Local germinal center reactions that persist in the lung after influenza infection are required for the generation of cross-reactive memory B cells.
Abstract
Respiratory influenza virus infection induces cross-reactive memory B cells targeting invariant regions of viral escape mutants. However, cellular events dictating the cross-reactive memory B cell responses remain to be fully defined. Here, we demonstrated that lung-resident memory compartments at the site of infection, rather than those in secondary lymphoid organs, harbor elevated frequencies of cross-reactive B cells that mediate neutralizing antibody responses to viral escape. The elevated cross-reactivity in the lung memory compartments was correlated with high numbers of VH mutations and was dependent on a developmental pathway involving persistent germinal center (GC) responses. The persistent GC responses were focused in the infected lungs in association with prolonged persistence of the viral antigens. Moreover, the persistent lung GCs supported the exaggerated B cell proliferation and clonal selection for cross-reactive repertoires, which served as the predominant sites for the generation of cross-reactive memory progenitors. Thus, we identified the distinct GC selection at local sites as a key cellular event for cross-reactive memory B cell response to viral escape, a finding with important implications for developing broadly protective influenza vaccines.
Protective memory responses provided by parental influenza vaccines primarily depend on neutralizing IgG antibodies (Abs) directed against hemagglutinin (HA), a major glycoprotein on the virus surface (Gerhard, 2001; Plotkin, 2013). The membrane distal region of the HA globular head is highly immunogenic and is the primary target of anti-HA Abs elicited by vaccination (Skehel and Wiley, 2000). However, the HA globular head undergoes continual antigenic evolution (Wiley et al., 1981), making vaccine-induced Abs less effective against drifted viruses. Moreover, new subtypes can emerge rapidly and unexpectedly, as experienced in the 2009 A/H1N1 pandemic virus and sporadic human infection with avian viruses such as H5N1 and H7N9. Thus, the evolving threats of influenza virus underscore the need for influenza vaccines that are more broadly protective.
HA conserved regions can be targeted by broadly cross-reactive Abs that exhibit potent virus-neutralizing activity in vitro and in vivo (Okuno et al., 1993; Throsby et al., 2008; Sui et al., 2009; Yoshida et al., 2009; Corti et al., 2010; Krause et al., 2011; Wrammert et al., 2011). Such cross-reactive Abs were observed in IgG and IgA fractions after respiratory exposure of viruses (Tamura et al., 1992; Tumpey et al., 2001; Margine et al., 2013). Of note, cross-reactive IgG Abs were higher in humans infected with influenza virus than in humans parentally boosted with vaccines (Moody et al., 2011; Wrammert et al., 2011; Li et al., 2012; Pica et al., 2012; Margine et al., 2013), suggesting that the cellular pathways for cross-reactive Ab responses are more active after respiratory virus infection.
Pulmonary-infected influenza virus initially primes virus-binding B cells in the lung-draining mediastinal LNs (MLNs; Coro et al., 2006). The infected lungs, albeit at delayed kinetics, also participate in the primary immune response, concordant with the ectopic formation of induced bronchus-associated lymphoid tissue (iBALT; Moyron-Quiroz et al., 2004; Halle et al., 2009). iBALTs are able to support germinal center (GC) formation (Moyron-Quiroz et al., 2004), suggesting intraorgan development of long-lived plasma cells and memory B cells, which are crucial cellular components for humoral memory responses (Joo et al., 2008; Onodera et al., 2012; Tarlinton and Good-Jacobson, 2013). Although immediate protection against homologous reinfection is mediated by preexisting neutralizing Abs from long-lived plasma cells, memory B cells serve as a reservoir of cross-reactive Ab repertoires in West Nile virus infection (Purtha et al., 2011). Therefore, it is now postulated that memory B cells are important for the broad protection against escape mutants, against which strain-specific Abs are no longer effective (Baumgarth, 2013). However, the memory B cell subset reserving cross-reactive repertoires and its developmental pathway has not been fully characterized.
Here, using two types of fluorochrome-labeled HA probes, we identified the cross-reactive memory B cell subset and dissected its developmental pathway after pulmonary influenza virus infection. Our data revealed a striking heterogeneity in the tissue localization, persistence, and selection for cross-reactivity among virus-specific GC responses. Among such heterogeneous GC responses, persistent GCs in the infected lungs profoundly selected and supplied cross-reactive memory repertoires into local sites, thereby potentiating the cross-protection at the site of infection.
RESULTS
Lung-resident memory B cells are enriched with highly mutated, cross-reactive Ab repertoires
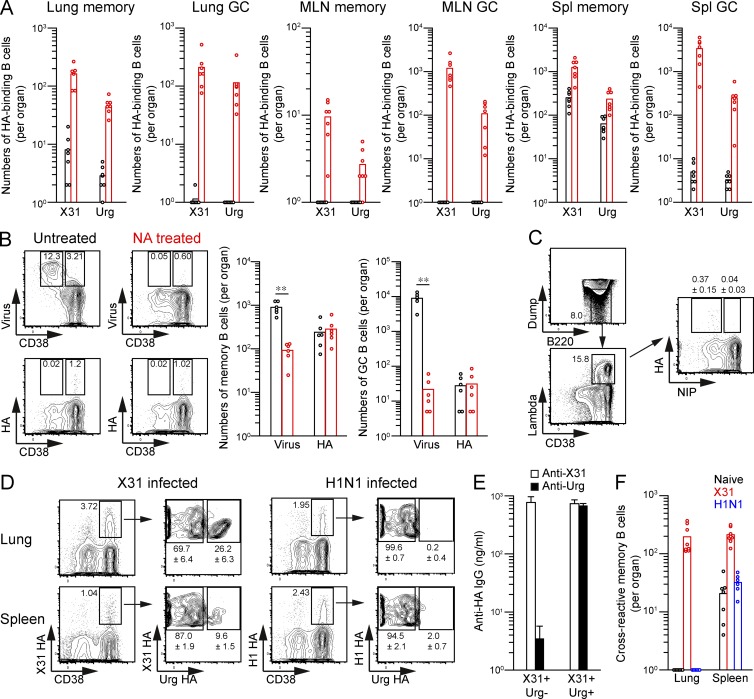
To identify HA-binding, cross-reactive B cell populations, we prepared recombinant HAs (rHAs) from two H3N2 virus strains, X31 and A/Uruguay/716/07, which share only 86.9% HA amino acid sequence similarity. The rHAs of these H3N2 strains were labeled with different fluorochromes for flow cytometric staining. Previous flow cytometric analysis has clearly identified HA-binding B cell populations in virus-primed mice; however, small numbers of HA-binding B cells were also detectable in unprimed mice (Doucett et al., 2005; Onodera et al., 2012). To assess the specificity of our HA probes, we first compared the staining profiles of naive and X31-infected mice (Fig. S1). Uruguay716-infected mice were excluded from the analysis, owing to insufficient pathogenicity and the lack of detectable immune responses in mice. After gating on IgM/D− isotype-switched B cells, the staining by both HA probes resulted in the clear visualization of HA-binding B cells in the X31-infected mice; however, small numbers of HA-binding CD38+ B cells were present in naive mice at 1/5 (spleen) or <1/10 (lung) the levels in the infected mice (Fig. 1 A). To determine the relative contribution of sialic acid and BCR to HA binding in naive splenocytes, we performed the following two experiments. First, splenocytes were pretreated with neuraminidase to inhibit sialic acid–mediated binding. Indeed, the neuraminidase treatment inhibited the binding of fluorochrome-labeled viruses to B cells, especially of the CD38dull GC phenotype, but failed to reduce the numbers of HA-binding CD38+ B cells in naive mice (Fig. 1 B), suggesting that the contribution of sialic acid–mediated binding is minor. Second, HA staining was performed using B cells from IgVH186.2-DFL16.1-JH2 gene-targeted mice (B1-8high) in which the B cells specifically bind to hapten NP (4-hydroxy-3-nitrophenylacetyl)/NIP (4-hydroxy-5-iodo-3-nitrophenylacetyl) in combination with an Igλ light chain (Shih et al., 2002). Therefore, we expected that the NIP-binding B cells from B1-8high mice would fail to bind HA if the HA binding is mediated by a specific BCR. We observed that both NIP-binding and -nonbinding B cell populations were present in the Igλ+CD38+IgD− B cell fraction in naive B1-8high splenocytes (Fig. 1 C); however, HA-binding B cells were more abundant in the NIP-nonbinding population, supporting that HA binds to B cells via a specific BCR rather than via sialic acid. Thus, although small numbers of HA-binding B cells are detected in naive mice, our HA probes identify HA-specific B cells, with low contamination of nonspecific B cells binding through sialic acid.
Figure 1.
Simultaneous staining by two HA probes segregates cross-reactive and strain-specific memory B cells. (A) The numbers of X31/Uruguay HA-binding CD38+ (memory) and CD38dull (GC) B cells among the PI-Dump−(IgM−IgD−Gr-1−CD3−CD5−CD11b−CD11c−CD43−CD90−CD93−TER-119−F4/80−CD117−CD138−PI−)B220+ fraction were counted from naive (black) and X31-infected (red) mice and plotted per organ. (B) Virus- and HA-binding memory phenotype B cells were enumerated in naive spleens with or without neuraminidase (NA) treatment. **, P < 0.01. (C) After gating on the dump−(IgD−Gr-1−CD3−CD5−CD11b−CD11c−CD43−CD90−CD93−TER-119−F4/80−CD117−CD138−PI−)B220+CD38+lambda+ fraction, HA-binding cells were compared between NIP-binding and -nonbinding populations in naive spleens from B1-8high mice. Over 20,000,000 events were analyzed from individual spleens, and the cells within enlarged B220 versus dump gate were collected for further analysis. The inset percentages represent the mean ± SD (n = 6). (D) X31 HA-binding lung and splenic memory B cells (PI-Dump−CD38+B220+) from X31- and H1N1-infected mice were separated by the cross-reactivity to Uruguay HA at day 40 after infection. The inset percentages represent the mean ± SD (n ≥ 6). (E) X31 HA-binding lung memory B cells with or without cross-reactivity to Uruguay HA were sorted and stimulated with LPS and IL-2/IL-4/IL-5 for 6 d on a layer of 3T3 fibroblasts. Anti-HA IgG Ab levels in culture supernatants from sorted memory B cells were determined by ELISA. The data represent the mean ± SD of three replicates and are representative of three independent experiments. (F) The numbers of cross-reactive memory B cells from naive, X31-, and H1N1-infected mice were plotted per organ. (A, B, and F) Each circle represents the result from an individual mouse. (A–C and F) The data are representative of two independent experiments.
Two distinct B cell populations were observed in the X31 HA-binding memory B cell compartment elicited in pulmonary X31 infection: one population bound to both X31 and Uruguay716 HAs, and the other bound solely to X31 HA (Fig. 1 D). The paucity of double-positive cells after infection with irrelevant H1N1 virus (A/Narita/1/09) confirms that this population emerges by antigen-induced events and is not simply the result of bystander effects of infection (Fig. 1 D). When single- and double-positive populations were separately stimulated in vitro, the double-positive population secreted cross-reactive IgG Abs that bound to both X31 and Uruguay HAs, and the single-positive population secreted strain-specific IgG Abs that bound only to X31 HA (Fig. 1 E), thus validating the flow cytometric results. Furthermore, we found that the cross-reactivity among lung memory B cells was significantly more frequent than that of splenic memory B cells from the same mice (Fig. 1 D). However, analysis of memory cross-reactivity in the MLNs was hampered by low cellularity of HA-binding memory B cells (Fig. 1 A). Again, although cross-reactive memory B cells were detected in naive or H1N1-infected spleens at 1/10 the level in X31-infected spleens, the number of such cells was below the detection limit in naive or H1N1-infected lungs (Fig. 1 F), supporting that the majority of cross-reactive memory B cells are induced by infection in an antigen-dependent manner. Together, these data reveal increased cross-reactivity of isotype-switched memory B cells at local sites, close to the viral entry point.
Intravascular staining has shown that lung T cells predominantly localize in the capillary vessels rather than in tissue (Anderson et al., 2012). To assess the tissue localization of lung memory B cells, we performed similar intravascular staining by anti-CD38 mAbs that bind to B cells other than GC B cells. Intravascular CD38 Ab clearly stained the major fraction of IgD+ B cells in the infected lung tissues (Fig. 2 A), supporting that lung naive B cells predominantly localize in capillary vessels, similar to T cells. In contrast, HA-binding memory B cells in the lungs were largely protected from intravascular CD38 staining (Fig. 2 A). Therefore, lung memory B cells mainly reside in tissue rather than in blood vessels, likely contributing to focused cross-reactivity at local sites.
Figure 2.
Highly mutated, cross-reactive memory B cells accumulate in the lungs. (A) After intravascular injection of CD38 mAbs, lung naive B cells (IgD+CD38+B220+) and HA-binding memory B cells (PI-Dump−CD38+B220+) were evaluated for intravascular staining. The flow cytometric gating used for analysis is shown, and the percentages of i.v. CD38+ cells are plotted. Each circle represents the result from an individual mouse. The data are representative of three independent experiments. ***, P < 0.001. (B–D) Cross-reactive and strain-specific memory B cells were sorted at days 40 and 160 after infection and subjected to VH repertoire and mutation analysis. Data are derived from VH sequences (day 40 strain, 18; day 40 cross, 14; day 160 strain, 23; day 160 cross, 30) that were obtained from three independent experiments. (B) VH usage was compared among strain-specific versus cross-reactive memory B cells. *, P < 0.05; **, P < 0.01. (C) Each circle represents the total numbers of VH mutations for an individual cell. **, P < 0.01; ***, P < 0.001. (A and C) Horizontal lines indicate mean. (D) Numbers of replacement and silent mutations in CDR1–2 and FWR1–3 were plotted. The data represent the mean ± SD.
IgVH mutational analysis has shown that human cross-reactive mAbs possess more mutations than strain-specific mAbs (Wrammert et al., 2011; Pappas et al., 2014). This led us to compare VH repertoires and the number of VH mutations in strain-specific memory B cells and cross-reactive memory B cells, which had been sorted using the gating scheme shown in Fig. 1 D. VH genes from memory B cells were subjected to single cell RT-PCR amplification using mouse pan-VH primers (Tiller et al., 2009). Intriguingly, we observed differential VH gene usage between cross-reactive and strain-specific memory B cells (Fig. 2 B) at days 40 and 160 after infection; VH1 family genes were more prevalent in cross-reactive memory B cells, along with lower representation of VH5 family genes. Moreover, cross-reactive memory B cells carried more VH gene mutations than strain-specific B cells (Fig. 2 C), although we could not find any significant bias for the location (complementarity-determining region [CDR] vs. framework region [FWR]) or the types (replacement vs. silent) of accumulated mutations at day 160 (Fig. 2 D).
Lung memory B cells mediate cross-reactive secondary IgG response
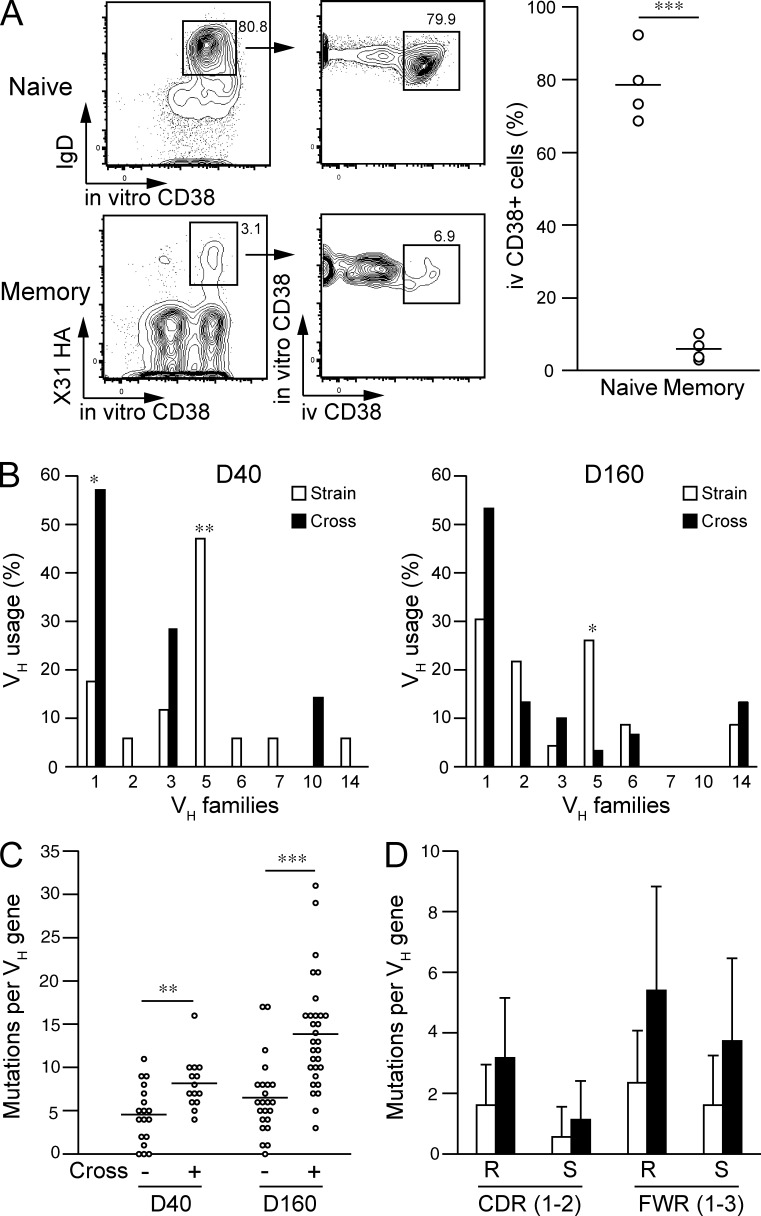
To address whether the increased cross-reactivity expanded the breadth of the secondary IgG response, we transferred X31 HA-binding lung and splenic memory B cells into scid mice together with primed CD4+ T cells. Although boosting with homologous X31 virus reactivated both strain-specific and cross-reactive memory B cells, boosting with drifted virus preferentially reactivated cross-reactive memory B cells (Fig. 3 A). Consistent with our previous observation (Onodera et al., 2012), lung and splenic memory B cells showed intrinsically similar abilities to generate IgG+ plasma cells upon homologous X31 stimulation; however, higher numbers of plasma cells were elicited from lung memory B cells than from splenic memory B cells after boosting with drifted Uruguay716 virus (Fig. 3 B). Thus, these data indicate that the increased cross-reactivity of lung memory B cells mediates a stronger cross-reactive secondary IgG response against a drifted virus in vivo.
Figure 3.
Lung memory B cells mediate cross-reactive secondary IgG response. Sorted X31 HA-binding memory B cells (PI-Dump−CD38+B220+) from lungs or spleens were transferred into CB17-scid mice (n = 4) with CD4+ T cells from infected mice and splenic B cells from naive mice. (A) At day 6 after boosting with X31 or Uruguay viruses, X31 HA-binding IgM−IgD−Gr-1−CD3−CD90−F4/80−PI−B220dull cells in splenocytes were gated and the cross-reactivity of restimulated memory B cells estimated. (B) Anti-HA IgG plasma cells in splenocytes were counted by ELISPOT using the same type of HA antigens for boosted virus strains. **, P < 0.01. (C) Serum Abs were recovered from memory cell–reconstituted mice and subjected to a microneutralization assay using MDCK cells after serial dilution (starting at a 1:10 dilution). The boosted virus strains were used for in vitro challenge. Each circle represents the result from an individual mouse. The data are representative of three independent experiments. *, P < 0.05.
Memory-derived Abs were then subjected to virus neutralization assays after serial dilution (starting at a 1:10 dilution; Fig. 3 C). Naive scid serum did not show any neutralization activity at a 1:10 dilution, and thus the detection limit was set to 10. X31-boosted Abs from restimulated lung and splenic memory B cells neutralized X31 virus to a comparable degree, but boosting with Uruguay716 virus elicited >10-fold higher titers of cross-neutralizing Abs from lung memory B cells. Given these results, we concluded that cross-reactive lung memory B cells are able to recognize virus-neutralizing epitopes in conserved regions of HAs and to secrete cross-neutralizing Abs upon restimulation with a drifted virus.
Monoclonal characterization of memory B cell–derived IgG Abs
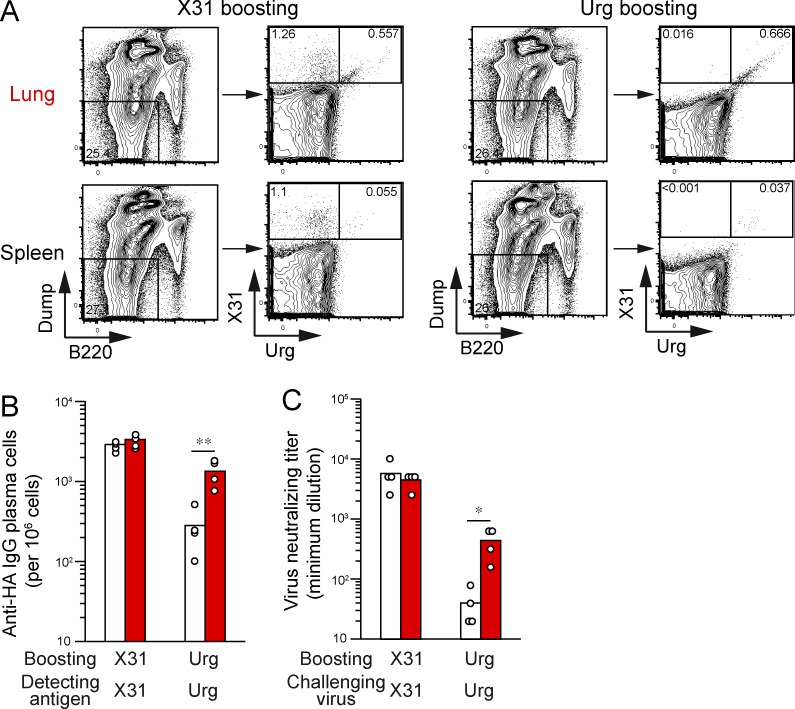
To further characterize the antigen specificity of lung memory B cells, we created recombinant monoclonal IgG Abs from single-sorted memory B cells in the lungs and spleens, as previously described (Smith et al., 2009; Tiller et al., 2009). In total, 23 mAbs were established from lungs (11) and spleens (12), and 11 and 10 mAbs, respectively, were found to bind to the X31 HA, confirming the specificity of our staining procedure using HA probes. Consistent with the flow cytometry data, lung-derived mAbs cross-reacted with the Uruguay716 virus to a greater extent than did their splenic counterparts (Fig. 4 A). Moreover, the conserved HA epitopes were included in the stem region: seven out of nine (lung) and one out of three (spleen) cross-reactive mAbs bound to the HA2 stem peptide, but none of nine strain-specific mAbs showed significant binding (Fig. 4 B). We then separated the panels of mAbs into two groups with (12 mAbs) or without (9 mAbs) detectable cross-reactivity to the Uruguay716 strain. We detected higher numbers of mutations in the cross-reactive mAbs (P = 0.0060; Fig. 4 C), confirming the previous polyclonal data (Fig. 2 C). To define the contribution of somatic hypermutations to the breadth of the reactivity, three cross-reactive mAbs (LM02, LM05, and LM09; Fig. 4 C, closed symbols) were reverted to the germline form, and the minimal concentrations for binding to the X31 and Uruguay viruses were determined (Fig. 4, D and E). We also prepared the germline form of three mAbs against H1 HA (clones NSP2, NPS27, and NSP29) as controls (Matsuzaki et al., 2014). All control mAbs showed no detectable binding to both X31 and Uruguay viruses below 50 µg/ml; therefore, the limit of detection in this analysis was set at 50 µg/ml. The mutated forms of the mAbs bound to both the X31 and drifted HAs at low concentrations, but the ability of germline-reverted mAbs to bind the X31 and Uruguay716 HAs was reduced (Fig. 4 E), consistent with previous findings in human heterosubtypic mAbs (Corti et al., 2011; Lingwood et al., 2012). However, in their germline form, all of the mAbs retained their cross-reactivity to drifted Uruguay716 virus. Thus, these results support the idea that somatic hypermutation is required for affinity maturation but is dispensable for the acquisition of cross-reactivity in influenza-infected mice, despite the correlation between mutational content and cross-reactivity.
Figure 4.
Cross-reactive mAbs target HA2 stem domain in a mutation-independent manner. HA-binding recombinant mAbs were generated from HA-binding memory B cells (PI-Dump−CD38+B220+) in lungs and spleens. (A) The minimal concentrations for binding to X31 and Uruguay716 were determined by ELISA. (B) Binding to HA2 peptide was analyzed by ELISA, and data are representative of three independent experiments. (C) Lung and splenic mAbs were subgrouped by their cross-reactivity to the Uruguay716 virus. The total number of mutations is plotted for each group. Closed symbols show the original clones used for germline reversion. Horizontal lines indicate mean. **, P < 0.01. (D and E) The mutated and germline forms of three mAbs (LM02, LM05, and LM09) were assessed by ELISA to determine the minimal concentrations for binding to X31 and Uruguay716. The detection limit of this assay was set at 50 µg/ml because of background signals from irrelevant germline mAbs. The data are representative of three independent experiments.
Pulmonary influenza virus infection gives rise to persistent GC responses
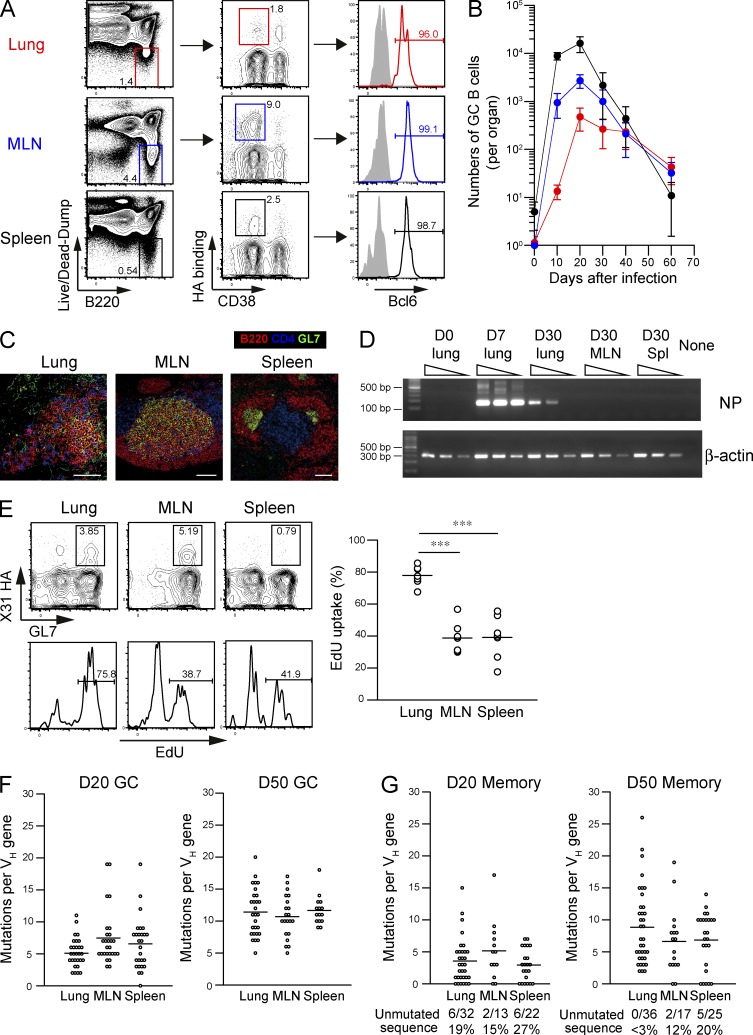
The higher number of mutations in cross-reactive Abs implies that the Abs originate from persistent GC responses because the mutational content of GC B cells increases in proportion to the period of time they spend in GCs (Takahashi et al., 2001; Kaji et al., 2012). Here, we evaluated the persistence of HA-binding GC responses in the indicated organs (Fig. 5, A and B). We first assessed the intracellular expression of the transcription factor Bcl-6, a reliable marker for GC B cells (Cattoretti et al., 1995). Indeed, >95% of CD38dull B cells from all organs were confirmed to express intracellular Bcl-6 at day 30 after infection (Fig. 5 A), thus defining CD38dull B cells as GC B cells even in the late time point. After the GC response peaked at day 20, a contraction phase followed in all organs (Fig. 5 B). Splenic GC B cells decreased 800-fold from day 20 to 60; however, lung GC B cells declined more slowly, with only an 11-fold reduction in the same period. MLN GCs exhibited intermediate reductions (85-fold) compared with lungs and spleens. Thus, GC responses persisted longer in local sites than in the spleen. Hereafter, we define HA-binding IgM/D−CD38dull B cells after day 20 as persistent GC B cells.
Figure 5.
Infection-induced persistent GCs generate highly mutated B cells. (A) HA-binding GC B cells [Live/Dead-Dump−(IgM−IgD−Gr-1−CD3−CD5−CD11b−CD11c−CD43−CD90−CD93−TER-119−F4/80−CD117−CD138−Live/Dead-Aqua−)CD38dullB220+] were gated, and the intracellular expression of Bcl-6 protein was evaluated by flow cytometry. The data are representative of three independent experiments. (B) The numbers of HA-binding GC B cells (PI-Dump−CD38dullB220+) in each organ are plotted at the indicated time points (mean ± SD; n = 4–9). Over 20,000,000 events were analyzed from individual spleens, and the cells within enlarged B220 versus dump gate were collected for further analysis at days 40 and 60 as shown in Fig. S1. The data are representative of two independent experiments. (C) Frozen sections (8 µm) were prepared from mice (n = 7) at day 30 after infection and stained with B220/CD4/GL7. In total, 569 (lungs), 272 (MLNs), and 1,439 (spleens) GL7+B220+ GC B cell clusters were scanned, and representative images on mean size are presented. Bars, 100 µm. (D) RNAs were extracted from the indicated organs (n = 6), serially diluted at threefold dilution, and subjected to one-step RT-PCR analysis for NP (40 cycles) and β-actin (23 cycles) genes. The data are representative of two independent experiments. (E) After intraperitoneal EdU injection (1 mg) at day 30 after infection, the EdU uptake of HA-binding GL7+ cells among Live/Dead-Dump−B220+ gate (as shown in A) in the indicated organs was analyzed 8 h later. Representative flow data for EdU labeling are presented, and the EdU+ ratios of HA-binding GC B cells are plotted. Each circle represents the result from an individual mouse. The data are representative of three independent experiments. ***, P < 0.001. (F and G) HA-binding GC B cells (PI-Dump−CD38dullB220+; F) and memory B cells (PI-Dump−CD38+B220+; G) were subjected to VH mutation analysis at days 20 and 50 after infection. Each circle represents the result from an individual cell. The combined data from three independent experiments are shown. (E–G) Horizontal lines indicate mean.
Persistent GC B cells express highly mutated immunoglobulin VH genes
We next tried to detect persistent GCs by immunohistochemistry using GL7 as a maker of GCs (Fig. 5 C). GL7+ B cell clusters were readily detectable in the MLN and spleen at a mean of 7.8 and 41.1 per section, respectively, at day 30 after infection. iBALT-like structures containing less dense GL7+ B cell clusters and CD4+ T cells, but lacking defined T cell area, were also observed in infected lungs around airways or blood vessels at a mean of 16.3 per section, confirming the presence of GL7+ GCs in these organs. In light of >1-mo persistence of viral antigen deposits near sites of infection (Kim et al., 2010), it is important to clarify whether the similar viral persistence is observed in our infection condition. Therefore, we performed RT-PCR analysis to detect viral RNA in the lungs, MLNs, and spleens at day 30 after infection (Fig. 5 D). Consistent with the results of the previous study (Kim et al., 2010), viral RNAs for nuclear protein were persistently detected in the lungs but not in the MLNs and spleens at day 30, even though the detected levels were much lower than those in the acute phase (day 7). Of note, the detection of viral antigens was associated with the exaggerated proliferation of lung GC B cells, as shown by the twofold higher frequencies in the labeling with 5-ethynyl-2′-deoxyuridine (EdU), which is a thymidine analogue that is incorporated into DNA during active DNA synthesis, compared with the frequencies in the other GC B cells (Fig. 5 E). We identified GC B cells as GL7+ cells in the EdU-labeling experiments because CD38 staining was unsuccessful in the EdU-staining condition. To determine whether the persistent viral antigens and exaggerated B cell proliferation affected the rate of somatic hypermutations, GC B cells were subjected to VH sequence analysis (Fig. 5 F). The majority of HA-binding GC B cells in all organs expressed mutated VH genes at day 20 after infection, and the number of mutations increased comparably over the next 30 d. The mean number of mutations reached 11.4 (lung), 10.7 (MLN), and 11.7 (spleen) per VH gene at day 50, roughly comparable with that in splenic GC B cells (10.0–15.2) at day 40 in mice primed with haptenated protein antigens (Takahashi et al., 2001; Kaji et al., 2012). Thus, although viral antigen deposits were persistently present in the infected lungs and the proliferative rates of the GC B cells were increased in this organ, persistent GC responses equally accumulated somatic hypermutations in all of the organs and generated highly mutated B cells at this late time point.
After proliferative expansion during the early response (<1–2 wk), memory B cells soon become resting and long-lived, undergoing slow turnover as the result of mutated precursors from persistent GCs (Schittek and Rajewsky, 1990; Kaji et al., 2012). In agreement with these observations, HA-binding memory B cell compartment in lung and spleen was first established by B cells with few mutations and then gradually replaced with repertoires possessing more mutations (lung D20 vs. D50 memory: P < 0.0001; spleen D20 vs. D50 memory: P = 0.005; Fig. 5 G). Of note, memory B cell replenishment appeared to be accelerated in the lung memory compartment, as reflected in the loss of unmutated memory B cells from the lungs at day 50 after infection (ratios of unmutated memory B cells at day 50; lung vs. MLN: P = 0.0359; lung vs. spleen: P = 0.0051; MLN vs. spleen: P = 0.4821; Fig. 5 G).
Cross-reactive memory B cells develop from persistent GCs
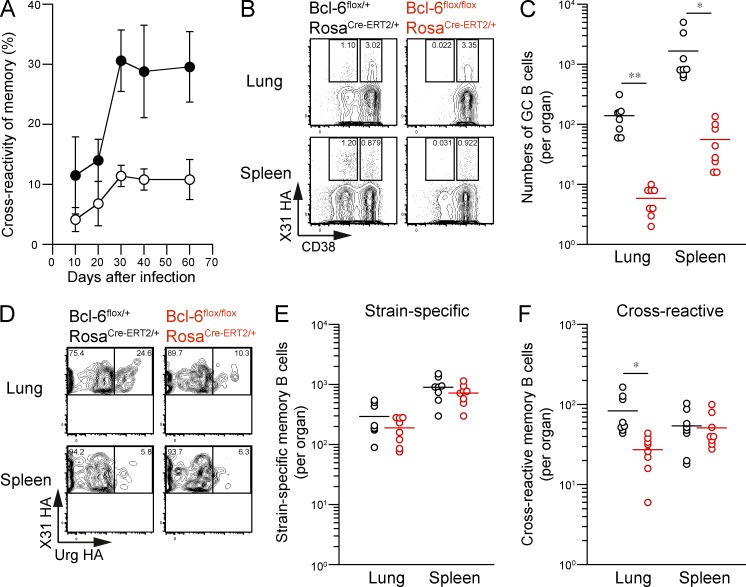
Mutational analysis of memory and GC B cell compartments has implied that cross-reactive memory B cells in the lungs originate from persistent GCs (Figs. 2 C and 5, F and G). Supporting this idea, we found that the prevalence of cross-reactive lung memory B cells increased from day 20 to 30, simultaneously with the beginning of the persistent GC phase (Fig. 6 A). To address whether persistent GC responses are required to establish cross-reactive memory B cells in the lungs, we used a tamoxifen-inducible Cre/loxP system to generate a mouse strain in which Bcl-6 can be conditionally deleted (Bcl-6flox/floxRosa26Cre-ERT2/+). As expected, tamoxifen treatment (days 14 and 15) resulted in a 10-fold reduction in the number of persistent GC B cells in flox/flox mice at day 28 after infection (Fig. 6, B and C); however, the treatment did not significantly affect the frequency of HA-binding memory B cells in either the lungs or spleens (Fig. 6 B). In contrast, we observed a specific reduction in the numbers of cross-reactive B cells among lung memory compartments after tamoxifen treatment (Fig. 6, D–F), whereas strain-specific memory B cells remained fairly intact. It is important to note that the absolute numbers of HA-binding memory B cells were smaller in flox/+ mice than in BALB/c mice, possibly because of the differential susceptibility of BALB/c and C57BL/6 mice to influenza virus infection and/or tamoxifen-dependent factors. However, we think the majority of cross-reactive memory B cells in spleens are again infection induced because naive flox/+ and flox/flox mice possessed only one-fifth the levels in the infected mice after the same tamoxifen treatment (flox/+: 10.3 ± 3.8 cells per organ; flox/flox: 6.3 ± 4.5 cells per organ; n = 7). These data clearly indicate that cross-reactive memory B cells in the lungs develop from persistent GCs, whereas strain-specific memory B cells are less dependent on persistent GCs for the development.
Figure 6.
Cross-reactive memory B cells develop from persistent GCs. (A) The cross-reactivity of splenic (open) and lung (closed) memory B cells (HA-binding PI-Dump−CD38+B220+) was assessed at the indicated time points (mean ± SD; n = 4–9). The data are representative of two independent experiments. (B) Mice with the indicated genotypes were orally treated with 2 mg tamoxifen at days 14 and 15 after infection, and lung and spleen cells were recovered at day 28 after infection. HA-binding GC (PI-Dump−CD38dullB220+) and memory B cells (PI-Dump−CD38+B220+) were gated. (C) The numbers of GC B cells per organ are plotted. *, P < 0.05; **, P < 0.01. (D) The cross-reactivity of memory B cells was compared for each mouse genotype. (E and F) The numbers of strain-specific (E) and cross-reactive (F) memory B cells are plotted. To detect the low frequency of splenic memory B cells, over 20,000,000 events were analyzed from individual spleens, and the cells within enlarged PI-Dump−B220+ gate were collected as shown in Fig. S1. *, P < 0.05. (C, E, and F) Each circle represents the result from an individual mouse. The data are representative of three independent experiments. Horizontal lines indicate mean.
Persistent GCs in the lungs select cross-reactive B cells with greater efficiency
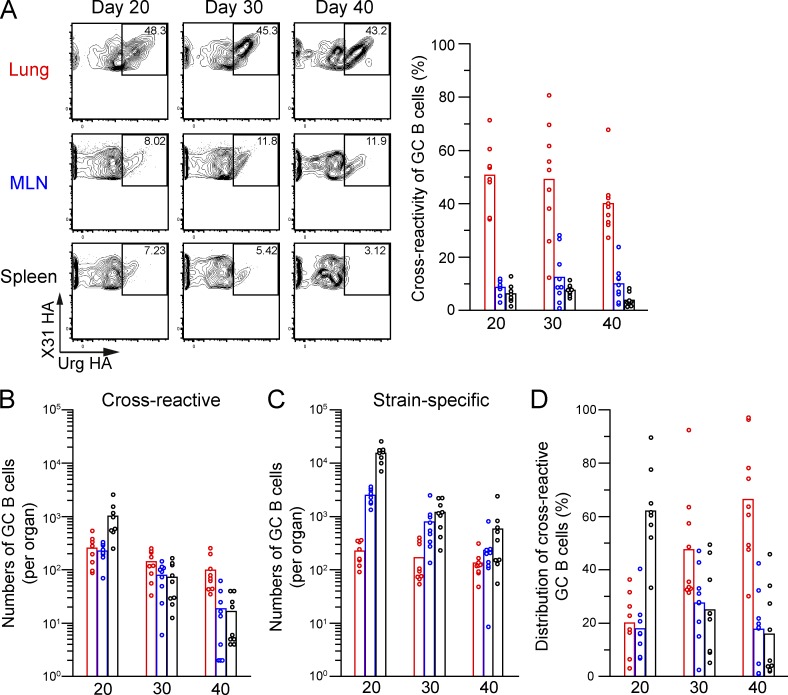
Although splenic GC B cells contracted more dramatically in the persistent GC phase, they retained cell numbers comparable with those in the lungs and MLNs, as a result of the high cellularity of the spleen (Fig. 5 B). To investigate where clonal selection for cross-reactivity occurs, we next assessed the numbers of strain-specific and cross-reactive GC B cells during the persistent GC phase (Fig. 7, A–C). Intriguingly, from day 20 to 40 after infection, cross-reactive B cells were significantly enriched in lung GCs but were scarce among splenic GC B cells (Fig. 7 A). Notably, lung GC B cells emerged with delayed kinetics (Fig. 5 B), and GC numbers reached to the peak at day 20, when the cross-reactivity of lung memory B cells was about to increase. Moreover, because lung GCs contracted to a lesser extent than other GCs (Fig. 7, B and C), they became the dominant sites harboring cross-reactive B cells at later time points (Fig. 7 D). Thus, compared with GC responses in other organs, lung GC responses select cross-reactive Ab repertoires more efficiently and persistently, implying that lung GC responses are the primary source of cross-reactive memory B cells in the lungs.
Figure 7.
Site-specific selection of cross-reactive GC B cells. (A) The cross-reactivity of HA-binding GC B cells (PI-Dump−CD38dullB220+) was analyzed in the lungs, MLNs, and spleens and plotted at several time points (n = 8–9). To reliably detect the low frequency of cross-reactive GC B cells in spleens, over 20,000,000 events were analyzed from individual spleens, and the cells within the PI-Dump−B220+ gate were collected at day 40. (B and C) The numbers of cross-reactive (B) and strain-specific (C) GC B cells in each organ were calculated using the flow cytometric data and the number of cells recovered for the organ. (D) The distribution of cross-reactive GC B cells in each organ was calculated from the cell numbers shown in B and C. (A–D) Each circle represents the result from an individual mouse. The data are representative of two independent experiments.
Lung GC B cells supply cross-reactive memory B cells at local sites
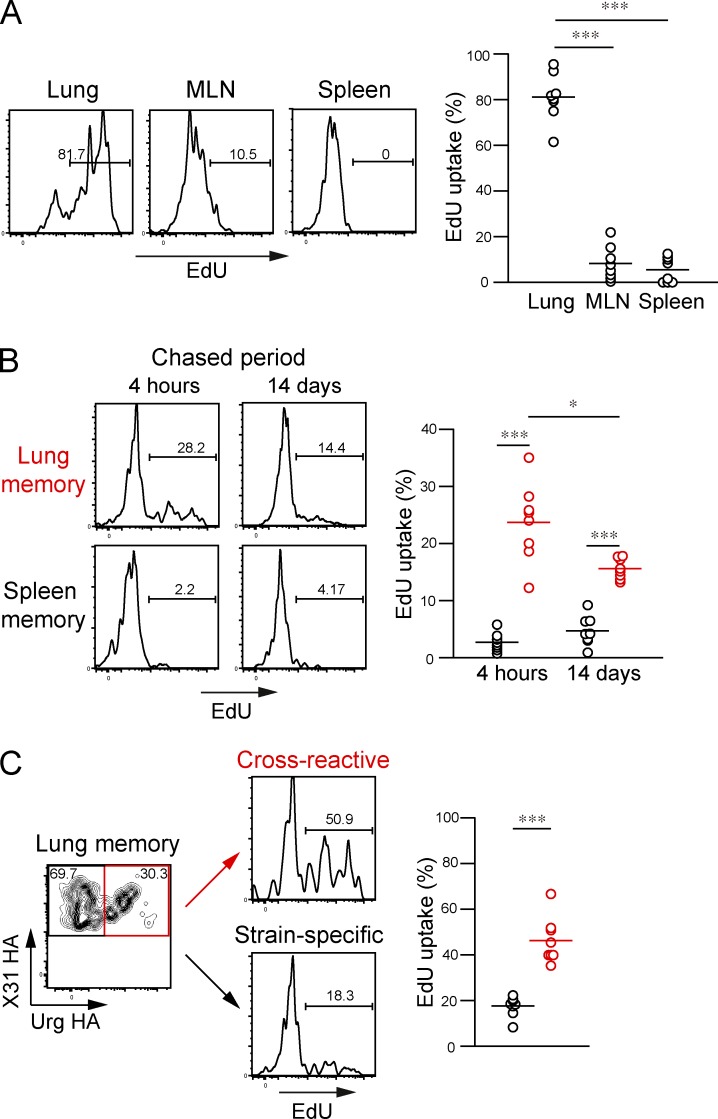
Intratracheal injection of small compounds can successfully label target cells near the lower respiratory tracts (lungs and MLNs), whereas cells in systemic organs remained unlabeled (Legge and Braciale, 2003; GeurtsvanKessel et al., 2009; Halle et al., 2009). Using a similar strategy, EdU was intratracheally injected on alternate days from day 18 to 30 for selective labeling of dividing GC B cells in local sites. Intratracheal EdU injection resulted in successful labeling of lung GC B cells (81.1% ± 10.5%, n = 8) at 4 h after the final EdU injection, but the majority of MLN and splenic GCs remained unlabeled with EdU (Fig. 8 A). Therefore, this labeling strategy allowed us to trace the progeny of lung GC B cells. Although EdU injection at a 2-d interval may be insufficient to label whole lung GC outputs from day 18 to 30, we used these conditions to avoid higher background labeling of other GC outputs caused by intensive EdU injection.
Figure 8.
Lung GCs supply cross-reactive memory B cells in the lungs. (A) After intratracheal EdU injection (100 µg in 100 µl PBS) on alternate days from day 18 to 30, the EdU uptake of HA-binding GL7+ cells among the Live/Dead-Dump−B220+ gate (as shown in Fig. 5, A and E) in the indicated organs was analyzed at day 30. Representative flow data for EdU staining are presented, and the EdU uptake of GC B cells in the indicated organs is plotted. ***, P < 0.001. (B) EdU uptake of memory B cells (HA-binding GL7− among the Live/Dead-Dump−B220+ gate) was analyzed at 4 h and 14 d after the final EdU injection. Representative flow data for EdU staining and EdU uptake of memory B cells are presented. *, P < 0.05; ***, P < 0.001. (C) Lung memory B cells (HA-binding GL7−) were divided into cross-reactive and strain-specific populations, and the EdU uptake of each population analyzed. The representative flow data and EdU uptake of each memory B cell population are presented. ***, P < 0.001. (A–C) Each circle represents the result from an individual mouse. The data are representative of two independent experiments. Horizontal lines indicate mean.
We analyzed the EdU uptake of HA-binding lung and splenic memory B cells at day 30, a time point when the cross-reactivity of lung memory B cells sharply increased (Fig. 6 A). At 4 h after the final EdU injection, we observed that 23.7 ± 6.8% of lung memory B cells incorporated EdU at this time point (Fig. 8 B), but this number dropped to 2.7 ± 1.6% in splenic memory B cells, supporting the preferential localization of lung GC–derived memory B cells in the same tissue. Consistent with the accelerated loss of unmutated memory B cells (Fig. 5 G), the percentages of EdU+ memory B cells gradually decreased from 23.7 to 15.6% in the lung compartment during 2-wk chase periods (Fig. 8 B), which resembles continual replenishment of lung-resident memory T cells (Ely et al., 2006; Zammit et al., 2006). This labeling strategy also revealed that 46.3 ± 10.1% of cross-reactive memory B cells in the lungs were EdU+ but that only 17.6 ± 4.4% of strain-specific memory B cells incorporated EdU under the same conditions (Fig. 8 C). Given the limited dosages and frequencies of EdU injection, it is likely that the EdU+ ratios of cross-reactive memory B cells were underestimated. Thus, these data demonstrate that lung GCs constitute an important cellular source of cross-reactive lung memory B cells. And also, combined with the results from Bcl-6–deficient mice, these data support the model that early GC responses in secondary lymphoid organs promptly generate strain-specific memory B cells, whereas lung-persistent GC responses are primarily dedicated to the generation of cross-reactive memory B cells in the same sites.
DISCUSSION
Serological analysis has established that the intranasal inoculation of influenza viruses or vaccines extends the breadth of Ab specificity and provides cross-protection against viral escape mutants at local sites (Tamura et al., 1992; Tumpey et al., 2001; Moody et al., 2011; Margine et al., 2013). Here, we have identified lung-persistent GCs as key induction sites of cross-reactive Ab responses, with a distinctive ability to select and supply cross-reactive memory progenitors in the late time points. The distinct GC selection in the lungs was associated with the persistence of viral antigens and exaggerated GC dynamics, suggesting a possible link between the viral antigen deposits in the local sites and the GC persistence and selection for cross-reactivity. Moreover, we observed preferential localization of lung GC–derived memory B cells as lung-resident cells, allowing cross-reactive repertoires to be focused at the site of infection. Thus, our findings reveal unique properties of GC-dependent memory B cell pathways at local sites that dictate cross-reactive Ab responses against respiratory virus reinfection.
By performing both polyclonal and monoclonal analyses, cross-reactive memory B cells were found to possess higher mutational content than strain-specific memory B cells, consistent with their origins from persistent GCs. Indeed, the persistent GC origin of cross-reactive memory B cells was clearly confirmed by the impaired development of cross-reactive memory B cells in mice lacking persistent GCs. Likewise, the fairly normal development of strain-specific memory B cells in the absence of persistent GCs underscored their origins via an early GC and/or GC-independent pathway by the beginning of the persistent GC phase. The heterogeneous pathways for memory B cell development have been elucidated in previous studies using protein antigens (Inamine et al., 2005; Tomayko et al., 2010; Kaji et al., 2012; Taylor et al., 2012). It is generally accepted that memory B cells develop in temporally distinct phases; the early phase accompanies the formation of a memory B cell repertoire possessing fewer mutations, and the late phase recruits a heavily mutated repertoire from persistent GC responses. Here, using influenza infection models, we successfully dissected distinct roles of the heterogeneous memory B cell pathways. The early pathway promptly generates strain-specific memory B cells, whereas the late pathway is dedicated to the generation of cross-reactive memory B cells. This temporal switch from strain specificity to cross-reactivity in memory B cell development appears to result from site-specific selection and persistence of cross-reactive GC B cells; to the best of our knowledge, these findings have not been documented to date in influenza virus infection. A temporal shift in memory B cell development would enable generation of rapid strain-specific responses for protection against the same virus and provide broader protection against antigenically divergent viruses during the later phase.
The augmented selection and persistence of cross-reactive B cells in lung GCs constitutes an important framework for the pronounced cross-reactivity of lung memory B cells and for broad protection against escape mutants in local sites. Persistent GC responses have been frequently observed after live virus infection or immunization with particulate viral antigens including Toll-like receptor agonists (Bachmann et al., 1996; Kasturi et al., 2011). Likewise, persistent GC responses have previously been observed in the lungs and MLNs after pulmonary influenza virus infection (Rothaeusler and Baumgarth, 2010; Boyden et al., 2012; Onodera et al., 2012). The survival and selection of GC B cells is tightly regulated by the BCR-mediated uptake of antigen and subsequent interactions with follicular helper T cells (Tfh cells; Victora and Nussenzweig, 2012; Gitlin et al., 2014). Therefore, we speculate that the site-specific fine-tuning of GC responses by antigens and Tfh cells is important for creating cross-reactive memory responses.
It is noteworthy that persistent viral antigens have been detected in infected lung tissue but not in other tissues at 1 mo after infection, despite the acute nature of influenza virus infection (Fig. 5 D; Kim et al., 2010). These data suggested that viral replication lasts in the infected lungs longer than previously thought. Because the Ab responses against influenza viruses peak at day 7 after primary infection (Rothaeusler and Baumgarth, 2010), influenza viruses in the lungs may have an increased chance to replicate in the presence of anti-HA Abs, which create the situation in which viral escape mutants are prone to emerge. If this is indeed the case, it is easily conceivable that local deposits of drifted HA antigens select cross-reactive repertoires with increased numbers of mutations, as has been observed in chronic virus infections with HIV (Liao et al., 2013; Doria-Rose et al., 2014; Wu et al., 2015). An alternative but not mutually exclusive possibility is that local-specific virus replication may somehow create HA antigens with different antigenic structures, resulting in the enhanced exposure of invariant HA regions to lung-resident B cells. Therefore, it is important to determine the exact nature of the persisting antigens and antigen-presenting cells to B cells in local sites. In addition, although GL7+ B cells and CD4+ T cells were observed in the histological examination of the infected lung, GL7+ B cell clusters were less dense, and there were no clearly segregated B cell follicles/T cell area, as seen in the secondary lymphoid organs. This implies that GC organization might be altered in the lungs, and this needs to be taken into account when assessing differential clonal selection in the lung GCs.
Site-restricted viral replication also generates a distinct cytokine milieu in infected lungs. Infection-induced cytokines, such as IFN-γ, IL-17, IL-27, and IL-6, along with Tfh-derived IL-21, profoundly influence GC responses through Tfh functions (Hsu et al., 2008; Batten et al., 2010; Karnowski et al., 2012; Lee et al., 2012). Given the heterogeneity of Tfh functions (Hsu et al., 2008; Bauquet at al., 2009; Johnston et al., 2009; Reinhardt et al., 2009), it is intriguing to speculate that persistent viral antigens may create a cytokine milieu that generates unique Tfh subsets in local sites, thereby sustaining functional GC responses over long periods.
A recent study using rapamycin, an immunosuppressive drug that inhibits the kinase mTOR, unveiled a novel B cell pathway for inducing cross-reactive Ab repertoires. Rapamycin-mediated mTOR inhibition suppressed B cell responses by reducing GC reactions and IgG switching, and, surprisingly, enhanced the induction of cross-reactive Ab repertoires (Keating et al., 2013). Although the exact nature (i.e., isotypes, mutational content, and B cell origin) of the rapamycin-induced cross-reactive Ab repertoires remains to be determined, this drug may potentiate T cell–dependent IgM Ab response that targets cross-reactive epitopes, as suggested by the authors. Therefore, at least two distinct pathways potentially elicit cross-reactive Ab repertoires to influenza viruses: the early IgM pathway, which can be enhanced by rapamycin-mediated immunosuppression, and the local GC-derived IgG pathway, which is exaggerated at the site of infection. Further comparative analysis of the breadth, persistence, and neutralizing activity of Abs may establish the functional compartmentalization of the two pathways. Moreover, elucidation of the heterogeneous pathways for cross-reactive Ab responses may allow us to prepare several types of broadly protective vaccines with differing modes of action, aiding efforts to cope with the large variability in pathogens and the immune histories of humans.
MATERIALS AND METHODS
Mice and virus.
BALB/c mice and CB17-scid mice were purchased from SLC and Clea, respectively. B1-8high mice were provided by M. Nussenzweig (The Rockefeller University, New York, NY). We independently generated Bcl-6 flox mice with loxP sites flanking exons 7–9 of the Bcl-6 gene using the procedures described previously (Kaji et al., 2012) and crossed with Rosa26-Cre-ERT2 mice (Taconic). All mice were maintained under specific pathogen–free conditions and used at 7–12 wk of ages. The influenza A viruses X31, A/Uruguay/716/07, and A/Narita/1/2009 (provided by T. Odagiri, M. Tashiro, and H. Hasegawa, National Institutes of Infectious Diseases, Tokyo, Japan) were grown in 10-d-old embryonated hen eggs or MDCK cells and purified through a 10–50% sucrose gradient as previously described (Takahashi et al., 2009). BALB/c mice, anesthetized by intraperitoneal injection with sodium pentobarbital, were inoculated intranasally with 0.1 lethal dose50 (LD50) in a volume of 20 µl. EdU (Life Technologies) was dissolved in PBS at 1 mg/ml and then inoculated intratracheally at 100 µl per injection into isoflurane-anesthetized mice using a 24-gauge indwelling needle (TOP Corporation). After intratracheal injection, the mice were kept in an upright position to allow sufficient spreading of the fluid throughout the lungs. Animal procedures were approved by Animal Ethics Committee of the National Institute of Infectious Diseases, Japan.
Abs and reagents.
Anti-FcγRII/III (2.4G2) mAbs were purified in our laboratory. rHA lacking C-terminal transmembrane regions was produced in a baculovirus expression system (Takara Bio Inc.) and conjugated with PE, APC, or PerCP in our laboratory (Onodera et al., 2012). NIP-BSA was conjugated with PE. Formalin-inactivated X31 viruses and anti-CD38 (CS/2) mAb were labeled with Alexa Fluor 488 in our laboratory. Anti-IgM (II/41)–biotin, anti-IgD (11-26c)–biotin, anti-B220/CD45R (RA3-6B2)–biotin, anti-CD93 (AA4.1)–biotin, anti-Thy1.2/CD90.2 (53-2.1)–biotin, anti-CD3 (145-2C11)–biotin, anti–Gr-1 (RB6-8C5)–biotin, anti-F4/80 (BM8)–biotin, anti-CD11b (M1/70)–biotin, anti-CD11c (N418)–biotin, anti-CD117 (2B8)–biotin, anti-CD8α (53-6.7)–biotin, anti–TER-119 (TER-119)–biotin, anti-CD5 (53-7.3)–biotin, anti-CD4 (GK1.5)–biotin, anti-IgD (11-26)–PE, and anti-CD4 (RM4-5)–APC were purchased from eBioscience. Anti-CD43 (S7)–biotin, anti-CD138 (281-2)–biotin, streptavidin (SA)-PE–Texas Red/PE-CF594, anti-GL7 (GL7)–PE, anti-lambda (R26-46)–FITC, and anti-Bcl6 (K112-91)–Alexa Fluor 488 were purchased from BD. Anti-CD38 (90)–FITC/Pacific Blue, anti-GL7 (GL7)–FITC, SA–Brilliant Violet510, anti-B220–Alexa Fluor 700/Pacific Blue/PE, and anti-CD19 (6D5)–biotin were purchased from BioLegend. Click-iT Plus EdU Alexa Fluor 488 kit and LIVE/DEAD Fixable Aqua Dead Cell Stain kit were purchased from Life Technologies. Goat anti–mouse IgG-HRP and goat anti–human IgG-HRP were purchased from SouthernBiotech.
Cell preparation.
Single-cell suspensions were prepared from the spleens and MLNs in DMEM (Sigma-Aldrich) containing 2% FCS, 2 mM l-glutamine, 100 IU/ml penicillin, 100 µg/ml streptomycin, and 5 × 10−5 M β-mercaptoethanol as described previously (Onodera et al., 2012). For isolation of lung cells, mice were perfused with PBS in the right ventricle to clear the blood. Lungs were minced and incubated at 37°C for 60 min in 2 mg/ml collagenase D (Roche) and 10 µg/ml DNase I (Roche) and then disrupted between the frosted ends of glass slides. After centrifugation in a 70%/44%/30% Percoll gradient, the cells at the 70%/44% interface were recovered. In some experiments, cells were treated with 16 mU neuraminidase (Roche) in FCS-free DMEM for 10 min at 37°C.
Flow cytometric analysis.
Flow cytometric analysis was performed as previously described (Onodera et al., 2012). In brief, cells were pretreated with anti-FcγRII/III mAb, incubated with mixtures of biotinylated mAbs (for memory/GC B cells, IgM, IgD, Gr-1, CD3, CD5, CD11b, CD11c, CD43, CD90, CD93, TER-119, F4/80, CD117, and CD138; for plasma cells, IgM, IgD, Gr-1, CD3, CD90, and F4/80) and labeled with anti-B220, anti-CD38, SA, propidium iodide (PI), or LIVE/DEAD Fixable Aqua, two types of rHA, and X31 virus. Bcl-6+ or EdU-labeled cells were detected by intracellular staining using anti–Bcl-6 or Click-iT Plus EdU kit according to the manufacturer’s instructions. Memory B cells were sorted from MACS-enriched populations using SA-microbeads (Miltenyi Biotech). Stained cells were analyzed or purified using FACSCanto II or FACSAria III (BD). A total of >1,000,000 events were collected, and data were analyzed with FlowJo software (Tree Star).
IgVH sequence analysis.
Single HA-binding IgM/D− memory and GC B cells were sorted directly into 10 µl water containing 50 ng carrier RNA (QIAGEN). RT reaction was performed using random hexamer (Invitrogen), and then cDNAs were subjected to two rounds of nested PCR as described previously (Tiller et al., 2009). Primers used for PCR were 5′ MsVHE, 3′ Cγ1 outer, and 3′ Cγ2a outer for first round and 5′ MSVHE, 3′ Cγ1 inner, and 3′ Cγ2a inner for second round. The PCR products were purified and directly sequenced using 3′ Cγ1 inner or 3′ Cγ2a inner primers. IgBlast was used to identify germline VDJ and the position of somatic hypermutations. To estimate the Taq-induced error rate, we sequenced 17,424 VDJ nucleotides from single-sorted HA-binding hybridomas and found one error. This error rate corresponds to 0.02 mutations per VH gene, indicating PCR error mutations are negligible in interpreting our results.
In vitro stimulation of memory B cells.
Purified HA-binding IgM/D− memory B cells (300 cells per well) were stimulated on a layer of mitomycin C–treated 3T3 fibroblasts in the presence of LPS, IL-2, IL-4, and IL-5 for 6 d as previously described (Takahashi et al., 2005).
Detection of anti-HA Ab titers and plasma cells.
For detection of anti-HA Ab titers and plasma cells, ELISA and ELISPOT were performed using rHA and HA2 peptide as coating antigens (Wang et al., 2010; Onodera et al., 2012). Virus-neutralization Ab titers were determined by microneutralization using MDCK cell lines (Takahashi et al., 2009).
Adoptive transfers.
B cells and CD4+ T cells were purified from pooled spleens using a MACS system with biotinylated mAbs against CD3, CD90, CD4, F4/80, Gr-1, CD11b, CD43, and CD138 (B cells) and CD19, B220, IgM, IgD, F4/80, Gr-1, CD11b, and CD8 (CD4+ T cells), followed by SA-microbeads (Miltenyi Biotech). B cells (2 × 106/head) from naive mice and CD4+ T cells (106/head) from infected mice were intravenously injected into CB17-scid mice with or without sorted memory B cells (3,000/head). On the next day, the recipient mice were intravenously boosted with inactivated viruses (20 µg), and the serum and spleens were collected at day 6 after boosting.
Generation of recombinant mAbs.
Mouse VH/Vk genes were PCR amplified from single-sorted memory B cells as described previously (Tiller et al., 2009). Recombinant mAbs were generated by transfecting both expression vectors into 293A cells (Invitrogen; Smith et al., 2009). In brief, the cells were grown to 80% confluency in 100-mm plates. Both expression vectors (3 µg each) were premixed with 50 µg polyethylenimine (Polysciences Inc.) and added to the cells. After 24-h incubation, the culture medium was replaced by a 50:50 mixture of RPMI/DMEM supplemented with antibiotic, 2 mM l-glutamine, and 1% Nutridoma-SP (Roche), and the medium was collected 4 d later. IgG1 Abs were purified from the culture supernatant using a protein G column (Thermo Fisher Scientific) and subjected to further analysis.
Histology.
Lung tissues were recovered after intratracheal injection of 50% Tissue-Tek OCT compound (Sakura Finetek Inc.) in PBS. All tissues were then embedded in Tissue-Tek OCT compound. After freezing, sections (8 µm) were deposited on slides and fixed in acetone. The sections were pretreated with anti-FcγRII/III mAb and then stained with anti-B220–PE, anti-CD4–APC, and anti-GL7–FITC. Stained sections were scanned under a confocal laser-scanning microscope LSM 700 (Carl Zeiss).
RT-PCR analysis.
Total RNA was extracted with an RNeasy Mini kit (QIAGEN), and the NP/β-actin genes were amplified by an OneStep RT-PCR kit (QIAGEN) according to the manufacturer’s instructions. The primers that were used for the amplification were Flu NP forward (5′-ATGCCATTCTGCCGCATTTG-3′) and Flu NP reverse (5′-CCTTATGGCCCAGTACCTGC-3′) for the influenza NP gene and β-actin forward (5′-ACTATTGGCAACGAGCGGTTC-3′) and β-actin reverse (5′-CCACCGATCCACACAGAGTA-3′) for the β-actin gene. After PCR amplification by 40 (NP) and 23 (β-actin) cycles, the PCR products were visualized on 2% agarose gel with EtBr staining.
Statistical analysis.
Statistical significance was determined using an unpaired, two-tailed Student’s t test. P-values <0.05 were considered significant and indicated by asterisks: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Accession numbers.
Complete sequence data are available from the DNA Data Bank of Japan (DDBJ), the EMBL Nucleotide Sequence Database, and GenBank under the following accession numbers: GC and memory B cells from lungs, MLNs, and spleens (AB774476–AB774768); cross-reactive and strain-specific memory B cells (AB774879–AB774931 and AB901108–AB901139).
Online supplemental material.
Fig. S1 shows the FACS gating strategy for HA-binding memory/GC B cells using two HA probes. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20142284/DC1.
Supplementary Material
Acknowledgments
We thank Drs. T. Odagiri, M. Tashiro, and H. Hasegawa for providing virus strains. We also thank Ms. E. Izumiyama, K. Fukuhara, and Mr. H. Miyadai for technical assistance.
This research was supported by grants to Y. Takahashi from the Ministry of Education, Culture, Sports, Science, and Technology in Japan, by grants to T. Kurosaki and Y. Takahashi from the Japan Science and Technology Agency, Core Research of Evolutional Science and Technology, by grants to K. Kobayashi and Y. Takahashi from the Ministry of Health, Labor, and Welfare in Japan (Emerging and Re-emerging Infectious Diseases and Regulatory Science of Pharmaceuticals and Medical Devices), and by grants to M. Ato and Y. Takahashi from the Japan Agency for Medical Research and Development.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- Ab
- antibody
- EdU
- 5-ethynyl-2′-deoxyuridine
- GC
- germinal center
- HA
- hemagglutinin
- iBALT
- induced bronchus-associated lymphoid tissue
- MLN
- mediastinal LN
- PI
- propidium iodide
- rHA
- recombinant HA
- Tfh cell
- follicular helper T cell
References
- Anderson K.G., Sung H., Skon C.N., Lefrancois L., Deisinger A., Vezys V., and Masopust D.. 2012. Cutting edge: intravascular staining redefines lung CD8 T cell responses. J. Immunol. 189:2702–2706. 10.4049/jimmunol.1201682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann M.F., Odermatt B., Hengartner H., and Zinkernagel R.M.. 1996. Induction of long-lived germinal centers associated with persisting antigen after viral infection. J. Exp. Med. 183:2259–2269. 10.1084/jem.183.5.2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batten M., Ramamoorthi N., Kljavin N.M., Ma C.S., Cox J.H., Dengler H.S., Danilenko D.M., Caplazi P., Wong M., Fulcher D.A., et al. 2010. IL-27 supports germinal center function by enhancing IL-21 production and the function of T follicular helper cells. J. Exp. Med. 207:2895–2906. 10.1084/jem.20100064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarth N. 2013. How specific is too specific? B-cell responses to viral infections reveal the importance of breadth over depth. Immunol. Rev. 255:82–94. 10.1111/imr.12094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauquet A.T., Jin H., Paterson A.M., Mitsdoerffer M., Ho I.C., Sharpe A.H., and Kuchroo V.K.. 2009. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat. Immunol. 10:167–175. 10.1038/ni.1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden A.W., Legge K.L., and Waldschmidt T.J.. 2012. Pulmonary infection with influenza A virus induces site-specific germinal center and T follicular helper cell responses. PLoS ONE. 7:e40733 10.1371/journal.pone.0040733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoretti G., Chang C.C., Cechova K., Zhang J., Ye B.H., Falini B., Louie D.C., Offit K., Chaganti R.S., and Dalla-Favera R.. 1995. BCL-6 protein is expressed in germinal-center B cells. Blood. 86:45–53. [PubMed] [Google Scholar]
- Coro E.S., Chang W.L., and Baumgarth N.. 2006. Type I IFN receptor signals directly stimulate local B cells early following influenza virus infection. J. Immunol. 176:4343–4351. 10.4049/jimmunol.176.7.4343 [DOI] [PubMed] [Google Scholar]
- Corti D., Suguitan A.L. Jr., Pinna D., Silacci C., Fernandez-Rodriguez B.M., Vanzetta F., Santos C., Luke C.J., Torres-Velez F.J., Temperton N.J., et al. 2010. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Invest. 120:1663–1673. 10.1172/JCI41902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Voss J., Gamblin S.J., Codoni G., Macagno A., Jarrossay D., Vachieri S.G., Pinna D., Minola A., Vanzetta F., et al. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 333:850–856. 10.1126/science.1205669 [DOI] [PubMed] [Google Scholar]
- Doria-Rose N.A., Schramm C.A., Gorman J., Moore P.L., Bhiman J.N., DeKosky B.J., Ernandes M.J., Georgiev I.S., Kim H.J., Pancera M., et al. NISC Comparative Sequencing Program. 2014. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 509:55–62. 10.1038/nature13036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucett V.P., Gerhard W., Owler K., Curry D., Brown L., and Baumgarth N.. 2005. Enumeration and characterization of virus-specific B cells by multicolor flow cytometry. J. Immunol. Methods. 303:40–52. 10.1016/j.jim.2005.05.014 [DOI] [PubMed] [Google Scholar]
- Ely K.H., Cookenham T., Roberts A.D., and Woodland D.L.. 2006. Memory T cell populations in the lung airways are maintained by continual recruitment. J. Immunol. 176:537–543. 10.4049/jimmunol.176.1.537 [DOI] [PubMed] [Google Scholar]
- Gerhard W. 2001. The role of the antibody response in influenza virus infection. Curr. Top. Microbiol. Immunol. 260:171–190. [DOI] [PubMed] [Google Scholar]
- GeurtsvanKessel C.H., Willart M.A., Bergen I.M., van Rijt L.S., Muskens F., Elewaut D., Osterhaus A.D., Hendriks R., Rimmelzwaan G.F., and Lambrecht B.N.. 2009. Dendritic cells are crucial for maintenance of tertiary lymphoid structures in the lung of influenza virus-infected mice. J. Exp. Med. 206:2339–2349. 10.1084/jem.20090410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin A.D., Shulman Z., and Nussenzweig M.C.. 2014. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 509:637–640. 10.1038/nature13300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halle S., Dujardin H.C., Bakocevic N., Fleige H., Danzer H., Willenzon S., Suezer Y., Hämmerling G., Garbi N., Sutter G., et al. 2009. Induced bronchus-associated lymphoid tissue serves as a general priming site for T cells and is maintained by dendritic cells. J. Exp. Med. 206:2593–2601. 10.1084/jem.20091472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.C., Yang P., Wang J., Wu Q., Myers R., Chen J., Yi J., Guentert T., Tousson A., Stanus A.L., et al. 2008. Interleukin 17-producing T helper cells and interleukin 17 orchestrate autoreactive germinal center development in autoimmune BXD2 mice. Nat. Immunol. 9:166–175. 10.1038/ni1552 [DOI] [PubMed] [Google Scholar]
- Inamine A., Takahashi Y., Baba N., Miyake K., Tokuhisa T., Takemori T., and Abe R.. 2005. Two waves of memory B-cell generation in the primary immune response. Int. Immunol. 17:581–589. 10.1093/intimm/dxh241 [DOI] [PubMed] [Google Scholar]
- Johnston R.J., Poholek A.C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A.L., Craft J., and Crotty S.. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 325:1006–1010. 10.1126/science.1175870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H.M., He Y., and Sangster M.Y.. 2008. Broad dispersion and lung localization of virus-specific memory B cells induced by influenza pneumonia. Proc. Natl. Acad. Sci. USA. 105:3485–3490. 10.1073/pnas.0800003105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji T., Ishige A., Hikida M., Taka J., Hijikata A., Kubo M., Nagashima T., Takahashi Y., Kurosaki T., Okada M., et al. 2012. Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J. Exp. Med. 209:2079–2097. 10.1084/jem.20120127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnowski A., Chevrier S., Belz G.T., Mount A., Emslie D., D’Costa K., Tarlinton D.M., Kallies A., and Corcoran L.M.. 2012. B and T cells collaborate in antiviral responses via IL-6, IL-21, and transcriptional activator and coactivator, Oct2 and OBF-1. J. Exp. Med. 209:2049–2064. 10.1084/jem.20111504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasturi S.P., Skountzou I., Albrecht R.A., Koutsonanos D., Hua T., Nakaya H.I., Ravindran R., Stewart S., Alam M., Kwissa M., et al. 2011. Programming the magnitude and persistence of antibody responses with innate immunity. Nature. 470:543–547. 10.1038/nature09737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating R., Hertz T., Wehenkel M., Harris T.L., Edwards B.A., McClaren J.L., Brown S.A., Surman S., Wilson Z.S., Bradley P., et al. 2013. The kinase mTOR modulates the antibody response to provide cross-protective immunity to lethal infection with influenza virus. Nat. Immunol. 14:1266–1276. 10.1038/ni.2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.S., Hufford M.M., Sun J., Fu Y.X., and Braciale T.J.. 2010. Antigen persistence and the control of local T cell memory by migrant respiratory dendritic cells after acute virus infection. J. Exp. Med. 207:1161–1172. 10.1084/jem.20092017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause J.C., Tsibane T., Tumpey T.M., Huffman C.J., Basler C.F., and Crowe J.E. Jr. 2011. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J. Virol. 85:10905–10908. 10.1128/JVI.00700-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.K., Silva D.G., Martin J.L., Pratama A., Hu X., Chang P.P., Walters G., and Vinuesa C.G.. 2012. Interferon-γ excess leads to pathogenic accumulation of follicular helper T cells and germinal centers. Immunity. 37:880–892. 10.1016/j.immuni.2012.10.010 [DOI] [PubMed] [Google Scholar]
- Legge K.L., and Braciale T.J.. 2003. Accelerated migration of respiratory dendritic cells to the regional lymph nodes is limited to the early phase of pulmonary infection. Immunity. 18:265–277. 10.1016/S1074-7613(03)00023-2 [DOI] [PubMed] [Google Scholar]
- Li G.M., Chiu C., Wrammert J., McCausland M., Andrews S.F., Zheng N.Y., Lee J.H., Huang M., Qu X., Edupuganti S., et al. 2012. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc. Natl. Acad. Sci. USA. 109:9047–9052. 10.1073/pnas.1118979109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.X., Lynch R., Zhou T., Gao F., Alam S.M., Boyd S.D., Fire A.Z., Roskin K.M., Schramm C.A., Zhang Z., et al. NISC Comparative Sequencing Program. 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 496:469–476. 10.1038/nature12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D., McTamney P.M., Yassine H.M., Whittle J.R., Guo X., Boyington J.C., Wei C.J., and Nabel G.J.. 2012. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 489:566–570. 10.1038/nature11371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margine I., Hai R., Albrecht R.A., Obermoser G., Harrod A.C., Banchereau J., Palucka K., García-Sastre A., Palese P., Treanor J.J., and Krammer F.. 2013. H3N2 influenza virus infection induces broadly reactive hemagglutinin stalk antibodies in humans and mice. J. Virol. 87:4728–4737. 10.1128/JVI.03509-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y., Sugawara K., Nakauchi M., Takahashi Y., Onodera T., Tsunetsugu-Yokota Y., Matsumura T., Ato M., Kobayashi K., Shimotai Y., et al. 2014. Epitope mapping of the hemagglutinin molecule of A/(H1N1)pdm09 influenza virus by using monoclonal antibody escape mutants. J. Virol. 88:12364–12373. 10.1128/JVI.01381-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody M.A., Zhang R., Walter E.B., Woods C.W., Ginsburg G.S., McClain M.T., Denny T.N., Chen X., Munshaw S., Marshall D.J., et al. 2011. H3N2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PLoS ONE. 6:e25797 10.1371/journal.pone.0025797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyron-Quiroz J.E., Rangel-Moreno J., Kusser K., Hartson L., Sprague F., Goodrich S., Woodland D.L., Lund F.E., and Randall T.D.. 2004. Role of inducible bronchus associated lymphoid tissue (iBALT) in respiratory immunity. Nat. Med. 10:927–934. 10.1038/nm1091 [DOI] [PubMed] [Google Scholar]
- Okuno Y., Isegawa Y., Sasao F., and Ueda S.. 1993. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol. 67:2552–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera T., Takahashi Y., Yokoi Y., Ato M., Kodama Y., Hachimura S., Kurosaki T., and Kobayashi K.. 2012. Memory B cells in the lung participate in protective humoral immune responses to pulmonary influenza virus reinfection. Proc. Natl. Acad. Sci. USA. 109:2485–2490. 10.1073/pnas.1115369109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas L., Foglierini M., Piccoli L., Kallewaard N.L., Turrini F., Silacci C., Fernandez-Rodriguez B., Agatic G., Giacchetto-Sasselli I., Pellicciotta G., et al. 2014. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature. 516:418–422. 10.1038/nature13764 [DOI] [PubMed] [Google Scholar]
- Pica N., Hai R., Krammer F., Wang T.T., Maamary J., Eggink D., Tan G.S., Krause J.C., Moran T., Stein C.R., et al. 2012. Hemagglutinin stalk antibodies elicited by the 2009 pandemic influenza virus as a mechanism for the extinction of seasonal H1N1 viruses. Proc. Natl. Acad. Sci. USA. 109:2573–2578. 10.1073/pnas.1200039109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin S.A. 2013. Complex correlates of protection after vaccination. Clin. Infect. Dis. 56:1458–1465. 10.1093/cid/cit048 [DOI] [PubMed] [Google Scholar]
- Purtha W.E., Tedder T.F., Johnson S., Bhattacharya D., and Diamond M.S.. 2011. Memory B cells, but not long-lived plasma cells, possess antigen specificities for viral escape mutants. J. Exp. Med. 208:2599–2606. 10.1084/jem.20110740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt R.L., Liang H.E., and Locksley R.M.. 2009. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat. Immunol. 10:385–393. 10.1038/ni.1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothaeusler K., and Baumgarth N.. 2010. B-cell fate decisions following influenza virus infection. Eur. J. Immunol. 40:366–377. 10.1002/eji.200939798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittek B., and Rajewsky K.. 1990. Maintenance of B-cell memory by long-lived cells generated from proliferating precursors. Nature. 346:749–751. 10.1038/346749a0 [DOI] [PubMed] [Google Scholar]
- Shih T.A., Roederer M., and Nussenzweig M.C.. 2002. Role of antigen receptor affinity in T cell-independent antibody responses in vivo. Nat. Immunol. 3:399–406. 10.1038/ni776 [DOI] [PubMed] [Google Scholar]
- Skehel J.J., and Wiley D.C.. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531–569. 10.1146/annurev.biochem.69.1.531 [DOI] [PubMed] [Google Scholar]
- Smith K., Garman L., Wrammert J., Zheng N.Y., Capra J.D., Ahmed R., and Wilson P.C.. 2009. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc. 4:372–384. 10.1038/nprot.2009.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Hwang W.C., Perez S., Wei G., Aird D., Chen L.M., Santelli E., Stec B., Cadwell G., Ali M., et al. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16:265–273. 10.1038/nsmb.1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Ohta H., and Takemori T.. 2001. Fas is required for clonal selection in germinal centers and the subsequent establishment of the memory B cell repertoire. Immunity. 14:181–192. 10.1016/S1074-7613(01)00100-5 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Inamine A., Hashimoto S., Haraguchi S., Yoshioka E., Kojima N., Abe R., and Takemori T.. 2005. Novel role of the Ras cascade in memory B cell response. Immunity. 23:127–138. 10.1016/j.immuni.2005.06.010 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Hasegawa H., Hara Y., Ato M., Ninomiya A., Takagi H., Odagiri T., Sata T., Tashiro M., and Kobayashi K.. 2009. Protective immunity afforded by inactivated H5N1 (NIBRG-14) vaccine requires antibodies against both hemagglutinin and neuraminidase in mice. J. Infect. Dis. 199:1629–1637. 10.1086/598954 [DOI] [PubMed] [Google Scholar]
- Tamura S., Ito Y., Asanuma H., Hirabayashi Y., Suzuki Y., Nagamine T., Aizawa C., and Kurata T.. 1992. Cross-protection against influenza virus infection afforded by trivalent inactivated vaccines inoculated intranasally with cholera toxin B subunit. J. Immunol. 149:981–988. [PubMed] [Google Scholar]
- Tarlinton D., and Good-Jacobson K.. 2013. Diversity among memory B cells: origin, consequences, and utility. Science. 341:1205–1211. 10.1126/science.1241146 [DOI] [PubMed] [Google Scholar]
- Taylor J.J., Pape K.A., and Jenkins M.K.. 2012. A germinal center-independent pathway generates unswitched memory B cells early in the primary response. J. Exp. Med. 209:597–606. 10.1084/jem.20111696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throsby M., van den Brink E., Jongeneelen M., Poon L.L., Alard P., Cornelissen L., Bakker A., Cox F., van Deventer E., Guan Y., et al. 2008. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE. 3:e3942 10.1371/journal.pone.0003942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T., Busse C.E., and Wardemann H.. 2009. Cloning and expression of murine Ig genes from single B cells. J. Immunol. Methods. 350:183–193. 10.1016/j.jim.2009.08.009 [DOI] [PubMed] [Google Scholar]
- Tomayko M.M., Steinel N.C., Anderson S.M., and Shlomchik M.J.. 2010. Cutting edge: Hierarchy of maturity of murine memory B cell subsets. J. Immunol. 185:7146–7150. 10.4049/jimmunol.1002163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumpey T.M., Renshaw M., Clements J.D., and Katz J.M.. 2001. Mucosal delivery of inactivated influenza vaccine induces B-cell-dependent heterosubtypic cross-protection against lethal influenza A H5N1 virus infection. J. Virol. 75:5141–5150. 10.1128/JVI.75.11.5141-5150.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora G.D., and Nussenzweig M.C.. 2012. Germinal centers. Annu. Rev. Immunol. 30:429–457. 10.1146/annurev-immunol-020711-075032 [DOI] [PubMed] [Google Scholar]
- Wang T.T., Tan G.S., Hai R., Pica N., Ngai L., Ekiert D.C., Wilson I.A., García-Sastre A., Moran T.M., and Palese P.. 2010. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc. Natl. Acad. Sci. USA. 107:18979–18984. 10.1073/pnas.1013387107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley D.C., Wilson I.A., and Skehel J.J.. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 289:373–378. 10.1038/289373a0 [DOI] [PubMed] [Google Scholar]
- Wrammert J., Koutsonanos D., Li G.M., Edupuganti S., Sui J., Morrissey M., McCausland M., Skountzou I., Hornig M., Lipkin W.I., et al. 2011. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 208:181–193. 10.1084/jem.20101352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Zhang Z., Schramm C.A., Joyce M.G., Kwon Y.D., Zhou T., Sheng Z., Zhang B., O’Dell S., McKee K., et al. NISC Comparative Sequencing Program. 2015. Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection. Cell. 161:470–485. 10.1016/j.cell.2015.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R., Igarashi M., Ozaki H., Kishida N., Tomabechi D., Kida H., Ito K., and Takada A.. 2009. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog. 5:e1000350 10.1371/journal.ppat.1000350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit D.J., Turner D.L., Klonowski K.D., Lefrançois L., and Cauley L.S.. 2006. Residual antigen presentation after influenza virus infection affects CD8 T cell activation and migration. Immunity. 24:439–449. 10.1016/j.immuni.2006.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.