Abstract
Chromatin immunoprecitation followed by high-throughput sequencing (ChIP-seq) is a powerful method that identifies protein-DNA binding sites in vivo. Recent studies have illustrated the value of ChIP-seq in studying transcription factor binding in various bacterial species under a variety of growth conditions. These results show that in addition to identifying binding sites, correlation of ChIP-seq data with expression data can reveal important information about bacterial regulons and regulatory networks. In this chapter, we provide an overview of the current state of knowledge about ChIP-seq methodology in bacteria, from sample preparation to raw data analysis. We also describe visualization and various bioinformatic analyses of processed ChIP-seq data.
1: Introduction
Transcriptional regulation of gene expression by transcription factors (TFs) is a common mechanism of regulation in bacteria [1]. Identifying all the genes directly regulated by a transcription factor can be challenging particularly for those that regulate a large number of genes and whose DNA binding sites might be less conserved and thus difficult to identify using bioinformatic approaches. By comparing RNA profiles of strains lacking the transcription factor to their parent strain, using either microarray technology or high-throughput sequencing technology, candidate genes controlled by a transcription factor can be identified. However, whole-genome expression analyses cannot reveal whether the influence of the TF on RNA levels is direct or indirect. This requires identification of TF binding within the appropriate promoter region. While it is possible to characterize binding to these regions using in vitro assays, recent advances in genome wide analysis, make identification of these binding sites possible in vivo, allowing identification of binding events under the same growth conditions that RNA levels were determined.
Chromatin immunoprecipitation (ChIP) followed by microarray analysis (ChIP-chip) or, more recently, high-throughput sequencing (ChIP-seq), which has a much higher resolution and signal-to-noise ratio than ChIP-chip, has been used to map genome-wide binding of many bacterial TFs [2-25]. While many of the studies have focused on TFs in Escherichia coli, recent studies have examined transcription factors in many diverse bacterial species, such as Vibrio cholerae, Vibrio harveyi, Rhodobacter sphaeroides, Salmonella enterica, Mycobacterium tuberculosis, and Caulobacter crescentus [2-25]. A subset of these studies also correlated occupancy data with expression data to investigate the regulons of certain TFs [2,3,5,7,8,13,16,21,22,25], and we predict the desire to use these assays to study other TFs will increase as the costs of the performing ChIP-seq decrease. Here, we provide an overview to using ChIP-seq to identify bacterial regulons, from sample preparation, data generation, data visualization, data analysis, and downstream bioinformatic and computational analyses.
We offer a general overview of each step and highlight specific analyses that have aided our work. It is important to note that analysis of ChIP-seq data in bacteria is an evolving pipeline and as such, new approaches and algorithms are being introduced frequently. It is beyond the scope of this review to provide a comprehensive description of known variations of ChIP-seq analysis, including the higher resolution variant, ChIP-exo. ChIP-exo has shown promise in the few bacterial studies where it has been used [26-29] but the analysis of ChIP-exo data has its own unique issues including sample preparation and data analysis. We encourage those interested in performing ChIP-seq experiments or variations on ChIP-seq to review other studies [2-25] if they want a broader sense of the experimental and analytical variables.
2: ChIP-seq Sample Preparation and Data Generation
2.1 General Overview of ChIP-seq Procedure
ChIP-seq reports on the DNA sequences that can be cross-linked by formaldehyde to a given transcription factor in actively growing cells and then enriched relative to genomic DNA when the DNA-protein complexes are precipitated by use of an antibody specific to the transcription factor. Aspects of the ChIP-seq method and resulting data analysis will be described in detail throughout this review (Figure 1). Several methods and review articles describe in detail how to perform ChIP-chip and ChIP-seq experiments [30-33], and we encourage readers to also review these publications. The methodology described in these articles differs and while each has been used to produced high-quality ChIP-seq data, we have found success following the method described by Davis et al [33]. Briefly, cells are grown in desired conditions to a designated growth phase and formaldehyde is added to crosslink proteins to DNA. Cells are pelleted, lysed, the lysate is sonicated to shear the DNA, and DNA-TF complexes of interest are enriched using an antibody specific to the TF being studied. The crosslinks are reversed by heating the sample to 65°C and the DNA that was bound to the TF of interest, the immunoprecipatant (IP), is isolated and sequenced along with a sample of DNA from un-enriched, non-antibody treated DNA (Input sample). The Input sample represents the background signal of available chromatin for IP, controlling for regions of genomic DNA that may be enriched for reasons other than TF binding. Areas of the genome bound by the TF are recovered in larger proportion in the IP fraction than unbound areas. Thus, areas of TF binding are regions of the genome that show enrichment in the IP fraction compared to the background Input fraction (Figure 1). In the next sections, we discuss what we consider “best practices” in designing ChIP-seq experiments.
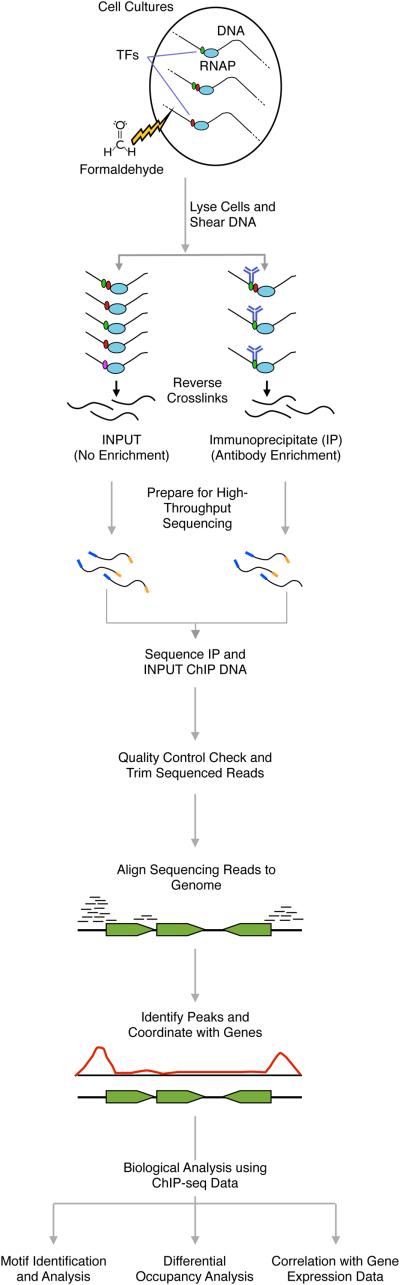
Figure 1. ChIP-seq Sample Preparation and Analysis.
ChIP-seq begins with cell cultures grown in defined conditions. When cultures reach the desired growth stage, they are treated with formaldehyde to crosslink proteins and DNA. The cells are lysed and sheared so the average DNA length is ~500 bp. The DNA-protein samples are split into two populations, the Immunoprecipitate (IP) fraction, which is enriched for a particular protein of interest using an antibody, and the Input fraction that is not enriched for any particular protein of interest. The crosslinks are reversed by heat treatment and the DNA fragments are subjected to high-throughput sequencing. After sequencing, the resulting sequencing reads are examined for quality and trimmed based on read quality. The trimmed reads are then aligned to a reference genome and algorithms and/or visual inspection identify ChIP-seq peaks. After peaks are associated with genes downstream, several bioinformatic analyses can be performed including motif identification and analysis, differential occupancy analysis, and correlation with expression data for in depth understanding of bacterial regulons.
2.2 Selection of TF and Experiment Design
The design of a ChIP-seq experiment for regulon analysis begins with the selection of the TF and designing a strategy for its expression. The use of transcription factor-specific antibodies is critical to the success of ChIP-seq experiments. The two primary methods to study TF binding involve using antibodies specific for the native protein or antibodies to a “tag” contained within a genetically engineered tagged variant of the TF. Both types of antibodies have been used to produce high quality ChIP-seq data and the choice depends on time, cost consideration, functionality of the tagged transcription factor and genetic tractability of the organism studied.
Since commercial antibodies are available for only a small subset of native bacterial TFs, it is necessary to raise antibodies against the TF of interest, which requires additional time and expenses associated with protein purification and antibody production. It is critical to test the specificity of the antiserum by performing western blot analysis using cell lysates prepared from wild-type and from a strain lacking the TF of interest. If cross-reactivity to other proteins is detected, additional purification of the antibody should be performed using affinity chromatography with the purified TF of interest immobilized to a column or by absorbing the antiserum with an acetone powder prepared from a strain lacking the protein of interest. Both methodologies have been successfully utilized in our laboratory to yield highly specific antibody solutions [2,3].
It is possible that antibodies recognizing the native protein are not specific enough for ChIP, even after purification. Many successful ChIP-seq studies have used affinity-tagged versions of the TF of interest, often using triple FLAG affinity tags [4,5,7,8,12,15,18,21-25] or myc tags [14]. Using an affinity-tagged protein allows the use of a commercial antibody that likely has little cross-reactivity with other proteins from the bacterium under study and negates the requirement of an antibody specific to the TF of interest. However, the addition of a tag to the TF may affect its ability to bind DNA, interact with RNA Polymerase (RNAP), or be regulated by another factor (e.g. protein or small molecule). Thus, use of tagged proteins requires additional in vitro and in vivo control experiments to ensure the activity of a tagged protein is similar to that of the native protein, such as comparing expression and DNA binding of tagged TFs to native TFs. However, another requirement of using a tagged version of the TF is that the method requires the ability to introduce a stable plasmid encoding the tagged transcription factor or to edit the genome to replace the wild-type transcription factor with the tagged variant. While this is relatively simple for some bacteria, other species lack genetically tractable systems. Therefore, for these bacteria, the only option is to raise antibodies against the TF of interest.
Finally, if the TF of interest is present at a low cellular copy number or if the environmental condition that activates a TF is unknown, it may be necessary to over-express the TF in order to detect a ChIP-signal. For example, many σ factors must be released from antisigma factors for activity [1]. If the environmental signal that causes release of the σ factor is unknown, over-expression of the σ factor may allow for detection of binding sites with ChIP-seq. This method of TF overexpression was used to map TF binding sites in Mycobacterium tuberculosis because the environmental signal required for activation was unknown [22]. Such over-expression can be accomplished by engineering a plasmid where the TF gene is expressed from an inducible promoter. However, over-expression may also result in binding to weaker affinity sites on the genome that may not be bound at wild-type concentrations of the TF. Therefore, care must be taken to determine which sites are bound under physiologically relevant conditions and to show that they are dependent on the inducing signal.
2.3 Checking the ChIP Procedure
In this section, we highlight specific steps that we have found important for successful ChIP-seq experiments. The method used for immunoprecipitation of the cross-linked DNA bound transcription was described by Davis et al [33]. Since library preparation and sequencing are still expensive and can take weeks to obtain data depending on access to sequencing cores, pilot experiments to examine the efficiency of ChIP before subjecting the samples to ChIP-seq are critical. The amount of antibody to add to cultures and the duration of antibody treatment will vary depending on the affinity of the antibody to the protein of interest and should be optimized empirically. ChIP-qPCR, wherein the isolated IP and Input DNA amount is measured using qPCR can be performed on samples where the amount and incubation time of the antibody are varied to evaluate optimal conditions for enrichment. If targets of the TF are known, amplifying targets of the TF of interest, including high-affinity targets, moderate-affinity targets, and low-affinity targets as well as regions of the genome not bound by the TF of interest should provide sufficient information to optimize conditions. By comparing the enrichment by qPCR of target sites to those not bound by the TF, the efficacy and efficiency of the ChIP experiment can be evaluated [33]. However, if the TF being studied lacks known target sites, one approach is to use the promoter region of the TF gene, since autoregulation is a common feature of bacterial TFs [34]. Many potential targets may need to be evaluated to obtain reliable ChIP-qPCR results.
Several controls are useful in curating the list of TF binding regions. First, the Input DNA is used in all ChIP-seq experiments to measure the background enrichment to eliminate nonspecific binding regions. Second, comparing the enrichment signal in a control strain lacking the TF or containing an untagged version of the TF is used to reveal any non-specific enrichment due to aberrant binding of the antibody, reducing the downstream analyses of false positive areas of enrichment. Alternatively, if a mutant strain is not available, it might be possible to perform ChIP-seq under growth conditions in which the protein of interest is inactive and shows no binding activity. However, the results of this approach should be interpreted with caution as the inactive TF may still exhibit residual DNA binding. Comparison of TF binding sites between different growth conditions is discussed in more detail in Section 3.3.
The ideal number of biological replicates for ChIP-seq experiments will depend on the desired downstream analyses. High quality and reproducible data using two biological replicates has shown to be sufficient for identification of highly enriched DNA binding locations [2,3]. However, at least three biological replicates will aid in identifying less enriched regions and also will provide greater statistical power for more robust analyses, such as differential binding analyses [35,36].
2.4 Sequencing ChIP DNA Samples
After ChIP, a sequencing library is constructed from the DNA samples - DNA is fragmented and size selected and sequencing adapters are ligated to the DNA fragments - and they are sequenced using a high-throughput sequencing platform, which is often accomplished at a sequencing facility. We have had success following the Illumina’s guidelines for library preparation, which is time intensive, involving many steps and reagents. A step-by-step protocol for ChIP-seq library construction is also described in a recent review [30]. Nevertheless, some samples may require additional troubleshooting during library preparation. Although our sequencing data have been generated using the Illumina technology, much of the procedure for analysis of sequencing results is independent of the technology. Illumina sequencing can be either single-end (sequenced from one end of the DNA fragment) or paired-end (sequenced from both ends of the DNA fragment) [37]. Successful ChIP-seq experiments have been performed using both methodologies. Furthermore, multiple biological samples can be sequenced together - multiplexed - by using unique DNA sequences, termed barcodes or indexes, when constructing the sequencing library [37]. These unique barcodes or indexes allow for the identification of the sequence data from each sample present in the sequencing experiment. Additionally, multiplexing can decrease the cost of the sequencing experiment, allowing additional biological replicates to be obtained. However, if multiplexing, total sequencing depth must be high enough to ensure enough reads for every experiment for proper analysis (at least 2-3 million reads per sample).
2.5 Processing Sequencing Results
The process of analyzing ChIP-seq data is currently an evolving pipeline and, as such, there is not a single best way to evaluate the data. We provide some examples here and in the following sections, but also encourage researchers to fully evaluate each algorithm used to determine if settings should be changed to match your particular experimental design. Furthermore, ChIP-seq data is publically available in NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/) and EMBL-EBI (http://www.ebi.ac.uk/), making it easy for all researchers to re-analyze data if potentially better analysis pipelines become available.
The first step in analysis of ChIP-seq data is to evaluate the quantity and quality of the ChIP-seq sequencing reads produced from the sequencing instrument. The number of reads in each experiment reflects the levels of coverage; downstream analyses may need to be normalized for any differences in total read number. For each read, the quality is measured as confidence of the base identity (quality score) at each position, which, for Illumina sequencing data, is reported in a FASTQC file associated with each experiment (Table 1). A low quality score for reads can affect the efficiency of alignment and thus the quality of the ChIP-seq data [37]. Generally, the quality scores within reads tends to decrease at both ends of the read. This initial examination is important to determine the quality and quantity of the raw data from the sequencing experiment before processing the data for biological analysis.
Table 1.
File formats used in ChIP-seq analysis.
| File Type | Brief Description | Use in Analysis |
|---|---|---|
| FASTQ | Illumina sequencing file from experimental run |
Raw ChIP-seq Data |
|
| ||
| FASTQC | Illumina quality control file for each Illumina sequencing run |
Evaluating ChIP-seq Sequencing Data |
|
| ||
| SAM | Alignment file from Bowtie2 or BWA |
Aligned ChIP-seq File |
|
| ||
| BAM | Binary SAM file | Aligned ChIP-seq File |
|
| ||
| wiggle (WIG) | File containing results of enumerating read hits at each base location |
Visualization File |
|
| ||
| ELAND | Another alignment file format, used as input in MOSAiCS |
For Peak Calling |
There are many tools and algorithms that can be used to process ChIP-seq data for biological analysis, and a full description and evaluation of each is beyond the scope of this review. We have utilized an on-line suite of tools called Galaxy [38]. The Galaxy system (www.usegalaxy.org) [38] is an open-source, user-friendly, web-based platform for data intensive biological research of next-generation sequencing data built from commonly used algorithms and tools. It also records the processes performed at each step, allowing full recall of every step performed in analysis. Many of the software packages present in Galaxy, along with many additional software packages, can be downloaded and run by the user individually, often on the command line. We will refer to these throughout this and the next sections. Regardless of the methods used, it is very important that careful records be kept on the analysis of ChIP-seq data. This includes the version of tools used as well as any changes made to the default settings for each algorithm. These records will allow future researchers to fully understand and, if necessary, repeat your analyses.
The first step is to process the sequencing reads to remove bases of low quality. This is accomplished by selectively measuring the quality of each base read and removing (trimming) bases from the reads based on read quality [39]. When trimming, it is important to ensure that reads are of a minimum length to ensure unambiguous alignment. After trimming, the sequencing reads are aligned to a reference genome to identify the genomic location of enrichment. A high-quality reference genome is required for optimal alignment, and many reference genomes can be obtained from NCBI (http://www.ncbi.nlm.nih.gov/genome/). Two common alignment algorithms are Bowtie2 [40] and BWA [41]. Both algorithms output the alignment data as a SAM file and/or a binary version of a SAM file called a BAM file (Table 1) [42]. Regardless of the algorithm used, the primary goal is to align as many of the sequence reads as possible to the reference genome as accurately as possible to ensure reliable downstream analysis.
3: Identifying Areas of Enrichment in ChIP-seq Experiments
After sequencing reads are aligned to the genome, there are many analyses that can be performed. In the following sections, we describe examples of these analyses that have proven useful in studying bacterial regulons. However, it is important to note that while many of these analyses are common to all ChIP-seq experiments (e.g. peak calling), some analyses are required for specific TFs or biological questions.
3.1 Use of Peak Calling Algorithms
After the sequencing reads are aligned to the genome, the next step is to identify areas of enrichment in the ChIP-seq data that represent where the TF binds throughout the genome, referred to as peaks. The robust identification of peaks is crucial for the success of a ChIP-seq analysis. Peak finding algorithms are intended to identify areas of enrichment in ChIP-seq data. These algorithms are important because they remove some subjectiveness from visual peak calling and provide a statistical basis for determining areas of enrichment. Examples of algorithms that have been used for bacterial ChIP-seq analysis include CisGenome [43], MACS [44], and MOSAiCS [45]. We have used MOSAiCS, which, unlike many other peak calling algorithms, was designed to analyze ChIP-seq data from bacteria as well as eukaryotes. MOSAiCS can also identify areas of enrichment for DNA binding proteins that display broad regions of enrichment, such as RNAP or nucleoid-like proteins like H-NS [45] rather than just symmetrically shaped peaks. However, it may be useful to analyze the ChIP-seq data with multiple algorithms and compare the enriched regions identified. While this would increase the computational and analysis time for your experiment, it would also provide multiple lines of evidence for enrichment at a particular genomic region.
It is important to note that as with any algorithm, the results provided by these tools may not include all possible peaks or could contain false positives. Therefore, care must be taken to ensure proper optimization of the algorithm for your experiment. For example, adjusting the false discovery rate (FDR) of a peak-calling algorithm will affect the number of peaks identified - a lower FDR will decrease the number of both total peaks identified as well as false-positive peaks identified. Furthermore, peak calling algorithms simply identify whether an area of the genome is enriched or not based on a statistical threshold - a binary identification. Thus, many peak calling algorithms may miss peaks with low enrichment above background. Additional visual inspection may be required to identify high value peaks in a ChIP-seq dataset, especially peaks with low enrichment (Section 4). The identification of peaks of low enrichment remains a weak aspect of ChIP-seq analysis and hopefully new algorithms will be developed that improve such peak calls.
3.2 Deconvolution of ChIP-seq Peaks
Some bacterial TFs bind to multiple, closely spaced locations within a promoter region (less than 100 bp separation) [46]. The ability to distinguish between these sites yields greater mechanistic insight into the transcriptional control mediated by the TF of interest. However, most standard peak finding algorithms are unable to resolve closely spaced binding sites, instead misidentifying these regions as a single binding site with the wrong genomic coordinates. To test for closely spaced binding sites, peak deconvolution algorithms such as CSDeconv [20,47], GEM [48], PICS [49] or dPeak [46] can be used. We found that dPeak was able to identify regions containing multiple TF binding sites, most of which were missed with standard peak finding algorithms [46]. Furthermore, dPeak was designed to leverage the greater resolution provided by paired-end sequencing data to provide high-resolution ChIP-seq binding site locations in bacteria [46]. The advantage of paired-end reads for distinguishing closely spaced binding sites stems from knowing the exact length of each read, allowing a single read to be unequivocally associated with one or both of two closely spaced binding sites. In contrast, for single-end sequencing, a single read cannot be assigned with certainty to one or both binding sites since the 3’ end must be approximated, typically by extending by the average library size. Thus, reads spanning only one binding event may be wrongly extended to cover both binding sites while long reads spanning both sites may not be under extended and cover only one binding site, leading to reduced sensitivity [46]. Therefore, if it is suspected that the protein of interest may have multiple binding sites located close together, then paired-end ChIP-seq data should be obtained to maximize the advantages of the dPeak algorithm.
3.3 Comparing ChIP-seq Data Between Biological Samples
It may be of interest to compare ChIP-seq data between growth conditions to study how different environments affect binding site occupancy or to compare wild type to the control lacking the TF of interest. When comparing biological replicates from different growth conditions, the differences in total read number must be normalized across samples. However, the best strategy for normalization is yet to be determined. Normalization to the background in a ChIP-seq experiment can be performed locally (surrounding only identified peaks) or on a global scale (normalize to all un-enriched genomic regions) [2]. Alternatively, spiking the DNA sample with synthetic DNA fragments will provide external reference sequences of known amounts for normalization [50].
After normalization, samples can then be compared for differential binding by the transcription factor. Due to the symmetrical, peak shape of enriched regions from most TFs, differential binding can be assessed by comparing the number of sequencing reads aligned around the peak summit. When comparing binding of other proteins, such as H-NS, which shows long regions of binding across the genome, it is better to compare the sequencing coverage under the entire enriched region [2]. In our studies, we have used the algorithm DBChIP [35], which was developed specifically to compare ChIP-seq enrichment of TF peaks between two experimental conditions, with multiple biological replicates for each condition. For example, we successfully used DBChip to identify oxygen-dependent differences in σ70 binding by analyzing σ70 ChIP-seq signal in E. coli under aerobic and anaerobic growth conditions (Figure 2) [2]. It is important to note that differential binding analysis is appropriate when comparing enrichment of the same site under different conditions. In contrast, we have found that ChIP-seq enrichment does not necessarily correlate with binding strength [2], thus preventing comparison of sites within the genome. Many factors likely determine the intensity of peaks across genomic sites, such as inherent cross-linking efficiency within a region of DNA [2]. We assume that these factors should remain constant across conditions.
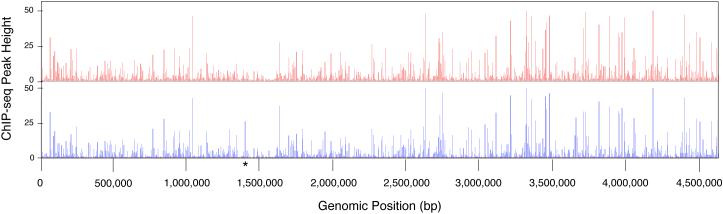
Figure 2. Comparing ChIP-seq data between growth conditions.
Shown are RNAP (σ70) ChIP-seq data traces collected from cultures grown under aerobic (red) or anaerobic (blue) conditions [2]. ChIP-seq IP/INPUT ratio is shown on the y-axis and genomic position is shown on the x-axis. The asterisk indicates an example of differential binding between growth conditions. This figure was generated in the MochiView browser [53].
4: Visualization of ChIP-seq Data
4.1 Importance of Visually Evaluating ChIP-seq Data
We have found that visual inspection is an important part of analyzing ChIP-seq data. Visual inspection is generally required to identify peaks with low enrichment above background, which current peak calling algorithms are unable to identify without introducing many false positives. Such low enrichment may be due to poor cross-linking efficiency or low sequencing coverage at that location in the genome. However, visual analysis of ChIP-seq data lacks statistical support and thus additional peaks identified only visually should be subjected to additional validation, which is discussed in Section 7.3. Visualization can also be useful in identifying false positive peaks. Canonical TF binding sites yield a bimodal enrichment profile where enrichment on the forward and reverse strands are staggered as a result of sequencing the 5′ portion of each DNA fragment [51,52]. Artifactual peaks typically lack this staggered peak signature exhibiting reads specific to only one strand, and thus often can be readily identified through visualization of reads mapping to both the forward and reverse strand. Algorithms such as QuEST [51] compares the enrichment profiles of both strands and can be used to filter out such artifactual peaks. Visual inspection of the data may also elucidate other trends that may have biological significance such as long binding regions, which might be overlooked when using the algorithmic results alone. Finally, visually evaluating ChIP-seq data can be important when considering the physiological context of binding events. The best way to visually inspect the data is to plot the ChIP-seq data on a genome browser and step through the genome.
4.2 Preparing Files for Visualization
The most common file format used for visualization of ChIP-seq data is the wiggle (or WIG) file (Table 1). Wiggle files are designed to display information dense data, such as ChIP-seq data throughout the genome. These files are generated by enumerating how often each read corresponds to each base in the genome. A composite wig file that incorporates data from both the forward and reverse strands is typically generated for visualization of ChIP-seq data; however, wig files for both the forward and reverse strand are useful for visualizing the staggered peak profiles described above. To generate composite wig files, we use the software package QuEST [51], that applies a peak shift estimation to generate a composite density profile for each enriched region. In addition to the wiggle file, visualization depends on an annotated genome. The more robust the annotation of the genome file, the more useful it will be for visualization analysis.
4.3 Visualization Using MochiView
There are many genome browsers available that can accept wiggle files and visualize ChIP-seq data. We use the genome browser MochiView [53] (Figure 2). MochiView was developed for use with yeast data, but functions very well to visualize multiple data types, including ChIP-seq, on a bacterial genome. MochiView is a fast browser that can accept many tracks of data without affecting performance. It is also highly customizable, allowing for easy generation of publication ready figures. All data are stored in a database in MochiView, which can be exported and shared, allowing easy transfer of data with other researchers.
In addition to visualizing ChIP-seq data, MochiView allows for easy import of ChIP-chip data, transcriptomic data, proteomic data, motif location data, and any other gene centric data set, allowing for simple comparison between multiple data types. It can also import the results of peak finding algorithms, deconvolution algorithms, and differential binding analysis. Finally, MochiView is written in Java, and therefore is operating system agnostic and runs equally well in Windows and Mac OS environments. One drawback of MochiView is that it is installed and run locally on your computer. So while you can easily share databases among users, MochiView does not lend itself to visualization via a web browser or server based method, where many users simultaneously use the same database. However, this drawback aside, MochiView has proven a robust and powerful genome browser.
4.4 Visualization Online
While local visualization with MochiView is our preferred way of analyzing ChIP-seq data, it is often advantageous to have a shared browser that anyone with a web browser can access. A commonly used genome browser is the UCSC Genome Browser (http://genome.ucsc.edu/index.html) [54]. This is built on the GBrowse genome browser [55], but has many tracks of annotation pre-loaded, making it easy to visualize ChIP-seq data quickly. Additionally, published tracks are available to be loaded into the browser, providing a platform to compare between different experiments from different research groups.
It is also possible to download and install an instance of GBrowse or another popular browser JBrowse [56] on a local server for which access can be controlled [55]. Tracks can be downloaded easily from UCSC for use in a personal instance of GBrowse. This can allow many users to access the same data and add to the data in a central location.
5: Associating ChIP-seq Peaks with Genes
After binding regions are identified, the next step in ChIP-seq analysis is to associate the peaks with the corresponding genes. Ideally, the peak could be algorithmically associated with genes based on proximity of the peak to the transcription start site (TSS). However, many genes in E. coli, the organism that we study, lack known transcription start sites, even with recent studies that have globally identified TSSs [57,58]. The translation start site, which is predicted for all known E. coli protein coding genes, can also be used in such an automated algorithm, but this may produce many false associations because the distance from the translation start site and the TSS is variable. Furthermore, such an automated analysis may ignore transcription start sites that are either far upstream of the translation start site or are located within genes or encode small RNAs.
We use visual inspection of each area of enrichment to determine what is the nearest operon as the most accurate way to ensure each peak is best associated with the corresponding operon. Such visual association also ensures that divergent promoters are identified and that sites located within genes are not overlooked. However, for a peak located within a region with divergently transcribed genes, it is impossible to associate the peak with either operon using ChIP-seq data alone, but the use of expression data can suggest which gene is regulated by the TF (Section 7). This will also potentially identify ChIP-seq peaks that do not associate with annotated genes, but may associate with known or unknown small RNAs [2].
6: Motif Analysis Using ChIP-seq Data
6.1 Value of ChIP-seq Data in Identification of Motifs
Since ChIP-seq data provides high-resolution (~10 bp [37]), location information of TF binding sites, the sequences bound by the TF can be analyzed to identify over-represented sequences. Many of motifs present in the literature have been generated from a small number of binding sites. The use of ChIP-seq data provides dozens to hundreds of binding site sequences, improving the power of motif identification. Updated motifs can aid in identifying false negative binding regions of the genome and can be used for further downstream biochemical and genetic experiments to examine binding efficiency [2].
6.2 Motif Identification from ChIP-seq Data
Many algorithms have been developed to search a given set of sequences and identify over-represented motifs. Regardless of the algorithm used, all require the sequences extracted from areas of enrichment. These areas may be the entire area of enrichment identified by MOSAiCS or specific genomic coordinates from a deconvoluted peak. Once the DNA sequences are obtained, they are submitted to an algorithm to search for a motif [59]. A commonly used algorithm is MEME [60] - or the related MEME-chip, which is designed for larger sequence inputs [61] - which can be run either online or locally via the command line [62]. In addition to MEME, other motif identification algorithms include info-gibbs [63] and consensus [64] which can be easily accessed at the Regulatory Sequence Analysis Tools (RSAT, http://rsat.ulb.ac.be) [65]. Regardless of the algorithm used, ChIP-seq data provide a valuable resource to better define the binding site sequences of transcription factors. Once identified, the location of motifs within the peak region can be measured using CentriMo [66]. The ease of binning ChIP-seq data and identifying motifs can allow for interesting biological comparisons, such as potential differences in motifs due to differential enrichment levels under various experimental conditions. It is important to note that, like all computational algorithms, motif-finding algorithms will always return results, even if those results are not biologically significant. Care must be taken to use the statistical results (E-value in MEME) to evaluate the results of these algorithms.
6.3 Searching Motifs Throughout the Genome and Within Peaks
After a motif has been identified, it is informative to search each area of enrichment for the occurrence of the motif to more accurately define the location of the TF binding site. This is done using a position weight matrix (PWM) that describes the frequency distribution of nucleotides at each position of the binding motif and is a common output of the aforementioned motif finders. In some cases, multiple binding sites may occur within a single enriched region. One significant challenge is the stringency of the cutoff to use in motif searches. Stringency parameters can be verified by coupling the results of the search with true binding sites identified from DNase I Footprinting or Electrophoretic Mobility Shift Assays (EMSA) experiments.
Further, the motif can be used to search the genome to identify potential binding sites missed by ChIP-seq analysis. Although motif searches have a high false positive rate, we have found that this analysis can provide important information regarding false negatives in the ChIP-seq data, that is locations with a high value motif but lacking a corresponding ChIP-seq peak [2]. Such locations have been shown to be predicted binding sites of TFs that are blocked by the binding of other proteins, such as nucleoid proteins H-NS [2]. There are many algorithms that can search a genome for motif binding, including MAST [67], PatSer [68], and the Delila suite [69]. We find the Delila software suite quite useful for fast, easily tunable genomic searches with motifs. A particularly useful and unique feature of the Delila suite is the ability to visualize graphically how a particular sequence was evaluated by a PWM using sequence walkers [69].
7: Correlation of ChIP-seq Data with Expression Data
7.1 Identification of Direct and Indirect Regulons in Bacteria
Because ChIP-seq data provides information about where transcription factors bind in vivo, correlation with expression data is crucial to identify regulons in bacteria. The source of expression data can be either from whole genome microarray analyses, RNA-seq analyses, or a combination of both. Many studies have demonstrated that correlation of a global transcription factor DNA binding with expression profiles of cells lacking that same TF compared to wild-type cells reveals important information about the regulon of the transcription factor [2,3,5,7,8,13,16,21,22,25]. Importantly, these studies allow for the global identification of the direct regulon - those operons that show a change in expression when the transcription factor is deleted and also have a corresponding transcription factor ChIP-seq peak upstream - and the indirect regulon - those operons that show a change in expression when the transcription factor is deleted but do not have a corresponding transcription factor ChIP-seq peak upstream. Correlation of ChIP-seq data with expression data also reveals the number of operons activated and repressed by the TF on a global scale. Along with other data, this type of analysis can reveal important features of transcription regulation and how bacteria respond to different environmental stimuli.
7.2 Visualizing Regulon Data on Metabolic Pathways
Once ChIP-seq and expression data are correlated, visualization of the data on metabolic pathways is a useful strategy to gain a physiological understanding of the TF’s function. Most visualization tools are gene centric, but can be used with ChIP-seq data once peaks are associated with genes. In a typical example, ChIP-seq occupancy and expression data are associated with each gene in the pathway [2]. The pathway tools function of MetaCyc (www.metacyc.org) and the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) are excellent tools for an initial genomic overview of the data [70-72]. Both MetaCyc and KEGG contain databases of gene and pathway information for many species and allow the simultaneous mapping of -omics data to annotated metabolic reactions within the cell. For E. coli there are specific on-line databases from which gene and pathway information can be obtained including EcoCyc [73], PortEco [74], and EcoGene [75]. Additionally, the metabolic pathways can be downloaded from the user-generated site WikiPathways [76]. To generate custom metabolic pathways, ProMetRa [77], GenMapp [78], or PathVisio [79] are good choices. However, it is important to ensure proper representation of the pathway and any specific changes that exist in the bacterial species being examined.
7.3 Validation of Regulon Analysis
While validation of all components of the direct and indirect regulon might not yet be realistic, it is valuable to complement the conclusions of regulon analysis using in vitro or in vivo methods. There are a variety of methods that can be used and here we will describe some common examples. Any well-established in vitro DNA binding assay (e.g. EMSA , DNase I footprinting, fluorescence polarization) can, in principle, be used to support TF binding to a specific sequence of DNA that was enriched in ChIP-seq experiments. DNAse I footprinting is advantageous because it determines the boundaries of the transcription factor binding site and is, therefore, very informative for evaluating or optimizing binding motif analysis [3]. However it is important to mention that the failure to observe binding in vitro does not rule out binding in vivo, and could signify an interesting regulatory mechanism. For example, some binding events require cooperative interactions with other TFs [2,80] and/or may require specific cellular conditions that are difficult to replicate in vitro.
To support direct transcriptional regulation, in vitro transcription assays or in vivo measurements of reporter gene fusions to the promoter of interest (e.g. ß-galactosidase activity assays or green fluorescent protein) or RNA levels by qRT-PCR can be performed with wild-type strains and strains that contain deletions or mutations of various TFs. These assays can be particularly informative for evaluating the effects of various growth conditions on transcriptional regulation of a particular operon. In nearly every regulon analysis study to date, a significant number of ChIP-seq binding sites are located upstream of genes that fail to exhibit differential expression under the growth conditions used in transcriptomic studies, leaving the biological function of the binding site unknown. In some cases, alternative growth conditions or the absence/inactivation of another TF may be required to observe an effect [2,3].
8: Summary and Perspectives
Here, we have described an overview of obtaining and analyzing ChIP-seq data. ChIP-seq analysis in bacteria is a powerful method of identifying in vivo binding sites of transcription factors and their regulons. To date, several studies have combined ChIP-seq data and expression data to gain a deep understanding of the regulon of transcription factors in multiple bacterial species grown under a small number of growth conditions. For example, we can combine data from our lab studying the oxygen or iron responsive TFs FNR, ArcA, IscR, and Fur to better define the regulation of oxygen-dependent gene expression in E. coli (Figure 3). As the cost of high-throughput sequencing decreases and more researchers have access to the technology, we are hopeful that more TF regulons will be defined in bacteria, providing new and exciting insights into bacterial regulons in a variety of species and growth conditions.
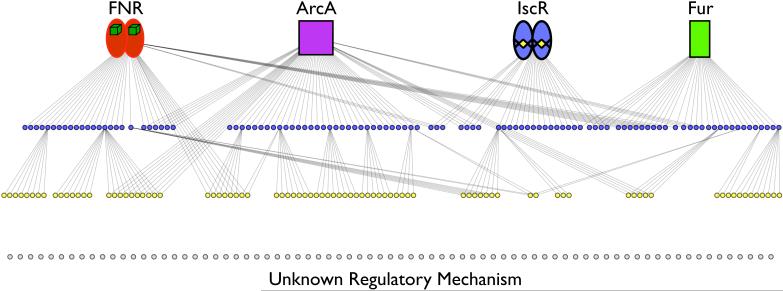
Figure 3. Example of combining bacterial regulons to better understand gene regulation in response to oxygen.
Shown is the proposed regulatory role of FNR (red ovals) [2], ArcA (purple square) [3], IscR (blue ovals) (Unpublished Data), and Fur (green rectangle) (Unpublished Data) on operons (blue, yellow, and gray circles) with a significant change in expression based on the presence or absence of O2. Lines indicate a regulatory role of a TF on expression from ChIP-chip/ChIP-seq and RNA expression data, either direct (blue circles) or indirect (yellow circles). Approximately 60% of the operons with an O2-dependent change in expression are regulated by FNR, ArcA, IscR, or Fur. The remaining 40% (gray circles) are regulated by other mechanisms not currently understood.
Highlights.
ChIP-seq is a powerful method for identifying transcription factor binding in vivo
Correlation of ChIP-seq and transcriptomic data can identify bacterial regulons
An overview of ChIP-seq methodology and data analysis in bacteria is provided
ChIP-seq sample preparation, data analysis, and computational analyses are reviewed
Acknowledgements
This work was funded by a grant from the NIH to PJK (GM045844). NAB was supported by the UW-Madison NIH Chemistry Biology Interface Training Grant (T32GM008505). This work was also funded in part by the DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kevin S. Myers, Laboratory of Genetics, University of Wisconsin – Madison; Madison, WI; USA; 53706 Great Lakes Bioenergy Research Center; University of Wisconsin – Madison; Madison, WI; USA; 53706.
Dan M. Park, Lawrence Livermore National Laboratory, Livermore, California, USA
Nicole A. Beauchene, Department of Biomolecular Chemistry; University of Wisconsin – Madison; Madison, WI; USA; 53706
Patricia J. Kiley, Department of Biomolecular Chemistry; University of Wisconsin – Madison; Madison, WI; USA; 53706 Great Lakes Bioenergy Research Center; University of Wisconsin – Madison; Madison, WI; USA; 53706.
References
- [1].Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat Rev Micro. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- [2].Myers KS, Yan H, Ong IM, Chung D, Liang K, Tran F, et al. Genome-scale analysis of Escherichia coli FNR reveals complex features of transcription factor binding. PLoS Genetics. 2013;9:e1003565. doi: 10.1371/journal.pgen.1003565. doi:10.1371/journal.pgen.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Park DM, Akhtar MS, Ansari AZ, Landick R, Kiley PJ. The bacterial response regulator ArcA uses a diverse binding site architecture to regulate carbon oxidation globally. PLoS Genetics. 2013;9:e1003839. doi: 10.1371/journal.pgen.1003839. doi:10.1371/journal.pgen.1003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Haycocks JRJ, Sharma P, Stringer AM, Wade JT, Grainger DC. The molecular basis for control of ETEC enterotoxin expression in response to environment and host. PLoS Pathog. 2015;11:e1004605. doi: 10.1371/journal.ppat.1004605. doi:10.1371/journal.ppat.1004605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kahramanoglou C, Seshasayee ASN, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, et al. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 2011;39:2073–2091. doi: 10.1093/nar/gkq934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Singh SS, Singh N, Bonocora RP, Fitzgerald DM, Wade JT, Grainger DC. Widespread suppression of intragenic transcription initiation by H-NS. Genes Dev. 2014;28:214–219. doi: 10.1101/gad.234336.113. doi:10.1101/gad.234336.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Prieto AI, Kahramanoglou C, Ali RM, Fraser GM, Seshasayee ASN, Luscombe NM. Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res. 2012;109:3524–3537. doi: 10.1093/nar/gkr1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Brown DR, Barton G, Pan Z, Buck M, Wigneshweraraj S. Nitrogen stress response and stringent response are coupled in Escherichia coli. Nat Comms. 2014;5:4115. doi: 10.1038/ncomms5115. doi:10.1038/ncomms5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fioravanti A, Fumeaux C, Mohapatra SS, Bompard C, Brilli M, Frandi A, et al. DNA binding of the cell cycle transcriptional regulator GcrA depends on N6-adenosine methylation in Caulobacter crescentus and other Alphaproteobacteria. PLoS Genetics. 2013;9:e1003541. doi: 10.1371/journal.pgen.1003541. doi:10.1371/journal.pgen.1003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fumeaux C, Radhakrishnan SK, Ardissone S, Théraulaz L, Frandi A, Martins D, et al. Cell cycle transition from S-phase to G1 in Caulobacter is mediated by ancestral virulence regulators. Nat Comms. 2014;5:4081. doi: 10.1038/ncomms5081. doi:10.1038/ncomms5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Solans L, Gonzalo-Asensio J, Sala C, Benjak A, Uplekar S, Rougemont J, et al. The PhoP-dependent ncRNA Mcr7 modulates the TAT secretion system in Mycobacterium tuberculosis. PLoS Pathog. 2014;10:e1004183. doi: 10.1371/journal.ppat.1004183. doi:10.1371/journal.ppat.1004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Perkins TT, Davies MR, Klemm EJ, Rowley G, Wileman T, James K, et al. ChIP-seq and transcriptome analysis of the OmpR regulon of Salmonella enterica serovars Typhi and Typhimurium reveals accessory genes implicated in host colonization. Mol Microbiol. 2013;87:526–538. doi: 10.1111/mmi.12111. doi:10.1111/mmi.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jones CJ, Newsom D, Kelly B, Irie Y, Jennings LK, Xu B, et al. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 2014;10:e1003984. doi: 10.1371/journal.ppat.1003984. doi:10.1371/journal.ppat.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Imam S, Noguera DR, Donohue TJ. Global analysis of photosynthesis transcriptional regulatory networks. PLoS Genetics. 2014;10:e1004837. doi: 10.1371/journal.pgen.1004837. doi:10.1371/journal.pgen.1004837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Crack JC, Munnoch J, Dodd EL, Knowles F, Al Bassam MM, Kamali S, et al. NsrR from Streptomyces coelicolor is a Nitric Oxide-Sensing [4Fe-4S] Cluster Protein with a Specialized Regulatory Function. J Biol Chem. 2015 doi: 10.1074/jbc.M115.643072. doi:10.1074/jbc.M115.643072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Davies BW, Bogard RW, Mekalanos JJ. Mapping the regulon of Vibrio cholerae ferric uptake regulator expands its known network of gene regulation. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1107894108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dong TG, Mekalanos JJ. Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res. 2012;40:7766–7775. doi: 10.1093/nar/gks567. doi:10.1093/nar/gks567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].van Kessel JC, Ulrich LE, Zhulin IB, Bassler BL. Analysis of activator and repressor functions reveals the requirements for transcriptional control by LuxR, the master regulator of quorum sensing in Vibrio harveyi. mBio. 2013;4 doi: 10.1128/mBio.00378-13. doi:10.1128/mBio.00378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Blasco B, Chen JM, Hartkoorn R, Sala C, Uplekar S, Rougemont J, et al. Virulence regulator EspR of Mycobacterium tuberculosis is a nucleoid-associated protein. PLoS Pathog. 2012;8:e1002621. doi: 10.1371/journal.ppat.1002621. doi:10.1371/journal.ppat.1002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lun DS, Sherrid A, Weiner B, Sherman DR, Galagan JE. A blind deconvolution approach to high-resolution mapping of transcription factor binding sites from ChIP-seq data. Genome Biol. 2009;10:R142. doi: 10.1186/gb-2009-10-12-r142. doi:10.1186/gb-2009-10-12-r142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Minch KJ, Rustad TR, Peterson EJR, Winkler J, Reiss DJ, Ma S, et al. The DNA-binding network of Mycobacterium tuberculosis. Nat Comms. 2015;6:5829. doi: 10.1038/ncomms6829. doi:10.1038/ncomms6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Galagan JE, Minch K, Peterson M, Lyubetskaya A, Azizi E, Sweet L, et al. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature. 2013;499:178–183. doi: 10.1038/nature12337. doi:10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fitzgerald DM, Bonocora RP, Wade JT. Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genetics. 2014;10:e1004649. doi: 10.1371/journal.pgen.1004649. doi:10.1371/journal.pgen.1004649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Petrone BL, Stringer AM, Wade JT. Identification of HilD-regulated genes in Salmonella enterica serovar Typhimurium. J Bacteriol. 2014;196:1094–1101. doi: 10.1128/JB.01449-13. doi:10.1128/JB.01449-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Stringer AM, Currenti S, Bonocora RP, Baranowski C, Petrone BL, Palumbo MJ, et al. Genome-scale analyses of Escherichia coli and Salmonella enterica AraC reveal noncanonical targets and an expanded core regulon. J Bacteriol. 2014;196:660–671. doi: 10.1128/JB.01007-13. doi:10.1128/JB.01007-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cho S, Cho YB, Kang TJ, Kim SC, Palsson B, Cho BK. The architecture of ArgR-DNA complexes at the genome-scale in Escherichia coli. Nucleic Acids Res. 2015;43:3079–3088. doi: 10.1093/nar/gkv150. doi:10.1093/nar/gkv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Carraro N, Matteau D, Luo P, Rodrigue S, Burrus V. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genetics. 2014;10:e1004714. doi: 10.1371/journal.pgen.1004714. doi:10.1371/journal.pgen.1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Poulin-Laprade D, Matteau D, Jacques P-É, Rodrigue S, Burrus V. Transfer activation of SXT/R391 integrative and conjugative elements: unraveling the SetCD regulon. Nucleic Acids Res. 2015;43:2045–2056. doi: 10.1093/nar/gkv071. doi:10.1093/nar/gkv071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Seo SW, Kim D, Latif H, O'Brien EJ, Szubin R, Palsson BØ. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat Comms. 2014;5:4910. doi: 10.1038/ncomms5910. doi:10.1038/ncomms5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bonocora RP, Wade JT. ChIP-Seq for Genome-Scale Analysis of Bacterial DNA-Binding Proteins. Methods Mol Biol. 2015;1276:327–340. doi: 10.1007/978-1-4939-2392-2_20. doi:10.1007/978-1-4939-2392-2_20. [DOI] [PubMed] [Google Scholar]
- [31].Galagan J, Lyubetskaya A, Gomes A. ChIP-Seq and the complexity of bacterial transcriptional regulation. Curr. Top. Microbiol. Immunol. 2013;363:43–68. doi: 10.1007/82_2012_257. doi:10.1007/82_2012_257. [DOI] [PubMed] [Google Scholar]
- [32].Jaini S, Lyubetskaya A, Gomes A, Peterson M, Park ST. Transcription Factor Binding Site Mapping Using ChIP-Seq. 2014 doi: 10.1128/microbiolspec.MGM2-0035-2013. doi:10.1128/microbiolspec.MGM2-0035-2013. [DOI] [PubMed] [Google Scholar]
- [33].Davis SE, Mooney RA, Kanin EI, Grass J, Landick R, Ansari AZ. Mapping E. coli RNA Polymerase and associated transcription factors and identifying promoters genome-wide. Meth Enzymol. 2011;498:449–471. doi: 10.1016/B978-0-12-385120-8.00020-6. [DOI] [PubMed] [Google Scholar]
- [34].Thieffry D, Huerta AM, Pérez-Rueda E, Collado-Vides J. From specific gene regulation to genomic networks: a global analysis of transcriptional regulation in Escherichia coli. Bioessays. 1998;20:433–440. doi: 10.1002/(SICI)1521-1878(199805)20:5<433::AID-BIES10>3.0.CO;2-2. doi:10.1002/(SICI)1521-1878(199805)20:5<433::AID-BIES10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- [35].Liang K, Keleş S. Detecting differential binding of transcription factors with ChIP-seq. Bioinformatics. 2012;28:121–122. doi: 10.1093/bioinformatics/btr605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liang K, Keleş S. Normalization of ChIP-seq data with control. BMC Bioinformatics. 2012;13:199. doi: 10.1186/1471-2105-13-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Goecks J, Nekruenko A, Talor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:1–38. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. doi:10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. doi:10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang Y, Liu T, Meyer C, Eeckhoute J, Johnson D, Bernstein B, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chung D, Kuan PF, Keleş S. Analysis of ChIP-seq Data with “mosaics” Package. 2012 [Google Scholar]
- [46].Chung D, Park D, Myers K, Grass J, Kiley P, Landick R, et al. dPeak: High resolution identification of transcription factor binding sites from PET and SET ChIP-Seq data. PLoS Comput Biol. 2013;9:e1003246. doi: 10.1371/journal.pcbi.1003246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gomes ALC, Abeel T, Peterson M, Azizi E, Lyubetskaya A, Carvalho L, et al. Decoding ChIP-seq with a double-binding signal refines binding peaks to single-nucleotides and predicts cooperative interaction. Genome Res. 2014;24:1686–1697. doi: 10.1101/gr.161711.113. doi:10.1101/gr.161711.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Guo Y, Mahony S, Gifford DK. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput Biol. 2012;8:e1002638. doi: 10.1371/journal.pcbi.1002638. doi:10.1371/journal.pcbi.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zhang X, Robertson G, Krzywinski M, Ning K, Droit A, Jones S, et al. PICS: Probabilistic Inference for ChIP-seq. Biometrics. 2010;67:151–163. doi: 10.1111/j.1541-0420.2010.01441.x. doi:10.1111/j.1541-0420.2010.01441.x. [DOI] [PubMed] [Google Scholar]
- [50].Bonhoure N, Bounova G, Bernasconi D, Praz V, Lammers F, Canella D, et al. Quantifying ChIP-seq data: a spiking method providing an internal reference for sample-to-sample normalization. Genome Res. 2014;24:1157–1168. doi: 10.1101/gr.168260.113. doi:10.1101/gr.168260.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wilbanks E, Facciotti M. Evaluation of algorithm performance in ChIP-seq peak detection. PLoS ONE. 2010 doi: 10.1371/journal.pone.0011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Homann OR, Johnson AD. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol. 2010;8:49. doi: 10.1186/1741-7007-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. doi:10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. doi:10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–1638. doi: 10.1101/gr.094607.109. doi:10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kim D, Hong JS-J, Qiu Y, Nagarajan H, Seo J-H, Cho B-K, et al. Comparative analysis of regulatory elements between Escherichia coli and Klebsiella pneumoniae by genome-wide transcription start site profiling. PLoS Genetics. 2012;8:e1002867. doi: 10.1371/journal.pgen.1002867. doi:10.1371/journal.pgen.1002867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Conway T, Creecy JP, Maddox SM, Grissom JE, Conkle TL, Shadid TM, et al. Unprecedented high-resolution view of bacterial operon architecture revealed by RNA sequencing. mBio. 2014;5:e01442–14. doi: 10.1128/mBio.01442-14. doi:10.1128/mBio.01442-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Das MK, Dai H-K. A survey of DNA motif finding algorithms. BMC Bioinformatics. 2007;8(Suppl 7):S21. doi: 10.1186/1471-2105-8-S7-S21. doi:10.1186/1471-2105-8-S7-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–73. doi: 10.1093/nar/gkl198. doi:10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. doi:10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Defrance M, van Helden J. info-gibbs: a motif discovery algorithm that directly optimizes information content during sampling. Bioinformatics. 2009;25:2715–2722. doi: 10.1093/bioinformatics/btp490. doi:10.1093/bioinformatics/btp490. [DOI] [PubMed] [Google Scholar]
- [64].Hertz GZ, Hartzell GW, Stormo GD. Identification of consensus patterns in unaligned DNA sequences known to be functionally related. Comput. Appl. Biosci. 1990;6:81–92. doi: 10.1093/bioinformatics/6.2.81. [DOI] [PubMed] [Google Scholar]
- [65].Thomas-Chollier M, Defrance M, Medina-Rivera A, Sand O, Herrmann C, Thieffry D, et al. RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res. 2011;39:W86–91. doi: 10.1093/nar/gkr377. doi:10.1093/nar/gkr377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Bailey TL, Machanick P. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 2012;40:e128. doi: 10.1093/nar/gks433. doi:10.1093/nar/gks433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. doi:10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- [69].Schneider TD, Stormo GD, Yarus MA, Gold L. Delila system tools. Nucleic Acids Res. 1984;12:129–140. doi: 10.1093/nar/12.1part1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2014;42:D459–71. doi: 10.1093/nar/gkt1103. doi:10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. doi:10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–205. doi: 10.1093/nar/gkt1076. doi:10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muñiz-Rascado L, et al. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39:D583–90. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Hu JC, Sherlock G, Siegele DA, Aleksander SA, Ball CA, Demeter J, et al. PortEco: a resource for exploring bacterial biology through high-throughput data and analysis tools. Nucleic Acids Res. 2014;42:D677–84. doi: 10.1093/nar/gkt1203. doi:10.1093/nar/gkt1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Zhou J, Rudd KE. EcoGene 3.0. Nucleic Acids Res. 2013;41:D613–24. doi: 10.1093/nar/gks1235. doi:10.1093/nar/gks1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:e184. doi: 10.1371/journal.pbio.0060184. doi:10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Neuweger H, Persicke M, Albaum SP, Bekel T, Dondrup M, Hüser AT, et al. Visualizing post genomics data-sets on customized pathway maps by ProMeTra - aeration-dependent gene expression and metabolism of Corynebacterium glutamicum as an example. BMC Syst Biol. 2009;3:82. doi: 10.1186/1752-0509-3-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. doi:10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- [79].Kutmon M, van Iersel MP, Bohler A, Kelder T, Nunes N, Pico AR, et al. PathVisio 3: an extendable pathway analysis toolbox. PLoS Comput Biol. 2015;11:e1004085. doi: 10.1371/journal.pcbi.1004085. doi:10.1371/journal.pcbi.1004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Lee DJ, Minchin SD, Busby SJW. Activating transcription in bacteria. Annu Rev Microbiol. 2012;66:125–152. doi: 10.1146/annurev-micro-092611-150012. doi:10.1146/annurev-micro-092611-150012. [DOI] [PubMed] [Google Scholar]