Significance
Complex I is a redox-driven proton pump, central for aerobic energy transduction. We show here by large-scale quantum and classical molecular simulations how reduction of quinone (Q) in the hydrophilic domain of complex I activates the proton pump in the membrane domain. Our simulations indicate that reduction of Q leads to local charge redistributions that trigger conformational changes via an array of alternating charged residues in the membrane domain, nearly 40 Å away. These mechanistic observations are supported by site-directed mutagenesis of a key residue triggering the activation process. The combined data provide molecular insight into how the long-range energy transduction is accomplished by complex I.
Keywords: NADH-quinone oxidoreductase, electron transfer, molecular dynamics simulations, QM/MM simulations, cell respiration
Abstract
Complex I functions as a redox-linked proton pump in the respiratory chains of mitochondria and bacteria, driven by the reduction of quinone (Q) by NADH. Remarkably, the distance between the Q reduction site and the most distant proton channels extends nearly 200 Å. To elucidate the molecular origin of this long-range coupling, we apply a combination of large-scale molecular simulations and a site-directed mutagenesis experiment of a key residue. In hybrid quantum mechanics/molecular mechanics simulations, we observe that reduction of Q is coupled to its local protonation by the His-38/Asp-139 ion pair and Tyr-87 of subunit Nqo4. Atomistic classical molecular dynamics simulations further suggest that formation of quinol (QH2) triggers rapid dissociation of the anionic Asp-139 toward the membrane domain that couples to conformational changes in a network of conserved charged residues. Site-directed mutagenesis data confirm the importance of Asp-139; upon mutation to asparagine the Q reductase activity is inhibited by 75%. The current results, together with earlier biochemical data, suggest that the proton pumping in complex I is activated by a unique combination of electrostatic and conformational transitions.
Complex I (NADH-quinone oxidoreductase) is the largest (550–980 kDa) and one of the most enigmatic enzymes of the electron transport chains of mitochondria and bacteria. It catalyzes electron transfer (eT) from reduced nicotinamide adenine dinucleotide (NADH) to quinone (Q) and couples the reaction to translocation of three to four protons across the membrane (1, 2). The established electrochemical proton gradient is further used to synthesize adenosine triphosphate (ATP) for active transport (3). Due to its central role in cellular respiration, elucidating the catalytic mechanism of complex I is crucial for understanding the molecular principles of biological energy transduction and for unveiling the origins of many mitochondrial disorders (4).
The electrons donated by NADH to complex I are transferred via flavin mononucleotide (FMN) to Q, bound at the lower edge of the hydrophilic domain at a distance of ∼80 Å from the FMN (Fig. 1). The eT process is mediated by seven to eight iron-sulfur (FeS) clusters, depending on the organism, and takes place in ∼100 μs (5). It is believed that the eT process does not couple to proton translocation, which is likely to occur on millisecond timescales (5, 6), but it is rather the oxidoreduction chemistry of the bound Q molecule that drives the proton pump (2, 5–9; cf. ref. 10).
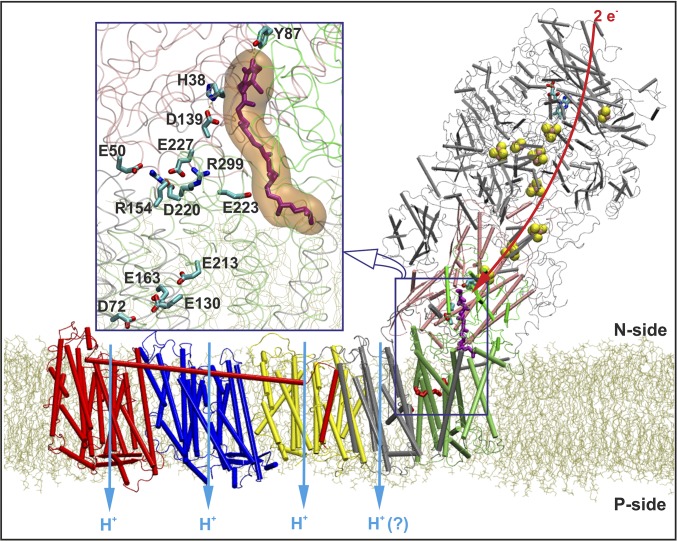
Fig. 1.
Complex I embedded in a lipid bilayer. The membrane bound subunit Nqo8 (in lime) is in close contact with the hydrophilic subunits Nqo4 (in pink) and Nqo6 (in green). The figure also shows the antiporter-like subunits Nqo14 (in yellow), Nqo13 (in blue), Nqo12 (in red), and subunits Nqo7/10/11 (in silver). Other hydrophilic subunits are shown with gray transparent ribbons. The eT path via FMN and FeS clusters to Q (in purple) is indicated with a red arrow, along with proton pumping in membrane-bound subunits (light blue arrows). (Inset) Q (purple) bound in a buried cavity, forming contacts with residues Asp-139, His-38, and Tyr-87 from Nqo4. A cluster of charged residues between the Q headgroup and residues comprising the E-channel are shown.
The proton-pumping machinery of complex I is located in the membrane domain of the enzyme (9) and is responsible for pumping three to four protons across the membrane (Fig. 1) (8, 11). Biochemical and structural studies suggest that the reduction of Q activates the proton pump via a conformational-driven coupling mechanism, accompanied by electrostatic gating (2, 6–8, 12–14). A recent study suggested that water-gated transitions and cooperative electrostatic couplings are also of importance in establishing a functional pump (15). Nevertheless, at the molecular level, the nature of this coupling still remains elusive, especially considering that the electron and proton transfer reactions are separated in both space and time; proton pumping takes place as far as 200 Å from the Q reduction site (1, 2, 9) and on a timescale three orders of magnitude slower than the eT process (5, 6).
In 2013, due to remarkable advances in membrane protein crystallography (16), the crystal structure of the entire complex I from Thermus thermophilus was resolved at a resolution of 3.3 Å (9). This was recently followed up by a crystal structure comprising core subunits of mitochondrial complex I from Yarrowia lipopytica at 3.6-Å resolution (17), revealing both similarities and differences in functionally critical locations (17).
The T. thermophilus structure revealed a membrane-bound Nqo8 subunit that links the hydrophilic eT domain to the remaining membrane subunits. Interestingly, parts of the Nqo8 subunit have structural similarity to the antiporter-like subunits Nqo12/13/14 of the membrane domain. It was therefore suggested that the Nqo8 subunit may participate in the proton translocation process, forming a fourth proton channel across the membrane (9). The bacterial complex I structure also revealed an unusual tunnel-like Q-binding cavity (Fig. 1, Inset), located ∼20 Å above the membrane plane, enclosed by subunits Nqo4, Nqo6, and Nqo8 (9). Although Q was not refined in the deposited structure, Baradaran et al. (9) cocrystallized complex I with Q analogs and suggested that the Q headgroup is located ∼12 Å from the terminal electron donating N2 cluster of the FeS chain, thus allowing for efficient eT (13, 18 and references therein). This finding is consistent with earlier biochemical data, suggesting that the Q headgroup is in hydrogen-bonding contact with Tyr-87 of subunit Nqo4 (19). Moreover, the structural data showed that the Q headgroup is also in hydrogen-bonding contact with His-38 of Nqo4, which in turn is further stabilized by an ion pair with Asp-139 (Fig. 1, the amino acid numbering is based on the structure of complex I from T. thermophilus, unless stated otherwise) (9).
Baradaran et al. (9) suggested that the coupling between the redox reactions at the Q-binding site and proton pumping might be established via a cluster of charged residues, located at the interface of the membrane and hydrophilic subunits. The charged cluster was found to connect to a network of acidic residues located at the center of the membrane subunits. This so-called E-channel, comprising residues Glu-130, Glu-213, and Glu-163 of Nqo8 and Asp-72 of Nqo7, may have an important role in the proton-pumping process (9). In addition, many of these residues are known to be involved in mitochondrial disorders (cf. ref. 9 and references therein), consistent with a central functional role.
Euro et al. (7) originally suggested that the negative charge of reduced quinone (Q-/• or Q2-) is the trigger of the proton-pumping mechanism (2, 6–9, 13, 20), but understanding how the coupling extends across large distances remains unexplained. Recent redox titrations performed on purified complex I from Escherichia coli revealed a tightly bound Q molecule with a midpoint potential (Em,7) of < −300 mV (21, 22). Because the Em,7 of NAD+/NADH is −320 mV, this suggests that the eT from NADH to the bound Q is isoenergetic and that the proton-pumping events are driven by subsequent reactions.
In this work, we combine large-scale computational simulations with biochemical experiments to shed light on the molecular mechanism of complex I. We show by combined hybrid quantum mechanics/molecular mechanics (QM/MM) calculations, large-scale classical molecular dynamics (MD) simulations, and electrostatic calculations that the reduction of bound Q is coupled to its local protonation by a His-Asp ion pair from subunit Nqo4 and that this reaction creates a significant charge imbalance in the Q-binding cavity. We further find that the charge imbalance is transmitted to the membrane subunits by long-range conformational transitions. The observed events are likely to comprise the initial charging steps of the proton pump and may thus provide mechanistic insight into the energy transduction process of complex I. We further test the proposed mechanistic picture by site-directed mutagenesis experiments of a key amino acid residue triggering the activation process. Our combined simulations and experimental results may explain findings from recent structural data (9, 17).
Results
Reduction-Induced Protonation of Q.
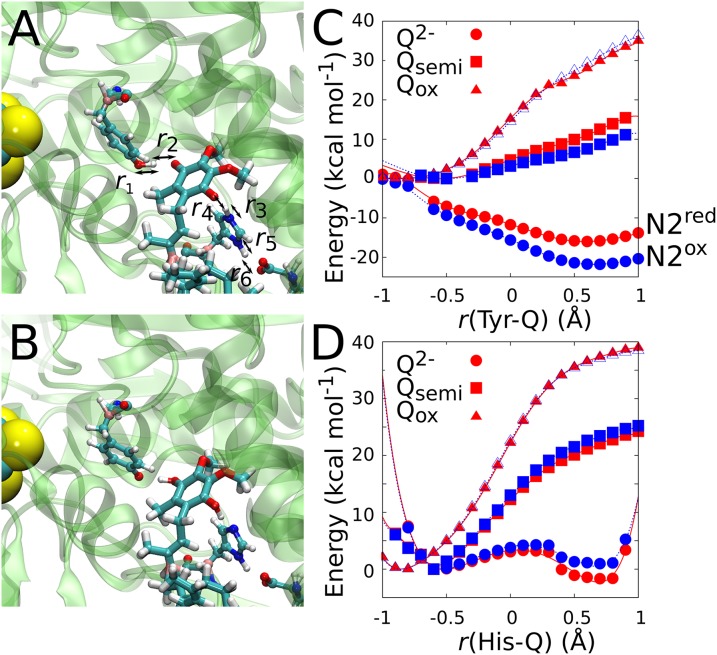
Figs. 2 and 3 show the dynamics of the quinone molecule in the Q-binding pocket obtained from QM/MM simulations with oxidized Q, a singly reduced semiquinone Q•/-, and doubly reduced Q2- states. In both the oxidized and semiquinone states, Tyr-87 remains protonated. The data also suggest that Asp-139 prefers to reside in a protonated state (Fig. 3C), although the proton was initially placed on His-38 (Figs. 2A and 3D). Our observed dynamics are consistent with proton transfer (pT) reaction profiles from these residues to Q, showing high reaction barriers of 12–40 kcal⋅mol−1 in the oxidized and semiquinone states (Fig. 2 C and D). The dynamics of the Q site drastically change in the doubly reduced Q2- state (Figs. 2 and 3), in which protons are rapidly transferred to Q from Tyr-87 and His-38, forming the protonated quinol species (QH2). This process links to local deprotonation of Asp-139 by His-38 through a proton relay process that further stabilizes the QH2 species. The behavior is consistent with the energetics obtained from pT reaction profiles, suggesting that in the Q2- state the pT from Tyr-87 and His-38 are exergonic by ∼15 and 5 kcal⋅mol−1, respectively. The weaker energetic driving on the latter is reflected in a transient backtransfer of the proton from QH2 to His-38 (Fig. 3B). Interestingly, the QM/MM single-point calculations further suggest that the redox state of the N2 cluster modulates the energetics of the pT processes (Fig. 2D), indicating that the protonation state of these residues may also affect the redox potential of the N2 cluster, and vice versa. This is consistent with previous experiments and may contribute to the unusually low Em values for bound Q (21). Our QM/MM data support the hypothesis by Baradaran et al. (9) that the double reduction of Q may lead to its local protonation by the nearby residues.
Fig. 2.
Reduction-triggered proton transfer in the Q-binding site. Snapshots of QM/MM MD simulations of (A) Qox and (B) Q2-. The reduction of Q to Q2- results in deprotonation of Tyr-87 and His-38, coupled to a pT from Asp-139. (C) QM/MM optimizations with r(Tyr-Q) = r1 − r2 and r(His-Q) = r3 − r4 as labeled in A, of the pT between Tyr-87 and Q, and (D) between His-38 and Q. The Q was modeled in the Qox (triangles), Q•/- (squares), and Q2- (circles) states with N2 in reduced (red) and oxidized states (blue).
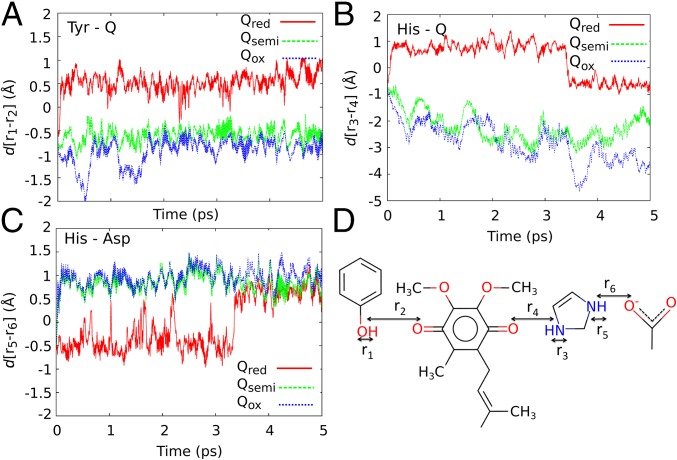
Fig. 3.
Reduction triggered dynamics of the Q binding. pT reactions resolved by pair distances, as labeled in D, in the Qox (blue), Q•/- (green), and Q2- (red) states obtained from QM/MM MD simulations between (A) Tyr-87 and Q; (B) between His-38 and Q; and (C) between His-38 and Asp-139. At ∼3.5 ps of the Q2- state, the proton is transiently transferred back from Q to His-38.
Conformational Changes in the Q-Binding Site.
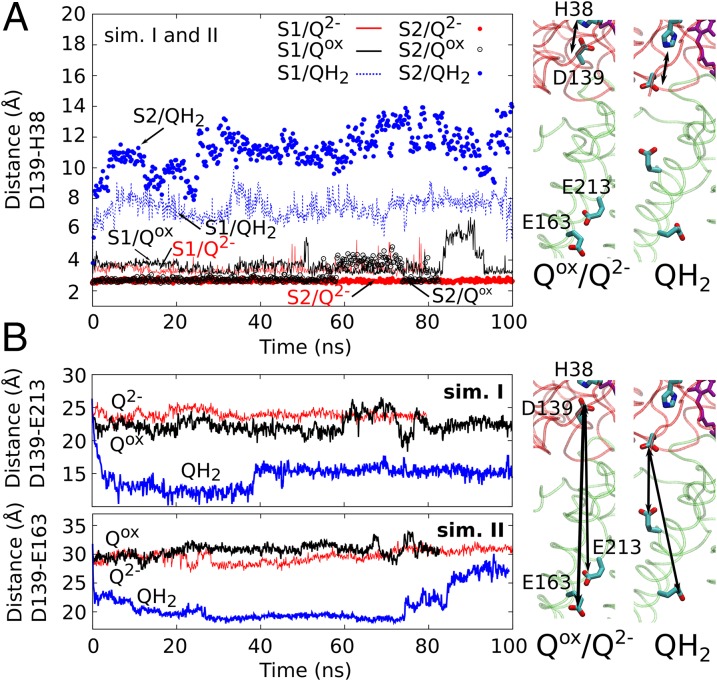
To probe the long timescale dynamics arising from the QH2 formation and the local deprotonation of Tyr-87 and Asp-139, we performed in total ∼1.5 μs of classical MD simulations in different redox and protonation states of the bound Q. Remarkably, in eight independent simulations of reduced and fully protonated QH2 with Asp-139 and Tyr-87 deprotonated, the anionic Asp-139 rapidly dissociates from His-38 (statistics collected from five independent simulations) and undergoes a large conformational change, bringing the side chain about 10–12 Å closer to the membrane domain (Fig. 4A). The observed dissociation of the His-Asp pair is also supported by the recently reported X-ray structure of complex I from Y. lipolytica (17), which is compared with the relaxed QH2 structure in SI Appendix, Fig. S1 (17).
Fig. 4.
Long-range coupling from Q site into membrane domain. (A) Asp-139 remains hydrogen-bonded to His-38 in the Qox and Q2- states, whereas the hydrogen bond dissociates in the QH2 state. (Right) MD snapshots of the Asp-His ion pair from simulations I. (B) Distances between Glu-163 and Glu-213 of Nqo8 and Asp-139 of Nqo4 with the Q. His-38, Asp-139, and Tyr-87 are modeled in the states Qox/HisH+/Asp−/TyrOH, Q2-/HisH+/Asp−/TyrOH, and QH2/His/Asp−/TyrO−. (Top and Bottom) Simulation setups I and II, respectively, with (Right) MD snapshots from simulation I.
Asp-139 resides on a four-helix bundle, which is known to be conformationally flexible based on structural and inhibitor-binding studies (12, 23). A likely reason for the observed behavior is the abstraction of the proton from the Asp-139/His-38 ion pair that links to the Q reduction. This dynamic behavior is found to be independent of the length of the Q isoprene chain, the lipid composition used in the simulations, and the classical charge parameterization model of the redox centers (Materials and Methods). Moreover, a similar dissociation is also observed in a 30-ns vacuum MD simulation of the complex I structure (SI Appendix, Fig. S2), suggesting that this behavior is robust.
To further probe the dissociation mechanism of the Asp-His ion pair, we performed MD simulations on a state corresponding to the situation before eT to the bound oxidized Q (Qox/HisH+/Asp−/TyrOH, where His, Asp, and Tyr refer to the respective residues 38, 139, and 87 of subunit Nqo4) and on a state that may form immediately after the reduction, but before pT from the surrounding residues (Q2-/HisH+/Asp−/TyrOH). In stark contrast to the rapid dissociation of the Asp-139 in the QH2/His/Asp−/TyrO− simulations discussed above, the Asp-139/His-38 ion pair remains intact for the complete simulation time in both alternative states (Fig. 4A). The findings suggest that the dissociation of the Asp-His ion pair and subsequent movement of Asp-139 toward the membrane domain result specifically from the charge imbalance created by the local proton abstraction in the Q-binding site and the associated local structural changes.
The rapid dissociation of the His-Asp ion pair has further interesting consequences. His-38, which is located ∼10 Å from Tyr-87 in the crystal structure (9), moves closer to the Tyr by 5–6 Å. This is in agreement with the recent X-ray structure of complex I from Y. lipolytica, where the analogous residues His-95 and Tyr-144 are within a distance of 4–5 Å (SI Appendix, Fig. S1). The movement of His-38 toward Tyr-87 takes place together with the dissociation of the hydrogen bond between Tyr-87 and the headgroup of the protonated quinol (QH2). Interestingly, these reported changes are not observed in the two other states, Qox/HisH+/Asp−/TyrOH and Q2-/HisH+/Asp−/TyrOH, presumably due to the stability of the HisH+-Asp− ion pair. The observed conformational changes are therefore likely to result from the deprotonation of the His-Asp ion pair induced by the reduction of Q.
The crystal structure of the bacterial complex I was resolved without a Q in its binding site near N2 (9), and, due to the relatively low resolution of the crystal structure, water molecules in the protein interior could not be identified (9). We performed additional classical MD simulations to assess the behavior of the Asp-His pair under such conditions (SI Appendix). When no Q is bound in the cavity, the His-Asp ion pair remains intact throughout the simulation trajectory, providing a good control for the Q-bound simulations described above. Simulations in the Q2-/HisH+/Asp−/TyrOH and QH2/His/Asp−/TyrO− states, with low-energy water molecules modeled in protein interior (SI Appendix), show that the His-Asp ion pair also remains intact in the former state, but rapidly dissociates in the latter state, in agreement with the longer simulations (Fig. 4A).
Site-Directed Mutagenesis Experiments of the His-Asp Ion Pair.
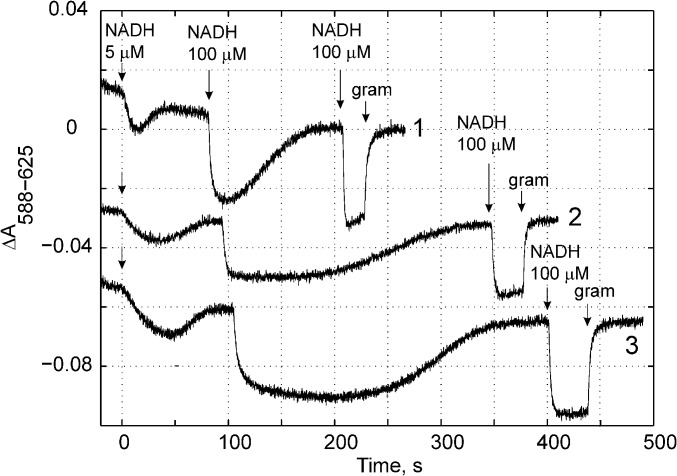
To probe the functional role of Asp-139 in triggering the redox-state–dependent activation of the proton pump (see above), we mutated the corresponding residue in E. coli, Asp-329, to an asparagine. Fig. 5 shows the membrane potential generation (Δψ) of the mutant and wild-type enzymes in liposomes reconstituted with purified complex I (24). For the wild-type enzyme, we find that the Δψ is accelerated after activation with the first NADH pulse (Fig. 5, trace 1), whereas inhibition by 80% of the NADH-ubiquionone oxidoreduction activity with rolliniastatin (Fig. 5, trace 2) decreases the amplitude of the Δψ formation and prolongs the time by which the membrane potential is sustained due to the lower rate of NADH oxidation. We find that the oxidoreduction activity of the D329N variant is significantly lowered to ∼25% of that of the wild-type enzyme (Fig. 5; see also ref. 25). Nevertheless, the maintained Δψ appears comparable to that of the wild-type enzyme, suggesting that the mutant is still able to pump protons across the membrane, but at a much reduced rate. The low activity of the mutant does not, however, allow us to establish the pumping stoichiometry, which may be lower than in the wild-type enzyme. Substantial inhibition of the oxidoreduction activity has also been observed by mutation of His-38 into alanine (26, 27). All these findings suggest that the His-Asp motif is central for the proton-coupled electron transfer function of complex I.
Fig. 5.
Electric potential (Δψ) generation monitored by absorbance changes at 588-625 nm of a potential-sensitive dye, Oxonol VI, in wild type (traces 1 and 2) and NuoCD D329N variant (trace 3) of complex I from E. coli. The Δψ generation was measured as described in ref. 24. The first addition of 5 µM NADH is required for complex I activation. Trace 2 shows the wild-type enzyme in the presence of 100 nM complex I inhibitor rolliniastatin, yielding 20% of wild-type activity. Addition of 2 µg/mL gramicidin (gram) dissipates Δψ. The second 100-µM NADH addition results in a fast Δψ generation followed by its dissipation due to NADH consumption. The generated Δψ can be completely dissipated by addition of gramicidin, after the third addition of NADH. The activity of the NuoCD D329N variant is 25% of the wild-type enzyme.
To understand the 25% residual activity of the mutant, we constructed an Asn-139 mutant in silico and performed MD simulations in the Q2-/His/Asn/TyrOH and QH−/His/Asn/TyrO− states. In contrast to the wild-type system, we modeled His-38 in the neutral state due to the absence of the negatively charged Asp-139. Our QM/MM MD simulations on the D139N mutant support the formation of QH− by deprotonation of Tyr-87 (SI Appendix, Fig. S3). We further find that in classical simulations of the mutant, the His-Asn pair rapidly dissociates in both states, in contrast to the results obtained for the wild-type simulations (SI Appendix, Fig. S4). Before proton transfer to Q, both His-38 and Tyr-87 remain strongly hydrogen-bonded to the Q2- species throughout the simulation trajectory, whereas after pT to Q, the QH− molecule drifts away from Tyr-87 toward the membrane by ∼15 Å (SI Appendix, Fig. S4), suggesting that there is a critical difference in the dynamics of the Q relative to the wild type. It is thus possible that, in the Asn-mutant enzyme, movement of the QH− species replaces the functional role of the anionic Asp-139 flip, resulting in the observed residual Q reductase and pumping activity.
Conformational Changes in the Membrane Subunit Nqo8.
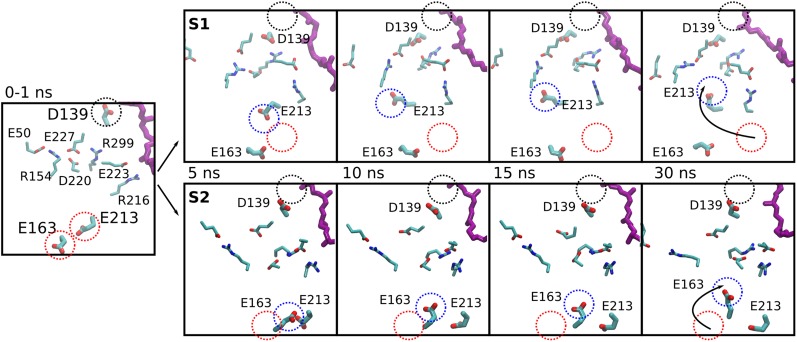
We find that the dissociation of Asp-139 from the Q-binding site leads to a large conformational rearrangement in the highly conserved E-channel region of the Nqo8 subunit, comprising a “quartet” of charged residues: Glu-130, Glu-213, Glu-163, and Asp-72 of Nqo7 (9) (Fig. 4B). This is a remarkable transition, considering that the region is located nearly 40 Å away from the Q site in the crystal structure. In two independent MD simulations, we observe that Glu-213 (in simulation S1) and Glu-163 (in simulation S2), which are located within ∼7 Å from each other in the X-ray structure and 35–40 Å from the Q headgroup, undergo large conformational changes; within 10 ns the residues flip toward Asp-139 and the N-side of the membrane, coming as close as 10 Å from Asp-139 (Fig. 4B). In contrast, in simulations of states with an oxidized Q or Q2- before its protonation, the distance between Glu-163/Glu-213 and Asp-139 remains close to the distances observed in the X-ray structure (∼30–35 Å, Fig. 4B).
To unveil how the conformational transitions in the Nqo8 subunit may lead to protonation changes of the E-channel residues, we performed continuum electrostatic calculations based on MD simulation snapshots. The pKa calculations suggest that the conformational changes lead to a drastic increase in the pKa of residues Glu-213/Glu-163, indicating a strong driving force for proton uptake at ∼25–40 ns in the two simulation trajectories (SI Appendix, Figs. S5 and S6). The calculations also suggest that residues Asp-72 and Glu-130 are likely to be protonated in the conformations sampled by the MD simulations. We therefore performed additional MD simulations with Glu-130 and Asp-72 protonated. The simulation shows that the residues in subunits Nqo4 and Nqo8 undergo similar conformational transitions as observed in longer simulations with all E-channel residues deprotonated (Fig. 4), suggesting that the observed redox-state–induced conformational changes in subunit Nqo8 results from the charge imbalance at the Q site.
Our simulations show that the conformational transitions in subunits Nqo4 and Nqo8 are reversible, which is critical to minimize energy dissipation in a thermodynamically efficient proton pump. As shown in Fig. 4B, after staying in an “up” conformation for nearly 50 ns, Glu-163 returns back to its crystallographic position at around 80 ns. In yet another simulation of the Q2-/HisH+/Asp−/TyrOH state (setup I), starting from a state with a dissociated HisH+-Asp− ion pair, the latter reversibly reforms within ∼15 ns of simulation time (SI Appendix, Fig. S7), further supporting the reversibility of the transition.
Electrostatic Coupling Elements.
The simulations of the alternative Q states reveal how the distant conformational transitions in Nqo4 near the Q headgroup and Nqo8 within the membrane could be linked. The simulations suggest that the dissociation of the anionic Asp-139 from the local Q-binding site disturbs a network of charged residues in subunit Nqo8, consisting of Arg-154, Asp-220, Glu-227, and Arg-299 (Figs. 1 and 6). The down-flip of the anionic Asp-139 causes Arg-154 and Arg-299 to rapidly dissociate from their initial crystallographic positions and to form ion pairs with Glu-50 in Nqo4 and Glu-223 in Nqo8, respectively. These structural perturbations involving rearrangement of Arg-299, Arg-216, and Arg-154 in subunit Nqo8 are likely to cause the glutamates in the E-channel to flip “up” and move closer to the N-side of the membrane (Fig. 6). A similar perturbation of this “dipolar cluster” is also observed in the Asp/Asn mutant simulation, in which the negatively charged QH− species is responsible for the transmitted conformational changes.
Fig. 6.
E-quartet and Q-site coupling. Simulation snapshots highlighting the long-range conformational transitions in complex I. Simulation snapshots are taken at 0–1, 5, 10, 15, and 30 ns from the 100-ns simulations (systems S1, Top; S2, Bottom) of the QH2 state. The initial position of Asp-139 is marked with a black dotted circle. Initial crystallographic positions and relaxed MD positions of Glu-213/Glu-163 are marked with red and blue dotted circles, respectively.
Together with the up-flip of the negatively charged E-channel residues, the anionic residues Asp-139 of Nqo4 and Glu-227 of Nqo8 create excess negative charge in this region, which may further lead to protonation of the E-channel residues. Indeed, electrostatic calculations on simulation snapshots show that, in conformations where E-channel residues are in the “up” conformation, the protonation probability of these residues increases drastically (SI Appendix, Table S1).
To assess how such protonation of the E-channel residues might alter their dynamics, we also performed MD simulations with Glu-163 protonated. We observe that when Glu-163 is protonated in its “up” conformation resulting from the Asp-139 dissociation, it flips “down” within a few nanoseconds of simulation time (SI Appendix, Fig. S8, Inset). In contrast, the residue retains its crystallographic conformation when the simulation is initiated with a protonated species (SI Appendix, Fig. S8), suggesting that the residue may be protonated in the likely resting state of the crystal structure. Overall, the protonation probability of the E-channel residues and the reversibility of the conformational transitions strongly suggest that these residues have an important role in proton pumping of complex I.
Discussion
We have used multiscale computational approaches and site-directed mutagenesis experiments of a key residue to address the structure, function, and dynamics of the respiratory complex I. Our combined findings suggest that the initial activation steps involve a charge imbalance arising from the Q reduction in the soluble domain that leads to a local proton-coupled electron transfer process in the Q-binding site. The effect of the excess charge is transmitted by concerted side-chain reorientations of charged residues at the interface of the soluble and membrane domains, leading to a contact between glutamates in the Nqo8 subunit with the N-side of the membrane and proton uptake from the bulk. The observed side-chain rotations have an interesting resemblance to the dynamics of a key acidic residue in the d-channel of cytochrome c oxidase, Glu-242 (28, 29). Previous site-directed mutagenesis experiments support the functional importance of the identified E-channel residues in the activation process (30–32) (SI Appendix, Table S2 and Fig. S9). Moreover, a recent computational study (15) suggested that the protonation state of buried carboxylates in the NuoN/K interface may control the transient formation of proton-conducting water wires from the bulk to buried titratable residues in NuoN. Interestingly, these residues are located only 15 Å from the conformationally active glutamate residues identified here and might thus control the protonation state of the residues near the NuoN/K interface. Such a coupling element could mediate the charge imbalance arising in the Q site all of the way to the antiporter-like subunits. Our findings suggest that unlike many energy-transducing enzymes that use primarily local electrostatic changes to drive proton pumping (33, 34), complex I may operate by a combination of electrostatic and conformational transitions.
Materials and Methods
MD Simulations.
The X-ray structure of complex I from T. thermophilus was obtained from the Protein Databank (PDB ID: 4HEA) (9). Two independent simulations setups were constructed: in model I, the protein was solvated in a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine membrane, TIP3P water molecules, and Na+ and Cl− ions, mimicking a 100-mM salt concentration. In model II, we used a mixed lipid bilayer model comprising 4-linoleic cardiolipin, 1,2-dilinoleyl-sn-glycero-3-phosphoethanolamine and -phosphatidylcholine lipids, in addition to TIP3P water molecules and Na+ and Cl− ions. The Q-binding site was modeled using Q10 and Q6 in the respective system setups I and II, comprising in total nearly 1 million (809,413–868,500) atoms. The Q headgroup was docked between Tyr-87 and His-38 of subunit Nqo4, and the isoprenoid tail was placed in the long tunnel-like cavity, identified using the HOLE program (35). Q was modeled in the quinol (QH2), doubly and singly anionic quinone (Q2- and QH−), and oxidized quinone (Qox) states. A short segment of the loop from subunit Nqo6 and two loops of subunit Nqo3 were modeled using MODELER (36). The MD simulations were performed using the CHARMM force field (37, 38) with parameters for the cofactors calculated using TURBOMOLE (39) or obtained from refs. 40–43. (SI Appendix, Tables S3 and S4). MD simulations with different redox and protonation states of the Q-binding site were performed using NAMD2 (44) at constant temperature (T = 310 K) and pressure (P = 1 atm) with 1 fs (model II) or 2 fs (model I) time steps and treating the long-range electrostatics with the Particle Mesh Ewald approach. The total simulation time for all studied systems was ∼1.5 µs. MD trajectories were analyzed in visual molecular dynamics (45). pKa values were estimated by continuum electrostatics calculations on structures obtained from the MD simulations using Karlsberg+ (46, 47), adaptive Poisson–Boltzmann solver (48), and Karlsberg 2 (49) with the protein modeled using explicit atoms and a polarizable medium with ε = 4. The details of the simulation setups are discussed in SI Appendix.
QM/MM Simulations.
Hybrid QM/MM simulations of the Qox, Q•/-, and Q2- states were performed with Nqo4 residues His-38, Tyr-87, Asp-139, and Thr-135 in addition to the Q headgroup up to the first isoprene unit in the QM region (SI Appendix, Table S5). Link atoms were introduced between the Cα and Cβ atoms of each residue and between atoms the C9 and C11 of Q. Starting structures were obtained from a 10-ns relaxed MD structure complex I, obtained by using harmonic restraints of 1 kcal⋅mol−1⋅Å-2 on the Cα atoms. The classical (MM) region was further trimmed for the QM/MM calculations to include subunits Nqo4-Nqo8. The QM region was described at the B3LYP/def2-SVP level (50–52), and the MM region using the CHARMM27 force field (37). Five picoseconds of unrestrained QM/MM MD simulations were performed at constant temperature (T = 310 K) using a 1-fs timestep in Q-Chem/CHARMM (53–55). The energetics of the pT process in different redox states was additionally studied using constrained QM/MM optimizations, with single-point energy evaluations using def2-TZVP basis sets (52).
Experimental Procedures.
Complex I from E. coli was isolated, purified, and reconstituted into liposomes as described in ref. (24). The conditions for the membrane potential assay were 50 mM Hepes–BTP [1,3-bis(Tris(hydroxymethyl)methylamino) propane], pH 7.0, 300 mM mannitol, 1 mM MgSO4, 5 mM, NH4Cl, 0.1 µM cytochrome bo3, 3 µM oxonol VI, proteoliposomes (∼40 µg complex I/mL), and 100 µM decylubiquinone. Mutagenesis of nuoH was performed using the QuikChange Lightning SiteDirected Mutagenesis Kits (Stratagene) and mutagenic oligonucleotides from Eurofins MWG Operon. The template plasmid contained the full-length nuoH gene with up- and downstream flanks and was used for restitution of the mutated gene to the chromosome of the nuoH-deficient E. coli strain using the pKO3 gene replacement system.
Supplementary Material
Acknowledgments
The authors are grateful for the computing time provided by the high-performance Biowulf cluster at the National Institutes of Health and the Center for Scientific Computing–the IT Center for Science Ltd., Finland. We acknowledge that part of the results of this research have been achieved using the PRACE-3IP project (FP7 RI-312763) resource Lindgren based in Sweden at Kungliga Tekniska Högskolan (KTH). V.S. acknowledges the Academy of Finland for postdoctoral researcher funding. This work was supported by the Max Planck Society (G.H.) and by grants from the Sigrid Jusélius Foundation (to M.W. and I.V.), the Biocentrum Helsinki (to M.W.), the Academy of Finland (to M.W., I.V., T.R.), the Center of Excellence in Biomembrane Research, Academy of Finland (to I.V., T.R., V.S.), European Research Council Advanced Grant CROWDED-PRO-LIPIDS (to I.V.), and the German Research Foundation and the Jane and Aatos Erkko Foundation (V.R.I.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1503761112/-/DCSupplemental.
References
- 1.Brandt U. Energy converting NADH:quinone oxidoreductase (complex I) Annu Rev Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 2.Verkhovskaya M, Bloch DA. Energy-converting respiratory Complex I: On the way to the molecular mechanism of the proton pump. Int J Biochem Cell Biol. 2013;45(2):491–511. doi: 10.1016/j.biocel.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 3.Yoshida M, Muneyuki E, Hisabori T. ATP synthase: A marvellous rotary engine of the cell. Nat Rev Mol Cell Biol. 2001;2(9):669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- 4.Mimaki M, Wang X, McKenzie M, Thorburn DR, Ryan MT. Understanding mitochondrial complex I assembly in health and disease. Biochim Biophys Acta. 2012;1817(6):851–862. doi: 10.1016/j.bbabio.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Verkhovskaya ML, Belevich N, Euro L, Wikström M, Verkhovsky MI. Real-time electron transfer in respiratory complex I. Proc Natl Acad Sci USA. 2008;105(10):3763–3767. doi: 10.1073/pnas.0711249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandt U. A two-state stabilization-change mechanism for proton-pumping complex I. Biochim Biophys Acta. 2011;1807(10):1364–1369. doi: 10.1016/j.bbabio.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Euro L, Belevich G, Verkhovsky MI, Wikström M, Verkhovskaya M. Conserved lysine residues of the membrane subunit NuoM are involved in energy conversion by the proton-pumping NADH:ubiquinone oxidoreductase (Complex I) Biochim Biophys Acta. 2008;1777(9):1166–1172. doi: 10.1016/j.bbabio.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Wikström M, Hummer G. Stoichiometry of proton translocation by respiratory complex I and its mechanistic implications. Proc Natl Acad Sci USA. 2012;109(12):4431–4436. doi: 10.1073/pnas.1120949109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature. 2013;494(7438):443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Vries S, Dörner K, Strampraad MJF, Friedrich T. Electron tunneling rates in respiratory complex I are tuned for efficient energy conversion. Angew Chem Int Ed Engl. 2015;54(9):2844–2848. doi: 10.1002/anie.201410967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wikström M. Two protons are pumped from the mitochondrial matrix per electron transferred between NADH and ubiquinone. FEBS Lett. 1984;169(2):300–304. doi: 10.1016/0014-5793(84)80338-5. [DOI] [PubMed] [Google Scholar]
- 12.Berrisford JM, Sazanov LA. Structural basis for the mechanism of respiratory complex I. J Biol Chem. 2009;284(43):29773–29783. doi: 10.1074/jbc.M109.032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohnishi ST, Salerno JC, Ohnishi T. Possible roles of two quinone molecules in direct and indirect proton pumps of bovine heart NADH-quinone oxidoreductase (complex I) Biochim Biophys Acta. 2010;1797(12):1891–1893. doi: 10.1016/j.bbabio.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pohl T, et al. Nucleotide-induced conformational changes in the Escherichia coli NADH:ubiquinone oxidoreductase (complex I) Biochem Soc Trans. 2008;36(Pt 5):971–975. doi: 10.1042/BST0360971. [DOI] [PubMed] [Google Scholar]
- 15.Kaila VRI, Wikström M, Hummer G. Electrostatics, hydration, and proton transfer dynamics in the membrane domain of respiratory complex I. Proc Natl Acad Sci USA. 2014;111(19):6988–6993. doi: 10.1073/pnas.1319156111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sazanov LA, Baradaran R, Efremov RG, Berrisford JM, Minhas G. A long road towards the structure of respiratory complex I, a giant molecular proton pump. Biochem Soc Trans. 2013;41(5):1265–1271. doi: 10.1042/BST20130193. [DOI] [PubMed] [Google Scholar]
- 17.Zickermann V, et al. Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I. Science. 2015;347(6217):44–49. doi: 10.1126/science.1259859. [DOI] [PubMed] [Google Scholar]
- 18.Moser CC, Farid TA, Chobot SE, Dutton PL. Electron tunneling chains of mitochondria. Biochim Biophys Acta. 2006;1757(9-10):1096–1109. doi: 10.1016/j.bbabio.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 19.Tocilescu MA, Fendel U, Zwicker K, Kerscher S, Brandt U. Exploring the ubiquinone binding cavity of respiratory complex I. J Biol Chem. 2007;282(40):29514–29520. doi: 10.1074/jbc.M704519200. [DOI] [PubMed] [Google Scholar]
- 20.Efremov RG, Sazanov LA. The coupling mechanism of respiratory complex I - a structural and evolutionary perspective. Biochim Biophys Acta. 2012;1817(10):1785–1795. doi: 10.1016/j.bbabio.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 21.Verkhovsky M, Bloch DA, Verkhovskaya M. Tightly-bound ubiquinone in the Escherichia coli respiratory complex I. Biochim Biophys Acta. 2012;1817(9):1550–1556. doi: 10.1016/j.bbabio.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 22.Verkhovskaya M, Wikström M. Oxidoreduction properties of bound ubiquinone in Complex I from Escherichia coli. Biochim Biophys Acta. 2014;1837(2):246–250. doi: 10.1016/j.bbabio.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Masuya T, Murai M, Ifuku K, Morisaka H, Miyoshi H. Site-specific chemical labeling of mitochondrial respiratory complex I through ligand-directed tosylate chemistry. Biochemistry. 2014;53(14):2307–2317. doi: 10.1021/bi500205x. [DOI] [PubMed] [Google Scholar]
- 24.Verkhovskaya M, Knuuti J, Wikström M. Role of Ca(2+) in structure and function of Complex I from Escherichia coli. Biochim Biophys Acta. 2011;1807(1):36–41. doi: 10.1016/j.bbabio.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Sinha PK, et al. Conserved amino acid residues of the NuoD segment important for structure and function of Escherichia coli NDH-1 (complex I) Biochemistry. 2015;54(3):753–764. doi: 10.1021/bi501403t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belevich G, Euro L, Wikström M, Verkhovskaya M. Role of the conserved arginine 274 and histidine 224 and 228 residues in the NuoCD subunit of complex I from Escherichia coli. Biochemistry. 2007;46(2):526–533. doi: 10.1021/bi062062t. [DOI] [PubMed] [Google Scholar]
- 27.Grgic L, Zwicker K, Kashani-Poor N, Kerscher S, Brandt U. Functional significance of conserved histidines and arginines in the 49-kDa subunit of mitochondrial complex I. J Biol Chem. 2004;279(20):21193–21199. doi: 10.1074/jbc.M313180200. [DOI] [PubMed] [Google Scholar]
- 28.Kaila VRI, Verkhovsky MI, Hummer G, Wikström M. Glutamic acid 242 is a valve in the proton pump of cytochrome c oxidase. Proc Natl Acad Sci USA. 2008;105(17):6255–6259. doi: 10.1073/pnas.0800770105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaila VRI, Verkhovsky MI, Hummer G, Wikström M. Mechanism and energetics by which glutamic acid 242 prevents leaks in cytochrome c oxidase. Biochim Biophys Acta. 2009;1787(10):1205–1214. doi: 10.1016/j.bbabio.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 30.Sinha PK, et al. Critical roles of subunit NuoH (ND1) in the assembly of peripheral subunits with the membrane domain of Escherichia coli NDH-1. J Biol Chem. 2009;284(15):9814–9823. doi: 10.1074/jbc.M809468200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurki S, Zickermann V, Kervinen M, Hassinen I, Finel M. Mutagenesis of three conserved Glu residues in a bacterial homologue of the ND1 subunit of complex I affects ubiquinone reduction kinetics but not inhibition by dicyclohexylcarbodiimide. Biochemistry. 2000;39(44):13496–13502. doi: 10.1021/bi001134s. [DOI] [PubMed] [Google Scholar]
- 32.Valentino ML, et al. The ND1 gene of complex I is a mutational hot spot for Leber’s hereditary optic neuropathy. Ann Neurol. 2004;56(5):631–641. doi: 10.1002/ana.20236. [DOI] [PubMed] [Google Scholar]
- 33.Kaila VRI, Verkhovsky MI, Wikström M. Proton-coupled electron transfer in cytochrome oxidase. Chem Rev. 2010;110(12):7062–7081. doi: 10.1021/cr1002003. [DOI] [PubMed] [Google Scholar]
- 34.Wikström M, Sharma V, Kaila VRI, Hosler JP, Hummer G. New perspectives on proton pumping in cellular respiration. Chem Rev. 2015;115(5):2196–2221. doi: 10.1021/cr500448t. [DOI] [PubMed] [Google Scholar]
- 35.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MSP. HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14(6):354–360, 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 36.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 37.MacKerell AD, Jr, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 38.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114(23):7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahlrichs R, Bär M, Häser M, Horn H, Kölmel C. Electronic structure calculations on workstation computers: The program system turbomole. Chem Phys Lett. 1989;162(3):165–169. [Google Scholar]
- 40.Postila PA, et al. Key role of water in proton transfer at the Qo-site of the cytochrome bc1 complex predicted by atomistic molecular dynamics simulations. Biochim Biophys Acta. 2013;1827(6):761–768. doi: 10.1016/j.bbabio.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 41.Chang CH, Kim K. Density functional theory calculation of bonding and charge parameters for molecular dynamics studies on [FeFe] hydrogenases. J Chem Theory Comput. 2009;5(4):1137–1145. doi: 10.1021/ct800342w. [DOI] [PubMed] [Google Scholar]
- 42.Freddolino PL, Dittrich M, Schulten K. Dynamic switching mechanisms in LOV1 and LOV2 domains of plant phototropins. Biophys J. 2006;91(10):3630–3639. doi: 10.1529/biophysj.106.088609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vagedes P, Rabenstein B, Åqvist J, Marelius J, Knapp EW. The deacylation step of acetylcholinestrase: Computer simulation studies. J Am Chem Soc. 2000;122(49):12254–12262. [Google Scholar]
- 44.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 46.Kieseritzky G, Knapp EW. Optimizing pKa computation in proteins with pH adapted conformations. Proteins. 2008;71(3):1335–1348. doi: 10.1002/prot.21820. [DOI] [PubMed] [Google Scholar]
- 47.Kieseritzky G, Knapp EW. Improved pK(a) prediction: Combining empirical and semimicroscopic methods. J Comput Chem. 2008;29(15):2575–2581. doi: 10.1002/jcc.20999. [DOI] [PubMed] [Google Scholar]
- 48.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rabenstein B, Knapp EW. Calculated pH-dependent population and protonation of carbon-monoxy-myoglobin conformers. Biophys J. 2001;80(3):1141–1150. doi: 10.1016/S0006-3495(01)76091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Becke AD. Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98(7):5648–5652. [Google Scholar]
- 51.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B Condens Matter. 1988;37(2):785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 52.Weigend F, Ahlrichs R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys Chem Chem Phys. 2005;7(18):3297–3305. doi: 10.1039/b508541a. [DOI] [PubMed] [Google Scholar]
- 53.Shao Y, et al. Advances in methods and algorithms in a modern quantum chemistry program package. Phys Chem Chem Phys. 2006;8(27):3172–3191. doi: 10.1039/b517914a. [DOI] [PubMed] [Google Scholar]
- 54.Woodcock HL, 3rd, et al. Interfacing Q-Chem and CHARMM to perform QM/MM reaction path calculations. J Comput Chem. 2007;28(9):1485–1502. doi: 10.1002/jcc.20587. [DOI] [PubMed] [Google Scholar]
- 55.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.