Plant sensing of bacterial pathogens induces ethylene, which is potentiated by salicylic acid and actively suppressed by effector(s) delivered to plant cells via the type III secretion system.
Abstract
Ethylene, a key phytohormone involved in plant-pathogen interaction, plays a positive role in plant resistance against fungal pathogens. However, its function in plant bacterial resistance remains unclear. Here, we report a detailed analysis of ethylene induction in Arabidopsis (Arabidopsis thaliana) in response to Pseudomonas syringae pv tomato DC3000 (Pst). Ethylene biosynthesis is highly induced in both pathogen/microbe-associated molecular pattern (PAMP)-triggered immunity and effector-triggered immunity (ETI), and the induction is potentiated by salicylic acid (SA) pretreatment. In addition, Pst actively suppresses PAMP-triggered ethylene induction in a type III secretion system-dependent manner. SA potentiation of ethylene induction is dependent mostly on MITOGEN-ACTIVATED PROTEIN KINASE6 (MPK6) and MPK3 and their downstream ACS2 and ACS6, two type I isoforms of 1-aminocyclopropane-1-carboxylic acid synthases (ACSs). ACS7, a type III ACS whose expression is enhanced by SA pretreatment, is also involved. Pst expressing the avrRpt2 effector gene (Pst-avrRpt2), which is capable of triggering ETI, induces a higher level of ethylene production, and the elevated portion is dependent on SALICYLIC ACID INDUCTION DEFICIENT2 and NONEXPRESSER OF PATHOGENESIS-RELATED GENE1, two key players in SA biosynthesis and signaling. High-order ACS mutants with reduced ethylene induction are more susceptible to both Pst and Pst-avrRpt2, demonstrating a positive role of ethylene in plant bacterial resistance mediated by both PAMP-triggered immunity and ETI.
Plants have multilayered defenses to ward off invading pathogens. The first line of defense is initiated by the detection of pathogen/microbe-associated molecular patterns (PAMPs) by plant cell surface pattern recognition receptors, which triggers a robust defense response known as PAMP-triggered immunity (PTI; Ausubel, 2005; Jones and Dangl, 2006; Boller and Felix, 2009; Spoel and Dong, 2012; Zipfel, 2014). To circumvent the immune responses of plants, pathogens are capable of delivering effector proteins into plant cells through the type III secretion system (TTSS) to suppress PTI to facilitate pathogenesis (Alfano and Collmer, 2004; Boller and He, 2009; Guo et al., 2009; Feng and Zhou, 2012). As a counter measure, plants evolved to possess a second line of defense in which RESISTANCE (R) proteins mediate the recognition of pathogen-derived effectors, initiating so-called effector-triggered immunity (ETI), which is stronger than PTI in general and frequently associated with hypersensitive response cell death (Ausubel, 2005; Glazebrook, 2005; Chisholm et al., 2006; Jones and Dangl, 2006; Dodds and Rathjen, 2010). Along with PAMPs and effectors, plant endogenous elicitors known as damage-associated molecular patterns (DAMPs), such as plant-derived peptides and cell wall fragments that are released upon wounding and infection, are also capable of inducing immune responses in plants (Boller and Felix, 2009).
Plant sensing of pathogen-originated PAMPs and effectors or plant-originated DAMPs activates multiple signal transduction pathways, including mitogen-activated protein kinase (MAPK) cascades (for review, see Zhang and Klessig, 2001; Pedley and Martin, 2005; Pitzschke et al., 2009; Rodriguez et al., 2010; Tena et al., 2011; Meng and Zhang, 2013). MAPK cascades are conserved eukaryotic signaling modules composed of three sequentially acting kinases with MAPKs at the bottom tier. MAPKs require phosphorylation activation by MAPK kinases (MAPKKs or MKKs), and MAPKKs in turn are activated by MAPKK kinases (MAPKKKs or MEKKs) via phosphorylation (Ichimura et al., 2002; Hamel et al., 2006). Among the 20 MAPKs in Arabidopsis (Arabidopsis thaliana), MPK3 and MPK6 share the highest homology and have a high level of functional redundancy. They also share the same upstream MAPKKs, MKK4 and MKK5 (Ren et al., 2002, 2008; Wang et al., 2007). In a MAPK cascade, the MAPKKK(s) receive signals from the sensors/receptors either directly or indirectly, and the outputs of a MAPK cascade are determined by the phosphorylation of MAPK substrates, which can be enzymes, transcription factors, and proteins with other biochemical functions (Liu and Zhang, 2004; Andreasson et al., 2005; Bethke et al., 2009; Mao et al., 2011; Meng et al., 2013; Guan et al., 2014; Xu and Zhang, 2015b).
MAPK cascades are primary signaling modules directly downstream of sensors/receptors. Identification of the first plant MAPK substrate revealed that MPK3/MPK6 positively regulate ethylene production through the phosphorylation-mediated stabilization of ACS2 and ACS6, two 1-aminocyclopropane-1-carboxylic acid synthase (ACS) isoforms (Liu and Zhang, 2004; Joo et al., 2008; Han et al., 2010; Xu and Zhang, 2014). ACS catalyzes the committing step of ethylene biosynthesis and is rate limiting in most vegetative tissues (Kende, 1993; Wang et al., 2002; Tsuchisaka et al., 2009; Xu and Zhang, 2015a). Arabidopsis has nine functional ACS isoforms that are classified into three groups based on their sequence homology and the presence/absence of phosphorylation sites in their C termini. They are type I (ACS1, ACS2, and ACS6), type II (ACS4, ACS5, ACS8, ACS9, and ACS11), and type III (ACS7; Chae and Kieber, 2005; Yoshida et al., 2005). In addition to the phosphorylation-mediated stabilization of ACS2/ACS6 proteins, MPK3 and MPK6 are also involved in the activation of ACS2 and ACS6 gene expression during plant immunity via WRKY33, another MPK3/MPK6 substrate (Mao et al., 2011; Li et al., 2012). The higher rate of de novo synthesis of ACS proteins resulted from ACS gene activation, which, coupled with their phosphorylation-induced stabilization by MPK3/MPK6, provides a vital supply of ACS enzymes to maintain a high rate of ethylene production in response to pathogen invasion (Li et al., 2012).
In addition to ethylene, other plant hormones, such as salicylic acid (SA) and jasmonic acid (JA), also function as key secondary signaling molecules in plant immunity. Their levels/activities are modulated by plant-pathogen interaction in response to the primary signaling pathways, either positively or negatively, and such changes can profoundly impact plant immunity (Glazebrook, 2005; Broekaert et al., 2006; van Loon et al., 2006; Katagiri and Tsuda, 2010; Pieterse et al., 2012; Kazan and Lyons, 2014). At present, the regulatory pathway(s) of SA and JA biosynthesis are mostly unknown. SA plays a central role in plant defense signaling in both local and systemic immunity, mainly through its downstream components NONEXPRESSER OF PATHOGENESIS-RELATED GENE1 (NPR1; for review, see Vlot et al., 2009; An and Mou, 2011; Spoel and Dong, 2012; Fu and Dong, 2013). A connection between SA and plant MAPKs was first made with the purification and identification of salicylic acid-induced protein kinase (SIPK), the tobacco (Nicotiana tabacum) ortholog of Arabidopsis MPK6 (Zhang and Klessig, 1997). However, the function of SIPK activation in the SA signaling pathway is still unknown. Recently, Arabidopsis MPK3, and to a lesser extent MPK6, were shown to play pivotal roles in SA-mediated priming of plant disease resistance (Beckers et al., 2009).
Antagonistic and synergistic interactions between SA, ethylene, and JA have been reported, which adds another layer of complexity (Broekaert et al., 2006; van Loon et al., 2006; Spoel and Dong, 2008; Robert-Seilaniantz et al., 2011; Pieterse et al., 2012). In general, it is considered that ethylene and SA are antagonistic in plant immunity (Glazebrook, 2005; Pieterse et al., 2012). Here, we report the PAMP- and effector-triggered ethylene induction and its potentiation by SA in Arabidopsis in response to Pseudomonas syringae pv tomato DC3000 (Pst) infection. The potentiation effect is mostly dependent on MPK3/MPK6 and involves ACS2 and ACS6, two ACS isoforms regulated by MPK3/MPK6 at both the transcriptional and posttranslational levels (Liu and Zhang, 2004; Han et al., 2010; Li et al., 2012). In addition, Pst is capable of inhibiting PAMP-triggered ethylene biosynthesis in a TTSS-dependent manner, suggesting an active suppression of plant ethylene production by Pst effector(s). The battle between Pst and Arabidopsis in controlling ethylene biosynthesis suggests a positive role of ethylene in bacterial resistance. Consistent with this, we found that the loss of ethylene biosynthesis in acs mutants leads to pathogen susceptibility. In summary, this research highlights a novel interaction between SA and ethylene and demonstrates that ethylene is a positive regulator in Arabidopsis immunity against a bacterial pathogen.
RESULTS
Pst Actively Suppresses Ethylene Induction in Arabidopsis during PTI
Although ethylene is recognized as an important plant hormone involved in plant disease resistance, no report has analyzed in detail its induction in Arabidopsis in response to Pst infection. To facilitate the measurement of ethylene, we grew Arabidopsis seedlings in gas chromatography (GC) vials similar to what we used for studying the Arabidopsis-Botrytis cinerea interaction (Han et al., 2010; Li et al., 2012), and Pst inoculation was simply done by the addition of inoculum to a final concentration of optical density at 600 nm (OD600) of 0.02. Ethylene accumulation in the headspace of the GC vials was monitored afterward. An advantage of this system in comparison with collecting leaves from soil-grown plants is that wounding-induced ethylene production can be avoided.
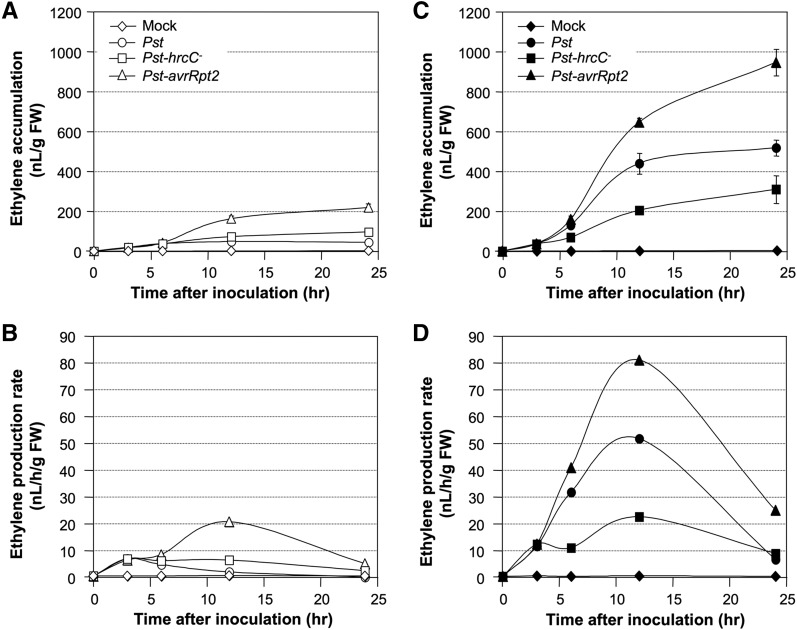
As shown in Figure 1A, ethylene gradually accumulated in the first 6 h after Pst inoculation and then reached a plateau. In contrast, ethylene accumulation continued to increase in seedlings inoculated with Pst-hrcC−, a Pst strain carrying a deletion mutation in hrcC gene that cannot deliver effectors into plant cells. After the conversion of ethylene accumulation to the average rates of ethylene production between the two adjacent time points (Fig. 1B), it became obvious that, within the first 3 h, ethylene induction rates were similar in Arabidopsis inoculated with Pst and Pst-hrcC−. After that, the ethylene production rate started to decline in Pst-inoculated plants, while that in Pst-hrcC−-inoculated Arabidopsis stayed at a relatively high level, which is consistent with the scenario that both Pst and Pst-hrcC− can trigger the PAMP-induced defense responses but only Pst can deliver effector proteins to suppress plant immunity and facilitate the pathogenesis process. As a result, we conclude that Pst is able to actively suppress ethylene induction by delivering effectors into Arabidopsis cells during PTI.
Figure 1.
Bacterial PAMP- and effector-triggered ethylene induction and its potentiation by SA in Arabidopsis. A, Fourteen-day-old seedlings grown in GC vials were inoculated with Pst, Pst-hrcC−, or Pst-AvrRpt2 (final OD600 = 0.02). Mock inoculation was used as a control. Ethylene accumulations in the headspace were determined at the indicated times. B, Replot of the data in A as the rates of ethylene production. Ethylene production rates were calculated as the average rates of ethylene production in the intervals of the two adjacent time points. C, Twelve-day-old seedlings grown in GC vials were treated with SA (final concentration of 100 µm). Two days later, they were inoculated with Pst, Pst-hrcC−, or Pst-avrRpt2 (final OD600 = 0.02). Mock inoculation was used as a control. Ethylene accumulations in the headspace were determined at the indicated times. D, Replot of the data in C as the average rates of ethylene production in the intervals of the two adjacent time points. All data were all collected side by side. Error bars indicate sd (n = 3). FW, Fresh weight.
avrRpt2 ETI Is Associated with a High Level of Ethylene Induction
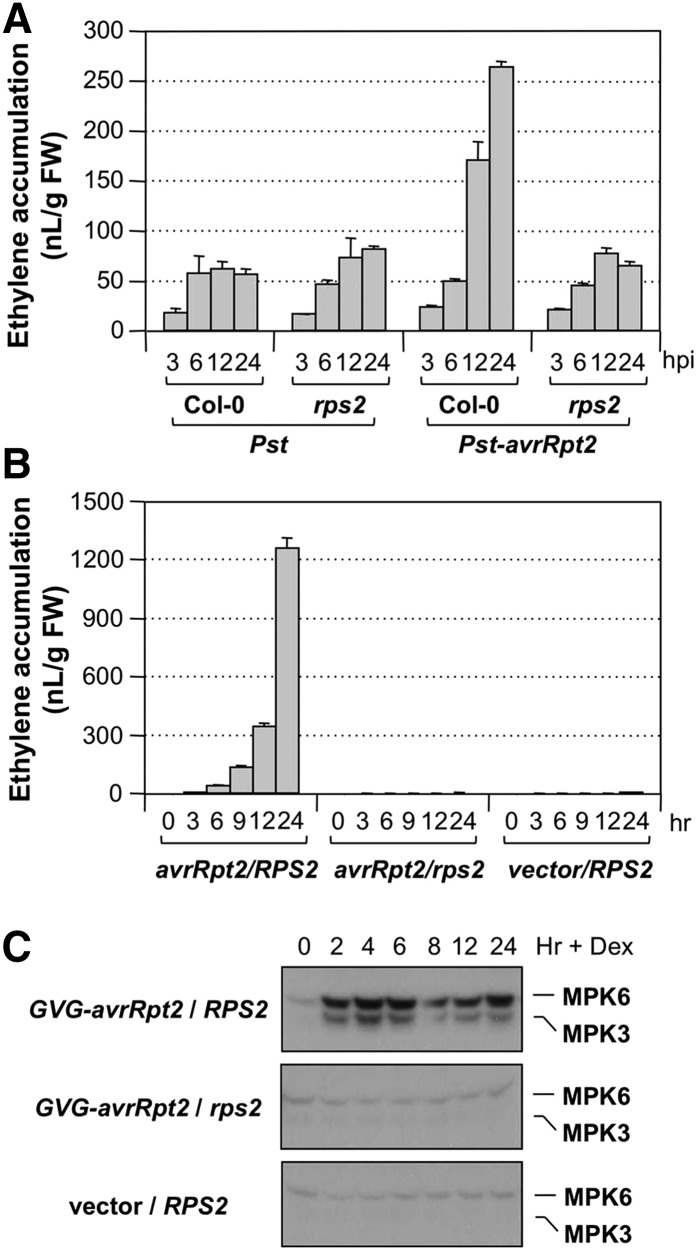
Seedlings inoculated with Pst expressing the avrRpt2 effector gene (Pst-avrRpt2) produced higher levels of ethylene. In the first 3 h, the ethylene production rate was similar to those inoculated with either Pst or Pst-hrcC− (Fig. 1, A and B). However, there was no sign of effector-mediated suppression of ethylene induction after 6 h; instead, the ethylene production rates surpassed that induced by Pst-hrcC− and continued to increase to a much higher level. The high-level ethylene production is likely a result of avrRpt2 effector-triggered activation of defense responses (i.e. ETI). In the end, seedlings inoculated with Pst-avrRpt2 produced much more ethylene than those inoculated with Pst-hrcC− or Pst (Fig. 1, A and B). In resistance to P. syringae2 (rps2) mutant seedlings that lack the R protein to sense the avrRpt2 effector, Pst-avrRpt2-induced ethylene was much lower, at a level similar to that in seedlings inoculated with Pst (Fig. 2A), confirming that the higher ethylene production is the result of an effector-triggered response.
Figure 2.
RPS2-dependent induction of ethylene biosynthesis in Arabidopsis triggered by the avrRpt2 effector. A, Higher levels of ethylene induction in Arabidopsis inoculated with Pst-avrRpt2 are dependent on the presence of RPS2, the corresponding R gene. Fourteen-day-old wild-type (Col-0) and rps2 mutant seedlings grown in GC vials were inoculated with Pst or Pst-avrRpt2 (final OD600 = 0.02). Ethylene accumulations in the headspace were determined at the indicated times. Error bars indicate sd (n = 3). FW, Fresh weight; hpi, hours post inoculation. B, Fourteen-day-old steroid-inducible avrRpt2 transgene (GVG-avrRpt2) in the wild-type Col-0 background (avrRpt2/RPS2), GVG-avrRpt2 in the rps2 mutant background (avrRpt2/rps2), and steroid-inducible vector control were treated with DEX (2 µm). Ethylene accumulations in the headspace were determined at the indicated times. Error bars indicate sd (n = 3). Seedlings were collected at the indicated times for in-gel kinase assays. C, RPS2-dependent activation of MPK3/MPK6 by the avrRpt2 effector. MAPK activities in samples collected in B were determined using 10 µg of total proteins by the in-gel kinase assay with myelin basic protein (MBP) as a substrate.
To demonstrate directly the induction of ethylene by the avrRpt2 effector, we utilized dexamethasone (DEX)-inducible promoter-driven avrRpt2 (GVG-avrRpt2 [GVG encodes a hybrid transcription factor consisting of one domain each from GAL4, VP16, and GR]) transgenic plants (McNellis et al., 1998). As shown in Figure 2B, treatment of GVG-avrRpt2 seedlings with DEX strongly induced ethylene biosynthesis. In the absence of RPS2, no induction of ethylene was observed. As another control, we also tested a vector control transgenic line and observed no ethylene production either. As a result, we can conclude that, in the absence of PTI, effector by itself is sufficient to trigger strong ethylene production. An in-gel kinase assay revealed the activation of MPK6 and MPK3 in GVG-avrRpt2 transgenic seedlings after DEX treatment (Fig. 2C). No activation of MPK3 and MPK6 was observed in GVG-avrRpt2 rps2 or vector control transgenic seedlings after DEX treatment, demonstrating the specificity of MPK3/MPK6 activation in the ETI.
SA Potentiates Pst-Induced Ethylene Production, a Process Dependent on NPR1
SA is known to enhance many defense responses triggered by Pst (Vlot et al., 2009; An and Mou, 2011; Fu and Dong, 2013). As a result, we examined ethylene production in SA-pretreated seedlings. In this experiment, the seedlings were treated with SA (100 µm) for 2 d before Pst inoculation. The normal SA response in the liquid-cultured seedling system was validated by the high-level PATHOGENESIS-RELATED1 (PR1) gene induction (Supplemental Fig. S1). Remarkably, pretreatment of seedlings with SA greatly enhanced the ethylene production induced by Pst inoculation (Fig. 1C). The maximal accumulation of ethylene in Pst-inoculated seedlings was approximately 50 nL g−1 fresh weight without SA pretreatment (Fig. 1A), whereas the ethylene induction in SA-pretreated seedlings reached greater than 500 nL g−1 fresh weight (Fig. 1C; the data in Fig. 1 were all collected side by side), an increase of about 10-fold.
SA pretreatment not only enhanced Pst-induced ethylene production but also promoted ethylene biosynthesis induced by Pst-hrcC− and Pst-avrRpt2 (Fig. 1, C and D), although the fold enhancement was not as high, at approximately 3-fold and approximately 4-fold for Pst-hrcC− and Pst-avrRpt2, respectively. SA treatment alone only weakly induced ethylene production (Supplemental Fig. S2), with a rate about twice the basal level without any treatment, which is very low in comparison with the pathogen-induced (Fig. 1, C and D, mock versus Pst inoculated) or flg22-induced (Supplemental Fig. S2) ethylene production. This provides strong support that increased ethylene biosynthesis after SA pretreatment is a potentiation (also known as priming) effect rather than a direct contribution to ethylene production.
SA potentiation of Pst-induced ethylene production was dependent on NPR1. In the npr1 mutant, Pst-induced ethylene production was similar to that in the wild-type (Columbia-0 [Col-0]) control. However, SA-potentiated ethylene induction was greatly reduced (Fig. 3), demonstrating that, similar to other SA-enhanced defense responses, SA-mediated potentiation of ethylene induction in response to Pst is also dependent on NPR1. In NahG transgenic plants expressing the bacterial salicylate hydroxylase (encoded by NahG gene) that converts SA to catechol, therefore effectively removing SA, the enhancement effect of SA pretreatment was also greatly inhibited (Fig. 3). These results demonstrate that (1) the liquid-cultured seedlings have a normal SA response and (2) the potentiation effect we observed is a bona fide SA-mediated response.
Figure 3.
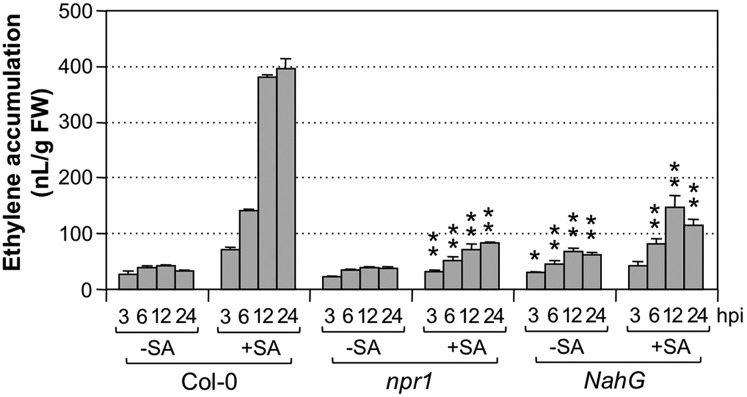
Potentiation of Pst-induced ethylene biosynthesis by SA pretreatment is dependent on functional NPR1, and the NahG transgene abolishes this SA effect. Twelve-day-old wild-type (Col-0), npr1, or NahG Arabidopsis seedlings grown in GC vials were treated with SA (+SA; final concentration of 100 µm) or a solvent of SA stock solution (−SA). Two days later, they were inoculated with Pst (final OD600 = 0.02). Ethylene accumulations in the headspace were determined at the indicated times. Error bars indicate sd (n = 3). Student’s t test was used to compare mutants and the wild type at the same time point after the same treatment (*, P ≤ 0.05; and **, P ≤ 0.01). FW, Fresh weight; hpi, hours post inoculation.
The Potentiation of Ethylene Induction by SA Is Associated with MAPK Activation
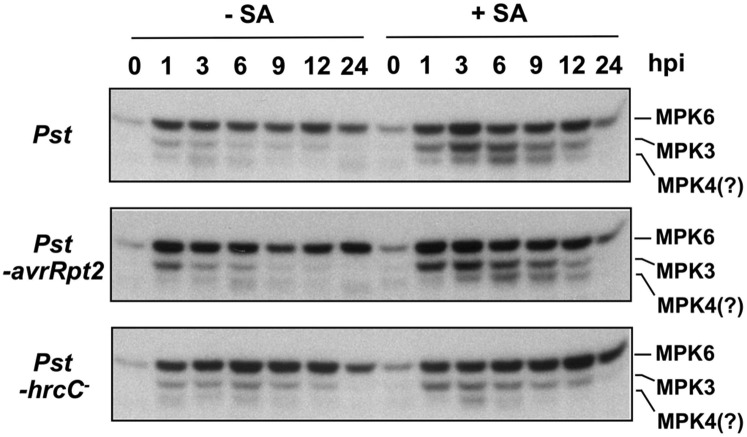
The MPK3/MPK6 cascade plays important roles in ethylene induction in response to B. cinerea at both the transcriptional and posttranslational levels (Han et al., 2010; Li et al., 2012). As a result, we examined the activation of MAPKs in seedlings inoculated with Pst, Pst-hrcC−, and Pst-avrRpt2 with and without SA pretreatment. We did not observe a major difference in the MAPK activation in Pst-, Pst-hrcC−-, or Pst-avrRpt2-inoculated seedlings (Fig. 4; Supplemental Fig. S3A). In mpk6 and mpk3 single mutants, their corresponding kinase activity bands disappeared (Supplemental Fig. S3A), demonstrating that the two larger MAPK activity bands are MPK6 and MPK3, respectively. The smallest and the weaker MAPK activity is likely to be MPK4 based on its size (Xu et al., 2014). It is also clear that, in the absence of MPK3, MPK6 activation was higher, a compensatory effect that we reported before (Wang et al., 2008). The lack of suppression of PAMP-triggered MAPK activation suggests that Pst does not possess effector(s) that directly target MPK3/MPK6 activation, and the suppression of ethylene production in Arabidopsis (Fig. 1) by Pst is likely a result of the suppression of other components in the ethylene induction process.
Figure 4.
SA pretreatment enhances MPK3/MPK6 activation by Pst. Twelve-day-old wild-type (Col-0) Arabidopsis seedlings grown in GC vials were treated with dimethyl sulfoxide (DMSO) solvent (−SA) or SA (+SA; final concentration of 100 µm). Two days later, they were inoculated with Pst, Pst-hrcC−, or Pst-avrRpt2 (final OD600 = 0.02). Seedlings were collected at the indicated times. MAPK activities were determined using 10 µg of total proteins by the in-gel kinase assay with MBP as a substrate. hpi, Hours post inoculation.
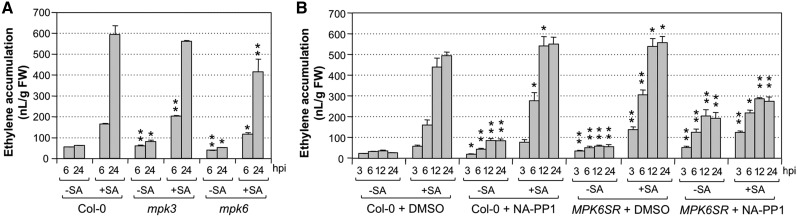
Seedlings pretreated with SA showed heightened MAPK activation, especially MPK3, after Pst, Pst-hrcC−, or Pst-avrRpt2 inoculation (Fig. 4; Supplemental Fig. S3). MPK4 activation was also stronger. MPK3 has been shown to play a key role in SA-induced priming (Beckers et al., 2009). To determine the role of MPK3 and MPK6 in Pst-induced ethylene production and its enhancement by SA pretreatment, we examined ethylene induction in mpk3, mpk6, and an mpk3 mpk6 double mutant rescued with an MPK6 variant (MPK6YG) that is sensitized to a 4-amino-1-tert-butyl-3-(1′-naphthyl)pyrazolo[3,4-d]pyrimidine (NA-PP1) inhibitor (genotype mpk3 mpk6 PMPK6:MPK6YG, MPK6SR for short; Xu et al., 2014). We first compared ethylene production in Col-0, mpk3 single mutant, and mpk6 single mutant after Pst inoculation with and without SA pretreatment. As shown in Figure 5A, loss of function of MPK6 slightly reduced ethylene biosynthesis, while no major difference was observed in the mpk3 mutant. In MPK6SR line 45 without NA-PP1 treatment, we observed normal SA potentiation of ethylene induction. In contrast, application of NA-PP1 almost completely blocked the potentiation effect of SA (Fig. 5B), demonstrating that MPK3/MPK6 are required for the full potentiation effect of SA. We also observed that, in the absence of SA pretreatment, Pst-induced ethylene production was higher in seedlings with NA-PP1 treatment. Since this was observed in both Col-0 and MPK6SR seedlings, we assumed that it might be caused by a nonspecific inhibition of some other unknown target(s) by NA-PP1. However, a stronger effect of NA-PP1 was observed in MPK6SR seedlings. Examination of another independently rescued mpk3 mpk6 double mutant line (MPK6SR line 58) gave similar results (Supplemental Fig. S4).
Figure 5.
Arabidopsis MPK3 and MPK6 play essential and overlapping roles in Pst-induced ethylene biosynthesis and SA potentiation. A, Twelve-day-old wild-type (Col-0), mpk3, and mpk6 seedlings grown in GC vials were treated with DMSO solvent (−SA) or SA (+SA; final concentration of 100 µm). Two days later, Pst was inoculated (final OD600 = 0.02), and ethylene accumulation in the headspace of the GC vials was measured. B, Twelve-day-old wild-type (Col-0) and chemical genetically rescued mpk3 mpk6 double mutant (MPK6SR line 45) seedlings grown in GC vials were treated with SA (+SA; final concentration of 100 µm) or DMSO, the solvent of SA stock solution (−SA). Two days later, they were inoculated with Pst (final OD600 = 0.02) after pretreatment with NA-PP1 (+NA-PP1; 5 µm final concentration) or solvent control (+DMSO) for 30 min. Ethylene accumulations in the headspace were determined at the indicated times. Error bars indicate sd (n = 3). Student’s t test was used to compare mutants and the wild type at the same time point after the same treatment (*, P ≤ 0.05; and **, P ≤ 0.01). FW, Fresh weight; hpi, hours post inoculation.
The Potentiation of Pst-Induced Ethylene Production by SA Is Dependent on ACS2, ACS6, and ACS7
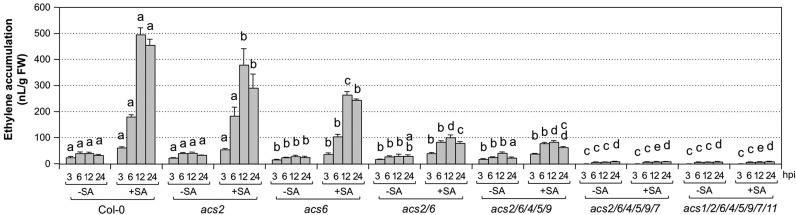
We next determined the contribution of different ACS isoforms using various acs mutants. In the acs2 single mutant, no significant reduction in ethylene induction was observed after Pst inoculation. Ethylene induction was slightly lower, but significant, in asc6 single, acs2 acs6 double, and acs2 acs6 acs4 acs5 acs9 quintuple mutants, and there was no significant difference between these four genotypes, suggesting that ACS6 also contributes to Pst-triggered ethylene biosynthesis. Ethylene induction was much lower in the acs2 acs6 acs4 acs5 acs9 acs7 sextuple mutant in comparison with the acs2 acs6 acs4 acs5 acs9 quintuple mutant (Fig. 6, −SA groups), demonstrating that Pst-induced ethylene production in Arabidopsis without SA pretreatment was mostly dependent on ACS7. The residual ethylene induction suggests the involvement of ACS8, since additional mutation of acs1 and acs11 in the acs2 acs6 acs4 acs5 acs9 acs7 mutant background did not further reduce the ethylene induction, and acs1 acs2 acs6 acs4 acs5 acs9 acs7 acs11 octuple mutant seedlings with only functional ACS8 still produced very low levels of ethylene. Alternatively, the residual ethylene production observed in octuple acs mutant seedlings could come from the bacteria. It was reported that P. syringae pathovars produce low levels of ethylene (Weingart and Volksch, 1997). Indeed, we found that Pst (with and without avrRpt2) produced low levels of ethylene (Supplemental Fig. S5). Without Arabidopsis seedlings, Pst does not grow much in Murashige and Skoog (MS) medium. As a result, we measured ethylene production by Pst of different concentrations from OD600 of 0.02 to 0.5. In the presence of Arabidopsis seedlings, Pst could grow from the initial concentration of 0.02 after inoculation to approximately 0.5 within 24 h. If we assume that Pst grew in a linear rate, its average concentration should be approximately 0.25. As shown in Supplemental Figure S5, GC vials with acs1 acs2 acs6 acs4 acs5 acs9 acs7 acs11 seedlings still accumulated more ethylene than the vials with only 0.25 OD600 Pst, suggesting that the octuple mutant seedlings still produced a small amount of ethylene. One caveat is that, in the presence of Arabidopsis seedlings, Pst might produce more ethylene due to its interaction with plants, which makes it difficult to draw a definitive conclusion about the involvement of ACS8. Either way, the contribution of ACS8 should be very small in comparison with ACS2, ACS6, or ACS7.
Figure 6.
SA potentiated ethylene induction in various acs mutants after Pst infection. Twelve-day-old wild-type (Col-0) seedlings and high-order acs mutants were treated with DMSO solvent (−SA) or SA (+SA; final concentration of 100 µm). Two days later, Pst was inoculated (final OD600 = 0.02), and ethylene accumulation in the headspace of the GC vials was measured at the indicated times. Error bars indicate sd (n = 3). One-way ANOVA was performed to compare acs mutants and the wild type at the same time point after the same treatment. Lowercase letters above the columns indicate statistically different groups. The two genotypes are considered to produce different amounts of ethylene when two or more time points are significantly different. The allele numbers are omitted for easy labeling. They are acs1-1, acs2-1, acs4-1, acs5-2, acs6-1, acs7-1, acs9-1, and acs11-1. FW, Fresh weight; hpi, hours post inoculation.
In contrast, SA potentiation of ethylene biosynthesis is mostly dependent on ACS2 and ACS6. In acs2 and asc6 single mutants, the enhanced ethylene production by SA pretreatment was reduced, and an additive effect was observed in the acs2 acs6 double mutant (Fig. 6, +SA groups). Instead of a 10-fold enhancement in the wild type, the acs2 acs6 double mutant only had an approximately 2-fold increase. There was no significant change in ethylene induction in the acs2 acs6 acs4 acs5 acs9 quintuple mutant in comparison with the acs2 acs6 double mutant, suggesting that ACS4, ACS5, and ACS9 played minimal roles in the process. Additional mutation of the ACS7 gene in the acs2 acs6 acs4 acs5 acs9 background reduced ethylene production further to a very low level that was no longer responsive to SA pretreatment. Again, the very low-level ethylene accumulation in acs1 acs2 acs6 acs4 acs5 acs9 acs7 acs11 seedlings inoculated with Pst could come either from Pst or Arabidopsis seedlings as a result of functional ACS8, as discussed above. In summary, we found that ACS7 is involved in Pst-induced ethylene induction, while ACS2 and ACS6 are more important to the SA-potentiated induction of ethylene biosynthesis.
The Potentiation of Ethylene Induction by SA Is Associated with Enhanced ACS Gene Expression
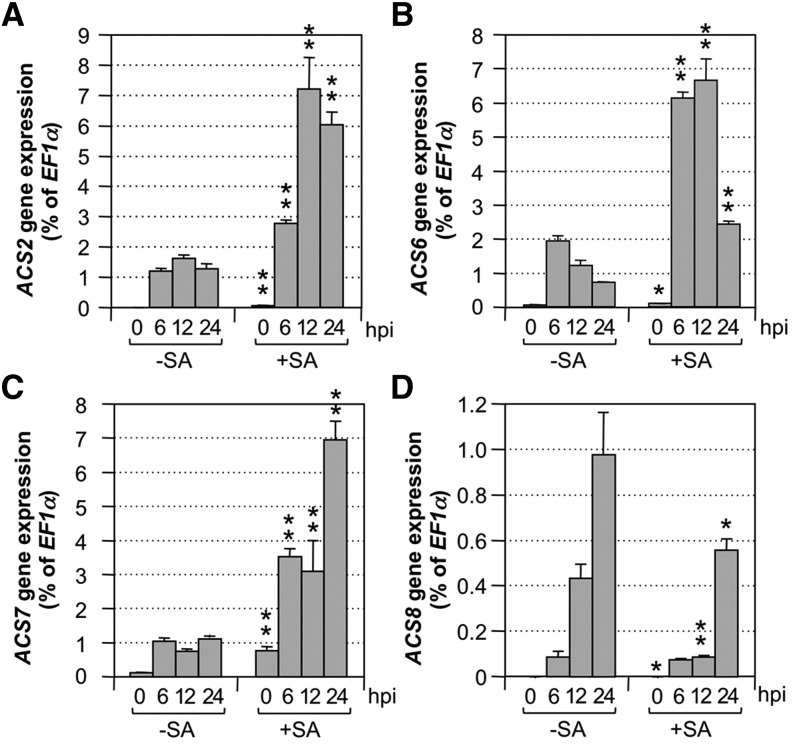
Without SA pretreatment, Pst-induced ethylene production was associated with ACS2, ACS6, ACS7, and ACS8 gene activation (Fig. 7), which is similar to B. cinerea-induced ethylene production (Li et al., 2012). SA pretreatment further enhanced the induction of ACS2, ACS6, and ACS7. Interestingly, SA pretreatment did not enhance ACS8 expression (Fig. 7D), which is consistent with the finding that ACS8 is not involved in SA-enhanced ethylene production (Fig. 6). As a result, SA-potentiated ethylene induction by Pst is likely a result of, at least partially, the enhanced gene activation of ACS2, ACS6, and ACS7. The higher level of ACS2/ACS6 expression in conjunction with MPK3/MPK6-mediated phosphorylation stabilization of ACS2/ACS6 proteins could lead to high cellular ACS activity levels and ethylene production. Pathway(s) that regulate ACS7 expression are unclear at present. Despite lacking a potential phosphorylation site in the C terminus, ACS7 was recently shown to be phosphorylated by a calcium-dependent protein kinase (CPK) activity at the N terminus, and phosphorylation could also lead to protein stabilization and ethylene production (Huang et al., 2013).
Figure 7.
SA potentiation of ethylene induction is associated with enhanced ACS2, ACS6, and ACS7 gene activation. Twelve-day-old wild-type (Col-0) seedlings grown in GC vials were treated with DMSO solvent (−SA) or SA (+SA; final concentration of 100 µm). Two days later, Pst was inoculated (final OD600 = 0.02), and seedlings were collected at the indicated times for total RNA preparation. After reverse transcription, transcript levels of ACS2 (A), ACS6 (B), ACS7 (C), and ACS8 (D) genes were quantified by real-time PCR. Expression of the ACS genes was calculated as the percentage of ELONGATION FACTOR-1α (EF1α) transcript. Error bars indicate sd (n = 3). Student’s t test was used to compare plants treated with SA (+SA) and DMSO solvent (−SA) at the same time point after the same treatment (*, P ≤ 0.05; and **, P ≤ 0.01). hpi, Hours post inoculation.
Even without SA pretreatment, we observed high levels of ACS2 and ACS6 gene activation at approximately 185- and 33-fold, respectively, at 6 h post inoculation, in response to Pst inoculation in Arabidopsis (Fig. 7, −SA groups). However, there is little/no contribution from ACS2 and ACS6 to Pst-induced ethylene production (Fig. 6, −SA groups). Using the in-gel kinase assay, we also detected similar levels of MPK3/MPK6 activation after Pst infection (Fig. 4; Supplemental Fig. S3A, top). Both should have led to an increase in ACS activity from ACS2/ACS6 isoforms and higher ethylene production. The lack of ACS2/ACS6 contribution suggests that Pst effector(s) might target these two ACS isoforms and impede their function either directly or indirectly.
The Elevated Ethylene Biosynthesis during ETI Is Dependent on ACS2/ACS6 Isoforms and SA Biosynthesis and Signaling
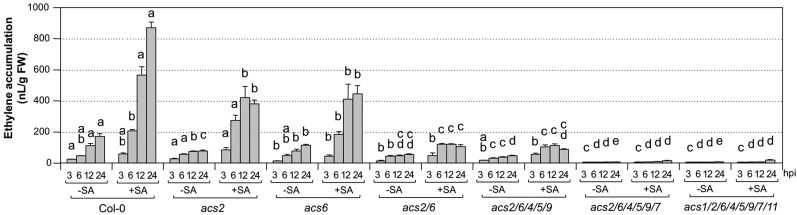
While ethylene induction by Pst in seedlings without SA pretreatment was mostly dependent on ACS7 (Fig. 6), ethylene induction by Pst-avrRpt2 without SA pretreatment was dependent on ACS2 and ACS6 in addition to ACS7 (Fig. 8, −SA groups). In the absence of ACS2 and ACS6 (acs2 acs6 double mutant), the level and kinetics of ethylene induction were almost identical to those induced by Pst (Figs. 6 and 8, −SA groups), suggesting that avrRpt2-triggered ethylene production is mostly dependent on ACS2 and ACS6. Ethylene induction by Pst-avrRpt2 in seedlings pretreated with SA was dependent on the same set of ACS genes (e.g. ACS2, ACS6, and ACS7) based on mutant analysis (Fig. 8, +SA groups). Again, the contribution of ACS8 to ethylene induction in response to Pst-avrRpt2 inoculation is minimal, if any. The low-level ethylene production by Pst-avrRpt2 (Supplemental Fig. S5) prevented us from drawing a definitive conclusion. In addition, the acs1 acs2 acs6 acs4 acs5 acs9 acs7 acs11 octuple mutant with only functional ACS8 did not show SA-enhanced ethylene production (Fig. 8), suggesting that ACS8 is not involved in the SA potentiation of ethylene biosynthesis in response to Pst-avrRpt2 inoculation either, which is similar to that in response to Pst inoculation.
Figure 8.
SA potentiated ethylene induction in high-order acs mutants after Pst-avrRpt2 inoculation. Twelve-day-old wild-type (Col-0) seedlings and acs mutants generated in Dr. A. Theologis’s laboratory were treated with DMSO solvent (−SA) or SA (+SA; final concentration of 100 µm). Two days later, Pst-avrRpt2 was inoculated (final OD600 = 0.02), and ethylene accumulation in the headspace of the GC vials was measured at the indicated times. Error bars indicate sd (n = 3). One-way ANOVA was performed to compare acs mutants and the wild type at the same time point after the same treatment. Lowercase letters above the columns indicate statistically different groups. The two genotypes are considered to produce different amounts of ethylene when two or more time points are significantly different. The allele numbers are omitted for easy labeling. They are acs1-1, acs2-1, acs4-1, acs5-2, acs6-1, acs7-1, acs9-1, and acs11-1. hpi, Hours post inoculation.
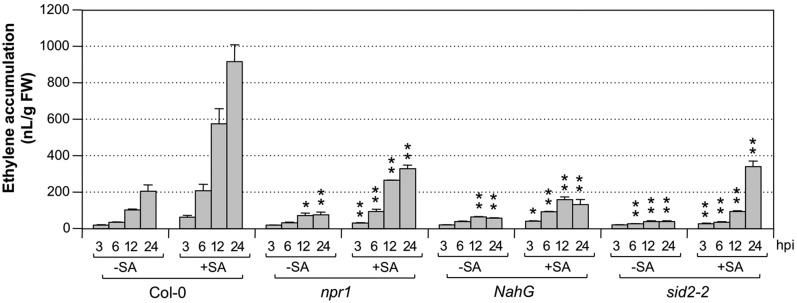
SA-potentiated ethylene production in seedlings inoculated with Pst-avrRpt2 was dependent on functional NPR1 (Fig. 9), similar to that inoculated with Pst (Fig. 3). It was interesting to see that the mutation of NPR1 also reduced ethylene induction by Pst-avrRpt2 in seedlings without SA pretreatment, suggesting that the SA signaling pathway is required for the effector-triggered ethylene production. Examination of NahG transgenic and sid2-2 mutant seedlings gave similar results (Fig. 9, −SA groups). The ethylene production in npr1, NahG, or sid2-2 seedlings inoculated with Pst-avrRpt2 was similar to that in seedlings inoculated with Pst (Fig. 3) in kinetics and magnitude. It is likely that the elevated ethylene induction triggered by effector is dependent on endogenously produced SA and signaling. We found that supplementation of SA cannot restore the potentiation of ethylene biosynthesis in sid2-2 mutant seedlings (Fig. 9; Supplemental Fig. S6), suggesting that SALICYLIC ACID INDUCTION DEFICIENT2 (SID2) may function more than simply as an SA biosynthetic enzyme. It is also possible that one-time application of exogenously supplied SA cannot fulfill the function of endogenously produced SA due to differences in the kinetics and magnitude of cellular SA levels.
Figure 9.
Elevated ethylene biosynthesis in response to Pst-avrRpt2 inoculation and SA pretreatment is dependent on functional NPR1 and SID2, and the NahG transgene abolishes this SA effect. Twelve-day-old wild-type (Col-0), npr1, sid2-2, or NahG Arabidopsis seedlings grown in GC vials were treated with SA (+SA; final concentration of 100 µm) or DMSO (−SA). Two days later, they were inoculated with Pst-avrRpt2 (final OD600 = 0.02). Ethylene accumulations in the headspace were determined at the indicated times. Error bars indicate sd (n = 3). Student’s t test was used to compare mutants and the wild type at the same time point after the same treatment (*, P ≤ 0.05; and **, P ≤ 0.01). FW, Fresh weight; hpi, hours post inoculation.
A Positive Role of Ethylene Biosynthesis in Plant Disease Resistance
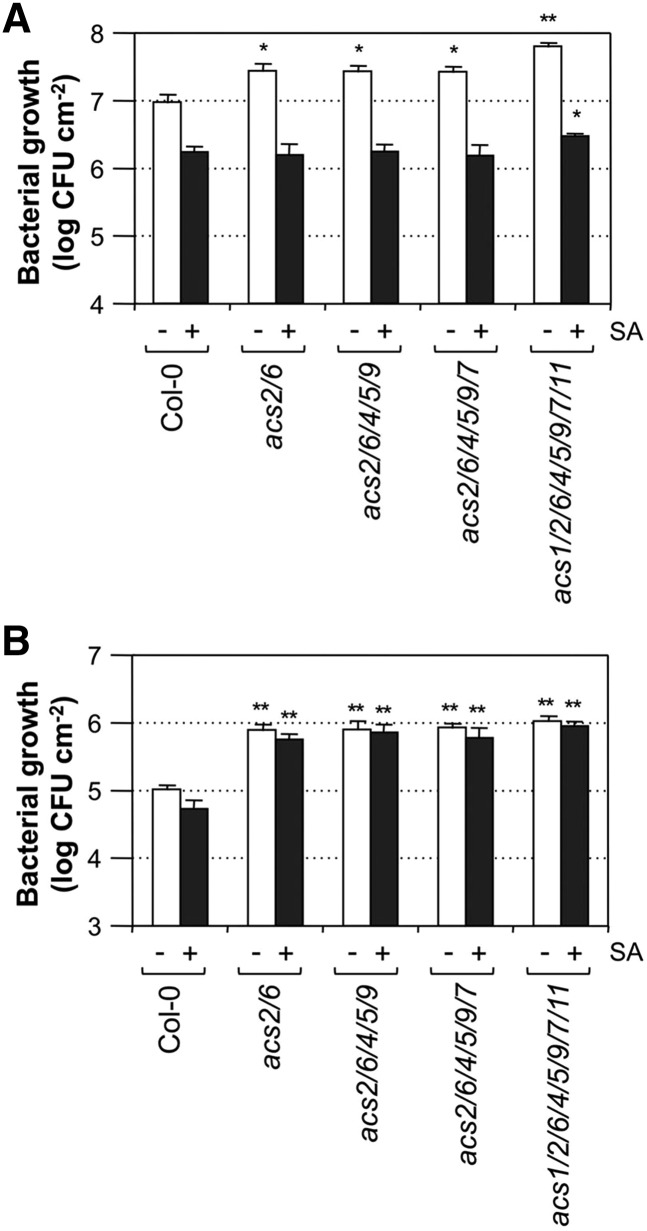
Enhanced ethylene biosynthesis is associated with both PTI and ETI in plant resistance against bacterial infection. In addition, Pst actively suppresses PAMP-triggered ethylene production via the TTSS-mediated process, and SA pretreatment potentiates Pst-induced ethylene biosynthesis, suggesting that ethylene might play an important role in plant immunity against bacterial infection. As a result, we examined the resistance of acs mutants infiltrated with Pst with and without SA pretreatment. As shown in Figure 10, loss of function of ACS2 and ACS6 genes, which are targets of the MPK3/MPK6-regulated ethylene production pathway, leads to higher pathogen susceptibility. Mutation of additional ACS genes, such as in acs2 acs6 acs4 acs5 acs9 quintuple, acs2 acs6 acs4 acs5 acs9 acs7 sextuple, and acs1 acs2 acs6 acs4 acs5 acs9 acs7 acs11 octuple mutants, did not further enhance the susceptibility (Fig. 10). The increase in Pst growth in acs mutants was small, less than 1 log, but the difference was significant and reproducible (Fig. 10A). In contrast, the growth of Pst-avrRpt2 was about 1 log higher in acs mutants than in the Col-0 control (Fig. 10B), suggesting that ethylene plays a positive role in both PTI and ETI in plant immunity against Pst infection.
Figure 10.
Ethylene is involved in plant resistance against both Pst and Pst-avrRpt2. Five-week-old wild-type (Col-0) and acs mutant Arabidopsis plants grown under a short-day light cycle were used for pathogen assays. Fully expanded leaves (three per plant) were infiltrated with either Pst (A) or Pst-avrRpt2 (B; OD600 = 0.001). SA pretreatment was performed by spraying the plants with SA (1 mm) and drenching the soil with SA (1 mm) at the same time for 24 h before pathogen infiltration. Bacterial growth was quantified 3 d post inoculation. Student’s t test was used to compare mutants and the wild type (*, P ≤ 0.05; and **, P ≤ 0.01). CFU, Colony-forming units.
SA pretreatment resulted in about a 1-log decrease in bacterial counts in Pst-inoculated plants (Fig. 10A). However, there was no significant difference in Pst counts in the wild-type control (Col-0) and various acs mutants after SA pretreatment. The loss of Pst susceptibility in acs mutants after SA pretreatment is likely due to a strong systemic acquired resistance (SAR) effect on other defense responses, which overshadowed the effect of ethylene on plant resistance against Pst. In contrast, SA pretreatment did not significantly affect the growth of Pst-avrRpt2 in Arabidopsis (Fig. 10B). Furthermore, even with SA pretreatment, acs mutants were still more susceptible to Pst-avrRpt2 (i.e. SA could not mask the requirement of ethylene biosynthesis during Arabidopsis resistance against Pst-avrRpt2). This result suggests that ethylene induction is more important to ETI and resistance against Pst-avrRpt2, which is consistent with the higher levels of ethylene induction during Arabidopsis-Pst-avrRpt2 interaction.
DISCUSSION
Our understanding of ethylene involvement in plant immunity is mostly based on analyses of ethylene sensing/signaling mutants, such as ethylene response1 and ethylene insensitive2 (ein2). Detailed measurement of ethylene induction in plant immunity and its functional analysis using ethylene biosynthetic mutants are lacking. In this study, we characterized the complex interplay between Arabidopsis and Pst in controlling ethylene biosynthesis (Fig. 11). Ethylene is highly induced after plant sensing of pathogen-derived PAMPs or effectors in Arabidopsis. It is interesting that Pst actively suppresses PAMP-triggered ethylene induction in Arabidopsis via a TTSS-dependent pathway. The battle between Arabidopsis and Pst over ethylene induction would suggest a positive role of ethylene in plant immunity. Consistent with this speculation, high-order acs mutants with greatly reduced ethylene induction are more susceptible to both Pst and Pst-avrRpt2, supporting a positive role of ethylene in plant immunity.
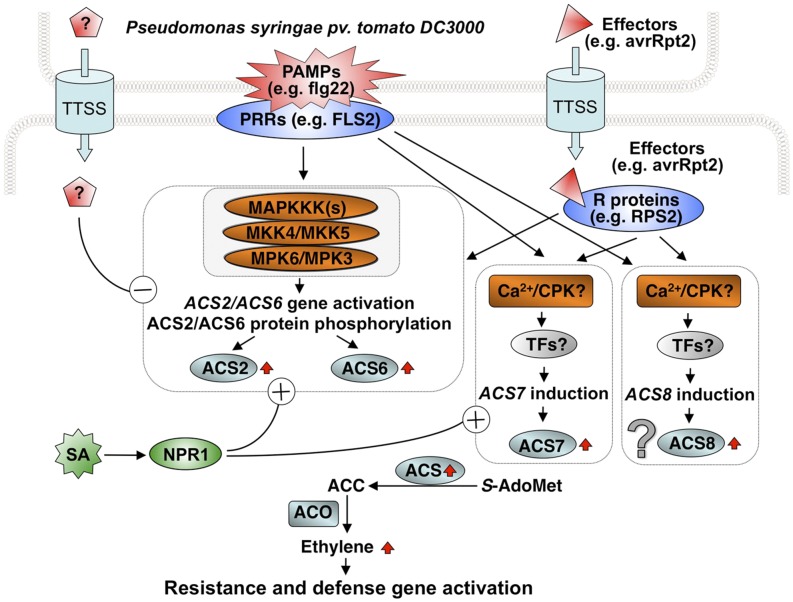
Figure 11.
Model depicting the regulation of ethylene biosynthesis in the Arabidopsis-Pst interaction, which involves PTI, effector-mediated suppression, ETI, and SA-induced potentiation. Three ACS members contribute to the Pst-induced ethylene biosynthesis. Among them, ACS2 and ACS6 are regulated by the stress-responsive MPK3/MPK6 cascade, while ACS7 is regulated by the unidentified signaling pathways. The contribution of ACS8 is uncertain due to the ethylene production by Pst. Even ACS8 is involved; its contribution should be minimal based on the low residual ethylene production in the acs1 acs2 acs6 acs4 acs5 acs9 acs7 acs11 mutant inoculated with Pst. All four ACS genes are activated at the transcriptional level. ACS2 and ACS6 are also regulated at the protein stability level by MPK3/MPK6 phosphorylation. Both the MAPK and unidentified pathways are activated in PTI and ETI. The MPK3/MPK6-regulated ACS2/ACS6 branch is also targeted by the unidentified effectors. This pathway is also the major target of the SA-induced potentiation of ethylene biosynthesis. In addition, induction of ACS7 expression by Pst is potentiated by SA pretreatment, which is similar to ACS2 and ACS6. In contrast, ACS8 is not involved in SA-potentiated ethylene production. ACC, 1-Aminocyclopropane-1-carboxylic acid; ACO, ACC oxidase; FLS2, FLAGELLIN SENSING2; PRRs, pattern-recognition receptors; S-AdoMet, S-adenosylmethionine; TFs, transcription factors.
Pretreatment of Arabidopsis with SA, a key hormone in plant SAR, greatly potentiates the ethylene induction in response to Pst inoculation. The SA potentiation effect is dependent mostly on MPK6 and MPK3, two pathogen-responsive MAPKs, and their downstream ACS2 and ACS6, two type I ACS isoforms. In addition, the sole member of type III ACS, ACS7, whose expression is induced by Pst infection and enhanced by SA pretreatment, also contributes to the pathogen-induced ethylene production. Pst-avrRpt2 induces a higher level of ethylene production, which is dependent on RPS2, the R protein corresponding to avrRpt2. Elevated ethylene induction after Pst-avrRpt2 inoculation is dependent on NPR1 and SID2, two key components in SA signaling and biosynthesis, respectively. This is consistent with a positive role of SA in both local immunity and the SAR response. Previously, it was reported that ethylene production in plants inoculated with Pst-avrRpt2 is partially compromised in NahG plants (Lieberherr et al., 2003). However, ethylene induction by Pst without the avrRpt2 effector was not characterized; therefore, it was unclear whether the SA-dependent ethylene induction is part of the PTI or ETI.
The Battle between Pst and Arabidopsis in Controlling Ethylene Biosynthesis during Plant Immunity
Ethylene induction during plant immunity is dependent on pathogen perception via pattern recognition receptors and R proteins, downstream signaling pathways such as the MPK3/MPK6 cascade, transcription factors involved in ACS gene activation, and phosphorylation stabilization of ACS proteins by MPK3/MPK6 and, possibly, CPKs (Fig. 11; Chae and Kieber, 2005; Xu and Zhang, 2014, 2015a). In addition, ethylene biosynthesis is targeted by Pst effectors, either directly or indirectly. Within the first 3 h of inoculation, the rates of ethylene production induced by Pst, Pst-hrcC−, and Pst-avrRpt2 were similar (Fig. 1B), which is a part of the PTI in response to all three Pst strains. After that, Pst-induced ethylene biosynthesis decreases in comparison with that induced by Pst-hrcC−, suggesting that PAMP-induced ethylene biosynthesis is suppressed by effectors delivered into plant cells. In contrast, Pst-avrRpt2 induces higher ethylene production as a result of ETI (Figs. 1B and 2). At this stage, the exact mechanism underlying the effector-mediated suppression of ethylene production is unclear. We can exclude a direct suppression of the pathogen-responsive MPK3/MPK6 in this case, because MPK3/MPK6 activation in seedlings inoculated with Pst is comparable to that inoculated with Pst-hrcC− (Fig. 4; Supplemental Fig. S3A), suggesting that Pst lacks effectors that negatively impact the activation of the MPK3/MPK6 cascade, a key signaling pathway in ethylene induction. We also observed high-level ACS2 and ACS6 gene activation in response to Pst inoculation in Arabidopsis (Fig. 7, −SA groups), which, together with MPK3/MPK6 activation, should have led to an increase in ACS activity and higher ethylene production. However, there is little/no contribution from ACS2 and ACS6 to Pst-induced ethylene production (Fig. 6, −SA groups), suggesting that Pst effectors might target these two ACS isoforms and impede their function directly or indirectly.
In SA-pretreated seedlings, Pst-induced ethylene biosynthesis is much higher than that induced by Pst-hrcC− (Fig. 1, C and D), suggesting that SA not only can reverse the suppression of Pst effectors but also can enhance ethylene biosynthesis beyond the maximal level that can be induced by PAMPs. It is likely that SA pretreatment induces additional sensing/signaling pathways that are otherwise absent in plant cells without SA pretreatment. These induced pathways can recognize additional PAMPs, DAMPs, and/or effectors upon bacterial infection. This speculation is consistent with the finding that flg22 treatment of SA-pretreated seedlings resulted in little increase in ethylene biosynthesis (Supplemental Fig. S7), indicating that SA does not elevate PAMP-triggered responses in general but rather may promote the recognition of additional PAMPs, DAMPs, and/or effectors, thus leading to more robust defense responses. Supporting this speculation, it was recently reported that SA affects Arabidopsis microbial pattern receptor kinase levels and signaling (Tateda et al., 2014).
Potentiation of Pst-Induced Ethylene Biosynthesis by SA Pretreatment in Plant Immunity
Plant immunity involves almost all plant hormones. Besides SA, ethylene, and JA, the three well-recognized stress/defense hormones, other plant hormones, including abscisic acid, GAs, auxins, cytokinins, and brassinosteroids, can all modulate plant immunity (for review, see Spoel and Dong, 2008; Pieterse et al., 2012; Kazan and Lyons, 2014). They function as secondary signaling molecules downstream of plant sensing of PAMPs, DAMPs, or effectors and can influence plant defense responses either positively or negatively (Glazebrook, 2005; Broekaert et al., 2006; van Loon et al., 2006; Pieterse et al., 2012). An increasing number of pathogen-derived effectors have been found to target/manipulate plant hormone biosynthesis or signaling to facilitate the pathogenesis process, illustrating the importance of plant hormones in plant immunity (Kazan and Lyons, 2014). Cross talk between different hormones, either antagonistically and synergistically, provides another layer of regulation to fine-tune plant immune responses to ward off pathogens.
Pretreatment of Arabidopsis with SA potentiates ethylene biosynthesis in plant immunity, revealing a novel cross talk between SA and ethylene. These enhanced ethylene levels might be important to not only ethylene-regulated defense gene expression and pathogen resistance but also to the maintenance of a balanced SA signaling. It was reported that loss of function of ethylene signaling leads to a higher level of SA biosynthesis and response (Chen et al., 2009). Loss of function of MPK3/MPK6 compromises the potentiation effect of SA pretreatment (Fig. 5; Supplemental Fig. S4). MPK3/MPK6 involvement in the process is likely a result of their regulation of ACS2 and ACS6, two major ACS isoforms contributing to the SA-potentiated ethylene induction (Figs. 6 and 8). ACS2 and ACS6 are downstream substrates of MPK3/MPK6 (Liu and Zhang, 2004; Han et al., 2010). In addition, ACS2 and ACS6 are also regulated at the transcriptional level by the MPK3/MPK6 cascade (Li et al., 2012). In-gel kinase assays revealed that SA pretreatment potentiates MPK3/MPK6 activation after Pst and Pst-avrRpt2 inoculation (Fig. 4; Supplemental Fig. S3). It is likely that enhanced phosphorylation stabilization of ACS2/ACS6 proteins by MPK3/MPK6, together with enhanced ACS2/ACS6 gene activation (Fig. 7), results in the great enhancement of ethylene biosynthesis in Arabidopsis after SA pretreatment. Consistent with this, SA potentiation of ethylene induction after both Pst and Pst-avrRpt2 inoculation is mostly dependent on ACS2 and ACS6. In the acs2 acs6 double mutant, more than 80% of the ethylene induction was blocked (Figs. 6 and 8).
Differential Regulation and Contribution of ACS Isoforms to Ethylene Biosynthesis in Plant Immunity
Based on mutant analysis, at least three isoforms of ACS, ACS2, ACS6, and ACS7, contribute to ethylene induction in plant immunity (Figs. 6 and 8). This conclusion is consistent with the activation of their gene expression in the process (Fig. 7). At this stage, whether ACS8 is also involved is hard to determine because of the lack of an acs8 mutant and the ability of Pst to produce low levels of ethylene (Supplemental Fig. S5). Based on their regulation by different signaling pathways, these four ACS isoforms can be classified into three different groups. The first group includes ACS2 and ACS6, two type I ACS that are regulated by the MPK3/MPK6 signaling pathway (Liu and Zhang, 2004; Han et al., 2010; Li et al., 2012). They are the major contributors to SA-potentiated ethylene biosynthesis (Figs. 6 and 8, +SA groups). In addition, they are targeted by Pst effectors, which results in their minimal contribution to ethylene induction by Pst (Fig. 6, −SA groups), despite (1) the high activation of MPK3/MPK6 (Fig. 4; Supplemental Fig. S3), which should phosphorylate ACS2 and ACS6 and stabilize the protein, and (2) ACS2/ACS6 gene activation (Fig. 7). Both should have led to their contribution to ethylene biosynthesis if they were not suppressed by Pst effectors. The second group includes ACS7, the sole member of type III ACS, which also contributes to the SA-potentiated ethylene induction (Figs. 6 and 8, +SA groups). The third group includes ACS8, a type II ACS. ACS8 might contribute to the low-level induction of ethylene by Pst, which is not responsive to SA pretreatment (Figs. 6 and 8, acs2 acs6 acs4 acs5 acs9 acs7 sextuple and acs1 acs2 acs6 acs4 acs5 acs9 acs7 acs11 octuple mutants with and without SA pretreatment). However, at least part, if not all, of this low-level ethylene is produced by Pst (Supplemental Fig. S5), which brings uncertainty to the involvement of ACS8 (Fig. 11).
Ethylene Is a Positive Regulator of Plant Immunity against Bacterial Pathogens
Ethylene is involved in diverse biological processes, including plant innate immunity. Based on analyses of ethylene signaling mutants, it is concluded that ethylene plays a positive role in plant resistance against necrotrophic pathogens but negatively influences plant resistance against bacterial pathogens (Glazebrook, 2005; Broekaert et al., 2006; van Loon et al., 2006; Pieterse et al., 2012). For instance, overexpression of Arabidopsis ETHYLENE RESPONSE FACTOR1, a transcription factor in the ethylene signaling pathway, results in increased resistance to B. cinerea but reduces SA-mediated resistance to Pst (Berrocal-Lobo et al., 2002). Analysis of the ein2 mutant revealed that ethylene signaling is not required for active resistance against bacterial pathogens but is involved in symptom development, and the lack of ethylene signaling makes plants more tolerant to the bacterial pathogens (Bent et al., 1992). In addition, EIN3 and EIN3-LIKE1 (EIL1) negatively impact the expression of SID2 and, therefore, SA biosynthesis and plant resistance against bacterial pathogens (Chen et al., 2009). However, recent studies also concluded that ethylene signaling is essential to the expression of genes encoding pattern-recognition receptors or DAMPs, such as ELICITOR PEPTIDE PRECURSORs, and therefore has a positive role in plant immunity against bacterial pathogens (Boutrot et al., 2010; Mersmann et al., 2010; Liu et al., 2013; Tintor et al., 2013; Zipfel, 2013). Furthermore, gene expression profiling analyses revealed that SA, JA, and ethylene all contribute positively to immunity to both biotrophic and necrotrophic pathogens (Glazebrook et al., 2003; Tsuda et al., 2009; Sato et al., 2010).
In this report, we conclude that ethylene plays a positive role in both PTI and ETI during plant immunity against bacterial pathogens based on an analysis of ethylene biosynthetic mutants. How to reconcile these different conclusions about the role of ethylene in plant bacterial resistance? One possible explanation is that key ethylene signaling components such as EIN2 and EIN3/EIL1 are critical to the functions of other plant hormones because of cross talk (Anderson et al., 2004; Chen et al., 2009; Pieterse et al., 2012). Their mutation may interfere with the functions of other plant hormones in plant immunity. For instance, loss of key ethylene signaling components, such as in the ein3 eil1 double mutant, leads to unchecked SA biosynthesis and/or signaling pathways. The exaggerated SA signaling leads to an enhanced resistance response in ethylene signaling mutants, giving the impression that ethylene has a negative role in plant bacterial resistance (Chen et al., 2009). The use of ethylene biosynthetic mutants in this study should alleviate this problem. Based on our results, ethylene plays a less important role in comparison with SA in plant basal resistance against Pst. The compromised resistance of the loss of ethylene biosynthetic mutants against Pst can be reversed by SA pretreatment (Fig. 10A), suggesting a dominant role of SA in the process. In contrast, SA pretreatment cannot mask the compromised resistance of ethylene biosynthesis mutants against Pst-avrRpt2 (Fig. 10B). Previously, ethylene biosynthesis was shown to be critical to plant resistance against B. cinerea, a necrotrophic pathogen (Tsuchisaka et al., 2009; Li et al., 2012). Together with the findings in this report, we can conclude that ethylene plays an important role in plant resistance against both fungal and bacterial pathogens.
MATERIALS AND METHODS
Plant Growth Conditions and Treatments
Soil-grown Arabidopsis (Arabidopsis thaliana) plants were maintained at 22°C in a growth chamber with a 14-h light cycle (100 µE m−2 s−1). For experiments, seeds were surface sterilized. After imbibition at 4°C for 3 to 5 d, seeds were sown in petri dishes with liquid one-half-strength MS medium and grown in a growth chamber at 22°C with continuous light (70 µE m−2 s−1). Six-day-old seedlings were transferred to 20-mL GC vials with 6 mL of liquid one-half-strength MS medium (seven seedlings per vial) and maintained under the same growth conditions. Twelve-day-old seedlings grown in GC vials were pretreated with SA (100 µm final concentration) by the addition of 100 mm stock solution prepared in DMSO solvent. Two days later, seedlings were inoculated with Pseudomonas syringae pv tomato DC3000 (Pst), Pst-hrcC−, or Pst-avrRpt2 or mock inoculated.
Pst strains were maintained as glycerol stocks at −80°C. For inoculum preparation, 20-µL stocks were spread on Difco Pseudomonas Agar F plates with appropriate antibiotics. After incubation at 28°C for approximately 24 h, bacteria were scraped off the plates and suspended in one-half-strength MS medium to OD600 of 1. Pst inoculation was simply done by adding 120 µL of inoculum to the GC vials with 6 mL of medium to a final OD600 of 0.02.
For experiments with multiple time points, at least two independent repetitions were performed. For single time point experiments, at least three independent repetitions were performed. Data from one of the independent repetitions with similar results are shown in the figures. Student’s t test was used for statistical analysis of most of the data. One and two asterisks above the columns are used to indicate differences that are statistically significant (P < 0.05) and very significant (P < 0.01), respectively. When different high-order acs mutants were compared, one-way ANOVA was performed. Lowercase letters above the columns are used to indicate statistically different groups. When two or more time points are significantly different, we conclude that the two genotypes produce statistically different amounts of ethylene.
Mutant Lines and Generation of Transgenic Plants
Arabidopsis ecotype Col-0 was used as the wild-type control. NahG, npr1, and sid2-2 seeds were kindly provided by Drs. Daniel Klessig and Xinnian Dong. DEX-inducible promoter-driven avrRpt2 transgenic plants (GVG-avrRpt2) were obtained from Dr. Brian Staskawicz’s laboratory (McNellis et al., 1998). Transfer DNA insertion mutant alleles of MPK3 (At3g45640) and MPK6 (At2g43790) were described previously (Liu and Zhang, 2004; Wang et al., 2007). High-order acs mutants generated in Dr. Athanasios Theologis’s laboratory (Tsuchisaka et al., 2009) were obtained from the Arabidopsis Biological Resource Center. The stock numbers are CS16564 (acs2), CS16569 (acs6), CS16581 (acs2 acs6), CS16644 (acs2 acs6 acs4 acs5 acs9), CS16649 (acs2 acs6 acs4 acs5 acs9 acs7), and CS16651 (acs1 acs2 acs6 acs4 acs5 acs9 acs7 acs11). The chemical genetically rescued mpk3 mpk6 double mutant (MPK6SR) was generated by using an NA-PP1 inhibitor-sensitized MPK6 variant, MPK6YG, driven by the MPK6 native promoter (PMPK6:MPK6YG). In the absence of NA-PP1 inhibitor, the functional PMPK6:MPK6YG is able to rescue the mpk3 mpk6 double mutant, resulting in plants with the mpk3 mpk6 PMPK6:MPK6YG genotype (named MPK6SR plants; Xu et al., 2014). Application of the NA-PP1 inhibitor (final concentration of 5 µm) can effectively inhibit the activity of MPK6YG, giving rise to the activity null mutants of both MPK3 and MPK6. Two independent transgenic lines were used, and similar results were obtained.
Ethylene Measurement
GC vials with Arabidopsis seedlings were flushed and capped immediately after treatment. At the indicated times, ethylene levels in the headspace of the GC vials were measured by GC as described previously (Liu and Zhang, 2004; Han et al., 2010). Ethylene production rates were calculated as the average rates of ethylene production in the intervals of the two adjacent time points. Seedlings were then collected, weighed, frozen in liquid nitrogen, and stored at −80°C for future analysis.
Protein Extraction and In-Gel Kinase Assay
Total proteins were extracted from treated seedlings as described previously (Liu and Zhang, 2004). The concentration of protein was determined by using the Bio-Rad protein assay kit with bovine serum albumin as the standard. MBP was used as the substrate for the in gel-kinase assay (Zhang and Klessig, 1997; Liu and Zhang, 2004).
RNA Extraction and Real-Time PCR Analysis
Total RNA was extracted using the Trizol reagent (Invitrogen). After DNase treatment, RNA (1 µg) was used for reverse transcription. Real-time PCR analysis was performed using an Opticon 2 real-time PCR machine as described previously (Ren et al., 2008). The transcript of the EF1α gene was used to normalize the samples. Gene expression was calculated as relative levels to that of the EF1α gene in the same sample. The primers used for real-time PCR were PR1 (At2g14610; 5′-CATACACTCTGGTGGGCCTTA-3′ and 5′-AGTGACCACAAACTCCATTGC-3′), ACS2 (At1g01480; 5′-GGATGGTTTAGGATTTGCTTTG-3′ and 5′-GCACTCTTGTTCTGGATTACCTG-3′), ACS6 (At4g11280; 5′-GTTCCAACCCCTTATTATCC-3′ and 5′-CCGTAATCTTGAACCCATTA-3′), ACS7 (At4g26200; 5′-ACGGTACGATACCATTGTGGA-3′ and 5′-GCTCGCCGTCTTTAGTTTTCT-3′), ACS8 (At4g37770; 5′-CCTTCCTTCCTTCAAGAATGC-3′ and 5′-GAGAGTCTCGTTAGCCGGAGT-3′), and EF1α (At5g60390; 5′-TGAGCACGCTCTTCTTGCTTTCA-3′ and 5′-GGTGGTGGCATCCATCTTGTTACA-3′).
Pathogen Resistance Assay
Five-week-old Col-0 and acs mutant plants grown under a short-day light cycle (10 h of light and 14 h of dark) were infiltrated with Pst or Pst-avrRpt2 (OD600 = 0.001). Eight leaves were collected 3 d post inoculation for quantification of bacterial growth. SA pretreatment was done by spraying the plants with SA (1 mm) and drenching soil with SA (1 mm) at the same time (Lawton et al., 1994; DeFraia et al., 2010). The flats were then covered with a dome for 24 h before bacterial infiltration. SA solution was prepared in water for drenching and in 0.01% (v/v) Silwet L-77 for spraying. The pH of the SA solutions was adjusted to 6.5 with KOH.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers MPK3 (At3g45640), MPK6 (At2g43790), EF1α (At5g60390), ACS1 (At3g61510), ACS2 (At1g01480), ACS4 (At2g22810), ACS5 (At5g65800), ACS6 (At4g11280), ACS7 (At4g26200), ACS8 (At4g37770), ACS9 (At3g49700), and ACS11 (At4g08040).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Validation of SA responsiveness in liquid-cultured Arabidopsis seedlings.
Supplemental Figure S2. SA treatment weakly induces ethylene biosynthesis in Arabidopsis.
Supplemental Figure S3. Enhanced activation of MPK3 and MPK6 by Pst in SA-pretreated Arabidopsis.
Supplemental Figure S4. MPK3 and MPK6 are required for SA-potentiated ethylene biosynthesis in Arabidopsis inoculated with Pst.
Supplemental Figure S5. Low-level ethylene production by Pst.
Supplemental Figure S6. Potentiation of Pst-induced ethylene biosynthesis by SA pretreatment is dependent on the function sid2-2.
Supplemental Figure S7. Weak potentiation of flg22-induced ethylene biosynthesis by SA pretreatment in Arabidopsis.
Supplementary Material
Acknowledgments
We thank Dr. Athanasios Theologis for the high-order acs mutant seeds donated to the Arabidopsis Biological Resource Center and the Arabidopsis Biological Resource Center for seed stocks.
Glossary
- PAMP
pathogen/microbe-associated molecular pattern
- PTI
pathogen/microbe-associated molecular pattern-triggered immunity
- TTSS
type III secretion system
- ETI
effector-triggered immunity
- DAMP
damage-associated molecular pattern
- SA
salicylic acid
- JA
jasmonic acid
- Pst
Pseudomonas syringae pv tomato DC3000
- GC
gas chromatography
- OD600
optical density at 600 nm
- DEX
dexamethasone
- Col-0
Columbia-0
- NA-PP1
4-amino-1-tert-butyl-3-(1′-naphthyl)pyrazolo[3,4-d]pyrimidine
- MS
Murashige and Skoog
- SAR
systemic acquired resistance
- DMSO
dimethyl sulfoxide
- CPK
calcium-dependent protein kinase
Footnotes
This work was supported by the Natural Science Foundation of China (grant nos. 91317303 and 31300244), the U.S. National Science Foundation (grant nos. IOS–0743957 and MCB–0950519), and the China Scholarship Council (grant no. CSC 201203250023 to R.G.).
Articles can be viewed without a subscription.
References
- Alfano JR, Collmer A (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu Rev Phytopathol 42: 385–414 [DOI] [PubMed] [Google Scholar]
- An C, Mou Z (2011) Salicylic acid and its function in plant immunity. J Integr Plant Biol 53: 412–428 [DOI] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson E, Jenkins T, Brodersen P, Thorgrimsen S, Petersen NHT, Zhu S, Qiu JL, Micheelsen P, Rocher A, Petersen M, et al. (2005) The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J 24: 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM. (2005) Are innate immune signaling pathways in plants and animals conserved? Nat Immunol 6: 973–979 [DOI] [PubMed] [Google Scholar]
- Beckers GJM, Jaskiewicz M, Liu Y, Underwood WR, He SY, Zhang S, Conrath U (2009) Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21: 944–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Innes RW, Ecker JR, Staskawicz BJ (1992) Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol Plant Microbe Interact 5: 372–378 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29: 23–32 [DOI] [PubMed] [Google Scholar]
- Bethke G, Unthan T, Uhrig JF, Pöschl Y, Gust AA, Scheel D, Lee J (2009) Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc Natl Acad Sci USA 106: 8067–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Boller T, He SY (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, Segonzac C, Chang KN, Qiao H, Ecker JR, Zipfel C, Rathjen JP (2010) Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc Natl Acad Sci USA 107: 14502–14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekaert WF, Delauré SL, De Bolle MFC, Cammue BPA (2006) The role of ethylene in host-pathogen interactions. Annu Rev Phytopathol 44: 393–416 [DOI] [PubMed] [Google Scholar]
- Chae HS, Kieber JJ (2005) Eto Brute? Role of ACS turnover in regulating ethylene biosynthesis. Trends Plant Sci 10: 291–296 [DOI] [PubMed] [Google Scholar]
- Chen H, Xue L, Chintamanani S, Germain H, Lin H, Cui H, Cai R, Zuo J, Tang X, Li X, et al. (2009) ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21: 2527–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- DeFraia CT, Zhang X, Mou Z (2010) Elongator subunit 2 is an accelerator of immune responses in Arabidopsis thaliana. Plant J 64: 511–523 [DOI] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Feng F, Zhou JM (2012) Plant-bacterial pathogen interactions mediated by type III effectors. Curr Opin Plant Biol 15: 469–476 [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Dong X (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64: 839–863 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Chen W, Estes B, Chang HS, Nawrath C, Métraux JP, Zhu T, Katagiri F (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J 34: 217–228 [DOI] [PubMed] [Google Scholar]
- Guan Y, Meng X, Khanna R, LaMontagne E, Liu Y, Zhang S (2014) Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet 10: e1004384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Tian F, Wamboldt Y, Alfano JR (2009) The majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol Plant Microbe Interact 22: 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, Ehlting J, Beaudoin N, Barbazuk B, Klessig D, Lee J, et al. (2006) Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci 11: 192–198 [DOI] [PubMed] [Google Scholar]
- Han L, Li GJ, Yang KY, Mao G, Wang R, Liu Y, Zhang S (2010) Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J 64: 114–127 [DOI] [PubMed] [Google Scholar]
- Huang SJ, Chang CL, Wang PH, Tsai MC, Hsu PH, Chang IF (2013) A type III ACC synthase, ACS7, is involved in root gravitropism in Arabidopsis thaliana. J Exp Bot 64: 4343–4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Shinozaki K, Tena G, Sheen J, Henry Y, Champion A, Kreis M, Zhang S, Hirt H, Wilson C, et al. (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Joo S, Liu Y, Lueth A, Zhang S (2008) MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J 54: 129–140 [DOI] [PubMed] [Google Scholar]
- Katagiri F, Tsuda K (2010) Understanding the plant immune system. Mol Plant Microbe Interact 23: 1531–1536 [DOI] [PubMed] [Google Scholar]
- Kazan K, Lyons R (2014) Intervention of phytohormone pathways by pathogen effectors. Plant Cell 26: 2285–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H. (1993) Ethylene biosynthesis. Annu Rev Plant Biol 44: 283–307 [Google Scholar]
- Lawton KA, Potter SL, Uknes S, Ryals J (1994) Acquired resistance signal transduction in Arabidopsis is ethylene independent. Plant Cell 6: 581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Meng X, Wang R, Mao G, Han L, Liu Y, Zhang S (2012) Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet 8: e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberherr D, Wagner U, Dubuis PH, Métraux JP, Mauch F (2003) The rapid induction of glutathione S-transferases AtGSTF2 and AtGSTF6 by avirulent Pseudomonas syringae is the result of combined salicylic acid and ethylene signaling. Plant Cell Physiol 44: 750–757 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, Tian X, Zhou JM (2013) BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci USA 110: 6205–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis TW, Mudgett MB, Li K, Aoyama T, Horvath D, Chua NH, Staskawicz BJ (1998) Glucocorticoid-inducible expression of a bacterial avirulence gene in transgenic Arabidopsis induces hypersensitive cell death. Plant J 14: 247–257 [DOI] [PubMed] [Google Scholar]
- Meng X, Xu J, He Y, Yang KY, Mordorski B, Liu Y, Zhang S (2013) Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25: 1126–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Zhang S (2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51: 245–266 [DOI] [PubMed] [Google Scholar]
- Mersmann S, Bourdais G, Rietz S, Robatzek S (2010) Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol 154: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley KF, Martin GB (2005) Role of mitogen-activated protein kinases in plant immunity. Curr Opin Plant Biol 8: 541–547 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28: 489–521 [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Schikora A, Hirt H (2009) MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Ren D, Liu Y, Yang KY, Han L, Mao G, Glazebrook J, Zhang S (2008) A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 105: 5638–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Yang H, Zhang S (2002) Cell death mediated by mitogen-activated protein kinase pathway is associated with the generation of hydrogen peroxide in Arabidopsis. J Biol Chem 277: 559–565 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JDG (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Rodriguez MCS, Petersen M, Mundy J (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61: 621–649 [DOI] [PubMed] [Google Scholar]
- Sato M, Tsuda K, Wang L, Coller J, Watanabe Y, Glazebrook J, Katagiri F (2010) Network modeling reveals prevalent negative regulatory relationships between signaling sectors in Arabidopsis immune signaling. PLoS Pathog 6: e1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Dong X (2008) Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3: 348–351 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Dong X (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12: 89–100 [DOI] [PubMed] [Google Scholar]
- Tateda C, Zhang Z, Shrestha J, Jelenska J, Chinchilla D, Greenberg JT (2014) Salicylic acid regulates Arabidopsis microbial pattern receptor kinase levels and signaling. Plant Cell 26: 4171–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena G, Boudsocq M, Sheen J (2011) Protein kinase signaling networks in plant innate immunity. Curr Opin Plant Biol 14: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintor N, Ross A, Kanehara K, Yamada K, Fan L, Kemmerling B, Nürnberger T, Tsuda K, Saijo Y (2013) Layered pattern receptor signaling via ethylene and endogenous elicitor peptides during Arabidopsis immunity to bacterial infection. Proc Natl Acad Sci USA 110: 6211–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A, Yu G, Jin H, Alonso JM, Ecker JR, Zhang X, Gao S, Theologis A (2009) A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 183: 979–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F (2009) Network properties of robust immunity in plants. PLoS Genet 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, Geraats BPJ, Linthorst HJM (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci 11: 184–191 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Wang H, Liu Y, Bruffett K, Lee J, Hause G, Walker JC, Zhang S (2008) Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell 20: 602–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KLC, Li H, Ecker JR (2002) Ethylene biosynthesis and signaling networks. Plant Cell (Suppl) 14: S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingart H, Volksch B (1997) Ethylene production by Pseudomonas syringae pathovars in vitro and in planta. Appl Environ Microbiol 63: 156–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Xie J, Yan C, Zou X, Ren D, Zhang S (2014) A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J 77: 222–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang S (2014) Regulation of ethylene biosynthesis and signaling by protein kinases and phosphatases. Mol Plant 7: 939–942 [DOI] [PubMed] [Google Scholar]
- Xu J, Zhang S (2015a) Ethylene biosynthesis and regulation in plants. In Wen CK, ed, Ethylene in Plants. Springer, Dordrecht, The Netherlands, pp 1–25 [Google Scholar]
- Xu J, Zhang S (2015b) Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci 20: 56–64 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Nagata M, Saito K, Wang KL, Ecker JR (2005) Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF (1997) Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9: 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF (2001) MAPK cascades in plant defense signaling. Trends Plant Sci 6: 520–527 [DOI] [PubMed] [Google Scholar]
- Zipfel C. (2013) Combined roles of ethylene and endogenous peptides in regulating plant immunity and growth. Proc Natl Acad Sci USA 110: 5748–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. (2014) Plant pattern-recognition receptors. Trends Immunol 35: 345–351 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.