Mehta et al. uncover a novel role for miR-212/132 as a regulator of early B cell development.
Abstract
MicroRNAs have emerged as key regulators of B cell fate decisions and immune function. Deregulation of several microRNAs in B cells leads to the development of autoimmune disease and cancer in mice. We demonstrate that the microRNA-212/132 cluster (miR-212/132) is induced in B cells in response to B cell receptor signaling. Enforced expression of miR-132 results in a block in early B cell development at the prepro–B cell to pro–B cell transition and induces apoptosis in primary bone marrow B cells. Importantly, loss of miR-212/132 results in accelerated B cell recovery after antibody-mediated B cell depletion. We find that Sox4 is a target of miR-132 in B cells. Co-expression of SOX4 with miR-132 rescues the defect in B cell development from overexpression of miR-132 alone, thus suggesting that miR-132 may regulate B lymphopoiesis through Sox4. In addition, we show that the expression of miR-132 can inhibit cancer development in cells that are prone to B cell cancers, such as B cells expressing the c-Myc oncogene. We have thus uncovered miR-132 as a novel contributor to B cell development.
B cells are the primary producers of immunoglobulin and play a critical role in adaptive immunity (Mauri and Bosma, 2012). The maintenance of proper B cell output from early hematopoietic progenitors, along with the production of an appropriate antibody repertoire, is critical to maintaining the balance between normal immune function and diseases such as autoimmunity and cancer. Therefore, B lymphopoiesis requires the intricate interplay of many different transcription factors in a complex gene regulatory network that controls lineage specification and commitment (Matthias and Rolink, 2005; Nutt and Kee, 2007; Mandel and Grosschedl, 2010). Antigen-independent B cell development begins with the differentiation of lymphoid primed multipotent progenitors to common lymphoid progenitors (CLPs), a process driven by the expression of PU.1 and Ikaros (Matthias and Rolink, 2005; Nutt and Kee, 2007), both of which may play a role in regulating Flt3 and IL-7R expression (DeKoter et al., 2002; Yoshida et al., 2006). These early progenitors also express Rag1 and Rag2, and thus begin the process of rearrangement of the Ig heavy chain (IgH) locus (Igarashi et al., 2002). Lineage specification to the next stages of B cell development, the prepro–B cell and pro–B cell, involves the up-regulation of several genes controlled by E2A and Ebf1 (O’Riordan and Grosschedl, 1999), including Pax5 (Cobaleda et al., 2007). Pax5 is essential for B cell lineage commitment, as it represses genes that are inappropriate for B cell development (Souabni et al., 2002). The transition to pre–B cells, the stage at which Ig light chain (IgL) rearrangement begins, and immature B cells, involves many factors, including Sox4 (Sun et al., 2013), which has also been implicated in regulating the expression of the Rag genes (Mallampati et al., 2014).
MicroRNAs, a class of small noncoding RNAs that negatively regulate gene expression, are key posttranscriptional regulators of hematopoietic cell fate decisions and immunity (O’Connell et al., 2010b). Several microRNAs regulate key checkpoints in B cell development, and the loss of a microRNA processing protein, Dicer, results in a block in the pro–B to pre–B cell transition (Koralov et al., 2008). In particular, both miR-150 and miR-34a regulate this transition by targeting c-Myb and Foxp1, respectively (Xiao et al., 2007; Zhou et al., 2007; Rao et al., 2010). Another example is miR-148a, which regulates plasma cell differentiation by targeting Bach2 (Porstner et al., 2015). In addition, miR-181 and miR-155 play an important role in B cell immune function by targeting AID to regulate class-switching and somatic hypermutation (Thai et al., 2007; de Yébenes et al., 2008; Teng et al., 2008). Importantly, deregulation of the expression of many microRNAs important in B cell development and function results in autoimmunity (Xiao et al., 2008) and the onset of B cell cancers (Eis et al., 2005; Costinean et al., 2006; Calin et al., 2008; Xiao et al., 2008; Puissegur et al., 2012).
Recently, the microRNA-212/132 cluster (miR-212/132) has emerged as an important regulator of hematopoietic stem cell function (Mehta et al., 2015), antiviral immunity (Lagos et al., 2010), macrophage and TH17 T cell immune function (Taganov et al., 2006; Shaked et al., 2009; Nahid et al., 2013; Nakahama et al., 2013), and inflammation and proliferation during wound healing (Li et al., 2015). In addition, it has been shown that miR-132 plays a role in the proliferation and invasion of certain solid tumors (Zhang et al., 2014; Jiang et al., 2015), as well as in pathological angiogenesis (Anand et al., 2010), thus making it a potential candidate for cancer therapeutics. Recently, several groups have also shown miR-212/132 to be deregulated in certain B cell cancers (Lawrie et al., 2008; Pede et al., 2013; Tavolaro et al., 2015). In this study, we uncover a novel role for miR-212/132 as a regulator of early B cell development by targeting the transcription factor Sox4. In addition, we find that miR-132 induces apoptosis in B cells and that this may be leveraged to inhibit the progression of B cell cancers, such as B cell lymphomas in mice expressing the c-Myc oncogene driven by the IgH enhancer (Langdon et al., 1986).
RESULTS
The microRNA-212/132 cluster is induced in B cells in response to BCR activation
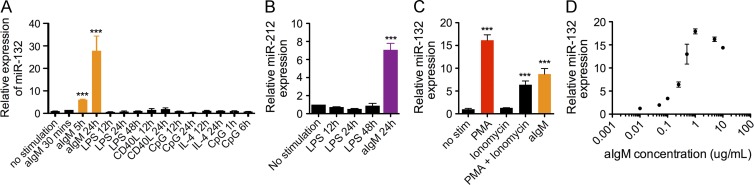
To investigate the role of the microRNA-212/132 cluster (miR-212/132) in B cells, we first examined the expression of miR-212/132 in various sorted splenic and bone marrow cell populations obtained from WT C57BL/6 mice. RNA was extracted from these cells, and then we performed TaqMan qPCR to profile microRNA expression. We initially found that miR-212/132 was not significantly enriched in either the splenic or bone marrow CD19+ B cell population; however, we found a significant up-regulation of miR-132 in these cells at 24 h in response to stimulation with mouse anti-IgM, which binds to and activates the BCR (Fig. 1 A). This induction peak could be replicated by stimulation with PMA and ionomycin, but not with activation of NF-κB through TLR ligands or anti-CD40, suggesting the induction requires multiple pathways downstream of BCR signaling, whereas NF-κB alone is insufficient (Fig. 1, A–C). The level of miR-132 induction demonstrated a strong dependence on the concentration of anti-IgM used for stimulation (Fig. 1 D).
Figure 1.
miR-132 is induced in B cells in response to BCR signaling. (A) Relative miR-132 expression in primary splenic B cells stimulated with anti-IgM (aIgM; 5 µg/ml), LPS (20 µg/ml), CD40L (2 µg/ml), IL-4 (10 ng/ml), or CpG (0.5 µM) for the indicated durations. miR-132 expression was detected using TaqMan RT-qPCR (n = 3). (B) Relative expression of miR-212 in splenic B cells stimulated with LPS (20 µg/ml) or anti-IgM (5 µg/ml) for the indicated times (n = 3). (C) Relative expression of miR-132 in splenic B cells stimulated for 8 h with a combination of PMA (15 ng/ml), ionomycin (0.5 µg/ml), and anti-IgM (5 µg/ml), as indicated (n = 3). (D) Relative expression of miR-132 in splenic B cells stimulated for 8 h with varying concentrations of anti-IgM as indicated (n = 3). Data represent two independent experiments and are represented as mean ± SEM. ***, P < 0.001, Student’s t test.
Enforced expression of microRNA-132 inhibits B cell development
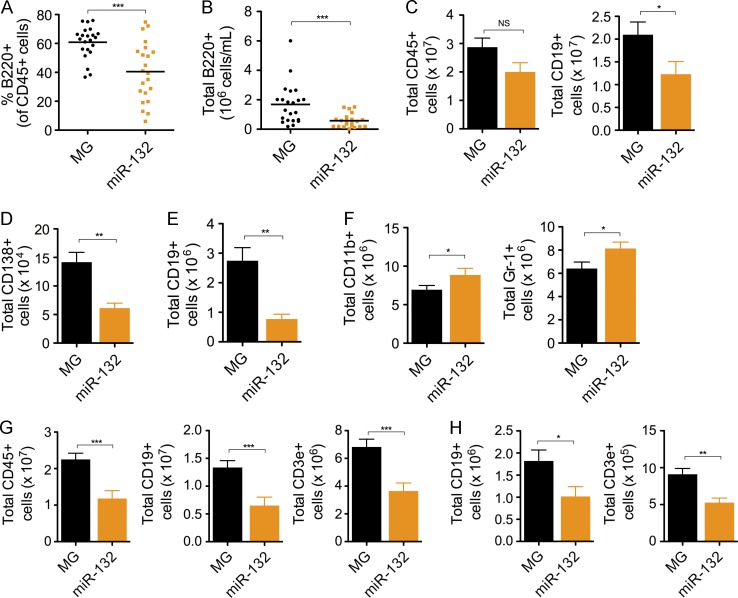
To understand the function of the miR-132/212 cluster on B cell development, we examined the effects of enforced expression of either miR-132 or miR-212 on B cell development. Bone marrow from WT C57BL/6 mice that was enriched for hematopoietic stem and progenitor cells (HSPCs) was retrovirally transduced with control vector (WTMG) or a miR-132 overexpression vector (WTmiR-132). These cells were then used to reconstitute the immune system of lethally irradiated C57BL/6 WT recipients, and we subsequently tracked the development of B cells. We found that WTmiR-132 mice had a significant depletion in the frequency and total number of peripheral blood B cells compared with WTMG controls at 3–4 mo after reconstitution (Fig. 2, A and B). This was accompanied by a decrease in the total number of leukocytes (CD45+), B cells (CD19+), and plasma cells (CD138+) in the spleen of WTmiR-132 mice (Fig. 2, C and D). The loss of B cells was dramatically mirrored in the bone marrow compartment (Fig. 2 E), where a minor increase in myeloid cells was also observed (Fig. 2 F). A similar depletion in total leukocytes, B cells, and T cells was observed in the spleen of mice at 9 mo after reconstitution (Fig. 2 G). The bone marrow compartment of these mice also demonstrated a loss in B cells and T cells, but no change in myeloid cells was apparent (Fig. 2 H).
Figure 2.
Enforced expression of miR-132 in mice alters B cell development. WT C57BL/6 mice were lethally irradiated and reconstituted with donor HPSCs expressing either a control (MG) or a miR-132–overexpressing (miR-132) vector. (A–F) Mice were analyzed at 4 mo after reconstitution (n = 10–12 mice per group). (A) Frequency of peripheral blood B cells (B220+) in MG and miR-132 mice. (B) Total number of peripheral blood B cells (B220+) in MG and miR-132 mice. (C) Total number of leukocytes (CD45+) and B cells (CD19+) in the spleen of MG and miR-132 mice. (D) Total number of plasma cells (CD138+) in the spleen of MG and miR-132 mice. (E) Total number of B cells (CD19+) in the bone marrow of MG and mIR-132 mice. (F) Total number of myeloid cells (CD11b+) and granulocytes (Gr-1+) in the bone marrow of MG and miR-132 mice. (G–H) miR-132 and MG mice analyzed at 9 mo after reconstitution. (G) Total number of leukocytes (CD45+), B cells (CD19+), and T cells (CD3e+) in the spleen of MG and miR-132 mice. (H) Total number of B cells (CD19+) and T cells (CD3e+) in the bone marrow of MG and miR-132 mice. Data represent at least two independent experiments and are represented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001, Student’s t test.
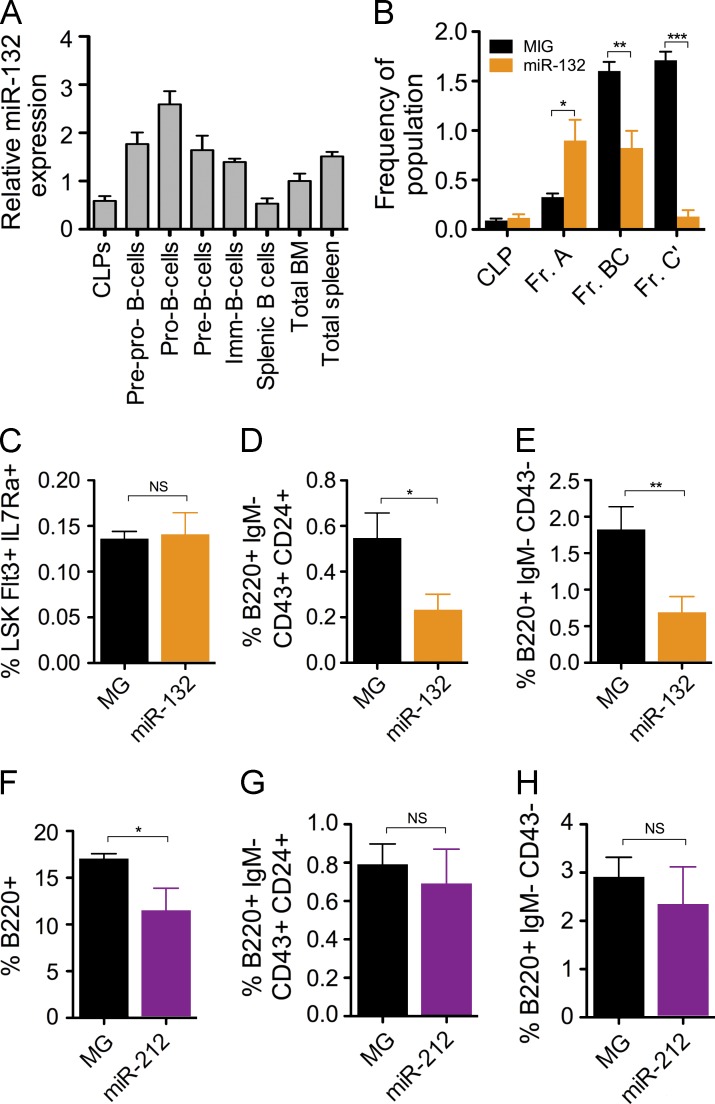
To better understand the influence of miR-212/132 in early B cell commitment, we profiled the expression of miR-132 in B cell progenitors. We found an enrichment in miR-132 in progenitor B (pro–B) cells and precursor B (pre–B) cells compared with total bone marrow cells and CLPs (Fig. 3 A). To investigate the consequence of this, we analyzed Hardy fractions in the bone marrow of WTmiR-132 and WTMG mice (Rumfelt et al., 2006). We found that WTmiR-132 mice had a comparable frequency of CLPs and an elevation in precursor–progenitor B (prepro–B) B cells (Hardy Fr. A) compared with WTMG mice (Fig. 3 B). However, we found a significant decrease in the frequency of both pre–B cells (Hardy Fr. BC) and pro–B cells (Hardy Fr. C’) in the bone marrow of WTmiR-132 mice (Fig. 3 B), suggestive of a developmental block between the prepro–B cell to pro–B cell transition. An alternative surface marker characterization of these B cell progenitor populations resulted in similar findings (Fig. 3, C–E). Enforced expression of miR-212 alone resulted in a decrease in B cells in the bone marrow compartment at 4 mo after reconstitution; however, no changes in the frequency of bone marrow B cell progenitors or splenic B cells was observed compared with controls (Fig. 3, F–H). Because miR-132 was more strongly induced in B cells, and its enforced expression in HSPCs produced a stronger effect on B cell development compared with that of miR-212, we chose to examine miR-132, among the two microRNAs in the miR-212/132 cluster, as a more relevant regulator of B cell biology.
Figure 3.
miR-132 causes a block in early B cell development. (A) miR-132 expression in various bone marrow B cell progenitor populations. Cells were sorted by FACS directly into RNA lysis buffer and miR-132 expression was detected using TaqMan RT-qPCR (n = 2). (B–E) Analysis of MG and miR-132 mice at 4 mo after reconstitution (n = 10–12 mice per group). (B) Hardy fractions for B cell progenitor populations in the bone marrow of MG and miR-132 mice. Populations were gated on lineage-negative cells (CD3, Ter119, Ly6C, Gr1, CD11b), and are presented as follows: CLP (lineage-CD19-CD24-CD43medCD93hiIgM-B220-cKit+IL7Ra+), Fr. A (prepro–B; lineage−B220+CD43+CD19−CD24−CD93+cKit+IL7Ra+), Fr. BC (pro–B; lineage−B220+CD43+CD19+CD24medCD93+), and Fr. C’ (pre–B; lineage−B220+CD43+CD19+CD24hiCD93+). (C) Frequency of CLPs using alternative surface markers (lineage−cKit+Sca1+Flt3+IL7Ra+) in the bone marrow MG and miR-132 mice. (D) Frequency of pro–B cells using alternative surface markers (B220+IgM−CD43+CD24+) in the bone marrow of MG and miR-132 mice. (E) Frequency of pre–B cells using alternative surface markers (B220+IgM−CD43−) in the bone marrow of MG and miR-132 mice. (F–H) WT C57BL/6 mice were lethally irradiated and reconstituted with donor HPSCs expressing either a control (MG) or a miR-212–overexpressing (miR-212) vector. Mice were analyzed at 4 mo after reconstitution (n = 4–5 mice per group). (F) Frequency of B cells (B220+) in the bone marrow of MG and miR-212 mice. (G) Frequency of pro–B cells (B220+IgM−CD43+CD24+) in the bone marrow of MG and miR-212 mice. (H) Frequency of pre–B cells (B220+IgM−CD43−) in the bone marrow of MG and miR-212 mice. Data represent at least two independent experiments and are represented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001, Student’s t test.
Loss of the microRNA-212/132 cluster enhances B cell development under nonhomeostatic conditions
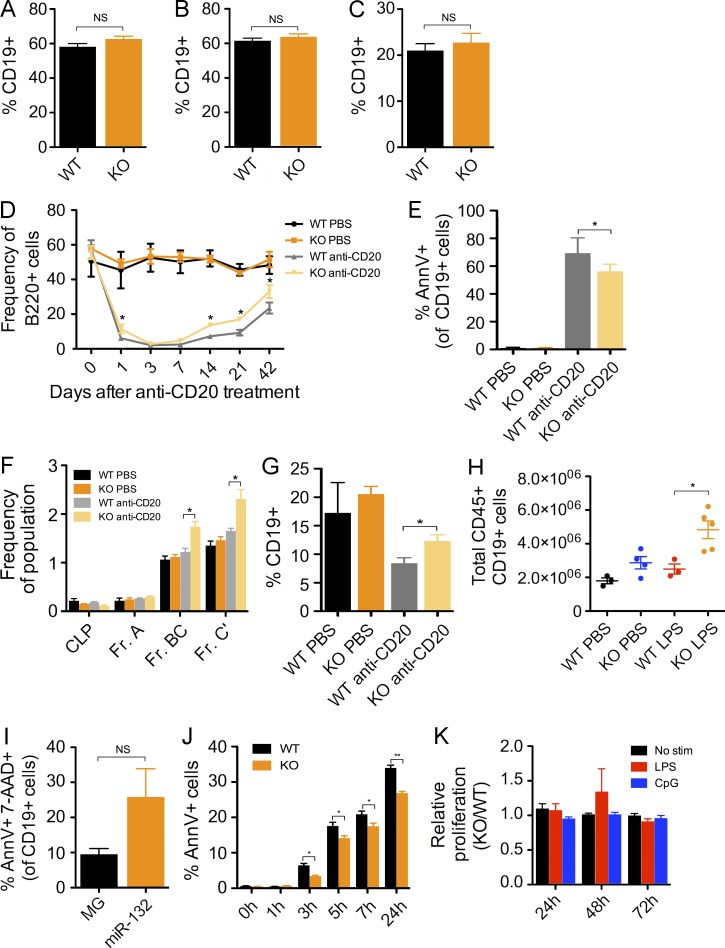
A close examination of the peripheral blood, spleen, and bone marrow of miR-212/132−/− and WT mice at 16-wk of age revealed no significant difference in the frequency of B cells present in these compartments (Fig. 4, A–C). To further understand the contribution of miR-212/132 to pro–B cell development, we challenged WT and miR-212/132−/− mice with an anti-CD20 antibody to deplete all mature B cells (Sarikonda et al., 2013) and track B cell development under nonhomeostatic conditions. We observed a rapid depletion of peripheral blood B cells in all mice treated with anti-CD20 compared with control mice treated with PBS (Fig. 4 D). We found, however, that KO mice had a slower depletion in B cells compared with WT mice, evidenced in the frequency of peripheral blood B cells 1 d after anti-CD20 treatment (Fig. 4 D). Consistent with this, peripheral blood B cells in KO mice had lower levels of apoptosis, measured by Annexin V expression, than WT mice 12 h after anti-CD20 treatment (Fig. 4 E). Within 3 d, the total number of B cells in both anti-CD20–treated cohorts was <1% of the total number of peripheral blood leukocytes. Interestingly, under these conditions, miR-212/132−/− mice demonstrated a more rapid rebound in the frequency of peripheral blood B cells compared with WT mice ∼42 d after anti-CD20 treatment (Fig. 4 D). A closer examination of the bone marrow compartment of these mice at 42 d revealed that KO mice had increased frequencies of pro–B cells (Hardy Fr. BC), pre–B cells (Hardy Fr. C’), and total B cells (CD19+) compared with WT mice after treatment with anti-CD20 (Fig. 4, F and G). This indicates a unique advantage of miR-212/132−/− B cell progenitors to replenish the B cell pool under conditions of stress. We further corroborated this result by treating miR-212/132−/− and WT mice with serial LPS injections to mimic aging under conditions of inflammatory stress (Esplin et al., 2011). Consistent with the aforementioned result, we found that miR-212/132−/− mice showed a large accumulation of B cells in the spleen compared with WT mice (Fig. 4 H).
Figure 4.
miR-212/132 regulates B cell apoptosis and the loss of miR-212/132 alters B cell development under nonhomeostatic conditions. (A–C) miR-212/132−/− (KO) and WT mice were analyzed at 12–16 wk of age to understand the physiological contribution of miR-212/132 on B cell output (n = 7–10 mice per group). (A) Frequency of B cells (CD19+) in the peripheral blood of KO and WT mice. (B) Frequency of B cells (CD19+) in the spleen of KO and WT mice. (C) Frequency of B cells (CD19+) in the bone marrow of KO and WT mice. (D–G) 16-wk old KO and WT mice were treated with either an anti-CD20 antibody (30 µg/mouse) or PBS (n = 3–4 mice per group). (D) Graph indicates the frequency of B cells (B220+) in the peripheral blood at each of the indicated time points after anti-CD20 antibody treatment. (E) Frequency of Annexin V+ cells within the peripheral blood B cell (CD19+) population 12 h after anti-CD20 treatment. (F) Frequency of cells in each Hardy fraction for each experimental group 42 d after anti-CD20 treatment. (G) Frequency of bone marrow B cells (CD19+) in each experimental group 42 d after anti-CD20 treatment. (H) 6-mo-old KO and WT mice were treated with 9 evenly spaced LPS (1 mg/kg of body weight) or PBS injections over one month. Graphs shows the total number of B cells (CD19+) in the spleen after the last injection. (I) Frequency of bone marrow CD19+ cells from MG and miR-132 mice that demonstrated positive staining for Annexin V and 7-AAD. (J) KO and WT splenic B cells were isolated and cultured in vitro for the indicated durations. Graph demonstrates the frequency of Annexin V+ cells at each time point. (K) KO or WT splenic B cells were cultured in the presence of no stimulation, or with LPS (20 µg/ml) or CpG (0.5 µM). Proliferation of these cells was measured at the indicated time points using the WST-1 cell proliferation assay. Data represents two independent experiments and is represented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001, Student’s t test.
The microRNA-212/132 cluster regulates B cell survival
We next sought to investigate whether miR-132 may influence B cell development by altering the survival or proliferation of B cells. We found that primary splenic B cells in WTmiR-132 at 2 mo after reconstitution demonstrated increased cell death compared with WTMG controls, as measured by Annexin V staining (Fig. 4 I), indicating that overexpression of miR-132 results in increased apoptosis in B cells in these mice. We next cultured primary splenic B cells from miR-212/132−/− and WT mice with or without anti-IgM, and found in both conditions miR-212/132−/− B cells were more resistant to apoptosis (Fig. 4 J), consistent with the potential role of miR-212/132 in regulating cell death. Importantly, we found no difference in proliferation of splenic B cells from miR-212/132−/− or WT mice in unstimulated conditions or in the presence of TLR ligands (Fig. 4 K).
Sox4 is a direct target of microRNA-132 in B cells
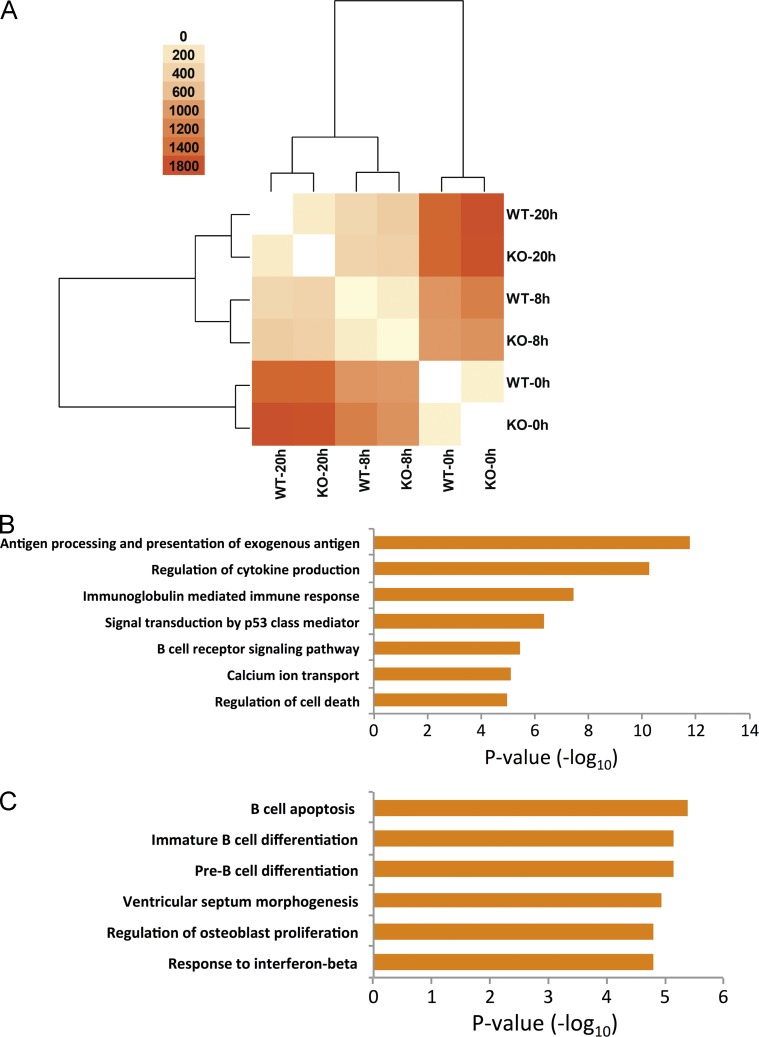
Seeking to better understand the underlying molecular mechanism for this defect in B cell development in WTmiR-132 mice, we began by performing RNA-seq analysis on immature B cells obtained from WT and miR-212/132−/− mice under steady-state conditions, and after 8 or 20 h of anti-IgM stimulation. We found several differentially expressed genes between samples (Fig. 5 A and Table 1). These were enriched for genes related to cell death, calcium ion transport, BCR signaling, and cytokine production when comparing anti-IgM–stimulated cells to unstimulated cells (Fig. 5 B). As expected, clustering analysis of differentially expressed genes revealed that WT and miR-212/132−/− cells from the same time points were more closely related to each other than to any other sample (Fig. 5 A). Among the genes that were differentially regulated between WT and miR-212/132−/− cells at the same time point, there was enrichment for genes related to B cell apoptosis and pre–B cell differentiation, as well as viral response genes (Fig. 5 C). These included genes such as Rag1, Sfrp1, Eif2ak2, Irf7, Mx1, and Rsad2. Importantly, we found enrichment in the expression of several miR-132 targets in miR-212/132−/− B cells compared with WT B cells, including the transcription factor Sox4, a known regulator of B cell development (Laurenti et al., 2013; Sun et al., 2013; Mallampati et al., 2014).
Figure 5.
Stimulation of miR-212/132−/− and WT B cells through the BCR results in dramatic gene expression changes. B cells were purified from miR-212/132−/− and WT mice and subsequently either left unstimulated or stimulated for 8 or 20 h with anti-IgM (5 µg/ml). RNA was harvested from the cells and processed for RNA-sequencing. (A) Heat map and clustering indicating the number of differentially expressed genes between each sample. (B) Functional annotations for differentially expressed genes between WT-stimulated B cells (8 or 20 h with anti-IgM) compared with WT unstimulated B cells. (C) Enriched functional annotations for genes differentially expressed between miR-212/132−/− and WT B cells at any of the time points.
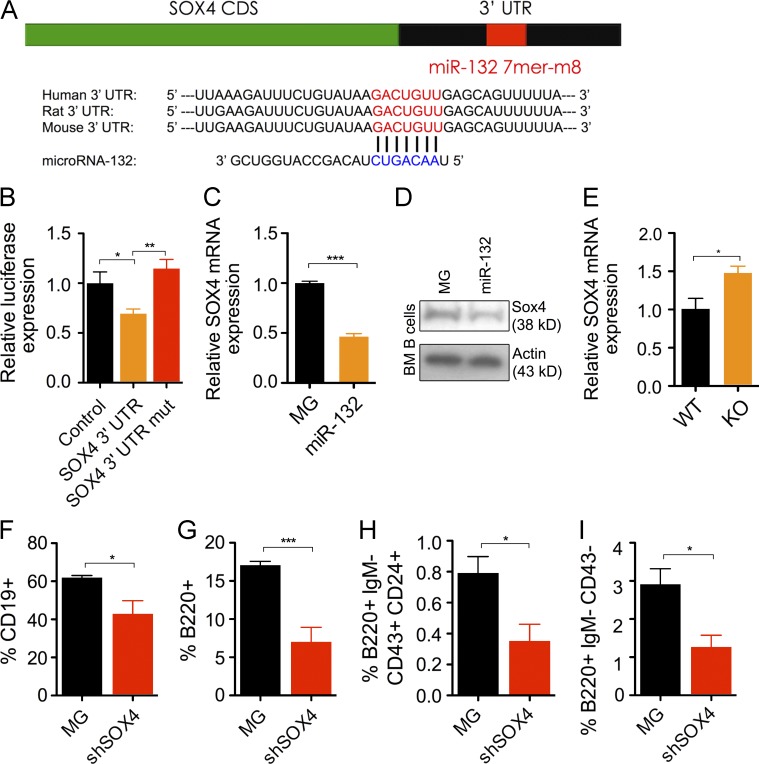
We have previously shown that Sox4 mRNA expression levels are decreased in the bone marrow of WTmiR-132 mice compared with WTMG controls (Mehta et al., 2015). Sox4 has a conserved, computationally predicted 7mer-m8 binding site for miR-132 (Fig. 6 A). We thus validated that it is a direct target of miR-132 and explored the possibility that miR-132 may regulate B cell development through Sox4. We transfected HEK293T cells with a construct that overexpressed miR-132 in the presence of a luciferase reporter immediately upstream of either the 3′UTR of Sox4, the 3′UTR of Sox4 with a mutated miR-132 binding site, or an irrelevant 3′UTR (control). Compared with the control vector, we found decreased expression of luciferase using the construct with an intact Sox4 3′UTR and restored luciferase expression with the mutated miR-132–binding site (Fig. 6 B). We found a significant decrease in Sox4 mRNA and protein expression levels in purified bone marrow B cells from WTmiR-132 mice when compared with WTMG controls (Fig. 6, C and D). Consistent with this, we found an elevation in Sox4 transcript expression in bone marrow B cells from miR-212/132−/− mice compared with WT mice after 42 d of anti-CD20 treatment (Fig. 6 E).
Figure 6.
Sox4 is a direct target of miR-132 and is a key regulator of B cell development. (A) Schematic of the predicted miR-132–binding site in the SOX4 3′ UTR. (B) Relative luciferase expression in HEK293T cells transfected with a miR-132 overexpression vector and either a vector containing luciferase only (control), luciferase and the Sox4 3′ UTR immediately downstream (Sox4 3′ UTR), or a vector containing luciferase and a Sox4 3′ UTR with a mutated miR-132–binding site (SOX4 3′ UTR mut; n = 5). (C) Sox4 transcript expression level, obtained by RT-qPCR, in B cells purified from the bone marrow of MG and mIR-132 mice (n = 3). (D) SOX4 protein expression levels, obtained by Western blot, in B cells purified from the bone marrow of MG and mIR-132 mice. (E) Sox4 transcript expression in B cells from WT and miR-212/132−/− (KO) mice 42 d after treatment with anti-CD20 (n = 5). (F–I) WT C57BL/6 mice were lethally irradiated and reconstituted with donor bone marrow cells transduced with either a control (MG) or SOX4 shRNA expressing (shSOX4) retroviral vector. Mice were analyzed at 4 mo after reconstitution (n = 4–5 per group). (F) Frequency of B cells (CD19+) in the spleen of MG and shSOX4 mice. (G) Frequency of B cells (B220+) in the bone marrow of MG and shSOX4 mice. (H) Frequency of pro–B cells in the bone marrow of MG and shSOX4 mice. (I) Frequency of pre–B cells in the bone marrow of MG and shSOX4 mice. Data represents two independent experiments and is represented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001, Student’s t test.
Although it is known that a complete loss of Sox4 has dramatic consequences on B cell development, we explored whether a partial decrease in Sox4 protein levels, as might be expected by up-regulation of a microRNA, may alter B cell fate. To do this, we reconstituted lethally irradiated C57BL/6 mice with HSPC-enriched bone marrow cells transduced with either a control vector (WTMG) or a vector expressing an shRNA knockdown for Sox4 (WTshSOX4) using a strategy previously described (Chaudhuri et al., 2012). WTshSOX4 mice, when compared with WTMG controls, demonstrated a significant depletion in the frequency of B cells in the spleen (Fig. 6 F) and bone marrow compartment (Fig. 6 G) at 4 mo after reconstitution. Although we observed no change in the frequency of prepro–B cells, there was a significant decrease in the frequency of pro–B cells (Fig. 6 H) and pre–B cells (Fig. 6 I) in the bone marrow of WTshSOX4 mice.
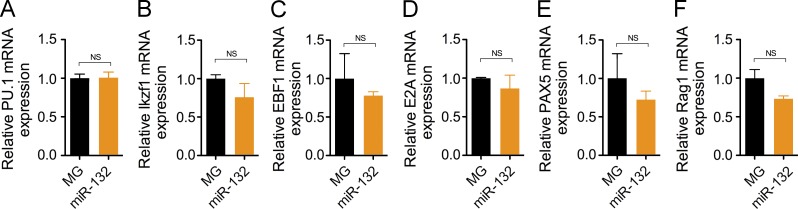
To determine if overexpression of miR-132 may alter B cell development genes upstream of SOX4, we looked at the expression of early regulators of B cell fate in bone marrow B cells from WTMG and WTmiR-132 mice. We found a decrease in Rag1 expression levels, though not statistically significant, in purified bone marrow B cells from WTmIR-132 when compared with WTMG cells, but observed no changes in the expression levels of other genes critical for B cell development, including PU.1, Ikxf1, Ebf11, E2A, and Pax5 (Fig. 7).
Figure 7.
Enforced expression of miR-132 does not alter expression of B cell development genes upstream of SOX4. Transcript expression levels of (A) PU.1, (B) Ikzf1, (C) EBF1, (D) E2A, (E) PAX5, and (F) Rag1 obtained by RT-qPCR from B cells purified from the bone marrow of MG and miR-132 mice (n = 3). Data represents two independent experiments and is represented as mean ± SEM.
Co-expression of Sox4 with microRNA-132 rescues defective B cell development with enforced expression of microRNA-132 alone
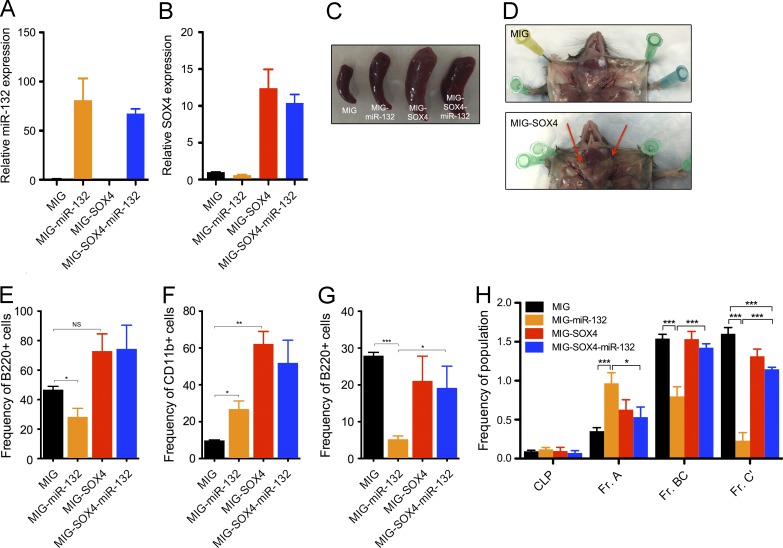
We next sought to investigate if Sox4 is the key target of miR-132 responsible for regulating B cell development. To do this, we attempted to rescue the defect in B cell development observed with enforced expression of miR-132 by coexpressing Sox4 that lacks the miR-132–binding site in the same cells. We reconstituted mice with HSPCs expressing an empty vector (WTMIG), or with a vector expressing either miR-132 only (WTmiR-132), Sox4 only (WTSOX4), or miR-132 and Sox4 together (WTmiR-132-SOX4) using a strategy previously described (Rao et al., 2010). We validated the expression of Sox4 and miR-132 in the respective samples by qPCR (Fig. 8, A and B). Surprisingly, we found that 38% of MIG-SOX4 mice and 31% of MIG-SOX4-miR-132 mice succumbed to an aggressive leukemia, characterized by markedly enlarged spleens and lymph nodes (Fig. 8, C and D). The cellular origin of these leukemias appeared variable, with 22% showing B cell infiltration in the lymph nodes (Fig. 8 E) and 78% showing a severe myeloid cell infiltration in the lymph nodes (Fig. 8 F). Importantly, we found that the cells in those mice with B cell infiltration did not express surface markers characteristic of early B cell progenitors.
Figure 8.
Co-expression of SOX4 rescues the defect in B cell development with expression of miR-132 alone. WT C57BL/6 mice were lethally irradiated and reconstituted with donor bone marrow cells expressing either a control (MIG), a miR-132–overexpressing (MIG-miR-132), a SOX4-overexpressing (MIG-SOX4), or a SOX4- and miR-132–overexpressing (MIG-SOX4-miR-132) retroviral vector (n = 5 mice per group). (A) Validation of miR-132 overexpression in donor HSPCs transduced with the labeled constructs. (B) Validation of SOX4 expression in donor HSPCs transduced with the labeled constructs. (C) Representative spleens from MIG, MIG-miR-132, MIG-SOX4, and MIG-SOX4-miR-132 mice at 4 mo after reconstitution. (D) Select LNs from MIG and MIG-SOX4 mice. Red arrows point to examples of enlarged LNs in MIG-SOX4 mice. (E) Frequency of B cells in LNs of MIG-SOX4 and MIG-SOX4-miR-132 mice with marked B cell expansion. (F) Frequency of myeloid cells in LNs of MIG-SOX4 and MIG-SOX4-miR-132 mice with marked myeloid cell expansion. (G) Frequency of bone marrow B cells in MIG, MIG-miR-132, MIG-SOX4, and MIG-SOX4-miR-132 mice at 4 mo after reconstitution. (H) Hardy fractions from the above cohorts of mice. Data represents the combination of two independent experiments and is represented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001, Student’s t test.
Analysis of the peripheral blood of mice lacking obvious disease at 3 mo after reconstitution revealed that co-expression of Sox4 partially rescues the loss of B cells observed with expression of miR-132 alone (Fig. 8 G), thus suggesting that miR-132 regulates B cell development in part by altering the expression of Sox4. An examination of early B cell progenitors by looking at Hardy fractions in the bone marrow of these mice was supportive of this finding. We found a partial normalization of the frequency of prepro–B cells (Fr. A), pro–B cells (Fr. BC), and pre–B cells (Fr. C’) in MIG-SOX4-mIR-132 mice compared with MIG-miR-132 mice (Fig. 8 H).
MicroRNA-132 protects Eμ-myc mice from B cell leukemia development
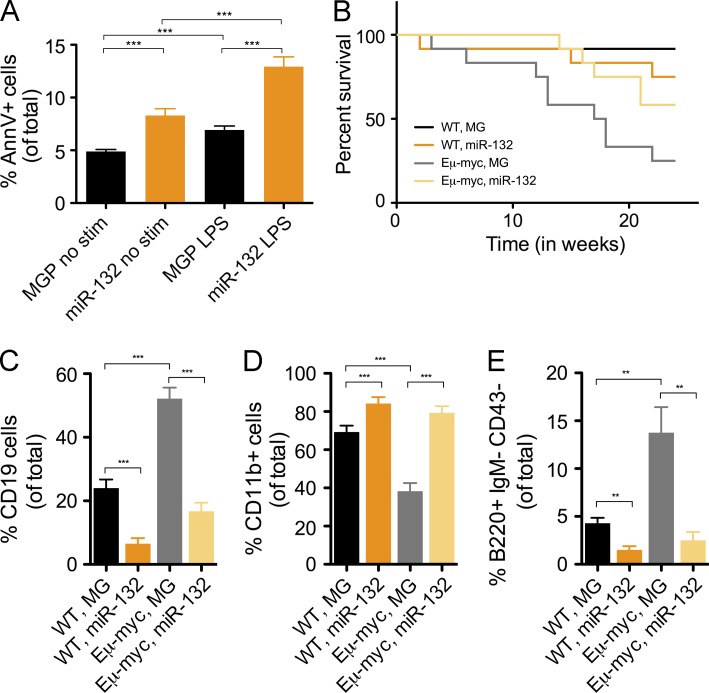
Several groups have shown that the microRNA-212/132 cluster is deregulated in certain leukemias of B cell origin (Pede et al., 2013; Tavolaro et al., 2015). Given our observation that miR-212/132 inhibits B cell development and induces B cell apoptosis, we asked if it could inhibit the growth of B cell leukemias/lymphomas. To this end, we overexpressed miR-132 in 70Z/3 cells, a pre–B cell lymphoblastic tumor cell line, and found increased Annexin V+ staining in cells expressing miR-132 compared with a control vector (Fig. 9 A). We next investigated whether delivering miR-132 to bone marrow cells from mice with a c-Myc transgene driven by an IgH enhancer (Eμ-myc mice; Langdon et al., 1986) could prevent the spontaneous occurrence of B cell cancers. To do this, we reconstituted lethally irradiated WT C57BL/6 mice with HSPC-enriched bone marrow cells from WT or Eμ-myc mice that were transduced with either a control vector or a miR-132–expressing vector. Over 6 mo, mice reconstituted with WT cells had no incidence of cancer. However, 75% of mice that received cells from Eμ-myc donors expressing the control vector developed spontaneous B cell lymphomas or leukemias and succumbed to disease (Fig. 9 B). Although 42% of the mice receiving Eμ-myc donor cells transduced with a miR-132 expression vector also succumbed, there was a significant improvement in survival compared with mice receiving Eμ-myc cells with the control vector (Fig. 9 B). Importantly, surviving mice that received Eμ-myc donor cells expressing miR-132 demonstrated a rescue in the frequency of CD19+ B cells, CD11b+ myeloid cells and pre–B cells in the bone marrow compartment compared with mice that received Eμ-myc donor cells with the control vector (Fig. 9, C–E). Thus, the expression of miR-132 had a protective effect on cancer occurrence in mice that were given Eμ-myc cells predisposed to forming B cell cancers.
Figure 9.
Enforced expression of miR-132 inhibits the development of spontaneous B cell cancers in cells with the Eμ-myc transgene. (A) Cells from a pre–B cell lymphoblastic leukemia line, 70Z/3, were transduced with a control vector (MGP) or a miR-132–overexpressing vector (miR-132) and cultured for 48 h with or without LPS (20 µg/ml). The frequency of cells expressing AnnV was then measured by FACS. (B–E) C57BL/6 mice were reconstituted with donor HSPCs from WT or Eμ-myc mice that were either transduced with a control vector (MG) or miR-132 overexpressing vector (miR-132). These mice were followed for 4 mo and harvested for analysis at this time point (n = 8–10 mice per group). (B) Survival curve for all experimental cohorts (P = 0.0037). Frequency of (C) bone marrow B cells (CD19+), (D) myeloid cells (CD11b+), and (E) pre–B cells (B220+IgM-CD43-) in all cohorts. Data represents two independent experiments and is represented as mean ± SEM. **, P < 0.01; ***, P < 0.001, Student’s t test.
DISCUSSION
A handful of microRNAs have been implicated in regulating hematopoietic cell fate decisions, including B cell lineage commitment (Xiao et al., 2007; O’Connell et al., 2010b; Rao et al., 2010). As posttranscriptional regulators of gene expression, these microRNAs primarily serve to fine-tune the expression of their targets, and in turn confer robustness to biological processes (Ebert and Sharp, 2012). When deregulated, however, microRNAs can tip the balance between normal and pathological function of a cell. In this study, we have uncovered a novel role for miR-212/132 in regulating the differentiation of prepro–B cells to pro–B cells by targeting the transcription factor Sox4. We found that deregulating miR-132 in vivo can affect survival of B cells and inhibit growth of certain B cell cancers.
We observed that miR-212/132 is highly induced in B cells in response to BCR activation, but not to activation of TLR receptors or CD40, which results in a strong NF-κB response. This highlights a potentially unique regulation of miR-212/132 in B cells, independent of what has been observed in macrophages, where a significant, but less dramatic induction of miR-212/132 has been observed with TLR4 stimulation (Taganov et al., 2006; Shaked et al., 2009). Importantly, this induction was replicated by stimulation with PMA, a DAG analogue, but not with ionomycin, which mimics Ca2+ influx into a cell, thus suggesting that miR-212/132 may be regulated by CREB or a host of other factors in B cells, as has been observed in neurons (Klein et al., 2007; Wen et al., 2010).
To investigate the developmental consequence of miR-212/132 up-regulation in B cells, we enforced expression of miR-132 in the bone marrow compartment of mice. We found that this enforced expression blocks the development of pro–B cells, which express the pre-BCR. Thus, it remains to be seen whether miR-212/132 may be induced by pre-BCR signaling and what role this may play in the developmental process. Our results are consistent with the critical role of microRNAs in the transition to pro–B and pre–B cells, as has been evidenced in mice with a genetic deletion of Dicer (Koralov et al., 2008). To date, this remains the earliest stage in B cell development at which a microRNA regulates B cell fate. Two other microRNAs, miR-150 and miR-34a, regulate the pro–B to pre–B cell transition by targeting c-Myb and Foxp1, respectively (Xiao et al., 2007; Zhou et al., 2007; Rao et al., 2010). Given the strong induction of miR-132 in response to BCR signaling, one may also hypothesize a potential role in B cell immune function. In fact, several microRNAs, such as miR-155, miR-181b, and miR-125b, have been shown to modulate immature and germinal center B cell function (de Yébenes et al., 2008; Teng et al., 2008; Porstner et al., 2015).
To investigate whether the role of miR-212/132 in B cell development is physiological, we analyzed miR-212/132−/− mice. We found that miR-212/132−/− B cells that were more resistant to apoptosis. In addition, miR-212/132−/− mice had an increased output of B cells compared with WT mice under nonhomeostatic conditions, such as when B cells were depleted with a mouse anti-CD20 antibody. This role of miR-212/132 in buffering B cell output under conditions of stress is consistent with the role of microRNAs in conferring robustness to many biological processes (Ebert and Sharp, 2012). We have recently shown that this microRNA cluster helps maintain normal hematopoietic stem cell output with age by buffering Foxo3 protein levels (Mehta et al., 2015). The current study demonstrates that miR-212/132 maintains B cell output through a different mechanism, serving more as a gatekeeper that modulates Sox4 protein levels when increased B cell output is required.
Aging the miR-212/132 mice did not reveal the occurrence of B cell cancers; however, it’s possible deregulation of miR-212/132 may aid in the progression of cancers once they have been established. In fact, it has been demonstrated that miR-212/132 is deregulated in cancers of various forms, including B cell leukemias, and that up-regulation of miR-212/132 in lung cancers inhibits proliferation and progression of the disease (Pede et al., 2013; Zhang et al., 2014; Jiang et al., 2015; Tavolaro et al., 2015). We found that enforced expression of miR-132 in donor cells from Eμ-myc mice, which are prone to develop pre–B and immature B cell lymphomas and leukemias, had a strong protective effect on cancer development. This model suggests that enforced expression of miR-132 decreases the number of cells susceptible to becoming cancerous, primarily by decreasing the number of pre–B cells in the bone marrow compartment. Consistent with this, previous studies have demonstrated that the incidence of B cell lymphomas can be decreased in Eμ-myc mice by enhancing the transition out of the pre–B cell stage to later stages of B cell development. This may be done by co-expression of membrane-bound IgM in the Eμ-myc background (Nussenzweig et al., 1988). Importantly, as was observed in primary B cells, the delivery of miR-132 to 70Z/3 cells, a B cell cancer line, induced apoptosis in these cells. We note, however, that this contrasts the role of miR-132 promoting pathological angiogenesis (Anand et al., 2010).
Our work also led to the discovery of Sox4 as a novel target for miR-132. We demonstrated that Sox4 is a key player in mediating the B cell development defect observed with miR-132 overexpression. A genetic deletion of Sox4 in mice has been shown to completely abolish the development of pro–B cells (Sun et al., 2013). Consistent with this, we found a dramatic loss of pro–B cells with miR-132 overexpression and a concomitant increase in B cell apoptosis. Importantly, we demonstrated that subthreshold levels of Sox4, either by miR-132 overexpression or by shRNA knockdown of Sox4, are sufficient to recreate the defect in pro–B cell development. We found that overexpression of miR-132 also affected downstream targets of Sox4 (Mallampati et al., 2014), particularly Rag1. Co-expression of Sox4 with miR-132 almost completely rescued the B cell developmental defect observed with expression of miR-132 alone. In particular, the frequency of prepro–B (Fr. BC) and pro–B (Fr. C’) cells was replenished in the bone marrow of mice overexpressing both Sox4 and miR-132 compared with mice overexpressing miR-132 alone. Although this may suggest that Sox4 is a critical target of miR-132 in regulating B cell development, the possibility exists that Sox4 replenishes the B cell pool through an indirect mechanism unrelated to the function of miR-132 in B cells. The role of Sox4 in B cell development remains poorly understood. By modulating the expression of miR-132, however, we have developed a new tool to study how graded expression of Sox4 can alter B cell fate decisions. Additionally, Sox4 is deregulated in several cancer cell lines, including being up-regulated in certain myelomas, and has been shown to promote the development of myeloid leukemias (Sandoval et al., 2012; Tiwari et al., 2013; Zhang et al., 2013). We found that enforced expression of Sox4, even in the presence of miR-132, led to myeloid and B cell lymphomas.
We have thus identified miR-212/132 as a novel player in regulating B cell development. Although much remains to be understood about the role of miR-212/132 in pro–B cell differentiation, we have identified a novel target for miR-212/132 in Sox4, and together these findings provide unique insight into the posttranscriptional mechanisms that govern B cell fate. Furthermore, our current work also suggests a potential role for miR-212/132 in combating cancers of B cell origin.
MATERIALS AND METHODS
DNA constructs.
For in vivo miR-132 expression and Sox4 shRNA experiments, we cloned either the mature miR-132 or Sox4 shRNA sequence in the microRNA-155 loop-and-arms format (O’Connell et al., 2010a) and into the MSCV-eGFP (MG) vector. For Sox4 rescue experiments, Sox4 lacking its 3′UTR was cloned into the MSCV-IRES-eGFP (MIG) vector using a previously described strategy (Rao et al., 2010). Sox4 shRNA target sequences are listed in Table S2.
For luciferase assays, the microRNA-132 expression cassette was subcloned into the pCDNA3 vector. The 3′UTR of Sox4 containing either the intact or mutated miR-132–binding site was cloned immediately downstream of luciferase in the pMiReport vector as previously described (Chaudhuri et al., 2012).
Cell culture.
Cells were cultured in a sterile incubator that was maintained at 37°C and 5% CO2. HEK293T cells were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin, and 100 U/ml streptomycin. Primary splenic B cells were cultured in complete RPMI supplemented with 10% FBS, 100 U/ml penicillin, 100 U/ml streptomycin, 50 µM β-mercaptoethanol, and the indicated stimulants. All stimulants, including anti-IgM (goat F(ab′)2 anti-mouse IgM; Southern Biotech), lipopolysaccharide (055:B5; Sigma-Aldrich), CpG (ODN 1826; Invivogen), mouse IL-4 (BioLegend), mouse CD40 ligand (eBioscience), phorbol-12-myristate-13-acetate (PMA; EMD Millipore), and ionomycin (EMD Millipore), were prepared as per manufacturer’s instructions and used at the concentration indicated in the text. For proliferation assays, the WST-1 cell proliferation reagent (Roche) was used as per manufacturer’s protocol.
Cell sorting for RNA extraction.
For miR-132 expression studies, we sorted cells either using MACS columns (Miltenyi Biotec) or by FACS. To isolate B cells, RBC-depleted bone marrow or spleen samples were sorted on MACS columns. In brief, bone marrow or spleen cells were incubated first with biotin-conjugated CD19 (positive selection) or CD43 (negative selection) antibodies (BioLegend) in MACS sorting buffer (Miltenyi Biotec), followed by incubation with streptavidin-conjugated magnetic beads (Miltenyi Biotec) as per manufacturer’s protocol, before being separated on the columns. For profiling studies on B cell progenitors, cells were sorted on a FACSAria IIu cell sorter (BD) at the Caltech Flow Cytometry Core Facility after surface staining with fluorescent antibodies for B220, CD19, CD43, CD24, IgM, CD11b, CD3e, Gr-1, Ter119, Nk1.1, Sca1, cKit, Flt3, and IL7Ra (BioLegend). RNA was then harvested from these cells using the miRNAeasy RNA prep kit (QIAGEN).
Expression profiling and qPCR.
RNA harvested from the respective cell populations was analyzed as previously described (Chaudhuri et al., 2012) by real-time qPCR (RT-qPCR) with a 7300 Real-Time PCR machine (Applied Biosystems). We used TaqMan MicroRNA Assays (Life Technologies) for miR-132, miR-212 and snoRNA-202 (control) to perform TaqMan qPCR as per manufacturer’s instructions. We performed SYBR Green–based RT-qPCR for mRNA expression of mouse Sox4, Ikzf1, Ebf1, E2A, Pax5, PU.1, and Rag1 after cDNA synthesis using qScript cDNA SuperMix (Quanta) and detection with PerfeCTa qPCR Fastmix with ROX (Quanta) as per manufacturer’s instructions. Table S3 lists primers used for qPCR.
Luciferase reporter assays.
We performed luciferase assays for the SOX4 3′UTR as has been previously described (Rao et al., 2010). 4 × 105 cells HEK293T cells, plated for 24 h, were transfected with either pCDNA or pCDNA-miR-132, a β-gal expression vector, and a pMiReport vector containing either no 3′UTR, the Sox4 3′UTR, or a Sox4 3′UTR with a mutant miR-132–binding site. 48 h later, cells were lysed using Reporter Lysis Buffer (Promega) and luciferase and β-gal expression was analyzed, respectively, using a Dual Luciferase kit (Promega) and a chemiluminescent β-gal reporter kit (Roche).
Immunoblotting.
Whole bone marrow samples and bone marrow B cells were sorted using the procedure described for RNA preparation using MACS columns (Miltenyi Biotec). Whole protein lysates were collected using RIPA lysis buffer (Sigma-Aldrich), and were subjected to gel-electrophoresis on a mini-PROTEAN TGX gradient (4–15%) gel (Bio-Rad Laboratories), and then transferred onto a PVDF membrane. Proteins were detected using the following antibodies: anti-Sox4 (154C4a; Abcam), actin-HRP (sc-1616; Santa Cruz Biotechnology, Inc.) and goat anti–mouse-IgG-HRP (sc-2005; Santa Cruz Biotechnology, Inc.).
Flow cytometry.
Respective tissue samples were collected from the appropriate mice as indicated in the text, RBC lysed (BioLegend), and processed as previously described (Chaudhuri et al., 2012). They were subsequently stained with a combination of fluorophore-conjugated antibodies (all from BioLegend), such as CD45, B220, CD19, CD43, CD24, IgM, CD11b, CD3e, Gr-1, Ter119, Nk1.1, Ly-6C, Sca1, cKit, Flt3, and IL7Ra. Surface markers were detected and analyzed using a MACSQuant10 Flow Cytometry machine (Miltenyi Biotec). Gating and analysis was performed using FlowJo software (Tree Star).
Animals.
All experiments were approved by the California Institute of Technology Institutional Animal Care and Use Committee. C57BL/6 WT and miR-212/132−/− mice were bred and housed in the Caltech Office of Laboratory Animal Resources (OLAR) facility. Bone marrow reconstitution experiments were performed as previously described (Chaudhuri et al., 2012) using donor cells from C57BL/6 WT mice or Eμ-myc mice, and with the appropriate retroviral vectors as described in the text. In vivo B cell depletion studies were performed by intraperitoneal delivery of 50 µg mouse anti-CD20 (5D2; Genentech) in 250 µl of PBS. Treated mice were subsequently bled at the indicated time points and cell populations in the peripheral blood were analyzed using FACS as described above.
RNA-seq data generation and analysis.
RNA-seq libraries were prepared from polyA+ selected RNA using the TruSeq RNA Sample Preparation kit at the Millard and Muriel Jacobs Genetics and Genomics Laboratory at Caltech. Libraries were sequenced on the Illumina HiSeq 2500 generating single-end 50 bp reads. The refSeq annotation for the mm9 version of the mouse genome was used to create a transcriptome Bowtie (version 0.12.7) (Langmead et al., 2009) index. Gene expression levels were estimated using eXpress (version 1.2.2; Roberts and Pachter, 2013), and DESeq (Anders and Huber, 2010) was used for evaluating differential expression.
Statistical tests.
All statistical analysis was done in GraphPad Prism software using an unpaired Student’s t test. Data were reported as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant.
Data access.
The RNA-seq data used in this study can be accessed from the Gene Expression Omnibus under the accession GSE66882.
Online supplemental material.
Table S1 shows the number of differentially expressed genes between WT and miR-212/132−/− splenic B cells with or without anti-IgM stimulation, as identified by RNA-seq analysis. Table S2 lists the sequences of the SOX4 shRNA knockdown constructs used in this study. Table S3 lists the primer sequences used for RT-qPCR analysis in this study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20150489/DC1.
Supplementary Material
Acknowledgments
We thank Diana Perez at the Caltech Flow Cytometry core facility, and Igor Antoshechkin and Vijaya Kumar at the Caltech Millard and Muriel Genetics and Genomics Laboratory. We also thank Michael T. Bethune for helpful discussions throughout the preparation of this manuscript.
This work was supported by funding from National Institutes of Health RO1AI079243 (D. Baltimore), National Research Service Award CA183220 (A. Mehta) and HL110691 (J.L. Zhao), the UCLA/Caltech Medical Scientist Training Program (A. Mehta and J.L. Zhao), and the Human Frontiers Science Foundation (M. Mann).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- CLP
- common lymphoid progenitor
- HSPC
- hematopoietic stem and progenitor cell
- MG
- MSCV-eGFP retroviral expression vector
- MIG
- MSCV-IRES-eGFP retroviral expression vector
- miR
- microRNA
- shRNA
- short-hairpin RNA
- shSOX4
- shRNA targeted against SOX4
References
- Anand S., Majeti B.K., Acevedo L.M., Murphy E.A., Mukthavaram R., Scheppke L., Huang M., Shields D.J., Lindquist J.N., Lapinski P.E., et al. 2010. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat. Med. 16:909–914. 10.1038/nm.2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., and Huber W.. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G.A., Cimmino A., Fabbri M., Ferracin M., Wojcik S.E., Shimizu M., Taccioli C., Zanesi N., Garzon R., Aqeilan R.I., et al. 2008. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc. Natl. Acad. Sci. USA. 105:5166–5171. 10.1073/pnas.0800121105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri A.A., So A.Y., Mehta A., Minisandram A., Sinha N., Jonsson V.D., Rao D.S., O’Connell R.M., and Baltimore D.. 2012. Oncomir miR-125b regulates hematopoiesis by targeting the gene Lin28A. Proc. Natl. Acad. Sci. USA. 109:4233–4238. 10.1073/pnas.1200677109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobaleda C., Schebesta A., Delogu A., and Busslinger M.. 2007. Pax5: the guardian of B cell identity and function. Nat. Immunol. 8:463–470. 10.1038/ni1454 [DOI] [PubMed] [Google Scholar]
- Costinean S., Zanesi N., Pekarsky Y., Tili E., Volinia S., Heerema N., and Croce C.M.. 2006. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl. Acad. Sci. USA. 103:7024–7029. 10.1073/pnas.0602266103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Yébenes V.G., Belver L., Pisano D.G., González S., Villasante A., Croce C., He L., and Ramiro A.R.. 2008. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J. Exp. Med. 205:2199–2206. 10.1084/jem.20080579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKoter R.P., Lee H.J., and Singh H.. 2002. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity. 16:297–309. 10.1016/S1074-7613(02)00269-8 [DOI] [PubMed] [Google Scholar]
- Ebert M.S., and Sharp P.A.. 2012. Roles for microRNAs in conferring robustness to biological processes. Cell. 149:515–524. 10.1016/j.cell.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eis P.S., Tam W., Sun L., Chadburn A., Li Z., Gomez M.F., Lund E., and Dahlberg J.E.. 2005. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA. 102:3627–3632. 10.1073/pnas.0500613102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esplin B.L., Shimazu T., Welner R.S., Garrett K.P., Nie L., Zhang Q., Humphrey M.B., Yang Q., Borghesi L.A., and Kincade P.W.. 2011. Chronic exposure to a TLR ligand injures hematopoietic stem cells. J. Immunol. 186:5367–5375. 10.4049/jimmunol.1003438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi H., Gregory S.C., Yokota T., Sakaguchi N., and Kincade P.W.. 2002. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 17:117–130. 10.1016/S1074-7613(02)00366-7 [DOI] [PubMed] [Google Scholar]
- Jiang X., Chen X., Chen L., Ma Y., Zhou L., Qi Q., Liu Y., Zhang S., Luo J., and Zhou X.. 2015. Upregulation of the miR-212/132 cluster suppresses proliferation of human lung cancer cells. Oncol. Rep. 33:705–712. 10.3892/or.2014.3637 [DOI] [PubMed] [Google Scholar]
- Klein M.E., Lioy D.T., Ma L., Impey S., Mandel G., and Goodman R.H.. 2007. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat. Neurosci. 10:1513–1514. 10.1038/nn2010 [DOI] [PubMed] [Google Scholar]
- Koralov S.B., Muljo S.A., Galler G.R., Krek A., Chakraborty T., Kanellopoulou C., Jensen K., Cobb B.S., Merkenschlager M., Rajewsky N., and Rajewsky K.. 2008. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 132:860–874. 10.1016/j.cell.2008.02.020 [DOI] [PubMed] [Google Scholar]
- Lagos D., Pollara G., Henderson S., Gratrix F., Fabani M., Milne R.S., Gotch F., and Boshoff C.. 2010. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat. Cell Biol. 12:513–519. 10.1038/ncb2054 [DOI] [PubMed] [Google Scholar]
- Langdon W.Y., Harris A.W., Cory S., and Adams J.M.. 1986. The c-myc oncogene perturbs B lymphocyte development in E-mu-myc transgenic mice. Cell. 47:11–18. 10.1016/0092-8674(86)90361-2 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S.L.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E., Doulatov S., Zandi S., Plumb I., Chen J., April C., Fan J.B., and Dick J.E.. 2013. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat. Immunol. 14:756–763. 10.1038/ni.2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie C.H., Saunders N.J., Soneji S., Palazzo S., Dunlop H.M., Cooper C.D., Brown P.J., Troussard X., Mossafa H., Enver T., et al. 2008. MicroRNA expression in lymphocyte development and malignancy. Leukemia. 22:1440–1446. 10.1038/sj.leu.2405083 [DOI] [PubMed] [Google Scholar]
- Li D., Wang A., Liu X., Meisgen F., Grünler J., Botusan I.R., Narayanan S., Erikci E., Li X., Blomqvist L., et al. 2015. MicroRNA-132 enhances transition from inflammation to proliferation during wound healing. J. Clin. Invest. 125:3008–3026. 10.1172/JCI79052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallampati S., Sun B., Lu Y., Ma H., Gong Y., Wang D., Lee J.S., Lin K., and Sun X.. 2014. Integrated genetic approaches identify the molecular mechanisms of Sox4 in early B-cell development: intricate roles for RAG1/2 and CK1ε. Blood. 123:4064–4076. 10.1182/blood-2013-12-543801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel E.M., and Grosschedl R.. 2010. Transcription control of early B cell differentiation. Curr. Opin. Immunol. 22:161–167. 10.1016/j.coi.2010.01.010 [DOI] [PubMed] [Google Scholar]
- Matthias P., and Rolink A.G.. 2005. Transcriptional networks in developing and mature B cells. Nat. Rev. Immunol. 5:497–508. 10.1038/nri1633 [DOI] [PubMed] [Google Scholar]
- Mauri C., and Bosma A.. 2012. Immune regulatory function of B cells. Annu. Rev. Immunol. 30:221–241. 10.1146/annurev-immunol-020711-074934 [DOI] [PubMed] [Google Scholar]
- Mehta A., Zhao J.L., Sinha N., Marinov G.K., Mann M., Kowalczyk M.S., Galimidi R.P., Du X., Erikci E., Regev A., et al. 2015. The MicroRNA-132 and MicroRNA-212 Cluster Regulates Hematopoietic Stem Cell Maintenance and Survival with Age by Buffering FOXO3 Expression. Immunity. 42:1021–1032. 10.1016/j.immuni.2015.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahid M.A., Yao B., Dominguez-Gutierrez P.R., Kesavalu L., Satoh M., and Chan E.K.. 2013. Regulation of TLR2-mediated tolerance and cross-tolerance through IRAK4 modulation by miR-132 and miR-212. J. Immunol. 190:1250–1263. 10.4049/jimmunol.1103060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahama T., Hanieh H., Nguyen N.T., Chinen I., Ripley B., Millrine D., Lee S., Nyati K.K., Dubey P.K., Chowdhury K., et al. 2013. Aryl hydrocarbon receptor-mediated induction of the microRNA-132/212 cluster promotes interleukin-17-producing T-helper cell differentiation. Proc. Natl. Acad. Sci. USA. 110:11964–11969. 10.1073/pnas.1311087110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig M.C., Schmidt E.V., Shaw A.C., Sinn E., Campos-Torres J., Mathey-Prevot B., Pattengale P.K., and Leder P.. 1988. A human immunoglobulin gene reduces the incidence of lymphomas in c-Myc-bearing transgenic mice. Nature. 336:446–450. 10.1038/336446a0 [DOI] [PubMed] [Google Scholar]
- Nutt S.L., and Kee B.L.. 2007. The transcriptional regulation of B cell lineage commitment. Immunity. 26:715–725. 10.1016/j.immuni.2007.05.010 [DOI] [PubMed] [Google Scholar]
- O’Connell R.M., Chaudhuri A.A., Rao D.S., Gibson W.S., Balazs A.B., and Baltimore D.. 2010a. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc. Natl. Acad. Sci. USA. 107:14235–14240. 10.1073/pnas.1009798107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell R.M., Rao D.S., Chaudhuri A.A., and Baltimore D.. 2010b. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 10:111–122. 10.1038/nri2708 [DOI] [PubMed] [Google Scholar]
- O’Riordan M., and Grosschedl R.. 1999. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 11:21–31. 10.1016/S1074-7613(00)80078-3 [DOI] [PubMed] [Google Scholar]
- Pede V., Rombout A., Vermeire J., Naessens E., Mestdagh P., Robberecht N., Vanderstraeten H., Van Roy N., Vandesompele J., Speleman F., et al. 2013. CLL cells respond to B-Cell receptor stimulation with a microRNA/mRNA signature associated with MYC activation and cell cycle progression. PLoS ONE. 8:e60275 10.1371/journal.pone.0060275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstner M., Winkelmann R., Daum P., Schmid J., Pracht K., Corte-Real J., Schreiber S., Haftmann C., Brandl A., Mashreghi M.F., et al. 2015. miR-148a promotes plasma cell differentiation and targets the germinal center transcription factors Mitf and Bach2. Eur. J. Immunol. 45:1206–1215 [DOI] [PubMed] [Google Scholar]
- Puissegur M.P., Eichner R., Quelen C., Coyaud E., Mari B., Lebrigand K., Broccardo C., Nguyen-Khac F., Bousquet M., and Brousset P.. 2012. B-cell regulator of immunoglobulin heavy-chain transcription (Bright)/ARID3a is a direct target of the oncomir microRNA-125b in progenitor B-cells. Leukemia. 26:2224–2232. 10.1038/leu.2012.95 [DOI] [PubMed] [Google Scholar]
- Rao D.S., O’Connell R.M., Chaudhuri A.A., Garcia-Flores Y., Geiger T.L., and Baltimore D.. 2010. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 33:48–59. 10.1016/j.immuni.2010.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., and Pachter L.. 2013. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods. 10:71–73. 10.1038/nmeth.2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumfelt L.L., Zhou Y., Rowley B.M., Shinton S.A., and Hardy R.R.. 2006. Lineage specification and plasticity in CD19- early B cell precursors. J. Exp. Med. 203:675–687. 10.1084/jem.20052444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval S., Kraus C., Cho E.C., Cho M., Bies J., Manara E., Accordi B., Landaw E.M., Wolff L., Pigazzi M., and Sakamoto K.M.. 2012. Sox4 cooperates with CREB in myeloid transformation. Blood. 120:155–165. 10.1182/blood-2011-05-357418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarikonda G., Sachithanantham S., Manenkova Y., Kupfer T., Posgai A., Wasserfall C., Bernstein P., Straub L., Pagni P.P., Schneider D., et al. 2013. Transient B-cell depletion with anti-CD20 in combination with proinsulin DNA vaccine or oral insulin: immunologic effects and efficacy in NOD mice. PLoS ONE. 8:e54712 10.1371/journal.pone.0054712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked I., Meerson A., Wolf Y., Avni R., Greenberg D., Gilboa-Geffen A., and Soreq H.. 2009. MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity. 31:965–973. 10.1016/j.immuni.2009.09.019 [DOI] [PubMed] [Google Scholar]
- Souabni A., Cobaleda C., Schebesta M., and Busslinger M.. 2002. Pax5 promotes B lymphopoiesis and blocks T cell development by repressing Notch1. Immunity. 17:781–793. 10.1016/S1074-7613(02)00472-7 [DOI] [PubMed] [Google Scholar]
- Sun B., Mallampati S., Gong Y., Wang D., Lefebvre V., and Sun X.. 2013. Sox4 is required for the survival of pro–B cells. J. Immunol. 190:2080-2089. 10.4049/jimmunol.1202736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov K.D., Boldin M.P., Chang K.J., and Baltimore D.. 2006. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 103:12481–12486. 10.1073/pnas.0605298103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavolaro S., Colombo T., Chiaretti S., Peragine N., Fulci V., Ricciardi M.R., Messina M., Bonina S., Brugnoletti F., Marinelli M., et al. 2015. Increased chronic lymphocytic leukemia proliferation upon IgM stimulation is sustained by the upregulation of miR-132 and miR-212. Genes Chromosomes Cancer. 54:222–234. 10.1002/gcc.22236 [DOI] [PubMed] [Google Scholar]
- Teng G., Hakimpour P., Landgraf P., Rice A., Tuschl T., Casellas R., and Papavasiliou F.N.. 2008. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 28:621–629. 10.1016/j.immuni.2008.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai T.H., Calado D.P., Casola S., Ansel K.M., Xiao C., Xue Y., Murphy A., Frendewey D., Valenzuela D., Kutok J.L., et al. 2007. Regulation of the germinal center response by microRNA-155. Science. 316:604–608. 10.1126/science.1141229 [DOI] [PubMed] [Google Scholar]
- Tiwari N., Tiwari V.K., Waldmeier L., Balwierz P.J., Arnold P., Pachkov M., Meyer-Schaller N., Schübeler D., van Nimwegen E., and Christofori G.. 2013. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 23:768–783. 10.1016/j.ccr.2013.04.020 [DOI] [PubMed] [Google Scholar]
- Wen A.Y., Sakamoto K.M., and Miller L.S.. 2010. The role of the transcription factor CREB in immune function. J. Immunol. 185:6413–6419. 10.4049/jimmunol.1001829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Calado D.P., Galler G., Thai T.H., Patterson H.C., Wang J., Rajewsky N., Bender T.P., and Rajewsky K.. 2007. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 131:146–159. 10.1016/j.cell.2007.07.021 [DOI] [PubMed] [Google Scholar]
- Xiao C., Srinivasan L., Calado D.P., Patterson H.C., Zhang B., Wang J., Henderson J.M., Kutok J.L., and Rajewsky K.. 2008. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 9:405–414. 10.1038/ni1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Ng S.Y., Zuniga-Pflucker J.C., and Georgopoulos K.. 2006. Early hematopoietic lineage restrictions directed by Ikaros. Nat. Immunol. 7:382–391. 10.1038/ni1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Alberich-Jorda M., Amabile G., Yang H., Staber P.B., Di Ruscio A., Welner R.S., Ebralidze A., Zhang J., Levantini E., et al. 2013. Sox4 is a key oncogenic target in C/EBPα mutant acute myeloid leukemia. Cancer Cell. 24:575–588. 10.1016/j.ccr.2013.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.G., Chen W.X., Wu Y.H., Liang H.F., and Zhang B.X.. 2014. MiR-132 prohibits proliferation, invasion, migration, and metastasis in breast cancer by targeting HN1. Biochem. Biophys. Res. Commun. 454:109–114. 10.1016/j.bbrc.2014.10.049 [DOI] [PubMed] [Google Scholar]
- Zhou B., Wang S., Mayr C., Bartel D.P., and Lodish H.F.. 2007. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc. Natl. Acad. Sci. USA. 104:7080–7085. 10.1073/pnas.0702409104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.