Abstract
Microsporidia comprise one of the largest groups of obligate intracellular pathogens and can infect virtually all animals, but host response to these fungal-related microbes has been poorly understood. Several new studies of the host transcriptional response to microsporidia infection have found infection-induced regulation of genes involved in innate immunity, ubiquitylation, metabolism, and hormonal signaling. In addition, microsporidia have recently been shown to exploit host recycling endocytosis for exit from intestinal cells, and to interact with host degradation pathways. Microsporidia infection has also been shown to profoundly affect behavior in insect hosts. Altogether, these and other recent findings are providing much-needed insight into the underlying mechanisms of microsporidia interaction with host animals.
Introduction
Microsporidia comprise a phylum of fungal-related, obligate intracellular parasites. This phylum contains species that parasitize almost all types of animals, including humans, fish, bees, and other insects. Over 1400 species of microsporidia have been described thus far and new species are being discovered each year [1-3]. Some species of microsporidia have a very narrow host range, while others have a relatively broad host range, including vertebrates and invertebrates. Transmissible microsporidia spores are often described as ubiquitous and have been detected in diverse environments ranging from deep sea vents [1] to intercontinental dust [4]. Microsporidia spores invade hosts with a polar tube to inject themselves directly into the host cell, where they undergo their entire replicative life cycle, and then ultimately differentiate back into spores to return to the environment. These microbes are widespread, but poorly understood, despite their importance to human health and agriculture.
The medical relevance of microsporidia was appreciated when they were found to be responsible for lethal diarrhea in AIDS patients, and death in transplant and immunocompromised patients. Microsporidia can infect any organ system, but predominantly infect the intestine in humans. There is a lack of drugs that are both safe and effective for treating microsporidiosis. For example, fumagillin is one of the few compounds that are effective in killing some species of microsporidia but unfortunately it is toxic to humans [5]. Some groups report that the prevalence of microsporidia infections in humans increasing, with many individuals carrying latent and asymptomatic infections [6-8]. For further details on the clinical relevance of microsporidia, we refer readers to a recent review of this topic [9].
Microsporidia also affect agriculturally relevant animals, predominantly through infections of fish and insects. Microsporidia have been responsible for the collapse of fisheries, and they have also been implicated in honey bee colony collapse disorder, a disease that is decimating the honey bees that pollinate many essential crops. Recently, progress has been made in developing vaccines and cell lines for study of fish infections by microsporidia [10-14]. Due to space limitations, we direct the reader to existing reviews of microsporidia infections and treatments in fish [15,16], and in honey bees [17].
Here we focus primarily on progress made in the basic research of microsporidia-host interactions. We review findings from genomic, transcriptional, cell biological, immunological, and behavioral studies published in the last two years that provide new insight into how hosts respond to these ubiquitous intracellular pathogens.
Analysis of microsporidian genomes and host-interacting proteins
There has been a rapid increase in the number of microsporidian genome sequences available, which has helped address questions of phylogeny, evolution and pathogenesis of the microsporidia. Microsporidia were originally classified as protists, but are now generally accepted to be a sister taxa to the fungi based on phylogenomic analysis [18,19]. Because of the challenges in manipulating microsporidia in the lab, it has been difficult to use this newly acquired microsporidian genome information to perform functional analysis. However, new findings have emerged that are providing insight into the microsporidian obligate intracellular lifestyle. A particular focus has been on the proteins secreted by microsporidia into the host cell, since these factors likely hold the key to microsporidia survival within the host cell. Importantly, two recent reports have experimentally verified secretion of some of these proteins from pathogen cells. The first example relates to the finding that many microsporidia genomes encode a secretion signal sequence on the enzyme hexokinase, which catalyzes the first step in glycolysis and the pentose phosphate pathway [19]. This secretion sequence was shown to be functional in a heterologous yeast expression system, where it could direct traffic through the yeast secretory system, supporting a model where microsporidia hexokinase could be directed into the host cell and perhaps promote anabolic metabolism in vivo [19]. Hexokinase secretion was recently verified experimentally using antibodies directed against hexokinase of Antonospora locustae, a species of microsporidia that infects locusts [20]. Interestingly, hexokinase localized to the nucleus in these studies, suggesting that it could alter host gene expression. This study also provided experimental confirmation for other pathogen proteins previously predicted to be secreted into host cells. A second set of microsporidia proteins that recently were experimentally verified as secreted came from genomic and proteomic analysis of Spraguea lophii, which infects Lophius monkfish [21]. The authors identified proteins released into the extracellular media from spores that were germinated in vitro, and found several microsporidia-specific proteins, as well as RICIN-B lectin-like proteins. The RICIN-B lectin-like proteins are also encoded in the genomes of other microsporidian species, and could possibly interact with carbohydrates found on host proteins. It will be exciting to functionally connect some of these secreted proteins with phenotypes long known to be caused by microsporidia infection, such as the dramatic ‘xenoma’ growths induced by many microsporidia infections in fish [22]. The past few years have seen many newly published microsporidia genomes, and these are covered in a recent review to which we direct readers for more information relating to progress in deciphering microsporidian biology using genomics [23].
Using this newly acquired genome information, several studies have focused on proteins that are unique to microsporidia, to learn more about the biology that characterizes these parasites and how they interact with their hosts. In particular, microsporidia-specific proteins such as spore wall proteins and polar tube proteins have received attention [24-29]. Some of these studies suggest a role for these unique proteins in promoting host cell entry. For example, it is thought that spore wall proteins may aid in adherence of spores to the host cell and thereby contribute to spore infectivity. The Zhang group recently reported that blocking either spore wall protein 16 (SWP16) or spore wall protein SWP11 using in vitro antibody treatments caused a 20% decrease in the adherence of Nosema bombycis spores to host cells in each case [24,26]. Further studies of proteins unique to microsporidia may provide insight into what underlies the unique properties of these parasites.
Host transcriptional response to microsporidia infection
Despite microsporidia being ubiquitous and significant parasites, very little was known about how host animals alter their gene expression in response to infection until just recently. This gap in our understanding has now been filled through analysis of the microsporidia-induced host response for several species, including insect hosts that have been studied for decades, as well as the nematode C. elegans, which has only recently been studied as a host for microsporidia infection. See Table 1 for a summary of pathways subjected to transcriptional regulation upon microsporidia infection in the host species that are discussed below.
Table 1.
Differentially regulated host pathways upon microsporidia infection.
| Host / microsporidia | |||
|---|---|---|---|
| Differentially regulated pathways | C. elegans / N. parisii | B. mori / N. bombycis | A. mellifera / N. ceranae |
| Autophagy | X | ||
| Ubiquitin proteasome system | X | X | X |
| Melanization | X | X | |
| Innate Immunity- Toll | X | ||
| Innate Immunity- IMD | |||
| Innate Immunity- JAK/STAT | X | ||
| C type lectins | X | X | |
| Antimicrobial peptide production | X | ||
| Metabolism |
X | X | X |
| References | [37] | [30,31] | [34,39] |
One of the first animals described as a host for microsporidia was the silkworm Bombyx mori, which can be infected by the microsporidia Nosema bombycis. Indeed, Louis Pasteur was one of the first scientists to describe microsporidia infection in silkworm, which causes a disease called pébrine. This disease is characterized by small larval size, delayed development, molting problems, and ‘prickly ash spots’. To understand more about the silk- worm response to microsporidia infection, the Zhou group recently analyzed changes in host transcription. They used a genome-wide (23K) microarray chip for B. mori and examined host transcriptional response to N. bombycis at 2, 4, 6, and 8 days post infection [30]. Then more recently, these transcriptional studies were extended by examining additional early timepoints with a more modern Digital Gene Expression (DGE) analysis method [31]. In both studies, the authors highlight the differential expression of many genes active in the synthesis and metabolism of a key regulator of silkworm development, juvenile hormone. These changes in gene expression are likely responsible for increases in juvenile hormone during infection [30], which in turn is likely responsible for the small body size and delayed development that are symptoms of silkworm pébrine disease [30,31]. Interestingly, juvenile hormone also accumulates upon infection in Nosema ceranae-infected honey bees. However, as opposed to causing stunted growth, in honey bees extra juvenile hormone may cause precocious foraging behaviors that are associated with microsporidia infection [32], although the cause of precocious foraging is still disputed [33]. The link between juvenile hormone regulation and symptoms of pébrine in silkworms is intriguing as a possible connection between changes in host gene expression and complex symptoms of disease. Additionally, the microarray study compared N. bombycis-induced transcriptional changes to changes resulting from infection by 4 non-microsporidian pathogens and found that 34/70 differentially regulated B. mori immune genes were uniquely regulated during infection by N. bombycis. Genes in the Toll and JAK/STAT pathways were found to be upregulated in expression, as well as several classes of anti-microbial peptides [30]. These findings were largely confirmed in the study using DGE [31].
In addition to the innate immune signaling pathways described above, many insects also use a melanization pathway to defend against microbes. The microarray study found genes of the serine protease cascade of the melanization pathway to be down-regulated upon infection with microsporidia. The authors postulate that secretion of serpins by the pathogen could be responsible for this down-regulation of host defense, and go on to show that hemolymph from N. bombycis-infected silkworms has slower rates of in vitro melanization than does uninfected silkworm hemolymph [30]. Interestingly, the serine protease cascade was also found to be downregulated in an RNA-seq study of Nosema ceranae-infected honey bees [34]. The DGE study of silkworms on the other hand found that lysozyme and lectins, key players in the melanization defense pathway were upregulated upon silkworm infection with N. bombycis [31]. However, lysozyme was downregulated in the honey bee system [34]. Taken together, these findings suggest that the melanization pathway may be a battleground for the ongoing arms race between host and pathogen, with each seeking to alter this important defense pathway to its own advantage.
A recently developed model host for studying microsporidia infection is the nematode C. elegans, which provides a tractable host with many genetic and molecular tools available for study. Nematocida parisii is a microsporidian species shown to naturally infect the intestines of C. elegans nematodes from around the world [35,36]. The transcriptional response of C. elegans to N. parisii microsporidia infection was measured using RNA-seq at 5 timepoints during infection and compared to transcriptional responses to other pathogens of C. elegans. Genes upregulated by N. parisii infection were largely distinct from those upregulated by infection with the extracellular pathogens Pseudomonas aeruginosa or Staphylococcus aureus, although there was extensive overlap in the set of genes downregulated by these distinct infections [37]. This finding is similar to the results of the B. mori microarray study described above, which found a high proportion of microsporidia-specific changes in gene induction compared to infection with other pathogens [30]. Interestingly, there was a striking similarity in the C. elegans host genes upregulated during N. parisii infection as compared to genes upregulated by viral infection, indicating a common host response to these very distinct intracellular pathogens. Many of the commonly upregulated genes contain F-box, FTH, and MATH domains that are associated with ubiquitin-mediated degradation [37]. The authors provide several additional lines of evidence to show that ubiquitin-mediated pathways are involved in the host response to microsporidia infection. In particular, they show that two downstream outputs of ubiquitin, the proteasome and autophagy, provide defense against infection. RNAi knock-down of proteasome subunits, as well as autophagy factors LGG-1 (Atg8/LC3 homolog) or ATG-18 led to increased pathogen load. Furthermore, they showed that ubiquitin as well as autophagy markers are targeted to parasite cells, and that the parasite may suppress that targeting [37]. Interestingly, in the silkworm model of microsporidia infection, DGE analysis of differentially expressed genes found that autophagy genes were regulated during infection by N. bombycis, particularly early in infection (6 hpi) [31]. Although autophagy genes were not induced by N. parisii infection, they did appear to play an important role in defense [37]. Thus, autophagy and other ubiquitin-mediated processes may be a common host response to intracellular infection by microsporidia.
A growing theme in host defense in C. elegans, as well as in other hosts, is that immune defense genes are induced when core host processes commonly targeted by pathogens are perturbed [38]. In keeping with this theme, C. elegans appears to induce intracellular defense genes in response to perturbation of proteasome function. In particular, E3 ubiquitin ligase components, which are induced by RNAi knock-down of proteasome subunits, as well as by pharmacological inhibitors of the proteasome, are also induced by microsporidia or viral infections [37]. Thus, microsporidia infection may be detected through the increased demand placed on the proteasome, although there are likely to be other cues as well. A recent study using a proteomics technique also supports the hypothesis that microsporidia counteracts host degradation pathways [39]. Proteomic analysis of infected and uninfected honey bee midguts identified 14 differentially expressed proteins, one of which was a proteasome subunit that was about half as abundant upon Nosema ceranae infection. Perhaps challenging the host proteasome is a common mechanism of pathogenesis employed by different species of microsporidia.
Microsporidia use host intracellular trafficking pathways for exit and remodel host cytoskeleton
A critical stage in the life cycle of any intracellular pathogen is to exit from the host cell and be transmitted to a new host, which requires the pathogen to navigate and interact with host pathways. Very little was known about microsporidia exit from host cells until recently. Previous analysis of microsporidia life cycles had assumed that microsporidia lyse host cells in order to exit. Indeed, several other intracellular pathogens such as Chlamydia have been shown to use such a strategy [40]. However, recent discoveries in C. elegans have found microsporidia can use a very well-orchestrated, multi-step exit strategy that does not lyse cells, but rather enables the host to live for a surprising length of time during prolific pathogen production, although microsporidia infection does eventually kill this host [35,41].
Earlier work in the C. elegans host system showed that host animals were alive while contagious, indicating that the C. elegans-infecting species of microsporidia, Nematocida parisii is able to exit from host intestinal cells and be excreted from the animal without causing death [35]. Additional studies of this host cell exit process indicated it to be non-lytic, because intestinal cells continued to exclude a small dye that enters perforated cells, even at timepoints when animals were actively excreting N. parisii spores [41]. More recently, it was shown by electron microscopy that intracellular spores are contained in a separate membrane compartment that can fuse with the host plasma membrane [42]. Importantly, N. parisii spores exit exclusively from the apical side of polarized intestinal cells, which allows them access to the lumen of the intestinal tract and therefore a route to excretion [41]. The recycling endosome regulator RAB-11 was identified by an RNAi screen to be instrumental in orchestrating the fusion of microsporidia-containing compartments with the host apical membrane. RNAi against RAB-11 decreased spore exit and reduced the transmission of infection from infected animals to their neighbors [42]. The authors propose that the action of this polarized smGTPase, RAB-11, may be responsible for the apical-only direction of parasite exit, an example of elegant repurposing of host trafficking machinery in response to infection.
Another interesting finding from studies of microsporidia in the C. elegans model system, is the degree to which the host cytoskeletal system is remodeled during infection. It was first noted that exit of N. parisii from host cells requires an intestinal-specific isoform of C. elegans actin, ACT-5. This protein is also mislocalized early during infection [41]. Interestingly, differential levels of host actin upon infection were identified in a proteomic study of the mosquito, Aedes aegypti, when it was co-infected with two species of microsporidia [43]. Although there are many possible roles actin could play during intracellular infection by parasites, perhaps use of actin during host cell exit is a common strategy employed by many different species of microsporidia. This finding also suggests that researchers should be cautious about using actin gene levels to normalize expression in transcriptional studies, since actin genes may not be have consistent levels of expression during infection [43].
Natural variation in host resistance and clearance of microsporidia
One recent study of microsporidia infection demonstrates how genetic variation in host responses to parasitic infections can affect host fitness across generations. In a study of the resistance of different strains of the roundworm C. elegans to its naturally occurring microsporidian parasite, N. parisii, the authors found that a strain of C. elegans from Hawaii had about 30-fold increased resistance to infection compared to the laboratory strain from England, as assessed by pathogen load. Furthermore, Hawaiian worms had more progeny than British worms after infection, indicating that the increased resistance could lead to a selective advantage. The enhanced resistance and fecundity of this strain when challenged with N. parisii appears to be due to the surprising ability of this strain to clear intracellular infection, but only during early larval stages of development [44]. Clearance of N. parisii from the intestinal epithelial cells of C. elegans in vivo is a striking finding, given that C. elegans does not have known professional immune cells. It would be interesting to analyze the mechanism of clearance in other hosts, to determine whether this epithelial clearance can also be used in hosts that have a professional immune system.
Changes in host behavior resulting from microsporidia infection
In addition to modulating host activities on a cellular level, microsporidia such as the honey bee-infecting species, Nosema ceranae, can also alter host behavior [45-48]. European honey bee populations have been decimated recently due to a phenomenon called ‘colony collapse disorder’ that may be at least partially due to infection by Nosema ceranae. A recent report suggests that “homing success”, a measure of the bees’ ability to return to the hive after kidnapping and being released from a far-away location, was significantly reduced in N. ceranae-infected bees compared to control animals [49]. This difference was largely due to decreased flight times and increased rest intervals of infected bees, rather than differences in navigation or other flight characteristics. The authors note that although this inability to return home can reduce colony size, it also can mitigate spread of infection throughout the colony, highlighting the complexity of factors at play in host response to microsporidia infections.
In another fascinating example of the complex behavioral changes that can occur upon microsporidia infection, Shi et al present data for a mechanism by which microsporidia infection can prevent locust swarming [50]. In their paper, the authors propose that the locust-infecting microsporidia, Antonospora (Nosema) locustae, acidifies the hindgut of the host locust during infection, which reduces growth of a particular commensal bacterial species that is responsible for producing pheromones that promote swarming behavior. Volatiles from the feces of infected locusts were less attractive to healthy locusts than volatiles from the feces of uninfected animals. In addition to reducing the onset of aggregating behaviors, RNAseq data shows that microsporidia infection suppresses synthesis of dopamine, a neurotransmitter that helps maintain swarming [50]. This study again illustrates how far-reaching the impact of microsporidia infection can be in a host, with changes in the host microbiome due to microsporidia adversely affecting important behaviors like locust swarming.
Conclusions
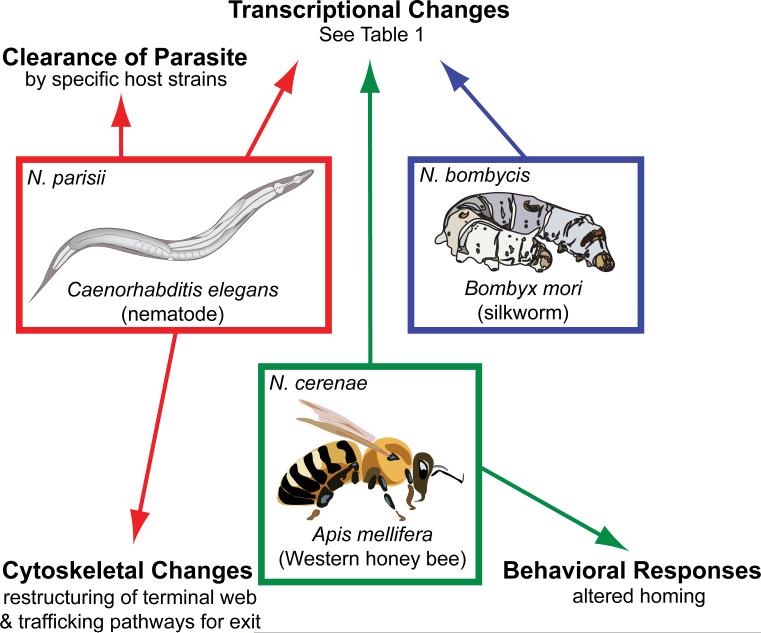
In conclusion, exciting progress has been made recently in investigating host responses to microsporidia infection. Many of the studies described above highlight the struggle between host and parasite for control of host defense pathways including innate and cellular immune pathways, cellular clearance and autophagy, and the proteasome. Interactions between host and pathogen manifest across many levels of host biology, ranging from transcriptional changes in the genome, to cytoskeletal and trafficking modifications within cells, and even to alterations in host behavior (Figure 1). Studying these interactions should help us understand infectious disease caused by microsporidia, and more generally the needs of both hosts and parasites.
Figure 1. Microsporidia-host interactions.
Recent studies conducted in nematodes, silkworms, and honeybees have enhanced our understanding of the basic biology of host-parasite interactions by examining how hosts respond to microsporidia infection. Examples of host responses studied in each host-parasite pair discussed in detail in this review are summarized above.
Highlights.
Hexokinase and lectin-like proteins are candidate effectors secreted by microsporidia
Microsporidia infection induces robust transcriptional changes in many host animals
Host ubiquitin and autophagy machinery target microsporidia cells in nematodes
Microsporidia exploit endocytic recycling of host nematode for directional exocytosis
Microsporidia infection regulates honey bee homing and locust swarming behavior
Acknowledgements
We thank Michael Botts and Lianne Cohen for helpful comments on the manuscript. This work was supported by NIH predoctoral training grant T32 GM008666 and a NSF Predoctoral Fellowship to S.C.S.; and NIAID R01 AI087528, the Searle Scholars Program, Packard Foundation and Burroughs Wellcome Fund fellowships to E.R.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sapir A, Dillman AR, Connon SA, Grupe BM, Ingels J, Mundo-Ocampo M, Levin LA, Baldwin JG, Orphan VJ, Sternberg PW. Microsporidia-nematode associations in methane seeps reveal basal fungal parasitism in the deep sea. Frontiers in microbiology. 2014;5:43. doi: 10.3389/fmicb.2014.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morsy K, Bashtar AR, Abdel-Ghaffar F, Al-Quraishy S. Morphological and phylogenetic description of a new xenoma-inducing microsporidian, Microsporidium aurata nov. sp., parasite of the gilthead seabream Sparus aurata from the Red Sea. Parasitology research. 2013;112:3905–3915. doi: 10.1007/s00436-013-3580-3. [DOI] [PubMed] [Google Scholar]
- 3.Andreadis TG, Takaoka H, Otsuka Y, Vossbrinck CR. Morphological and molecular characterization of a microsporidian parasite, Takaokaspora nipponicus n. gen., n. sp. from the invasive rock pool mosquito, Ochlerotatus japonicus japonicus. J Invertebr Pathol. 2013;114:161–172. doi: 10.1016/j.jip.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Favet J, Lapanje A, Giongo A, Kennedy S, Aung YY, Cattaneo A, Davis-Richardson AG, Brown CT, Kort R, Brumsack HJ, et al. Microbial hitchhikers on intercontinental dust: catching a lift in Chad. The ISME journal. 2013;7:850–867. doi: 10.1038/ismej.2012.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desoubeaux G, Maakaroun-Vermesse Z, Lier C, Bailly E, Morio F, Labarthe F, Bernard L, Chandenier J. Successful treatment with fumagillin of the first pediatric case of digestive microsporidiosis in a liver-kidney transplant. Transplant infectious disease : an official journal of the Transplantation Society. 2013;15:E250–259. doi: 10.1111/tid.12158. [DOI] [PubMed] [Google Scholar]
- 6.Kotkova M, Sak B, Kvetonova D, Kvac M. Latent microsporidiosis caused by Encephalitozoon cuniculi in immunocompetent hosts: a murine model demonstrating the ineffectiveness of the immune system and treatment with albendazole. PLoS One. 2013;8:e60941. doi: 10.1371/journal.pone.0060941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sak B, Brady D, Pelikanova M, Kvetonova D, Rost M, Kostka M, Tolarova V, Huzova Z, Kvac M. Unapparent microsporidial infection among immunocompetent humans in the Czech Republic. J Clin Microbiol. 2011;49:1064–1070. doi: 10.1128/JCM.01147-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sak B, Kvac M, Kucerova Z, Kvetonova D, Sakova K. Latent microsporidial infection in immunocompetent individuals - a longitudinal study. PLoS neglected tropical diseases. 2011;5:e1162. doi: 10.1371/journal.pntd.0001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashfaq A, White AC., Jr. Microsporidiasis. Handbook of clinical neurology. 2013;114:183–191. doi: 10.1016/B978-0-444-53490-3.00012-1. [DOI] [PubMed] [Google Scholar]
- 10.Mc CS, Sheppard J, Wright GM, Speare DJ. Development of the microsporidian parasite, Loma salmonae, in a rainbow trout gill epithelial cell line (RTG-1): evidence of xenoma development in vitro. Parasitology. 2014:1–6. doi: 10.1017/S0031182014001620. [DOI] [PubMed] [Google Scholar]
- 11.Saleh M, Kumar G, Abdel-Baki AA, El-Matbouli M, Al-Quraishy S. In vitro growth of the microsporidian Heterosporis saurida in the eel kidney EK-1 cell line. Dis Aquat Organ. 2014;108:37–44. doi: 10.3354/dao02690. [DOI] [PubMed] [Google Scholar]
- 12.Kumar G, Saleh M, Abdel-Baki AA, Al-Quraishy S, El-Matbouli M. In vitro cultivation model for Heterosporis saurida (Microsporidia) isolated from lizardfish, Saurida undosquamis (Richardson). Journal of fish diseases. 2014;37:443–449. doi: 10.1111/jfd.12123. [DOI] [PubMed] [Google Scholar]
- 13.Saleh M, Kumar G, Abdel-Baki AA, Dkhil M, El-Matbouli M, Al-Quraishy S. Development of a novel in vitro method for drug development for fish; application to test efficacy of antimicrosporidian compounds. The Veterinary record. 2014;175:561. doi: 10.1136/vr.102604. [DOI] [PubMed] [Google Scholar]
- 14•.Harkness JE, Guselle NJ, Speare DJ. Demonstrated efficacy of a pilot heterologous whole-spore vaccine against Microsporidial gill disease in rainbow trout. Clinical and vaccine immunology : CVI. 2013;20:1483–1484. doi: 10.1128/CVI.00340-13. [This study suggests that vaccines against some microsporidia dieseases may be feasible in the fishery industry.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stentiford GD, Feist SW, Stone DM, Bateman KS, Dunn AM. Microsporidia: diverse, dynamic, and emergent pathogens in aquatic systems. Trends in parasitology. 2013;29:567–578. doi: 10.1016/j.pt.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Sanders JL, Watral V, Kent ML. Microsporidiosis in zebrafish research facilities. ILAR journal / National Research Council, Institute of Laboratory Animal Resources. 2012;53:106–113. doi: 10.1093/ilar.53.2.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Heever JP, Thompson TS, Curtis JM, Ibrahim A, Pernal SF. Fumagillin: an overview of recent scientific advances and their significance for apiculture. Journal of agricultural and food chemistry. 2014;62:2728–2737. doi: 10.1021/jf4055374. [DOI] [PubMed] [Google Scholar]
- 18.Capella-Gutierrez S, Marcet-Houben M, Gabaldon T. Phylogenomics supports microsporidia as the earliest diverging clade of sequenced fungi. BMC biology. 2012;10:47. doi: 10.1186/1741-7007-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuomo CA, Desjardins CA, Bakowski MA, Goldberg J, Ma AT, Becnel JJ, Didier ES, Fan L, Heiman DI, Levin JZ, et al. Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth. Genome research. 2012;22:2478–2488. doi: 10.1101/gr.142802.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Senderskiy IV, Timofeev SA, Seliverstova EV, Pavlova OA, Dolgikh VV. Secretion of Antonospora (Paranosema) locustae proteins into infected cells suggests an active role of microsporidia in the control of host programs and metabolic processes. PLoS One. 2014;9:e93585. doi: 10.1371/journal.pone.0093585. [The authors use antibodies directed against microsporidia proteins to experimentally demonstrate that these proteins are secreted into host cytoplasm.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell SE, Williams TA, Yousuf A, Soanes DM, Paszkiewicz KH, Williams BA. The genome of Spraguea lophii and the basis of host-microsporidian interactions. PLoS Genet. 2013;9:e1003676. doi: 10.1371/journal.pgen.1003676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lom J, Dykova I. Microsporidian xenomas in fish seen in wider perspective. Folia parasitologica. 2005;52:69–81. [PubMed] [Google Scholar]
- 23.Corradi N, Selman M. Latest progress in microsporidian genome research. J Eukaryot Microbiol. 2013;60:309–312. doi: 10.1111/jeu.12030. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Dang X, Ma Q, Liu F, Pan G, Li T, Zhou Z. Characterization of a novel spore wall protein NbSWP16 with proline-rich tandem repeats from Nosema bombycis (microsporidia). Parasitology. 2014:1–9. doi: 10.1017/S0031182014001565. [DOI] [PubMed] [Google Scholar]
- 25.Meng X, Zheng J, Gao Y, Zhang Y, Jia H. Evaluation of spore wall protein 1 as an alternative antigen for the diagnosis of Encephalitozoon cuniculi infection of farmed foxes using an enzyme-linked immunosorbent assay. Vet Parasitol. 2014;203:331–334. doi: 10.1016/j.vetpar.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 26•.Yang D, Dang X, Peng P, Long M, Ma C, Qin JJ, Wu H, Liu T, Zhou X, Pan G, et al. NbHSWP11, a microsporidia Nosema bombycis protein, localizing in the spore wall and membranes, reduces spore adherence to host cell BME. The Journal of parasitology. 2014;100:623–632. doi: 10.1645/13-286.1. [The authors show how spore wall proteins may contribute to infectivity.] [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Geng L, Long M, Li T, Li Z, Yang D, Ma C, Wu H, Ma Z, Li C, et al. Identification of a novel chitin-binding spore wall protein (NbSWP12) with a BAR-2 domain from Nosema bombycis (microsporidia). Parasitology. 2013;140:1394–1402. doi: 10.1017/S0031182013000875. [DOI] [PubMed] [Google Scholar]
- 28.Zhu F, Shen Z, Hou J, Zhang J, Geng T, Tang X, Xu L, Guo X. Identification of a protein interacting with the spore wall protein SWP26 of Nosema bombycis in a cultured BmN cell line of silkworm. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2013;17:38–45. doi: 10.1016/j.meegid.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 29.Dang X, Pan G, Li T, Lin L, Ma Q, Geng L, He Y, Zhou Z. Characterization of a subtilisin-like protease with apical localization from microsporidian Nosema bombycis. J Invertebr Pathol. 2013;112:166–174. doi: 10.1016/j.jip.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 30••.Ma Z, Li C, Pan G, Li Z, Han B, Xu J, Lan X, Chen J, Yang D, Chen Q, et al. Genome-wide transcriptional response of silkworm (Bombyx mori) to infection by the microsporidian Nosema bombycis. PLoS One. 2013;8:e84137. doi: 10.1371/journal.pone.0084137. [This transcriptional study of the silkworm response to microsporidia infection describes regulation of genes involved in immunity, hormonal signaling and metabolism.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yue YJ, Tang XD, Xu L, Yan W, Li QL, Xiao SY, Fu XL, Wang W, Li N, Shen ZY. Early responses of silkworm midgut to microsporidium infection - A Digital Gene Expression analysis. J Invertebr Pathol. 2015;124:6–14. doi: 10.1016/j.jip.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Goblirsch M, Huang ZY, Spivak M. Physiological and behavioral changes in honey bees (Apis mellifera) induced by Nosema ceranae infection. PLoS One. 2013;8:e58165. doi: 10.1371/journal.pone.0058165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonnell CM, Alaux C, Parrinello H, Desvignes JP, Crauser D, Durbesson E, Beslay D, Le Conte Y. Ecto- and endoparasite induce similar chemical and brain neurogenomic responses in the honey bee (Apis mellifera). BMC ecology. 2013;13:25. doi: 10.1186/1472-6785-13-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aufauvre J, Misme-Aucouturier B, Vigues B, Texier C, Delbac F, Blot N. Transcriptome analyses of the honeybee response to Nosema ceranae and insecticides. PLoS One. 2014;9:e91686. doi: 10.1371/journal.pone.0091686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Troemel ER, Felix MA, Whiteman NK, Barriere A, Ausubel FM. Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans. PLoS Biol. 2008;6:2736–2752. doi: 10.1371/journal.pbio.0060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felix MA, Barkoulas M. Robustness and flexibility in nematode vulva development. Trends in genetics : TIG. 2012;28:185–195. doi: 10.1016/j.tig.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 37••.Bakowski MA, Desjardins CA, Smelkinson MG, Dunbar TA, Lopez-Moyado IF, Rifkin SA, Cuomo CA, Troemel ER. Ubiquitin-mediated response to microsporidia and virus infection in C. elegans. PLoS Pathog. 2014;10:e1004200. doi: 10.1371/journal.ppat.1004200. [The authors use RNA-seq, RNAi knock-down and cell biological analysis to demonstrate a role for ubiquitin and autophagy in the C. elegans response to microsporidia.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen LB, Troemel ER. Microbial pathogenesis and host defense in the nematode C. elegans. Curr Opin Microbiol. 2014;23C:94–101. doi: 10.1016/j.mib.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vidau C, Panek J, Texier C, Biron DG, Belzunces LP, Le Gall M, Broussard C, Delbac F, El Alaoui H. Differential proteomic analysis of midguts from Nosema ceranae-infected honeybees reveals manipulation of key host functions. J Invertebr Pathol. 2014;121:89–96. doi: 10.1016/j.jip.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Friedrich N, Hagedorn M, Soldati-Favre D, Soldati T. Prison break: pathogens’ strategies to egress from host cells. Microbiology and molecular biology reviews : MMBR. 2012;76:707–720. doi: 10.1128/MMBR.00024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Estes KA, Szumowski SC, Troemel ER. Non-lytic, actin-based exit of intracellular parasites from C. elegans intestinal cells. PLoS Pathog. 2011;7:e1002227. doi: 10.1371/journal.ppat.1002227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42••.Szumowski SC, Botts MR, Popovich JJ, Smelkinson MG, Troemel ER. The small GTPase RAB-11 directs polarized exocytosis of the intracellular pathogen N. parisii for fecal-oral transmission from C. elegans. Proc Natl Acad Sci U S A. 2014;111:8215–8220. doi: 10.1073/pnas.1400696111. [This study used an RNAi screen to identify a key role for the small GTPase RAB-11 in microsporidia exocytosis from host intestinal cells.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duncan AB, Agnew P, Noel V, Demettre E, Seveno M, Brizard JP, Michalakis Y. Proteome of Aedes aegypti in response to infection and coinfection with microsporidian parasites. Ecology and evolution. 2012;2:681–694. doi: 10.1002/ece3.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44•.Balla KM, Andersen EC, Kruglyak K, Troemel ER. A wild C. elegans strain has enhanced epithelial immunity to a natural microsporidian parasite. PLoS Pathog. 2015 doi: 10.1371/journal.ppat.1004583. [The authors identify a wild strain of C. elegans that can clear microsporidia infection from intestinal epithelial cells, but only during early life.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cepero A, Ravoet J, Gomez-Moracho T, Bernal JL, Del Nozal MJ, Bartolome C, Maside X, Meana A, Gonzalez-Porto AV, de Graaf DC, et al. Holistic screening of collapsing honey bee colonies in Spain: a case study. BMC research notes. 2014;7:649. doi: 10.1186/1756-0500-7-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Botias C, Martin-Hernandez R, Barrios L, Meana A, Higes M. Nosema spp. infection and its negative effects on honey bees (Apis mellifera iberiensis) at the colony level. Veterinary research. 2013;44:25. doi: 10.1186/1297-9716-44-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Natsopoulou ME, McMahon DP, Doublet V, Bryden J, Paxton RJ. Interspecific competition in honeybee intracellular gut parasites is asymmetric and favours the spread of an emerging infectious disease. Proceedings. Biological sciences / The Royal Society. 2015;282:20141896. doi: 10.1098/rspb.2014.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naug D, Gibbs A. Behavioral changes mediated by hunger in honeybees infected with Nosema ceranae. Apidologie. 2009;40:595–599. [Google Scholar]
- 49.Wolf S, McMahon DP, Lim KS, Pull CD, Clark SJ, Paxton RJ, Osborne JL. So near and yet so far: harmonic radar reveals reduced homing ability of Nosema infected honeybees. PLoS One. 2014;9:e103989. doi: 10.1371/journal.pone.0103989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Shi W, Guo Y, Xu C, Tan S, Miao J, Feng Y, Zhao H, St Leger RJ, Fang W. Unveiling the mechanism by which microsporidian parasites prevent locust swarm behavior. Proc Natl Acad Sci U S A. 2014;111:1343–1348. doi: 10.1073/pnas.1314009111. [This study shows how microsporidia infection reduces hindgut bacteria that produce locust-swarming pheromones, and also performs RNA-seq analysis.] [DOI] [PMC free article] [PubMed] [Google Scholar]