Activated phytochrome is sufficient to sustain robust circadian rhythms in the dark even in the absence of exogenous sugars, revealing the importance of light signaling pathways in clock function.
Abstract
The sensitivity of the circadian system to light allows entrainment of the clock, permitting coordination of plant metabolic function and flowering time across seasons. Light affects the circadian system via both photoreceptors, such as phytochromes and cryptochromes, and sugar production by photosynthesis. In the present study, we introduce a constitutively active version of phytochrome B-Y276H (YHB) into both wild-type and phytochrome null backgrounds of Arabidopsis (Arabidopsis thaliana) to distinguish the effects of photoreceptor signaling on clock function from those of photosynthesis. We find that the YHB mutation is sufficient to phenocopy red light input into the circadian mechanism and to sustain robust rhythms in steady-state mRNA levels even in plants grown without light or exogenous sugars. The pace of the clock is insensitive to light intensity in YHB plants, indicating that light input to the clock is constitutively activated by this allele. Mutation of YHB so that it is retained in the cytoplasm abrogates its effects on clock function, indicating that nuclear localization of phytochrome is necessary for its clock regulatory activity. We also demonstrate a role for phytochrome C as part of the red light sensing network that modulates phytochrome B signaling input into the circadian system. Our findings indicate that phytochrome signaling in the nucleus plays a critical role in sustaining robust clock function under red light, even in the absence of photosynthesis or exogenous sources of energy.
The circadian system has evolved as an endogenous time-keeping mechanism that confines many biochemical and physiological processes to specific parts of the day and allows plants to accurately measure seasonal transitions (for review, see Song et al., 2013; Hsu and Harmer, 2014). To remain synchronized with the regular diurnal cycle, the plant circadian system is exquisitely sensitive to environmental changes in light and temperature (Fankhauser and Staiger, 2002; Jones, 2009). Although specific temperature sensors have remained elusive, light input to the circadian system occurs primarily via phytochromes, cryptochromes, and the ZEITLUPE family of photosensory F-box proteins (Somers et al., 1998, 2000; Devlin and Kay, 2000; Pudasaini and Zoltowski, 2013).
Phytochromes (phys) are the primary red and far-red light photoreceptors in plants (Bae and Choi, 2008) and consist of a five-member protein family in Arabidopsis (Arabidopsis thaliana; Clack et al., 1994; Franklin and Quail, 2010). Phys reversibly switch between Pr and Pfr forms upon absorption of red or far-red light, respectively (Rockwell et al., 2006), and the ratio of these forms within the cell controls the shade avoidance response and contributes to light perception (Casal, 2013). PhyA is the most divergent phy, with a specialized role as a far-red light sensor (Casal et al., 2014), whereas phyB is the predominant red light sensor in Arabidopsis (Whitelam and Devlin, 1997). Although oscillation of circadian transcripts is dampened under constant far-red light, phyA retains photoregulatory control of these and other genes under these conditions (Wenden et al., 2011). By contrast, phyA, phyB, and phyD each appear to contribute to maintenance of circadian rhythms under constant red light (Rc; Somers et al., 1998; Devlin and Kay, 2000). PhyD single mutants have a wild-type circadian phenotype but have an additive effect when introgressed into a phyB background (Devlin and Kay, 2000). Although recent studies on temperate grasses have established a direct role for phyC in photoperiod sensing (Chen et al., 2014; Woods et al., 2014), phyC has not been formally described as part of the circadian system in Arabidopsis. However, phyC is presumed to act similarly to phyE as a modulator of phyB activity since neither phyC nor phyE is capable of forming the homodimers necessary for signaling activity in Arabidopsis (Clack et al., 2009) or in rice (Oryza sativa; Xie et al., 2014), and instead, both function as heterodimers with other phys.
The circadian system is typically conceptualized as a core molecular oscillator reset by light and temperature stimuli that regulates the expression of multiple output pathways (Harmer, 2009). Outputs are easily defined as processes under circadian control that do not feed back into the circadian system; however, the distinction between input and core components of the circadian system has become increasingly blurred with our expanding knowledge of the circadian system. For example, phys transduce light signals from the red and far-red portion of the spectrum into the circadian system, but the transcription of this photoreceptor family is concurrently regulated by the clock (Somers et al., 1998; Bognár et al., 1999; Devlin and Kay, 2000; Tóth et al., 2001; Wenden et al., 2011). Despite this complication, the core oscillator is generally considered to consist of a complex web of interacting feedback loops that are sufficient to generate a self-sustaining oscillation of transcripts and proteins (Fogelmark and Troein, 2014; Hsu and Harmer, 2014). Although the relative effect of light on each core circadian component has not been systematically determined, several reports have identified transcripts that are acutely induced by light (Wang and Tobin, 1998; Ito et al., 2003; Locke et al., 2005). CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) are morning-phased, light-inducible transcription factors that induce expression of PSEUDO-RESPONSE REGULATOR9 (PRR9; Schaffer et al., 1998; Wang and Tobin, 1998; Farré et al., 2005). PRR9, whose transcription is itself light induced (Ito et al., 2003; Jones et al., 2012), represses expression of CCA1 and LHY in partnership with the later-phased PRR7, PRR5, and TIMING OF CAB1 (TOC1; Matsushika et al., 2000; Nakamichi et al., 2010). Later in the subjective day, TOC1 and REVEILLE8 act as negative and positive transcription factors, respectively, that act to reinforce the robust oscillations of circadian genes (Farinas and Más, 2011; Gendron et al., 2012; Huang et al., 2012; Hsu et al., 2013). In addition to acute photoreceptor-mediated effects on the clock, it is well established that increasing fluence rates of light under constant conditions quickens circadian pace in plants, as well as many other species (Aschoff, 1960). For plants, the shorter period length seen at higher fluence rates of light may be due in part to increased photosynthesis, a process that is strongly regulated by phys and other photoreceptors (Haydon et al., 2013). However, Arabidopsis seedlings lacking all five phytochromes exhibit a shorter circadian period than the wild type under dim light, despite being deficient in photosynthetic light capture (Strasser et al., 2010; Hu et al., 2013).
The ability of phyB-E to form homo- and heterodimers introduces complexity into the interpretation of individual phy mutant phenotypes (Sharrock and Clack, 2004; Clack et al., 2009; Hu et al., 2013). In addition, period length in phyABCDE mutants is modestly sensitive to different fluence rates of red light (Hu et al., 2013), suggesting that metabolic processes may complicate the assessment of the role of Phys in light input to the circadian clock. To better understand phy-dependent and phy-independent light signaling inputs to the circadian system, we now exploit the phytochrome B-Y276H (YHB) mutant allele that induces constitutive phyB signaling in the absence of light (Su and Lagarias, 2007; Hu et al., 2009). By introducing the YHB allele into the phyABCDE background, we resolve the direct roles of red light from those of phyB activation on clock output/function. We demonstrate that nuclear-localized but not cytosolic YHB is sufficient to maintain circadian rhythmicity in constant darkness in the absence of endogenous photoreceptor activation or photosynthesis. Our studies also identify a regulatory role for phyC within the circadian system to enhance phyB signaling input under dim red light.
RESULTS
YHB Sustains Core Clock Transcript Cycling in Light-Grown Plants Transferred to Constant Darkness
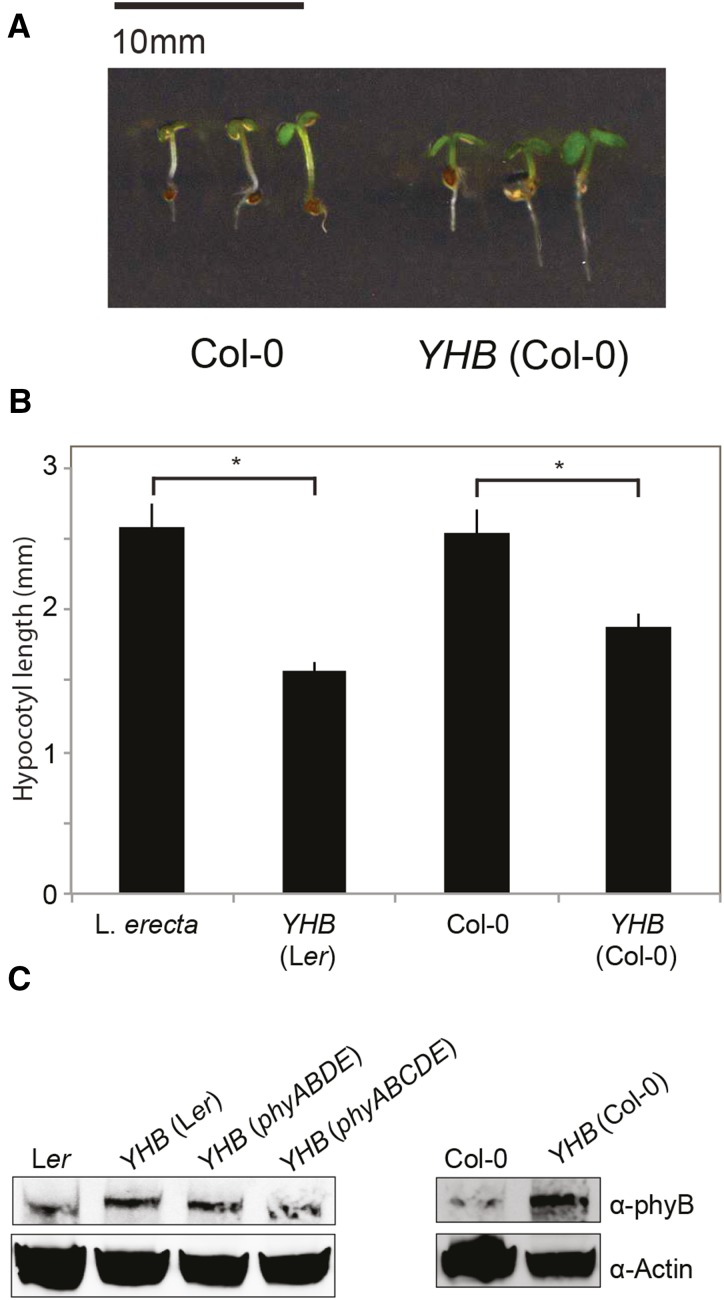
Previous work has demonstrated that YHB is sufficient to induce photomorphogenesis and to initiate transcriptional cascades that mimic red light-induced phy signaling in the absence of light (Su and Lagarias, 2007; Hu et al., 2009). The circadian system is exquisitely sensitive to changes in day length and light intensity (Salomé et al., 2008; Jones, 2009), so we were curious whether the YHB allele would mimic phyB-mediated light input into the circadian system. We therefore crossed the transgenic genomic YHB allele into Arabidopsis plants carrying a clock-regulated bioluminescent reporter. Previous screens have used a COLD, CIRCADIAN RHYTHM AND RNA BINDING2 (CCR2)::LUCIFERASE (LUC) reporter (Columbia accession [Col]) to measure circadian rhythms in the dark (Martin-Tryon et al., 2007), and so we generated Col plants containing both the YHB allele and the CCR2::LUC reporter in addition to introducing a CCA1::LUC2 reporter into existing YHB (Landsberg erecta [Ler]) lines (Hu et al., 2009). Introduction of the YHB allele confers shortened hypocotyls and expanded cotyledons in Ler seedlings grown in darkness or in constant light (Su and Lagarias, 2007; Hu et al., 2009). Similar short hypocotyls were observed in our newly generated YHB (Col) lines when compared with wild-type Col controls grown under white light (P < 0.05; Fig. 1, A and B). YHB was moderately overexpressed compared with endogenous phyB in these lines (Fig. 1C).
Figure 1.
Characterization of an additional YHB allele. A, Morphology of seedlings grown under 30 μmol m−2 s−1 white light in 12:12 L/D cycles. Wild-type (Col-0) and Col-0 seedlings transformed with a PHYB::PHYB-Y276H construct (YHB) were grown on Murashige and Skoog (MS) medium for 6 d. B, Quantification of hypocotyl lengths of Col-0, YHB (Col-0), Ler, and YHB (Ler) seedlings grown as described in (A). Previously described Ler seedlings similarly transformed with YHB are presented for comparison (Su and Lagarias, 2007). sem is presented, n > 20. *, Significant difference for the indicated comparison (P < 0.001, Student’s t test). C, Immunoblot analysis of phyB/YHB protein levels showing relative phyB accumulation in Col-0 and YHB (Col-0) seedlings. Ler wild type and various transgenic plants harboring the Ler YHB transgenic allele derived from YHB/phyA201phyB-5 line #5 (Su and Lagarias, 2007) are presented for comparison.
We initially tested whether constitutively active YHB protein would be sufficient to maintain robust luciferase rhythms in light/dark (L/D)-entrained plants transferred to constant darkness (Fig. 2, A–D). Assessments of circadian rhythms have historically used Suc as a media supplement to enhance bioluminescence in transgenic plants (Millar et al., 1992), although recent work has demonstrated that this exogenous Suc can itself act as an entrainment signal (Dalchau et al., 2011; Haydon et al., 2013). We consequently compared luciferase activity in our YHB lines in either the presence or absence of exogenous Suc to facilitate comparison with historical and more recent data sets. We observed that rhythms of CCR2-driven luciferase activity in YHB seedlings grown with Suc had increased amplitude throughout the experiment compared with wild-type controls (P < 0.001; Fig. 2A), although there was no significant difference in period length between the two populations (τ = 25.4 ± 0.21 and 24.93 ± 0.08 for wild-type and YHB, respectively; P = 0.1). CCA1-driven luciferase activity was similarly increased in YHB (Ler) lines in the presence of Suc (Fig. 2B). In the absence of Suc, we observed that rhythmic bioluminescence in both Col wild-type and YHB seedlings containing a CCR2:LUC reporter dampened considerably and became arrhythmic following 1 d of constant darkness (Fig. 2C). By contrast, robust circadian rhythms were retained in YHB CCA1::LUC2 lines grown in the absence of Suc (τ = 26.0 ± 0.23; Fig. 2D). Since rhythmic luminescence activity was not observed from the CCA1::LUC2 reporter control in control plants in the absence of Suc (Fig. 2D), YHB helps maintain robust CCA1::LUC2 cycling in darkness. Taken together, these studies show that YHB enhances cycling amplitudes of both clock output (CCR2) and clock gene (CCA1) reporters and sustains rhythmic CCA1-regulated luciferase activity in the absence of light and Suc.
Figure 2.
Effect of YHB on bioluminescence rhythms in constant darkness. A and C, Bioluminescence of Col-0 (solid) and YHB (dashed) seedlings containing a CCR2::LUC reporter grown on MS medium with (A) or without (C) exogenous Suc; n > 9. B and D, Bioluminescence of Ler (solid) and YHB (Ler; dashed) seedlings containing a CCA1::LUC2 reporter grown on MS medium with (B) or without (D) exogenous Suc; n > 10. YHB (Ler) is presented on a secondary axis in D for clarity. E and F, Abundance of CCR2 transcripts as measured by quantitative reverse transcription (qRT)-PCR after transfer of seedlings to constant darkness. Plants were grown with (F) or without (E) exogenous Suc as described in A and C. G and H, Abundance of CCA1 transcripts as measured by qRT-PCR after transfer of seedlings to constant darkness. Plants were grown with (H) or without (G) exogenous Suc as described in A and C. Plants were entrained under 60 μmol m−2 s−1 white light in 12:12 L/D cycles for 6 d and then transferred to constant darkness at ZT12 on day 6. Gray bars indicate subjective day, whereas black bars indicate subjective night. CCR2 and CCA1 mRNA levels were normalized to PP2a. sem is shown.
To better understand the role of exogenous Suc in the maintenance of clock-regulated gene expression in darkness, we examined the steady-state levels of CCR2 and CCA1 transcripts in both YHB (Col) and wild-type control lines (Fig. 2, E–H). In the absence of Suc, CCR2 transcript accumulation in the wild type mirrored the luciferase activity data, with one rhythmic peak of transcript accumulation before dampening toward arrhythmia (Fig. 2E). Rhythms were more robust in YHB plants grown without Suc, exhibiting two obvious peaks of transcript abundance. Similarly, the abundance of CCA1 transcripts dampened very quickly in wild-type plants in constant darkness, regardless of the presence of exogenous Suc, whereas rhythmicity was retained in YHB-expressing lines (Fig. 2, G and H).
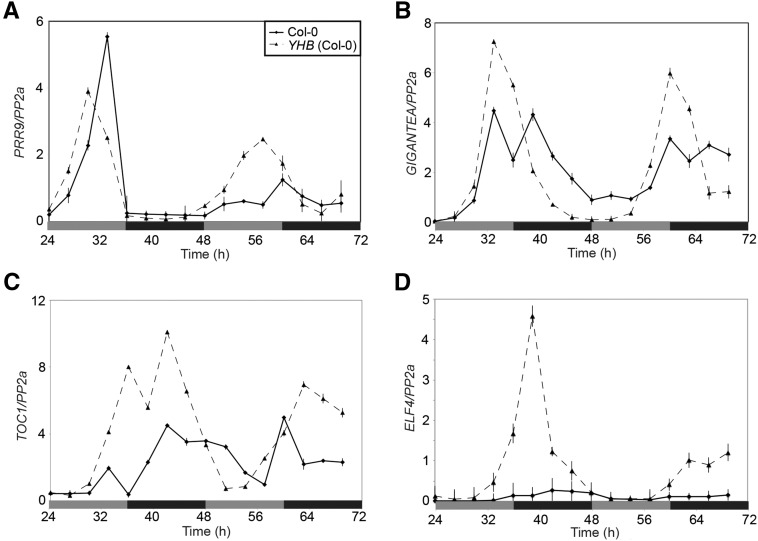
We next performed qRT-PCR analysis of additional core clock transcripts in dark-adapting Col wild-type and YHB plants. Following L/D entrainment and transfer to constant darkness at Zeitgeber Time (ZT) 12, we assessed transcript levels of several core circadian clock genes (Hsu and Harmer, 2014). The presence of YHB was sufficient to maintain rhythms in transcript levels of the morning-phased genes CCA1 and PRR9 as well as the evening-phased genes GIGANTEA (GI), TOC1, and EARLY FLOWERING4 (ELF4; Figs. 2G and 3). In all cases, transcript accumulation rhythms dampened more significantly in wild-type seedlings than in YHB seedlings. These results indicate that, independent of the presence of Suc, YHB acts to sustain rhythmic expression of core clock transcripts in constant darkness, a phenomenon not seen in wild-type seedlings.
Figure 3.
YHB sustains core clock transcript cycling in L/D entrained plants transferred to constant darkness. Abundance of circadian transcripts as measured by qRT-PCR after transfer of seedlings (grown on Suc-free medium) to constant darkness. Levels of PRR9 (A), GI (B), TOC1 (C), and ELF4 (D) mRNA were assessed. Plants were grown on 0.5× MS medium and entrained to 12:12 L/D cycles with 60 μ mol m−2 s−1 white light for 10 d before transfer to constant darkness at ZT12. mRNA levels for each gene were normalized to PP2a; sem is shown. Gray bars indicate subjective day, whereas black bars indicate subjective night.
Circadian Period of YHB Plants Is Insensitive to Fluence Rate
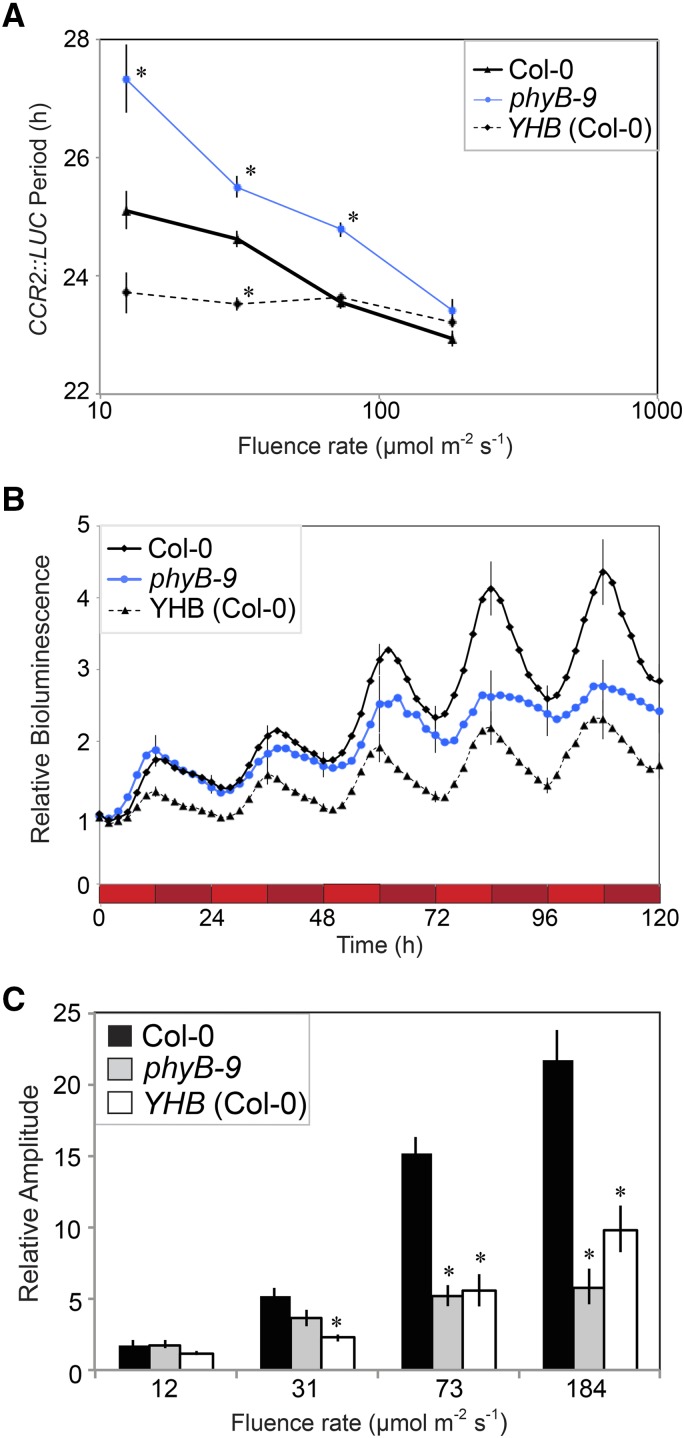
We next examined YHB influence on circadian rhythms under a range of intensities of Rc. Following an entrainment period, wild-type, YHB, and phyB-9 seedlings grown on Suc-free medium were released into Rc, with circadian period and amplitude measured via activity of the CCR2::LUC reporter (Fig. 4). Similar to previous reports (Somers et al., 1998; Palágyi et al., 2010), the circadian period of wild-type plants shortened from 25.1 ± 0.33 h to 22.9 ± 0.13 h as the fluence rate increased from 12 to 184 μmol m−2 s−1 (Fig. 4A). PhyB-9 seedlings exhibited a similar, albeit more exaggerated response over the fluence rates tested, as was reported previously (Fig. 4A; Somers et al., 1998; Palágyi et al., 2010). By contrast, YHB seedlings were essentially unresponsive to increasing fluence rates of Rc (up to 184 μmol m−2 s−1), with period length remaining approximately 23.5 h at all fluence rates tested (Fig. 4A). This unresponsiveness resulted in the greatest period difference between YHB and the wild type at the lowest fluence rate of Rc (12 μmol m−2 s−1) tested, where a approximately 1.5-h shorter period was observed in the transgenic plant (Fig. 4A). By contrast, under high-intensity Rc, the period lengths of all genotypes were nearly identical. Taken together, these results indicate that period length in YHB plants is nearly insensitive to increasing fluence rates of red light.
Figure 4.
YHB suppresses the clock's response to increasing fluence rates of red light. A, Fluence rate response curve to measure free-running circadian period under red light. phyB-9 (blue) and YHB (dashed) alleles were crossed into a Col-0 background (solid line) carrying a CCR2::LUC luciferase reporter. Homozygous lines were grown under 60 μmol m−2 s−1 white light in 12:12 L/D cycles on 0.5× MS medium for 6 d before transfer to Rc at the indicated fluence rate on day 6. B, Example of circadian rhythms observed in A. Seedlings were entrained as described in A before being transferred to 184 μ mol m−2 s−1 Rc. Shaded red bars indicate subjective night. C, Amplitude of luciferase rhythms reported in A. sem is shown. *, Significant difference compared with the wild type, Bonferroni adjusted Student’s t test.
Differences in the amplitude of bioluminescence rhythms were also detected for YHB and wild-type plants after transfer to high fluence rates of Rc. Although similar for all genotypes under 12 μmol m−2 s−1 Rc, the amplitude of bioluminescence was greatly enhanced in the wild type with increasing fluence rate of Rc, whereas the responsiveness of the YHB plants was reduced. Indeed, the discrepancy between the amplitude of these genotypes was most pronounced at the highest fluence rates examined, where YHB seedlings were half as bright as wild-type controls (Fig. 4, B and C). Similar to the period phenotype, these results indicate that clock amplitude in YHB plants is less responsive to increasing fluence rates of Rc than in the wild type.
YHB Influences Clock Gene Expression in the Absence of Other Phys
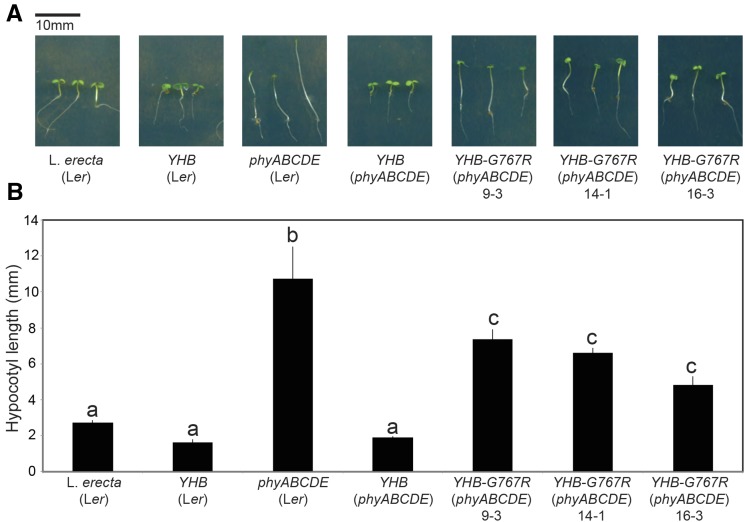
Type II phys (phyB-E) form homo- and heterodimers that complicate interpretation of phenotypes of loss-of-function phy mutants and of gain-of-function YHB transgenics (Sharrock and Clack, 2004; Clack et al., 2009; Hu et al., 2009). The additive circadian defect of phyABD mutants compared with phyAB has been assumed to indicate an ability of phyD to provide light input into the circadian system in the absence of phyB (Devlin and Kay, 2000). However, the loss of phyD potentially also alters the amount of phyC-phyE heterodimers in the two genotypes, providing an alternative explanation for their distinct phenotypes. Similarly, introduction of the YHB allele would alter the amounts of the homo- and heterodimeric species of endogenous phyB-E proteins. To better understand how YHB and by extension phyB influence the circadian system, we introduced the YHB allele into the recently isolated phyABCDE quintuple mutant (Hu et al., 2013). In contrast to the photomorphogenesis-challenged phenotypes of the phyABCDE parental line under 12:12 L/D cycles, YHB(phyABCDE) plants looked similar to wild-type seedlings with short hypocotyls and expanded cotyledons (Fig. 5).
Figure 5.
Morphology of seedlings transformed with YHB and YHB-G767R alleles. A, Seedlings were grown on 0.5× MS medium without Suc for 6 d under 30 μmol m−2 s−1 white light in 12:12 L/D cycles. B, Quantification of hypocotyl lengths of seedlings shown in A. sem is presented, n > 20. Letters indicate significantly different populations (P < 0.001, Tukey’s honestly significant difference test).
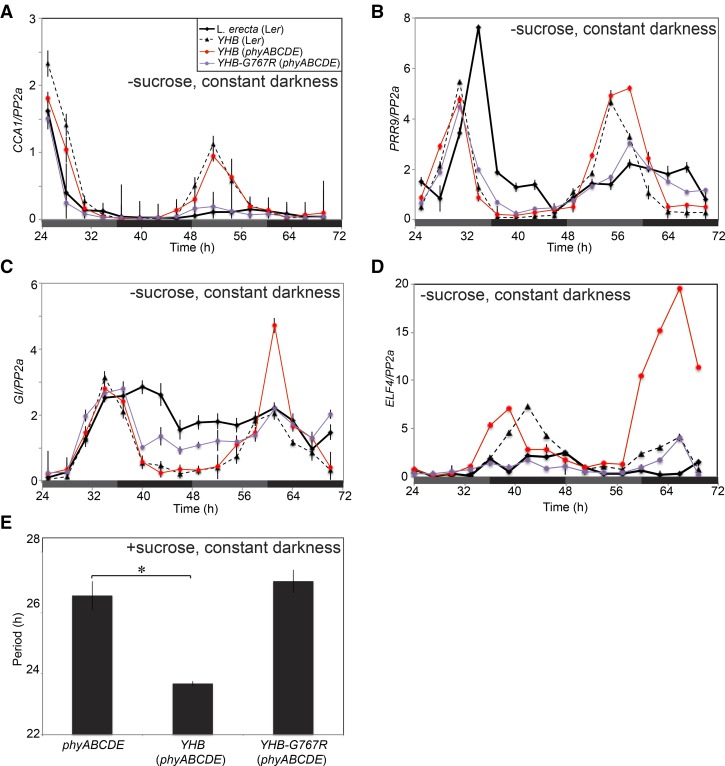
To further explore the effect of YHB in the absence of other phys, we assessed the accumulation of core clock gene transcripts in the phyABCDE mutant background using qRT-PCR. The phyABCDE mutant was generated in the Ler background; hence, we used a previously reported YHB line [YHB(Ler); Su and Lagarias, 2007; Hu et al., 2009] as a control. As observed for YHB(Ler) plants, YHB(phyABCDE) seedlings transferred to constant darkness displayed robustly rhythmic CCA1 accumulation, a response that is strongly damped in the Ler wild type (Fig. 6A). YHB(phyABCDE) also sustained rhythmic expression of PRR9, GI, and ELF4 transcripts in prolonged darkness (Fig. 6, B–D). All of these clock genes displayed dampened oscillations in the dark-adapting Ler wild type, similar to our results in the Col accession (Figs. 2 and 3). These data indicate that endogenous phys are not required for YHB-mediated maintenance of robust circadian rhythms in darkness.
Figure 6.
YHB in the absence of other phys influences clock gene expression. A to D, Abundance of circadian transcripts under constant darkness in YHB seedlings in the presence or absence of native phys using qRT-PCR. Levels of CCA1 (A), PRR9 (B), GI (C), and ELF4 (D) mRNA were assessed. Plants were entrained for 10 d in 12:12 L/D cycles on Suc-free MS media with 60 μmol m−2 s−1 white light before transfer to constant darkness. mRNA levels for each gene were normalized to PP2a; sem is shown. E, Circadian periodicity of phyABCDE, YHB(phyABCDE), and YHB-G767R(phyABCDE) seedlings expressing a CCA1::LUC2 reporter when grown on MS + Suc plates. Plants were entrained for 6 d under 60 μmol m−2 s−1 white light in 12:12 L/D cycles before being transferred to constant darkness at ZT12. Bioluminescence from groups of five seedlings was pooled for each data point, n > 8. sem is shown. *, Significant difference (P = 0.0016, Student’s t test).
Cytosolic YHB Has Little Effect on Clock Gene Expression or Circadian Pace
In contrast to endogenous phy, YHB does not require light activation to migrate from the cytoplasm to the nucleus and is instead constitutively targeted to the nucleus (Su and Lagarias, 2007). In the nucleus, YHB acts similarly to the Pfr form of phyB by binding PHYTOCHROME INTERACTING FACTOR (PIF) basic Helix-Loop-Helix (bHLH) transcription factors and targeting them for degradation (for review, see Bae and Choi, 2008). More recently, signaling roles for Pfr in the cytoplasm have been reported (Paik et al., 2012; Hughes, 2013). To evaluate the contribution of cytoplasmic Pfr signaling into the circadian system, we introduced the G767R mutation into the YHB allele (YHB-G767R). Phys containing the G767R mutation are retained in the cytoplasm (Wagner and Quail, 1995; Ni et al., 1999; Matsushita et al., 2003). This has been attributed to the inability of the G767R mutant to interact with PIF3 and then be imported into the nucleus (Pfeiffer et al., 2012). Surprisingly, the double phy mutant partially complemented the phyABCDE null mutant: light-grown YHB-G767R(phyABCDE) lines exhibited expanded cotyledons and shorter hypocotyls compared with the parental phyABCDE seedlings (Fig. 5). However, these results contrast with the strong hyperactivity of YHB in both null and wild-type backgrounds (Fig. 5). Thus, it is clear that the G757R mutation largely suppresses the gain-of-function activity of YHB, presumably by retaining it in the cytosol.
To assess the role of cytoplasmic YHB-G767R within the circadian system, we used qRT-PCR to assess its effects on transcript levels of genes with poor cycling in wild-type plants maintained in constant darkness in the absence of exogenous Suc (Fig. 6, A–D). Whereas YHB-expressing plants demonstrated robust rhythms in steady-state mRNA levels, dark-adapting YHB-G767R(phyABCDE) lines grown in the absence of Suc showed rapidly damping rhythms very similar to those observed in dark-adapting wild-type Ler.
We next examined the effects of activated phy containing the G767R mutation on circadian pace. The YHB or YHB-G767R transgenes were crossed into phyABCDE mutants expressing the CCA1::LUC2 reporter, and rhythms in bioluminescence activity were assessed in dark-adapting plants grown in the presence of exogenous Suc. Whereas YHB(phyABCDE) plants had a shorter period of 23.68 ± 0.08 h compared with the parental phyABCDE (26.57 ± 0.48 h), the period length of YHB-G767R(phyABCDE) plants was indistinguishable from the control (27.04 ± 0.37 h). These data indicate that both the shortening of circadian period by YHB and its enhancement of rhythms in transcript abundance are dependent upon its nuclear localization.
PhyC Modulates Light Input to the Circadian System
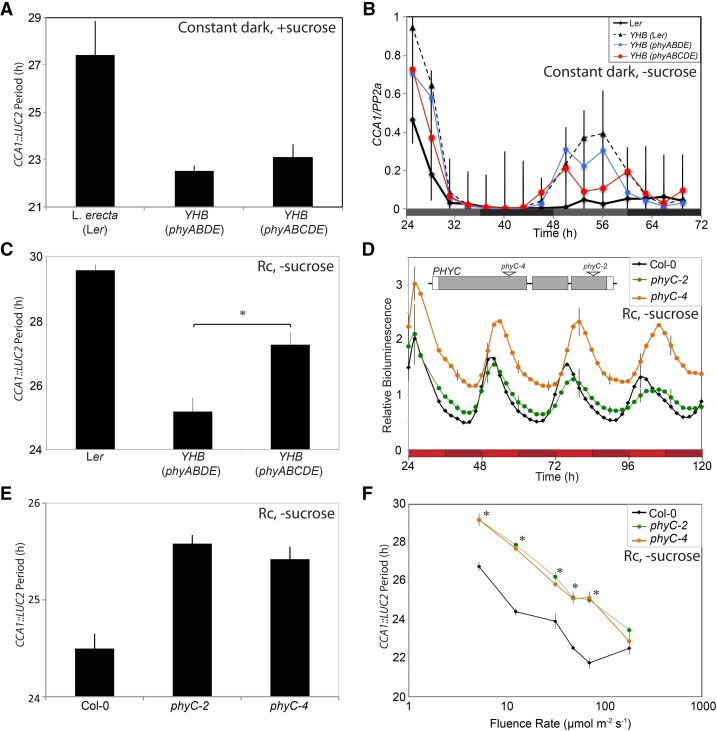
PhyC protein does not accumulate in phyABDE seedlings (which therefore phenocopy phyABCDE plants; Hu et al., 2013), and thus Arabidopsis phyC function depends upon other phys. Since phyC forms heterodimers with phyB or phyD (Clack et al., 2009), we were curious whether the presence of phyC was able to alter the activity of YHB. We therefore introduced the YHB allele into a phyABDE background to compare the clock phenotype of these YHB(phyABDE) seedlings with that of YHB(phyABCDE) plants (Fig. 7, A–C). In these two dark-adapting lines grown in the presence of Suc, the rhythms of CCA1::LUC2 expression were indistinguishable (Fig. 7A), with periods of 22.51 ± 0.24 and 23.13 ± 0.54 h in YHB(phyABDE) and YHB(phyABCDE), respectively. Similarly, in the absence of supplemental Suc, no significant difference in the accumulation of the clock gene transcripts CCA1 (Fig. 7B), PRR9, GI, or TOC1 (Supplemental Fig. S1) was detected between these two lines. Nevertheless, YHB(phyABDE) seedlings did exhibit a significantly shorter period than YHB(phyABCDE) under dim red light in the absence of Suc (τ = 25.18 ± 0.42 and 27.26 ± 0.38, respectively; P = 0.02; Fig. 7C), suggesting that phyC can enhance YHB signaling into the circadian system.
Figure 7.
PhyC modulates light input into the circadian system. A, Circadian periodicity of Ler), YHB(phyABDE), and YHB(phyABCDE) seedlings transformed with CCA1::LUC2 after transfer to constant darkness. Plants were grown under 60 μmol m−2 s−1 white light in 12:12 L/D cycles for 6 d with supplemental Suc before being transferred to constant darkness at ZT12. Bioluminescence from groups of five seedlings was pooled for each data point, n > 9. B, Abundance of CCA1 transcripts under constant darkness in YHB(ABDE) and YHB(ABCDE) seedlings using qRT-PCR. Plants were entrained to 12:12 L/D cycles on Suc-free MS medium under 60 μmol m−2 s−1 white light for 10 d before transfer to constant darkness at ZT12. mRNA levels for each gene were normalized to PP2a; sem is shown. C, Circadian periodicity of Ler, YHB(phyABDE), and YHB(phyABCDE) seedlings transferred to dim red light (1 μmol m−2 s−1). Seedlings were grown on 0.5× Suc-free MS medium and entrained for 6 d in 12:12 L/D cycles under 60 μmol m−2 s−1 white light before being transferred to Rc. Bioluminescence from groups of five seedlings was pooled for each data point, n > 7. D, Period estimates of Col-0, phyC-2, and phyC-4 seedlings under Rc. Plants were entrained in 12:12 L/D cycles for 6 d before transfer to 20 μmol m−2 s−1 Rc. The insert shows a schematic illustration of the PHYC locus indicating transfer DNA (T-DNA) insertion locations for phyC-2 and phyC-4. 5′ and 3′ untranslated regions are shown in white boxes, and exons are shown in gray. T-DNA insertion points are indicated with white triangles. E, Period estimates of seedlings transformed with a CCA1::LUC2 reporter. Wild-type (Col-0), phyC-2, and phyC-4 seedlings were entrained as described in (D) before being transferred to 20 μ mol m−2 s−1 Rc. F, Fluence rate response curve to evaluate the effect of phyC on the free-running period of the circadian system. Wild-type (Col-0, black line), phyC-2 (green), and phyC-4 (orange) were entrained as described in D before being transferred to Rc at the indicated fluence rate. sem is shown. *, Significant difference from the wild type (Bonferroni-adjusted Student’s t test).
PhyC mutants have hypocotyl growth defects (Franklin et al., 2003; Monte et al., 2003), implying an important role of phyC in modulating the activity of other phys (Franklin et al., 2003; Monte et al., 2003; Hu et al., 2013). Our data show that phyC influences the activity of YHB under Rc (Fig. 7C), supporting the hypothesis that phyC acts as a light input into the circadian system. To more directly test this hypothesis, we introduced a CCA1::LUC2 reporter into phyC-2 (Monte et al., 2003) and phyC-4, two independent T-DNA insertion lines in the Col accession (Fig. 7D). Both phyC-2 and phyC-4 mutants had a circadian period approximately 1.5 h longer than wild-type controls under Rc (Fig. 6, D and E), although the amplitude of these rhythms appeared unaffected (Fig. 7D). Similar results were obtained from multiple independent T2 lines transformed with the CCA1::LUC2 reporter (Supplemental Fig. S2). Both phyC-2 and phyC-4 seedlings exhibited longer circadian periods than the wild type across a broad range of fluence rates (Fig. 7F). Such data led us to conclude that phyC modulates red light input into the circadian system in a manner similar to that of phyB (Devlin and Kay, 2000).
DISCUSSION AND CONCLUSION
YHB Mimics Continuous Light Input into the Circadian System in Darkness
We have assessed circadian clock function in YHB-expressing seedlings, allowing us to evaluate the effects of a single active phy species on the circadian system independently from light effects on photosynthesis. The YHB mimic of light-activated phyB was sufficient to sustain high-amplitude, rhythmic accumulation of CCA1, PRR9, TOC1, and GI transcripts in constant darkness in the absence of exogenous sugar in both Col and Ler accessions (Figs. 2, E and G, 3, and 6). This may be due to the increased expression of ELF4 in YHB plants, which shows a robust peak of expression on the first subjective day of free run in this genotype but not in the wild type (Figs. 3D and 6D). This difference precedes the first observed difference in cyclic amplitude in CCA1 and PRR9 transcripts in YHB and control plants, which is not seen until the morning of the second subjective day in free run (Figs. 2G, 3A, and 6, A and B). ELF4 forms part of the Evening Complex (Nusinow et al., 2011), which directly represses expression of clock genes such as PRR7, PRR9, GI, and ARRHYTHMO (LUX; Herrero et al., 2012; Mizuno et al., 2014; Box et al., 2015). Intriguingly, ELF4 also is necessary for red light-mediated induction of CCA1 and LHY (Kikis et al., 2005). We therefore suggest that sustained high-amplitude expression of Evening Complex components contributes to the maintenance of transcriptional rhythms in YHB plants in the dark.
Although luciferase and transcript oscillations were observed in YHB lines in constant darkness, the activity of YHB was not sufficient to prevent lengthening of the circadian period under these conditions when compared with even dim red light. We observed periods of 26 h in YHB CCA1::LUC2 reporter lines (Fig. 2D), and the later phase of peak CCA1 transcript accumulation also suggests a longer-than-24-h period in YHB seedlings in both Col and Ler accessions (Figs. 2G and 6A). CCA1::LUC2 activity rhythms in dark-adapting YHB plants grown with Suc are shorter (τ = 24.8 ± 0.3 h; Fig. 2B) than in YHB plants grown in the absence of Suc (26.0 ± 0.23 h; Fig. 2D), although the mechanism underlying this difference in period remains unclear. Similarly, PRR9 transcript oscillation was also sustained by YHB in darkness and displayed an advanced phase of peak accumulation compared with the wild-type control (Figs. 3A and 6B). Whether these PRR9 phase advances are due to underlying differences in periodicity rather than phase will be the subject of future investigations. Notably, YHB enhances rhythms in transcript abundance even in the absence of other phys (Fig. 6). Taken together, our data establish that YHB sustains robust clock function in the absence of photosynthesis and in the absence of light activation of other photoreceptors, including phys.
Dissecting the Role of Phys as Light Inputs to the Circadian System
The circadian period lengths of many diurnal species, including plants, are shortened in response to higher fluence rates of constant light, a phenomenon known as Aschoff’s rule (Aschoff, 1960; Somers et al., 1998; Devlin and Kay, 2000). This pattern is apparent in the red light fluence rate response curves presented here (Figs. 4A and 7F). Rhythmic amplitude of luciferase activity tended to increase with fluence rate (Fig. 4C), similar to the enhancement caused by added Suc (Fig. 2, A and C; Dalchau et al., 2011). Even low fluence rates of Rc caused maximal period shortening in YHB plants, suggesting full activation of phy signaling pathways to the clock even under dim light conditions (Fig. 4A).
Our analysis clearly indicates that YHB activity sustains phy-signaling input into the circadian system in darkness regardless of the presence of exogenous Suc. However, in continuous light, it is also clear that the clock receives additional red light-derived signaling cues from other phys, from the effects of light-driven chlorophyll synthesis, and/or from metabolic changes induced by photosynthesis itself (Hu et al., 2013). PhyA, phyB, and phyD have each been shown to contribute to light perception by the circadian system (Somers et al., 1998; Devlin and Kay, 2000). Recent studies reveal that phyABDE and phyABCDE mutants have indistinguishable circadian phenotypes (Hu et al., 2013), consistent with the evidence that phyC protein is unstable in the absence of other phys (Clack et al., 2009). The current study defines a role for phyC within the circadian system by demonstrating both a circadian phenotype in phyC mutants and modulation of YHB activity by phyC (Fig. 7). The long-period phenotype of phyC mutants across a range of fluence rates (Fig. 7F) suggests that phyC also contributes to red light signaling into the clock, consistent with previous reports describing the altered morphology of phyC mutants (Franklin et al., 2003; Monte et al., 2003). The shorter periods of YHB(phyABDE) compared with YHB(phyABCDE) under Rc (Fig. 7C) strongly suggest that phyC activation, presumably as the phyC(Pfr):YHB heterodimer, is responsible for the shorter circadian period in the YHB(phyABDE) line. In this regard, the slightly shorter period of dark-adapting YHB(phyABDE) compared with YHB(phyABCDE; Fig. 7A) could reflect the influence of residual phyC(Pfr) that had not fully reverted to phyC(Pr) at the onset of darkness. These results illustrate one consequence of the many interactions between phys that underlie the complex regulation of the circadian system by red light (Sharrock and Clack, 2004).
Mechanistic Hypothesis for the Regulatory Role of YHB in the Circadian System
Sugars, either produced via photosynthesis or applied exogenously, can both affect the pace of the clock and act as a time-of-day cue (Dalchau et al., 2011; Haydon et al., 2013). Thus, it can be difficult to distinguish photoreceptor-mediated and metabolic effects of light on circadian clock function. The ability of the constitutively active YHB allele of phyB to maintain high-amplitude transcriptional rhythms in the dark in the absence of exogenous sugars (Figs. 3 and 6) demonstrates that phy signaling alone is sufficient to maintain robust clock function. Recent studies implicate light-regulated interactions of PhyB with a subset of nuclear clock proteins, including CCA1, LHY, GI, TOC1, LUX, and ELF3 (Yeom et al., 2014). Under red light, the relative strength of some of these interactions is altered in planta, with binding to LUX increasing while interactions with CCA1 and TOC1 diminish (Yeom et al., 2014). Since all of the clock components function in the nucleus, yet require synthesis and transit through the cytosol, it is possible that interactions with phys could occur in both the nucleus and the cytosol. However, our analyses indicate that cytosolic YHB-G767R is unable to sustain circadian rhythms seen in YHB lines in constant darkness, nor does it shorten the clock period as measured by CCA1::LUC-dependent luminescence (Fig. 6). These activities thus appear dependent on the nuclear localization of YHB. However, YHB-G767R seems to evoke an advance in the phase of PRR9 expression during the early stages of free run, suggesting a modest cytoplasmic role for YHB at least within this subloop of the circadian system (Fig. 6B). We speculate that this response could be due to cytosolic retention of Pfr-interacting factors such as TOC1 that inhibit expression of PRR9 in the nucleus (Huang et al., 2012), an intriguing possibility that we will explore in future studies.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The pJM63-YHBg construct, genomic YHB sequence including approximately 2.3 kb native PHYB promoter (Su and Lagarias, 2007), was transformed into Col-0 wild type by the floral dip method. The resultant YHBg/Col line #1 was crossed with pCCR2::LUC/Col (Martin-Tryon et al., 2007) to obtain the YHBg/CCR2::LUC line. The phyB-9/CCR2::LUC line was generated by crossing CCR2::LUC plants with phyB-9 obtained from the Arabidopsis Biological Resource Center (line CS6217). CCA1::LUC2/Ler, CCA1::LUC2/YHBg/phyABDE, CCA1::LUC2/phyABDE, and CCA1::LUC2/phyABCDE were described previously (Hu et al., 2013). CCA1::LUC2/YHBg/phyABCDE was also obtained from the cross between CCA1::LUC2/YHBg/phyABDE and the phyABCDE quintuple mutant (Hu et al., 2013). CCA1::LUC2/YHBg was obtained from the cross between CCA1::LUC2/Ler and the previously reported YHBg/Ler line (Su and Lagarias, 2007). The pJM63-YHBg-G767R construct was created by site-directed mutagenesis and then transformed into CCA1::LUC2/phyABCDE, resulting in multiple genetically single insertion lines of YHB-G767R/CCA1::LUC2/phyABCDE. The phyC-2 mutant (Monte et al., 2003) was provided by Dr. Peter Quail (Plant Gene Expression Center, Albany, CA). The phyC-4 mutant (Salk_007004 line) was newly isolated; it was PCR genotyped using oligonucleotides described in Supplemental Table S1. The pEarleyGate301-pCCA1::LUC2 construct (Hu et al., 2013) was transformed into Col-0, phyC-2, and phyC-4 to obtain corresponding transgenic lines. Unless otherwise stated, all plants were grown under 60 μmol m−2 s−1 white light with 12:12 L/D photoperiods for 6 d before transfer to the constant conditions described for each assay.
Luciferase Imaging Assays
Plants were entrained for 6 d in 12:12 L/D cycles under white light on MS medium with or without supplemental 3% (w/v) Suc before being sprayed with 3 mm d-luciferin in 0.01% (v/v) Triton X-100. Plants were then transferred to free-running conditions under red light-emitting diodes of indicated fluence rate or held in constant darkness at ZT12 of day 6 as previously described (Jones et al., 2010). Bioluminescence from groups of 10 seedlings was pooled for each data point where seedlings were transferred to constant darkness. Imaging was completed over 5 d, and data were processed using Metamorph software (Molecular Devices). Patterns of luciferase activity were fitted to cosine waves using Fourier fast transform-nonlinear least squares (Plautz et al., 1997) to estimate circadian period length.
qRT-PCR
RNA was isolated and qRT-PCR performed as previously described (Jones et al., 2010). Samples were run in triplicate, with starting quantity estimated from critical thresholds using the standard curve of amplification. Data for each sample were normalized to PROTEIN PHOSPHATASE 2A (PP2a) expression as an internal control. Primer sets used are described in Supplemental Table S1.
Protein Extraction and Immunoblot Analysis
Dark-grown, 4-d-old seedlings were harvested for protein extraction as previously described (Su and Lagarias, 2007). After quantifying the total protein concentrations with the Pierce BCA protein assay kit (Thermo Scientific), equal amounts of proteins were separated on 4% to 20% ExpressPlus PAGE gels (GenScript) and then semidry transferred onto an Immobilon-FL PVDF membrane (EMD Millipore). PhyB and actin were immunodetected by anti-phyB B1 (gift from Dr. Peter Quail, 1:300 dilution) and anti-actin (#MA1-744, 1:1,000, Thermo Scientific) monoclonal antibodies, respectively. The IRDye 800CW goat-anti-mouse IgG (H+L) secondary antibody (LI-COR) was used to detect the primary antibodies. Immunoreactive bands were recorded by scanning the membrane with the Odyssey infrared imaging system (LI-COR).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Analysis of YHB function in the absence of other phytochromes.
Supplemental Figure S2. Circadian periodicity of phyC mutants.
Supplemental Table S1. Oligos used in this study.
Glossary
- Rc
constant red light
- Ler
Landsberg erecta
- L/D
light/dark
- MS
Murashige and Skoog
- qRT
quantitative reverse transcription
- T-DNA
transfer DNA
- ZT
Zeitgeber Time
Footnotes
This work was supported by the National Institutes of Health (grant nos. GM069418 to S.L.H. and GM068552 to J.C.L.), U.S. Department of Agriculture National Institute of Food and Agriculture (Hatch Project CA-D*–MCB–4126-H to J.C.L.), the Leverhulme Trust (grant no. ECF–2012–358 to M.A.J.), the Royal Society (grant no. RG130746 to M.A.J.), the Oppenheimer Memorial Trust (PhD studentship to S.L.), and the University of Essex (to M.A.J. and S.L.). M.A.J. is a Leverhulme Early Career Fellow.
Articles can be viewed without a subscription.
References
- Aschoff J. (1960) Exogenous and endogenous components in circadian rhythms. Cold Spring Harb Symp Quant Biol 25: 11–28 [DOI] [PubMed] [Google Scholar]
- Bae G, Choi G (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59: 281–311 [DOI] [PubMed] [Google Scholar]
- Bognár LK, Hall A, Adám E, Thain SC, Nagy F, Millar AJ (1999) The circadian clock controls the expression pattern of the circadian input photoreceptor, phytochrome B. Proc Natl Acad Sci USA 96: 14652–14657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box MS, Huang BE, Domijan M, Jaeger KE, Khattak AK, Yoo SJ, Sedivy EL, Jones DM, Hearn TJ, Webb AAR, et al. (2015) ELF3 controls thermoresponsive growth in Arabidopsis. Curr Biol 25: 194–199 [DOI] [PubMed] [Google Scholar]
- Casal JJ. (2013) Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol 64: 403–427 [DOI] [PubMed] [Google Scholar]
- Casal JJ, Candia AN, Sellaro R (2014) Light perception and signalling by phytochrome A. J Exp Bot 65: 2835–2845 [DOI] [PubMed] [Google Scholar]
- Chen A, Li C, Hu W, Lau MY, Lin H, Rockwell NC, Martin SS, Jernstedt JA, Lagarias JC, Dubcovsky J (2014) Phytochrome C plays a major role in the acceleration of wheat flowering under long-day photoperiod. Proc Natl Acad Sci USA 111: 10037–10044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol 25: 413–427 [DOI] [PubMed] [Google Scholar]
- Clack T, Shokry A, Moffet M, Liu P, Faul M, Sharrock RA (2009) Obligate heterodimerization of Arabidopsis phytochromes C and E and interaction with the PIF3 basic helix-loop-helix transcription factor. Plant Cell 21: 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalchau N, Baek SJ, Briggs HM, Robertson FC, Dodd AN, Gardner MJ, Stancombe MA, Haydon MJ, Stan GB, Gonçalves JM. , et al. (2011) The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc Natl Acad Sci USA 108: 5104–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Kay SA (2000) Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12: 2499–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Staiger D (2002) Photoreceptors in Arabidopsis thaliana: light perception, signal transduction and entrainment of the endogenous clock. Planta 216: 1–16 [DOI] [PubMed] [Google Scholar]
- Farinas B, Más P (2011) Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J 66: 318–329 [DOI] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Fogelmark K, Troein C (2014) Rethinking transcriptional activation in the Arabidopsis circadian clock. PLoS Comput Biol 10: e1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Davis SJ, Stoddart WM, Vierstra RD, Whitelam GC (2003) Mutant analyses define multiple roles for phytochrome C in Arabidopsis photomorphogenesis. Plant Cell 15: 1981–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Quail PH (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL. (2009) The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Haydon MJ, Mielczarek O, Robertson FC, Hubbard KE, Webb AAR (2013) Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502: 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, et al. (2012) EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Devisetty UK, Harmer SL (2013) Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife 2: e00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Harmer SL (2014) Wheels within wheels: the plant circadian system. Trends Plant Sci 19: 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Franklin KA, Sharrock RA, Jones MA, Harmer SL, Lagarias JC (2013) Unanticipated regulatory roles for Arabidopsis phytochromes revealed by null mutant analysis. Proc Natl Acad Sci USA 110: 1542–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Su Y-S, Lagarias JC (2009) A light-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcriptional networks. Mol Plant 2: 166–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79 [DOI] [PubMed] [Google Scholar]
- Hughes J. (2013) Phytochrome cytoplasmic signaling. Annu Rev Plant Biol 64: 377–402 [DOI] [PubMed] [Google Scholar]
- Ito S, Matsushika A, Yamada H, Sato S, Kato T, Tabata S, Yamashino T, Mizuno T (2003) Characterization of the APRR9 pseudo-response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol 44: 1237–1245 [DOI] [PubMed] [Google Scholar]
- Jones MA. (2009) Entrainment of the Arabidopsis circadian clock. J Plant Biol 52: 202–209 [Google Scholar]
- Jones MA, Covington MF, DiTacchio L, Vollmers C, Panda S, Harmer SL (2010) Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc Natl Acad Sci USA 107: 21623–21628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Williams BA, McNicol J, Simpson CG, Brown JWS, Harmer SL (2012) Mutation of Arabidopsis SPLICEOSOMAL TIMEKEEPER LOCUS1 causes circadian clock defects. Plant Cell 24: 4066–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis EA, Khanna R, Quail PH (2005) ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J 44: 300–313 [DOI] [PubMed] [Google Scholar]
- Locke J, Southern M, Kozma-Bognar L, Hibberd V, Brown P, Turner M, Millar A (2005) Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol 1: 2005 0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Tryon EL, Kreps JA, Harmer SL (2007) GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol 143: 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Mizuno T (2000) Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol 41: 1002–1012 [DOI] [PubMed] [Google Scholar]
- Matsushita T, Mochizuki N, Nagatani A (2003) Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424: 571–574 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Short SR, Chua NH, Kay SA (1992) A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell 4: 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Nomoto Y, Oka H, Kitayama M, Takeuchi A, Tsubouchi M, Yamashino T (2014) Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol 55: 958–976 [DOI] [PubMed] [Google Scholar]
- Monte E, Alonso JM, Ecker JR, Zhang Y, Li X, Young J, Austin-Phillips S, Quail PH (2003) Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. Plant Cell 15: 1962–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua N-H, Sakakibara H (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1999) Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400: 781–784 [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I, Yang S, Choi G (2012) Phytochrome regulates translation of mRNA in the cytosol. Proc Natl Acad Sci USA 109: 1335–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palágyi A, Terecskei K, Adám E, Kevei E, Kircher S, Mérai Z, Schäfer E, Nagy F, Kozma-Bognár L (2010) Functional analysis of amino-terminal domains of the photoreceptor phytochrome B. Plant Physiol 153: 1834–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A, Nagel MK, Popp C, Wüst F, Bindics J, Viczián A, Hiltbrunner A, Nagy F, Kunkel T, Schäfer E (2012) Interaction with plant transcription factors can mediate nuclear import of phytochrome B. Proc Natl Acad Sci USA 109: 5892–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA (1997) Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms 12: 204–217 [DOI] [PubMed] [Google Scholar]
- Pudasaini A, Zoltowski BD (2013) Zeitlupe senses blue-light fluence to mediate circadian timing in Arabidopsis thaliana. Biochemistry 52: 7150–7158 [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC (2006) Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57: 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, Xie Q, McClung CR (2008) Circadian timekeeping during early Arabidopsis development. Plant Physiol 147: 1110–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Sharrock RA, Clack T (2004) Heterodimerization of type II phytochromes in Arabidopsis. Proc Natl Acad Sci USA 101: 11500–11505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA (1998) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490 [DOI] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Song YH, Ito S, Imaizumi T (2013) Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci 18: 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser B, Sánchez-Lamas M, Yanovsky MJ, Casal JJ, Cerdán PD (2010) Arabidopsis thaliana life without phytochromes. Proc Natl Acad Sci USA 107: 4776–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YS, Lagarias JC (2007) Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell 19: 2124–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth R, Kevei E, Hall A, Millar AJ, Nagy F, Kozma-Bognár L (2001) Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol 127: 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Quail PH (1995) Mutational analysis of phytochrome B identifies a small COOH-terminal-domain region critical for regulatory activity. Proc Natl Acad Sci USA 92: 8596–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Wenden B, Kozma-Bognár L, Edwards KD, Hall AJW, Locke JCW, Millar AJ (2011) Light inputs shape the Arabidopsis circadian system. Plant J 66: 480–491 [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Devlin PF (1997) Roles of different phytochromes in Arabidopsis photomorphogenesis. Plant Cell Environ 20: 752–758 [Google Scholar]
- Woods DP, Ream TS, Minevich G, Hobert O, Amasino RM (2014) PHYTOCHROME C is an essential light receptor for photoperiodic flowering in the temperate grass, Brachypodium distachyon. Genetics 198: 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Kagawa T, Takano M (2014) The phytochrome B/phytochrome C heterodimer is necessary for phytochrome C-mediated responses in rice seedlings. PLoS One 9: e97264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom M, Kim H, Lim J, Shin AY, Hong S, Kim JI, Nam HG (2014) How do phytochromes transmit the light quality information to the circadian clock in Arabidopsis? Mol Plant 7: 1701–1704 [DOI] [PubMed] [Google Scholar]