Abstract
The ability of microbial cells to exist in multiple states is a ubiquitous property that promotes adaptation and survival. This phenomenon has been extensively studied in the opportunistic pathogen Candida albicans, which can transition between multiple phenotypic states in response to environmental signals. C. albicans normally exists as a commensal in the human body, but can also cause debilitating mucosal infections or life-threatening systemic infections. The ability to switch between cellular forms contributes to C. albicans’ capacity to infect different host niches, and strictly regulates the program of sexual mating. We review the unique properties associated with different phenotypic states, as well as how interactions between cells in different states can further augment microbial behavior.
The white-opaque switch governs multiple aspects of C. albicans biology
C. albicans exists as a harmless commensal in the gastrointestinal tract and is also commonly isolated from the skin and oral cavity of healthy individuals [1]. However, it can transition to an infectious colonizer of virtually any organ in the body, particularly in immunocompromised patients. C. albicans is currently the most common cause of life-threatening fungal infections in these individuals, with an estimated mortality rate of 35–67% [2,3].
C. albicans exhibits several forms of phenotypic switching in which cells undergo a reversible and heritable transition between alternative cell states. The best characterized of these epigenetic transitions is the white-opaque switch, named for the appearance of colonies on agar media. White cells are round and form brighter, dome-shaped colonies, whereas opaque cells are elliptical and form flatter, duller colonies [4]. Miller and Johnson demonstrated that the switch to the opaque state is necessary for efficient sexual mating [5**], stimulating interest into how phenotypic diversity impacts important physiological traits in C. albicans. Although refractory to mating, white cells were subsequently shown to respond to mating pheromones resulting in ‘sexual’ biofilms [6**], as further discussed below. White and opaque states exhibit many other distinct properties as outlined in Table 1. For example, white cells are predicted to have a fermentative metabolism, whereas opaque cells express genes associated with an oxidative metabolism [7]. Opaque cells are better colonizers of the skin, whereas white cells are more virulent in a bloodstream model of infection [8,9]. Furthermore, white cells are more susceptible to phagocytosis by host macrophages [10] and secrete a leukocyte chemoattractant [11], whereas opaque cells do not. These findings raise the possibility that the switch to the opaque state could promote immune evasion, although opaque cells are also more sensitive to reactive oxygen species, a common defense mechanism of host phagocytes [12].
Table 1. Properties of phenotypic states in C. albicans.
Characteristic properties of the heritable cell states in C. albicans. Cells in each state have unique phenotypic properties. MTL, mating-type locus. Predicted metabolic preferences based on gene expression studies.
| State | MTL Configuration | Major Regulator | Cues supporting formation of each state | Metabolism | Preferred niche | Pheromone response |
|---|---|---|---|---|---|---|
| White | Heterozygous or homozygous | Efg1 [13,14,19] | Stable at 25°C or 37° C [35], aerobic conditions [37] | Fermentative [7] | Commensal, Bloodstream [8] | Biofilm formation [6] |
| Opaque | Usually homozygous [5,28] but can be heterozygous [30] | Wor1 [16–18] | 25°C [35], CO2 [36], anaerobiosis [37], N-acetyl glucosamine [38] | Oxidative [7] | Skin [9] | Mating [5] |
| Gray | Heterozygous or homozygous [44] | N/A | Stable at 25°C or 37°C on rich media (YPD) [44] | Unique carbohydrate metabolism [44] | Skin [44] | Mating at low efficiency [44] |
| GUT | Heterozygous [45] | Requires Wor1 over-expression [45] | Passage through the mammalian gastrointestinal tract [45] | Adapted to the gastro-intestinal tract, high iron [45] | Commensal in gastro-intestinal tract [45] | No response [45] |
Regulation of Phenotypic Switching
C. albicans strains are typically isolated in the white phenotypic state, in which the regulatory circuit is dependent on the transcription factor Efg1 [13–15]. Conversely, expression of the Wor1 transcription factor is necessary and sufficient for stable transitioning to the opaque state [16–18]. Wor1 acts in concert with at least five other transcription factors as part of a network of positive and negative feedback loops to regulate bi-stability in C. albicans [15,19]. Chromatin-level changes control the switching frequency, including a role for hyperacetylation of histone H3K56 in promoting opaque cell formation [20–23] and deposition of H2A.Z in promoting the white state [24]. Mediator complex also regulates white-opaque switching, as deletion of certain mediator components can destabilize the white or opaque states [25]. More recently, experiments demonstrated that the long 5′ untranslated region (5′ UTR) of WOR1 regulates switching by reducing translational efficiency on the WOR1 transcript [26]. These experiments reveal complex transcriptional, post-transcriptional and chromatin-mediated regulation of the switch, with parallels to other heritable circuits including embryonic stem cell differentiation [27].
The white-opaque switch is also controlled by transcription factors encoded at the MTL (mating-type locus). Switching from white to opaque typically requires that diploid strains are homozygous at this locus (i.e., either “MTLa/a” or “MTLα/α”) [5] [28], as expression of Wor1 is repressed by a complex of a1 and α2 transcription factors that is present in MTLa/α cells [29]. However, recent work has shown that certain a/α isolates can also switch to the opaque state under specific environmental conditions [30]. A similar white-opaque switch has been discovered in the related species Candida tropicalis [31**,32] and Candida dubliniensis [33]. In the case of C. tropicalis, white-opaque switching appears to be independent of MTL regulation [32,34]. The fact that the switch exists exclusively in three commensal fungi that co-evolved with the mammalian host suggests that this transition provides an advantage for survival in vivo.
Multiple environmental factors influence C. albicans switching between white and opaque states. Conditions that affect switching in vitro include temperature [35], carbon dioxide [36], anaerobiosis [37], N-acetylglucosamine [38] and genotoxic and oxidative stress [39]. In vivo, opaque-to-white switching occurs en masse during systemic infection [40], whereas increased switching in the opposite direction, from white to opaque, was observed in one strain during gastrointestinal colonization [41]. The sensitivity of the switch to a diverse array of environmental factors is reflective of multiple pathways impinging on white-opaque signaling. Some of these have been defined and include the pheromone MAPK cascade [42**], the cAMP/PKA pathway [38], and the Hog1 stress-activated protein kinase pathway [43].
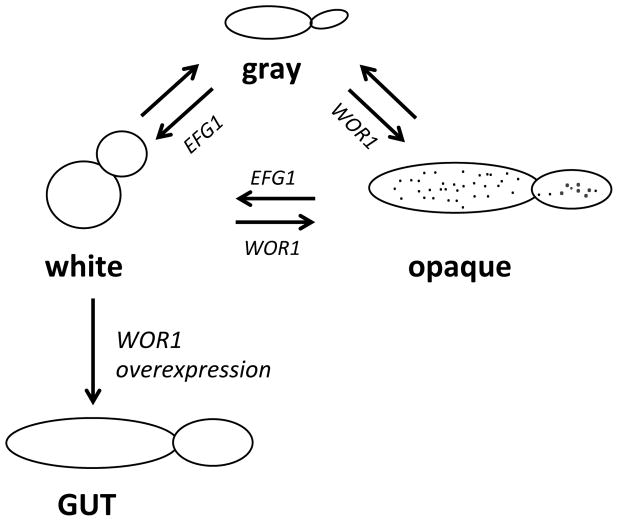
Recently, C. albicans was shown to stably exist in two additional phenotypic states, termed the “gray” and “GUT” states, that are related to white and opaque forms (Figure 1). Gray cells displayed a distinct metabolic profile from white and opaque cells, and were more successful than either of these states in an ex vivo mouse tongue infection model [44**]. Cells in the GUT state were observed following gastrointestinal colonization using a strain that overexpressed the WOR1 gene [45*]. GUT cells were hypercompetitive when retested in this niche, indicating a potential role for this phenotypic state in promoting commensalism [45]. The metabolic reprogramming of cells can therefore generate specialized forms that enable colonization and infection of different host niches.
Figure 1. Candida albicans cells can exist in four related heritable states.
The white state is positively regulated by the transcription factor Efg1, while the opaque state requires expression of the transcription factor Wor1. Other transcription factors that co-regulate the white-opaque switch with Wor1 and Efg1 are not shown. Opaque cells often display characteristic ‘pimples’ on the cell surface when analyzed by scanning electron microscopy. Deletion of both of Wor1 and Efg1 can promote formation of the grey state. White cells overexpressing the Wor1 transcription factor can be induced to adopt the GUT (gastrointestinally induced transition) phenotypic state after passage through the murine gastrointestinal tract. GUT cells were characterized in a/α cell types and appear similar in shape to opaque cells, but unlike opaque cells they show increased fitness in colonizing the gastrointestinal tract.
C. albicans mating necessitates switching to the opaque state
The most striking difference between white and opaque cells is their sexual fecundity. Opaque cells are the mating competent form of C. albicans, mating a million times more efficiently than white cells [5]. Conventional heterothallic mating occurs between diploid a and α opaque cells and is driven by the secretion and response to sex-specific pheromones (Figure 2A). A conserved MAPK cascade transduces the pheromone signal resulting in activation of the transcription factor Cph1 (ortholog of Saccharomyces cerevisiae Ste12). This triggers polarized growth, cell fusion and karyogamy, resulting in the formation of tetraploid mating products [46–49]. Meiosis has not been observed in C. albicans and instead tetraploid cells return to the diploid state by a series of reductional mitotic divisions [50–52]. It was revealed that C. albicans diploid cells can reduce their ploidy to that of true haploids by a similar parasexual process, further extending the range of ploidy states that can be adopted by this species (also see Gerstein and Berman review, this issue). Haploids are homozygous at the MTL locus and can readily switch to the opaque state and mate to reform diploids, or can undergo spontaneous diploidization [53]. The transient formation of haploid a and α cells could potentially support mating events even in individuals that appear to be carrying a single a/α diploid strain.
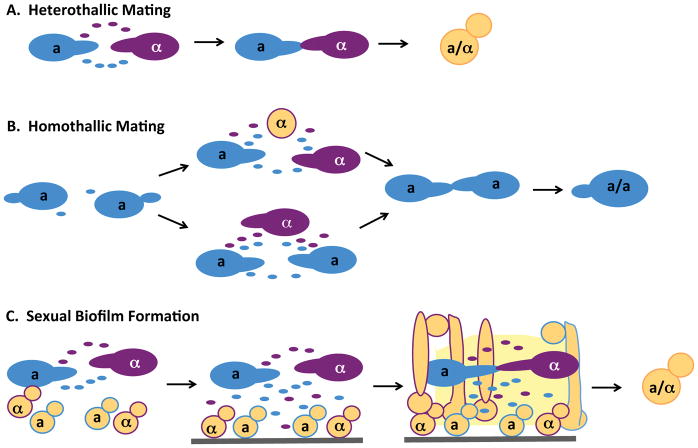
Figure 2. Interactions between white and opaque cells in a population.
A. Conventional heterothallic mating occurs between diploid opaque C. albicans cells of opposite mating types. Pheromone signaling stimulates mating projection formation and cell fusion to generate a/α tetraploid products.
B. Same-sex opaque cells can undergo mating in the presence of an opposite-sex white cell (top) or opaque cell (bottom). Pheromone signaling between these cell types leads to amplification of the pheromone response and conjugation between cells of the same sex.
C. Pheromone signaling from opaque cells can stimulate responding white a or α cells to adhere to a surface and undergo formation of a ‘sexual biofilm’ that includes filamentous cells and secretion of an extracellular matrix. Within this immobilized structure, pheromone gradients between opaque cells are stabilized thereby leading to more efficient chemotropism and conjugation.
The population structure of C. albicans is predominantly clonal [54,55], although there is evidence of a mixed evolutionary history consistent with a sexually or parasexually reproducing species [56,57]. In addition to heterothallic mating, experiments have shown that C. albicans is capable of unisexual, homothallic mating [58*]. In particular, C. albicans opaque a cells lacking the α pheromone-degrading protease Bar1 experience autocrine pheromone signaling, resulting in same-sex a-a mating. Same-sex mating was also detected in ménage-à-trois matings, where conjugation between cells of the same sex was induced by pheromone secreted by cells of the opposite sex (see Figure 2B) [58]. Subsequent experiments established that activation of the pheromone response was sufficient to cause opaque cells to undergo same-sex conjugation [59].
Despite being unable mate, C. albicans white cells can respond to pheromones produced by opposite-sex opaque cells, upregulating production of their own pheromones in response. Synergistic signaling between the two cell types can enhance mating between rare opaque cells due to the amplification of pheromone levels [60**]. White cells can thereby facilitate the mating of opaque cells, despite being essentially sterile themselves (Figure 2B). Interestingly, the role of such “helper” white cells promotes both opposite-sex and same-sex mating between minority opaque cells [60]. Thus, cooperation between white and opaque cells can support mating even in populations where only one cell type is in the “mating-competent” opaque state.
Pheromone signaling drives biofilm formation
Biofilms are communities of cells that adhere to inert or biological surfaces. They are often associated with an extracellular matrix and communal cells exhibit distinct phenotypic properties from free-floating planktonic cells [61]. C. albicans causes a variety of device-associated infections that involve the formation of biofilms on catheters, heart valves, pacemakers, dentures, and prosthetic joints [62]. Biofilm formation is initiated when a basal layer of unicellular yeast adheres to the substrate, upon which a layer of pseudohyphal and hyphal cells grows during maturation [63]. These microbial communities are often drug resistant due to the occlusion of antifungal drugs, the upregulation of drug efflux pumps, and the existence of persister cells within the biofilm [64,65]. Conventional studies of biofilm formation in C. albicans have utilized MTLa/α cell types; these make up the majority of clinical isolates whereas less than 10% of strains are naturally homozygous for MTLa or MTLα [56,66].
More recently, it has been demonstrated that pheromone signaling in C. albicans can promote the formation of ‘sexual’ biofilms. These were first observed in white a and α cells responding to pheromones secreted by opaque cells of the opposite sex [6]. Responding white cells were induced to undergo substrate adhesion, filamentation, and development of a complex biofilm structure. Subsequently, it was shown that white cells also respond to pheromones secreted by opaque cells of the same sex, using a self-activation mechanism similar to that that drives same-sex mating [67]. Sexual biofilms contain a basal layer of unicellular yeast cells upon which a layer of filamentous cells is formed, but appear to lack the drug resistance and small-molecule impermeability characteristic of ‘conventional’ biofilms generated by MTLa/α cells [68]. Interestingly, the permeability of sexual biofilms supports the build-up and maintenance of pheromone gradients within the biofilm. Thus, pheromones secreted by minority opaque cells form stable gradients within this structure, facilitating chemotropism between opaque a and α partners [6]. Presumably as a result of this mechanism, mating between minority opaque cells is 5- to 100-fold higher when these cells are seeded in a sexual biofilm compared to a conventional biofilm [69*]. These findings suggest that C. albicans has evolved two types of biofilms that are influenced by their MTL configuration; conventional a/α biofilms that are ‘pathogenic’ based on their general impermeability and resistance to antifungals, and sexual biofilms that may provide an appropriate setting for mating to occur in vivo [68,70,71]. It is also hypothesized that the white-opaque switch evolved to generate sexual biofilms that facilitate mating of Candida strains in the host [71].
The transcriptional regulation of both conventional and sexual biofilms has been examined in C. albicans. In conventional a/α biofilms, a network of six major transcription factors regulates expression of ~2,200 genes [72*]. A comparison between the transcriptional control of sexual biofilms and conventional biofilms revealed that four of the six transcriptional regulators involved in conventional biofilms are also required for sexual biofilms, indicating partial overlap of the two regulatory circuits [73*]. Upstream signaling in conventional biofilms involves the Ras1/cAMP pathway, which activates the master transcriptional regulator Efg1 [68,74]. However, there have been conflicting reports concerning how the pheromone signal is transduced via the MAPK pathway to activate sexual biofilm formation. Experiments in our lab and others have shown that pheromone signaling in white cells requires the Cph1 transcription factor [42,60,73], and that this factor is essential for the transcriptional response and adhesive properties of pheromone-treated white cells [73]. In contrast, a separate set of studies indicated that pheromone signaling acts through the Tec1 transcription factor, and that Cph1 is not required for sexual biofilm formation [75]. Differences in experimental conditions may explain some of these differences [71], although all groups agree that Tec1 makes a significant contribution to both conventional and sexual biofilm formation [42,73,75,76], which reflects, at least in part, a key role for Tec1 in promoting hyphal growth [77,78]. Overall, it is clear that questions remain concerning the precise signaling mechanisms contributing to the establishment of sexual biofilms, as well as the role of these biofilms during infection of the host.
Recent studies in C. tropicalis have revealed striking differences with C. albicans with respect to the properties of white and opaque cells. For example, while switching between states is influenced by multiple environmental conditions in C. albicans, only N-acetylglucosamine has been shown to also affect the transition in C. tropicalis [31,32]. Furthermore, while pheromone signaling induces the formation of sexual biofilms in C. tropicalis as in C. albicans, it is responding opaque cells, not white cells, that contribute to biofilm development in the former [79*]. Thus, opaque cells are both the mating competent form and the biofilm forming cell type in C. tropicalis. These observations raise intriguing questions as to the evolution of the white-opaque switch in Candida species. At face value, they suggest that the switch did not simply evolve so that biofilms generated by white cells could facilitate mating between opaque cells, as white cells do not form sexual biofilms in C. tropicalis. It is possible that this mechanism represents a further refinement of the mating system in C. albicans that occurred after it diverged from C. tropicalis. There is also an incomplete understanding of the in vivo niches supporting the growth of white and opaque cells in the mammalian host. Given recent advances in our understanding of the spatial and temporal heterogeneity of the human mycobiome (see review by Xu and Dongari-Bagtzoglou, this issue), it is likely that we will soon learn more about the niches favored by white and opaque forms in the natural host. Distinguishing cells in different phenotypic states is likely be driven by emerging techniques such as RNA FISH (fluorescence in situ hybridization) to quantify the in vivo expression of state-specific gene transcripts.
Conclusions
C. albicans generates diversity by undergoing heritable and reversible transitions between multiple alternative states. The creation of phenotypic diversity is an established mechanism to ensure the stability and health of microbial populations, providing an ‘insurance policy’ against changing environmental conditions [80,81]. Morphological plasticity is a common attribute of both pathogenic and non-pathogenic species. In the model yeast S. cerevisiae, different phenotypic states can be generated by changes in chromosomal copy number or by prions [82,83], while phenotypic variation has been observed in multiple human fungal pathogens [84](see also review by Wang and Lin, this issue). In the case of C. albicans, not only do different states offer potential fitness advantages, but also interactions between cells in different states can provide additional benefits, such as increasing mating efficiency or induction of sexual biofilm formation. The result of this synergy is that the ‘whole is greater than the sum of its parts’ when it comes to evaluating phenotypic diversity. Further investigation of different phenotypes are certain to lead to greater understanding of C. albicans biology, as well as providing new insights into cell specialization in microbial species.
Highlights.
Phenotypic switching between states occurs in the fungal pathogen C. albicans.
Switching provides an adaptive strategy for different host environments.
Interactions between cells in different states can promote unique responses.
These include facilitating mating and formation of sexual biofilms.
Acknowledgments
We thank Iuliana Ene, Emily Mallick and anonymous reviewers for comments on the manuscript. Work in the author’s laboratory is supported by National Institutes of Health grants AI081704 and AI112362, and by a PATH award from the Burroughs Wellcome Fund. CMS is supported by National Institutes of Health fellowship F31DE024036.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Christine M. Scaduto, Email: Christine_Scaduto@brown.edu.
Richard J. Bennett, Email: Richard_Bennett@brown.edu.
References
- 1.Cassone A, Cauda R. Candida and candidiasis in HIV-infected patients: where commensalism, opportunistic behavior and frank pathogenicity lose their borders. AIDS. 2012;26:1457–1472. doi: 10.1097/QAD.0b013e3283536ba8. [DOI] [PubMed] [Google Scholar]
- 2.Brown GD, Denning DG, Gow NA, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 3.Kett DH, Azoulay E, Echeverria PM, Vincent JL. Candida bloodstream infections in intensive care units: analysis of the extended prevalence of infection in intensive care unit study. Crit Care Med. 2011;39:665–670. doi: 10.1097/CCM.0b013e318206c1ca. [DOI] [PubMed] [Google Scholar]
- 4.Slutsky BSM, Anderson J, Risen L, Pfaller M, Soll DR. White-opaque transition”: a second high frequency switching system in Candida albicans. Journal of Bacteriology. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **5.Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. This landmark paper showed that the white-opaque switch is the key to efficient mating in C. albicans, and that the switch is regulated by the MTL locus. [DOI] [PubMed] [Google Scholar]
- **6.Daniels KJ, Srikantha T, Lockhart SR, Pujol C, Soll DR. Opaque cells signal white cells to form biofilms in Candida albicans. EMBO J. 2006;25:2240–2252. doi: 10.1038/sj.emboj.7601099. This work was the first to demonstrate that pheromone signaling could lead to sexual biofilm formation in C. albicans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lan CY, Newport G, Murillo LA, Jones T, Scherer S, Davis RW, Agabian N. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc Natl Acad Sci U S A. 2002;99:14907–14912. doi: 10.1073/pnas.232566499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kvaal CA, Srikantha T, Soll DR. Misexpression of the white-phase-specific gene WH11 in the opaque phase of Candida albicans affects switching and virulence. Infect Immun. 1997;65:4468–4475. doi: 10.1128/iai.65.11.4468-4475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kvaal C, Lachke SA, Srikantha T, Daniels K, McCoy J, Soll DR. Misexpression of the opaque phase specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect Immun. 1999;67:6652–6662. doi: 10.1128/iai.67.12.6652-6662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lohse MB, Johnson AD. Differential phagocytosis of white versus opaque Candida albicans by Drosophila and mouse phagocytes. PLoS One. 2008;3:e1473. doi: 10.1371/journal.pone.0001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geiger J, Wessels D, Lockhart SR, Soll DR. Release of a potent polymorphonuclear leukocyte chemoattractant is regulated by white-opaque switching in Candida albicans. Infection and Immunity. 2004;72:667–677. doi: 10.1128/IAI.72.2.667-677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolotila MP, Diamond RD. Effects of neutrophils and in vitro oxidants on survival and phenotypic switching of Candida albicans WO-1. Infect Immun. 1990;58:1174–1179. doi: 10.1128/iai.58.5.1174-1179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sonnerborn A, Tebarth B, Ernst JF. Control of white-opaque switching in Candida albicans by the Efg1p morphogenetic regulator. Infect Immun. 1999;67:4655–4660. doi: 10.1128/iai.67.9.4655-4660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srikantha T, Tsai LK, Daniels K, Soll DR. EFG1 null mutants of Candida albicans switch but cannot express the complete phenotype of white-phase budding cells. J Bacteriol. 2000;182:1580–1591. doi: 10.1128/jb.182.6.1580-1591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernday AD, Lohse MB, Fordyce PM, Nobile CJ, DeRisi JL, Johnson AD. Structure of the transcriptional network controlling white-opaque switching in Candida albicans. Mol Microbiol. 2013;90:22–35. doi: 10.1111/mmi.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang G, Wang H, Chou S, Nie X, Chen J, Liu H. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci U S A. 2006;103:12813–12818. doi: 10.1073/pnas.0605270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srikantha T, Borneman AR, Daniels KJ, Pujol C, Wu W, Seringhaus MR, Gerstein M, Yi S, Snyder M, Soll DR. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot Cell. 2006;5:1674–1687. doi: 10.1128/EC.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zordan RE, Galgoczy DJ, Johnson AD. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci U S A. 2006;103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hnisz D, Majer O, Frohner IE, Komnenovic V, Kuchler K. The Set3/Hos2 histone deacetylase complex attenuates cAMP/PKA signaling to regulate morphogenesis and virulence of Candida albicans. PLoS Pathog. 2010;6:e1000889. doi: 10.1371/journal.ppat.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hnisz D, Schwarzmuller T, Kuchler K. Transcriptional loops meet chromatin: a dual-layer network controls white-opaque switching in Candida albicans. Mol Microbiol. 2009;74:1–15. doi: 10.1111/j.1365-2958.2009.06772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stevenson JS, Liu H. Regulation of white and opaque cell-type formation in Candida albicans by Rtt109 and Hst3. Mol Microbiol. 2011;81:1078–1091. doi: 10.1111/j.1365-2958.2011.07754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevenson JS, Liu H. Nucleosome assembly factors CAF-1 and HIR modulate epigenetic switching frequencies in an H3K56 acetylation-associated manner in Candida albicans. Eukaryot Cell. 2013;12:591–603. doi: 10.1128/EC.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guan Z, Liu H. Overlapping functions between SWR1 Deletion and H3K56 acetylation in Candida albicans. Eukaryot Cell. 2015;14:578–587. doi: 10.1128/EC.00002-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang A, Liu Z, Myers LC. Differential regulation of white-opaque switching by individual subunits of Candida albicans mediator. Eukaryot Cell. 2013;12:1293–1304. doi: 10.1128/EC.00137-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan Z, Liu H. The WOR1 5′ untranslated region regulates white-opaque switching in Candida albicans by reducing translational efficiency. Mol Microbiol. 2015;97:125–138. doi: 10.1111/mmi.13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuch BB, Mitrovich QM, Homann OR, Hernday AD, Monighetti CK, De La Vega FM, Johnson AD. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet. 2010;6:e1001070. doi: 10.1371/journal.pgen.1001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lockhart SR, Pujol C, Daniels KJ, Miller MG, Johnson AD, Pfaller MA, Soll DR. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics. 2002;162:737–745. doi: 10.1093/genetics/162.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- 30.Xie J, Tao L, Nobile CJ, Tong Y, Guan G, Sun Y, Cao C, Hernday AD, Johnson AD, Zhang L, et al. White-opaque switching in natural MTLa/alpha isolates of Candida albicans: evolutionary implications for roles in host adaptation, pathogenesis, and sex. PLoS Biol. 2013;11:e1001525. doi: 10.1371/journal.pbio.1001525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **31.Porman AM, Alby K, Hirakawa MP, Bennett RJ. Discovery of a phenotypic switch regulating sexual mating in the opportunistic fungal pathogen Candida tropicalis. Proc Natl Acad Sci U S A. 2011;108:21158–21163. doi: 10.1073/pnas.1112076109. This work established that Candida troplicalis, previously thought to reproduce only asexually, can undergo a white-opaque switch that regulates mating. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie J, Du H, Guan G, Tong Y, Kourkoumpetis TK, Zhang L, Bai FY, Huang G. N-acetylglucosamine induces white-to-opaque switching and mating in Candida tropicalis, providing new insights into adaptation and fungal sexual evolution. Eukaryot Cell. 2012;11:773–782. doi: 10.1128/EC.00047-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pujol C, Daniels KJ, Lockhart SR, Srikantha T, Radke JB, Geiger J, Soll DR. The closely related species Candida albicans and Candida dubliniensis can mate. Eukaryot Cell. 2004;3:1015–1027. doi: 10.1128/EC.3.4.1015-1027.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porman AM, Hirakawa MP, Jones SK, Jr, Wang N, Bennett RJ. MTL-independent phenotypic switching in Candida tropicalis and a dual role for Wor1 in regulating switching and filamentation. PLoS Genet. 2013;9:e1003369. doi: 10.1371/journal.pgen.1003369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rikkerink EHA, Magee BB, Magee PT. Opaque-white phenotype transition: a programmed morphological transition in Candida albicans. J Bacteriol. 1988;170:895–890. doi: 10.1128/jb.170.2.895-899.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang G, Srikantha T, Sahni N, Yi S, Soll DR. CO2 regulates white-to-opaque switching in Candida albicans. Curr Biol. 2009;19:330–334. doi: 10.1016/j.cub.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dumitru R, Navarathna DH, Semighini CP, Elowsky CG, Dumitru RV, Dignard D, Whiteway M, Atkin AL, Nickerson KW. In vivo and in vitro anaerobic mating in Candida albicans. Eukaryot Cell. 2007;6:465–472. doi: 10.1128/EC.00316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang G, Yi S, Sahni N, Daniels KJ, Srikantha T, Soll DR. N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 2010;6:e1000806. doi: 10.1371/journal.ppat.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alby K, Bennett RJ. Stress-induced phenotypic switching in Candida albicans. Mol Biol Cell. 2009;20:3178–3191. doi: 10.1091/mbc.E09-01-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kvaal CA, Srikantha T, Soll DR. Misexpression of the white-phase-specific gene WH11 in the opaque phase of Candida albicans affects switching and virulence. Infect Immun. 1997;65:4468–4475. doi: 10.1128/iai.65.11.4468-4475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramirez-Zavala B, Reuss O, Park YN, Ohlsen K, Morschhauser J. Environmental induction of white-opaque switching in Candida albicans. PLoS Pathog. 2008;4:e1000089. doi: 10.1371/journal.ppat.1000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **42.Ramirez-Zavala B, Weyler M, Gildor T, Schmauch C, Kornitzer D, Arkowitz R, Morschhauser J. Activation of the Cph1-dependent MAP kinase signaling pathway induces white-opaque switching in Candida albicans. PLoS Pathog. 2013;9:e1003696. doi: 10.1371/journal.ppat.1003696. This work establishes that the pheromone MAPK cascade which induces mating also induces cells to switch from the white to opaque state. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang SH, Cheng JH, Deng FS, Tsai PA, Lin CH. A novel function for Hog1 stress-activated protein kinase in controlling white-opaque switching and mating in Candida albicans. Eukaryot Cell. 2014;13:1557–1566. doi: 10.1128/EC.00235-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **44.Tao L, Du H, Guan G, Dai Y, Nobile CJ, Liang W, Cao C, Zhang Q, Zhong J, Huang G. Discovery of a “white-gray-opaque” tristable phenotypic switching system in Candida albicans: roles of non-genetic diversity in host adaptation. PLoS Biol. 2014;12:e1001830. doi: 10.1371/journal.pbio.1001830. This work describes the discovery of an additonal phenotypic state, the “gray” state, which has unique virulence and physiological properties. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *45.Pande K, Chen C, Noble SM. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet. 2013;45:1088–1091. doi: 10.1038/ng.2710. These authors identified the ‘GUT’ phenotype in cells overexpressing the Wor1 transcription factor upon passage through the murine gastrointestinal tract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Magee BB, Legrand M, Alarco AM, Raymond M, Magee PT. Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Mol Microbiol. 2002;46:1345–1351. doi: 10.1046/j.1365-2958.2002.03263.x. [DOI] [PubMed] [Google Scholar]
- 47.Chen J, Chen J, Lane S, Liu H. A conserved mitogen-activated protein kinase pathway is required for mating in Candida albicans. Mol Microbiol. 2002;46:1335–1344. doi: 10.1046/j.1365-2958.2002.03249.x. [DOI] [PubMed] [Google Scholar]
- 48.Bennett RJ, Uhl MA, Miller MG, Johnson AD. Identification and characterization of a Candida albicans mating pheromone. Molecular and Cellular Biology. 2003;23:8189–8201. doi: 10.1128/MCB.23.22.8189-8201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bennett RJ, Johnson AD. Mating in Candida albicans and the search for a sexual cycle. Annu Rev Microbiol. 2005;59:233–255. doi: 10.1146/annurev.micro.59.030804.121310. [DOI] [PubMed] [Google Scholar]
- 50.Bennett RJ, Johnson AD. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 2003;22:2505–2515. doi: 10.1093/emboj/cdg235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forche A, Alby K, Schaefer D, Johnson AD, Berman J, Bennett RJ. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 2008;6:e110. doi: 10.1371/journal.pbio.0060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hickman MA, Paulson C, Dudley AM, Berman J. Parasexual ploidy reduction drives population heterogeneity through random and transient aneuploidy in Candida albicans. Genetics. 2015 doi: 10.1534/genetics.115.178020. Early online as 10.1534/genetics.115.178020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hickman MA, Zeng G, Forche A, Hirakawa MP, Abbey D, Harrison BD, Wang YM, Su CH, Bennett RJ, Wang Y, et al. The ‘obligate diploid’ Candida albicans forms mating-competent haploids. Nature. 2013;494:55–59. doi: 10.1038/nature11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gräser Y, Volovsek M, Arrington J, Schönian G, Presber W, Mitchell TG, Vilgalys R. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc Natl Acad Sci U S A. 1996;93:12473–12477. doi: 10.1073/pnas.93.22.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson JB, Wickens C, Khan M, Cowen LE, Federspiel N, Jones T, Kohn LM. Infrequent genetic exchange and recombination in the mitochondrial genome of Candida albicans. J Bacteriol. 2001;183:865–872. doi: 10.1128/JB.183.3.865-872.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Odds FC, Bougnoux ME, Shaw DJ, Bain JM, Davidson AD, Diogo D, Jacobsen MD, Lecomte M, Li SY, Tavanti A, et al. Molecular phylogenetics of Candida albicans. Eukaryot Cell. 2007;6:1041–1052. doi: 10.1128/EC.00041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tavanti A, Gow NA, Maiden MC, Odds FC, Shaw DJ. Genetic evidence for recombination in Candida albicans based on haplotype analysis. Fungal Genet Biol. 2004;41:553–562. doi: 10.1016/j.fgb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- *58.K, Schaefer D, Bennett RJ. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature. 2009;460:890–893. doi: 10.1038/nature08252. This paper is the first demonstration that C. albicans cells can undergo same-sex, homothallic mating. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alby K, Bennett RJ. Interspecies pheromone signaling promotes biofilm formation and same sex mating in Candida albicans. Proc Natl Acad Sci U S A. 2011;108:2510–2515. doi: 10.1073/pnas.1017234108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **60.Tao L, Cao C, Liang W, Guan G, Zhang Q, Nobile CJ, Huang G. White cells facilitate opposite- and same-sex mating of opaque cells in Candida albicans. PLoS Genet. 2014;10:e1004737. doi: 10.1371/journal.pgen.1004737. These authors demonstrate that pheromone signaling can occur between white and opaque cells, leading to increased mating efficiency, particularly in same-sex mating. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clinical Microbiology Reviews. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fox EP, Nobile CJ. A sticky situation: untangling the transcriptional network controlling biofilm development in Candida albicans. Transcription. 2012;3:315–322. doi: 10.4161/trns.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Finkel JS, Mitchell AP. Genetic control of Candida albicans biofilm development. Nat Rev Microbiol. 2011;9:109–118. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.LaFleur MD, Kumamoto CA, Lewis K. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob Agents Chemother. 2006;50:3839–3846. doi: 10.1128/AAC.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramage G, Bachmann S, Patterson TF, Wickes BL, Lopez-Ribot JL. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J Antimicrob Chemother. 2002;49:973–980. doi: 10.1093/jac/dkf049. [DOI] [PubMed] [Google Scholar]
- 66.Lockhart SR, Pujol C, Daniels KJ, Miller MG, Johnson AD, Pfaller MA, Soll DR. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics. 2002;162:737–745. doi: 10.1093/genetics/162.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yi S, Sahni N, Daniels KJ, Lu KL, Huang G, Srikantha T, Soll DR. Self-induction of a/a or alpha/alpha biofilms in Candida albicans is a pheromone-based paracrine system requiring switching. Eukaryot Cell. 2011;10:753–760. doi: 10.1128/EC.05055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yi S, Sahni N, Daniels KJ, Lu KL, Srikantha T, Huang G, Garnaas AM, Soll DR. Alternative mating type configurations (a/alpha versus a/a or alpha/alpha) of Candida albicans result in alternative biofilms regulated by different pathways. PLoS Biol. 2011;9:e1001117. doi: 10.1371/journal.pbio.1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *69.Park YN, Daniels KJ, Pujol C, Srikantha T, Soll DR. Candida albicans forms a specialized “sexual” as well as “pathogenic” biofilm. Eukaryot Cell. 2013;12:1120–1131. doi: 10.1128/EC.00112-13. This work shows that sexual biofilims formed by white cells can increase the mating efficiency between minority opaque cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Soll DR. Why does Candida albicans switch? FEMS Yeast Res. 2009;9:973–989. doi: 10.1111/j.1567-1364.2009.00562.x. [DOI] [PubMed] [Google Scholar]
- 71.Soll DR. The evolution of alternative biofilms in an opportunistic fungal pathogen: an explanation for how new signal transduction pathways may evolve. Infect Genet Evol. 2014;22:235–243. doi: 10.1016/j.meegid.2013.07.013. [DOI] [PubMed] [Google Scholar]
- *72.Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–138. doi: 10.1016/j.cell.2011.10.048. This work established the transcriptonal network regulating ‘conventional’ biofilms formed by a/α cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *73.Lin CH, Kabrawala S, Fox EP, Nobile CJ, Johnson AD, Bennett RJ. Genetic control of conventional and pheromone-stimulated biofilm formation in Candida albicans. PLoS Pathog. 2013;9:e1003305. doi: 10.1371/journal.ppat.1003305. These authors examined the transcriptional regulation of sexual biofilms and shows that the transcription factor Cph1 mediates pheromone signaling in white cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramage G, VandeWalle K, Lopez-Ribot JL, Wickes BL. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol Lett. 2002;214:95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- 75.Sahni N, Yi S, Daniels KJ, Huang G, Srikantha T, Soll DR. Tec1 mediates the pheromone response of the white phenotype of Candida albicans: insights into the evolution of new signal transduction pathways. PLoS Biol. 2010;8:e1000363. doi: 10.1371/journal.pbio.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol. 2005;15:1150–1155. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 77.Schweizer A, Rupp S, Taylor BN, Rollinghoff M, Schroppel K. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol Microbiol. 2000;38:435–445. doi: 10.1046/j.1365-2958.2000.02132.x. [DOI] [PubMed] [Google Scholar]
- 78.Daniels KJ, Srikantha T, Pujol C, Park YN, Soll DR. Role of Tec1 in the development, architecture, and integrity of sexual biofilms of Candida albicans. Eukaryot Cell. 2015;14:228–240. doi: 10.1128/EC.00224-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *79.Jones SK, Jr, Hirakawa MP, Bennett RJ. Sexual biofilm formation in Candida tropicalis opaque cells. Mol Microbiol. 2014;92:383–398. doi: 10.1111/mmi.12565. This work establishes that, unlike C. albicans, only opaque cells form sexual biofilms in the related fungal pathogen C. tropicalis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boles BR, Thoendel M, Singh PK. Self-generated diversity produces “insurance effects” in biofilm communities. Proc Natl Acad Sci U S A. 2004;101:16630–16635. doi: 10.1073/pnas.0407460101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kolter R, Greenberg EP. Microbial sciences: the superficial life of microbes. Nature. 2006;441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 82.Tan Z, Hays M, Cromie GA, Jeffery EW, Scott AC, Ahyong V, Sirr A, Skupin A, Dudley AM. Aneuploidy underlies a multicellular phenotypic switch. Proc Natl Acad Sci U S A. 2013;110:12367–12372. doi: 10.1073/pnas.1301047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482:363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jain N, Fries BC. Antigenic and phenotypic variations in fungi. Cell Microbiol. 2009;11:1716–1723. doi: 10.1111/j.1462-5822.2009.01384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]