Abstract
The oral mycobiota is an important component of the oral microbiota that has only recently received increased attention. The diversity and complexity of the oral mycobiota in healthy humans is greater than any other body site. Dysbiotic imbalance of indigenous fungal communities in immunosuppressed hosts has been proposed to lead to oropharyngeal fungal infections. As in other body sites, to survive and thrive in the oral cavity fungi have to maintain mutually beneficial relationships with the resident bacterial microbiota and the host. Here we review our current understanding of the composition of the oral mycobiota and how it may be influenced by oral commensal bacteria and the host environment.
Keywords: oral mycobiota, bacterial interactions, host immunity
Graphical Abstract
Introduction
The oral microbiota is a complex ecosystem primarily represented by the bacterial and fungal Kingdoms. Although not yet fully established, it has long been assumed that fungi are a minor component of the oral microbiota, compared to prokaryotes. Shotgun sequencing studies are needed to confirm this assumption. With the advent of new high-throughput sequencing methodologies, global analyses of the oral bacterial Kingdom received much more attention over the past few years compared to fungi. This is because sophisticated 16S rRNA gene sequencing pipelines and a comprehensive, curated sequence database that facilitates accurate oral bacterial taxonomic assignment have been available to investigators [1]. As a result, the complexity and biodiversity of the bacterial component of the oral microbiota in health, and community shifts in common oral diseases such as periodontitis and caries have been well characterized [2–6]. However, despite the possibility that bacterial community shifts in these diseases may be influenced by fungal shifts, only one study included simultaneous analysis of fungi [6].
The mycobiota is a medically important component of the oral microbiota since opportunistic fungal infections commonly afflict the oral mucosa of immunocompromised hosts. Most infections are triggered by the genus Candida and are assumed to result from an overgrowth of indigenous species in a permissive host environment [7]. However, because only recently a universal DNA barcode was described for fungal identification [8], no studies have explored the role of global fungal population shifts during oral fungal infection and the ecological determinants of these shifts. As fungal genomic technologies are developing, explorations of the human mycobiome in different body sites have started to shed some light on the complexity and heterogeneity of fungal communities at these sites [9–12]. Recently, two studies describing the oral mycobiota have also emerged [13,14]. These studies are important because they gave new insights on the complexity of the core oral mycobiota in health. However, they did not contribute significantly to a deeper understanding of the relationships between the mycobiota and the resident bacteria or the host in the healthy state.
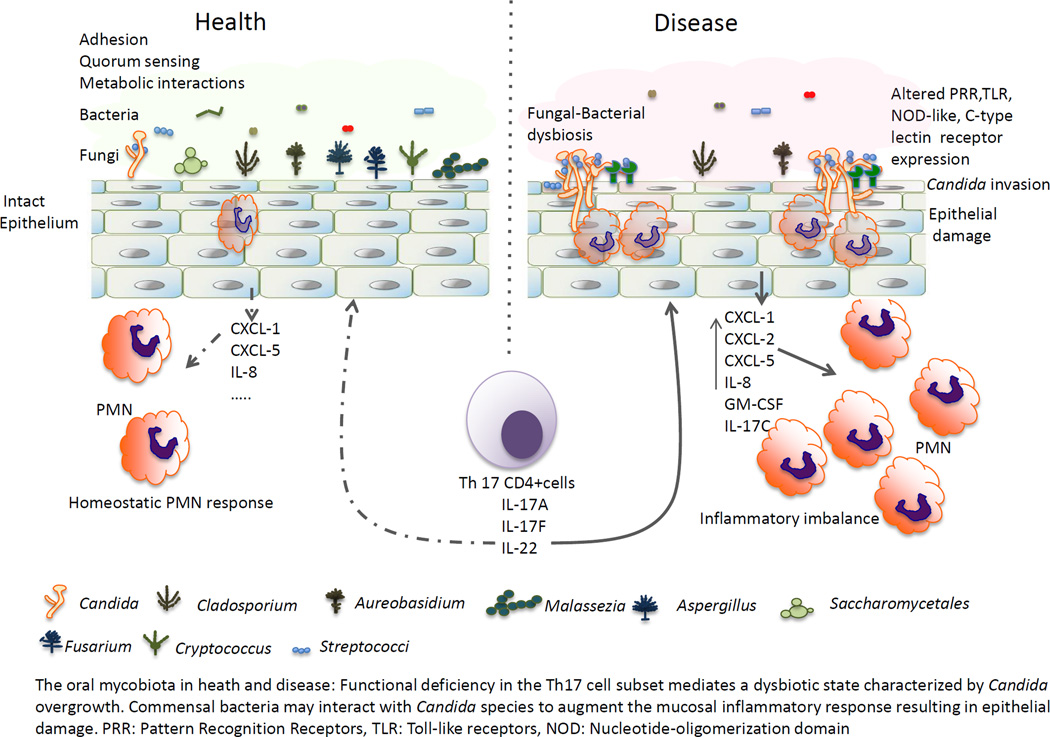
As in other mucosal sites, to survive and thrive in the oral cavity fungi have to develop mutualistic relationships both with the indigenous bacterial microbiota and the host. Mutualistic interactions of fungi with commensal bacteria involve physical binding, communication via signaling molecules, and metabolic exchange during co-adaptation in the multiple oral micro-environmental niches [15]. In addition, alterations in the host environment are essential in shaping the fungal microbiota composition and in the development of fungal diseases [15–17]. Thus, fungi, bacteria and host form complex and dynamic ecological relationships in the oral cavity. In this review, we summarized the current state in our understanding of the influence of commensal bacteria and host environment on colonization patterns and virulence of oral opportunistic fungi.
Core oral fungal microbiota in health
Assembling accurate information on the diversity and composition of the healthy state or core mycobiota is important for subsequent studies of fungal community shifts in oral diseases. The first insight into the diversity and composition of the oral mycobiome in health came from Ghannoum and colleagues [14]. This group utilized a novel multitag pyrosequencing approach to investigate the fungal taxa in the oral cavity of 20 healthy individuals. The diversity of the oral mycobiota was exemplified by the discovery of 85 fungal genera, including 74 culturable and 11 non-culturable [14]. Compared to the fungal diversity in skin and other mucosal sites this represents significantly greater diversity [17]. Increased diversity may be due to the constant exposure to environmental fungi via food intake and mouth breathing, and the diverse micro-environments present in the oral cavity which allow different taxa with unique nutritional requirements to thrive. This study used oral rinse samples, thus the diversity reflects the diversity in oral ecological niches such as the tongue, buccal mucosa and supragingival plaque. However, it is possible that the oral diversity was still underestimated since fungi forming tenacious biofilms with bacteria in anaerobic environments within gingival sulci or in periodontal pockets [18] are not well represented in rinse samples. As expected, Candida species were most frequently identified (sequences detected in 75% of participants) and included one or more of the following species: C. albicans, C. parapsilosis, C. tropicalis, C. khmerensis and C. metapsilosis. Other oral fungal genera included Cladosporium, Aureobasidium, Saccharomycetales, Aspergillus, Fusarium, and Cryptococcus. Thirteen taxa were overall identified as core components of the “basal” oral mycobiome found in >20% of individuals sampled [14]. Interestingly, the true pathogenic fungi Aspergillus, Fusarium, and Cryptococcus, were identified as healthy oral colonizers for the first time. It is reasonable to hypothesize that these fungi may be under continuous surveillance and control by healthy immune systems. In this study low abundance (<1%) fungal sequences were not analyzed, potentially omitting low abundance organisms that may overgrow in disease states. Finally, the type of taxonomic analysis used may have led to the identification of a large percentage of sequences (36.1%) as non-culturable fungi [14].
More recently, the core oral mycobiota was revisited in a pyrosequencing analysis of internal transcribed spacer 1 amplicons in saliva samples of 6 healthy subjects [8]. In order to capture low abundance genera that might be present in a relatively large percentage of healthy individuals, this study did not apply a 1% abundance threshold for further taxonomic analysis. Also, although the sampling method was similar to the earlier study of the oral mycobiome [14], taxonomic assignments were generated using a different database that excludes “uncultured” reference sequences. Despite the differences in methodology between the two studies there was substantial overlap in defining the core oral mycobiota. Seven consensus members of the core oral mycobiome, Candida/Pichia, Cladosporium/Davidiella, Alternaria/Lewia, Aspergillus/Emericella/Eurotium, Fusarium/Gibberella, Cryptococcus/Filobasidiella, and Aureobasidium were identified [13,14]. Five genera that were identified in all healthy individuals only in the second study [13], were Malassezia, Irpex, Cytospora/Valsa, Lenzites/Trametes, and Sporobolomyces/Sporidiobolus. Although Malassezia is a bona fide human skin commensal and pathogen [19], it was also identified in the sputum of all cystic fibrosis patients sampled by Hogan and co-workers [12], suggesting that the oral cavity may be a portal of entry for this organism into the respiratory tract under compromising host conditions. The other four genera are common soil and/or plant pathogens, raising the possibility that they are transient and not stable colonizers of the oral cavity. Longitudinal sampling of the same individuals is required to resolve this issue.
Several challenges remain in interpreting and integrating data on the composition of the core oral mycobiota using fungal genomic or metagenomic approaches from different groups. First, there is lack of uniformity in the utilization of curated databases among studies. Since fungal taxonomic nomenclature differs in each database, and in fact is continuously evolving as more sequences become available, interpretation of sequence data from different groups becomes a formidable task [17,20]. Although further curation is needed, a recently updated ITS1 sequence database for use in the Visualization and Analysis of Microbial Population Structures website, may provide a useful tool for more standardized metagenomic analyses of human samples by different groups [12]. Second, a consensus abundance level in each sample that “qualifies” sequences for further taxonomic analysis is lacking. This may affect our ability to discriminate between transient environmental passers-by and low abundance but stable colonizers. Thirdly, because of large subject to subject variability in sequencing data [13,14], the number of healthy individuals sampled may have a great impact on which fungal genera are identified as members of the core fungal microbiota. The identification of a large proportion of sequences as unculturable fungi [14] and challenge in utilizing culture methods to verify sequence data, sometimes even for culturable species [21], raises the question whether these data should be verifiable with culture approaches before new organisms are “fully vetted” as bona fide members of the core mycobiota.
Influence of host environment on the oral mycobiota
Several core components of the oral mycobiota are stable intra-individually over time but variable between healthy individuals [22]. However, virtually nothing is known about host factors affecting the composition of the oral mycobiota in health that could explain the inter-individual variability. Ghannoum and co-workers suggested that such differences may be associated with gender or ethnicity, but evidence is weak due to limited sample size and lack of consistency across all gender groups and ethnicities [14].
Systemic host health alteration is associated with most oral fungal diseases, regardless of whether they result from overgrowth of indigenous species as in the case of candidiasis, or from exposure to environmental pathogens, as in the case of oral histoplasmosis. This underpins the universal importance of a permissive host environment in influencing colonization or overgrowth of fungal organisms in the oral cavity. Although alterations of host immunity have been hypothesized to directly impact the core oral mycobiota composition leading to a disease-promoting dysbiotic state, there are limited studies to date that have examined global oral fungal community shifts in immunosuppressed humans. A global analysis using 454 pyrosequencing showed a significant shift in the oral mycobiome, with Epicoccum and Alternaria abundantly colonizing HIV-infected patients but not healthy individuals, whereas Candida was abundant in both groups [16]. In pharmacologically immunosuppressed solid organ transplant recipients both culture and pyrosequencing studies showed the oral mycobiota to be dominated by Candida species [23–25]. Candida species load and diversity were positively correlated with the dose of mycophenolate mofetil in a renal transplant population, suggesting a causal link [24].
Several types of genetic disorders are also associated with Candida overgrowth and increased risk of infection, such as chronic mucocutaneous candidiasis and the autoimmune polyendocrine syndrome type I [17]. However, global changes in other members of the oral mycobiota have not been examined in these conditions. A common underlying link between all known host systemic conditions associated with oral Candida overgrowth is functional immunodeficiency in the Th17 CD4+ cell subset, confirming their central role in mucosal protection.
The innate oral epithelial fungal recognition systems and subsequent responses that drive a protective Th17 immunity in the oral mucosa are presently unclear. Toll-like receptors, NOD-like receptors, and/or C-type lectin receptors may survey fungi that come in contact with the superficial oral epithelial cell layers and trigger appropriate responses controlling the growth of certain fungal species, while sparing others, to maintain a homeostatic balance. Along these lines an analysis of the intestinal mycobiota in Dectin-1 knout-out mice with experimentally-induced colitis showed that the proportion of opportunistic pathogenic fungi including Candida and Trichosporon increases, whereas nonpathogenic Saccharomyces decrease [26], which suggests that dectin-1 has role in maintaining a homeostatic fungal community balance in the gut. Whether the oral mycobiota is similarly affected by a functional deficiency in recognition receptors is unknown, although C-type lectin receptors such as CLEC6A have been proposed as suitable candidates [17]. On the other hand, a fungal-bacterial dysbiotic state associated with overexpression of certain oral epithelial pattern recognition receptors, such as TLR2, may lead to an aggravated inflammatory response to fungal opportunistic pathogens, that promotes neutrophil-mediated oral pathology [27].
Influence of oral bacteria on the mycobiota
There is a growing understanding that bacterial and fungal communities are integrally associated in the oral cavity, since they occupy the same micro-environmental niches. However, only two studies to date have examined both the bacterial and fungal component of the microbiota in the same oral samples using next generation sequencing [16, 23]. The two studies tested the hypothesis that in immunosuppressed states oral fungal community shifts are accompanied by shifts in bacterial communities. However, a study of HIV+ individuals reported that fungal community shifts occurred in the absence of significant bacterial shifts [16]. Despite this general observation, some data in this study suggested that positive associations between certain fungal and bacterial taxa identified in health, diminished and even became negative in the HIV+ host background [16]. A study conducted in lung transplant recipients showed distinct shifts in both the bacterial and fungal oropharyngeal communities associated with immunosuppression [23]. However this study did not perform statistical analyses to evaluate the strength of associations between specific bacterial and fungal genera. Interestingly, the vast majority of lung transplant recipients were co-colonized with an increased abundance of Streptococci and Candida species [23].
Because Candida species are the most amenable to isolation, identification and culture, culture studies have concentrated on the effects of bacteria on this genus. Changes in bacterial diversity or abundance by antibiotic treatments may increase oral Candida growth in humans, albeit with lower frequencies or intensities compared to other mucosal sites [28]. In immunocompromised patients on long-term prophylactic antibiotics erythematous candidiasis is common [29], but the combined effects of antibiotics and immunosuppression may be required for fungal infection. There are no high-throughput sequencing studies on the effect of antibiotics on the oral mycobiota in humans or animals.
Interestingly, a triple antibiotic combination treatment that significantly diminished gut bacterial diversity in mice, resulted in sustained, high-level GI colonization with C. albicans, a species not indigenous to mice. Even more interesting was the fact that high-level C. albicans colonization in antibiotics-treated mice was associated with high probability of increased relative abundance of Streptococcus [21]. These results are in agreement with the reduced bacterial diversity and high levels of oropharyngeal colonization with Streptococci and Candida in lung transplant recipients receiving antibiotic prophylaxis [23]. Taken together, these reports suggest that Streptococci and C. albicans may interact through several molecular mechanisms to promote synergistic upper and/or lower GI tract colonization in mammalian hosts.
Because C. albicans and viridans Streptococci are dominant oral commensals in humans, the molecular mechanisms of their interaction have been extensively studied and include physical adhesion, signaling molecules and metabiotic molecules that may influence fungal growth, gene expression and pathogenicity [reviewed in 15]. Early studies showed viridans species to form biofilm communities with C. albicans in vitro in which the hyphal biomass was enhanced [30,31]. Co-aggregation interactions between C. albicans and oral Streptococci are mediated by adhesins that are multifunctional proteins [32–34]. Co-aggregation also requires fungal O-Mannosylation and benefits from the synthesis of bacterial and fungal exopolymers such as soluble α- and β-glucans [35–38]. Oral Streptococci can have a positive effect on hyphal growth via quorum sensing molecules, such as autoinducer-2, and small metabolic molecules, such as hydrogen peroxide [8,39]. Gene expression analysis of C. albicans forming polymicrobial biofilms with oral bacteria in vitro showed that known virulence genes encoding certain secreted aspartyl-proteinases (SAP4/SAP6) were up-regulated [40]. Altering virulence gene expression patterns as a result of cell-cell signaling in polymicrobial biofilms may play a significant role in disease development.
It is possible that positive fungal-bacterial interactions result in enhanced fungal colonization of sites favored by bacteria that may otherwise not be populated by fungi, as in the case of the mouse gut or human tooth surfaces by C. albicans [18,21]. Increased fungal colonization of ectopic sites combined with increased virulence gene expression may promote disease development in these sites. Although strong evidence of pathogenic synergy in humans is lacking, organotypic model and animal studies showed that certain oral streptococcal species display synergistic virulence with C. albicans on oral mucosal or tooth surfaces [27, 37, 40, 41].
Future directions
The roles that individual members of the core oral mycobiota may play in the community dynamics that sustain health or promote disease remain to be elucidated. Characterizing the interactions of newly recognized members of the core oral mycobiota with dominant Candida and Streptococcal species and assessing their impact on community development is an important first step in our understanding of the role of these fungi in health or disease. A reappraisal of the pathoecology of common oral fungal diseases such as oropharyngeal candidiasis using metagenomic sequencing is currently needed. Shotgun sequencing approaches may not only reveal new fungal genera associated with fungal disease but may also unveil previously unrecognized roles for oral bacteria or the host response in pathobiology. Improved statistical models to more accurately estimate the effect size of host and bacterial parameters have to be employed in such high-throughput surveys [21].
The representation of unculturable fungi in healthy and disease states needs to be accurately assessed and progress needs to be made in their culturability. Microbial loads in addition to mere presence of certain species have to be assessed, as there is considerable overlap in species community membership in oral health and disease for both fungal and bacterial organisms [3,15]. However, even when coupled with qPCR quantification [12], next generation sequencing data cannot differentiate between live and dead organisms. Until these methods are optimized to prevent genomic amplification in non-viable cells, microscopic, culture and/or biochemical approaches will be needed for definitive species identification or quantification. In addition, coupling phenotypic heterogeneity with genomic information (reviewed by Scaduto and Bennett, this issue), as in white, opaque, or grey cell morphologies associated with ploidy in Candida (reviewed by Gerstein and Berman, this issue), holds great promise in identifying novel community-based traits associated with health or disease states. Finally, development of appropriate infection models to include fastidious fungal organisms in the study of microbial community-level interactions with the host, coupled with integrated systems-based approaches, are needed to improve our understanding of the transition to dysbiosis and loss of homeostasis in the oral mucosa.
Highlights.
The membership of the core oral mycobiota in health has been defined
Global community shifts in disease have not been identified yet
Host influences composition and pathogenic activity of the oral mycobiota
Commensal bacteria may contribute to a fungal dysbiosis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mark Welch JL, Utter DR, Rossetti BJ, Mark Welch DB, Eren AM, Borisy GG. Dynamics of tongue microbial communities with single-nucleotide resolution using oligotyping. Front Microbiol. 2014;5:568. doi: 10.3389/fmicb.2014.00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gross EL, Beall CJ, Kutsch SR, Firestone ND, Leys EJ, Griffen AL. Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One. 2012;7:e47722. doi: 10.1371/journal.pone.0047722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aas JA, Barbuto SM, Alpagot T, Olsen I, Dewhirst FE, Paster BJ. Subgingival plaque microbiota in HIV positive patients. J Clin Periodontol. 2007;34:189–195. doi: 10.1111/j.1600-051X.2006.01034.x. *This study examined both bacterial and fungal community membership in oral samples.
- 7.Lalla RV, Patton LL, Dongari-Bagtzoglou A. Oral candidiasis: pathogenesis, clinical presentation, diagnosis and treatment strategies. J Calif Dent Assoc. 2013;41:263–268. [PubMed] [Google Scholar]
- 8.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding C Fungal Barcoding Consortium Author L. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang J, Iliev ID, Brown J, Underhill DM, Funari VA. Mycobiome: Approaches to analysis of intestinal fungi. J Immunol Methods. 2015 doi: 10.1016/j.jim.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen LD, Viscogliosi E, Delhaes L. The lung mycobiome: an emerging field of the human respiratory microbiome. Front Microbiol. 2015;6:89. doi: 10.3389/fmicb.2015.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung WH, Croll D, Cho JH, Kim YR, Lee YW. Analysis of the nasal vestibule mycobiome in patients with allergic rhinitis. Mycoses. 2015;58:167–172. doi: 10.1111/myc.12296. [DOI] [PubMed] [Google Scholar]
- 12.Willger SD, Grim SL, Dolben EL, Shipunova A, Hampton TH, Morrison HG, Filkins LM, O'Toole GA, Moulton LA, Ashare A, et al. Characterization and quantification of the fungal microbiome in serial samples from individuals with cystic fibrosis. Microbiome. 2014;2:40. doi: 10.1186/2049-2618-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dupuy AK, David MS, Li L, Heider TN, Peterson JD, Montano EA, Dongari-Bagtzoglou A, Diaz PI, Strausbaugh LD. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One. 2014;9:e90899. doi: 10.1371/journal.pone.0090899. **This study amended the core oral mycobiota membership to include the fastidious organism Malassezia, previously known as skin colonizer.
- 14. Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6:e1000713. doi: 10.1371/journal.ppat.1000713. **The first study to examine the oral mycobiota in healthy humans using pyrosequencing.
- 15.Xu H, Jenkinson HF, Dongari-Bagtzoglou A. Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol Oral Microbiol. 2014;29:99–116. doi: 10.1111/omi.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mukherjee PK, Chandra J, Retuerto M, Sikaroodi M, Brown RE, Jurevic R, Salata RA, Lederman MM, Gillevet PM, Ghannoum MA. Oral mycobiome analysis of HIV-infected patients: identification of Pichia as an antagonist of opportunistic fungi. PLoS Pathog. 2014;10:e1003996. doi: 10.1371/journal.ppat.1003996. *This study targeted both 16S and 18S rRNA genes in the same oral rinse samples using pyrosequencing.
- 17.Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014;14:405–416. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zijnge V, van Leeuwen MB, Degener JE, Abbas F, Thurnheer T, Gmur R, Harmsen HJ. Oral biofilm architecture on natural teeth. PLoS One. 2010;5:e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saunders CW, Scheynius A, Heitman J. Malassezia fungi are specialized to live on skin and associated with dandruff, eczema, and other skin diseases. PLoS Pathog. 2012;8:e1002701. doi: 10.1371/journal.ppat.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diaz PI, Strausbaugh LD, Dongari-Bagtzoglou A. Fungal-bacterial interactions and their relevance to oral health: linking the clinic and the bench. Front Cell Infect Microbiol. 2014;4:101. doi: 10.3389/fcimb.2014.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shankar J, Solis NV, Mounaud S, Szpakowski S, Liu H, Losada L, Nierman WC, Filler SG. Using Bayesian modelling to investigate factors governing antibiotic-induced Candida albicans colonization of the GI tract. Sci Rep. 2015;5:8131. doi: 10.1038/srep08131. **This study utilized improved regression models to more accurately assess strength of associations between fungal colonization, and bacterial and host parameters in the mouse gut. It is also the first study to report synergy between C. albicans and streptococci in colonization.
- 22.Monteiro-da-Silva Filipa, Araujo Ricardo, Sampaio-Maia Benedita. Interindividual variability and intraindividual stability of oral fungal microbiota over time. Med Mycol. 2014;52:496–503. doi: 10.1093/mmy/myu027. [DOI] [PubMed] [Google Scholar]
- 23. Charlson ES, Diamond JM, Bittinger K, Fitzgerald AS, Yadav A, Haas AR, Bushman FD, Collman RG. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. 2012;186:536–545. doi: 10.1164/rccm.201204-0693OC. **One of two studies to examine oral samples for both bacterial and fungal sequences using next generation techniques. The study reported that immunoisuppressed transplant recipients on prohylactic antibiotics have abundant oral colonization with Candida and streptococcal species.
- 24. Diaz PI, Hong BY, Frias-Lopez J, Dupuy AK, Angeloni M, Abusleme L, Terzi E, Ioannidou E, Strausbaugh LD, Dongari-Bagtzoglou A. Transplantation-associated long-term immunosuppression promotes oral colonization by potentially opportunistic pathogens without impacting other members of the salivary bacteriome. Clin Vaccine Immunol. 2013;20:920–930. doi: 10.1128/CVI.00734-12. *The study examined the relationship between the dose of immunosuppressants and oral colonization by Candida species as determined by culture methods.
- 25.Dongari-Bagtzoglou A, Dwivedi P, Ioannidou E, Shaqman M, Hull D, Burleson J. Oral Candida infection and colonization in solid organ transplant recipients. Oral Microbiol Immunol. 2009;24:249–254. doi: 10.1111/j.1399-302X.2009.00505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. : Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. **Elegant demonstration that a host fungal recognition receptor can shape the composition of the mouse intestinal mycobiota.
- 27. Xu H, Sobue T, Thompson A, Xie Z, Poon K, Ricker A, Cervantes J, Diaz PI, Dongari-Bagtzoglou A. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol. 2014;16(2):214–231. doi: 10.1111/cmi.12216. **First demonstration of pathogenic synergy between C. albicans and a streptococcal commensal in the oral mucosa.
- 28.Maraki S, Margioris AN, Orfanoudaki E, Tselentis Y, Koumantakis E, Kontoyiannis DP, Rovithi M, Samonis G. Effects of doxycycline, metronidazole and their Page 15 of 16 combination on Candida species colonization of the human oropharynx, intestinal lumen and vagina. J Chemother. 2003;15:369–373. doi: 10.1179/joc.2003.15.4.369. [DOI] [PubMed] [Google Scholar]
- 29.Fotos PG, Vincent SD, Hellstein JW. Oral candidosis. Clinical, historical, and therapeutic features of 100 cases. Oral Surg Oral Med Oral Pathol. 1992;74:41–49. doi: 10.1016/0030-4220(92)90213-a. [DOI] [PubMed] [Google Scholar]
- 30.Hsu LY, Minah GE, Peterson DE, Wingard JR, Merz WG, Altomonte V, Tylenda CA. Coaggregation of oral Candida isolates with bacteria from bone marrow transplant recipients. J Clin Microbiol. 1990;28:2621–2626. doi: 10.1128/jcm.28.12.2621-2626.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkinson HF, Lala HC, Shepherd MG. Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect Immun. 1990;58:1429–1436. doi: 10.1128/iai.58.5.1429-1436.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrian E, Qi G, Wang J, Halperin SA, Lee SF. Role of surface proteins SspA and SspB of Streptococcus gordonii in innate immunity. Microbiology. 2012;158:2099–2106. doi: 10.1099/mic.0.058073-0. [DOI] [PubMed] [Google Scholar]
- 33.Jenkinson HF, Demuth DR. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol Microbiol. 1997;23:183–190. doi: 10.1046/j.1365-2958.1997.2021577.x. [DOI] [PubMed] [Google Scholar]
- 34.Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun. 2010;78:4644–4652. doi: 10.1128/IAI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dutton LC, Nobbs AH, Jepson K, Jepson MA, Vickerman MM, Aqeel Alawfi S, Munro CA, Lamont RJ, Jenkinson HF. O-mannosylation in Candida albicans enables development of interkingdom biofilm communities. MBio. 2014;5:e00911. doi: 10.1128/mBio.00911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ricker A, Vickerman M, Dongari-Bagtzoglou A. Streptococcus gordonii glucosyltransferase promotes biofilm interactions with Candida albicans. J Oral Microbiol. 2014;6 doi: 10.3402/jom.v6.23419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai CH, Gonzalez- Begne M, Watson G, Krysan DJ, Bowen WH, et al. : Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun. 2014;82:1968–1981. doi: 10.1128/IAI.00087-14. **The study provides strong evidence for C. albicans-S. mutans pathogenic synergy in an animal model of caries.
- 38.Gregoire S, Xiao J, Silva BB, Gonzalez I, Agidi PS, Klein MI, Ambatipudi KS, Rosalen PL, Bauserman R, Waugh RE, et al. : Role of glucosyltransferase B in interactions of Candida albicans with Streptococcus mutans and with an experimental pellicle on hydroxyapatite surfaces. Appl Environ Microbiol. 2011;77:6357–6367. doi: 10.1128/AEM.05203-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bamford CV, d'Mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun. 2009;77:3696–3704. doi: 10.1128/IAI.00438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cavalcanti YW, Morse DJ, da Silva WJ, Del-Bel-Cury AA, Wei X, Wilson M, Milward P, Lewis M, Bradshaw D, Williams DW. Virulence and pathogenicity of Candida albicans is enhanced in biofilms containing oral bacteria. Biofouling. 2015;31:27–38. doi: 10.1080/08927014.2014.996143. [DOI] [PubMed] [Google Scholar]
- 41.Diaz PI, Xie Z, Sobue T, Thompson A, Biyikoglu B, Ricker A, Ikonomou L, Dongari- Bagtzoglou A. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun. 2012;80:620–632. doi: 10.1128/IAI.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]