Sekiya et al. demonstrate that deletion of the nuclear orphan receptor Nr4a in T reg cells results in a fatal systemic immunopathology due to abrogated suppressive capability in limiting Th2 and Tfh conversion.
Abstract
Regulatory T (T reg) cells are central mediators of immune suppression. As such, T reg cells are characterized by a distinct pattern of gene expression, which includes up-regulation of immunosuppressive genes and silencing of inflammatory cytokine genes. Although an increasing number of transcription factors that regulate T reg cells have been identified, the mechanisms by which the T reg cell–specific transcriptional program is maintained and executed remain largely unknown. The Nr4a family of nuclear orphan receptors, which we recently identified as essential for the development of T reg cells, is highly expressed in mature T reg cells as well, suggesting that Nr4a factors play important roles even beyond T reg cell development. Here, we showed that deletion of Nr4a genes specifically in T reg cells caused fatal systemic immunopathology. Nr4a-deficient T reg cells exhibited global alteration of the expression of genes which specify the T reg cell lineage, including reduction of Foxp3 and Ikzf4. Furthermore, Nr4a deficiency abrogated T reg cell suppressive activities and accelerated conversion to cells with Th2 and follicular helper T (Tfh) effector-like characteristics, with heightened expression of Th2 and Tfh cytokine genes. These findings demonstrate that Nr4a factors play crucial roles in mature T reg cells by directly controlling a genetic program indispensable for T reg cell maintenance and function.
T reg cells are a distinct CD4+ T cell subset with immunosuppressive activities. Targets of T reg cell–mediated suppression include a variety of immune cells in both the innate and adaptive arms of immunity, and T reg cells act at both the priming and effector phases of immune reactions. Thus, T reg cells enforce proper control of self-reactive and pathogen-specific immune responses (Miyara and Sakaguchi, 2007; Sakaguchi et al., 2008; Vignali et al., 2008). For example, repression of germinal center (GC) B cell reactions is exerted through several targets. T reg cells can suppress the initial priming of naive CD4+ T (T naive) cells by dendritic cells, thus preventing differentiation of Tfh cells. In addition, T reg cells, especially those localized in GCs, can repress the ability of Tfh cell to activate GC B cells (Chung et al., 2011; Linterman et al., 2011). The T reg subset residing in the GC, called follicular regulatory T (Tfr) cells, is also thought to directly suppress GC B cell activities (Lim et al., 2004; Iikuni et al., 2009).
T reg cells exert suppressive functions through cell contact–dependent and –independent mechanisms by expressing cell surface molecules including CTLA-4 and CD25, as well as the antiinflammatory cytokines IL-10 and TGF-β1 (Takahashi et al., 1998; Hori et al., 2003; Khattri et al., 2003). Importantly, the expression of proinflammatory cytokines including IFN-γ, IL-2, and IL-4 is strictly repressed in T reg cells (Thornton and Shevach, 1998; Thornton and Shevach, 2000; Fontenot et al., 2003; Hori et al., 2003). As expected based on the wide variety of immunosuppressive functions of T reg cells, various transcription factors (TFs) have been reported to regulate their immunosuppressive genetic program (Luo and Li, 2013). The contribution of each TF ranges from global to specific, as revealed by the variety of phenotypes induced upon disruption of each transcription factor in T reg cells. For example, deletion of Foxp3 in T reg cells results in the acute development of autoimmune diseases. The severity of these diseases is similar to the severity of those observed in T reg cell–deficient mice, revealing the central role of Foxp3 as a lineage-specifying TF in T reg cells (Kim et al., 2007). However, disruption of Foxo1 or Eos does not globally disturb gene expression in T reg cells, but instead leads to dysregulation of a set of inflammatory genes, which includes Ifng (Pan et al., 2009; Ouyang et al., 2012). Although an increasing number of TFs that regulate T reg cells have been identified, the mechanisms by which the T reg cell–specific transcriptional program is maintained and/or executed remain largely unknown. For example, the fact that Foxp3 has been suggested to amplify or stabilize rather than to initiate the T reg cell–transcriptional program during T reg cell development implies the existence of other TFs that also globally regulate the T reg cell genetic program (Gavin et al., 2007; Lin et al., 2007). In addition, how T reg cells repress the expression of cytokines IL-4 and IL-21, whose aberrant activation potentially compromises the ability of T reg cells to control GC reactions, remains largely unknown as well.
We recently demonstrated that members of the Nr4a family of nuclear orphan receptors, via their ability to induce Foxp3, play crucial roles in T reg cell differentiation (Sekiya et al., 2011, 2013). T cell–specific deletion of all Nr4a family members (Nr4a1, Nr4a2, and Nr4a3) results in complete loss of T reg cells and development of severe systemic autoimmunity (Sekiya et al., 2013). However, because all Nr4a family members are up-regulated in T reg cells, it is likely that they play roles in mature T reg cells as well (Hill et al., 2007; Lin et al., 2007; Wei et al., 2009; Moran et al., 2011; Sekiya et al., 2011). To test this, we deleted Nr4a factors specifically in T reg cells to elucidate their roles in this T cell subset. Our findings reveal crucial roles for Nr4a factors in T reg cells as shown by the various immunological abnormalities occurring upon their deletion in T reg cells. We also found that Nr4a factors globally regulate a T reg cell–transcriptional program, including sustained expression of the key T reg cell effectors Foxp3, Ikzf4, and Il2ra, as well as repression of Il4 and Il21. Thus, Nr4a factors maintain T reg cell–lineage stability and T reg cell suppressive activities.
RESULTS
Development of systemic immunopathology in mice lacking Nr4a factors in T reg cells
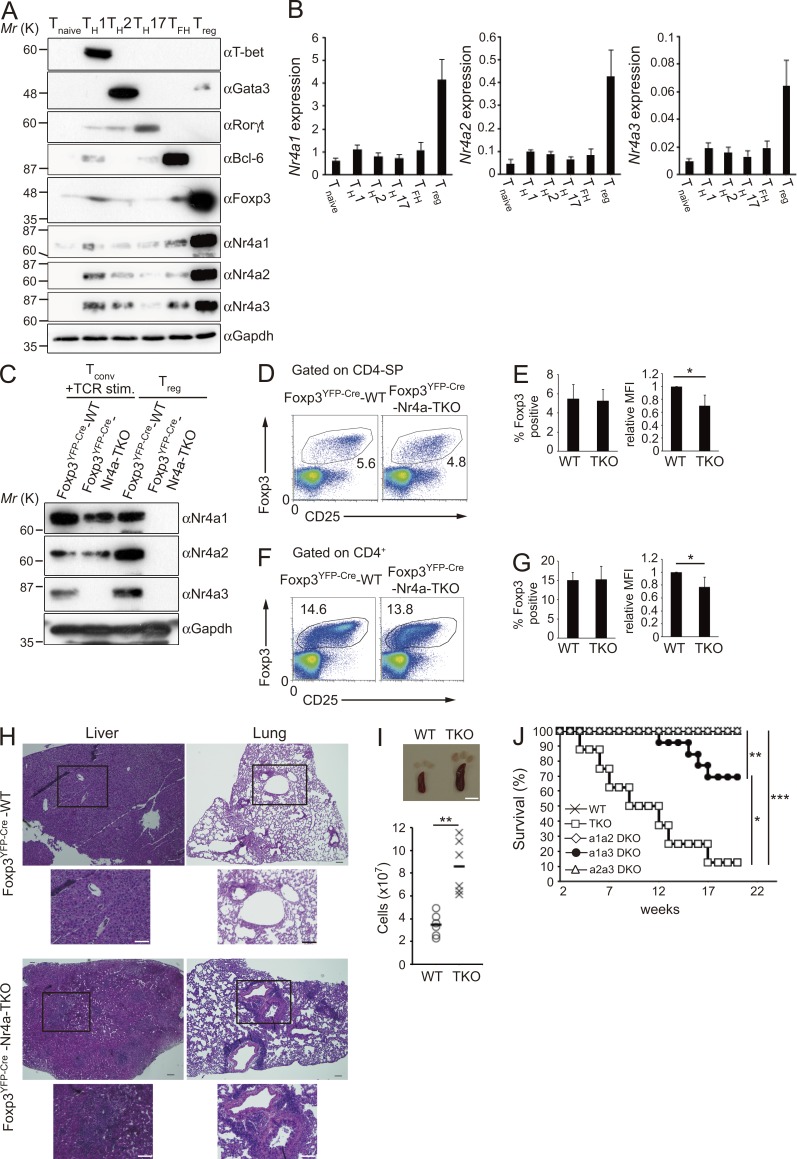
As expression levels of all Nr4a family members have been reported to be elevated in T reg cells, it was expected that Nr4a factors play important roles in mature T reg cells. First, we confirmed higher expression of all Nr4a family members in T reg cells compared with other CD4+ T cell subsets, at both mRNA and protein levels (Fig. 1, A and B). Next, because the complete absence of T reg cells upon T cell–specific deletion of Nr4a genes using CD4-Cre hampered analysis of their function in T reg cells, we conditionally deleted all Nr4a genes specifically in T reg cells, by crossing Foxp3YFP-Cre mice (Rubtsov et al., 2008) with Nr4a1f/fNr4a2f/fNr4a3−/− (Sekiya et al., 2013; called Foxp3YFP-Cre Nr4a-triple knockout [Foxp3YFP-Cre-Nr4a-TKO] herein) mice. We confirmed specific ablation of Nr4a1 and Nr4a2 in T reg cells, but not in conventional CD4+ T cells, as well as ubiquitous deletion of Nr4a3 (Fig. 1 C).
Figure 1.
Loss of Nr4a expression in T reg cells induces multiorgan autoimmunity. (A) Immunoblot analysis of CD4+ T cell subset markers and Nr4a factors in the indicated CD4+ T cell subsets. (B) qRT-PCR analysis of mRNA expression of Nr4a factors in the indicated CD4+ T cell subsets. Results are presented relative to expression of the control gene Hprt. Data are representative of three independent experiments (mean and SD of triplicates). Protein and mRNA samples, used in A and B, respectively, were prepared from sorted T naive, Tfh, and T reg cells from mice, and from in vitro–differentiated Th1, Th2, and Th17 cells, as described in Materials and methods. (C) Immunoblot analysis of Nr4a factors in sorted conventional CD4+ T (CD4+CD25low) cells and T reg (CD4+CD25hiYFP+) cells from Foxp3YFP-Cre-WT and Foxp3YFP-Cre-Nr4a-TKO mice. Conventional T cells were stimulated with anti-CD3 and anti-CD28 antibodies for 2 h to induce Nr4a proteins to detectable levels. (D) Flow cytometry profiles of thymic CD4-SP cells from 4-wk-old mice. Numbers indicate the percentage of cells in the indicated regions. (E) Quantification of the results in D. (left) Frequency of Foxp3+ cells in CD4-SP cell populations in the thymus. (right) Mean fluorescence intensities (MFI) of Foxp3 in T reg cells. MFI of WT T reg cells are set as one. (F) Flow cytometry profiles of CD4+ T cells from pooled spleens and lymph nodes from 4-wk-old mice. Numbers indicate the percentage of cells in the indicated regions. (G) Quantification of the results in F. (left) Frequency of Foxp3+ cells in CD4+ T cells. (right) MFI of Foxp3 in T reg cells. MFI of WT T reg cells are set as one (n = 5 mice per genotype, pooled from 4 independent cohorts per genotype, mean, and SD). (H) Hematoxylin and eosin staining of tissue sections from 5-wk-old mice. Outlined area indicates area shown enlarged below. Bars, 100 µm. (I) Spleens and lymph nodes from 5-wk-old mice (left), and total cellularity of spleens in 5–7-wk old mice (n = 6 mice per genotype, pooled from 4 independent cohorts per genotype). Each symbol represents an individual mouse; small horizontal lines indicate the mean. Bar, 5 mm. (J) Survival of Foxp3YFP-Cre-WT and Foxp3YFP-Cre-Nr4a-TKO mice, and of mice doubly deficient in Nr4a1 and Nr4a2 (a1a2-DKO), Nr4a1 and Nr4a3 (a1a3-DKO), or Nr4a2 and Nr4a3 (a2a3-DKO) in T reg cells, monitored for 20 wk (n = 8–10 per group). *, P < 0.05; **, P < 0.01; ***, P < 0.005 (Mann-Whitney U tests [E, G, and I], Kaplan-Meier analysis with Mantel-Cox log-rank tests [J]).
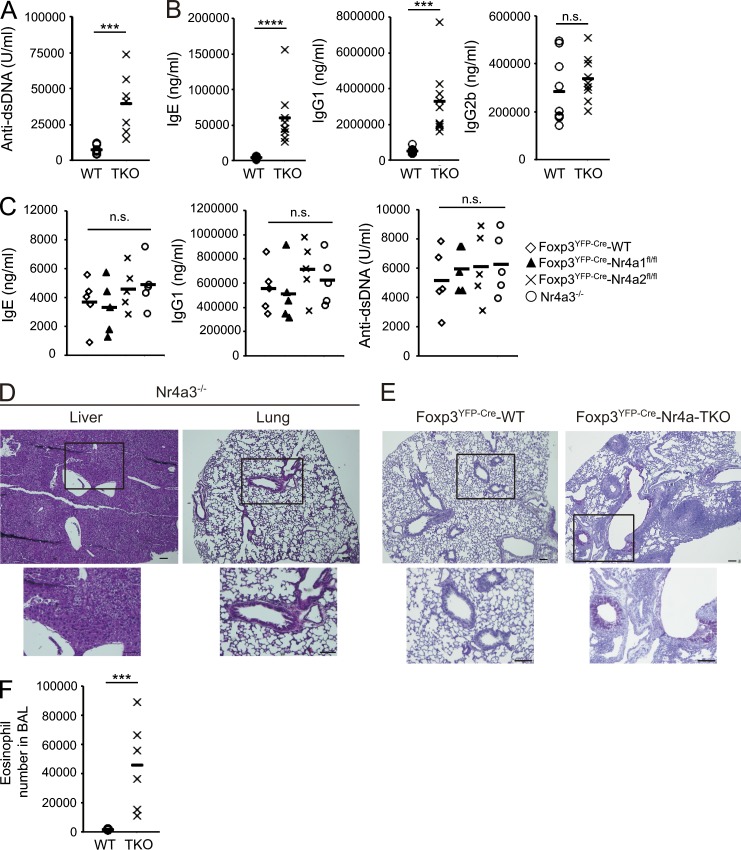
In Foxp3YFP-Cre-Nr4a-TKO mice, T reg cells were observed at proportions comparable to those in Foxp3YFP-CreNr4a1+/+Nr4a2+/+Nr4a3+/+ (called Foxp3YFP-Cre-WT herein) mice. However, expression levels of Foxp3 in T reg cells were significantly lower in Foxp3YFP-Cre-Nr4a-TKO mice (Fig. 1, D–G). Massive lymphoproliferation was observed in Foxp3YFP-Cre-Nr4a-TKO mice, and the mice developed fatal multiorgan autoimmunity (Fig. 1, H–J). The life span of Foxp3YFP-Cre-Nr4a-TKO mice was much shorter than those of each combination of double-knockouts (DKOs), indicating redundancy among Nr4a family members (Fig. 1 J). Among the DKO combinations, only Nr4a1a3-DKO mice showed autoimmune phenotypes and shorter life spans, showing that Nr4a1 and Nr4a3 made stronger contributions than Nr4a2 in T reg cells. Levels of autoreactive antibodies, and of IgG1 and IgE, were significantly elevated in Foxp3YFP-Cre-Nr4a-TKO mice (Fig. 2, A and B). Levels of IgG1 and IgE and levels of autoreactive antibodies were not elevated in Nr4a single-knockouts, showing again the functional redundancy among Nr4a factors in T reg cells (Fig. 2 C). In addition, as Nr4a3 was ubiquitously deleted, T reg cell–extrinsic effects may be a concern in the interpretation of the phenotypes of Foxp3YFP-Cre-Nr4a-TKO mice. However, as no signs of inflammation were detected in Nr4a3−/− mice, such T reg cell–extrinsic effects of Nr4a3 deletion can be ignored in Foxp3YFP-Cre-Nr4a-TKO mice (Fig. 2, C and D).
Figure 2.
Th2/IgE-type immune reactions are evident in Foxp3YFP-Cre-Nr4a-TKO mice. (A) Titers of antibodies to double-stranded DNA in sera from 5–7-wk-old mice (n = 7 mice per genotype, pooled from 5 independent cohorts per genotype). (B) Titers of IgE, IgG1, and IgG2b in serum from 5–7-wk-old mice (n = 9 mice per genotype, pooled from 7 independent cohorts per genotype). (C) Titers of IgE, IgG1, and anti–double-stranded DNA antibodies in serum from 5–7-wk-old mice (n = 5 mice per genotype, pooled from 4 independent cohorts per genotype). (D) Hematoxylin and eosin staining of tissue sections from 5-wk-old Nr4a3−/− mice. Outlined area indicates area shown enlarged below. Scale bars, 100 µm. (E) Periodic acid-Schiff (PAS) staining of mucus-producing cells (pink) in the lungs from 5-wk-old Foxp3YFP-Cre-WT and Foxp3YFP-Cre-Nr4a-TKO mice. Outlined area indicates area shown enlarged below. Bars, 100 µm. (F) Total eosinophil counts in the bronchoalveolar lavage (BAL) fluids from 5–7-wk-old Foxp3YFP-Cre-WT and Foxp3YFP-Cre-Nr4a-TKO mice (n = 6 mice per genotype, pooled from 5 independent cohorts per genotype). Each symbol in (A, B, C, and F) represents an individual mouse; small horizontal lines indicate the mean. ***, P < 0.005; ****, P < 0.001 (Mann-Whitney tests [A, B, F], one-way ANOVA with Bonferroni multiple comparisons test [C]).
Accelerated mucus production by lung epithelia, as well as intense infiltration of eosinophils into the airways, was observed in Foxp3YFP-Cre-Nr4a-TKO mice (Fig. 2, E and F). These results indicated that Nr4a family members play important roles in T reg cells in a redundant manner, and their deletion results in severe Th2/IgE-type inflammation.
Global alteration of the T reg cell transcriptional program in Nr4a-deficient T reg cells
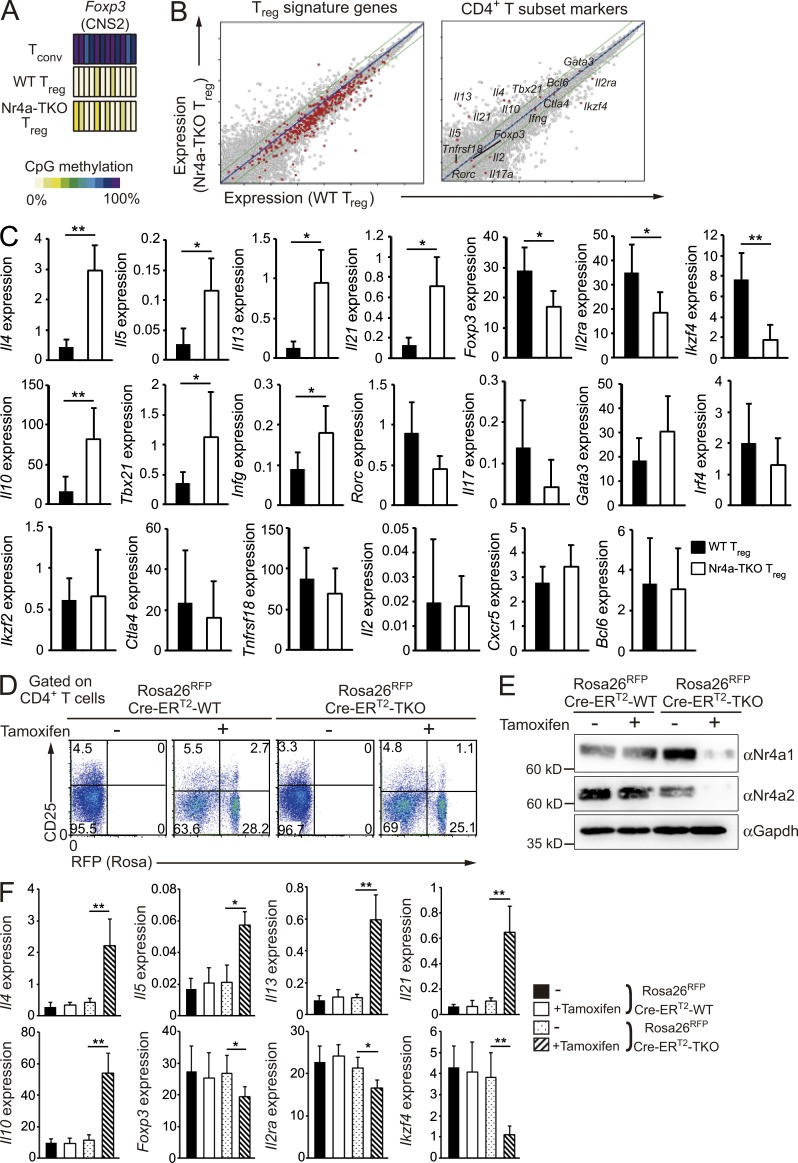
Next, we characterized T reg cells isolated from Foxp3YFP-Cre-Nr4a-TKO mice at a molecular level. We first investigated the methylation pattern at the CNS2 enhancer region of the Foxp3 locus; this region is demethylated specifically in T reg cells (Floess et al., 2007; Kim and Leonard, 2007; Bailey-Bucktrout et al., 2013). Fig. 3 A shows that demethylation of the CNS2 enhancer region was normal in Nr4a-TKO T reg cells. This result showed that Nr4a-TKO T reg cells, sorted as the CD4+YFP+ population from Foxp3YFP-Cre-Nr4a-TKO mice, were truly composed of T reg cells and not conventional CD4+ T cells transiently expressing Foxp3, as this region is not demethylated in such cells (Miyao et al., 2012).
Figure 3.
Dysregulation of T reg cell–associated transcription in Nr4a-TKO T reg cells. (A) CpG methylation profiles of T reg cell–specific demethylated sites at the Foxp3 CNS2 enhancer. (B) Expression profiles of WT T reg cells (horizontal axis) and Nr4a-TKO T reg cells (vertical axis) of the indicated groups of genes. (C) qRT-PCR analysis of the indicated genes in WT and Nr4a-TKO T reg cells from 4–6-wk-old mice. Results are presented relative to expression of the control gene Hprt (n = 10 mice per genotype, pooled from 7 independent cohorts per genotype). (D) Flow cytometry profiles of CD4+ cells from the peripheral bloods of Rosa26RFPCre-ERT2-WT and Rosa26RFPCre-ERT2-Nr4a-TKO mice, before and after tamoxifen treatment. Numbers indicate the percentage of cells in the indicated regions. (E) Immunoblot analysis of Nr4a1 and Nr4a2 in sorted T reg cells (CD4+CD25hiRFP+) from mice treated with tamoxifen, or in sorted T reg cells (CD4+CD25hi) from littermates treated with vehicle. (F) qRT-PCR analysis of mRNA for the indicated genes in T reg cells (CD4+CD25hiRFP+) from mice treated with tamoxifen, or in sorted T reg cells (CD4+CD25hi) from littermate mice treated with vehicle. Results are presented relative to expression of the control gene Hprt (n = 6 mice per genotype, pooled from 2 independent cohorts per genotype). Error bars denote mean ± SD. *, P < 0.05; **, P < 0.01 (Mann-Whitney tests [C], one-way ANOVA with Bonferroni test [F]).
Next, we comprehensively characterized mRNA expression in Nr4a-TKO T reg cells. Notably, genes reported to be exclusively expressed in T reg cells were globally down-regulated in Nr4a-TKO T reg cells (Fig. 3 B, T reg signature genes; Hill et al., 2007). Both microarray analysis and quantitative reverse transcription PCR (qRT-PCR) showed that T reg effector genes, including Ikzf4 and Il2ra, which encode Eos and CD25, respectively, were substantially down-regulated in Nr4a-TKO T reg cells (Fig. 3, B and C). qRT-PCR analysis also showed a significant reduction of Foxp3 in Nr4a-TKO T reg cells (Fig. 3 C). Investigation of other CD4+ T cell subset markers revealed aberrant up-regulation of Th2 and Tfh cytokine genes, including Il4 and Il21, in Nr4a-TKO T reg cells (Fig. 3, B and C).
To confirm that similar mRNA changes occur after acute deletion of Nr4a factors in mature T reg cells, we crossed WT mice or Nr4a1fl/flNr4a2fl/flNr4a3−/− mice with Rosa26-loxP-Stop-loxP, Cre-ERT2 mice (called Rosa26RFPCre-ERT2-WT, and Rosa26RFPCre-ERT2-Nr4a-TKO mice herein). In the Rosa26RFPCre-ERT2-Nr4a-TKO mice, Nr4a1 and Nr4a2 genes are deleted under the control of tamoxifen, and the cells in which Cre was activated are permanently marked by RFP expression (Fig. 3, D and E; Seibler et al., 2003; Luche et al., 2007). As shown in Fig. 3 F, Foxp3, Ikzf4, and Il2ra levels were reduced and Th2 and Tfh cytokine genes were up-regulated in T reg cells (CD4+CD25hi) after acute deletion of Nr4a factors. Thus, these data confirmed that Nr4a factors continuously maintain gene expression in mature T reg cells.
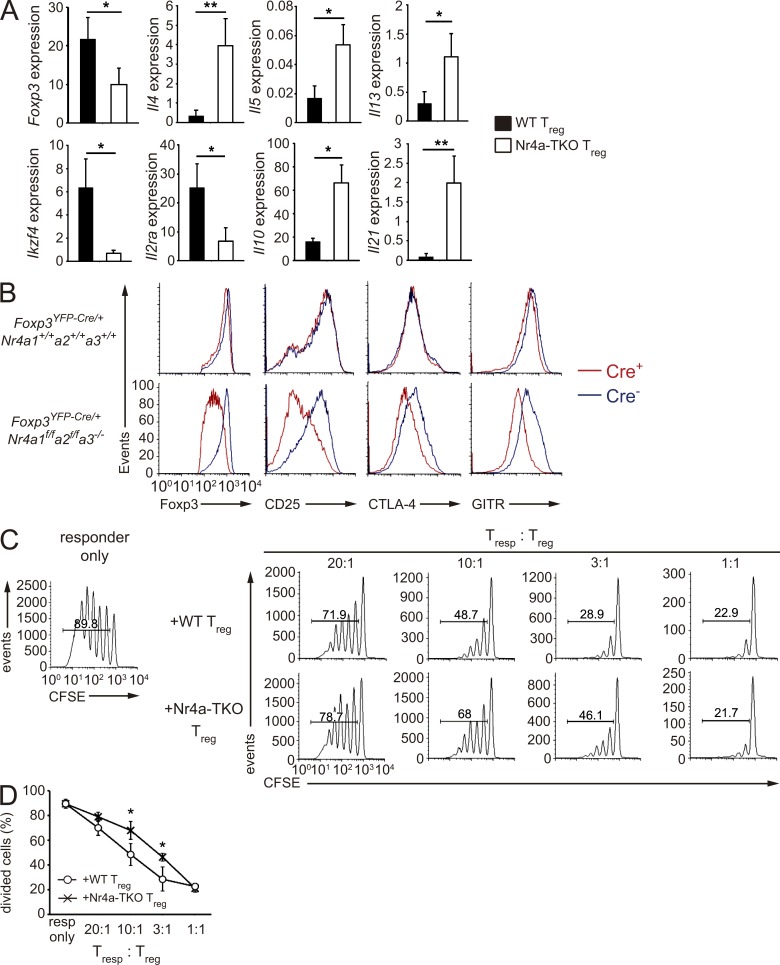
As inflammatory environments potentially disrupt Foxp3 expression, as well as T reg cell lineage integrity (Zhou et al., 2009; Bailey-Bucktrout et al., 2013; Komatsu et al., 2014), the observed abnormality in Nr4a-TKO T reg cells could be attributed to the severe inflammation observed in Foxp3YFP-Cre-Nr4a-TKO mice. To address this issue, we analyzed Foxp3YFP-Cre/+Nr4a1f/fNr4a2f/fNr4a3−/− X-chromosomal Foxp3 gene heterozygous females, in which WT and Nr4a-TKO T reg cells coexist and no overt inflammation was observed at the time of analysis. As shown in Fig. 4 (A and B), reduction of Foxp3 and CD25 levels were specific to the Nr4a-TKO counterparts at both mRNA and protein levels. Reduction of Ikzf4, as well as up-regulation of cytokine genes, was also observed in Nr4a-TKO T reg cells in Foxp3YFP-Cre/+Nr4a1f/fNr4a2f/fNr4a3−/− mice (Fig. 4 A).
Figure 4.
Nr4a-deficient T reg cells show attenuated suppressive activity. (A) qRT-PCR analysis of mRNA for the indicated genes in CD4+YFP+ T reg cells sorted either from 4–6-wk-old Foxp3YFP-Cre/+Nr4a1+/+Nr4a2+/+Nr4a3+/+ or Foxp3YFP-Cre/+Nr4a1f/fNr4a2f/fNr4a3−/− heterozygous female mice. In these mice, Cre+ T reg cells and Cre− T reg cells theoretically coexist at a 1:1 ratio in an individual mouse because Foxp3 is on the X-chromosome and a single X-chromosome is randomly inactivated in each somatic cell in females. Results are presented relative to expression of the control gene Hprt (n = 8 per group). (B) Flow cytometry profiles of the indicated proteins in YFP-Cre+Foxp3+ and YFP-Cre−Foxp3+ T reg cells from 5-wk-old Foxp3YFP-Cre/+Nr4a1+/+Nr4a2+/+Nr4a3+/+ and Foxp3YFP-Cre/+Nr4a1f/fNr4a2f/fNr4a3−/− heterozygous female mice. (C) In vitro suppression assay of WT naive CD4+ T cells (responding T cells, T resp), labeled with the cytosolic dye carboxyfluorescein diacetate succinimidyl ester (CFSE), in the presence of WT or Nr4a-TKO T reg cells. Numbers indicate the percentage of cells in the indicated area. (D) Quantification of results in (C). T resp cell division was analyzed by CFSE dilution at the indicated ratios of T resp and T reg cells. Data are representative of three independent experiments. Error bars denote mean ± SD (A and D). *, P < 0.05; **, P < 0.01 (Mann-Whitney tests [A], two-way ANOVA with Sidak multiple comparisons test [D]).
Next, we functionally characterized Nr4a-TKO T reg cells using an in vitro suppression assay. As shown in Fig. 4 (C and D), when tested on proliferation of responder T naive cells, Nr4a-TKO T reg cells showed attenuated suppression activity. Collectively, these results suggest that Nr4a factors globally regulate T reg cell–specific transcriptional programs and support the full suppressive activity of T reg cells.
Nr4a factors directly regulate T reg cell transcriptional programs
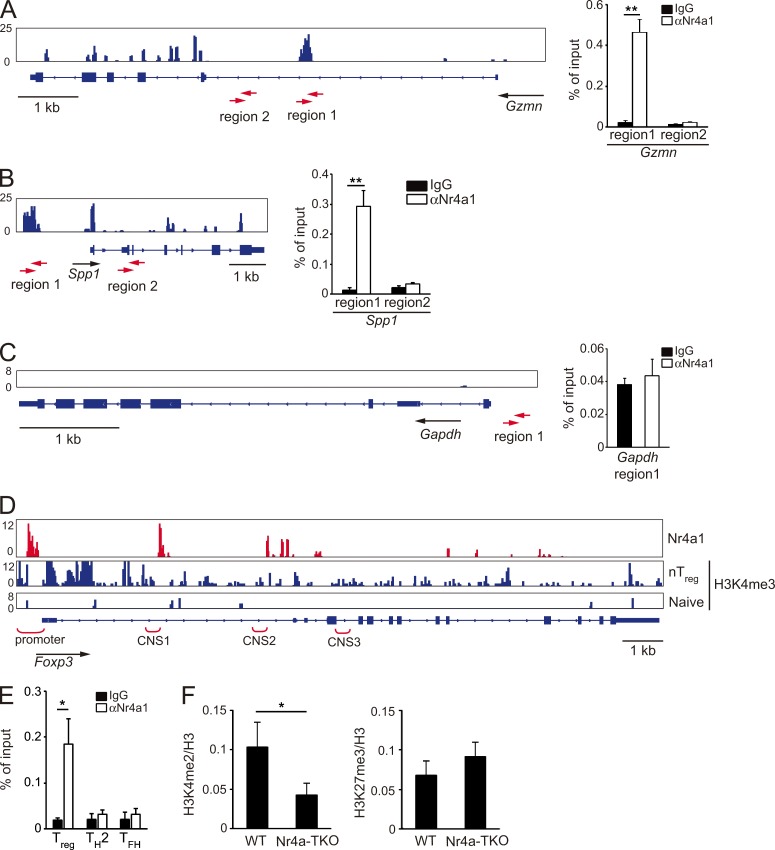
As members of a nuclear receptor super family, Nr4a factors exert their biological activities mainly by regulating transcription (Hamers et al., 2013). We performed ChIP followed by high-throughput sequencing (ChIP-seq) to reveal T reg cell transcriptional programs directly regulated by Nr4a factors. First, the result of the ChIP-seq analysis was evaluated by ChIP-qPCR. Regions in intron 1 of the Gzmn gene and 2 kb upstream of the Spp1 gene, each of which corresponds to randomly selected ChIP peaks with high fidelity in the ChIP-seq analysis, also showed enrichment in anti-Nr4a1 immunoprecipitates compared with control-IgG precipitates in ChIP-qPCR analysis. Regions that did not show peaks in ChIP-seq analysis also did not show binding in ChIP-qPCR analysis (Fig. 5, A–C).
Figure 5.
Direct regulation of T reg cell–associated genes by Nr4a factors. (A–C) Genome browser view of anti-Nr4a1 ChIP-seq profile maps. Distributions of Nr4a1 binding on Gzmn (A), Spp1 (B), and Gapdh (C) loci are shown for WT T reg cells (left). Vertical scales are noted. (right) ChIP-qPCR analysis with anti-Nr4a1 or control IgG antibodies in WT T reg cells, at the indicated regions in Gzmn (A), Spp1 (B), and Gapdh (C) loci. Regions analyzed are shown below the genome browser views (red arrows). Values are presented as the percentage of corresponding input. (D) Genome browser view of ChIP-seq profile maps. The distribution of Nr4a1-binding sites on the Foxp3 locus is shown for WT T reg cells. The distribution of H3K4me3 modifications on Foxp3 (Wei et al., 2009) is shown for nT reg and for T naive cells. Vertical scales are noted. (E) ChIP-qPCR analysis of the enrichment of Nr4a1 at the Foxp3 promoter in WT T reg, Th2, and Tfh cells. Values are presented as the percentage of corresponding input. (F) ChIP-qPCR analysis of the Foxp3 promoter region using chromatin extracts from WT and Nr4a-TKO T reg cells immunoprecipitated with anti-H3K4me2 or anti-H3K27me3 antibodies. Results are presented relative to those obtained with antihistone H3 antibodies, which recognize both modified and unmodified histone H3. Shown are representative results of three biological replicates (right in A–C, E, and F), each performed in triplicate, means ± SD. *, P < 0.05; **, P < 0.01 (Student’s t test).
We found Nr4a1 bound to the promoter, as well as to regions proximal to the CNS1 and CNS2 enhancers of the Foxp3 gene, recapitulating our previous findings by ChIP-qPCR (Sekiya et al., 2011, 2013; Fig. 5 D). ChIP-qPCR analysis revealed that binding of Nr4a1 to the Foxp3 promoter is specific to T reg cells, as this binding was not observed in Th2 and Tfh cells (Fig. 5 E). We also observed down-regulation of an active chromatin modification, lysine 4 dimethylation of histone H3 (H3K4me2), at the Foxp3 promoter in Nr4a-TKO T reg cells compared with WTT reg cells (Fig. 5 F).
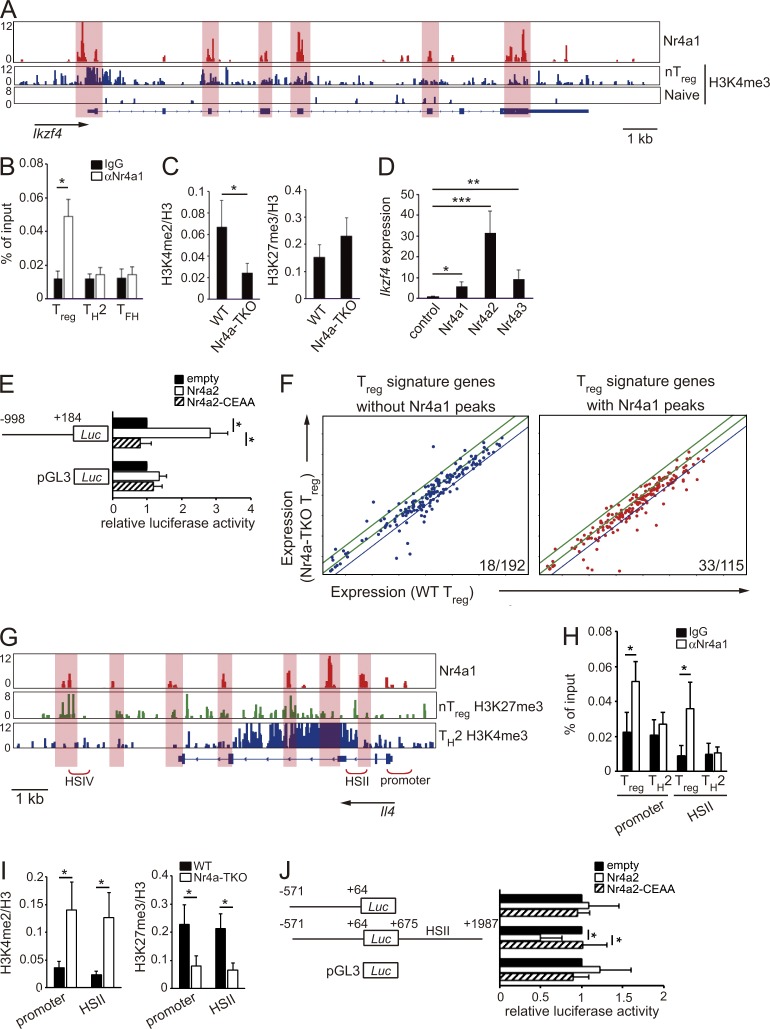
In addition, we found several peaks of Nr4a1 binding at the Ikzf4 locus (Fig. 6 A). Importantly, through a parallel investigation of previous genome-wide studies for histone modifications in CD4+ T cell subsets (Wei et al., 2009), we found that Nr4a1-binding peaks substantially overlapped with peaks of lysine 4 trimethylation of histone H3 (H3K4me3) in T reg cells, an active chromatin modification (Fig. 6 A, highlighted areas). These results suggest that Nr4a factors directly bind to the Ikzf4 locus and incorporate activating histone modifications. By ChIP-qPCR, we observed binding of Nr4a1 to the Ikzf4 promoter in T reg cells, not in Th2 and Tfh cells (Fig. 6 B). H3K4me2 modification at the Ikzf4 promoter was reduced in Nr4a-TKO T reg cells compared with that in WTT reg cells (Fig. 6 C). Furthermore, forced expression of Nr4a factors strongly induced Ikzf4 expression in T naive cells (Fig. 6 D). To further determine whether Nr4a factors transactivate Ikzf4 expression, we expressed an Ikzf4 promoter-luciferase reporter construct in Jurkat T lymphoma cells. As shown in Fig. 6 E, WT Nr4a2, but not a DNA binding–deficient form of Nr4a2 (Saijo et al., 2009), transactivated the Ikzf4 promoter. Collectively, these results suggest that Nr4a factors directly transactivate Ikzf4 expression in T reg cells.
Figure 6.
Direct repression of Il4 by Nr4a factors. (A) Genome browser view of ChIP-seq profile maps. The distribution of Nr4a1 bind sites on the Ikzf4 locus is shown for WT T reg cells. The distribution of H3K4me3 modification on Ikzf4 (Wei et al., 2009) are shown for nT reg and for T naive cells. Vertical scales are noted. (B) ChIP-qPCR analysis of the enrichment of Nr4a1 binding at the Ikzf4 promoter in WT T reg, Th2, and Tfh cells. Values are presented as the percentage of corresponding input. (C) ChIP-qPCR analysis of the Ikzf4 promoter region using chromatin extracts from WT and Nr4a-TKO T reg cells after immunoprecipitation with anti-H3K4me2 and anti-H3K27me3 antibodies. Results are presented relative to those obtained with antihistone H3 antibodies. (D) qRT-PCR analysis of Ikzf4 mRNA expression in naive CD4+ T cells transduced with a control retrovirus or Nr4a1-, Nr4a2-, or Nr4a3-expressing retroviruses. Expression was normalized to Hprt levels. Expression levels of control virus-transduced cells are set as one. (E) Luciferase reporter assays of a pGL3-control plasmid or an Ikzf4 reporter plasmid constructed from the Ikzf4 promoter region (−998 to +184). Promoter activity was analyzed in Jurkat cells transfected with empty plasmids or plasmids encoding WT Nr4a2 or a DNA binding-deficient form of Nr4a2 (Nr4a2-CEAA). Promoter activities are shown relative to those of empty plasmid-transfected cells. (F) Expression profiles of T reg signature genes in WT T reg cells (horizontal axis) and Nr4a-TKO T reg cells (vertical axis). The same microarray datasets that are shown in Fig. 3 B are highlighted for T reg signature genes, which were further divided according to the presence (right) or absence (left) of Nr4a1 peaks determined in the ChIP-seq analysis. Blue lines in each panel show the border of twofold down-regulation in Nr4a-TKO T reg cells, compared with WT T reg cells. Numbers in the each panel show the ratio of genes that showed more than twofold down-regulation in Nr4a-TKO T reg compared with WT T reg cells. (G) Nr4a1 occupancy in WT T reg cells, deposition of H3K27me3 in nT reg cells and H3K4me3 in Th2 cells (Wei et al., 2009) across the Il4 locus. Vertical scales are noted. (H) ChIP-qPCR with anti-Nr4a1 or control IgG antibodies in WT T reg and Th2 cells. Immunoprecipitated DNA was amplified with primers corresponding to the Il4 promoter and HSII regions. Values are presented as the percentage of corresponding input. (I) ChIP-qPCR of WT and Nr4a-TKO T reg cells; immunoprecipitation with antibodies for H3K4me2 or H3K27me3 was followed by qPCR analysis with primer sets corresponding to the indicated regions. Results are presented relative to those obtained with antihistone H3 antibodies. (J) Luciferase reporter assays. Reporter constructs of pGL3-control, and pGL3 linked with the Il4 promoter (−571 to +64) with or without a region encompassing HSII (+675 to +1987), were transfected into Jurkat cells, together with empty plasmids or plasmids encoding WT Nr4a2 or Nr4a2-CEAA. Promoter activities are shown relative to those of empty plasmid–transfected cells. Shown are representative results of three (B, C, E, and H–J) or four (D) biological replicates, each performed in triplicate, means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.005, (Student’s t test [B, C, H, and I] one-way ANOVA with Bonferroni test [E and J]).
Among the 307 genes comprising the T reg signature gene set, 115 genes, including Foxp3, Ikzf4, and Il2ra, showed Nr4a1-binding peaks in ChIP-seq analysis (Fig. 6 F). A parallel examination of ChIP-seq and microarray data revealed that expression levels of 33 out of the 115 genes with Nr4a1 binding peaks were down-regulated >2-fold in Nr4a-TKO T reg cells compared with those in WT T reg cells. 18 out of 192 genes without Nr4a1-binding peaks were down-regulated >2-fold in Nr4a-TKO T reg cells (Fig. 6 F). These results strongly indicate that Nr4a factors directly regulate a substantial fraction of the T reg cell–transcriptional program.
Nr4a factors directly suppress IL-4 expression
We investigated the molecular mechanisms by which Nr4a factors repress Th2 cytokine genes in T reg cells. ChIP-seq data showed several Nr4a1 binding peaks at the Il4 gene locus, including the HSII site, which is thought to regulate Il4 gene expression (Agarwal and Rao, 1998; Zhu et al., 2003; Fig. 6 G). Furthermore, we found partial overlaps of Nr4a1-binding peaks with peaks of either lysine 27 trimethylation of histone H3 (H3K27me3), a repressive histone modification in T reg cells, or H3K4me3 in Th2 cells (Fig. 6 G, highlighted areas). By ChIP-qPCR, we observed binding of Nr4a1 to the Il4 promoter and HSII sites in T reg cells, but not in Th2 cells (Fig. 6 H). We also observed augmented H3K4me2 modifications, as well as attenuated H3K27me3 modifications, at the aforementioned regions in Nr4a-TKO T reg cells (Fig. 6 I). By luciferase reporter assay, we found that Nr4a2 significantly suppressed Il4 promoter activity in the presence of the region containing the HSII site (Fig. 6 J). Altogether, these results strongly suggest that Nr4a factors directly suppress IL-4 expression in T reg cells.
Nr4a factors maintain lineage stability in T reg cells
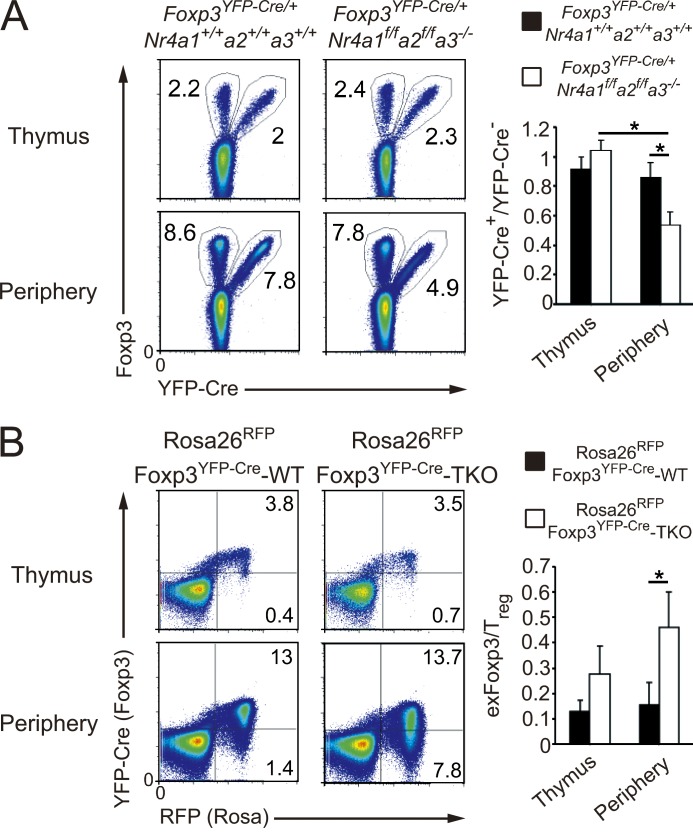
T reg cells are a stable cell lineage, particularly under noninflammatory, homeostatic conditions (Rubtsov et al., 2010; Miyao et al., 2012). To address the contribution of Nr4a factors to the lineage stability of T reg cells, we first compared the homeostatic characteristics of WT and Nr4a-TKO T reg cells in a single individual mouse by analyzing Foxp3YFP-Cre/+ heterozygous females. Although the ratio of YFP-Cre+ and YFP-Cre− T reg cells was close to 1:1 in the thymus, the proportion of peripheral YFP-Cre+ T reg cells was significantly smaller in Foxp3YFP-Cre/+Nr4a1f/fNr4a2f/fNr4a3−/− mice than in Foxp3YFP-Cre/+Nr4a1+/+Nr4a2+/+Nr4a3+/+ mice (Fig. 7 A). These results indicate that Nr4a-deficient T reg cells had attenuated competitive fitness in the presence of Nr4a-sufficient T reg cells.
Figure 7.
Accelerated loss of Foxp3 expression and conversion to exFoxp3 cells of Nr4a-TKO T reg cells. (A, left) Flow cytometry of CD4-SP thymocytes and CD4+ T cells from pooled spleens and lymph nodes (Periphery) in 4-wk-old Foxp3YFP-Cre/+Nr4a1+/+Nr4a2+/+Nr4a3+/+ and Foxp3YFP-Cre/+Nr4a1f/fNr4a2f/fNr4a3−/− mice. (right) Quantification of the flow cytometry results. Ratios of YFP+/YFP− in total Foxp3+ cells are shown (n = 4 mice per genotype, pooled from 3 independent cohorts per genotype, mean and SD). (B, left) Flow cytometry of CD4-SP thymocytes and peripheral CD4+ T cells from 4-wk-old Rosa26RFPFoxp3YFP-Cre-WT and Rosa26RFPFoxp3YFP-Cre-Nr4a-TKO mice. (right) Quantification of the flow cytometry results. The ratios of exFoxp3/T reg cells within CD4+RFP+ cells from the thymus and periphery of 4–5-wk-old mice were quantified (n = 4 mice per genotype, pooled from 3 independent cohorts per genotype, mean and SD). *, P < 0.05 (one-way ANOVA with Bonferroni test).
We tracked the fate of Nr4a-TKO T reg cells, using Foxp3YFP-Cre x Rosa26-loxP-Stop-loxP mice, in which any cell that has once expressed Foxp3 is permanently marked by RFP expression. In these mice, YFP-Cre+RFP+ cells are T reg cells, whereas the YFP-Cre−RFP+ cells are considered to be exFoxp3 cells that had once expressed Foxp3 but then extinguished its expression (Zhou et al., 2009). By analyzing the ratio of exFoxp3 to T reg cells, we found a larger population of exFoxp3 cells in Rosa26RFPFoxp3YFP-CreNr4a1f/fNr4a2f/fNr4a3−/− (called Rosa26RFPFoxp3YFP-Cre-Nr4a-TKO herein) mice than in Rosa26RFPFoxp3YFP-CreNr4a1+/+Nr4a2+/+Nr4a3+/+ (called Rosa26RFPFoxp3YFP-Cre-WT herein) mice, indicating an accelerated conversion of T reg cells to exFoxp3 cells in the absence of Nr4a factors (Fig. 7 B).
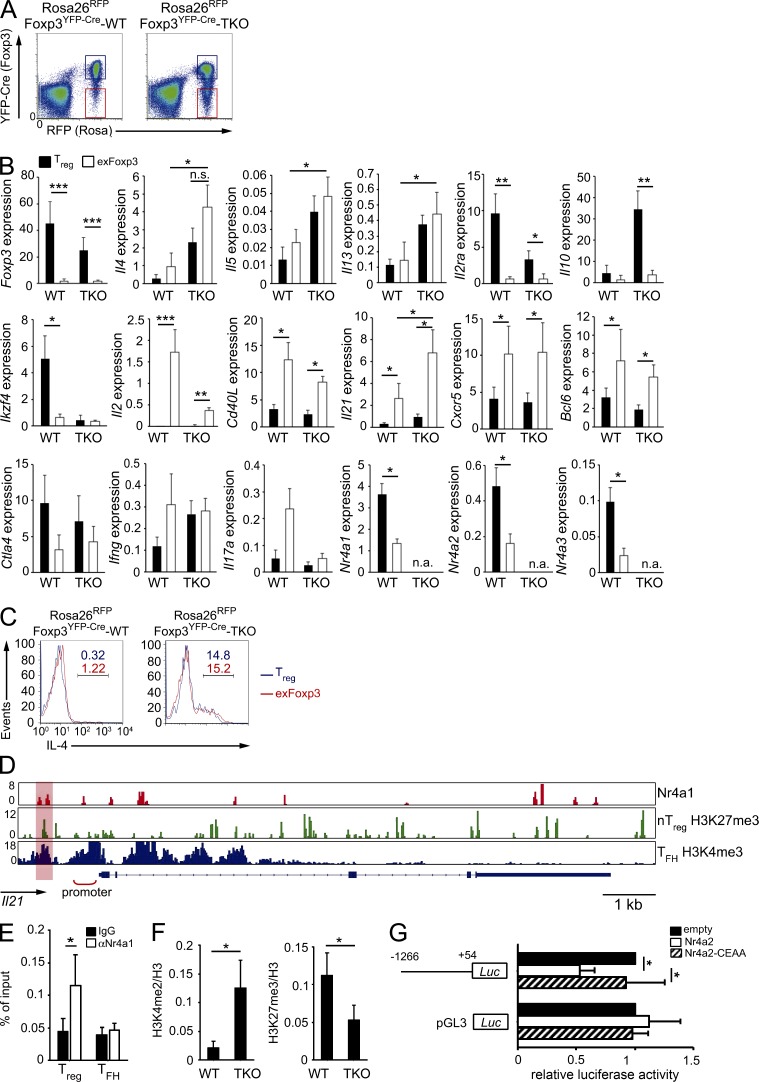
We next characterized the Nr4a-TKO exFoxp3 cells using qRT-PCR. Cells were obtained from 6–8-wk-old mice, at which age exFoxp3 cells emerged as a separate population from parental T reg cells (Fig. 8 A). We found that Il2ra and Il10, two genes directly related to the immune-suppressive effects of T reg cells, were down-regulated in exFoxp3 cells compared with those in T reg cells in both WT and Nr4a-TKO backgrounds (Fig. 8 B). Co-stimulatory effector genes, including Cd40L and Il2, were up-regulated in exFoxp3 cells compared with those in T reg cells, also regardless of WT or Nr4a-TKO backgrounds (Fig. 8 B). Investigation of Tfh-associated genes revealed higher levels of Il21, Cxcr5, and Bcl6 in exFoxp3 cells compared with T reg cell–counterparts in both WT and Nr4a-TKO backgrounds. However, as for Il21, its expression levels were higher in Nr4a-TKO exFoxp3 cells than in WT exFoxp3 cells (Fig. 8 B). Expression of Th2-cytokines was higher in Nr4a-TKO exFoxp3 cells than in WT exFoxp3 cells at both mRNA and protein levels (Fig. 8, B and C). Importantly, we observed that expression levels of all Nr4a factors were reduced in exFoxp3 cells, compared with those in T reg cells (Fig. 8 B). This may explain the elevated expression of Il4 and Il21 in WT exFoxp3 cells.
Figure 8.
Nr4a-deficient exFoxp3 cells acquire Th2 and Tfh effector-like gene expression patterns. (A) FACS profiles of peripheral CD4+ T cells from 7-wk-old Rosa26RFPFoxp3YFP-Cre-WT and Rosa26RFPFoxp3YFP-Cre-Nr4a-TKO mice. Cell populations denoted with blue and red lines were sorted as T reg cells and exFoxp3 cells, respectively, and further analyzed in experiments in B. (B) qRT-PCR analysis of the indicated mRNAs in T reg and exFoxp3 cells from 6–8-wk-old Rosa26RFPFoxp3YFP-Cre-WT and Rosa26RFPFoxp3YFP-Cre-Nr4a-TKO mice (n = 7 mice per genotype, pooled from 5 independent cohorts per genotype). n.a., not available. (C) Flow cytometric analysis of IL-4 expression in T reg (blue) and exFoxp3 (red) cells from 5-wk-old Rosa26RFPFoxp3YFP-Cre-WT and Rosa26RFPFoxp3YFP-Cre-Nr4a-TKO mice after 5 h stimulation with PMA plus ionomycin in the presence Brefeldin A. Blue (T reg) and red (exFoxp3) numbers indicate the percentage of cells in the indicated area. (D) Nr4a1 occupancy in WT T reg cells, deposition of H3K27me3 in nT reg cells (Wei et al., 2009) and H3K4me3 in Tfh cells (Lu et al., 2011) across the Il21 locus. Vertical scales are noted. (E) ChIP-qPCR analysis of Nr4a1 binding 1,200 bp upstream of the Il21 transcriptional start site (highlighted region in D) in WT T reg and Tfh cells. (F) ChIP-qPCR analysis of the region 1,200 bp upstream of the Il21 transcriptional start site using chromatin extracts obtained from WT and Nr4a-TKO T reg cells immunoprecipitated with anti-H3K4me2 and anti-H3K27me3 antibodies. Results are presented relative to those obtained with antihistone H3 antibodies. Shown are means ± SD of three independent experiments, each performed in triplicate. (G) Luciferase reporter assays of a pGL3-control plasmid or an Il21 reporter plasmid constructed from the Il21 promoter region (−1266 to +54). Promoter activity was analyzed in Jurkat cells transfected with empty plasmids or plasmids encoding WT Nr4a2 or a DNA-binding deficient form of Nr4a2 (Nr4a2-CEAA). Promoter activities are shown relative to those of empty plasmid-transfected cells. Shown are a representative results of three (E and F) or four (G) biological replicates, each performed in triplicate, means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.005 (Student’s t test [E and F], one-way ANOVA with Bonferroni test [B and G]).
Because IL-21 is an important mediator of Tfh cell function, we investigated the molecular mechanisms by which Nr4a factors repress Il21 expression. Analysis of our ChIP-seq data revealed several Nr4a1 binding peaks at the Il21 gene locus (Fig. 8 D). We also found partial overlaps of Nr4a1 binding peaks with peaks of either H3K27me3 in T reg cells, or H3K4me3 in Tfh cells (Lu et al., 2011; Fig. 8 D, highlighted regions). By ChIP-qPCR, we observed binding of Nr4a1 to a region 1.2 kb upstream of the Il21 transcriptional start site in T reg cells, but not in Tfh cells (Fig. 8 E). We also observed enhanced H3K4me2 modifications, as well as reduced H3K27me3 modifications at the same region in Nr4a-TKO T reg cells, compared with WT T reg cells (Fig. 8 F). By luciferase reporter assay, we found that Nr4a2 significantly suppressed Il21 promoter activity (Fig. 8 G).
Collectively, the aforementioned results suggest that Nr4a factors maintain lineage stability in T reg cells and prevent T reg cells from converting to exFoxp3 cells with Th2- and Tfh-effector–like characteristics.
Nr4a-deficient T reg cells acquire an atypical ability to elicit GC reactions
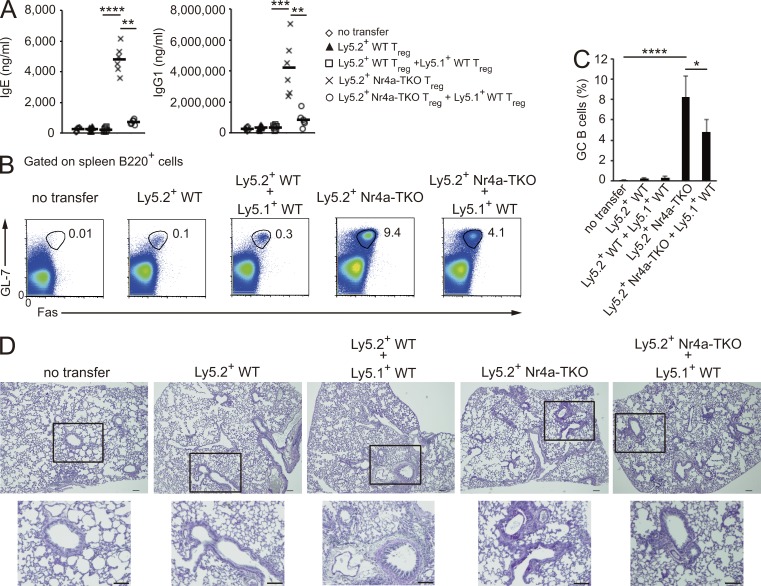
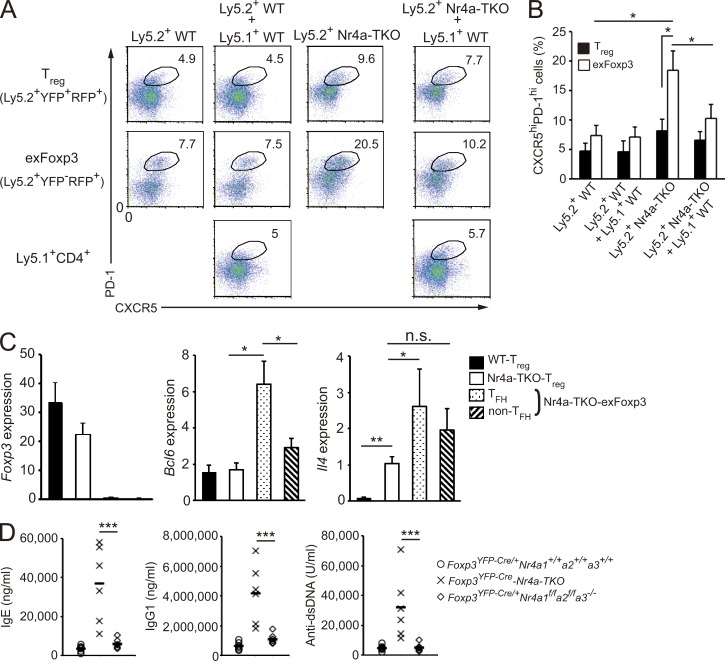
Elevated levels of autoreactive IgE and IgG1 were detected in Foxp3YFP-CreNr4a-TKO mice (Fig. 2, A and B). As Th2- and Tfh-cytokine genes were up-regulated in Nr4a-TKO T reg cells, we suspected that T reg cells aberrantly gained the ability to elicit GC reactions upon loss of Nr4a factors. To address this issue, we transferred isolated T reg cells into T cell–deficient CD3ε−/− mice, without immunization, and monitored GC reactions in the recipient mice. WT T reg and Nr4a-TKO T reg cells were isolated from Ly5.2+Rosa26RFPFoxp3YFP-Cre-WT mice and Ly5.2+Rosa26RFPFoxp3YFP-CreNr4a-TKO mice, respectively, and transferred with or without congenic Ly5.1+ WT T reg cells into recipient mice (Fig. S1 A). 5 wk after transfer, mice receiving Nr4a-TKO T reg cells showed significantly elevated levels of IgE and IgG1, as well as induction of GC B cells and lung inflammation (Fig. 9, A–D). By analyzing T reg cells and exFoxp3 cells individually, we found that Nr4a-TKO exFoxp3 cells contained a larger CXCR5hiPD-1hi Tfh population than T reg cells did, indicating that it was mainly the exFoxp3 cells that contributed to the GC reactions in the recipient mice (Fig. 10, A and B; Fig. S1, B and C). Analysis of the Tfh and non-Tfh fractions of Nr4a-TKO exFoxp3 cells revealed that Il4 expression is elevated in both (Fig. 10 C). This showed that Nr4a-TKO exFoxp3 cells are composed of Th2 cells and cells with a Tfh/Th2 hybrid phenotype.
Figure 9.
Nr4a-TKO T reg cells induce GC reactions. (A) Total IgE and IgG1 levels in recipient CD3ε-deficient mice transferred with no cells, Ly5.2+WT T reg cells (4 × 105), or Ly5.2+Nr4a-TKO T reg cells (4 × 105), with or without Ly5.1+ WT T reg cells (2 × 105). Each symbol represents an individual mouse; small horizontal lines indicate the mean. (B) Flow cytometry profiles of B220+ spleen cells from A. Numbers adjacent to outlined areas indicate the percentage of cells in each. (C) Quantification of results in B. Frequencies of GL-7+Fas+ GC B cells among B220+ cells are shown (n = 5 mice per group, pooled from 2 independent experiments). (D) PAS staining of mucus-producing cells (pink) in the lungs of CD3ε-deficient mice from A. Outlined area is enlarged below. Bars, 100 µm. Error bars in C denote mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001 (one-way ANOVA with Bonferroni test).
Figure 10.
GC reaction–inducing ability of Nr4a-deficient T reg cells is suppressed by coexisting WT T reg cells. (A) Flow cytometry of spleen T reg (CD4+YFP+RFP+) and exFoxp3 (CD4+YFP−RFP+) cells from Fig. 9 A. Co-transferred Ly5.1 total CD4+ T cells are also shown. The definition of the CXCR5hiPD-1hi Tfh gating is shown in Fig. S1 C. (B) Quantification of results in A. Frequencies of CXCR5hiPD-1hi cells among T reg and exFoxp3 cells (n = 5 mice per group, pooled from 2 independent experiments). Numbers adjacent to outlined areas indicate the percentage of cells in each. (C) qRT-PCR analysis of Foxp3, Bcl-6, and Il4 mRNA in the indicated cells. WT T reg cells were isolated from mice that had received Ly5.2+ WT T reg cells. Nr4a-TKO T reg (Ly5.2+YFP+RFP+), Nr4a-TKO exFoxp3 cells in the Tfh fraction (Ly5.2+YFP−RFP+CXCR5hiPD-1hi), and Nr4a-TKO exFoxp3 cells in the non-Tfh fraction (Ly5.2+YFP−RFP+CXCR5lowPD-1low) were all isolated from the mice that had received only Ly5.2+ Nr4a-TKO T reg cells. Expression was normalized to Hprt levels. Shown are a representative result of two biological replicates, each performed in triplicate. (D) Titers of total IgE, IgG1, and anti–double-stranded DNA antibodies in sera from 5–7-wk-old Foxp3YFP-Cre/+Nr4a1+/+Nr4a2+/+Nr4a3+/+, Foxp3YFP-Cre-Nr4a-TKO, and Foxp3YFP-Cre/+Nr4a1f/fNr4a2f/fNr4a3−/− mice (n = 6 mice per genotype, pooled from 4 independent cohorts per genotype). Each symbol represents an individual mouse; small horizontal lines indicate the mean. Error bars (B and C) denote mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.005 (one-way ANOVA with Bonferroni test).
The finding that a substantial fraction (∼65%) of transferred Nr4a-TKO T reg cells retained Foxp3 expression in the recipient mice at the time of analysis (Fig. S1 B) suggested that Nr4a-deficient T reg cells do not have the ability to suppress GC reactions elicited by Nr4a-TKO T reg cell–transfer. This hypothesis was supported by the observation that cotransferred WT T reg cells were able to suppress induction of IgE and IgG1 as well as the lung inflammation induced by Nr4a-TKO T reg cell–transfer (Fig. 9, A and D). The induction of GC B cells and Tfh cells was also partially, but significantly, suppressed by cotransferred WT T reg cells (Figs. 9, B and C, and 10, A and B). Supporting these observations, production of IgE, IgG1, and self-reactive antibodies was much lower in female Foxp3YFP-Cre/+Nr4a1f/fNr4a2f/fNr4a3−/− mice in which Nr4a-sufficient T reg and Nr4a-deficient T reg cells coexist than production in Foxp3YFP-Cre-Nr4a-TKO mice (Fig. 10 D).
Altogether, these results showed that T reg cells abnormally obtained the capacity to elicit GC reactions upon loss of Nr4a factors. Furthermore, the ability to suppress GC responses was impaired in Nr4a-TKO T reg cells, although WT T reg cells elicited suppression of GC responses upon transfer with Nr4a-TKO T reg cells.
DISCUSSION
Our studies revealed that Nr4a factors play crucial roles in mature T reg cells. We showed that Nr4a factors contribute to a large fraction of a T reg cell–specific transcriptional program, as revealed by a global reduction of T reg signature gene expression in Nr4a-defficient T reg cells. Apparently, not all of the genes in this group are directly regulated by Nr4a factors, however; instead, Nr4a factors directly regulate several key T reg cell lineage–specifying factors, including Foxp3, Ikzf4, and Il2ra. Because the products of these genes (Foxp3, Eos, and CD25) are crucial for maintenance of T reg cell lineage (Fontenot et al., 2005; Setoguchi et al., 2005; Kim et al., 2007; Pan et al., 2009), the observed global reduction of T reg signature gene expression in Nr4a-TKO T reg cells can be attributed to both the impairment of expression of genes directly regulated by Nr4a factors and the secondary effects of down-regulation of those key T reg regulators. In addition, because the Foxp3 CNS2 enhancer was properly demethylated in Nr4a-TKO T reg cells, Nr4a factors maintain Foxp3 expression independent of demethylation of this locus. Instead, Nr4a factors may positively contribute at the promoter to the sustained transcription of Foxp3. This speculation is supported by the ChIP data in this study, as well as our previous studies (Sekiya et al., 2011, 2013).
Nr4a family members play redundant roles in T reg cells, as revealed by the severity of disease phenotypes in Foxp3YFP-CreNr4a-TKO mice compared with those in double-knockouts. Contributions from Nr4a1 and Nr4a3 are stronger than those of Nr4a2. The fundamental basis of this functional redundancy should reside in their shared binding profiles, which are speculated from the highly homologous DNA-binding domains shared among Nr4a family members. Indeed, replacement of the DNA-binding domain of Nr4a2 with that of Nr4a1 does not affect the Foxp3-inducing ability of Nr4a2 (Sekiya et al., 2013). In Nr4a2-single-knockout T reg cells, however, there is some reduction of Foxp3 or CD25 levels but no dysregulation of proinflammatory cytokine genes (Sekiya et al., 2011). This suggests that the functional redundancy among Nr4a family members is not completely equivalent. Also related to this issue, attenuated maintenance of Foxp3 expression in Nr4a2-single-knockout T reg cells does not seem to be substantially exacerbated in Nr4a-TKO T reg cells (Sekiya et al., 2011). This may be due to the enhanced expression of IL-10 in Nr4a-TKO T reg cells, which was not observed in Nr4a2-single-knockout T reg cells. As IL-10 maintains Foxp3 expression in T reg cells (Murai et al., 2009), the elevated IL-10 expression may partially compensate for the accelerated loss of Foxp3 expression in Nr4a-TKO T reg cells.
In contrast to the complete loss of T reg cell development in CD4Cre-Nr4a-TKO mice, down-regulation of Foxp3 in Nr4a-TKO T reg cells seemed to be gradual, rather than an acute cessation. This can be explained by the epigenetic memory of T reg cells, including demethylation of the Foxp3 CNS2 enhancer, which stably maintains Foxp3 expression once T reg cells have matured (Miyao et al., 2012). The effectiveness of the T reg cell epigenetic memory is exemplified by the recent unexpected observation by Levine et al. (2014) who reported that even removal of T cell receptor signaling, a crucial component of T reg cell development, did not result in an acute loss of Foxp3 expression in T reg cells. Although the details of the roles of Nr4a factors in this epigenetic memory of T reg cells remain unknown, the fact that they are important for the maintenance of Foxp3 expression in T reg cells is revealed by several observations in this study: localization of their binding sites at the Foxp3 promoter and enhancers, the accelerated conversion of Nr4a-deficient T reg cells to exFoxp3 cells, and the reduction of Foxp3 expression in Nr4a-TKO T reg cells even under noninflammatory conditions.
Depletion of T reg cells from adult mice induces severe autoimmunity, resulting in death within several weeks (Kim et al., 2007). However, the autoimmunity observed in Foxp3YFP-Cre-Nr4a-TKO mice is relatively milder and the majority of mice survive for more than a month. In addition, the suppressive activity of Nr4a-TKO T reg cells was not completely abrogated in vitro. Considering the normal or even elevated levels of some of the central effectors of T reg cell–mediated suppression, including CTLA-4 and IL-10, in Nr4a-TKO T reg cells, it is possible that the roles of Nr4a factors in T reg cells are selective. Indeed, Foxp3Cre-CTLA4fl/fl mice die at early ages (100% death within ∼10 wk) and Foxp3Cre-IL10fl/fl mice develop colitis, phenotypes that are not observed in Foxp3YFP-Cre-Nr4a-TKO mice (Rubtsov et al., 2008; Wing et al., 2008). However, elevated IgE, IgG1, and autoreactive antibody levels in Foxp3YFP-Cre-Nr4a-TKO mice seemed much higher than those observed in other Foxp3-Cre-knockouts reported so far. Therefore, among the pleiotropic roles of T reg cells in immunosuppression, Nr4a factors seem to be particularly important in suppression of Th2/Tfh reactions. In addition, acquisition of Th2- and Tfh-effector functions by Nr4a-TKO T reg and exFoxp3 cells should exacerbate Th2 and Tfh reactions in Foxp3YFP-Cre-Nr4a-TKO mice.
Antibody production is stringently controlled by T reg cells (Lim et al., 2004; Iikuni et al., 2009; Chung et al., 2011; Linterman et al., 2011). As IL-4 and IL-21 positively regulate GC reactions (Abbas et al., 1990; King, 2009; Crotty, 2011), expression of both cytokines must be silenced in T reg cells. Indeed, expression levels of both Il4 and Il21 are tightly repressed in Tfr cells (Linterman et al., 2011), the GC-residing subset of T reg cells with suppressive activity against GC reactions. In this study, we demonstrated that Nr4a factors directly suppress both Il4 and Il21 expression in T reg cells. In addition, deletion of Nr4a factors aberrantly induced self-reactive IgG1 and IgE. Although we did not specifically analyze the Tfr subset in this study, it seems highly likely that Nr4a factors also repress IL-4 and IL-21 expression in Tfr cells, thus mediating normal Tfr cell functions.
Nr4a-TKO T reg cells aberrantly acquired the ability to induce GC reactions, particularly after their conversion to exFoxp3 cells. In agreement with these findings, conversion of T reg cells to Tfh cells upon loss of Foxp3 expression was reported previously (Tsuji et al., 2009), who reported that WT T reg cells lost Foxp3 expression and simultaneously up-regulated expression of Tfh-associated genes, including CXCR5 and PD-1, upon transfer into Cd3ε−/− mice. They also found that those CXCR5hiPD-1hi WT exFoxp3 cells activate production of IgA by GC B cells in Peyer’s patches, but not in the spleen or lymph nodes. We also observed up-regulation of Tfh-associated genes both in WT and Nr4a-TKO exFoxp3 cells. These concordant observations by others and us suggest that the acquisition of Tfh cell phenotypes is a general characteristic of exFoxp3 cells that is programmed in T reg cells, presumably to mediate some aspects of their protective roles in certain environments, including gut. However, in this study, we detected a substantial induction of GC B cells even in spleens of Cd3ε−/− mice after transfer of Nr4a-TKO T reg cells, resulting in elevated production of IgE, as well as lung inflammation. Collectively, such abnormal induction of GC reactions can be considered as an aberrant and/or ectopic execution of the Tfh developmental program that intrinsically resides in T reg cells. This abnormal program is driven in Nr4a-TKO T reg cells by their accelerated conversion to exFoxp3 cells, enhanced expression of Th2 and Tfh cytokines, and impaired suppressive activity.
Altogether, we revealed that Nr4a factors play crucial roles in T reg cells, mediating the two defining characteristics of T reg cells: stability of cell lineage (Rubtsov et al., 2010; Miyao et al., 2012) and immunosuppressive functions (Sakaguchi et al., 2008). Upon loss of Nr4a genes, T reg cells tend to become exFoxp3 cells, unleashing aberrant Tfh and Th2 effector genetic programs. Furthermore, although WT T reg cells can suppress GC reactions, this ability is diminished in Nr4a-TKO T reg cells. As a result of these combined abnormalities, severe systemic autoimmune disease develops in Foxp3YFP-Cre-Nr4a-TKO mice. The Nr4a family of nuclear receptors, as essential components of T reg cell biology, thus present attractive therapeutic targets for treatment of various disorders, particularly those related to GC reactions, such as atopic allergies, asthma, and chronic allograft rejection (Lloyd and Hawrylowicz, 2009; Wehner et al., 2010).
MATERIALS AND METHODS
Mice.
All research involving animals was performed in accordance with the Guidelines for Animal Care approved by Keio University. Animals were maintained in specific pathogen–free conditions. A license for research-use of Nr4a3−/− mice (TF0937) was purchased from Taconic. Rosa26RFP mice (Luche et al., 2007) were as previously described (Muto et al., 2013). The Nr4a1-floxed and Nr4a2-floxed mice from H. Ichinose (Tokyo Institute of Technology, Yokohama, Kanagawa, Japan) have been described elsewhere (Kadkhodaei et al., 2009; Sekiya et al., 2011). Foxp3YFP-Cre knock-in mice were provided by A. Rudensky (Memorial Sloan-Kettering Cancer Center, New York, NY). Cre-ERT2 mice have been described previously (Seibler et al., 2003). All mouse strains were maintained on a C57BL/6 genetic background.
Antibodies.
FITC-, phycoerythrin (PE)-, PerCP-Cy5.5-, allophycocyanin (APC)-, or APC-Cy7–conjugated monoclonal antibodies for CD4 (L3T4), CD8 (53–6.7), CD25 (PC61.5), Foxp3 (FJK-16s), IL-4 (11B11), and CD45.1 (A20) antibodies were purchased from eBioscience. Anti-Nr4a1 monoclonal antibody (H1648) was purchased from Perseus Proteomics, and monoclonal antihistone H3 (ab1791), polyclonal anti-di-methylated-histone H3 lys4 (Y47), and polyclonal anti-tri-methylated-histone H3 lys27 (ab6002) were purchased from Abcam.
Western blotting.
T naive cells (CD4+CD25lowCD62Lhigh) were sorted from the spleen and lymph node cells. T reg (CD4+CD25highFoxp3+) cells were freshly prepared from spleen and lymph node cells of Foxp3YFP-Cre mice by sorting. Tfh cells (CD4+CXCR5highPD-1high) were collected from splenocytes of OT-II mice that were immunized s.c. in the scruff with 50 µg of OVA (Sigma-Aldrich) mixed in complete Freund’s adjuvant (CFA), at day 8 after immunization. Th1, Th2, and Th17 cells were differentiated in vitro for 4 d in RPMI-1640 medium supplemented with 1 µg/ml plate-coated anti-CD3ε (2C11), 0.5 µg/ml anti-CD28 (57.31), 55 µM 2-Mercaptoethanol (Invitrogen), and 10% FBS, supplemented with the following: for Th1, 5 µg/ml anti–IL-4 (11B11), 5 ng/ml IL-2 (PeproTech), 10 ng/ml IL-12 (PeproTech); for Th2, 5 µg/ml anti–IFN-γ (XMG1.2), 10 ng/ml IL-2 (PeproTech), 10 ng/ml IL-4 (PeproTech); for Th17, 5 µg/ml anti–IL-4 (11B11), 5 µg/ml anti–IFN-γ (XMG1.2), 5 µg/ml anti–IL-2 (JES6-1A12) antibodies, 10 ng/ml human IL-6 (R&D Systems), and 1 ng/ml recombinant human TGF-β1 (R&D Systems). Proteins were dissolved in SDS-PAGE sample buffer, and then separated by SDS-PAGE and electrotransferred to an Immobilon-P PDVF membrane (Millipore). Membranes were hybridized with the following antibodies: anti-GAPDH mAb (3H12; MBL), anti–T-bet mAb (4B10; Santa Cruz Biotechnology, Inc.), anti-Gata3 mAb (HG3-31; Santa Cruz Biotechnology, Inc.), anti-Rorγt mAb (AFKJS-9; eBioscience), anti-Bcl6 mAb (K112-91; BD), anti-Foxp3 mAb (D608R; Cell Signaling Technology), anti-Nr4a1 mAb (H1648), anti-Nr4a2 mAb (N1404), and anti-Nr4a3 mAb (H7833), and then visualized using an ImmunoStar-LD detection system (WAKO).
Cre-ERT2–mediated acute deletion of Nr4a genes in adult mice.
For activation of Cre-ERT2 (fusion of Cre recombinase with a mutant human estrogen receptor ligand-binding domain), Rosa26RFPCre-ERT2-WT, or Rosa26RFPCre-ERT2-Nr4a-TKO mice were administrated i.p. with Tamoxifen (Sigma-Aldrich) dissolved in corn oil for 4 consecutive days (2 mg/d) from day 0. At day 11, mice were sacrificed, and CD4+CD25highRFP+ cells were isolated from pooled spleen and lymph node cells as T reg cells, in which Cre recombinase was activated. For control samples, CD4+CD25high cells were sorted from vehicle-treated littermates.
Isolation of T reg cells and exFoxp3 cells.
Isolation of T reg cells from Foxp3YFP-Cre mice was performed by sorting the CD4+YFP+ population from pooled spleen and lymph node cells using PerCP-Cy5.5–conjugated monoclonal antibodies for CD4 (RM4-5; Tonbo Biosciences). Isolation of T reg cells and exFoxp3 cells from Rosa26RFPFoxp3YFP-Cre mice was done by sorting CD4+YFP+RFP+ cells and CD4+YFP-RFP+ cells from pooled spleen and lymph node cells, respectively.
Plasmid construction.
For transient expression of Nr4a2, a cDNA comprising the open reading frame (ORF) of human NR4A2 (595 of 598 aa are identical to mouse Nr4a2) was inserted into the pCMV plasmid. The cDNA that encodes a DNA-binding–deficient form of Nr4a2 (Nr4a2-CEAA) was generated by site-directed mutagenesis performed by PCR-based methods.
Histopathological examination.
Tissues were fixed in 4% formalin, embedded in paraffin, and cut into 5-µm sections. Tissue sections were then stained with hematoxylin and eosin, or periodic acid Schiff (PAS).
Chromatin immunoprecipitation (ChIP).
WT and Nr4a-TKO T reg were isolated from Foxp3YFP-Cre-WT and Foxp3YFP-Cre-Nr4a-TKO mice, respectively, by sorting the CD4+YFP+ population from pooled spleen and lymph node cells. Tfh cells (CD4+CXCR5highPD-1high) were collected from splenocytes of OT-II mice, which were immunized s.c. in the scruff with 50 µg of OVA (Sigma-Aldrich) mixed in CFA at day 8 after immunization. Th2 cells were differentiated for 4 d in RPMI-1640 medium supplemented with 1 µg/ml plate-coated anti-CD3ε (2C11), 0.5 µg/ml anti-CD28 (57.31), 55 µM 2-Mercaptoethanol (Invitrogen), and 10% FBS, supplemented with 5 µg/ml anti–IFN-γ (XMG1.2), 10 ng/ml IL-2 (PeproTech), and 10 ng/ml IL-4 (PeproTech). For histone immunoprecipitations, freshly isolated T reg cells were fixed and, for Nr4a1 immunoprecipitations, isolated T reg cells were restimulated with anti-CD28 antibodies (clone PV1.17, 0.5 µg/ml) and plate-coated anti-CD3 antibodies (clone 2C11, 2 µg/ml) for 2 h to reinduce Nr4a1 protein, whose expression is acutely reduced during ex vivo handling of T reg cells. 5 × 106 cells were fixed with 1 ml of 1% formaldehyde for 10 min with occasional swirling at room temperature. Cross-linking was stopped by addition of glycine to 125 mM. Cells were then lysed in 100 µl of lysis buffer (20 mM Tris, pH 7.5, 5 mM EDTA, and 0.5% SDS supplemented with a protease inhibitor cocktail [Nacalai tesque]) for 10 min while rotating at 4°C. Genomic fragments were sonicated to a mean size of ∼200 bp using an Acoustic Solubilizer (Covaris) with a 200-bp shearing protocol. After insoluble material was removed by centrifugation at 14,000 rpm for 10 min, 5 µl of the supernatants were taken as input, and the remainder was diluted fivefold with dilution buffer (1% Triton X-100, 1 mM EDTA, 150 mM NaCl, and 20 mM Tris, pH 8.0, supplemented with protease inhibitor cocktail). Immunoprecipitation was performed with 2 µg anti-Nr4a1 monoclonal antibody (H1648; Perseus Proteomics), monoclonal antihistone H3 (ab1791; Abcam), polyclonal anti–di-methylated-histone H3 lys4 (Y47; Abcam), polyclonal anti–tri-methylated-histone H3 lys27 (ab6002; Abcam), or control mouse IgG for 4 h at 4°C with rotation. Complexes were recovered by incubation with 15 µl Protein G-DynaBeads (Invitrogen), which had been preincubated in 100 µl of blocking buffer (500 µg/ml BSA and 200 µg/ml sheared salmon sperm DNA in dilution buffer) for 2 h at 4°C. Precipitates were washed serially with 100 µl RIPA buffer (50 mM, Tris pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% Na-deoxycholate, 1% NP-40, and 1 mM EDTA), 100 µl high salt buffer (50 mM Tris, pH 8.0, 500 mM NaCl, 0.1% SDS, 0.5% Na-deoxycholate, 1% NP-40, and 1 mM EDTA), 100 µl LiCl buffer (50 mM Tris, pH 8.0, 1 mM EDTA, 250 mM LiCl, 1% NP-40, and 0.5% Na-deoxycholate), and then twice with 100 µl TE. Chromatin samples were eluted from the beads three times with 70 µl elution buffer (1% SDS and 0.1 M NaHCO3) for 30 min at room temperature with constant agitation. Next, cross-linking was reversed by overnight incubation at 65°C. After the reversal of cross-linking, proteinase K was added to the samples at 0.2 mg/ml, and the samples incubated for 5 h at 56°C. After phenol-chloroform extraction, the aqueous phase was ethanol-precipitated and dissolved in 50 µl TE buffer. Products were analyzed either by quantitative PCR analysis (ChIP-qPCR) or by next-generation sequencing (ChIP-seq). Primer sets used in ChIP-qPCR are shown in Table S1.
ChIP-seq.
Chromatin DNA were prepared as described in the Chromatin immunoprecipitation section. 10 ng immunoprecipitated DNA fragments, which were pooled from several independent preparations, were blunt-ended, and then ligated to the Illumina adaptors. After purification of the DNAs with 250–350 bp length on gel electrophoresis, they were amplified using a TruSeq ChIP Sample Prep kit (Illumina) according to the manufacture’s protocol and sequenced with a HiSeq 1500 sequencer (Illumina). The obtained single-end 50-nt reads were base-called by using the Illumina Analysis Pipeline. All uniquely matching reads were mapped to the mouse genome (mm10) using the CLC Genomics Workbench software (Filgen). After removal of redundant reads, candidate binding peaks were predicted using ChIP-Seq analysis tool equipped in CLC Genomics Workbench. Around 50,000 peaks were detected at a p-value level of <10−5 and false discovery rate of <1%. Binding of a gene was defined as having a peak of ±5 kb of gene bodies. Raw ChIP-seq sequences of H3K4me3 in nT reg, T naive, and Th2 cells, H3K27me3 in nT reg cells were obtained from Short Read Archive database (SRP000706). Raw ChIP-seq sequences of H3K4me3 in Tfh (described as ex vivo Tfh cells in Lu et al., 2011) were obtained from Gene Expression Omnibus (GEO) database (GSE32864). The sequence reads were mapped to mouse genome (mm10) using CLC Genomics Workbench software (Filgen).
Bisulfite sequencing.
Genomic DNA extracted from ∼2 × 105 cells from male mice was digested with BamHI. After phenol-chloroform extraction, the aqueous phases were ethanol-precipitated and dissolved in dH2O. DNA concentrations were adjusted to a range of 500 ng–2 µg in 19 µl dH2O, then 1 µl of 6 N NaOH was added; the mixture was then incubated at 37°C for 15 min. Next, 120 µl of reaction reagent (3.6 M sodium bisulfite, 0.57 mM hydroquinone, and 0.3 N NaOH) were added to the samples. Samples were then treated with 15 cycles of 95°C for 30 s/50°C for 15 min. The reactions were then desalted using the Wizard DNA Clean-up System (Promega), and eluted with 50 µl of TE. 3 µl of 6 N NaOH were added to the samples, followed by incubation for 5 min at room temperature. The products were then ethanol-precipitated and dissolved in 20 µl of TE buffer. The desired regions were PCR amplified with primer sets shown in Table S1. Amplified fragments were then T/A cloned into the pGEM-T Easy Vector (Promega). More than 10 independent clones in each group were sequenced. A primer set used in bisulfite sequencing is shown in Table S2.
Transient transfections and luciferase assays.
5 × 105 Jurkat T cell lymphoma cells were transiently transfected with 1 µg of the indicated pGL3-luciferase plasmids and 8 µg of the Nr4a2-expressing or empty pCMV plasmids, using NEPA21 Type II electroporator (NEPAGENE). For each transfection, 1 µg of β-galactosidase-expressing plasmid was added for internal control. Luciferase activities were analyzed by Luciferase Reporter Assay System (Promega), at 36 h after transfection.
Detection of Tfh cells and germinal center B cells in splenocytes.
Total splenocytes were isolated and stained with APC-Cy7–conjugated antibody to CD4 (RM4-5; Tonbo Biosciences), APC-conjugated monoclonal antibody to PD-1 (RMP1-30; BioLegend), and biotinylated antibody to CXCR5 (2G8; BD), followed by PerCP-Cy5.5-conjugated streptavidin for Tfh detection. For control staining of Tfh cell detection, cells were stained with APC-Cy7–conjugated antibody to CD4 (RM4-5; Tonbo Biosciences), APC-conjugated rat IgG2a (eBR2a), and biotinylated rat IgG2a (eBR2a) followed by PerCP-Cy5.5–conjugated streptavidin. For GC B cell detection, splenocytes were stained with PE-conjugated antibody to GL-7 (GL-7; eBioscience), PerCP-Cy5.5-conjugated antibody to B220 (RA3-6B2; Tonbo Biosciences), and APC-conjugated antibody to Fas (15A7; eBioscience).
ELISA.
For analysis of total immunoglobulin, sera were analyzed with a mouse Ig ELISA Quantitation kit (Bethyl Laboratories), according to the manufacturer’s protocol. Anti–double-strand DNA antibodies were analyzed with a mouse Anti-dsDNA Ig ELISA kit (Alpha Diagnostic), according to the manufacturer’s protocol.
qRT-PCR and microarray analysis of mRNA.
Total RNA was extracted from freshly prepared T naive cells, T reg cells, Tfh cells, and exFoxp3 cells, or from in vitro–differentiated Th1, Th2, and Th17 cells, using RNAiso PLUS (Takara Bio Inc.). Differentiation conditions for Th1, Th2, and Th17 cells were described in the Western blotting section. For qRT-PCR analysis, samples were subjected to reverse transcription using a High Capacity cDNA Synthesis kit (Applied Biosystems). PCR analysis was performed with an iCycler iQ multicolor real-time PCR detection system (Bio-Rad Laboratories) and SsoFast EvaGreen Supermix (Bio-Rad Laboratories). Sequences for primers used in this study are available upon request. For microarray analysis, samples were further cleaned using an NucleoSpin RNA Clean-up XS (MACHEREY-NAGEL), labeled with Cyanine 3-CTP using a Low Input Quick Amp Labeling kit (Agilent), and hybridized to a 4 × 44K Whole Mouse Genome Oligo Microarray (Agilent). Expression values for each probe set were calculated using the RMA method with GeneSpring GX 11.0 software (Agilent).
In vitro suppression assay.
Responder cells (Ly5.1+CD4+CD25−CD62Lhi; 2 × 104 cells) from Ly5.1+ WT C57/BL6 mice were labeled with 5 µM CFSE (Invitrogen), and then cultured with or without sorted T reg cells (CD4+YFP+) from Foxp3YFP-Cre-Nr4a-TKO mice or Foxp3YFP-Cre-WT mice at the indicated ratio for 96 h at 37°C in 200 µl of RPMI-1640 medium plus 10% FBS, supplemented with 1 µg/ml anti-CD3 antibodies (clone 2C11) and 55 µM β-mercaptoethanol, in the presence of 2 × 104 T cell–depleted splenocytes irradiated at 20 Gy, in round-bottomed 96-well culture plates. The CFSE dilution of Ly5.1+ responder cells was analyzed by flow cytometry.
Adoptive transfer of T reg cells.
Sorted WT or Nr4a-TKO T reg cells were intravenously transferred into 6–8-wk-old CD3ε-deficient recipient mice without immunization. 5 wk after transfer, the sera, BAL fluid, lymphoid organs, and lungs were collected for analysis.
Accession nos.
Gene expression data and high-throughput sequencing datasets have been deposited in the NCBI Gene Expression Omnibus with accession nos. GSE70306 and GSE70417, respectively.
Statistical analysis.
P-values were calculated with GraphPad Prism software. P-values < 0.05 were considered significant. All error bars in graphs represent SEM calculated at least three replicates.
Online supplemental material.
Fig. S1 shows the sorted cell population before and after adoptive transfer in the experiment shown in Fig. 9. Tables S1 and Table S2 list the gene-specific primers used for ChIP-qPCR analysis and bisulfite sequencing, respectively. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20142088/DC1.
Supplementary Material
Acknowledgments
We thank N. Shiino, Y. Noguchi, H. Yamane, R. Komine, and M. Asakawa for technical assistance and Y. Ushijima for manuscript preparation. We thank O. Ohara and Y. Hasegawa for ChIP-seq analysis.
This work was supported by Japan Society for the Promotion of Science KAKENHI Grant-in-Aid for Young Scientists (A) 26713019, Grant-in-Aid for Scientific Research (S) 25221305, the Takeda Science Foundation, the Uehara Memorial Foundation, Mochida Memorial Foundation for Medical and Pharmaceutical Research, Kanae Foundation, and the SENSHIN Medical Research Foundation, Keio Gijuku Academic Developmental Funds.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- BAL
- bronchoalveolar lavage
- CFA
- complete Freund’s adjuvant
- GC
- germinal center
- TF
- transcription factor
- Tfh
- T follicular helper
- Tfr
- follicular regulatory T
- T reg cell
- regulatory T cell
References
- Abbas A.K., Urioste S., Collins T.L., and Boom W.H.. 1990. Heterogeneity of helper/inducer T lymphocytes. IV. Stimulation of resting and activated B cells by Th1 and Th2 clones. J. Immunol. 144:2031–2037. [PubMed] [Google Scholar]
- Agarwal S., and Rao A.. 1998. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 9:765–775. 10.1016/S1074-7613(00)80642-1 [DOI] [PubMed] [Google Scholar]
- Bailey-Bucktrout S.L., Martinez-Llordella M., Zhou X., Anthony B., Rosenthal W., Luche H., Fehling H.J., and Bluestone J.A.. 2013. Self-antigen-driven activation induces instability of regulatory T cells during an inflammatory autoimmune response. Immunity. 39:949–962. 10.1016/j.immuni.2013.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y., Tanaka S., Chu F., Nurieva R.I., Martinez G.J., Rawal S., Wang Y.H., Lim H., Reynolds J.M., Zhou X.H., et al. 2011. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat. Med. 17:983–988. 10.1038/nm.2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663. 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- Floess S., Freyer J., Siewert C., Baron U., Olek S., Polansky J., Schlawe K., Chang H.D., Bopp T., Schmitt E., et al. 2007. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 5:e38 10.1371/journal.pbio.0050038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot J.D., Gavin M.A., and Rudensky A.Y.. 2003. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4:330–336. 10.1038/ni904 [DOI] [PubMed] [Google Scholar]
- Fontenot J.D., Rasmussen J.P., Gavin M.A., and Rudensky A.Y.. 2005. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat. Immunol. 6:1142–1151. 10.1038/ni1263 [DOI] [PubMed] [Google Scholar]
- Gavin M.A., Rasmussen J.P., Fontenot J.D., Vasta V., Manganiello V.C., Beavo J.A., and Rudensky A.Y.. 2007. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 445:771–775. 10.1038/nature05543 [DOI] [PubMed] [Google Scholar]
- Hamers A.A., Hanna R.N., Nowyhed H., Hedrick C.C., and de Vries C.J.. 2013. NR4A nuclear receptors in immunity and atherosclerosis. Curr. Opin. Lipidol. 24:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J.A., Feuerer M., Tash K., Haxhinasto S., Perez J., Melamed R., Mathis D., and Benoist C.. 2007. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 27:786–800. 10.1016/j.immuni.2007.09.010 [DOI] [PubMed] [Google Scholar]
- Hori S., Nomura T., and Sakaguchi S.. 2003. Control of regulatory T cell development by the transcription factor Foxp3. Science. 299:1057–1061. 10.1126/science.1079490 [DOI] [PubMed] [Google Scholar]
- Iikuni N., Lourenço E.V., Hahn B.H., and La Cava A.. 2009. Cutting edge: regulatory T cells directly suppress B cells in systemic lupus erythematosus. J. Immunol. 183:1518–1522. 10.4049/jimmunol.0901163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadkhodaei B., Ito T., Joodmardi E., Mattsson B., Rouillard C., Carta M., Muramatsu S., Sumi-Ichinose C., Nomura T., Metzger D., et al. 2009. Nurr1 is required for maintenance of maturing and adult midbrain dopamine neurons. J. Neurosci. 29:15923–15932. 10.1523/JNEUROSCI.3910-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattri R., Cox T., Yasayko S.A., and Ramsdell F.. 2003. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 4:337–342. 10.1038/ni909 [DOI] [PubMed] [Google Scholar]
- Kim H.P., and Leonard W.J.. 2007. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J. Exp. Med. 204:1543–1551. 10.1084/jem.20070109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., Rasmussen J.P., and Rudensky A.Y.. 2007. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8:191–197. 10.1038/ni1428 [DOI] [PubMed] [Google Scholar]
- King C. 2009. New insights into the differentiation and function of T follicular helper cells. Nat. Rev. Immunol. 9:757–766. 10.1038/nri2644 [DOI] [PubMed] [Google Scholar]
- Komatsu N., Okamoto K., Sawa S., Nakashima T., Oh-hora M., Kodama T., Tanaka S., Bluestone J.A., and Takayanagi H.. 2014. Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nat. Med. 20:62–68. 10.1038/nm.3432 [DOI] [PubMed] [Google Scholar]
- Levine A.G., Arvey A., Jin W., and Rudensky A.Y.. 2014. Continuous requirement for the TCR in regulatory T cell function. Nat. Immunol. 15:1070–1078. 10.1038/ni.3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H.W., Hillsamer P., and Kim C.H.. 2004. Regulatory T cells can migrate to follicles upon T cell activation and suppress GC-Th cells and GC-Th cell-driven B cell responses. J. Clin. Invest. 114:1640–1649. 10.1172/JCI200422325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Haribhai D., Relland L.M., Truong N., Carlson M.R., Williams C.B., and Chatila T.A.. 2007. Regulatory T cell development in the absence of functional Foxp3. Nat. Immunol. 8:359–368. 10.1038/ni1445 [DOI] [PubMed] [Google Scholar]
- Linterman M.A., Pierson W., Lee S.K., Kallies A., Kawamoto S., Rayner T.F., Srivastava M., Divekar D.P., Beaton L., Hogan J.J., et al. 2011. Foxp3+ follicular regulatory T cells control the germinal center response. Nat. Med. 17:975–982. 10.1038/nm.2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C.M., and Hawrylowicz C.M.. 2009. Regulatory T cells in asthma. Immunity. 31:438–449. 10.1016/j.immuni.2009.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K.T., Kanno Y., Cannons J.L., Handon R., Bible P., Elkahloun A.G., Anderson S.M., Wei L., Sun H., O’Shea J.J., and Schwartzberg P.L.. 2011. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity. 35:622–632. 10.1016/j.immuni.2011.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche H., Weber O., Nageswara Rao T., Blum C., and Fehling H.J.. 2007. Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur. J. Immunol. 37:43–53. 10.1002/eji.200636745 [DOI] [PubMed] [Google Scholar]
- Luo C.T., and Li M.O.. 2013. Transcriptional control of regulatory T cell development and function. Trends Immunol. 34:531–539. 10.1016/j.it.2013.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao T., Floess S., Setoguchi R., Luche H., Fehling H.J., Waldmann H., Huehn J., and Hori S.. 2012. Plasticity of Foxp3+ T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity. 36:262–275. 10.1016/j.immuni.2011.12.012 [DOI] [PubMed] [Google Scholar]
- Miyara M., and Sakaguchi S.. 2007. Natural regulatory T cells: mechanisms of suppression. Trends Mol. Med. 13:108–116. 10.1016/j.molmed.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Moran A.E., Holzapfel K.L., Xing Y., Cunningham N.R., Maltzman J.S., Punt J., and Hogquist K.A.. 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208:1279–1289. 10.1084/jem.20110308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai M., Turovskaya O., Kim G., Madan R., Karp C.L., Cheroutre H., and Kronenberg M.. 2009. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat. Immunol. 10:1178–1184. 10.1038/ni.1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto G., Kotani H., Kondo T., Morita R., Tsuruta S., Kobayashi T., Luche H., Fehling H.J., Walsh M., Choi Y., and Yoshimura A.. 2013. TRAF6 is essential for maintenance of regulatory T cells that suppress Th2 type autoimmunity. PLoS ONE. 8:e74639 10.1371/journal.pone.0074639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W., Liao W., Luo C.T., Yin N., Huse M., Kim M.V., Peng M., Chan P., Ma Q., Mo Y., et al. 2012. Novel Foxo1-dependent transcriptional programs control T(reg) cell function. Nature. 491:554–559. 10.1038/nature11581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F., Yu H., Dang E.V., Barbi J., Pan X., Grosso J.F., Jinasena D., Sharma S.M., McCadden E.M., Getnet D., et al. 2009. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 325:1142–1146. 10.1126/science.1176077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov Y.P., Rasmussen J.P., Chi E.Y., Fontenot J., Castelli L., Ye X., Treuting P., Siewe L., Roers A., Henderson W.R. Jr., et al. 2008. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 28:546–558. 10.1016/j.immuni.2008.02.017 [DOI] [PubMed] [Google Scholar]
- Rubtsov Y.P., Niec R.E., Josefowicz S., Li L., Darce J., Mathis D., Benoist C., and Rudensky A.Y.. 2010. Stability of the regulatory T cell lineage in vivo. Science. 329:1667–1671. 10.1126/science.1191996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo K., Winner B., Carson C.T., Collier J.G., Boyer L., Rosenfeld M.G., Gage F.H., and Glass C.K.. 2009. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 137:47–59. 10.1016/j.cell.2009.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S., Yamaguchi T., Nomura T., and Ono M.. 2008. Regulatory T cells and immune tolerance. Cell. 133:775–787. 10.1016/j.cell.2008.05.009 [DOI] [PubMed] [Google Scholar]
- Seibler J., Zevnik B., Küter-Luks B., Andreas S., Kern H., Hennek T., Rode A., Heimann C., Faust N., Kauselmann G., et al. 2003. Rapid generation of inducible mouse mutants. Nucleic Acids Res. 31:e12 10.1093/nar/gng012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T., Kashiwagi I., Inoue N., Morita R., Hori S., Waldmann H., Rudensky A.Y., Ichinose H., Metzger D., Chambon P., and Yoshimura A.. 2011. The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat. Commun. 2:269 10.1038/ncomms1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T., Kashiwagi I., Yoshida R., Fukaya T., Morita R., Kimura A., Ichinose H., Metzger D., Chambon P., and Yoshimura A.. 2013. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat. Immunol. 14:230–237. 10.1038/ni.2520 [DOI] [PubMed] [Google Scholar]
- Setoguchi R., Hori S., Takahashi T., and Sakaguchi S.. 2005. Homeostatic maintenance of natural Foxp3+ CD25+ CD4+ regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J. Exp. Med. 201:723–735. 10.1084/jem.20041982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Kuniyasu Y., Toda M., Sakaguchi N., Itoh M., Iwata M., Shimizu J., and Sakaguchi S.. 1998. Immunologic self-tolerance maintained by CD25+CD4+ naturally anergic and suppressive T cells: induction of autoimmune disease by breaking their anergic/suppressive state. Int. Immunol. 10:1969–1980. 10.1093/intimm/10.12.1969 [DOI] [PubMed] [Google Scholar]
- Thornton A.M., and Shevach E.M.. 1998. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J. Exp. Med. 188:287–296. 10.1084/jem.188.2.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton A.M., and Shevach E.M.. 2000. Suppressor effector function of CD4+CD25+ immunoregulatory T cells is antigen nonspecific. J. Immunol. 164:183–190. 10.4049/jimmunol.164.1.183 [DOI] [PubMed] [Google Scholar]
- Tsuji M., Komatsu N., Kawamoto S., Suzuki K., Kanagawa O., Honjo T., Hori S., and Fagarasan S.. 2009. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer’s patches. Science. 323:1488–1492. 10.1126/science.1169152 [DOI] [PubMed] [Google Scholar]
- Vignali D.A., Collison L.W., and Workman C.J.. 2008. How regulatory T cells work. Nat. Rev. Immunol. 8:523–532. 10.1038/nri2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner J.R., Fox-Talbot K., Halushka M.K., Ellis C., Zachary A.A., and Baldwin W.M. III. 2010. B cells and plasma cells in coronaries of chronically rejected cardiac transplants. Transplantation. 89:1141–1148. 10.1097/TP.0b013e3181d3f271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G., Wei L., Zhu J., Zang C., Hu-Li J., Yao Z., Cui K., Kanno Y., Roh T.Y., Watford W.T., et al. 2009. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 30:155–167. 10.1016/j.immuni.2008.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing K., Onishi Y., Prieto-Martin P., Yamaguchi T., Miyara M., Fehervari Z., Nomura T., and Sakaguchi S.. 2008. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 322:271–275. 10.1126/science.1160062 [DOI] [PubMed] [Google Scholar]
- Zhou X., Bailey-Bucktrout S.L., Jeker L.T., Penaranda C., Martínez-Llordella M., Ashby M., Nakayama M., Rosenthal W., and Bluestone J.A.. 2009. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat. Immunol. 10:1000–1007. 10.1038/ni.1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Cote-Sierra J., Guo L., and Paul W.E.. 2003. Stat5 activation plays a critical role in Th2 differentiation. Immunity. 19:739–748. 10.1016/S1074-7613(03)00292-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.