Identification and analysis of novel miRNAs in diploid strawberry revealed mechanisms of miRNA evolution and miRNA-mediated regulation of large gene families.
Abstract
The wild strawberry (Fragaria vesca) has recently emerged as an excellent model for cultivated strawberry (Fragaria × ananassa) as well as other Rosaceae fruit crops due to its short seed-to-fruit cycle, diploidy, and sequenced genome. Deep sequencing and parallel analysis of RNA ends were used to identify F. vesca microRNAs (miRNAs) and their target genes, respectively. Thirty-eight novel and 31 known miRNAs were identified. Many known miRNAs targeted not only conserved mRNA targets but also developed new target genes in F. vesca. Significantly, two new clusters of miRNAs were found to collectively target 94 F-BOX (FBX) genes. One of the miRNAs in the new cluster is 22 nucleotides and triggers phased small interfering RNA production from six FBX genes, which amplifies the silencing to additional FBX genes. Comparative genomics revealed that the main novel miRNA cluster evolved from duplications of FBX genes. Finally, conserved trans-acting siRNA pathways were characterized and confirmed with distinct features. Our work identified novel miRNA-FBX networks in F. vesca and shed light on the evolution of miRNAs/phased small interfering RNA networks that regulate large gene families in higher plants.
The wild strawberry (Fragaria vesca) is an emerging model for the commercial strawberry (Fragaria × ananassa) as well as other Rosaceae fruit crops, including apple (Malus domestica), pear (Pyrus communis), peach (Prunus persica), and cherry (Prunus avium). While many of the rosaceous fruit trees require several years of juvenile growth before flowering, F. vesca takes only 3.5 to 5 months from seed to flower and fruit. The small genome size (240 Mb), a sequenced genome, extensive tissue- and stage-specific transcriptomes, diploidy, and ease of transformation make F. vesca an ideal model system for investigations into fundamental biological questions and identifications of the regulatory pathways of important economic traits (Shulaev et al., 2011; Darwish et al., 2013; Kang et al., 2013; Pantazis et al., 2013).
MicroRNAs (miRNAs) represent an important class of regulatory molecules found in plants and animals. Processed from fold-back hairpin precursor RNAs, miRNAs are 20 to 22 nucleotides in length and negatively regulate target genes through homology-directed cleavage or translational inhibition of mRNAs (Bartel, 2004; Mallory and Vaucheret, 2006). Certain miRNAs, in addition, can trigger the biogenesis of another type of regulatory small RNA called trans-acting small interfering RNA (tasiRNA). In Arabidopsis (Arabidopsis thaliana), the noncoding precursor RNA of tasiRNA is first cleaved by a trigger miRNA. One of the cleavage products is then converted into a double-stranded RNA by RNA-DEPENDENT RNA POLYMERASE6. This double-stranded RNA is subsequently cleaved, in progressive 21-nucleotide phased intervals, by DICER-LIKE4 (DCL4) into 21-nucleotide tasiRNAs (Allen et al., 2005; Yoshikawa et al., 2005). Recently, genome-wide small RNA sequencing and analyses of an ever-expanding number of plant species have revealed the production of 21-nucleotide secondary small interfering RNAs (siRNAs) from hundreds of genes through a mechanism like tasiRNAs. These siRNAs may function in trans (tasiRNAs) or in cis (cis-acting siRNAs) to cleave their target genes (Johnson et al., 2009; Zhai et al., 2011; Xia et al., 2012, 2013; Fei et al., 2013). Collectively, these tasiRNAs and cis-acting siRNAs, which are produced via progressive 21-nucleotide cleavage from the trigger miRNA cleavage site, are termed phased small interfering RNAs (phasiRNAs). Accordingly, phasiRNA-producing loci are termed PHAS loci (Zhai et al., 2011). In dicots, phasiRNAs have been found to be generated from and presumably to regulate large and conserved gene families such as those coding for the NUCLEOTIDE-BINDING AND LEUCINE-RICH REPEAT (NB-LRR) proteins, MYB transcription factors, and PENTATRICOPEPTIDE REPEAT (PPR) proteins (Zhai et al., 2011; Xia et al., 2012, 2013). The vast phasiRNA networks in higher plant genomes are just beginning to be recognized and discovered, and the functional roles of these phasiRNA networks are yet to be illuminated.
One of the most interesting and unique developmental processes in strawberry is its fruit. Unlike other conventional fruits, which usually develop from the ovary of a flower, the fleshy fruit of strawberry develops from the stem tip, the receptacle. The ovary of the strawberry flower (called the achene) instead dries up and dots the surface of the receptacle. Each achene contains a single seed and can be easily removed by manual manipulation. Nitsch (1950) showed that removing all of achenes from the receptacle prevented receptacle fruit enlargement. Furthermore, exogenous auxin application acted as a substitute for achenes and stimulated receptacle fruit growth. This experiment established auxin as an essential phytohormone that promotes strawberry receptacle enlargement and suggested achenes as the source of auxin (Nitsch, 1950). Despite these important early discoveries, not much is known about the underlying molecular mechanisms of strawberry fruit development. Recently, we comprehensively profiled mRNA levels during F. vesca flower and fruit development and identified stage- and tissue-specific transcripts, in particular transcripts unique to the receptacle (Kang et al., 2013; Hollender et al., 2014). Analysis of the F. vesca fruit transcriptome revealed the endosperm and seed coat as the primary tissues for fertilization-induced auxin and GA biosynthesis. However, it remains unknown whether small RNAs play a role in regulating flower and fruit development and if small RNAs contribute to the success of receptacle fruit formation.
Here, we collected tissues of various developmental stages from F. vesca and dissected the fruit into separate fine parts to profile their small RNA population by deep sequencing and computational analyses. We identified two novel miRNA clusters, one bearing eight different miRNAs in an approximately 8-kb genomic region and the other with four copies of the same miRNA. All nine miRNAs were found to target a large number of F-BOX (FBX) genes, with the majority of them targeting at a conserved region coding for the F-box domain. One of the nine miRNAs is of 22 nucleotides in length, targets at least six FBX genes, and initiates secondary phasiRNA production, leading to amplification of the silencing effect. Hence, combining the ability to initiate phasiRNAs with target pairing at the highly conserved F-box domain, these novel miRNAs are particularly effective in regulating large gene families. Moreover, the main miRNA cluster was shown to arise only recently from duplications of the target FBX genes in the F. vesca genome. Our work defines a new miRNA-FBX regulatory network in F. vesca and provides novel insights into the evolution of miRNAs and their mRNA targets.
RESULTS
Identification of Novel and Known miRNA Families in F. vesca
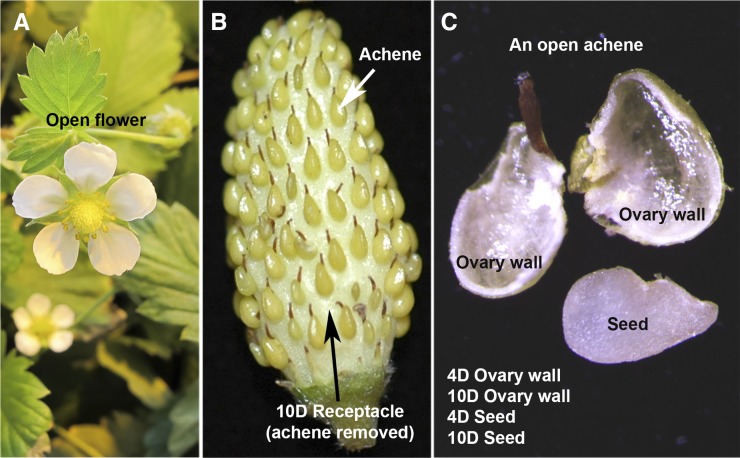
To maximize our chances of identifying F. vesca miRNAs, we isolated RNA from vegetative tissues (young leaves and seedlings), flowers (flower buds and newly opened flowers), and fruit tissues that separated achenes from the receptacle (Fig. 1). We further dissected each achene to harvest the ovary wall and the seed separately (Fig. 1C). Since fruit initiation occurs only after successful fertilization, the ovary wall and seed were harvested at 4 days post anthesis (DPA) as well as 10 DPA. Together, small RNAs from nine different tissues/libraries were sequenced to yield 10 to 60 million small RNA sequence reads per library (Supplemental Table S1).
Figure 1.
Photographs of F. vesca floral and fruit tissues used as the source of small RNAs. A, An open flower. B, An image of a developing fruit, illustrating the relationship between achene, the botanical fruit, and receptacle, the accessary fruit. C, Photograph showing a dissected achene, revealing ovary wall and the white seed, normally enclosed inside the ovary wall. 4D and 10D indicate samples harvested at 4 and 10 DPA, respectively.
miRNA identification followed an established pipeline (Supplemental Fig. S1). First, we identified 91 MIRNA genes, encoding 59 unique miRNA sequences and belonging to 31 known miRNA families (Supplemental Table S2). In general, known miRNA families are conserved across multiple plant species, have many family members, and are of higher abundance. For example, miR156 has eight variants (i.e. eight unique sequences) derived from 15 distinct loci in the genome. To identify novel miRNAs not previously described, all small RNAs with length of 20 to 22 nucleotides were subjected to stringent filtering criteria (Meyers et al., 2008; Supplemental Fig. S1). A total of 33 miRNA loci were identified that had greater than 75% processing accuracy as well as an miRNA star (miRNA*) present in the same library. After moving the three recently reported F. × ananassa miRNAs, fve-miR11283, fve-miR11284, and fve-miR11285 (Supplemental Table S2; Li et al., 2013), from our novel list, we have identified 30 novel miRNAs from F. vesca. They were submitted to miRBase and named fve-miR11286 to fve-miR11314 (Table I; Supplemental Table S3). Unlike the conserved miRNA families, most of the novel fve-miRNAs are coded by a single MIRNA locus and present at a lower abundance (Supplemental Table S3). Eleven of the 30 novel miRNAs are 22 nucleotides in length, which is a typical feature of miRNAs capable of initiating phasiRNA production (Chen et al., 2010; Cuperus et al., 2010).
Table I. Novel miRNAs identified in F. vesca.
| Novel miRNA Name | Mature Sequence | Length | Strand | Scaffold | Position |
|---|---|---|---|---|---|
| fve-miR11286 |
UUGGAGAGAGAGUAGACAAUG |
21 |
+ |
scf0513102 |
292,158 |
| fve-miR11287 |
UCAGGGAUUGUUUCAUAGACC |
21 |
− |
scf0512986 |
31,505 |
| fve-miR11288a(miRFBX1) |
UGGGAUUUGGCGAAUUGUGGU |
21 |
+ |
scf0513142 |
48,824 |
| fve-miR11288c2*(miRFBX4*) |
UUCGUUCGGAUUCCAAUUCAAA |
22 |
+ |
scf0513142 |
50,334 |
| fve-miR11289 |
UCGUCGACAUCAAAGGGCACC |
21 |
− |
scf0513044 |
1,313,006 |
| fve-miR11290 |
UGCUUCAAGUCUGGCCAAUACU |
22 |
− |
scf0513192 |
861,001 |
| fve-miR11291 |
UUGCGGUCUUGUCUCUUCCAAU |
22 |
− |
scf0513113 |
286,610 |
| fve-miR11292 |
UUGUAGUUCAGCGCCUCCGCC |
21 |
− |
scf0513157 |
951,427 |
| fve-miR11293 |
UUCUUCCUCAGGAACCUCCACC |
22 |
+ |
scf0513111 |
73,845 |
| fve-miR11294 |
UUCACCUGGACCAUAACUGACC |
22 |
− |
scf0513110 |
746,561 |
| fve-miR11295 |
CUCAUUCAAUUUCGGUAUUCAG |
22 |
− |
scf0513024 |
360,589 |
| fve-miR11296 |
UUUUUGAUGGCUGGAAUCCAGU |
22 |
− |
scf0513153 |
13,050 |
| fve-miR11297 |
CAGACAAGAUCGAUCUCGCCU |
21 |
+ |
scf0513124 |
126,841 |
| fve-miR11298 |
UUGAGGGGCUUAACGAUUACC |
21 |
− |
scf0512969 |
462,110 |
| fve-miR11299 |
CAAAUAGGGUUGGCUGAUACU |
21 |
− |
scf0513061 |
229,454 |
| fve-miR11300 |
CAACAUCACUGUUCUCUUCCU |
21 |
− |
scf0513136 |
542,507 |
| fve-miR11301 |
UCAGAGUUGUAAUAUAUUGAU |
21 |
+ |
scf0513131 |
1,429,968 |
| fve-miR11302 |
AGGACCGCCAUCACGUUUUGG |
21 |
+ |
scf0513002 |
843,149 |
| fve-miR11303 |
UCAAACAUCACUGCAGCUGUA |
21 |
+ |
scf0513134 |
1,902,196 |
| fve-miR11304 |
UAGUGUUUCAGCUUGACAAG |
20 |
+ |
scf0513172 |
207,257 |
| fve-miR11305 |
UUUUGGUCCGAAUCCGAGCUCC |
22 |
− |
scf0513080 |
37,252 |
| fve-miR11306 |
CUACCGAAGAACUUUGCAAAAG |
22 |
+ |
scf0512935 |
232,516 |
| fve-miR11307 |
UAAGCGACGGACUCCAAUCGC |
21 |
+ |
scf0513152 |
2,200,024 |
| fve-miR11308 |
UAAGUUAGGAUUCUAGUUACC |
21 |
− |
scf0513177 |
3,815,717 |
| fve-miR11309 |
UUUGUUUGGCAUGCAGUUGGC |
21 |
+ |
scf0513081 |
504,742 |
| fve-miR11310 |
AGGGAUCACAACUCCAGUGCU |
21 |
− |
scf0513138 |
426,264 |
| fve-miR11311 |
UGAGAAUGAUGUGGAUCUCAGC |
22 |
+ |
scf0513105 |
1,393,548 |
| fve-miR11312 |
UGGAGCUGUUGGGGAGAGUUA |
21 |
+ |
scf0513098 |
2,538,938 |
| fve-miR11313 |
GAAAAGAAUGGACUCUCCGGGG |
22 |
+ |
scf0513158 |
5,649,065 |
| fve-miR11314 |
AGAGUUGUGGAUGCUAUGAAU |
21 |
+ |
scf0513111 |
544,059 |
| fve-miR11288b(miRFBX2) |
UGGGAUUGGGCGAAUUUUGGU |
21 |
+ |
scf0513142 |
49,251 |
| fve-miR11288c1(miRFBX3) |
UCCAUCGUUUCGACACGCAGG |
21 |
+ |
scf0513142 |
50,416 |
| fve-miR11288c2(miRFBX4) |
UGAAUUGGGAUUUGUCGAAUU |
21 |
+ |
scf0513142 |
50,374 |
| fve-miR11288d1(miRFBX5) |
UCCAUCGUUUUGAGACACAGG |
21 |
+ |
scf0513142 |
52,546 |
| fve-miR11288d2(miRFBX6) |
UGAAUUGGGAUUUGGCGAAUU |
21 |
+ |
scf0513142 |
52,504 |
| fve-miR11288e1(miRFBX7) |
UGAUGGGUAGUCUGGAGAGGAU |
22 |
+ |
scf0513142 |
55,908 |
| fve-miR11288e2(miRFBX8) |
UGAAGUGGGAUUUGGCGAAUU |
21 |
+ |
scf0513142 |
55,825 |
| fve-miR11315(miRFBX9) | CUAGUCAUUGGUCAUAGCAUC | 21 | − | scf0513106 | 456,307 |
Newly Evolved Target Genes of Conserved miRNAs
A high-throughput approach, called parallel analysis of RNA ends (PARE), or degradome sequencing, or genome-wide mapping of uncapped and cleaved transcripts (Addo-Quaye et al., 2008; German et al., 2008; Willmann et al., 2014) can experimentally validate target transcripts of miRNAs by capturing mRNA cleavage products. This approach has the advantage of concurrent identification of all cleavage products in the genome as well as a high detection sensitivity, compared with older techniques such as northern blotting or 5′ RACE. To maximize the identification of miRNA targets, we dissected F. vesca tissues, mirroring the tissues used for small RNA sequencing; we then pooled RNA from these dissected tissues into three different PARE libraries: flower, fruit, and vegetative tissues. In total, approximately 86 million to approximately 108 million high-quality reads from each PARE library were mapped to the F. vesca transcriptome (Supplemental Table S1). Cleavage targets of 27 (of the 31) known miRNA families were found, totaling 280 target genes (Supplemental Table S4). As expected, these known miRNA families mainly targeted canonical conserved target genes, most of which encode transcription factors.
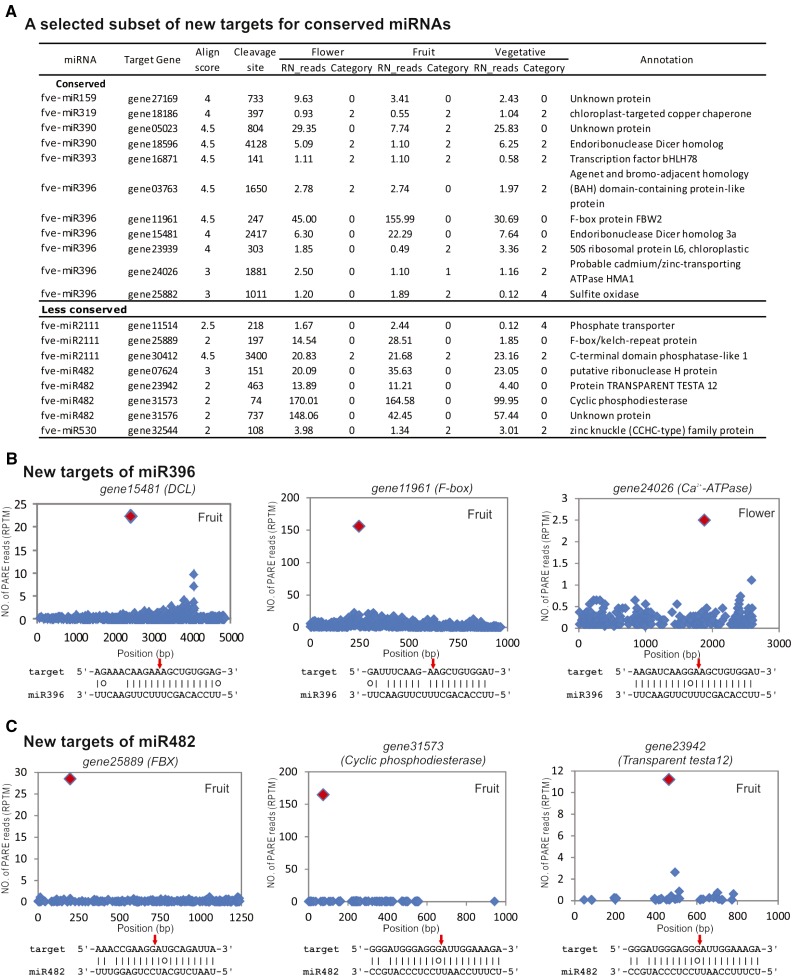
Unexpectedly, we found that many known miRNA families gained new targets, perhaps species or lineage specific (Supplemental Table S4). For example, apart from the previously known target genes encoding the growth regulation factor and the basic helix-loop-helix transcription factors (Rodriguez et al., 2010; Debernardi et al., 2012), fve-miR396 appeared to cleave 19 new target genes (Fig. 2A; Supplemental Table S4). The PARE tags validating the miRNA cleavage sites are shown in T-plots (Fig. 2B). Interestingly, miR396 and miR390 acquired as targets DCL genes (gene18596 and gene15481 in F. vesca, respectively; Fig. 2, A and B). Given the prior instances of miRNA regulation of miRNA processing or effector machineries in higher plants (Vaucheret et al., 2004; Zhai et al., 2011), it is not surprising that F. vesca also involves miRNAs in potential small RNA feedback loops.
Figure 2.
Novel target genes of conserved and less conserved miRNAs. A, List of selected novel target genes of conserved and less conserved miRNAs based on PARE data from flower, fruit, and vegetative tissues. Repeat normalized reads (RN_reads) are derived by dividing PARE read abundance by the number of loci in the genome to which the reads align. Alignment score (between 0 and 4.5) is calculated based on how well a miRNA pairs with its target sequence; a lower score indicates a better alignment between the miRNA and its target. The category score (between 0 and 4) considers the ratio of the PARE tag reads mapped to the corresponding miRNA-mediated cleavage site to the total number of PARE tags in a given target mRNA. The lower the category score, the higher the confidence of the cleaved targets. B, T-plots for selected target genes of miR396. The y axis indicates the abundance of cleavage PARE reads (reads per 10 million [RPTM]). The x axis indicates the coding sequence nucleotide position (bp), with 1 marking the first nucleotide of the start codon. The red diamond denotes the cleavage site by the corresponding miRNA in the fruit or flower PARE library. The base pairing between miRNA and the target transcript is indicated beneath each plot, with the exact cleavage site indicated by a red arrow. C, T-plots for selected target genes of miR482.
Two different 22-nucleotide miRNAs or families, miR482/miR2118 and miR2109, were previously reported to target a large number of NB-LRR disease resistance genes in tomato (Solanum lycopersicum) and Medicago truncatula (Zhai et al., 2011; Shivaprasad et al., 2012). Most M. truncatula NB-LRR transcripts produce phasiRNAs, triggering additional layers of NB-LRR transcript degradation (Zhai et al., 2011). In F. vesca, four different variants of fve-miR482 (miR482a, miR482b, miR482c, and miR482d) were identified; together, they targeted 50 F. vesca genes, the majority of which code for NB-LRRs (Supplemental Table S4). However, 18 were novel targets unrelated to NB-LRRs (Fig. 2, A and C; Supplemental Table S4). Similarly, miR2109 targeted 40 different NB-LRR genes. Our results indicate that even conserved miRNAs may rapidly acquire new family- or species-specific targets.
Two Clusters of Novel fve-miRNAs Targeting FBX Genes
Previous work hypothesized that most newly formed young miRNAs were neutrally evolving and evolutionarily transient loci. These new loci may not harbor any function but could serve as the source of novel regulatory variations, which could be captured into existing networks (Cuperus et al., 2011; Nozawa et al., 2012). Therefore, young miRNAs may not necessarily have targets or may have targets that are weakly or imprecisely cleaved. However, through analysis of the PARE data, we identified 112 target genes for 25 of the 30 novel miRNAs (Supplemental Table S5). This ratio (25 of 30) is, surprisingly, similar to that of conserved miRNAs with identifiable targets (27 of 31). In contrast, fewer target genes were identified for each of the novel miRNAs, and there was relatively low cleavage tag abundance for these targets (Supplemental Table S5). In addition, unlike conserved miRNAs that often targeted multiple members of a gene family, most novel miRNAs targeted genes from different gene families. Similar observations were made in other plant species, reflecting the properties of young miRNAs (Pantaleo et al., 2010; Xia et al., 2012; Zhu et al., 2012).
One of the most striking observations from the novel miRNA-target analyses was the unusually large number of FBX target genes for miR11288a, which targeted 11 FBX genes with high cleavage signals (category 0-2 in Supplemental Table S5). Further computational target prediction revealed up to 27 FBX genes with an fve-miR11288a target site (Supplemental Table S6), suggesting the existence of a previously uncharacterized yet extensive miRNA-FBX pathway in F. vesca.
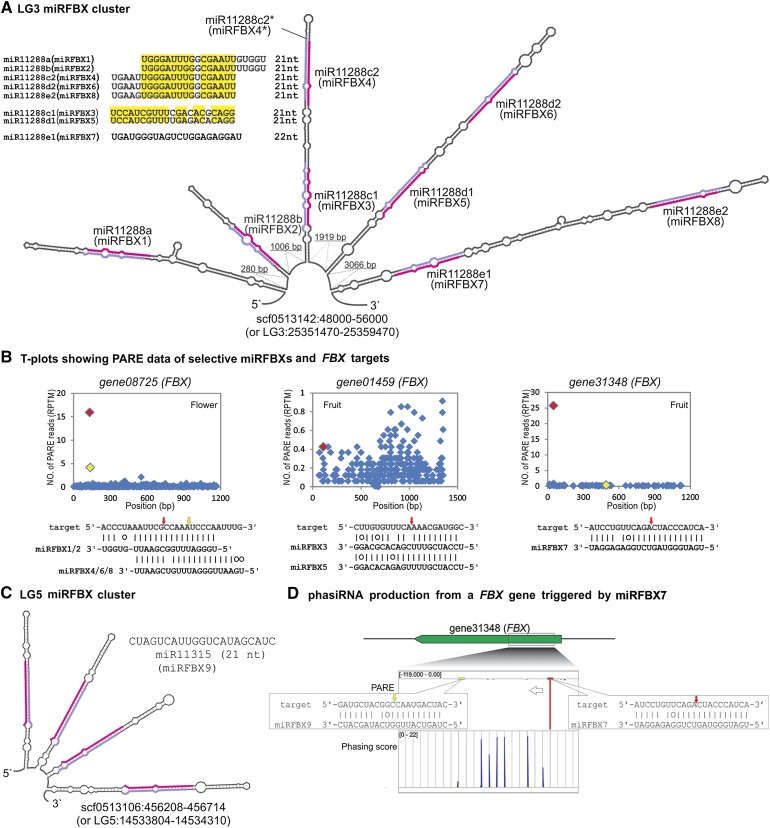
Interestingly, manual examination of the fve-MIR11288a locus at Linkage Group 3 (LG3) revealed seven potential miRNAs located within 8 kb downstream of fve-MIR11288a (scf0513142:48000-56000 or LG3:25351470-25359470; Fig. 3A). These seven potential MIRNAs reside within four good stem-loop structures and could generate miRNAs precisely (Fig. 3A). Therefore, these seven miRNAs are considered as novel miRNAs. Because of their sequence similarity to each other (Fig. 3A; Supplemental Fig. S2), miRBase named them as fve-miR11288a, fve-miR11288b, fve-miR11288c1, fve-miR11288c2, fve-miR11288d1, fve-miR11288d2, fve-miR11288e1, and fve-miR11288e2 (Fig. 3A; Table I; Supplemental Table S3). However, these eight related miRNAs are more easily remembered as miRFBX1, miRFBX2, miRFBX3, miRFBX4, miRFBX5, miRFBX6, miRFBX7, and miRFBX8 (Fig. 3A) because of their targeting at a large number of FBX genes (see below). Within this 8-kb region, the star sequence of miRFBX4 was annotated as a novel miRNA in earlier computational identification (Fig. 3A; Supplemental Fig. S3).
Figure 3.
Two new fve-miRNA clusters in the F. vesca genome. A, Computationally predicted multiple stem-loop structure from an 8-kb region in scf0513142 located in LG3. Eight pairs of miRNA and miRNA* are marked in pink and light purple, respectively. Their names as given by miRBase (miR11288a, miR11288b, miR11288c1, miR11288c2, miR11288d1, miR11288d2, miR11288e1, and miR11288e2) are indicated; an easier to remember name, miRFBX1 to miRFBX8, is also given for each miRNA in the cluster. Seven of these eight miRNAs are related in sequence, as shown in the top left corner; conserved nucleotides are highlighted in yellow. B, T-plots for selected fve-miRFBX genes and their targeted FBX transcripts. The red and yellow diamonds denote the cleavage sites of miRFBXs. Yellow refers to a second cleavage event by a second miRFBX. C, Another stem-loop structure in LG5 codes for a cluster of four identical miRNAs (miR11315/miRFBX9). The miRFBX9 sequence is indicated above the structure. D, Primary and secondary cleavages of gene31348 (FBX) and phasiRNA production. gene31348 shown on the top is cleaved by miRFBX7 (red arrow) and miRFBX9 (yellow arrow). In the PARE track, cleavage tag abundance is shown as a red down-pointing peak; the negative value indicates antisense transcript (left-pointing arrow); numbers in brackets indicate the range of PARE read abundance. In the phasing score track, phasiRNA peaks are evenly distributed and upward pointing. The phasing score inside the bracket was calculated from the flower small RNA library.
This cluster of eight miRNAs exhibited additional features. First, while fve-miFBX1 and fve-miRFBX2 are each derived from an independent stem-loop precursor, the other three pairs, fve-miRFBX3 and fve-miRFBX4, fve-miRFBX5 and fve-miRFBX6, and fve-miRFBX7 and fve-miRFBX8, are derived from three stem-loop precursors. The derivation of multiple miRNAs from the same stem-loop precursor was observed previously in other plants (Zhang et al., 2010). Second, seven of the eight matured fve-miRNAs (except fve-miRFBX7) show sequence similarities, with a minimum identity of 15 nucleotides, to one another (Fig. 3A). Strikingly, all eight fve-miRNAs were predicted to collectively target 94 FBX genes (Supplemental Table S6). Cleavage products of 51 of the 94 FBX genes were confirmed by PARE (Fig. 3B; Supplemental Table S6). Many of these FBX genes could be cleaved by two or more miRNAs (Fig. 3B; Supplemental Table S6). For simplicity, we refer to this cluster of eight fve-miRNAs as the LG3 miRFBX cluster.
To identify FBX-targeting miRNAs elsewhere in the genome, we conducted target gene prediction for all unique small RNAs with good stem-loop precursor structures (derived from earlier analyses) and found another novel miRNA, fve-miR11315 (miRFBX9). This miRNA is encoded by a cluster of four stem-loop structures (Fig. 3C) located within an approximately 500-bp region (scf0513106: 456,208–456,714; or LG5: 14,533,804–14,534,310) and bears no sequence similarity to miRNAs at the LG3 miRFBX cluster (Fig. 3, A and C). This miRNA (fve-miR11315 or miRFBX9) was predicted to target 12 FBX genes, and the PARE data confirmed the cleavage of seven of the 12 FBX genes (Supplemental Table S6). We refer to this second cluster as the LG5 miRFBX cluster. Collectively, these nine miRNAs (miRFBX1–miRFBX9) at the LG3 and LG5 miRFBX clusters are termed fve-miRFBXs.
fve-miRFBX7 Cleaves FBX Transcripts and Initiates phasiRNA Production
Among the nine fve-miRFBXs, only fve-miRFBX7 is 22 nucleotides in length and possesses an initial uridine. In plants, 22-nucleotide uridine-initiated miRNAs are capable of triggering phasiRNAs from cleaved mRNAs of PHAS loci (Chen et al., 2010; Cuperus et al., 2010). We assessed whether fve-miRFBX7 can initiate phasiRNA production. Using our PHAS profiling algorithm described previously (Xia et al., 2013) and the nine small RNA libraries, we identified a total of 24 PHAS FBX genes for which phased peaks of phasiRNAs could be detected (Supplemental Table S7). However, only six of these 24 PHAS FBX genes are directly cleaved by fve-miRFBX7, as indicated by solid alignment scores between fve-miRFBX7 and the respective FBX genes (Supplemental Table S7).
In addition, fve-miRFBX7 appears to possess common FBX targets with miRFBX9, gene31348, gene31347, gene31349, gene13786, and gene27192 (Supplemental Table S6), indicating that fve-miRFBX9 and fve-miRFBX7 may participate in a two-hit cleavage to delimit the boundary of a region that gives rise to phasiRNAs. For example, fve-miRFBX9 cleaves FBX gene31348 at a site approximately 220 bp downstream of the fve-miRFBX7 cleavage site (Fig. 3D). Indeed, abundant and progressively cleaved phasiRNAs were produced from the region immediately after the fve-miRFBX7 cleavage site (Fig. 3D).
miRFBXs Mostly Target the Conserved F-Box Domain
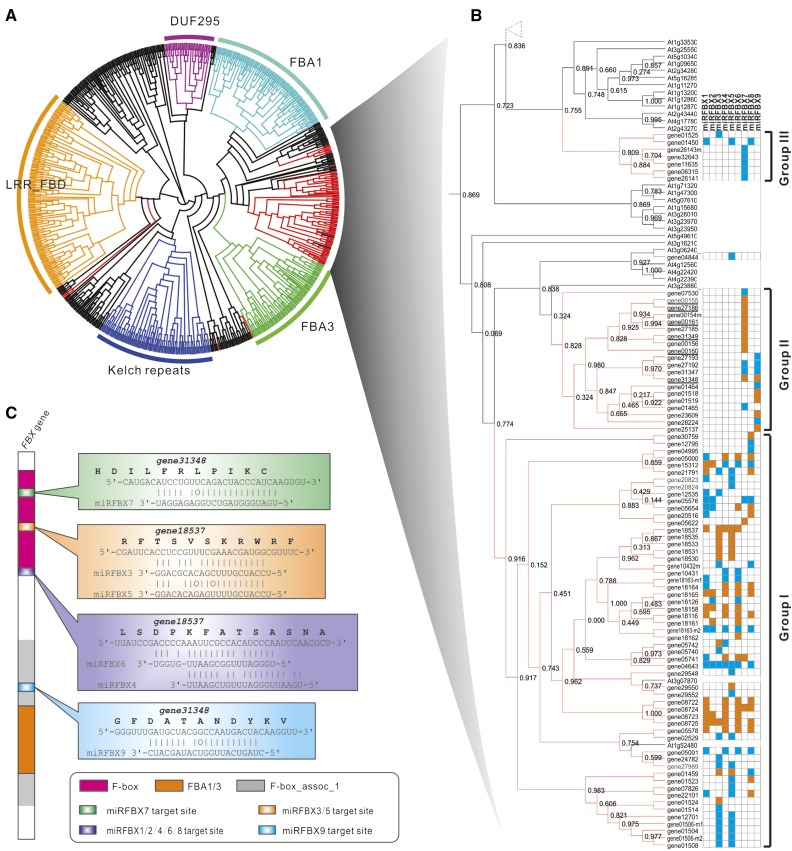
Plant FBX genes constitute a large family of genes (approximately 800 in F. vesca and approximately 690 in Arabidopsis) and can be classified into several distinct subfamilies based on domain structures (Kuroda et al., 2002; Xu et al., 2009). To investigate which specific FBX gene subfamilies are regulated by the clustered miRFBXs and deduce their potential functions, we performed phylogenetic analysis using all the FBX genes from Arabidopsis (691 genes; Xu et al., 2009) and the set of 96 F. vesca FBX genes targeted by the clustered miFBXs (Supplemental Table S6). As shown in Figure 4A, the Arabidopsis FBX genes are grouped into several distinct subfamilies, including FBA1, FBA3, LRR-FBD, Kelch repeats, and Domain Unknown Function295. Almost all the F. vesca FBX genes (except five) targeted by the fve-miRFBXs favorably cluster with the FBA1 and FBA3 subfamilies (Fig. 4A). The subtree for the F. vesca FBX genes could be further divided into three groups. Group I is targeted mainly by the LG3 miRFBX cluster (except miRFBX7). Group II is targeted by fve-miRFBX7 and fve-miRFBX9 and encompasses many PHAS FBX genes. Group III is preferentially regulated by miRFBX7 (Fig. 4B).
Figure 4.
Analysis of FBX genes targeted by the nine fve-miRFBXs. A, Phylogenetic tree incorporating all Arabidopsis FBX genes and miRFBX-targeted FBX genes in F. vesca. Five major classes are marked with distinct colors and indicated by group names (minor groups are in black). B, Enlarged F. vesca FBX genes targeted by the nine miRFBXs shown as orange lines. These genes are divided into three groups (I, II, and III) according to positions on the tree and the targeting miRNAs. Blue-filled grids indicate FBX genes bearing a predicted target site (align score ≤ 5) for a given miRNA; orange-filled grids indicate the detection of cleavage products by PARE. PHAS FBX genes directly targeted by fve-miRFBX7 are underlined. C, Diagram of the FBX protein with typical domains characteristic of the FBA1/3 subfamilies. The positions of various miRFBX target sites are indicated, and examples of miRNA pairing to target FBX sequences are included on the right. Not all FBX genes harbor all the miRNA target sites.
Next, we examined the distribution and conservation pattern of miRFBX target sites within the FBX coding sequence. As the fve-miRFBX-targeted FBX genes are more similar to FBA1/3 subfamilies, we modeled the structure of an FBX gene with three typical domains, the F-box, FBA1/3, and the F-box_assoc_1 domain (Fig. 4C). The target sites of fve-miRFBX3, fve-miRFBX5, and fve-miRFBX7 are located within the encoded F-box domain (Fig. 4C). In contrast, the target sites of fve-miRFBX1, fve-miRFBX2, fve-miRFBX4, fve-miRFBX6, and fve-miRFBX8 are located at the C-terminal end of the F-box domain. The fve-miRFBX9 target site is in the F-box_assoc_1 domain in some FBX genes (Fig. 4C). Therefore, targeting at the more conserved region of the FBX genes endows these fve-miRFBXs with the ability to regulate a large number of FBX family members simultaneously. However, not every FBX gene harbors the target sites of all members of the miRFBXs; therefore, each FBX gene may only be targeted by a subset of the miRFBXs as illustrated in Figure 4B.
The LG3 Cluster of miRFBXs Recently Evolved in the F. vesca Genome from Gene Duplication
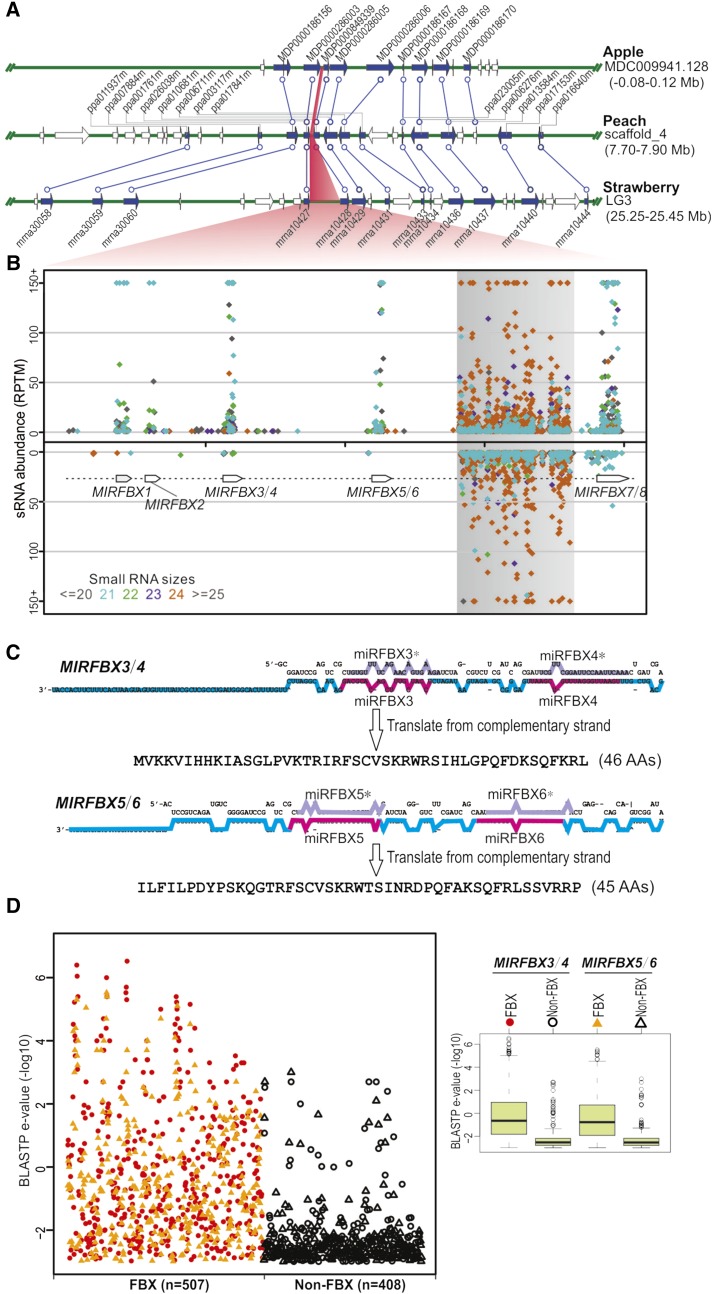
Does this regulatory network of miRFBX-FBX exist in other higher plants? Similar miRNA pathways have not been reported even in related species such as apple and peach (Xia et al., 2012; Zhu et al., 2012). Therefore, we searched and then identified syntenic regions of the LG3 miRFBX locus in many plant species (Supplemental Fig. S4). Strikingly, this region is significantly expanded in F. vesca (Fig. 5A) when compared with even the closely related Rosaceae species apple (in the genome between MDP0000286003 and MDP000349339) and peach (between ppa026038m and ppa010681m), suggesting that the miRFBX cluster may have recently evolved in F. vesca from a common ancestor of the Rosaceae. Small RNA mapping data at the LG3 miRFBX region revealed abundant miRNAs produced from the five putative MIRNA precursor loci, all in the same transcriptional orientation (Fig. 5B). Between MIRFBX5/6 and MIRFBX7/8 is a region with repetitive sequences that gave rise to large quantities of 24-nucleotide small RNAs in both directions (Fig. 5B).
Figure 5.
The LG3 miRFBX cluster likely evolved recently from FBX genes. A, Alignment of syntenic regions among apple, peach, and F. vesca. Genes are shown as arrows, with syntenic genes as filled blue arrows. Each syntenic gene pair is connected by a blue line. The LG3 miRFBX cluster region in strawberry and the corresponding syntenic regions in apple and peach are shaded in red. B, Small RNA distribution within the LG3 miRFBX region in F. vesca. Small RNAs of different length are denoted with different color dots. A region of repetitive DNA that yielded abundant 24-nucleotide small RNAs is shaded in gray. C, Stem-loop structures for MIRFBX3/4 and MIRFBX5/6 precursor RNAs. miRNA and miRNA* are highlighted in pink and light purple, respectively. The reverse-complementary sequence of the longer arm of the stem-loop structure (the cyan- and pink-colored region) codes for a peptide of 46 (for MIRFBX3/4) or 45 (for MIRFBX5/6) amino acids (AAs). D, Summary of BLASTP results for the two short peptides in C. BLASTP search was conducted against F. vesca proteins with an e-value cutoff of 1,000. All the hits (915 proteins) were classified into two groups: FBX proteins (507) and non-FBX proteins (408). Many more FBX proteins were obtained with significantly lower BLASTP e-values (show in −log10 scale) than the non-FBX proteins. The e-value differences between FBX and non-FBX proteins for the two peptides are shown in the box plot on the right.
To investigate from where these LG3 MIRFBX genes could originate, BLASTX was conducted with the LG3 MIRFBX DNA sequence (approximately 8 kb) as a query against public protein collections. The MIRFBX3/4 and MIRFBX5/6 regions showed significant similarity to protein-coding genes; the reverse complement strands of MIRFBX3/4 and MIRFBX5/6 loci can be translated into appreciable peptides of 45 or 46 amino acids, respectively (Fig. 5C). When BLASTP was conducted against all F. vesca protein-coding genes, the top hits for either of these two peptides were FBX proteins (Fig. 5D), and these FBX homologs showed significantly lower e-values than the non-FBX hits (Student’s t test, 2.2e-16; Fig. 5D). Our analysis strongly suggests that MIRFBX3/4 and MIRFBX5/6 were derived from FBX genes. Considering the high level of sequence similarity among the five stem-loop sequences within the LG3 miRFBX cluster (Supplemental Fig. S2), it is likely that these five stem loops at the LG3 evolved from duplications of a common precursor (see “Discussion”).
Expression of Precursor Transcripts of miRFBXs and Their Target FBX Genes in Various Tissues
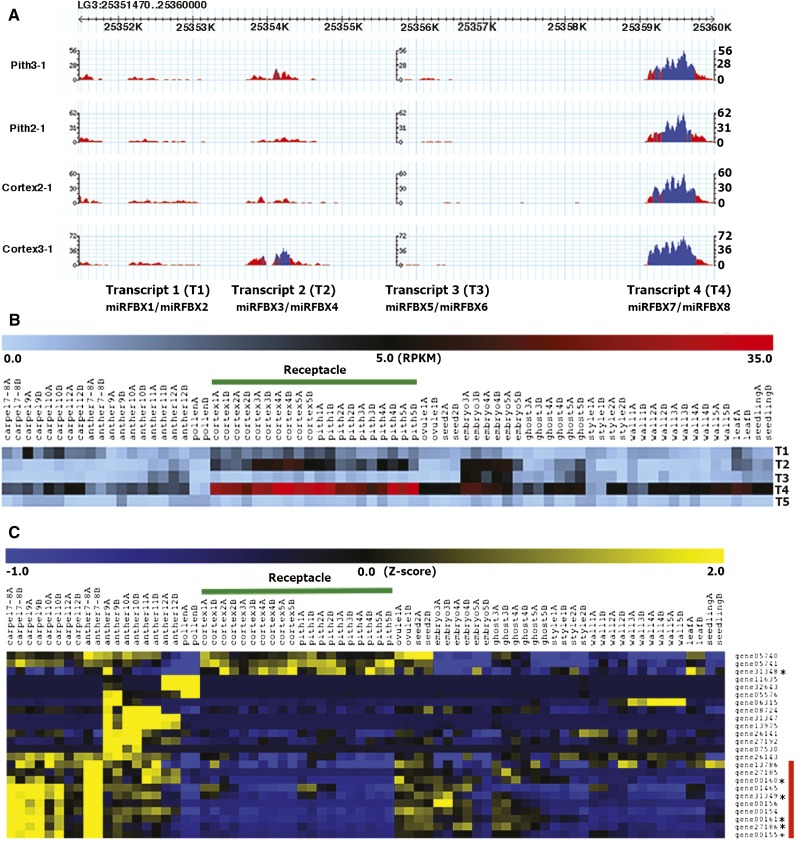
Since our initial small RNA libraries were made for the sole purpose of discovering small RNAs in F. vesca, the expression of each miRNA could not be quantitatively assessed in the absence of replicates for each of the nine small RNA libraries. However, we previously generated RNA-seq data for 37 different floral, fruit, and vegetative tissues in F. vesca with two biological replicates for each tissue type (Kang et al., 2013; Hollender et al., 2014). The levels of miRNA precursor transcripts could be examined. Figure 6A illustrates the RNA-seq data in the LG3 fve-miRFBX cluster region, which corresponds to at least four different precursor transcripts (transcripts 1–4). Of particular interest to us is transcript 4, which is the precursor for both fve-miRFBX7 and miRFBX8. Transcript 4 (Fig. 6B, row 4) is highly abundant and specific to both the pith and the cortex of the receptacle fruit at all five fruit developmental stages (pith 1–5 and cortex 1–5). Consistent with the precursor RNA expression, the small RNA read abundance for miRFBX7 and miRFBX8 also showed highest expression in the receptacle among the nine tissues sampled (Supplemental Fig. S5). fve-miRFBX7/8 may thus be involved in regulating receptacle fruit development. The RNA-seq read abundance for the precursor transcript of the MIRFBX9 cluster (transcript 5) was also examined, which showed extremely low levels in all floral and fruit tissues (Fig. 6B). This is consistent with the low expression of mature fve-miRFBX9 in flower and fruit tissues (Supplemental Fig. S5).
Figure 6.
miRFBX7/8 precursor transcript is specifically expressed in the receptacle fruit. A, RNA-seq read abundance displayed via GBrowse at the Genome Database for Rosaceae (GDR) on the LG3 miRFBX cluster region (LG3: 25351470–25359470). There appear to be four distinct transcription units: miRFBX1/2 (T1, transcript 1), miRFBX3/4 (T2, transcript 2), miRFBX5/6 (T3, transcript 3), and miRFBX7/8 (T4, transcript 4). Blue color indicates greater than 20× read coverage, while red color is 20× or less coverage. B, Heat map showing read abundance (reads per kilobase per million [RPKM]) of the four LG3 precursor transcripts (T1, T2, T3, and T4) shown in A and the LG5 miRFBX9 precursor transcript (labeled as T5). RNA-seq data of 37 different tissues and developmental stages (labeled by number) are shown. The two biological replicates are labeled as A and B. The receptacle (highlighted by the green line) is divided into pith and cortex at fruit developmental stages 1 to 5. C, Hierarchical clustering (Pearson correlation) showing the relative expression level of 24 FBX genes targeted by miRFBX7. Mapped reads across the same 37 tissues/stages were normalized via the Normalize Genes/Rows function in MeV4.8. The phasiRNA-producing FBX genes (triggered by miRFBX7) are marked by asterisks. The red bar on the right side donates the cluster showing reduced expression in pith and cortex of the receptacle.
To test if there is an inverse relationship in transcript abundance between miRFBX7 and its FBX targets, we investigated the transcript abundance of the 24 FBX genes, either predicted or proven by PARE analysis to be targets of miRFBX7 (Supplemental Table S6), using the same comprehensive flower and fruit RNA-seq data sets (Kang et al., 2013; Hollender et al., 2014). Hierarchical clustering of Z-score normalized mapped reads across 37 tissues was shown for each of the 24 FBX genes (Fig. 6C). Interestingly, 10 FBX genes clustered together (at the bottom of the heat map) and showed dramatic reduction of transcript abundance in the pith and cortex tissues (Fig. 6C). This is consistent with possible degradation of these 10 FBX transcripts by miRFBX7, whose precursor specifically accumulates in the pith and cortex (transcript 4 in Fig. 6B) and whose mature miRNAs were most abundant in the receptacle tissue (Supplemental Fig. S5). Among these 10 FBX genes, five are known to produce phasiRNAs triggered by miRFBX7 (Fig. 6C; Supplemental Table S7). Hence, this analysis identified a miRFBX7-FBX-phasiRNA network operating in the receptacle fruit of F. vesca.
Conserved tasiRNA Pathways in F. vesca
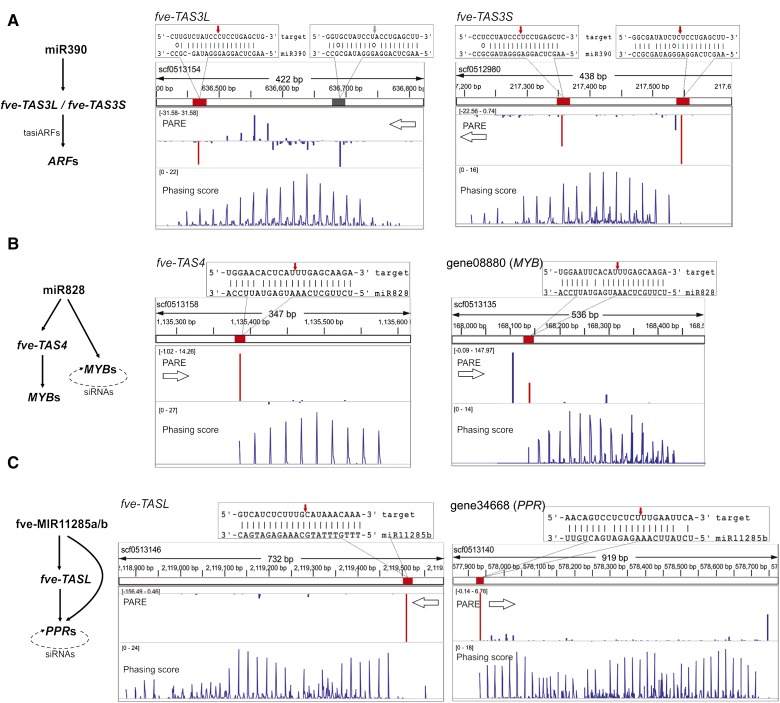
Three major conserved tasiRNA pathways have been characterized in plants; their biogenesis is initiated by three distinct miRNAs: miR390, miR828, and the miR7122 superfamilies (Fei et al., 2013; Xia et al., 2013). All three tasiRNA pathways are also found in F. vesca (Fig. 7). The miR390-TAS3 pathway is one of the most well-characterized pathways in plants and was identified in mosses as well as higher plants (Axtell et al., 2007; Howell et al., 2007). In Arabidopsis, miR390 cleaves TAS3 transcripts using a two-hit mechanism, generating tasiARFs that guide the degradation of transcripts encoding AUXIN RESPONSE FACTOR2 (ARF2), ARF3, and ARF4. tasiARFs regulate many important developmental processes ranging from leaf patterning and lateral root initiation to developmental timing (Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006; Marin et al., 2010). F. vesca has two TAS3 genes that we named TAS3L (L for long) and TAS3S (S for short; Fig. 7A). fve-TAS3L is processed similarly to Arabidopsis TAS3, resulting in two fve-tasiARFs. In contrast, fve-TAS3S is cleaved at both 3′ and 5′ sites (Fig. 7A). The phasiRNAs, including one tasiARF, are produced downstream of both cleavage sites. The demonstrated cleavage at both 3′ and 5′ sites of fve-TAS3S by the PARE data argues against an absolute requirement for a noncleavable 5′ site in the two-hit mode of tasiARF biogenesis (Axtell et al., 2006; Allen and Howell, 2010).
Figure 7.
Conserved tasiRNA pathways in F. vesca. Diagrams on the left illustrate conserved trans-acting pathways in F. vesca. On the right are genomic configurations of TAS genes showing three tracks: position of the miRNA target site (red or gray rectangular boxes) within the scaffold, PARE data (middle track), and phasing score (bottom track) viewed in the Integrative Genomics Viewer. Base pairing between the miRNA and corresponding transcript is enlarged above the tracks, with the exact cleavage site marked with a red or gray arrow. The gray box indicates a miR390 target site that is not cleaved. The PARE track and the phasing score track are as described in Figure 3D. The values in brackets in the PARE track and the phasing score track were derived from the flower PARE library and the flower small RNA library, respectively. A higher phasing score indicates a higher confidence of the PHAS loci. A, fve-TAS3L and fve-TAS3S are homologous genes located on different scaffolds. Each has two miR390-binding sites, indicated by the red and gray boxes in the top track. Cleavages by miR390 are shown by downward (antisense) or upward (sense) peaks. Red peaks indicate cleavage products by the indicated miRNA. Blue peaks indicate other cleavage products. B, fve-TAS4 and MYB genes are both cleaved by miR828 in typical one-hit mode. The dotted arrow in the diagram at left indicates self-cleavage by phasiRNAs. C, fve-TASL and PPR genes are each cleaved once by fve-miR11285a/b (previously named as fve-PPRtri1/2; Li et al., 2013; Xia et al., 2013). This initiates phasiRNAs revealed by the phasiRNA peaks. The dotted arrow in the diagram at left indicates self-cleavage by phasiRNAs.
miR828 is conserved in eudicots (Cuperus et al., 2011); although it was not identified in our list of known or conserved miRNAs due to its extremely low expression level, its cleavage of fve-TAS4 and tasiRNA production were confirmed by the PARE data and phasing analyses, respectively (Fig. 7B). In addition to targeting fve-TAS4, fve-miR828 directly cleaved several MYB genes such as gene08880 (Fig. 7B; Supplemental Table S8) leading to phasiRNAs, some of which target transcripts from cognate MYB genes. Copies of fve-MIR11285a/b (previously named as fve-PPRtri1/2) were described to target PPR genes directly or via cleavage of an intergenic noncoding locus named as a TAS-LIKE gene (TASL; Xia et al., 2013). This was confirmed by our PARE data, and the ensuing phasiRNAs were detected in many small RNA libraries (Fig. 7C).
DISCUSSION
F. × ananassa is a hybrid octoploid and poses tremendous challenges in genetic and genomic studies. Past studies of cultivated strawberry, including miRNA identification (Ge et al., 2013; Li et al., 2013; Xu et al., 2013), were mostly focused on the ripening stage fruit. In this work, we focused our analysis on F. vesca, a diploid wild strawberry whose genome has been sequenced (Shulaev et al., 2011). Based on stringent criteria (Meyers et al., 2008), we identified 31 known and 38 novel miRNAs from F. vesca. Our identification and analyses of miRNAs in F. vesca benefited from the sequenced F. vesca genome (Shulaev et al., 2011), allowing accurate mapping of sequenced reads, a well-described developmental morphology of reproductive development (Hollender et al., 2012) that allowed accurate tissue and stage identification, and the availability of extensive transcriptome data sets in F. vesca (Kang et al., 2013; Hollender et al., 2014), which enabled us to examine the expression patterns of miRNA precursors and miRNA target genes. Importantly, the established transformation protocol and the ease of gene knockout in a diploid background will enable future functional studies.
A Majority of Conserved fve-miRNAs Have Evolved Novel Targets
One surprising finding is that 20 of the 31 known miRNAs exhibited not only conserved canonical gene targets but also previously unreported, presumably novel, gene targets. For example, among the 30 target genes of miR396, 11 are conserved targets (Supplemental Table S4, genes highlighted in boldface) but 19 are novel targets (Supplemental Table S4, genes not in boldface). Previous PARE analysis carried out in apple revealed that, among the 19 conserved miRNAs, two miRNAs (miR319 and miR396) were found to have new gene targets (Xia et al., 2012). By comparing our F. vesca list with that of apple (Xia et al., 2012), we found that the new gene targets of miR319 and miR396 in apple are distinct from those in F. vesca. Therefore, new miRNA-target pairs may form relatively easily and change rapidly in evolutionary time, underscoring the dynamic nature of miRNA-based regulation and its continuous evolution.
phasiRNA Production in Combination with miRNA-Target Pairing at Conserved Protein Domains Can Efficiently and Simultaneously Switch Off Members of Large Gene Families
More significantly, our work identified a novel miRNA-FBX network in F. vesca. Nine novel miRFBXs were found to cluster at two chromosomal regions: eight at LG3 within an 8-kb region and four at LG5 within a 500-bp region (Fig. 3, A and C). MIRNA genes are generally scattered in plant genomes. Only 17% of plant MIRNA genes were found to reside within clusters, and usually fewer than three MIRNA genes are clustered together (Nozawa et al., 2012). This is in general much lower than the clustered MIRNAs in human and Drosophila spp. (approximately 40%; Altuvia et al., 2005; Nozawa et al., 2010). The two miRFBX clusters found in F. vesca demonstrated that the relatively large size of the MIRNA gene family could arise quickly from tandem duplications, even though this is rather rare in plants.
In legumes, the miR482/2118 superfamily regulates hundreds of NB-LRR genes by targeting at the conserved P-loop region and triggering profuse secondary phasiRNA production (Zhai et al., 2011; Shivaprasad et al., 2012). In apple, three conserved miRNAs, miR828, miR858, and miR159, were shown to potentially target up to 81 different MYB genes (Xia et al., 2012). Target sites of miR858 and miR828 are within the conserved R3 domain of MYBs. In this study, we discovered that nine novel miRFBXs target a total of 94 FBX genes and that the target sites of eight miRNAs fall either partially or fully within the region coding for the conserved F-box domain (Fig. 4C). Our work, combined with recent findings mentioned above, reveals that miRNAs targeting at conserved protein domains can be particularly powerful in coordinately regulating large gene families.
Of increasing significance are recent discoveries of 22-nucleotide miRNAs capable of initiating phasiRNAs from protein-coding PHAS genes in higher plants. The 22-nucleotide miRNAs were reported in legumes to trigger phasiRNA production from more than 74 loci coding for the NB-LRRs (Zhai et al., 2011; Arikit et al., 2014). The resulting phasiRNAs further coregulate en masse, in trans or cis, NB-LRR transcripts. Similarly, 22-nucleotide miR828 reinforces its silencing of the MYB family of genes by instigating phasiRNAs from MYB transcripts (Xia et al., 2012). The superfamily of miR7122, conserved in eudicots, was also found to induce phasiRNAs from PPR genes in a wide variety of plants (Xia et al., 2013). In this study, we found a miRNA-phasiRNA pathway specifically targeting FBX genes in F. vesca. Although the function of these various phasiRNA networks is unknown, these prior findings together with the work reported here suggest that phasiRNA production, in combination with miRNA-target pairing at conserved protein domains, is a highly efficient and sustainable strategy evolved in higher plants to simultaneously switch off members of large gene families. With increasing analyses of plant genomes, more such miRNA-phasiRNA pathways may be discovered.
The LG3 miRFBX Cluster Recently Evolved from FBX Genes
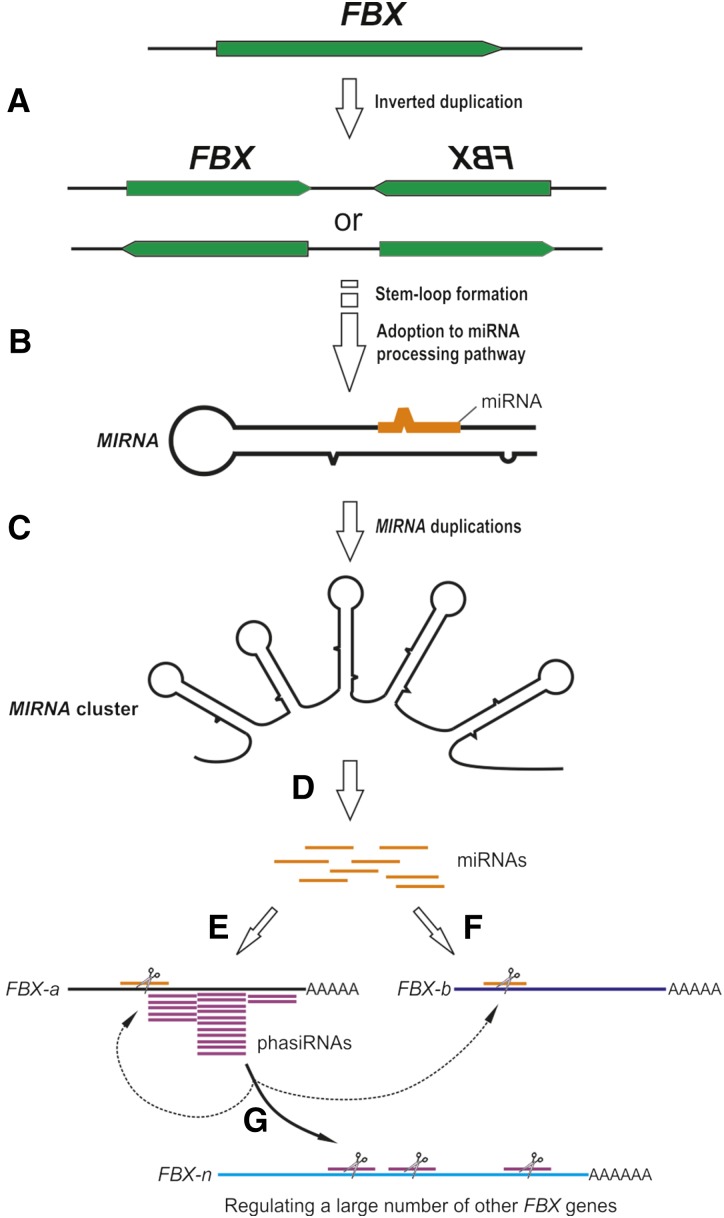
To our knowledge, the miRFBX-FBX network has never been reported in other plant species; we are curious about how this novel miRFBX-FBX network evolved. Prior publications indicated that new MIRNAs could evolve from target genes, preexisting MIRNA genes, repetitive elements, or random sequences (Allen et al., 2004; Felippes et al., 2008; Piriyapongsa and Jordan, 2008; Xia et al., 2013). We found that the LG3 miRFBX cluster region is expanded significantly in the F. vesca genome when compared with its syntenic region in closely related species (Fig. 5A; Supplemental Fig. S4). Furthermore, the MIRFBX3/4 and MIRFBX5/6 in the LG3 cluster showed significant similarity to the FBX genes (Fig. 5, C and D), suggesting that the LG3 miRFBXs likely evolved from duplications of FBX genes. We propose a model to illustrate the evolution of miRFBXs and the miRFBX-regulated network of FBX genes (Fig. 8). First, inverted duplication of an FBX gene (or gene fragment) led to transcripts capable of forming stem-loop structures (Fig. 8A). Gradually, the stem-loop structure was adopted by the miRNA processing machinery to yield miRNA and the locus became the MIRNA gene (Fig. 8B). Through subsequent duplications, a single MIRNA became a cluster of MIRNAs (Fig. 8C). Over time, the clustered MIRNAs diverged from one another and yielded a family of miRNAs (Fig. 8D) that target different FBX genes (Fig. 8, E and F). The 22-nucleotide miRNAs (i.e. miRFBX7) not only cleave their target FBX genes (FBX-a) but also primed their target genes (FBX-a) for phasiRNA production (Fig. 8E). phasiRNAs subsequently amplify the silencing effect by targeting additional FBX genes in cis to regulate their cognate gene (FBX-a) or in trans to regulate other FBX genes (FBX-b and FBX-n; Fig. 8G).
Figure 8.
A model illustrating the evolution and mode of action of LG3 miRFBXs in F. vesca. A, The stem-loop structure of a MIRNA gene came from inverted duplication of an FBX gene (or gene fragment). B, The stem-loop structure was gradually adopted by the miRNA processing machinery to yield a miRNA, and the locus became the MIRNA gene. C, Subsequent local duplication of the initial MIRNA gene led to the MIRNA cluster consisting of five stem loops. D, Each MIRNA gene diverges over time to give rise to a family of related miRNAs (orange), which were capable of regulating a large number of FBX genes. E, The 22-nucleotide miRNAs (orange) produced from MIRFBX7 guided the initial cleavage of its FBX target gene (FBX-a), which is followed by phasiRNA (purple) production. F, The 21-nucleotide miRNAs guided the cleavage of their target FBX genes (e.g. FBX-b). G, The phasiRNAs (purple) derived from E further amplified the silencing effect by guiding the degradation of additional FBX genes (FBX-a, FBX-b, and FBX-n) in cis and trans.
The miRFBX7-FBX-phasiRNA Network May Function in the Receptacle of Strawberry
FBX genes encode the substrate-recognition subunits of the SCF (for S-phase kinase associated protein1-Cullin-F Box) ubiquitin ligases and constitute one of the largest gene families, with 820 members in F. vesca (Xu et al., 2009; Hollender et al., 2014). FBX proteins regulate diverse biological processes and were grouped into two general types based on whether they function in conserved processes, such as embryogenesis and circadian rhythms, or relatively specialized processes, such as pollen recognition and pathogen response (Xu et al., 2009). Xu et al. (2009) noted that those with functions in conserved processes experienced little expansion in gene numbers during the evolution from the common ancestor of eudicots and monocots, while those in specialized processes experienced frequent gene duplications, in particular tandem duplications that may be critical to the evolution of specialized functions. Indeed, most of the FBX genes targeted by fve-miRFBXs reported here are more closely grouped with FBA1/3, which belongs to the second group with specialized functions. This is consistent with our finding that miRFBX-targeted FBX genes experienced significant gene duplications, as shown by their adjacent chromosomal locations in the F. vesca genome (Fig. 6C). This observation indicates that the FBX-targeting miRFBXs may have evolved to regulate strawberry-specific processes, which is further supported by the expression pattern of precursor RNAs for miRFBX7/8. Specifically, the precursor RNA for miRFBX7/8 (transcript 4) was highly expressed in cortex and pith (Fig. 6B). The pith and cortex are the inner and outer tissues of the receptacle, which is a unique part of the strawberry flower that enlarges rapidly to form the edible fleshy fruit. Hence, the miRFBX7-FBX-phasiRNA network may function in the receptacle and be responsible for the unique properties of the strawberry fruit. Most of the FBX genes in the miRFBX7-FBX-phasiRNA network are homologous to the Arabidopsis Constitutive expresser of PR1 (CPR1)/CPR30 (At4g12560; Supplemental Table S6). The Arabidopsis CPR1/CPR30 FBX proteins were shown to negatively regulate the accumulation of a Resistance protein, Suppressor of npr1-1, Constitutive1, via the SCF complex (Gou et al., 2012). By cleaving CPR1/CPR30-type FBX transcripts, miRFBX could act to promote Resistance protein accumulation and enhance disease resistance in the developing receptacle fruit.
MATERIALS AND METHODS
Plant Materials
A seventh generation inbred line of Fragaria vesca, Yellow Wonder 5AF7 (Slovin et al., 2009), was grown in soil in growth chambers under 12 h of light as described previously (Hollender et al., 2012). Young unexpanded leaves of 6-month-old plants, unopened flowers between stages 8 and 12 (staging according to Hollender et al. [2012]), open flowers (within 1 d of opening), and receptacles (10 DPA) were harvested with hand dissection to remove the achenes from each receptacle (Fig. 1). Achenes (fertilized ovaries) were dissected open to separate the ovary wall from the seed inside. The ovary wall and seeds were harvested at 4 and 10 DPA, respectively.
For seedlings, seeds were surface sterilized, plated on Murashige and Skoog medium, stratified at 4°C for 1 week in darkness, and transferred to a growth chamber with 12 h of light. Four-week-old seedlings were collected.
RNA Extraction and Sequencing
For small RNA sequencing, total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen) with certain modifications. Specifically, the supernatant, passed through a QIAshredder spin column, was precipitated in 1.5 volumes of ethanol and then centrifuged for 2 min at 12,000 rpm. The pellet was dissolved in 50 µL of water and mixed with 500 µL of Qiazol and then 140 μL of chloroform. After centrifugation, the supernatant was transferred to an RNeasy mini spin column for further purification following the instructions from the RNeasy Plant Mini Kit. The quality of total RNA was checked by the Experion Automated Electrophoresis System (Bio-Rad). For library preparation following the Illumina TruSeq Small RNA Sample Preparation protocol, 10 to 20 µg of total RNA per sample with RNA integrity number above 7.8 was sent to the Weill Cornell Medical College Genomics Resources Core Facility. The libraries were sequenced on the Illumina HiSeq 2000 platform.
For PARE analysis, total RNA from different tissues (described above) was extracted using the RNeasy Plant Mini Kit following the manufacturer’s instructions. Equal amounts of RNA from different tissues were pooled to form three samples: vegetative (leaf and seedling), flower (open and unopened), and fruit (achene and receptacles). For the achene tissue, both 4- and 10-d-old achenes were scraped off the receptacle. Approximately 100 to 150 μg of total RNA per sample was sent to the Beijing Genomics Institute for PARE library construction and sequencing on the Illumina HiSeq 2000 platform. Prior to trimming, 50-bp single-end-sequence reads were obtained (Supplemental Table S1).
miRNA Identification
miRNA identification was based on previously well-established criteria (Meyers et al., 2008; Xia et al., 2012) and is summarized in Supplemental Figure S1. The raw reads were processed by first discarding low-quality reads, then removing adaptors, and finally collapsing identical small RNA reads into one using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Reads homologous to other noncoding RNAs, like tRNA and ribosomal RNAs, were removed by BLASTN alignment against Rfam 10, allowing at most two mismatches. The remaining small RNAs were subjected to miRNA identification. The prescreen for miRNAs relied on several criteria: (1) reads had to map exactly to the genome at no more than 16 loci; (2) reads had to be 20 to 22 nucleotides in length; (3) raw read counts had to be 10 or higher in at least one of the nine libraries; and (4) the novel miRNA had to derive from a well-supported stem-loop structure (Supplemental Fig. S3), defined as four or fewer mispairings and one or fewer central bulge in the region of miRNA:miRNA* pairing. Afterward, conserved miRNAs were identified using BLASTN against mature miRNAs in miRBase (release 20; http://www.mirbase.org/), allowing up to two base differences. Those small RNAs, with no hit in the miRBase and combined miRNA/miRNA* reads accounting for more than 75% of reads in the precursor locus (spanning 150 bp upstream to 150 bp downstream of the small RNA), were defined as novel miRNAs. The putative precursor sequences were folded with the Vienna RNA package (Hofacker, 2003).
The total number of reads that perfectly match the F. vesca genome in a given library was used for the normalization of read abundance. Each miRNA was normalized to 10 million reads. F. vesca genome sequences (fvesca_v1.0_scaffolds.fna.gz) were downloaded from the GDR (http://www.rosaceae.org/; Shulaev et al., 2011).
miRNA-Target Analysis
Targetfinder 1.6 (Fahlgren et al., 2007) was used to predict miRNA targets, and CleaveLand version 2.0 (Addo-Quaye et al., 2009) was then applied to analyze the PARE data after adaptor trimming and genomic mapping. The F. vesca transcriptome (GeneMark hybrid coding sequence) was retrieved from the GDR (http://www.rosaceae.org/; Shulaev et al., 2011). The alignment penalty score, assigned by Targetfinder 1.6, was used in target prediction. Only matches with scores of 5 or less were retained. Two parameters, the alignment score and the category, were used to evaluate the reliability of target genes (Supplemental Tables S4 and S5). An alignment score up to 5 was used in target prediction (Li et al., 2010; Xia et al., 2012); a lower score indicates a better alignment between miRNA and its target. The category score (between 0 and 4), on the other hand, considers the ratio of the PARE tag reads mapped to the corresponding miRNA-mediated cleavage site to the total number of PARE tags in a given target mRNA (Addo-Quaye et al., 2009). The lower the category score, the higher the confidence of the cleaved targets.
RNA-seq Analysis
Seventy-four RNA-seq libraries from 37 different F. vesca flower, fruit, leaf, and seedling tissues (i.e. two replicates per tissue) were generated in our earlier work (Kang et al., 2013; Hollender et al., 2014). RNA-seq reads in the LG3 fve-miRFBX cluster region were visualized from GBrowse hosted at the GDR (www.rosaceae.org/gb/gbrowse/fragaria_vesca_v1.1-lg/). A screenshot from this was incorporated into Figure 6A. Furthermore, RNA-seq mapped reads (RPKM) of the precursor transcripts at the LG3 and LG5 clusters as well as the 24 predicted FBX target genes of miRFBX7 (Supplemental Table S6) were extracted from the RNA-seq libraries (Kang et al., 2013; Hollender et al., 2014) and imported into MultiExperimental Viewer MeV4.8 (Saeed et al., 2003), respectively. RPKM values across the 37 tissues of the precursor transcripts at the LG3 and LG5 clusters are shown as a heat map (Fig. 6B). The Normalize Genes/Rows function in MeV4.8 was used to normalize each of the 24 FBX genes across all 74 libraries. Hierarchical clustering with Pearson correlation was then used to display the relative expression trend of the 24 FBX genes targeted by miRFBX7 (Fig. 6C).
TAS Gene and PHAS F-Box Gene Identification
Phasing analysis of secondary siRNAs was conducted as described previously (Xia et al., 2013). The same algorithms were used for P value and phasing score calculation. Genes with P < 0.001 were considered as TAS or PHAS genes. The Integrative Genomics Viewer (Thorvaldsdóttir et al., 2013) was used to view the phasing score.
F-Box Gene Classification and Phylogenetic Tree Construction
The 691 Arabidopsis (Arabidopsis thaliana) FBX gene sequences were retrieved from the data of Xu et al. (2009), and the classification of Arabidopsis FBX genes was based on the domain annotation in the same work. For the 94 F. vesca FBX genes targeted by miRFBXs, de novo domain structure annotation was performed using CD-search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Among the 94 genes, the gene bodies of three genes (gene26143, gene00154, and gene10432) were corrected from the original sequences due to inaccurate annotations, and two genes (gene18163 and gene01506) were separated into two additional genes. In total, 787 FBX genes (697 from Arabidopsis and 96 from F. vesca) were used for the construction of phylogenetic trees. First, their full-length amino acid sequences were aligned using the multiple alignment tool MUSCLE (Edgar, 2004). Then, the tree was constructed by FastTree using default settings (Price et al., 2009). The resulting tree was viewed and annotated using Dendroscope 3 (Huson and Scornavacca, 2012).
Synteny Analysis of the F. vesca LG3 miRFBX Region
Syntenic regions among plants and their alignments were retrieved from the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/index/home). BLASTX and BLASTP analyses were conducted locally with an e-value cutoff of 1,000, and plots of BLASTP e-value distribution were generated using the R package.
Raw and processed reads of all nine small RNA libraries and three PARE libraries are available from the Gene Expression Omnibus at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo). With the exception of the open flower small RNA library (accession no. GSE44930), the eight small RNA libraries and three PARE libraries are deposited under the accession number GSE61798.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Informatics pipeline for predicting miRNAs from F. vesca.
Supplemental Figure S2. Alignment of miRFBX precursors of the LG3 miRFBX cluster.
Supplemental Figure S3. Secondary structures of 32 novel miRNA precursors.
Supplemental Figure S4. Syntenic analysis in several plant species for the region containing the LG3 miRFBX cluster.
Supplemental Figure S5. Read abundance of mature miRFBXs.
Supplemental Table S1. Summary of sequence reads of nine small RNA libraries and three PARE libraries from F. vesca (in millions).
Supplemental Table S2. Known miRNAs in F. vesca.
Supplemental Table S3. Novel miRNAs identified in F. vesca.
Supplemental Table S4. Target genes of known miRNAs in F. vesca.
Supplemental Table S5. Target genes of novel miRNAs in F. vesca.
Supplemental Table S6. FBX target genes of miRFBX1-9.
Supplemental Table S7. A list of 24 PHAS FBX genes targeted by miRFBX7.
Supplemental Table S8. PHAS MYB genes targeted by miR828.
Supplementary Material
Acknowledgments
We thank Dr. Chunying Kang for the photographs shown in Figure 1 and help with Figure 6B and Drs. Chunying Kang and Julie Caruana for comments and edits.
Glossary
- miRNA
microRNA
- tasiRNA
trans-acting small interfering RNA
- siRNA
small interfering RNA
- phasiRNA
phased small interfering RNA
- PARE
parallel analysis of RNA ends
- RNA-seq
RNA sequencing
- GDR
Genome Database for Rosaceae
Footnotes
This work was supported by the National Science Foundation (grant no. MCB0923913 to Zh.L. and IOS1257869 to B.C.M.) and by the Maryland Agricultural Experiment Station Hatch Project (grant no. MD–CBMG–0738 to Zh.L).
Articles can be viewed without a subscription.
References
- Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18: 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo-Quaye C, Miller W, Axtell MJ (2009) CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25: 130–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouché N, Gasciolli V, Vaucheret H (2006) DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol 16: 927–932 [DOI] [PubMed] [Google Scholar]
- Allen E, Howell MD (2010) miRNAs in the biogenesis of trans-acting siRNAs in higher plants. Semin Cell Dev Biol 21: 798–804 [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC (2004) Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet 36: 1282–1290 [DOI] [PubMed] [Google Scholar]
- Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H (2005) Clustering and conservation patterns of human microRNAs. Nucleic Acids Res 33: 2697–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikit S, Xia R, Kakrana A, Huang K, Zhai J, Yan Z, Valdes-Lopez O, Prince S, Musket TA, Nguyen HT, et al. (2014) An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. Plant Cell 26: 4584–4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP (2006) A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Snyder JA, Bartel DP (2007) Common functions for diverse small RNAs of land plants. Plant Cell 19: 1750–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH (2010) 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA 107: 15269–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC (2010) Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol 17: 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwish O, Slovin JP, Kang C, Hollender CA, Geretz A, Houston S, Liu Z, Alkharouf NW (2013) SGR: an online genomic resource for the woodland strawberry. BMC Plant Biol 13: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi JM, Rodriguez RE, Mecchia MA, Palatnik JF (2012) Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet 8: e1002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16: 939–944 [DOI] [PubMed] [Google Scholar]
- Fei Q, Xia R, Meyers BC (2013) Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25: 2400–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippes FF, Schneeberger K, Dezulian T, Huson DH, Weigel D (2008) Evolution of Arabidopsis thaliana microRNAs from random sequences. RNA 14: 2455–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Collier SA, Byrne ME, Martienssen RA (2006) Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol 16: 933–938 [DOI] [PubMed] [Google Scholar]
- Ge A, Shangguan L, Zhang X, Dong Q, Han J, Liu H, Wang X, Fang J (2013) Deep sequencing discovery of novel and conserved microRNAs in strawberry (Fragaria × ananassa). Physiol Plant 148: 387–396 [DOI] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26: 941–946 [DOI] [PubMed] [Google Scholar]
- Gou M, Shi Z, Zhu Y, Bao Z, Wang G, Hua J (2012) The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J 69: 411–420 [DOI] [PubMed] [Google Scholar]
- Hofacker IL. (2003) Vienna RNA secondary structure server. Nucleic Acids Res 31: 3429–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollender CA, Geretz AC, Slovin JP, Liu Z (2012) Flower and early fruit development in a diploid strawberry, Fragaria vesca. Planta 235: 1123–1139 [DOI] [PubMed] [Google Scholar]
- Hollender CA, Kang C, Darwish O, Geretz A, Matthews BF, Slovin J, Alkharouf N, Liu Z (2014) Floral transcriptomes in woodland strawberry uncover developing receptacle and anther gene networks. Plant Physiol 165: 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC (2007) Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Scornavacca C (2012) Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol 61: 1061–1067 [DOI] [PubMed] [Google Scholar]
- Johnson C, Kasprzewska A, Tennessen K, Fernandes J, Nan GL, Walbot V, Sundaresan V, Vance V, Bowman LH (2009) Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res 19: 1429–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C, Darwish O, Geretz A, Shahan R, Alkharouf N, Liu Z (2013) Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell 25: 1960–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Takahashi N, Shimada H, Seki M, Shinozaki K, Matsui M (2002) Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol 43: 1073–1085 [DOI] [PubMed] [Google Scholar]
- Li H, Mao W, Liu W, Dai H, Liu Y, Ma Y, Zhang Z (2013) Deep sequencing discovery of novel and conserved microRNAs in wild type and a white-flesh mutant strawberry. Planta 238: 695–713 [DOI] [PubMed] [Google Scholar]
- Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R (2010) Transcriptome-wide identification of microRNA targets in rice. Plant J 62: 742–759 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Vaucheret H (2006) Functions of microRNAs and related small RNAs in plants. Nat Genet (Suppl) 38: S31–S36 [DOI] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A (2010) miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, et al. (2008) Criteria for annotation of plant microRNAs. Plant Cell 20: 3186–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch JP. (1950) Growth and morphogenesis of the strawberry as related to auxin. Am J Bot 37: 211–215 [Google Scholar]
- Nozawa M, Miura S, Nei M (2010) Origins and evolution of microRNA genes in Drosophila species. Genome Biol Evol 2: 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M, Miura S, Nei M (2012) Origins and evolution of microRNA genes in plant species. Genome Biol Evol 4: 230–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaleo V, Szittya G, Moxon S, Miozzi L, Moulton V, Dalmay T, Burgyan J (2010) Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J 62: 960–976 [DOI] [PubMed] [Google Scholar]
- Pantazis CJ, Fisk S, Mills K, Flinn BS, Shulaev V, Veilleux RE, Dan Y (2013) Development of an efficient transformation method by Agrobacterium tumefaciens and high throughput spray assay to identify transgenic plants for woodland strawberry (Fragaria vesca) using NPTII selection. Plant Cell Rep 32: 329–337 [DOI] [PubMed] [Google Scholar]
- Piriyapongsa J, Jordan IK (2008) Dual coding of siRNAs and miRNAs by plant transposable elements. RNA 14: 814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF (2010) Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA, Baulcombe DC (2012) A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24: 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, et al. (2011) The genome of woodland strawberry (Fragaria vesca). Nat Genet 43: 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slovin JP, Schmitt K, Folta KM (2009) An inbred line of the diploid strawberry Fragaria vesca f. semperflorens for genomic and molecular genetic studies in the Rosaceae. Plant Methods 5: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crété P, Bartel DP (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann MR, Berkowitz ND, Gregory BD (2014) Improved genome-wide mapping of uncapped and cleaved transcripts in eukaryotes: GMUCT 2.0. Methods 67: 64–73 [DOI] [PubMed] [Google Scholar]
- Xia R, Meyers BC, Liu Z, Beers EP, Ye S, Liu Z (2013) MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA biogenesis in eudicots. Plant Cell 25: 1555–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Zhu H, An YQ, Beers EP, Liu Z (2012) Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol 13: R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Ma H, Nei M, Kong H (2009) Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proc Natl Acad Sci USA 106: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Yin L, Ying Q, Song H, Xue D, Lai T, Xu M, Shen B, Wang H, Shi X (2013) High-throughput sequencing and degradome analysis identify miRNAs and their targets involved in fruit senescence of Fragaria ananassa. PLoS ONE 8: e70959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, Li Y, González AJ, Yan Z, Kitto SL, Grusak MA, et al. (2011) MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev 25: 2540–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Gao S, Zhou X, Xia J, Chellappan P, Zhou X, Zhang X, Jin H (2010) Multiple distinct small RNAs originate from the same microRNA precursors. Genome Biol 11: R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Xia R, Zhao B, An YQ, Dardick CD, Callahan AM, Liu Z (2012) Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC Plant Biol 12: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.