A gibberellin oxidase exposes the regulatory association between gibberellic acid and cellulose biosynthesis during cell expansion.
Abstract
Here, we show a mechanism for expansion regulation through mutations in the green revolution gene gibberellin20 (GA20)-oxidase and show that GAs control biosynthesis of the plants main structural polymer cellulose. Within a 12,000 mutagenized Sorghum bicolor plant population, we identified a single cellulose-deficient and male gametophyte-dysfunctional mutant named dwarf1-1 (dwf1-1). Through the Sorghum propinquum male/dwf1-1 female F2 population, we mapped dwf1-1 to a frameshift in GA20-oxidase. Assessment of GAs in dwf1-1 revealed ablation of GA. GA ablation was antagonistic to the expression of three specific cellulose synthase genes resulting in cellulose deficiency and growth dwarfism, which were complemented by exogenous bioactive gibberellic acid application. Using quantitative polymerase chain reaction, we found that GA was positively regulating the expression of a subset of specific cellulose synthase genes. To cross reference data from our mapped Sorghum sp. allele with another monocotyledonous plant, a series of rice (Oryza sativa) mutants involved in GA biosynthesis and signaling were isolated, and these too displayed cellulose deficit. Taken together, data support a model whereby suppressed expansion in green revolution GA genes involves regulation of cellulose biosynthesis.
In all higher plants, development of upright plant growth is advanced by a highly regulated process of cell division, cell fate determination, and cell expansion (Xie et al., 2011). Cell shape and morphogenesis are largely acquired through anisotropic expansion, during which the plant cell wall provides the structural integrity needed to resist internal turgor pressure and simultaneously extend in a controlled and organized manner (Cosgrove and Jarvis, 2012). Cellulose is the main load-bearing component on the plant cell wall, and cellulose is synthesized in a highly organized manner according to the expansion state and growth pattern (Tsekos, 1999). A number of phytohormones are involved in regulating expansion, including auxin (Paque et al., 2014), brassinosteroid (Xie et al., 2011), the rapid alkalinization factor peptide hormone (Haruta et al., 2014), cytokinin (Downes and Crowell, 1998), and GAs (Keyes et al., 1989). A complex and still poorly understood interplay exists among phytohormones to precisely regulate cellular expansion machinery.
Sorghum bicolor is one of the most agriculturally important grains produced worldwide (Paterson, 2008). It is largely cultivated in Africa, India, China, and the United States for both grain and biomass. An advantage that Sorghum sp. holds over many other grain crops is drought tolerance, an important trait in low-rainfall conditions and anticipated climate change shifts (Schmidhuber and Tubiello, 2007). The Sorghum sp. genome (730 Mb; Paterson et al., 2009) is much smaller than maize (Zea mays; 2,500 Mb) and most grasses (e.g. wheat [Triticum aestivum], 15,000 Mb; barley [Hordeum vulgare], 5,100 Mb), and genetic tools are available (Paterson et al., 2009; Liu and Godwin, 2012). Here, we aimed to isolate a cellulose-deficient mutant from within a chemical mutagenesis population, which we named dwarf1-1 (dwf1-1). From here, we aimed to characterize the genetic mutation underlying severe cellulose deficiency and aberrant expansion in a dwf1-1. Most cellulose-deficient mutants have been mapped to cellulose synthase (CESA) genes (Arioli et al., 1998; Taylor et al., 2003; Sethaphong et al., 2013). dwf1-1 did not map to a CESA gene but rather, mapped to a GA20-oxidase involved in GA biosynthesis. We then sought to explore the feedback between dwarfism and cellulose deficiency and used rice (Oryza sativa), because many of the GA biosynthetic mutants are characterized as green revolution genes. Chemical inhibition, exogenous GA complementation, transcriptional analyses, and phenotypic data were used to support our findings.
RESULTS
dwf1-1 Displays Dwarfed Morphology and Reduced Cellulose Content
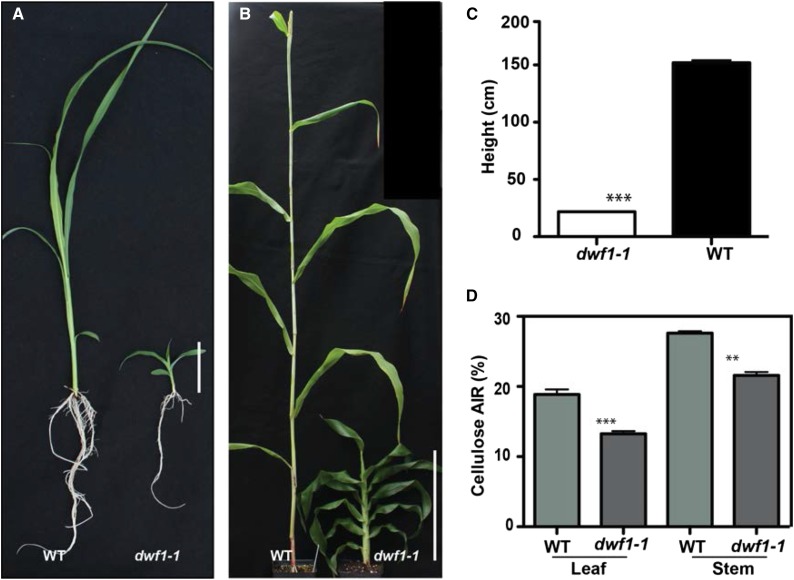
The Sorghum sp. dwf1-1 mutant arose from an M2 mutagenesis population that we developed (as described in “Materials and Methods”). The mutant was immediately identifiable compared with the wild type because of a lack of expansion and extremely dwarfed morphology (Fig. 1, A and B). Expansion defects and dwarfed plant form persisted until reaching maturity (Fig. 1B). The mutant displayed a reduction in total plant height (Fig. 1C) equivalent to one-tenth the size of mature wild-type plants. Cellulose content was determined in M2 dwf1-1 plants and found to be around 30% lower than that in the wild type. Leaf and stem cellulose content was measured and found to be significantly lower than that in the wild type in both phenological growth stages assessed (Fig. 1D; P < 0.001 and P < 0.01, Student’s t test).
Figure 1.
dwf1-1 displays dwarfed plant form and cellulose deficit. A, Wild-type (WT) and dwf1-1 plants grown in natural light conditions for 2 weeks in the greenhouse (24°C). Bar = 5 cm. B, The fully expanded mature wild type and dwf1-1. Bar = 25 cm. C, Height measurement between dwf1-1 and the wild type evaluated at maturity (n = 100). D, Cellulose content was measured as a proportion of AIR and determined on leaves and stems of dwf1-1 and the wild type (n = 4; error bars indicate sem). Asterisks indicate significance by Student’s t test. **, P = 0.01; ***, P = 0.001.
Infertile Pollen in dwf1-1 and Inheritance Analysis Using Wild-Type Pollen
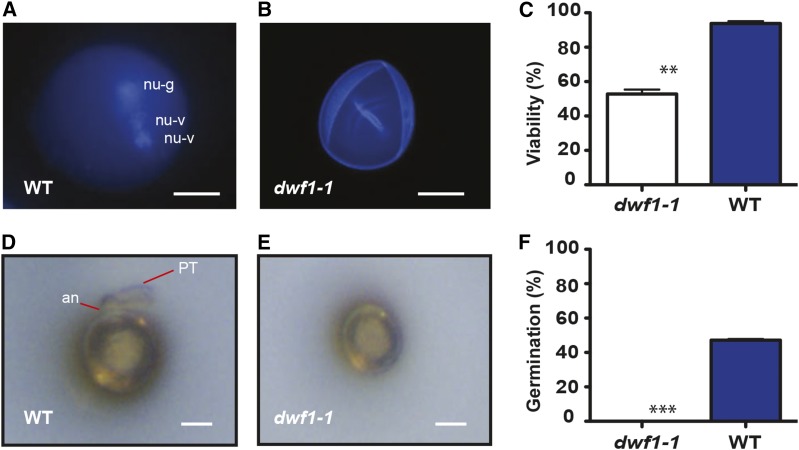
Inflorescence from dwf1-1 was unable to produce seed. A plausible explanation was that severe cellulose-deficient mutants, such as Arabidopsis (Arabidopsis thaliana) cesa1 and Atcesa3, were determined to be male gametophytic lethal (Persson et al., 2007), and this may also be the case for dwf1-1. We therefore characterized pollen viability and germination (Fig. 2). It was found that the dwf1-1 pollen did not germinate compared with robust germination in the wild type (Fig. 2, D–F). Germination in the wild type was approximately 45%, and this rate is in agreement with previous reports indicating the germination rates in Sorghum sp. (Lansac et al., 1994; Tuinstra and Wedel, 2000; Burke et al., 2007). Additional histochemical staining with aniline blue, 4′,6-diamidino-2-phenylindole, and Fluorescein Diacetate also showed differences in the health of dwf1-1 pollen. Approximately one-half of the dwf1-1 pollen grains were found to be nonviable according to a modified Alexander stain method (Peterson et al., 2010; Fig. 2, A–C). To test for female gametophyte fertility, we crossed wild-type pollen onto the stigma of dwf1-1, and the resulting cross was fertile and capable of producing viable seed.
Figure 2.
Pollen viability and germination are dysfunctional in dwf1-1. A, Example of wild-type (WT) pollen that has been stained with 4′,6-diamidino-phenylindole to show the three-nucleated pollen grain and spherical shape. nu-g, Germinative nuclei; nu-v, vegetative nuclei. Bar = 10 μm. B, Representative example of dwf1-1 pollen highlighting internally collapsed surface, absence of nuclei, and aberrant spherical shape. Bar = 10 μm. C, Viability of pollen grains determined by a modified Alexander stain (n = 100; error bar is sem of three technical replicates). Asterisks indicate significance by Student’s t test. **, P = 0.01. Light micrographs of pollen germination determined in the wild type (D) and dwf1-1 (E). In the wild type, evidence of the protruding annulus (an) and the germinating pollen tube (PT) is noted, but they are absent in dwf1-1. Bar = 10 μm. F, Germination rate determined as the percentage of pollen that developed the pollen tube (error bars represent sem; n = 3 independent replications [100 counts each]). Asterisks indicate significant difference. ***, P = 0.001.
For trait inheritance analysis, pollen from the wild type was used and crossed into dwf1-1. Because the mutant arose from the M2, we anticipated it to be a recessive trait. This was quantitatively verified in the F2 dwf1-1 and wild-type cross. Here, 440 seedlings were assessed, and it was determined that 346 were phenotypically equivalent to the wild type and that 94 were dwf1-1, which was statistically a 3:1 ratio (χ2 = 1.634, degrees of freedom = 1, P > 0.05). Additional populations were analyzed, and they verified a consistent 3:1 inheritance of the trait. Thus, we observed dwf1-1 inheritance that was consistent with a single recessive gene inherited in a Mendelian manner.
The dwf1-1 Mutation Maps to a GA20-oxidase and Not to a CESA Gene
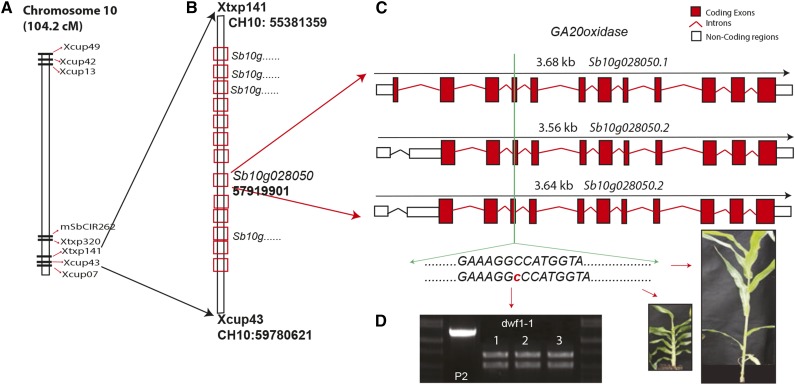
The parental lines used for map-based cloning were dwf1-1 (parent 1) as the female receptor and wild-type species Sorghum propinquum (parent 2) as the pollen donor, and the segregating population in the F2 generation was assessed. Genetic markers for S. bicolor × S. propinquum were derived from the works by Bhattramakki et al. (2000) and Billot et al. (2013). Bulked segregant analysis allowed for a linkage of dwf1-1 to the lower arm of chromosome 10. Additional markers from chromosome 10 were obtained (Xu et al., 1995; Bhattramakki et al., 2000; Kong et al., 2000, 2013; Menz et al., 2002; Billot et al., 2013), and we determined that the mutation was associated to a region approximately 8-cm long between the genome landmark of Xtxp141 and Xcup3 (Fig. 3, A and B). Selected Cleaved Amplified Polymorphic Sequences (CAPS) markers and chromosome walking linked the mutation to a group of GA20-oxidases. Cloning and sequencing (all primers in Supplemental Table S1) of the GA20-oxidase identified a frameshift mutation, which caused the loss of 60 amino acids within the first splice variant and 40 amino acids in the remaining two alternative splicing forms of the gene (Fig. 3C; Sb10g28050). CAPS marker within the region containing the mutation verified the mapping population, with all but the parental S. propinquum containing the mutation (Fig. 3D).
Figure 3.
Map-based cloning of dwf1-1. A, Physical map of chromosome 10 with relevant SSR markers (Kong et al., 2013). B, Genomic region between SSR markers Xtxp141 and Xcup43 with the location of the genetic locus for the GA20-oxidase (Sb10g028050). C, The three alternative gene models (splice variant) determined for Sb10g028050 with the position of the frameshift mutation highlighted. D, CAPS marker for the area containing the mutation observed in the dwf1-1 population (examples 1–3) but not in wild-type S. propinquum (P2; plant specimen indicated by red arrows).
Undetectable Levels of GAs in dwf1-1
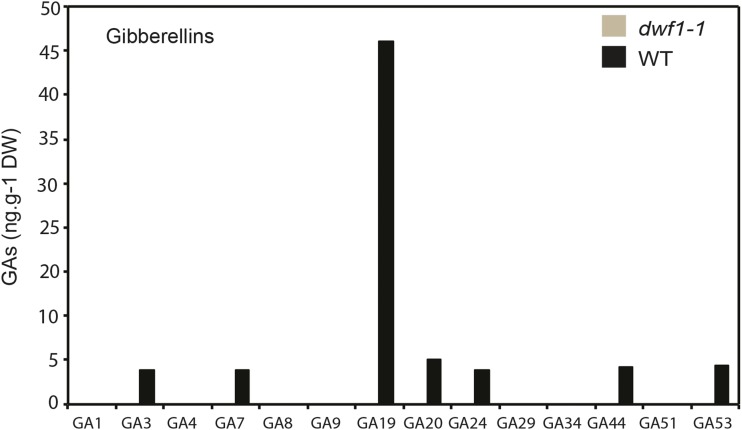
A metabolic investigation of plant hormones was performed (Plant Hormone Analysis Laboratory; National Research Council of Canada). Numerous GAs were present in wild-type tissue, including GA3, GA7, GA19, GA20, GA24, GA44, and GA53 (Fig. 4). By contrast, no detectable GAs were measured in dwf1-1 (Fig. 4). These data implied that the early 13 hydroxylation pathway conducive to formation of bioactive GA1 and GA8 (not bioactive GA; GA53 → GA44 → GA19 → GA20 → GA29 → GA1 → GA8) was dysfunctional in dwf1-1. In the assessment of hormones (Plant Hormone Analysis Laboratory; National Research Council of Canada), GA12 was not included, and GA53 was detected at 4 ng g−1 dry weight of wild-type tissue, which was not represented graphically (Fig. 4). The absence of any significant levels of GAs in the dwf1-1 was consistent with a metabolic block in the biosynthetic pathway and the genetic dysfunction in the GA pathway. Additionally, sampling time may have been important to reveal the abundance of different GAs. For example, in our analysis, we did not detect GA1, GA4, or GA8, but the precursors were evident in wild-type tissue: GA19 to GA20 to GA4 to GA1. GAs are highly regulated by light and photoperiodism, and a prior study (Lee et al., 1998) showed that the highest level of GA53 corresponds to the lowest levels of GA1 and GA8. It was, therefore, plausible that our sampling focused on a proportionally high point in production of GA53 and GA19, which correspondingly overlapped with a low sampling point for GA1 and GA8. Importantly, transcriptional analysis of early genes in GA biosynthesis (stages 1 and 2; Hedden and Kamiya, 1997; Hedden and Phillips, 2000) revealed a lack of differential expression (Supplemental Fig. S1). These data were consistent with the downstream position of GA20-oxidase in the GA pathway. Other hormones analyzed (auxins and cytokinins) failed to display notable differences in accumulation (Supplemental Fig. S2). Taken together, data were consistent with a nonredundant role for the GA20-oxidase encoded by dwf1-1 in biosynthesis of GA.
Figure 4.
Analyses of GAs. dwf1-1 and the wild type grown were examined for the occurrence of 12 key metabolic GAs. No GAs were detected in the dwf1-1, whereas the main GAs in the 13 early hydroxylation pathway were quantified for the wild type (WT). DW, Dry weight.
dwf1-1 Phenotype Is Rescued by the Application of Exogenous GA3
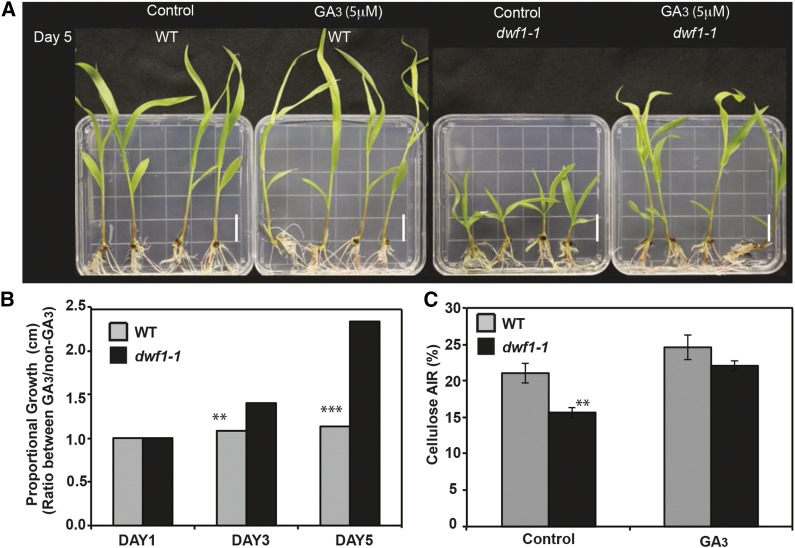
In prior studies, phenotypes linked to GA deficit could be complemented with exogenous GA (Hedden and Phillips, 2000). Furthermore, recent chemical complementation experiments of GA mutants in Sorghum sp. used application of exogenous GA3 (Ordonio et al., 2014), consistent with the work by Lee et al. (1998). Therefore, we hypothesized that the genetic block leading to no bioactive GAs in dwf1-1 could be chemically complemented by application of exogenous GA3. Importantly, this was a two-tiered question to establish whether either or both (1) cellulose deficiency and (2) dwarf phenotype could be complemented using GA3 applied to dwf1-1. Because of male gametophytic lethality, the homozygous dwf1-1 seedlings were selected from a segregating F2 population. Upon transplanting homozygous dwf1-1 to GA3-supplemented media, the dwarfed growth phenotype was fully rescued (Fig. 5, A and B). On a timescale, within 2 d of GA3 application, the dwf1-1 (height) grew 60% in length compared with the dwf1-1-control. Wild-type plants also reached a greater height (7.6% increase) compared with the untreated wild-type control. After 5 d of exposure, wild-type plants exposed to GA3 displayed a 12% increase compared with untreated controls. However, the dwf1-1 was 120% longer than dwf1-1 controls after 5 d and phenotypically indistinguishable from the wild type. These data support chemical complementation of the dwf1-1 by GA3, which is in agreement with the lack of GAs. Also, cellulose deficit in dwf1-1 was rescued with exogenous GA3. Here, after 7 d exposure to 5 µm GA3, the cellulose content in dwf1-1 was almost identical to that in the wild type, and although marginally lower, it was found not to be significantly different (P = 0.1246, Student’s t test; Fig. 5C). Thus, cellulose and dwarfism phenotypes in dwf1-1 were linked to a GA deficit as expected from the genetic linkage data.
Figure 5.
Chemical complementation of dwf1-1. A, Visual assessment of dwf1-1 and the wild type (WT) grown for 5 d in media supplemented with exogenous GA3. Bar = 1 cm. B, Proportional growth observed on the GA3- versus the non-GA3-treated wild type and dwf1-1. Proportions are presented. Statistical assessment was performed on expansion capacity of aerial plant tissue with n = 4 with five technical replicates. C, Bars indicate cellulose content (as a proportion of AIR). Control is untreated compared with the GA3-treated wild-type and dwf1-1 samples (n = 4 with five technical replicates; error bars represent sem). Asterisks indicate significance by Student’s t test. **, P = 0.01; ***, P = 0.001.
In addition to dwarfism and cellulose deficiency, the dwf1-1 line was characterized by male sterility. Therefore, we sought to determine if the application of GA3 could revert male infertility. After GA3 application, it was found that the inflorescences were able to set seeds, albeit in a small percentage (5%–10%). The plantlets arising from seeds were phenotypically equivalent to the maternal dwf1-1 line.
GA Inhibition Reduces Cellulose Synthesis in the Wild Type But Not in dwf1-1
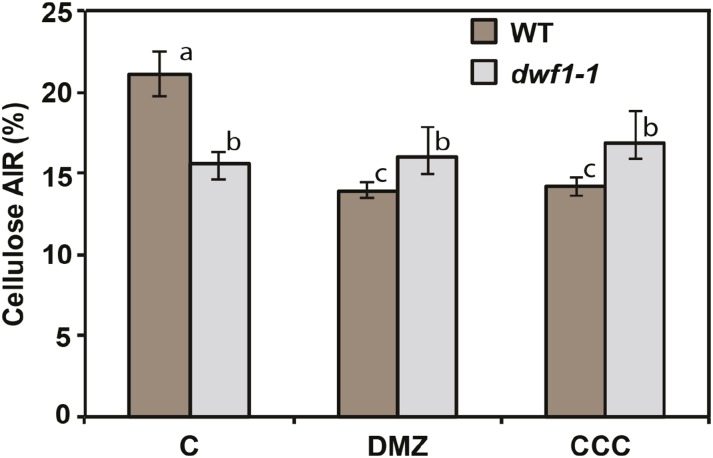
By theory, pharmacological inhibition of GA biosynthesis should phenocopy genetic dysfunction with respect to causing reduced cellulose synthesis. This experiment was also important to pharmacologically mimic the dwf1-1 mutant. To test this, after plants were sufficiently past seedling emergence stage and the first true leaves began to expand, they were transplanted to media supplemented with 0.2 or 0.4 mm chlorocholine chloride (CCC) and 0.2 or 0.4 mm daminozide (DMZ), both inhibitors of GA biosynthesis. In the wild type, a substantial drop in cellulose was observed after inhibition of GA (Fig. 6; P = 0.001), consistent with the genetic data earlier presented. By contrast, in the dwf1-1 mutant background, there was no significant change in the cellulose content after pharmacological inhibition of GA (Fig. 6). Therefore, GA regulation, both genetically and pharmacologically, may account for a significant capacity of cellulose biosynthesis in Sorghum sp. However, our observations do not exclude contributions to cellulose regulation from other mechanisms.
Figure 6.
Pharmacological inhibition of GA causes cellulose deficit. The result of applying two different GA inhibitors (DMZ and CCC) was assessed compared with untreated controls (C). Cellulose content was measured as the proportion of AIR in wild-type (WT) and dwf1-1 plantlets (aerial tissue) after 10 d of exposure compared with C. ANOVA was used to established significance between and across groups (n = 4 with five technical replicates). Different letters indicate significance established at P ≤ 0.05.
Rice GA Mutants Phenocopy Sorghum sp. dwf1-1 in Cellulose Deficiency
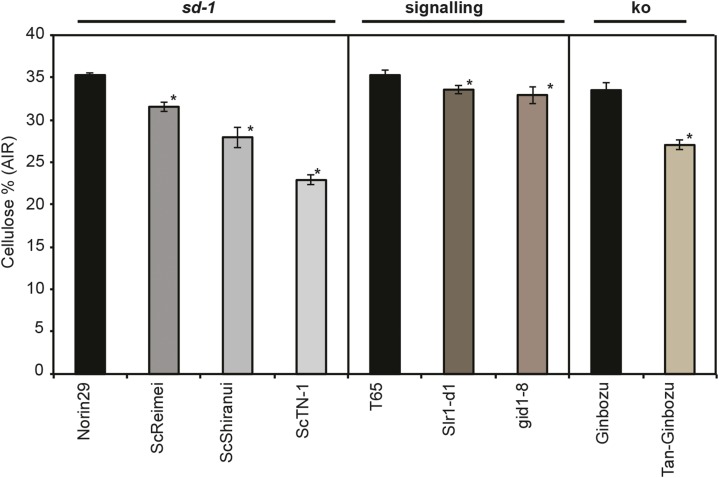
In lieu of our map-based cloning of a single mutant allele identified from within our 12,000 Sorghum sp. population and the difficulties in transformation of the parental sweet Sorghum sp. variety for single-gene complementation, we sought to identify alleles in another monocotyledonous species: rice. To evaluate how GA dysfunction impacted cellulose biosynthesis, six GA-related mutant lines with semidwarf or dwarf phenotypes were examined (Fig. 7). Two groups of semidwarf mutants defective in GA biosynthesis were investigated. Mutant alleles of semidwarf-1 (sd-1; ga20-oxidase), including two sd-1 intermediate alleles (scReimei and scShiranui) and one sd-1 null allele (scTN-1; Xia et al., 1991; Ogi et al., 1993; Irie et al., 2008), were grown alongside and compared with their parent wild-type Norin 29 (Fig. 7). Here, scReimei is the weakest allele, and scTN-1 is the most severe (Okuno et al., 2014). Corresponding, mutations all induced a reduction in cellulose biosynthesis (milligrams per gram cell wall acid-insoluble residue [AIR]).
Figure 7.
Analysis of cellulose biosynthesis in rice semidwarf varieties with GA dysfunction. The cellulose content of AIR from rice seedlings with genetic mutations in the GA pathway was determined on 10-d-old seedlings (10 d postgermination). For cellulose analysis, three composite biological replicates consisting of five seedlings each were used along with three technical replicates. Error bars indicate sem of three replicates. *, Significance (P = 0.01) using a Student’s t test from the mean of the appropriate control.
To further explore the link between GA and cellulose biosynthesis, we examined cellulose content in a mild mutation in ent-kaurene oxidase (KO; Tan-Ginbozu), which catalyzed an early step in GA biosynthesis (Itoh et al., 2004; Okuno et al., 2014) compared with its parental Ginbozu. This class of mutation in the GA biosynthetic pathway also resulted in less cellulose content (milligrams per gram AIR) than that in parental plants (Fig. 7).
We questioned whether dysfunctional GA signaling may also cause reduced cellulose content. Therefore, we examined mutations in SLENDER1 (SLR1) and GIBBERELLIN INSENSITIVE DWARF1 (GID1). SLR1-d1 is a mutant encoding a constitutively active form of a suppressor protein for GA signaling: SLR1 (Ueguchi-Tanaka et al., 2007; Asano et al., 2009). By contrast, GID1 encodes a soluble GA receptor that positively regulates GA signaling (Ueguchi-Tanaka et al., 2007), and thus, the above two mutants both show suppressed GA signaling. Interestingly, the reduction in cellulose content was modest and relatively consistent for GA signaling mutants compared with more severe GA biosynthetic mutants (Fig. 7).
Specific CESA Transcripts Were Down-Regulated in dwf1-1 Mutant and Conversely Activated by Exogenous GA Application, Suggesting Regulatory Specificity
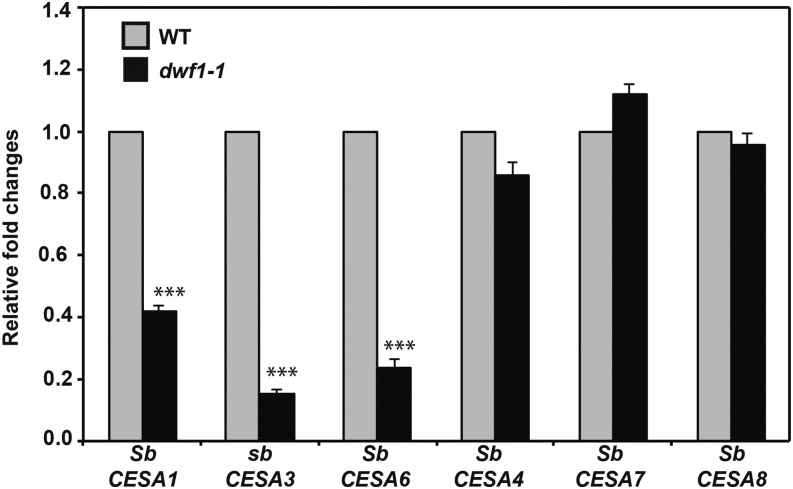
CESA genes were bioinformatically annotated based on prior phylogenetic studies that included Arabidopsis, setaria (Setaria viridis), and maize along with Sorghum sp. (Petti et al., 2013). Based on bioinformatic data available, we annotated a total of eight Sorghum sp. genes as CESA; they were Sb09g005280, Sb03g004310, Sb01g004210, Sb02g006290, Sb02g007810, Sb01g019720, Sb02g025020, and Sb02g010110. Little is known about specific properties of the CESA complex in Sorghum sp., but by contrast, we know that primary and secondary cell wall CESA complexes acquire a heterotrimeric configuration in Arabidopsis (Desprez et al., 2007; Persson et al., 2007) with a probable stoichiometry of 1:1:1 (Gonneau et al., 2014), which displays coexpression at the transcriptional level (Brown et al., 2005; Persson et al., 2005). Although we cannot safely overlay data from the Arabidopsis system on monocotyledons because of notable differences in vascular development and physiological adaptations, we examined homology to functionally characterized CESA from Arabidopsis with the Sorghum sp. accessions. Results were Sb09g005280 (CESA1-like or CESA10-like), Sb03g004310 (CESA1-like or CESA10-like), Sb01g004210 (CESA3-like), Sb02g006290 (CESA3-like), Sb02g007810 (CESA6-like), Sb01g019720 (CESA4-like), Sb02g025020 (CESA7-like), and Sb02g010110 (CESA81-like). For our study, we sought to ask whether any differential expression occurred in dwf1-1. The transcripts that we annotated as primary cell wall-related Sb09g005280 (CESA1-like or CESA10-like), Sb01g004210 (CESA3-like), and Sb02g007810 (CESA6-like) were differentially down-regulated in a similar manner (Fig. 8; P < 0.001). All other genes were not differentially expressed. These included putative CESA genes annotated as most homologous to the Arabidopsis cluster of three secondary cell wall CESA genes (CESA4, CESA7, and CESA8; P > 0.05; Fig. 8). Although this does not rule out GA-mediated regulation of secondary cell wall CESAs, it does illustrate a clustering of putative CESA genes that were down-regulated in the GA-deficient dwf1-1. To query transcriptional regulatory features, we examined the promoter region (4,000 bp upstream) of all CESA genes for the presence of cis-trans acting elements, particularly those related to hormone response in prior studies. The promoters of both primary and secondary CESA genes contained putative GA-, cytokinin-, and auxin-responsive elements (Supplemental Table S2). No clear pattern was evident, suggesting that additional work will be needed to decipher the transcriptional regulatory machinery involved.
Figure 8.
Transcriptional analysis of Sorghum sp. CESAs. The relative expressions of primary and secondary cell wall SbCESA genes (Petti et al., 2013) were determined by quantitative reverse transcription-PCR in 14-d-old plantlets (aerial tissue) from the wild type (WT) and dwf1-1. Error bars indicate sem of three replicates. ***, Significance (P = 0.001).
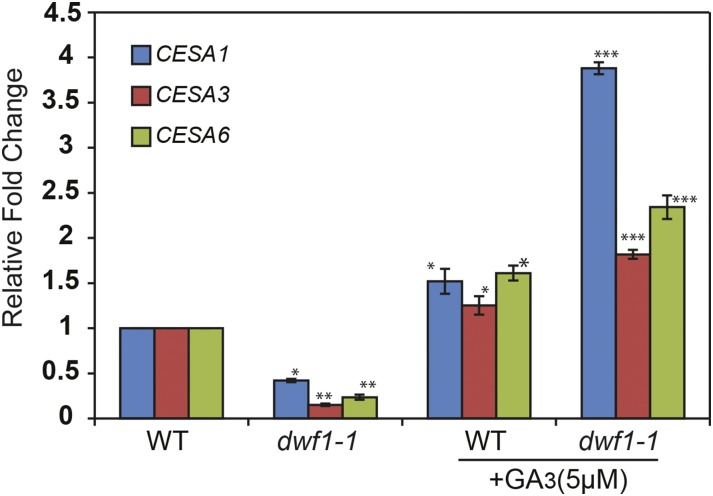
To advance the question of whether GA was capable of positively regulating CESA transcription, we quantified transcript abundance for the CESA genes that were differentially down-regulated in dwf1-1 in wild-type Sorghum sp. after the addition of exogenous bioactive GA3. The experiment was performed by exogenously applying GA3 to 10-d-old light-grown (16-h-light/8-h-dark cycle) Sorghum sp. seedlings for 1 h. Normalized against wild-type expression, the addition of GA3 caused modest but significant increased gene expression for all three putative CESA genes (Fig. 9). In the dwf1-1 mutant, we also examined gene expression before and after the addition of GA3. In contrast with data from wild-type plants, the expression of CESA genes in dwf1-1 was highly increased (5-fold) after GA3 addition (Fig. 9).
Figure 9.
Transcriptional analysis of Sorghum sp. CESAs after GA application. The relative expressions of primary cell wall SbCESA genes were determined by quantitative reverse transcription-PCR in 7- to 10-d-old plantlets (aerial tissue) from the wild type (WT) and dwf1-1 with and without GA3 addition. Error bars indicate sem of three replicates. Asterisks indicate significant difference. *, P = 0.05; **, P = 0.01; ***, P = 0001.
DISCUSSION
Our forward genetics screen was in Sorghum sp. because of prior genetic screens for altered cellulose being performed in Arabidopsis. There are prominent differences between dicot and monocot cell walls. Arabidopsis has a type I cell wall, whereas most economically important biomass and grain crops are grasses with divergent type II cell walls (Carpita and Gibeaut, 1993). Our primary screen was for (1) dwarfed plant form and (2) cellulose deficit, which revealed dwf1-1. Map-based cloning of dwf1-1 to SbGA20-oxidase was in contrast to both our expectations. Certainly, dwarfed and fertility-compromised mutant phenotypes were highly consistent with GA mutants, such as ga1-3 (Cheng et al., 2004) and gid1 (Aya et al., 2009) and also, show similar growth defects in rice (Sasaki et al., 2002) and Arabidopsis (Plackett et al., 2012) GA20-oxidase mutants. In fact, the rice GA20-oxidase mutant was isolated as a green revolution locus (Sasaki et al., 2002). However, current studies have mapped cellulose-deficient mutants to only a handful of genes, and most of these are CESAs (Arioli et al., 1998; Fagard et al., 2000; Desprez et al., 2002; Daras et al., 2009) or a small handful of CESA accessory proteins, including POM2/CELLULOSE SYNTHASE INTERACTING1 (Lei et al., 2012), COBRA (Roudier et al., 2005), and KORRIGAN (Lane et al., 2001). With a genetic screen for cellulose deficit linked to GA20-oxidase, we uncovered a GA-mediated mechanism for regulating cellular expansion. Importantly, this result was supported by pharmacological inhibition of GA, which also led to cellulose deficit (Fig. 6). Correspondingly, in both wild-type parental lines and the dwf1-1 mutant CESA, transcript abundance was significantly greater after exogenous GA3 addition (Fig. 9), suggesting positive regulation. Thus, an obvious question from these results is whether cellulose deficit could be the feature of GA deficit that leads to poor pollen viability and dwarfed growth form. Several logical points are consistent with this possibility. Both Atcesa1 and Atcesa3 mutants are pollen lethal and severely dwarfed (Persson et al., 2007) as is dwf1-1. Dwarfed growth is a common feature of cellulose deficiency (Fagard et al., 2000; Desprez et al., 2002) and GA deficit (Koornneef and van der Veen, 1980). Furthermore, transcriptional feedback was observed between GA deficit in dwf1-1 and primary cell wall CESAs (Fig. 7), which are known to be coregulated (Brown et al., 2005; Persson et al., 2005).
Primary and secondary cell wall cellulose biosyntheses have quite different roles in plant development. Transcriptional control of secondary cell wall CESAs has been studied, and several members of the NAC (for NAM [No Apical Meristem], ATAF1-2, and CUC2 [Cup-Shaped Cotyledon]) family of transcription factors have been identified as key regulators (Yamaguchi et al., 2010). In primary cell wall cellulose synthesis, posttranscriptional (Held et al., 2008) and posttranslational control (Chen et al., 2010) features have been observed. Our results support the role of GA as a positive regulator of CESA initiated transcriptionally. However, an additional question is how other hormones needed for expansion fit with proposed GA regulation of cellulose biosynthesis. For example, auxin and brassinosteroid are also critical for controlled expansion (Frigerio et al., 2006; Xie et al., 2011; Wolf et al., 2012; Paque et al., 2014). Prior evidence suggests cross talk between auxin and GA metabolism in Arabidopsis to regulate expansion (Frigerio et al., 2006), which in some circumstance, can involve auxin signaling upstream of the GA (Paque et al., 2014). Furthermore, dysfunction in brassinosteroid levels also seems to influence both primary and secondary cell wall cellulose biosyntheses. This signal either exists divergent of GA as a developmentally or environmentally induced signal or interacts with GA. Deciphering such a regulatory signaling pattern will be important for our understanding of anisotropic expansion in plants. GA responsive elements (Chen et al., 2006) were identified within the promoter of Sorghum sp. CESA genes, suggesting the possibility for direct regulatory features. During review of this article, Huang et al. (2015) published a causal association between GA signaling and the secondary cellulose biosynthetic process in rice. Huang et al. (2015) propose a model whereby GA-mediated DELLA-NAC (SLR1) signaling of an MYB transcription factor directly regulates CESA transcription, which is consistent with prior studies (Yamaguchi et al., 2010). Huang et al. (2015) also illustrate a vehicle for possible interaction with auxin-mediated development (Frigerio et al., 2006; Paque et al., 2014), which is where SLR1 (NAC) has been previously characterized in Arabidopsis lateral root formation (Fukaki et al., 2002). It will be interesting to examine whether the rice pathway is conserved in Arabidopsis and other species and how GA responses intersect with other hormonal signals during the regulation of plant expansion.
Classically involved in stem elongation, a mechanism for expansion regulation through GA involves antagonistic signaling between light and GAs (de Lucas et al., 2008). Interestingly, recent evidence suggests that PHYTOCHROME INTERACTING FACTOR4 (PIF4) influences the cellulose biosynthetic process (Bischoff et al., 2011) in Arabidopsis. PIF4 positively regulates genes involved in cellular expansion, and this is negatively regulated by the light photoreceptor phytochrome B (Huq and Quail, 2002). The transcriptional activity of PIF4 is also blocked during light signaling by DELLA that binds to the recognition sites for target genes (de Lucas et al., 2008). Taken with the findings by Huang et al. (2015), here, the switch to photomorphogenesis upon exposure to light inhibits hypocotyl expansion through PIF4. Understanding whether interplay exists between GA-mediated photomorphogenesis and the regulation of the DELLA-NAC transcriptional machinery (Huang et al., 2015) will be interesting future work.
Because cellulose was significantly reduced but not ablated in dwf1-1, the possibility exists that GA regulation provides a necessary biosynthetic cue to maximize cellulose production during cell wall development. We postulate that high-density organization of cellulose and a requirement for biosynthesis during key times of cellular expansion likely need regulatory mechanism(s) to adequately allocate resources. GA seems to directly fulfill one such role in regulating cellulose biosynthetic processes in Sorghum sp. and rice.
MATERIALS AND METHODS
Generating Ethyl Methanesulfonate Mutagenesis Population in Sorghum sp.
Ethyl methanesulfonate (EMS; Sigma) causes preferential alkylation of guanidine and thus, a shift of G:C base paring to A:T and was used to generate an mutagenesis population in Sorghum bicolor (the parental wild type). Approximately 12,000 seeds were exposed to a lethality-based saturation assay to establish the required dose of EMS needed (0.25%–1.5% [w/v] with a stepwise increase of 0.25% [w/v]), the optimal EMS concentration being estimated as 0.75% (w/v). Seeds were then exposed to 0.75% (w/v) EMS for 12 h followed by neutralization in sodium thiosulfate (0.2 m) and sequentially washed (M0 generation). They were air dried and sown on soilless media (MetroMix 360; SunGro Industries) supplemented with 3 g of Osmocote (The Scotts Company) at field water conditions. Seedlings were grown in a temperature-controlled greenhouse before being transplanted to the field after 6 weeks. Plants were phenotypically scored for the occurrence of aberrant expansion and reduced cellulose content. One such line named dwf1-1 was identified. For height estimation, 100 plants that were greenhouse grown were used for the mutant and the wild type. Plants of interest had their inflorescence bagged upon emergence to promote self-pollination. When this failed in the case of dwf1-1, the reciprocal cross between the wild type and dwf1-1 was performed. Viable progeny and inheritance of the trait were ultimately established using pollen from the wild type and dwf1-1 as the pollen receptor. The F2 population resulting from this cross was analyzed for the inheritance of the dwf1-1 trait.
Additional Plant Material
For the rice (Oryza sativa) sd-1 mutants, three isogenic lines (two intermediate alleles [scReimei and scShiranui] and one null allele [scTN-1]) were used with their recurrent parent, Norin 29 (Xia et al., 1991; Ogi et al., 1993; Irie et al., 2008). For the rice KO mutant, Tan-Ginbozu was used with its original cv Ginbozu (Itoh et al., 2004). For the rice GA-signaling mutants, gid1-8 and Slr1-d1 were used with their original cv Taichung 65 (Ueguchi-Tanaka et al., 2007; Asano et al., 2009). Briefly, seeds of each rice variety were surface sterilized and planted on one-half-strength Murashige and Skoog medium agar plates with and without GA3 (5 µm) supplementation. Seeds were germinated and grown in a vertical position in an environmentally controlled growth chamber at 24°C with a 16-h-light/8-h-dark cycle. Biomass from the first internodes was used for the analysis of cellulose content of GA-deficient mutant rice varieties along with the respective wild-type controls. For cellulose analysis (described below), three composite biological replicates consisting of five seedlings each were used along with three technical replicates. Total seedling lengths were recorded for areal tissue 10 d postgermination.
Cellulose Quantification
Cell walls were prepared according to the work by Reiter et al. (1993), and cellulose estimation was completed on 5 mg of sample according to the work by Updegraff (1969). To standardize the comparison, tissue was staged (Petti et al., 2013) and used for biological and technical replicates. Whole stems were used, and leaves of identical position and age were collected from the mutant and the wild type.
Pollen Viability and Germination Test
Viability of the pollen grains for both the mutant and the wild type was tested according to the work by Peterson et al. (2010) in triplicate and at different times to account for the intrinsic variability of pollen viability (Lansac et al., 1994; Tuinstra and Wedel, 2000; Burke et al., 2007). Pollen and germinability of the dwf1-1 and wild-type pollen were tested according to the works by Lansac et al. (1994) and Tuinstra and Wedel (2000) (n = 3; 100 counts each).
Mapping of the dwf1-1 Mutant
Bulked segregant analysis was used to determine the mutation underlying the dwarf phenotype. The parental lines used were dwf1-1 (parent 1) as the female receptor and the wild-type species Sorghum propinquum (parent 2) as the pollen donor. Genetic markers used were simple sequence repeats (SSRs) characterized and described for a similar cross type (S. bicolor × S. propinquum) in the works by Bhattramakki et al. (2000) and Billot et al. (2013). SSRs were tested for polymorphism in the parental lines P1 and P2 and a heterozygous F1. The SSRs displaying polymorphism were used for the analysis of the two bulks comprised of 25 individuals in each bulk. Equal amounts of purified DNA (4 μg) were used to generate the bulked DNA, and the final concentration used for the PCR analysis was 50 ng μL−1. PCR products were visualized on a 3.5% (w/v) agarose gel and scored as P1, P2, or heterozygous. After chromosomal linkage, SSRs from published resources (Bhattramakki et al., 2000; Kong et al., 2000, 2013; Billot et al., 2013) and CAPS markers were developed to obtain a close association to a genetic marker on a population of 200 segregants of dwf1-1. Chromosome walking was used to identify targets genes, which were then PCR amplified and sequenced. Sequences were compared with the reference S. bicolor genomes sequence (Paterson et al., 2009) and S. propinquum (Mace et al., 2013).
Hormone Analysis
GAs (GA1, GA3, GA7, GA8, GA19, GA20, GA24, GA29, GA34, GA44, GA51, and GA53), auxin, and cytokinins were examined as described in the works by Abrams et al. (2003), Ross et al. (2004), and Zaharia et al. (2005). These analyses were commercially determined as a fee for service product by the National Research Council of Canada. Arial tissue samples for both dwf1-1 and the wild type were light-grown 10-d-old seedlings.
Chemicals and Calibration Curves
A number of compounds, namely dihydroxyphaseic acid, abscisic acid (ABA)-glycosyl ester (GE), phaseic acid (PA), 7′- OH-ABA, neoPA, trans-ABA, and indole-3-acetic acid (IAA)-Glu, were synthesized and prepared at the National Research Council of Canada. ABA, IAA-Leu, IAA-Ala, IAA-Asp, IAA, zeatin (Z), zeatin-riboside (ZR), isopentenyladenosine riboside, and isopentenyladenosine were purchased from Sigma-Aldrich. dhZ and dhZR were purchased from Olchemim Ltd., and GA1, GA3, GA4, GA7, GA8, GA9, GA19, GA20, GA24, GA29, GA44, and GA53 were purchased from the Research School of Chemistry, Australian National University. Deuterated forms of the hormones that were used as internal standards included d3-dihydroxyphaseic acid, d5-ABA-GE, d3-PA, d4-7′-OH-ABA, d3-neoPA, d4-ABA, d4-trans-ABA, d3-IAA-Leu, d3-IAA-Ala, d3-IAA-Asp, and d3-IAA-Glu, and they were synthesized and prepared at the National Research Council of Canada according to works by Abrams et al. (2003) and Zaharia et al. (2005). d5-IAA was purchased from Cambridge Isotope Laboratories. d3-dhZ, d3-dhZR, d5-Z-O-Glu, d6-iPR, and d6-iP were purchased from Olchemim Ltd. d2-GA1, d2-GA3, d2-GA4, d2-GA7, d2-GA8, d2-GA9, d2-GA19, d2-GA20, d2-GA24, d2-GA29, d2-GA34, d2-GA44, d2-GA51, and d2-GA53 were purchased from the Research School of Chemistry, Australian National University. Calibration curves were created for all compounds of interest. Quality control (QC) samples were run along with the tissue samples.
Instrumentation
Analysis was performed with ultraperformance liquid chromatography (UPLC)/electrospray ionization-mass spectrometry (MS)/MS using a Waters ACQUITY UPLC System equipped with a binary solvent delivery manager and a sample manager coupled to a Waters Micromass Quattro Premier XE Quadrupole Tandem Mass Spectrometer through a Z-spray interface. MassLynx and QuanLynx (Micromass) were used for data acquisition and data analysis.
Extraction and Purification
An aliquot (100 μL) containing all of the internal standards, each at a concentration of 0.2 ng μL−1, was added to homogenized sample (approximately 50 mg); 3 mL of isopropanol:water:glacial acetic acid (80:19:1, v/v/v) was further added, and the samples were agitated in the dark for 14 to 16 h at 4°C. Samples were then centrifuged, and the supernatant was isolated and dried on a Büchi Syncore Polyvap. Furthermore, they were reconstituted in 100 μL of acidified methanol, adjusted to 1 mL with acidified water, and then, partitioned against 2 mL of hexane. After 30 min, the aqueous layer was isolated and dried as above. Dry samples were reconstituted in 800 μL of acidified methanol and adjusted to 1 mL with acidified water. The reconstituted samples were passed through equilibrated Sep-Pak C18 Cartridges (Waters Inc.), and the final eluate was split in two equal portions. One portion was dried completely (and stored), whereas the other portion was dried down to the aqueous phase on a LABCONCO Centrivap Concentrator (Labconco Corporation). The second portion was partitioned against ethyl acetate (2 mL) and further purified using an Oasis WAX Cartridge (Waters Inc.). The GA-enriched fraction in the second portion was eluted with 2 mL of acetonitrile:water (80:20, v/v) and then dried on a centrivap as described above. An internal standard blank was prepared with 100 μL of deuterated internal standards mixture. A QC standard was prepared by adding 100 μL of a mixture containing all of the analytes of interest, each at a concentration of 0.2 ng μL−1, to 100 μL of internal standard mix. Finally, both portions, blanks, and QCs were reconstituted in a solution of 40% (v/v) methanol containing 0.5% (v/v) acetic acid and 0.1 ng μL−1 of each of the recovery standards.
Hormone Quantification by HPLC-ESI-MS/MS
Performed as a fee for service product by the National Research Council of Canada, samples were injected onto an ACQUITY UPLC HSS C18 SB Column (2.1 × 100 mm, 1.8 μm) with an inline filter and separated by a gradient elution of water containing 0.02% (v/v) formic acid against an increasing percentage of a mixture of acetonitrile:methanol (50:50, v/v). Briefly, the analysis uses the Multiple Reaction Monitoring function of the MassLynx v4.1 (Waters Inc.) control software. The resulting chromatographic traces are quantified offline by the QuanLynx v4.1 software (Waters Inc.), wherein each trace is integrated, and the resulting ratio of signals (nondeuterated to internal standard) is compared with a previously constructed calibration curve to yield the amount of analyte present (nanograms per sample). Calibration curves were generated from the Multiple Reaction Monitoring signals obtained from standard solutions based on the ratio of the chromatographic peak area for each analyte to that of the corresponding internal standard as described by Ross et al. (2004). The QC samples, internal standard blanks, and solvent blanks were also prepared and analyzed along each batch of tissue samples.
Pharmacological Inhibition of GA in the Wild Type
Two GAs inhibitors, CCC and DMZ, were exogenously applied to wild-type plant material at the concentrations of 0.2 and 0.4 mm. The two inhibitors were known to be targeting early and late metabolic positions of the GA biosynthesis, CCC and DMZ (Rademacher, 2000). Plantlets exposed to these inhibitors were harvested, freeze dried, ground up, subjected to cellulose estimation as previously described, and compared with control plantlets.
Chemical Complementation of dwf1-1 Phenotypes with Exogenous GA
An F2 population (dwf1-1 and the wild type) was used to select homozygous dwf1-1 alongside the wild type. Each genotype was transferred to plates containing one-half-strength Murashige and Skoog medium supplemented with 5 μm GA3 (Lo et al., 2008) and mock control plates. Plantlets were grown vertically with approximately four plants per plate (n = 10). The effect of GA3 on expansion was evaluated visually by seedling length, and cellulose content was measured according to the work by Updegraff (1969).
RNA Extraction, Reverse Transcription-PCR, and Promoter Analyses
The pathway option of GRAMENE (www.gramene.org) was used to select candidate genes from the GA biosynthetic genes in S. bicolor. Sorghum sp. CESA genes were previously identified (Petti et al., 2013), and a list of target genes and primers was compiled (Supplemental Table S1). Total mRNA was extracted as per the work by Petti et al. (2013) and converted to single-strand complementary DNA (AB Applied Biosystem). Transcript abundance was quantified by SyBr Green semiquantitative assay and completed on each biological replicate (n = 3 biological and technical replicates) on a StepOne Real-Time System (AB Applied Biosystem). The ∆∆ method was used for quantifying the relative expression levels normalized against the actin gene (Supplemental Table S1). Promoters were defined as 4,000 bp upstream of the initiation codon and examined using the PLACE Database for cir-acting regulatory DNA elements (Higo et al., 1999).
Statistical Analyses
Statistical analysis used ANOVA, Student’s t test, and Tukey’s post hoc test. Analyses were carried out on Graphpad PRISM5. Significance was established at α = 0.05.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Transcriptional analysis of Sorghum sp. early GA biosynthetic genes.
Supplemental Figure S2. Analyses of auxins and cytokinins.
Supplemental Table S1. List of primers and sequences.
Supplemental Table S2. Promoter analyses data.
Glossary
- ABA
abscisic acid
- AIR
acid-insoluble residue
- CCC
chlorocholine chloride
- DMZ
daminozide
- EMS
ethyl methanesulfonate
- IAA
indole-3-acetic acid
- MS
mass spectrometry
- QC
quality control
Footnotes
This work was supported by the Japan Society for the Promotion of Science (KAKENHI grant no. 26660278 to K.H.), the U.S. Department of Energy (grant no. Department of Energy-Funding Opportunity Announcement 10–0000368 to S.D.), and the National Science Foundation (grant nos. 1256029 and 1355438 to S.D.).
Articles can be viewed without a subscription.
References
- Abrams SR, Nelson K, Ambrose SJ (2003) Deuterated abscisic acid analogs for mass spectrometry and metabolism studies. J Labelled Comp Radiopharm 46: 273–283 [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R, et al. (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720 [DOI] [PubMed] [Google Scholar]
- Asano K, Hirano K, Ueguchi-Tanaka M, Angeles-Shim RB, Komura T, Satoh H, Kitano H, Matsuoka M, Ashikari M (2009) Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Mol Genet Genomics 281: 223–231 [DOI] [PubMed] [Google Scholar]
- Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M (2009) Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21: 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattramakki D, Dong J, Chhabra AK, Hart GE (2000) An integrated SSR and RFLP linkage map of Sorghum bicolor (L.) Moench. Genome 43: 988–1002 [PubMed] [Google Scholar]
- Billot C, Ramu P, Bouchet S, Chantereau J, Deu M, Gardes L, Noyer JL, Rami JF, Rivallan R, Li Y, et al. (2013) Massive sorghum collection genotyped with SSR markers to enhance use of global genetic resources. PLoS One 8: e59714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff V, Desprez T, Mouille G, Vernhettes S, Gonneau M, Höfte H (2011) Phytochrome regulation of cellulose synthesis in Arabidopsis. Curr Biol 21: 1822–1827 [DOI] [PubMed] [Google Scholar]
- Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke IC, Wilcut JW, Allen NS (2007) Viability and in vitro germination of Johnsongrass (Sorghum halepense) pollen. Weed Technol 21: 23–29 [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Chen PW, Chiang CM, Tseng TH, Yu SM (2006) Interaction between rice MYBGA and the gibberellin response element controls tissue-specific sugar sensitivity of alpha-amylase genes. Plant Cell 18: 2326–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ehrhardt DW, Somerville CR (2010) Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc Natl Acad Sci USA 107: 17188–17193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Qin L, Lee S, Fu X, Richards DE, Cao D, Luo D, Harberd NP, Peng J (2004) Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131: 1055–1064 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Jarvis MC (2012) Comparative structure and biomechanics of plant primary and secondary cell walls. Front Plant Sci 3: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daras G, Rigas S, Penning B, Milioni D, McCann MC, Carpita NC, Fasseas C, Hatzopoulos P (2009) The thanatos mutation in Arabidopsis thaliana cellulose synthase 3 (AtCesA3) has a dominant-negative effect on cellulose synthesis and plant growth. New Phytol 184: 114–126 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Vernhettes S, Fagard M, Refrégier G, Desnos T, Aletti E, Py N, Pelletier S, Höfte H (2002) Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol 128: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes BP, Crowell DN (1998) Cytokinin regulates the expression of a soybean beta-expansin gene by a post-transcriptional mechanism. Plant Mol Biol 37: 437–444 [DOI] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio M, Alabadí D, Pérez-Gómez J, García-Cárcel L, Phillips AL, Hedden P, Blázquez MA (2006) Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol 142: 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Gonneau M, Desprez T, Guillot A, Vernhettes S, Höfte H (2014) Catalytic subunit stoichiometry within the cellulose synthase complex. Plant Physiol 166: 1709–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Kamiya Y (1997) Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol 48: 431–460 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Held MA, Penning B, Brandt AS, Kessans SA, Yong W, Scofield SR, Carpita NC (2008) Small-interfering RNAs from natural antisense transcripts derived from a cellulose synthase gene modulate cell wall biosynthesis in barley. Proc Natl Acad Sci USA 105: 20534–20539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Wang S, Zhang B, Shang-Guan K, Shi Y, Zhang D, Liu X, Wu K, Fu X, Zhou Y (2015) A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 27: 1681–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie K, Hua C, Tun YT, Kikuchi F, Fujimaki H, Kikuchi F, Nagamine T (2008) Genetic analysis of sterility caused by anther indehiscence gene linked with a semidwarfing gene locus, sd1 in rice. J Agric Sci 53: 69–74 [Google Scholar]
- Itoh H, Tatsumi T, Sakamoto T, Otomo K, Toyomasu T, Kitano H, Ashikari M, Ichihara S, Matsuoka M (2004) A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol Biol 54: 533–547 [DOI] [PubMed] [Google Scholar]
- Keyes GJ, Paolillo DJ, Sorrels ME (1989) The effects of dwarfing genes Rht1 and Rht2 on cellular dimensions and rate of leaf elongation in wheat. Ann Bot 64: 683–690 [Google Scholar]
- Kong L, Dong J, Hart GE (2000) Characteristics, linkage-map positions, and allelic differentiation of Sorghum bicolor (L.) Moench DNA simple-sequence repeats (SSRs). Theor Appl Genet 101: 438–448 [Google Scholar]
- Kong W, Jin H, Franks CD, Kim C, Bandopadhyay R, Rana MK, Auckland SA, Goff VH, Rainville LK, Burow GB, et al. (2013) Genetic analysis of recombinant inbred lines for Sorghum bicolor × Sorghum propinquum. G3 (Bethesda) 3: 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH (1980) Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theor Appl Genet 58: 257–263 [DOI] [PubMed] [Google Scholar]
- Lane DR, Wiedemeier A, Peng L, Höfte H, Vernhettes S, Desprez T, Hocart CH, Birch RJ, Baskin TI, Burn JE, et al. (2001) Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol 126: 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansac AR, Sullivan CY, Johnson BE, Lee KW (1994) Viability and germination of the pollen of sorghum [Sorghum bicolor (L.) Moench]. Ann Bot (Lond) 74: 27–33 [DOI] [PubMed] [Google Scholar]
- Lee I, Foster KR, Morgan PW (1998) Effect of gibberellin biosynthesis inhibitors on native gibberellin content, growth and floral initiation in Sorghum bicolor. J Plant Growth Regul 17: 185–195 [DOI] [PubMed] [Google Scholar]
- Lei L, Li S, Gu Y (2012) Cellulose synthase interactive protein 1 (CSI1) mediates the intimate relationship between cellulose microfibrils and cortical microtubules. Plant Signal Behav 7: 714–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Godwin ID (2012) Highly efficient sorghum transformation. Plant Cell Rep 31: 999–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SF, Yang SY, Chen KT, Hsing YI, Zeevaart JAD, Chen LJ, Yu SM (2008) A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20: 2603–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace ES, Tai S, Gilding EK, Li Y, Prentis PJ, Bian L, Campbell BC, Hu W, Innes DJ, Han X, et al. (2013) Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat Commun 4: 2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz MA, Klein RR, Mullet JE, Obert JA, Unruh NC, Klein PE (2002) A high-density genetic map of Sorghum bicolor (L.) Moench based on 2926 AFLP, RFLP and SSR markers. Plant Mol Biol 48: 483–499 [DOI] [PubMed] [Google Scholar]
- Ogi Y, Kato H, Maruyama K, Kikuchi F (1993) The effects of culm length and other agronomic characters caused by semidwarfing genes at the sd-1 locus in rice. Jpn J Breeding 43: 267–275 [Google Scholar]
- Okuno A, Hirano K, Asano K, Takase W, Masuda R, Morinaka Y, Ueguchi-Tanaka M, Kitano H, Matsuoka M (2014) New approach to increasing rice lodging resistance and biomass yield through the use of high gibberellin producing varieties. PLoS One 9: e86870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonio RL, Ito Y, Hatakeyama A, Ohmae-Shinohara K, Kasuga S, Tokunaga T, Mizuno H, Kitano H, Matsuoka M, Sazuka T (June 13, 2014) Gibberellin deficiency pleiotropically induces culm bending in sorghum: an insight into sorghum semi-dwarf breeding. Sci Rep http://dx.doi.org/10.1038/srep05287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paque S, Mouille G, Grandont L, Alabadí D, Gaertner C, Goyallon A, Muller P, Primard-Brisset C, Sormani R, Blázquez MA, et al. (2014) AUXIN BINDING PROTEIN1 links cell wall remodeling, auxin signaling, and cell expansion in Arabidopsis. Plant Cell 26: 280–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH. (2008) Genomics of sorghum. Int J Plant Genomics 2008: 362451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R, Slovin JP, Chen C (2010) A simplified method for differential staining of aborted and non-aborted pollen grains. Int J Plant Biol 1: e13 [Google Scholar]
- Petti C, Shearer A, Tateno M, Ruwaya M, Nokes S, Brutnell T, Debolt S (2013) Comparative feedstock analysis in Setaria viridis L. as a model for C4 bioenergy grasses and Panicoid crop species. Front Plant Sci 4: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plackett AR, Powers SJ, Fernandez-Garcia N, Urbanova T, Takebayashi Y, Seo M, Jikumaru Y, Benlloch R, Nilsson O, Ruiz-Rivero O, et al. (2012) Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell 24: 941–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher W. (2000) Growth retardants: effects on gibberellin biosynthesis and other metabolic pathways. Annu Rev Plant Physiol Plant Mol Biol 51: 501–531 [DOI] [PubMed] [Google Scholar]
- Reiter WD, Chapple CC, Somerville CR (1993) Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science 261: 1032–1035 [DOI] [PubMed] [Google Scholar]
- Ross AR, Ambrose SJ, Cutler AJ, Feurtado JA, Kermode AR, Nelson K, Zhou R, Abrams SR (2004) Determination of endogenous and supplied deuterated abscisic acid in plant tissues by high-performance liquid chromatography-electrospray ionization tandem mass spectrometry with multiple reaction monitoring. Anal Biochem 329: 324–333 [DOI] [PubMed] [Google Scholar]
- Roudier F, Fernandez AG, Fujita M, Himmelspach R, Borner GHH, Schindelman G, Song S, Baskin TI, Dupree P, Wasteneys GO, et al. (2005) COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17: 1749–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, et al. (2002) Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416: 701–702 [DOI] [PubMed] [Google Scholar]
- Schmidhuber J, Tubiello FN (2007) Global food security under climate change. Proc Natl Acad Sci USA 104: 19703–19708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethaphong L, Haigler CH, Kubicki JD, Zimmer J, Bonetta D, DeBolt S, Yingling YG (2013) Tertiary model of a plant cellulose synthase. Proc Natl Acad Sci USA 110: 7512–7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsekos I. (1999) The sites of cellulose synthesis in algae: diversity and evolution of cellulose-synthesizing enzyme complexes. J Phycol 35: 635–655 [Google Scholar]
- Tuinstra MR, Wedel J (2000) Estimation of pollen viability in grain sorghum contribution no. 99-212-J from the Kansas Agric. Exp. Stn. Crop Sci 40: 968–970 [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Katoh E, Ohmiya H, Asano K, Saji S, Hongyu X, Ashikari M, Kitano H, Yamaguchi I, et al. (2007) Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19: 2140–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Wolf S, Hématy K, Höfte H (2012) Growth control and cell wall signaling in plants. Annu Rev Plant Biol 63: 381–407 [DOI] [PubMed] [Google Scholar]
- Xia B, Hanada K, Kikuchi F (1991) Character expression of the semidwarfing gene sd-1 in rice (Oryza sativa L.): 1. Effects of nitrogen levels on the expression of some agronomic characteristics. Jpn J Crop Sci 60: 36–41 [Google Scholar]
- Xie L, Yang C, Wang X (2011) Brassinosteroids can regulate cellulose biosynthesis by controlling the expression of CESA genes in Arabidopsis. J Exp Bot 62: 4495–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YL, Li L, Wu K, Peeters AJ, Gage DA, Zeevaart JA (1995) The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc Natl Acad Sci USA 92: 6640–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Ohtani M, Mitsuda N, Kubo M, Ohme-Takagi M, Fukuda H, Demura T (2010) VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22: 1249–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharia LI, Galka MM, Ambrose SJ, Abrams SR (2005) Preparation of deuterated abscisic acid metabolites for use in mass spectrometry and feeding studies. J Labelled Comp Radiopharm 48: 435–445 [Google Scholar]