Significance
The molecular organization of eukaryotic mechanosensory channels is largely uncharacterized. This characterization, having the correct parts list, is a necessary beginning step toward understanding how the channel transduces mechanical signals. Here we investigate the organization of the degenerin/epithelial sodium channel (DEG/ENaC) mechanosensory transduction channel in the six touch receptor neurons of Caenorhabditis elegans. Previous work has suggested that four membrane proteins formed a channel complex: the DEG/ENaC proteins MEC-4 and MEC-10 form the channel pore and the stomatin-like protein MEC-2 and paraoxonase-like protein MEC-6 act as auxiliary proteins. Using single molecule imaging, we find that the transduction complex is simpler, being a MEC-4:MEC-4:MEC-10 trimer. In contrast to the previous suggestion, this trimer does not appear to associate substantially with MEC-2 or MEC-6.
Keywords: mechanosensory channels, DEG/ENaC proteins, channel stoichiometry
Abstract
Caenorhabditis elegans senses gentle touch in the six touch receptor neurons (TRNs) using a mechanotransduction complex that contains the pore-forming degenerin/epithelial sodium channel (DEG/ENaC) proteins MEC-4 and MEC-10. Past work has suggested these proteins interact with the paraoxonase-like MEC-6 and the cholesterol-binding stomatin-like MEC-2 proteins. Using single molecule optical imaging in Xenopus oocytes, we found that MEC-4 forms homotrimers and MEC-4 and MEC-10 form 4:4:10 heterotrimers. MEC-6 and MEC-2 do not associate tightly with these trimers and do not influence trimer stoichiometry, indicating that they are not part of the core channel transduction complex. Consistent with the in vitro data, MEC-10, but not MEC-6, formed puncta in TRN neurites that colocalize with MEC-4 when MEC-4 is overexpressed in the TRNs.
Few of the sensory transduction molecules needed to detect touch, sound, and other mechanical stimuli are known (1–4), and the molecular organization of most of those that are known has not been studied. In Caenorhabditis elegans, for example, gentle touch is transduced in the six touch receptor neurons (TRNs) by a channel formed from the degenerin/epithelial sodium channel (DEG/ENaC) proteins MEC-4 and MEC-10 (5, 6; MEC derives from “mechanosensory abnormal,” the name of the gene class), but the exact nature of the transduction complex is not known.
Previous experiments suggested that two other membrane proteins, the stomatin-like protein MEC-2 (7) and the paraoxonase-like protein MEC-6 (8), contributed to the transduction complex. First, MEC-2, MEC-6, and MEC-4 are essential for the production of the transduction current, whereas MEC-10 has relatively minor effects on it (5, 6). Second, MEC-2 and MEC-6 increased the activity of MEC-4(d) channels 5 d after injection of their cRNAs into Xenopus oocytes without apparently changing the amount of surface-localized MEC-4(d) protein (8–11). [The dominant mec-4(d) mutation, which results in an A713T substitution, produces a hyperactive channel that is constitutively active (9, 12, 13) and leads to TRN degeneration.] Third, MEC-2 and MEC-6 coimmunoprecipitated with MEC-4(d), MEC-10, and each other in heterologous cells (8–10). Fourth, antibodies to MEC-2 and FLAG-tagged MEC-6 labeled puncta in vivo, which appeared to colocalize with MEC-4::YFP (8, 10).
Recently, however, other observations made us reinvestigate this model. First, although MEC-6 did not affect MEC-4 abundance in Xenopus oocytes, loss of mec-6 in vivo led to a drastic reduction in MEC-4::YFP expression in the TRNs (8). Second, studies in Drosophila melanogaster found that, although DEG/ENaC proteins needed for mechanosensation were present (14–18), obvious MEC-6-like proteins were not (19). Third, we wondered about the importance of the puncta and the apparent colocalization of the proteins in them, because in vivo electrophysiological studies of the TRNs (5) showed that the estimated number of active channels equaled the number of puncta, but single channels were unlikely to be visible by fluorescence microscopy. Moreover, MEC-10::GFP did not form puncta in vivo (20).
Here we study the interactions and stoichiometry of the proteins of the TRN transduction channel complex. Our results suggest that the ENaC proteins form both a MEC-4 homotrimer and a MEC-4:MEC-4:MEC-10 heterotrimer and that this core channel complex does not form a high-affinity complex with either MEC-2 or MEC-6.
Results
Association and Stoichiometry of the MEC-4 Channel Complex in Xenopus Oocytes.
We examined the association and stoichiometry of the proteins of the MEC-4 channel complex using single molecule optical imaging (21, 22) in Xenopus oocytes. We tagged the MEC-2, 4, 6, and 10 proteins at their N or C termini with EGFP and/or mCherry. Because MEC-4 does not form an open channel in oocytes, we used tagged-MEC-4(d), which forms a constitutively open channel (9) to test the functionality of these proteins. The tagged proteins and untagged proteins functioned similarly, with MEC-4(d) inducing current, coexpressed MEC-2 and MEC-6 boosting the current and MEC-10 inhibiting the current (Fig. S1), as shown earlier (8, 9). In addition, the MEC-4::TagRFP and MEC-4::GFP each restored touch sensitivity to the mec-4 null mutant, mec-4(u253) (SI Materials and Methods).
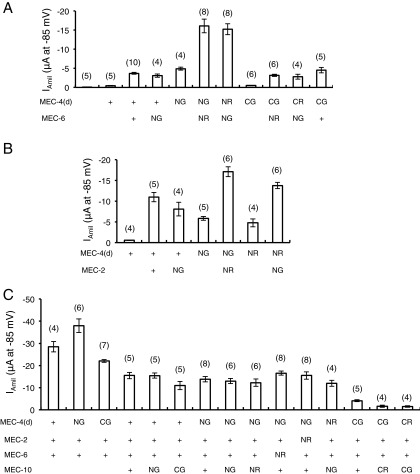
Fig. S1.
The effect of N- or C-terminal EGFP/mCherry-tagged MEC-4(d) (A–C), MEC-6 (A and C), MEC-2 (B and C), and MEC-10 (C) on the amiloride-sensitive current (IAmil) at −85 mV in Xenopus oocytes (mean ± SEM). The number of tested oocytes is given in parentheses. G, EGFP; R, mCherry; N and C, whether the tag is at the N or C terminus of the MEC protein, respectively; +, untagged MEC proteins.
To examine the molecular composition of the MEC-4 channel complex, we expressed the EGFP-tagged MEC-4 at low density on the plasma membrane of oocytes so that each fluorescent spot represented a single complex. We photobleached an area of membrane containing 50–200 spots and counted the number of photobleaching steps in individual spots to determine the number of EGFP-tagged molecules in each complex. Total internal reflection fluorescence microscopy (TIRF) imaging does not discriminate between functional and nonfunctional proteins or protein complexes. Nonetheless, most, if not all, of the observed spots represent functional proteins because they have been transported to the cell surface. In addition, electrophysiological evidence (Fig. S1) suggests that the surface proteins are functional.
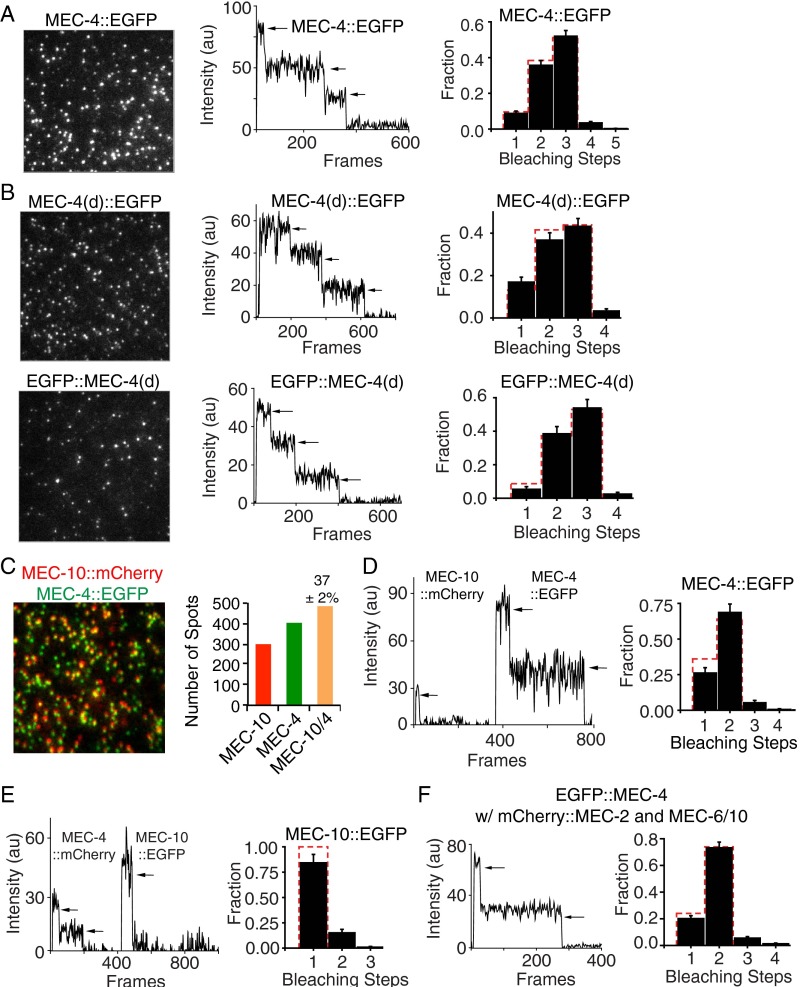
When MEC-4 is expressed alone, both N- and C-terminally tagged versions of MEC-4 were immobile and yielded photobleaching steps that fit the expected binomial distribution for a homotrimer (Fig. 1A, Fig. S2A, and Movie S1). Similarly, tagged MEC-4(d) also formed a homotrimeric channel (Fig. S2B). Because both MEC-4 WT and MEC-4(d) had the same stoichiometry, we used the tagged MEC-4 WT for rest of the single molecule optical imaging experiments in oocytes. The stoichiometry of MEC-10 expressed on its own could not be determined because it was too mobile on the oocyte surface (Movie S2). In contrast, coexpression of MEC-10 along with MEC-4 produced immobile spots of MEC-10 (Movie S3). We analyzed the relative localization of MEC-4 and MEC-10 and found a high level of colocalization, whether the proteins were N- or C-terminally tagged (Fig. 1B and Fig. S2C). We counted photobleaching steps in the spots that showed MEC-4/MEC-10 colocalization and found that they contained two molecules of MEC-4 and one of MEC-10 (Fig. 1C and Fig. S2 D and E). Thus, the total number of subunits in the complex remains three even in heteromers.
Fig. 1.
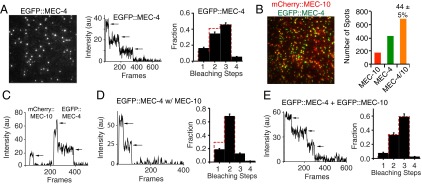
The stoichiometry and colocalization of the MEC-4/MEC-10 channel complex on the plasma membrane of Xenopus oocytes. (A) Single-molecule irreversible photobleaching indicates EGFP::MEC-4 forms trimers on its own. (Left) TIRF image of EGFP::MEC-4 (here and in all other TIRF images, the field is 13 × 13 μm). (Center) Fluorescence traces of a EGFP::MEC-4 complex yielding three bleaching steps (arrows). (Right) Observed frequency distributions of the number of bleaching steps (black bars) and the predicted binomial distribution (red dotted bars) for trimers here and in Figs. 1E and 2 D and G and Fig. S2 A and B (for dimers in Figs. 1D and 2F and Fig. S2 D and F). The errors in the subunit counting data are given by I/N*√n (n = total number of spots for each step; N = total number of spots for all steps). (B) The colocalization of mCherry::MEC-10 and EGFP::MEC-4. (Left) Representative image. (Right) Number of spots with MEC-10 alone (red), MEC-4 alone (green), and both proteins (orange). Here and in Figs. 2 A–C and Fig. S2C, the colocalization fraction after subtracting random colocalization is given as the mean ± SEM. (C) Fluorescence traces showing the photobleaching of a MEC-4:MEC-4:MEC-10 heterotrimer. (D) An example (Left) and quantification (Right) of the photobleaching of EGFP::MEC-4 in the presence of untagged MEC-10. (E) An example (Left) and quantification (Right) of the photobleaching of EGFP::MEC-4/10 heterotrimer.
Fig. S2.
The stoichiometry and colocalization of MEC-4 and MEC-10 in Xenopus oocytes. (A and B) MEC-4::EGFP, MEC-4(d)::EGFP, and EGFP::MEC-4(d) forms trimers on its own. (Left) TIRF image of MEC-4::EGFP, MEC-4(d)::EGFP, or EGFP::MEC-4(d). (Center) Example of a MEC-4::EGFP, MEC-4(d)::EGFP, or EGFP::MEC-4(d) complex yielding three bleaching steps. (Right) Quantification of their photobleaching steps. (C) The colocalization of MEC-10::mCherry and MEC-4::EGFP. (Left) Representative image. (Right) Number of spots with MEC-10 alone (red), MEC-4 alone (green), and both proteins (orange). The colocalization fraction after subtracting random colocalization is given as the mean ± SEM. (D and E) An example (Left) and quantification (Right) of the photobleaching of the 4:4:10 heterotrimer. (F) An example (Left) and quantification (Right) of the photobleaching of EGFP::MEC-4 in the presence of mCherry::MEC-2, untagged MEC-10, and MEC-6.
Further evidence for a 4:4:10 heterotrimer combination came from two experiments. First we found that, in contrast to the three subunit counts from EGFP-tagged MEC-4 alone (Fig. 1A and Fig. S2A), coexpression of EGFP-tagged MEC-4 with an untagged MEC-10 resulted in spots containing only two EGFPs (Fig. 1D). Second, when we coexpressed MEC-4 and MEC-10 that were both tagged with EGFP, the spots contained three EGFP molecules (Fig. 1E). These results are consistent with the trimeric stoichiometry of the related ASIC channels and their ability to form heterotrimers (23, 24).
Prior work suggested that MEC-4 colocalizes with MEC-2 and MEC-6 in vivo (8, 10). We asked whether these proteins can also be part of the core channel complex. We found that colocalization of tagged versions of MEC-4 and MEC-10 was not affected by coexpression of untagged MEC-2 and MEC-6 (Fig. 2A). Moreover, N-terminally tagged MEC-6 did not associate with tagged MEC-4, either with or without MEC-10 and MEC-2 (Fig. 2 B and C). In addition, the untagged MEC-6 did not change the stoichiometry of MEC-4 alone and MEC-4/MEC-10 complexes (Fig. 2 D–F and Fig. S2F). These results indicate that MEC-6 is not a core part of the channel complex.
Fig. 2.
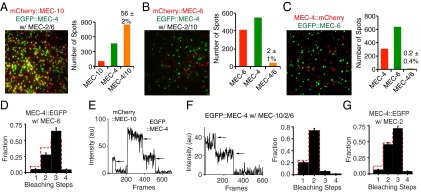
MEC-2, MEC-6, and MEC-4 do not colocalize on the plasma membrane of Xenopus oocytes. (A) The colocalization of mCherry::MEC-10 and EGFP::MEC-4 in the presence of untagged MEC-2 and MEC-6. (B) mCherry::MEC-6 and EGFP::MEC-4 failed to colocalize in the presence of untagged MEC-2 and MEC-10. (C) MEC-4::mCherry and EGFP::MEC-6 fail to colocalize. (D) The trimeric stoichiometry of MEC-4::EGFP in the presence MEC-6. (E) An example of the photobleaching of a MEC-4:MEC-4:MEC-10 heterotrimer in the presence of untagged MEC-2 and MEC-6. (F) An example (Left) and quantification (Right) of the photobleaching of EGFP::MEC-4 in the presence of untagged MEC-10, MEC-2, and MEC-6. (G) The trimeric stoichiometry of MEC-4::EGFP in the presence MEC-2.
We also asked whether MEC-2 colocalized with MEC-4. Both N- and C-terminally tagged MEC-2 proteins were highly mobile even in the presence of MEC-4 or MEC-4/MEC-10 and MEC-6 (Movies S4–S6). In addition, MEC-2 did not change the stoichiometry of the MEC-4 and MEC-4/10 trimers (Fig. 2 E–G and Fig. S2F). These results indicate that, like MEC-6, MEC-2 is not a core part of the channel complex.
MEC-4 Interacts Weakly with MEC-2 and MEC-6 Even at High Density in Xenopus Oocytes.
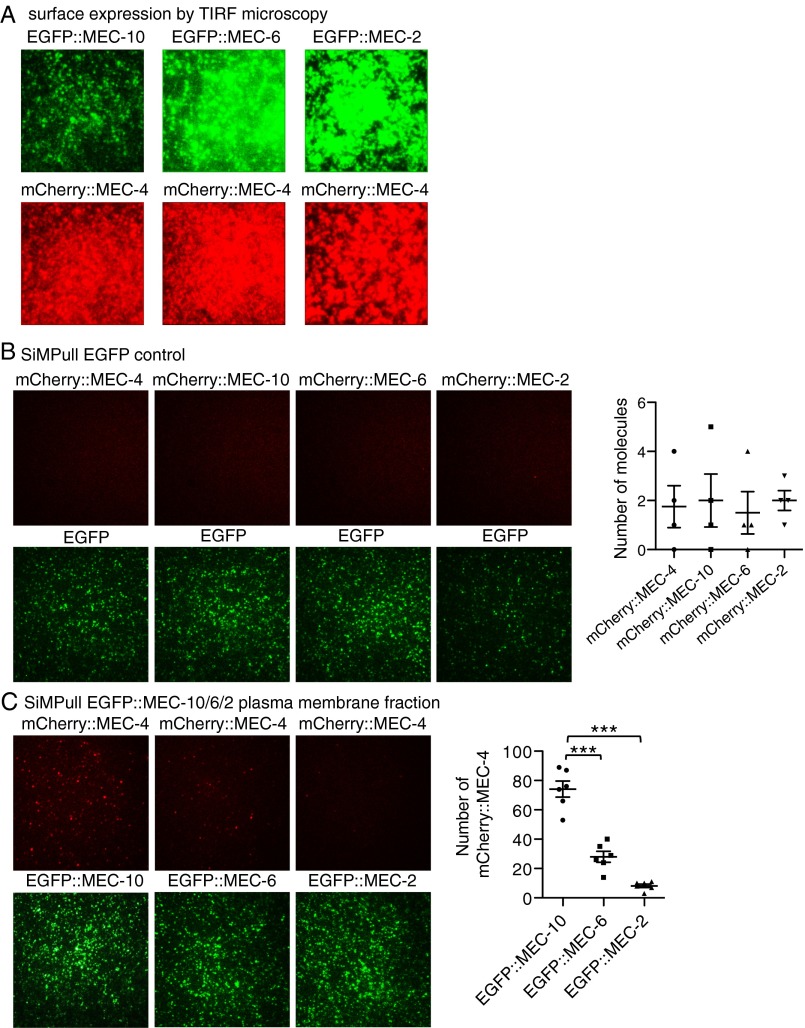
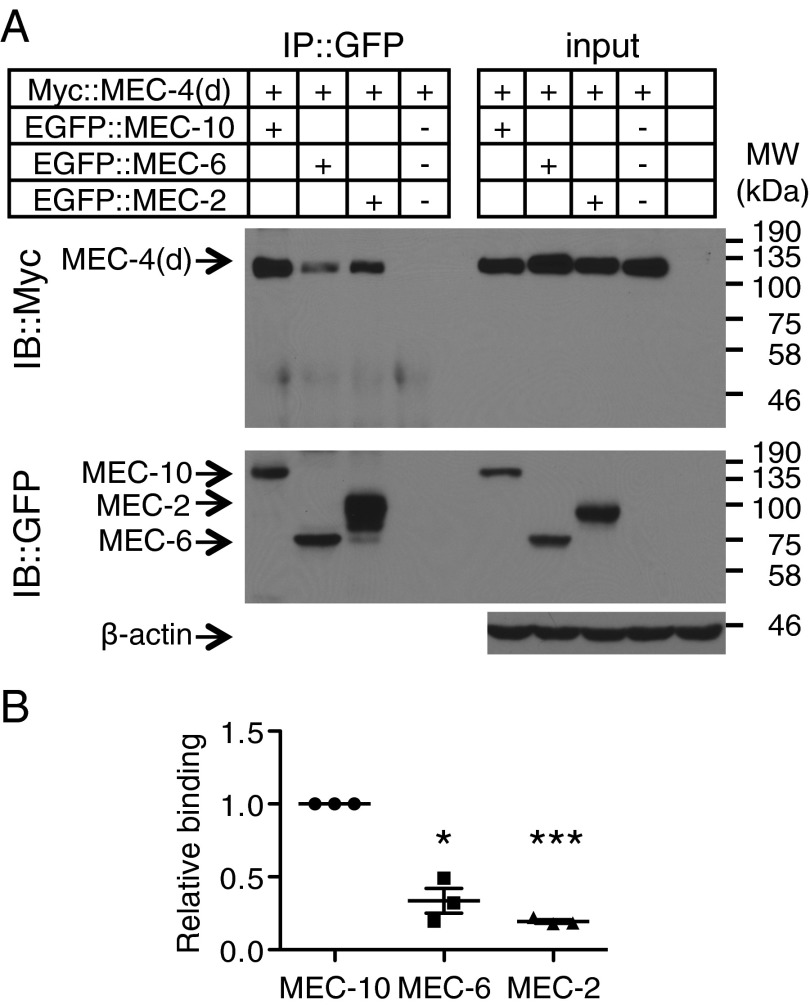
Because single molecule experiments are done at very low density and may not detect weak interactions between the MEC proteins, we performed coimmunoprecipitation and single-molecule pull-down [SiMPull (25)] at high expression density (Fig. S3A). Coimmunoprecipitation used MEC-4(d) that was tagged with the Myc epitope [Myc::MEC-4(d)] and N-terminally EGFP-tagged versions of MEC-10, MEC-6, and MEC-2. More MEC-4 was precipitated by MEC-10 than by either MEC-6 or MEC-2 (Fig. 3). Similarly, in SiMPull (performed either on whole lysate or on a plasma membrane fraction), MEC-10 pulled down more molecules of MEC-4 than either MEC-6 or MEC-2 (Fig. 4A and Fig. S3 B and C) and conversely, MEC-4 pulled down more molecules of MEC-10 than of either MEC-6 or MEC-2 (Fig. 4B and Fig. S3B). Hence, even at high expression density, we found that MEC-4 interacted relatively weakly with MEC-6 and MEC-2.
Fig. S3.
SiMPull between MEC-4 and MEC-10, MEC-6, and MEC-2 in Xenopus oocytes. (A) EGFP or mCherry-tagged MEC-4, MEC-10, MEC-6, and MEC-2 were highly expressed on oocytes surface as seen by TIRF imaging before SiMPull experiments. (B) EGFP control did not pull down the mCherry-tagged MEC-4, MEC-10, MEC-6, and MEC-2. (Left) Images of pulled-down mCherry::MEC-4/10/6/2 (Upper) and EGFP (Lower). (Right) Average number of pulled-down mCherry-tagged proteins per image area. (C) Images (Left) and quantification (Right) of pulled-down mCherry::MEC-4 (Upper) and EGFP::MEC-10/6/2 (Lower) from the isolated plasma membrane fraction. Each dot in the plot represents one experiment using the plasma membrane fraction collected from 30 oocytes. Each image area has ∼700 EGFP-tagged molecules. ***P < 0.001, one-way ANOVA with Tukey post hoc.
Fig. 3.
Immunoprecipitation (IP) of Myc::MEC-4(d) by EGFP-tagged MEC-10, MEC-6, and MEC-2 from the total lysate of Xenopus oocytes. (A) Western blot, which is representative of three independent experiments. Negative (−) controls replaced the EGFP-tagged MEC proteins with EGFP::HA. The rightmost lane represents the total lysate of uninjected oocytes. (Top) Immunoblot (IB) probed with an anti-Myc antibody. (Middle) Same membrane that had been stripped using the Restore Western Blot Stripping Buffer (Thermo Scientific) and reprobed with an anti-GFP antibody. (Bottom) Total lysates probed with an anti–β-actin antibody. Molecular weights (kDa) of the protein markers used in the experiments are indicated on the right. (B) Relative binding of MEC-10, MEC-6, and MEC-2 to MEC-4(d) (Materials and Methods). Data were normalized and compared with that of MEC-10. Each dot in the plot represents an independent experiment. Statistical significance (*P < 0.05, ***P < 0.001) was determined using one-sample t test with Bonferroni correction.
Fig. 4.
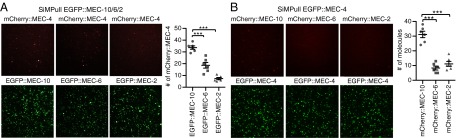
Physical interaction between MEC-4, MEC-10, MEC-6, and MEC-2 in oocytes by SiMPull. (A) Images (Left) and quantification (number of fluorescent molecules per imaging area; Right) of pulled-down mCherry::MEC-4 (Upper) and EGFP::MEC-10/6/2 (Lower) from the total lysate. Here and in B and Fig. S3, the image field is 13 × 13 μm. Each dot in the plot represents one experiment using the total lysate of oocytes (and in B and Fig. S3B). (B) Images (Left) and quantification (Right) of pulled-down mCherry::MEC-10/6/2 (Upper) and EGFP::MEC-4 (Lower), respectively, from the total lysate. Each image field has ∼400 EGFP-tagged molecules. Statistical difference (***P < 0.001) was analyzed using one-way ANOVA with Tukey post hoc.
MEC-6 Does Not Tightly Associate with the MEC-4/MEC-10 Channel Complex in C. elegans.
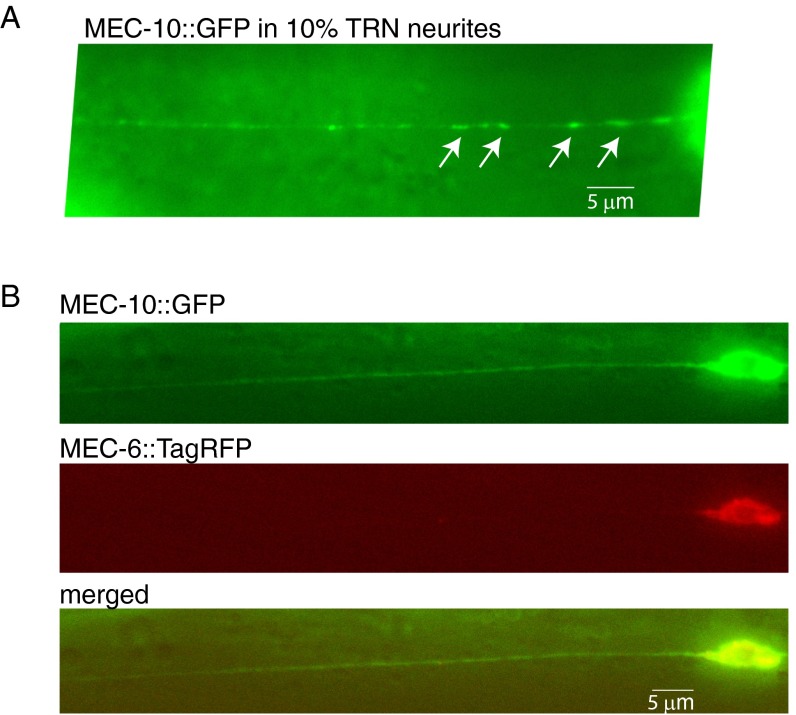
Our single molecule analysis (above) showed that MEC-4 can form homotrimers on its own or 4:4:10 heterotrimers when coexpressed with MEC-10. In vivo, MEC-10::GFP was previously shown to be predominantly localized in the TRN cell body and to have diffuse expression in the neurite (20, 26). We confirmed this (Fig. 5A, upper image; 90% of 40 TRNs), although we found that in 10% of the TRNs MEC-10::GFP formed MEC-4-like puncta in the proximal neurite (Fig. S4A). However, coexpression in vivo of MEC-10::GFP and MEC-4::TagRFP resulted in their reliable colocalization into puncta throughout the TRN neurite (Fig. 5B and Fig. S5; 50 of 50 TRNs examined). Moreover, coexpression of multiple copies of untagged MEC-4 resulted in MEC-10::GFP puncta in the TRN neurite (Fig. 5A). Coexpression of multiple copies of MEC-6 did not have this effect (Fig. S4). In contrast, expression of multiple copies of C-terminally GFP-tagged MEC-4 did not induce colocalizing puncta of MEC-6::3XFLAG (Fig. 5 C and D). These observations are consistent with our finding in oocytes that MEC-4 can form homotrimers and prior work that showed that the formation of MEC-4::YFP puncta does not require MEC-10 (6), and indicate that, although MEC-4 and MEC-10 can coassemble in vivo, MEC-10 is not a major component of the MEC-4 channel puncta in the TRN neurite.
Fig. 5.
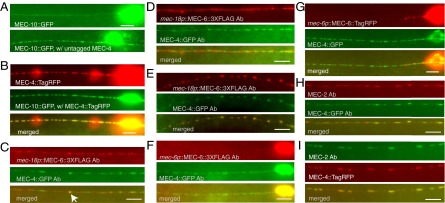
The colocalization of MEC-4, MEC-10, MEC-6, and MEC-2 in vivo. (Scale bar, 5 μm.) (A) The expression of MEC-10::GFP without (Upper) and with extra MEC-4 (Lower) in the TRNs. (B) MEC-10::GFP puncta in the presence of MEC-4::TagRFP and their colocalization in the TRN neurite; 10 ng/μL mec-10:gfp DNA was injected into TU4353 [uIs146 (mec-4::tagrfp)]. All 50 animals from three stable transgenic lines had GFP puncta that colocalized with MEC-4::TagRFP. (C–E) Puncta of MEC-6::3XFLAG expressed from the mec-18 promoter (mec-18p) and MEC-4::GFP expressed from the mec-4 promoter did not colocalize in most TRNs (C and D), only with occasional overlap seen in some TRN neurites (arrow in C), but colocalized well in ∼30% TRN neurites (E). MEC-6::3XFLAG (F) and MEC-6::TagRFP (G) expressed from the mec-6 promoter (mec-6p) were rarely observed in TRN neurite and did not colocalize with MEC-4::GFP. (H and I) The colocalization of MEC-2 with MEC-4::GFP (H) and MEC-4::TagRFP (I). Proteins indicated by Ab were detected by immunofluorescence using antibodies to FLAG, GFP, and MEC-2.
Fig. S4.
Expression of MEC-10::GFP in C. elegans TRNs. (A) MEC-10::GFP formed puncta (indicated by arrows) in proximal neurites of about 10% TRNs. (B) MEC-10::GFP had diffuse expression in neurites in the presence of MEC-6::TagRFP. Images are representative of 20 TRNs examined.
Fig. S5.
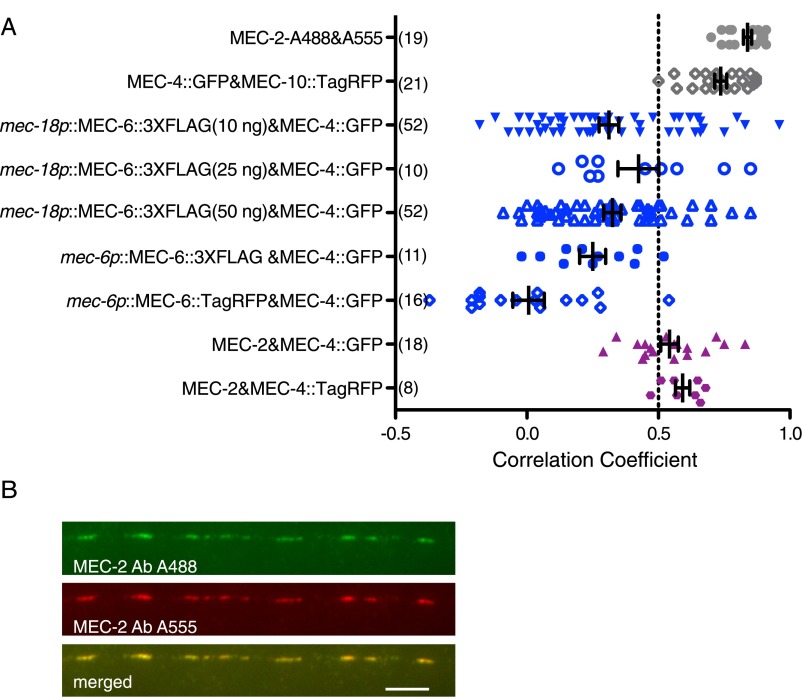
Colocalization of MEC-4, MEC-10, MEC-2, and MEC-6 in the TRN neurites. (A) Quantification of the colocalization (correlation coefficient). The number of TRN neurites examined is indicated in parentheses. The amount of injected mec-18p::mec-6::3xflag DNA (10, 25, or 50 ng/μL) is also given in parentheses. Except for MEC-4::TagRFP and MEC-10::GFP, and MEC-6::TagRFP and MEC-4::GFP (which directly imaged fluorescent proteins), all proteins were detected by immunofluorescence (Fig. 5). Different concentration of injected DNA had no effect on correlation of MEC-6::3xFLAG (expressed from mec-18 promoter) and MEC-4::GFP by one-way ANOVA with Tukey post hoc. The correlation coefficient of MEC-6 [expressed from either mec-18 (injected with 10 ng/μL DNA) or mec-6 promoter] and MEC-4::GFP was significantly different from that of MEC-2 and MEC-4::GFP, as well as MEC-4::GFP and MEC-10::TagRFP (P < 0.0001 by Student's t test). (B) MEC-2 detected by an anti–MEC-2 antibody and an Alexa 488 (A488)-conjugated anti-rabbit antibody and an Alexa 555 (A555)-conjugated anti-rabbit antibody (secondary antibodies) as a positive control for colocalization. (Scale bar, 5 μm.)
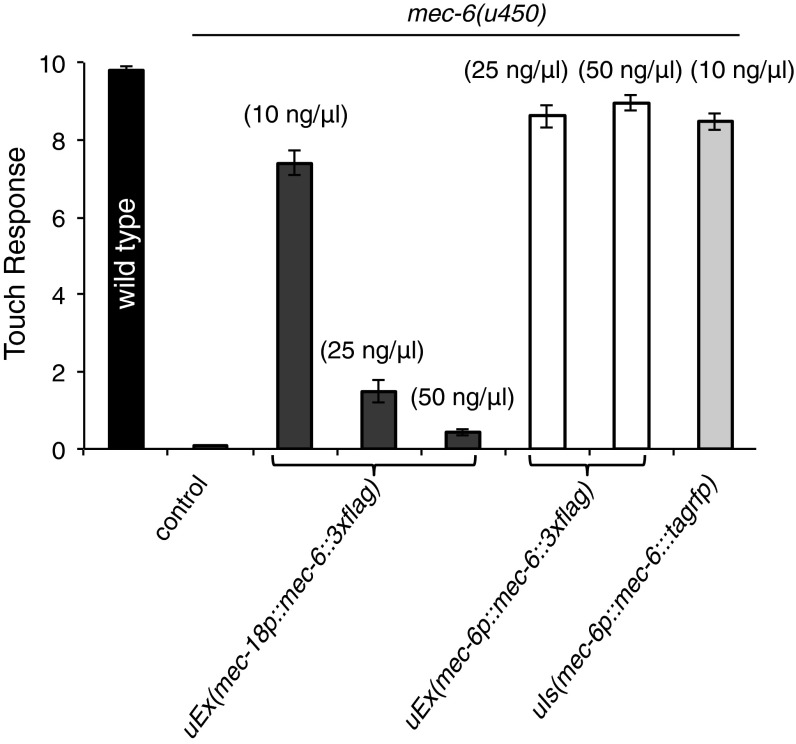
Our previous work suggested that C-terminally FLAG-tagged MEC-6 expressed under the TRN specific promoter of mec-18 colocalized with MEC-4::YFP puncta (8). Because the strains and plasmid used in this previous study had been lost, we generated a new plasmid and new strains to repeat those experiments. We injected mec-18p::mec-6::3xflag DNA at three different concentrations (10, 25, and 50 ng/μL), and although the intensity of immunostaining was not significantly different, transformation with 10 ng/μL, but not the higher concentrations, restored touch sensitivity to the mec-6 null mutant, mec-6(u450) (Fig. S6). In contrast to the strong colocalization between MEC-4 and MEC-10, MEC-4 and MEC-6 was widely scattered, with ∼30% of the TRNs neurites showing a correlation (Pearson correlation coefficient > 0.5; Fig. 5E and Fig. S5).
Fig. S6.
Rescue of touch sensitivity by MEC-6 fusion proteins in vivo. The MEC-6 fusion proteins were expressed from the mec-18 promoter (mec-18p) or the mec-6 promoter (mec-6p). Touch response indicates the number of times animals responded to 10 touches delivered alternately near the head and tail. The concentration of injected DNA is given in parentheses. All values are given as mean ± SEM. For wild type and mec-6(u450), 30 animals were tested; for the rescuing experiment, 25–60 animals from two to four transgenic lines or one integrated line (MEC-6::TagRFP) were tested.
Because the mec-18 promoter is a very strong promoter, we also examined MEC-6::3xFLAG and MEC-6::TagRFP expression from the mec-6 promoter itself and found both proteins to be primarily located to the cell body, with few puncta visible in the TRN neurite (Fig. 5 F and G). The little MEC-6::3XFLAG and MEC-6::TagRFP signal found in neurite was confined to the ∼40-μm proximal neurite of only 20–30% TRNs (50 TRNs examined for each). As with the mec-18 promoter both of the MEC-6 fusions from the mec-6 promoter rescued mec-6(u450) touch insensitivity (Fig. S6). Unlike the results with the mec-18 promoter, there was virtually no colocalization of MEC-6::3XFLAG or MEC-6::TagRFP with MEC-4::GFP (Fig. 5 F and G and Fig. S5). Together these results suggest that MEC-6 does not associate with MEC-4 in TRN neurites at normal expression levels. In contrast, as found previously (10), MEC-2 puncta colocalized well with tagged MEC-4 in 80% of the TRNs examined (n = 26; Fig. 5 H and I and Fig. S5).
SI Materials and Methods
Electrophysiology and Immunoprecipitation in Xenopus Oocytes.
To test the function of C- and/or N-terminally EGFP and mCherry-tagged MEC-4(d), MEC-10, MEC-6, and MEC-2 proteins, we injected Xenopus laevis oocytes (Xenopus I, Dexter or Ecocyte) with 10 ng of cRNA for untagged or tagged MEC-4(d), MEC-2, and 1 ng MEC-6 cRNA or 2 ng tagged MEC-6 cRNA. For coexpression of MEC-4(d), MEC-2, and MEC-6 with or without MEC-10, we injected oocytes with 7.5 ng cRNA for tagged or untagged MEC-4(d), MEC-2, MEC-10, and 0.75 ng MEC-6 cRNA or 1.5 ng tagged MEC-6 cRNA, because oocytes were healthier and produced more robust results when less cRNA was injected. Oocytes were maintained as previously described (6) Membrane current was measured 4–6 d after cRNA injection using a two-electrode voltage clamp as described previously (9).
Immunoprecipitation was performed 5 d after injection of 10 ng cRNA of N-terminally Myc-tagged MEC-4(d), N-terminally EGFP-tagged MEC-10 and MEC-2, or 2 ng cRNA of N-terminally EGFP-tagged MEC-6. Oocytes homogenates were prepared using 10 μL lysis buffer (20 mM Tris⋅HCl, pH = 7.6, 100 mM NaCl, and 1% Nonidet P-40) with proteinase inhibitor (Roche) per oocyte. The total lysate was diluted in lysis buffer 1:10 and incubated with anti-GFP antibodies (rabbit polyclonal, ab290; Abcam) at 4 °C overnight. Then, Protein A/G PLUS-Agarose (Santa Cruz Biotechnology) was added and incubated at 4 °C for 2 h to precipitate target protein complex. Protein samples were subjected to SDS/PAGE. The total lysate of one oocyte equivalent was loaded per lane for input, and five oocyte equivalents were loaded per lane for immunoprecipitation. Protein was detected by Western blot using antibodies against GFP (mouse monoclonal, sc-9996; Santa Cruz Biotechnology), Myc (mouse monoclonal 9E10; Sigma), and HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories). HRP was detected using the ECL Western Blotting reagent (Amersham).
cDNA and Plasmids Used in C. elegans.
Fluorescence fusion plasmids were generated from pPD95.75 (www.addgene.org/static/cms/files/Vec95.pdf). mec-10::gfp (TU#1176) was made by cloning the 1,925-bp sequence upstream of the start codon and the 4,232 bp of coding sequence (without the stop codon) of the mec-10 genomic DNA into pPD95.75. mec-6::tagrfp (TU#1174) and mec-4::tagrfp (TU#1175) were generated by removing yfp::unc-54 3′UTR from mec-6::yfp and mec-4::yfp (8), respectively, using KpnI and ApaI and replacing it with tagrfp::unc-54 3′UTR from TU#930. mec-4::gfp (TU#1192) were generated by removing yfp::unc-54 3′UTR from mec-4::yfp using KpnI and ApaI and replacing it with gfp::unc-54 3′UTR from pPD95.75. The mec-18p::mec-6::3Xflag (TU#1193) construct was generated using the Three-Fragment Vector Construction Kit (Invitrogen). The mec-18 promoter (400-bp genomic sequence upstream and the start codon) was cloned into pDONRP4P1R. The mec-6 coding sequence without the start codon and the stop codon was cloned from the genomic sequence into pDONR221. The sequences encoding 3XFLAG and unc-54 3′UTR were cloned into pDONRP2RP3. TU#1190 was made by replacing GFP in pPD95.75 with 3xFLAG from p3XFLAG-CMV-14 (Sigma-Aldrich). mec-6::3Xflag (TU1191) was made by cloning the 3,787-bp sequence upstream of the start codon and 2,872-bp coding sequence (without the stop codon) of mec-6 genomic DNA into TU#1190.
C. elegans Strains.
The following strains were generated during this study: TU3971 [uIs146 (mec-4::tagrfp)], TU4352 [uIs146 (mec-4::tagrfp); uEx871(mec-10::gfp, myo-2p::mCherry)], TU4353 [uIs146 (mec-4::tagrfp); uEx872(mec-10::gfp, myo-2p::mCherry)], TU4630 [uIs193(mec-10::gfp, myo-2p::mCherry); uEx914 (mec-4p::mec-4, inx-20p::gfp)], TU4631 [uIs193; uEx915(mec-4p::mec-4, inx-20p::gfp)], TU4643-4651 [mec-6(u450); uEx931-939(mec-18p::mec-6::3xflag; myo-2p::mCherry)], TU4720-TU4722 [uEx940-942 (mec-4::gfp, mec-6p::mec-6::3xflag; myo-2p::mCherry)], TU4723-4731 [uEx943-951(mec-4::gfp, mec-18p::mec-6::3xflag; myo-2p::mCherry)], and TU4739 [uIs148 (mec-6p::mec-6::tagrfp), uEx852 (mec-4::gfp)].
Transgenic animals were prepared by microinjection, and integrated transgenes were generated by UV irradiation (37). For all expression experiments, we microinjected WT (N2) animals with 5–50 ng/μL relevant GFP or TagRFP fusion plasmid (unless otherwise indicated) and 2 ng/μL myo-2p::mCherry plasmid (PCFJ90; Addgene) and/or 20 ng/μL inx-20p::gfp (TU#1196) plasmid and pBluescript SK plasmid to bring the total DNA to 100 ng/μL.
For the rescuing experiments, we usually microinjected 1–50 ng/μL relevant plasmid, 2 ng/μL myo-2p::mCherry (PCFJ90; Addgene), 40 ng/μL lin-15(+) plasmid, and pBluescript SK to bring the total DNA to 100 ng/μL. When we rescued mec-4(u253) with mec-4::tagrfp and mec-4::gfp, we injected 0.2 ng/μL mec-4::tagrfp or 0.1 ng/μL mec-4::gfp linearized by HindIII, 2.5 ng/μL inx-20p::gfp linearized by SphI, and 125 ng/μL genomic DNA linearized by EcoRI and KpnI from OP50 E.coli. Extragenic expression of MEC-4::TagRFP and MEC-4::GFP restored touch sensitivity to mec-4(u253) null mutants in C. elegans (responses to 10 touches applied alternately near the head and the tail (mean ± SEM): control, 0 ± 0, n = 30; uEx(mec-4::tagrfp), 7.1 ± 0.3, n = 33 from three stable lines; uEx(mec-4::gfp), 4.7 ± 0.7, n = 20 from two stable lines.
C. elegans Immunofluorescence and Microscopy.
The following antibodies were used for immunostaining: anti–MEC-2 N terminus (10), anti-FLAG (mouse, F1804; Sigma), and anti-GFP (rabbit polyclonal A11122 and mouse monoclonal 3E6; Life Technologies) diluted 1:200, Rhodamine Red-X-conjugated and Alexa Fluor 488-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories), and Alexa Fluor 488/555-conjugated goat anti-rabbit/mouse IgG (Life Technologies) diluted 1:700. Cross-reactions between secondary antibodies and primary antibodies from different species have been examined and found to be minimal.
Fluorescence and immunofluorescence were observed using a Zeiss Axio Observer Z1 inverted microscope equipped with a 100×, NA 1.40 oil immersion objective and a Photometrics CoolSnap HQ2 camera (Photometrics). Live animals were anesthetized using 100 mM 2,3-butanedione monoxime in 10 mM Hepes, pH 7.4.
Discussion
MEC-4 Forms a Homotrimeric or Heterotrimeric Channel with MEC-10.
This study demonstrates that MEC-4 and MEC-10 can form 4:4:4 homotrimers and 4:4:10 heterotrimers. This arrangement is consistent with the homotrimer channels formed by the chicken DEG/ENaC protein ASIC1 as seen by X-ray crystallography and the heterotrimeric channels formed by rat ASIC1a and ASIC2a as detected by single molecule photobleaching (23, 24) and the findings from immunoelectron microscopy that half of the membrane-associated MEC-4 exists as doublets (27). Together these data suggest that the trimer is the functional form of the channel in vivo.
In contrast to MEC-4, which formed immobile homotrimers on the oocyte surface, MEC-10 was very mobile on its own, so we could not determine its stoichiometry. Because MEC-10(d), the equivalent to MEC-4(d), does not form a functional channel in Xenopus oocytes (9), it may not be able to form trimers on its own. If so, the presence of MEC-10 in the 4:4:10 trimer may require MEC-4 to enable its incorporation. Moreover, we never observed any 4:10:10 complexes, again suggesting that MEC-10 subunits do not interact. Thus, MEC-4 and MEC-10 differ from rat ASIC channels which can form both 1a:1a:2a and 1a:2a:2a heterotrimers (24).
The differences between MEC-4 and MEC-10 may explain several genetic results: (i) loss of MEC-4 causes complete touch insensitivity (28), whereas loss of MEC-10 does not (6); (ii) MEC-4(d) causes nearly 100% TRN cell death and its toxicity does not require MEC-10, whereas MEC-10(d) only causes about 30% TRN cell death and its toxicity requires MEC-4 (29); and (iii) MEC-10::GFP localization needs MEC-4, whereas MEC-4::YFP localization as puncta does not need MEC-10 (6).
Other members of the DEG/ENaC family also differ in their ability to form functional homomeric channels. The mammalian epithelial sodium channel has three subunits (α-, β-, and γ-ENaC), but only α-ENaC forms a functional channel on its own (30). In contrast, ASIC channels (including ASIC1, ASIC2, and ASIC3) can form functional homomeric and heterotrimer channels with distinct physiological properties (31).
The ability of MEC-4 to form homotrimers and MEC-4 and MEC-10 to form 4:4:10 heterotrimers raises the question about the physiological function of each of these forms. Electrophysiological evidence predicts that the number of functional channels is about the same as the number of puncta (5), yet each punctum clearly contains many channels and we could not detect single channels in vivo with standard confocal microscopy, using either fluorescent protein tags or fluorescent antibodies. This result suggests that most of the MEC-4 homotrimeric (and possibly the MEC-4/MEC-10 heterotrimeric) channels in the TRN neurite puncta are inactive. Because MEC-4 puncta can be seen with anti–MEC-4 antibodies (27), the puncta are not artifacts of fluorescent protein expression. One possibility is that the puncta are intracellular reservoirs for MEC-4 (and possibly other proteins). Indeed, Cueva et al. (27) found about half of the MEC-4 immunogold labeling in electron micrographs to be associated with the plasma membrane of TRNs, but the rest to be associated with 15-protofilament microtubules, and Butterworth et al. (32) suggested that mammalian ENaC channels can enter the plasma membrane from a recycling pool.
A Simplified Mechanosensory Channel Complex.
The data presented here simplify the model of the mechanotransduction complex in the TRNs. We confirmed the stable association of MEC-4 and MEC-10, but suggest that the other membrane proteins, MEC-2 (and perhaps UNC-24, which we did not test) and MEC-6, whose expression boosts the activity of MEC-4–containing channels (5, 8, 9), have transient interactions with MEC-4 and MEC-10. MEC-2 binds cholesterol and we have hypothesized that it may be necessary for touch sensitivity via modulation of the lipid microenvironment of the MEC-4/MEC-10 channel (33). The finding that MEC-2 and a similar protein, podocin, form multimeric complexes in HEK293T1/2 cells (33), but that MEC-2 has only a weak interaction with MEC-4, suggests that their colocalization to the TRN neurite puncta (10, 27) reflects not direct interaction but their clustering into large common complexes by other protein factors, perhaps to affect the local cholesterol environment.
Because MEC-6 seems to be confined to the TRN cell body, it may play its main role there to boost MEC-4 channel expression. Our data indicate that MEC-6 and MEC-2 are not integral components of the channel transduction complex. Our results, however, cannot exclude the possibility that these proteins directly affect, albeit transiently, the function of the transduction complex.
Materials and Methods
Single Molecule Imaging.
For stoichiometry and colocalization experiments, DNA constructs for C- and/or N-terminally EGFP and mCherry-tagged proteins were generated in vector pGEMHE-X-EGFP/mCherry and/or pGEMHE-EGFP-X (21). pGEMHE-mCherry-X was generated by removing EGFP sequence from pGEMHE-EGFP-X by BamHI and BsrGI and replacing it with mCherry from pGEMHE-X-mCherry. Plasmids propagation and cRNA synthesis were done as before (9). The function of C- and/or N-terminally EGFP and mCherry-tagged proteins was tested electrophysiologically in Xenopus laevis oocytes (SI Materials and Methods).
For single molecule imaging by TIRF microscopy, cRNA injection for EGFP/mCherry-tagged proteins was optimized for expression. The following amount of cRNA was injected: 1.25 ng cRNA for EGFP::MEC-4, 0.5 ng cRNA for mCherry::MEC-10, 0.1 ng cRNA for mCherry::MEC-6, 0.05 ng cRNA for mCherry::MEC-2, and 0.5 ng cRNA for untagged MEC-2, 6, and 10. For MEC-4::EGFP expression alone, 5–10 ng cRNA was injected. Imaging of individual protein complex on Xenopus oocyte membrane by TIRF microscopy was performed as previously described (21, 22, 34). Briefly, oocytes were manually devitellinized after 1–2 d of expression at 16 °C and placed on high refractive index coverglass (n = 1.78; Olympus America) and imaged using Olympus 100×, NA 1.65 oil immersion objective at room temperature.
EGFP and mCherry-tagged MEC proteins were excited using a phoxX 488 (60 mW) laser and a 593-nm diode-pumped solid-state laser, respectively. For subunit counting with EGFP tags, a 495-nm long-pass dichroic mirror was used at excitation in combination with a 525/50-nm band-pass filter at emission. For colocalization experiments where both EGFP and mCherry were excited sequentially, a z488/594 rpc polychroic (Chroma) was used at excitation, and 525/50- and 629/53-nm band-pass filters for EGFP and mCherry, respectively, were used at emission. Five hundred to 800 frames at the rate of 20 Hz were acquired for subunit counting, whereas 1,000 frames (∼200 for mCherry and ∼800 for EGFP, sequentially) were acquired at the same rate for colocalization using an EMCCD (Andor iXon DV887) camera.
Only single, immobile, and diffraction-limited spots were analyzed. The number of bleaching steps was determined manually for each single spot included in the analysis; 200–800 spots from 5 to 10 oocytes from three to five different batches were analyzed for most of the constructs. Observed frequency distribution of photobleaching steps for each construct was plotted and compared with the expected binomial distributions for a dimer, trimer, tetramer, and pentamer that were calculated using a fixed probability of 80% of mEGFP being fluorescent.
Single-molecule colocalization of red (mCherry) and green (EGFP) spots were analyzed as previously described (34). Random colocalization was 1–6% for EGFP and mCherry-tagged MEC-4 and MEC-10 and 1–2% between EGFP and mCherry-tagged MEC-4 and MEC-6.
SiMPull.
SiMPull experiments were performed 2–3 d after injecting either 25 ng cRNA for EGFP::MEC-4 and mCherry::MEC-10/2/6, or 10 ng cRNA for EGFP::MEC-2/10 and mCherry::MEC-4, and 1 ng cRNA for EGFP::MEC-6 into Xenopus oocytes. SiMPull was performed on total lysate or only plasma membrane [mechanically isolated from oocytes as previously described (35)] lysate as described in Jain et al. (25). In the plasma membrane experiments, 1 ng cRNA for untagged MEC-6 was also added. Briefly, channels/flow chambers were prepared on coverslips passivated with monofunctional and biotinylated polyethylene glycol. Biotinylated anti-EGFP antibody (goat polyclonal, ab6658; Abcam) was then immobilized by incubating 40 nM of antibody on Neutravidin (Thermofisher) coated channels. Sample lysate was passed through the channels and imaged by TIRF microscopy as described above. The specificity of SiMPull was confirmed by showing that EGFP did not pull down mCherry-tagged MEC proteins (Fig. S3B).
Immunoprecipitation in Xenopus Oocytes.
Immunoprecipitation were performed 5 d after cRNA injection as described previously (9). One oocyte equivalent was loaded per lane for lysate input, and five oocyte equivalents were loaded per lane for immunoprecipitation. Proteins were detected by Western blot using HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) and the ECL Western Blotting reagent (Amersham). Band density was measured from the autoradiography films using Image J (National Institutes of Health) and was used to calculate the relative binding of EGFP::MEC-10/6/2 and Myc::MEC-4(d): [IP complex detected by the anti-Myc antibody]/([EGFP::MECs input] × [Myc::MEC-4(d) input]). Details are given in SI Materials and Methods.
The specificity of the immunoprecipitation was confirmed in two ways. First, 1 ng cRNA encoding EGFP::HA was used as a negative control for EGFP::MEC-10/6/2 immunoprecipitation of Myc::MEC-4(d); none of the proteins were immunoprecipitated. Second, we probed the immuno-complexes for a Xenopus oocyte membrane protein, β-integrin, by using a monoclonal antibody (8C8; Developmental Studies Hybridoma Bank, University of Iowa) and did not detect the β-integrin.
C. elegans Procedures.
Strains were maintained and studied at 20 °C on the OP50 strain of Escherichia coli according to Brenner (36). All of the translational fusions were based on pPD95.75 (www.addgene.org/static/cms/files/Vec95.pdf). Transgenic animals were generated by microinjection and integrated transgenes were generated by UV irradiation (37). Details about strains and plasmids are given in SI Materials and Methods.
Immunostaining of larvae and adults was performed according to Miller and Shakes (38). Antibodies and microscopy used for immunofluorescence are given in SI Materials and Methods. To quantify colocalization, we selected the best-focused images (from images taken at several focal planes) that contained at least 25 μm of the TRN neurite and correlated the fluorescence intensities of the puncta (after subtracting the background) from the two color channels using Coloc 2 (fiji.sc/Coloc2). Correlation coefficient was represented by the Pearson’s R value above the automatic threshold (39, 40). MEC-2 immunofluorescence with the protein colabeled with Alexa 488 and Alexa 555 was used as a positive control for colocalization.
Statistics.
Data are presented with their mean ± SEM. Error bars indicate SEM, unless noted. Statistical significance was determined using the Student t test (with Welch’s correction when data being compared do not have equal variances), one-way ANOVA (for multiple samples), and one-sample t test (for Western blot) using GraphPad Prism5 software (www.graphpad.com/scientific-software/prism/).
Supplementary Material
Acknowledgments
We thank Jian Yang (Columbia University) and Yong Yu (St. Johns University) for providing the Xenopus laevis oocytes and Oliver Hobert, Elizabeth Miller, and members of the M.C. laboratory for discussion. This work was supported by National Institutes of Health Grants GM30997 (to M.C.) and NS35549 (to E.Y.I.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515968112/-/DCSupplemental.
References
- 1.Chalfie M. Neurosensory mechanotransduction. Nat Rev Mol Cell Biol. 2009;10(1):44–52. doi: 10.1038/nrm2595. [DOI] [PubMed] [Google Scholar]
- 2.Geffeney SL, Goodman MB. How we feel: Ion channel partnerships that detect mechanical inputs and give rise to touch and pain perception. Neuron. 2012;74(4):609–619. doi: 10.1016/j.neuron.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ranade SS, et al. Piezo2 is the major transducer of mechanical forces for touch sensation in mice. Nature. 2014;516(7529):121–125. doi: 10.1038/nature13980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu Z, et al. SWELL1, a plasma membrane protein, is an essential component of volume-regulated anion channel. Cell. 2014;157(2):447–458. doi: 10.1016/j.cell.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Hagan R, Chalfie M, Goodman MB. The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals. Nat Neurosci. 2005;8(1):43–50. doi: 10.1038/nn1362. [DOI] [PubMed] [Google Scholar]
- 6.Arnadóttir J, O’Hagan R, Chen Y, Goodman MB, Chalfie M. The DEG/ENaC protein MEC-10 regulates the transduction channel complex in Caenorhabditis elegans touch receptor neurons. J Neurosci. 2011;31(35):12695–12704. doi: 10.1523/JNEUROSCI.4580-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang M, Gu G, Ferguson EL, Chalfie M. A stomatin-like protein necessary for mechanosensation in C. elegans. Nature. 1995;378(6554):292–295. doi: 10.1038/378292a0. [DOI] [PubMed] [Google Scholar]
- 8.Chelur DS, et al. The mechanosensory protein MEC-6 is a subunit of the C. elegans touch-cell degenerin channel. Nature. 2002;420(6916):669–673. doi: 10.1038/nature01205. [DOI] [PubMed] [Google Scholar]
- 9.Goodman MB, et al. MEC-2 regulates C. elegans DEG/ENaC channels needed for mechanosensation. Nature. 2002;415(6875):1039–1042. doi: 10.1038/4151039a. [DOI] [PubMed] [Google Scholar]
- 10.Zhang S, et al. MEC-2 is recruited to the putative mechanosensory complex in C. elegans touch receptor neurons through its stomatin-like domain. Curr Biol. 2004;14(21):1888–1896. doi: 10.1016/j.cub.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 11.Brown AL, Liao Z, Goodman MB. MEC-2 and MEC-6 in the Caenorhabditis elegans sensory mechanotransduction complex: Auxiliary subunits that enable channel activity. J Gen Physiol. 2008;131(6):605–616. doi: 10.1085/jgp.200709910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Driscoll M, Chalfie M. The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration. Nature. 1991;349(6310):588–593. doi: 10.1038/349588a0. [DOI] [PubMed] [Google Scholar]
- 13.Brown AL, Fernandez-Illescas SM, Liao Z, Goodman MB. Gain-of-function mutations in the MEC-4 DEG/ENaC sensory mechanotransduction channel alter gating and drug blockade. J Gen Physiol. 2007;129(2):161–173. doi: 10.1085/jgp.200609672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L, et al. Contribution of Drosophila DEG/ENaC genes to salt taste. Neuron. 2003;39(1):133–146. doi: 10.1016/s0896-6273(03)00394-5. [DOI] [PubMed] [Google Scholar]
- 15.Zhong L, Hwang RY, Tracey WD. Pickpocket is a DEG/ENaC protein required for mechanical nociception in Drosophila larvae. Curr Biol. 2010;20(5):429–434. doi: 10.1016/j.cub.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mauthner SE, et al. Balboa binds to pickpocket in vivo and is required for mechanical nociception in Drosophila larvae. Curr Biol. 2014;24(24):2920–2925. doi: 10.1016/j.cub.2014.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Y, Wang Y, Wang Q, Wang Z. The role of PPK26 in Drosophila larval mechanical nociception. Cell Reports. 2014;9(4):1183–1190. doi: 10.1016/j.celrep.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 18.Gorczyca DA, et al. Identification of Ppk26, a DEG/ENaC Channel Functioning with Ppk1 in a Mutually Dependent Manner to Guide Locomotion Behavior in Drosophila. Cell Reports. 2014;9(4):1446–1458. doi: 10.1016/j.celrep.2014.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hicks MA, et al. The evolution of function in strictosidine synthase-like proteins. Proteins. 2011;79(11):3082–3098. doi: 10.1002/prot.23135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chatzigeorgiou M, et al. Spatial asymmetry in the mechanosensory phenotypes of the C. elegans DEG/ENaC gene mec-10. J Neurophysiol. 2010;104(6):3334–3344. doi: 10.1152/jn.00330.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4(4):319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulbrich MH, Isacoff EY. Rules of engagement for NMDA receptor subunits. Proc Natl Acad Sci USA. 2008;105(37):14163–14168. doi: 10.1073/pnas.0802075105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 2007;449(7160):316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- 24.Bartoi T, Augustinowski K, Polleichtner G, Gründer S, Ulbrich MH. Acid-sensing ion channel (ASIC) 1a/2a heteromers have a flexible 2:1/1:2 stoichiometry. Proc Natl Acad Sci USA. 2014;111(22):8281–8286. doi: 10.1073/pnas.1324060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain A, et al. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473(7348):484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamat S, Yeola S, Zhang W, Bianchi L, Driscoll M. NRA-2, a nicalin homolog, regulates neuronal death by controlling surface localization of toxic Caenorhabditis elegans DEG/ENaC channels. J Biol Chem. 2014;289(17):11916–11926. doi: 10.1074/jbc.M113.533695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cueva JG, Mulholland A, Goodman MB. Nanoscale organization of the MEC-4 DEG/ENaC sensory mechanotransduction channel in Caenorhabditis elegans touch receptor neurons. J Neurosci. 2007;27(51):14089–14098. doi: 10.1523/JNEUROSCI.4179-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chalfie M, Sulston J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol. 1981;82(2):358–370. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- 29.Huang M, Chalfie M. Gene interactions affecting mechanosensory transduction in Caenorhabditis elegans. Nature. 1994;367(6462):467–470. doi: 10.1038/367467a0. [DOI] [PubMed] [Google Scholar]
- 30.Canessa CM, et al. Amiloride-sensitive epithelial Na+ channel is made of three homologous subunits. Nature. 1994;367(6462):463–467. doi: 10.1038/367463a0. [DOI] [PubMed] [Google Scholar]
- 31.Benson CJ, et al. Heteromultimers of DEG/ENaC subunits form H+-gated channels in mouse sensory neurons. Proc Natl Acad Sci USA. 2002;99(4):2338–2343. doi: 10.1073/pnas.032678399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butterworth MB, Edinger RS, Johnson JP, Frizzell RA. Acute ENaC stimulation by cAMP in a kidney cell line is mediated by exocytic insertion from a recycling channel pool. J Gen Physiol. 2005;125(1):81–101. doi: 10.1085/jgp.200409124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huber TB, et al. Podocin and MEC-2 bind cholesterol to regulate the activity of associated ion channels. Proc Natl Acad Sci USA. 2006;103(46):17079–17086. doi: 10.1073/pnas.0607465103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abuin L, et al. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69(1):44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivanina T, et al. Phosphorylation by protein kinase A of RCK1 K+ channels expressed in Xenopus oocytes. Biochemistry. 1994;33(29):8786–8792. doi: 10.1021/bi00195a021. [DOI] [PubMed] [Google Scholar]
- 36.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chelur DS, Chalfie M. Targeted cell killing by reconstituted caspases. Proc Natl Acad Sci USA. 2007;104(7):2283–2288. doi: 10.1073/pnas.0610877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller DM, Shakes DC. Immunofluorescence microscopy. Methods Cell Biol. 1995;48:365–394. [PubMed] [Google Scholar]
- 39.Costes SV, et al. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J. 2004;86(6):3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young HM, Furness JB, Shuttleworth CW, Bredt DS, Snyder SH. Co-localization of nitric oxide synthase immunoreactivity and NADPH diaphorase staining in neurons of the guinea-pig intestine. Histochemistry. 1992;97(4):375–378. doi: 10.1007/BF00270041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.