An evolutionarily conserved gene regulates resistance to a devastating necrotrophic fungal crop plant pathogen by controlling major antifungal defense pathways.
Abstract
Although Sclerotinia sclerotiorum is a devastating necrotrophic fungal plant pathogen in agriculture, the virulence mechanisms utilized by S. sclerotiorum and the host defense mechanisms against this pathogen have not been fully understood. Here, we report that the Arabidopsis (Arabidopsis thaliana) Mediator complex subunit MED16 is a key component of basal resistance against S. sclerotiorum. Mutants of MED16 are markedly more susceptible to S. sclerotiorum than mutants of 13 other Mediator subunits, and med16 has a much stronger effect on S. sclerotiorum-induced transcriptome changes compared with med8, a mutation not altering susceptibility to S. sclerotiorum. Interestingly, med16 is also more susceptible to S. sclerotiorum than coronatine-insensitive1-1 (coi1-1), which is the most susceptible mutant reported so far. Although the jasmonic acid (JA)/ethylene (ET) defense pathway marker gene PLANT DEFENSIN1.2 (PDF1.2) cannot be induced in either med16 or coi1-1, basal transcript levels of PDF1.2 in med16 are significantly lower than in coi1-1. Furthermore, ET-induced suppression of JA-activated wound responses is compromised in med16, suggesting a role for MED16 in JA-ET cross talk. Additionally, MED16 is required for the recruitment of RNA polymerase II to PDF1.2 and OCTADECANOID-RESPONSIVE ARABIDOPSIS ETHYLENE/ETHYLENE-RESPONSIVE FACTOR59 (ORA59), two target genes of both JA/ET-mediated and the transcription factor WRKY33-activated defense pathways. Finally, MED16 is physically associated with WRKY33 in yeast and in planta, and WRKY33-activated transcription of PDF1.2 and ORA59 as well as resistance to S. sclerotiorum depends on MED16. Taken together, these results indicate that MED16 regulates resistance to S. sclerotiorum by governing both JA/ET-mediated and WRKY33-activated defense signaling in Arabidopsis.
Sclerotinia sclerotiorum is one of the most devastating necrotrophic fungal plant pathogens in agriculture. It infects over 400 plant species worldwide and causes annual losses of more than $200 million in the United States (Boland and Hall, 1994; Bolton et al., 2006). S. sclerotiorum depends on several virulence mechanisms to successfully attack the broad range of host plants. One mechanism is to produce the non-host-selective toxin oxalic acid, which inhibits plant defense responses, modulates host redox environment, suppresses autophagy, and activates cell wall-degrading enzymes (Marciano et al., 1983; Godoy et al., 1990; Cessna et al., 2000; Rollins and Dickman, 2001; Kim et al., 2008; Williams et al., 2011; Kabbage et al., 2013). Secretion of cell wall-degrading enzymes is another virulence mechanism of S. sclerotiorum, which facilitates penetration, tissue maceration, and plant cell wall depolymerization (Lumsden, 1979; Riou et al., 1991, 1992). S. sclerotiorum may also secrete effector proteins, such as S. sclerotiorum INTEGRIN-LIKE and S. sclerotiorum CHORISMATE MUTASE1, to diminish plant defense responses (Kabbage et al., 2013; Zhu et al., 2013). A recent bioinformatic study revealed that the S. sclerotiorum genome encodes a large set of candidate effector proteins (Guyon et al., 2014), which may have functions in Sclerotinia spp. pathogenesis.
Compared with the virulence mechanisms, resistance in host plants against S. sclerotiorum is less well understood. Microarray results indicate that S. sclerotiorum infection induces the expression of genes encoding components of diverse biological processes, including the jasmonic acid (JA) and ethylene (ET) signaling pathways (Zhao et al., 2007, 2009). Recent studies of Arabidopsis (Arabidopsis thaliana) mutants revealed that JA, ET, auxin, abscisic acid, nitric oxide, and reactive oxygen species all contribute to basal resistance against S. sclerotiorum (Guo and Stotz, 2007; Perchepied et al., 2010; Stotz et al., 2011). Conclusions in these studies about the role of the salicylic acid (SA) signaling pathway in resistance to S. sclerotiorum are contradictory (Guo and Stotz, 2007; Perchepied et al., 2010; Wang et al., 2012; Nováková et al., 2014). Nevertheless, these results suggest that the Arabidopsis basal resistance against S. sclerotiorum is complex and involves multiple signaling pathways.
JA and ET are well known to cooperate in resistance against necrotrophic fungal pathogens (Thomma et al., 2001; Kunkel and Brooks, 2002; Glazebrook, 2005). They synergistically induce the pathogen-defense gene PLANT DEFENSIN1.2 (PDF1.2) and antagonize each other’s specific responses (Penninckx et al., 1998; Schenk et al., 2000). For instance, JA reduces ET-induced expression of the apical hook-regulating gene HOOKLESS1 (HLS1; Turner et al., 2002), whereas ET suppresses JA-mediated activation of wound-responsive genes, including VEGATATIVE STORAGE PROTEIN1 (VSP1), VSP2, and JASMONATE RESPONSIVE1 (JR1; Rojo et al., 1999; Lorenzo et al., 2004). Two transcription factors, ETHYLENE INSENSITIVE3 (EIN3) and ETHYLENE INSENSITIVE3-LIKE1 (EIL1), which regulate most, if not all, of the ET responsiveness, are signaling hubs of JA/ET cooperation (Chao et al., 1997; Solano et al., 1998; Alonso et al., 2003b; An et al., 2010). JA and ET signaling converge at EIN3/EIL1, inducing genes encoding APETALA2/ETHYLENE-RESPONSIVE FACTORs (AP2/ERFs), such as OCTADECANOID-RESPONSIVE ARABIDOPSIS APETALA2/ETHYLENE-RESPONSIVE FACTOR59 (ORA59) and ERF1, which in turn activate PDF1.2 expression (Solano et al., 1998; Lorenzo et al., 2003; Pré et al., 2008; Zarei et al., 2011; Zhu et al., 2011). On the other hand, EIN3 and EIL1 interact with and repress the activity of MYC2, a transcription factor responsible for the activation of JA-mediated wound responses. Conversely, MYC2 attenuates HLS1 expression by promoting EIN3/EIL1 proteolysis and repressing their activity (Song et al., 2014; Zhang et al., 2014a). Such cooperation between JA and ET allows plants to prioritize defense against invading necrotrophic pathogens over development and other responses.
The transcription factor WRKY33 is an important regulator of resistance to necrotrophic fungal pathogens. Expression of the WRKY33 gene is highly inducible by Botrytis cinerea infection (AbuQamar et al., 2006). Mutations in WRKY33 cause enhanced susceptibility to B. cinerea and Alternaria brassicicola, whereas overexpression of WKRY33 leads to increased resistance to these pathogens and elevated basal expression of PDF1.2 (Zheng et al., 2006). Moreover, expression of the Brassica napus WRKY33 (BnWRKY33) gene is highly inducible by S. sclerotiorum infection, and overexpression of BnWRKY33 in B. napus results in constitutive expression of BnPDF1.2 and markedly enhanced resistance to S. sclerotiorum (Wang et al., 2014), suggesting that WRKY33 may be a positive regulator of resistance against S. sclerotiorum. WRKY33 has been shown to directly control the expression of ORA59 during the later stages of pathogen infection (Birkenbihl et al., 2012), but how WRKY33 activates ORA59 transcription is unclear.
In eukaryotes, RNA POLYMERASE II (RNAPII) catalyzes the transcription of protein-encoding genes (Woychik and Hampsey, 2002). A highly conserved multiprotein complex named Mediator plays an essential role in RNAPII-mediated transcription (Kim et al., 1994; Kornberg, 2005; Takagi and Kornberg, 2006; Conaway and Conaway, 2011a). Mediator exists in multiple functionally distinct forms and serves as either a transcriptional activator or a repressor, depending on its associated protein partners (Conaway and Conaway, 2011b). The Mediator core contains more than 20 subunits organized into head, middle, and tail modules (Guglielmi et al., 2004; Chadick and Asturias, 2005). This core associates with the RNAPII complex to form the holoenzyme, stimulating basal transcription and supporting the activation of transcription by specific transcription activators (Mittler et al., 2001; Baek et al., 2002; Zhu et al., 2006; Ansari et al., 2009). Individual Mediator subunits converge diverse signals to the RNAPII transcription complex via interaction with a particular or a class of transcription activators, leading to pathway-specific gene transcription (Balamotis et al., 2009; Kagey et al., 2010; Takahashi et al., 2011). The Mediator core also interacts with a kinase module, which prevents its binding to the RNAPII complex, resulting in transcriptional repression (Holstege et al., 1998; Akoulitchev et al., 2000; Knuesel et al., 2009). The distinct forms of Mediator thus underlie various pathway-specific transcription activation or suppression (Balamotis et al., 2009).
The Arabidopsis Mediator complex contains 27 conserved subunits and six additional subunits whose positions in the complex are unassigned (Bäckström et al., 2007; Mathur et al., 2011). A number of Arabidopsis Mediator subunits have been implicated in signaling pathways related to plant development and abiotic responses. For instance, the Arabidopsis MEDIATOR SUBUNIT14 (MED14)/STRUWWELPETER is a key regulator of cell proliferation (Autran et al., 2002). MED25/PHYTOCHROME AND FLOWERING TIME1 was first identified as a key regulator of flowering (Cerdán and Chory, 2003) and later found to regulate final organ size and light signaling (Xu and Li, 2011; Klose et al., 2012). MED18 was found to control flowering time and floral organ identity (Zheng et al., 2013). MED5a/MED33a/REDUCED EPIDERMAL FLUORESCENCE4 (REF4)-RELATED1 and MED5b/MED33b/REF4 are required for phenylpropanoid homeostasis (Bonawitz et al., 2012). The Mediator kinase module subunits CYCLIN-DEPENDENT KINASE8 (CDK8)/HUA ENHANCER3, MED12/CRYPTIC PRECOCIOUS, and MED13/MACCHI-BOU2 regulate the specification of floral organ identity, early embryo patterning/flowering, and embryo patterning/cotyledon organogenesis, respectively (Wang and Chen, 2004; Gillmor et al., 2010; Ito et al., 2011; Imura et al., 2012). MED17, MED18, and MED20a play roles in small and long noncoding RNA production (Kim et al., 2011). Moreover, MED2/MED32, MED14, and MED16/SENSITIVE TO FREEZING6 regulate cold-responsive genes (Knight et al., 1999, 2008, 2009; Hemsley et al., 2014). MED16 also modulates iron uptake and homeostasis (Yang et al., 2014; Zhang et al., 2014b).
Mediator is emerging as a master regulator of plant immune responses. MED14, MED15, MED16, and MED19a have been shown to regulate the SA-triggered immunity against biotrophic and hemibiotrophic pathogens (Canet et al., 2012; Zhang et al., 2012, 2013; Caillaud et al., 2013), whereas MED8, MED12, MED13, MED16, MED21, MED25, and CDK8 have been found to function in JA/ET-mediated immunity against necrotrophic pathogens (Dhawan et al., 2009; Kidd et al., 2009; Zhang et al., 2012; Zhu et al., 2014). MED18 also functions in resistance to necrotrophic pathogens, but the resistance is independent of the JA/ET signaling (Lai et al., 2014). Interestingly, the Arabidopsis oomycete pathogen Hyaloperonospora arabidopsidis RXLR effector44 (HaRxL44) interacts with MED19a and promotes its degradation, which shifts the balance of defense transcription from SA to JA/ET signaling (Caillaud et al., 2013). This result suggests that, while Mediator positively regulates plant immunity, pathogens may have evolved virulence mechanisms to interfere with Mediator function.
In this study, we identified MED16 as a central regulator of basal resistance against S. sclerotiorum. We show that MED16 is required not only for JA/ET-mediated defense responses but also for ET-induced suppression of JA-mediated wound responses. We demonstrate that MED16 associates with WRKY33 and mediates WRKY33-activated defense gene expression and resistance to S. sclerotiorum. Our results indicate that MED16 is a key regulator of both JA/ET-mediated and the transcription factor WRKY33-activated defense signaling in Arabidopsis.
RESULTS
MED16 Is a Critical Mediator Subunit for Basal Resistance against S. sclerotiorum
Since mutations in the Arabidopsis Mediator subunits MED8, MED16, and MED25 compromise basal resistance against the necrotrophic fungal pathogens B. cinerea and A. brassicicola (Kidd et al., 2009; Zhang et al., 2012), they may also affect resistance to S. sclerotiorum. To test this hypothesis, we inoculated med8, med16-1, sensitive to freezing6-2 (sfr6-2), med25-1, med25-2, and wild-type plants with S. sclerotiorum. Interestingly, while med16/sfr6 and med25 mutants exhibited enhanced susceptibility to S. sclerotiorum, med8 did not show altered susceptibility compared with the wild type (Fig. 1, A–D). At 48 h post inoculation (hpi), the average lesion sizes on med8 and wild-type plants were approximately 7.8 mm, and those on med16/sfr6 and med25 were approximately 16.3 mm and approximately 11.9 mm, respectively (Fig. 1A). At 5 d post inoculation (dpi), approximately 27.7% of the inoculated med8 and wild-type plants were decayed, whereas 100% and approximately 62.5% of the inoculated med16/sfr6 and med25 plants, respectively, were decayed (Fig. 1C). These results indicate that both MED16 and MED25, but not MED8, contribute to basal resistance against S. sclerotiorum and that MED16 plays a more important role than MED25 in this resistance.
Figure 1.
Susceptibility of 14 Mediator subunit mutants to S. sclerotiorum. A, Size of the necrotic lesions formed on the S. sclerotiorum-infected wild-type (WT), med8, med16-1, sfr6-2, med25-1, and med25-2 plants at 24 and 48 hpi. Data represent means of lesion sizes on 24 leaves (on 24 plants) with sd. Different letters above the bars indicate significant differences (P < 0.05, one-way ANOVA). B, Disease symptoms on the S. sclerotiorum-infected wild-type, med8, med16-1, sfr6-2, med25-1, and med25-2 leaves. Photographs were taken at 24 and 48 hpi. C, Decay percentages of S. sclerotiorum-infected wild-type, med8, med16-1, sfr6-2, med25-1, and med25-2 plants at 3, 4, and 5 dpi. Data represent means of three groups (12 plants per group) of S. sclerotiorum-infected plants with sd. Different letters above the bars indicate significant differences (P < 0.05, one-way ANOVA). D, Disease symptoms on the S. sclerotiorum-infected wild-type, med8, med16-1, sfr6-2, med25-1, and med25-2 plants. Photographs were taken at 5 dpi. E, Size of the necrotic lesions formed on the S. sclerotiorum-infected wild-type, med8, med13, med14, med16, med17, med18, med20a, med23, med25, med31, med32, med33b, med34, and med36 plants at 48 hpi. Data represent means of lesion sizes on 24 leaves (on 24 plants) with sd. Different letters above the bars indicate significant differences (P < 0.05, one-way ANOVA). F, Disease symptoms on the S. sclerotiorum-infected wild-type, med8, med13, med14, med16, med17, med18, med20a, med23, med25, med31, med32, med33b, med34, and med36 leaves. Photographs were taken at 48 hpi. The statistical comparisons in A and C were performed among the wild type, med8, med16-1, sfr6-2, med25-1, and med25-2 for each time point. Experiments in A, C, and E were repeated three independent times with similar trends. Results from a representative experiment are presented. Photographs in B, D, and F represent typical disease symptoms on S. sclerotiorum-infected leaves or plants.
As Mediator is a multisubunit complex, other Mediator subunits may also function in basal resistance against S. sclerotiorum. To test this, we identified transfer DNA (T-DNA) insertion homozygous lines for the Mediator subunits MED13, MED17, MED18, MED20a, MED23, MED31, MED32, MED33b, MED34, and MED36 and tested their susceptibility to S. sclerotiorum. The T-DNA lines for MED13, MED18, MED32, and MED33b have been characterized previously (Supplemental Table S1), and those for MED17, MED20a, MED23, MED31, MED34, and MED36 are either knockout or knockdown mutants (Supplemental Fig. S1). The wild type, med8, med16-1, med25-1, and the previously characterized med14-1 T-DNA insertion mutant were also included in the experiment (Supplemental Table S1). At 48 hpi, the average lesion sizes on med8, med13, med17, med18, med20a, med23, med31, med32, med33b, med34, and med36 were not significantly different from that on the wild type, whereas the average lesion sizes on med14, med16, and med25 were significantly larger than that on the wild type (Fig. 1, E and F). Among all the tested Mediator subunit mutants, med16 exhibited the highest susceptibility to S. sclerotiorum, indicating that MED16 is a critical Mediator subunit regulating basal resistance against S. sclerotiorum.
MED16 and MED8 Differentially Regulate S. sclerotiorum-Induced Transcriptional Reprogramming
Although Mediator is a multisubunit complex, not all subunits are required for a specific biological process (Balamotis et al., 2009; Hemsley et al., 2014). For basal resistance against S. sclerotiorum, MED16 appears to play a more important role than other Mediator subunits (Fig. 1, E and F). To uncover the molecular mechanisms underlying this unique requirement of MED16 for basal resistance against S. sclerotiorum, we performed a microarray experiment to monitor S. sclerotiorum-induced transcriptome changes in med16-1, med8, and the wild type (National Center for Biotechnology Information Gene Expression Omnibus series no. GSE65165). We chose med8 over other Mediator subunit mutants because med8 is susceptible to B. cinerea and A. brassicicola but not to S. sclerotiorum (Kidd et al., 2009; Fig. 1). We inoculated med16-1, med8, and wild-type plants with S. sclerotiorum. Leaf tissues were collected as controls at 0 dpi, the inoculated leaves were collected as local tissues at 1 dpi, and the upper uninoculated leaves were collected as systemic tissues at 2 and 4 dpi. Triplicate experiments were performed independently, and the data were analyzed to identify genes that were induced or suppressed in med16-1, med8, and the wild type. We used q values to identify induced or suppressed genes. Genes that showed a 2-fold or higher induction or suppression with a low q value (q ≤ 0.05) in med16-1, med8, and the wild type were chosen for further analysis. We found that all three genotypes exhibited dramatic transcriptional reprogramming upon S. sclerotiorum inoculation and that both med16-1 and med8 significantly shifted their transcriptome profiles (Fig. 2, A and B). The numbers of genes up- or down-regulated in med16-1 were larger than those in the wild type in local tissues at 1 dpi and systemic tissues at 4 dpi and smaller in systemic tissues at 2 dpi (Fig. 2A). In med8, except for the number of genes up-regulated in local tissues at 1 dpi, all others were smaller than those in the wild type (Fig. 2A).
Figure 2.
S. sclerotiorum-induced transcriptome changes in med16 and med8. Three independent RNA samples per genotype at each time point were used for the microarray experiment, and data were analyzed to identify genes that showed a 2-fold or higher induction or suppression with a low q value (q ≤ 0.05) in med16-1, med8, and the wild type or genes that showed a 2-fold or larger difference in their expression levels with a low q value (q ≤ 0.05) between med16-1 or med8 and the wild type. A, Dynamic changes in the number of genes that are up- or down-regulated in the wild type (WT), med16, and med8 after S. sclerotiorum infection. B, Overlapping circles indicating the number of genes that are commonly, partially commonly, or uniquely up- or down-regulated at 1 and 4 dpi in the wild type, med16, and med8. C, Dynamic changes in the number of genes that are differentially expressed between med16 and the wild type and between med8 and the wild type after S. sclerotiorum infection. D, Overlapping circles indicating the number of genes that are commonly or uniquely differentially expressed at 1 and 4 dpi between med16 or med8 and the wild type.
We then queried the microarray data and identified genes that showed a 2-fold or larger difference in their expression levels with a low q value (q ≤ 0.05) between med16-1 or med8 and the wild type. As shown in Figure 2C, considerably more genes were differentially expressed between med16-1 and the wild type than between med8 and the wild type. A total of 493, 2,101, 921, and 2,561 genes were differentially expressed between med16-1 and the wild type at 0, 1, 2, and 4 dpi, respectively, whereas only 236, 97, 259, and 453 genes were differentially expressed between med8 and the wild type (Fig. 2C; Supplemental Data Set S1). We compared the differentially expressed genes in med16-1 and med8 and found that approximately 43.6% to 61.9% of the genes that were differentially expressed in med8 were also differentially expressed in the med16-1 mutant (Fig. 2D), suggesting that MED16 and MED8 regulate some common genes during S. sclerotiorum infection, although these genes may not contribute to basal resistance against S. sclerotiorum. Taken together, these results indicate that the med16-1 mutation has a much broader impact than med8 on S. sclerotiorum-induced transcriptome changes.
Since MED16 positively regulates both SA and JA/ET signaling (Wathugala et al., 2012; Zhang et al., 2012), we wondered how SA and JA/ET pathway genes were influenced by the med16-1 mutation during S. sclerotiorum infection. To this end, we further explored the microarray data and identified SA and JA/ET pathway genes that showed a 2-fold or larger difference in their expression levels with a low q value (q ≤ 0.05) between med16-1 and the wild type (Table I). Interestingly, the JA biosynthesis genes LIPOXYGENASE3 (LOX3), ALLENE OXIDE CYCLASE3 (AOC3), and OPR3, the ET biosynthesis genes 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE2 (ACS2) and ACS8, as well as the SA biosynthesis genes ISOCHORISMATE SYNTHASE1 (ICS1) and ENHANCED DISEASE SUSCEPTIBILITY5 (EDS5) were up-regulated in med16-1 in local tissues at 1 dpi and/or systemic tissues at 4 dpi. Surprisingly, however, while a number of SA pathway genes (EDS1, PHYTOALEXIN DEFICIENT4 [PAD4], NONEXPRESSER OF PATHOGENESIS-RELATED GENES1 [NPR1], WRKY18, WRKY38, WRKY53, PATHOGENESIS-RELATED GENE1 [PR1], and PR2) and several JA-regulated wound-responsive genes (MYC2, JAR1, VSP1, and VSP2) were up-regulated in med16-1, a group of JA/ET-regulated defense genes (ORA59, ERF1, ERF14, PDF1.2, PDF1.2b, PDF1.2c, PDF1.3, PR4, and Chitinase B [ChiB]) and PR5 were down-regulated. On the other hand, only a few of these genes were differentially expressed between med8 and the wild type (Table I). These results indicate that, during S. sclerotiorum infection, SA signaling and JA-mediated wound signaling were enhanced, whereas JA/ET-controlled defense signaling was inhibited in the med16-1 mutant.
Table I. Defense genes that are differentially expressed between med16-1 or med8 and the wild type during S. sclerotiorum infection.
AGI, Arabidopsis Genome Initiative; FC, fold change.
| AGI Locus | Gene Name |
med16-1/Wild Type |
med8/Wild Type |
AGI Description | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Local, 1 dpi |
Systemic, 4 dpi |
Local, 1 dpi |
Systemic, 4 dpi |
||||||||
| Log2(FC) | q Value | Log2(FC) | q Value | Log2(FC) | q Value | Log2(FC) | q Value | ||||
| JA/ET pathway genes | |||||||||||
| At1g17420 | LOX3 | 2.105 | 0.003 | 1.131 | 0.007 | LIPOXYGENASE3 | |||||
| At3g25780 | AOC3 | 1.846 | 0.007 | 2.49 | 0 | 2.06 | 0.003 | ALLENE OXIDE CYCLASE3 | |||
| At2g06050 | OPR3 | 1.037 | 0.033 | 12-OXOPHYTODIENOATE-REDUCTASE3 | |||||||
| At1g32640 | MYC2 | 1.293 | 0.005 | JASMONATE INSENSITIVE1 | |||||||
| At2g46370 | JAR1 | 1.069 | 0.008 | JASMONATE RESISTANT1 | |||||||
| At5g24780 | VSP1 | 3.062 | 0.002 | 2.224 | 0 | VEGETATIVE STORAGE PROTEIN1 | |||||
| At5g24770 | VSP2 | 2.642 | 0.002 | 2.684 | 0 | VEGETATIVE STORAGE PROTEIN2 | |||||
| At1g01480 | ACS2 | 1.628 | 0.004 | 2.583 | 0.001 | 1-AMINO-CYCLOPROPANE-1-CARBOXYLATE SYNTHASE2 | |||||
| At4g37770 | ACS8 | 1.779 | 0.002 | 1-AMINO-CYCLOPROPANE-1-CARBOXYLATE SYNTHASE8 | |||||||
| At1G06160 | ORA59 | −3.72 | 0.001 | −4.616 | 0 | Ethylene-responsive factor | |||||
| At3g23240 | ERF1 | −1.021 | 0.004 | ETHYLENE-RESPONSIVE FACTOR1 | |||||||
| At1G04370 | ERF14 | −1.823 | 0.004 | ETHYLENE-RESPONSIVE FACTOR14 | |||||||
| At5g44420 | PDF1.2 | −4.234 | 0.004 | −5.712 | 0 | PLANT DEFENSIN1.2 | |||||
| At2G26020 | PDF1.2b | −7.437 | 0.002 | −9.356 | 0 | PLANT DEFENSIN1.2b | |||||
| At5g44430 | PDF1.2c | −6.647 | 0.002 | −8.894 | 0 | PLANT DEFENSIN1.2c | |||||
| At2g26010 | PDF1.3 | −5.289 | 0.007 | −7.961 | 0.001 | PLANT DEFENSIN1.3 | |||||
| At3g04720 | PR4/HEL | −2.933 | 0.009 | −3.998 | 0.001 | PATHOGENESIS-RELATED4 | |||||
| At3G12500 | ChiB | −4.521 | 0.001 | −2.588 | 0 | −1.559 | 0.006 | BASIC CHITINASE | |||
| SA pathway genes | |||||||||||
| At3g48090 | EDS1 | 1.055 | 0.001 | ENHANCED DISEASE SUSCEPTIBILITY1 | |||||||
| At3g52430 | PAD4 | 2.989 | 0 | 2.011 | 0 | PHYTOALEXIN DEFICIENT4 | |||||
| At1g74710 | ICS1 | 1.912 | 0.001 | 2.551 | 0 | ISOCHORISMATE SYNTHASE1 | |||||
| At4g39030 | EDS5 | 2.019 | 0.003 | 2.513 | 0 | 1.171 | 0.006 | ENHANCED DISEASE SUSCEPTIBILITY5 | |||
| At1g64280 | NPR1 | 1.187 | 0.002 | NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 | |||||||
| At4g31800 | WRKY18 | 2.041 | 0.002 | WRKY DNA-BINDING PROTEIN18 | |||||||
| At5g22570 | WRKY38 | 3.528 | 0.002 | 3.613 | 0 | 1.695 | 0.007 | WRKY DNA-BINDING PROTEIN38 | |||
| At4g23810 | WRKY53 | 2.111 | 0.001 | 2.7 | 0 | 2.84 | 0.001 | WRKY DNA-BINDING PROTEIN53 | |||
| At2g14610 | PR1 | 1.348 | 0.006 | 1.743 | 0.017 | 1.115 | 0.018 | PATHOGENESIS-RELATED GENE1 | |||
| At3g57260 | PR2 | 2.166 | 0.004 | 3.841 | 0 | 1.099 | 0.036 | 1.499 | 0.005 | PATHOGENESIS-RELATED GENE2 | |
| At1g75040 | PR5 | −1.88 | 0.001 | −1.222 | 0.001 | −1.025 | 0.013 | PATHOGENESIS-RELATED GENE5 | |||
MED16 Is a Key Regulator of Basal Resistance against S. sclerotiorum
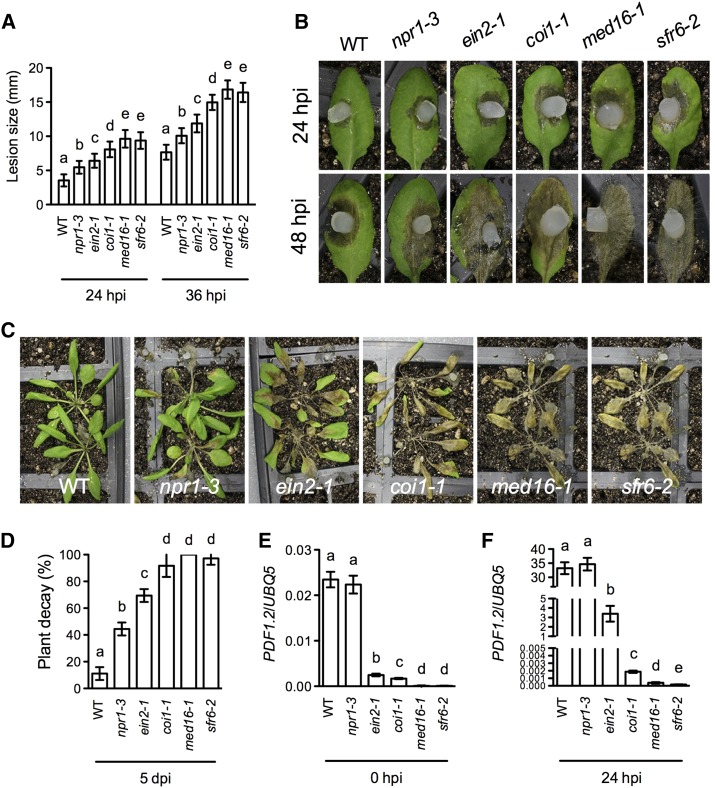
It has been shown that the SA, JA, and ET signaling mutants, npr1, coi1, and ein2, respectively, have enhanced susceptibility to S. sclerotiorum (Guo and Stotz, 2007). To compare the susceptibility between these mutants and med16, we inoculated npr1-3, ein2-1, coi1-1, med16-1, sfr6-2, and wild-type plants with S. sclerotiorum. We used the coi1-1 null allele because it is extremely susceptible to S. sclerotiorum compared with other mutants (Perchepied et al., 2010). As shown in Figure 3, A to D, npr1-3, ein2-1, and coi1-1 were all more susceptible to S. sclerotiorum than the wild type. We consistently observed enhanced susceptibility in the npr1-3 mutant, supporting the previous conclusion that NPR1-mediated SA signaling plays a role in resistance to S. sclerotiorum (Guo and Stotz, 2007). In our experiments, the coi1-1 mutant was more susceptible than ein2-1, which in turn was more susceptible than npr1-3. Interestingly, the med16/sfr6 mutants exhibited even higher susceptibility than coi1-1 to S. sclerotiorum. At 36 hpi, the average lesion sizes (approximately 16.6 mm) on med16/sfr6 plants were slightly but significantly larger than that (approximately 14.9 mm) on coi1-1 (Fig. 3A). At 5 dpi, approximately 91.6% and approximately 98.6% of inoculated coi1-1 and med16/sfr6 plants, respectively, were decayed, even though these percentages were not significantly different (Fig. 3D). These results together demonstrate that MED16 is a key regulator of basal resistance against S. sclerotiorum in Arabidopsis.
Figure 3.
S. sclerotiorum-induced defense responses in med16, npr1, ein2, and coi1. A, Size of the necrotic lesions formed on the S. sclerotiorum-infected wild type (WT), npr1-3, ein2-1, coi1-1, med16-1, and sfr6-2 at 24 and 36 hpi. Data represent means of lesion sizes on 24 leaves (on 24 plants) with sd. Different letters above the bars indicate significant differences (P < 0.05, one-way ANOVA). The statistical comparisons were performed among the wild type, npr1-3, ein2-1, coi1-1, med16-1, and sfr6-2 for each time point. B, Disease symptoms on the S. sclerotiorum-infected wild-type, npr1-3, ein2-1, coi1-1, med16-1, and sfr6-2 leaves. Photographs were taken at 24 and 48 hpi. C, Disease symptoms on the S. sclerotiorum-infected wild-type, npr1-3, ein2-1, coi1-1, med16-1, and sfr6-2 plants. Photographs were taken at 5 dpi. D, Decay percentages of S. sclerotiorum-infected wild-type, npr1-3, ein2-1, coi1-1, med16-1, and sfr6-2 plants at 5 dpi. Data represent means of three groups (12 plants per group) of S. sclerotiorum-infected plants with sd. Different letters above the bars indicate significant differences (P < 0.05, one-way ANOVA). E, PDF1.2 expression levels in the wild type, npr1-3, ein2-1, coi1-1, med16-1, and sfr6-2 at 0 hpi. Expression of PDF1.2 was normalized against the constitutively expressed UBIQUITIN5 (UBQ5). Data represent means of three biological replicates (samples taken from different plants during the same experiment) with sd. Different letters above the bars indicate significant differences (P < 0.05, one-way ANOVA). F, PDF1.2 expression levels in the wild type, npr1-3, ein2-1, coi1-1, med16-1, and sfr6-2 at 24 hpi. Expression of PDF1.2 was normalized against the constitutively expressed UBQ5. Data represent means of three biological replicates with sd. Different letters above the bars indicate significant differences (P < 0.05, one-way ANOVA). Experiments in A, D, E, and F were repeated three independent times with similar trends. Results from a representative experiment are presented. Photographs in B and C represent typical disease symptoms on S. sclerotiorum-infected leaves or plants.
We have shown previously that mutations in MED16 block B. cinerea-induced expression of PDF1.2, a marker gene of the JA/ET-mediated defense responses (Zhang et al., 2012). Since COI1 is known to be essential for PDF1.2 expression (Penninckx et al., 1996; Lorenzo et al., 2003; Pré et al., 2008), we compared S. sclerotiorum-induced PDF1.2 expression in the coi1-1 and med16/sfr6 mutants. The wild type, npr1-3, and ein2-1 were included in the experiment as controls. As shown in Figure 3, E and F, and Supplemental Figure S2, PDF1.2 was drastically induced in npr1-3 and the wild-type plants and also significantly induced in ein2-1, but it was not induced in the coi1-1 and med16/sfr6 plants. The expression levels of PDF1.2 in the med16/sfr6 plants were even lower than those in coi1-1. Therefore, like the COI1 protein, MED16 is essential for S. sclerotiorum-induced PDF1.2 expression.
MED16 Regulates ET-Activated Inhibition of JA-Mediated Wound Signaling
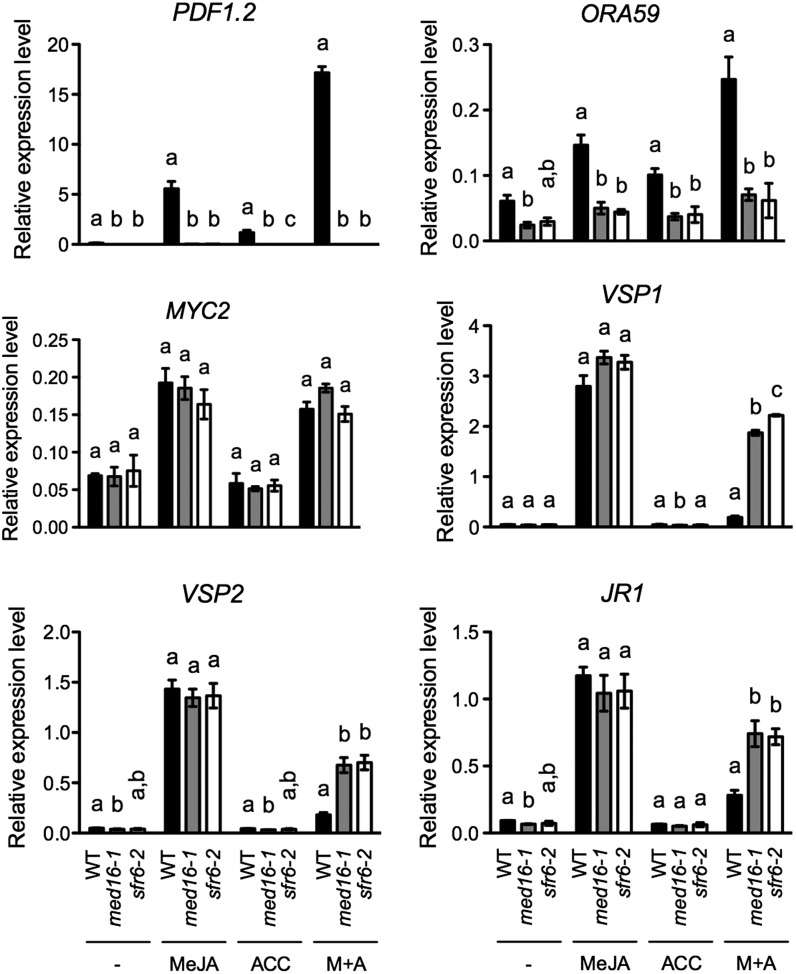
JA and ET signaling are well established to interact both synergistically and antagonistically (Glazebrook, 2005; Pieterse et al., 2009). JA and ET independently and synergistically induce pathogen-responsive genes, such as PDF1.2, to support resistance against necrotrophic pathogens, whereas ET inhibits JA-induced wound-responsive genes, such as VSP1, VSP2, and JR1. Since JA/ET-regulated pathogen-responsive genes and JA-mediated wound-responsive genes are differentially regulated in med16-1 during S. sclerotiorum infection (Table I), MED16 may modulate the cross talk between JA and ET signaling. To test this hypothesis, we treated med16-1, sfr6-2, and wild-type plants with methyl jasmonate (MeJA), 1-aminocyclopropane-1-carboxylic acid (ACC), or their combination and tested the induction of the pathogen-responsive genes PDF1.2 and ORA59 as well as the wound-responsive genes VSP1, VSP2, and JR1. As shown in Figure 4, JA, ET, and their combination induced PDF1.2 and ORA59 expression in the wild-type plants but not in med16-1 and sfr6-2, confirming the requirement of MED16 for induction of these two genes. On the other hand, JA-induced expression of VSP1, VSP2, and JR1 was not altered by the med16/sfr6 mutations, indicating that the JA-mediated wound signaling pathway is intact in these mutants. As expected, ET significantly inhibited JA-induced expression of VSP1, VSP2, and JR1 in the wild-type plants, but the inhibition was largely alleviated in the med16-1 and sfr6-2 mutants, indicating that MED16 is required for ET to maximally suppress JA-mediated wound responses.
Figure 4.
MeJA-, ACC-, and their combination-induced pathogen- and wound-responsive genes in med16. Ten-day-old wild-type (WT), med16-1, and sfr6-2 seedlings grown on one-half-strength Murashige and Skoog medium were treated with 0.1 mm MeJA, 0.1 mm ACC, or 0.1 mm MeJA plus 0.1 mm ACC (M+A). Seedlings for the negative control (−) were treated with water. Total RNA was extracted from plant tissues collected 24 h after the treatment and subjected to quantitative PCR (qPCR) analysis. Expression of ORA59, PDF1.2, MYC2, VSP1, VSP2, and JR1 was normalized against the constitutively expressed UBQ5. Data represent means of three biological replicates with sd. Different letters above the bars indicate significant differences (P < 0.05, one-way ANOVA). The statistical comparisons were performed among the wild type, med16-1, and sfr6-2 for each treatment. The experiment was repeated three independent times with similar trends. Results from a representative experiment are presented.
MED16 Mediates the Recruitment of RNAPII to the PDF1.2 and ORA59 Genes
Recruitment of RNAPII is a critical step for the transcription of protein-encoding genes in eukaryotic cells (Woychik and Hampsey, 2002). Since MED16 is required for JA/ET-induced transcription of PDF1.2 and ORA59 (Fig. 4), it may mediate the recruitment of RNAPII to these genes. To test this hypothesis, we performed chromatin immunoprecipitation with an RNAPII-specific antibody to analyze RNAPII occupancy on the coding regions of the PDF1.2 and ORA59 genes in med16-1 and wild-type plants treated with or without MeJA plus ACC. The CYP79B3 gene was included as a control, since MeJA plus ACC treatment does not drastically induce this gene (Mikkelsen et al., 2003). As shown in Figure 5, in the wild-type plants, MeJA plus ACC treatment dramatically increased RNAPII occupancy on three different sites in the coding regions of PDF1.2 and ORA59 but not on those of CYP79B3, indicating that the treatment induced gene-specific enrichment of RNAPII. In the med16-1 mutant, on the other hand, MeJA plus ACC treatment did not induce considerable increases in RNAPII occupancy on PDF1.2 and ORA59, indicating that MED16 is a key Mediator subunit for JA/ET-induced recruitment of RNAPII to PDF1.2 and ORA59.
Figure 5.
MeJA/ACC-induced recruitment of RNAPII to PDF1.2 and ORA59 in med16. Chromatin immunoprecipitation-qPCR results show that the med16-1 mutation prevented MeJA/ACC-induced recruitment of NRPB2 (DNA-directed RNA polymerase II subunit RPB2) to the coding regions of PDF1.2 and ORA59. The CYP79B3 gene was included as a control. Chromatin was extracted from wild-type (WT) and med16-1 seedlings treated with (+) or without (−) 0.1 mm MeJA plus 0.1 mm ACC (MeJA+ACC) for 24 h and then precipitated with anti-RPB2 antibody. The amount of precipitated DNA corresponding to a specific gene region was determined by qPCR and normalized to input DNA. Data represent means of three biological replicates with sd. Asterisks indicate significant differences between the wild type and med16 (P < 0.05, Student’s t test). The experiment was repeated three independent times with similar trends. Results from a representative experiment are presented.
MED16 Is Physically Associated with WRKY33
The transcription factor WRKY33 has been shown to bind to two distinct W box-containing regions in the ORA59 promoter and to regulate B. cinerea-induced prolonged expression of ORA59 (Zheng et al., 2006; Birkenbihl et al., 2012). Since MED16 is required for the recruitment of RNAPII to the ORA59 gene (Fig. 5), WRKY33 may activate ORA59 transcription by physically interacting with MED16. To test this hypothesis, we subcloned the coding regions of MED16 and WRKY33 into the yeast two-hybrid bait vector pGBKT7 and the prey vector pGADT7, respectively, and cotransformed the resulting plasmids pGBKT7-MED16 and pGADT7-WRKY33 into the yeast (Saccharomyces cerevisiae) strain AH109. The empty bait and prey vectors, pGBKT7 and pGADT7, were used as negative controls and cotransformed with pGADT7-WRKY33 and pGBKT7-MED16, respectively. As shown in Figure 6A, yeast cells harboring pGBKT7-MED16 and pGADT7-WRKY33 grew on quadruple synthetic dextrose (SD) dropout medium (Ade-His-Leu-Trp) supplemented with 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (X-α-Gal) and formed blue colonies, whereas those harboring either pGBKT7 and pGADT7-WRKY33 or pGBKT7-MED16 and pGADT7 did not grow and change color. These results indicate that MED16 and WRKY33 physically interact in yeast.
Figure 6.
Physical association between MED16 and WRKY33. A, Growth and color of yeast cells carrying pGBKT7/pGADT7-WRKY33, pGBKT7-MED16/pGADT7, or pGBKT7-MED16/pGADT7-WRKY33. Yeast cells carrying pGBKT7-MED16/pGADT7-WRKY33 grew on quadruple dropout medium supplemented with X-α-Gal (SD-Ade-His-Leu-Trp+X-α-Gal) and formed blue colonies, whereas those harboring either pGBKT7/pGADT7-WRKY33 or pGBKT7-MED16/pGADT7 did not grow and change color, indicating that MED16 interacts with WRKY33 in yeast. B, Morphology (top row) of and S. sclerotiorum disease symptoms (middle and bottom rows) on the wild type (WT), med16-1, and the MED16pro:MED16-3×FLAG med16-1 transgenic line. Representative photographs were taken 4 weeks after germination (top row), at 48 hpi (middle row), and 5 dpi (bottom row). C, The MED16-3×FLAG fusion protein was detected in MED16pro:MED16-3×FLAG med16-1 plants but not in the wild type and med16-1, indicating the specificity of anti-FLAG antibody for immunoblot analysis of MED16-3×FLAG. The asterisks show two nonspecific bands present in all genotypes. D, Nuclear protein extracts of MED16pro:MED16-3×FLAG/35Spro:4×Myc-WRKY33 and MED16pro:MED16-3×FLAG plants were immunoprecipitated with anti-Myc antibody. The precipitated proteins were separated by SDS-PAGE, transferred onto a nitrocellulose membrane, and probed with anti-FLAG antibody (right image). Inputs were analyzed with the anti-FLAG and anti-Myc antibodies, showing the presence or absence of MED16-3×FLAG (left image) and 4×My-WRKY33 (middle image). IP, Immunoprecipitation; W, western blotting. The experiment was repeated four independent times with the same results.
In order to test whether MED16 is physically associated with WRKY33 in planta, we created transgenic Arabidopsis plants coexpressing functional epitope-tagged MED16 and WRKY33 proteins. We first generated transgenic med16-1 plants expressing a MED16-3×FLAG fusion driven by its native promoter. The MED16pro:MED16-3×FLAG transgene complemented all of the med16-1 mutant phenotypes, including pale green leaf color, lack of inducible PDF1.2 expression, and enhanced pathogen susceptibility (Fig. 6B; Supplemental Fig. S3, A–C). When the transgenic plants were subjected to immunoblot analysis with an anti-FLAG antibody, two protein bands corresponding to MED16-3×FLAG were detected (Fig. 6C). These bands were not present in med16-1 and wild-type plants, confirming the specificity of the anti-FLAG immunoblot analysis. The MED16pro:MED16-3×FLAG transgene was then crossed into the previously characterized 35S:4×Myc-WRKY33 transgenic line (Mao et al., 2011). We chose this transgenic line because the fusion protein has been demonstrated to be functional and can be easily detected using an anti-Myc antibody (Mao et al., 2011). Additionally, overexpression of WRKY33 has been shown to elevate basal levels of PDF1.2 (Zheng et al., 2006), which would allow us to detect the interaction between MED16 and WRKY33 in the absence of any treatment.
The transgenic plants coexpressing MED16-3×FLAG and 4×Myc-WRKY33 were subjected to a coimmunoprecipitation experiment. The MED16pro:MED16-3×FLAG transgenic line was used as a negative control in this experiment. As shown in Figure 6D, MED16-3×FLAG was coimmunoprecipitated with 4×Myc-WRKY33 using the anti-Myc antibody from the nuclear protein extract of the MED16pro:MED16-3×FLAG/35S:4×Myc-WRKY33 plants but not from that of the MED16pro:MED16-3×FLAG plants. Moreover, MED16-3×FLAG was not coimmunoprecipitated with 6×Myc using the anti-Myc antibody from protein extract of Nicotiana benthamiana leaf tissues transiently coexpressing MED16-3×FLAG and 6×Myc (Supplemental Fig. S4). These results clearly indicate that WRKY33 physically associates with MED16 in planta.
MED16 Is Required for WRKY33-Activated Transcription of PDF1.2 and ORA59 and Resistance to S. sclerotiorum
Previous work has shown that WRKY33 is required for B. cinerea-induced expression of PDF1.2 and ORA59 and that overexpression of WRKY33 leads to elevated basal expression of PDF1.2 (Zheng et al., 2006; Birkenbihl et al., 2012). Since WRKY33 associates with MED16 and MED16 is required for the recruitment of RNAPII to the PDF1.2 and ORA59 genes (Figs. 5 and 6), the constitutive expression of PDF1.2 in the WRKY33-overexpressing plants may depend on MED16. To test this hypothesis, we crossed the previously characterized 35S:4×Myc-WRKY33 transgene (Mao et al., 2011) into the med16-1 mutant background and compared the basal expression levels of PDF1.2 and ORA59 in the wild-type, med16-1, 35S:4×Myc-WRKY33, and 35S:4×Myc-WRKY33 med16-1 plants. Confirming the previous observation (Zheng et al., 2006), basal transcription levels of PDF1.2 were elevated in the 35S:4×Myc-WRKY33 plants (Fig. 7A). Similarly, basal expression of ORA59 was also enhanced in the 35S:4×Myc-WRKY33 plants (Fig. 7A), which supports the conclusion that the ORA59 gene is a direct target of WRKY33 (Birkenbihl et al., 2012). Importantly, the enhancement effects of the overexpression of WRKY33 on PDF1.2 and ORA59 expression were completely blocked by the med16-1 mutation (Fig. 7A), indicating that WRKY33-activated transcription of PDF1.2 and ORA59 requires MED16.
Figure 7.
Dependence of WRKY33-activated defense responses on MED16. A, Expression levels of PDF1.2 and ORA59 in the wild type (WT), med16-1, 35S:4×Myc-WRKY33 (oeWRKY33), and 35S:4×Myc-WRKY33 med16-1 plants. Leaves of 4-week-old soil-grown plants were collected and subjected to total RNA extraction and qPCR analysis. Expression of PDF1.2 and ORA59 was normalized against the constitutively expressed UBQ5. Data represent means of three biological replicates with sd. Different letters above the bars indicate significant differences (P < 0.05, one-way ANOVA). B, Disease symptoms on the S. sclerotiorum-infected wild-type, wrky33-1, med16-1, 35S:4×Myc-WRKY33, and 35S:4×Myc-WRKY33 med16-1 leaves. Photographs were taken at 24 and 48 hpi. C, Disease symptoms on the S. sclerotiorum-infected wild-type, wrky33-1, med16-1, 35S:4×Myc-WRKY33, and 35S:4×Myc-WRKY33 med16-1 plants. Photographs were taken at 5 dpi. D, Size of the necrotic lesions formed on the S. sclerotiorum-infected wild type, wrky33-1, med16-1, 35S:4×Myc-WRKY33, and 35S:4×Myc-WRKY33 med16-1 at 36 and 48 hpi. Data represent means of lesion sizes on 24 leaves with sd. Different letter above the bars indicate significant differences (P < 0.05, one-way ANOVA). The statistical comparisons were performed among the wild type, wrky33-1, med16-1, 35S:4×Myc-WRKY33, and 35S:4×Myc-WRKY33 med16-1 for each time point. E, Decay percentages of S. sclerotiorum-infected wild-type, wrky33-1, med16-1, 35S:4×Myc-WRKY33, and 35S:4×Myc-WRKY33 med16-1 plants at 5 dpi. Data represent means of three groups (12 plants per group) of S. sclerotiorum-infected plants with sd. Different letters above the bars indicate significant differences (P < 0.05, one-way ANOVA). Experiments in A, D, and E were repeated three independent times with similar trends. Results from a representative experiment are presented. Photographs in B and C represent typical disease symptoms on S. sclerotiorum-infected leaves or plants.
A recent report indicates that overexpression of BnWRKY33 in B. napus confers markedly enhanced resistance to S. sclerotiorum (Wang et al., 2014), suggesting that WRKY33 may be a positive regulator of resistance against S. sclerotiorum. Indeed, the wrky33-1 mutant was slightly but significantly more susceptible than the wild type to S. sclerotiorum (Fig. 7, B–E), and overexpression of WRKY33 provided marginally but significantly increased resistance, which could be seen before 36 hpi (Fig. 7D). Intriguingly, the increased resistance disappeared as disease progressed. Nevertheless, the resistance conferred by the overexpression of WRKY33 was completed inhibited by the med16-1 mutation, indicating that WRKY33-mediated resistance against S. sclerotiorum also depends on MED16.
DISCUSSION
Using genetic, molecular, and biochemical approaches, here we demonstrate that (1) MED16 is a central regulator of basal resistance against S. sclerotiorum in Arabidopsis; (2) MED16 is required for ET-promoted suppression of JA-mediated wound responses; (3) MED16 is required for the recruitment of RNAPII to both PDF1.2 and ORA59; and (4) MED16 is physically associated with WRKY33 and is required for WKRY33-activated transcription of PDF1.2 and ORA59 and resistance to S. sclerotiorum.
Mediator is a multisubunit complex, and the requirement for each subunit depends on specific biological processes (Balamotis et al., 2009; Hemsley et al., 2014). Among the 14 Mediator subunit mutants tested in this study, med14, med16, and med25 exhibited significantly enhanced susceptibility to S. sclerotiorum (Fig. 1). Although med8, med13, and med18 have been shown to be more susceptible than the wild type to the necrotrophic fungal pathogens B. cinerea and A. brassicicola (Kidd et al., 2009; Lai et al., 2014; Zhu et al., 2014), they did not show enhanced susceptibility to S. sclerotiorum. This is probably because MED8, MED13, and MED18 do not play a major role in regulating S. sclerotiorum-induced transcriptome changes (Fig. 2), suggesting that the molecular mechanisms underlying susceptibility/resistance against various necrotrophic pathogens are different. On the other hand, the med16 mutant is considerably more susceptible than med14 and med25 to S. sclerotiorum (Fig. 1, E and F), indicating that MED16 plays a more important role than MED14 and MED25 in regulating basal resistance against S. sclerotiorum.
Surprisingly, the med16/sfr6 mutants are even more susceptible to S. sclerotiorum than the coi1-1 null mutant, and the transcription levels of PDF1.2 are also lower in med16 than in coi1-1 (Fig. 3; Supplemental Fig. S2). Since coi1-1 has been shown to be the most susceptible Arabidopsis mutant to S. sclerotiorum and the coi1-1 mutation completely blocks JA/ET-induced PDF1.2 expression (Pré et al., 2008; Perchepied et al., 2010), the med16 mutation may either have a stronger effect than coi1-1 on JA/ET-mediated defense signaling or compromise both JA/ET-dependent and -independent defense pathways.
The function of MED16 in JA/ET-mediated defense signaling has recently been revealed. Zhang et al. (2012) and Wathugala et al. (2012) have shown that mutations in MED16 block JA/ET- and B. cinerea-induced defense gene expression and compromise resistance to B. cinerea and A. brassicicola. It has been well documented that expression of the defense gene PDF1.2 is regulated by a group of AP2/ERF domain transcription factors including ORA59 and ERF1 (Lorenzo et al., 2003; Pré et al., 2008) and that genes encoding the AP2/ERF factors are, in turn, controlled by the transcription factors EIN3 and EIL1 (Solano et al., 1998; Zhu et al., 2011). Here, we show that MED16 is required for the recruitment of RNAPII to both ORA59 and PDF1.2 (Fig. 5), suggesting that MED16 may directly participate in both EIN3/EIL1- and AP2/ERF-mediated transcription. Consistent with this hypothesis, elevated expression of PDF1.2 activated by the overexpression of ERF5 depends on MED16 (Wathugala et al., 2012). Furthermore, EIN3, EIL1, ORA59, and ERF1 have all been shown to interact with MED25, a Mediator subunit physically associating with MED16 (Çevik et al., 2012; Yang et al., 2014). Taken together, it could be concluded that MED16 plays an essential role in relaying defense signals of the JA/ET pathway to the RNAPII transcription machinery. Whether any of the EIN3/EIL1 and AP2/ERF transcription factors interact directly with MED16 needs further investigation.
Unlike COI1 and MED25, which are required for both branches of the JA signaling pathway, namely, the wound response branch and the defense response branch (Xie et al., 1998; Pré et al., 2008; Kidd et al., 2009; Chen et al., 2012), MED16 is only required for the defense response branch (Fig. 4). In fact, MED16 positively contributes to ET-induced suppression of JA-mediated wound responses (Fig. 4), suggesting either a direct or indirect role for MED16 in JA-ET cross talk (Dong, 1998; Pieterse et al., 2009).
MED16 is required for the transcription factor WRKY33-activated defense signaling. WRKY33 is an important regulator of defense responses against necrotrophic fungal pathogens (Zheng et al., 2006; Lai et al., 2011). Mutations in WRKY33 compromise B. cinerea-induced defense gene expression and enhance susceptibility to both B. cinerea and A. brassicicola. Expression of the WRKY33 gene is highly inducible by B. cinerea infection (AbuQamar et al., 2006), but the induction does not require the JA and ET signaling components COI1 and EIN2, respectively (Zheng et al., 2006), suggesting that WRKY33 may be activated by a JA/ET-independent defense pathway. Recently, Wang et al. (2014) reported that expression of the BnWRKY33 gene is highly inducible by S. sclerotiorum infection and that overexpression of BnWRKY33 in B. napus leads to increased basal expression of BnPDF1.2 and resistance to S. sclerotiorum. Consistent with that report, we found that the Arabidopsis wrky33-1 mutant is more susceptible to S. sclerotiorum than the wild type and that overexpression of WRKY33 confers a low level of resistance to S. sclerotiorum during early time points of infection (Fig. 7). Based on the microarray data, WKRY33 is also inducible by S. sclerotiorum in Arabidopsis and MED16 is not required for the induction. However, our further study revealed that WRKY33-activated defense responses depend on MED16. We found that MED16 physically associates with WRKY33 and is required for WRKY33-activated PDF1.2 and ORA59 expression and resistance to S. sclerotiorum (Figs. 6 and 7). These results indicate that MED16 is required not only for JA/ET-mediated defense responses but also for WRKY33-activated defense signaling. Blocking the WRKY33-mediated defense signaling in med16 probably contributes to the reduced PDF1.2 expression and enhanced susceptibility to S. sclerotiorum.
Although results from previous studies about the role of NPR1 in resistance against S. sclerotiorum are contradictory (Guo and Stotz, 2007; Perchepied et al., 2010), we consistently observed that the npr1-3 mutant is more susceptible than the wild type to this pathogen (Fig. 3, A–D). The npr1-3 mutation does not affect S. sclerotiorum-induced PDF1.2 expression (Fig. 3, E and F), which is consistent with NPR1 being an SA signaling component (Cao et al., 1997). Since the SA biosynthesis mutants, SA induction-deficient1 (sid1)/eds5 and sid2/eds16, as well as nahG (encoding a salicylate hydroxylase) transgenic plants, which are impaired in SA accumulation, do not show enhanced susceptibility to S. sclerotiorum (Perchepied et al., 2010), how NPR1 plays a positive role in resistance to this necrotrophic fungal pathogen needs further investigation. Nevertheless, because we have previously shown that mutations in MED16 reduce NPR1 protein accumulation (Zhang et al., 2012), the reduced NPR1 protein levels in the med16 mutant might contribute to the enhanced susceptibility to S. sclerotiorum.
In summary, our study revealed that MED16 is a central component of basal resistance against S. sclerotiorum. We demonstrate that MED16 is essential not only for JA/ET-mediated defense pathways but also for the transcription factor WRKY33-activated defense signaling. Further investigations on the molecular mechanisms underlying the function of MED16 in resistance to S. sclerotiorum would help in the design of strategies for controlling this broad-host-range necrotrophic fungal pathogen.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The wild-type Arabidopsis (Arabidopsis thaliana) Columbia ecotype was utilized. The med mutants used in this study are listed in Supplemental Table S1, and wrky33-1 (SALK_006603), npr1-3, ein2-1, and coi1-1 were described previously (Cao et al., 1997; Xie et al., 1998; Alonso et al., 1999; Zheng et al., 2006). The T-DNA insertion lines were obtained from either the Arabidopsis Biological Resource Center at Ohio State University or the European Arabidopsis Stock Centre at the University of Nottingham. Homozygous mutant plants of the T-DNA insertion lines were confirmed with primers flanking the T-DNA insertions (Supplemental Table S2) and the left border primers LBa1 and LB3 (Sessions et al., 2002; Alonso et al., 2003a). MED16pro:MED16-3×FLAG/35Spro:4×My-WRKY33 plants were generated by crossing MED16pro:MED16-3×FLAG med16-1 with the previously characterized 35Spro:4×My-WRKY33 transgenic line (Mao et al., 2011). The 35Spro:4×My-WRKY33 transgene was also crossed into the med16-1 mutant background to generate 35Spro:4×My-WRKY33 med16-1 plants. The Arabidopsis seeds were sown on autoclaved soil (Sunshine MVP; Sun Gro Horticulture) and cold treated at 4°C for 3 d. Plants were germinated and grown at 23°C to 25°C under a 16-h-light/8-h-dark regime.
Pathogen Infection
Sclerotinia sclerotiorum inoculation was performed as described by Guo and Stotz (2007) with minor modifications. Briefly, sclerotia of S. sclerotiorum isolate 1980 were germinated at room temperature on potato dextrose agar medium (213400; Becton-Dickinson). About 3 d later, a small piece of agar containing mycelia was transferred onto minimal medium (1 g of NaOH, 3 g of dl-malic acid, 2 g of NH4NO3, 0.1 g of MgSO4·7H2O, and 39 g of Bacto-agar per liter; Cruickshank, 1983) and cultured for about 3 d prior to inoculation to reduce the pathogen aggressiveness (Guo and Stotz, 2007). An agar plug (2 mm in diameter) containing the advancing edge of S. sclerotiorum mycelia was removed to inoculate Arabidopsis leaves. One rosette leaf per plant was inoculated for basal resistance assessment. Lengths and widths of lesions were measured with a caliper before disease symptoms expanded beyond inoculated leaves, and the average of the length and the width was used to represent the size of a lesion. The decay percentage of S. sclerotiorum-infected plants was scored to assess disease development. A plant was considered to be decayed when the lesion expanded beyond the inoculated leaf to the center of the plant.
RNA Analysis
RNA extraction, reverse transcription, and qPCR analyses were carried out as described by DeFraia et al. (2010). Primers used for qPCR in this study are listed in Supplemental Table S3.
Microarray Analysis
Four-week-old soil-grown plants were inoculated with the S. sclerotiorum isolate 1980. Total RNA samples extracted from leaf tissues collected at the indicated time points after S. sclerotiorum inoculation were subjected to microarray analysis. Briefly, RNA concentration was determined on a NanoDrop Spectrophotometer (Thermo Fisher Scientific), and sample quality was assessed using the 2100 Bioanalyzer (Agilent Technologies). Complementary DNA (cDNA) was synthesized from 200 ng of total RNA and used as a template for in vitro transcription in the presence of T7 RNA Polymerase and cyanine-labeled CTPs using the Quick Amp Labeling kit (Agilent Technologies) according to the manufacturer’s protocol. The amplified, labeled copy RNA was purified using the RNeasy Mini kit (Qiagen). For each array, 1,650 ng of Cy3-labeled copy RNA was fragmented and hybridized with rotation at 65°C for 17 h. Samples were hybridized to Arabidopsis 4 × 44k arrays (Agilent Technologies). The arrays were washed according to the manufacturer’s protocol and then scanned on a G2505B scanner (Agilent Technologies). Data were extracted using Feature Extraction 10.1.1.1 software (Agilent Technologies).
Data (individual signal intensity values) obtained from the microarray probes were background corrected using the normexp+offset method, in which a small positive offset (k = 50) was added to move the corrected intensities away from zero (Ritchie et al., 2007). The resulting data were log transformed (using 2 as the base) and normalized between individual samples by scaling the individual log-transformed signal intensities so that all data sets had comparable lower quartile, median, and upper quartile values (Smyth, 2005). After normalization, Student’s t test was performed considering a probe-by-probe comparison between different genotypes at the same time point using wild-type Columbia as the reference sample and between different time points of the same genotype using the 0-h sample as the reference. In each comparison, a P value and fold change were computed for each gene locus. The gene expression fold changes were computed based on the normalized log-transformed signal intensity data. To control false discovery rate and correct multiple hypothesis testing, a q value was calculated and used to assess the significance of each test (Storey and Tibshirani, 2003). The comparison results were further explored to obtain numbers of overlapped genes between/among different comparisons.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed as described by Saleh et al. (2008) with minor modifications. Briefly, approximately 2 g of 2-week-old seedlings was submerged in 50 mL of cross-linking buffer (10 mm Tris-HCl, pH 8, 0.4 m Suc, 1 mm phenylmethylsulfonyl fluoride [PMSF], 1 mm EDTA, and 1% [v/v] formaldehyde) and vacuum infiltrated three times for 3 to 4 min each at room temperature. The cross-linking reaction was stopped by adding 2.5 mL of 2 m Gly to a final concentration of 100 mm and vacuum infiltration for 5 min. Plant tissues were washed three times with cold sterile deionized water. After removing water, plant tissues were submerged in liquid nitrogen, ground to a fine powder, and resuspended in 20 to 25 mL of cold nuclei isolation buffer (15 mm PIPES, pH 6.8, 0.25 m Suc, 5 mm MgCl2, 60 mm KCl, 15 mm NaCl, 1 mm CaCl2, 0.9% [v/v] Triton X-100, 1 mm PMSF, 2 μg mL−1 pepstatin A, and 2 μg mL−1 aprotinin). After brief vortex and incubation, the homogenized slurry was filtered through one layer of Miracloth. After centrifugation at 3,220g for 20 min, the pellet (nuclei) was resuspended in 1.5 mL of cold nuclei lysis buffer (50 mm HEPES, pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.1% [w/v] SDS, 0.1% [w/v] sodium deoxycholate, 1% [v/v] Triton X-100, 1 μg mL−1 pepstatin A, and 1 μg mL−1 aprotinin). DNA was sheared into approximately 500-bp (200–1,000 bp) fragments by 6 to 10 min of 3-s pause sonication at 40% to 43% amplitude using a TM-100 sonic disruptor (TekMar). After centrifugation at 13,800g for 10 min, the supernatant (200 μL) was diluted 5-fold with nuclei lysis buffer and precleared by adding 50 μL of salmon (Oncorhynchus keta) sperm DNA/protein A agarose beads. After removing the agarose beads, 5 μL of anti-RPB2 antibody (ab10338; Abcam) was added, and the mixture was incubated at 4°C for 5 h to overnight with gentle rotation, and then 60 to 75 μL of salmon sperm DNA/protein A agarose beads was added and the incubation was continued for 2 to 3 h. After centrifugation at 3,800g for 2 min, the agarose beads were sequentially washed with low-salt wash buffer (20 mm Tris-HCl, pH 8, 150 mm NaCl, 0.2% [w/v] SDS, 0.5% [v/v] Triton X-100, and 2 mm EDTA), high-salt wash buffer (20 mm Tris-HCl, pH 8, 500 mm NaCl, 0.2% [w/v] SDS, 0.5% [v/v] Triton X-100, and 2 mm EDTA), LiCl wash buffer (10 mm Tris-HCl, pH 8, 0.25 m LiCl, 1% [w/v] sodium deoxycholate, 1% [v/v] Nonidet P-40, and 1 mm EDTA), and TE buffer (twice; 1 mm EDTA and 10 mm Tris-HCl, pH 8). The immunocomplexes were eluted with freshly prepared elution buffer (0.1 m NaHCO3 and 0.5% [w/v] SDS) and incubated at 65°C for 15 min with gentle rotation. Twenty microliters of 5 m NaCl was added to 500 μL of the immunocomplex solution, the mixture was incubated at 65°C for 4 h to overnight to reverse cross linking, then 20 μL of 1 m Tris-HCl, pH 6.5, 10 μL of 0.5 m EDTA, and 2 μL of proteinase K (10 mg mL−1) were added, and the mixture was incubated at 45°C for 1.5 h to digest the proteins. Immunoprecipitated DNA was purified using a mixture of phenol:chloroform:isoamyl alcohol (25:24:1), and the resulting DNA was used for qPCR analysis. The amount of precipitated DNA corresponding to a specific gene region was determined by qPCR and normalized to input DNA (Haring et al., 2007). Primers used for chromatin immunoprecipitation-qPCR are listed in Supplemental Table S4.
Plasmid Construction and Plant Transformation
The 3×FLAG fragment was removed with SpeI and XbaI from pCR8GW-XB3New-3×FLAG (Wang et al., 2006) and ligated into the corresponding sites of pBluescript SK+. The primers EcoRI-MED16F and SpeI-MED16R (Supplemental Table S5) were used to amplify the coding region of MED16 from Arabidopsis cDNAs. The PCR products were digested with EcoRI and SpeI and ligated into the corresponding sites of pBluescript SK+-3×FLAG, resulting in the plasmid pBluescript SK−MED16-3×FLAG. The primers SalI-MED16PF and EcoRI-MED16PR (Supplemental Table S5) were used to amplify the promoter region of MED16 from the Arabidopsis genomic DNA. The PCR products were digested with SalI and EcoRI and ligated into the corresponding sites of pBluescript SK+-MED16-3×FLAG, resulting in the plasmid pBluescript SK+-MED16pro:MED16-3×FLAG. The MED16pro:MED16-3×FLAG fragment was then recovered using SalI and XbaI and subcloned into the corresponding sites of pBI101-Luc (Zhang et al., 2012). The resulting plasmid pBI101-MED16pro:MED16-3×FLAG was introduced into the Agrobacterium tumefaciens strain GV3101(pMP90) by electroporation and transformed into med16-1 following the floral dip method (Clough and Bent, 1998).
Chemical Treatment
Ten-day-old seedlings grown on one-half-strength Murashige and Skoog medium were treated with 0.1 mm MeJA, 0.1 mm ACC, or their combination. Seedlings for the negative control were treated with water. Plant tissues except roots were collected and subjected to total RNA extraction.
Coimmunoprecipitation
The coimmunoprecipitation assay was performed as described by Qiu et al. (2008) with minor modifications. Briefly, nuclei were isolated according to Gendrel et al. (2002) and Nelson et al. (2006). Nuclei were resuspended in coimmunoprecipitation buffer (100 mm Tris-HCl, pH 7.5, 75 mm NaCl, 1 mm EDTA, 0.1% [v/v] Triton X-100, 0.05% [w/v] SDS, 10% [v/v] glycerol, 2.5 mm dithiothreitol, 50 μg mL−1 protease inhibitors tosyl-l-phenylalanyl chloromethyl keton and tosyl-l-lysyl-chloromethane hydrochloride, and 0.6 mm PMSF). Ten units of Universal Nuclease (88700; Pierce Biotechnology) was added into the suspension, and the mixture was incubated on ice for 1 h and centrifuged at 16,000g for 30 min. The supernatant was incubated with anti-Myc antibody (sc-789; Santa Cruz Biotechnology) overnight at 4°C followed by precipitation with protein G Plus-Agarose (sc-2002; Santa Cruz Biotechnology) for 4 h. After washing four times with coimmunoprecipitation buffer, proteins were eluted by boiling in 40 μL of 2× Laemmli sample buffer for 10 min. The eluates were separated by 8% (w/v) SDS-PAGE, transferred onto a nitrocellulose membrane (1215458; ME Manufacturing), and probed with anti-FLAG antibody (3165; Sigma) to detect coimmunoprecipitated MED16-3×FLAG protein.
Yeast Two-Hybrid Assay
The full-length coding sequence of MED16 was amplified from Arabidopsis cDNAs with primers SalI-MED16F and EcoRI-MED16R (Supplemental Table S5). SalI/EcoRI-digested PCR products were cloned into the corresponding site of the bait vector pGBKT7. Full-length WRKY33 coding sequence was amplified from Arabidopsis cDNAs with primers BamHI-WRKY33F and EcoRI-WRKY33R (Supplemental Table S5), digested with BamHI and EcoRI, and cloned into the prey vector pGADT7. The resulting plasmids pGBKT7-MED16 and pGADT7-WRKY33 were cotransformed into the yeast (Saccharomyces cerevisiae) strain AH109. The bait vector pGBKT7 and prey vector pGADT7 were cotransformed with pGADT7-WRKY33 and pGBKT7-MED16, respectively, to generate negative controls. The presence of the transgenes was confirmed by growth on an SD-Leu-Trp agar plate. To assess protein interactions, the transformed yeast cells were suspended in liquid SD-Leu-Trp medium to an optical density at 600 nm of 1. Five microliters of suspended yeast cells was dropped onto an SD-Ade-His-Leu-Trp+X-α-Gal (4 mg mL−1) agar plate. The resulting agar plate was incubated at 30°C and observed for yeast growth and color changes.
Statistical Methods
Except for those used in microarray analysis, statistical analyses were performed using the one-way ANOVA in Prism 5.0b (GraphPad Software) and the data analysis tools in Excel of Microsoft Office 2004 for Macintosh (Student’s t test: two samples assuming unequal variances).
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MED8 (At2g03070), MED13 (At1g55325), MED14 (At3g04740), MED16 (At4g04920), MED17 (At5g20170), MED18 (At2g22370), MED20a (At3g28230), MED23 (At1g23230), MED25 (At1g25540), MED31 (At5g19910), MED32 (At1g11760), MED33b (At2g48110), MED34 (At1g31360), MED36 (AT4g25630), NPR1 (At1g64280), EIN2 (At5g03280), COI1 (At2g39940), WRKY33 (At2g38470), ORA59 (At1g06160), PDF1.2 (At5g44420), MYC2 (At1g32640), VSP1 (At5g24780), VSP2 (At5g24770), JR1 (At3g16470), and UBQ5 (At3g62250), and the National Center for Biotechnology Information Gene Expression Omnibus series number GSE65165 (microarray data).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. MED gene expression in the corresponding T-DNA insertion mutants.
Supplemental Figure S2. Comparison of PDF1.2 expression in med16 and coi1-1.
Supplemental Figure S3. Complementation of med16 with MED16pro:MED16-3×FLAG.
Supplemental Figure S4. Evidence that 6×Myc does not interact with MED16-3×FLAG.
Supplemental Table S1. Mediator mutants used in this study.
Supplemental Table S2. Primers used for the identification of homozygous T-DNA insertion mutant plants.
Supplemental Table S3. Primers used for qPCR analysis of gene expression.
Supplemental Table S4. Primers used for chromatin immunoprecipitation-qPCR analysis.
Supplemental Table S5. Primers used for plasmid construction.
Supplemental Data Set S1. Comparison of gene expression between med16-1 and the wild type.
Supplementary Material
Acknowledgments
We thank Dr. Zhixiang Chen (Purdue University) for the 35S:4×My-WRKY33 seeds, Dr. Wen-Yuan Song (University of Florida) for the pCR8GW-XB3New-3×FLAG plasmid, the Arabidopsis Biological Resource Center at Ohio State University for SALK_006603, SALK_018156, SALK_111977, SAIL_889_C08, SALK_119080, SALK_035522, SALK_028490, SALK_022477, SALK_087178, and SALK_093373 seeds, and the European Arabidopsis Stock Centre at the University of Nottingham for GABI_507F08 seeds.
Glossary
- JA
jasmonic acid
- ET
ethylene
- SA
salicylic acid
- hpi
hours post inoculation
- dpi
days post inoculation
- T-DNA
transfer DNA
- MeJA
methyl jasmonate
- ACC
1-aminocyclopropane-1-carboxylic acid
- SD
synthetic dextrose
- qPCR
quantitative PCR
- cDNA
complementary DNA
- PMSF
phenylmethylsulfonyl fluoride
Footnotes
This work was supported by the National Science Foundation (grant no. IOS–0842716 to Z.M.), the National Sclerotinia Initiative (grant no. 58–5442–3–029 to J.A.R. and Z.M.), and the China Scholarship Council (scholarship to X.D.).
Articles can be viewed without a subscription.
References
- AbuQamar S, Chen X, Dhawan R, Bluhm B, Salmeron J, Lam S, Dietrich RA, Mengiste T (2006) Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J 48: 28–44 [DOI] [PubMed] [Google Scholar]
- Akoulitchev S, Chuikov S, Reinberg D (2000) TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407: 102–106 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003a) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Solano R, Wisman E, Ferrari S, Ausubel FM, Ecker JR (2003b) Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc Natl Acad Sci USA 100: 2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y, et al. (2010) Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari SA, He Q, Morse RH (2009) Mediator complex association with constitutively transcribed genes in yeast. Proc Natl Acad Sci USA 106: 16734–16739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autran D, Jonak C, Belcram K, Beemster GT, Kronenberger J, Grandjean O, Inzé D, Traas J (2002) Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J 21: 6036–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S (2007) Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell 26: 717–729 [DOI] [PubMed] [Google Scholar]
- Baek HJ, Malik S, Qin J, Roeder RG (2002) Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol Cell Biol 22: 2842–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamotis MA, Pennella MA, Stevens JL, Wasylyk B, Belmont AS, Berk AJ (2009) Complexity in transcription control at the activation domain-mediator interface. Sci Signal 2: ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Diezel C, Somssich IE (2012) Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol 159: 266–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland GJ, Hall R (1994) Index of plant hosts of Sclerotinia sclerotiorum. Can J Plant Pathol 16: 93–108 [Google Scholar]
- Bolton MD, Thomma BP, Nelson BD (2006) Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol Plant Pathol 7: 1–16 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Soltau WL, Blatchley MR, Powers BL, Hurlock AK, Seals LA, Weng JK, Stout J, Chapple C (2012) REF4 and RFR1, subunits of the transcriptional coregulatory complex mediator, are required for phenylpropanoid homeostasis in Arabidopsis. J Biol Chem 287: 5434–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud MC, Asai S, Rallapalli G, Piquerez S, Fabro G, Jones JD (2013) A downy mildew effector attenuates salicylic acid-triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biol 11: e1001732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canet JV, Dobón A, Tornero P (2012) Non-recognition-of-BTH4, an Arabidopsis mediator subunit homolog, is necessary for development and response to salicylic acid. Plant Cell 24: 4220–4235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Cerdán PD, Chory J (2003) Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Cessna SG, Sears VE, Dickman MB, Low PS (2000) Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell 12: 2191–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çevik V, Kidd BN, Zhang P, Hill C, Kiddle S, Denby KJ, Holub EB, Cahill DM, Manners JM, Schenk PM, et al. (2012) MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol 160: 541–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadick JZ, Asturias FJ (2005) Structure of eukaryotic Mediator complexes. Trends Biochem Sci 30: 264–271 [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chen R, Jiang H, Li L, Zhai Q, Qi L, Zhou W, Liu X, Li H, Zheng W, Sun J, et al. (2012) The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW (2011a) Function and regulation of the Mediator complex. Curr Opin Genet Dev 21: 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW (2011b) Origins and activity of the Mediator complex. Semin Cell Dev Biol 22: 729–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank RH. (1983) Distinction between Sclerotinia species by their pectic zymograms. Trans Br Mycol Soc 80: 117–119 [Google Scholar]
- DeFraia CT, Zhang X, Mou Z (2010) Elongator subunit 2 is an accelerator of immune responses in Arabidopsis thaliana. Plant J 64: 511–523 [DOI] [PubMed] [Google Scholar]
- Dhawan R, Luo H, Foerster AM, Abuqamar S, Du HN, Briggs SD, Mittelsten Scheid O, Mengiste T (2009) HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. (1998) SA, JA, ethylene, and disease resistance in plants. Curr Opin Plant Biol 1: 316–323 [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA (2002) Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871–1873 [DOI] [PubMed] [Google Scholar]
- Gillmor CS, Park MY, Smith MR, Pepitone R, Kerstetter RA, Poethig RS (2010) The MED12-MED13 module of Mediator regulates the timing of embryo patterning in Arabidopsis. Development 137: 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Godoy G, Steadman JR, Dickman MB, Dam R (1990) Use of mutants to demonstrate the role of oxalic acid in pathgenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris. Physiol Mol Plant Pathol 37: 179–191 [Google Scholar]
- Guglielmi B, van Berkum NL, Klapholz B, Bijma T, Boube M, Boschiero C, Bourbon HM, Holstege FC, Werner M (2004) A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res 32: 5379–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Stotz HU (2007) Defense against Sclerotinia sclerotiorum in Arabidopsis is dependent on jasmonic acid, salicylic acid, and ethylene signaling. Mol Plant Microbe Interact 20: 1384–1395 [DOI] [PubMed] [Google Scholar]
- Guyon K, Balagué C, Roby D, Raffaele S (2014) Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genomics 15: 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M (2007) Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley PA, Hurst CH, Kaliyadasa E, Lamb R, Knight MR, De Cothi EA, Steele JF, Knight H (2014) The Arabidopsis mediator complex subunits MED16, MED14, and MED2 regulate mediator and RNA polymerase II recruitment to CBF-responsive cold-regulated genes. Plant Cell 26: 465–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- Imura Y, Kobayashi Y, Yamamoto S, Furutani M, Tasaka M, Abe M, Araki T (2012) CRYPTIC PRECOCIOUS/MED12 is a novel flowering regulator with multiple target steps in Arabidopsis. Plant Cell Physiol 53: 287–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Sono T, Tasaka M, Furutani M (2011) MACCHI-BOU 2 is required for early embryo patterning and cotyledon organogenesis in Arabidopsis. Plant Cell Physiol 52: 539–552 [DOI] [PubMed] [Google Scholar]
- Kabbage M, Williams B, Dickman MB (2013) Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog 9: e1003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K (2009) The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, Min JY, Dickman MB (2008) Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol Plant Microbe Interact 21: 605–612 [DOI] [PubMed] [Google Scholar]
- Kim YJ, Björklund S, Li Y, Sayre MH, Kornberg RD (1994) A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77: 599–608 [DOI] [PubMed] [Google Scholar]
- Kim YJ, Zheng B, Yu Y, Won SY, Mo B, Chen X (2011) The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J 30: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C, Büche C, Fernandez AP, Schäfer E, Zwick E, Kretsch T (2012) The mediator complex subunit PFT1 interferes with COP1 and HY5 in the regulation of Arabidopsis light signaling. Plant Physiol 160: 289–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Mugford SG, Ulker B, Gao D, Thorlby G, Knight MR (2009) Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J 58: 97–108 [DOI] [PubMed] [Google Scholar]
- Knight H, Thomson AJ, McWatters HG (2008) Sensitive to freezing6 integrates cellular and environmental inputs to the plant circadian clock. Plant Physiol 148: 293–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Veale EL, Warren GJ, Knight MR (1999) The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell 11: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ (2009) The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev 23: 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. (2005) Mediator and the mechanism of transcriptional activation. Trends Biochem Sci 30: 235–239 [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5: 325–331 [DOI] [PubMed] [Google Scholar]
- Lai Z, Li Y, Wang F, Cheng Y, Fan B, Yu JQ, Chen Z (2011) Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23: 3824–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Schluttenhofer CM, Bhide K, Shreve J, Thimmapuram J, Lee SY, Yun DJ, Mengiste T (2014) MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat Commun 5: 3064. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden RD. (1979) Histology and physiology of pathogenesis in plant diseases caused by Sclerotinia species. Phytopathology 69: 890–896 [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciano P, Di Lenna P, Nelson BD (1983) Oxalic acid, cell wall-degrading enzymes and pH in pathogensis and their significance in the virulence of two Sclerotinia sclerotiorum isolates on sunflower. Physiol Plant Pathol 22: 339–345 [Google Scholar]
- Mathur S, Vyas S, Kapoor S, Tyagi AK (2011) The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol 157: 1609–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Petersen BL, Glawischnig E, Jensen AB, Andreasson E, Halkier BA (2003) Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol 131: 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler G, Kremmer E, Timmers HT, Meisterernst M (2001) Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep 2: 808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Sova P, Bomsztyk K (2006) Fast chromatin immunoprecipitation assay. Nucleic Acids Res 34: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková M, Sašek V, Dobrev PI, Valentová O, Burketová L (2014) Plant hormones in defense response of Brassica napus to Sclerotinia sclerotiorum: reassessing the role of salicylic acid in the interaction with a necrotroph. Plant Physiol Biochem 80: 308–317 [DOI] [PubMed] [Google Scholar]
- Penninckx IAMA, Eggermont K, Terras FRG, Thomma BPHJ, De Samblanx GW, Buchala A, Métraux JP, Manners JM, Broekaert WF (1996) Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8: 2309–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IAMA, Thomma BPHJ, Buchala A, Métraux JP, Broekaert WF (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10: 2103–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perchepied L, Balagué C, Riou C, Claudel-Renard C, Rivière N, Grezes-Besset B, Roby D (2010) Nitric oxide participates in the complex interplay of defense-related signaling pathways controlling disease resistance to Sclerotinia sclerotiorum in Arabidopsis thaliana. Mol Plant Microbe Interact 23: 846–860 [DOI] [PubMed] [Google Scholar]
- Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Pré M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink J (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol 147: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Fiil BK, Petersen K, Nielsen HB, Botanga CJ, Thorgrimsen S, Palma K, Suarez-Rodriguez MC, Sandbech-Clausen S, Lichota J, et al. (2008) Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J 27: 2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou C, Freyssinet G, Fevre M (1991) Production of cell wall-degrading enzymes by the phytopathogenic fungus Sclerotinia sclerotiorum. Appl Environ Microbiol 57: 1478–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou C, Freyssinet G, Fevre M (1992) Purification and characterization of extracellular pectinolytic enzymes produced by Sclerotinia sclerotiorum. Appl Environ Microbiol 58: 578–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK (2007) A comparison of background correction methods for two-colour microarrays. Bioinformatics 23: 2700–2707 [DOI] [PubMed] [Google Scholar]
- Rojo E, León J, Sánchez-Serrano JJ (1999) Cross-talk between wound signalling pathways determines local versus systemic gene expression in Arabidopsis thaliana. Plant J 20: 135–142 [DOI] [PubMed] [Google Scholar]
- Rollins JA, Dickman MB (2001) pH signaling in Sclerotinia sclerotiorum: identification of a pacC/RIM1 homolog. Appl Environ Microbiol 67: 75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97: 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2005) Limma: linear models for microarray data. In Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, eds, Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer, New York, pp 397–420 [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Huang H, Gao H, Wang J, Wu D, Liu X, Yang S, Zhai Q, Li C, Qi T, et al. (2014) Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26: 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz HU, Jikumaru Y, Shimada Y, Sasaki E, Stingl N, Mueller MJ, Kamiya Y (2011) Jasmonate-dependent and COI1-independent defense responses against Sclerotinia sclerotiorum in Arabidopsis thaliana: auxin is part of COI1-independent defense signaling. Plant Cell Physiol 52: 1941–1956 [DOI] [PubMed] [Google Scholar]
- Takagi Y, Kornberg RD (2006) Mediator as a general transcription factor. J Biol Chem 281: 80–89 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, et al. (2011) Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146: 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Penninckx IA, Broekaert WF, Cammue BP (2001) The complexity of disease signaling in Arabidopsis. Curr Opin Immunol 13: 63–68 [DOI] [PubMed] [Google Scholar]
- Turner JG, Ellis C, Devoto A (2002) The jasmonate signal pathway. Plant Cell (Suppl) 14: S153–S164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Chen X (2004) HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development 131: 3147–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YS, Pi LY, Chen X, Chakrabarty PK, Jiang J, De Leon AL, Liu GZ, Li L, Benny U, Oard J, et al. (2006) Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 18: 3635–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Fang H, Chen Y, Chen K, Li G, Gu S, Tan X (2014) Overexpression of BnWRKY33 in oilseed rape enhances resistance to Sclerotinia sclerotiorum. Mol Plant Pathol 15: 677–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Tan X, Zhang Z, Gu S, Li G, Shi H (2012) Defense to Sclerotinia sclerotiorum in oilseed rape is associated with the sequential activations of salicylic acid signaling and jasmonic acid signaling. Plant Sci 184: 75–82 [DOI] [PubMed] [Google Scholar]
- Wathugala DL, Hemsley PA, Moffat CS, Cremelie P, Knight MR, Knight H (2012) The Mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol 195: 217–230 [DOI] [PubMed] [Google Scholar]
- Williams B, Kabbage M, Kim HJ, Britt R, Dickman MB (2011) Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog 7: e1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik NA, Hampsey M (2002) The RNA polymerase II machinery: structure illuminates function. Cell 108: 453–463 [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xu R, Li Y (2011) Control of final organ size by Mediator complex subunit 25 in Arabidopsis thaliana. Development 138: 4545–4554 [DOI] [PubMed] [Google Scholar]
- Yang Y, Ou B, Zhang J, Si W, Gu H, Qin G, Qu LJ (2014) The Arabidopsis Mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. Plant J 77: 838–851 [DOI] [PubMed] [Google Scholar]
- Zarei A, Körbes AP, Younessi P, Montiel G, Champion A, Memelink J (2011) Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol Biol 75: 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang C, Zhang Y, Sun Y, Mou Z (2012) The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24: 4294–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yao J, Zhang Y, Sun Y, Mou Z (2013) The Arabidopsis Mediator complex subunits MED14/SWP and MED16/SFR6/IEN1 differentially regulate defense gene expression in plant immune responses. Plant J 75: 484–497 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhu Z, An F, Hao D, Li P, Song J, Yi C, Guo H (2014a) Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell 26: 1105–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wu H, Wang N, Fan H, Chen C, Cui Y, Liu H, Ling HQ (2014b) Mediator subunit 16 functions in the regulation of iron uptake gene expression in Arabidopsis. New Phytol 203: 770–783 [DOI] [PubMed] [Google Scholar]
- Zhao J, Buchwaldt L, Rimmer SR, Sharpe A, McGregor L, Bekkaoui D, Hegedus D (2009) Patterns of differential gene expression in Brassica napus cultivars infected with Sclerotinia sclerotiorum. Mol Plant Pathol 10: 635–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Wang J, An L, Doerge RW, Chen ZJ, Grau CR, Meng J, Osborn TC (2007) Analysis of gene expression profiles in response to Sclerotinia sclerotiorum in Brassica napus. Planta 227: 13–24 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Guan H, Leal F, Grey PH, Oppenheimer DG (2013) Mediator subunit18 controls flowering time and floral organ identity in Arabidopsis. PLoS ONE 8: e53924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Qamar SA, Chen Z, Mengiste T (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48: 592–605 [DOI] [PubMed] [Google Scholar]
- Zhu W, Wei W, Fu Y, Cheng J, Xie J, Li G, Yi X, Kang Z, Dickman MB, Jiang D (2013) A secretory protein of necrotrophic fungus Sclerotinia sclerotiorum that suppresses host resistance. PLoS ONE 8: e53901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Wirén M, Sinha I, Rasmussen NN, Linder T, Holmberg S, Ekwall K, Gustafsson CM (2006) Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol Cell 22: 169–178 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Schluttenhoffer CM, Wang P, Fu F, Thimmapuram J, Zhu JK, Lee SY, Yun DJ, Mengiste T (2014) CYCLIN-DEPENDENT KINASE8 differentially regulates plant immunity to fungal pathogens through kinase-dependent and -independent functions in Arabidopsis. Plant Cell 26: 4149–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, An F, Feng Y, Li P, Xue L, A M, Jiang Z, Kim JM, To TK, Li W, et al. (2011) Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA 108: 12539–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.