ABSTRACT
The life cycle of hepatitis C virus (HCV) is highly dependent on host cellular proteins for virus propagation. In order to identify the cellular factors involved in HCV propagation, we performed protein microarray assay using the HCV nonstructural 5A (NS5A) protein as a probe. Of ∼9,000 human cellular proteins immobilized in a microarray, approximately 90 cellular proteins were identified as NS5A interactors. Of these candidates, Pim1, a member of serine/threonine kinase family composed of three different isoforms (Pim1, Pim2, and Pim3), was selected for further study. Pim kinases share a consensus sequence which overlaps with kinase activity. Pim kinase activity has been implicated in tumorigenesis. In the present study, we verified the physical interaction between NS5A and Pim1 by both in vitro pulldown and coimmunoprecipitation assays. Pim1 interacted with NS5A through amino acid residues 141 to 180 of Pim1. We demonstrated that protein stability of Pim1 was increased by NS5A protein and this increase was mediated by protein interplay. Small interfering RNA (siRNA)-mediated knockdown or pharmacological inhibition of Pim kinase abrogated HCV propagation. By employing HCV pseudoparticle entry and single-cycle HCV infection assays, we further demonstrated that Pim kinase was involved in HCV entry at a postbinding step. These data suggest that Pim kinase may represent a new host factor for HCV entry.
IMPORTANCE Pim1 is an oncogenic serine/threonine kinase. HCV NS5A protein physically interacts with Pim1 and contributes to Pim1 protein stability. Since Pim1 protein expression level is upregulated in many cancers, NS5A-mediated protein stability may be associated with HCV pathogenesis. Either gene silencing or chemical inhibition of Pim kinase abrogated HCV propagation in HCV-infected cells. We further showed that Pim kinase was specifically required at an early entry step of the HCV life cycle. Thus, we have identified Pim kinase not only as an HCV cell entry factor but also as a new anti-HCV therapeutic target.
INTRODUCTION
Hepatitis C virus (HCV) is a major etiological agent of chronic liver disease, including cirrhosis and hepatocellular carcinoma (1). Approximately 170 million people are chronically infected with HCV, and HCV-related disease leads to 350,000 deaths annually (2). Although recent development of direct-acting antivirals (DAAs) displayed significant progress in HCV treatment regimens, there are still many issues, including unaffordable high cost of drugs, genotypic efficacy, and occasional occurrence of resistance-associated variants. HCV is an enveloped virus with a positive-sense, single-stranded RNA that belongs to the genus Hepacivirus within the family Flaviviridae (3). The 9.6-kb HCV genome encodes a single polyprotein of 3,010 amino acids, which is sequentially processed into 3 structural proteins (core, E1, and E2) and 7 nonstructural proteins (p7 and NS2 to NS5B) (1, 2). Nonstructural 5A (NS5A) is a multifunctional protein consisting of 447 amino acid residues. NS5A protein exists in two different sizes of polypeptide (p56 and p58), which is phosphorylated mainly at serine residues by cellular kinase (3). NS5A protein interacts with many cellular and viral proteins and regulates viral replication and host cellular signaling pathways (4, 5). We have previously reported that NS5A modulates tumor necrosis factor alpha (TNF-α) signaling of the host cells through the interaction with TRAF2 (6) and also regulates TGF-β signaling (7), which are implicated in HCV-associated liver pathogenesis. In addition, we showed that NS5A modulated β-catenin signaling that might play a crucial role in HCV pathogenesis (8). More recently, we reported that NS5A interacted with cellular Pin1 (9) and PI4KIIIα (10), and regulated HCV replication. All these data firmly support the idea that NS5A not only plays an important role in HCV replication but also contributes to HCV-mediated liver pathogenesis.
The provirus integration site for Moloney murine leukemia virus (Pim1) was first identified as an activated gene in Molony murine leukemia virus-induced T cell lymphoma (11). Pim1 belongs to an oncogenic serine/threonine kinase family with two other members, Pim2 and Pim3. Pim1 shares sequence homologies of 71% with Pim2 and 61% with Pim3. Pim1 is a proto-oncogene whose activation promotes the development of cancer in animals (12, 13). Pim kinases are involved in various cellular processes, including cell cycle regulation, proliferation, apoptosis, and signal transduction pathways (11). Overexpression of Pim contributes to malignant transformation and tumorigenesis (14, 15) and the expression levels of Pim proteins are associated with their actual activities. Indeed, it has been previously reported that Pim kinases are upregulated in solid tumors and hepatoma cells (16–18).
Using protein microarray analysis, we have identified approximately 90 NS5A-interacting cellular proteins. Here we show that NS5A physically interacts with Pim1. Protein interaction was verified by both in vitro pulldown and coimmunoprecipitation assays. Moreover, NS5A increased protein stability of Pim1 through downregulation of the polyubiquitination process. Silencing of Pim kinases abrogated HCV propagation. This was further verified by a Pim kinase inhibitor. We further showed that Pim kinases are involved in the entry step of HCV infection. These data suggest that Pim protein may be a legitimate target for anti-HCV therapy.
MATERIALS AND METHODS
Plasmids and DNA transfection.
Total RNAs were isolated from Huh7 cells by using RiboEx (GeneAll), and full-length Pim1, Pim2, Pim3, and BAD were amplified from cDNA synthesized by using a cDNA synthesis kit (Toyobo) according to the manufacturer's instructions. PCR products were inserted into the corresponding enzyme sites of the plasmid pCMV10-3x Flag (Sigma-Aldrich). Pim1 was subcloned into either the plasmid pGEX-4T-1 (Amersham Biosciences AB, Uppsala, Sweden) or pGFP-C1. pEF6B Myc-tagged wild-type and mutant forms of NS5A were described previously (10). A small interfering RNA (siRNA)-resistant Pim2 mutant was constructed by introducing three silent mutations at the siRNA binding site with PCR-based mutagenesis. All DNA transfections were performed by using polyethyleneimine (Sigma-Aldrich) as we described previously (19).
Cell culture.
All cell lines were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin in 5% CO2 at 37°C. Huh6 cells harboring subgenomic replicon derived from genotype 2a were grown as reported previously (19). Primary human hepatocytes were grown in medium supplemented with 5% fetal bovine serum, 1% penicillin-streptomycin, and 1% hepatocyte growth factor in 5% CO2 at 37°C.
GST pulldown assay.
The glutathione S-transferase (GST)-Pim1 fusion protein was expressed in Escherichia coli BL21 and purified with glutathione-Sepharose 4B beads (Amersham Biosciences) according to the manufacturer's instructions. HEK293T cells were transfected with the pEF6B Myc-tagged NS5A plasmid. At 24 h after transfection, cells were harvested in lysis buffer. The cell lysate was centrifuged at 13,500 rpm for 15 min, and then protein concentration was determined by the Bradford assay (Bio-Rad). For the in vitro binding assay, Myc-tagged NS5A was incubated with either GST or GST-Pim1 fusion protein for 2 h at 4°C in cell lysis buffer. The samples were washed four times in lysis buffer, and then bound protein was detected by immunoblot assay.
Immunoprecipitation.
HEK293T cells were cotransfected with Flag-tagged Pim1 and Myc-tagged NS5A plasmid. Total amounts of DNA were adjusted by adding an empty vector. At 48 h after transfection, cell lysates were centrifuged at 13,500 rpm for 15 min. The supernatant was incubated at 4°C overnight with the appropriate antibody. The samples were further incubated with 30 μl of protein A beads (Sigma-Aldrich) for 1 h. The beads were washed five times in washing buffer, and then bound protein was detected by immunoblot assay as we described previously (10).
Protein stability assay.
Huh7.5 cells were transiently transfected with various plasmids. At 24 h after transfection, cells were treated with 20 μg/ml cycloheximide (Sigma-Aldrich) for various times. The resulting cell lysates were immunoblotted with the appropriate antibodies to monitor protein turnover.
RNA interference.
siRNAs targeting Pim1 (sense, 5′-ACAUUUACAACUCAUUCCA-3′; antisense, 5′-UGGAAUGAGUUGUAAAUGU-3′), Pim2 (sense, 5′-GUGGAGUUGUCCAUCGUGACA-3′; antisense, 5′-CUCCTCUUCUGGTUGCUCTGT-3′), and Pim3 (sense, 5′-GGCGTGCTTCTCTACGATA-3′; antisense, 5′-CCGCACGAAGAGATGCTAT-3′), as well as Pim3 #2 for the siRNA resistance experiment (sense, 5′-CCCUGGGUGGAUACUUGAA-3′; antisense, 5′-UUCAAGUAUCCACCCA GGG-3′) and the universal negative-control siRNA, were purchased from Bioneer. siRNA transfection was performed using a Lipofectamine RNAiMax reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Immunoblot assay.
Immunoblot analysis was performed as we described previously (19) using the following antibodies: rabbit anti-NS5A, rabbit anti-core, and rabbit anti-NS3 from Byung-Yoon Ahn (Korea University), rabbit anti-Pim1 (Cell signaling), anti-Pim2 (Cell Signaling), anti-Pim3 (Cell Signaling), mouse anti-β actin (Sigma-Aldrich), mouse anti-Myc (Santa Cruz), mouse anti-ubiquitin (Santa Cruz), and mouse anti-Flag (Sigma-Aldrich). Either horseradish peroxidase-conjugated goat anti-rabbit antibody or goat anti-mouse antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) was used as a secondary antibody.
HCV pseudoparticle entry assay.
HCV pseudoparticles (HCVpp) with E1 and E2 glycoproteins derived from genotype 1a (H77) or genotype 2a (JFH-1) and vesicular stomatitis virus pseudoparticles (VSVpp) were generated as previously described (20). Briefly, HEK293T cells were transfected with the glycoprotein-expressing plasmid, Gag-Pol (polymerase)-packaging plasmid, and transfer vector encoding the firefly luciferase reporter protein by using polyethyleneimine (Sigma-Aldrich). Supernatants containing HCVpp or VSVpp were collected 48 h after transfection. For the infection assay, Huh7.5 cells or primary human hepatocyte were either transfected with siRNAs for 48 h or treated with Pim kinase inhibitor either for 2 h or 36 h. Cells were then infected with either HCVpp or VSVpp for 6 h. Unbound viruses were removed and cells were replaced with respective culture media. At 72 h postinfection, cells were harvested and luciferase activity was analyzed.
Single-cycle HCV infection.
Single-round infectious HCV (HCVsc) was generated from a replicon trans-packaging system as previously described (21, 22). Briefly, Huh7.5 cells were cotransfected with pHH/SGR-Luc plasmid, which carries a bicistronic HCV subgenomic (NS3-5) replicon firefly luciferase reporter with a Pol I promoter/terminator, and an HCV core-NS2 expression plasmid by using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Culture medium containing HCV-like infectious particles (HCV-LP) was collected at day 4 after transfection. For the single-cycle HCV infection assay, Huh7.5 cells were treated with either siRNAs or Pim kinase inhibitor before infection with HCVsc. At day 2 postinfection, cells were harvested and luciferase activity was analyzed.
Luciferase reporter assay.
For the dual-luciferase reporter assay, Huh7.5 cells were transfected with pRL-HL plasmid and pCH110 reference plasmid as we described previously (19). The cells were then incubated with various dosages of Pim kinase inhibitor. Following incubation for 48 h, cells were harvested and dual-luciferase assays were performed according to the manufacturer's instructions (Promega). For the HCV-LP reporter assay, Huh7.5 cells were pretreated with either dimethyl sulfoxide (DMSO) (vehicle) or Pim inhibitor for 2 h or treated with various siRNAs for 48 h and then inoculated with HCV-LP. At 48 h postinfection, cells were harvested, and then luciferase activity was determined.
Immunofluorescence assay.
Huh 7.5 cells seeded on cover slides were infected with Jc1 for 48 h, and then green fluorescent protein (GFP)-tagged Pim1 was transfected for 16 h. Cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min and then permeabilized with 0.1% Triton X-100 in PBS for 10 min at 37°C. After three washes with PBS, fixed cells were blocked with 1% bovine serum albumin (BSA) in PBS for 1 h at room temperature. The cells were then incubated with a rabbit anti-NS5A antibody overnight at 4°C. After three washes with PBS, cells were incubated with either tetramethylrhodamine isothiocyanate (TRITC)-conjugated donkey anti-rabbit IgG for 1 h at room temperature. Cells were further incubated with BODIPY (439/503) (Invitrogen) to detect lipid droplets (green). Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to label nuclei. After three washes with PBS, cells were analyzed using the Zeiss LSM 700 laser confocal microscopy system (Carl Zeiss, Inc., Thornwood, NY).
WST assay.
Approximately 1.5 × 104 cells seeded on 24-well plates were either transfected with various siRNAs or treated with Pim inhibitor. Cell viability was measured by using water-soluble tetrazolium salt (WST) (Dail Lab) according to the manufacturer's protocol.
Pim inhibitor assay.
Pim-kinase inhibitor IX, SGI-1776 (Merck Millipore), was dissolved in DMSO, and then aliquots were stored at −20°C. Huh7.5 cells were infected with Jc1 for 4 h and then incubated in complete medium containing various amounts of Pim inhibitor. At 48 h or 72 h postinfection, cells were harvested, and both protein and RNA levels were analyzed as described above.
Statistical analysis.
Data are presented as means and standard deviations (SDs). Student's t test was used for statistical analysis.
RESULTS
Identification of Pim1 as an NS5A interactor in protein array.
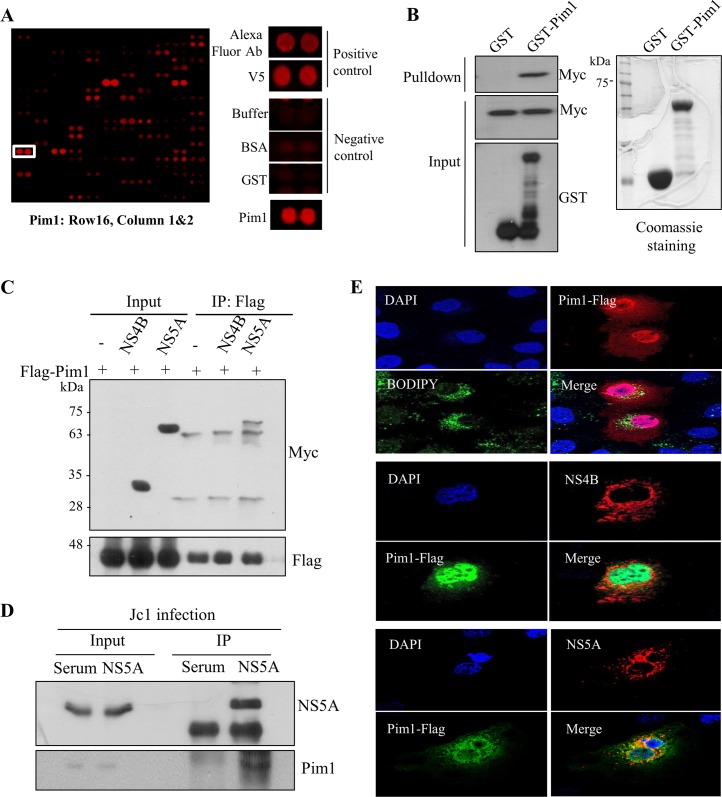
To identify cellular proteins interacting with HCV NS5A protein, we performed protein microarray assays using the HCV NS5A protein as a probe. Statistically significant candidate interactors were identified by using methods we reported previously (23). Approximately 90 cellular proteins were identified as HCV NS5A interactors (Table 1). Pim1 was identified as one of the candidate hits, and both positive- and negative-control hits are shown in Fig. 1A. Since overexpression of Pim contributes to tumorigenesis and progression of a variety of malignancies (24), we selected Pim1 for further characterization. First, to verify the protein array data, we performed an in vitro GST pulldown assay using GST-Pim1 purified from E. coli and cell lysates expressing Myc-tagged NS5A. Figure 1B shows that NS5A selectively interacted with Pim1. Coimmunoprecipitation data further demonstrated that NS5A specifically interacted with Pim1 (Fig. 1C). Next, we investigated whether Pim1 interacted with NS5A in the context of HCV infection. Huh7.5 cells were infected with HCV Jc1. Cell lysates harvested at 72 h after HCV infection were immunoprecipitated with either control serum or an anti-NS5A antibody, and then bound proteins were analyzed by immunoblotting with an anti-Pim1 antibody. Indeed, endogenous Pim1 protein interacted with NS5A in Jc1-infected cells (Fig. 1D). These data suggest that Pim1 may colocalize with NS5A in HCV-infected cells. To investigate this possibility, Huh7 cells were either transfected with Myc-tagged NS4B or infected with Jc1, and then cells were analyzed by an immunofluorescence assay. Pim1 was widely expressed in both the nucleus and the cytoplasm as reported previously (11). As shown in Fig. 1E, both Pim1 and NS5A were colocalized in the cytoplasm, as indicated by the yellow fluorescence in the merged image. However, neither the lipid droplet nor NS4B was colocalized with Pim1. Collectively, these data suggest that Pim1 specifically interacts with NS5A both in vitro and in vivo.
TABLE 1.
Candidate NS5A interactors identified by protein microarray screening
| Category | Gene name | Protein header/functiona | Database ID | Z-score |
|---|---|---|---|---|
| Enzyme activity | BIRC4 | Baculoviral IAP repeat-containing 4 | NM_001167.2 | 20.67644 |
| SMYD2 | SET and MYND domain-containing protein 2 | NM_020197.1 | 12.68368 | |
| NUDT16L1 | Nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 1 | NM_032349.1 | 8.37184 | |
| DYNLRB2 | Dynein, light chain, roadblock type 2 | BC054892.1 | 6.37752 | |
| PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol specific) | BC007565.1 | 6.27688 | |
| AHCYL1 | S-Adenosyl homocysteine hydrolase-like 1 (AHCYL1) | NM_006621.3 | 5.77758 | |
| SMYD2 | SET and MYND domain-containing protein 2 | BC094747.1 | 5.72049 | |
| LENG1 | Leukocyte receptor cluster (LRC) member 1 | NM_024316.1 | 5.87047 | |
| PRO1853 | Protein MidA homolog, mitochondrial | NM_144736.3 | 4.7122 | |
| BAZ2B | Bromodomain adjacent to zinc finger domain, 2B, mRNA | BC012576.1 | 4.16548 | |
| FOXN3 | Forkhead box protein N3 | NM_005197.2 | 4.0929 | |
| BIRC3 | Baculoviral IAP repeat-containing 3, transcript variant 1 | NM_001165.3 | 4.29417 | |
| ERH | Enhancer of rudimentary homolog (Drosophila) | NM_004450.1 | 3.97872 | |
| APEX2 | APEX nuclease (apurinic/apyrimidinic endonuclease) 2 | NM_014481.2 | 3.53651 | |
| AFF4 | AF4/FMR2 family, member 4 | BC025700.1 | 3.5936 | |
| LMX1A | LIM homeobox transcription factor 1, alpha | BC066353.1 | 3.57715 | |
| AHCYL2 | Putative adenosyl homocysteinase 2 | 3.52102 | ||
| MED4 | Mediator of RNA polymerase II transcription subunit 4 | NM_014166.2 | 3.5665 | |
| PTK6 | PTK6 protein tyrosine kinase6 | NM_005975.2 | 3.37491 | |
| NR1D1 | Nuclear receptor subfamily 1, group D, member 1 | BC056148.1 | 3.31879 | |
| MSL3L1 | Male-specific lethal 3-like 1 (Drosophila) (MSL3L1), transcript variant 2 | NM_078630.1 | 3.20073 | |
| ATP5O | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | NM_001697.1 | 3.13397 | |
| Kinase activity | MOBKL1A | Mps one binder kinase activator-like 1A (yeast) | NM_173468.2 | 8.7318 |
| PIM1 | pim-1 oncogene | NP_002639 | 7.79512 | |
| CASS4 | Cas scaffolding protein family member 4 | BC027951.1 | 5.99433 | |
| STK40 | Serine/threonine kinase 40 | NM_032017.1 | 5.61888 | |
| PBK | PDZ binding kinase | NP_060962 | 5.18151 | |
| FGFR2 | Fibroblast growth factor receptor 2 | NP_075261 | 4.65511 | |
| BMX | BMX nonreceptor tyrosine kinase, transcript variant 2 | NP_001712 | 4.47222 | |
| CDC42BPA | CDC42 binding protein kinase alpha (DMPK-like) | NP_055641 | 4.49835 | |
| CRK | v-crk sarcoma virus CT10 oncogene homolog (avian), transcript variant II | NM_016823.2 | 4.25547 | |
| CRKL | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | NM_016823.2 | 4.17322 | |
| NTRK2 | Neurotrophic tyrosine kinase, receptor, type 2 (NTRK2), transcript variant b | NM_024946.1 | 4.15096 | |
| PACSIN1 | Protein kinase C and casein kinase substrate in neurons 1 | NM_020804.2 | 4.64833 | |
| KDR | Kinase insert domain receptor (a type III receptor tyrosine kinase) | NP_002244 | 3.94292 | |
| BCKDK | Branched chain keto acid dehydrogenase kinase | BC007363.1 | 3.91099 | |
| CAMK2D | Calcium/calmodulin-dependent protein kinase II delta, transcript variant 3 | NP_742113 | 4.00775 | |
| PLK1 | Polo-like kinase 1 (Drosophila) (PLK1) | NM_005030.2 | 3.58005 | |
| PLK1 | Serine/threonine-protein kinase PLK1 | BC002369.1 | 3.41265 | |
| Phosphatidylinositol binding | SNX10 | Sorting nexin 10 | NM_013322.2 | 3.74939 |
| SNX9 | Sorting nexin-9 | NM_016224.3 | 3.77455 | |
| SNX11 | Sorting nexin 11, transcript variant 2 | NM_013323.1 | 3.64875 | |
| Protein binding | LASP1 | LIM and SH3 domain protein 1 | NM_006148.1 | 33.05845 |
| CTTN | Cortactin (CTTN), transcript variant 2 | NM_138565.1 | 22.69205 | |
| ABI1 | Abl-interactor 1 | BC024254.1 | 16.00561 | |
| MOBK1B | Mps one binder kinase activator-like 1B (yeast) | BC003398.1 | 9.54172 | |
| NCK2 | NCK adaptor protein 2 | BC000103.1 | 9.12951 | |
| LOC100134934 | cDNA clone MGC:54300 IMAGE:6500495, complete CDS | BC047782.1 | 8.00897 | |
| BIRC7 | Baculoviral IAP repeat-containing 7 (livin) | BC014475.1 | 5.61211 | |
| SRI | Sorcin, transcript variant 1 | NM_003130.1 | 5.75242 | |
| C10orf63 | Chromosome 10 open reading frame 63 | NM_145010.1 | 4.65994 | |
| NOSTRIN | Nitric oxide synthase trafficker | NM_001039724.1 | 4.61737 | |
| RSRC1 | Arginine/serine-rich coiled-coil protein 1 | BC010357.1 | 4.25257 | |
| ATPAF2 | ATP synthase mitochondrial F1 complex assembly factor 2 | NM_145691.3 | 3.67391 | |
| RPH3AL | Rabphilin 3A-like (without C2 domains) | BC005153.1 | 3.64295 | |
| RTF1 | Rtf1, Paf1/RNA polymerase II complex component, homolog (Saccharomyces cerevisiae) | NM_015138.2 | 3.47942 | |
| MAPRE1 | Microtubule-associated protein, RP/EB family, member 1 | NM_012325.1 | 3.47942 | |
| PRC1 | Protein regulator of cytokinesis 1, transcript variant 1 | NM_003981.2 | 3.44361 | |
| APTX | Aprataxin, transcript variant 4 | NM_017692.1 | 3.18041 | |
| Receptor signaling complex scaffold | BIN1 | Bridging integrator 1 | BC004101.1 | 31.31669 |
| BIN1 | Bridging integrator 1, transcript variant 6 | NM_139348.1 | 25.38501 | |
| NCK1 | NCK adaptor protein 1 | NM_006153.3 | 18.70147 | |
| PSTPIP1 | Proline-serine-threonine phosphatase interacting protein 1 | NM_003978.2 | 15.95336 | |
| OSTF1 | Osteoclast stimulating factor 1 | NM_012383.2 | 19.18626 | |
| PACSIN2 | Protein kinase C and casein kinase substrate in neuron 2 | BC008037.2 | 16.05689 | |
| TRAF2 | TNF receptor-associated factor 2 | NM_021138.2 | 13.22362 | |
| AMPH | Amphiphysin | BC034376.1 | 11.85537 | |
| BAIAP2 | BAI1-associated protein 2, transcript variant 2 | NM_017451.1 | 3.4949 | |
| RNA binding | ZRANB2 | Zinc finger, RAN-binding domain containing 2, transcript variant 1 | NM_203350.1 | 7.76416 |
| DDX49 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 | NM_019070.1 | 5.74855 | |
| HSPC148 | Spliceosome-associated protein homolog (S. cerevisiae) | BC040946.1 | 5.90144 | |
| PPIE | Peptidylprolyl isomerase E (cyclophilin E) (PPIE), transcript variant 3 | NM_203457.1 | 4.34933 | |
| RAD51AP1 | RAD51-associated protein 1 | BC016330.1 | 3.49199 | |
| EIF4E2 | Eukaryotic translation initiation factor 4E family member 2 | NM_004846.1 | 3.32846 | |
| Other/unknown | C7orf49 | Chromosome 7 open reading frame 49 | NM_024033.1 | 8.27604 |
| PCLO | Piccolo (presynaptic cytomatrix protein) | BC001304.1 | 6.43267 | |
| C22orf33 | Chromosome 22 open reading frame 33 | NM_178552.2 | 5.93821 | |
| C6orf60 | Family with sequence similarity 184, member A | BC009055.1 | 5.74081 | |
| MGC24125 | Hypothetical protein MGC24125 | NM_173796.2 | 5.7021 | |
| MAB21L2 | Protein MAb-21-like 2 | NM_006439.3 | 4.98314 | |
| SH3YL1 | SH3 domain containing, Ysc84-like 1 (S. cerevisiae) | BC008374.1 | 4.57286 | |
| NIP30 | NEFA-interacting nuclear protein NIP30 | NM_024946.1 | 4.13742 | |
| ODF2L | Outer dense fiber of sperm tails 2-like | BC009779.1 | 3.99614 | |
| CCDC149 | Coiled-coil domain containing 149 | 3.88679 | ||
| SNX10 | Sorting nexin 10 | NM_013322.2 | 3.74939 | |
| C9orf19 | Chromosome 9 open reading frame 19 | BC017918.1 | 3.74358 | |
| SH3YL1 | SH3 domain containing, Ysc84-like 1 (S. cerevisiae), mRNA (cDNA clone MGC:12275 IMAGE:3996120), complete CDS | BC008375.1 | 3.86647 | |
| CCDC55 | Coiled-coil domain containing 55, transcript variant 1 | NM_032141.1 | 3.51135 | |
| FAM104A | Family with sequence similarity 104, member A, transcript variant 2 | NM_032837.1 | 3.46587 | |
| MAGEB4 | Melanoma antigen family B, 4 | BC032852.2 | 3.22202 |
CDS, coding sequence; IAP, inhibitor of apoptosis; MAB; male abnormal.
FIG 1.
(A) Identification of Pim1 in a protein microarray. Both positive and negative controls are shown. (B) Pim1 interacts with HCV NS5A protein. HEK293T cells were transfected with Myc-tagged NS5A, and total cell lysates harvested at 24 h after transfection were incubated with either GST or GST-Pim1. After pulldown by GST beads, bound protein was detected by immunoblot analysis with an anti-Myc antibody. (C) HEK293T cells were cotransfected with Myc-tagged NS5A and Flag-tagged Pim1. At 36 h after transfection, total cell lysates were immunoprecipitated with an anti-Flag antibody, and bound protein was detected by immunoblot analysis with an anti-Myc antibody. (D) Huh7.5 cells were infected with Jc1 for 4 h. At 48 h postinfection, cell lysates were immunoprecipitated with either control serum or an anti-NS5A antibody. Bound proteins were analyzed by immunoblotting with an anti-Pim1 antibody. (E) Huh7 cells seeded on glass coverslips were either transfected with Myc-tagged NS4B or infected with Jc1. At 48 h postinfection, cells were transfected with Flag-tagged Pim1. For lipid droplet staining, Huh7 cells were transfected with Flag-tagged Pim1. At 16 h after transfection, cells were fixed in 4% paraformaldehyde and were further incubated with BODIPY to detect lipid droplets (green). Immunofluorescence staining was performed by using the indicated antibodies. Dual staining showed colocalization of Pim1 and NS5A in the cytoplasm as yellow fluorescence in the merged image. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to label nuclei (blue).
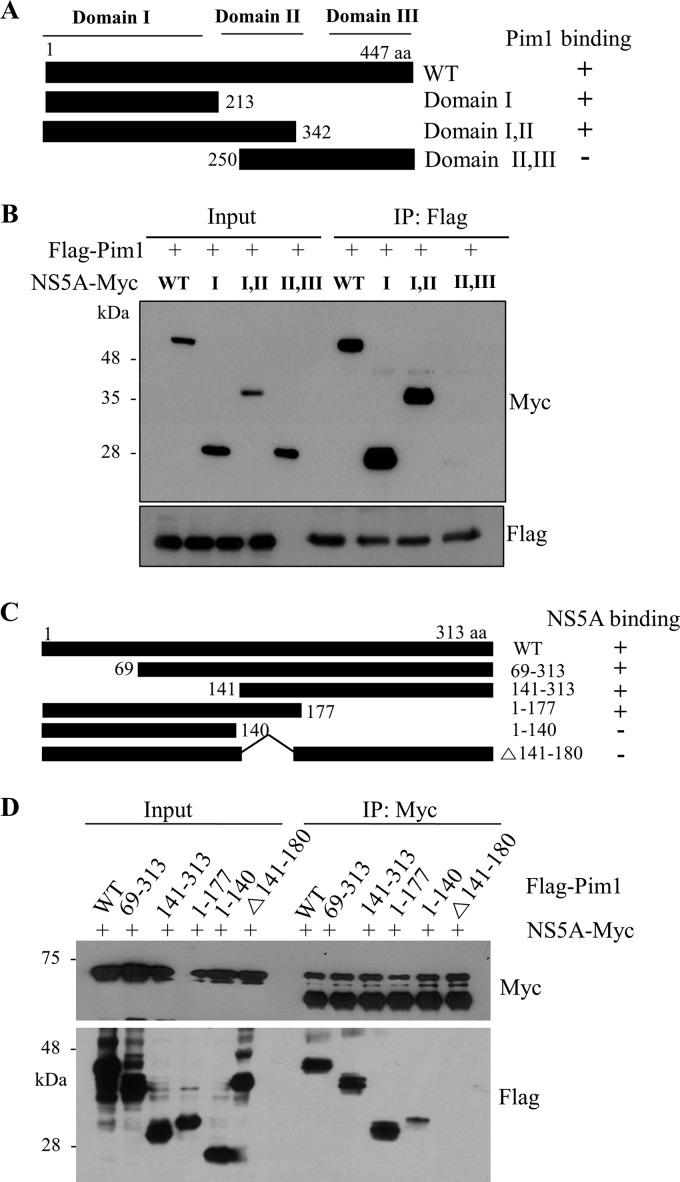
HCV NS5A protein interacts with Pim1 through domain I of NS5A and amino acid residues 141 to 180 of Pim1.
To determine the region in NS5A responsible for Pim1 binding, the interaction between Pim1 and various truncated mutants of NS5A (Fig. 2A) was determined by a transfection-based coimmunoprecipitation assay. As shown in Fig. 2B, Pim1 interacted with domain I and the region spanning domains I and II but not with the region spanning domains II and III. This result indicated that domain I was responsible for binding with Pim1. Next, we determined the region in Pim1 for NS5A binding. We constructed various deletion mutants of Pim1 (Fig. 2C) based on a previous report (25). Figure 2D demonstrated that NS5A interacted with mutants 1–177 and 141–313 but not with mutants 1–140 and Δ141–180, indicating that NS5A interacted with Pim1 through the region encompassing amino acids (aa) 141 to 180 of Pim1. In fact, this sequence contains aspartic acid, which is essential for the activity of Pim kinase. This amino acid is well conserved in all Pim kinase family members (13).
FIG 2.
HCV NS5A interacts with Pim1 through the domain I of NS5A and amino acid residues 141 to 177 of Pim1. (A) Schematic illustration of both wild-type and mutant forms of the NS5A expression plasmid. (B) Pim1 interacts with the domain I of NS5A. HEK293T cells were cotransfected with Flag-tagged Pim1 and Myc-tagged NS5A expression plasmids. At 36 h after transfection, cell lysates were immunoprecipitated with an anti-Flag antibody, and bound proteins were immunoblotted with an anti-Myc antibody. Expression of Myc-tagged NS5A and Flag-tagged Pim1 was verified by immunoblotting with an anti-Myc or anti-Flag antibody using the same cell lysates. (C) Schematic illustration of both wild-type and mutant Pim1expression plasmids. (D) NS5A interacts with amino acid residues 141 to 177 of Pim1. HEK293T cells were cotransfected with Myc-tagged NS5A and Flag-tagged Pim1 expression plasmids. At 36 h after transfection, cell lysates were immunoprecipitated with an anti-Myc antibody, and bound proteins were immunoblotted with an anti-Flag antibody. Protein expressions of Myc-tagged NS5A and Flag-tagged Pim1 were verified by immunoblotting with an anti-Myc or anti-Flag antibody using the same cell lysates.
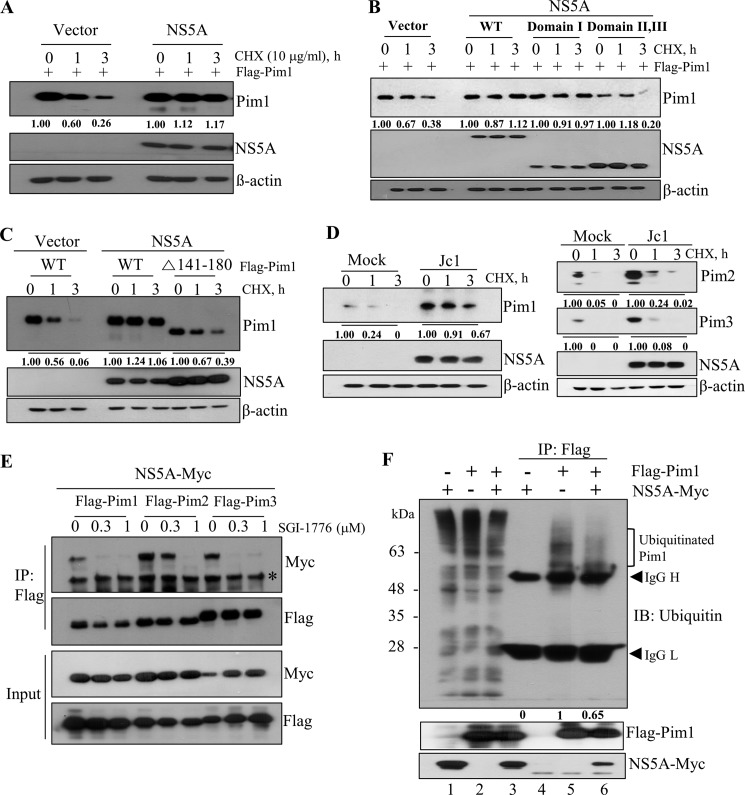
NS5A stabilizes the Pim1 protein.
Unlike other kinases, Pim kinases have no regulatory domain. Therefore, once Pim proteins are expressed, they constitutively maintain an active state, implying that expression of Pim protein itself correlated with its kinase activity (11, 12). We postulated that NS5A might regulate Pim1 protein stability to maintain its kinase activity. To verify this possibility, Huh7.5 cells were cotransfected with Flag-tagged Pim1 and Myc-tagged NS5A expression plasmid. Cells were then treated with cycloheximide (CHX), and protein stability was analyzed. As shown in Fig. 3A, the level of Pim1 was gradually decreased in CHX-treated vector control cells, whereas the Pim1 level was unchanged in the presence of NS5A protein. To investigate whether protein interplay was required for protein stability, we analyzed Pim1 protein stability by using a binding-defective mutant of NS5A. As shown in Fig. 3B, protein stability of Pim1 was unaltered by both wild-type NS5A and domain I of NS5A, whereas Pim1 protein was gradually degraded in the presence of a binding-defective domain II,III mutant of NS5A. To further verify this result, we performed a protein stability assay by using binding-defective Pim1. Figure 3C shows that the level of wild-type Pim1 was stable in the presence of NS5A, whereas the level of the binding-defective Δ141–180 mutant was gradually decreased, indicating that protein interplay between NS5A and Pim1 contributes to the stability of Pim1. Since Pim kinase family members share sequence homology with overlapping functions (12), we hypothesized that stabilities of other Pim kinases might also be regulated by NS5A protein in Jc1-infected cells. To explore this possibility, Huh7.5 cells infected with Jc1 for 72 h were treated with CHX, and protein levels of Pim1, Pim2, and Pim3 were determined by immunoblot analysis. As expected, Pim kinases were highly stabilized in Jc1-infected cells compared to mock-infected cells (Fig. 3D). We verified that stabilities of Pim kinases were prominently stabilized by NS5A (data not shown). This raised the possibility that NS5A might interact with other Pim kinases. Indeed, we demonstrated that NS5A interacted with both endogenous Pim2 and Pim3 kinases (data not shown). Taken together, these data show that protein interplay between NS5A and Pim kinases contributes to protein stability of Pim kinases. We then asked whether kinase activity was required for protein interaction between NS5A and Pim proteins. HEK293T cells were treated with increasing amounts of SGI-1776 and Pim kinase inhibitor and then cotransfected with Myc-tagged NS5A together with each Flag-tagged Pim kinase plasmid. At 36 h after transfection, cell lysates were immunoprecipitated with an anti-Flag antibody, and then bound proteins were detected by immunoblot analysis using an anti-Myc antibody. As shown in Fig. 3E (bottom), NS5A protein expression levels were unaffected by Pim kinase inhibitor. However, coimmunoprecipitated NS5A protein levels were gradually decreased with increasing amounts of SGI-1776, indicating that protein interplay between NS5A and Pim required kinase activity of Pim. To investigate how the ubiquitination pattern of Pim1 was altered by NS5A, HEK293T cells were cotransfected with Myc-tagged NS5A and Flag-tagged Pim1. Cell lysates harvested at 36 h after transfection were immunoprecipitated with an anti-Flag antibody, and then bound proteins were immunoblotted using an antiubiquitin antibody to detect ubiquitination. As demonstrated in Fig. 3F, the level of ubiquitylated Pim1 was decreased in the presence of NS5A (lane 6 versus lane 5). These data clearly show that NS5A promotes Pim1 stability through protein interplay.
FIG 3.
NS5A stabilizes the Pim1 protein. (A) Huh7.5 cells were cotransfected with Flag-tagged Pim1 and either vector control or Myc-tagged NS5A expression plasmid. At 30 h after transfection, cells were treated with 10 μg/ml of CHX for the indicated times, and protein expression levels were analyzed by immunoblot analysis with the indicated antibodies. (B) Huh7.5 cells were cotransfected with Flag-tagged Pim1 and various Myc-tagged NS5A expression plasmids. Cells were treated as described for panel A, and protein expression levels were analyzed. (C) Huh7.5 cells were cotransfected with the wild-type or mutant form of Pim1 in the absence (vector) or presence of NS5A. At 30 h after transfection, cells were treated with 10 μg/ml of CHX for the indicated times, and protein expression levels were determined by immunoblot analysis with the indicated antibodies. (D) Huh7.5 cells were infected with Jc1 for 72 h. Cells were treated with 10 μg/ml CHX for the indicated times, and protein expression levels were determined by immunoblot analysis with the indicated antibodies. (E) HEK293T cells were treated with increasing amounts of Pim kinase inhibitor for 1 h and then cotransfected with each of Flag-tagged Pim kinase and Myc-tagged NS5A plasmids. At 36 h after transfection, cell lysates were immunoprecipitated with an anti-Flag antibody, and then bound proteins were detected by immunoblot analysis using an anti-Myc antibody. (F) Ubiquitination of Pim1 is decreased by NS5A protein. HEK293T cells were cotransfected with Myc-tagged NS5A and Flag-tagged Pim1. At 36 h after transfection, total cell lysates were immunoprecipitated with anti-Flag antibody, and then bound proteins were immunoblotted with an antiubiquitin antibody.
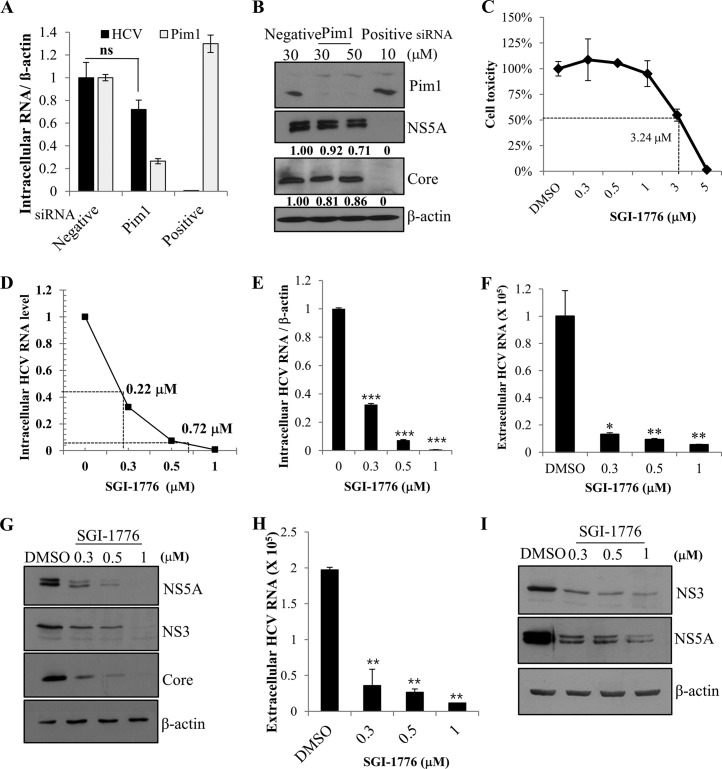
Kinase activity of Pim protein is required for HCV propagation.
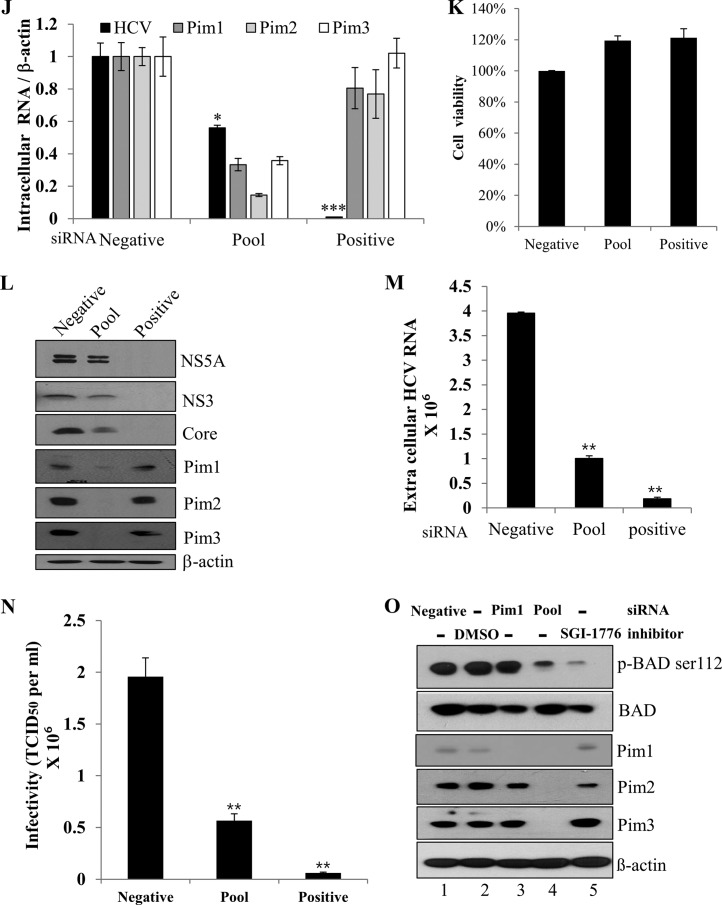
To investigate the functional involvement of Pim1 in HCV propagation, Huh7.5 cells transfected with siRNAs were infected with Jc1. At 48 h postinfection, both intracellular HCV RNA and Pim1 RNA levels were determined. We showed that silencing of Pim1 had little effect on intracellular HCV RNA level (Fig. 4A). Similarly, HCV protein levels were unaffected by knockdown of Pim1 (Fig. 4B). Since Pim family proteins share a high sequence homology and functionally overlap in various cellular processes, we investigated the functional role of Pim proteins in HCV propagation by using SGI-1776. SGI-1776 specifically inhibits all Pim kinase activities. It binds the ATP-binding pocket of Pim and has been proven to have antitumor activity (26). We first determined a 50% cytotoxic concentration for SGI-1776 by a water-soluble-tetrazolium assay and showed that treatment of Pim inhibitor did not induce any cell toxicity at concentrations up to 1 μM (Fig. 4C). We also established a dose response curve of the antiviral activity of SGI-1776 (Fig. 4D). The 50% and 90% inhibitory concentrations (IC50 and IC90) for SGI-1776 were 0.22 μM and 0.72 μM, respectively. To investigate the effects of Pim inhibitor on HCV propagation, Huh7.5 cells were pretreated with various concentrations of SGI-1776 and then infected with Jc1. The cells were further incubated with culture medium containing either DMSO or SGI-1776 for 48 h. We demonstrated that intracellular HCV RNA level (Fig. 4E), extracellular HCV RNA level (Fig. 4F), and HCV protein levels (Fig. 4G) were dramatically decreased by Pim kinase inhibitor and that inhibition occurred in a dose-dependent manner. To further verify these results, naive Huh7.5 cells were infected with culture supernatant harvested from the experiment whose results are shown in Fig. 4E. As shown in Fig. 4H and I, both extracellular HCV RNA and HCV infectivity levels were significantly decreased in kinase inhibitor-treated cells. Since HCV propagation was impaired by Pim kinase inhibitor but not by knockdown of Pim1 alone, we speculated that other Pim kinases might be involved in HCV propagation. We therefore investigated the effect of knockdown of all Pim kinases on HCV propagation by using a pool of siRNA constructs targeting all Pim kinases. As shown in Fig. 4J, intracellular HCV RNA level was significantly decreased in all Pim kinase knockdown cells without causing any cell toxicity (Fig. 4K). Similarly, HCV protein expression levels (Fig. 4L) and extracellular HCV RNA levels (Fig. 4M) were significantly reduced by knockdown of all three Pim kinases. To further verify these results, the effects of knockdown of all Pim kinases on viral infectivity were determined by using 50% tissue culture infective doses (TCID50). As shown in Fig. 4N, HCV titer was significantly decreased in all Pim kinase knockdown cells. To determine whether siRNA-mediated knockdown or the use of a kinase inhibitor was more potent to suppress Pim kinase activity, we analyzed phosphorylation status of BAD, which is one of the substrates of Pim kinase. For this purpose, Huh7.5 cells were either transfected with siRNAs (Fig. 4O, lanes 1, 3, and 4) or treated with Pim kinase inhibitor (lanes 2 and 5). Cells were then transfected with Flag-tagged BAD plasmid. At 24 h after transfection, Pim kinase activity was determined by using an anti-phospho-BAD antibody. As shown in Fig. 4O, knockdown of Pim1 had no perceptible effect on phosphorylation of BAD in Huh7.5 cells (lane 3), whereas knockdown of all Pim kinases showed a drastic decrease in phosphorylation of BAD protein (lane 4). It is worth noting that the level of phosphorylation of BAD was more decreased in kinase inhibitor treated cells than in Pim knockdown cells (Fig. 4O, lane 5 versus lane 4), indicating that pharmacological inhibition was more effective than siRNA-mediated silencing of Pim kinases.
FIG 4.
Kinase activity of Pim is required for HCV propagation. (A) Huh7.5 cells were transfected with the indicated siRNA constructs for 48 h and then infected with Jc1 for 4 h. At 48 h postinfection, intracellular HCV RNA and Pim1 RNA level were analyzed by qRT-PCR. (B) Huh7.5 cells were treated as described for panel A, and total cell lysates were immunoblotted with the indicated antibodies. “Negative” indicates irrelevant negative siRNA; “positive” indicates HCV-specific siRNA. (C) Huh7.5 cells were treated with either DMSO or various doses of SGI-1776 for 48 h and then cell toxicity together, and 50% cytotoxic concentrations were determined by using a water-soluble-tetrazolium assay. (D) Huh7.5 cells were pretreated with either DMSO or various doses of SGI-1776 for 1 h and then infected with Jc1 for 4 h. After 48 h of incubation, the IC50 and IC90 for SGI-1776 were determined in a graph. (E and F) Huh7.5 cells were pretreated with either DMSO or various doses of SGI-1776 for 1 h and then infected with Jc1 for 4 h. Following 48 h cultivation in the presence of either DMSO or SGI-1776, both intracellular HCV RNA levels (E) and extracellular HCV RNA levels (F) were analyzed by qRT-PCR. The asterisks indicate significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (G) Total cell lysates harvested from the set of experiment described for panel E were immunoblotted with the indicated antibodies. (H) Cell culture supernatants were harvested from the experiment whose results are shown in panel E, and extracellular HCV RNAs were analyzed by qRT-PCR. The asterisks indicate significant differences (**, P < 0.01). (I) Naive Huh7.5 cells were infected with culture supernatants harvested from the experiment whose results are shown in panel E for 48 h, and protein expression was analyzed by immunoblotting with the indicated antibodies. (J to M) Huh7.5 cells were transfected with a 30 nM Pim siRNA pool targeting Pim1, Pim2, and Pim3 mRNA. At 48 h after transfection, cells were infected with Jc1 for 4 h. At 48 h postinfection, intracellular RNA levels (J), protein levels (L), and extracellular HCV RNA levels (M) were analyzed. The asterisks indicate significant differences (**, P < 0.01; ***, P < 0.001). “Pool” indicates siRNAs targeting mRNAs of Pim1, Pim2, and Pim3. (K) Huh7.5 cells were treated as described for panel J, and cell viability was determined by WST assay. (N) Huh7.5 cells were transfected with the indicated siRNAs for 48 h and then GFP-tagged Jc1 cells were infected for 72 h. HCV infectivity was determined by limiting dilution assays. Infected cells were assessed with a fluorescent cell analyzer microscope. (O) Huh7.5 cells were either transfected with the indicated siRNAs (lanes 1, 3, and 4) for 24 h or treated with DMSO or Pim inhibitor (lanes 2 and 5) for 2 h. Cells were then transfected with Flag-tagged BAD plasmid. At 24 h after transfection, total cell lysates were analyzed for protein expression, and Pim kinase activity was indirectly determined by immunoblot analysis using an anti-phospho-BAD antibody.
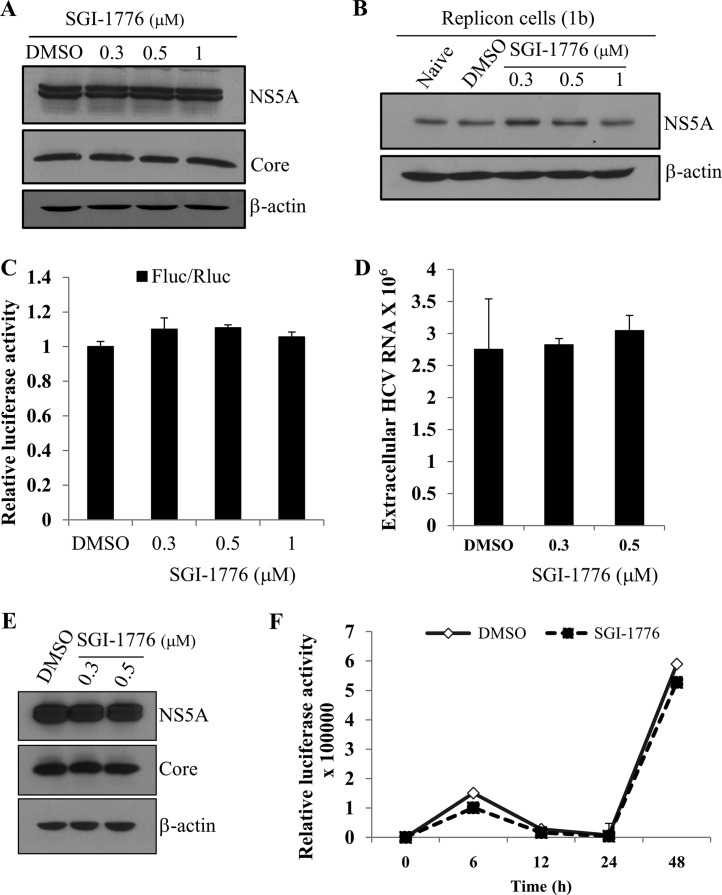
Pim kinases are not involved in the replication, translation, and virion production steps in the HCV life cycle.
To gain insight into the role of Pim kinases in HCV life cycle, we analyzed which steps of the HCV life cycle were required for Pim kinases. Huh7.5 cells were infected with Jc1 for 48 h and then treated with either DMSO or Pim kinase inhibitor. As shown in Fig. 5A, treatment of Pim kinases inhibitor after viral infection had no effect on HCV protein levels. We further confirmed that Pim kinases inhibitor displayed no discernible effect on viral protein expression level in HCV subgenomic replicon cells (Fig. 5B). We next asked if Pim kinases are involved in HCV internal ribosome entry site (IRES)-mediated translation. To address this question, Huh7.5 cells were treated with various concentrations of SGI-1776 and then transfected with pRL-HL and β-galactosidase plasmid as we reported previously (19), and then luciferase activity was determined. We demonstrated that treatment of Pim kinase inhibitor displayed no effects on HCV IRES-dependent translation (Fig. 5C). These data indicated that Pim kinase might be involved in the virion production step of the HCV life cycle. To explore this possibility, Huh7.5 cells infected with Jc1 were treated with either DMSO or SGI-1776. At 48 h after inhibitor treatment, extracellular HCV RNAs were analyzed by quantitative reverse transcription-PCR (qRT-PCR). Figure 5D showed that Pim kinases were not involved in the viral production step of the HCV life cycle. To further confirm this result, naive Huh7.5 cells were infected with culture supernatant harvested from the experiment whose results are shown in Fig. 5D, and protein expression levels were determined. As shown in Fig. 5E, viral protein expression levels were unaffected by Pim kinase inhibitor, suggesting that Pim might be involved in other steps of the HCV life cycle. To further investigate the possible involvement of Pim kinase in the initiation of replication step, Huh7.5 cells were treated with either DMSO or SGI-1776 for 36 h and electroporated with JFH1-luc RNA, and then luciferase activity was determined. As shown in Fig. 5F, luciferase activity was exponentially increased in DMSO-treated cells due to HCV RNA replication. However, luciferase activity was not decreased in Pim inhibitor-treated cells compared to DMSO-treated cells. This result showed that Pim kinase inhibitor displayed no inhibitory effect on HCV replication and thus that Pim is important for HCV entry independent of the interaction with NS5A.
FIG 5.
Pim kinases are not involved in the replication, translation, and virion production steps in the HCV life cycle. (A) Huh7.5 cells were infected with Jc1 for 4 h. At 48 h postinfection, cells were treated with either DMSO (vehicle) or various doses of SGI-1776. At 2 days after inhibitor treatment, total cellular extracts were immunoblotted with the indicated antibodies. (B) Huh7 cells harboring HCV subgenomic replicon were treated with either DMSO or various concentrations of SGI-1776. At 48 h after inhibitor treatment, total cellular extracts were immunoblotted with the indicated antibodies. (C) Huh7.5 cells were treated with various concentrations of SGI-1776 for 2 h. Cells were then transfected with 0.5 μg of pRL-HL and 0.5 μg of β-galactosidase plasmid (21). Cell lysates harvested at 48 h after transfection were used to determine luciferase activity and normalized using β-galactosidase activity. (D) Huh7.5 cells infected with Jc1 were treated with either DMSO or various doses of SGI-1776. At 2 days after inhibitor treatment, extracellular HCV RNA levels were analyzed by qRT-PCR. (E) Naive Huh7.5 cells were infected with culture supernatant harvested from the experiment whose results are shown in panel D, and protein expression levels were analyzed by immunoblotting with the indicated antibodies. (F) Huh7.5 cells were treated with either DMSO or SGI-1776 for 36 h and then electroporated with 10 μg of JFH1-luc RNA. Cells were harvested at the indicated time points, and then luciferase activities were determined.
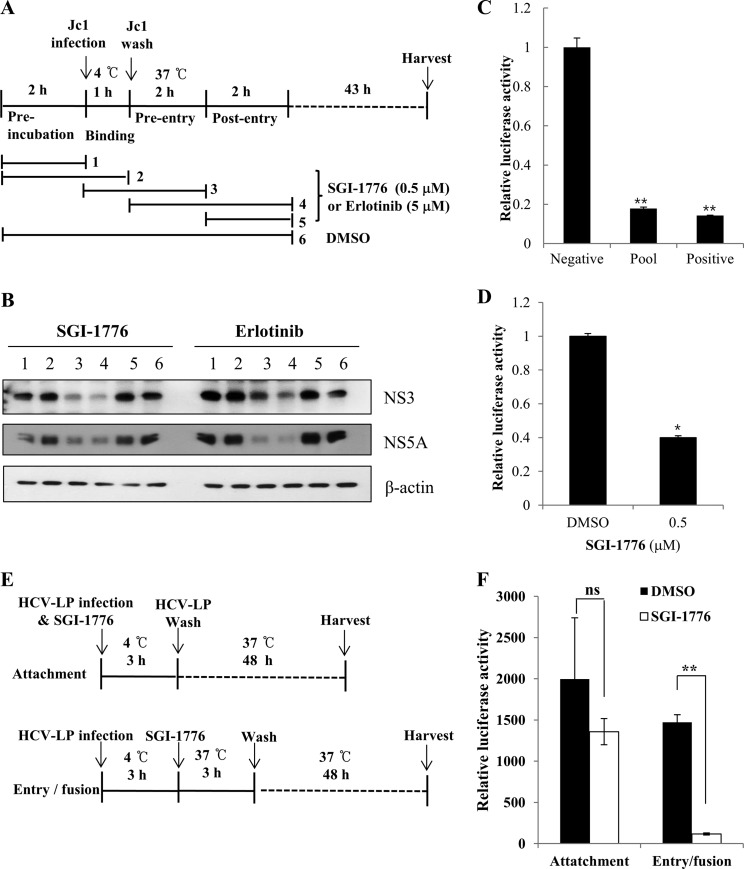
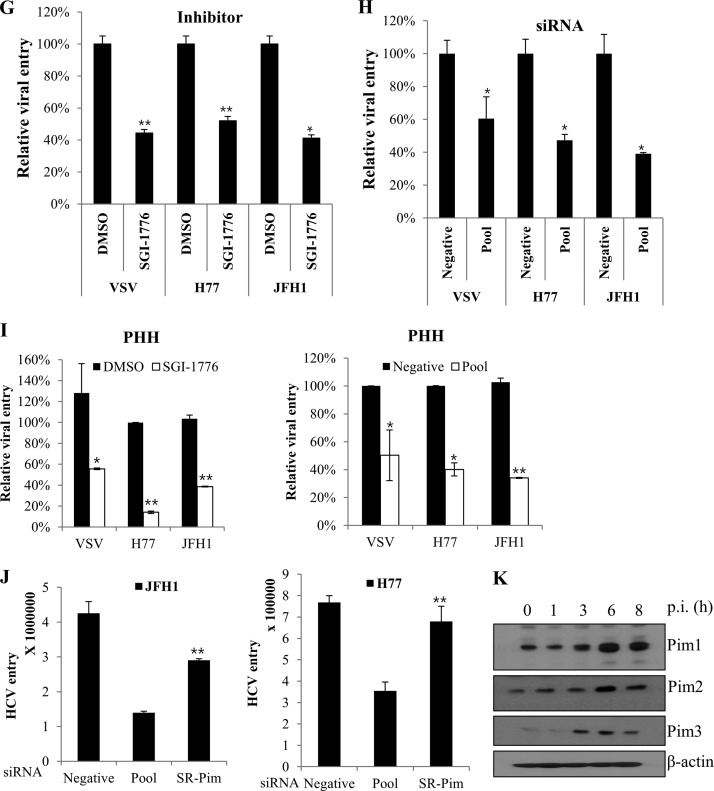
Pim kinases are required during the entry step of the viral infection.
To investigate if Pim kinases were required at an early step of HCV infection, we performed time-of-addition infection assays using Pim kinase inhibitor (Fig. 6A). Huh7.5 cells were incubated with Jc1 viral inoculum to assess the effects of Pim kinase inhibitor over the time course of HCV infection. Cells were treated with an inhibitor for various time periods, and then viral protein expressions were assessed. Figure 6B shows that viral protein expressions were unaffected by the Pim kinase inhibitor during both binding and postentry steps of the HCV infection. However, viral protein expressions were almost undetectable at the entry step in the presence of SGI-1776, indicating that Pim kinases were required during the entry stage of the HCV life cycle. Consistently, we verified that erlotinib, an epidermal growth factor receptor (EGFR) inhibitor which is a well-known inhibitor of HCV entry, specifically inhibited the entry step without affecting either binding or postentry steps of the HCV infection (Fig. 6B). To verify whether Pim kinases were required for entry, we performed viral entry assays using HCV-like particles (HCV-LP). We performed viral entry assays using HCV-LP as described previously (21, 22). In this system, HCV-LP can undergo single-cycle infection without producing virions. Figure 6C shows that HCV-LP luciferase activity was significantly reduced in Pim kinase knockdown cells compared to the negative control. Similarly, luciferase activity was significantly decreased in inhibitor-treated cells compared to DMSO-treated cells (Fig. 6D), indicating that Pim kinases are required for viral entry. To further confirm these results, we performed time-of-addition entry assays using HCV-LP. Huh7.5 cells were treated with SGI-1776 at two different time points to distinguish between the attachment and the entry/fusion steps (Fig. 6E). As shown in Fig. 6F, HCV-LP luciferase activity was significantly suppressed at the viral entry step but not at the attachment step. To further verify if the Pim kinases were required for the entry step, we performed viral entry assays using both HCVpp and VSVpp. Figure 6G shows that not only HCVpp entry but also VSVpp entry was significantly decreased in Pim kinase inhibitor-treated cells. Similarly, both HCVpp and VSVpp entries were significantly reduced in Pim kinase knockdown cells (Fig. 6H), suggesting that Pim kinase may be a common cofactor required for various viral entries. To verify this entry effect in primary cells, primary human hepatocytes were either treated with SGI-1776 or transfected with pooled Pim siRNAs and then infected with various pseudoparticles. Viral entry was determined by luciferase activity at 72 h postinfection. As shown in Fig. 6I, HCVpp entries in primary human hepatocytes were also significantly reduced in both SGI-1776 and siRNA-treated cells, further confirming that Pim is required for HCV entry. To investigate if Pim-NS5A interaction plays a role in HCV entry, we performed recovery experiments for HCV entry using an siRNA-resistant Pim mutant in Pim knockdown cells. As shown in Fig. 6J, HCV entry levels were rescued by siRNA-resistant Pim in both HCV genotypes. Since HCVpp is lacking NS5A, these data indicate that Pim kinase itself regulates HCV entry irrespective of Pim-NS5A interaction. To investigate if Pim kinase expression was also regulated at an early stage of HCV infection, Huh7.5 cells were infected with Jc1, and then protein expression of each Pim kinase was analyzed at various time points by immunoblot assay. Indeed, expression of all Pim proteins was abruptly elevated at 3 to 6 h postinfection (Fig. 6K). This may be due to the fact that Pim kinases are required at the entry step of HCV infection.
FIG 6.
Pim kinases are required during the entry step of the HCV life cycle. (A) Schematic illustration of the experimental design. SGI-1776 was treated for the indicated time periods. DMSO was treated as a vehicle control. (B) Huh7.5 cells were incubated with Jc1 for binding at 4°C for 1 h in the absence or presence of either SGI-1776 or erlotinib. After the cells had been washed in PBS, the temperature was shifted to 37°C to allow cell entry in the absence or presence of either SGI-1776 or erlotinib. At 48 h postinfection, protein expression levels were determined by immunoblot analysis using the indicated antibodies. (C) Huh7.5 cells were transfected with the indicated siRNA constructs and then infected with HCV-LP for 4 h. At 48 h postinfection, cells were harvested and firefly luciferase activity was determined. The data for each experiment are averages of triplicate values, with error bars showing standard deviations. (D) Huh7.5 cells were treated with either DMSO or SGI-1776 for 2 h and then infected with HCV-LP for 4 h. At 48 postinfection, firefly luciferase activity in total cell lysates was analyzed. (E) Schematic illustration of the experimental design. (Top) Huh7.5 cells were incubated with HCV-LP in the presence of SGI-1776 at 4°C for 3 h (viral attachment step) and then washed in PBS. The temperature was shifted to 37°C, and cells were further cultured for 48 h. (Bottom) Huh7.5 cells were incubated with HCV-LP at 4°C for 3 h and treated with SGI-1776, and the temperature was shifted to 37°C for 3 h (viral entry/fusion step). Cells were washed in PBS and further cultured for 48 h. (F) Huh7.5 cells treated as described for panel E were analyzed to discriminate between the attachment step and the entry/fusion step by measuring luciferase activity. (G) Huh7.5 cells were treated with Pim kinase inhibitor for 2 h. Cells were then infected with either VSVpp or HCVpp derived from genotype 1a (H77) and 2a (JFH1) for 6 h. At 72 h postinfection, cells were harvested and viral entry was determined by luciferase activity. (H) Huh7.5 cells were transfected with siRNAs for 48 h. Cells were then infected with either VSVpp or HCVpp, and viral entry was determined by a luciferase activity assay at 72 h postinfection. (I) Primary human hepatocytes were either treated with SGI-1776 (left) for 36 h or transfected with the indicated siRNAs (right) for 48 h and then infected with either VSVpp or HCVpp for 6 h. At 72 h postinfection, cells were harvested and viral entry was determined by luciferase activity. (J) Huh7.5 cells were transfected with a pool of Pim siRNAs for 24 h. Cells were further transfected with siRNA-resistant Pim for 24 h and then infected with HCVpp (right, genotype 1a; left, genotype 2a) for 6 h. At 24 h postinfection, cells were harvested, and viral entry was determined by a luciferase activity assay. (K) Huh7.5 cells were infected with Jc1. At the indicated times postinfection (p.i.), protein expression levels of Pim kinases were analyzed by immunoblotting with the indicated antibodies. Asterisks in all graphs show statistical signficance: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
DISCUSSION
HCV encodes 10 functional proteins. Among these proteins, NS5A has been shown to interact with numerous cellular proteins to modulate host signaling pathways and HCV-induced pathogenesis (6–10, 27). By employing protein array screening, we identified Pim1 as the HCV NS5A interactor. Since Pim1 has been implicated in tumorigenesis and progression of a variety of malignancies (24), we explored the possible involvement of Pim1 in HCV propagation. Pim1 belongs to the active serine/threonine kinase family. However, it has been reported previously that NS5A is not a substrate for Pim1 kinase (28). Pim1 shares a common high sequence homology with other members, Pim2 and Pim3. All Pim kinases have overlapping functions in various cellular processes, including cell cycle, cell survival, apoptosis, and signal transduction pathways (29–32). Pim1 functions as an oncogene whose activation promotes cancer in animals (33, 34). Overexpression of Pim1 contributes to tumorigenosis (33), and the levels of Pim proteins are associated with their actual activities. Indeed, elevated levels of Pim kinases have been associated with solid tumors and hematological malignancies (14, 16). Although Pim1 but not Pim2 or Pim3 was initially identified as an NS5A interactor in our protein array assay, we also demonstrated that both Pim2 and Pim3 interacted with NS5A by coimmunoprecipitation assay. We showed that knockdown of Pim1 displayed little effect on HCV propagation, whereas knockdown of all Pim proteins impaired HCV propagation. These results suggest that Pim family proteins may either function in a cooperative manner or act redundantly in HCV propagation, and thus, functional inactivation of Pim1 could be compensated for by other Pim family proteins, as previously reported for Pim-1−/− mice (34).
Protein interaction between Pim1 and NS5A was confirmed by both in vitro binding and coimmunoprecipitation assays. We further verified that both Pim1 and NS5A proteins were colocalized in the cytoplasm of Jc1-infected cells. Protein interaction between Pim1 and NS5A is mediated by domain I of NS5A and aa 141 to 180 of Pim1. It is worth noting that Pim kinase inhibitor abrogated protein interaction in a dose-dependent manner, implying that kinase activity of Pim protein is required for protein interplay between Pim and NS5A. Since Pim proteins become autonomously activated once they are expressed, we explored the possible involvement of NS5A in Pim protein stability. Pim protein levels in CHX-treated cells were monitored in the absence or presence of NS5A protein. We showed that Pim protein level was stably maintained in the presence of NS5A protein. Importantly, Pim protein level was decreased in the presence of a binding-defective mutant of NS5A. Likewise, the level of a binding-defective mutant of Pim1 was also decreased even in the presence of wild-type NS5A. In addition, we demonstrated that NS5A modulated protein stability of Pim1 by suppressing the ubiquitylation process. Since Pim1 is an immediate early gene and its half-life is short, all these data suggest that NS5A contributes to Pim1 protein stability via protein interplay.
We evaluated the functional role of Pim1 in HCV propagation. Knockdown of Pim1 displayed little effect on intracellular HCV RNA and HCV protein levels. However, pharmacological inhibition of Pim kinases with SGI-1776 suppressed HCV RNA, HCV protein, and extracellular HCV RNA levels. Since SGI-1776 inhibits all Pim kinase activities, this raised the question of whether other Pim kinases might be involved in HCV propagation. Using a pool of siRNA constructs targeting all Pim kinases, we demonstrated that intracellular HCV RNA, HCV protein, and extracellular HCV RNA levels were significantly decreased in Pim knockdown cells. This may be due to the fact that all Pim kinases share a consensus sequence and are functionally redundant (11, 12, 34, 35).
Finally, we investigated which step of the HCV life cycle required for Pim kinases. We demonstrated that Pim kinases were not involved in replication, IRES-mediated translation, or virion production steps in the HCV life cycle. We therefore employed time-of-addition infection assays using a Pim kinase inhibitor and showed that Pim kinases are required during the entry step of the HCV life cycle. In fact, protein expressions of all Pim kinases surged at 3 h after Jc1 infection, which overlaps the time of viral entry. Using HCV-LP and HCVpp assays, we further verified that Pim kinases are involved in the entry step but not the attachment step of the HCV infection.
To date, Pim kinases have been shown to be implicated in cell survival, apoptosis, modulation of signal transduction, and tumorigenesis. Here, we provide the first evidence that HCV coopts Pim kinases for viral entry via unknown mechanisms. It was reported previously that HCV transiently activates the PI3K-AKT pathway during virus entry (36). Growing evidence suggests that Pim kinases cross talk with PI3K-AKT kinases to produce synergetic effects on cell proliferation and survival signaling pathways (37–39). Therefore, we are tempted to speculate that Pim kinase might regulate HCV entry by promoting CD81 and claudin-1 receptor complex formation via PI3K-AKT signaling pathway. However, additional studies are needed to better understand the implication of Pim kinase in HCV entry. Since Pim kinase has been considered a novel target for anticancer therapy (12, 26, 40), it is also possible that HCV may regulate Pim kinase activity to modulate other oncogenic signal transduction pathways which can cause HCV-mediated liver pathogenesis. Taken together, Pim protein may represent a novel therapeutic target for HCV.
ACKNOWLEDGMENTS
We are indebted to Ralf Bartenschlager (University of Heidelberg) for the Jc1 construct and Charles Rice for Huh7.5 cells.
This work was supported by the Basic Science Research Program (2012026351) from the Ministry of Science, ICT and Future Planning, South Korea, and by the Korean Health Technology R&D Project (HI13C1746), Ministry of Health and Welfare, South Korea.
We have no conflicts of interests to declare.
REFERENCES
- 1.Joyce MA, Tyrrell DL. 2010. The cell biology of hepatitis C virus. Microbes Infect 12:263–271. doi: 10.1016/j.micinf.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki T, Ishii K, Aizaki H, Wakita T. 2007. Hepatitis C viral life cycle. Adv Drug Deliv Rev 59:1200–1212. doi: 10.1016/j.addr.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee A, Ray RB, Ray R. 2010. Oncogenic potential of hepatitis C virus proteins. Viruses 2:2108–2133. doi: 10.3390/v2092108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nanda SK, Herion D, Liang TJ. 2006. The SH3 binding motif of HCV NS5A protein interacts with Bin1 and is important for apoptosis and infectivity. Gastroenterology 130:794–809. doi: 10.1053/j.gastro.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 5.Tripathi LP, Kambara H, Chen Y-A, Nishimura Y, Moriishi K, Okamoto T, Morita E, Abe T, Mori Y, Matsuura Y, Mizuguchi K. 2013. Understanding the biological context of NS5A-host interactions in HCV infection: a network-based approach. J Proteome Res 12:2537–2551. doi: 10.1021/pr3011217. [DOI] [PubMed] [Google Scholar]
- 6.Park KJ, Choi SH, Choi DH, Park JM, Yie SW, Lee SY, Hwang SB. 2003. Hepatitis C virus NS5A protein modulates c-Jun N-terminal kinase through interaction with tumor necrosis factor receptor-associated factor 2. J Biol Chem 278:30711–30718. doi: 10.1074/jbc.M209623200. [DOI] [PubMed] [Google Scholar]
- 7.Choi SH, Hwang SB. 2006. Modulation of the transforming growth factor-beta signal transduction pathway by hepatitis C virus nonstructural 5A protein. J Biol Chem 281:7468–7478. doi: 10.1074/jbc.M512438200. [DOI] [PubMed] [Google Scholar]
- 8.Park CY, Choi SH, Kang SM, Kang JI, Ahn BY, Kim H, Jung G, Choi KY, Hwang SB. 2003. Nonstructural 5A protein activates beta-catenin signaling cascades: implication of hepatitis C virus-induced liver pathogenesis. J Hepatol 51:853–864. doi: 10.1016/j.jhep.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 9.Lim YS, Tran HT, Park SJ, Yim SA, Hwang SB. 2011. Peptidyl-prolyl isomerase Pin1 is a cellular factor required for hepatitis C virus propagation. J Virol 85:8777–8788. doi: 10.1128/JVI.02533-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim YS, Hwang SB. 2011. Hepatitis C virus NS5A protein interacts with phosphatidylinositol 4-kinase type IIIalpha and regulates viral propagation. J Biol Chem 286:11290–11298. doi: 10.1074/jbc.M110.194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nawijn MC, Alendar A, Berns A. 2011. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer 11:23–34. doi: 10.1038/nrc2986. [DOI] [PubMed] [Google Scholar]
- 12.Narlik-Grassow M, Blanco-Aparicio C, Carnero A. 2014. The PIM family of serine/threonine kinases in cancer. Med Res Rev 34:136–159. doi: 10.1002/med.21284. [DOI] [PubMed] [Google Scholar]
- 13.Fujii C, Nakamoto Y, Lu P, Tsuneyama K, Popivanova BK, Kaneko S, Mukaida N. 2005. Aberrant expression of serine/threonine kinase Pim-3 in hepatocellular carcinoma development and its role in the proliferation of human hepatoma cell lines. Int J Cancer 114:209–218. doi: 10.1002/ijc.20719. [DOI] [PubMed] [Google Scholar]
- 14.Ren K, Gou X, Xiao M, Wang M, Liu C, Tang Z, He W. 2013. The over-expression of Pim-2 promote the tumorigenesis of prostatic carcinoma through phosphorylating eIF4B. Prostate 73:1462–1469. doi: 10.1002/pros.22693. [DOI] [PubMed] [Google Scholar]
- 15.Cibull TL, Jones TD, Li L, Eble JN, Ann Baldridge L, Malott SR, Luo Y, Cheng L. 2006. Overexpression of Pim-1 during progression of prostatic adenocarcinoma. J Clin Pathol 59:285–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y, Wang YY, Nakamoto Y, Li YY, Baba T, Kaneko S, Fujii C, Mukaida N. 2010. Accelerated hepatocellular carcinoma development in mice expressing the Pim-3 transgene selectively in the liver. Oncogene 29:2228–2237. doi: 10.1038/onc.2009.504. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Anderson PD, Luo W, Gius D, Roh M, Abdulkadir SA. 2012. Pim1 kinase is required to maintain tumorigenicity in MYC-expressing prostate cancer cells. Oncogene 14:1794–1803. doi: 10.1038/onc.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong J, Wang J, Ren K, Liu C, Li B, Shi Y. 2009. Serine/threonine kinase Pim-2 promotes liver tumorigenesis induction through mediating survival and preventing apoptosis of liver cell. J Surg Res 153:17–22. doi: 10.1016/j.jss.2008.03.033. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen LN, Lim YS, Pham LV, Shin HY, Kim YS, Hwang SB. 2014. Stearoyl coenzyme a desaturase 1 is associated with hepatitis C virus replication complex and regulates viral replication. J Virol 88:12311–12325. doi: 10.1128/JVI.01678-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sung PS, Murayama A, Kang W, Kim MS, Yoon SK, Fukasawa M, Kondoh M, Kim JS, Kim H, Kato T, Shin EC. 2014. Hepatitis C virus entry is impaired by claudin-1 downregulation in diacylglycerol acyltransferase-1-deficient cells. J Virol 88:9233–9244. doi: 10.1128/JVI.01428-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masaki T, Suzuki R, Saeed M, Mori K, Matsuda M, Aizaki H, Ishii K, Maki N, Miyamura T, Matsuura Y, Wakita T, Suzuki T. 2010. Production of infectious hepatitis C virus by using RNA polymerase I-mediated transcription. J Virol 84:5824–5835. doi: 10.1128/JVI.02397-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q, Zhang YY, Chiu S, Hu Z, Lan KH, Cha H, Sodroski C, Zhang F, Hsu CS, Thomas E, Liang TJ. 2014. Integrative functional genomics of hepatitis C virus infection identifies host dependencies in complete viral replication cycle. PLoS Pathog 10:e1004163. doi: 10.1371/journal.ppat.1004163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ngo HT, Pham LV, Kim JW, Lim YS, Hwang SB. 2013. Modulation of mitogen-activated protein kinase-activated protein kinase 3 by hepatitis C virus core protein. J Virol 87:5718–5731. doi: 10.1128/JVI.03353-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pang W, Tian X, Bai F, Han R, Wang J, Shen H, Zhang X, Liu Y, Yan X, Jiang F, Xing L. 2014. Pim-1 kinase is a target of miR-486-5p and eukaryotic translation initiation factor 4E, and plays a critical role in lung cancer. Mol Cancer 13:240. doi: 10.1186/1476-4598-13-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma J, Arnold HK, Lilly MB, Sears RC, Kraft AS. 2007. Negative regulation of Pim-1 protein kinase levels by the subunit of PP2A. Oncogene 35:5145–5153. [DOI] [PubMed] [Google Scholar]
- 26.Blanco-Aparicio C, Carnero A. 2013. Pim kinases in cancer: diagnostic, prognostic and treatment opportunities. Biochem Pharmacol 85:629–643. doi: 10.1016/j.bcp.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 27.Ross-Thriepland D, Harris M. 2015. Hepatitis C virus NS5A: enigmatic but still promiscuous 10 years on! J Gen Virol 96:727–738. doi: 10.1099/jgv.0.000009. [DOI] [PubMed] [Google Scholar]
- 28.Masaki T, Matsunaga S, Takahashi H, Nakashima K, Kimura Y, Ito M, Matsuda M, Murayama A, Kato T, Hirano H, Endo Y, Lemon SM, Wakita T, Sawasaki T, Suzuki T. 2014. Involvement of hepatitis C virus NS5A hyperphosphorylation mediated by casein kinase I-α in infectious virus production. J Virol 88:7541–7555. doi: 10.1128/JVI.03170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bachmann M, Hennemann H, Xing PX, Hoffmann I, Möröy T. 2004. The oncogenic serine/threonine kinase Pim-1 phosphorylates and inhibits the activity of Cdc25C-associated kinase 1 (C-TAK1): a novel role for Pim-1 at the G2/M cell cycle checkpoint. J Biol Chem 279:48319–48328. [DOI] [PubMed] [Google Scholar]
- 30.Nihira K, Ando Y, Yamaguchi T, Kagami Y, Miki Y, Yoshida K. 2010. Pim-1 controls NF-jB signalling by stabilizing RelA/p65. Cell Death Differ 17:689–698. doi: 10.1038/cdd.2009.174. [DOI] [PubMed] [Google Scholar]
- 31.Aho TL, Sandholm J, Peltola KJ, Mankonen HP, Lilly M, Koskinen PJ. 2004. Pim-1 kinase promotes inactivation of the pro-apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS Lett 571:43–49. doi: 10.1016/j.febslet.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 32.Gu JJ, Wang Z, Reeves R, Magnuson NS. 2009. PIM1 phosphorylates and negatively regulates ASK1-mediated apoptosis. Oncogene 28:4261–4271. doi: 10.1038/onc.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hogan C, Hutchison C, Marcar L, Milne D, Saville M, Goodlad J, Kernohan N, Meek D. 2008. Elevated levels of oncogenic protein kinase Pim-1 induce the p53 pathway in cultured cells and correlate with increased Mdm2 in mantle cell lymphoma. J Biol Chem 283:18012–18023. doi: 10.1074/jbc.M709695200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Lugt NM, Domen J, Verhoeven E, Linders K, van der Gulden H, Allen J, Berns A. 1995. Proviral tagging in E mu-myc transgenic mice lacking the Pim-1 proto-oncogene leads to compensatory activation of Pim-2. EMBO J 11:2536–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan B, Zemskova M, Holder S, Chin V, Kraft A, Koskinen PJ, Lilly M. 2003. The PIM-2 kinase phosphorylates BAD on serine 112 and reverses BAD-induced cell death. J Biol Chem 278:45358–45367. [DOI] [PubMed] [Google Scholar]
- 36.Liu Z, Tian Y, Machida K, Lai MM, Luo G, Foung SK, Ou JH. 2012. Transient activation of the PI3K-AKT pathway by hepatitis C virus to enhance viral entry. J Biol Chem 287:41922–41930. doi: 10.1074/jbc.M112.414789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo G, Qiu X, Wang S, Chen Y, Rothman PB, Wang Z, Chen Y, Wang G, Chen JL. 2010. Oncogenic E17K mutation in the pleckstrin homology domain of AKT1 promotes v-Abl-mediated pre-B-cell transformation and survival of Pim-deficient cells. Oncogene 29:3845–3853. doi: 10.1038/onc.2010.149. [DOI] [PubMed] [Google Scholar]
- 38.Mondello P, Cuzzocrea S, Mian M. 2014. Pim kinases in hematological malignancies: where are we now and where are we going? J Hematol Oncol 7:95. doi: 10.1186/s13045-014-0095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warfel NA, Kraft AS. 2015. PIM kinase (and Akt) biology and signaling in tumors. Pharmacol Ther 15:1–9. doi: 10.1016/j.pharmthera.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magnuson NS, Wang Z, Ding G, Reeves R. 2010. Why target PIM1 for cancer diagnosis and treatment? Future Oncol 6:1461–1478. doi: 10.2217/fon.10.106. [DOI] [PMC free article] [PubMed] [Google Scholar]