Allelic variation in a blue-light photoreceptor modulates temperature sensitivity.
Abstract
Wild strains of Arabidopsis (Arabidopsis thaliana) exhibit extensive natural variation in a wide variety of traits, including response to environmental changes. Ambient temperature is one of the major external factors that modulates plant growth and development. Here, we analyze the genetic architecture of natural variation in thermal responses of Arabidopsis. Exploiting wild accessions and recombinant inbred lines, we reveal extensive phenotypic variation in response to ambient temperature in distinct developmental traits such as hypocotyl elongation, root elongation, and flowering time. We show that variation in thermal response differs between traits, suggesting that the individual phenotypes do not capture all the variation associated with thermal response. Genome-wide association studies and quantitative trait locus analyses reveal that multiple rare alleles contribute to the genetic architecture of variation in thermal response. We identify at least 20 genomic regions that are associated with variation in thermal response. Further characterizations of temperature sensitivity quantitative trait loci that are shared between traits reveal a role for the blue-light receptor CRYPTOCHROME2 (CRY2) in thermosensory growth responses. We show the accession Cape Verde Islands is less sensitive to changes in ambient temperature, and through transgenic analysis, we demonstrate that allelic variation at CRY2 underlies this temperature insensitivity across several traits. Transgenic analyses suggest that the allelic effects of CRY2 on thermal response are dependent on genetic background suggestive of the presence of modifiers. In addition, our results indicate that complex light and temperature interactions, in a background-dependent manner, govern growth responses in Arabidopsis.
Temperature is a critical environmental factor that has major effects on the growth, development, and distribution of plants across the globe (Fitter and Fitter, 2002; Samach and Wigge, 2005, 2013; Kotak et al., 2007; Penfield, 2008). With the predicted increase in global temperatures, and their potential impact on agricultural productivity, there are efforts to understand the genetic basis of temperature responses in plants. Traditionally, temperature effects have been studied in the context of extreme stress responses such as heat shock or cold shock (Kotak et al., 2007; Barrero-Gil and Salinas, 2013; Song et al., 2013; Storey and Storey, 2013). In recent times, there has been an interest in analyzing the response of plants to changes in their growth temperature within the nonstress range of l6°C to 27°C, as even small changes in temperature can have major impacts on plant growth and development (Wigge, 2013; Franklin et al., 2014). In this study, we refer to various phenotypic responses in plants that are attributable to small changes in ambient temperature as temperature/thermal response.
A few specific phenotypic responses are often used to uncover the genetic and molecular basis of thermal response in plants. These include temperature-induced changes in hypocotyl elongation, flowering time, and circadian clock as well as molecular phenotypes such as the expression levels of At3g12580, one of the 14 genes encoding HEAT SHOCK PROTEIN70 (HSP70; Gray et al., 1998; Blázquez et al., 2003; Halliday et al., 2003; Michael et al., 2003; Lempe et al., 2005; Balasubramanian et al., 2006b; Edwards et al., 2006; Koini et al., 2009; Kumar and Wigge, 2010; Kumar et al., 2012). In addition, cryptic phenotypes, which are often revealed in higher temperatures, are also used to uncover genes associated with variation in thermal response in plants (Queitsch et al., 2002, 2012; Sureshkumar et al., 2009).
Higher temperatures result in elongated hypocotyls, early flowering, and an increased HSP70 expression (Gray et al., 1998; Blázquez et al., 2003; Balasubramanian et al., 2006b). Studies in the last decade have demonstrated that there is a genetic basis for most of the phenotypic responses observed upon temperature changes (Blázquez et al., 2003; Halliday et al., 2003; Balasubramanian et al., 2006b; Kumar and Wigge, 2010; Wigge, 2013). The temperature-induced hypocotyl elongation has been shown to be mediated by an increase in auxin levels, resulting from an increased expression of the YUCCA genes (e.g. YUCCA8), which encode enzymes involved in their biosynthesis (Gray et al., 1998; Koini et al., 2009; Franklin et al., 2011). The expression of the transcription factor PHYTOCHROME INTERACTING FACTOR4 (PIF4) is induced at higher temperatures, which in turn leads to the increase in the expression of the YUCCA genes (Franklin et al., 2011). Recently, a role has been demonstrated for DE-ETIOLATED1 (DET1) and ELONGATED HYPOCOTYL5 (HY5) in controlling the hypocotyl elongation response by modulating PIF4 expression according to the ambient temperature (Delker et al., 2014). PIF4 has also been suggested to underlie temperature-induced early flowering in short days (Kumar et al., 2012).
Mutations in the ACTIN-RELATED PROTEIN6 (ARP6) gene result in hyperactive thermal response even at lower temperatures (Kumar and Wigge, 2010). ARP6 is part of the SWR1 complex that plays a role in the incorporation/eviction of different histone variants (Deal et al., 2007). Based on this, it has been suggested that the incorporation/eviction dynamics of the histone variant H2A.Z on to the nucleosomes provide a direct mechanism for the perception of temperature (Kumar and Wigge, 2010). However, it has also been demonstrated that the presence of H2A.Z correlates with responsive genes and is not necessarily specific to temperature (Coleman-Derr and Zilberman, 2012).
On the other hand, the analysis of natural variation in flowering responses identified FLOWERING LOCUS M (FLM), which encodes a MADS domain containing floral repressor, to confer variation in thermosensory flowering response (Balasubramanian et al., 2006b). The function of FLM appears to be compromised at higher temperatures by changes in splicing patterns, resulting in derepression of FLOWERING LOCUS T (FT), and therefore the induction of flowering (Balasubramanian et al., 2006b; Balasubramanian and Weigel, 2006; Posé et al., 2013). Another MADS domain transcription factor SHORT VEGETATIVE PHASE (SVP), which interacts with FLM has also been shown to play a role in temperature-induced flowering, with the SVP protein being degraded at higher temperatures contributing to the derepression of FT (Lee et al., 2013). While there is agreement as to the thermal response in hypocotyl elongation, the genetic interaction between FLM and PIF4 is yet to be analyzed (Verhage et al., 2014).
Here, we decipher and reveal extensive natural variation in thermal responses of Arabidopsis (Arabidopsis thaliana) by analyzing hundreds of accessions and recombinant inbred lines (RILs) across several traits. We show that the thermosensitivity measured in one trait is only weakly correlated with thermosensitivity in another trait. We use genome-wide association studies (GWASs) and quantitative trait locus (QTL) analysis to map loci for thermal response and show that the genetic architecture of thermal response comprises multiple genomic regions consisting of potentially rare alleles. We show allelic variation at CRYPTOCHROME2 (CRY2) is associated with variation in thermal response. We identify Cvi-0 to be less responsive to temperature and demonstrate that the CRY2-Cvi-0 allele underlies this response. In addition to highlighting a role for CRY2 in thermal response, our results reveal complex genetic as well as genotype × environment interactions that link light and temperature response in plants.
RESULTS
Wild Strains of Arabidopsis Display Extensive Variation in Temperature Responses
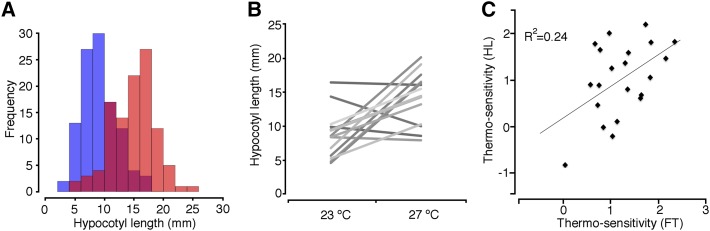
Previous analyses indicated that there is substantial genetically controlled variation in thermal response to flowering time (Lempe et al., 2005; Balasubramanian et al., 2006b). To assess whether there is variation in thermal response in other phenotypes, we analyzed hypocotyl elongation in more than 5,000 plants representing 139 accessions at two different temperatures (23°C and 27°C in short days; Supplemental Table S1). There was substantial variation in hypocotyl lengths, with most accessions displaying longer hypocotyls at higher temperatures (Fig. 1A). ANOVA with the accessions as a factor of random effect and hypocotyl length as response revealed high heritability in hypocotyl elongation, with the genotypes accounting for 84% and 78% of the variation at 23°C and 27°C, respectively (P < 0.0001; Supplemental Table S2). This suggests that much of the observed variation is genetically controlled. When we combined the 23°C and 27°C data sets and analyzed the effects of temperature and genotype as well as genotype × temperature interactions through ANOVA with hypocotyl length as response, we found that 44% of variation could be attributed to the genotype (P < 0.0001). Consistent with the observed reduction in heritability (44% as opposed to 83% or 78% when data sets from different temperature were analyzed separately) with the combined data set, temperature accounted for 32% of the variation in hypocotyl lengths (P < 0.0001). Analysis of the reaction norms of accessions revealed that the response of the accessions to changes in temperature differed, suggestive of genotype × environment interactions (Fig. 1B; Supplemental Fig. S1). Consistent with this, through ANOVA, we quantified the effect of genotype × temperature interaction, which accounted for approximately 13% of the phenotypic variation (P < 0.0001). These findings suggested that there is extensive natural variation in thermal response in hypocotyl elongation.
Figure 1.
Natural variation in hypocotyl elongation response to temperature. A, Distribution of average hypocotyl lengths for accessions grown at 23°C (blue) or 27°C (red). The overlap is shown in a different color. B, Reaction norms of Arabidopsis accessions displaying differential thermal response in hypocotyl elongation. Only a small subset of accessions is shown. Crossing reaction norms indicate genotype × environment interactions. The reaction norms for all accessions are provided in Supplemental Figure S1. C, Correlation between temperature sensitivities in flowering time (FT) or hypocotyl elongation (HL). Temperature sensitivities were calculated from the slopes of the reaction norms of flowering time and hypocotyl elongation at 23°C and 27°C.
To assess whether common factors modulate temperature sensitivity, we calculated the temperature sensitivity as the slope of the reaction norms for each of the accessions and compared the temperature sensitivity in hypocotyl elongation with that of flowering time. We used the data from our current study and/or from our previous analysis (Supplemental Table S1; Balasubramanian et al., 2006b). There is a significant yet relatively small (R2 = 0.24, P < 0.0001) correlation between thermosensitivities in flowering time and hypocotyl elongation (Fig. 1C). This suggests, while there are some common factors, phenotype-specific processes contribute to differential responses to temperature for different traits.
GWAS for Temperature-Induced Hypocotyl Elongation
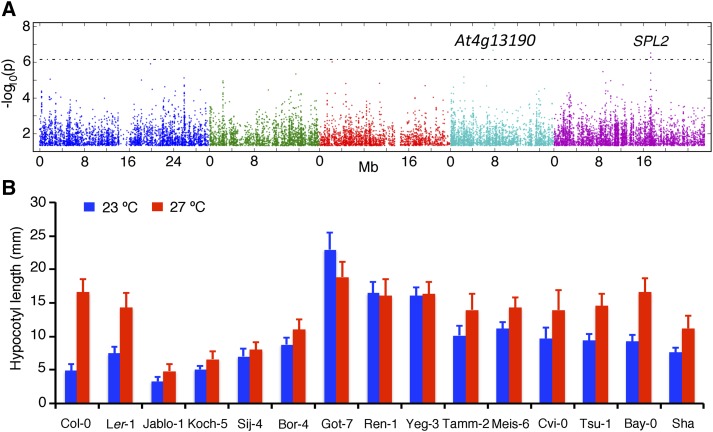
To decipher whether there are common allelic variants that could explain the variation in thermal response in hypocotyl elongation, we took advantage of the genotypic information available through the 1,001 genomes project (http://www.1001genomes.org) and carried out association analysis for temperature sensitivity. The GWAS identified two regions that appear to be associated with temperature sensitivity (Fig. 2A; Supplemental Fig. S2). In chromosome 5, the Single Nucleotide Polymorphism (SNP) with the strongest association and the SNPs that are in linkage disequilibrium fell on the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE2 (SPL2) gene (Fig. 2A). SPL2 belongs to a family of transcription factors, which are targets for the temperature-responsive microRNA, miR156 (Lee et al., 2010). Recently, it has also been shown that mutations in SPL2 can modulate sensitivity to extreme temperature conditions, thus making SPL2 a potential candidate gene for thermal response (Stief et al., 2014). Sequence analysis of SPL2 using the 1,001 genomes project revealed several deletions in both the SPL2 coding region as well as in the promoter, although further experiments are required to analyze the potential causal polymorphisms.
Figure 2.
Genome-wide association analysis and the identification of strains with reduced thermal response in hypocotyl elongation. A, Manhattan plots for genetic association for temperature sensitivity in hypocotyl elongation generated through the GWAS Web Application. The plot generated through linear regression is shown here. For other models, refer to Supplemental Figure S1. The position of the candidate gene SPL2 is shown. B, Hypocotyl elongation in accessions identified to be less responsive to ambient temperature changes including the parents of the RILs analyzed in this study.
The SNPs in the second region with significant association fell on a gene encoding a protein kinase (At4g13190), which is yet to be characterized (Fig. 2A). Association analysis using residuals of the regression of hypocotyl elongation at 23°C on to 27°C as a measure of temperature sensitivity provided similar results. In addition, the use of different statistical methods (Kruskal-Wallis analysis or approximate mixed model [AMM]) did not alter the Manhattan plots, although the significance of the association decreased in the AMM analysis (Supplemental Fig. S2). We did not find any additional SNPs with significant association. Thus, in contrast to flowering time where multiple common alleles have been found through GWAS (Atwell et al., 2010), there are fewer common allelic variants that appear to be associated with variation in temperature-induced hypocotyl elongation.
Temperature Sensitivity in Hypocotyl Elongation Segregates as a Multigenic Complex Trait
Because GWAS analysis identified only a couple of candidate regions, we looked for strains with reduced thermal response. To identify such accessions, we calculated the percentage of variance in hypocotyl elongation that can be attributed to temperature for each of the strains (Supplemental Table S1). For the majority (75%) of the strains, temperature accounted for more than 50% of the variation, with the median variance explained by temperature being 70.87%. We concentrated on the bottom quartile of the strains, with reduced temperature sensitivity, which included two different sets of behaviors (Fig. 2B). First, there were strains with impaired temperature-induced hypocotyl elongation in which the hypocotyl length was relatively short even at higher temperatures (e.g. Koch-5, Sij-4, Jablo-1, and Bor-4). Second, there were strains in which the hypocotyl length was much higher than the average at 23°C, suggesting that they exhibit a high temperature response even at lower temperatures (e.g. Got-7, Ren-1, Yeg-3, Meise-6, Tamm-2, and Cape Verde Islands [Cvi-0]). Among these strains, Sij-4 also displays a strong temperature-dependent growth defect, in addition to the absence of temperature-induced hypocotyl elongation. Further analysis of Sij-4 revealed that allelic variation at the ICARUS1 locus, encoding a tRNAHis guanylyl transferase, underlies this phenotype, indicating a role for fundamental processes in conferring variation in thermal response (Zhu et al., 2015). To decipher the genetic architecture of these accession-specific responses, we analyzed three F2 populations derived from the relatively temperature-insensitive strains (Tsushima [Tsu-0], Meise-6, and Yeg-3) with the reference strain Columbia (Col-0), which displays relatively higher sensitivity (Supplemental Fig. S3). All three analyzed F2 populations displayed a continuous distribution and lacked any bimodal distributions at either temperature, suggesting that multiple loci contribute to quantitative thermal responses in these strains.
Genetic Architecture of Temperature Response in RILs
Because our analysis suggested that thermal response is likely to be multigenic and quantitative, we chose to carry out QTL analysis using the RILs. We selected the Tsu-0 × Col-0, Cvi-0 × Col-0, and Bayreuth (Bay-0) × Shahdara (Sha) RILs in which the parental lines differed in their thermal response (Fig. 2B). To maximize capturing the variation associated with thermal response, we phenotyped all three RILs for several temperature sensitive traits (i.e. hypocotyl elongation, root elongation, and flowering time) across two different temperatures (23°C and 27°C). We observed high broad-sense heritabilities for all traits (Supplemental Table S2). Consistent with earlier observations, we observed longer roots, longer hypocotyls, and early flowering at higher temperatures across all RILs (Supplemental Tables S3–S5). The genetic correlations between the different phenotypes were weak, suggesting that the underlying genetic factors are only partially shared between the phenotypes (Supplemental Table S6).
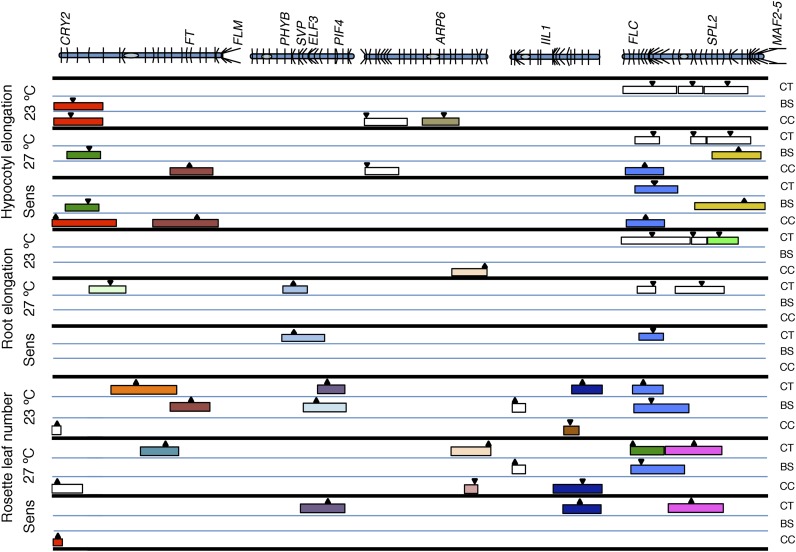
To identify the loci associated with thermal response, we applied two approaches. First, we identified QTLs that are detected only in one temperature, which indicates its temperature dependence. Second, we calculated temperature sensitivity for each of the three traits in the different RIL populations, used the same as phenotype, and carried out QTL analysis. We compiled the QTLs detected through these methods (Fig. 3; Table I; Supplemental Fig. S4; Supplemental Table S7). Among a total of 53 QTLs detected for a variety of traits, we observed 19 genomic regions that contribute to temperature response (Fig. 3; Table I). While some of the regions were detected as QTLs for multiple traits across populations, a majority of the QTLs were specific for individual traits, confirming that the analysis of multiple traits is required to capture variation in temperature response (Supplemental Table S7). Most QTLs (approximately 66%) for all the traits showed large effect explaining more than 10% of the phenotypic variance, while the remaining loci showed a moderate effect (5%–10%). Our analysis lacked the power to capture QTLs that accounted for less than 5% of phenotypic variation.
Figure 3.
Comparative map positions of the QTLs. QTLs detected for hypocotyl elongation, root elongation, and rosette leaf number at two different temperatures (23°C and 27°C) as well as temperature sensitivity (Sens) for all three traits in three different RILs is depicted. The QTLs representing potentially the same loci are shaded in the same color, with the colored box representing two LOD confidence intervals. The arrow represents the position with the highest LOD threshold. The upward or downward arrows indicate that the Col-0 and Sha alleles increase and decrease the trait value, respectively. The positions of genes known to be associated with ambient temperature response are shown above. CT, Col-0 × Tsu-0; BS, Bay-0 × Sha; CC, Col-0 × Cvi-0; MAF2-5, MADS AFFECTING FLOWERING2-5.
Table I. Summary of temperature-sensitive QTLs detected in this study.
ColTsu, Col-0 × Tsu-0 RILs; ColCvi, Col-0 × Cvi-0 RILs; BaySha, Bay-0 × Sha RILs; Chr, chromosome; Position, genomic position of the marker with highest LOD threshold; Interval, 2× LOD confidence interval for the QTL; RLN, rosette leaf number.
| RIL | Chr | Position | Interval | Trait | Category | QTL |
|---|---|---|---|---|---|---|
| CviCol | 1 | 0.59 | 0.59–1.62 | RLN | Sensitivity | 1 |
| CviCol | 1 | 0.59 | 0–5.0 | Hypocotyl length | Sensitivity | 1 |
| CviCol | 1 | 2.99 | 0–9.19 | Hypocotyl length | 23Specific | 1 |
| BaySha | 1 | 3.21 | 0–8.6 | Hypocotyl length | 23Specific | 1 |
| BaySha | 1 | 4.99 | 2.00–8.19 | Hypocotyl length | Sensitivity | 2 |
| BaySha | 1 | 6.37 | 2–8.19 | Hypocotyl length | 27Specific | 2 |
| ColTsu | 1 | 10.57 | 6.53–13.27 | Root length | 27Specific | 3 |
| ColTsu | 1 | 15.04 | 10.57–22.79 | RLN | 23Specific | 4 |
| ColTsu | 1 | 19.79 | 15.4–22.79 | RLN | 27Specific | 5 |
| CviCol | 1 | 25.1 | 17.8–29.3 | Hypocotyl length | Sensitivity | 6 |
| CviCol | 1 | 25.1 | 21.59–29.3 | Hypocotyl length | 27Specific | 6 |
| BaySha | 1 | 25.82 | 20.63–29.01 | RLN | 23Specific | 6 |
| ColTsu | 2 | 9.65 | 7.6–11.84 | Root length | 27Specific | 7 |
| ColTsu | 2 | 9.65 | 6.93–14.66 | Root length | Sensitivity | 7 |
| BaySha | 2 | 12.98 | 10–19.42 | RLN | 23Specific | 8 |
| ColTsu | 2 | 14.66 | 9.65–18.0 | RLN | Sensitivity | 9 |
| ColTsu | 2 | 14.66 | 11.84–18 | RLN | 23Specific | 9 |
| CviCol | 3 | 14.92 | 11–17.99 | Hypocotyl length | 23Specific | 10 |
| CviCol | 3 | 20.19 | 18.19–20.72 | RLN | 27Specific | 11 |
| CviCol | 3 | 22.14 | 16.67–22.14 | Root length | 23Specific | 12 |
| ColTsu | 3 | 23.04 | 16–23 | RLN | 27Specific | 12 |
| CviCol | 4 | 11.87 | 10.6–13.17 | RLN | 23Specific | 13 |
| ColTsu | 4 | 14.17 | 10.0–17.0 | RLN | Sensitivity | 14 |
| ColTsu | 4 | 14.17 | 12.53–17.04 | RLN | 23Specific | 14 |
| CviCol | 4 | 14.8 | 8.91–17.67 | RLN | 27Specific | 14 |
| ColTsu | 5 | 1.01 | 0–7.99 | RLN | 27Specific | 15 |
| BaySha | 5 | 2.77 | 1.5–12.1 | RLN | 27Specific | 16 |
| ColTsu | 5 | 3.44 | 1.01–7.99 | RLN | 23Specific | 16 |
| BaySha | 5 | 4.64 | 1.47–14 | RLN | 23Specific | 16 |
| CviCol | 5 | 4.74 | 0–9.85 | Hypocotyl length | Sensitivity | 16 |
| CviCol | 5 | 4.74 | 0–7.99 | Hypocotyl length | 27Specific | 16 |
| ColTsu | 5 | 5.31 | 2.33–7.99 | Root length | Sensitivity | 16 |
| ColTsu | 5 | 5.31 | 2.33–10 | Hypocotyl length | Sensitivity | 16 |
| ColTsu | 5 | 13.04 | 5–18.74 | RLN | Sensitivity | 17 |
| ColTsu | 5 | 13.04 | 7.99–18.74 | RLN | 27Specific | 17 |
| ColTsu | 5 | 18.74 | 15.79–21.84 | Root length | 23Specific | 18 |
| BaySha | 5 | 23.87 | 17.25–25.92 | Hypocotyl length | 23Specific | 19 |
| BaySha | 5 | 23.87 | 13.96–25.92 | Hypocotyl length | Sensitivity | 19 |
Allelic Variation at CRY2 Modulates Temperature Sensitivity
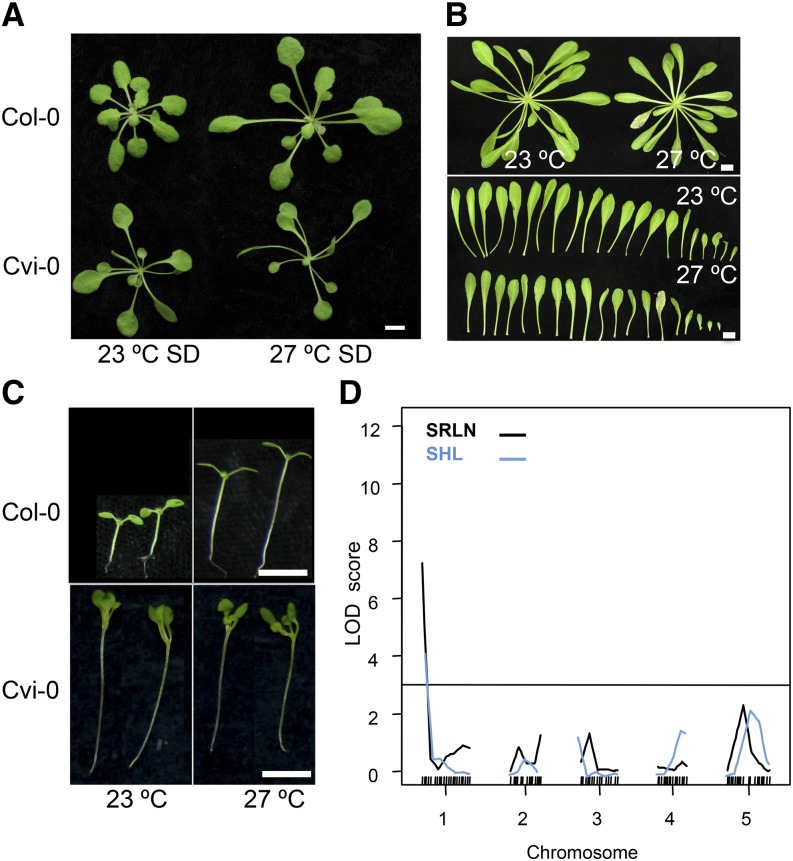
Among the parental strains used in our analysis, Cvi-0 plants appeared very similar at 23°C and 27°C in several ways, suggesting that they had a reduced response to this change in temperature (Fig. 4, A–C). In contrast to Col-0, Cvi-0 displays high temperature (27°C)-associated phenotypes such as longer hypocotyls, elongated petioles, early flowering, and altered plant architecture at lower temperatures (23°C) as well (Fig. 4, A–C). QTL analysis for temperature sensitivity in both flowering time and hypocotyl elongation in the Col-0 × Cvi-0 RILs mapped to the top of chromosome 1 (Fig. 4D). RILs that carry the Cvi-0 allele at this QTL displayed reduced temperature sensitivity due to longer hypocotyls and early flowering at lower temperatures.
Figure 4.
Reduced sensitivity to temperature in Cvi-0 maps to the top of chromosome 1. A, Architecture of Col-0 or Cvi-0 plants grown at 23°C or 27°C. B, Petiole elongation in Cvi-0 at 23°C compared with 27°C. C, Temperature-induced hypocotyl elongation in Col-0 compared with Cvi-0. D, QTL analysis of temperature sensitivity in hypocotyl elongation (light blue) or flowering time (black) in the Col-0 × Cvi-0 RILs. Bars = 6 mm (A, C, and D) and 10 mm (B). SRLN and SHL represent temperature sensitivity in rosette leaf number (SRLN) and hypocotyl length (SHL).
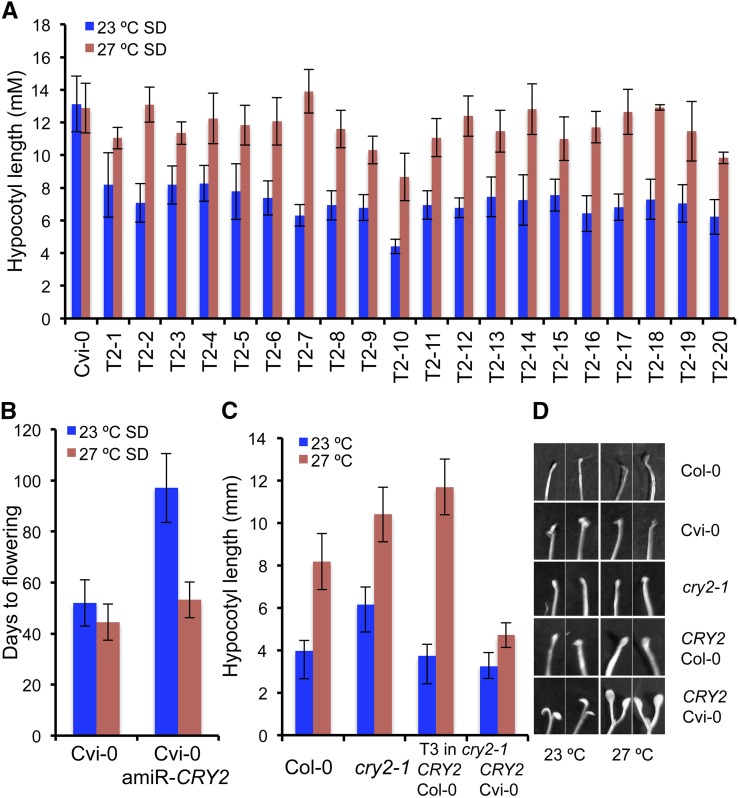
Within the confidence intervals for these QTLs, Cvi-0 is known to carry a hyperactive gain-of-function allele of CRY2 that leads to early flowering, and this phenotype has been attributed to the function of CRY2 as a blue-light receptor in light response (El-Din El-Assal et al., 2001). A hyperactive photoreceptor would increase sensitivity to light and thus lead to a shorter hypocotyl. However, Cvi-0 exhibits longer hypocotyls at both temperatures. To test whether the CRY2-Cvi-0 allele is the underlying locus for the longer hypocotyls and reduced temperature sensitivity of Cvi-0, we generated artificial microRNAs against CRY2 in the Cvi-0 background (35S::amiRCRY2) in which the CRY2 expression levels were severely reduced (Supplemental Fig. S5A). We assessed the transgenic plants for hypocotyl elongation and flowering time at varied temperatures (Fig. 5). 35S::amiRCRY2 lines displayed shorter hypocotyls compared with Cvi-0, suggesting that Cvi-CRY2 contributes to the longer hypocotyls observed in Cvi-0 (Fig. 5A). However, this inhibition of hypocotyl elongation was observed only at 23°C, and the 35S::amiRCRY2 lines displayed temperature-induced hypocotyl elongation and restored temperature sensitivity. This suggests that the CRY2-Cvi-0 allele contributes to the longer hypocotyls observed at lower temperatures and CRY2 is one of the genes underlying the compromised ambient thermal response observed in the Cvi-0 accession (Fig. 5A). Consistent with this, the 35S::amiRCRY2-Cvi-0 lines also flowered later, again, only at lower temperatures, thus restoring temperature sensitivity in flowering time as well (Fig. 5B). In addition, 35S::amiRCRY2-Cvi-0 plants also displayed short petioles and altered plant architecture and appeared similar to Col-0 plants grown at lower temperatures, suggesting that CRY2-Cvi-0 allele underlies the high-temperature phenotype exhibited by Cvi-0 even under lower thermal regimes (Supplemental Fig. S5, B and C).
Figure 5.
Allelic variation at CRY2 modulates thermal responses in Arabidopsis. A, Restoration of temperature sensitivity in hypocotyl elongation in plants in which CRY2 function is compromised via artificial microRNA against CRY2. Hypocotyl elongation at two different temperatures in 20 independent T2 families of 35S::amiRCRY2 lines in the Cvi-0 background compared with Cvi-0. B, Restoration of temperature sensitivity in flowering time in 35S::amiRCRY2 in the Cvi-0 background, measured as days to flowering. C, Hypocotyl elongation at different temperatures in T3 transgenic plants carrying either the Col-0 or the Cvi-0 allele in the cry2-1 background compared with Col-0 and cry2-1. D, Cotyledon opening response in Col-0, Cvi-0, cry2-1, and transgenic plants carrying either the Col-0 (CRY2pro::CRY2-Col-0) or the Cvi-0 allele (CRY2pro::CRY2-Cvi-0) in the Col-0 background in dark. SD, Short-day condition.
To test whether allelic variation at CRY2 modulates temperature sensitivity, we generated transgenic plants expressing CRY2 alleles from Col-0 (pCRY2Col::CRY2Col-0) or Cvi-0 (pCRY2Cvi::CRY2Cvi-0) in the cry2-1 mutant background (Guo et al., 1998) and compared the transgenic lines for their temperature sensitivity. While the pCRY2Col::CRY2Col-0 plants displayed temperature sensitivity in hypocotyl elongation, pCRY2Cvi::CRY2Cvi-0 plants were insensitive to temperature (Fig. 5C). In addition, the temperature insensitivity was also observed in flowering time, petiole elongation, and plant architecture, confirming that allelic differences at CRY2 underlie variation in temperature sensitivity between the Col-0 and Cvi-0 accessions of Arabidopsis (Fig. 5C; Supplemental Fig. S6, A–C).
Genotype × Environment Interactions Modify the Effects of CRY2-Cvi-0
The effects of the CRY2-Cvi-0 allele on hypocotyl elongation differed between the Cvi-0 and Col-0 backgrounds. While displaying temperature insensitivity in both backgrounds, the CRY2-Cvi-0 allele conferred longer hypocotyls in the Cvi-0 background and shorter hypocotyls in the Col-0 background (Fig. 5, A and C), suggesting the presence of background-specific modifiers of CRY2 function in this response. In addition, F1 plants derived from a Col-0 × Cvi-0 cross exhibited temperature-induced hypocotyl elongation, indicating that the temperature sensitivity conferred by the CRY2-Cvi-0 is recessive in nature (Supplemental Fig. S7A). This is in contrast to flowering time, where the CRY2-Cvi-0 allele acts as a dominant allele, which is attributed to the increased stability of the CRY2-Cvi-0 protein (El-Din El-Assal et al., 2001). However, temperature does not seem to affect the stability of the CRY2 protein, as previous studies found no obvious differences between different temperatures (Gould et al., 2013). Therefore, we wondered whether differential light × temperature interactions might modulate the discrepancy in hypocotyl elongation observed in both backgrounds.
The CRY2-Cvi-0 has been previously shown to confer enhanced cotyledon unfolding during photomorphogenesis (Botto et al., 2003). Therefore, we hypothesized that a short hypocotyl may result from enhanced cotyledon opening, which may inhibit hypocotyl elongation. To test this, we compared the effects of CRY2-Cvi-0 and CRY2-Col-0 alleles on cotyledon opening in the cry2-1 background. The cotyledon unfolding was significantly higher in the plants carrying the CRY2-Cvi-0 allele compared with the plants harboring the CRY2-Col-0 allele in the cry2-1 mutant background across all the conditions tested (Fig. 5D; Supplemental Fig. S6E). In addition, the pCRY2Cvi::CRY2Cvi-0 plants in the cry2-1 mutant background displayed cotyledon opening very early compared with pCRY2Col::CRY2Col-0 plants at both temperatures (Fig. 5D). This is consistent with the hypothesis that the hypocotyl elongation stops after cotyledon opening, resulting in the shorter hypocotyls. This effect was seen in both Col-0 (carrying the endogenous CRY2-Col-0 allele) as well as the cry2-1 mutants, suggesting that this photomorphogenetic phenotype conferred by the CRY2-Cvi-0 is dominant in the Col-0 background (Fig. 5D; Supplemental Fig. S6D). However, this phenotype was not that obvious in the Cvi-0 accession, which harbors the CRY2-Cvi-0 allele (Fig. 5D). While the Cvi-0 plants did display mild cotyledon opening, it was quite weak compared with the pCRY2Cvi::CRY2Cvi-0 plants in the Col-0 background (Fig. 5D; Supplemental Fig. S6D). Therefore, it appears that the early cotyledon opening in the Col-0 background results in short hypocotyls at both temperatures, contrasting its effect of longer hypocotyls in the Cvi-0 background at both temperatures. Thus, genetic background modifies the effect of the CRY2-Cvi-0 allele on light response, which in turn alters the temperature-induced hypocotyl elongation response.
To assess the underlying mechanism for this background-dependent effect, we analyzed the expression levels of CRY2 in Col-0, Cvi-0, and the different transgenic lines (Supplemental Fig. S8). We found the pCRY2Col::CRY2Col-0 plants to be expressing CRY2 similar to Col-0 or Cvi-0, while there was a major increase in the expression levels of the CRY2-Cvi-0 allele in the Col-0 background (Supplemental Fig. S8). This suggests that the observed phenotypic differences could be attributed at least partially to the differential activity of the CRY2-Cvi-0 promoter in the Col-0 and Cvi-0 backgrounds.
Because the original CRY2 allelic variation was defined through the analysis of Landsberg erecta (Ler) and Cvi-0 accessions, we analyzed the F1 plants derived from the Ler × Cvi-0 cross, which revealed that the temperature insensitivity segregated as a recessive trait in this cross as well (Supplemental Fig. S7A). In addition, the near-isogenic line EDI-NIL (El-Din El-Assal et al., 2001), which harbors the genomic region encompassing the CRY2 locus from Cvi-0 in the otherwise Ler background, also displayed temperature sensitivity, suggesting the presence of modifiers in the Ler background that suppress the effects of the CRY2-Cvi-0 allele. Consistently, transgenic plants in the Ler background carrying the Ler or Cvi-0 alleles for CRY2 also displayed temperature-induced hypocotyl elongation (Supplemental Fig. S7B). Thus, genetic modifiers in different backgrounds modulate the light × temperature interactions conferring the differential responses seen in Arabidopsis accessions.
DISCUSSION
Multiple Rare Alleles Rather Than Common Alleles Underlie Most of the Variation in Thermal Responses in Arabidopsis
In this study, we have uncovered the genetic architecture of temperature-dependent growth responses in Arabidopsis. Thermal response can be measured using a variety of phenotypes. While we observed correlation between temperature sensitivities for flowering time and hypocotyl elongation among accessions, there was almost no correlation between thermal responses measured through different traits in the RILs (Supplemental Table S6), suggesting that the underlying molecular mechanisms modulating thermosensitivity differ between distinct traits. Consistent with this, out of the 12 QTLs detected for temperature sensitivity, only two QTLs were shared between traits and RILs. Nevertheless, we have identified several genomic regions that modulate temperature responses in specific traits. Therefore, our results show that analysis of temperature response using individual traits fails to capture the majority of variation in thermal response.
We have carried out GWAS for hypocotyl elongation and temperature sensitivity in this response. Unlike flowering time, for which several strong associations have been reported (Atwell et al., 2010), we found no associations with hypocotyl length and detected only two regions associated with temperature sensitivity in hypocotyl elongation. This suggests that the genetic architecture of thermal response in hypocotyl elongation is quite different from the genetic architecture of flowering time (Salomé et al., 2011). However, there is extensive genetically controlled phenotypic variation, as the heritabilities for hypocotyl elongation at both temperatures were quite high. Therefore, it appears that multiple loci contribute to thermal responses in hypocotyl elongation. Consistent with this, QTL analysis uncovered multiple genomic regions that display temperature-dependent effects. This suggests that most of the variation in thermal response is attributable to alleles that occur at relatively low frequency in populations, which are not easily detectable through association analysis. This is consistent with the findings that often genes with demonstrated allelic effects in hypocotyl elongation have been identified only in a subset of strains, with common allelic variation having been reported only for the PHYTOCHROME B (PHY B) and PHYC, although rare alleles of PHYA, PHYD, and CRY2 have been described (Aukerman et al., 1997; El-Din El-Assal et al., 2001; Maloof et al., 2001; Balasubramanian et al., 2006a; Filiault et al., 2008). Thermal response, a complex trait, modulated both by genetic and genotype × environment interactions, may benefit from multiple loci/alleles, which may allow incremental changes in a smaller scale that could suit adaptations to multiple environments. Overall, our findings suggest that most of the variation in thermal response may be conferred through rare alleles rather than common alleles in natural populations.
GWAS and QTL Analysis Identifies Loci Associated with Thermal Response
Several earlier studies have shown that the expression level of miR156 changes in response to growth temperature (Lee et al., 2010; An et al., 2011; May et al., 2013; Stief et al., 2014). miR156 levels regulate developmental transitions in plants, including the floral transition (Wang, 2014). Most of the miR156-related responses have been explained through its role in down-regulating the SPL family of transcription factors (Wang, 2014). Among the SPL family of transcription factors, SPL2 is relatively less explored, and recently, it has been linked with heat shock memory and thermo tolerance (Stief et al., 2014). Our studies have identified a strong association between allelic variations at SPL2 and temperature sensitivity in hypocotyl elongation, making SPL2 an obvious candidate gene for ambient temperature response. The SNPs with strongest associations were within the promoter of the SPL2 gene. However, the promoter region of SPL2 appears to harbor several deletions, and thus further analysis is required to confirm a role for SPL2 and to work out the potential underlying mechanisms for this response.
The QTL mapping revealed several regions that appear to modulate sensitivity to temperature, which include known as well as additional candidate genes for thermal response. Consistent with earlier findings, a QTL for flowering time in the Bay-0/Sha population based around the FLM gene was detected only at 23°C but not at 27°C (Supplemental Table S7), suggesting that real differences are captured in our approach. One of the QTL for thermosensitive growth responses across multiple traits/RILs mapped to the top of chromosome 5, which includes the candidate gene FLOWERING LOCUS C (FLC). FLC, in addition to playing a key role in vernalization response (Michaels and Amasino, 1999; Sheldon et al., 1999), has also been linked to pleiotropic effects, including temperature compensation of the circadian clock, thermosensitivity in flowering time, and plant architecture (Michaels and Amasino, 1999; Sheldon et al., 1999; Poduska et al., 2003; Edwards et al., 2005, 2006; Balasubramanian et al., 2006b; Sibout et al., 2008). Therefore, it is conceivable that FLC could be the underlying locus for this QTL for thermal response.
We found several genes for which natural variation has been reported to colocalize with some of the QTLs that we detected for thermal response. The region around the FT gene, which has been previously implicated in thermal response, was identified as a region for temperature sensitivity in hypocotyl elongation (Balasubramanian et al., 2006b; Schwartz et al., 2009; Liu et al., 2014). We found QTLs encompassing the EARLY FLOWERING3 (ELF3) gene as a QTL for temperature sensitivity in flowering time in the Col-0/Tsu-0 and Bay/Sha RILs (Thines and Harmon, 2010; Undurraga et al., 2012; Mizuno et al., 2014; Box et al., 2015). Similarly, a QTL around PIF4 was found as a QTL for temperature sensitivity in flowering time in the Col-0/Tsu-0 population (Brock et al., 2010; Kumar et al., 2012; Delker et al., 2014). The MADS domain transcription factor SVP plays a role in thermal response, it has also been identified as a candidate conferring natural variation through both association mapping and QTL analysis, and its effect is also dependent on the genetic background (Lee et al., 2007, 2013; Atwell et al., 2010; Méndez-Vigo et al., 2013). We found a QTL for thermosensitivity in root length mapping to this region. Further analysis is required to verify any of these genes are the causal genes for the identified QTLs. There were some regions that appeared in more than one RIL/trait, for which we did not find any obvious candidate genes. These include the bottom of chromosome 4, which was detected as a QTL for temperature sensitivity in flowering in the Col-0/Tsu-0 population and as a QTL for flowering time only at 27°C in the Col-0/Cvi-0 population.
Cvi-0 Is Less Sensitive to Small Changes in Ambient Temperature
Several studies have revealed that the accession Cvi-0 is unusual in both phenotypic as well as genotypic characteristics (Alonso-Blanco et al., 1998; Rao and Davis, 1999; Borsani et al., 2001; El-Din El-Assal et al., 2001; Nordborg et al., 2005; Tessadori et al., 2009; Vasseur et al., 2011, 2014). Our findings show that within the tested ambient temperature range (23°C–27°C), Cvi-0 exhibits high temperature-associated phenotypes such as early flowering, longer hypocotyls, and elongated petioles even at lower temperatures. Cvi-0 was collected from the Cape Verde islands, where the fluctuations in average temperatures throughout the year is fairly minimal (23°C–27°C for most months) when compared with other natural habitats for Arabidopsis. Therefore, our findings might reflect a relaxed sensitization to temperature within this range, which may suit local environmental conditions. Our observations are also consistent with previous studies, which have shown that Cvi-0 is less sensitive to stomatal opening in response to environmental changes (Monda et al., 2011; Vasseur et al., 2014). Leaf temperature measurements in response to CO2 revealed that, unlike Col-0, Cvi-0 displayed minimal changes in leaf temperature (Monda et al., 2011). Furthermore, it has been shown that the heat-induced leaf hyponasty response is reduced in the Cvi-0 accession (van Zanten et al., 2009), consistent with our findings. A recent study by Suter and Widmer (2013) also found the Cvi-0 accession to be less responsive to a 3-d heat shock treatment. However, our studies do not rule out differential thermal responses in Cvi-0 in other temperature ranges.
We have shown that the CRY2-Cvi-0 allele is at least partially responsible for this reduced temperature sensitivity. Knocking down CRY2 in the Cvi-0 background increased sensitivity to temperature across a variety of traits supporting this hypothesis. The restoration of temperature sensitivity in the amiRCRY2-Cvi-0 lines is mostly due to a suppression of hypocotyl elongation at lower temperature conditions, suggesting that CRY2-Cvi-0 confers the high-temperature response at lower temperatures. Previous studies have shown that the CRY2-Cvi-0 allele inhibits hypocotyl elongation in response to an increase in blue light (El-Din El-Assal et al., 2001). Therefore, this role of CRY2-Cvi-0 in inducing hypocotyl elongation is likely to be independent of its role in light signaling. However, the exact mechanism through which CRY2-Cvi-0 confers temperature insensitivity remains unknown. Previous studies have shown that the stability of the CRY2 protein does not change in response to a change in temperature (Gould et al., 2013). While CRY2-Cvi-0 is more stable than CRY2-Ler, as shown earlier (El-Din El-Assal et al., 2001), it is still unclear how a stable CRY2-Cvi-0 modulates temperature perception and response. It is interesting to note that although CRY2 is known very well for its role in blue-light perception, it has also been implicated in sensing redox status and magnetism as well as entrainment of the circadian clock by temperature (Kaushik et al., 2007; Gegear et al., 2008; Yoshii et al., 2010; Fogle et al., 2011). Therefore, it is conceivable that CRY2-Cvi-0, due to its increased stability, may also modulate thermal response, although further studies are required to explore the underlying mechanism.
Thermal Responses Are Modified through Light × Temperature Interactions in a Background-Dependent Manner
Plant response to environmental changes is often a combinatorial response to multiple cues. Our study provides a nice example for how light and temperature interact to elicit a phenotypic response. We have shown that the CRY2-Cvi-0 allele leads to cotyledon opening even in darkness. This is consistent with earlier findings (Botto et al., 2003; Vasseur et al., 2014), where it has been demonstrated that the cotyledon unfolding to hourly pulses of far-red light is stronger in the Cvi-0 accession compared with Col-0 and CRY2-Cvi-0 may be contributing to altered thermal responses. However, in contrast to earlier studies, we have analyzed the effects of the alleles in a common loss-of-function background (cry2-1 in the Col-0 background), which allows us to decipher some of these interactions precisely. The CRY2-Cvi-0 allele is a gain-of-function allele with respect to its blue-light response and acts in a dominant manner. The dominant gain-of-function in blue-light signaling leads to an early cotyledon opening even in dark. Once cotyledons are open, the hypocotyl elongation stops. Thus, in the Col-0 background, we observed abolition of the temperature-induced hypocotyl elongation, because the cotyledons are opened due to the hyperactive light signaling conferred by CRY2-Cvi-0. The increased expression levels of CRY2-Cvi-0 specifically in the Col-0 background could possibly be the underlying mechanism for the same. While analysis of an individual phenotype would suggest that CRY2-Cvi-0 in the Col-0 background is insensitive in temperature-induced hypocotyl elongation, the underlying mechanism appears to the hyperactive light signaling conferred by the CRY2-Cvi-0, which leads to early cotyledon opening. However, it remains unclear as to how the CRY2-Cvi-0 confers differential response in petiole elongation in the Cvi-0 and Col-0 backgrounds. Nevertheless, the thermal response phenotype is an indirect effect of light × temperature interactions. On the other hand, we observed that these effects of this allele are highly attenuated in the Cvi-0 background, demonstrating how light and temperature interactions in a background-dependent manner modulate phenotypic responses in Arabidopsis.
Our findings support that unlike flowering time, the genetic architecture of the natural variation in thermal response likely comprises multiple small-effect alleles that occur at low frequency across global populations. We have uncovered several potential genomic regions, which can now be explored further to identify the causal genes that modulate natural variation in thermal response. We have characterized one of the underlying QTLs, which reveal a role for allelic variation at CRY2 in mediating natural variation in thermal response. Our findings with CRY2 demonstrate the complexity of light and temperature interactions and the effect of various genetic backgrounds on this interaction. Future studies would uncover the modifiers that regulate the light × temperature interactions, which lead to differential growth responses in plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All the accessions used in this study are listed with additional information in Supplemental Table S1. Seeds for accessions and the mutants were obtained from the Arabidopsis Biological Resource Center, the Institut National de la Research Agronomique, or RIKEN. The EDI-NIL and the CRY2 transgenic lines in the Ler background were gifted by Dr. Carlos Alonso-Blanco. The cry2 mutants in the Ler background (fha-1–fha-3) and the Col-0 background (cry2-1, CS3732) have been previously described (Koornneef et al., 1991; Guo et al., 1998). The transgenic lines harboring the CRY2-Col-0 and CRY2-Cvi-0 alleles were generated in this study (see below). The plants were grown in controlled growth chambers (Percival Scientific) or growth rooms at 23°C or 27°C under short-day (16-h-dark/8-h-light) conditions. Plants for transformation were grown in long day (16-h-light/8-h-dark) conditions. Relative humidity was maintained at 65% in all conditions. The plants were watered periodically.
Scoring of Flowering Time, Hypocotyl Elongation, and Primary Root Length
Flowering time was measured using either days to flowering or rosette leaf number, as described previously (Lempe et al., 2005). Hypocotyl elongation was measured either in plates or in soil. For the plate measurements, seeds were sterilized either by washing in 70% (v/v) ethanol with 0.1% (v/v) Triton X-100 for 3 to 5 min followed by 95% (v/v) ethanol for 1 to 2 min or by washing in bleach for 20 min followed by a water wash. Twenty seeds for each genotype (accession or RIL) were spotted onto 0.5× or 1× Murashige and Skoog media (pH 5.7) with or without Suc. Plates were placed in light- and humidity-controlled Percival chambers. For measurements in soil, seeds were stratified for 4 d in 0.1% (w/v) agarose and sown in soil, and the plants were cultivated in growth rooms at 23°C or 27°C in short days. Ten-day-old seedlings from soil were transferred to acetate sheets and scanned on a flatbed scanner. Plates were photographed along with a scale. Hypocotyl length was measured with ImageJ64 for Mac (National Institutes of Health). We found the heritability for this trait to be substantially higher in soil when compared with the plates across multiple experiments. For root length, plates were photographed on day 5, and the root length was measured through ImageJ64 as described above. To analyze photomorphogenesis/skotomorphogenesis, different genotypes were sown on Murashige and Skoog plates, covered with aluminum foil, and then kept vertically in Percival growth chambers for 10 d at the appropriate temperatures and light conditions. Plates were scanned after 10 d, and the plants were analyzed for their growth response, including cotyledon opening. Cotyledon opening was analyzed by measuring the angle between the embryonic leaves through ImageJ. To analyze allelic effects of CRY2, plants representing three independently derived stable transgenic lines at T3 harboring the pCRY2Cvi::CRY2Cvi-0 and pCRY2Col::CRY2Col-0 transgenes were analyzed twice, but, for comparison, data from a single transgenic line used for the phenotypic analysis is shown.
Statistical Analyses
Statistical analyses were done using JMP or SPSS (SAS Institute), Microsoft Excel, and R. The Col-0/Tsu-0, Col-0/Cvi-0, and Bay-0/Sha RILs have been previously described (Loudet et al., 2002; Simon et al., 2008). For hypocotyl elongation, at least 20 plants per RIL per temperature (23°C and 27°C short day) were measured. For flowering time analysis, six plants per RIL per temperature were grown in growth rooms in a completely randomized design. Broad-sense heritability was calculated as between-line variance divided by total variance. The total variance was partitioned into between-line variance and the residuals in a one-way ANOVA model using the genotype as a single factor of random effect and the phenotype as the response. Genetic correlations between the traits were calculated as described previously (Lempe et al., 2005). Temperature sensitivity was measured as described previously using the slope of the reaction norms (Lempe et al., 2005). To assess the effects of genotype, environment, and genotype × environment interactions, the entire data set (i.e. hypocotyl length for all plants across both temperature conditions) was analyzed using ANOVA, and the variance was partitioned. All the phenotypic data used for the analyses of various statistics are provided in Supplemental Table S2.
GWAS and QTL Analyses
The GWAS analyses were conducted using the GWAS Web Application online tool that harbors the genotypic information from the 1,001 genomes project (http://www.1001genomes.org; Seren et al., 2012). GWAS was conducted using the linear regression model/Kruskal-Wallis or AMM approach. Significant associations were found for temperature sensitivity in hypocotyl elongation. Temperature sensitivity measured either through the slope of the reaction norm or the residuals of the regression of hypocotyl length at 23°C on to 27°C gave similar results. The phenotypic data used for GWAS are provided in Supplemental Table S1. After initial analysis with AMM, we used the SNP with the highest association in the SPL2 gene as a cofactor and reran the association analyses. We analyzed the sampling bias on GWAS by systematically removing sets of eight accessions multiple times and found that it did not make any difference to the detected associations. QTL mapping was performed using the Multiple QTL modeling in R/qtl (http://www.biostat.jhsph.edu/kbroman/qtl/). Empirical log of the odds (LOD) threshold was determined through analysis of 5,000 permutations, which typically resulted in LOD scores around 2.5 for most of the phenotypes. As a conservative measure, we considered QTLs whose LOD score was above 3.
Generation of Transgenic Plants
A CRY2 genomic DNA fragment containing 2 kb of promoter sequence was PCR amplified from Col-0 and Cvi-0 using Phusion DNA polymerase with primers described in the Supplemental Table S8. The fragment was then cloned into a Gateway vector pDONR207 and then moved to destination vector pFK202 and used for Col-0 and cry2-1 mutant plant transformation. An artificial microRNA against CRY2 was constructed using the primers described in Supplemental Table S8, cloned under the Cauliflower mosaic virus 35S promoter (35S::amiRCRY2), and transformed into the Cvi-0 plants. Due to the requirement of long dormancy for germination of the Cvi-0 strain, transgenic seeds were sown after 3 to 4 months of storage. The presence of the CRY2 transgene was confirmed by genotyping using primers described in Supplemental Table S8 (El-Din El-Assal et al., 2001). Multiple independent T1 lines were analyzed for each of the transgenic plants and for specific analysis; independent T2 or T3 lines were used as specified. The specific mutations representing different CRY2 alleles were confirmed in transgenic plants by sequencing. The expression of CRY2 in transgenic plants was quantified by quantitative real-time PCR with SYBR Green on a LightCycler system (LightCycler 480, Roche) using primers described in Supplemental Table S8.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Reaction norms that represent thermal response across all the analyzed strains.
Supplemental Figure S2. Genome-wide association analysis of temperature sensitivity in hypocotyl elongation.
Supplemental Figure S3. Frequency distributions of hypocotyl lengths for F2 populations.
Supplemental Figure S4. QTL maps for multiple traits in different RIL populations.
Supplemental Figure S5. Cvi-0 plants in which CRY2 function is compromised display temperature sensitivity in petiole elongation.
Supplemental Figure S6. Allelic variation at CRY2 confers variation in temperature-associated architectural phenotypes.
Supplemental Figure S7. The effect of CRY2-Cvi-0 allele is dependent on the genetic background.
Supplemental Figure S8. Comparison of expression levels of CRY2 in transgenic lines.
Supplemental Table S1. Descriptions and the phenotypes of accessions used in the GWAS analysis.
Supplemental Table S2. Summary statistics for accessions and the recombinant inbred lines.
Supplemental Table S3. Phenotypic data from the Col-0 × Tsu-0 RIL population.
Supplemental Table S4. Phenotypic data from the Col-0 × Cvi-0 RIL population.
Supplemental Table S5. Phenotypic data from the Bay-0 × Sha RIL population.
Supplemental Table S6. Genetic correlations between various traits.
Supplemental Table S7. QTL identified in this work using different RILs.
Supplemental Table S8. Oligos used in this study.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center, the Institut National de la Recherche Agronomique, RIKEN, Carlos-Alonso Blanco, and Kemal Kazan for seeds and Richard Burke, Tim Connallon, and members of the Balasubramanian laboratory for discussions and critical comments on the article. The authors would also like to thank the anonymous reviewers for their comments, which helped improve this article.
Glossary
- RIL
recombinant inbred line
- GWAS
genome-wide association study
- QTL
quantitative trait locus
- AMM
approximate mixed model
- Col-0
Columbia
- Ler
Landsberg erecta
- Sha
Shahdara
- LOD
log of the odds
Footnotes
This work was supported by an International Postgraduate Research Scholarship (to W.Z.), by the Australian Research Council (Discovery Early Career Researcher Award DE130100320 to C.T., Discovery grants DP110100964 to S.S. and DP0983875 to S.B., and Future Fellowship FT100100377 to S.B.), and by Monash University (Larkins Fellowship to S.B.).
Articles can be viewed without a subscription.
References
- Alonso-Blanco C, Peeters AJ, Koornneef M, Lister C, Dean C, van den Bosch N, Pot J, Kuiper MT (1998) Development of an AFLP-based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J 14: 259–271 [DOI] [PubMed] [Google Scholar]
- An FM, Hsiao SR, Chan MT (2011) Sequencing-based approaches reveal low ambient temperature-responsive and tissue-specific microRNAs in phalaenopsis orchid. PLoS One 6: e18937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA (1997) A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell 9: 1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Agrawal M, Michael TP, Wessinger C, Maloof JN, Clark R, Warthmann N, Chory J, Weigel D (2006a) The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat Genet 38: 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Lempe J, Weigel D (2006b) Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet 2: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Weigel D (2006) Temperature Induced Flowering in Arabidopsis thaliana. Plant Signal Behav 1: 227–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero-Gil J, Salinas J (2013) Post-translational regulation of cold acclimation response. Plant Sci 205-206: 48–54 [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Ahn JH, Weigel D (2003) A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet 33: 168–171 [DOI] [PubMed] [Google Scholar]
- Borsani O, Valpuesta V, Botella MA (2001) Evidence for a role of salicylic acid in the oxidative damage generated by NaCl and osmotic stress in Arabidopsis seedlings. Plant Physiol 126: 1024–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto JF, Alonso-Blanco C, Garzarón I, Sánchez RA, Casal JJ (2003) The Cape Verde Islands allele of cryptochrome 2 enhances cotyledon unfolding in the absence of blue light in Arabidopsis. Plant Physiol 133: 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box MS, Huang BE, Domijan M, Jaeger KE, Khattak AK, Yoo SJ, Sedivy EL, Jones DM, Hearn TJ, Webb AA, et al. (2015) ELF3 controls thermoresponsive growth in Arabidopsis. Curr Biol 25: 194–199 [DOI] [PubMed] [Google Scholar]
- Brock MT, Maloof JN, Weinig C (2010) Genes underlying quantitative variation in ecologically important traits: PIF4 (phytochrome interacting factor 4) is associated with variation in internode length, flowering time, and fruit set in Arabidopsis thaliana. Mol Ecol 19: 1187–1199 [DOI] [PubMed] [Google Scholar]
- Coleman-Derr D, Zilberman D (2012) Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet 8: e1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Topp CN, McKinney EC, Meagher RB (2007) Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19: 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C, Sonntag L, James GV, Janitza P, Ibañez C, Ziermann H, Peterson T, Denk K, Mull S, Ziegler J, et al. (2014) The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Reports 9: 1983–1989 [DOI] [PubMed] [Google Scholar]
- Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JC, Lynn JR, Straume M, Smith JQ, Millar AJ (2006) FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18: 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KD, Lynn JR, Gyula P, Nagy F, Millar AJ (2005) Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics 170: 387–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Din El-Assal S, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M (2001) A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet 29: 435–440 [DOI] [PubMed] [Google Scholar]
- Filiault DL, Wessinger CA, Dinneny JR, Lutes J, Borevitz JO, Weigel D, Chory J, Maloof JN (2008) Amino acid polymorphisms in Arabidopsis phytochrome B cause differential responses to light. Proc Natl Acad Sci USA 105: 3157–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter AH, Fitter RS (2002) Rapid changes in flowering time in British plants. Science 296: 1689–1691 [DOI] [PubMed] [Google Scholar]
- Fogle KJ, Parson KG, Dahm NA, Holmes TC (2011) CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science 331: 1409–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Toledo-Ortiz G, Pyott DE, Halliday KJ (2014) Interaction of light and temperature signalling. J Exp Bot 65: 2859–2871 [DOI] [PubMed] [Google Scholar]
- Gegear RJ, Casselman A, Waddell S, Reppert SM (2008) Cryptochrome mediates light-dependent magnetosensitivity in Drosophila. Nature 454: 1014–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould PD, Ugarte N, Domijan M, Costa M, Foreman J, Macgregor D, Rose K, Griffiths J, Millar AJ, Finkenstädt B, et al. (2013) Network balance via CRY signalling controls the Arabidopsis circadian clock over ambient temperatures. Mol Syst Biol 9: 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Halliday KJ, Salter MG, Thingnaes E, Whitelam GC (2003) Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J 33: 875–885 [DOI] [PubMed] [Google Scholar]
- Kaushik R, Nawathean P, Busza A, Murad A, Emery P, Rosbash M (2007) PER-TIM interactions with the photoreceptor cryptochrome mediate circadian temperature responses in Drosophila. PLoS Biol 5: e146. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10: 310–316 [DOI] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA (2010) H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Lee H, Yoo SJ, Lee JH, Kim W, Yoo SK, Fitzgerald H, Carrington JC, Ahn JH (2010) Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res 38: 3081–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Ryu HS, Chung KS, Posé D, Kim S, Schmid M, Ahn JH (2013) Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342: 628–632 [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH (2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev 21: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempe J, Balasubramanian S, Sureshkumar S, Singh A, Schmid M, Weigel D (2005) Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet 1: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Adrian J, Pankin A, Hu J, Dong X, von Korff M, Turck F (2014) Induced and natural variation of promoter length modulates the photoperiodic response of FLOWERING LOCUS T. Nat Commun 5: 4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F (2002) Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet 104: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Maloof JN, Borevitz JO, Dabi T, Lutes J, Nehring RB, Redfern JL, Trainer GT, Wilson JM, Asami T, Berry CC, et al. (2001) Natural variation in light sensitivity of Arabidopsis. Nat Genet 29: 441–446 [DOI] [PubMed] [Google Scholar]
- May P, Liao W, Wu Y, Shuai B, McCombie WR, Zhang MQ, Liu QA (2013) The effects of carbon dioxide and temperature on microRNA expression in Arabidopsis development. Nat Commun 4: 2145. [DOI] [PubMed] [Google Scholar]
- Méndez-Vigo B, Martínez-Zapater JM, Alonso-Blanco C (2013) The flowering repressor SVP underlies a novel Arabidopsis thaliana QTL interacting with the genetic background. PLoS Genet 9: e1003289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Salome PA, McClung CR (2003) Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc Natl Acad Sci USA 100: 6878–6883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Nomoto Y, Oka H, Kitayama M, Takeuchi A, Tsubouchi M, Yamashino T (2014) Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol 55: 958–976 [DOI] [PubMed] [Google Scholar]
- Monda K, Negi J, Iio A, Kusumi K, Kojima M, Hashimoto M, Sakakibara H, Iba K (2011) Environmental regulation of stomatal response in the Arabidopsis Cvi-0 ecotype. Planta 234: 555–563 [DOI] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al. (2005) The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S. (2008) Temperature perception and signal transduction in plants. New Phytol 179: 615–628 [DOI] [PubMed] [Google Scholar]
- Poduska B, Humphrey T, Redweik A, Grbić V (2003) The synergistic activation of FLOWERING LOCUS C by FRIGIDA and a new flowering gene AERIAL ROSETTE 1 underlies a novel morphology in Arabidopsis. Genetics 163: 1457–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posé D, Verhage L, Ott F, Yant L, Mathieu J, Angenent GC, Immink RG, Schmid M (2013) Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503: 414–417 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Carlson KD, Girirajan S (2012) Lessons from model organisms: phenotypic robustness and missing heritability in complex disease. PLoS Genet 8: e1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S (2002) Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624 [DOI] [PubMed] [Google Scholar]
- Rao MV, Davis KR (1999) Ozone-induced cell death occurs via two distinct mechanisms in Arabidopsis: the role of salicylic acid. Plant J 17: 603–614 [DOI] [PubMed] [Google Scholar]
- Salomé PA, Bomblies K, Laitinen RA, Yant L, Mott R, Weigel D (2011) Genetic architecture of flowering-time variation in Arabidopsis thaliana. Genetics 188: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Wigge PA (2005) Ambient temperature perception in plants. Curr Opin Plant Biol 8: 483–486 [DOI] [PubMed] [Google Scholar]
- Schwartz C, Balasubramanian S, Warthmann N, Michael TP, Lempe J, Sureshkumar S, Kobayashi Y, Maloof JN, Borevitz JO, Chory J, et al. (2009) Cis-regulatory changes at FLOWERING LOCUS T mediate natural variation in flowering responses of Arabidopsis thaliana. Genetics 183: 723–732, 1SI–7SI [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seren Ü, Vilhjálmsson BJ, Horton MW, Meng D, Forai P, Huang YS, Long Q, Segura V, Nordborg M (2012) GWAPP: a web application for genome-wide association mapping in Arabidopsis. Plant Cell 24: 4793–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Plantegenet S, Hardtke CS (2008) Flowering as a condition for xylem expansion in Arabidopsis hypocotyl and root. Curr Biol 18: 458–463 [DOI] [PubMed] [Google Scholar]
- Simon M, Loudet O, Durand S, Bérard A, Brunel D, Sennesal FX, Durand-Tardif M, Pelletier G, Camilleri C (2008) Quantitative trait loci mapping in five new large recombinant inbred line populations of Arabidopsis thaliana genotyped with consensus single-nucleotide polymorphism markers. Genetics 178: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Irwin J, Dean C (2013) Remembering the prolonged cold of winter. Curr Biol 23: R807–R811 [DOI] [PubMed] [Google Scholar]
- Stief A, Altmann S, Hoffmann K, Pant BD, Scheible WR, Bäurle I (2014) Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26: 1792–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey KB, Storey JM (2013) Molecular biology of freezing tolerance. Compr Physiol 3: 1283–1308 [DOI] [PubMed] [Google Scholar]
- Sureshkumar S, Todesco M, Schneeberger K, Harilal R, Balasubramanian S, Weigel D (2009) A genetic defect caused by a triplet repeat expansion in Arabidopsis thaliana. Science 323: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Suter L, Widmer A (2013) Phenotypic effects of salt and heat stress over three generations in Arabidopsis thaliana. PLoS One 8: e80819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessadori F, van Zanten M, Pavlova P, Clifton R, Pontvianne F, Snoek LB, Millenaar FF, Schulkes RK, van Driel R, Voesenek LA, et al. (2009) Phytochrome B and histone deacetylase 6 control light-induced chromatin compaction in Arabidopsis thaliana. PLoS Genet 5: e1000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Harmon FG (2010) Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc Natl Acad Sci USA 107: 3257–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Undurraga SF, Press MO, Legendre M, Bujdoso N, Bale J, Wang H, Davis SJ, Verstrepen KJ, Queitsch C (2012) Background-dependent effects of polyglutamine variation in the Arabidopsis thaliana gene ELF3. Proc Natl Acad Sci USA 109: 19363–19367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten M, Voesenek LA, Peeters AJ, Millenaar FF (2009) Hormone- and light-mediated regulation of heat-induced differential petiole growth in Arabidopsis. Plant Physiol 151: 1446–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasseur F, Bontpart T, Dauzat M, Granier C, Vile D (2014) Multivariate genetic analysis of plant responses to water deficit and high temperature revealed contrasting adaptive strategies. J Exp Bot 65: 6457–6469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasseur F, Pantin F, Vile D (2011) Changes in light intensity reveals a major role for carbon balance in Arabidopsis responses to high temperature. Plant Cell Environ 34: 1563–1576 [DOI] [PubMed] [Google Scholar]
- Verhage L, Angenent GC, Immink RG (2014) Research on floral timing by ambient temperature comes into blossom. Trends Plant Sci 19: 583–591 [DOI] [PubMed] [Google Scholar]
- Wang JW. (2014) Regulation of flowering time by the miR156-mediated age pathway. J Exp Bot 65: 4723–4730 [DOI] [PubMed] [Google Scholar]
- Wigge PA. (2013) Ambient temperature signalling in plants. Curr Opin Plant Biol 16: 661–666 [DOI] [PubMed] [Google Scholar]
- Yoshii T, Hermann C, Helfrich-Förster C (2010) Cryptochrome-positive and -negative clock neurons in Drosophila entrain differentially to light and temperature. J Biol Rhythms 25: 387–398 [DOI] [PubMed] [Google Scholar]
- Zhu W, Ausin I, Seleznev A, Méndez-Vigo B, Picó FX, Sureshkumar S, Sundaramoorthi V, Bulach D, Powell D, Seemann T, et al. (2015) Natural variation identifies ICARUS1, a universal gene required for cell proliferation and growth at high temperatures in Arabidopsis thaliana. PLoS Genet 11: e1005085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.