Function gain to promote carotenoid overaccumulation affects chromoplast biogenesis.
Abstract
Carotenoids are crucial for plant growth and human health. The finding of ORANGE (OR) protein as a pivotal regulator of carotenogenesis offers a unique opportunity to comprehensively understand the regulatory mechanisms of carotenoid accumulation and develop crops with enhanced nutritional quality. Here, we demonstrated that alteration of a single amino acid in a wild-type OR greatly enhanced its ability to promote carotenoid accumulation. Whereas overexpression of OR from Arabidopsis (Arabidopsis thaliana; AtOR) or from the agronomically important crop sorghum (Sorghum bicolor; SbOR) increased carotenoid levels up to 2-fold, expression of AtORHis (R90H) or SbORHis (R104H) variants dramatically enhanced carotenoid accumulation by up to 7-fold in the Arabidopsis calli. Moreover, we found that AtORAla (R90A) functioned similarly to AtORHis to promote carotenoid overproduction. Neither AtOR nor AtORHis greatly affected carotenogenic gene expression. AtORHis exhibited similar interactions with phytoene synthase (PSY) as AtOR in posttranscriptionally regulating PSY protein abundance. AtORHis triggered biogenesis of membranous chromoplasts in the Arabidopsis calli, which shared structures similar to chromoplasts found in the curd of the orange cauliflower (Brassica oleracea) mutant. By contrast, AtOR did not cause plastid-type changes in comparison with the controls, but produced plastids containing larger and electron-dense plastoglobuli. The unique ability of AtORHis in mediating chromoplast biogenesis is responsible for its induced carotenoid overproduction. Our study demonstrates ORHis/Ala as powerful tools for carotenoid enrichment in plants, and provides insights into the mechanisms underlying ORHis-regulated carotenoid accumulation.
Carotenoids are widely distributed in nature and give colors varying from light yellow and orange to dark red. As a group of the most abundant pigments, carotenoids are of great importance to both plants and animals (Fraser and Bramley, 2004; Cazzonelli and Pogson, 2010). In plants, carotenoids function as light-harvesting pigments and photoprotectants in photosynthesis (Niyogi, 1999; Domonkos et al., 2013). They serve as precursors for the biosynthesis of phytohormones, abscisic acid, and strigolactones, and for the production of flower and fruit flavor and aroma (Auldridge et al., 2006; Walter and Strack, 2011). In animals such as fish, birds, and butterflies, the vivid colors produced by carotenoids affect reproduction and survival through courtship behavior and protective color (Cazzonelli, 2011). Most importantly, carotenoids are crucial for human nutrition and health by providing precursors for vitamin A biosynthesis and by reducing the risk of certain chronic diseases, such as cancers, cardiovascular diseases, and age-related eye diseases (Rao and Rao, 2007).
Carotenoids are synthesized de novo in plants and algae as well as in some bacteria and fungi (Moise et al., 2014; Nisar et al., 2015). The first committed step in the carotenoid biosynthesis pathway is the condensation of two geranylgeranyl diphosphate molecules to phytoene catalyzed by phytoene synthase (PSY). Following four steps of desaturation and isomerization, phytoene is turned into lycopene. Lycopene can be cyclized to produce α- and β-carotene. Upon oxygenation and epoxidation, α-carotene is converted into lutein and β-carotene into zeaxanthin and other xanthophylls (Hirschberg, 2001; Zhu et al., 2010; Ruiz-Sola and Rodríguez-Concepción, 2012; Moise et al., 2014; Yuan et al., 2015). Carotenoids can be synthesized in nearly all plastids and stored at high concentrations in chloroplasts of green tissues and in chromoplasts of nongreen organs, such as fruits, flowers, and tubers (Lu and Li, 2008; Li and Yuan, 2013).
Significant efforts have been made to increase carotenoid content in food crops due to the importance of carotenoids to human nutrition and health (Giuliano et al., 2008; Farré et al., 2011). The common approach is to alter the catalytic activity of the pathway enzymes. For example, overexpression of a minipathway of β-carotene biosynthesis results in the production of Golden Rice (Oryza sativa), golden potato (Solanum tuberosum), and yellow corn (Zea mays; Paine et al., 2005; Diretto et al., 2007; Naqvi et al., 2009). Alteration of PSY and endogenous gene expression leads to a redirect of metabolic flux into and between branches of carotenoid biosynthesis in several crop species (Fraser et al., 2002; Ducreux et al., 2005; Yan et al., 2010). A different strategy that builds a strong sink to enhance carotenoid accumulation has arisen following the cloning of ORANGE (OR) from the cauliflower (Brassica oleracea) orange curd mutant (Lu et al., 2006).
OR encodes a DnaJ Cys-rich zinc finger domain containing protein, which is highly conserved among divergent plant species (Lu et al., 2006). The mutant allele of OR with a large retrotransposon insertion in cauliflower (BoORMUT) causes the normal white curd tissue to accumulate high levels of β-carotene. Expression of the BoORMUT transgene in both white cauliflower and potato tubers leads to the production of orange tissues with enhanced carotenoid content (Lu et al., 2006; Lopez et al., 2008). The increased carotenoid accumulation in the BoORMUT plants is found to be associated with the formation of carotenoid sequestration structures, which are suggested to serve as an effective sink to facilitate sequestration and storage of carotenoids (Lopez et al., 2008; Li et al., 2012).
Although BoORMUT has been demonstrated to be an effective molecular tool in enhancing carotenoid content in plants, it contains a large retrotransposon insertion and causes a dwarf phenotype in the homozygous mutant in cauliflower (Li et al., 2001; Lu et al., 2006), which limits its potential application in crop nutritional improvement. In addition, the insertion of a large retrotransposon in BoORMUT produces three alternatively spliced transcripts, but none of them, alone, induces β-carotene accumulation at a level comparable with BoORMUT (Lu et al., 2006), thus hindering the mechanistic study of the OR function.
To further explore the function of OR and its utilization as an effective genetic tool either through traditional breeding or biotechnology approaches for developing crops with enhanced carotenoid content, we mutagenized wild-type OR from Arabidopsis (Arabidopsis thaliana; AtOR) along with the agronomically important crop sorghum (Sorghum bicolor; SbOR). The mutations were based on the recent study of the melon (Cucumis melo) OR protein, where a single Arg to His substitution is responsible for the orange-flesh melon fruit phenotype (Tzuri et al., 2015). Our data show that AtORHis (R90H) and SbORHis (R104H) were able to greatly increase carotenoid content. In addition, AtORAla (R90A) also promoted a high level of carotenoid accumulation. Both AtOR and AtORHis were found to posttranscriptionally regulate PSY protein level. Transmission electron microscopy (TEM) analysis revealed that AtORHis exhibited additional function and induced the formation of membranous chromoplasts in Arabidopsis calli, which was not observed in the calli of wild-type and AtOR-overexpressing lines. Thus, the induction of chromoplast biogenesis with enhanced sink strength is responsible for the AtORHis- or AtORAla-induced carotenoid overaccumulation.
RESULTS
AtORHis (R90H) Promotes Carotenoid Accumulation in Arabidopsis Callus
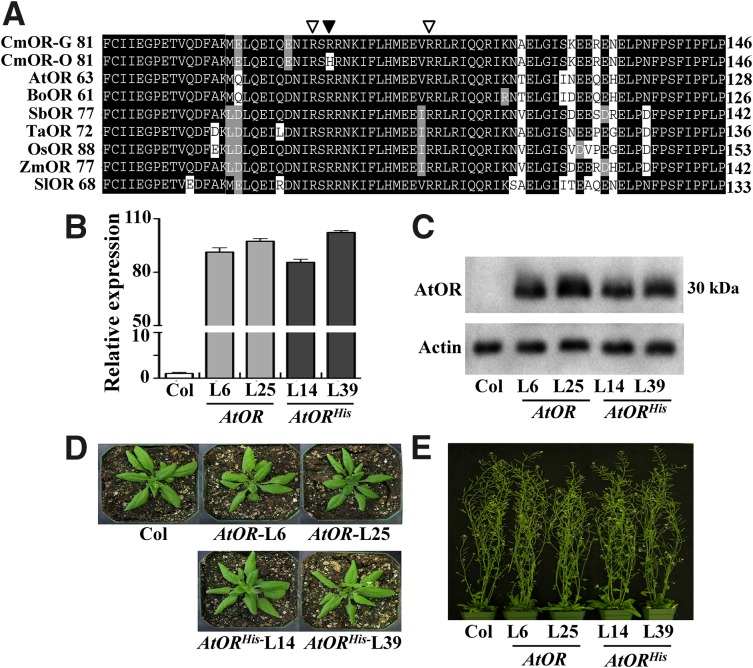
Cauliflower BoORMUT regulates high levels of β-carotene accumulation (Lu et al., 2006). Our recent genetic and functional studies of a melon OR homologous gene (CmOR) reveal that CmOR controls the inheritance of fruit flesh color in melon (Tzuri et al., 2015). A single nucleotide polymorphism in CmOR was found to be linked with the orange fruit phenotype. This single nucleotide polymorphism results in an Arg to His change in the CmOR 108th amino acid. Alignment of the OR protein sequences showed that this Arg is highly conserved among various plant species (Fig. 1A; Supplemental Fig. S1).
Figure 1.
Characterization of AtOR- and AtORHis-overexpressing transgenic lines. A, Alignment of partial OR protein sequences from melon (CmOR) with green-flesh (G) and orange-flesh (O) fruit, respectively, Arabidopsis (AtOR), cauliflower (BoOR), sorghum (SbOR), wheat (Triticum aestivum) (TaOR), rice (OsOR), maize (Zea mays; ZmOR), and tomato (Solanum lycopersicum;SlOR). Protein accession numbers are given in the Supplemental Figure S1 legend. The black triangle shows the position of Arg to His mutation, and the white triangle indicates the 88th or 102nd Arg (R) in AtOR. B, Real-time quantitative reverse transcription (qRT)-PCR analysis of the relative expression level of AtOR in the calli of Columbia (Col) control, AtOR lines L6 and L25, and AtORHis lines L14 and L39. C, Western-blot analysis of AtOR protein levels in the calli of Col, AtOR, and AtORHis lines. Actin protein levels were analyzed as loading control. D, Phenotype of 22-d-old plants of Col, AtOR, and AtORHis lines grown in soil. E, Phenotype of 35-d-old plants grown in soil.
To investigate whether alteration of this conserved Arg to His in other OR proteins induced carotenoid accumulation, site-directed mutagenesis (SDM) was carried out with AtOR to generate AtORHis (R90H) mutation. The nucleotides 269 and 270 of AtOR were changed from GA to AT by SDM, resulting in the codon CGA to CAT alteration and changing Arg to His in AtOR (Supplemental Table S1). AtORHis along with AtOR was overexpressed in Arabidopsis. Two T3 homozygous AtOR or AtORHis transgenic lines with comparable expression levels were selected (Fig. 1B). They showed a correlated increase of the OR protein levels (Fig. 1C). The AtORHis and AtOR transgenic lines grew normally, as did the wild-type plant (Fig. 1, D and E), showing no growth inhibition.
Carotenoid composition and content are highly conserved in leaf tissues for optimal photosynthesis (Domonkos et al., 2013). Overexpression of PSY, the gene encoding the rate-limiting enzyme in carotenogenesis, exerts no effect on leaf carotenoid level in Arabidopsis (Maass et al., 2009). Similarly, the carotenoid content as well as chlorophyll level in the adult leaves of AtOR and AtORHis transgenic Arabidopsis lines was similar to that of the wild-type control (Supplemental Fig. S2), as that in the leaves of the wild type and the BoORMUT cauliflower plants (Li et al., 2001).
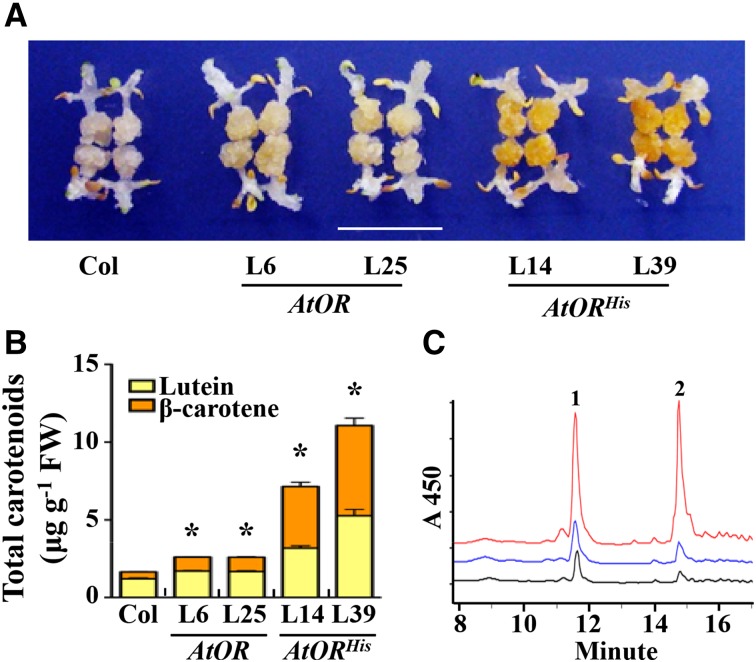
In contrast to leaves, nongreen tissues, such as callus, are frequently responsive to increased pathway flux with enhanced carotenoid accumulation (Maass et al., 2009; Cao et al., 2012). Indeed, callus has been established as an effective system for rapid functional characterization of genes involved in carotenoid biosynthesis and accumulation (Bai et al., 2014). To examine whether AtORHis caused carotenoid accumulation, calli were induced directly from the seeds of these T3 homozygous AtOR and AtORHis transgenic lines. The seed-derived Arabidopsis calli of the wild type were nearly white in color, the AtOR-overexpressing calli had a light-yellow color, and the AtORHis lines exhibited a dark-orange hue (Fig. 2A).
Figure 2.
AtORHis-overexpressing transgenic lines accumulate high levels of total carotenoids in the calli. A, The calli of Col, AtOR, and AtORHis lines. Bar = 1 cm. B, Total carotenoid levels of Col, AtOR, and AtORHis lines measured by HPLC. Data are the average ± se of three biological replicates.*, Significant difference when compared with Col (P ≤ 0.05, n = 3); FW, fresh weight. C, HPLC elute profiles of carotenoids from the calli. The black line is for Col, the blue line is for AtOR-L6, and the red line is for AtORHis-L39. Peak 1 represents lutein, and peak 2 represents β-carotene.
To see whether the color difference was associated with carotenoid levels, carotenoid content in these calli was quantified by HPLC. The wild-type calli accumulated low levels of carotenoids with lutein and β-carotene as the main compounds (Fig. 2B). Consistent with the visible callus color difference, the two AtOR-overexpressing lines showed slightly but significantly higher levels of total carotenoids than the wild type. The AtORHis-overexpressing lines, however, accumulated much higher levels of carotenoids (Fig. 2B). The total carotenoid content in the AtORHis-overexpressing lines was up to 6.7 times that in the wild type, whereas the level in AtOR-overexpressing lines was up to 1.6 times. Noticeably, both AtOR and AtORHis promoted β-carotene biosynthesis (Fig. 2, B and C). In comparison with a ratio of 26% of β-carotene in the wild-type calli, AtOR increased the β-carotene ratio to 35%, whereas AtORHis caused the calli to accumulate β-carotene at approximately 50% of the total carotenoids (Fig. 2, B and C). The stimulation of β-carotene accumulation by AtORHis was similar to that observed with BoORMUT in the transgenic potato tubers (Li et al., 2012). The results suggest that AtORHis confers carotenoid accumulation similar to BoORMUT.
To further explore the implication of ORHis as a new genetic tool for crop nutritional quality improvement, a SbOR was mutagenized with an R104H change (Supplemental Table S1) and overexpressed. Two pairs of SbOR and SbORHis homozygous transgenic lines with comparable expression levels were obtained (Supplemental Fig. S3). Whereas the calli from the SbOR lines had a slight yellow color compared with the untransformed Arabidopsis calli, the SbORHis lines showed intense orange calli (Supplemental Fig. S3). Much higher levels of carotenoids were detected in the SbORHis lines than the SbOR lines (Supplemental Fig. S3). Similar to the cases found in the AtORHis lines, SbORHis induced carotenoid accumulation, with β-carotene composing approximately 50% of the total carotenoids (Supplemental Fig. S3). These results indicate a conserved function of His substitution in an OR protein in inducing carotenoid accumulation among plant species.
Both Amino Acid Type and Position Are Important, and AtORAla Functions Similarly to AtORHis
To investigate whether His substitution at the conserved Arg position in an OR protein was essential for its role in promoting high levels of carotenoid accumulation, additional mutagenesis of AtOR was carried out. To see the amino acid type effect, a number of nucleotide substitutions were introduced into AtOR to replace the codon for the 90th Arg by SDM (Supplemental Table S1). Phe, Lys, and Ala were chosen due to their amino acid properties. His is a polar aromatic amino acid with a very unique chemical property, which makes it difficult to find a proper amino acid to replace it (Betts and Russell, 2003). Phe is also an aromatic acid and has some structural similarity to His. Lys was chosen for its similarity to Arg, as these two amino acids normally could substitute each other without affecting the protein function. Ala was chosen for its small size and nonpolar property (Pogliani, 1994).
The AtORPhe, AtORLys, and AtORAla constructs generating R90F, R90K, and R90A changes, respectively, were overexpressed in Arabidopsis. Approximately 20 independent transgenic lines for each construct were obtained. Calli from over 20 T2 transgenic seedlings for each line were induced and visually screened for the color phenotype. None of the AtORPhe lines showed the same intense orange calli as the AtORHis lines (Supplemental Fig. S4), indicating that Phe did not function as His in AtOR. AtORLys containing Lys substitution also did not give the intensive orange calli (Supplemental Fig. S4). By contrast, 7 out of 17 AtORAla positive transgenic lines showed orange calli (Supplemental Fig. S4). Analysis of these transgenic lines by real-time qRT-PCR showed that AtOR in some of the AtORAla, AtORPhe, and AtORLys transgenic lines was expressed at comparable levels (Supplemental Fig. S4), indicating that the nonorange calli of AtORPhe and AtORLys were not caused by low expression of the transgenes.
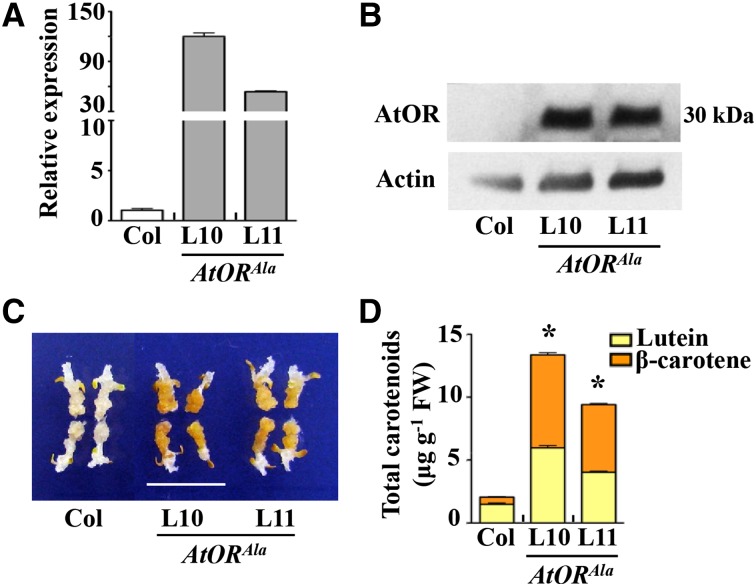
Two T3 homozygous AtORAla lines with high AtOR transcript and AtOR protein levels were generated (Fig. 3, A and B). The calli of these two homozygous AtORAla lines exhibited a dark-orange color similar to those from the AtORHis lines (Fig. 3C). They accumulated high levels of carotenoids, with β-carotene accounting for approximately 55% of the total carotenoids (Fig. 3D). Both the total carotenoid level increase and the β-carotene-to-lutein ratio in the AtORAla lines were similar to those in the AtORHis transgenic lines. These findings suggest that the His substitution (R90H) in AtOR was not unique in inducing carotenoids accumulation. Ala, with very different physiochemical properties from His, was also able to change AtOR function.
Figure 3.
AtORAla-overexpressing transgenic lines accumulate high levels of total carotenoids in the calli. A, Relative expression levels of AtOR tested by qRT-PCR in the calli of Col and AtORAla lines. B, AtOR protein levels in the calli of Col and AtORAla lines. C, The callus color of Col and AtORAla lines. Bar = 1 cm. D, Total carotenoid levels in the calli of Col and AtORAla lines, measured by HPLC. *, Significant difference when compared with Col (P ≤ 0.05, n = 3).
In addition to the amino acid type, the amino acid position effect on the AtOR function was also tested. Both AtORHis and BoORMUT mutations cause a carotenoid overproduction phenotype. The sequence alterations were in the N-terminal region of OR proteins at amino acid position 90 in AtOR and 104 in BoOR. Therefore, two more His mutations of AtOR in this N-terminal region were produced to replace the 88th Arg (R88H) and 102nd Arg (R102H; Fig. 1A). None of the Arabidopsis transgenic lines overexpressing AtORHis-88 and AtORHis-102 showed orange calli as AtORHis and AtORAla calli despite similar transgene expression levels in some lines (Supplemental Fig. S4). This result indicates that His at the 90th position is much more important than that at the 88th or 102nd for AtORHis function. Together, these findings suggest that both the amino acid position and type are crucial for OR in conferring high levels of carotenoid accumulation.
Plant genomes contain another OR family protein named OR-Like (Zhou et al., 2015). In Arabidopsis, AtOR-Like (At5g06130) shares 69% amino acid sequence identity with AtOR. AtOR-Like also has the conserved Arg at the 97th position (Supplemental Fig. S5). The Arg in AtOR-Like was mutated to His (R97H), and the AtOR-LikeHis construct was overexpressed in Arabidopsis. T2 seeds of 30 independent transgenic lines were induced for callus formation. None of these AtOR-LikeHis-overexpressing lines showed orange calli as AtORHis and AtORAla (data not shown). It appears that the promotion of carotenoid overaccumulation by the His substitution only works with OR but not with the OR-Like gene.
AtORHis Does Not Dramatically Affect Carotenoid Metabolic Gene Expression or Alter Plastid Localization
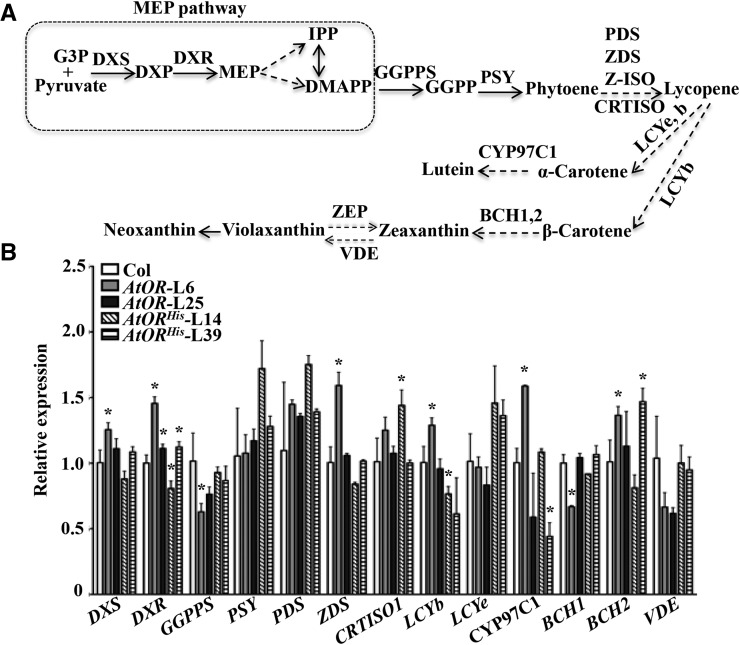
Since both AtORHis and AtORAla shared a similar effect on carotenoid accumulation, we focused the following studies on AtORHis lines. To investigate whether AtOR and AtORHis enhanced carotenoid accumulation by affecting carotenogenic gene expression, the transcript levels of carotenoid biosynthesis genes (Fig. 4A) as well as homologous genes of pepper plastid fusion and/or translocation factor (Pfts; Hugueney et al., 1995) in the calli of these transgenic lines in comparison with the wild type were measured. RNA from the same callus samples used for carotenoid measurement was extracted. qRT-PCR analysis revealed that, in general, no dramatic difference in the expression of the core carotenoid biosynthetic genes was observed in the AtOR- or AtORHis-overexpressing lines in comparison with the wild-type control, although some genes in one line showed a significant difference (Fig. 4B). The result was consistent with previous studies showing that the transcript levels of carotenoid biosynthetic genes were similar between white and orange cauliflower (Li et al., 2001). Thus, the AtORHis-induced carotenoid accumulation appeared not to be related to transcriptional regulation of carotenogenic genes. Moreover, examination of the homolog of pepper Pfts (ATP-dependent zinc metalloprotease [FtsH2]) along with FtsH homologs in Arabidopsis revealed that the His substitution in AtORHis did not significantly alter the Pfts homologous gene expression (Supplemental Fig. S6).
Figure 4.
Expressions of carotenoid biosynthetic genes are not greatly affected in the calli of AtOR and AtORHis transgenic lines. A, Outline of carotenoid biosynthetic pathway. Solid arrows indicate one step, and dashed arrows indicate multiple steps. G3P, Glyceraldehyde-3-phosphate; DXP, 1-deoxy-d-xylulose-5-phosphate; MEP, 2-C-methyl-d-erythritol-4-phosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate, GGPP, geranylgeranyl diphosphate; DXS, 1-deoxy-d-xylulose-5-phosphate synthase; DXR, 1-deoxy-d-xylulose 5-phosphate reductoisomerase; GGPPS, geranylgeranyl diphosphate synthase; PDS, phytoene desaturase; ZDS, ζ-carotene desaturase; Z-ISO, ζ-carotene isomerase; CRTISO, carotenoid isomerase; LCYe, lycopene ε-cyclase; LCYb, lycopene β-cyclase; CYP97C1, cytochrome P450-type monooxygenase 97C; BCH1/2, β-carotenoid hydroxylase 1/2; ZEP, zeaxanthin epoxidase; VDE, violaxanthin deepoxidase. B, Relative expression level of carotenogenic genes in Col and AtOR and AtORHis lines. Data are the average ± se of three biological replicates with two technical trials. *, Significant difference when compared with Col (P ≤ 0.05, n = 3).
AtOR has been shown to localize in plastids (Zhou et al., 2015). To find out the basis of AtORHis action in regulating carotenoid accumulation, we first checked whether this Arg to His substitution affected its subcellular localization. AtORHis fused with GFP was expressed in Arabidopsis. The AtORHis-GFP transgenic lines also had orange calli (Supplemental Fig. S7), indicating that the AtORHis-GFP was functional. Examination of the GFP signals in the AtORHis-GFP and AtOR-GFP transgenic lines showed that AtORHis exhibited the same subcellular location in chloroplast as AtOR (Supplemental Fig. S7). This result revealed that the His substitution in the AtORHis mutant did not affect the plastidial location of the OR protein.
AtORHis Has Similar Function as AtOR in Posttranscriptional Regulation of PSY Protein Level
Our recent study discovered that AtOR physically interacts with PSY and posttranscriptionally regulates PSY protein level (Zhou et al., 2015). To find out whether AtORHis enhanced carotenoid accumulation with strong regulation of PSY, we examined the interaction between AtORHis and PSY, and compared the effect of AtOR and AtORHis in regulating PSY protein level and activity.
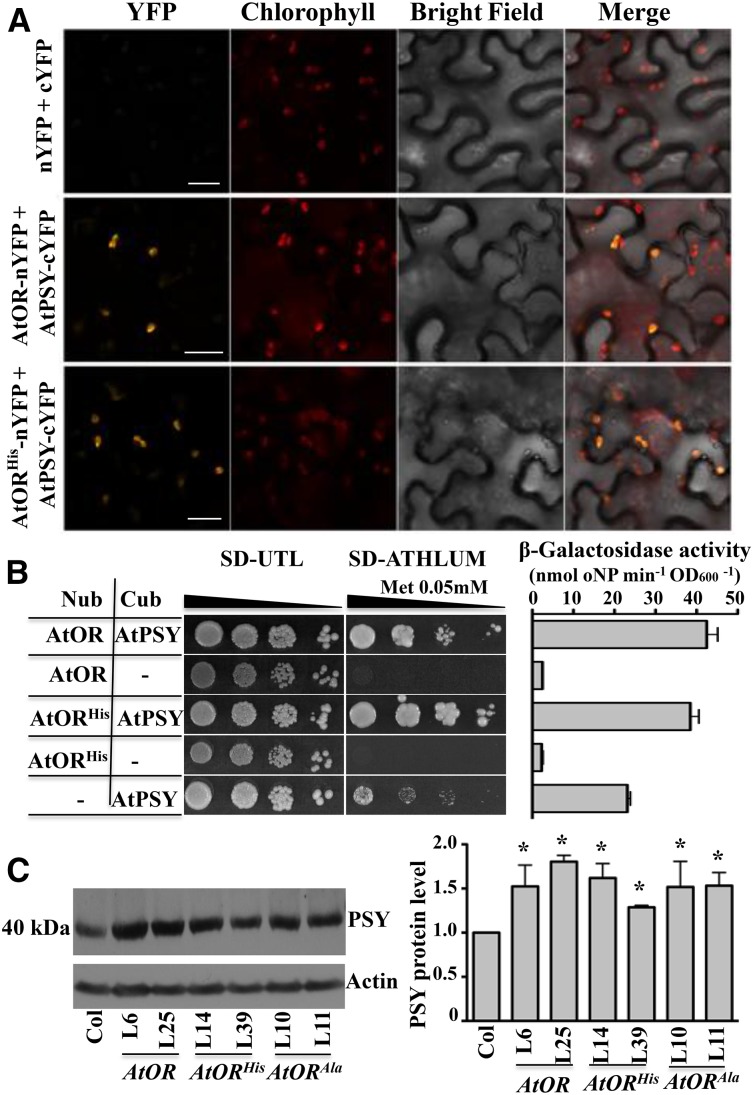
To test the interaction between AtORHis and PSY, bimolecular fluorescence complementation (BiFC) assays in Nicotiana benthamiana leaves were performed to investigate whether AtORHis and PSY interaction occurred in vivo and the subcellular localization of their interaction. Yellow fluorescent protein (YFP) signals were observed when AtORHis-N terminus of YFP (AtORHis-nYFP) and PSY-C terminus of YFP (PSY-cYFP) were coexpressed in tobacco (Nicotiana tabacum) leaf epidermal cells, similar to the interaction between AtOR-nYFP and PSY-cYFP (Fig. 5A). Such interactions occurred in chloroplasts, consistent with the plastidial localizations of these proteins (Supplemental Fig. S7; Shumskaya et al., 2012). In addition, a split-ubiquitin membrane-based yeast two-hybrid (Y2H) system was used to check their interaction in vitro, as OR contains two transmembrane domains (Lu et al., 2006). Both AtORHis and AtOR were found to interact directly with PSY in the Y2H assay and showed similar interactions by quantification of the reporter gene LacZ encoding β-galactosidase via o-nitrophenyl-β-d-galactopyranoside activity measurements (Fig. 5B). These results demonstrate that AtORHis was also physically associated with PSY, showing similar interaction with PSY as AtOR.
Figure 5.
AtORHis and AtOR share a similar interaction with AtPSY and the same effect on regulating AtPSY protein level. A, BiFC of AtOR or AtORHis interaction with AtPSY. Vectors with N terminus of YFP (nYFP) and C terminus of YFP (cYFP), AtOR-nYFP and AtPSY-cYFP, or AtORHis-nYFP and AtPSY-cYFP were coexpressed in tobacco leaves. The fluorescence signals were detected with a confocal microscope. Bar = 20 μm. B, Yeast two-hybrid (Y2H) analysis of AtOR and AtORHis interactions with AtPSY. Combinations of vectors with N terminus of ubiquitin (Nub), C terminus of ubiquitin (Cub), Nub-AtOR, Nub-AtORHis, or AtPSY-Cub were coexpressed in yeast (Saccharomyces cerevisiae) cells. Yeast was grown on nonselective medium (synthetic defined premix/Uri-Thr-Leu [SD/UTL]) and selective medium (synthetic defined premix/Ala-Thr-His-Leu-Uri-Met [SD/ATHLUM]), with 0.05 mm Met with four series of 10 times dilution. For the β-galactosidase activity assay, yeast was cultured in liquid medium overnight and tested. OD600, absorbance at wavelength 600 nm; oNPG, o-nitrophenyl-β-d-galactopyranoside. C, Western blot of AtPSY in the calli of Col, AtOR, AtORHis, and AtORAla lines. Actin protein level was used as a loading control. Relative PSY protein levels were calculated from quantification of western band signals. *, Significant difference when compared with Col (P ≤ 0.05, n = 3).
To see whether AtORHis mediated the enhanced carotenoid production by promoting high accumulation or activity of PSY, PSY protein levels and activity in the calli of the AtOR and AtORHis as well as the AtORAla transgenic lines were compared by western-blot analysis and activity assay. Consistent with previous observation of enhanced PSY protein levels in leaves of the AtOR transgenic lines (Zhou et al., 2015), increased PSY protein abundance was observed in the calli of the AtOR lines in comparison with the wild type (Fig. 5C). The AtORHis and AtORAla transgenic lines also contained increased levels of PSY similar to the AtOR lines (Fig. 5C). The result indicated that AtORHis and AtORAla posttranscriptionally regulated PSY protein abundance but did not promote higher accumulation than AtOR. No enhanced PSY activities were observed in these transgenic lines (Supplemental Fig. S8). Thus, the dramatic differences in promoting carotenoid accumulation between AtOR and AtORHis suggest that the mutations might gain additional function to confer high levels of carotenoid production.
AtORHis Promotes Chromoplast Biogenesis
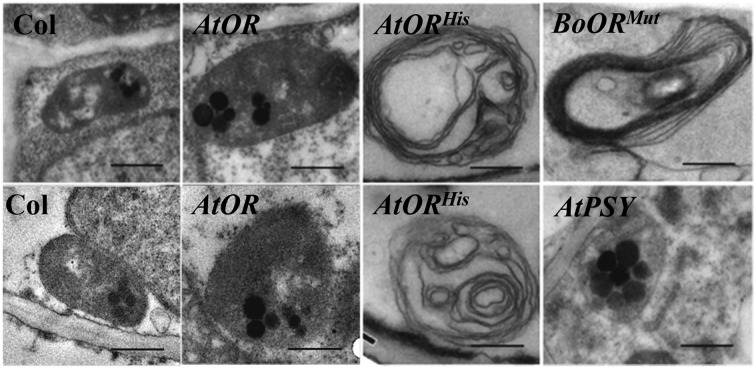
BoORMUT is known to trigger chromoplast formation in cauliflower (Lu et al., 2006). To see whether AtORHis promoted chromoplast biogenesis, TEM analysis was carried out to examine plastids in the calli of the wild type as well as in the AtOR and AtORHis transgenic lines. In addition, plastids were compared with those from orange curd cauliflower mutant and calli of an Arabidopsis PSY-overexpressing line (Maass et al., 2009). Plastids and plastoglobuli within these plastids were clearly seen in the calli of wild-type samples. Similar plastid structures generally harboring larger plastoglobuli with a higher electron density were observed in the calli expressing AtOR compared with wild-type calli (Fig. 6). Overexpression of AtPSY has been shown to result in carotenoid accumulation in the calli of Arabidopsis transgenic lines (Maass et al., 2009). Plastid structures similar to those in the calli of AtOR lines but with much larger plastoglobuli and a higher electron density were observed in the calli of AtPSY lines (Fig. 6). By contrast, the AtORHis callus cells contained large plastids with dark sinuous membrane structures that generally lacked plastoglobuli (Fig. 6). The AtORHis-induced plastids shared the typical membranous chromoplasts from the orange cauliflower mutant (Fig. 6) as shown previously (Paolillo et al., 2004). These findings suggest that AtORHis induced biogenesis of the same membranous chromoplasts as BoORMUT. Unlike AtORHis, the similar plastid structures in cells of wild-type and AtOR calli indicate that AtOR did not confer chromoplast differentiation.
Figure 6.
TEM images of plastids in the Arabidopsis calli of Col, AtOR, AtORHis, and AtPSY transgenic lines as well as in the orange curd of BoORMut cauliflower mutant. Plastids with plastoglobuli were observed in the calli of Col, AtOR, and AtPSY transgenic lines. Characteristic membranous chromoplasts found in the curd of BoORMut cauliflower mutant were observed in the AtORHis transgenic lines. Bar = 0.5 μm.
DISCUSSION
OR represents a unique class of regulatory genes in mediating carotenoid accumulation (Li and Yuan, 2013). Two natural mutations of OR are found to promote high levels of β-carotene accumulation in plants. The first one is BoORMUT from orange curd cauliflower mutant. BoORMUT contains a large retrotransposon insertion and produces three alternatively spliced transcripts, but none of them alone induces massive carotenoid accumulation (Lu et al., 2006). The second one is the recently discovered CmOR in melon. The CmOR protein with a single amino acid difference of His or Arg distinguishes orange-flesh melon from white- or green-flesh melon (Tzuri et al., 2015). In this study, we further demonstrated that this single Arg to His substitution was effective in changing the functionality of a wild-type OR in promoting carotenoid overaccumulation. In addition, we discovered that His was not the only amino acid that promoted carotenoid overproduction, as Ala substitution also functioned similarly in enhancing carotenoid level. AtORHis was shown to posttranscriptionally regulate PSY protein abundance. However, its specific role in stimulating carotenoid overaccumulation relied on its unique ability to promote chromoplast biogenesis, which was documented in Arabidopsis calli. Our findings firmly imply the feasibility of creating a functional allele of OR in a crop species for nutritional quality improvement. In addition, since Arabidopsis normally does not contain a tissue with chromoplasts, the demonstration of chromoplast formation establishes the Arabidopsis calli as a unique and effective system for functional study of genes involved in chromoplast biogenesis and carotenoid accumulation.
His/Ala Substitution of the Conserved Arg in an OR Protein Promotes High Levels of Carotenoid Accumulation
Both AtORHis and AtORAla were found to mediate high levels of carotenoid accumulation in the Arabidopsis callus system, and especially promoted the overproduction of β-carotene, the most potent provitamin A carotenoid. Clearly, the conserved Arg position in an OR protein is crucial for the ORHis or ORAla function in enhancing carotenoid accumulation. The His substitutions of Arg at nearby sites, such as at the 88th or 102nd position of AtOR, were unable to induce carotenoid accumulation. Although His substitution at the conserved Arg site of OR was important, His was not the only amino acid that altered carotenoid levels in the OR mutants. AtORAla had a functional role similar to AtORHis.
His is one of the most common amino acids found in the active or binding site of proteins (Betts and Russell, 2003). His mutation in an OR may change or create a new active center or give new interaction ability with some unknown proteins to this region, which is under investigation. Although Ala has quite different chemical properties from His, Ala may cause the same change in the active center or structure of AtOR as His. Unfortunately, there are presently no related three-dimensional structures available to allow for protein modeling that could shed light on these structural changes. Future protein three-dimensional structure analysis of the OR protein could be helpful in the prediction of active center and important amino acid sites. The information could help explain the basis of the His and Ala substitutions and produce new candidates for OR mutations to mediate carotenoid metabolism. Indeed, the BoORMUT mutation does not affect the conserved Arg site.
Recent reports show that the wild-type OR proteins also have the capacity to induce carotenoid biosynthesis. Overexpression of a sweet potato (Ipomoea batatas) OR gene produces light-yellow calli and increases carotenoid content in the transgenic sweet potato roots (Kim et al., 2013; Park et al., 2015). Similarly, expression of AtOR produces a clear color difference with approximately 2-fold increased carotenoid levels in the AtOR ZmPSY transgenic rice calli in comparison with the ZmPSY-only transgenic lines (Bai et al., 2014). Consistent with these reports, significantly increased carotenoid levels with a clearly visible color difference were also observed in this study when AtOR and SbOR were overexpressed. However, the AtORHis, AtORAla, and SbORHis transgenic lines produced much large quantities of carotenoids with intense orange calli in comparison with the wild-type OR lines.
Dual Function of AtORHis in Regulating PSY Protein Abundance and Chromoplast Biogenesis
PSY catalyzes the rate-limiting step in the carotenoid biosynthetic pathway. The transcript levels of PSY along with other carotenogenesis genes were not dramatically affected by the overexpression of either AtOR or AtORHis, as shown in the other studies (Li et al., 2001; Lopez et al., 2008; Kim et al., 2013). OR was recently discovered to serve as a major posttranscriptional regulator of PSY protein level and activity (Zhou et al., 2015), providing an explanation for a wild-type OR-mediated carotenoid biosynthesis. Here, we found that AtORHis also directly interacted with PSY in plastids, the place where carotenoids are synthesized. Similar interactions of AtOR or AtORHis with PSY were observed in Y2H and BiFC assays, showing that the His substitution in OR did not alter its interaction with PSY. A similar up-regulation of PSY protein levels was observed in the AtOR and AtORHis transgenic lines, suggesting that AtORHis does not promote higher accumulation of PSY protein abundance than AtOR. Interestingly, no increased PSY enzyme activity was detected between the callus samples of wild-type and transgenic lines, which might be due to low sensitivity of the activity assay for the OR callus samples. The fact that there was no dramatic difference in PSY protein level and activity between the AtORHis and AtOR lines indicates that AtORHis gains additional function to mediate carotenoid overproduction.
Our previous studies show that BoORMUT confers carotenoid accumulation in chromoplasts (Li et al., 2001; Lu et al., 2006). Expression of the BoORMUT transgene in both white cauliflower and potato tubers leads to the biogenesis of chromoplasts with enhanced carotenoid content (Lu et al., 2006; Lopez et al., 2008). Consistent with these studies, we demonstrated that AtORHis also functioned in inducing chromoplast biogenesis. BoORMUT is known to induce the formation of membranous chromoplasts containing sinuous or scattered membranes in orange cauliflower mutant (Paolillo et al., 2004). The typical and similar membranous chromoplasts were only observed in the AtORHis samples, but not seen in the AtOR transgenic lines. In the AtOR samples, a different type of plastids containing plastoglobuli was observed, which shared the same structures as those found in the wild-type controls. These findings support the specific function of AtORHis in conferring chromoplast biogenesis.
Chromoplasts are the plastids that possess unique mechanisms to synthesize and store large quantities of carotenoids (Lu and Li, 2008; Egea et al., 2010; Li and Yuan, 2013). Biogenesis of chromoplasts enhances sink strength for carotenoid accumulation by effectively sequestering the newly synthesized carotenoids to stimulate continuous biosynthesis and stable storage within the plastids (Rabbani et al., 1998; Li et al., 2012; Kilambi et al., 2013). Increases in either chromoplast size or number have been found to contribute to enhanced carotenoid content in tomato pigment mutants (Liu et al., 2004; Kolotilin et al., 2007; Galpaz et al., 2008). As both AtOR and AtORHis lines shared similar levels of OR expression, and AtORHis did not promote higher PSY activity than AtOR to control carotenoid biosynthesis, the much higher level of carotenoid accumulation in the AtORHis transgenic lines was likely due to the specific capacity of AtORHis in triggering chromoplast biogenesis, which provides enhanced plastid sink strength for carotenoid biosynthesis and storage.
The mechanism underlying AtORHis-induced chromoplast biogenesis remains to be elucidated. Chromoplast biogenesis is associated with membrane proliferation, and remodeling of the internal membrane represents the most prominent change during chromoplast differentiation (Egea et al., 2010; Li and Yuan, 2013). Stacks of membranes in the chromoplasts of the OR mutants are evidenced by TEM from this and previous studies (Li et al., 2001; Paolillo et al., 2004). In addition, our ongoing study reveals that AtORHis physically interacts with a number of proteins involved in the plastid protein import apparatus. Based on these studies, a possible mechanism of AtORHis in controlling chromoplast biogenesis for carotenoid accumulation is postulated to modulate plastid membrane proliferation and/or chromoplast preprotein import via regulating proteins in the plastid protein import machinery. Future elucidation of the specific mechanism underlying OR-regulated chromoplast biogenesis could also open up a new area of plastid biology.
An Effective Arabidopsis Callus System for Functional Study of Genes Involved in Chromoplast Biogenesis and Carotenoid Accumulation
Arabidopsis normally does not contain chromoplasts in its tissues, which hinders progress in the study of genes, signaling, and metabolic processes involved in chromoplast biogenesis. Previously, we observed that calli derived from cauliflower BoORMUT plants accumulate β-carotene with an intense orange color (Li et al., 2006), and BoORMUT confers chromoplast formation in different plant species (Lu et al., 2006; Lopez et al., 2008). The current study produced Arabidopsis ORHis transgenic lines that contained intense orange color with carotenoid accumulation in the calli. Electron microscopy revealed the presence of membranous chromoplasts similar to chromoplasts found in the curd of the orange cauliflower mutant (Li et al., 2001; Paolillo et al., 2004). Thus, we generated Arabidopsis lines that contain a tissue with a chromoplast development program.
The callus is very responsive to genes involved in carotenoid biosynthesis with different color phenotype and has been used as a model plant system to study the gene functions in carotenoid metabolism. Maize callus transformed with PSY from different plant species shows marked differences in color intensity and was used to identify the most efficacious PSY for Golden Rice 2 production (Paine et al., 2005). Arabidopsis callus expressing AtPSY gives an intense orange color and is used to investigate the effect of PSY on carotenoid sequestration (Maass et al., 2009). Expression of a bacterial PSY in different citrus genotypes yields calli of various colors with diverse natural carotenoid patterns and provides information to interpret the common appearance of a favored β,β-pathway following an increased expression of PSY in plants (Cao et al., 2012). Recently, rice embryogenic callus was developed for rapid functional characterization of genes involved in carotenoid biosynthesis following validation with known and unknown function genes (Bai et al., 2014).
The Arabidopsis ORHis or ORAla callus system could be used as a unique model for rapid functional study of genes involved in chromoplast biogenesis in addition to carotenoid metabolism. By overexpressing or suppressing the expression of associated genes in these lines, their effects on chromoplast development and carotenoid accumulation can be examined in the callus system following quick visual screening of callus phenotypes. As Arabidopsis can be easily transformed with a large number of gene constructs, this Arabidopsis callus system likely provides an effective and relatively high throughput approach to functionally characterize candidate genes for their involvement in chromoplast biogenesis and carotenoid metabolism.
ORHis and ORAla Serve as New Genetic Tools for Crop Nutritional Quality Improvement
Carotenoids, especially β-carotene, are indispensable to human nutrition and provide the primary dietary source for vitamin A biosynthesis. Low levels of carotenoids in major food crops contribute to the global prevalence of vitamin A deficiency, which leads to significant efforts to generate carotenoid-enriched crops either through biotechnology or traditional breeding. By expression of PSY or a minipathway of carotenoid biosynthesis, numerous transgenic crops with enhanced carotenoid levels are produced (Fraser et al., 2002; Ducreux et al., 2005; Paine et al., 2005; Diretto et al., 2007; Naqvi et al., 2009, Welsch et al., 2010). Selection of favorable alleles that alter carotenoid metabolic flux toward β-carotene results in the breeding of orange maize with high β-carotene content (Harjes et al., 2008; Yan et al., 2010). In contrast to modification of catalytic activity of the pathway, the discovery of OR provides an alternative and complementary approach for carotenoid enhancement in food crops via enhancing the storage sink strength (Li and Van Eck, 2007). Moreover, OR promotes continuously increased carotenoid accumulation in the BoORMUT transgenic potato tubers during postharvest storage (Li et al., 2012).
The demonstration that ORHis and ORAla promoted high levels of carotenoid accumulation suggests their potential as new genetic tools for crop carotenoid enhancement. In contrast to BoORMUT that contains a large retrotransposon insertion, with none of the alternatively spliced transcripts showing an effect in enhancing carotenoid level comparable with the BoORMUT allele (Lu et al., 2006), ORHis or ORAla contains one or two nucleotide changes required for the His or Ala substitution in a wild-type OR gene. Such a change is sufficient to alter OR function for carotenoid accumulation. In addition, whereas homozygous BoORMUT causes a dwarf phenotype in cauliflower (Li et al., 2001), ORHis exhibits no deteriorating effects on plant growth and development in melon as well as in the transgenic Arabidopsis.
OR is highly conserved among diverse plant species. Genome editing has emerged as a potent new tool that enables site-specific modifications of a genome (Sander and Joung, 2014). As one or two nucleotide changes alter the OR function, the rapidly developing genome-editing techniques can be utilized to induce the site-specific mutagenesis of a crop OR gene. Moreover, genetic diversity among crop species provides an additional resource for discovering the functional variants of OR. Indeed, the variable sites of OR mutations in ORHis/Ala and BoORMUT suggest the potential to identify new functional alleles from genetic variation of crops. Thus, in addition to the biotechnology approach, there is potential to introduce functional alleles of OR into staple crops through traditional breeding to improve crop nutritional quality.
MATERIALS AND METHODS
Plant Materials and Callus Induction
Arabidopsis (Arabidopsis thaliana) plants were grown in a computer-controlled growth chamber under 14 h of light and 10 h of dark at 23°C. To induce calli, Arabidopsis seeds were surface sterilized in 20% (v/v) bleach for 15 min and sown on plates of seed-derived callus medium (4.30 g L−1 Murashige and Skoog basal salt, 0.1% [v/v] Gamborg's vitamin, 1.0 g L−1 MES, 3% [w/v] Suc, 0.5 mg L−1 2,4-dichlorophenoxyacetic acid, 2 mg L−1 indole-3-acetic acid, 0.5 mg L−1 2-isopentenyladenine, adjusted pH to 5.8 by 1 m KOH, and 0.4% [w/v] Phytagel [Sigma]) as described (Maass et al., 2009). Following stratification for 2 d in dark at 4°C, the seeds were germinated under light (16 h of light/8 h of dark) at 23°C for 5 d and grown in dark for 2 weeks in a growth incubator. Calli were collected, frozen in liquid nitrogen, and stored at −80°C until use.
SDM and Arabidopsis Transformation
To identify the conserved Arg in the OR proteins, the protein sequences of AtOR (At5g61670), AtOR-like (At5g06130), and SbOR (Sb04g033280; http://www.plantgdb.org/SbGDB/) were aligned with melon (Cucumis melo) CmOR (Tzuri et al., 2015). SDM primers specific to change Arg to other amino acids in these OR proteins were designed using the Agilent QuickChange Primer Design program (http://www.genomics.agilent.com/primerDesignProgram.jsp; Supplemental Table S2). AtOR, AtOR-like, and SbOR in pCR2.1 vector were mutagenized using the QuikChange II XL Site-Directed Mutagenesis Kit according to the instruction manual of manufacturer (Agilent Technologies, catalog number 200521). The OR mutant variants along with their wild-type versions were cloned into pCAMBIA1300S with Cauliflower mosaic virus 35S promoter (Zhou et al., 2011) to generate constructs producing AtOR R90A, R90H, R90K, R90F, R88H, and R102H as well as SbOr R104H.
The plasmid constructs were transferred into Agrobacterium tumefaciens GV3101 by electroporation and introduced into Arabidopsis by floral dip. The transformed T1 seeds were screened for positive transformants on plates of Murashige and Skoog medium containing 1% (w/v) Suc, 30 mg L−1 hygromycin, 50 mg L−1 carbenicillin, and 0.6% (w/v) agar. Several independent homologous T3 lines for each construct were selected and used for phenotype and further analysis.
Carotenoid Extraction and Analysis
Carotenoids from Arabidopsis calli (200 mg) were extracted and analyzed essentially following the method as described by Lopez et al. (2008). Carotenoids were identified by their characteristic absorption spectra and their retention times in comparison with standards and published spectra, and quantified using a β-carotene calibration curve (Li et al., 2012). Samples were analyzed in triplicate with three biological replicates.
Protein Extraction, Western-Blot Analysis, and PSY Activity Assay
Total proteins from Arabidopsis calli were extracted in a cold room with a phenol-based protein extraction protocol (Yang et al., 2007). Powdered calli (200 mg) was added with 600 μL of extraction buffer (0.5 m Tris-HCl [pH 7.5], 50 mm EDTA, and 0.1 m KCl with 10 mm dithiothreitol and 1 mm phenylmethylsulfonyl fluoride added just before use) and 600 μL of Tris-EDTA saturated phenol (pH 6.7). The sample mixtures were vigorously shaken for 30 min and centrifuged at 12,000 rpm for 30 min. Proteins in the upper phenolic phase were collected, precipitated with 5 volumes of 0.1 m NH4AOC dissolved in methanol overnight, and centrifuged at 4,000 rpm for 40 min at 4°C. Pellets were washed with prechilled methanol, air dried, and dissolved in 100 μL of solution containing 7 m urea and 2 m thiourea. Protein concentration was measured by adding 1 μL of sample to 499 μL of protein assay buffer (Bio-Rad).
For western-blot analysis, 50 μg of total proteins was separated by 10% SDS-PAGE gels and blotted onto a polyvinylidene difluoride membrane (EMD Millipore). Membranes were blocked overnight and included with primary anti-OR antibody (Lu et al., 2006) or anti-PSY antibody followed by secondary goat anti-rabbit IgG antibody (Bio-Rad). Signals were detected using the ECL Prime Western Blotting Detection Reagent following the manufacturer’s manual (GE Healthcare). AtActin was detected in the same membrane using anti-Actin antibody (Sigma-Aldrich, A0848) for loading control. PSY western signal was quantified with Actin as an internal control using ImageJ (http://www.di.uq.edu.au/sparqimagejblots).
The in vitro PSY activity assay was performed with 400 µg of plastid membrane proteins isolated from callus samples as detailed (Zhou et al., 2015). Samples were analyzed with two technical trials and two biological replicates.
RNA Extraction and qRT-PCR
Total RNA was extracted from 100 mg of calli using Trizol reagent (Life Technologies). The complementary DNA was synthesized from 1 μg of RNA with oligo(dT) using Superscript III reverse transcriptase (Invitrogen). Real-time qRT-PCR was performed using SYBR Green Supermix (Bio-Rad) and gene-specific primers (Supplemental Table S2) in an ABI 7500 Real-Time PCR system (Applied Biosystems). The Arabidopsis ubiquitin extension protein1 gene was used as a nuclear gene internal control, and relative expression levels were calculated using the cycle threshold method (Lyi et al., 2007). Values reported represent the averages of three biological replicates with two technical trials.
Y2H and BiFC Assay
The Y2H assay was done using the split-ubiquitin system (Zhou et al., 2015). Coding sequence (CDS) of AtPSY without its transit peptide and stop codon was cloned into pMetYCgate vector to produce a fusion protein with Cub, while AtOR and AtORHis CDSs without their transit peptides were cloned into pNXgate to generate a fusion protein with Nub. The pMetYCgate constructs were transformed into yeast strain THY.AP5, and pNXgate to strain THY.AP4. Mating of these two strains brought these two vectors to the diploid yeast cells, which were then inoculated to a selective medium (synthetic defined premix/Ala-Thr-His-Leu-Uri-Met with 0.05 mm Met) for protein interaction tests. β-galactosidase activity in yeast was assayed in triplicate and calculated as nmol o-nitrophenyl-β-d-galactopyranoside min−1 absorbance at wavelength 600 nm−1 as described, with minor modification (20 mg L−1 Met added to the medium; Zhou et al., 2015).
BiFC was performed as described (Zhou et al., 2015). In brief, AtPSY CDS without stop codon was cloned to the vector pUC-SPYCE to make a fusion protein with cYFP. AtOR and AtORHIS CDSs without stop codons were cloned to pUC-SPYNE to produce a fusion protein with nYFP. Plasmids in Agrobacterium sp. GV3101 were used to inject 4-week-old tobacco (Nicotiana tabacum) leaves. Two days after injection, the leaves were observed under confocal microscope (Leica TCS SP5 Laser Scanning Confocal Microscope). The YFP and chlorophyll fluorescence was detected with excitation light wavelength at 488 nm and emission filter for YFP at 520 to 560 nm and for chlorophyll at 620 to 680 nm. Images were taken and processed using Leica LAS AF software.
TEM
Arabidopsis calli of Col, AtOR, AtORHis, and AtPSY were analyzed by TEM following the protocol described previously (Cao et al., 2012). The calli were fixed, followed by dehydration and embedding. The embedded samples were sectioned with Leica UC6 ultramicrotome to obtain ultrathin sections, which were then stained and analyzed by a transmission electron microscope (HITACHI H-7650).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of full length OR proteins.
Supplemental Figure S2. Leaf chlorophyll and total carotenoid contents of the AtOR and AtORHis lines.
Supplemental Figure S3. SbORHis callus and carotenoid content.
Supplemental Figure S4. Phenotype and AtOR expression level in T2 calli of AtOR mutation lines.
Supplemental Figure S5. Alignment of the protein sequences of AtOR and AtOR-Like.
Supplemental Figure S6. Relative expression levels of Arabidopsis Pftf homologous genes FtsH1, FtsH2, FtsH5, and FtsZ1 in AtOR and AtORHis calli.
Supplemental Figure S7. AtORHis-GFP calli and AtOR-GFP and AtORHis-GFP subcellular localization.
Supplemental Figure S8. PSY activity in Col, AtOR, AtORHis, and AtORAla calli.
Supplemental Table S1. Site-directed mutagenesis sites in AtOR and SbOR.
Supplemental Table S2. Primer sequences used in this study.
Glossary
- TEM
transmission electron microscopy
- qRT
quantitative reverse transcription
- SDM
site-directed mutagenesis
- BiFC
bimolecular fluorescence complementation
- nYFP
N terminus of YFP
- cYFP
C terminus of YFP
- Y2H
yeast two-hybrid
- Nub
N terminus of ubiquitin
- Cub
C terminus of ubiquitin
- YFP
yellow fluorescent protein
- CDS
coding sequence
Footnotes
This work was supported by the United States-Israel Binational Agricultural Research and Development Fund (grant no. BARD US–4423–11) and by Harvestplus Research Consortium (to R.W.).
Articles can be viewed without a subscription.
References
- Auldridge ME, McCarty DR, Klee HJ (2006) Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr Opin Plant Biol 9: 315–321 [DOI] [PubMed] [Google Scholar]
- Bai C, Rivera SM, Medina V, Alves R, Vilaprinyo E, Sorribas A, Canela R, Capell T, Sandmann G, Christou P. , et al. (2014) An in vitro system for the rapid functional characterization of genes involved in carotenoid biosynthesis and accumulation. Plant J 77: 464–475 [DOI] [PubMed] [Google Scholar]
- Betts MJ, Russell RB (2003) Amino acid properties and consequences of substitutions. In Barnes MR, Gray IC, eds, Bioinformatics for Geneticists. John Wiley & Sons, Ltd., Chichester, UK, pp 289–316 [Google Scholar]
- Cao H, Zhang J, Xu J, Ye J, Yun Z, Xu Q, Xu J, Deng X (2012) Comprehending crystalline β-carotene accumulation by comparing engineered cell models and the natural carotenoid-rich system of citrus. J Exp Bot 63: 4403–4417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli CI. (2011) Carotenoids in nature : insights from plants and beyond. Funct Plant Biol 38: 833–847 [DOI] [PubMed] [Google Scholar]
- Cazzonelli CI, Pogson BJ (2010) Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci 15: 266–274 [DOI] [PubMed] [Google Scholar]
- Diretto G, Welsch R, Tavazza R, Mourgues F, Pizzichini D, Beyer P, Giuliano G (2007) Silencing of beta-carotene hydroxylase increases total carotenoid and beta-carotene levels in potato tubers. BMC Plant Biol 7: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domonkos I, Kis M, Gombos Z, Ughy B (2013) Carotenoids, versatile components of oxygenic photosynthesis. Prog Lipid Res 52: 539–561 [DOI] [PubMed] [Google Scholar]
- Ducreux LJM, Morris WL, Hedley PE, Shepherd T, Davies HV, Millam S, Taylor MA (2005) Metabolic engineering of high carotenoid potato tubers containing enhanced levels of beta-carotene and lutein. J Exp Bot 56: 81–89 [DOI] [PubMed] [Google Scholar]
- Egea I, Barsan C, Bian W, Purgatto E, Latché A, Chervin C, Bouzayen M, Pech JC (2010) Chromoplast differentiation: current status and perspectives. Plant Cell Physiol 51: 1601–1611 [DOI] [PubMed] [Google Scholar]
- Farré G, Bai C, Twyman RM, Capell T, Christou P, Zhu C (2011) Nutritious crops producing multiple carotenoids--a metabolic balancing act. Trends Plant Sci 16: 532–540 [DOI] [PubMed] [Google Scholar]
- Fraser PD, Bramley PM (2004) The biosynthesis and nutritional uses of carotenoids. Prog Lipid Res 43: 228–265 [DOI] [PubMed] [Google Scholar]
- Fraser PD, Romer S, Shipton CA, Mills PB, Kiano JW, Misawa N, Drake RG, Schuch W, Bramley PM (2002) Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc Natl Acad Sci USA 99: 1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galpaz N, Wang Q, Menda N, Zamir D, Hirschberg J (2008) Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J 53: 717–730 [DOI] [PubMed] [Google Scholar]
- Giuliano G, Tavazza R, Diretto G, Beyer P, Taylor MA (2008) Metabolic engineering of carotenoid biosynthesis in plants. Trends Biotechnol 26: 139–145 [DOI] [PubMed] [Google Scholar]
- Harjes CE, Rocheford TR, Bai L, Brutnell TP, Kandianis CB, Sowinski SG, Stapleton AE, Vallabhaneni R, Williams M, Wurtzel ET. , et al. (2008) Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319: 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg J. (2001) Carotenoid biosynthesis in flowering plants. Curr Opin Plant Biol 4: 210–218 [DOI] [PubMed] [Google Scholar]
- Hugueney P, Bouvier F, Badillo A, d’Harlingue A, Kuntz M, Camara B (1995) Identification of a plastid protein involved in vesicle fusion and/or membrane protein translocation. Proc Natl Acad Sci USA 92: 5630–5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilambi HV, Kumar R, Sharma R, Sreelakshmi Y (2013) Chromoplast-specific carotenoid-associated protein appears to be important for enhanced accumulation of carotenoids in hp1 tomato fruits. Plant Physiol 161: 2085–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Ahn YO, Ahn MJ, Jeong JC, Lee HS, Kwak SS (2013) Cloning and characterization of an Orange gene that increases carotenoid accumulation and salt stress tolerance in transgenic sweetpotato cultures. Plant Physiol Biochem 70: 445–454 [DOI] [PubMed] [Google Scholar]
- Kolotilin I, Koltai H, Tadmor Y, Bar-Or C, Reuveni M, Meir A, Nahon S, Shlomo H, Chen L, Levin I (2007) Transcriptional profiling of high pigment-2dg tomato mutant links early fruit plastid biogenesis with its overproduction of phytonutrients. Plant Physiol 145: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lu S, Cosman KM, Earle ED, Garvin DF, O’Neill J (2006) β-Carotene accumulation induced by the cauliflower Or gene is not due to an increased capacity of biosynthesis. Phytochemistry 67: 1177–1184 [DOI] [PubMed] [Google Scholar]
- Li L, Paolillo DJ, Parthasarathy MV, Dimuzio EM, Garvin DF (2001) A novel gene mutation that confers abnormal patterns of β-carotene accumulation in cauliflower (Brassica oleracea var. botrytis). Plant J 26: 59–67 [DOI] [PubMed] [Google Scholar]
- Li L, Van Eck J (2007) Metabolic engineering of carotenoid accumulation by creating a metabolic sink. Transgenic Res 16: 581–585 [DOI] [PubMed] [Google Scholar]
- Li L, Yang Y, Xu Q, Owsiany K, Welsch R, Chitchumroonchokchai C, Lu S, Van Eck J, Deng XX, Failla M. , et al. (2012) The Or gene enhances carotenoid accumulation and stability during post-harvest storage of potato tubers. Mol Plant 5: 339–352 [DOI] [PubMed] [Google Scholar]
- Li L, Yuan H (2013) Chromoplast biogenesis and carotenoid accumulation. Arch Biochem Biophys 539: 102–109 [DOI] [PubMed] [Google Scholar]
- Liu Y, Roof S, Ye Z, Barry C, van Tuinen A, Vrebalov J, Bowler C, Giovannoni J (2004) Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc Natl Acad Sci USA 101: 9897–9902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez AB, Van Eck J, Conlin BJ, Paolillo DJ, O’Neill J, Li L (2008) Effect of the cauliflower Or transgene on carotenoid accumulation and chromoplast formation in transgenic potato tubers. J Exp Bot 59: 213–223 [DOI] [PubMed] [Google Scholar]
- Lu S, Li L (2008) Carotenoid metabolism: biosynthesis, regulation, and beyond. J Integr Plant Biol 50: 778–785 [DOI] [PubMed] [Google Scholar]
- Lu S, Van Eck J, Zhou X, Lopez AB, O’Halloran DM, Cosman KM, Conlin BJ, Paolillo DJ, Garvin DF, Vrebalov J, et al. (2006) The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 18: 3594–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyi SM, Zhou X, Kochian LV, Li L (2007) Biochemical and molecular characterization of the homocysteine S-methyltransferase from broccoli (Brassica oleracea var. italica). Phytochemistry 68: 1112–1119 [DOI] [PubMed] [Google Scholar]
- Maass D, Arango J, Wüst F, Beyer P, Welsch R (2009) Carotenoid crystal formation in Arabidopsis and carrot roots caused by increased phytoene synthase protein levels. PLoS One 4: e6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moise AR, Al-Babili S, Wurtzel ET (2014) Mechanistic aspects of carotenoid biosynthesis. Chem Rev 114: 164–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi S, Zhu C, Farre G, Ramessar K, Bassie L, Breitenbach J, Perez Conesa D, Ros G, Sandmann G, Capell T. , et al. (2009) Transgenic multivitamin corn through biofortification of endosperm with three vitamins representing three distinct metabolic pathways. Proc Natl Acad Sci USA 106: 7762–7767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisar N, Li L, Lu S, Khin NC, Pogson BJ (2015) Carotenoid metabolism in plants. Mol Plant 8: 68–82 [DOI] [PubMed] [Google Scholar]
- Niyogi KK. (1999) Photoprotection revisited: Genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol 50: 333–359 [DOI] [PubMed] [Google Scholar]
- Paine JA, Shipton CA, Chaggar S, Howells RM, Kennedy MJ, Vernon G, Wright SY, Hinchliffe E, Adams JL, Silverstone AL. , et al. (2005) Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat Biotechnol 23: 482–487 [DOI] [PubMed] [Google Scholar]
- Paolillo DJ Jr, Garvin DF, Parthasarathy MV (2004) The chromoplasts of Or mutants of cauliflower (Brassica oleracea L. var. botrytis). Protoplasma 224: 245–253 [DOI] [PubMed] [Google Scholar]
- Park SC, Kim SH, Park S, Lee HU, Lee JS, Park WS, Ahn MJ, Kim YH, Jeong JC, Lee HS. , et al. (2015) Enhanced accumulation of carotenoids in sweetpotato plants overexpressing IbOr-Ins gene in purple-fleshed sweetpotato cultivar. Plant Physiol Biochem 86: 82–90 [DOI] [PubMed] [Google Scholar]
- Pogliani L. (1994) Structure property relationships of amino acids and some dipeptides. Amino Acids 6: 141–153 [DOI] [PubMed] [Google Scholar]
- Rabbani S, Beyer P, Lintig J, Hugueney P, Kleinig H (1998) Induced β-carotene synthesis driven by triacylglycerol deposition in the unicellular alga Dunaliella bardawil. Plant Physiol 116: 1239–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao AV, Rao LG (2007) Carotenoids and human health. Pharmacol Res 55: 207–216 [DOI] [PubMed] [Google Scholar]
- Ruiz-Sola MÁ, Rodríguez-Concepción M (2012) Carotenoid biosynthesis in Arabidopsis: a colorful pathway. Arabidopsis Book 10: e0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK (2014) CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32: 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumskaya M, Bradbury LMT, Monaco RR, Wurtzel ET (2012) Plastid localization of the key carotenoid enzyme phytoene synthase is altered by isozyme, allelic variation, and activity. Plant Cell 24: 3725–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzuri G, Zhou X, Chayut N, Yuan H, Portnoy V, Meir A, Sa’ar U, Baumkoler F, Mazourek M, Lewinsohn E, et al. (2015) A ‘golden’ SNP in CmOr governs the fruit flesh color of melon (Cucumis melo). Plant J 82: 267–279 [DOI] [PubMed] [Google Scholar]
- Walter MH, Strack D (2011) Carotenoids and their cleavage products: biosynthesis and functions. Nat Prod Rep 28: 663–692 [DOI] [PubMed] [Google Scholar]
- Welsch R, Arango J, Bär C, Salazar B, Al-Babili S, Beltrán J, Chavarriaga P, Ceballos H, Tohme J, Beyer P (2010) Provitamin A accumulation in cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. Plant Cell 22: 3348–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Kandianis CB, Harjes CE, Bai L, Kim EH, Yang X, Skinner DJ, Fu Z, Mitchell S, Li Q, et al. (2010) Rare genetic variation at Zea mays crtRB1 increases beta-carotene in maize grain. Nat Genet 42: 322–327 [DOI] [PubMed] [Google Scholar]
- Yang Y, Thannhauser TW, Li L, Zhang S (2007) Development of an integrated approach for evaluation of 2-D gel image analysis: impact of multiple proteins in single spots on comparative proteomics in conventional 2-D gel/MALDI workflow. Electrophoresis 28: 2080–2094 [DOI] [PubMed] [Google Scholar]
- Yuan H, Zhang J, Nageswaran D, Li L (2015) Carotenoid metabolism and regulation in horticultural crops. Hortic Res (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, McQuinn R, Fei Z, Wolters AM, Van Eck J, Brown C, Giovannoni JJ, Li L (2011) Regulatory control of high levels of carotenoid accumulation in potato tubers. Plant Cell Environ 34: 1020–1030 [DOI] [PubMed] [Google Scholar]
- Zhou X, Welsch R, Yang Y, Álvarez D, Riediger M, Yuan H, Fish T, Liu J, Thannhauser TW, Li L (2015) Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc Natl Acad Sci USA 112: 3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Bai C, Sanahuja G, Yuan D, Farré G, Naqvi S, Shi L, Capell T, Christou P (2010) The regulation of carotenoid pigmentation in flowers. Arch Biochem Biophys 504: 132–141 [DOI] [PubMed] [Google Scholar]