Significance
Sex determination is a fundamental biological problem faced by all metazoans. To understand the sex determination pathway, it is important to identify all the genes involved in this process. In this study, we have identified a novel gene, spenito (nito), which is required for sex determination in Drosophila melanogaster. Loss of nito function in the soma transforms female tissues to male, and loss of nito function in female germ-line stem cells changes their sexual identity and prevents them from proper differentiation. We show that nito is a cofactor for Sex-lethal (Sxl) auto regulation, a process that remains an important textbook model for regulated alternative splicing.
Keywords: germ-line stem cell, sex determination, alternative splicing
Abstract
Sex-lethal (Sxl) encodes the master regulator of the sex determination pathway in Drosophila and acts by controlling sex identity in both soma and germ line. In females Sxl maintains its own expression by controlling the alternative splicing of its own mRNA. Here, we identify a novel sex determination gene, spenito (nito) that encodes a SPEN family protein. Loss of nito activity results in stem cell tumors in the female germ line as well as female-to-male somatic transformations. We show that Nito is a ubiquitous nuclear protein that controls the alternative splicing of the Sxl mRNA by interacting with Sxl protein and pre-mRNA, suggesting that it is directly involved in Sxl auto-regulation. Given that SPEN family proteins are frequently mutated in cancers, our results suggest that these factors might be implicated in tumorigenesis through splicing regulation.
Sex determination in Drosophila is under the control of the master regulatory gene Sex-lethal (Sxl) (1). Sxl acts downstream of the X-chromosome counting mechanism and encodes a female-specific RNA binding protein. Once activated, Sxl maintains its own expression by regulating the alternative splicing of its pre-mRNA. Sxl controls female fate by controlling somatic and germ-line sex identity as well as dosage compensation (2). In female somatic cells, Sxl controls the alternative splicing of transformer (tra), which together with transformer2 (tra2) controls the alternative splicing of doublesex (dsx) and fruitless (fru). dsx and fru in turn encode sex-specific transcription factors that control male versus female morphology, physiology, and behavior (3, 4). In addition, Sxl represses the male-specific dosage compensation system by regulating male-specific lethal 2 (msl-2) both at the level of alternative splicing and translational control (5).
In the female germ-line Sxl regulates sex identity by a different mechanism, as tra, tra2, msl-2 have no roles in the germ line (2). In the ovary, germ-line stem cells (GSCs) located at the anterior tip of the germarium divide to produce another GSC and a cystoblast (CB) that is committed to differentiate. Sxl protein accumulates to high levels in the GSCs/CBs and is required for the proper differentiation of the germ cells (6). Germ cells lacking Sxl cannot differentiate and instead produce stem cell tumors. The identity of Sxl target genes in the germ line is not well characterized; however, a recent study indicates that nanos, a gene required for GSC maintenance, is a Sxl target (7). Indeed, Sxl has been proposed to promote the differentiation of GSCs by downregulating Nanos levels in CBs by binding to the nanos 3′ UTR (7). In addition, Sxl is also important for repressing the expression of testis-specific genes, including Phf7, a male germ-line identity gene (8). In the absence of Sxl, Phf7 is mis-expressed leading to germ-line tumors (9).
Sxl does not act alone to control splicing. Several genes, including sans fille (snf), virilizer (vir), female-lethal-2-d (fl(2)d), SPF45, U1-70K, U2af38, U2af50, and protein partner of sans-fille (pps) facilitate Sxl splicing autoregulation (10–18). Except for pps, these genes encode either general splicing factors or proteins associated with spliceosomes. They all act to maintain the Sxl autoregulatory splicing loop by interacting with Sxl itself. In addition, some of them are involved in the splicing of other Sxl splicing targets such as tra or msl-2 (1). Interestingly, these genes have essential functions besides Sxl regulation and null mutations are associated with zygotic lethality in both sexes. Therefore, the roles of these factors in sex determination were revealed from genetic interactions (snf, U1-70K, U2af38) (11, 17), temperature-sensitive mutation (vir) (15), clonal analysis (fl(2)d) (10), or biochemical studies (SPF45, U2af50 and pps) (12, 14, 17).
Here, we characterize spenito (nito), a novel regulator of Sxl, which is required to maintain sex identity and Sxl levels in both the female germ-line and somatic tissues. Nito is required for the proper alternative splicing of the Sxl pre-mRNA in both germ line and soma, and forms a complex with Sxl protein and its pre-mRNA, thus identifying an important component of the sex determination pathway.
Results
nito Is an Essential Gene Required for Ovarian GSC Differentiation.
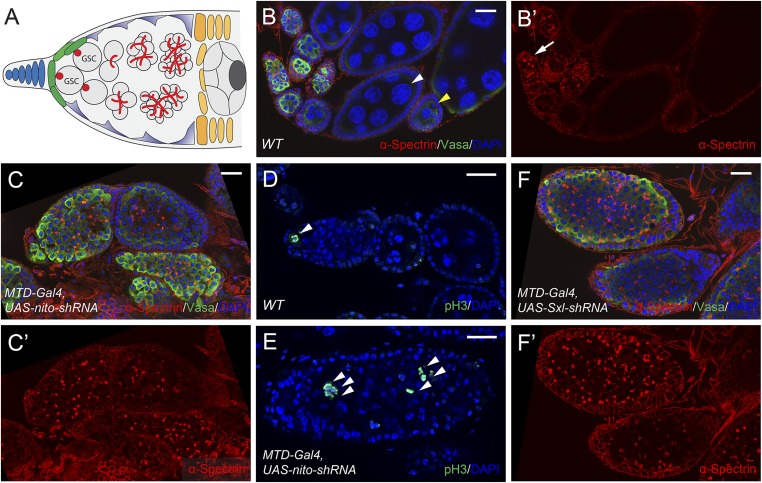
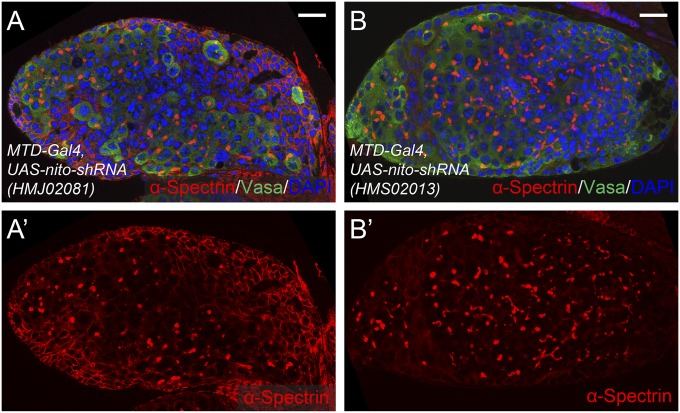
nito was identified from our previous RNAi screen in Drosophila GSCs (19). Specifically, RNAi knockdown of nito driven by the germ-line-specific MTD-Gal4 driver resulted in complete sterility in females. In wild-type ovarioles, two or three GSCs are located in the anterior tip of the germarium (Fig. 1 A–B′). Strikingly, nito shRNA ovarioles are filled with undifferentiated stem-cell-like cells, and nurse cells and oocytes are not formed (Fig. 1 C and C′, compared with WT in Fig. 1 B and B′). Further, stem-cell-like cells associated with nito shRNA ovaries retain their proliferative potential as shown by staining with the mitotic marker phosphorylated histone H3 (pH3) (Fig. 1E, compared with WT in Fig. 1D). Note that the same stem-cell tumor phenotype was observed with three independent nito shRNAs and two long dsRNA RNAi lines (Methods) (Fig. S1 A–B′), indicating that nito is an essential gene required for GSC differentiation.
Fig. 1.
Nito is essential for ovarian GSC differentiation. (A) Diagram showing the structure of a wild type germarium. (B–B′) WT ovarioles stained for α-Spectrin, Vasa and DAPI. The α-Spectrin antibody labels the round spectrosomes in GSCs (arrow); the Vasa antibody labels all germ cells and DAPI labels nuclei to monitor oocyte (yellow arrowhead) and nurse cell formation (white arrowhead). (C–C′) Egg chambers expressing nito shRNA by MTD-Gal4 were stained for α-Spectrin, Vasa and DAPI. Note the numerous stem-cell-like cells labeled by α-Spectrin and the absence of differentiated nurse cells. (D–E) pH3 staining in WT egg chambers and egg chambers expressing nito shRNA. In WT egg chambers, pH3-positive cells were restricted to the anterior tip of the germarium but were detected throughout nito shRNA egg chambers (arrowheads). (F–F′) Egg chambers expressing Sxl shRNA stained for α-Spectrin, Vasa and DAPI. (Scale bars: 20 μm.)
Fig. S1.
Two independent nito shRNAs result in similar stem-cell-tumor in the germ-line. (A–B′) Egg chambers expressing shRNAs targeting nito (HMJ02081) or nito (HMS02013) using MTD-Gal4 stained for α-Spectrin, Vasa and DAPI. (Scale bars: 20 μm.)
nito Is Required for Sex Determination in the Soma.
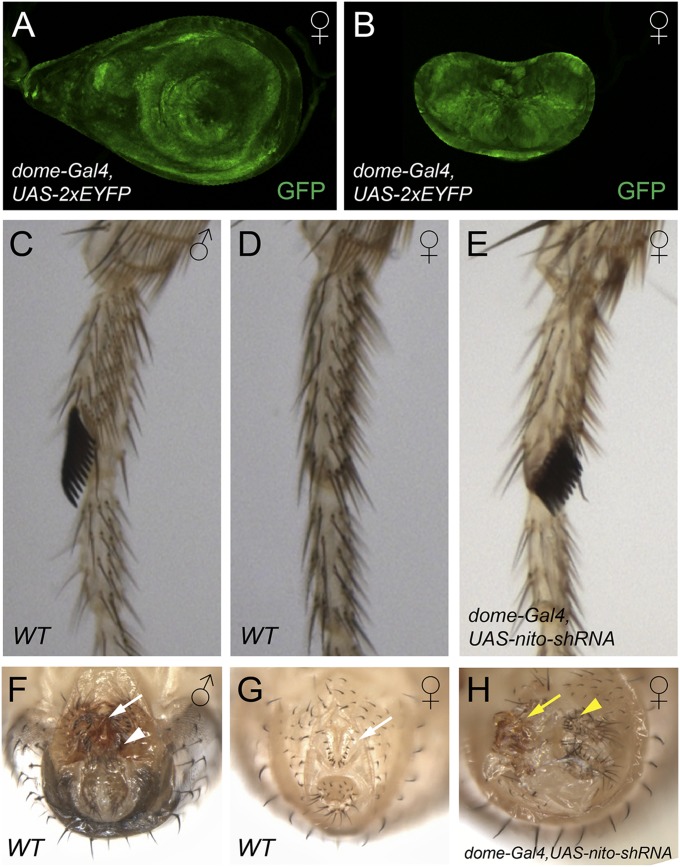
Because the germ-line phenotype of nito could reflect perturbations in a number of developmental processes affecting either germ-line proliferation or differentiation, we examined nito loss-of-function phenotypes in somatic tissues. Strikingly, expression of nito shRNA using dome-Gal4, that drives expression in both the leg and genital discs (Fig. 2 A and B), led to the transformation of female tissues into that of males. This is evidenced by the appearance of dark, thickened bristles, the male sex combs, in the forelegs of nito shRNA females (Fig. 2E, compare with WT in Fig. 2 C and D). This phenotype is almost fully penetrant and occurs in 97% (n = 78) of females examined. In addition, there are strong abnormalities in the genitalia of these female flies. First, a rotation defect has occurred in 71% (n = 78) of dome-Gal4/nito-shRNA females (Fig. 2H). Second, typical female external structures, such as vaginal bristles (Fig. 2G, white arrow), are absent in the genitalia (Fig. 2H). Third, structures resembling those of males, such as penis apparatus and claspers can be identified (Fig. 2 F and H). These transformations suggest that Nito is a component of the Drosophila sex determination pathway in the soma. Because Sxl shRNA generates a stem-cell-tumor phenotype in the germ line similar to that of nito (Fig. 1 F and F′), the nito germ-line phenotype therefore could be due to sex determination defects associated with Sxl (see below).
Fig. 2.
Nito is required for sex determination in somatic tissues. (A and B) dome-Gal4 drives expression in the first pair of leg discs (A) and genital discs (B), as shown by UAS-2xEYFP. (C) Foreleg of a WT male with the dark thickened sex comb bristles. (D) Foreleg of a WT female. Note the absence of sex combs. (E) Foreleg of a female fly expressing nito shRNA driven by dome-Gal4. Some bristles are transformed into male sex combs. (F and G) Genitalia of wild-type male (F) and female (G) flies showing distinct morphology, such as penis apparatus and claspers in male (F, arrow and arrowhead, respectively) and vaginal bristles in female (G, arrow). (H) nito shRNA driven by dome-Gal4 transforms female genital morphology into male-like, as evidenced by the absence of vaginal bristles and appearance of structures resembling penis apparatus (arrow) and claspers (arrowhead).
Nito Is a Ubiquitously Expressed Nuclear Protein That Is Crucial in Both Sexes.
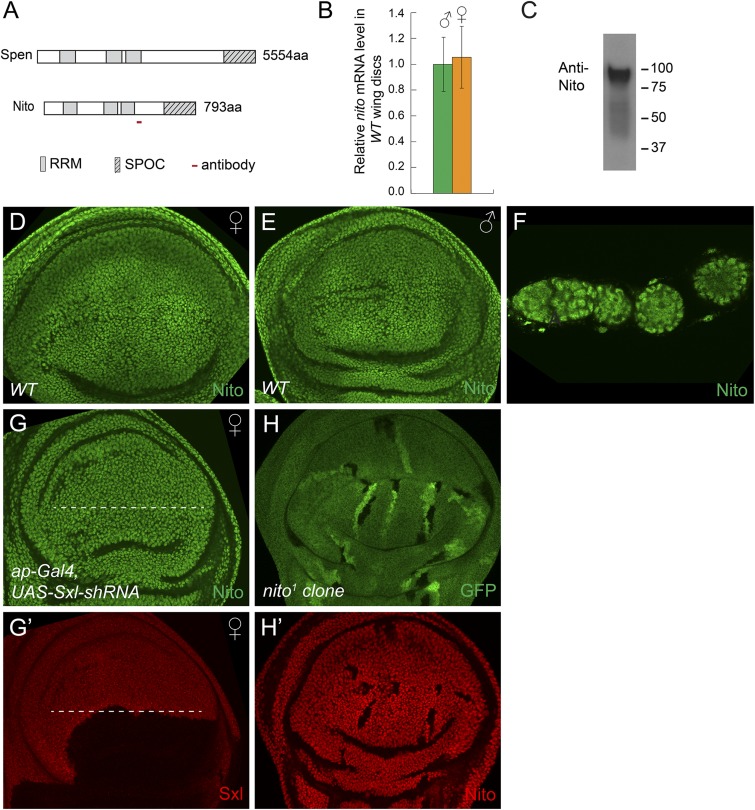
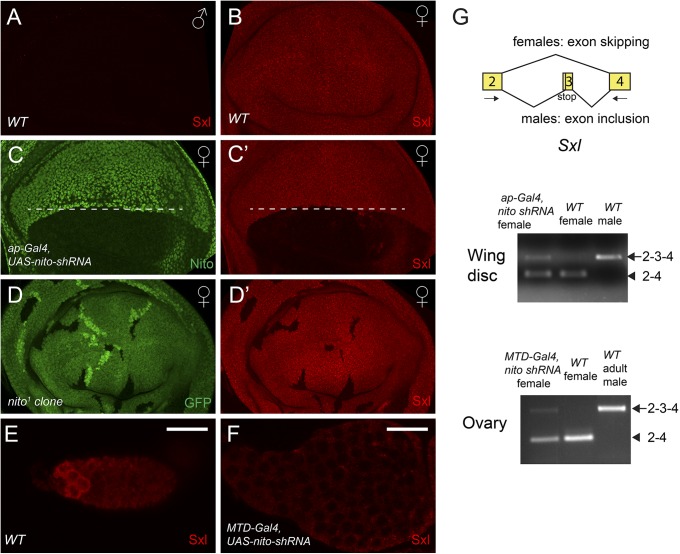
Nito, together with Split ends (Spen), are members of the SPEN protein family characterized by three N-terminal RNA recognition motifs (RRMs) and a C-terminal SPOC (Spen paralog and ortholog C-terminal) domain (Fig. 3A) (20, 21). To analyze Nito expression, we raised a polyclonal antibody against a 22 amino acid peptide (Methods). A Western blot showed that this antibody recognizes a protein of the expected ∼89 kDa size in Drosophila S2 cell lysates (Fig. 3C). Nito is ubiquitously expressed in all tissues examined, including imaginal discs and ovaries, and localizes to the nucleus (Fig. 3 D and F). Furthermore, expression of nito shRNA using ap-Gal4 led to almost complete depletion of the Nito protein in the dorsal half of wing discs demonstrating the specificity of the antibody (Fig. 4C and Fig. S2 A and B).
Fig. 3.
Nito is a ubiquitous nuclear protein that shows no sex-biased expression. (A) Schematic diagram showing domain structures of Spen and Nito proteins and the peptide used to generate the Nito antibody. (B) nito mRNA levels in male and female WT wing discs were measured by qRT-PCR. Error bars represent SDs. (C) Nito antibody recognizes a single band of the predicted ∼89 kDa size in S2 cell lysates. (D–F) Nito antibody stainings in WT female (D) or male (E) wing discs and ovarioles (F). (G–G′) Expression of Sxl shRNA in the dorsal half of the wing disk (below the dashed line) using ap-Gal4 leads to depletion of Sxl protein, but has no effect on Nito protein levels. (H–H′) Nito antibody staining in wing discs containing nito1 mutant clones, marked by the absence of GFP. Note the absence of Nito staining in nito1 clones.
Fig. 4.
Nito is required for Sxl levels and regulates Sxl mRNA splicing. (A and B) Sxl stainings in WT male (A) and female (B) wing discs. (C–C′) Expressing nito shRNA in the dorsal half of the disk (below the dashed line) using ap-Gal4 leads to strong reduction of Nito (C) and Sxl (C′) stainings. (D–D′) Sxl antibody staining (D′) in wing discs containing nito1 mutant clones, which are marked by the absence of GFP (D). Note the absence of Nito and Sxl staining in nito1 clones. (E and F) Sxl stainings in WT egg chambers (E) or in egg chambers expressing nito shRNA by MTD-Gal4 (F). Scale bars: 20 μm. (G) Diagram showing the alternative splicing event that produces the male- or female-specific Sxl transcripts. The arrows indicate the primers used for RT-PCR. Sxl splicing was analyzed by RT-PCR using RNA extracted from wing discs or ovaries. Male-specific bands: 2–3-4. Female-specific bands: 2–4.
Fig. S2.
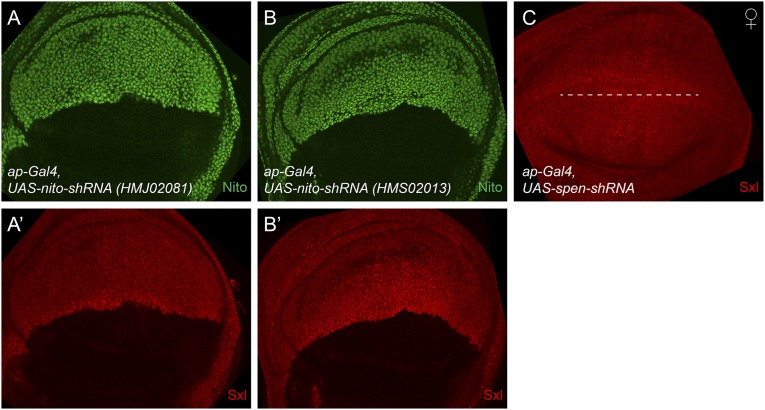
nito, but not spen, regulates Sxl levels in wing discs. (A–B′) Expression of nito shRNA (HMJ02081 or HMS02013) in the dorsal half of the wing disk using ap-Gal4 leads to a strong reduction of both Nito (A and B) and Sxl (A′ and B′) stainings. (C) Expression of spen shRNA in the dorsal half of the disk (below the dashed line) by ap-Gal4 does not affect Sxl protein levels. spen shRNA generates embryonic lethality with cuticle and head defects when expressed using MTD-Gal4 (47), which resembles the phenotype of the spen mutant, indicating that the shRNA is functional.
Because nito affects sex determination, we tested whether its expression level is biased in females versus males. To exclude the maternal contribution from ovaries, we compared nito mRNA levels in wing discs. As shown in Fig. 3B, nito mRNA levels were similar in female and male wing discs. In addition, Nito antibody staining showed similar protein levels in female and male discs (Fig. 3 D and E). Further, Nito is not regulated by Sxl as its protein level is not affected in Sxl RNAi discs (Fig. 3 G and G′). Together, these data indicate that nito is not differentially expressed in males versus females.
We generated a null allele of nito by imprecise P-element excision (referred to as nito1) to test whether nito is an essential gene. nito1 homozygous animals die during larval stages and homozygous mutant clones show the absence of Nito protein indicating that nito1 is a null mutation (Fig. 3 H and H′). Interestingly, nito1 causes lethality in both females and males, indicating that nito is an essential gene. Further, nito1 is lethal over a deficiency of the nito locus indicating that the lethality is likely due to the nito mutation. Consistent with this, nito RNAi driven by a ubiquitous Gal4 such as actin-Gal4 or tubulin-Gal4 is associated with larval lethality.
Nito Regulates Sxl Levels by Controlling Sxl Alternative Splicing.
The phenotype associated with loss of Nito function in both soma and germ line suggests that Nito may regulate Sxl activity. In ovaries, Sxl is enriched in GSCs and their immediate daughter cells (Fig. 4E) (6), whereas in somatic tissues such as wing discs Sxl is expressed ubiquitously in females but absent in males (Fig. 4 A and B). We expressed nito shRNA in the germ line using MTD-Gal4 and in the dorsal half of the wing disk using ap-Gal4. Strikingly, knockdown of nito in both tissues led to a significant reduction of Sxl levels (Fig. 4 F, C, and C′). Note that similar results were obtained using two additional nito shRNA lines as well as in homozygous nito1 mutant clones (Fig. 4 D and D′ and Fig. S2 A′ and B′). However, the level of Sxl is not affected when Split-ends (Spen), another member of the SPEN family, was knocked down by shRNA in the wing disk (Fig. S2C), indicating a specific role of Nito in the sex determination pathway. Altogether, we have identified a new component of the Drosophila sex determination pathway that acts in both the germ line and soma by affecting Sxl levels.
Sxl transcripts are alternatively spliced, with exon 3 containing a stop codon that is included in males but skipped in females, leading to truncated Sxl forms in males but functional proteins in females (Fig. 4G) (22). Because Nito has three RRMs, we asked whether Nito has a potential role in Sxl splicing and used a pair of primers that detects the small female and large male spliced Sxl products (Fig. 4G) (12) to analyze Sxl splicing by RT-PCR in the absence of Nito. In nito shRNA female wing discs or ovaries, a large band corresponding to the male-specific spliced form was clearly detected (Fig. 4G). Note that in wing discs and ovaries, female-specific transcripts are detected due to contributions from remaining WT disk cells or somatic follicle cells, respectively, as nito was knocked down in half of the discs or in the germ line only. Together, our results indicate that Nito regulates Sxl levels by controlling its alternative splicing.
Nito Interacts with Sxl Protein and its Pre-mRNA in S2 Cells.
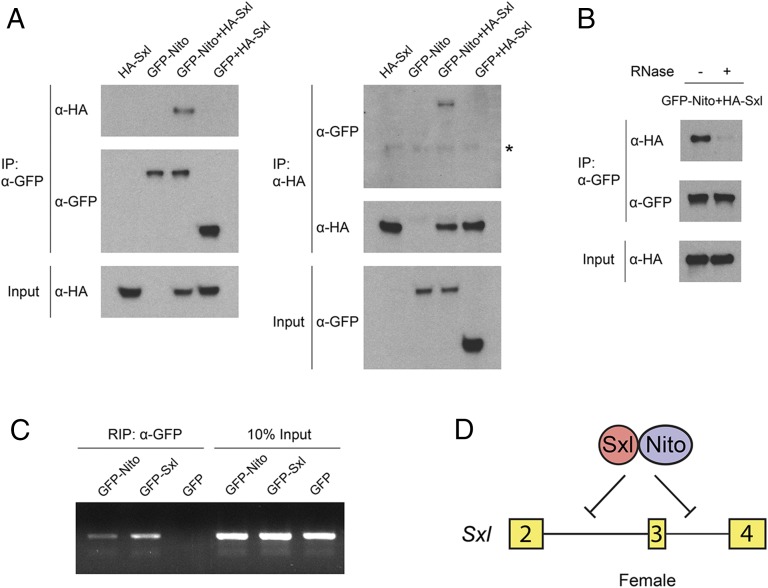
We then analyzed how Nito controls Sxl alternative splicing. Because the key protein that binds Sxl pre-mRNA and inhibits splicing of male-specific exon 3 is Sxl itself (2), we examined whether Nito interacts with Sxl using a coimmunoprecipitation (co-IP) assay in Drosophila S2 cells. GFP-Nito and HA-Sxl were expressed either individually or in combination in S2 cells, and GFP alone was used as a control (Fig. 5A). HA-Sxl was detected in the precipitate obtained using anti-GFP nanobody from GFP-Nito cells, but not from GFP-expressing cells. Similarly, GFP-Nito, but not GFP, was pulled down by HA-Sxl. Further, we tested whether the interaction between Nito and Sxl is dependent on the presence of RNA. Interestingly, the amount of Sxl-HA pulled down by GFP-Nito is strongly reduced in the presence of RNase, suggesting that the Nito/Sxl interaction is mediated or stabilized by RNA (Fig. 5B). Finally, we performed RNA immunoprecipitation (RIP) experiments in S2 cells to analyze whether Nito can interact with Sxl pre-mRNA, which was detected by RT-PCR using an intron 3-exon 4 primer pair (12). As shown in Fig. 5C, GFP-Sxl, GFP-Nito, but not GFP alone, can pull down Sxl pre-mRNA from cell lysates. Together, the specific interactions between Nito, Sxl, and Sxl pre-mRNA support the model that Nito forms a complex with Sxl and that they together regulate alternative splicing of Sxl mRNA (Fig. 5D).
Fig. 5.
Nito interacts with Sxl and Sxl pre-mRNA in S2 cells. (A) HA-Sxl, GFP-Nito or GFP expression vectors were transfected individually or together into Drosophila S2 cells. Cell lysates were immunoprecipitated using GFP nanobody or anti-HA antibody and analyzed by Western blot. GFP alone is used as a control. Asterisk indicates IgG heavy chain. (B) S2 cells were transfected with GFP-Nito and HA-Sxl, and Co-IP was performed using GFP nanobody in the absence or presence of RNase A and RNase T1. (C) S2 cells were transfected with GFP-Nito, GFP-Sxl or GFP and immunoprecipitated with GFP nanobody. The presence of Sxl pre-mRNA was detected by RT-PCR using an intron 3/exon 4 primer pair. GFP-Sxl was used as a positive control and GFP alone as a negative control. (D) Model: Nito forms a complex with Sxl and together they repress the splicing of Sxl exon 3 in female tissues.
Screen for Additional Splicing Genes Involved in Sex Determination.
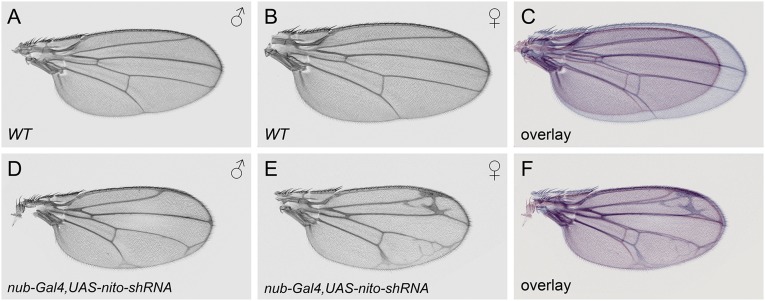
Because we identified nito as an important gene involved in sex determination by regulating Sxl splicing, we asked whether there are other unidentified genes in the Drosophila genome acting in this pathway. Because most known Sxl-autoregulatory proteins are associated with spliceosomes (2), we screened a collection of 316 RNAi lines representing 247 splicing-associated genes and RNA-binding proteins (Dataset S1), using MTD-Gal4 for the germ line and dome-Gal4 for somatic phenotypes. In addition, we also included in our screen a wing-specific driver, nub-Gal4, as many RNAi lines exhibit lethality with dome-Gal4 and prevent the detection of potential sex determination phenotypes. Because Sxl is responsible for the larger wing size in females, examination of wing size using nub-Gal4 allows detection of potential genes involved in sex determination, as shown in the case of nito (Fig. S3 A–F).
Fig. S3.
Nito shRNA affects wing growth more strongly in females than in males. (A) WT male wing. (B) WT female wing. (C) Overlay of the images in A (red) and B (blue) shows that a WT female wing is about 30% larger than a WT male wing. Male (D) and female (E) wings in which nito shRNA was expressed using the nub-Gal4 driver. (F) Overlay of D and E showing that both male and female wings reach about the same size upon nito knockdown.
Our screen successfully identified known components of the sex determination pathway: tra and tra2 RNAi showed strong female-to-male transformation in sex combs and genitalia when induced by dome-Gal4; and fl(2)d and vir were identified using the nub-Gal4 driver as both have a stronger effect in female wings than in male wings. Strikingly, besides these genes we did not identify any other genes that showed a sex-related phenotype. However, we did characterize GSC differentiation phenotypes associated with tsu, mago, RnpS1, Rbp9 RNAi lines, as well as wing growth and pattern defects with kul, CG7879, ASPP, tst and Syp RNAi lines. These data provide a valuable resource of phenotypes associated with splicing-related genes (data available at www.flyrnai.org/RSVP.html).
Discussion
We describe the characterization of Nito as a novel component of the Drosophila sex determination pathway. Nito loss-of-function results in stem-cell tumor phenotypes in the germ-line and sexual transformations in the soma. Interestingly, Nito affects Sxl protein levels in both GSCs and somatic tissues by regulating Sxl pre-mRNA alternative splicing, most likely directly as Nito interacts with the Sxl protein and pre-mRNA. The role of Nito is reminiscent of the previously reported roles of splicing factors in Sxl auto regulation, such as both subunits of U2AF (17), U1-70K (17, 18), Fl(2)d (10, 23), SPF45 (13, 14), Vir (15, 24), and Snf (11, 16). Our data support earlier reports that Sxl physically interacts with components of the spliceosome to simultaneously block utilization of the 3′ and 5′ splice sites of the male exon.
Nito and Spen are members of the SPEN protein family that are evolutionarily conserved from plants, worms, flies to mice and humans (25). Both proteins contain three N-terminal RRM domains and one C-terminal SPOC domain. The sequence similarity between these domains is low and there is no conservation outside these motifs, suggesting that they have evolved specific functions following a duplication event (20), as indicated by our observation that spen is not required for Sxl regulation (Fig. S2C). In Drosophila, spen was first identified in several genetic screens looking for components of the receptor tyrosine kinase (RTK) signaling pathway (26). Subsequent studies found that spen is implicated in a variety of cellular and developmental processes including neuronal cell fate specification, axon guidance, cell cycle, Hox gene regulation, and cell death (27–30). These pleiotropic effects are likely due to the involvement of spen in multiple signaling pathways (31–33). However, the molecular mechanisms underlying the function of Spen in these pathways are not understood.
Genetic studies in Drosophila have shown that nito overexpression results in a rough eye phenotype (20) and that it plays a redundant role with spen in Wnt signaling (21), but how Nito is involved in these processes is not known. Biochemical studies indicate that Nito, like its human ortholog, copurify with the precatalytic spliceosome (complex B) (34). In addition, nito, as well as many other splicing factors, was identified in an RNAi screen for RAS/MAPK signaling components (35). Consistent with these findings, we find that nito is required for the alternative splicing of the master sex-determination gene Sxl. Previously, both Spen and Nito were thought to act mainly as transcription factors through their SPOC domains, our findings however clearly indicate that Nito is involved in mRNA splicing. It is intriguing to note that PPS, another important factor required for Sxl splicing, also has a SPOC domain (12). Similar to Nito, PPS also forms a complex with Sxl protein and its pre-mRNA (12). In the future it will be crucial to dissect how different protein domains contribute to the function of SPEN family proteins.
Then what is the “main” role of nito? On one hand, the phenotypes in the sex comb, genitalia and germ line appear specific to Sxl and such phenotypes do not depend on the genetic interaction with other genes in the sex determination pathway. On the other hand, nito clearly has other non-sex-specific functions, as revealed by the lethality, rough eye, and wing phenotype observed in both sexes (Fig. S3 A–F). Because a null allele of nito is associated with zygotic lethality, the RNAi knockdown approach is a powerful method to reveal sex-related phenotypes. Interestingly, our RNAi screen targeting splicing factors did not identify any new additional sex determination genes, indicating that there are a limited number of genes yet to be identified in this pathway. Finally, intriguingly, three recent studies have identified SPEN and Rbm15 (the mouse and human ortholog of Nito) as factors interacting with Xist, the long noncoding RNA that is essential for dosage compensation in mammals (36–38). Clearly, future experiments such as RNA-seq will be necessary to elucidate the mechanism and logic of Nito-mediated signaling events.
Rbm15, also known as OTT, was originally identified from infants with acute megakaryoblastic leukemia (AMKL) (39, 40). The t(1, 22) chromosomal translocation results in fusion of RBM15 and MKL1, and the fusion protein is responsible for AMKL development as shown in a mouse model (41). In addition to this chromosome translocation, recent cancer genome sequencing projects have found that RBM15 and SPEN (also known as SHARP) are mutated in many different types of cancers, such as adenoid cystic carcinomas and bladder cancers (42). Given that SPEN family proteins are frequently mutated or deleted in cancers, they have been proposed to act as potential tumor suppressors (42). Studies of Spen and Nito in Drosophila will provide mechanistic insights to our understanding of this important family of proteins.
Methods
Details on the fly strains used in this study, as well as how the null nito mutation was isolated and how nito clones were generated can be found in SI Methods. Protocols used for antibody staining, reagents, how Nito antibodies were generated, coimmunoprecipitation protocols, RT-PCR, and information on primers and RNA immunoprecipitation (RIP), can be found in SI Methods.
SI Methods
Fly Strains.
The following stocks were used in this study: w1118 (used as wild-type, WT), MTD-Gal4, ap-Gal4, nub-Gal4, dome-Gal4, UAS-2xEYFP (43), Sxl shRNA (HMS00609), nito shRNA (HMS00166), nito shRNA (HMS02013), nito shRNA (HMJ02081), nito dsRNA (VDRC 20942), nito dsRNA (VDRC 114704). Experiments presented in Figs. 1, 2, and 4 and Fig. S3 were done using nito shRNA (HMS00166).
To generate a null nito mutation, we used the homozygous viable P-element insertion nitoHP25329 located in the 5′UTR of the nito gene. After mobilization of the HP25329 P-element, we screened for homozygous lethal lines and recovered a null allele, nito1, that deletes 1,357 bp and is lethal over Df(2R)Exel6055 (43F1 to 44A4) that uncovers the nito locus. The Nito antibody, raised against amino acids 479–500, cannot detect any Nito antigens in nito1 mutant clones (Fig. 3 H and H′).
To generate mutant clones, nito1 was recombined with FRTG13 and crossed to y w hsflp; ubiGFP FRTG13 flies. The progeny were heat-shocked at 37 °C for 1 h twice at first- and second-instar larval stage.
Antibody Stainings in Discs and Ovaries.
Larval wing discs and female ovaries were stained as described (19). Briefly, tissues were dissected in PBS and fixed in 4% formaldehyde in PBST (PBS + 0.1% Triton X-100). After blocking in 1% normal donkey serum in PBST for 1 h, the samples were incubated with the primary antibody in the same solution at 4 °C overnight. After three washes in PBST, samples were incubated with the secondary antibody for 2 h at room temperature, washed in PBST three times, and subsequently mounted in Vectashield. All images were taken on a Zeiss LSM 780 microscope.
The following antibodies were used: mouse anti-α-Spectrin (1:10) (3A9, DSHB), rabbit anti-Vasa (1:250) (Santa Cruz Biotechnology), mouse anti-Sxl (1:10) (M18, DSHB), rabbit anti-Nito (1:500), rabbit anti-phospho-Histone H3 (1:1,000) (Millipore), rabbit anti-GFP (1:1,000) (Molecular Probes), mouse anti-GFP (1:200) (Molecular Probes), Alexa 488- or 555- conjugated secondary antibodies (1:1,000) (Molecular Probes) and DAPI (1:1,000) (Molecular Probes).
Nito antibodies were generated in rabbits against a peptide containing amino acids 479–500 (KSSKPPYDESALEYRRPEYDPY) and affinity-purified at YenZym Antibodies. Polyclonal antisera were raised in two rabbits, YZ3137 and YZ3138, and gave similar staining patterns. All of the experiments described in the paper were performed with antiserum from YZ3137.
Adult legs and wings were mounted in a 1:1 (vol/vol) mixture of Permount (Fisher Scientific) and xylene. The genitalia images were taken in stacks and rendered with HeliconFocus software.
Coimmunoprecipitation.
To generate the GFP-Nito plasmid, a nito full-length cDNA (GH11110) was cloned into the Drosophila Gateway vector pAGW. HA-Sxl and GFP-Sxl were constructed following PCR of Sxl (the MS3 isoform) from UAS-Sxl flies (44) and cloned into pAHW and pAGW, respectively. GFP was cloned into pAWM as a control.
Drosophila S2 cells were maintained at 25 °C in Schneider’s medium supplemented with 10% FBS. One microgram of total DNA was transfected into S2 cells in a single well of six-well plates with Effectene (QIAGEN). After 48 h, cells were lysed in IP lysis buffer (Pierce) with Halt Protease Inhibitor (Thermo Scientific). Lysates were incubated with anti-GFP nanobody agarose beads (Allele Biotechnology) or anti-HA agarose (Sigma) for 2 h at 4 °C. The beads were washed 3–4 times with 1 mL lysis buffer. Protein complexes were eluted and detected by Western blotting using anti-GFP antibody (A6455, Molecular Probes) or anti-HA antibody (3F10, Roche). For RNase treatment experiment, 100 μL of RNase A (10 mg/mL, Thermo Scientific) and 5 μL of RNase T1 (1,000 U/μL, Thermo Scientific) were added to 1 mL of lysate and incubated for 30 min at 30 °C, then overnight at 4 °C with beads (12, 17).
RT-PCR.
Total RNA was extracted from dissected wing discs or ovaries using TRIzol (Invitrogen), digested with DNase I (Qiagen) and purified using the RNeasy Mini kit (Qiagen). cDNA was generated from 1 μg of purified RNA using the iScript cDNA Synthesis lit (Bio-Rad). For nonreal time PCR, TaKaRa Taq polymerase was used. For qPCR, iQ SYBR Green Supermix (Bio-Rad) was used and reactions were measured in a CFX96 Real-Time PCR detection system (Bio-Rad). qPCR results for nito expression in male and female wing discs (Fig. 3B) were normalized to the reference gene αTubulin84B. Sxl primers used in Fig. 4G are described in ref. 12. nito primers in Fig. 3B and αTubulin84B primers are listed below.
| nito_PP17280_f | GGCTACAAGGTACTTTGCGTC |
| nito_PP17280_r | TACTCGCGGTACAGTGTCTCC |
| Sxl-f | GTGGTTATCCCCCATATGGC |
| Sxl-r | GATGGCAGAGAATGGGAC |
| αTubulin84B-f | CAACCAGATGGTCAAGTGCG |
| αTubulin84B-r | ACGTCCTTGGGCACAACATC |
RNA Immunoprecipitation (RIP).
RIP experiments were performed following previous protocols (45, 46). One microgram of DNA was transfected into S2 cells in 60-mm plates with Effectene (QIAGEN). After 48 h, cells were lysed in IP lysis buffer (Pierce) with Halt Protease Inhibitor (Thermo Scientific) and RNasin plus (40 U/mL, Promega). After centrifugation, 10% of the supernatant were saved as the input and the rest were incubated with anti-GFP nanobody agarose beads (Allele Biotechnology) for 2 h at 4 °C. The beads were washed 3–4 times with 1 mL of lysis buffer. To elute RNA from the RNA/protein complexes, the beads were treated with proteinase K solution for 10 min at 55 °C. Total RNA from the input and the beads were extracted by using RNeasy Micro kit (QIAGEN). cDNA was synthesized using SuperScript III First-Strand Synthesis System (Life Technologies) with four of the eluted RNA and random hexamers. TaKaRa Taq polymerase was used for two rounds of PCR amplification with primers and conditions described in ref. 12.
| Sxl-intron | GAGGGTCAGTCTAAGTTATATTCG |
| Sxl-r | GATGGCAGAGAATGGGAC |
Supplementary Material
Acknowledgments
We thank Ken Cadigan, Jamila Horabin, Ilaria Rebay, and Helen Salz for fly stocks and antibodies. We thank Mitzi Kuroda, Leonard Rabinow, Stephanie Mohr, Richard Binari, Benjamin Housden, and Ben Ewen-Campen for comments on the manuscript. D.Y. is supported by Damon Runyon Cancer Research and Charles A. King Trust Postdoctoral Fellowship. N.P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515891112/-/DCSupplemental.
References
- 1.Cline TW, Meyer BJ. Vive la différence: Males vs females in flies vs worms. Annu Rev Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- 2.Salz HK, Erickson JW. Sex determination in Drosophila: The view from the top. Fly (Austin) 2010;4(1):60–70. doi: 10.4161/fly.4.1.11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clough E, et al. Sex- and tissue-specific functions of Drosophila doublesex transcription factor target genes. Dev Cell. 2014;31(6):761–773. doi: 10.1016/j.devcel.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryner LC, et al. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87(6):1079–1089. doi: 10.1016/s0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- 5.Camara N, Whitworth C, Van Doren M. The creation of sexual dimorphism in the Drosophila soma. Curr Top Dev Biol. 2008;83:65–107. doi: 10.1016/S0070-2153(08)00403-1. [DOI] [PubMed] [Google Scholar]
- 6.Chau J, Kulnane LS, Salz HK. Sex-lethal facilitates the transition from germline stem cell to committed daughter cell in the Drosophila ovary. Genetics. 2009;182(1):121–132. doi: 10.1534/genetics.109.100693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chau J, Kulnane LS, Salz HK. Sex-lethal enables germline stem cell differentiation by down-regulating Nanos protein levels during Drosophila oogenesis. Proc Natl Acad Sci USA. 2012;109(24):9465–9470. doi: 10.1073/pnas.1120473109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang SY, Baxter EM, Van Doren M. Phf7 controls male sex determination in the Drosophila germline. Dev Cell. 2012;22(5):1041–1051. doi: 10.1016/j.devcel.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shapiro-Kulnane L, Smolko AE, Salz HK. Maintenance of Drosophila germline stem cell sexual identity in oogenesis and tumorigenesis. Development. 2015;142(6):1073–1082. doi: 10.1242/dev.116590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Granadino B, Campuzano S, Sánchez L. The Drosophila melanogaster fl(2)d gene is needed for the female-specific splicing of Sex-lethal RNA. EMBO J. 1990;9(8):2597–2602. doi: 10.1002/j.1460-2075.1990.tb07441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliver B, Perrimon N, Mahowald AP. Genetic evidence that the sans fille locus is involved in Drosophila sex determination. Genetics. 1988;120(1):159–171. doi: 10.1093/genetics/120.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson ML, Nagengast AA, Salz HK. PPS, a large multidomain protein, functions with sex-lethal to regulate alternative splicing in Drosophila. PLoS Genet. 2010;6(3):e1000872. doi: 10.1371/journal.pgen.1000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaouki AS, Salz HK. Drosophila SPF45: A bifunctional protein with roles in both splicing and DNA repair. PLoS Genet. 2006;2(12):e178. doi: 10.1371/journal.pgen.0020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lallena MJ, Chalmers KJ, Llamazares S, Lamond AI, Valcárcel J. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell. 2002;109(3):285–296. doi: 10.1016/s0092-8674(02)00730-4. [DOI] [PubMed] [Google Scholar]
- 15.Hilfiker A, Amrein H, Dübendorfer A, Schneiter R, Nöthiger R. The gene virilizer is required for female-specific splicing controlled by Sxl, the master gene for sexual development in Drosophila. Development. 1995;121(12):4017–4026. doi: 10.1242/dev.121.12.4017. [DOI] [PubMed] [Google Scholar]
- 16.Flickinger TW, Salz HK. The Drosophila sex determination gene snf encodes a nuclear protein with sequence and functional similarity to the mammalian U1A snRNP protein. Genes Dev. 1994;8(8):914–925. doi: 10.1101/gad.8.8.914. [DOI] [PubMed] [Google Scholar]
- 17.Nagengast AA, Stitzinger SM, Tseng CH, Mount SM, Salz HK. Sex-lethal splicing autoregulation in vivo: Interactions between SEX-LETHAL, the U1 snRNP and U2AF underlie male exon skipping. Development. 2003;130(3):463–471. doi: 10.1242/dev.00274. [DOI] [PubMed] [Google Scholar]
- 18.Salz HK, et al. The Drosophila U1-70K protein is required for viability, but its arginine-rich domain is dispensable. Genetics. 2004;168(4):2059–2065. doi: 10.1534/genetics.104.032532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan D, et al. A regulatory network of Drosophila germline stem cell self-renewal. Dev Cell. 2014;28(4):459–473. doi: 10.1016/j.devcel.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jemc J, Rebay I. Characterization of the split ends-like gene spenito reveals functional antagonism between SPOC family members during Drosophila eye development. Genetics. 2006;173(1):279–286. doi: 10.1534/genetics.106.055558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang JL, Lin HV, Blauwkamp TA, Cadigan KM. Spenito and Split ends act redundantly to promote Wingless signaling. Dev Biol. 2008;314(1):100–111. doi: 10.1016/j.ydbio.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 22.Bell LR, Horabin JI, Schedl P, Cline TW. Positive autoregulation of sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell. 1991;65(2):229–239. doi: 10.1016/0092-8674(91)90157-t. [DOI] [PubMed] [Google Scholar]
- 23.Penn JK, et al. Functioning of the Drosophila Wilms’-tumor-1-associated protein homolog, Fl(2)d, in Sex-lethal-dependent alternative splicing. Genetics. 2008;178(2):737–748. doi: 10.1534/genetics.107.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultt C, Hilfiker A, Nothiger R. virilizer regulates Sex-lethal in the germline of Drosophila melanogaster. Development. 1998;125(8):1501–1507. doi: 10.1242/dev.125.8.1501. [DOI] [PubMed] [Google Scholar]
- 25.Sánchez-Pulido L, Rojas AM, van Wely KH, Martinez-A C, Valencia A. SPOC: A widely distributed domain associated with cancer, apoptosis and transcription. BMC Bioinformatics. 2004;5:91. doi: 10.1186/1471-2105-5-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rebay I, et al. A genetic screen for novel components of the Ras/Mitogen-activated protein kinase signaling pathway that interact with the yan gene of Drosophila identifies split ends, a new RNA recognition motif-containing protein. Genetics. 2000;154(2):695–712. doi: 10.1093/genetics/154.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuang B, Wu SC, Shin Y, Luo L, Kolodziej P. split ends encodes large nuclear proteins that regulate neuronal cell fate and axon extension in the Drosophila embryo. Development. 2000;127(7):1517–1529. doi: 10.1242/dev.127.7.1517. [DOI] [PubMed] [Google Scholar]
- 28.Chen F, Rebay I. split ends, a new component of the Drosophila EGF receptor pathway, regulates development of midline glial cells. Curr Biol. 2000;10(15):943–946. doi: 10.1016/s0960-9822(00)00625-4. [DOI] [PubMed] [Google Scholar]
- 29.Wiellette EL, et al. spen encodes an RNP motif protein that interacts with Hox pathways to repress the development of head-like sclerites in the Drosophila trunk. Development. 1999;126(23):5373–5385. doi: 10.1242/dev.126.23.5373. [DOI] [PubMed] [Google Scholar]
- 30.Mace K, Tugores A. The product of the split ends gene is required for the maintenance of positional information during Drosophila development. BMC Dev Biol. 2004;4:15. doi: 10.1186/1471-213X-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doroquez DB, Orr-Weaver TL, Rebay I. Split ends antagonizes the Notch and potentiates the EGFR signaling pathways during Drosophila eye development. Mech Dev. 2007;124(9-10):792–806. doi: 10.1016/j.mod.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin HV, et al. Splits ends is a tissue/promoter specific regulator of Wingless signaling. Development. 2003;130(14):3125–3135. doi: 10.1242/dev.00527. [DOI] [PubMed] [Google Scholar]
- 33.Querenet M, Goubard V, Chatelain G, Davoust N, Mollereau B. Spen is required for pigment cell survival during pupal development in Drosophila. Dev Biol. 2015;402(2):208–215. doi: 10.1016/j.ydbio.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 34.Herold N, et al. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol Cell Biol. 2009;29(1):281–301. doi: 10.1128/MCB.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashton-Beaucage D, et al. A functional screen reveals an extensive layer of transcriptional and splicing control underlying RAS/MAPK signaling in Drosophila. PLoS Biol. 2014;12(3):e1001809. doi: 10.1371/journal.pbio.1001809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moindrot B, et al. A Pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Reports. 2015;12(4):562–572. doi: 10.1016/j.celrep.2015.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monfort A, et al. Identification of Spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Reports. 2015;12(4):554–561. doi: 10.1016/j.celrep.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma Z, et al. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat Genet. 2001;28(3):220–221. doi: 10.1038/90054. [DOI] [PubMed] [Google Scholar]
- 40.Mercher T, et al. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc Natl Acad Sci USA. 2001;98(10):5776–5779. doi: 10.1073/pnas.101001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mercher T, et al. The OTT-MAL fusion oncogene activates RBPJ-mediated transcription and induces acute megakaryoblastic leukemia in a knockin mouse model. J Clin Invest. 2009;119(4):852–864. doi: 10.1172/JCI35901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su H, et al. Split end family RNA binding proteins: Novel tumor suppressors coupling transcriptional regulation with RNA processing. Cancer Translational Medicine. 2015;1(1):21–25. [Google Scholar]
- 43.Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461(7263):537–541. doi: 10.1038/nature08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horabin JI. Splitting the Hedgehog signal: sex and patterning in Drosophila. Development. 2005;132(21):4801–4810. doi: 10.1242/dev.02054. [DOI] [PubMed] [Google Scholar]
- 45.Stitzinger SM, Conrad TR, Zachlin AM, Salz HK. Functional analysis of SNF, the Drosophila U1A/U2B” homolog: identification of dispensable and indispensable motifs for both snRNP assembly and function in vivo. RNA. 1999;5(11):1440–1450. doi: 10.1017/s1355838299991306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang F, et al. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012;151(4):871–884. doi: 10.1016/j.cell.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staller MV, et al. Depleting gene activities in early Drosophila embryos with the “maternal-Gal4-shRNA” system. Genetics. 2013;193(1):51–61. doi: 10.1534/genetics.112.144915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.