The three GTP-binding proteins function within the heterotrimeric G protein complex, increasing the diversity of possible heterotrimer combinations.
Abstract
Heterotrimeric G proteins, consisting of Gα, Gβ, and Gγ subunits, are a conserved signal transduction mechanism in eukaryotes. However, G protein subunit numbers in diploid plant genomes are greatly reduced as compared with animals and do not correlate with the diversity of functions and phenotypes in which heterotrimeric G proteins have been implicated. In addition to GPA1, the sole canonical Arabidopsis (Arabidopsis thaliana) Gα subunit, Arabidopsis has three related proteins: the extra-large GTP-binding proteins XLG1, XLG2, and XLG3. We demonstrate that the XLGs can bind Gβγ dimers (AGB1 plus a Gγ subunit: AGG1, AGG2, or AGG3) with differing specificity in yeast (Saccharomyces cerevisiae) three-hybrid assays. Our in silico structural analysis shows that XLG3 aligns closely to the crystal structure of GPA1, and XLG3 also competes with GPA1 for Gβγ binding in yeast. We observed interaction of the XLGs with all three Gβγ dimers at the plasma membrane in planta by bimolecular fluorescence complementation. Bioinformatic and localization studies identified and confirmed nuclear localization signals in XLG2 and XLG3 and a nuclear export signal in XLG3, which may facilitate intracellular shuttling. We found that tunicamycin, salt, and glucose hypersensitivity and increased stomatal density are agb1-specific phenotypes that are not observed in gpa1 mutants but are recapitulated in xlg mutants. Thus, XLG-Gβγ heterotrimers provide additional signaling modalities for tuning plant G protein responses and increase the repertoire of G protein heterotrimer combinations from three to 12. The potential for signal partitioning and competition between the XLGs and GPA1 is a new paradigm for plant-specific cell signaling.
The classical heterotrimeric G protein consists of a GDP/GTP-binding Gα subunit with GTPase activity bound to an obligate dimer formed by Gβ and Gγ subunits. In the signaling paradigm largely elucidated from mammalian systems, the plasma membrane-associated heterotrimer contains Gα in its GDP-bound form. Upon receiving a molecular signal, typically transduced by a transmembrane protein (e.g. a G protein-coupled receptor), Gα exchanges GDP for GTP and dissociates from the Gβγ dimer. Both Gα and Gβγ interact with intracellular effectors to initiate downstream signaling cascades. The intrinsic GTPase activity of Gα restores Gα to the GDP-bound form, which binds Gβγ, thereby reconstituting the heterotrimer (McCudden et al., 2005; Oldham and Hamm, 2008).
Signal transduction through a heterotrimeric G protein complex is an evolutionarily conserved eukaryotic mechanism common to metazoa and plants, although there are distinct differences in the functional intricacies between the evolutionary branches (Jones et al., 2011a, 2011b; Bradford et al., 2013). The numbers of each subunit encoded within genomes, and therefore the potential for combinatorial complexity within the heterotrimer, is one of the most striking differences between plants and animals. For example, the human genome encodes 23 Gα (encoded by 16 genes), five Gβ, and 12 Gγ subunits (Hurowitz et al., 2000; McCudden et al., 2005; Birnbaumer, 2007). The Arabidopsis (Arabidopsis thaliana) genome, however, only encodes one canonical Gα (GPA1; Ma et al., 1990), one Gβ (AGB1; Weiss et al., 1994), and three Gγ (AGG1, AGG2, and AGG3) subunits (Mason and Botella, 2000, 2001; Chakravorty et al., 2011), while the rice (Oryza sativa) genome encodes one Gα (Ishikawa et al., 1995), one Gβ (Ishikawa et al., 1996), and either four or five Gγ subunits (Kato et al., 2004; Chakravorty et al., 2011; Botella, 2012). As expected, genomes of polyploid plants have more copies due to genome duplication, with the soybean (Glycine max) genome encoding four Gα, four Gβ (Bisht et al., 2011), and 10 Gγ subunits (Choudhury et al., 2011). However, Arabidopsis heterotrimeric G proteins have been implicated in a surprisingly large number of phenotypes, which is seemingly contradictory given the relative scarcity of subunits. Arabidopsis G proteins have been implicated in cell division (Ullah et al., 2001; Chen et al., 2006) and morphological development in various tissues, including hypocotyls (Ullah et al., 2001, 2003), roots (Ullah et al., 2003; Chen et al., 2006; Li et al., 2012), leaves (Lease et al., 2001; Ullah et al., 2001), inflorescences (Ullah et al., 2003), and flowers and siliques (Lease et al., 2001), as well as in pathogen responses (Llorente et al., 2005; Trusov et al., 2006; Cheng et al., 2015), regulation of stomatal movement (Wang et al., 2001; Coursol et al., 2003; Fan et al., 2008) and development (Zhang et al., 2008; Nilson and Assmann, 2010), cell wall composition (Delgado-Cerezo et al., 2012), responses to various light stimuli (Warpeha et al., 2007; Botto et al., 2009), responses to multiple abiotic stimuli (Huang et al., 2006; Pandey et al., 2006; Trusov et al., 2007; Zhang et al., 2008; Colaneri et al., 2014), responses to various hormones during germination (Ullah et al., 2002), and postgermination development (Ullah et al., 2002; Pandey et al., 2006; Trusov et al., 2007). Since the Gγ subunit appeared to be the only subunit that provides diversity in heterotrimer composition in Arabidopsis, it was proposed that all functional specificity in heterotrimeric G protein signaling was provided by the Gγ subunit (Trusov et al., 2007; Chakravorty et al., 2011; Thung et al., 2012, 2013). This allowed for only three heterotrimer combinations to account for the wide range of G protein-associated phenotypes.
In addition to the above typical G protein subunits, the plant kingdom contains a conserved protein family of extra-large GTP-binding proteins (XLGs). XLGs differ from typical Gα subunits in that they possess a long N-terminal extension of unknown function, but they are similar in that they all have a typical C-terminal Gα-like region, with five semiconserved G-box (G1–G5) motifs. The XLGs also possess the two sequence features that differentiate heterotrimeric G protein Gα subunits from monomeric G proteins: a helical region between the G1 and G2 motifs and an Asp/Glu-rich loop between the G3 and G4 motifs (Lee and Assmann, 1999; Ding et al., 2008; Heo et al., 2012). The Arabidopsis XLG family comprises XLG1, XLG2, and XLG3, and all three have demonstrated GTP-binding and GTPase activities, although they differ from GPA1 in exhibiting a much slower rate of GTP hydrolysis, with a Ca2+ cofactor requirement instead of an Mg2+ requirement, as for canonical Gα proteins (Heo et al., 2012). All three Arabidopsis XLGs were observed to be nuclear localized (Ding et al., 2008). Although much less is known about XLGs than canonical Gα subunits, XLG2 positively regulates resistance to the bacterial pathogen Pseudomonas syringae and was immunoprecipitated with AGB1 from tissue infected with P. syringae (Zhu et al., 2009). xlg3 mutants, like agb1 mutants, are impaired in root-waving and root-skewing responses (Pandey et al., 2008). During the preparation of this report, Maruta et al. (2015) further investigated XLG2, particularly focusing on the link between XLG2 and Gβγ in pathogen responses. Based on symptom progression in xlg mutants, they found that XLG2 is a positive regulator of resistance to both bacterial and fungal pathogens, with a minor contribution from XLG3 in resistance to Fusarium oxysporum. XLG2 and XLG3 are also positive regulators of reactive oxygen species (ROS) production in response to pathogen-associated molecular pattern elicitors. The resistance and pathogen-associated molecular pattern-induced ROS phenotypes of the agg1 agg2 and xlg2 xlg3 double mutants were not additive in an agg1 agg2 xlg2 xlg3 quadruple mutant, indicating that these two XLGs and the two Gγ subunits function in the same, rather than parallel, pathways. Unfortunately, the close proximity of XLG2 and AGB1 on chromosome 4 precluded the generation of an agb1 xlg2 double mutant; therefore, direct genetic evidence of XLG2 and AGB1 interaction is still lacking, but physical interactions between XLG2 and the Gβγ dimers were shown by yeast (Saccharomyces cerevisiae) three-hybrid and bimolecular fluorescence complementation (BiFC) assays (Maruta et al., 2015). Localization of all three XLGs was also reexamined, indicating that XLGs are capable of localizing to the plasma membrane in addition to the nucleus (Maruta et al., 2015).
Interestingly, several other plant G protein-related phenotypes, in addition to pathogen resistance, have been observed only in Gβ and Gγ mutants, with opposite phenotypes observed in Gα (gpa1) mutants. Traditionally, the observation of opposite phenotypes in Gα versus Gβγ mutants in plants and other organisms has mechanistically been attributed to signaling mediated by free Gβγ, which increases in abundance in the absence of Gα. However, an intriguing alternative is that XLG proteins fulfill a Gα-like role in forming heterotrimeric complexes with Gβγ and function in non-GPA1-based G protein signaling processes. If XLGs function like Gα subunits, the corresponding increase in subunit diversity could potentially account for the diversity of G protein phenotypes. In light of this possibility, we assessed the heterotrimerization potential of all possible XLG and Gβγ dimer combinations, XLG localization and its regulation by Gβγ, and the effect of xlg mutation on selected known phenotypes associated with heterotrimeric G proteins. Our results provide compelling evidence for the formation of XLG-Gβγ heterotrimers and reveal that plant G protein signaling is substantially more complex than previously thought.
RESULTS
XLG-Gβγ Interaction in Vivo
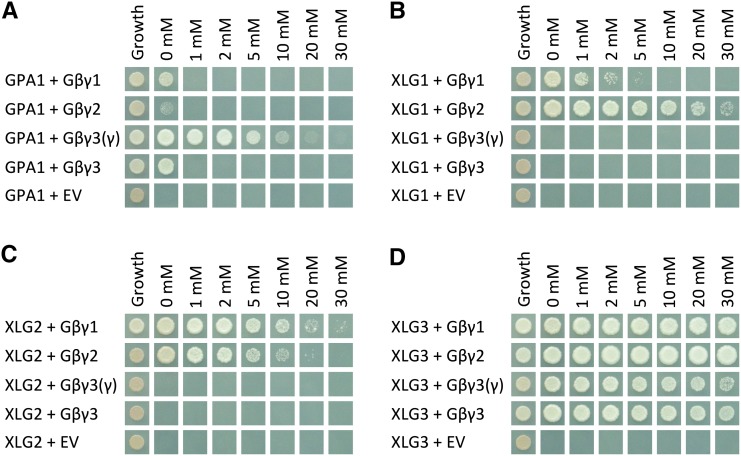
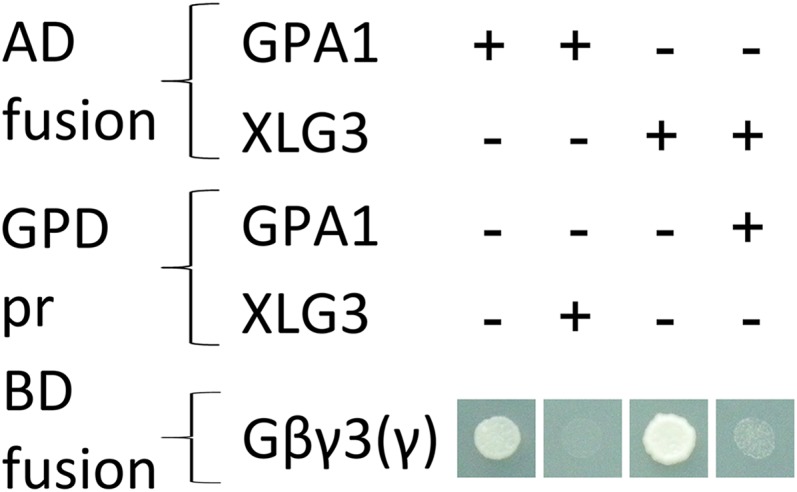
Maruta et al. (2015) previously reported that XLG2 interacts with AGB1/AGG1, AGB1/AGG2, and AGB1/AGG3 in yeast three-hybrid assays, but they did not test the ability of XLG1 or XLG3 to interact with any of the Gβγ dimers. We tested all three XLGs against all three Gβγ dimers. Evidence for a transmembrane domain in AGG3, which would place the C terminus of AGG3 outside of the cell (Wolfenstetter et al., 2015), was taken into consideration by including an additional AGB1/AGG3 construct [designated Gβγ3(γ)], in which only the Gγ-like domain (residues 1–112) of AGG3 was fused to the GAL4 binding domain. We also investigated interaction strength within the complexes, wherein growth on higher concentrations of 3-amino-1,2,4-triazole (3-AT) is an accepted indicator of a stronger or more stable interaction (Durfee et al., 1993; Ursic et al., 2004).
The interaction between GPA1 and Gβγ dimers containing AGG1 or AGG2 was quite weak, with yeast growth observed only in the absence of 3-AT. The only dimer with which GPA1 showed a strong interaction was AGB1/AGG3(γ) (Fig. 1A). XLG1 and XLG2 interaction with Gβγ dimers containing AGG1 or AGG2 (Fig. 1, B and C) was stronger than that of GPA1. XLG1 and XLG2 did not interact in yeast with Gβγ dimers containing AGG3 (Fig. 1, B and C); however, XLG3 interacted strongly with all Gβγ dimers, including those containing AGG3 (Fig. 1D). Therefore, we found strong evidence from this approach for the interaction of eight of the 12 heterotrimeric complexes tested.
Figure 1.
Yeast three-hybrid assays demonstrate the specificity of GPA1 for AGB1/AGG3, XLG1 and XLG2 for AGB1/AGG1 and AGB1/AGG2, and XLG3 for all three Gβγ dimers. Yeast three-hybrid assays tested the interactions between GPA1 (A), XLG1 (B), XLG2 (C), and XLG3 (D; GAL4 activation domain fusions) with the AGB1/AGG1 (Gβγ1), AGB1/AGG2 (Gβγ2), and AGB1/AGG3 (Gβγ3) Gβγ dimers (Gγ fused to the GAL4 binding domain, with Gβ [AGB1] as the bridge protein). A truncated γ3(γ) was also tested, consisting of only the Gγ-like domain of AGG3 (residues 1–112), lacking the implicated transmembrane domain. Yeast growth on synthetic complete (SC)-Trp-Leu confirmed transformation and cell viability. Interactions were assayed on SC-Trp-Leu-Met-His supplemented with the indicated concentrations of 3-AT (0–30 mm). EV, Empty vector.
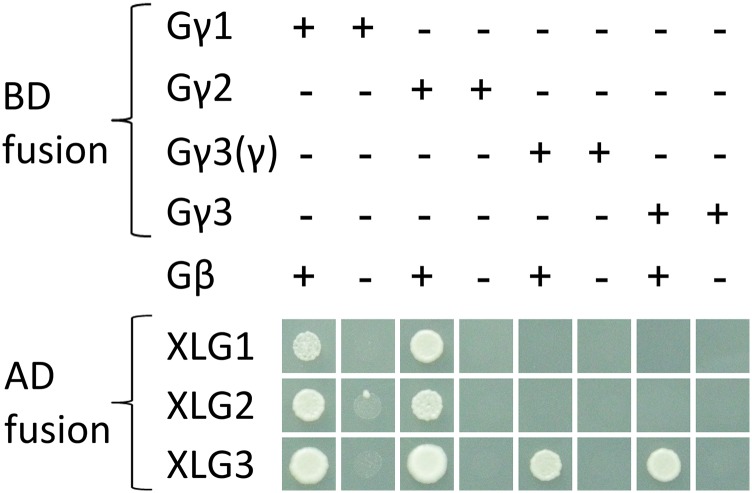
In the above assays, the XLGs or GPA1 were fused to the GAL4 activation domain, the Gγ subunits (AGG1, AGG2, or AGG3) were fused to the GAL4 binding domain (multiple cloning site 1 [MCS1] of the yeast three-hybrid vector pBridge), and AGB1 was used as the bridge protein, expressed from MCS2 of pBridge. Therefore, it was possible that the yeast reporter genes were activated by direct interaction between each XLG and each Gγ subunit. To assess whether the yeast three-hybrid interactions do in fact involve a heterotrimeric interaction (i.e. with a requirement for AGB1), the AGB1 bridge protein was not included, and the assay was repeated. As shown in Figure 2, coexpression of the Gβ subunit, AGB1, was required for interaction. These results raised the hypothesis that GPA1 and the XLGs may compete with each other for Gβγ binding. Since XLG1 and XLG2 did not bind the same Gβγ dimers as GPA1 in yeast (Fig. 1), XLG3 and AGB1/AGG3(γ) were selected to test this hypothesis. Either GPA1 or XLG3 was expressed under the control of the strong GPD promoter (Bitter and Egan, 1984; Partow et al., 2010), in competition with the GAL4:XLG3 or GAL4:GPA1 activation domain fusion, respectively. XLG3 was able to inhibit GPA1-AGB1/AGG3 interaction, and GPA1 was able to inhibit XLG3-AGB1/AGG3 interaction (Fig. 3).
Figure 2.
The interaction of XLGs with Gγ subunits is AGB1 dependent. Yeast three-hybrid assays tested the interactions of GPA1, XLG1, XLG2, and XLG3 with the AGG1 (Gγ1), AGG2 (Gγ2), and AGG3 (Gγ3) subunits. Interactions were assayed in the presence and absence of AGB1. A truncated γ3(γ) subunit was also included, which includes only the Gγ-like domain of AGG3 (residues 1–112). All interactions were assayed by growth on SC-Trp-Leu-Met-His supplemented with 2 mm 3-AT. Two millimolars of 3-AT was used because the pBridge-AGG1 construct exhibited autoactivation (i.e. growth in combination with the empty vector) on SC-Trp-Leu-Met-His supplemented with 0 or 1 mm 3-AT.
Figure 3.
GPA1 and XLG3 compete with each other for binding of AGB1/AGG3 in yeast. Yeast three-hybrid competition assays tested the interaction between GPA1 and AGB1/AGG3(γ) [Gβγ3(γ)] with or without additional expression of XLG3 and the interaction between XLG3 and AGB1/AGG3(γ) [Gβγ3(γ)] with or without additional expression of GPA1. The fourth protein (XLG3 or GPA1, respectively) was expressed under the control of the strong GPD promoter (pr). All interactions were assayed by growth on SC-Trp-Leu-Met-His supplemented with 2 mm 3-AT.
XLG Three-Dimensional Structure Analysis in Silico
The ability of all three XLGs to interact with Gβγ dimers, and of XLG3 to compete with GPA1 in yeast, suggests that all four G proteins are structurally similar. As seen in the alignment of Ding et al. (2008), primary sequence conservation between GPA1 and the XLG proteins is moderate, with percentage identities of GPA1 and the XLG Gα domains of 26.1%, 23.2%, and 28.5% for XLG1446-888, XLG2435-861, and XLG3396-848, respectively. Therefore, we performed an in silico analysis of each XLG using Phyre2 (Kelley and Sternberg, 2009), which generates a protein fold profile for subsequent analysis by the secondary structure predictor Psi-Pred (Jones, 1999b). Phyre2 then identifies similar tertiary structures in a fold library via profile-profile alignments (Kelley and Sternberg, 2009). The top 20 in silico structural matches for all three XLGs corresponded to heterotrimeric G protein Gα subunits, including GPA1 (Supplemental Table S1), with 99.9% to 100% confidence, and the conserved secondary structure patterning of the XLGs and GPA1 is evident when the Phyre2 data are superimposed over an alignment of their Gα regions (Supplemental Fig. S1). As an independent secondary confirmation, the genTHREADER software was used to conduct additional fold recognition analyses (Jones, 1999a). genTHREADER returned 13 hits for each of the three XLGs that were classed as either certain (P < 0.0001) or high (P = 0.0001–0.001), all of which corresponded to the same 13 heterotrimeric G protein Gα structures, including that of GPA1 (Supplemental Table S2). In contrast to the high-confidence matching of the C-terminal domains of the XLGs with canonical Gα subunit structures, there were no high-ranking hits that matched the N-terminal domains of the XLGs.
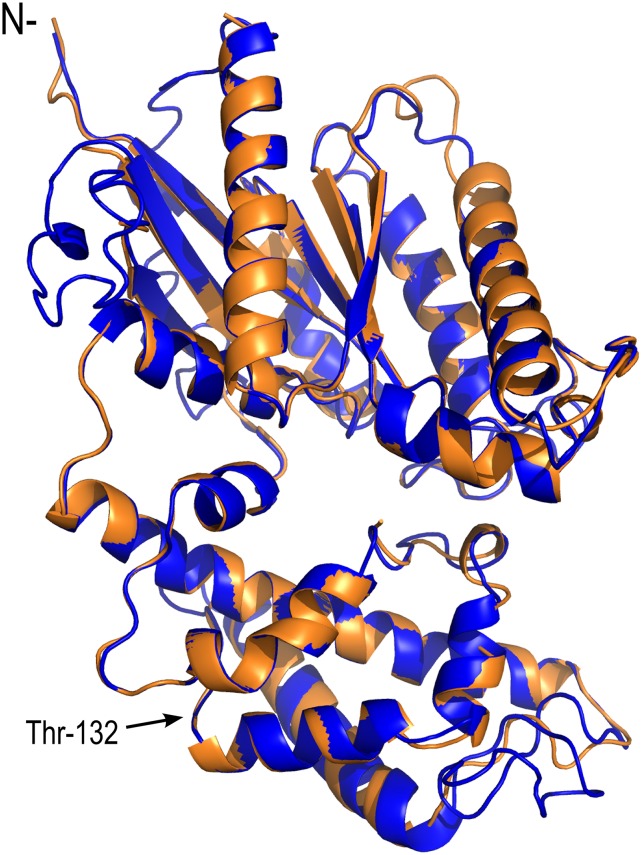
We next examined the Gα region of XLG3 in greater detail by utilizing the I-TASSER server for protein structure modeling, assigning the empirically resolved crystal structure of GPA1 (Protein Data Bank no. 2XTZ:A; Jones et al., 2011a) as a template. For reference, the XLG3 Gα region encompasses residues 429 to 849, with residue 429 aligning to GPA1 residue 37 (Supplemental Fig. S1). To allow direct correlation, the exogenous MSGI tetrapeptide found at the N terminus of the GPA1 2XTZ crystal structure (Jones, 1999a) was computationally affixed to the XLG3 Gα domain; therefore, the first XLG3 native residue corresponds to residue 5 in the XLG3alpha.pdb structure (Supplemental File S1). The predicted XLG3 structure aligned closely to the crystal structure of GPA1 (Fig. 4; Supplemental Fig. S2). The theoretical model of the XLG3 Gα domain was validated as a reasonable approximation of the as yet undescribed XLG3 crystal structure, with a passing self-compatibility score (82.55% of residue 3D–1D [three dimension-one dimension] scores ≥ 0.2) from Verify-3D (Eisenberg et al., 1997), a ProSA Z-score of −7.2 (Supplemental Fig. S3; Wiederstein and Sippl, 2007), and a plausible overall quality score of 83.894 (Supplemental Fig. S4) from ERRAT (Colovos and Yeates, 1993), which is designed to verify protein structures empirically determined from crystallography data. Together, the Phyre2 and genTHREADER identification of structural homology to metazoan and plant Gα subunits, and the close alignment of the XLG3 and GPA1 Gα regions by I-TASSER, support XLG function as a component of the plant heterotrimeric G protein.
Figure 4.
The XLG3 Gα-like region aligns closely to the crystal structure of GPA1. Superimposition of computationally derived XLG3 structural features on the empirically derived GPA1 crystal structure demonstrates their shared three-dimensional characteristics. Blue indicates the XLG3 Gα-like region, orange indicates the GPA1 Gα-like region, and N- indicates the N terminus of the Gα regions. Thr-132 marks the NVen210 insertion site in the αB-αC loop of GPA1 used for BiFC (Gookin and Assmann, 2014): each of the XLGs was modified at the analogous residue.
XLG-Gβγ Interaction in Planta
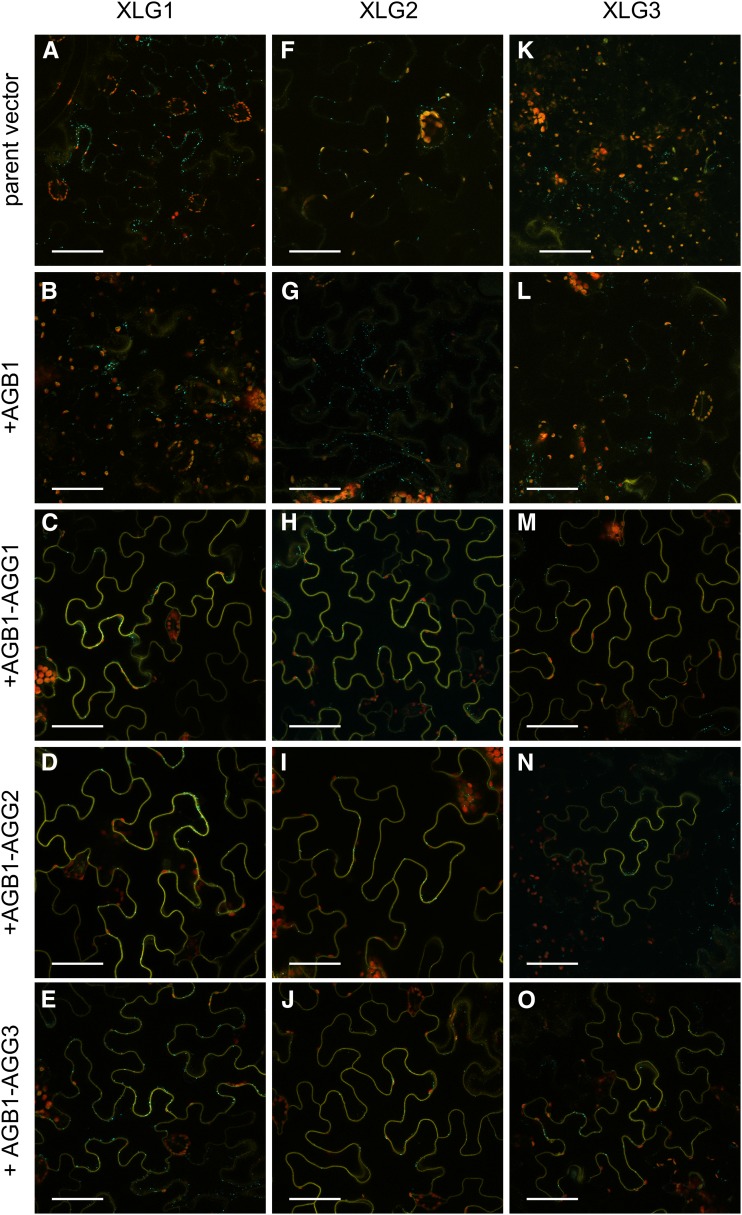
To further test the hypothesis that XLGs function as Gα subunits, we utilized our recently developed multicassette pDOE vector BiFC system (Gookin and Assmann, 2014) with reduced nonspecific signal from self-assembly (Ohashi et al., 2012; Gookin and Assmann, 2014) to evaluate the XLG-Gβγ interactions in planta and assess subcellular localization. GPA1 interacts with AGB1-AGG dimers (Gookin and Assmann, 2014) when tagged internally, in a configuration consistent with a mammalian heterotrimer crystal structure (McCudden et al., 2005). Given the results of our yeast three-hybrid competition assay, we hypothesized that the XLGs and GPA1 would interact similarly with AGB1-AGG dimers. Accordingly, we inserted the NmVenus210 fragment into the middle of each XLG protein at a position analogous to the αB-αC loop of GPA1 (Fig. 4) to generate XLG1L, XLG2L, and XLG3L parent vectors, to which AGB1 was added to create three XLGL-CVen210:AGB1 test vectors. Negative control assays with the XLGL parent vectors (with an unfused CVen210 fragment in the second cassette) did not show any nonspecific BiFC signal (Fig. 5, A, F, and K). The XLGL-CVen210:AGB1 test constructs also showed zero to near zero signal (Fig. 5, B, G, and L). In all cases, positive transformation was confirmed by a vector-integrated XT-Golgi-mTq2 marker. Positive interaction between each XLGL fusion and AGB1 was only obtained when exogenous untagged AGG1 (Fig. 5, C, H, and M), AGG2 (Fig. 5, D, I, and N), or AGG3 (Fig. 5, E, J, and O) subunits were provided via cotransformation with a second vector. Population level images show the reproducibility of these assays (Supplemental Fig. S5).
Figure 5.
XLG1, XLG2, and XLG3 interact with AGB1 at the plasma membrane in an AGG-dependent manner. In all assays, positive transformation is confirmed by Golgi-localized mTurquoise2 (mTq2) fluorescence from the pDOE XT-Golgi-mTq2 marker. A, The XLG1L-CVen210 parent vector does not produce nonspecific BiFC signal. B, The XLG1L-CVen210:AGB1 construct shows zero to near-zero signal in the absence of a coexpressed Gγ subunit. C, The XLG1L-CVen210:AGB1 construct produces BiFC signal in the presence of coexpressed AGG1. D, The XLG1L-CVen210:AGB1 construct produces BiFC signal in the presence of coexpressed AGG2. E, The XLG1L-CVen210:AGB1 construct produces BiFC signal in the presence of coexpressed AGG3. F, The XLG2L-CVen210 parent vector does not produce nonspecific BiFC signal. G, The XLG2L-CVen210:AGB1 construct shows zero to near-zero signal in the absence of a coexpressed Gγ subunit. H, The XLG2L-CVen210:AGB1 construct produces BiFC signal in the presence of coexpressed AGG1. I, The XLG2L-CVen210:AGB1 construct produces BiFC signal in the presence of coexpressed AGG2. J, The XLG2L-CVen210:AGB1 construct produces BiFC signal in the presence of coexpressed AGG3. K, The XLG3L-CVen210 parent vector does not produce nonspecific BiFC signal. L, The XLG3L-CVen210:AGB1 construct shows zero to near-zero signal in the absence of a coexpressed Gγ subunit. M, The XLG3L-CVen210:AGB1 construct produces BiFC signal in the presence of coexpressed AGG1. N, The XLG3L-CVen210:AGB1 construct produces BiFC signal in the presence of coexpressed AGG2. O, The XLG3L-CVen210:AGB1 construct produces BiFC signal in the presence of coexpressed AGG3. Vectors were agroinfiltrated into N. benthamiana leaves at an optical density at 600 nm (OD600) of 0.0075 to 0.01, and images were acquired 57 to 60 h post infiltration. Yellow indicates mVenus BiFC, blue indicates mTq2, and red indicates chlorophyll autofluorescence. Bars = 50 μm.
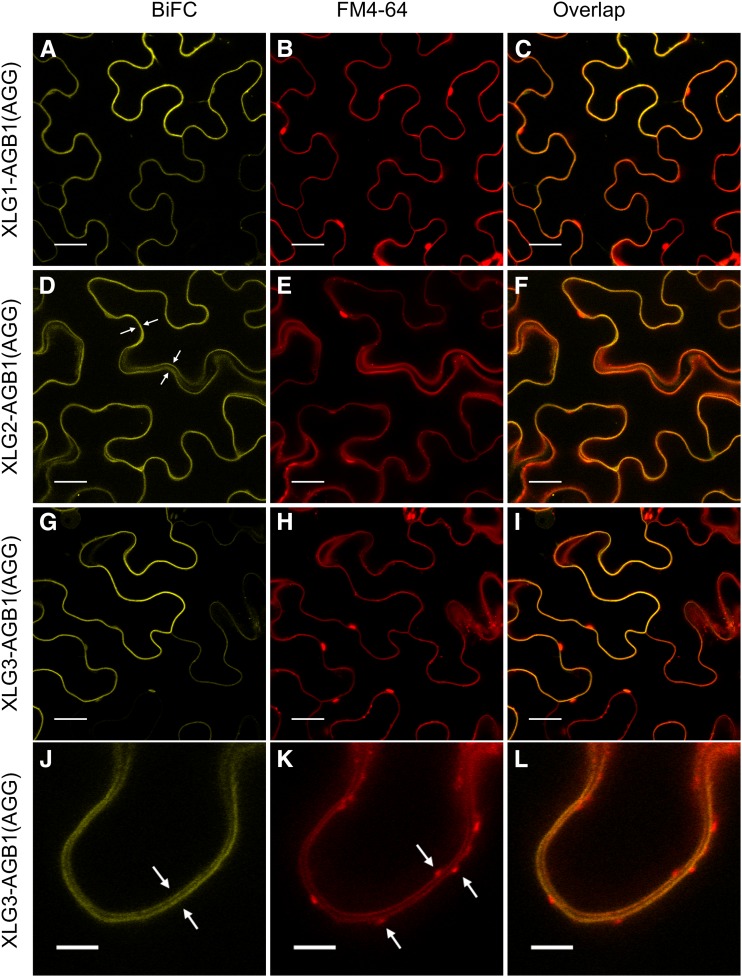
To identify the subcellular localization of the XLG-AGB1-AGG heterotrimers, we performed high-magnification (63× and 95×) colocalization experiments with the plasma membrane staining dye FM4-64 (Bolte et al., 2004) and found that the interaction occurs specifically at the plasma membrane (Fig. 6). Furthermore, the XLG-Gβγ heterotrimers do not colocalize with internalizing FM4-64-labeled vesicles (Fig. 6, J, K, and L). These results complement our yeast data and show a dependency on all three subunits for a positive XLG-AGB1 interaction at the plasma membrane in planta.
Figure 6.
XLG-based heterotrimers localize specifically to FM4-64-marked plasma membranes. The UBIQUITIN10 promoter-driven XLG-AGB1 test constructs were cotransformed with the 35S-driven pDOE-γ1γ2 vector (Gookin and Assmann, 2014) as a source of untagged AGG1 and AGG2. A, XLG1 interacts with AGB1. B, FM4-64 stains the plasma membrane. C, XLG1-AGB1 BiFC signal overlaps with the FM4-64-marked plasma membrane. D, XLG2 interacts with AGB1. The four white arrows mark the plasma membranes of two adjacent transformed cells traversing in and out of the 1.1-μm focal plane. E, FM4-64 marks the plasma membrane. F, XLG2-AGB1 BiFC signal overlaps with the FM4-64-marked plasma membrane. G, XLG3 interacts with AGB1. H, FM4-64 stains the plasma membrane. I, XLG3-AGB1 BiFC signal overlaps with the FM4-64-marked plasma membrane. J, High-resolution/magnification image of the XLG3-AGB1 BiFC signal visible at the two distinct plasma membranes of two adjacent cells, marked with white arrows. K, FM4-64 specifically labels the two distinct plasma membranes. White arrows mark FM4-64-labeled vesicles. L, XLG3-AGB1 BiFC signal overlaps with the two distinct FM4-64-marked plasma membranes but not with the FM4-64-marked vesicles. Test vectors were agroinfiltrated into N. benthamiana leaves at an OD600 of 0.0075, and images were acquired 62 to 67 h post infiltration. FM4-64 was infiltrated at 50 μm just prior to imaging. Images in A to I were acquired at 63× magnification with a 1.1-μm optical slice; bars = 20 μm. Images in J to L were acquired at 95× (63× magnification plus a 1.5× zoom of the scan area during image acquisition; 0.09 μm per pixel resolution); bars = 5 μm. Yellow indicates mVenus BiFC, and red indicates FM4-64.
XLG Subcellular Localization
All three Arabidopsis XLG proteins were originally described as nuclear resident proteins (Ding et al., 2008), but Maruta et al. (2015) showed XLG1 localization at the plasma membrane and XLG2 and XLG3 localization at both the plasma membrane and nucleus.
To reassess XLG subcellular localization patterns, we created UBIQUITIN10 promoter-driven XLG:mVenus fusions. We reasoned that the use of the optimized and bright monomeric mVenus fluoroprotein, in combination with the slower kinetics of the UBIQUITIN10 promoter-driven expression, would allow for enhanced temporal resolution (Gookin and Assmann, 2014). Analysis of XLG:mVenus fusions transiently expressed in Nicotiana benthamiana leaves showed that XLG1 is primarily extranuclear, with nuclear signal ranging from not detectable to weak (Fig. 7, A and B). XLG2:mVenus localization varied from predominantly nuclear (Fig. 7C) to an even distribution between nucleus and the cytoplasm (Fig. 7D). XLG3:mVenus consistently localized to both the nucleus and cytoplasm (Fig. 7, E and F) in every agroinfiltration experiment. The nuclear accumulation patterns of the three XLGs were confirmed in high-magnification (63×) colocalization experiments, with mTq2-tagged nuclear specific histone 2B (Heidstra et al., 2004) colocalizing with the XLGs in the nucleus (Supplemental Fig. S6). In comparison, our BiFC results show that all three XLGs interact with all three Gβγ dimers at the cell periphery (Fig. 5; Supplemental Fig. S5), suggesting that coexpression of partner subunits is a key determinant of XLG intracellular location.
Figure 7.
Subcellular localization of Arabidopsis XLG proteins in N. benthamiana leaves. A to D, UBIQUITIN10 promoter-driven XLG:mVenus fusions show differing localization patterns. A and B, XLG1 is predominantly extranuclear with nearly undetectable (A) to very weak (B) nuclear signal. C and D, XLG2 varies between strong nuclear signal (C) and signal evenly divided between the nucleus and cytoplasm (D). E and F, XLG3 consistently localizes to the nucleus and the cytoplasm. Agroinfiltrations of N. benthamiana leaves were performed at an OD600 of 0.0075 to 0.01 and imaged at 60 to 68 h, except for F, which was imaged at 6 d post infiltration. Yellow indicates mVenus, blue indicates mTq2, and red indicates chlorophyll autofluorescence. Bars = 50 μm.
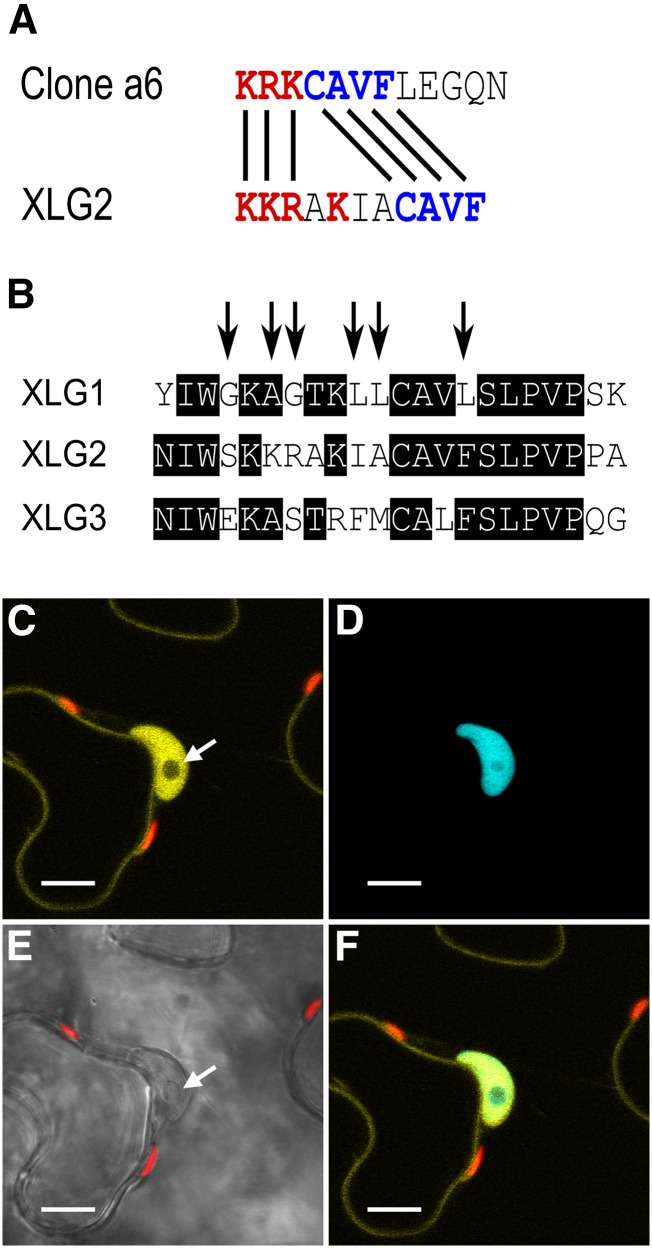
Sequence analysis of the three XLGs using the PredictProtein software suite (Yachdav et al., 2014) only identified the classic monopartite nuclear localization signal (NLS) present in XLG3 (RRKKKKK), previously identified by Ding et al. (2008). We next used computational regular expression to search the set of NLSs identified and functionally characterized in cv BY-2 tobacco (Nicotiana tabacum) cells by Kosugi et al. (2009). Our search identified a potential noncanonical NLS in XLG2 (KKRAKiaCAVF) through a partial match with CAVF (Fig. 8A). This potential XLG2 NLS resides within a small stretch of sequence that is semiconserved among the XLGs (Fig. 8B). Accordingly, we assessed the NLS functionality of this region. We used residue substitution to modify the XLG1 sequence to mirror XLG2 in this stretch (Fig. 8B, arrows) and expressed the resultant XLG1m1:mVenus fusion in N. benthamiana leaves. In contrast to XLG1 (Fig. 7, A and B; Supplemental Fig. S6A), XLG1m1 produced strong nuclear signal (Fig. 8C), specifically colocalizing with the nucleus-specific signal of mTq2:histone 2B (Fig. 8, D and F) and providing evidence that the atypical NLS of XLG2 is functional.
Figure 8.
XLG2 has a functional NLS. A, The XLG2 NLS has a similar basic-positive patch of residues (red) and a CAVF motif (blue) identical to the empirically validated clone a6 sequence (Kosugi et al., 2009). B, The XLG2 NLS resides in a semiconserved stretch of sequence in the XLG family starting at XLG2 residue 424, and the arrows show the XLG1 residues mutated to mirror XLG2 to create the XLG1m1 construct. XLG1m1:mVenus fusions localized to the nucleus in N. benthamiana leaves, demonstrating that the XLG2 NLS is a functional regulatory domain. (Compare with XLG1:mVenus localization in Figure 7, A and B.) C to F, Agroinfiltration of XLG1m1:mVenus into N. benthamiana leaves at an OD600 of 0.0075 to 0.01 and imaged 48 h later at 63× magnification. C, XLG1m1:mVenus localizes to the interior of the nucleus and at the plasma membrane; the nucleolus (white arrow) does not appreciably accrue XLG1m1. D, The nucleus is specifically marked by mTq2:histone 2B. E, The nucleus and nucleolus (white arrow) are clearly visible in the bright-field channel. F, XLG1m1:mVenus and mTq2:histone 2B colocalization is specific to the nucleus. Yellow indicates mVenus, blue indicates mTq2, and red indicates chlorophyll autofluorescence. Bars = 10 μm.
Since XLG2 and XLG3 also show both nuclear and extranuclear localization, we analyzed these sequences in search of a nuclear export signal (NES). We did not find a direct match to the classic consensus pattern for a Leu-rich NES (Xu et al., 2012); therefore, we used computational regular expression to search the set of validated NESs identified by Kosugi et al. (2008). Our analyses based on hydrophobic residue spacing identified a potential unconventional class 2 NES in XLG3 (LELRILKL; Fig. 9A). Two of these residues (I and the final L) were also weakly identified by NetNES (la Cour et al., 2004), suggesting the presence of a NES. We created two residue substitution mutants to map the domain, in which conserved hydrophobic residues that are functionally important to the motif were substituted with Gly (lowercase g): XLG3m1:mVenus (gEgRILKL) and XLG3m2:mVenus (LELRggKg; Fig. 9A). Both XLG3m1 and XLG3m2 produced strong nuclear signal (Fig. 9, B–D), demonstrating that the identified NES regulates XLG3 subcellular localization.
Figure 9.
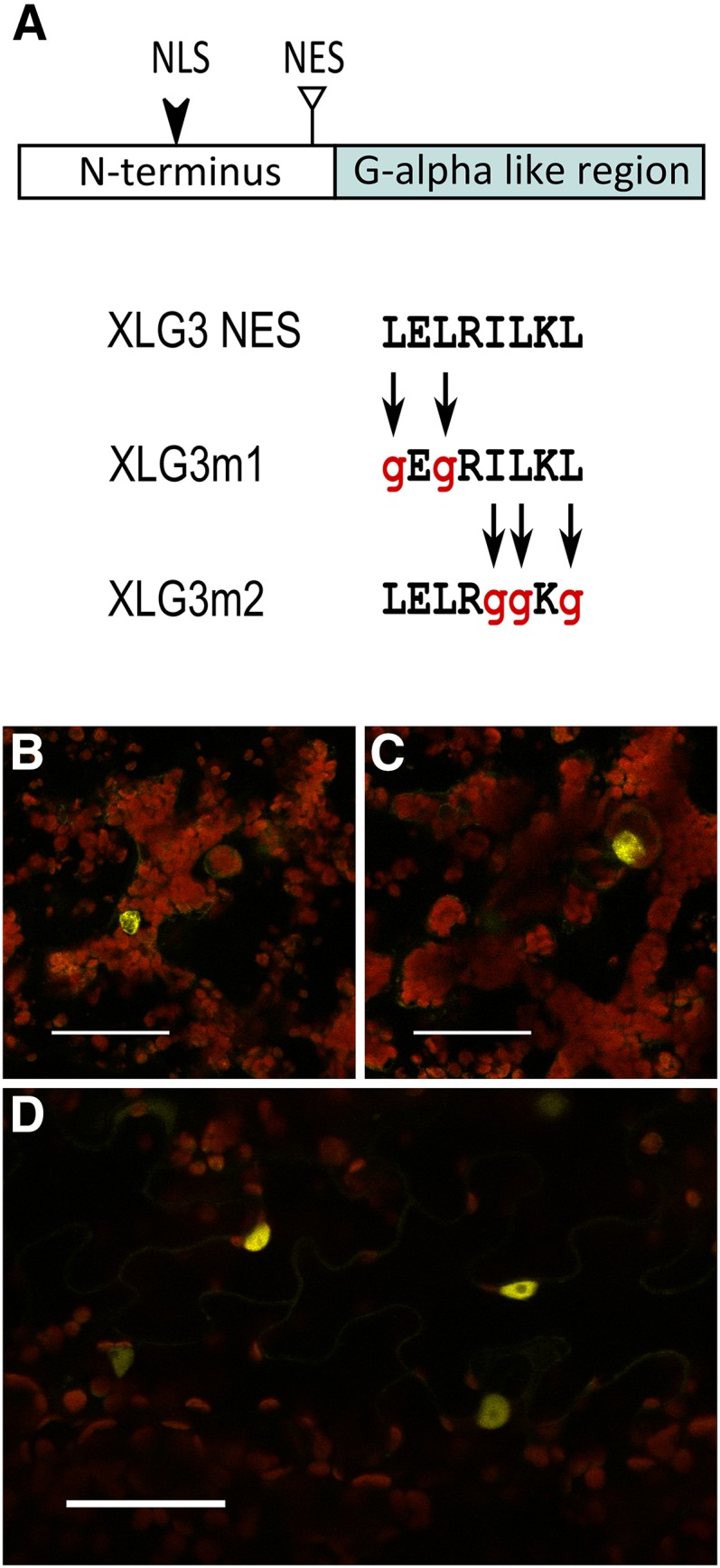
XLG3 has a functional NES. A, The N-terminal domain of XLG3 has an unconventional NES (triangle) located just upstream of the Gα-like region. The XLG3 NES was mutated by substituting hydrophobic residues at the N terminus (XLG3m1) or the C terminus (XLG3m2) of the domain with Gly residues. Both mutants showed nearly exclusive nuclear localization. B and C, XLG3m1 nuclear localization in two focal planes of the same mesophyll cells. D, XLG3m2 nuclear localization in epidermal pavement cells. Agroinfiltrations of N. benthamiana leaves were performed at an OD600 of 0.0075 to 0.01 and imaged 50 to 55 h later. Yellow indicates mVenus, blue indicates mTq2, and red indicates chlorophyll autofluorescence. Bars = 50 μm.
We next investigated XLG3:mVenus localization upon the coexpression of 35S-driven untagged AGB1-AGG1 and AGB1-AGG2 dimers. In both assays, XLG3 was sequestered at the plasma membrane, and in many cases, the nucleus was devoid of fluorescent signal (Fig. 10). These results provide evidence that XLG3 localization is dynamic and, in conjunction with our BiFC results, demonstrate that the plasma membrane is the site of XLG-Gβγ interaction.
Figure 10.
Overexpression of Gβγ dimers sequesters XLG3 at the plasma membrane. XLG3::mVenus driven by the UBIQUITIN10 promoter is retained at the plasma membrane when coexpressed with 35S-driven AGB1-AGG1 (A) or AGB1-AGG2 (B) dimer. (Compare with localization of the XLG3::mVenus fusion protein in the absence of additional Gβγ in Figure 7, E and F.) Note the lack of nuclear signal in two nuclei (white arrows) visible in the bright-field channel. The plasma membrane retention was consistent over time; these images were acquired 6 d post infiltration of N. benthamiana leaves at a final OD600 of 0.0075 for XLG3 and 0.025 for Gβγ dimers. Yellow indicates mVenus, blue indicates mTq2, and red indicates chlorophyll autofluorescence. Bars = 50 μm.
xlg Mutants Recapitulate agb1/agg1 and agb1/agg2 Phenotypes
The majority of G protein-related phenotypes have been characterized for gpa1 and agb1 mutants; however, for a growing but not yet comprehensive number of phenotypes, details have been described for Gγ subunit mutants as well. Comparison of published phenotypes suggests that while AGB1 (the only Gβ subunit) participates in all G protein-mediated processes, the majority of gpa1 phenotypes are restricted to recapitulation in agg3 mutants. For example, hypersensitivity to abscisic acid during germination (Ullah et al., 2002; Chakravorty et al., 2011), decreased etiolated hypocotyl elongation (Ullah et al., 2001; Chakravorty et al., 2011), round leaf morphology (Ullah et al., 2001; Chakravorty et al., 2011), hyposensitivity to abscisic acid inhibition of stomatal opening (Wang et al., 2001; Chakravorty et al., 2011), shortened silique morphology (Trusov et al., 2008; Chakravorty et al., 2011), and round seed (Chakravorty et al., 2011) phenotypes are all similar in gpa1 and agg3 mutants. The recapitulation of gpa1 phenotypes in agg3 mutants correlates well with the GPA1-AGB1/AGG3 binding specificity demonstrated in Figure 1. Conversely, a number of agg1 and agg2 phenotypes are recapitulated in agb1, but opposite or wild-type phenotypes are observed in gpa1 mutants (Table I). Our yeast three-hybrid results, showing that AGB1/AGG1 and AGB1/AGG2 interact preferentially with XLGs over GPA1, raise the intriguing possibility that XLGs form signaling complexes with Gβγ and may especially be involved in agb1/agg1 and agb1/agg2 phenotypes. We selected several phenotypes present in agb1 mutants, but absent from (or not yet assessed in) gpa1 mutants, for examination in xlg mutants. The four phenotypes we selected were NaCl, tunicamycin, and d-Glc hypersensitivity and increased stomatal density.
Table I. Summary of phenotypic differences between gpa1 and agb1 mutants, and participation of the XLGs in these phenotypes.
G protein phenotypes from the literature in which gpa1 and agb1 mutants display opposite or different phenotypes are listed. Phenotypes of gpa1, agb1, xlg, agg1/agg2, and agg3 mutants are annotated as hypersensitive, hyposensitive, or wild type for responses. Morphological phenotypes are annotated as increased, decreased, or wild type. New results from this article are included in boldface. Phenotypes were often found to be similar or aligned between xlg, agb1, and agg1/agg2 mutants. References, in square brackets, are as follows: [1] Llorente et al. (2005); [2] Trusov et al. (2006); [3] Zhu et al. (2009); [4] Maruta et al. (2015); [5] Trusov et al. (2007); [6] Chakravorty et al. (2011); [7] Ding et al. (2008); [8] Chen et al. (2006); [9] Li et al. (2012); [10] Ullah et al. (2003); [11] Pandey et al. (2008); [12] Colaneri et al. (2014); [13] Chen and Brandizzi (2012); [14] Wang et al. (2006); [15] Zhang et al. (2008); and [16] Nilson and Assmann (2010).
| Phenotype | Genotypes |
||||
|---|---|---|---|---|---|
| gpa1 | agb1 | xlgs | agg1/2 | agg3 | |
| Susceptibility to pathogens | ↓ Decreased [1,2] | ↑ Increased [1,2,3,4] | ↑ Increased [3,4] | ↑ Increased [5] | ─ Wild type [6] |
| Primary root growth rate/length | ─ Wild type [7,8] | ↑ Increased [7,8] | ↑ Increased [7] | ─ Wild type [9] | |
| Lateral root proliferation | ↓ Decreased [10] | ↑ Increased [10] | ↑ Increased [7] | ↑ Increased [5] | ─ Wild type [6] |
| Root waving and skewing | ↓ Wild type/minor decrease [11] | ↓ Decreased [11] | ↓ Decreased [11] | ||
| Sensitivity to salt post germination | ↓ Hyposensitive [12] | ↑ Hypersensitive [12] | ↑ Hypersensitive | ↑ Hypersensitive | ─ Wild type |
| Sensitivity to tunicamycin post germination | ─ Wild type | ↑ Hypersensitivea [13] | ↑ Hypersensitive | ↑ Hypersensitive | ─ Wild type |
| Sensitivity to d-Glc post germination | ─ Wild type | ↑ Hypersensitive [14] | ↑ Hypersensitive | ↑ Hypersensitive | ─ Wild type |
| Stomatal density/index | ↓ Decreased [15,16] | ↑ Increased [15] | ↑ Increased | ↑ Increased | ─ Wild type [6] |
Wang et al. (2007) described the opposite, with agb1 hyposensitive.
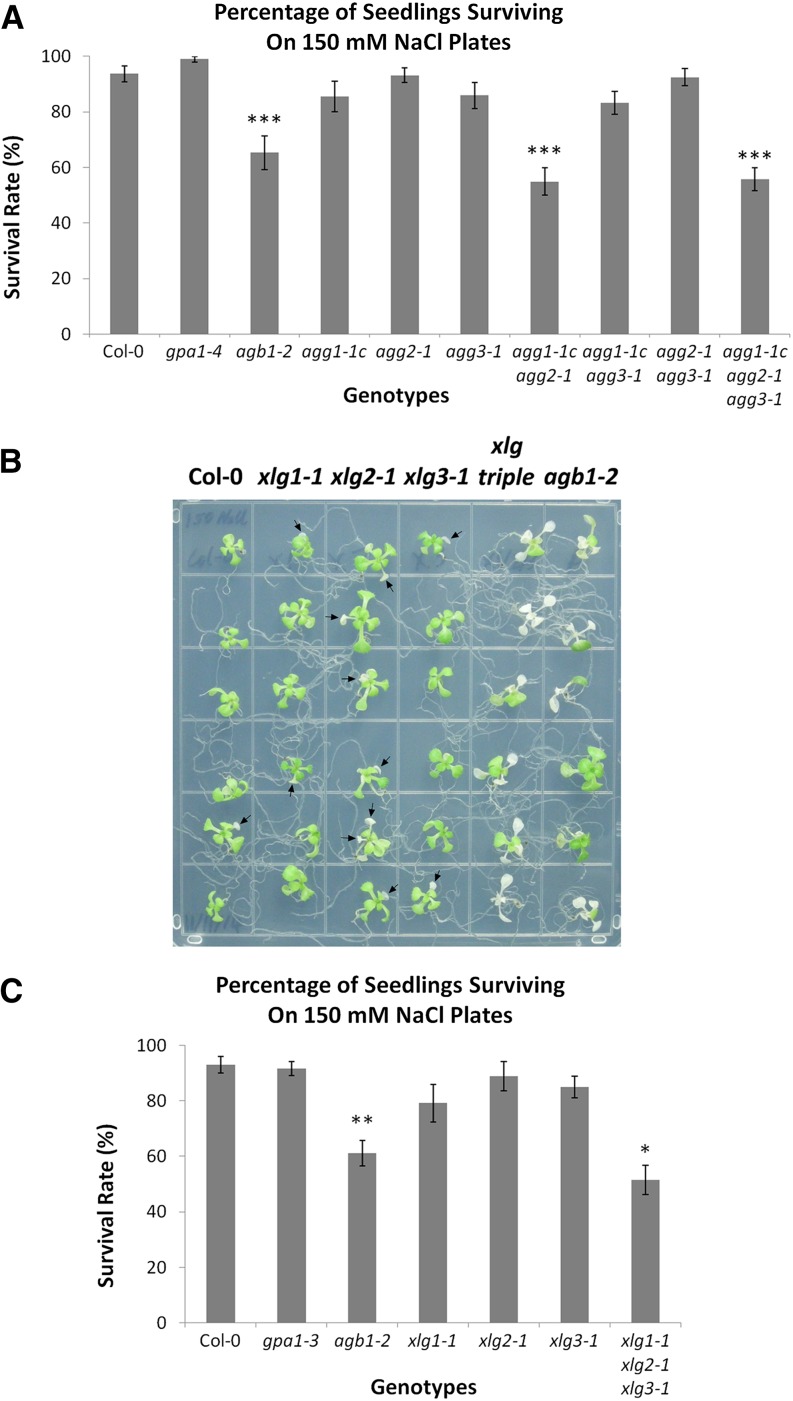
Colaneri et al. (2014) showed that Arabidopsis agb1 mutants are hypersensitive to NaCl, while gpa1 is hyposensitive to NaCl. Urano et al. (2014) examined Gα mutants available in rice and maize (Zea mays) and found that the Gα mutants were, much like gpa1 mutants in Arabidopsis, hyposensitive to salt treatment. The hypersensitivity of Gβ mutants to NaCl, and the hyposensitivity of Gα mutants, suggested that NaCl sensitivity is a candidate for a potential xlg phenotype. Colaneri et al. (2014) found that agg1 and agg2 single mutants were not hypersensitive to NaCl; however, the agg1-1c agg2-1 agg3-1 triple mutant appeared hypersensitive to NaCl.
As we hypothesize that agg1- and/or agg2-mediated phenotypes are a potential indicator of XLG involvement, we first clarified Gγ subunit involvement in NaCl sensitivity by testing all single, double, and triple mutant combinations of Gγ mutants and deduced that AGG1 and AGG2 play redundant roles in NaCl sensitivity: agg1-1c agg2-1 double mutants displayed a similar NaCl hypersensitivity to the agb1-2 mutant or the agg triple mutant (Fig. 11A). AGG3 is not involved in salt sensitivity, as all agg3-containing genotypes, aside from the agg triple mutant, displayed wild-type sensitivity (Fig. 11A). We next investigated the salinity phenotype of the xlg single and triple mutants. An increased number of xlg single mutant seedlings, as compared with wild-type seedlings, displayed slight chlorosis, indicated by the arrows in Figure 11B, on 150 mm NaCl plates. The chlorosis was generally limited to one or two leaves in the single xlg mutants and was much more pronounced in both the agb1-2 mutant and the xlg triple mutant (Fig. 11B). Similarly, when compared with the wild type, the agb1-2 and xlg triple mutants each displayed a clear decrease in seedling survival (Fig. 11C). Therefore, the agb1-2, agg1-1c agg2-1 double, and xlg triple mutants displayed similar NaCl hypersensitivity phenotypes, suggesting the possibility that up to six heterotrimer combinations, XLG1/AGB1/AGG1, XLG1/AGB1/AGG2, XLG2/AGB1/AGG1, XLG2/AGB1/AGG2, XLG3/AGB1/AGG1, and XLG3/AGB1/AGG2, function in salt sensitivity.
Figure 11.
agb1, agg1 agg2, and the xlg triple mutant are hypersensitive to salt. Seeds were sown and germinated on 0.5× Murashige and Skoog (MS) plates (1% [w/v] Suc and 1% [w/v] agar). After 9 d of growth, seedlings were transferred to 0.5× MS plates (1% [w/v] Suc and 1% [w/v] agar) supplemented with 150 mm NaCl. Seedling survival was scored once seedling death was apparent in the most severely affected genotypes (14–21 d). Assays were conducted with mutants of the Gγ subunits (A) and xlg mutants (B and C). A representative image of the results with the xlg mutants and agb1-2 is shown in B, and quantification is shown in C. Columbia-0 (Col-0), agb1, and gpa1 controls were included in both assays. In B, xlg triple refers to the xlg1-1 xlg2-1 xlg3-1 mutant. Significant differences from Col-0 (Student’s t test) are indicated: *, P < 0.05 to 0.01; **, P < 0.01 to 0.001; and ***, P < 0.001. All values are means ± se.
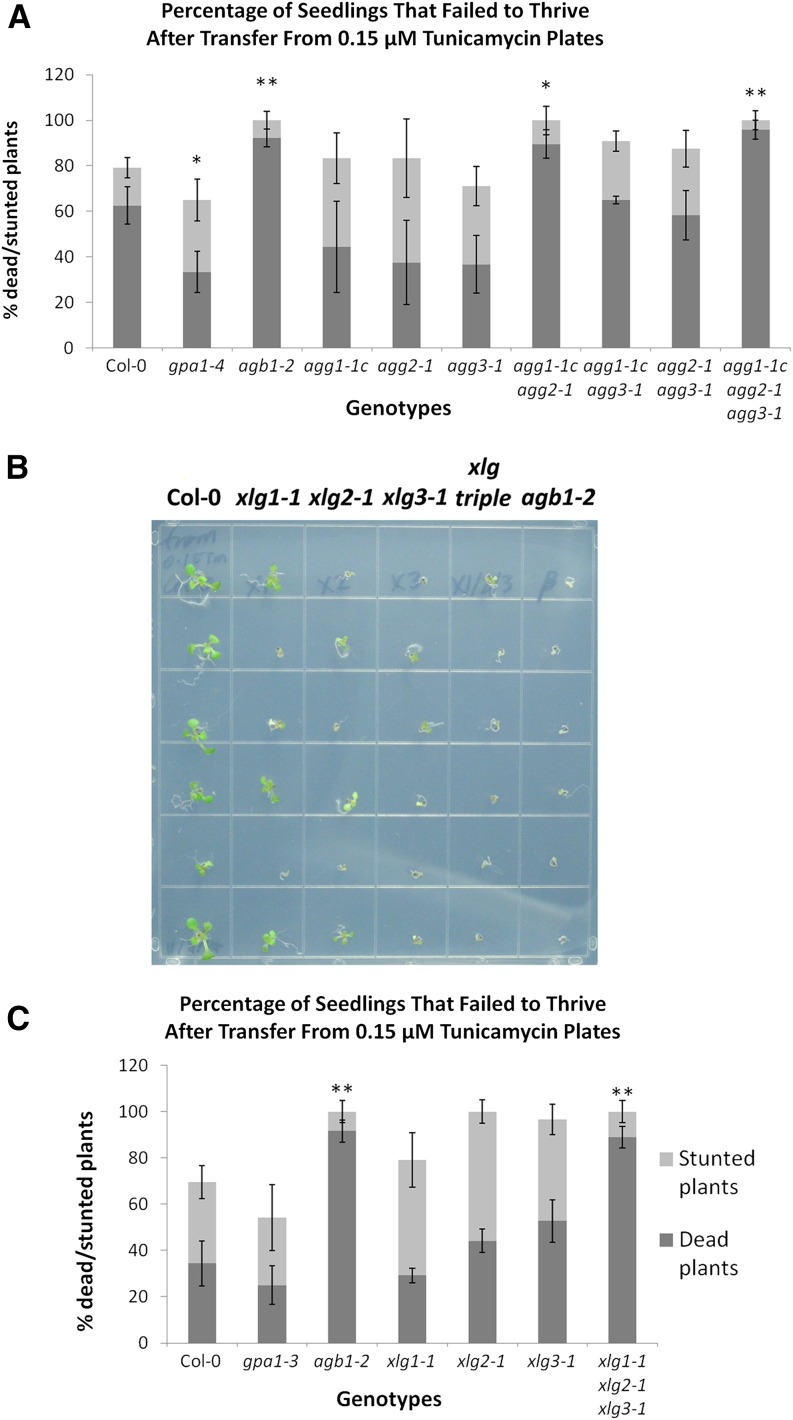
Another potential xlg phenotype is hypersensitivity to tunicamycin, a mixture of homologous compounds that block nitrogen-linked glycosylation, thereby inducing the unfolded protein response (UPR; Schröder and Kaufman, 2005), a stress response initiated in the endoplasmic reticulum when the cellular protein production and folding machinery is overcome, for example by inhibited posttranslational modification (Koizumi et al., 2001; Schröder and Kaufman, 2005). Wang et al. (2007) initially reported that the agb1-2 mutant was hyposensitive to tunicamycin, while gpa1-4 displayed wild-type sensitivity. Contrary to the initial report, Chen and Brandizzi (2012) reported that multiple agb1 mutants, including agb1-2, were hypersensitive to tunicamycin in a plate-based seedling assay assessing recovery from tunicamycin. Similar to the report from Chen and Brandizzi (2012), in our hands, agb1-2 was more sensitive than the wild type to tunicamycin, with 0.15 µm tunicamycin the optimal concentration for differentiating agb1-2 from the wild type.
We then investigated the Gγ subunit mutants in the tunicamycin recovery assay. The agg1-1c agg2-1 double mutant and agg triple mutant, like agb1-2, were particularly hypersensitive to tunicamycin. As with the salt assay, the gpa1 and single Gγ mutants were not hypersensitive to tunicamycin (Fig. 12A), suggesting that AGG1 and AGG2 play redundant roles and that tunicamycin sensitivity is a candidate xlg phenotype. As seen in Figure 12B, the xlg single mutants, particularly xlg2-1 and xlg3-1, were indeed hypersensitive to tunicamycin, showing a clear increase in the number of stunted seedlings as compared with the wild type (Fig. 12, B and C). The xlg triple mutant fully recapitulated the agb1-2 hypersensitivity phenotype, with approximately 90% of seedlings of both genotypes unable to recover from the treatment (Fig. 12B) and scored as dead (Fig. 12C). The similarity of tunicamycin hypersensitivity phenotypes suggests that the XLGs may function in a heterotrimer with AGB1/AGG1 and AGB1/AGG2.
Figure 12.
agb1, agg1 agg2, and the xlg triple mutant are hypersensitive to tunicamycin. Seeds were sown and germinated on 0.5× MS plates (1% [w/v] Suc and 1% [w/v] agar) supplemented with 0.15 µm tunicamycin. After 6 d of growth, seedlings were transferred to 0.5× MS plates (1% [w/v] Suc and 1% [w/v] agar) and allowed to recover for an additional 10 d before being scored, as outlined in “Materials and Methods.” Seedlings that failed to thrive (stunted + dead seedlings) are presented for mutants of the Gγ subunits (A) and xlg mutants (B and C). A representative image of the results with the xlg mutants and agb1-2 is shown in B, and quantification is shown in C. Col-0, agb1, and gpa1 controls were included in both assays. In B, xlg triple refers to the xlg1-1 xlg2-1 xlg3-1 mutant. Significant differences from Col-0 (Student’s t test) are indicated: *, P < 0.05 to 0.01; and **, P < 0.01 to 0.001. All values are means ± se.
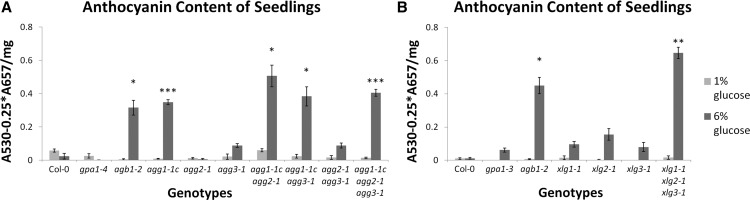
A third potential xlg phenotype is the sensitivity of agb1 to d-Glc in postgermination development. It has been demonstrated that agb1-2 is hypersensitive to d-Glc when grown on plates (Wang et al., 2006). Additionally, the ARABIDOPSIS REGULATOR OF G PROTEIN SIGNALING protein (AtRGS1) has been implicated in d-Glc signaling, as rgs1 seedlings are hyposensitive to d-Glc (Chen and Jones, 2004), and AtRGS1 internalizes upon d-Glc treatment in an AGB1- and AGG1-/AGG2-dependent manner (Urano et al., 2012b). We germinated seeds of the G protein mutants on medium supplemented with 1% or 6% (w/v) d-Glc under high-light conditions to synchronize germination (Trusov et al., 2007) and then allowed seedlings to grow under low-light conditions, to replicate the assay conducted by Wang et al. (2006). However, in our hands, even 6% (w/v) d-Glc was not sufficient to severely repress the development of agb1-2 seedlings. Instead, the increased stress placed upon the agb1-2 seedlings was manifested in increased anthocyanin production at higher d-Glc concentrations; therefore, we quantitatively assessed the d-Glc stress phenotype by measuring anthocyanin content (Mancinelli et al., 1991).
The anthocyanin content of all genotypes was similarly negligible when grown on medium supplemented with 1% (w/v) d-Glc (Fig. 13). However, anthocyanin content in 6% (w/v) d-Glc was elevated in agb1-2 and all agg1-1c-containing genotypes (agg1-1c, agg1-1c agg2-1, agg1-1c agg3-1, and the agg triple mutant) but not in gpa1-4 (Fig. 13A). Therefore, d-Glc sensitivity fits the criteria of an agb1/agg1 or agb1/agg2 phenotype that lacks GPA1 involvement. When we repeated the assay with xlg mutants, the xlg triple mutant reproduced the agb1-2 phenotype (Fig. 13B). These results suggest that XLG/AGB1/AGG1 heterotrimers function in postgermination d-Glc sensitivity.
Figure 13.
agb1, agg1, and the xlg triple mutant are hypersensitive to d-Glc. Seeds were sown on 0.5× MS plates (1% [w/v] agar) supplemented with 1% or 6% (w/v) d-Glc. Seeds were germinated under 120 µmol m−2 s−1 white light for 1 d, then light intensity was dimmed to 60 µmol m−2 s−1. After 24 d of growth in long-day conditions, anthocyanins were extracted and quantified for mutants of the Gγ subunits (A) and xlg mutants (B). Col-0, agb1, and gpa1 controls were included in both assays. Significant differences from Col-0 (Student’s t test) are indicated: *, P < 0.05 to 0.01; **, P < 0.01 to 0.001; and ***, P < 0.001. All values are means ± se.
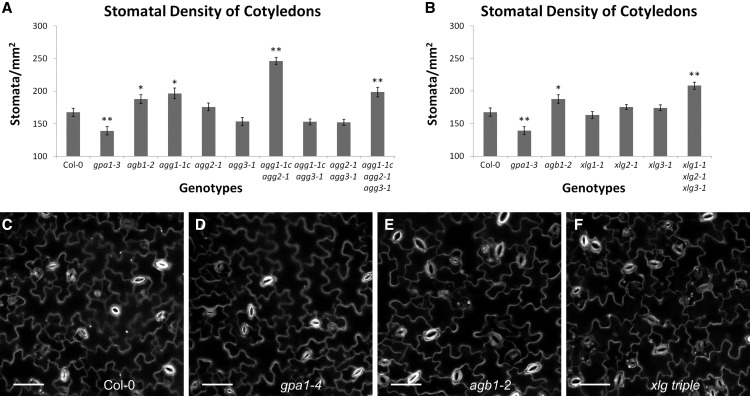
Finally, we investigated the stomatal density phenotype of the G protein mutants. Zhang et al. (2008) found that gpa1 and agb1 mutants displayed a lower and higher stomatal density than the wild type, respectively, in cotyledons. Chakravorty et al. (2011) showed that the agg3 mutants resembled the wild type in cotyledon stomatal density. These results suggested that stomatal density is another candidate phenotype for XLG involvement.
The cotyledons of gpa1 and agb1 mutants displayed lower and higher stomatal density than the wild type (Fig. 14, A, C, D, and E), respectively, in agreement with the results of Zhang et al. (2008). As seen in Figure 14A, the agg1-1c mutant also displayed an increase in stomatal density, as did the agg1-1c agg2-1 double mutant and the agg triple mutant. Of the Gγ mutants, the agg1-1c agg2-1 double mutant displayed the largest increase in stomatal density, higher even than agb1 or the agg triple mutant. All agg3-containing genotypes displayed a slight, but not statistically significant, decrease in stomatal density as compared with the wild type. Like the agb1 mutant, the xlg triple mutant displayed an increase in stomatal density (Fig. 14, B, C, E, and F), indicating that XLG/AGB1/AGG1,2 heterotrimers repress stomatal development. In fact, the stomatal density of the xlg triple mutant was greater than that of the agb1 mutant (P = 0.02, Student’s t test). These results may indicate a dual function for AGB1, with GPA1/AGB1/AGG3 heterotrimers stimulating stomatal development and XLG/AGB1/AGG1 and XLG/AGB1/AGG2 heterotrimers repressing stomatal development. This would explain the greater severity of the agg1-1c agg2-1 (Fig. 14A) and xlg triple (Fig. 14B) mutants when compared with agb1 and the agg triple mutant.
Figure 14.
agb1, agg1 agg2, and the xlg triple mutants display increased stomatal density. Propidium iodide-stained cotyledons of 9-d-old seedlings were imaged using a confocal microscope, and images were used to quantify stomatal density for mutants of the Gγ subunits (A) and xlg mutants (B). The assay was conducted for all genotypes simultaneously, and the Col-0, agb1-2, and gpa1-3 controls represent the same data in A and B. Significant differences from Col-0 (Student’s t test) are indicated: *, P < 0.05 to 0.01; and **, P < 0.01 to 0.001. All values are means ± se. Representative images of propidium iodide-stained cotyledons are shown for Col-0 (C), gpa1-4 (D), agb1-2 (E), and the xlg triple mutant (F). White indicates propidium iodide stain. Bars = 50 µm.
DISCUSSION
XLGs as Noncanonical Gα Proteins
Here, we make a case that the three XLGs are noncanonical Gα subunits that function within the heterotrimeric G protein complex. The sole canonical Gα protein subunit in Arabidopsis is GPA1, originally identified by Ma et al. (1990). In 1999, we reported the identification of the first noncanonical Gα-like protein in plants, Arabidopsis XLG1, and we demonstrated that the XLG1 protein binds GTP (Lee and Assmann, 1999). Analysis by Maruta et al. (2015) suggests that XLGs evolved from canonical Gα subunits. Furthermore, our in silico structural analysis shows that the XLG3 Gα domain and GPA1 are strikingly similar (Fig. 4; Supplemental Figs. S1 and S2), suggesting that GPA1 and XLG3 may share a common binding surface on Gβγ. The intermolecular arrangement of canonical Gα with Gβγ in the heterotrimer is well established (Wall et al., 1998), and all three XLG family members show strong structural conservation of their Gα region with the empirically derived GPA1 structure (Supplemental Tables S1 and S2). In our yeast three-hybrid assays, we observed that GPA1 and XLG3 are capable of competing with each other for binding of the AGB1/AGG3 dimer (Fig. 3). Competition between GPA1 and XLG3 for Gβγ-binding also suggests that GPA1 and XLG3 share a common binding surface on Gβγ, supporting the hypothesis that XLGs and GPA1 bind Gβγ in a similar conformation. This idea is further supported by the efficient interaction between XLGs and Gβγ dimers that is observed (Fig. 5) upon insertion of the NmVenus210 fragment into a position analogous to the αB-αC loop insertion, a position that also enables the observation of GPA1 interaction with AGB1-AGG dimers in BiFC (Gookin and Assmann, 2014). Our protein-protein interaction data and phenotypic analyses, in combination with the genetic evidence provided by Maruta et al. (2015), suggest that the recapitulation of agb1 phenotypes that we observe in the xlg triple mutant is likely due to the XLGs, AGB1, and AGG1/AGG2 functioning in the same pathways. In short, we provide evidence from several different approaches that all three XLG proteins participate within a heterotrimeric G protein complex.
One impediment to denoting XLGs as true Gα subunits relates to the fact that bona fide Gα subunits display a GTP/GDP switch during the activation/inactivation cycle, and while XLGs are GTP-/GDP-binding and GTPase-capable proteins (Heo et al., 2012), this switch has not been shown to control activation/inactivation. However, in truth, the single canonical Arabidopsis Gα subunit, GPA1, also falls short of fulfilling this stringent criterion. The activation/inactivation dynamics of GPA1 are poorly understood, due to unusual GTP-binding and hydrolysis kinetics (Johnston et al., 2007, 2008; Urano et al., 2012a), regulation by the atypical seven transmembrane domain containing AtRGS1 (Chen et al., 2003; Jones et al., 2011b; Urano et al., 2012b), and functional interactions between receptor-like kinases and G proteins (Bommert et al., 2013; Liu et al., 2013; Ishida et al., 2014). Indeed, the in planta GTP-/GDP-binding kinetics of GPA1, as well as those of the XLGs, are unknown. Because Gβγ availability may alter the ratio of GTP-bound versus GDP-bound Gα subunits, the addition of XLGs as potentially competing Gβγ interactors may have implications for the in planta activity of the Gα pool, including GPA1.
It should be noted that we provide direct evidence for GPA1 and XLG3 competition for Gβγ binding only in yeast; it would be valuable to perform analogous experiments in planta as well. Such experiments may be technically challenging, given previously noted difficulties in expressing tagged XLG protein at adequate levels for such assessments (Ding et al., 2008). While competition between GPA1 and XLGs for Gβγ could be a common theme, it is also possible that XLG- and GPA1-based heterotrimers modulate shared molecular signaling pathways. GPA1 interacts with GCR1, the only putative Arabidopsis G protein-coupled receptor based on sequence and structural homology (Plakidou-Dymock et al., 1998; Pandey and Assmann, 2004; Warpeha et al., 2007; Taddese et al., 2014), GTG1/GTG2, the two Arabidopsis G protein-coupled receptor-type G proteins (Pandey et al., 2009), and RGS1, the sole demonstrated Gα GTPase-activating protein in Arabidopsis (Chen et al., 2003). The potential interactions of the XLGs with these signaling elements remain to be explored.
Interaction of XLG Proteins with Gβγ Dimers in Vivo
The interaction of one of the XLGs, XLG2, with Gβγ was shown by yeast three-hybrid analysis and confirmed by BiFC by Maruta et al. (2015), but interaction strength was not investigated. We now demonstrate that all three XLGs bind Gβγ dimers, with distinct specificity, and with differences in binding strength in the yeast three-hybrid system. The canonical Gα subunit, GPA1, displayed a preference for AGB1/AGG3(γ) over AGB1/AGG1 or AGB1/AGG2 (Fig. 1A). Similarly strong, or even stronger, interactions than that of GPA1 with AGB1/AGG3(γ) were observed for several of the XLG-Gβγ interactions (Fig. 1). The fact that XLG interactions with Gβγ dimers are as strong as or stronger than the Gβγ interactions of the canonical GPA1 is evidence that XLGs function in the heterotrimer.
There were some slight discrepancies between the yeast three-hybrid results of Maruta et al. (2015) and our yeast three-hybrid data. For example, Maruta et al. (2015) obtained positive results using XLG2, AGB1, and an untagged AGG3 subunit, whereas our results were negative using XLG2 and AGG3, with untagged AGB1 as the bridge protein (Fig. 1C). One explanation is that the XLG2-AGB1/AGG3 interaction is extremely weak and fell below the detectable threshold in our conditions and assay configuration. It also should be noted that negative yeast three-hybrid results are not always definitive for lack of interaction. For example, we did not see strong evidence for GPA1/AGB1/AGG1 or GPA1/AGB1/AGG2 heterotrimers in yeast, yet gpa1, agb1, and agg1-1c agg2-1 mutants all display hypersensitivity to d-Glc repression of germination (Ullah et al., 2002; Pandey et al., 2006; Trusov et al., 2007), functionally implicating complexes of these subunits in planta. As GPA1/AGB1/AGG1 and GPA1/AGB1/AGG2 heterotrimers have been observed in planta by BiFC (Gookin and Assmann, 2014), the formation of these heterotrimers likely requires plant-specific cofactors.
Maruta et al. (2015) previously showed that XLG2 interacted with all three Arabidopsis Gβγ dimers by BiFC. However, the use of the sticky N-173/C-155 overlap-based split enhanced yellow fluorescent protein (eYFP) fragments (Kodama and Hu, 2012), coupled with strong expression driven by the double 35S promoter present in the pSAT vectors and a relatively long protoplast incubation time of 16 to 24 h, could confound the results (Maruta et al., 2015). To assess XLG-Gβγ heterotrimer formation, we performed BiFC for all three XLGs using the pDOE low-background BiFC system (Gookin and Assmann, 2014). Our results show that all three XLGs interact with AGB1/AGG1, AGB1/AGG2, and AGB1/AGG3 at the plasma membrane (Figs. 5 and 6), with a signal localization pattern reminiscent of the GPA1-Gβγ BiFC signal (Gookin and Assmann, 2014). Importantly, no BiFC signal was observed in the absence of a coexpressed Gγ subunit, demonstrating, as in our yeast three-hybrid results, that the XLG-Gβ interaction is not bipartite and that true heterotrimer formation occurs. This contrasts with the data from Maruta et al. (2015), which showed BiFC signal between XLG2 and the three Gγ subunits in agb1 protoplasts. One possible explanation is that XLG2 and the Gγ subunits exist in a complex strong and stable enough (without AGB1) to allow detectable BiFC. Alternatively, the BiFC signal observed by Maruta et al. (2015) could have arisen from nonspecific self-assembly of the BiFC fragments, as outlined above.
XLG Localization
The previously described subcellular localization of XLGs in the nucleus and Gβγ dimers at the plasma membrane could provide an impediment to the hypothesis that XLGs function as heterotrimeric G protein subunits, although AGB1 localization to the nucleus has also been reported, albeit without an equivalently expressed Gγ subunit that might provide a membrane anchor for the Gβγ dimer (Anderson and Botella, 2007). We reassessed the subcellular localization of the XLGs using Agrobacterium tumefaciens-mediated transient transformation in N. benthamiana, as opposed to biolistic expression in Vicia faba guard cells (Ding et al., 2008). Our assays indicate that the three XLGs have variable preferences for nuclear localization (Fig. 7; Supplemental Fig. S6). The extranuclear localization of all three proteins at the cell periphery is consistent with the recent report of XLG localization at the plasma membrane (Maruta et al., 2015). Our BiFC data show XLG heterotrimer localization is restricted to the cell periphery, specifically at the plasma membrane (Figs. 5 and 6). However, in BiFC, the two tagged interacting proteins are physically coupled by the irreversible association of the split fluoroprotein and are not free to move independently. Therefore, we examined the influence of untagged Gβγ dimers on XLG localization, choosing XLG3:mVenus as an example, since it reliably showed nearly even localization in the cytosol and nucleus. When provided in excess, untagged AGB1/AGG1 or AGB1/AGG2 dimers were able to sequester XLG3 at the plasma membrane, in many cases rendering the nucleus devoid of XLG3 signal (Fig. 10). In the absence of cosupplied Gβγ, the diverse localization patterns are also supported by our bioinformatic analysis and residue substitution mutants, which show that XLG2 has a noncanonical NLS (Fig. 8) and that XLG3 has both a classical NLS (Ding et al., 2008) and an unconventional NES (Fig. 9). It is intriguing to speculate that, upon activation and dissociation from Gβγ, the XLGs may act to directly transfer signal from the plasma membrane to the nucleus. This idea is lent credence by the interaction of XLG with RELATED TO VERNALIZATION1 (RTV1), a DNA-binding protein with a plant-specific B3-like DNA-binding domain (Heo et al., 2012). RTV1 plays an important role in flowering time control, likely by regulating floral integrator genes such as SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 and FLOWERING LOCUS T. The XLG2-RTV1 interaction occurs in the nucleus and stimulates RTV1 chromatin binding in chromatin immunoprecipitation assays (Heo et al., 2012).
XLGs Function in G Protein-Related Phenotypes
If XLGs function with Gβγ dimers in a Gα-like manner, XLG mutation should result in G protein-associated phenotypes. Some indicators that XLGs participate in AGB1 signaling are known (Table I). Based on the analysis of G protein phenotypes in the literature, we identified four other agb1 phenotypes in which gpa1 mutants behaved like the wild type, displayed an opposite phenotype to agb1, or had not been assigned. Salt, tunicamycin, and d-Glc are abiotic stresses that agb1 mutants are hypersensitive to during postgermination development. In the absence of similar phenotypes in gpa1 mutants, we examined the xlg mutants and identified agb1-like phenotypes in the xlg triple mutant (Figs. 11–14). Our results thereby indicate that XLG1,2,3/AGB1/AGG1,2 heterotrimers play a role in suppressing responses to abiotic stress. Salt has recently been implicated in stimulation of the UPR (Liu and Howell, 2010; Wang et al., 2011), much as tunicamycin traditionally has been. Therefore, G proteins can be hypothesized to function as suppressors of the UPR downstream of the convergence of different stimuli such as salt and tunicamycin. Similarly, AGB1, AGG1 and AGG2, but not GPA1, have been implicated in cell death and ROS production downstream of receptor-like kinases such as BAK1-INTERACTING RECEPTOR-LIKE KINASE1 (BIR1), FLAGELLIN-SENSITIVE2, EF-Tu RECEPTOR, and CHITIN ELICITOR RECEPTOR KINASE1 (Liu et al., 2013). Additionally, Respiratory burst oxidase protein D- and F-mediated P. syringae resistance and ROS production are dependent on AGB1 but not GPA1 (Torres et al., 2013), and an increased rate of powdery mildew entry into epidermal cells was observed upon surface inoculation of agb1 but not gpa1 mutant leaves (Lorek et al., 2013). Indeed, Maruta et al. (2015) demonstrated that disruption of either XLG2 or AGB1 was able to suppress the cell death phenotype of bir1 and resulted in impaired pathogen defense responses, including ROS production. These results illustrate that G proteins modulate multiple cell death pathways, either suppressing (Warpeha et al., 2008; Wei et al., 2008; Chen and Brandizzi, 2012; Colaneri et al., 2014; Yu and Assmann, 2015) or stimulating (Liu et al., 2013; Maruta et al., 2015) cell death in a stimuli-specific manner. XLGs also function in developmental pathways. xlg mutants display increased primary root growth (Ding et al., 2008) and increased root waving and skewing (Pandey et al., 2008), both of which are also agb1 phenotypes (Chen et al., 2006; Pandey et al., 2008). We demonstrate that XLGs are also involved in the development of aerial tissues, with xlg mutants exhibiting an increase in stomatal density (Fig. 14).
The subunit phenotypes, including those summarized in Table I and those we analyzed in Figures 11 to 14, correlate well with the heterotrimer configurations we observed in yeast (Fig. 1). We found significant overlap of agg1 and agg2 phenotypes with xlg phenotypes. With the exception of hypersensitivity to d-Glc during germination (Pandey et al., 2006; Trusov et al., 2007), few overlapping phenotypes between gpa1 and agg1/agg2 mutants have been identified to date. This observation, in combination with our yeast data, suggests that there are eight higher affinity heterotrimer combinations that are responsible for the majority of G protein signaling: GPA1/AGB1/AGG3, XLG1/AGB1/AGG1, XLG1/AGB1/AGG2, XLG2/AGB1/AGG1, XLG2/AGB1/AGG2, XLG3/AGB1/AGG1, XLG3/AGB1/AGG2, and XLG3/AGB1/AGG3. Our criteria for choosing potential xlg phenotypes to investigate centered on agg1 and/or agg2 phenotypes that were not recapitulated in gpa1 mutants. Therefore, it should be noted that our study was deliberately biased toward the investigation of potential xlg/agb1/agg1-agg2 phenotypes and was not expected to identify additional gpa1/agb1/agg1-agg2 phenotypes.
The distinct phenotypes displayed by gpa1 and xlg mutants suggest that GPA1 and the XLGs mediate distinct molecular pathways, at least in some cases, and that competition between GPA1 and the XLGs for Gβγ binding could be important for molecular function. Additionally, GPA1 and the XLGs would be expected to bind and regulate different sets of effectors. A small number of GPA1 interactors have been described that could be evaluated for interaction with XLGs, including PHOSPHOLIPASE Dα1 (Zhao and Wang, 2004), PREPHENATE DEHYDRATASE1 (Warpeha et al., 2006), PIRIN1 (Warpeha et al., 2007; Orozco-Nunnelly et al., 2014), THYLAKOID FORMATION1 (Huang et al., 2006), and those identified in the interactome of Klopffleisch et al. (2011). There may also be effector specificity within the XLG family, as the majority of the G protein-related pathogen responses have been attributed to XLG2 (Zhu et al., 2009; Maruta et al., 2015). However, significant overlap among XLG effectors is also expected, due to the redundancy displayed by the XLGs in our salinity (Fig. 11), tunicamycin (Fig. 12), d-Glc (Fig. 13), and stomatal density (Fig. 14) assays.
Extending G Protein Heterotrimer Diversity
The lack of G protein diversity in plants such as Arabidopsis has been an unexpected finding in the postgenomic era. The initial identification of GPA1, AGB1 (Gβ), and AGG1/AGG2 (Gγ) allowed for only two different heterotrimer combinations, which was assumed from 2001 to 2011 to be the full complement of Arabidopsis G protein subunits, until the identification of the atypical Gγ subunit, AGG3 (Chakravorty et al., 2011). Even the characterization of AGG3 only expanded the number of heterotrimer combinations to three and, surprisingly, suggested that Gγ was the only subunit conferring isoform diversity in Arabidopsis. Here, we provide evidence from several complementary approaches that all three XLGs function as components of the heterotrimeric G protein complex. XLGs and GPA1 bind selected Gβγ dimers with similar or greater strength (Fig. 1), and XLG3 and GPA1 compete with each other for the binding of AGB1/AGG3 in our yeast results (Fig. 3). Furthermore, in silico analysis suggests that the three XLG proteins structurally resemble GPA1 (Fig. 4; Supplemental Figs. S1 and S2; Supplemental Tables S1 and S2). All three XLGs interact with Gβγ dimers in planta (Fig. 5), XLG3 is sequestered at the plasma membrane by both AGB1/AGG1 and AGB1/AGG2 (Fig. 10), and xlg triple mutants phenocopy several agb1 phenotypes (Figs. 11–14). Therefore, it appears that the Gα family in Arabidopsis comprises four members: GPA1, XLG1, XLG2, and XLG3. The addition of the XLGs as Gα subunits expands the repertoire of heterotrimer combinations from three to 12.
MATERIALS AND METHODS
Plant Lines and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) transfer DNA insertions used have been described previously and were isolated in the Col-0 background, with the exception of agg1-1c, which was identified in the Wassilewskija ecotype but introgressed into Col-0 by Trusov et al. (2007). Mutants used were gpa1-3 and gpa1-4 (Jones et al., 2003), agb1-2 (Ullah et al., 2003), xlg1-1, xlg2-1, xlg3-1, and the xlg1-1 xlg2-1 xlg3-1 triple mutant (Ding et al., 2008), agg1-1c and agg2-1 (Trusov et al., 2007), and agg3-1 (Chakravorty et al., 2011). Aside from the agg1-1c agg2-1 double mutant (Trusov et al., 2007), the Gγ double and triple mutants have not previously been described but are combinations of the above published alleles. Arabidopsis plants were grown in Metro Mix 360 soil (Sun Gro Horticulture), and Nicotiana benthamiana plants were grown in a 1:1 mix of Metro Mix 360 and Sunshine Mix LC1 (Sun Gro Horticulture), in growth chambers with an 8-h-light/16-h-dark cycle, 120 µmol photons m−2 s−1 light, and 20°C.
Yeast Three-Hybrid Experiments
XLG clones were mobilized from pCR8/GW/TOPO (Life Technologies) into pDEST-GADT7 (Uetz et al., 2006) by Gateway cloning methods (Life Technologies). Gγ subunit genes and truncations of the Gγ subunits were cloned into the EcoRI/SalI restriction sites of MCS1 of pBridge. AGB1 was cloned into the NotI/BglII restriction sites of MCS2 to generate yeast (Saccharomyces cerevisiae) three-hybrid constructs that express the Gβ and Gγ subunits. Competition assay constructs were made by cloning XLG3 or GPA1 into the SpeI/EcoRI sites of p416GPD under the control of the strong yeast promoter GPD. The GPDpr::XLG3::CYC1 terminator and GPDpr::GPA1::CYC1 terminator expression cassettes were excised from the vectors by PCR, which added flanking NotI and BspEI restriction sites. The PCR fragments were cloned into the NotI/BspEI sites in the backbone of pDEST-GADT7-GPA1 and pDEST-GADT7-XLG3 vectors, yielding pDEST-GADT7-GPA1-GPDpr::XLG3 and pDEST-GADT7-XLG3-GPDpr::GPA1 vectors, respectively, for competition assays.
Yeast assays were performed in the Y2HGold yeast strain background (Clontech). Diploid cells from mating of Y2HGold bait strains with Y187 (Clontech) prey strains were selected on SC-Trp-Leu plates. To test for the presence and strength of interactions, diploid cells were spotted onto SC-Trp-Leu-Met-His medium supplemented with a range of 3-AT concentrations (0, 1, 2, 5, 10, 20, and 30 mm) to suppress His synthesis at increasing 3-AT levels (Durfee et al., 1993). Interaction was assessed by activation of the His synthesis reporter gene, which was scored by yeast growth, and the strength of interaction was assessed by growth on plates supplemented with progressively higher 3-AT concentrations.
BiFC and Fluorescent Protein Analyses
BiFC and localization analysis was performed essentially as described previously (Gookin and Assmann, 2014). Protocols for agroinfiltration, protoplast transformation, and confocal microscopy, as well as a description of the improved pDOE vector set used here, can be found in a recent publication (Gookin and Assmann, 2014). Cloning was performed using the strategies outlined in detail by Gookin and Assmann (2014), Methods S1 and S2. Briefly, XLGL protein fusions with internal NVen210 fragments were created by overlap PCR and inserted into the pDOE-10 MCS1 (simultaneously removing the vector-encoded tag) to create parent vectors. AGB1 was inserted into MCS3 to create the three UBIQUITIN10 promoter-driven XLGL-CVen210:AGB1 test vectors. UBIQUITIN10-driven XLG and residue-substituted XLG:mVenus fusions were created similarly by bridging pDOE-17 MCS1 and MCS3 in one cloning step (i.e. the 5′ end of the open reading frame was cloned into MCS1 and the 3′ end into MCS3). Likewise, the mTq2:histone 2B construct was created by bridging MCS1 and MCS3 of pDOE-19. The 35S promoter-driven untagged AGB1-AGG and single AGG constructs were made similarly and utilized previously (Gookin and Assmann, 2014). Agroinfiltration at low optical densities is critical for avoiding artifacts; therefore, test vectors were infiltrated at a final OD600 of 0.0075 to 0.01. When utilized, additional subunits were added in presumed excess by infiltrating at an OD600 of 0.025 to 0.03. The FM4-64 dye (Life Technologies), which marks the plasma membrane, was diluted to 50 µm and infiltrated into the transiently transformed N. benthamiana leaves just prior to imaging. Population level images were acquired at 20× magnification. Finer detail images were collected using a 40× magnification water-immersion objective. High-resolution/magnification images showing the specificity of histone 2B and XLG colocalization, and the specific overlap of XLG-Gβγ heterotrimer formation with the plasma membrane marker dye FM4-64, were obtained using a 63× oil-immersion lens and a 1.1-μm optical slice. The specific localization of XLG-Gβγ heterotrimer formation at the plasma membrane was further demonstrated at 95× magnification by the additional application of a 1.5× zoom of the confocal scan area during image acquisition (0.09 μm per pixel resolution). Images were acquired using the Zeiss LSM-510 confocal microscope and analyzed using the Zen 2009/2012 software.
Plate Assays
Plate assays were performed on standard 0.5× MS medium (Sigma) plates with 1% (w/v) agar (Sigma) supplemented with 1% (w/v) Suc (Calbiochem) for all assays except the d-Glc sensitivity assay. After plating, seeds were stratified at 4°C for 2 d before plates were transferred to short-day growth chamber conditions, as described above. Assays were performed with plates oriented horizontally. In the salt sensitivity assay, seeds were plated as described above and grown for 9 d, and then the seedlings were transferred to medium supplemented with 150 mm NaCl (EMD Millipore). Seedlings were grown for an additional 14 to 21 d. Tunicamycin assays were performed similarly to the assay of Wang et al. (2006). Briefly, seeds were initially plated on medium supplemented with 0.15 µm tunicamycin (Sigma). Seedlings were grown for 6 d, transferred to standard medium, and allowed to recover for an additional 10 d before scoring. Seedlings fell into three categories in our scoring system: (1) seedlings that did not grow after transfer from the tunicamycin plates and were clearly chlorotic and brown (dead); (2) seedlings that grew and often greened after transfer but were visibly impaired in their growth (stunted); and (3) seedlings that thrived after transfer (healthy). d-Glc sensitivity assays were performed using medium supplemented with 1% or 6% (w/v) d-Glc (Sigma) instead of Suc, with seedlings grown under a long-day 16-h-light/8-h-dark cycle. Seeds were germinated under high-light conditions (120 µmol photons m−2 s−1) for 1 d before lights were dimmed to 60 µmol photons m−2 s−1 as per the method of Wang et al. (2006), who described their assay as run under dim light. Seedlings were grown for 24 d. All plate assays were repeated at least three times with similar results.
Stomatal Density Assay
Seeds were plated and stratified as above. Seedlings were grown horizontally for 9 d in growth chambers with an 8-h-light/16-h-dark cycle, 90 to 100 µmol photons m−2 s−1 light, and 20°C. Cotyledons were excised, stained with 1.5 μm propidium iodide (Sigma) for 30 min in the dark, and mounted on coverslips with the abaxial side facing down. Images of abaxial epidermes were taken using an inverted Zeiss LSM-510 confocal microscope with a 20× objective lens. At least 10 seedlings were used for each genotype. Stomatal density was quantified from the images.
Anthocyanin Extraction
Anthocyanins were extracted according to the method of Mancinelli et al. (1991) with minor modifications. Briefly, intact aerial tissue of three to five seedlings was weighed, placed in acidic methanol (1% [v/v] HCl), and stored at 4°C in the dark overnight. Absorbance of the buffer with the leached pigments was measured in duplicate at A530 and A657 for three independent samples per genotype per condition using a Nanodrop 2000 (Thermo Scientific). The peak absorbance of anthocyanins occurs at 530 nm; however, chlorophyll also absorbs light at 530 nm, at approximately 25% of its peak A657. Therefore, as per the procedure of Mancinelli et al. (1991), comparative anthocyanin content was estimated by calculating (A530 − 0.25 × A657) per mg of aerial tissue.
Arabidopsis Genome Initiative (AGI) locus identifiers for the heterotrimeric G protein subunits outlined in this article are: GPA1 (AT2G26300), XLG1 (AT2G23460), XLG2 (AT4G34390), XLG3 (AT1G31930), AGB1 (AT4G34460), AGG1 (AT3G63420), AGG2 (AT3G22942), AGG3 (AT5G20635), and RGS1 (AT3G26090).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Predicted secondary structure elements of the Gα regions of the XLGs.
Supplemental Figure S2. Rotated view of the predicted XLG3 structure in Figure 4.
Supplemental Figure S3. ProSA validation of the XLG3 Gα theoretical structure model.
Supplemental Figure S4. ERRAT analysis of the XLG3 Gα theoretical structure model.
Supplemental Figure S5. Population level images of XLG-AGB1 BiFC.
Supplemental Figure S6. High-magnification colocalization of XLGs and histone 2B.
Supplemental Table S1. Summary of structural template matches for the XLG proteins, obtained using Phyre2.
Supplemental Table S2. Summary of structural template matches for the XLG proteins, obtained using genTHREADER.
Supplemental File S1. Protein Data Bank file of the computationally derived XLG3α structure.
Supplementary Material
Glossary
- ROS
reactive oxygen species
- BiFC
bimolecular fluorescence complementation
- 3-AT
3-amino-1,2,4-triazole
- NLS
nuclear localization signal
- NES
nuclear export signal
- UPR
unfolded protein response
- Col-0
Columbia-0
- MS
Murashige and Skoog
- OD600
optical density at 600 nm
Footnotes
This work was supported by the National Science Foundation (grant no. MCB–1121612 to S.M.A.) and by the U.S. Department of Agriculture/Agriculture and Food Research Initiative (postdoctoral grant no. 2011–67012–30722 to M.J.M.).
Articles can be viewed without a subscription.
References
- Anderson DJ, Botella JR (2007) Expression analysis and subcellular localization of the Arabidopsis thaliana G-protein β-subunit AGB1. Plant Cell Rep 26: 1469–1480 [DOI] [PubMed] [Google Scholar]
- Birnbaumer L. (2007) Expansion of signal transduction by G proteins: the second 15 years or so. From 3 to 16 α subunits plus βγ dimers. Biochim Biophys Acta 1768: 772–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht NC, Jez JM, Pandey S (2011) An elaborate heterotrimeric G-protein family from soybean expands the diversity of plant G-protein networks. New Phytol 190: 35–48 [DOI] [PubMed] [Google Scholar]
- Bitter GA, Egan KM (1984) Expression of heterologous genes in Saccharomyces cerevisiae from vectors utilizing the glyceraldehyde-3-phosphate dehydrogenase gene promoter. Gene 32: 263–274 [DOI] [PubMed] [Google Scholar]
- Bolte S, Talbot C, Boutte Y, Catrice O, Read ND, Satiat-Jeunemaitre B (2004) FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J Microsc 214: 159–173 [DOI] [PubMed] [Google Scholar]
- Bommert P, Je BI, Goldshmidt A, Jackson D (2013) The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502: 555–558 [DOI] [PubMed] [Google Scholar]
- Botella JR. (2012) Can heterotrimeric G proteins help to feed the world? Trends Plant Sci 17: 563–568 [DOI] [PubMed] [Google Scholar]
- Botto JF, Ibarra S, Jones AM (2009) The heterotrimeric G-protein complex modulates light sensitivity in Arabidopsis thaliana seed germination. Photochem Photobiol 85: 949–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford W, Buckholz A, Morton J, Price C, Jones AM, Urano D (2013) Eukaryotic G protein signaling evolved to require G protein-coupled receptors for activation. Sci Signal 6: ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty D, Trusov Y, Zhang W, Acharya BR, Sheahan MB, McCurdy DW, Assmann SM, Botella JR (2011) An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana. Plant J 67: 840–851 [DOI] [PubMed] [Google Scholar]
- Chen JG, Gao Y, Jones AM (2006) Differential roles of Arabidopsis heterotrimeric G-protein subunits in modulating cell division in roots. Plant Physiol 141: 887–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Jones AM (2004) AtRGS1 function in Arabidopsis thaliana. Methods Enzymol 389: 338–350 [DOI] [PubMed] [Google Scholar]
- Chen JG, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP (2003) A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301: 1728–1731 [DOI] [PubMed] [Google Scholar]
- Chen Y, Brandizzi F (2012) AtIRE1A/AtIRE1B and AGB1 independently control two essential unfolded protein response pathways in Arabidopsis. Plant J 69: 266–277 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Li JF, Niu Y, Zhang XC, Woody OZ, Xiong Y, Djonović S, Millet Y, Bush J, McConkey BJ, et al. (2015) Pathogen-secreted proteases activate a novel plant immune pathway. Nature 521: 213–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury SR, Bisht NC, Thompson R, Todorov O, Pandey S (2011) Conventional and novel Gγ protein families constitute the heterotrimeric G-protein signaling network in soybean. PLoS ONE 6: e23361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaneri AC, Tunc-Ozdemir M, Huang JP, Jones AM (2014) Growth attenuation under saline stress is mediated by the heterotrimeric G protein complex. BMC Plant Biol 14: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2: 1511–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coursol S, Fan LM, Le Stunff H, Spiegel S, Gilroy S, Assmann SM (2003) Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423: 651–654 [DOI] [PubMed] [Google Scholar]
- Delgado-Cerezo M, Sánchez-Rodríguez C, Escudero V, Miedes E, Fernández PV, Jordá L, Hernández-Blanco C, Sánchez-Vallet A, Bednarek P, Schulze-Lefert P, et al. (2012) Arabidopsis heterotrimeric G-protein regulates cell wall defense and resistance to necrotrophic fungi. Mol Plant 5: 98–114 [DOI] [PubMed] [Google Scholar]
- Ding L, Pandey S, Assmann SM (2008) Arabidopsis extra-large G proteins (XLGs) regulate root morphogenesis. Plant J 53: 248–263 [DOI] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ (1993) The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev 7: 555–569 [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Luthy R, Bowie JU (1997) VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol 277: 396–404 [DOI] [PubMed] [Google Scholar]
- Fan LM, Zhang W, Chen JG, Taylor JP, Jones AM, Assmann SM (2008) Abscisic acid regulation of guard-cell K+ and anion channels in Gβ- and RGS-deficient Arabidopsis lines. Proc Natl Acad Sci USA 105: 8476–8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gookin TE, Assmann SM (2014) Significant reduction of BiFC non-specific assembly facilitates in planta assessment of heterotrimeric G-protein interactors. Plant J 80: 553–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra R, Welch D, Scheres B (2004) Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev 18: 1964–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S, Assmann SM (2012) Ca2+-dependent GTPase, extra-large G protein 2 (XLG2), promotes activation of DNA-binding protein related to vernalization 1 (RTV1), leading to activation of floral integrator genes and early flowering in Arabidopsis. J Biol Chem 287: 8242–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Taylor JP, Chen JG, Uhrig JF, Schnell DJ, Nakagawa T, Korth KL, Jones AM (2006) The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell 18: 1226–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurowitz EH, Melnyk JM, Chen YJ, Kouros-Mehr H, Simon MI, Shizuya H (2000) Genomic characterization of the human heterotrimeric G protein α, β, and γ subunit genes. DNA Res 7: 111–120 [DOI] [PubMed] [Google Scholar]
- Ishida T, Tabata R, Yamada M, Aida M, Mitsumasu K, Fujiwara M, Yamaguchi K, Shigenobu S, Higuchi M, Tsuji H, et al. (2014) Heterotrimeric G proteins control stem cell proliferation through CLAVATA signaling in Arabidopsis. EMBO Rep 15: 1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa A, Iwasaki Y, Asahi T (1996) Molecular cloning and characterization of a cDNA for the β subunit of a G protein from rice. Plant Cell Physiol 37: 223–228 [DOI] [PubMed] [Google Scholar]
- Ishikawa A, Tsubouchi H, Iwasaki Y, Asahi T (1995) Molecular cloning and characterization of a cDNA for the α subunit of a G protein from rice. Plant Cell Physiol 36: 353–359 [DOI] [PubMed] [Google Scholar]
- Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen JG, Siderovski DP, Jones AM, Willard FS (2007) GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc Natl Acad Sci USA 104: 17317–17322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CA, Willard MD, Kimple AJ, Siderovski DP, Willard FS (2008) A sweet cycle for Arabidopsis G-proteins: recent discoveries and controversies in plant G-protein signal transduction. Plant Signal Behav 3: 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Ecker JR, Chen JG (2003) A reevaluation of the role of the heterotrimeric G protein in coupling light responses in Arabidopsis. Plant Physiol 131: 1623–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT. (1999a) GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J Mol Biol 287: 797–815 [DOI] [PubMed] [Google Scholar]
- Jones DT. (1999b) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292: 195–202 [DOI] [PubMed] [Google Scholar]
- Jones JC, Duffy JW, Machius M, Temple BRS, Dohlman HG, Jones AM (2011a) The crystal structure of a self-activating G protein α subunit reveals its distinct mechanism of signal initiation. Sci Signal 4: ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Temple BRS, Jones AM, Dohlman HG (2011b) Functional reconstitution of an atypical G protein heterotrimer and regulator of G protein signaling protein (RGS1) from Arabidopsis thaliana. J Biol Chem 286: 13143–13150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato C, Mizutani T, Tamaki H, Kumagai H, Kamiya T, Hirobe A, Fujisawa Y, Kato H, Iwasaki Y (2004) Characterization of heterotrimeric G protein complexes in rice plasma membrane. Plant J 38: 320–331 [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJE (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Klopffleisch K, Phan N, Augustin K, Bayne RS, Booker KS, Botella JR, Carpita NC, Carr T, Chen JG, Cooke TR, et al. (2011) Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol Syst Biol 7: 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y, Hu CD (2012) Bimolecular fluorescence complementation (BiFC): a 5-year update and future perspectives. Biotechniques 53: 285–298 [DOI] [PubMed] [Google Scholar]
- Koizumi N, Martinez IM, Kimata Y, Kohno K, Sano H, Chrispeels MJ (2001) Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol 127: 949–962 [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, Yanagawa H (2009) Six classes of nuclear localization signals specific to different binding grooves of importin α. J Biol Chem 284: 478–485 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H (2008) Nuclear export signal consensus sequences defined using a localization-based yeast selection system. Traffic 9: 2053–2062 [DOI] [PubMed] [Google Scholar]
- la Cour T, Kiemer L, Mølgaard A, Gupta R, Skriver K, Brunak S (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel 17: 527–536 [DOI] [PubMed] [Google Scholar]
- Lease KA, Wen J, Li J, Doke JT, Liscum E, Walker JC (2001) A mutant Arabidopsis heterotrimeric G-protein β subunit affects leaf, flower, and fruit development. Plant Cell 13: 2631–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YRJ, Assmann SM (1999) Arabidopsis thaliana ‘extra-large GTP-binding protein’ (AtXLG1): a new class of G-protein. Plant Mol Biol 40: 55–64 [DOI] [PubMed] [Google Scholar]
- Li S, Liu Y, Zheng L, Chen L, Li N, Corke F, Lu Y, Fu X, Zhu Z, Bevan MW, et al. (2012) The plant-specific G protein γ subunit AGG3 influences organ size and shape in Arabidopsis thaliana. New Phytol 194: 690–703 [DOI] [PubMed] [Google Scholar]
- Liu J, Ding P, Sun T, Nitta Y, Dong O, Huang X, Yang W, Li X, Botella JR, Zhang Y (2013) Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. Plant Physiol 161: 2146–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH (2010) Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22: 2930–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente F, Alonso-Blanco C, Sánchez-Rodriguez C, Jorda L, Molina A (2005) ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J 43: 165–180 [DOI] [PubMed] [Google Scholar]
- Lorek J, Griebel T, Jones AM, Kuhn H, Panstruga R (2013) The role of Arabidopsis heterotrimeric G-protein subunits in MLO2 function and MAMP-triggered immunity. Mol Plant Microbe Interact 26: 991–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM (1990) Molecular cloning and characterization of GPA1, a G protein α subunit gene from Arabidopsis thaliana. Proc Natl Acad Sci USA 87: 3821–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancinelli AL, Rossi F, Moroni A (1991) Cryptochrome, phytochrome, and anthocyanin production. Plant Physiol 96: 1079–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta N, Trusov Y, Brenya E, Parekh U, Botella JR (2015) Membrane-localized extra-large G proteins and Gβγ of the heterotrimeric G proteins form functional complexes engaged in plant immunity in Arabidopsis. Plant Physiol 167: 1004–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Botella JR (2000) Completing the heterotrimer: isolation and characterization of an Arabidopsis thaliana G protein γ-subunit cDNA. Proc Natl Acad Sci USA 97: 14784–14788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Botella JR (2001) Isolation of a novel G-protein γ-subunit from Arabidopsis thaliana and its interaction with Gβ. Biochim Biophys Acta 1520: 147–153 [DOI] [PubMed] [Google Scholar]
- McCudden CR, Hains MD, Kimple RJ, Siderovski DP, Willard FS (2005) G-protein signaling: back to the future. Cell Mol Life Sci 62: 551–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilson SE, Assmann SM (2010) The α-subunit of the Arabidopsis heterotrimeric G protein, GPA1, is a regulator of transpiration efficiency. Plant Physiol 152: 2067–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K, Kiuchi T, Shoji K, Sampei K, Mizuno K (2012) Visualization of cofilin-actin and Ras-Raf interactions by bimolecular fluorescence complementation assays using a new pair of split Venus fragments. Biotechniques 52: 45–50 [DOI] [PubMed] [Google Scholar]
- Oldham WM, Hamm HE (2008) Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol 9: 60–71 [DOI] [PubMed] [Google Scholar]
- Orozco-Nunnelly DA, Muhammad D, Mezzich R, Lee BS, Jayathilaka L, Kaufman LS, Warpeha KM (2014) Pirin1 (PRN1) is a multifunctional protein that regulates quercetin, and impacts specific light and UV responses in the seed-to-seedling transition of Arabidopsis thaliana. PLoS ONE 9: e93371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Assmann SM (2004) The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein α subunit GPA1 and regulates abscisic acid signaling. Plant Cell 16: 1616–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Chen JG, Jones AM, Assmann SM (2006) G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol 141: 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pandey S, Monshausen GB, Ding L, Assmann SM (2008) Regulation of root-wave response by extra large and conventional G proteins in Arabidopsis thaliana. Plant J 55: 311–322 [DOI] [PubMed] [Google Scholar]
- Pandey S, Nelson DC, Assmann SM (2009) Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136: 136–148 [DOI] [PubMed] [Google Scholar]
- Partow S, Siewers V, Bjørn S, Nielsen J, Maury J (2010) Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast 27: 955–964 [DOI] [PubMed] [Google Scholar]
- Plakidou-Dymock S, Dymock D, Hooley R (1998) A higher plant seven-transmembrane receptor that influences sensitivity to cytokinins. Curr Biol 8: 315–324 [DOI] [PubMed] [Google Scholar]
- Schröder M, Kaufman RJ (2005) ER stress and the unfolded protein response. Mutat Res 569: 29–63 [DOI] [PubMed] [Google Scholar]
- Taddese B, Upton GJG, Bailey GR, Jordan SRD, Abdulla NY, Reeves PJ, Reynolds CA (2014) Do plants contain G protein-coupled receptors? Plant Physiol 164: 287–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thung L, Chakravorty D, Trusov Y, Jones AM, Botella JR (2013) Signaling specificity provided by the Arabidopsis thaliana heterotrimeric G-protein γ subunits AGG1 and AGG2 is partially but not exclusively provided through transcriptional regulation. PLoS ONE 8: e58503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thung L, Trusov Y, Chakravorty D, Botella JR (2012) Gγ1+Gγ2+Gγ3=Gβ: the search for heterotrimeric G-protein γ subunits in Arabidopsis is over. J Plant Physiol 169: 542–545 [DOI] [PubMed] [Google Scholar]
- Torres MA, Morales J, Sánchez-Rodríguez C, Molina A, Dangl JL (2013) Functional interplay between Arabidopsis NADPH oxidases and heterotrimeric G protein. Mol Plant Microbe Interact 26: 686–694 [DOI] [PubMed] [Google Scholar]
- Trusov Y, Rookes JE, Chakravorty D, Armour D, Schenk PM, Botella JR (2006) Heterotrimeric G proteins facilitate Arabidopsis resistance to necrotrophic pathogens and are involved in jasmonate signaling. Plant Physiol 140: 210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y, Rookes JE, Tilbrook K, Chakravorty D, Mason MG, Anderson D, Chen JG, Jones AM, Botella JR (2007) Heterotrimeric G protein γ subunits provide functional selectivity in Gβγ dimer signaling in Arabidopsis. Plant Cell 19: 1235–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y, Zhang W, Assmann SM, Botella JR (2008) Gγ1 + Gγ2 ≠ Gβ: heterotrimeric G protein Gγ-deficient mutants do not recapitulate all phenotypes of Gβ-deficient mutants. Plant Physiol 147: 636–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Dong YA, Zeretzke C, Atzler C, Baiker A, Berger B, Rajagopala SV, Roupelieva M, Rose D, Fossum E, et al. (2006) Herpesviral protein networks and their interaction with the human proteome. Science 311: 239–242 [DOI] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Temple B, Boyes DC, Alonso JM, Davis KR, Ecker JR, Jones AM (2003) The β-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell 15: 393–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Wang S, Jones AM (2002) Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol 129: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Young JC, Im KH, Sussman MR, Jones AM (2001) Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292: 2066–2069 [DOI] [PubMed] [Google Scholar]
- Urano D, Colaneri A, Jones AM (2014) Gα modulates salt-induced cellular senescence and cell division in rice and maize. J Exp Bot 65: 6553–6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Jones JC, Wang H, Matthews M, Bradford W, Bennetzen JL, Jones AM (2012a) G protein activation without a GEF in the plant kingdom. PLoS Genet 8: e1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Phan N, Jones JC, Yang J, Huang J, Grigston J, Taylor JP, Jones AM (2012b) Endocytosis of the seven-transmembrane RGS1 protein activates G-protein-coupled signalling in Arabidopsis. Nat Cell Biol 14: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursic D, Chinchilla K, Finkel JS, Culbertson MR (2004) Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res 32: 2441–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall MA, Posner BA, Sprang SR (1998) Structural basis of activity and subunit recognition in G protein heterotrimers. Structure 6: 1169–1183 [DOI] [PubMed] [Google Scholar]
- Wang HX, Weerasinghe RR, Perdue TD, Cakmakci NG, Taylor JP, Marzluff WF, Jones AM (2006) A Golgi-localized hexose transporter is involved in heterotrimeric G protein-mediated early development in Arabidopsis. Mol Biol Cell 17: 4257–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Xu Q, Yuan M (2011) The unfolded protein response induced by salt stress in Arabidopsis. Methods Enzymol 489: 319–328 [DOI] [PubMed] [Google Scholar]
- Wang S, Narendra S, Fedoroff N (2007) Heterotrimeric G protein signaling in the Arabidopsis unfolded protein response. Proc Natl Acad Sci USA 104: 3817–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XQ, Ullah H, Jones AM, Assmann SM (2001) G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292: 2070–2072 [DOI] [PubMed] [Google Scholar]
- Warpeha KM, Gibbons J, Carol A, Slusser J, Tree R, Durham W, Kaufman LS (2008) Adequate phenylalanine synthesis mediated by G protein is critical for protection from UV radiation damage in young etiolated Arabidopsis thaliana seedlings. Plant Cell Environ 31: 1756–1770 [DOI] [PubMed] [Google Scholar]
- Warpeha KM, Lateef SS, Lapik Y, Anderson M, Lee BS, Kaufman LS (2006) G-protein-coupled receptor 1, G-protein Gα-subunit 1, and prephenate dehydratase 1 are required for blue light-induced production of phenylalanine in etiolated Arabidopsis. Plant Physiol 140: 844–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warpeha KM, Upadhyay S, Yeh J, Adamiak J, Hawkins SI, Lapik YR, Anderson MB, Kaufman LS (2007) The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol 143: 1590–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Zhou W, Hu G, Wei J, Yang H, Huang J (2008) Heterotrimeric G-protein is involved in phytochrome A-mediated cell death of Arabidopsis hypocotyls. Cell Res 18: 949–960 [DOI] [PubMed] [Google Scholar]
- Weiss CA, Garnaat CW, Mukai K, Hu Y, Ma H (1994) Isolation of cDNAs encoding guanine nucleotide-binding protein β-subunit homologues from maize (ZGB1) and Arabidopsis (AGB1). Proc Natl Acad Sci USA 91: 9554–9558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35: W407–W410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenstetter S, Chakravorty D, Kula R, Urano D, Trusov Y, Sheahan MB, McCurdy DW, Assmann SM, Jones AM, Botella JR (2015) Evidence for an unusual transmembrane configuration of AGG3, a class C Gγ subunit of Arabidopsis. Plant J 81: 388–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Grishin NV, Chook YM (2012) NESdb: a database of NES-containing CRM1 cargoes. Mol Biol Cell 23: 3673–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachdav G, Kloppmann E, Kajan L, Hecht M, Goldberg T, Hamp T, Hönigschmid P, Schafferhans A, Roos M, Bernhofer M, et al. (2014) PredictProtein: an open resource for online prediction of protein structural and functional features. Nucleic Acids Res 42: W337–W343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Assmann SM (May 12, 2015) The heterotrimeric G-protein β subunit, AGB1, plays multiple roles in the Arabidopsis salinity response. Plant Cell Environ http://dx.doi.org/10.1111/pce.12542 [DOI] [PubMed] [Google Scholar]
- Zhang L, Hu G, Cheng Y, Huang J (2008) Heterotrimeric G protein α and β subunits antagonistically modulate stomatal density in Arabidopsis thaliana. Dev Biol 324: 68–75 [DOI] [PubMed] [Google Scholar]
- Zhao J, Wang X (2004) Arabidopsis phospholipase Dα1 interacts with the heterotrimeric G-protein α-subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. J Biol Chem 279: 1794–1800 [DOI] [PubMed] [Google Scholar]
- Zhu H, Li GJ, Ding L, Cui X, Berg H, Assmann SM, Xia Y (2009) Arabidopsis extra large G-protein 2 (XLG2) interacts with the Gβ subunit of heterotrimeric G protein and functions in disease resistance. Mol Plant 2: 513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.