A negative feedback loop involving two Ethylene Response Factors fine-tunes growth inhibition and stress tolerance activation under mannitol-induced stress.
Abstract
Leaf growth is a tightly regulated and complex process, which responds in a dynamic manner to changing environmental conditions, but the mechanisms that reduce growth under adverse conditions are rather poorly understood. We previously identified a growth inhibitory pathway regulating leaf growth upon exposure to a low concentration of mannitol and characterized the ETHYLENE RESPONSE FACTOR (ERF)/APETALA2 transcription factor ERF6 as a central activator of both leaf growth inhibition and induction of stress tolerance genes. Here, we describe the role of the transcriptional repressor ERF11 in relation to the ERF6-mediated stress response in Arabidopsis (Arabidopsis thaliana). Using inducible overexpression lines, we show that ERF6 induces the expression of ERF11. ERF11 in turn molecularly counteracts the action of ERF6 and represses at least some of the ERF6-induced genes by directly competing for the target gene promoters. As a phenotypical consequence of the ERF6-ERF11 antagonism, the extreme dwarfism caused by ERF6 overexpression is suppressed by overexpression of ERF11. Together, our data demonstrate that dynamic mechanisms exist to fine-tune the stress response and that ERF11 counteracts ERF6 to maintain a balance between plant growth and stress defense.
Plants are constantly challenged to survive and maintain growth in changing environments. In natural environments, as well as in laboratories, growth conditions are rarely optimal, generating a weak but continuous stress. In such suboptimal conditions, the equilibrium between sustained plant growth and activation of stress defense mechanisms is defied and needs to be continuously rebalanced and fine-tuned (Claeys and Inzé, 2013).
To unravel these growth- and defense-related mechanisms in Arabidopsis (Arabidopsis thaliana), researchers commonly use in vitro setups in which different growth inhibitory compounds are added to the growth medium (Verslues et al., 2006; Lawlor, 2013; Claeys et al., 2014). Mannitol, for example, is a frequently applied compound to induce mild stress because it results in both inhibition of leaf growth and activation of stress-responsive genes (Kreps et al., 2002; Skirycz et al., 2010, 2011; Dubois et al., 2013; Claeys et al., 2014; Trontin et al., 2014). Two putative receptor-like kinases, ENHANCED GROWTH ON MANNITOL1 (EGM1) and EGM2, are presumably involved in the detection of mannitol and further downstream activation of the growth and tolerance responses (Trontin et al., 2014). Previously, we have shown that mannitol-induced responses are specific to the different stages of Arabidopsis leaf development (Skirycz et al., 2010). In very young Arabidopsis leaves, in which cells are not yet expanding but still actively dividing, exposure to mannitol triggers the accumulation of the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) and the transcriptional induction of ethylene-related genes. Interestingly, these responses are extremely fast, with several ETHYLENE RESPONSE FACTORs (ERFs; ERF1, ERF2, ERF5, ERF6, and ERF11) being induced in growing leaves within hours upon sudden exposure of roots to mannitol (Skirycz et al., 2011).
The ERFs are transcription factors belonging to the large APETALA2 (AP2)/ERF family of plant-specific transcription factors (Riechmann and Meyerowitz, 1998; Nakano et al., 2006). They all possess a single repeat of the well-conserved AP2 domain, which is important for DNA binding. Additionally, other small domains are conserved between several but not all ERFs, enabling a detailed classification in 12 ERF subgroups (Sakuma et al., 2002; Nakano et al., 2006). For example, the mannitol-induced ERF11 belongs to subgroup VIIIa, the members of which contain a C-terminal ERF amphiphilic repression (EAR) motif enabling transcriptional repression of the downstream targets (Ohta et al., 2001; Yang et al., 2005; Nakano et al., 2006; Li et al., 2011). The other mannitol-induced ERFs are members of the subgroups IXa (ERF1 and ERF2) or IXb (ERF5 and ERF6). These contain an N-terminally located conserved stretch of acidic amino acids (called the 2nd Conserved Motif of group IX [CMIX-2]), which might function as a transcriptional activator domain. The transcriptional activators ERF5 and ERF6 additionally harbor a conserved C-terminal motif (CMIX-5) identified as a putative phosphorylation site by mitogen-activated protein kinases (MPKs), which distinguishes group IXa from group IXb (Fujimoto et al., 2000; Nakano et al., 2006).
ERF6 is an activating transcription factor with documented roles in the response to various abiotic and biotic stress conditions, such as oxidative stress (Sewelam et al., 2013; Wang et al., 2013; Vermeirssen et al., 2014), high light (Vogel et al., 2014), cold (Lee et al., 2005; Xin et al., 2007), and biotic stress induced by biotrophic and necrotrophic pathogens (AbuQamar et al., 2006; Dombrecht et al., 2007; Hu et al., 2008; Moffat et al., 2012; Son et al., 2012; Meng et al., 2013). We have previously unraveled the molecular and biological function of ERF6 and its close homolog ERF5 in the mannitol-induced stress response, specifically in actively growing young Arabidopsis leaves (Dubois et al., 2013). We propose the following model: upon sudden exposure to mannitol, MPK3 and MPK6 could posttranslationally activate ERF5/ERF6 through phosphorylation, as has previously been reported under reactive oxygen species-mediated oxidative stress (Popescu et al., 2009; Son et al., 2012; Wang et al., 2013). Active ERF6 proteins transcriptionally induce ERF6 gene expression and act as early and central regulators of the mannitol-induced stress response. On the one hand, ERF6 transcriptionally activates the GA2-OXIDASE6 (OX6) gene, encoding an enzyme degrading bioactive GAs and thus reducing cellular GA levels (Rieu et al., 2008). Through activation of GA degradation, ERF5/ERF6 stimulate the stabilization of DELLA proteins, which in turn inhibit growth of young leaves by pushing dividing cells out of the mitotic cell cycle (Claeys et al., 2012). Under mannitol stress, ERF5/ERF6 thus induce leaf growth inhibition, and consequently, plants overexpressing ERF6 show extreme dwarfism. On the other hand, ERF6 was found to rapidly activate 332 putative target genes, highly enriched for stress-responsive genes and for genes involved in transcriptional regulation, suggesting that ERF6 is situated upstream of a stress-related network of transcription factors. ERF6 was, for example, shown to directly activate the expression of genes encoding the stress tolerance-related transcription factors MYB DOMAIN PROTEIN51 (MYB51), Salt Tolerance Zinc Finger (STZ), and WRKY33. Thus, on top of inhibiting Arabidopsis leaf growth, ERF6 also induces, independently of the GA/DELLA pathway, genes involved in stress tolerance (Dubois et al., 2013).
The transcriptional repressor ERF11 has been much less studied but has been identified in several gene expression studies following different kinds of stresses. For example, ERF11 expression is altered upon several abiotic stresses, such as K+ depletion (Hampton et al., 2004), cold exposure (Lee et al., 2005; Vergnolle et al., 2005), H2O2 treatment (Wang et al., 2006), and exposure to chronic UV-B (Hectors et al., 2007) and 3-(3,4-dichlorophenyl)-1,1-dimethylurea, an inhibitor of photosynthetic electron transport (Vogel et al., 2012). ERF11 is also known to be responsive to several hormones, especially jasmonic acid and abscisic acid (ABA), and to infection with the oomycete Phytophthora parasitica and Alternaria brassicicola (McGrath et al., 2005; Dombrecht et al., 2007; Eulgem and Somssich, 2007; Libault et al., 2007). ERF11 is further known to be a negative regulator of ethylene biosynthesis upon increased ABA levels by directly repressing the expression of the ACC synthases ACS2 and ACS5, explaining the known ABA-ethylene antagonism (Li et al., 2011). However, despite its induction by various and numerous stresses, the biological and molecular role of ERF11 in biotic and abiotic stress responses is still poorly understood.
In this study, we demonstrate that the transcriptional repressor ERF11 antagonizes the activator ERF6, providing a mechanism to maintain the balance between plant growth and stress tolerance upon stress exposure.
RESULTS
ERF6 and ERF11 Are Induced Simultaneously by Mannitol-Mediated Stress and by ACC
Previously, we have developed a mild stress assay in which young Arabidopsis seedlings were exposed to a low concentration of mannitol (25 mm), which induces a rapid leaf growth inhibition without affecting developmental timing. Using this assay, the transcription factors ERF6 and ERF11 have been shown to be transcriptionally induced by mannitol within hours upon exposure, specifically in actively growing Arabidopsis leaves (Skirycz et al., 2011). To get a better time resolution on this expression pattern, we performed a detailed time-course experiment and measured the expression of ERF6 and ERF11 within minutes upon exposure to mannitol using the following experimental setup: Arabidopsis seedlings are grown in vitro on one-half-strength Murashige and Skoog (MS) medium overlaid with a nylon mesh until 9 d after stratification (DAS). At this moment, the third true leaf is still mainly composed of proliferating cells. The mesh with plants is subsequently transferred to growth medium supplemented with 25 mm mannitol, and the young Arabidopsis seedlings are harvested on mRNA-stabilizing solution after 5, 10, 15, 30, 45, 60, 120, and 180 min of stress exposure, followed by microdissection of the third true leaf (less than 0.1 mm2 in size). By measuring ERF6 and ERF11 expression levels by quantitative reverse transcription (qRT)-PCR, we show that in young Arabidopsis leaves, both genes were induced simultaneously by mannitol stress already after 60 min (PERF6 = 0.003 and PERF11 = 0.002, ANOVA; Fig. 1A). Consistent with our previous observation that mannitol rapidly induces ACC accumulation (Skirycz et al., 2011), transfer of young seedlings to 5 μm ACC-containing growth medium triggered a simultaneous up-regulation of ERF6 and ERF11 within 45 min of exposure (PERF6 = 0.002 and PERF11 = 0.037, ANOVA; Fig. 1B). We also measured the expression level of STZ, MYB51, and GA2-OX6, which are target genes of ERF6 (Dubois et al., 2013). Interestingly, the increased expression of these genes upon mannitol treatment occurred simultaneously with the ERF6 and ERF11 induction (Supplemental Fig. S1). The transcriptional induction of ERF6 at the same time as its own target genes is consistent with our previous observations that stress-activated ERF6 proteins are able to induce both their own expression as well as that of STZ, MYB51, and GA2-OX6. Thus, ERF11, ERF6, and its target genes are induced at the same time upon treatment with mannitol in actively growing Arabidopsis leaves.
Figure 1.
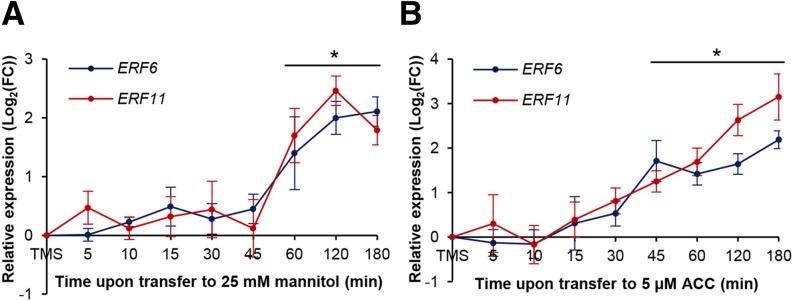
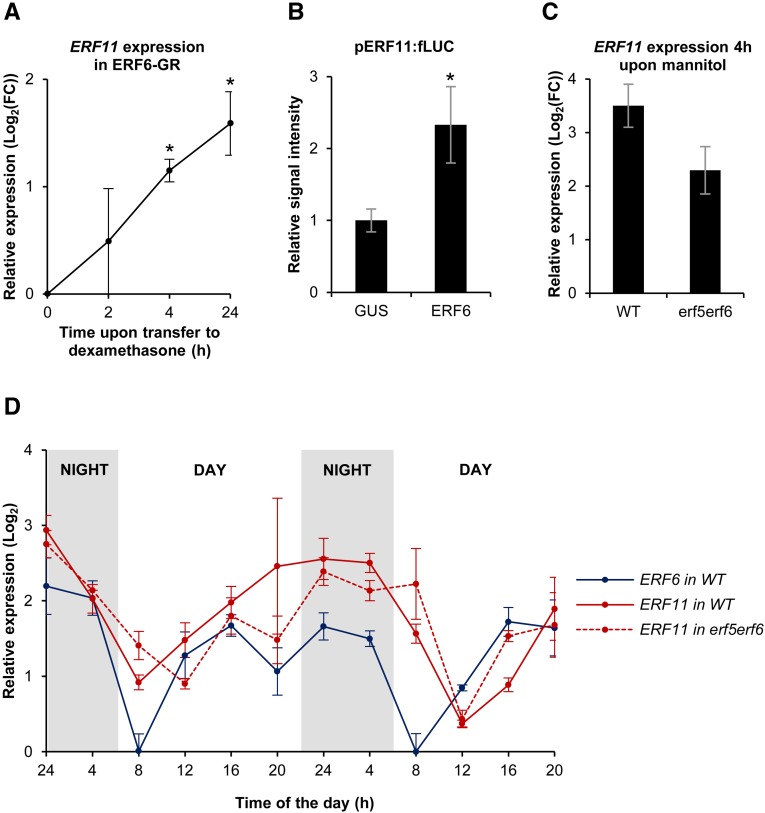
Transcriptional induction of ERF6 and ERF11 upon short-term exposure to mannitol or ACC. Expression levels of ERF6 and ERF11 in proliferating leaf tissue measured by qRT-PCR upon exposure to mild mannitol-induced stress (25 mm; A) or 5 μm ACC (B). Represented values are means of three biological repeats with their se relative to the expression value upon transfer to MS (TMS) medium as a control. *, P < 0.05 (ANOVA) for both genes. FC, Fold change.
ERF6 Is an Activator of ERF11 Expression under Mannitol-Mediated Stress
The expression pattern of ERF11 upon mannitol exposure is similar to that of known ERF6 target genes, raising the question whether ERF11 is an ERF6 target gene as well. To further investigate this possibility, we first determined whether ERF6 is able to activate ERF11 expression. To this end, a 35S:ERF6-GR (ERF6-GR) line, containing a fusion between ERF6 and the glucocorticoid receptor, was used (Dubois et al., 2013). If ERF11 is a target gene of ERF6, up-regulation of ERF11 transcripts is expected to occur rapidly upon induction of ERF6. We therefore transferred ERF6-GR plants to dexamethasone-containing growth medium (see “Materials and Methods”) and measured ERF11 transcript levels by qRT-PCR at multiple time points following induction (Fig. 2A). ERF11 transcript levels gradually increased within 4 h upon dexamethasone treatment to up to 2-fold (P = 0.02, ANOVA), suggesting that ERF11 is a direct target of ERF6.
Figure 2.
Transcriptional activation of ERF11 by ERF6 under mannitol-induced stress. A, Expression levels of ERF11 in the growing third leaf (15 DAS) of ERF6-GR plants, relative to the wild type, measured by qRT-PCR at several time points (h) upon transfer to dexamethasone-containing medium. *, P < 0.05 (ANOVA). B, Luciferase transactivation assay with cotransformation of 35S:ERF6 and pERF11:fLUC showing the activation of the ERF11 promoter by ERF6. Values are signal intensities normalized to the cotransformed normalization plasmid rLUC and relative to the negative control (35S:GUS + pERF11:fLUC). *, P < 0.05. C, Expression levels of ERF11 in wild-type (WT) plants and erf5erf6 mutants 4 h upon exposure to mannitol. D, Expression levels of ERF6 and ERF11 at different times of the day in actively growing leaves (15 DAS) of wild-type and erf5erf6 plants grown in soil. For all graphs, the represented values are the means of three biological repeats with their se. FC, Fold change.
Next, activation of the ERF11 promoter (pERF11) by ERF6 was analyzed using a luciferase transactivation assay in tobacco (Nicotiana tabacum) protoplasts by cotransformation of 35S:ERF6 and pERF11:fLUC (for firefly luciferase) constructs. The observed increase in light signal emitted by fLUC suggests that ERF6 is able to activate the promoter of ERF11 (P < 2E-16; Fig. 2B). To further confirm this, the mannitol-mediated induction of ERF11 was analyzed in an erf5erf6 double mutant background (Dubois et al., 2013), compared with wild-type plants. After 4 h of exposure to mannitol, ERF11 was 11.3-fold higher expressed in mannitol-treated wild-type plants, compared with the controls transferred to MS medium (Fig. 2C). This clear induction of ERF11 expression was less pronounced (P = 0.07) in erf5erf6 mutants, where the induction by mannitol was 4.9-fold (Fig. 2C). Together, these data show that ERF5/ERF6 are involved in the rapid induction of ERF11 upon exposure to low concentrations of mannitol.
The expression of the mannitol-induced ERFs was previously thought to be low under control conditions (Andriankaja et al., 2012; Dubois et al., 2013; Meng et al., 2013). However, in growing Arabidopsis leaves of plants grown in soil, we observed that this is not the case throughout the entire day but that instead, ERF6 and ERF11 transcript profiles show a similar diurnal pattern (Fig. 2D), suggesting coregulation of both genes. ERF6 and ERF11 transcript levels were low in the morning and gradually accumulated until the evening. During the night, transcript levels remained stable and abruptly decreased again in the early morning. To test if the regulation of ERF11 expression throughout the day is mediated by ERF6, the diurnal ERF11 pattern was measured in erf5erf6 mutants under nonstress conditions, showing a similar ERF11 expression pattern as in the wild type (Fig. 2D). We thus conclude that ERF6 is an activator of ERF11 under mannitol-induced stress but that it is not responsible for the diurnal expression pattern of ERF11 expression under control conditions.
Overexpression of ERF11 Negatively Affects Leaf Growth
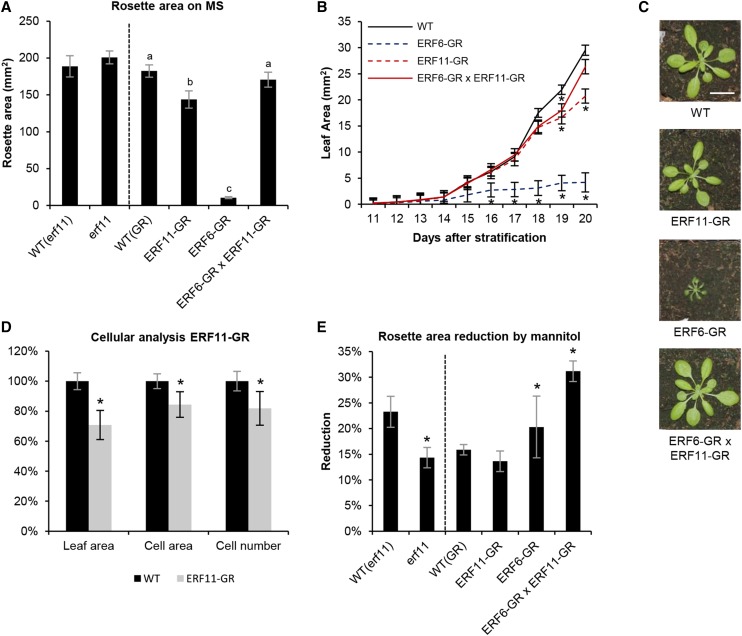
Because, under growth-repressive conditions, ERF11 is induced in actively growing leaves of plants, we speculated that ERF11 might be involved in the regulation of leaf growth. To test this hypothesis, we first measured rosette growth of the homozygous line erf11 (SALK_116053; Alonso et al., 2003) knocked out for ERF11 due to a transfer DNA insertion in the 5′ untranslated region of ERF11 and showing clearly reduced ERF11 expression levels (residual ERF11 expression, on average, 5%; Supplemental Fig. S2). At 22 DAS, the rosettes of erf11 mutants were indistinguishable from wild-type rosettes (P = 0.96, ANOVA; Fig. 3A). Because the absence of a phenotype in the knockout line might be due to low ERF11 activity under control conditions, we also phenotyped transgenic plants overexpressing the dexamethasone-inducible ERF11-GR construct. Rosettes of ERF11-GR plants grown in vitro on dexamethasone-containing medium were significantly smaller than wild-type plants (Fig. 3A), with an average size reduction of 21% (P = 2E-5, ANOVA; Fig. 3, A and B). The ERF11-mediated growth reduction was also visible on every individual leaf of the rosette of soil-grown plants watered with a dexamethasone solution (P = 2E-5, ANOVA; Fig. 3C; Supplemental Fig. S3A).
Figure 3.
ERF11 regulates Arabidopsis leaf growth under mannitol-induced stress conditions. A, Projected rosette area at 22 DAS of the erf11 mutant and the dexamethasone-inducible overexpression lines of ERF6 (ERF6-GR) and ERF11 (ERF11-GR) and of both in ERF6-GR × ERF11-GR double homozygous plants, grown under control conditions (medium supplemented with dexamethasone for the GR lines). Represented values are the means of three biological repeats with their se. *, P < 0.05, compared with the wild type (WT). B, Size of the third true leaf over time of the GR lines described in A upon exposure to dexamethasone from 9 DAS onward. *, P < 0.05 (ANOVA), compared with the wild type. C, Rosettes of the GR lines described in A grown in soil for 22 d and watered with a dexamethasone-containing solution. D, Cellular measurements of the abaxial side of the third leaf of the wild type and ERF11-GR at 20 DAS from the plants shown in B. *, P < 0.05, compared with the wild type. E, Reduction in rosette area caused by growth for 22 d on medium supplemented with 25 mm mannitol, compared with the rosette area of the same line grown under control conditions. Error bars represent the se of three biological repeats. *, P < 0.05 (ANOVA), compared with the wild type. Bar = 2 cm.
To investigate the effect of overexpressing ERF11 on leaf development in more detail, 35S:ERF11-GR and wild-type plants were grown on MS medium and dexamethasone treatment was started at 9 DAS, when all cells of the third leaf are actively proliferating, and the size of the third leaf was measured every day until 20 DAS. Leaf growth inhibition started to be visible at 18 DAS, i.e. after 9 d of overexpression of ERF11 (Fig. 3B). To unravel the cellular mechanism behind this growth reduction, cell number and cell area of the harvested leaves were measured at 20 DAS. Leaves of plants overexpressing ERF11 showed a 16% decrease in cell area (P = 0.01, ANOVA) and an 18% reduction in cell number (P = 0.01, ANOVA; Fig. 3D). Together, these data show that ERF11 has the capacity to repress leaf growth by negatively affecting cell area and number.
Finally, because ERF6 and ERF11 are coregulated and because the ERF6-GR line has previously been reported to inhibit leaf growth as well (Dubois et al., 2013), we further explored whether both transcription factors could act together to regulate leaf growth. We therefore crossed the 35S:ERF6-GR line with the 35S:ERF11-GR line and selected plants that were homozygous for both constructs and that expressed the respective constructs to a comparable level as the single homozygous lines (Fig. 4A). Although both individual lines showed, respectively, strong and mild growth inhibition when grown on dexamethasone, surprisingly, when both ERF6 and ERF11 are overexpressed simultaneously, the extreme dwarfism induced by ERF6 is almost completely abolished and the rosette area is again comparable with the wild type (Fig. 3A). This rescue of the phenotype was observed both in experiments conducted in vitro (Fig. 3A) and in soil (Fig. 3C) under long-term (Fig. 3A) and short-term (Fig. 3B) dexamethasone treatment and holds for all rosette leaves (Supplemental Fig. S3B). Taken together, these results demonstrate that ERF6 and ERF11 are able to negatively affect leaf growth individually, while simultaneous overexpression of both transcription factors releases this growth inhibition.
Figure 4.
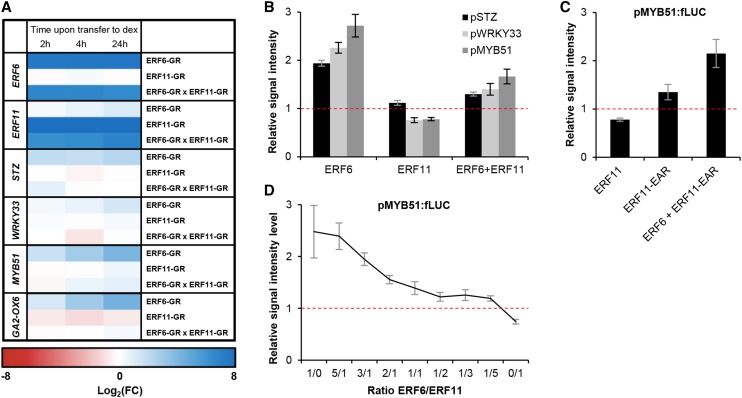
ERF6 and ERF11 antagonistically regulate common target genes. A, Expression levels of ERF6, ERF11, and the ERF6 target genes in dexamethasone-inducible overexpression lines of ERF6 (ERF6-GR), ERF11 (ERF11-GR), and both in ERF6-GR × ERF11-GR double homozygous plants. Expression values are the average of at least three biological repeats. Values are relative to the expression level in wild-type plants subjected to the same treatment. B, Protoplast activation assay with pSTZ:fLUC, pWRKY33:fLUC, and pMYB51:fLUC for binding and effect of ERF6, ERF11, and a combination of both in a 1:1 ratio. C, Protoplast activation assay on the promoter of MYB51 for the native ERF11 protein, a truncated ERF11 in which the repressive EAR domain has been removed, and a combination in a 1:1 ratio of ERF6 and the truncated ERF11. D, Titration protoplast activation assay on the promoter of MYB51 with multiple concentrations of ERF6 and ERF11. For B to D, values are signal intensities normalized to the cotransformed normalization plasmid rLUC and relative to the negative control (35S:GUS + pTARGET:fLUC, horizontal line). Values represent the average of at least two biological repeats with their se. FC, Fold change.
ERF11 Is Involved in the Regulation of Leaf Growth under Mild Stress
To explore whether ERF11 is involved in leaf growth inhibition under stress, we phenotyped ERF11 loss- and gain-of-function lines under growth-limiting conditions mediated by low concentrations of mannitol. After 22 d of growth, the rosette size on mannitol was reduced by 14% in erf11 and by 23% in the wild type compared with the rosette area of the corresponding line on control conditions (Fig. 3E). Thus, erf11 mutants are more tolerant to mannitol (P = 0.02, ANOVA), indicating that ERF11 plays a role in leaf growth regulation under mannitol-induced growth-limiting conditions. For the ERF11-overexpressing plants, however, exposure to mild stress caused a growth inhibition comparable with the mannitol-induced growth reduction observed in wild-type plants (14% and 16%, respectively; P = 0.7, ANOVA; Fig. 3E). This shows that ERF11-overexpressing plants are equally sensitive to mannitol-induced stress and that overexpression of ERF11 does not mimic mannitol-induced growth inhibition. By contrast, ERF6-overexpressing plants were previously shown, using a weaker ERF6 overexpression line with a milder phenotype, to be hypersensitive to mannitol (Dubois et al., 2013). Interestingly, when 35S:ERF6-GR × 35S:ERF11-GR plants were grown on medium supplemented with mannitol, the reduction in leaf growth was also more pronounced than in wild-type plants (31% versus 16% for the wild type; P = 0.02, ANOVA). Taken together, these data demonstrate that ERF11 and ERF6 are both involved in the regulation of leaf growth inhibition under stress.
At the Molecular Level, ERF6 and ERF11 Compete for Common Target Genes
Because ERF6 is a transcriptional activator and ERF11 is a transcriptional repressor, we reasoned that simultaneous induction of both transcription factors by stress could generate a possible antagonism and competition for common target genes. We therefore transferred the double homozygous ERF6-GR × ERF11-GR line and the appropriate controls to dexamethasone-containing medium and measured, in the actively growing third leaf, the expression level of the ERF6 target genes: GA2-OX6, MYB51, WRKY33, and STZ (Dubois et al., 2013). In the ERF6-GR line, these target genes were induced within 4 h upon dexamethasone treatment (P < 0.05; Fig. 4A). By contrast, overexpression of only ERF11 (ERF11-GR) did not affect the expression of these genes. Interestingly, when ERF11 was cooverexpressed with ERF6 (ERF6-GR × ERF11-GR), the expression of the target genes was not induced by dexamethasone, and thus these genes were no longer induced by ERF6 within the measured time frame (Fig. 4A).
Multiple molecular models can explain this antagonistic relationship between ERF6 and ERF11. First, ERF11 could directly repress the expression of ERF6 and thereby indirectly repress the induction of the ERF6 target genes. However, neither expression analyses of ERF6 in the ERF11-GR line upon dexamethasone treatment nor promoter-binding assays of 35S:ERF11 on pERF6:fLUC support this hypothesis (Supplemental Fig. S4). Second, ERF6 and ERF11 could form heterodimers, which would be responsible for the repressive function on the promoters of the genes targeted by ERF6. Third, ERF11 could physically withhold ERF6 proteins from their targets, indirectly inhibiting their induction. Both hypotheses imply physical interaction at the protein level between ERF6 and ERF11. However, despite successive attempts using quantitative yeast (Saccharomyces cerevisiae) two-hybrid (Y2H) assays to measure putative interactions between ERF6 and ERF11, no protein-protein interactions between both transcription factors could be detected (Supplemental Fig. S5). Finally, ERF11 could directly compete with ERF6 for binding on the promoter of target genes. To test this hypothesis, we first performed multiple protoplast activation assays using fusions of the promoters of the known ERF6 target genes with the gene encoding fLUC (pSTZ:fLUC, pMYB51:fLUC, and pWRKY33:fLUC) and cotransformed each of them with either 35S:ERF6, or 35S:ERF11, or a combination of both in a 1:1 ratio. As expected, transformation with ERF6 alone caused an increase in signal intensity (Fig. 4B), strongly suggesting activation of the promoters of the three target genes. By contrast, upon transformation with ERF11 alone, the signal intensity did not increase and even showed a weak but significant and reproducible reduction for pMYB51 and pWRKY33. Upon cotransformation with both transcription factors, only a slight activation of the three reporter constructs was observed. To further confirm these indications that ERF11 would bind and repress the promoters of the ERF6 target genes rather than withholding ERF6 from activating the targets, we generated a truncated variant of ERF11, in which the EAR domain was removed. The signal intensities using ERF11-EAR were always higher than the signal obtained with native ERF11, and the truncated ERF11 was unable to inhibit the activation by ERF6 when cotransformed in a 1:1 ratio (Fig. 4C). Finally, to further quantify the strength of the transcriptional activation and repression by ERF6 and ERF11, respectively, we performed a titration assay using pMYB51:fLUC (the promoter on which ERF6 and ERF11 had the most pronounced effect) in combination with multiple ERF6/ERF11 ratios (Fig. 4D). The ability of ERF6 to activate the expression of pMYB51:fLUC gradually decreased with rising ERF11 concentrations. The decrease holds until a 1:1 ratio was reached, when protoplasts were transfected with an equal concentration of 35S:ERF6 and 35S:ERF11. Further increase in ERF11 concentration did then no longer dramatically decrease the ability of ERF6 to activate the pMYB51:fLUC signal, which reached a plateau. Only when no ERF6 was present did absolute repression of the pMYB51 by ERF11 become visible. Based on these experiments, we conclude that ERF11 antagonizes ERF6 function most likely through direct competition for the same promoters and plausibly the same cis-regulatory elements. The resulting effect on target gene expression depends on the abundance of each of the transcription factors, and the activation by ERF6 appears to be stronger than the repression by ERF11, which occurs through the presence of the EAR domain.
At Least Two Parallel Pathways Are Upstream of ERF6 and ERF11
Recently, two receptor-like kinases, EGM1 and EGM2, were shown to be involved in the mannitol-induced growth inhibition and were suggested to act upstream of the ERF5/ERF6 pathway (Trontin et al., 2014). The latter study analyzed egm1 and egm2 mutants on higher concentrations of mannitol (60 mm), which not only restrict plant growth, but also induce clear stress symptoms (narrow and curling leaves). Therefore, we first phenotypically characterized the egm1 and egm2 mutant on 25 mm mannitol. Because EGM1 and EGM2 are tandem-duplicated genes, no double egm1egm2 mutant is available, but both genes are not fully redundant (Trontin et al., 2014). Surprisingly, although egm1 and egm2 did, as reported, tolerate high mannitol concentrations (60 mm; respectively, 43% and 39% larger rosettes than the wild type; P < 0.05) significantly better, they were not significantly more tolerant to the mild mannitol concentration (25 mm; Supplemental Fig. S6A). By contrast, the erf5erf6 mutant grew significantly better than the wild type under low mannitol stress conditions (P < 0.05, ANOVA; Dubois et al., 2013) but not on higher mannitol concentrations (Supplemental Fig. S6B). These differential growth responses to mannitol suggest that the EGMs and ERF5/ERF6 are not directly involved in the same linear pathway. The observation that both egm mutants show a growth reduction of, on average, 21% and 49% under 25 and 60 mm mannitol stress, respectively, compared with growth on MS, further indicates that EGM1 and EGM2 are not solely responsible for leaf growth reduction by mannitol, but that instead, multiple pathways are controlling the mannitol-induced leaf growth inhibition.
EGM1 is known to be transcriptionally induced by 60 mm mannitol in growing leaves (Trontin et al., 2014), thus in the same tissue as in which ERF5, ERF6, and ERF11 are induced. Therefore, we further explored the exact timing of the induction of EGM1 upon mannitol stress and measured the EGM1 transcript levels upon short-term exposure to 25 mm mannitol (as above). Surprisingly, the short-term mild mannitol treatment did not increase the EGM1 transcript level within the measured time frame (from 5 to 180 min; Supplemental Fig. S7A). This is consistent with the short-term expression data from Skirycz et al. (2011), where the EGM transcript is only significantly induced 12 h upon mannitol (2-fold induction at 24 h) but not at 1.5 h nor at 3 h (Supplemental Fig. S7B). Thus, these results demonstrate that EGM genes are not transcriptionally induced by mannitol in actively growing leaves within the same time frame as ERF6 and ERF11.
Finally, we analyzed whether ERF5, ERF6, or ERF11 expression was modified in the egm1 and egm2 mutant backgrounds. To this end, egm1 and egm2 mutants were exposed to long-term mannitol (25 and 60 mm) treatment, and ERF5, ERF6, and ERF11 transcript levels were measured by qRT-PCR. Under long-term severe mannitol stress, the expression level of the selected ERF genes was significantly lower (on average, 40%) in the egm1 mutant than in the wild type, and the same tendency was found for the egm2 mutant (Supplemental Fig. S8C). Surprisingly, under control conditions, the ERF6 expression level in the egm1 mutant was slightly, but significantly, higher (34%) than in the wild type (Supplemental Fig. S8A). Moreover, the expression of ERF6 and ERF11 in the egm1 mutant under low mannitol concentrations were down-regulated to levels lower than under control conditions (25% and 66%, respectively; P < 0.05), whereas levels similar to control conditions would be expected in the case of a simple linear activating EGM/ERF pathway (Supplemental Fig. S8B). Thus, although these results suggest that EGMs are involved in the regulation of ERF expression under severe mannitol stress, the EGM/ERF pathway is not linear and might act in parallel with other pathways.
DISCUSSION
As demonstrated before, low concentrations of mannitol trigger a dual response in young Arabidopsis leaves. Mannitol induces, on the one hand, a rapid inhibition of leaf growth and, on the other hand, the transcriptional activation of a plethora of stress-responsive genes (Skirycz et al., 2010, 2011; Dubois et al., 2013; Trontin et al., 2014). This transcriptional response, involving more than 1,500 genes in growing leaves, is thought to be established through transcriptional cascades. In a simplified view, one or a few early transcription factors activate multiple other regulators, which, in turn, activate their own target genes. To unravel the sequential steps of these cascades and to distinguish the early players from late-induced stress-responsive genes, we opted in our previous research and in the presented article for time-course experiments following sudden exposure to mannitol (Skirycz et al., 2011; Claeys et al., 2012). As described previously, ERF5 and ERF6 are among the few early regulators induced by mannitol and regulate more than one-half of the early mannitol-induced genes (Skirycz et al., 2011; Dubois et al., 2013). Here, we show that ERF11 is induced in growing leaves at the same timing as ERF6, within 1 h upon sudden exposure to mannitol.
ERF6 Induces ERF11 under Mannitol Stress in Growing Leaves
Although the mechanisms that activate the ERF6 protein under stress start to be unraveled (Son et al., 2012; Dubois et al., 2013; Wang et al., 2013), much less is known about how ERF11 is induced by mannitol. Because ERF11 and the known ERF6 target genes are induced simultaneously by mannitol, we tested the hypothesis that ERF11 could be a target gene of ERF6 by expression analysis with qRT-PCR and a luciferase assay and confirmed the induction of ERF11 by ERF6. Because it is rather counterintuitive that the activator (ERF6) and its targets are induced simultaneously, it is important to emphasize that ERF6 is first activated posttranslationally upon stress and subsequently activates its downstream target genes, as well as its own expression. Thus, in growing leaves of plants exposed to mannitol, ERF11 and ERF6 are induced simultaneously, because both transcripts are activated by ERF6. We further used this coexpression as a characteristic to explore whether ERF6 could activate ERF11 under other stress conditions. A clear coexpression (Pearson correlation coefficient = 0.73) was observed between ERF6 and ERF11 under numerous biotic and abiotic stress conditions (Supplemental Fig. S9; Dombrecht et al., 2007; Hruz et al., 2008). Interestingly, a fast and simultaneous induction of ERF6 and ERF11, similar to our findings upon mannitol-induced stress, has been reported during short-term cold treatment, where ERF6 and ERF11 were identified among the very few early (74 genes after 3-h treatment) cold-responsive genes (Lee et al., 2005) and after 15-min treatment with the chitin-mimicking compound chitooctaose (Libault et al., 2007). In general, a meta-analysis of available stress data sets showed that ERF5, ERF6, and ERF11, as well as STZ and WRKY33, all belong to a confined group of 197 genes differentially expressed in more than 80% of the stress-related data sets, suggesting that they belong to a core set of general stress-responsive genes (Ma and Bohnert, 2007).
The Regulators Upstream of ERF11 Are Diverse and Context Dependent
Several other transcription factors have been proposed to transcriptionally regulate ERF11 in different developmental contexts. The MULTIPROTEIN BRIDGE FACTOR1c (MBF1c), a transcriptional coactivator, has been suggested to act upstream of ERF11 expression, because ERF11 transcript levels were increased in MBF1c overexpression lines, although no evidence of direct regulation was provided (Suzuki et al., 2005). Among the genes induced upon MBF1c overexpression (steady state), ERF6 and several ERF6 target genes, such as GA2-OX6 and WRKY33, were found, so it cannot be excluded that ERF6 also induced these genes as a secondary effect of MBF1c overexpression. Another identified candidate for transcriptional activation of ERF11 is the ELONGATED HYPOCOTYL5 (HY5), a basic leucine zipper (bZIP) transcriptional activator involved in hypocotyl growth (Lee et al., 2007; Song et al., 2008; Li et al., 2011). HY5 has been shown to bind the ERF11 promoter in vivo through its preferential CACGTG binding sequence, present in the 1-kb region upstream of the ERF11 coding sequence. The ERF11 transcript levels have also been shown to be decreased in the hy5 mutant, which might be consistent with the more pronounced hypocotyl growth of this mutant, because ERF11 is able to negatively affect growth. Hypocotyl growth is primarily driven by cell expansion (Boron and Vissenberg, 2014; Ragni and Hardtke, 2014), a process that ERF11 is able to control. Finally, the MYC2/JASMONATE-INSENSITIVE1 (JIN1) basic helix-loop-helix (bHLH) transcriptional repressor has been proposed to be a negative regulator of ERF11 expression during the jasmonic acid-mediated response by binding the same CACGTG box in the ERF11 promoter (Dombrecht et al., 2007). Thus, multiple transcription factors have been proposed as regulators of ERF11 expression in different developmental or environmental contexts, and for HY5, MYC2/JIN1, and ERF6, direct regulation has been found. In contrast to ERF6, neither MYC2/JIN1 nor HY5 are induced in growing leaves by low concentrations of mannitol, and under stress, ERF11 is less coexpressed with these regulators than with ERF6 (Supplemental Fig. S9). Therefore, we conclude that ERF11 is transcriptionally regulated by different transcription factors depending on the context and that ERF6 might be one of the activators of ERF11 under several stresses and particularly under mannitol-induced stress.
Competition for the Same Promoters at the Molecular Basis of the ERF6-ERF11 Antagonism
ERF11 is known as a transcriptional repressor, because it possesses an EAR domain (Ohta et al., 2001; Yang et al., 2005; Nakano et al., 2006; Li et al., 2011). The simultaneous induction of a transcriptional activator and repressor suggests the presence of a regulatory network in which the repressor could attenuate the response induced by the activator. Our experimental data support this hypothesis: in a transgenic line overexpressing both transcription factors, ERF11 is able to suppress the ERF6-mediated activation of the downstream genes. In a transactivation assay, competition for the promoter of common target genes was shown to occur, resulting in either activation by ERF6 or repression by ERF11 in a concentration-dependent way. Furthermore, the growth inhibitory pathway induced by ERF6 was no longer activated when both ERF6 and ERF11 were overexpressed, and the plants overexpressing both transcription factors did no longer show the ERF6-induced dwarfism. Similar antagonistic relationships between two or more regulatory proteins controlling growth and stress defense have been described in relation to both biotic and abiotic stress. Generally, the activator and repressor are both transcriptionally induced by the stress, and this also holds true for ERF6 and ERF11. For example, upon infection with Pseudomonas syringae, effector-triggered immunity mechanisms induce both activators of defensive mechanisms, i.e. resistance to P. syringae4/RCC1 (CHC1)-LIKE DOMAIN1 (rps4-RLD1) and several TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors, as well as the repressor SUPPRESSOR OF rps4-RLD1 (SRFR1; Kim et al., 2014). In a similar way, exposure to UV-B-induced stress induces both HY5 and UV-B RESISTANCE8 (UVR8) as activators and REPRESSOR OF UV-B-INDUCED PHOTOMORPHOGENESIS1 (RUP1) and RUP2 to antagonize UVR8, although it should be noted that in this example, RUP1 and RUP2 are not transcription factors (Gruber et al., 2010). In both examples, however, the antagonism was established through heterodimerization of the activating and repressing regulatory proteins, resulting in a repressive complex that blocks further stress response. Despite several attempts, we did not detect heterodimerization between ERF6 and ERF11. Moreover, our data showed that ERF11 uses its EAR domain to repress gene expression and that the truncated ERF11 without EAR is unable to counteract the activation by ERF6, demonstrating that ERF6 sequestration by ERF11 is unlikely to be at the basis of the competition. Heterodimerization between ERFs is, however, known to exist and has previously been reported for ERF5 and ERF6 (Son et al., 2012). This interaction was therefore included as a positive control in our assays but could not be validated. Thus, antagonistic relationships between an activating and a repressing transcription factor occur both under biotic and abiotic stress to temper the stress response and can be conferred at the molecular level both through protein-protein interaction and through competition for shared target genes.
Multiple Players Act Upstream of the ERF6-ERF11 Regulon
Upstream of the ERFs, multiple pathways have previously been identified in different contexts and might therefore induce the transcriptional cascade under mannitol stress. First, a phosphorylation cascade working downstream of the ethylene receptors and mediated through MPK3 and MPK6 has been shown to phosphorylate and thereby activate ERF6 in other abiotic stress contexts (Popescu et al., 2009; Son et al., 2012; Wang et al., 2013). Supporting the involvement of this ethylene-mediated pathway in actively growing leaves of plants exposed to mannitol, the time-course experiments showed that ACC induced ERF6 and ERF11 more rapidly than mannitol, which is consistent with our previously proposed model in which mannitol first triggers the accumulation of ACC, which, in turn, activates several ERFs (Skirycz et al., 2011). However, a recent study identified two putative receptor-like kinases, EGM1 and EGM2, as possible activators upstream of the mannitol-induced ERFs (Trontin et al., 2014). If so, our results suggest that their activation by mannitol should occur at the posttranscriptional level, because it was shown that their transcriptional induction was much slower than the up-regulation of the ERFs. Furthermore, phenotypic analysis of egm1, egm2, and erf5erf6 mutants under control conditions and mild and severe mannitol stress demonstrated that the egm mutants did not phenocopy the erf5erf6 mutant under stress: the egm mutants were more tolerant to severe stress, and the erf5erf6 mutant was more tolerant to mild stress. Based on these results, we speculate that ERF5 and ERF6 are induced by low mannitol concentrations and regulate growth under these conditions, while EGM proteins are more likely to be involved in tolerance against more severe stress levels. Thus, although EGM proteins and ERF5 and ERF6 are all involved in the mannitol-induced stress response, it is unlikely that they act in a same linear pathway. The reduced ERF5, ERF6, and ERF11 expression levels in egm mutants grown on high mannitol concentrations nonetheless indicate that the EGM proteins are involved in the activation of these ERFs. Thus, although the way in which EGMs might regulate ERF expression under mannitol stress still remains highly elusive, our experiments demonstrate that several pathways interact at multiple levels, probably in a concentration-dependent manner, rendering the stress response extremely complex.
A Model for the Antagonism between ERF6 and ERF11 in the Regulation of Leaf Growth under Stress
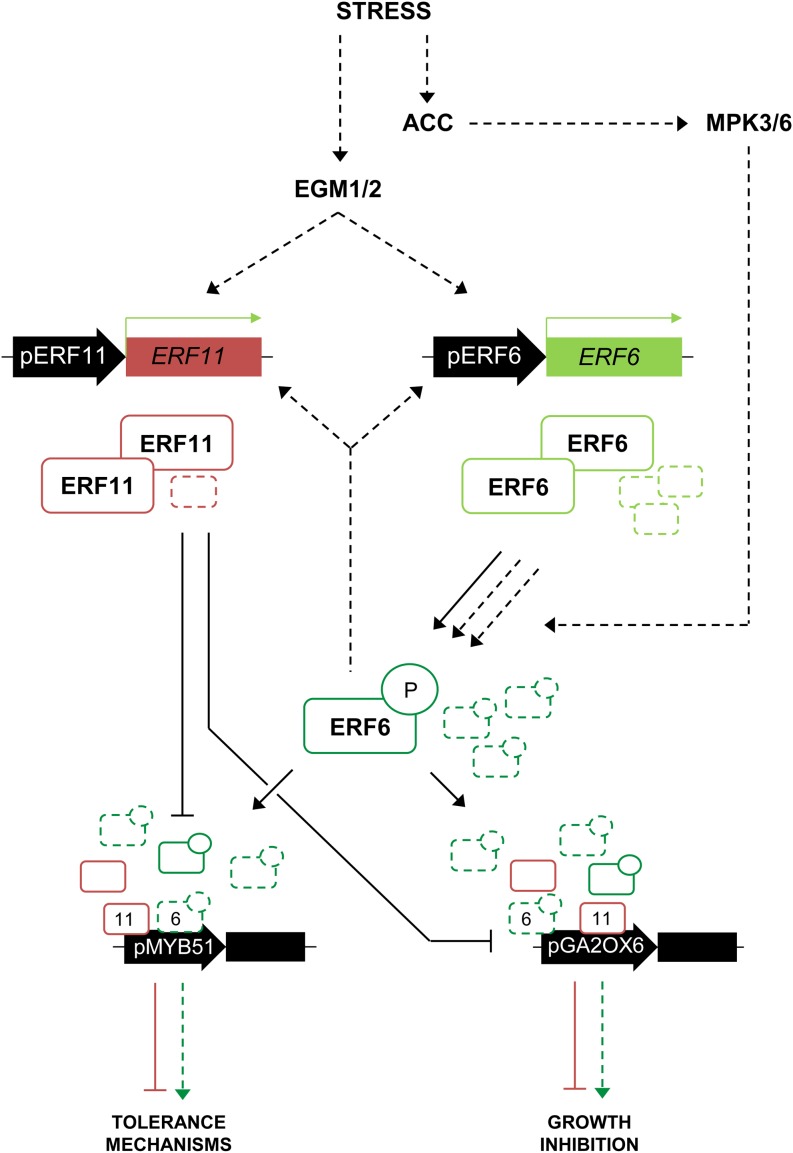
Based on our results, we propose the following model for the role of ERF6 and ERF11 in the regulation of leaf growth under stress (Fig. 5). Prior to stress, ERF11 and ERF6 are lowly expressed and their expression varies throughout the day. ERF11, which, to our knowledge, does not possess any putative phosphorylation site for posttranslational regulation (Nakano et al., 2006), is likely to be active and restricts the expression of GA2-OX6 and the stress-responsive genes such as MYB51. ERF6, by contrast, is known to need activation through phosphorylation by MPK3 and MPK6 to activate downstream targets (Son et al., 2012; Meng et al., 2013; Wang et al., 2013). However, because growth conditions are never optimal, low levels of ERF6 proteins are probably sporadically phosphorylated. In this low level of stress context, the sporadically activated ERF6 proteins cannot compete with the ERF11 repressive proteins. Plant growth is sustained, because the ERF6/ERF11 balance is equilibrated. Disturbing this balance by, for example, overexpressing only ERF6 or ERF11 clearly affects leaf growth, while overexpression of both restores the balance and thereby growth. When plants gradually perceive stress, rising, but still mild, stress levels cause phosphorylation of ERF6, and the ratio of active ERF6 is likely to dramatically increase, resulting in out-competition of ERF11 by ERF6 on the promoters of their target genes and in a net activation of the targets by ERF6. As demonstrated by luciferase assays, activation of targets by ERF6 was stronger than repression by ERF11 when ERF6 levels exceeded ERF11 levels. Thereby, growth is blocked and defense mechanisms are activated to safeguard plant survival. However, higher amounts of active ERF6 proteins also generate the production of more ERF11 proteins. When stress levels are again declining, this relatively high amount of ERF11 proteins is able to rapidly block the stress response when ERF6 activity levels are decreasing to rapidly switch off the stress response when it becomes unnecessary.
Figure 5.
Schematic overview of the putative roles of ERF6 and ERF11 under stress. Under basal levels of stress (plain arrows and proteins), ERF11 and ERF6 are lowly expressed. Low levels of ERF11 proteins are sufficient to repress the activation of the stress response mechanisms, as they are likely more abundant than the active, phosphorylated ERF6 proteins. Under higher stress levels (dashed arrows and proteins), ACC accumulates and MPK3/MPK6 phosphorylates and thereby activates ERF6. ERF6 now outcompetes ERF11 proteins on the promoter of the target genes, activating tolerance mechanisms and growth inhibition. However, ERF6 also transcriptionally activates ERF11, increasing ERF11 protein levels, which enables a rapid inhibition of the stress response when the stress levels decrease. P, Phosphorylation residue.
The ERF6-ERF11 Loop May Be a General Module to Fine-Tune Stress Responses
The literature also contains many examples of studies, mainly biotic stress related, in which ERF6 and ERF11 were found among the differentially expressed genes, although the experiments were not conducted on growing leaves but rather on mature leaf tissue or complete seedlings (McGrath et al., 2005; AbuQamar et al., 2006; Dombrecht et al., 2007; Eulgem and Somssich, 2007; Libault et al., 2007; Ma and Bohnert, 2007; Hu et al., 2008; Moffat et al., 2012; Son et al., 2012; Meng et al., 2013; Vermeirssen et al., 2014). In brief, ethylene and the described ERFs are generally induced in response to necrotrophic pathogens such as Botrytis cinerea and control the expression of the plant defensive proteins PDF1.1 and PDF1.2. Moreover, the described target genes of ERF6 and ERF11, i.e. STZ, WRKY33, and MYB51, were reported to be involved in tolerance against biotic stress as well. This suggests that the presented regulatory network might be active in a broader context than growing leaf tissue. Thus, we speculate that the described balance might also be involved in mature leaves to avoid uncontrolled activation of the defensive response upon biotic stress exposure. Because mannitol is a molecule secreted by fungal pathogens during the infection process (Trontin et al., 2014), it might mimic such a biotic stress response in vitro. At low concentrations, mannitol was found to induce the presented regulatory network only in actively growing leaves, but at higher concentrations, the ERFs were also induced in complete seedlings. Thus, the presented ERF-centered network might be functional during different developmental stages of leaf growth, depending on the severity of the stress. For this reason, future studies to elucidate the role of ERF6 and ERF11 in different stress contexts and different tissues should take development, stress levels, and timing into account, not confounding growing leaves and, in seedlings, low stress levels and severe stress, and the short-term stress response and the long-term adaptive response, because it is likely that the subtle balance between plant growth and stress defense is regulated in a specific way in these different contexts.
CONCLUSION
In this study, we uncovered the presence of a negative feedback mechanism to balance growth and defense upon exposure to mild mannitol-induced stress. We demonstrated that the transcriptional repressor ERF11 is able to counteract the action of the activator ERF6 by inhibiting their common target genes in a dose-dependent manner. Further investigations are, however, necessary to elucidate the exact mechanism occurring in leaves and should include analysis for competition on the same cis-regulatory elements and in planta protein-protein interaction studies. We speculated that inhibition of the downstream responses by ERF11 ensures sustained growth under low-stress conditions. Upon sudden exposure to mannitol, the expression of both transcription factors is induced simultaneously in growing Arabidopsis leaves, activated upstream by both an ethylene-mediated cascade and through a mannitol receptor-mediated pathway. In these stress conditions, the balance is shifted toward extensive activation of ERF6, the levels of which increase exponentially through an autoactivation loop, causing the activation of the growth inhibitory pathway and the induction of downstream tolerance-related transcription factors. In parallel, the activation of the repressive factor ERF11 functions as a brake to counteract the ERF6-driven activation and to ensure proper restoration of the balance when the stress levels are again decreasing. Consistently, the generation of transgenic plants in which the balance is disturbed in one or the other direction generates plants with pronounced growth penalties, while plants overexpressing both the activator and the repressor have a reequilibrated balance and therefore normal growth phenotypes. The described balance likely occurs in all shoot tissues and conditions where ERF6 and ERF11 are expressed but might fulfill different functions and therefore result in a specific outcome depending on the exact context. In actively growing Arabidopsis leaves, it provides a tightly regulated, but flexible, system to control leaf growth in a dynamic way upon continuously changing environmental stress conditions.
MATERIALS AND METHODS
Plant Lines
The single erf5, erf6, and erf11 mutants were obtained from the SALK collection (references SALK_076967 [erf5], SALK_030723 [erf6], and SALK_116053 [erf11]). The egm1 and egm2 mutants were described by Trontin et al. (2014) and were kindly provided by Olivier Loudet. All lines used are in Columbia background.
Soil Plant Growth Conditions and Individual Leaf Area Measurements
Plants were grown in Gottinger pots (8 × 8 × 8.5 cm) filled with soil (Saniflor, without osmocote) at 21°C under a 16-h-day (110 μmol m–2 s–1) and 8-h-night regime. Pots were covered with transparent plastic foil for 4 d to stimulate germination. Watering with 15 mL of water was applied at 9, 13, 16, and 20 DAS. In experiments in which GR lines were used, plants were watered with 15 mL of a 15 µm dexamethasone-containing solution. For leaf size measurements, 12 plants were grown per line, the transgenic lines were always grown together with the appropriate control on the same tray, and randomization was done between the genotypes. At 22 DAS, plants were harvested, and leaf series were made by cutting each individual leaf of the rosette and ranking them from old to young on a square agar plate. Plates were photographed, and pictures were subsequently analyzed using ImageJ version 1.45 (National Institutes of Health; http://rsb.info.nih.gov/ij/) to measure the size of each individual leaf.
In Vitro Plant Growth Conditions and Experiments
Seedlings were grown in vitro on one-half-strength MS medium (Murashige and Skoog, 1962) containing 1% (w/v) Suc at 21°C under a 16-h-day (110 μmol m–2 s–1) and 8-h-night regime. For long-term experiments without transfer, 9 g L–1 agar was added to the medium. To facilitate transfer for the short-term experiments, 6.5 g L–1 agar was used, and the growth medium was overlaid with a nylon mesh (Prosep) of 20-µm pore size. For leaf growth experiments and expression analyses, 32 and 64 seeds, respectively, were distributed equally on a 14-cm-diameter petri dish. To enable correct comparisons, the different lines were always grown together on one plate with the appropriate control.
Exposure to Long-Term Mannitol Stress and Rosette Growth Analysis
For long-term exposure to mannitol stress, plants were grown on one-half-strength MS medium containing 9 g L–1 agar and 25 or 60 mm mannitol, and plates were photographed at 22 DAS. The pictures were subsequently analyzed using ImageJ version 1.45 to measure the size of each individual rosette.
Exposure to Short-Term Mannitol Stress or ACC- and Dexamethasone-Induced Expression of ERF6 and/or ERF11
Arabidopsis (Arabidopsis thaliana) seedlings were grown in vitro on one-half-strength MS medium overlaid with a nylon mesh until the third leaf had completely emerged from the shoot apical meristem but was still in a fully proliferative stage, at 9 DAS (except for the experiments performed for Fig. 4A, where the transfer was done at 15 DAS). At this time point, the mesh with plants was transferred to plates with one-half-strength MS medium containing 25 mm mannitol (plant culture tested; Sigma), 5 µm ACC (Sigma), or 5 µm dexamethasone (Sigma).
For expression analysis performed for Figure 1, the young Arabidopsis seedlings were harvested after 5, 10, 15, 30, 45, 60, 120, and 180 min of stress exposure, followed by microdissection of the third true leaf for leaf growth analysis (less than 0.1 mm2 in size).
Leaf Growth Analysis
Leaf growth analysis over time was performed on the third true leaf harvested at different time points after transfer to dexamethasone. After clearing with 70% ethanol, leaves were mounted in lactic acid on microscopic slides. For each time point, about 15 to 20 leaves per genotype were photographed with a binocular, and abaxial epidermal cells (100–200) were drawn for three representative leaves with a DMLB microscope (Leica) fitted with a drawing tube and a differential interference contrast objective. Photographs of leaves and drawings were used to measure leaf area and cell size, respectively, using ImageJ version 1.45, and average cell numbers were calculated by dividing the leaf area by the drawn area followed by multiplication of this factor with the number of drawn pavement cells. Calculated cell areas were ln transformed prior to all subsequent analyses.
Expression Analysis (qRT-PCR)
RNA samples were obtained from three independent experiments and from multiple plates within each experiment. For the experiments depicted in Figure 1, whole seedlings were harvested rapidly, submerged in 6 mL of the mRNA-stabilizing RNAlater solution (Ambion), and after overnight storage at 4°C, dissected under a binocular microscope on a cooling plate with precision microscissors. Dissected leaves were transferred to a new tube, frozen in liquid nitrogen, and ground with a Retsch machine and 3-mm metal balls. For the experiments depicted in Figures 2D and 4A, the third true leaf was harvested, frozen immediately in liquid nitrogen, and ground. The harvesting of samples during the night was performed under green light. RNA was subsequently extracted with TriZol (Invitrogen) and further purified with the RNeasy Mini Kit (Qiagen). DNA digestion was done on columns with RNase-free DNase I (Invitrogen).
For complementary DNA synthesis, the iScript cDNA Synthesis Kit (Bio-Rad) was used according to the manufacturer’s instructions using 1 μg of RNA. qRT-PCR was done on a LightCycler 480 (Roche Diagnostics) in 384-well plates with LightCycler 480 SYBR Green I Master Mix (Roche) according to the manufacturer’s instructions. Melting curves were analyzed to check primer specificity. Normalization was done against the average of housekeeping genes AT1G13320, AT2G32170, AT2G28390: ΔCt = Ct (gene) – Ct (mean [housekeeping genes]) and ΔΔCt = ΔCt (control line) – ΔCt (line of interest), where Ct refers to the number of cycles at which SYBR Green fluorescence reaches an arbitrary value during the exponential phase of amplification. Primers were designed with the QuantPrime Web site (Arvidsson et al., 2008; Skirycz et al., 2010). Primers used in this study are as follows: ERF5, AAATTCGCGGCGGAGATTCGTG and TCAAACGTCCCAAGCCAAACGC; ERF6, TCGAATCCTCCTCGCGTTACTG and TTCGGTGGTGCGATCTTCAACG; ERF11, ATGGCACCGACAGTTAAAAC and TCAGTTCTCAGGTGGAGGA; EGM, TGGCTCATGTGTGGTCAATCTGG and TCATTAGCAGCGTCTTGCACAC; GA2-OX6, TGGATCCCAATCCCATCTGACC and TCTCCCATTCGTCAATGCCTGAAG; MYB51, GCCCTTCACGGCAACAAATG and GGTTATGCCCTTGTGTGTAACTGG; STZ, TCACAAGGCAAGCCACCGTAAG and TTGTCGCCGACGAGGTTGAATG; and WRKY33, CTTCCACTTGTTTCAGTCCCTCTC and CTGTGGTTGGAGAAGCTAGAACG.
Protoplast Activation Assay
The protoplast activation assay was performed as previously described (De Sutter et al., 2005; Pauwels et al., 2010). All transformation constructs were obtained using the Gateway cloning system, and all liquid handlings were done on the Tecan Genesis automated platform (De Sutter et al., 2005). The protoplast activation assay was performed in a 3-d-old Bright Yellow-2 (BY-2) tobacco (Nicotiana tabacum) cell culture subcultured from a 6- to 10-d-old culture. BY-2 cells were protoplasted using a 1% Cellulase (Kyowa Chemical Products) and 0.1% Pectolyase (Kyowa Chemical Products) enzyme solution in a 0.4% mannitol (Sigma) buffer. Protoplasts were then washed, counted, and diluted to 500,000 mL–1. For every transcription factor-promoter combination, 100 µL (50,000 protoplasts) was used. To confirm the activating and repressing regulatory effect of ERF6 or ERF11, respectively, on the promoters of ERF6, ERF11, STZ, MYB51, and WRKY33, protoplasts were cotransfected with 35S:ERF6 or/and 35S:ERF11 (in p2GW7) and pERF6:fLUC, pERF11:fLUC, pSTZ:fLUC, pMYB51:fLUC, or pWRKY33:fLUC (in pM42GW7). Promoters were defined as the 2,000 bp upstream of the start codon. fLUC encodes the firefly luciferase enzyme. Every protoplast sample was transfected with 2 µg per construct as well as with 2 µg of normalization construct expressing the Renilla luciferase (rLUC) enzyme. For the experiments depicted in Figure 5C, the used amounts of 35S:ERF6 and 35S:ERF11 constructs were, respectively, 4 and 0 µg (1:0 ratio), 3 and 1 µg (3:1), 2.68 and 1.32 µg (2:1), 2 and 2 µg (1:1), 1.32 and 2.68 µg (1:2), 1 and 3 µg (1:3), and 0 and 4 µg (0:1). Transformed protoplasts were further grown by gentle shaking overnight in the dark in BY-2 medium to allow expression of the constructs. The next day, the BY-2 medium was removed and protoplasts were lysed in Cell Culture Lysis Reagent (Promega). Protoplast content was transferred to Nunc plates (Thermo Scientific), and fLUC and rLUC activities were measured using the Dual Luciferase Assay (Promega) and the LumiStar Galaxy (De Sutter et al., 2005). Measured fLUC activities were then normalized to rLUC activities.
Quantitative Y2H
For the protein-protein interaction analysis with quantitative Y2H, the ERF6 or ERF11 coding sequence was fused N-terminally to either the GAL4BD (DNA-binding domain of GAL4, bait) or the GAL4AD (activation domain, prey). Each transcription factor was used both as a bait and as a prey, generating four possible construct combinations. For each combination, the two constructs were cotransformed in competent yeast (Saccharomyces cerevisiae) cells of the PJ69-4A strain using polyethylene glycol lithium acetate-mediated transformation. As a control, each construct was also cotransformed with an empty vector to detect autoactivation. For the quantitative assay, yeast cultures were grown overnight in liquid synthetic defined (Clontech) minimal medium supplemented with an amino acid mix without Leu and Trp (-L-W dropout supplement, Clontech). The next day, the optical density at 600 nm was determined and used to start new cultures with equal amounts of yeast cells, either in nonselective medium (synthetic defined -L-W) or in selective medium (without Leu, Trp, and His, Clontech). The selective cultures were supplemented with 5, 25, or 50 mm 3-amino-1,2,4-triazole (Sigma) to distinguish autoactivation from protein-protein interaction. Upon 24 h of growth, the optical density at 600 nm was measured to quantify the growth, and the nonselective cultures were measured as a control.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Transcriptional induction of ERF6 target genes upon short-term exposure to mannitol.
Supplemental Figure S2. Expression level of ERF11 in the erf11 T-DNA line.
Supplemental Figure S3. Rosette phenotype of ERF11-overexpressing plants grown in soil.
Supplemental Figure S4. Effect of ERF11 on ERF6 expression.
Supplemental Figure S5. Quantitative Y2H assay with ERF6 and ERF11.
Supplemental Figure S6. Growth measurements of egm1, egm2, and erf5erf6 on mild and severe mannitol stress.
Supplemental Figure S7. Expression analysis of EGM in young Arabidopsis leaves upon short-term mannitol treatment.
Supplemental Figure S8. Expression levels of ERF5, ERF6, and ERF11 in the egm1 and egm2 mutants.
Supplemental Figure S9. Coexpression analysis of ERF11 and its putative regulators under multiple stress-related conditions.
Supplementary Material
Acknowledgments
We thank the whole Systems Biology of Yield group for the stimulating environment and constructive scientific discussions, Katrien Maleux for the practical help, and Dr. Annick Bleys for help with improving the article.
Glossary
- ACC
1-aminocyclopropane-1-carboxylic acid
- ABA
abscisic acid
- DAS
days after stratification
- qRT
quantitative reverse transcription
- MS
Murashige and Skoog
- Y2H
yeast two-hybrid
- BY-2
Bright Yellow-2
Footnotes
This work was supported by the Interuniversity Attraction Poles Program (IUAP P7/29 MARS) initiated by the Belgian Science Policy Office, Ghent University (Bijzonder Onderzoeksfonds Methusalem project no. BOF08/01M00408 and Multidisciplinary Research Partnership Biotechnology for a Sustainable Economy project no. 01MRB510W), the Agency for Innovation by Science and Technology (predoctoral fellowship to L.V.d.B.), and the Research Foundation Flanders (predoctoral fellowship to H.C.).
Articles can be viewed without a subscription.
References
- AbuQamar S, Chen X, Dhawan R, Bluhm B, Salmeron J, Lam S, Dietrich RA, Mengiste T (2006) Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J 48: 28–44 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GTS, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22: 64–78 [DOI] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B (2008) QuantPrime: a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boron AK, Vissenberg K (2014) The Arabidopsis thaliana hypocotyl, a model to identify and study control mechanisms of cellular expansion. Plant Cell Rep 33: 697–706 [DOI] [PubMed] [Google Scholar]
- Claeys H, Inzé D (2013) The agony of choice: how plants balance growth and survival under water-limiting conditions. Plant Physiol 162: 1768–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H, Skirycz A, Maleux K, Inzé D (2012) DELLA signaling mediates stress-induced cell differentiation in Arabidopsis leaves through modulation of anaphase-promoting complex/cyclosome activity. Plant Physiol 159: 739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H, Van Landeghem S, Dubois M, Maleux K, Inzé D (2014) What is stress? Dose-response effects in commonly used in vitro stress assays. Plant Physiol 165: 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sutter V, Vanderhaeghen R, Tilleman S, Lammertyn F, Vanhoutte I, Karimi M, Inzé D, Goossens A, Hilson P (2005) Exploration of jasmonate signalling via automated and standardized transient expression assays in tobacco cells. Plant J 44: 1065–1076 [DOI] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Skirycz A, Claeys H, Maleux K, Dhondt S, De Bodt S, Vanden Bossche R, De Milde L, Yoshizumi T, Matsui M, et al. (2013) ETHYLENE RESPONSE FACTOR6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol 162: 319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber H, Heijde M, Heller W, Albert A, Seidlitz HK, Ulm R (2010) Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci USA 107: 20132–20137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton CR, Bowen HC, Broadley MR, Hammond JP, Mead A, Payne KA, Pritchard J, White PJ (2004) Cesium toxicity in Arabidopsis. Plant Physiol 136: 3824–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hectors K, Prinsen E, De Coen W, Jansen MAK, Guisez Y (2007) Arabidopsis thaliana plants acclimated to low dose rates of ultraviolet B radiation show specific changes in morphology and gene expression in the absence of stress symptoms. New Phytol 175: 255–270 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Barlet X, Deslandes L, Hirsch J, Feng DX, Somssich I, Marco Y (2008) Transcriptional responses of Arabidopsis thaliana during wilt disease caused by the soil-borne phytopathogenic bacterium, Ralstonia solanacearum. PLoS ONE 3: e2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Son GH, Bhattacharjee S, Kim HJ, Nam JC, Nguyen PDT, Hong JC, Gassmann W (2014) The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J 78: 978–989 [DOI] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor DW. (2013) Genetic engineering to improve plant performance under drought: physiological evaluation of achievements, limitations, and possibilities. J Exp Bot 64: 83–108 [DOI] [PubMed] [Google Scholar]
- Lee BH, Henderson DA, Zhu JK (2005) The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17: 3155–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang L, Yu Y, Quan R, Zhang Z, Zhang H, Huang R (2011) The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J 68: 88–99 [DOI] [PubMed] [Google Scholar]
- Libault M, Wan J, Czechowski T, Udvardi M, Stacey G (2007) Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol Plant Microbe Interact 20: 900–911 [DOI] [PubMed] [Google Scholar]
- Ma S, Bohnert HJ (2007) Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol 8: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Xu J, He Y, Yang KY, Mordorski B, Liu Y, Zhang S (2013) Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25: 1126–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat CS, Ingle RA, Wathugala DL, Saunders NJ, Knight H, Knight MR (2012) ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS ONE 7: e35995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, Chico JM, Bossche RV, Sewell J, Gil E, et al. (2010) NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Gerstein M, Snyder M, Dinesh-Kumar SP (2009) MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev 23: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni L, Hardtke CS (2014) Small but thick enough: the Arabidopsis hypocotyl as a model to study secondary growth. Physiol Plant 151: 164–171 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM (1998) The AP2/EREBP family of plant transcription factors. Biol Chem 379: 633–646 [DOI] [PubMed] [Google Scholar]
- Rieu I, Eriksson S, Powers SJ, Gong F, Griffiths J, Woolley L, Benlloch R, Nilsson O, Thomas SG, Hedden P, et al. (2008) Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell 20: 2420–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Sewelam N, Kazan K, Thomas-Hall SR, Kidd BN, Manners JM, Schenk PM (2013) Ethylene response factor 6 is a regulator of reactive oxygen species signaling in Arabidopsis. PLoS ONE 8: e70289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Claeys H, De Bodt S, Oikawa A, Shinoda S, Andriankaja M, Maleux K, Eloy NB, Coppens F, Yoo SD, et al. (2011) Pause-and-stop: the effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell 23: 1876–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, De Bodt S, Obata T, De Clercq I, Claeys H, De Rycke R, Andriankaja M, Van Aken O, Van Breusegem F, Fernie AR, et al. (2010) Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol 152: 226–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son GH, Wan J, Kim HJ, Nguyen XC, Chung WS, Hong JC, Stacey G (2012) Ethylene-responsive element-binding factor 5, ERF5, is involved in chitin-induced innate immunity response. Mol Plant Microbe Interact 25: 48–60 [DOI] [PubMed] [Google Scholar]
- Song YH, Yoo CM, Hong AP, Kim SH, Jeong HJ, Shin SY, Kim HJ, Yun DJ, Lim CO, Bahk JD, et al. (2008) DNA-binding study identifies C-box and hybrid C/G-box or C/A-box motifs as high-affinity binding sites for STF1 and LONG HYPOCOTYL5 proteins. Plant Physiol 146: 1862–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Rizhsky L, Liang H, Shuman J, Shulaev V, Mittler R (2005) Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c. Plant Physiol 139: 1313–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trontin C, Kiani S, Corwin JA, Hématy K, Yansouni J, Kliebenstein DJ, Loudet O (2014) A pair of receptor-like kinases is responsible for natural variation in shoot growth response to mannitol treatment in Arabidopsis thaliana. Plant J 78: 121–133 [DOI] [PubMed] [Google Scholar]
- Vergnolle C, Vaultier MN, Taconnat L, Renou JP, Kader JC, Zachowski A, Ruelland E (2005) The cold-induced early activation of phospholipase C and D pathways determines the response of two distinct clusters of genes in Arabidopsis cell suspensions. Plant Physiol 139: 1217–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeirssen V, De Clercq I, Van Parys T, Van Breusegem F, Van de Peer Y (2014) Arabidopsis ensemble reverse-engineered gene regulatory network discloses interconnected transcription factors in oxidative stress. Plant Cell 26: 4656–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45: 523–539 [DOI] [PubMed] [Google Scholar]
- Vogel MO, Gomez-Perez D, Probst N, Dietz KJ (2012) Combinatorial signal integration by APETALA2/Ethylene Response Factor (ERF)-transcription factors and the involvement of AP2-2 in starvation response. Int J Mol Sci 13: 5933–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel MO, Moore M, König K, Pecher P, Alsharafa K, Lee J, Dietz KJ (2014) Fast retrograde signaling in response to high light involves metabolite export, MITOGEN-ACTIVATED PROTEIN KINASE6, and AP2/ERF transcription factors in Arabidopsis. Plant Cell 26: 1151–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Du Y, Zhao X, Miao Y, Song CP (2013) The MPK6-ERF6-ROS-responsive cis-acting Element7/GCC box complex modulates oxidative gene transcription and the oxidative response in Arabidopsis. Plant Physiol 161: 1392–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PC, Du YY, An GY, Zhou Y, Miao C, Song CP (2006) Analysis of global expression profiles of Arabidopsis genes under abscisic acid and H2O2 applications. J Integr Plant Biol 48: 62–74 [Google Scholar]
- Xin Z, Mandaokar A, Chen J, Last RL, Browse J (2007) Arabidopsis ESK1 encodes a novel regulator of freezing tolerance. Plant J 49: 786–799 [DOI] [PubMed] [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K (2005) Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol 58: 585–596 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.