Abstract
Objective
The purpose of this study is to evaluate the ability of machine learning to discriminate between magnetic resonance images (MRI) of normal and pathological human articular cartilage obtained under standard clinical conditions.
Method
An approach to MRI classification of cartilage degradation is proposed using pattern recognition and multivariable regression in which image features from MRIs of histologically scored human articular cartilage plugs were computed using weighted neighbor distance using compound hierarchy of algorithms representing morphology (WND-CHRM). The WND-CHRM method was first applied to several clinically available MRI scan types to perform binary classification of normal and osteoarthritic osteochondral plugs based on the OARSI histological system. In addition, the image features computed from WND-CHRM were used to develop a multiple linear least-squares regression model for classification and prediction of an OARSI score for each cartilage plug.
Results
The binary classification of normal and osteoarthritic plugs yielded results of limited quality with accuracies between 36% and 70%. However, multiple linear least-squares regression successfully predicted OARSI scores and classified plugs with accuracies as high as 86%. The present results improve upon the previously-reported accuracy of classification using average MRI signal intensities and parameter values.
Conclusion
MRI features detected by WND-CHRM reflect cartilage degradation status as assessed by OARSI histologic grading. WND-CHRM is therefore of potential use in the clinical detection and grading of osteoarthritis.
Keywords: Osteoarthritis, Human articular cartilage, MRI, Pattern recognition, Classification
Introduction
Osteoarthritis (OA) is a highly prevalent and potentially debilitating joint disease characterized at least in part by the degradation of articular cartilage1–3. One impediment to the development of effective therapeutics for OA is the difficulty of early diagnosis. Therefore, there is great interest attached to the development of noninvasive MRI techniques that will permit early detection of OA4. This can be regarded as a classification problem, with perhaps the most natural approach being detection and grading based on physically-meaningful individual MRI parameter measurements such as relaxation times.
However, MR parameters overlap substantially between different degrees of cartilage degradation5–7. Therefore, a difference in group mean values of a given parameter between individuals with differing OA status does not necessarily indicate clinical utility of this parameter; classification according to even a statistically significantly varying parameter may result in a clinical test of limited sensitivity (SE) and specificity (SP)8. Examples of this were seen in the work of Lukas et al., who performed univariate classification of Osteoarthritis Research Society International (OARSI) graded human osteochondral plugs, using the Mahalanobis distance metric9. While statistically significant differences in the T1-weighted (T1W) images and magnetization transfer ratios were seen with respect to degradation status, the corresponding classification accuracies were only 0.60 and 0.55.
As an alternative to univariate classification, analysis of content descriptors, or features, extracted from images, can be used for classification. These methods discriminate between groups based on inherent patterns and textures within images; the application of such techniques to identifying early OA in human subjects has recently been explored10–12. Urish et al. calculated four categories of MRI features, including histogram, gray level co-occurrence matrix, gray level run length matrix, and z-score from T2 parameter maps obtained as part of the Osteoarthritis Initiative (OAI)12, 13. Using the support-vector machine approach, they achieved 71% accuracy in predicting the symptomatic progression of OA as defined by the Western Ontario and McMaster Universities Arthritis (WOMAC) questionnaire14. Shamir et al. 2009 applied a more extensive pattern recognition algorithm, using over 2,900 features, weighted neighbor distance using compound hierarchy of algorithms representing morphology (WND-CHRM), to X-ray images classified by Kellgren and Lawrence (KL) grades10, 15. WND-CHRM was able to classify subjects with early OA (KL = 2) as compared to controls (KL = 0) with 80% accuracy. However, plain radiography is limited in its potential to detect early disease and to monitor subtle changes over time16. In contrast, MRI provides excellent delineation of musculoskeletal tissues and sensitivity to both morphologic features and matrix characteristics of joint cartilage17–19, making it a much more appealing imaging modality for early detection and monitoring of OA.
Here, we extend the previous work of Lukas et al. on univariate classification using individual MR parameters9 by examining the ability of WND-CHRM to assign osteochondral plugs to either a normal or OA group based on analysis of MR images. Each plug was separately classified based on OARSI histological score20 to provide the comparison gold standard with WND-CHRM. We applied WND-CHRM to widely available clinical MRI scan types, which are sensitive to cartilage matrix changes associated with OA, including T1, T2, T2*, ADC and MT weighted images. We also explored several weightings corresponding to those acquired as part of the OAI13, and to a T2 parameter map.
In addition to binary classification, the image features detected by WND-CHRM were used to develop a multiple linear least-squares regression model to predict the OARSI score for each plug. This provides an additional approach to classification that, in effect, makes use of all OARSI score information rather than discretizing the plugs into two groups and may accordingly provide more robust results.
Materials and Methods
Sample Preparation and Histology
Sample preparation for the present study has been previously described9. Briefly, human tissue was obtained from knee joints in an Institutional Review Board approved protocol from subjects undergoing elective arthroplasty. Two adjacent osteochondral plugs (5 mm dia, n = 36 pairs) were harvested from standardized sites within the femur (lateral anterior, LA, lateral posterior, LP, medial anterior, MA, and medial posterior, MP), as depicted by Figure 1. The samples allotted for histology were immediately fixed in formalin after harvest, while the samples allotted for imaging were flash frozen and stored at −80°C until analyzed.
Figure 1.
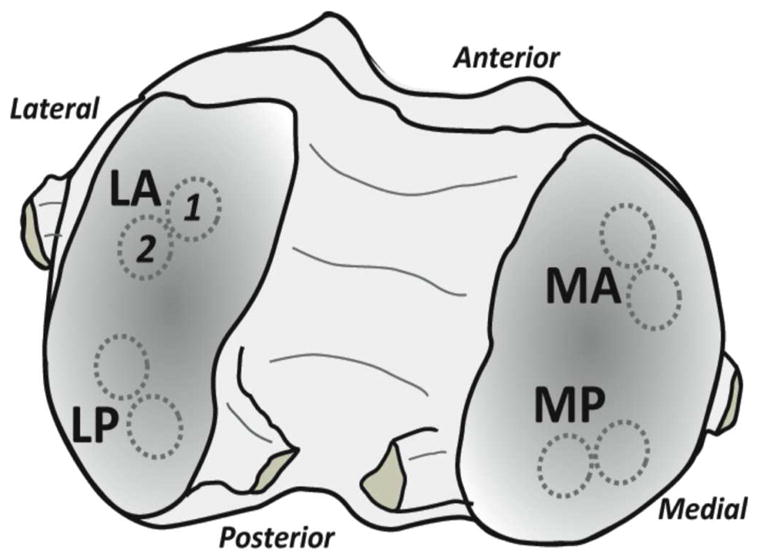
Paired osteochondral samples were harvested from defined locations on the femoral condyles and used for histology and MRI measurements. Samples were obtained from medial anterior (MA), medial posterior (MP), lateral anterior (LA) and lateral posterior (LP) locations. Illustration based on Figure 1 of Reference 9.
Histology was performed by two independent observers for OA severity21 using the OARSI histopathology assessment system20. OARSI histological scores provide an overall assessment of cartilage OA status, including the depth of progression, or grade, and the extent of surface area affected, or stage. OARSI scores range from 0–24 and are calculated as the product of the grade (0–6) and the stage (0–4). The intraclass correlation coefficient (ICC) was calculated to assess the level of agreement between the two observers for the grade, stage and score, for each plug.
MRI measurements
The MRI protocol for the present study has been previously described9. Briefly, samples were inserted into a susceptibility-matched polyetherimide (ULTEM) sample holder containing Fluorinert® FC-77 (Sigma-Aldrich, St. Louis, MO) and heated to 37.0 ± 0.1 °C prior to data acquisition. The ULTEM holder also contained a Dulbecco’s phosphate-buffered saline (DPBS; Invitrogen) standard for normalizing weighted images. Imaging was performed using a 3T Philips Achieva system equipped with an 8-channel SENSE knee coil with sample temperature maintained at 37.0 ± 0.1 °C for the duration of the acquisition.
T1W measurements
A 2D Look-Locker sequence with echo planar imaging (EPI) readout (echo time, TE = 5 ms, repetition time, TR = 6 s, 55 inversion times (TI) ranging from 18 ms to 2757 ms, flip angle, FA, = 14°, EPI factor = 3) was used to acquire two 4 mm thick slices with band width (BW) = 17.5 kHz, field of view, (FOV) = 75 × 44.5 mm2 (vertical x horizontal), matrix size, (MTX) = 120 × 66 pixels, and number of signal averages, (NSA) = 2.
T2-weighted measurements (T2W)
A 3D multi-echo spin echo sequence (TE = 12 ms, TR = 767 ms, echo train length, ETL, = 30) was used with BW = 28.2 kHz, FOV = 75 × 45 × 23 mm, MTX = 188 × 78 × 7 pixels, and NSA = 1.
T2 map
A T2 parameter map was generated voxel-by-voxel using a two-parameter monoexponential function from all 30 magnitude MR images from the 3D multi-echo spin echo sequence at TE ranging from 12 ms to 360 ms.
T2*-weighted measurements (T2*W)
A 2D gradient echo sequence (TE = 1.5 ms, ΔTE = 4.2 ms, TR = 2 s, FA = 25°, ETL = 30) was used to acquire two 3.5 mm thick slices with BW = 98.9 kHz, FOV =75 x 45 mm, MTX = 152 × 73 pixels, and NSA = 2.
Diffusion weighted imaging (DWI) measurements
A 2D spin echo sequence with EPI readout (TE = 62 ms, TR = 2 s, EPI factor = 3) was used to acquire two 4 mm thick slices at b-values of 0, 333, 666, 1000, 1333, 1666, and 2000 s/mm2, applied in three orthogonal gradient directions, Δ = 25.3 ms, δ = 12.4 ms, BW = 12 kHz, FOV = 75 × 43.75 mm, MTX = 96 × 43 pixels, and NSA = 1.
MTW measurements
A 2D FFE (fast field echo) sequence (TE = 2.4 ms, TR = 517 ms, FA = 25°) preceded by 50 ms sinc-shaped presaturation pulses with 1 kHz offset and B1 = 2.15 μT was used to acquire two 5 mm slices, BW = 98.9 kHz, FOV = 75 × 45 mm, MTX = 152 × 73 pixels, and NSA = 2.
T1W measurements as per the OAI
A 3D FFE sequence (TE = 7.57 ms, TR = 20 ms, FA = 13°, partial Fourier factor = 0.75) was used with BW = 31.1 kHz, FOV = 75 × 45 × 24 mm, MTX = 240 × 144 × 16 pixels, and NSA = 1.
T2W measurements as per the OAI
A 2D multi-slice multi-echo turbo spin echo sequence (TE = 10 ms, TR = 2700 ms, ETL = 7) was used to acquire two 3 mm thick slices with BW = 59.8 kHz, FOV = 75 × 45 mm (vertical × horizontal), MTX = 240 × 101 pixels, and NSA = 1.
ROI selection
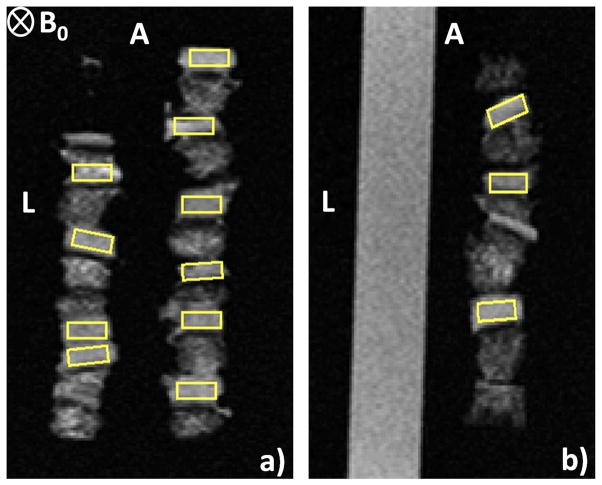
All weighted images were normalized to the average signal intensity of the Dulbecco’s phosphate buffered saline (DPBS) contained within the sample holder. ROIs centered on the full thickness cartilage cross-sectional area, excluding subchondral bone, were drawn using ImageJ software (National Institutes of Health, Bethesda, MD). ROIs of uniform size (15 x 7 pixels) were identified in all images (Fig. 2). Images with insufficient cartilage for analysis were discarded and 36 ROIs for each MR contrast modality remained for classification. Figure 2 depicts 11 ROIs within a T2W (2D spin-echo) image.
Figure 2.
T2 W (2D spin-echo) of TE = 20 ms images of cartilage samples with ROIs (yellow color). The 4-well ULTEM sample holder contained six plugs per well in three wells, and DPBS in the fourth well. The ‘anterior’ and ‘left’ sample positions are indicated by the letters ‘A’ and ‘L’, respectively, while the static magnetic field, B0, is oriented into the image plane. (a) Slice 1, intersecting two of the wells containing cartilage. (b) slice 2, intersecting the third cartilage-containing well and the DPBS-containing well.
Feature extraction
We elected to use overall OARSI score rather than OARSI grade for group assignment because 1) we analyzed ROIs of full-thickness cartilage and 2) score provides a larger range of values for regression. Plugs with sufficient pixels were assigned to one of two groups: normal or early OA changes, defined as OARSI score ≤ 6.0 (n = 18), and henceforth referred to as “normal”, and more established OA, with OARSI score > 6.0 (n = 18), henceforth referred to as “OA”.
WND-CHRM image features were computed for each weighting of each scan type. The WND-CHRM algorithm extracts a generic set of numerical image content descriptors, including textures, statistical distribution of pixel values, and factors from polynomial decomposition of the raw image, as well as several image transforms of the raw image as previously described22, 23. Image transforms used for feature extraction include wavelet (Symlet 5, level 1), Fourier, Chebyshev and an edge transform, as well as several combinations of transforms performed in tandem (Chebyshev-Fourier, wavelet-Fourier, Fourier-wavelet, Fourier-Chebyshev, Chebyshev-wavelet, Fourier-edge, and wavelet-edge)23. This results in a final set of 2,919 features (WND-CHRM version 1.50). The features are then ranked by their Fisher discriminant, defined here as the ratio of variance of class means from the pooled mean to the mean of within-class variances11. Because not all features are equally informative and those that are less informative may primarily represent noise, only the top 15% of the features are used for binary univariate classification as below11, 23.
Binary univariate classification analysis
Two methods were used to classify this dataset: an analysis to measure separability of the two classes within the dataset itself and a standard leave-one-out cross validation to estimate predictive capability. Separability in this sense provides an estimate of maximum predictive capability.
In the separability analysis, all images were used to train the model. Then, each image in the set was selected as a test image in turn to compute class probabilities using the weighted neighbor distance classifier (WND5)22. In the WND5 classifier, the feature vectors from the training images are arranged in a (weighted) feature space, where each feature value is multiplied by its corresponding Fisher discriminant. The relative probability of a test image belonging to a certain class is calculated as the mean of the inverse fifth power of the Fisher-score weighted distances, r, between the test image and all training images of that class. The test image is then assigned to the class with the highest probability. However, since the distance from a test image to itself is r = 0, calculation of 1/r 5 would result in division by zero. Thus in this separability analysis, the test image contributes to the weights used to construct the feature space but is left out of the class probability calculation.
In the standard leave-one-out analysis, the accuracy of the model was obtained by selecting a test image at random 100 times with replacement, and classifying test images as normal or OA using the WND5 classifier trained on the remaining 35 images in the dataset.
Multivariable prediction and classification by regression
A multiple linear least-squares regression model for predicting OARSI scores was constructed using the WND-CHRM feature set and SciPy24, a mathematical toolkit written in the Python programming language. The independent variables, or predictors, were the descriptive values assigned to particular features, while the dependent variable, or outcome, was the histologic OARSI score for each sample.
The number of features used in the regression model was taken as a parameter to be optimized by minimizing the root-mean square (RMS) difference between the model-predicted OARSI score and each sample’s actual OARSI score. In contrast to the Fisher discriminants used for feature ranking and weighting in the binary univariate classification analysis, Pearson coefficients for correlation between each feature and the images’ corresponding OARSI score were used for ranking and weighting in linear regression. Features were added to form successive regression models in rank order until the RMS no longer decreased.
The predicted OARSI score for each of the 36 samples were calculated using standard linear regression provided by the scipy.linalg.lstsq function, where the coefficients for the regression were the Pearson-weighted feature values from the training images and the corresponding ordinates were the OARSI scores corresponding to the training images. The Shapiro-Wilk test for normality was performed, which showed that the residuals followed a normal distribution. The predicted score for the test image was determined from the inner product of the test image’s features and the least squares solution returned by lstsq. In other words, predicted OARSI score was calculated as the sum of feature values, weighted by least squares coefficients. Each of the 36 images was tested in this way, with the other 35 acting as the training set.
After the optimal number of features for regression was determined by minimizing the RMS, the predicted OARSI scores were plotted against the actual OARSI scores. The coefficient of determination (R2) between these two scores was determined and used to evaluate each MR scan type. The SE was calculated as the proportion of correctly predicted OA samples (OARSI > 6), the SP was calculated as the proportion of correctly predicted normal samples (OARSI ≤ 6.0), and the accuracy was determined as the average of the SE and SP.
Results
The normal and OA groups had average scores of 3.66 ± 2.07 and 9.60 ± 3.75, respectively, with 18 being the maximum OARSI score in the latter group, out of a possible maximum score of 24. The normal and OA groups had average grades of 1.60 ± 0.37 and 2.9 ± 0.79, respectively, with 4.5 being the maximum OARSI grade in the latter group, indicating mild degradation. Thus, all samples evaluated displayed considerable structural integrity. The inter-observer agreement was moderate; the ICCs of the grade, stage and score were 0.45, 0.63 and 0.55 respectively, with a mean difference of only 1.46 between the overall OARSI scores.
Table 1 shows the results of the separability analysis for the best performing weighted images out of each series of image weightings, as well as for the T2 map, and for all of the T1W and T2W images corresponding to those used in the OAI. The accuracy among these was at least 0.78, but was generally considerably greater, indicating that the classifiers are able to detect subtle differences between the normal and OA images. However, despite the separability of the two groups under WND-CHRM analysis, the algorithm was largely unsuccessful at classifying the images using a standard leave-one-out analysis, with accuracies generally in the range of 50% (Table 2).
Table 1.
WND-CHRM-derived mean and error of SE, SP and accuracy of the separability test. The measurements listed represent the weighting at which the highest classification accuracy was achieved, representing the highest degree of separation between the classes.
| MR Measurement | SE | SP | Accuracy |
|---|---|---|---|
| T1W (TI = 982 ms) | 0.94 ± 0.01 | 0.94 ± 0.01 | 0.94 ± 0.00 |
| T2W (TE = 72 ms) | 0.85 ± 0.02 | 0.85 ± 0.02 | 0.85 ± 0.01 |
| T2 (ms) | 0.78 ± 0.02 | 0.78 ± 0.02 | 0.78 ± 0.01 |
| T2*W (TE = 1.44 ms) | 0.94 ± 0.02 | 1.0 ± 0.01 | 0.97 ± 0.01 |
| DWI (b = 999 s/mm2) | 0.78 ± 0.02 | 0.83 ± 0.02 | 0.81 ± 0.01 |
| MTW (Tsat = 200 ms) | 0.94 ± 0.01 | 0.89 ± 0.02 | 0.92 ± 0.01 |
| OAI weightings | |||
|---|---|---|---|
| T1W (TR = 7.57 ms) | 0.83 ± 0.02 | 0.94 ± 0.02 | 0.89 ± 0.01 |
| T2W (TE = 10 ms) | 0.63 ± 0.02 | 1.0 ± 0.00 | 0.82 ± 0.01 |
| T2W (TE = 20 ms) | 0.78 ± 0.02 | 1.0 ± 0.00 | 0.89 ± 0.01 |
| T2W (TE = 30 ms) | 0.83 ± 0.02 | 0.89 ± 0.01 | 0.86 ± 0.01 |
| T2W (TE = 40 ms) | 0.78 ± 0.02 | 0.94 ± 0.02 | 0.86 ± 0.01 |
| T2W (TE = 50 ms) | 0.78 ± 0.02 | 0.94 ± 0.01 | 0.86 ± 0.01 |
| T2W (TE = 60 ms) | 0.89 ± 0.02 | 0.78 ± 0.02 | 0.83 ± 0.01 |
| T2W (TE = 70 ms) | 0.83 ± 0.02 | 0.83 ± 0.02 | 0.83 ± 0.01 |
Table 2.
WND-CHRM-derived mean and error of SE, SP and accuracy reported from the standard leave-one-out cross validation as a conventional test for assessing the quality of the classifier. The MR measurements listed correspond to those in Table 1. Note that the separability of the dataset as shown in Table 1 did not translate into predictive accuracy as shown here.
| MR Measurement | SE | SP | Accuracy |
|---|---|---|---|
| T1W (TI = 982 ms) | 0.63 ± 0.1 | 0.61 ± 0.1 | 0.62 ± 0.07 |
| T2W (TE = 72 ms) | 0.53 ± 0.1 | 0.58 ± 0.09 | 0.56 ± 0.07 |
| T2 (ms) | 0.47 ± 0.1 | 0.74 ± 0.09 | 0.61 ± 0.07 |
| T2*W (TE = 1.44 ms) | 0.52 ± 0.1 | 0.56 ± 0.1 | 0.54 ± 0.07 |
| DWI (b = 999 s/mm2) | 0.68 ± 0.09 | 0.72 ± 0.09 | 0.70 ± 0.06 |
| MTW (Tsat = 200 ms) | 0.38 ± 0.1 | 0.68 ± 0.09 | 0.53 ± 0.07 |
| OAI weightings | |||
|---|---|---|---|
| T1W (TR = 7.57 ms) | 0.42 ± 0.1 | 0.40 ± 0.1 | 0.41 ± 0.07 |
| T2W (TE = 10 ms) | 0.35 ± 0.09 | 0.69 ± 0.09 | 0.52 ± 0.07 |
| T2W (TE = 20 ms) | 0.31 ± 0.09 | 0.67 ± 0.09 | 0.49 ± 0.07 |
| T2W (TE = 30 ms) | 0.70 ± 0.09 | 0.72 ± 0.09 | 0.71 ± 0.06 |
| T2W (TE = 40 ms) | 0.64 ± 0.09 | 0.61 ± 0.1 | 0.63 ± 0.07 |
| T2W (TE = 50 ms) | 0.41 ± 0.09 | 0.31 ± 0.09 | 0.36 ± 0.07 |
| T2W (TE = 60 ms) | 0.52 ± 0.1 | 0.57 ± 0.1 | 0.54 ± 0.07 |
| T2W (TE = 70 ms) | 0.56 ± 0.09 | 0.42 ± 0.1 | 0.49 ± 0.07 |
The MR imaging contrast modalities of each scan type that demonstrated the greatest degree of separability between classes (Table 1) were used to generate a multiple linear regression model using image features and gold standard OARSI scores. The most informative features for all scan types were the ones computed using the polynomial decomposition of the raw image, including the Zernike and Chebyshev statistics. Additional informative features were those computed from the image transforms, such as the Haralick textures computed from the wavelet transform and the Tamura textures computed from the Fourier transform of the image. Overall, there were varying numbers of informative features for each scan type. The decrease in the Pearson correlation coefficients in the set of rank ordered features typically followed a power law, rather than exponential, distribution; a small number of features had high coefficients, followed by a large number of features with relatively low coefficients. The optimal number of informative features was defined as the value for which the prediction of OARSI score no longer improves upon the inclusion of additional features, as determined by the RMS error.
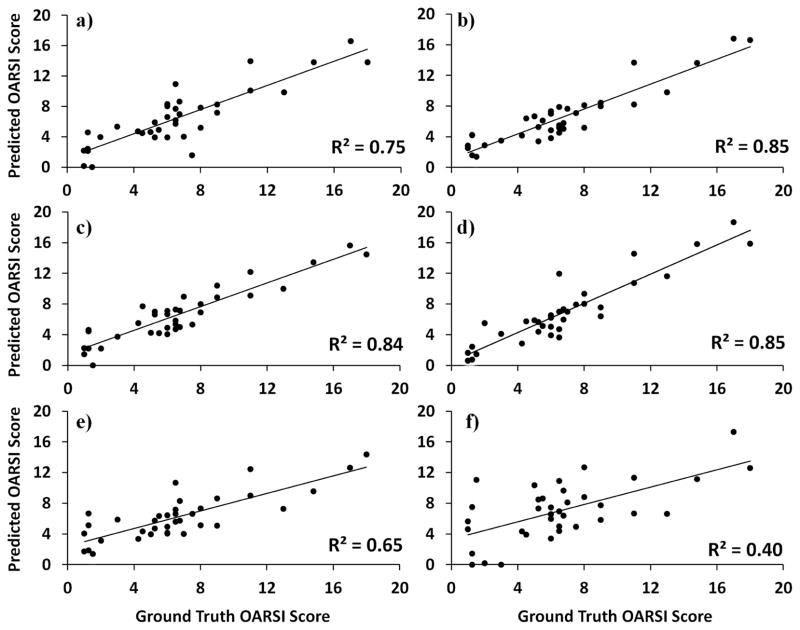
The optimal RMS values and corresponding percentage of informative features for each scan type are listed in Table 3. None of the scan types provided optimal OARSI score prediction using less than 1.9% or more than 8.4% of the features. Of the conventional MR modalities, T2W with TE = 72 ms resulted in the lowest RMS error value (1.62) using 6.1% of the features (Table 3). A plot of predicted versus actual OARSI scores for each MR scan type is shown in Figures 3 and 4 using the feature set defined by the optimal percentage of features indicated in Table 3. The prediction with T2W of TE = 72 ms exhibited one of the highest R2 values (0.85), yet the lowest binary classification accuracy (0.69) (Table 3).
Table 3.
Results from the multiple linear regression. The lowest RMS value for each contrast is listed, as well as the corresponding percentage of features used to optimize the model. Classification by regression results are reported as R2, indicating the coefficient of determination between the actual OARSI score and the model-predicted score. Calculated SE, SP and accuracy of the model are also reported.
| MR Measurement | RMS | Percentage of features | R2 | SE | SP | Accuracy |
|---|---|---|---|---|---|---|
| T1W (TI = 982 ms) | 2.13 | 3.2 % | 0.75 | 0.78 | 0.83 | 0.81 |
| T2W (TE = 72 ms) | 1.62 | 6.1 % | 0.85 | 0.67 | 0.72 | 0.69 |
| T2 (ms) | 3.57 | 5.7 % | 0.40 | 0.83 | 0.56 | 0.70 |
| T2*W (TE = 1.44 ms) | 1.74 | 8.4 % | 0.83 | 0.72 | 0.72 | 0.72 |
| DWI (b = 999 s/mm2) | 1.70 | 1.9 % | 0.85 | 0.83 | 0.89 | 0.86 |
| MTW (Tsat = 200 ms) | 2.53 | 6.6 % | 0.65 | 0.72 | 0.83 | 0.78 |
| OAI weightings | ||||||
|---|---|---|---|---|---|---|
| T1W (TR = 7.57 ms) | 2.05 | 4.4 % | 0.76 | 0.72 | 0.67 | 0.69 |
| T2W (TE = 10 ms) | 2.10 | 7.3 % | 0.76 | 0.78 | 0.72 | 0.75 |
| T2W (TE = 20 ms) | 2.40 | 5.6 % | 0.67 | 0.89 | 0.78 | 0.83 |
| T2W (TE = 30 ms) | 2.59 | 7.3 % | 0.63 | 0.89 | 0.72 | 0.81 |
| T2W (TE = 40 ms) | 2.91 | 7.9 % | 0.57 | 0.83 | 0.61 | 0.72 |
| T2W (TE = 50 ms) | 2.19 | 3.6 % | 0.73 | 1.0 | 0.72 | 0.86 |
| T2W (TE = 60 ms) | 2.38 | 6.4 % | 0.68 | 0.83 | 0.67 | 0.75 |
| T2W (TE = 70 ms) | 2.76 | 3.0 % | 0.60 | 0.83 | 0.83 | 0.83 |
Figure 3.
Predicted OARSI score versus actual OARSI score for each sample as determined by the multiple regression for a) T1W (TI = 982 ms), b) T2W (TE = 72 ms), c) T2*W (TE = 1.44 ms), d) DWI (b = 999 s/mm2), e) MTW (Tsat = 200 ms), f) T2 (ms). R2 values for each scan type are listed on the respective graphs.
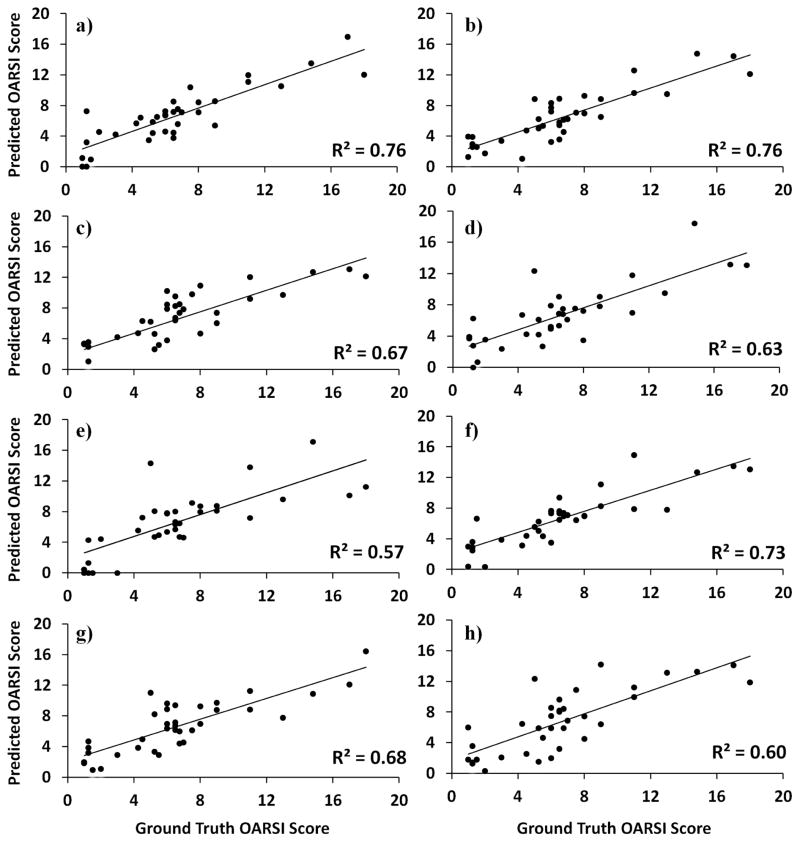
Figure 4.
Predicted OARSI score versus actual OARSI score for each sample as determined by the multiple regression for a) T1W (TR = 7.57 ms), b) T2W (TE = 10 ms), c) T2W (TE = 20 ms), d) T2W (TE = 30 ms), e) T2W (TE = 40 ms), f) T2W (TE = 50 ms), g) T2W (TE = 60 ms), h) T2W (TE = 70 ms). R2 values for each scan type are listed on the respective graphs.
Using standard leave-one-out analysis, DWI of b = 999 s/mm2 achieved a classification accuracy of 0.70, with a SE and SP of 0.68 and 0.72, respectively (Table 2). In addition, the regression based on this weighted image was able to predict OARSI scores with an RMS of 1.70 and an R2 of 0.85, and to classify the samples with an accuracy of 0.86 (Table 3, Fig. 3).
As indicated by the best-fit line graphs (Figs. 3 and 4), the actual and predicted OARSI scores for all of the weighted images exhibited correlations with R2 between 0.65 and 0.85, while the regression based on the T2 map was much less satisfactory, with R2 = 0.40, consistent with the fact that this regression exhibited the largest RMS value of any scan type (3.57), and likewise exhibited one of the lowest regression-based binary classification accuracies (0.70).
Discussion
Arthroscopic and radiographic evaluations provide the current diagnostic standards for characterizing cartilage matrix loss associated with OA15, 25, 26. Arthroscopy is an invasive procedure and provides only qualitative or semi-quantitative evaluation of cartilage. Radiographic evaluation based on KL or OARSI-OMERACT27 scoring focuses on later stages of disease and is non-specific and insensitive to detect change. Whole organ MR evaluation based on semi-quantitative expert assessment28, 29 and 3D MRI quantitative evaluation30 focus on apparent morphological changes in early or later disease stages. There is a need for more sensitive MRI methods that can detect subtle changes associated with pre-radiographic OA and that may potentially serve as biomarkers for disease onset and progression.
We found that application of an emerging machine learning algorithm, WND-CHRM, provided accuracy of up to 86% in classifying MR images of normal and early OA cartilage. This compares favorably with other analyses9, which also specifically addressed discrimination between these histologically-graded ostechondral plugs from human subjects with idiopathic non-advanced OA. The present approach relies upon the analysis of a large set of image features and provides compelling evidence that feature analysis yields information that may serve as a surrogate for histology.
Histological scoring systems that assess the structural composition of cartilage matrix and determine the severity of degradation31 can be used as a reference standard to establish a classification scheme using image features32–34. The OARSI histopathology assessment has been shown to be reliable and reproducible35 and is an informative gold standard for cartilage classification using quantitative MR measurements9. Feature analysis of cartilage matrix-sensitive MR images can provide information about underlying patterns and textures. We found that WND-CHRM may be an efficient method for the classification of cartilage MRIs based on OARSI scores. WND-CHRM differs from traditional pattern recognition classification techniques in that it computes over 2,900 features based on both the raw image and on multiple image transforms. WND-CHRM showed very limited ability to provide an accurate binary classification of these images using a standard leave-one-out cross-validation analysis. However, the separability results (Table 1) indicate that such classification may be successful with a larger number of training images; a larger database would result in more stable feature weights with less variation from training set to training set.
In contrast to discrete classification, multiple least-squares regression based on image features was able to predict an OARSI score for each plug and successfully discriminate between MRIs of normal and OA cartilage. We found that image features, such as the Zernicke polynomials and Chebyshev statistics, computed on the wavelet transforms of the image were the most informative. In contrast to bulk pixel statistics, these types of features are sensitive to various types of textures in the image, such as spatial relationships between pixel intensities. This indicates that the classifier exhibits sensitivity to structural changes in cartilage at the spatial resolution afforded by MR imaging.
The T2W image with TE = 50 ms was the least successful in discrete classification according to the standard leave-one-out analysis (Table 2), but through use of regression was able to predict OARSI scores with one of the highest classification accuracies of the scan types evaluated (Table 3). This indicates the power of relationships established through use of the entire range of OARSI scores to build a classification model. In terms of the actual measurement, transverse relaxation is influenced by several factors, all of which are affected by the process of degradation, including matrix macromolecular composition, collagen fiber orientation and cartilage water content36. In addition, DWI also exhibited highly favorable classification accuracy through regression, which is consistent with the classification results reported by Lukas et al.9. The ability for features to detect subtle pattern differences associated with OA in DWI may imply that DWI is particularly sensitive to early degenerative changes, which can affect both magnitude and anisotropy of diffusion37.
In previous work using the same dataset as explored in the present analysis, cartilage samples were classified using individual MR parameter values and signal intensities using the univariate Mahalanobis distance; this accounts for the possibility of different variances within the normal and OA groups. In that study, the T2W image of TE = 60 ms from the OAI protocol demonstrated the best performance with an accuracy of 0.759. The WND-CHRM analysis performed in the present study improved upon this (Table 3) and may further indicate the utility of T2 weighting in characterizing OA. However, classifiers derived from the 3D T2W images and the T2 parameter map did not perform as well as the T2W measurements from the OAI protocol. This is consistent with the low accuracy in the separability analysis (Table 1). Further, the RMS error of the T2 regression model was the highest of the scan types tested, indicating poor prediction of OARSI scores; this led to poor SP when the predicted scores were used for classification. The poor performance of T2 in this this study is in contrast to its relatively good performance in classifying cartilage in other studies5, 9. Of the 3D imaging protocols, the T2W image with TE = 72 ms provided the best separability and was found also to exhibit the maximum classification accuracy of the 3D weightings (Tables 1 and 3). The T2 parameter map, derived from all 30 T2 weightings, demonstrated substantially worse accuracy, presumably because the data incorporated into the monoexpontial fit for T2 included weightings with poor diagnostic performance.
There are certain limitations to our study. Given the success of multiparametric analysis of cartilage matrix in other contexts38–41, the extension of the present approach to analysis of images obtained using two or more MR image scan types would likely be of substantial benefit. In effect, this would increase the dimensionality of parameter space and may be of particular benefit in regression analysis. The features themselves are derived from independent image transforms and in many cases represent orthogonal components of these transforms. While they are therefore non-redundant, given the number of features, a certain degree of near-multi-collinearity is possible. In addition, the small physical size of the samples in this work resulted in a relatively limited number of pixels available for analysis. This number was further reduced by our selection of ROI’s consisting of only 15 x 7 pixels in order to avoid edge and partial volume effects. It is expected that this would reduce the overall reliability of the method due to statistical variation, as well as systematically limiting the accuracy of larger-scale, lower spatial frequency image and transform features. While larger physical samples would therefore be desirable, we note that in potential medical applications, the physical size of the region available for image analysis will be intrinsically limited and defined by the size of the structures under investigation. Further, we note that the MR measurements and histologic scores were obtained on different, though adjacent, plugs. This allowed us to avoid the potential effects on histology of sample degradation during the lengthy imaging sessions as well as potential tissue swelling, although it does not account for the small potential variability in tissue characteristics, or even in OARSI score, between adjacent sites. We also note that this study was restricted to analysis of imaging modalities that are relatively routinely available in the clinical setting. Other contrast modalities, including rotating frame techniques, such as T1ρ42, would also be of great value to explore.
Based on the success of the WND-CHRM algorithm in analysis of articular cartilage matrix as described here, its further application to more heterogeneous images consisting of a greater range of joint structures affected by OA may be productive, as previously demonstrated in X-ray images10, 11. Thus, performance of WND-CHRM in the classification of MR images may be substantially improved through the inclusion of additional joint features, such as subchondral bone and meniscus. To be fully effective, this may require use of specialized imaging techniques such as ultrashort TE, sweep imaging with Fourier Transform or near-zero TE43–45 so that these more rigid structures are more readily visualized. Nevertheless, we achieved substantial success through analysis of cartilage matrix using MR measures that are routinely available in clinical settings. The analysis was performed on samples from individuals across a range of early pathologic changes. Thus, our results indicate the ability to non-invasively assign individual subjects to a degree of OA pathology. One essential difference between our in vitro study of explants and analyses performed in the in vivo setting is the requirement for reproducible tissue segmentation in the latter. While challenging, great progress has been made in the registration and segmentation of joint cartilage in clinical images, with several automated methods having been developed46–48. These advances would permit WND-CHRM or related analyses to be performed on joint structures in addition to cartilage.
In conclusion, we have demonstrated that image and image transform features derived from MR images obtained using standard clinical sequences under clinical imaging conditions may be useful for the detection and staging of OA in human articular cartilage.
Acknowledgments
This work was supported by the Intramural Research Program of the NIH, National Institute on Aging and by NIH R01 AR063712 (C.P. Neu) and R21 AR066230 (C.P. Neu).
Role of Funding Source
This work was supported in part by the Intramural Research Program of the NIH, National Institute on Aging, by NIH R01 AR063712 and by R21 AR066230. The study sponsors had no involvement in in study design, collection and interpretation of data, writing of the manuscript, or manuscript submission.
Footnotes
Conflict of Interest
The authors have no conflicts of interest to declare.
Author Contributions
Conception and design: BGA, VAL, DAR, IGG, RGS
Analysis and interpretation of the data: BGA, CEC, MB, JMB, IGG, RGS
Drafting of the article: BGA, CEC, IGG, RGS
Critical revision of the article for important intellectual content: All authors
Final approval of the article: All authors
Statistical expertise: BGA, CEC, MB, IGG, RGS
Obtaining of funding: CPN, IGG, RGS
Administrative, technical, or logistic support: BGA, CEC, MB, VAL, JMB, IGG
Collection and assembly of data: BGA, MB, VAL, DAR, CPN
BA and RGS take full responsibility for the integrity of this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Beth G. Ashinsky, Email: beth.ashinsky@nih.gov.
Christopher E. Coletta, Email: christopher.coletta@nih.gov.
Mustapha Bouhrara, Email: mustapha.bouhrara2@nih.gov.
Vanessa A. Lukas, Email: vannylukas@gmail.com.
Julianne M. Boyle, Email: jmboyle@oakland.edu.
David A. Reiter, Email: reiterda@nia.nih.gov.
Corey P. Neu, Email: cpneu@purdue.edu.
Ilya G. Goldberg, Email: goldbergil@helix.nih.gov.
Richard G. Spencer, Email: spencer@helix.nih.gov.
References
- 1.Davis MA. Epidemiology of osteoarthritis. Clin Geriatr Med. 1988;4:241–255. [PubMed] [Google Scholar]
- 2.Shapiro LM, McWalter EJ, Son MS, Levenston M, Hargreaves BA, Gold GE. Mechanisms of osteoarthritis in the knee: MR imaging appearance. J Magn Reson Imaging. 2014;39:1346–1356. doi: 10.1002/jmri.24562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swedberg JA, Steinbauer JR. Osteoarthritis. Am Fam Physician. 1992;45:557–568. [PubMed] [Google Scholar]
- 4.Burstein D, Gray ML. Is MRI fulfilling its promise for molecular imaging of cartilage in arthritis? Osteoarthritis Cartilage. 2006;14:1087–1090. doi: 10.1016/j.joca.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Lin PC, Reiter DA, Spencer RG. Sensitivity and specificity of univariate MRI analysis of experimentally degraded cartilage. Magn Reson Med. 2009;62:1311–1318. doi: 10.1002/mrm.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen CT, Fishbein KW, Torzilli PA, Hilger A, Spencer RG, Horton WE., Jr Matrix fixed-charge density as determined by magnetic resonance microscopy of bioreactor-derived hyaline cartilage correlates with biochemical and biomechanical properties. Arthritis Rheum. 2003;48:1047–1056. doi: 10.1002/art.10991. [DOI] [PubMed] [Google Scholar]
- 7.Laurent D, Wasvary J, O’Byrne E, Rudin M. In vivo qualitative assessments of articular cartilage in the rabbit knee with high-resolution MRI at 3T. Magn Reson Med. 2003;50:541–549. doi: 10.1002/mrm.10566. [DOI] [PubMed] [Google Scholar]
- 8.Spencer RG, Pleshko N. How do statistical differences in matrix-sensitive magnetic resonance outcomes translate into clinical assignment rules? J Am Acad Orthop Surg. 2013;21:438–439. doi: 10.5435/JAAOS-21-07-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lukas VA, Fishbein KW, Lin PC, Schar M, Schneider E, Neu CP, et al. Classification of Histologically Scored Human Knee Osteochondral Plugs by Quantitative Analysis of Magnetic Resonance Images at 3T. J Orthop Res. 2015 doi: 10.1002/jor.22810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shamir L, Ling SM, Scott W, Hochberg M, Ferrucci L, Goldberg IG. Early detection of radiographic knee osteoarthritis using computer-aided analysis. Osteoarthritis Cartilage. 2009;17:1307–1312. doi: 10.1016/j.joca.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shamir L, Ling SM, Scott WW, Jr, Bos A, Orlov N, Macura TJ, et al. Knee x-ray image analysis method for automated detection of osteoarthritis. IEEE Trans Biomed Eng. 2009;56:407–415. doi: 10.1109/TBME.2008.2006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urish KL, Keffalas MG, Durkin JR, Miller DJ, Chu CR, Mosher TJ. T2 texture index of cartilage can predict early symptomatic OA progression: data from the osteoarthritis initiative. Osteoarthritis Cartilage. 2013;21:1550–1557. doi: 10.1016/j.joca.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peterfy CG, Schneider E, Nevitt M. The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthritis Cartilage. 2008;16:1433–1441. doi: 10.1016/j.joca.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellamy N, Buchanan WW, Goldsmith CH, Campbell J, Stitt LW. Validation study of WOMAC: a health status instrument for measuring clinically important patient relevant outcomes to antirheumatic drug therapy in patients with osteoarthritis of the hip or knee. J Rheumatol. 1988;15:1833–1840. [PubMed] [Google Scholar]
- 15.Kellgren JH, Lawrence JS. Radiological assessment of osteo-arthrosis. Ann Rheum Dis. 1957;16:494–502. doi: 10.1136/ard.16.4.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y, Gunther KP, Brenner H. Reliability of radiographic grading of osteoarthritis of the hip and knee. Scand J Rheumatol. 1997;26:155–165. doi: 10.3109/03009749709065675. [DOI] [PubMed] [Google Scholar]
- 17.Braun HJ, Gold GE. Advanced MRI of articular cartilage. Imaging Med. 2011;3:541–555. doi: 10.2217/iim.11.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braun HJ, Gold GE. Diagnosis of osteoarthritis: imaging. Bone. 2012;51:278–288. doi: 10.1016/j.bone.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi JA, Gold GE. MR imaging of articular cartilage physiology. Magn Reson Imaging Clin N Am. 2011;19:249–282. doi: 10.1016/j.mric.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pritzker KP, Gay S, Jimenez SA, Ostergaard K, Pelletier JP, Revell PA, et al. Osteoarthritis cartilage histopathology: grading and staging. Osteoarthritis Cartilage. 2006;14:13–29. doi: 10.1016/j.joca.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Neu CP, Reddi AH, Komvopoulos K, Schmid TM, Di Cesare PE. Increased friction coefficient and superficial zone protein expression in patients with advanced osteoarthritis. Arthritis Rheum. 2010;62:2680–2687. doi: 10.1002/art.27577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orlov N, Shamir L, Macura T, Johnston J, Eckley DM, Goldberg IG. WND-CHARM: Multi-purpose image classification using compound image transforms. Pattern Recognit Lett. 2008;29:1684–1693. doi: 10.1016/j.patrec.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shamir L, Orlov N, Eckley DM, Macura T, Johnston J, Goldberg IG. Wndchrm - an open source utility for biological image analysis. Source Code Biol Med. 2008;3:13. doi: 10.1186/1751-0473-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Walt S, Colbert SC, Varoquaux G. Computing in Science & Engineering. Vol. 13. IEEE; Mar-Apr. 2011. The NumPy Array: A Structure for Efficient Numerical Computation; pp. 22–30. [Google Scholar]
- 25.Noyes FR, Stabler CL. A system for grading articular cartilage lesions at arthroscopy. Am J Sports Med. 1989;17:505–513. doi: 10.1177/036354658901700410. [DOI] [PubMed] [Google Scholar]
- 26.Oakley SP, Portek I, Szomor Z, Appleyard RC, Ghosh P, Kirkham BW, et al. Arthroscopy -- a potential “gold standard” for the diagnosis of the chondropathy of early osteoarthritis. Osteoarthritis Cartilage. 2005;13:368–378. doi: 10.1016/j.joca.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Ornetti P, Brandt K, Hellio-Le Graverand MP, Hochberg M, Hunter DJ, Kloppenburg M, et al. OARSI-OMERACT definition of relevant radiological progression in hip/knee osteoarthritis. Osteoarthritis Cartilage. 2009;17:856–863. doi: 10.1016/j.joca.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterfy CG, Guermazi A, Zaim S, Tirman PF, Miaux Y, White D, et al. Whole-Organ Magnetic Resonance Imaging Score (WORMS) of the knee in osteoarthritis. Osteoarthritis Cartilage. 2004;12:177–190. doi: 10.1016/j.joca.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Runhaar J, Schiphof D, van Meer B, Reijman M, Bierma-Zeinstra SM, Oei EH. How to define subregional osteoarthritis progression using semi-quantitative MRI Osteoarthritis Knee Score (MOAKS) Osteoarthritis Cartilage. 2014;22:1533–1536. doi: 10.1016/j.joca.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 30.Trattnig S, Domayer S, Welsch GW, Mosher T, Eckstein F. MR imaging of cartilage and its repair in the knee--a review. Eur Radiol. 2009;19:1582–1594. doi: 10.1007/s00330-009-1352-3. [DOI] [PubMed] [Google Scholar]
- 31.Pauli C, Whiteside R, Heras FL, Nesic D, Koziol J, Grogan SP, et al. Comparison of cartilage histopathology assessment systems on human knee joints at all stages of osteoarthritis development. Osteoarthritis Cartilage. 2012;20:476–485. doi: 10.1016/j.joca.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bittersohl B, Mamisch TC, Welsch GH, Stratmann J, Forst R, Swoboda B, et al. Experimental model to evaluate in vivo and in vitro cartilage MR imaging by means of histological analyses. Eur J Radiol. 2009;70:561–569. doi: 10.1016/j.ejrad.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 33.Gahunia HK, Babyn P, Lemaire C, Kessler MJ, Pritzker KP. Osteoarthritis staging: comparison between magnetic resonance imaging, gross pathology and histopathology in the rhesus macaque. Osteoarthritis Cartilage. 1995;3:169–180. doi: 10.1016/s1063-4584(05)80051-2. [DOI] [PubMed] [Google Scholar]
- 34.Saadat E, Jobke B, Chu B, Lu Y, Cheng J, Li X, et al. Diagnostic performance of in vivo 3-T MRI for articular cartilage abnormalities in human osteoarthritic knees using histology as standard of reference. Eur Radiol. 2008;18:2292–2302. doi: 10.1007/s00330-008-0989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Custers RJ, Creemers LB, Verbout AJ, van Rijen MH, Dhert WJ, Saris DB. Reliability, reproducibility and variability of the traditional histologic/histochemical grading system vs the new OARSI osteoarthritis cartilage histopathology assessment system. Osteoarthritis Cartilage. 2007;15:1241–1248. doi: 10.1016/j.joca.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 36.Burstein D, Bashir A, Gray ML. MRI techniques in early stages of cartilage disease. Invest Radiol. 2000;35:622–638. doi: 10.1097/00004424-200010000-00008. [DOI] [PubMed] [Google Scholar]
- 37.Meder R, de Visser SK, Bowden JC, Bostrom T, Pope JM. Diffusion tensor imaging of articular cartilage as a measure of tissue microstructure. Osteoarthritis Cartilage. 2006;14:875–881. doi: 10.1016/j.joca.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Lin PC, Reiter DA, Spencer RG. Classification of degraded cartilage through multiparametric MRI analysis. J Magn Reson. 2009;201:61–71. doi: 10.1016/j.jmr.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin PC, Irrechukwu O, Roque R, Hancock B, Fishbein KW, Spencer RG. Multivariate analysis of cartilage degradation using the support vector machine algorithm. Magn Reson Med. 2012;67:1815–1826. doi: 10.1002/mrm.23189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reiter DA, Irrechukwu O, Lin PC, Moghadam S, Von Thaer S, Pleshko N, et al. Improved MR-based characterization of engineered cartilage using multiexponential T2 relaxation and multivariate analysis. NMR Biomed. 2012;25:476–488. doi: 10.1002/nbm.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griebel AJ, Trippel SB, Emery NC, Neu CP. Noninvasive assessment of osteoarthritis severity in human explants by multicontrast MRI. Magn Reson Med. 2013;71:807–814. doi: 10.1002/mrm.24725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh A, Haris M, Cai K, Kogan F, Hariharan H, Reddy R. High resolution T1rho mapping of in vivo human knee cartilage at 7T. PLoS One. 2014;9:e97486. doi: 10.1371/journal.pone.0097486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rautiainen J, Lehto LJ, Tiitu V, Kiekara O, Pulkkinen H, Brunott A, et al. Osteochondral repair: evaluation with sweep imaging with fourier transform in an equine model. Radiology. 2013;269:113–121. doi: 10.1148/radiol.13121433. [DOI] [PubMed] [Google Scholar]
- 44.Gatehouse PD, Thomas RW, Robson MD, Hamilton G, Herlihy AH, Bydder GM. Magnetic resonance imaging of the knee with ultrashort TE pulse sequences. Magn Reson Imaging. 2004;22:1061–1067. doi: 10.1016/j.mri.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 45.Lu A, Barger AV, Grist TM, Block WF. Improved spectral selectivity and reduced susceptibility in SSFP using a near zero TE undersampled three-dimensional PR sequence. J Magn Reson Imaging. 2004;19:117–123. doi: 10.1002/jmri.10435. [DOI] [PubMed] [Google Scholar]
- 46.Ashinsky B, Bouhrara M, Urish KL, Coletta CE, Goldberg IG, Spencer RG. Predicting Early Osteoarthritis in the Human Knee: Pattern Recognition and Machine Learning Classification of Magnetic Resonance Images. 2015 Annual Meeting of the Orthopaedic Research Society; Las Vegas, NV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Folkesson J, Dam EB, Olsen OF, Pettersen PC, Christiansen C. Segmenting articular cartilage automatically using a voxel classification approach. IEEE Trans Med Imaging. 2007;26:106–115. doi: 10.1109/TMI.2006.886808. [DOI] [PubMed] [Google Scholar]
- 48.Fripp J, Crozier S, Warfield SK, Ourselin S. Automatic segmentation and quantitative analysis of the articular cartilages from magnetic resonance images of the knee. IEEE Trans Med Imaging. 2010;29:55–64. doi: 10.1109/TMI.2009.2024743. [DOI] [PMC free article] [PubMed] [Google Scholar]