Brassinosteroid signaling is central to controlling auricle development and establishing the blade-sheath boundary in maize leaf.
Abstract
Brassinosteroids (BRs) are plant hormones involved in various growth and developmental processes. The BR signaling system is well established in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) but poorly understood in maize (Zea mays). BRASSINOSTEROID INSENSITIVE1 (BRI1) is a BR receptor, and database searches and additional genomic sequencing identified five maize homologs including duplicate copies of BRI1 itself. RNA interference (RNAi) using the extracellular coding region of a maize zmbri1 complementary DNA knocked down the expression of all five homologs. Decreased response to exogenously applied brassinolide and altered BR marker gene expression demonstrate that zmbri1-RNAi transgenic lines have compromised BR signaling. zmbri1-RNAi plants showed dwarf stature due to shortened internodes, with upper internodes most strongly affected. Leaves of zmbri1-RNAi plants are dark green, upright, and twisted, with decreased auricle formation. Kinematic analysis showed that decreased cell division and cell elongation both contributed to the shortened leaves. A BRASSINOSTEROID INSENSITIVE1-ETHYL METHANESULFONATE-SUPPRESSOR1-yellow fluorescent protein (BES1-YFP) transgenic line was developed that showed BR-inducible BES1-YFP accumulation in the nucleus, which was decreased in zmbri1-RNAi. Expression of the BES1-YFP reporter was strong in the auricle region of developing leaves, suggesting that localized BR signaling is involved in promoting auricle development, consistent with the zmbri1-RNAi phenotype. The blade-sheath boundary disruption, shorter ligule, and disrupted auricle morphology of RNAi lines resemble KNOTTED1-LIKE HOMEOBOX (KNOX) mutants, consistent with a mechanistic connection between KNOX genes and BR signaling.
Brassinosteroids (BRs) are ubiquitous plant hormones that promote plant growth by regulating cell elongation and division (Clouse, 1996; Clouse et al., 1996). BRs have other diverse roles, including enhancing tracheary element differentiation, stimulating ATPase activity, controlling microtubule orientation, and controlling flowering time, fertility, and leaf development (Iwasaki and Shibaoka, 1991; Clouse et al., 1996; Li et al., 1996; Schumacher et al., 1999; Catterou et al., 2001; Oh et al., 2011). BRs also function in tolerance to both biotic and abiotic stresses such as extreme temperatures, drought, and pathogens (Krishna, 2003).
Deficiencies in BR biosynthesis or signaling produce characteristic dwarf plant phenotypes (Clouse et al., 1996; Szekeres et al., 1996; Fujioka et al., 1997). Plant height is an important agricultural trait, as seen in the Green Revolution, where semidwarf mutants contributed to increased yields in small-grain crops (Salas Fernandez et al., 2009). BR-deficient dwarf rice (Oryza sativa) produced increased grain and biomass yields because the erect leaf habit allowed higher planting densities under field conditions (Sakamoto et al., 2006). In fact, Green Revolution Uzu barley (Hordeum vulgare) is based on a mutation of the UZU1 gene, which encodes a homolog of BRASSINOSTEROID INSENSITIVE1 (BRI1), a BR receptor (Chono et al., 2003).
Genes functioning in BR pathways have been identified by the analysis of dwarf mutants in several species, including Arabidopsis (Arabidopsis thaliana) and rice. Arabidopsis bri1 mutants are shortened, have reduced apical dominance, and are male sterile (Clouse et al., 1996). BRI1 encodes a Leu-rich repeat (LRR) receptor-like kinase that is located in the plasma membrane and contains an extracellular domain responsible for BR binding, a transmembrane sequence, and a cytoplasmic protein kinase domain (Li and Chory, 1997; Vert et al., 2005; Belkhadir and Chory, 2006). The island domain and subsequent LRR 22 are critical for BR binding (Kinoshita et al., 2005; Hothorn et al., 2011; She et al., 2011). Phosphorylation of the conserved residues Ser-1044 and Thr-1049 in the kinase activation loop activates the BRI1 kinase (Wang et al., 2005), while dephosphorylation of BRI1 by PROTEIN PHOSPHATASE2A inhibits its function (Wu et al., 2011).
BRI1 is partially redundant in BR signaling with related BRASSINOSTEROID INSENSITIVE1-LIKE RECEPTOR KINASE (BRL) paralogs, both in Arabidopsis and rice. In Arabidopsis, even though null alleles of brl1 or brl3 did not show obvious phenotypic defects in shoots, they enhanced the developmental defects of a weak bri1-5 mutant. In contrast to ubiquitously expressed BRI1, BRL1, BRL2, and BRL3 are tissue specific, mostly expressed in vascular tissues, while BRL1 and BRL3 are also expressed in root apices (Caño-Delgado et al., 2004; Zhou et al., 2004; Fàbregas et al., 2013). Both BRL1 and BRL3 can bind brassinolide (BL; Caño-Delgado et al., 2004). In rice, OsBRI1 is similar to the Arabidopsis BRI1 gene, and phenotypes of OsBRI1 rice mutants include dwarf plants with shortened internodes, erect leaves that are twisted and dark green, and photomorphogenesis in the dark (Yamamuro et al., 2000). There are three BR receptors in rice as well, and while OsBRI1 is universally expressed in all organs, OsBRL1 and OsBRL3 are expressed mostly in roots (Nakamura et al., 2006).
To date, two mutant genes of the BR biosynthetic pathway have been reported in maize (Zea mays). A classic dwarf mutant, nana plant1 (na1), has a mutation in a DE-ETIOLATED2 homologous gene, which encodes a 5α-reductase enzyme in the BR biosynthesis pathway (Hartwig et al., 2011), while the brassinosteroid-dependent1 (brd1) gene encodes brassinosteroid C-6 oxidase (Makarevitch et al., 2012). The maize BR-deficient mutants have shortened internodes, twisted, dark green, erect leaves, and feminized male flowers (Hartwig et al., 2011; Makarevitch et al., 2012). However, no genes in BR signaling have yet been reported in maize. Understanding BR signaling in maize might help improve this important crop for the production of biofuels, biomass, and grain yield. Here, we took a transgenic RNA interference (RNAi) approach to generate maize plants partially deficient for BRI1. These knockdown lines demonstrate that BRI1 functions are generally conserved in maize compared with other plant species, but they also exhibit unique phenotypes, suggesting either that maize possesses novel BR-regulated developmental processes or that aspects of maize morphology reveal processes not evident in other plants.
RESULTS
BRI1 Homologs in Maize
Database searches were conducted to determine whether maize possessed the potential to encode all the core components of the canonical BR signal transduction pathway. As reported in Supplemental Data Set S1, high-confidence homologs were identified for BRI1, BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1, BRASSINOSTEROID INSENSITIVE1 KINASE INHIBITOR1, BRASSINOSTEROID-SIGNALING KINASE1, CONSTITUTIVE DIFFERENTIAL GROWTH1, BRASSINOSTEROID INSENSITIVE1 SUPPRESSOR1, BRASSINOSTEROID-INSENSITIVE2, and BRASSINOSTEROID INSENSITIVE1-ETHYL METHANESULFONATE-SUPPRESSOR1/BRASSINAZOLE-RESISTANT1 (BES1/BZR1). As such, it appears highly likely that the core BR signaling system is conserved in maize. Interestingly, an unequivocal homolog of rice DWARF AND LOW-TILLERING could not be identified. Whether this reflects a gap in the genome assembly or a biological difference between rice and maize remains to be determined.
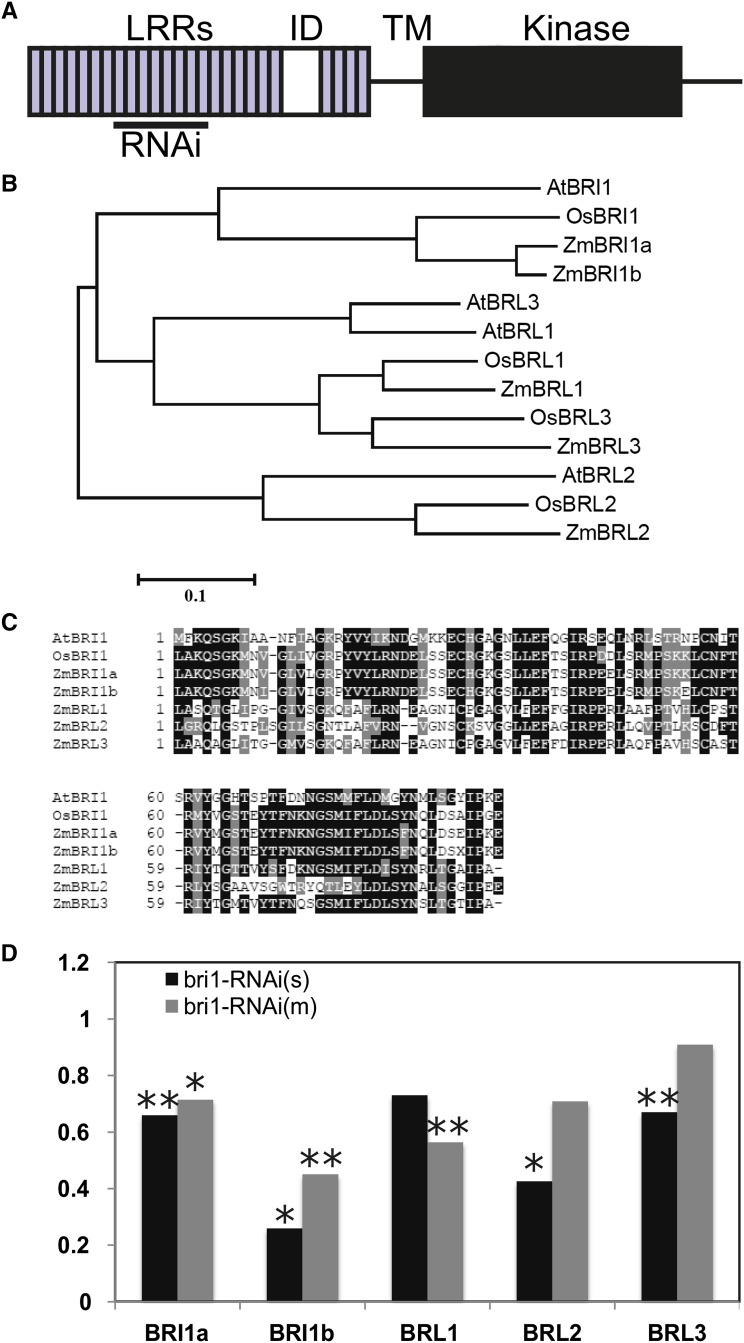
BRI1 belongs to a family of LRR receptor-like kinases, and at least two BRI1-like genes are involved in BR signaling in Arabidopsis (Caño-Delgado et al., 2004; Zhou et al., 2004). Maize BRI1 homologs were identified by using the Arabidopsis and rice BRI1 amino acid sequences to search the maize genome. A phylogenetic analysis was carried out to identify relationships among maize BRI1 homologs. Protein products were aligned (Supplemental Fig. S1), and a phylogenetic tree was obtained by the neighbor-joining method (Fig. 1). Five maize genes encode proteins belonging to the BRI1 family (Table I). Figure 1B shows that two maize gene products are most closely related to Arabidopsis and rice BRI1, while three additional genes encode proteins similar to BRL1, BRL2, and BRL3. The top hit, herein designated zmbri1a, corresponds to GRMZM2G048294 on chromosome 8. The ZmBRI1a protein product shows 54% amino acid identity and 69% similarity to Arabidopsis BRI1 (AtBRI1) and 79% amino acid identity and 88% similarity to rice BRI1 (OsBRI1). A second gene, GRMZM2G449830, located on chromosome 5, contained only a partial sequence in the genome assembly (cv B73 version 3) and was annotated as a transposable element. However, it was found through cloning and sequencing to contain a complete gene, designated zmbri1b (GenBank accession no. KP099562). The predicted ZmBRI1b protein shows 93% amino acid identity and 95% similarity to maize ZmBRI1a, 79% amino acid identity and 88% similarity to OsBRI1, and 54% amino acid identity and 69% similarity to AtBRI1. The island domain or insertion domain between LRRs 21 and 22 is a distinguishing characteristic of the BRI1 family of receptor kinases and the site of BR binding in BRI1 (Kinoshita et al., 2005; Hothorn et al., 2011; She et al., 2011). All five maize BRI1/BRL homologs contained a conserved island domain and LRR 22 sequences involved in BR binding (Fig. 1C).
Figure 1.
Characterization of BRI1 homologs in maize. A, BRI1 protein domains. The region of BRI1a used for RNAi is indicated. ID, Island domain; TM, transmembrane domain. B, Phylogenetic analysis showed that there are five BRI1 homologs in maize, including three BRL genes and a maize-specific duplication of the BRI1 gene. C, Island domain and LRR 22 domain alignment of BRI1 homologs show that all maize homologs belong to the BRI1 family. D, Quantitative reverse transcription (qRT)-PCR expression analysis of BRI1 homologs in zmbri1-RNAi plants. Shoot tissue was collected from plants showing characteristic zmbri1-RNAi phenotypes at the approximately eight-leaf stage. bri1-RNAi(s) is the strong event, and bri1-RNAi(m) is a mild event. Bars show proportional expression of the genes in RNAi lines relative to wild-type siblings, with wild-type expression set to 1. ANOVA P values: *, P < 0.05; and **, P < 0.01.
Table I. Sequence homology comparisons among maize BRI1-related proteins.
| Gene Model/GenBank Accession No. | Maize BRI | AtBRI1 | OsBRI1 | ZmBRI1a | ZmBRI1b | ZmBRL1 | ZmBRL2 |
|---|---|---|---|---|---|---|---|
| % identity (% similarity) | |||||||
| GRMZM2G048294/XM_008658585 | ZmBRI1a | 54 (68) | 79 (88) | 100 | |||
| GRMZM2G449830/KP099562 | ZmBRI1b | 55 (69) | 79 (88) | 93 (95) | 100 | ||
| GRMZM2G092604/XM_008654300 | ZmBRL1 | 51 (64) | 47 (60) | 50 (64) | 50 (65) | 100 | |
| GRMZM2G002515/XM_008660956 | ZmBRL2 | 45 (60) | 44 (58) | 43 (58) | 45 (60) | 45 (60) | 100 |
| GRMZM2G438007/XR_565429 | ZmBRL3 | 46 (62) | 44 (57) | 44 (59) | 49 (64) | 70 (82) | 46 (62) |
The expression patterns of these five BRI1/BRL homologs were examined by querying publicly available gene expression data sets. According to the Maize Gene Expression Atlas (Sekhon et al., 2011), the zmbri1a and zmbri1b transcripts are widely expressed in most tissues sampled, with zmbri1a showing somewhat higher expression levels and a more ubiquitous pattern than zmbri1b (Supplemental Fig. S2). Notably, both are expressed in shoot apices, developing leaves, internodes, and roots. The zmbrl genes all show generally lower steady-state transcript levels than the zmbri1 genes except in a couple instances, such as roots and germinating seeds, where zmbrl transcripts are present at higher levels than zmbri1b. The zmbrl1 and zmbrl3 transcripts are more highly expressed than zmbrl2 except in roots, where all three genes are expressed at comparable levels. In most tissues, the zmbrl2 transcript was below reliable detection levels.
Expression along the developmental gradient of a growing seedling leaf was also examined (Wang et al., 2014). As shown in Supplemental Figure S3, zmbri1a and zmbri1b expression was high at the base of the leaf in the zones of active cell division and cell elongation, gradually declining as cells differentiate and mature. Expression of the zmbrl genes was generally very low except for a brief increase in zmbrl1 transcript levels near the region of the leaf where tissues change from acting as carbon sinks and become photosynthetically active as source tissue.
zmbri1-RNAi Lines Show Compromised BR Signaling
To generate loss-of-function RNAi suppression of maize BRI1-related genes, a 498-bp fragment encoding LRRs 6 to 12 of the extracellular domain in the protein product was cloned in forward and reverse orientations into a pMCG1005 vector, controlled by the maize ubiquitin1 promoter, and transformed into maize. Among 17 independent bri1-RNAi T0 transgenic events, 13 showed clear dwarf phenotypes similar to those reported in rice dwarf61 or maize na1 seedlings (Yamamuro et al., 2000; Hartwig et al., 2011). Two events were selected for further analysis, one producing strong phenotypes and one producing intermediate phenotypes. Each was backcrossed three or more generations to a cv B73 inbred background.
Because BRI1 belongs to a gene family, it is likely that multiple members participate in BR signaling, perhaps redundantly, and that multiple members were targeted by the RNAi construct. To determine which genes in the maize genome were likely to be targeted by the zmbri1-RNAi construct, nucleotide database searches were conducted using the zmbri1a complementary DNA (cDNA) fragment contained in the RNAi construct. The results of this search were consistent with the phylogeny shown in Figure 1B. The two top hits included the original zmbri1a and zmbri1b, with 93% nucleotide identity. The zmbril homologs showed somewhat lower homology, ranging from 43.9% to 50.4% nucleotide identity, although localized stretches of nucleotides showed higher levels of homology (alignments are shown in Supplemental Fig. S4). The expression of these five genes was examined using RNA obtained from young developing shoot tissue, which included shoot apical meristems as well as nodes, internodes, and growing regions of leaves of approximately the seven youngest plastochrons, from the upper, most strongly affected nodes of the plant. Both zmbri1-RNAi lines showed decreased expression of all five BRI1 homologs compared with nontransgenic siblings (Fig. 1D).
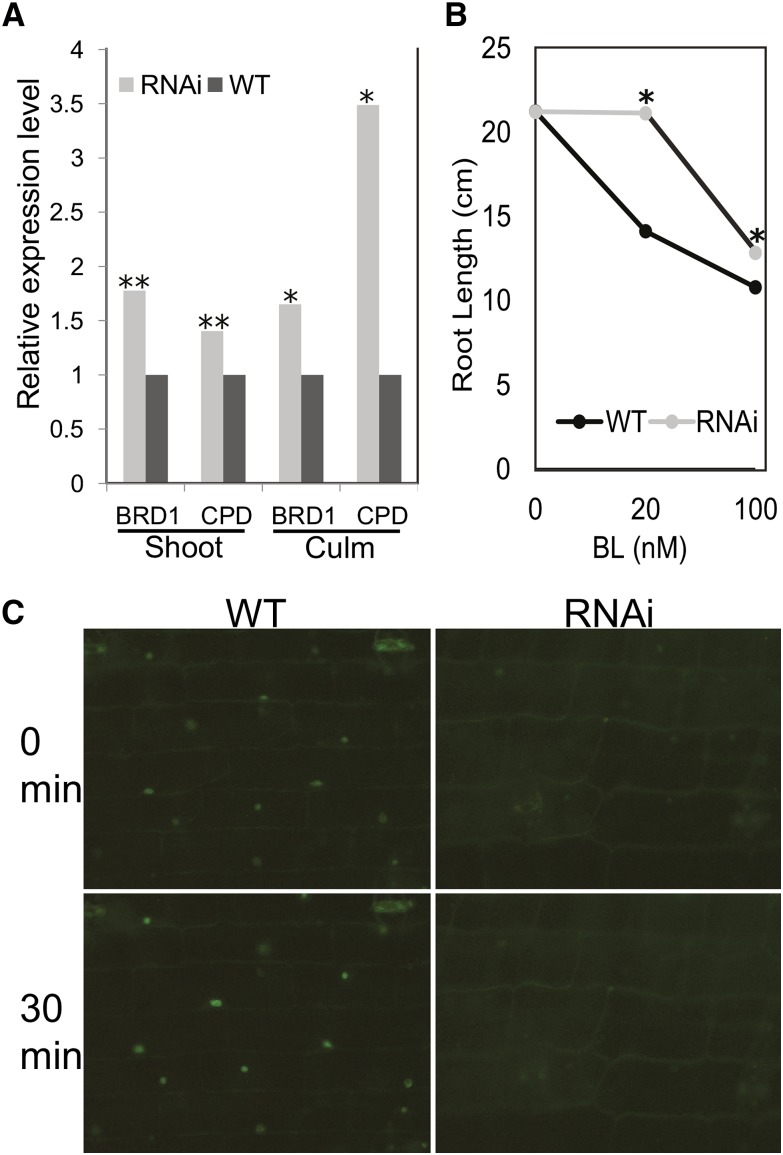
The expression of the BR marker genes brd1 and constitutive photomorphogenic dwarf (cpd), encoding enzymes involved in BR biosynthesis, was examined by qRT-PCR. These genes are negatively feedback regulated by BR signaling in Arabidopsis and rice, causing increased expression in mutants with decreased BR signaling (Mathur et al., 1998; Hong et al., 2002; Bai et al., 2007). As seen in Figure 2, both shoot and stalk tissues from strongly affected regions of the plants have increased expression of cpd and brd1, consistent with decreased BR signaling activity in strong zmbri1-RNAi lines compared with nontransgenic siblings.
Figure 2.
zmbri1-RNAi plants have disrupted BR signaling. A, BR marker gene expression in strong zmbri1-RNAi plants. Tissue was collected from 40-d-old plants when phenotypes were clearly exhibited and the most strongly affected portions of the plants were developing. Shoot tissue included culm, meristem, and growing regions of leaves from approximately the seven youngest plastochrons. Culm tissue included node and internode tissue from the same stage. Bars show proportional expression of genes in RNAi lines over wild-type (WT) siblings, with gene expression set to 1 in the wild type. Student’s t test P values: *, P < 0.05; and **, P < 0.01. B, BL root growth inhibition assay. Points show root length in cm of BL-treated roots versus mock-treated roots. The asterisk indicates a significant difference between wild-type and RNAi seedlings at P < 0.05 by Student’s t test. C, BES1-YFP expression pattern in wild-type and zmbri1-RNAi leaf sheath tissue. In the wild type, BES1-YFP accumulation in nuclei of leaf sheath cells could be induced to higher levels by BL application. The zmbri1-RNAi cells showed decreased BES1-YFP nuclear expression in untreated tissue and a slower response to exogenous BL treatment.
To further analyze BR signaling, we generated a BES1-yellow fluorescent protein (YFP) reporter for BR activity. BES1 is a transcription factor that is posttranslationally stabilized and translocated into the nucleus to regulate gene expression in response to BR signaling (Yin et al., 2002). As such, BR signaling is expected to increase nuclear fluorescence due to BES1-YPF accumulation (Ryu et al., 2010). The maize genome was searched for BES1 homologs, and GRMZM2G102514 was clearly the top candidate. The predicted protein encoded by this gene model contains 313 amino acids and shows 50% overall identity and 62% similarity to Arabidopsis BES1 and 78% identity and 82% similarity to OsBZR1. To create a stable BES1-YFP maize line, the YFP coding sequence was translationally fused to the 3′ end of the maize BES1 coding region within the native genomic context of the gene, including 5′ and 3′ regions as well as introns (Mohanty et al., 2009). As expected, nuclear fluorescence in leaf sheath cells is BR inducible in transgenic BES1-YFP lines, indicating that the fusion protein responds to BRs and can serve as a BR reporter (Fig. 2C).
Since BES1-YFP responds to BRs, a decrease in BR signaling should cause less BES1-YFP accumulation in the nucleus. To test the hypothesis that zmbri1-RNAi disrupts BR signaling, we examined several tissues in zmbri1-RNAi; BES1-YFP lines. As seen in Figure 2C, there is decreased nuclear fluorescence in zmbri1-RNAi leaf sheath cells compared with the wild-type cells, consistent with decreased levels of BR signaling and BES1-YFP accumulation. To see if these transgenic cells were able to respond to BR, we exposed both transgenic and nontransgenic leaf sheath cells to 1 μm BL. Figure 2C shows that wild-type cells are more responsive to BL than RNAi cells, confirming that BR signaling is impaired in zmbri1-RNAi lines.
To further confirm disrupted BR signaling in zmbri1-RNAi lines, we performed a BR root growth inhibition assay, a commonly used bioassay for BR sensitivity (Clouse, 1996; Chono et al., 2003; Müssig et al., 2003; Kim et al., 2007; Wang et al., 2007; Hartwig et al., 2012). Germinated seeds were treated with distilled, deionized water (mock), 20 nm BL, or 100 nm BL. After 10 d, wild-type plants showed a 2-fold greater inhibition of root growth compared with zmbri1-RNAi seedlings, indicating decreased BL sensitivity in transgenic seedlings (Fig. 2B; Supplemental Fig. S5).
zmbri1-RNAi Alters Maize Plant Architecture
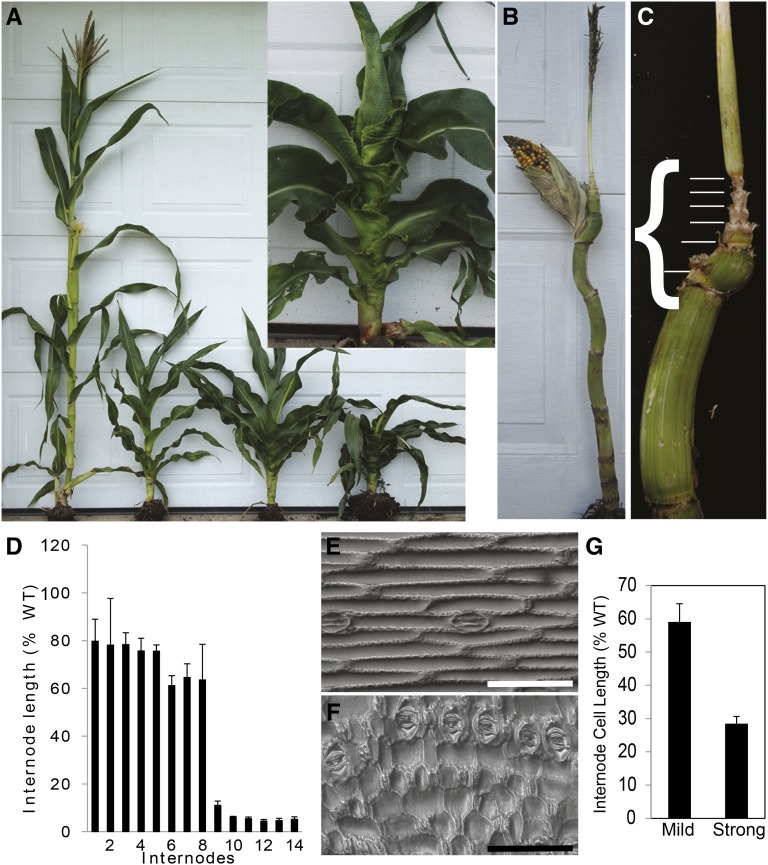
In both mild and strong zmbri1-RNAi lines, plant height is decreased compared with nontransgenic siblings (Fig. 3; Table II). Internodes were analyzed to determine the basis of the shorter plant height. While the number of internodes was not significantly changed (Table II), internode length was dramatically shorter in zmbri1-RNAi lines than in their nontransgenic siblings. In the milder lines, internodes are shortened by similar proportions throughout the plant (Supplemental Fig. S6), while in strong lines, internodes between the ear and tassel nodes were most dramatically affected (Fig. 3, B–D). Epidermal cell lengths in the strongly affected ninth internodes were shortened an average of 40% in the mild, and approximately 70% in the strong, zmbri1-RNAi lines compared with their nontransgenic counterparts, suggesting that decreased cell elongation is a major contributing factor to the decreased internode length (Fig. 3, E–G).
Figure 3.
zmbri1-RNAi plant architecture. A, zmbri1-RNAi plants with a wild-type (WT) sibling. The inset shows a closeup image of a zmbri1-RNAi plant. B and C, Dissected zmbri1-RNAi plants show extremely shortened internodes clustered between the ear and tassel nodes. D, Internode length measurements of zmbri1-RNAi plants. Bars show proportional internode lengths of the strong zmbri1-RNAi plants over wild-type siblings. While early internodes are moderately shortened, later internodes are strongly affected; the x axis represents internodes, and the y axis represents the proportional internode length of transgenic lines. E and F, Epidermal cells from internode 9 of the wild type (E) and zmbri1-RNAi (F). Bars = 100 μm. G, Internode 9 epidermal cells were shortened in both mild and strong zmbri1-RNAi lines. Bars show proportional cell length of zmbri1-RNAi plants over wild-type siblings.
Table II. Phenotypic analyses of RNAi lines.
| Line | Node Count | Blade Length | Blade Width | Leaf Sheath Length | Plant Height | Flowering Time |
|---|---|---|---|---|---|---|
| cm | d | |||||
| RNAi-strong lines | ||||||
| RNAi | 16.25 ± 0.3 | 62.13 ± 1.7 | 8.5 ± 0.2 | 4.75 ± 0.4 | 93.4 ± 1.4 | n.d.a |
| Wild type | 16.5 ± 0.3 | 75.25 ± 0.9 | 8.53 ± 0.1 | 14.15 ± 0.2 | 230.7 ± 7.4 | n.d. |
| Pb | 0.27 | 0.00067 | 0.45 | 2.95E-05 | 0.008 | n.d. |
| RNAi-mild lines | ||||||
| RNAi | 15 ± 0 | 67.25 ± 2 | 7.975 ± 0.4 | 7.725 ± 0.1 | 160.9 ± 5.3 | 60.5 ± 0.2 |
| Wild type | 15.25 ± 0.3 | 75.25 ± 0.5 | 8.05 ± 0.1 | 12.55 ± 0.2 | 220.5 ± 2.4 | 59.4 ± 0.4 |
| P | 0.19 | 0.01 | 0.43 | 2.16E-05 | 3.17E-06 | 0.015 |
Since most of the mutants have enclosed tassels, accurate data could not be obtained for this analysis.
P values are based on Student’s t test.
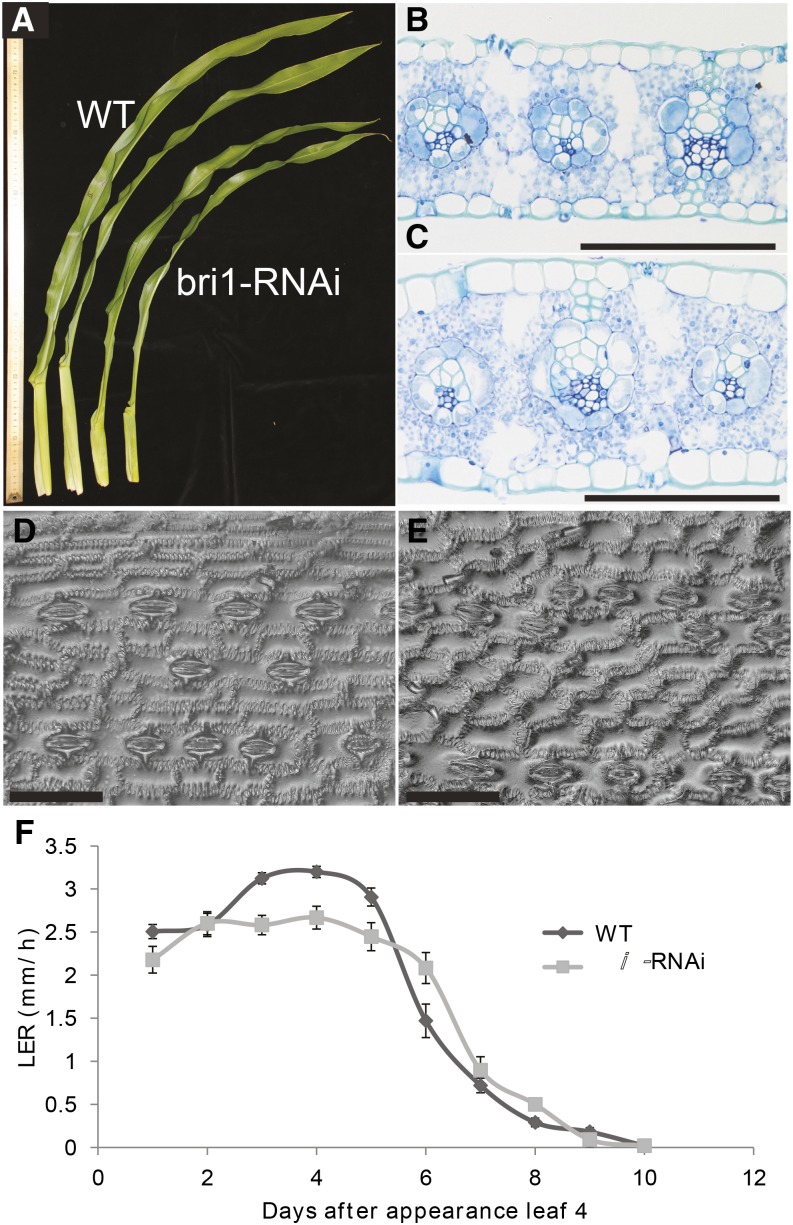
Leaf blades and sheathes were both shorter in RNAi lines (Fig. 4; Table II). Adult leaves in zmbri1-RNAi transgenic lines were dark green, upright, thickened, and had a wrinkled surface, compared with nontransgenic lines (Figs. 3A and 4C; Supplemental Fig. S7). These phenotypes were more apparent in adult leaves than in seedlings and stronger in field-grown plants than in the greenhouse. Like internode epidermal cells, leaf epidermal cells of zmbri1-RNAi lines were also shortened compared with nontransgenic siblings (Fig. 4, D and E). In the first leaf above the ear node, epidermal pavement cells sampled halfway along the blade length and midway between the midrib and margin had a mean length of 85.8 ± 3.4 μm in the wild type, while zmbri1-RNAi cells averaged 46.3 ± 2.7 μm (Student’s t test, P < 0.0001). Interestingly, cells showed increased depth (Fig. 4, B and C), even though their lengths were decreased, indicating a role for BR signaling in regulating directional cell growth. In the lines with the strongest phenotype, upper leaves strongly enclasped the tassel, necessitating manual unfurling to allow pollen release (Fig. 3A). Transgenic lines with mild phenotypes showed a delay in flowering (Table II), but the enclosed tassels of strong zmbri1-RNAi lines prevented accurate determination of anthesis onset.
Figure 4.
zmbri1-RNAi inhibits leaf growth. A, Wild-type (WT; left two leaves) versus zmbri1-RNAi leaves of greenhouse-grown plants. B and C, Mature leaf blade transverse sections. B, The wild type. C, zmbri1-RNAi leaves are thickened compared with the wild type. zmbri1-RNAi leaf epidermal cells are enlarged in cross section, even though they are shorter in length. Tissue was sampled halfway along the blade length and midway between the midrib and margin of the first leaf above the ear node. D, Leaf epidermal cells of the wild type (pavement cell mean, 85.8 ± 3.4 μm). E, zmbri1-RNAi shows decreased cell elongation (pavement cell mean, 46.3 ± 2.7 μm; Student’s t test P < 0.0001). Bars = 100 μm (B–E). F, Kinematic analysis showed that leaf elongation rate (LER) is decreased in zmbri1-RNAi during steady-state growth (days 1–5 after leaf emergence).
BRI1 Regulates Cell Division and Cell Expansion in Maize Leaf Growth
To understand the cellular basis of the zmbri1-RNAi shortened leaf phenotype, we performed a kinematic growth analysis on the fourth seedling leaf (Nelissen et al., 2013). This analysis quantifies growth parameters, including cell size, cell division rates, cell expansion rates, and the extents of the cell division and cell expansion zones on the leaf, thus allowing the number of cells undergoing each process to be calculated. After backcrossing to cv B73, seedling leaves did not display the dramatic corkscrew appearance or other morphological disturbances displayed by adult leaves. Nonetheless, the final length of seedling leaf 4 was decreased (Table III). Following the growth of the fourth leaf over time showed that the average growth rate or leaf elongation rate during steady-state growth was significantly reduced for zmbri1-RNAi (Fig. 4F). At the cellular level, both cell expansion, as measured by the final cell size, and cell production were reduced, by 7.6% and 7.9%, respectively (Table III). The kinematic analysis further showed that zmbri1-RNAi impacted cell division by affecting both the number of dividing cells and the rate at which they divide (Table III). The wild-type siblings had 26% more dividing cells, while the zmbri1-RNAi leaves showed cell division rate increased by 15.5% (cell cycle duration decreased by 16.03%), and the size of the zone of cell division and the number of dividing cells were decreased by 38.12% and 26.02%, respectively. In conclusion, our data demonstrate that the reduced leaf size of the zmbri1-RNAi lines is due to decreased cell number and cell size, indicating that BRI1 signaling regulates both cell division and cell expansion.
Table III. Kinematic growth analysis of leaf 4.
| Parameter | Wild Type | zmbri1-RNAi | Difference | P |
|---|---|---|---|---|
| % | ||||
| Size division zone (cm) | 1.98 ± 0.08 | 1.43 ± 0.08 | −38.12 | 0.009 |
| Size dividing cells (μm) | 26.77 ± 0.31 | 23.39 ± 0.87 | −14.43 | 0.047 |
| No. of dividing cells | 799.54 ± 32.45 | 634.47 ± 50.37 | −26.02 | 0.061 |
| Leaf elongation rate (mm h−1) | 2.81 ± 0.09 | 2.41 ± 0.07 | −16.79 | 0.004 |
| Cell production | 19.51 ± 0.31 | 18.08 ± 0.98 | −7.9 | 0.281 |
| Cell division rate (cell cell−1 h−1) | 0.024 ± 0.001 | 0.029 ± 0.003 | 15.52 | 0.314 |
| Cell cycle duration (h) | 28.48 ± 1.51 | 24.54 ± 2.70 | −16.03 | 0.289 |
| Mature cell length (μm) | 144.38 ± 2.35 | 134.14 ± 7.71 | −7.63 | 0.315 |
| Final leaf length (mm) | 606.5 ± 11.02 | 554 ± 17.58 | −9.48 | 0.068 |
zmbri1-RNAi Plants Show Leaf Auricle Phenotypes That Resemble KNOTTED1-LIKE HOMEOBOX Mutants
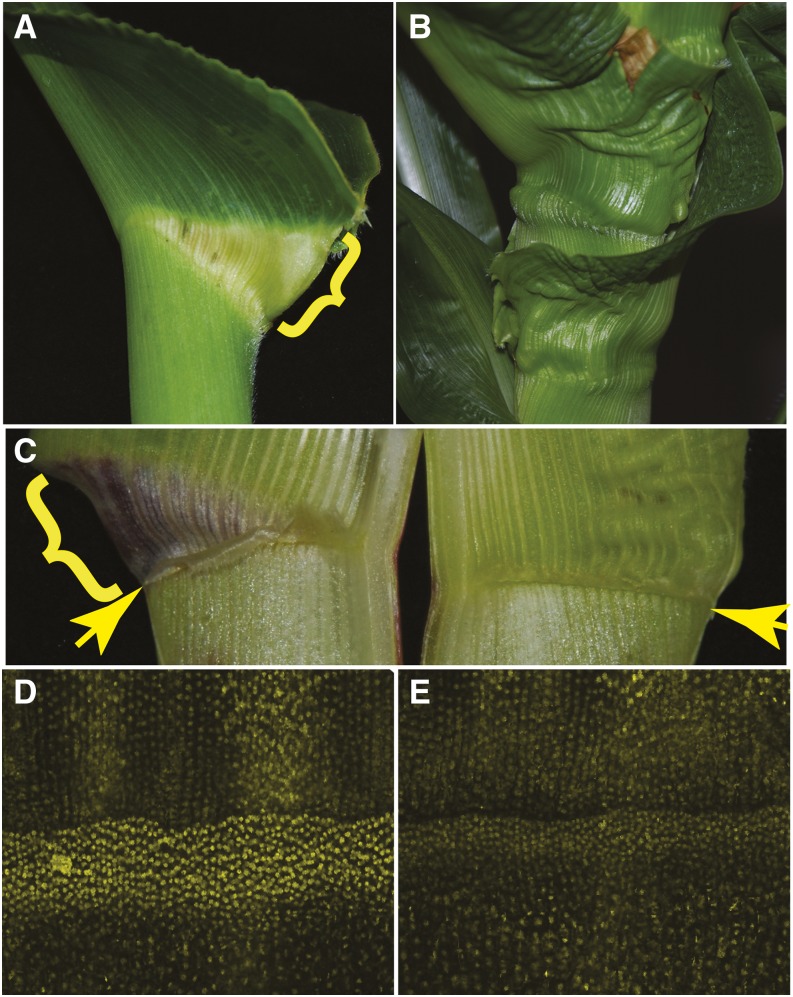
There was also a notable effect on auricle development in zmbri1-RNAi plants, as reported for the maize brd1 mutant (Fig. 5; Makarevitch et al., 2012). Like the internode shortening, upper nodes of the plants were more prone to displaying auricle disruption. In strongly affected leaves, auricles are largely missing and the ligules are decreased in length (Fig. 5, B and C). Sometimes only a rudimentary ligule is present. The auricle regions of zmbri1-RNAi leaves are thickened compared with nontransgenic leaves, even more so than in the lamina (Supplemental Fig. S7). In grass leaves, the auricle is a hinge-like structure located between the blade and sheath, which allows the blade to attain its normal angle (Moreno et al., 1997). In rice, BR mutants cause a decreased leaf angle and erect leaf habit compared with the wild type (Yamamuro et al., 2000; Morinaka et al., 2006), whereas BR gain-of-function mutants show increased lamina joint bending (Li et al., 2009). The altered auricle morphology of zmbri1-RNAi plants led us to hypothesize that BR signaling might have a specific function in maize auricle development. Consistent with these morphological changes, a striking accumulation of the BES1-YFP marker for BR activity was observed in nuclei of cells in the ligule-auricle region of developing maize leaves (Fig. 5, D and E). The band of increased BES1-YFP expression is sharply delineated at the distal end, consistent with a proposed boundary function separating the sheath and blade (Tsuda et al., 2014). The width of the auricle band was narrower, and nuclear BES1-YFP fluorescence intensity was significantly lower, in the auricle region of zmbri1-RNAi leaves than in the wild type (Fig. 5, D and E; Supplemental Table S1).
Figure 5.
zmbri1-RNAi disrupted leaf auricle formation. A, Auricle (bracket) of a wild-type leaf. B, zmbri1-RNAi leaves lacking well-defined auricles. C, Adaxial view of leaves. The wild type is on the left showing a normal auricle (bracket) and ligule (arrow). The zmbri1-RNAi leaf (right) lacks a normal auricle and has a reduced ligule (arrow). D and E, Confocal images showing nuclear BES1-YFP protein accumulation in a band of cells at the ligule-auricle region in developing plastochron 7 leaves. Signal intensity in the auricle band of the wild type (D) was 47% higher than that in the band of zmbri1-RNAi cells (E). The auricle band of zmbri1-RNAi (E) was narrower and nuclear fluorescence intensity was less bright compared with the wild-type band (D).
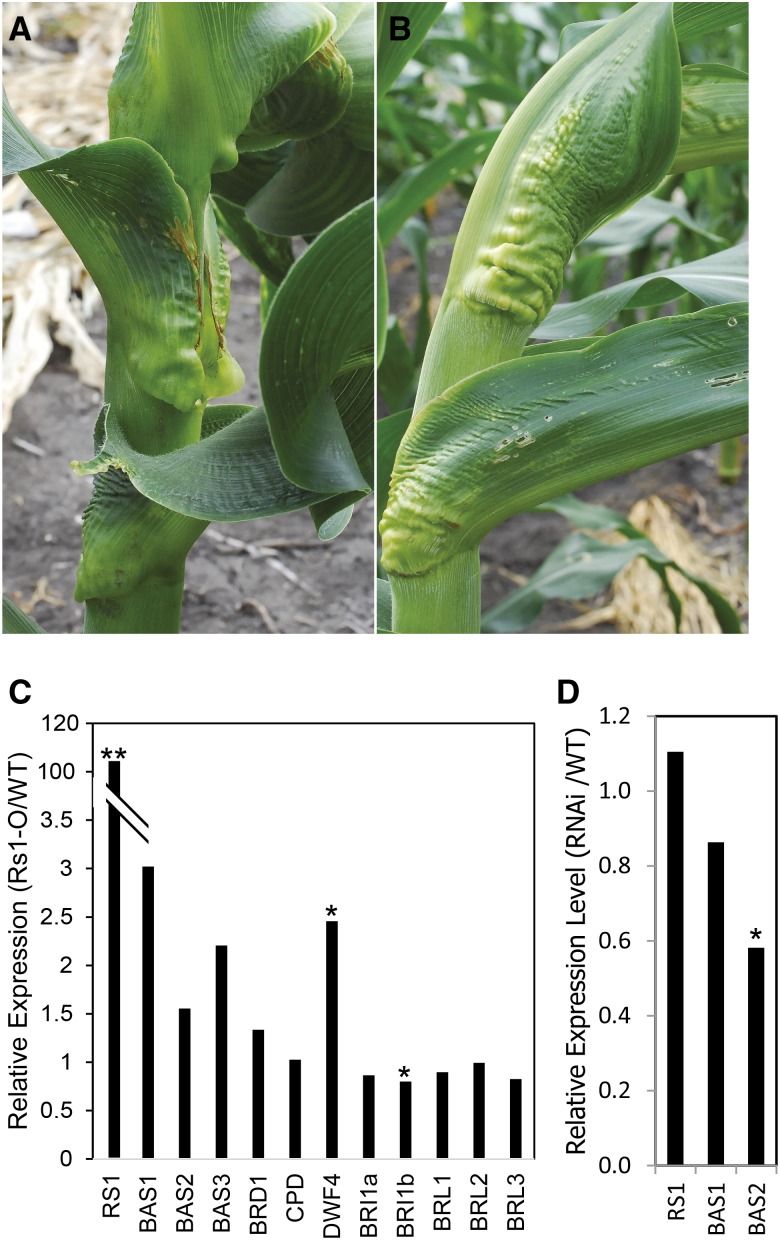
The leaf phenotype of the zmbri1-RNAi lines bears striking similarity to KNOTTED1-LIKE HOMEOBOX (KNOX) mutants, particularly Rough Sheath1-O (Rs1-O; Fig. 6; Becraft and Freeling, 1994; Schneeberger et al., 1995). Consistent with these phenotypes, the rice KNOX gene rice homeobox1 (OSH1) was recently shown to promote the expression of BR catabolic genes (Tsuda et al., 2014). To test whether a similar interaction exists between KNOX genes and BR signaling in maize, qRT-PCR was performed on RNA isolated from the auricle regions of developing leaves. As shown in Figure 6, Rs1-O mutants showed a dramatic increase in rs1 transcript, consistent with the mutant resulting from ectopic expression in leaves (Schneeberger et al., 1995). Three maize PHYB-4 ACTIVATION-TAGGED SUPPRESSOR1 (BAS1) homologs most similar to those up-regulated by OSH1 in rice all showed increased expression in Rs1-O, although not statistically significant. Among the BRI1 homologs, zmbri1b showed a significant decrease in Rs1-O. An increase in dwarf4 (dwf4) transcript abundance was also observed, consistent with derepression due to decreased feedback from BR signaling; however, no such response was observed for the other BR reporter genes cpd and brd1. In the auricle region of zmbri1-RNAi leaves, no change was observed for rs1 transcript levels, while two of the BAS1 homologs were decreased. Taken together, these results provide evidence of slight BR perturbations in Rs1-O leaves but no evidence of BR regulating rs1 expression.
Figure 6.
zmbri1-RNAi plants resemble Rs1-O mutants. A, zmbri1-RNAi. B, Rs1-O. C, BR-related gene expression in the auricle region of developing Rs1-O mutant leaves. The dwf4 and zmbri1b genes show altered expression in the mutant. D, KNOX rs1 gene expression was not altered in the auricle region of developing zmbri1-RNAi leaves. qRT-PCR was performed on RNA isolated from auricle regions of wild-type (WT) versus mutant or RNAi lines. Two millimeters of leaf tissue was dissected from the auricle region of plastochron 7 to 10 leaves from 10-leaf stage plants. *, P < 0.05; and **, P < 0.01 by Student’s t test.
DISCUSSION
While BR functions are well studied in Arabidopsis and rice (Clouse, 1996, 2011; Krishna, 2003), to our knowledge, this is the first report on BR signaling components in maize. We took a transgenic approach by targeting BRI1, the BR receptor, with RNAi. The zmbri1-RNAi lines show consistent phenotypes after more than four generations of backcrosses to cv B73 or other inbred lines. As expected, knockdown of zmbri1 caused decreased plant height (Fig. 3A; Table II), consistent with other species (Li and Chory, 1997; Yamamuro et al., 2000). This dwarf phenotype results from reduced internode length due in large part to decreased cell elongation (Fig. 3, D–G), consistent with BR control of cell expansion (Friedrichsen and Chory, 2001; Hartwig et al., 2011). Given the results in leaves, it is likely that decreased cell division also contributes to the shortened internodes.
The strong event showed differential effects on specific internodes, with those above the ear node most strongly affected. It is currently unknown whether this reflects some aspect of BR hormone biology. That the mild event showed nearly uniform effects throughout the plant suggests that this could be a specific effect of this particular event. Interestingly, Rs1 mutants also show allele-specific regional differences in their phenotypic expression; Rs1-O (Fig. 6) has the most pronounced effects on upper nodes, similar to strong zmbri1-RNAi, while Rs1-Z effects are more prevalent in earlier leaves (Becraft and Freeling, 1994). Thus, there might be some aspect of plant phase progression interacting with these genes.
A novel function for BRs in maize sex determination was reported for BR-deficient na1 mutants (Hartwig et al., 2011). These dwarves exhibit a tasselseed phenotype, where female floral structures fail to abort in male inflorescences. Fertilization of these perfect flowers results in kernels forming on tassels. Tasselseed phenotypes were never observed on our zmbri1-RNAi lines. It remains to be determined whether this is due to insufficient knockdown of BRI1 function or to unknown activities of biosynthetic products of the 5α-reductase enzyme encoded by na1.
Leaves of zmbri1-RNAi plants were short, thick, upright, twisted, and dark green in color. Even though the phenotypes of zmbri1-RNAi leaves become stronger in adult plants, a kinematic growth analysis showed that seedling leaf 4 was impacted and revealed important information about the functions of BR signaling in maize leaf development. Most cellular growth parameters were affected, including both cell division and elongation, similar to Arabidopsis (Szekeres et al., 1996; Azpiroz et al., 1998; Oh et al., 2011; Zhiponova et al., 2013). Decreased cell length indicated that zmbri1-RNAi inhibited cell elongation. While cell length was decreased, cell diameter was increased in the dorsiventral axis, contributing to the thickened blade phenotype and suggesting that the effect of BRs on cell expansion is directional (Fig. 4, B and C). Kinematic analysis showed that leaf elongation rate was significantly reduced (Fig. 4F). The extent of the cell division zone and the number of dividing cells were decreased. Cell division rate and cell cycle duration were also impacted, although these parameters were not statistically significant (Table III).
Several plant hormones affect both cell division and cell expansion (Takatsuka and Umeda, 2014). For example, GA can promote both cell division and expansion (Gonzalez et al., 2010; Band and Bennett, 2013) and was shown to function at the transition between these processes (Nelissen et al., 2012). In Arabidopsis leaves, BR has also been shown to impact both cell number and cell size (Gonzalez et al., 2010; Zhiponova et al., 2013). In addition, it was shown in the maize leaf that BR levels are high at the base of the leaf, where the actual growth processes of cell division and cell expansion take place (Nelissen et al., 2012). The high expression levels of zmbri1a and zmbri1b, but not the other brl genes, suggest that the BRI1 receptors encoded by these two genes are of primary importance for regulating leaf growth. The concomitant high BR hormone levels, high expression levels of zmbri1a and zmbri1b (Wang et al., 2014), and the observed phenotypes of the zmbri1-RNAi lines reported here suggest that the BR/BRI signaling cascade has a focus of action in the growth zones at the leaf base to regulate leaf size.
The auricle is a hinge-like structure at the blade-sheath junction that determines leaf angle. A striking phenotype of BR mutants in rice and barley is the upright leaf habit (Yamamuro et al., 2000; Chono et al., 2003), and application of BR to the leaf joint in rice can increase leaf angle by promoting auricle expansion (Tsuda et al., 2014). Leaf angle is an important agricultural trait because upright leaves increase sunlight penetration into a crop canopy (Sinclair and Sheehy, 1999). Breeding improvements to corn yields have arisen not from increases in individual plant yields but primarily from increased tolerance to higher cropping densities, enabled in large part by changes in shoot architecture (Duvick et al., 2004; Duvick, 2005). Upright leaf habit is a key component of the maize plant ideotype (Mock and Pearce, 1975) and has been correlated with increased yields at high planting densities (Pendleton et al., 1968; Pepper et al., 1977; Lambert and Johnson, 1978). Indeed, the BR-deficient rice d4 mutant showed increased grain yield due to improved tolerance to higher planting density (Sakamoto et al., 2006).
Our results demonstrate a key role for BR signaling in auricle development. The zmbri1-RNAi plants showed disrupted auricle morphology, and the BES1-YFP reporter suggests a localized concentration of BR signaling activity in the developing auricle (Fig. 5). The disrupted auricle phenotype was highly reminiscent of dominant KNOX mutants such as Rs1-O (Fig. 6; Becraft and Freeling, 1994; Schneeberger et al., 1995). A disrupted ligule was reported in maize brd1 mutants, but the KNOX similarity was not noted (Makarevitch et al., 2012). KNOX genes are important in shoot apical meristems for cells to maintain their pluripotent identities (Hay and Tsiantis, 2009). Down-regulation of KNOX genes is required for leaf initiation; therefore, the expression of these genes is normally repressed in leaf primordia (Hay and Tsiantis, 2010). Altered cell fate and disrupted organ shape are two basic consequences when KNOX genes are ectopically expressed in leaves (Hake et al., 2004). One of the abnormalities caused by mutations that derepress KNOX genes during leaf development is proximalization, a transformation of blade identity to sheath (Schneeberger et al., 1998; Foster et al., 1999). This phenotype is seen in several maize KNOX gene mutants and is most prevalent in the auricle/ligule region (Schneeberger et al., 1998; Foster et al., 1999).
The phenotypic resemblance between zmbri1-RNAi plants and KNOX mutants suggests a mechanistic relation between BRs and KNOX genes in auricle and/or leaf development and is consistent with recent reports of a regulatory connection between BR and KNOX genes in shoot apical meristems (Tsuda et al., 2014). It was recently reported that the KNOTTED1 and OSH1 proteins bind several BR metabolic genes in maize and rice, respectively, and that OSH1 promotes the expression of BAS1 homologs (Bolduc et al., 2012; Tsuda et al., 2014). BAS1 encodes a cytochrome P450 enzyme that catabolizes BRs in Arabidopsis (Tanaka et al., 2005; Turk et al., 2005). Overexpression of OSH1 caused decreased sensitivity of rice auricle tissue to BR application, suggesting that KNOX mutants act at least in part by modulating BR levels. Our results suggested the possibility of a mild impact on BR signaling in the auricle region of Rs1-O leaves but no evidence for BR signaling regulating rs1 expression. It is possible that KNOX genes and BR signaling pathways converge on a similar set of genes that control auricle development. It also remains possible that similar growth defects are produced by independent mechanisms.
Arabidopsis and rice each contain one gene, BRI1, encoding the major BR receptor, as well as three BRL genes, at least two of which contribute to BR signaling (Caño-Delgado et al., 2004; Zhou et al., 2004; Kim and Wang, 2010). Maize contains two close BRI1 homologs and three BRLs. One of the BRI1 homologs was not complete in the cv B73 version 2 genome assembly but was found to be intact and expressed. The phylogeny indicates that BRI1 duplicated specifically in maize, while the BRL1 and BRL3 pair resulted from independent duplications, one in the Arabidopsis lineage and another in the cereal lineage prior to the divergence of maize and rice. Given the high level of sequence similarity and the similar expression patterns between the maize bri1 homologs, it is highly likely that they function redundantly. The RNAi lines reported here were designed using the extracellular domain and decreased the expression of all five family members. The relatively low level of nucleotide similarity between the zmbri1a fragment used for RNAi and the BRL genes makes it questionable whether they are direct targets of suppression or whether decreased expression could be an indirect consequence of zmbri1 suppression. A topic for future studies will be sorting out the various contributions of individual receptor genes to BR signaling and different phenotypic effects in maize.
In summary, we show that BR signaling affects plant height, cell expansion, leaf morphology, and auricle development in maize. Similarities between zmbri1-RNAi plants and KNOX mutants support the relation between BR signaling and KNOX genes. Future studies should help reveal mechanisms of the interactions between these two pathways in leaf development, and that knowledge can be used to help optimize maize architecture for crop production.
MATERIALS AND METHODS
Bioinformatic Analyses
To survey and identify candidate BR signaling genes in maize (Zea mays), the canonical Arabidopsis (Arabidopsis thaliana) genes were identified at The Arabidopsis Information Resource, and protein amino acid sequences were copied. In the case of multiple isoforms, the one listed as the representative gene model was chosen. The amino acid sequence was used in a BLAST search of the National Center for Biotechnology Information (NCBI) maize reference protein database to identify GenPept accessions. These amino acid sequences were then used in BLAST searches of Gramene (http://ensembl.gramene.org/) to search translated gene models from the cv B73 reference genome AGPv3 (Proteins).
To identify BRI1 and BRL sequences, BLAST was used to search maize sequences in the NCBI databases, including Nonredundant protein sequences, Nucleotide collection, Reference RNA sequences, and High throughput genomic sequences (Altschul et al., 1990). With full-length Arabidopsis BRI1 amino acid sequences, 479 BLAST hits were returned with scores ranging from 276 to 1,105. Rice BRI1 amino acid sequences generated 331 BLAST hits with scores ranging from 284 to 1,699. All alignments were generated with ClustalW, and phylogenetic trees were obtained using default parameters of the Mega5.1 neighbor-joining method with Poisson model (Larkin et al., 2007; Tamura et al., 2011). Sequences belonging to the same clades as BRI1 or BRL proteins were selected for further analysis.
zmbri1b Cloning
The GRMZM2G449830 gene model, corresponding to the zmbri1b gene, contained 903 bp of transcribed sequence in the genome assembly (cv B73 version 2). The portion of zmbri1b contained in this gene model corresponded to a protein with nine LRR domains and no kinase domain. A combination of PCR cloning and database searching was used to identify the remaining sequence of the zmbri1b homolog. All primers and sequences used for cloning zmbri1b are listed in Supplemental Table S2. Since there was high similarity among the other four BRI1 homologs, initial attempts to identify the 3′ region of zmbri1b were made via PCR with primers based on conserved domains of these related genes. The template consisted of cDNA prepared from cv B73 seedling leaves using the Invitrogen SuperScript III reverse transcriptase kit. Using the Kinase5-R primer, located on kinase domains of other BRI1 homologs, as the reverse primer with a zmbri1b specific forward primer, an additional 1,393-bp sequence of zmbri1b homolog was found. By designing more primers (chr5-R1, chr5-R2, chr5-R3, and chr5-R4) based on 3′ sequences of zmbri1a, another 476 bp of this gene was identified. From this newly identified sequence, 55 nucleotides (sequence 1; Supplemental Table S2) were used to search maize in the NCBI Expressed Sequence Tags (EST) database and identified three ESTs (gi|32834046, gi|14243881, and gi|211181084) that encompassed the remaining 3′ coding region and untranslated region (UTR) of zmbri1b. No specific ESTs for the 5′ end of zmbri1b could be found in available databases. With the conserved domain approach for the N terminus of zmbri1b, another additional 196-bp sequence was found by PCR amplification using forward primers (chr5W-F7 and chr5W-F8) based on the first LRR domain of zmbri1a, in combination with the zmbri1b-specific qbri1-5R reverse primer. From this part, a 115-bp sequence (sequence 2; Supplemental Table S2) specific to brib was used for database searches and identified an NCBI Genome Survey Sequence record (gi|34267396) that included the start codon and 5′ UTR. To confirm that all these various fragments originated from a single gene, the entire zmbri1b cDNA was amplified with FideliTaq PCR Master Mix (2×; Affymetrix) proofreading enzyme from cv B73 seedling leaf cDNA via primers located on the 5′ UTR (Chr5-N1) and 3′ UTR (Chr5-UR). This nearly full-length cDNA was cloned into a TA vector and sequenced at the Iowa State University DNA Facility, and the sequence was deposited in GenBank (accession no. KP099562).
Production of Transgenic Lines
Because bioinformatic analyses indicated that GRMZM2G048294 (zmbri1a) encodes a likely maize ortholog of BRI1, a full-length cDNA [(ZM_BFb-Clone) Zea mays ‘B73’ 0034N07] was obtained from the Arizona Genomics Institute. The zmbri1-RNAi construct was generated by amplifying the region of the cDNA coding for the extracellular domain (bases 611–1,108 beginning from the start codon). Appropriate restriction sites were added to the primers 5′-TGGACCTCTCCGGGAACAAGAT-3′ and 5′-TGGTGCAGTTGGAGATTGAC-3′, and the fragment was cloned into the pMCG1005 RNAi vector. AvrII and AscI enzymes were used to put the BRI1 fragment in the reverse orientation between the ALCOHOL DEHYDROGENASE1 intron and rice Waxy-a intron, while XmaI and SpeI enzymes were used to put the BRI1 fragment in the forward orientation between OCTOPINE SYNTHASE 3′ and the rice Waxy-a intron. Transgenic lines were identified by glufosinate resistance and the expected BR-like mutant seedling phenotype.
BES1-YFP Construction
The citrine YFP-tagged construct, BES1-YFP, was made by fusing YFP on the C terminus of BES1 using the MultiSite Gateway method (Invitrogen). Briefly, the native 3,139-bp promoter and genomic coding sequence of zmbes1 (GRMZM2G102514; XP_008653282) was amplified with primers BES1_3GWp1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTgcattgatgctatcggagat-3′, where upper case letters represent Gateway [TM] cloning sequence and lowercase letters represent BES1 gene-specific sequence) and BES1_3GWp4 (5′-GGGGACAACTTTGTATAGAAAAGTTGGGTGcttggcgccgacgccga-3′), and the 1,300-bp native terminator of bes1 was amplified with primers BES1_3GWp3 (5′-GGGGACAACTTTGTATAATAAAGTTGAGtgaagcgattggcagcaa-3′) and BES1_3GWp2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTAcgttcgttccatctcctctc-3′), where uppercase letters represent Gateway cloning sequence and lowercase letters represent BES1 gene-specific sequence, and they were cloned into MultiSite Gateway donor vectors. The resulting two entry vectors were then recombined into the binary vector pAM1006 (a derivative of pTF101.1 carrying the Gateway cassette) with the entry vector containing YFP, yielding BES1-YFP. BES1-YFP was introduced into Agrobacterium tumefaciens EHA101 and transformed into maize line HiII at the Iowa State University Plant Transformation Facility. Transgenic lines were identified by glufosinate resistance and evaluated by YFP fluorescence for BES1-YFP expression.
qRT-PCR
Wild-type cv B73 and segregating RNAi lines were grown in the greenhouse for 40 d until the RNAi phenotypes were readily discernible, typically around the eight-leaf seedling stage, at which point the actively growing tissues were dissected out. For shoots, this included the nodes, internodes, and growing regions of leaves from approximately the seven youngest plastochrons as well as the shoot apical meristems. For culms, this included node and internode tissue from plastochrons 3 through 7, corresponding to nodes 9 to 13. Tissues were frozen immediately in liquid nitrogen, ground, and kept in −80°C until RNA purification. Auricle tissue was isolated from plants at the 10-leaf stage. Leaf tissues were isolated from the area within approximately 2 mm around the auricle, and ligule was dissected from plastochron 7 to 10 leaves immersed in RNAlater (Qiagen). Leaf tissues were also preserved in RNAlater overnight at 4°C until RNA purification.
RNA was purified using the Qiagen RNeasy Mini Kit according to the manufacturer’s protocol. RNA concentrations were measured by a NanoDrop ND-1000 spectrophotometer, samples were DNase treated (Promega RQ1 RNase-free DNase), and 1 to 2 µg of RNA was used per experiment. A total of 1 µL of DNase and 1.2 µL of RQ1 buffer were used, and volumes were adjusted to 12 µL with nuclease-free water. Samples were incubated at 37°C for 30 min, after which 1 µL of RQ1 DNase stop solution was added and samples were kept at 65°C for 10 min. A total of 11 µL of this volume was used directly for reverse transcription-PCR. Reverse transcription-PCR was performed with Invitrogen SuperScript III reverse transcriptase using the manufacturer’s procedure. From the 20-µL cDNA reaction, 1 µL was used for further PCRs. qRT-PCR was performed using iQ SYBR Green Supermix (Bio-Rad) and gene-specific primers (Supplemental Table S2) on a Stratagene MX4000 instrument located at the Iowa State University DNA Facility. A ubiquitin-conjugating enzyme gene (GRMZM2G027378) was used as an internal standard. A minimum of four biological and two technical replicates were used for each analysis. Statistical comparisons with ANOVA were made using JMP 11.0 statistical software.
Phenotypic Analyses
Each transgenic event was backcrossed a minimum of three times into a cv B73 inbred genetic background. All phenotypic analyses were done on plants grown under field conditions in Ames, Iowa. Plant height was measured from the bottom of the plant (the soil surface) to the top of the tassel. For internode length measurements, fully mature plants of segregating families were dug from the ground and dissected. Three wild-type and three RNAi plants were sampled for each comparison. Nodes and internodes were numbered and counted starting from the first seedling leaf node at the bottom of the plant, progressing to the upper parts. Two or more impressions of epidermal cells were obtained from the middle of the ninth internode by painting the culm surface with clear nail polish. After drying, the nail polish was peeled and examined with an Olympus BX60 microscope using differential interference contrast. Impressions were digitally photographed with a Jenoptik C5 camera, and cell lengths were measured with PROGRES 2.0 image-analysis software. Between 72 and 107 cells were counted per impression.
Tissue samples from mature leaf blades and auricle tissues were prepared for histological examination by cutting into small pieces and fixing in 5% (v/v) formalin, 50% (v/v) ethyl alcohol, and 10% (v/v) glacial acetic acid. Each leaf blade was sampled halfway along the blade length and midway between the midrib and margin. Auricles were also sampled midway between the midrib and margin. The dehydration and paraffin-embedding protocol was as described (Berlyn and Miksche, 1976). Paraffin blocks were sectioned at 7 to 10 μm with a Leica RM2235 manual microtome, and sections were affixed on glass slides, dewaxed, stained with 0.1% (w/v) Toluidine Blue, and mounted with Permount.
Kinematic Analysis of Leaf Growth
The kinematic analysis was performed as described by Nelissen et al. (2013). Leaf 4 was measured daily from appearance until growth stopped. By doing so, the leaf elongation rate was calculated (in mm h−1). The first days of linear increase are considered as steady-state growth (Ben-Haj-Salah and Tardieu, 1995). Leaf 4 was sampled during steady-state growth (2 d after appearance) for 4′,6-diamidino-phenylindole staining and differential interference microscopy to determine the cell length profiles. The growth analyses (leaf elongation rate and final leaf length) were done on at least five plants per genotype, and the kinematic analysis was performed on three plants per genotype.
BES1-YFP Response to BL in Maize Tissues
The behavior of the BES1-YFP reporter in response to BL was examined in several tissues from maize lines expressing the transgene. These included leaf sheath, mature and developing auricle, and leaf blade. Tissues were exogenously treated with varying concentrations of pure BL prepared in 1× phosphate-buffered saline buffer. A droplet of BL solution was applied to maize tissue (tissue was not submerged in solution), and BES1-YFP expression was checked at various intervals over a time course with a fluorescence microscope. While all these tissues could be observed to respond, the most favorable for photomicrography was sheath tissue. Nuclear fluorescence within the preauricle band of developing leaves was measured using ImageJ as described (McCloy et al., 2014).
BR Root Inhibition Assay
Seeds from segregating ears were surface sterilized (70% [v/v] ethanol followed by 5% [v/v] bleach) and soaked in sterilized wet paper towels for 2 to 3 d until germination. Germinated seeds were transferred to treatment containers containing paper towels soaked with the BL treatment solution, 0, 20, or 100 nm pure BL in distilled, deionized water. Paper towels at the bottom of the containers were rewet with the appropriate BL solution every other day. Seedlings were grown under continuous light at room temperature for 8 d, at which time each plant was genotyped for the presence of the BAR gene (marker for the transgene) and the primary root length was measured. Seedlings were PCR genotyped after root measurements. Ten to 18 seedlings were used for each treatment group. Statistical analyses were done with the JMP statistical software package.
Image Analysis
Images used for fluorescence quantification were captured by confocal microscopy at the Iowa State University Confocal and Multiphoton Facility. Image analysis was done using ImageJ V:2.0.0-rc-23 software (Fiji Is Just; Schindelin et al., 2012). For each nucleus, total YFP signal was calculated via the corrected total cell fluorescence method (McCloy et al., 2014).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number KP099562.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Amino acid sequence alignment of BRI1 homologs from Arabidopsis, rice, and maize.
Supplemental Figure S2. Transcript levels of maize bri1 homologs in cv B73 maize tissues.
Supplemental Figure S3. Transcript levels of maize BRI1 homologs in developing cv B73 maize seedling leaf 3 and relative transcript levels in 1-cm increments along maize seedling leaf 4 as determined by qRT-PCR.
Supplemental Figure S4. Nucleotide sequence of the zmbri1a cDNA used for the RNAi construct aligned with BRI1 homologs.
Supplemental Figure S5. BR root growth assay.
Supplemental Figure S6. Internode lengths of mild zmbri1-RNAi plants.
Supplemental Figure S7. Leaf traits of zmbri1-RNAi plants.
Supplemental Table S1. Fluorescence intensity of nuclei in the auricle bands of wild-type versus zmbri1-RNAi leaves.
Supplemental Table S2. Primers and sequences used.
Supplemental Data Set S1. Maize BR signaling genes.
Supplementary Material
Acknowledgments
We thank the Iowa State University Bessey Microscopy and Nanoimaging Facility and the Confocal and Multiphoton Facility for assistance with microscopy and Maradi Pho for extensive technical support.
Glossary
- BR
brassinosteroid
- BL
brassinolide
- LRR
leucine-rich repeat
- RNAi
RNA interference
- cDNA
complementary DNA
- qRT
quantitative reverse transcription
- NCBI
National Center for Biotechnology Information
- UTR
untranslated region
Footnotes
This work was supported by the Iowa State University Plant Sciences Institute and the Genetics, Development, and Cell Biology Department and by the Ministry of National Education, Republic of Turkey (education fellowship to G.K.).
Present address: Department of Genetics and Bioengineering, Faculty of Architecture and Engineering, Ahi Evran University, 40100 Kirsehir, Turkey.
Articles can be viewed without a subscription.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Azpiroz R, Wu Y, LoCascio JC, Feldmann KA (1998) An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell 10: 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY (2007) Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA 104: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band LR, Bennett MJ (2013) Mapping the site of action of the Green Revolution hormone gibberellin. Proc Natl Acad Sci USA 110: 4443–4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW, Freeling M (1994) Genetic analysis of Rough sheath1 developmental mutants of maize. Genetics 136: 295–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Chory J (2006) Brassinosteroid signaling: a paradigm for steroid hormone signaling from the cell surface. Science 314: 1410–1411 [DOI] [PubMed] [Google Scholar]
- Ben-Haj-Salah H, Tardieu F (1995) Temperature affects expansion rate of maize leaves without change in spatial distribution of cell length (analysis of the coordination between cell division and cell expansion). Plant Physiol 109: 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlyn GP, Miksche JP (1976) Botanical Microtechnique and Cytochemistry. Iowa State University Press, Ames, IA [Google Scholar]
- Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O’Connor D, Grotewold E, Hake S (2012) Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev 26: 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caño-Delgado A, Yin Y, Yu C, Vafeados D, Mora-García S, Cheng JC, Nam KH, Li J, Chory J (2004) BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131: 5341–5351 [DOI] [PubMed] [Google Scholar]
- Catterou M, Dubois F, Schaller H, Aubanelle L, Vilcot B, Sangwan-Norreel BS, Sangwan RS (2001) Brassinosteroids, microtubules and cell elongation in Arabidopsis thaliana. II. Effects of brassinosteroids on microtubules and cell elongation in the bul1 mutant. Planta 212: 673–683 [DOI] [PubMed] [Google Scholar]
- Chono M, Honda I, Zeniya H, Yoneyama K, Saisho D, Takeda K, Takatsuto S, Hoshino T, Watanabe Y (2003) A semidwarf phenotype of barley uzu results from a nucleotide substitution in the gene encoding a putative brassinosteroid receptor. Plant Physiol 133: 1209–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD. (1996) Molecular genetic studies confirm the role of brassinosteroids in plant growth and development. Plant J 10: 1–8 [DOI] [PubMed] [Google Scholar]
- Clouse SD. (2011) Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC (1996) A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol 111: 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick DN. (2005) Genetic progress in yield of United States maize (Zea mays L.). Maydica 50: 193–202 [Google Scholar]
- Duvick DN, Smith JSC, Cooper M (2004) Long-term selection in a commercial hybrid maize breeding program. In Janick J, ed, Plant Breeding Reviews 24 (Part 2). Long Term Selection: Crops, Animals, and Bacteria. John Wiley & Sons, New York, pp 109–151 [Google Scholar]
- Fàbregas N, Li N, Boeren S, Nash TE, Goshe MB, Clouse SD, de Vries S, Caño-Delgado AI (2013) The brassinosteroid insensitive1-like3 signalosome complex regulates Arabidopsis root development. Plant Cell 25: 3377–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T, Yamaguchi J, Wong BC, Veit B, Hake S (1999) Gnarley1 is a dominant mutation in the knox4 homeobox gene affecting cell shape and identity. Plant Cell 11: 1239–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen D, Chory J (2001) Steroid signaling in plants: from the cell surface to the nucleus. BioEssays 23: 1028–1036 [DOI] [PubMed] [Google Scholar]
- Fujioka S, Li J, Choi YH, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J, et al. (1997) The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 9: 1951–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, Jikumaru Y, Chae E, Dhondt S, Van Daele T, De Milde L, Weigel D, Kamiya Y, et al. (2010) Increased leaf size: different means to an end. Plant Physiol 153: 1261–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S, Smith HMS, Holtan H, Magnani E, Mele G, Ramirez J (2004) The role of knox genes in plant development. Annu Rev Cell Dev Biol 20: 125–151 [DOI] [PubMed] [Google Scholar]
- Hartwig T, Chuck GS, Fujioka S, Klempien A, Weizbauer R, Potluri DP, Choe S, Johal GS, Schulz B (2011) Brassinosteroid control of sex determination in maize. Proc Natl Acad Sci USA 108: 19814–19819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig T, Corvalan C, Best NB, Budka JS, Zhu JY, Choe S, Schulz B (2012) Propiconazole is a specific and accessible brassinosteroid (BR) biosynthesis inhibitor for Arabidopsis and maize. PLoS ONE 7: e36625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Tsiantis M (2009) A KNOX family TALE. Curr Opin Plant Biol 12: 593–598 [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M (2010) KNOX genes: versatile regulators of plant development and diversity. Development 137: 3153–3165 [DOI] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, Inukai Y, Fujioka S, Shimada Y, Takatsuto S, Agetsuma M, Yoshida S, Watanabe Y, et al. (2002) Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J 32: 495–508 [DOI] [PubMed] [Google Scholar]
- Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, Wilson IA, Chory J (2011) Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki T, Shibaoka H (1991) Brassinosteroids act as regulators of tracheary-element differentiation in isolated Zinnia mesophyll cells. Plant Cell Physiol 32: 1007–1014 [Google Scholar]
- Kim TW, Lee SM, Joo SH, Yun HS, Lee Y, Kaufman PB, Kirakosyan A, Kim SH, Nam KH, Lee JS, et al. (2007) Elongation and gravitropic responses of Arabidopsis roots are regulated by brassinolide and IAA. Plant Cell Environ 30: 679–689 [DOI] [PubMed] [Google Scholar]
- Kim TW, Wang ZY (2010) Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol 61: 681–704 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Caño-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J (2005) Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Krishna P. (2003) Brassinosteroid-mediated stress responses. J Plant Growth Regul 22: 289–297 [DOI] [PubMed] [Google Scholar]
- Lambert RJ, Johnson RR (1978) Leaf angle, tassel morphology, and the performance of maize hybrids. Crop Sci 18: 499–502 [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li D, Wang L, Wang M, Xu YY, Luo W, Liu YJ, Xu ZH, Li J, Chong K (2009) Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol J 7: 791–806 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J (1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401 [DOI] [PubMed] [Google Scholar]
- Makarevitch I, Thompson A, Muehlbauer GJ, Springer NM (2012) Brd1 gene in maize encodes a brassinosteroid C-6 oxidase. PLoS ONE 7: e30798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Molnár G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C, et al. (1998) Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J 14: 593–602 [DOI] [PubMed] [Google Scholar]
- McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A (2014) Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle 13: 1400–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock JJ, Pearce RB (1975) An ideotype of maize. Euphytica 24: 613–623 [Google Scholar]
- Mohanty A, Luo A, DeBlasio S, Ling X, Yang Y, Tuthill DE, Williams KE, Hill D, Zadrozny T, Chan A, et al. (2009) Advancing cell biology and functional genomics in maize using fluorescent protein-tagged lines. Plant Physiol 149: 601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco MK, Sen TZ, Dharmawardhana PD, Ren L, Schaeffer M, Naithani S, Amarasinghe V, Thomason J, Harper L, Gardiner J, et al. (2013) Maize metabolic network construction and transcriptome analysis. Plant Genome http://dx.doi.org/10.3835/plantgenome2012.09.0025
- Moreno MA, Harper LC, Krueger RW, Dellaporta SL, Freeling M (1997) liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev 11: 616–628 [DOI] [PubMed] [Google Scholar]
- Morinaka Y, Sakamoto T, Inukai Y, Agetsuma M, Kitano H, Ashikari M, Matsuoka M (2006) Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol 141: 924–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müssig C, Shin GH, Altmann T (2003) Brassinosteroids promote root growth in Arabidopsis. Plant Physiol 133: 1261–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Fujioka S, Sunohara H, Kamiya N, Hong Z, Inukai Y, Miura K, Takatsuto S, Yoshida S, Ueguchi-Tanaka M, et al. (2006) The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol 140: 580–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Rymen B, Coppens F, Dhondt S, Fiorani F, Beemster GTS (2013) Kinematic analysis of cell division in leaves of mono- and dicotyledonous species: a basis for understanding growth and developing refined molecular sampling strategies. Methods Mol Biol 959: 247–264 [DOI] [PubMed] [Google Scholar]
- Nelissen H, Rymen B, Jikumaru Y, Demuynck K, Van Lijsebettens M, Kamiya Y, Inzé D, Beemster GTS (2012) A local maximum in gibberellin levels regulates maize leaf growth by spatial control of cell division. Curr Biol 22: 1183–1187 [DOI] [PubMed] [Google Scholar]
- Oh MH, Sun J, Oh DH, Zielinski RE, Clouse SD, Huber SC (2011) Enhancing Arabidopsis leaf growth by engineering the BRASSINOSTEROID INSENSITIVE1 receptor kinase. Plant Physiol 157: 120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton JW, Smith GE, Winter SR, Johnston TJ (1968) Field investigation of the relationships of leaf angle in corn (Zea mays L.) to grain yield and apparent photosynthesis. Agron J 60: 422–424 [Google Scholar]
- Pepper GE, Pearce RB, Mock JJ (1977) Leaf orientation and yield of maize. Crop Sci 17: 883–886 [Google Scholar]
- Ryu H, Kim K, Cho H, Hwang I (2010) Predominant actions of cytosolic BSU1 and nuclear BIN2 regulate subcellular localization of BES1 in brassinosteroid signaling. Mol Cells 29: 291–296 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, et al. (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24: 105–109 [DOI] [PubMed] [Google Scholar]
- Salas Fernandez MG, Becraft PW, Yin Y, Lübberstedt T (2009) From dwarves to giants? Plant height manipulation for biomass yield. Trends Plant Sci 14: 454–461 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger R, Tsiantis M, Freeling M, Langdale JA (1998) The rough sheath2 gene negatively regulates homeobox gene expression during maize leaf development. Development 125: 2857–2865 [DOI] [PubMed] [Google Scholar]
- Schneeberger RG, Becraft PW, Hake S, Freeling M (1995) Ectopic expression of the knox homeo box gene rough sheath1 alters cell fate in the maize leaf. Genes Dev 9: 2292–2304 [DOI] [PubMed] [Google Scholar]
- Schumacher K, Vafeados D, McCarthy M, Sze H, Wilkins T, Chory J (1999) The Arabidopsis det3 mutant reveals a central role for the vacuolar H+-ATPase in plant growth and development. Genes Dev 13: 3259–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66: 553–563 [DOI] [PubMed] [Google Scholar]
- She J, Han Z, Kim TW, Wang J, Cheng W, Chang J, Shi S, Wang J, Yang M, Wang ZY, et al. (2011) Structural insight into brassinosteroid perception by BRI1. Nature 474: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair TR, Sheehy JE (1999) Erect leaves and photosynthesis in rice. Science 283: 1455 [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Takatsuka H, Umeda M (2014) Hormonal control of cell division and elongation along differentiation trajectories in roots. J Exp Bot 65: 2633–2643 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Asami T, Yoshida S, Nakamura Y, Matsuo T, Okamoto S (2005) Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol 138: 1117–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Kurata N, Ohyanagi H, Hake S (2014) Genome-wide study of KNOX regulatory network reveals brassinosteroid catabolic genes important for shoot meristem function in rice. Plant Cell 26: 3488–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Wang H, Torres QI, Ward JM, Murthy G, et al. (2005) BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J 42: 23–34 [DOI] [PubMed] [Google Scholar]
- Vert G, Nemhauser JL, Geldner N, Hong F, Chory J (2005) Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol 21: 177–201 [DOI] [PubMed] [Google Scholar]
- Wang L, Czedik-Eysenberg A, Mertz RA, Si Y, Tohge T, Nunes-Nesi A, Arrivault S, Dedow LK, Bryant DW, Zhou W, et al. (2014) Comparative analyses of C4 and C3 photosynthesis in developing leaves of maize and rice. Nat Biotechnol 32: 1158–1165 [DOI] [PubMed] [Google Scholar]
- Wang L, Xu YY, Li J, Powell RA, Xu ZH, Chong K (2007) Transgenic rice plants ectopically expressing AtBAK1 are semi-dwarfed and hypersensitive to 24-epibrassinolide. J Plant Physiol 164: 655–664 [DOI] [PubMed] [Google Scholar]
- Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD (2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Wang X, Li X, Kamiya Y, Otegui MS, Chory J (2011) Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci Signal 4: ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Zhiponova MK, Vanhoutte I, Boudolf V, Betti C, Dhondt S, Coppens F, Mylle E, Maes S, González-García MP, Caño-Delgado AI, et al. (2013) Brassinosteroid production and signaling differentially control cell division and expansion in the leaf. New Phytol 197: 490–502 [DOI] [PubMed] [Google Scholar]
- Zhou A, Wang H, Walker JC, Li J (2004) BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J 40: 399–409 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.