Summary
The mechanisms by which tumour cells metastasize and the role cell polarity proteins play in this process are not well understood. We report that Partitioning defective protein 3 (Par3) is dysregulated in metastasis in human breast cancer, and is associated with higher tumour grade and ErbB2-positive status. Downregulation of Par3 cooperated with ErbB2 to induce cell invasion and metastasis in vivo. Interestingly, the metastatic behaviour was not associated with an overt mesenchymal phenotype. However, loss of Par3 inhibited E-cadherin junction stability, disrupted membrane and actin dynamics at cell-cell junctions and decreased cell-cell cohesion in a Tiam1/Rac-GTP pathway dependent manner. Inhibition of this pathway restored E-cadherin junction stability and blocked invasive behaviour of cells lacking Par3 suggesting that loss of Par3 promotes metastatic behaviour of ErbB2-induced tumour epithelial cells by decreasing cell-cell cohesion.
Introduction
The Partitioning defective (Par) family of cell polarity proteins, Par1, Par3 (Pard3), Par4, Par5 and Par6, were identified in Caenorhabditis elegans as regulators of asymmetric cell division during early embryonic development1–3. In mammals, Par proteins including Par1b (EMK1/MARK2), Par3, Par4 (LKB1), 14-3-3 (Par5), Par6 and protein kinase C ι/ζ (atypical PKC, aPKC), facilitate generation of intracellular asymmetry and regulate various processes such as establishment of apical-basal polarity, directional cell migration, and asymmetric cell division4. Although the precise mechanisms by which Par proteins regulate these biological remains to be understood, it involves a coordinated regulation of protein localization and actin/microtubule cytoskeletal dynamics5.
Very little is known about the pathways that regulate loss of cell and tissue architecture in carcinoma, however, emerging evidence points to a direct role for Par proteins as important regulators6. We demonstrated that erythroblastic leukaemia viral oncogene homolog 2 (ErbB2) (also referred to as human epidermal growth factor receptor 2, HER2) interacts with Par6/aPKC complex and that this interaction is required during ErbB2-induced disruption of apical-basal polarity and transformation of polarized epithelial cells7. Par6 is also a critical mediator of transforming growth factor-β (TGF-β)-induced epithelial to mesenchymal transition (EMT) and metastasis6. Others and we have demonstrated that Par6 and aPKC are overexpressed and/or amplified in breast, ovarian and lung cancers8–10 and the genomic region containing the Pard3 gene is deleted in lung, head and neck and oesophageal squamous cell carcinoma cell lines11, 12.
The Par complex member Par3, is a signalling scaffold that contains three PSD-95/Discs-large/ZO-1 (PDZ) domains, an N-terminal dimerization domain and a C-terminal aPKC interaction domain4, 13. The PDZ domains interact with cell surface proteins such as Junctional Adhesion Molecules, (JAM)14 and Nectin15, Par66, phospholipids (PIP2) and phosphatase and tensin homolog (PTEN)16–18. The C-terminal domain of Par3 interacts with aPKC to inhibit its kinase activity19, 20 and with a Rac1 GTPase-specific GTP exchange factor Tiam1 (T Lymphoma invasion and metastasis) to inhibit its exchange activity21. Whereas the Par3-aPKC interaction regulates establishment of the apical membrane, Par3-Tiam1 interaction is sufficient for tight junction biogenesis22. A similar Par3-Tiam1-dependent, but aPKC-independent, interaction is also required during dendritic spine morphogenesis23. Expression of dominant negative Rac or downregulation of Tiam1 rescued Par3 loss phenotype, demonstrating that the ability of Par3 to inhibit Rac activation plays an important role during dendritic spine morphogenesis. Here, we report our findings on the role Par3 plays during metastasis of breast cancer.
Results
Loss of Par3 cooperates with ErbB2 to induce invasive behaviour in mammary epithelial cells
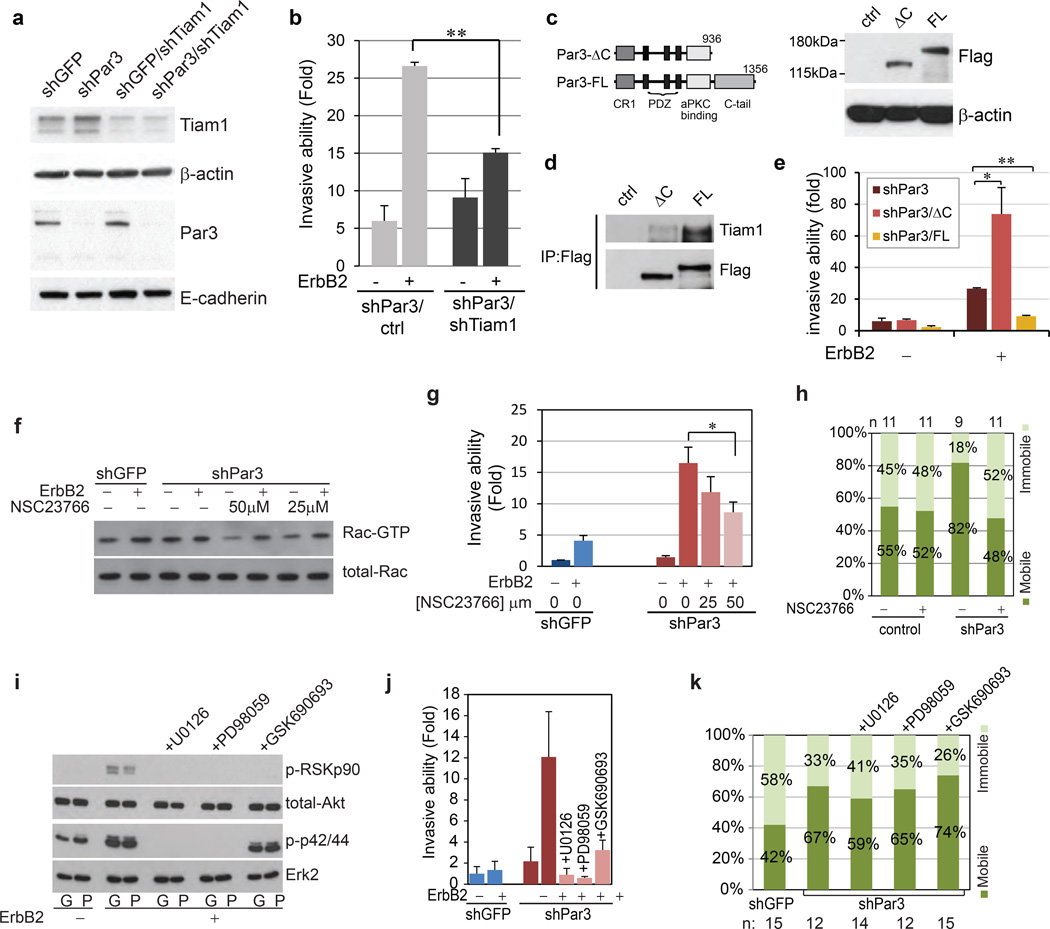
We designed two independent short hairpin RNAs (shRNAs) targeting Par3 (Fig. 1a) and generated stable populations of MCF-10A cells expressing an inducible form of the oncogenic receptor tyrosine kinase ErbB2 (10A.B2)24. Par3 shRNA-B was shown to be the most efficient shRNA and was used in the further experiments.
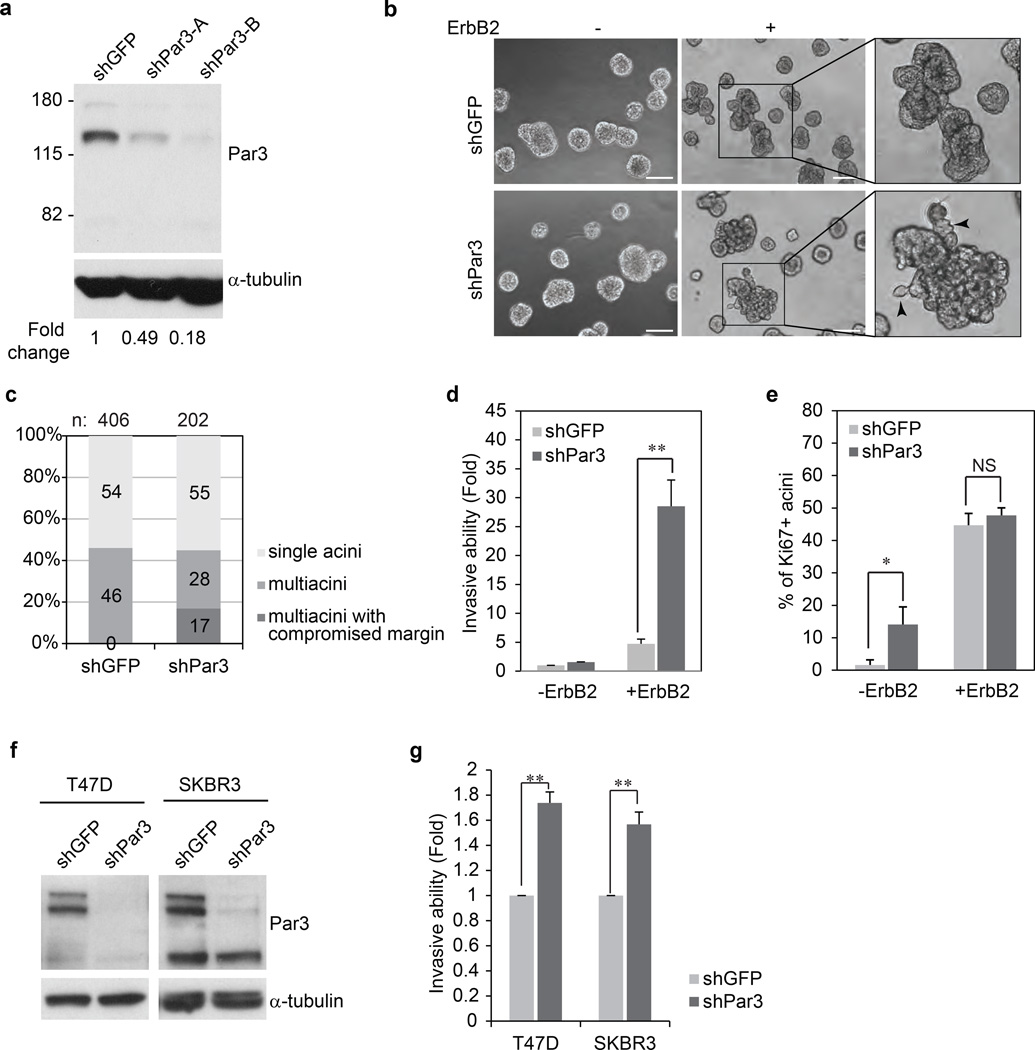
Figure 1. Loss of Par3 cooperates with ErbB2 to induce invasive behaviour in mammary epithelial cells.
(a) Par3 protein level in 10A.B2 cells expressing GFP shRNA and two shRNAs targeting Par3. (b) Phase contrast images of day 12 acinar structures unstimulated (−) or stimulated (+) with dimerizer AP1510, a small molecule ligand to activate ErbB2, for four days. Right panels show a higher magnification of the multiacinar structures. Arrows point to the protrusions from the multiacinar. (Scale bar = 100 µm) (c) ErbB2 activated acini were classified according to their microscopic morphology. Multiacinar structures were identified as those having three or more acinar structures and those with invasive protrusions were categorized as those with compromised margin. Percentages were determined by scoring 406 acini for shGFP and 2002 for shPar3. (d) 10A.B2 cells stably expressing shGFP or shPar3 were seeded for transwell invasion assays, with or without activation of ErbB2 and incubated for 48 hours. Results are represented as fold change in invasion compared to shGFP cells without ErbB2 stimulation. The data are presented as mean ± SEM collected from three independent experiments; +ErbB2 **P=007 by Student’s t-test. (e) Percentage of Ki67 positive acini. Day 16 acini from 10A.B2 shGFP, 10A.B2 shPar3 cells were grown with or without ErbB2 stimulation for 4 days and immunostained for Ki67. The data are presented as mean ± SEM collected from four independent experiments, −ErbB2 *P=0.069, +ErbB2 P=0.510 by Student’s t-test. (NS) (f) Lysates from shGFP or shPar3 T47D and SKBR3 cells were analysed for Par3 protein levels. (g) T47D and SKBR3 cells were seeded for transwell invasion assays and incubated for 24 hours and 72 hours respectively. Invaded cells were quantified represented as fold change in invasion compared to shGFP cells. The data are presented as mean ± SEM collected from three independent experiments; T47D **P=0.002, SKBR3 **P=0.009 by Student’s t-test.
Activation of ErbB2 in both shGFP and shPar3 cells disrupted normal three-dimensional (3D) acinar morphogenesis and induced formation of multiacinar structures when grown in Matrigel (Fig. 1b, 1c). Interestingly, 17% of multiacinar structures derived from shPar3 cells showed evidence of loose cell-cell adhesions and invasive protrusions (Fig. 1b, lower panel, 1c). Consistent with invasive protrusions (Fig. S1), activation of ErbB2 induced a significant increase in invasion of shPar3 cells when compared to shGFP cells (30 fold versus 5 fold increase respectively) (Fig. 1d) in transwell invasion assays. Both shGFP and shPar3 cells showed an increase in proliferation in response to ErbB2 activation as monitored by Ki-67 expression (Fig. 1e). Acini derived from shPar3 cells, in the absence of ErbB2 activation, showed increased proliferation compared to shGFP cells (Fig. 1e), consistent with recent observations in mouse mammary epithelial cells25 where loss of Par3 induced an increase in cell proliferation. However, loss of Par3, in the absence of ErbB2 activation, was not sufficient to induce multiacinar structures (Fig. 1b).
In addition to MCF-10A cells, we knocked down Par3 in T47D and SKBR3 cells (Fig. 1f). Both T47D and SKBR3 cells expressing shPar3 showed a significant increase in invasive ability in transwell invasion assays (Fig. 1g). Thus, loss of Par3 induced invasion both in ErbB2-transformed MCF-10A cells and in human breast tumour-derived cell lines.
Downregulation of Par3 promotes metastasis of primary mouse mammary tumours in vivo
To determine if downregulation of Par3 induces metastasis in vivo, we generated lentiviral shRNAs targeting mouse Par3 (Fig. 2a). Primary tumour epithelial cells were isolated from transgenic mice expressing Neu/ErbB2 deletion mutant (NDL) (an extracellular domain mutant that promotes constitutive receptor dimerization) under the control of mouse mammary tumour virus (MMTV) long terminal repeat (LTR)26. Cells were infected with a control virus or mPar3 shRNA co-expressing enhanced green fluorescent protein (EGFP) as a reporter for infected cells. Infected, but not selected, cells were transplanted into pre-cleared mammary fat pads of NOD/SCID mice. Both vector and mPar3 shRNA-infected cells developed GFP-positive tumours with the same latency. One cohort of mice was sacrificed 12 weeks after transplantation when the tumour size reached 1.5 cm diameter. In the second cohort of mice, the primary tumour nodes were removed at week six to relieve the tumour burden and the mice were maintained for 14 to 18 more weeks (Fig. 2b). The primary tumours generated by mPar3 shRNA-infected cells or vector-infected cells did not differ significantly in total mass or their histology (Fig. 2c, 2d). mPar3 shRNA-infected cells developed significantly more GFP-positive lung metastases (5 out of 10) compared to the lung metastases in vector-infected group (1 out of 10) (Fig. 2e, p = 0.008). Primary tumours and metastases from both vector and shmPar3 had epithelial morphology and expressed E-cadherin at cell-cell contacts and shmPar3-derived primary tumours and metastases had downregulation of Par3 expression (n= 3; Fig. 2d, 2e, 2f). In addition, loss of Par3 also promoted local invasion of transplanted tumour cells (See Fig S2 for details). Thus loss of Par3 was sufficient to initiate metastatic behaviour in epithelial cells derived from ErbB2-induced primary mouse mammary tumours.
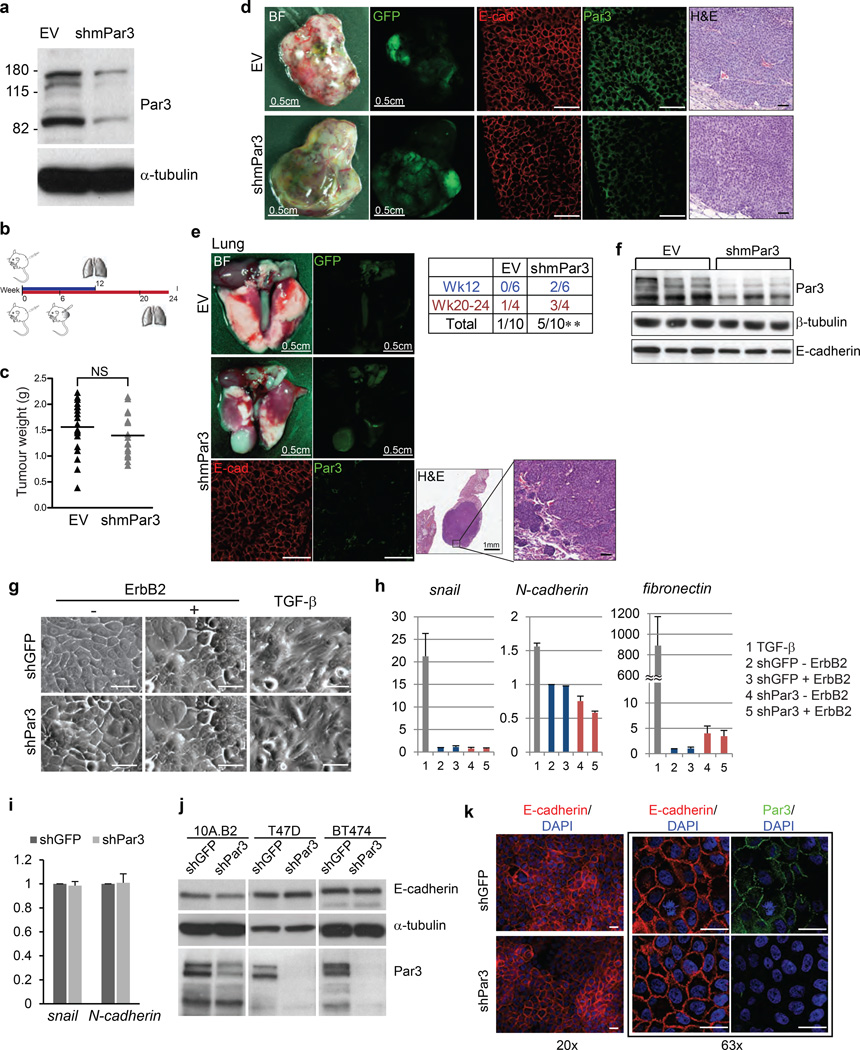
Figure 2. Downregulation of Par3 promotes metastasis without EMT.
(a) Validation of shRNA against mouse Par3 (shmPar3) in mouse mammary epithelial Comma1-D cells by immunoblot. (b) Schematic illustration of the transplantation experiment. (c) Weight of tumours grown from MMTV/NDL empty vector (EV) or mPar3 shRNA (shmPar3) transplantation at week12. The solid lines represent the mean value. n=18 (EV) and 17 (shmPar3). P=0.290 (NS) by unpaired t-test. Bright field and GFP images of (d) primary tumours collected 12 weeks after transplantation or (e) the representative lungs collected 24 weeks after transplantation are shown. Tissue sections from the primary tumours or the lung metastasis in shmPar3 transplantation or primary tumours were stained with immunoflorescent labels against E-cadherin and Par3 or H&E. (Scale bar = 50 µm) (e) The incidence of lung metastasis is shown in the table. EV P=0.341 (NS), shmPar3 **P=0.008 by one sample t-test with test value=0. (f) Cell lysates from primary tumours grown from transplantation were analysed for Par3 and E-cadherin and β-tubulin was served as loading control by immunoblot. (g) Phase contrast images of 10A.B2 cells untreated or treated with ErbB2 dimerizer or TGF-β (5.0 ng/ml) for four days. (Scale bar = 100 µm) (h) Expression of mesenchymal markers in 10A.B2 cells was determined by quantitative PCR using primers against snail, N-cadherin and fibronectin. The data were normalized to β-actin mRNA levels, TGF-β stimulation was used as positive control. The data were presented as mean ± SEM collected from three independent experiments. (i) mRNA from T47D cells was examined for snail and N-cadherin levels by quantitative PCR. The data were normalized to β-actin and fold changes are shown as mean ± SEM collected from 3 independent experiments. (j) Protein lysates from shGFP or shPar3 10A.B2, T47D and BT474 cells were analysed for changes in E-cadherin protein levels. (k) Confluent 10A.B2 cells were immunostained for E-cadherin and the nuclei. Boxed images show the immunostaining for Par3 and E-cadherin at higher magnification. (Scale bar = 20 µm)
Acquisition of invasive behaviour is frequently associated with EMT. However, activation of ErbB2 did not induce morphological changes associated with EMT in either shGFP or shPar3 cells (Fig. 2g). However, stimulation of both shGFP and shPar3 cells with TGF-β, a known promoter of EMT, induced a loss of epithelial morphology and acquisition of spindle-shaped mesenchymal morphology (Fig. 2g, right panel). Consistent with the lack of changes in cell morphology, the expression of mesenchymal markers such as snail, N-cadherin and fibronectin was not altered by loss of Par3 in either 10A.B2 or T47D cells (Fig.2h and 2i). We note that there was a modest increase in fibronectin expression (less than 5 fold), which was low compared to the more than 800 fold increase in expression observed in response to TGF-β (Fig. 2h). In addition, loss of Par3 did not decrease expression of epithelial marker E-cadherin in 10A.B2, T47D or BT474 cells (Fig. 2j). We also did not observe obvious differences in E-cadherin localization in confluent monolayers of epithelial cells (Fig. 2k) or tumours derived from shPar3 cells (Fig. 2d). These observations suggest that loss of Par3 cooperates with ErbB2 to induce invasive behaviour in epithelial cells without being associated with an overt EMT.
Loss of Par3 decreases cell-cell cohesion
To understand the mechanism by which loss of Par3 induces invasion, we monitored ErbB2-induced changes in MCF-10A cell behaviour in monolayers. Dividing shPar3 daughter cells failed to stay cohesive within a colony whereas shGFP daughters remained proximal to each other within an epithelial island (Fig. 3a) suggesting a decrease in cell-cell cohesion. To test if this decrease in cohesiveness is due to increased motility promoted by integrin-mediated signalling27, shGFP and shPar3 cells were plated on Matrigel-coated coverslips in the absence or presence of ErbB2 activation (Fig. 3b). The cells did not differ in their ability to adhere and spread on a Matrigel coated surface demonstrating that loss of Par3 did not activate integrin-mediated functions. To test if loss of Par3 affects cell-cell interactions, we performed a cell aggregation assay by suspending cells in drops of media hanging from the lid of a culture dish. Activation of ErbB2 in shGFP cells induced a decrease number of cell clumps (Fig. 3c, 3d), indicating weakened cell cohesiveness, which is consistent with previous reports showing that activation of ErbB2 can induce a decrease in E-cadherin function28. ShPar3 cells, in the absence of ErbB2 activation, decreased the number of clumps and activation of ErbB2 induced dispersal of all large clumps in shPar3 cells (Fig. 3c, 3d).
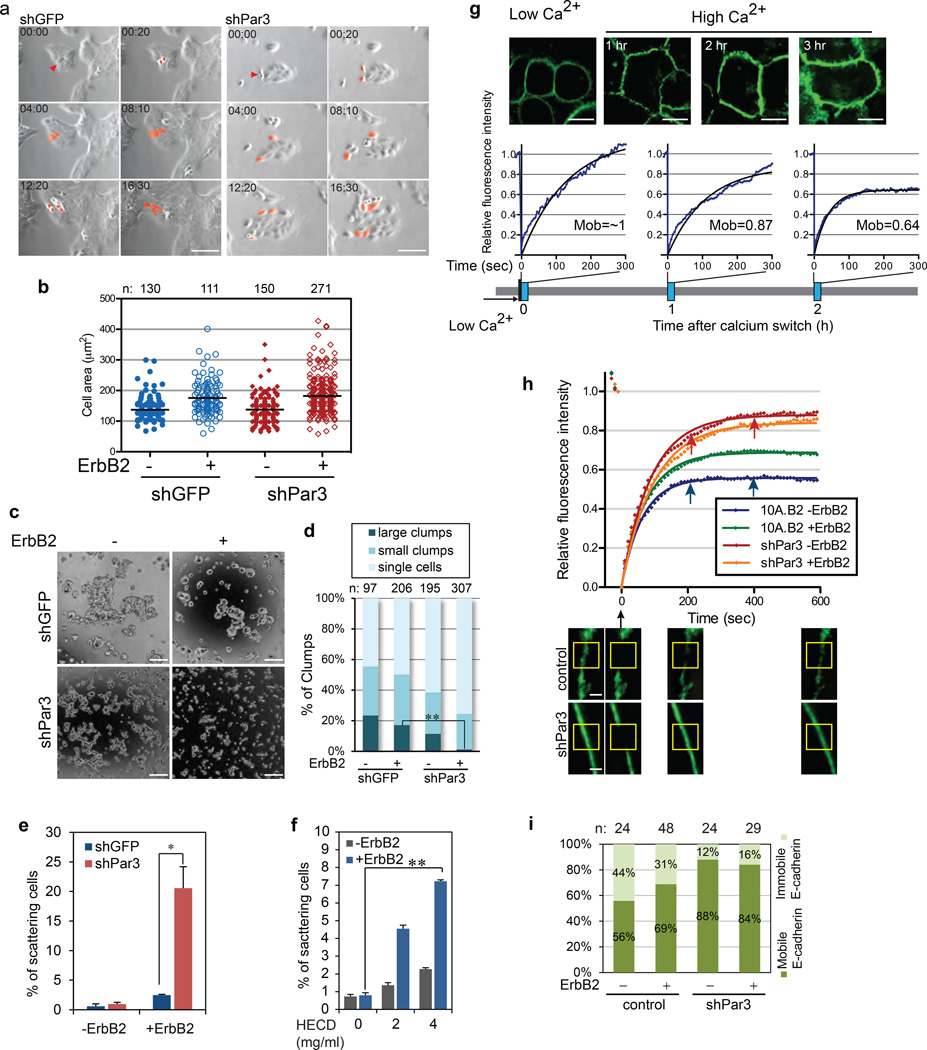
Figure 3. Loss of Par3 weakens cell-cell adhesions and inhibits E-cadherin junction stability.
(a) 10A.B2 cells were grown on plastic dishes with ErbB2 stimulation and imaged every 10 minutes for 18 hours. Still snapshots are shown. In 00:00 images, the red arrows point to a mother cell. In subsequent images, the red colour tracks the progeny of the mother cells. (Scale bar = 100 µm) (b) Cell spreading assay with 10A.B2 cells. The graph shows the scatter plot of the cell area. The solid lines represent the mean value. (c) 10A.B2 cells were cultured in hanging drops as indicated for 20 hours, and then imaged by phase contrast microscopy. Representative images from each condition are shown. (Scale bar = 100 µm) (d) The clump size was measured and categorized into single cell, small clumps (2~10 cells) and large clumps (more than 10 cells). The percentage of clumps was presented as a bar graph on the right. **P=0.0045 by Student’s t-test. (e) Quantification of cell scattering of 10A.B2 shGFP or shPar3 cells grown on plastic dishes, without (−) or with (+) ErbB2 stimulation for 24 hours. A cell was scored as a scattered cell if it lost contact with its neighbours. Results are presented as mean ± SEM from three independent experiments. (f) Quantification of cell scattering of 10A.B2 cells grown in the presence of HECD-1 or control mIgG for 24 hour. (n=3, mean ± SEM). (g) Confluent monolayer of control or shPar3 10A.B2 cells expressing E-cadherin-GFP were incubated in low calcium medium for four hours and switched to high calcium medium. FRAP analysis was performed at indicated time points. The black lines indicate best fitting curves of nonlinear regression analysis. E-cadherin mobility was calculated from the fitting curves. Sample size n=8, 9 and 8 respectively. (Scale bar = 10 µm) (h) FRAP was conducted on control and shPar3 cells expressing E-cadherin-GFP. The bottom panel shows the representative confocal sections of the cells before and after various time points after photobleaching. The yellow-boxes mark the bleached regions. (Scale bar = 2 µm) (i) Quantification of E-cadherin immobile and mobile fractions from Fig 3g. Sample sizes are shown above the graph.
E-cadherin-based adherens junctions (herein referred to as E-cadherin junctions) are the primary mediators of cell-cell interaction in epithelial cells and disruption of E-cadherin junctions is known to induce cell scattering. Activation of ErbB2 induced scattering of shPar3, but not the control shGFP cells (Fig. 3e, phase images in Fig. S3a) and neither shGFP nor shPar3 cells scattered in the absence of ErbB2 activation. Interestingly, shGFP cells exposed to E-cadherin neutralizing antibody (HECD-1)29 scattered upon ErbB2 activation (Fig 3f, S3b), demonstrating that cell scattering in the Par3-depleted cells is due to a combined effect of weakened E-cadherin junctions and activation of ErbB2.
Par3 is required for E-cadherin junction stability
We investigated if Par3 regulates functional properties of E-cadherin junctions. During formation of E-cadherin junctions, nascent junctions are unstable and have E-cadherin molecules that constantly recycle at junctions. In contrast, mature junctions are stable and have a high fraction of immobile E-cadherin in MDCK cell monolayers30, 31. 10A.B2 cells expressing E-cadherin-GFP was subject to calcium switch and E-cadherin at cell-cell contacts was monitored by fluorescence recovery after photobleaching (FRAP). 100% of E-cadherin was recovered within five minutes after photobleaching at t=0, whereas, only 87% and 64% of E-cadherin was recovered at 60 min and 120 min after calcium switch respectively, demonstrating that the percentage of the immobile fraction of E-cadherin increased as cell-cell junctions mature (Fig. 3g).
FRAP analysis in established junctions revealed that only 56% of the GFP signal was recovered in control cells, whereas, 88% of the GFP signal was recovered in shPar-3 demonstrating that only 12% of the E-cadherin junctions were immobile in cells lacking Par3 (Fig.3h and 3i). These observations demonstrate that E-cadherin junctions were labile in mammary epithelial cells lacking Par3.
Loss of Par3 activates Tiam1-Rac signalling
Par3 is known to interact with Tiam1 and inhibit its guanine nucleotide exchange activity towards Rac GTPase22. While Rac activity is required for E-cadherin junction formation32, constitutive activation of Rac induces disruption of E-cadherin junctions by promoting aberrant actin remodelling33, 34. To monitor changes in Tiam1 activation and Rac-GTP levels in shPar3 cells we used Rac1 G15A, a nucleotide-free Rac1 mutant that interacts with active Tiam135, 36, and p21-activated kinase (PAK) binding domain (PAK-PBD) pull down assays37. Cells lacking Par3 had a dramatic increase in the levels of active Tiam1 (Fig. 4a) and high Rac-GTP levels under unstimulated conditions compared to the levels present in shGFP cells (Fig. 4b, comparing lanes 1 and 5). Activation of ErbB2 induced a four-fold increase in Rac-GTP levels in shGFP cells (Fig. 4b), similar to the levels observed in shPar3 irrespective of ErbB2 activation. In contrast, loss of Par3 did not have any effect on either basal or ErbB2-induced Ras/mitogen activated protein kinase (MAPK) activation or ErbB2-activated AKT pathway (Fig 4c).
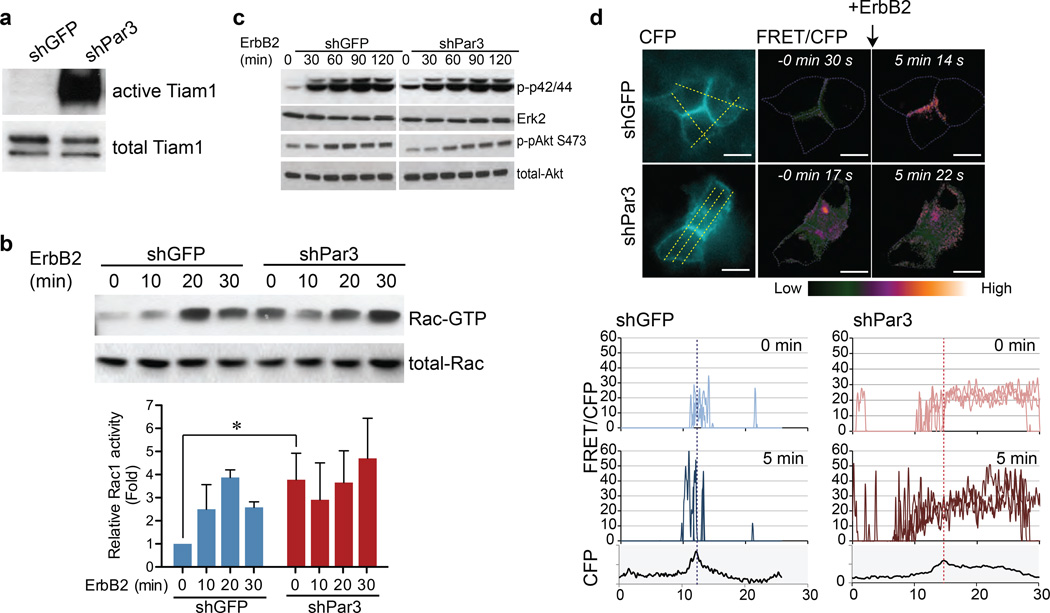
Figure 4. Loss of Par3 induces aberrant activation of Tiam1-Rac signalling.
(a) Cell lysates from shGFP or shPar3 10A.B2 cells were subjected to Tiam1 Rac-GEF activity assay. The level of active Tiam1 pulled down by Rac1 G15A agarose beads (upper panel) and total Tiam1 (lower panel) were monitored by immunoblotting with Tiam1 antibody. (b) Cell lysates, from confluent monolayers stimulated with ErbB2 dimerizer for indicated times, were incubated with GST-PAK1 agarose beads and bound active Rac (Rac-GTP) was monitored by immunoblotting with anti-Rac antibody. Total Rac levels were monitored using one tenth of the input lysate. Quantification of relative levels of Rac-GTP of three independent experiments is shown in the graph below (mean ± SEM). (c) Cell lysates from confluent monolayers stimulated with ErbB2 for indicated time were analysed for phospho-p42/44, total Erk2, phospho-Akt S473 and total Akt levels by immunoblot analysis. (d) Confluent 10A.B2 cells expressing Raichu-Rac were treated with ErbB2 dimerizer. CFP and FRET images were obtained before stimulation and five minutes after stimulation. Representative FRET/CFP ratio images after background subtraction are shown. The ratio is represented by LUT colours from green to orange. All images were normalized using the same threshold and ratio range. The CFP donor images are shown on the left. (Scale bar = 20 µm)The change in Rac activity was presented by plotting the histogram of the indicated lines across the cells (yellow lines) in the normalized ratio pixel images. The dotted vertical lines in the graphs represent the cell-cell junction determined by the maximum CFP intensity.
To monitor changes in spatial distribution of Rac-GTP activation in shPar3 cells, we used Raichu-Rac biosensor by Förster resonance energy transfer (FRET). The localization of Raichu-Rac fusion monitored as CFP image is different from endogenous Rac and is known to be related to local GEF activity38. Raichu-Rac was mainly localized to cell-cell junctions in shGFP cells, whereas it showed a diffuse distribution in the cytosol in shPar3 cells (Fig. 4d). Upon ErbB2 activation, Rac-GTP loading was focally increased at cell-cell junctions in shGFP cells, whereas Rac-GTP loading was diffusely elevated throughout the cytosol in shPar3 cells (Fig. 4d, S4a). These observations demonstrate that Par3 is required for spatial control of Rac-GTP activation in mammary epithelial cells.
Contribution of Tiam1-Rac and ErbB2 signalling to Par3 loss-induced cell invasion and E-cadherin junction stability
Downregulation of Tiam1 (Fig 5a) efficiently inhibited ErbB2-induced invasion in shPar3 cells (Fig. 5b), demonstrating that increased Tiam1 activity in shPar3 cells is required for cell invasion. In addition, we generated Flag-tagged RNAi-resistant Par3 full length (Par3-FL) and a truncated mutation lacking the C-terminus, which cannot bind to Tiam122 (Par3-ΔC) (Fig. 5c, 5d). Expression of Par3-FL was able to inhibit Par3 loss-induced cell invasion, while Par3-ΔC failed to rescue the Par3-loss phenotype (Fig. 5e). Furthermore, stable expression of dominant-negative Tiam1 (Fig. S4b), a mutant of Tiam1 that contains N-terminal pleckstrin homology (PH) domain, the proximal coiled coil (CC) and adjacent Ex domain (Tiam1-PHn-CC-Ex)39 (Fig. S4b), inhibited the formation of shPar3 acini with compromised margins (Fig. S4c) and decreased the ability of shPar3 cells to invade through Matrigel (Fig. S4d). Lastly, we tested the effects of the compound NSC23766, an inhibitor that interferes with binding of Rac to Tiam1 or TrioN GEFs40. Treatment of shPar3 cells with NSC23766 at concentrations sufficient to decrease the high basal levels of Rac-GTP observed in shPar3 cells (Fig. 5f) inhibited ErbB2-induced cell invasion (Fig. 5g) and cell scattering (Fig. S4e). Thus, the Tiam1-Rac pathway was required for shPar3-induced cell invasion.
Figure 5. Contribution of Tiam1-Rac and ErbB2 signalling to Par3 loss-induced phenotype.
(a) Immunoblot of Tiam1 level in 10A.B2 shGFP or shPar3 cells expressing shRNA targeting Tiam1. (b) 10A.B2 shPar3 cells expressing control vector or shTiam1 were subjected to transwell invasion assays as indicated. Invaded cells were quantified and presented as fold change compared to shGFP cells without ErbB2 stimulation. Data are shown as mean ± SEM collected from three independent experiments;. **P=0.001 by Student’s t-test. (c) Left Panel: Schematic representation of Par3 full length (FL) and ΔC constructs used in this study. Abbreviation: CR1, conserved region 1. Right Panel: Cell lysates from 10A.B2 shPar3 expressing RNAi-resistant Flag-tagged Par3-FL and Par3-ΔC were immunoblotted using antibody against Flag. (d) Lysates from cells expressing control vector, Flag-Par3-FL or Flag-Par3-ΔC were immunoprecipitated with anti-Flag antibody and immunoblotted for Tiam1. (e) shPar3 cells expressing control vector, Flag-Par3-FL or Flag-Par3-ΔC, without or with ErbB2 stimulation, were subjected to transwell invasion assay. (mean ± SEM from 3 independent experiments. Student’s t-test: *P=0.08, **P<4.15E-05) (f) Cell lysates from 10A.B2 cells stimulated with ErbB2 dimerizer for 15 minutes in the absence (−) or presence (+) of two different concentrations of NSC23766 were subjected to Rac activity assay. (g) 10A.B2 shGFP or shPar3 cells were were subjected to transwell invasion assay as described above. The Rac inhibitor, NSC23766, was added at the indicated concentrations at the same time as ErbB2 activation. (mean ± SEM from 3 independent experiments, Student’s t-test: *P=0.051). (h) FRAP analysis was conducted on the E-cadherin-GFP expressing 10A.B2 cells in the absence or presence of 50 µM NSC23766 compound. The E-cadherin immobile and mobile fractions were calculated and plotted. Sample sizes are listed above the graph. (i) Cell lysates form shGFP (G) or shPar3 (P) cells stimulated with ErbB2 dimerizer for 30 minutes in the absence or presence of 1 µM U0126, 50 µM PD98056 or 300 nM GSK690693 were analysed for phospho-RSKp90, total-Akt, phospho-p42/44 and total Erk2 by Western blot. The cells treated with inhibitors were subjected to (j) transwell invasion assay (mean ± SEM, n=3) and (k) E-cadherin FRAP analysis (sample sizes are listed below the graph) as described above.
To determine if dysregulation of Tiam1-Rac.GTP is required for changes in E-cadherin junction, shPar3/E-cadherin-GFP cells were treated with NSC23766 and subjected to FRAP analysis. While treatment of the parental 10A.B2 cells with NSC23766 did not change the percentage of immobile fraction of E-cadherin, treatment of shPar3 cells induced a more than two fold increase in the immobile E-cadherin fraction (Fig. 5h) demonstrating that the increase in Rac activation is required for shPar3-induced inhibition of E-cadherin junction stability.
To understand how ErbB2 signalling cooperates with shPar3 to promote unstable E-cadherin junctions and cell invasion, we blocked ErbB2-induced MAPK activation using MEK inhibitors U0126 and PD98059, or AKT using GSK690693 (Fig. 5i). Suppression of MEK or AKT activity inhibited ErbB2-induced invasion of shPar3 cells (Fig. 5j). Interestingly, none of these inhibitors restored shPar3-induced defects in E-cadherin junction turnover (Fig. 5k).
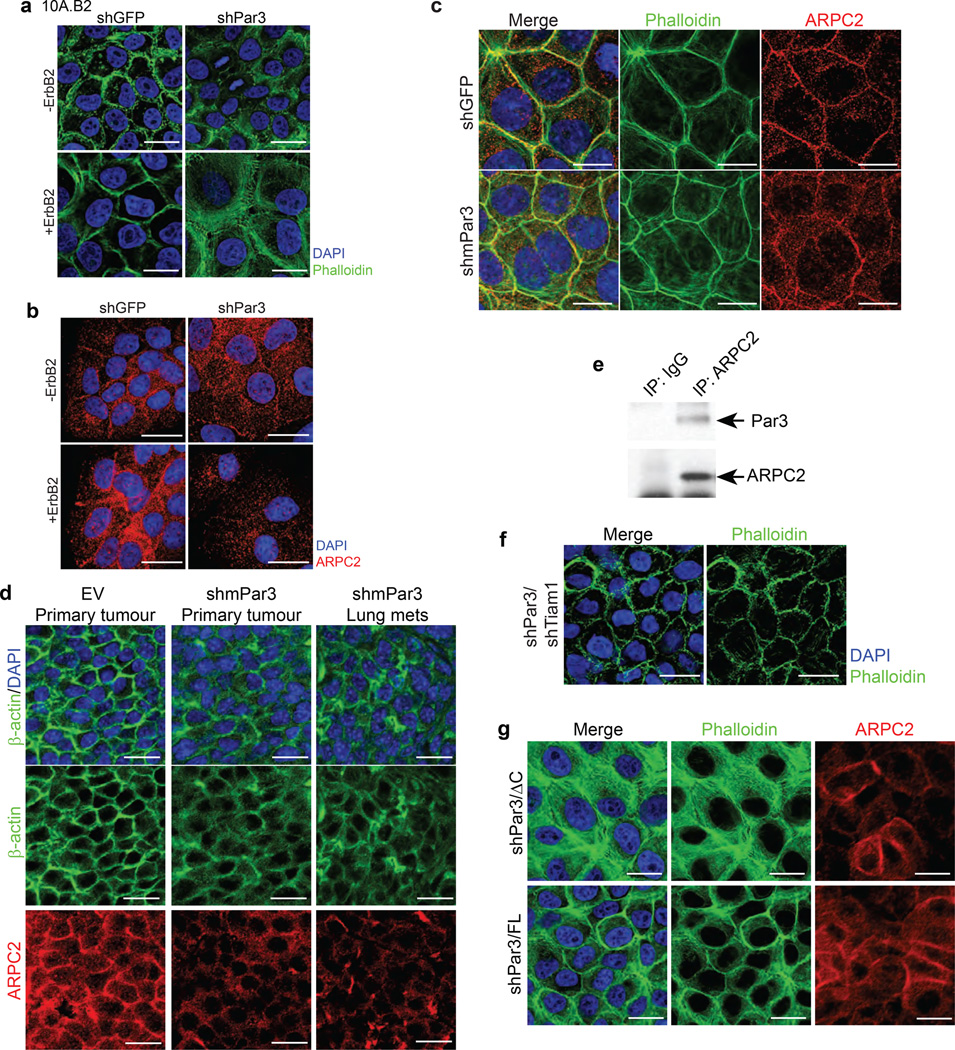
Loss of Par3 induces changes in actin cytoskeleton organization and ARPC2 localization
The actin cytoskeleton undergoes major changes during E-cadherin junction maturation. Since Rac activity is known to regulate actin dynamics41, 42 by affecting actin nucleators including the Arp2/3 complex, we investigated whether actin cytoskeleton organization or Arp2/3 complex localization is altered in Par3 knockdown cells. In a confluent layer of shGFP 10A.B2 epithelial cells, F-actin was organized at cell-cell junctions to form a compact cortical actin belt (Fig. 6a, left panel), whereas, shPar3 cells had loosely organized F-actin (Fig. 6a, right panel). ARPC2, (also known as p34-ARC)43, a component of the Arp2/3 complex was localized to cell-cell junctions in shGFP cells but was mislocalized throughout the cell in shPar3 cells (Fig. 6b). ErbB2 activation induced a modest loosening of the cortical actin in shGFP cells and a severe disruption of cortical actin and ARPC2 localization in shPar3 cells (Fig. 6a, 6b). Loss of Par3-induced changes in cortical actin and ARPC2 were observed in Eph4, a normal mouse mammary cell line, stably expressing shPar3 (Fig. 6c), as well as in the T47D breast cancer cell line (Fig S5a), demonstrating that the shPar3-induced changes in cortical actin and ARPC2 localization were not specific for MCF-10A cells.
Figure 6. Loss of Par3 induces changes in actin organization and mislocalizes Arp2/3 complex.
Confluent monolayers of 10A.B2 cells expressing shGFP or shPar3, without or with ErbB2 activation, were (a) stained with phalloidin for F-actin (green) and nuclei (DAPI, blue), or (b) immunostained with ARPC2 (red). (c) Eph4 mammary epithelial cells expressing shGFP or shmPar3 were stained with phalloidin for actin cytoskeleton (green) and immunostained for ARPC2 (red). (d) Tissue sections of tumours generated using MMTV-NDL tumour cells infected with vector or mPar3 RNAi lentivirus were immunostained for actin cytoskeleton (β-actin, green) or ARPC2 (red). (e) Cell lysates from confluent monolayers of 10A.B2 cells were immunoprecipitated using anti-ARPC2 and immunoblotted as indicated. Rabbit IgG was used as non-specific control in the experiments. (f) shPar3 10A.B2 cells expressing Tiam1 shRNA were stained with phalloidin for actin cytoskeleton. (g) shPar3 10A.B2 cells expressing Par3-FL and Par3-ΔC were stained with phalloidin for actin cytoskeleton (green) and immunostained for ARPC2 (red). (Scale bar = 20 µm above)
Interestingly, primary tumour generated from empty vector-infected NDL tumour cells (Fig. 2d) had well-organized cortical actin and ARPC2 localization to cell-cell junctions, whereas the tumour and metastasis generated from shPar3-infected cells showed loose cortical actin and mislocalization of ARPC2 from cell-cell junctions (Fig. 6d, S5b). In addition, the tumour epithelia in the spontaneous lung metastasis of MMTV-NDL mice had a dramatic disruption of cortical actin organization and ARPC2 localization compared to that observed in cells within primary tumours from the same mice (Fig. S5c). Furthermore, we observe a biochemical interaction between ARPC2 and Par3 by co-immunoprecipitation analysis (Fig. 6e, S5d). These observations demonstrate that loss of Par3 relates to disruption of cortical actin and mislocalization of ARPC2 in epithelial cells in culture and in vivo.
Given that Par3 loss-induced cell invasion requires Tiam1, we investigated the role played by Tiam1 on actin organization and ARPC2 localization. Knockdown of Tiam1 in shPar3 cells or inhibition of Tiam1-Rac activation using NSC23766 lead to an increase in cortical actin organization (Fig 6f, S5e). Whereas expression of Par3-FL in shPar3 cells restored cortical actin organization, expression of Par3-ΔC failed to do so (Fig. 6g). Interestingly, both Par3-FL and Par3-ΔC restored ARPC2 localization to cell-cell junctions (Fig. 6g) suggesting that ARPC2 localization can be uncoupled from Tiam1-Rac signalling and that cell-cell junction localization of ARPC2 was not sufficient to restore cortical actin organization.
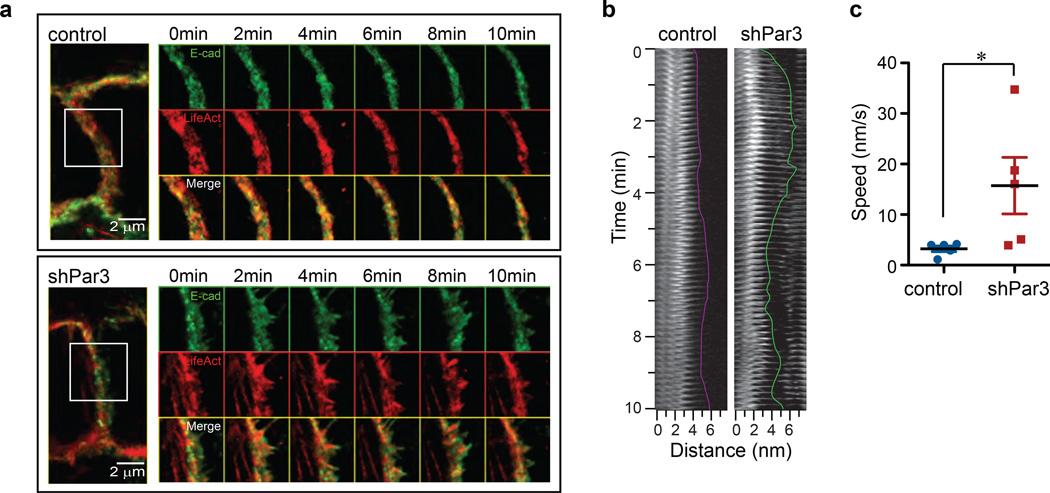
Loss of Par3 promotes aberrant actin and E-cadherin dynamics at cell-cell junctions
The decrease in cell-cell cohesion may be due to aberrant actin and cadherin dynamics at cell-cell contact. To test this directly, we performed time-lapse imaging of cells coexpressing E-cadherin-GFP and red fluorescent protein TagRFP fused to Lifeact, a 17-amino acid peptide that labels F-actin44. In the control cells, E-cadherin (green) colocalized with F-actin (red) to form a continuous, smooth junctional structure of even thickness (Fig. 7a). During the 10 minutes of time-lapse imaging, we did not observe any noticeable change in the overall organization of F-actin or E-cadherin (Fig. 7a, upper box). However, in shPar3 cells F-actin organization was complex with thin actin filaments adjacent to the cortical belt, similar to the structures reported in immature cell-cell contacts. During the 10 minutes of time-lapse imaging, E-cadherin and F-actin at cell-cell junctions of shPar3 cells were highly dynamic with oscillating protrusions and retractions (Fig. 7a, lower box, see also Supplementary Video 1). Kymograph analysis of Lifeact-TagRFP from representative regions in control and shPar3 cells revealed that the average speed of actin protrusions at cell-cell junctions in shPar3 cells was 4–5 fold higher compared to that observed in control cells (Fig. 7b, 7c). Thus, loss of Par3 promotes dynamic remodelling of F-actin and E-cadherin at cell-cell junctions.
Figure 7. Loss of Par3 introduces changes in actin and E-cadherin dynamics at cell-cell junctions.
(a) Confluent monolayers of control and shPar3 10A.B2 cells stably co-expressing E-cadherin-GFP and TagRFP-Lifeact were imaged every 5 seconds for 10 minutes. Representative images of cell-cell junctions are shown. The right panels show time-lapse montages at 2 minute intervals of an area delineated by a white box in the cell-cell junction images in the left panel. (b) Kymograph of a representative region of TagRFP-Lifeact at the cell-cell junction in control cells (left) or shPar3 cells (right). (c) The mean speed of five protrusions was measured from kymographs and shown in the graph. *P=0.056 by Student’s t-test .
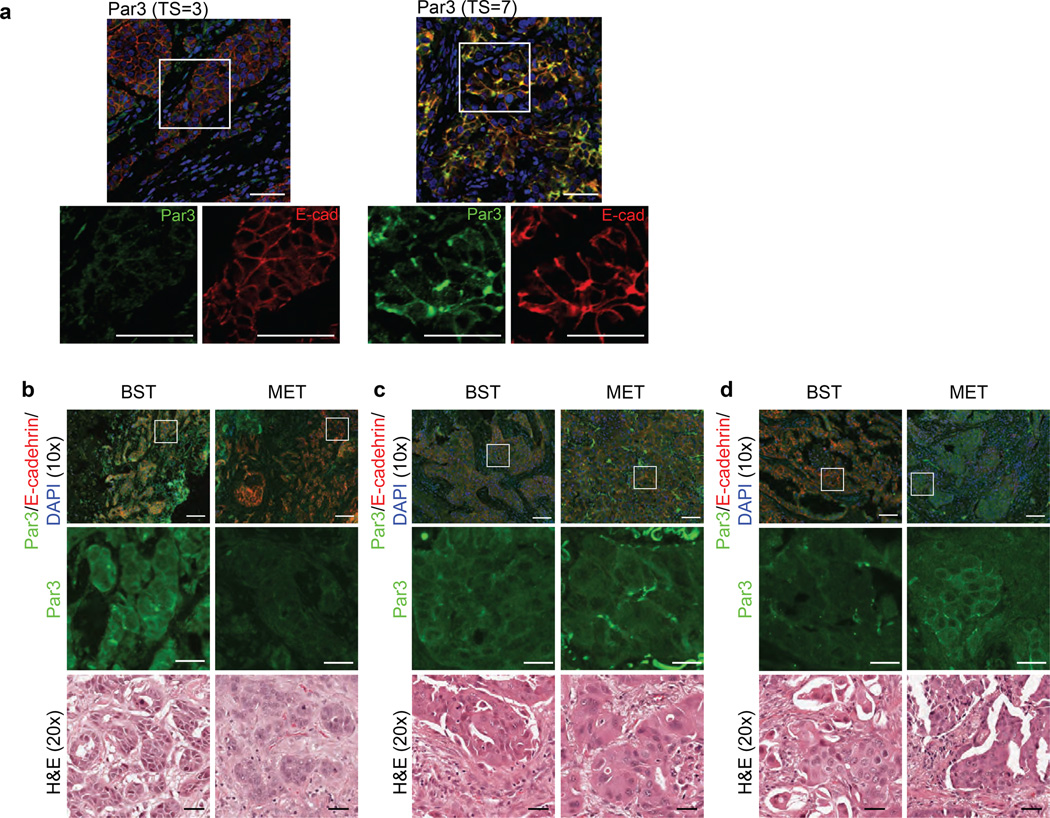
Dysregulation of Par3 in human breast cancer
To understand the relationship between Par3 expression and tumour characteristics in invasive breast cancer (IBC), we analysed formalin-fixed paraffin-embedded tissue (FFPET) samples of human IBCs (n=98) assembled on tissue microarrays45. Par3 expression was evaluated by indirect immunofluorescence using anti-Par3 antibody. Change in membrane-localized Par3 was studied using the Allred Score (Total Score (TS) range 0–8) (See Fig. S for more details). An Allred score of 3 corresponds to as few as 10% cells with weak membrane signal and an Allred score 7 indicates strong membrane Par3 in 66% of cells (Fig. 8a). Par3 membrane localization was significantly decreased in IBCs associated with clinically aggressive prognostic factors, including higher nuclear grade, negative ER status, and positive HER2 status (Suppl. Table 1); indicators of poor clinical prognosis. We have also observed Par3 mislocalized in some human breast cancer samples analysed (Fig. S6), however, the cause and consequence of Par3 mislocalization is still unclear.
Figure 8. Dysregulation of Par3 in human breast cancer.
(a) IHC for Par3 protein expression representing a low (Total Score, TS=3) and high (TS=7) Allred score (Par3: green, E-cadherin: red, nuclei: DAPI). The lower panels represent a zoomed image of the samples. (Scale bar = 50 µm) (b), (c) and (d) Primary human breast tumours (BST) and matched metastasis (MET) were immunostained for Par3 (green) and E-cadherin (red). (Scale bar = 100 µm) Middle inserts are zoomed images of the boxed region. Bottom inserts correspond to H&E stained images of the tumour or metastasis. (Scale bar = 20 µm) (b) shows the representative pair in which the metastasis has a decrease in the level of membrane Par3 expression compared to the matched primary tumour. (c) shows the representative pair in which Par3 protein expression is at the same level in BST and MET. (d) shows the representative pair in which the metastasis has an decrease in the level of membrane Par3 expression compared to the matched primary tumour.
To address if changes in Par3 expression are associated with breast cancer metastasis, we identified 14 pairs of primary breast tumour (BST) and matched metastasis (MET) from the same patient and analysed them for expression of Par3 by immunohistochemistry and quantified using Allred score. The results demonstrate that eight out of 14 metastases have a decrease in the level of membrane Par3 expression compared to matched primary tumours (Fig. 8b), two out of 14 metastases show no change in levels of Par3 (Fig. 8c), and four out 14 metastases show an increase in the levels of Par3 when compared to the Par3 levels in matched primary tumours (Fig. 8d). Statistical analysis using binomial test suggests a significant probability (p= 0.0174) for BST>MET scenario (Fig. 8b), but not the other two groups. These observations provide direct evidence to suggest a trend of downregulation of Par3 in the metastases. We acknowledge that analysis of a larger cohort of primary and metastasis samples could provide additional insight. However, our current analysis of this 14 matched pair cohort of samples provides statistically significant evidence in support of our conclusion that downregulation of Par3 is associated with metastasis in breast cancer.
Discussion
We demonstrate that downregulation of Par3 expression is frequently observed in human breast cancer and promotes metastasis of ErbB2-induced mouse mammary tumours. Downregulation of Par3 promoted invasive behaviour of epithelial cells by inducing actin and E-cadherin dynamics at cell-cell contacts and inhibiting cell-cell cohesion in a Tiam1 dependent manner. Thus, we identify Par3 polarity protein as a novel metastasis suppressor in breast cancer.
Although ErbB2 is overexpressed in more than 60% of breast cancer patients with ductal carcinoma in situ (DCIS), only 15% of IDC express ErbB2 suggesting that expression of ErbB2 by itself is not sufficient to drive metastatic disease. Consistently, loss of 14-3-3σ and overexpression of 14-3-3ζ cooperates with ErbB2 to induce invasion and metastasis in cell culture and animal models of ErbB2-positive breast cancers46, 47. 14-3-3 (Par5) interacts with Par3 to regulate cell polarity during early divisions of C. elegans embryos and in mammalian epithelial cells48, 49. Our findings, in conjunction with previous reports, identify the Par3-14-3-3 signalling module as an important regulator of metastasis in ErbB2-overexpressing breast cancers.
We find that the aberrant increase in Tiam1 mediated Rac-GTP loading was required for Par3-loss induced effects on cortical actin and E-cadherin junction dynamics. While shPar3 cells also show mislocalization of ARPC2, restoring ARPC2 localization to cell-cell junctions was not sufficient to restore cortical actin organization or inhibit invasion. Thus, loss of Par3 promotes spatially dysregulated, Tiam1-mediated, Rac activation, which leads to uncontrolled Arp2/3 activity and actin dynamics. This aberrant actin dynamics likely compromises E-cadherin junctions and decreases cohesion between tumor epithelial cells and promote metastasis.
We speculate that, in addition to loss of Par3, unregulated activation of Arp2/3 can promote aberrant actin remodelling and compromise cell-cell cohesion to promote metastatic behaviours of epithelial cells. Consistent with this possibility, coexpression of Arp2 and Arp2/3 activator WAVE2 predicts poor outcome in invasive breast cancers50 and dysregulation of proteins that regulate actin nucleation is frequently observed in multiple cancers51. Elevated expression of both Cdc42 and Arp2/3 have been observed in invasive cells52. Loss of Arp2/3 inhibitors may also promote aggressive cancer. Consistent with this possibility, the genetic region containing α-catenin, an inhibitor of Arp2/353, was deleted in two out of three metastases but retained in the matched primary basal breast cancers54. Understanding the mechanisms of actin regulation during tumour progression will provide us new insights into tumour cell behaviour.
Recently, much evidence has suggested that the EMT process, typically associated with normal development, may be a critical process in tumour progression55, 56. Although we do not observe EMT, a role for partial EMT cannot be rule out. However, it is also possible that loss of Par3-induced metastasis does not require EMT, which is consistent with a concurrent study demonstrating that loss of Par cooperates with Notch intracellular domain or Ras61L-induced metastasis to promote metastasis of mammary tumors without showing EMT57. Our observations provide a novel perspective to our understanding of the cancer metastasis process.
Metastatic tumours are known to have significant evidence for loss of polarity, disordered differentiation and loss of lineage-specific markers in carcinomas. Although these changes can be highly pleomorphic, cells within the primary tumour as well as cells that are migrating away from the primary tumour tend to retain significant epithelial characteristics such as expression of epithelial keratins, mucins and E-cadherin58. While tumour cells that have undergone EMT during metastasis are thought to regain epithelial characteristics by undergoing a mesenchymal to epithelial transition55, the molecular mechanisms by which these transitions occur are poorly understood. Our observations on decrease in cell-cell cohesion and acquisition of a ‘metastatic-epithelial’ phenotype provides an additional perspective and identifies regulation of E-cadherin junction maturation, changes in cortical actin organization and cell-cell cohesion as mechanisms used by tumour epithelium to metastasize. Deeper and broader understanding of this process will not only identify novel predictive biomarkers for tumours with metastatic potential, but also identify novel strategies to inhibit metastasis.
METHODS
Plasmids
The lentiviral shRNA constructs against GFP (## RHS4459), human PARD3 (TRCN0000118134, mature antisense sequence 5’-ATCATAAGATTTGTCGATGGC-3’) and mouse Pard3 (TRCN0000094399, mature antisense sequence 5’-AAAGTGATAGAGATAAACTGG-3’) were obtained from The RNAi consortium (TRC) lentiviral shRNA library (Open Biosystems, Thermo Scientific). pLKO.3G vector was obtain from Christophe Benoist and Diane Mathis through Addgene (Addgene plasmid 14748). Tiam1 shRNA, targeting Homo sapiens Tiam1 (1305–1325bp) was generated by amplifying a 97-mer oligonucleotide (5’-TGCTGTTGACAGTGAGCGAAAGAGACTCCTCCGTACAGTATAGTGAAGCCACAGATGTATACTGTACGGAGGAGTCTCTTCTGCCTACTGCCTCGGA-3’) and cloning into retroviral pMSCV-LTR-hygromycin vector59. Preparation of virus, infection and selection were performed as previously described3. LifeAct-TagRFP was purchased from ibidi and cloned into XhoI/HpaI sites of pMSCV-blasticidin vector. RNAi-resistant Par3 construct was generated by mutating the RNAi recognition sequence (2782–2803bp) of wild type Par3 (kindly provided by Ziqiang Yang and Ben Neel, Ontario Cancer Institute) to GCTATTGATAAGAGTTACGAT encoding the same protein by Quikchange site-directed mutagenesis. The full length and ΔC (1–936aa) cDNA was amplified by PCR and cloned into pMSCV-hygro-3×Flag vector using an HpaI site. Tiam1-PHn-Ex-CC was generated by PCR amplification of pK-Tiam1-PHn-CC-Ex (kindly provided by Ian G. Macara, University of Virginia School of Medicine, VA) as an EcoRV/MfeI fragment and cloned into HpaI/EcoRI sites of pMSCV-PIG-Myc vector (kindly provided by S. Lowe, Cold Spring Harbor Laboratory, NY).
Antibodies and reagents
Antibodies were listed in Supplemental Table 2. Growth factor reduced Matrigel (BD Transduction Laboratories) was used for 3D culture and transplantation experiments. Small molecule ligand AP1510 (ARIAD Pharmaceuticals) was used to activate the chimeric ErbB2 receptor in 10A.B2 cells as previously described24. Recombinant human TGF-β (Abcam) and Rac inhibitor NSC23766 (Calbiochem), MEK inhibitor U0126 (EMD Millipore) and PD98059 (Calbiochem) and AKT inhibitor GSK690693 (Tocris Bioscience) were used as described.
Cell culture
10A.B2 cells were generated and cultured as previously described60. 3D culture of 10A.B2 cells, inducible ErbB2 activation and indirect immunofluorescence of 3D acini were performed as described earlier. T47D, BT474 and SKBR3 cells were cultured according to the protocol provided by ATCC.
Hanging Drop Assay
Drops (20 µl) of 10A.B2 cells (2.5×105 cells/ml) were pipetted onto the inner surface of the lid of a 12 well culture plate. The lid was quickly flipped over and then placed on the plate so that the drops were hanging from the lid with the cells suspended within them. To prevent evaporation, 2.0 ml serum-free culture medium was placed in the well. After 20 hours at 37 °C, the lid of the plate was inverted and photographed using a ZEISS inverted tissue culture microscope at 20× magnification. At least 5 drops were analysed per experiment. Clump areas were measured using ImageJ.
Transwell invasion assay
Transwell invasion assay were performed as previously described61. 1×105 cells were seeded on top of the 0.8µm pore size, Growth Factor Reduced Matrigel Invasion Chambers (BD Biosciences, Cat# 354483). Cells were incubated for 24 hours (for T47D cells) or 48 hours (for 10A.B2 cells) and fixed and counted.
Cell spreading assay
Cell spreading assay were performed on 10A.B2 cells as previously described62. Cells were plated on Matrigel-coated coverslips and allowed to adhere for 2 hours.
Rac-GTP pulldown
Confluent cell monolayers were starved with assay medium overnight before ErbB2 activation/stimulation. Assays were performed using the Rac1 activation assay kit (Cell Biolabs) according to the manufacturer’s instructions.
Tiam1 activity assay
Assays were performed using the active Rac-GEF assay kit (Cell Biolabs) according to the manufacturer’s instructions.
Immunoprecipitation
10A.B2 cells were lysed in in TNE buffer (25 mM Hepes, 150 mM NaCl, 0.5 mM EDTA, 5 mM MgCl2, 1% Triton X-100, 1 mM Na3VO4, 100 µg/mL aprotinin, 2.5 µg/mL leupeptin, and 1 µg/mL pepstatin). Lysates were collected by scraping the plates and were centrifuged at 15,000×g for 15 minutes at 4 °C. Protein concentration was measured using the Biorad protein assay. 1mg total protein were incubate with indicated antibodies for 4 hours at 4°C and the immune complex was then captured by Protein G Sepharose beads (GE Healthcare Life Science). The immunoprecipitated proteins were further analysed by SDS-PAGE and immunoblotting to determine protein interactions60.
Raichu-Rac Förster Resonance Energy Transfer (FRET) assay
Cells were transiently transfected with pRaichu-Rac plasmid by Amaxa electroporation (Lonza). 20 hours after transfection, cells were starved in assay medium overnight. Cells were imaged on a Zeiss Axiovert 200M wide-field microscope at 63× magnification maintained at 37°C, 5.0 % CO2. For dual-emission ratio, we used an ET436/20× excitation filter, a T515LP diachronic mirror, an ET520LP emission filter for FRET channel, and an ET636/20× excitation filter, a T455LP dichroic mirror and an ET480/40m emission filter for CFP channel (Chroma Technology Crop). FRET, CFP and phase contrast channels were sequentially collected before adding the ErbB2 stimulation and every 5 minutes after ErbB2 activated. The FRET channel and CFP channel exposure time was 80 milliseconds when the digital gain was set to 24. After background extraction, the ratio image was created with ImageJ PixFRET plugin63.
Fluorescence Recovery After Photobleaching (FRAP) analysis
FRAP experiments were performed using spinning disk microscopy (Perkin Elmer UltraVIEW Vox) with an APON 60×, 1.49NA oil immersion lens (Olympus). Excitation of GFP was carried out at a 488nm laser line of an argon laser. Before photobleaching, three images were acquired. A selected region (10.0 µm×10.0 µm) was bleached by a 5.0 milliseconds single laser pulse (UltraVIEW PK Device) at 80% laser transmission power. Recovery images were collected using 20% laser transmission power for 10 minutes at a scanning rate of 12 fpm. The average intensity was determined in the photobleaching region and normalized to the intensity before bleaching using ImageJ software.
Microscopy
Live image were performed by spinning disk microscopy (Perkin Elmer UltraVIEW Vox) with an PL APO 100×, 1.40 NA oil immersion objective (Olympus), Perfect Focus (Nikon) and automatic thermal stage and CO2 supply. Images were captured at the rate of 12 fpm using Volocity Software. Images were processed using ImageJ software.
Primary mammary tumour cell transplantation
Single primary mammary tumour cells from MMTV-NDL mouse were prepared as described previously64. Isolated cells were maintained in DMEM/F12 medium containing 2.0 % BSA, MEGM Singlequot (Lonza), 20 ng/ml basic fibroblast growth factor (Sigma-Aldrich), 4.0 µg/ml heparin (Sigma-Aldrich). Isolated cells were infected with concentrated shRNA lentivirus (MOI=5) in the ultra-low attachment plates (Corning). Infected cells were grown at 37 °C in the tissue culture incubator for two days and dissociated into single cells using dispase. Cells were resuspended at 2,500 cell/10 µl in RPMI and 1:1 mixed with Matrigel and injected into cleared mammary fat pads of three week old NOD/SCID mice following established procedures65. Mice were palpated weekly for tumour onset, starting two weeks after the operation. Tumours were collected as described in Fig. 2b. All tumours were imaged using stereo fluorescence microscope (Leica) and collected for IHC. The images were all taken using 50ms exposure time. Brightness between 25–255 was score as GFP positive.
Quantitative PCR
Total RNA was isolated under indicated conditions using TRIzol reagent following the manufacturer’s protocol (Invitrogen). Quantitative PCR assays were performed as described previously66. Primer sequences used are as follows:
fibronectin forward: 5’- GAG GGG ACC TGC AGC CAC AA -3’
fibronectin reverse: 5’- TTC GCA ACC TGC GGG AAA AA-3’
human β-actin forward: 5’-TTC AAC ACC CCA GCC ATG-3’
human β-actin reverse: 5’-GCC AGT GGT ACG GCC AGA-3’
snail forward: 5’-TGC AGG ACT CTA ATC CAA GTT TAC C-3’
snail reverse: 5’-GTG GGA TGG CTG CCA GC-3’
N-cadherin forward: 5’-GAC GGT TCG CCA TCC AGA C-3’
N-cadherin reverse: 5’-TCG ATT GGT TTG ACC ACG G-3’
Tumour immunohistochemistry (IHC) and image analysis
Immunohistochemistry of Formalin-fixed paraffin-embedded tissue (FFPET) samples of human invasive breast cancers was performed as previously described66. Microscopy images were acquired using Aperio Scanscope. Identical microscope parameters were applied to paired primary tumours and metastasis.
Tumour characterization
In human tumour microarray samples, Histological grade of the IBCs was determined utilizing the method of Scarff-Bloom-Richardson as modified by Elston-Ellis (grades 1–3; well to poorly differentiated)67. ER (nuclear) was quantified using the Allred Score (range 0–8; positive >2)68. HER2 was quantified using the Dako HercepTest Score (0–3+; negative = 0/1+; indeterminate = 2+; positive = 3+)69. Membrane Par 3 was also quantified using the Allred Score (range 0–8) for each sample. Total Score (TS) = Proportion score (range 0–5) + Intensity Score (range 0–3)70
Statistical analysis
The specific statistical tests used are indicated in the figure legends alongside the P values.
Supplementary Material
Acknowledgements
The authors would like to thank William J. Muller for the NDL transgenic mice, Ian Macara for Tiam1 cDNA, Michiyuki Matsuda for Raichu-Rac plasmid, Dr. Diane Barber for sharing anti-ARPC2 antibody. We thank Jennifer Haynes for critical reading of the manuscript and Mikala Egeblad, and Raffaella Sordella for helpful comments. We thank Stephen Hearn for assistance with microscopy and Charles Camarda for assistance with the graphic abstract. We like to thank members of the Muthuswamy laboratory for critical comments on the manuscripts and insightful discussions. This work was supported by CA098830, BC075024, Era of Hope Scholar award from DOD Breast Cancer Research Program; Rita Allen Foundation, Lee K Margaret Lau Chair for breast cancer research and Campbell Family Institute for Breast cancer research to SKM. This was also funded in part by the Ontario Ministry of Health and Long Term Care. The views expressed do not necessarily reflect those of the OMOHLTC.
References
- 1.Gomes JE, Bowerman B. Caenorhabditis elegans par genes. Curr Biol. 2002;12:R444. doi: 10.1016/s0960-9822(02)00939-9. [DOI] [PubMed] [Google Scholar]
- 2.Kirby C, Kusch M, Kemphues K. Mutations in the par genes of Caenorhabditis elegans affect cytoplasmic reorganization during the first cell cycle. Dev Biol. 1990;142:203–215. doi: 10.1016/0012-1606(90)90164-e. [DOI] [PubMed] [Google Scholar]
- 3.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. J Cell Sci. 2006;119:979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 5.Mellman I, Nelson WJ. Coordinated protein sorting, targeting and distribution in polarized cells. Nat Rev Mol Cell Biol. 2008;9:833–845. doi: 10.1038/nrm2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aranda V, Nolan ME, Muthuswamy SK. Par complex in cancer: a regulator of normal cell polarity joins the dark side. Oncogene. 2008;27:6878–6887. doi: 10.1038/onc.2008.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aranda V, et al. Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat Cell Biol. 2006;8:1235–1245. doi: 10.1038/ncb1485. [DOI] [PubMed] [Google Scholar]
- 8.Eder AM, et al. Atypical PKCiota contributes to poor prognosis through loss of apical-basal polarity and cyclin E overexpression in ovarian cancer. Proc Natl Acad Sci U S A. 2005;102:12519–12524. doi: 10.1073/pnas.0505641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nolan ME, et al. The polarity protein Par6 induces cell proliferation and is overexpressed in breast cancer. Cancer Res. 2008;68:8201–8209. doi: 10.1158/0008-5472.CAN-07-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Regala RP, et al. Atypical protein kinase C iota is an oncogene in human non-small cell lung cancer. Cancer Res. 2005;65:8905–8911. doi: 10.1158/0008-5472.CAN-05-2372. [DOI] [PubMed] [Google Scholar]
- 11.Rothenberg SM, et al. A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers. Cancer Res. 70:2158–2164. doi: 10.1158/0008-5472.CAN-09-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zen K, et al. Defective expression of polarity protein PAR-3 gene (PARD3) in esophageal squamous cell carcinoma. Oncogene. 2009;28:2910–2918. doi: 10.1038/onc.2009.148. [DOI] [PubMed] [Google Scholar]
- 13.Etemad-Moghadam B, Guo S, Kemphues KJ. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell. 1995;83:743–752. doi: 10.1016/0092-8674(95)90187-6. [DOI] [PubMed] [Google Scholar]
- 14.Ebnet K, et al. The cell polarity protein ASIP/PAR-3 directly associates with junctional adhesion molecule (JAM) EMBO J. 2001;20:3738–3748. doi: 10.1093/emboj/20.14.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takekuni K, et al. Direct binding of cell polarity protein PAR-3 to cell-cell adhesion molecule nectin at neuroepithelial cells of developing mouse. J Biol Chem. 2003;278:5497–5500. doi: 10.1074/jbc.C200707200. [DOI] [PubMed] [Google Scholar]
- 16.Feng W, Wu H, Chan LN, Zhang M. Par-3-mediated junctional localization of the lipid phosphatase PTEN is required for cell polarity establishment. J Biol Chem. 2008;283:23440–23449. doi: 10.1074/jbc.M802482200. [DOI] [PubMed] [Google Scholar]
- 17.Nagai-Tamai Y, Mizuno K, Hirose T, Suzuki A, Ohno S. Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells. 2002;7:1161–1171. doi: 10.1046/j.1365-2443.2002.00590.x. [DOI] [PubMed] [Google Scholar]
- 18.Wu H, et al. PDZ domains of Par-3 as potential phosphoinositide signaling integrators. Mol Cell. 2007;28:886–898. doi: 10.1016/j.molcel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 19.Izumi Y, et al. An atypical PKC directly associates and colocalizes at the epithelial tight junction with ASIP, a mammalian homologue of Caenorhabditis elegans polarity protein PAR-3. J Cell Biol. 1998;143:95–106. doi: 10.1083/jcb.143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamanaka T, et al. PAR-6 regulates aPKC activity in a novel way and mediates cell-cell contact-induced formation of the epithelial junctional complex. Genes Cells. 2001;6:721–731. doi: 10.1046/j.1365-2443.2001.00453.x. [DOI] [PubMed] [Google Scholar]
- 21.Mertens AE, Rygiel TP, Olivo C, van der Kammen R, Collard JG. The Rac activator Tiam1 controls tight junction biogenesis in keratinocytes through binding to and activation of the Par polarity complex. J Cell Biol. 2005;170:1029–1037. doi: 10.1083/jcb.200502129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol. 2005;7:262–269. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Macara IG. The PAR-6 polarity protein regulates dendritic spine morphogenesis through p190 RhoGAP and the Rho GTPase. Dev Cell. 2008;14:216–226. doi: 10.1016/j.devcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muthuswamy SK, Gilman M, Brugge JS. Controlled dimerization of ErbB receptors provides evidence for differential signaling by homo- and heterodimers. Mol Cell Biol. 1999;19:6845–6857. doi: 10.1128/mcb.19.10.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCaffrey LM, Macara IG. The Par3/aPKC interaction is essential for end bud remodeling and progenitor differentiation during mammary gland morphogenesis. Genes Dev. 2009;23:1450–1460. doi: 10.1101/gad.1795909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrechek ER, et al. Amplification of the neu/erbB-2 oncogene in a mouse model of mammary tumorigenesis. Proc. Natl Acad. Sci. USA. 2000;97:3444–3449. doi: 10.1073/pnas.050408497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Rooij J, Kerstens A, Danuser G, Schwartz MA, Waterman-Storer CM. Integrin-dependent actomyosin contraction regulates epithelial cell scattering. J Cell Biol. 2005;171:153–164. doi: 10.1083/jcb.200506152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D'Souza B, Taylor-Papadimitriou J. Overexpression of ERBB2 in human mammary epithelial cells signals inhibition of transcription of the E-cadherin gene. Proc Natl Acad Sci U S A. 1994;91:7202–7206. doi: 10.1073/pnas.91.15.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimoyama Y, et al. Cadherin cell-adhesion molecules in human epithelial tissues and carcinomas. Cancer Res. 1989;49:2128–2133. [PubMed] [Google Scholar]
- 30.Cavey M, Rauzi M, Lenne P-F, Lecuit T. A two-tiered mechanism for stabilization and immobilization of E-cadherin. Nature. 2008;453:751–756. doi: 10.1038/nature06953. [DOI] [PubMed] [Google Scholar]
- 31.Baum B, Georgiou M. Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J Cell Biol. 2011;192:907–917. doi: 10.1083/jcb.201009141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braga VM, Machesky LM, Hall A, Hotchin NA. The small GTPases Rho and Rac are required for the establishment of cadherin-dependent cell-cell contacts. J Cell Biol. 1997;137:1421–1431. doi: 10.1083/jcb.137.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu YS, et al. Force measurements in E-cadherin-mediated cell doublets reveal rapid adhesion strengthened by actin cytoskeleton remodeling through Rac and Cdc42. J Cell Biol. 2004;167:1183–1194. doi: 10.1083/jcb.200403043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraemer A, Goodwin M, Verma S, Yap AS, Ali RG. Rac is a dominant regulator of cadherin-directed actin assembly that is activated by adhesive ligation independently of Tiam1. Am J Physiol Cell Physiol. 2007;292:C1061–C1069. doi: 10.1152/ajpcell.00073.2006. [DOI] [PubMed] [Google Scholar]
- 35.Arthur WT, Ellerbroek SM, Der CJ, Burridge K, Wennerberg K. XPLN, a Guanine Nucleotide Exchange Factor for RhoA and RhoB, But Not RhoC. Journal of Biological Chemistry. 2002;277:42964–42972. doi: 10.1074/jbc.M207401200. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Mata R, et al. Analysis of activated GAPs and GEFs in cell lysates. Methods in enzymology. 2006;406:425–437. doi: 10.1016/S0076-6879(06)06031-9. [DOI] [PubMed] [Google Scholar]
- 37.Benard V, Bokoch GM. Assay of Cdc42, Rac, and Rho GTPase activation by affinity methods. Methods in enzymology. 2002;345:349–359. doi: 10.1016/s0076-6879(02)45028-8. [DOI] [PubMed] [Google Scholar]
- 38.Itoh RE, et al. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol. 2002;22:6582–6591. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stam JC, et al. Targeting of Tiam1 to the plasma membrane requires the cooperative function of the N-terminal pleckstrin homology domain and an adjacent protein interaction domain. J Biol Chem. 1997;272:28447–28454. doi: 10.1074/jbc.272.45.28447. [DOI] [PubMed] [Google Scholar]
- 40.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci U S A. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamazaki D, Oikawa T, Takenawa T. Rac-WAVE-mediated actin reorganization is required for organization and maintenance of cell-cell adhesion. J Cell Sci. 2007;120:86–100. doi: 10.1242/jcs.03311. [DOI] [PubMed] [Google Scholar]
- 42.Yamada S, Nelson WJ. Localized zones of Rho and Rac activities drive initiation and expansion of epithelial cell-cell adhesion. J Cell Biol. 2007;178:517–527. doi: 10.1083/jcb.200701058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ. The Human Arp2/3 Complex Is Composed of Evolutionarily Conserved Subunits and Is Localized to Cellular Regions of Dynamic Actin Filament Assembly. The Journal of Cell Biology. 1997;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riedl J, et al. Lifeact: a versatile marker to visualize F-actin. Nature methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lampkin SR, Allred DC. Preparation of Paraffin Blocks and Sections Containing Multiple Tissue Samples Using a Skin Biopsy Punch. Journal of Histotechnology. 1990;13:121–122. [Google Scholar]
- 46.Ling C, Zuo D, Xue B, Muthuswamy S, Muller WJ. A novel role for 14-3-3sigma in regulating epithelial cell polarity. Genes Dev. 2010;24:947–956. doi: 10.1101/gad.1896810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu J, et al. 14-3-3zeta Cooperates with ErbB2 to promote ductal carcinoma in situ progression to invasive breast cancer by inducing epithelial-mesenchymal transition. Cancer cell. 2009;16:195–207. doi: 10.1016/j.ccr.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nance J. PAR proteins and the establishment of cell polarity during C. elegans development. Bioessays. 2005;27:126–135. doi: 10.1002/bies.20175. [DOI] [PubMed] [Google Scholar]
- 49.Hurd TW, et al. Phosphorylation-dependent binding of 14-3-3 to the polarity protein Par3 regulates cell polarity in mammalian epithelia. Curr Biol. 2003;13:2082–2090. doi: 10.1016/j.cub.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 50.Iwaya K, Norio K, Mukai K. Coexpression of Arp2 and WAVE2 predicts poor outcome in invasive breast carcinoma. Mod Pathol. 2007;20:339–343. doi: 10.1038/modpathol.3800741. [DOI] [PubMed] [Google Scholar]
- 51.Nurnberg A, Kitzing T, Grosse R. Nucleating actin for invasion. Nat Rev Cancer. 2011;11:177–187. doi: 10.1038/nrc3003. [DOI] [PubMed] [Google Scholar]
- 52.Mizutani K, Miki H, He H, Maruta H, Takenawa T. Essential role of neural Wiskott-Aldrich syndrome protein in podosome formation and degradation of extracellular matrix in src-transformed fibroblasts. Cancer Research. 2002;62:669–674. [PubMed] [Google Scholar]
- 53.Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell. 2005;123:903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–2004. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 56.Rhim AD, et al. EMT and Dissemination Precede Pancreatic Tumor Formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCaffrey LM, Montalbano J, Mihai C, Macara IG. Loss of the par3 polarity protein promotes breast tumorigenesis and metastasis. Cancer cell. 2012;22:601–614. doi: 10.1016/j.ccr.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson EW, Newgreen DF, Tarin D. Carcinoma invasion and metastasis: a role for epithelial-mesenchymal transition? Cancer Res. 2005;65:5991–5995. doi: 10.1158/0008-5472.CAN-05-0616. discussion 5995. [DOI] [PubMed] [Google Scholar]
- 59.Paddison PJ, et al. Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nature methods. 2004;1:163–167. doi: 10.1038/nmeth1104-163. [DOI] [PubMed] [Google Scholar]
- 60.Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin G, Aranda V, Muthuswamy SK, Tonks NK. Identification of PTPN23 as a novel regulator of cell invasion in mammary epithelial cells from a loss-of-function screen of the 'PTP-ome'. Gene Dev. 2011;25:1412–1425. doi: 10.1101/gad.2018911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang XA, et al. Function of the tetraspanin CD151-alpha6beta1 integrin complex during cellular morphogenesis. Mol Biol Cell. 2002;13:1–11. doi: 10.1091/mbc.01-10-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feige JN, Sage D, Wahli W, Desvergne B, Gelman L. PixFRET, an ImageJ plug-in for FRET calculation that can accommodate variations in spectral bleed-throughs. Microscopy research and technique. 2005;68:51–58. doi: 10.1002/jemt.20215. [DOI] [PubMed] [Google Scholar]
- 64.Zhang M, et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 2008;68:4674–4682. doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ehmann UK, et al. Cultured mouse mammary epithelial cells: normal phenotype after implantation. J Natl Cancer Inst. 1987;78:751–757. [PubMed] [Google Scholar]
- 66.Zhan L, et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. C. W. Elston & I. O. Ellis. Histopathology 1991; 19; 403–410. Histopathology. 2002;41:151–152. discussion 152–153. [PubMed] [Google Scholar]
- 68.Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 69.Wolff AC, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Archives of pathology & laboratory medicine. 2007;131:18–43. doi: 10.5858/2007-131-18-ASOCCO. [DOI] [PubMed] [Google Scholar]
- 70.Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155–168. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.