NOS1−/− mice show reduced inflammatory responses and tissue damage in experimental sepsis models. Baig et al show that NOS1-derived NO production in macrophages leads to proteolysis of SOCS1 to alleviate its repression of NFkB transcriptional activity in response to TLR4-mediated responses.
Abstract
The NF-κB pathway is central to the regulation of inflammation. Here, we demonstrate that the low-output nitric oxide (NO) synthase 1 (NOS1 or nNOS) plays a critical role in the inflammatory response by promoting the activity of NF-κB. Specifically, NOS1-derived NO production in macrophages leads to proteolysis of suppressor of cytokine signaling 1 (SOCS1), alleviating its repression of NF-κB transcriptional activity. As a result, NOS1−/− mice demonstrate reduced cytokine production, lung injury, and mortality when subjected to two different models of sepsis. Isolated NOS1−/− macrophages demonstrate similar defects in proinflammatory transcription on challenge with Gram-negative bacterial LPS. Consistently, we found that activated NOS1−/− macrophages contain increased SOCS1 protein and decreased levels of p65 protein compared with wild-type cells. NOS1-dependent S-nitrosation of SOCS1 impairs its binding to p65 and targets SOCS1 for proteolysis. Treatment of NOS1−/− cells with exogenous NO rescues both SOCS1 degradation and stabilization of p65 protein. Point mutation analysis demonstrated that both Cys147 and Cys179 on SOCS1 are required for its NO-dependent degradation. These findings demonstrate a fundamental role for NOS1-derived NO in regulating TLR4-mediated inflammatory gene transcription, as well as the intensity and duration of the resulting host immune response.
Orchestration of the severity, duration, and location of inflammation is critical to achieving sterilizing immunity while minimizing tissue injury. The NF-κB pathway is central to establishing this balance. NF-κB promotes transcription of genes involved in every aspect of the immune response, from differentiation and homeostatic regulation of immune cells, to modulation of barrier function and leukocyte recruitment during acute activation and the deployment of immune effector mechanisms that mediate antimicrobial activity (Xia et al., 1997; Badrichani et al., 1999; Jeon et al., 1999; Alcamo et al., 2001; Tiruppathi et al., 2008; Jacobson and Birukov, 2009; Lawrence, 2009; Baltimore, 2011; Li et al., 2011; Pittet et al., 2011; Ruland, 2011; Sadik et al., 2011; Fieren, 2012; Koelink et al., 2012; Summers deLuca and Gommerman, 2012; Sun and Karin, 2012; Takizawa et al., 2012). Disrupted NF-κB signaling leads to defective pathogen clearance, autoimmunity, and tissue injury (Bowie and O’Neill, 2000; Javaid et al., 2003; Aktan, 2004; Dixon, 2004; Block and Hong, 2005; Parsons et al., 2005; Dombrovskiy et al., 2007; Panettieri et al., 2008; Jacobson and Birukov, 2009; Liu, 2011; Nacher and Hidalgo, 2011; Rossi et al., 2011; Ruland, 2011; Edgley et al., 2012; Ma and Malynn, 2012).
NF-κB signaling is complex, leading to multiple distinct transcription complexes whose formation and stability are known to be regulated by a wide array of protein modifications, including phosphorylation, ubiquitination, and glutathiolation, as well as modifications by reactive oxygen and nitrogen species (Bowie and O’Neill, 2000; Marshall and Stamler, 2001; Ben-Neriah, 2003; Bubici et al., 2006; Mattioli et al., 2006; Nicholas et al., 2007; Geng et al., 2009; Lin et al., 2012; Sabatel et al., 2012). Nitric oxide (NO) has long been appreciated to promote feedback inhibition of NF-κB because of the important role of the high output NO synthase 2 (NOS) in inflammation. However, mammals encode three distinct NOS isozymes: neuronal (nNOS or NOS1), inducible (iNOS or NOS2), and endothelial (eNOS or NOS3; Knowles and Moncada, 1994). To determine the precise roles for each of these in inflammatory signaling, we analyzed animals with targeted mutations in each of the three NOS isoforms in two models of sepsis. We demonstrate that, unique among the three NOS enzymes, NO produced by the low-output NOS1 plays a critical role in promoting NF-κB–mediated proinflammatory cytokine transcription in animals and in isolated macrophages.
The canonical NF-κB complex is comprised of p65 and p50 subunits held inactive in the cytoplasm by IκB. The targeted proteolysis of IκB results in nuclear translocation of the p65:p50 complex, and subsequent transcriptional activation. IκB degradation is promoted by a large number of proinflammatory signaling pathways, and its preservation is the target of many antiinflammatory pathways. (Jacobs and Harrison, 1998; Vanden Berghe et al., 1999; Basak et al., 2007; Baltimore, 2011; Ruland, 2011). We therefore determined its status in the NOS1−/− macrophages challenged with LPS, but found no defect in IκB degradation. Instead, we found that p65 protein levels were not maintained in the knockout cells. Suppressor of cytokine signaling (SOCS1) has been reported to promote the degradation of DNA-bound p65 protein leading to the suppression of NF-κB activity and inflammation (Kinjyo et al., 2002; Nakagawa et al., 2002; Ben-Neriah, 2003; Park et al., 2003; Ryo et al., 2003; Baetz et al., 2004; Gingras et al., 2004; Strebovsky et al., 2011; Linossi and Nicholson, 2012). We show that NOS1-derived NO promotes the S-nitrosation of SOCS1, and that this ablates its capacity to bind and target p65 for degradation. Therefore, we show that NO from NOS1 permits the full transcriptional activation of NF-κB in macrophages by suppressing SOCS1.
RESULTS
NOS1−/− animals are protected from model septic injury and mortality, whereas NOS2−/− and NOS3−/− animals demonstrate greater susceptibility
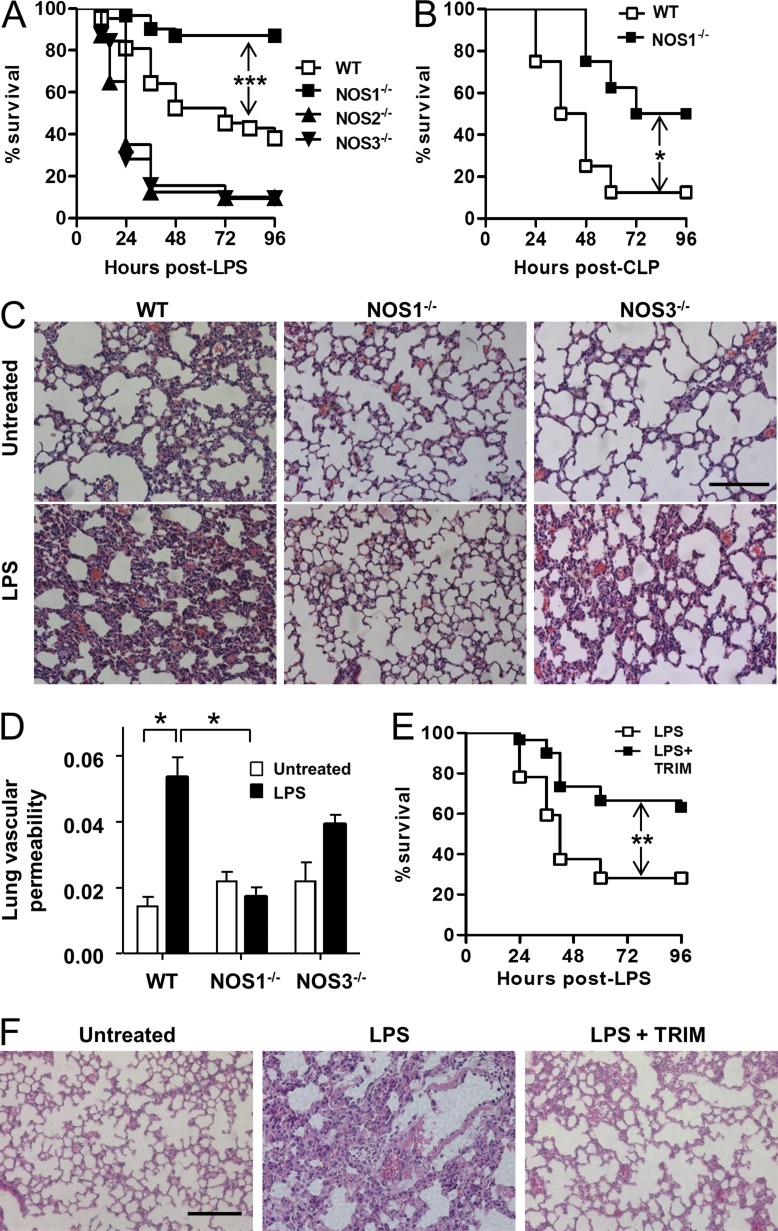
We analyzed the response of NOS1−/−, NOS2−/−, and NOS3−/− animals with an experimental model of sepsis: i.p. LPS injection. Similar to earlier studies, we observed increased mortality in NOS2−/− and NOS3−/− animals (Hickey et al., 1997; Wang et al., 2004; Miki et al., 2005; Fig. 1 A). In contrast, NOS1−/− mice showed increased survival when compared with control animals (Fig. 1 A). We tested whether compensatory expression changes in NOS1 could help to explain the susceptible NOS2−/− or NOS3−/− phenotypes, but we detected comparable levels of NOS1 expression in WT, NOS2−/−, and NOS3−/− (unpublished data). We also tested responses to a polymicrobial sepsis model, cecal ligation and puncture (CLP). Again, NOS1−/− animals were protected compared with WT (Fig. 1 B). Acute lung injury is a major cause of morbidity and mortality associated with acute systemic inflammation (Matthay and Zemans, 2011). Although WT and NOS3−/− lungs demonstrated marked inflammatory remodeling, as well as increased vascular permeability assessed by capillary filtration analysis (Kfc), NOS1−/− lungs were protected in both assays (Fig. 1, C and D). To determine if these effects were caused by the loss of NOS1 and not by any compensatory effects of gene deletion, we also treated WT mice with the NOS1/NOS2 inhibitor 1-(2-Trifluoromethylphenyl) imidazole, TRIM (Handy et al., 1995). In agreement with many previous studies, we find that NOS2 is not significantly expressed until hours after the activation of inflammation, and expect that TRIM’s mechanism of action at this time should be solely through NOS1 (see also Fig. 5, F and G). Consistent with this, TRIM treatment recapitulated the results of genetic ablation of NOS1 for both mortality and histological correlates of injury (Fig. 1, E and F). Although NOS2 and NOS3 play protective roles in septic injury, we demonstrate that NOS1, uniquely, promotes the injury associated with model sepsis.
Figure 1.
NOS1 deficiency protects against model septic injury. (A) Survival of WT (n = 42), NOS1−/− (n = 31), NOS2−/− (n = 40), and NOS3−/− (n = 32) mice injected with LPS (30 mg/kg, i.p.), compiled from at least 3 independent experiments using 10 mice per group, or (B) WT (n = 8) and NOS1−/− (n = 8) mice, compiled from 2 independent experiments, were subjected to CLP and survival was monitored for the indicated time. (C) Lungs of WT, NOS3−/−, and NOS1−/− mice were harvested 8 h after i.p. LPS challenge and sections were stained with H&E. Representative images from three mice per phenotype analyzed in three independent experiments are shown. Bar, 100 µM. (D) Lung vascular permeability measurements (Kfc) of WT, NOS3−/−, and NOS1−/−, mice before and 8 h after LPS challenge. Three mice per phenotype were analyzed in three independent experiments. *, P < 0.05, Student’s t test. (E) Survival of WT mice treated with LPS alone (n = 32) or pretreated for 2 h with TRIM (25 mg/kg, i.p.) before LPS (n = 30), compiled from at least 3 independent experiments with 10 mice per group (F). Lung histological analyses were performed on WT mice as in C, either control or with pretreatment with TRIM, as in E. Significance in A–C was determined using Fisher’s exact test. *, P < 0.02 (B); **, P < 0.001 (E); ***, P < 0.0005 (A).
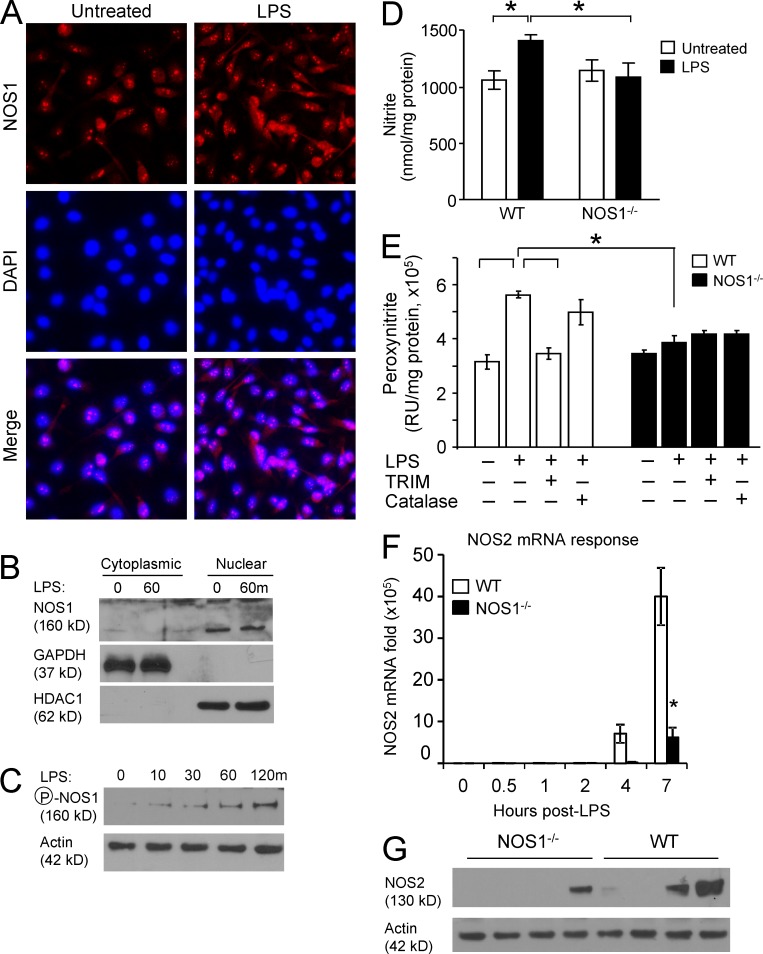
Figure 5.
NOS1 localizes to the nuclei of macrophages and is required for rapid NO production after LPS treatment. (A) Nuclear localization of NOS1 was demonstrated in BMDM after fixation and immunostaining for NOS1 (red), nuclei were counterstained with DAPI and representative images from 2 independent experiments are shown (bar, 25 µM). (B) BMDM cytoplasmic and nuclear fractions were immunoblotted to demonstrate the subcellular location of NOS1. GAPDH and HDAC1 serve as controls for subcellular fractionation. (C) NOS1 Serine 1412 phosphorylation, which correlates with enzymatic activation, was assayed by immunoblot of BMDM stimulated with LPS (100 ng/ml) for the indicated time points. Data are representative of three independent experiments. (D) Nitrite accumulation, as an indirect measure of NO production, was detected in the supernatants of WT and NOS1−/− BMDM using a Sievers 280i Nitric Oxide Analyzer, before or after LPS (100 ng/ml for 1 h). *, P < 0.05. (E) Peroxynitrite production, another indirect measure of NO production, was assayed by incubating WT or NOS1−/− BMDM with coumarin-7-boronic acid (10 µM) for 30 min, with or without LPS (100 ng/ml). Fluorescence measurement was performed using HPLC. *, P < 0.0003. (F) Quantitative RT-PCR analysis of LPS-induced NOS2 mRNA and (G) immunoblot analysis of NOS2 protein, from WT and NOS1−/− BMDM treated with LPS (250 ng/ml) demonstrates no detection of the inducible NOS (NOS2) at early time points after LPS in BMDM of either genotype. P < 0.01 (Student’s t test). Data are representative of six (D and E) and three (F) independent experiments; all are presented as mean ± SD. Data in B, C, and G are representative of three independent experiments.
NOS1 is required for TLR4-mediated cytokine production and NF-κB activation, in vitro and in vivo
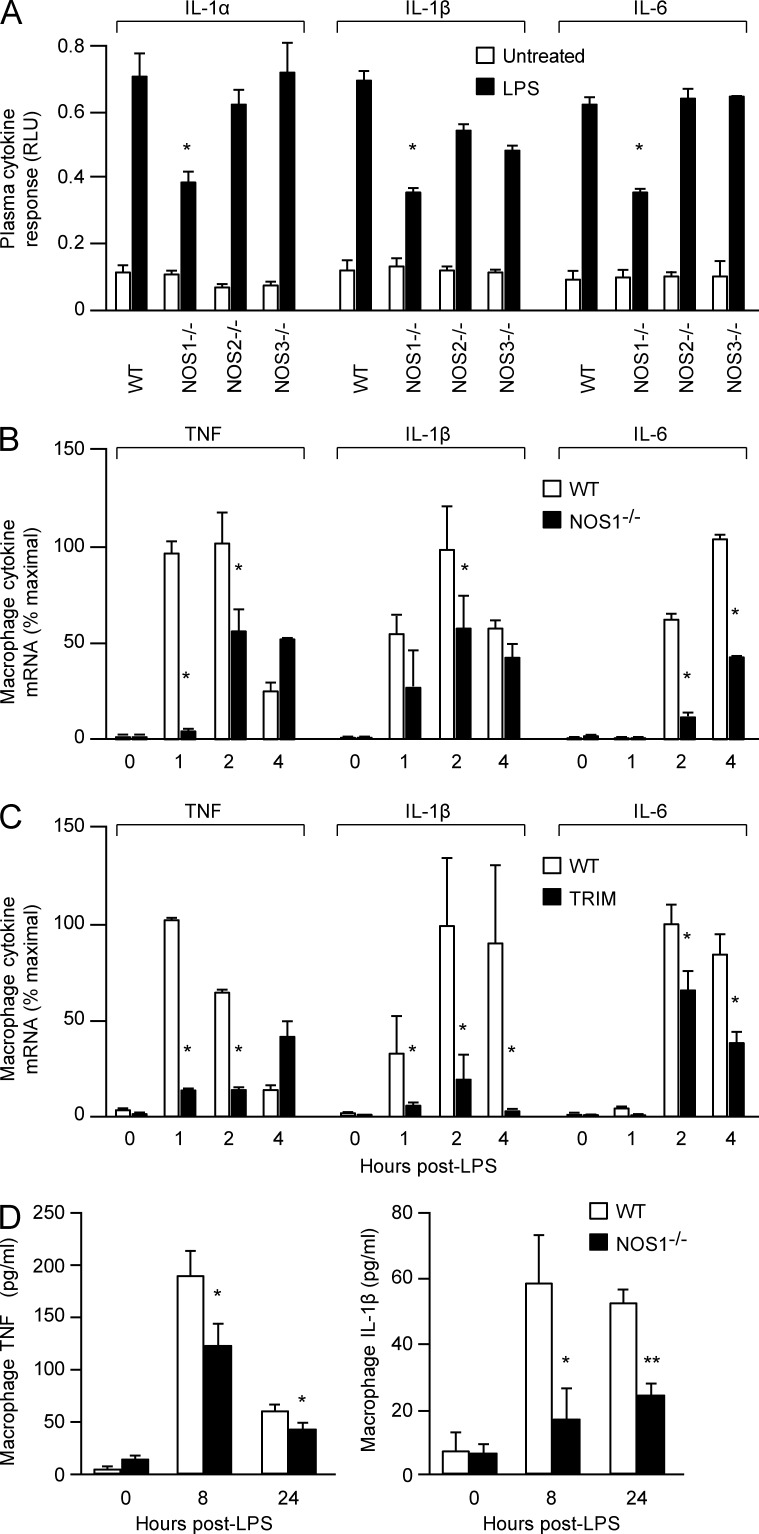
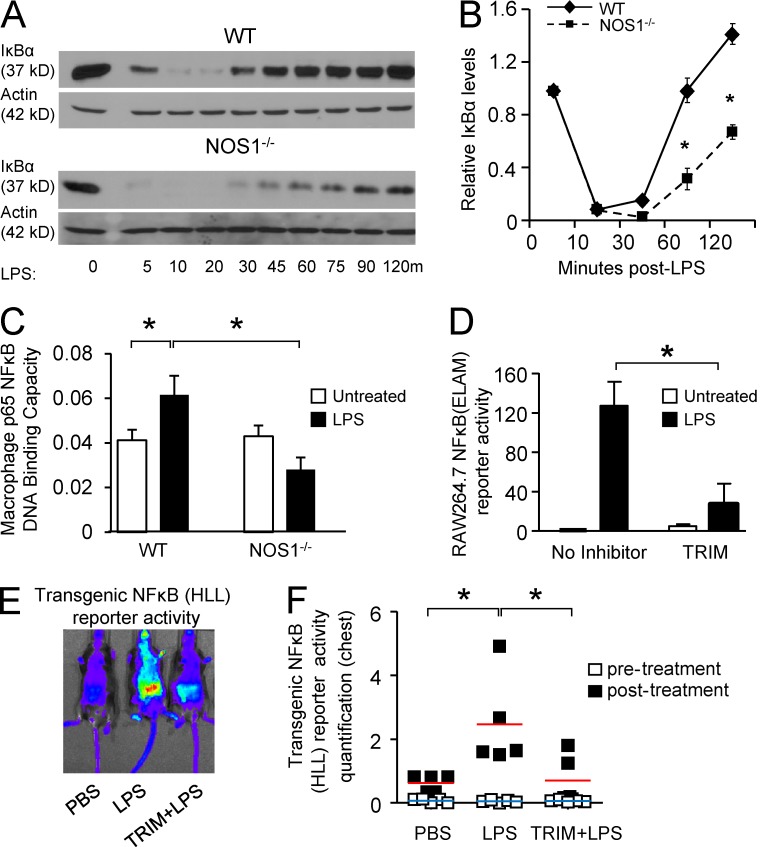
De novo transcription and translation of cytokines plays a critical role in promoting inflammatory tissue injury during sepsis. In accordance with their protection from septic injury, we observed that NOS1−/− mice displayed significantly decreased plasma cytokine responses 8 h after i.p. LPS treatment compared with WT, whereas NOS2−/− and NOS3−/− mice mounted normal cytokine responses (Fig. 2 A). Macrophages are critical cellular mediators of inflammation in vivo (MacMicking, et al., 1997). We analyzed NOS1 function in these cells and found that they recapitulated the in vivo phenotype of NOS1 deficiency. BM-derived macrophages (BMDMs) prepared from NOS1−/− mice demonstrated significant reductions in mRNA and protein accumulation for an array of cytokines (TNF, IL1β, IL6, and MIP2; Fig. 2, B–D). Acute inhibition of NOS1 in WT BMDM by pretreatment with TRIM also reduced cytokine transcriptional responses to LPS treatment (Fig. 2 C). These broad defects in proinflammatory cytokine transcription led us to investigate the signaling and transcriptional pathways downstream of TLR4. NF-κB activation is critical for expression of a wide variety of inflammatory cytokines, so we interrogated this pathway in the NOS1−/− cells. Early signaling events in the pathway converge on the degradation of IκB, which liberates the transcription factor complex to the nucleus. NOS1−/− BMDMs responded to LPS treatment by degrading IκB similarly to WT cells, suggesting that the earliest signaling events triggered by TLR4 are intact. Despite this, we found that reexpression of IκB after degradation is significantly diminished (Fig. 3, A and B). Because IκB is one of its major targets, we analyzed NF-κB transcriptional activation downstream of IκB degradation. Despite detecting an increase in nuclear p65 DNA-binding activity in LPS-treated WT BMDM, we saw no increase in NOS1−/− BMDM (Fig. 3 C). Consistent with this, a mouse macrophage cell line (RAW264.7) stably transfected with an NF-κB–driven luciferase reporter (endothelial cell–leukocyte adhesion molecule [ELAM] cells; Gantner et al., 2003) and treated with LPS for 6 h demonstrated significantly reduced luciferase reporter activity after pretreatment with TRIM (Fig. 3 D). Mice expressing another NF-κB–driven luciferase reporter ubiquitously as a transgene (HLL-mice) demonstrated that TRIM treatment could also significantly dampen systemic NF-κB activation in vivo, 24 h after LPS treatment (Sadikot et al., 2001; Fig. 3, E and F). Our findings demonstrate a novel and important role for NOS1 in promoting a broad array of proinflammatory cytokine expression by driving NF-κB DNA binding and transcriptional activation.
Figure 2.
Proinflammatory cytokine responses to LPS are diminished in NOS1−/− animals and cultured macrophages. (A) IL-1α, IL-1β, and IL-6 in the plasma of WT, NOS1−/−, NOS2−/−, and NOS3−/− mice before and 8 h after LPS (30 mg/kg, i.p.) injection. Cytokine levels in plasma were assessed using a Luminex panel and expressed as relative light units (RLU). Nine mice for each treatment condition and genotype were analyzed and data are compiled from three independent experiments, mean ± SEM. *, P < 0.005 (Student’s t test). (B) Quantitative RT-PCR analysis of cytokine mRNA expression of TNF, IL-6, and IL-1β in BMDMs isolated from WT and NOS1-/− mice and stimulated with LPS (250 ng/ml). *, P < 0.0005. (C) mRNA expression analysis of TNF, IL-6, and IL-1β in WT macrophages (no inhibitor) and WT macrophages treated 2 h with TRIM (100 nM) before LPS (250 ng/ml) stimulation. *, P < 0.003 (Student’s t test). (D) ELISA measurements of TNF and IL-1β cytokine levels in culture media of LPS-activated BMDM (250 ng/ml). *, P < 0.02; **, P < 0.001 (Student’s t test). Representative data are shown from three (B–D) independent experiments, and are expressed as mean ± SD.
Figure 3.
NOS1 is required for NF-κB transcriptional activation but not upstream signaling. (A) Representative immunoblots for IκB degradation after LPS treatment (250 ng/ml) from WT and NOS1−/− BMDMs. (B) Densitometry analysis of IκB levels as in (A), normalized to actin, presented as mean ± SEM of quantitation of three independent experiments. *, P < 0.02 (Student’s t test). (C) p65 DNA-binding activity was assayed from the isolated nuclei of WT and NOS1−/− BMDM before and after LPS (250 ng/ml) for 1 h. *, P < 0.001, Student’s t test. (D) RAW264.7 macrophage cell line stably expressing an NF-κB–driven luciferase reporter (ELAM) were pretreated with TRIM (50 µM) for 2 h and then stimulated with LPS (100 ng/ml) for 6 h. *, P < 0.0005, Student’s t test. C and D are representative of three independent experiments, presented as mean ± SD of three replicates. Representative digital images of (E) and quantitation of (F) NF-κB transcriptional activity from mice expressing a transgenic luciferase reporter (HLL) 24 h after treatment with PBS (control), LPS or LPS after 2 h pretreatment with TRIM (50 mg/kg, i.p.) to inhibit NOS1. Luminescence was evaluated by IVIS 10 min after i.p. injection of 30 mg/kg luciferin. Quantitation focused on activity in the chest (n = 6 animals per group compiled from two separate experiments using 3 mice per treatment), and includes basal reporter activity before LPS or PBS treatment for comparison. *, P < 0.05 (Student’s t test). Averages are shown as horizontal bars, red for treated, and blue for control values.
NOS1 is required to block proteolytic degradation of p65 in LPS-activated macrophages
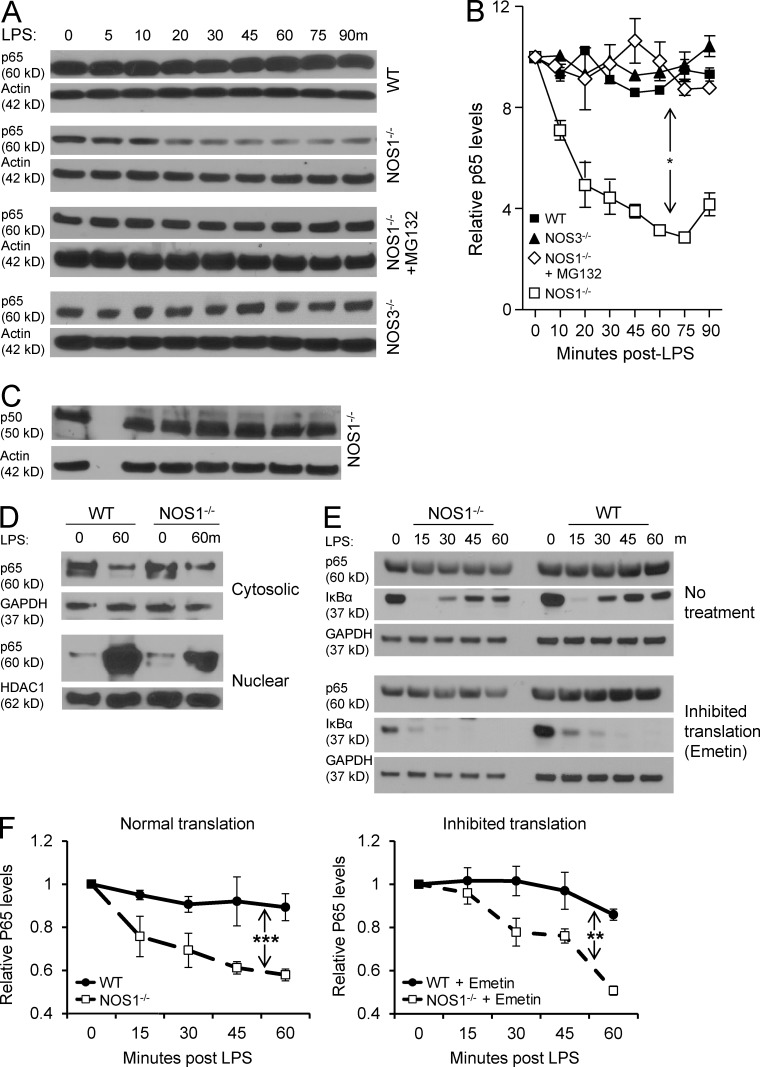
Unexpectedly, NOS1−/− BMDM showed a rapid decrease in p65 protein levels after LPS stimulation, despite comparable expression of p65 in unstimulated cells. This was in clear contrast with WT or NOS3−/− cells, where p65 protein levels remained stable (Fig. 4, A and B). NF-κB p50, the canonical binding partner for p65, was not degraded in NOS1−/− or WT BMDM (Fig. 4 C and not depicted). p65 is central to canonical NF-κB transcription, so a reduction in its protein level could account for the cytokine transcriptional defects in the absence of NOS1 (Alcamo et al., 2001; Vallabhapurapu and Karin, 2009; Baltimore, 2011; Fig. 2). To determine whether the loss of p65 was caused by synthesis or degradation, we treated NOS1−/− macrophages with the proteasome inhibitor MG132, and were able to rescue p65 protein levels after LPS treatment (Fig. 4, A and B). Consistent with this, we compared p65 protein levels in NOS1−/− and WT BMDMs after blocking de novo protein synthesis with emetin (Grollman, 1968) and found that knockout cells still lost p65 protein after LPS treatment (Fig. 4, E and F). These results support the hypothesis that NOS1 blocks proteasomal degradation of p65 in macrophages, rather than affecting p65 synthesis. We prepared cytosolic and nuclear fractions from BMDMs and analyzed p65 protein abundance. Although little difference was detected in cytoplasmic p65, nuclear p65 protein was greatly depleted from NOS1−/− BMDMs (Fig. 4 D). This led us to determine the subcellular localization of NOS1 in macrophages. Using immunofluorescence imaging and nuclear fractionation of BMDMs, we found that NOS1 was mainly in the nucleus, regardless of the LPS-activation state of the cell (Fig. 5, A and B). This finding is consistent with other reports that NOS1 can function in the nucleus and further supports the hypothesis that this enzyme plays an important role in regulating gene expression (Yuan et al., 2004; Saluja et al., 2010; Villanueva and Giulivi, 2010). We then undertook functional characterization of NOS1 to ensure that it is enzymatically active in BMDMs. Consistent with previous observations that NOS1 is phosphorylated on Ser1412 on activation (Huang et al., 2012), we found that LPS treatment induced NOS1 phosphorylation on this site within minutes in BMDMs (Fig. 5 C). This correlated with two indirect measures of NO. We detected very early pulses of nitrite (60 min) and peroxynitrite (30 min) production in BMDMs. However, NOS1−/− BMDMs failed to produce either of these products of NO within this time frame (Fig. 5, D and E). Similarly, TRIM pretreatment blocked production of either product in WT cells (Fig. 5 E and not depicted). Consistent with NOS1 activity reported in other cell types, calcium ionophore treatment drove nitrite production in WT, but not in NOS1−/− BMDMs (unpublished data). To exclude the possibility that NOS2 was contributing to early NO activity, we examined NOS2 mRNA and protein at time points after LPS treatment. We found no detectable expression in WT or knockout cells until 4 h after LPS treatment, well after the early LPS-mediated NOS activity we detected (Fig. 5, F and G). Our data demonstrate that LPS stimulation activates mechanisms that promote p65 degradation, and that uniquely among the NOS family enzymes, NOS1 is capable of blocking this, and thereby, of maintaining p65 protein levels.
Figure 4.
NOS1−/− BMDM fail to maintain p65 protein levels after exposure to LPS. Representative immunoblots (A) and densitometric quantitation (B) of NF-κB p65 protein levels in BMDM stimulated with LPS (250 ng/ml), normalized to actin, for the indicated times and genotypes. Where indicated, cells were pretreated with MG132 (50 µM, proteasome inhibitor). *, P < 0.01. (C) Immunoblot analysis of total NF-κB p50 levels in NOS1−/− macrophages after LPS (250 ng/ml). (D) Isolated cytoplasmic and nuclear fractions of WT and NOS1−/− BMDM were probed for p65 by immunoblot before and after LPS (250 ng/ml) for 60 min, GAPDH and HDAC1 are loading controls for cytoplasmic or nuclear protein, respectively. p65 protein degradation dynamics was assessed by (E) immunoblot and (F) densitometric quantitation (normalized to GAPDH) after inhibition of de novo protein synthesis by treatment of BMDM with emetin (100 µg/ml) or control, followed by LPS (250 ng/ml) for the indicated times. The loss of IκBα re-expression demonstrates the effective inhibition of protein translation. For all of the above experiments, representative blots are shown from 3 independent experiments and quantitation, based on all experiments, are presented as mean ± SEM. P-values were determined by Anova two-way test with Bonferroni post-test to compare replicates: *, P < 0.02; **, P < 0.001; and ***, P < 0.0001.
NOS1-derived NO mediates S-nitrosation of SOCS1, blocking its ability to target p65 for degradation
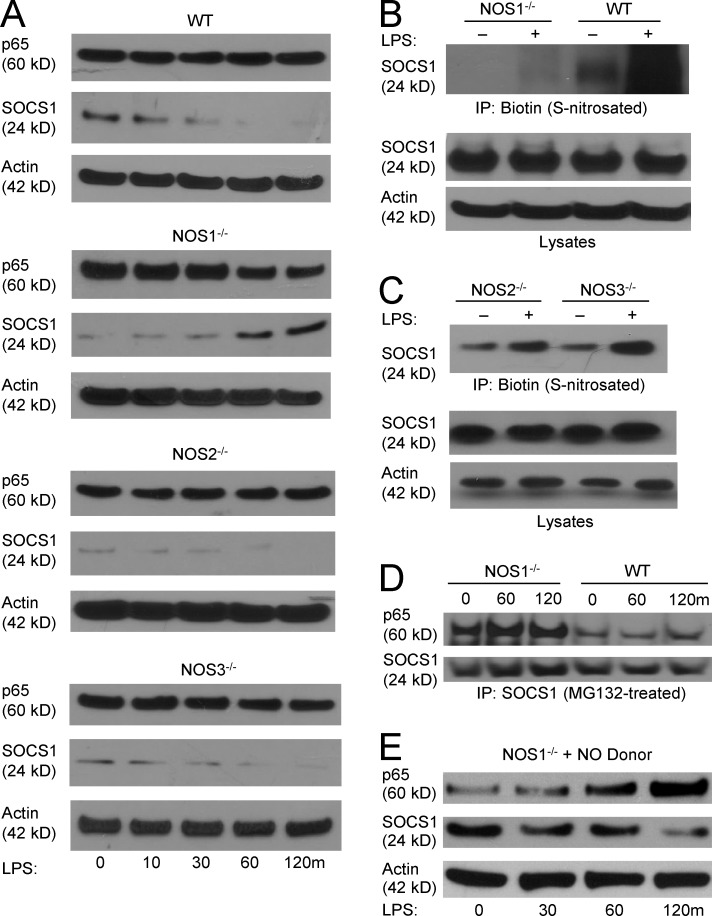
One mechanism reported to direct p65 protein degradation involves the antiinflammatory molecule SOCS1. SOCS1 is reported to mediate ubiquitination of DNA-bound p65 leading to its proteasomal lysis (Ben-Neriah, 2003; Ryo et al., 2003; Strebovsky et al., 2011). To determine if SOCS1 could be involved in NOS1-dependent signaling, we analyzed its protein expression in macrophages. WT, NOS2−/−, and NOS3−/− BMDM demonstrate decreasing SOCS1 protein abundance for the time period examined. In contrast, NOS1−/− cells alone demonstrated accumulation of SOCS1 protein that tracked with the drop in p65 protein levels after LPS stimulation (Fig. 6 A). To determine if SOCS1 is a direct target of NO, we analyzed protein S-nitrosation. By performing biotin switch assays on MG132-treated cells (to retain protein that would otherwise be proteasomally degraded) we found that in WT BMDM, SOCS1 demonstrates low basal S-nitrosation that increases dramatically after 1-h LPS treatment. In stark contrast, NOS1−/− cells demonstrate much lower basal nitrosation and no detectable increase with LPS treatment (Fig. 6 B). NOS2−/− and NOS3−/− BMDM exhibited low basal and robust LPS-induced S-nitrosation, demonstrating that these enzymes are not required to modify SOCS1 during the time course we studied (Fig. 6 C). In addition to the effect on protein stability, we also found that NOS1 regulates the ability of SOCS1 to physically interact with p65. Immunoprecipitates of SOCS1 from MG132-treated BMDM show greater binding to p65 in NOS1−/− than in WT cells (Fig. 6 D). Finally, we tested the ability of exogenous NO to rescue the defects seen in the knockout macrophages by using the fast-releasing NO donor, diethylamine-nonoate (DEANO). Concurrent treatment with DEANO and LPS reversed the degradation of p65 seen in NOS1−/− cells, while also promoting the degradation of SOCS1 (Fig. 6 E). These observations demonstrate that a rapid pulse of NO from NOS1 facilitates SOCS1 degradation, leading to stabilization of p65 protein and sustained NF-κB–dependent transcription. To our knowledge, this is the first demonstration of a posttranslational modification of SOCS1 that regulates its ability to suppress NF-κB nuclear activity.
Figure 6.
NOS1-derived NO mediates S-nitrosation of SOCS1 and prevents SOCS1-mediated proteasomal degradation of p65. (A) Immunoblot analysis of p65 and SOCS1 in WT, NOS1−/−, NOS2−/−, and NOS3−/− BMDM treated with LPS (250 ng/ml) for the indicated times. (B) SOCS1 S-nitrosation was detected using the biotin switch method on protein from WT and NOS1−/− BMDMs pretreated with MG132 (50 µM) for 1 h before treatment with LPS (250 ng/ml) for the indicated times, followed by immunoblotting for SOCS1. (C) NOS2−/− and NOS3−/− BMDMs analyzed by biotin switch method, as in B. (D) Co-immunoprecipitation of SOCS1 and p65 in WT and NOS1−/− BMDMs, pretreated with MG132 (50 µM, 1 h) before LPS (250 ng/ml) for the indicated time intervals. (E) Immunoblot analysis of p65 and SOCS1 total protein levels in NOS1−/− BMDM treated with DEANO (NO donor, 5 µM) and LPS (250 ng/ml). All blots shown are representative of at least two (C and D) or three (A, B, and E) independent experiments.
Cys147 and Cys179 are essential for SOCS1 nitrosation and proteasomal degradation
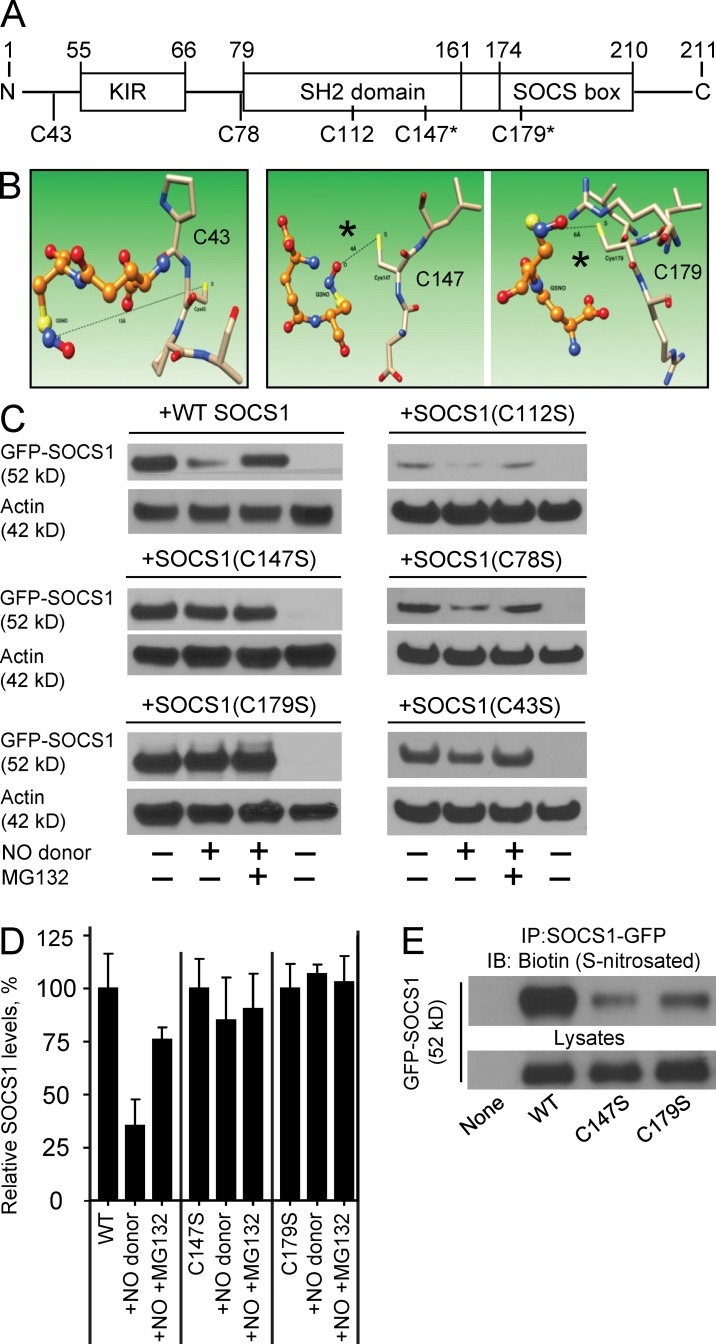
We next sought to determine the exact residues on SOCS1 that are modified by NOS1-derived NO. We used both predictive molecular modeling techniques, followed by functional analysis of individual point mutants of the candidate residues. SOCS1 contains 5 cysteine residues, and we assessed their likelihood for S-nitrosation by in silico docking models (Fig. 7 A). We analyzed the protein sequence and structure of SOCS1 and modeled the predicted accessibility of each cysteine sulfur atom to the nitrogen atom in S-nitrosoglutathione (GSNO) using minimal energy calculations (Fig. 7 B and not depicted). We chose GSNO because it is the likely physiological carrier of reactive NO (Sun et al., 2011; Green et al., 2012). This approach predicted the most susceptible residues to nitrosation were Cys147 and Cys179, which are within the SH2 domain (at the vicinity of the SOCS box) and the SOCS box domain itself (the p65 binding domain), respectively (Fig. 7, A and B). Modification to these domains could explain our observation that NOS1 decreases the binding between SOCS1 and p65 even in the absence of proteasomal degradation. To empirically verify these in silico predictions, we transiently expressed either WT or individual cysteine to serine point mutant constructs of SOCS1 in HEK293 cells. Transfected cells were exposed to exogenous NO using DEANO, with or without MG132, to assess the susceptibility of each protein to NO-directed proteasomal degradation. In agreement with our in silico predictions, we determined that although WT SOCS1 protein was degraded upon exposure to NO, C147S and C179S mutants were only minimally affected (Fig. 7, C and D). In contrast, mutations of the three cysteines predicted not to be targets of S-nitrosation (C43S, C78S, and C112S) demonstrated degradation similar to WT SOCS1 (Fig. 7 C). To confirm that this effect was directly correlated with S-nitrosation, we performed a biotin switch assay. Although WT SOCS1 was robustly modified in the presence of the NO donor, S-nitrosation was dampened in the C147S and C179S mutants (Fig. 7 E). These findings demonstrate that the two cysteine residues in SOCS1 closest to the SOCS box are critical for regulating the stability of SOCS1 and its capacity to repress p65-mediated inflammatory transcription. Our results demonstrate that modifications of both Cys147 and Cys179 are required for full S-nitrosation and subsequent degradation of SOCS1.
Figure 7.
Molecular modeling and functional testing demonstrate that Cys147 and Cys179 of SOCS1 are the targets for S-nitrosation. (A) Protein domain structure of SOCS1 illustrating the positions of all 5 candidate cysteine residues. (B) Molecular modeling was performed to predict the accessibility of cysteine residues of SOCS1 (represented as colored stick atomic models) to the likely transnitrosation donor, GSNO (represented as ball and stick atomic models). The shorter distances between the cysteine sulfur atom and the nitrogen of GSNO are predicted to permit S-nitrosation for Cys147 (4 Å, middle) and Cys179 (6 Å, right), whereas Cys43 (13 Å, left), Cys112 (11 Å, not depicted), and Cys78 (11 Å, not depicted) are predicted to be too far apart to permit the reaction. (C) Representative immunoblots of GFP-tagged constructs of SOCS1 or point mutations of each cysteine (C147S, C179S, C112S, C78S, and C43S) were transfected into HEK293 cells, which were then treated with a 15-min pulse of 20 ng/ml IL-1β. Some of the cells were pretreated with 50 µM proteasome inhibitor for 1 h (MG132 +) and some were treated with 10 µM DEANO for 1 h after IL-1β (NO donor +). Stability of SOCS1 protein levels was determined by immunoblot and (D) quantified relative to β-actin from three independent experiments (mean ± SEM). (E) The capacity of SOCS1 mutants C147S, and C179S for S-nitrosation was compared with WT SOCS1 in transiently transfected HEK293 cells using the biotin switch method. Immunoprecipitates of GFP-SOCS1 variants were assayed for biotin (S-nitrosation), and lysates were immunoblotted for total SOCS1 as control. Data are representative of two independent experiments.
DISCUSSION
Immunoregulatory functions for NO have long been recognized, owing to the potent induction of the high output, inducible NOS2 isoform associated with the inflammatory response. NO from NOS2 is known to mediate feedback inhibition of proinflammatory signaling pathways, including NF-κB (Peng et al., 1995; Matthews et al., 1996; Togashi et al., 1997; delaTorre et al., 1999; Marshall and Stamler, 2001; Marshall et al., 2004; Reynaert et al., 2004; Kelleher et al., 2007). Although some studies have suggested that lower concentrations of NO from NOS1 can promote inflammation (Porras et al., 2006; Kanwar et al., 2009; Lange et al., 2010; Duma et al., 2011), the mechanism of action for NOS1 has remained obscure. In this study, we demonstrate that NO from NOS1 is critical for NF-κB activity, specifically through the S-nitrosation–mediated degradation of SOCS1, which blocks the targeted proteolysis of p65, permitting sustained proinflammatory gene transcription.
We confirmed a physiologically important role for NOS1 in two models of sepsis, a sterile inflammatory model induced by i.p. treatment with LPS, as well as polymicrobial sepsis resulting from CLP. Both treatments resulted in significantly reduced mortality in NOS1−/− animals (Fig. 1). This is unique to NOS1 because, consistent with the findings of others, we demonstrate that NOS2−/− and NOS3−/− mice show increased mortality (Hickey et al., 1997; Wang et al., 2004; Miki et al., 2005; Duma et al., 2011). Inflammatory cytokines are critical for septic injury, and NOS1−/− mice and NOS1−/− BMDM both exhibit diminished cytokine responses to LPS treatment (Fig. 2). Indeed, pharmacological inhibition of NOS1 with TRIM replicates these NOS1−/− phenotypes. Although TRIM is able to inhibit the activity of NOS2, as well as NOS1, we do not detect any expression of NOS2 at time points where we can detect and inhibit NOS1-dependent, NO-derived products (Fig. 5). Collectively, these findings demonstrate that NOS enzymatic activity, specific to NOS1, is required for the inflammatory injury caused by sepsis.
The variety of the cytokines affected hinted at a defect in the inflammatory signaling pathways downstream of TLR4. We were surprised by the observation that whereas IκB degradation was normal in NOS1−/− macrophages, NF-κB–dependent transcription was significantly reduced (Fig. 3). Our findings that other proximal signaling pathways activated by TLR4, ERK1/2, and JNK MAPK activation were intact in NOS1−/− cells led us to scrutinize the NF-κB pathway in more detail (unpublished data). We were surprised to find that the levels of p65 protein were significantly decreased in activated NOS1−/− macrophages, in clear distinction with WT or NOS3−/− cells, which maintained p65 protein (Fig. 4, A and B). We observed that proteasomal inhibition with MG132 could rescue p65 protein levels in NOS1−/− cells, and that inhibition of protein synthesis with emetin did not abolish the difference between WT and knockout p65 responses. In addition, we observed no changes in protein levels for the canonical p65 binding partner, NF-κB p50 (Fig. 4 C). Collectively, these findings demonstrate that NOS1-derived NO regulates the specific proteasomal targeting of p65. p50 homodimers can mediate distinct antiinflammatory transcriptional responses compared with the p65–p50 complex (Driessler et al., 2004). Our studies therefore suggest that in addition to modulating the intensity of the inflammatory response, NOS1-derived NO may influence the quality of the inflammatory response to TLR4 stimulation, as well.
SOCS1, a signaling molecule best known for negative regulation of JAK/STAT signaling, has been reported to mediate ubiquitination of DNA-bound p65, leading to its proteasomal lysis (Ben-Neriah, 2003; Ryo et al., 2003; Strebovsky et al., 2011). This mechanism was consistent with our observations of p65, and indeed, we did find that SOCS1 protein levels were higher in LPS-treated NOS1−/− cells (Fig. 6 A). Consistent with this, we used the biotin switch method to find that SOCS1 was S-nitrosated in WT macrophages within 60 min of LPS treatment. Unlike NOS2−/− or NOS3−/− cells, S-nitrosation at these time points was completely absent in NOS1−/− cells (Fig. 6, B and C). Our predictive modeling and functional mutational analysis of SOCS1 identified Cysteine 147 and Cysteine 179 as being required for S-nitrosation and SOCS1 degradation (Fig. 7). We also observed that in the absence of proteasomal degradation, NOS1−/− macrophages display reduced binding of SOCS1 and p65 compared with WT (Fig. 6 D). It is possible, therefore, that SOCS1 inhibition may be transient and reversible, or more durable once the SOCS1 protein is degraded. This notion is supported by the finding that although the nitroso modification of both Cys147 or Cy179 residues was required to direct SOCS1 proteolysis, the individual mutants retained a reduced ability to bind to p65 (Fig. 7, C and E; and not depicted).
Unlike other reports that have demonstrated NO can reduce the activity of NF-κB by direct nitrosation as a result of NOS2 activity (Kelleher et al., 2007), we observed that NO generation from NOS1, within 60 min after TLR4 activation, led to stabilization of the p65 transcription factor. This is in agreement with previously reported in vitro observations that NOS1 regulates the DNA binding ability of p65 (Peng et al., 1995; Matthews et al., 1996). We have identified direct nitrosation of SOCS1 as the molecular mechanism for this effect. This extends the importance of SOCS1-mediated regulation of p65, and suggests that the ability to modulate the balance of SOCS1 and p65 through pharmacological targeting of NOS1 could provide important therapeutic interventions to shape the inflammatory responses in a wide range of clinically relevant diseases. Collectively, we find that the anti-inflammatory proteolytic targeting of NF-κB p65 by SOCS1 is an active mechanism during LPS stimulation of macrophages, and that NO, derived uniquely from NOS1, is required to suppress SOCS1 and drive sustained inflammatory transcription.
MATERIALS AND METHODS
Animals and cell culture.
Animal handling and experiments were conducted in accordance with the policies of the Animal Care Facility of the University of Illinois at Chicago and the National Institute of Environmental Health Sciences (NIEHS). All animals were acquired from The Jackson Laboratory. NOS1 knockout mice, NOS2 knockout mice, and NOS3 knockout mice were obtained from JAX. WT C57BL/6 animals were used as control. Strain-specific genotyping was performed by Transnetyx Inc. In all experiments, we used 8–12-wk-old male animals. BMDMs were prepared from C57/BL6, NOS1−/−, NOS2−/−, or NOS3−/− mice (The Jackson Laboratory) by culturing single-cell suspensions of BM for 5–7 d in Dulbecco’s minimal essential medium (DMEM) from Invitrogen or RPMI (Gibco) made complete by addition of 10% heat-inactivated FBS (Atlanta Biologicals or Gibco), 100 U/ml penicillin and 100 µg/ml streptomycin from CellGro by Mediatech or 1% (vol/vol) Antibiotic-Antimycotic mix from Life Technologies, with 10–20% L929 cell-conditioned (ATCC) complete DMEM medium. Macrophages were washed and lifted by spraying cells with 1 mM EDTA in 1× PBS from an 18-gauge needle (BD), and then plated for experiments 1–3 d later. RAW264.7 cells stably transfected with the ELAM luciferase reporter (Hume et al., 2001) were cultured in RPMI (Life Technologies) made complete as above. Luciferase expression was assessed by lysing cells (Promega), adding Luciferase Assay Reagent (Promega) and detection using a Wallac Victor2 (PerkinElmer).
In vivo studies.
Septic inflammation was induced in mice using two methods. i.p. LPS animals received an intraperitoneal injection of 30 mg/kg LPS (Escherichia coli, 0111:B4; Sigma-Aldrich), a dose demonstrating a mortality rate of >80% in C57/BL6 animals. CLP was performed under ketamine/xylazine anesthesia as described previously (Rittirsch et al., 2009). The distal 20% (below the ileocecal valve, ∼1 cm from the tip) of the cecum was ligated with a 6–0 suture. The cecum was punctured 4 times with a 20-gauge needle. For survival studies, lethal endpoints were observed for 96 h, at which point all remaining animals were sacrificed. For histopathological observation, animals were sacrificed and lungs were removed 8 h after treatment. Lungs were fixed in 10% neutral buffered formalin, dehydrated, embedded in paraffin, sectioned, and then stained with hematoxylin and eosin (at the facilities of Research Histology and Tissue Imaging Core [RHTIC], University of Illinois, Chicago, IL). Microvessel Kfc was measured to determine the pulmonary microvascular permeability to liquids, as described previously (Vogel et al., 2000). In brief, after standard 30 min equilibration perfusion, the outflow pressure was rapidly elevated by 13 cm H2O for 20 min and then returned to normal. The changes of lung wet weight reflect net fluid extravasation. At the end of each experiment, lungs were dissected free of nonpulmonary tissue, and lung dry weight was determined. Kfc (ml/min/cmH2O/dry weight [g]) was calculated from the slope of the recorded weight change normalized to the pressure change and to lung dry weight. For some experiments, the NOS1/NOS2 dual inhibitor, 1-(2-trifluoromethylphenyl) imidazole (TRIM; 25 mg/kg, i.p.) was administered 2 h before LPS administration.
For cytokine production analysis, blood samples from 6 mice per phenotype were clotted and centrifuged 2,000 g for 20 min, and serum was stored at −80°C until assay. The concentrations of IL-1α, IL-1β, and IL-6 were detected by ELISA Luminex xMAP detection, according to the manufacturer’s instructions (EMD Millipore).
HLL transgenic mice (C57B6/DBA background) express Photinus luciferase cDNA under the control of proximal 5′ HIV-LTR promoter. Only 8–12-wk-old males were used in the experiment. Bioluminescence intravital imaging was performed using an IVIS charge-coupled camera (PerkinElmer). 24 h after the indicated treatment, mice were anesthetized by isoflurane, fur was removed from chest and abdomen, 100 µl luciferin (300 mg/ml) was injected i.p. (Caliper Life Sciences), and imaging was performed 10–15 min later. Quantitative analysis was performed by selecting signal over the chest area only, and data were expressed as photons per second.
Nitrite and peroxynitrite measurements.
Nitrite, a stable metabolite of NO, was measured in culture supernatants by chemiluminescence. BMDMs were washed twice with HBSS and incubated in serum-free HBSS (Life Technologies) containing 1 mM L-arginine (Sigma-Aldrich) at 37°C, and treated with LPS (E. coli, B4:111) for 2 h. After treatment, media was collected, kept ice cold, and centrifuged to remove floating cells. NO concentration in the culture medium was assessed indirectly by measuring NO2− accumulation using a 280i Nitric Oxide Analyzer (Sievers Instruments) and reported as nmol NO per mg protein. Measurement of peroxynitrite production was done using Coumarin-7-boronate (CBA; Sigma-Aldrich). BMDM plated in 6-well plates, either treated with 100 µM TRIM (Sigma-Aldrich) for 1 h or control, were washed twice with DPBS, and incubated in serum-free HBSS buffer containing 20 µM CBA. Cells were then stimulated with LPS (250 ng/ml). Catalase (10 U/ml; Sigma-Aldrich) was also added in the negative control group. After 60 min, media was collected and centrifuged at 3,000 rpm for 5 min. Fluorescence analysis of the oxidation product 7-OH-coumarin (COH) was performed on a Beckman-Coulter HPLC system. Samples (20 µl) were separated on a Synergi-Fusion (250 × 4.6 mm; Phenomenex) using an isocratic elution with 35% vol/vol acetonitrile, at a flow rate of 1 ml/min. Fluorescence was measured using 350 nm (excitation) and 450 nm (emission). Authentic COH solution was injected onto the HPLC to verify the retention time and generate a standard curve.
Confocal analysis.
For SOCS1 and p65 staining, BMDMs were plated on coverslips (19-mm diam) and stimulated in 12-well plates. Cells were fixed with 2% paraformaldehyde for 15 min at room temperature and permeabilized for 10 min with ice-cold 100% methanol slowly added to prechilled cells at −20°C. All subsequent steps were conducted at room temperature. The cells were blocked with 5% BSA in 1× PBS for 1 h, and then stained with primary antibodies (mouse anti-SOCS1 and rabbit anti-p65 antibody from Cell Signaling Technology; 1/200 in blocking buffer) for 1 h. After washing with PBS (×5), the cells were stained with Alexa Fluor 568–conjugated goat anti–rabbit IgG and Alexa Fluor 488–conjugated donkey anti–mouse IgG (1/500 in PBS) for 1 h. DAPI was used for nuclear counterstaining according to the manufactorer’s protocol. The coverslips were mounted onto glass slides with VECTASHIELD Mounting Media (Vector Laboratories) and analyzed on a LSM 510 META confocal laser scanning microscope (Carl Zeiss) fitted with: 25 mW diode UV laser (DAPI, blue), 1 mW HeNe laser (Alexa Fluor 546, red), 30 mW tunable Ar laser (Alexa Fluor 488, green), and 63×/1.2 Water DIC C-Apochromat objective (UIC imaging core facility). Images were all taken with the same set exposure and processed using LSM software (Carl Zeiss).
mRNA isolation, cDNA synthesis, and real-time PCR.
Total RNA was isolated from BMDMs using TRIzol reagent according to the manufacturer’s specifications (Invitrogen). cDNA was synthesized using TaqMan RT-PCR kit (Life Technology) according to the manufacturer’s specifications. qPCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) on a 7500 Real-Time PCR System (Applied Biosystems). The following primer sets (each 5 pmol) were used to amplify cDNA fragments: TNF forward primer 5′-TTCGGCTACCCCAAGTTCAT-3′ and reverse primer 5′-CGCACGTAGTTCCGCTTTC-3′; IL-1β forward primer 5′-CCATGGCACATTCTGTTCAAA-3′ and reverse primer 5′-GCCCATCAGAGGCAAGGA-3′; IL6 forward primer 5′-CCACGGCCTTCCCTACTTC-3′ and reverse primer 5′-TTGGGAGTGGTATCCTCTGTGA-3′; GAPDH forward primer 5′-GCACAGTCAAGGCCGAGAAT-3′ and reverse primer 5′-GCCTTCTCCATGGTGGTGAA-3′. Cycle threshold (Ct) values were transformed to linear scale (1/2Ct), and the resulting data were normalized to GAPDH values for each sample. Data are presented as the percentage of the maximal mRNA response for each dataset. Analysis was performed with Microsoft Excel. To analyze levels of NOS2 mRNA in BMDM, total mRNA was isolated from 1.2–1.4 × 106 BMDM with RNA STAT-60 (Tel-Test, Inc.) according to manufacturer’s protocol. 4.6 µg of total mRNA was converted to cDNA using high capacity cDNA RT kit (Applied Biosystems), and 0.46 µg of resulting cDNA was further analyzed by qPCR using Fast SYBR Green Master mix (Applied Biosystems), AB7500 cycler, and the following primers: NOS2 forward 5′-CAGCTGGGCTGTACAAACCTT-3′; NOS2 reverse 5′-CATTGGAAGTGAAGCGTTTCG-3′ (de Souza et al., 2013; Shi et al., 2003); GAPDH forward 5′-AACGACCCCTTCATTGAC-3′; GAPDH reverse 5′-TCCACGACATACTCAGCAC-3′. Ct values were analyzed as above. All values were normalized to the values obtained for GAPDH for the same sample to correct for differences in cell number. Results are presented as fold change compared with untreated controls.
Nuclear/cytoplasmic fractionation, cytokine, and p65 DNA binding measurements.
The separation of nuclear extracts from cytoplasmic fractions was performed using a Nuclear/Cytosol Extraction kit (BioVision,), Nuclear Extraction kit (Cayman Chemical Company), or NE-PER Nuclear Protein Extraction kit (Thermo Fisher Scientific). Efficient cytoplasmic and nuclear fractionation was confirmed by Western blotting analysis using anti-GAPDH antibody (Cell Signaling Technology) for cytoplasmic fraction and anti-HDAC1 antibody (Santa Cruz Biotechnology, Inc.) for nuclear fraction. NF-κB p65 activity (nuclear DNA binding) was measured with NF-κB (p65) Transcription Factor Assay kit (Cayman Chemical Company) according to the manufacturer’s protocol. ∼1 µg of nuclear proteins were analyzed for each condition. Results were obtained with Spectra Max M5e (Molecular Devices) plate reader (absorbance at 450 nm). The same pools of BMDM were used to measure cytokine levels in parallel to confirm the phenotype, analyzing TNF and IL-1β cytokines with ELISA kits obtained from Thermo Fisher Scientific. Measurements were performed with a Spectra Max M5e (Molecular Devices) plate reader at 450 and 550 nm.
Immunoblotting, co-immunoprecipitation, and biotin switch assays.
For immunoblot analysis, cells were lysed in RIPA buffer (Santa Cruz Biotechnology, Inc.) containing protease and phosphatase inhibitors (Sigma-Aldrich), resolved on NuPAGE 4–12% gels (Novex; Life Technologies), and blotted and probed with the following antibodies against phospho-nNOS-Ser1412, p50, IκBα, GAPDH (Santa Cruz Biotechnology, Inc.), SOCS1 (Abcam), HDAC1, NOS1, GAPDH, β-actin, and anti-p65 (Cell Signaling Technology). After secondary HRP staining (Cell Signaling Technology or Jackson ImmunoResearch Laboratories), chemiluminescence was detected using SuperSignal (Thermo Fisher Scientific) or a luminol-coumeric acid-H2O2 chemiluminescence solution, and film (Denville Scientific Inc.) Images were digitally scanned (HP Officejet J4550 scanner) and analyzed using ImageJ (National Institutes of Health; Schneider et al., 2012). For coimmunoprecipitation, cells were lysed in NP-40 lysis buffer (20 mM Tris-HCL, pH 8.0, 137 mM NaCl, 10% glycerol, 1% NP-40, 2 mM EDTA) with protease inhibitors. Protein concentration was normalized using the BCA assay (Bio-Rad Laboratories). 100 µg of protein lysate was incubated with mouse anti-SOCS1 antibody (Invitrogen) plus protein A/G agarose beads (Santa Cruz Biotechnology, Inc.) overnight, rotating at 4°C. Beads were washed 4× with lysis buffer and protein was analyzed by immunoblot. Biotin-switch assays were conducted using the S-nitrosylated Protein Detection kit (Cayman Chemicals) according to the manufacturer’s instructions. Differentiated BMDM were pretreated with MG132 (50 µM) for 1 h and stimulated with LPS (250 ng/ml) for 30 min. S-nitrosated proteins, isolated from an equal number of cells, were precipitated using streptavidin Dynabeads (M-280 Streptavidin; Invitrogen), resolved by NuPAGE 4–12% gels (Novex; Life Technologies) and immunoblotted with either peroxidase-conjugated anti-SOCS1 antibody (1:100; Cell Signaling Technology) or rabbit anti-SOCS1 antibody (1:1,000; Abcam), followed by HRP-conjugated secondary antibody (anti–rabbit; Jackson ImmunoResearch Laboratories).
Expression of GFP-tagged SOCS1 and cysteine mutants in HEK293 cells.
HEK293 cells were maintained in complete DMEM. Cells were transiently transfected with pCMV6-AC-GFP Vector (OriGene), which expresses SOCS1 (mouse; available from GenBank under accession no. NM_009896.2) driven by the constitutive CMV promoter, using Lipofectamine 2000 reagent according to the manufacturer’s protocol. Cysteine mutations (C147S, C179S, C43S, C78S and C112S) were introduced in SOCS1 ORF (GenScript USA Inc.). HEK293 cells were stimulated with a 15 min pulse of 20 ng/ml IL-1β (Prospec Inc.) and treated with 10 µM DEA NONOate (DEANO) for 1 h (Alexis Biochemicals). Diethylamine NO (DEANO) is a member of the NONOate class NO donors. At 37°C, pH 7.4, it generates NO spontaneously with a half-life of 2.1 min (Schödel et al., 2009). Total SOCS1 and corresponding p65 protein level was also determined in the presence and absence of 50 µM MG132 (Sigma-Aldrich) to inhibit proteasomal lysis.
Molecular modeling and computational docking.
The 3-dimensional structure of SOCS1 region (residues 65–212) was available from Protein Model Port, while a model of the N terminus (residues 1–64) was constructed using the I-Tasser modeling server. Ramachandran plot analysis was done using SAVES server to test the model quality. The 3-dimensional conformer of the substrate S-nitrosylated glutathione (GSNO; CID_104858) was initially retrieved from Pubchem compound search. The Accelrys discovery studio program (Discovery Studio Modeling Environment; Release 4.0; Accelrys Software Inc.) was used to energy minimize the structure using CHARMM force field parameters. Charges on the NO-Cys were then manually implemented as reported in a recently published study (Han, 2008). GSNO was then exported to Mcule docking for docking calculations. Mcule utilizes the docking calculations based on program AutoDock Vina. The preparation of protein and substrate file was done according to the standard AutoDock procedure. In brief, including conversion of input ligands defined by mcule IDs, SMILES, or InChI strings to 2D MOL, generation of defined stereoisomers, conversion of ligand 2D MOL to 3D MOL, conversion of ligand 3D MOL to PDBQT, preparation of docking targets and finally docking. Out of the best four docking poses the top scoring one was analyzed further. Castp server was used for protein pocket detection. The hydrophobicity index for the pocket was calculated according to the Kyte-Doolittle scale (Kyte and Doolittle, 1982) using the Protein GRAVY module of the Sequence Manipulation Suite server.
Statistical analyses.
Statistical analyses were performed using Microsoft Excel or Prism (GraphPad). Significance was assessed using two-tailed Student’s t test, Anova two-way test with Bonferroni post-test, or Fisher’s exact test. All data are representative of at least two independent experiments and error refers to standard deviation unless otherwise stated.
Acknowledgments
The authors are indebted to Ms. Mary Maliakal for preliminary work on NOS1 regulation of p65 phosphorylation.
This work was sponsored by the American Heart Association, Scientist Development Grants nos. 09SDG2250933 and 13GRNT16400010; and Department of Defense grant W911NF-12-1-0493/61758-LS to M.G. Bonini.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- BMDM
- BM-derived macrophage
- CLP
- cecal ligation and puncture
- ELAM
- endothelial cell-leukocyte adhesion molecule
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- GSNO
- S-nitrosoglutathione
- HDAC
- histone deacetylase
- NO
- nitric oxide
- NOS
- NO synthase
- SOCS
- suppressor of cytokine signaling
- TRIM
- 1-(2-Trifluoromethylphenyl) imidazole
References
- Aktan F. 2004. iNOS-mediated nitric oxide production and its regulation. Life Sci. 75:639–653. 10.1016/j.lfs.2003.10.042 [DOI] [PubMed] [Google Scholar]
- Alcamo E., Mizgerd J.P., Horwitz B.H., Bronson R., Beg A.A., Scott M., Doerschuk C.M., Hynes R.O., and Baltimore D.. 2001. Targeted mutation of TNF receptor I rescues the RelA-deficient mouse and reveals a critical role for NF-kappa B in leukocyte recruitment. J. Immunol. 167:1592–1600. 10.4049/jimmunol.167.3.1592 [DOI] [PubMed] [Google Scholar]
- Badrichani A.Z., Stroka D.M., Bilbao G., Curiel D.T., Bach F.H., and Ferran C.. 1999. Bcl-2 and Bcl-XL serve an anti-inflammatory function in endothelial cells through inhibition of NF-kappaB. J. Clin. Invest. 103:543–553. 10.1172/JCI2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetz A., Frey M., Heeg K., and Dalpke A.H.. 2004. Suppressor of cytokine signaling (SOCS) proteins indirectly regulate toll-like receptor signaling in innate immune cells. J. Biol. Chem. 279:54708–54715. 10.1074/jbc.M410992200 [DOI] [PubMed] [Google Scholar]
- Baltimore D. 2011. NF-κB is 25. Nat. Immunol. 12:683–685. 10.1038/ni.2072 [DOI] [PubMed] [Google Scholar]
- Basak S., Kim H., Kearns J.D., Tergaonkar V., O’Dea E., Werner S.L., Benedict C.A., Ware C.F., Ghosh G., Verma I.M., and Hoffmann A.. 2007. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 128:369–381. 10.1016/j.cell.2006.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Neriah Y. 2003. Pinning NF-kappaB to the nucleus. Mol. Cell. 12:1344–1345. 10.1016/S1097-2765(03)00493-3 [DOI] [PubMed] [Google Scholar]
- Block M.L., and Hong J.S.. 2005. Microglia and inflammation-mediated neurodegeneration: multiple triggers with a common mechanism. Prog. Neurobiol. 76:77–98. 10.1016/j.pneurobio.2005.06.004 [DOI] [PubMed] [Google Scholar]
- Bowie A., and O’Neill L.A.. 2000. Oxidative stress and nuclear factor-kappaB activation: a reassessment of the evidence in the light of recent discoveries. Biochem. Pharmacol. 59:13–23. 10.1016/S0006-2952(99)00296-8 [DOI] [PubMed] [Google Scholar]
- Bubici C., Papa S., Dean K., and Franzoso G.. 2006. Mutual cross-talk between reactive oxygen species and nuclear factor-kappa B: molecular basis and biological significance. Oncogene. 25:6731–6748. 10.1038/sj.onc.1209936 [DOI] [PubMed] [Google Scholar]
- de Souza K.P., Silva E.G., de Oliveira Rocha E.S., Figueiredo L.B., de Almeida-Leite C.M., Arantes R.M., de Assis Silva Gomes J., Ferreira G.P., de Oliveira J.G., Kroon E.G., and Campos M.A.. 2013. Nitric oxide synthase expression correlates with death in an experimental mouse model of dengue with CNS involvement. Virol. J. 10:267 10.1186/1743-422X-10-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- delaTorre A., Schroeder R.A., Punzalan C., and Kuo P.C.. 1999. Endotoxin-mediated S-nitrosylation of p50 alters NF-kappa B-dependent gene transcription in ANA-1 murine macrophages. J. Immunol. 162:4101–4108. [PubMed] [Google Scholar]
- Dixon B. 2004. The role of microvascular thrombosis in sepsis. Anaesth. Intensive Care. 32:619–629. [DOI] [PubMed] [Google Scholar]
- Dombrovskiy V.Y., Martin A.A., Sunderram J., and Paz H.L.. 2007. Rapid increase in hospitalization and mortality rates for severe sepsis in the United States: a trend analysis from 1993 to 2003. Crit. Care Med. 35:1244–1250. 10.1097/01.CCM.0000261890.41311.E9 [DOI] [PubMed] [Google Scholar]
- Driessler F., Venstrom K., Sabat R., Asadullah K., and Schottelius A.J.. 2004. Molecular mechanisms of interleukin-10-mediated inhibition of NF-kappaB activity: a role for p50. Clin. Exp. Immunol. 135:64–73. 10.1111/j.1365-2249.2004.02342.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duma D., Fernandes D., Bonini M.G., Stadler K., Mason R.P., and Assreuy J.. 2011. NOS-1-derived NO is an essential triggering signal for the development of systemic inflammatory responses. Eur. J. Pharmacol. 668:285–292. 10.1016/j.ejphar.2011.05.065 [DOI] [PubMed] [Google Scholar]
- Edgley A.J., Krum H., and Kelly D.J.. 2012. Targeting fibrosis for the treatment of heart failure: a role for transforming growth factor-β. Cardiovasc Ther. 30:e30–e40. 10.1111/j.1755-5922.2010.00228.x [DOI] [PubMed] [Google Scholar]
- Fieren M.W. 2012. The local inflammatory responses to infection of the peritoneal cavity in humans: their regulation by cytokines, macrophages, and other leukocytes. Mediators Inflamm. 2012:976241 10.1155/2012/976241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner B.N., Simmons R.M., Canavera S.J., Akira S., and Underhill D.M.. 2003. Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J. Exp. Med. 197:1107–1117. 10.1084/jem.20021787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H., Wittwer T., Dittrich-Breiholz O., Kracht M., and Schmitz M.L.. 2009. Phosphorylation of NF-kappaB p65 at Ser468 controls its COMMD1-dependent ubiquitination and target gene-specific proteasomal elimination. EMBO Rep. 10:381–386. 10.1038/embor.2009.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras S., Parganas E., de Pauw A., Ihle J.N., and Murray P.J.. 2004. Re-examination of the role of suppressor of cytokine signaling 1 (SOCS1) in the regulation of toll-like receptor signaling. J. Biol. Chem. 279:54702–54707. 10.1074/jbc.M411043200 [DOI] [PubMed] [Google Scholar]
- Green L.S., Chun L.E., Patton A.K., Sun X., Rosenthal G.J., and Richards J.P.. 2012. Mechanism of inhibition for N6022, a first-in-class drug targeting S-nitrosoglutathione reductase. Biochemistry. 51:2157–2168. 10.1021/bi201785u [DOI] [PubMed] [Google Scholar]
- Grollman A.P. 1968. Inhibitors of protein biosynthesis. V. Effects of emetine on protein and nucleic acid biosynthesis in HeLa cells. J. Biol. Chem. 243:4089–4094. [PubMed] [Google Scholar]
- Han S. 2008. Force field parameters for S-nitrosocysteine and molecular dynamics simulations of S-nitrosated thioredoxin. Biochem. Biophys. Res. Commun. 377:612–616. 10.1016/j.bbrc.2008.10.017 [DOI] [PubMed] [Google Scholar]
- Handy R.L., Wallace P., Gaffen Z.A., Whitehead K.J., and Moore P.K.. 1995. The antinociceptive effect of 1-(2-trifluoromethylphenyl) imidazole (TRIM), a potent inhibitor of neuronal nitric oxide synthase in vitro, in the mouse. Br. J. Pharmacol. 116:2349–2350. 10.1111/j.1476-5381.1995.tb15078.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey M.J., Sharkey K.A., Sihota E.G., Reinhardt P.H., Macmicking J.D., Nathan C., and Kubes P.. 1997. Inducible nitric oxide synthase-deficient mice have enhanced leukocyte-endothelium interactions in endotoxemia. FASEB J. 11:955–964. [DOI] [PubMed] [Google Scholar]
- Huang Z., Hoffmann F.W., Fay J.D., Hashimoto A.C., Chapagain M.L., Kaufusi P.H., and Hoffmann P.R.. 2012. Stimulation of unprimed macrophages with immune complexes triggers a low output of nitric oxide by calcium-dependent neuronal nitric-oxide synthase. J. Biol. Chem. 287:4492–4502. 10.1074/jbc.M111.315598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume D.A., Underhill D.M., Sweet M.J., Ozinsky A.O., Liew F.Y., and Aderem A.. 2001. Macrophages exposed continuously to lipopolysaccharide and other agonists that act via toll-like receptors exhibit a sustained and additive activation state. BMC Immunol. 2:11 10.1186/1471-2172-2-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M.D., and Harrison S.C.. 1998. Structure of an IkappaBalpha/NF-kappaB complex. Cell. 95:749–758. 10.1016/S0092-8674(00)81698-0 [DOI] [PubMed] [Google Scholar]
- Jacobson J.R., and Birukov K.G.. 2009. Activation of NFkB and coagulation in lung injury by hyperoxia and excessive mechanical ventilation: one more reason “low and slow” is the way to go? Transl. Res. 154:219–221. 10.1016/j.trsl.2009.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaid K., Rahman A., Anwar K.N., Frey R.S., Minshall R.D., and Malik A.B.. 2003. Tumor necrosis factor-alpha induces early-onset endothelial adhesivity by protein kinase Czeta-dependent activation of intercellular adhesion molecule-1. Circ. Res. 92:1089–1097. 10.1161/01.RES.0000072971.88704.CB [DOI] [PubMed] [Google Scholar]
- Jeon Y.J., Han S.H., Kang J.S., Koh W.S., and Yang K.H.. 1999. Acetylaminofluorene inhibits nitric oxide production in LPS-stimulated RAW 264.7 cells by blocking NF-kappa B/Rel activation. Toxicol. Lett. 104:195–202. 10.1016/S0378-4274(98)00372-5 [DOI] [PubMed] [Google Scholar]
- Kanwar J.R., Kanwar R.K., Burrow H., and Baratchi S.. 2009. Recent advances on the roles of NO in cancer and chronic inflammatory disorders. Curr. Med. Chem. 16:2373–2394. 10.2174/092986709788682155 [DOI] [PubMed] [Google Scholar]
- Kelleher Z.T., Matsumoto A., Stamler J.S., and Marshall H.E.. 2007. NOS2 regulation of NF-kappaB by S-nitrosylation of p65. J. Biol. Chem. 282:30667–30672. 10.1074/jbc.M705929200 [DOI] [PubMed] [Google Scholar]
- Kinjyo I., Hanada T., Inagaki-Ohara K., Mori H., Aki D., Ohishi M., Yoshida H., Kubo M., and Yoshimura A.. 2002. SOCS1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity. 17:583–591. 10.1016/S1074-7613(02)00446-6 [DOI] [PubMed] [Google Scholar]
- Knowles R.G., and Moncada S.. 1994. Nitric oxide synthases in mammals. Biochem. J. 298:249–258. 10.1042/bj2980249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelink P.J., Overbeek S.A., Braber S., de Kruijf P., Folkerts G., Smit M.J., and Kraneveld A.D.. 2012. Targeting chemokine receptors in chronic inflammatory diseases: an extensive review. Pharmacol. Ther. 133:1–18. 10.1016/j.pharmthera.2011.06.008 [DOI] [PubMed] [Google Scholar]
- Kyte J., and Doolittle R.F.. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132. 10.1016/0022-2836(82)90515-0 [DOI] [PubMed] [Google Scholar]
- Lange M., Nakano Y., Traber D.L., Hamahata A., Esechie A., Jonkam C., Bansal K., Traber L.D., and Enkhbaatar P.. 2010. Role of different nitric oxide synthase isoforms in a murine model of acute lung injury and sepsis. Biochem. Biophys. Res. Commun. 399:286–291. 10.1016/j.bbrc.2010.07.071 [DOI] [PubMed] [Google Scholar]
- Lawrence T. 2009. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 1:a001651 10.1101/cshperspect.a001651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Grivennikov S.I., and Karin M.. 2011. The unholy trinity: inflammation, cytokines, and STAT3 shape the cancer microenvironment. Cancer Cell. 19:429–431. 10.1016/j.ccr.2011.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.C., Huang G.D., Hsieh C.W., and Wung B.S.. 2012. The glutathionylation of p65 modulates NF-κB activity in 15-deoxy-Δ¹²,¹⁴-prostaglandin J₂-treated endothelial cells. Free Radic. Biol. Med. 52:1844–1853. 10.1016/j.freeradbiomed.2012.02.028 [DOI] [PubMed] [Google Scholar]
- Linossi E.M., and Nicholson S.E.. 2012. The SOCS box-adapting proteins for ubiquitination and proteasomal degradation. IUBMB Life. 64:316–323. 10.1002/iub.1011 [DOI] [PubMed] [Google Scholar]
- Liu Y. 2011. Cellular and molecular mechanisms of renal fibrosis. Nat. Rev. Nephrol. 7:684–696. 10.1038/nrneph.2011.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A., and Malynn B.A.. 2012. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nat. Rev. Immunol. 12:774–785. 10.1038/nri3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J., Xie Q.W., and Nathan C.. 1997. Nitric oxide and macrophage function. Annu. Rev. Immunol. 15:323–350. 10.1146/annurev.immunol.15.1.323 [DOI] [PubMed] [Google Scholar]
- Marshall H.E., and Stamler J.S.. 2001. Inhibition of NF-kappa B by S-nitrosylation. Biochemistry. 40:1688–1693. 10.1021/bi002239y [DOI] [PubMed] [Google Scholar]
- Marshall H.E., Hess D.T., and Stamler J.S.. 2004. S-nitrosylation: physiological regulation of NF-kappaB. Proc. Natl. Acad. Sci. USA. 101:8841–8842. 10.1073/pnas.0403034101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthay M.A., and Zemans R.L.. 2011. The acute respiratory distress syndrome: pathogenesis and treatment. Annu. Rev. Pathol. 6:147–163. 10.1146/annurev-pathol-011110-130158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews J.R., Botting C.H., Panico M., Morris H.R., and Hay R.T.. 1996. Inhibition of NF-kappaB DNA binding by nitric oxide. Nucleic Acids Res. 24:2236–2242. 10.1093/nar/24.12.2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli I., Geng H., Sebald A., Hodel M., Bucher C., Kracht M., and Schmitz M.L.. 2006. Inducible phosphorylation of NF-kappa B p65 at serine 468 by T cell costimulation is mediated by IKK epsilon. J. Biol. Chem. 281:6175–6183. 10.1074/jbc.M508045200 [DOI] [PubMed] [Google Scholar]
- Miki S., Takeyama N., Tanaka T., and Nakatani T.. 2005. Immune dysfunction in endotoxicosis: role of nitric oxide produced by inducible nitric oxide synthase. Crit. Care Med. 33:716–720. 10.1097/01.CCM.0000159200.69314.3A [DOI] [PubMed] [Google Scholar]
- Nacher M., and Hidalgo A.. 2011. Fire within the vessels: interactions between blood cells and inflammatory vascular injury. Front Biosci (Schol Ed). 3:1089–1100. 10.2741/213 [DOI] [PubMed] [Google Scholar]
- Nakagawa R., Naka T., Tsutsui H., Fujimoto M., Kimura A., Abe T., Seki E., Sato S., Takeuchi O., Takeda K., et al. 2002. SOCS-1 participates in negative regulation of LPS responses. Immunity. 17:677–687. 10.1016/S1074-7613(02)00449-1 [DOI] [PubMed] [Google Scholar]
- Nicholas C., Batra S., Vargo M.A., Voss O.H., Gavrilin M.A., Wewers M.D., Guttridge D.C., Grotewold E., and Doseff A.I.. 2007. Apigenin blocks lipopolysaccharide-induced lethality in vivo and proinflammatory cytokines expression by inactivating NF-kappaB through the suppression of p65 phosphorylation. J. Immunol. 179:7121–7127. 10.4049/jimmunol.179.10.7121 [DOI] [PubMed] [Google Scholar]
- Panettieri R.A. Jr., Kotlikoff M.I., Gerthoffer W.T., Hershenson M.B., Woodruff P.G., Hall I.P., and Banks-Schlegel S.. National Heart, Lung, and Blood Institute. 2008. Airway smooth muscle in bronchial tone, inflammation, and remodeling: basic knowledge to clinical relevance. Am. J. Respir. Crit. Care Med. 177:248–252. 10.1164/rccm.200708-1217PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.H., Kim K.E., Hwang H.Y., and Kim T.Y.. 2003. Regulatory effect of SOCS on NF-kappaB activity in murine monocytes/macrophages. DNA Cell Biol. 22:131–139. 10.1089/104454903321515931 [DOI] [PubMed] [Google Scholar]
- Parsons P.E., Eisner M.D., Thompson B.T., Matthay M.A., Ancukiewicz M., Bernard G.R., and Wheeler A.P.. NHLBI Acute Respiratory Distress Syndrome Clinical Trials Network. 2005. Lower tidal volume ventilation and plasma cytokine markers of inflammation in patients with acute lung injury. Crit. Care Med. 33:1–6, discussion :230–232. 10.1097/01.CCM.0000149854.61192.DC [DOI] [PubMed] [Google Scholar]
- Peng H.B., Libby P., and Liao J.K.. 1995. Induction and stabilization of I kappa B alpha by nitric oxide mediates inhibition of NF-kappa B. J. Biol. Chem. 270:14214–14219. 10.1074/jbc.270.23.14214 [DOI] [PubMed] [Google Scholar]
- Pittet L.A., Quinton L.J., Yamamoto K., Robson B.E., Ferrari J.D., Algül H., Schmid R.M., and Mizgerd J.P.. 2011. Earliest innate immune responses require macrophage RelA during pneumococcal pneumonia. Am. J. Respir. Cell Mol. Biol. 45:573–581. 10.1165/rcmb.2010-0210OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porras M., Martín M.T., Torres R., and Vergara P.. 2006. Cyclical upregulated iNOS and long-term downregulated nNOS are the bases for relapse and quiescent phases in a rat model of IBD. Am. J. Physiol. Gastrointest. Liver Physiol. 290:G423–G430. 10.1152/ajpgi.00323.2005 [DOI] [PubMed] [Google Scholar]
- Reynaert N.L., Ckless K., Korn S.H., Vos N., Guala A.S., Wouters E.F., van der Vliet A., and Janssen-Heininger Y.M.. 2004. Nitric oxide represses inhibitory kappaB kinase through S-nitrosylation. Proc. Natl. Acad. Sci. USA. 101:8945–8950. 10.1073/pnas.0400588101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittirsch D., Huber-Lang M.S., Flierl M.A., and Ward P.A.. 2009. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat. Protoc. 4:31–36. 10.1038/nprot.2008.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi B., Angiari S., Zenaro E., Budui S.L., and Constantin G.. 2011. Vascular inflammation in central nervous system diseases: adhesion receptors controlling leukocyte-endothelial interactions. J. Leukoc. Biol. 89:539–556. 10.1189/jlb.0710432 [DOI] [PubMed] [Google Scholar]
- Ruland J. 2011. Return to homeostasis: downregulation of NF-κB responses. Nat. Immunol. 12:709–714. 10.1038/ni.2055 [DOI] [PubMed] [Google Scholar]
- Ryo A., Suizu F., Yoshida Y., Perrem K., Liou Y.C., Wulf G., Rottapel R., Yamaoka S., and Lu K.P.. 2003. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell. 12:1413–1426. 10.1016/S1097-2765(03)00490-8 [DOI] [PubMed] [Google Scholar]
- Sabatel H., Di Valentin E., Gloire G., Dequiedt F., Piette J., and Habraken Y.. 2012. Phosphorylation of p65(RelA) on Ser(547) by ATM represses NF-κB-dependent transcription of specific genes after genotoxic stress. PLoS ONE. 7:e38246 10.1371/journal.pone.0038246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadik C.D., Kim N.D., and Luster A.D.. 2011. Neutrophils cascading their way to inflammation. Trends Immunol. 32:452–460. 10.1016/j.it.2011.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadikot R.T., Jansen E.D., Blackwell T.R., Zoia O., Yull F., Christman J.W., and Blackwell T.S.. 2001. High-dose dexamethasone accentuates nuclear factor-kappa b activation in endotoxin-treated mice. Am. J. Respir. Crit. Care Med. 164:873–878. 10.1164/ajrccm.164.5.2008059 [DOI] [PubMed] [Google Scholar]
- Saluja R., Saini R., Mitra K., Bajpai V.K., and Dikshit M.. 2010. Ultrastructural immunogold localization of nitric oxide synthase isoforms in rat and human eosinophils. Cell Tissue Res. 340:381–388. 10.1007/s00441-010-0947-y [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schödel J., Padmapriya P., Marx A., Huang P.L., Ertl G., and Kuhlencordt P.J.. 2009. Expression of neuronal nitric oxide synthase splice variants in atherosclerotic plaques of apoE knockout mice. Atherosclerosis. 206:383–389. 10.1016/j.atherosclerosis.2009.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Jung Y.J., Tyagi S., Gennaro M.L., and North R.J.. 2003. Expression of Th1-mediated immunity in mouse lungs induces a Mycobacterium tuberculosis transcription pattern characteristic of nonreplicating persistence. Proc. Natl. Acad. Sci. USA. 100:241–246. 10.1073/pnas.0136863100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebovsky J., Walker P., Lang R., and Dalpke A.H.. 2011. Suppressor of cytokine signaling 1 (SOCS1) limits NFkappaB signaling by decreasing p65 stability within the cell nucleus. FASEB J. 25:863–874. 10.1096/fj.10-170597 [DOI] [PubMed] [Google Scholar]
- Summers deLuca L., and Gommerman J.L.. 2012. Fine-tuning of dendritic cell biology by the TNF superfamily. Nat. Rev. Immunol. 12:339–351. 10.1038/nri3193 [DOI] [PubMed] [Google Scholar]
- Sun B., and Karin M.. 2012. Obesity, inflammation, and liver cancer. J. Hepatol. 56:704–713. 10.1016/j.jhep.2011.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Wasley J.W., Qiu J., Blonder J.P., Stout A.M., Green L.S., Strong S.A., Colagiovanni D.B., Richards J.P., Mutka S.C., et al. 2011. Discovery of s-nitrosoglutathione reductase inhibitors: potential agents for the treatment of asthma and other inflammatory diseases. ACS Med Chem Lett. 2:402–406. 10.1021/ml200045s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa H., Boettcher S., and Manz M.G.. 2012. Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood. 119:2991–3002. 10.1182/blood-2011-12-380113 [DOI] [PubMed] [Google Scholar]
- Tiruppathi C., Shimizu J., Miyawaki-Shimizu K., Vogel S.M., Bair A.M., Minshall R.D., Predescu D., and Malik A.B.. 2008. Role of NF-kappaB-dependent caveolin-1 expression in the mechanism of increased endothelial permeability induced by lipopolysaccharide. J. Biol. Chem. 283:4210–4218. 10.1074/jbc.M703153200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togashi H., Sasaki M., Frohman E., Taira E., Ratan R.R., Dawson T.M., and Dawson V.L.. 1997. Neuronal (type I) nitric oxide synthase regulates nuclear factor kappaB activity and immunologic (type II) nitric oxide synthase expression. Proc. Natl. Acad. Sci. USA. 94:2676–2680. 10.1073/pnas.94.6.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S., and Karin M.. 2009. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 27:693–733. 10.1146/annurev.immunol.021908.132641 [DOI] [PubMed] [Google Scholar]
- Vanden Berghe W., De Bosscher K., Boone E., Plaisance S., and Haegeman G.. 1999. The nuclear factor-kappaB engages CBP/p300 and histone acetyltransferase activity for transcriptional activation of the interleukin-6 gene promoter. J. Biol. Chem. 274:32091–32098. 10.1074/jbc.274.45.32091 [DOI] [PubMed] [Google Scholar]
- Villanueva C., and Giulivi C.. 2010. Subcellular and cellular locations of nitric oxide synthase isoforms as determinants of health and disease. Free Radic. Biol. Med. 49:307–316. 10.1016/j.freeradbiomed.2010.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel S.M., Gao X., Mehta D., Ye R.D., John T.A., Andrade-Gordon P., Tiruppathi C., and Malik A.B.. 2000. Abrogation of thrombin-induced increase in pulmonary microvascular permeability in PAR-1 knockout mice. Physiol. Genomics. 4:137–145. [DOI] [PubMed] [Google Scholar]
- Wang W., Mitra A., Poole B., Falk S., Lucia M.S., Tayal S., and Schrier R.. 2004. Endothelial nitric oxide synthase-deficient mice exhibit increased susceptibility to endotoxin-induced acute renal failure. Am. J. Physiol. Renal Physiol. 287:F1044–F1048. 10.1152/ajprenal.00136.2004 [DOI] [PubMed] [Google Scholar]
- Xia Y., Pauza M.E., Feng L., and Lo D.. 1997. RelB regulation of chemokine expression modulates local inflammation. Am. J. Pathol. 151:375–387. [PMC free article] [PubMed] [Google Scholar]
- Yuan Z., Liu B., Yuan L., Zhang Y., Dong X., and Lu J.. 2004. Evidence of nuclear localization of neuronal nitric oxide synthase in cultured astrocytes of rats. Life Sci. 74:3199–3209. 10.1016/j.lfs.2003.10.037 [DOI] [PubMed] [Google Scholar]