Evolution of an alpine genome involved an unusually high number of pericentric inversions, plausibly facilitating the evolution of heavy metal-related traits.
Abstract
Noccaea caerulescens (formerly known as Thlaspi caerulescens), an extremophile heavy metal hyperaccumulator model plant in the Brassicaceae family, is a morphologically and phenotypically diverse species exhibiting metal tolerance and leaf accumulation of zinc, cadmium, and nickel. Here, we provide a detailed genome structure of the approximately 267-Mb N. caerulescens genome, which has descended from seven chromosomes of the ancestral proto-Calepineae Karyotype (n = 7) through an unusually high number of pericentric inversions. Genome analysis in two other related species, Noccaea jankae and Raparia bulbosa, showed that all three species, and thus probably the entire Coluteocarpeae tribe, have descended from the proto-Calepineae Karyotype. All three analyzed species share the chromosome structure of six out of seven chromosomes and an unusually high metal accumulation in leaves, which remains moderate in N. jankae and R. bulbosa and is extreme in N. caerulescens. Among these species, N. caerulescens has the most derived karyotype, with species-specific inversions on chromosome NC6, which grouped onto its bottom arm functionally related genes of zinc and iron metal homeostasis comprising the major candidate genes NICOTIANAMINE SYNTHASE2 and ZINC-INDUCED FACILITATOR-LIKE1. Concurrently, copper and organellar metal homeostasis genes, which are functionally unrelated to the extreme traits characteristic of N. caerulescens, were grouped onto the top arm of NC6. Compared with Arabidopsis thaliana, more distal chromosomal positions in N. caerulescens were enriched among more highly expressed metal homeostasis genes but not among other groups of genes. Thus, chromosome rearrangements could have facilitated the evolution of enhanced metal homeostasis gene expression, a known hallmark of metal hyperaccumulation.
Noccaea caerulescens (formerly known as Thlaspi caerulescens) is a diploid (2n = 14) biennial or short-living perennial plant from the family Brassicaceae. N. caerulescens is native to Europe, with a patchy distribution from the United Kingdom and France to Slovakia and from Germany and Poland southward to northern Spain and Italy. The widespread occurrence in Scandinavia is thought to represent naturalized populations over the past few hundred years (Koch et al., 1998, and refs. therein). N. caerulescens is one of the 120 Noccaea spp., which, together with two other genera, constitute the tribe Coluteocarpeae (approximately 127 species; Al-Shehbaz, 2012). However, the generic treatment of the tribe is far from settled, and up to 12 genera are recognized in Coluteocarpeae by F.K. Meyer (Meyer, 2001, 2006a, 2006b; Koch and German, 2013).
Together with the metal hyperaccumulator species Arabidopsis halleri, N. caerulescens is among the most prominent plant model systems for the study of heavy metal hyperaccumulation and associated hypertolerance (Krämer, 2010; Hanikenne and Nouet, 2011; Pollard et al., 2014). N. caerulescens is a hyperaccumulator of zinc (Zn) on metalliferous as well as nonmetalliferous soils and of cadmium (Cd) on metalliferous soils (Reeves et al., 2001; Krämer, 2010), and its populations on serpentine soils are known to hyperaccumulate nickel (Ni; Reeves and Brooks, 1983; Reeves, 1988; Visioli et al., 2012; Maestri et al., 2013). N. caerulescens is a morphologically highly diverse species comprising at least two, but up to eight (http://www.gbif.org), recognized subspecies with partly overlapping distribution ranges (the simplest treatment includes ssp. caerulescens and sylvestris, the latter including populations formerly described as Thlaspi calaminare or N. caerulescens ssp. calaminare). As the morphological variation often has a clinal character, the taxonomic and biological value of various intraspecific taxa is questionable, and detailed studies will be needed to resolve this issue (for discussion, see Koch and German, 2013). In addition to pervasive morphological variation (Koch and German, 2013) apparently reinforced by geographical constraints (Besnard et al., 2009), there is also pronounced phenotypic variation in metal tolerance and accumulation (Escarré et al., 2000; Reeves et al., 2001; Assunção et al., 2006; Xing et al., 2008; Krämer, 2010; Tuomainen et al., 2010).
Although N. caerulescens is the most commonly studied metal hyperaccumulator model species, with more than 210 studies published on the subject (Pollard et al., 2014), the detailed genome structure of N. caerulescens remains unresolved. Assunção et al. (2006) published the first amplified fragment-length polymorphism-based genetic linkage map and identified quantitative trait loci for Zn accumulation in roots. Another amplified fragment-length polymorphism-based genetic map based on a cross between two accessions with differential Cd accumulation and tolerance was used to identify quantitative trait loci associated with the accumulation of Cd and Zn (Deniau et al., 2006). Both maps comprised the expected seven linkage groups with dense clusters of linked markers located on each linkage group, most likely corresponding to centromeric regions with suppressed recombination rates. Apart from the tentative identification of centromeres, small numbers of orthologous markers shared with Arabidopsis thaliana and A. halleri did not allow the establishment of chromosome collinearity between these genomes for inference of the genome structure of N. caerulescens.
More recently, Mandáková and Lysak (2008) reconstructed karyotype evolution in eight Brassicaceae species of tribes in extended lineage II (Franzke et al., 2011) by comparative chromosome painting (CCP) using chromosome-specific bacterial artificial chromosome (BAC) contigs of A. thaliana. They concluded that genomes of all the analyzed species with seven or 14 chromosome pairs (n = 7/14) were derived from the eight chromosomes of the Ancestral Crucifer Karyotype (ACK; n = 8) via an ancestral n = 7 genome named the proto-Calepineae Karyotype (PCK). The genome of N. caerulescens (accession Korenec), analyzed as a representative of the tribe Coluteocarpeae (formerly Noccaeeae) by Mandáková and Lysak (2008), has descended from a PCK-like ancestor but showed a remarkably high number of secondary chromosome rearrangements. By comparison with the ancestral PCK, in N. caerulescens six of the seven ancestral chromosomes were reshuffled by inversions encompassing pericentromeric regions. However, that study did not establish the detailed structure of the N. caerulescens genome, including precise positions of chromosome break points. Consequently, evolutionary steps leading to the origin of the inversion chromosomes were reconstructed only approximately, and gene content could not be estimated.
Our study here provides a detailed comparative genome structure of N. caerulescens and relates the genome structure to the evolution of heavy metal-related extreme traits. Based on a more precise definition of ancestral genomic blocks within the ACK (Cheng et al., 2013) and using PCK as the most probable ancestral genome of N. caerulescens, we carried out a detailed analysis of the entire karyotype, including inversions, by means of CCP. Furthermore, considering the unusually high incidence of inversions, we were intrigued to find out whether this variation is unique to the reference accession (Mandáková and Lysak, 2008) or fixed in populations on both nonmetalliferous and metalliferous soils enriched for different heavy metal elements. Toward this aim, we constructed detailed comparative karyotypes for 13 populations of N. caerulescens from metalliferous and nonmetalliferous soils throughout the European species distribution range, including, in particular, populations from southern France known for the extraordinarily high concentrations of Zn and Cd in their leaves (St. Félix de Pallières and Viviez; Reeves et al., 2001). Next, we asked whether the inversions are specific to N. caerulescens or shared by other Coluteocarpeae species by deducing the chromosome structure and evolution of the N. caerulescens genome in comparison with two other species from the tribe Coluteocarpeae, Noccaea jankae and Raparia bulbosa. Finally, we addressed the question of whether chromosome inversions and rearrangements might have affected the physical positions of metal homeostasis candidate genes that were proposed to act in naturally selected metal hyperaccumulation and associated hypertolerance of N. caerulescens. We tested for the clustering of functionally related metal homeostasis genes in closer proximity on the chromosome toward supergene formation as well as for the relation between changes in chromosomal position and gene expression among metal homeostasis genes.
RESULTS
Before detailed cytogenetic analyses were carried out, we verified chromosome numbers in all species under study and accessions thereof (Table I; Supplemental Fig. S1). Except for the tetraploid N. jankae (2n = 4x = 32), all remaining accessions were diploid (2n = 2x = 14). We estimated nuclear genome size in our reference accession of N. caerulescens (Korenec; CZ1) at 267 Mb by flow cytometry.
Table I. Populations analyzed in this study.
Soil types are as follows: N, nonmetalliferous;, C, calamine; Cu, copper; and U, ultramafic/serpentine. Asterisks indicate populations from which plant material was sampled in the wild; plants from the remaining populations were grown from seeds. Daggers indicate populations analyzed by Besnard et al. (2009). n/a, Not applicable.
| Code | Population | Soil Type | Altitude | Latitude | Longitude |
|---|---|---|---|---|---|
| m | N | E | |||
| N. caerulescens | |||||
| S1 | Sweden: Kungs-Husby | N | 17 | 59° 30′ | 17° 20′ |
| S2 | Sweden: Sularp | N | 76 | 55° 42′ | 13° 20′ |
| CH1† | Switzerland: Mont d’Amin | N | 1,300 | 47° 04′ 55′′ | 6° 54′ 13′′ |
| CH2† | Switzerland: Haslital | N | 1,200 | 46° 39′ 22′′ | 8° 18′ 01′′ |
| CH3† | Switzerland: Vallée de Saas | N | 1,600 | 46° 09′ 50′′ | 7° 55′ 27′′ |
| CZ1* | Czech Republic: Kořenec | N | 640 | 49° 31′ 38.84′′ | 16° 45′ 31.76′′ |
| CZ2* | Czech Republic: Mohelno | U | 300 | 49° 06.403′ | 16° 11.279′ |
| SK1* | Slovakia: Špania Dolina | Cu | 750 | 48° 48.563′ | 19° 07.962′ |
| F1* | France: Bergenbach | U | 480 | 47° 54.366′ | 6° 57.676′ |
| F2* | France: Viviez | C | 200 | 44° 33.887′ | 2° 12.796′ |
| F3* | France, Firmi: Puy de Wolf | U | 260 | 44° 32′ 07.15′′ | 2° 17′ 46.25′′ |
| F4* | France: St. Félix de Pallières | C | 340 | 44° 02′ 40.23′′ | 3° 56′ 23.92′′ |
| F5* | France: St. Laurent le Minier | C | 250 | 43° 55.959′ | 3° 39.740′ |
| N. jankae | |||||
| Slovakia: Tribeč Mountains, Zobor Mountain* | N | 250 | 48°20′ 17.03′′ | 18° 07′ 39.51′′ | |
| R. bulbosa | |||||
| Endemic to Greece; seeds provided by the botanical garden of the University of Heidelberg | U, N | n/a | n/a | n/a | |
Comparative Structure of the N. caerulescens Genome
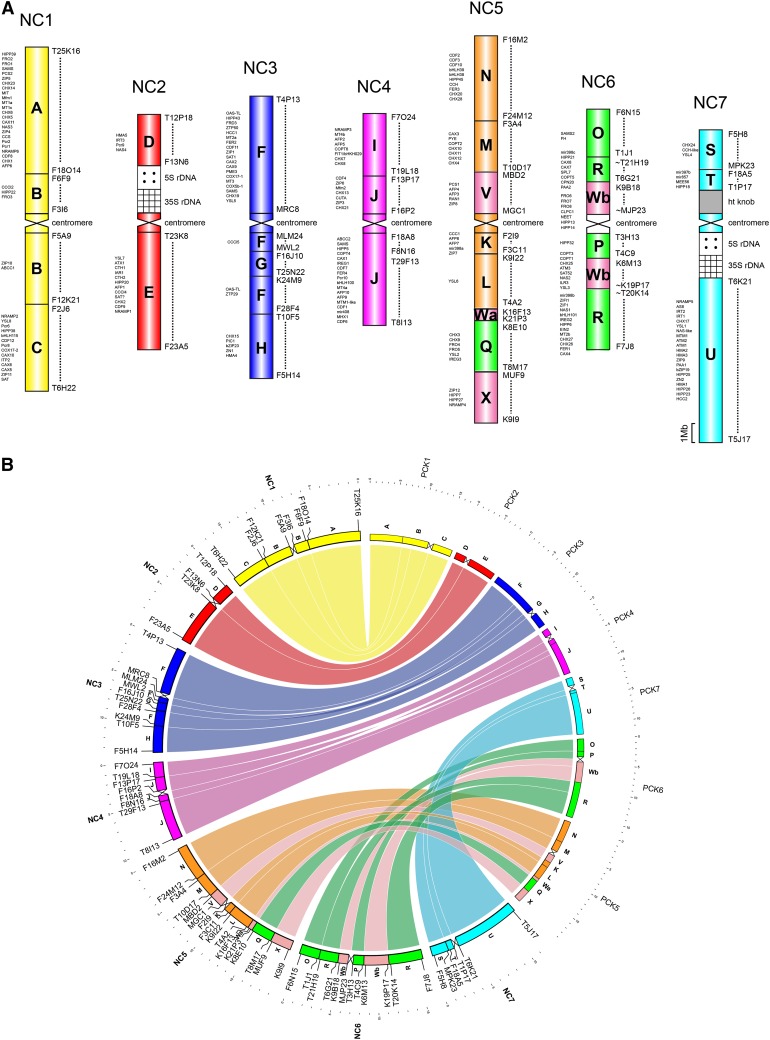
A comparative cytogenomic map of N. caerulescens (Fig. 1A) was constructed by multicolor CCP using chromosome-specific BAC contigs of A. thaliana (for examples, see Fig. 2). Differentially labeled BAC contigs were used as painting probes to identify all 24 ancestral genomic blocks (Schranz et al., 2006; refined in Cheng et al., 2013) in the genomes of N. caerulescens and two other Coluteocarpeae species. Our experiments unambiguously identified the positions of all 24 genomic blocks in the analyzed chromosome complements (Fig. 1A). The karyotype of N. caerulescens consists of metacentric chromosomes without terminal nucleolar organizer regions. Instead, the 35S and 5S ribosomal DNA (rDNA) loci are adjacent to each other and located at the pericentromere of chromosomes NC2 and NC7. A heterochromatic knob is located on the short arm of NC7. The N. caerulescens genome is largely collinear with the ancestral PCK genome (Fig. 1B); thus, the genome of N. caerulescens has most likely descended from the PCK, like all other taxa in extended lineage II (Mandáková and Lysak, 2008). In comparison with PCK, centromeric regions of six N. caerulescens chromosomes were rearranged by pericentric and paracentric inversions; chromosome NC2 has the ancestral PCK2-like structure (Fig. 1).
Figure 1.
Structure of the N. caerulescens genome. Cytomolecular comparative map based on chromosome painting analyses (A) and Circos diagram (B) show the extent of collinearity between the N. caerulescens genome (NC1–NC7) and the ancestral karyotype, designated as PCK (PCK1–PCK7). Uppercase A to X mark 24 conserved genomic blocks defined here by A. thaliana BAC clones (www.arabidopsis.org). In A, genes of heavy metal homeostasis are listed on the left side (Supplemental Data Set S1). Chromosome in situ localizations of 5S and 35S rDNA probes are shown in Supplemental Figure S4.
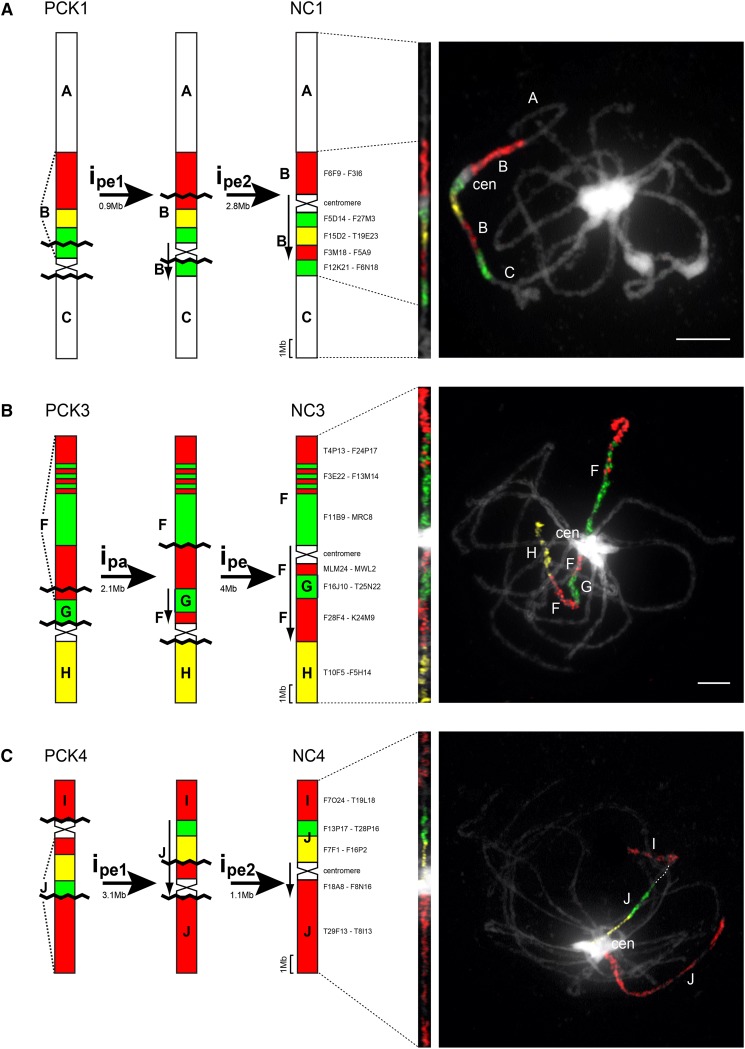
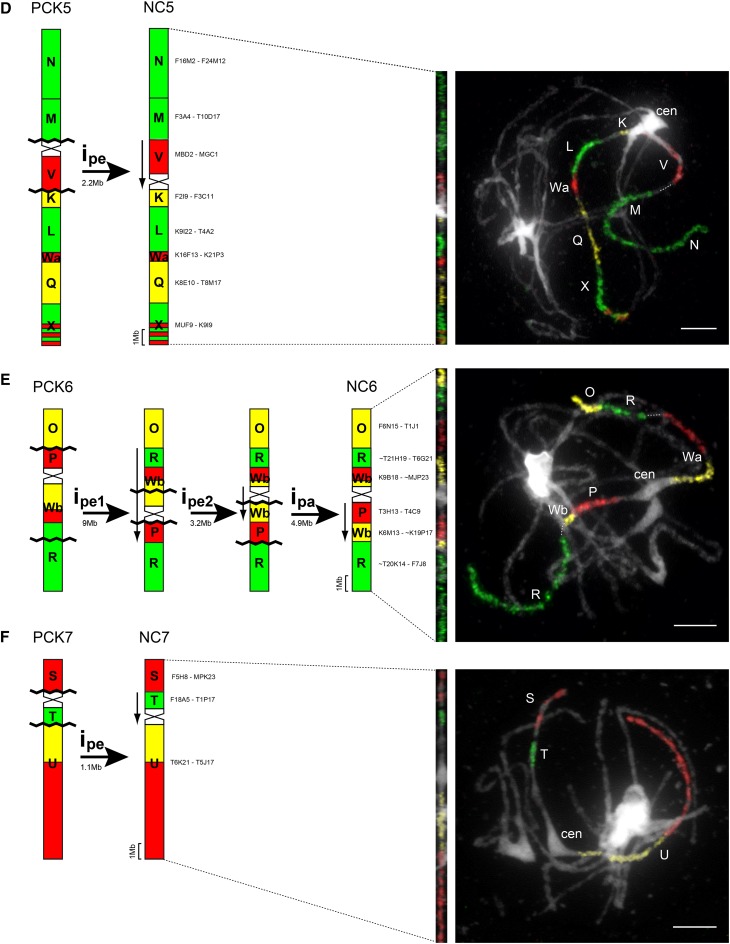
Figure 2.
Inference of chromosome rearrangements during evolution of the N. caerulescens genome from the ancestral PCK genome. Shown are schematic summaries (left) and examples of CCP analysis (right). NC2 retaining the ancestral structure is not shown. ipa, Paracentric inversion; ipe, pericentric inversion. Downward-pointing arrows denote the inverse orientation of chromosome regions after inversions. The approximate size of each inversion was estimated based on the A. thaliana genome sequence (www.arabidopsis.org); rDNA loci were not considered. Chromosomes were painted with A. thaliana BAC contigs labeled by biotin-dUTP (red), digoxigenin-dUTP (green), and Cy3-dUTP (yellow); BAC clone coordinates of each contig used are given. Chromosomes were counterstained by 4′,6-diamidino-2-phenylindole (DAPI). Bars = 5 µm.
Inversion Rearrangements
A reconstruction of chromosome rearrangements shuffling six PCK-like chromosomes to extant chromosomes of N. caerulescens is shown in Figure 2. The current pattern of genomic blocks on chromosomes NC1 and NC4 is most parsimoniously explained by two subsequent pericentric inversions reshuffling blocks B and J, respectively, and changing the centromere position (Fig. 2, A and C). The relative extent of inversion rearrangements based on the A. thaliana collinear genomic sequence (inversion size [Mb]/total chromosome length [Mb] × 100) is 22.1% for NC1 and 30.6% for NC4. One paracentric and one pericentric inversion explain the origin of NC3 from its precursor PCK3; inversions rearranged 29.2% of NC3 (Fig. 2B). The extant structures of chromosomes NC5 and NC7 can be explained by single pericentric inversions (Fig. 2, D and F), and the two chromosomes are least rearranged (11.1% for NC5 and 9.2% for NC7). The origin of NC6 most probably required three inversion events, one paracentric and two pericentric ones (Fig. 2E), reshuffling more than half (56.7%) of the ancestral chromosome. In total, at least nine pericentric and two paracentric inversions occurred during the evolution of the N. caerulescens karyotype. Not considering centromeres, the size of inverted regions varied from 0.9 Mb (NC1: pericentric inversion 1) to 9 Mb (NC6: pericentric inversion 1), with the average inversion size of 3.1 Mb. From 22 inversion break points, 10 localize to pericentromeric heterochromatin (46%), eight within a genomic block (36%), and four between two genomic blocks (18%; Fig. 2).
Intraspecific Karyotype Stasis in N. caerulescens
Revealing the high incidence of inversions in our reference accession of N. caerulescens (population CZ1; Mandáková and Lysak, 2008; this study), we questioned whether these rearrangements are present in other N. caerulescens populations across its European distribution area (Table I; Supplemental Fig. S1). Toward this aim, we sampled 13 populations of N. caerulescens from Sweden to southern France (a transect of approximately 3,000 km) following three main variable criteria: (1) geography, (2) altitude, and (3) soil type (populations on nonmetalliferous versus metalliferous soils).
To assess the influence of metalliferous soils, we collected plants from different types of soils based on literature records (Reeves et al., 2001; Banásová et al., 2008) and verified the reported values by the determination of soil and leaf metal concentrations (Table II). Populations Mohelno (CZ2), Bergenbach (F1), and Puy de Wolf (F3) are located on natural ultramafic/serpentine soils characterized by an unusually high magnesium-calcium ratio and high levels of Ni, cobalt, and chromium and sometimes of Cd and Zn (population F3; Table II). As expected, the Czech population growing on serpentinites exhibited Ni hyperaccumulation (average leaf Ni concentration of 1,800 µg g−1 dry biomass). Our measurements for the two French populations were in accordance with those reported by Reeves et al. (2001), confirming the hyperaccumulation of Ni in Bergenbach and of Ni, Cd, and Zn in Puy de Wolf. The population of Špania Dolina (SK1), an old copper (Cu) mining area with high soil Cu concentrations, had leaf Zn concentrations in the hyperaccumulator range (Table II; Banásová et al., 2008). Finally, the remaining three French populations are located on calamine soils of former Zn/lead mining sites (St. Félix de Pallières [F4] and St. Laurent le Minier [F5]) or at a smelter site (Viviez [F2]) with Zn ore having been brought in from a variety of sources, reflected by hyperaccumulation of both Zn and Cd in shoots (Table II; Reeves et al., 2001). Our data confirmed that these seven analyzed populations occur on a diverse set of metalliferous soils, spanning elevated to extreme concentrations of heavy metals. All of these populations can be classified as metal hyperaccumulating, with considerable between-population variation in hyperaccumulation properties.
Table II. Soil and leaf metal concentrations in populations of N. caerulescens analyzed in this study.
Leaf metal concentrations corresponding to metal hyperaccumulation (greater than 100, 3,000, or 1,000 mg kg−1 for Cd, Zn, and Ni, respectively) are highlighted in boldface (Krämer, 2010). n.d., Below detection limits. Values given are arithmetic means of between one and three replicate samples.
| Code | Population | Soil Concentration |
Leaf Concentration |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Cd | Zn | Cu | Ni | Cd | Zn | Cu | Ni | ||
| mg kg−1 dry mass | mg kg−1 dry biomass | ||||||||
| CZ2 | Czech Republic: Mohelno | n.d. | 58 | 7.9 | 170 | 17 | 1,600 | 3.5 | 1,800 |
| SK1 | Slovakia: Špania Dolina | 0.50 | 120 | 320 | 11 | 16 | 7,400 | 12 | 14 |
| F1 | France: Bergenbach | n.d. | 82 | 10 | 1,800 | 5.3 | 2,100 | 3.9 | 10,000 |
| F2 | France: Viviez | 51 | 5,100 | 425 | 36 | 390 | 6,100 | 12 | 3.1 |
| F3 | France, Firmi: Puy de Wolf | 0.55 | 210 | 31 | 2,600 | 110 | 4,000 | 5.7 | 13,000 |
| F4 | France: St. Félix de Pallières | 98 | 22,000 | 60 | 12 | 720 | 12,000 | 15 | 3.0 |
| F5 | France: St. Laurent le Minier | 220 | 94,000 | 12 | 37 | 750 | 13,000 | 6.1 | 1.4 |
We reconstructed the entire karyotype by CCP in randomly chosen plants from each of the seven populations on metalliferous soils and from six other populations on nonmetalliferous soils (Table I; Supplemental Fig. S1). Our analysis was particularly centered on the potential incidence of inversion polymorphism among populations originating from different parts of the distribution range or contrasting edaphic soil types. On the basis of CCP employing a set of individual A. thaliana BAC clones, all populations analyzed showed the same karyotype structure and inversion break points as described for population CZ1 (Figs. 1 and 2).
Are Inversions Unique to N. caerulescens?
After establishing the consensus genome structure of N. caerulescens, we analyzed the genomes of two other species from the tribe Coluteocarpeae using painting probes specific for the seven chromosomes of N. caerulescens. The tetraploid N. jankae is endemic to southern Slovakia, northern Hungary (Meyer, 2006b), and Romania (Király et al., 2013) and is closely related to N. caerulescens (Supplemental Fig. S2; Koch and Al-Shehbaz, 2004). The diploid R. bulbosa (formerly Thlaspi bulbosum) is endemic to Greece (Meyer, 2006a) and is a sister taxon to genus Noccaea (Supplemental Fig. S2; Koch and Mummenhoff, 2001; Koch and Al-Shehbaz, 2004; Kiefer et al., 2014). Whereas N. jankae occurs on limestones, andesites, or sandy soils (Meyer, 2006b) and can be classified as an accumulator, but not a hyperaccumulator, of Ni and Zn (Reeves and Brooks, 1983), R. bulbosa grows on various substrates, including serpentines (Reeves and Brooks, 1983; Artelari, 2002; P. Trigas, personal communication), and is able to hyperaccumulate both Ni and Zn at significantly lower levels than N. caerulescens (Reeves and Brooks, 1983).
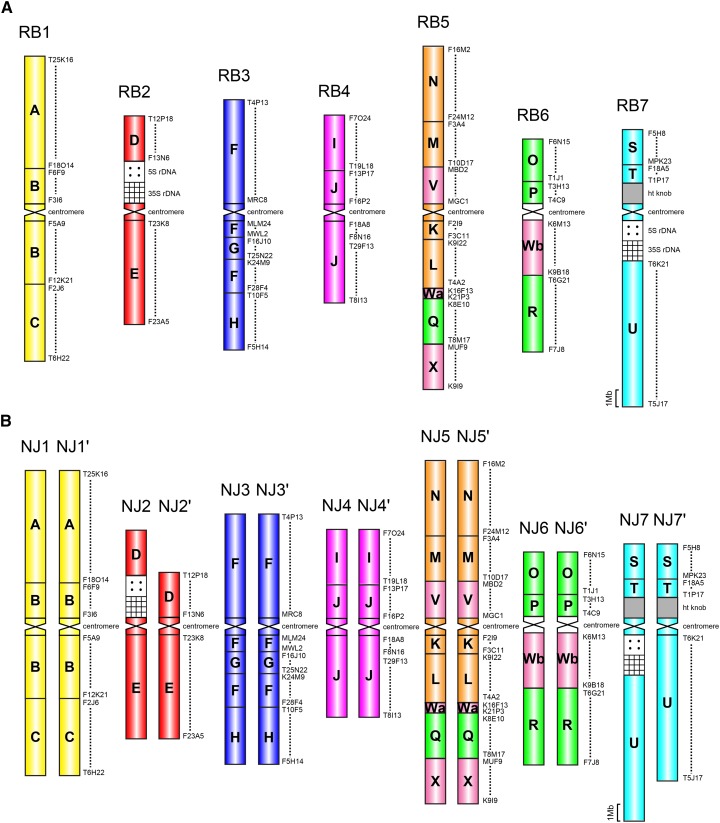
Our analysis showed that six out of the seven chromosomes including rDNA loci and the knob in N. jankae and R. bulbosa (Fig. 3; Supplemental Fig. S3) have the same structure as in N. caerulescens (Fig. 1A). The two species differ from N. caerulescens in the arrangement of genomic blocks on chromosomes NJ6 and RB6, respectively. Whereas the N. caerulescens NC6 chromosome was rearranged by three inversions (Fig. 2E), chromosomes NJ6 and RB6 retained their ancestral PCK6-like structure (Fig. 3). This suggests that genomes of N. jankae and R. bulbosa have a more ancestral character than the genome of N. caerulescens and that the PCK6 homologue was reshuffled only in this species or a clade of very closely related taxa. Moreover, we could conclude that both subgenomes of N. jankae do not differ by structural chromosome rearrangements (Fig. 3B); thus, the tetraploid genome originated through autopolyploidy or hybridization between two taxa with a highly similar genome structure.
Figure 3.
Cytomolecular comparative maps of the diploid genome of R. bulbosa (A) and the tetraploid genome of N. jankae (B) based on CCP analysis. Uppercase A to X mark 24 conserved genomic blocks defined here by A. thaliana BAC clones (www.arabidopsis.org). Chromosome in situ localizations of 5S and 35S rDNA probes are shown in Supplemental Figure S4.
Pericentric Inversions Are Increasing the Karyotype Symmetry
The predominance of pericentric inversions during genome evolution of the three Coluteocarpeae species was associated with the reduction of ancestral karyotype asymmetry of PCK (Figs. 1 and 3). This tendency is corroborated by the centromeric index (CI; length of the short arm to the total chromosome length × 100) calculated for the ancestral PCK genome and the three extant species using A. thaliana sequence data as a proxy. Whereas the mean CI of the ancestral PCK genome is 28%, the CI value equals 39% in N. jankae and R. bulbosa, and N. caerulescens has the most symmetric karyotype of all (CI = 41%). This means that in Noccaea and Raparia spp., the ancestral acrocentric to submetacentric chromosomes have become more symmetric, with arms roughly equal in length.
Genome Structure in the Context of the Metal Homeostasis Network
Metal-related extreme traits of N. caerulescens are also found, yet in much weaker form, in R. bulbosa and N. jankae, but not in the vast majority of Brassicaceae species, including A. thaliana. Thus, we tested for the consequences of changes in genome structure on chromosomal positions of candidate genes that were proposed to functionally contribute to metal-related extreme traits. Characteristically, steady-state transcript levels of metal homeostasis candidate genes are known to be far higher in the metal hyperaccumulators N. caerulescens and A. halleri than in closely related nontolerant nonaccumulator reference species such as A. thaliana (Krämer, 2010). Moreover, the contribution to metal hyperaccumulation and hypertolerance was demonstrated for a subset of these genes by RNA interference in A. halleri (Hanikenne et al., 2008; Deinlein et al., 2012). The gene content of each of the 24 conserved genomic blocks is known in the A. thaliana reference genome. This information was used to approximate chromosomal positions of metal homeostasis genes, as well as their changes, in the PCK and N. caerulescens genomes based on the chromosomal position of each conserved genomic block. By comparison with the ancestral PCK (Fig. 1B), rearrangements that occurred in the N. caerulescens lineage removed regions containing a low density of metal homeostasis genes away from one arm of chromosomes PCK3 and PCK7, which are otherwise very rich in metal homeostasis genes (Figs. 1A and 2, B and F; Supplemental Data Set S1). Moreover, a small rearrangement in NC4 places the major candidate plasma membrane Zn uptake-related genes ZINC-REGULATED TRANSPORTER, IRON-REGULATED TRANSPORTER-LIKE PROTEIN3 (ZIP3) and ZIP6 in closer proximity to two very important iron homeostasis-related genes. One of these genes is the candidate gene NATURAL RESISTANCE ASSOCIATED MACROPHAGE PROTEIN3 encoding a membrane transporter involved in metal mobilization from the vacuole, and the second one is FE-DEFICIENCY INDUCED TRANSCRIPTION FACTOR1, which encodes an iron-regulatory transcription factor of central importance (Figs. 1A and 2C). In this context, it is important to note that the molecular homeostasis of the metals hyperaccumulated in N. caerulescens is tightly connected with the homeostasis of the chemically similar, important micronutrient iron (Fe; Krämer and Sinclair, 2012). All these rearrangements in N. caerulescens by comparison with PCK are shared with N. jankae and R. bulbosa, which are either Ni and Zn accumulating or hyperaccumulating taxa (see above). Furthermore, a significant enrichment of genes contributing to the homeostasis of Cd was found in block A and is common to all karyotypes (11% metal homeostasis genes overall versus 24% in block A; P < 0.05, Fisher’s exact test; Supplemental Data Set S2). Note that these are predictions based on the known gene content of conserved genomic bocks in A. thaliana.
Whereas linkage group 6 in N. jankae and R. bulbosa share the ancestral PCK-like chromosome structure (Fig. 3), in N. caerulescens, genomic block R on NC6, originally on the bottom arm of PCK6, was split into two segments (Fig. 1A). One segment of block R now resides in inverted orientation on the other, top arm of the chromosome NC6 in N. caerulescens (Fig. 2E). Importantly, the ancestral block R is enriched in genes contributing to Cu homeostasis (Fig. 1A; 10 Cu homeostasis genes out of 21 metal homeostasis genes in total; P < 0.05, Fisher’s exact test; Supplemental Data Set S2), of which a segment containing five Cu homeostasis genes out of a total of eight metal homeostasis genes now resides in the novel position on the top arm. These genes include the Cu-regulatory transcription factor gene SQUAMOSA PROMOTER BINDING PROTEIN-LIKE7 alongside the Cu-regulatory miRNA398c gene, the chloroplast thylakoid membrane P1B-type Cu(I)-ATPase-encoding gene PAA2, as well as the vacuolar membrane Cu-mobilizing transporter-encoding gene COPT5 (Fig. 1A). By contrast, the portion of block R that remains on the bottom arm of NC6, as in the ancestral karyotype, mostly contains genes related to Fe and Zn homeostasis.
Sixty percent of all metal homeostasis genes present in block Wb of PCK6 are related to Fe, whereas only 26% of genome-wide metal homeostasis genes contribute to Fe homeostasis, indicating that Fe homeostasis genes are enriched in block Wb (P < 0.01, Fisher’s exact test; Supplemental Data Set S2). Compared with block Wb on the bottom arm of PCK6, chromosome NC6 contains the ancestral block Wb in two subsegments. One segment of Wb has been relocated in inverted orientation to the other, top arm of chromosome NC6, and the other segment forms part of an inversion on the bottom arm of NC6 (Fig. 2E). In this fashion, several Fe homeostasis genes with additional roles in Zn homeostasis of block Wb were brought in far closer physical proximity to the general Fe homeostasis genes on the stationary segment of block R on the bottom arm of NC6. These major candidate genes are characterized by enhanced expression in N. caerulescens and by their status as candidate genes for metal hyperaccumulation and hypertolerance. They include the NICOTIANAMINE SYNTHASE-encoding gene NAS2 (van de Mortel et al., 2006; Deinlein et al., 2012), the putative cytoplasmic metal cation importer gene IAA-LEUCINE RESISTANT3 (van de Mortel et al., 2006), and the putative metal-nicotianamine complex transporter gene YELLOW STRIPE-LIKE3 (YSL3; Gendre et al., 2007) in block Wb and ZINC-INDUCED FACILITATOR1 (ZIF1) and its paralogue ZIFL1 (van de Mortel et al., 2006) in block R (bottom arm of NC6). On the other, top arm of NC6, genes involved in the organellar Fe/metal homeostasis of block Wb are positioned in far closer genomic proximity to the set of Cu homeostasis genes on the relocated segment of block R (Fig. 1).
These structural rearrangements have the potential of resulting in reduced rates of recombination within each of two groups of functionally related metal homeostasis genes. For each of these groups, genes located in segments of different genomic blocks are now placed in much closer physical proximity on either arm of N. caerulescens chromosome NC6 than in the ancestral karyotype. Moreover, recombination rates between the two separated segments of blocks R and Wb could possibly be enhanced. Thus, a lineage-specific linkage resorting can be observed on N. caerulescens chromosome NC6, grouping Cu and chloroplast-related metal homeostasis functions on one side of the centromere, whereas Fe- and Zn-related functions, as well as metal hypertolerance- and hyperaccumulation-related functions, are grouped on the other side of the centromere. Overall, as compared with the ancestral state, the inversions that occurred toward the extant NC6 chromosome resulted in a more even distribution of metal homeostasis genes across both arms of that chromosome.
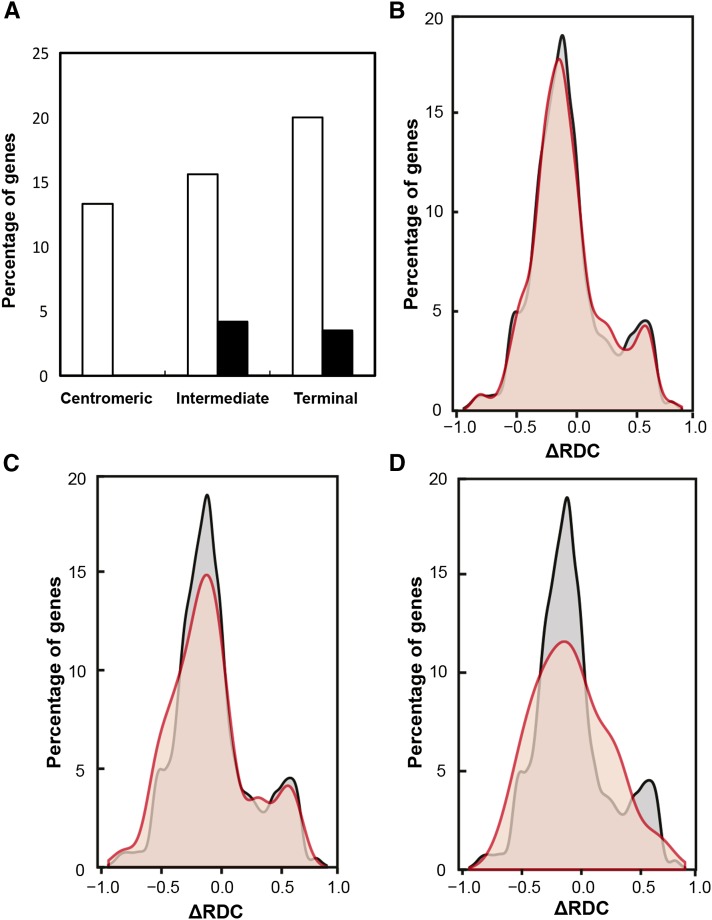
Because the evolution of metal hyperaccumulation and hypertolerance appears to have involved increased expression of metal homeostasis genes (Becher et al., 2004; Weber et al., 2004; Talke et al., 2006; Hanikenne et al., 2008), we tested for relationships between predicted chromosome positions and gene expression. Out of the metal homeostasis genes positioned in the centromeric, intermediate, and terminal chromosome regions in N. caerulescens, respectively, 13.3%, 15.6%, and 20% are more highly expressed in N. caerulescens than in A. thaliana, based on published data (Fig. 4A; van de Mortel et al., 2006, 2008). Moreover, none, 4.2%, and 3.5% of metal homeostasis genes located in centromeric, intermediate, and distal chromosome positions, respectively, exhibit a different chromosome position in N. caerulescens by comparison with the ancestral karyotype (PCK), in addition to higher expression in N. caerulescens than in A. thaliana (Fig. 4A). Interestingly, all of these genes are located in a more distal chromosome position in N. caerulescens than in the PCK (Fig. 4A).
Figure 4.
Relationships between genomic position and metal homeostasis gene expression in N. caerulescens. A, Each white bar shows the proportion of genes expressed at higher levels in N. caerulescens than in A. thaliana, out of all metal homeostasis genes located in the respective chromosome region in N. caerulescens (centromeric, intermediate, or terminal; van de Mortel et al., 2006, 2008). For these genes, black bars show the subset of genes positioned in a different chromosome region in N. caerulescens than in the ancestral karyotype. The length of each chromosome arm was divided into three approximately equally sized regions (centromeric, intermediate, or terminal) based on the assembly physical ruler approach according to Figure 1A. B to D, Relative frequency distribution of chromosome position ΔRDC between N. caerulescens and A. thaliana for different groups of genes. Data are given in black/gray for all genes (B–D, for reference) and in red/orange for genes that are more highly expressed in N. caerulescens than in A. thaliana (B), for all metal homeostasis genes (C), and for metal homeostasis genes that are more highly expressed in N. caerulescens than in A. thaliana (D). Shown are percentages of genes, plotted against the ΔRDC between N. caerulescens and A. thaliana. The relative distance of a gene from the centromere (RDC) is defined as the distance of the 5′ end of a given gene from the centromere divided by the total length of the chromosome arm on which the gene is located. For example, ΔRDC of 0 corresponds to identical relative chromosome positions in both species, ΔRDC of 1 corresponds to a centromeric position in A. thaliana and a terminal position in N. caerulescens, and ΔRDC of −1 corresponds to a terminal position in A. thaliana and a centromeric position in N. caerulescens.
Next, we attempted a more quantitative approximation of chromosome positional differences between the N. caerulescens genome and that of A. thaliana for all A. thaliana genes. We used the information provided here (Fig. 1A) and assembled pseudochromosomes of N. caerulescens from genomic blocks taken from the A. thaliana genome assembly in order to obtain approximate physical chromosome positions for all genes in N. caerulescens. We were then able to compute, for each gene, an approximated difference in the relative distance from the centromere between N. caerulescens and A. thaliana. The distribution of genes that are more highly expressed in N. caerulescens than in A. thaliana was only slightly skewed toward more distal chromosome positions in N. caerulescens than in A. thaliana, when compared with all genes (Fig. 4B; change in relative distance of a gene from the centromere [ΔRDC] between 0.15 and 0.4). In relation to all genes, metal homeostasis genes in general showed an overall trend toward alterations in chromosome positions, with predominantly more proximal positions in N. caerulescens than in A. thaliana (Fig. 4C; ΔRDC between −0.3 and −0.75). In contrast, metal homeostasis genes that are more highly expressed in N. caerulescens were overall positioned more distally on N. caerulescens chromosomes than on those of A. thaliana and also more distally than all metal homeostasis genes and all genes (Fig. 4D; ΔRDC between 0.05 and 0.45 and between 0.7 and 0.9; Fig. 4, A–C).
Across all genes of the N. caerulescens genome regardless of their expression, chromosome positions are predominantly slightly more proximal when compared with A. thaliana (Fig. 4, B–D, peak of ΔRDC at slightly negative values). There was an additional, minor secondary peak of gene frequency at increased distance from the centromere in N. caerulescens (Fig. 4, B–D). Structural rearrangements on the bottom arms of chromosomes NC1 and NC3 predominated among the genes constituting this secondary peak (Fig. 2; Supplemental Fig. S5). The four inversions transforming the acrocentric chromosomes PCK1 and PCK3 into metacentrics NC1 and NC3, respectively, put blocks C (NC1) and H (NC3) into more distal chromosome positions in relation to the respective centromeres (Fig. 2).
Finally, we addressed whether, by comparison with the PCK, a changed genomic environment is associated with altered gene expression in N. caerulescens. Out of all genomic regions that were subject to a change in genomic environment during the evolutionary history of N. caerulescens, block M (NC5) and part of block B (F12K21–F6N18; NC1) showed a significant enrichment for genes more highly expressed in N. caerulescens than in A. thaliana (P < 0.001 and 0.05, respectively; Fisher’s exact test; Supplemental Data Set S3). Both regions were positioned adjacent to the centromere in the ancestral genome and are in a more distal chromosome position in N. caerulescens (Figs. 1B and 2).
DISCUSSION
Navigating a Whole-Genome Assembly Using the N. caerulescens Comparative Genome Map
The Zn/Cd/Ni hyperaccumulator N. caerulescens is one of the most prominent model systems in which to study the uptake and handling of heavy metals in land plants. Here, we provide the detailed structure of the N. caerulescens genome through cross-species in situ hybridization of A. thaliana BAC contigs to N. caerulescens chromosomes. The resulting cytogenetic map of all 14 chromosome arms can navigate a scaffold assembly when the 267-Mb genome of N. caerulescens is sequenced. This approach has been successfully applied for genome assembly in Schrenkiella parvula (Dassanayake et al., 2011), Thellungiella salsuginea (Wu et al., 2012), and Arabis alpina (Willing et al., 2015). In the absence of high-resolution comparative genetic maps for N. caerulescens, our cytogenetic map enables the direct comparison between collinear chromosomes and genome regions among N. caerulescens, A. thaliana, A. halleri, and other crucifer species at a precision of approximately 100 kb.
The N. caerulescens Genome Lacks Large-Scale Structural Intraspecific Variation
Upon divergence from the proto-Calepineae ancestor, all N. caerulescens chromosomes, except NC2, have undergone inversions within or close to the centromere, changing the chromosome symmetry and ancestral orientation of affected genomic regions (Fig. 1). Cytogenetic analysis of 13 different populations of N. caerulescens from contrasting soil types throughout its European distribution range showed a remarkable stasis of inversion rearrangements among the analyzed populations, regardless of the geographic origin, altitude, and soil type. Therefore, the origin of the extant N. caerulescens genome predates the recolonization of large parts of Europe from refugia in southern Europe after the Last Glacial Maximum (Koch et al., 1998; Besnard et al., 2009). However, a comprehensive phylogeographic and genomic study of N. caerulescens and its closest relatives, elucidating the extent of intraspecific genetic variation and historical colonization routes, is much needed (Besnard et al., 2009; M. Koch, personal communication). The intraspecific karyotype stability in N. caerulescens further implies that the adaptation to different metalliferous soils was not associated with large-scale chromosome rearrangements. However, the existing genomic makeup can have an adaptive significance in species-wide traits, in particular the pronounced metal hyperaccumulation and the ability to colonize diverse habitats and edaphic soil types, whereby underdominant de novo inversion rearrangements are probably selected against.
Inversions on Five Chromosomes Predated the Speciation in Genus Noccaea and Coluteocarpeae and Corroborate the Common Ancestry of These Taxa
As the chromosome rearrangements in N. caerulescens appeared to be unique in comparison with five tribes of the extended lineage II (Mandáková and Lysak, 2008), we analyzed karyotypes of two other members of the tribe Coluteocarpeae. We found that the altered structure of five of the seven ancestral chromosomes is shared among N. caerulescens, N. jankae, and R. bulbosa. For PCK6 homologues, the ancestral structure was retained in N. jankae and R. bulbosa, whereas it was altered by three inversions in N. caerulescens. These results suggest that inversion rearrangements on homologues PCK1, PCK3 to PCK5, and PCK7 occurred already in a common ancestor of the genera Noccaea/Raparia and that inversions on the PCK6 homologue are specific to the N. caerulescens lineage. It is noteworthy that the inversion rearrangements on the five chromosomes remained unaltered during the speciation process in the Coluteocarpeae. Moreover, the cytogenomic data corroborate the close relationship between the genera Noccaea and Raparia, shown by the analysis of plastid and internal transcribed spacer sequences (Koch and Al-Shehbaz, 2004), and are not in conflict with the proposed inclusion of the genus Raparia in the genus Noccaea (Al-Shehbaz, 2014).
Pericentric Inversions Have an Impact on the Local Genetic Structure
Our reconstruction of genome evolution from the ancestral PCK karyotype in the three Coluteocarpeae species showed that pericentric inversions were the dominating type of rearrangements in this group. In N. caerulescens, 46% of inversion break points localized into the immediate proximity of centromeres (Fig. 2) and thus into pericentromeric repeat-rich regions that are mainly occupied by tandem repeats, transposons, retrotransposons, pseudogenes, and 5S and 35S rDNA (Arabidopsis Genome Initiative, 2000; Hall et al., 2006). Repeat-rich genomic regions are generally prone to nonallelic homologous recombination, mediating the observed inversion events (Schubert and Lysak, 2011).
Pericentric inversions with one break point close to or within the pericentromere can have a 2-fold consequence. Genes within euchromatic regions are brought into the immediate proximity of pericentromeric heterochromatin, whereas parts of pericentromeric heterochromatin are relocated into juxtaposition with gene-rich regions. It is well established that heterochromatin spreading into neighboring euchromatic regions can cause gene silencing or modify gene expression (Talbert and Henikoff, 2006; Eichten et al., 2012). The opposite process (i.e. relocation of genes from the vicinity of pericentromeric heterochromatin into euchromatic context) may conversely enhance the expression of the relocated genes. Our data here are in agreement with this model. Specifically, this is exemplified by genes involved in metal homeostasis, a process that is strongly altered in N. caerulescens as a result of adaptive evolution. Among metal homeostasis genes, enhanced gene expression in N. caerulescens, especially that of metal homeostasis genes, was associated with overall more distal chromosome positions of genes, or the movement to a more distal chromosome position, when compared with the ancestral karyotype (Fig. 4). Additionally, this is exemplified by the ancestral block M on NC5 and a segment of block B on the bottom arm of chromosome NC1. Both regions show a significantly larger number of genes that display higher expression in a euchromatic region of N. caerulescens than in A. thaliana, where both regions are pericentromeric. Further analysis of changes in the chromosome positions of genes highlighted blocks C and H on the bottom arms of chromosomes NC1 and NC3, respectively, as blocks that are shifted distally by comparison with the PCK genome. Block H contains the HEAVY METAL ATPASE4 (HMA4) gene, which is the functionally most important gene in metal hyperaccumulation and associated hypertolerance of A. halleri known to date (Hanikenne et al., 2008). Ó Lochlainn et al. (2011) showed that HMA4, which is very highly expressed in both A. halleri and N. caerulescens, has undergone similar mutations in N. caerulescens as in A. halleri HMA4 (i.e. cis-activating promoter mutations and gene copy number expansion). In addition to permitting enhanced gene expression, a change in chromosomal position can alter recombination rates, which are known to be nonrandom along chromosomes and usually suppressed in centromeric regions (Mercier et al., 2015). The increased chromosome symmetry due to multiple pericentric inversions might generally contribute to recombination of low-recombining regions when they are brought to more distal, heterochromatin-free positions. Concerted evolution of the three tandem HMA4 gene copies was reported among A. halleri HMA4 gene copies (Hanikenne et al., 2013). This was proposed to result from homologous recombination-mediated repair of somatic double-strand breaks, but illegitimate meiotic recombination (e.g. through single or double crossovers) can also contribute to mutations altering copy number or to sequence exchange between paralogous gene copies, and the tandem HMA4 gene copies of N. caerulescens have not been examined in this regard. Generally, increased recombination rates would also increase the likelihood of gene copy number expansion or contraction through illegitimate recombination.
In the adaptation to changing environments and new ecological niches, a tight linkage of genes into one so-called supergene can become advantageous (Schwander et al., 2014). As the lowered rate of recombination within a supergene is required to ensure its integrity, recombination suppression within inversions reinforces supergene maintenance (Schwander et al., 2014). Examples of supergenes associated with inversions include a plumage polymorphism in the white-throated sparrow (Thomas et al., 2008) and annual and perennial ecotypes in the yellow monkeyflower (Mimulus guttatus; Lowry and Willis, 2010). In the latter case, the inversion polymorphism can be directly implicated in the reproductive isolation and morphological differentiation between the two ecotypes. A candidate region for supergene formation by inversion is block J on NC4, including the Zn transporter-encoding genes ZIP3 and ZIP6. To date, ZIP3 transcript was reported as undetected in N. caerulescens (Assunção et al., 2001), and ZIP6 transcript levels were reported as higher in N. caerulescens than in the nonaccumulator Thlaspi arvense (Hammond et al., 2006). This region is common to all Coluteocarpeae species (Figs. 1 and 3). The two inversions on NC6, leading to supergenes encompassing Cu and organellar metal homeostasis genes on the top chromosome arm, and Zn/Fe homeostasis likely related to metal hyperaccumulation and hypertolerance on the bottom chromosome arm, are specific to N. caerulescens (Figs. 1 and 3). Remarkably, the latter group of genes encompass NAS2, YSL3, and ZIFL1, involved in the biosynthesis of the low-Mr metal chelator nicotianamine (Deinlein et al., 2012) and transmembrane transport of metal-nicotianamine complexes (Gendre et al., 2007) or of nicotianamine alone (Haydon and Cobbett, 2007; Haydon et al., 2012), respectively. Transcript levels of all three genes are elevated in N. caerulescens by comparison with closely related nonaccumulator species (van de Mortel et al., 2006, 2008), and NAS2 has been demonstrated to contribute to Zn hyperaccumulation in A. halleri (Deinlein et al., 2012).
Underdominance of Pericentric Inversions and Their Fixation and Role in Speciation
In both plants and animals (the latter usually represented by Drosophila spp.), paracentric inversions were observed more frequently than pericentric ones (Coyne et al., 1991, 1993; Kirkpatrick, 2010; Auger and Sheridan, 2012). One reason is that small pericentric inversions frequently formed within heterochromatin-rich and low-recombining pericentromeric regions are difficult to detect by genetic mapping or conventional chromosomal analyses (Anderson et al., 2010). Despite being less frequently reported, many pericentric inversions do not need to be underdominant if recombination is suppressed in inverted regions, as shown in Drosophila spp. (Coyne et al., 1991, 1993). Such inversions do not reduce the fertility significantly and become fixed in a population, particularly if inverted regions are small. In the field bean (Vicia faba), a pericentric inversion comprising 20% of the total chromosome length did not compromise pollen fertility, presumably due to the lack of recombination within the inverted region (Sjödin, 1971). As regions rearranged by one or two inversions in N. caerulescens comprise only up to 30% of the total chromosome length, it is possible that the fertility of inversion heterozygotes remained high. Moreover, underdominant chromosome rearrangements can be fixed more easily in short-living plants (annuals and biennials) and particularly in selfers (Charlesworth, 1992; Coyne and Orr, 2004). As inversions impede chromosome pairing and suppress recombination in heterokaryotypes, they are important in restricting gene flow and in promoting lineage sorting, divergence, and speciation (Noor et al., 2001; Rieseberg, 2001; Lowry and Willis, 2010; Nosil and Feder, 2013, and refs. therein). Along the same line, pericentric inversions could have contributed to the reproductive isolation and genetic divergence of the Coluteocarpeae ancestor from its PCK-like predecessor and possibly had a role in environmental adaptation.
Structural rearrangements during the evolutionary history of metal-hyperaccumulating and hypertolerant N. caerulescens predate the origin of this species and are partly shared with closely related species. Rearrangements are unusually rich in inversions known to propel lineage sorting and divergence. The genome of N. caerulescens has undergone global resorting of metal homeostasis functions within linkage groups. As an outcome, recombination between functionally distinct metal homeostasis genes can be predicted to be enhanced, and recombination between functionally related metal homeostasis genes is likely reduced. Furthermore, distal chromosome positions are associated with increased gene expression levels, in particular for metal homeostasis genes. The chromosome rearrangements reported here are outcomes of natural selection, which have occurred during the evolution of the N. caerulescens lineage in a stepwise fashion.
MATERIALS AND METHODS
Plant Material
Noccaea caerulescens, Noccaea jankae, and Raparia bulbosa plants were either collected in the wild or grown from seeds in a growth chamber and cultivated in a greenhouse. Table I shows the origin of all populations analyzed. To induce flowering in plants grown from seed, a cold treatment (3 months at 4°C, 16-h-day/8-h-night cycle) was applied to 1-month-old plants. Young inflorescences from all sampled plants were collected and fixed in freshly prepared fixative (ethanol:acetic acid, 3:1) for several hours or overnight. The fixative was exchanged for 70% (v/v) ethanol, and the material was stored at −20°C until use. For the analysis of heavy metal content in plants collected in the wild, healthy rosette leaves were sampled and desiccated with cobalt-free silica gel, and soil samples from under the same plants were acquired (diameter, approximately 15 cm; depth, approximately 15–20 cm).
Chromosome Preparations, Chromosome Counts, and Genome Size Estimation
Chromosome spreads from fixed young flower buds containing immature anthers were prepared according to the published protocol (Lysak and Mandáková, 2013). Ready-to-use chromosome spreads were checked under a phase contrast for suitable chromosome figures and the amount of cytoplasm. When appropriate, preparations were treated with 100 µg mL−1 RNase (AppliChem) in 2× SSC (20× SSC = 3 m sodium chloride and 300 mm trisodium citrate, pH 7) for 60 min and 0.1 mg mL−1 pepsin (Sigma) in 0.01 m HCl at 37°C for 5 min, postfixed in 4% (v/v) formaldehyde in 2× SSC for 10 min, washed in 2× SSC twice for 5 min, and dehydrated in an ethanol series (70%, 90%, and 100% [v/v], 2 min each). Both mitotic and meiotic (pachytene) chromosome figures were observed under a phase contrast and Olympus BX-61 epifluorescence microscope after counterstaining chromosomes with Vectashield containing DAPI (2 µg mL−1). The genome size of N. caerulescens was estimated by flow cytometry using the protocol described by Doležel et al. (2007) with Solanum pseudocapsicum (2C = 2.59 pg; Temsch et al., 2010) as an internal reference standard.
DNA Probes
The Arabidopsis thaliana BAC clone T15P10 (AF167571) bearing 35S ribosomal RNA gene repeats was used for chromosome in situ localization of nucleolar organizer regions, and the A. thaliana clone pCT4.2 (M65137), corresponding to a 500-bp 5S rDNA repeat, was used for the localization of 5S rDNA loci. The A. thaliana-type telomere repeat (TTTAGGG)n was prepared according to Ijdo et al. (1991). For CCP in species of the Coluteocarpeae, A. thaliana chromosome-specific BAC clones grouped into contigs according to 24 genomic blocks of the ACK (Schranz et al., 2006) were used. BAC contigs followed the limits of genomic blocks as reported by Cheng et al. (2013), except for blocks I and J (I, F7O24 [AC007142]–T19L18 [AC004747]; J, F18A8 [AC003105]–T8I13 [AC002337]). All DNA probes were labeled with Cy3-, biotin-, or digoxigenin-dUTP via nick translation as described by Lysak and Mandáková (2013). Labeled probes were pooled, ethanol precipitated, desiccated, and dissolved in 20 µL of 50% (v/v) formamide and 10% (v/v) dextran sulfate in 2× SSC per slide.
Comparative Chromosome Painting and Microscopy
Twenty microliters of the probe was pipetted on chromosome-containing spread and immediately denatured on a hot plate at 80°C for 2 min. Hybridization was carried out in a moist chamber at 37°C overnight, followed by posthybridization washing in 20% (v/v) formamide in 2× SSC at 42°C. The immunodetection of hapten-labeled probes was performed as described earlier (Lysak and Mandáková, 2013). Chromosomes were counterstained with DAPI in Vectashield. Fluorescence signals were analyzed and photographed with an Olympus BX-61 epifluorescence microscope and AxioCam CCD camera (Zeiss). Monochromatic images were pseudocolored and merged using the Adobe Photoshop CS5 software (Adobe Systems). Pachytene chromosomes in Figure 2 were straightened using the plugin Straighten Curved Objects (Kocsis et al., 1991) in ImageJ (National Institutes of Health).
Multielement Analysis
Aliquots of 1 mL of analytical-grade concentrated HNO3 (65%, w/w) and 3 mL of concentrated HCl (37%, w/w) were added to each subsample of 0.25 g of air-dried soil (sieved to less than 2-mm particle size) in an acid-washed perfluoroalkoxy (TFM) microwave vessel. The suspension was mixed gently by hand and kept at room temperature 20 min before closing the vessels. Digestion was carried out by heating from room temperature to 160°C in 15 min, holding at 160°C for 15 min, and cooling samples down over 45 min in sealed containers in a microwave-assisted chemical digestion system (MARS Xpress; CEM). Samples were filled up to a 10-mL final volume with ultrapure water and filtered through Whatman No. 5951/2 paper before analysis. Air-dried leaf tissues were ground using an acid-washed pestle and mortar. Three milliliters of concentrated HNO3 (65%, w/w) was gently mixed with between 13 and 33 mg of tissue powder in perfluoroalkoxy microwave vessels. Digests were heated from room temperature to 190°C in 20 min, held at 190°C for 15 min, and cooled down over 60 min in sealed containers in a microwave-assisted chemical digestion system (MARS Xpress; CEM). After cooling to room temperature in ambient air, digests were filled up with ultrapure water to a total volume of 10 mL. Multielement analysis was carried out by inductively coupled optical emission spectrometry using the ICAPDuo 6500 system (Thermo Scientific), calibrated with five multielement standard solutions, pipetted from single-element standard solutions (AAS Standards; Bernd Kraft), and one blank solution (Cd, 228.8 nm; Cu, 327.3 nm; Ni, 231.6 nm; and Zn, 213.8 nm for most samples; Zn, 334.5 nm for high-Zn soils). Before and after each set of samples, one blank, one multielement standard solution, and one digest of certified reference material (Virginia tobacco [Nicotiana tabacum] leaves CTA-VTL 2) were measured for quality control.
Genome-Wide in Silico Analysis
Given a list of known metal homeostasis genes and their positions in genomic blocks (Supplemental Data Set S1), we calculated the percentage of genes assigned to each individual metal, for example Cu, out of all metal homeostasis genes in a given genomic block. We then compared this percentage with the overall genome-wide percentage of Cu-related genes out of all metal homeostasis genes, applying Fisher’s exact test to identify statistically significant differences. Similarly, we calculated the percentage of genes that are more highly expressed in N. caerulescens than in A. thaliana (referred to as differentially expressed genes) out of all genes in a given genomic block, and we compared this with the percentage of differentially expressed genes summed over all genomic blocks of the genome. The differentially expressed genes list was obtained from previously published microarray data (van de Mortel et al., 2006, 2008). Because of variation in sequence divergence among genes of N. caerulescens, cross-species microarray hybridization data cannot be employed to reliably identify genes that are expressed at lower levels in N. caerulescens than in A. thaliana.
To calculate the RDC, the number of nucleotides between the 5′ end of the gene and the centromere was divided by the total length of the chromosome arm on which the gene is located. The total lengths of NC (N. caerulescens) and PCK chromosome arms were estimated as the sum of their constituent A. thaliana genomic blocks as shown in Figure 2. Subtracting the RDC value of a gene in A. thaliana from its RDC value in NC/PCK provided the ΔRDC of a gene in NC/PCK relative to A. thaliana, as shown in Figure 4, B to D, and Supplemental Figure S5.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Geographic locations of N. caerulescens populations analyzed in this study.
Supplemental Figure S2. Phylogenetic relationships between Coluteocarpeae taxa analyzed in this study.
Supplemental Figure S3. Comparative structure of R. bulbosa and N. jankae chromosomes as inferred by CCP.
Supplemental Figure S4. Fluorescence in situ localization of 5S (green) and 35S (red) rDNA loci on chromosomes in N. caerulescens, N. jankae, and R. bulbosa.
Supplemental Figure S5. Distribution of genes and chromosome position differences with respect to A. thaliana along chromosomes of N. caerulescens and the ancestral PCK genome.
Supplemental Data Set S1. List of heavy metal homeostasis genes.
Supplemental Data Set S2. Distribution of heavy metal homeostasis genes among 24 genomic blocks in the N. caerulescens genome.
Supplemental Data Set S3. Differentially expressed genes in N. caerulescens as compared with the A. thaliana genome.
Supplementary Material
Acknowledgments
We thank Roger D. Reeves for helpful information on French populations of N. caerulescens and valuable comments on the article, Guillaume Besnard and Torbjörn Säll for providing seeds of Swiss and Swedish populations of N. caerulescens, respectively, Marcus A. Koch for seeds of R. bulbosa and insightful comments, Petra Düchting for carrying out sample preparation and measurements for multielement analyses, Petra Hloušková for Circos visualizations, and Pavel Trávníček for genome size estimation.
Glossary
- Zn
zinc
- Cd
cadmium
- Ni
nickel
- CCP
comparative chromosome painting
- BAC
bacterial artificial chromosome
- ACK
ancestral crucifer karyotype
- PCK
proto-Calepineae karyotype
- rDNA
ribosomal DNA
- Cu
copper
- CI
centromeric index
- Fe
iron
- RDC
relative distance of a gene from the centromere
- ΔRDC
change in relative distance of a gene from the centromere
- DAPI
4′,6-diamidino-2-phenylindole
- PCK
proto-Calepineae Karyotype
- ACK
Ancestral Crucifer Karyotype
Footnotes
This work was supported by the Czech Science Foundation (excellence cluster grant no. P501/12/G090 to M.A.L.), the European Social Fund projects (grant no. CZ.1.07/2.3.00/30.0037 to T.M. and grant no. CZ.1.07/2.3.00/20.0189 to M.A.L.), and the Deutsch Forschungsgemeinschaft (SPP1529 ADAPTOMICS grant no. Kr1967/10–1 to V.S. and U.K.).
Articles can be viewed without a subscription.
References
- Al-Shehbaz IA. (2012) A generic and tribal synopsis of the Brassicaceae (Cruciferae). Taxon 61: 931–954 [Google Scholar]
- Al-Shehbaz IA. (2014) A synopsis of the genus Noccaea (Coluteocarpeae, Brassicaceae). Harv Pap Bot 19: 25–51 [Google Scholar]
- Anderson LK, Covey PA, Larsen LR, Bedinger P, Stack SM (2010) Structural differences in chromosomes distinguish species in the tomato clade. Cytogenet Genome Res 129: 24–34 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796. [DOI] [PubMed] [Google Scholar]
- Artelari R. (2002) Thlaspi L. In Strid A, Tan K, eds, Flora Hellenica, Vol 2 ARG Gantner Verlag, Koenigstein, Germany, pp 253–261 [Google Scholar]
- Assunção AGL, Da Costa Martins P, De Folter S, Vooijs R, Schat H, Aarts MGM (2001) Elevated expression of metal transporter genes in three accessions of the metal hyperaccumulator Thlaspi caerulescens. Plant Cell Environ 24: 217–226 [Google Scholar]
- Assunção AGL, Pieper B, Vromans J, Lindhout P, Aarts MG, Schat H (2006) Construction of a genetic linkage map of Thlaspi caerulescens and quantitative trait loci analysis of zinc accumulation. New Phytol 170: 21–32 [DOI] [PubMed] [Google Scholar]
- Assunção AGL, Ten Bookum WM, Nelissen HJM, Vooijs R, Schat H, Ernst WHO (2003) Differential metal-specific tolerance and accumulation patterns among Thlaspi caerulescens populations originating from different soil types. New Phytol 159: 411–419 [DOI] [PubMed] [Google Scholar]
- Auger DL, Sheridan WF (2012) Plant chromosomal deletions, insertions, and rearrangements. In Bass HW, Birchler JA, eds, Plant Cytogenetics. Springer, New York, pp 3–36 [Google Scholar]
- Banásová V, Horak O, Nadubinská M, Čiamporová M, Lichtscheidl I (2008) Heavy metal content in Thlaspi caerulescens J. et C. Presl growing on metalliferous and non-metalliferous soils in central Slovakia. Int J Environ Pollut 33: 133–145 [Google Scholar]
- Becher M, Talke IN, Krall L, Krämer U (2004) Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J 37: 251–268 [DOI] [PubMed] [Google Scholar]
- Besnard G, Basic N, Christin PA, Savova-Bianchi D, Galland N (2009) Thlaspi caerulescens (Brassicaceae) population genetics in western Switzerland: is the genetic structure affected by natural variation of soil heavy metal concentrations? New Phytol 181: 974–984 [DOI] [PubMed] [Google Scholar]
- Charlesworth B. (1992) Evolutionary rates in partially self-fertilizing species. Am Nat 140: 126–148 [DOI] [PubMed] [Google Scholar]
- Cheng F, Mandáková T, Wu J, Xie Q, Lysak MA, Wang X (2013) Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell 25: 1541–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Aulard S, Berry A (1991) Lack of underdominance in a naturally occurring pericentric inversion in Drosophila melanogaster and its implications for chromosome evolution. Genetics 129: 791–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Meyers W, Crittenden AP, Sniegowski P (1993) The fertility effects of pericentric inversions in Drosophila melanogaster. Genetics 134: 487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA (2004) Speciation. Sinauer, Sunderland, MA [Google Scholar]
- Dassanayake M, Oh DH, Haas JS, Hernandez A, Hong H, Ali S, Yun DJ, Bressan RA, Zhu JK, Bohnert HJ, et al. (2011) The genome of the extremophile crucifer Thellungiella parvula. Nat Genet 43: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinlein U, Weber M, Schmidt H, Rensch S, Trampczynska A, Hansen TH, Husted S, Schjoerring JK, Talke IN, Krämer U, et al. (2012) Elevated nicotianamine levels in Arabidopsis halleri roots play a key role in zinc hyperaccumulation. Plant Cell 24: 708–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniau AX, Pieper B, Ten Bookum WM, Lindhout P, Aarts MGM, Schat H (2006) QTL analysis of cadmium and zinc accumulation in the heavy metal hyperaccumulator Thlaspi caerulescens. Theor Appl Genet 113: 907–920 [DOI] [PubMed] [Google Scholar]
- Doležel J, Greilhuber J, Suda J (2007) Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc 2: 2233–2244 [DOI] [PubMed] [Google Scholar]
- Eichten SR, Ellis NA, Makarevitch I, Yeh CT, Gent JI, Guo L, McGinnis KM, Zhang X, Schnable PS, Vaughn MW, et al. (2012) Spreading of heterochromatin is limited to specific families of maize retrotransposons. PLoS Genet 8: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escarré J, Lefèbvre C, Gruber W, Leblanc M, Lepart J, Rivière Y, Delay B (2000) Zinc and cadmium hyperaccumulation by Thlaspi caerulescens from metalliferous and nonmetalliferous sites in the Mediterranean area: implications for phytoremediation. New Phytol 145: 429–437 [DOI] [PubMed] [Google Scholar]
- Franzke A, Lysak MA, Al-Shehbaz IA, Koch MA, Mummenhoff K (2011) Cabbage family affairs: the evolutionary history of Brassicaceae. Trends Plant Sci 16: 108–116 [DOI] [PubMed] [Google Scholar]
- Gendre D, Czernic P, Conéjéro G, Pianelli K, Briat JF, Lebrun M, Mari S (2007) TcYSL3, a member of the YSL gene family from the hyper-accumulator Thlaspi caerulescens, encodes a nicotianamine-Ni/Fe transporter. Plant J 49: 1–15 [DOI] [PubMed] [Google Scholar]
- Hall AE, Kettler GC, Preuss D (2006) Dynamic evolution at pericentromeres. Genome Res 16: 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Bowen HC, White PJ, Mills V, Pyke KA, Baker AJ, Whiting SN, May ST, Broadley MR (2006) A comparison of the Thlaspi caerulescens and Thlaspi arvense shoot transcriptomes. New Phytol 170: 239–260 [DOI] [PubMed] [Google Scholar]
- Hanikenne M, Kroymann J, Trampczynska A, Bernal M, Motte P, Clemens S, Krämer U (2013) Hard selective sweep and ectopic gene conversion in a gene cluster affording environmental adaptation. PLoS Genet 9: e1003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanikenne M, Nouet C (2011) Metal hyperaccumulation and hypertolerance: a model for plant evolutionary genomics. Curr Opin Plant Biol 14: 252–259 [DOI] [PubMed] [Google Scholar]
- Hanikenne M, Talke IN, Haydon MJ, Lanz C, Nolte A, Motte P, Kroymann J, Weigel D, Krämer U (2008) Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature 453: 391–395 [DOI] [PubMed] [Google Scholar]
- Haydon MJ, Cobbett CS (2007) A novel major facilitator superfamily protein at the tonoplast influences zinc tolerance and accumulation in Arabidopsis. Plant Physiol 143: 1705–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon MJ, Kawachi M, Wirtz M, Hillmer S, Hell R, Krämer U (2012) Vacuolar nicotianamine has critical and distinct roles under iron deficiency and for zinc sequestration in Arabidopsis. Plant Cell 24: 724–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijdo JW, Wells RA, Baldini A, Reeders ST (1991) Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res 19: 4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer M, Schmickl R, German DA, Mandáková T, Lysak MA, Al-Shehbaz IA, Franzke A, Mummenhoff K, Stamatakis A, Koch MA (2014) BrassiBase: introduction to a novel knowledge database on Brassicaceae evolution. Plant Cell Physiol 55: e3. [DOI] [PubMed] [Google Scholar]
- Király G, Ferakova V, Mereďa P, Hodálová I, Eliáš P (2013) Thlaspi jankae: the IUCN red list of threatened species, version 2014.2. www.iucnredlist.org. (September 10, 2014)
- Kirkpatrick M. (2010) How and why chromosome inversions evolve. PLoS Biol 8: e1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M, Al-Shehbaz IA (2004) Taxonomic and phylogenetic evaluation of the American “Thlaspi” species: identity and relationship to the Eurasian genus Noccaea (Brassicaceae). Syst Bot 29: 375–384 [Google Scholar]
- Koch M, Mummenhoff K (2001) Thlaspi s.str. (Brassicaceae) versus Thlaspi s.l.: morphological and anatomical characters in the light of ITS nrDNA sequence data. Plant Syst Evol 227: 209–225 [Google Scholar]
- Koch M, Mummenhoff K, Hurka H (1998) Systematics and evolutionary history of heavy metal tolerant Thlaspi caerulescens in western Europe: evidence from genetic studies based on isozyme analysis. Biochem Syst Ecol 26: 823–838 [Google Scholar]
- Koch MA, German DA (2013) Taxonomy and systematics are key to biological information: Arabidopsis, Eutrema (Thellungiella), Noccaea and Schrenkiella (Brassicaceae) as examples. Front Plant Sci 4: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocsis E, Trus BL, Steer CJ, Bisher ME, Steven AC (1991) Image averaging of flexible fibrous macromolecules: the clathrin triskelion has an elastic proximal segment. J Struct Biol 107: 6–14 [DOI] [PubMed] [Google Scholar]
- Krämer U. (2010) Metal hyperaccumulation in plants. Annu Rev Plant Biol 61: 517–534 [DOI] [PubMed] [Google Scholar]
- Krämer U, Sinclair SA (2012) The zinc homeostasis network of land plants. Biochim Biophys Acta 1823: 1553–1567 [DOI] [PubMed] [Google Scholar]
- Lowry DB, Willis JH (2010) A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol 8: e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Mandáková T (2013) Analysis of plant meiotic chromosomes by chromosome painting. Methods Mol Biol 990: 13–24 [DOI] [PubMed] [Google Scholar]
- Maestri E, Pirondini A, Visioli G, Marmiroli N (2013) Trade-off between genetic variation and ecological adaptation of metallicolous and non-metallicolous Noccaea and Thlaspi species. Environ Exp Bot 96: 1–10 [Google Scholar]
- Mandáková T, Lysak MA (2008) Chromosomal phylogeny and karyotype evolution in x=7 crucifer species (Brassicaceae). Plant Cell 20: 2559–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M (2015) The molecular biology of meiosis in plants. Annu Rev Plant Biol 66: 5.1–5.31 [DOI] [PubMed] [Google Scholar]
- Meyer FK. (2001) Kritische Revision der “Thlaspi”: Arten Europas, Afrikas und Vorderasiens. Spezieller Teil. I. Thlaspi L. Haussknechtia 8: 3–42 [Google Scholar]
- Meyer FK. (2006a) Kritische Revision der “Thlaspi”: Arten Europas, Afrikas und Vorderasiens: Spezieller Teil. VIII. Raparia F.K. Mey. Haussknechtia 11: 196–206 [Google Scholar]
- Meyer FK. (2006b) Kritische Revision der “Thlaspi”: Arten Europas, Afrikas und Vorderasiens. Spezieller Teil. IX. Noccaea Moench. Haussknechtia 12: 1–343 [Google Scholar]
- Noor MAF, Grams KL, Bertucci LA, Reiland J (2001) Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci USA 98: 12084–12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Feder JL (2013) Genome evolution and speciation: toward quantitative descriptions of pattern and process. Evolution 67: 2461–2467 [DOI] [PubMed] [Google Scholar]
- Ó Lochlainn S, Bowen HC, Fray RG, Hammond JP, King GJ, White PJ, Graham NS, Broadley MR (2011) Tandem quadruplication of HMA4 in the zinc (Zn) and cadmium (Cd) hyperaccumulator Noccaea caerulescens. PLoS ONE 6: e17814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard AJ, Reeves RD, Baker AJ (2014) Facultative hyperaccumulation of heavy metals and metalloids. Plant Sci 217-218: 8–17 [DOI] [PubMed] [Google Scholar]
- Reeves RD. (1988) Nickel and zinc accumulation by species of Thlaspi L., Cochlearia L. and other genera of the Brassicaceae. Taxon 37: 309–318 [Google Scholar]
- Reeves RD, Brooks RR (1983) European species of Thlaspi L. (Cruciferae) as indicators of nickel and zinc. J Geochem Explor 18: 275–283 [Google Scholar]
- Reeves RD, Schwartz C, Morel JL, Edmonson J (2001) Distribution and metal-accumulating behaviour of Thlaspi caerulescens and associated metallophytes in France. Int J Phytoremediation 3: 145–172 [Google Scholar]
- Rieseberg LH. (2001) Chromosomal rearrangements and speciation. Trends Ecol Evol 16: 351–358 [DOI] [PubMed] [Google Scholar]
- Schranz ME, Lysak MA, Mitchell-Olds T (2006) The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci 11: 535–542 [DOI] [PubMed] [Google Scholar]
- Schubert I, Lysak MA (2011) Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet 27: 207–216 [DOI] [PubMed] [Google Scholar]
- Schwander T, Libbrecht R, Keller L (2014) Supergenes and complex phenotypes. Curr Biol 24: R288–R294 [DOI] [PubMed] [Google Scholar]
- Sjödin J. (1971) Induced paracentric and pericentric inversions in Vicia faba L. Hereditas 67: 39–54 [Google Scholar]
- Talbert PB, Henikoff S (2006) Spreading of silent chromatin: inaction at a distance. Nat Rev Genet 7: 793–803 [DOI] [PubMed] [Google Scholar]
- Talke IN, Hanikenne M, Krämer U (2006) Zinc-dependent global transcriptional control, transcriptional deregulation, and higher gene copy number for genes in metal homeostasis of the hyperaccumulator Arabidopsis halleri. Plant Physiol 142: 148–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temsch EM, Greilhuber J, Krisai R (2010) Genome size in liverworts. Preslia 82: 63–80 [Google Scholar]
- Thomas JW, Cáceres M, Lowman JJ, Morehouse CB, Short ME, Baldwin EL, Maney DL, Martin CL (2008) The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotrichia albicollis) is a complex rearrangement and suppressor of recombination. Genetics 179: 1455–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomainen M, Tervahauta A, Hassinen V, Schat H, Koistinen KM, Lehesranta S, Rantalainen K, Häyrinen J, Auriola S, Anttonen M, et al. (2010) Proteomics of Thlaspi caerulescens accessions and an inter-accession cross segregating for zinc accumulation. J Exp Bot 61: 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Mortel JE, Almar Villanueva L, Schat H, Kwekkeboom J, Coughlan S, Moerland PD, Ver Loren van Themaat E, Koornneef M, Aarts MGM (2006) Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol 142: 1127–114716998091 [Google Scholar]
- van de Mortel JE, Schat H, Moerland PD, Ver Loren van Themaat E, van der Ent S, Blankestijn H, Ghandilyan A, Tsiatsiani S, Aarts MGM (2008) Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant Cell Environ 31: 301–324 [DOI] [PubMed] [Google Scholar]
- Visioli G, Vincenzi S, Marmiroli M, Marmiroli N (2012) Correlation between phenotype and proteome in the Ni hyperaccumulator Noccaea caerulescens subsp. caerulescens. Environ Exp Bot 77: 156–164 [Google Scholar]
- Weber M, Harada E, Vess C, Roepenack-Lahaye E, Clemens S (2004) Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyperaccumulation factors. Plant J 37: 269–281 [DOI] [PubMed] [Google Scholar]
- Willing EM, Rawat V, Mandáková T, Maumus F, James GV, Nordström KJV, Becker C, Warthmann N, Chica C, Szarzynska B, et al. (2015) Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nature Plants 1: 14023 [DOI] [PubMed] [Google Scholar]
- Wu HJ, Zhang Z, Wang JY, Oh DH, Dassanayake M, Liu B, Huang Q, Sun HX, Xia R, Wu Y, et al. (2012) Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc Natl Acad Sci USA 109: 12219–12224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing JP, Jiang RF, Ueno D, Ma JF, Schat H, McGrath SP, Zhao FJ (2008) Variation in root-to-shoot translocation of cadmium and zinc among different accessions of the hyperaccumulators Thlaspi caerulescens and Thlaspi praecox. New Phytol 178: 315–325 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.