Reducing ethylene sensitivity by modifying the expression of a negative regulator of ethylene signal transduction improves grain yield in maize under drought stress environments.
Abstract
Lack of sufficient water is a major limiting factor to crop production worldwide, and the development of drought-tolerant germplasm is needed to improve crop productivity. The phytohormone ethylene modulates plant growth and development as well as plant response to abiotic stress. Recent research has shown that modifying ethylene biosynthesis and signaling can enhance plant drought tolerance. Here, we report novel negative regulators of ethylene signal transduction in Arabidopsis (Arabidopsis thaliana) and maize (Zea mays). These regulators are encoded by the ARGOS gene family. In Arabidopsis, overexpression of maize ARGOS1 (ZmARGOS1), ZmARGOS8, Arabidopsis ARGOS homolog ORGAN SIZE RELATED1 (AtOSR1), and AtOSR2 reduced plant sensitivity to ethylene, leading to enhanced drought tolerance. RNA profiling and genetic analysis suggested that the ZmARGOS1 transgene acts between an ethylene receptor and CONSTITUTIVE TRIPLE RESPONSE1 in the ethylene signaling pathway, affecting ethylene perception or the early stages of ethylene signaling. Overexpressed ZmARGOS1 is localized to the endoplasmic reticulum and Golgi membrane, where the ethylene receptors and the ethylene signaling protein ETHYLENE-INSENSITIVE2 and REVERSION-TO-ETHYLENE SENSITIVITY1 reside. In transgenic maize plants, overexpression of ARGOS genes also reduces ethylene sensitivity. Moreover, field testing showed that UBIQUITIN1:ZmARGOS8 maize events had a greater grain yield than nontransgenic controls under both drought stress and well-watered conditions.
There is an increasing demand for food and feed due to global population growth, urbanization, and rapid middle-class emergence. Lack of water limits crop yields worldwide; Bot et al. (2000) estimated that 45% of agricultural lands are subject to continuous or frequent drought conditions. Drought-tolerant varieties can reduce the impact of drought on crop productivity. The phytohormone ethylene regulates many aspects of plant growth and development, from seed germination, leaf expansion, and floral transition to organ senescence, fruit ripening, and the response to abiotic stresses, such as drought, high temperature, freezing, shading, and nutrient deficiency. Ethylene is one of the most widely used hormones in agriculture to increase yield and reduce production costs. For example, ethylene can reduce lodging in wheat (Triticum aestivum) and barley (Hordeum vulgare) by shortening the stem, therefore improving grain yield and quality. Studies have shown that inhibitors of ethylene biosynthesis and perception can mitigate yield loss by enhancing plant tolerance to abiotic stresses, such as drought, heat, and a combination of both (Hays et al., 2007; Kawakami et al., 2010, 2013; Huberman et al., 2014). This study explores the potential to improve crop performance by modifying ethylene sensitivity.
At the molecular level, ethylene responses in Arabidopsis (Arabidopsis thaliana) are initiated by the binding of ethylene to a family of endoplasmic reticulum (ER)- and Golgi membrane-localized receptors, including ETHYLENE RESPONSE1 (ETR1), ETR2, ETHYLENE RESPONSE SENSOR1 (ERS1), ERS2, and ETHYLENE-INSENSITIVE4 (EIN4; Chang et al., 1993; Hua and Meyerowitz, 1998). The ethylene signal is transduced from the receptors to the nuclear protein EIN3 via CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) and EIN2 (Ju et al., 2012; Qiao et al., 2012). CTR1 is a Raf-like kinase and interacts physically with the receptors (Huang et al., 2003). EIN3 and ETHYLENE-INSENSITIVE3-LIKE1 (EIL1) are the master transcription factors controlling ethylene-responsive gene expression (Chao et al., 1997). EIN2, an ER-tethered protein, functions as a shuttle to transduce the signal from the membrane to the nucleus via its cleavable C-terminal domain (Ju et al., 2012; Qiao et al., 2012). Proteins that modulate ethylene perception include REVERSION-TO-ETHYLENE SENSITIVITY1 (RTE1) and RESPONSIVE-TO-ANTAGONIST1, the former promoting the activity of ETR1 (Resnick et al., 2006, 2008; Dong et al., 2010) and the latter, a transporter, providing the ethylene cofactor copper and also playing a role in the biogenesis of active ethylene receptors (Hirayama et al., 1999; Binder et al., 2010). Many aspects of the ethylene signaling pathway and the sequences of the proteins involved are conserved between dicots and monocots (Guillaume and Sauter, 2008).
Beltrano et al. (1999) reported that exogenous application of the ethylene biosynthesis inhibitor aminoethoxyvinylglycine reversed a drought stress syndrome in wheat. Transgenic maize (Zea mays) plants with reduced ethylene biosynthesis, via silencing 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE6, have shown enhanced yields compared with nontransgenic controls in water-deficit and low-nitrogen environments (Habben et al., 2014). Here, we report novel, negative regulators of ethylene signal transduction from maize and Arabidopsis. These regulators are encoded by the ARGOS gene family, whose first member was identified in Arabidopsis (Hu et al., 2003). ARGOS genes encode a predicted integral membrane protein (Supplemental Fig. S1) and are known to promote plant organ growth by increasing cell number and/or cell size when overexpressed in Arabidopsis (Hu et al., 2003, 2006; Wang et al., 2009; Feng et al., 2011). Previously, the phenotype of enlarged leaves in AtARGOS overexpression Arabidopsis was interpreted as a result of the prolonged expression of AINTEGUMENTA and CYCLIN D3;1, and AtARGOS was proposed to function downstream of AUXIN-RESISTANT1 as a signaling component in the auxin signal transduction pathway (Hu et al., 2003). An earlier study indicates that Arabidopsis ARGOS-LIKE (AtARL) also regulates lateral organ size but does not affect cell proliferation in Arabidopsis. Instead, AtARL promotes cell expansion. AtARL was hypothesized to act downstream of BRASSINOSTEROID-INSENSITIVE1 in the brassinosteroid signaling pathway (Hu et al., 2006). However, in both cases, the function of the overexpressed genes was not established. We found that ARGOS genes, when overexpressed, reduce plant (Arabidopsis and maize) sensitivity to ethylene. Overexpressed ARGOS targets the ethylene receptor complex, possibly affecting ethylene perception as well as ethylene signal transduction. We also found that overexpression of ARGOS in maize results in improved grain yields under both drought stress and well-watered conditions.
RESULTS
Overexpression of ZmARGOS1 Confers Ethylene Insensitivity in Arabidopsis
Eight family members of ARGOS have been identified in maize and four in Arabidopsis (Supplemental Fig. S1). We initially focused our attention on one of these members: ZmARGOS1. To determine its molecular function, ZmARGOS1 was overexpressed in Arabidopsis under the control of the cauliflower mosaic virus 35S promoter (35S). Transgene expression was confirmed in eight events by northern blotting (Supplemental Fig. S2). The 35S:ZmARGOS1 Arabidopsis plants had wider and longer leaves than wild-type plants at bolting time, and flowering time was delayed 3 to 10 d dependent on growth conditions (Fig. 1A), similar to Arabidopsis overexpressing AtARGOS genes (Hu et al., 2003, 2006; Feng et al., 2011) and rice ARGOS (Wang et al., 2009). In wild-type plants, perianth organs in flowers abscised soon after pollination, and inflorescences generally had three to five opened flowers. In contrast, petals and sepals of the 35S:ZmARGOS1 plants remained turgid and intact for a longer time, and the inflorescences had seven to 10 opened flowers (Supplemental Fig. S3). These phenotypes of enlarged leaves, delayed flowering time, and delayed flower senescence were also reported in the ethylene-insensitive mutants etr1-1 and ein2-1 (Guzmán and Ecker, 1990; Ogawara et al., 2003; Patterson and Bleecker, 2004), suggesting that ARGOS may be involved in the ethylene pathway.
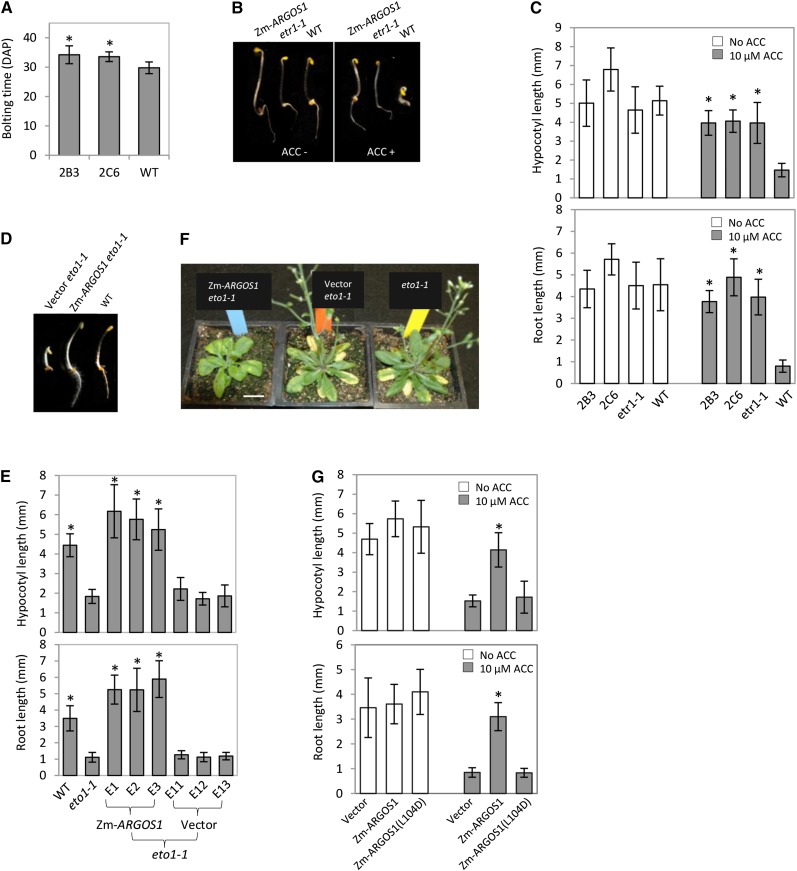
Figure 1.
Overexpression of ZmARGOS1 reduces ethylene responses in Arabidopsis plants. A, Bolting time is delayed in transgenic Arabidopsis plants overexpressing ZmARGOS1 relative to wild-type plants. Two 35S:ZmARGOS1 transgenic events (2B3 and 2C6) and wild-type (WT) controls were grown under a regime of 16 h of light (approximately 120 mE m−2 s−1) at 24°C and 8 h of darkness at 23°C. Means are shown for bolting time, days after planting (DAP). Error bars indicate sd; n = 18. Student’s t test was performed to compare the transgenic events and the wild-type control (*, P < 0.01). B, 35S:ZmARGOS1 Arabidopsis plants are insensitive to the ethylene precursor ACC. Seedlings were germinated in the dark in the presence or absence of 10 μm ACC for 3 d. Composite images of representative seedlings of 35S:ZmARGOS1 transgenic, etr1-1 mutant, and wild-type Columbia-0 (Col-0) plants are shown. C, Hypocotyl and root lengths of 3-d-old etiolated Arabidopsis seedlings. The triple response assay was conducted with 10 μm ACC. The data represent means of 10 to 20 seedlings from two 35S:ZmARGOS1 events (2B3 and 2C6), an etr1-1 mutant, and wild-type controls. Error bars indicate sd. Student’s t test was performed to compare the transgenic events and the etr1-1 mutant with the wild-type control (*, P < 0.01). D, Overexpression of ZmARGOS1 in the Arabidopsis eto1-1 mutant background. The 3-d-old, etiolated 35S:ZmARGOS1 eto1-1 seedlings lack the ethylene triple response phenotype in the absence of exogenously supplied ethylene. A composite image shows representative seedlings of wild-type plants and the transgenic Arabidopsis eto1-1 plants carrying ZmARGOS1 and empty vector. E, Hypocotyl and root lengths of etiolated Arabidopsis eto1-1 mutant plants overexpressing ZmARGOS1. Seeds were germinated under dark in the absence of exogenously supplied ethylene for 3 d. Three 35S:ZmARGOS1 events (ZmARGOS1 E1, E2, and E3) and three empty vector events (Vector E11, E12, and E13) in the eto1-1 mutant background are shown. Wild-type Arabidopsis and the eto1-1 mutant served as controls. The hypocotyls and roots in the 35S:ZmARGOS1 eto1-1 plants are significantly (*, P < 0.01) longer than those in the vector eto1-1 control events and the eto1-1 mutant. Error bars indicate sd; n = 20. F, Flowering time was delayed in the 35S:ZmARGOS1 eto1-1 Arabidopsis plants relative to the eto1-1 mutant. The delayed flowering time was observed in all 10 independent events tested. Representative 34-d-old transgenic eto1-1 plants carrying 35S:ZmARGOS1 and empty vector as well as the eto1-1 mutant plants are shown. Bar = 20 mm. G, ZmARGOS1(L104D), a mutated version of ZmARGOS1, was unable to confer ethylene insensitivity in Arabidopsis plants. Root and hypocotyl lengths were determined in transgenic Arabidopsis plants carrying empty vector, 35S:ZmARGOS1, and 35S:ZmARGOS1(L104D). Twelve T1 seeds per construct, each representing an independent event, were randomly selected based on the yellow fluorescent protein (YFP) marker and germinated in the dark in the presence or absence of 10 µm ACC. The means of the hypocotyl and root lengths are shown for 3-d-old seedlings. Error bars indicate sd. Student’s t test was performed to compare the transgenic events and empty vector controls (*, P < 0.01).
To investigate the effect of 35S:ZmARGOS1 on Arabidopsis responses to ethylene, seeds were germinated in the presence of ethylene or the precursor 1-aminocyclopropane-1-carboxylic acid (ACC; Fig. 1, B and C; Supplemental Fig. S4). Etiolated wild-type seedlings showed inhibition of hypocotyl and root growth, exaggerated curvature of the apical hook, and excessive radial swelling of the hypocotyl (Fig. 1B), which is the typical triple response of Arabidopsis to ethylene exposure (Guzmán and Ecker, 1990). However, the triple response phenotype was absent in the etiolated 35S:ZmARGOS1 seedlings (Fig. 1, B and C), demonstrating that the 35S:ZmARGOS1 plants are insensitive to exogenous ethylene.
To determine the response to endogenous ethylene, the ethylene overproducer1-1 (eto1-1) mutant (Chae et al., 2003) was transformed with 35S:ZmARGOS1 and transgene expression was confirmed by reverse transcription (RT)-PCR (Supplemental Fig. S5). In the three independent events examined, overexpression of ZmARGOS1 overrode the constitutive ethylene response in the etiolated eto1-1 seedlings (Fig. 1, D and E). The light-grown eto1-1 mutant plants flowered earlier than wild-type plants, but the floral transition in 35S:ZmARGOS1 eto1-1 plants was delayed (Fig. 1F) relative to eto1-1, similar to that of 35S:ZmARGOS1 in a wild-type background. The delayed flowering time was observed in all 10 independent events tested. These results confirmed that the 35S:ZmARGOS1 plants are insensitive to ethylene.
To further verify the activity of ZmARGOS1 in conferring ethylene insensitivity, a mutant version, ZmARGOS1(L104D), was generated by substituting Leu-104 with Asp in the highly conserved Pro-rich motif (Supplemental Fig. S1). Western analysis showed that the Leu substitution did not negatively affect the protein expression level in transgenic plants (Supplemental Fig. S6). The 35S:ZmARGOS1(L104D) Arabidopsis plants were sensitive to ACC (Fig. 1G), and flowering time was equivalent to nontransgenic controls (data not shown), indicating that the ethylene insensitivity in the 35S:ZmARGOS1 plants is dependent on the ZmARGOS1 protein.
Overexpression of ZmARGOS8 and Arabidopsis ARGOS Genes Also Decreases Ethylene Sensitivity in Arabidopsis
To determine if other ARGOS genes can modulate ethylene responses as well, ZmARGOS8, Arabidopsis ARGOS homolog ORGAN SIZE RELATED1 (AtOSR1), and AtOSR2 (Qin et al., 2014) were overexpressed in Arabidopsis. The 35S:AtOSR1 and 35S:AtOSR2 etiolated seedlings displayed the ethylene-insensitive phenotype in the triple response assay, as did the 35S:ZmARGOS1 plants (Fig. 2). Overexpression of ZmARGOS8 significantly reduced the ethylene-induced triple response as well, but the phenotype was weaker than that of the 35S:ZmARGOS1 plants (Fig. 2; Supplemental Fig. S7). Like ZmARGOS1(L104D), a mutated version of ZmARGOS8, ZmARGOS8(L67D), was not able to confer ACC insensitivity (Fig. 2).
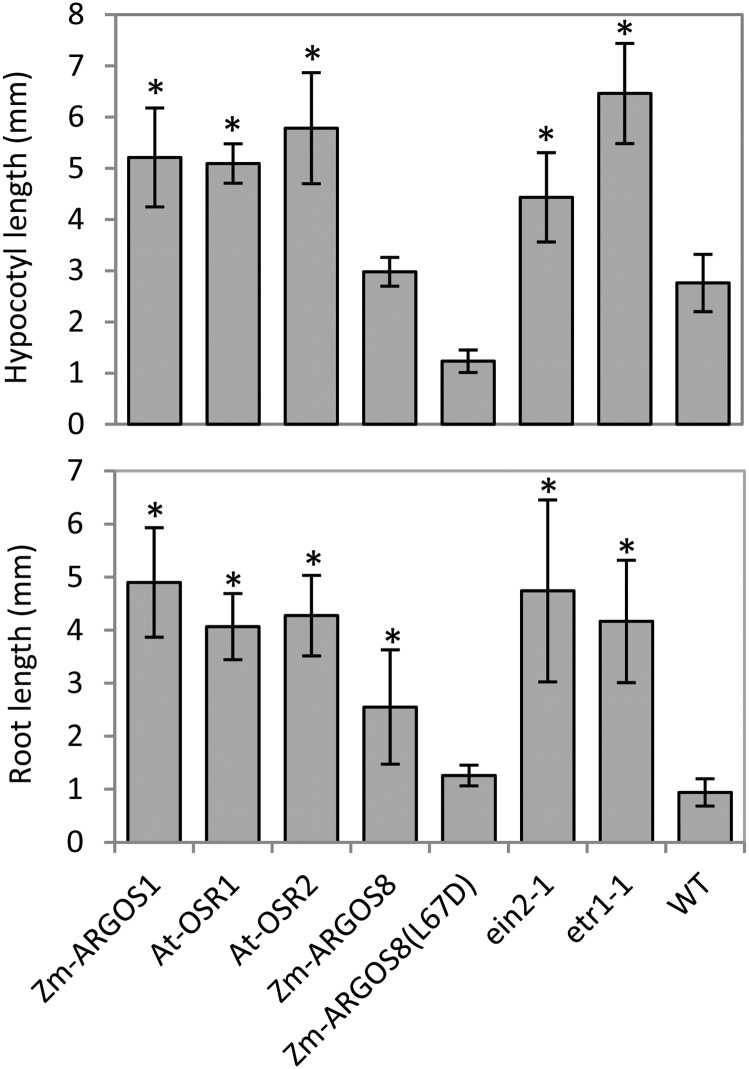
Figure 2.
The 35S:AtOSR1, 35S:AtOSR2, and 35S:ZmARGOS8 transgenic Arabidopsis plants have reduced ethylene sensitivity. Root and hypocotyl lengths are shown for 3-d-old etiolated Arabidopsis T1 seedlings germinated in the presence of 10 µm ACC. The data represent means of 12 to 20 independent events randomly selected for each construct based on YFP marker expression in T1 seeds. The 35S:ZmARGOS1 events, ethylene-insensitive mutants etr1-1 and ein2-1, and wild-type (WT) plants served as controls. Error bars indicate sd. Student’s t test was performed to compare the transgenic plants and the mutants with the wild-type control (*, P < 0.01).
Ethylene Biosynthesis Is Increased But the Expression of Ethylene-Inducible Genes Is Down-Regulated in ZmARGOS1 Arabidopsis Plants
Previous research has shown that the etr1-1 and ein2-1 mutants have increased ethylene biosynthesis (Guzmán and Ecker, 1990). Therefore, we determined ethylene emission in 35S:ZmARGOS1 leaves and found that they released 5- to 7-fold more ethylene than the vector control and wild-type plants (Fig. 3A). We would predict concomitant induced expression of ethylene-inducible genes if the transgenic plant had sensed ethylene normally. Northern analysis, however, showed that the steady-state levels of mRNA for the ethylene-inducible ETHYLENE-INSENSITIVE3-BINDING F-BOX PROTEIN2 (EBF2) and ETHYLENE RESPONSE FACTOR5 (ERF5) were decreased in the 35S:ZmARGOS1 plants relative to the vector control (Fig. 3B). In the aerial tissues (rosette leaves and apical meristem) of 19-d-old plants, transcript levels of the EIN3/EIL1-activated EBF2, PROTOCHLOROPHYLLIDE OXIDOREDUCTASE A (PORA), PORB, FLAGELLIN-SENSITIVE2 (FLS2), and ETHYLENE RESPONSE DNA BINDING FACTOR (EDF) genes (Alonso et al., 2003; Konishi and Yanagisawa, 2008; Zhong et al., 2009; Boutrot et al., 2010) were down-regulated in the 35S:ZmARGOS1 plants, while the EIN3/EIL1-repressed SALICYLIC ACID INDUCTION DEFICIENT2 (SID2; Chen et al., 2009) was up-regulated, as revealed by RNA sequencing analysis (Table I). Expression of nine ERF genes was reduced at least 50% relative to the vector control. Among these genes, ERF1, ERF2, ERF4, ERF5, ERF9, ERF11, and ERF72 were reported to be inducible by ethylene (Büttner and Singh, 1997; Solano et al., 1998; Fujimoto et al., 2000). ERF3 is not responsive to ethylene treatments (Fujimoto et al., 2000), and we found that its expression was not changed in the 35S:ZmARGOS1 plants. As predicted, expression of the ERF-regulated defensin genes PLANT DEFENSIN1.2 (PDF1.2), CHITINASE, BASIC CHITINASE (CHI-B), and PATHOGENESIS-RELATED4 (PR4; Solano et al., 1998; Fujimoto et al., 2000) was reduced (Table I). These results confirmed that the 35S:ZmARGOS1 transgenic plants were unable to properly sense endogenous elevated ethylene levels and suggested that ZmARGOS1 may act on the ethylene signaling components upstream of EIN3/EIL1.
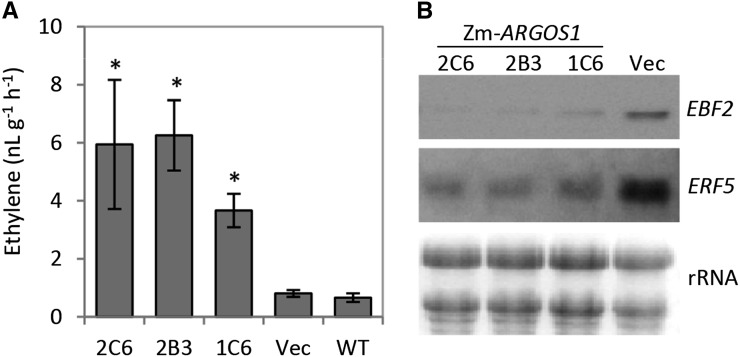
Figure 3.
Increased ethylene production and reduced expression of ethylene-inducible genes in Arabidopsis overexpressing ZmARGOS1. A, Ethylene production in rosette leaves of 20-d-old Arabidopsis plants is shown for three 35S:ZmARGOS1 events (2C6, 2B3, and 1C6), vector controls (Vec), and wild-type (WT) Col-0 plants. Ethylene was collected for a period of 22 h and subsequently measured using a gas chromatograph. Error bars indicate sd; n = 4. Student’s t test was performed to compare the transgenic events and empty vector controls with wild-type plants (*, P < 0.01). B, Reduced expression of ethylene-inducible genes in transgenic Arabidopsis plants overexpressing ZmARGOS1. Total RNA was extracted from rosette leaves of 3-week-old plants. Northern-blot analysis of three 35S:ZmARGOS1 events (2C6, 2B,3 and 1C6) and the vector control (Vec) was performed using 10 µg of RNA per lane and probed with the ethylene-inducible genes AtEBF2 and AtERF5. A gel stained with ethidium bromide is shown at bottom (rRNA) as a control for loading.
Table I. Ethylene-responsive gene expression in the aerial tissues (rosette leaves and apical meristem) of 19-d-old 35S:ZmARGOS1 Arabidopsis plants, as measured by RNA sequencing analysis.
Sequence reads were normalized to RPKtM. Values are means ± sd, with three replications for transgenic plants (TR) and four replications for vector controls (Ve). P represents the Student’s t test statistic (two-sided) P value.
| Gene | Locus | TR | Ve | TR:Ve Ratio | P |
|---|---|---|---|---|---|
| RPKtM | |||||
| EBF2 | At5g25350 | 305.8 ± 25.2 | 737.8 ± 43.0 | 0.41 | 0.0000 |
| PORA | At5g54190 | 2.2 ± 0.5 | 9.3 ± 5.1 | 0.23 | 0.0091 |
| PORB | At4g27440 | 1,618 ± 124.5 | 2,172.7 ± 169.6 | 0.74 | 0.0038 |
| FLS2 | At5g46330 | 113 ± 17.7 | 298 ± 13.3 | 0.38 | 0.0001 |
| SID2 | At1g74710 | 10.2 ± 0.9 | 6.5 ± 2.2 | 1.57 | 0.0475 |
| EDF1 | At1g25560 | 416.5 ± 29.7 | 733.3 ± 37.6 | 0.57 | 0.0001 |
| EDF2 | At1g68840 | 490.3 ± 34.8 | 1,200.1 ± 36.0 | 0.41 | 0.0000 |
| EDF4 | At1g13260 | 795.6 ± 15.8 | 1,339.5 ± 34.6 | 0.59 | 0.0000 |
| ERF1 | At4g17500 | 112.5 ± 8.7 | 211.3 ± 13.2 | 0.53 | 0.0001 |
| ERF2 | At5g47220 | 186.1 ± 8.8 | 347.9 ± 24.2 | 0.53 | 0.0000 |
| ERF3 | At1g50640 | 481.9 ± 14.4 | 478.0 ± 19.2 | 1.01 | 0.7744 |
| ERF4 | At3g15210 | 419.7 ± 19.9 | 649.9 ± 31.5 | 0.65 | 0.0001 |
| ERF5 | At5g47230 | 69.4 ± 4.6 | 270.5 ± 33.0 | 0.26 | 0.0000 |
| ERF6 | At4g17490 | 88.7 ± 10.2 | 236.9 ± 17.0 | 0.37 | 0.0000 |
| ERF9 | At5g44210 | 17.4 ± 4.9 | 53.9 ± 11.9 | 0.32 | 0.0019 |
| ERF11 | At1g28370 | 30.2 ± 4.2 | 74.9 ± 13.6 | 0.40 | 0.0010 |
| ERF13 | At2g44840 | 11.7 ± 5.8 | 26.4 ± 7.4 | 0.45 | 0.0524 |
| ERF72 | At3g16770 | 1,079.2 ± 196.3 | 2,541.1 ± 263.7 | 0.42 | 0.0004 |
| ERF104 | At5g61600 | 233.6 ± 8.6 | 556.1 ± 50.1 | 0.42 | 0.0000 |
| PDF1.2 | At5g44420 | 147.7 ± 51.5 | 564.9 ± 77.7 | 0.26 | 0.0009 |
| PDF1.2c | At5g44430 | 31.7 ± 15.1 | 222.0 ± 43.5 | 0.14 | 0.0005 |
| PDF1.2b | At2g26020 | 26.1 ± 8.8 | 209.8 ± 26.8 | 0.12 | 0.0001 |
| CHITINASE | At2g43590 | 52.6 ± 9.3 | 127.5 ± 40.8 | 0.41 | 0.0109 |
| CHI-B | At3g12500 | 37.2 ± 5.7 | 57.8 ± 11.8 | 0.64 | 0.0376 |
| PR4 | At3g04720 | 779.0 ± 44.8 | 1,175.1 ± 117.0 | 0.66 | 0.0014 |
ZmARGOS1 Targets the Upstream Components of the Ethylene Signal Transduction Pathway
The ARGOS family is composed of small integral membrane proteins containing unconserved N- and C-terminal regions and two transmembrane helices that flank a highly conserved Pro-rich motif of eight amino acids (Supplemental Fig. S1). As an example, AtOSR2 has only 67 amino acid residues. Protein sequence analysis did not reveal any catalytic sites in ARGOS proteins. There is no evidence indicating that ARGOS is one of the signaling cascade steps to relay the ethylene signal. Therefore, we hypothesized that ARGOS may play a regulatory role by directly, or indirectly, modifying the ethylene signaling components. To determine where ZmARGOS1 acts in the pathway, the ctr1-1 Arabidopsis mutant was transformed with 35S:ZmARGOS1 and transgene expression was confirmed by RT-PCR (Supplemental Fig. S5). Of 15 independent events examined, the light-grown 35S:ZmARGOS1 ctr1-1 plants all displayed the characteristic constitutive ethylene response phenotype (Fig. 4A), as did the ctr1-1 mutant (Kieber et al., 1993). In addition, the event C2 overexpressing the FLAG-HA epitope-tagged ZmARGOS1 (Supplemental Fig. S6) in a wild-type background was crossed with the ctr1-1 mutant to generate 35S:ZmARGOS1 ctr1-1 plants. Like those events produced by directly transforming ctr1-1, the 35S:ZmARGOS1 ctr1-1 plants also showed the same phenotype as the ctr1-1 mutant (Fig. 4B).
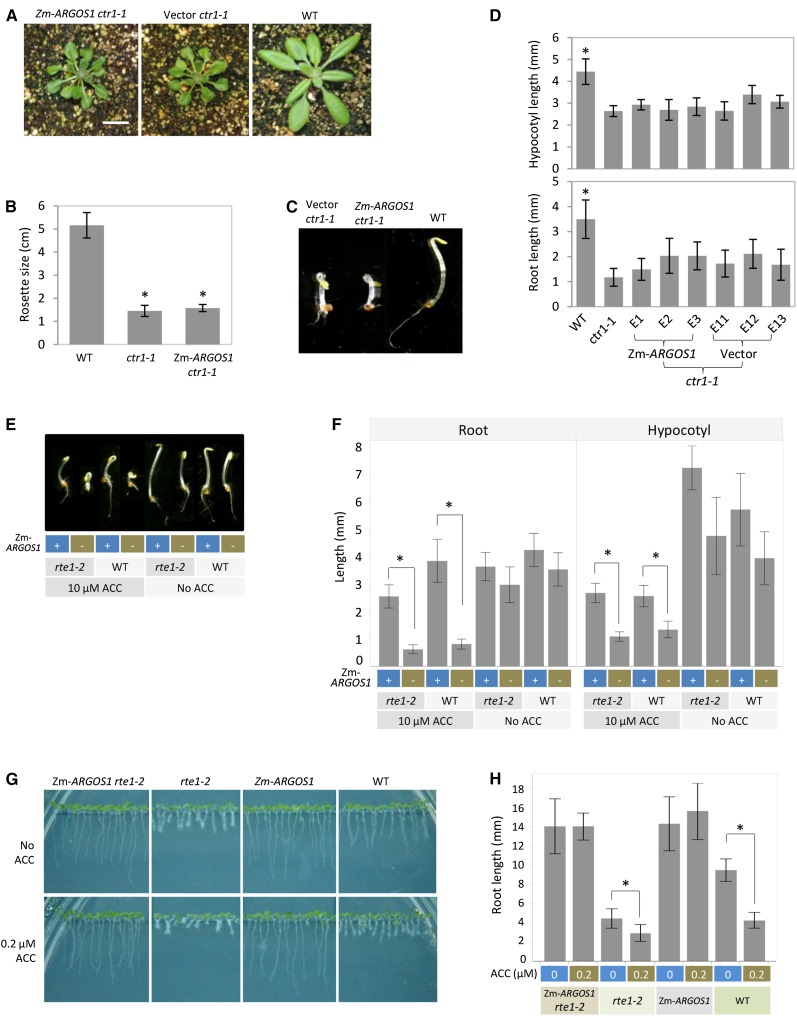
Figure 4.
Overexpression of ZmARGOS1 in Arabidopsis ctr1-1 and rte1-2 mutant plants. A, Constitutive ethylene response phenotypes in transgenic Arabidopsis ctr1-1 plants carrying 35S:ZmARGOS1 (ZmARGOS1 ctr1-1) and empty vector (Vector ctr1-1). Wild-type (WT) Col-0 plants grown in the same conditions served as controls. Representative 23-d-old plants are shown. Bar = 10 mm. B, Rosette sizes measured in the 35S:ZmARGOS1 ctr1-1 Arabidopsis plants, the ctr1-1 mutant, and the wild-type control. The 35S:ZmARGOS1 ctr1-1 plants were generated by crossing the 35S:ZmARGOS1-FLAG-HA event C2 with the ctr1-1 mutant followed by self-pollination. The plants were grown for 20 d. The data represent means of 13 to 17 plants. Error bars indicate sd. Student’s t test was performed to compare the transgenic plants and the ctr1-1 mutant with wild-type plants (*, P < 0.01). No significant difference was found between the 35S:ZmARGOS1 ctr1-1 plants and the ctr1-1 mutant (P > 0.05, Student’s t test). C, Overexpression of ZmARGOS1 in the Arabidopsis ctr1-1 mutant background. Etiolated ctr1-1 seedlings overexpressing ZmARGOS1 (ZmARGOS1 ctr1-1) display the constitutive triple response in the absence of exogenously supplied ethylene, similar to the transgenic ctr1-1 plants carrying empty vector (Vector ctr1-1). Wild-type Col-0 seedlings grown in the same conditions served as controls. A composite image of representative 3-d-old etiolated seedlings is shown. D, Hypocotyl and root lengths of etiolated Arabidopsis ctr1-1 mutant plants overexpressing ZmARGOS1. Seeds were germinated under dark conditions in the absence of exogenously supplied ethylene for 3 d. Three 35S:ZmARGOS1 events (ZmARGOS1 E1, E2, and E3) and three empty vector events (Vector E11, E12, and E13) in the ctr1-1 mutant background are shown. Wild-type Arabidopsis and the ctr1-1 mutant served as controls. Error bars indicate sd; n = 20. Student’s t test was performed to compare the 35S:ZmARGOS1 and empty vector constructs, and no difference was found. The hypocotyl and root lengths in the wild type are significantly longer than those in the ctr1-1 transgenic plants (*, P < 0.05). E, Overexpressing ZmARGOS1 in the Arabidopsis rte1-2 mutant background. The 35S:ZmARGOS1 rte1-2 plants were generated by crossing the ZmARGOS1 events 2B3 and 2C6 with the rte1-2 mutant followed by self-pollination. A composite image of representative 3-d-old etiolated seedlings is shown. The triple response assay was conducted in the presence or absence of 10 µm ACC. F, Root and hypocotyl lengths of 3-d-old, etiolated Arabidopsis seedlings are presented for the 35S:ZmARGOS1 rte1-2 plants, rte1-2 mutant, 35S:ZmARGOS1 transgenic plants, and wild-type controls. The 35S:ZmARGOS1 rte1-2 plants were generated by crossing the ZmARGOS1 events 2B3 and 2C6 with the rte1-2 mutant followed by self-pollination. The triple response assay was conducted in the presence and absence of 10 µm ACC. Measurements are shown for the event 2B3 cross. Error bars indicate sd; n = 10. Student’s t test was performed to compare the plants with or without the ZmARGOS1 transgene. *, P < 0.01. G, Root phenotypes are shown for the 35S:ZmARGOS1 rte1-2 Arabidopsis plants, the rte1-2 mutant, 35S:ZmARGOS1 transgenic plants, and wild-type controls. Plants were grown for 5 d in agar that contained one-half-strength Murashige and Skoog medium with 0 or 0.2 µm ACC and were set vertically in a growth chamber under a regime of 16 h of light (approximately 120 mE m−2 s−1) at 24°C and 8 h of darkness at 23°C. H, Root lengths of the 5-d-old Arabidopsis seedlings are shown for the 35S:ZmARGOS1 rte1-2 plants, rte1-2 mutant, 35S:ZmARGOS1 transgenic plants, and wild-type controls grown in the light in the presence of 0 or 0.2 µm ACC. Error bars indicate sd; n = 15. Student’s t test was performed to compare the plants in the presence and absence of ACC within each genotype. *, P < 0.01.
Under dark conditions, the etiolated seedlings of the 35S:ZmARGOS1 ctr1-1 Arabidopsis exhibited the exaggerated curvature of the apical hook and inhibited the growth of hypocotyls and roots in the absence of exogenously supplied ethylene (Fig. 4C), similar to the ctr1-1 mutant (Kieber et al., 1993). In the three independent events tested, no difference in hypocotyl or root lengths was detected between the 35S:ZmARGOS1 ctr1-1 Arabidopsis plants and the ctr1-1 plants carrying empty vector (Fig. 4D). Since CTR1 acts as a suppressor in the ethylene signal transduction pathway, these results suggested that the signaling pathway downstream of CTR1 functions properly even in the presence of overexpressed ZmARGOS1.
The 35S:ZmARGOS1 plants in a wild-type background from two events, 2B3 and 2C6, were crossed with the ETR1 null allele etr1-7 and the RTE1 loss-of-function mutant rte1-2. Both mutants have increased ethylene sensitivity (Hua and Meyerowitz, 1998; Resnick et al., 2006). The triple response assay showed that the 35S:ZmARGOS1 rte1-2 Arabidopsis plants had reduced ethylene sensitivity relative to the wild type and rte1-2 (Fig. 4, E and F). When seedlings were grown vertically on agar in the presence of ACC under light (Růzicka et al., 2007), the rte1-2 mutant had shorter roots and more pronounced root hairs due to increased ethylene sensitivity relative to the wild type (Fig. 4G). Roots of the 35S:ZmARGOS1 rte1-2 plants were longer than those of wild-type and rte1-2 plants in the absence and presence of 0.2 µm ACC (Fig. 4, G and H), confirming reduced ethylene sensitivity in the rte1-2 mutant overexpressing ZmARGOS1. Similarly, the 35S:ZmARGOS1 etr1-7 plants showed reduced ethylene sensitivity relative to the wild type and etr1-7 in both assays (Supplemental Figs. S8 and S9).
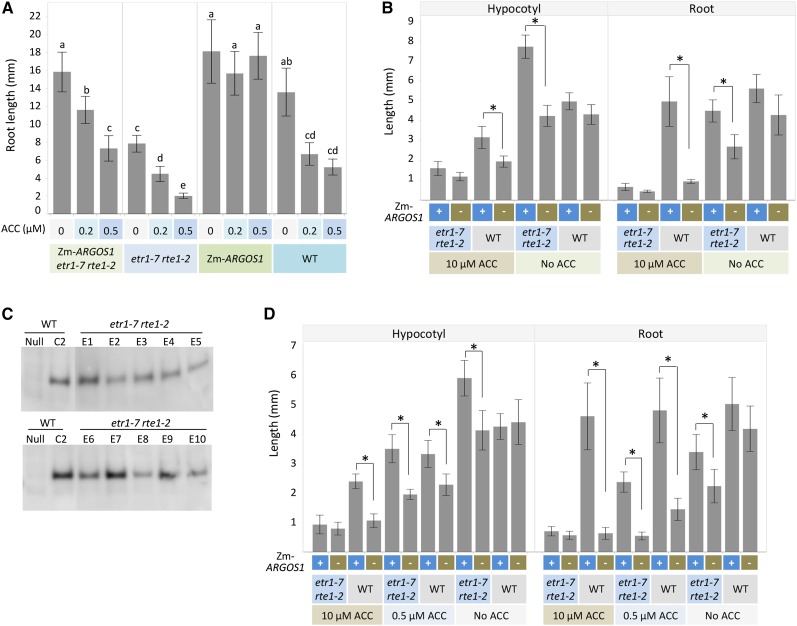
Because RTE1 interacts physically with ETR1 in Arabidopsis and modifies the activity of the ethylene receptor (Resnick et al., 2006, 2008; Dong et al., 2010), we tested ZmARGOS1 overexpression in an etr1-7 rte1-2 double mutant of Arabidopsis. The 35S:ZmARGOS1 etr1-7 rte1-2 plants were generated by crossing the 35S:ZmARGOS1 event 2B3 with the double mutant followed by self-pollination. Under light growth conditions, the 35S:ZmARGOS1 etr1-7 rte1-2 plants were less sensitive to 0.2 and 0.5 µm ACC relative to the double mutant (Fig. 5A; Supplemental Fig. S10). However, the ethylene-insensitive phenotype caused by ZmARGOS1 overexpression is weaker in the double mutant background than in the wild-type background (Fig. 5A). In addition, the root inhibition in the 35S:ZmARGOS1 etr1-7 rte1-2 plants was responsive to ACC concentrations, but it was not in the wild-type plants overexpressing ZmARGOS1 (Fig. 5A). These results indicate that the etr1-7 rte1-2 double mutation partially suppresses the ethylene-insensitive phenotype of ZmARGOS1 overexpression.
Figure 5.
Overexpression of ZmARGOS1 in the Arabidopsis etr1-7 rte1-2 double mutant. A, Root lengths in the 35S:ZmARGOS1 etr1-7 rte1-2 Arabidopsis plants compared with the etr1-7 rte1-2 double mutant. The 35S:ZmARGOS1 etr1-7 rte1-2 plant was generated by crossing the ZmARGOS1 event 2B3 and the etr1-7 rte1-2 double mutant followed by self-pollination. Plants were grown for 5 d in agar that contained one-half-strength Murashige and Skoog medium with 0, 0.2, or 0.5 µm ACC and were set vertically in a growth chamber under a regime of 16 h of light (approximately 120 mE m−2 s−1) at 24°C and 8 h of darkness at 23°C. The root lengths of the 35S:ZmARGOS1 plants in the wild-type (WT) background showed no significant difference under 0, 0.2, and 0.5 µm ACC (P > 0.05, ANOVA), but the 35S:ZmARGOS1 etr1-7 rte1-2 plants were significantly different (P < 0.05, ANOVA). Significant differences are denoted by different letters. Data are means ± sd; n = 10 to 20. B, Hypocotyl and root lengths of 3-d-old, etiolated Arabidopsis seedlings are presented for the 35S:ZmARGOS1 etr1-7 rte1-2 plants and the etr1-7 rte1-2 double mutant. The 35S:ZmARGOS1 etr1-7 rte1-2 plant was generated by crossing the ZmARGOS1 event 2B3 and the etr1-7 rte1-2 double mutant followed by self-pollination. The triple response assay was conducted in the presence and absence of 10 µm ACC in the dark. The wild-type Arabidopsis transgenic plants carrying 35S:ZmARGOS1 and nontransgenic wild-type plants served as controls. Error bars indicate sd; n = 10. Student’s t test was performed to compare the plants with or without the ZmARGOS1 transgene. *, P < 0.01. C, Immunoblot analysis of ZmARGOS1 overexpression in the etr1-7 rte1-2 Arabidopsis mutant plants. A FLAG-HA epitope-tagged ZmARGOS1 was overexpressed in the double mutant (etr1-7 rte1-2) or the wild-type background under the control of the cauliflower mosaic virus 35S promoter. Ten events (E1–E10) are shown. Event C2 of 35S:ZmARGOS1-FLAG-HA in the wild-type background served as a positive control. Western-blot analysis was performed with anti-FLAG antibodies. D, Hypocotyl and root lengths of 3-d-old, etiolated Arabidopsis seedlings are presented for the 35S:ZmARGOS1 etr1-7 rte1-2 plants and the etr1-7 rte1-2 double mutant. The 35S:ZmARGOS1 etr1-7 rte1-2 plant was generated by transforming the etr1-7 rte1-2 double mutant plants with 35S:ZmARGOS1-FLAG-HA. The triple response assay was conducted with four independent events (E1, E4, E7, and E9 in C) in the presence of 0, 0.5, or 10 µm ACC in the dark. Results from event E1 are shown. The wild-type Arabidopsis transgenic plants carrying 35S:ZmARGOS1 and nontransgenic wild-type plants served as controls. Error bars indicate sd; n = 10. Student’s t test was performed to compare the plants with or without the ZmARGOS1 transgene. *, P < 0.01.
The ethylene response of the 35S:ZmARGOS1 etr1-7 rte1-2 plants was also tested in the triple response assay under dark conditions. Although the etiolated 35S:ZmARGOS1 etr1-7 rte1-2 seedlings had reduced sensitivity to 0.5 µm ACC relative to the wild type and the double mutant (data not shown), they displayed the typical triple response phenotype under 10 µm ACC (Fig. 5B). To make sure ZmARGOS1 is expressed in the transgenic plants and to test additional independent events, we used the FLAG-HA epitope-tagged ZmARGOS1 construct to transform the etr1-7 rte1-2 double mutant. Transgene expression in 10 events was determined by western blotting (Fig. 5C), and four events (E1, E4, E7, and E9) were chosen for phenotyping. In the triple response assay under dark conditions, etiolated 35S:ZmARGOS1 etr1-7 rte1-2 seedlings showed reduced sensitivity to 0.5 µm ACC relative to the wild type and the double mutant (Fig. 5D). However, they displayed the typical triple response phenotype under 10 µm ACC (Fig. 5D), as the 2B3 event in the double mutant background did (Fig. 5B), confirming that ZmARGOS1 is not able to properly function in rte1-2 mutant seedlings when ETR1 is absent and the concentration of exogenously supplied ACC is high. A normal interaction between RTE1 and ETR1 likely is required for ZmARGOS1 conferring ethylene insensitivity in Arabidopsis seedlings. Taken together, these results suggested that ZmARGOS1 may act between the ethylene receptor and CTR1, affecting ethylene perception or the early stages of ethylene signal transduction.
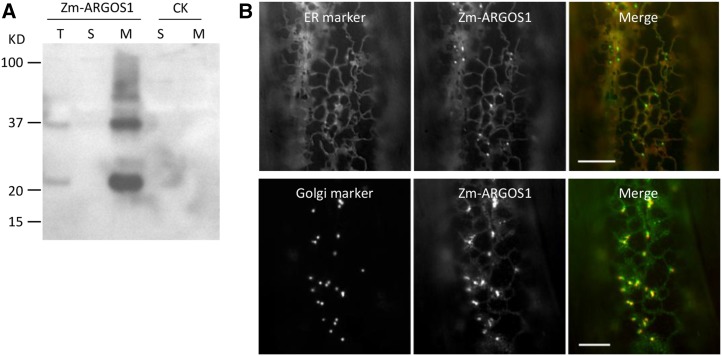
ZmARGOS1 Is Localized in the ER and Golgi Membranes
Sequence analysis with PRODIV-TMHMM (Viklund and Elofsson, 2004) predicted that ZmARGOS1 and other family members contain two transmembrane α-helices. Cell fractionation analysis showed that the FLAG-HA-tagged ZmARGOS1 was present in the microsomal fraction but nondetectable in the soluble fraction (Fig. 6A), confirming that ZmARGOS1 is a membrane protein. To determine the subcellular localization, the Aequorea coerulescens GFP was fused to the C terminus of ZmARGOS1 and the construct was transfected into Arabidopsis. In stable transgenic plants, the GFP tag did not affect ZmARGOS1 function in conferring ethylene insensitivity (data not shown). Fluorescence microscopy of the hypocotyl cells showed that the ZmARGOS1-GFP fusion protein was localized to moderately fluorescing threads that form a loose web-like pattern within the cell (Supplemental Fig. S11). Also, small bright GFP-positive bodies, in close association with the threads, were observed within the lumen of each cell. The localization pattern of the fusion protein in hypocotyl cells of stable transgenic Arabidopsis is similar to that observed in onion (Allium cepa) epidermal cells transiently expressing the ZmARGOS1-GFP fusion protein (Fig. 6B). In onion epidermal cells, coexpression of the fusion protein and a fluorescently tagged ER marker, ER-ck CD3-953 (Nelson et al., 2007), showed strong colocalization to a web-like pattern throughout the lumen of the cell (Fig. 6B). Additionally, coexpression of the ZmARGOS1-GFP fusion protein and a fluorescently tagged Golgi marker, G-ck CD3-961 (Nelson et al., 2007), indicated that there was a strong association between the strongly fluorescent GFP-positive bodies and the Golgi. These ZmARGOS1-GFP fusion protein bodies were less than 1 µm in diameter and were closely associated with the ER. Expression of the fluorescent ARGOS1 fusion protein was not observed in nuclei, plastids, vacuoles, cytoplasm, vacuolar membranes, and the plasma membranes in the transgenic Arabidopsis hypocotyl or onion epidermal cells.
Figure 6.
ER and Golgi membrane localization of overexpressed ZmARGOS1. A, Immunoblot analysis of cellular fractions of Arabidopsis plants overexpressing FLAG-HA epitope-tagged ZmARGOS1 (Zm-ARGOS1) and an untagged ZmARGOS1 control (CK). Total (T) homogenates were ultracentrifuged to separate the soluble (S) and microsomal membrane (M) fractions. Western-blot analysis was performed with anti-FLAG antibodies. B, Overexpressed ZmARGOS1 protein is localized to the ER and Golgi in transiently transformed onion epidermal cells. The top row shows fluorescence microscopy images of the interior of single cells displaying colocalization of the GFP-tagged ZmARGOS1 (middle) with an ER marker, the cyan fluorescent protein-tagged ER CD3-953 (left). At right is the merged image of ZmARGOS1 (green) and ER CD3-953 (red). The bottom row shows images of GFP-tagged ZmARGOS1 (middle) and a Golgi marker, the cyan fluorescent protein-tagged Golgi CD3-961 (left). The merged image of ZmARGOS1 (green) and Golgi CD3-961 (red) at right indicates that there is a strong association between the strongly fluorescent ZmARGOS1-GFP fusion bodies and the Golgi marker. Bars = 10 µm.
ARGOS Transgenic Arabidopsis Plants Have Increased Drought Tolerance
ARGOS transgenic Arabidopsis plants were tested along with the etr1-1 and ein2-1 mutants for drought tolerance. Drought stress was applied by withholding water at 14 d after germination. Since Arabidopsis leaves wilt during drought stress, maintenance of the leaf area was used as a criterion for evaluating the drought tolerance of transgenic plants. LemnaTec HTSBonitUV software was used to capture and segment red-green-blue images. Estimates of the leaf area of the Arabidopsis plants were obtained in terms of the number of green pixels. The data for each image were averaged to obtain estimates of mean and sd for the green pixel counts for transgenic and wild-type plants. Parameters for a noise function were obtained by straight line regression of the squared deviation versus the mean pixel count using data for all images in a batch. Error estimates for the mean pixel count data were calculated using the fit parameters for the noise function. The mean pixel counts for transgenic and wild-type plants were summed to obtain an assessment of the overall leaf area for each image. The 4-d interval with maximal wilting was obtained by selecting the interval that corresponds to the maximum difference in plant growth. The individual wilting responses of the transgenic and wild-type plants were obtained by normalization of the data using the value of the green pixel count of the first day in the interval. The drought tolerance of the transgenic plant compared with the wild-type plant was scored by summing the weighted difference between the wilting responses of transgenic plants and wild-type plants over days 2 through 4; the weights were estimated by propagating the error in the data. A positive drought tolerance score corresponds to a transgenic plant with slower wilting compared with the wild-type plant. An effect size statistic for the difference in wilting response between transgenic and wild-type plants is obtained from the weighted sum of the squared deviations. When the transgenic replicates show a significant difference (score of greater than 2) from the control replicates, the line is then considered a validated drought-tolerant line. In the drought assay, the 35S:ZmARGOS1, 35S:ZmARGOS8, 35S:AtOSR1, and 35S:AtOSR2 plants showed a significant delay in leaf area loss relative to wild-type controls (Fig. 7). The increased drought tolerance was also observed in the etr1-1 and ein2-1 mutants. However, the transgenic plants overexpressing the mutated, loss-of-function ARGOS, ZmARGOS1(L104D) and ZmARGOS8(L67D), were not significantly different from wild-type plants (Fig. 7B). After establishing the role of ARGOS in Arabidopsis growth and development, we next tested its functionality in maize.
Figure 7.
Overexpression of ARGOS genes in Arabidopsis increased drought tolerance in a wilting assay. A, Increased drought tolerance in the ARGOS transgenic Arabidopsis plants. A representative phenotype is shown for the 35S:AtOSR1 plants and wild-type (WT) controls 15 d after the last watering. B, Overexpression of multiple ARGOS genes increases drought tolerance in Arabidopsis as well as that of etr1-1 and ein2-1 mutants. Maintenance of the green leaf area under drought stress was used as a criterion for evaluating the drought tolerance of transgenic plants. A drought score greater than 2 indicates drought-tolerant plants.
Overexpressing ZmARGOS Genes Reduces Ethylene Responses in Transgenic Maize Plants
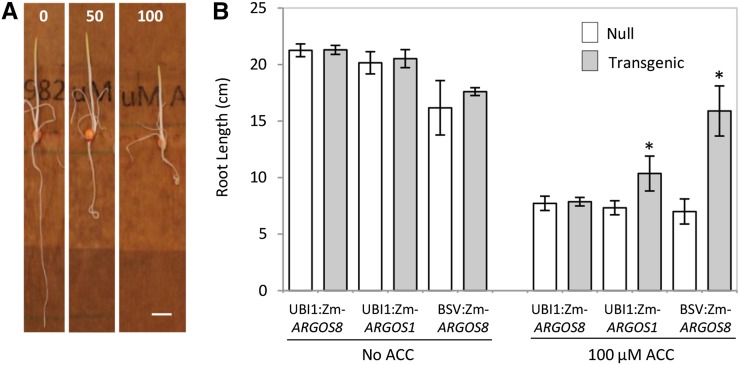
ZmARGOS1 and ZmARGOS8 were overexpressed in maize under the control of the maize UBIQUITIN1 promoter (UBI1) and the banana streak virus promoter (BSV; Schenk et al., 2001). Both promoters drive constitutive expression, with BSV being stronger than UBI1. To determine the transgenic maize response to exogenously supplied ACC, seeds were germinated in the presence of the ethylene precursor. The ACC treatment reduced root elongation and affected root gravitropism in nontransgenic seedlings (Fig. 8A) but did so to a lesser extent in the UBI1:ZmARGOS1 events (Fig. 8B). The inhibition of root growth was detectable at 50 μm ACC, and the severity of the phenotype intensified with an increase in ACC concentration. The BSV:ZmARGOS8 plants were insensitive to ACC, with no obvious root inhibition observed at 100 μm (Fig. 8B). However, when ZmARGOS8 expression was driven by the relatively weaker UBI1, the root length of the transgenic and nontransgenic plants was indistinguishable in this assay (Fig. 8B). In the absence of exogenously supplied ACC, no difference in root growth was detected among the ARGOS transgenic and nontransgenic seedlings (Fig. 8B). The reduced ethylene response suggested that ARGOS overexpression affects ethylene sensitivity in maize, similar to that in transgenic Arabidopsis.
Figure 8.
Overexpression of ARGOS genes reduced ethylene sensitivity in maize. A, The ethylene precursor ACC inhibits root growth and affects gravitropism in maize seedlings. Representative 5-d-old seedlings are shown for the wild-type maize plants grown in filter paper rolls set vertically in the dark in the presence of 0, 50, and 100 µm ACC. Bar = 2 cm. B, Overexpressing ARGOS genes reduces the ethylene response in maize seedlings. Four UBI1:ZmARGOS1 and UBI1:ZmARGOS8 events and three BSV:ZmARGOS8 events were germinated in the dark in the presence of 0 and 100 µm ACC. The data represent means of the root lengths of 5-d-old seedlings. Student’s t test was performed to compare the transgenic and nontransgenic (null) segregants (*, P < 0.05). Error bars indicate sd.
Maize Events Overexpressing ZmARGOS8 Have Increased Grain Yield under Field Conditions
Guo et al. (2014) reported that UBI1:ZmARGOS1 has a positive effect on maize grain yield, but only in particular environments. The ZmARGOS1 construct reduces ethylene sensitivity in maize (Fig. 8B) and has potential in yield improvement under dry and high-temperature conditions. However, yield reduction as revealed in humid and low-temperature conditions limits its practical applications. The strength of ARGOS activity and expression levels of a transgene likely influence its capability in regulating ethylene sensitivity in transgenic plants. ZmARGOS8, whose activity is weaker than ZmARGOS1, is able to reduce ethylene sensitivity when overexpressed in Arabidopsis (Fig. 2) and maize (Fig. 8B) and to enhance drought tolerance in Arabidopsis (Fig. 7B). Aiming at developing maize hybrids that have yield advantage under drought-stressed environments with no yield loss in optimal growing conditions, we elected to determine the functionality of ZmARGOS8 and, therefore, tested eight single-copy UBI1:ZmARGOS8 events in a hybrid background over a 2-year period at multiple locations throughout the United States. At the end of each growing season, locations were categorized into either a high-drought-stress or low-drought-stress environment based on several drought stress parameters (Loffler et al., 2005). Grain yield was analyzed using a mixed model via ASReml (Gilmour et al., 2009; Habben et al., 2014), and Table II shows that plants grown under a high-drought-stress condition had, on average, a 56% decrease in yield compared with those grown under a low-stress condition, indicating the severity of water limitation on grain yield. Under the high-stress condition, all events showed a statistically significant increase in yield relative to the bulk null comparator (Table II). Interestingly, when these same events were grown in low-stress, high-yielding conditions, they also showed a significant increase in yield relative to the comparator. Thus, events overexpressing the UBI1:ZmARGOS8 transgenic cassette showed yield efficacy not only under drought stress but also under well-watered conditions.
Table II. Grain yield of maize UBI1:ZmARGOS8 transgenic events and the bulk null in a hybrid under high- and low-drought-stress conditions.
Data are from eight individual transgenic maize events (plus construct mean) and the bulk null at high- and low-drought-stress locations in 2012 and 2013. The predicted difference for each transgenic entry is compared with the bulk null. All analyses were implemented using ASReml with output of the model presented as best linear unbiased predictions (see “Materials and Methods”). Asterisks indicate predicted differences significant at P < 0.05.
| Entry | High-Drought Stress |
Low-Drought Stress |
||
|---|---|---|---|---|
| Yield Prediction | Predicted Difference | Yield Prediction | Predicted Difference | |
| Mg ha−1(bushel acre−1) | ||||
| DP-E3.10 | 7.83 (124.4) | 0.32 (5.1)* | 13.55 (215.5) | 0.26 (4.1)* |
| DP-E3.12 | 7.83 (124.6) | 0.33 (5.3)* | 13.55 (215.4) | 0.25 (4.1)* |
| DP-E4.13 | 7.83 (124.5) | 0.33 (5.3)* | 13.55 (215.4) | 0.25 (4.0)* |
| DP-E4.15 | 7.84 (124.6) | 0.33 (5.3)* | 13.54 (215.3) | 0.24 (3.9)* |
| DP-E4.16 | 7.83 (124.4) | 0.32 (5.2)* | 13.55 (215.5) | 0.26 (4.1)* |
| DP-E4.17 | 7.84 (124.6) | 0.33 (5.3)* | 13.55 (215.5) | 0.26 (4.1)* |
| DP-E5.10 | 7.83 (124.5) | 0.33 (5.2)* | 13.55 (215.4) | 0.25 (4.0)* |
| DP-E5.03 | 7.83 (124.5) | 0.33 (5.2)* | 13.55 (215.5) | 0.26 (4.1)* |
| Constructa | 7.83 (124.5) | 0.33 (5.2)* | 13.55 (215.4) | 0.26 (4.1)* |
| Bulk null | 7.50 (119.3) | – | 13.29 (211.4) | – |
Construct, The mean of transgene positive events.
To enhance our understanding of transgene efficacy in other genetic backgrounds, seven of the eight ARGOS8 events were converted into two inbreds, top crossed to two testers, and field yield tested. Similar to the results described above, locations were grouped into high- and low-drought-stress environments. In both hybrids, all events with the exception of one (DP-E5.03; hybrid 2) showed a statistically significant increase in grain yield under high-drought stress (Table III). At the construct level, events averaged a 6.6 and 4.5 bu ac−1 increase in hybrid 1 and hybrid 2, respectively. Yield increases of the best event (DP-E4.17) ranged from 5 to 10.9 bu ac−1 across the two hybrids under a high-drought-stress condition (Table III). In the low-stress environment, all events showed either a positive or neutral grain yield increase in both hybrids, with there being an overall significant increase in yield at the construct level. To determine the basis of the enhanced grain yield, we measured several ear parameters in both hybrids (Table IV). There was a significant increase in kernel number per ear, with both hybrids averaging more than 20 extra kernels. The ear length was also increased significantly relative to the comparator, but there was no change in ear width (Table IV). Based on these results collected in high-yielding germplasm, UBI1:ZmARGOS8 events consistently demonstrated increased yield under both drought stress and well-watered conditions.
Table III. Grain yield of maize UBI1:ZmARGOS8 transgenic events and the bulk null in two hybrids under high- and low-drought-stress conditions.
Data are from eight individual transgenic maize events (plus construct mean) and the bulk null converted into two hybrids and grown at high- and low-drought-stress locations in 2013. The predicted difference for each transgenic entry is compared with the bulk null. All analyses were implemented using ASReml with output of the model presented as best linear unbiased predictions (see “Materials and Methods”). Asterisks indicate predicted differences significant at P < 0.05.
| Entry |
High-Drought Stress |
Low-Drought Stress |
||
|---|---|---|---|---|
| Yield Prediction | Predicted Difference | Yield Prediction | Predicted Difference | |
| Mg ha−1(bushel acre−1) | ||||
| Hybrid 1 | ||||
| DP-E3.10 | 8.87 (141.0) | 0.52 (8.3)* | 14.34 (228.0) | 0.29 (4.6)* |
| DP-E3.12 | 8.83 (140.5) | 0.49 (7.8)* | 14.29 (227.2) | 0.24 (3.8)* |
| DP-E4.13 | 8.75 (139.1) | 0.41 (6.5)* | 14.19 (225.6) | 0.14 (2.3) |
| DP-E4.15 | 8.81 (140.1) | 0.47 (7.4)* | 14.24 (226.5) | 0.20 (3.1) |
| DP-E4.16 | 8.61 (136.8) | 0.26 (4.2)* | 14.08 (223.9) | 0.03 (0.5) |
| DP-E4.17 | 9.03 (143.6) | 0.69 (10.9)* | 14.49 (230.4) | 0.44 (7.0)* |
| DP-E5.03 | 8.81 (140.0) | 0.46 (7.4)* | 14.29 (227.2) | 0.24 (3.8)* |
| Constructa | 8.76 (139.3) | 0.42 (6.6)* | 14.22 (226.1) | 0.17 (2.7)* |
| Bulk null | 8.34 (132.7) | – | 14.05 (223.4) | – |
| Hybrid 2 | ||||
| DP-E3.10 | 7.62 (121.2) | 0.21 (3.4)* | 12.80 (203.5) | 0.14 (2.3) |
| DP-E3.12 | 7.63 (121.3) | 0.22 (3.6)* | 12.80 (203.6) | 0.15 (2.4) |
| DP-E4.13 | 7.80 (124.0) | 0.39 (6.3)* | 12.94 (205.8) | 0.29 (4.6)* |
| DP-E4.15 | 7.63 (121.3) | 0.22 (3.5)* | 12.78 (203.1) | 0.12 (1.9) |
| DP-E4.16 | 7.86 (125.1) | 0.46 (7.3)* | 13.03 (207.2) | 0.38 (6.0)* |
| DP-E4.17 | 7.72 (122.8) | 0.32 (5.0)* | 12.89 (204.9) | 0.23 (3.7)* |
| DP-E5.03 | 7.39 (117.5) | −0.02 (-0.3) | 12.56 (199.7) | −0.10 (-1.5) |
| Constructa | 7.69 (122.2) | 0.28 (4.5)* | 12.85 (204.3) | 0.20 (3.1)* |
| Bulk null | 7.41 (117.8) | – | 12.65 (201.2) | – |
Construct, The mean of transgene positive events.
Table IV. Ear parameters of maize UBI1:ZmARGOS8 transgenic events and the bulk null in two hybrids.
Data are presented at the construct level compared with the bulk null in two hybrids at a low-drought-stress location in 2013. All analyses were implemented using ASReml with output of the model presented as best linear unbiased predictions (see “Materials and Methods”). Asterisks indicate predicted differences significant at P < 0.05.
| Entry | Hybrid 1 |
Hybrid 2 |
||||
|---|---|---|---|---|---|---|
| Kernels per Ear | Ear Length | Ear Width | Kernels per Ear | Ear Length | Ear Width | |
| n | cm (inches) | n | cm (inches) | |||
| Constructa | 559* | 19.4 (7.6)* | 5.3 (2.1) | 559* | 19.5 (7.7)* | 5.3 (2.1) |
| Bulk null | 538 | 19.0 (7.5) | 5.3 (2.1) | 537 | 19.0 (7.5) | 5.3 (2.1) |
Construct, The mean of transgene positive events.
DISCUSSION
It has been reported that overexpression of ARGOS genes promotes lateral organ growth by regulating cell number and/or size in Arabidopsis (Hu et al., 2003, 2006; Feng et al., 2011). However, the molecular mechanism of this enhanced organ growth is not known. We found that ARGOS negatively modulates the response to ethylene when overexpressed in Arabidopsis and maize. The enhanced ethylene production and reduced expression of a large number of ethylene-inducible genes in 35S:ZmARGOS1 Arabidopsis events supports the concept of reduced ethylene sensitivity in transgenic plants. Overexpressing ZmARGOS1 in the Arabidopsis ctr1-1 mutant suggested that ARGOS may not target the ethylene signaling pathway downstream of CTR1. Further genetic analysis showed that ZmARGOS1 can reduce ethylene sensitivity in the loss-of-function mutant rte1-2 or the null mutant etr1-7. However, ZmARGOS1 was not able to properly function in Arabidopsis rte1-2 mutant plants when the ethylene receptor ETR1 was absent and the exogenously supplied ACC concentration was high, suggesting that ZmARGOS1 likely targets ethylene perception or the initial stage of ethylene signal transduction. Cell fractionation and microscopy results indicated that overexpressed ZmARGOS1 was localized to the ER and Golgi membrane, where the ethylene receptor complex resides. This subcellular localization is consistent with its role in regulating ethylene signaling.
According to the current model of ethylene perception and signal transduction (Ju et al., 2012; Qiao et al., 2012), in the absence of ethylene, the ethylene receptors are in an active form that activates CTR1. The activated CTR1 represses downstream signaling by phosphorylating EIN2. The membrane-localized RTE1 plays a regulatory role by interacting with the ethylene receptor ETR1 (Dong et al., 2008, 2010; Resnick et al., 2008). Overexpression of Arabidopsis RTE1 and the tomato (Solanum lycopersicum) homolog Green-Ripe (Barry and Giovannoni, 2006) likely enhances the ethylene receptor activity, reducing plant sensitivity to ethylene. In the presence of ethylene, ethylene binding by receptors causes a decrease in CTR1 kinase activity, subsequently resulting in cleavage and nuclear translocation of the EIN2 C terminus, which activates the EIN3/EIL1-dependent ethylene response (Ju et al., 2012; Qiao et al., 2012). ZmARGOS1 overexpression confers ethylene insensitivity, possibly by enhancing the activity of ethylene receptors or by keeping CTR1 active even in the presence of ethylene. As a result, the expression of the EIN3/EIL1-activated genes is down-regulated while the EIN3/EIL1-suppressed genes are up-regulated in the Arabidopsis plants overexpressing ZmARGOS1 (Table I).
We tested an Arabidopsis ARGOS transfer DNA insertion knockdown line (GK-627B07) and an AtOSR1 knockout mutant (GK-436G04). No obvious phenotype was observed in the single mutants and the argos osr1 double mutant (data not shown). However, the triple mutant of the AtARGOS knockdown, AtOSR1 knockout, and rte1-2 showed an increase in ethylene sensitivity relative to the rte1-2 mutant (J. Shi and R.L. Archibald, unpublished data). Similar results were obtained when the double mutant was moved into an etr1-7 mutant background (J. Shi and R.L. Archibald, unpublished data). There are four ARGOS genes in Arabidopsis (Supplemental Fig. S1), and functional redundancy is evident (Feng et al., 2011). It would be interesting to see how a quadruple knockout Arabidopsis line would respond to ethylene.
Arabidopsis plants overexpressing various ARGOS members were found to be tolerant to drought in a wilting assay. This tolerance is likely a consequence of reduced ethylene sensitivity, because the transgenic plants were insensitive to ethylene and the mutant versions of both ZmARGOS1 and ZmARGOS8, which cannot reduce ethylene sensitivity when overexpressed, were unable to confer drought tolerance. In addition, the ethylene-insensitive mutant etr1-1 and ein2-1 also showed increased drought tolerance in the wilting assay. In water-deficient conditions, the reduced leaf-wilting phenotype is possibly a result of reduced water loss through transpiration in ethylene-insensitive plants. Tholen et al. (2008) reported that stomatal conductance of the etr1-1 mutant was 44% lower than in wild-type plants. The finding of ethylene-inhibiting abscisic acid-induced stomatal closure in drought-stressed Arabidopsis (Tanaka et al., 2005) also supports the concept of a water conservation effect in ethylene-insensitive Arabidopsis. Whether this occurs in Arabidopsis plants overexpressing ARGOS genes remains to be determined.
Maize plants overexpressing ZmARGOS8 also had ethylene sensitivity reduced and concomitant better grain yield relative to nontransgenic controls in water-deficient environments, indicating enhanced drought tolerance. This result is consistent with our earlier observation that reduced ethylene production improves drought tolerance in maize (Habben et al., 2014). Grain yield is a complex trait affected by numerous molecular and physiological processes during vegetative and reproductive growth. It is widely recognized that drought has a negative effect on plant growth and that ethylene plays an important role in this phenotype. Maintenance of plant growth under stressed conditions could mitigate the yield loss. The UBI1:ZmARGOS8 transgenic plants had longer ears and produced more kernels, suggesting that their growth was less affected by drought relative to nontransgenic controls. Previous research also showed that overexpressing ARGOS promotes growth in leaves, flowers, and seeds in maize (Guo et al., 2014), Arabidopsis (Hu et al., 2003, 2006; Feng et al., 2011), and tobacco (Nicotiana tabacum; Kuluev et al., 2011).
Field yield trials have shown that UBI1:ZmARGOS8 events enhance maize grain yields under both drought-stress and well-watered conditions. A yield increase in multiple genetic backgrounds suggests the broader applicability of ZmARGOS8 in crop production. Although overexpression of ZmARGOS1 can improve maize yields in certain drought environments, a negative effect on yields was observed in cool and high-humidity conditions (Guo et al., 2014). This performance difference between the two ARGOS genes may reflect a magnitude variation in activity between ZmARGOS1 and ZmARGOS8, with the former being stronger in reducing ethylene sensitivity (Figs. 2 and 8). Given the effect of ethylene on myriad cellular and developmental processes, it is expected that a drastic modification of ethylene signal transduction would not be beneficial in maximizing grain yields. Instead, obtaining an optimal ethylene level and/or ethylene sensitivity may be more desirable.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) mutants eto1-1, etr1-7, rte1-2, and ctr1-1 are in the Col-0 ecotype. The eto1-1 and ctr1-1 mutants were obtained from the Arabidopsis Biological Resource Center, and rte1-2 and ctr1-1 were a gift from Caren Chang (University of Maryland). Plants were grown under fluorescent lamps supplemented with incandescent lights (approximately 120 mE m−2 s−1) in growth chambers with a 16-h light period at 24°C and an 8-h dark period at 23°C and 50% relative humidity.
For the Arabidopsis triple response assay, surface-sterilized and stratified seeds were germinated in the presence of ethylene gas (Praxair) in an air-tight container or on medium (one-half-strength Murashige and Skoog salts with 1% [w/v] Suc and 0.8% [w/v] agar) containing ACC (Calbiochem) at the stated concentrations. Hypocotyl and root lengths were measured by photographing the seedlings using a dissection microscope with a digital camera and using image-analysis software (ImageJ; National Institutes of Health).
For assaying the maize (Zea mays) seedling response to ACC, seeds were placed in a row between two layers of filter papers wetted with an ACC aqueous solution at the stated concentrations. The filter paper was rolled up with a piece of waxed paper on the outside and set vertically in a beaker containing 1 inch (2.5 cm) of the same solution. The beaker was covered with plastic wrap to prevent evaporation and placed at 24°C in the dark. Seedling phenotypes were scored 5 d after seeding.
Transgene Constructs and Plant Transformation
Open reading frames of maize or Arabidopsis genes were PCR amplified, cloned into pENTR/D-TOPO vector (Invitrogen), and confirmed by sequencing. The mutated versions of ARGOS genes were created by the PCR method using primers containing the desired mutations. To generate the FLAG-HA epitope-tagged ZmARGOS1, PCR primers was designed to include the ZmARGOS1-specific sequence and the coding sequence for the FLAG and HA epitope (DYKDDDDKVKLYPYDVPDYAAA). Using Gateway technology (Invitrogen), the genes in the pENTR/D entry vector were mobilized into the binary vector pBC.Yellow (de la Luz Gutiérrez-Nava et al., 2008), which contains the cauliflower mosaic virus 35S promoter and the phaseolin terminator. The binary vector has two selectable markers: YFP under the control of the desiccation-responsive AtRd29 promoter for color selection and the BIALAPHOS RESISTANCE gene for herbicide selection. The Agrobacterium tumefaciens strain GV3101 was used to transform Arabidopsis Col-0 with the flower-dipping method (Clough and Bent, 1998).
GFP and ZmARGOS1 fusions were created by joining the PCR product of the Aequorea coerulescens GFP and ZmARGOS1 in a vector containing Gateway attL4/3 recombination sites, the cauliflower mosaic virus 35S promoter, and the nopaline synthase terminator. A linker sequence encoding for GGGSGGGS was placed between the two genes. The recombinant gene was integrated into a binary vector containing Gateway attR4/3 recombination sites and the selectable marker UBI1 PRO:UBI1 INTRON1:MoPAT:PinII Term (Unger et al., 2001; Cigan et al., 2005). The A. tumefaciens strain LBA4404 that harbors the construct was used to transform the Arabidopsis ecotype as stated above.
For maize transformation, the complementary DNA (cDNA) sequences of ZmARGOS1 and ZmARGOS8 were integrated between the maize UBI1 or BSV and the PinII terminator using Gateway technology and cointegrated with plant transformation vectors as described previously (Unger et al., 2001; Cigan et al., 2005). Plasmids were introduced into A. tumefaciens strain LBA4404 and used to transform maize embryos from a proprietary inbred. Multiple independent events were generated for each construct. Single-copy transfer DNA integration events that expressed the transgene were selected and advanced for crosses to wild-type plants and further characterization.
For subcellular colocalization of ZmARGOS1, binary plasmids carrying cyan fluorescent protein-tagged ER and Golgi markers (ER-ck CD3-953 and G-ck CD3-961; Nelson et al., 2007) were obtained from the Arabidopsis Biological Resource Center. Onion (Allium cepa) inner epidermal cell transformation was performed as described previously (Scott et al., 1999).
Ethylene Measurements
Whole leaves were excised from 3-week-old Arabidopsis plants. After letting the wound-induced ethylene burst subside for 2 h, the leaves then were placed in 9.77-mL amber vials containing a filter paper disc wetted with 50 μL of distilled water and sealed with aluminum crimp seals. After a 20-h incubation period, 1-mL samples were taken from the headspace of each sealed vial. The ethylene content was quantified by gas chromatography as described (Habben et al., 2014). Ethylene production rate was expressed as nL h−1 g−1 fresh weight.
Gene Expression Analysis by RNA Sequencing
Total RNAs were isolated from aerial tissues of 19-d-old Arabidopsis plants by use of the Qiagen RNeasy kit for total RNA isolation (Qiagen). Sequencing libraries from the resulting total RNAs were prepared using the TruSeq mRNA-Seq kit according to the manufacturer’s instructions (Illumina). Briefly, mRNAs were isolated via attachment to oligo(dT) beads, fragmented to a mean size of 150 nucleotides, reverse transcribed into cDNA using random primers, end repaired to create blunt end fragments, 3′ A-tailed, and ligated with Illumina indexed TruSeq adapters. Ligated cDNA fragments were PCR amplified using Illumina TruSeq primers, and purified PCR products were checked for quality and quantity on the Agilent Bioanalyzer DNA 7500 chip (Agilent Technologies). Ten nanomolar pools made up of three samples with unique indices were generated. Pools were sequenced using TruSeq Illumina GAIIx-indexed sequencing. Each pool of three was hybridized to a single flow-cell lane and was amplified, blocked, linearized, and primer hybridized using the Illumina cBot. Fifty base pairs of insert sequence and 6 bp of index sequence were generated on the Illumina GAIIx. Sequences were trimmed based on quality scores and deconvoluted based on the index identifier. The resulting sequences were bowtie aligned (Langmead et al., 2009) to the Arabidopsis gene set and normalized to relative parts per kilobase per 10 million (RPKtM; Mortazavi et al., 2008). The generated RPKtM data matrix was visualized and analyzed in GeneData Analyst software (Genedata).
RNA Analysis
For northern-blot analysis, total RNA was extracted from Arabidopsis leaf tissues. Ten micrograms of RNA was separated by electrophoresis on a 1% (w/v) agarose/formaldehyde/MOPS gel and blotted to a nylon membrane. Probe labeling, hybridization, and washing were carried out according to the manufacturer’s instructions. To determine transgene expression in Arabidopsis with RT-PCR, cDNA was synthesized with oligo(dT) primers using SuperScript II RNase H− reverse transcriptase (Invitrogen Life Technology). PCR was conducted using the Advantage-GC 2 PCR kit (Clontech). The primers used were PF1 (5′-GACACCCAGCAGCTGATCAACAG-3′) and PR2 (5′-ATGTAGGTCGGTCCGGTTCCACCG-3′).
Cell Fractionation and Immunoblotting
Microsomal membranes and soluble fraction were isolated from 3-week-old Arabidopsis plants according to Chen et al. (2002). Protein was separated by SDS-PAGE, blotted to a polyvinylidene difluoride membrane, and probed with monoclonal anti-FLAG (Sigma-Aldrich) antibodies. The primary antibodies were detected with the Pierce Fast Western Blot Kit, ECL Substrate (Thermo Fisher Scientific). Monoclonal anti-HA antibodies (Thermo Fisher Scientific) were also used to analyze the expression of FLAG-HA epitope-tagged ARGOS proteins.
Fluorescence Microscopy
Hypocotyls of Arabidopsis seedlings and onion inner epidermal peels were harvested and immediately placed in phosphate-buffered saline (pH 7.2) on glass slides for microscopic observations. Observations and images were taken with a Leica DMRXA epifluorescence microscope with a mercury light source. Two different fluorescent filter sets were used to monitor Ac-GFP1 fluorescence: Alexa 488 MF-105 (excitation, 486–500 nm; dichroic, 505LP; emission, 510–530) and Red-Shifted GFP 41001 (excitation, 460–500 nm; dichroic, 505LP; emission, 510–560). Cyan fluorescence was monitored using an Aqua 31036v2 filter set (excitation, 426–446 nm; dichroic, 455LP; emission, 465–495). All filter sets were from Chroma Technology. Images were captured with a Photometrics CoolSNAP HQ CCD. The camera and microscope were controlled, and the images manipulated, by Molecular Devices MetaMorph imaging software.
Quantitative Drought Assay for Arabidopsis Plants
YFP-positive transgenic seed and YFP-negative nontransgenic sibs from a segregating T2 population were sown in a single flat on Scotts Metro-Mix 360 soil supplemented with Peters fertilizer and Osmocote. Flats were configured with eight square pots each. Each of the square pots was filled with soil. Each pot (or cell) was sown to produce nine seedlings in a 3 × 3 array. Within a flat, four pots consisted of transgenic plants and four pots consisted of nontransgenic control plants. The soil was watered to saturation, and then plants were grown in conditions of 16 h of light/22°C and 8 h of dark/20°C at approximately 65% relative humidity. Plants were given a normal watering regime until day 14 after germination. At day 14, plants were given a final saturating watering. Digital images of the plants were taken once per day at the same time of day, starting at the onset of visible drought stress symptoms, approximately 14 d after the last watering, until the plants appeared desiccated. Typically, 4 consecutive days of data were captured.
Maize Hybrid Yield Testing
To evaluate the grain yield of the UBI::ZmARGOS8 transgenic events, field trials were conducted with a hybrid over a range of environments in 2 years. Using standard backcrossing techniques, the insertion was backcrossed from a donor inbred line into two inbred lines, one in each of two major complementary heterotic groups. Two hybrids originating from these converted inbred lines were evaluated over a similar range of environments. Hybrid seed for these trials was generated in a winter nursery and sent back to North America for the subsequent growing season. Hybrid seed segregated 1:1 for the transgene, and selectable markers linked to the transgene were used to identify and separate the transgene-positive F1 seed from the transgene-negative F1 seed (event nulls). A subsample of each of the event nulls was combined to create a bulk null control. All subsequent yield comparisons were made between the F1 transgene-positive hybrid and the bulk null. In some cases, other transgenic deregulated traits, including herbicide tolerance and/or insect protection, were already included in these hybrids. When this occurred, the same deregulated genes were present in both the experimental and bulk null entries of a hybrid. Multiple individual events were backcrossed to determine the effect of insertion site on efficacy. Experimental events and bulk null controls were grown in field environments at research centers in Woodland, California; Garden City, Kansas; Plainview, Texas; York, Nebraska; Fruitland, Iowa; Marion, Iowa; Johnston, Iowa; Windfall, Indiana; Princeton, Indiana; Sciota, Illinois; and San Jose, Illinois. Some environments were managed to impose various levels of drought stress, while others were managed for optimum yield/nonstress conditions.
Experimental designs were set up as a randomized complete block or split-plot arrangement with hybrids as main plots and transgene status (positive or wild type) as the split plot. Three to four replicates were established at each location, and the plant population density used was typical for growers in that particular region. Harvest weight and grain moisture from each plot were used to calculate yield per area at a constant moisture. Statistical analysis was conducted within an environment to eliminate plot-level outliers before analyzing across environments. Individual growing locations were classified as stressed or low stressed based on management practices and yield levels attained at those locations (Loffler et al., 2005). Statistical models accounting for environment effects were used to eliminate within-location spatial variation. Best linear unbiased predictions were generated at the individual event level as well as across events (construct level) for both the stress and low-stress environmental groupings (Gilmour et al., 2009; Habben et al., 2014). At one low-stress environment, 10 preselected and consecutive plants within each plot had ears removed before combine harvest in order to evaluate individual ear characteristics such as ear length, ear width, and kernels per ear via image analysis. The 10 ears were imaged and shelled for grain weight, and a moisture sample was taken. After combine harvest, the grain weight of the imaged ears was adjusted to the combine moisture for that plot and added to the grain weight of the remaining plot. Average ear length, ear width, and kernels per ear values were calculated from the 10 ears in each plot and submitted as one plot value per trait.
Sequence data for the genes described in this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ZmARGOS1 (JN252297), ZmARGOS8 (JN252302), AtOSR1 (At2g41230), and AtOSR2 (At2g41225).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Schematic diagram of the structure of ARGOS proteins and sequence alignment of maize and Arabidopsis homologs.
Supplemental Figure S2. Northern blot analysis of transgene expression in the transgenic Arabidopsis plants.
Supplemental Figure S3. Delayed senescence of perianth organs of the flower in the 35S:ZmARGOS1 transgenic Arabidopsis plants.
Supplemental Figure S4. Overexpression of ZmARGOS1 confers ethylene insensitivity in Arabidopsis.
Supplemental Figure S5. RT-PCR analysis of the 35S:ZmARGOS1 transgene expression in an eto1-1 or ctr1-1 mutant background of Arabidopsis.
Supplemental Figure S6. Overexpression of a mutated version of ZmARGOS1 in Arabidopsis plants.
Supplemental Figure S7. Reduced ethylene sensitivity in the 35S:ZmARGOS8 transgenic Arabidopsis plants.
Supplemental Figure S8. Overexpression of ZmARGOS1 in an etr1-7 mutant background reduces plant sensitivity to ethylene.
Supplemental Figure S9. Overexpression of ZmARGOS1 in Arabidopsis etr1-7 mutant.
Supplemental Figure S10. Overexpression of ZmARGOS1 in Arabidopsis etr1-7 rte1-2 mutant.
Supplemental Figure S11. Subcellular localization of ZmARGOS1 protein in transgenic Arabidopsis plants.
Note Added in Press
An article describing some aspects of the function of AtARGOS and AtARL in Arabidopsis was published recently (Rai MI, Wang X, Thibault DM, Kim HJ, Bombyk MM, Binder BM, Shakeel SN, Schaller GE [2015] BMC Plant Biol 15: 157).
Acknowledgments
We thank Caren Chang for kindly providing rte1-2 and etr1-7 Arabidopsis mutants; Kathleen Schellin, Hongyu Wang, Jim Saylor, and Jason Brothers for assistance with phenotyping and genotyping; Karen Kratky for measuring ethylene; Wally Marsh for onion transformation; Mary Beatty and Gina Zastrow-Hayes for RNA sequencing analysis; Brooke Peterson-Burch and Stanley Luck for bioinformatics and statistics support; Salim Hakimi, Mary Trimnell, Hua Mo, and Weiguo Cai for excellent contributions to the field studies; our colleagues at outlying breeding stations who conducted first-rate yield trials; and Tom Greene, Mike Lassner, and Mark Cooper for organizational leadership and helpful input.
Glossary
- ER
endoplasmic reticulum
- ACC
1-aminocyclopropane-1-carboxylic acid
- RT
reverse transcription
- BSV
banana streak virus promoter
- Col-0
Columbia-0
- cDNA
complementary DNA
- RPKtM
relative parts per kilobase per ten million
- GFP
green fluorescent protein
Footnotes
Articles can be viewed without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Barry CS, Giovannoni JJ (2006) Ripening in the tomato Green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signaling. Proc Natl Acad Sci USA 103: 7923–7928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrano J, Ronco MG, Montaldi ER (1999) Drought stress syndrome in wheat is provoked by ethylene evolution imbalance and reversed by rewatering, aminoethoxyvinylglycine, or sodium benzoate. J Plant Growth Regul 18: 59–64 [DOI] [PubMed] [Google Scholar]
- Binder BM, Rodríguez FI, Bleecker AB (2010) The copper transporter RAN1 is essential for biogenesis of ethylene receptors in Arabidopsis. J Biol Chem 285: 37263–37270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bot AJ, Nachtergaele FO, Young A (2000) Land Resource Potential and Constraints at Regional and Country Levels: World Soil Resources Reports 90, Land and Water Development Division. FAO, Rome [Google Scholar]
- Boutrot F, Segonzac C, Chang KN, Qiao H, Ecker JR, Zipfel C, Rathjen JP (2010) Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc Natl Acad Sci USA 107: 14502–14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner M, Singh KB (1997) Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc Natl Acad Sci USA 94: 5961–5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae HS, Faure F, Kieber JJ (2003) The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15: 545–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chen H, Xue L, Chintamanani S, Germain H, Lin H, Cui H, Cai R, Zuo J, Tang X, Li X, et al. (2009) ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21: 2527–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Randlett MD, Findell JL, Schaller GE (2002) Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J Biol Chem 277: 19861–19866 [DOI] [PubMed] [Google Scholar]
- Cigan AM, Unger-Wallace E, Haug-Collet K (2005) Transcriptional gene silencing as a tool for uncovering gene function in maize. Plant J 43: 929–940 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de la Luz Gutiérrez-Nava M, Aukerman MJ, Sakai H, Tingey SV, Williams RW (2008) Artificial trans-acting siRNAs confer consistent and effective gene silencing. Plant Physiol 147: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CH, Jang M, Scharein B, Malach A, Rivarola M, Liesch J, Groth G, Hwang I, Chang C (2010) Molecular association of the Arabidopsis ETR1 ethylene receptor and a regulator of ethylene signaling, RTE1. J Biol Chem 285: 40706–40713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CH, Rivarola M, Resnick JS, Maggin BD, Chang C (2008) Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. Plant J 53: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Qin Z, Yan J, Zhang X, Hu Y (2011) Arabidopsis ORGAN SIZE RELATED1 regulates organ growth and final organ size in orchestration with ARGOS and ARL. New Phytol 191: 635–646 [DOI] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box–mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour AR, Gogel BJ, Cullis BR, Thompson R (2009) ASReml User Guide, Release 30. VSN International, Hemel Hempstead, UK [Google Scholar]
- Guillaume RG, Sauter M (2008) Ethylene biosynthesis and signaling in rice. Plant Sci 175: 32–42 [Google Scholar]
- Guo M, Rupe MA, Wei J, Winkler C, Goncalves-Butruille M, Weers BP, Cerwick SF, Dieter JA, Duncan KE, Howard RJ, et al. (2014) Maize ARGOS1 (ZAR1) transgenic alleles increase hybrid maize yield. J Exp Bot 65: 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habben JE, Bao X, Bate NJ, DeBruin JL, Dolan D, Hasegawa D, Helentjaris TG, Lafitte RH, Lovan N, Mo H, et al. (2014) Transgenic alteration of ethylene biosynthesis increases grain yield in maize under field drought-stress conditions. Plant Biotechnol J 12: 685–693 [DOI] [PubMed] [Google Scholar]
- Hays DB, Do JH, Mason RE, Morgan G, Finlayson SA (2007) Heat stress induced ethylene production in developing wheat grains induces kernel abortion and increased maturation in a susceptible cultivar. Plant Sci 172: 1113–1123 [Google Scholar]
- Hirayama T, Kieber JJ, Hirayama N, Kogan M, Guzman P, Nourizadeh S, Alonso JM, Dailey WP, Dancis A, Ecker JR (1999) RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis. Cell 97: 383–393 [DOI] [PubMed] [Google Scholar]
- Hu Y, Poh HM, Chua NH (2006) The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J 47: 1–9 [DOI] [PubMed] [Google Scholar]
- Hu Y, Xie Q, Chua NH (2003) The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15: 1951–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271 [DOI] [PubMed] [Google Scholar]
- Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J 33: 221–233 [DOI] [PubMed] [Google Scholar]
- Huberman M, Riov J, Goldschmidt EE, Apelbaum A, Goren R (2014) The novel ethylene antagonist, 3-cyclopropyl-1-enyl-propanoic acid sodium salt (CPAS), increases grain yield in wheat by delaying leaf senescence. Plant Growth Regul 73: 249–255 [Google Scholar]
- Ju C, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML, et al. (2012) CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci USA 109: 19486–19491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami EM, Oosterhuis DM, Snider J (2013) High temperature and the ethylene antagonist 1-methylcyclopropene alter ethylene evolution patterns, antioxidant responses, and boll growth in Gossypium hirsutum. Am J Plant Sci 4: 1400–1408 [Google Scholar]
- Kawakami EM, Oosterhuis DM, Snider JL (2010) Physiological effects of 1-methylcyclopropene on well-watered and water-stressed cotton plants. J Plant Growth Regul 29: 280–288 [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S (2008) Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J 55: 821–831 [DOI] [PubMed] [Google Scholar]
- Kuluev BR, Knyazev AV, Iljassowa AA, Chemeris AV (2011) Constitutive expression of the ARGOS gene driven by dahlia mosaic virus promoter in tobacco plants. Russ J Plant Physiol 58: 507–515 [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffler CM, Wei J, Fast T, Gogerty J, Langton S, Bergman M, Merrill B, Cooper M (2005) Classification of maize environments using crop simulation and geographic information systems. Crop Sci 45: 1708–1716 [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Ogawara T, Higashi K, Kamada H, Ezura H (2003) Ethylene advances the transition from vegetative growth to flowering in Arabidopsis thaliana. J Plant Physiol 160: 1335–1340 [DOI] [PubMed] [Google Scholar]
- Patterson SE, Bleecker AB (2004) Ethylene-dependent and -independent processes associated with floral organ abscission in Arabidopsis. Plant Physiol 134: 194–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Shen Z, Huang SS, Schmitz RJ, Urich MA, Briggs SP, Ecker JR (2012) Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338: 390–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Zhang X, Zhang X, Feng G, Hu Y (2014) The Arabidopsis ORGAN SIZE RELATED 2 is involved in regulation of cell expansion during organ growth. BMC Plant Biol 14: 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick JS, Rivarola M, Chang C (2008) Involvement of RTE1 in conformational changes promoting ETR1 ethylene receptor signaling in Arabidopsis. Plant J 56: 423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick JS, Wen CK, Shockey JA, Chang C (2006) REVERSION-TO-ETHYLENE SENSITIVITY1, a conserved gene that regulates ethylene receptor function in Arabidopsis. Proc Natl Acad Sci USA 103: 7917–7922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růzicka K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E (2007) Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Remans T, Sági L, Elliott AR, Dietzgen RG, Swennen R, Ebert PR, Grof CP, Manners JM (2001) Promoters for pregenomic RNA of banana streak badnavirus are active for transgene expression in monocot and dicot plants. Plant Mol Biol 47: 399–412 [DOI] [PubMed] [Google Scholar]
- Scott A, Wyatt S, Tsou PL, Robertson D, Allen NS (1999) Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques 26: 1125–1132 [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sano T, Tamaoki M, Nakajima N, Kondo N, Hasezawa S (2005) Ethylene inhibits abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol 138: 2337–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholen D, Pons TL, Voesenek LACJ, Poorter H (2008) The role of ethylene perception in the control of photosynthesis. Plant Signal Behav 3: 108–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger E, Betz S, Xu R, Cigan AM (2001) Selection and orientation of adjacent genes influences DAM-mediated male sterility in transformed maize. Transgenic Res 10: 409–422 [DOI] [PubMed] [Google Scholar]
- Viklund H, Elofsson A (2004) Best α-helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Sci 13: 1908–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Sang Y, Song J, Gao XQ, Zhang X (2009) Expression of a rice OsARGOS gene in Arabidopsis promotes cell division and expansion and increases organ size. J Genet Genomics 36: 31–40 [DOI] [PubMed] [Google Scholar]
- Zhong S, Zhao M, Shi T, Shi H, An F, Zhao Q, Guo H (2009) EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci USA 106: 21431–21436 [DOI] [PMC free article] [PubMed] [Google Scholar]