Ethylene Response Factors are transcription factors that act as a key regulatory hub in plant response to abiotic stresses, integrating ethylene, ABA, jasmonate, and redox signaling.
Abstract
Ethylene is essential for many developmental processes and a key mediator of biotic and abiotic stress responses in plants. The ethylene signaling and response pathway includes Ethylene Response Factors (ERFs), which belong to the transcription factor family APETALA2/ERF. It is well known that ERFs regulate molecular response to pathogen attack by binding to sequences containing AGCCGCC motifs (the GCC box), a cis-acting element. However, recent studies suggest that several ERFs also bind to dehydration-responsive elements and act as a key regulatory hub in plant responses to abiotic stresses. Here, we review some of the recent advances in our understanding of the ethylene signaling and response pathway, with emphasis on ERFs and their role in hormone cross talk and redox signaling under abiotic stresses. We conclude that ERFs act as a key regulatory hub, integrating ethylene, abscisic acid, jasmonate, and redox signaling in the plant response to a number of abiotic stresses.
Environmental stresses, including drought, salinity, high light, and extreme temperatures, influence plant growth and productivity. These abiotic stresses result in reductions in growth, stomatal and nonstomatal limitations on photosynthesis, and alterations in both hormonal balance and reduction/oxidation (redox) processes, potentially leading to enhanced lipid peroxidation, protein oxidation, and DNA damage (Munns and Tester, 2008; Mittler et al., 2011; Munné-Bosch et al., 2013). Plant responses and adaptation to abiotic stresses are controlled by molecular signal transduction cascades. In these cascades, plant hormones as a part of the signal network function as central integrators that link and reprogram the complex stress-adaptive signaling cascades (Ma et al., 2006; Golldack et al., 2014).
Several plant hormones, such as ethylene (Zhao and Schaller, 2004; Cheng et al., 2013), abscisic acid (ABA; Wu et al., 2007), jasmonates (Cela et al., 2011; Cheng et al., 2013), salicylic acid (Jayakannan et al., 2015), GAs (Magome et al., 2008), cytokinins (Wu et al., 2014b), auxin (He et al., 2005), and brassinosteroids (Divi et al., 2010), have been reported to be involved in stress signaling. Despite its simple two-carbon structure, the plant hormone ethylene serves as a key mediator of biotic and abiotic stress factors. Transcription factors (TFs) control the majority of stress response genes, and of more than 1,800 TFs identified in Arabidopsis (Arabidopsis thaliana; Arabidopsis Genome Initiative, 2000; Riechmann et al., 2000; Gong et al., 2004), the APETALA2 (AP2)/Ethylene Response Factor (ERF) superfamily plays a pivotal role in adaptation to biotic and abiotic stresses, such as those caused by pathogens, wounding, cold and heat stress, UV light, drought, and salinity (Mizoi et al., 2012). A genome-wide analysis of plant species has identified, for instance, the following numbers of members of the AP2/ERF superfamily: Arabidopsis, 147 (Nakano et al., 2006); Populus spp., 200 (Zhuang et al., 2008); Brassica spp., 291 (Song et al., 2013); Citrus spp., 108 (Ito et al., 2014); Vitis spp., 149 (Licausi et al., 2010); Solanum spp., 155 (Charfeddine et al., 2015); and Oryza spp., 180 (Nakano et al., 2006; Sharoni et al., 2011).
Sakuma et al. (2002) classified AP2/ERF TFs into five subfamilies: AP2, related to ABSCISIC ACID INSENSITIVE3 (ABI3)/VIVIPAROUS1 (VP1), dehydration-responsive element (DRE) binding protein, ERF, and others according to the number and similarity of the DNA binding domains. ERFs have been extensively reported to be involved in the response to pathogen attack by binding to sequences containing AGCCGCC motifs (the GCC box), a cis-acting ethylene response element (Solano et al., 1998; Berrocal-Lobo et al., 2002; Lorenzo et al., 2002). However, recent research has shown that several ERFs also bind to DREs and play a regulatory role in plant responses to abiotic stresses (Cheng et al., 2013). Although the role of AP2/ERF TFs as mediators of stress responses and development programs has been reviewed recently (Xu et al., 2011; Mizoi et al., 2012; Licausi et al., 2013), little emphasis has been put on the role of ERFs in abiotic stress tolerance. Here, we focus on the role of ERFs in plant tolerance to abiotic stresses, with an emphasis on hormone cross talk, redox regulation, and abiotic stress signaling.
ERFs IN ETHYLENE RESPONSE
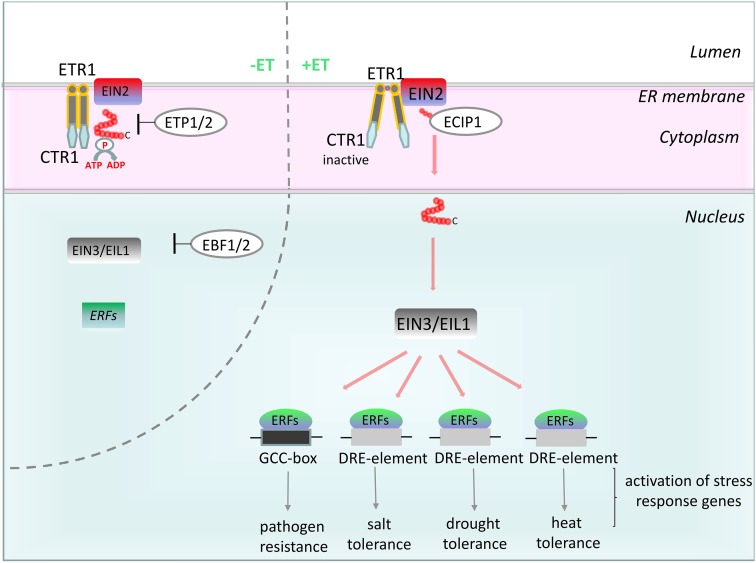
The ethylene signaling and response pathway to the cell nucleus, where the transcription of hundreds of genes is altered, was revealed as a result of the analysis of the model system Arabidopsis (Fig. 1). Ethylene is sensed by five receptors localized at the endoplasmic reticulum membrane that are divided into subfamily I (Ethylene Response1 [ETR1] and Ethylene Response Sensor1 [ERS1]) and subfamily II (ETR2, ERS2, and Ethylene Insensitive4 [EIN4]; Chen et al., 2005; Lacey and Binder, 2014). The ethylene signaling and response pathway also includes the downstream components Constitutive Triple Response1 (CTR1), EIN2, EIN3/Ethylene Insensitive-Like Protein1 (EIL1), and ERFs (Kendrick and Chang, 2008; Stepanova and Alonso, 2009). CTR1 is a negative regulator of ethylene signaling. In the absence of ethylene, the receptors promote CTR1 kinase activity, which phosphorylates the C-terminal domain of EIN2 and thereby, prevents its nucleus localization. However, in the presence of ethylene, the receptors and CTR1 are inactive (Kieber et al., 1993; Gao et al., 2003; Huang et al., 2003; Ju et al., 2012). In contrast, EIN2, which is localized at the ER membrane along with the ethylene receptors and CTR1, positively regulates ethylene signaling (Bisson et al., 2009; Bisson and Groth, 2010). In the absence of ethylene, EIN2 protein levels are reduced because of the interaction with two F-box proteins: Ethylene Insensitive2-Targeting Protein1 (ETP1) and ETP2 (Qiao et al., 2009). In the presence of ethylene, the inactivation of the receptors and CTR1 results in the dephosphorylation and cleavage of the EIN2 C terminus and translocation to the nucleus, where it directly or indirectly regulates EIN3/EIL1 activation (Ju et al., 2012). An MA3 domain-containing protein (ECIP1) interacts with the EIN2 C terminus, leading to enhanced ethylene response (Lei et al., 2011). In the absence of ethylene, the EIN3/EIL1 protein levels are extremely low because of the protein turnover involving the F-box proteins Ethylene Insensitive3-Binding F-Box Protein1 (EBF1) and EBF2. The presence of ethylene down-regulates EBF1 and EBF2 and leads to the accumulation of EIN3/EIL1 proteins, which initiates a transcriptional cascade that results in the activation and repression of hundreds of genes (An et al., 2010). In Arabidopsis, one of the direct targets of EIN3 is the ERF genes (Solano et al., 1998). AtERF and ERF will be used in this review interchangeably.
Figure 1.
Model of the ethylene (ET) signaling pathway to ERFs. In the absence of ethylene (left), the ethylene receptors promote CTR1 kinase activity, resulting in the phosphorylation of the C-terminal domain of EIN2. Because of the protein turnover involving the F-box proteins ETP1/2 and EBF1/2, the protein levels of both EIN2 and EIN3/EIL1 are extremely low. In the presence of ethylene (right), the inactivation of the ethylene receptors and CTR1 results in the dephosphorylation and cleavage of the EIN2 C terminus and translocation to the nucleus, where they regulate EIN3/EIL1 activation directly or indirectly. The direct targets of EIN3 are the TF genes ERFs, such as ERF1, which activates, depending on the stress conditions (either biotic [pathogen infection] or abiotic [e.g. dehydration, salinity, or heat shock] stress), a specific set of stress response genes by binding to the specific cis-acting GCC box and DRE elements. ER, Endoplasmic reticulum; ECIP1, EIN2 C-TERMINUS INTERACTING PROTEIN1.
Recent studies revealed that ERFs are key regulators in abiotic stress tolerance in several species. Enhanced ERF expression has been reported after drought, salinity, light stress, and cold and heat treatments among other abiotic stresses (Table I). Several ERF genes have been reported to be induced by salt stress (38 study cases), drought (27 study cases), low temperature (18 study cases), heat stress (3 study cases), and changes in light availability (14 study cases). It should be noted that ERF gene expression is common to various abiotic stresses, including salt, drought, and cold stress treatment (12 study cases), salt and drought (8 study cases), and others. The effects of overexpressing ERF genes in plant response to abiotic stress have been studied in various plant systems (Table II).
Table I. Expression of genes from the ERF subfamily under abiotic stress.
—, Not studied; JERF1, Jasmonate and Ethylene Response Factor1; OPBP1, Osmotin Promoter Binding Protein1; BIERF3, BTH-Induced ERF Transcriptional Factor3; TSRF1, Tomato Stress-Responsive Factor1; TERF, Tomato Ethylene Response Factor; SlERF, Tomato Ethylene Response Factor; SHN1, ethylene-responsive transcription factor WIN1.
| Species | ERFs | Up-Regulation under Stress |
References | ||||
|---|---|---|---|---|---|---|---|
| Salt | Drought | Cold | Heat | Light | |||
| Arabidopsis | AtERF1a | Yes | Yes | — | Yes | — | Cheng et al., 2013 |
| Arabidopsis | AtERF4 | Yes | — | — | — | — | Seo et al., 2010 |
| Arabidopsis | AtERF5 | No | Yes | — | — | — | Dubois et al. (2013) |
| Arabidopsis | AtERF6 | Yes | Yes | Yes | No | Yes | Dubois et al. (2013); Sewelam et al. (2013); Wang et al. (2013a); Vogel et al. (2014) |
| Arabidopsis | AtERF98 | Yes | — | — | — | — | Zhang et al. (2012) |
| Arabidopsis | AtERF104 | — | — | — | — | Yes | Vogel et al. (2014) |
| Arabidopsis | AtERF105 | — | — | — | — | Yes | Vogel et al. (2014) |
| Brassica rapa | BrERF4 | No | — | — | — | — | Seo et al. (2010) |
| Pepper (Capsicum annuum) | CaERFLP1a | Yes | No | No | — | — | Lee et al. (2004) |
| Pepper | CaPF1a | Yes | Yes | Yes | — | — | Yi et al. (2004) |
| Chickpea (Cicer arietinum) | CarERF116 | Yes | Yes | Yes | Yes | — | Deokar et al. (2015) |
| Chrysanthemum nankingense | CnERF1 | No | — | Yes | — | — | Gao et al. (2015) |
| Camellia reticulata | CitERF | Yes | Yes | Yes | — | — | Yang et al. (2011) |
| Citrus sinensis | CsERF | Yes | Yes | Yes | — | — | Ma et al. (2014) |
| Gossypium hirsutum | GhERF1 | Yes | Yes | Yes | — | — | Qiao et al. (2008) |
| G. hirsutum | GhERF2 | Yes | Yes | Yes | — | — | Jin et al. (2010) |
| G. hirsutum | GhERF3 | Yes | Yes | Yes | — | — | Jin et al. (2009) |
| G. hirsutum | GhERF5 | Yes | Yes | Yes | — | — | Jin et al. (2010) |
| G. hirsutum | GhERF6 | Yes | Yes | Yes | — | — | Jin et al. (2010) |
| Soybean (Glycine max) | GmERF3a | Yes | Yes | No | — | — | Zhang et al. (2009) |
| Jatropha curcas | JcERF1 | Yes | Yes | — | — | — | Yang et al. (2014) |
| Chinese boxthorn (Lycium chinense) | LchERF | Yes | Yes | — | — | — | Wu et al. (2014a, 2014b) |
| Common bird's-foot trefoil Lotus corniculatus | LcERF054 | Yes | No | — | — | — | Sun et al. (2014a) |
| L. corniculatus | LcERF080 | Yes | — | — | — | — | Sun et al. (2014b) |
| Alfala (Medicago sativa) | MsERF8 | Yes | Yes | — | — | — | Chen et al. (2012) |
| Tobacco (Nicotiana tabacum) | JERF1a | Yes | Yes | Yes | — | — | Wu et al. (2007) |
| Tobacco | JERF3a | Yes | Yes | Yes | — | — | Wang et al. (2004) |
| Tobacco | OPBP1 | Yes | — | — | — | — | Guo et al. (2004) |
| Tobacco | OsBIERF3 | Yes | — | — | — | — | Cao et al. (2006) |
| Tobacco | Tsi1a | Yes | — | — | — | — | Park et al. (2001) |
| Tomato (Lycopersicon esculentum) | LeERF3b | Yes | Yes | Yes | No | Yes | Chen et al. (2008); Severo et al. (2015) |
| Tomato | TSRF1a | Yes | Yes | — | — | — | Zhang et al. (2007) |
| Tomato | TERF1a | Yes | Yes | — | — | — | Gao et al. (2008) |
| Tomato | LeERF1 | Yes | — | — | — | — | Hu et al. (2014) |
| Tomato | LeERF2 | Yes | — | — | — | Yes | Hu et al. (2014); Severo et al. (2015) |
| Tomato | SlERF5 | Yes | Yes | Yes | Yes | — | Pan et al. (2012) |
| Tomato | ERF A.1 | — | — | — | — | Yes | Severo et al. (2015) |
| Tomato | ERF A.3 (Pti4) | — | — | — | — | Yes | Severo et al. (2015) |
| Tomato | ERF B.1 | — | — | — | — | Yes | Severo et al. (2015) |
| Tomato | ERF B.2 | — | — | — | — | Yes | Severo et al. (2015) |
| Tomato | ERF B.3 (LeERF4) | — | — | — | — | Yes | Severo et al. (2015) |
| Tomato | ERF C.6 (ERF5) | — | — | — | — | Yes | Severo et al. (2015) |
| Tomato | ERF D.1 | — | — | — | — | Yes | Severo et al. (2015) |
| Tomato | ERF D.3 | — | — | — | — | Yes | Severo et al. (2015) |
| Tomato | ERF G.2 (Pti6) | — | — | — | — | Yes | Severo et al. (201) |
| Tomato | TERF2/LeERF2 | — | No | Yes | — | — | Zhang and Huang (2010) |
| Sugarcane (Saccharum officinarum) | SodERF3 | Yes | — | — | — | — | Trujillo et al. (2008) |
| Wheat (Triticum aestivum) | TaERF1a | Yes | Yes | Yes | — | — | Xu et al. (2007) |
| T. aestivum | TaERF3 | Yes | Yes | — | — | — | Rong et al. (2014) |
| Tamarix hispida | ThERF1a | Yes | Yes | — | — | — | Wang et al. (2014); Liu et al. (2014) |
| T. hispida | ThERF5 | Yes | Yes | — | — | — | Liu et al. (2014) |
| T. hispida | ThERF12 | Yes | No | — | — | — | Liu et al. (2014) |
| Triticum turgidum | TdSHN1a | Yes | Yes | Yes | — | — | Djemal and Khoudi (2015) |
ERFs described as capable of binding to GCC box and DRE elements.
Table II. Abiotic stress response studied in transgenic plants overexpressing ERFs.
BIERF3, BTH-induced ERF transcriptional Factor3; TERF, Tomato Ethylene Response Factor; SlERF, Tomato Ethylene Response Factor.
| Gene | Transgenic Plants | Effect | References |
|---|---|---|---|
| AtERF1 | Arabidopsis | Salt, drought, and heat stress tolerance | Cheng et al. (2013) |
| AtERF4 | Arabidopsis | Hypersensitive to salt stress | Yang et al. (2005) |
| AtERF5 | Arabidopsis | Hypersensitive to osmotic stress | Dubois et al. (2013) |
| AtERF6 | Arabidopsis | Hypersensitive to osmotic stress | Dubois et al. (2013) |
| AtERF98 | Arabidopsis | Salt tolerance | Zhang et al. (2012) |
| BrERF4 | Arabidopsis | Salt and drought tolerance | Seo et al. (2010) |
| CarERF116 | Arabidopsis | Osmotic and freezing tolerance | Deokar et al. (2015) |
| LcERF054 | Arabidopsis | Salt tolerance | Sun et al. (2014a) |
| CaPF1 | Arabidopsis | Freezing tolerance | Yi et al. (2004) |
| CaPF1 | Virginia pine (Pinus virginiana) | Heat and heavy metal tolerance | Tang et al. (2005) |
| CaPF1 | Potato (Solanum tuberosum) | Drought, freezing, heat, and heavy metal tolerance | Youm et al. (2008) |
| CaERFLP1 | Tobacco | Salt tolerance | Lee et al. (2004) |
| CsERF | Tobacco | Cold tolerance | Ma et al. (2014) |
| JERF1 | Rice (Oryza sativa) | Drought tolerance | Zhang et al. (2010) |
| JERF3 | Tobacco | Salt, drought, and freezing tolerance | Wang et al. (2004); Wu et al. (2008) |
| Tsi1 | Tobacco | Salt tolerance | Park et al. (2001) |
| OPBP1 | Tobacco | Salt tolerance | Guo et al. (2004) |
| TaERF1 | Arabidopsis | Salt, drought, and freezing tolerance | Xu et al. (2007) |
| OsBIERF3 | Tobacco | Salt tolerance | Cao et al. (2006) |
| SodERF3 | Tobacco | Salt and drought tolerance | Trujillo et al. (2008) |
| TERF1 | Rice | Salt and drought tolerance | Gao et al. (2008) |
| TERF1 | Tobacco | Salt tolerance | Huang et al. (2004) |
| MsERF8 | Tobacco | Salt tolerance | Chen et al. (2012) |
| GmERF8 | Tobacco | Salt and drought tolerance | Zhang et al. (2009) |
| JcERF1 | Tobacco | Salt tolerance | Yang et al. (2014) |
| LchERF | Tobacco | Salt tolerance | Wu et al. (2014a) |
| TSRF1 | Tobacco | Negative regulator of salt stress | Zhang et al. (2007) |
| TSRF1 | Zea mays | Salt tolerance | Wang et al. (2013a, 2013b) |
| TSRF1 | Rice | Drought tolerance | Quan et al. (2010) |
| LeERF1 | Tomato | Salt tolerance | Hu et al. (2014) |
| LeERF2 | Tomato | Salt tolerance | Hu et al. (2014) |
| SlERF5 | Tomato | Salt and drought tolerance | Pan et al. (2012) |
| ThERF1 | Arabidopsis | Negative regulator of salt and drought stress | Wang et al. (2014) |
| TERF2/LeERF2 | Tobacco | Freezing tolerance | Zhang and Huang (2010) |
Several TFs from Arabidopsis and other plant species that belong to the ERF subfamily have been reported to be capable of binding to both GCC box and DRE elements (Table I). For instance, AtERF1 binds specifically to GCC boxes in the promoter regions of the ethylene- and jasmonate-responsive plant defensin (PDF1.2) and basic-chitinase (b-CHI) genes (Solano et al., 1998) and DRE elements in the promoters of the Δ1-Pyrroline-5-Carboxylate Synthetase1 (P5CS1), Germin-Like Protein9 (GLP9), osmotin34 (OSM34), similar to RCD one5 (SRO5), responsive to desiccation29B (RD29B), Early Response to Dehydration7 (ERD7), and RD20 genes, thus conferring not only resistance to pathogen attack but also, tolerance to several abiotic stresses, including drought, salt, and heat stress (Cheng et al., 2013). Interestingly, the affinity of ERF1 for the DRE elements in the promoter of the P5CS1 was much higher than it was for the GCC box in the promoters of b-CHI and PDF1.2 (Cheng et al., 2013). Other ERFs also specifically bind to both GCC box and DRE elements (Table I). The pepper ethylene-responsive factor-like protein1 (ERFLP1) gene was identified from Xanthomonas spp.-infected plants and encodes the CaERFLP1 protein. CaERFLP1 showed enhanced expression under salt stress but not cold or drought stress. Overexpression of CaERFLP1 led to increased salt stress tolerance in tobacco plants (Lee et al., 2004; Table II). Expression of pathogen and freezing tolerance-related protein1 (CaPF1) was induced in plant response to various abiotic stresses, including cold, salt, and drought stress in Arabidopsis. Overexpression of CaPF1 resulted in enhanced resistance to freezing in Arabidopsis (Yi et al., 2004), and in potato, it resulted in enhanced resistance to freezing, heat, heavy metal, and oxidative stress (Youm et al., 2008). Other examples of ERFs binding to both GCC box and DRE elements include ERF3 from soybean (Zhang et al., 2009); JERF1, JERF3, and tobacco stress-induced protein1 (Tsi1) from tobacco plants (Park et al., 2001; Wang et al., 2004; Wu et al., 2007); TSRF1 from tomato plants (Zhang et al., 2007); and SHN1 from wheat (Triticum durum; Djemal and Khoudi, 2015), which are all induced by drought, salt, and/or cold stress. A complete list of the TF genes that encode proteins from the ERF subfamily, including those that bind to both GCC box and DRE elements, and are involved in abiotic stress tolerance is given in Table I.
ERFs AND HORMONE CROSS TALK
Plant response and adaptation to environmental stresses require the coordinated interaction of hormone signaling pathways to regulate the expression of TF genes that allows the plant to fine tune specific stress responses. Expression of ERFs as downstream components of the ethylene signaling and response pathway can be induced by ethylene as well as biotic and abiotic stresses. Jasmonic acid and ABA have also been reported to be involved in the regulation of ERFs under abiotic stresses. Moreover, ethylene signaling interacts with other plant hormone pathways, such as those regulated by salicylic acid, gibberellins, and brassinosteroids, during plant adaptation to abiotic stresses. Indeed, exogenous application of these phytohormones has led to induced expression of a number of ERF genes (Table III). However, the molecular transduction mechanisms underlying pathway cross talk are still only partly understood.
Table III. Hormonal effects on the transcription of ERF genes.
BRs, Brassinosteroids; GAs, gibberellins; —, not studied; TERF, Tomato Ethylene Response Factor; SlERF, Tomato Ethylene Response Factor.
| Gene | Induced Expression |
References | ||||||
|---|---|---|---|---|---|---|---|---|
| Ethylene | ABA | Jasmonates | Salicylic Acid | GAs | Auxin | BRs | ||
| AtERF1a | Yes | Yes | Yes | — | — | — | — | Cheng et al. (2013) |
| AtERF4 | Yes | Yes | Yes | — | — | — | — | Yang et al. (2005) |
| BrERF4 | Yes | No | Yes | — | — | — | — | Seo et al. (2010) |
| CaPF1a | Yes | Yes | Yes | — | — | — | — | Yi et al. (2004) |
| CarERF116 | Yes | Yes | — | Yes | Yes | — | — | Deokar et al. (2015) |
| CsERF | Yes | Yes | Yes | Yes | No | No | No | Ma et al. (2014) |
| GhERF1 | Yes | Yes | — | — | — | — | — | Qiao et al. (2008) |
| GhERF2 | Yes | Yes | — | — | — | — | — | Jin et al. (2010) |
| GhERF3 | Yes | Yes | — | — | — | — | — | Jin et al. (2009) |
| GhERF5 | Yes | Yes | — | — | — | — | — | Jin et al. (2010) |
| GhERF6 | Yes | Yes | — | — | — | — | — | Jin et al. (2010) |
| GmERF3a | Yes | Yes | Yes | Yes | — | — | — | Zhang et al. (2009) |
| JcERF1 | Yes | Yes | — | — | — | — | — | Yang et al. (2014) |
| LchERF | Yes | — | — | — | — | — | — | Wu et al. (2014a) |
| LcERF054 | Yes | Yes | Yes | No | — | — | — | Sun et al. (2014a) |
| TSRF1 | Yes | — | — | Yes | — | — | — | Zhang et al. (2007) |
| MsERF8 | Yes | Yes | Yes | Yes | Yes | Yes | — | Chen et al. (2012) |
| JERF1a | Yes | Yes | Yes | — | — | — | — | Wu et al. (2007) |
| JERF3a | Yes | Yes | Yes | — | — | — | — | Wang et al. (2004) |
| OPBP1 | Yes | — | Yes | No | — | — | — | Guo et al. (2004) |
| Tsi1a | Yes | No | Yes | Yes | — | — | — | Park et al. (2001) |
| TERF1a | Yes | — | — | — | — | — | — | Gao et al. (2008) |
| SlERF5 | Yes | Yes | No | No | — | — | — | Pan et al. (2012) |
| SodERF3 | Yes | Yes | Yes | No | — | — | — | Trujillo et al. (2008) |
| TaERF1a | Yes | Yes | — | Yes | — | — | — | Xu et al. (2007) |
ERFs described as capable of binding to GCC box and DRE elements.
Jasmonates
Lorenzo et al. (2002) have reported that ERF1 is a downstream component of not only the ethylene but also, the jasmonate signaling pathway in Arabidopsis. Lorenzo et al. (2002) observed that ERF1 expression can be induced rapidly by ethylene and jasmonic acid as well as synergistically by both hormones, and they suggested that ERF1 acts as a key element in the regulation of ethylene/jasmonic acid-dependent defense response genes (Lorenzo et al., 2002). Recent studies using the jasmonic acid-insensitive mutant jasmonic acid amido-synthetase1-1 exposed to drought, salt, and heat stress revealed blocked ERF1 expression, indicating that ERF1 induction requires jasmonic acid as well as ethylene signaling under a number of abiotic stresses (Cheng et al., 2013). Ethylene/jasmonic acid signaling is also required in the induction of other ERFs to a number of abiotic stresses; examples of these ERFs include ERF6 (Sewelam et al., 2013), JERF1 (Wu et al., 2007, 2008), JERF3 (Wang et al., 2004), Tsi1 (Park et al., 2001), OPBP1 (Guo et al., 2004) and GmERF3 (Zhang et al., 2009).
ABA
ABA plays an important role in the response of plants to abiotic stresses, such as drought, salinity, and extreme temperatures. ERF1 overexpression has been observed to enhance drought, salt, and heat stress resistance in Arabidopsis plants accompanied with increased levels of ABA and Pro (Cheng et al., 2013). As an osmolite, Pro contributes to stress tolerance because its accumulation may prevent water loss. ABA has been reported to partially modulate Pro accumulation (Sharma et al., 2011). However, ABA negatively regulates ERF1 induction as shown in the ABA-hypersensitive abi1 and abi2 knockout mutants. Nevertheless, in the constitutive ethylene signaling mutant ctr1, ERF1 expression was even higher than in wild-type plants after ABA treatment. This indicates that ethylene/jasmonic acid signaling could not be blocked by the negative effect of ABA (Cheng et al., 2013). ABA treatment also repressed the expression of ERF6 in Arabidopsis (Sewelam et al., 2013).
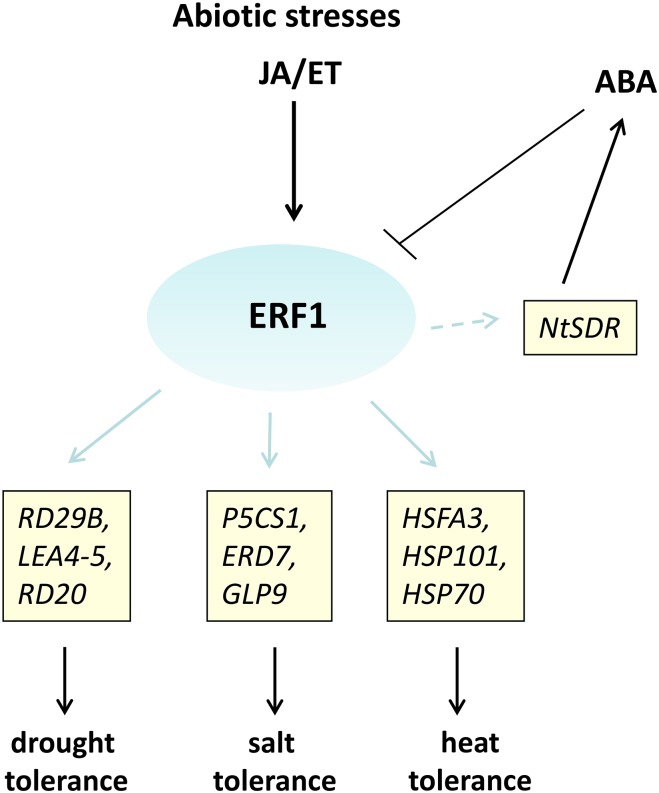
In contrast to ERF1 and ERF6, other ERF genes have been reported to be induced by ABA, including CsERF (Ma et al., 2014), GmERF3 (Zhang et al., 2009), LchERF (Wu et al., 2014a), JERF3 (Wu et al., 2008), TSRF1 (Quan et al., 2010), TaERF1 (Xu et al., 2007), and JERF1 (Wu et al., 2007). In addition, plants overexpressing TaERF1 were found to be highly sensitive to exogenous ABA treatment, resulting in rapid stomatal closure (Xu et al., 2007). Interestingly, JERF1 overexpression also increased leaf and root growth of tobacco significantly under salinity and low temperature accompanied by increased ABA levels (Wu et al., 2007). JERF1 was found to interact with multiple cis-acting elements and may activate both stress-responsive and ABA biosynthesis-related genes (such as tobacco short-chain dehydrogenase/reductase [NtSDR]), resulting in enhanced tolerance to salinity and cold stress in tobacco (Wu et al., 2007). Also, TSRF1 overexpression in tobacco enhanced expression of the ABA biosynthesis-related gene NtSDR, resulting in increased ABA contents. Moreover, overexpression of TSRF1 in tobacco plants resulted in enhanced drought tolerance and increased Pro contents (Quan et al., 2010; Cheng et al., 2013). Complex interaction signaling between ABA, ethylene, and ERF proteins, such as ERF1, JERF1, and TSRF1, seems to regulate ABA biosynthesis; however, ABA also acts as a negative regulator of ERF1 gene induction (Cheng et al., 2013). Interestingly, all three ERF proteins can bind to both GCC box and DRE elements and induce salt, heat, drought, and cold tolerance (Table I). A model for ethylene, jasmonic acid, and ABA cross talk through ERF1 under abiotic stress is shown in Figure 2.
Figure 2.
Proposed model for ethylene (ET), jasmonic acid (JA), and ABA cross talk through ERFs under abiotic stress. ERF1 induces expression of genes involved in abiotic stress tolerance. It has been postulated that, through the activation of JERF1 and TSRF1 (ERFs from the same ERF subfamily), ERF1 activates expression of NtSDR, an ABA biosynthesis-related gene. In turn, ABA might down-regulate ERF1 expression under abiotic stress. However, the negative effect of ABA does not seem to block ET/JA signaling. LEA4-5, Late-Embryogenesis Abundant Protein4-5; HSFA3, Heat-Shock Transcription Factor A3; HSP101, Heat-Shock Protein101.
Some proteins from the ERF subfamily can also act as negative regulators, such as AtERF4 and AtERF7, which are localized in the nuclear bodies and modulate ABA responses. Induced AtERF4 expression has been reported to make plants less sensitive to ABA, inhibit the expression of genes that are responsive to ABA, and confer hypersensitivity to salt stress in Arabidopsis (Yang et al., 2005). It has been suggested that AtERF7 activation inhibits the expression of genes induced by ABA, thereby decreasing tolerance to drought stress (Song et al., 2005).
Other Hormones
Salicylic acid has long been known to play a role in the induction of defense mechanisms in plants; however, recent studies revealed that it participates in abiotic stress signaling (Stevens et al., 2006; Horváth et al., 2007). It has been revealed that salicylic acid signaling enhances salt and oxidative stress tolerance in Arabidopsis by the induction of the NONEXPRESSER OF PATHOGENESIS RELATED1 (NPR1) gene (Jayakannan et al., 2015). Upon pathogen infection, ethylene is known to enhance salicylic acid/NPR1-dependent defenses through the ethylene signaling and response pathway (Leon-Reyes et al., 2009). Salicylic acid treatment induced expression of the ethylene TF genes AtERF6 (Sewelam et al., 2013), TaERF1 (Xu et al., 2007), TSRF1 (Huang et al., 2004), MsERF8 (Chen et al., 2012), GmERF3 (Zhang et al., 2009), and CarERF116 (Deokar et al., 2015), whereas expression of SodERF3 (Trujillo et al., 2008) and CsERF (Ma et al., 2014) was reduced by salicylic acid.
Recently, ERF6 has been reported to be involved in cross talk between the ethylene and gibberellin/DELLA pathway. ERF6 expression inhibits leaf growth by activating the transcription of the GIBBERELN2-OXIDASE6 gene, resulting in inactivation of gibberellins by DELLA stabilization under osmotic stress conditions in Arabidopsis (Dubois et al., 2013). However, the rapid ERF6 activation was found to be independent of EIN3/EIL1. It has been shown that ERF6 is activated by a mitogen-activated protein kinase cascade, including MITOGEN-ACTIVATED PROTEIN KINASE3 (MPK3)/MPK6. Phosphorylation of ERF6 by MPK3/MPK6 in gain-of-function transgenic plants increases ERF6 protein stability in vivo (Dubois et al., 2013; Meng et al., 2013).
Brassinosteroids have been found to mediate thermotolerance and salt tolerance and induce expression of several hormone-responsive genes, such as PDF1.2, suggesting cross talk between brassinosteroids and the ethylene, jasmonic acid, ABA, and salicylic acid signaling pathways (Divi et al., 2010). In Citrus spp. plants, expression of the CsERF gene was neither induced nor reduced after treatment with brassinosteroids, auxin, and gibberellin 3 (Ma et al., 2014). In contrast, gibberellin 3 treatment induced expression of both the CaERF116 and MsERF8 genes (Chen et al., 2012; Deokar et al., 2015).
ERFs AND REDOX SIGNALING
The reactive oxygen species (ROS) signaling network controls a broad range of biological processes, including biotic and abiotic stress responses, by activating defense genes (Mittler et al., 2011). ROS, such as 1O2, hydrogen peroxide, O2−, and •HO, are molecules that are considered to be both signaling and potentially damaging molecules (Iqbal et al., 2014). Salinity, drought, and cold stresses enhance ROS production, which results in an imbalance between ROS production and ROS scavenging (Miller et al., 2010). Antioxidants, such as ascorbic acid, glutathione, carotenoids, and tocopherols, as well as enzymes, such as superoxide dismutase, ascorbate peroxidase, catalase, and glutathione peroxidase, play an essential role in ROS scavenging mechanisms (Apel and Hirt, 2004; Munné-Bosch et al., 2013). Moreover, Pro plays a potential role in ROS detoxification because it is typically accumulated in response to osmotic stress (Sharma et al., 2011). Extracellular ROS, which are produced by peroxidases and NADPH oxidases, can transmit intracellular signals rapidly to the nucleus and/or amplify signals passing from the chloroplast to the cell nucleus through the action of secondary messengers, such as mitogen-activated protein kinases and plant hormones. Therefore, the TFs activated by ROS result in the transcription of a large number of genes (Miller et al., 2010; Munné-Bosch et al., 2013).
Oxidative stress treatment induced ERF1 expression (Sewelam et al., 2013), and overexpression of ERF1 leads to Pro accumulation and induces expression of P5CS1 (Cheng et al., 2013). This, in turn, catalyzes the first step of Pro synthesis, resulting in enhanced drought tolerance in Arabidopsis (Cheng et al., 2013). This result suggests, on the one hand, that ERF1 might regulate ROS signaling and on the other hand, that Pro accumulation seems to be a common response of ERFs to stress. This is, indeed, documented for the majority of TFs from the ERF subfamily that enhance abiotic stress tolerance, such as JERF1 (Zhang et al., 2010), TSRF1 (Quan et al., 2010), GmERF3 (Zhang et al., 2009), Tomato Ethylene-Response Factor5 (Pan et al., 2012), CsERF (Ma et al., 2014), JcERF1 (Yang et al., 2014), LeERF1, LeERF2 (Hu et al., 2014), MsERF8 (Chen et al., 2012), LcERF054, LcERF080 (Sun et al., 2014a, 2014b), LchERF (Wu et al., 2014a), and TaERF3 (Rong et al., 2014). Overexpression of LeERF1, LeERF2, and MsERF8 has been reported to elevate Pro accumulation and reduce malondialdehyde levels, an indicator of lipid peroxidation, in tomato and tobacco plants under salt stress (Cheng et al., 2013; Hu et al., 2014). Tang et al. (2005) have reported that overexpression of CaPF1 enhances biotic and abiotic stress tolerance in transgenic Virginia pine by regulating antioxidant enzyme activities. This result coincides with that for transgenic potato plants that overexpress CaPF1 and were more tolerant after oxidative stress treatment than control plants (Youm et al., 2008). AtERF98 has been reported to enhance salt tolerance by regulating the expression of ascorbate biosynthesis genes, resulting in reduced ROS levels in Arabidopsis (Zhang et al., 2012). JERF3 regulates the expression of ROS-related genes, such as SUPEROXIDE DISMUTASE, ASCORBATE PEROXIDASE1 (APX1), NtAPX2, and GLUTATHIONE PEROXIDASE in tobacco plants, resulting in decreased accumulation of ROS and enhancing tolerance to drought, salt, and freezing (Wu et al., 2008). ERF6 shows highly induced expression under oxidative stress treatment, such as hydrogen peroxide, and high light stress. Up-regulation of ROS-responsive gene expression analyzed in erf6 mutants revealed that ERF6 seems to be a negative regulator of ROS-responsive gene expression. In contrast, several antioxidant enzymes, such as MONODEHYDROASCORBATE REDUCTASE3, CATALASE3, and VITAMIN C DEFECTIVE2, showed down-regulation in erf6 mutants, indicating that ERF6 is a positive antioxidant regulator under biotic and abiotic stresses (Sewelam et al., 2013). Wang et al. (2013a) reported that ERF6 can bind specifically to the ROS-responsive cis-acting element7 (ROSE7)/GCC box, and enhances high light tolerance given that ROSE7-type genes showed no activation in erf6-1 mutants under high light stress.
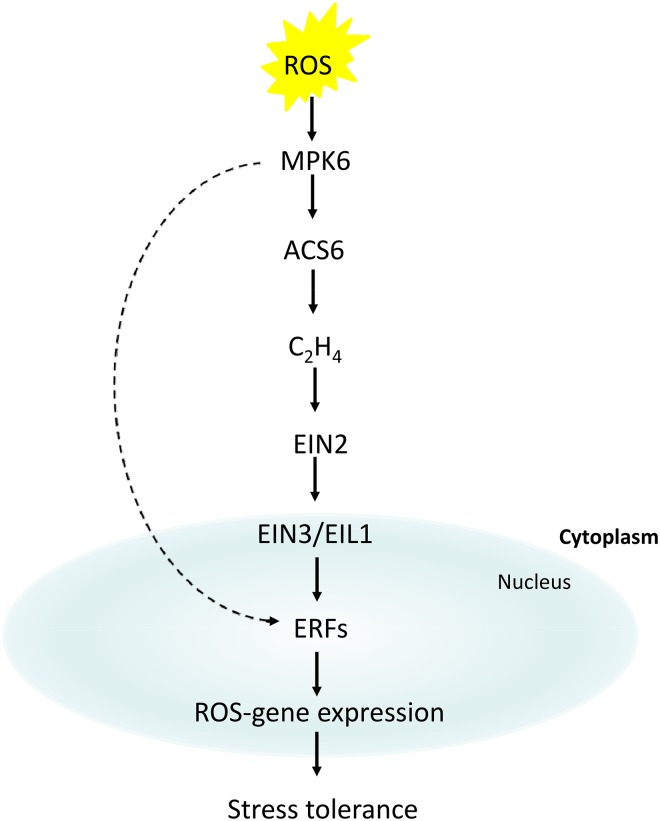
So far, little is known about the involvement of ROS signaling in the activation of ERFs. The MAPK cascade is well known to play an important role in ROS signaling. Ethylene synthesis is known to be positively regulated by the MPK6-mediated phosphorylation of ACC SYNTHASE2 (ACS2) and ACS6 (Liu and Zhang, 2004). Recently, several studies revealed that ERF6 is activated independently of EIN3/EIL1 by MPK6 phosphorylation, resulting in ROS-responsive gene expression in Arabidopsis (Dubois et al., 2013; Meng et al., 2013; Sewelam et al., 2013; Wang et al., 2013b). It is hypothesized that inactive ERF6 is kept at a basal level and can be rapidly phosphorylated, thereby reducing the time lag in transcriptional activation (Dubois et al., 2013; Fig. 3).
Figure 3.
Proposed model for ROS signaling to ERFs. Biotic and abiotic stresses enhance ROS production, resulting in the activation of MPK6, which activates ethylene biosynthesis by phosphorylation of ACS6. Then, EIN2, EIN3/EIL1, and finally, ERF1 are activated, which could result in the activation of ROS gene expression that enhances stress tolerance. Recently, it has been suggested that ERF6 is activated by MPK6 phosphorylation independently of EIN3/EIL1 under oxidative stress.
ERFs may also play a role in linking redox and hormonal regulation in plant responses to abiotic stresses. Tocopherols, which belong to the vitamin E group of compounds, are lipid-soluble antioxidants found in the chloroplasts. Photosynthetic tissues accumulate α-tocopherol in chloroplasts and to a lesser extent, its immediate precursor γ-tocopherol. ERF1 expression in the γ-tocopherol methyltransferase (vte4) mutant, which is deficient in α-tocopherol but accumulates γ-tocopherol, was reduced in parallel with lower jasmonic acid levels in the vte4 mutant compared with the wild type (Cela et al., 2011). These results indicate that γ-tocopherol represses jasmonic acid and ethylene signaling and response pathways in salt-stressed vte4 plants, thus suggesting a link between redox and hormonal signaling in the regulation of ERF1 expression in Arabidopsis. In another example of retrograde signaling from the chloroplast to the nucleus, Vogel et al. (2014) found that ERF6, ERF104, and ERF105 expression was rapidly (within 1 min) up-regulated upon exposure to high light in Arabidopsis. This response was deregulated in triose phosphate translocator (tpt) mutants. Similarly, activation of MPK6 was up-regulated after 1 min in the wild type but not in the tpt mutant (Vogel et al., 2014). Vogel et al. (2014) propose that metabolite export through the tpt in the chloroplast leads to subsequent MPK6 activation and ERF gene expression in the nucleus, therefore representing an additional mechanism of chloroplast to nucleus retrograde signaling.
CONCLUSION AND PROSPECTS
Molecular genetic studies have been pivotal in dissecting the ethylene signaling and response pathway. New insights from recent years have revealed that ERFs regulate not only biotic but also, abiotic stress responses. Thus, overexpression of a number of ERFs enhances salt, drought, light stress, and cold and heat tolerance as well as pathogen resistance in Arabidopsis plants. Ethylene, jasmonic acid, and ABA have been reported to be involved in the regulation of ERFs under abiotic stresses. For instance, ERF1 is involved in both ethylene and jasmonic acid signaling pathways. Moreover, plants that overexpress ERF1 enhance ABA levels under drought stress, indicating that ERF1 may regulate ABA biosynthesis. However, ABA negatively regulates ERF1 induction. Furthermore, induced ERF expression under oxidative stress suggests that ERFs might regulate ROS-responsive gene expression, thereby conferring stress tolerance. Because of the fact that many stresses act hand in hand with each other and not in isolation, it is clear that there is cross talk between biotic and abiotic stress responses. The specific binding activity of several ERFs to both GCC box and DRE elements depending on the stress conditions supports this hypothesis. There are still, however, many gaps in our knowledge on ERFs and hormonal cross talk, and the answers to remaining questions are important to increase our understanding of stress adaptation. Alongside the cross talk with jasmonic acid and ABA, the cross talk between ERFs and auxin, cytokinin, gibberellin, salicylic acid, and brassinosteroid responses should be studied in more detail. ERFs seem to regulate ROS-responsive gene expression, but more evidence of synergistic ERFs- and ROS-responsive genes is needed.
Glossary
- ABA
abscisic acid
- ROS
reactive oxygen species
- TF
transcription factor
Footnotes
This work was supported by the Spanish Government (grant no. BFU2012–32057) and the Generalitat de Catalunya (Catalan Institution for Research and Advanced Studies Academia Award to S.M.-B.).
References
- An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y, et al. (2010) Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29: 23–32 [DOI] [PubMed] [Google Scholar]
- Bisson MM, Bleckmann A, Allekotte S, Groth G (2009) EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochem J 424: 1–6 [DOI] [PubMed] [Google Scholar]
- Bisson MM, Groth G (2010) New insight in ethylene signaling: autokinase activity of ETR1 modulates the interaction of receptors and EIN2. Mol Plant 3: 882–889 [DOI] [PubMed] [Google Scholar]
- Cao Y, Wu Y, Zheng Z, Song F (2006) Overexpression of the rice EREBP-like gene OsBIERF3 enhances disease resistance and salt tolerance in transgenic tobacco. Physiol Mol Plant Pathol 67: 202–211 [Google Scholar]
- Cela J, Chang C, Munné-Bosch S (2011) Accumulation of γ- rather than α-tocopherol alters ethylene signaling gene expression in the vte4 mutant of Arabidopsis thaliana. Plant Cell Physiol 52: 1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charfeddine M, Saïdi MN, Charfeddine F, Hammami A, Bouzid RG (2015) Genome-wide analysis and expression profiling of the ERF transcription factor family in potato (Solanum tuberosum L.). Mol Biotechnol 57: 348–358 [DOI] [PubMed] [Google Scholar]
- Chen G, Hu Z, Grierson D (2008) Differential regulation of tomato ethylene responsive factor LeERF3b, a putative repressor, and the activator Pti4 in ripening mutants and in response to environmental stresses. J Plant Physiol 165: 662–670 [DOI] [PubMed] [Google Scholar]
- Chen T, Yang Q, Gruber M, Kang J, Sun Y, Ding W, Zhang T, Zhang X (2012) Expression of an alfalfa (Medicago sativa L.) ethylene response factor gene MsERF8 in tobacco plants enhances resistance to salinity. Mol Biol Rep 39: 6067–6075 [DOI] [PubMed] [Google Scholar]
- Chen YF, Etheridge N, Schaller GE (2005) Ethylene signal transduction. Ann Bot (Lond) 95: 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MC, Liao PM, Kuo WW, Lin TP (2013) The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol 162: 1566–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deokar AA, Kondawar V, Kohli D, Aslam M, Jain PK, Karuppayil SM, Varshney RK, Srinivasan R (2015) The CarERF genes in chickpea (Cicer arietinum L.) and the identification of CarERF116 as abiotic stress responsive transcription factor. Funct Integr Genomics 15: 27–46 [DOI] [PubMed] [Google Scholar]
- Divi UK, Rahman T, Krishna P (2010) Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol 10: 151–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djemal R, Khoudi H (February 17, 2015) Isolation and molecular characterization of a novel WIN1/SHN1 ethylene-responsive transcription factor TdSHN1 from durum wheat (Triticum turgidum. L. subsp. durum). Protoplasma 10.1007/s00709-015-0775-8 [DOI] [PubMed] [Google Scholar]
- Dubois M, Skirycz A, Claeys H, Maleux K, Dhondt S, De Bodt S, Vanden Bossche R, De Milde L, Yoshizumi T, Matsui M, et al. (2013) ETHYLENE RESPONSE FACTOR6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol 162: 319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Li P, Song A, Wang H, Wang Y, Ren L, Qi X, Chen F, Jiang J, Chen S (2015) Isolation and characterization of six AP2/ERF transcription factor genes in Chrysanthemum nankingense. Int J Mol Sci 16: 2052–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Zhang H, Tian Y, Li F, Zhang Z, Lu X, Chen X, Huang R (2008) Expression of TERF1 in rice regulates expression of stress-responsive genes and enhances tolerance to drought and high-salinity. Plant Cell Rep 27: 1787–1795 [DOI] [PubMed] [Google Scholar]
- Gao Z, Chen YF, Randlett MD, Zhao XC, Findell JL, Kieber JJ, Schaller GE (2003) Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J Biol Chem 278: 34725–34732 [DOI] [PubMed] [Google Scholar]
- Golldack D, Li C, Mohan H, Probst N (2014) Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci 5: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W, Shen YP, Ma LG, Pan Y, Du YL, Wang DH, Yang JY, Hu LD, Liu XF, Dong CX, et al. (2004) Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol 135: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo ZJ, Chen XJ, Wu XL, Ling JQ, Xu P (2004) Overexpression of the AP2/EREBP transcription factor OPBP1 enhances disease resistance and salt tolerance in tobacco. Plant Mol Biol 55: 607–618 [DOI] [PubMed] [Google Scholar]
- He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44: 903–916 [DOI] [PubMed] [Google Scholar]
- Horváth E, Szalai G, Janda T (2007) Induction of abiotic stress tolerance by salicylic acid signaling. J Plant Growth Regul 26: 290–300 [Google Scholar]
- Hu N, Tang N, Yan F, Bouzayen M, Li Z (2014) Effect of LeERF1 and LeERF2 overexpression in the response to salinity of young tomato (Solanum lycopersicum cv. Micro-Tom) seedlings. Acta Physiol Plant 36: 1703–1712 [Google Scholar]
- Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J 33: 221–233 [DOI] [PubMed] [Google Scholar]
- Huang Z, Zhang Z, Zhang X, Zhang H, Huang D, Huang R (2004) Tomato TERF1 modulates ethylene response and enhances osmotic stress tolerance by activating expression of downstream genes. FEBS Lett 573: 110–116 [DOI] [PubMed] [Google Scholar]
- Iqbal N, Umar S, Khan NA, Khan MIR (2014) A new perspective of phytohormones in salinity tolerance: regulation of proline metabolism. Environ Exp Bot 100: 34–42 [Google Scholar]
- Ito TM, Polido PB, Rampim MC, Kaschuk G, Souza SGH (2014) Genome-wide identification and phylogenetic analysis of the AP2/ERF gene superfamily in sweet orange (Citrus sinensis). Genet Mol Res 13: 7839–7851 [DOI] [PubMed] [Google Scholar]
- Jayakannan M, Bose J, Babourina O, Shabala S, Massart A, Poschenrieder C, Rengel Z (2015) The NPR1-dependent salicylic acid signalling pathway is pivotal for enhanced salt and oxidative stress tolerance in Arabidopsis. J Exp Bot 66: 1865–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin LG, Huang B, Li H, Liu JY (2009) Expression profile and transactivation analysis of a novel ethylene-responsive transcription factor gene GhERF5 from cotton. Prog Nat Sci 19: 563–572 [Google Scholar]
- Jin LG, Li H, Liu JY (2010) Molecular characterization of three ethylene responsive element binding factor genes from cotton. J Integr Plant Biol 52: 485–495 [DOI] [PubMed] [Google Scholar]
- Ju C, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML, et al. (2012) CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci USA 109: 19486–19491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick MD, Chang C (2008) Ethylene signaling: new levels of complexity and regulation. Curr Opin Plant Biol 11: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Lacey RF, Binder BM (2014) How plants sense ethylene gas—the ethylene receptors. J Inorg Biochem 133: 58–62 [DOI] [PubMed] [Google Scholar]
- Lee JH, Hong JP, Oh SK, Lee S, Choi D, Kim WT (2004) The ethylene-responsive factor like protein 1 (CaERFLP1) of hot pepper (Capsicum annuum L.) interacts in vitro with both GCC and DRE/CRT sequences with different binding affinities: possible biological roles of CaERFLP1 in response to pathogen infection and high salinity conditions in transgenic tobacco plants. Plant Mol Biol 55: 61–81 [DOI] [PubMed] [Google Scholar]
- Lei G, Shen M, Li ZG, Zhang B, Duan KX, Wang N, Cao YR, Zhang WK, Ma B, Ling HQ, et al. (2011) EIN2 regulates salt stress response and interacts with a MA3 domain-containing protein ECIP1 in Arabidopsis. Plant Cell Environ 34: 1678–1692 [DOI] [PubMed] [Google Scholar]
- Leon-Reyes A, Spoel SH, De Lange ES, Abe H, Kobayashi M, Tsuda S, Millenaar FF, Welschen RAM, Ritsema T, Pieterse CMJ (2009) Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol 149: 1797–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, Giorgi FM, Zenoni S, Osti F, Pezzotti M, Perata P (2010) Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genomics 11: 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P (2013) APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol 199: 639–649 [DOI] [PubMed] [Google Scholar]
- Liu W, Wang Y, Gao C (2014) The ethylene response factor (ERF) genes from Tamarix hispida respond to salt, drought and ABA treatment. Trees 28: 317–327 [Google Scholar]
- Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R (2002) ETHYLENE RESPONSE FACTOR1 integrates signal from ethylene and jasmonate pathway in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Gong Q, Bohnert HJ (2006) Dissecting salt stress pathways. J Exp Bot 57: 1097–1107 [DOI] [PubMed] [Google Scholar]
- Ma Y, Zhang L, Zhang J, Chen J, Wu T, Zhu S, Yan S, Zhao X, Zhong G (2014) Expressing a Citrus ortholog of Arabidopsis ERF1 enhanced cold-tolerance in tobacco. Sci Hortic 174: 64–76 [Google Scholar]
- Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K (2008) The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J 56: 613–626 [DOI] [PubMed] [Google Scholar]
- Meng X, Xu J, He Y, Yang KY, Mordorski B, Liu Y, Zhang S (2013) Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25: 1126–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33: 453–467 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F (2011) ROS signaling: the new wave? Trends Plant Sci 16: 300–309 [DOI] [PubMed] [Google Scholar]
- Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819: 86–96 [DOI] [PubMed] [Google Scholar]
- Munné-Bosch S, Queval G, Foyer CH (2013) The impact of global change factors on redox signaling underpinning stress tolerance. Plant Physiol 161: 5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Seymour GB, Lu C, Hu Z, Chen X, Chen G (2012) An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep 31: 349–360 [DOI] [PubMed] [Google Scholar]
- Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH (2001) Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13: 1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Chang KN, Yazaki J, Ecker JR (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev 23: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao ZX, Huang B, Liu JY (2008) Molecular cloning and functional analysis of an ERF gene from cotton (Gossypium hirsutum). Biochim Biophys Acta 1779: 122–127 [DOI] [PubMed] [Google Scholar]
- Quan R, Hu S, Zhang Z, Zhang H, Zhang Z, Huang R (2010) Overexpression of an ERF transcription factor TSRF1 improves rice drought tolerance. Plant Biotechnol J 8: 476–488 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rong W, Qi L, Wang A, Ye X, Du L, Liang H, Xin Z, Zhang Z (2014) The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol J 12: 468–479 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Seo YJ, Park JB, Cho YJ, Jung C, Seo HS, Park SK, Nahm BH, Song JT (2010) Overexpression of the ethylene-responsive factor gene BrERF4 from Brassica rapa increases tolerance to salt and drought in Arabidopsis plants. Mol Cells 30: 271–277 [DOI] [PubMed] [Google Scholar]
- Severo J, Tiecher A, Pirrello J, Regad F, Latché, Pech JC, Bouzayen M, Rombaldi CV (2015) UV-C radiation modifies the ripening and accumulation of ethylene response factor (ERF) transcripts in tomato fruits. Postharvest Biol Technol 102: 9–16 [Google Scholar]
- Sewelam N, Kazan K, Thomas-Hall SR, Kidd BN, Manners JM, Schenk PM (2013) Ethylene response factor 6 is a regulator of reactive oxygen species signaling in Arabidopsis. PLoS One 8: e70289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Villamor JG, Verslues PE (2011) Essential role of tissue-specific proline synthesis and catabolism in growth and redox balance at low water potential. Plant Physiol 157: 292–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharoni AM, Nuruzzaman M, Satoh K, Shimizu T, Kondoh H, Sasaya T, Choi IR, Omura T, Kikuchi S (2011) Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol 52: 344–360 [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17: 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Li Y, Hou X (2013) Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics 14: 573–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Alonso JM (2009) Ethylene signaling and response: where different regulatory modules meet. Curr Opin Plant Biol 12: 548–555 [DOI] [PubMed] [Google Scholar]
- Stevens J, Senaratna T, Sivasithamparam K (2006) Salicylic acid induces salinity tolerance in tomato (Lycopersico nesculentum cv. Roma): associated changes in gas exchange, water relations and membrane stabilization. Plant Growth Regul 49: 77–83 [Google Scholar]
- Sun ZM, Zhou ML, Xiao XG, Tang YX, Wu YM (2014a) Genome-wide analysis of AP2/ERF family genes from Lotus corniculatus shows LcERF054 enhances salt tolerance. Funct Integr Genomics 14: 453–466 [DOI] [PubMed] [Google Scholar]
- Sun ZM, Zhou ML, Xiao XG, Tang YX, Wu YM (2014b) Overexpression of a Lotus corniculatusAP2/ERF transcription factor gene, LcERF080, enhances tolerance to salt stress in transgenic Arabidopsis. Plant Biotechnol Rep 8: 315–324 [Google Scholar]
- Tang W, Charles TM, Newton RJ (2005) Overexpression of the pepper transcription factor CaPF1 in transgenic Virginia pine (Pinus Virginiana Mill.) confers multiple stress tolerance and enhances organ growth. Plant Mol Biol 59: 603–617 [DOI] [PubMed] [Google Scholar]
- Trujillo LE, Sotolongo M, Menéndez C, Ochogavía ME, Coll Y, Hernández I, Borrás-Hidalgo O, Thomma BP, Vera P, Hernández L (2008) SodERF3, a novel sugarcane ethylene responsive factor (ERF), enhances salt and drought tolerance when overexpressed in tobacco plants. Plant Cell Physiol 49: 512–525 [DOI] [PubMed] [Google Scholar]
- Vogel MO, Moore M, König K, Pecher P, Alsharafa K, Lee J, Dietz KJ (2014) Fast retrograde signaling in response to high light involves metabolite export, MITOGEN-ACTIVATED PROTEIN KINASE6, and AP2/ERF transcription factors in Arabidopsis. Plant Cell 26: 1151–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Huang Z, Chen Q, Zhang Z, Zhang H, Wu Y, Huang D, Huang R (2004) Ectopic overexpression of tomato JERF3 in tobacco activates downstream gene expression and enhances salt tolerance. Plant Mol Biol 55: 183–192 [DOI] [PubMed] [Google Scholar]
- Wang L, Qin L, Liu W, Zhang D, Wang Y (2014) A novel ethylene-responsive factor from Tamarix hispida, ThERF1, is a GCC-box- and DRE-motif binding protein that negatively modulates abiotic stress tolerance in Arabidopsis. Physiol Plant 152: 84–97 [DOI] [PubMed] [Google Scholar]
- Wang M, Liu C, Li S, Zhu D, Zhao Q, Yu J (2013a) Improved nutritive quality and salt resistance in transgenic maize by simultaneously overexpression of a natural lysine-rich protein gene, SBgLR, and an ERF transcription factor gene, TSRF1. Int J Mol Sci 14: 9459–9474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Du Y, Zhao X, Miao Y, Song CP (2013b) The MPK6-ERF6-ROS-responsive cis-acting Element7/GCC box complex modulates oxidative gene transcription and the oxidative response in Arabidopsis. Plant Physiol 161: 1392–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Ji J, Wang G, Guan C, Jin C (2014a) LchERF, a novel ethylene-responsive transcription factor from Lycium chinense, confers salt tolerance in transgenic tobacco. Plant Cell Rep 33: 2033–2045 [DOI] [PubMed] [Google Scholar]
- Wu L, Chen X, Ren H, Zhang Z, Zhang H, Wang J, Wang XC, Huang R (2007) ERF protein JERF1 that transcriptionally modulates the expression of abscisic acid biosynthesis-related gene enhances the tolerance under salinity and cold in tobacco. Planta 226: 815–825 [DOI] [PubMed] [Google Scholar]
- Wu L, Zhang Z, Zhang H, Wang XC, Huang R (2008) Transcriptional modulation of ethylene response factor protein JERF3 in the oxidative stress response enhances tolerance of tobacco seedlings to salt, drought, and freezing. Plant Physiol 148: 1953–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, He J, Chen J, Yang S, Zha D (2014b) Alleviation of exogenous 6-benzyladenine on two genotypes of eggplant (Solanum melongena Mill.) growth under salt stress. Protoplasma 251: 169–176 [DOI] [PubMed] [Google Scholar]
- Xu ZS, Chen M, Li LC, Ma YZ (2011) Functions and application of the AP2/ERF transcription factor family in crop improvement. J Integr Plant Biol 53: 570–585 [DOI] [PubMed] [Google Scholar]
- Xu ZS, Xia LQ, Chen M, Cheng XG, Zhang RY, Li LC, Zhao YX, Lu Y, Ni ZY, Liu L, et al. (2007) Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol Biol 65: 719–732 [DOI] [PubMed] [Google Scholar]
- Yang H, Yu C, Yan J, Wang X, Chen F, Zhao Y, Wei W (2014) Overexpression of the Jatropha curcas JcERF1 gene coding an AP2/ERF-type transcription factor increase tolerance to salt in transgenic tobacco. Biochemistry 79: 1226–1236 [DOI] [PubMed] [Google Scholar]
- Yang XY, Xie JX, Lu XP, Liu YZ, Peng SA (2011) Isolation of a citrus ethylene-responsive element binding factor gene and its expression in response to abiotic stress, girdling and shading. Sci Hortic 127: 275–281 [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K (2005) Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol 58: 585–596 [DOI] [PubMed] [Google Scholar]
- Yi SY, Kim JH, Joung YH, Lee S, Kim WT, Yu SH, Choi D (2004) The pepper transcription factor CaPF1 confers pathogen and freezing tolerance in Arabidopsis. Plant Physiol 136: 2862–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youm JW, Jeon JH, Choi D, Yi SY, Joung H, Kim HS (2008) Ectopic expression of pepper CaPF1 in potato enhances multiple stresses tolerance and delays initiation of in vitro tuberization. Planta 228: 701–708 [DOI] [PubMed] [Google Scholar]
- Zhang G, Chen M, Li L, Xu Z, Chen X, Guo J, Ma Y (2009) Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot 60: 3781–3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li W, Chen J, Yang Y, Zhang Z, Zhang H, Wang XC, Huang R (2007) Transcriptional activator TSRF1 reversely regulates pathogen resistance and osmotic stress tolerance in tobacco. Plant Mol Biol 63: 63–71 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Huang R (2010) Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol Biol 73: 241–249 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li F, Li D, Zhang H, Huang R (2010) Expression of ethylene response factor JERF1 in rice improves tolerance to drought. Planta 232: 765–774 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wang J, Zhang R, Huang R (2012) The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J 71: 273–287 [DOI] [PubMed] [Google Scholar]
- Zhao XC, Schaller GE (2004) Effect of salt and osmotic stress upon expression of the ethylene receptor ETR1 in Arabidopsis thaliana. FEBS Lett 562: 189–192 [DOI] [PubMed] [Google Scholar]
- Zhuang J, Cai B, Peng RH, Zhu B, Jin XF, Xue Y, Gao F, Fu XY, Tian YS, Zhao W, et al. (2008) Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem Biophys Res Commun 371: 468–474 [DOI] [PubMed] [Google Scholar]