Abstract
Background
Accumulating studies reveal that aberrant microRNA (miRNA) expression can affect the development of chemotherapy drug resistance by modulating the expression of relevant target proteins. The aim of this study was to investigate the role of miR-133b in the development of drug resistance in ovarian cancer cells.
Methods
We examined the levels of miR-133b expression in ovarian carcinoma tissues and the human ovarian carcinoma cell lines (A2780, A2780/DDP and A2780/Taxol, respectively). We determined the cell viability of these cell lines treated with cisplatin or paclitaxel in the presence or absence of miR-133b or anti-miR-133b transfection using the MTT assay. Reverse transcription polymerase chain reaction and Western blotting were used to assess the mRNA and protein expression levels of two drug-resistance-related genes: glutathione S-transferase (GST)-π and multidrug resistance protein 1 (MDR1). The dual-luciferase reporter assay was used to detect the promoter activity of GST-π in the presence and absence of miR-133b.
Results
The expression of miR-133b was significantly lower in primary resistant ovarian carcinomas than in the chemotherapy-sensitive carcinomas (P<0.05), and the same results were found in primary resistant ovarian cell lines (A2780/Taxol and A2780/DDP) compared to the chemotherapy-sensitive cell line (A2780; P<0.05). Following miR-133b transfection, four cell lines showed increased sensitivity to paclitaxel and cisplatin, while anti-miR-133b transfection reduced cell sensitivity to paclitaxel and cisplatin. Dual-luciferase reporter assay showed that miR-133b interacted with the 3′-untranslated region of GST-π. Compared to controls, the mRNA and protein levels of MDR1 and GST-π were downregulated after miR-133b transfection and upregulated after anti-miR-133b transfection.
Conclusion
MicroRNA-133b may reduce ovarian cancer drug resistance by silencing the expression of the drug-resistance-related proteins, GST-π and MDR1. In future, the combination of miR-133b with chemotherapy agents may prevent the development of drug resistance in ovarian cancers.
Keywords: cisplatin, drug resistance, ovarian cancer cells, paclitaxel
Introduction
Epithelial ovarian cancer is the leading cause of death from gynecological cancers worldwide, and is the fifth leading cause of cancer death in women.1 The 5-year survival rate for all stages of ovarian cancer has been estimated to be 45.6%.2 Primary treatment of epithelial ovarian cancer is surgical resection of visible disease, followed by adjuvant chemotherapy. Conventional chemotherapy drugs, such as cisplatin and paclitaxel, have long-term clinical application in many cancers, including ovarian cancer.3 Unfortunately, numerous ovarian cancer patients who initially respond to cisplatin or paclitaxel therapy become drug resistant and relapse. Therefore, the identification of new biomarkers, treatments, and therapeutic targets for human epithelial ovarian cancer is required.
MicroRNAs (miRNAs) are promising diagnostic and prognostic molecular biomarkers in cancer.4,5 They are endogenous, noncoding RNAs that silence gene expression by inhibiting mRNA stability or translation, and frequently target the 3′-untranslated regions (3′-UTRs).6 Typically, miRNAs function in a variety of cellular events (eg, proliferation, differentiation, and apoptosis) and have been implicated in tumorigenesis.7–9 In recent years, more attention has been paid to the role of miRNAs in drug resistance, with accumulating evidence that aberrant miRNA expression can affect drug resistance by modulating the expression of relevant target proteins, including drug transporters, cell apoptosis mediators, and cell-cycle-related components.10 For example, miR-27a,11 miR-106a,12 and miR-48713 have been implicated in the development of drug resistance by regulating gene expression of a number of different target genes.
In this study, we examined the role of miR-133b in drug resistance. miR-133b modulates cell proliferation, cell apoptosis, cell migration, and invasion, and is downregulated in gastric,14 bladder,15 and colon16 cancers. Guo et al showed that miR-133b has a tumor suppressor role in gastric cancer; it decreases fascin actin-bundling protein 1 expression, a protein that increases cell motility and cancer metastasis.14 Similarly, Chen et al reported that miR-133b decreases bladder cancer cell proliferation and apoptosis by reducing expression of Bcl-w and Akt1.15 On the other hand, Qin et al reported that miR-133b acts as an oncogene in cervical cancer, and promotes carcinoma development by activating the ERK and AKT1 signaling pathways.17 Therefore, miR-133b may play multiple roles in cancer proliferation and metastasis.
Despite the importance of miR-133b in tumorigenesis and the potential function of miRNAs in drug resistance, there are no studies investigating the role of miR-133b in drug-resistant human epithelial ovarian cancer cells to date. In this study, we investigated whether miR-133b is involved in the development of cisplatin and paclitaxel drug resistance in an ovarian cancer cell line.
Materials and methods
Cell culture
The serous cystic adenocarcinoma ovarian cell line A2780 and the paclitaxel-resistant A2780 (A2780/Taxol) cell line were kindly given to us by Dr Biao Wang (College of Basic Medical Sciences, China Medical University, Shenyang, People’s Republic of China). The cisplatin-resistant A2780 (A2780/DDP) cell line was purchased from the China Center for Type Culture Collection (Shanghai, People’s Republic of China), and the OVCAR3 cell line was purchased from American Type Culture Collection (ATCC) (Manassas, VA, USA). Cells were maintained in RPMI 1640 (A2780/DDP, A2780/Taxol, OVCAR3) or Dulbecco’s Modified Eagle’s Medium (A2780) supplemented with 10% fetal bovine serum, 100 units/mL penicillin, and 100 μg/mL streptomycin. Cell lines were kept in a humidified atmosphere of 5% CO2 at 37°C with or without cisplatin (0, 37.5, 150, 600, 2,400, and 9,600 ng/mL) or paclitaxel (0, 6.25, 25, 100, 400, and 1,600 ng/mL) treatment. Transfection with 50 nM miR-133b, anti-miR-133b (Biomics Biotech, Jiangsu, People’s Republic of China), or a scrambled miRNA sequence (miR-C, Anti-C) was performed using the Lipofectamine 2000 in accordance with the manufacturer’s guidelines (Thermo Fisher Scientific, Waltham, MA, USA). The sequences were as follows: miR-133b, 5′-UUU GGU CCC CUU CAA CCA GCU A-3′; miR-C, 5′-UUC UCC GAA CGU GUC ACG UTT-3′; anti-miR-133b, 5′-UAG CUG GUU GAA GGG GAC CAA A-3′; and anti-C, 5′-CAG UAC UUU UGU GUA GUA CAA-3′. No ethics statement was required from the institutional review board for the use of these cell lines.
Ovarian carcinoma specimens
Forty-four ovarian carcinomas were collected from patients who underwent surgical resection at the Department of Gynecology of the First Affiliated Hospital of China Medical University (Shenyang, Liaoning, People’s Republic of China). The tumor specimens were independently confirmed by two pathologists. Samples were frozen immediately in liquid nitrogen and stored at −80°C until use. None of the patients had preoperative chemotherapy or radiotherapy. Informed consent was obtained from all subjects, the study was approved by the China Medical University Ethics Committee, and all specimens were handled and made anonymous according to ethical and legal standards.
Cell viability assay
MTT assay using the tetrazolium 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide dye (Beyotime, Jiangsu, People’s Republic of China) was employed to determine cell viability. Briefly, approximately 2.5×103 cells/well were seeded into a 96-well plate and allowed to adhere overnight. At 0, 24, 48, and 72 hours, 20 μL of MTT solution was added into each well of the plate, and the plates was subsequently incubated for 4 hours in the incubator. Then, the liquid was removed from the plate, and 150 μL of dimethyl sulfoxide was added to each well. The plate was measured at 490 nm.
Dual-luciferase reporter assay
The miR-133b binding site in glutathione S-transferase (GST)-π, which is abundantly expressed in cancer cells, was predicted using a web-based miRNA program (http://www.microrna.org). The target sequence of the wild-type 3′-UTR of GST-π was cloned into a luciferase vector containing the Renilla luciferase gene. In addition, a mutant 3′-UTR of GST-π was cloned by Genethem (Shanghai Genechem Co., Ltd, Shanghai, People’s Republic of China). HEK-293T cells and A2780 cells were cotransfected with miR-C or miR-133b using Lipofectamine 2000 (Thermo Fisher Scientific). Cells were collected 48 hours after transfection and analyzed using the Dual-Luciferase Reporter Assay System (Promega Corporation, Fitchburg, WI, USA). Luciferase activity values were normalized relative to that of the Renilla luciferase internal control. Each reporter plasmid was transfected at least three times, and each sample was assayed in triplicate. The wild sequence for GST-π (NM_000852) 3′ UTR was: GGGTTGGGGGGACTCTGAGCGGGAGGCAGAGTTTGCCTTCCTTTCTCCAGGACCAATAAAATTTCTAAGAGAGCT; while mutant sequence was GGGTTGGGGGGACTCTGAGCGGGAGGCAGAGTTTGCCTTCCTTTCTCCATCGGTCGTAAAATTTCTAAGAGAGCT.
Real-time reverse transcription polymerase chain reaction
Total RNA was extracted from the ovarian carcinoma cell lines using TRIzol® (Takara, Kyoto, Japan). Real-time reverse transcription (RT) polymerase chain reaction (PCR) was performed from 2 μg of total RNA using avian myeloblastosis virus reverse transcriptase and random primers (Takara). PCR primers were designed according to the sequences in GenBank (Table S1). Amplification of cDNA was performed according to the manufacturer’s protocol using an SYBR Premix Ex Taq II kit (Takara). All the PCR experiments were accompanied with a no-template control and 18S as an internal control. The relative gene expression level (the amount of target gene normalized to the endogenous control gene) was calculated using the comparative CT method: 2−ΔΔCt.
Western blot analysis
Protein quantification was performed according to the Bradford method using the Bio-Rad protein assay kit (Bio-Rad Laboratories Inc., Hercules, CA, USA). A total of 40 μg of denatured proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 12% acrylamide gels, and then transferred to Hybond™ membranes (Amersham Biosciences, Freiburg, Germany). The membranes were blocked overnight in 5% skim milk in Tris-buffered saline with Tween 20. For immunoblotting, the membranes were incubated at 4°C overnight with the multidrug resistance 1 (MDR1) antibody (1:500; Bioss, Peking, People’s Republic of China) and the GST-π antibody (1:500; Proteintech Group, Chicago, USA). Then, they were rinsed with Tris-buffered saline with Tween 20 and incubated with the anti-rabbit IgG antibodies conjugated to horseradish peroxidase (Dako, Carpinteria CA, USA) at a dilution of 1:5,000. After applying electrochemiluminescent plus detection reagents (Santa Cruz Biotechnology Inc., Dallas, TX, USA), the protein bands were visualized using an X-ray film (Fujifilm, Tokyo, Japan). The immunoblots were washed with Western blot stripping buffer (pH 2–3; Nacalai, Tokyo, Japan) and probed using a monoclonal antibody specific for glyceraldehyde-3-phosphate (1:2,000; Proteintech Group).
Statistical analysis
Statistical evaluation was performed using the Student’s t-test to differentiate the means of different groups. A P-value of <0.05 was considered statistically significant. SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) was employed to analyze all data.
Results
miR-133b expression is lower in drug resistant ovarian carcinomas and cell lines
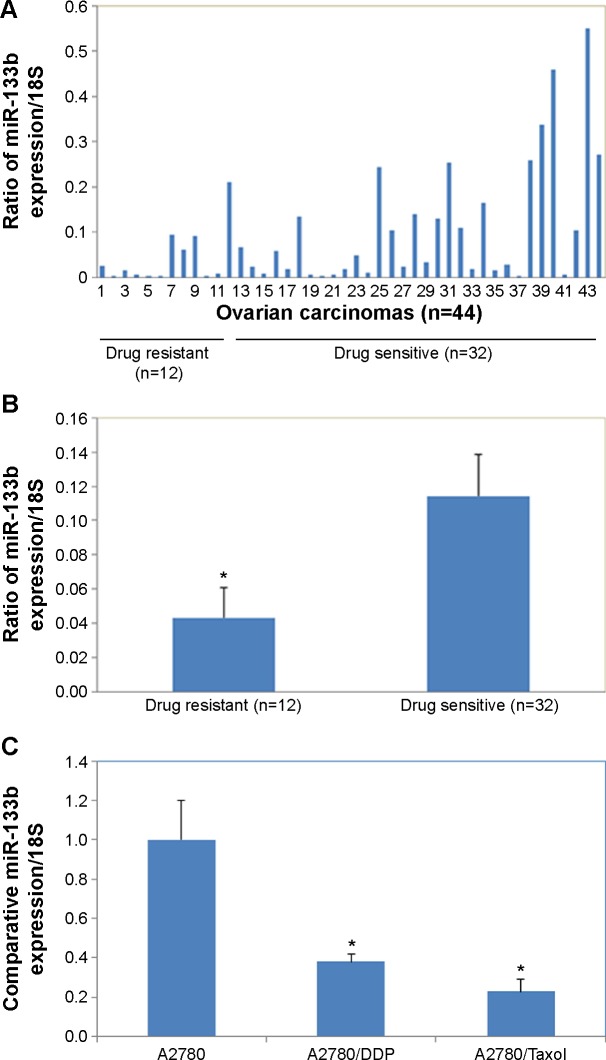
miR-133b expression was significantly lower in primary resistant ovarian carcinomas compared to the chemotherapy-sensitive carcinomas (Figure 1A and B, P<0.05). miR-133b expression was significantly lower in primary resistant ovarian cell lines (A2780/Taxol and A2780/DDP) compared to the chemotherapy sensitive (A2780) cell line (Figure 1C, P<0.05).
Figure 1.
MiR-133b expression is lower in drug-resistant ovarian carcinomas and cell lines.
Notes: (A and B) miR-133b expression was significantly lower in primary resistant ovarian carcinomas compared to the chemotherapy-sensitive carcinomas. (C) Comparison of miR-133b expression relative to 18S expression in the nonresistant ovarian cancer cell line (A2780), the cisplatin-resistant ovarian cell line (A2780/DDP), and the paclitaxel-resistant ovarian cell line (A2780/Taxol) determined by reverse transcription polymerase chain reaction. Data are expressed as the mean ± standard deviation; *P<0.05.
miR-133b transfection increases the sensitivity of ovarian carcinoma cell lines to chemotherapy drugs
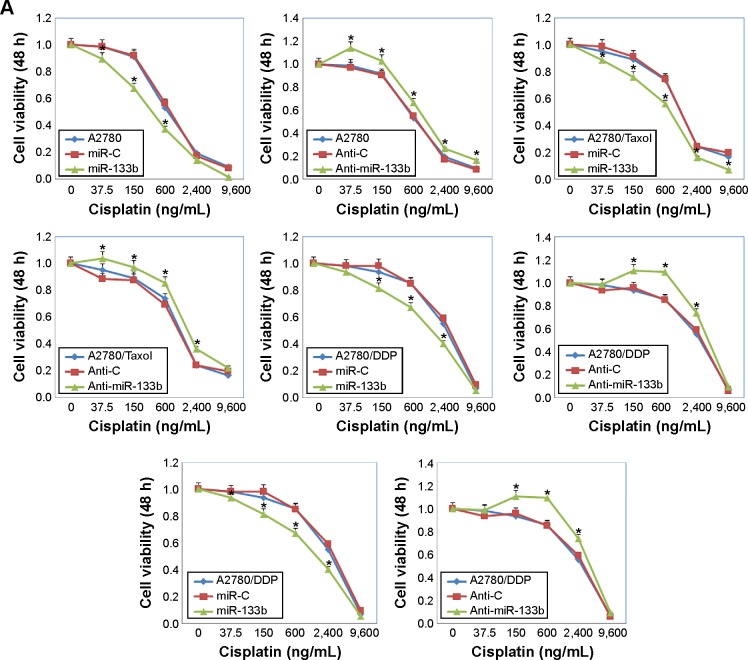
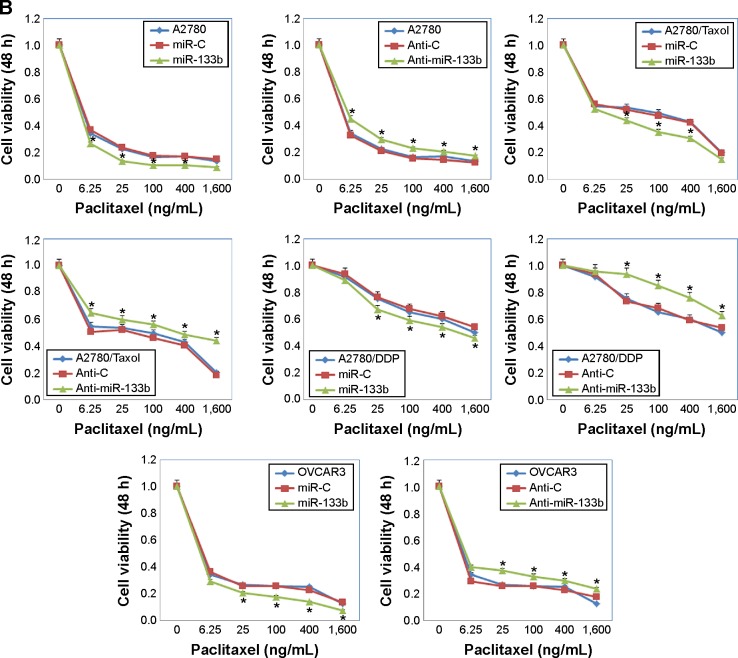
Following miR-133b transfection, the sensitivity of A2780 cells to cisplatin was increased compared with negative control cells or miR-C cells; however, anti-miR-133b transfection decreased the sensitivity to cisplatin (Figure 2A, P<0.05). Similar results were observed with the A2780/DDP, A2780/Taxol, and OVCAR3 cells lines. The sensitivity to paclitaxel was also increased after miR-133b transfection, while it was decreased after anti-miR-133b transfection in all cell lines (Figure 2B, P<0.05). Details can be found in Table S2.
Figure 2.
MiR-133b transfection increases the sensitivity of ovarian carcinoma cell lines to chemotherapy drugs.
Notes: Cell viability after 48 hours of the A2780, A2780/DPP, A2780/Taxol, and OVCAR3 cells transfected with a scrambled miRNA sequence (miR-C) or miR-133b, and scrambled anti-miR-133b (Anti-C) or anti-miR-133b. Cells were treated with increasing doses of (A) cisplatin (0, 37.5, 150, 600, 2,400, and 9,600 ng/mL) or (B) paclitaxel (0, 6.25, 25, 100, 400, and 1,600 ng/mL). Results are representative of three separate experiments; data are expressed as the mean ± standard deviation; *P<0.05 compared with miR-C or Anti-C group.
Abbreviation: h, hours.
Predicted binding region and transcriptional effects of miR-133b on its target gene, GST-π
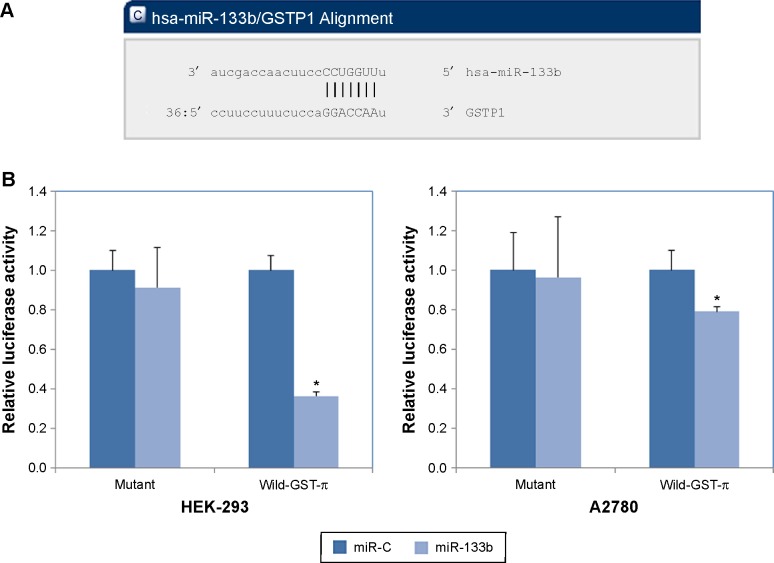
To gain a greater understanding of the role of miR-133b in the drug resistance of human ovarian cancer, we looked for its potential downstream targets using a web-based miRNA program (http://www.microrna.org). Based on the predicted binding region of miR-133b in the 3′-UTR of GST-π, we propose that GST-π is a direct target of miR-133b (Figure 3A). Furthermore, using the dual-luciferase reporter assay, we found that miR-133b significantly decreased the luciferase activity of the GST-π 3′-UTR in both HEK-293 T and A2780 cells (Figure 3B, P<0.05 for both), but not mutant sequences of the 3′-UTR of GST-π. These results indicate that miR-133b directly binds to the 3′-UTR of wild type GST-π.
Figure 3.
Predicted binding region and transcriptional effects of miR-133b on its target gene, GST-π.
Notes: (A) The predicted seed region of miR-133b (hsa-miRNA-133b) in the 3′-UTR of GST-π (GST-P1) determined using a miRNA target program (http://www.microrna.org). (B) Effect of miR-133b on GST-π promoter activity using a Mutant and a Wild-GST-π. Luciferase activity (gene promoter activity) is unchanged when using a scrambled miRNA (miR-C) sequence or miR-133b with the Mutant. However, when the Wild-GST-π is used, the promoter activity is significantly reduced by miR-133b compared to miR-C. Each reporter plasmid was transfected at least three times, and each sample was assayed in triplicate; data are expressed as the mean ± standard deviation; *P<0.05.
Abbreviations: 3′-UTR, 3′-untranslated region; Mutant, mutant 3′-UTR of GST-π; Wild-GST-π, wild-type 3′-UTR of GST-π; GST, glutathione S-transferase.
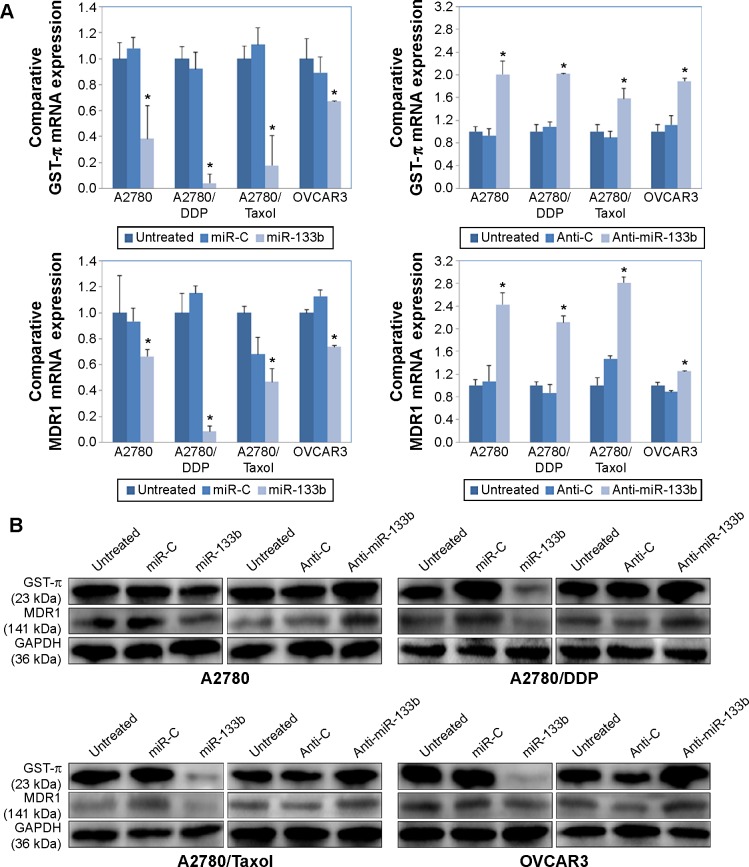
miR-133b downregulates MDR1 and GST-π expression levels
Compared to controls, miR-133b transfection decreased the mRNA expression levels of MDR1 and GST-π in all four cell lines, while anti-miR-133b transfection upregulated MDR1 and GST-π mRNA expression (Figure 4A, P<0.05). At the same time, protein expression levels of MDR1 and GST-π were also lower in miR-133b-transfected cells and were higher in Anti-miR-133b-transfected cells compared to controls (Figure 4B).
Figure 4.
miR-133b downregulates GST-π, MDR1 mRNA, and protein expression levels.
Notes: (A) Comparison of GST-π and MDR1 mRNA levels relative to 18S mRNA levels in in the A2780, A2780/DDP, A2780/Taxol, and OVCAR3 cells alone (untreated); or transfected with a scrambled miRNA sequence (miR-C) or miR-133b, or anti-miR-133b or scrambled anti-miRNA sequence (Anti-C), determined by reverse transcription polymerase chain reaction. Results are representative of three separate experiments; data are expressed as the mean ± standard deviation; *P<0.05. (B) Comparison of GST-π and MDR1 protein levels in A2780, A2780/DDP, A2780/Taxol, and OVCAR3 cells alone (untreated); or transfected with a scrambled miRNA sequence (miR-C) or miR-133b, or anti-miR-133b or scrambled anti-miRNA sequence (Anti-C), determined by Western blotting. GAPDH expression is used as a control.
Abbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GST, glutathione S-transferase; MDR, multidrug resistance 1.
Discussion
This study investigated whether miR-133b contributes to the development of drug resistance in ovarian cancer. We showed that miR-133b expression was significantly lower in primary resistant ovarian carcinomas compared to the chemotherapy-sensitive carcinomas. In addition, miR-133b expression was lower in cisplatin-resistant and paclitaxel-resistant ovarian cancer cell lines compared to the chemotherapy-sensitive cells. Moreover, after miR-133b transfection, all examined cell lines showed an increased sensitivity to paclitaxel and cisplatin, while anti-miR-133b transfection decreased sensitivity to paclitaxel and cisplatin. Together, these results suggest that miR-133b expression is important for chemotherapy drug sensitivity.
The role of miRNAs in the development of chemotherapy drug resistance in ovarian tumors has recently attracted attention. For example, van Jaarsveld et al reported that miR-141 induces cisplatin resistance by directly targeting the Kelch-Like ECH-Associated Protein 1 gene,18 while miR-21-3p induces cisplatin resistance in ovarian tumors by targeting the neuron navigator 3 gene.19 On the other hand, overexpression of some miRNAs will improve drug sensitivity rather than induce drug resistance. In particular, He et al reported that miR-152 sensitizes ovarian cancer cells to cisplatin;20 while Zhou et al reported that miR-449a enhances cisplatin-induced cytotoxicity in ovarian cancer cells by decreasing NOTCH1 expression.21 Thus different miRNAs may have different functions in A2780 cells through targeting different genes.
We predicted that GST-π was a direct target of miR-133b. Moreover, we confirmed that miR-133b could target GST-π expression using a dual-luciferase reporter assay. GSTs are associated with the detoxification of cytostatic drugs, and previous studies indicate that GST polymorphisms may affect drug metabolism and influence chemotherapy and cancer survival.22,23 For example, Beeghly et al showed that reduced GST function may improve ovarian cancer survival after postoperative chemotherapy and that evaluating GST functional polymorphisms may help to predict ovarian cancer prognosis.24 Indeed, many drug-resistant cancers express high levels of the π isoform of GST. Therefore, as increased miR-133b expression can reduce GST-π expression, it may prevent development of drug resistance and improve ovarian cancer prognosis. Our results showed that miR-133b transfection decreased GST-π expression at both the mRNA and protein level, and therefore may reduce chemotherapy drug resistance.
Drug resistance of tumor cells involves several mechanisms. For example, another important protein involved in drug resistance is MDR1. MDR1, also known as the P-glycoprotein 1 or the ATP-binding cassette subfamily B member 1, is important for transporting drugs across the cell membrane. Increased MDR1 expression can enhance drug resistance as the protein pumps drugs out of the cells and decreases the intracellular drug concentration.25–30 In our study, miR-133b transfection decreased MDR1 expression at both mRNA and protein levels.
Therefore, we demonstrated that overexpression of miR-133b increases ovarian cancer cell sensitivity to cisplatin and paclitaxel by decreasing GST-π and MDR1 expression. We suggest that the combination of miR 133b with chemotherapy agents may prevent the development of drug resistance in ovarian cancers. However, the precise molecular mechanisms of miR-133b, and the opportunities for its clinical manipulation, require further investigation.
Conclusion
In summary, we demonstrated that overexpression of miR-133b increases ovarian cancer cell sensitivity to cisplatin and paclitaxel by decreasing GST-π and MDR1 expression. We suggest that the combination of miR-133b with chemotherapy agents may prevent the development of drug resistance in ovarian cancers. However, the precise molecular mechanisms of miR-133b, and the opportunities for its clinical manipulation, require further investigation.
Supplementary materials
Table S1.
Primers for reverse transcription polymerase chain reaction
| Gene | Primer sequence | Annealing temperature (°C) | Product size (bp) | Extension time (seconds) |
|---|---|---|---|---|
| GST-π | F: 5′-TTTCGCCGCCGCAGTCT-3′ | 60 | 135 | 34 |
| R: 5′-TCCACGGTCACCACCTCCTC-3′ | ||||
| MDR1 | F: 5′-AGGCTATCATTACTCTTTACC-3′ | 60 | 161 | 34 |
| R: 5′-TCTGGCTTCCGTTGC-3′ | ||||
| miR-133b | F: 5′-CCTCAGAAGAAAGATGCC-3′ | 60 | 85 | 34 |
| R: 5′-GCTGGTTGAAGGGGAC-3′ | ||||
| 18S | F: 5′-ACGGACAGGATTGACAGATT-3′ | 60 | 288 | 34 |
| R: 5′-GGCGTAGGGTAGGCACA-3′ |
Table S2.
Comparative cell viability result
| Cisplatin (ng/mL) | 0 | 37.5 | 150 | 600 | 2,400 | 9,600 |
|---|---|---|---|---|---|---|
| A2780 | ||||||
| Untreated | 1 | 0.9888 | 0.9124 | 0.5321 | 0.1901 | 0.0911 |
| miR-C | 1 | 0.9870 | 0.9230 | 0.5670 | 0.1700 | 0.0807 |
| miR-133b | 1 | 0.8978 | 0.6791 | 0.3747 | 0.1414 | 0.0114 |
| Anti-C | 1 | 0.9687 | 0.9023 | 0.5467 | 0.1710 | 0.0812 |
| Anti-miR-133b | 1 | 1.1375 | 1.0291 | 0.6687 | 0.2645 | 0.1625 |
| A2780/Taxol | ||||||
| Untreated | 1 | 0.9512 | 0.8930 | 0.7392 | 0.2390 | 0.1678 |
| miR-C | 1 | 0.9850 | 0.9138 | 0.7453 | 0.2436 | 0.1973 |
| miR-133b | 1 | 0.8838 | 0.7597 | 0.5607 | 0.1620 | 0.0701 |
| Anti-C | 1 | 0.9038 | 0.8760 | 0.6902 | 0.2436 | 0.1973 |
| Anti-miR-133b | 1 | 1.0389 | 0.9736 | 0.8559 | 0.3623 | 0.2236 |
| A2780/DDP | ||||||
| Untreated | 1 | 0.9798 | 0.9344 | 0.8533 | 0.5512 | 0.0666 |
| miR-C | 1 | 0.9775 | 0.9842 | 0.8479 | 0.5891 | 0.0935 |
| miR-133b | 1 | 0.9349 | 0.8157 | 0.6748 | 0.4039 | 0.0534 |
| Anti-C | 1 | 0.9491 | 0.9570 | 0.8492 | 0.5910 | 0.0543 |
| Anti-miR-133b | 1 | 0.9842 | 1.1026 | 1.0899 | 0.7376 | 0.0915 |
| OVCAR3 | ||||||
| Untreated | 1 | 0.8269 | 0.6313 | 0.4059 | 0.1387 | 0.1360 |
| miR-C | 1 | 0.8024 | 0.6137 | 0.4370 | 0.1392 | 0.1453 |
| miR-133b | 1 | 0.6518 | 0.4720 | 0.2361 | 0.1038 | 0.1016 |
| Anti-C | 1 | 0.8024 | 0.6124 | 0.4357 | 0.1423 | 0.1430 |
| Anti-miR-133b | 1 | 0.9473 | 0.7267 | 0.5712 | 0.2828 | 0.2097 |
|
| ||||||
| Paclitaxel (ng/mL) | 0 | 6.25 | 25 | 100 | 400 | 1,600 |
|
| ||||||
| A2780 | ||||||
| Untreated | 1 | 0.3446 | 0.2263 | 0.1647 | 0.1698 | 0.1354 |
| miR-C | 1 | 0.3670 | 0.2344 | 0.1756 | 0.1701 | 0.1481 |
| miR-133b | 1 | 0.2652 | 0.1348 | 0.1044 | 0.1041 | 0.0912 |
| Anti-C | 1 | 0.3267 | 0.2134 | 0.1576 | 0.1470 | 0.1248 |
| Anti-miR-133b | 1 | 0.4507 | 0.2973 | 0.2313 | 0.2069 | 0.1730 |
| A2780/Taxol | ||||||
| Untreated | 1 | 0.5472 | 0.5348 | 0.4954 | 0.4298 | 0.1996 |
| miR-C | 1 | 0.5599 | 0.5191 | 0.4745 | 0.4244 | 0.1973 |
| miR-133b | 1 | 0.5270 | 0.4398 | 0.3540 | 0.3051 | 0.1523 |
| Anti-C | 1 | 0.5060 | 0.5112 | 0.4575 | 0.4024 | 0.1797 |
| Anti-miR-133b | 1 | 0.6459 | 0.5975 | 0.5588 | 0.4864 | 0.4415 |
| A2780/DDP | ||||||
| Untreated | 1 | 0.9153 | 0.7517 | 0.6538 | 0.6006 | 0.4989 |
| miR-C | 1 | 0.9349 | 0.7644 | 0.6769 | 0.6241 | 0.5400 |
| miR-133b | 1 | 0.8895 | 0.6712 | 0.5906 | 0.5382 | 0.4586 |
| Anti-C | 1 | 0.9391 | 0.7346 | 0.6848 | 0.5891 | 0.5343 |
| Anti-miR-133b | 1 | 0.9570 | 0.9367 | 0.8479 | 0.7597 | 0.6279 |
| OVCAR3 | ||||||
| Untreated | 1 | 0.3430 | 0.2667 | 0.2569 | 0.2509 | 0.1239 |
| miR-C | 1 | 0.3629 | 0.2566 | 0.2586 | 0.2281 | 0.1374 |
| miR-133b | 1 | 0.2936 | 0.2036 | 0.1759 | 0.1382 | 0.0739 |
| Anti-C | 1 | 0.2947 | 0.2570 | 0.2558 | 0.2284 | 0.1739 |
| Anti-miR-133b | 1 | 0.3966 | 0.3721 | 0.3297 | 0.2975 | 0.2345 |
Acknowledgments
This work was supported by the Liaoning Science and Technology Grant (2013021077) and the Natural Scientific Foundation of China (81202049, 81472400, and 81472502).
Footnotes
Author contributions
YZ, SC, JWJ, KXS and ZHZ conceived the study, carried out the acquisition, analysis, and interpretation of data, and took part in drafting/revising the article. All authors read and approved the final manuscript, and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Cress RD, Chen YS, Morris CR, Petersen M, Leiserowitz GS. Characteristics of Long-Term Survivors of Epithelial Ovarian Cancer. Obstet Gynecol. 2015;126(3):491–497. doi: 10.1097/AOG.0000000000000981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma A, Raina V, Lokeshwar N, Deo SV, Shukla NK, Mohanti BK. Phase II study of cisplatin, etoposide and paclitaxel in locally advanced or metastatic adenocarcinoma of gastric/gastroesophageal junction. Indian J Cancer. 2006;43(1):16–19. doi: 10.4103/0019-509x.25770. [DOI] [PubMed] [Google Scholar]
- 4.Allegra A, Alonci A, Campo S, et al. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (review) Int J Oncol. 2012;41(6):1897–1912. doi: 10.3892/ijo.2012.1647. [DOI] [PubMed] [Google Scholar]
- 5.Sotillo E, Thomas-Tikhonenko A. Shielding the messenger (RNA): microRNA-based anticancer therapies. Pharmacol Ther. 2011;131:18–32. doi: 10.1016/j.pharmthera.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 9.Babu JM, Prathibha R, Jijith VS, Hariharan R, Pillai MR. A miR-centric view of head and neck cancers. Biochim Biophys Acta. 2011;1816(1):67–72. doi: 10.1016/j.bbcan.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 10.To KK, Zhan Z, Litman T, Bates SE. Regulation of ABCG2 expression at the 3′ untranslated region of its mRNA through modulation of transcript stability and protein translation by a putative microRNA in the S1 colon cancer cell line. Mol Cell Biol. 2008;28(17):5147–5161. doi: 10.1128/MCB.00331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Hu S, Wang J, et al. MiR-27a modulates MDR1/P-glycoprotein expression by targeting HIPK2 in human ovarian cancer cells. Gynecol Oncol. 2010;119:125–130. doi: 10.1016/j.ygyno.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Xu H, Shen H, Li H. microRNA-106a modulates cisplatin sensitivity by targeting PDCD4 in human ovarian cancer cells. Oncol Lett. 2014;7(1):183–188. doi: 10.3892/ol.2013.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma MT, He M, Wang Y, et al. MiR-487a resensitizes mitoxantrone (MX)-resistant breast cancer cells (MCF-7/MX) to MX by targeting breast cancer resistance protein (BCRP/ABCG2) Cancer Lett. 2013;339(1):107–115. doi: 10.1016/j.canlet.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 14.Guo L, Bai H, Zou D, et al. The role of microRNA-133b and its target gene FSCN1 in gastric cancer. J Exp Clin Cancer Res. 2014;33:99. doi: 10.1186/s13046-014-0099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen XN, Wang KF, Xu ZQ, et al. MiR-133b regulates bladder cancer cell proliferation and apoptosis by targeting Bcl-w and Akt1. Cancer Cell Int. 2014;14:70. doi: 10.1186/s12935-014-0070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin CW, Li XR, Zhang Y, et al. TAp63 suppress metastasis via miR-133b in colon cancer cells. Br J Cancer. 2014;110(9):2310–2320. doi: 10.1038/bjc.2014.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin W, Dong P, Ma C, et al. MicroRNA-133b is a key promoter of cervical carcinoma development through the activation of the ERK and AKT1 pathways. Oncogene. 2012;31(36):4067–4075. doi: 10.1038/onc.2011.561. [DOI] [PubMed] [Google Scholar]
- 18.van Jaarsveld MT, Helleman J, Boersma AW, et al. miR-141 regulates KEAP1 and modulates cisplatin sensitivity in ovarian cancer cells. Oncogene. 2013;32(36):4284–4293. doi: 10.1038/onc.2012.433. [DOI] [PubMed] [Google Scholar]
- 19.Pink RC, Samuel P, Massa D, Caley DP, Brooks SA, Carter DR. The passenger strand, miR-21-3p, plays a role in mediating cisplatin resistance in ovarian cancer cells. Gynecol Oncol. 2015;137(1):143–151. doi: 10.1016/j.ygyno.2014.12.042. [DOI] [PubMed] [Google Scholar]
- 20.He J, Yu JJ, Xu Q, et al. Downregulation of ATG14 by EGR1-MIR152 sensitizes ovarian cancer cells to cisplatin-induced apoptosis by inhibiting cyto-protective autophagy. Autophagy. 2015;11(2):373–384. doi: 10.1080/15548627.2015.1009781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y, Chen Q, Qin R, Zhang K, Li H. MicroRNA-449a reduces cell survival and enhances cisplatin-induced cytotoxicity via downregulation of NOTCH1 in ovarian cancer cells. Tumour Biol. 2014;35(12):12369–12378. doi: 10.1007/s13277-014-2551-3. [DOI] [PubMed] [Google Scholar]
- 22.Popęda M, Płuciennik E, Bednarek AK. Proteins in cancer multidrug resistance. Postepy Hig Med Dosw (Online) 2014;68:616–632. doi: 10.5604/17322693.1103268. Polish. [DOI] [PubMed] [Google Scholar]
- 23.Schnekenburger M, Karius T, Diederich M. Regulation of epigenetic traits of the glutathione S-transferase P1 gene: from detoxification toward cancer prevention and diagnosis. Front Pharmacol. 2014;5:170. doi: 10.3389/fphar.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beeghly A, Katsaros D, Chen H, et al. Glutathione S-transferase polymorphisms and ovarian cancer treatment and survival. Gynecol Oncol. 2006;100(2):330–337. doi: 10.1016/j.ygyno.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 25.Zhang T, Guan M, Jin HY, Lu Y. Reversal of multidrug resistance by small interfer ing double-stranded RNAs in ovarian cancer cells. Gynecol Oncol. 2005;97:501–507. doi: 10.1016/j.ygyno.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 26.Chen J, Ding Z, Peng Y, et al. HIF-1α inhibition reverses multidrug resistance in colon cancer cells via downregulation of MDR1/P-glycoprotein. PLoS One. 2014;9(6):e98882. doi: 10.1371/journal.pone.0098882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andorfer P, Rotheneder H. Regulation of the MDR1 promoter by E2F1 and EAPP. FEBS Lett. 2013;587:1504–1509. doi: 10.1016/j.febslet.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 28.Wang Q, Wang Z, Chu L, et al. The Effects and Molecular Mechanisms of MiR-106a in Multidrug Resistance Reversal in Human Glioma U87/DDP and U251/G Cell Lines. PLoS One. 2015;10(5):e0125473. doi: 10.1371/journal.pone.0125473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Januchowski R, Wojtowicz K, Sujka-Kordowska P, Andrzejewska M, Zabel M. MDR gene expression analysis of six drug-resistant ovarian cancer cell lines. Biomed Res Int. 2013;2013:241763. doi: 10.1155/2013/241763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xing AY, Shi DB, Liu W, et al. Restoration of chemosensitivity in cancer cells with MDR phenotype by deoxyribozyme, compared with ribozyme. Exp Mol Pathol. 2013;94:481–485. doi: 10.1016/j.yexmp.2013.03.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Primers for reverse transcription polymerase chain reaction
| Gene | Primer sequence | Annealing temperature (°C) | Product size (bp) | Extension time (seconds) |
|---|---|---|---|---|
| GST-π | F: 5′-TTTCGCCGCCGCAGTCT-3′ | 60 | 135 | 34 |
| R: 5′-TCCACGGTCACCACCTCCTC-3′ | ||||
| MDR1 | F: 5′-AGGCTATCATTACTCTTTACC-3′ | 60 | 161 | 34 |
| R: 5′-TCTGGCTTCCGTTGC-3′ | ||||
| miR-133b | F: 5′-CCTCAGAAGAAAGATGCC-3′ | 60 | 85 | 34 |
| R: 5′-GCTGGTTGAAGGGGAC-3′ | ||||
| 18S | F: 5′-ACGGACAGGATTGACAGATT-3′ | 60 | 288 | 34 |
| R: 5′-GGCGTAGGGTAGGCACA-3′ |
Table S2.
Comparative cell viability result
| Cisplatin (ng/mL) | 0 | 37.5 | 150 | 600 | 2,400 | 9,600 |
|---|---|---|---|---|---|---|
| A2780 | ||||||
| Untreated | 1 | 0.9888 | 0.9124 | 0.5321 | 0.1901 | 0.0911 |
| miR-C | 1 | 0.9870 | 0.9230 | 0.5670 | 0.1700 | 0.0807 |
| miR-133b | 1 | 0.8978 | 0.6791 | 0.3747 | 0.1414 | 0.0114 |
| Anti-C | 1 | 0.9687 | 0.9023 | 0.5467 | 0.1710 | 0.0812 |
| Anti-miR-133b | 1 | 1.1375 | 1.0291 | 0.6687 | 0.2645 | 0.1625 |
| A2780/Taxol | ||||||
| Untreated | 1 | 0.9512 | 0.8930 | 0.7392 | 0.2390 | 0.1678 |
| miR-C | 1 | 0.9850 | 0.9138 | 0.7453 | 0.2436 | 0.1973 |
| miR-133b | 1 | 0.8838 | 0.7597 | 0.5607 | 0.1620 | 0.0701 |
| Anti-C | 1 | 0.9038 | 0.8760 | 0.6902 | 0.2436 | 0.1973 |
| Anti-miR-133b | 1 | 1.0389 | 0.9736 | 0.8559 | 0.3623 | 0.2236 |
| A2780/DDP | ||||||
| Untreated | 1 | 0.9798 | 0.9344 | 0.8533 | 0.5512 | 0.0666 |
| miR-C | 1 | 0.9775 | 0.9842 | 0.8479 | 0.5891 | 0.0935 |
| miR-133b | 1 | 0.9349 | 0.8157 | 0.6748 | 0.4039 | 0.0534 |
| Anti-C | 1 | 0.9491 | 0.9570 | 0.8492 | 0.5910 | 0.0543 |
| Anti-miR-133b | 1 | 0.9842 | 1.1026 | 1.0899 | 0.7376 | 0.0915 |
| OVCAR3 | ||||||
| Untreated | 1 | 0.8269 | 0.6313 | 0.4059 | 0.1387 | 0.1360 |
| miR-C | 1 | 0.8024 | 0.6137 | 0.4370 | 0.1392 | 0.1453 |
| miR-133b | 1 | 0.6518 | 0.4720 | 0.2361 | 0.1038 | 0.1016 |
| Anti-C | 1 | 0.8024 | 0.6124 | 0.4357 | 0.1423 | 0.1430 |
| Anti-miR-133b | 1 | 0.9473 | 0.7267 | 0.5712 | 0.2828 | 0.2097 |
|
| ||||||
| Paclitaxel (ng/mL) | 0 | 6.25 | 25 | 100 | 400 | 1,600 |
|
| ||||||
| A2780 | ||||||
| Untreated | 1 | 0.3446 | 0.2263 | 0.1647 | 0.1698 | 0.1354 |
| miR-C | 1 | 0.3670 | 0.2344 | 0.1756 | 0.1701 | 0.1481 |
| miR-133b | 1 | 0.2652 | 0.1348 | 0.1044 | 0.1041 | 0.0912 |
| Anti-C | 1 | 0.3267 | 0.2134 | 0.1576 | 0.1470 | 0.1248 |
| Anti-miR-133b | 1 | 0.4507 | 0.2973 | 0.2313 | 0.2069 | 0.1730 |
| A2780/Taxol | ||||||
| Untreated | 1 | 0.5472 | 0.5348 | 0.4954 | 0.4298 | 0.1996 |
| miR-C | 1 | 0.5599 | 0.5191 | 0.4745 | 0.4244 | 0.1973 |
| miR-133b | 1 | 0.5270 | 0.4398 | 0.3540 | 0.3051 | 0.1523 |
| Anti-C | 1 | 0.5060 | 0.5112 | 0.4575 | 0.4024 | 0.1797 |
| Anti-miR-133b | 1 | 0.6459 | 0.5975 | 0.5588 | 0.4864 | 0.4415 |
| A2780/DDP | ||||||
| Untreated | 1 | 0.9153 | 0.7517 | 0.6538 | 0.6006 | 0.4989 |
| miR-C | 1 | 0.9349 | 0.7644 | 0.6769 | 0.6241 | 0.5400 |
| miR-133b | 1 | 0.8895 | 0.6712 | 0.5906 | 0.5382 | 0.4586 |
| Anti-C | 1 | 0.9391 | 0.7346 | 0.6848 | 0.5891 | 0.5343 |
| Anti-miR-133b | 1 | 0.9570 | 0.9367 | 0.8479 | 0.7597 | 0.6279 |
| OVCAR3 | ||||||
| Untreated | 1 | 0.3430 | 0.2667 | 0.2569 | 0.2509 | 0.1239 |
| miR-C | 1 | 0.3629 | 0.2566 | 0.2586 | 0.2281 | 0.1374 |
| miR-133b | 1 | 0.2936 | 0.2036 | 0.1759 | 0.1382 | 0.0739 |
| Anti-C | 1 | 0.2947 | 0.2570 | 0.2558 | 0.2284 | 0.1739 |
| Anti-miR-133b | 1 | 0.3966 | 0.3721 | 0.3297 | 0.2975 | 0.2345 |