The product of a termination factor promotes maturation of a transfer RNA in Arabidopsis plastids.
Abstract
Plastid gene expression is crucial for organelle function, but the factors that control it are still largely unclear. Members of the so-called mitochondrial transcription termination factor (mTERF) family are found in metazoans and plants and regulate organellar gene expression at different levels. Arabidopsis (Arabidopsis thaliana) mTERF6 is localized in chloroplasts and mitochondria, and its knockout perturbs plastid development and results in seedling lethality. In the leaky mterf6-1 mutant, a defect in photosynthesis is associated with reduced levels of photosystem subunits, although corresponding messenger RNA levels are unaffected, whereas translational capacity and maturation of chloroplast ribosomal RNAs (rRNAs) are perturbed in mterf6-1 mutants. Bacterial one-hybrid screening, electrophoretic mobility shift assays, and coimmunoprecipitation experiments reveal a specific interaction between mTERF6 and an RNA sequence in the chloroplast isoleucine transfer RNA gene (trnI.2) located in the rRNA operon. In vitro, recombinant mTERF6 bound to its plastid DNA target site can terminate transcription. At present, it is unclear whether disturbed rRNA maturation is a primary or secondary defect. However, it is clear that mTERF6 is required for the maturation of trnI.2. This points to an additional function of mTERFs.
Originally derived from a cyanobacterium-like ancestor (Raven and Allen, 2003), chloroplasts have retained a reduced genome that predominantly encodes proteins involved in photosynthesis and organellar gene expression (OGE), while much of its genetic heritage now resides in the nucleus (for review, see Leister and Kleine, 2011). Hence, tight coordination of nuclear and plastid gene expression is required to ensure the development and maintenance of chloroplasts and their functionality. Although chloroplasts preserve features of prokaryotic genome organization, their gene expression system is far more complex than that of its cyanobacterial progenitor (for review, see Liere et al., 2011), as the maturation of chloroplast RNAs and their translation into proteins require a plethora of nucleus-encoded proteins. This machinery now includes additional RNA polymerases and σ-factors as well as monospecific or merospecific RNA maturation factors that promote RNA transcription, splicing, editing, end formation, or translation (for review, see Stern et al., 2010; Lerbs-Mache, 2011; Hammani et al., 2014; Tiller and Bock, 2014; Börner et al., 2015; Schmitz-Linneweber et al., 2015; Shikanai, 2015).
Families of proteins with similar modular architectures comprising repeated helical motifs play important roles in OGE (Hammani et al., 2014). One of these families is the mitochondrial transcription termination factor family (mTERF), members of which have been identified in both metazoans and plants (Linder et al., 2005). The family founder member, human mTERF1, is one of four mammalian mTERF proteins and was identified one-quarter of a century ago as a factor that promotes transcription termination in human mitochondrial extracts (Kruse et al., 1989). The assumed function of human mTERF1 as a transcription terminator (of heavy-strand transcripts) gave the whole family its name. Lately, a model has been suggested in which the major function of human mTERF1 is not termination of transcription but termination of antisense transcription to prevent light-strand transcripts from proceeding around the mitochondrial DNA (mtDNA) circle, thus avoiding transcriptional interference at the light-strand promoter from which they originated (Terzioglu et al., 2013).
The mouse mTERF2 coimmunoprecipitates with mTERF1 and mTERF3, and it was suggested that this possibly occurs by binding to the same mtDNA region (Wenz et al., 2009). In contrast, another report showed that the DNA-binding activity of mTERF2 is not sequence specific (Pellegrini et al., 2009). Thus, the molecular function of mouse mTERF2 remains unclear. Knockout Mterf3 mice are embryo lethal (Park et al., 2007). In the conditional Mterf3 heart knockout, levels of the 39S mitochondrial ribosomal subunit are reduced and ribosomal assembly is perturbed (Wredenberg et al., 2013); thereby, a novel role for mouse mTERF3 in the biogenesis of metazoan mitochondrial ribosomes was identified. Like Mterf3 knockout mice (Park et al., 2007), Mterf4 knockout mice are embryo lethal (Cámara et al., 2011). Loss of mTERF4 in the mouse heart increases steady-state levels of mtDNA transcripts, including 12S and 16S ribosomal RNA (rRNA), by activating de novo transcription (Cámara et al., 2011). Moreover, two different immunoprecipitation strategies have identified NSUN4 (for NOL1/NOP2/Sun domain family member4), a mitochondrial rRNA methyltransferase, as an interaction partner of mTERF4 (Cámara et al., 2011). Interestingly, human mTERF4 forms a complex with NSUN4 that is required to assemble the small and large ribosomal subunits to form a monosome (Metodiev et al., 2014).
The fact that mammalian mTERF1, mTERF3, and mTERF4 share a common fold and are structurally similar to each other, even though mTERF3 was crystallized in the absence of its substrate (Spåhr et al., 2010) and mTERF4 in complex with another protein (Spåhr et al., 2012), supports the conclusion that mTERF proteins have evolved to bind nucleic acids (Byrnes and Garcia-Diaz, 2011). However, it is becoming increasingly clear that the nucleic acid need not necessarily be double-stranded DNA (dsDNA); RNAs can also serve as targets of mTERFs. Intriguingly, while the function for mTERF2 has not been clarified yet, three of the four mammalian mTERFs do not actually terminate transcription, as their designation suggests, but appear to function in antisense transcription termination and ribosome biogenesis.
In spite of their even greater diversity in plants (for review, see Kleine, 2012), knowledge of the functions of mTERFs in photosynthetic organisms remains sparse. Most of the 35 Arabidopsis (Arabidopsis thaliana) mTERF proteins are targeted to mitochondria and/or chloroplasts (Babiychuk et al., 2011), and mTERF proteins have been shown to be associated with the plastid transcriptionally active chromosome in Arabidopsis and maize (Zea mays; Pfalz et al., 2006; Majeran et al., 2012). This nucleoid-enriched proteome includes proteins involved in DNA organization, replication, and repair as well as in transcription, mRNA processing, splicing, and editing, supporting a pivotal role for mTERFs in OGE (Majeran et al., 2012). The importance of mTERF proteins in plants is demonstrated by their loss-of-function phenotypes. Arabidopsis mutants defective in the mTERF proteins SINGLET OXYGEN-LINKED DEATH ACTIVATOR10 (SOLDAT10)/mTERF1 (Meskauskiene et al., 2009) and BELAYA SMERT (BSM)/RUGOSA2 (RUG2)/mTERF4 (Babiychuk et al., 2011; Quesada et al., 2011) exhibit developmental arrest in the embryo. The maize BSM ortholog ZmmTERF4 is responsible for group II intron splicing, and this function in chloroplast RNA splicing might be conserved in BSM/RUG2 (Hammani and Barkan, 2014). In light of the role of BSM in chloroplast intron splicing (Hammani and Barkan, 2014), Hsu et al. (2014) investigated splicing events in mitochondria from mterf15 plants. Indeed, their results strongly suggest that mTERF15 is also involved in intron splicing, namely splicing of intron 3 in the mitochondrial NADH dehydrogenase2 (nad2) transcript. In vitro studies have shown that the mTERF-like gene of Chlamydomonas reinhardtii (MOC1) binds specifically to a sequence within the mitochondrial rRNA-coding module S3. Because knockout of MOC1 alters antisense transcription around this site, it was concluded that MOC1 acts as a transcription terminator in vivo (Wobbe and Nixon, 2013).
In this study, we describe the identification and characterization of mTERF6, a protein that is crucial for chloroplast development. mTERF6 interacts in vitro and in vivo with a sequence found in the chloroplast Ile transfer RNA gene trnI.2 [tRNAIle(GAU)] located 3′ of rrn16 (16S rRNA). Lack of this interaction is associated with the reduced accumulation of plastid ribosomes in knockdown (mterf6-1) and knockout (mterf6-2) mutants. Furthermore, T7 RNA polymerase-dependent transcription of synthetic genes containing the mTERF6 target sequence can be terminated in vitro by the addition of recombinant mTERF6. At present, it is not resolved whether altered rRNA maturation in mterf6 mutants is due to a primary or a secondary effect. However, lack of mTERF6 leads to strongly reduced aminoacylation of trnI.2; thus, mTERF6 is most likely a factor involved in the modification of pretrnI.2.
RESULTS
Identification and Phenotypic Analysis of Mutants for the MTERF6 Locus
Screening of the Arabidopsis GABI_KAT transfer DNA (T-DNA) insertion collection (Rosso et al., 2003) for lines that show alterations in the effective quantum yield of PSII, designated ΦII (Varotto et al., 2000), resulted in the recovery of a set of mutants with defects in photosynthesis. In the line photosynthesis altered mutant48-1 (pam48-1; GABI_152G06), the ΦII was significantly reduced compared with the wild type (Table I). Furthermore, the maximum quantum yield of PSII (variable fluorescence [Fv]/maximum fluorescence [Fm]) was drastically reduced (wild type, 0.81 ± 0.01; pam48-1, 0.54 ± 0.03), implying a defect in energy transfer within PSII. Moreover, an increase in the excitation pressure of PSII, as indicated by the magnitude of 1-qP, was observed (Table I). When grown on soil, pam48-1 mutants displayed a reduced growth rate. The pale-green coloration of true leaves (Fig. 1A) reflects their lower overall chlorophyll content, and the chlorophyll a/b ratio was slightly decreased (Table I).
Table I. Basic photosynthetic parameters and chlorophyll content of wild-type (Col-0), pam48-1 (mterf6-1), and 35S:PAM48.1 pam48-1 (35S:mTERF6.1 mterf6-1) plants at the eight-leaf rosette stage.
Photosynthetic parameters are derived from measurements on at least five leaves from different plants. Actinic light intensity was 95 µE m−2 s−1. Acetone-extracted leaf pigments were measured photometrically, and chlorophyll concentrations were calculated according to Lichtenthaler (1987) and are reported in μg mg−1 fresh weight. Mean values ± sd are provided.
| Parameter | Col-0 | mterf6-1 | 35S:mTERF6.1 mterf6-1 |
|---|---|---|---|
| Fv/Fm | 0.81 ± 0.01 | 0.54 ± 0.03a | 0.82 ± 0.01 |
| ΦII | 0.70 ± 0.03 | 0.43 ± 0.02a | 0.69 ± 0.04 |
| Nonphotochemical quenching | 0.14 ± 0.03 | 0.11 ± 0.04a | 0.16 ± 0.02 |
| 1-qP | 0.11 ± 0.03 | 0.57 ± 0.04a | 0.14 ± 0.03 |
| Chlorophyll a + b | 1.15 ± 0.09 | 0.81 ± 0.17a | 1.12 ± 0.13 |
| Chlorophyll a/b | 3.01 ± 0.03 | 2.75 ± 0.12 | 3.05 ± 0.06 |
Values significantly different from Col-0 (P < 0.05).
Figure 1.
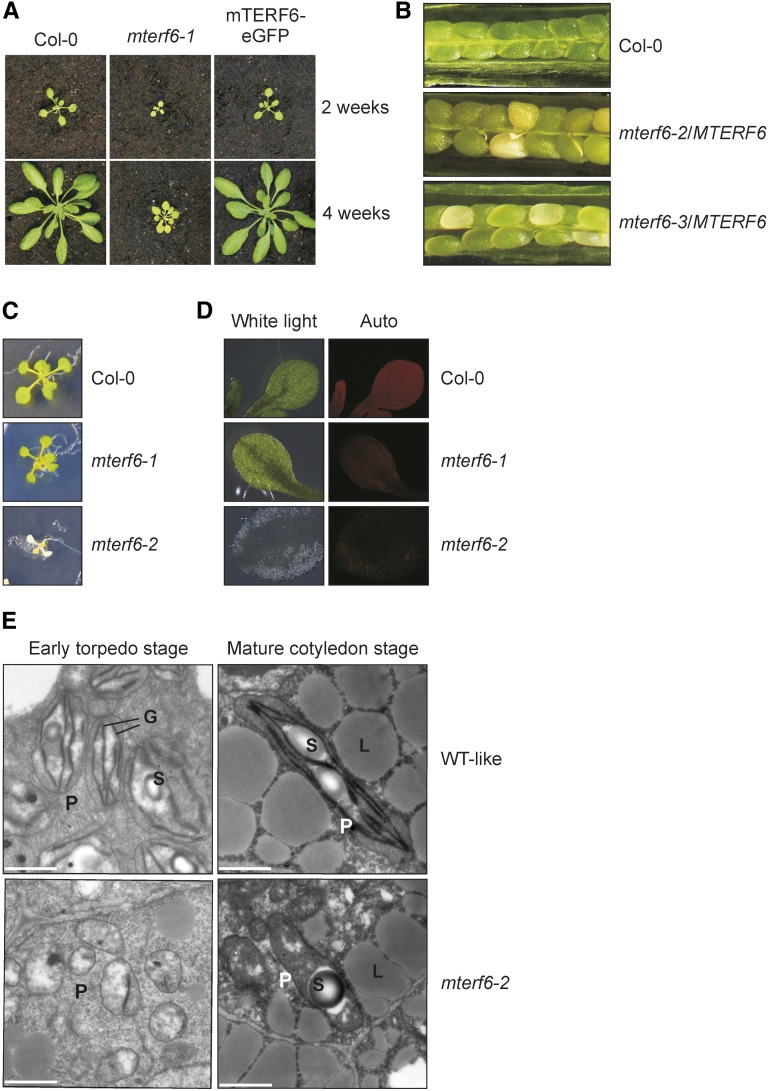
Phenotypic characterization of mterf6 T-DNA insertion mutants. A, Phenotypes of 2- and 4-week-old wild-type (Col-0) plants, mterf6-1, and mterf6-1 complemented with the first splice variant of the AT4G38160 gene (mTERF6-eGFP). B, In siliques of mterf6-2/MTERF6 and mterf6-3/MTERF6 plants, white ovules are found interspersed with normal green ovules. White ovules account for about 25% (122 of 523 for mterf6-2/MTERF6 and 70 of 276 for mterf6-3/MTERF6) of all ovules analyzed. C, When grown on Murashige and Skoog (MS) medium supplemented with Suc, 18-d-old mterf6-2 seedlings show significant growth retardation. They display white cotyledons and have just developed the first pair of true leaves, while wild-type (Col-0) and mterf6-1 seedlings of the same age present three pairs of true leaves. D, White light (left) and chlorophyll fluorescence (Auto; right) images of cotyledons of wild-type, mterf6-1, and mterf6-2 seedlings. E, Transmission electron micrographs of ultrathin sections of heterozygous and homozygous mterf6-2 embryos. Early torpedo and mature cotyledon stage embryos were isolated from green (homozygous wild type [WT] and heterozygous wild type like) and white mutant ovules of mterf6-2/MTERF6 siliques. The green embryos show well-developed chloroplasts with thylakoid membranes beginning to form grana. Plastids in the mutant embryo remain in the proplastid stage. Bar = 1 μm. G, Grana; L, lipid bodies; P, plastids; S, starch grain.
The Leaky Mutation pam48-1 Is Due to T-DNA Insertion in the 5′ Untranslated Region of MTERF6
The photosynthetic phenotype of pam48-1 segregated as a monogenic, recessive trait, and cosegregation of the sulfadiazine resistance marker on the T-DNA with the mutant phenotype indicated that the mutation was due to the T-DNA insertion. Isolation of the genomic sequence flanking the left border of the T-DNA enabled identification of the insertion site in the 5′ untranslated region of the gene AT4G38160 (also known as PIGMENT DEFECTIVE191; Tzafrir et al., 2004) or MTERF6 (Kleine, 2012) at position −115 relative to the start codon (Supplemental Fig. S1A). According to The Arabidopsis Information Resource genome annotation 10, AT4G38160 is a single-copy gene with three predicted transcript splice forms, AT4G38160.1, AT4G38160.2, and AT4G38160.3, which differ only at their 3′ ends (Supplemental Fig. S1A). In order to experimentally test the three gene models, reverse transcription (RT)-PCR was performed on head-to-tail ligated mRNA (circular RT-PCR; Supplemental Fig. S1B). Sequencing showed 5′ and 3′ ends that mapped close to those expected for gene model AT4G38160.1. Additional RT-PCRs did not amplify products corresponding to AT4G38160.2, AT4G38160.3, or alternative gene models, indicating that only AT4G38160.1 transcripts are made in the tissues analyzed here (Supplemental Fig. S1B). Furthermore, RT-PCR and real-time PCR analysis showed that the T-DNA insertion reduces levels of the AT4G38160.1 transcript by about 90% compared with the wild type (Supplemental Fig. S1, C and D).
To confirm that the altered activity of AT4G38160 was responsible for the mutant phenotype of pam48-1, the pam48-1 line was genetically complemented with the full-length splice variant 1 (AT4G38160.1) fused upstream of the enhanced GFP reporter gene under the control of the cauliflower mosaic virus 35S promoter (mTERF6-eGFP). Subsequent phenotypic and chlorophyll fluorescence analyses showed that the wild-type function had been restored in the complemented mutant (Fig. 1; Table I). This indicates that the pam48-1 phenotype is indeed caused by knockdown of the AT4G38160 gene. Therefore, we refer to pam48-1, pam48-2, and pam48-3 (see below) in the following as mterf6-1, mterf6-2, and mterf6-3 and to AT4G38160/PAM48 as mTERF6.
Complete Loss of mTERF6 Function Results in Seedling Lethality and Impairment of Chloroplast Development
Because the mterf6-1 mutation does not completely prevent transcription of the AT4G38160 gene (Supplemental Fig. S1C), further mutant lines were sought. The SAIL_360_H09 and SGT1851-3-3 lines were identified in the SIGnAL T-DNA Express Arabidopsis Gene Mapping Tool (http://signal.salk.edu/cgi-bin/tdnaexpress) and in the Dissociation line list (Parinov et al., 1999), respectively. In these lines, designated mterf6-2 (previously designated as pde191-2; www.seedgenes.org; Tzafrir et al., 2004) and mterf6-3, the pCSA110 T-DNA and the Dissociation transposon were inserted in exon 1 of MTERF6 at positions 706 and 511 relative to the start codon, respectively (Supplemental Fig. S1A). In both mterf6-2 and mterf6-3, MTERF6.1 transcripts were undetectable (Supplemental Fig. S1C). Heterozygous mterf6-2/MTERF6 and mterf6-3/MTERF6 plants were phenotypically indistinguishable from wild-type plants. However, in siliques of mterf6-2/MTERF6 and mterf6-3/MTERF6 plants, approximately 25% of ovules (122 out of 523 ovules for mterf6-2/MTERF6 and 70 out of 276 ovules for mterf6-3/MTERF6) turned white (Fig. 1B; segregation significant with χ2 = 0.82 or χ2 = 0.019, respectively), indicating that chloroplast development is disrupted in embryos homozygous for the mutant alleles.
Homozygous mterf6-2 mutant seeds were smaller than wild-type seeds and failed to germinate on soil. Moreover, in seedlings grown on Suc-containing medium, mterf6-2 seedlings displayed an albino phenotype and generally stopped growing after 2 weeks, while the mterf6-1 seedlings appeared almost like the wild type under these conditions (Fig. 1C). Additionally, chlorophyll autofluorescence was barely detectable in mterf6-2 seedlings (Fig. 1D). This, together with the white mterf6-2 ovules (Fig. 1B), indicates that chloroplast development was already significantly disturbed during embryogenesis in mutants homozygous for this allele. To further investigate chloroplast development in mterf6-2 embryos, early torpedo stage and mature cotyledon stage embryos isolated from green (homozygous wild type and heterozygous wild type like) and white (homozygous mutant) ovules from mterf6-2/MTERF6 plants were fixed, embedded, sectioned, and examined for the presence and morphology of embryo plastids using transmission electron microscopy. In the green embryos, the thylakoid membrane system developed and began to stack into grana (Fig. 1E), a significant indicator that plastids had begun to differentiate into chloroplasts. In contrast, only early, protoplast-like plastids were detected in mutant embryos at the early torpedo stage (Fig. 1E). In contrast, grana began to develop in only a minor fraction of the mterf6-2 plastids but did not proceed to become mature cotyledon-stage plastids (Fig. 1E). Altogether, the data suggest that mTERF6 is essential for plastid development.
mTERF6 Is Targeted to Both Chloroplasts and Mitochondria
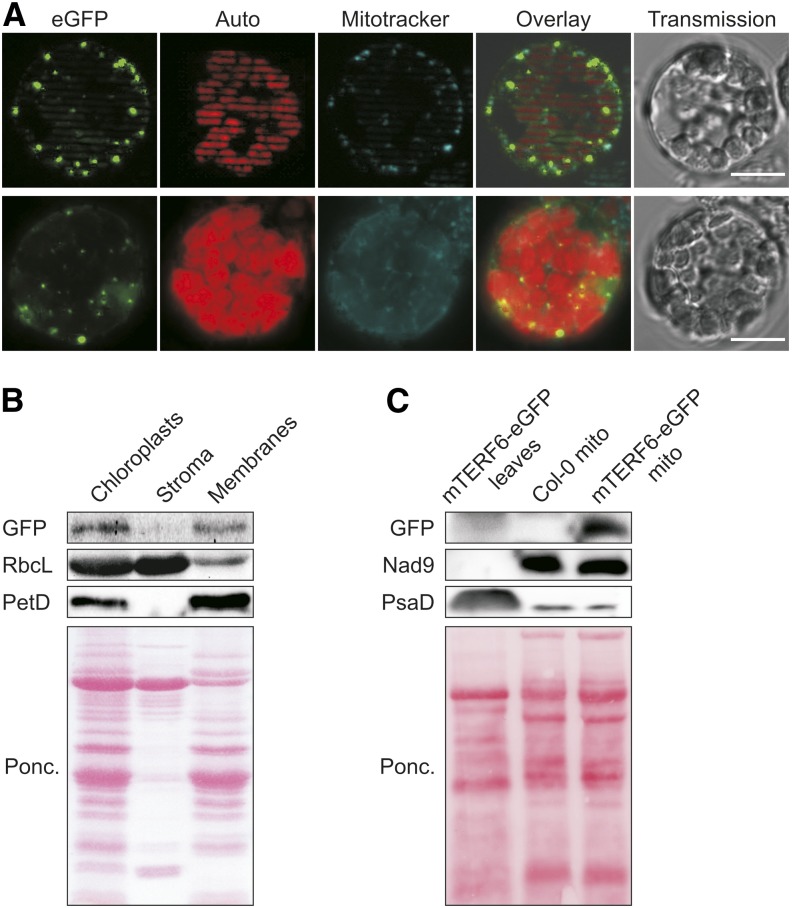
GFP fusions of all members of the Arabidopsis mTERF family have recently been monitored by fluorescence microscopy in protoplasts produced from transient expression assays and from guard cells of transgenic plants (Babiychuk et al., 2011). These data indicated that mTERF6 is targeted to mitochondria (Babiychuk et al., 2011). However, the altered morphology of mterf6-2 chloroplasts, the coexpression analysis, and the aforementioned sorting of mTERF6 to the chloroplast cluster (Kleine, 2012) all argue in favor of a function of mTERF6 in chloroplasts. Furthermore, computational prediction of the subcellular localization of mTERF6 by the various algorithms implemented in SUBA3 (http://suba.plantenergy.uwa.edu.au; Tanz et al., 2013) yielded a plastid localization with maximum score. Therefore, the subcellular location of mTERF6 was experimentally revisited by examining protoplasts of mterf6-1 plants that overexpressed the mTERF6-eGFP fusion with a fluorescence microscope. Localization of the fusion protein to mitochondria was confirmed (Fig. 2A), but in addition, dotted fluorescence signals were detected that colocalized with the chlorophyll autofluorescence (Fig. 2A), suggesting that mTERF6 is targeted to both chloroplasts and mitochondria. To further explore the issue, chloroplasts from mTERF6-eGFP plants were fractionated into stroma and membranes. The fractions were subjected to immunoblot analysis, and the purity of fractions was tested by monitoring the stromal Rubisco, large subunit (RbcL) and thylakoid Photosynthetic electron transfer D (PetD) proteins (Fig. 2B). Using this approach, the mTERF6 fusion protein was found specifically in the chloroplast membrane fraction (Fig. 2B). Furthermore, to confirm the mitochondrial localization of mTERF6 based on an analysis of intracellular GFP fluorescence distribution in mTERF6-eGFP plants (Fig. 2A), a leaf extract from 4-week-old mTERF6-eGFP plants and mitochondria-enriched fractions from Columbia-0 (Col-0) and mTERF6-eGFP plants was subjected to immunoblot analysis (Fig. 2C). The mitochondrial Nad9 protein was only detected in the mitochondria-enriched fractions, whereas the chloroplast PsaD signal was strong in the leaf extract and (very) weak in both mitochondria-enriched fractions, corroborating the enrichment of mitochondria and the depletion of chloroplasts in these two preparations. However, the GFP-tagged protein was not detected in the mTERF6-eGFP leaf extract but only in the mitochondria-enriched fraction of mTERF6-eGFP plants, confirming mitochondrial localization of mTERF6 (Fig. 2C). In summary, mTERF6 is dually targeted to mitochondria and chloroplasts.
Figure 2.
The mTERF6 protein is targeted to chloroplasts and mitochondria. A, The mterf6-1 mutant was stably transformed to express an mTERF6-eGFP fusion protein, and expression of the fusion was monitored in protoplasts of positive transformants using fluorescence microscopy. The eGFP signal reveals the distribution of the fusion protein, autofluorescence of chlorophyll (red) pinpoints chloroplasts (Auto), and Mitotracker signals (cyan) visualize mitochondria (Mitotracker). Bars = 10 µm. B, The mTERF6 protein is targeted to chloroplasts. Chloroplast, stroma, and membrane fractions were isolated from complemented mterf6-1 (mTERF6-eGFP) plants, subjected to SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and exposed to antibodies raised against GFP (to detect the mTERF6-GFP fusion protein), RbcL (as a control for stromal proteins), or PetD (as a control for the membrane fraction). C, The mTERF6 protein is targeted to mitochondria. A whole-leaf extract from complemented 4-week-old mterf6-1 (mTERF6-eGFP) plants and mitochondria-enriched fractions (mito) from Col-0 and mTERF6-eGFP plants were subjected to SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and exposed to antibodies raised against GFP (to detect the mTERF6-GFP fusion protein), Nad9 (as a control for mitochondrial proteins), or PsaD (as a control for chloroplast proteins). Ponc., Ponceau Red.
Impairment of mTERF6 Affects Chloroplast Gene Expression
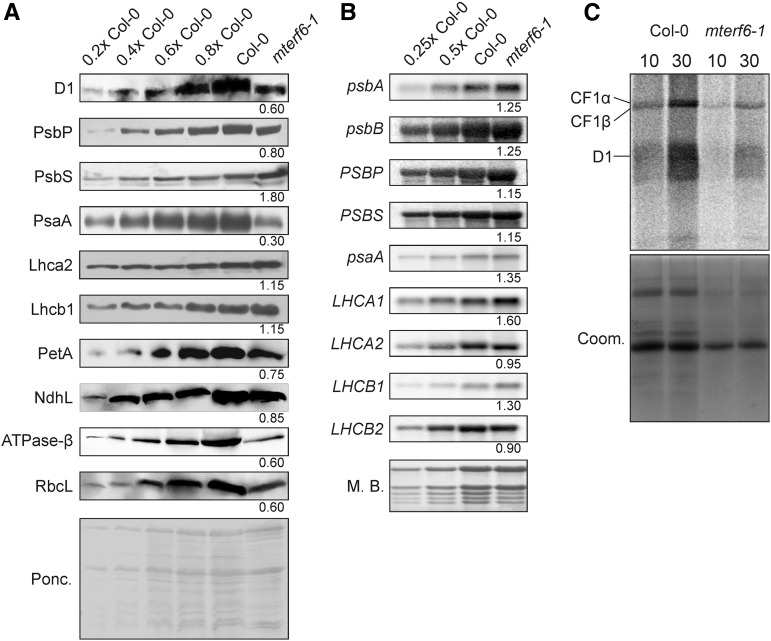
To study the effects of decreased mTERF6 activity, the leaky mterf6-1 allele was used in the following experiments. Immunoblot analysis was performed on total protein extracts from wild-type (Col-0) and mterf6-1 leaves to investigate whether the defect in photosynthetic activity found in the mterf6-1 mutant was a consequence of reduced amounts of photosynthetic proteins. In the mterf6-1 mutant, representative chloroplast-encoded subunits of PSII and PSI, the cytochrome b6f complex, and the chloroplast ATP synthase accumulated to lower levels than in the wild type (Fig. 3A). On the contrary, nucleus-encoded proteins like PsbP (a subunit of the oxygen-evolving complex), NdhL [a subunit of the chloroplast NAD(P)H dehydrogenase complex], and Lhca2 and Lhcb1 (subunits of light-harvesting complex I [LHCI] and LHCII, respectively) accumulated to or nearly to wild-type levels, whereas PsbS showed a 1.8-fold increase relative to the wild type (Fig. 3A).
Figure 3.
The accumulation of chloroplast-encoded proteins is disrupted at the posttranscriptional level in mterf6-1. A, Immunoblot analysis of representative thylakoid proteins. Total leaf protein extracts from wild-type (Col-0) and mterf6-1 plants were fractionated by SDS-PAGE, and blots were probed with antibodies raised against individual subunits of photosynthetic complexes. Increasing levels of wild-type proteins were loaded in the lanes marked 0.2× Col-0, 0.4× Col-0, 0.6× Col-0, 0.8× Col-0, and Col-0. Loading was adjusted to the fresh weights of leaf tissue. The Ponceau Red (Ponc.)-stained blot served as the loading control. Quantification of signals relative to the wild type (=1) is provided below each mterf6-1 lane. B, Steady-state levels of the transcripts of photosynthetic genes in wild-type (Col-0) and mterf6-1 plants. Total RNA was isolated from fresh leaf material obtained from wild-type and mterf6-1 plants, and aliquots (3.75, 7.5, and 15 µg from the wild type; 15 µg from mterf6-1) were fractionated on a formaldehyde-containing denaturing gel, transferred onto a nylon membrane, and probed with [α-32P]dCTP-labeled complementary DNA (cDNA) fragments specific for transcripts encoding individual subunits of PSII (psbA, psbB, PSBP, and PSBS), PSI (psaA), LHCI (LHCA1 and LHCA2), and LHCII (LHCB1 and LHCB2). rRNA was visualized by staining the membrane with Methylene Blue (M. B.) and served as a loading control. Quantification of signals relative to the wild type (=1) is provided below each mterf6-1 lane. C, In vivo pulse labeling of thylakoid membrane proteins with [35S]Met in the presence of cycloheximide indicates that translation occurs at reduced rates in mterf6-1 chloroplasts. Proteins were resolved by SDS-PAGE after pulse labeling for 10 and 30 min and visualized by autoradiography. CF1α and CF1β, α- and β-subunits of the ATP synthase; Coom, Coomassie Blue.
The reduced accumulation of plastid-encoded thylakoid proteins in the mterf6-1 mutant could be due to a translational defect or might be the consequence of impaired accumulation of their transcripts. To assess the latter possibility, RNA gel-blot analysis was performed to examine levels of the plastid-encoded transcripts for PSI and PSII. Our results indicated that even slightly higher amounts of transcripts of psaA (encoding a reaction center protein of PSI) and psbA and psbB (encoding the D1 and CP47 proteins of PSII, respectively) accumulated in mterf6-1 and wild-type plants (Fig. 3B). However, other chloroplast transcripts are affected by an impairment of mTERF6. For example, as determined by real-time PCR, RNA polymerase β subunit-1 (rpoC1), ribosomal proteins S12 (rps12), ribosomal protein L16 (rpl16), rpl2, and ycf3 (for hypothetical chloroplast open reading frame3) are up-regulated while petB, petD, and NADH dehydrogenase ND1 are down-regulated in knockout mterf6 mutants (Supplemental Fig. S8A). The levels of transcripts of nuclear photosynthesis genes were comparable to wild-type levels. Together, these findings suggest that levels of nucleus-encoded photosynthetic proteins are not diminished in the mterf6-1 mutant. Moreover, the reduced amounts of chloroplast-encoded photosynthetic proteins seen in the mterf6-1 mutant are not associated with lower levels of transcripts of the corresponding chloroplast genes.
To determine whether a defect in translation is responsible for the reduced accumulation of chloroplast proteins, the synthesis of plastid-encoded thylakoid membrane proteins was studied by pulse labeling of wild-type and mterf6-1 mutant leaves in the presence of cycloheximide, which inhibits the translation of nucleus-encoded proteins. After pulse labeling for 10 and 30 min, de novo synthesis of the D1 protein and the α- and β-subunits of the ATP synthase were found to be strongly reduced in mterf6-1 relative to the wild type (Fig. 3C). Thus, the synthesis of chloroplast proteins is indeed reduced in the mterf6-1 mutant.
Accumulation of Plastid Ribosomes Is Reduced in mterf6 Mutants
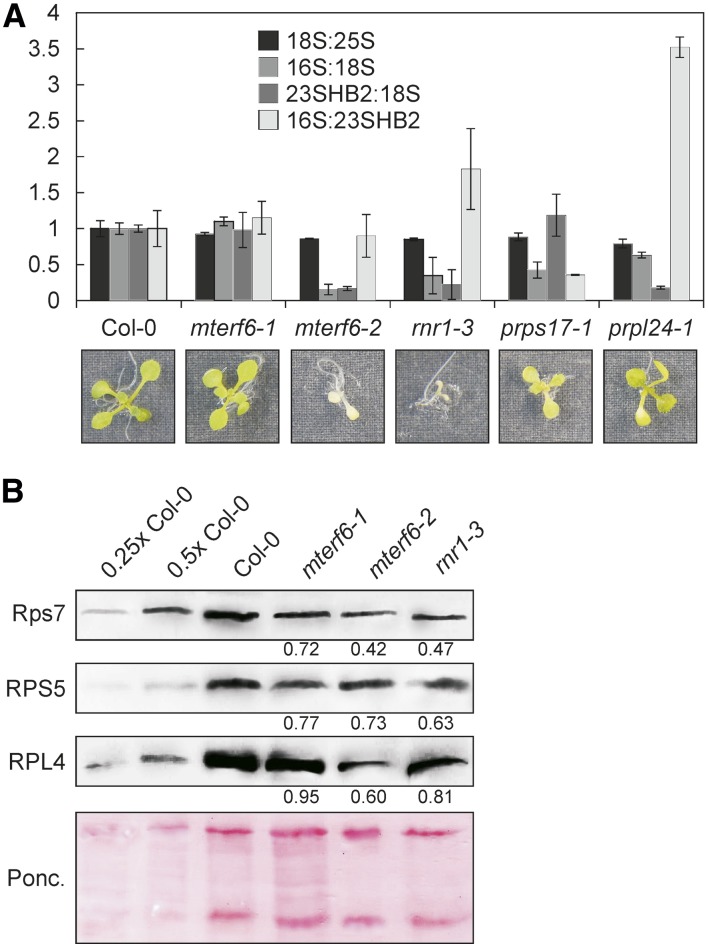
Because rRNAs do not stably accumulate unless they are incorporated into ribosomal subunits, the quantification of 16S and 23S rRNA levels can serve as a proxy for the accumulation of the 30S and 50S ribosomal subunits, respectively. Moreover, the small and large ribosomal subunits can accumulate independently from each other, because they only associate during the translation process. Thus, rRNA quantifications can serve to distinguish between defects in biogenesis of the large versus the small subunit of the ribosome (Tiller et al., 2012). Amounts of chloroplast rRNAs were decreased in soil-grown mterf6-1 plants (Fig. 3B; Supplemental Fig. S2A) and even more in mterf6-2 (grown on MS medium; Supplemental Fig. S2), as determined by Methylene Blue staining of membranes. To quantify those reductions accurately, total RNA preparations of the wild type and mterf6-1 and mterf6-2 mutants grown on MS medium were analyzed with a Bioanalyzer, whereby 16S rRNA, 23S rRNA, and 18S rRNA levels were quantified as means for the plastid small and large and the cytosolic small ribosomal subunit, respectively. Total RNA preparations of the ribonucleotide reductase1-3 (rnr1-3) mutant (Bollenbach et al., 2005) and the chloroplast ribosomal mutants plastid ribosomal protein S17 (prps17-1) and plastid ribosomal protein L24 (prpl24-1; Romani et al., 2012) served as controls for altered rRNA accumulation (Supplemental Fig. S2). Indeed, as shown before (Tiller et al., 2012), the prps17-1 mutant showed a decreased 16S-18S rRNA ratio and no significant change in the 23S-18S rRNA ratio (Fig. 4A), indicating a specific reduction of the small plastid ribosomal subunit. In prpl24-1, the large plastid ribosomal subunit was specifically reduced (indicated as the 23S-18S rRNA ratio in Fig. 4A). Interestingly, rRNA levels were wild type like in mterf6-1 seedlings grown on MS medium (Fig. 4A), although rRNA levels were decreased in soil-grown mterf6-1 plants and the more sensitive 23S rRNA detection by northern-blot analysis revealed a reduction of spliced 23S rRNA levels to 63% in mterf6-1 seedlings. In mterf6-2 and rnr1-3, already Bioanalyzer profiles revealed the reduction of both the 16S-18S rRNA and the 23S-18S rRNA ratios, indicating a reduced accumulation of the small and the large plastid ribosomal subunits (Fig. 4A). This finding was corroborated by immunoblot analysis of total protein extracts from wild-type, mterf6-1, mterf6-2, and rnr1-3 seedlings. In the mterf6-1 mutant, representative proteins of the small (chloroplast-encoded Rps7 and nucleus-encoded RPS5) and large (nucleus-encoded RPL4) plastid ribosomal subunits accumulated to lower levels, as in the wild type (Fig. 4B). This reduction was even more pronounced in mterf6-2 and rnr1-3 seedlings (Fig. 4B). In sum, the accumulation of ribosomal subunits is reduced.
Figure 4.
The accumulation of plastid ribosomes is reduced in mterf6 mutants. A, Total RNA preparations of MS medium-grown wild-type (Col-0), mterf6-1, mterf6-2, rnr1-3, prps17-1, and prpl24-1 seedlings were run on a Bioanalyzer (Supplemental Fig. S2C), and changes in rRNA ratios were calculated with ImageJ (Col-0 set to 1). Error bars indicate sd. 23SHB2, HIDDEN BREAK2 product of 23S rRNA. B, Immunoblot analysis of representative plastid ribosome subunits. Total leaf protein extracts from wild-type (Col-0), mterf6-1, mterf6-2, and rnr1-3 seedlings were fractionated by SDS-PAGE, and blots were probed with antibodies raised against the plastid-encoded Rps7 and the nucleus-encoded RPS5 and RPL4 proteins. Increasing levels of wild-type proteins were loaded in the lanes marked 0.25× Col-0, 0.5× Col-0, and Col-0. Loading was adjusted to the fresh weights of seedlings. The Ponceau Red (Ponc.)-stained blot served as the loading control. Quantification of signals relative to the wild type (=1) is provided below each mutant lane.
mTERF6 Binds to Chloroplast DNA Sequences
Because some mammalian mTERF proteins bind to specific mtDNA sequences in vitro and in vivo (Kruse et al., 1989; Roberti et al., 1991; Daga et al., 1993; Fernandez-Silva et al., 1997; Terzioglu et al., 2013), we asked whether mTERF6 might similarly interact with plant organellar DNAs.
We first created a structural model of mTERF6 to study whether the protein is, in principle, capable of binding to nucleic acids. To this end, we examined the recently published crystal structures of human mTERF1 bound to its double-stranded mtDNA target (Jiménez-Menéndez et al., 2010; Yakubovskaya et al., 2010). The human mTERF1 protein folds into a half-doughnut structure (Yakubovskaya et al., 2010) that wraps around the DNA helix (Fig. 5A). mTERF6 was aligned with the human mTERF1 structure, and the model obtained by I-TASSER (Zhang, 2008) suggests that mTERF6 consists of a central core that is structurally homologous to human mTERF1, flanked by short N- and C-terminal extensions (Fig. 5A). In fact, the mTERF fold has evolved to mediate protein-nucleic acid (especially dsDNA) interactions (Spåhr et al., 2010; Yakubovskaya et al., 2010). In this respect, the model predicts that mTERF6 should also be capable of binding dsDNA in a similar fashion to that observed for human mTERF1.
Figure 5.
The mTERF6 protein is capable of in vitro binding to both DNA and RNA. A, Structural homology modeling of mTERF6. The tertiary structure of human mTERF1 (hMTERF1; deposited as 3MVA) was downloaded from the National Center for Biotechnology Information structure database (http://www.ncbi.nlm.nih.gov/structure) and matched to the predicted structure of mTERF6. The three-dimensional structure of mTERF6 was calculated by I-TASSER (see “Materials and Methods”), and the molecular graphics images were produced using the UCSF Chimera package (see “Materials and Methods”). B, SDS-PAGE showing the purity of recombinant His6-tagged mTERF6 (mTERF6) expressed in E. coli and purified on nickel-nitrilotriacetic acid agarose. C, Southwestern and northwestern analyses to demonstrate the in vitro binding of His6-tagged mTERF6 (mTERF6) to dsDNA and RNA, respectively. Shown is the Ponceau Red-stained membrane blotted with increasing concentrations of recombinant mTERF6 protein and a maltose binding protein-tagged control protein. The left and right parts of the membrane were incubated with a radiolabeled total DNA or RNA preparation, respectively.
To experimentally test for the nucleic acid-binding activity of mTERF6, full-length His6-tagged mTERF6 (His6-mTERF6) was expressed in Escherichia coli (for purity of the recombinant protein, see Fig. 5B) and used for southwestern and northwestern analyses on membrane blots bearing the reconstituted His6-mTERF6 protein. Indeed, recombinant mTERF6 is capable of binding both DNA and RNA (Fig. 5C).
To further investigate the DNA-binding activity of mTERF6 and to identify its target sequences, a B1H system was employed that was originally designed to identify DNA sequences recognized by transcription factors (Meng and Wolfe, 2006). The B1H site-selection system employs as key components a transcription factor expression vector, a reporter vector that contains a library of randomized binding sites, and a bacterial selection strain. Full-length mTERF6.1 was expressed as a C-terminal fusion to the α-subunit of RNA polymerase RpoA, hereafter referred to as α-mTERF6. Transcription of this α-mTERF6 fusion was controlled by the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible lppC/lacUV5 promoter. Furthermore, a library of randomized 15-bp oligonucleotides was cloned upstream of a weak lac promoter that drives the expression of HISTIDINE3 and URACIL REQUIRING3. If mTERF6 recognizes a DNA sequence within a library member, the RNA polymerase will be recruited to the promoter, activating transcription of the reporter genes. After setting up the selection conditions as described in “Materials and Methods,” 53 randomly selected colonies were sequenced. Among the sequences obtained, three 15-bp motifs were found to be overrepresented (e-values are as follows: motif 1 = 3.6e-029, motif 2 = 1.7e-016, and motif 3 = 3.0e+001). These were identified in 11, 19, and eight of the 53 sequences, respectively (Supplemental Fig. S3) by the de novo motif discovery tool Multiple Em for Motif Elicitation (MEME; http://meme.sdsc.edu/meme/cgi-bin/meme.cgi; Bailey and Elkan, 1994) using default settings. The sequences in each motif were used to construct a sequence logo (http://weblogo.berkeley.edu/logo.cgi; Crooks et al., 2004) that shows the level of conservation at each position as a function of its base frequency (Supplemental Fig. S3). The motif files were submitted to the Motif Alignment and Search Tool (MAST) implemented in MEME and searched against the mitochondrial and chloroplast genome sequences. Intriguingly, significant sequence hits were only found in or adjacent to six chloroplast genes, referred to as target I (trnV.1), target II (trnI.2/trnI.3), target III (rps7.1/rps7.2), target IV (psbA), target V (trnT.1), and target VI (ndhH and trnT.1; Table II; Supplemental Fig. S3). Note that the complete chloroplast rRNA operon (containing the trnI.2/trnI.3 genes) is located in an inverted repeat of the chloroplast genome, and for simplicity, we will refer in the following to only one of these isoforms. Furthermore, some of the targets are associated with genes that code for isoacceptor tRNAs (Sato et al., 1999). For instance, the inverted repeat tRNA genes trnV.2 and trnV.3 as well as trnV.1 all code for Val tRNAs.
Table II. Putative target sequences of mTERF6 identified by bacterial one-hybrid (B1H) screening.
The consensus sequences derived from the sequences identified in the B1H screen (Supplemental Fig. S4) were used to query the nuclear, mitochondrial, and chloroplast genome sequences.
| Name | Locus Identifier | Gene Name | Description | Identified Sequence (5′–3′) | P |
|---|---|---|---|---|---|
| target I | ATCG00450 | trnV.1 | 3′ of tRNA-Val (trnV.1) | CATTAAAATCAAAAT | 3.2e-05 |
| target II | ATCG00930/ATCG01200 | trnI.2/trnI.3 | Intron of tRNA-Ile (trnI.2 and trnI.3) | ATTGCGTCGTTGTGC | 9.5e-06 |
| target III | ATCG00900/ATCG01240 | rps7.1/rps7.2 | Exon of ribosomal proteins s7.1 and s7.2 (rps7.1 and rps7.2) | CAGTAAAAGCAAGAC | 1.1e-05 |
| target IV | ATCG00020 | psbA | Exon of psbA | ATGGTATTCGTGAAC | 4.5e-05 |
| target V | ATCG00260 | trnT.1 | 3′ of tRNA-Thr (trnT.1) | ATCATGACTATATCC | 6.4e-0 |
| target VI | ATCG01110/ATCG00260 | ndhH/ trnT.1 | Exon of NAD(P)H dehydrogenase subunit H (ndhH)/3′ of tRNA-Thr (trnT.1) | GAATTGAGTCGTAT | 3.0e-05 |
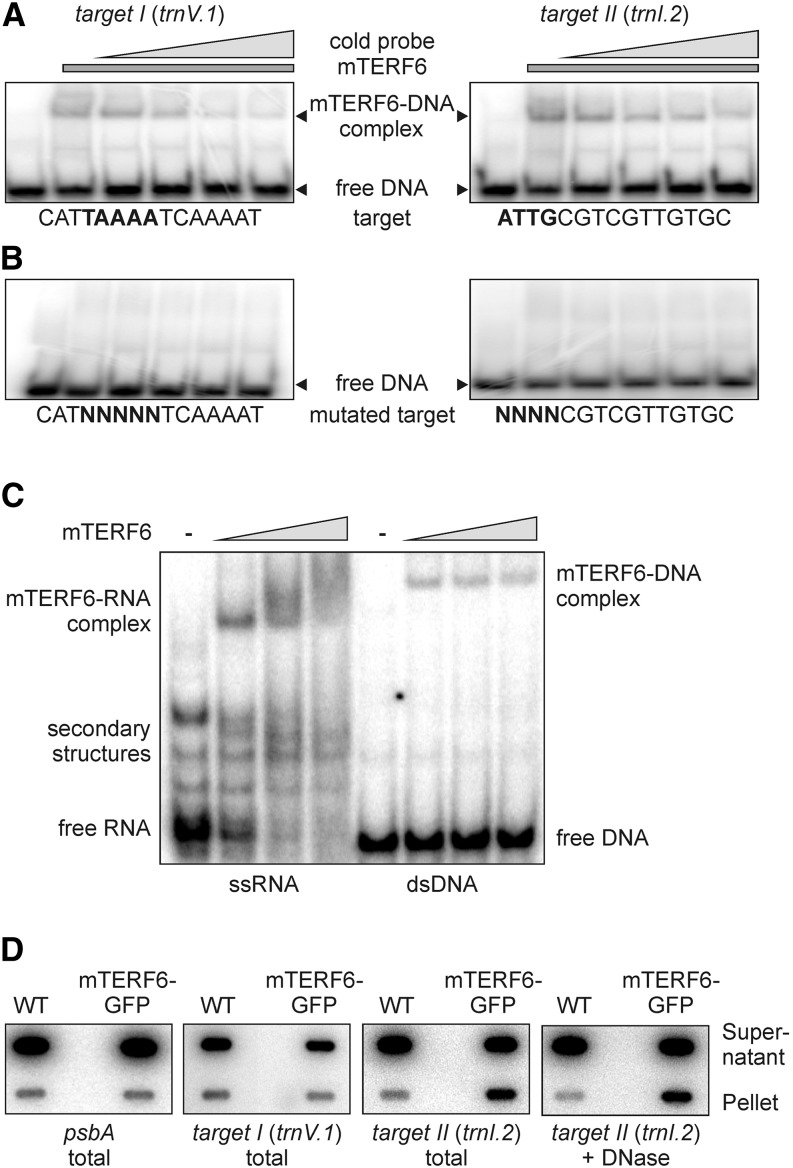
To experimentally corroborate the nucleic acid-binding activity of mTERF6 to these specific sites, His6-mTERF6 was used for electrophoretic mobility shift assays (EMSAs; Fig. 6, A and B; Supplemental Fig. S4). To this end, six different double-stranded 60-bp oligonucleotides were designed, representing each of the six tentative binding sites identified in the chloroplast genome. When 1 µg of His6-mTERF6 was added to the different 32P-labeled probes and the mixtures were electrophoresed, band shifts were observed with the target I (trnV.1) and target II (trnI.2) probes, indicating that these sequences formed complexes with the protein (Fig. 6A). However, protein-DNA complexes were not produced upon incubation with the other probes tested: psbA, trnT.1, rps7.1, and ndhH (Supplemental Fig. S4). Furthermore, the intensity of the shifted target I (trnV.1) and target II (trnI.2) bands decreased progressively when increasing concentrations of the respective unlabeled dsDNA (Fig. 6A) were added to incubation mixtures. Moreover, EMSA experiments with a 32P-labeled probe in which the core region of the target I (trnV.1) and target II (trnI.2) binding sequences was mutated did not result in shifted bands (Fig. 6B), suggesting that mTERF6 specifically binds in vitro to the target I (trnV.1) and target II (trnI.2) dsDNA sequences (Table II).
Figure 6.
The mTERF6 protein interacts with a nucleic acid sequence located in the plastid trnI.2 gene (target II). A, EMSAs were performed with recombinant His6-tagged mTERF6. Aliquots (1 µg) of recombinant protein were incubated with 100 ng of [γ-32P]ATP-labeled dsDNA probes representing the target I and target II sequences of mTERF6 in the presence of increasing concentrations (up to 10-fold relative to the labeled probe; indicated by the gray triangles) of the same unlabeled probe as competitor. Binding reactions were then subjected to electrophoresis on nondenaturing TBE-polyacrylamide gels (see “Materials and Methods”). EMSAs for the other four putative target sequences are shown in Supplemental Figure S4. B, Demonstration of specific sequence binding. EMSA reactions were performed as in A with probes in which 4 or 5 bp within the target I or target II sequence had been replaced by random nucleotides (mutated target). C, EMSA showing that mTERF6 binds ssRNA matching the trnI.2/3 target (target II). Increasing concentrations (50, 250, and 500 nm; indicated by the gray triangles) of recombinant His6-tagged mTERF6 protein were incubated with 50 pm [γ-32P]ATP-labeled ssRNA and dsDNA probes corresponding to the target II sequence in the plastid trnI.2 gene. Binding reactions were fractionated on nondenaturing TBE-polyacrylamide gels. D, Coimmunoprecipitation analysis to investigate the in vivo binding properties of mTERF6 to target I and target II sequences. The detergent-treated stromal extracts of complemented mterf6-1 (35S:MTERF6.1 mterf6-1) and wild-type (WT; Col-0) plants were subjected to immunoprecipitation with GFP antibodies. Nucleic acids from the supernatant and the immunoprecipitation pellets were recovered (total) and for probing with target II in addition DNase treated (+ DNase). Nucleic acids were slot blotted onto a nylon membrane and probed with [α-32P]dCTP-labeled cDNA fragments specific for target I, target II, and psbA.
Because southwestern and northwestern analyses indicated that recombinant mTERF6 is capable of binding both DNA and RNA (Fig. 5C), we tested whether mTERF6 can also bind to the target II (trnI.2) RNA sequence in vitro. In fact, in an EMSA experiment, mTERF6 was found to bind more strongly to single-stranded RNAs (ssRNAs) spanning the same sequence as the target II (trnI.2) dsDNA (Fig. 6C). Furthermore, coimmunoprecipitation experiments with chloroplast extracts isolated from the complemented mutant line (mTERF6-eGFP) showed that mTERF6 binds in vivo to a sequence bearing target II (trnI.2) but not target I (trnV.1) and also not the psbA control sequence (Fig. 6D). The strength of the target II (trnI.2) signal was essentially the same after DNase treatment (Fig. 6D), demonstrating that mTERF6 binds in vivo to target II (trnI.2) RNA.
mTERF6 Terminates Transcription in Vitro
Human mTERF1 was originally identified as a DNA-binding protein that protected a specific site in the Leu tRNA gene located 3′ of the rrn16S gene (Kruse et al., 1989) in mitochondria and was later shown to be sufficient to mediate transcriptional termination in vitro (Asin-Cayuela et al., 2005). Strikingly, although the mTERF1 target and mTERF6 target II sequences are not related, their relative positions in the organelle genomes are similar (Supplemental Fig. S5A). Because mTERF6 can bind target II (trnI.2) dsDNA (although weaker than its corresponding RNA; Fig. 6C), we investigated whether mTERF6 can terminate transcription in vitro. For this purpose, an assay for transcription termination activity was adapted from Prieto-Martín et al. (2004). The pGEM plasmid containing either the intact or a mutated target II (trnI.2) site (Supplemental Fig. S5) was linearized downstream of the T7 promoter and the target II (trnI.2) site and then transcribed with T7 RNA polymerase in the presence of recombinant mTERF6. When the termination activity was tested with the intact target site, two transcripts were generated: a longer run-off transcript and a shorter truncated transcript that corresponds to the terminated transcript (Supplemental Fig. S5C). Substituting the wild-type target site with a mutated target II site resulted in a prominent run-off transcript and a shorter faint band, which did not correspond in size to the terminated transcript. Moreover, when the termination assay is carried out with the intact binding site and increasing concentrations of the recombinant mTERF6 protein, increased amounts of the terminated transcript are produced, while the omission of mTERF6 prevented the formation of this terminated transcript (Supplemental Fig. S5D). Thus, the recombinant mTERF6 protein is not only capable of binding to the target II (trnI.2) sequence, its attachment promotes the in vitro termination of transcription within that sequence.
Reduced mTERF6 Levels Lead to Altered rRNA Maturation
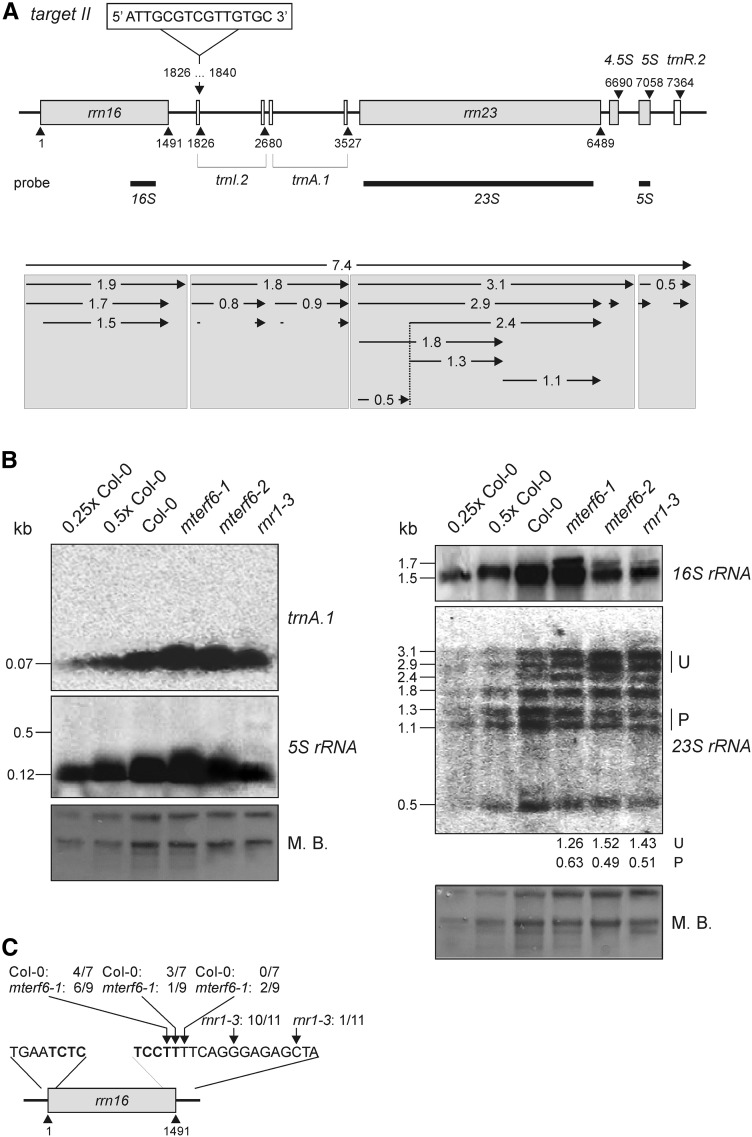
The chloroplast Ile tRNA gene (trnI.2) that harbors the target II (trnI.2) site is transcribed within the rrn operon. In the chloroplasts of land plants, the rRNAs incorporated into the 30S and 50S ribosomal subunits are encoded in the rrn operon, which contains the genes for 16S, 23S, 4.5S, and 5S rRNAs together with tRNAs for Ile, Ala, and Arg (Fig. 7A). This gene cluster is transcribed as a single molecule and processed via multiple endonucleolytic cleavage and exonucleolytic trimming events, as also occurs in bacteria (Strittmatter and Kössel, 1984).
Figure 7.
Expression and processing of chloroplast rRNAs and trnA.1 in wild-type (Col-0), mterf6-1, mterf6-2, and rnr1-3 seedlings grown on MS medium, and mapping of 16S rRNA ends in Col-0 and mterf6-1 plants. A, Schematic representation of the chloroplast rrn operon and the mTERF6 target site located in the trnI.2 gene (target II). Gray and white boxes indicate genes encoding rRNA and tRNA, respectively; introns are depicted as solid lines. The positions of the probes used in northern-blot analysis are marked by black rectangles under the operon. B, Analysis of transcript levels and splice forms of rrn genes and trnA.1. Total RNA was isolated as in Figure 2B, resolved on a denaturing gel, transferred onto a nitrocellulose membrane, and probed with [α-32P]dCTP-labeled cDNA fragments specific for rrn16, rrn23, and rrn5 as indicated in A and an [γ-32P]ATP end-labeled oligonucleotide probe specific for trnA.1. rRNA was visualized by staining the membrane with Methylene Blue (M. B.) as a loading control. Quantification of unprocessed (U) and processed (P) 23S rRNA relative to the wild type (=1) is provided below the mterf6-1, mterf6-2, and rnr1-3 lanes. C, Mapping of 16S rRNA 3′ ends by circular RT-PCR. The arrows point to the 3′ ends detected, and the numbers of clones obtained at the indicated positions are given. The rnr1-3 data are taken from Bollenbach et al. (2005). The 5′ and 3′ ends of the mature 16S rRNA sequence are depicted in boldface.
As shown above, the accumulation of plastid ribosomes is reduced in mterf6 plants (Fig. 4). To assess whether and to what extent the binding of mTERF6 to the target II (trnI.2) site 3′ of the rrn16S gene might influence the expression or processing of the chloroplast rrn operon, total RNA was analyzed by RNA gel-blot analysis (Fig. 7). The rnr1-3 mutant, which shows plastid ribosome deficiency of a similar magnitude to that seen in the mterf6-2 mutant (Fig. 3), was included as a control. Our results indicate that in mterf6 mutants, but not in the rnr1-3 mutant, levels of the mature trnA.1 and 5S rRNA transcripts were increased relative to the wild type (Fig. 7B). In contrast, the levels of mature 23S rRNA were decreased to 63% in mterf6-1 and to approximately 50% of the wild-type levels in mterf6-2 and rnr1-3 seedlings, and an unprocessed precursor overaccumulated (Fig. 7B). Moreover, in mterf6 and rnr1-3 mutants, a higher Mr product that is not seen in the wild type was detected by the rrn16 probe (Fig. 7B). Such overaccumulation of 16S and 23S rRNAs has been found before in the rnr1-3 mutant (Bollenbach et al., 2005) as well as in differentiation and greening-like1 (dal1; Bisanz et al., 2003) and several other Arabidopsis mutants defective in plastid ribosomal proteins (Tiller and Bock, 2014). Remarkably, genes encoding proteins involved in plastid rRNA processing, like the 16S rRNA processing protein RimM (AT5G46420), DAL1 (AT2G33430; Bisanz et al., 2003), and RNR1, are coexpressed with MTERF6 (as determined with the Arabidopsis Co-Response Database (http://csbdb.mpimp-golm.mpg.de/csbdb/dbcor/ath/ath_tsgq.html; Steinhauser et al., 2004). RNR1 is one of the few proteins for which a direct role in rRNA processing has been established, and its activity is essential for rRNA 3′ end maturation (Bollenbach et al., 2005). Therefore, we tested whether a relative lack of mTERF6 has similar effects on rRNA maturation to the rnr1 mutation by mapping the 5′ and 3′ ends of 16S rRNA in mterf6-1 and wild-type plants by means of circular RT-PCR (Fig. 7C). All clones analyzed had mature 5′ ends in wild-type and mterf6-1 plants. Furthermore, all wild-type-derived clones and seven out of nine mterf6-1-derived clones displayed mature 3′ ends. The remaining two mterf6-1-derived clones contained only a one-nucleotide 3′ extension (Fig. 7C). Consequently, mterf6-1 plants do not exhibit the same molecular phenotype as rnr1 plants, in which no mature 16S rRNA 3′ ends are detected: Bollenbach et al. (2005) found that 10 of their 11 clones contained five-nucleotide 3′ extensions; the 11th bore a 12-nucleotide 3′ extension.
Overall, it is not clear at present whether altered rRNA maturation in mterf6 mutants is due to a direct effect resulting from a lack of binding of mTERF6 at the rrn operon or results from a partial loss of translation and therefore represents a secondary effect of the reduction of mTERF6 levels.
Relative Lack of mTERF6 Leads to the Overaccumulation of tRNAIle(GAU) (trnI.2) RNAs
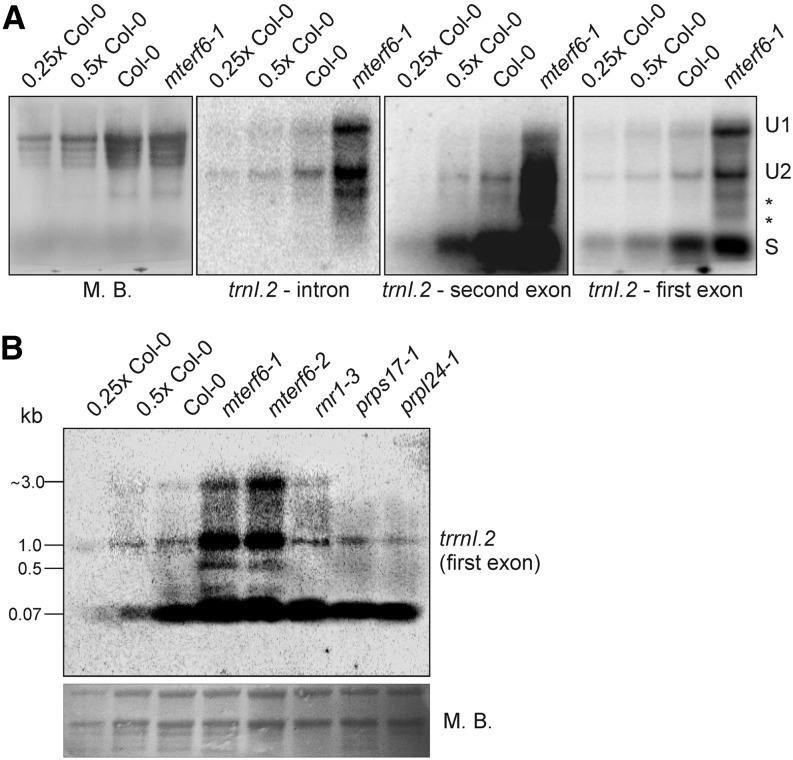
The determined target II (trnI.2) site covers the last nucleotide of the first exon and the very beginning of the trnI.2 intron. Therefore, we asked whether mTERF6 might act as a splicing factor of trnI.2. To determine how mTERF6 affects trnI.2 expression and maturation, RNA gel-blot hybridizations were carried out using probes targeting both exons of trnI.2 as well as its intron (Fig. 8). Both unspliced and mature trnI.2 RNAs accumulated to much higher levels in mterf6-1 compared with the wild type (Fig. 8A). Furthermore, two additional unprocessed forms were found in mterf6-1 mutant plants (Fig. 8A, asterisks); however, these might also be represented in wild-type plants, as indicated by the faint bands obtained by probing with trnI.2 second exon antisense primers (Fig. 8A). To investigate whether enhanced trnI.2 accumulation might be solely a consequence of pleiotropic effects of impaired plastid translation, RNA gel-blot hybridization using a probe targeting the first exon of trnI.2 was performed for the control mutants rnr1-3, prps17-1, and prpl24-1. In prps17-1 and prpl24-1, trnI.2 accumulated to wild-type levels, while trnI.2 accumulation was slightly enhanced in rnr1-3, although not to the extent seen in the mterf6 mutants (Fig. 8B). At all events, the relative lack of mTERF6 leads to an overaccumulation of trnI.2 levels and seems not to preclude the splicing of trnI.2.
Figure 8.
Expression and processing of chloroplast trnI.2 in wild-type (Col-0) and mterf6 plants. A, Analysis of the levels and splicing patterns of the trnI.2/trnI.3 (trnI.2) transcript in wild-type (Col-0) and mterf6-1 soil-grown plants. Total RNA was isolated as in Figure 2B, resolved on a denaturing gel, transferred onto a nylon membrane, and probed with [γ-32P]ATP end-labeled oligonucleotide probes specific for the intron and first and second exons. rRNA was visualized by staining the membrane with Methylene Blue (M. B.) as a loading control. S, Spliced; U1, unprocessed 1; U2, unprocessed 2. B, Analysis of the levels and processing patterns of the trnI.2/trnI.3 (trnI.2) transcript in wild-type (Col-0), mterf6-1, and mterf6-2 seedlings and, as controls, rnr1-3, prps17-1, and prpl24-1 seedlings grown on MS medium. Total RNA was treated as in A and probed with [γ-32P]ATP end-labeled oligonucleotide probes specific for the first exon of trnI.2. rRNA was visualized by staining the membrane with Methylene Blue as a loading control.
The trnI.2 binding site lies next to one of the origins of DNA replication (oriA), as determined in the tobacco (Nicotiana tabacum) plastid genome (Kunnimalaiyaan and Nielsen, 1997). Real-time PCR was performed to compare copy numbers of chloroplast DNA surrounding the trnI.2 binding site and also the psbA gene in the mterf6-1 mutant compared with the wild type. However, plastid DNA levels were not significantly altered in mterf6-1 (Supplemental Fig. S6A). The overaccumulation of trnI.2 transcripts could also be caused by an accelerated trnI.2 transcription rate in mterf6 mutants. However, run-on analysis showed that the transcription rates of trnI.2, rrn16 (5′ of the target II binding site), trnA.1, and rrn23 (both 3′ of the target II binding site) were similar in mterf6-1 and wild-type plants (Supplemental Fig. S6B).
The mTERF4 protein of maize (ZmmTERF4) was recently identified as a protein that coimmunoprecipitates with many chloroplast introns. Moreover, the splicing of some of these introns (group II) is disrupted in Zmmterf4 mutants, which accounts for the loss of plastid ribosomes in such plants (Hammani and Barkan, 2014). To find out whether mTERF6 associates with more plastid RNA-binding sites in vivo, RNA immunoprecipitation and hybridization to gene chips (RIP-chip) was performed as described (Schmitz-Linneweber et al., 2005). We failed to identify more RNA targets of mTERF6 (Supplemental Fig. S7; Supplemental Table S1), although we had observed binding of mTERF6 to target II (trnI.2) RNA both in vitro (Fig. 6C) and in vivo (Fig. 6D). The absence of target II RNA binding in the RIP-chip assay could result from the prominent membrane localization of mTERF6. Therefore, to completely rule out a tentative splicing defect in mterf6 plants, splicing of chloroplast transcripts was investigated by real-time PCR analyses in knockout mterf6 mutants (Supplemental Fig. S8). To account for pleiotropic effects on the splicing of plastid introns, the mutants chloroplast biogenesis19 (clb19) and plastid transcriptionally active2 (ptac2) were used as controls. In these mutants, transcription (ptac2) or transcript editing and transcription (clb19) in chloroplasts is impaired, such that they display, like the mterf6 mutants, severe impairment of chloroplast development and reduced 16S and 23S rRNA levels (Pfalz et al., 2006; Chateigner-Boutin et al., 2008). The real-time assay revealed an increased ratio of unspliced to spliced mRNA for some intron-containing plastid mRNAs in the mterf6 mutants (Supplemental Fig. S8). However, the same extent of unspliced-to-spliced ratios was also seen in the clb19 and ptac2 mutants. Taken together, these results exclude a specific primary splicing defect in mterf6 mutant plants.
Reduced mTERF6 Levels Result in trnI.2 Maturation Deficiency
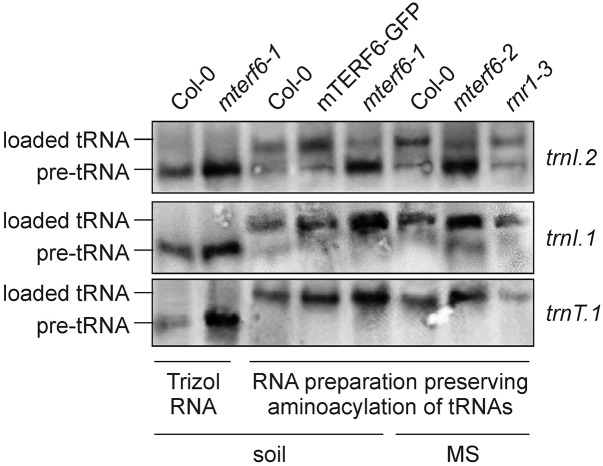
In human, an A-to-G transition in the middle of the mTERF1-protected DNA region, which has been associated with the MELAS syndrome (mitochondrial myopathy, encephalopathy lactic acidosis, and stroke-like episodes), drastically reduces the affinity of mTERF1 for its target sequence (for review, see Kleine and Leister, 2015). Strikingly, this point mutation in the tRNALeu(UUR) gene causes aminoacylation deficiency of tRNALeu(UUR) and also reduced association of mRNA with ribosomes (Chomyn et al., 2000). This prompted us to investigate the aminoacylation status of trnI.2 in mterf6 mutants. The proportion of tRNA that is charged with its amino acid in vivo was determined for plastid trnI.2 [coding for tRNAIle(GAU)] and as controls for trnI.1 [coding for tRNAIle(CAU)] and trnT.1 [coding for tRNAThr(GGU)] using acid conditions for RNA extraction and gel electrophoresis. Our results indicated a marked reduction in the efficiency of charging of trnI.2 with its amino acid Ile in mterf6-1 plants (Fig. 9); this defect could be rescued in complemented mterf6-1 plants. Moreover, charging of trnI.2 was not abolished in rnr1-3 seedlings but was strongly reduced in mterf6-2 seedlings (Fig. 9). In contrast, trnI.1 and trnT.1 were charged at normal levels in both mterf6 mutants and the rnr1-3 mutant (Fig. 9). Together, these findings strongly suggest that mTERF6 is required for the maturation of trnI.2.
Figure 9.
Aminoacylation of trnI.2 is reduced in mterf6 plants. Analysis of trnI.2 and trnI.1 aminoacylation in soil-grown wild-type (Col-0), mterf6-1, and complemented mterf6-1 (35S:MTERF6.1 mterf6-1) plants and, furthermore, in MS medium-grown Col-0, mterf6-2, and, as a control, rnr1-3 seedlings. Northern-blot analyses were carried out with total RNA isolated with the Trizol method or isolated under conditions that preserve the aminoacylation of tRNAs. In the gel system used here (see “Materials and Methods”), the aminoacylated tRNA (loaded tRNA) migrates slower than its corresponding deacylated pre-tRNA. To visualize the difference in electrophoretic mobility, aliquots of Trizol-prepared wild-type and mterf6-1 RNA were included on the gel. Samples of 5 µg of RNA were separated by electrophoresis, blotted, and probed with [γ-32P]ATP end-labeled oligonucleotide probes specific for trnI.2, trnI.1, and trnT.1.
DISCUSSION
In this study, we show that the nucleus-encoded mTERF6 protein binds to an RNA sequence located in the tRNA for Ile(GAU). To our knowledge, our results provide the first evidence for the notion that the activity of an mTERF protein might be a critical component in tRNA maturation distinct from the splicing process. Overall, the relative deficit of mature chloroplast ribosomal RNAs in mterf6 mutants leads to a reduction in the number of functional ribosomes, compromising chloroplast protein synthesis and consequently perturbing chloroplast development and plant growth.
A Relative Lack of mTERF6 Disturbs Chloroplast rRNA Maturation
Reduced amounts of mTERF6 in the mterf6-1 mutant are associated with a relative lack of mature 23S rRNA and an overaccumulation of 23S rRNA precursors and a 16S rRNA precursor (Fig. 7). Chloroplast rRNA genes are cotranscribed as part of the rrn operon, which also encodes tRNAIle(GAU), tRNAAla(UGC), and tRNAArg(ACG) (Fig. 7). The 7.4-kb rRNA precursor is thought to be cleaved by unidentified endonuclease(s), releasing pre-tRNAs for Ile and Ala and pre-rRNAs for 16S rRNA, as well as the dicistronic intermediates rrn23-rrn4.5 and rrn5-tRNR.2 (Bollenbach et al., 2007). The pre-tRNAs and pre-rRNAs are subsequently processed at their 5′ and 3′ ends by chloroplast homologs of the bacterial RNases P, Z, and R1 and a polynucleotide phosphorylase, PNPase (Bollenbach et al., 2007). In contrast to their bacterial counterparts, pre-rrn16 and pre-rrn5 rRNAs are not processed close to their mature termini; therefore, they accumulate long 3′ tails, which require 3′ to 5′ exonucleolytic processing by RNR1 and/or PNPase (Yamamoto et al., 2000; Walter et al., 2002; Bollenbach et al., 2005). Among the numerous mutants that accumulate rrn16 and rrn23 precursors are dal1 (Bisanz et al., 2003) and rnr1 (Bollenbach et al., 2005). The genes DAL1 and RNR1 are coexpressed with MTERF6. DAL1 and RNR1 are ribonucleases and bind RNA, and mTERF6 also possesses RNA-binding capacity (Fig. 6). However, and in contrast to the rnr1 mutant, the mterf6-1 mutant does not interfere with 3′ end maturation of rrn16 (Fig. 7).
Molecular Functions of mTERFs
As in the case of the mterf6-1 mutant, chloroplast transcript accumulation in the soldat10 mutant, another of the few characterized Arabidopsis mterf mutants, is not generally impaired, but levels of rRNAs seem to be down-regulated specifically (Meskauskiene et al., 2009). However, rRNA maturation was not investigated in soldat10 mutants, and as yet, nothing is known about the molecular function of SOLDAT10. Another Arabidopsis mTERF protein, BSM (Babiychuk et al., 2011), which was also named RUG2 (Quesada et al., 2011), is indeed required for the maintenance of normal levels of transcripts in mitochondria and chloroplasts. In the absence of BSM/RUG2, most mitochondrial genes are down-regulated, whereas the majority of the chloroplast genes are up-regulated (Quesada et al., 2011). Moreover, deficiency of BSM/RUG2 (Babiychuk et al., 2011) or of its maize ortholog ZmmTERF4 (Hammani and Barkan, 2014) leads to incorrect processing of plastid transcripts. Data published by Hammani et al. (2014) demonstrate a role for ZmmTERF4 in the splicing of chloroplast group II introns, the first molecular function assigned to a plant mTERF so far. Hammani and Barkan (2014) suggest that other mTERFs, especially Arabidopsis BSM/RUG2, might execute similar functions. Indeed, in Arabidopsis mutants for the mitochondria-localized mTERF15 protein, splicing of nad2 intron 3 is significantly reduced (Hsu et al., 2014). However, our data indicate that the primary function of mTERF6 is not intron splicing (Fig. 8; Supplemental Fig. S8).
Transcription initiated at the human mitochondrial HEAVY STRAND1 promoter is terminated at a specific site in the gene for tRNALeu located 3′ of the rrn16 gene. Human mTERF1 was originally identified in mitochondrial extracts as a factor that binds to this site (Kruse et al., 1989; Supplemental Fig. S5), and it was later shown to be capable of mediating transcription termination in vitro (Asin-Cayuela et al., 2005). In this context, it is remarkable that mTERF6 binds to a sequence located in the trnI.2 intron (target II) in the chloroplast genome, which also lies 3′ of the rrn16 gene. We have also demonstrated that recombinant mTERF6 mediates transcription termination in vitro (Supplemental Fig. S5). Furthermore, the closest mTERF6 homolog in human is mTERF1: the two proteins share 25%/46% identity/similarity over a stretch of 194 amino acids. In principle, partial loss of human mTERF1 or Arabidopsis mTERF6 should lead to reduced occupancy of its target sites and, thus, to increased read-through transcription at these sites. However, manipulation of human mTERF1 expression levels has minimal effects on steady-state levels of sense-strand transcripts (Hyvärinen et al., 2010). In vivo studies in Mterf1 knockout mice led to a model in which the major function of mTERF1 is to prevent light strand transcripts from proceeding around the mtDNA circle, thus avoiding transcriptional interference at the light strand promoter from which they originated (Terzioglu et al., 2013).
In contrast to the above-mentioned studies (Hyvärinen et al., 2010; Terzioglu et al., 2013), knockdown of mTERF6 clearly affects the maturation of chloroplast rRNA, and translational capacity is reduced (Figs. 3 and 7). Like the mterf6 mutants, mouse heart cells that lack mTERF3 (Wredenberg et al., 2013) or mTERF4 (Cámara et al., 2011) are impaired in translation. Interestingly, human mTERF4 forms a complex with NSUN4, a mitochondrial rRNA methyltransferase, which is required to assemble the small and large ribosomal subunits to form a monosome (Metodiev et al., 2014). Therefore, it emerges that the physiological function of this evolutionarily conserved protein family is not restricted to transcription termination. Undoubtedly, the functions of mTERFs, including mTERF6, are essential for the development of plant (Tzafrir et al., 2004; Meskauskiene et al., 2009; Babiychuk et al., 2011) and animal (Park et al., 2007; Cámara et al., 2011) embryos. However, we are just beginning to understand the mechanistic details of their specific functions in these processes.
Why Is trnI.2 Aminoacylation Reduced in mter6 Mutants?
In mterf6-1 and even more in mterf6-2, the level of aminoacylated trnI.2 is strongly reduced (Fig. 9). The plastid isoleucyl-tRNA synthetase was identified as OVULE ABORTION2 (Berg et al., 2005). Furthermore, both trnI.1 and trnI.2 are essential, at least in tobacco (Alkatib et al., 2012). However, the knockout mterf6-2 mutant does not display an aborted ovule phenotype but survives on sugar-supplemented MS medium until the seedling stage and dies with the emergence of the first true leaves (Fig. 1C). This difference in survival is likely to be explained by the facts that (1) aminoacylation of trnI.2 is not completely abolished in the mterf6 mutants and (2) trnI.1 is charged to wild-type levels in mterf6 mutants (Fig. 9).
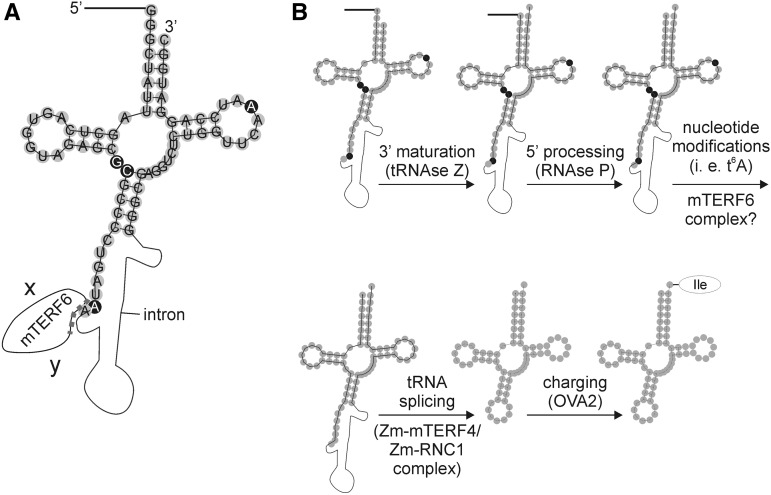
Before a precursor tRNA becomes functional and will be finally loaded with its amino acid, it has to undergo several maturation steps (Fig. 10). Thus, tRNA biogenesis involves the synthesis of the initial transcript, processing to remove the 5′ leader, trimming the 3′ trailer, adding CCA, splicing introns that may be present (as is the case for trnI.2), and modification of multiple nucleotides (for review, see Hopper et al., 2010). The chronological order of these events is still under debate and might vary in various tRNAs. In Figure 10, the order of tRNA processing events is derived from tRNA maturation steps in yeast (for review, see Hopper et al., 2010) and defining the maturation status of Methanopyrus kandleri tRNAs with the help of RNA sequencing data (Su et al., 2013). The hypothetical order of the observed tRNA processing events is here as follows: (1) 3′ maturation, (2) 5′ processing, (3) nucleotide modifications, (4) splicing, and (5) charging with its amino acid. The responsible enzymes for events 1, 2, and 5 have been identified (Schiffer et al., 2002; Berg et al., 2005; Evans et al., 2006). For event 4, ZmmTERF4 (Hammani and Barkan, 2014) and ZmRNC1 (Watkins et al., 2007) have been identified to be required for tRNAIle(GAU) intron splicing in maize. ZmmTERF4 is the ortholog to Arabidopsis BSM (mTERF4), which is involved in intron splicing of clpP (Babiychuk et al., 2011). Because the determined target II (trnI.2) site of mTERF6 covers the last nucleotide of the first exon and the beginning of the trnI.2 intron, we speculated whether mTERF6 might act as a splicing factor of trnI.2. However, both precursor and mature trnI.2 transcripts overaccumulate in mterf6 mutants (Fig. 8), arguing against a function of mTERF6 as a trnI.2 splicing factor. Theoretically, this would leave a function in event 3, nucleotide modifications of trnI.2. Interestingly, the identified interaction partner NSUN4 of mammalian mTERF4 (Cámara et al., 2011) is an rRNA nucleotide modification enzyme. Currently, nothing is known about nucleotide modifications of plant tRNAIle(GAU). However, some of these modifications can be surely derived from the known ones in mammalian mitochondrial tRNAIle(GAU). Nucleotides that are posttranscriptionally modified in mammalian mitochondrial tRNAIle(GAU) are located at positions 9, 26, 27, 37, and 58 of the tRNA (Suzuki and Suzuki, 2014). The modification of nucleotide A-37 is of particular interest, because the identified binding site of mTERF6 lies in close proximity to this nucleotide (Fig. 10). Because the largest part of the identified mTERF6 binding site covers the beginning of the trnI.2 intron, it is important to note that the tRNA is spliced after nucleotides have been posttranscriptionally modified. A-37 is threonyl-carbamoylated, leading to N6-threonyl-carbamoyl-adenosine (t6A). Thus, mTERF6 might be involved in introducing a t6A modification to trnI.2, and consequently, the incorrectly modified pre-trnI.2 in mterf6 mutants would not be loaded with its amino acid Ile. This, in turn, would result in an elongation defect and explain the reduced translation rate in mterf6-1 mutants (Fig. 3C), as observed for Arabidopsis mutants defective for the chloroplast tRNA adenosine deaminase Arg (Delannoy et al., 2009).
Figure 10.
Hypothetical maturation steps of Arabidopsis tRNAIle(GAU) (trnI.2). A, Schematic representation of Arabidopsis plastid pre-trnI.2. The mature trnI.2 was drawn with the RNAfold Webserver (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi), and pre-tRNA properties were added manually. Filled gray circles represent exon nucleotides, and the black line and the thin black curve represent the 5′ leader and the intron, respectively. Nucleotides that are posttranscriptionally modified in human mitochondrial tRNAIle(GAU) (Suzuki and Suzuki, 2014) are indicated by white letters in black circles. In human mitochondrial tRNAIle(GAU), nucleotide A-37 is threonyl-carbamoylated, leading to t6A. The identified mTERF6 binding site (marked with the dotted line) spans the last nucleotide (A-38) of the first exon and the very beginning of the trnI.2 intron. The letters x and y symbolize yet unidentified interaction partners of mTERF6. B, Schematic representation of the tentative maturation process. Precursor tRNAs are transcribed with additional sequences at the 5′ and 3′ ends of the tRNA (not shown here), and some tRNA genes contain introns, as is the case for trnI.2. The tRNA has to undergo several maturation steps before it is functional and can be charged with its amino acid. The order of tRNA processing events is derived from tRNA maturation steps in yeast (for review, see Hopper et al., 2010) and defining the maturation status of M. kandleri tRNAs with the help of RNA sequencing data (Su et al., 2013). The hypothetical order of observed tRNA processing events is as follows: (1) 3′ processing, (2) 5′ processing, (3) nucleotide modifications, (4) splicing, and (5) charging with its amino acid. However, the exact chronological order might deviate for various tRNAs. ZmmTERF4 and ZmRNC1 are proteins that are required for tRNAIle(GAU) intron splicing in maize.
Outlook
Future work will have to identify the interaction partners of mTERF6 and will show whether mTERF6 indeed is involved in introducing t6A modifications. Further studies should address the possible multiple functions of mTERF6, including a function in mitochondria and a transcription termination function that might be masked in vivo by redundancy with other mTERF family members.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The mterf6-1 mutant (GABI_152G06) was identified in the GABI-KAT collection (Rosso et al., 2003) based on alterations in ΦII. The mutants mterf6-2 (SAIL_360_H09), mterf6-4 (SALK_098509), and ptac2 (GK_015F09) were identified in the SIGnAL database (Alonso et al., 2003), and mterf6-3 (SGT1851-3-3) was found in the database described by Parinov et al. (1999). The mutant clb19-3 (SALK_123752; Chateigner-Boutin et al., 2008) has been described previously, as have prps17-1 and prpl24-1 (Romani et al., 2012) and rnr1-3 (Bollenbach et al., 2005). With the exception of mterf6-3 (which is a Landsberg erecta strain), all mutants are in the Col-0 background.
Arabidopsis (Arabidopsis thaliana) plants were routinely grown on potting soil (Stender) under controlled greenhouse conditions (daylight was supplemented with illumination from HQI Powerstar 400W/D, providing a total fluence of approximately 180 µmol photons m−2 s−1 on leaf surfaces and an 8/16-h light/dark cycle). Wuxal Super fertilizer (8% [w/w] nitrogen, 8% [w/w] P2O5, and 6% [w/w] K2O; MANNA) was used according to the manufacturer’s instructions. Where indicated, seedlings were grown on agar (Sigma-Aldrich) containing 1.5% (w/v) Suc and 0.3% (w/v) Gelrite (Roth) at 22°C under 100 µmol photons m−2 s−1 provided by white fluorescent lamps.
Complementation of the mterf6-1 Mutant and Intracellular Localization of eGFP Fusions
For complementation of the mterf6-1 mutant, a cDNA encompassing the AT4G38160.1 coding region was amplified by PCR (for primer information, see Supplemental Table S2). The PCR product was cloned with Gateway technology and subcloned into the plant expression vector pB7FWG to generate a fusion with eGFP (Clontech Laboratories) under the control of the cauliflower mosaic virus 35S promoter. The construct was introduced into Arabidopsis Col-0 and mterf6-1 plants by floral dip (Clough and Bent, 1998). Plants were then grown in the greenhouse, and seeds were collected. Individual transgenic plants were selected on the basis of their resistance to BASTA. The success of the complementation was confirmed by PCR as well as phenotypic and chlorophyll fluorescence analyses.
For eGFP visualization, sterile cotyledons of 3-week-old transformed plants were cut into small pieces and incubated for 16 h at 24°C in the dark in a protoplasting solution (10 mm MES, 20 mm CaCl2, 0.5 m mannitol, pH 5.8, 0.1 g mL−1 macerozyme [Duchefa], and 0.1 g mL−1 cellulase [Duchefa]). Following the isolation of protoplasts as described (Dovzhenko et al., 2003), preparations were viewed with a Fluorescence Axio Imager microscope in ApoTome mode (Zeiss). Fluorescence was excited with the X-Cite Series 120 fluorescence lamp (EXFO), and images were collected in the 500- to 550-nm (eGFP fluorescence), 570- to 640-nm (Mitotracker fluorescence), and 670- to 750-nm (chlorophyll autofluorescence) ranges.
Chlorophyll a Fluorescence Measurements
Five plants of each genotype were analyzed, and average values and sd were calculated. In vivo chlorophyll a fluorescence of single leaves was measured using the Dual-PAM 100 (Walz). Pulses (0.5 s) of red light (5,000 μmol photons m−2 s−1) were used to determine the maximum fluorescence and the ratio (Fm − F0)/Fm = Fv/Fm, where F0 is the minimum fluorescence. A 15-min exposure to red light (37 μmol photons m−2 s−1) was used to drive electron transport before measuring ΦII and 1-qP.
Chlorophyll Concentration Measurements
For chlorophyll extraction, approximately 30 mg of leaf tissue from 4-week-old plants was ground in liquid nitrogen in the presence of 80% (v/v) acetone. After the removal of cell debris by centrifugation, absorption was measured with the Ultrospec 3100 pro spectrophotometer (GE Healthcare). Chlorophyll concentrations were calculated after Lichtenthaler (1987).
Transmission Electron Microscopy
Embryos were isolated from their seeds and fixed immediately with 2.5% (v/v) glutaraldehyde in fixative buffer (75 mm sodium cacodylate and 2 mm MgCl2, pH 7) for 1 h at room temperature, rinsed several times in fixative buffer, and postfixed for 2 h with 1% (w/v) osmium tetroxide in fixative buffer at room temperature. After two washing steps in distilled water, the cells were stained en bloc with 1% (w/v) uranyl acetate in 20% (v/v) acetone for 30 min. Dehydration was performed with a graded acetone series. Samples were infiltrated and embedded in Spurr’s low-viscosity resin (Spurr, 1969). After polymerization, ultrathin sections with a thickness between 50 and 70 nm were cut with a diamond knife and mounted on uncoated copper grids. The sections were poststained with aqueous lead citrate (100 mm, pH 13). All micrographs were taken with an EM 912 electron microscope (Zeiss) equipped with an integrated OMEGA energy filter operated in the zero-loss mode.
Protein Isolation, Western Blotting, and RIP-chip Analyses
Proteins were homogenized in 2× SDS sample buffer (62.5 mm Tris-HCl, pH 6.8, 20% [v/v] glycerol, 4% [w/v] SDS, 100 mm dithiothreitol [DTT], and 0.05% [w/v] Bromophenol Blue), incubated for 7 min at 75°C, and centrifuged for 15 min. The protein concentration in the supernatant was quantified by staining with Amido Black (Schaffner and Weissmann, 1973). Proteins were fractionated by vertical electrophoresis on a 12% (w/v) SDS-polyacrylamide gel and transferred to polyvinylidene difluoride membranes (Millipore). Filters were than incubated with antibodies specific for Nad9 (Lamattina et al., 1993), the PSII subunit D1 (obtained from Jürgen Soll, University of Munich), and other photosynthesis proteins (Agrisera). Antibodies recognizing plastid ribosomal proteins were obtained from Uniplastomic. Signals were detected by enhanced chemiluminescence (ECL kit; Amersham Bioscience) using an ECL reader system (Fusion FX7; PeqLab) and quantified using ImageJ (http://rsbweb.nih.gov/ij).
Chloroplasts from 5-week-old Arabidopsis plants were prepared as described by Kupsch et al. (2012). Stroma was separated from membranes by centrifugation at 40,000g for 10 min. Membranes were washed twice, and equal volumes of total chloroplasts, stroma, and membranes were loaded onto an SDS-PAGE system and investigated by western blot as described (Kupsch et al., 2012).
The RIP-chip procedures have been described previously (Kupsch et al., 2012) and were performed on 5-week-old 35S:MTERF6.1:eGFP mterf6-1 plants and, as a control, FERREDOXIN-NADP(+)-OXIDOREDUCTASE:eGFP plants (Marques et al., 2004). Protein-RNA complexes were immunoprecipitated using 2.5 µg of monoclonal anti-GFP antibody (Invitrogen).
Mitochondria from 4-week-old Arabidopsis plants were prepared as described by Meyer et al. (2009).
Nucleic Acid Extraction
For DNA isolation, leaf tissue was homogenized in extraction buffer containing 200 mm Tris-HCl, pH 7.5, 25 mm NaCl, 25 mm EDTA, and 0.5% (w/v) SDS. After centrifugation, DNA was precipitated from the supernatant by adding isopropyl alcohol. After washing with 70% (v/v) ethanol, the DNA was dissolved in distilled water.
For RNA isolation, frozen tissue was ground in liquid nitrogen. Following the addition of Trizol (Invitrogen) and chloroform according to the manufacturer’s instructions, RNA was precipitated from the aqueous phase with isopropyl alcohol, washed with 70% (v/v) ethanol, and dissolved in RNase-free water. The concentration and purity of RNA samples were determined spectroscopically in the GeneQuant pro RNA/DNA Calculator (GE Healthcare Europe). Isolated RNA was stored at −80°C until further use.
RNA Gel-Blot Analysis
Northern blotting and hybridization of probes were performed using standard procedures. Aliquots (2.5–10 µg) of total RNA were denatured and fractionated on a 1.2% (w/v) agarose gel and blotted onto a nylon membrane (Roche). Blots were stained with 0.04% (w/v) Methylene Blue in 0.5 m sodium acetate (pH 5.2). Probes complementary to psbA, psbB, PSBP, PSBS, psaA, LHCA1, LHCA2, LHCB1, LHCB2, rbcL, ndhC, rrn5S, rrn16S, and rrn23 were amplified from cDNA and labeled with [α-32P]dCTP. Probes for tRNA detection were generated by end labeling corresponding primers with [γ-32P]ATP using polynucleotide T4 kinase (Fermentas; for primer information, see Supplemental Table S2). Hybridizations were performed for 16 h at 65°C (detection of photosynthetic and rrn transcripts) or at 44°C (detection of trn transcripts). After washing, the filters were exposed to a phosphorimager screen and analyzed with the Typhoon Variable Mode Imager (GE Healthcare).
Aminoacylation Analysis of tRNAs
For RNA isolation, which preserves aminoacylation of tRNAs, frozen tissue was ground in liquid nitrogen. After the addition of 300 µL of 0.3 m sodium acetate (pH 4.5) and 10 mm Na2EDTA, RNA was isolated according to Varshney et al. (1991). RNA was fractionated as described (Jester et al., 2003), the gel was electroblotted onto a nylon membrane, and the detection was done as described above.
Circular RT-PCR
For the determination of 16S rRNA end sequences, total RNA (5 µg) was treated with 40 units of T4 RNA ligase (New England Biolabs) in a total volume of 50 µL. RNA was precipitated by the addition of 5 µL of 3 m sodium acetate (pH 5.2) and 100 µL of 99% (v/v) ethanol, and the pellet was washed in 70% (v/v) ethanol. cDNA was then synthesized using SuperScript II RNaseH reverse transcriptase (Invitrogen) and the reverse primer 16S-R1 (Supplemental Table S2). The region of the circularized 16S rRNAs containing the junction between the original 5′ and 3′ ends was then amplified by RT-PCR using 16S-R2 and 16S-F1 primers. The circular RT-PCR products were cloned using the pGEM-Easy Kit and sequenced. For the determination of MTERF6 transcript ends, capped transcripts were converted to 5′-monophosphorylated transcripts by treating 1 µg of total RNA with 2 units of tobacco (Nicotiana tabacum) acid pyrophosphatase (Epicentre) at 37°C for 1 h in the presence of 40 units of RNase inhibitor (Thermo Scientific) in the appropriate buffer. In the control reaction, pyrophosphatase was omitted. The positions of MTERF6 gene-specific primers used for circular RT-PCR are indicated in Supplemental Figure S1. For primer sequence information, see Supplemental Table S2.
cDNA Synthesis and Real-Time PCR Analysis
cDNA synthesis and real-time PCR analysis were generally done as described previously (Voigt et al., 2010). All reactions were performed in triplicate on at least two biological replicates. The genes investigated, and the corresponding primers, are listed in Supplemental Table S2. Real-time PCR analysis to detect the splice forms of chloroplast transcripts was performed as described (de Longevialle et al., 2008).
In Vivo Labeling of Thylakoid Proteins
Leaf discs of 0.4-cm diameter from plants at the eight-leaf rosette stage were vacuum infiltrated in a syringe containing 20 µg mL–1 cycloheximide in 10 mL of 1 mm K2HPO4-KH2PO4 (pH 6.3) and 0.1% (w/v) Tween 20 and incubated for 30 min to block cytosolic translation. Then, leaves were again infiltrated with the same solution containing 0.1 mCi mL–1 [35S]Met and transferred into the light (20 µmol photons m–2 s–1), and four leaf discs were collected after 10 and 30 min. Chloroplast isolation and thylakoid extraction were performed with buffers as described (Kunst, 1998), but the step gradient was omitted. Thylakoid proteins were separated by Tris-Gly SDS-PAGE (12% [w/v] polyacrylamide). Signals were detected with the Typhoon phosphorimager (GE Healthcare).
B1H Assay
The B1H assay was performed according to a previously described protocol (Meng and Wolfe, 2006). AT4G38160.1 was amplified from Col-0 cDNA (for primer information, see Supplemental Table S2), and the PCR product was cloned into the KpnI and XbaI sites of the pB1H2-pr2w2 vector using enzymes from New England Biolabs. This vector was then transformed into the Escherichia coli strain USOΔhisBΔpyrFΔrpoZ (Meng and Wolfe, 2006).
The 18-nucleotide random library was generated by cloning the 5′-ACTGCGGCCGCTATCAGNNNNNNNNNNNNNNNNNNGAATTCATACTACTA-3′ sequence into the pH3U3-mcs vector. Self-activating sequences were eliminated by negative selection on 5-fluoroorotic acid. The library vector was then introduced into the E. coli strain USOΔhisBΔpyrFΔrpoZ containing the pB1H-pr2w2-mTERF6.1 vector, and the selection screen was performed on selective medium without His, containing appropriate antibiotics (100 µg mL–1 ampicillin, 50 µg mL–1 kanamycin, and 10 µg mL–1 tetracycline), 10 µm IPTG, and increasing concentrations (0, 1, 2, and 4 mm) of 3-aminotriazole. Surviving colonies were screened via colony PCR using the primers 5′-CAAATATGTATCCGCTCATGAC-3′ and 5′-GGGCTTTCTGCTCTGTCATAG-3′ flanking the random region in the pH3U3 vector, and the PCR products were sequenced. The resulting sequences were analyzed with MEME (http://meme.sdsc.edu/meme/cgi-bin/meme.cgi; Bailey and Elkan, 1994) using default settings to obtain a consensus binding sequence that was subsequently submitted to the MAST implemented in MEME and used to query the mitochondrial and chloroplast genome sequences.
mTERF6.1 Expression in E. coli and Purification
AT4G38160.1 was amplified from Col-0 cDNA (for primer information, see Supplemental Table S2), and the PCR product was cloned into the pET151 vector using TOPO cloning technology (Invitrogen). E. coli BL21-CodonPlus (DE3)-RIPL cells harboring the pET151-mTERF6.1 construct were induced with 1 mm IPTG, incubated in a shaker for 3 h at 30°C, and harvested at 7,500g for 10 min. For purification of inclusion bodies, all the following steps were performed at 4°C. The cell pellet was lysed for 30 min in lysis buffer (50 mm NaH2PO4 [pH 8], 350 mm NaCl, 10 mm imidazole, and 1 mg mL–1 lysozyme [Sigma-Aldrich]) and then sonicated 10 times for 20 s. The resulting pellet after centrifugation for 30 min at 16,000g contained the inclusion bodies. Inclusion bodies were solubilized in Strep-tag washing buffer (IBA) supplemented with 10% (v/v) N-laurylsarcosine and Complete Protease Inhibitor Cocktail tablets (Roche Applied Science). The postcentrifugation supernatant was diluted 1:5 with Strep-tag washing buffer supplemented with 4% (v/v) Triton X-100 and 1% (w/v) CHAPS and dialyzed against dialysis buffer (50 mm Tris-HCl [pH 8], 300 mm NaCl, 2% [v/v] Triton X-100, and 1% [w/v] CHAPS). After the addition of nickel-nitrilotriacetic acid agarose (Qiagen), the dialysate was incubated for 2 h with rotation. Washing steps were performed with washing buffer (50 mm Tris-HCl [pH 8], 300 mm NaCl, and 20 mm imidazole). The recombinant mTERF6.1-His protein was eluted with elution buffer (50 mm Tris-HCl [pH 8], 300 mm NaCl, 500 mm imidazole, and 30% [v/v] glycerol). Protein concentration was determined with the Bradford assay (Bio-Rad) using bovine serum albumin (BSA) as the standard. Protein purity was checked by SDS-PAGE and subsequent Coomassie Blue staining.
EMSAs
EMSAs were performed either with dsDNA generated with the universal primer described by Badis et al. (2009) and primers containing the 15-bp binding sequence and a common 20-bp sequence at the 3′ end with which to anneal the universal primer (Supplemental Table S2) or with ssRNA produced by in vitro transcription. For the EMSAs shown in Figure 6 and Supplemental Figure S4, 2 µg of mTERF6.1-His protein was incubated for 30 min at 25°C in binding buffer (10 mm Tris-HCl [pH 7.5], 1 mm EDTA, 0.1 m KCl, 0.1 mm DTT, 5% [v/v] glycerol, and 0.01 mg mL–1 BSA) with 100 ng of 32P-labeled dsDNA probes and increasing concentrations (0, 100, 250, 500, and 1,000 ng) of the corresponding unlabeled dsDNA probe (competition). Samples were then subjected to electrophoresis on a vertical native 8% (w/v) polyacrylamide gel in TBE buffer (89 mm Tris-borate, 89 mm boric acid, and 2 mm EDTA). For the EMSAs shown in Supplemental Figure S4, DNA fragments were generated by PCR using a primer containing a T7 promoter sequence (Supplemental Table S2) and the M13 primer, with a pGEM plasmid containing the trnI2.3 binding site as a template. PCR products were then either labeled by random priming using the Klenow fragment (New England Biolabs) or used for in vitro transcription in the presence of [α-32P]UTP according to the T7 in vitro transcription protocol (Fermentas). Binding was performed in 25-µL reactions in binding buffer (40 mm Tris-HCl [pH 7.5], 100 mm NaCl, 0.1 mg mL–1 BSA, 4 mm DTT, and 0.5 mg mL–1 heparin) at 25°C for 20 min. Protein concentrations of 50, 250, and 500 nm and a nucleic acid concentration of approximately 50 pm were used. Samples were separated on a nondenaturing 5% (w/v) polyacrylamide (29:1 acrylamide:bisacrylamide) gel prepared in 0.5× TBE buffer (45 mm Tris-borate, 45 mm boric acid, and 1 mm EDTA). The dried gels were exposed to a phosphorimager screen and analyzed with the Typhoon Variable Mode Imager (GE Healthcare).
In Vitro Transcription Termination Assay
Oligonucleotides representing the intact or mutated trnI.2/3 binding site were annealed to their reverse complement (for oligonucleotide information, see Supplemental Table S2) and cloned into the pGEM-T easy vector (Promega). The vectors were cut with SalI and gel purified using the Wizard SV Gel and PCR Clean-Up System (Promega). In vitro transcription was performed in 20-µL reactions containing 100 ng of each vector, 1 µm mTERF6.1, ATP, CTP, and GTP (0.5 mm each), 12 µm UTP, 50 µCi of [α-32P]UTP, 20 units of T7 RNA polymerase (Fermentas), 4 µL of 5× transcription buffer, and 20 units of RiboLock RNase Inhibitor (Fermentas). Reactions were incubated for 1 h at 25°C, denatured by heating in the presence of 80% (w/v) formaldehyde, and analyzed on 8% (w/v) denaturing polyacrylamide gels containing 8 m urea and 1× TBE. Gels were visualized as above.
Southwestern and Northwestern Analyses
Southwestern and northwestern experiments were performed as described (Uyttewaal et al., 2008). One and 2 µg of recombinant mTERF6.1 and 1.5 µg of maltose binding protein were separated by SDS-PAGE and transferred onto a polyvinylidene difluoride Immobilon-P membrane (Millipore). Filters were incubated with total Arabidopsis DNA or RNA, which were end labeled with [γ-32P]ATP using polynucleotide T4 kinase (Fermentas).
Nucleic Acid Immunoprecipitation
Chloroplasts were isolated from wild-type and complemented mterf6-1 (35S:mTERF6.1 mterf6-1) plants as described previously (Stoppel et al., 2012) and ruptured in coimmunoprecipitation buffer (20 mm Tris-HCl, pH 8, 150 mm NaCl, 1 mm EDTA, 2 mm DTT, 0.5% [v/v] Nonidet P-40, and Complete Protease Inhibitor Cocktail tablets [Roche]) by incubation on ice for 15 min interrupted by occasional short vortexing. The lysates were cleared by centrifugation (4°C, 46,000g, and 30 min). One milligram of chloroplast extract was incubated with GFP-Trap_M (Chromotek) at 4°C for 1 h with rotation. After three washes with coimmunoprecipitation buffer, nucleic acids were isolated by phenol/chloroform extraction followed by ethanol precipitation. One-tenth of the supernatant nucleic acids and one-half of the nucleic acids recovered from the pellet were spotted onto a nylon membrane using a slot-blot device (Bio-Rad). Filters were then hybridized with 32P-labeled PCR probes specific for the indicated genomic regions in the figures (for primer information, see Supplemental Table S2).
Run-On Transcription
Chloroplasts from wild-type and mterf6-1 plants were isolated as described above. Intact chloroplasts were separated from broken ones by centrifugation (4°C, 15 min, and 3,200g) on top of a 40%/80% Percoll step gradient. A total of 2.5 × 107 chloroplasts were resuspended in 100 µL of run-on transcription buffer (30 mm HEPES-KOH, pH 7.7, 10 mm MgCl2, 25 mm KOAc, 1 µg µL–1 heparin, 10 mm DTT, 150 µm each ATP, GTP, and CTP, 50 mCi of [32P]UTP, and 40 units of RNase inhibitor [Fermentas]) by pipetting up and down several times. Reactions were incubated for 30 min at 25°C, and then RNA was extracted using Trizol (Invitrogen) and subsequent ethanol precipitation. Quantities of 250 ng and 1 µg of PCR-amplified fragments (rrn16 and rrn23) and 200 ng and 2 µg of a 70-mer (trnI.2 and trnA.1) were spotted onto a nylon membrane using a slot-blot device (Bio-Rad) and hybridized with the recovered RNAs.
Computational Analyses
Plotting the intensity of lanes was done using ImageJ (http://rsbweb.nih.gov/ij) according to the manual’s recommendations.
Protein sequences were retrieved from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and The Arabidopsis Information Resource (http://www.arabidopsis.org). Putative chloroplast transit peptides were predicted by ChloroP (http://www.cbs.dtu.dk/services/ChloroP; Emanuelsson et al., 1999). Amino acid sequences were aligned using the ClustalW program (www.ebi.ac.uk/clustalw; Chenna et al., 2003), and alignments were shaded according to sequence similarity using the Boxshade server 3.21 (www.ch.embnet.org/software/BOX_form.html).
The mTERF6 three-dimensional protein structure was predicted by I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER; Zhang, 2008) using the crystal structures of human mTERF1 bound to dsDNA (Jiménez-Menéndez et al., 2010; Yakubovskaya et al., 2010) as templates. These structures are deposited in the RCSB Protein Data Bank (http://www.rcsb.org/pdb/home/home.do) as 3N6S (resolution of 3.1 Å) and 3MVA (resolution of 2.2 Å). Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (http://www.cgl.ucsf.edu/chimera).
The sequences isolated from a binding-site selection experiment using the B1H system were analyzed with the de novo motif discovery tool MEME (http://meme.sdsc.edu/meme/cgi-bin/meme.cgi; Bailey and Elkan, 1994) using default settings. The sequences of each motif identified were used to construct a sequence logo (http://weblogo.berkeley.edu/logo.cgi; Crooks et al., 2004) that shows the level of conservation at each position as a function of its information content and base frequency. The motif files were submitted to MAST implemented in MEME and searched against mitochondrial and chloroplast genome sequences.
Sequence identifiers for mTERF6 homologs are as follows: Arabidopsis lyrata (GI:297797802), Ricinus communis (GI:255546666), Populus trichocarpa (GI:224132470), Vitis vinifera (GI:225428362), Glycine max (GI:255638191), Oryza sativa (GI:115477278), Phyllostachys edulis (GI:284434657), and Sorghum bicolor (GI:242079975).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification and characterization of mterf6 T-DNA insertion mutants.
Supplemental Figure S2. Chloroplast rRNAs are underrepresented in mterf6 mutants.
Supplemental Figure S3. Target sequences of mTERF6 identified with a B1H screen.
Supplemental Figure S4. mTERF6 does not bind to the designation sequences of rps7.1, psbA, trnT.1, and ndhH.
Supplemental Figure S5. In vitro transcription termination activity of mTERF6.
Supplemental Figure S6. DNA copy number and the transcription rate around the target II sequence are not altered.
Supplemental Figure S7. No significant association of mTERF6 with RNAs could be detected by RIP-chip analysis.
Supplemental Figure S8. Lack of mTERF6 does not lead to specific splicing defects in chloroplast transcripts.
Supplemental Table S1. RIP-chip data for Supplemental Figure S7.
Supplemental Table S2. Primers used in this study.
Acknowledgments
We thank Joachim Kurth for the initial isolation of the pam48-1 mutant; Vincent Schulz for assistance, Ralf Bernd Klösgen and Martin Schattat for providing the FERREDOXIN-NADP(+)-OXIDOREDUCTASE:eGFP seeds, and Catherine Colas des Francs-Small and Ian Small for providing the clb19-3 seeds; David B. Stern for providing the rnr1-3 seeds; Paul Hardy for critical reading of the article; and Elisabeth Gerick for technical assistance.
Glossary
- OGE
organellar gene expression
- mtDNA
mitochondrial DNA
- rRNA
ribosomal RNA
- dsDNA
double-stranded DNA
- ΦII
effective quantum yield of photosystem II
- Fv/Fm
maximum quantum yield of photosystem II
- T-DNA
transfer DNA
- RT
reverse transcription
- Col-0
Columbia-0
- MS
Murashige and Skoog
- B1H
bacterial one-hybrid
- IPTG
isopropylthio-β-galactoside
- MEME
Multiple Em for Motif Elicitation
- MAST
Motif Alignment and Search Tool
- EMSA
electrophoretic mobility shift assay
- ssRNA
single-stranded RNA
- RIP-chip
RNA immunoprecipitation and hybridization to gene chips
- cDNA
complementary DNA
- DTT
dithiothreitol
- BSA
bovine serum albumin
- t6A
N6-threonyl-carbamoyl-adenosine
References
- Alkatib S, Fleischmann TT, Scharff LB, Bock R (2012) Evolutionary constraints on the plastid tRNA set decoding methionine and isoleucine. Nucleic Acids Res 40: 6713–6724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Asin-Cayuela J, Schwend T, Farge G, Gustafsson CM (2005) The human mitochondrial transcription termination factor (mTERF) is fully active in vitro in the non-phosphorylated form. J Biol Chem 280: 25499–25505 [DOI] [PubMed] [Google Scholar]
- Babiychuk E, Vandepoele K, Wissing J, Garcia-Diaz M, De Rycke R, Akbari H, Joubès J, Beeckman T, Jänsch L, Frentzen M, et al. (2011) Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc Natl Acad Sci USA 108: 6674–6679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. (2009) Diversity and complexity in DNA recognition by transcription factors. Science 324: 1720–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Berg M, Rogers R, Muralla R, Meinke D (2005) Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J 44: 866–878 [DOI] [PubMed] [Google Scholar]
- Bisanz C, Bégot L, Carol P, Perez P, Bligny M, Pesey H, Gallois JL, Lerbs-Mache S, Mache R (2003) The Arabidopsis nuclear DAL gene encodes a chloroplast protein which is required for the maturation of the plastid ribosomal RNAs and is essential for chloroplast differentiation. Plant Mol Biol 51: 651–663 [DOI] [PubMed] [Google Scholar]
- Bollenbach TJ, Lange H, Gutierrez R, Erhardt M, Stern DB, Gagliardi D (2005) RNR1, a 3′-5′ exoribonuclease belonging to the RNR superfamily, catalyzes 3′ maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana. Nucleic Acids Res 33: 2751–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollenbach TJ, Schuster G, Portnoy V, Stern DB (2007) Processing, degradation, and polyadenylation of chloroplast transcripts. In Bock R, ed, Topics in Current Genetics, Vol 19. Springer, Berlin, pp 175–211 [Google Scholar]
- Börner T, Aleynikova AY, Zubo YO, Kusnetsov VV (2015) Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim Biophys Acta 1847: 761–769 [DOI] [PubMed] [Google Scholar]
- Byrnes J, Garcia-Diaz M (2011) Mitochondrial transcription: how does it end? Transcription 2: 32–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cámara Y, Asin-Cayuela J, Park CB, Metodiev MD, Shi Y, Ruzzenente B, Kukat C, Habermann B, Wibom R, Hultenby K, et al. (2011) MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab 13: 527–539 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Ramos-Vega M, Guevara-García A, Andrés C, de la Luz Gutiérrez-Nava M, Cantero A, Delannoy E, Jiménez LF, Lurin C, Small I, et al. (2008) CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J 56: 590–602 [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomyn A, Enriquez JA, Micol V, Fernandez-Silva P, Attardi G (2000) The mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episode syndrome-associated human mitochondrial tRNALeu(UUR) mutation causes aminoacylation deficiency and concomitant reduced association of mRNA with ribosomes. J Biol Chem 275: 19198–19209 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga A, Micol V, Hess D, Aebersold R, Attardi G (1993) Molecular characterization of the transcription termination factor from human mitochondria. J Biol Chem 268: 8123–8130 [PubMed] [Google Scholar]
- Delannoy E, Le Ret M, Faivre-Nitschke E, Estavillo GM, Bergdoll M, Taylor NL, Pogson BJ, Small I, Imbault P, Gualberto JM (2009) Arabidopsis tRNA adenosine deaminase arginine edits the wobble nucleotide of chloroplast tRNAArg(ACG) and is essential for efficient chloroplast translation. Plant Cell 21: 2058–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Longevialle AF, Hendrickson L, Taylor NL, Delannoy E, Lurin C, Badger M, Millar AH, Small I (2008) The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J 56: 157–168 [DOI] [PubMed] [Google Scholar]
- Dovzhenko A, Dal Bosco C, Meurer J, Koop HU (2003) Efficient regeneration from cotyledon protoplasts in Arabidopsis thaliana. Protoplasma 222: 107–111 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D, Marquez SM, Pace NR (2006) RNase P: interface of the RNA and protein worlds. Trends Biochem Sci 31: 333–341 [DOI] [PubMed] [Google Scholar]
- Fernandez-Silva P, Martinez-Azorin F, Micol V, Attardi G (1997) The human mitochondrial transcription termination factor (mTERF) is a multizipper protein but binds to DNA as a monomer, with evidence pointing to intramolecular leucine zipper interactions. EMBO J 16: 1066–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Barkan A (2014) An mTERF domain protein functions in group II intron splicing in maize chloroplasts. Nucleic Acids Res 42: 5033–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Bonnard G, Bouchoucha A, Gobert A, Pinker F, Salinas T, Giegé P (2014) Helical repeats modular proteins are major players for organelle gene expression. Biochimie 100: 141–150 [DOI] [PubMed] [Google Scholar]
- Hopper AK, Pai DA, Engelke DR (2010) Cellular dynamics of tRNAs and their genes. FEBS Lett 584: 310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YW, Wang HJ, Hsieh MH, Hsieh HL, Jauh GY (2014) Arabidopsis mTERF15 is required for mitochondrial nad2 intron 3 splicing and functional complex I activity. PLoS ONE 9: e112360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvärinen AK, Kumanto MK, Marjavaara SK, Jacobs HT (2010) Effects on mitochondrial transcription of manipulating mTERF protein levels in cultured human HEK293 cells. BMC Mol Biol 11: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jester BC, Levengood JD, Roy H, Ibba M, Devine KM (2003) Nonorthologous replacement of lysyl-tRNA synthetase prevents addition of lysine analogues to the genetic code. Proc Natl Acad Sci USA 100: 14351–14356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Menéndez N, Fernández-Millán P, Rubio-Cosials A, Arnan C, Montoya J, Jacobs HT, Bernadó P, Coll M, Usón I, Solà M (2010) Human mitochondrial mTERF wraps around DNA through a left-handed superhelical tandem repeat. Nat Struct Mol Biol 17: 891–893 [DOI] [PubMed] [Google Scholar]
- Kleine T. (2012) Arabidopsis thaliana mTERF proteins: evolution and functional classification. Front Plant Sci 3: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Leister D (2015) Emerging functions of mammalian and plant mTERFs. Biochim Biophys Acta 1847: 786–797 [DOI] [PubMed] [Google Scholar]
- Kruse B, Narasimhan N, Attardi G (1989) Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell 58: 391–397 [DOI] [PubMed] [Google Scholar]
- Kunnimalaiyaan M, Nielsen BL (1997) Fine mapping of replication origins (ori A and ori B) in Nicotiana tabacum chloroplast DNA. Nucleic Acids Res 25: 3681–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L. (1998) Preparation of physiologically active chloroplasts from Arabidopsis. Methods Mol Biol 82: 43–48 [DOI] [PubMed] [Google Scholar]
- Kupsch C, Ruwe H, Gusewski S, Tillich M, Small I, Schmitz-Linneweber C (2012) Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps. Plant Cell 24: 4266–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamattina L, Gonzalez D, Gualberto J, Grienenberger JM (1993) Higher plant mitochondria encode an homologue of the nuclear-encoded 30-kDa subunit of bovine mitochondrial complex I. Eur J Biochem 217: 831–838 [DOI] [PubMed] [Google Scholar]
- Leister D, Kleine T (2011) Role of intercompartmental DNA transfer in producing genetic diversity. Int Rev Cell Mol Biol 291: 73–114 [DOI] [PubMed] [Google Scholar]
- Lerbs-Mache S. (2011) Function of plastid sigma factors in higher plants: regulation of gene expression or just preservation of constitutive transcription? Plant Mol Biol 76: 235–249 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Liere K, Weihe A, Börner T (2011) The transcription machineries of plant mitochondria and chloroplasts: composition, function, and regulation. J Plant Physiol 168: 1345–1360 [DOI] [PubMed] [Google Scholar]
- Linder T, Park CB, Asin-Cayuela J, Pellegrini M, Larsson NG, Falkenberg M, Samuelsson T, Gustafsson CM (2005) A family of putative transcription termination factors shared amongst metazoans and plants. Curr Genet 48: 265–269 [DOI] [PubMed] [Google Scholar]
- Majeran W, Friso G, Asakura Y, Qu X, Huang M, Ponnala L, Watkins KP, Barkan A, van Wijk KJ (2012) Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol 158: 156–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JP, Schattat MH, Hause G, Dudeck I, Klösgen RB (2004) In vivo transport of folded EGFP by the DeltapH/TAT-dependent pathway in chloroplasts of Arabidopsis thaliana. J Exp Bot 55: 1697–1706 [DOI] [PubMed] [Google Scholar]
- Meng X, Wolfe SA (2006) Identifying DNA sequences recognized by a transcription factor using a bacterial one-hybrid system. Nat Protoc 1: 30–45 [DOI] [PubMed] [Google Scholar]
- Meskauskiene R, Würsch M, Laloi C, Vidi PA, Coll NS, Kessler F, Baruah A, Kim C, Apel K (2009) A mutation in the Arabidopsis mTERF-related plastid protein SOLDAT10 activates retrograde signaling and suppresses 1O2-induced cell death. Plant J 60: 399–410 [DOI] [PubMed] [Google Scholar]
- Metodiev MD, Spåhr H, Loguercio Polosa P, Meharg C, Becker C, Altmueller J, Habermann B, Larsson NG, Ruzzenente B (2014) NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet 10: e1004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EH, Tomaz T, Carroll AJ, Estavillo G, Delannoy E, Tanz SK, Small ID, Pogson BJ, Millar AH (2009) Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol 151: 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, Ye D, Yang WC, Kumaran M, Sundaresan V (1999) Analysis of flanking sequences from dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11: 2263–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CB, Asin-Cayuela J, Cámara Y, Shi Y, Pellegrini M, Gaspari M, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, et al. (2007) MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell 130: 273–285 [DOI] [PubMed] [Google Scholar]
- Pellegrini M, Asin-Cayuela J, Erdjument-Bromage H, Tempst P, Larsson NG, Gustafsson CM (2009) MTERF2 is a nucleoid component in mammalian mitochondria. Biochim Biophys Acta 1787: 296–302 [DOI] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R (2006) pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18: 176–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Martín A, Montoya J, Martínez-Azorín F (2004) Phosphorylation of rat mitochondrial transcription termination factor (mTERF) is required for transcription termination but not for binding to DNA. Nucleic Acids Res 32: 2059–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Sarmiento-Mañús R, González-Bayón R, Hricová A, Pérez-Marcos R, Graciá-Martínez E, Medina-Ruiz L, Leyva-Díaz E, Ponce MR, Micol JL (2011) Arabidopsis RUGOSA2 encodes an mTERF family member required for mitochondrion, chloroplast and leaf development. Plant J 68: 738–753 [DOI] [PubMed] [Google Scholar]
- Raven JA, Allen JF (2003) Genomics and chloroplast evolution: what did cyanobacteria do for plants? Genome Biol 4: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberti M, Mustich A, Gadaleta MN, Cantatore P (1991) Identification of two homologous mitochondrial DNA sequences, which bind strongly and specifically to a mitochondrial protein of Paracentrotus lividus. Nucleic Acids Res 19: 6249–6254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani I, Tadini L, Rossi F, Masiero S, Pribil M, Jahns P, Kater M, Leister D, Pesaresi P (2012) Versatile roles of Arabidopsis plastid ribosomal proteins in plant growth and development. Plant J 72: 922–934 [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S (1999) Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res 6: 283–290 [DOI] [PubMed] [Google Scholar]
- Schaffner W, Weissmann C (1973) A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal Biochem 56: 502–514 [DOI] [PubMed] [Google Scholar]
- Schiffer S, Rösch S, Marchfelder A (2002) Assigning a function to a conserved group of proteins: the tRNA 3′-processing enzymes. EMBO J 21: 2769–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Lampe MK, Sultan LD, Ostersetzer-Biran O (2015) Organellar maturases: a window into the evolution of the spliceosome. Biochim Biophys Acta 1847: 798–808 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier R, Barkan A (2005) RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 17: 2791–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T. (2015) RNA editing in plants: machinery and flexibility of site recognition. Biochim Biophys Acta 1847: 779–785 [DOI] [PubMed] [Google Scholar]
- Spåhr H, Habermann B, Gustafsson CM, Larsson NG, Hallberg BM (2012) Structure of the human MTERF4-NSUN4 protein complex that regulates mitochondrial ribosome biogenesis. Proc Natl Acad Sci USA 109: 15253–15258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spåhr H, Samuelsson T, Hällberg BM, Gustafsson CM (2010) Structure of mitochondrial transcription termination factor 3 reveals a novel nucleic acid-binding domain. Biochem Biophys Res Commun 397: 386–390 [DOI] [PubMed] [Google Scholar]
- Spurr AR. (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 26: 31–43 [DOI] [PubMed] [Google Scholar]
- Steinhauser D, Usadel B, Luedemann A, Thimm O, Kopka J (2004) CSB.DB: a comprehensive systems-biology database. Bioinformatics 20: 3647–3651 [DOI] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR (2010) Chloroplast RNA metabolism. Annu Rev Plant Biol 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Stoppel R, Manavski N, Schein A, Schuster G, Teubner M, Schmitz-Linneweber C, Meurer J (2012) RHON1 is a novel ribonucleic acid-binding protein that supports RNase E function in the Arabidopsis chloroplast. Nucleic Acids Res 40: 8593–8606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter G, Kössel H (1984) Cotranscription and processing of 23S, 4.5S and 5S rRNA in chloroplasts from Zea mays. Nucleic Acids Res 12: 7633–7647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AA, Tripp V, Randau L (2013) RNA-Seq analyses reveal the order of tRNA processing events and the maturation of C/D box and CRISPR RNAs in the hyperthermophile Methanopyrus kandleri. Nucleic Acids Res 41: 6250–6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Suzuki T (2014) A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res 42: 7346–7357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanz SK, Castleden I, Hooper CM, Vacher M, Small I, Millar HA (2013) SUBA3: a database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Res 41: D1185–D1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzioglu M, Ruzzenente B, Harmel J, Mourier A, Jemt E, López MD, Kukat C, Stewart JB, Wibom R, Meharg C, et al. (2013) MTERF1 binds mtDNA to prevent transcriptional interference at the light-strand promoter but is dispensable for rRNA gene transcription regulation. Cell Metab 17: 618–626 [DOI] [PubMed] [Google Scholar]
- Tiller N, Bock R (2014) The translational apparatus of plastids and its role in plant development. Mol Plant 7: 1105–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N, Weingartner M, Thiele W, Maximova E, Schöttler MA, Bock R (2012) The plastid-specific ribosomal proteins of Arabidopsis thaliana can be divided into non-essential proteins and genuine ribosomal proteins. Plant J 69: 302–316 [DOI] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyttewaal M, Mireau H, Rurek M, Hammani K, Arnal N, Quadrado M, Giegé P (2008) PPR336 is associated with polysomes in plant mitochondria. J Mol Biol 375: 626–636 [DOI] [PubMed] [Google Scholar]
- Varotto C, Pesaresi P, Maiwald D, Kurth J, Salamini F, Leister D (2000) Identification of photosynthetic mutants of Arabidopsis by automatic screening for altered effective quantum yield of photosystem 2. Photosynthetica 38: 497–504 [Google Scholar]
- Varshney U, Lee CP, RajBhandary UL (1991) Direct analysis of aminoacylation levels of tRNAs in vivo: application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J Biol Chem 266: 24712–24718 [PubMed] [Google Scholar]
- Voigt C, Oster U, Börnke F, Jahns P, Dietz KJ, Leister D, Kleine T (2010) In-depth analysis of the distinctive effects of norflurazon implies that tetrapyrrole biosynthesis, organellar gene expression and ABA cooperate in the GUN-type of plastid signalling. Physiol Plant 138: 503–519 [DOI] [PubMed] [Google Scholar]
- Walter M, Kilian J, Kudla J (2002) PNPase activity determines the efficiency of mRNA 3′-end processing, the degradation of tRNA and the extent of polyadenylation in chloroplasts. EMBO J 21: 6905–6914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins KP, Kroeger TS, Cooke AM, Williams-Carrier RE, Friso G, Belcher SE, van Wijk KJ, Barkan A (2007) A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell 19: 2606–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T, Luca C, Torraco A, Moraes CT (2009) mTERF2 regulates oxidative phosphorylation by modulating mtDNA transcription. Cell Metab 9: 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wobbe L, Nixon PJ (2013) The mTERF protein MOC1 terminates mitochondrial DNA transcription in the unicellular green alga Chlamydomonas reinhardtii. Nucleic Acids Res 41: 6553–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wredenberg A, Lagouge M, Bratic A, Metodiev MD, Spåhr H, Mourier A, Freyer C, Ruzzenente B, Tain L, Grönke S, et al. (2013) MTERF3 regulates mitochondrial ribosome biogenesis in invertebrates and mammals. PLoS Genet 9: e1003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakubovskaya E, Mejia E, Byrnes J, Hambardjieva E, Garcia-Diaz M (2010) Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell 141: 982–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Puente P, Deng XW (2000) An Arabidopsis cotyledon-specific albino locus: a possible role in 16S rRNA maturation. Plant Cell Physiol 41: 68–76 [DOI] [PubMed] [Google Scholar]
- Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]