Auxin regulates a progressive cell cycle during adult maize lateral root formation.
Abstract
Plants have evolved a unique plasticity of their root system architecture to flexibly exploit heterogeneously distributed mineral elements from soil. Local high concentrations of nitrate trigger lateral root initiation in adult shoot-borne roots of maize (Zea mays) by increasing the frequency of early divisions of phloem pole pericycle cells. Gene expression profiling revealed that, within 12 h of local high nitrate induction, cell cycle activators (cyclin-dependent kinases and cyclin B) were up-regulated, whereas repressors (Kip-related proteins) were down-regulated in the pericycle of shoot-borne roots. In parallel, a ubiquitin protein ligase S-Phase Kinase-Associated Protein1-cullin-F-box proteinS-Phase Kinase-Associated Protein 2B-related proteasome pathway participated in cell cycle control. The division of pericycle cells was preceded by increased levels of free indole-3-acetic acid in the stele, resulting in DR5-red fluorescent protein-marked auxin response maxima at the phloem poles. Moreover, laser-capture microdissection-based gene expression analyses indicated that, at the same time, a significant local high nitrate induction of the monocot-specific PIN-FORMED9 gene in phloem pole cells modulated auxin efflux to pericycle cells. Time-dependent gene expression analysis further indicated that local high nitrate availability resulted in PIN-FORMED9-mediated auxin efflux and subsequent cell cycle activation, which culminated in the initiation of lateral root primordia. This study provides unique insights into how adult maize roots translate information on heterogeneous nutrient availability into targeted root developmental responses.
Roots have developed adaptive strategies to reprogram their gene expression and metabolic activity in response to heterogeneous soil environments (Osmont et al., 2007). By this way, local environmental stimuli can be integrated into the developmental program of roots (Forde, 2014; Giehl and von Wirén, 2014). In resource-depleted environments, an important heterogeneously distributed soil factor is nutrient availability, which then directs lateral root growth preferentially into nutrient-rich patches (Zhang and Forde, 1998; Lima et al., 2010; Giehl et al., 2012). Such directed lateral root development depends on regulatory networks that integrate both local and systemic signals to coordinate them with the overall plant nutritional status (Ruffel et al., 2011; Guan et al., 2014). As shown by the impact of the N status-dependent regulatory module CLAVATA3/EMBRYO-SURROUNDING REGION-related peptides-CLAVATA1 leucine-rich repeat receptor-like kinase, economizing the costs for root development is pivotal for a resource-efficient strategy in nutrient acquisition (Araya et al., 2014). In recent years, strategies on yield and efficiency improvement have been developed that are primarily based on the manipulation of root system architecture (Gregory et al., 2013; Lynch, 2014; Meister et al., 2014). A common imperative of these strategies is to develop crops that use water and nutrients more efficiently, allowing the reduction of fertilizer input and potentially hazardous environmental contamination.
Maize (Zea mays) plays an eminent role in global food, feed, and fuel production, which is also a consequence of its unique root system (Rogers and Benfey, 2015). The genetic analysis of maize root architecture revealed a complex molecular network coordinating root development during the whole lifecycle (for review, see Hochholdinger et al., 2004a, 2004b). Identification of root type-specific lateral root mutants in maize emphasized the existence of regulatory mechanisms involved in the branching of embryonic roots, which are distinct from those in postembryonic roots (Hochholdinger and Feix, 1998; Woll et al., 2005). Under heterogeneous nutrient supplies, nitrate-rich patches increased only the length of lateral roots in primary and seminal roots, whereas they increased both length and density of lateral roots from shoot-borne roots of adult maize plants (Yu et al., 2014a). Remarkably, modulation of the extensive postembryonic shoot-borne root stock has a great potential to improve grain yield and nutrient use efficiency (Hochholdinger and Tuberosa, 2009).
Lateral root branching is critical to secure anchorage and ensure adequate uptake of water and nutrients. In maize, these roots originate from concentric single-file layers of pericycle and endodermis cells (Fahn, 1990; Jansen et al., 2012). Lateral root initiation is the result of auxin-dependent cell cycle progression (Beeckman et al., 2001; Jansen et al., 2013a). Most of the molecular changes during the cell cycle like, for instance, the induction of positive regulators, such as cyclins (CYCs) and cyclin-dependent kinases (CDKs), and the repression of Kip-related proteins (KRPs), thus account for a reactivation of the cell cycle (Beeckman et al., 2001; Himanen et al., 2002, 2004). In eukaryotes, ubiquitin-mediated degradation of cell cycle proteins plays a critical role in the regulation of cell division (Hershko, 2005; Jakoby et al., 2006). Conjugation of ubiquitin to a substrate requires the sequential action of three enzymes: ubiquitin-activating enzyme, ubiquitin-conjugating enzyme, and ubiquitin-protein ligase (E3). The E3 enzymes are responsible for the specificity of the pathway, and several classes of E3 enzymes have been implicated in cell cycle regulation, including the S-Phase Kinase-Associated Protein1-cullin-F-box protein (SCF) and Really Interesting New Gene (RING) finger-domain ubiquitin ligases (Del Pozo and Manzano, 2014). The F-box protein S-Phase Kinase-Associated Protein 2B (SKP2B) encodes an F-box ubiquitin ligase, which plays an important role in the cell cycle by regulating the stability of KRP1 and pericycle founder cell division during lateral root initiation (Ren et al., 2008; Manzano et al., 2012).
It has been shown that auxin is involved in long-distance signaling to adjust root growth in response to local nutrient availability (Giehl et al., 2012), and it is likely to serve in long-distance signaling for local nutrient responses as well (for review, see Rubio et al., 2009; Krouk et al., 2011; Saini et al., 2013; Forde, 2014). Polar auxin transport is instrumental for the generation of local auxin maxima, which guide these cells to switch their developmental program (Vanneste and Friml, 2009; Lavenus et al., 2013). In Arabidopsis (Arabidopsis thaliana), the PIN-FORMED (PIN) family of auxin efflux carrier proteins controls the directionality of auxin flows to maximum formation at the tip or pericycle cells (Benková et al., 2003; Laskowski et al., 2008; Marhavý et al., 2013). Auxin responses in protoxylem or protophloem cells of the basal meristem coincide with the site of lateral root initiation (De Smet et al., 2007; Jansen et al., 2012). In these defined pericycle cells, the phloem pole pericycle founder cells are primed before auxin accumulation occurs (De Smet et al., 2007; Jansen et al., 2012, 2013a). In contrast to dicots, the larger PIN family in monocots has a more divergent phylogenetic structure (Paponov et al., 2005). It is likely that monocot-specific PIN genes regulate monocot-specific morphogenetic processes, such as the development of a complex root system (Wang et al., 2009; Forestan et al., 2012).
The molecular control of lateral root initiation of the root system to heterogeneous nitrate availabilities is not yet understood in maize. In this study, the plasticity of lateral root induction in adult shoot-borne roots of maize in response to local high concentration of nitrate was surveyed in an experimental setup that simulated patchy nitrate distribution. RNA-sequencing (RNA-Seq) experiments and cell type-specific gene expression analyses showed that local nitrate triggers progressive cell cycle control during pericycle cell division. In addition, tissue-specific determination of indole-3-acetic acid (IAA) and its metabolites combined with auxin maxima determination by DR5 supported a role of basipetal auxin transport during lateral root initiation in shoot-borne roots. Thereby, this study provides unique insights in how auxin orchestrates cell cycle control under local nitrate stimulation in the shoot-borne root system of maize.
RESULTS
Local High Nitrate Promotes Early Pericycle Cell Divisions and Lateral Root Formation
Heterogeneous nitrate environments were simulated in a split-root system in which different nitrate levels were supplied. To determine how local high nitrate contributes to lateral root formation, we examined emerging lateral roots from 2 d on after local high-nitrate stimulation. Striking differences in length and density of lateral roots were observed 6 d after treatment (Fig. 1, A–C). The local effect on the promotion of lateral root density (136% increase) was more significant than that of lateral root length (55% increase; Fig. 1, B and C). Subsequently, early stages of lateral root initiation were monitored in pericycle cells to determine the distinct developmental stages of lateral root primordium formation at different time points and increasing distances from the root tip. Transverse sections of paraffin-embedded root fragments were analyzed, and early pericycle cell divisions were considered as anticlinal or periclinal by Safranin O and Fast Green staining (Supplemental Fig. S1A). At 24 h after treatment (Fig. 1D), peak differences in pericycle cell divisions were detected between homogeneous low-nitrate and local high nitrate treatments in the region between 5 and 25 mm from the root tip compared with 12 (Supplemental Fig. S1B) and 36 h (Supplemental Fig. S1C) after treatment.
Figure 1.
Effect of homogeneous low nitrate (homo LN) and local high nitrate (local HN) supplies on lateral root development in shoot-borne roots of maize. A, Shoot-borne roots were grown hydroponically in low nitrate (0.5 mm NO3−). Subsequently, two shoot-borne roots were split supplied with low nitrate (0.5 mm NO3−; left) or high nitrate (4 mm NO3−; right) for a period of 6 d. Representative roots are displayed. Bar = 1 cm. Average length (in millimeters; B) and density (millimeters−1; C) of lateral roots initiated from the whole shoot-borne roots. DAT, Days after treatment. D, Early pericycle cell divisions in shoot-borne roots 24 h after local HN stimulation. E, Frequency of lateral root primordia (LRP) in the region between 5 and 25 mm from the root tip at 24 h after treatment (HAT). Stages I to III, IV and V, VI to VII, and VIII are indicated according to Supplemental Figure S2. B to E, Error bars represent se for four biological replicates. Asterisks denote a significant difference according to paired Student’s t test. *, P ≤ 0.05; **, P ≤ 0.01.
To obtain a comprehensive view of lateral root primordium development, microscopic analyses were conducted to determine the developmental stages and number of emerged lateral roots in the region of 5 to 25 mm from the tip of shoot-borne roots (Fig. 1E). Stages I to VIII are defined in Supplemental Figure S2. Consistent with the observation of more emerged lateral roots (Fig. 1C), early divisions at stages I to III were significantly induced by local high nitrate stimulation (Fig. 1E).
RNA-Seq Analyses of the Stele of Maize Shoot-Borne Roots in Response to Local High Nitrate Stimulation
The transcriptome of stele tissue extracted from the region between 5 and 25 mm of shoot-borne roots of the maize inbred line B73 (Fig. 2A) was subjected to RNA-Seq to identify genes associated with lateral root initiation in response to the previously determined 24-h local high nitrate stimulation (Fig. 1D). Each treatment was analyzed in four biological replicates with 20 to 24 individual roots per replicate (Supplemental Data Set S1). On average, RNA-Seq experiments yielded approximately 24 million 90-bp paired-end reads per sample (Supplemental Data Set S1). After quality trimming and removal of duplicate reads, approximately 87% of all sequences were mapped to the maize reference genome (ZmB73_RefGen_v2; Supplemental Data Set S1). After removal of redundant reads, 66% of the remaining reads mapped uniquely to the filtered gene set of maize (FGSv2, release 5b.60; Supplemental Data Set S1), which comprises 39,656 high-confidence gene models. A gene was declared as expressed if five or more reads mapped in all four replicates of a sample. In total, 22,796 genes were expressed in at least one nitrate treatment (Supplemental Data Set S2). A complete list of expressed genes with normalized expression values is provided in Supplemental Data Set S2.
Figure 2.
Stele-specific transcriptome analysis and differentially expressed cell cycle genes in response to local high nitrate (local HN) stimulation. A, Shoot-borne roots were manually dissected in three tissues: 5 mm of root tip (5 mm), stele covering all central cell types covered by the pericycle (5–25 mm of stele), and cortex covering all radial tissues between endodermis and epidermis (5–25 mm). B, Frequency of differentially expressed genes normalized according to log2 Fc with FDR ≤ 5% and │Fc│ ≥ 2. Homo LN, Homogeneous low nitrate. C, RNA-Seq revealed differentially expressed cell cycle genes. CDKB and CYCB genes (G2/M transition; left) were induced by local HN, whereas KRP genes were inhibited by local HN (G1/S transition; right). D, Heat map of selected cell cycle genes (CDKB1;1, CDKB2;1, CYCB1;1, CYCB1;3, CYCB2;1, CYCB2;2, KRP1, KRP2, KRP3, and KRP4) in the stele determined by qRT-PCR after a time-course experiment of 36 h. Transcript abundance was normalized to myosin and expressed in base log2 values (GenBank accession no. 486090G09.x1). E, Expression of cell cycle genes (CDKB1;1, CDKB2;1, KRP1, KRP2, CYCB1;1, and CYCB2;1) in pericycle cells at the phloem poles determined by qRT-PCR 24 h after local HN stimulation. Error bars represent se for four biological replicates. Asterisks denote a significant difference according to paired Student’s t test. *, P ≤ 0.05; **, P ≤ 0.01.
Pairwise comparisons were performed to determine genes differentially expressed between local high nitrate and homogeneous low nitrate treatment (control). To specifically focus on genes with a strong response to local high nitrate stimulation, only genes with a false discovery rate (FDR) ≤5% and a fold change (Fc) ≥2 were considered in subsequent analyses. In total, 582 genes were differentially expressed in response to local high nitrate stimulation, among which 508 genes (87%) were up-regulated and 74 genes (13%) were down-regulated (Fig. 2B).
Overrepresentation of Differentially Expressed Genes Involved in Cell Cycle Control and Ubiquitin-Dependent Protein Degradation
Differentially expressed genes were functionally classified according to Gene Ontology (GO) terms using agriGO (http://bioinfo.cau.edu.cn/agriGO/analysis.php). To identify significantly overrepresented (FDR ≤ 1%) functional categories, a singular enrichment analysis (SEA) was performed. SEA compares each annotated gene with all annotated expressed genes in a given nitrate treatment. Among the 582 differentially expressed genes (Fig. 2B), 513 genes were annotated, and 26 GO terms displayed overrepresentation (Supplemental Table S1). Among those, several GO terms related to cell cycle regulation were overrepresented (GO: 0006950, 0007050, 0022402, 0004861, and 0016538).
Differentially expressed genes were further assigned to MapMan functional categories to compare the distribution of over- and underrepresented functional classes between nitrate treatments (Supplemental Table S2). The expected number of genes for each functional group was calculated based on all expressed genes and subsequently compared with the detected number of genes in this group. In the overrepresented group protein, 24 genes related to protein degradation (proteolysis) were enriched in bin 29 of MapMan outputs (Supplemental Fig. S3 shows a simplified illustration according to MapMan; Supplemental Table S3 has a complete gene list extracted from MapMan). Protein degradation is partially dependent on proteasomes in cooperation with SKP, Cullin, the F box-containing complex (SCF), and RING-E3 activities. In total, nine genes that encode F-BOX proteins that belong to the SCF E3 ligase complex were identified in this study (Supplemental Fig. S3; Supplemental Table S3). In detail, we identified an SKP2B subunit (GRMZM2G138176) and a subunit of Cullin (GRMZM2G551108) that belongs to an SCF E3 ligase complex. Moreover, 13 subunits (Supplemental Table S3) of the RING E3 ligase complex and 1 subunit (GRMZM2G428119) of the Broad complex, Tramtrak, Bric à brac/Pox virus and zinc finger Cullin E3 ligase complex were found (Supplemental Fig. S3). Finally, a subunit (GRMZM2G038126) related to a 26S proteasome regulatory protein was found. The detailed features of ubiquitin-dependent degradation are summarized in Supplemental Table S3.
Expression Dynamics Profiling on Cell Cycle Progression
RNA-Seq results suggested differential expression of numerous cell cycle genes in the overrepresented group cell and enriched GO term 0007050 that control cell cycle arrest upon induction with local high nitrate. Among those, transcript levels of six cell cycle repressors of the KRP family (GRMZM2G037926, GRMZM2G084570, GRMZM2G101613, GRMZM2G116885, GRMZM2G157510, and GRMZM5G854731) that arrest progressive cell cycle activity were down-regulated. In contrast, three cell cycle activators of the CDKB class (CDKB1;1: GRMZM2G495626; CDKB2;1: GRMZM2G068193; and CDKB2;2: GRMZM2G070115) and eight cell cycle activators of the CYCB family (GRMZM2G005619, GRMZM2G034647, GRMZM2G062453, GRMZM2G073003, GRMZM2G073671, GRMZM2G138886, and GRMZM2G310115) were up-regulated (Fig. 2C; Supplemental Table S4).
To further validate the effects of local high nitrate on cell cycle regulation, expression patterns of selected maize cell cycle genes were examined by quantitative reverse transcription (qRT)-PCR in stele tissue. After clustering, two diametrically opposed expression patterns were observed compared with the homogeneous low nitrate treatment: CYCB genes were up-regulated, whereas KRP genes were down-regulated under local high nitrate conditions (Fig. 2C). Repression of KRP genes and activation of CYCB genes were observed as early as 12 h after local high nitrate induction (Fig. 2D). These results are in line with early pericycle cell divisions after 24 h of high nitrate stimulation (Fig. 1D; Supplemental Fig. S1A). Pericycle cells at the phloem poles were isolated by laser-capture microdissection (LCM) to determine cell-specific expression of cell cycle genes by qRT-PCR. The results of these experiments showed that local high nitrate induced the expression of CYCB and CDKB genes, whereas the expression of KRP genes was inhibited (Fig. 2E).
Auxin Redistribution and Basipetal Auxin Transport
RNA-Seq results suggested progressive cell cycle activation and a higher demand for SCF ubiquitin-ligase-mediated protein degradation. Both processes are typical for auxin responses in lateral root initiation and emergence. A maize marker line expressing red fluorescent protein (RFP) under the control of a DR5 auxin-responsive promoter was used to analyze the spatial characteristics of the auxin response in shoot-borne roots. To study the effect of deficient auxin transport on the formation of lateral roots and the localization of the auxin response in maize, shoot-borne roots were split incubated in the presence of 1-N-naphthylphthalamic acid (NPA) after they were initiated (Supplemental Fig. S4). In shoot-borne roots treated for 4 d with NPA, no lateral roots emerged compared with either nitrate treatment (Supplemental Fig. S4A). Moreover, auxin maxima were observed in differentiated metaxylem vessels of roots subjected to local high nitrate (Supplemental Fig. S4B), whereas no maxima were found in NPA-treated roots (Supplemental Fig. S4C). Instead, at a distance of 6 to 8 mm from the root tip, an auxin maximum was established in pericycle cells of local high nitrate-treated roots (Supplemental Fig. S4D), whereas a diffuse signal was detected and no auxin response maximum was formed in the stele tissue of NPA-treated shoot-borne roots (Supplemental Fig. S4E).
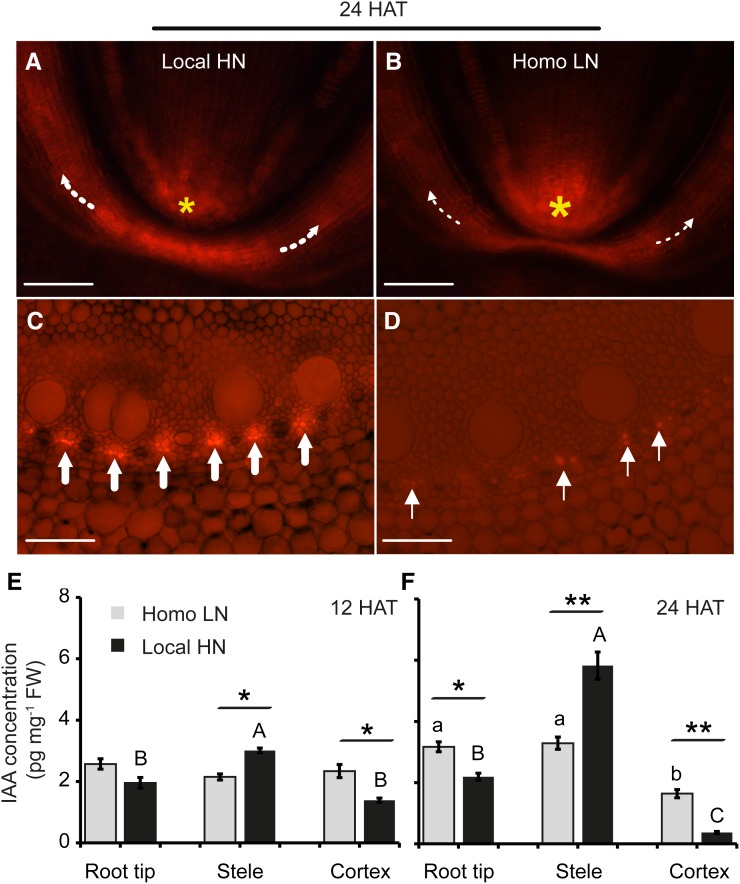
To survey the effects of local high nitrate on polar auxin transport, shoot-borne roots grown for 4 d in NPA were transferred to high or low nitrate. After 24 h, local high nitrate-treated shoot-borne root tissues expressed the RFP signal in both columella and lateral root cap cells (Fig. 3A). In contrast, incubation in homogeneous low nitrate enhanced the RFP signal exclusively within root apical meristem cells (Fig. 3B). Typically, a strong RFP signal was observed at the phloem poles of roots grown under local high nitrate in the lateral root initiation zone (Fig. 3C). In contrast, weak signals were monitored after homogeneous low-nitrate treatment (Fig. 3D). Moreover, IAA and its metabolites concentrations were determined to address the effect of nitrate on IAA distribution and its biosynthesis and degradation within root tissues (root tip, stele, and cortex) in shoot-borne roots (Fig. 3, E and F; Supplemental Fig. S5). Significantly higher IAA levels in the root tip and relatively uniform levels across tissues were detected when roots were grown in homogeneous low nitrate conditions (Fig. 3, E and F). IAA was distributed asymmetrically across three distinct tissues when induced by local high nitrate, with the highest level in the stele followed by a significant decrease in cortex and root tip (Fig. 3, E and F). In contrast, methylindole-3-acetic acid (MeIAA), an inactive form of IAA, was not induced by local high nitrate (Supplemental Fig. S5, A and B). This observation was supported by spatiotemporal IAA activity measurements. These experiments showed that auxin was transported from root tip and cortex to the stele as soon as 12 h after stimulation. Hence, measured auxin was not generated by local biosynthesis or degradation, because the concentrations of indole-3-acetonitrile (IAN), a putative IAA precursor, and 2-oxoindole-3-acetic acid (OxIAA), a degradation product of IAA, remained unchanged when induced by local high nitrate (Supplemental Fig. S5, C–F). In addition, the other two conjugated forms IAA-Ala and IAA-Glu were also not induced by local high nitrate application (Supplemental Fig. S5, G–J).
Figure 3.
Effect of local high nitrate (local HN) on tissue-specific auxin distribution. The initiated shoot-borne roots were incubated in NPA solution for 4 d and then followed by low or high nitrate treatment for 24 h. DR5 reporter activity in DR5::RFP-expressing transgenic maize lines in longitudinal orientation close to the root tip (A and B) or in cross sections (C and D) of the lateral root initiation zone was detected after high and low nitrate stimuli, respectively. White dotted arrows indicate the basipetal auxin transport direction, and yellow asterisks mark the differences of auxin accumulation in columella. White arrows indicate auxin accumulation in the phloem poles. HAT, hours after treatment; homo LN, homogeneous low nitrate. Bars = 150 μm. IAA concentrations in root tip, stele, and cortex of shoot-borne roots induced by homo LN and local HN 12 (E) and 24 HAT (F). Error bars represent se for four biological replicates. Different letters indicated significant differences among means when given a single homo LN (lowercase letters) or local HN (capital letters) treatment (P ≤ 0.05 by one-way ANOVA). Asterisks denote a significant difference between homo LN and local HN treatments according to paired Student’s t test. FW, Fresh weight. *, P ≤ 0.05; **, P ≤ 0.01.
Tissue- and Cell-Specific Expression Dynamics of ZmPINs
RNA-Seq revealed that ZmPIN1a, ZmPIN1c, and ZmPIN9 were significantly induced in the shoot-borne root stele by local high nitrate treatment (Fig. 4A). To further study potential roles of ZmPIN genes in auxin transport between different tissues, tissue-specific expression of the three ZmPIN genes was further monitored by qRT-PCR. These experiments independently confirmed and extended the RNA-Seq results (Fig. 4B). ZmPIN1a and ZmPIN1c transcript levels were induced in the root tip (Fig. 4C). In cortex tissue, only ZmPIN1a was up-regulated (Fig. 4D). ZmPIN9 was the only one of these genes that was exclusively expressed in the stele and subjected to a linear time-dependent induction by local high nitrate (Fig. 4, B–D). Transcript levels of ZmPIN9 closely correlated with those of CYCB and CDKB (Fig. 4E) but displayed reciprocal correlation to those of the KRP genes (Fig. 4F). Taken together, tissue-specific expression of ZmPIN genes and the local distribution of DR5 reporter activity indicated that auxin transport might directly or indirectly affect cell cycle processes by regulation of the SCF ubiquitin-ligase complex.
Figure 4.
Tissue-specific transcript abundance of ZmPIN genes. A, RNA-Seq revealed three ZmPIN genes significantly up-regulated in the root stele when induced by local high nitrate (local HN). The expression values were normalized by fragments per kilobase of transcript per million reads (FPKM) of local HN versus homogeneous low nitrate (homo LN). **, FDR-corrected P ≤ 1%. Expression patterns of ZmPIN1a, ZmPIN1c, and ZmPIN9 in stele (B), root tip (C), and cortex (D) as determined by qRT-PCR after local HN induction in a time-course experiment. Different letters indicate significant levels at P ≤ 0.05 after ANOVA. E, Correlation between transcript levels of CYCB and CDKB genes and ZmPIN9 in the stele (circles). F, Correlation between transcript levels of KRP genes and ZmPIN9 in the stele (triangles). The data points were extracted from B and Figure 2D at 0, 6, 12, and 24 h of local HN induction. Different letters indicated significant differences among means (P ≤ 0.05 by one-way ANOVA).
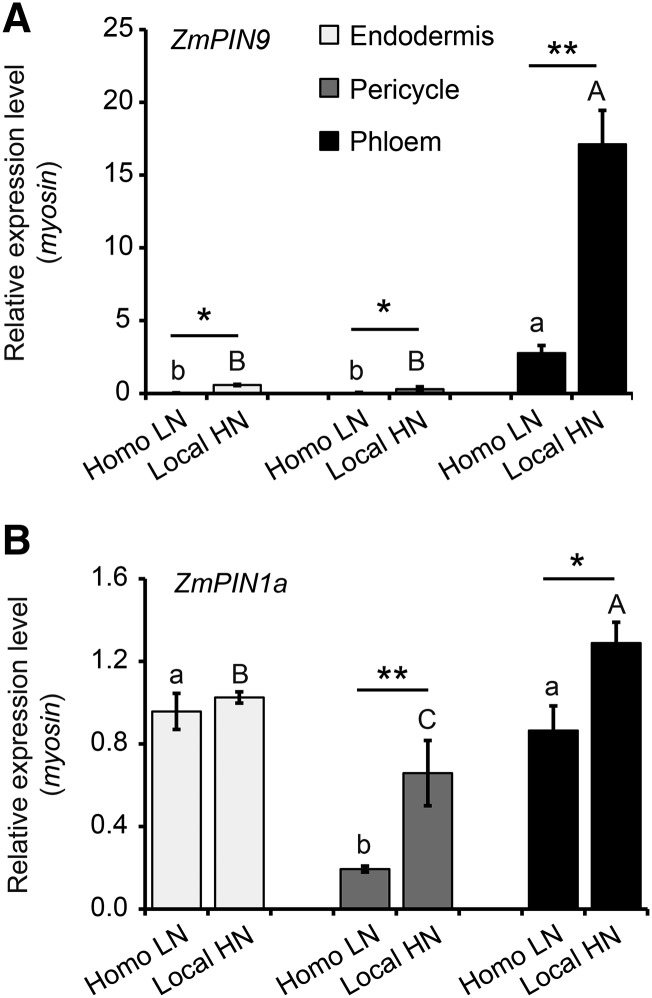
Pericycle founder cells related to lateral root initiation were captured by LCM from shoot-borne roots to further explore the effects of cell-specific auxin efflux on auxin maxima formation. Epifluorescence microscopy of unstained transverse sections revealed smaller xylem pole pericycle cells with thick autofluorescent cell walls, whereas the large pericycle cells at the phloem poles had thinner cell walls and lacked autofluorescence (Supplemental Fig. S6A). In maize, pericycle cells and endodermis cells at the phloem poles contribute to lateral root formation (Supplemental Fig. S6, B and C). These cell types were isolated by LCM from transverse root sections at distances of 5 and 10 mm from the root tip (Supplemental Fig. S6, D and E). Excellent RNA quality was obtained from these samples (RNA integrity number [RIN] values > 6.7; Supplemental Fig. S7). Cell type-specific expression analyses of ZmPIN1a, ZmPIN1c, and ZmPIN9 revealed that ZmPIN9 (Fig. 5A; Supplemental Fig. S8) transcripts were much more abundant under local high nitrate treatment in phloem pole cells compared with pericycle and endodermis cells, whereas ZmPIN1a and ZmPIN1c were not (Fig. 5B; Supplemental Fig. S8). The cell type-specific up-regulation of ZmPIN9 under local nitrate indicated that this transporter contributes to auxin provision of adjacent pericycle cells. Taken together, net auxin efflux direction of local high nitrate stimulation navigated by ZmPIN9 is illustrated in Figure 6A.
Figure 5.
Cell type-specific localization of ZmPIN9 (A) and ZmPIN1a (B) expressions as determined in three cell types related to lateral root initiation. Cells were isolated by LCM 24 h after transfer to local high nitrate (local HN). Paired Student’s t test was performed for homogeneous low nitrate (homo LN) and local HN treatments, and significance levels are indicated by asterisks. Different letters indicate significant differences among means when given a single homo LN (lowercase letters) or local HN (capital letters) treatment (P ≤ 0.05 by one-way ANOVA). *, P ≤ 0.05; **, P ≤ 0.01.
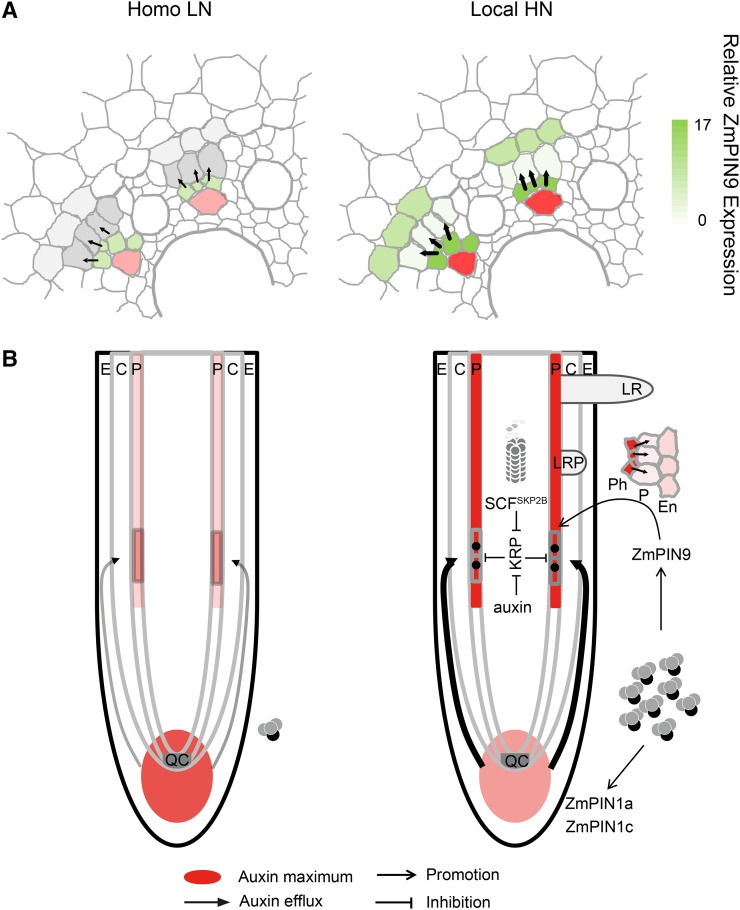
Figure 6.
Model of the molecular mechanisms underlying local high nitrate (local HN)-induced lateral root initiation in shoot-borne roots of maize. A, Schematic illustration of cell type-specific relative ZmPIN9 expression (grey to green gradient). Auxin maxima in phloem poles have been highlighted (red). Black arrows indicate suggested net auxin flux directions. Homo LN, Homogeneous low nitrate. B, At low nitrate concentrations, auxin accumulates in the root apical meristem (Fig. 3, B and F). Low auxin efflux capacity fails to induce basipetal auxin transport. In the presence of local HN, basipetal auxin transport (thick arrows) is facilitated by ZmPIN1a and ZmPIN1c (Fig. 3, A and C). Increased auxin in the lateral root initiation zone is transported into competent pericycle cells by ZmPIN9 (Figs. 3C, 4B, and 5A). Lateral root initiation is mediated by either auxin directly or the auxin-mediated ubiquitin-dependent protein degradation machinery (SCFSKP2B complex; Supplemental Fig. S3), either of which repress KRPs to facilitate for cell cycle activation (Fig. 2). C, Cortex; E, epidermis; En, endodermis; LR, lateral root; LRP, lateral root primordium; P, pericycle; Ph, phloem; QC, quiescent center.
DISCUSSION
Lateral root branching is a pivotal developmental process of root systems to extract nutrients from heterogeneous soil environments. Local nitrate remodels lateral root patterning by local signaling and in dependence of systemic regulation (Ruffel et al., 2011; Forde, 2014; Guan et al., 2014). Impressive progress has been made in understanding the mechanisms regulating lateral root development in roots of Arabidopsis (Giehl and von Wirén, 2014) and crops (von Behrens et al., 2011; Smith and De Smet, 2012; Orman-Ligeza et al., 2013). However, little is known about the mechanisms controlling lateral root branching in adult postembryonic shoot-borne roots of cereals. The extensive shoot-borne root system of maize substantially contributes to grain yield and nutrient use efficiency (for review, see Hochholdinger et al., 2004a, 2004b; Hochholdinger and Tuberosa, 2009). Through our studies of nitrate on lateral root patterning in maize, we identified a dramatic and highly cell type-specific response of postembryonic shoot-borne roots to local nitrate. With the nitrate-dependent regulation of auxin maxima in phloem pole cells and downstream cell cycle control, this study unravels a unique link between the availability of heterogeneous nitrate and the developmental programs of lateral root formation.
Local High Nitrate Promotes Auxin-Mediated Cell Cycle Activation during Lateral Root Initiation
A foraging response of postembryonic shoot-borne roots and subsequently altered lateral root morphology in adult maize roots was simulated in a split-root system (Yu et al., 2014a, 2014b). There, the local availability of nitrate exerted a higher number of lateral root primordia in stages I to III in shoot-borne roots relative to those grown under low nitrate (Fig. 1E). This observation suggested that local high nitrate supply significantly affects the first anticlinal and periclinal divisions of pericycle founder cells. In Arabidopsis, regulation of CDKs and a class of CDK-inhibitory proteins has been proposed as regulatory links between the control of cell division and morphological plasticity (De Veylder et al., 2001; Himanen et al., 2002, 2004). The increased frequency of dividing pericycle cells and emerged lateral roots suggested that local high nitrate activates competent but previously inactive pericycle cells (Fig. 1, C and E). This notion is supported by the observation that the gene expression levels of six KRP inhibitors (GRMZM2G037926, GRMZM2G084570, GRMZM2G101613, GRMZM2G116885, GRMZM2G157510, and GRMZM5G854731) that suppress the division of competent pericycle cells were down-regulated (Fig. 2). Cell cycle genes are differentially expressed in activated and nonactivated pericycle cells in Arabidopsis (de Almeida Engler et al., 2009). KRP1, KRP2, and KRP4 have been proposed as primary candidates for controlling the activation of divisions in nondividing cells and to specifically prevent cell cycle induction for formative divisions of lateral roots in the pericycle (Himanen et al., 2002, 2004; Ren et al., 2008; de Almeida Engler et al., 2009). Accordingly, KRP1 overexpression inhibited auxin-mediated cell division of pericycle cells, resulting in a dramatic decrease in the number of lateral roots (Ren et al., 2008). KRP2 regulates early lateral root initiation by blocking the G1-to-S transition and was down-regulated transcriptionally after 4 h of auxin induction (Himanen et al., 2002). KRP4 responded less strongly and showed a weak negative response upon auxin induction (Himanen et al., 2002; de Almeida Engler et al., 2009). The response of KRP3 deviated significantly from that of the other KRPs and was induced upon transfer to auxin, when it was highly expressed in actively dividing cells in Arabidopsis (De Veylder et al., 2001; Himanen et al., 2002). In contrast, in the stele of maize, the orthologous KRP3 gene showed a similar transcript pattern as the other KRP genes in response to auxin or nitrate (Jansen et al., 2013a; Fig. 2D). This suggests divergent cell cycle control mechanisms between dicots and monocots. Thus, these results show that the homeostasis of KRP proteins, which actively regulate the G1-to-S transition in the pericycle cells, is auxin dependent. During spontaneous lateral root initiation, competent pericycle cells proceed to the G2 phase of the cell cycle, whereas the remaining pericycle cells stay in the G1 phase (Beeckman et al., 2001). In this study, a set of CYCB genes (GRMZM2G005619, GRMZM2G034647, GRMZM2G062453, GRMZM2G073003, GRMZM2G073671, GRMZM2G138886, and GRMZM2G310115), CDKB1;1 (GRMZM2G495626), CDKB2;1 (GRMZM2G068193), and CDKB2;2 (GRMZM2G070115) were up-regulated and likely act in G2- to M-phase regulation (Fig. 2; Supplemental Table S4; Himanen et al., 2002, 2004). In Arabidopsis, CYCB proteins stimulate cell division and tissue growth (Lee et al., 2003). Moreover, B1- and B2-type CDK proteins display a maximum of kinase activity at the G2-to-M transition and during mitosis (Inzé and De Veylder, 2006). In addition, CYCB2;2 promotes mitotic cell cycle and cell divisions probably through association with CDKB2 in the root meristem (Lee et al., 2003; Sabelli et al., 2014). In this study, qRT-PCR-based time-course experiments on stele-specific gene expression have been implemented to survey the effects of local high nitrate stimulation on the cell cycle. The expression patterns of selected CDKB, CYCB, and KRP genes are distinctively clustering in line with their phase-specific control of the cell cycle (Fig. 2, D and E). These nitrate-inducible patterns are thoroughly analogous to the expression profiles observed in Arabidopsis during lateral root initiation induced by auxin (Himanen et al., 2002; Jansen et al., 2013a; Fig. 2D). Stability of KRP2 is controlled by CDKB1;1 phosphorylation (Verkest et al., 2005a), although KRP1 does not interact with CDKB1;1 and CYCB proteins (De Veylder et al., 2001). In this model, the CDKB1;1 kinase inhibits KRP2 activity, resulting in a higher activity at the G2-to-M transition and consequently, more mitotic cell divisions (Verkest et al., 2005a). In view of these results, we conclude that, in maize, local high nitrate supply promotes early pericycle cell divisions by modulating mitosis-specific cell cycle progression.
A Role of the Ubiquitin-Proteasome System in Nitrate-Dependent Cell Cycle Control
It has been shown that ubiquitin-dependent degradation of key regulatory proteins of the cell cycle plays a crucial role in a sequence of events leading to cell division (for review, see Del Pozo and Manzano, 2014). In addition to CDK protein phosphorylation, KRP1 and KRP2 are also known to be degraded by the 26S proteasome during the G1- to S-phase transition of the cell cycle (Verkest et al., 2005a, 2005b; Jakoby et al., 2006). SKP2B, an F-box protein, controls cell cycle by targeting KRP1 protein turnover in Arabidopsis (Ren et al., 2008). In the stele-specific RNA-Seq analysis presented here, genes contributing to the ubiquitin-proteasome system SCFSKP2B complex were strikingly enriched as illustrated by MapMan visualization (Supplemental Fig. S3; Supplemental Table S3). In Arabidopsis, KRP1 degradation is dependent on the SCFSKP2B complex that consists of CULLIN (GRMZM2G428119), SKP2B (GRMZM2G138176), and the 26S proteasome (GRMZM2G038126; Ren et al., 2008; Supplemental Fig. S3). Recently, we have also shown that the rootless with undetectable meristem1 mutant, which is defective in lateral root formation, displays significant inhibition of ubiquitin-dependent protein degradation and cell cycle/division processes (Zhang et al., 2014). Thus, our RNA-Seq results, together with the finding that SKP2B overexpression promotes KRP1 degradation in Arabidopsis (Ren et al., 2008), suggest that the ubiquitin-dependent SCFSKP2B complex might target KRP protein degradation for reactivation of the mitotic cell cycle in maize (Fig. 6). This does not, however, exclude the possibility that the cell cycle is directly regulated by auxin at the transcriptional level (Himanen et al., 2002, 2004; Fig. 6).
Basipetal Auxin Transport Modulated by Local High Nitrate
Auxin is a key regulator of lateral root formation (De Smet et al., 2007; Dubrovsky et al., 2008; Rubio et al., 2009; Krouk et al., 2011; Saini et al., 2013) but also, involved in lateral root development in response to local nutrient availability (Krouk et al., 2011; Giehl et al., 2012). Induction of genes specific for G1/S cell cycle transition and activation of protein degradation is controlled by auxin signal perception and transduction (Himanen et al., 2002, 2004). As early as 3 or 6 h but not later than 12 h after stimulation with local high nitrate, the genes CYCB1;1 and CYCB2;1 involved in the G2-to-M transition showed simultaneous induction by auxin (Himanen et al., 2002; Jansen et al., 2013a; Fig. 2D). This observation supports the notion that phloem pole pericycle cells gain mitotic activity, which synchronizes with the induction of G2/M phase-specific genes at the same time (Figs. 1D and 2E; Supplemental Fig. S1A). In contrast, early changes of cell division induced by nitrate are at least 12 h delayed compared with auxin induction (Fig. 2D; Himanen et al., 2002). Based on this observation, we hypothesize that local high nitrate modulates auxin homeostasis required for pericycle cell divisions rather than directly acting on the cell cycle genes. In fact, transcriptome and reverse genetic studies have shown that the auxin status and auxin transporters are strongly modulated by nitrogen availability (for review, see Krouk et al., 2011).
The basipetal flow of auxin in the lateral root cap and epidermis and its subsequent accumulation in pericycle founder cells are thought to drive the first formative divisions in pericycle cells (Dubrovsky et al., 2001; De Smet et al., 2006). Disruption of this process inhibits lateral root outgrowth in Arabidopsis (Casimiro et al., 2001; Himanen et al., 2002). To address the effects of local high nitrate stimulation on basipetal auxin transport in shoot-borne roots, lateral root initiation was synchronized experimentally by NPA before induction by different nitrate levels (Supplemental Fig. S4). NPA treatment keeps pericycle cells in an undifferentiated or cell division-competent state (Jansen et al., 2012, 2013b). Indeed, early vascular differentiation was inhibited at a distance of 6 to 8 mm from the root tip (Supplemental Fig. S4, B and C). Visualization of the activity of the auxin-responsive DR5 reporter indicated that the auxin signal was elevated in those lateral founder cells that were subjected to local high nitrate (Fig. 3A) but not in roots grown in homogeneous low nitrate (Fig. 3B). Accordingly, higher auxin maxima were formed in the phloem poles and their neighboring cells in the lateral root initiation zone in response to nitrate (Fig. 3C). In contrast, addition of NPA caused accumulation of auxin in root apical meristem cells (Fig. 3B), which decreased auxin levels to a suboptimal level for the induction of founder cell division in the lateral root initiation zone (Casimiro et al., 2001; Fig. 3D). These results indicated that local high nitrate may partly restore basipetal auxin transport and release the inhibition from NPA. Auxin maxima in the phloem poles could be a major trigger for the priming of neighboring pericycle cells and subsequent anticlinal divisions (Jansen et al., 2012; Supplemental Fig. S5C). Tissue-specific determination of active IAA and its metabolites validated possible auxin transport from root tip and cortex to root stele tissue as soon as 12 h after treatment in response to local nitrate supply (Figs. 3, E and F and 4, C and D; Supplemental Fig. S5), which is in agreement with the release of KRP proteins at the same time (Fig. 2D). The concentration of MeIAA was not induced by local high nitrate, although it has a stronger capacity to induce lateral roots than IAA (Supplemental Fig. S5, A and B). In addition, both the precursor (IAN; Supplemental Fig. S5, C and D) and the degradation product (OxIAA; Supplemental Fig. S5, E and F) of IAA were not induced by local high nitrate. Furthermore, conjugated forms of IAA (IAA-Ala and IAA-Glu) presented here might not contribute to the auxin homeostasis (Supplemental Figs. S5, G–J). Based on these results, local nitrate-enhanced basipetal auxin transport may contribute to early auxin gradients in pericycle cells in the lateral root initiation zone.
Cell Type-Specific Expression Dynamics of PIN Genes Navigate Auxin Efflux for Pericycle Priming
Polar auxin transport, partially mediated by PIN auxin efflux carriers, is crucial for generating auxin gradients between cells (Benková et al., 2003; Friml et al., 2003; Blilou et al., 2005). Despite their functional redundancy, each PIN protein has been implicated in particular developmental processes in Arabidopsis (Friml et al., 2002a, 2002b, 2003; Blilou et al., 2005; Marhavý et al., 2013). In contrast to dicots, the phylogenetic structure of the larger PIN families in monocots is more divergent (Paponov et al., 2005; Wang et al., 2009; Forestan et al., 2012). ZmPIN1a, ZmPIN1b, and ZmPIN1c, which are orthologs of AtPIN1, may perform the functions of AtPIN3, AtPIN4, and AtPIN7, for which no orthologs have been identified in maize (Forestan et al., 2012; Villiers and Kwak, 2012). ZmPIN1-mediated transport of auxin and the related auxin fluxes during maize axillary meristem, kernel, and ligule development have been studied in detail (Carraro et al., 2006; Gallavotti et al., 2008; Forestan et al., 2010; Johnston et al., 2014). These studies have significantly extended the understanding of auxin transport on the initiation of shoot organs in maize. On top of this, the results of this study add that ZmPIN1 is likely involved in shootward auxin flows to be channeled through the lateral root cap (Band et al., 2014; Figs. 3A and 4C). It is important to note that basipetal auxin flow results in a considerable increase in auxin levels in phloem pole cells at approximately the site where pericycle cells start to divide. This observation supports the notion that local nitrate-mediated basipetal auxin transport plays a role in the activation of pericycle cells.
PIN-mediated auxin efflux further promotes periclinal auxin transport, enabling auxin to move between epidermal, cortical, and endodermal cells (Band et al., 2014). Auxin movement into founder cells has been reinforced by PIN3 localized laterally in the inner membrane in endodermis cells. This provides a local auxin reflux pathway important for additional lateral root initiation (Marhavý et al., 2013). Auxin treatment induces PIN3-GFP in endodermal and cortical cell files of the Arabidopsis root (Marhavý et al., 2013). In maize, ZmPIN1a is exclusively induced by local high nitrate stimulation in the cortex. Hence, ZmPIN1a may contribute to auxin export from the cortex to the stele (Fig. 4D). This conclusion is further supported by the expression profile of ZmPIN1a, which displayed significantly higher transcript levels in the endodermis compared with the pericycle (Supplemental Fig. S8). The up-regulation of ZmPIN1a supports a regulatory function for the endodermis during auxin maximum formation. Moreover, this indicates that the interaction between pericycle cells and the adjacent endodermis cells defines a development-specific auxin reflux pathway, which supports pericycle founder cells to reach auxin threshold levels required to initiating lateral roots. Monocot root-specific PIN9 genes have been identified in maize, rice (Oryza sativa), and wheat (Triticum aestivum; Paponov et al., 2005; Wang et al., 2009; Forestan et al., 2012). In rice, a higher expression level of OsPIN9 was found in the root and the stem base compared with other tissues (Wang et al., 2009). Likewise, ZmPIN9 is exclusively expressed in shoot nodes and roots of maize (Forestan et al., 2012). Moreover, PIN9 was highly expressed in lateral root primordia, pericycle cells, and vascular tissues in rice (Wang et al., 2009) as well as in the endodermis, pericycle, and phloem of maize (Forestan et al., 2012). Taken together, the divergent structure and phylogeny and the expression of the monocot PIN9 genes indicate a subfunctionalization associated with monocot-specific morphogenetic processes (Paponov et al., 2005; Forestan et al., 2012). In this study, we revealed that ZmPIN9 expression in the stele is induced in parallel with the transcriptional up-regulation of CYCB proteins and repression of KRP proteins after local supply of high nitrate (Figs. 2D and 4, B, E, and F). Taken together, these results suggest a major role of ZmPIN9 in auxin efflux from phloem to pericycle cells (Figs. 5 and 6A), and they suggest that, in maize, the dividing potential of phloem pericycle cells rather than that of xylem pericycle cells relies on auxin efflux by the monocot-specific ZmPIN9 protein.
In summary, this study supports a model for PIN-dependent auxin efflux and cell cycle activation under local nitrate availability (Fig. 6B). Lateral roots are initiated from the pericycle, a process mediated by auxin-related cell cycle progression. Before cell cycle activation, pericycle cells need to be specified as the founder cells by auxin. ZmPIN1-mediated basipetal auxin transport to the lateral root initiation zone and ZmPIN9-dependent redistribution of auxin to pericycle cells are orchestrated by local high nitrate supply. Auxin relieves the inhibition from KRP proteins for progressive cell cycle either directly or by induction of the SCFSKP2B-regulated ubiquitin-dependent proteasome. Thus, this study uncovered a direct regulatory pathway between heterogeneous nitrate stimulation and auxin transport dynamics that underpins cell cycle decisions before lateral root initiation.
MATERIALS AND METHODS
Plant Material and Growth Condition
Seeds of the maize (Zea mays) inbred line B73 were surface sterilized and germinated in paper rolls in distilled water as previously described (Hetz et al., 1996). Seven days after germination, the endosperm of each seedling was excised, and uniform seedlings with two visible leaves and four embryonic roots were transferred to low nitrate [0.5 mm NO3−, Ca(NO3)2] nutrient solution and grown as previously described (Yu et al., 2014a; long-term experiment). Initiation of shoot-borne roots was recorded in a time-course experiment until the emergence of the seventh whorl of shoot-borne roots, which were split treated by high- [4 mm NO3−, Ca(NO3)2] and low nitrate solution as control. The experiment was conducted in a growth chamber in a 16-h-light (25°C)/8-h-dark (21°C) cycle for 2 months.
Synchronization of Lateral Root Initiation
Synchronization of lateral root initiation was performed according to a modified protocol from the works by Himanen et al. (2002) and Jansen et al. (2013b). Plants were grown in low nitrate solution until the seventh whorl of shoot-borne roots started to emerge (approximately 60 d after germination). Then, plants were transferred into a 25 μm solution of the polar auxin transport inhibitor NPA (Sigma-Aldrich) in 0.5 mm NO3− for 4 d. Lateral roots were then synchronously induced by transferring the plants to low- (0.5 mm NO3−) or local high nitrate (4 mm NO3−) solution without NPA (Supplemental Fig. S4).
Morphological and Anatomical Analyses of Lateral Roots Initiation
To determine lateral root length and density, nitrate-treated shoot-borne roots were harvested, scanned using ScanMaker 9800XL (Microtek), and subsequently analyzed by WinRHIZO (www.regent.qc.ca). Lateral root density was calculated by the number of emerged lateral roots divided by the length of the region where lateral roots emerged. Microscopic analysis was conducted to determine the number of lateral root primordia and emerged lateral roots. For this purpose, the total number of lateral root initiation events was counted, and lateral root primordia were classified according to their developmental stages: I to III, IV and V, V to VII, and VIII (Malamy and Benfey, 1997; Supplemental Fig. S2).
Histochemical, Histological, and Microscopic Analyses
Visualization of Cell Divisions by Safranin O and Counterstaining by Fast Green
Maize shoot-borne roots were fixed in 4% (w/v) paraformaldehyde for 24 h at 4°C as described by Lim et al. (2000); 10-μm sections were prepared by an RM2125RTS Microtome (Leica; www.leicabiosystems.com). Staining of deparaffinized specimens with Safranin O (Roth) and Fast Green FCF (Sigma-Aldrich) was performed according to standard histological procedures (Johansen, 1940). Stained specimens were analyzed under an AxioCam MRc Microscope (Carl Zeiss Microimaging) and documented with Axio-Imager software (Carl Zeiss Microimaging).
Localization of Auxin Signal Response
Transgenic lines carrying a DR5::RFP auxin response reporter with a synthetic auxin promoter were subjected to histological analysis (Gallavotti et al., 2008). Roots were fixed in 4% (w/v) paraformaldehyde (1× phosphate-buffered saline [PBS] buffer, pH 7.4) under vacuum (500 MPa) for 10 min at 4°C. The solution was replaced with fresh fixative after two vacuum infiltration steps. Subsequently, the infiltrated samples were incubated at 4°C for >1 h. Tissues were then rinsed at 4°C several times with 1× PBS buffer. Root samples were then embedded in 8% (w/v) low-melting agarose (peqGOLD Universal Agarose; peQLab Biotechnologie) with 0.5% (w/v) gelatin (AppliChem). Trimmed agarose blocks with properly oriented samples were sectioned in 100-μm increments by a VT1200 Vibratome (Leica), mounted with distilled water, and immediately observed under an Axio-Imager Epifluorescence Microscope (Carl Zeiss) and a confocal laser-scanning microscope (Carl Zeiss LSM 780). Autofluorescence was detected with an AxioCam MRc Microscope (Carl Zeiss Microimaging), and images were taken with an Axio-Imager (Carl Zeiss Microimaging). A filter set (Carl Zeiss Filter Set 43) for rhodamine was used for visualization of the RFP signal. Another two filter sets for the cyan fluorescent protein signal (Carl Zeiss Filter Set 55) were used, and nucleus staining with 4′,6-diamidino-phenylindole (Carl Zeiss Filter Set 38) was performed.
IAA Detection by Ultraperformance Liquid Chromatography-Electrospray Ionization-Tandem Mass Spectrometry
Based on the histological and DR5 fluorescent results, the most apical 5 mm of the shoot-borne root were excised and defined as root tip. Subsequently, the stele was separated from the cortical parenchyma and epidermis by peeling off the outer tissue layers (Saleem et al., 2009). For each biological replicate, 10 individual shoot-borne roots were pooled for root tip, cortical parenchyma, and stele tissues and immediately frozen in liquid nitrogen before storage at −80°C and freeze drying.
Solid Phase Extraction of Auxins
The phytohormone extraction procedure was adapted and modified from the works by Kojima et al. (2009) and Seo et al. (2011). Frozen tissues were crushed to fine powder using a TissueLyser (Qiagen) with two steel balls in a 2-mL Eppendorf tube and then mixed with 180 µL of extraction solution in water (Milli-Q Reference System; Merck):methanol (Th. Geyer; 50:50) containing 0.5% (w/v) formic acid (Biosolve Chimie) for 30 s. The mixture was sonicated for 5 min and then extracted with an overhead shaker for 15 min. Next, 720 µL of water was added, and the complete extraction solution was again mixed in the overhead shaker for 1 h. After centrifugation at 14,000 rpm for 10 min, the supernatant was transferred to a new Eppendorf tube. All steps were performed at 4°C. Afterward, the procedure was repeated, and the supernatants were combined.
Internal standards were dissolved in methanol and added to each combined supernatant (Supplemental Table S5). The methanol of the extracted solution was then evaporated in a vacuum centrifuge for 20 min, and the residue was then used for solid phase extraction cleanup. The supernatant was then added to a 1-mL/30-mg HLB Cartridge (Waters), which was conditioned before with 1 mL of methanol containing 0.1% (w/v) formic acid and equilibrated with 2× 1 mL of water containing 0.1% (w/v) formic acid. The sample was then washed two times with 1 mL of water containing 0.1% (w/v) formic acid and eluted with 2× 600 µL of 90% (v/v) methanol with 0.1% (w/v) formic acid. Now, the methanol was evaporated with a vacuum centrifuge, and the water residue was filled up to 1 mL with water containing 0.1% (w/v) formic acid. Then, all was mixed for 30 s and sonicated for 2 min at 4°C. In the second SPE step, a 1-mL/30-mg MCX Cartridge (Waters) was used. The MCX cartridge was conditioned with 1 mL of methanol containing 0.1% (w/v) formic acid and equilibrated with 2× 1 mL of water containing 0.1% (w/v) formic acid. Samples were then added to the column and washed two times with 1 mL of water containing 0.1% (w/v) formic acid. Auxin was then eluted with 2× 600 µL of 100% methanol. Samples were then evaporated in a vacuum centrifuge to dryness, resolved in 10 µL of 50% (v/v) methanol containing 0.5% (w/v) formic acid, mixed for 30 s, sonicated for 2 min, filled up to 50 µL with 40 µL of water, and transferred to a Ultraperformance Liquid Chromatography (UPLC) vial for analysis.
Liquid Chromatography-Tandem Mass Spectrometry
Ten-microliter extracted samples were then injected into an ACQUITY Ultra-Performance LC System (Waters) coupled with a Xevo TQ MS Mass Spectrometer (Waters). The sample analytes were separated on an ACQUITY UPLC BEH C18 (Waters) coupled with a VanGuard precolumn BEH C18 (Waters). For quantification of the analytes, one fragment ion was used for quantification, two fragment ions were used for qualification, and two fragment ions were used as internal standards (Supplemental Table S5). Mass spectrometry data were processed by using TargetLynx V4.1 SCN 904 (Waters; Supplemental Text S1). The peak area of the diagnostic product ion was used for quantification.
qRT-PCR
qRT-PCR was performed in a CFX 384TM Real-Time System (Bio-Rad). RNA was isolated from the three root tissues described above using the RNeasy Mini Kit (Qiagen). All RNA samples were quantified and qualified using a NanoDrop Spectrophotometer (Thermo Scientific; PeqLab) and agarose gel electrophoresis, respectively. Complementary DNA (cDNA) for qRT-PCR analysis was synthesized from 1 μg of total RNA with the qScript cDNA SuperMix (Quanta Biosciences). Each PCR reaction contained 4 μL of MESA Blue qPCR Mastermix Plus for SYBR Assay No ROX (Eurogentec), 1 μL of cDNA sample, and 100 nm gene-specific oligonucleotide primers to a final volume of 8 μL. The primer efficiency of each oligonucleotide was calculated using the following dilution series: 1, 1/2, 1/4, 1/8, 1/16, 1/32, 1/64, and 1/128. The relative expression levels of the transcripts were calculated with reference to the housekeeping gene myosin (GenBank accession no. 486090G09.x1) previously used in the laboratory to quantify gene expression in maize roots (Dembinsky et al., 2007). Significant differences in gene expression levels were determined by paired Student’s t test or one-way ANOVA. Cell cycle and PIN gene expression analyses were performed with primer pairs as previously described (Rymen et al., 2007; Forestan et al., 2010, 2012).
Root Stele-Specific RNA-Seq Experiment
RNA Isolation and cDNA Library Preparation for RNA-Seq
The steles of shoot-borne roots were separated from cortical parenchyma between 5 and 25 mm from the root tip. Dissected stele tissues were pooled and ground in liquid nitrogen. RNA was extracted by the RNeasy Plus Universal Mini Kit (Qiagen). RNA quality was assessed by agarose gel electrophoresis and an Agilent RNA 6000 Nano LabChipon Agilent 2010 Bioanalyzer (Agilent Technologies). All samples had an excellent quality as documented by RIN values >9.7. During the quality control steps, an Agilent DNA 1000 LabChip (Agilent Technologies) and an ABI StepOnePlus Real-Time PCR System (Applied Biosystems) were used for quantification and qualification of the sample libraries. The cDNA libraries for RNA-Seq were constructed according to the TruSeq RNA Sample Prep Kit (Illumina). For sequencing, eight libraries were pooled in one lane of a flow cell. Cluster preparation and paired-end read sequencing were performed according to the HiSeq 2000 guidelines (Illumina).
Processing, Mapping of RNA-Seq Reads, and Statistical Procedures for Analyzing Differentially Expressed Genes
Raw sequencing reads generated by the Illumina HiSEq 2000 System were initially processed and quality trimmed with SHORE (http://1001genomes.org/software/shore.html). At least 20 million clean reads were obtained by removing reads containing adapters, reads containing >10% unknown bases, and low-quality reads containing >50% low-quality bases. Read mapping was performed with CLC Genomics Workbench (http://www.clcbio.com/products/clc-genomics-workbench/). All high-quality reads were mapped to the maize B73 reference genome (RefGen_v2; ftp://ftp.gramene.org/pub/gramene/maizesequence.org/), allowing large gaps of up to 50 kb to span introns. In addition, redundant reads mapping at the same starting coordinate and mapping orientation (stacked reads) were removed from the set of uniquely mapping RNA-Seq reads. The remaining reads of all samples were projected to the filtered gene set (FGSv2, release 5b.60; http://ftp.maizesequence.org/release-5b/filtered-set/) of the B73 reference genome derived from the maize genome sequencing project. Only those reads uniquely mapping to the reference data set were subsequently used for analyses. Differential gene expression was determined using the normalized fragments per kilobase of transcript per million reads values in conjunction with empirical analysis of differential gene expression (Robinson et al., 2010) implemented in the CLC Genomics Workbench using an FDR ≤ 5% cutoff (Benjamini and Hochberg, 1995) and a log2 Fc of ≥1.
GO and Metabolic Pathway Analyses
GO functional categories were assigned to differentially expressed genes using the web-based agriGO software (http://bioinfo.cau.edu.cn/agriGO/analysis.php). SEA was used to compute enriched categories by comparing a list of differentially expressed genes to all expressed genes. According to the work by Benjamini and Yekutieli (2001), multiple comparison correction was performed, and FDR was controlled at ≤1%.
The MapMan bin classification system (Thimm et al., 2004) was performed for functional annotation of differentially expressed genes and subsequently visualized based on the functional annotated file ZmB73_5b_FGS_cds_2012 (http://mapman.gabipd.org). To determine if specific functional groups were overrepresented among the differentially expressed genes with reference to all genes represented in the whole genome, the expected number of genes for each functional category was calculated based on the distribution of functional categories among all genes on the genome. To determine if significantly more or fewer genes than expected were detected for each individual category, a χ2 test for independence with Yates’ continuity correction (Yates, 1934) was performed.
Cell-Specific Gene Expression Analysis by LCM
Fixation, Protection, and Embedding of Samples
Target cell types were isolated from frozen sections according to improved modified protocol from the work by Nakazono et al. (2003). Root segments from 5 to 10 mm from the root tip were dissected and fixed immediately in a freshly prepared mixture with 75% (v/v) ethanol and 25% (v/v) acetic acid by vacuum infiltration (300 MPa) for 15 min on ice, and afterward, they were swirled gently for 1 h on ice at 4°C. The solution was replaced with fresh fixative, and vacuum infiltration/swirl steps were repeated two times. To minimize the formation of ice crystals, the fixative was replaced by fresh 15% (w/v) Suc (Sigma-Aldrich) in 0.01 m diethyl pyrocarbonate (Sigma-Aldrich)-treated PBS buffer (137 mm NaCl, 8.01 mm Na2HPO4, 2.68 mm KCl, and 1.47 mm KH2PO4, pH 7.3) and infiltrated as described before. The 15% (w/v) Suc vacuum infiltration/swirl step was repeated again. Subsequently, fresh 40% (w/v) Suc (in 0.01 m diethyl pyrocarbonate-treated PBS buffer) was used to replace 15% Suc, and then, the vacuum infiltration/swirl step was conducted as described above and repeated with fresh 40% Suc solution three times. The root segments were embedded in 7 × 7 × 5-mm disposable molds (Polysciences Europe GmbH) with PolyFreeze-Clear Tissue Freezing Medium (Polysciences Europe GmbH). Then, molds with oriented segments were held close to the surface of liquid nitrogen until the surface freezing medium of molds froze. Samples were then immediately immersed into liquid nitrogen and stored at −80°C until cryosectioning. The procedure of fixation/protection/embedding was performed within 1 d.
Cryosectioning and Isolation of Pericycle Founder Cells by LCM
Before cryosectioning, the blocks with root segments were equilibrated in a precooled Cryostat at −28°C for about 15 min and then trimmed into 20-μm-thick sections using a CM1850 Cryotome (Leica) at −28°C. Serial frozen sections were mounted on PEN Membrane Slides (Carl Zeiss Microscopy) precoated with Poly-l-Lys (0.1% [w/v]; Sigma-Aldrich). Subsequently, sections were incubated in ice-cold 70% (v/v) ethanol for several minutes to reduce RNase activity. Before LCM, the freezing medium was removed, and sections were dehydrated. Sequential washes were performed in staining jars as follows: 50% (v/v) ethanol at room temperature (RT) for 5 min, 70% (v/v) ethanol at RT for 30 s, 95% (v/v) ethanol at RT for 30 s, absolute ethanol at RT for 1 min, and dehydration of samples for 2 min in xylene two times at RT. After dehydration, slides were air dried and then kept in a vacuum desiccator with silica gel (Sigma-Aldrich). Laser microdissection was performed with a PALM MicroBeam Platform (Carl Zeiss Microimaging; www.zeiss.com). At least 40 pericycle founder cells at the phloem poles were isolated from individual cross sections. In total, approximately 2,000 cells (40 cells × 50 sections) were collected per adhesive cap (Carl Zeiss Microscopy) within 1 h and pooled for downstream analyses.
Cell-Specific RNA Isolation and Linear Amplification for qRT-PCR
RNA was extracted from microdissected cells with 10 μL of lysis buffer from the Arcturus PicoPure Frozen RNA Isolation Kit (Life Technologies). Reaction tubes were incubated upside down on a 42°C block for 30 min; subsequently, they were briefly centrifuged at 800g for 2 min, and then, RNA extraction was performed according to the manufacturer’s protocol. RNA quality was evaluated using an RNA 6000 Pico LabChip Kit (Agilent Technologies) on an Agilent 2100 Bioanalyzer (Agilent Technologies). Then, total RNA (>3 ng) was linearly amplified by the RiboAmp HS PLUS RNA Amplification Kit (Applied Biosystems) according to the manufacturer’s instructions, and RNA quality was determined by an RNA 6000 Nano LabChip Kit on an Agilent 2100 Bioanalyzer (Agilent Technologies). cDNAs were synthesized from linear-amplified RNA using qScript cDNA SuperMix (Quanta Biosciences), and qRT-PCR was performed as described previously.
Raw sequencing data from this article are stored at the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP058076.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Early pericycle cell divisions in shoot-borne roots.
Supplemental Figure S2. Different stages of lateral root primordium formation are illustrated by representative images.
Supplemental Figure S3. Schematic overview of the components of the ubiquitin-dependent degradation system (SCFSKP2B complex) induced in response to local high nitrate.
Supplemental Figure S4. Inhibition of lateral root formation and auxin response by NPA treatment.
Supplemental Figure S5. MeIAA, IAN, OxIAA, IAA-Ala, and IAA-Glu concentrations in root tip, stele, and cortex of shoot-borne roots induced by homogeneous low and local high nitrate 12 and 24 h after treatment.
Supplemental Figure S6. Pericycle cells dividing at the phloem poles and illustration of LCM-microdissected cell types.
Supplemental Figure S7. RNA quality of LCM-dissected cell types evaluated by the RIN.
Supplemental Figure S8. Relative expression level of ZmPIN1c in three cell types related to lateral root initiation.
Supplemental Table S1. Comprehensively enriched GO terms obtained by SEAs among differentially expressed genes in pairwise comparisons between local high nitrate and homogeneous low nitrate treatments.
Supplemental Table S2. Determination of over- and underrepresented functional classes among differentially expressed genes (Fc ≥ 2; FDR ≤ 5%) that were functionally annotated by MapMan.
Supplemental Table S3. Gene list of the ubiquitin-dependent protein degradation pathway extracted from bin 29 of the MapMan analysis.
Supplemental Table S4. Gene list of cell cycle-related genes extracted from bin 31 of the MapMan analysis.
Supplemental Table S5. Internal standards used for the analysis of IAA and its metabolites by UPLC-ESI-Tandem Mass Spectrometry.
Supplemental Data Set S1. Overview of sample collection, RNA-Seq output, mapping results, and alignments to the B73 reference genome.
Supplemental Data Set S2. Complete list of 22,796 expressed genes.
Supplemental Text S1. Detailed procedure of MS data processing using TargetLynx V4.1 SCN 904.
Supplementary Material
Acknowledgments
We thank David Jackson (Cold Spring Harbor Laboratory) for sharing DR5::RFP transgenic maize seeds, Andreas Meyer (University of Bonn) for support in confocal laser-scanning microscopy, and Mikio Nakazono (Nagoya University) for stimulating discussions on the optimization of LCM.
Glossary
- cDNA
complementary DNA
- Fc
fold change
- FDR
false discovery rate
- GO
Gene Ontology
- IAA
indole-3-acetic acid
- IAN
indole-3-acetonitrile
- LCM
laser-capture microdissection
- MeIAA
methylindole-3-acetic acid
- MS
mass spectrometry
- NPA
1-N-naphthylphthalamic acid
- OxIAA
2-oxoindole-3-acetic acid
- PBS
phosphate-buffered saline
- qRT
quantitative reverse transcription
- RIN
RNA integrity number
- RNA-Seq
RNA sequencing
- RT
room temperature
- SEA
singular enrichment analysis
Footnotes
This work was supported by the Chinese Universities Scientific Fund (grant no. 2012YJ039 to P.Y.), the China Scholarship Council (Post-Graduate Study Abroad Program no. 201306350120 to P.Y.), the State Key Basic Research and Development Plan of China (grant no. 2013CB127402 to C.L.), the National Natural Science Foundation of China (grant no. 31272232 to C.L. and Innovative Group Grant no. 31421092 to C.L.), and the Deutsche Forschungsgemeinschaft (grants for root research to the laboratory of F.H.).
Articles can be viewed without a subscription.
References
- Araya T, Miyamoto M, Wibowo J, Suzuki A, Kojima S, Tsuchiya YN, Sawa S, Fukuda H, von Wirén N, Takahashi H (2014) CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc Natl Acad Sci USA 111: 2029–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band LR, Wells DM, Fozard JA, Ghetiu T, French AP, Pound MP, Wilson MH, Yu L, Li W, Hijazi HI, et al. (2014) Systems analysis of auxin transport in the Arabidopsis root apex. Plant Cell 26: 862–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman T, Burssens S, Inzé D (2001) The peri-cell-cycle in Arabidopsis. J Exp Bot 52: 403–411 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300 [Google Scholar]
- Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29: 1165–1188 [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Carraro N, Forestan C, Canova S, Traas J, Varotto S (2006) ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize. Plant Physiol 142: 254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inzé D, Sandberg G, Casero PJ, et al. (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida Engler J, De Veylder L, De Groodt R, Rombauts S, Boudolf V, De Meyer B, Hemerly A, Ferreira P, Beeckman T, Karimi M, et al. (2009) Systematic analysis of cell-cycle gene expression during Arabidopsis development. Plant J 59: 645–660 [DOI] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B. , Frei dit Frey N, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al (2007) Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690 [DOI] [PubMed] [Google Scholar]
- De Smet I, Vanneste S, Inzé D, Beeckman T (2006) Lateral root initiation or the birth of a new meristem. Plant Mol Biol 60: 871–887 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inzé D (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Pozo JC, Manzano C (2014) Auxin and the ubiquitin pathway. Two players-one target: the cell cycle in action. J Exp Bot 65: 2617–2632 [DOI] [PubMed] [Google Scholar]
- Dembinsky D, Woll K, Saleem M, Liu Y, Fu Y, Borsuk LA, Lamkemeyer T, Fladerer C, Madlung J, Barbazuk B, et al. (2007) Transcriptomic and proteomic analyses of pericycle cells of the maize primary root. Plant Physiol 145: 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Rost TL, Colón-Carmona A, Doerner P (2001) Early primordium morphogenesis during lateral root initiation in Arabidopsis thaliana. Planta 214: 30–36 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benková E (2008) Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci USA 105: 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahn A. (1990) Plant Anatomy, Ed 4 Pergamon Press, New York [Google Scholar]
- Forde BG. (2014) Nitrogen signalling pathways shaping root system architecture: an update. Curr Opin Plant Biol 21: 30–36 [DOI] [PubMed] [Google Scholar]
- Forestan C, Farinati S, Varotto S (2012) The maize PIN gene family of auxin transporters. Front Plant Sci 3: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestan C, Meda S, Varotto S (2010) ZmPIN1-mediated auxin transport is related to cellular differentiation during maize embryogenesis and endosperm development. Plant Physiol 152: 1373–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Benková E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jürgens G, et al. (2002a) AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K (2002b) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Yang Y, Schmidt RJ, Jackson D (2008) The relationship between auxin transport and maize branching. Plant Physiol 147: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl RF, Lima JE, von Wirén N (2012) Localized iron supply triggers lateral root elongation in Arabidopsis by altering the AUX1-mediated auxin distribution. Plant Cell 24: 33–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl RF, von Wirén N (2014) Root nutrient foraging. Plant Physiol 166: 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PJ, Atkinson CJ, Bengough AG, Else MA, Fernández-Fernández F, Harrison RJ, Schmidt S (2013) Contributions of roots and rootstocks to sustainable, intensified crop production. J Exp Bot 64: 1209–1222 [DOI] [PubMed] [Google Scholar]
- Guan P, Wang R, Nacry P, Breton G, Kay SA, Pruneda-Paz JL, Davani A, Crawford NM (2014) Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc Natl Acad Sci USA 111: 15267–15272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A. (2005) The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ 12: 1191–1197 [DOI] [PubMed] [Google Scholar]
- Hetz W, Hochholdinger F, Schwall M, Feix G (1996) Isolation and characterization of rtcs, a maize mutant deficient in the formation of nodal roots. Plant J 10: 845–857 [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Vuylsteke M, Vanneste S, Vercruysse S, Boucheron E, Alard P, Chriqui D, Van Montagu M, Inzé D, Beeckman T (2004) Transcript profiling of early lateral root initiation. Proc Natl Acad Sci USA 101: 5146–5151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F, Feix G (1998) Early post-embryonic root formation is specifically affected in the maize mutant lrt1. Plant J 16: 247–255 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Park WJ, Sauer M, Woll K (2004a) From weeds to crops: genetic analysis of root development in cereals. Trends Plant Sci 9: 42–48 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Tuberosa R (2009) Genetic and genomic dissection of maize root development and architecture. Curr Opin Plant Biol 12: 172–177 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Woll K, Sauer M, Dembinsky D (2004b) Genetic dissection of root formation in maize (Zea mays) reveals root-type specific developmental programmes. Ann Bot (Lond) 93: 359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D, De Veylder L (2006) Cell cycle regulation in plant development. Annu Rev Genet 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Jakoby MJ, Weinl C, Pusch S, Kuijt SJ, Merkle T, Dissmeyer N, Schnittger A (2006) Analysis of the subcellular localization, function, and proteolytic control of the Arabidopsis cyclin-dependent kinase inhibitor ICK1/KRP1. Plant Physiol 141: 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen L, Hollunder J, Roberts I, Forestan C, Fonteyne P, Van Quickenborne C, Zhen RG, McKersie B, Parizot B, Beeckman T (2013a) Comparative transcriptomics as a tool for the identification of root branching genes in maize. Plant Biotechnol J 11: 1092–1102 [DOI] [PubMed] [Google Scholar]
- Jansen L, Parizot B, Beeckman T (2013b) Inducible system for lateral roots in Arabidopsis thaliana and maize. Methods Mol Biol 959: 149–158 [DOI] [PubMed] [Google Scholar]
- Jansen L, Roberts I, De Rycke R, Beeckman T (2012) Phloem-associated auxin response maxima determine radial positioning of lateral roots in maize. Philos Trans R Soc Lond B Biol Sci 367: 1525–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen DA. (1940) Plant Microtechnique. McGraw-Hill, New York [Google Scholar]
- Johnston R, Wang M, Sun Q, Sylvester AW, Hake S, Scanlon MJ (2014) Transcriptomic analyses indicate that maize ligule development recapitulates gene expression patterns that occur during lateral organ initiation. Plant Cell 26: 4718–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima M, Kamada-Nobusada T, Komatsu H, Takei K, Kuroha T, Mizutani M, Ashikari M, Ueguchi-Tanaka M, Matsuoka M, Suzuki K, et al. (2009) Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol 50: 1201–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Ruffel S, Gutiérrez RA, Gojon A, Crawford NM, Coruzzi GM, Lacombe B (2011) A framework integrating plant growth with hormones and nutrients. Trends Plant Sci 16: 178–182 [DOI] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CA, Hogeweg P, Marée AF, Scheres B (2008) Root system architecture from coupling cell shape to auxin transport. PLoS Biol 6: e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Roberts I, Guyomarc’h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L (2013) Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci 18: 450–458 [DOI] [PubMed] [Google Scholar]
- Lee J, Das A, Yamaguchi M, Hashimoto J, Tsutsumi N, Uchimiya H, Umeda M (2003) Cell cycle function of a rice B2-type cyclin interacting with a B-type cyclin-dependent kinase. Plant J 34: 417–425 [DOI] [PubMed] [Google Scholar]
- Lim J, Helariutta Y, Specht CD, Jung J, Sims L, Bruce WB, Diehn S, Benfey PN (2000) Molecular analysis of the SCARECROW gene in maize reveals a common basis for radial patterning in diverse meristems. Plant Cell 12: 1307–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima JE, Kojima S, Takahashi H, von Wirén N (2010) Ammonium triggers lateral root branching in Arabidopsis in an AMMONIUM TRANSPORTER1;3-dependent manner. Plant Cell 22: 3621–3633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP. (2014) Root phenes that reduce the metabolic costs of soil exploration: opportunities for 21st century agriculture. Plant Cell Environ 10.1111/pce.12451 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Manzano C, Ramirez-Parra E, Casimiro I, Otero S, Desvoyes B, De Rybel B, Beeckman T, Casero P, Gutierrez CC, Del Pozo J (2012) Auxin and epigenetic regulation of SKP2B, an F-box that represses lateral root formation. Plant Physiol 160: 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavý P, Vanstraelen M, De Rybel B, Zhaojun D, Bennett MJ, Beeckman T, Benková E (2013) Auxin reflux between the endodermis and pericycle promotes lateral root initiation. EMBO J 32: 149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister R, Rajani MS, Ruzicka D, Schachtman DP (2014) Challenges of modifying root traits in crops for agriculture. Trends Plant Sci 19: 779–788 [DOI] [PubMed] [Google Scholar]
- Nakazono M, Qiu F, Borsuk LA, Schnable PS (2003) Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissues of maize. Plant Cell 15: 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman-Ligeza B, Parizot B, Gantet PP, Beeckman T, Bennett MJ, Draye X (2013) Post-embryonic root organogenesis in cereals: branching out from model plants. Trends Plant Sci 18: 459–467 [DOI] [PubMed] [Google Scholar]
- Osmont KS, Sibout R, Hardtke CS (2007) Hidden branches: developments in root system architecture. Annu Rev Plant Biol 58: 93–113 [DOI] [PubMed] [Google Scholar]
- Paponov IA, Teale WD, Trebar M, Blilou I, Palme K (2005) The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci 10: 170–177 [DOI] [PubMed] [Google Scholar]
- Ren H, Santner A, del Pozo JC, Murray JA, Estelle M (2008) Degradation of the cyclin-dependent kinase inhibitor KRP1 is regulated by two different ubiquitin E3 ligases. Plant J 53: 705–716 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers ED, Benfey PN (2015) Regulation of plant root system architecture: implications for crop advancement. Curr Opin Biotechnol 32: 93–98 [DOI] [PubMed] [Google Scholar]
- Rubio V, Bustos R, Irigoyen ML, Cardona-López X, Rojas-Triana M, Paz-Ares J (2009) Plant hormones and nutrient signaling. Plant Mol Biol 69: 361–373 [DOI] [PubMed] [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM (2011) Nitrogen economics of root foraging: transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc Natl Acad Sci USA 108: 18524–18529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymen B, Fiorani F, Kartal F, Vandepoele K, Inzé D, Beemster GTS (2007) Cold nights impair leaf growth and cell cycle progression in maize through transcriptional changes of cell cycle genes. Plant Physiol 143: 1429–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Dante RA, Nguyen HN, Gordon-Kamm WJ, Larkins BA (2014) Expression, regulation and activity of a B2-type cyclin in mitotic and endoreduplicating maize endosperm. Front Plant Sci 5: 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini S, Sharma I, Kaur N, Pati PK (2013) Auxin: a master regulator in plant root development. Plant Cell Rep 32: 741–757 [DOI] [PubMed] [Google Scholar]
- Saleem M, Lamkemeyer T, Schützenmeister A, Fladerer C, Piepho HP, Nordheim A, Hochholdinger F (2009) Tissue specific control of the maize (Zea mays L.) embryo, cortical parenchyma, and stele proteomes by RUM1 which regulates seminal and lateral root initiation. J Proteome Res 8: 2285–2297 [DOI] [PubMed] [Google Scholar]
- Seo M, Jikumaru Y, Kamiya Y (2011) Profiling of hormones and related metabolites in seed dormancy and germination studies. Methods Mol Biol 773: 99–111 [DOI] [PubMed] [Google Scholar]
- Smith S, De Smet I (2012) Root system architecture: insights from Arabidopsis and cereal crops. Philos Trans R Soc Lond B Biol Sci 367: 1441–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Vanneste S, Friml J (2009) Auxin: a trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Verkest A, Manes CLDO, Vercruysse S, Maes S, Van Der Schueren E, Beeckman T, Genschik P, Kuiper M, Inzé D, De Veylder L (2005a) The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17: 1723–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A, Weinl C, Inzé D, De Veylder L, Schnittger A (2005b) Switching the cell cycle: Kip-related proteins in plant cell cycle control. Plant Physiol 139: 1099–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villiers F, Kwak JM (2012) Comparative genomics and molecular characterization of the maize PIN family proteins. Front Plant Sci 3: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Behrens I, Komatsu M, Zhang Y, Berendzen KW, Niu X, Sakai H, Taramino G, Hochholdinger F (2011) Rootless with undetectable meristem 1 encodes a monocot-specific AUX/IAA protein that controls embryonic seminal and post-embryonic lateral root initiation in maize. Plant J 66: 341–353 [DOI] [PubMed] [Google Scholar]
- Wang JR, Hu H, Wang GH, Li J, Chen JY, Wu P (2009) Expression of PIN genes in rice (Oryza sativa L.): tissue specificity and regulation by hormones. Mol Plant 2: 823–831 [DOI] [PubMed] [Google Scholar]
- Woll K, Borsuk LA, Stransky H, Nettleton D, Schnable PS, Hochholdinger F (2005) Isolation, characterization, and pericycle-specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1. Plant Physiol 139: 1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates F. (1934) Contingency tables involving small numbers and the χ2 test. Suppl J Roy Stat Soc 1: 217–235 [Google Scholar]
- Yu P, Li X, Yuan L, Li C (2014a) A novel morphological response of maize (Zea mays) adult roots to heterogeneous nitrate supply revealed by a split-root experiment. Physiol Plant 150: 133–144 [DOI] [PubMed] [Google Scholar]
- Yu P, White PJ, Hochholdinger F, Li C (2014b) Phenotypic plasticity of the maize root system in response to heterogeneous nitrogen availability. Planta 240: 667–678 [DOI] [PubMed] [Google Scholar]
- Zhang H, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Paschold A, Marcon C, Liu S, Tai H, Nestler J, Yeh CT, Opitz N, Lanz C, Schnable PS, et al. (2014) The Aux/IAA gene rum1 involved in seminal and lateral root formation controls vascular patterning in maize (Zea mays L.) primary roots. J Exp Bot 65: 4919–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.