Sequences flanking the degron accelerate or decelerate the rate of auxin-induced degradation of repressor proteins.
Abstract
Ubiquitin-mediated protein degradation is a common feature in diverse plant cell signaling pathways; however, the factors that control the dynamics of regulated protein turnover are largely unknown. One of the best-characterized families of E3 ubiquitin ligases facilitates ubiquitination of auxin (aux)/indole-3-acetic acid (IAA) repressor proteins in the presence of auxin. Rates of auxin-induced degradation vary widely within the Aux/IAA family, and sequences outside of the characterized degron (the minimum region required for auxin-induced degradation) can accelerate or decelerate degradation. We have used synthetic auxin degradation assays in yeast (Saccharomyces cerevisiae) and in plants to characterize motifs flanking the degron that contribute to tuning the dynamics of Aux/IAA degradation. The presence of these rate motifs is conserved in phylogenetically distant members of the Arabidopsis (Arabidopsis thaliana) Aux/IAA family, as well as in their putative Brassica rapa orthologs. We found that rate motifs can act by enhancing interaction between repressors and the E3, but that this is not the only mechanism of action. Phenotypes of transgenic plants expressing a deletion in a rate motif in IAA28 resembled plants expressing degron mutations, underscoring the functional relevance of Aux/IAA degradation dynamics in regulating auxin responses.
Regulated turnover of diverse proteins by E3 ubiquitin ligases is critical for cell signaling in eukaryotes, yet we know remarkably little about what controls the rate of protein turnover. The ubiquitin system is among the most highly conserved across eukaryotes, and provides an intuitive platform for testing the links between the dynamics of signaling events and nearly every aspect of cell biology. The Arabidopsis (Arabidopsis thaliana) genome encodes nearly 700 predicted F-box proteins (Hua and Vierstra, 2011). TRANSPORT INHIBITOR RESPONSE1 (TIR1) is one of the best-characterized F-box proteins and acts as the receptor for the hormone auxin (for review, see Pierre-Jerome et al., 2013). In the presence of auxin, TIR1 acts within an S-Phase Kinase-Associated Protein1 (Skp1), Cullin, F-box complex (SCF)-type E3 ubiquitin ligase (SCFTIR1) that is responsible for ubiquitination and subsequent proteasome-mediated degradation of auxin (Aux)/indole-3-acetic-acid (IAA) transcriptional repressors (Gray et al., 2001; Ramos et al., 2001; Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005; Maraschin Fdos et al., 2009). Under low auxin conditions, Aux/IAA repressors restrict the activity of auxin response factor (ARF) transcription factors (Ulmasov et al., 1997; Tiwari et al., 2001, 2004; Overvoorde et al., 2005). When auxin levels increase, the hormone itself acts as molecular glue between the Aux/IAA and TIR1 (Tan et al., 2007). Efficient auxin binding requires both TIR1 and an Aux/IAA, leading to the conclusion that Aux/IAAs act as coreceptors (Calderón Villalobos et al., 2012).
The rate of auxin-induced degradation varies widely among members of the large multigene Aux/IAA family, and sequences outside of the degron (defined as the minimum region required for auxin-induced degradation) accelerate or decelerate degradation in a substrate-specific manner (Worley et al., 2000; Ramos et al., 2001; Dreher et al., 2006; Calderón Villalobos et al., 2012; Havens et al., 2012). A recapitulation of auxin response in yeast (Saccharomyces cerevisiae) demonstrated that Aux/IAA degradation rate is the major determinant of the rate of transcriptional activation by ARFs (Pierre-Jerome et al., 2014). As Aux/IAAs are among the strongest and earliest auxin response genes, they also contribute negative feedback within the system (for review, see Del Bianco and Kepinski, 2011). Additional layers of complexity include coexpression of several Aux/IAAs in tissue and development-specific contexts and cross regulation by other hormone signaling pathways (for review, see Pierre-Jerome et al., 2013). This complexity has made interrogation of dynamics within the auxin pathways challenging.
In this work, we utilized synthetic auxin degradation systems in yeast and in plants to identify and characterize two motifs outside the Aux/IAA degron that were required to maintain wild-type rates of degradation. The presence of these rate motifs were conserved in phylogenetically distant members of the Arabidopsis Aux/IAA family, as well as in Aux/IAAs from Brassica rapa. Whereas some of the sequences we identified as rate motifs could modulate the strength of interaction between the Aux/IAA repressor and the auxin receptor, other motifs had no effect on this interaction. This finding points to additional, currently unknown levels of control on degradation dynamics. Finally, we determined that Aux/IAA rate motifs are functionally relevant, as plants expressing an IAA28 variant with a deleted rate motif displayed similar developmental defects as plants expressing an IAA28 variant with a mutated degron sequence.
RESULTS
A Small Domain with Conserved Boundaries Can Recapitulate the Degradation Dynamics of Full-Length Aux/IAA Proteins
Aux/IAAs share several conserved domains critical for their function as transcriptional repressors (Fig. 1A; for review, see Pierre-Jerome et al., 2013). Domain I is required for interaction with the TOPLESS corepressor, whereas domains III and IV mediate interactions with ARF transcription factors and with other Aux/IAAs. Domain II contains the canonical degron and is necessary and sufficient to confer rapid auxin-induced degradation (Worley et al., 2000; Gray et al., 2001; Ramos et al., 2001; Zenser et al., 2001). To locate the critical regions within the Aux/IAA proteins required for tuning auxin-induced degradation rates, we first identified the minimal fragment required for wild-type degradation behavior.
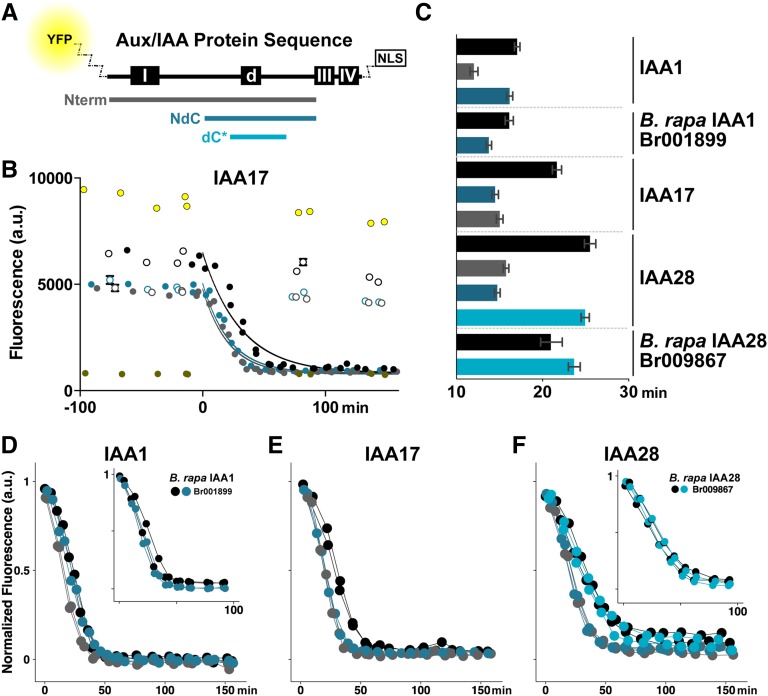
Figure 1.
Sequences flanking the degron are required to recapitulate degradation rates of full-length (FL) Aux/IAAs. A, Schematic of the domain structure of a FL Aux/IAA indicating the location of domain I (repression), the conserved degron (d), and domains III/IV (dimerization). Colored bars indicate the size and position of peptide fragments tested for degradation dynamics in relation to FL protein (black). All peptide fragments and FL proteins were expressed as fusions with an N-terminal fluorescent protein (YFP or VENUS) and a C-terminal 2xSV40 nuclear localization sequence (NLS). B, Raw IAA17 degradation as captured by time course fluorescence flow cytometry. Diploid yeast strains coexpressing TIR1 and YFP-IAA17 fragments were treated with 10 μm auxin (filled circles) or mock (white circles) at time = 0 min. Data from two independent replicates are plotted together, and each point represents 1,000 to 3,000 individual events recorded in the cytometer. Error bars representing the sem are shown for each point, but in most cases the marker size is larger than the error. Bright-yellow circles are data from cells expressing YFP with no protein fusion, whereas dark-yellow circles are data from cells expressing an untagged Aux/IAA. Data were fit to a one-phase decay nonlinear regression with a band representing the 95% confidence intervals across the fit. C, Half-lives for all fusion proteins presented in B and D to F. For all data, half-lives with error were calculated from nonlinear regression fits such as those as shown in B. Error bars represent 95% confidence intervals. D to F, Short peptides (either NdC or dC*) of Arabidopsis IAA1 (D), IAA17 (E), and IAA28 (F) are able to degrade at least as rapidly as their respective FL proteins as assessed by time lapse fluorescence flow cytometry. To facilitate comparison of degradation dynamics, mean fluorescence values were normalized to starting fluorescence. All data represent two independent experiments. NdC and dC* fragments of putative B. rapa orthologs of IAA1 and IAA28 (insets) similarly recapitulated FL degradation dynamics.
We selected a subset of Arabidopsis Aux/IAAs (IAA1, IAA3, IAA17, and IAA28) from diverse points within the Aux/IAA phylogeny (Overvoorde et al., 2005; Dreher et al., 2006) with a range of degradation rates (Dreher et al., 2006; Havens et al., 2012). We used a synthetic auxin signaling system in yeast (Havens et al., 2012) to characterize auxin-induced degradation dynamics of these Aux/IAAs. This system enables precise control of auxin input levels and can isolate the dynamics of user-defined Aux/IAA and TIR1/AUXIN SIGNALING F-BOX (AFB) pairs, parameters that complicate interpretation of similar assays in plants. Aux/IAA degradation rate as measured in the synthetic auxin system assays is an amalgamated parameter, integrating the rates of a number of distinct biochemical mechanisms, including multiple protein interactions, protein modification, and protein synthesis. Serial truncations of IAA1, IAA17, and IAA28 (Fig. 1A) were fused to yellow fluorescent protein (YFP) and an NLS. An NLS was included in all constructs to minimize potential differences in subcellular localization. Aux/IAA fragments or their FL counterparts were then coexpressed in yeast with the TIR1 auxin receptor, and Aux/IAA levels were measured by time lapse fluorescence flow cytometry (Fig. 1B). Degradation was quantified by calculating auxin-induced half-lives for each Aux/IAA fragment, and significance is indicated by nonoverlapping 95% confidence intervals (equivalent to P < 0.05; Fig. 1C). To easily compare degradation dynamics of several different fragments, fluorescence data were normalized as fold initial (Fig. 1, D–F).
Removal of the conserved domain III/IV to generate the amino terminus (Nterm) fragment (Fig. 1A, gray bar) did not slow auxin-induced Aux/IAA degradation when compared with FL proteins (Fig. 1, C–F). This is in agreement with previous observations that the N-terminal portion of Pisum sativum IAA6 (PSIAA6) and IAA17 contains the minimal region necessary to target a luciferase fusion for rapid degradation in plant assays (Worley et al., 2000; Ramos et al., 2001; Dreher et al., 2006). For IAA28, removal of domains III/IV exerted modestly accelerated degradation (Fig. 1, C and F), a finding we have observed before (Havens et al., 2012). This same trend was also observed for IAA17 (Fig. 1, C and E) and, more weakly, for IAA1 (Fig. 1, C and D). These results imply that dimerization or higher order complex formation mediated by domains III/IV negatively affects Aux/IAA degradation dynamics, but the magnitude of this effect is highly repressor dependent.
Further truncation analysis identified a smaller fragment (N-terminal-degron-C-terminal [NdC]; Fig. 1A, dark-blue bar) that fully recapitulated FL degradation dynamics in IAA1 (Fig. 1, C and D) as well as the closely related IAA3 (Supplemental Fig. S1). For IAA17 (Fig. 1E) and IAA28 (Fig. 1F), the NdC fragment degraded faster than FL (Fig. 1C). The NdC fragment lacks domain I, which had been shown previously to be dispensable for rapid PSIAA6 and IAA17 degradation (Worley et al., 2000; Dreher et al., 2006). In the case of IAA28, additional truncations were able to further reduce the fragment to 28 amino acids (degron-C-terminal-short [dC*]; Fig. 1A, bright-blue bar) that exhibited auxin-induced degradation dynamics equivalent to that of FL IAA28 (Fig. 1, C and F).
The previously characterized domains of Aux/IAAs are well conserved across all land plants (Lau et al., 2008; Prigge et al., 2010). We predicted that the degradation behavior of NdC or dC* fragments would be similarly conserved and could be isolated using truncations analogous to those applied to Arabidopsis Aux/IAAs. To test this theory, we focused on B. rapa, a crop species relative of Arabidopsis estimated to have diverged 14.5 to 20.4 million years ago (Bowers et al., 2003). We identified one putative IAA1 ortholog (Br001899) and two putative IAA28 orthologs (Br009867 and Br036557; Supplemental Fig. S1). When coexpressed in yeast with Arabidopsis TIR1, the FL B. rapa Aux/IAA orthologs exhibited auxin-induced degradation rates that were quite similar to those of their Arabidopsis counterparts (Supplemental Fig. S1), with the exception of one of the B. rapa IAA28 orthologs (Br036557) where the degradation half-life was faster but turnover was not as complete. NdC fragments (Supplemental Fig. S1) were then compared with FL proteins. For the IAA1 ortholog (Br001899), the FL and NdC fragment had nearly identical auxin-induced degradation profiles (Fig. 1, C and D inset). The dC* fragment of one of the putative BrIAA28 orthologs (Br009867) similarly recapitulated its respective FL degradation profile (Fig. 1, C and F inset). The dC* fragment of the second IAA28 ortholog (Br036557) degraded more slowly and completely than FL (Supplemental Fig. S1). Thus, at least within the Brassicaceae, the region between domains I and III encodes the critical information for setting the rate of auxin-induced Aux/IAA turnover.
The KR Motif N-terminal of the Degron Accelerates Auxin-Induced Degradation Rate
Further truncations were next used to determine specific residues controlling degradation dynamics within the NdC and dC* fragments (Fig. 2A; Supplemental Fig. S2). For IAA1 (Fig. 2B), removal of the sequences N-terminal of the degron (dC, light blue) strongly slowed degradation dynamics but maintained degradation rates higher than the degron alone (d, green). Removing N-terminal residues had mild effects on IAA3 (Supplemental Fig. S2) and IAA17 (Fig. 2C) degradation dynamics, but no effect on IAA28 (Fig. 2D). We predicted that Aux/IAAs not strongly affected by removal of N-terminal regions might be more strongly influenced by removal of C-terminal regions. To assess this, we generated Nd fragments of each Aux/IAA (Fig. 2A, orange). As expected, IAA28.Nd degradation dynamics slowed to the level of the degron alone (Fig. 2D; Supplemental Fig. S2). Similar removal of C-terminal residues had moderate effects on IAA3 (Supplemental Fig. S2) and IAA17 (Fig. 2C), but only a weak effect on IAA1 (Fig. 2B).
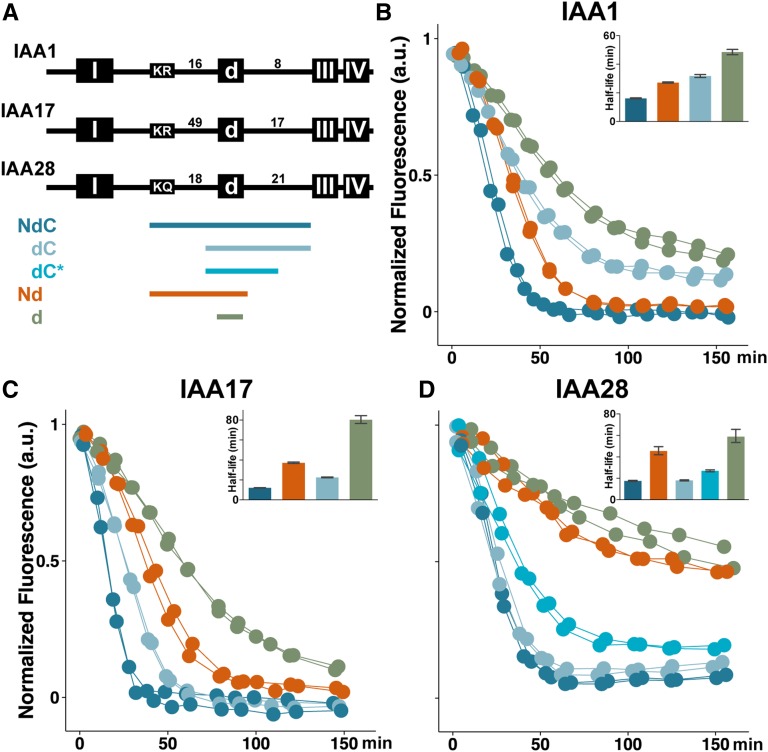
Figure 2.
The impact of the N- and C-terminal rate motifs on degradation dynamics is Aux/IAA dependent. A, Colored bars indicate the size and position of Aux/IAA peptide fragments tested for degradation dynamics; colors are used consistently in all sections of this figure and in all following figures. Numbers indicate how many amino acids are found between conserved domains. B to D, Regions N- and C-terminal of the degron influence IAA1 (B) and IAA17 (C) degradation dynamics, whereas only C-terminal regions of the IAA28 (D) degron impact degradation. Auxin-induced degradation in the presence of TIR1 was captured as before; half-lives (inset) are presented with 95% confidence intervals.
To ensure that other factors were not confounding these degradation rate comparisons, we performed a number of controls. We found that all YFP-IAA1-2xSV40 fusion proteins localized to the nucleus (Supplemental Fig. S2), and that there was no correlation between auxin-induced degradation half-life and basal YFP-IAA expression level (Supplemental Fig. S2). Moreover, YFP-degron linker length did not contribute to degradation speed (Supplemental Fig. S2).
The NdC and Nd fragments are flanked at the N terminus by a conserved KR motif (Abel et al., 1994) previously shown to be important for stability of IAA17 under endogenous, but not exogenous, auxin conditions in whole seedlings (Dreher et al., 2006). Although the KR residues are highly conserved among many of the Arabidopsis Aux/IAAs, the number and identity of the residues between the KR and the degron are highly variable (Fig. 2A). To test whether the KR residues themselves may be acting as an N-terminal rate motif, we mutated the KR within the NdC fragments (Fig. 3, A–C and G). As expected for IAA17 (Dreher et al., 2006), Ala substitution of the KR resulted in slowed auxin-induced degradation similar to that observed when the KR was completely removed in the dC fragment (Fig. 3A). This effect was even more pronounced in IAA1 (Fig. 3B). Substitutions of the K or R alone or both residues together slowed IAA1.NdC degradation to the equivalent degree (Fig. 3B), and the degradation half-lives were identical to deletion of the entire N-terminal sequence in the dC fragment (Fig. 3G). Mutating the KR in the context of the entire N-terminal half of IAA1 or IAA17 similarly slowed the rate of auxin-induced degradation (Supplemental Fig. S3). These KR alterations had no obvious effect on subcellular localization of IAA1.NdC (Supplemental Fig. S2).
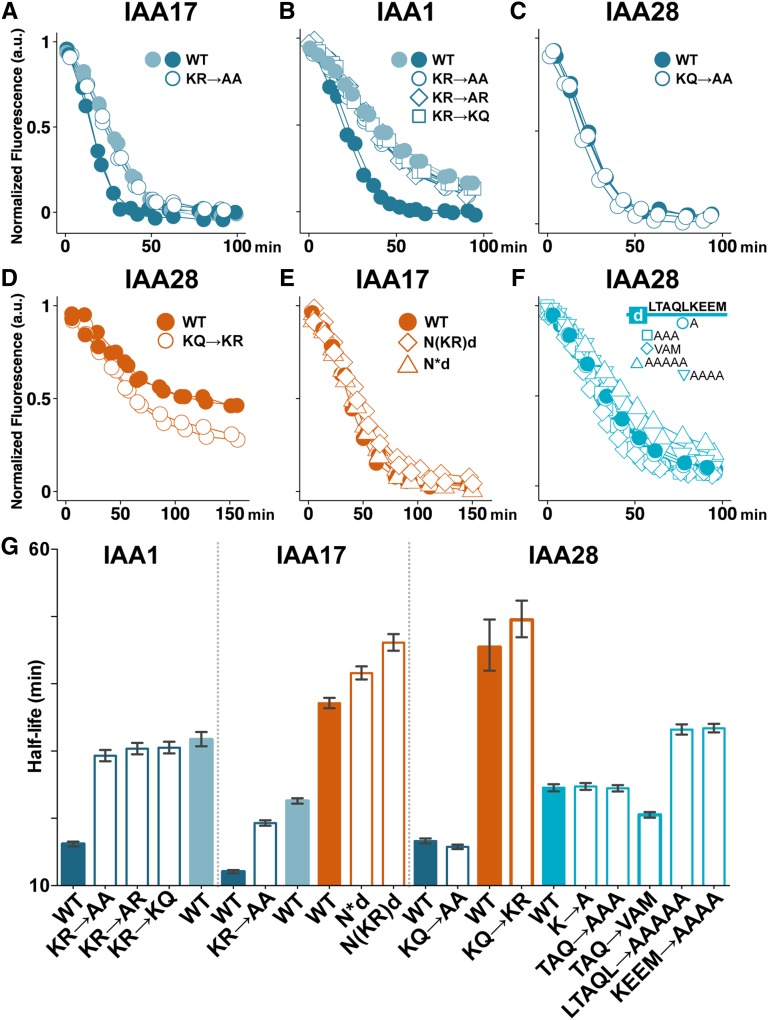
Figure 3.
Specific residues in the N terminus contribute to degradation rate, whereas rate determinants in the C terminus are not necessarily sequence specific. A to C, Ala substitution (white circles) of the KR rate motif residues within the NdC fragment (dark-blue) in IAA17 (A) and IAA1 (B) slowed degradation similar to the dC fragment (light-blue), whereas Ala substitution of the KQ residues of IAA28 (C) had no effect on degradation dynamics. D, Replacing the IAA28 KQ with KR (white circles) did not affect half-life but promoted more complete degradation of the Nd fragment (orange). E, Shortening the distance between the IAA17 degron and the KR rate motif by either moving the KR forward 25 residues (diamonds) or deleting 25 residues (triangles) did not lead to faster degradation of IAA17.Nd. F, The C-terminal rate motif of IAA28 displayed no obvious sequence specificity. Ala substitutions and polar→nonpolar mutations (white symbols) of the nine residues immediately terminal of the degron in the IAA28.dC* fragment (bright-blue) had weak effects on degradation. G, Half-lives with 95% confidence intervals for data in A to F. WT, Wild type.
IAA28 belongs to a small subfamily of Aux/IAAs that did not retain the KR motif (Dreher et al., 2006). Instead, IAA28 has a KQ that can be aligned in a similar position. Given that the N-terminal region of the degron was dispensable to retain rapid IAA28 degradation (Fig. 2D, dC and dC*), and that the R residue in the IAA1 KR motif was critical to the function of the KR rate motif (Fig. 3B), we hypothesized that IAA28 was a naturally occurring loss-of-function variant. Consistent with this hypothesis, Ala substitution of the KQ had no effect on degradation dynamics (Fig. 3, C and G; Supplemental Fig. S3). We then predicted that replacement of the divergent KQ with KR would restore N-terminal rate motif activity. When the KQ→KR substitution was generated in the IAA28.Nd fragment, we observed no significant difference in degradation half-lives, although the substitution did result in a reproducible enhancement of the degree of turnover compared with wild-type IAA28.Nd (Fig. 3D).
The intervening sequence between the KR and the degron (Fig. 2A) is much longer in IAA17 (49 amino acids) than in IAA1 and IAA3 (16 and 25 amino acids, respectively). Additionally, mutating the KR had a more modest effect on IAA17 degradation when compared with the effect of these mutations on IAA1 (Fig. 3, A, B, and G). We hypothesized that the distance between the KR and the degron might impact its function. To test this theory, we altered the KR-to-degron distance in the slowly degrading IAA17.Nd fragment to examine whether this would accelerate degradation. We moved the KR motif closer by either removing one-half of the intervening amino acids (IAA17.N*d) or shifting the KR into the middle of the intervening sequence [IAA17.N(KR)d]. Neither of these changes accelerated auxin-induced degradation (Fig. 3E); instead, both resulted in slightly longer half-lives compared with the IAA17.Nd wild type (Fig. 3G). Similar alterations of KR-to-degron distance in IAA3 had no effect or again slightly slowed IAA3.Nd degradation (Supplemental Fig. S3). Thus, distance alone did not explain the differential impact of the KR rate motif on degradation rate among Aux/IAAs.
The Residues Immediately C-Terminal of the Degron Accelerate Auxin-Induced Degradation Rate
We next looked at how residues C-terminal of the degron were influencing degradation dynamics. For all IAAs tested in our studies, removal of these residues significantly impacted degradation dynamics (Nd versus NdC; Fig. 2B–D; Supplemental Fig. S2). This effect was greatest for IAA28, so we focused our studies on the IAA28 C-terminal residues for further analysis. Comparison of auxin-induced degradation of IAA28.dC and IAA28.dC* revealed that removing the last 11 C-terminal residues only modestly slowed degradation (Fig. 2D). This suggested that the remaining 10 residues immediately C-terminal of the degron (LTAQLKEEM) were most important. These residues had little sequence conservation across the Aux/IAAs, apart from enrichment for polar residues (Supplemental Fig. S2), making it difficult to identify potential motifs. Ala substitution of the single K (a possible ubiquitination target) in IAA28.dC* had no effect on degradation (Fig. 3, F and G). This result is consistent with recent data demonstrating that Lys residues are not required for ubiquitination of IAA1 (Gilkerson et al., 2015). Expanded Ala substitutions within the LTAQL or KEEM residues had no effect or modestly slowed degradation (Fig. 3F, G). Conversion of the two polar residues in this region to similar nonpolar residues (TAQ→VAM) slightly accelerated degradation (Fig. 3, F and G). Thus, we did not identify specific amino acids within this region that were critical for its function in determining degradation dynamics. However, among the Aux/IAAs, there is variation in the number of residues between the degron and domains III/IV. In our small sampling, the longer the C-terminal domain was, the stronger the influence on degradation dynamics was (IAA1 < IAA3 = IAA17 < IAA28). We hypothesized that this trend may indicate a structural or linker role for this region in facilitating protein-protein interactions. To address this, we replaced the C-terminal rate motif of IAA28.dC with a flexible linker of identical length (IAA28.dC-Flex) and assessed whether turnover dynamics were maintained. Our data instead revealed that IAA28.dC-Flex behaved like the degron alone (Supplemental Fig. S3), exhibiting much slower turnover than IAA28.dC. Thus, the residues within the C-terminal rate motif seem likely to act as more than a simple flexible linker between the degron and domains III/IV, but the rules of their activity remain to be elucidated.
Rate Motifs Tune Aux/IAA Degradation Rate in Plant Roots
The KR rate motif was previously known to affect Aux/IAA protein stability in plants (Dreher et al., 2006); however, it was unclear whether the C-terminal rate motif would have the same importance in plant cells as it did in yeast. To test this, we generated transgenic Arabidopsis lines expressing heat shock-inducible fusions of IAA28 fragments with the bright YFP variant VENUS (Guseman et al., 2015). Following heat shock, seedlings were treated with auxin, and turnover of VENUS-IAA28 fragments was measured in root tips (Fig. 4A). As predicted by the yeast experiments, the IAA28.Nd fragment lacking the C-terminal rate motif degraded more slowly than the IAA28.dC fragment (Fig. 4B; Supplemental Fig. S4). The IAA28.dC* fragment lacking part of the C-terminal domain exhibited slightly slower degradation than IAA28.dC, as was observed in yeast. VENUS-only control plant lines showed very little change in fluorescence in the presence of auxin. These findings are consistent with our previous work showing that domain II-VENUS (equivalent to our IAA28.Nd) has significantly higher steady-state levels than IAA28.dC when expressed constitutively from the 35S promoter (Havens et al., 2012).
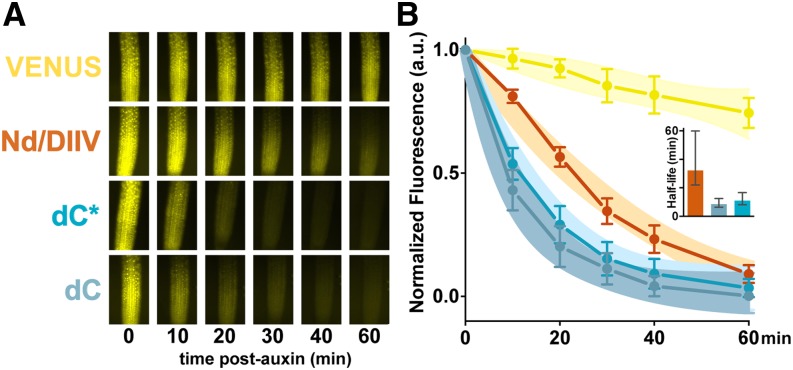
Figure 4.
Aux/IAA rate motifs are functional in plant roots. The C-terminal rate motif predictably influenced IAA28 degradation dynamics in plants. A, When expressed in Arabidopsis roots, an IAA28.Nd fragment (orange) lacking the C-terminal rate motif degrades more slowly in the presence of exogenous auxin than IAA28 fragments with a full (dC, light-blue) or partial (dC*, bright-blue) C-terminal rate motif. Root tips of 7-d-old Arabidopsis seedlings expressing heat shock-induced VENUS-tagged IAA28 reporters or VENUS alone (yellow) were treated with 5 μm auxin and imaged periodically for 1 h to capture degradation profiles. B, Quantification of fluorescence microscopy data from two replicates. Between one and five individuals representing two to four independent insertions were analyzed for each genotype. Mean fluorescence intensity values ±sem are plotted. Data were fit by one-phase decay nonlinear regression, and 95% confidence bands for the fit are shown. Auxin-induced degradation half-lives were calculated through nonlinear regression, and means were plotted with 95% confidence intervals (inset).
The KR Rate Motif Does Not Alter IAA Affinity for the TIR1 F-Box Protein
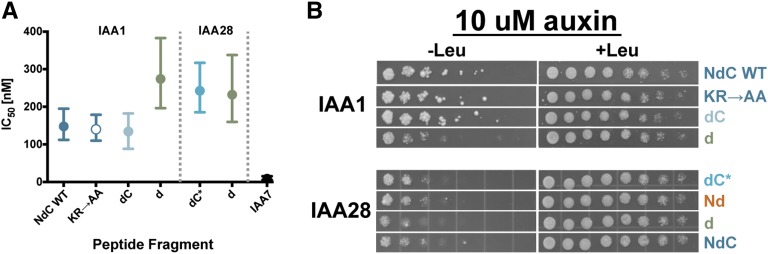
Enhancing TIR1 affinity for Aux/IAAs can accelerate Aux/IAA degradation rate (Yu et al., 2013). To test whether rate motifs act by influencing Aux/IAA binding to TIR1, we used two measures of interaction strength: in vitro competition binding experiments and yeast two-hybrid assays. Data from both assays indicated that the KR rate motif does not influence Aux/IAA affinity for TIR1 (Fig. 5; Supplemental Fig. S5). The IC50 values for IAA1.NdC, IAA1.NdC-KR→AA, and IAA1.dC were all within 100 to 200 nm (Fig. 5A). This indicates that IAA1 peptides with a mutated KR or peptides lacking the KR entirely were equally efficient binding competitors as wild-type IAA1.NdC. Yeast two-hybrid data further corroborated this finding, with the same IAA1 fragments showing no differences in interaction strength with TIR1 (Fig. 5B; Supplemental Fig. S5). The IAA1.d, missing both the N- and C-terminal rate motifs, interacted more weakly with TIR1 than the wild-type or mutant IAA1.NdC fragments in both assays (Fig. 5, A and B). Thus, the C-terminal rate motif of IAA1 influenced degradation at least in part by altering interaction strength with TIR1.
Figure 5.
Binding affinity between auxin repressor and receptor is not always correlated with degradation rate. Aux/IAAs with wild-type, mutant, or deleted rate motifs show no differences in ability to interact with TIR1. A, Aux/IAA peptide competition binding assays were used to assess strength of interaction with TIR1. Inhibitory concentration 50% (IC50) values were measured by an AlphaScreen assay using purified TIR1 and IAA7.Nterm conjugated to donor and acceptor beads, respectively. Data shown represent one experiment with n = 6 and are typical of results obtained in additional experiments. B, Interaction between TIR1 and indicated Aux/IAA peptides was assessed by yeast two-hybrid assay in the presence of 10 μm auxin. WT, Wild type.
The IAA28.dC* peptide fragment that contains part of the C-terminal rate motif has a similar IC50 value as IAA28.d (Fig. 5A), despite large differences in degradation rates (Fig. 2D). Yeast two-hybrid analysis demonstrated that IAA28.dC* and IAA28.d showed similar interaction strength with TIR1, with the degron alone exhibiting only a slightly weaker interaction (Fig. 5B; Supplemental Fig. S5). Auxin binding assays in the presence of TIR1 indicate that IAA28.dC* has a larger dissociation constant than IAA1.NdC (Supplemental Fig. S5; Supplemental Protocol S1), consistent with the peptide competition assays (Fig. 5A). IAA28 fragments either completely lacking the C-terminal rate motif (Nd) or with only a partial motif (dC*) showed similar interactions with TIR1 when compared with fragments with an intact C-terminal rate motif (NdC; Fig. 5B). These results indicated that the C-terminal rate motif of IAA28, unlike the equivalent region of IAA1, did not influence interaction with TIR1. Furthermore, these experiments revealed that receptor:substrate interaction strength can be decoupled from substrate degradation rate.
Degradation Rate Is Critical for Aux/IAA Function in Plants
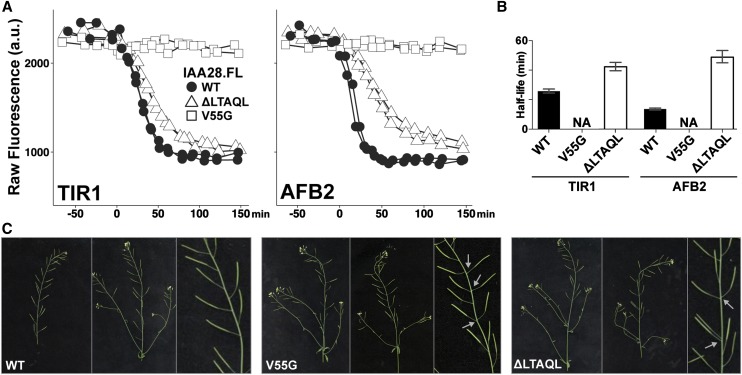
We next investigated the impact of rate motifs on Aux/IAA function, focusing on the C-terminal rate motif of IAA28. As with other Aux/IAAs, mutations within the IAA28 degron stabilize the resulting protein and lead to a range of auxin-resistant phenotypes (Ramos et al., 2001; Rogg et al., 2001). For iaa28-1, these phenotypes include defective lateral roots, diminished adult size, and loss of apical dominance (Rogg et al., 2001). When assayed with either TIR1 or AFB2 in yeast, a mutation in the degron of a FL IAA28 (V55G) completely abolished auxin-induced degradation (Fig. 6, A and B). In contrast, removing part of the C-terminal rate motif (ΔLTAQL) resulted in only a partial slowdown of IAA28 degradation (Fig. 6, A and B).
Figure 6.
Rate motif deletions slow degradation of FL IAA28 and result in phenotypes similar to stabilized IAA28 when expressed in plants. A, Degradation profiles of IAA28 variants were measured in yeast expressing either TIR1 or AFB2 auxin receptors. Partial deletion of the IAA28 C-terminal rate motif (ΔLTAQL, white triangles) in the context of FL IAA28 substantially slowed degradation when compared with the wild-type protein (black circles). As expected, a mutation in the degron (V55G, white squares) completely abolished auxin-induced degradation. Raw mean fluorescence values from two independent experiments are shown. B, Half-lives with 95% confidence intervals for data in A. Half-lives for IAA28.FL-V55G were not applicable (NA) as no auxin-induced degradation was observed. C, Plants expressing IAA28.FL-ΔLTAQL or IAA28.FL-V55G from the IAA28 promoter showed a range of phenotypes, including disruptions in phyllotaxy (gray arrows) and reduced fertility, when compared with plants expressing wild-type (WT) IAA28. Phenotypes of plants expressing IAA28.FL-V55G were more severe than those of plants expressing IAA28.FL-ΔLTAQL, consistent with a stronger effect of the respective mutations on auxin-induced degradation.
We then generated transgenic plants expressing wild-type, V55G, and ΔLTAQL versions of FL IAA28 from the native IAA28 promoter (Rogg et al., 2001). When compared with plants expressing wild-type IAA28, plants expressing IAA28-V55G or IAA28-ΔLTAQL exhibited a range of phenotypes (Fig. 6C), including disruptions in phyllotaxy (gray arrows) and reduced fertility. Overall, the transgenic plants displayed mild phenotypes. This is consistent with previous reports that expression of iaa28-1 from its native promoter confers milder phenotypes in the Columbia-0 background than what is observed in transgenics made in Wassilewskija, the background of the original mutant (Rogg et al., 2001). Phenotypes of plants expressing IAA28-V55G were more severe than those of plants expressing IAA28-ΔLTAQL, consistent with a stronger effect of the respective mutations on auxin-induced degradation rates in yeast. These results confirm the regulatory role that Aux/IAA rate motifs play in auxin responses. Moreover, the observed aberration in phyllotactic pattern caused by a slowed turnover of IAA28, likely reflecting an altered rate of primordia production at the shoot apex, supports the recently proposed model that Aux/IAAs are auxin-initiated timers that synchronize developmental processes (Guseman et al., 2015).
DISCUSSION
Induced turnover of repressors is an effective and sensitive determinant of signaling dynamics. In this work, we used a synthetic approach to identify and characterize multiple motifs that tune degradation dynamics of the Aux/IAA repressor proteins in Arabidopsis and B. rapa. This work supports broader use of synthetic systems for rapid interrogation of hormone signaling dynamics, as it both successfully resulted in de novo identification of the KR motif, previously uncovered through highly intensive and complex assays in seedlings (Dreher et al., 2006), as well as identified, to our knowledge, new regions previously not associated with auxin-induced degradation. In addition, we discovered that although some rate motifs enhanced substrate interaction with receptor-auxin complexes, this was not universally true. This decoupling of Aux/IAA turnover dynamics from the hormone-induced interaction of Aux/IAAs and TIR1 points to currently unknown regulatory factors, a search for which could be greatly aided by a combination of synthetic and in-plant assays.
The conserved KR motif had been previously shown to influence IAA17 turnover in Arabidopsis seedlings (Dreher et al., 2006). Here, we show that the KR acts as an auxin-responsive rate motif in several Aux/IAA proteins (Fig. 3, A and B), and that the magnitude of this effect was correlated with the proximity of the KR rate motif to the degron (Fig. 2A; Supplemental Fig. S2). However, moving the KR sequence closer to the degron was not sufficient to accelerate degradation rate (Fig. 3E; Supplemental Fig. S3). IAA28 is a natural KR variant. It is part of a divergent clade, including IAA18 and IAA26, all of which have a KQ or KK rather than a KR (Rogg et al., 2001; Dreher et al., 2006). Although the KQ was dispensable for maintenance of wild-type IAA28 degradation dynamics, replacement of the KQ with KR enhanced the degree of IAA28 turnover (Fig. 3D). It remains to be seen whether the auxin-induced degradation rates of other members of this divergent clade are similarly insensitive to deletions N-terminal to the degron.
The second rate motif identified was immediately C-terminal of the Aux/IAA degron (Fig. 2B–D; Supplemental Fig. S2). As shown previously for the KR motif (Dreher et al., 2006), we found that the C-terminal motif was important for degradation rate in plants as well as yeast (Fig. 4). The sequence of the C-terminal region is highly variable across the Aux/IAA family, and the influence of this motif on TIR1 interaction was Aux/IAA dependent (Fig. 5). Although the C-terminal motif enhanced affinity of IAA1 for TIR1 in the presence of auxin, it had no effect on complex affinity for IAA28. Because the C-terminal region is not included in the TIR1-auxin-Aux/IAA structure (Tan et al., 2007), additional structural studies may be needed to elucidate the function of this motif.
Binding experiments indicated that the KR rate motif does not influence Aux/IAA affinity for TIR1 (Fig. 5), strongly suggesting an additional unknown mechanism for increasing efficiency of SCFTIR1/AFB substrate ubiquitination. Several recent reports suggest attractive models for this effect, including enhancing interaction with E2 ubiquitin-conjugating enzymes (for review, see Chang and Barford, 2014). Several substrates of the anaphase-promoting complex/cyclosome-type ubiquitin ligase were shown to possess initiation motifs containing positively charged residues that work in conjunction with a specific E2 enzyme to promote efficient ubiquitin chain initiation and substrate degradation (Williamson et al., 2011). As E2 enzymes involved in the auxin signaling pathway are identified (Wen et al., 2014), it will be interesting to assess whether the Aux/IAA rate motifs may be acting as initiation motifs, or if they are mediating interactions with other components of the ubiquitin-proteasome system (for review, see Komander and Rape, 2012; Callis, 2014).
The highly conserved domains III/IV in the C-terminal half of Aux/IAAs and ARFs have recently been identified as a phagocyte oxidase/bud emergence protein (PB1) domain (Korasick et al., 2014; Nanao et al., 2014; Dinesh et al., 2015; Han et al., 2015; Korasick et al., 2015). These studies have demonstrated that isolated ARF and Aux/IAA PB1 domains are capable of mediating the formation of higher order complexes. We observed that removal of the PB1 domain in several different Aux/IAAs resulted in varying degrees of accelerated auxin-induced turnover (Fig. 1), suggesting that IAA-IAA interactions mediated through the PB1 might negatively influence Aux/IAA turnover.
Unraveling the mechanisms guiding Aux/IAA degradation dynamics may provide deeper insight into the evolution of dynamic behaviors within this pathway. Changing the rate motif would allow for tweaking transcriptional dynamics in an auxin signaling circuit without altering the ability of the Aux/IAA to interact with either the signal (auxin) or the receptor. The Aux/IAA family has undergone large expansion in flowering plants, and Arabidopsis Aux/IAAs possess a diverse range of degradation rates (Dreher et al., 2006; Havens et al., 2012). Differential degradation rates among Aux/IAAs could enable exploration of a range of transcriptional dynamics (Pierre-Jerome et al., 2014) and contribute to complex behaviors, such as the transcriptional oscillations that are observed in lateral roots (De Smet, 2012). A recent study of engineered IAA14 rate variants provided a strong link between Aux/IAA degradation rate with the rate of organ initiation (Guseman et al., 2015). Our results with IAA28 rate variants and phyllotactic pattern further support a direct connection between the rate of cell level auxin response and coordination of carefully timed, larger scale developmental events.
MATERIALS AND METHODS
Yeast Methods
Yeast (Saccharomyces cerevisiae) transformations were performed using a standard lithium acetate protocol (Gietz and Woods, 2002) into mating type locus a (Mata) W303-1A or mating type locus α (Matα) W814-29B (gifts from the Gottschling Laboratory). Yeast peptone dextrose (YPD) and synthetic complete (SC) medium were made according to standard protocols and supplemented with 80 mg mL−1 adenine. MATa and MATα strains were mated by coinoculating strains at low density into YPD medium, growing overnight at 30°C, streaking out to single colonies on SC-Leu-Trp to select for diploids, and isostreaking on YPD plates before glycerol stocking.
Plasmid Construction
FL Aux/IAAs and truncations were fused to an N-terminal enhanced YFP and C-terminal SV40 NLS as previously described (Havens et al., 2012). Truncated Aux/IAAs were synthesized (www.idtdna.com) with partial enhanced YFP and NLS and then cloned into the pGP4GY vector backbone via Gibson assembly (Gibson et al., 2009). Yeast two-hybrid vectors were generated by Gateway cloning. TIR1 was cloned into the pGILDA-GW backbone and Aux/IAAs into pB42-AD-GW (vectors provided by Jianping Hu, Michigan State University).
Putative Brassica rapa orthologs of IAA1 were identified by searching the corresponding Arabidopsis (Arabidopsis thaliana) gene locus (AT4G14560) in the Brassica Database (BRAD; http://brassicadb.org/brad/; Cheng et al., 2011). The amino acid sequences of Arabidopsis Aux/IAAs and putative B. rapa orthologs were then compared using the BRAD BLAST. No IAA1 matches were found using the BRAD ortholog search for AtIAA1. However, two putative IAA1 orthologs were identified by BLAST scores: Br039732 and Br001899. We abandoned Br039732 because it is syntenic to IAA14 and expressed poorly in yeast. We pursued study of Br001899 because it is a nonsyntenic ortholog of AtIAA1 and has synteny with the closely related AtIAA2. The two putative B. rapa IAA28 orthologs (Br009867 and Br036557) matched three BRAD selection criteria: both the ortholog and synteny search features, and top BLAST alignment score.
Heat shock (HS)::VENUS-IAA-NLS plant expression vectors were constructed by fusing Aux/IAA truncations to an N-terminal VENUS and C-terminal SV40 NLS repeat using Gly-Ala linkers (GAGAGAGAGAGP and GAGA, respectively; Nishimura et al., 2009) via Gibson cloning (Gibson et al., 2009). The resulting fragments were restriction cloned into pGREEN vectors containing the soybean (Glycine max) heat shock promoter HS6871 (Gray et al., 2001).
Yeast YFP-IAA Degradation Assays
Yeast degradation assays were carried out as previously described (Havens et al., 2012). In brief, yeast strains coexpressing stably-integrated TIR1 variants and YFP-tagged Aux/IAA proteins were prepared by transferring a freshly grown colony from YPD plates into SC medium. Flow cytometry was used to estimate the cell density (in events μL−1) and dilute cells such that cultures were in log phase 16 h later and for the duration of the experiment. All cultures were grown at 30°C with shaking. Preauxin measurements were taken to ascertain baseline expression, followed by addition of auxin (10 μm indole-3-acetic acid) or mock treatment (95% [v/v] ethanol). Measurements were acquired over the course of 120 to 150 min following auxin treatment, with intervals ranging from 10 min early in the YFP-IAA degradation phase to 20 min later in the degradation phase. Controls were measured every hour for the duration of the experiment. At least two independent replicates were performed and plotted for each assay. All data were normalized by subtracting background autofluorescence and normalizing to preauxin fluorescence levels for each strain. Nonlinear regression analysis (plateau followed by one-phase decay) was performed in Prism6 (GraphPad) using raw data to calculate degradation half-lives and basal fluorescence levels. We also confirmed that all YFP-IAA1-2xSV40 fusion proteins localized to the nucleus (Supplemental Fig. S2) and that there was no correlation between auxin-induced degradation half-life and basal YFP-IAA expression level (Supplemental Fig. S2).
Generation of Transgenic Plants
A 3-kb IAA28 promoter fragment (Rogg et al., 2001) was cloned into pGREEN vectors, and IAA28 variant coding DNA sequences were cloned downstream. HS::VENUS-IAA-NLS constructs were generated by fusing IAA28 coding DNA sequences to an N-terminal VENUS and C-terminal SV40 NLS repeat using Gly-Ala linkers via Gibson cloning. The resulting fragments were cloned into pGREEN vectors containing the soybean heat shock promoter HS6871 (Gray et al., 2001).
Columbia ecotype Arabidopsis plants were transformed with HS::VENUS-IAA-NLS or pIAA28::IAA28.FL variant plant expression vectors using the floral dip method (Clough and Bent, 1998). Plants were selected on 0.5× Linsmaier-Skoog (LS) agar plates containing 50 mg/mL kanamycin. Two or three independent T2 lines for each construct were used for heat shock degradation assays. Three independent T2 lines were grown on selection for 8 d before transfer to soil to examine adult phenotypes.
Heat Shock Induction and VENUS-IAA Degradation Assays
Seedlings were stratified at 4°C for 2 d on plates containing 0.5× LS and 0.8% (w/v) agar, and then grown in 16-h-light:8-h-dark conditions for 7 d. Plates were placed on a slide warmer set at 37°C for 2 h to induce expression. Seedlings were then arranged on agar blocks containing either 5 µM IAA or mock treatment and sprayed with liquid 0.5× LS containing either 5 µM IAA or vehicle. A coverslip was placed over the agar blocks, and plants were imaged at 0, 10, 20, 30, 40, and 60 min posttreatment.
Plants were imaged using a Leica DMI 3000B microscope fitted with a Leica long-working 40x HCX PL FLUORSTAR objective and illuminated with a Lumencor SOLA light source. Images were captured using Leica LAS AF version 2.6.0 software and a Leica DFC 345FX camera. Fiji software was used to quantify fluorescence in a region of interest centered on each image. Nonfluorescent siblings were used to calculate background levels. Fluorescence for each plant was analyzed by subtracting the background level of fluorescence and then normalizing to the initial time point. Half-lives were calculated in Prism6 (GraphPad) using one-phase exponential decay fit.
Aux/IAA Competition Binding Assays
Arabidopsis IAA7(1-125) DNA was amplified by PCR with primers designed to incorporate the Flag epitope at the Nterm and was then ligated into a pET15b vector containing an N-terminal hexa-His tag. His-Flag-IAA7 was purified from BL21(DE3 [Novagen]) cells by nickel affinity chromatography. Expression and purification of FL Arabidopsis TIR1 in complex with the FL Arabidopsis SKP1-LIKE1 were described previously (Tan et al., 2007). IAA1 and IAA28 peptides were synthesized at EZBioLab as 5 to 10 mg of 95% (w/w) HPLC purified peptide.
Aux/IAA peptide competition was assayed by measuring IC50 values in an AlphaScreen assay (PerkinElmer) with purified TIR1 and IAA7.Nterm conjugated to donor and acceptor beads, respectively. Competitor Aux/IAA peptides were initially dissolved in water at approximately 1.0 mm. Concentrations were determined using extinction coefficients and absorbance at 280 nm values. Competition binding experiments were carried out by diluting the unlabeled IAA1, IAA7, and IAA28 peptides in the range of 0.5 to 5,000 nm in binding buffer [20 mm Tris, 200 mm NaCl, 1 mm tris(2-carboxyethyl)phosphine, 0.02 mg mL−1 ovalbumin, pH 8.0, 4 µm Arabidopsis SKP1-LIKE1/TIR1-glutathione S-transferase, 20 nm Flag-IAA7.Nterm, and 50 µm auxin] to a final volume of 50 µL. The samples were then incubated at room temperature for 2 h prior to addition of 2 µL alpha bead mixture (binding buffer containing 0.3% [v/v] Tween 20, 6 µL of 5 mg mL−1 AlphaScreen Glutathione Donor, and 3 µL of 5 mg mL−1 AlphaLisa anti-FLAG Acceptor beads in 100 µL final volume). The entire mixture was covered, placed in the dark, and allowed to incubate at 4°C over night. The plate was warmed to room temperature, and chemiluminescent signal was read on an Enspire 230 multilabel reader (PerkinElmer). In the presence of 50 μm auxin, the TIR1 and IAA7.Nterm associate and energy transfer from donor to acceptor beads produces a luminescent/fluorescent signal. Adding increasing concentrations of unlabeled Aux/IAA fragments to compete with the IAA7.Nterm substrate disrupts association between the beads, leading to a decrease in luminescent signal. This signal disruption was used to calculate the IC50 value for each peptide. The competition binding data were analyzed and plotted with Prism software (GraphPad). The experiment was done using white 384-well plates. All conditions were replicated six times.
Yeast Two-Hybrid Assays
The pGILDA-GW-TIR1 vector was transformed into the EGY48 yeast strain and selected on synthetic defined media (SD)-Ura-Trp. The pB42-AD-GW-Aux/IAA plasmids were transformed into the EGY48 pGILDA-TIR1 yeast strain and selected on SD-Ura-Trp-His. To assess interaction between TIR1 and the Aux/IAA fragments, the double-transformed strains were serially diluted onto SD+Gal-Ura-Trp-His-Leu plates supplemented with 10 μm auxin or mock treatment. For growth controls, the same strains were also serially diluted onto SD+Gal-Ura-Trp-His+Leu plates supplemented with 10 μm auxin or mock treatment. Plates were grown at 30°C for up to 1 week. Yeast transformed with pGILDA-TIR1 and pB42-AD-IAA7 (provided by Mark Estelle, University of California, San Diego) were used as a positive control strain, and yeast transformed with pGILDA-GW-TIR1 × pB42-GW-AD-stop codon were used as a negative control strain.
Western-Blot Analyses
Yeast grown overnight in liquid SC at 30°C with shaking were diluted to optical density = 0.6 into fresh SC medium and grown for 3 to 4 more hours until optical density = 0.8 to 1.0. Cells were then treated with auxin (10 μm) or mock (95% [v/v] ethanol), and 2.5-mL aliquots of cells were harvested at several time points up to 1 h after auxin treatment. Cells were lysed by glass bead beating, and protein samples were loaded into a 4% to 20% (v/v) TGX gel (Bio-Rad). Anti-GFP (Sigma-Aldrich) was used to probe for YFP-tagged IAA28, and anti-Phosphoglycerate Kinase1 (PGK1; Abcam) was used as a loading control. YFP band intensity was normalized to PGK1 band intensity and then fold initialized to the t = 0 time point.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequences flanking the degron are required to recapitulate degradation rates of FL IAA3 and are conserved in B. rapa orthologs.
Supplemental Figure S2. The influence of N- and C-terminal rate motifs on degradation dynamics is IAA dependent and is not confounded by differences in nuclear localization, basal expression, or linker length.
Supplemental Figure S3. The N-terminal KR residues contribute to Aux/IAA degradation rate, whereas rate determinants in the C terminus cannot be replaced with a flexible linker.
Supplemental Figure S4. Degradation of VENUS-IAA28 fragments in plant roots.
Supplemental Figure S5. Binding affinity between IAA28 and IAA1 fragments and the TIR1 auxin receptor is not always correlated with degradation rate.
Supplemental Protocol S1. Materials and methods.
Supplementary Material
Acknowledgments
We thank Eric Klavins and members of the Nemhauser and Zheng laboratories for helpful discussions.
Glossary
- NLS
nuclear localization sequence
- FL
full length
- IC50
inhibitory concentration 50%
- PB1
Phox/Bem1P
- YPD
yeast peptone dextrose
- SC
synthetic complete
Footnotes
This work was supported by the Paul G. Allen Family Foundation (to J.L.N.), National Science Foundation (grant nos. MCB–1411949 to J.L.N. and DBI–0115870 to N.Z.), and National Institutes of Health (grant nos. R01–GM107084 to J.L.N. and R01–CA076584 to N.Z. and fellowship F32CA180514 to B.L.M.). N.Z. is a Howard Hughes Medical Institute investigator.
Articles can be viewed without a subscription.
References
- Abel S, Oeller PW, Theologis A (1994) Early auxin-induced genes encode short-lived nuclear proteins. Proc Natl Acad Sci USA 91: 326–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J. (2014) The ubiquitination machinery of the ubiquitin system. Arabidopsis Book 12: e0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Barford D (2014) Insights into the anaphase-promoting complex: a molecular machine that regulates mitosis. Curr Opin Struct Biol 29: 1–9 [DOI] [PubMed] [Google Scholar]
- Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Li P, Hua W, Wang X (2011) BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol 11: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Del Bianco M, Kepinski S (2011) Context, specificity, and self-organization in auxin response. Cold Spring Harb Perspect Biol 3: a001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I. (2012) Lateral root initiation: one step at a time. New Phytol 193: 867–873 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005a) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M (2005b) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dinesh DC, Kovermann M, Gopalswamy M, Hellmuth A, Calderón Villalobos LI, Lilie H, Balbach J, Abel S (2015) Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response. Proc Natl Acad Sci USA 112: 6230–6235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Gilkerson J, Kelley DR, Tam R, Estelle M, Callis J (2015) Lysine residues are not required for proteasome-mediated proteolysis of the Auxin/indole acidic acid protein IAA1. Plant Physiol 168: 708–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Guseman JM, Hellmuth A, Lanctot A, Feldman TP, Moss BL, Klavins E, Calderón Villalobos LI, Nemhauser JL (2015) Auxin-induced degradation dynamics set the pace for lateral root development. Development 142: 905–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Park Y, Kim I, Kim EH, Yu TK, Rhee S, Suh JY (2015) Correction for Han et al., Structural basis for the auxin-induced transcriptional regulation by Aux/IAA17. Proc Natl Acad Sci USA 112: E60225617363 [Google Scholar]
- Havens KA, Guseman JM, Jang SS, Pierre-Jerome E, Bolten N, Klavins E, Nemhauser JL (2012) A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol 160: 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Vierstra RD (2011) The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol 62: 299–334 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81: 203–229 [DOI] [PubMed] [Google Scholar]
- Korasick DA, Chatterjee S, Tonelli M, Dashti H, Lee SG, Westfall CS, Fulton DB, Andreotti AH, Amarasinghe GK, Strader LC. , et al. (2015) Defining a Two-pronged Structural Model for PB1 (Phox/Bem1p) Domain Interaction in Plant Auxin Responses. J Biol Chem 290: 12868–12878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick DA, Westfall CS, Lee SG, Nanao MH, Dumas R, Hagen G, Guilfoyle TJ, Jez JM, Strader LC (2014) Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci USA 111: 5427–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S, Jürgens G, De Smet I (2008) The evolving complexity of the auxin pathway. Plant Cell 20: 1738–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraschin Fdos S, Memelink J, Offringa R (2009) Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J 59: 100–109 [DOI] [PubMed] [Google Scholar]
- Nanao MH, Vinos-Poyo T, Brunoud G, Thévenon E, Mazzoleni M, Mast D, Lainé S, Wang S, Hagen G, Li H, et al. (2014) Structural basis for oligomerization of auxin transcriptional regulators. Nat Commun 5: 3617. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M (2009) An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6: 917–922 [DOI] [PubMed] [Google Scholar]
- Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Liu A, Onodera C, Quach H, et al. (2005) Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17: 3282–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre-Jerome E, Jang SS, Havens KA, Nemhauser JL, Klavins E (2014) Recapitulation of the forward nuclear auxin response pathway in yeast. Proc Natl Acad Sci USA 111: 9407–9412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre-Jerome E, Moss BL, Nemhauser JL (2013) Tuning the auxin transcriptional response. J Exp Bot 64: 2557–2563 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Lavy M, Ashton NW, Estelle M (2010) Physcomitrella patens auxin-resistant mutants affect conserved elements of an auxin-signaling pathway. Curr Biol 20: 1907–1912 [DOI] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser O, Callis J (2001) Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13: 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogg LE, Lasswell J, Bartel B (2001) A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13: 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderón-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen R, Wang S, Xiang D, Venglat P, Shi X, Zang Y, Datla R, Xiao W, Wang H (2014) UBC13, an E2 enzyme for Lys63-linked ubiquitination, functions in root development by affecting auxin signaling and Aux/IAA protein stability. Plant J 80: 424–436 [DOI] [PubMed] [Google Scholar]
- Williamson A, Banerjee S, Zhu X, Philipp I, Iavarone AT, Rape M (2011) Regulation of ubiquitin chain initiation to control the timing of substrate degradation. Mol Cell 42: 744–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J (2000) Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J 21: 553–562 [DOI] [PubMed] [Google Scholar]
- Yu H, Moss BL, Jang SS, Prigge M, Klavins E, Nemhauser JL, Estelle M (2013) Mutations in the TIR1 auxin receptor that increase affinity for auxin/indole-3-acetic acid proteins result in auxin hypersensitivity. Plant Physiol 162: 295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenser N, Ellsmore A, Leasure C, Callis J (2001) Auxin modulates the degradation rate of Aux/IAA proteins. Proc Natl Acad Sci USA 98: 11795–11800 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.