A cytoplamic calcineurin-like-dependent protein kinase affects salt and osmotic stress responses by preferentially localizing to the vacuolar membrane under stress.
Abstract
The role of calcium-mediated signaling has been extensively studied in plant responses to abiotic stress signals. Calcineurin B-like proteins (CBLs) and CBL-interacting protein kinases (CIPKs) constitute a complex signaling network acting in diverse plant stress responses. Osmotic stress imposed by soil salinity and drought is a major abiotic stress that impedes plant growth and development and involves calcium-signaling processes. In this study, we report the functional analysis of CIPK21, an Arabidopsis (Arabidopsis thaliana) CBL-interacting protein kinase, ubiquitously expressed in plant tissues and up-regulated under multiple abiotic stress conditions. The growth of a loss-of-function mutant of CIPK21, cipk21, was hypersensitive to high salt and osmotic stress conditions. The calcium sensors CBL2 and CBL3 were found to physically interact with CIPK21 and target this kinase to the tonoplast. Moreover, preferential localization of CIPK21 to the tonoplast was detected under salt stress condition when coexpressed with CBL2 or CBL3. These findings suggest that CIPK21 mediates responses to salt stress condition in Arabidopsis, at least in part, by regulating ion and water homeostasis across the vacuolar membranes.
Drought and salinity cause osmotic stress in plants and severely affect crop productivity throughout the world. Plants respond to osmotic stress by changing a number of cellular processes (Xiong et al., 1999; Xiong and Zhu, 2002; Bartels and Sunkar, 2005; Boudsocq and Lauriére, 2005). Some of these changes include activation of stress-responsive genes, regulation of membrane transport at both plasma membrane (PM) and vacuolar membrane (tonoplast) to maintain water and ionic homeostasis, and metabolic changes to produce compatible osmolytes such as Pro (Stewart and Lee, 1974; Krasensky and Jonak, 2012). It has been well established that a specific calcium (Ca2+) signature is generated in response to a particular environmental stimulus (Trewavas and Malhó, 1998; Scrase-Field and Knight, 2003; Luan, 2009; Kudla et al., 2010). The Ca2+ changes are primarily perceived by several Ca2+ sensors such as calmodulin (Reddy, 2001; Luan et al., 2002), Ca2+-dependent protein kinases (Harper and Harmon, 2005), calcineurin B-like proteins (CBLs; Luan et al., 2002; Batistič and Kudla, 2004; Pandey, 2008; Luan, 2009; Sanyal et al., 2015), and other Ca2+-binding proteins (Reddy, 2001; Shao et al., 2008) to initiate various cellular responses.
Plant CBL-type Ca2+ sensors interact with and activate CBL-interacting protein kinases (CIPKs) that phosphorylate downstream components to transduce Ca2+ signals (Liu et al., 2000; Luan et al., 2002; Batistič and Kudla, 2004; Luan, 2009). In several plant species, multiple members have been identified in the CBL and CIPK family (Luan et al., 2002; Kolukisaoglu et al., 2004; Pandey, 2008; Batistič and Kudla, 2009; Weinl and Kudla, 2009; Pandey et al., 2014). Involvement of specific CBL-CIPK pair to decode a particular type of signal entails the alternative and selective complex formation leading to stimulus-response coupling (D’Angelo et al., 2006; Batistič et al., 2010).
Several CBL and CIPK family members have been implicated in plant responses to drought, salinity, and osmotic stress based on genetic analysis of Arabidopsis (Arabidopsis thaliana) mutants (Zhu, 2002; Cheong et al., 2003, 2007; Kim et al., 2003; Pandey et al., 2004, 2008; D’Angelo et al., 2006; Qin et al., 2008; Tripathi et al., 2009; Held et al., 2011; Tang et al., 2012; Drerup et al., 2013; Eckert et al., 2014). A few CIPKs have also been functionally characterized by gain-of-function approach in crop plants such as rice (Oryza sativa), pea (Pisum sativum), and maize (Zea mays) and were found to be involved in osmotic stress responses (Mahajan et al., 2006; Xiang et al., 2007; Yang et al., 2008; Tripathi et al., 2009; Zhao et al., 2009; Cuéllar et al., 2010).
In this report, we examined the role of the Arabidopsis CIPK21 gene in osmotic stress response by reverse genetic analysis. The loss-of-function mutant plants became hypersensitive to salt and mannitol stress conditions, suggesting that CIPK21 is involved in the regulation of osmotic stress response in Arabidopsis. These findings are further supported by an enhanced tonoplast targeting of the cytoplasmic CIPK21 through interaction with the vacuolar Ca2+ sensors CBL2 and CBL3 under salt stress condition.
RESULTS
CIPK21 Is Ubiquitously Expressed in Arabidopsis Tissues and Up-Regulated under Stress Conditions
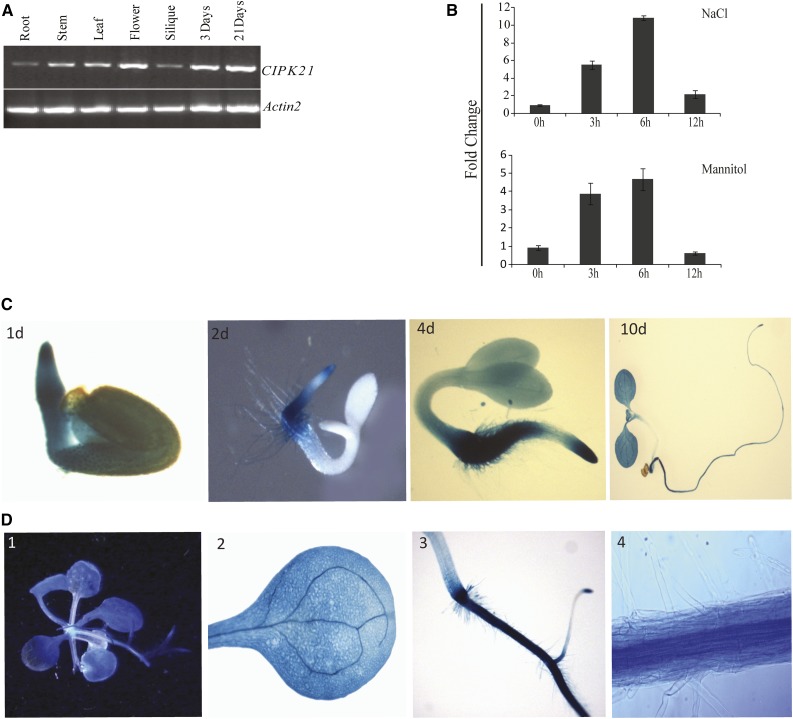
To investigate the function of CIPK21 in planta, the expression profile of CIPK21 was determined by semiquantitative reverse transcription (RT)-PCR analysis in the vegetative growth phase during which stress-related phenotypic assays are usually performed (Fig. 1A). A ubiquitous expression pattern was observed in all examined tissues including root, stem, silique, and flower of 21-d-old Arabidopsis plants and during early seedling development in 3-d-old seedlings (Fig. 1A). In addition, quantitative RT-PCR revealed severalfold increase in CIPK21 transcript in seedlings exposed to NaCl or mannitol compared with water-treated controls (Fig. 1B), indicating that CIPK21 might play a vital role in responses to salinity and osmotic stress. Further, CIPK21 transcript was also highly induced by treatment with polyethylene glycol, abscisic acid (ABA), cold, and drought stress conditions (Supplemental Fig. S1).
Figure 1.
Expression analysis of CIPK21. A, RT-PCR analysis of CIPK21 expression in different organs and during seed germination of Arabidopsis plants. Total RNA was isolated from various tissues (root, stem, leaf, flower, and silique) of the wild-type (Col-0) plants growing under long-day conditions or from germinating seeds and young seedlings (3 and 21 d after sowing). RT-PCR was performed with CIPK21-specific primers and ACTIN2-specific primers. B, Quantitative RT-PCR of CIPK21 in wild-type seedlings under salt and mannitol treatments. Salt (300 mm) and mannitol (400 mm) treatments were given to 3-week-old MS-grown wild-type seedlings in the Petri plate. Tissue was collected at different time intervals, and RNA was isolated to make first-strand cDNA. Quantitative PCR was done using gene-specific forward and reverse primers. ACTIN2 was used to normalize the variance among the samples. Change in transcript level was calculated as fold change with respect to water control, indicated at y axis. ses in the replicates are indicated by error bars. C, Expression of the CIPK21-promoter-β-glucuronidase (uidA) fusion seedlings 1, 2, 4, and 10 d after germination. D, Histochemical GUS activity of transgenic plants expressing uidA reporter gene driven by CIPK21 promoter. Expression pattern in 3-week-old seedlings (1) and individual organ of adult plant (2–4), mature leaves (2), and root (3 and 4, enlarged view).
To further investigate the expression profile of CIPK21, we generated transgenic Arabidopsis plants harboring CIPK21::GUS (CIPK21 promoter::β-glucuronidase) reporter construct and performed GUS staining to determine the promoter activity. During early seedling development, GUS activity was detected in embryos and most of the vegetative tissues (Fig. 1C). Expression in 1-d-old seedling exhibited a strong and uniformly distributed pattern (Fig. 1C, 1d). In 10-d-old seedlings, GUS activity was detected in open cotyledons, less strong in roots, and undetectable in the hypocotyl (Fig. 1C, 10d). Ubiquitous expression was also observed in leaves, and a high expression was detected in vascular tissues (Fig. 1D, 2). In roots, GUS staining was strong in the root tip (Fig. 1D, 3) and vascular tissues (Fig. 1D, 4). The expression pattern of the Arabidopsis CIPK21 gene was described in the publicly available microarray data (Winter et al., 2007; Supplemental Fig. S2), and that supported the RT-PCR and GUS analysis results.
Figure 2.
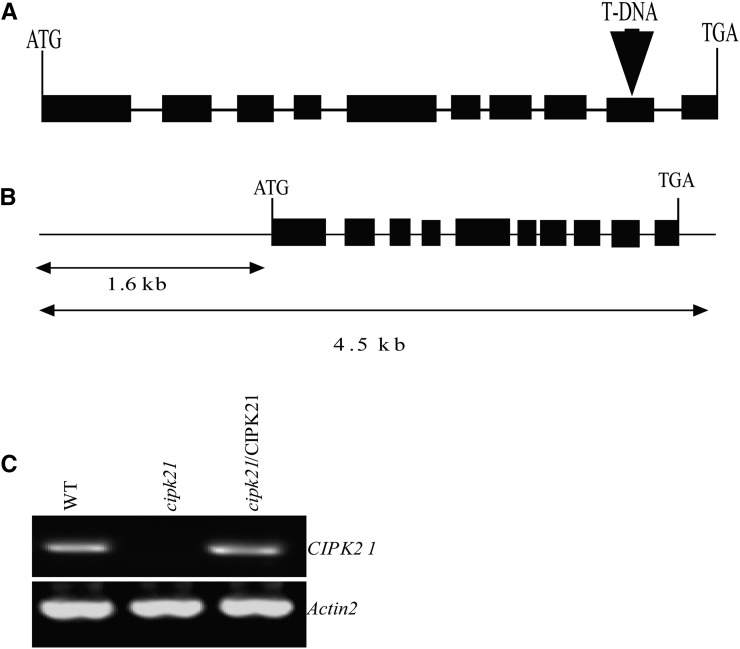
Isolation and complementation of the cipk21 T-DNA insertional mutant. A, Scheme representing the Arabidopsis CIPK21 gene. Exons (black boxes) and introns (lines) are indicated. The position of the T-DNA insertion is indicated by a triangle (not represented to scale). B, Genomic DNA fragment used for complementation. A 4.5-kb fragment including the CIPK21 coding region and 1.6 kb of the 5′ flanking DNA upstream of the start codon and 3′ untranslated region was amplified by PCR and cloned into the pCAMBIA 1300 for plant transformation. C, RT-PCR analysis of CIPK21 gene expression from wild-type (WT), cipk21, and cipk21/CIPK21 plants. Expression of ACTIN2 was analyzed as a loading control.
Figure 3.
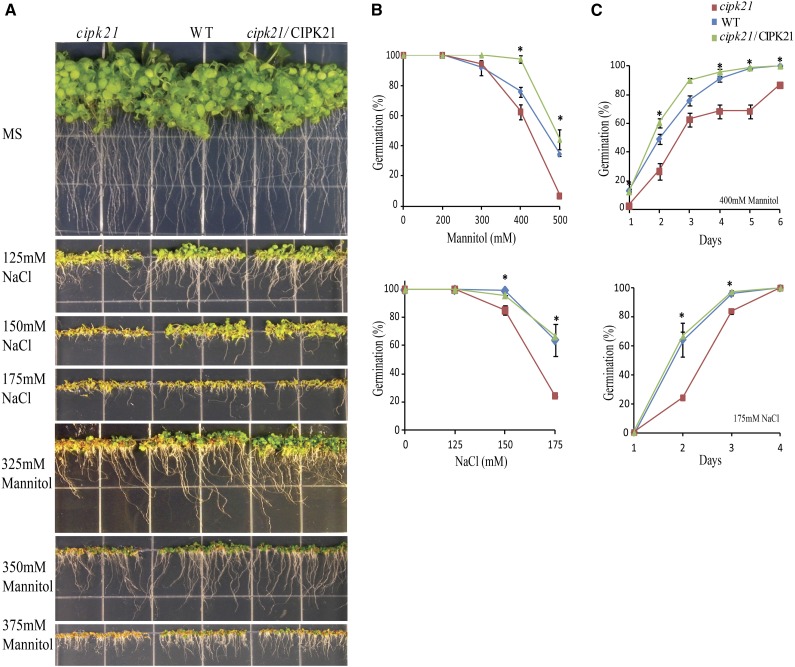
Phenotypic analysis of cipk21 mutant. A, Inhibition of germination and growth of young seedlings in wild-type (WT), cipk21, and cipk21/CIPK21 plants grown on normal MS agar and MS agar containing different concentration of NaCl and mannitol. Seeds were incubated at 4°C for 6 d before transfer to 23°C for germination. The photographs were taken on day 12 after transfer to 23°C. B, Seed germination rate of the wild type, cipk21, and cipk21/CIPK21 in different concentrations of NaCl and mannitol. Germination was scored at 2 d (for NaCl) and 3 d (for mannitol) after incubation at 23°C. C, Kinetics of seed germination for wild-type, cipk21, and cipk21/CIPK21 plants on medium containing 175 mm NaCl or 400 mm mannitol. Results in B and C are presented as average values along with ses from three experiments. The wild type has been indicated by rhombuses, cipk21 by squares, and cipk21/CIPK21 by triangles. *, Significant differences (one-way ANOVA) between the wild type and cipk21 (P < 0.05) in B and C.
Figure 4.
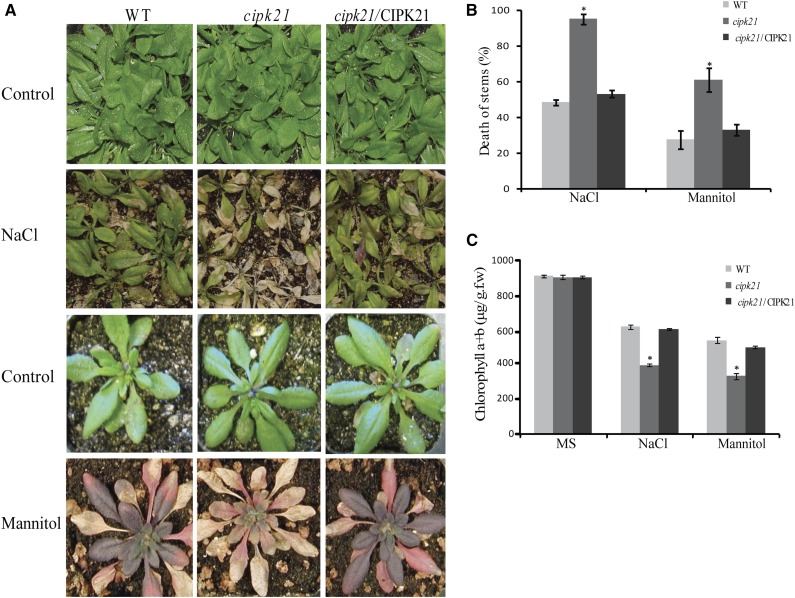
Adult cipk21 mutant plants are hypersensitive to NaCl and mannitol. A, Rosette leaf stage of mature plants of wild-type (WT), cipk21, and cipk21/CIPK21 plants treated with water (top and third rows), 400 mm mannitol (bottom row), or 300 mm NaCl (second row). B, Quantification of death of stem after 300 mm NaCl and 400 mm mannitol treatment. C, Chlorophyll content of plants treated with NaCl and mannitol. Results depicted in B and C are average values ± se of three independent experiments. Three-week-old plants were treated with 300 mm NaCl and 400 mm mannitol three times after every 3 d. Photographs were taken on the 10th day after treatment. *, Significant differences (one-way ANOVA) between the wild type and cipk21 (P < 0.05) in B and C.
Disruption of CIPK21 Gene Expression Renders Plants Hypersensitive to Salt and Osmotic Stresses
For functional characterization of CIPK21, a transfer DNA (T-DNA) insertion mutant allele, GABI-Kat (GK)-838H08-025702 (http://www.GABI-Kat.de), of the CIPK21 (At5g57630) gene was isolated. A wild-type CIPK21 gene contains 10 exons and nine introns, whereas the mutant contained a T-DNA insertion after nucleotide 1,137 within the ninth exon (Fig. 2A). RT-PCR analysis showed that the insertion disrupted the expression of the full-length CIPK21 transcript in the homozygous cipk21 mutant (Fig. 2C). Transgenic complementation lines were produced by introducing the genomic fragment containing 1.6 kb upstream of the start codon (ATG), the complete coding region, and the 3′ untranslated region of CIPK21 into the cipk21 mutant background (Fig. 2B). The expression of CIPK21 was restored in these transgenic plants (Fig. 2C).
The cipk21 mutant and wild-type plants grew well under the normal greenhouse conditions. Abrogation of CIPK21 did not result in any significant morphological difference from the wild type, suggesting that CIPK21 might not affect plant growth and development under normal conditions. The up-regulation of CIPK21 transcript under osmotic stress prompted us to perform the phenotypic analysis of cipk21 mutant under several abiotic stress conditions. These assays indicated hypersensitivity of cipk21 mutant in medium containing high salt or mannitol (Fig. 3A). We observed inhibition of both root growth and emergence of cotyledons in the cipk21 mutant under salt and mannitol stress, where germination of cipk21 seeds was inhibited to a greater extent than that of wild-type seeds or seeds of complemented lines (Fig. 3B). In growth medium with 175 mm NaCl, 64% of wild-type seeds germinated within 2 d, whereas only 24% of cipk21 seeds germinated (Fig. 3B). The germination rate of cipk21 seeds was also reduced (<7%) compared to the wild type (34%) in 500 mm mannitol (Fig. 3B). Likewise, kinetics of seed germination in the presence of 175 mm NaCl and 400 mm mannitol also showed similar difference (Fig. 3C).
To determine the role of CIPK21 in stress regulation in adult plants, analysis of cipk21 mutant, the wild type, and complemented transgenic lines was done under various stress conditions. In the comparative analysis of stress tolerance, cipk21 plants exhibited hypersensitivity to salt and mannitol treatment compared with the wild type and the complemented line (Fig. 4A).
Quantitative analysis showed that the stems of cipk21 plant collapsed more frequently than those in both wild-type and complemented lines (Fig. 4B). Salt and osmotic stress also led to more severe chlorosis in cipk21 mutant leaves compared with the wild type. Measurement of total chlorophyll content indicated that cipk21 mutant leaves retained less chlorophyll than wild-type and complemented lines (Fig. 4C). Taken together, these data support the hypothesis that CIPK21 functions as a positive regulator of salt and drought responses.
CIPK21 Interacts with CBL2 and CBL3
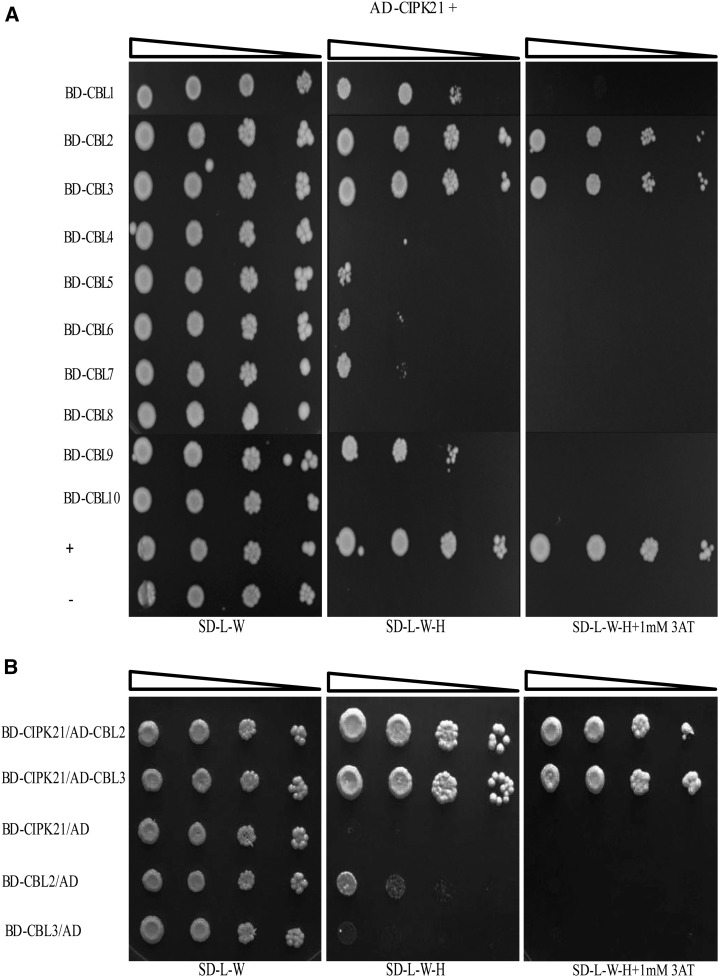
In the CBL-CIPK-mediated Ca2+-signaling pathway, a single CBL or multiple CBLs might decode the Ca2+ signature generated during a particular stress and subsequently target the interacting CIPK. To determine which CBL targets CIPK21, yeast (Saccharomyces cerevisiae) two-hybrid analysis was performed between CIPK21 (in activation domain [AD] vector) and all 10 Arabidopsis CBLs (in binding domain [BD] vector). The yeast cells coexpressing CIPK21 and several CBLs proliferated on the selection medium SD-H-L-W containing 1 mm 3-AT. Stronger interaction was detected with CBL2 and CBL3, and weaker interaction was shown with CBL1 and CBL9 (Fig. 5A). To confirm the interaction, we also performed vector-swapping analysis by generating the AD.CBL2 and AD.CBL3 and BD.CIPK21 constructs. The result further validated the interaction between CBL2/CBL3 with CIPK21 regardless of the vectors expressing the two proteins (Fig. 5B).
Figure 5.
CIPK21 interacts with CBL2 and CBL3 in yeast two-hybrid assay. A, Dilution series of yeast AH109 strains transformed with AD-CIPK21 and all CBLs proteins in BD vectors. Yeast two-hybrid analysis identified CBL2 and CBL3 as predominant CIPK21 interactors. In addition, a weak interaction was also observed with CBL1 and CBL9. Interaction between the full-length AD-CIPK8/BD-CBL1 and AD-CIPK21/BD used as positive and negative controls, respectively. B, Yeast two-hybrid analysis showing vector swapping of CBL2 and CBL3 interaction with CIPK21. Dilution series of yeast AH109 strains transformed with BD-CIPK21 and CBL2 and CBL3 proteins in AD vectors. The combination of plasmids in A and B is indicated on the left, and decreasing cell densities in the dilution series are illustrated by narrowing triangles. Yeast was grown on SD-L-W medium (first column), SD-L-W-H medium (second column), or SD-L-W-H medium containing 1 mm 3-AT (third column).
CIPK21 Colocalizes to the Vacuolar Membrane with CBL2 and CBL3
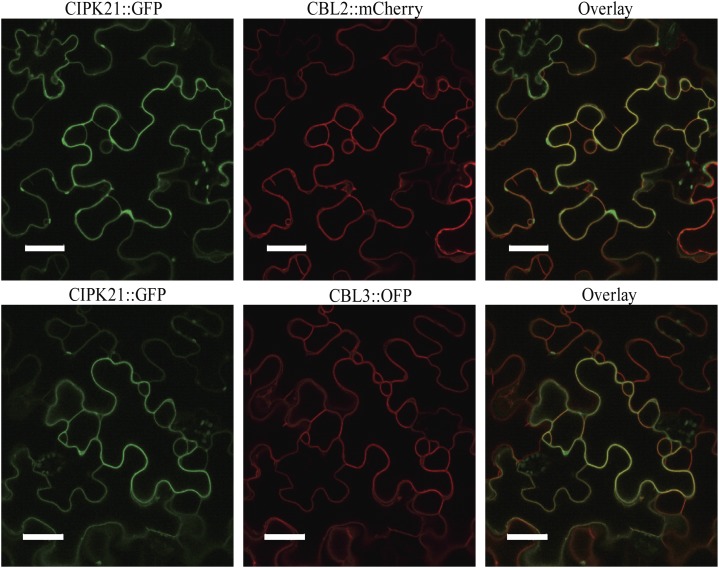
Subcellular localization of a given protein provides a clue for its potential function. To understand the function of CIPK21, we examined subcellular localization of CIPK21 using CIPK21:GFP fusion construct through transient expression in Nicotiana benthamiana leaves. CIPK21:GFP fusion protein exhibited cytosolic and nuclear fluorescence (Supplemental Fig. S3; Batistič et al., 2010), similar to other CIPK members reported earlier (D’Angelo et al., 2006; Cheong et al., 2007; Kim et al., 2007; Batistič et al., 2010). CIPK21 interaction with CBL2 and CBL3 observed in yeast two-hybrid assays was validated by subcellular colocalization of CBL2/CBL3 and CIPK21 in plant cells using CIPK21:GFP cotransformed with either CBL2:mCherry or CBL3:orange fluorescent protein (OFP). CBL2 and CBL3 are known to be localized to tonoplast in both N. benthamiana and Arabidopsis cells (Batistič et al., 2010; Tang et al., 2012). CIPK21, when coexpressed with CBL2 or CBL3, exhibited cytoplasmic and tonoplastic localization, in contrast to cytosolic and nuclear expression of CIPK21 individually (Figs. 6 and 7). In a parallel experiment, no change in subcellular localization of CIPK21:GFP was seen when cotransformed with CBL1n:OFP (PM marker) or two-pore channel1 (TPC1; tonoplast marker; Supplemental Fig. S4).
Figure 6.
Colocalization of CIPK21 with CBL2 and CBL3 proteins in the epidermal peel cells of N. benthamiana. CIPK21:GFP coexpressed with either CBL2:mCherry (top row, red) or CBL3:OFP (bottom row, red) display the formation of CBL2/CIPK21 and CBL3/CIPK21 complexes preferentially at the vacuolar membrane, as indicated by the yellow color in the overlay image (right column). Bar = 40 µm.
Figure 7.
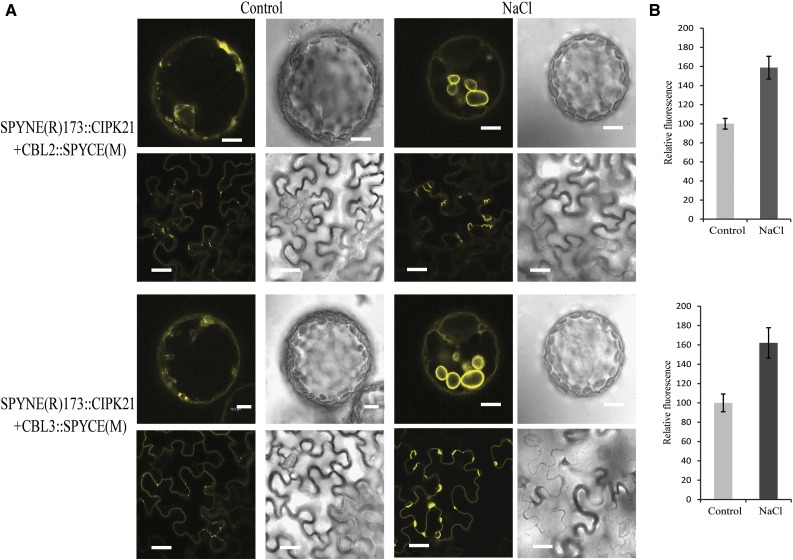
Effects of salt stress on the CBL2/3-CIPK21 BiFC complexes. A, Investigation of interaction of CBL2 and CBL3 with CIPK21 by BiFC in epidermal cells of N. benthamiana. CBL2/CBL3-CIPK21 complexes formed mostly at the vacuolar membrane as reported for individual CBL2 and CBL3 (left columns). Upon stress treatment with 125 mm NaCl, large tonoplastic vesicles were observed compared with control for both CBL2-CIPK21 and CBL3-CIPK21 (right columns) as shown in protoplasts (top row; bars = 10 µm) and intact epidermal cells (bottom row; bars = 40 µm). Plasmid combinations are indicated on the left. B, Quantification of the relative fluorescence intensity to monitor the effects of salt stress treatment on BiFC complexes formed by N-terminal of YFP (YN)-CIPK21/CBL2-C-terminal of YFP (YC; above) and YN-CIPK21/CBL3-YC (below), which were incubated with 125 mm NaCl or control (10 mm MES, pH 5.6, and 10 mm MgCl2) medium. Results are presented as average values along with ses from three experiments.
Differential Localization of CBL2/CBL3-CIPK21 Complexes under Salt Stress
To further elucidate the in vivo interaction of CIPK21 with CBL2 or CBL3, CIPK21 was fused to the N-terminal fragment of yellow fluorescent protein (YFP), and CBL2 and CBL3 were fused to the C-terminal fragment of YFP for bimolecular fluorescence complementation (BiFC) analysis. The BiFC constructs were coinfiltrated in the N. benthamiana leaves followed by confocal microscopy, which revealed that both CBL2:CIPK21 and CBL3:CIPK21 complexes were mostly present on the vacuolar membranes. In addition to this, we also observed BiFC signal for the CBL6 and CIPK21 pair. However, no fluorescence was observed upon coexpression of CBL5 or CBL7 with CIPK21 (Supplemental Fig. S5). Even though the expression pattern and phenotypic analysis of CIPK21 suggested its possible role in osmotic and salt stress, it was important to understand the function of CBL2- or CBL3-CIPK21 complex formation under specific abiotic stress conditions. To address the relevance of these interactions in a stress-signaling pathway, we determined the subcellular localization of the CBL:CIPK complex under salt stress. Upon salt stress treatment, increase in relative fluorescence intensity of CBL2:CIPK21 or CBL3:CIPK21 was observed on tonoplast compared with the untreated controls as depicted in the BiFC signal in epidermal cells and the protoplast of N. benthamiana (Fig. 7). The control pGPTV-II-BAR:35S::mVENUS vector showed no change in the fluorescence intensities under salt stress (Supplemental Fig. S6).
DISCUSSION
The role of cellular Ca2+ in stress signal transduction in plants is well established (Knight et al., 1996, 1997; Sanders et al., 1999, 2002; Knight and Knight, 2000; Rudd and Franklin-Tong, 2001; Luan et al., 2002; Luan, 2009; Steinhorst and Kudla, 2014). CBLs and CIPKs form complexes that decode the Ca2+ changes in plant responses to stress signals (Liu et al., 2000; Chikano et al., 2001; Gong et al., 2002; Kim et al., 2003; Pandey, 2008; Luan, 2009; Weinl and Kudla, 2009; Kudla et al., 2010). Although forward and reverse genetic studies have advanced our understanding of the role of these components in Arabidopsis (Pandey et al., 2004, 2007; D’Angelo et al., 2006; Cheong et al., 2007; Qin et al., 2008; Tripathi et al., 2009; Tang et al., 2012, 2015; Drerup et al., 2013), many of the CIPKs are still not functionally characterized. This study is centered on the characterization of CIPK21 to decipher its possible role in the regulation of osmotic and salt stress responses in Arabidopsis. CIPK21 is possibly connected with intracellular Ca2+ changes by the Ca2+ sensors CBL2 and CBL3 that interact physically with CIPK21 based on yeast two-hybrid assay, in vivo localization, and BiFC interaction analysis (Figs. 5–7).
The ubiquitous expression of CIPK21 in different tissues matched a pattern observed for CBL2 and CBL3 (Tang et al., 2012). The expression pattern detected by histochemical staining of promoter-CIPK21:GUS plants (Fig. 1) is more similar to promoter-CBL3-GUS (Tang et al., 2012). Differential expression of several other CBL and CIPK genes has provided significant directions to characterize them functionally under one or multiple abiotic stress conditions (Cheong et al., 2003; Kim et al., 2003; Pandey et al., 2004, 2007; D’Angelo et al., 2006). CIPK21 expression was up-regulated by salt, mannitol, ABA, cold, and drought conditions, and this served as an important clue to undertake a detailed functional analysis of this gene. It is quite interesting to note that CIPK21 appears to contribute to osmotic stress responses induced by salt and mannitol, which is very similar to CIPK1 function in plants (D’Angelo et al., 2006). In contrast to cipk21 plants, cipk1 mutant did not respond to salt (NaCl) stress. Interestingly, CIPK1, unlike CIPK21, forms alternate complexes with CBL1 and CBL9 and hence mediates osmotic and ABA responses at the PM.
In response to changing environmental conditions or developmental stages, some regulatory proteins relocate themselves to different subcellular compartments to execute their designated functions. Previously, most of the CIPK family members including CIPK21 were reported to be located in the cytoplasm and the nucleus (Cheong et al., 2007; Kim et al., 2007; Batistič et al., 2010). Importantly, it has been shown that some of the CIPKs such as CIPK1, CIPK14, CIPK23, and CIPK24 are targeted to different locations in the cell through interaction with CBL partners (D’Angelo et al., 2006; Xu et al., 2006; Cheong et al., 2007; Kim et al., 2007; Waadt et al., 2008; Batistič et al., 2010; Tang et al., 2012, 2015). Coexpressing CIPK21 with CBL2 or CBL3 targets CIPK21 preferentially to the tonoplast (Fig. 6), probably in a manner similar to CIPK1 and CIPK23, both of which are targeted to the PM on interaction with CBL1 and CBL9. Amino acid sequence analysis showed the presence of N-terminal myristoylation and acylation sites in both CBL1 and CBL9, known for targeting proteins to cellular membranes. These sites are responsible for anchoring the CBLs along with the interacting CIPKs to the PM, where the CBL-CIPK complex can phosphorylate the downstream target proteins (D’Angelo et al., 2006; Cheong et al., 2007). This hypothesis is supported by experiments where the NAF (CBL-interacting domain) motif is deleted from CIPKs, leading to abolition of localization of CIPK1 and CIPK23 to the PM (D’Angelo et al., 2006; Cheong et al., 2007; Hashimoto et al., 2012). Subcellular localization of CBL2 and CBL3 to the vacuolar membrane is attributed to S-acylation of three Cys residues at the N terminus as shown for CBL2 (Batistič et al., 2012). Localization of CBL2 and CBL3 to the tonoplast suggests a possible function of CBL2- or CBL3-CIPK21 complexes at the vacuolar membrane in regulating the osmotic stress responses (Batistič et al., 2008, 2010). However, a role for CBL2 and CBL3 has not yet been identified in salt and osmotic stress, as cbl2, cbl3, or cbl2/cbl3 double mutants do not exhibit hypersensitivity to salt stress compared with cipk21, supporting a possible involvement of other CBLs in the functional regulation of CIPK21 (Tang et al., 2012). In addition to CBL2 and CBL3, in the yeast two-hybrid analysis, CIPK21 interacts weakly with CBL1, CBL5, CBL6, CBL7, and CBL9. These weaker interacting CBLs might also form complex with CIPK21 in planta but possibly require higher Ca2+ concentration. The yeast system either may not maintain the high Ca2+ concentration required by a CBL or may not be able to generate transient Ca2+ spikes, a hallmark in plant stress response. In both scenarios, there is possibility of alternative CBLs regulating CIPK21 function. Our hypothesis is supported by the fact that the CBL6:CIPK21 complex yields a strong BiFC signal, even though the yeast two-hybrid shows a very weak interaction. In a recent report, tonoplast-localized protein S-acyl transferase10 (PAT10) was reported to be critical for development and salt tolerance in Arabidopsis (Zhou et al., 2013), evident by salt hypersensitivity of pat10 mutant plants. Moreover, CBL2/CBL3/CBL6 were speculated to be the putative substrates of PAT10 (Zhou et al., 2013). PAT10 regulates the tonoplast localization of CBL2, CBL3, and CBL6 (whose membrane association depends on palmitoylation; Zhou et al., 2013). This possibly suggests the role of targeting of the three CBLs to tonoplast during salt stress.
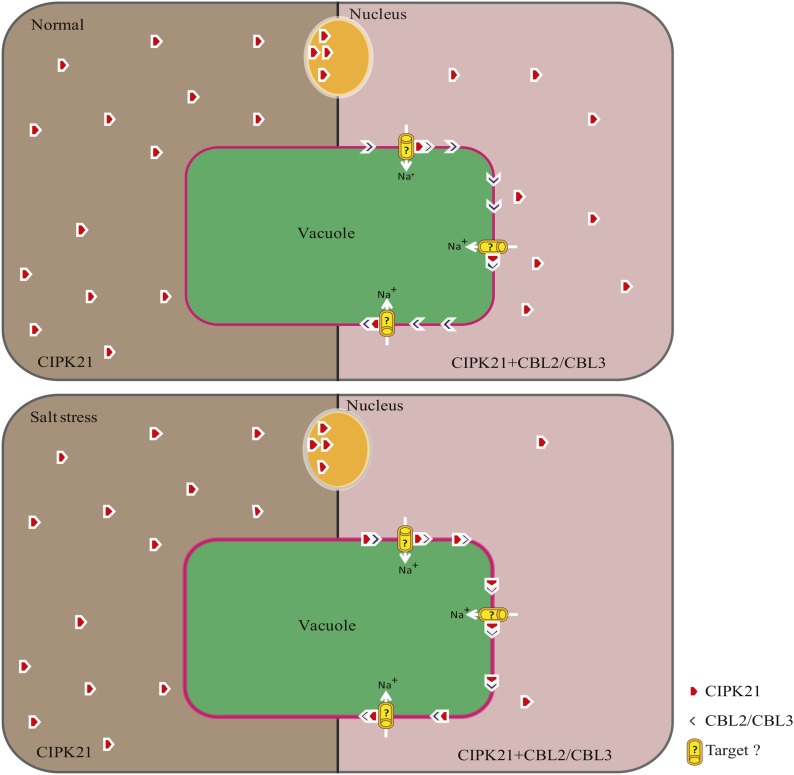
To conclude, although our data here support the notion that, under salt stress, Ca2+ sensor proteins CBL2 or CBL3 might target CIPK21 to the vacuolar membrane to enhance salt tolerance (Fig. 8), we speculate that CIPK21 might also interact with other CBLs that target the kinase possibly to other locations, including PM, under various conditions. Getting targeted to the tonoplast by CBL2 and CBL3 may not be exclusively responsible for CIPK21 function under stress conditions. Identification and characterization of downstream target(s) will certainly enhance our understanding of the mechanism of CIPK21 gene functions and its involvement in stress adaptability.
Figure 8.
Hypothetical model for CIPK21 function in osmotic and salt stress signaling. Under normal (top left) and salt stress conditions (bottom left), CIPK21 is localized in the cytoplasm and the nucleus. Upon coexpression of CBL2 or CBL3, CIPK21-CBL2/CBL3 complex formation initiates in the vacuolar membrane (top right), which is enhanced under salt stress (bottom right). Possibly under salt stress, CBL2/CBL3 detects the rise in cytoplasmic Ca2+, activates CIPKs (CIPK21), and targets them to the tonoplast. Possible targets of CIPK21 may be vacuolar membrane Na+-channels/transporters facilitating Na+ sequestration from cytosol into the vacuole under salt stress, which also might be responsible for the osmoregulation in plant cell.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Stress Treatments
Arabidopsis (Arabidopsis thaliana) ecotype Columbia 0 (Col-0) seeds were treated with isopropanol for 5 min and with 50% (v/v) bleach solution for 15 min, washed five times with sterile water, and plated on one-half-strength Murashige and Skoog (MS) basal medium (Murashige and Skoog, 1962) solidified with 0.8% (w/v) agar for aseptic growth and stratified at 4°C for 4 d under dark conditions. Seeds of Arabidopsis wild-type Col-0 and cipk21 mutant were grown under long-day (16-h-light/8-h-dark cycle) conditions in a growth room until flowering stage for plant transformation and generation of seeds. Three-week-old seedlings grown on one-half-strength (MS) basal medium were subjected to different stress treatments. Salinity and osmotic treatments were given to 3-week-old seedlings (wild type, cipk21, and cipk21/CIPK21) by applying 300 mm NaCl and 400 mm mannitol solution, respectively, on MS plates. For control, sterile water was added to the MS plates, parallel to treated samples. Tissue was harvested at different time points (0, 3, 6, and 12 h) from treated as well as control MS plates and immediately frozen in liquid nitrogen. For ABA treatment, 100 µm (±)-cis, trans-ABA (Sigma) was sprayed onto seedlings to ensure total coverage of the foliage area. The plants were incubated at room temperature under white light. For polyethylene glycol treatment, 30% (w/v) polyethylene glycol 6000 was added to the seedlings on MS plates, and the seedlings were incubated at room temperature under white light. For cold treatment, seedlings were transferred to 4°C conditions in the cold room under white light. For drought treatment, 4-d-old seedlings grown on MS media were exposed in the laminar flow hood for dehydration as described previously (Pandey et al., 2005).
RT-PCR Analysis
Total RNA was isolated with Tripure isolation reagent (Roche Diagnostics). To analyze the gene expression by RT-PCR, total RNA (0.2 μg) from plants was heated to 65°C for 7 min and then subjected to RT reaction using SuperScript II RNase H reverse transcriptase (200 units per reaction; Invitrogen) with oligo(dT) primer for 50 min at 42°C. PCR amplification was performed as follows: initial denaturation at 94°C for 2 min followed by 25 cycles of incubations at 94°C for 20 s, 55°C for 40 s, and 72°C for 1 min, with a final extension at 72°C for 10 min with AmpliTaq polymerase (two units per reaction; Perkin-Elmer). ACTIN2 expression level was used as a quantitative control. Aliquots of individual PCR products were resolved by agarose gel electrophoresis and visualized with ethidium bromide under UV light. All experiments were repeated at least three times, and results from one representative experiment are shown.
For quantitative expression analysis by real-time PCR, total RNA was isolated using TriReagent (Sigma Aldrich) from samples harvested at different time points, both from treated and control MS plates. Two micrograms of DNase-treated total RNA was used to synthesize the first-strand complementary DNA (cDNA) in 50 µL of reaction volume using the High-Capacity cDNA Archive Kit (Applied Biosystems). Primers were synthesized for CIPK21 preferentially, from the 3′ end, employing Primer Express (PE Applied Biosystems) with default settings (Supplemental Table S1). Primers were analyzed using the BLAST tool of National Center for Biotechnology Information for their specificity to the respective gene and were also confirmed by dissociation curve analysis after the PCR reaction (Supplemental Table S1). KAPA SYBR FAST Master Mix (Kapa Biosystems) was used to determine the expression levels of the genes in the ABI Prism 7000 sequence detection system (Applied Biosystems). To normalize the variance among samples, ACTIN2 was used as the endogenous control. Relative expression values were calculated employing the ΔΔ cycle threshold method.
Isolation and Complementation of cipk21 T-DNA Insertion Mutant
The cipk21 mutant (GK-838H08-025702) was isolated from the GABI-Kat Simple Search T-DNA insertion collections (http://www.GABI-Kat.de). The T-DNA insertion was confirmed by CIPK21 gene-specific PCR. For complementation of the cipk21 mutant, a 4.5-kb fragment including the CIPK21 coding region and 1.6-kb 5′-upstream region of the start codon (ATG) was amplified by PCR from Arabidopsis genomic DNA inclusive of restriction sites EcoRI and BamHI. The PCR product was cloned into the binary vector pCAMBIA1300 (CAMBIA). The construct was transformed into Agrobacterium tumefaciens strain GV3103 and introduced into cipk21 mutant plants by the floral dip method (Clough and Bent, 1998). Briefly, A. tumefaciens cells were grown in Luria-Bertani broth for 24 h at 30°C. The cells were collected by centrifugation and resuspended in infiltration medium (one-half-strength MS medium, 5% [w/v] Suc, 1× Gamborg’s vitamins, 0.044 μm benzylaminopurine, and 0.04% [v/v] Silwet L77) to an optical density at 600 nm of 1.5 to 2.0. Plants were dipped into this suspension for 30 s and then transferred to growth room conditions. Transgenic seeds were plated on one-half-strength MS medium containing 0.8% (w/v) agar, 112 mg L–1 Gamborg’s B5 vitamin mixture and 15 mg mL–1 hygromycin. Resistant seedlings were transplanted to soil and grown in the greenhouse to produce seeds.
Analysis of CIPK21 Promoter-GUS Expression in Transgenic Plants
To generate the CIPK21 promoter-GUS construct, the 5′ flanking DNA of CIPK21 coding region was amplified. The 1.6-kb PCR fragment was cloned in pBI101.1 vector (Clontech) using XbaI and BamHI sites. The construct was used to transform wild-type Arabidopsis plants, and transformants were selected on 50 µg mL–1 of kanamycin. T2 transgenic seedlings were stained with 5-bromo-4-chloro-3-indolyl-d-glucuronide for 12 h followed by incubation in 80% (v/v) ethanol to remove chlorophyll (Jefferson et al., 1987).
Yeast Two-Hybrid Interaction Analysis
To examine whether there was a direct physical interaction between the CIPK21 and CBLs, the yeast (Saccharomyces cerevisiae) two-hybrid system was employed. The CIPK21 cDNA was cloned into the activation domain vector (pGAD.GH), and 10 CBL genes were cloned into the DNA-binding domain vector (pGBT9.BS). AD-CIPK21 and each of the BD-CBL plasmids were cotransformed into yeast strain AH109, and cotransformants were selected on synthetic complete medium lacking Trp and Leu (SD-L-W). Spot dilution assays were performed by inoculating transformed yeast cells on a medium lacking Trp and Leu (SD-L-W) and allowed to grow at 28°C (at 200 rpm overnight). These cultures were diluted to obtain an optical density at 600 nm of 0.5. This culture was 10-fold serially diluted, and 5 µL of each of these dilutions was spotted on synthetic complete medium lacking Trp, Leu, and His and supplemented with 1 mm 3-amino-1,2,4-triazole (Sigma) to score growth as an indicator of the protein-protein interaction. Plates were incubated at 28°C for 4 d. The sequences of primers used are listed in Supplemental Table S2, and previously published constructs are mentioned in Supplemental Table S3.
Germination and Phenotype Assay
Approximately 100 seeds each from the wild-type, cipk21 mutant, and cipk21/CIPK21 complemented lines were planted in triplicate on MS medium and MS medium with different concentrations of NaCl and mannitol and incubated at 4°C for 4 d before transferring to 23°C under long-day conditions. Germination (emergence of radicles) was scored daily for 12 d. The percentage value of salt- or mannitol-treated plants versus plants grown under normal conditions was used to represent the data. Phenotypic assays shown in Figure 3A were performed in a similar manner except that the plates were placed vertically on a rack. Plant growth was monitored and photographed after 12 d of a 16-h of light and 8 h of dark cycle.
Salt and Drought Tolerance Analysis
For the salt and drought tolerance assays, 3-week-old plants were subjected to 300 mm NaCl and 400 mm mannitol solution, respectively, for every 3 d and subsequently monitored for chlorosis/bleaching phenotype regularly for the next 2 weeks. Photographs were recorded on the 10th day. All experiments were repeated at least three times, and results from one representative experiment are shown in Figure 4A. For quantitative assessment of stress tolerance assays, chlorophyll was extracted using dimethyl sulfoxide as described earlier (Barnes et al., 1992). Arnon’s equations were used to calculate the chlorophyll contents (Arnon, 1949). In addition, drooping of inflorescence and subsequent drying were considered to be indicators of the deaths of the stems (Kim et al., 2007). Deaths of the stems were also calculated as quantitative measure from the results of three independent experiments. Statistical significance was determined using one-way ANOVA (Excel version 2007, Microsoft).
Subcellular Localization, BiFC, and Quantification of BiFC Signal
All the constructs used in this study were cloned into the pGPTVII backbone vectors (Walter et al., 2004). The complete protein-encoding region of CBL2, CBL3, and CIPK21 were amplified by PCR using a Phusion-Polymerase (Finnzymes) and fused to the coding region of mCherry, OFP, and GFP, respectively. Additionally, for BiFC analysis, CBL2 and CBL3 were fused to the C-terminal fragment of YFP in pSPYCE(M), and CIPK21 was fused to the N-terminal fragment of YFP in pSPYNE173 and pSPYNE(R)173 (Walter et al., 2004; Waadt et al., 2008). All the constructs were verified by sequencing. Primers used in this work are included in Supplemental Table S2. A list of constructs generated previously is included in Supplemental Table S3. Plasmids were transformed into A. tumefaciens (GV3101 pMP90) and transfected to Nicotiana benthamiana leaves as described (Walter et al., 2004; Waadt and Kudla, 2008). For NaCl treatments, the leaf discs of N. benthamiana after 3 d of post-A. tumefaciens infiltration were incubated in control (10 mm MES, pH 5.6, and 10 mm MgCl2) and different doses of salt (i.e. 75, 125, and 175 mm NaCl) for 16 h in growth room conditions. For most of the representation in figures, 125 mm NaCl treatment was used in all the BiFC experiment involving salt stress. The control and salt-treated leaf discs were analyzed under confocal microscope for YFP fluorescence in BiFC assays. Fluorescence microscopy was performed with an inverted microscope (Leica DMIRE2) equipped with the Leica TCS SP2 laser-scanning device (Leica Microsystems). Detection of fluorescence was performed as follows: GFP, excitation at 488 nm, scanning at 500 to 535 nm; YFP (BiFC), excitation at 514 nm, scanning at 525 to 600 nm; mCherry, excitation at 543 nm, scanning at 600 to 630 nm; and OFP, excitation at 543 nm, scanning at 565 to 595 nm. Autofluorescence of plastids was detected at 650 to 720 nm. All images were acquired using a 633/1.20 water immersion objective (HCX PL Apo CS) from Leica. Protoplast preparations were performed according to D’Angelo et al. (2006).
For quantification of BiFC signals of CBL2-CIPK21 or CBL3-CIPK21 interactions under salt stress, a total of 12 independent leaves from different plants of the same age were coinfiltrated for one experimental condition. One separate leaf of each plant was also infiltrated with 0.3 optical density culture having p19, which serves as a negative control. The infiltrated plants were kept under the regime of 16- and 8-h light and dark cycles, respectively, at room temperature conditions. After 2 d of incubation, uniform-size leaf discs from the same leaf were floated in control (buffer: 10 mm MES, pH 5.6, and 10 mm MgCl2) and salt stress conditions (buffer plus 75, 125, and 175 mm NaCl) in a 24-well microtiter plate. All the solutions used for the treatments were prepared in activation buffer without acetosyringone. The microtiter plate was incubated for a 16-h regime of light and dark cycle for 16 and 8 h, respectively, at growth room conditions. Ten random areas from each leaf disc were photographed using an inverted fluorescence microscope (Leica Microsystems) equipped with a Hamamatsu Orca AG camera (Hamamatsu Photonics), which was guided by OPENLAB software (Improvision). All photographs were recorded at the same OPENLAB software setting (i.e. 20× objectives, 1× binning, 80 gain, 0 offset, 1 digital gain, and 500-ms exposure time) for YFP and GFP. The mean intensity of each image was subtracted with that of p19 control, and then relative change in the interaction strength was calculated with respect to control. Absolute fluorescence from YFP signal (BiFC) under control and the salt-treated sample was analyzed in three different experiments in which the p19 signal was deducted to confirm the specificity of the detected signals. Relative fluorescence intensities were calculated by mean values and ses from the results of three independent experiments.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Elevated expression of CIPK21 gene in the wild-type (Col-0) plants under various conditions such as ABA (100 µm), cold (4°C), drought, or polyethylene glycol (30% [w/v]) treatments.
Supplemental Figure S2. Expression of the CIPK21 gene in the Arabidopsis eFP Browser.
Supplemental Figure S3. Subcellular localization of the CIPK21 in the epidermal peel cells of N. benthamiana.
Supplemental Figure S4. CIPK21:GFP coexpressed either with CBL1n:OFP or TPC1:OFP in the epidermal cells of N. benthamiana.
Supplemental Figure S5. Investigation of interaction of CBLs with CIPK21 by BiFC in the epidermal peel cells of N. benthamiana.
Supplemental Figure S6. Determination of the relative fluorescence produced to monitor the effects of salt stress treatment on pGPTV-II-BAR-35S::mVENUS vector, which was incubated with 125 mm NaCl or control (10 mm MES, pH 5.6, and 10 mm MgCl2) medium.
Supplemental Table S1. List of primers used for quantitative PCR/RT-PCR.
Supplemental Table S2. List of primers used for generation of constructs.
Supplemental Table S3. Published constructs used in this work.
Supplementary Material
Acknowledgments
We thank Dr. Oliver Batistič and Lena Schmidt (Universität Münster) for providing pGPTV-II-BAR-35S-CBL02::mCHERRY and pGPTV-II-HYG-35S-SPYNE-173::CIPK21 and Dr. Mool Chand Tyagi (Indian Agricultural Research Institute) and Dr. Tapasya Srivastava (Department of Genetics, University of Delhi South Campus) for proofreading the article.
Glossary
- PM
plasma membrane
- RT
reverse transcription
- ABA
abscisic acid
- T-DNA
transfer DNA
- BiFC
bimolecular fluorescence complementation
- Col-0
ecotype Columbia 0
- MS
Murashige and Skoog
- cDNA
complementary DNA
- AD
activation domain
- BD
binding domain
- OFP
orange fluorescent protein
- YFP
yellow fluorescent protein
Footnotes
This work was supported by the Department of Science and Technology, Ministry of Science and Technology, Government of India (to G.K.P.), the Deutsche Forschungsgemeinschaft (grant no. FOR964 to J.K.), a grant from the National Science Foundation (to S.L.), and research fellowships from the Council for Scientific and Industrial Research, India (to P.K., A.S., A.K.Y., and I.T.).
References
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes JD, Balaguer L, Manrique E, Elvira S, Davison AW (1992) A reappraisal of the use of DMSO for the extraction and determination of chlorophylls a and b in lichens and higher plants. Environ Exp Bot 32: 85–100 [Google Scholar]
- Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24: 23–58 [Google Scholar]
- Batistič O, Kudla J (2004) Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 219: 915–924 [DOI] [PubMed] [Google Scholar]
- Batistič O, Kudla J (2009) Plant calcineurin B-like proteins and their interacting protein kinases. Biochim Biophys Acta 1793: 985–992 [DOI] [PubMed] [Google Scholar]
- Batistič O, Rehers M, Akerman A, Schlücking K, Steinhorst L, Yalovsky S, Kudla J (2012) S-acylation-dependent association of the calcium sensor CBL2 with the vacuolar membrane is essential for proper abscisic acid responses. Cell Res 22: 1155–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistič O, Sorek N, Schültke S, Yalovsky S, Kudla J (2008) Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell 20: 1346–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistič O, Waadt R, Steinhorst L, Held K, Kudla J (2010) CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J 61: 211–222 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Laurière C (2005) Osmotic signaling in plants: multiple pathways mediated by emerging kinase families. Plant Physiol 138: 1185–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Kim KN, Pandey GK, Gupta R, Grant JJ, Luan S (2003) CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 15: 1833–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Pandey GK, Grant JJ, Batistič O, Li L, Kim BG, Lee SC, Kudla J, Luan S (2007) Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J 52: 223–239 [DOI] [PubMed] [Google Scholar]
- Chikano H, Ogawa M, Ikeda Y, Koizumi N, Kusano T, Sano H (2001) Two novel genes encoding SNF-1 related protein kinases from Arabidopsis thaliana: differential accumulation of AtSR1 and AtSR2 transcripts in response to cytokinins and sugars, and phosphorylation of sucrose synthase by AtSR2. Mol Gen Genet 264: 674–681 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cuéllar T, Pascaud F, Verdeil JL, Torregrosa L, Adam-Blondon AF, Thibaud JB, Sentenac H, Gaillard I (2010) A grapevine Shaker inward K+ channel activated by the calcineurin B-like calcium sensor 1-protein kinase CIPK23 network is expressed in grape berries under drought stress conditions. Plant J 61: 58–69 [DOI] [PubMed] [Google Scholar]
- D’Angelo C, Weinl S, Batistič O, Pandey GK, Cheong YH, Schültke S, Albrecht V, Ehlert B, Schulz B, Harter K, et al. (2006) Alternative complex formation of the Ca-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J 48: 857–872 [DOI] [PubMed] [Google Scholar]
- Drerup MM, Schlücking K, Hashimoto K, Manishankar P, Steinhorst L, Kuchitsu K, Kudla J (2013) The calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF. Mol Plant 6: 559–569 [DOI] [PubMed] [Google Scholar]
- Eckert C, Offenborn JN, Heinz T, Armarego-Marriott T, Schültke S, Zhang C, Hillmer S, Heilmann M, Schumacher K, Bock R, et al. (2014) The vacuolar calcium sensors CBL2 and CBL3 affect seed size and embryonic development in Arabidopsis thaliana. Plant J 78: 146–156 [DOI] [PubMed] [Google Scholar]
- Gong D, Guo Y, Jagendorf AT, Zhu JK (2002) Biochemical characterization of the Arabidopsis protein kinase SOS2 that functions in salt tolerance. Plant Physiol 130: 256–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JF, Harmon A (2005) Plants, symbiosis and parasites: a calcium signalling connection. Nat Rev Mol Cell Biol 6: 555–566 [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Eckert C, Anschütz U, Scholz M, Held K, Waadt R, Reyer A, Hippler M, Becker D, Kudla J (2012) Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases (CIPKs) is required for full activity of CBL-CIPK complexes toward their target proteins. J Biol Chem 287: 7956–7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held K, Pascaud F, Eckert C, Gajdanowicz P, Hashimoto K, Corratgé-Faillie C, Offenborn JN, Lacombe B, Dreyer I, Thibaud JB, et al. (2011) Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res 21: 1116–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BG, Waadt R, Cheong YH, Pandey GK, Dominguez-Solis JR, Schültke S, Lee SC, Kudla J, Luan S (2007) The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J 52: 473–484 [DOI] [PubMed] [Google Scholar]
- Kim KN, Cheong YH, Grant JJ, Pandey GK, Luan S (2003) CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15: 411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR (1996) Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8: 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Knight MR (2000) Imaging spatial and cellular characteristics of low temperature calcium signature after cold acclimation in Arabidopsis. J Exp Bot 51: 1679–1686 [DOI] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR (1997) Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J 12: 1067–1078 [DOI] [PubMed] [Google Scholar]
- Kolukisaoglu U, Weinl S, Blazevic D, Batistič O, Kudla J (2004) Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol 134: 43–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasensky J, Jonak C (2012) Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot 63: 1593–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Batistič O, Hashimoto K (2010) Calcium signals: the lead currency of plant information processing. Plant Cell 22: 541–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97: 3730–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S. (2009) The CBL-CIPK network in plant calcium signaling. Trends Plant Sci 14: 37–42 [DOI] [PubMed] [Google Scholar]
- Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W (2002) Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell (Suppl) 14: S389–S400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S, Sopory SK, Tuteja N (2006) Cloning and characterization of CBL-CIPK signalling components from a legume (Pisum sativum). FEBS J 273: 907–925 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Pandey GK. (2008) Emergence of a novel calcium signaling pathway in plants: CBL-CIPK signaling network. Physiol Mol Biol Plants 14: 51–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, Cheong YH, Kim BG, Grant JJ, Li L, Luan S (2007) CIPK9: a calcium sensor-interacting protein kinase required for low-potassium tolerance in Arabidopsis. Cell Res 17: 411–421 [DOI] [PubMed] [Google Scholar]
- Pandey GK, Cheong YH, Kim KN, Grant JJ, Li L, Hung W, D’Angelo C, Weinl S, Kudla J, Luan S (2004) The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16: 1912–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S (2005) ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol 139: 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, Grant JJ, Cheong YH, Kim BG, Li G, Luan S (2008) Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Mol Plant 1: 238–248 [DOI] [PubMed] [Google Scholar]
- Pandey GK, Kanwar P, Pandey A (2014) Global Comparative Analysis of CBL-CIPK Gene Families in Plants, Ed 1. Springer International Publishing, New York [Google Scholar]
- Qin Y, Li X, Guo M, Deng K, Lin J, Tang D, Guo X, Liu X (2008) Regulation of salt and ABA responses by CIPK14, a calcium sensor interacting protein kinase in Arabidopsis. Sci China C Life Sci 51: 391–401 [DOI] [PubMed] [Google Scholar]
- Reddy AS. (2001) Calcium: silver bullet in signaling. Plant Sci 160: 381–404 [DOI] [PubMed] [Google Scholar]
- Rudd JJ, Franklin-Tong VE (2001) Unraveling response-specificity in Ca2+ signaling pathways in plant cells. New Phytol 151: 7–33 [DOI] [PubMed] [Google Scholar]
- Sanders D, Brownlee C, Harper JF (1999) Communicating with calcium. Plant Cell 11: 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF (2002) Calcium at the crossroads of signaling. Plant Cell (Suppl) 14: S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal SK, Pandey A, Pandey GK (May 7, 2015) The CBL-CIPK signaling module in plants: a mechanistic perspective. Physiol Plant http://dx.doi.org/10.1111/ppl.12344 [DOI] [PubMed] [Google Scholar]
- Scrase-Field SA, Knight MR (2003) Calcium: just a chemical switch? Curr Opin Plant Biol 6: 500–506 [DOI] [PubMed] [Google Scholar]
- Shao HB, Song WY, Chu LY (2008) Advances of calcium signals involved in plant anti-drought. C R Biol 331: 587–596 [DOI] [PubMed] [Google Scholar]
- Steinhorst L, Kudla J (2014) Signaling in cells and organisms: Calcium holds the line. Curr Opin Plant Biol 22: 14–21 [DOI] [PubMed] [Google Scholar]
- Stewart GR, Lee JA (1974) The role of proline accumulation in halophytes. Planta 120: 279–289 [DOI] [PubMed] [Google Scholar]
- Tang RJ, Liu H, Yang Y, Yang L, Gao XS, Garcia VJ, Luan S, Zhang HX (2012) Tonoplast calcium sensors CBL2 and CBL3 control plant growth and ion homeostasis through regulating V-ATPase activity in Arabidopsis. Cell Res 22: 1650–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang RJ, Zhao FG, Garcia VJ, Kleist TJ, Yang L, Zhang HX, Luan S (2015) Tonoplast CBL-CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc Natl Acad Sci USA 112: 3134–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewavas AJ, Malhó R (1998) Ca2+ signalling in plant cells: the big network! Curr Opin Plant Biol 1: 428–433 [DOI] [PubMed] [Google Scholar]
- Tripathi V, Parasuraman B, Laxmi A, Chattopadhyay D (2009) CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. Plant J 58: 778–790 [DOI] [PubMed] [Google Scholar]
- Waadt R, Kudla J (2008) In Planta Visualization of Protein Interactions Using Bimolecular Fluorescence Complementation (BiFC). CSH Protocols, New York [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistič O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Weinl S, Kudla J (2009) The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol 184: 517–528 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Huang Y, Xiong L (2007) Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol 144: 1416–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Zhu JK (1999) Interaction of osmotic stress, temperature, and abscisic acid in the regulation of gene expression in Arabidopsis. Plant Physiol 119: 205–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Zhu JK (2002) Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ 25: 131–139 [DOI] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360 [DOI] [PubMed] [Google Scholar]
- Yang W, Kong Z, Omo-Ikerodah E, Xu W, Li Q, Xue Y (2008) Calcineurin B-like interacting protein kinase OsCIPK23 functions in pollination and drought stress responses in rice (Oryza sativa L.). J Genet Genomics 35: 531–543, S1–S2 [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun Z, Zheng J, Guo X, Dong Z, Huai J, Gou M, He J, Jin Y, Wang J, et al. (2009) Cloning and characterization of a novel CBL-interacting protein kinase from maize. Plant Mol Biol 69: 661–674 [DOI] [PubMed] [Google Scholar]
- Zhou LZ, Li S, Feng QN, Zhang YL, Zhao X, Zeng YL, Wang H, Jiang L, Zhang Y (2013) PROTEIN S-ACYL TRANSFERASE10 is critical for development and salt tolerance in Arabidopsis. Plant Cell 25: 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.