Abstract
The transcription factor XBP1 has been linked to the development of highly secretory tissues such as plasma cells and Paneth cells, yet its function in granulocyte maturation has remained unknown. Here we discovered an unexpectedly selective and absolute requirement for XBP1 in eosinophil differentiation without an effect on the survival of basophils or neutrophils. Progenitors of myeloid cells and eosinophils selectively activated the endoribonuclease IRE1α and spliced Xbp1 mRNA without inducing parallel endoplasmic reticulum (ER) stress signaling pathways. Without XBP1, nascent eosinophils exhibited massive defects in the post-translational maturation of key granule proteins required for survival, and these unresolvable structural defects fed back to suppress critical aspects of the transcriptional developmental program. Hence, we present evidence that granulocyte subsets can be distinguished by their differential reliance on secretory-pathway homeostasis.
Introduction
The endoplasmic reticulum (ER) is a critical regulator of calcium storage and signaling, lipid biosynthesis, and the proper folding and post-translational modification of secreted and transmembrane proteins. This organelle functions in a highly integrated manner to support these fundamental and interconnected biological processes, and disruptions in specific ER tasks are often counterbalanced by compensatory modulation of parallel ER abilities. ER dysfunction, or stress, can be caused by the intraluminal accumulation of misfolded proteins. If the influx of new protein substrates into the ER overwhelms its steady-state protein-folding capacity, a multi-pronged response known as the ‘unfolded protein response’ (UPR) is triggered to ameliorate cellular ER stress.
The UPR is driven by the combined action of the ER membrane–localized kinase-endoribonuclease IRE1α (encoded by Ern1), the transcription factor ATF6 and the kinase PERK. When ER stress occurs, IRE1α splices out 26 nucleotides from cytoplasmic Xbp1 mRNA and thereby induces a shift in the reading frame that leads to the translation of a highly active transcription factor involved in the UPR1. PERK induces translational repression by phosphorylating the translation-initiation factor eIF2α2, which subsequently activates the transcription factors ATF4 and CHOP (encoded by Ddit3)3. Simultaneously, ATF6, together with XBP1, regulates various genes encoding stress-response factors4, 5. This tripartite response reduces ER stress by upregulating the expression of genes encoding protein chaperones, redox enzymes and glycosylases to enhance protein folding within the ER while simultaneously reducing the overall translation rate to slow the influx of new proteins. Successful resolution of ER stress results in cellular adaptation and protection from future proteotoxic insults, while failure to ameliorate the offending stimulus leads to cell death.
XBP1 is best known as a major developmental regulator of highly secretory cells such as plasma cells6, Paneth cells7 and pancreatic acinar cells8. These cell types are generally equipped with extensive ER networks to cope with the burden of constitutive synthesis of secreted proteins upon terminal maturation. In the absence of Xbp1, a condition that can sensitize cells to ER stress by diminishing adaptive protein-folding capacity, such highly secretory cells generally demonstrate a disorganized ultrastructure of the endomembrane pathway and increased cell death8. Notably, XBP1 also supports the differentiation and survival of certain cell types that are not thought to engage in constitutively abundant production of secreted proteins, such as resting splenic dendritic cells9. We therefore postulated that developmental and cell-survival requirements for the UPR, and XBP1 in particular, might extend beyond ‘professional’ secretory cells due to the need to cope with transient but intense bursts of protein production encountered during development. To this end, we chose to study granulocyte differentiation as a model system for investigating the physiological importance of the UPR during intense, developmentally restricted demands on the secretory pathway.
Neutrophils, eosinophils and basophils constitute the three main types of granulocytes. Large quantities of neutrophils are produced and stored in the bone marrow, poised for rapid deployment to sites of infection and injury to destroy invading microorganisms and help coordinate type 1 immune responses10. In contrast, the comparatively rare eosinophils11 and basophils12 are typically associated with type 2 immune responses, allergy and parasitic infection. However, published reports have shown that these ‘enigmatic’ cells, particularly eosinophils, can serve important roles in the regulation of insulin sensitivity13, adipose tissue development14, regenerative responses to injury15, tumor burden16 and the survival of memory plasma cells17.
The earliest stages of granulocyte differentiation are morphologically similar across lineages and are marked by rapid upregulation and intense production of granulocyte-specific effector proteins18. As differentiation progresses over several days, granule-protein production is gradually extinguished and components of the secretory pathway, including the ER, are substantially downsized18. Thus, granulocyte differentiation represents a tractable model system for investigating how the UPR, and XBP1 in particular, regulates survival and adaptation during physiologically relevant, temporally restricted states requiring high secretory-pathway functionality.
Here we report a highly selective and indispensable role for XBP1 and its upstream activator IRE1α in the development of mouse eosinophils but not of other granulocyte lineages. Hematopoietic ablation of Xbp1 resulted in complete, cell-intrinsic loss of mature eosinophils and progenitors of eosinophils without affecting upstream precursors. Unbiased transcriptome analyses of hematopoietic progenitor populations along the eosinophil developmental continuum revealed that diminished basal ER protein-folding capacity actively prevented terminal maturation after the commitment of progenitor cells to the eosinophil lineage, in part by downregulating expression of Gata1, which encodes the master eosinophil lineage–commitment factor GATA-1, and genes encoding indispensable granule proteins. Collectively our findings identify an unexpected interplay between cellular stress and a major lineage-determining transcription factor and indicate that the IRE1α-XBP1 signaling axis is a potential therapeutic target for eosinophil-mediated diseases.
Results
XBP1 is indispensable for eosinophil production
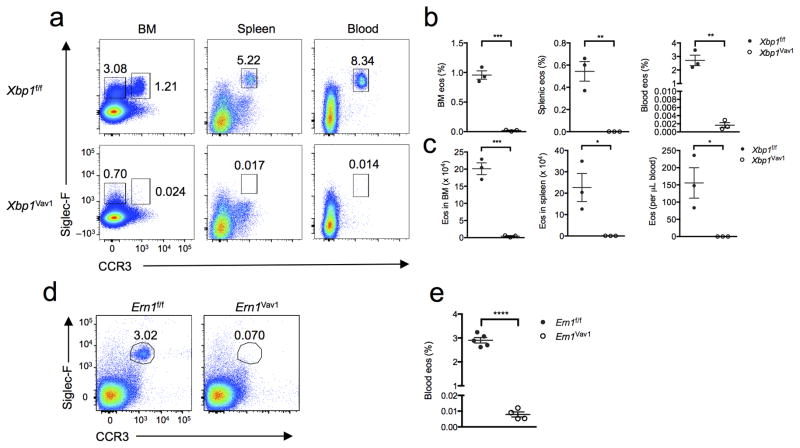
To investigate a possible role for XBP1 in granulocyte development, we crossed mice expressing Cre recombinase from the hematopoietic compartment–specific Vav1 promoter (Vav1-Cre mice) to mice with loxP-flanked Xbp1 alleles (Xbp1f/f mice) to generate Xbp1f/fVav1-Cre mice (called ‘Xbp1Vav1 mice’ here) for targeted deletion of Xbp1 in the hematopoietic compartment. Total bone marrow cellularity was unaffected by loss of Xbp1 (Supplementary Fig. 1), and the frequency of splenic T cells, B cells, macrophages, neutrophils and basophils was essentially equivalent in Xbp1f/f and Xbp1Vav1 mice (Supplementary Fig. 2a–i), which indicated that Xbp1 deficiency was generally well tolerated across the immune system. Similar to results in published reports9, Xbp1Vav1 mice exhibited a reduction of approximately twofold in CD11c+ splenic dendritic cells relative to their abundance in Xbp1f/f mice (Supplementary Fig. 2j,k). Unexpectedly, we observed highly selective and complete ablation of the eosinophil lineage in Xbp1Vav1 bone marrow, spleen and blood compared with the abundance of this lineage in Xbp1f/f bone marrow (Fig. 1a–c). Cre expression alone had no effect on the number of peripheral eosinophils (Supplementary Fig. 3). The persistence of neutrophils and basophils was not due to the escape and population expansion of rare Xbp1-sufficient progenitor cells, as sorted bone marrow granulocytes exhibited robust deletion of Xbp1 (Supplementary Fig. 4a,b). Because Xbp1 mRNA is directly spliced by IRE1α, we sought to determine whether Ern1f/fVav1-Cre (Ern1Vav1) mice were similarly deficient in eosinophils. Ern1Vav1 mice were a full phenocopy of the Xbp1Vav1 mice (Fig. 1d,e), which indicated that physiological eosinophil differentiation in mice was wholly dependent on the spliced form of Xbp1.
Figure 1.
XBP1 is required for eosinophil differentiation. (a) Flow cytometry of cells from the bone marrow (BM), spleen and blood of Xbp1f/f and Xbp1Vav1 mice, gated on non-B, non-T cells (spleen and blood only). Numbers adjacent to outlined areas indicate percent Siglec-F+CCR3+ mature eosinophils (top right) or Siglec-F+CCR3− immature bone marrow eosinophils (top left, far left column). (b) Frequency of mature (Siglec-F+CCR3+) eosinophils (eos) in the bone marrow, spleen and blood of Xbp1f/f and Xbp1Vav1 mice (n = 3 per genotype). (c) Absolute number of mature eosinophils in the bone marrow (two tibias and two femurs per mouse), spleen and blood of Xbp1f/f and Xbp1Vav1 mice (n = 3 per genotype). (d) Flow cytometry of cells from the blood of Ern1f/f and Ern1Vav1 mice. Numbers adjacent to outlined areas indicate percent Siglec-F+CCR3+ mature eosinophils. (e) Frequency of mature eosinophils in blood of Ern1f/f mice (n = 5) and Ern1Vav1 mice (n = 4). Each symbol (b,c,e) represents an individual mouse; small horizontal lines indicate the mean (± s.e.m.). *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 (Student’s t-test). Data are representative of at least three independent experiments with at least three mice per group in each.
Eosinophilopoiesis involves physiological ER stress
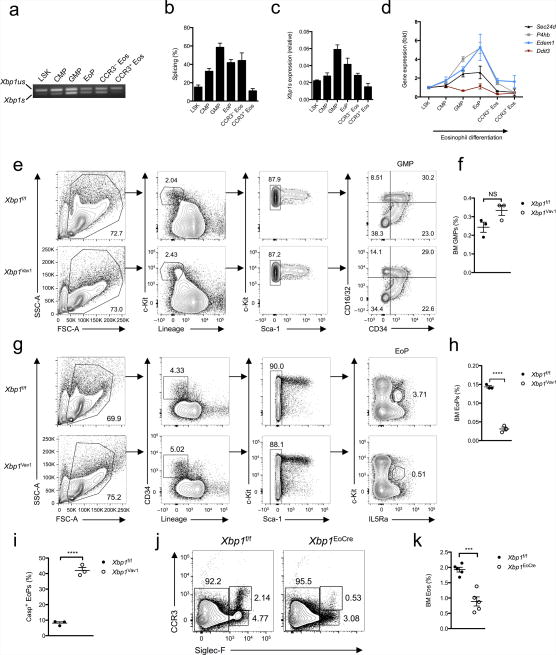
Since XBP1 was an absolute requirement for eosinophil development, we next sought to determine whether the IRE1α-XBP1 pathway is temporally regulated during eosinophil differentiation. Hematopoietic stem cells identified as lineage marker–negative (Lin−) Sca-1+ c-Kit+ (LSK) cells progressively commit to the eosinophil lineage after passing through the stages of Lin−Sca-1−c-Kit+CD34+CD16/32− common myeloid progenitor (CMP), Lin−Sca-1−c-Kit+CD34+CD16/32+ granulocyte-macrophage progenitor (GMP) and IL-5Rα+Lin−Sca-1− c-KitdimCD34+ eosinophil progenitor (EoP)19. Immature eosinophils upregulate their expression of the sialoadhesin Siglec-F, while fully mature eosinophils additionally express the chemokine receptor CCR3 (ref. 20). We sorted cells along the eosinophil-differentiation spectrum and evaluated the ratio of full-length Xbp1 mRNA to active, spliced Xbp1 mRNA by quantitative PCR, after validating Actb as a suitably stable housekeeping reference gene across multiple cell lineages (data not shown). Notably, Xbp1 mRNA was progressively spliced during differentiation, with the greatest activation in GMPs and EoPs (Fig. 2a–c). Splicing of Xbp1 mRNA correlated with the induction of numerous downstream genes that are targets of XBP1, such as P4hb, Edem1 and Sec24d (Fig. 2d). Upon final cellular maturation, protein-synthetic demands drop considerably, which probably explains why terminally differentiated eosinophils no longer spliced Xbp1 mRNA. In contrast, Ddit3 was not upregulated during eosinophil differentiation (Fig. 2d), which suggested that the PERK axis was not induced. We were unable to detect expression of PERK by immunoblot analysis in any cell type examined except CCR3+ eosinophils (data not shown), which again suggested that this UPR signaling branch was minimally active during eosinophil differentiation. However, we were unable to rule out the possibility that small amounts of PERK are phosphorylated during eosinophil differentiation. Collectively these results suggested that developing eosinophils underwent a branch-specific UPR characterized by activation of IRE1α without activation of PERK. Similar cases of selective branch use have been observed in both macrophages and plasma cells, although why branch selectivity occurs, in a teleological sense, remains poorly understood21,22.
Figure 2.
XBP1 is potently activated during eosinophil differentiation in vivo and is required upon commitment to the eosinophil lineage. (a) PCR analysis of spliced (Xbp1s) and unspliced (Xbp1us) Xbp1 mRNA in LSK cells, CMPs, GMPs, EoPs and CCR3− or CCR3+ eosinophils purified by flow cytometry. (b) Frequency of Xbp1s mRNA among total Xbp1 mRNA in sorted LSK cells, CMPs, GMPs, EoPs, CCR3− eosinophils, and CCR3+ eosinophils (n = 3 mice per cell type). (c) Quantitative PCR analysis of the Xbp1s isoform in cells as in a (n = 3 mice per cell type); results were normalized to those of Actb. (d) Quantitative PCR analysis of Sec24d, P4hb, Edem1 and Ddit3 in cells as in a (n = 3 mice per cell type); results (normalized as in c) are presented relative to those of LSK cells. (e) Gating strategy for flow cytometry of bone marrow GMPs from Xbp1f/f and Xbp1Vav1 mice. Numbers adjacent to outlined areas indicate percent cells with leukocyte side scatter (SSC-A) or forward scatter (FSC-A) (far left), c-Kit+Lin− cells (middle left) or c-Kit+Sca-1− cells (middle right); numbers in quadrants indicate percent GMPs in each (far right). (f) Frequency of GMPs in the bone marrow of Xbp1f/f and Xbp1Vav1 mice (n = 3 per genotype). (g) Gating strategy for flow cytometry of bone marrow EoPs from Xbp1f/f and Xbp1Vav1 mice. Numbers adjacent to outlined areas indicate percent cells with leukocyte side or forward scatter (far left), CD34+Lin− cells (middle left), c-Kitpos–negSca-1− cells (middle right) or c-KitintIL-5Rα+ cells (far right). (h) Frequency of EoPs in bone marrow from Xbp1f/f and Xbp1Vav1 mice (n = 3 per genotype). (i) Frequency of EoPs exhibiting caspase activity (Casp+), assessed by flow cytometry (n = 3 mice per genotype). (j) Flow cytometry of eosinophils in bone marrow from Xbp1f/f and Xbp1eoCRE mice. Numbers adjacent to outlined areas indicate percent CCR3pos–negSiglec-F− cells (left), CCR3+Siglec-F+ cells (top right) or CCR3−Siglec-F+ cells (bottom right). (k) Frequency of eosinophils in bone marrow from Xbp1f/f and Xbp1eoCRE mice (n = 5 mice per genotype). Each symbol (f,h,i,k) represents an individual mouse; small horizontal lines indicate the mean (± s.e.m.). NS, not significant (P > 0.05); *P < 0.001 and **P < 0.0001 (Student’s t-test). Data are from one experiment representative of three experiments with more than three independent biological replicates (a) or are representative of at least three independent experiments with at least three mice per group in each (b–k; mean and s.e.m. in b–d,f,h,i,k).
XBP1 specifically supports eosinophil lineage survival
Given that Xbp1 mRNA was maximally spliced in GMPs, we next sought to determine whether XBP1 was required for the differentiation and survival of GMPs. The frequency of bone marrow GMPs in Xbp1Vav1 mice was similar to that in Xbp1f/f mice (Fig. 2e,f), despite robust deletion of Xbp1 (Supplementary Fig. 4c). Therefore, despite a high frequency of Xbp1 mRNA splicing in GMPs, XBP1 did not substantially affect their differentiation and steady-state survival. In contrast, the EoP compartment was much smaller in Xbp1Vav mice than in Xbp1f/f mice (Fig. 2g,h), which indicated that XBP1 was required at the earliest identifiable eosinophil-committed developmental stage. As the in vivo roles of XBP1 in cellular differentiation often correlate with defects in cell survival8, we sought to determine whether Xbp1Vav1 EoPs exhibited signs of enhanced cell death. As expected, residual EoPs from Xbp1Vav1 mice demonstrated increased staining of active caspases (Fig. 2i), which indicated that XBP1 sustained the viability of EoPs.
To determine whether XBP1 was required for commitment to the eosinophil lineage, we bred the Xbp1f/f mice onto the Epx-Cre (eoCRE) strain23 to generate ‘Xbp1eoCRE’ mice. The eoCRE knock-in strain expresses Cre from the open reading frame of the endogenous gene encoding eosinophil peroxidase (Epx) and thus expresses Cre only after commitment to the eosinophil lineage and is exceedingly specific for eosinophils rather than cells of other hematopoietic lineages23. Xbp1eoCRE mice had a significantly smaller bone marrow eosinophil population than Xbp1f/f mice had (Fig. 2j,k) and, notably, the surviving peripheral eosinophils had escaped deletion of Xbp1 (data not shown). Thus, XBP1 was needed to sustain viability of the eosinophil-committed progenitor.
Eosinophil development requires XBP1S cell-intrinsically
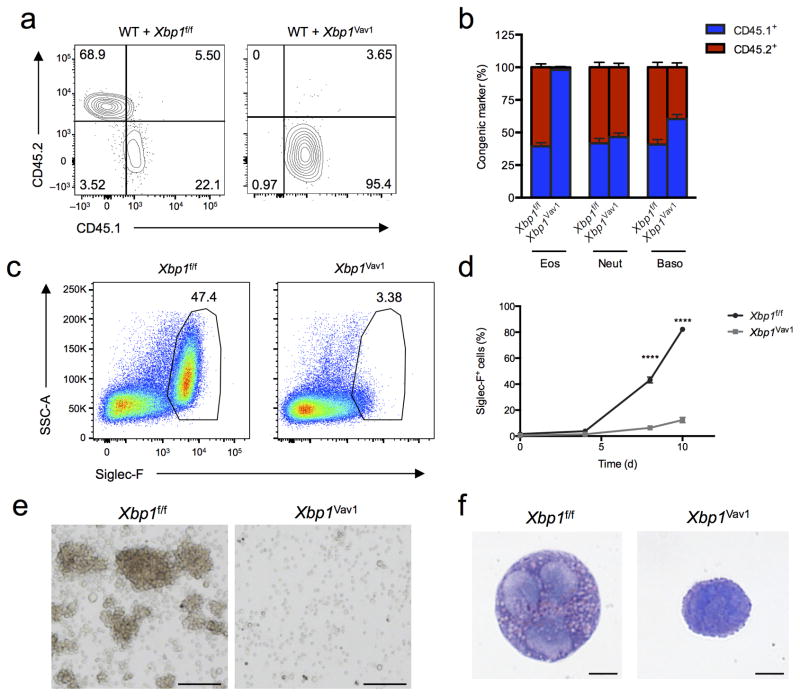
Eosinophil development relies on many transcription factors, such as GATA-1 and PU.1, but is also regulated by extracellular cues, such as interferon-γ24. To address whether the observed phenotype stemmed from cell-intrinsic or cell-extrinsic defects, we generated mixed–bone marrow chimeras by injecting a 1:1 mixture of congenically marked CD45.1+ wild-type cells and CD45.2+ Xbp1f/f or Xbp1Vav1 cells into lethally irradiated CD45.1+ recipient mice. We found that Xbp1Vav1 CD45.2+ cells were completely incapable of generating mature eosinophils, although their production of neutrophils and basophils was normal (Fig. 3a,b), which indicated that XBP1 functioned in a cell-intrinsic way to sustain eosinophil development. In further support of those findings, our efforts to generate Xbp1Vav1 bone marrow–derived eosinophils (BMDEs)25 were unsuccessful, even in cultures containing interleukin 5 (IL-5) supplemented with additional pro-survival cytokines, such as IL-3 and GM-CSF (Fig. 3c and data not shown). The frequency of Siglec-F+ cells was significantly lower in Xbp1Vav1 cultures than in Xbp1f/f cultures at days 8 and 10 (Fig. 3d), when eosinophil populations are normally rapidly expanding. Xbp1Vav1 cells failed to acquire the characteristic eosinophil granule-associated high-side-scatter profile or to form proliferating colonies (Fig. 3c,e). Furthermore, cytospin analysis of Siglec-F+ BMDEs at day 8 that were negative for the DNA-binding dye DAPI (DAPI−), which had been purified by flow cytometry, revealed that nascent eosinophils derived from Xbp1Vav1 cells initiated nuclear segmentation but were incapable of generating mature eosinophilic granules (Fig. 3f), indicative of arrested development. Notably, blocking IRE1α with the potent small-molecule inhibitor 4μ8C26 had no effect on the survival of fully differentiated eosinophils from the bone marrow ( had no effect on survival of fully differentiated eosinophils from the bone marrow (Supplementary Fig. 5). We noted similar effects on splenic eosinophils purified from mice with transgenic IL-5 expression27 and in mature BMDEs incubated with concentrations of 4μ8C as high as 64 μM (data not shown). The function of XBP1, therefore, mapped in a cell-intrinsic way to a developmental window after commitment to the eosinophil lineage but before terminal differentiation.
Figure 3.
XBP1 is a cell-intrinsic requirement for eosinophil development. (a) Flow cytometry assessing expression of the congenic markers CD45.2 (Xbp1f/f or Xbp1Vav1) and CD45.1 (wild type) on peripheral blood eosinophils from chimeras generated with a mixture of wild-type bone marrow plus either Xbp1f/f bone marrow (WT + Xbp1f/f) or Xbp1Vav1 bone marrow (WT + Xbp1Vav1). Numbers in quadrants indicate percent cells in each. (b) Reconstitution efficiency of eosinophils, neutrophils (Neut) and basophils (Baso) in blood from mixed-bone marrow chimeras as in a (n = 6 host mice per chimera type). (c) Flow cytometry analyzing Siglec-F expression Xbp1f/f and Xbp1Vav1 BMDEs at day 8 of culture, gated on DAPI− singlets. Numbers adjacent to outlined areas indicate percent Siglec-F+ cells. (d) Frequency of Siglec-F+ cells among Xbp1f/f and Xbp1Vav1 BMDEs at days 0, 4, 8 and 10 of culture (n = 3 independent cultures per genotype). (e) Microscopy of hematopoietic colonies from Xbp1f/f and Xbp1Vav1 BMDEs at day 8 of culture. Scale bars, 100 μm. (f) Microscopy of cytospin analysis of DAPI−Siglec-F+ singlets sorted from Xbp1f/f and Xbp1Vav1 BMDEs at day 8 of culture. Scale bars, 5 μm. *P < 0.0001 (two-way analysis of variance (ANOVA) with the Šidák correction for multiple comparisons). Data are from two independent experiments with six mice per group (a,b; mean and s.e.m. in b) or more than three independent experiments with at least three mice per group (c–f; mean and s.e.m. in d).
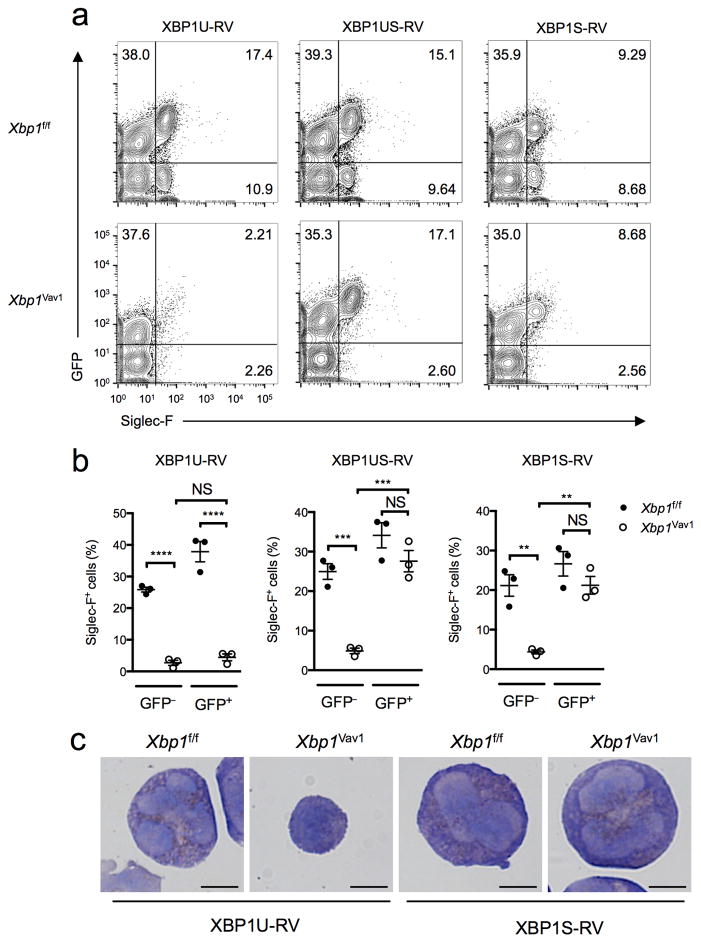
We next sought to determine whether ectopic overexpression of various Xbp1 mRNA isoforms could ‘rescue’ the developmental defects in eosinophils. Retroviral overexpression of full-length, unspliced but spliceable Xbp1 mRNA (Xbp1us) or its shorter, spliced isoform (Xbp1s) fully restored eosinophil development in vitro (Fig. 4a,b). However, Xbp1Vav1 cells were not ‘rescued’ by the over-expression of a full-length, unspliceable Xbp1 mRNA variant (Xbp1u) (Fig. 4a,b), which further established the requirement for the Xbp1s isoform in eosinophilopoiesis. Of note, the ‘rescued’ cells displayed full morphological recovery (Fig. 4c). Thus, the substantial eosinophil developmental defects observed in Xbp1Vav1 mice could be readily corrected in vitro but only when the spliced Xbp1s RNA isoform was supplied.
Figure 4.
Xbp1s can restore eosinophil development in Xbp1-deficient bone marrow cultures, but Xbp1u cannot. (a) Flow cytometry analyzing the expression of GFP and Siglec-F in Xbp1f/f and Xbp1Vav1 BMDEs transduced with retrovirus expressing Xbp1u mRNA (XBP1U-RV), Xbp1us mRNA (XBP1US-RV) or Xbp1s mRNA (XBP1S-RV) assessed at day 8 of culture. Numbers in quadrants indicate percent cells in each. (b) Frequency of Siglec-F+ eosinophils among total GFP− and GFP+ cells from Xbp1f/f and Xbp1Vav1 BMDE cultures transduced as in a. Each symbol represents an individual mouse (n = 3 per genotype); small horizontal lines indicate the mean (± s.e.m.). (c) Microscopy of cytospin analyses of GFP+Siglec-F+ cells sorted from Xbp1f/f and Xbp1Vav1 BMDEs transduced as in a (below images), assessed at day 8 of culture. Scale bars, 5 μm. *P < 0.01, **P < 0.001 and ***P < 0.0001 (one-way ANOVA with the Holm-Šidák correction for multiple comparisons). Data are representative of three independent experiments with three biological replicates in each.
Developmental networks are XBP1-independent in GMPs
Beyond its functions in maintaining ER homeostasis, XBP1 can control cell-type-specific transcriptional networks, including major cell-identity determinants, such as Mist1 in myoblasts28. Therefore, we sought to determine whether XBP1 might influence the expression of known eosinophil developmental regulators in GMPs. GATA-1, GATA-2, C/EBPα, C/EBPβ, CEBPε, IRF8, PU.1, Id2 and TRIB1 positively regulate eosinophil development, while FOG1 and Id1 act as negative regulators29,30. Despite the presence of robust splicing of Xbp1 mRNA, all known regulators of eosinophil development were expressed normally in Xbp1-deficient GMPs (Fig. 5a), which indicated that XBP1 was dispensable for the baseline expression of canonical regulators of eosinophil differentiation.
Figure 5.
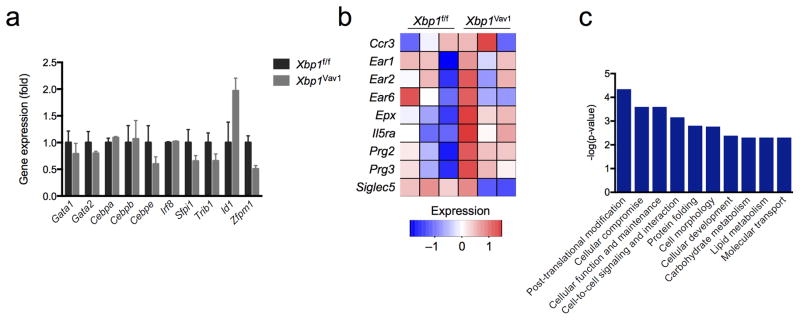
The GMP-differentiation capacity is not affected by Xbp1 deficiency. (a) Quantitative PCR analysis of genes encoding products known to regulate eosinophil differentiation, in sorted GMPs from Xbp1f/f and Xbp1Vav1 mice (n = 3 per genotype); results were normalized to those of Actb and are presented relative to those of Xbp1f/f GMPs. (b) RNA-seq analysis of selected genes encoding eosinophil markers, in Xbp1f/f and Xbp1Vav1 GMPs (n = 3 mice per genotype). (c) Pathway analysis of the ten molecular and cellular gene-ontology functions most significantly dysregulated in Xbp1Vav1 GMPs relative to their regulation in Xbp1f/f GMPs, ranked by P value from most significant (left) to least significant (right); results are presented as raw P values. Data are representative of three independent experiments (a; mean and s.e.m.) or one experiment with three independent biological replicates per group (b,c).
Attempts to isolate the few remaining CD34+ EoPs from Xbp1Vav1 mice were unsuccessful due to technical limitations (data not shown). Therefore, to delineate the effect of Xbp1 deficiency after eosinophil commitment, we used a published strategy to enforce eosinophil differentiation on GMPs via retroviral overexpression of GATA-2 (ref. 31). Normally only ~1% of GMPs have the potential to develop into eosinophils, but overexpression of GATA-2 induces commitment to the eosinophil lineage31. We purified GMPs from Xbp1f/f and Xbp1Vav1 mice by flow cytometry and infected the cells with control retrovirus expressing green fluorescent protein (GFP) alone or retrovirus expressing GATA-2 and GFP; after 36 h, we re-sorted GFP+ (infected) cells and cultured the cells for various times31. While control retrovirus–transduced Xbp1f/f cells gave rise almost exclusively to macrophages and neutrophils, GATA-2-transduced cells demonstrated robust formation of eosinophilic granules and had high expression of genes encoding canonical markers of the eosinophil lineage (Supplementary Fig. 6a,b). However, similar to their maturation defects in BMDEs, secretory granules in GATA-2-transduced Xbp1Vav1 GMPs were immature compared with those in GATA-2-transduced Xbp1f/f GMPs (Supplementary Fig. 6c). Hence, GATA-2-transduced GMPs represented an effective phenocopy of critical aspects of eosinophil differentiation interrupted by loss of Xbp1.
After validating GATA-2 overexpression as a model for early eosinophil differentiation, we performed high-throughput sequencing for cDNA (RNA-seq) on freshly sorted GMPs and GATA-2- transduced GMPs from Xbp1f/f and Xbp1Vav1 mice to identify stage-specific, XBP1-dependent processes. Of the 153 genes differentially expressed in Xbp1f/f GMPs versus Xbp1Vav1 GMPs, none encoded terminal eosinophil effectors (Fig. 5b and Supplementary Table 1), which suggested that defects in eosinophil development had yet to manifest. Additionally, pathway analysis identified alterations in canonical secretory pathway–associated processes, such as post-translational modifications, protein folding, carbohydrate metabolism, lipid metabolism, and organization of the Golgi apparatus (Fig. 5c and Supplementary Table 2). Despite the finding of reduced expression of genes encoding factors involved in multiple networks responsible for maintaining ER homeostasis, cellular viability and differentiation pathways were unaffected in GMPs, which indicated that deletion of Xbp1 before commitment to the eosinophil lineage impaired the adaptive capacity of the ER without causing acute cellular impairment.
XBP1 sustains eosinophil development after commitment
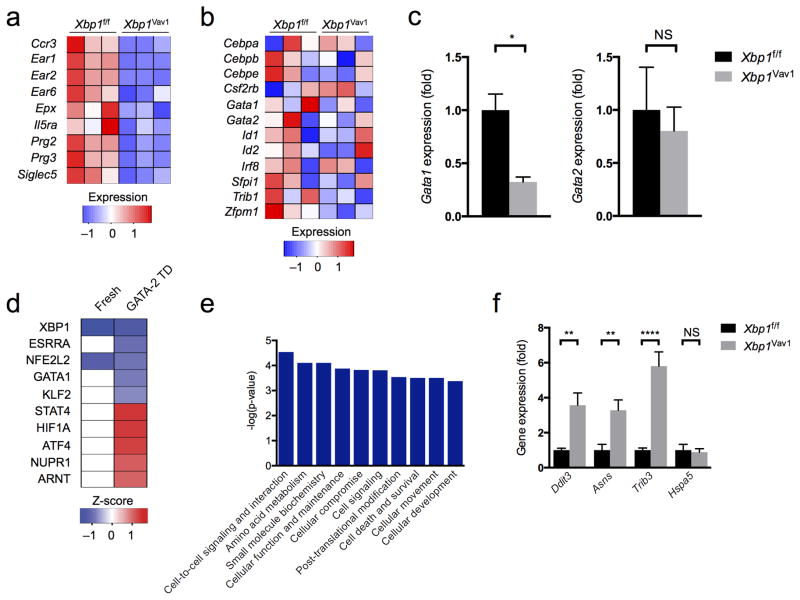
Notably, in GATA-2-transduced GMPs cultured for 48 h, we observed pronounced downregulation of genes encoding several markers of terminal eosinophil differentiation (Fig. 6a and Supplementary Table 3). Genes encoding most known transcriptional regulators of eosinophil development generally had equivalent expression in GATA-2-transduced Xbp1f/f and Xbp1Vav1 GMPs, although the gene encoding GATA-1, a key transcription factor for eosinophil development, was modestly downregulated in GATA-2-transduced Xbp1f/f GMPs relative to its expression in GATA-2-transduced Xbp1Vav1 GMPs (Fig. 6b,c). Expression of the gene encoding GATA-2 was similar in Xbp1f/f and Xbp1Vav1 cells (Fig. 6c), which indicated that the effects on Gata1 were not due to unequal ectopic overexpression of Gata2. Additionally, pathway analysis of potential upstream transcriptional regulators predicted that GATA-1 signaling was inhibited in Xbp1Vav1 cells, on the basis of downregulation of classic targets of GATA-1, such as Cdkn2c, Abcb10 and Prg2, but only after commitment to the eosinophil lineage (Fig. 6d and Supplementary Table 2). Inefficient GATA-1-mediated signaling might have contributed to the loss of expression of genes encoding many eosinophil markers. Of the 200 genes differentially expressed in GATA-2-overexpressing Xbp1f/f GMPs versus their Xbp1Vav1 counterparts, less than 20% overlapped with genes known to be upregulated during BMDE differentiation32, which indicated that loss of Xbp1 inhibited limited and specific facets of the eosinophil developmental program.
Figure 6.
Loss of XBP1-mediated protein-quality-control mechanisms interferes with eosinophil transcriptional identity. (a,b) RNA-seq analysis of genes encoding eosinophil markers (a) or regulators of eosinophil development (b), in GATA-2-transduced Xbp1f/f and Xbp1Vav1 GMPs (n = 3 independent cultures per genotype). (c) Quantitative PCR analysis of Gata1 and Gata2 in GATA-2-transduced Xbp1f/f and Xbp1Vav1 GMPs (n = 3 independent cultures per genotype) (presented as in Fig. 5a). (d) Pathway analysis–predicted top five activated or inhibited upstream transcriptional regulators (ranked by activation z-score) most responsible for the transcriptional differences between GATA-2-transduced (GATA-2 TD) Xbp1f/f GMPs and GATA-2-transduced Xbp1Vav1 GMPs, showing functional repression (blue) or activation (red) in Xbp1Vav1 cells compared with Xbp1f/f cells, and predicted influence on freshly sorted GMPs (Fresh). (e) Pathway analysis–predicted ten most significantly dysregulated molecular and cellular gene-ontology functions in GATA-2-transduced Xbp1f/f and Xbp1Vav1 GMPs (presented as in Fig. 5b). (f) Quantitative PCR analysis of Ddit3, Asns, Trib3 and Hspa5 in GATA-2-transduced Xbp1f/f and Xbp1Vav1 GMPs (n = 3 independent cultures per genotype) (presented as in Fig. 5a). *P < 0.05, **P < 0.01 and ***P < 0.0001 (Student’s t-test (c) or one-way ANOVA with the Holm-Šidák correction for multiple comparisons (f)). Data are representative of one experiment (a,b,d,e) or two independent experiments with three independent biological replicates per group (c,f; mean and s.e.m.).
We additionally observed that genes encoding products involved in amino-acid metabolism and cell-death pathways were markedly induced in GATA-2-transduced Xbp1Vav1 GMPs relative to their expression in GATA-2-transduced Xbp1f/f GMPs (Fig. 6e), which indicated that eosinophil differentiation without XBP1 was cytotoxic. The PERK signaling axis is considered to be pro-apoptotic and regulates amino-acid metabolism via ATF4 and CHOP33, and we therefore sought to determine whether dysregulated gene expression reflected the activation of alternative UPR signaling branches. Consistent with that possibility, pathway analysis predicted ATF4 to be a strong mediator of the observed transcriptional changes (Fig. 6d). By quantitative PCR, we confirmed that the expression of several genes that are targets of ATF4 (Ddit3, Asns and Trib3) was upregulated in GATA-2-transduced Xbp1Vav1 GMPs compared with their expression in GATA-2-transduced Xbp1f/f GMPs (Fig. 6f). Unexpectedly, Hspa5 expression was not higher in GATA-2-transduced Xbp1Vav1GMPs than in GATA-2-transduced Xbp1f/f GMPs (Fig. 6f), which suggested that Xbp1 deficiency in eosinophil-committed cells activated an atypical, ATF4-dominated UPR.
The activation of PERK-ATF4 may have been driven by the accumulation of misfolded or incorrectly processed eosinophil-specific proteins. To investigate this possibility, we analyzed the posttranslational maturation of the eosinophil granule proteins PRG2 and EPX. Strikingly, we observed substantial accumulation of the ‘pre-pro-forms’ and/or ‘pro-forms’ of both granule proteins in Xbp1Vav1 cells after 96 h of culture (Fig. 7a–d), which indicated a nearly complete block in terminal post-translational maturation. Furthermore, ultrastructural analyses revealed substantial ER swelling and failure to nucleate granule packing (Fig. 7e), suggestive of ER stress and severe granule dysfunction upon commitment to the eosinophil lineage. Together these data suggested close coupling among ER homeostasis, eosinophil transcriptional identity, granule formation and cell death.
Figure 7.
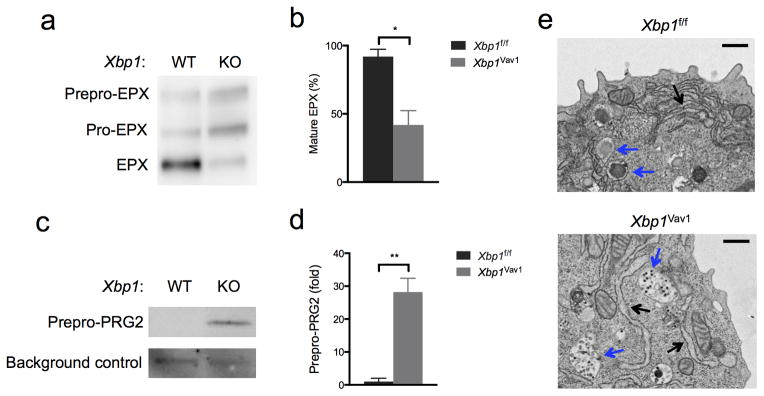
Xbp1 deficiency causes defects in granule-protein maturation and secretory-pathway ultrastructure. (a) Immunoblot analysis of EPX maturation (left margin) in GATA-2-transduced Xbp1f/f (f/f) and Xbp1Vav1 (Vav1) GMPs. (b) Frequency of mature EPX in GATA-2-transduced Xbp1f/f and Xbp1Vav1 GMPs (n = 3 independent cultures per genotype). (c) Immunoblot analysis of the maturation of pre-pro-PRG2 in GATA-2-transduced Xbp1f/f and Xbp1Vav1 GMPs; below (Ctrl), nonspecific background band (loading control). (d) Abundance of immature pro-PRG2 (pre-pro-PRG2) in GATA-2-transduced Xbp1f/f and Xbp1Vav1 GMPs (n = 3 independent cultures per genotype); results were normalized to those of the nonspecific background band and are presented relative those of Xbp1f/f GMPs. (e) Transmission electron microscopy of GATA-2-transduced Xbp1f/f and Xbp1Vav1 GMPs: blue arrows indicate developing secretory granules; black arrows indicate the ER. Scale bars, 500 nm. *P < 0.05 and **P < 0.01 (Student’s t-test). Data are representative of (a,c,e) or from (b,d) one experiment with three independent biological replicates per group (mean and s.e.m. in b,d).
XBP1 links granulogenesis to Gata1 expression
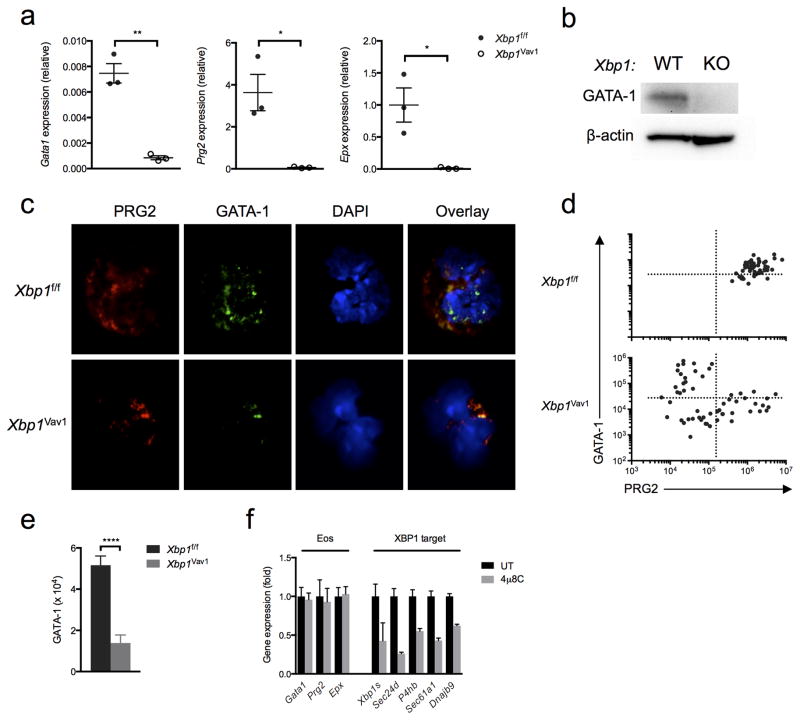
To verify the connection between XBP1 and the expression of granule proteins, we cultured BMDEs from Xbp1f/f and Xbp1Vav1 mice for 8 d, sorted DAPI−Siglec-F+ cells and quantified expression of the gene encoding GATA-1 and genes encoding the key eosinophil effectors PRG2 and EPX. After 8 d of culture, Xbp1f/f BMDEs robustly express genes encoding key transcription factors as well as numerous genes encoding eosinophil markers25. All three genes (Gata1, Prg2 and Epx) were markedly downregulated in Xbp1Vav1 BMDEs relative to their expression in Xbp1f/f BMDEs (Fig. 8a), which suggested that the expression of Gata1 and terminal differentiation markers was ‘titrated’ by the ‘health’ of the secretory pathway. The transcriptional defects were more severe in Siglec-F+ BMDEs than in GATA-2-transduced GMPs (Fig. 8a), possibly because GATA-2 can upregulate Gata1 expression34. Similarly, GATA-1 was essentially undetectable by immunoblot analysis in Xbp1Vav1 BMDE cultures at day 8 (Fig. 8b).
Figure 8.
Xbp1 deficiency prevents the accumulation of GATA-1 in eosinophils through an indirect mechanism. (a) Quantitative PCR analysis of Gata1, Prg2 and Epx in sorted DAPI−Siglec-F+ BMDEs from Xbp1f/f and Xbp1Vav1 mice (n = 3 independent cultures per genotype), assessed at day 8 of culture; results were normalized to those of Actb. (b) Immunoblot analysis of GATA-1 and β-actin (loading control) in Xbp1f/f and Xbp1Vav1 BMDEs at day 8 of culture. (c) Immunofluorescence microscopy of developing eosinophils among Xbp1f/f and Xbp1Vav1 BMDEs at day 8 of culture, stained for PRG2, GATA-1 and DAPI. (d) Scatter plots of PRG2 fluorescence versus GATA-1 fluorescence in Xbp1f/f BMDEs (n = 50) and Xbp1Vav1 BMDEs (n = 56) at day 8 of culture. (e) Staining intensity of GATA-1 in developing PRG2+ eosinophils among Xbp1f/f BMDEs (n = 47) and Xbp1Vav1 BMDEs (n = 30) at day 8 of culture. (f) Quantitative PCR analysis of the canonical eosinophil genes Gata1, Prg2 and Epx, as well as Xbp1s and the XBP1 target genes Sec24d, P4hb, Sec61a1 and Dnajb9, in BMDE cultures treated for 8 h with the vehicle dimethyl sulfoxide (DMSO) or 4μ8C (20 μM) (n = 3 independent cultures per condition); results were normalized to those of Actb and are presented relative to those of vehicle-treated cells. *P < 0.05 and **P < 0.01 and ***P < 0.0001 (Student’s t-test). Data are representative of at least two independent experiments (a,f; mean and s.e.m.), two independent experiments (b) or two experiments with three independent biological replicates per genotype (c) or are from one experiment with three biological replicates pooled per genotype (d,e; mean and s.e.m. in e).
To further confirm that downregulation of Gata1 occurred in eosinophil-committed cells, we performed immunofluorescence analysis of GATA-1 and PRG2 in Xbp1f/f and Xbp1Vav1 BMDE cultures. Xbp1f/f BMDEs co-expressed GATA-1 and PRG2, while the rare Xbp1Vav1 BMDEs, identified by granulocytic nuclear morphology and PRG2+ granules, completely lacked staining of GATA-1 (Fig. 8c). We were able to detect occasional GATA-1+ cells in Xbp1Vav1 BMDE cultures, but these cells never had eosinophil granules (Fig. 8d,e), which suggested that Gata1 expression was mutually exclusive with granule formation under Xbp1-deficient conditions. To determine whether Gata1 and genes encoding eosinophil granule proteins were direct targets of XBP1, we blocked XBP1 signaling for 8 h in developing BMDEs with 4μ8C. Unlike the expression of targets of XBP1 such as Sec24d, Sec61a1, Dnajb9 and P4hb, the expression of Gata1, Prg2 and Epx was unaffected by acute blockade of IRE1α (Fig. 8f), which indicated that these genes were not directly regulated by IRE1α-XBP1 but instead were regulated by other downstream processes. Given our collective results, we propose that Xbp1 deficiency damages the adaptive capacity of the ER, which, after the commitment of progenitors to the eosinophil lineage, results in pronounced defects in the maturation of granule proteins and secretory-pathway function that feed back to downregulate the expression of Gata1 and genes encoding eosinophil granule proteins in a futile attempt to allow survival of irremediable cellular stress.
DISCUSSION
In this study we have identified an unexpectedly selective requirement for the transcription factor XBP1 in eosinophil development. Through the use of multiple genetic and cellular models, we found that XBP1 was potently and selectively activated during eosinophil differentiation and was required in a cell-intrinsic way for the survival of committed EoPs. Loss of Xbp1 resulted in the downregulation of a network of genes encoding products dedicated to preserving ER protein folding and homeostasis of secretory pathways in hematopoietic progenitors. After commitment of progenitors to the eosinophil lineage, those secretory-pathway vulnerabilities were overwhelmed, which resulted in critical defects in secretory granule formation, downregulation of expression of the gene encoding the major lineage determinant GATA-1 and loss of expression of genes encoding key eosinophil granule proteins. Collectively our data revealed that granulocytes could be distinguished by their reliance on maintaining homeostasis of the secretory pathway via the stress-response transcription factor XBP1.
Eosinophil development results from carefully coordinated activity of transcription factors such as GATA-1, PU.1, C/EBPα, C/EBPε and IRF8. Notably, all factors known to be involved in eosinophil development are also required by one or more alternative granulocyte lineages, which attests to the intimate transcriptional and ontological relatedness among neutrophils, basophils and eosinophils. We observed a complete loss of Xbp1Vav1 eosinophils starting at the EoP stage, with no effects on peripheral basophils or neutrophils, which established XBP1 as a factor exclusive for eosinophil development among granulocytes. The residual Xbp1Vav1 EoPs exhibited increased caspase activity relative to such activity in Xbp1f/f EoPs, which indicated that Xbp1Vav1 EoPs were eliminated by cell death. Furthermore, reduced eosinophil counts in Xbp1eoCRE mice demonstrated that XBP1 operated after lineage commitment. As nascent granules are present in the earliest identifiable committed EoPs19, these experiments traced the function of XBP1 to eosinophil granulogenesis.
We detected potent splicing of Xbp1 mRNA in several myeloidprogenitor cell types along the eosinophil developmental continuum, which suggested that ER stress is a characteristic of steady-state hematopoiesis. This finding was consistent with published reports demonstrating that myelopoiesis is marked by increased protein synthesis and oxidative stress35. Unexpectedly, the phenotype of Xbp1Vav1 mice was exceedingly specific for eosinophils, which revealed that most cells did not require XBP1 to survive developmental stressors. XBP1 is required for immunoglobulin production by plasma cells, although Xbp1-deficient B220intCD138+ intermediate forms of plasma cells are found in normal numbers in vivo36. Notably, we were unable to identify similar residual populations of immature eosinophils, which suggested that unlike plasma cells, committed eosinophils cannot exist in a partially mature state.
Eosinophils are singular among granulocytes in that they must safely store highly basic granule proteins (isoelectric point, >10) such as PRG2, EPX and the eosinophil-associated RNases, many of which are cytotoxic and/or exhibit membrane-destabilizing properties37,38. Additionally, eosinophils uniquely store certain granule proteins (PRG2) as crystalline depots39,40 and are histologically identifiable by Congo red staining41, suggestive of tightly regulated, physiological intracellular amyloid formation. Given these nuances of eosinophil granule development and function, it is notable that inefficient granule formation itself, although not excessive production of granule proteins, appears to be a major regulator of the survival of EoPs. Mice with heterozygous or homozygous deletion of various genes encoding products involved in pathways for the processing and disposal of secreted protein (such as Galc, Atp6v0a1, Lyst and Txndc5) are deficient in eosinophils42. Additionally, it has been demonstrated that eosinophil development crucially relies on production of the lineage-specific granule proteins PRG2 and EPX, which cumulatively occupy more than 50% of specific granules by volume43. That finding and the granule-formation defects observed in Xbp1Vav1 cells committed to the eosinophil lineage suggest that eosinophil development may require finely tuned expression of granule proteins, wherein too little or too much corrupts terminal differentiation. However, we found that wild-type BMDEs developed normally upon retroviral overexpression of PRG2 and/or EPX (data not shown), which suggested that eosinophil development is acutely sensitive to the scarcity and processing of granule proteins but can adapt to the excessive production of granule proteins.
Immunofluorescence analysis revealed that Xbp1Vav1 cells were able to produce eosinophil-specific granule proteins to a limited degree. Additionally, both PRG2 and EPX were consistently located within granule-like punctate structures in BMDEs (data not shown), which challenged the possibility of leakage of cytotoxic proteins due to granule destabilization. By driving GMPs into the eosinophil lineage via overexpression of GATA-2, we were able to detect severe blockades in the maturation of both EPX and PRG2, along with severe ultrastructural defects in granule nucleation. The lack of observable eosinophilic Wright-Giemsa staining in Xbp1Vav1 primary BMDEs confirmed the defect in eosinophil granule-protein maturation, as the acidic ‘pre-pro-’ portions block eosinophilic staining. The accumulation of these immature species correlated with transcriptional downregulation of genes encoding eosinophil granule proteins and the induction of ER stress–mediated PERK-ATF4 activity, in support of a model in which crucial components of the eosinophil developmental program are modulated on the basis of ER and granule ‘health’. It is possible that PERK, which is normally inactive during eosinophil differentiation, may mediate some of this feedback. Given the observed severity of the blockade, it is likely that Xbp1Vav1 cells never adequately resolved ER and granule stress, which resulted in sustained activation of PERK, permanent repression of the eosinophil developmental program and eventual cell death. Whether a complementary feedback loop exists to signal poor granule filling, as probably occurs in Prg2−/−Epx−/− mice, remains to be determined.
We showed that both Xbp1Vav1 primary BMDEs and GATA-2-transduced Xbp1Vav1 GMPs substantially downregulated numerous genes encoding factors involved in the terminal differentiation of eosinophils, such as Prg2 and Epx, relative to the expression of these genes in their respective Xbp1f/f counterparts, yet these same genes were unaffected by short-term blockade of IRE1α-XBP1 signaling in BMDE cultures. Although spliced XBP1 protein itself has a relatively short half-life of just 22 min (ref. 1), the half-lives of the proteins encoded by its transcriptional target genes are considerably longer44. It is likely that persistent inhibition of XBP1-dependent signaling is needed to deplete the expression of target genes below the threshold necessary to maintain homeostasis of the ER and granules. Although the mechanism remains unclear, we have shown that this perturbation fed back to downregulate key elements of the eosinophil developmental program, including the gene encoding GATA-1, potentially as an adaptive mechanism to allow time for the disposal of accumulated misfolded proteins.
So far, XBP1 is the only transcription factor to our knowledge that distinguishes the development of eosinophils from that of other granulocytes, which suggests that subtle differences in cellular biological processes provide a previously unappreciated ‘handle’ for fine-tuning the production of different granulocyte subsets. Furthermore, our work has demonstrated that the function of XBP1 in cellular development could not be accurately predicted solely on the basis of the abundance of secreted protein production. Additional studies are needed to explore whether this selectivity derives from stress-mediated cell type–specific relationships between XBP1 and key developmental regulators such as GATA-1 or derives instead from particularly complex requirements for the folding of granule proteins. It is also possible that downregulation of the production of granule proteins is a common feature of granulocyte progenitors undergoing ER stress, but that eosinophils are selectively eliminated because the production of granule proteins itself is as important to survival as that of canonical lineage-determining factors43. Collectively, these results indicate that the IRE1α-XBP1 signaling pathway represents an additional pharmacologically tractable signaling axis for modulating eosinophil development with therapeutic potential for the treatment of a wide variety of eosinophil-mediated disorders.
METHODS
Materials
The IRE1α inhibitor 4μ8C was from Millipore. Antibody to (anti-) GATA-1 (D52H6), anti-β-actin (4967S) and anti-PERK (D11A8) were from Cell Signaling Technologies. J.J. Lee and N.A. Lee provided the rat monoclonal antibody to mouse PRG2 (MT-14.7) and the mouse monoclonal antibody to mouse EPX (MM25-82.2). Unless indicated otherwise, all antibodies for flow cytometry were from BioLegend. All cytokines were from Peprotech. Retroviral plasmids MSCV-GFP, MSCV-XBP1U/S-IRES-GFP, MSCV-XBP1S-IRES-GFP and MSCV-XBP1U-IRES-GFP have been described45. Gata2 and Prg2 were cloned into the plasmid MSCV-IRES-GFP by PCR with high-fidelity DNA polymerase (Kapa Biosystems) to generate MSCV-GATA-2-IRES-GFP and MSCV-PRG2-IRES-GFP. Epx was cloned by PCR into plasmid MSCV-IRES-hCD4 with high-fidelity DNA polymerase (Kapa Biosystems) to generate MSCV-EPX-IRES-hCD4.
Mice
Mice were housed in the specific pathogen–free facility of Weill Cornell Medical College and were handled in accordance with guidelines from the Research Animal Resource Center of the Center of Comparative Medicine and Pathology at Weill Cornell Medical College (protocol 2012-0004). Mice with hematopoietic system–specific or eosinophil-specific deletion of Xbp1 were generated by mating of Xbp1f/f mice with the B6.Cg-Tg(Vav1-cre)A2Kio/J strain (Jackson Laboratory) or the eoCRE strain23, respectively. Ern1f/f mice have been described46 and were backcrossed with a ‘speed congenic strategy’ to a C57BL/6 background of greater than 98%, then were mated with Vav1-Cre mice to generate Ern1Vav1 mice. Congenic CD45.1+ C57BL/6 mice were from Jackson Laboratories. Il5-transgenic BALB/c mice were provided by C. Gerard. Sex- and age-matched littermates were used as controls throughout the study.
Cell culture
Bone marrow eosinophil cultures were prepared as described25, with slight modification. Bone marrow cells were harvested from femurs, tibias and pelvic bones. After lysis of red blood cells, c-Kit+ hematopoietic&epsi progenitor cells were enriched by positive selection with anti-c-Kit MACS beads (Miltenyi Biotec) and were resuspended to a density of 1 × 106 cells per ml in RPMI-1640 medium (VWR) supplemented with 20% FBS (Atlanta Biologicals), 2 mM l-glutamine (Cellgro), 25 mM HEPES, pH 7.2–7.6 (Cellgro), non-essential amino acids (Cellgro), 1 mM sodium pyruvate, 100 U penicillin, 100 μg/ml streptomycin (Corning Cellgro), 50 μM β-mercaptoethanol (Sigma) and the cytokines SCF (100 ng/ml) and Flt3L (100 ng/ml), followed by culture for 4 d. After day 4, the cells were washed and then were cultured in the presence of IL-5 (10 ng/ml) for the duration of the culture, with medium changes on days 8 and 10. For certain experiments, BMDE cultures were supplemented with 10 ng/ml IL-3 and/or 10 ng/ml GM-CSF. Eosinophils from Il5-transgenic mice were isolated by mechanical disruption of the spleen as described47. For overnight incubation with the IRE1α inhibitor 4μ8C, bone marrow cells (after lysis of red blood cells) were cultured at a density of 1 × 106 cells per ml in complete RPMI medium supplemented with 10 ng/ml IL-5 to enhance eosinophil survival in vitro.
Sorted GMPs for retroviral transduction were cultured as described31 with slight modification. Sorted cells were infected with retrovirus containing the plasmid MSCV-GATA-2-IRES-GFP and then were grown for 36 h in Iscove’s modified Dulbecco’s medium (Corning) containing 20% FBS, 100 U penicillin, 100 μg/ ml streptomycin and 50 μM β-mercaptoethanol supplemented with SCF (20 ng/ml), IL-3 (20 ng/ml), IL-5 (50 ng/lL), IL-6 (20 ng/ml), IL-7 (20 ng/ml), IL-11 (10 ng/ml) and GM-CSF (10 ng/ml). GFP+ cells were then re-sorted and then grown in SCF (20 ng/ml), IL-3 (20 ng/ml), IL-5 (50 ng/ml), and GM-CSF (10 ng/ml) for the remainder of the culture period (up to 5 d). Time in culture reported for all GATA-2-transduction experiments was measured starting after the second sort. Where indicated, cell morphology was assessed by cytospin followed by modified staining with Wright-Giemsa (WG16) according to the manufacturer’s protocol (Sigma).
Retroviral infection
Retroviruses were produced by co-transfection of retroviral vectors with gag-pol and VSV-G expression plasmids into 293T cells through the use of Effectene reagent according to the manufacturer’s instructions (Qiagen). Retroviral supernatants were harvested and then were concentrated 100× by ultracentrifugation for 2 h at 112,398g with a Sorvall SW-28 rotor. Concentrated viruses were used for ‘spin-infection’ of target primary cell cultures at 1,000g for 1.5 h at 32 °C in the presence of 8 μg/ml polybrene.
RNA extraction, RT-qPCR and XBP1 splicing assay
Total RNA was prepared with TRIzol according to the manufacturer’s instructions (Invitrogen). cDNA was synthesized with a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative RT-PCR was performed with SYBR Green fluorescent reagent on the Mx3005P QPCR System (Strategene). The amount of mRNA was normalized with Actb as a control by the comparative threshold cycle method. Efficiency of Cre recombinase– mediated deletion of Xbp1 with loxP-flanked exon 2 was assessed by quantitative PCR as described48. Primer sequences used for quantitative PCR in this study are in Supplementary Table 4. The Xbp1 mRNA–splicing assay was performed as described21. Primers surrounding the splice site in the Xbp1 mRNA (5′-ACACGCTTGGGAATGGACAC-3′ and 5′-CCA TGGGAAGATGTTCTGGG-3′) were used for amplification by PCR, after which the products were separated by electrophoresis through a 2.5% agarose gel and visualized by staining with ethidium bromide. The frequency of Xbp1 mRNA splicing was calculated by ImageJ software via densitometry as follows: intensity of Xbp1s band / ([intensity of Xbp1s band] + [intensity of Xbp1us band]) × 100.
Flow cytometry
Flow cytometry of bone marrow cells, splenocytes, blood cells and BMDEs was conducted using the following antibodies (all from BioLegend unless stated otherwise): anti-Gr-1 (RB6-8C5), anti-F4/80 (BM8), anti-Ly6G (1A8), anti-B220 (RA3-6B2), anti-CD11c (N418), anti-CD45.1 (A20), anti-CD45.2 (104), anti-Siglec-F (E50-2440; BD Pharmingen), anti-CCR3 (J073E5), anti-FcεRI (MAR-1), anti-CD4 (RM4-5), anti-CD8α (53-6.7), anti-CD16/32 (93), anti-CD11b (M1/70), antibody to IL-5 receptor-α (T21; BD Pharmingen), anti-CD34 (RAM34; eBioscience), anti-Ter119 (TER-119), anti-Sca-1 (D7), anti-CD3 (145-2C11) and anti-c-Kit (2B8). Events were acquired on an LSR II flow cytometer system (BD Biosciences) and data were analyzed with FlowJo software (TreeStar). Cell sorting was performed on a FACSAria II SORP cell sorter (Becton Dickinson) at Weill Cornell Medical College, with exclusion of DAPI+ cells and doublets. Before sorting, stem and progenitor populations underwent initial enrichment by staining of cells with biotin-conjugated antibodies to lineage markers, followed by incubation with streptavidin MACS beads (Miltenyi Biotec) or by positive selection of c-Kit+ cells with anti-c-Kit MACS beads (Miltenyi Biotec). Lineage markers used for GMP staining were defined as Gr-1, CD11c, CD11b, CD8α, CD4, CD3, B220 and Ter119, while lineage markers used for EoP staining were defined as Gr-1, CD11c, CD8α, CD4, CD3, B220 and Ter119. Caspase activity was measured with a CaspGLOW Fluorescein Active Caspase Staining Kit according to the manufacturer’s instructions (eBioscience). Staining with annexin V and propidium iodide for analysis of cell death was measured with an Annexin V Apoptosis Detection Kit APC according to the manufacturer’s instructions (eBioscience).
Bone marrow chimeras
Recipient CD45.1+ C57BL/6 mice were switched to a Sulfatrim diet 7 d before irradiation. Mice were then exposed to a single lethal full-body irradiation of 1,000 rads. 24 h after irradiation, 5 × 106 total bone marrow cells from a 1:1 mixture of wild-type C57BL/6 (CD45.1+) bone marrow cells and Xbp1f/f or Xbp1Vav1 (CD45.2+) bone marrow cells were injected into the irradiated hosts. The mice were maintained on a Sulfatrim diet for a minimum of 12 weeks, after which the proportion of engraftment from each donor was measured by flow cytometry.
Immunofluorescence
Suspension cells from BMDE cultures were transferred onto glass slides by cytospin and were fixed for 10 min at 25 °C with 4% paraformaldehyde. Cells were repeatedly washed with PBS, then nonspecific binding was blocked and cells were permeabilized by incubation for 30–60 min with 3% goat serum, 1% BSA and 0.1% Triton X-100 in PBS. Cells were then incubated overnight at 4 °C with anti-GATA-1 and anti-PRG2 (identified above) in PBS with 1% BSA and 0.1% Triton-X100. Primary antibodies were visualized by staining with secondary antibodies Alexa Fluor 555–conjugated goat anti-rat (A21434) and Alexa Fluor 488–conjugated goat anti-rabbit (A11029) (both from Molecular Probes), and cells were counterstained with 1 μg/ml DAPI (4,6-diamidino-2-phenylindole) and were mounted with Slowfade Gold (Life Technologies). Digital confocal images for quantification by ImageJ software were captured on an Olympus IX81. High-quality images were captured on an AxioVert LSM710 confocal microscope (Zeiss).
For image quantification, ImageJ software was used for determination of the average pixel intensity of 16-bit greyscale images from each channel individually. The fluorescence intensity of GATA-1 and PRG2 was calculated with ImageJ software as follows: corrected total cell fluorescence = region of interest integrated density – ([area of selected cell region of interest] × [mean fluorescence of background]).
Immunoblots
Cell pellets were lysed in RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP40, 0.5% deoxycholate, 0.1% SDS and 50 mM NaF) supplemented with a protease inhibitor tablet. Homogenates were centrifuged at 12,000g for 10 min at 4 °C, and the supernatants were collected. Protein concentration in lysates was measured with a Pierce BCA Protein Assay Kit (Thermo Scientific) and equivalent amounts of protein were used for immunoblot analysis of GATA-1, PRG2, EPX and β-actin.
RNA-seq
Total RNA from GMPs and GATA-2-transduced GMPs was extracted with TRIzol according to the manufacturer’s instructions. For GMPs, poly(A)+ RNA-seq libraries were prepared with an Illumina TruSeq RNA Sample Preparation Kit V2 according to the manufacturer’s instructions (Illumina). For GATA-2-transduced GMPs, libraries for Illumina sequencing were obtained by the generation of double-stranded cDNA with a SMARTer UltraTMLow Input RNA Kit for Illumina sequencing (634826; Clontech Laboratories), followed by library preparation with a TruSeq sample preparation kit (IP-202-1012) according to the manufacturer’s instructions (Illumina). Sequence data from Illumina’s HiSeq2000 sequencer were ‘de-multiplexed’ to generate FASTQ files for each sample with Illumina’s CASAVA pipeline (version 1.8.2).
The ‘reads’ that passed Illumina’s quality and purity filter were aligned to the mouse genome (Illumina iGenomes mm9 build) with the STAR aligner program (version 2.3.0) with default parameters. The resulting SAM alignment files were then converted to the BAM file format, then were sorted and indexed with SAMtools (version 0.1.14). The HTSeq python package49 was used for summarization of the aligned sequence reads into counts. Differential gene expression was analyzed with the DESeq Bioconductor package50. Transcripts were considered differentially expressed with a difference in expression of 1.5-fold or more and an adjusted P value (false-discovery rate) < 0.1. Pathway analysis of enriched gene ontology and predicted upstream transcriptional regulators was performed with Ingenuity Pathway Analysis software (Qiagen). For heat maps, variance-stabilized data from DESeq RNA-seq analyses were exported, manually curated for specific genes in each category, and the gene-expression z-score was visualized with the Heatmap.2 function within the gplots R library.
All differentially expressed transcripts identified by RNA-seq analyses of GATA-2-transduced GMPs were also compared with the 623 unique upregulated transcripts (P < 0.05; difference in expression of twofold or more) identified in a published study of BMDE maturation that examined IL-5-mediated transcriptional changes after 4 d in culture31 (GEO accession code GSE55386).
Transmission electron microscopy
Cells were fixed with a modified Karmovsky’s fix and a secondary fixation in reduced osmium tetroxide. Following dehydration, samples were embedded in an epon analog resin. Ultrathin sections (65 nm in thickness) were contrasted with lead citrate and were viewed on a JEM 1400 electron microscope (JEOL USA) operated at 120 kV. Digital images were captured on a Veleta 2K x 2K charge-coupled device camera (Olympus-SIS).
Statistical analyses
For comparison of two groups, data were analyzed by Student’s t-test. When more than two groups were compared, data were analyzed by one-way or two-way ANOVA with the Šidák correction for multiple comparisons. All statistical analyses were performed with Graphpad Prism 6, with P values of <0.05 considered statistically significant.
Supplementary Material
Acknowledgments
We thank L. Cohen-Gould for assistance with electron microscopy; J. McCormick for assistance with cell sorting; the Starr Foundation Tri-Institutional Stem Cell Derivation Laboratory and Flow Cytometry and Microscopy Core facility for technical assistance, the Weill Cornell Epigenomics Core facility for library preparation and RNA sequencing; J. Cubillos-Ruiz and C. Tan for technical assistance; A. Espinosa, S. Hai and members of the Glimcher laboratory for suggestions and critical reading of this manuscript; A. Doyle for conversations; J.J. Lee and N.A. Lee (Mayo Clinic) for eoCRE mice, antibody to PRG2 and antibody to EPX; C. Gerard (Children’s Hospital Medical, Boston) for Il5-transgenic BALB/c mice; and T. Iwawaki (Gunma University) for Ern1f/f mice. Supported by the US National Institutes of Health (R01DK082448 to L.H.G., R01HL095699 to L.A.S. and R37AI020241 to P.F.W.).
Footnotes
Accession codes. GEO: Illumina RNA-seq raw sequence reads and read counts, GSE65753. Referenced gene expression analysis accession GEO: GSE55386
AUTHOR CONTRIBUTIONS
S.E.B. and L.H.G. designed and analyzed the experiments; S.E.B. conducted experiments and wrote the manuscript; R.L. performed high-resolution immunofluorescence microscopy imaging. S.A. contributed to the design of certain experiments; A.H.L. provided reagents, L.H.G., L.A.S., and P.F.W. supervised the research and edited the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests. L.H.G. is on the Board of Directors and holds equity in Bristol Myers Squibb Pharmaceutical Company.
References
- 1.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 2.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 3.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shoulders MD, et al. Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 2013;3:1279–1292. doi: 10.1016/j.celrep.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reimold AM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 7.Kaser A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwakoshi NN, Pypaert M, Glimcher LH. The transcription factor XBP-1 is essential for the development and survival of dendritic cells. J Exp Med. 2007;204:2267–2275. doi: 10.1084/jem.20070525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. Neutrophil function: from mechanisms to disease. Annu Rev Immunol. 2012;30:459–489. doi: 10.1146/annurev-immunol-020711-074942. [DOI] [PubMed] [Google Scholar]
- 11.Furuta GT, Atkins FD, Lee NA, Lee JJ. Changing roles of eosinophils in health and disease. Ann Allergy Asthma Immunol. 2014;113:3–8. doi: 10.1016/j.anai.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karasuyama H, Mukai K, Obata K, Tsujimura Y, Wada T. Nonredundant roles of basophils in immunity. Annu Rev Immunol. 2011;29:45–69. doi: 10.1146/annurev-immunol-031210-101257. [DOI] [PubMed] [Google Scholar]
- 13.Wu D, et al. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science. 2011;332:243–247. doi: 10.1126/science.1201475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu Y, et al. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell. 2014;157:1292–1308. doi: 10.1016/j.cell.2014.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heredia JE, et al. Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell. 2013;153:376–388. doi: 10.1016/j.cell.2013.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simson L, et al. Regulation of carcinogenesis by IL-5 and CCL11: a potential role for eosinophils in tumor immune surveillance. J Immunol. 2007;178:4222–4229. doi: 10.4049/jimmunol.178.7.4222. [DOI] [PubMed] [Google Scholar]
- 17.Chu VT, et al. Eosinophils are required for the maintenance of plasma cells in the bone marrow. Nat Immunol. 2011;12:151–159. doi: 10.1038/ni.1981. [DOI] [PubMed] [Google Scholar]
- 18.Bainton DF, Farquhar MG. Segregation and packaging of granule enzymes in eosinophilic leukocytes. J Cell Biol. 1970;45:54–73. doi: 10.1083/jcb.45.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwasaki H, et al. Identification of eosinophil lineage-committed progenitors in the murine bone marrow. J Exp Med. 2005;201:1891–1897. doi: 10.1084/jem.20050548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voehringer D, van Rooijen N, Locksley RM. Eosinophils develop in distinct stages and are recruited to peripheral sites by alternatively activated macrophages. J Leukoc Biol. 2007;81:1434–1444. doi: 10.1189/jlb.1106686. [DOI] [PubMed] [Google Scholar]
- 21.Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma Y, Shimizu Y, Mann MJ, Jin Y, Hendershot LM. Plasma cell differentiation initiates a limited ER stress response by specifically suppressing the PERKdependent branch of the unfolded protein response. Cell Stress Chaperones. 2010;15:281–293. doi: 10.1007/s12192-009-0142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyle AD, et al. Homologous recombination into the eosinophil peroxidase locus generates a strain of mice expressing Cre recombinase exclusively in eosinophils. J Leukoc Biol. 2013;94:17–24. doi: 10.1189/jlb.0213089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Bruin AM, et al. Eosinophil differentiation in the bone marrow is inhibited by T cell-derived IFN-γ. Blood. 2010;116:2559–2569. doi: 10.1182/blood-2009-12-261339. [DOI] [PubMed] [Google Scholar]
- 25.Dyer KD, et al. Functionally competent eosinophils differentiated ex vivo in high purity from normal mouse bone marrow. J Immunol. 2008;181:4004–4009. doi: 10.4049/jimmunol.181.6.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cross BC, et al. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci USA. 2012;109:E869–E878. doi: 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tominaga A, et al. Transgenic mice expressing a B cell growth and differentiation factor gene (interleukin 5) develop eosinophilia and autoantibody production. J Exp Med. 1991;173:429–437. doi: 10.1084/jem.173.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Acosta-Alvear D, et al. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 29.Uhm TG, Kim BS, Chung IY. Eosinophil development, regulation of eosinophilspecific genes, and role of eosinophils in the pathogenesis of asthma. Allergy Asthma Immunol Res. 2012;4:68–79. doi: 10.4168/aair.2012.4.2.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satoh T, et al. Critical role of Trib1 in differentiation of tissue-resident M2-like macrophages. Nature. 2013;495:524–528. doi: 10.1038/nature11930. [DOI] [PubMed] [Google Scholar]
- 31.Iwasaki H, et al. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20:3010–3021. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fulkerson PC, Schollaert KL, Bouffi C, Rothenberg ME. IL-5 triggers a cooperative cytokine network that promotes eosinophil precursor maturation. J Immunol. 2014;193:4043–4052. doi: 10.4049/jimmunol.1400732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15:481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujiwara T, et al. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Todd DJ, et al. XBP1 governs late events in plasma cell differentiation and is not required for antigen-specific memory B cell development. J Exp Med. 2009;206:2151–2159. doi: 10.1084/jem.20090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Acharya KR, Ackerman SJ. Eosinophil granule proteins: form and function. J Biol Chem. 2014;289:17406–17415. doi: 10.1074/jbc.R113.546218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kroegel C, Costabel U, Matthys H. Mechanism of membrane damage mediated by eosinophil major basic protein. Lancet. 1987;1:1380–1381. doi: 10.1016/s0140-6736(87)90686-6. [DOI] [PubMed] [Google Scholar]
- 39.Barker RL, Gleich GJ, Pease LR. Acidic precursor revealed in human eosinophil granule major basic protein cDNA. J Exp Med. 1988;168:1493–1498. doi: 10.1084/jem.168.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soragni A, et al. Toxicity of eosinophil MBP is repressed by intracellular crystallization and promoted by extracellular aggregation. Mol Cell. 2015;57:1011–1021. doi: 10.1016/j.molcel.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grouls V, Helpap B. Selective staining of eosinophils and their immature precursors in tissue sections and autoradiographs with Congo red. Stain Technol. 1981;56:323–325. doi: 10.3109/10520298109067335. [DOI] [PubMed] [Google Scholar]
- 42.Brown SD, Moore MW. The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mammalian Genome. 2012;23:632–640. doi: 10.1007/s00335-012-9427-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doyle AD, et al. Expression of the secondary granule proteins major basic protein 1 (MBP-1) and eosinophil peroxidase (EPX) is required for eosinophilopoiesis in mice. Blood. 2013;122:781–790. doi: 10.1182/blood-2013-01-473405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 46.Wang HB, Ghiran I, Matthaei K, Weller PF. Airway eosinophils: allergic inflammation recruited professional antigen-presenting cells. Journal of immunology. 2007;179(11):7585–7592. doi: 10.4049/jimmunol.179.11.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jurczak MJ, Lee AH, Jornayvaz FR, Lee HY, Birkenfeld AL, Guigni BA, et al. Dissociation of inositol-requiring enzyme (IRE1alpha)-mediated c-Jun N-terminal kinase activation from hepatic insulin resistance in conditional X-box-binding protein-1 (XBP1) knock-out mice. The Journal of biological chemistry. 2012;287(4):2558–2567. doi: 10.1074/jbc.M111.316760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.