Translationally-controlled tumor protein binds with a class of ethylene receptors at the endoplasmic reticulum and affects protein degradation.
Abstract
Ethylene is an important phytohormone in the regulation of plant growth, development, and stress response throughout the lifecycle. Previously, we discovered that a subfamily II ethylene receptor tobacco (Nicotiana tabacum) Histidine Kinase1 (NTHK1) promotes seedling growth. Here, we identified an NTHK1-interacting protein translationally controlled tumor protein (NtTCTP) by the yeast (Saccharomyces cerevisiae) two-hybrid assay and further characterized its roles in plant growth. The interaction was further confirmed by in vitro glutathione S-transferase pull down and in vivo coimmunoprecipitation and bimolecular fluorescence complementation assays, and the kinase domain of NTHK1 mediates the interaction with NtTCTP. The NtTCTP protein is induced by ethylene treatment and colocalizes with NTHK1 at the endoplasmic reticulum. Overexpression of NtTCTP or NTHK1 reduces plant response to ethylene and promotes seedling growth, mainly through acceleration of cell proliferation. Genetic analysis suggests that NtTCTP is required for the function of NTHK1. Furthermore, association of NtTCTP prevents NTHK1 from proteasome-mediated protein degradation. Our data suggest that plant growth inhibition triggered by ethylene is regulated by a unique feedback mechanism, in which ethylene-induced NtTCTP associates with and stabilizes ethylene receptor NTHK1 to reduce plant response to ethylene and promote plant growth through acceleration of cell proliferation.
Although well known as an aging phytohormone for the acceleration of fruit ripening, organ senescence, and abscission, ethylene also regulates many aspects of vegetative growth and development (Abeles et al., 1992; Vandenbussche et al., 2012). In general, ethylene tends to function as an inhibitor of vegetative growth but also, promotes reproductive growth, accelerating the lifecycle, especially under environmental stress. Over a century ago, Neljubow (1901) first discovered that ethylene inhibits hypocotyl elongation of pea (Pisum sativum) seedlings, enhances horizontal growth, and leads to radial swelling in the dark, which is known as the triple response. Based on this discovery, a series of mutants was identified in Arabidopsis (Arabidopsis thaliana), and the classical signaling transduction model was established by genetic analysis. In the absence of ethylene, negative regulator Constitutive Triple Response1 (CTR1) tightly interacts with receptor complex (Clark et al., 1998), phosphorylates the central factor Ethylene-Insensitive2 (EIN2), and prevents its translocation into nucleus, thus blocking ethylene signaling transduction. In the presence of ethylene, receptors and CTR1 are inactivated, leading to the dephosphorylation and cleavage of EIN2. The C terminus of EIN2 is then translocated into the nucleus to activate downstream transcription factors EIN3/Ethylene-Insensitive3-Like (EIL)s followed by expression of target genes (Ju et al., 2012; Qiao et al., 2012; Wen et al., 2012). In this pathway, the ethylene receptors EIN2 and EIN3 are regulated by ubiquitination and proteasome-mediated degradation (Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004; Binder et al., 2007; Chen et al., 2007; Kevany et al., 2007; Qiao et al., 2009; An et al., 2010).
As for the role of ethylene in the regulation of vegetative growth, one of the most prominent pieces of evidence is the variant seedling morphology of signaling mutant plants. In Arabidopsis, mutant plants with enhanced ethylene response, such as ethylene response1-6 (etr1-6) and ctr1-1, often show retarded growth, leading to dwarfed seedlings and smaller leaves compared with the wild type (Kieber et al., 1993; Hua and Meyerowitz, 1998). On the contrary, etr1-1, ein2-1, ein3-1, and other ethylene-insensitive or low-sensitivity mutants often show accelerated seedling growth (Chang et al., 1993; Chao et al., 1997; Alonso et al., 1999). Unlike ctr1-1, ein2-1, or ein3-1, mutants of individual ethylene receptors show only weak effects on seedling growth because of the functional redundancy of different receptors (Hua and Meyerowitz, 1998; Hall and Bleecker, 2003). However, each receptor seems to have its own features of protein structure and expression pattern and therefore, may play unique roles in addition to the general role of ethylene perception (Shakeel et al., 2013). In Arabidopsis, mutation and transgenic analysis illustrate that subfamily I receptors play more significant roles in ethylene-regulated growth responses than subfamily II receptors do, probably because of the more efficient activation of CTR1 by subfamily I receptors (Hua and Meyerowitz, 1998; Hall and Bleecker, 2003; Wang et al., 2003; Qu et al., 2007). Additionally, the kinase activity of ETR1 may play a role in the activation of CTR1 (Hall et al., 2012).
Moreover, subfamily I receptors have stronger associations with CTR1 than subfamily II receptors (Clark et al., 1998; Cancel and Larsen, 2002). A similar preference is observed for the associations between ethylene receptors and CTRs in tomato (Solanum lycopersicum; Lin et al., 2008a, 2009b). In addition, ctr1 loss-of-function mutants continue to exhibit a residual ethylene response (Larsen and Chang, 2001; Larsen and Cancel, 2003), and the quadruple ethylene receptor loss-of-function mutant etr1 etr2 ein4 ethylene response sensor2 (ers2) and the etr1 ers1 double mutant display a more severe phenotype than the ctr1 loss-of-function mutants (Hua and Meyerowitz, 1998; Hall and Bleecker, 2003). Therefore, other than the classical CTR1-dependent pathway, other pathways might exist, possibly through unique receptor-interacting factors (Ju et al., 2012). One possible pathway is the two-component signaling pathway involving the phosphorelay transduction (Hass et al., 2004; Mason et al., 2005; Scharein et al., 2008; Scharein and Groth, 2011), but direct evidence is still lacking. Gel filtration analysis of ethylene receptor complexes in Arabidopsis further indicates that isoform-specific interacting proteins may exist for different receptors to mediate ethylene signaling (Chen et al., 2010). Indeed, Arabidopsis RTE1 was identified as an ETR1-specific interacting protein to mediate CTR1-independent ethylene response (Resnick et al., 2006, 2008; Zhou et al., 2007; Dong et al., 2010; Qiu et al., 2012). Expression of the N-terminal fragment of ETR1 (1-349) in ctr1-1 partially suppresses its constitutive ethylene-response phenotype, and this effect is dependent on the Ethylene Sensitivity1 (RTE1; Qiu et al., 2012). Another example is that Arabidopsis Tetatricopeptide Repeat Protein1 and tomato Tetratricopeptide Repeat Protein1 were identified as interacting factors with subfamily I ethylene receptors to modulate ethylene response and plant development (Lin et al., 2008b, 2009a). We recently showed that tobacco Histidine Kinase1 (NTHK1) Ethylene Receptor-interacting Protein2 (NEIP2), an ankyrin domain protein, interacts with subfamily II receptors to regulate plant response to abiotic stresses in tobacco (Nicotiana tabacum; Cao et al., 2015).
Previously, we studied roles of tobacco subfamily II ethylene receptor NTHK1 in plant growth and stress response. The transcripts of NTHK1 were inducible under salt stress and wounding (Zhang et al., 1999, 2001). Overexpression of NTHK1 in both tobacco and Arabidopsis exhibited reduced ethylene sensitivity, accelerated growth, and increased salt sensitivity of transgenic plants (Xie et al., 2002; Cao et al., 2006, 2007). The kinase domain and Ser/Thr kinase activity of NTHK1 were differentially required for salt response and seedling growth (Zhou et al., 2006; Chen et al., 2009). Surprisingly, NTHK1 seems to play more prominent roles in these responses than the subfamily I member NtETR1 (Chen et al., 2009). Characterized downstream factors involved in NTHK1-regulated stress response and plant growth processes include two transcription factors AtNAC2 and AtMYB59 (He et al., 2005; Mu et al., 2009), an NIMA-related kinase NEK6 (Zhang et al., 2011), and an ankyrin domain protein NEIP2 (Cao et al., 2015). To further elucidate the mechanism underlying the function of NTHK1, we initiated a yeast (Saccharomyces cerevisiae) two-hybrid assay to screen components that interact with NTHK1. Finally, a translationally controlled tumor protein (TCTP) was identified and further investigated. NtTCTP protein accumulation is induced by ethylene treatment. Overexpression of NtTCTP promotes seedling growth, mainly through the control of cell proliferation. Genetic analysis reveals that NtTCTP is required for NTHK1-mediated ethylene response and seedling growth.
RESULTS
Screening and Identification of NTHK1-Interacting Proteins
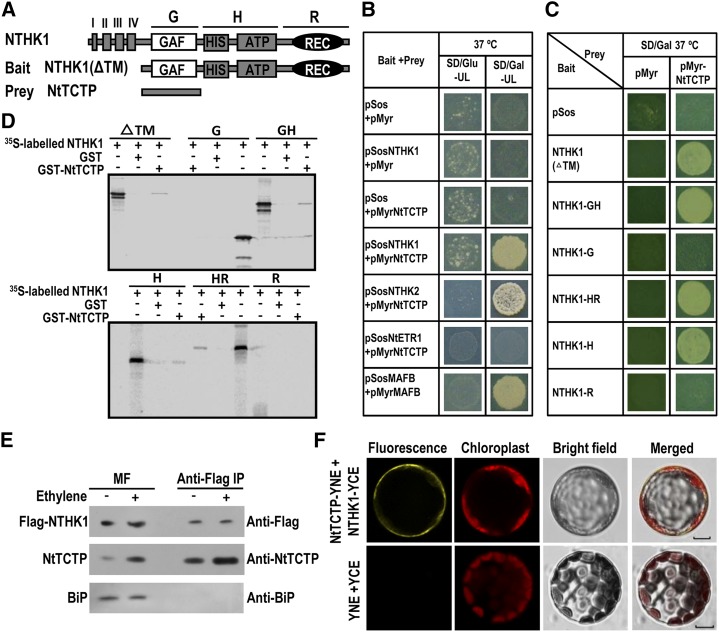
To unravel the mechanism underlying the role of NTHK1 in regulating ethylene response and plant growth, we screened NTHK1-interacting proteins using a yeast two-hybrid assay. The NTHK1 coding region lacking the transmembrane domains (145–762 amino acids) was used as the bait in a yeast CytoTrap two-hybrid system (Fig. 1A). Two TCTPs that had been screened out for several times were selected for additional investigation (Fig. 1, A–C). The two TCTPs, named NtTCTP1 and NtTCTP2, are highly similar, with only two amino acid variations. Sequence blast in all three tobacco databases (http://solgenomics.net/) showed that there are four TCTPs (NtTCTP1, NtTCTP2, NtTCTP3, and NtTCTP4) in the tobacco genome. NtTCTP3 and NtTCTP4 are highly homologous to each other but less similar to NtTCTP1 and NtTCTP2 (Supplemental Fig. S1). Compared with TCTPs in Arabidopsis, NtTCTP1 and NtTCTP2 are more similar to AtTCTP1 (AT3G16640), whereas NtTCTP3 and NtTCTP4 are more similar to AtTCTP2 (AT3G05540), which was supposed to be a nonfunctional pseudo-gene (Berkowitz et al., 2008; Supplemental Fig. S1). For convenience, NtTCTP is used to represent NtTCTP1, whereas NtTCTP3 and NtTCTP4 together are represented by tobacco protein translationally controlled tumor protein-like (NtTCTPL) in this study.
Figure 1.
NtTCTP interacts with tobacco ethylene receptor NTHK1. A, Schematic representation showing the structure of NTHK1, the bait, and the prey NtTCTP. Gray box I shows the putative signal peptide; gray boxes II to IV show transmembrane regions. B, NtTCTP interacts with NTHK1 and NTHK2 but not with NtETR1 in the yeast two-hybrid assay. Transformants grown on SD/Gal-UL plate but not on SD/Glu-UL plate at 37°C indicate positive interactions. The combination of pSosMAFB plus pMyrMAFB was used as the positive interaction control; combinations of both empty plasmids, empty pSos plus pMyrNtTCTP, and empty pMyr plus pSosNTHK1 were used as negative interaction controls. C, The kinase domain of NTHK1 is indispensable for interaction with NtTCTP in the yeast two-hybrid assay. Combination of empty pSos plus pMyrNtTCTP and combinations of empty pMyr plus different NTHK1 truncated versions were used as negative interaction controls. D, NTHK1 kinase domain mediates interaction with NtTCTP by GST pull-down assay. Equal amounts of GST-NtTCTP or GST were incubated with 35S-labeled different NTHK1-truncated versions, and 35S-labeled NTHK1 truncations were loaded as input controls. E, Identification of the interaction between NTHK1 and NtTCTP by Co-IP assay. Flag-NTHK1 was expressed in tobacco leaves through Agrobacterium spp.-mediated transformation. After expression for 2 d, seedlings were treated with 100 μL L−1 ethylene for 3 h. Treatment without ethylene was set simultaneously. MF was prepared from total cell extracts using ultracentrifugation and then incubated with anti-Flag antibody-linked agarose beads for immunoprecipitation of NTHK1-interacting protein complexes. MFs without immunoprecipitation were used as controls. As a negative control, anti-BiP antibody was used to detect the ER marker BiP. F, BiFC assay of the NTHK1-NtTCTP interaction. NtTCTP-YNE and NTHK1-YCE were coexpressed in tobacco leaf protoplasts, and yellow fluorescence signal was detected by an LCSM. YNE and YCE were coexpressed as a negative interaction control. MAFB, V-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B; Co-IP, coimmunoprecipitation; BiP, immunoglobulin-binding protein.
NtTCTP encodes a small protein containing 168 amino acids, with a predicted pI of 4.33 and an Mr of 18.8 kD (http://web.expasy.org/compute/pi/). Both National Center for Biotechnology Information CD search and Sanger Pfam search showed that NtTCTP possesses a TCTP superfamily domain (1–165 amino acids). Compared with TCTPs from other plants and animals, NtTCTP is more similar to plant TCTP proteins (AtTCTP, soybean [Glycine max] TCTP, rice [Oryza sativa] TCTP, and maize [Zea mays] TCTP), with many diverged amino acid substitutions in previously characterized functional domains of nonplant TCTPs (Supplemental Fig. S2). NtTCTP has more than 75% identity with TCTP homologs from Arabidopsis, soybean, rice, maize, and some other plants but less than 37% identity with those from fruit fly, mouse, human, and other animals. All of these data imply that plant TCTPs may evolve divergently from animals and have different functions.
There are several ethylene receptors in tobacco, among which NTHK1 and NTHK2 belong to subfamily II receptors, and NtETR1 is a subfamily I receptor (Xie et al., 2003; Zhang et al., 2004; Chen et al., 2009). To test the interactions between NtTCTP and different ethylene receptors, the coding regions of these receptors without transmembrane domains were cloned into pSos vector and cotransformed, respectively, with the prey pMyr-NtTCTP plasmid into yeast cdc25H cells. Transformants harboring either pSos-NTHK1 or pSos-NTHK2 plus pMyr-NtTCTP could grow on SD/Gal-UL but not SD/Glu-UL medium at 37°C, whereas transformants harboring pSos-NtETR1 plus pMyr-NtTCTP could not grow on SD/Gal-UL medium at 37°C (Fig. 1B). This result indicates that NtTCTP could interact with the two subfamily II receptors NTHK1 and NTHK2 but not with the subfamily I receptor NtETR1.
NTHK1 has four hydrophobic regions (I–IV), a GAF domain, a kinase domain (HIS plus ATP), and a receiver domain (REC) from the N terminus to the C terminus (Fig. 1A). To examine which domain was critical for the interaction with NtTCTP, the coding regions of various truncated versions of NTHK1 were cloned into the pSos vector as baits for the yeast two-hybrid assay. As shown in Figure 1C, transformants harboring pSos-NTHK1(ΔTM) (−GAF-HisATP [−GH], −HisATP-RD [−HR], and − HisATP [−H]) plus pMyr-NtTCTP grew well on selective SD/Gal-UL medium at 37°C, whereas transformants containing NTHK1 (−GAF [−G] and −RD [−R]) plus pMyr-NtTCTP could not grow. This result indicates that the kinase domain of NTHK1 is necessary for the interaction with NtTCTP. To confirm the interaction between NtTCTP and NTHK1, a glutathione S-transferase (GST) pull-down assay was carried out. Both GST-NtTCTP fusion proteins and [35S]Met-labeled NTHK1 proteins were incubated with GST affinity resin. As a result, GST-NtTCTP could pull down ΔTM, GH, H, and HR truncated versions of NTHK1 but not G or R truncated version. As a negative control, GST could not pull down any versions of NTHK1 (Fig. 1D). These results indicate that the kinase domain of NTHK1 is the major interaction domain with NtTCTP.
We further used the coimmunoprecipitation (Co-IP) assay to examine in vivo interactions between NTHK1 and NtTCTP. Flag-NTHK1 was expressed in tobacco leaves for immunoprecipitation. As shown in Figure 1E, NtTCTP could be coimmunoprecipitated with the Flag-NTHK1 protein, and this confirmed the interaction between NtTCTP and NTHK1. Interestingly, ethylene treatment enhances the expression of NtTCTP and promotes its interaction with NTHK1 (Fig. 1E). In addition, ethylene treatment enhanced the localization of NtTCTP to the membrane system (Supplemental Fig. S3). A bimolecular fluorescence complementation (BiFC) assay was also performed to confirm the interaction of NtTCTP and NTHK1. A yellow florescence signal was detected using confocal microscopy when NTHK1-C terminus of yellow fluorescence protein (YCE) and NtTCTP-N terminus of yellow fluorescence protein (YNE) were coexpressed in tobacco protoplasts (Fig. 1F). However, no yellow florescence was observed in YCE- and YNE-coexpressed protoplasts under the same condition, indicating the specificity of the interaction between NTHK1 and NtTCTP in tobacco protoplasts.
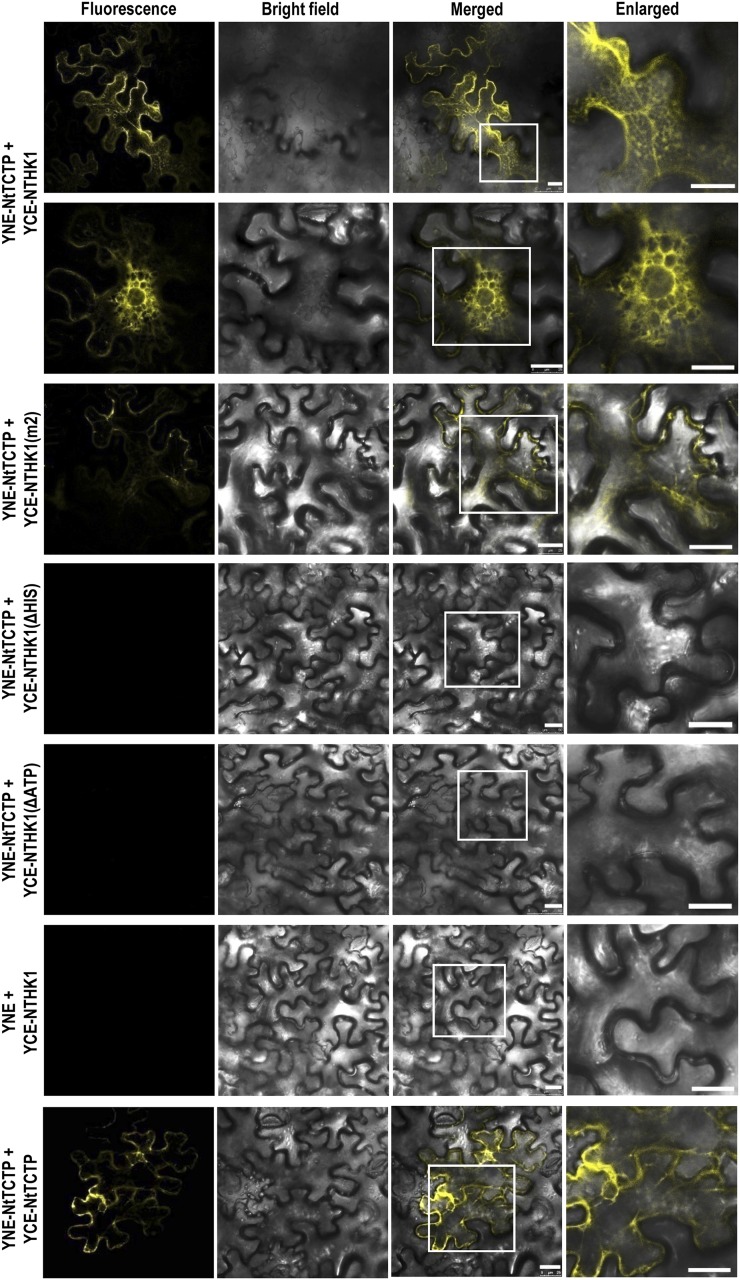
To obtain more substantial information on the interaction between NtTCTP and NTHK1, additional BiFC assays in Nicotiana benthamiana leaves were carried out. The results showed that a yellow fluorescent protein (YFP) signal could be detected at endoplasmic reticulum (ER)-like structures when YNE-NtTCTP and YCE-NTHK1 were coexpressed in tobacco leaf cells (Fig. 2), suggesting their interaction at the ER, which is consistent with the ER localization of the NTHK1 (Cao et al., 2015). Moreover, the mutation of NTHK1 (m2), which eliminates its kinase activity (Chen et al., 2009), reduced the interaction remarkably, and the deletion of His or ATP-binding subdomain completely disrupted the interaction between NTHK1 and NtTCTP (Fig. 2). This indicates that NtTCTP interacts with NTHK1 at the ER and that the kinase domain of NTHK1 is required for their interaction. Moreover, NtTCTP could possibly form a dimer and/or even a polymer by interacting with itself (Fig. 2).
Figure 2.
The kinase domain of NTHK1 is indispensable for the interaction with NtTCTP. Fusing with the C-terminal one-half of YFP, the full-length kinase-inactivated mutation (m2), His-truncated (ΔHIS), and ATP-truncated (ΔATP) types of NTHK1 were coexpressed separately with the YNE-NtTCTP in N. benthamiana leaves through Agrobacterium spp.-mediated transformation. Coexpression of YNE-NtTCTP and YCE-NtTCTP was used as a positive interaction control, and coexpression of YNE and YCE-NTHK1 was used as a negative interaction control. Fluorescence signals were detected using a confocal microscope. Column 4 shows enlargements of the parts of the ER area marked by white frames. Bar = 25 μm.
Expression Pattern and Subcellular Localization of NtTCTP
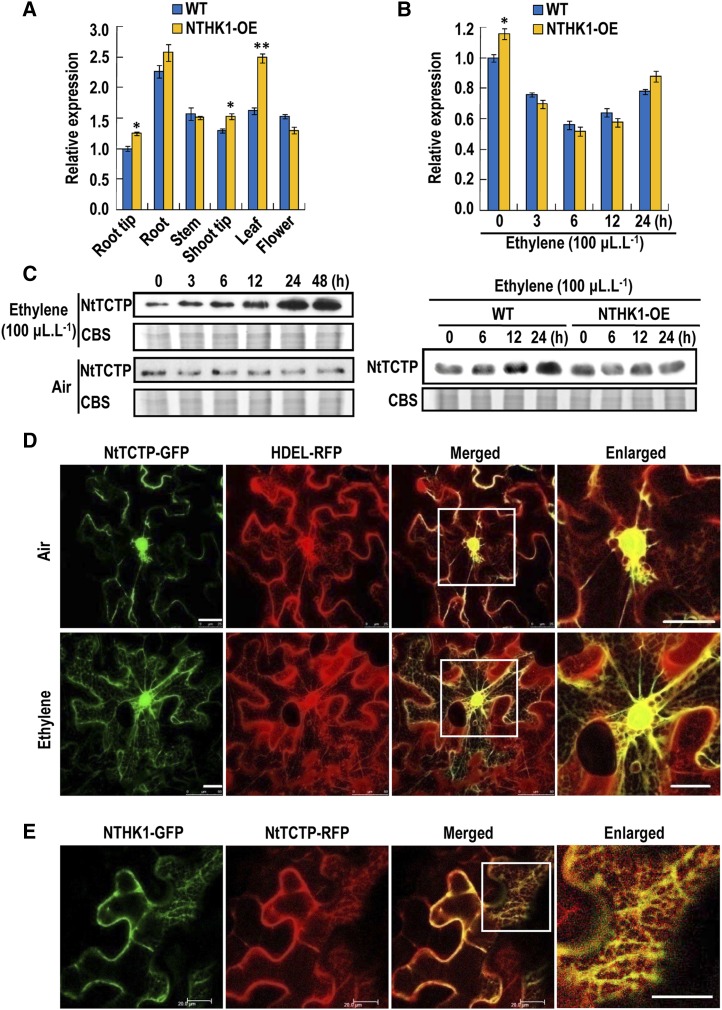
NtTCTP transcript levels were examined in different plant organ/tissues by quantitative real-time (qRT)-PCR. Figure 3A shows that transcripts of NtTCTP were more abundant in root than in other organs of wild-type tobacco plants. In NTHK1-overexpressing tobacco plants (NTHK1-OE; Cao et al., 2006), the NtTCTP transcript levels were more abundant in root and leaf than in the other organs tested. Specifically, there was a significant increase of NtTCTP transcript level in leaf compared with the wild type (Fig. 3A).
Figure 3.
NtTCTP expression pattern and subcellular localization. A, NtTCTP transcript level in different organs by qRT-PCR. Root tip, root, stem, shoot tip, and leaf were all from 48-DAS tobacco plants grown in soil; root tip was 1-cm long from the primary root, and flower indicates only open flowers. Transcript levels of NtTCTP were normalized to NttubA1 and relative to the root tips of wild-type (WT) plants. The data are expressed as the means ± sd from three samples. Student’s t tests between wild-type and NTHK1-overexpressing (NTHK1-OE) plants were performed. Three biological replicates were performed with similar results, and one of them was shown. *, P < 0.05; **, P < 0.01. B, Expression pattern of NtTCTP at transcript level under ethylene treatment by qRT-PCR. The 12-DAS seedlings grown on one-half-strength MS were treated with 100 μL L−1 ethylene for different durations, and total RNA was extracted for qRT-PCR. Transcript levels of NtTCTP were normalized to NttubA1 and relative to the untreated wild-type plants. The data are expressed as the means ± sd from three independently treated samples. Student’s t tests between wild-type and NTHK1-overexpressing plants at each time point were performed. Three biological replicates were performed with similar results, and one of them was shown. *, P < 0.05. C, NtTCTP protein expression pattern under ethylene treatment by immunoblotting. Ethylene treatment was the same as in B. Anti-NtTCTP mouse polyclonal antibody was used for NtTCTP protein detection. D, Subcellular localization of NtTCTP. Both NtTCTP-GFP and HDEL-RFP (an ER marker protein) were expressed in N. benthamiana leaves through Agrobacterium spp.-mediated transformation. Fluorescence signals were detected using a confocal microscope. For ethylene treatment, transformed N. benthamiana plants were treated with 100 μL L−1 ethylene for 3 h before observation. Column 4 shows the enlargements of the nuclear areas marked by the white frames. Bar = 25 μm. E, Colocalization of NTHK1 and NtTCTP at the ER. Both NTHK1-GFP and NtTCTP-RFP were expressed in N. benthamiana leaves through Agrobacterium spp.-mediated transformation. Fluorescence signals were detected using a confocal microscope. Column 4 shows the enlargement of the part of the ER area marked by the white frame. Bar = 20 μm. CBS, Coomassie Blue staining of the total proteins; NttubA1, N. tabacum tubulin alpha-1.
Because NtTCTP is an ethylene receptor-interacting protein, we examined whether NtTCTP is responsive to ethylene at both transcript and protein levels. Under ethylene treatment, NtTCTP transcript level declined continuously for 12 h in both the wild type and NTHK1-overexpressing plants and then tended to recover gradually (Fig. 3B). In contrast, the NtTCTP protein was induced in wild-type plants over a time course of ethylene treatment (Fig. 3C). However, in NTHK1-overexpressing plants, ethylene treatment could not induce high accumulation of NtTCTP like in wild-type tobacco plants (Fig. 3C), suggesting that overexpression of NTHK1 led to reduced ethylene sensitivity.
To characterize the subcellular localization of NtTCTP, NtTCTP-GFP fusion protein was expressed in tobacco protoplasts or N. benthamiana leaf cells. The GFP signal could be detected in the nucleus, cytoplasm, and ER (Supplemental Fig. S4). Furthermore, NtTCTP colocalized with the ER marker HDEL (Fig. 3D). Ethylene treatment enhanced the accumulation of NtTCTP at the ER (Fig. 3D; Supplemental Fig. S4B). This result is consistent with results from the Co-IP assay (Fig. 1E; Supplemental Fig. S3). Considering the interaction between NtTCTP and NTHK1, we examined whether NTHK1 and NtTCTP could colocalize in cells. When NTHK1-GFP and NtTCTP-red fluorescent protein (RFP) were coexpressed in N. benthamiana leaf cells, yellow signals merged by the green and red fluorescence were detected at the ER (Fig. 3E), indicating that NtTCTP colocalizes with NTHK1 at the ER.
Taking together the results from the Co-IP and BiFC assays (Figs. 1E and 2; Supplemental Fig. S3), we conclude that NtTCTP interacts with NTHK1 mainly at the ER. Ethylene treatment could induce the accumulation of NtTCTP proteins and enhance its interaction with NTHK1.
NtTCTP and NTHK1 Participate in Regulation of Plant Growth
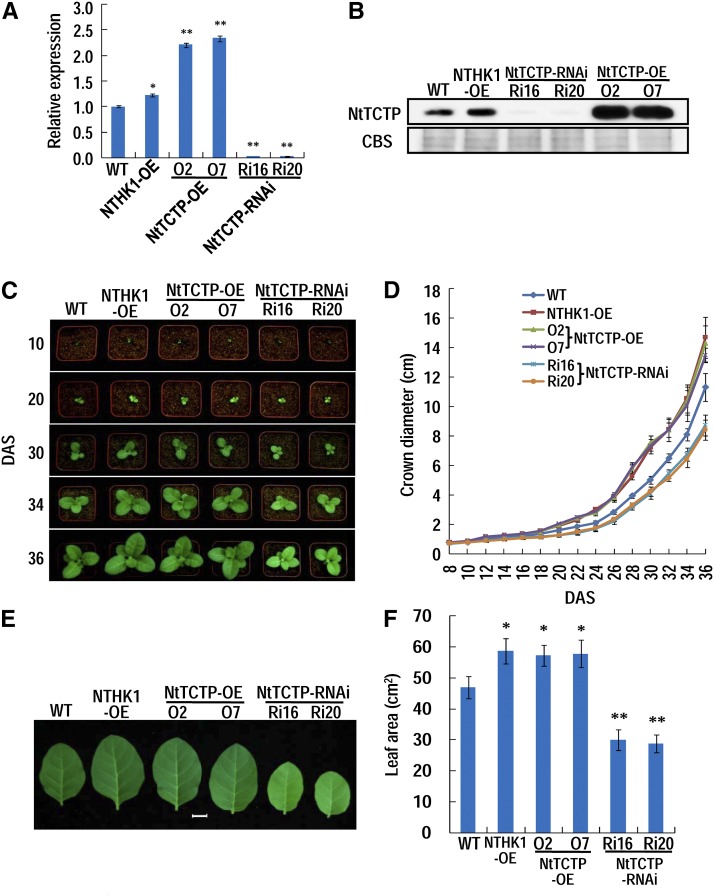
As an important phytohormone, ethylene is indispensable for the fine tuning of plasticity during plant growth and development (Abeles et al., 1992). When ethylene biosynthesis and/or signaling are disturbed, plants often grow abnormally, appearing as promoted vegetative growth but delayed reproductive growth or the opposite (Kieber et al., 1993; Chao et al., 1997; Hua and Meyerowitz, 1998). Our previous work showed that overexpression of NTHK1 reduced ethylene sensitivity and promoted seedling growth both in tobacco and Arabidopsis (Cao et al., 2006, 2007). As an NTHK1-interacting protein, NtTCTP was further investigated for its role in regulating plant growth. Transgenic tobacco lines overexpressing or silencing NtTCTP were generated, and expression levels were examined by northern-blotting analysis (Supplemental Fig. S5A). From these lines, two NtTCTP-overexpressing lines (NtTCTP-OE: O2 and O7) and two NtTCTP-silencing lines (NtTCTP-RNA interference [RNAi]: Ri16 and Ri20) were selected for the following experiments, and expression levels of NtTCTP were reconfirmed by qRT-PCR and immunoblotting assay (Fig. 4, A and B). It should be noted that, in the selected NtTCTP-silencing lines, the expression levels of both NtTCTP1 and NtTCTP2 were inhibited. However, the transcript level of NtTCTPL (NtTCTP3 and NtTCTP4) was only slightly down-regulated compared with the dramatic down-regulation of NtTCTP (Supplemental Fig. S5B). The NTHK1-overexpressing line (NTHK1-OE) 16-4 was also used for comparative analysis (Cao et al., 2006).
Figure 4.
NtTCTP and NTHK1 promote plant growth. A, NtTCTP expression levels in NtTCTP-overexpressing (NtTCTP-OE) and RNAi transgenic lines were detected by qRT-PCR. The 12-DAS tobacco seedlings were used for total RNA extraction and qRT-PCR. Expression levels of NtTCTP were normalized to NttubA1 and relative to the wild-type (WT) seedlings. The data are expressed as the means ± sd from three samples. Student’s t tests between the wild type and each transgenic line were performed. Three biological replicates were performed with similar results, and one of them was shown. *, P < 0.05; **, P < 0.01. B, NtTCTP protein level in different transgenic lines. NtTCTP protein level was detected by immunoblotting with anti-NtTCTP mouse polyclonal antibody. C, Comparison of the growth of different tobacco lines. Plants at various stages are shown. The pot size is 8 × 8 cm. D, Measurement of seedling size at different stages. E, Photographs of the sixth true leaves from 48-DAS tobacco plants. Bar = 2 cm. F, Measurement of the sixth true leaf area from 48-DAS tobacco plants. For D and F, each data point is the mean ± se from three biological replicates (each replicate has 10 seedlings). Asterisks indicate a significant difference from the wild type (Student’s t test). CBS, Coomassie Blue staining of the total proteins; NttubA1, N. tabacum tubulin alpha-1; *, P < 0.05; **, P < 0.01.
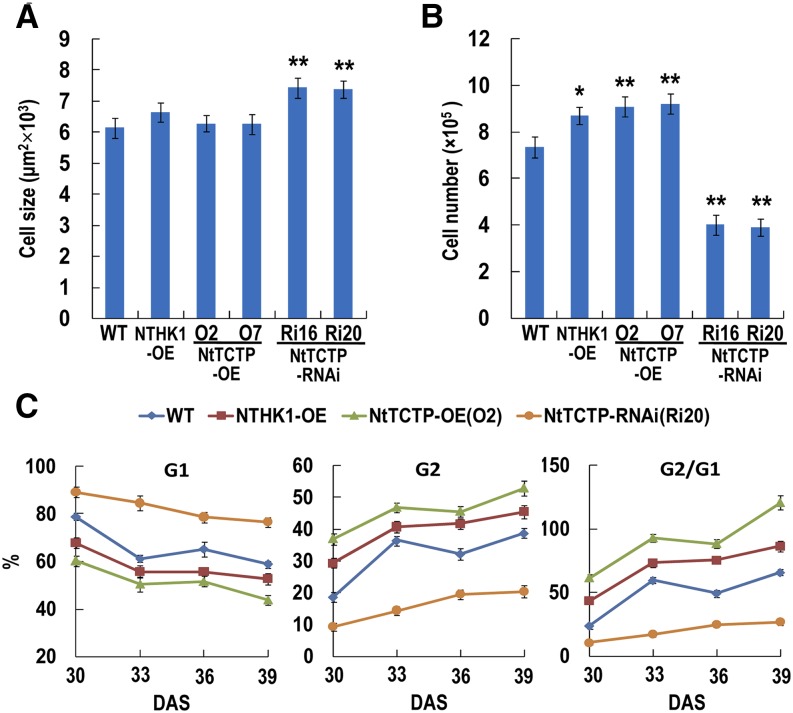
Morphologically, both NtTCTP- and NTHK1-overexpressing seedlings were larger, whereas NtTCTP-silencing seedlings were smaller than those of the wild type, especially at the approximately 30-d after stratification (DAS) stage (Fig. 4, C and D). The leaves of NtTCTP- and NTHK1-overexpressing seedlings were also larger than those of the wild type. In contrast, the leaves of NtTCTP-silencing seedlings were smaller than those of the wild type (Fig. 4, E and F). Two NTHK1-silencing lines also showed retarded growth and smaller leaves compared with the wild-type seedlings (Supplemental Fig. S6). These results indicate that both NtTCTP and NTHK1 play positive roles in vegetative growth. Because leaf area is mainly determined by cell number and cell size, we further used the sixth true leaves from 48-DAS tobacco plants (the growth of six true leaves was nearly stopped at this stage in our experimental condition) to measure the two parameters of the upper epidermis. The epidermal cell sizes of NtTCTP-silencing lines were significantly larger than, whereas the epidermal cell sizes of the NTHK1- and NtTCTP-overexpressing lines were similar to those of the wild type at this stage (Fig. 5A; Supplemental Fig. S7A). In contrast, the leaf upper epidermal cell numbers of NTHK1- and NtTCTP-overexpressing lines were larger, whereas the cell numbers of NtTCTP-silencing lines were smaller than those of the wild type (Fig. 5B). These results indicate that the final large leaf phenotype of NTHK1- and NtTCTP-overexpressing plants is mainly caused by increases in cell numbers.
Figure 5.
NtTCTP and NTHK1 control cell cycle and cell proliferation. A, Quantification of epidermal cell size from different tobacco lines. The sixth true leaves from 48-DAS tobacco plants were collected for SEM analysis of leaf upper epidermal cells. The data are expressed as the means ± se from three independent experimental groups (three leaves per group, four independent areas per leaf, and at least 30 cells per each area were measured using ImageJ 1.38X). **, Significant difference from the wild type (WT; Student’s t test; P < 0.01). B, Comparison of epidermal cell numbers in each leaf. The data are expressed as the means ± se of three independent experimental groups (three leaves per group). Asterisks indicate significant differences from the wild type (Student’s t test). *, P < 0.05; **, P < 0.01. C, Percentage of G1 and G2 cells and G2 to G1 ratio in the sixth true leaves of various tobacco lines. G1 and G2 indicate different phase of the cell cycle. The data are expressed as the means ± se from three independent experimental groups (eight leaves per group).
To investigate the mechanisms underlying significant cell number variation, we examined DNA ploidy distribution in cells of the sixth true leaves from different lines by flow cytometry. During the rapid expanding period, G1 cells of both NtTCTP-overexpressing (O2) and NTHK1-overexpressing (16-4) lines were less than those of the wild type, and the G1 cells of the NtTCTP-overexpressing line were even less than the NTHK1-overexpressing line (Fig. 5C; Supplemental Fig. S7B). In contrast, the G1 cells of NtTCTP-silencing line (Ri16) were significantly more than those of the wild type. The situations of G2 cells were just the opposite in all tested tobacco lines (Fig. 5C; Supplemental Fig. S7B). Along with the leaf expanding, the ratio of G2 to G1 steadily increased in all tobacco lines, with those in NtTCTP- and NTHK1-overexpressing lines being higher and those in the NtTCTP-silencing line being lower than those in the wild type (Fig. 5C). These results indicate that NTHK1 and NtTCTP may promote cell cycle and cell proliferation for the enhancement of leaf and seedling growth.
Effects of 1-Aminocyclopropane-1-Carboxylic Acid on NtTCTP- and NTHK1-Regulated Leaf Growth
As stated above, both NtTCTP and NTHK1 play positive roles in seedling growth, mainly through promotion of cell proliferation. Because NTHK1 is a member of subfamily II ethylene receptors, we examined whether ethylene could influence the roles of NtTCTP and NTHK1 in regulation of seedling growth. Tobacco seeds were germinated on one-half-strength Murashige and Skoog medium (MS) for continuous growth. The third true leaves of all tobacco lines began to sprout at the 12-DAS stage, and seedlings at this stage were transplanted to fresh one-half-strength MS and treated with 1-aminocyclopropane-1-carboxylic acid (ACC). Then, the third true leaves were collected every day until 22 DAS for measurements of leaf area, upper epidermal cell size, and cell number (Supplemental Fig. S8).
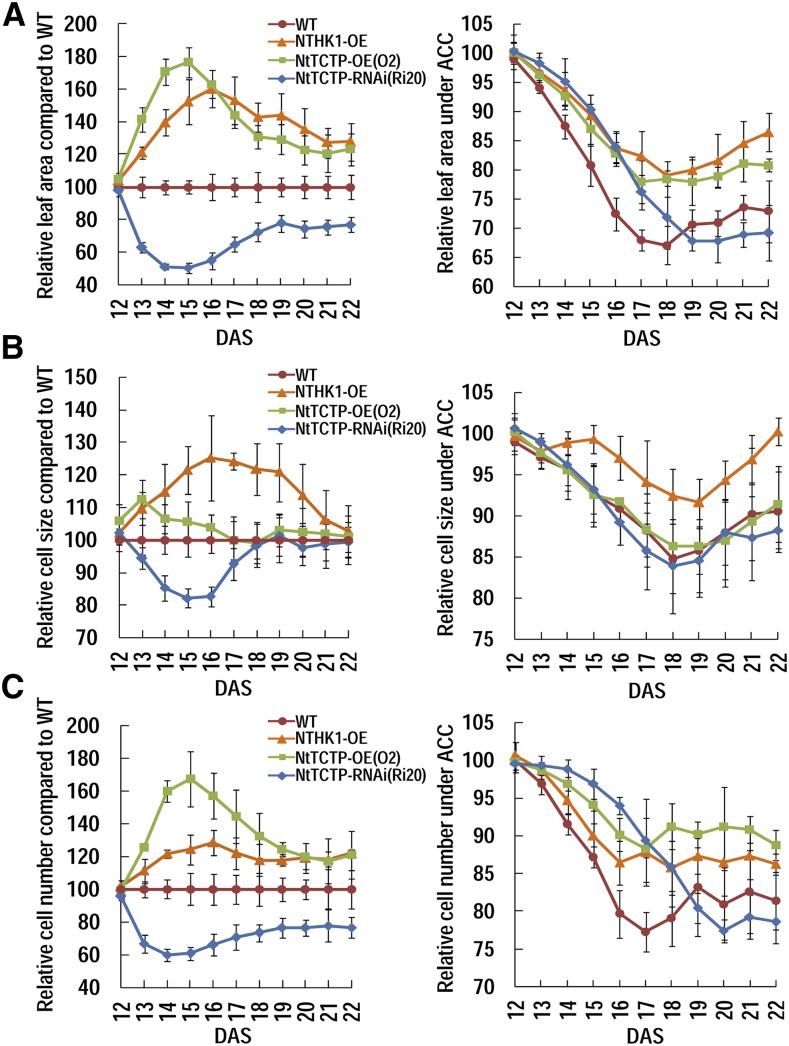
The leaf areas, upper epidermal cell sizes, and cell numbers of NTHK1-overexpressing, NtTCTP-overexpressing (O2), and NtTCTP-silencing (Ri20) tobacco lines were calculated as relative values compared with those of the wild type (Fig. 6, left). Overall, under normal growth conditions, the leaf areas of NTHK1- and NtTCTP-overexpressing lines were larger than those of the wild type, and the peaks were at 16 and 15 DAS, respectively. The leaf area of the NtTCTP-silencing line was smaller than that of the wild type and exhibited a peak of decrease at 14 to 15 DAS, just opposite to the pattern of the NtTCTP-overexpressing line, indicating that NtTCTP is essential for leaf growth (Fig. 6A, left). In the NTHK1-overexpressing line, the peak leaf size increase occurred 1 d later than that in the NtTCTP-overexpressing line and showed a slightly different growth pattern (Fig. 6A, left). The variation of cell size was also compared with that of the wild type. The relative leaf cell size of the NtTCTP-overexpressing line showed only a small increase at early days, whereas the relative leaf cell size of the NtTCTP-silencing line decreased evidently before 17 DAS and then returned to the wild-type level, suggesting that NtTCTP is required for cell expansion at the early stage of leaf growth (Fig. 6B, left). In the NTHK1-overexpressing plants, leaf cells were larger than those of the wild type during the whole period, with a peak of increase at 16 DAS, and finally returned to the wild-type level at 22 DAS (Fig. 6B, left).
Figure 6.
Comparison of the leaf growing pattern between different tobacco lines in the presence or absence of the ethylene precursor ACC. At the stage of 12 DAS, the third true leaves of all tobacco lines begin to sprout. Then, seedlings were transplanted to control or ACC-containing medium for further growth, and the third true leaves were collected every day for measurements of leaf area, cell size, and cell numbers. A, Relative leaf area of each transgenic line compared with the wild type (WT) and relative leaf area under ACC treatment compared to the mock. B, Relative cell size of each transgenic line compared with the wild type and relative cell size under ACC treatment compared with the mock. C, Relative cell number of each transgenic line compared with the wild type and relative cell number under ACC treatment compared with the mock. The data are expressed as the means ± se from three independent experimental groups. For leaf area, eight leaves per group were measured. For cell size and number calculation, the upper epidermis areas of four leaves from each group were photomicrographed and analyzed using ImageJ 1.38X; four independent areas averaged between the apical and basal positions were examined, and at least 30 cells per area were measured.
The leaf cell numbers were further compared with those of the wild type. The relative cell numbers of all transgenic tobacco lines showed similar patterns with their leaf areas, except that the increase in the NTHK1-overexpressing line was relatively less than that in the NtTCTP-overexpressing line (Fig. 6C, left). Together, at early stages, the leaf growth of the NtTCTP-overexpressing/silencing line is correlated with the cell number more largely than the cell size, whereas the NTHK1-overexpressing line depends on both the cell size and cell number. However, the final leaf areas of both the NtTCTP- and NTHK1-overexpressing lines seem to be substantially dependent on leaf cell numbers (Figs. 4F and 5, A and B).
In the presence of ACC, all of the relative parameters (leaf area, cell size, and cell number under ACC treatment compared with the nontreatment control at the corresponding time points) decreased during the treatment until the period of 17 to 20 DAS and then remained at stable levels or increased to different levels (Fig. 6, right). Compared with the wild type, the relative leaf areas and cell numbers of both the NtTCTP- and NTHK1-overexpressing lines were at higher levels, suggesting that the NtTCTP- and NTHK1-overexpressing lines show reduced sensitivity to ACC in the two parameters (Fig. 6, A, right and C, right). The two parameters were at lower levels for the NtTCTP-silencing lines, especially at the later stage of the treatment period, possibly reflecting slightly enhanced ACC sensitivity. It should be noted that the reduction of relative cell number in the NtTCTP-overexpressing line was even less than in the NTHK1-overexpressing line (Fig. 6C, right). As for relative cell size, the reduction pattern of both the NtTCTP-overexpressing and NtTCTP-silencing lines was similar to that of the wild type, whereas the relative cell size of the NTHK1-overexpressing line was only mildly decreased and then recovered to the level before ACC treatment, indicating a reduced sensitivity to ACC for cell size, which was specific for the NTHK1-overexpressing line (Fig. 6B, right). All of these results reveal that, although both NtTCTP and NTHK1 confer reduced sensitivities to ACC on leaf growth, NtTCTP functioned mainly through the regulation of cell number, and NTHK1 functioned through regulation of both cell number and cell size. It should be noted that the relative leaf area and cell number of the NtTCTP-silencing line decreased more slowly than those of the wild type before 18 to 19 DAS and then decreased to lower levels than the wild type (Fig. 6, A, right and C, right). However, the absolute leaf area and cell numbers of the NtTCTP-silencing line were always smaller than those of the wild type with or without ACC treatment (Supplemental Fig. S8).
NtTCTP and NTHK1 Regulate Root Growth and Lateral Root Formation
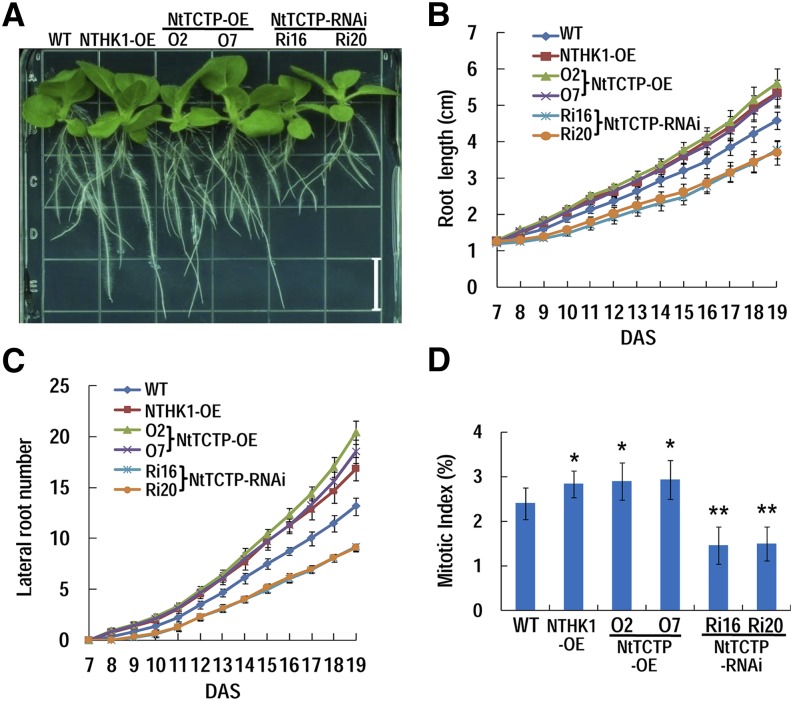
In addition to regulating leaf growth, the roles of NtTCTP and NTHK1 on root growth were also investigated (Fig. 7); 7-DAS tobacco seedlings were transplanted to fresh one-half-strength MS for continuous growing and placed vertically for convenient observation. NtTCTP-silencing lines showed retarded root growth, with significantly reduced primary root length and lateral root number (Fig. 7, A–C), whereas the performances of the NTHK1- and NtTCTP-overexpressing lines were just the opposite (Fig. 7). Then, the lateral root densities were calculated. The lateral root density of the NtTCTP-silencing lines was significantly lower than that of the wild type during the whole observation period. In contrast, the NtTCTP-overexpressing lines showed significantly increased lateral root density compared with that of the wild type (Supplemental Fig. S9). The lateral root density of the NTHK1-overexpressing line also increased significantly before 12 DAS (Supplemental Fig. S9).
Figure 7.
NtTCTP and NTHK1 regulate root growth and development. A, Growth of different tobacco lines on vertical one-half-strength MS plates for 20 DAS. Bar = 1.5 cm. B, Primary root length of various lines at the early stage of root development; 7-DAS seedlings were transferred to new one-half-strength MS plates and placed vertically for continuous growth. Seedlings were photographed every day until 20 DAS for examination of root parameters. The data are expressed as the means ± se from three independent experimental groups (n = 12). C, Lateral root numbers in different plant lines. Others are the same as in B. D, Measurement of the mitotic index of root tips from 7-DAS seedlings. The data are expressed as the means ± se from three independent experimental groups (n = 6). Asterisks indicate significant differences from the wild type (WT; Student’s t test). *, P < 0.05; **, P < 0.01.
Considering that both NTHK1 and NtTCTP promoted cell proliferation in leaves (Figs. 5 and 6), we further examined whether cell proliferation in roots was affected. The mitotic indices of root tips from 7-DAS seedlings were measured. The mitotic indices of the NTHK1- and NtTCTP-overexpressing lines were significantly higher than those of the wild type, whereas this parameter in the NtTCTP-silencing lines was significantly lower than that of the wild type (Fig. 7D). These results suggest that both NtTCTP and NTHK1 promote root growth through controlling cell proliferation.
NtTCTP and NTHK1 Confer Reduced Sensitivity to Ethylene
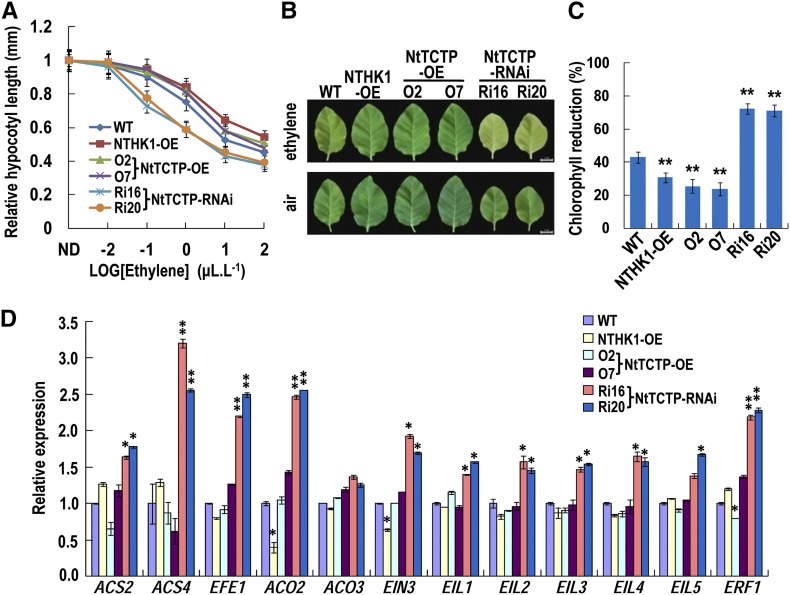
Our previous work showed that seedlings of the NTHK1-overexpressing lines were less sensitive to ACC treatment (Xie et al., 2002; Cao et al., 2007). We further analyzed whether the NTHK1-interacting protein NtTCTP affects the ethylene response. Under treatments with different concentrations of ethylene, hypocotyl lengths of 5-DAS tobacco etiolated seedlings were measured. The relative hypocotyl length of the NtTCTP- and NTHK1-overexpressing lines was significantly longer than that of the wild type under 1.0 μL L−1 or higher concentration of ethylene treatments (Fig. 8A). On the contrary, the NtTCTP-silencing lines showed significantly shortened hypocotyls under 0.1 μL L−1 or higher concentration of ethylene treatments (Fig. 8A). These results indicate that both NtTCTP and NTHK1 confer mildly reduced ethylene sensitivity in etiolated seedlings.
Figure 8.
NtTCTP and NTHK1 confer reduced ethylene sensitivity. A, Relative hypocotyl length of etiolated seedlings in response to ethylene treatments. The data are expressed as the means ± se (n > 30). Relative hypocotyls of NTHK1-overexpressing (NTHK1-OE) and NtTCTP-overexpressing (NtTCTP-OE) lines were significantly longer than the wild type (WT) at 1.0 μL L−1 or higher concentrations of ethylene; the relative hypocotyls of NtTCTP-silencing lines were significantly shorter than the wild type at ethylene concentrations of 0.1 μL L−1 or higher (Student’s t test; P < 0.01). B, Ethylene-induced leaf senescence of different tobacco lines. The sixth true leaves from 60-DAS plants were treated with 100 μL L−1 ethylene in sealed boxes for 5 d to induce senescence. Treatment in air was used as a control. Bar = 2 cm. C, Percentage of chlorophyll reduction compared with the control after ethylene treatment. The data are expressed as the means ± se from three independent experimental groups (six leaves per group). **, Significant difference from the wild type (Student’s t test; P < 0.01). D, Expressions of genes related to ethylene biosynthesis and signaling in various tobacco lines. The 12-DAS tobacco seedlings were used for total RNA extraction and qRT-PCR. Expression levels of all genes were normalized to NttubA1 and relative to wild-type seedlings. The data are expressed as the means ± sd from three samples. Student’s t tests between the wild type and each transgenic line were performed. Three biological replicates were performed with similar results, and one of them is shown. ND, Nondetected ethylene; NttubA1, N. tabacum tubulin alpha-1; *, P < 0.05; **, P < 0.01.
Because ethylene induces leaf senescence, we further examined this ethylene effect on tobacco lines. The sixth true leaves from 60-d-old tobacco plants (at approximately this stage, the sixth true leaves of tobacco matured and became senescent in our experimental condition) were detached and placed on wet filter paper in sealed boxes for ethylene treatment. After 5 d, the leaf senescence of the NTHK1- and NtTCTP-overexpressing lines was less severe than that of the wild type in the presence or absence of ethylene, whereas the leaf senescence of the NtTCTP-silencing lines was more severe than that of the wild type (Fig. 8B). After ethylene treatment, the chlorophyll reduction in the NTHK1- and NtTCTP-overexpressing lines was significantly lower than that in the wild type, whereas this parameter in NtTCTP-silencing lines was significantly higher (Fig. 8C; Supplemental Fig. S10), suggesting that both NTHK1 and NtTCTP confer reduced sensitivity to ethylene during the leaf senescent process.
Then, the transcript levels of genes involved in ethylene biosynthesis and signaling were examined by qRT-PCR. Transcript levels of four ethylene biosynthesis-related genes (1-Aminocyclopropane-1-Carboxylate Synthase2 [ACS2], ACS4, Ethylene Forming Enzyme [EFE], and (1-Aminocyclopropane-1-Carboxylate Oxidase2 [ACO2]), EIN3, EIL1 to EIL5, and ERF1 were significantly up-regulated in the NtTCTP-silencing lines, suggesting that NtTCTP is probably required for suppression of genes related to ethylene biosynthesis and/or signaling (Fig. 8D). However, overexpression of NtTCTP only slightly reduced the transcript level of the ACS4 gene but not the other genes listed above, and overexpression of NTHK1 reduced the transcript levels of ACO2 and EIN3 genes but not the others examined (Fig. 8D).
NtTCTP Is Necessary for NTHK1 in Regulation of Seedling Growth and Ethylene Response
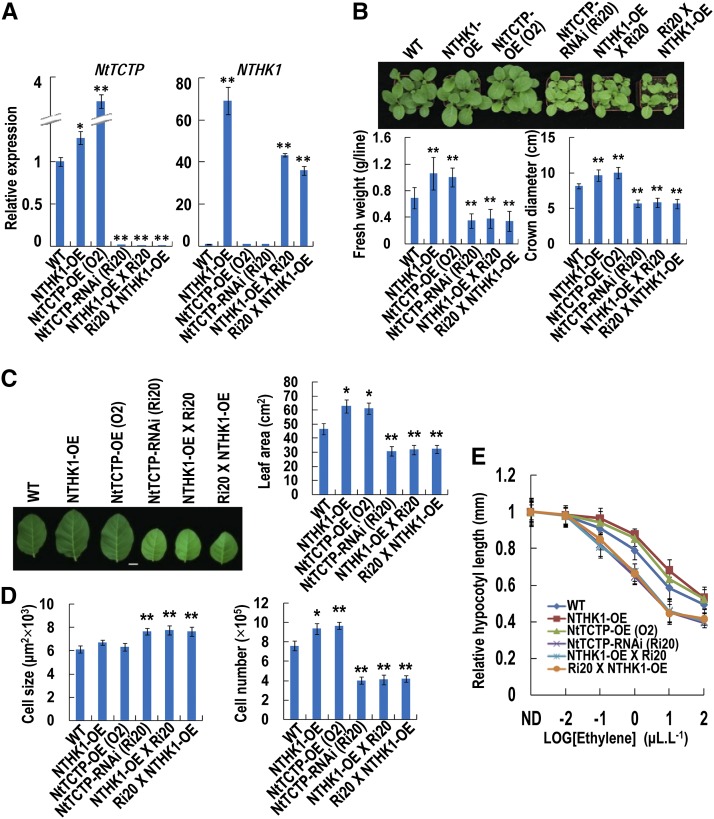
As an NTHK1-interacting protein, NtTCTP has a similar function to NTHK1 in reducing ethylene sensitivity and promoting plant growth (Figs. 4–8). To further elucidate their genetic interaction, we generated hybrids between the NTHK1-overexpressing line (16-4) and the NtTCTP-silencing line (Ri20; including NTHK1-OE × Ri20 and Ri20 × NTHK1-OE). In the two hybrids, the expression level of NTHK1 was increased approximately 40 times, whereas the expression level of NtTCTP was decreased more than 50 times compared with that of the wild type (Fig. 9A). Both hybrids showed significantly decreased crown diameter, fresh weight, and leaf area compared with the wild type but were indistinguishable from each other and the NtTCTP-silencing line (Fig. 9, B and C). The cell size and cell number of both hybrids were also very similar to those of the NtTCTP-silencing line (Fig. 9D; Supplemental Fig. S11). Ethylene response was further examined, and both hybrids showed enhanced sensitivity to ethylene, like the NtTCTP-silencing line, with significantly shorter hypocotyls than those of the wild type (Fig. 9E). These results illustrated that the silencing of NtTCTP eliminated the roles of NTHK1 in regulation of plant growth and ethylene response, suggesting that NtTCTP is required for the normal function of NTHK1.
Figure 9.
NtTCTP is required for NTHK1 to regulate plant growth and ethylene response. A, The expression levels of NtTCTP and NTHK1 in the F1 generation of the hybrids between the NTHK1-overexpressing (NTHK1-OE) line and the NtTCTP-silencing (NtTCTP-RNAi) line Ri20 were detected by qRT-PCR. Expression levels were normalized to NttubA1 and relative to the wild-type (WT) seedlings. The data are expressed as the means ± sd from three samples. Student’s t tests between the wild type and each tobacco line were performed. Three biological replicates were performed with similar results, and one of them is shown. *, P < 0.05; **, P < 0.01. B, Phenotypes of the F1 hybrids compared with wild-type and other tobacco lines. The F1 hybrids showed similar phenotypes on seedling size as the NtTCTP-silencing line. The 34-DAS seedlings of different tobacco lines growing in soil were photographed, and parameters (crown diameter and fresh weight) were measured. The data are expressed as the means ± se from three independent experiments (n = 12). **, Significant difference from the wild type (Student’s t test; P < 0.01). C, Comparison of leaf area in various plant lines. The sixth true leaves from 48-DAS tobacco plants were photographed, and leaf areas were measured. The data are expressed as the means ± se from three independent experiments (n = 10). Asterisks indicate a significant difference from the wild type (Student’s t test). Bar = 2 cm. *, P < 0.05; **, P < 0.01. D, Comparison of leaf cell size and cell number in different tobacco lines. The sixth true leaves from 48-DAS tobacco plants were collected for SEM analysis of leaf upper epidermal cells. The data are expressed as the means ± se from three independent experimental groups (three leaves per group, four independent areas per leaf, and at least 30 cells per each area were measured using ImageJ 1.38X). Asterisks indicate a significant difference from the wild type (Student’s t test). *, P < 0.05; **, P < 0.01. E, Relative hypocotyl length of 5-DAS etiolated seedlings after ethylene treatment. The data are expressed as the means ± se (n > 50). Relative hypocotyls of the F1 seedlings were significantly shorter than the wild type when the ethylene concentration was at 0.1 μL L−1 or higher (Student’s t test; P < 0.01). ND, Nondetected ethylene; NttubA1, N. tabacum tubulin alpha-1.
NtTCTP Protects NTHK1 from 26S Proteasome-Mediated Degradation
NtTCTP interacts with ethylene receptor NTHK1 to regulate plant growth and ethylene response. However, the molecular mechanism and biological function underlying this interaction were unknown. Previous research revealed that several ethylene receptors were dynamically controlled by 26S proteasome-mediated degradation (Chen et al., 2007; Kevany et al., 2007), and TCTPs were found to participate in regulating proteins stability in humans and yeast (Liu et al., 2005; Rinnerthaler et al., 2013). We then investigated whether NTHK1 protein was also regulated by the similar mechanism and whether the NTHK1-interacting protein NtTCTP participated in this process. Flag-tagged NTHK1 proteins were expressed in tobacco leaf cells and treated with the protein synthesis inhibitor cycloheximide (CHX) for different durations.
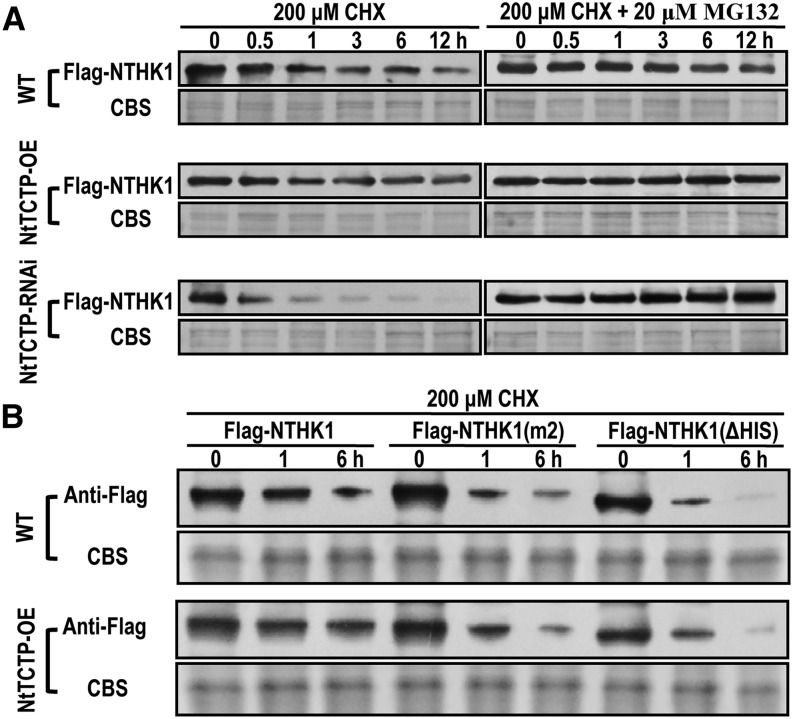
Then, the protein level was examined by immunoblotting assay using the anti-Flag antibody. In wild-type tobacco leaves, the NTHK1 protein level declined gradually under CHX treatment (Fig. 10A). In the leaves of the NtTCTP-overexpressing line (O2), CHX treatment did not alter the NTHK1 protein levels. On the contrary, in the leaves of the NtTCTP-silencing line (Ri20), the NTHK1 protein was reduced apparently and finally, disappeared (Fig. 10A). Moreover, inclusion of the proteasome inhibitor MG132 in the above tests suppressed the decrease of NTHK1 protein in all tobacco leaves (Fig. 10A). These results indicate that NTHK1 is controlled by the 26S proteasome-mediated protein degradation system and that NtTCTP can stabilize the NTHK1 protein.
Figure 10.
NtTCTP protects NTHK1 from proteasome-mediated degradation. A, Flag-NTHK1 was expressed in tobacco leaves of the wild type (WT), NtTCTP-overexpressing line (O2), and NtTCTP-silencing line (Ri20) through Agrobacterium spp.-mediated transformation. After expression for 2 d, leaves were treated with 200 μm CHX or 200 μm CHX plus 20 μm MG132 for different times, and the flag-NTHK1 protein was detected by immunoblotting with anti-Flag antibody. B, The direct interaction is necessary for NtTCTP to stabilize NTHK1. Kinase-inactivated mutation (m2; reducing the interaction between NTHK1 and NtTCTP) and His-truncated (ΔHIS; eliminating the interaction) types of NTHK1 were used for the protein stability assay. Agrobacterium spp.-mediated transformation, CHX treatment, and immunoblotting were performed as described in A. CBS, Coomassie Blue staining of the protein.
To unravel the function of NtTCTP-NTHK1 interaction in stabilizing the NTHK1 protein from degradation, the protein stabilities of two mutational types of NTHK1 were also examined (Fig. 10B). Although the NTHK1 is more stable in NtTCTP-overexpressing plants than in wild-type plants, the m2 mutational type of NTHK1, which has a weak interaction with NtTCTP (Fig. 2), showed more rapid degradation in both wild-type and NtTCTP-overexpressing tobacco plants (Fig. 10B). Moreover, truncated NTHK1 with deletion of His domain (ΔHIS), which could not interact with NtTCTP (Fig. 2), showed a much faster degradation under CHX treatment in wild-type plants (Fig. 10B). Overexpression of NtTCTP turned out to be nonfunctional for stabilizing the NTHK1 (ΔHIS) protein. All of these data indicate that the association of NtTCTP protects NTHK1 from proteasome-mediated degradation.
DISCUSSION
As for the role of ethylene in regulating plant growth, many studies have been focused on the regulation of cell elongation and expansion. Here, we identified NtTCTP as an interacting protein and stabilizer of the ethylene receptor NTHK1 to reduce plant ethylene sensitivity and promote plant growth through the acceleration of cell proliferation.
In animal systems, TCTP interacts with many proteins, including microtubules, actin filaments, the mitotic spindle, checkpoint protein Chfr, nuclear proteins nucleophosmin and nucleolin, transcription factor octamer-binding transcription factor4, Na+-K+-ATP synthase, antiapoptotic proteins of the B-cell lymphoma2 family, the ataxia-telangiectasia-mutated kinase, and p53 protein (Bommer, 2012; Zhang et al., 2012). TCTP may stabilize microtubules and the spindle, and it acts as a general mitotic regulator during cell cycle progression (Bommer, 2012). In tobacco plants, NtTCTP associates with the ethylene receptor NTHK1, an His kinase-like protein with Ser/Thr kinase activity, to stabilize this protein (Figs. 1, 2, and 10). Truncation of the His/ATP subdomain in NTHK1 disrupted the interaction of NTHK1 with NtTCTP. Mutation at NTHK1 without kinase activity has a weak interaction with NtTCTP (Fig. 2). Our results suggest that the kinase domain of NTHK1 is essential for the interaction with NtTCTP, whereas the kinase activity, although important, might not be required for this association. However, the failure of truncation of the His/ATP subdomain in NTHK1 to interact with NtTCTP could be caused by protein structural changes by deletion of a whole subdomain instead of a residue substitution. Additionally, because NTHK1 has Ser/Thr kinase activity and can autophosphorylate and phosphorylate other proteins (Xie et al., 2003; Chen et al., 2009; Cao et al., 2015), it is also possible that loss of kinase activity and/or phosphorylation would cause conformational changes and modulate interactions among proteins. In Arabidopsis, the phosphorylation state of ETR1 might affect its affinity for EIN2 and CTR1 (Bisson and Groth, 2010; Hall et al., 2012). Which part of the NtTCTP is required for its interaction with NTHK1 has yet to be investigated.
Subcellular colocalization is required for protein interaction in vivo. In this study, we discovered that NtTCTP localized to the cytoplasm and the nucleus and partially, at the ER and colocalized with NTHK1, mainly at the ER (Fig. 3; Supplemental Fig. S4B). Partial localization of NtTCTP at the ER can also be observed from fractionation analysis (Supplemental Fig. S3). Partial localization at the ER was also observed for human TCTP, which possibly colocalized with Eukaryotic Translation Elongation Factor 1B (Cans et al., 2003). Interestingly, ethylene treatment increased the protein level of NtTCTP at the ER (Figs. 1 and 3; Supplemental Fig. S3), probably through enhancing the interaction with NTHK1. In Arabidopsis, the interaction with the ethylene receptor is also required for the localization of CTR1 to the ER (Gao et al., 2008; Zhong et al., 2008). However, potential interactions with some other unknown factors at the ER could not be excluded. Expression induction by ethylene may also contribute to the increase of the NtTCTP protein in the membrane fraction (MF). It is interesting to note that, when coexpressed with NTHK1, the nuclear localization of NtTCTP seems to be altered (Fig. 3E), leading to a possible translocation from nucleus to ER. Additional analysis of the subcellular localization of NtTCTP in the NTHK1-knockout background may corroborate the translocation and its biological significance.
Both NTHK1- and NtTCTP-overexpressing plants exhibited larger leaves/seedlings than the wild type, and the NtTCTP-silencing plants showed smaller leaves/seedlings (Figs. 4 and 7), indicating that both NTHK1 and NtTCTP play positive roles in regulating plant growth. This promotion likely occurs through the activation of cell cycle progression and cell proliferation, which was seen from the increase of cell numbers and G2 cells in leaves of the NTHK1- and NtTCTP-overexpressing plants and the elevation of the mitotic index in root tips of these plants (Figs. 4 and 7). It should be noted that, although both the NTHK1- and NtTCTP-overexpressing plants had large leaf/seedling size, the NTHK1-overexpressing plants had a lower percentage of G2 cells and fewer cell numbers in their leaves than those in the leaves of the NtTCTP-overexpressing plants (Figs. 4 and 5). This is likely correlated with the expression level of NtTCTP, because the elevation of the NtTCTP expression level in the NTHK1-overexpressing plants was relatively lower than that in the NtTCTP-overexpressing plants (Figs. 3A and 4, A and B). Two transgenic NTHK1-silencing RNAi lines with the lowest transcript levels showed slightly smaller leaves/seedlings than those of the wild type (Supplemental Fig. S6), supporting that NTHK1 plays a positive role in regulation of plant growth.
During the early stage of seedling growth (12–22 DAS), NTHK1 seems to control leaf growth through the regulation of both cell numbers and cell size, whereas NtTCTP controls leaf area mainly through the regulation of cell numbers (Fig. 6). This divergence may be caused by the existence of some additional NTHK1-interacting proteins functioning in the early stage of leaf growth. Recently, another NTHK1-interacting protein NEIP2 was also identified and characterized (Cao et al., 2015). NEIP2 functions in regulating the plant growth at early stages and is responsive to salt stress (Cao et al., 2015). It should be noted that, in the NtTCTP-silencing lines, cell size decreased at the early stages but increased significantly at the mature stage compared with the wild type. In Arabidopsis, silencing of TCTP also resulted in large epidermal cells in petals and leaves (Brioudes et al., 2010). This phenomenon is supposed to be the result of growth compensation for significantly reduced cell numbers (Truernit and Haseloff, 2008; Tsukaya, 2008) or because of possibly elevated endoreduplication in these cells (Melaragno et al., 1993; Gendreau et al., 1997). In contrast to the large leaf/seedlings of the NtTCTP-overexpressing tobacco lines in this study, the AtTCTP-overexpressing lines showed no obvious phenotypic changes in Arabidopsis, possibly because of the limited increase in protein level (approximately 50% increase; Berkowitz et al., 2008). In fact, overexpression of AtTCTP accelerates plant growth, although the development of the adult plants was not altered. In addition, the overexpression of AtTCTP also increased leaf epidermal cell numbers through the positive regulation of cell cycle and proliferation (Brioudes et al., 2010).
Although roles of ethylene receptors in ethylene perception and signal transduction have been studied extensively, more and more diverged functions of different receptors have been discovered, such as the opposite role of ETR1 and other receptors in controlling ethylene-induced nutation (Binder et al., 2006; Kim et al., 2011), the opposite role of ETR1 and EIN4 in regulating responses to fumonisin B1 (Knoester et al., 1998), the contrasting roles for ETR1 and ERS1 in regulation of plant growth (Liu et al., 2010b), the contrasting roles for ETR1 and ETR2 in seed germination control under salt stress (Wilson et al., 2014b), the unique role of ETR1 in modulating the effects of far-red light on seed germination (Wilson et al., 2014a), the special role of ETR2 in trichome development (Plett et al., 2009), and roles of receiver domain-containing receptors (ETR1, ETR2, and EIN4) in growth recovery after ethylene treatment (Kim et al., 2011). As a subfamily II ethylene receptor, tobacco NTHK1 showed a more obvious role in the promotion of seedling growth than subfamily I ethylene receptors NtETR1 and AtETR1 (Cao et al., 2007; Chen et al., 2009). The divergent functions of a specific receptor may depend on these interacting proteins. NtTCTP interacts with NTHK1 but not with NtETR1, suggesting a special role of NTHK1 (Fig. 1B). Additional genetic analysis through crosses between the NTHK1-overexpressing line and the NtTCTP-silencing line reveals that NtTCTP is likely required for NTHK1 to regulate the ethylene response and plant growth, mainly through controlling cell proliferation (Fig. 9). Considering the basic role of TCTP in regulating cell proliferation in a widespread species, the inhibition of the NTHK1-overexpressing phenotype in NtTCTP-silencing background could be caused by a general disruption of cell proliferation. Additional transgenic analysis with mutational types of NTHK1 to show the relationship of physical interaction between NTHK1 and NtTCTP with genetic consequences would be valuable. Together, we propose that NTHK1 plays an important role in constraining ethylene response in light to maintain normal growth, likely through specific interaction with NtTCTP and the regulation of cell proliferation. This conclusion is further strengthened by the fact that NTHK1 needs NtTCTP to maintain its stabilization (Fig. 10).
The degradation of 26S proteasome-mediated protein is a widespread and efficient mechanism for the fine tuning of phytohormone signaling, including ethylene (Vierstra, 2009; Santner and Estelle, 2010). Currently, several type II ACSs, EIN2 and EIN3, have been found to be regulated by the ubiquitin-proteasome-mediated degradation system (Guo and Ecker, 2003; Potuschak et al., 2003; Wang et al., 2004; Joo et al., 2008; Qiao et al., 2009; Lyzenga et al., 2012). Moreover, protein levels of Arabidopsis ethylene receptor AtETR2 and tomato ethylene receptors LeETR4 and LeETR6 are also regulated through a 26S proteasome-mediated degradation pathway (Chen et al., 2007; Kevany et al., 2007). Recent work in Arabidopsis further revealed that the protein levels of all five ethylene receptors are posttranscriptionally regulated under high-ethylene concentration, possibly through the proteasome-mediated pathway (Shakeel et al., 2015). In addition, CTR1 serves to protect ETR1 from ethylene-induced protein turnover (Shakeel et al., 2015). In this study, tobacco ethylene receptor NTHK1 was also degraded rapidly when treated with CHX, and this degradation was mediated by a 26S proteasome-dependent pathway. It is interesting to note that the overexpression of NtTCTP reduced the degradation of NTHK1, whereas the silencing of NtTCTP led to more rapid degradation (Fig. 10A). Moreover, when the interaction between NTHK1 and NtTCTP was disrupted by mutations or truncations in NTHK1, NTHK1 protein became more unstable, even in the presence of overexpressed NtTCTP (Fig. 10B). These results imply that the interaction of NtTCTP with NTHK1 is required for the protection of NTHK1 from degradation, suggesting a sophisticated mechanism for tight control of ethylene receptor stability. Human TCTP and its homolog from a parasite may act as a molecular chaperone under heat shock conditions, protecting proteins from denaturation (Gnanasekar et al., 2009). Yeast TCTP colocalized with several proteins involved in deubiquitination and seemed to act as an inhibitor of proteasome for the protection of proteins involved in stress-induced cell death (Rinnerthaler et al., 2013). Therefore, TCTP may adopt unique roles in protecting some important proteins.
To maintain normal lifecycles for survival and reproduction, rigorous self-control of ethylene signaling by feedback regulations can occur at both biosynthesis and signaling processes, including both positive and negative feedback (Vandenbussche et al., 2012). A typical case is that EIN3 promotes the transcription of EIN3 Binding F-box Protein2 and then induces 26S proteasome-mediated degradation of itself to restrain ethylene signaling and maintain normal plant growth (Guo and Ecker, 2003; Potuschak et al., 2003; Konishi and Yanagisawa, 2008). In this work, ethylene-induced TCTP proteins in tobacco are likely translocated to ER and associated with ethylene receptor NTHK1 to protect it from degradation. This action will strengthen the roles of NTHK1 probably in activating CTR1 and the inhibition of EIN2, allowing the desensitization of ethylene signaling and response. Simultaneously, increased TCTP proteins promote cell proliferation to maintain normal plant growth. Moreover, the stabilized NTHK1 may also recruit other factors, such as NEIP2, to promote cell expansion at the early stage of growth (Cao et al., 2015). Our study reveals a unique negative feedback mechanism of tightly restraining excessive ethylene response and promoting growth recovery from inhibited growth. In this model, NtTCTP acts as a brake of ethylene signaling and an enhancer of cell proliferation to coordinate normal vegetative growth and ethylene response.
MATERIALS AND METHODS
Yeast Two-Hybrid Screen
The yeast (Saccharomyces cerevisiae) two-hybrid screening was performed according to the instruction manual of the CytoTrap XR Library Construction Kit (Stratagene). The NTHK1 coding region without the transmembrane domains (amino acids 145–762) was amplified from the original plasmid (Zhang et al., 2001) by PCR with primers BaitF and BaitR (Supplemental Table S1) and cloned into pSos vector, which is the bait construct for screening target proteins, at BamHI and SalI sites. Total RNAs from 4-week-old tobacco (Nicotiana tabacum; variety Xanthi) seedlings were extracted, and the mRNA was further isolated with the PolyATract mRNA Isolation System IV (Promega). The complementary DNA (cDNA) library was constructed with the Yeast CytoTrap XR Library Construction Kit (Stratagene). Then, the yeast cells (cdc25H) were cotransformed with bait pSos-NTHK1(ΔTM) and cDNA library pMyr plasmids and examined for their growth on selection medium at 24°C and 37°C, respectively. Cotransformants that could grow normally at 37°C on SD/Gal-UL but not on SD/Glu-UL were considered as positive colonies showing possible interactions between NTHK1 and corresponding proteins expressed in pMyr plasmids. These positive colonies were picked out, and inserted cDNAs were amplified by PCR and sequenced, among which repeatedly appearing insertions were selected as candidates. To verify the interaction, complete coding sequence (CDS) of candidates was amplified by PCR from cDNA, cloned into the pMyr vector, cotransformed with pSos-NTHK1(ΔTM), and examined as described above.
NTHK1 contains several domains in addition to transmembrane domains. To detect which domain is necessary for the interaction, sequences encoding G, GH, H, HR, and R domains were amplified by PCR with primer pairs of BaitF and GR, BaitF and HR, HF and HR, HF and BaitR, and RF and BaitR, respectively (Supplemental Table S1), and cloned into pSos vectors at BamHI and SalI sites. pMyr plasmids containing full-length CDS of the target gene and each of these pSos plasmids were each cotransformed into yeast cells and examined as described above.
GST Pull-Down Assay
For expression and purification of GST-NtTCTP fusion protein from Escherichia coli, the complete CDS of NtTCTP was digested from pMyr-NtTCTP vector with BamHI and SalI and ligated into the pGEX-6P-1 vector. The plasmids were transformed into the BL21(DE3) strain by heat shock for protein expression. Isopropylthio-β-galactoside with a final concentration of 0.15 mm was added to the culture for protein induction. The GST-NtTCTP fusion protein was purified with Glutathione Sepharose 4B (Amersham). The GST protein was also expressed using empty pGEX-6P-1 plasmids and purified as described above.
For expression of [35S]Met-labeled protein, sequences encoding NTHK1 (ΔTM), G domain, GH domain, H domain, HR domain, and R domain were amplified by PCR with primer pairs of TNTK1F and TNTK1R, TNTK1F and TNTGR, TNTK1F and TNTHR, TNTHF and TNTHR, TNTHF and TNTK1R, and TNTRF and TNTK1R, respectively (Supplemental Table S1), and cloned into pTNT vector at XhoI and SalI for in vitro translation. Then, [35S]Met-labeled different domains of NTHK1 were synthesized by using the TNT Quick Coupled Transcription/Translation Systems (Promega) according to the instruction manual.
GST pull-down assay was performed by adding 10 μg of GST or GST-NtTCTP fusion protein, 50 μL of Glutathione Sepharose 4B (Amersham), 20 μL of [35S]Met-labeled protein in binding buffer (10 mm Na2HPO4, 2 mm KH2PO4, 140 mm NaCl, 2.7 mm KCl, and 10% [v/v] glycerol), and 5 μL of 200 mg mL−1 bovine serum albumin (dissolved in binding buffer) into round-bottom Eppendorf tubes and supplementing with binding buffer to a final volume of 100 μL. Samples were mixed gently, rotated for 2 h at 4°C, and then centrifuged at 12,000 rpm for 2 min at 4°C. The supernatants were discarded, and the resins were washed three times with precold binding buffer. Then, the proteins were eluted with 30 μL of elution buffer (10 mm reduced glutathione and 50 mm Tris-HCl, pH 8.0) and separated by 10% (w/v) SDS-PAGE. After fixation in 10% (v/v) acetic acid and 20% (v/v) methanol for 2 h, gels were immersed sequentially in 7% (v/v) acetic acid, 7% (v/v) methanol, and 10% (v/v) glycerol for 5 min, dried for 2 h, and then exposed to phosphor screen (Amersham).
Co-IP
Complete CDS of NTHK1 was amplified by PCR with NTHK1-GWF1 and NTHK1-GWR1 primers (Supplemental Table S1), ligated into the ImpGWB2 TOPO Vector (Life Technologies), and then translocated into the pGWB412 vector through homologous recombination. pGWB412-NTHK1 plasmids plus P19 (to avoid gene silencing) were transferred into tobacco leaves through agrobacterium (EHA105)-mediated transformation (Liu et al., 2010a). After growth at 25°C under a 14-h-light/10-h-dark photoperiod for 2 d, tobacco seedlings were treated with 100 to 0 μL L−1 ethylene for 3 h in a sealed box, and then transformed leaves were collected for the Co-IP assay.
For the Co-IP assay, tobacco leaves were homogenized in extraction buffer (30 mm Tris-HCl, pH 8.0, 150 mm NaCl, 20% [v/v] glycerol, 2 mm EDTA, and 1 mm dithiothreitol) with 1× protease and phosphatase inhibitors mixture (Thermo Fisher) on ice and then centrifuged at 5,000g for 30 min at 4°C. The supernatant was filtered through miracloth (Calbiochem) and then centrifuged at 100,000g for 1 h at 4°C. The pellet (MF) was suspended in ice-cold membrane resuspension buffer (20 mm Tris-HCl, pH 7.5, 10% [v/v] glycerol, 1.5% [v/v] Triton X-100, 3 mm MgCl2, and 1 mm EDTA) with 1× protease and phosphatase inhibitors mixture (Thermo Fisher) and incubated on ice for 1 h. Then, 200 μg of MF was diluted to 500 μL with immunoprecipitation buffer (30 mm Tris-HCl, pH 7.5, 150 mm NaCl, 3 mm MgCl2, 1 mm EDTA, and 1× protease and phosphatase inhibitors), and 100 μL of mAnti-DDDDK-Tag mAb-Magnetic Beads (MBL) were added. The mixture was incubated at 4°C for 6 h with gentle agitation. The beads were washed five times with ice-cold IP buffer. Then, protein was eluted from the beads using 40 μL of Laemmli sample buffer; 20 μL of eluted protein was separated by SDS-PAGE followed by an immunoblotting assay. For immunoblotting, anti-Flag antibody (1:5,000; MBL) and anti-NtTCTP antibody (1:2,000) were used to detect Flag-NTHK1 and NtTCTP, respectively. Horseradish peroxidase-conjugated goat anti-mouse secondary antibody (EarthOx) was used at 1:30,000 dilution, and signal was detected using the SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher). Anti-NtTCTP antibody was produced from mice immunized with GST-NtTCTP protein (Beijing ABT Genetic Engineering Technology). To examine the solubility of Flag-NTHK1 and NtTCTP proteins in membrane resuspension buffer, 200 μg of resuspended MF was centrifuged at 150,000g for 1 h. Then, 5% of the supernatant and 10% of the resuspended pellet with the additions of 10 μg of MF and 10 μg of soluble fraction were separated by SDS-PAGE followed by an immunoblotting assay (Supplemental Fig. S3).
BiFC
BiFC assay of the interaction between NTHK1 and NtTCTP was performed using the tobacco protoplast transformation system. The coding region of NTHK1 was amplified by PCR with NTHK1-BiFCF1 and NTHK1-BiFCR1 primers (Supplemental Table S1) from the original plasmid (Zhang et al., 2001) and ligated into the pSPYCE vector (Walter et al., 2004) at BamHI and SalI. The complete CDS of NtTCTP was amplified by PCR with primers NtTCTP-BiFCF1 and NtTCTP-BiFCR1 from pMyr-NtTCTP plasmids and cloned into pSPYNE vector. pSPYCE-NTHK1 and pSPYNE-NtTCTP plasmids were cotransformed into tobacco protoplasts. Simultaneously, cotransformation of empty pSPYNE and pSPYCE plasmids was set as a negative interaction control. After incubation at 23°C for 16 h, YFP signals were detected using a laser confocal scanning microscope (LCSM). Preparation and transformation of tobacco protoplasts were performed according to the protocol by Yoo et al. (2007) with little modification.
The Agrobacterium spp.-mediated Nicotiana benthamiana leaf transformation system was used to elucidate which domain of NTHK1 was necessary for its interaction with NtTCTP. The mutational type of NTHK1 (m2) that eliminated the kinase activity (Chen et al., 2009), two truncated types of NTHK1 (ΔHIS and ΔATP), and the full-length CDS of NTHK1 were amplified with NTHK1-BiFCF2 and NTHK1-BiFCR2 primers (Supplemental Table S1) and ligated into the pSPYCE (MR) vector (Waadt et al., 2008). The CDS of NtTCTP was amplified with NtTCTP-BiFCF2 and NtTCTP-BiFCR2 primers (Supplemental Table S1) and ligated into the pSPYNE(R)173 and pSPYCE(MR) vectors, respectively (Waadt et al., 2008). YNE-NtTCTP and YCE fused with each type of NTHK1 were coexpressed in tobacco leaves through the Agrobacterium spp.-mediated tobacco leaf transformation system (Liu et al., 2010a). YNE and YCE-NTHK1 were coexpressed as a negative interaction control, whereas coexpression of YNE-NtTCTP and YCE-NtTCTP was set as a positive interaction control. Then, the YFP signals were detected using LCSM.
Plant Growth Conditions and Treatments
Seeds of wild-type tobacco (variety Xanthi) and NTHK1-overexpressing tobacco line 16-4 (Cao et al., 2006) were sterilized in 1% (w/v) sodium hypochlorite for 12 min, washed with sterilized water four times, and plated on one-half-strength MS. After stratification at 4°C in the dark for 3 d, plates were kept at 25°C under a 14-h-light/10-h-dark photoperiod for plant growth. Seedlings growing on one-half-strength MS for 8 DAS were transplanted to soil for additional growth at 25°C under a 16-h-light/8-h-dark photoperiod. During the entire lifecycle, root tips, roots, stems, shoot tips, leaves, and flowers were collected for detection of the NtTCTP transcript level.
For ethylene treatment, 12-DAS seedlings were placed in sealed boxes with 100 μL L−1 ethylene at 25°C under a 14-h-light/10-h-dark photoperiod and collected at different times for detection of the NtTCTP transcript and protein levels. Treatments with air were used as controls.
Immunoblotting Assay
To detect the NtTCTP protein level, collected materials were thoroughly ground to powder in liquid nitrogen, and proteins were extracted by using the Plant Protein Extraction Kit (CWBIO). Protein extracts were separated by 12% (w/v) SDS-PAGE and transferred onto polyvinylidene difluoride membrane. Then, immunoblotting was carried out by using anti-NtTCTP antibody (1:2,000) and detected as described in the Co-IP assay.
Subcellular Localization Analysis
To observe the subcellular localization of NTHK1 and NtTCTP, tobacco protoplasts and N. benthamiana leaves were used for expression of GFP- or RFP-fused NTHK1 and NtTCTP proteins, and florescence was detected using an LCSM.
For observation of subcellular localization in tobacco protoplasts, the coding region of NtTCTP was amplified by PCR as described in the BiFC assay and ligated into the pBI221-GFP vector at BamHI and SalI sites. Purified pBI221-NtTCTP-GFP plasmids were transformed into tobacco protoplasts by the polyethylene glycol-CaCl2 method. The pBI221-GFP plasmids were also transformed as a control. After incubation at 25°C for 16 h, green florescence signals were detected using an LCSM.
For observation of subcellular localization in tobacco leaf cells, the coding region of NTHK1 and NtTCTP in the ImpGWB2 TOPO vector was translocated to the pGWB405 and pGWB454 vectors, respectively, through homologous recombination. Purified pGWB405-NtTCTP plasmids were then transformed into tobacco leaves as described in the Co-IP assay. HDEL-RFP was coexpressed with NtTCTP-GFP in tobacco leaves to identify the ER localization of NtTCTP. To observe colocalization of NTHK1 and NtTCTP, both pGWB405-NTHK1 and pGWB454-NtTCTP plasmids were cotransformed into tobacco leaves. After incubation at 25°C under a 16-h-light/8-dark-h photoperiod for 3 d, florescence signals were detected using an LCSM. For ethylene treatment, transformed seedlings were placed in a sealed box with 100 μL L−1 ethylene for 3 h.
Generation of Transgenic Plants and Analysis of Growth Phenotype
To obtain NtTCTP-overexpressing tobacco lines, complete CDS of NtTCTP was cut from the pMyr-NtTCTP vector with BamHI and SalI and ligated into the pBIN438 vector under the control of a 35S promoter plus a translation enhancer-Ω (Wuriyanghan et al., 2009). For RNAi analysis, the CDS of NtTCTP was amplified with NtTCTPRIF and NtTCTPRIR primers (Supplemental Table S1) and then ligated two times into RNAi vector pZH01 in an inversely oriented manner. Two transcript regions of NTHK1 (type 1, 2,125–2,630 bp; type 2, 1,382–1,904 bp) were used to form two different pZH01 constructs for the NtTCTP-silencing transgene (the primers used for vector construction are listed in Supplemental Table S1). These constructs were transformed into tobacco (variety Xanthi) plants using the Agrobacterium spp.-mediated leaf disc transformation method. All NtTCTP transgenic lines were first screened by reverse transcription-PCR and then confirmed by northern blotting. Two NtTCTP-overexpressing lines (O2 and O7) and two NtTCTP-silencing lines (Ri16 and Ri20) were used in this study. NtTCTP protein levels were detected by immunoblotting with anti-NtTCTP antibody. Transcript levels of NtTCTPL in the two NtTCTP-silencing lines were detected by qRT-PCR with the qRT-NtTCTPLF and qRT-NtTCTPLR primers (Supplemental Table S2). All NTHK1-silencing lines were screened by qRT-PCR, and five lines (2-1, 2-2, 2-7, 1-8, and 1-36) were used for phenotypic analysis.
For plant growth analysis, seeds of wild-type tobacco, all selected transgenic lines, and the NTHK1-overexpressing line 16-4 (Cao et al., 2006) were sown on one-half-strength MS as described above; 8-DAS seedlings were transplanted to soil for additional growth at 25°C under a 16-h-light/8-h-dark photoperiod. Seedling crown diameters were measured every 2 d, and photographs were taken. The sixth true leaves of 48-DAS plants were photographed, and leaf areas were measured with ImageJ 1.38X software. To investigate leaf epidermal cells, the sixth true leaves of 48-DAS plants were detached for scanning electron microscope (SEM) analysis. Epidermal cell size was then measured using ImageJ 1.38X, and the cell number per leaf was calculated. For detection of DNA ploidy distribution, the sixth true leaves from different lines were chopped in the chopping buffer (Galbraith et al., 1983) and then passed through nylon filters (pore size of 37 μm). Then, the nuclei were stained with 4′,6′-diaminophenylindole and analyzed by flow cytometry. To investigate the growth of the root system, 7-DAS seedlings with simultaneous germination were transferred to fresh one-half-strength MS plates and placed vertically. Seedlings were photographed every 1 d until 20 DAS for primary root length measure and lateral root number counting. To measure the mitotic index of root tips, the root tips of 7-DAS seedlings were cut, stabilized in Carnoy’s fluid for 24 h at 4°C, and then dehydrated sequentially in 90%, 80%, and 70% (v/v) ethanol for 30 min. Fixed root tips were dissociated in 1 mol L−1 HCl (60°C) for 10 min and washed two times with distilled water. The slide was squashed so that only the meristematic zone was encased for observation of cell division phases. Chromosomes were stained with carbolfuchsin staining solution and observed using optical microscopy. Dividing cell numbers and total cell numbers were counted in each view field.
For dynamic analysis of the growth of different tobacco lines under ACC treatment, 12-DAS seedlings were transplanted to fresh one-half-strength MS with or without 10 μm ACC. The third true leaves were harvested daily from 12 to 22 DAS for measurements of leaf area and cell size through micrographs of the upper epidermis, and the cell number was then calculated (Skirycz et al., 2011).
Observation and Analysis of Ethylene Responses
To investigate phenotypes under ethylene treatment, the classical triple response was examined as before (Cao et al., 2007) with some modification. Seeds of different tobacco lines were plated on one-half-strength MS. After stratification at 4°C in the dark for 3 d, plates were placed in sealed boxes with different concentrations of ethylene. After treatment at 25°C in the dark for 5 d, the hypocotyl length was measured. In addition, for examination of ethylene-induced leaf senescence, the sixth true leaves from 60-DAS tobacco plants were treated with 100 μL L−1 ethylene in sealed boxes for 5 d. Treatments without exogenous ethylene were set as controls. After treatments, leaves were photographed, and the chlorophyll content was measured.
qRT-PCR
qRT-PCR was used to detect transcript levels of all genes, including NTHK1, NtTCTPs, and genes related to ethylene synthesis and signaling. For qRT-PCR, total RNA was extracted by using TRIzol Reagent (Life Technologies). First-strand cDNA was synthesized with the ReverTra Ace qPCR RT Kit (TOYOBO). PCR reactions were performed using the SYBR Green Realtime PCR Master Mix (TOYOBO) with a LightCycler 480 II (Roche). All primers used for qRT-PCR are listed in Supplemental Table S2. The PCR results were calculated using LightCycler 480 software release 1.5.0 SP3. Three replicates were used for each data point, and all experiments were repeated at least two times.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NtTCTP, KM507327; AtTCTP, NP_188286.1; GmTCTP, NP_001237819.1; OsTCTP, NP_001068405.1; ZmTCTP, NP_001105104.1; DmTCTP, NP_650048.1; MmTCTP, NP_033455.1; HsTCTP, NP_003286.1; NTHK1, AF026267; ACS2, EU123522; ACS4, EU123523.1; EFE1, Z29529; ACO2, Z46349; ACO3, X83229; EIN3, AF247568; EIL1, AY248903; EIL2, AY248904; EIL3, AY248905; EIL4, AY248906; EIL5, AY248907; ERF1, D38123.1; and NttubA1, AJ421411.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence comparison among TCTPs from tobacco and Arabidopsis.
Supplemental Figure S2. Sequence comparison of NtTCTP with other TCTPs from plants and animals.
Supplemental Figure S3. Examination of the solubility of Flag-NTHK1 and NtTCTP proteins the membrane resuspension buffer used for the Co-IP assay.
Supplemental Figure S4. Subcellular localization of NtTCTP was revealed by GFP fusion in different expression systems.
Supplemental Figure S5. Expression analysis of NtTCTPs in transgenic tobacco lines.
Supplemental Figure S6. RNA interference analysis of NTHK1 in tobacco.
Supplemental Figure S7. SEM and flow cytometric analyses of leaf cells from different tobacco lines.
Supplemental Figure S8. Leaf area, cell size, and cell number of different tobacco lines in the presence or absence of ethylene precursor ACC.
Supplemental Figure S9. Lateral root density in different tobacco lines.
Supplemental Figure S10. Chlorophyll contents in leaves of various plants treated with ethylene or air.
Supplemental Figure S11. SEM of leaf upper epidermal cells from different tobacco lines.
Supplemental Table S1. Primers used for gene cloning and vector construction.
Supplemental Table S2. Primers used for qRT-PCR analysis.
Supplementary Material
Glossary
- ACC
1-aminocyclopropane-1-carboxylic acid
- BiFC
bimolecular fluorescence complementation
- cDNA
complementary DNA
- CHX
cycloheximide
- Co-IP
coimmunoprecipitation
- DAS
days after stratification
- ER
endoplasmic reticulum
- LCSM
laser confocal scanning microscope
- MF
membrane fraction
- MS
Murashige and Skoog medium
- qRT
quantitative real-time
- SEM
scanning electron microscope
Footnotes
This work is supported by the National Natural Science Foundation of China (grant no. 91317306), the National Key Basic Research Project (grant nos. 2015CB755702 and 2012CB114202), and the State Key Lab of Plant Genomics.
Articles can be viewed without a subscription.
References
- Abeles FB, Morgan PW, Saltveit ME Jr (1992) Ethylene in Plant Biology. Academic Press, San Diego, CA [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y, et al. (2010) Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz O, Jost R, Pollmann S, Masle J (2008) Characterization of TCTP, the translationally controlled tumor protein, from Arabidopsis thaliana. Plant Cell 20: 3430–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BM, O’Malley RC, Wang W, Zutz TC, Bleecker AB (2006) Ethylene stimulates nutations that are dependent on the ETR1 receptor. Plant Physiol 142: 1690–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BM, Walker JM, Gagne JM, Emborg TJ, Hemmann G, Bleecker AB, Vierstra RD (2007) The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 19: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson MM, Groth G (2010) New insight in ethylene signaling: autokinase activity of ETR1 modulates the interaction of receptors and EIN2. Mol Plant 3: 882–889 [DOI] [PubMed] [Google Scholar]
- Bommer UA. (2012) Cellular function and regulation of the translationally controlled tumour protein TCTP. Open Allergy J 5: 19–32 [Google Scholar]
- Brioudes F, Thierry AM, Chambrier P, Mollereau B, Bendahmane M (2010) Translationally controlled tumor protein is a conserved mitotic growth integrator in animals and plants. Proc Natl Acad Sci USA 107: 16384–16389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancel JD, Larsen PB (2002) Loss-of-function mutations in the ethylene receptor ETR1 cause enhanced sensitivity and exaggerated response to ethylene in Arabidopsis. Plant Physiol 129: 1557–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cans C, Passer BJ, Shalak V, Nancy-Portebois V, Crible V, Amzallag N, Allanic D, Tufino R, Argentini M, Moras D, et al. (2003) Translationally controlled tumor protein acts as a guanine nucleotide dissociation inhibitor on the translation elongation factor eEF1A. Proc Natl Acad Sci USA 100: 13892–13897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao WH, Liu J, He XJ, Mu RL, Zhou HL, Chen SY, Zhang JS (2007) Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol 143: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao WH, Liu J, Zhou QY, Cao YR, Zheng SF, Du BX, Zhang JS, Chen SY (2006) Expression of tobacco ethylene receptor NTHK1 alters plant responses to salt stress. Plant Cell Environ 29: 1210–1219 [DOI] [PubMed] [Google Scholar]
- Cao YR, Chen HW, Li ZG, Tao JJ, Ma B, Zhang WK, Chen SY, Zhang JS (2015) Tobacco ankyrin protein NEIP2 interacts with ethylene receptor NTHK1 and regulates plant growth and stress responses. Plant Cell Physiol 56: 803–818 [DOI] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chen T, Liu J, Lei G, Liu YF, Li ZG, Tao JJ, Hao YJ, Cao YR, Lin Q, Zhang WK, et al. (2009) Effects of tobacco ethylene receptor mutations on receptor kinase activity, plant growth and stress responses. Plant Cell Physiol 50: 1636–1650 [DOI] [PubMed] [Google Scholar]
- Chen YF, Gao Z, Kerris RJ III, Wang W, Binder BM, Schaller GE (2010) Ethylene receptors function as components of high-molecular-mass protein complexes in Arabidopsis. PLoS ONE 5: e8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Shakeel SN, Bowers J, Zhao XC, Etheridge N, Schaller GE (2007) Ligand-induced degradation of the ethylene receptor ETR2 through a proteasome-dependent pathway in Arabidopsis. J Biol Chem 282: 24752–24758 [DOI] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C (1998) Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA 95: 5401–5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CH, Jang M, Scharein B, Malach A, Rivarola M, Liesch J, Groth G, Hwang I, Chang C (2010) Molecular association of the Arabidopsis ETR1 ethylene receptor and a regulator of ethylene signaling, RTE1. J Biol Chem 285: 40706–40713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JM, Smalle J, Gingerich DJ, Walker JM, Yoo SD, Yanagisawa S, Vierstra RD (2004) Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci USA 101: 6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051 [DOI] [PubMed] [Google Scholar]
- Gao Z, Wen CK, Binder BM, Chen YF, Chang J, Chiang YH, Kerris RJ III, Chang C, Schaller GE (2008) Heteromeric interactions among ethylene receptors mediate signaling in Arabidopsis. J Biol Chem 283: 23801–23810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Höfte H (1997) Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanasekar M, Dakshinamoorthy G, Ramaswamy K (2009) Translationally controlled tumor protein is a novel heat shock protein with chaperone-like activity. Biochem Biophys Res Commun 386: 333–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Hall AE, Bleecker AB (2003) Analysis of combinatorial loss-of-function mutants in the Arabidopsis ethylene receptors reveals that the ers1 etr1 double mutant has severe developmental defects that are EIN2 dependent. Plant Cell 15: 2032–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BP, Shakeel SN, Amir M, Ul Haq N, Qu X, Schaller GE (2012) Histidine kinase activity of the ethylene receptor ETR1 facilitates the ethylene response in Arabidopsis. Plant Physiol 159: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass C, Lohrmann J, Albrecht V, Sweere U, Hummel F, Yoo SD, Hwang I, Zhu T, Schäfer E, Kudla J, Harter K (2004) The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J 23: 3290–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44: 903–916 [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271 [DOI] [PubMed] [Google Scholar]
- Joo S, Liu Y, Lueth A, Zhang S (2008) MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J 54: 129–140 [DOI] [PubMed] [Google Scholar]
- Ju C, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML, et al. (2012) CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci USA 109: 19486–19491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevany BM, Tieman DM, Taylor MG, Cin VD, Klee HJ (2007) Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J 51: 458–467 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Kim H, Helmbrecht EE, Stalans MB, Schmitt C, Patel N, Wen CK, Wang W, Binder BM (2011) Ethylene receptor ETHYLENE RECEPTOR1 domain requirements for ethylene responses in Arabidopsis seedlings. Plant Physiol 156: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoester M, van Loon LC, van den Heuvel J, Hennig J, Bol JF, Linthorst HJ (1998) Ethylene-insensitive tobacco lacks nonhost resistance against soil-borne fungi. Proc Natl Acad Sci USA 95: 1933–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S (2008) Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J 55: 821–831 [DOI] [PubMed] [Google Scholar]
- Larsen PB, Cancel JD (2003) Enhanced ethylene responsiveness in the Arabidopsis eer1 mutant results from a loss-of-function mutation in the protein phosphatase 2A A regulatory subunit, RCN1. Plant J 34: 709–718 [DOI] [PubMed] [Google Scholar]
- Larsen PB, Chang C (2001) The Arabidopsis eer1 mutant has enhanced ethylene responses in the hypocotyl and stem. Plant Physiol 125: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Alexander L, Hackett R, Grierson D (2008a) LeCTR2, a CTR1-like protein kinase from tomato, plays a role in ethylene signalling, development and defence. Plant J 54: 1083–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Arciga-Reyes L, Zhong S, Alexander L, Hackett R, Wilson I, Grierson D (2008b) SlTPR1, a tomato tetratricopeptide repeat protein, interacts with the ethylene receptors NR and LeETR1, modulating ethylene and auxin responses and development. J Exp Bot 59: 4271–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Ho CW, Grierson D (2009a) AtTRP1 encodes a novel TPR protein that interacts with the ethylene receptor ERS1 and modulates development in Arabidopsis. J Exp Bot 60: 3697–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Zhong S, Grierson D (2009b) Recent advances in ethylene research. J Exp Bot 60: 3311–3336 [DOI] [PubMed] [Google Scholar]
- Liu H, Peng HW, Cheng YS, Yuan HS, Yang-Yen HF (2005) Stabilization and enhancement of the antiapoptotic activity of mcl-1 by TCTP. Mol Cell Biol 25: 3117–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang Y, Tang S, Zhao Q, Zhang Z, Zhang H, Dong L, Guo H, Xie Q (2010a) An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J 61: 893–903 [DOI] [PubMed] [Google Scholar]
- Liu Q, Xu C, Wen CK (2010b) Genetic and transformation studies reveal negative regulation of ERS1 ethylene receptor signaling in Arabidopsis. BMC Plant Biol 10: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyzenga WJ, Booth JK, Stone SL (2012) The Arabidopsis RING-type E3 ligase XBAT32 mediates the proteasomal degradation of the ethylene biosynthetic enzyme, 1-aminocyclopropane-1-carboxylate synthase 7. Plant J 71: 23–34 [DOI] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE (2005) Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melaragno JE, Mehrotra B, Coleman AW (1993) Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5: 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu RL, Cao YR, Liu YF, Lei G, Zou HF, Liao Y, Wang HW, Zhang WK, Ma B, Du JZ, et al. (2009) An R2R3-type transcription factor gene AtMYB59 regulates root growth and cell cycle progression in Arabidopsis. Cell Res 19: 1291–1304 [DOI] [PubMed] [Google Scholar]
- Neljubov D. (1901) Ueber die horizontale Nutation der Stengel von Pisum sativum und einiger Anderen Pflanzen. Beih Bot Zentralbl 10: 128–139 [Google Scholar]
- Plett JM, Mathur J, Regan S (2009) Ethylene receptor ETR2 controls trichome branching by regulating microtubule assembly in Arabidopsis thaliana. J Exp Bot 60: 3923–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Qiao H, Chang KN, Yazaki J, Ecker JR (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev 23: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Shen Z, Huang SS, Schmitz RJ, Urich MA, Briggs SP, Ecker JR (2012) Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338: 390–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Xie F, Yu J, Wen CK (2012) Arabidopsis RTE1 is essential to ethylene receptor ETR1 amino-terminal signaling independent of CTR1. Plant Physiol 159: 1263–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Hall BP, Gao Z, Schaller GE (2007) A strong constitutive ethylene-response phenotype conferred on Arabidopsis plants containing null mutations in the ethylene receptors ETR1 and ERS1. BMC Plant Biol 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick JS, Rivarola M, Chang C (2008) Involvement of RTE1 in conformational changes promoting ETR1 ethylene receptor signaling in Arabidopsis. Plant J 56: 423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick JS, Wen CK, Shockey JA, Chang C (2006) REVERSION-TO-ETHYLENE SENSITIVITY1, a conserved gene that regulates ethylene receptor function in Arabidopsis. Proc Natl Acad Sci USA 103: 7917–7922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinnerthaler M, Lejskova R, Grousl T, Stradalova V, Heeren G, Richter K, Breitenbach-Koller L, Malinsky J, Hasek J, Breitenbach M (2013) Mmi1, the yeast homologue of mammalian TCTP, associates with stress granules in heat-shocked cells and modulates proteasome activity. PLoS ONE 8: e77791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Estelle M (2010) The ubiquitin-proteasome system regulates plant hormone signaling. Plant J 61: 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharein B, Groth G (2011) Phosphorylation alters the interaction of the Arabidopsis phosphotransfer protein AHP1 with its sensor kinase ETR1. PLoS ONE 6: e24173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharein B, Voet-van-Vormizeele J, Harter K, Groth G (2008) Ethylene signaling: identification of a putative ETR1-AHP1 phosphorelay complex by fluorescence spectroscopy. Anal Biochem 377: 72–76 [DOI] [PubMed] [Google Scholar]
- Shakeel S, Gao Z, Amir M, Chen YF, Rai MI, Ul Haq N, Schaller GE (2015) Ethylene regulates levels of ethylene-receptor/CTR1 signaling complexes in Arabidopsis thaliana. J Biol Chem 290: 12415–12424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakeel SN, Wang X, Binder BM, Schaller GE (2013) Mechanisms of signal transduction by ethylene: overlapping and non-overlapping signalling roles in a receptor family. AoB Plants 5: plt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Claeys H, De Bodt S, Oikawa A, Shinoda S, Andriankaja M, Maleux K, Eloy NB, Coppens F, Yoo SD, et al. (2011) Pause-and-stop: the effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell 23: 1876–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Haseloff J (2008) Arabidopsis thaliana outer ovule integument morphogenesis: ectopic expression of KNAT1 reveals a compensation mechanism. BMC Plant Biol 8: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H. (2008) Controlling size in multicellular organs: focus on the leaf. PLoS Biol 6: e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Vaseva I, Vissenberg K, Van Der Straeten D (2012) Ethylene in vegetative development: a tale with a riddle. New Phytol 194: 895–909 [DOI] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang KL, Yoshida H, Lurin C, Ecker JR (2004) Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428: 945–950 [DOI] [PubMed] [Google Scholar]
- Wang W, Hall AE, O’Malley R, Bleecker AB (2003) Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc Natl Acad Sci USA 100: 352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Zhang C, Ji Y, Zhao Q, He W, An F, Jiang L, Guo H (2012) Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res 22: 1613–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RL, Bakshi A, Binder BM (2014a) Loss of the ETR1 ethylene receptor reduces the inhibitory effect of far-red light and darkness on seed germination of Arabidopsis thaliana. Front Plant Sci 5: 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RL, Kim H, Bakshi A, Binder BM (2014b) The ethylene receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 have contrasting roles in seed germination of Arabidopsis during salt stress. Plant Physiol 165: 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuriyanghan H, Zhang B, Cao WH, Ma B, Lei G, Liu YF, Wei W, Wu HJ, Chen LJ, Chen HW, et al. (2009) The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice. Plant Cell 21: 1473–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Zhang JS, Zhou HL, Li J, Zhang ZG, Wang DW, Chen SY (2003) Serine/threonine kinase activity in the putative histidine kinase-like ethylene receptor NTHK1 from tobacco. Plant J 33: 385–393 [DOI] [PubMed] [Google Scholar]
- Xie C, Zhang ZG, Zhang JS, He XJ, Cao WH, He SJ, Chen SY (2002) Spatial expression and characterization of a putative ethylene receptor protein NTHK1 in tobacco. Plant Cell Physiol 43: 810–815 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang B, Chen HW, Mu RL, Zhang WK, Zhao MY, Wei W, Wang F, Yu H, Lei G, Zou HF, et al. (2011) NIMA-related kinase NEK6 affects plant growth and stress response in Arabidopsis. Plant J 68: 830–843 [DOI] [PubMed] [Google Scholar]
- Zhang J, de Toledo SM, Pandey BN, Guo G, Pain D, Li H, Azzam EI (2012) Role of the translationally controlled tumor protein in DNA damage sensing and repair. Proc Natl Acad Sci USA 109: E926–E933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JS, Xie C, Liu F, Liu FH, Chen SY (1999) A novel tobacco gene coding for a product similar to bacterial two-component regulators. Chin Sci Bull 44: 1025–1029 [Google Scholar]
- Zhang JS, Xie C, Shen YG, Chen SY (2001) A two-component gene (NTHK1) encoding a putative ethylene-receptor homolog is both developmentally- and stress-regulated in tobacco. Theor Appl Genet 102: 815–824 [Google Scholar]
- Zhang ZG, Zhou HL, Chen T, Gong Y, Cao WH, Wang YJ, Zhang JS, Chen SY (2004) Evidence for serine/threonine and histidine kinase activity in the tobacco ethylene receptor protein NTHK2. Plant Physiol 136: 2971–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Lin Z, Grierson D (2008) Tomato ethylene receptor-CTR interactions: visualization of NEVER-RIPE interactions with multiple CTRs at the endoplasmic reticulum. J Exp Bot 59: 965–972 [DOI] [PubMed] [Google Scholar]
- Zhou HL, Cao WH, Cao YR, Liu J, Hao YJ, Zhang JS, Chen SY (2006) Roles of ethylene receptor NTHK1 domains in plant growth, stress response and protein phosphorylation. FEBS Lett 580: 1239–1250 [DOI] [PubMed] [Google Scholar]
- Zhou X, Liu Q, Xie F, Wen CK (2007) RTE1 is a Golgi-associated and ETR1-dependent negative regulator of ethylene responses. Plant Physiol 145: 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.