Abscisic acid-activated protein kinases interact with each other and with protein phosphatases that modulate abscisic acid responses.
Abstract
The plant hormone abscisic acid (ABA) controls growth and development and regulates plant water status through an established signaling pathway. In the presence of ABA, pyrabactin resistance/regulatory component of ABA receptor proteins inhibit type 2C protein phosphatases (PP2Cs). This, in turn, enables the activation of Sucrose Nonfermenting1-Related Protein Kinases2 (SnRK2). Open Stomata1 (OST1)/SnRK2.6/SRK2E is a major SnRK2-type protein kinase responsible for mediating ABA responses. Arabidopsis (Arabidopsis thaliana) expressing an epitope-tagged OST1 in the recessive ost1-3 mutant background was used for the copurification and identification of OST1-interacting proteins after osmotic stress and ABA treatments. These analyses, which were confirmed using bimolecular fluorescence complementation and coimmunoprecipitation, unexpectedly revealed homo- and heteromerization of OST1 with SnRK2.2, SnRK2.3, OST1, and SnRK2.8. Furthermore, several OST1-complexed proteins were identified as type 2A protein phosphatase (PP2A) subunits and as proteins involved in lipid and galactolipid metabolism. More detailed analyses suggested an interaction network between ABA-activated SnRK2-type protein kinases and several PP2A-type protein phosphatase regulatory subunits. pp2a double mutants exhibited a reduced sensitivity to ABA during seed germination and stomatal closure and an enhanced ABA sensitivity in root growth regulation. These analyses add PP2A-type protein phosphatases as another class of protein phosphatases to the interaction network of SnRK2-type protein kinases.
Land plants adapted the molecule abscisic acid (ABA) as a hormone to control plant water status and to regulate developmental processes in response to limited water conditions (Cutler et al., 2010; Raghavendra et al., 2010; Hauser et al., 2011). In particular, ABA regulates seed dormancy (Finkelstein et al., 2008), root growth and development (De Smet et al., 2006; Duan et al., 2013), and stomatal movements (Kim et al., 2010) in response to environmental cues, including drought and salinity.
ABA is perceived by a family of pyrabactin resistance/regulatory component of abscisic acid receptor (PYR/RCAR) ABA receptors (Ma et al., 2009; Park et al., 2009). In complex with ABA, PYR/RCAR proteins interact with and negatively regulate type 2C protein phosphatases (PP2Cs; Ma et al., 2009; Park et al., 2009; Santiago et al., 2009; Nishimura et al., 2010; Szostkiewicz et al., 2010). Inhibition of PP2Cs enables the activation of Sucrose Nonfermenting1-Related Protein Kinases2 (SnRK2; Fujii et al., 2009; Melcher et al., 2009) through a release of dephosphorylation and steric inhibition (Umezawa et al., 2009; Vlad et al., 2009; Soon et al., 2012; Xie et al., 2012).
In Arabidopsis (Arabidopsis thaliana), the SnRK2-type protein kinase family consists of 10 members (Hrabak et al., 2003). Although salt and osmotic stress activate nine of these family members (Boudsocq et al., 2004, 2007; Umezawa et al., 2004; Yoshida et al., 2006), the major kinases activated by ABA are SnRK2.2/SRK2D, SnRK2.3/SRK2I, and Open Stomata1 (OST1)/SnRK2.6/SRK2E (Mustilli et al., 2002; Yoshida et al., 2002, 2006; Boudsocq et al., 2004, 2007; Fujii et al., 2007; Fujii and Zhu, 2009; Nakashima et al., 2009; Umezawa et al., 2009). In addition, OST1 is activated by low humidity (Yoshida et al., 2002, 2006) and required for guard cell CO2 signal transduction (Xue et al., 2011; Merilo et al., 2013). SnRK2-type protein kinase activation requires the phosphorylation of the activation loop within the kinase domain and its stabilization by the DI domain/SnRK2 box located C-terminal to the activation loop (Belin et al., 2006; Boudsocq et al., 2007; Umezawa et al., 2009; Vlad et al., 2010; Yunta et al., 2011). Phosphorylation of the activation loop can occur through trans(auto)phosphorylation (Belin et al., 2006; Ng et al., 2011) or potentially through not yet identified kinases. The C-terminal DII domain/ABA box is required for PP2C-type protein phosphatase interaction and ABA activation (Belin et al., 2006; Yoshida et al., 2006). However, the deletion of this domain did not affect the activation of OST1 in response to osmotic stress and low humidity, suggesting ABA-dependent and -independent activation mechanisms of OST1 (Yoshida et al., 2006).
Substrates or interacting proteins of SnRK2-type protein kinases include abscisic acid responsive elements (ABRE)-binding factors (AREB/ABF)- and abscisic acid insensitive5 (ABI5)-type transcription factors (Kobayashi et al., 2005; Furihata et al., 2006; Sirichandra et al., 2010), the basic helix-loop-helix (bHLH) transcription factor inducer of C-repeat-binding factor (CBF) expression1 (ICE1; Ding et al., 2015), the ion channels slow anion channel-associated1 (SLAC1), quick anion channel (QUAC1)/aluminium-activated malate transporter12 (ALMT12), and potassium channel in Arabidopsis (KAT1; Geiger et al., 2009; Lee et al., 2009; Sato et al., 2009; Sasaki et al., 2010; Brandt et al., 2012; Imes et al., 2013), the anion/proton exchanger chloride channel a (CLCa; Wege et al., 2014), the NADPH oxidase resporatory burst oxidase homolog F (RbohF; Sirichandra et al., 2009), 14-3-3 proteins (Shin et al., 2007), the SnRK2-interacting calcium sensor (SCS; Bucholc et al., 2011), and the functionally unknown protein SnRK2-substrate1 (SNS1; Umezawa et al., 2013). In addition, putative substrates were identified through mapping peptide phosphorylation preferences (Vlad et al., 2008; Sirichandra et al., 2010) and phosphoproteomics approaches (Shin et al., 2007; Umezawa et al., 2013; Wang et al., 2013). However, validation and functional analyses of such putative SnRK2-type protein kinase substrates or interacting proteins are incomplete.
Consistent with the environmental conditions in which SnRK2-type protein kinases are activated, mutations in SnRK2-type protein kinases rendered Arabidopsis hypersensitive to drought, osmotic stress, and low humidity and insensitive to elevated CO2 (Mustilli et al., 2002; Yoshida et al., 2002, 2006; Umezawa et al., 2004; Xie et al., 2006; Fujii et al., 2007, 2011; Fujii and Zhu, 2009; Fujita et al., 2009; Xue et al., 2011; Merilo et al., 2013; Tian et al., 2015). These defects were linked to altered expression of stress-responsive genes (Umezawa et al., 2004; Fujii et al., 2007, 2011; Fujii and Zhu, 2009; Fujita et al., 2009; Nakashima et al., 2009) and impaired stomatal responses (Merlot et al., 2002; Mustilli et al., 2002; Yoshida et al., 2002; Xie et al., 2006; Vahisalu et al., 2010; Xue et al., 2011; Merilo et al., 2013). Of the ABA-activated SnRK2-type protein kinases, SnRK2.2 and SnRK2.3 function together predominantly during seed germination and young seedling development (Fujii et al., 2007; Fujii and Zhu, 2009; Fujita et al., 2009; Nakashima et al., 2009). OST1 plays a major role in guard cells (Merlot et al., 2002; Mustilli et al., 2002; Yoshida et al., 2002; Xie et al., 2006; Xue et al., 2011). Disruption of all three ABA-activated SnRK2-type protein kinases resulted in a significant decrease in ABA sensitivity compared with single and double mutants (Fujii and Zhu, 2009; Fujita et al., 2009; Nakashima et al., 2009). Consistently, OST1 was found to also be expressed in the leaf vasculature, roots, and seeds, and in-gel kinase assays showed OST1 activity in roots and seeds (Mustilli et al., 2002; Nakashima et al., 2009).
Type 2A protein phosphatases (PP2As) are heterotrimeric holoenzyme complexes consisting of regulatory PP2AA and PP2AB and catalytic PP2AC subunits (Xu et al., 2006b; Shi, 2009). The Arabidopsis genome encodes 3 PP2AA, 17 PP2AB (2 B, 9 B′, and 6 B″ subunits), and 5 PP2AC subunits (DeLong, 2006; Farkas et al., 2007). Forward and reverse genetics approaches revealed functional roles of PP2A-type protein phosphatases in ABA signaling (Kwak et al., 2002; Pernas et al., 2007; Saito et al., 2008; Charpentier et al., 2014), regulation of auxin flux (Fisher et al., 1996; Garbers et al., 1996; Rashotte et al., 2001; Michniewicz et al., 2007; Ballesteros et al., 2013; Dai et al., 2012), brassinosteroid signaling (Tang et al., 2011; Wu et al., 2011), ethylene signaling and biosynthesis (Larsen and Cancel, 2003; Skottke et al., 2011), methyl jasmonate signaling (Saito et al., 2008; Trotta et al., 2011), blue-light signaling (Tseng and Briggs, 2010), plant microbe interactions (Charpentier et al., 2014; Segonzac et al., 2014), and microtubule organization (McClinton and Sung, 1997; Camilleri et al., 2002; Kirik et al., 2012).
PP2AA subunits consist of 15 tandem Huntingtin, elongation factor3 (EF3), PP2A, and the yeast (Saccharomyces cerevisiae) kinase TOR1 (HEAT) repeats forming a hook-like structure for binding to the PP2AB and PP2AC subunits (Xu et al., 2006b; Farkas et al., 2007). Severe developmental defects were observed when pp2aa1/roots curl in NPA1 (rcn1) was combined with either pp2aa2 or pp2aa3, indicating that RCN1 plays a major role in the regulation of PP2A activity (Zhou et al., 2004; Michniewicz et al., 2007). In the Arabidopsis Wassilewskija (Ws) accession, RCN1 mutation results in an ABA hyposensitivity in seed germination and stomatal closure (Kwak et al., 2002; Saito et al., 2008). In contrast, the catalytic subunit mutant pp2ac2 exhibited ABA hypersensitivity in seed germination, root growth, and seedling development (Pernas et al., 2007). Previous pharmacological studies have suggested that both positively regulating and negatively regulating PP2A-type protein phosphatases function in ABA signaling (Schmidt et al., 1995; Hey et al., 1997), leading to the question of the identity of the underlying genes.
The goal of this study was to identify and characterize OST1-interacting proteins (OIPs). Using in planta OST1 protein complex isolations, we identified family members of the SnRK2-type protein kinases, PP2A-type protein phosphatases, and proteins involved in lipid and galactolipid metabolism as OIPs. Additional analyses revealed that regulatory PP2AA and PP2AB′ subunits form an interaction network with ABA-activated SnRK2-type protein kinases. Phenotypically, pp2a double-mutant combinations were ABA hyposensitive during seed germination and stomatal closure and hypersensitive to ABA in root growth assays. Together, our data add PP2A-type protein phosphatases as another family of protein phosphatases into the interaction network of SnRK2-type protein kinases.
RESULTS
Generation and Functional Characterization of OST1-HF Lines
The SnRK2-type protein kinase OST1 was fused at its C terminus to a 6xHis-3xFLAG (HF) tag, resulting in the OST1-HF construct. As a control, a green fluorescent protein (GFP)-HF fusion was generated. Furthermore, the kinase inactive versions OST1DA-HF, harboring the D140A mutation in the proton acceptor site (http://www.uniprot.org/uniprot/Q940H6), and OST1SA-HF, harboring the S175A mutation of the major phosphorylation site in the activation loop (Belin et al., 2006; Umezawa et al., 2009; Vlad et al., 2010), were generated. In addition, the OST1ΔC-HF construct, with deletion of the DII domain/ABA box (amino acids P319–M362; Belin et al., 2006; Yoshida et al., 2006), was constructed. All constructs, driven by the ubiquitin10 (pUBQ10) promoter, were transformed into the ost1-3 mutant in the Arabidopsis Columbia-0 (Col-0) background (SALK_008068; Yoshida et al., 2002). Expression of these constructs was verified by western blot and anti-FLAG immunodetection (Supplemental Fig. S1A).
To confirm the functionality of the OST1-HF constructs, initial coimmunoprecipitation (co-IP) experiments were performed using the PP2C-type protein phosphatase Abscisic Acid Insensitive1 (ABI1) as the positive control (Yoshida et al., 2006; Umezawa et al., 2009; Nishimura et al., 2010) and the yellow fluorescent protein mVenus (Kremers et al., 2006) as the negative control (Supplemental Fig. S1B). In these experiments, OST1-HF was probed for interaction with mVenus-ABI1 or mVenus, and HF-ABI1 was probed for interaction with mVenus-OST1 or mVenus (Supplemental Fig. S1B). After transient expression in Nicotiana benthamiana leaves, purification of either OST1-HF or HF-ABI1 did not copurify the control protein mVenus. In purified OST1-HF fractions, mVenus-ABI1 was hardly detected, whereas HF-ABI1 copurified mVenus-OST1 very efficiently (Supplemental Fig. S1B). These results suggest that the HF tag fused adjacent to the C-terminal DII domain/ABA box of OST1 might interfere with PP2C-type protein phosphatase interaction (see “Discussion”).
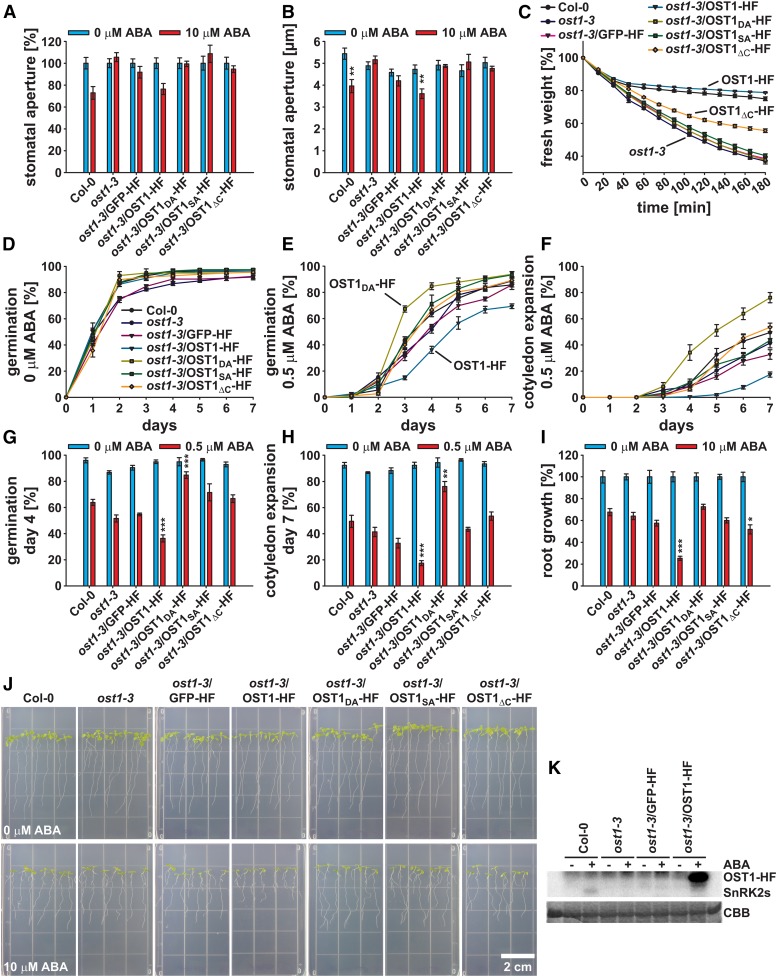
OST1 loss of function mutants are characterized by reduced leaf temperature, impairment of stomatal closure in response to ABA and CO2, and an enhanced water loss under low humidity and drought conditions (Merlot et al., 2002; Mustilli et al., 2002; Yoshida et al., 2002; Xie et al., 2006; Xue et al., 2011). ABA response analyses were performed to investigate whether the different OST1-HF constructs could complement the ost1-3 mutant phenotype (Fig. 1). Investigations of 10 µm ABA-induced stomatal closure revealed that only the OST1-HF wild-type construct could complement the ost1-3 mutant phenotype (Fig. 1, A and B), consistent with Col-0 wild type-like wilting kinetics after rosette leaves were detached from the roots (Supplemental Movie S1). To describe the leaf wilting kinetics more quantitatively, rosette fresh weight was measured every 15 min for a time period of 3 h (Fig. 1C). The results show that only OST1-HF fully complemented the ost1-3 phenotype (Fig. 1C). OST1ΔC-HF partially complemented the ost1-3 wilting phenotype, which was consistent with previous reports (Yoshida et al., 2006).
Figure 1.
OST1-HF constructs affect ABA responses. A and B, Stomatal apertures of 22- to 26-d-old one-half-strength MS agar-grown seedlings 2 h after incubation in 0 (blue bars) or 10 μm ABA (red bars; means ± sem [n = 3–4] with ≥17 stomata per n) normalized to the 0 μm ABA control conditions (A) and raw stomatal aperture values given in micrometers (B). C, Fresh weight time course of detached rosettes (n = 4). Values were normalized to the initial t = 0 min time point. D and E, Time-dependent seed germination in presence of 0 (D) or 0.5 μm ABA (E). F, Time-dependent cotyledon expansion in presence of 0.5 μm ABA. G and H, Seed germination on day 4 after stratification (G) and cotyledon expansion on day 7 after stratification (H) in the presence of 0 (blue bars) or 0.5 μm ABA (red bars). D to H, Means ± sem (n = 4) with 49 seeds per n and normalized to the seed count. I, Root growth of seedlings shown in J in the presence of 0 (blue bars) or 10 μm ABA (red bars; means ± sem [n = 5] with seven seedlings per n) normalized to the 0 μm ABA control conditions. J, Representative images of the root growth assay in I. K, In-gel kinase assay of indicated lines after 1 h treatment with 0 (−) or 10 μm ABA (+). Upper, 32P autoradiograph of kinase activity. Lower, Coomassie Brilliant Blue (CBB) control protein staining. Statistical values for differences between the Col-0 wild type and the indicated lines were calculated using one-way ANOVA (B) and two-way ANOVA (in G–I). *, P < 0.05; **, P < 0.01; ***, P < 0.001. See also Supplemental Movie S1.
In seed germination and cotyledon expansion assays, ost1-3/OST1-HF exhibited an enhanced sensitivity to 0.5 µm ABA compared with the Col-0 wild type, whereas the kinase inactive ost1-3/OST1DA-HF exhibited a reduced sensitivity to ABA (Fig. 1, D–H), indicating a dominant negative effect of this kinase version.
When 4-d-old seedlings were transferred to one-half-strength Murashige and Skoog (MS) medium supplemented with 10 μm ABA, root growth of ost1-3/OST1-HF was strongly inhibited compared with the other investigated lines (Fig. 1, I and J). Also, ost1-3/OST1ΔC-HF exhibited a slight but significant ABA hypersensitivity in the root growth assay (Fig. 1, I and J).
The phenotypical analyses clearly show the in planta functionality of OST1-HF. To biochemically validate these data, in-gel kinase assays were performed with protein extracts of 15-d-old seedlings (Fig. 1K). SnRK2-type protein kinase activation was detected in Col-0 wild-type plants 1 h after 10 μm ABA application, which was strongly reduced in the absence of ABA (Fig. 1K, left), consistent with previous reports (Fujii et al., 2007; Fujii and Zhu, 2009). This activation was not detected in ost1-3 and ost1-3/GFP-HF plants. Compared with ABA activation of SnRK2-type protein kinases in Col-0 wild-type background, OST1-HF was strongly activated only in presence of ABA (Fig. 1K, right). Note the shift in band size because of the HF tag fusion.
Taken together, OST1-HF is functional in plants (Fig. 1) but might exhibit a reduced interaction with PP2C-type protein phosphatases (Supplemental Fig. S1B). Hence, through protein complex isolations, OST1-HF can be used for the identification of previously unreported OIPs.
OST1-HF Protein Complex Isolations and Validations of Interactions
Protein complex isolations of transgenic Arabidopsis lines were conducted on 15-d-old seedlings as described in “Materials and Methods.” Liquid chromatography (LC)-tandem mass spectrometry (MS/MS) data (Supplemental Table S1, A–N) were collected from seven experiments treated with 200 mm sorbitol for 5, 30, or 60 min and seven experiments treated with 50 μm ABA for 30 or 60 min. In total, three independent protein isolations were performed on ost1-3/OST1-HF and ost1-3/GFP-HF (as background control) after sorbitol or ABA treatments. In addition, one experiment was conducted on ost1-3/OST1ΔC-HF treated with 200 mm sorbitol, and one experiment was conducted on ost1-3/OST1DA-HF treated with 50 μm ABA. From all experiments, in total, 120,299 peptides were identified and probed against The Arabidopsis Information Resource 10 (TAIR10) Protein Database.
To identify potential OIPs, protein scores were calculated by the number of peptides identified from OST1-HF purifications minus the number of peptides identified from GFP-HF purifications. Experiment scores were calculated by the number of OST1-HF purifications minus the number of GFP-HF purifications in which a given protein was identified. We included data on the OST1DA-HF and OST1ΔC-HF purifications in the OST1-HF score calculations. OIPs were identified through either unique peptides that specifically match to only one protein of the Arabidopsis proteome (unique score in Table I; Supplemental Table S2) or any peptide that matches to a respective protein sequence (total score in Table I; Supplemental Table S3). Note that Supplemental Tables S2 and S3 list only the proteins that were absent in the respective ost1-3/GFP-HF background control experiments.
Table I. List of potential OIPs selected for additional analyses.
Displayed are the Arabidopsis Gene Identifier (AGI) codes of the respective OIPs with information on detected phosphorylation status, experiment scores (shown in parentheses), and protein scores. Experiment scores were calculated as the number of OST1(DA, ΔC)-HF purification experiments in which the protein was identified minus the number of GFP-HF purification experiments in which the protein was identified. Protein scores were calculated as the number of protein-matching peptides from OST1(DA, ΔC)-HF purification experiments minus the number of protein-matching peptides from GFP-HF purification experiments. Scores were calculated according to the treatment (200 mm sorbitol or 50 µm ABA) from unique peptides, which match only that specific Arabidopsis protein (unique score), and all peptides, which match to that protein (total score). The sums of scores from sorbitol and ABA treatment experiments are also displayed. −, Not detected; ECIP1, EIN2 C terminus interacting protein1.
| AGI | OIP | Phosphorylated | Unique Score Sorbitol | Unique Score ABA | Sum Unique Score | Total Score Sorbitol | Total Score ABA | Sum Total Score |
|---|---|---|---|---|---|---|---|---|
| AT4G33950.1 | OST1 | Yes | 971 (4) | 917 (4) | 1,888 (8) | 3,671 (1) | 2,856 (3) | 6,527 (4) |
| AT4G23920.1 | UGE2 | No | 16 (4) | 11 (4) | 27 (8) | 23 (4) | 19 (4) | 42 (8) |
| AT3G44690.1 | OIP1 | No | 26 (4) | 57 (2) | 83 (6) | 26 (4) | 57 (2) | 83 (6) |
| AT3G03530.1 | NPC4 | No | 24 (4) | 53 (2) | 77 (6) | 34 (4) | 57 (2) | 91 (6) |
| AT3G09880.1 | PP2AB′beta | No | 12 (4) | 15 (2) | 27 (6) | 19 (4) | 23 (2) | 42 (6) |
| AT5G14720.1 | OIP4 | No | 4 (1) | 31 (4) | 35 (5) | 4 (1) | 31 (4) | 35 (5) |
| AT3G09980.1 | OIP6 | No | 3 (1) | 16 (4) | 19 (5) | 3 (1) | 17 (4) | 20 (5) |
| AT5G66880.1 | SnRK2.3 | No (yes) | 5 (2) | 5 (3) | 10 (5) | 2,609 (1) | 1,872 (3) | 4,481 (4) |
| AT1G78290.2 | SnRK2.8 | No | 3 (2) | 7 (3) | 10 (5) | 126 (4) | 81 (4) | 207 (8) |
| AT4G24800.1 | ECIP1 | Yes | 4 (2) | 7 (3) | 11 (5) | 4 (2) | 7 (3) | 11 (5) |
| AT2G46610.1 | RS31a | No | 7 (1) | 21 (3) | 28 (4) | 7 (1) | 21 (3) | 28 (4) |
| AT3G06510.2 | SFR2 | No | — | 16 (4) | 16 (4) | — | 16 (4) | 16 (4) |
| AT5G03470.1 | PP2AB′alpha | No | 5 (2) | 8 (2) | 13 (4) | 5 (2) | 8 (2) | 13 (4) |
| AT1G08050.1 | OIP20 | Yes | — | 29 (3) | 29 (3) | — | 29 (3) | 29 (3) |
| AT1G13320.1 | PP2AA3 | No | 2 (1) | 9 (2) | 11 (3) | 4 (3) | 10 (1) | 14 (4) |
| AT1G70770.1 | OIP26 | Yes | — | 8 (3) | 8 (3) | — | 8 (3) | 8 (3) |
| AT3G50500.2 | SnRK2.2 | No (yes) | 4 (2) | 1 (1) | 5 (3) | 2,508 (1) | 1,809 (3) | 4,317 (4) |
| AT1G59830.1 | PP2AC1 | No | 2 (1) | 2 (1) | 4 (2) | 5 (2) | 4 (1) | 9 (3) |
| AT3G25800.1 | PP2AA2 | No | 1 (1) | 0 (0) | 1 (1) | 5 (3) | 6 (0) | 11 (3) |
| AT3G26030.1 | PP2AB′delta | No | — | — | — | 6 (3) | 5 (3) | 11 (6) |
| AT1G10430.1 | PP2AC2 | No | — | — | — | 3 (2) | 2 (1) | 5 (3) |
Potential OIPs, which were identified in at least three experiments or with at least 10 peptides, were selected for additional validation. These proteins are labeled in red in Supplemental Tables S2 and S3 and listed in Table I. Note that proteins detected by only nonunique peptides (PP2AB′delta and PP2AC2; Table I) are not unequivocally identified in the purifications. PP2C-type protein phosphatases were not detected in our analyses, most likely because of a reduced interaction of OST1-HF with PP2C-type protein phosphatases (e.g. ABI1; Supplemental Fig. S2B). Also, other known OIPs or substrates (see introduction) were not detected. Apart from OST1, our analyses identified three SnRK2-type protein kinases by unique peptides and five members of this kinase family by nonunique peptides (Table I; Supplemental Tables S2 and S3). Furthermore, five PP2A subunits were identified by unique peptides, and two additional putative subunits were identified by nonunique peptides (Table I). Note that four of the seven identified PP2A-type protein phosphatase subunits were detected only at low abundance (Table I), and thus, additional research is required to substantiate these interactions. The highest scoring OIP was UDP-d-Glc 4 epimerase2 (UGE2), which was identified in all OST1-HF purifications (Table I). The proteins OIP4, OIP6, Arg/Ser-rich splicing factor 31A (RS31a), sensitive to freezing2 (SFR2), and OIP20 exhibited higher scores from ABA-treated samples compared with sorbitol-treated samples (Table I).
Subcellular localization analyses (Supplemental Fig. S2A), protein-protein interaction analyses using bimolecular fluorescence complementation (BiFC; Supplemental Fig. S2, B and C), and co-IP analyses (Supplemental Fig. S2D) were performed on proteins listed in Table I. OIPs were localized in various subcellular compartments, including plasma membrane and tonoplast (nonspecific phospholipase C4 [NPC4]), nucleus and nuclear bodies (OIP1 and RS31a), chloroplast envelope membrane (SFR2), or punctuate structures (OIP6 and OIP26; Supplemental Fig. S2A).
To visualize OST1 BiFC complexes with OIPs, high-resolution images were acquired (Supplemental Fig. S2B). Semiquantitative BiFC analyses were performed at low resolution using identical imaging settings to quantify BiFC emissions from multiple cells (“Materials and Methods”; Supplemental Fig. S2C). In general, the subcellular localization of the OIP determined the subcellular localization of its complex with OST1 as indicated by BiFC (Supplemental Fig. S2, B and C). In BiFC analyses, UGE2 and NPC4 exhibited the strongest BiFC emission, which was comparable with the interaction of OST1 with the PP2C-type protein phosphatase ABI1 (Supplemental Fig. 2C, upper). Only OIP26, which located in punctuate structures (Supplemental Fig. S2B, lower right), did not form complexes with OST1 in BiFC analyses (Supplemental Fig. S2, B, lower right and C, lower). In co-IP analyses, OST1 was fused to the C terminus of mVenus or the monomeric cyan fluorescent protein (mT)urquoise to prevent unwanted effects as observed for the OST1-HF and mVenus-ABI1 interaction (Supplemental Fig. S1B). mVenus-OST1 and mT-OST1 were detected by anti-GFP western blot after coexpression and purification of HF-tagged OIPs (Supplemental Fig. S2D), confirming their interactions in planta. Note that OIP1-HF, OIP4-HF, and the galactolipid-remodeling enzyme SFR2-HF proteins were not detected by anti-FLAG detection after immunoprecipitation (Supplemental Fig. S2D, right), indicating a low expression level or an inefficient purification of these proteins. Although mT-OST1 was detected in these co-IP analyses, interaction of OST1 with OIP1, OIP4, and SFR2 requires additional validation (Supplemental Fig. S2D, right).
Transfer DNA (T-DNA) lines of selected OIP genes were isolated and compared with Col-0 and ost1-3 in terms of their ABA responses (Supplemental Fig. S3). npc4-1, oip4-2, and sfr2-3 exhibited a slight ABA hypersensitivity during seed germination and cotyledon expansion (Supplemental Fig. S3, A–D). In addition, uge2-2 exhibited a slight ABA hypersensitivity for the cotyledon expansion (Supplemental Fig. S3D). None of the investigated oip mutants exhibited altered ABA response in root growth and stomatal closure assays (Supplemental Fig. S3, E–G).
Because the in vivo protein complex isolations of OST1-HF identified various SnRK2-type protein kinases and PP2A-type protein phosphatase subunits (Table I), we chose to further investigate these interactions.
OST1 Interacts with SnRK2-Type Protein Kinases in BiFC and Co-IP Analyses
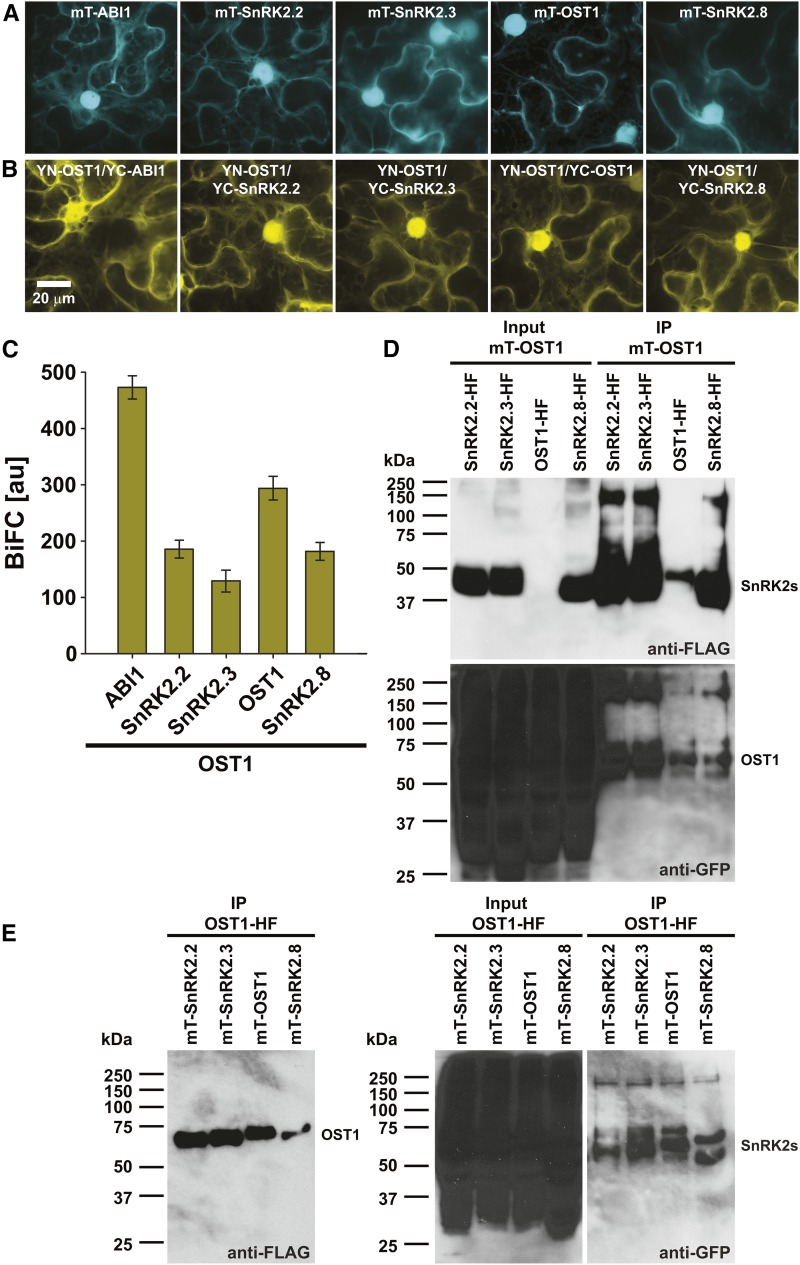
An unexpected finding from the in vivo OST1 protein complex isolations was that three to five other SnRK2-type protein kinases copurified with OST1-HF (Table I; Supplemental Tables S2 and S3). Subcellular localization analyses using (mT)urquoise fusion proteins in transiently transformed N. benthamiana revealed similar expression levels of SnRK2.2, SnRK2.3, OST1, and SnRK2.8 and localization in the cytoplasm and nucleus, similar to the PP2C-type protein phosphatase ABI1 (Fig. 2A). BiFC analyses indicated OST1 interactions with the respective SnRK2-type protein kinases in the cytoplasm and nucleus as well as with ABI1, which was used as a positive control (Fig. 2B). Quantification of the BiFC emissions indicated a stronger OST1-OST1 homodimer versus OST1-SnRK2 heterodimer formation (Fig. 2C). In co-IP analyses, SnRK2-HF and (mT)urquoise-OST1 (Fig. 2D) or OST1-HF and (mT)urquoise-SnRK2 interactions were investigated (Fig. 2E). These analyses confirmed OST1-OST1 homomerizations and OST1-SnRK2 heteromerizations (Fig. 2, D and E). Note that, in western-blot analyses of the raw protein extracts, indicated as input in Figure 2, D and E, OST1-HF was not always detected. However, after HF purification (immunoprecipitation), OST1-HF was enriched and clearly detected (Fig. 2 D, upper and E, left).
Figure 2.
The SnRK2-type protein kinase OST1 forms homo- and heteromers with other SnRK2-type protein kinases. A, (mT)urquoise-ABI1 and mT-SnRK2s are localized in the cytoplasm and the nucleus. B, BiFC analyses indicate OST1 interaction with ABI1 and SnRK2s in the cytoplasm and the nucleus. YC, Yellow fluorescent protein C-terminal fragment; YN, yellow fluorescent protein N-terminal fragment. A and B, Maximum projections of 32-plane z stacks acquired at high magnification with optimized brightness and contrast. C, BiFC quantifications measured from entire low-magnification images, which were acquired using identical settings for all investigated construct combinations (means ± sem; n = 10 images). D, mT-OST1 copurifies with SnRK2-HF fusion proteins. Western blots of HF-tagged SnRK2s (anti-FLAG; upper) and mT-OST1 (anti-GFP; lower) after coexpression in N. benthamiana (input) and anti-FLAG immunoprecipitation (IP) of HF-tagged SnRK2s. E, mT-SnRK2 fusion proteins copurify with OST1-HF. Western blot of OST1-HF (anti-FLAG; left) and mT-SnRK2s (anti-GFP; right) after coexpression in N. benthamiana (input) and anti-FLAG IP of OST1-HF.
SnRK2-Type Protein Kinases Form an Interaction Network with Regulatory PP2AA and PP2AB′ Subunits
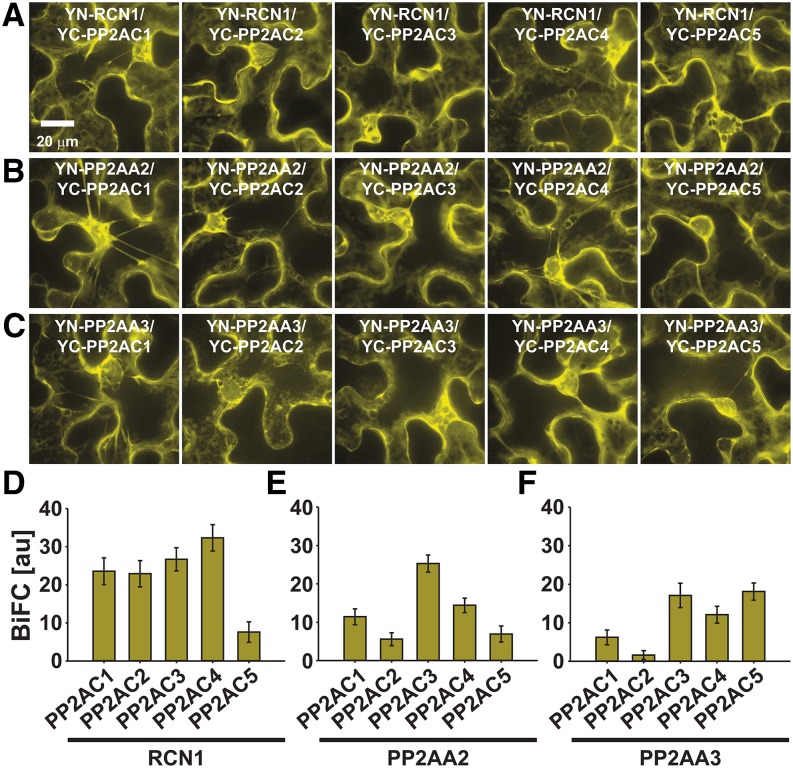
Interaction of the SnRK2-type protein kinase OST1 with the PP2A-type protein phosphatase regulatory B′ subunit PP2AB′beta (Supplemental Fig. S2) and the identification of five to seven PP2A subunits in the OST1-HF protein complex isolations (Table I) raised the question of whether OST1 generally interacts with PP2A-type protein phosphatase subunits. Therefore, BiFC analyses were conducted to investigate interactions of OST1 with all three regulatory PP2AA subunits, three regulatory PP2AB′ subunits (PP2AB′alpha, PP2AB′beta, and PP2AB′delta), which were identified from the OST1-HF protein complex isolations (Table I), and all five catalytic PP2AC subunits (Supplemental Fig. S4, A and D). In these analyses, BiFC emissions were observed for all OST1 and PP2A-type protein phosphatase subunit combinations. However, very low BiFC emissions were observed from OST1 combined with the catalytic PP2AC subunits compared with the PP2AA regulatory subunit interactions (Supplemental Fig. S4D). Note that OST1 interaction with PP2AC5 was at almost background levels, and thus, additional research is needed to substantiate this putative interaction (Supplemental Fig. S4D, inset). In general, OST1 and PP2A subunit complexes were localized in the cytoplasm and nucleus, with the exception of OST1 and the regulatory subunit PP2AB′delta, which was not detected in the nucleus (Supplemental Fig. S4A, upper right). In addition, OST1 interaction with PP2AB′beta, PP2AB′delta, and the catalytic PP2AC subunits was also observed in punctuate structures (Supplemental Fig. S4A, arrows).
To further investigate whether PP2A-type protein phosphatase subunits form an interaction network with SnRK2-type protein kinases, the regulatory PP2A subunits PP2AA1 (RCN1) and PP2AB′beta were probed for interactions with SnRK2.2, SnRK2.3, and OST1 in BiFC analyses (Supplemental Fig. S4, B, C, E, and F). The results of these assays indicated interactions of RCN1 (Supplemental Fig. S4B) and PP2AB′beta (Supplemental Fig. S4C) with all three SnRK2-type protein kinases in the cytoplasm and nucleus. Compared with the OST1 and ABI1 BiFC emission, the RCN1 and SnRK2 BiFC emission was 41% to 67% (Supplemental Fig. S4E), and the PP2AB′beta and SnRK2 BiFC emission was 9% to 11% (Supplemental Fig. S4F).
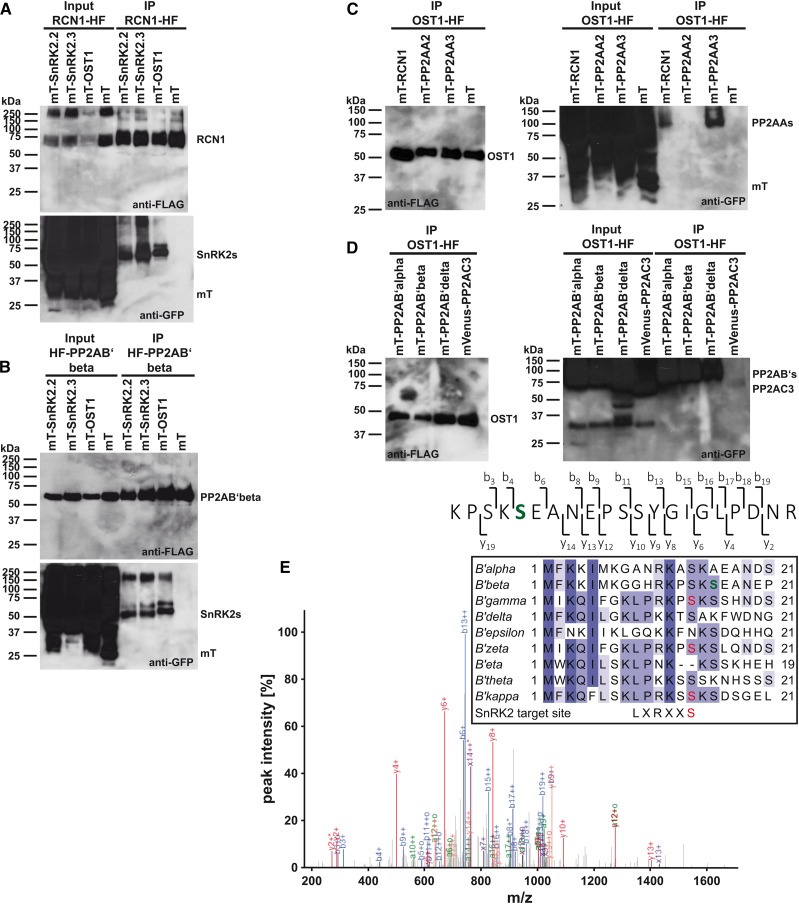
The SnRK2-type protein kinase and PP2A-type protein phosphatase subunit interaction network was further investigated using co-IP analyses. In these analyses, the regulatory PP2A subunits PP2AA1 (RCN1) and PP2AB′beta were fused to the HF tag and coexpressed with (mT)urquoise-SnRK2s or (mT)urquoise alone, which was used as a negative control (Fig. 3, A and B). The results show that purification of RCN1-HF (Fig. 3A) and HF-PP2AB′beta (Fig. 3B) could efficiently copurify the SnRK2-type protein kinases SnRK2.2, SnRK2.3, and OST1 but not the control protein (mT)urquoise (anti-GFP blots in Fig. 3, A and B). In additional experiments, OST1-HF was coexpressed with (mT)urquoise- or mVenus-PP2As or (mT)urquoise alone (Fig. 3, C and D). Purification of OST1-HF efficiently copurified the regulatory PP2AA subunits RCN1 and PP2AA3 but not PP2AA2 and the control protein (mT)urquoise (Fig. 3C). Also, the regulatory B′ subunits PP2AB′alpha, PP2AB′beta, and PP2AB′delta were efficiently copurified with OST1-HF (Fig. 3D), consistent with data of the protein complex isolations (Table I). Copurification of the catalytic C-subunit PP2AC3 with OST1-HF was barely detected (Fig. 3D). These results were consistent with the very low BiFC fluorescence emission of OST1 and PP2AC subunits in BiFC analyses (Supplemental Fig. S4D, inset).
Figure 3.
SnRK2-type protein kinases interact with PP2A-type protein phosphatase regulatory subunits in co-IP analyses. A, Co-IP analyses of the regulatory PP2AA-subunit RCN1-HF with (mT)urquoise-SnRK2s and mT. B, Co-IP analyses of the regulatory PP2AB′-subunit HF-PP2AB′beta with mT-SnRK2s and mT. C, Co-IP analyses of OST1-HF with mT-PP2AA regulatory A subunits and mT. D, Co-IP analyses of OST1-HF with mT-PP2AB′ regulatory B′ subunits and mVenus-PP2AC3 catalytic C subunit. A to D, Western blots of HF-tagged proteins (anti-FLAG) and mT- or mVenus-tagged proteins (anti-GFP) after coexpression in N. benthamiana (input) and anti-FLAG immunoprecipitation (IP) of HF-tagged proteins. E, MS/MS spectrum of an HF-PP2AB′beta phosphopeptide after copurification with mVenus-OST1. The high-intensity fragment ion peaks from b6 onward indicate that one of the first two Ser residues is phosphorylated. Fragment ion b3, observed as low-intensity peak, localizes the site of modification to S16 (green). E, inset, MuscleWS alignment of PP2AB′ subunits. Conserved amino acids are highlighted by different tones of purple. PP2AB′beta S16 (green) is conserved in six of nine PP2AB′ subunits. PP2AB′gamma, PP2AB′zeta, and PP2AB′kappa harbor an SnRK2-type protein kinase consensus target site (red; Vlad et al., 2008; Sirichandra et al., 2010).
Phosphopeptide enrichment and LC-MS/MS analyses after co-IP of HF-PP2AB′beta with mVenus-OST1 (Supplemental Fig. S2D, left) indicated phosphorylation of Ser-16 in PP2AB′beta (Fig. 3E). This Ser-16 residue is conserved in six of nine PP2AB′ subunits. Investigations of the N-terminal sequences of PP2AB′ subunits revealed that PP2AB′gamma, PP2AB′zeta, and PP2AB′kappa exhibit a consensus SnRK2-type protein kinase target site at Ser-14 (Fig. 3E, inset; Vlad et al., 2008; Sirichandra et al., 2010). In summary, BiFC and co-IP analyses revealed strong evidence for the formation of a protein-protein interaction network between SnRK2-type protein kinases and PP2A-type protein phosphatase regulatory A and B′ subunits. PP2AB′ subunits might be target proteins of SnRK2-type protein kinases in planta.
PP2A-Type Protein Phosphatase Subunit Mutants Exhibit Altered ABA Responses
PP2A-type protein phosphatases are known to be involved in ABA responses. In the Arabidopsis Ws accession, the regulatory PP2AA subunit mutant rcn1-1 exhibited ABA-hyposensitive phenotypes, whereas the single catalytic PP2AC subunit mutant pp2ac2 was ABA hypersensitive (Kwak et al., 2002; Pernas et al., 2007). In our analyses, we used T-DNA insertion lines in the Col-0 accession. Currently, it is unknown which regulatory PP2AB subunits function in ABA responses. Because the Arabidopsis genome encodes 17 PP2AB subunits, including 9 PP2AB′ subunits (Farkas et al., 2007), we focused on functional characterizations of regulatory PP2AA subunits and catalytic PP2AC subunits.
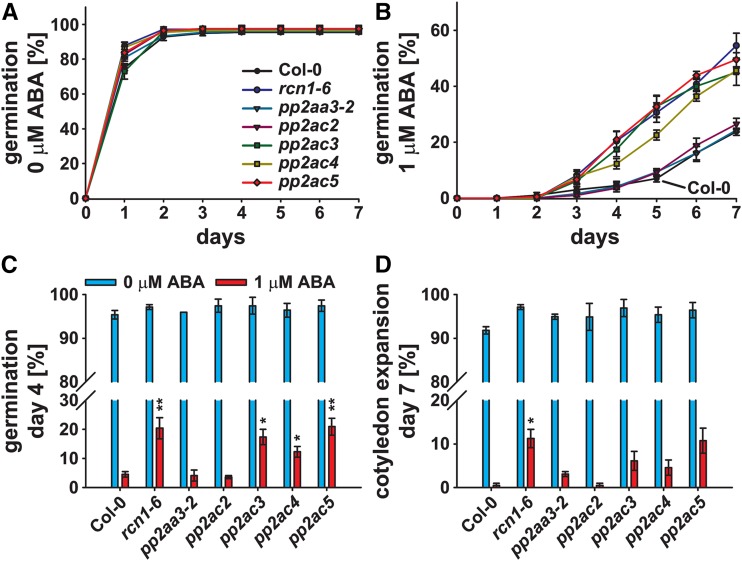
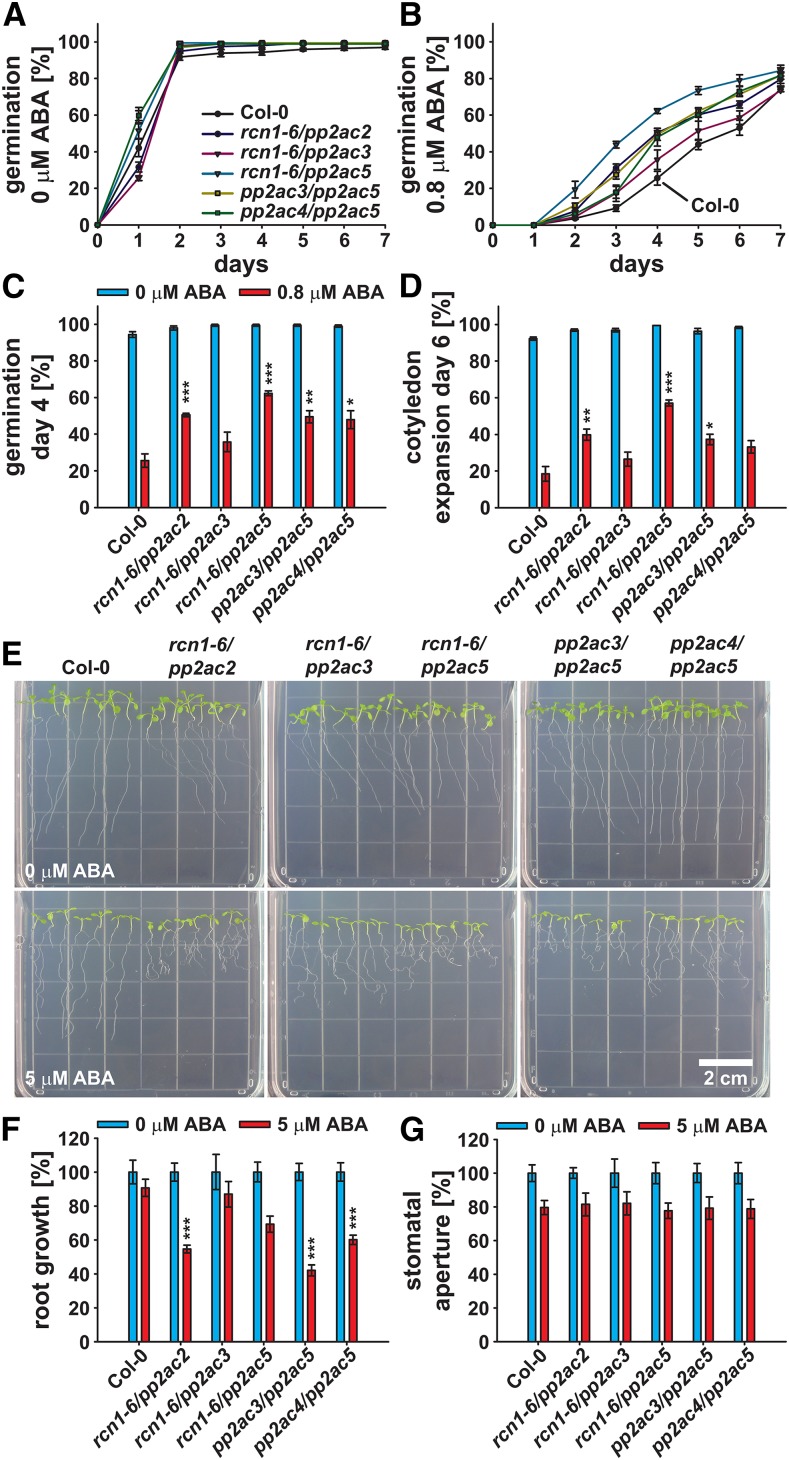
Seed germination analyses in response to 1 μm ABA revealed a reduced ABA sensitivity of the regulatory PP2AA subunit mutant rcn1-6 and the catalytic PP2AC subunit mutants pp2ac3, pp2ac4, and pp2ac5 (Fig. 4, A and B). Seed germination of these lines was significantly increased when grown on one-half-strength MS supplemented with 1 μm ABA compared with the Col-0 wild type on day 4 after stratification (Fig. 4C). Also, a significantly higher percentage of rcn1-6 mutant seedlings was green with expanded cotyledons on day 7 after stratification (Fig. 4D). ABA sensitivities of the mutant lines pp2aa3-2 and pp2ac2 were similar to the Col-0 wild type (Fig. 4).
Figure 4.
PP2A-type protein phosphatase subunit single mutants exhibit a reduced ABA sensitivity during seed germination. A and B, Time-dependent seed germination of Col-0 and indicated pp2a single mutants in the presence of 0 (A) or 1 μm ABA (B). C and D, Seed germination on day 4 after stratification (C) and cotyledon expansion on day 7 after stratification (D) in the presence of 0 (blue bars) or1 μm ABA (red bars). Means ± sem (n = 4) with 49 seeds per n normalized to the seed count. Statistical values for differences between the Col-0 wild type and the pp2a single-mutant lines were calculated using two-way ANOVA. *, P < 0.05; **, P < 0.01.
Analysis of seedling growth on one-half-strength MS supplemented with or without 5 μm ABA was performed and compared with the ABA-insensitive abi1-1 mutant in the Col-0 background (Supplemental Fig. S5). In contrast to abi1-1, which was insensitive to ABA, the pp2a mutants exhibited a root growth reduction similar to the Col-0 wild type in response to ABA (Supplemental Fig. S5B). Note that rcn1-6 exhibited an agravitropic root growth on control medium, which was further enhanced in the presence of 5 μm ABA (Supplemental Fig. S5A).
In Arabidopsis, PP2A ternary complexes potentially consist of 255 different PP2A-subunit combinations. We further investigated ABA responses of regulatory PP2AA-subunit and catalytic PP2AC-subunit double-mutant combinations, in which only a limited number of potential PP2A-type protein phosphatase ternary complexes can be formed. The regulatory A subunit PP2AA1 (RCN1) plays a major role in PP2A-type protein phosphatase regulation in the Ws accession (Zhou et al., 2004; Michniewicz et al., 2007). Therefore, rcn1-6 and pp2ac double-mutant combinations were also generated and functionally analyzed.
In seed germination and cotyledon expansion/greening assays on one-half-strength MS control medium, the investigated double-mutant combinations exhibited germination rates and seedling growth similar to the Col-0 wild type (Fig. 5, A, C, and D). However, in response to 0.8 μm ABA, all investigated rcn1-6/pp2ac and pp2ac double-mutant combinations exhibited an increased seed germination rate when compared with the Col-0 wild type (Fig. 5B). On day 4 after stratification, seed germination of rcn1-6/pp2ac2, rcn1-6/pp2ac5, pp2ac3/pp2ac5, and pp2ac4/pp2ac5 was significantly increased and ≥2-fold compared with the Col-0 wild type (Fig. 5C). The number of seedlings with green and expanded cotyledons on day 6 after stratification was also significantly higher for rcn1-6/pp2ac2, rcn1-6/pp2ac5, and pp2ac3/pp2ac5 (Fig. 5D). The rcn1-6/pp2ac3 double mutant had no significant effect on ABA inhibition of seed germination and cotyledon expansion in these experimental conditions (Fig. 5, C and D).
Figure 5.
rcn1-6/pp2ac and pp2ac double mutants exhibit altered ABA responses during seed germination and seedling growth. A and B, Time-dependent seed germination of Col-0 and indicated pp2a double mutants in the presence of 0 (A) or 0.8 μm ABA (B). C and D, Seed germination on day 4 after stratification (C) and cotyledon expansion on day 6 after stratification (D) in the presence of 0 (blue bars) or 0.8 μm ABA (red bars). A to D, Means ± sem (n = 4) with 49 seeds per n normalized to the seed count. E, Four-day-old seedlings were transferred to one-half-strength MS agar plates supplemented with 0 (upper) or 5 μm ABA (lower) and grown for an additional 5 d. F, Root growth of seedlings shown in E in the presence of 0 (blue bars) and 5 μm ABA (red bars; means ± sem [n = 5] with seven seedlings per n) normalized to the 0 μm ABA control conditions. G, Stomatal apertures 2 h after incubation in 0 (blue bars) or 5 μm ABA (red bars; means ± sem [n = 4] with ≥19 stomata per n) normalized to the 0 μm ABA control conditions. Statistical values for differences between the Col-0 wild type and the pp2a double-mutant lines were calculated using two-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In contrast to the reduced ABA sensitivity during seed germination, the rcn1-6/pp2ac2, pp2ac3/pp2ac5, and pp2ac4/pp2ac5 double mutants exhibited a significantly enhanced sensitivity to 5 μm ABA in root growth assays (Fig. 5, E and F). Root growth of these mutants was inhibited to 42% to 60% of control conditions compared with 70% of rcn1-6/pp2ac5 and 91% of the Col-0 wild type (Fig. 5F). Interestingly, on control medium, all investigated rcn1-6/pp2ac double mutants exhibited an rcn1-6-like agravitropic root growth phenotype (Fig. 5E, upper; Supplemental Fig. S5A). Such agravitropic root growth was also observed for pp2ac3/pp2ac5 (Fig. 5E, upper).
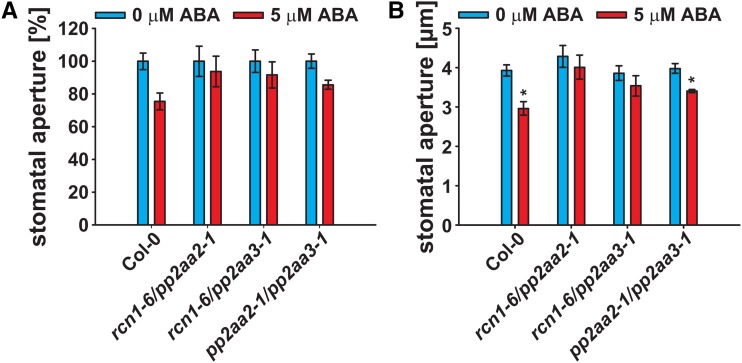
None of the investigated rcn1-6/pp2ac and pp2ac double-mutant combinations exhibited altered responses for ABA-induced stomatal closure (Fig. 5G). Therefore, we investigated stomatal responses of regulatory PP2AA-subunit double-mutant combinations (Fig. 6). rcn1-6/pp2aa2-1 and rcn1-6/pp2aa3-1 double mutants exhibited strongly reduced A-subunit expression (Supplemental Fig. S6A) and consequently, reduced growth compared with the Col-0 wild type and the pp2aa2-1/pp2aa3-1 double mutant (Supplemental Fig. S6B). These data are consistent with a previous report on rcn1-1/pp2aa2-1 and rcn1-1/pp2aa3-1 double mutants (Zhou et al., 2004).
Figure 6.
PP2AA-subunit double mutants exhibit a reduced ABA sensitivity in stomatal closure. Stomatal apertures of 29- to 33-d-old plants 2 h after incubation in 0 (blue bars) or 5 μm ABA (red bars; means ± sem [n = 3–6] with ≥30 stomata per n) normalized to the 0 μm ABA control conditions (A) and real aperture values (micrometers; B). Statistical values for differences between the control and 5 μm ABA treatments were calculated using one-way ANOVA. *, P < 0.05.
Stomata of the Col-0 wild type and the pp2aa2-1/pp2aa3-1 double mutant significantly responded to 5 μm ABA and closed to 75% and 86% of apertures, respectively, when compared with control conditions (Fig. 6). In contrast, stomata of the rcn1-6/pp2aa2-1 and rcn1-6/pp2aa3-1 double mutants did not respond significantly to 5 μm ABA (Fig. 6B). In summary, disruption of PP2A activity renders Arabidopsis Col-0 less sensitive to ABA during seed germination and stomatal closure but hypersensitive to ABA during seedling development.
Regulatory PP2AA Subunits and Catalytic PP2AC Subunits Interact in BiFC and Yeast Two-Hybrid Analyses
Because of the large number of potential PP2A-type protein phosphatase ternary complexes, knowledge of PP2A-subunit expression, subcellular localization, and interaction patterns is essential for understanding PP2A-type protein phosphatase function. BiFC, subcellular localization, and yeast two-hybrid analyses were performed to investigate whether PP2AA- and PP2AC-subunit interactions and PP2A-subunit localizations correlate with the observed differences in ABA responses of pp2a single and double mutants (Figs. 4 and 5). BiFC emission was observed for all possible regulatory PP2AA-subunit and PP2AC-subunit combinations (Fig. 7). Protein complexes of PP2AA1 (RCN1; Fig. 7A), PP2AA2 (Fig. 7B), and PP2AA3 (Fig. 7C) with PP2AC subunits were predominantly located in the cytoplasm. Note that the single PP2AA- and PP2AC-subunit proteins fused to (mT)urquoise or mVenus localized in the cytoplasm and nucleus (Supplemental Fig. S7, A and B). Regulatory PP2AB subunits were differentially distributed within the cell (Supplemental Fig. S7, C–E). Quantification of BiFC emissions indicated a reduced interaction of RCN1 with PP2AC5 (Fig. 7D). PP2AA2 interacted most strongly with PP2AC3 (Fig. 7E), and PP2AA3 interacted most strongly with PP2AC3-PP2AC5 (Fig. 7F). Yeast two-hybrid analyses confirmed the broad interaction spectrum of PP2AA subunits with PP2AC subunits (Supplemental Fig. S8). However, in our experimental condition, PP2AA3 did not interact with PP2AC4 (Supplemental Fig. S8C). Taken together, regulatory PP2AA and catalytic PP2AC subunits form a broad interaction network, which is consistent with partial functional overlap of PP2A subunits in ABA responses.
Figure 7.
Regulatory PP2AA subunits interact with catalytic PP2AC subunits in BiFC analyses. A and D, BiFC analyses of YN-RCN1 with YC-PP2AC1-C5. B and E, BiFC analyses of YN-PP2AA2 with YC-PP2AC1-C5. C and F, BiFC analyses of YN-PP2AA3 with YC-PP2AC1-C5. A to C, High-magnification 32-plane z-stack maximum projections with optimized brightness and contrast showing PP2AA- and PP2AC-subunit complex formations in the cytoplasm. YC, Yellow fluorescent protein C-terminal fragment; YN, yellow fluorescent protein N-terminal fragment. D to F, BiFC quantifications measured from entire low-magnification images, which were acquired using identical settings for each BiFC experiment (means ± sem [n = 10 images]).
Analyses of PP2A-Type Protein Phosphatase and SnRK2-Type Protein Kinase Cross Regulation
The physical interactions of ABA-activated SnRK2-type protein kinases with PP2A-type protein phosphatases, their functional involvement in ABA responses, and the identification of a potential phosphorylation site in regulatory PP2AB′ subunits raised the question of whether such kinase and phosphatase pairs directly regulate the activity of each other.
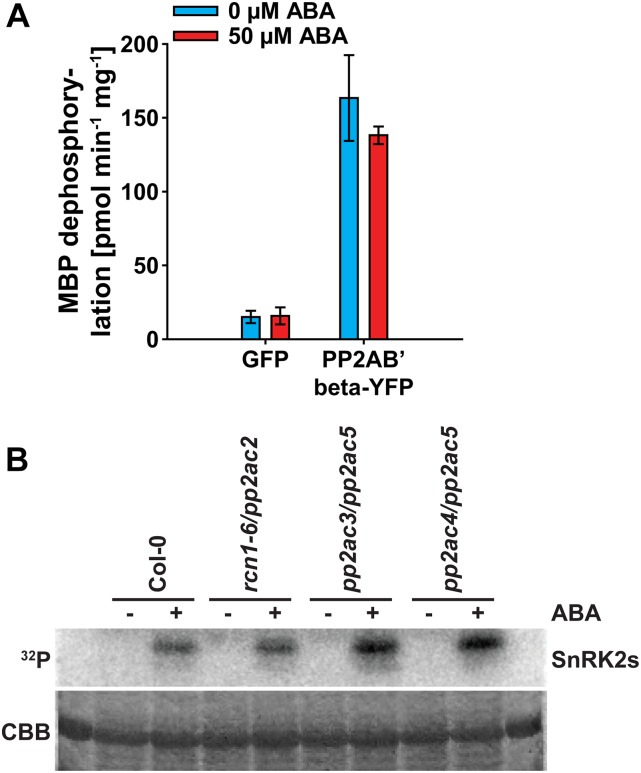
PP2A-type protein phosphatase activity was investigated after PP2A complex isolations of 2-week-old seedlings expressing PP2AB′beta fused to yellow fluorescent protein (PP2AB′beta-YFP) or GFP as control (Fig. 8A; Tang et al., 2011). Using myelin basic protein (MBP) as substrate, isolated PP2A complexes exhibited PP2A activity, which was clearly reduced in the GFP control purifications (Fig. 8A). However, compared with control conditions, 1 h of 50 µm ABA treatment did not alter PP2A activity (Fig. 8A).
Figure 8.
Analyses of PP2A-type protein phosphatase and SnRK2-type protein kinase cross regulation. A, PP2A-type protein phosphatase activity of 2-week-old GFP control seedlings and PP2AB′beta-YFP using MBP as a substrate. Activities were measured after GFP complex isolations 1 h after 0 μm ABA (blue bars) or 50 μm ABA (red bars) treatments. Data represent means ± sd of n = 3 experiments. B, In-gel kinase assay of 1-week-old Col-0 wild type and indicated pp2a double mutants in response to 1 h of 0 μm ABA (−) or 10 μm ABA (+) treatment. Coomassie Brilliant Blue (CBB) staining indicates the protein loading.
In-gel kinase assays were performed on protein extracts of 1-week-old Col-0, rcn1-6/pp2ac2, pp2ac3/pp2ac5, and pp2ac4/pp2ac5 double-mutant seedlings, with the most severe root growth phenotype in response to ABA (Figs. 5, E and F and 8B). Compared with the Col-0 wild type, pp2a double mutants did not exhibit any drastically altered ABA activation of SnRK2-type protein kinases in response to 1 h of 10 µm ABA treatment (Fig. 8B). In summary, we did not resolve any cross regulation of SnRK2-type protein kinase and PP2A-type protein phosphatase activity within a 1-h time period of ABA treatment.
DISCUSSION
Design and ABA Responses of OST1-HF Lines
The SnRK2-type protein kinase OST1 is one of the major ABA signaling components in guard cells, but it also functions in roots and seeds (Merlot et al., 2002; Mustilli et al., 2002; Yoshida et al., 2002; Xie et al., 2006; Fujii and Zhu, 2009; Fujita et al., 2009; Acharya et al., 2013). Our goal was to identify OIPs using in planta protein complex isolations followed by LC-MS/MS analyses.
We ubiquitously expressed OST1-HF constructs, including inactive D140A and S175A mutations (Belin et al., 2006; Boudsocq et al., 2007; Vlad et al., 2010), and the C-terminally truncated OST1ΔC-HF construct lacking the DII domain/ABA box in the ost1-3 mutant background (Supplemental Fig. S1A). OST1ΔC is activated by osmotic stress and low humidity but not by ABA, indicating that it can function in an ABA-independent manner (Yoshida et al., 2006). From these constructs, only OST1-HF could fully complement guard cell-related phenotypes of ost1-3 (Fig. 1, A–C; Supplemental Movie S1), indicating that these responses require the active kinase and the DII domain/ABA box (Yoshida et al., 2006). These results were consistent with previous reports, which analyzed the ABA activation of OST1 and complementation of ost1 phenotypes using similar constructs (Belin et al., 2006; Yoshida et al., 2006; Boudsocq et al., 2007; Acharya et al., 2013)
Ectopic expression of OST1-HF rendered seeds and roots hypersensitive to ABA (Fig. 1, D–J). Similarly, overexpression of the ABA receptors pyrabactin resistance1 (PYR1), PYR1-like2 (PYL2), and PYL5 or OST1, as positive regulators of ABA signaling, enhances ABA sensitivity (Santiago et al., 2009; Mosquna et al., 2011; Acharya et al., 2013; Waadt et al., 2014a), whereas overexpression of the PP2C-type protein phosphatases ABI1 and HAB1, as negative regulators, decreases ABA sensitivity (Santiago et al., 2009; Nishimura et al., 2010).
The OST1ΔC-HF construct partially complemented the enhanced leaf water loss of ost1-3, which was consistent with a previous report (Yoshida et al., 2006). ost1-3/OST1ΔC-HF plants additionally exhibited a slight ABA hypersensitivity in root growth assays (Fig. 1, I and J). Such a phenotype might be explained by the previously reported low basal activity of OST1ΔC, which might target and activate downstream ABA signaling components (Yoshida et al., 2006).
Expression of the inactive OST1DA-HF construct resulted in a reduced ABA sensitivity during seed germination and young seedling development (Fig. 1, D–H). OST1DA-HF might compete with endogenous ABA-activated SnRK2-type protein kinases for substrate binding, thereby inhibiting ABA responses. These data are consistent with the inhibition of ABA responses in Vicia faba guard cells through overexpression of the ABA-activated protein kinase (AAPK), which was inactivated through a mutation in the ATP-binding site (Li et al., 2000).
In contrast to OST1DA-HF, the inactive activation loop mutant OST1SA-HF did not affect any ABA responses in the ost1-3 mutant background (Fig. 1). It is possible that phosphorylation of the activation loop might be required for substrate binding.
Identification of OIPs
LC-MS/MS analyses of proteins copurified with OST1-HF identified several potential OIPs (Table I; Supplemental Tables S2–S4). However, none of the known OST1 substrates (see introduction) were identified in our analyses. Three possible reasons could explain this result. (1) Low protein abundance of known OST1-interacting proteins in total protein extracts due to tissue-specific expression. (2) Reduced interactions of OST1-HF with PP2C-type protein phosphatases due to the HF-tag fusion adjacent to the DII-domain/ABA-box (Supplemental Fig. S1B). This feature may enable an enriched copurification of OST1-interacting proteins that do not require interaction with the DII-domain/ABA-box. (3) OST1 interactions with substrates can be transient and may occur only during signaling. The present analyses demonstrate interactions with various proteins previously not known to interact with SnRK2-type protein kinases. These interactions have been confirmed using co-IP and BiFC analyses (Supplemental Fig. S2). Below, we discuss some interesting aspects of OIPs.
The OIPs UGE2, NPC4, and SFR2 Are Implicated in Lipid Metabolism
UGE2, NPC4, and SFR2 were confirmed to interact with OST1 (Table I; Supplemental Fig. S2). NPC4 hydrolyzes phosphatidylcholine or phosphatidylethanolamine to produce inorganic phosphate-containing head groups and diacylglycerol and has been implicated in phosphate recycling from phospholipids during phosphate deprivation (Nakamura et al., 2005). Diacylglycerol produced by NPC4 can be converted to phosphatidic acid, which acts as a signaling molecule and is also involved in ABA signaling (Jacob et al., 1999; Testerink and Munnik, 2005; Peters et al., 2010). Analyses of npc4 mutants revealed its role in salt, osmotic stress, and ABA responses (Peters et al., 2010; Kocourková et al., 2011).
UGE activity has been implicated to provide UDP-Gal for cell wall and galactolipid biosynthesis (Dörmann and Benning 1998; Rösti et al., 2007). Galactolipids are synthesized at the chloroplast envelope membranes from UDP-Gal and diacylglycerol to produce monogalactosyldiacylglycerol, which can be further processed to digalactosyldiacylglycerol (Moellering and Benning, 2011). Both galactolipids are substrates of SFR2, which produces oligogalactolipids and diacylglycerol (Benning and Ohta, 2005; Moellering et al., 2010; Moellering and Benning, 2011). SFR2 was originally identified through a screen for mutants sensitive to freezing (Warren et al., 1996; Thorlby et al., 2004). Interestingly, SFR2 activity was also induced by osmotic stress and dehydration (Moellering et al., 2010), and the modulation of lipid compositions is known to occur in response to abiotic stresses (Moellering and Benning, 2011). Recently, OST1 has also been implicated in modulating freezing tolerance (Ding et al., 2015).
NPC4, UGE2, and SFR2 might be involved in ABA-mediated responses to abiotic stresses through membrane lipid remodeling, which may be regulated through OST1. Initial ABA response analyses revealed that T-DNA insertions in these genes rendered plants ABA hypersensitive during seed germination and young seedling development (Supplemental Fig. S3, A–D). Note that NPC4 and UGE2 belong to larger gene families. Therefore, higher order mutants would be required for additional investigations in the future.
OST1 Interacts with Other SnRK2-Type Protein Kinases
One major finding of the in planta protein complex isolations was that OST1 formed OST1-OST1 homomer and OST1-SnRK2-type protein kinase heteromer complexes (Table I), which was confirmed for SnRK2.2, SnRK2.3, and SnRK2.8 in BiFC and co-IP analyses (Fig. 2). Analyses of recombinant OST1 protein suggested a monomeric status of OST1 (Yunta et al., 2011). However, this study provides evidence that SnRK2-type protein kinases form oligomers in planta (Fig. 2; Table I). This might indicate that other plant components are required for SnRK2-type protein kinase oligomerization.
SnRK2-type protein kinase complexes could contribute to the amplification of the SnRK2 phosphorylation status through trans(auto)phosphorylation. In vitro trans(auto)phosphorylation of OST1 has been reported (Ng et al., 2011). Phosphorylation of the activation loop is required for SnRK2-type protein kinase activity (Belin et al., 2006; Boudsocq et al., 2007; Umezawa et al., 2009; Vlad et al., 2010). Trans(auto)phosphorylation could potentially contribute to the activation status in osmotic and salt stress conditions, where 9 of 10 SnRK2-type protein kinases are activated (Boudsocq et al., 2004). However, this potential mechanism may not hold true for ABA that mainly activates OST1, SnRK2.2, and SnRK2.3 (Mustilli et al., 2002; Yoshida et al., 2002; Boudsocq et al., 2004; Fujii et al., 2007; Fujii and Zhu, 2009; Nakashima et al., 2009).
SnRK2-Type Protein Kinases Interact with PP2A-Type Protein Phosphatase Regulatory Subunits
Among the potential OIPs identified from the in planta protein complex isolations were five to seven PP2A-type protein phosphatase subunits (Table I). More detailed analyses revealed that OST1 interacted with regulatory PP2AA subunits and PP2AB′ subunits in co-IP and BiFC analyses (Fig. 3; Supplemental Fig. S4). Interactions were also confirmed for the regulatory subunits PP2AA1 (RCN1) and PP2AB′beta with the three ABA-activated SnRK2-type protein kinases (Fig. 3; Supplemental Fig. S4). These data suggest a broad interaction network between SnRK2-type protein kinases and PP2A-type protein phosphatases other than the well-established interactions of SnRK2-type protein kinases with PP2Cs (Yoshida et al., 2006; Umezawa et al., 2009; Vlad et al., 2009; Nishimura et al., 2010).
PP2A-Type Protein Phosphatases Function in ABA Responses
Studies on PP2A-type protein phosphatase subunit mutants in the Arabidopsis Ws accession revealed a reduced ABA sensitivity of the regulatory PP2AA-subunit mutant rcn1-1 in seed germination and stomatal responses (Kwak et al., 2002; Saito et al., 2008) and an ABA hypersensitivity of the catalytic PP2AC-subunit mutant pp2ac2 in seed germination and root growth (Pernas et al., 2007). rcn1-6, rcn1-6/pp2ac2, rcn1-6/pp2ac5, and pp2ac3/pp2ac5 mutants in Arabidopsis Col-0 exhibited reduced ABA sensitivity during seed germination and cotyledon expansion (Figs. 4, B–D and 5, B–D). The catalytic C-subunit single mutant pp2ac2 did not exhibit any altered ABA sensitivity under our experimental conditions (Fig. 4; Supplemental Fig. S5). Reduced ABA sensitivity during stomatal closure was observed for the PP2AA-subunit double mutants rcn1-6/pp2aa2-1 and rcn1-6/pp2aa3-1 (Fig. 6) but not for rcn1-6/pp2ac and pp2ac double-mutant combinations (Fig. 5G). Differences between ABA responses in Ws and Col-0 may result from altered expression levels between natural accessions of Arabidopsis. For instance, PP2A-type protein phosphatase activity in Ws was 4-fold higher compared with that in Col-0 (Hu et al., 2014). Gene dosages of regulatory PP2AA subunits were discussed to be important for PP2A-type protein phosphatase function (DeLong, 2006).
The regulatory PP2AA-subunit mutant rcn1-6 exhibited enhanced root curling in the presence of ABA (Supplemental Fig. S5A), and the ABA sensitivity of root growth and/or curling was found to be strongly increased in the double mutants rcn1-6/pp2ac2, rcn1-6/pp2ac5, pp2ac3/pp2ac5, and pp2ac4/pp2ac5 (Fig. 5, E and F). Our ABA response analyses indicate that PP2A-type protein phosphatases function as positive regulators of ABA responses in seeds and guard cells and as negative regulators in roots. Previous studies on stomatal ABA responses using the PP2A-type protein phosphatase inhibitor okadaic acid provided evidence that okadaic acid-sensitive protein phosphatases function as negative regulators of ABA responses in Pisum sativum and V. faba (Schmidt et al., 1995; Hey et al., 1997; Schwarz and Schroeder, 1998) and as positive regulators in Arabidopsis (Pei et al., 1997; Kwak et al., 2002). Moreover, research in P. sativum suggested that okadaic acid-sensitive protein phosphatases function as negative and positive regulators of ABA responses depending on the conditions (Hey et al., 1997). PP2A-type protein phosphatases function not only in ABA responses but also, in other plant hormone and light responses (Lillo et al., 2014). PP2A-subunit mutants have been characterized by enhanced ethylene levels and ethylene responses (Larsen and Cancel, 2003; Muday et al., 2006; Skottke et al., 2011), altered auxin transport (Garbers et al., 1996; Rashotte et al., 2001; Muday et al., 2006; Michniewicz et al., 2007; Ballesteros et al., 2013), and an enhanced blue light-induced stomatal opening (Tseng and Briggs, 2010). The observed differences in ABA responses of PP2A-type protein phosphatase mutants might be a result of their direct interactions with ABA-activated SnRK2-type protein kinases and cross talk with auxin, ethylene, and/or blue-light responses.
We did not resolve a strong direct link of biochemical cross regulation between SnRK2-type protein kinases and PP2A-type protein phosphatases after a short 1-h ABA treatment in planta (Fig. 8). Future analyses will require investigations of SnRK2-type protein kinase activities in higher order pp2a mutants and in specific tissues and cell types and activity measurements of PP2A-type protein phosphatases after prolonged ABA treatments. Note that a 1-d ABA treatment drastically reduced PP2A-type phosphatase activity in germinating seeds (Hu et al., 2014).
Regulatory PP2AA Subunits and Catalytic PP2AC Subunits Interact in Vivo, and Regulatory PP2AB Subunits Are Differentially Localized
Information about the subcellular distributions and interaction patterns of PP2A subunits is essential for understanding their cellular functions. There is limited knowledge about interaction patterns among PP2A subunits (Haynes et al., 1999), and subcellular localizations are known for the regulatory PP2AA subunits and six PP2AB′ subunits (Blakeslee et al., 2008; Matre et al., 2009; Tang et al., 2011; Trotta et al., 2011; Tran et al., 2012). We performed comprehensive interaction analyses of all three regulatory PP2AA subunits and all five catalytic PP2AC subunits and detected a broad interaction spectrum using BiFC (Fig. 7) and yeast two-hybrid analyses (Supplemental Fig. S8). Regulatory PP2AB subunits are thought to be necessary for the subcellular targeting of PP2A holoenzyme complexes (DeLong, 2006; Farkas et al., 2007). This hypothesis is supported by diverse localization patterns of the 17 PP2AB subunits (Supplemental Fig. S7, C–E).
CONCLUSION
The identification of interacting proteins or substrates of ABA-activated SnRK2-type protein kinases is crucial for unraveling the ABA signaling network and its communication with other signaling pathways. We performed in vivo protein complex isolations of the SnRK2-type protein kinase OST1, identified OIPs using LC-MS/MS, and confirmed their interactions with OST1 using co-IP and BiFC analyses. Our major findings were (1) OST1 unexpectedly forms homo- and heteromers with other SnRK2-type protein kinases, (2) OST1 interacts with enzymes involved in lipid metabolism, (3) SnRK2-type protein kinases form complexes with regulatory subunits of PP2A-type protein phosphatases, and (4) PP2A-type protein phosphatase double mutants exhibit ABA response phenotypes. Our findings provide additional insights into the roles of regulatory PP2AA subunits and catalytic PP2AC subunits in ABA responses, which might be linked to SnRK2-type protein kinase interactions and potentially, cross talk with other signaling pathways.
MATERIALS AND METHODS
T-DNA Lines and Genotyping
T-DNA lines were obtained from the Arabidopsis Biological Resource Center and the Nottingham Arabidopsis Stock Centre. T-DNA insertions were confirmed by PCR on genomic DNA and sequencing of the left and right borders. Genomic DNA was isolated using the cetyl-trimethyl-ammonium bromide method (Stacey and Isaac, 1994). RNA was isolated using the RNeasy Plant Mini Kit (Quiagen) and reverse transcribed using the First-Strand cDNA Synthesis Kit and NotI d(T)18 primers (GE Healthcare). Mutant status was confirmed by PCR using cDNA as template (38 cycles) including actin2 as expression control.
The following T-DNA lines in Col-0 accession were used in this work: ost1-3 (SALK_008068; Yoshida et al., 2002), rcn1-6 (SALK_059903; Blakeslee et al., 2008), pp2aa2-1 (SALK_042724; Zhou et al., 2004), pp2aa3-1 (SALK_014113; Zhou et al., 2004), pp2aa3-2 (SALK_099550; Zhou et al., 2004), pp2ac2 (GK-072G03), pp2ac3 (SALK_035009), pp2ac4 (GK-089F04), pp2ac5 (SALK_139822; Tang et al., 2011), npc4-1 (SALK_046713C; Peters et al., 2010), oip1-1 (SALK_022011C), oip4-2 (SALK_065417C), sfr2-3 (SALK_106253C; Moellering et al., 2010), and uge2-2 (SALK_024044C; Rösti et al., 2007).
Oligonucleotides and Plasmids
Detailed lists of oligonucleotides and plasmids used in this work are provided in Supplemental Tables S5 and S6, respectively.
Plant Growth and ABA Response Assays
Arabidopsis (Arabidopsis thaliana) seeds were surface sterilized in 70% (v/v) ethanol and 0.04% (v/v) SDS followed by three washes in 100% ethanol and sown on one-half-strength MS (pH 5.8; Sigma) supplemented with 0.8% (w/v) Phyto Agar (RPI). After 4 d of stratification at 4°C in the dark, plants were grown in a growth room in long-day conditions (16-h-light/8-h-dark cycle) with 50 to 80 μE m−2 s−1 light intensity at 25°C to 27°C and 25% to 30% relative humidity. Six-day-old seedlings were transferred to pots and grown in either the growth room or a Conviron CMP3244 Plant Growth Chamber with a 16-h-d (22°C)/8-h-night (18°C) cycle, 50 to 100 μE m−2 s−1, and 40% to 50% relative humidity.
ABA response assays were performed as described previously (Waadt et al., 2014a). Seed germination assays were performed in the growth room on one-half-strength MS agar supplemented with 0.5 or 0.8 μm (+)-ABA (TCI), 1 μm (±)-ABA (Sigma), or the respective concentration of ethanol as solvent control. Analyses represent mean values ± sem of four technical replicates normalized to the seed count of each experiment. Root growth assays were performed on 4-d-old seedlings, which were transferred to one-half-strength MS agar supplemented with 5 to 10 μm ABA or the respective concentration of ethanol as solvent control and grown vertically in the growth room for an additional 5 d. Root length was measured using Fiji (http://fiji.sc/Fiji; Schindelin et al., 2012) or the Root Detection software (http://www.labutils.de/rd.html), analyzed as means of seven seedlings ± sem of four to five technical replicates, and normalized to the one-half-strength MS control conditions.
ABA-induced stomatal closure analyses were performed with detached leaves of 3- to 5-week-old plants, which were floated in stomatal assay buffer (5 mm KCl, 50 μm CaCl2, and 10 mm MES-Tris, pH 6.15 [pp2a mutants] or pH 5.6 [ost1-3 and OST1-HF lines]) for 2 h. Subsequently, 5 to 10 μm (+)-ABA or the respective concentration of ethanol as solvent control was added to the opening buffer followed by an additional 2 h of incubation. Leaf epidermal tissue was isolated using the blending method, and images were acquired using an inverted light microscope. Stomatal apertures were measured using Fiji (Schindelin et al., 2012). Data represent mean stomatal apertures ± sem of three to six experiments.
For the dry down experiment, rosettes of 29-d-old plants grown in soil in the growth chamber were cut, placed immediately on dry Whatman paper, and imaged every 0.5 min for 30 min. Quantitative data on 25-d-old plants (n = 4) were determined by rosette fresh weight measurements at intervals of 15 min for a time period of 3 h. Data were normalized to the initial (t = 0 min) fresh weight.
Significance of data was analyzed using one-way ANOVA for stomatal assays or two-way ANOVA for every other assay.
Subcellular Localization and BiFC Analyses
For subcellular localization analyses, coding sequences were inserted into plant expression vectors harboring a pGPTVII.bar or pGPTVII.hyg backbone (Walter et al., 2004) and an expression cassette consisting of the pUBQ10 promoter (AT4G05310; Norris et al., 1993; Krebs et al., 2012) or the pUBQ10-driven β-estradiol inducible system (Schlücking et al., 2013) and the tHSP18.2 terminator (AT5G59720; Nagaya et al., 2010) and fused to the fluorescent proteins mTurquoise (Goedhart et al., 2010) or (m)Venus (Nagai et al., 2002; Kremers et al., 2006). Constructs for BiFC analyses were generated by ligation of coding sequences into hygII-SPYNE(R) and kanII-SPYCE(M) or kanII-SPYCE(MR) plasmids (Waadt et al., 2008). Plasmids were transformed into Agrobacterium tumefaciens GV3101 (pMP90; Koncz and Schell, 1986) and transiently expressed in leaves of 5- to 6-week-old Nicotiana benthamiana plants together with the p19 silencing suppressor as described (Waadt et al., 2014b). Subcellular localizations and BiFC analyses were performed by confocal microscopy using an Eclipse TE2000-U Microscope (Nikon) as described previously (Waadt et al., 2014a, 2014b). Subcellular localizations of fluorescent protein fusions and BiFC complexes were acquired using a Plan Apo 60×/1.20 WI ∞/0.15 to 0.18 WD 0.22 objective and displayed as maximum projections of 32-plane z stacks with optimized brightness and contrast. Semiquantitative BiFC analyses were performed using low magnification (Plan 20×/0.40 ∞/0.17 WD 1.3). Images were acquired using identical settings (exposure time and gain), thus enabling comparisons of fluorescence intensities among parallel investigated construct combinations. BiFC emissions were quantified from entire images using Fiji.
In Vivo Protein Complex Isolations and Co-IP Analyses
Coding sequences were inserted into the pGPTVII.bar plant expression vector harboring either a pUBQ10-NosT expression cassette (see above; Walter et al., 2004) and an HF tag (HHHHHHDYDIPTTASENLYFQGELDYKDHDGDYKDHDIDYKDDDDK) located 3′ of the multiple cloning site or a pUBQ10-tHSP18.2 expression cassette and the HF tag located 5′ of the multiple cloning site and transformed into A. tumefaciens GV3101 (pMP90; Koncz and Schell, 1986).
For in vivo protein complex isolations, GFP-HF and OST1(DA, SA, ΔC)-HF constructs were transformed into the ost1-3 mutant background (Yoshida et al., 2002) by the floral dip method (Clough and Bent, 1998). Transformants were selected on one-half-strength MS agar plates supplemented with 10 μg mL−1 glufosinate and further selected by western blot and anti-FLAG immunodetection. Homozygous T4 lines were sown on one-half-strength MS agar plates (12 plates with 24 seedlings per plate) and grown vertically in the growth room for 15 d. For transient expression, A. tumefaciens harboring HF constructs or mTurquoise- or mVenus-tagged constructs and the p19 strain were cotransfected into one N. benthamiana leaf and incubated for 3 to 4 d. Transient expression in N. benthamiana was performed as described (Waadt et al., 2014b).
Plant tissue was harvested and incubated for 1 h in one-half-strength MS (liquid). Subsequently, medium was exchanged with one-half-strength MS (liquid) and 200 mm sorbitol, or (+)-ABA was added to a final concentration of 50 μm followed by an incubation for 5, 30, or 60 min. Plant tissue was dried briefly using paper towels, frozen in liquid N2, and extracted in SII buffer (100 mm Na2HPO4/NaH2PO4, pH 8.0, 150 mm NaCl, 5 mm EDTA, 5 mm EGTA, and 0.1% [v/v] Triton X-100) supplemented with protease inhibitor (Roche), phosphatase inhibitors 2 and 3 (Sigma), and 1 mm phenylmethylsulfonyl fluoride. After 10 min of rotation at 4°C, raw extracts were sonicated (20 times for 0.5 s on ice) followed by a 20-min centrifugation at 20,000g and filtration through a 0.45 μm syringe filter. Protein extracts were mixed with (1:100, v/v) anti-FLAG M2 magnetic beads (Sigma) equilibrated in SII buffer and rotated for 4 h at 4°C. Proteins bound to the anti-FLAG M2 magnetic beads were concentrated using the DynaMag-2 Magnetic Particle Concentrator (Invitrogen) and washed three to four times in ≥20 bead volumes of SII buffer followed by two washes in FLAG-to-His buffer (100 mm Na2HPO4/NaH2PO4, pH 8.0, 150 mm NaCl, and 0.05% [v/v] Triton X-100). Proteins were eluted four times in one bead volume of FLAG-to-His buffer supplemented with 500 μg mL−1 3xFLAG peptide (Sigma) and stored at −80°C. Western blot and immunodetection were performed using a mouse anti-FLAG M2 antibody (Sigma) or rabbit monoclonal anti-GFP antibody (Invitrogen) followed by a secondary goat anti-mouse or anti-rabbit IgG (H + L) horseradish peroxidase-conjugate antibody (BioRad) as described (Waadt et al., 2014b).
For LC-MS/MS analyses, proteins were precipitated by addition of 1:3 (v/v) cold 100% trichloracetic acid (TCA), followed by 30-min incubation on ice and 30-min centrifugation at 20,000g and 4°C. Protein pellets were washed two times with 100% ice-cold acetone, centrifuged for 10 min at 20,000g and 4°C, air dried, and stored at −80°C.
Mass Spectrometry and Data Analysis
Protein pellets were resolubilized in 50 μL of 0.2% (v/v) ProteaseMAX surfactant prepared in 50 mm ammonium bicarbonate and adjusted to 100 μL of volume with 8 m urea. Samples were reduced and alkylated before overnight digestion with trypsin. The protein digest was pressure loaded onto a biphasic trapping column packed with 2.5 cm of 5 μm Partisphere Strong Cation Exchanger (Hichrom) followed by an additional 2.5 cm of 5 μm Aqua C18 Resin (Phenomenex). Resin-bound peptides were desalted with buffer A (5% [v/v] acetonitrile and 0.1% [v/v] formic acid) by flow through the trap column. The trap and analytical columns (100-μm i.d. capillary with a 5-μm pulled tip packed with 15 cm of 3 μm Aqua C18 Resin; Phenomenex) were assembled using a zero-dead volume union (Upchurch Scientific). LC-MS/MS analysis of the samples was performed on LTQ OrbitrapVelos (Thermo Scientific) interfaced at the front end with a quaternary HP 1100 Series HPLC Pump (Agilent Technology) using MudPIT technology (Washburn et al., 2001). Tandem mass spectrometry spectra were collected in a data-dependent fashion, and resulting spectra were extracted from the Xcalibur data system format into MS2 format using RawXtract. Protein identification was done with Integrated Proteomics Pipeline (IP2) by searching against the TAIR10 database (downloaded January of 2011) and filtering to 1% false positive at the spectrum level using the DTASelect2.0 program (Tabb et al., 2002). Carbamidomethylation on Cys was defined as fixed modification, and phosphorylation on Ser, Thr, and Tyr was included as variable modification in the database search criteria.
Identified peptides were blasted against the TAIR10 protein database (http://www.arabidopsis.org/index.jsp) and used to calculate protein and experiment scores for each identified protein using an in house-designed Dismemberer Java application. Protein scores were calculated as the number of protein-matching peptides identified in the OST1(DA, ΔC)-HF purification experiments minus the number of the protein-matching peptides identified in the GFP-HF purification experiments. Experiment scores were calculated as the number of OST1(DA, ΔC)-HF purification experiments in which the protein was identified minus the number of GFP-HF purification experiments in which the protein was identified. Scores were calculated from unique peptides, of which their sequence matches only one single TAIR10 annotated protein, or any protein-matching peptides, of which their sequence matches any TAIR10 annotated protein. Proteins with experiment scores ≥3 or protein scores ≥10 were considered as potential OIPs and used for additional validation.
Phosphopeptide Enrichment
Phosphopeptides were enriched using the hydroxyapatite (HAP) method (Fonslow et al., 2012) with some modifications. An HAP column of 5 cm in length was prepared from ceramic HAP (Biorad) suspended in methanol and packed into a 250-µm i.d. and 360-µm o.d. fritted capillary. The HAP column was equilibrated using 20 mm Tris (pH 7.2) for 15 min. Tryptic digests were adjusted to pH 7.4 and loaded on the HAP column. The flow through containing unbound peptides was collected for additional LC-MS/MS analysis. Nonspecifically bound peptides and salts were washed off the HAP column using 250 µL of 20 mm Tris and 60% (v/v) acetonitrile (pH 7.2). Enriched phosphopeptides were eluted from the HAP column using 250 µL of 10 mm KH2PO4 (pH 7.8). The elution was repeated with 250 µL of 100 mm KH2PO4 (pH 7.8). Both elution fractions were pooled, acidified, and loaded on a biphasic trap column for MudPIT analysis.
Immunodetection of PP2AA Subunits
For immunodetection of PP2AA subunits, seedlings were grown for 6 d in constant light on vertical one-half-strength MS agar plates containing 1% (w/v) Suc. Double-mutant seedlings in segregating families were identified by their short root phenotype. Whole seedlings were harvested and ground in liquid N2. Plant extract preparation, SDS-PAGE, immunoblotting techniques, and probing with anti-RCN1 antiserum were described previously (Deruère et al., 1999).
PP2A Activity Assays
Two-week-old PP2AB′β-YFP (Tang et al., 2010) and GFP control seedlings grown vertically on one-half-strength MS agar medium were incubated for 1 h in liquid one-half-strength MS medium followed by an additional incubation in liquid one-half-strength MS medium supplemented with 50 µm ABA or EtOH as solvent control. 33P-Labeled MBP was prepared using the Protein Ser/Thr Phosphatase Assay System according to the manufacturer's instructions. Plant tissue was frozen and ground in liquid N2, then suspended in IP buffer (50 mm Tris, 100 mm NaCl, 0.3 m Suc, 0.2% [v/v] Triton X-100, 2 µg mL−1 aprotinin, 1 µg mL−1 leupeptin, and 0.2 mm phenylmethylsulfonyl fluoride) and centrifuged at 16,000g for 10 min at 4°C to pellet debris. Two-hundred fifty micrograms of total protein was added to 300 µL Protein A Sepharose resin slurry and beads were washed twice with IP buffer followed by a wash with PP2A buffer (50 mm Tris-HCl, pH 8.5, 0.1 mm EDTA, 0.01% [v/v] Brij 35; Skottke et al., 2011). Beads were resuspended in 100 µL PP2A buffer and the slurry dispensed into 30-µL aliquots. Labeled substrate (5 µL) was added to each aliquot to start the reaction. After 15 min at 30°C, each reaction was stopped with 35 µL 2× stop solution (100 mm NaF, 20 mm EDTA, 2 mm Na-pyrophosphate) followed by 200 µL of ice-cold 25% [v/v] TCA and centrifuged at 16,000g. Radioactivity in 50 µL of the supernatant was measured in a scintillation counter. Assays were performed in triplicate; similar results were obtained in a separate experiment and in assays using phosphorylated 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE6 peptide (Skottke et al., 2011) as substrate.
In-Gel Kinase Assays
In-gel kinase assays were performed as previously described (Ichimura et al., 2000; Kim et al., 2012) with slight modifications. Proteins of 1-week-old Arabidopsis seedlings, grown on vertical one-half-strength MS agar plates, were transferred to liquid one-half-strength MS for 1 h followed by 1 h of treatment with or without 10 µm ABA. Proteins were extracted by homogenization in liquid N2 and resuspension in extraction buffer (100 mm HEPES, pH 7.5, 5 mm EDTA, 5 mm EGTA, 0.5% [v/v] Triton X-100, 150 mm NaCl, 0.5 mm dithiothreitol [DTT], 10 mm NaF, 0.5% [v/v] protease inhibitor [Sigma-Aldrich], 0.5% [v/v] phosphatase inhibitor [Sigma-Aldrich], 5 mm Na3VO4, and 5 mm β-glycerophosphate 2Na). Cell debris was removed by 30 min of centrifugation at 20,000g at 4°C, and supernatant was transferred to new tubes. Protein concentration was determined by the bicinchoninic acid method (Thermo Fisher Scientific). Twenty micrograms of protein was subjected to electrophoresis on an SDS polyacrylamide gel that contained 0.25 to 0.5 mg mL−1 Histone III-S (Sigma-Aldrich). After electrophoresis, the gel was washed three times in washing buffer (5 mm Tris-HCl, pH 8.0, 0.5 mm DTT, 0.1 mm Na3VO4, 5 mm NaF, 0.5 mg mL−1 bovine serum albumin, and 0.1% [v/v] Triton X-100) for 30 min at room temperature followed by additional two washes in renaturation buffer (25 mm Tris-HCl, pH 8.0, 1 mm DTT, 0.1 mm Na3VO4, and 5 mm NaF) and one wash in renaturation buffer at 4°C overnight. The gel was equilibrated in reaction buffer (25 mm HEPES, pH 7.5, 2 mm EGTA, 12 mm MgCl2, 1 mm DTT, and 0.1 mm Na3VO4) for 30 min at room temperature and then incubated in reaction buffer supplemented with 50 μCi of [γ-32P]ATP (10 μCi μL−1; 3,000 Ci mmol−1; PerkinElmer) for 90 min at room temperature. The reaction was stopped by washing with wash solution (5% [v/v] TCA and 1% [v/v] sodium pyrophosphate) until no radioactivity was detected in the wash solution. The gel was stained with Coomassie Brilliant Blue, dried on a paper, and exposed to an autoradiography film for 3 to 5 d at −80°C. Data shown are a representative of two experiments.
PP2AB′-Subunit Alignment
PP2AB′-subunit sequences were downloaded from TAIR10 (https://www.arabidopsis.org/) according to the annotated AGIs (Farkas et al., 2007). Protein sequences were aligned by the Jalview software (Waterhouse et al., 2009) using the MuscleWS algorithm.
Yeast Two-Hybrid Analyses
Coding sequences were inserted into pGBT9.BS and pGAD.GH vectors (Elledge et al., 1991). The PJ69-4A yeast (Saccharomyces cerevisiae) strain (James et al., 1996) grown at 28°C in YPD agar medium (2% [w/v] bactopeptone, 1% [w/v] yeast extract, 2% [w/v] Glc, and 2% [w/v] bactoagar) was transformed using the polyethylene glycol/lithium acetate method (Gietz et al., 1992), plated on complete supplement mixture (CSM) agar medium (3.35% [w/v] Yeast Nitrogen Base/(NH4)2SO4, 0.32% [w/v] CSM-Leu-Trp, 2% [w/v] Glc, and 2% [w/v] bactoagar), and incubated for 2 to 3 d at 28°C. Successfully transformed yeast colonies were restreaked to new CSM-Leu-Trp agar medium and incubated 1 to 2 d at 28°C; 5 μL of a 10-fold dilution series (optical density at 600 nm of 100 to 10−4 in 2% [w/v] Glc) of transformants was spotted on CSM-Leu-Trp agar medium as a control and CSM-Leu-Trp-His agar medium and supplemented with 2.5 mm 3-amino-1,2,4-triazole for selection of positive interaction. Yeast transformants were incubated for 5 to 7 d at 28°C.
AGIs of loci investigated in this study are provided in Table I and Supplemental Tables S1 to S6.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Western blot of HF lines and control co-IP experiments.
Supplemental Figure S2. Subcellular localizations and interaction analyses of OST1 with OIPs.
Supplemental Figure S3. OIP mutants exhibit a reduced ABA sensitivity during seed germination.
Supplemental Figure S4. SnRK2-type protein kinases interact with PP2A-type protein phosphatase subunits in BiFC analyses.
Supplemental Figure S5. ABA induces an enhanced root curling of rcn1-6.
Supplemental Figure S6. Reduced expression of regulatory PP2AA subunits affects plant growth.
Supplemental Figure S7. Subcellular localizations of PP2A-subunit fluorescent protein fusions.
Supplemental Figure S8. Regulatory PP2AA subunits interact with catalytic PP2AC subunits in yeast two-hybrid analyses.
Supplemental Table S1. LC-MS/MS data from protein complex isolations.
Supplemental Table S2. List of proteins identified by unique peptides and found only in ost1-3/OST1(DA, ΔC)-HF purifications.
Supplemental Table S3. List of proteins identified by any matching peptide and found only in ost1-3/OST1(DA, ΔC)-HF purifications.
Supplemental Table S4. Summarized list of potential OIPs selected for additional analyses (related to Table I).
Supplemental Table S5. Oligonucleotides used in this work.
Supplemental Table S6. Plasmids used and generated in this work.
Supplemental Movie S1. A 30 min time-lapse movie of wilting detached rosettes.
Supplementary Material
Acknowledgments
We thank James Moresco (The Scripps Research Institute), Felix Hauser and Cun Wang (University of California, San Diego) for initial help with data analyses, and Jörg Kudla (University of Münster) for providing various plasmids.
Glossary
- ABA
abscisic acid
- BiFC
bimolecular fluorescence complementation
- co-IP
coimmunoprecipitation
- Col-0
Columbia-0
- DTT
dithiothreitol
- HAP
hydroxyapatite
- LC
liquid chromatography
- MS
Murashige and Skoog medium
- TAIR10
The Arabidopsis Information Resource 10
- T-DNA
transfer DNA
- Ws
Wassilewskija
- MS/MS
tandem mass spectrometry
Footnotes
This work was supported by the Alexander von Humboldt Foundation (Feodor Lynen Fellowship to R.W.), the National Institutes of Health (grant no. GM060396 to J.I.S.; the development of the 6xHIS-3xFLAG tag used was supported by grant nos. F32GM083585 to D.A.N., R01GM056006 to S.A.K., and R01GM067837 to S.A.K.; and grant no. P41 GM103533 to J.R.Y. for proteomics analyses), the National Science Foundation (grant no. MCB1414339 to J.I.S. and grant no. IOS–1145585 to A.D. for biochemical analyses of type 2A protein phosphatase-type protein phosphatases), and the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy (grant no. DE–FG02–03ER15449 to J.I.S. for root growth analyses).
Articles can be viewed without a subscription.
References
- Acharya BR, Jeon BW, Zhang W, Assmann SM (2013) Open Stomata 1 (OST1) is limiting in abscisic acid responses of Arabidopsis guard cells. New Phytol 200: 1049–1063 [DOI] [PubMed] [Google Scholar]
- Ballesteros I, Domínguez T, Sauer M, Paredes P, Duprat A, Rojo E, Sanmartín M, Sánchez-Serrano JJ (2013) Specialized functions of the PP2A subfamily II catalytic subunits PP2A-C3 and PP2A-C4 in the distribution of auxin fluxes and development in Arabidopsis. Plant J 73: 862–872 [DOI] [PubMed] [Google Scholar]
- Belin C, de Franco PO, Bourbousse C, Chaignepain S, Schmitter JM, Vavasseur A, Giraudat J, Barbier-Brygoo H, Thomine S (2006) Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol 141: 1316–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C, Ohta H (2005) Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J Biol Chem 280: 2397–2400 [DOI] [PubMed] [Google Scholar]
- Blakeslee JJ, Zhou HW, Heath JT, Skottke KR, Barrios JA, Liu SY, DeLong A (2008) Specificity of RCN1-mediated protein phosphatase 2A regulation in meristem organization and stress response in roots. Plant Physiol 146: 539–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Barbier-Brygoo H, Laurière C (2004) Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem 279: 41758–41766 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Droillard MJ, Barbier-Brygoo H, Laurière C (2007) Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol Biol 63: 491–503 [DOI] [PubMed] [Google Scholar]
- Brandt B, Brodsky DE, Xue S, Negi J, Iba K, Kangasjärvi J, Ghassemian M, Stephan AB, Hu H, Schroeder JI (2012) Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc Natl Acad Sci USA 109: 10593–10598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholc M, Ciesielski A, Goch G, Anielska-Mazur A, Kulik A, Krzywińska E, Dobrowolska G (2011) SNF1-related protein kinases 2 are negatively regulated by a plant-specific calcium sensor. J Biol Chem 286: 3429–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri C, Azimzadeh J, Pastuglia M, Bellini C, Grandjean O, Bouchez D (2002) The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell 14: 833–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M, Sun J, Wen J, Mysore KS, Oldroyd GE (2014) Abscisic acid promotion of arbuscular mycorrhizal colonization requires a component of the PROTEIN PHOSPHATASE 2A complex. Plant Physiol 166: 2077–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Dai M, Zhang C, Kania U, Chen F, Xue Q, McCray T, Li G, Qin G, Wakeley M, Terzaghi W, et al. (2012) A PP6-type phosphatase holoenzyme directly regulates PIN phosphorylation and auxin efflux in Arabidopsis. Plant Cell 24: 2497–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Zhang H, Inzé D, Beeckman T (2006) A novel role for abscisic acid emerges from underground. Trends Plant Sci 11: 434–439 [DOI] [PubMed] [Google Scholar]
- DeLong A. (2006) Switching the flip: protein phosphatase roles in signaling pathways. Curr Opin Plant Biol 9: 470–477 [DOI] [PubMed] [Google Scholar]
- Deruère J, Jackson K, Garbers C, Söll D, Delong A (1999) The RCN1-encoded A subunit of protein phosphatase 2A increases phosphatase activity in vivo. Plant J 20: 389–399 [DOI] [PubMed] [Google Scholar]
- Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S (2015) OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev Cell 32: 278–289 [DOI] [PubMed] [Google Scholar]
- Dörmann P, Benning C (1998) The role of UDP-glucose epimerase in carbohydrate metabolism of Arabidopsis. Plant J 13: 641–652 [DOI] [PubMed] [Google Scholar]
- Duan L, Dietrich D, Ng CH, Chan PM, Bhalerao R, Bennett MJ, Dinneny JR (2013) Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25: 324–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ, Mulligan JT, Ramer SW, Spottswood M, Davis RW (1991) Lambda YES: a multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia coli mutations. Proc Natl Acad Sci USA 88: 1731–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas I, Dombrádi V, Miskei M, Szabados L, Koncz C (2007) Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci 12: 169–176 [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59: 387–415 [DOI] [PubMed] [Google Scholar]
- Fisher RH, Barton MK, Cohen JD, Cooke TJ (1996) Hormonal studies of fass, an Arabidopsis mutant that is altered in organ elongation. Plant Physiol 110: 1109–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonslow BR, Niessen SM, Singh M, Wong CC, Xu T, Carvalho PC, Choi J, Park SK, Yates JR III (2012) Single-step inline hydroxyapatite enrichment facilitates identification and quantitation of phosphopeptides from mass-limited proteomes with MudPIT. J Proteome Res 11: 2697–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK (2011) Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc Natl Acad Sci USA 108: 1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu JK (2009) Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA 106: 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, et al. (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 50: 2123–2132 [DOI] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2006) Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA 103: 1988–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbers C, DeLong A, Deruére J, Bernasconi P, Söll D (1996) A mutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis. EMBO J 15: 2115–2124 [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, et al. (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhart J, van Weeren L, Hink MA, Vischer NO, Jalink K, Gadella TW Jr (2010) Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nat Methods 7: 137–139 [DOI] [PubMed] [Google Scholar]
- Hauser F, Waadt R, Schroeder JI (2011) Evolution of abscisic acid synthesis and signaling mechanisms. Curr Biol 21: R346–R355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes JG, Hartung AJ, Hendershot JD III, Passingham RS, Rundle SJ (1999) Molecular characterization of the B′ regulatory subunit gene family of Arabidopsis protein phosphatase 2A. Eur J Biochem 260: 127–136 [DOI] [PubMed] [Google Scholar]
- Hey SJ, Bacon A, Burnett E, Neill SJ (1997) Abscisic acid signal transduction in epidermal cells of Pisum sativum L. Argenteum: both dehydrin mRNA accumulation and stomatal responses require protein phosphorylation and dephosphorylation. Planta 202: 85–92 [Google Scholar]
- Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, et al. (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Zhu Y, Shen G, Zhang H (2014) TAP46 plays a positive role in the ABSCISIC ACID INSENSITIVE5-regulated gene expression in Arabidopsis. Plant Physiol 164: 721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K (2000) Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J 24: 655–665 [DOI] [PubMed] [Google Scholar]
- Imes D, Mumm P, Böhm J, Al-Rasheid KA, Marten I, Geiger D, Hedrich R (2013) Open stomata 1 (OST1) kinase controls R-type anion channel QUAC1 in Arabidopsis guard cells. Plant J 74: 372–382 [DOI] [PubMed] [Google Scholar]
- Jacob T, Ritchie S, Assmann SM, Gilroy S (1999) Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc Natl Acad Sci USA 96: 12192–12197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI (2010) Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Kunz HH, Bhattacharjee S, Hauser F, Park J, Engineer C, Liu A, Ha T, Parker JE, Gassmann W, et al. (2012) Natural variation in small molecule-induced TIR-NB-LRR signaling induces root growth arrest via EDS1- and PAD4-complexed R protein VICTR in Arabidopsis. Plant Cell 24: 5177–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik A, Ehrhardt DW, Kirik V (2012) TONNEAU2/FASS regulates the geometry of microtubule nucleation and cortical array organization in interphase Arabidopsis cells. Plant Cell 24: 1158–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T (2005) Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J 44: 939–949 [DOI] [PubMed] [Google Scholar]
- Kocourková D, Krcková Z, Pejchar P, Veselková S, Valentová O, Wimalasekera R, Scherer GF, Martinec J (2011) The phosphatidylcholine-hydrolysing phospholipase C NPC4 plays a role in response of Arabidopsis roots to salt stress. J Exp Bot XX: 3753–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Krebs M, Held K, Binder A, Hashimoto K, Den Herder G, Parniske M, Kudla J, Schumacher K (2012) FRET-based genetically encoded sensors allow high-resolution live cell imaging of Ca²⁺ dynamics. Plant J 69: 181–192 [DOI] [PubMed] [Google Scholar]
- Kremers GJ, Goedhart J, van Munster EB, Gadella TW Jr (2006) Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry 45: 6570–6580 [DOI] [PubMed] [Google Scholar]
- Kwak JM, Moon JH, Murata Y, Kuchitsu K, Leonhardt N, DeLong A, Schroeder JI (2002) Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14: 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PB, Cancel JD (2003) Enhanced ethylene responsiveness in the Arabidopsis eer1 mutant results from a loss-of-function mutation in the protein phosphatase 2A A regulatory subunit, RCN1. Plant J 34: 709–718 [DOI] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S (2009) A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA 106: 21419–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang XQ, Watson MB, Assmann SM (2000) Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science 287: 300–303 [DOI] [PubMed] [Google Scholar]
- Lillo C, Kataya AR, Heidari B, Creighton MT, Nemie-Feyissa D, Ginbot Z, Jonassen EM (2014) Protein phosphatases PP2A, PP4 and PP6: mediators and regulators in development and responses to environmental cues. Plant Cell Environ 37: 2631–2648 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Matre P, Meyer C, Lillo C (2009) Diversity in subcellular targeting of the PP2A B’eta subfamily members. Planta 230: 935–945 [DOI] [PubMed] [Google Scholar]
- McClinton RS, Sung ZR (1997) Organization of cortical microtubules at the plasma membrane in Arabidopsis. Planta 201: 252–260 [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, et al. (2009) A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merilo E, Laanemets K, Hu H, Xue S, Jakobson L, Tulva I, Gonzalez-Guzman M, Rodriguez PL, Schroeder JI, Broschè M, et al. (2013) PYR/RCAR receptors contribute to ozone-, reduced air humidity-, darkness-, and CO2-induced stomatal regulation. Plant Physiol 162: 1652–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Mustilli AC, Genty B, North H, Lefebvre V, Sotta B, Vavasseur A, Giraudat J (2002) Use of infrared thermal imaging to isolate Arabidopsis mutants defective in stomatal regulation. Plant J 30: 601–609 [DOI] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Moellering ER, Benning C (2011) Galactoglycerolipid metabolism under stress: a time for remodeling. Trends Plant Sci 16: 98–107 [DOI] [PubMed] [Google Scholar]
- Moellering ER, Muthan B, Benning C (2010) Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science 330: 226–228 [DOI] [PubMed] [Google Scholar]
- Mosquna A, Peterson FC, Park SY, Lozano-Juste J, Volkman BF, Cutler SR (2011) Potent and selective activation of abscisic acid receptors in vivo by mutational stabilization of their agonist-bound conformation. Proc Natl Acad Sci USA 108: 20838–20843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muday GK, Brady SR, Argueso C, Deruère J, Kieber JJ, DeLong A (2006) RCN1-regulated phosphatase activity and EIN2 modulate hypocotyl gravitropism by a mechanism that does not require ethylene signaling. Plant Physiol 141: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A (2002) A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20: 87–90 [DOI] [PubMed] [Google Scholar]
- Nagaya S, Kawamura K, Shinmyo A, Kato K (2010) The HSP terminator of Arabidopsis thaliana increases gene expression in plant cells. Plant Cell Physiol 51: 328–332 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Awai K, Masuda T, Yoshioka Y, Takamiya K, Ohta H (2005) A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J Biol Chem 280: 7469–7476 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, et al. (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50: 1345–1363 [DOI] [PubMed] [Google Scholar]
- Ng LM, Soon FF, Zhou XE, West GM, Kovach A, Suino-Powell KM, Chalmers MJ, Li J, Yong EL, Zhu JK, et al. (2011) Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases. Proc Natl Acad Sci USA 108: 21259–21264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, et al. (2010) PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J 61: 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris SR, Meyer SE, Callis J (1993) The intron of Arabidopsis thaliana polyubiquitin genes is conserved in location and is a quantitative determinant of chimeric gene expression. Plant Mol Biol 21: 895–906 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei ZM, Kuchitsu K, Ward JM, Schwarz M, Schroeder JI (1997) Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9: 409–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernas M, García-Casado G, Rojo E, Solano R, Sánchez-Serrano JJ (2007) A protein phosphatase 2A catalytic subunit is a negative regulator of abscisic acid signalling. Plant J 51: 763–778 [DOI] [PubMed] [Google Scholar]
- Peters C, Li M, Narasimhan R, Roth M, Welti R, Wang X (2010) Nonspecific phospholipase C NPC4 promotes responses to abscisic acid and tolerance to hyperosmotic stress in Arabidopsis. Plant Cell 22: 2642–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA perception and signalling. Trends Plant Sci 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Rashotte AM, DeLong A, Muday GK (2001) Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. Plant Cell 13: 1683–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösti J, Barton CJ, Albrecht S, Dupree P, Pauly M, Findlay K, Roberts K, Seifert GJ (2007) UDP-glucose 4-epimerase isoforms UGE2 and UGE4 cooperate in providing UDP-galactose for cell wall biosynthesis and growth of Arabidopsis thaliana. Plant Cell 19: 1565–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito N, Munemasa S, Nakamura Y, Shimoishi Y, Mori IC, Murata Y (2008) Roles of RCN1, regulatory A subunit of protein phosphatase 2A, in methyl jasmonate signaling and signal crosstalk between methyl jasmonate and abscisic acid. Plant Cell Physiol 49: 1396–1401 [DOI] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Márquez JA, Cutler SR, Rodriguez PL (2009) Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 60: 575–588 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Mori IC, Furuichi T, Munemasa S, Toyooka K, Matsuoka K, Murata Y, Yamamoto Y (2010) Closing plant stomata requires a homolog of an aluminum-activated malate transporter. Plant Cell Physiol 51: 354–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Sato Y, Fukao Y, Fujiwara M, Umezawa T, Shinozaki K, Hibi T, Taniguchi M, Miyake H, Goto DB, et al. (2009) Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem J 424: 439–448 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlücking K, Edel KH, Köster P, Drerup MM, Eckert C, Steinhorst L, Waadt R, Batistic O, Kudla J (2013) A new β-estradiol-inducible vector set that facilitates easy construction and efficient expression of transgenes reveals CBL3-dependent cytoplasm to tonoplast translocation of CIPK5. Mol Plant 6: 1814–1829 [DOI] [PubMed] [Google Scholar]
- Schmidt C, Schelle I, Liao YJ, Schroeder JI (1995) Strong regulation of slow anion channels and abscisic acid signaling in guard cells by phosphorylation and dephosphorylation events. Proc Natl Acad Sci USA 92: 9535–9539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz M, Schroeder JI (1998) Abscisic acid maintains S-type anion channel activity in ATP-depleted Vicia faba guard cells. FEBS Lett 428: 177–182 [DOI] [PubMed] [Google Scholar]
- Segonzac C, Macho AP, Sanmartín M, Ntoukakis V, Sánchez-Serrano JJ, Zipfel C (2014) Negative control of BAK1 by protein phosphatase 2A during plant innate immunity. EMBO J 33: 2069–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139: 468–484 [DOI] [PubMed] [Google Scholar]
- Shin R, Alvarez S, Burch AY, Jez JM, Schachtman DP (2007) Phosphoproteomic identification of targets of the Arabidopsis sucrose nonfermenting-like kinase SnRK2.8 reveals a connection to metabolic processes. Proc Natl Acad Sci USA 104: 6460–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C, Davanture M, Turk BE, Zivy M, Valot B, Leung J, Merlot S (2010) The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PLoS One 5: e13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C, Gu D, Hu HC, Davanture M, Lee S, Djaoui M, Valot B, Zivy M, Leung J, Merlot S, et al. (2009) Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett 583: 2982–2986 [DOI] [PubMed] [Google Scholar]
- Skottke KR, Yoon GM, Kieber JJ, DeLong A (2011) Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet 7: e1001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon FF, Ng LM, Zhou XE, West GM, Kovach A, Tan MH, Suino-Powell KM, He Y, Xu Y, Chalmers MJ, et al. (2012) Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335: 85–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey J, Isaac PG (1994) Isolation of DNA from plants. Methods Mol Biol 28: 9–15 [DOI] [PubMed] [Google Scholar]
- Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, Assaad FF, Christmann A, Grill E (2010) Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J 61: 25–35 [DOI] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR III (2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1: 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim TW, Zhou HW, Deng Z, Gampala SS, et al. (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerink C, Munnik T (2005) Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci 10: 368–375 [DOI] [PubMed] [Google Scholar]
- Thorlby G, Fourrier N, Warren G (2004) The SENSITIVE TO FREEZING2 gene, required for freezing tolerance in Arabidopsis thaliana, encodes a beta-glucosidase. Plant Cell 16: 2192–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Hou C, Ren Z, Pan Y, Jia J, Zhang H, Bai F, Zhang P, Zhu H, He Y, et al. (2015) A molecular pathway for CO₂ response in Arabidopsis guard cells. Nat Commun 6: 6057. [DOI] [PubMed] [Google Scholar]
- Tran HT, Nimick M, Uhrig RG, Templeton G, Morrice N, Gourlay R, DeLong A, Moorhead GB (2012) Arabidopsis thaliana histone deacetylase 14 (HDA14) is an α-tubulin deacetylase that associates with PP2A and enriches in the microtubule fraction with the putative histone acetyltransferase ELP3. Plant J 71: 263–272 [DOI] [PubMed] [Google Scholar]
- Trotta A, Wrzaczek M, Scharte J, Tikkanen M, Konert G, Rahikainen M, Holmström M, Hiltunen HM, Rips S, Sipari N, et al. (2011) Regulatory subunit B′gamma of protein phosphatase 2A prevents unnecessary defense reactions under low light in Arabidopsis. Plant Physiol 156: 1464–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng TS, Briggs WR (2010) The Arabidopsis rcn1-1 mutation impairs dephosphorylation of Phot2, resulting in enhanced blue light responses. Plant Cell 22: 392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, Shinozaki K (2013) Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signal 6: rs8. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K (2004) SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 17306–17311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T, Puzõrjova I, Brosché M, Valk E, Lepiku M, Moldau H, Pechter P, Wang YS, Lindgren O, Salojärvi J, et al. (2010) Ozone-triggered rapid stomatal response involves the production of reactive oxygen species, and is controlled by SLAC1 and OST1. Plant J 62: 442–453 [DOI] [PubMed] [Google Scholar]
- Vlad F, Droillard MJ, Valot B, Khafif M, Rodrigues A, Brault M, Zivy M, Rodriguez PL, Merlot S, Laurière C (2010) Phospho-site mapping, genetic and in planta activation studies reveal key aspects of the different phosphorylation mechanisms involved in activation of SnRK2s. Plant J 63: 778–790 [DOI] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S (2009) Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Turk BE, Peynot P, Leung J, Merlot S (2008) A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J 55: 104–117 [DOI] [PubMed] [Google Scholar]
- Waadt R, Hitomi K, Nishimura N, Hitomi C, Adams SR, Getzoff ED, Schroeder JI (2014a) FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. eLife 3: e01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Schlücking K, Schroeder JI, Kudla J (2014b) Protein fragment bimolecular fluorescence complementation analyses for the in vivo study of protein-protein interactions and cellular protein complex localizations. Methods Mol Biol 1062: 629–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang P, Xue L, Batelli G, Lee S, Hou YJ, Van Oosten MJ, Zhang H, Tao WA, Zhu JK (2013) Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc Natl Acad Sci USA 110: 11205–11210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G, McKown R, Marin AL, Teutonico R (1996) Isolation of mutations affecting the development of freezing tolerance in Arabidopsis thaliana (L.) Heynh. Plant Physiol 111: 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR III (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 19: 242–247 [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege S, De Angeli A, Droillard MJ, Kroniewicz L, Merlot S, Cornu D, Gambale F, Martinoia E, Barbier-Brygoo H, Thomine S, et al. (2014) Phosphorylation of the vacuolar anion exchanger AtCLCa is required for the stomatal response to abscisic acid. Sci Signal 7: ra65. [DOI] [PubMed] [Google Scholar]
- Wu G, Wang X, Li X, Kamiya Y, Otegui MS, Chory J (2011) Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci Signal 4: ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Ren R, Zhang YY, Pang Y, Yan C, Gong X, He Y, Li W, Miao D, Hao Q, et al. (2012) Molecular mechanism for inhibition of a critical component in the Arabidopsis thaliana abscisic acid signal transduction pathways, SnRK2.6, by protein phosphatase ABI1. J Biol Chem 287: 794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Wang Y, Williamson L, Holroyd GH, Tagliavia C, Murchie E, Theobald J, Knight MR, Davies WJ, Leyser HM, et al. (2006) The identification of genes involved in the stomatal response to reduced atmospheric relative humidity. Curr Biol 16: 882–887 [DOI] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH (2006a) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360 [DOI] [PubMed] [Google Scholar]
- Xu Y, Xing Y, Chen Y, Chao Y, Lin Z, Fan E, Yu JW, Strack S, Jeffrey PD, Shi Y (2006b) Structure of the protein phosphatase 2A holoenzyme. Cell 127: 1239–1251 [DOI] [PubMed] [Google Scholar]
- Xue S, Hu H, Ries A, Merilo E, Kollist H, Schroeder JI (2011) Central functions of bicarbonate in S-type anion channel activation and OST1 protein kinase in CO2 signal transduction in guard cell. EMBO J 30: 1645–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K (2002) ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol 43: 1473–1483 [DOI] [PubMed] [Google Scholar]
- Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K (2006) The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem 281: 5310–5318 [DOI] [PubMed] [Google Scholar]
- Yunta C, Martínez-Ripoll M, Zhu JK, Albert A (2011) The structure of Arabidopsis thaliana OST1 provides insights into the kinase regulation mechanism in response to osmotic stress. J Mol Biol 414: 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HW, Nussbaumer C, Chao Y, DeLong A (2004) Disparate roles for the regulatory A subunit isoforms in Arabidopsis protein phosphatase 2A. Plant Cell 16: 709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.