A Physcomitrella patens protein regulates both ethylene and abscisic acid signaling, and this dual function was subsequently lost during evolution.
Abstract
Land plants have evolved adaptive regulatory mechanisms enabling the survival of environmental stresses associated with terrestrial life. Here, we focus on the evolution of the regulatory CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) component of the ethylene signaling pathway that modulates stress-related changes in plant growth and development. First, we compare CTR1-like proteins from a bryophyte, Physcomitrella patens (representative of early divergent land plants), with those of more recently diverged lycophyte and angiosperm species (including Arabidopsis [Arabidopsis thaliana]) and identify a monophyletic CTR1 family. The fully sequenced P. patens genome encodes only a single member of this family (PpCTR1L). Next, we compare the functions of PpCTR1L with that of related angiosperm proteins. We show that, like angiosperm CTR1 proteins (e.g. AtCTR1 of Arabidopsis), PpCTR1L modulates downstream ethylene signaling via direct interaction with ethylene receptors. These functions, therefore, likely predate the divergence of the bryophytes from the land-plant lineage. However, we also show that PpCTR1L unexpectedly has dual functions and additionally modulates abscisic acid (ABA) signaling. In contrast, while AtCTR1 lacks detectable ABA signaling functions, Arabidopsis has during evolution acquired another homolog that is functionally distinct from AtCTR1. In conclusion, the roles of CTR1-related proteins appear to have functionally diversified during land-plant evolution, and angiosperm CTR1-related proteins appear to have lost an ancestral ABA signaling function. Our study provides new insights into how molecular events such as gene duplication and functional differentiation may have contributed to the adaptive evolution of regulatory mechanisms in plants.
Modern-day land plants are derived from an ancient aquatic ancestor. Following colonization of the land, terrestrial plants evolved a diverse range of adaptive mechanisms that enabled survival of the various adversities associated with terrestrial life. Many of these acclimations are regulated by the phytohormone-mediated mechanisms that coordinate plant development and growth with changes in environmental conditions. While many studies have revealed that the major phytohormonal signaling pathways, such as those associated with auxin, abscisic acid (ABA), and ethylene, have remained fundamentally conserved during land-plant evolution, some variations have been identified as individual signaling pathways and have been studied in depth (Marella et al., 2006; Rensing et al., 2008; Eklund et al., 2010; Prigge et al., 2010; Sakata et al., 2010; Yasumura et al., 2012; Komatsu et al., 2013). For example, phylogenetic analysis has indicated that the auxin response factors of Arabidopsis (Arabidopsis thaliana) and Physcomitrella patens have undergone independent processes of expansion and divergence (Paponov et al., 2009; Romanel et al., 2009). In contrast, the GA signaling pathway appears to have arisen during land-plant evolution, sometime following the divergence of the bryophytes from the main land-plant lineage (Hirano et al., 2007; Yasumura et al., 2007).
The ethylene signaling pathway is of particular relevance to plant survival of water-related stresses (Voesenek and Sasidharan, 2013). In this article, we describe investigations of the evolutionary trajectory of this pathway, with particular focus on the role of gene duplication and divergence in that trajectory. Much of our current understanding of ethylene signaling in angiosperms is derived from genetic analyses in Arabidopsis (Stepanova and Alonso, 2009; Yoo et al., 2009; Wang et al., 2013). Essentially, ethylene binds to membrane-spanning ethylene receptors (e.g. ETR1; Bleecker et al., 1988; Chang et al., 1993; Hua and Meyerowitz, 1998). The binding of ethylene to ethylene receptors inactivates these receptors and, consequently, inactivates the downstream CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) protein (Kieber et al., 1993; Clark et al., 1998). Inactivation of CTR1 permits the activation of the downstream ETHYLENE INSENSITIVE2 (EIN2) protein and the consequent activation of a transcriptional regulatory cascade consisting first of EIN3 and then of ETHYLENE RESPONSE FACTOR (ERF)-type transcription factors (Chao et al., 1997; Solano et al., 1998; Alonso et al., 1999). There is already evidence that gene duplication and subsequent divergence have played important roles in the modulation of ethylene-regulated adaptive responses. For example, different rice (Oryza sativa) varieties have evolved two distinct strategies for combating the adverse effects of submergence. These distinct strategies evolved independently through the recruitment of different ERFs (SUBMERGENCE 1A in the one case and SNORKEL1 and SNORKEL2 in the other; Xu et al., 2006; Hattori et al., 2009; Voesenek and Bailey-Serres, 2013). These independent recruitments resulted from separate gene duplication events and illustrate how diversity can evolve through gene duplication and the subsequent novel functional specialization of duplicated genes (Flagel and Wendel, 2009; Zou et al., 2009). In addition, these observations are consistent with the previous suggestion that once a stress-regulated mechanism has been established, plants may increase the range of stresses that they can deal with through the acquisition of duplicate genes that gain new responses to different stresses (Zou et al., 2009). More recently, subfunctionalization of multiple ethylene receptors has been reported in Arabidopsis, with different receptor molecules regulating seed germination differentially during salt stress by modulating ABA signaling (Wilson et al., 2014), again suggesting that the ethylene signaling pathway is highly susceptible to plastic change related to stress adaptation. In order to further our understanding of the molecular events that have enabled adaptive evolution in land plants, we compared the CTR1 component of the ethylene signaling pathways of a bryophyte (P. patens, an extant relative of early land plants) and of an angiosperm (Arabidopsis, a relatively recent land plant).
Ethylene, together with other phytohormones such as ABA, plays major roles in plant adaptive responses to environmental stresses, including salinity, drought, and submergence (Bailey-Serres and Voesenek, 2008; Cao et al., 2008; Hattori et al., 2009; Wilkinson and Davies, 2010; Fukao et al., 2011). Ethylene signaling has been studied mostly in angiosperms, and many known ethylene responses are specific to vascular plants, including effects on seed germination, etiolated seedlings, and fruit development (Abeles et al., 1992). However, ethylene also has important functions in bryophytes. We previously showed that ethylene regulates the growth of P. patens, particularly in conditions of water-related stress (Yasumura et al., 2012). First, we found that the P. patens genome encodes proteins similar to components of the angiosperm ethylene signaling pathway, including the ethylene receptors PpETR1 to PpETR7, a CTR1-like protein, EIN3, and ERF proteins. Next, by altering the activity of an ethylene receptor, we showed that ethylene plays a regulatory role in modifying the P. patens growth pattern in response to submergence and osmotic stress (Yasumura et al., 2012). These observations suggest that ethylene signaling has functioned in the regulation of plant growth in response to water-related stresses since the earliest phases of evolution of the land-plant lineage. Here, we investigate the possibility that the diversification of adaptive responses to different water-related stresses that occurred during land-plant evolution may have been driven by the diversification of the function of the ethylene signaling component CTR1.
Studies with Arabidopsis have shown that CTR1 plays a major regulatory role in the ethylene signaling pathway (Stepanova and Alonso, 2009; Yoo et al., 2009). AtCTR1 represses the ethylene response in the absence of ethylene by phosphorylating EIN2, thus preventing EIN2 from entering the nucleus (where it activates EIN3 and downstream ERF transcription factors; Ju et al., 2012; Qiao et al., 2012). AtCTR1 interacts directly with the ethylene receptors, and this interaction maintains AtCTR1 in an active state (Kieber et al., 1993; Clark et al., 1998). However, when the ethylene receptors bind ethylene, AtCTR1 is deactivated. As a result, EIN2 is no longer subject to CTR1-dependent phosphorylation, and a C-terminal portion of EIN2 consequently moves to the nucleus, thus inducing ethylene responses via the stabilization of EIN3 (Ju et al., 2012; Qiao et al., 2012).
Although the Arabidopsis genome contains several genes related in sequence to AtCTR1, AtCTR1 itself is the only one of these known to be involved in ethylene signaling (Adams-Phillips et al., 2004). Here, we show that the inclusion of related bryophyte sequences in phylogenetic analyses reveals the likely occurrence of a gene duplication event in the lineage leading to the angiosperms and following the divergence of bryophytes. The resultant second gene, At4g24480, was previously reported to be the most closely related gene to AtCTR1 in the Arabidopsis genome (Adams-Phillips et al., 2004). Given the importance of CTR1 in the ethylene signaling pathway and the possibility that this gene duplication event occurred after the ethylene signaling function was established, we hypothesized that duplication of CTR1-like genes may have contributed to the evolution of ethylene-regulated adaptive responses in angiosperms. In the case of tomato (Solanum lycopersicum), different CTR1 homologs were shown to have distinct expression patterns, indicating the potential drive for functional divergence when multiple copies are available (Adams-Phillips et al., 2004), although the precise nature of this proposed functional differentiation or redundancy remains unclear (Zhong et al., 2008). In this study, we consider the possibility of functional divergence of CTR1-related proteins during land-plant evolution. First, we investigate the function of a CTR1-related protein in P. patens, thus identifying possible ancestral functions of CTR1. Second, we compare the functional characteristics of AtCTR1 and related proteins in Arabidopsis.
When studying the regulation of plant adaptive responses to water-related stress, it is important to consider another plant stress hormone, ABA. The ABA signaling pathway is activated by the binding of ABA to the ABA receptors REGULATORY COMPONENTS OF ABSCISIC ACID RECEPTOR/PYRABACTIN RESISTANCE (PYR)/PYRABACTIN RESISTANCE-LIKE (PYL) family proteins, which leads to the inactivation of type 2C protein phosphatases such as ABSCISIC ACID INSENSITIVE1 (ABI1; Raghavendra et al., 2010). Ethylene signaling is known to interact with ABA signaling in Arabidopsis to regulate seed germination, root and shoot growth, and hyponastic growth of petioles in angiosperms (Sharp and LeNoble, 2002; Benschop et al., 2007; McManus, 2012). Several studies have dissected the molecular mechanisms underlying the interactions between ethylene and ABA signaling (Wilkinson and Davies, 2010; Arc et al., 2013). For example, the reduced ethylene signaling characteristic of the Arabidopsis etr1 and ein2 mutants was shown to result in enhanced ABA sensitivity of seed germination, while this ABA sensitivity was shown to be reduced in ctr1 and ethylene overproducing3 mutants displaying constitutive ethylene responses (Subbiah and Reddy, 2010). An additional study found that ethylene and ABA signaling act antagonistically in seeds but synergistically in root growth and that such interactions involve major signaling components such as ETR1, CTR1, EIN2, and ABI1 (Beaudoin et al., 2000). However, while cross talk between ethylene and ABA signaling pathways has been studied in angiosperms, the nature of this cross talk in bryophytes is poorly understood. Therefore, here, we also sought to elucidate how ethylene signaling interacts with ABA signaling in the bryophyte species P. patens and to understand the possible involvement of the P. patens CTR1-related protein in that interaction.
In summary, our investigations have revealed a possible ancestral role for CTR1 proteins in both ethylene and ABA signaling. While ethylene signaling functions are retained by some members of the angiosperm CTR1 family, a role in ABA signaling was not detected, suggesting that this function has been lost by CTR1 family proteins during the course of land-plant evolution. Our observations provide important information concerning ancestral relationships between the ethylene and ABA signaling pathways.
RESULTS
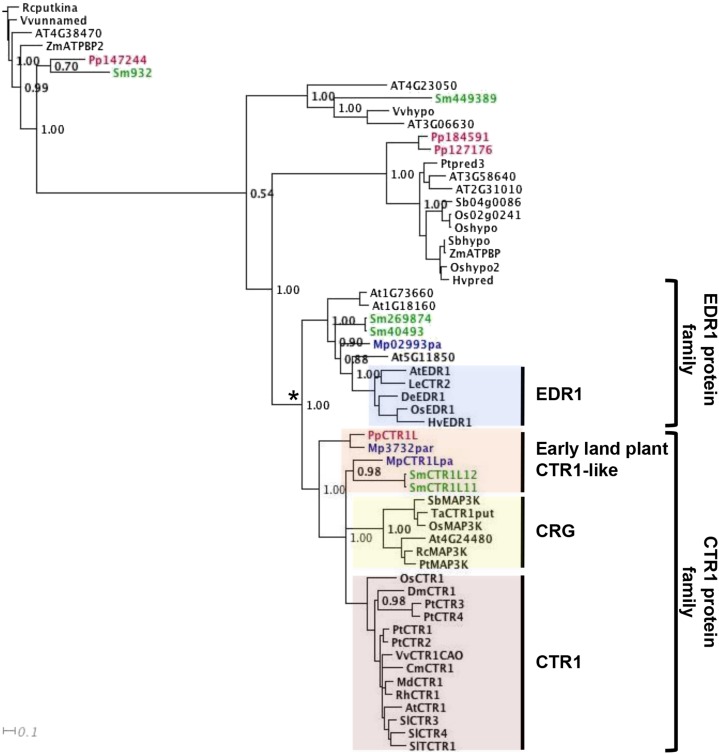
Phylogenetic Analysis Indicates That the P. patens Genome Encodes Only a Single CTR1-Like Protein and Identifies a CTR1-Encoding Gene Duplication Prior to the Angiosperm Divergence
The CTR1 protein was first identified in Arabidopsis (Kieber et al., 1993) and has subsequently been studied in a number of different species. To analyze the phylogenetic relationships between CTR1 proteins and related proteins from various species, CTR1 and related sequences were collected from databases containing angiosperm, lycophyte, and bryophyte sequences and used to construct a phylogenetic tree (Fig. 1). This tree identifies a group of bryophyte and lycophyte protein sequences that are the most closely related to angiosperm CTR1 proteins and that fall within a region marked as early land-plant CTR1-like proteins (Fig. 1). Interestingly, only a single protein sequence from the fully sequenced P. patens genome (PpCTR1L, encoded by the PpCTR1L gene) is included within this group, suggesting that PpCTR1L is one of the relatively few single-copy genes found in the P. patens genome (which is the product of a recent genome duplication event; Rensing et al., 2008). The relationship of these proteins was further supported by an additional tree (Supplemental Fig. S1) that was built on more broadly selected amino acid sequences using an independent method of tree inference and was also consistent with our previous phylogenetic analysis (within which algal sequences were considered; Yasumura et al., 2012).
Figure 1.
PpCTR1L is related to CTR1 homologs and CTR1-like proteins. AtCTR1, AtEDR1, and their related sequences form a monophyletic group, which is further subdivided into the EDR1 and CTR1 gene families. Within the CTR1 gene family, angiosperm sequences form two distinctive subfamilies labeled as CTR1 and CRG, while early land-plant CTR1-like sequences are placed outside of these two subfamilies. The tree was constructed using the maximum likelihood method on aligned amino acid sequences listed in Supplemental Table S1. Bootstrap support values are indicated at internal nodes. The scale bar indicates the number of changes per site. Sequences from P. patens are shown in red, those from Marchantia polymorpha in blue, and those from S. moellendorffii in green. The asterisk indicates the node at which the EDR1 and CTR1 protein families join together.
Moreover, our analyses placed early land-plant CTR1-like sequences within a monophyletic group that includes not only angiosperm CTR1 proteins but also another group of angiosperm proteins labeled CRG for CTR1-related group (Fig. 1). This observation suggests the occurrence of a gene duplication event after the divergence of the bryophytes and lycophytes and before that of the angiosperms. An alternative possibility could be that the duplication had occurred earlier in evolution and that the duplicate copy was subsequently lost in the lineage that contains P. patens. However, since none of multiple CTR1 homolog sequences from the lycophyte Selaginella moellendorffii were included in either of the CTR1 or CRG families (Fig. 1), it is more likely that the divergence of these two families occurred subsequent to the divergence of the lycophytes. In addition, our analyses identified the ENHANCED DISEASE RESISTANCE1 (EDR1) protein family as the most closely related group to the CTR1 protein family, with these protein families together forming a monophyletic group (marked with an asterisk in Fig. 1) and, thus, sharing the same ancestry. EDR1 has been studied previously in Arabidopsis and rice and shown to be involved in the regulation of disease resistance (Frye and Innes, 1998; Shen et al., 2011). In rice, EDR1 also plays a role in the regulation of ethylene production (Shen et al., 2011). The functions of the Arabidopsis CTR1, CRG, and EDR1 proteins are further considered below.
The P. patens CTR1-Like Protein Interacts with Ethylene Receptors
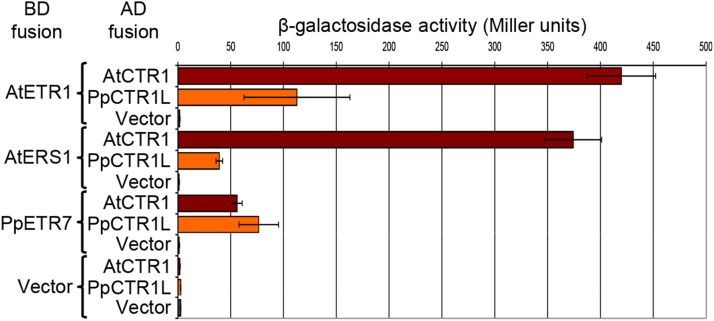
We next studied the functions of the CTR1-like protein we had previously identified in the bryophyte P. patens (Fig. 1), in the expectation that such analyses might enable elucidation of the ancestral function of CTR1 gene family proteins. PpCTR1-like (PpCTR1L) is encoded by a single copy gene in the genome of P. patens and was placed in a basal position in the CTR1 subfamily in phylogenetic analyses (Fig. 1). First, we tested the ability of PpCTR1L to interact with ethylene receptors from Arabidopsis and P. patens (Fig. 2; Supplemental Fig. S2). Yeast two-hybrid assays revealed that PpCTR1L can bind to the Arabidopsis ethylene receptors AtETR1 and ETHYLENE RESPONSE SENSOR1 (AtERS1) and P. patens PpETR7, although the strength of the affinity of PpCTR1L for AtETR1 and AtERS1 was much less than that of AtCTR1 (Fig. 2). Because PpCTR1L is a bryophyte protein, these data suggest that CTR1 family proteins had acquired the ability to interact with ethylene receptors prior to the bryophyte divergence. Because it has also been reported that ethylene receptor and CTR1 homologs from the charophacean alga Spirogyra pratensis interact with one another (Ju et al., 2015), this interaction appears to have existed prior to the colonization of the land and to have been subsequently conserved in land-plant lineages.
Figure 2.
PpCTR1L can bind to ethylene receptors. A quantitative yeast two-hybrid assay for interactions between AtCTR1, PpCTR1L, and ethylene receptors from moss and Arabidopsis is shown. The transmembrane domains of the ethylene receptors and the kinase domains of CTR1-related proteins were excluded from the constructs to avoid these domains interfering with the assay. Results are expressed as means ± sd (n = 3), and the experiment was conducted twice. AD, Activation domain; BD, DNA binding domain.
A Ppctr1l Knockout Line Is Defective in Responding to Ethylene and Exhibits Constitutive Ethylene Responses
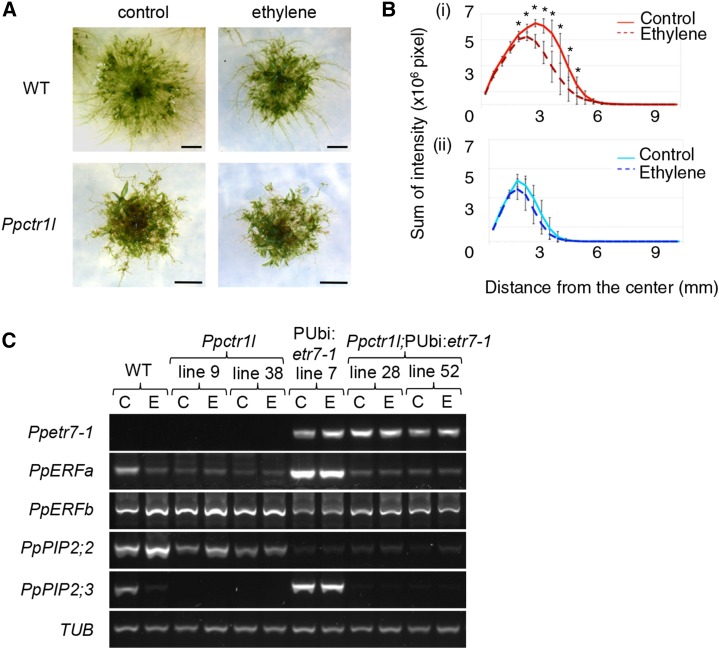
AtCTR1 is involved in various aspects of the growth and developmental regulation of Arabidopsis, as suggested by the Atctr1-1 mutant phenotype. For example, in addition to displaying the triple response in the absence of ethylene, the Atctr1-1 mutant also exhibits various growth defects, including reduced leaf size, hyponastic leaves, reduced cauline shoot height, and delayed flowering (Kieber et al., 1993). We next determined the function of PpCTR1L in P. patens, taking a mutant analysis approach. We removed PpCTR1L from the P. patens genome via homologous recombination, making multiple independent Ppctr1l deletion mutant lines (Supplemental Fig. S3). First, Ppctr1l was tested for altered ethylene sensitivity or responses (Fig. 3). The morphological response to ethylene of P. patens was previously defined in terms of altered filamentous growth architecture: ethylene-treated plants display enhanced growth of peripheral colonizing filaments and reduced growth density in central parts of the moss plant (Yasumura et al., 2012). While wild-type plants responded to ethylene treatment as described, no such ethylene-inducible changes were observed in Ppctr1l colonies (Fig. 3, A and B), suggesting that the Ppctr1l line is defective in responding to ethylene.
Figure 3.
Ppctr1l knockout moss plants exhibit constitutive ethylene responses. A, Fourteen-day-old wild-type (WT) and Ppctr1l plants, previously transferred on day 5 following inoculation to an airflow chamber filled with air (control) or with 3 µL L−1 ethylene (ethylene) and incubated for a further 10 d. Bars = 5 mm. B, Radial intensity was measured, and the sum of intensity was plotted against distance from the center of the plant for ethylene-treated (dotted lines) and control (solid lines) 14-d-old wild-type (i) and Ppctr1l (ii) plants. n = 7 for each graph. Asterisks indicates data points where the differences between air-treated and ethylene-treated plants were statistically significant (error bars represent sd). C, Semiquantitative reverse transcription (RT)-PCR showing levels of transgene transcripts, of putative PpERF transcripts (PpERFa and PpERFb), and of transcripts encoding P. patens aquaporins (PpPIP2;2 and PpPIP2;3) in the wild type, Ppctrl1 lines, pUbi:Ppetr7-1, and Ppctrl1; pUbi:Ppetr7-1 lines, using tubulin (PpTUB) as a control. RNA was harvested from 7-d-old protonema treated in an air-flow chamber with air (C) or air containing 3 µL L−1 ethylene (E) for 3 d.
Further molecular studies revealed that Ppctr1l confers constitutive ethylene-responsive gene expression (Fig. 3C). These experiments included the previously described P. patens transgenic line pUbi:Ppetr7-1. This line expresses a dominant mutant version of the PpETR7 ethylene receptor (Ppetr7-1) under the control of a maize (Zea mays) ubiquitin promoter (Ubi) from pBract211(http://www.bract.org). Ppetr7-1 is unable to bind ethylene and, hence, confers insensitivity to ethylene, presumably because, as in the case of the Arabidopsis ethylene receptors, it activates downstream signaling to repress ethylene responses (Yasumura et al., 2012). To determine whether PpCTR1L functionally interacts with PpETR7, the PpCTR1L gene was removed from pUbi:Ppetr7-1 via homologous recombination, thus generating multiple independent lines that lacked PpCTR1L and expressed Ppetr7-1 (Ppctr1l; pUbi:Ppetr7-1; Supplemental Fig. S3). As shown in Figure 3C, the expression of transgene Ppetr7-1 was maintained in pUbi:Ppetr7-1 lines and Ppctr1l; pUbi:Ppetr7-1 lines throughout the experiment. Expression levels were tested for PpERFa and PpERFb, which are members of a large gene family phylogenetically shown to be homologous to the angiosperm ERF family, and for Plasma Membrane Intrinsic Protein2;2 (PpPIP2;2) and PpPIP2;3, both of which encode ethylene-regulated aquaporins. PpERFa and PpPIP2;3 transcripts are down-regulated by ethylene and are expressed at elevated levels in ethylene-insensitive pUbi:Ppetr7-1 lines. The expression analysis revealed that the expression of PpERFa and PpPIP2;3 remained low or absent in Ppctr1l mutants regardless of the presence or absence of ethylene in the PpETR7 or pUbi:Ppetr7-1 backgrounds (Fig. 3C). These observations indicate that the Ppctr1l mutant constitutively displays an ethylene-induced state with respect to PpERFa and PpPIP2;3, suggesting that PpCTR1L acts as a negative regulator of P. patens ethylene responses that acts downstream of PpETR7, with a function analogous to that of AtCTR1 in Arabidopsis.
In contrast, PpERFb is up-regulated by ethylene and down-regulated in the pUbi:Ppetr7-1 background. While the expression of PpERFb was up-regulated in the Ppctr1l mutant, an observation consistent with the role for PpCTR1L proposed above, PpERFb expression remained at low levels in the Ppctr1l; pUbi:Ppetr7-1 lines, albeit somewhat higher than in pUbi:Ppetr7-1. The reasons for the reduced expression in Ppctr1l; pUbi:Ppetr7-1 compared with Ppctr1l remain unclear. In addition, PpPIP2;2, another ethylene-inducible gene, was not detectably up-regulated in Ppctr1l or Ppctr1l; pUbi:Ppetr7-1 lines. These observations may suggest that regulatory factors additional to the canonical ETR1-CTR1 pathway are involved in the regulation of ethylene-responsive gene expression in P. patens.
PpCTR1L Regulates P. patens Growth and Ethylene Responses through Functional Interaction with Ethylene Receptors
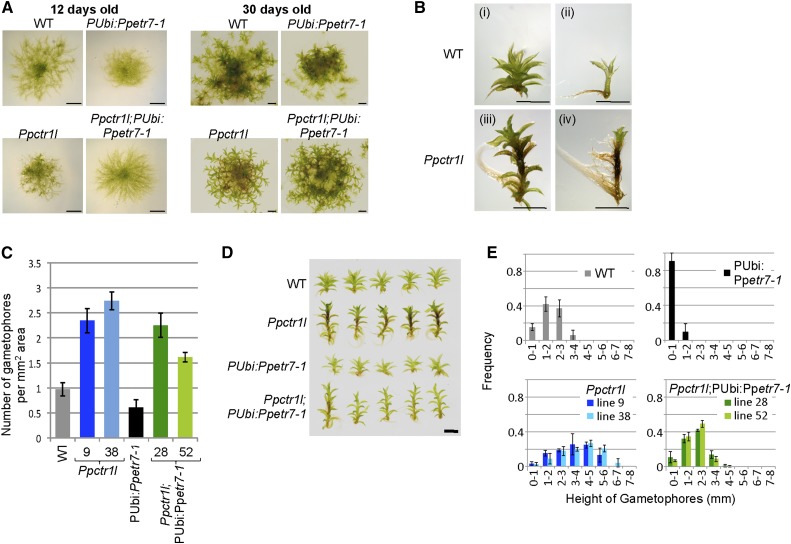
To further investigate the function of PpCTR1L, growth was compared between the wild type and Ppctr1l on day 12 after inoculation, when filamentous structure normally predominates colonies, and again on day 30, when colonies grow gametophores (or leafy shoots; Fig. 4). It was observed that filamentous growth was reduced in Ppctr1l compared with the wild type, given the difference in colony size on day 12, while Ppctr1l showed more pronounced gametophore growth than the wild type on day 30 (Fig. 4A). In addition, Ppctr1l gametophores were much taller than wild-type gametophores (Fig. 4B, i and iii), and rhizophore induction was also altered (Fig. 4B, ii and iv). While rhizophore growth is normally restricted to the basal regions of wild-type gametophores, Ppctr1l gametophores grew rhizophores throughout most of their length, from the base to the tip. Taken together, these Ppctr1l mutant phenotypes indicate that PpCTR1L is involved in the regulation of growth at various stages of development. More precisely, PpCTR1L promotes filamentous growth, inhibits gametophore growth, and inhibits rhizophore development in P. patens.
Figure 4.
Functional interactions between PpETR7 and PpCTR1L. A, Loss of PpCTR1L altered the morphology of the moss colony and the effect of pUbi:Ppetr7-1 expression. Bars = 2 mm. B, Ppctr1l gametophores (iii and iv) are taller than wild-type (WT) gametophores (i and ii) and grow rhizoids in an uncharacteristic way, as revealed when leaf-like structures were removed (ii and iv). Bars = 2 mm. C, Mean number of gametophores, calculated per mm2 of colony area for the wild type, Ppctrl1 lines, pUbi:Ppetr7-1, and Ppctrl1; pUbi:Ppetr7-1 lines. D, The tallest gametophore was collected from five different plants each for the wild type, Ppctrl1, pUbi:Ppetr7-1, and Ppctrl1; pUbi:Ppetr7-1. Bar = 2 mm. E, Frequency distribution of gametophore heights for the wild type, Ppctrl1 lines, pUbi:Ppetr7-1, and Ppctrl1; pUbi:Ppetr7-1 lines. The results are expressed as means ± sd (n > 120).
We next determined if PpCTR1L functions downstream of ethylene receptors, using the above-described Ppctr1l; pUbi:Ppetr7-1 lines (which lack PpCTR1L and express the mutant Ppetr7-1 ethylene receptor). In a functionally equivalent experiment in Arabidopsis, an etr1-1 ctr1-1 double mutant exhibits the constitutive ethylene responses characteristic of ctr1-1, indicating that CTR1 acts downstream of Arabidopsis ethylene receptors in ethylene signaling (Hua et al., 1995; Roman et al., 1995). pUbi:Ppetr7-1 in the wild-type background confers, as a result of its ethylene insensitivity, a number of morphological alterations, including a densely packed filamentous colony structure and reduced gametophore development (Fig. 4A; Yasumura et al., 2012). The absence of PpCTR1L, in turn, conferred upon pUbi:Ppetr7-1 plants morphological changes in filamentous growth, overall colony size, and altered gametophore development (Fig. 4A). Close examination of the number and size of gametophores (Fig. 4, C–E) revealed that expression of Ppetr7-1 and loss of PpCTR1L had opposite effects. In other words, the growth phenotype of Ppctr1l is the reverse of the effect of ethylene insensitivity and likely represents a constitutive ethylene response phenotype. In addition, the effect of Ppctr1l masked the effect of pUbi:Ppetr7-1, as the phenotype of Ppctr1l; pUbi:Ppetr7-1 resembles the Ppctr1l phenotype more than the phenotype conferred by pUbi:Ppetr7-1 alone. This observation suggests that PpCTR1L indeed functions downstream of the ethylene receptor as a negative regulator of ethylene signaling. For example, ethylene insensitivity caused a reduction in the number of gametophores per unit colony area of pUbi:Ppetr7-1 to half that of the wild type (Fig. 4C), suggesting that ethylene promotes gametophore growth. In contrast, Ppctr1l grew more than twice as many gametophores as the wild type, and the likely explanation is that it exhibited an enhanced ethylene response. The removal of PpCTR1L from pUbi:Ppetr7-1 brought the number of gametophores up to the level seen in Ppctr1l mutant lines (Fig. 4C), suggesting that the effect of Ppctr1l is epistatic to that of pUbi:Ppetr7-1. Furthermore, when the tallest gametophores were compared (Fig. 4D), gametophores from Ppctr1l lines and Ppctr1l; pUbi:Ppetr7-1 lines were observed to be morphologically similar to each other but distinct from those of wild-type and pUbi:Ppetr7-1 lines. When the height of gametophores was compared globally across colonies (Fig. 4E), it was found that Ppctr1l; pUbi:Ppetr7-1 lines had a similar distribution of gametophore height to the wild type, which was distinctively different from that of pUbi:Ppetr7-1 lines. Thus, removing PpCTR1L shifted the frequency curve of gametophore height of pUbi:Ppetr7-1 lines toward that of the wild type, although not quite as far as that of Ppctr1l lines. These results suggest that PpETR7 and PpCTR1L are both involved in the same aspects of developmental regulation. It should be noted, however, that the colony morphology of Ppctr1l; pUbi:Ppetr7-1 lines was not identical to that of Ppctr1l, which may suggest that pUbi:Ppetr7-1 also exerts its effect through components other than PpCTR1L. Nonetheless, the results obtained in the above experiments provide sufficient data to suggest that PpCTR1L likely functions downstream of ethylene receptors through direct interaction and that PpCTR1L plays a role in regulating ethylene responses as a negative regulator.
PpCTR1L Modulates Both ABA and Ethylene Signaling
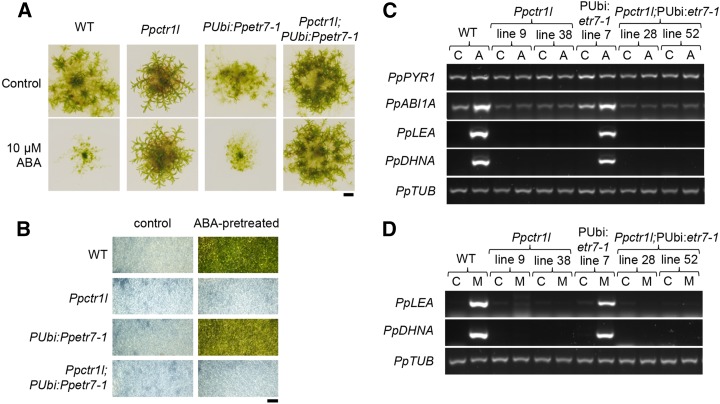
ABA is a stress hormone that regulates plant adaptive responses to environmental change. In P. patens, ABA confers desiccation tolerance (Khandelwal et al., 2010; Komatsu et al., 2013; Yotsui et al., 2013). Because ethylene also regulates water relation responses in P. patens (Yasumura et al., 2012), we sought to determine if the ABA and ethylene signaling pathways interact with one another (Fig. 5). Because ABA causes dramatic growth reductions in P. patens, we first compared the growth of the wild-type, pUbi:Ppetr7-1, and Ppctrl1 mutant lines with or without ABA (Fig. 5A). While the expression of pUbi:Ppetr7-1 did not alter ABA sensitivity, ABA responses were completely absent in Ppctr1l mutants (Fig. 5A). This lack of ABA response was further shown to be physiologically relevant in P. patens plants subjected to desiccation stress. Pretreatment of P. patens with ABA usually confers a degree of desiccation tolerance that allows survival of the complete desiccation that would otherwise be lethal, as shown for the wild-type and pUbi:Ppetr7-1 lines (Fig. 5B). In contrast, Ppctr1l mutants do not display desiccation tolerance, even following ABA pretreatment, consistent with the above described insensitivity to ABA.
Figure 5.
PpCTR1L function is required for ABA signaling in P. patens. A, Fourteen-day-old wild-type (WT), Ppctr1l, pUbi:etr7-1, and Ppctr1l; pUbi:etr7-1 plants grown on medium lacking ABA (control) or containing 10 μm ABA. Bar = 2 mm. B, Seven-day-old protonemal tissue of wild-type, Ppctr1l, pUbi:etr7-1, and Ppctr1l; pUbi:etr7-1 plants, previously transferred to medium lacking ABA (control) or containing 10 μm ABA (ABA-pretreated) for 1 d, were subjected to desiccation treatment and tested for survival. Bar = 2 mm. C, Semiquantitative RT-PCR showing levels of transcripts encoding ABA signaling components (PpPYR1 and PpABI1A) and of ABA-regulated transcripts (PpLEA and PpDHNA) in the wild type, Ppctrl1 lines, pUbi:Ppetr7-1, and Ppctrl1; pUbi:Ppetr7-1 lines, using PpTUB as a control. RNA was harvested from 8-d-old protonema that was transferred to medium containing 0 μm (C) or 10 μm (A) ABA on day 7 following inoculation. D, Semiquantitative RT-PCR showing levels of ABA-regulated transcripts (PpLEA and PpDHNA) in the wild type, Ppctrl1 lines, pUbi:Ppetr7-1, and Ppctrl1; pUbi:Ppetr7-1 lines, using PpTUB as a control. RNA was harvested from 8-d-old protonema that was transferred to medium containing 0 mm (C) or 300 mm (M) mannitol on day 7 following inoculation.
In further experiments, we showed that ABA receptors and signaling components are likely to be expressed in Ppctr1l plants, because RT-PCR analyses detected normal levels of PpPYR1 transcripts (encoding a putative ABA receptor; Umezawa et al., 2010; Chater et al., 2011) and normal or only slightly reduced levels of PpABI1A transcripts (encoding an ABA signaling component; Komatsu et al., 2009) in Ppctr1l mutants (Fig. 5C). Nevertheless, Ppctr1l mutants failed to up-regulate the expression of ABA-inducible genes such as PpABI1A, Late Embryogenesis Abundant (PpLEA; Kamisugi and Cuming, 2005), and Dehydrin-like A (PpDHNA; Saavedra et al., 2006) when treated with ABA (Fig. 5C). Furthermore, when P. patens plants were subjected to osmotic stress by the addition of mannitol to the growth medium, the expression of PpLEA and PpDHNA was up-regulated in the wild type and in the pUbi:Ppetr7-1 line through ABA signaling, but not in Ppctrll mutants (Fig. 5D). Thus, PpCTR1L is an essential component of the ABA signaling mechanism in P. patens. Because expression of the mutant ethylene receptor Ppetr7-1 did not affect ABA signaling, PpCTR1L may serve as a point of intersection of the ethylene and ABA signaling pathways in P. patens.
Ethylene inactivates AtCTR1, and this system is likely conserved for PpCTR1L in P. patens. However, the addition of ethylene had little effect on ABA responses in P. patens (Supplemental Fig. S4), suggesting that inactivation of PpCTR1L by ethylene did not prevent ABA signaling. Thus, PpCTR1L may have a dual function in transducing ethylene and ABA signals and modulating ABA signaling independently of ethylene signaling.
The Ability to Bind Ethylene Receptors Is Shared by All Members of the CTR1 Protein Family, Including the CTR1-Related Arabidopsis Protein AtCRG
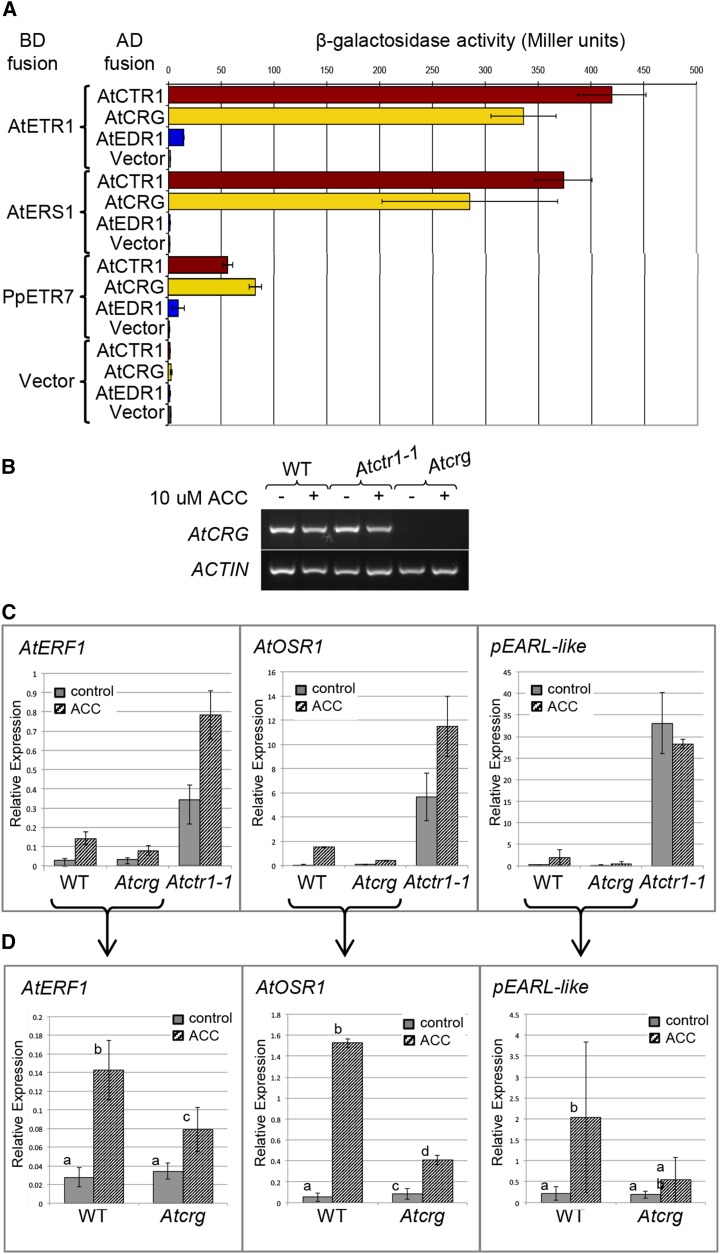
Given that PpCTR1L shares the same ancestry with Arabidopsis AtCTR1 and related proteins, At4g24480 and AtEDR1 (Fig. 1), we sought to find out to what extent the function is conserved or diversified among the CTR1-related proteins. In Arabidopsis, while AtCTR1 and AtEDR1 have been functionally characterized, very little is known about At4g24480, which was included in the CRG (Fig. 1; At4g24480 is hereafter referred to as AtCRG). We compared the functional properties of these proteins, first by testing their abilities to interact with ethylene receptors. CTR1 proteins interact physically with ethylene receptors to repress ethylene signaling, and this interaction is key to the function of CTR1. Here, we found that AtCTR1 and AtCRG both interacted strongly with two of the Arabidopsis ethylene receptors, AtETR1 and AtERS1, and with a P. patens ethylene receptor, PpETR7, in yeast two-hybrid assays. AtEDR1, on the other hand, showed minimal binding with AtETR1 and no binding with AtERS1 (Fig. 6; Supplemental Fig. S2). Previously, LeCTR2, a member of the EDR1 subfamily in tomato, was reported to interact with two of the three subfamily I ethylene receptors (Lin et al., 2008; Zhong et al., 2008). However, given that the data were produced using a qualitative β-Gal plate assay and that AtEDR1 also showed minimal binding activity using this same assay (Supplemental Fig. S2), it is possible that AtEDR1 and LeCTR2 actually have comparatively similar binding abilities. Therefore, the ability to interact with ethylene receptors, an ability that arose prior to the bryophyte divergence, appears to have been retained as a property characteristic of the CTR1 protein family throughout subsequent land-plant evolution.
Figure 6.
The AtCRG protein interacts with ethylene receptors and may play a role in ethylene responses. A, Quantitative yeast two-hybrid assay for interactions between AtCTR1, related proteins, and ethylene receptors. The data shown for AtCTR1 in combination with AtETR1, AtERS1, and BD vector are identical to those presented in Figure 2. The transmembrane domains of the ethylene receptors and the kinase domains of CTR1-related proteins were excluded from the constructs to avoid these domains interfering with the assay. The results are expressed as means ± sd (n = 9), and the experiment was conducted twice. B, Semiquantitative RT-PCR determination of AtCRG transcript levels, using ACTIN transcripts as a control. RNA was harvested from wild-type (WT) plants and mutant lines of Atctr1-1 and Atcrg T-DNA insertion, all of which were treated (+) or untreated (−) with 10 μm 1-aminocyclopropane-1-carboxylic acid (ACC). C, Quantitative RT-PCR determination of AtERF, AtOSR1, and pEARL-LIKE transcript levels by quantitative PCR, using UBIQUITIN C as a reference. RNA was extracted from wild-type plants and mutant lines of Atcrg T-DNA insertion and Atctr1-1, all of which were sprayed with water (gray bars) or 0.5 mm ACC control (shaded bars) at 24 h before samples were harvested. The results are expressed as means ± sd (n = 6), and two independent biological replicates were included in the experiments. D, An extract of the data for wild-type and Atcrg T-DNA insertion plants from the data shown in C. Means with different letters in each graph are significantly different (Student’s t test, P < 0.05; n = 6).
In addition, our observations suggest that there has been an increase in the binding affinity of CTR1 and ethylene receptors during land-plant evolution, with the more recent Arabidopsis proteins (ethylene receptors, AtCTR1, and AtCRG) having relatively high affinities for one another when compared with the interaction between PpETR7 and PpCTR1L (Fig. 2). Thus, there seems to have been a trend toward intraspecific optimization of the binding efficiency of CTR1 and ethylene receptors during the course of evolution.
Ethylene-Inducible Gene Expression Is Differentially Altered in Atctr1 and Atcrg Mutants, Suggesting a Functional Differentiation of AtCTR1 and AtCRG
The fact that AtCRG retained the ability to interact with ethylene receptors suggested a possible involvement of AtCRG in ethylene signaling. To find out about the role of AtCRG in ethylene signaling, we identified a SALK transfer DNA (T-DNA) insertion line, SO25685 (hereafter Atcrg), which contains a T-DNA insertion into the AtCRG locus and lacks the expression of AtCRG (Fig. 6B). First, we determined if the Atcrg mutant seedlings express the constitutive triple response phenotype that is characteristic of mutants lacking AtCTR1. While the Atctr1-1 mutant constitutively displayed the triple response, the Atcrg mutant exhibited the triple response only in the presence of ACC, as did the wild type and Atedr1-1 mutant plants (Supplemental Fig. S5). This lack of a constitutive ethylene response in the Atcrg mutant suggests that AtCRG is functionally distinct from AtCTR1. Moreover, while the Atctr1-1 mutant exhibits various mutant phenotypes, including dwarfism and delayed flowering, the Atcrg mutant is not detectably different from the wild type in growth habit (data not shown), further suggesting a functional diversification between these two duplicate genes.
Nevertheless, AtCRG is likely to play a role in ethylene signaling because its absence affects the ethylene-inducible expression of the following genes: ERF1, ORGAN SIZE RELATED1 (OSR1), and a pEARL1-like gene, which contains similarity to early Arabidopsis aluminium-induced (pEARL) genes (At4g12470; Hall et al., 2012). The expression levels of these three genes were examined in the absence or presence of ACC treatment in the wild type and Atcrg and Atctr1-1 mutants, as shown in Figure 6C. The comparison between wild-type and Atcrg plants is shown more clearly in Figure 6D (in which the scale is set in the appropriate range). This result indicates that AtERF1 and AtOSR1 transcripts are expressed at basal levels in wild-type plants grown in the absence of ACC but induced to higher levels when plants are sprayed with ACC. These genes are expressed at elevated levels in Atctr1-1 mutant plants and at even higher levels in these plants upon ACC application (Fig. 6C). On the other hand, ACC-dependent induction was observed, but to a reduced extent in Atcrg mutants (Fig. 6D). Although the pEARL-like gene was shown to be highly ethylene inducible in 4-d-old dark-grown seedlings by Hall et al. (2012), its ACC responsiveness appeared to be highly variable (although statistically significant) in the rosette leaves of the 4-week-old plants that were used in this experiment (Fig. 6D). This variable ACC-dependent induction of the pEARL-like gene was possibly reduced in Atcrg mutants, but this observation was not statistically supported (Fig. 6D). Nevertheless, while the pEARL-like gene was overexpressed in Atctr1-1 mutants, such a degree of altered expression pattern was not observed in Atcrg mutants (Fig. 6C). Because the ways in which these transcript levels were misregulated differ between the Atctr1-1 and Atcrg mutants, it seems likely that, although AtCTR1 and AtCRG mediate ethylene signaling through interaction with ethylene receptors, they have evolved to play different roles as a result of the functional diversification that followed gene duplication. It might even suggest that while AtCTR1 negatively regulates ethylene signaling, AtCRG has a positive effect on ethylene signaling with respect to the regulation of downstream gene expression.
Because we had shown that PpCTR1L has dual functions in the ethylene and ABA signaling pathways, we next tested whether ABA signaling might be affected in the Atctr1-1, Atcrg, and Atedr1 mutants. However, we could discern no detectable differences between any of these mutants and the wild type with respect to ABA-responsive transcript induction (Supplemental Fig. S6), suggesting that an ancestral function for CTR1-like proteins in mediating ABA signaling (as seen in P. patens) has been lost during subsequent land-plant evolution. This may have been due to changes in the functions of AtCTR1 and AtCRG themselves, or alternatively, due to modifications in downstream components (with AtCTR1 and AtCRG retaining the ability to mediate ABA signaling). This latter possibility could be tested in future studies by determining if AtCTR1 or AtCRG can complement the Ppctr1l mutant (restore ABA sensitivity).
DISCUSSION AND CONCLUSION
CTR1-Related Proteins Are Ancient Regulatory Proteins That Played a Key Role in Growth Regulation in Response to Ethylene in Land Plants
Although the AtCTR1 protein was previously thought to be functionally unique (Adams-Phillips et al., 2004; Yin et al., 2008), our phylogenetic analyses suggest that both AtCTR1 and AtCRG proteins share ancestry with a P. patens protein, PpCTR1L, and are likely to be the products of a gene duplication event that occurred after the divergence of the bryophytes and the lycophytes (Fig. 1). We also conclude that PpCTR1L is likely to function as a negative regulator of ethylene signaling and to regulate the growth and development of P. patens, just like AtCTR1 in Arabidopsis, because the following results were obtained. (1) PpCTR1L interacts with ethylene receptors (Fig. 2). (2) The removal of PpCTR1L abolishes the P. patens response to ethylene and confers a constitutive ethylene response phenotype (Figs. 3 and 4). (3) PpCTR1L functionally interacts with (a mutated version of) the ethylene receptor Ppetr7-1 to regulate the development and growth of P. patens (Fig. 4). (4) Morphological and RT-PCR data from Ppctr1l and pUbi:Ppetr7-1 transgenic lines suggests that PpETR7 and PpCTR1L have antagonistic effects (Fig. 4). Moreover, PpCTR1L has a dual function, playing roles in both ethylene and ABA signaling pathways (Fig. 5). This dual function is apparently not a property of AtCTR1 or AtCRG (Supplemental Fig. S6). Finally, the ability to interact with ethylene receptors is shared by proteins of the CTR1 family, since all of AtCTR1, AtCRG, and PpCTR1L retain the ability to interact with ethylene receptors, an ability not shared with the sister group of EDR1 proteins (Fig. 6; Supplemental Fig. S2). Thus, CTR1 proteins constitute an ancient mechanism of growth regulation by ethylene by acting directly downstream of ethylene receptors, which is likely to have been present in the land-plant ancestor 450 million years ago. This conclusion is consistent with the recent finding that the ethylene signaling pathway is present in extant charophytic algae and, thus, is likely to have evolved prior to the colonization of the land (Ju et al., 2015).
While PpCTR1L and AtCTR1 were both shown to function downstream of ethylene perception, this study revealed that these homologs differ in a number of ways, allowing us to speculate an evolutionary history of the CTR1 protein family, as illustrated in Figure 7. First, PpCTR1L is a single unique gene in the P. patens genome, whereas two homologs have been identified in Arabidopsis, AtCTR1 and AtCRG. There are indications that AtCTR1 and AtCRG have functionally diverged or specialized, as the dwarf mutant phenotype of Atctr1-1 is not shared by the Atcrg mutant (whose phenotype is indistinguishable from that of the wild type in appearance). The absence of an obvious mutant phenotype in Atcrg mutants may suggest that AtCRG plays a minor role and that its loss can be complemented by AtCTR1 (whereas loss of AtCTR1 cannot be complemented by endogenous AtCRG in Atctr1-1 mutants). However, the fact that the Atcrg mutant showed the altered expression patterns of ethylene-regulated genes (ERF1 and OSR1) may indicate that AtCRG has its own specific function. Future studies will reveal the function of AtCRG and provide further insights into the nature of functional specialization between AtCTR1 and AtCRG.
Figure 7.
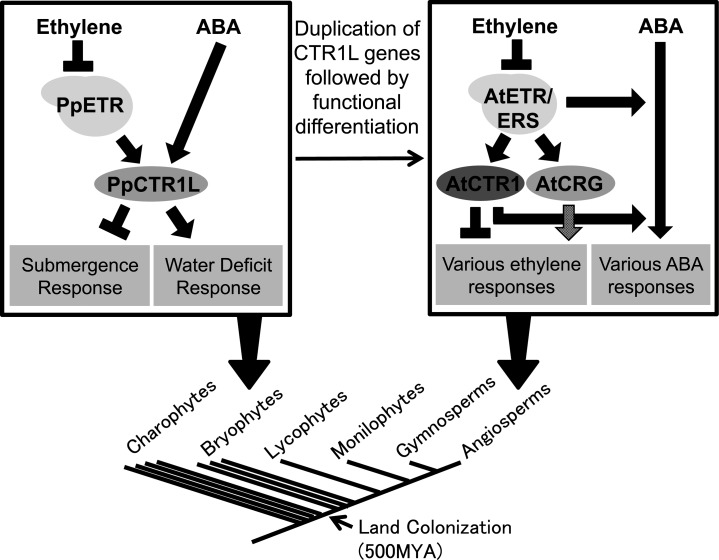
Schematic diagram to explain the hypothesized evolutionary changes. In bryophytes, which diverged at an early stage of land-plant evolution, a single copy of PpCTR1L regulates environmental stress responses by mediating both ethylene and ABA signals. In angiosperm plants, the ethylene signaling pathway diversified through the duplication of signaling components, including CTR1 homologs, followed by their functional differentiation. The way in which the ethylene signaling pathway cross talks with the ABA signaling pathway appears to be different from that in bryophytes. Such evolutionary changes involving the CTR1 protein family may have contributed to optimizing the regulatory mechanisms for stress responses. MYA, Million years ago.
Second, another difference was found in the affinity of CTR1 proteins to ethylene receptors. In the yeast two-hybrid assay, AtCTR1 and AtCRG appeared to have much higher affinities to Arabidopsis ethylene receptors than PpCTR1L to PpETR7. On the other hand, there was also an indication that AtCTR1 and AtCRG had a much reduced affinity toward PpETR7, suggesting that there might have been intraspecific optimization for the Arabidopsis pairs. It should also be noted that the ethylene receptors have diversified independently in the bryophytes and in the angiosperms. While there are two distinct classes (class I and class II) of ethylene receptors in angiosperms, only class I ethylene receptors are present in the P. patens genome (Yasumura et al., 2012). Therefore, there may have been changes in both CTR1 proteins and ethylene receptors of Arabidopsis to optimize protein-protein interactions.
Third, although alternative ethylene signaling pathways that operate independently of AtCTR1 have been suggested in Arabidopsis (Qiu et al., 2012), it is largely thought that the function of AtCTR1 covers the known ethylene response of Arabidopsis. In this study, we observed that the Ppetr7-1 mutation exerted substantial effects through PpCTR1L but some of its effect without involving PpCTR1L. This result indicates that, in P. patens, ethylene receptors may interact with other downstream components as well as with PpCTR1L. It may be possible to speculate that there are multiple signaling pathways that ethylene receptors can regulate in land plants.
In addition, Atctr1-1 mutant phenotypes include dwarfism, reduction in the size of leaves, reduction in shoot height, and delayed flowering (Kieber et al., 1993), which could be summarized as a prolonged life cycle. On the other hand, Ppctr1l mutant lines have an increased induction rate of gametophores, which might be interpreted as an accelerated life cycle, and increased length of gametophores. These mutant phenotypes appeared to be, at least superficially, opposite. Since ethylene was shown to regulate adaptive responses on water relations in both species, such differences may reflect the different strategies they evolved in order to cope with water-related environmental stress.
The most significant finding of this study, however, was that PpCTR1L was shown to be essential for ABA signaling in P. patens (Fig. 5). Intriguingly, while Ppctr1 mutants are completely insensitive to ABA, Atctr1-1 or Atcrg Arabidopsis mutants exhibit ABA responses that are not detectably different from those of the wild type (Supplemental Fig. S6).
Modification of the Ethylene and ABA Signaling Mechanisms Has Accompanied Developmental and Physiological Changes during Land-Plant Evolution
Previously, we proposed that ethylene signaling may have been essential for land colonization by plants (Yasumura et al., 2012). This was because ethylene is involved in adaptive responses to drought and submergence in bryophytes. Similarly, ABA signaling was also suggested to have played a vital role in the evolution of land plants, as it confers bryophyte desiccation tolerance (Komatsu et al., 2013). Both ethylene and ABA have retained these ancestral roles in adaptive responses to water relations throughout land-plant evolution. For example, ethylene was shown to confer rice submergence tolerance (Xu et al., 2006; Hattori et al., 2009; Voesenek and Bailey-Serres, 2013), while ABA confers Arabidopsis seed desiccation tolerance (Maia et al., 2014) and drought tolerance (Finkelstein and Rock, 2002; Verslues and Zhu, 2005; Cutler et al., 2010). On the other hand, following the divergence of the bryophytes, the land plants evolved various adaptive structures specialized for dealing with water-related stress, including xylem for long-distance transport of water, roots and root hairs for water absorption, and stomata in leaves for regulating water loss. Since such evolutionary changes would have required accompanying regulatory changes, they may have led to the modification or acquisition of new pathways in ethylene and ABA signaling. For example, ABA signaling recruited a new branch to its regulatory pathway to regulate seed germination and stomatal closure to minimize water loss after the divergence of the bryophytes (Komatsu et al., 2013). Such an addition of new regulatory branches appeared to be brought about by recruiting new components, ABI5 and SLOW ANION CHANNEL-ASSOCIATED1 (Komatsu et al., 2013). Notably, the mechanism of stomatal closing in response to ABA and increased CO2 concentration was shown to be conserved between bryophytes and angiosperms (Chater et al., 2011), which may suggest that it would have required only a small tweak in a regulatory pathway to link in the water availability, ABA, and stomatal closing responses.
Another possible scenario would be that a signaling pathway diversified through the duplication of signaling components was followed by subsequent functional differentiation. This may have been a way in which the evolution of the CTR1 protein family contributed to optimizing the regulatory mechanisms for stress responses. In the bryophyte P. patens, the single-copy PpCTR1L gene regulates environmental stress responses by mediating both ethylene and ABA signals (Fig. 7). In Arabidopsis, on the other hand, loss-of-function mutations in either AtCTR1 or AtCRG did not lead to abolished ABA responses, unlike those of PpCTR1L in P. patens. Another study, however, has shown that Atctr1-10 or Atctr1-1 mutants displayed reduced ABA sensitivity in seed germination responses (Beaudoin et al., 2000). In addition, a recent study has suggested the involvement of ethylene receptors in modulating ABA signaling in Arabidopsis (Wilson et al., 2014). Taken together, these data suggest that the ethylene pathway cross talks with the ABA pathway through ethylene receptors and AtCTR1 in Arabidopsis (Fig. 7), although their effects on this cross talk are not as strong as is that of their homologs in P. patens. Further investigation, such as a study of Atctr1;crg double mutants, will determine the extent to which AtCTR1 and/or AtCRG influences ABA signaling (as well as ethylene signaling). Together with the fact that AtCTR1 is involved in various ethylene responses that are not applicable in bryophytes, such as root development and fruit ripening, the mechanisms through which the CTR1 proteins exert their effects are different in Arabidopsis and in P. patens. Thus, although at the protein level, PpCTR1L and the two Arabidopsis homologs AtCTR1 and AtCRG share the ancestral function of interaction with ethylene receptors, their roles in development and physiology have diversified during the course of land-plant evolution. Such diversification would be consistent with, and provide a mechanistic understanding of, the highly diverse effects of ethylene, depending on species and environment (Pierik et al., 2006).
By revealing the dual function of PpCTR1L in the ethylene and ABA signaling pathways, this study provides a new insight into the molecular mechanism and evolutionary history of the interaction between ethylene and ABA. In addition, this study adds to our understanding of the molecular mechanisms via which plants have evolved a variety of adaptive responses in dealing with water-related environmental stresses.
MATERIALS AND METHODS
Plant Strains and Growth Conditions
Physcomitrella patens ssp. patens (Gransden2004 strain) was provided by Yasuko Kamisugi (University of Leeds). P. patens cultures were grown at 25°C with continuous light at a light intensity of 50 to 60 µmol m–2 s–1 on the minimum growth medium specified by Ashton and Cove (1977), with or without supplements (10 µm ABA, 3 µL L−1 ethylene, or 0.3 m mannitol) as indicated. Ethylene was applied at 3 µL L−1 with continuous airflow at 1 L m−1 in cuvettes (24 L). Ethylene concentrations in the cuvette were verified by analyzing air samples with a gas chromatograph with a photoionization detector (Syntech Spectras Analyzer GC955-100; Synspec). The growth chamber was kept at 21°C with continuous light of 50 to 60 µmol m–2 s–1. Moss plants were grown from spot inocula of filamentous tissue (1 mm diameter) for morphological observation, and tissues for DNA/RNA extraction were grown from fragmented protonema on a cellophane disc placed on medium. Arabidopsis (Arabidopsis thaliana) plants were either germinated on soil or on one-half-strength Murashige and Skoog (MS) medium following a seed sterilization step and kept at 21°C with a 16-h/8-h light/dark cycle using 100 to 120 µmol m–2 s–1 light. For phytohormone treatment, ACC was added to one-half-strength MS medium at 20 µm and ABA was added at 10 µm, and soil-grown plants were sprayed with 0.5 mm ACC. The SALK T-DNA insertion line SO25685 (Atcrg) was obtained from the Nottingham Arabidopsis Stock Center.
Gametophore Measurement
All leafy shoots were individually picked with forceps at the base from each moss plant and laid down on 1% (w/v) agarose for observation, measurement of the length (height), and counting of the total number of gametophores. Measurements were made using the program ImageJ (http://rsbweb.nih.gov/ij/) on photographs of the leafy shoots. At least four colonies in which 30 to 60 gametophores were found were used to generate data for each plant line. Student’s t test was applied for the statistical test, and differences with P < 0.05 were taken as statistically significant.
Quantification of Filamentous Growth
Images of P. patens were used to segment the whole plant structure and measure tissue density. A shading correction was used to compensate for any uneven illumination across the field, and the image was inverted to provide a positive estimate of tissue density. The plant structure was accentuated using contrast-limited histogram equalization (Zuiderveld, 1994) and automatically segmented using an intensity threshold that minimizes interclass variance between the object and background (Otsu, 1979). The intensity-weighted centroid was used to determine the center of the plant, and the integrated intensity and pixel count were determined in expanding 500-µm rings following background subtraction of the original intensity image or the segmented image, respectively, to provide a profile of the tissue density and surface coverage with radius. For statistical comparison, these values were integrated to give a single measure for the whole colony and subjected to Student’s t test, and differences with P < 0.05 were taken as statistically significant.
Sequence Identification
CTR1-related sequences were obtained from P. patens, Selaginella moellendorffii, and various angiosperm species through BLAST search of the Physcomitrella genome Web site (http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html), the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), and the Selaginella Genomics (http://selaginella.genomics.purdue.edu/) databases. Marchantia polymorpha sequences were obtained from EST collections held by the Kohchi laboratory (University of Kyoto) by BLAST search with the help of Katsuyuki Yamato and Kimitsune Ishizaki.
Phylogenetic Analysis
For the phylogenetic tree shown in Figure 1, the amino acid sequences of the listed CTR1-related sequences (Supplemental Table S1) were aligned using MergeAlign (Collingridge and Kelly, 2012) and subject to phylogenetic inference using maximum likelihood implemented in the FastTree algorithm (Price et al., 2010). A 100-bootstrap replicate maximum likelihood tree was inferred utilizing the Jones-Taylor-Thornton model of amino acid substitution and CAT rate heterogeneity. Bootstrap support values are indicated at internal nodes. Scale bars indicate the number of changes per site. Trees were viewed using the program Dendroscope (Huson et al., 2007). A DendroBLAST phylogenetic tree (Supplemental Fig. S1) was inferred using a wider selection of amino acid sequences (Supplemental Table S2) with a phylogenetic analysis tool, Dendroblast (http://www.dendroblast.com), using the default program parameters (Kelly and Maini, 2013).
Yeast Two-Hybrid Assay
Expression constructs were made with the plasmid pB42AD for the activation domain-fusion proteins with CTR1-related sequences and with the plasmid pLEXA for the DNA binding domain-fusion proteins with ethylene receptor sequences. The kinase domain of the CTR1-related proteins and the transmembrane domain of the ethylene receptor proteins were excluded from the constructs by amplifying the part of these proteins using the primers as specified in Supplemental Table S3. The constructs were introduced to the yeast strain EGY48+pSH18.2. β-Galactosidase assays were performed following the instructions in the Yeast Protocols Handbook (PT3024-1; http://www.clontech.com).
Transformation of P. patens
Construction of the plasmid containing the construct pUbi:Ppetr7-1 and the moss plant pUbi:Ppetr7-1 was as described by Yasumura et al. (2012). The Ppctr1l knockout construct was prepared using a 1,585-bp upstream sequence of PpCTR1L and a 1,580-bp downstream sequence of PpCTR1L, which flanked the p35S:nptII:t35S cassette from 35S-Kan (http://www.pgreen.ac.uk) for selecting transformants using kanamycin or G418 sulfate. Approximately 10 µg of the linearized plasmids containing the Ppctr1l knockout construct or the construct pUbi:Ppetr7-1 was used for each transformation experiment. P. patens was transformed after polyethylene glycol-mediated DNA uptake into protoplasts, essentially according to Schaefer et al. (1991). Protoplasts were regenerated for 5 d on BCD medium and then transferred to fresh medium containing 50 µg mL−1 G418 sulfate or hygromycin B at 16 µg mL−1. After 2 weeks, regenerants were transferred to nonselective BCD medium for another 2 weeks. Stable transformants were subsequently selected by plating once again on selective medium. Transformants were screened by PCR to check for construct integration followed by DNA gel-blot analysis to determine whether they contained multiple copies of the construct (Supplemental Fig. S3).
DNA and RNA Analyses
DNA and RNA were extracted from P. patens essentially according to Langdale et al. (1988). P. patens protonemal tissue was ground in liquid N2 and added to extraction buffer (100 mm Tris, pH 8.6, 1% [v/v] sarkosyl, 4 m guanidine thiocyanate, 25 mm EDTA, pH 8, 25 mm EGTA, pH 8, and 100 mm β-mercaptoethanol) followed by phenol/chloroform treatment. Nucleic acid was precipitated through isopropanol precipitation, and RNA and DNA were separated by lithium chloride precipitation and collected through ethanol precipitation. The detailed protocol is available at http://dps.plants.ox.ac.uk/langdalelab/protocols/RNA/RNA.html. Gel blots (Supplemental Fig. S3) were prepared and hybridized as described by Langdale et al. (1988) using a 1,574-bp fragment of PpCTR1L flanking sequence at the 3′ region, amplified using the primers PpCTR1L3flanF and PpCTR1L3flanR (Supplemental Table S3), as a probe. RNA was extracted from Arabidopsis using Trizol (Life Technologies), according to the manufacturer’s manual.
RT-PCR
Complementary DNA (cDNA) was generated using SuperScript II reverse transcriptase (Life Technologies) from 2 µg of DNaseI (amplification grade; Life Technologies)-treated total RNA with an oligo(dT) primer. For semiquantitative PCR, subsequent PCR was set up with diluted samples of cDNA and Taq polymerase (Life Technologies) according to the manufacturer’s instructions. Primers used in the subsequent PCR are listed in Supplemental Table S3, and PCR conditions were as follows: 94°C for 120 s; 17 to 28 cycles of 94°C for 30 s, 54°C to 57°C for 30 s, and 72°C for 70 s; and then 72°C for 10 min. PCR products were visualized using SYBR Green (Life Technologies), and the cycle number was adjusted to obtain data during the log phase of PCR amplification. The cycle numbers of the log phase were confirmed by checking the amplification status at three different cycle numbers (plus or minus two cycles of the cycle at which the data were taken), as shown in Supplemental Figure S7. The experiments were repeated using biological replicates to confirm that the results were reproducible. For quantitative PCR, PCR was set up with diluted samples of cDNA and KOD SYBR qPCR Mix (Toyobo) according to the manufacturer’s instruction and carried out using the 7300 Applied Biosystem thermocycler. Primers used are listed in Supplemental Table S3, and PCR conditions were as follows: 94°C for 120 s; 45 cycles of 98°C for 10 s, 54°C for 10 s, and 68°C for 30s; followed by a dissociation step. Three technical replicates were prepared for each reaction, and the result was analyzed using the software LinRegPCR version 2012.0 (Ruijter et al., 2009). The computed N0 values, which reflect the initial amounts of template in the reaction mixes, were normalized by the reference transcript UBIQUITIN C. The result was also normalized by another transcript, GAPDH, which encodes glyceraldehyde 3-P, to ensure the suitability of the reference gene.
P. patens Desiccation Test
Six-day-old moss filamentous tissue grown on a cellophane disc was transferred to medium containing 0 or 10 µm ABA. At 24 h after the transfer, the moss tissue was transferred to and maintained in a petri dish containing no growth medium or water source for 3 d. The dried tissue was transferred back to growth medium and observed for growth recovery at day 14 after the transfer.
Triple Response of Arabidopsis
Arabidopsis seeds were surface sterilized with 5% (v/v) bleach and 70% (v/v) ethanol, followed by a water wash. Seeds were placed on one-half-strength MS medium containing 0 or 10 µm ACC and incubated first at 4°C in the dark for 4 d and then at 21°C in the dark for 3 d. Seedlings were observed with a dissection microscope.
Sequence data from this article can be found in the GenBank database under the accession numbers listed in Supplemental Tables S1 and S2 and under the following accession numbers: AtERF1, NM_113225.2; PpERFa, XP_001782760; PpERFb, XP_001767185; PpPIP2;2, AY494192.1; PpPIP2;3, DQ018113.1; PpTUB, AB096719.1; putative PpPYR1, XP_001762113.1; PpABI1A, AB369256.1; PpLEA, AY870926; PpDHNA, AY365466; COLD REGULATED47, NM_101894; AtCOR78, NM_124610; AtOSR1, AY091149; AtpEARL-LIKE, AY093032; AtCOR15a, NM_129815; AtEDH14, NM_001198479; LOW TEMPERATURE INDUCED29, NM_180616; AtLTI30, NM_114957; and ACTIN2, AK317305.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic analysis of CTR1- and EDR-related proteins.
Supplemental Figure S2. Yeast two-hybrid plate assay.
Supplemental Figure S3. Characterization of Ppctr1l lines.
Supplemental Figure S4. Ethylene does not alter ABA response in P. patens.
Supplemental Figure S5. Triple response assay.
Supplemental Figure S6. ABA-inducible gene expression.
Supplemental Figure S7. Full result of semiquantitative RT-PCR analysis with amplification status at +/−2 cycles.
Supplemental Table S1. Accession numbers and description of the sequences used in the phylogenetic analysis shown in Figure 1.
Supplemental Table S2. Accession numbers and description of the sequences used in the phylogenetic analysis shown in Supplemental Figure S1.
Supplemental Table S3. Primers used in this study (5′→3′).
Supplementary Material
Acknowledgments
We thank Yasuko Kamisugi (University of Leeds) for P. patens; Rob Welschen (Utrecht University) and John Baker (University of Oxford) for technical assistance; and Kimitsune Ishizaki (Kobe University), Takayuki Kohchi (University of Kyoto), and Katsuyuki Yamato (Kinki University) for help with BLAST search and sharing of M. polymorpha EST data.
Glossary
- ABA
abscisic acid
- RT
reverse transcription
- T-DNA
transfer DNA
- ACC
1-aminocyclopropane-1-carboxylic acid
- MS
Murashige and Skoog
- cDNA
complementary DNA
Footnotes
This work was supported by the Biotechnological and Biological Science Research Council (grant no. BB/F020759/1) and by St. John’s College, University of Oxford (research support to N.P.H.).
Y.Y. and N.P.H. designed the research; Y.Y. performed most of the experiments; R.P., S.K., M.S., L.A.C.J.V., and N.P.H. supervised the experiments and analyzed the data; Y.Y. and N.P.H. wrote the article with contributions from all the authors.
Articles can be viewed without a subscription.
References
- Abeles FB, Morgan PW, Saltveit MEJ (1992) Ethylene in Plant Biology, Ed 2 Academic Press, San Diego [Google Scholar]
- Adams-Phillips L, Barry C, Kannan P, Leclercq J, Bouzayen M, Giovannoni J (2004) Evidence that CTR1-mediated ethylene signal transduction in tomato is encoded by a multigene family whose members display distinct regulatory features. Plant Mol Biol 54: 387–404 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Arc E, Sechet J, Corbineau F, Rajjou L, Marion-Poll A (2013) ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front Plant Sci 4: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton NW, Cove DJ (1977) The isolation and preliminary characterisation of auxotrophic and analogue resistant mutants of the moss, Physcomitrella patens. Mol Gen Genet 154: 87–95 [Google Scholar]
- Bailey-Serres J, Voesenek LA (2008) Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339 [DOI] [PubMed] [Google Scholar]
- Beaudoin N, Serizet C, Gosti F, Giraudat J (2000) Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12: 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop JJ, Millenaar FF, Smeets ME, van Zanten M, Voesenek LA, Peeters AJ (2007) Abscisic acid antagonizes ethylene-induced hyponastic growth in Arabidopsis. Plant Physiol 143: 1013–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H (1988) Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241: 1086–1089 [DOI] [PubMed] [Google Scholar]
- Cao YR, Chen SY, Zhang JS (2008) Ethylene signaling regulates salt stress response: an overview. Plant Signal Behav 3: 761–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chater C, Kamisugi Y, Movahedi M, Fleming A, Cuming AC, Gray JE, Beerling DJ (2011) Regulatory mechanism controlling stomatal behavior conserved across 400 million years of land plant evolution. Curr Biol 21: 1025–1029 [DOI] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C (1998) Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA 95: 5401–5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingridge PW, Kelly S (2012) MergeAlign: improving multiple sequence alignment performance by dynamic reconstruction of consensus multiple sequence alignments. BMC Bioinformatics 13: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Eklund DM, Thelander M, Landberg K, Ståldal V, Nilsson A, Johansson M, Valsecchi I, Pederson ER, Kowalczyk M, Ljung K, et al. (2010) Homologues of the Arabidopsis thaliana SHI/STY/LRP1 genes control auxin biosynthesis and affect growth and development in the moss Physcomitrella patens. Development 137: 1275–1284 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Rock CD (2002) Abscisic acid biosynthesis and response. The Arabidopsis Book 1: e0058, doi/10.1199/tab.0058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF (2009) Gene duplication and evolutionary novelty in plants. New Phytol 183: 557–564 [DOI] [PubMed] [Google Scholar]
- Frye CA, Innes RW (1998) An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Yeung E, Bailey-Serres J (2011) The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23: 412–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BP, Shakeel SN, Amir M, Ul Haq N, Qu X, Schaller GE (2012) Histidine kinase activity of the ethylene receptor ETR1 facilitates the ethylene response in Arabidopsis. Plant Physiol 159: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, et al. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Hirano K, Nakajima M, Asano K, Nishiyama T, Sakakibara H, Kojima M, Katoh E, Xiang H, Tanahashi T, Hasebe M, et al. (2007) The GID1-mediated gibberellin perception mechanism is conserved in the lycophyte Selaginella moellendorffii but not in the bryophyte Physcomitrella patens. Plant Cell 19: 3058–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM (1995) Ethylene insensitivity conferred by Arabidopsis ERS gene. Science 269: 1712–1714 [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271 [DOI] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R (2007) Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8: 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C, Van de Poel B, Cooper ED, Thierer JH, Gibbons TR, Delwiche CF, Chang C (2015) Conservation of ethylene as a plant hormone over 450 million years of evolution. Nat Plant 1: 14004. [DOI] [PubMed] [Google Scholar]
- Ju C, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML, et al. (2012) CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci USA 109: 19486–19491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisugi Y, Cuming AC (2005) The evolution of the abscisic acid-response in land plants: comparative analysis of group 1 LEA gene expression in moss and cereals. Plant Mol Biol 59: 723–737 [DOI] [PubMed] [Google Scholar]
- Kelly S, Maini PK (2013) DendroBLAST: approximate phylogenetic trees in the absence of multiple sequence alignments. PLoS ONE 8: e58537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal A, Cho SH, Marella H, Sakata Y, Perroud PF, Pan A, Quatrano RS (2010) Role of ABA and ABI3 in desiccation tolerance. Science 327: 546. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Komatsu K, Nishikawa Y, Ohtsuka T, Taji T, Quatrano RS, Tanaka S, Sakata Y (2009) Functional analyses of the ABI1-related protein phosphatase type 2C reveal evolutionarily conserved regulation of abscisic acid signaling between Arabidopsis and the moss Physcomitrella patens. Plant Mol Biol 70: 327–340 [DOI] [PubMed] [Google Scholar]
- Komatsu K, Suzuki N, Kuwamura M, Nishikawa Y, Nakatani M, Ohtawa H, Takezawa D, Seki M, Tanaka M, Taji T, et al. (2013) Group A PP2Cs evolved in land plants as key regulators of intrinsic desiccation tolerance. Nat Commun 4: 2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale JA, Rothermel BA, Nelson T (1988) Cellular pattern of photosynthetic gene expression in developing maize leaves. Genes Dev 2: 106–115 [DOI] [PubMed] [Google Scholar]
- Lin Z, Alexander L, Hackett R, Grierson D (2008) LeCTR2, a CTR1-like protein kinase from tomato, plays a role in ethylene signalling, development and defence. Plant J 54: 1083–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia J, Dekkers BJ, Dolle MJ, Ligterink W, Hilhorst HW (2014) Abscisic acid (ABA) sensitivity regulates desiccation tolerance in germinated Arabidopsis seeds. New Phytol 203: 81–93 [DOI] [PubMed] [Google Scholar]
- Marella HH, Sakata Y, Quatrano RS (2006) Characterization and functional analysis of ABSCISIC ACID INSENSITIVE3-like genes from Physcomitrella patens. Plant J 46: 1032–1044 [DOI] [PubMed] [Google Scholar]
- McManus MT. (2012) Annual Plant Reviews, The Plant Hormone Ethylene, Vol 44 Wiley-Blackwell, Hoboken, NJ [Google Scholar]
- Otsu N. (1979) A threshold selection method from gray-level histograms. IEEE Trans Systems Man Cybernetics 9: 62–66 [Google Scholar]
- Paponov IA, Teale W, Lang D, Paponov M, Reski R, Rensing SA, Palme K (2009) The evolution of nuclear auxin signalling. BMC Evol Biol 9: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, Tholen D, Poorter H, Visser EJ, Voesenek LA (2006) The Janus face of ethylene: growth inhibition and stimulation. Trends Plant Sci 11: 176–183 [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP (2010) FastTree 2: approximately maximum-likelihood trees for large alignments. PLoS ONE 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Lavy M, Ashton NW, Estelle M (2010) Physcomitrella patens auxin-resistant mutants affect conserved elements of an auxin-signaling pathway. Curr Biol 20: 1907–1912 [DOI] [PubMed] [Google Scholar]
- Qiao H, Shen Z, Huang SS, Schmitz RJ, Urich MA, Briggs SP, Ecker JR (2012) Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338: 390–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Xie F, Yu J, Wen CK (2012) Arabidopsis RTE1 is essential to ethylene receptor ETR1 amino-terminal signaling independent of CTR1. Plant Physiol 159: 1263–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA perception and signalling. Trends Plant Sci 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Roman G, Lubarsky B, Kieber JJ, Rothenberg M, Ecker JR (1995) Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: five novel mutant loci integrated into a stress response pathway. Genetics 139: 1393–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanel EA, Schrago CG, Couñago RM, Russo CA, Alves-Ferreira M (2009) Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS ONE 4: e5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra L, Svensson J, Carballo V, Izmendi D, Welin B, Vidal S (2006) A dehydrin gene in Physcomitrella patens is required for salt and osmotic stress tolerance. Plant J 45: 237–249 [DOI] [PubMed] [Google Scholar]
- Sakata Y, Nakamura I, Taji T, Tanaka S, Quatrano RS (2010) Regulation of the ABA-responsive Em promoter by ABI3 in the moss Physcomitrella patens: role of the ABA response element and the RY element. Plant Signal Behav 5: 1061–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer D, Zryd JP, Knight CD, Cove DJ (1991) Stable transformation of the moss Physcomitrella patens. Mol Gen Genet 226: 418–424 [DOI] [PubMed] [Google Scholar]
- Sharp RE, LeNoble ME (2002) ABA, ethylene and the control of shoot and root growth under water stress. J Exp Bot 53: 33–37 [PubMed] [Google Scholar]
- Shen X, Liu H, Yuan B, Li X, Xu C, Wang S (2011) OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis. Plant Cell Environ 34: 179–191 [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Alonso JM (2009) Ethylene signaling and response: where different regulatory modules meet. Curr Opin Plant Biol 12: 548–555 [DOI] [PubMed] [Google Scholar]
- Subbiah V, Reddy KJ (2010) Interactions between ethylene, abscisic acid and cytokinin during germination and seedling establishment in Arabidopsis. J Biosci 35: 451–458 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K (2010) Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol 51: 1821–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Zhu JK (2005) Before and beyond ABA: upstream sensing and internal signals that determine ABA accumulation and response under abiotic stress. Biochem Soc Trans 33: 375–379 [DOI] [PubMed] [Google Scholar]
- Voesenek LA, Bailey-Serres J (2013) Flooding tolerance: O2 sensing and survival strategies. Curr Opin Plant Biol 16: 647–653 [DOI] [PubMed] [Google Scholar]
- Voesenek LA, Sasidharan R (2013) Ethylene—and oxygen signalling—drive plant survival during flooding. Plant Biol (Stuttg) 15: 426–435 [DOI] [PubMed] [Google Scholar]
- Wang F, Cui X, Sun Y, Dong CH (2013) Ethylene signaling and regulation in plant growth and stress responses. Plant Cell Rep 32: 1099–1109 [DOI] [PubMed] [Google Scholar]
- Wilkinson S, Davies WJ (2010) Drought, ozone, ABA and ethylene: new insights from cell to plant to community. Plant Cell Environ 33: 510–525 [DOI] [PubMed] [Google Scholar]
- Wilson RL, Kim H, Bakshi A, Binder BM (2014) The ethylene receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 have contrasting roles in seed germination of Arabidopsis during salt stress. Plant Physiol 165: 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
- Yasumura Y, Crumpton-Taylor M, Fuentes S, Harberd NP (2007) Step-by-step acquisition of the gibberellin-DELLA growth-regulatory mechanism during land-plant evolution. Curr Biol 17: 1225–1230 [DOI] [PubMed] [Google Scholar]
- Yasumura Y, Pierik R, Fricker MD, Voesenek LA, Harberd NP (2012) Studies of Physcomitrella patens reveal that ethylene-mediated submergence responses arose relatively early in land-plant evolution. Plant J 72: 947–959 [DOI] [PubMed] [Google Scholar]
- Yin XR, Chen KS, Allan AC, Wu RM, Zhang B, Lallu N, Ferguson IB (2008) Ethylene-induced modulation of genes associated with the ethylene signalling pathway in ripening kiwifruit. J Exp Bot 59: 2097–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho Y, Sheen J (2009) Emerging connections in the ethylene signaling network. Trends Plant Sci 14: 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yotsui I, Saruhashi M, Kawato T, Taji T, Hayashi T, Quatrano RS, Sakata Y (2013) ABSCISIC ACID INSENSITIVE3 regulates abscisic acid-responsive gene expression with the nuclear factor Y complex through the ACTT-core element in Physcomitrella patens. New Phytol 199: 101–109 [DOI] [PubMed] [Google Scholar]
- Zhong S, Lin Z, Grierson D (2008) Tomato ethylene receptor-CTR interactions: visualization of NEVER-RIPE interactions with multiple CTRs at the endoplasmic reticulum. J Exp Bot 59: 965–972 [DOI] [PubMed] [Google Scholar]
- Zou C, Lehti-Shiu MD, Thomashow M, Shiu SH (2009) Evolution of stress-regulated gene expression in duplicate genes of Arabidopsis thaliana. PLoS Genet 5: e1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuiderveld K. (1994) Contrast limited adaptive histograph equalization. In Heckbert PS, ed, Graphics Gems IV. Academic Press Professional, San Diego, CA, pp 474–485 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.