A major plant plasma membrane-targeted bacterial virulence protein is linked to reduced expression of an Arabidopsis gene required for innate immunity.
Abstract
Many bacterial pathogens of plants and animals deliver effector proteins into host cells to promote infection. Elucidation of how pathogen effector proteins function not only is critical for understanding bacterial pathogenesis but also provides a useful tool in discovering the functions of host genes. In this study, we characterized the Pseudomonas syringae pv tomato DC3000 effector protein Avirulence Protein E (AvrE), the founding member of a widely distributed, yet functionally enigmatic, bacterial effector family. We show that AvrE is localized in the plasma membrane (PM) and PM-associated vesicle-like structures in the plant cell. AvrE contains two physically interacting domains, and the amino-terminal portion contains a PM-localization signal. Genome-wide microarray analysis indicates that AvrE, as well as the functionally redundant effector Hypersensitive response and pathogenicity-dependent Outer Protein M1, down-regulates the expression of the NONRACE-SPECIFIC DISEASE RESISTANCE1/HARPIN-INDUCED1-LIKE13 (NHL13) gene in Arabidopsis (Arabidopsis thaliana). Mutational analysis shows that NHL13 is required for plant immunity, as the nhl13 mutant plant displayed enhanced disease susceptibility. Our results defined the action site of one of the most important bacterial virulence proteins in plants and the antibacterial immunity function of the NHL13 gene.
As a common mechanism of pathogenesis, many animal and plant pathogenic bacteria utilize the type III secretion system (T3SS) to deliver effector proteins into the host cell (Galán and Collmer, 1999; He et al., 2004). For the past decade, Pseudomonas syringae pv tomato (Pst) strain DC3000 has been employed as a model pathogen to define the virulence functions of type III secretion system effectors (T3Es). Study of T3Es has led to significant advances not only in the understanding of the fundamental mechanisms underlying bacterial pathogenesis but also in the discovery and characterization of functions of relevant plant genes (Block and Alfano, 2011; Dou and Zhou, 2012; Xin and He, 2013). However, despite much progress in this area, the virulence functions of most T3Es from Pst DC3000 and other plant pathogenic bacteria remain undefined, illustrating substantial potential in using pathogen virulence factors as probes in the characterization of the functions of plant genes.
Among the most crucial T3Es of plant pathogenic bacteria is the Avirulence Protein E (AvrE) family of effectors, of which AvrE from Pst DC3000 is the founding member (Lorang and Keen, 1995; Degrave et al., 2015). The AvrE effector family is present in diverse plant pathogenic bacteria that belong to the genera Pseudomonas, Pantoea, Erwinia, Dickeya, and Pectobacterium. Importantly, mutation of avrE orthologs has been shown to cause a dramatic decrease in the virulence of a number of bacteria (Gaudriault et al., 1997; Bogdanove et al., 1998; Frederick et al., 2001; DebRoy et al., 2004; Boureau et al., 2006). In P. syringae, AvrE is functionally redundant to another effector protein, Hypersensitive response and pathogenicity-dependent Outer Protein M1 (HopM1; Alfano et al., 2000; DebRoy et al., 2004; Badel et al., 2006). Mutation of either avrE or hopM1 alone does not strongly affect Pst DC3000 virulence, but the avrE hopM1 double mutant is severely impaired in virulence (Badel et al., 2006). Because of the crucial virulence role of AvrE family effectors and the wide distribution of this effector family in diverse plant pathogenic bacteria, an understanding of the virulence functions of AvrE family effectors is expected to have a substantial impact on our understanding of bacterial diseases in plants. Similarly, because the virulence functions of AvrE family effectors are not understood at the molecular level, study of AvrE family effectors may lead to new insights into the functions of relevant plant genes.
For technical reasons, AvrE family effectors have been very challenging to study due to their extremely large size (approximately 200 kD) and high toxicity to plant and yeast (Saccharomyces cerevisiae) cells. Nevertheless, some progress has been made in the characterization of AvrE family effectors. For example, Ham et al. (2008, 2009) identified two sequence motifs in AvrE family effectors: the WxxxE motif (where x stands for any amino acid other than Trp or Glu) at the N-terminal one-half and a putative endoplasmic reticulum membrane retention/retrieval-like signal (ERMRS) at the C terminus. Some WxxxE effectors from human pathogenic bacteria function as guanine nucleotide-exchange factors to activate small GTPases. In these effectors, the WxxxE motif appears to play a structural role in properly positioning the catalytic loop and is required for the guanine nucleotide-exchange factor activity (Alto et al., 2006; Ohlson et al., 2008; Huang et al., 2009c). However, whether the WxxxE motif in the AvrE family has a similar or a different structural function has yet to be determined. Similarly, it is not known whether the C-terminal ERMRS motif indeed targets AvrE family effectors to the endoplasmic reticulum (ER) in the plant cell.
All AvrE family effectors examined, including AvrE from Pst DC3000, Water-Soaking E (WtsE) from Pantoea stewartii, Disease-specific protein A (DspA)/E from Erwinia amylovora, and DspE from Pectobacterium carotovorum, are strong inducers of water soaking and/or cell death when expressed in host or nonhost plants (Frederick et al., 2001; Boureau et al., 2006; Ham et al., 2006, 2008, 2009; Degrave et al., 2008; Kim et al., 2011; Hogan et al., 2013). Conversely, deletion of avrE family genes delays or abolishes this ability (Bogdanove et al., 1998; Frederick et al., 2001; Badel et al., 2006; Ham et al., 2006). In addition, AvrE family effectors have been shown to suppress plant defense responses, such as callose deposition and expression of the defense gene PATHOGENESIS RELATED1 (DebRoy et al., 2004; Boureau et al., 2006; Ham et al., 2008, 2009), and DspA/E was reported to interact with several putative receptor kinases from apple (Malus domestica; Meng et al., 2006). Intererstingly, DspA/E affects actin dynamics and vesicle trafficking in yeast (Siamer et al., 2011), and a genetic screen identified sphingolipid intermediates and protein phosphatase 2A regulatory subunits important for its toxicity in yeast (Siamer et al., 2014). Transcriptional profiling of the host maize (Zea mays) plants shows that WtsE induces genes involved in secondary metabolism and the suppression of genes involved in photosynthesis (Asselin et al., 2015). In the nonhost plant Arabidopsis (Arabidopsis thaliana), transgenic expression of E. amylovora DspA/E results in the induction of a large suite of salicylic acid (SA)-dependent defense genes (Degrave et al., 2013). However, overall, our understanding of AvrE family effectors remains fragmentary. In particular, we do not know their sites of action in the host cell or the specific host genes that are required for their virulence functions, presenting a significant roadblock to our general understanding of bacterial pathogenesis in plants.
In this study, we initiated efforts to define (1) the subcellular localization of AvrE inside the plant cell and (2) the host gene expression associated with the virulence function of AvrE in the host plant Arabidopsis. These experiments led to the findings that (1) AvrE, although containing no known plasma membrane (PM)-targeting signal, is localized to the host PM and PM-associated vesicle-like structures, (2) AvrE contains two physically interacting domains with the N-terminal portion containing a PM-targeting signal, and (3) importantly, the virulence function of AvrE is linked to down-regulation of the expression of a specific member of the NONRACE-SPECIFIC DISEASE RESISTANCE1/HARPIN-INDUCED1-LIKE (NHL) gene family, NHL13, which we found to be required for antibacterial immunity in Arabidopsis. Together, these results defined the site of action of one of the most important bacterial virulence proteins in plants and the antibacterial immunity function of the NHL13 gene.
RESULTS
Transgenic Expression of avrE in Arabidopsis Restores the Virulence of the CONSERVED EFFECTOR LOCUS DELETION Mutant of Pst DC3000
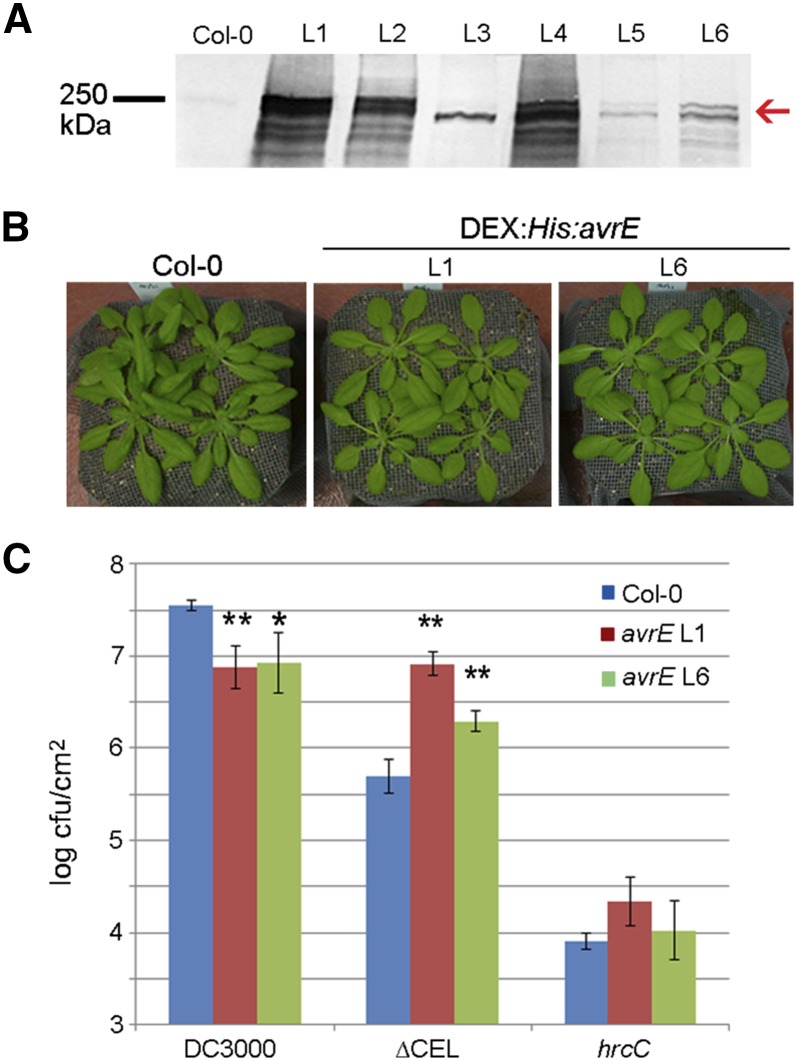
AvrE has been shown to be translocated into the host cell (Guttman et al., 2002; Badel et al., 2006). To study the action of AvrE inside plant cells, we transgenically expressed the Pst DC3000 avrE gene as a 6× His fusion under the control of the dexamethasone (DEX)-inducible promoter (DEX:His:avrE). Six independent lines that expressed His:AvrE at different levels upon 30 µm DEX treatment were obtained (Fig. 1A). We found that application of DEX induced tissue necrosis in all the transgenic lines, which is consistent with the involvement of AvrE family effectors in triggering cell death in host plants. In the absence of DEX treatment, high-expression lines (e.g. L1) showed slightly smaller plant size, but no other obvious morphological or developmental difference, compared with the wild-type Columbia-0 (Col-0) plants, was observed (Fig. 1B).
Figure 1.
Basal expression of DEX:His:avrE in transgenic Arabidopsis complemented the growth of the ∆CEL mutant. A, Western-blot analysis of His:AvrE proteins in transgenic Arabidopsis plants 6 h after 30 µm DEX treatment. A polyclonal AvrE antibody was used to detect AvrE. The red arrow indicates the predicted size of AvrE. B, Representative images of 4-week-old Col-0 and transgenic DEX:His:avrE plants. C, Basal expression of DEX:His:avrE in transgenic Arabidopsis lines L1 and L6 is sufficient to substantially enhance the growth of the ∆CEL mutant. DC3000, ∆CEL, and hrcC were hand infiltrated at 1 × 106 colony-forming units (cfu) mL−1. Bacterial numbers were counted 3 d after infiltration. Asterisks indicate significant differences between Col-0 and L1 or L6: **, P < 0.01; and *, P < 0.05.
For in-depth analysis, we selected lines 1 and 6, which showed high and low levels, respectively, of His:AvrE expression upon DEX induction (Fig. 1A). To determine whether the expression of His:AvrE in each of these lines could complement the virulence defect of the CONSERVED EFFECTOR LOCUS DELETION (∆CEL) mutant, which contains genomic deletions in the avrE and hopM1 genes (Alfano et al., 2000), we inoculated the two lines with Pst DC3000, the ∆CEL mutant, or the hrp gene conserved C (hrcC) mutant, which is defective in type III secretion and therefore nonpathogenic (Yuan and He, 1996). Pretreatment with DEX at concentrations as low as 0.3 µm 24 h before bacterial inoculation, however, caused tissue necrosis within 1 d in both lines, making it impossible to perform standard disease assays. To circumvent this, we tested the possibility that a basal level of His:AvrE expression (even though undetectable by immunoblot) might be sufficient to complement the ∆CEL mutant. As shown in Figure 1C, despite a slightly enhanced resistance that the His:avrE lines showed to Pst DC3000 infection, a substantial increase of the growth of the ∆CEL mutant was observed in both lines, although line 6 (i.e. low AvrE expression) showed a less dramatic disease phenotype. The hrcC mutant grew similarly in Col-0 versus line 1/6 (Fig. 1C). This result suggests that a very low level of His:AvrE inside the host cell, which likely mimics the physiological level delivered by bacteria during infection, is sufficient to provide a transkingdom complementation of the ∆CEL mutant.
AvrE Is Targeted to the PM and PM-Associated Vesicle-Like Structures in Arabidopsis Leaf Cells
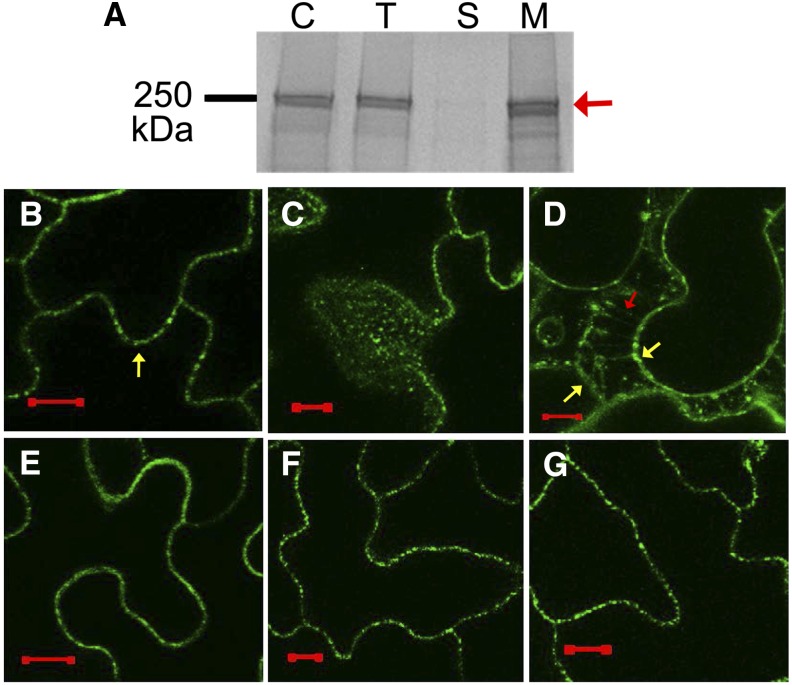
Computer analysis of the AvrE protein sequence suggests a putative C-terminal ERMRS motif in AvrE family effector proteins (Ham et al., 2008, 2009). To determine whether AvrE is localized in the ER as predicted or in another organelle, we conducted subcellular fractionation experiments. Total protein extract of DEX:His:avrE line 1 after DEX treatment was fractionated into soluble and membrane (microsomal) fractions by ultracentrifugation. As shown in Figure 2A, AvrE was found almost exclusively in the membrane fraction, as opposed to the soluble fraction.
Figure 2.
AvrE is localized at the PM and PM-associated vesicle-like structures in the plant cell. A, Subcellular fractionation of His:AvrE expressed in transgenic Arabidopsis plants. C, Crude plant extract; T, total plant protein; S, soluble protein fraction; M, total membrane protein fraction. The red arrow indicates the His:AvrE band. B and C, Confocal images of His:YFP:AvrE expressed in leaf cells of transgenic Arabidopsis plants. The yellow arrow indicates the PM. C is a z-stacked image. D, Confocal image (z-stack) of His:YFP:AvrE expressed in leaf cells of transgenic Arabidopsis plants after plasmolysis using 0.5 m NaCl. The red arrow indicates PM Hechtian strands, and the yellow arrows indicate the PM. E, Confocal image (z-stack) of AvrE:VFP expressed in N. benthamiana leaf cells. F and G, Confocal images of His:YFP:AvrE-ee (F) and His:YFP:AvrE-kk (G) expressed in leaf cells of transgenic Arabidopsis plants. Bars = 10 µm.
To visualize to which membrane(s) AvrE is targeted in the plant cell, we made an expression construct in which yellow fluorescent protein (YFP) was fused to the N-terminal end of AvrE. The YFP-AvrE fusion protein was expressed transiently in Nicotiana benthamiana as well as stably in transgenic Arabidopsis under the control of a DEX-inducible promoter (i.e. DEX:His:YFP:avrE). In addition, we created a second construct in which Venus fluorescent protein (VFP) was fused to the C terminus of AvrE and expressed transiently in N. benthamiana under the control of a β-estradiol-inducible promoter (i.e. β-estradiol:avrE:VFP). DEX or β-estradiol treatment of His:YFP:AvrE and AvrE:VFP induced tissue necrosis, indicating that the fusion proteins are active (Supplemental Fig. S1A). We performed confocal imaging at early time points (around 4 h after DEX or 6 h after β-estradiol treatment) prior to AvrE-induced cell death. Contrary to the predicted ER localization of AvrE family effector proteins (Ham et al., 2008, 2009), both His:YFP:AvrE and AvrE-VFP were found at the cell periphery (Fig. 2, B and E). After plasmolysis, His:YFP:AvrE was found in Hechtian strands, which are characteristic of PM-associated proteins (Oparka, 1994), thus confirming the PM localization of His:YFP:AvrE (Fig. 2D). In addition, we also observed prominent His:YFP:AvrE signals as PM-associated vesicle-like structures (Fig. 2C).
Previous work has defined the requirement for two WxxxE motifs common among AvrE family effector proteins (Ham et al., 2009); however, the specific functions of these motifs remain elusive. To determine if the ERMRS or WxxxE motifs are involved in PM targeting, transgenic Arabidopsis plants expressing YFP-tagged AvrE-ee (defective in the WxxxE motif; Ham et al., 2009) or AvrE-kk (defective in the ERMRS motif; Ham et al., 2009) variants under the control of a DEX-inducible promoter (i.e. DEX:His:YFP:avrE-ee and DEX:His:YFP:avrE-kk, respectively) were generated. As shown in Figure 2, F and G, His:YFP:AvrE-ee and His:YFP:AvrE-kk proteins showed localization patterns similar to that of His:YFP:AvrE, suggesting that neither the WxxxE motif nor the ERMRS motif is important for targeting AvrE to the PM and PM-associated vesicles in the host cell.
AvrE Contains Two Interactive Domains, and the N-Terminal One-Half Contains a PM-Targeting Signal
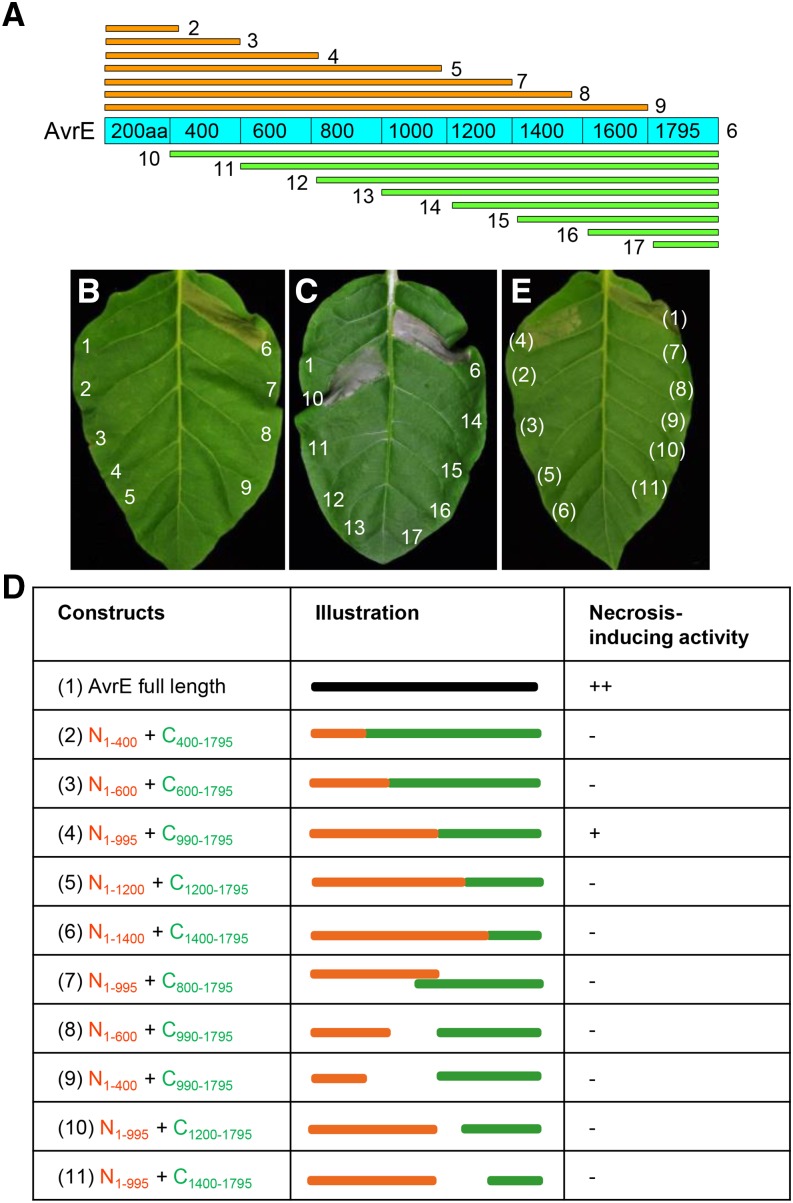
To define which portion(s) of the AvrE protein is required for its PM localization and/or virulence activity, we conducted a systematic deletion analysis. As shown in Figure 3A, serial 200-amino acid deletions from the N or C terminus of AvrE were constructed, and the truncated AvrE variants were transiently expressed in N. benthamiana and tobacco (Nicotiana tabacum) leaves. Induction of plant tissue necrosis was used as a proxy of AvrE’s function in these experiments. We found that the results in N. benthamiana and tobacco were identical; therefore, only the results from experiments using tobacco are shown (Fig. 3, B and C). Whereas full-length AvrE and, at a slower rate, AvrE200-1795aa induced necrosis, all other truncated versions of AvrE lost the ability to induce necrosis (Fig. 3, B and C). We examined AvrE protein expression from these constructs and found that all AvrE constructs produced the expected AvrE proteins (Supplemental Fig. S2). Taken together, these data suggest that AvrE’s necrosis-inducing activity requires almost the full-length protein.
Figure 3.
Serial deletion analysis reveals a putative two-domain structure of AvrE. A, Diagram showing the AvrE deletion constructs used in this study. B and C, Necrosis-inducing activity of AvrE proteins transiently expressed in tobacco leaves. Numbering is as follows: 1, empty vector; 2, AvrE1-200aa; 3, AvrE1-400aa; 4, AvrE1-596aa; 5, AvrE1-995aa; 6, His:AvrE; 7, AvrE1-1200aa; 8, AvrE1-1400aa; 9, AvrE1-1600aa; 10, AvrE200-1795aa; 11, AvrE400-1795aa; 12, AvrE600-1795aa; 13, AvrE800-1795aa; 14, AvrE990-1795aa; 15, AvrE1200-1795aa; 16, AvrE1400-1795aa; and 17, AvrE1600-1795aa. Necrosis is characterized by tissue collapse and discoloration (e.g. gray color) in the infiltrated area. D, Combinations of AvrE deletion derivatives used for transient coexpression experiments in tobacco. For each combination, two Agrobacterium tumefaciens strains (optical density at 600 nm [OD600] = 0.1) were mixed at a 1:1 ratio and infiltrated into tobacco leaves. E, Necrosis (indicated by tissue collapse with gray color) induced by combinations of truncated AvrE proteins. Photographs were taken 2 d after 10 µm DEX spray. Numbering is the same as shown in D.
Our serial deletion from the N and C terminus of AvrE does not rule out the possibility that AvrE contains discrete functional domains, each of which alone is not sufficient to confer a virulence phenotype but, when combined with another domain, is. To test this possibility, we coexpressed various combinations of two truncated AvrE proteins that make up the full-length AvrE equivalent and examined whether they could reconstitute the necrosis-inducing activity in tobacco. As shown in Figure 3, D and E, coexpression of AvrE1-995aa (AvrE-N) and AvrE990-1795aa (AvrE-C) restored the necrosis-inducing activity, while all other tested combinations did not. These data suggest that AvrE contains two functional domains, one in the N-terminal one-half and the other in the C-terminal one-half.
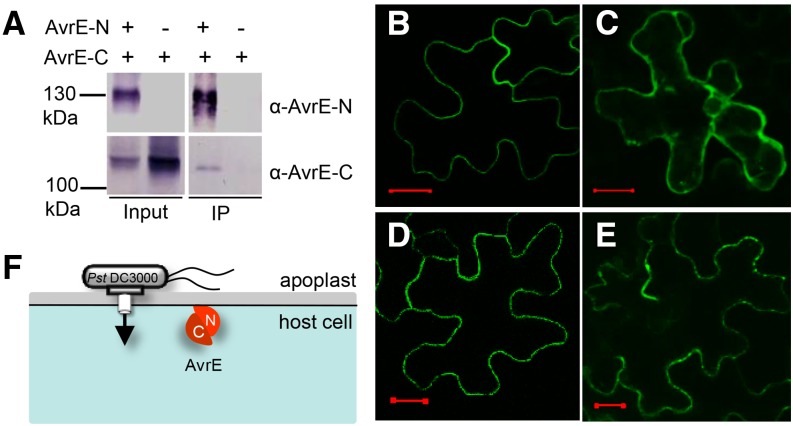
The requirement for a specific combination of AvrE-N and AvrE-C, expressed separately in planta, in restoring the necrosis-inducing activity raises the possibility that the two halves of AvrE may physically interact in vivo. This possibility was tested by coimmunoprecipitation of AvrE-N and AvrE-C expressed transiently in N. benthamiana leaves. As shown in Figure 4A, AvrE-C could be pulled down by an antibody against AvrE-N, confirming the physical interaction of the N- and C-terminal halves in planta.
Figure 4.
The two domains of AvrE physically interact, and the N-terminal domain contains a PM localization signal. A, Coimmunoprecipitation shows that AvrE-N and AvrE-C interact in N. benthamiana. The affinity-purified AvrE-N antibody was used to pull down AvrE-N. AvrE-N and AvrE-C were detected by AvrE-N and AvrE-C antibodies, respectively. IP, Immunoprecipitate. B to E, Confocal images of GFP:AvrE-N in N. benthamiana leaf cells (B), GFP:AvrE-C in N. benthamiana leaf cells (z-stack; C), GFP:AvrE-N coexpressed with AvrE-C in N. benthamiana leaf cells (D), and GFP:AvrE-C protein coexpressed with AvrE-N in N. benthamiana leaf cells (z-stack; E). Expression of GFP:AvrE-N and GFP:AvrE-C was under the control of the cauliflower mosaic virus 35S promoter (pSITEII vector). A. tumefaciens containing GFP:avrE-N/C and DEX:avrE-C/N constructs was adjusted to OD600 = 0.1 and mixed at a ratio of 1:2 (GFP:avrE-N/C versus DEX:avrE-C/N) for infiltration. Twenty-four hours after A. tumefaciens infiltration, 10 µm DEX was sprayed to induce the expression of untagged AvrE-N and AvrE-C proteins. Confocal images were collected 5 h after DEX treatment. Bars = 20 µm. F, A simplified model showing the two physically interacting domains of AvrE and their localization in the plant cell. C, AvrE-C; N, AvrE-N.
To determine whether the PM-targeting signal is localized within the N- or C-terminal one-half of AvrE, we constructed GFP-tagged AvrE-N and AvrE-C proteins and investigated the subcellular localization of AvrE-N and AvrE-C in N. benthamiana leaf cells by confocal fluorescence microscopy. Similar to full-length AvrE, GFP:AvrE-N was mainly localized at the cell periphery (Fig. 4B) and, after plasmolysis, to Hechtian strands (Supplemental Fig. S1B). Conversely, GFP:AvrE-C showed more diffuse signal in the cytosol and on the periphery of the nucleus (Fig. 4C). Interestingly, however, when coexpressed with nontagged AvrE-N, GFP-AvrE-C showed a shift of signal toward the cell periphery (Fig. 4E). In contrast, when coexpressed with nontagged AvrE-C, GFP-AvrE-N remained at the cell periphery (Fig. 4D). The AvrE-N-dependent shift in the subcellular localization of AvrE-C, together with the observed physical interaction between AvrE-N and AvrE-C (Fig. 4A), suggest that AvrE-N contains a PM-targeting signal that could drive the entire AvrE protein to the host PM (Fig. 4F).
Genome-Wide Host Gene Expression Analysis Uncovers a Link between AvrE Virulence Function and Down-Regulation of the NHL13 Gene in Arabidopsis
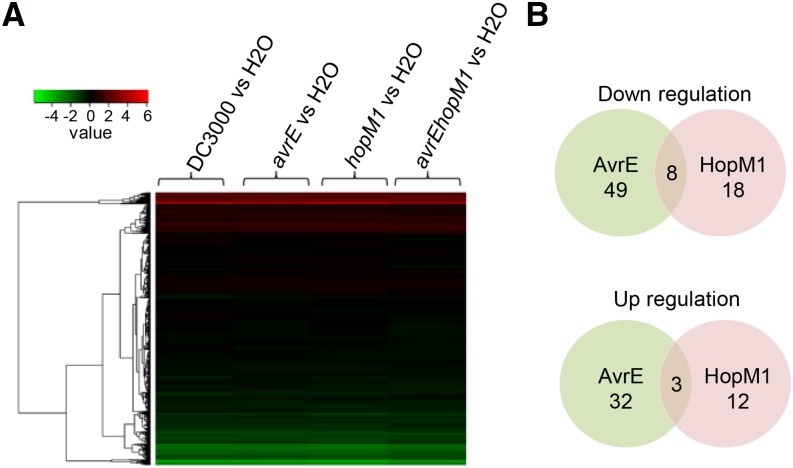
Despite the importance of the AvrE family effectors in plant-bacterial interactions, to date, no specific plant gene has been identified, directly or indirectly, to be functionally associated with their virulence functions. To address this important question, we conducted a genome-wide microarray analysis of Arabidopsis during Pst DC3000 infection. Because AvrE and HopM1 in Pst DC3000 are functionally redundant (DebRoy et al., 2004; Badel et al., 2006), we used the following bacterial strains to discern the shared and specific contributions of AvrE and HopM1 to host gene expression: Pst DC3000 (avrE+ and hopM1+), the avrE mutant (hopM1+), the hopM1 mutant (avrE+), and the avrE hopM1 double mutant. Arabidopsis leaves were inoculated with 1 × 108 cfu mL−1 bacteria or water (as a control), total RNA was extracted, and genome-wide gene expression was analyzed (data submitted to the Gene Expression Omnibus; GSE67179). A Pearson correlation analysis was performed, and a good correlation was found among biological replicates of each treatment (Supplemental Table S1). Surprisingly, Arabidopsis plants infected by the four bacterial strains showed remarkably similar gene expression profiles (Fig. 5A). Neither Gene Ontology analysis (using the DAVID Bioinformatics Resources; Huang et al., 2009a, 2009b) nor manual examination revealed global alteration of gene expression that is associated with a canonical defense signaling pathway, including the SA, jasmonate, or pathogen-associated molecular pattern-triggered immunity (PTI) pathway. However, we were able to identify small subsets of Arabidopsis genes that were specifically regulated (2-fold or greater) by AvrE, HopM1, or both (Fig. 5B; Supplemental Table S2). Interestingly, a group of eight Arabidopsis genes is down-regulated by both AvrE and HopM1 (Table I). Among them is NHL13, a novel member of the large NDR1/HIN1-LIKE gene family (Supplemental Fig. S3). We also found an enrichment of genes encoding PM-localized proteins among the genes regulated by AvrE and/or HopM1 (Table I; Supplemental Table S2), suggesting a possible link of AvrE/HopM1 function to host PM protein dynamics.
Figure 5.
Microarray analysis of the virulence effects of bacterium-delivered AvrE and HopM1 on Arabidopsis gene expression in leaves. A, Heat map of global gene expression changes in response to the different bacterial strains indicated: Pst DC3000 (avrE+ and hopM1+), the avrE mutant (hopM1+), the hopM1 mutant (avrE+), and the avrE hopM1 double mutant. Bacteria (1 × 108 cfu mL−1) were vacuum infiltrated into Arabidopsis leaves, and leaf samples were collected 7 h after inoculation to allow sufficient time for type III effector delivery and, at the same time, to avoid bacterial population differences between the strains, especially between the avrE hopM1 double mutant and Pst DC3000 (Nomura et al., 2011). B, Venn diagram showing the numbers of genes up- or down-regulated by AvrE (comparing infection by the hopM1 mutant with infection by the avrE hopM1 mutant), HopM1 (comparing infection by the avrE mutant with infection by the avrE hopM1 mutant), or both.
Table I. List of Arabidopsis genes regulated by both AvrE and HopM1 during Pst DC3000 infection.
For log2 fold change, down-regulated genes are indicated by the minus symbol and up-regulated genes are indicated by the plus symbol. Log2 fold change indicates the gene expression change caused by AvrE (by comparison between hopM1 and avrE hopM1 strains) or HopM1 (by comparison between avrE and avrE hopM1 strains). P < 0.1; log2 > 1 or < −1.
| Gene Annotation | Gene Identifier | Log2 Fold Change |
PM | |||
|---|---|---|---|---|---|---|
| AvrE | P | HopM1 | P | |||
| Universal stress protein (USP) family protein | AT3G62550 | −1.973 | 0.010 | −1.933 | 0.045 | |
| NDR1/HIN1-LIKE13; NHL13 | AT2G27080 | −1.947 | 0.006 | −2.048 | 0.044 | Yes |
| Unknown protein | AT5G11070 | −1.654 | 0.005 | −1.503 | 0.006 | |
| Octicosapeptide/Phox/Bem1p (PB1) domain-containing protein | AT3G26510 | −1.500 | 0.026 | −1.072 | 0.019 | |
| Arabinogalactan protein20; AGP20 | AT3G61640 | −1.459 | 0.059 | −1.655 | 0.044 | Yes |
| Plasma-membrane associated cation-binding protein1; PCAP1 | AT4G20260 | −1.219 | 0.050 | −1.125 | 0.053 | Yes |
| Cys-rich receptor-like kinase10 | AT4G23180 | −1.166 | 0.075 | −1.004 | 0.071 | Yes |
| Calcineurin B-like-interacting protein kinase9 | AT1G01140 | −1.042 | 0.038 | −1.072 | 0.020 | |
| Glutathione S-transferase24; GSTU6 | AT2G29440 | +2.206 | 0.048 | +1.604 | 0.062 | |
| NAC (for no apical meristem [NAM], Arabidopsis transcription activation factor [ATAF], cup-shaped cotyledon [CUC]) domain-containing protein25 | AT1G61110 | +1.509 | 0.064 | +1.031 | 0.088 | |
| Xyloglucan endotransglycosylase-related8; XTR8 | AT3G44990 | +1.025 | 0.020 | +1.290 | 0.049 | |
NHL13 Is Required for Antibacterial Immunity
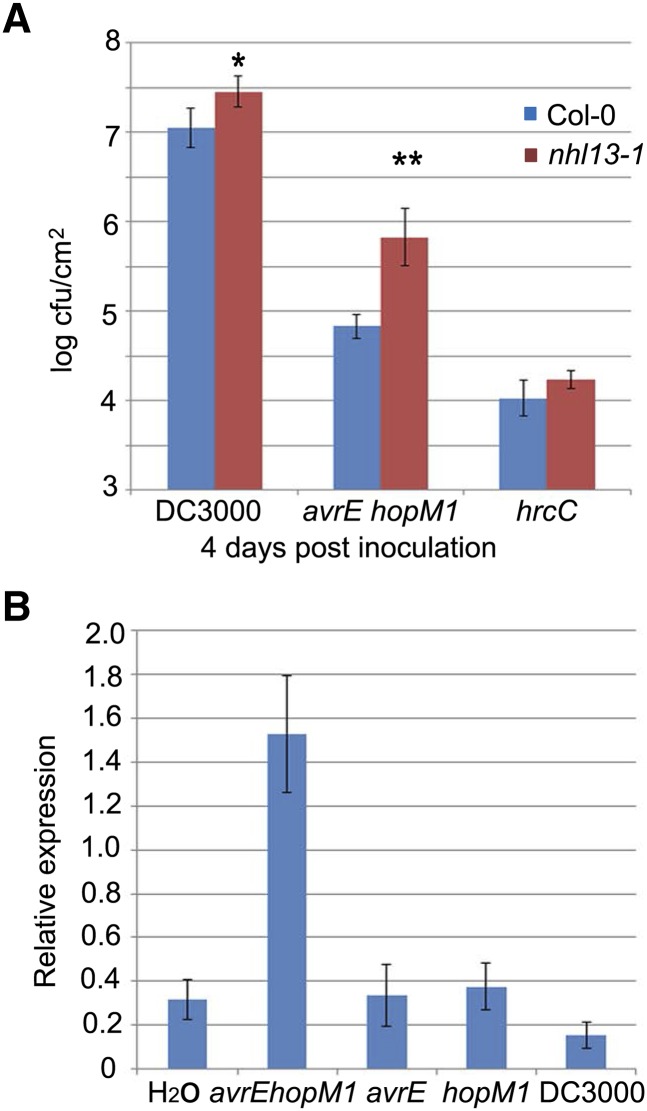
To determine whether the Arabidopsis genes down-regulated by both AvrE and HopM1 are biologically relevant to the function of AvrE/HopM1 in Pst DC3000, we performed disease assays on confirmed gene knockout plants (Supplemental Table S3). An initial screen-of-disease assay using the ∆CEL strain was performed with all the lines. Most Arabidopsis mutant lines examined did not show a change in disease susceptibility, with the exception of SALK_016726C (NHL13) plants, which showed a robust level of enhanced disease susceptibility (Supplemental Fig. S4). In addition, SALK_060447 (AT1G61110) plants showed slightly enhanced disease susceptibility (Supplemental Fig. S4). However, an independent knockout line of AT1G61110, SALK_060459, did not show any enhanced disease susceptibility (Supplemental Fig. S4). Therefore, we focused our further analyses on the NHL13 gene and carried out further disease assays with nhl13-1 and nhl13-2 plants using DC3000, the avrE hopM1 mutant, and the hrcC mutant (Fig. 6A; Supplemental Fig. S5). These experiments showed that the enhanced susceptibility of the nhl13 mutants was most obvious in the context of infection by the avrE hopM1 double mutant compared with infection by Pst DC3000 or the hrcC mutant (Fig. 6A; Supplemental Fig. S5C), suggesting specificity to the AvrE/HopM1 function.
Figure 6.
NHL13 is required for Arabidopsis immunity. A, The nhl13-1 mutant plant showed enhanced susceptibility to the avrE hopM1 mutant. Five-week-old Col-0 and nhl13-1 plants were dip inoculated with Pst DC3000, the avrE hopM1 mutant, or the hrcC mutant at 1 × 108 cfu mL−1. The bacterial population was counted 4 d after inoculation. Asterisks indicate significant differences in bacterial multiplication when compared with that in Col-0 plants: **, P < 0.01; and *, P < 0.05. B, Results of qRT-PCR to measure the levels of the NHL13 transcript in response to inoculation with Pst DC3000, the avrE mutant, the hopM1 mutant, the avrE hopM1 mutant, or water. The level of NHL13 was normalized to that of PROTEIN PHOSPHATASE 2A SUBUNIT A3 (PP2AA3; At1G13320).
We also conducted quantitative reverse transcription (qRT)-PCR analysis of the NHL13 transcript during Pst DC3000 infection to verify the microarray results. NHL13 gene expression was found to be induced by the avrE hopM1 mutant compared with mock (water) treatment but was suppressed by Pst DC3000, the avrE mutant, and the hopM1 mutant (Fig. 6B). These results are consistent with the microarray results and show that AvrE and HopM1 could redundantly suppress the induction of the NHL13 gene by the avrE hopM1 mutant. These results, together with the disease assay data, identify NHL13, to our knowledge, as the first biologically relevant downstream transcriptional output of the redundant virulence functions of AvrE and HopM1.
We conducted further experiments to examine NHL13 expression in response to the type III secretion-defective hrcC mutant and the avirulent strain DC3000 (avrRpt2). We found that there was almost no induction of the NHL13 transcript in response to the hrcC mutant (Supplemental Fig. S6), suggesting that NHL13 expression during infection is likely dependent on the function of the T3SS and/or certain type III effectors. DC3000 (avrRpt2) induced the expression of the NHL13 gene (Supplemental Fig. S6). This result provides further evidence that NHL13 is an immunity-associated gene and shows that effector-triggered immunity could counter DC3000 (i.e. AvrE/HopM1-mediated) suppression of NHL13 gene expression.
DISCUSSION
In this study, we defined the subcellular localization, functional domains, and associated host gene expression of the founding member of an important family of bacterial effectors, which, to date, have been recalcitrant to functional characterization. We demonstrated that AvrE has two physically interactive halves that are separable and that AvrE resides in the plant PM and PM-associated vesicle-like structures, with the N-terminal one-half of AvrE containing the PM-targeting signal. AvrE contributes to the down-regulation of a specific subset of Arabidopsis genes during Pst DC3000 infection, including a member of the NDR1/HIN1 gene family, NHL13, which we found to be required for antibacterial immunity.
AvrE family effectors were previously predicted to contain a putative C-terminal ERMRS motif (Ham et al., 2008, 2009). Therefore, it is both surprising and interesting that our membrane fractionation and confocal microscopy experiments show a clear PM localization of AvrE. AvrE does not contain an obvious transmembrane domain, and computer programs fail to predict a known PM-targeting signal in AvrE. Therefore, we predict that AvrE likely contains a novel PM-targeting signal or employs a novel mechanism to reach the host PM. Ham et al. (2009) previously also identified a conserved amino acid motif, WxxxE, in AvrE family effectors. However, we found that this motif, like the ERMRS, is not involved in PM targeting of AvrE. Instead, our analysis showed that the N-terminal one-half of AvrE (AvrE-N) is efficiently targeted to the plant PM and, furthermore, when coexpressed with the C-terminal one-half of AvrE (AvrE-C), can shift cytosol- and nucleus-localized AvrE-C to the PM (Fig. 4, B–E). These observations strongly suggest that a novel PM-targeting signal is located within the N-terminal one-half of AvrE and that this signal is sufficiently strong to target the entire AvrE protein to the host PM. Sphingolipids, which are perturbed by DspA/E in yeast cells (Siamer et al., 2014), are a significant component of the PM lipids in plants (Markham et al., 2013). Therefore, it is of interest in the future to determine whether AvrE affects the PM-associated sphingolipid pathway in plants.
Our serial 200-amino acid deletion analysis led to a novel finding regarding the domain structure of AvrE. We found that, while the N-terminal and C-terminal halves of AvrE (i.e. AvrE-N and AvrE-C) by themselves could not trigger plant cell death, coexpression of the two halves could reconstitute the cell death activity in N. benthamiana. Furthermore, AvrE-N and AvrE-C fragments physically interact in planta. Thus, AvrE appears to contain two interacting, functional domains, both of which are required for the full activity of AvrE. Although it is well known that many plant disease resistance proteins contain discrete functional domains that could physically interact with each other (Qi and Innes, 2013), to our knowledge, this is the first time a bacterial effector has been found to have this property.
Our microarray analysis showed that the virulence functions of AvrE and HopM1 are not associated with the global down-regulation of canonical defense signaling pathways (including the SA, jasmonate, and PTI pathway). This finding is in contrast to other studies in which bacterial effectors were found to globally down-regulate PTI-associated gene expression (Block and Alfano, 2011; Dou and Zhou, 2012; Xin and He, 2013). For example, AvrPto and AvrPtoB from Pst DC3000 target the pattern recognition receptor FLAGELLIN INSENSITIVE2 and/or coreceptor BRI1-ASSOCIATED RECEPTOR KINASE1, and their functions are linked to suppression of the expression of a suite of host genes (e.g. FLG22-INDUCED RECEPTOR-LIKE KINASE1) associated with PTI (He et al., 2006; Shan et al., 2008; Xiang et al., 2008). Our results, therefore, support the redundant effector group model proposed by Kvitko et al. (2009), in which AvrE/HopM1 function either at a step downstream of pathogen-associated molecular pattern signaling or through a distinct mechanism to promote pathogenesis.
We found that the effect of AvrE, when delivered by bacteria during infection, on the host Arabidopsis gene expression is very different from that of E. amylovora DspA/E, when transgenically expressed in the nonhost Arabidopsis, which results in the induction of a large suite of SA-dependent defense genes (Degrave et al., 2013). This difference is likely due to host versus nonhost responses to the AvrE family of effectors. Instead, our microarray analysis shows that NHL13 is one of the most down-regulated genes by AvrE and HopM1 in the context of Pst DC3000 infection (Table I). We found this result particularly interesting because the NHL family of genes, with approximately 40 members in Arabidopsis (Supplemental Fig. S3), has been reported to be associated with plant defense responses, although the specific defense signaling pathway(s) that regulates the expression of NHL genes has not been defined (Varet et al., 2002, 2003; Zheng et al., 2004). One of the founding members of this gene family, NDR1, is a PM protein required for immunity as well as abiotic stress responses (Knepper et al., 2011). Another member of the NHL gene family, HIN1, was shown to be induced by harpins (Gopalan et al., 1996), which are the extracellular components of the T3SS (Kvitko et al., 2007; Bocsanczy et al., 2008; Boureau et al., 2011). In our study, we not only found that NHL13 is down-regulated by AvrE and HopM1 but also found that the nhl13 mutants are compromised in disease resistance, especially to the avrE hopM1 mutant (Fig. 6). Our results, therefore, uncovered, to our knowledge, the first example of host PM-targeting bacterial effectors whose virulence functions are linked to the down-regulation of a member of the NHL gene family that is required for antibacterial immunity in plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants used in this study were all in Col-0 background. SALK transfer DNA insertion lines were ordered from the Arabidopsis Biological Resource Center (www.arabidopsis.org). Arabidopsis was grown in an environmentally controlled chamber under a 12-h-day/12-h-night cycle (120 μmol m−2 s−1 cool-white fluorescent light, 22°C). Nicotiana benthamiana and tobacco (Nicotiana tabacum) plants were grown in a growth chamber with a 12-h-day/12-h-night cycle (approximately 200 μmol m−2 s−1 fluorescent light, 24°C).
Molecular Cloning
To make the DEX:His:avrE construct, chromosomal DNA purified from Pst DC3000 was used as a template to clone the avrE gene by PCR. The avrE primers AvrE-F1 and AvrE-R1 were used (Supplemental Table S4). avrE with a 5′ sequence extension coding for the 6× His tag was first cloned into the pGEM-T Easy vector (Promega) for sequencing. The correct insert was retrieved by cutting with XhoI and XbaI and was ligated into the pBUD-DEX (pBD) vector. To create the DEX:His:YFP:avrE fusion construct, the gene encoding YFP was inserted into the DEX:His:AvrE construct at the XhoI and KpnI sites. Constructs for the expression of truncated AvrE proteins, DEX:avrE-N/C, DEX:His:YFP:avrE-ee, and DEX:His:YFP:avrE-kk, were cloned into the pBD vector using a similar strategy. For DEX:His:YFP:avrE-ee and DEX:His:YFP:avrE-kk, templates containing avrE-ee or avrE-kk (Ham et al., 2009) were used for PCR, and primers AvrE-F1/AvrE-R1 and AvrE-F1/AvrE-kk R1 were used, respectively (Supplemental Table S4).
The constructs β-estradiol:avrE:VFP and GFP:avrE-N/C were cloned by Gateway cloning (Life Technologies). Primers used for making β-estradiol:avrE:VFP were AvrE F2 and AvrE R2A; primers used for making 35S:GFP:avrE-N were AvrE F2 and AvrE-N R1; primers used for making 35S:GFP:avrE-C were AvrE-C F1 and AvrE-C R1 (Supplemental Table S4). A pENTR/D-TOPO entry clone was first generated, and avrE was then recombined into pER8-DEST (for β-estradiol:avrE:VFP) or pSITEII-GFP vector (for GFP:avrE-N/C; Martin et al., 2009).
Transient Expression in N. benthamiana and Tobacco and Production of Transgenic Arabidopsis
Plant transformation constructs expressing avrE were mobilized into Agrobacterium tumefaciens GV3850 or GV3101 by electroporation, and the resultant clones were used to transiently transform tobacco or N. benthamiana cells by syringe infiltration. For transient expression of AvrE deletion constructs (Fig. 3), A. tumefaciens GV3850 containing each construct was infiltrated into tobacco leaves at OD600 = 0.1. Ten micromolar DEX was sprayed 1 d after infiltration. Photographs were taken 2 or 3 d after DEX treatment. Total protein was extracted 8 h (for the AvrE200-1975 construct) or 24 h (for the other constructs) after DEX treatment for western blot (Supplemental Fig. S2).
Stable transgenic Arabidopsis expressing DEX:His:avrE and DEX:His:YFP:avrE/-ee/-kk were generated by floral dipping of Col-0 plants (Bent, 2006). Multiple T1 lines were selected for each genotype and propagated to produce the T2 generation for experiments.
Microsomal Membrane Fraction Isolation and Western Blotting
Four-week-old transgenic Arabidopsis plants expressing His:AvrE were sprayed with 30 µm DEX to induce protein production. One gram of plant leaf tissue was collected 6 h after induction. Microsomal fraction isolation was performed as described previously (Nomura et al., 2006). Proteins from different fractions were subjected to SDS-PAGE separation and transferred onto Immobilon-P membranes (Millipore) by semidry transfer. A rabbit anti-AvrE polyclonal antibody was used to detect His:AvrE. Coimmunoprecipitation of AvrE-N and AvrE-C was conducted following the procedures described previously (Yang et al., 2012).
Confocal Fluorescence Microscopy
For AvrE localization, 4-week-old DEX:His:YFP:avrE, DEX:His:YFP:avrE-ee, and DEX:His:YFP:avrE-kk transgenic Arabidopsis plants were sprayed with 3 µm DEX. Four hours after treatment, leaves were randomly selected for confocal imaging. For AvrE:VFP localization, A. tumefaciens containing the β-estradiol:avrE:VFP construct was infiltrated into the leaves of N. benthamiana. One day after A. tumefaciens infiltration, leaves were infiltrated with 20 µm β-estradiol, and confocal imaging was conducted at approximately 6 h after treatment. Imaging was performed using an Olympus FluoView FV1000 Laser-Scanning Confocal Microscope for YFP/VFP visualization at the excitation wavelength of 515 nm or GFP visualization at 488 nm. Images were collected with a 40× oil-immersion objective and processed with Olympus Fluoview Viewer software, version 2.0B.
Microarray Analysis of Arabidopsis Gene Expression
Leaves of 5-week-old Arabidopsis Col-0 plants were vacuum infiltrated with Pst DC3000, the avrE mutant, the hopM1 mutant, or the avrE hopM1 double mutant at 1 × 108 cfu mL−1. Tissue samples were collected by snap freezing in liquid nitrogen 7 h after bacterial infiltration, and RNA was extracted using RNeasy plant mini kits (Qiagen). The Gene Chip Arabidopsis ATH1 genome array (Affymetrix) was used for microarray analysis in the Research Technology Support Facility at Michigan State University.
qRT-PCR Analysis of the NHL13 Gene
The experimental conditions and RNA isolation methods used were the same as those described for the microarray experiments (see above). The complementary DNA were synthesized from total RNA by Moloney murine leukemia virus reverse transcriptase (Invitrogen). The gene PP2AA3 (At1G13320) was used as an internal control. Primers used in qRT-PCR are described in Supplemental Table S4.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Transient expression of His:YFP:AvrE and AvrE:VFP proteins.
Supplemental Figure S2. Detection of truncated AvrE proteins in N. benthamiana leaves.
Supplemental Figure S3. Phylogenetic tree of the NHL gene family.
Supplemental Figure S4. Disease phenotypes of mutants of AvrE/HopM1-regulated Arabidopsis genes.
Supplemental Figure S5. Characterization of nhl13 mutants.
Supplemental Figure S6. Measurement of NHL13 transcript levels during infection.
Supplemental Table S1. Pearson correlation matrix of microarray data.
Supplemental Table S2. Arabidopsis genes regulated by AvrE or HopM1.
Supplemental Table S3. A list of Arabidopsis mutants analyzed in disease assays.
Supplemental Table S4. Primers used in this study.
Supplementary Material
Acknowledgments
We thank John-Scott Craig and Bethany Huot for editing the article and members of the S.Y.H. laboratory for insightful discussions throughout this work.
Glossary
- T3SS
type III secretion system
- T3Es
type III secretion system effectors
- Pst
Pseudomonas syringae pv tomato
- ERMRS
reticulum membrane retention/retrieval-like signal
- ER
endoplasmic reticulum
- SA
salicylic acid
- PM
plasma membrane
- DEX
dexamethasone
- Col-0
Columbia-0
- cfu
colony-forming units
- PTI
pathogen-associated molecular pattern-triggered immunity
- qRT
quantitative reverse transcription
- OD600
optical density at 600 nm
Footnotes
This work was supported by the National Institutes of Health (grant nos. AI060761 and GM109928 to S.Y.H.), the Gordon and Betty Moore Foundation (grant no. GBMF3037 to S.Y.H.), the U.S. Department of Energy, Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science (grant no. DE–FG02–91ER20021 to S.Y.H. for infrastructural support), and Michigan State University (dissertation completion fellowship to X.-F.X.).
Articles can be viewed without a subscription.
References
- Alfano JR, Charkowski AO, Deng WL, Badel JL, Petnicki-Ocwieja T, van Dijk K, Collmer A (2000) The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc Natl Acad Sci USA 97: 4856–4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alto NM, Shao F, Lazar CS, Brost RL, Chua G, Mattoo S, McMahon SA, Ghosh P, Hughes TR, Boone C, et al. (2006) Identification of a bacterial type III effector family with G protein mimicry functions. Cell 124: 133–145 [DOI] [PubMed] [Google Scholar]
- Asselin JA, Lin J, Perez-Quintero AL, Gentzel I, Majerczak D, Opiyo SO, Zhao W, Paek SM, Kim MG, Coplin DL, et al. (2015) Perturbation of maize phenylpropanoid metabolism by an AvrE family type III effector from Pantoea stewartii. Plant Physiol 167: 1117–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badel JL, Shimizu R, Oh HS, Collmer A (2006) A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol Plant Microbe Interact 19: 99–111 [DOI] [PubMed] [Google Scholar]
- Bent A. (2006) Arabidopsis thaliana floral dip transformation method. Methods Mol Biol 343: 87–103 [DOI] [PubMed] [Google Scholar]
- Block A, Alfano JR (2011) Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr Opin Microbiol 14: 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocsanczy AM, Nissinen RM, Oh CS, Beer SV (2008) HrpN of Erwinia amylovora functions in the translocation of DspA/E into plant cells. Mol Plant Pathol 9: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove AJ, Kim JF, Wei Z, Kolchinsky P, Charkowski AO, Conlin AK, Collmer A, Beer SV (1998) Homology and functional similarity of an hrp-linked pathogenicity locus, dspEF, of Erwinia amylovora and the avirulence locus avrE of Pseudomonas syringae pathovar tomato. Proc Natl Acad Sci USA 95: 1325–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boureau T, ElMaarouf-Bouteau H, Garnier A, Brisset MN, Perino C, Pucheu I, Barny MA (2006) DspA/E, a type III effector essential for Erwinia amylovora pathogenicity and growth in planta, induces cell death in host apple and nonhost tobacco plants. Mol Plant Microbe Interact 19: 16–24 [DOI] [PubMed] [Google Scholar]
- Boureau T, Siamer S, Perino C, Gaubert S, Patrit O, Degrave A, Fagard M, Chevreau E, Barny MA (2011) The HrpN effector of Erwinia amylovora, which is involved in type III translocation, contributes directly or indirectly to callose elicitation on apple leaves. Mol Plant Microbe Interact 24: 577–584 [DOI] [PubMed] [Google Scholar]
- DebRoy S, Thilmony R, Kwack YB, Nomura K, He SY (2004) A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci USA 101: 9927–9932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrave A, Fagard M, Perino C, Brisset MN, Gaubert S, Laroche S, Patrit O, Barny MA (2008) Erwinia amylovora type three-secreted proteins trigger cell death and defense responses in Arabidopsis thaliana. Mol Plant Microbe Interact 21: 1076–1086 [DOI] [PubMed] [Google Scholar]
- Degrave A, Moreau M, Launay A, Barny MA, Brisset MN, Patrit O, Taconnat L, Vedel R, Fagard M (2013) The bacterial effector DspA/E is toxic in Arabidopsis thaliana and is required for multiplication and survival of fire blight pathogen. Mol Plant Pathol 14: 506–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrave A, Siamer S, Boureau T, Barny MA (January 13, 2015) The AvrE superfamily: ancestral type III effectors involved in suppression of pathogen-associated molecular pattern-triggered immunity. Mol Plant Pathol http://dx.doi.org/10.1111/mpp.12237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou D, Zhou JM (2012) Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12: 484–495 [DOI] [PubMed] [Google Scholar]
- Frederick RD, Ahmad M, Majerczak DR, Arroyo-Rodríguez AS, Manulis S, Coplin DL (2001) Genetic organization of the Pantoea stewartii subsp. stewartii hrp gene cluster and sequence analysis of the hrpA, hrpC, hrpN, and wtsE operons. Mol Plant Microbe Interact 14: 1213–1222 [DOI] [PubMed] [Google Scholar]
- Galán JE, Collmer A (1999) Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284: 1322–1328 [DOI] [PubMed] [Google Scholar]
- Gaudriault S, Malandrin L, Paulin JP, Barny MA (1997) DspA, an essential pathogenicity factor of Erwinia amylovora showing homology with AvrE of Pseudomonas syringae, is secreted via the Hrp secretion pathway in a DspB-dependent way. Mol Microbiol 26: 1057–1069 [DOI] [PubMed] [Google Scholar]
- Gopalan S, Wei W, He SY (1996) hrp gene-dependent induction of hin1: a plant gene activated rapidly by both harpins and the avrPto gene-mediated signal. Plant J 10: 591–600 [DOI] [PubMed] [Google Scholar]
- Guttman DS, Vinatzer BA, Sarkar SF, Ranall MV, Kettler G, Greenberg JT (2002) A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295: 1722–1726 [DOI] [PubMed] [Google Scholar]
- Ham JH, Majerczak D, Ewert S, Sreerekha MV, Mackey D, Coplin D (2008) WtsE, an AvrE-family type III effector protein of Pantoea stewartii subsp. stewartii, causes cell death in non-host plants. Mol Plant Pathol 9: 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham JH, Majerczak DR, Arroyo-Rodriguez AS, Mackey DM, Coplin DL (2006) WtsE, an AvrE-family effector protein from Pantoea stewartii subsp. stewartii, causes disease-associated cell death in corn and requires a chaperone protein for stability. Mol Plant Microbe Interact 19: 1092–1102 [DOI] [PubMed] [Google Scholar]
- Ham JH, Majerczak DR, Nomura K, Mecey C, Uribe F, He SY, Mackey D, Coplin DL (2009) Multiple activities of the plant pathogen type III effector proteins WtsE and AvrE require WxxxE motifs. Mol Plant Microbe Interact 22: 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nürnberger T, Sheen J (2006) Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125: 563–575 [DOI] [PubMed] [Google Scholar]
- He SY, Nomura K, Whittam TS (2004) Type III protein secretion mechanism in mammalian and plant pathogens. Biochim Biophys Acta 1694: 181–206 [DOI] [PubMed] [Google Scholar]
- Hogan CS, Mole BM, Grant SR, Willis DK, Charkowski AO (2013) The type III secreted effector DspE is required early in Solanum tuberosum leaf infection by Pectobacterium carotovorum to cause cell death, and requires Wx(3-6)D/E motifs. PLoS ONE 8: e65534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA (2009a) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA (2009b) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Huang Z, Sutton SE, Wallenfang AJ, Orchard RC, Wu X, Feng Y, Chai J, Alto NM (2009c) Structural insights into host GTPase isoform selection by a family of bacterial GEF mimics. Nat Struct Mol Biol 16: 853–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Thammarat P, Lommel SA, Hogan CS, Charkowski AO (2011) Pectobacterium carotovorum elicits plant cell death with DspE/F but the P. carotovorum DspE does not suppress callose or induce expression of plant genes early in plant-microbe interactions. Mol Plant Microbe Interact 24: 773–786 [DOI] [PubMed] [Google Scholar]
- Knepper C, Savory EA, Day B (2011) Arabidopsis NDR1 is an integrin-like protein with a role in fluid loss and plasma membrane-cell wall adhesion. Plant Physiol 156: 286–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitko BH, Park DH, Velásquez AC, Wei CF, Russell AB, Martin GB, Schneider DJ, Collmer A (2009) Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog 5: e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitko BH, Ramos AR, Morello JE, Oh HS, Collmer A (2007) Identification of harpins in Pseudomonas syringae pv. tomato DC3000, which are functionally similar to HrpK1 in promoting translocation of type III secretion system effectors. J Bacteriol 189: 8059–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang JM, Keen NT (1995) Characterization of avrE from Pseudomonas syringae pv. tomato: a hrp-linked avirulence locus consisting of at least two transcriptional units. Mol Plant Microbe Interact 8: 49–57 [DOI] [PubMed] [Google Scholar]
- Markham JE, Lynch DV, Napier JA, Dunn TM, Cahoon EB (2013) Plant sphingolipids: function follows form. Curr Opin Plant Biol 16: 350–357 [DOI] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59: 150–162 [DOI] [PubMed] [Google Scholar]
- Meng X, Bonasera JM, Kim JF, Nissinen RM, Beer SV (2006) Apple proteins that interact with DspA/E, a pathogenicity effector of Erwinia amylovora, the fire blight pathogen. Mol Plant Microbe Interact 19: 53–61 [DOI] [PubMed] [Google Scholar]
- Nomura K, Debroy S, Lee YH, Pumplin N, Jones J, He SY (2006) A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223 [DOI] [PubMed] [Google Scholar]
- Nomura K, Mecey C, Lee YN, Imboden LA, Chang JH, He SY (2011) Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc Natl Acad Sci USA 108: 10774–10779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlson MB, Huang Z, Alto NM, Blanc MP, Dixon JE, Chai J, Miller SI (2008) Structure and function of Salmonella SifA indicate that its interactions with SKIP, SseJ, and RhoA family GTPases induce endosomal tubulation. Cell Host Microbe 4: 434–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka KJ. (1994) Plasmolysis: new insights into an old process. New Phytol 126: 571–591 [Google Scholar]
- Qi D, Innes RW (2013) Recent advances in plant NLR structure, function, localization, and signaling. Front Immunol 4: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nürnberger T, Martin GB, Sheen J (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siamer S, Guillas I, Shimobayashi M, Kunz C, Hall MN, Barny MA (2014) Expression of the bacterial type III effector DspA/E in Saccharomyces cerevisiae down-regulates the sphingolipid biosynthetic pathway leading to growth arrest. J Biol Chem 289: 18466–18477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siamer S, Patrit O, Fagard M, Belgareh-Touzé N, Barny MA (2011) Expressing the Erwinia amylovora type III effector DspA/E in the yeast Saccharomyces cerevisiae strongly alters cellular trafficking. FEBS Open Bio 1: 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varet A, Hause B, Hause G, Scheel D, Lee J (2003) The Arabidopsis NHL3 gene encodes a plasma membrane protein and its overexpression correlates with increased resistance to Pseudomonas syringae pv. tomato DC3000. Plant Physiol 132: 2023–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varet A, Parker J, Tornero P, Nass N, Nürnberger T, Dangl JL, Scheel D, Lee J (2002) NHL25 and NHL3, two NDR1/HIN1-1ike genes in Arabidopsis thaliana with potential role(s) in plant defense. Mol Plant Microbe Interact 15: 608–616 [DOI] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, et al. (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol 18: 74–80 [DOI] [PubMed] [Google Scholar]
- Xin XF, He SY (2013) Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol 51: 473–498 [DOI] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, He SY (1996) The Pseudomonas syringae Hrp regulation and secretion system controls the production and secretion of multiple extracellular proteins. J Bacteriol 178: 6399–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng MS, Takahashi H, Miyazaki A, Hamamoto H, Shah J, Yamaguchi I, Kusano T (2004) Up-regulation of Arabidopsis thaliana NHL10 in the hypersensitive response to Cucumber mosaic virus infection and in senescing leaves is controlled by signalling pathways that differ in salicylate involvement. Planta 218: 740–750 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.