ABSTRACT
Transcription and replication of influenza A virus are carried out in the nuclei of infected cells in the context of viral ribonucleoproteins (RNPs). The viral polymerase responsible for these processes is a protein complex composed of the PB1, PB2, and PA proteins. We previously identified a set of polymerase-associated cellular proteins by proteomic analysis of polymerase-containing intracellular complexes expressed and purified from human cells. Here we characterize the role of NXP2/MORC3 in the infection cycle. NXP2/MORC3 is a member of the Microrchidia (MORC) family that is associated with the nuclear matrix and has RNA-binding activity. Influenza virus infection led to a slight increase in NXP2/MORC3 expression and its partial relocalization to the cytoplasm. Coimmunoprecipitation and immunofluorescence experiments indicated an association of NXP2/MORC3 with the viral polymerase and RNPs during infection. Downregulation of NXP2/MORC3 by use of two independent short hairpin RNAs (shRNAs) reduced virus titers in low-multiplicity infections. Consistent with these findings, analysis of virus-specific RNA in high-multiplicity infections indicated a reduction of viral RNA (vRNA) and mRNA after NXP2/MORC3 downregulation. Silencing of NXP2/MORC3 in a recombinant minireplicon system in which virus transcription and replication are uncoupled showed reductions in cat mRNA and chloramphenicol acetyltransferase (CAT) protein accumulation but no alterations in cat vRNA levels, suggesting that NXP2/MORC3 is important for influenza virus transcription.
IMPORTANCE Influenza virus infections appear as yearly epidemics and occasional pandemics of respiratory disease, with high morbidity and occasional mortality. Influenza viruses are intracellular parasites that replicate and transcribe their genomic ribonucleoproteins in the nuclei of infected cells, in a complex interplay with host cell factors. Here we characterized the role of the human NXP2/MORC3 protein, a member of the Microrchidia family that is associated with the nuclear matrix, during virus infection. NXP2/MORC3 associates with the viral ribonucleoproteins in infected cells. Downregulation of NXP2/MORC3 reduced virus titers and accumulations of viral genomic RNA and mRNAs. Silencing of NXP2/MORC3 in an influenza virus CAT minireplicon system diminished CAT protein and cat mRNA levels but not genomic RNA levels. We propose that NXP2/MORC3 plays a role in influenza virus transcription.
INTRODUCTION
Influenza viruses cause an acute respiratory disease that annually affects millions of people worldwide (Global Influenza Surveillance and Response System [GISRS] [http://www.who.int/influenza/gisrs_laboratory/en/]). The genome of influenza A viruses is about 13 kb long and consists of eight single-stranded negative-sense RNA segments. The viral proteome includes 10 viral proteins that have been studied extensively (1) and another 8 proteins, probably accessory proteins, that were identified more recently (reviewed in reference 2). The transcription and replication of influenza viruses occur in the nuclei of infected cells and are mediated by the viral polymerase, a heterotrimer composed of the PB1, PB2, and PA subunits, in the context of viral ribonucleoprotein complexes (RNPs) (3; reviewed in references 4 to 7). The virus recruits host cell factors to help carry out these processes, and in some specific cases, their roles in virus replication have been determined (reviewed in references 4 and 8 to 10). In one such study, we identified the nuclear matrix NXP2 protein as a factor associated with influenza virus polymerase in vivo by proteomic analysis of recombinant purified polymerase complexes (11). Since influenza virus RNA synthesis is connected to the nuclear matrix (12, 13), we decided to further characterize the role of NXP2 in the virus infection cycle.
The NXP2 protein (also called MORC3, ZCW5, ZCWCC3, or KIAA0136) belongs to the Microrchidia (MORC) family. This is a relatively uncharacterized nuclear protein family with highly conserved ancestors in prokaryotic cells (14, 15). Five members of the MORC family (MORC1 to MORC4 and the more divergent SMCHD1 protein) have been identified in humans. They contain three conserved domains, including (i) a GHL (gyrase B, Hsp90, and MutL) ATPase domain at the N terminus (16), (ii) a CW-type zinc finger domain containing four conserved cysteine and two tryptophan residues in the middle portion (17), and (iii) a coiled-coil dimerization domain at the C terminus (15, 18). The MORC proteins show tissue-specific expression patterns and a wide range of biological functions, such as transcription regulation, chromatin condensation and remodeling, and DNA break repair (reviewed in reference 14). More specifically, NXP2/MORC3 was shown to bind SUMO-2 to repress transcription. In addition, Takahashi et al. reported that the NXP2/MORC3 protein localized to PML nuclear bodies (PML-NBs) in an ATPase-dependent manner, recruited p53 to PML-NBs, and activated p53 to induce cellular senescence in normal human and mouse fibroblasts (19). More recent studies demonstrated a two-step mechanism involved in the colocalization of NXP2/MORC3 with PML-NBs (20). Note that it has been described previously that PML-NBs play a role in antiviral defense, including defense against influenza virus infections (21; reviewed in reference 22).
Here we show that NXP2/MORC3 associates with the influenza virus polymerase and viral RNPs in infected cells. NXP2/MORC3-silencing experiments indicated that this protein is necessary for efficient influenza virus replication. Downregulation of NXP2/MORC3 led to diminished virus genomic and mRNA levels. However, only transcription of viral RNPs, not RNA replication, was slightly affected in a recombinant minireplicon system, indicating that this host factor is required for virus transcription.
MATERIALS AND METHODS
Viruses and cells.
The A549 and HEK293T human cell lines were obtained from J. A. Melero and J. C. de la Torre, respectively. The MDCK and BHK21 cell lines were purchased from ATCC. Cell culture was carried out as described previously (23). The influenza virus strains A/WSN/33 (H1N1) (WSN) and A/Victoria/3/75 (H3N2) (Vic) were used and titrated in MDCK cells as described previously (24). Vesicular stomatitis virus (VSV) titers were determined in BHK21 cells. Adenovirus 5 (Ad5), kindly provided by P. Fortes, was grown and titrated in HEK293T cells as described previously (25).
Plasmids.
The mission plasmid expressing short hairpin RNA 1 (shRNA1; CCGGTgcggagcatgttgttgttcTTCAAGAGAgaacaacaacatgctccgcTTTTTG) targeting NXP2/MORC3 was purchased from Sigma-Aldrich (St. Louis, MO). The plasmid expressing short hairpin RNA 2 (shRNA2; CCGGTgaaggggtacaagaagcagTTCAAGAGActgcttcttgtaccccttcTTTTTG) targeting NXP2/MORC3 was designed using previously published data (19) (the target sense and antisense sequences are shown in lowercase). A control shRNA plasmid expressing a sequence from Thermotoga maritima (CCGGTaattctccgaacgtgtcacTTCAAGAGAgtgacacgttcggagaattTTTTTG) was kindly provided by A. Rodriguez-Frandsen. Plasmids used to generate lentiviral particles, i.e., the lentiviral vector pLKO.1-Puro (Addgene), the packaging vector psPAX2 (Addgene), and the envelope vector pMD2.G (Addgene) (26, 27), were kindly provided by P. Gastaminza. Plasmids pCMVPB1, pCMVPB2, pCMVPA, and pCMVNP, expressing the influenza virus polymerase subunits and the NP, were described previously (28). Plasmid pHHCAT, which transcribes a virus-like cat gene in negative polarity (29), was provided by A. Rodriguez-Frandsen.
Lentiviral particle production and cell transduction.
Lentiviral particles were produced in HEK293T cells by cotransfection of plasmids psPAX2 and pMD2.G with each of the pLKO-based shRNA vectors as described previously (26, 27). Supernatants were collected at 40 to 48 h posttransfection, filtered through a 0.45-μm filter, and used to transduce A549 or HEK293T cells. Since the lentiviral vectors confer resistance to puromycin, the minimum amount of supernatant necessary to confer resistance to puromycin (5 μg/ml) to 100% of the cells was used. Silencing was tested by Western blotting, typically at 3 days posttransduction. The viability of transduced cells was determined by the 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay (30).
Western blotting.
Protein samples were separated by SDS-PAGE and electrotransferred to Immobilon filters. Western blotting was carried out essentially as described previously (31). The membranes were saturated with 5% nonfat milk for 1 h and then incubated with the primary antibodies for 1 h at room temperature or overnight at 4°C. The filters were washed with phosphate-buffered saline (PBS)–0.25% Tween 20 and incubated with the appropriate secondary antibody conjugated to horseradish peroxidase. After further washing as described above, the filters were developed by enhanced chemiluminescence. Nonsaturated films were scanned, and relative density values were measured using ImageJ software (32). The following primary antibodies were used: for NXP2/MORC3, a rabbit polyclonal antibody kindly provided for Norimitsu Inoue; for PA, monoclonal antibodies 2 and 9 (33, 34); for PB2, monoclonal antibodies 8 and 22 (33); for PB1, a rabbit polyclonal antibody (35); for NP, a rabbit polyclonal antibody (36), and for β-tubulin, a mouse monoclonal antibody (T5076; Sigma).
Immunoprecipitation assay.
Immunoprecipitation was performed as described previously (37). Briefly, mock-infected or infected cells were collected and lysed in a buffer containing 150 mM NaCl, 5 mM EDTA, 1.5 mM MgCl2, 50 mM Tris-HCl, pH 8.5, 0.5% IGEPAL, 1 mM dithiothreitol (DTT), 1 U/ml human placental ribonuclease inhibitor (HPRI), and protease inhibitors containing EDTA. The lysate was centrifuged at 10,000 × g, and the supernatant was immunoprecipitated with a polyclonal anti-NXP2/MORC3 antibody (ab76813; Abcam) or an unrelated antibody. The immunocomplexes were washed 10 times with lysis buffer, and the immunoprecipitated proteins were analyzed by Western blotting.
Confocal immunofluorescence microscopy.
A549 cells were grown on glass coverslips and infected at a high multiplicity of infection (MOI) (3 PFU/cell) with strain WSN. At various times postinfection, the cells were fixed with methanol for 20 min at −20°C, washed twice with PBS, and incubated with a buffer containing 2% bovine serum albumin (BSA) in PBS for 1 h. The coverslips were incubated with antibodies diluted in PBS-0.1% BSA for 1 h, washed with PBS, and subsequently incubated with a 1:400 dilution of a goat anti-mouse antibody conjugated to Alexa 488, 594, or 694 (Invitrogen, Carlsbad, CA). The following primary antibodies were used: rabbit anti-NP polyclonal antibody (38), rat anti-NP polyclonal antibody (39), rabbit anti-PML polyclonal antibody (H-238; Santa Cruz), or mouse anti-NXP2/MORC3 polyclonal antibody (76813; Abcam). Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole). The coverslips were washed with PBS and mounted with Prolong (Invitrogen, Carlsbad, CA). Confocal microscopy was performed with a Leica TCS SP5 laser scanning system (Leica Microsystems). Images of 1,024 by 1,024 pixels at an 8-bit gray-scale depth were acquired sequentially every 0.13 to 0.3 μm through a 63%, 1.40-numerical-aperture (NA) oil-immersion lens by employing LAS AF v2.6.0 software (Leica Microsystems). Image analysis was performed using ImageJ (32) and the Jacop software plug-in (40) to determine Manders's coefficient. The statistical significance of NXP2/MORC3 and NP colocalization was analyzed using randomization (41) and the method of Van Steensel et al. (42), which confirmed that colocalization occurred in a nonrandom manner. Colocalized pixel maps were generated using the “colocalization threshold” tool of the ImageJ package.
Minireplicon assay.
The recombinant minireplicon assay was performed essentially as described previously (43). In brief, cultures of HEK293T cells (2.5 × 106 cells) were transfected with a mixture of plasmids expressing the RNP components (pCMVPA, 2.5 ng; pCMVPB1, 12.5 ng; pCMVPB2, 12.5 ng; and pCMVNP, 500 ng) and a genomic plasmid expressing a viral RNA (vRNA)-like cat reporter gene (pHHCAT, 500 ng), using the calcium phosphate technique (44). At 20 h posttransfection, total cell extracts were prepared, and chloramphenicol acetyltransferase (CAT) accumulation was determined by enzyme-linked immunosorbent assay (ELISA) (GE Healthcare), using purified CAT enzyme as the standard. For RNA extraction, cell pellets were resuspended in 1 ml of TRIzol reagent (Invitrogen), and the RNA was purified as recommended by the manufacturer. The RNA was digested with RNase-free DNase (1 U/mg) for 1 h at 37°C, extracted with phenol-chloroform-isoamyl alcohol, and precipitated with ethanol. For dot blot hybridization, aliquots of purified RNAs were denatured for 15 min at 55°C in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 7.5% formaldehyde, and fixed on nylon filters by UV cross-linking. Hybridization was carried out overnight at 40°C in 6× SSC, 0.5% SDS, 5× Denhardt's mixture, and 26 to 47% formamide, using gene-specific oligonucleotides as probes. After washing with 0.5× SSC-0.5% SDS at 40°C, the signals were quantified in a phosphorimager. At this point, the probes were eliminated by heating at 70°C in H2O, and the filters were rehybridized with a ribosomal 5S probe to control RNA loading. Synthetic oligonucleotides specific to the different viral RNA species were labeled with [γ-32P]ATP and polynucleotide kinase and used as probes.
RNA isolation and quantification of viral NP RNAs.
Quantification of virus-specific RNA was carried out by reverse transcription–real-time quantitative PCR (RT-qPCR) essentially as described previously (45). Briefly, total RNA was extracted from infected or mock-infected A549 cells by using TRIzol reagent (Invitrogen) as described above. cDNAs complementary to the mRNA or vRNA type of influenza virus NP RNA segment were synthesized with tagged primers (45). A 5.5-μl mixture containing 100 ng of total RNA and 10 pmol of tagged primer was heated for 10 min at 65°C, chilled immediately on ice for 5 min, and then heated again at 60°C. After 5 min, 14.5 μl of a preheated reaction mixture containing 4 μl of 5× First Strand buffer (Invitrogen), 1 μl of 0.1 M dithiothreitol, 1 μl of deoxynucleoside triphosphate (dNTP) mix (10 mM concentration of each dNTP), 1 μl of Superscript III reverse transcriptase (200 U/μl; Invitrogen), 1 μl RNasin Plus RNase inhibitor (40 U/μl; Promega), and 6.5 μl saturated trehalose was added and incubated for 1 h. qPCR was performed with SYBR GreenER qPCR SuperMix on a Prism 7000 sequence detection system (Applied Biosystems). Four microliters of a 10-fold dilution of cDNA was added to the qPCR mixture. The cycling conditions for qPCR were 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min.
RESULTS AND DISCUSSION
We previously reported a proteomic analysis of the intracellular complexes formed by a recombinant influenza virus polymerase (11). This analysis revealed a series of human proteins that were stably associated with the viral enzyme. One such associated factor was NXP2/MORC3, a nuclear matrix-associated protein with RNA-binding activity (15).
NXP2/MORC3 interacts with the influenza virus polymerase complex during viral infection.
In the present study, we wanted to determine whether NXP2/MORC3 interacts with the viral polymerase complex in infected cells. Cultures of HEK293T cells were infected with influenza virus A/WSN/33 (H1N1) (WSN) or A/Victoria/3/75 (H3N2) (Vic) at an MOI of 3 PFU/cell. At 6 h postinfection (hpi), total cell extracts from infected or mock-infected cells were collected and immunoprecipitated with an anti-NXP2/MORC3 antibody, using total mouse IgG as a control. The immunoprecipitated proteins were analyzed by Western blotting with antibodies specific for NXP2/MORC3, PA, and PB1. The results showed that the PA and PB1 polymerase subunits associated with NXP2/MORC3, whereas none of the proteins were immunoprecipitated with the control mix of antibodies (Fig. 1).
FIG 1.
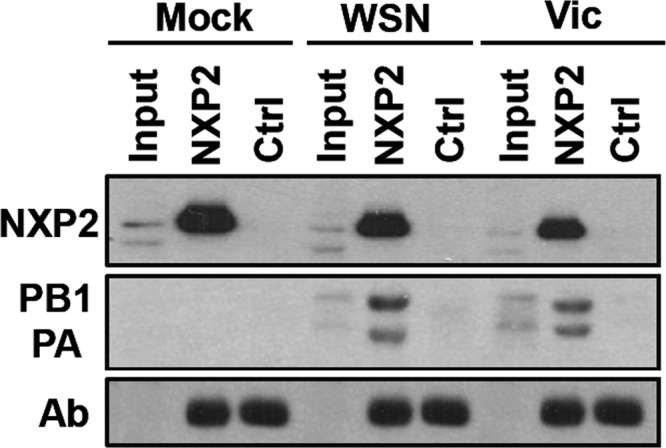
Interaction of NXP2/MORC3 with influenza virus polymerase. Cultures of HEK293T cells were infected with the WSN or Victoria (Vic) influenza virus strain at an MOI of 3 PFU/cell or mock infected (Mock). At 6 hpi, total cell extracts were prepared and immunoprecipitated with anti-NXP2/MORC3 or control antibodies. The presence of NXP2/MORC3, PB1, and PA was examined by Western blotting with an antibody specific for NXP2/MORC3 or a mixture of anti-PB1 and anti-PA antibodies. Input, total cell extracts; NXP2, immunoprecipitates obtained with NXP2/MORC3-specific antibodies; Ctrl, immunoprecipitates obtained with unspecific antibodies. The positions of NXP2/MORC3, PB1, PA, and the IgG heavy chain (Ab) are indicated to the left. Changes in electrophoretic mobility for the PB1 and PA proteins are a consequence of the immunoprecipitation process.
Cellular accumulation and localization of NXP2/MORC3 change during influenza virus infection.
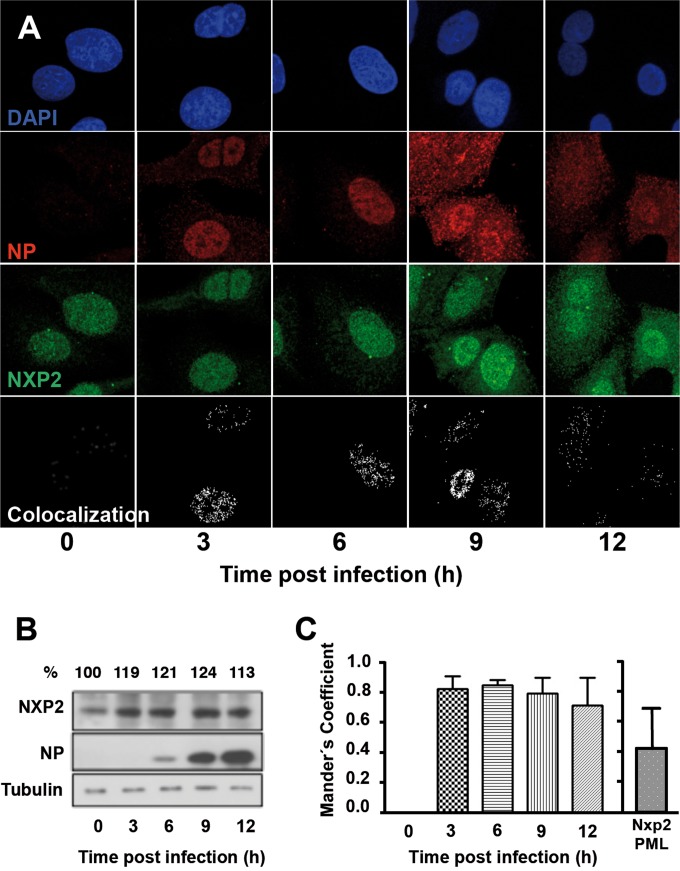
We next examined whether the intracellular distribution or expression level of NXP2/MORC3 was altered during infection. The localization of NXP2/MORC3 was analyzed by confocal immunofluorescence microscopy, and the results are presented in Fig. 2A. Cultures of A549 cells were mock infected or infected with the WSN strain at an MOI of 3 PFU/cell and at various times after infection were fixed and analyzed by immunofluorescence with antibodies specific for NXP2/MORC3 or NP. The NXP2/MORC3 protein was found to be distributed throughout the nucleus but excluded from nucleoli in uninfected cells, as reported previously (15), as well as in early infected cells, but the staining was slightly stronger and the signal could be detected in the cytoplasm at later times of infection (Fig. 2A, 9 to 12 hpi). This change in cellular distribution is specific for NXP2/MORC3, since the localization of other nuclear proteins that interact with viral RNPs, such as RNA polymerase II (Pol II), the CHD6 chromatin remodeler, or the SFPQ/PSF splicing factor, was not affected by the infection (37, 46, 47). The apparent overexpression detected by immunofluorescence assay was verified by Western blotting. Cultures of A549 cells were infected with the influenza virus WSN strain at an MOI of 3 PFU/cell or left uninfected, and total cell extracts were prepared at various times after infection. The accumulation of NXP2/MORC3 was determined using an antibody specific for NXP2/MORC3, with an antibody for NP used as a marker of infection and one for β-tubulin used as a loading marker (Fig. 2B). An increase in the accumulation of NXP2/MORC3 was consistently observed at various times after influenza virus infection.
FIG 2.
Accumulation and intracellular localization of NXP2/MORC3 during influenza virus infection. (A) Cultures of A549 cells were infected with the WSN influenza virus at an MOI of 3 PFU/cell. At the indicated times after infection, cells were fixed and processed for confocal immunofluorescence microscopy. Representative optical sections from three independent experiments are shown. Nuclei are presented in blue, NP in red, and NXP2/MORC3 in green. Colocalization masks are shown in white. (B) Total cell extracts of parallel cultures were analyzed by Western blotting with antibodies specific for NXP2/MORC3, NP, and β-tubulin (loading marker). Representative data from one of three independent experiments are presented. Numbers at the top indicate the quantification of the NXP2/MORC3 signal, using the value obtained at the time of virus inoculation as 100%. (C) Quantification of NP-NXP2/MORC3 colocalization. ImageJ software was used to calculate Manders's coefficient for colocalization. Averages for the values from a representative experiment are shown. Error bars indicate standard deviations. As a specificity control, the panel on the right shows Manders's coefficient for colocalization of NXP2 with PML, as an example of an alternative nuclear protein.
To analyze the potential colocalization of NXP2/MORC3 with influenza virus RNPs, we used NP as a surrogate marker. The results of applying a colocalization mask showing only overlapping NXP2/MORC3-NP pixels as white spots are presented in Fig. 2A, lower panels, and show a nuclear colocalization pattern. Manders's coefficient was determined as described in Materials and Methods, and the results are presented in Fig. 2C. These values were shown to be significant as ascertained by randomization (41, 42), confirming that NXP2/MORC3-NP colocalization occurred in a nonrandom manner. The specificity of such colocalization was verified by performing a similar analysis with NXP2-PML proteins, and no significant colocalization was observed with this control protein pair in human A549 cells (Fig. 2C). These results are in agreement with the coimmunoprecipitation data and confirm that NXP2/MORC3 associates with the viral RNP complex, most probably through interaction with the polymerase (11), during influenza virus infection.
NXP2/MORC3 downregulation affects influenza virus multiplication.
The results presented above suggest that NXP2/MORC3 may be important for influenza virus infection. To investigate the potential relevance of the NXP2/MORC3 protein in the virus life cycle, we generated A549 cell populations in which NXP2/MORC3 was downregulated by transduction with lentiviral vectors expressing two alternative NXP2/MORC3-specific shRNAs (shRNA1 and shRNA2 cells) or an irrelevant sequence (shRNA C cells), as a control. As shown in Fig. 3A, the expression of the NXP2/MORC3 protein was highly reduced in shRNA1 or shRNA2 cells compared with shRNA C cells (around 80% reduction). Both shRNA1 and shRNA2 cells showed consistent and prolonged NXP2/MORC3 downregulation, without any overt impact on cell viability (Fig. 3A and B). In humans, influenza virus infections occur in the epithelium of the upper respiratory tract under low-MOI conditions. Thus, to establish the potential significance of NXP2/MORC3, we infected shRNA C, shRNA1, and shRNA2 cells with influenza virus at an MOI of 0.001 PFU/cell and determined the viral titers in the supernatants during infection by plaque assay on MDCK cells. The results of kinetic experiments run in triplicate are presented in Fig. 3C and show a significant reduction in viral titer, of around 1 log unit, for both NXP2/MORC3-deficient cell lines compared with control cells. These results indicate that the NXP2/MORC3 protein is important for influenza virus replication. To determine whether NXP2/MORC3 is specifically relevant for influenza virus infection, the multiplication of the following two additional viruses was studied in NXP2/MORC3-downregulated cells: VSV, as a further example of a negative-strand RNA virus; and adenovirus 5 (Ad5), as a nuclear DNA virus that is fully dependent on the cell transcriptional machinery. Cultures of A549 shRNA1, shRNA2, or shRNA C cells were infected with either VSV or Ad5, and the amount of virus accumulated in the culture supernatant (VSV) or infected cells (Ad5) was determined by a plaque assay on BHK21 cells (VSV) or a focus-forming assay on HEK293T cells (Ad5). As presented in Fig. 4, the multiplication of none of these viruses was affected by downregulation of NXP2/MORC3, indicating that this human protein is a host factor specifically important for influenza virus replication.
FIG 3.
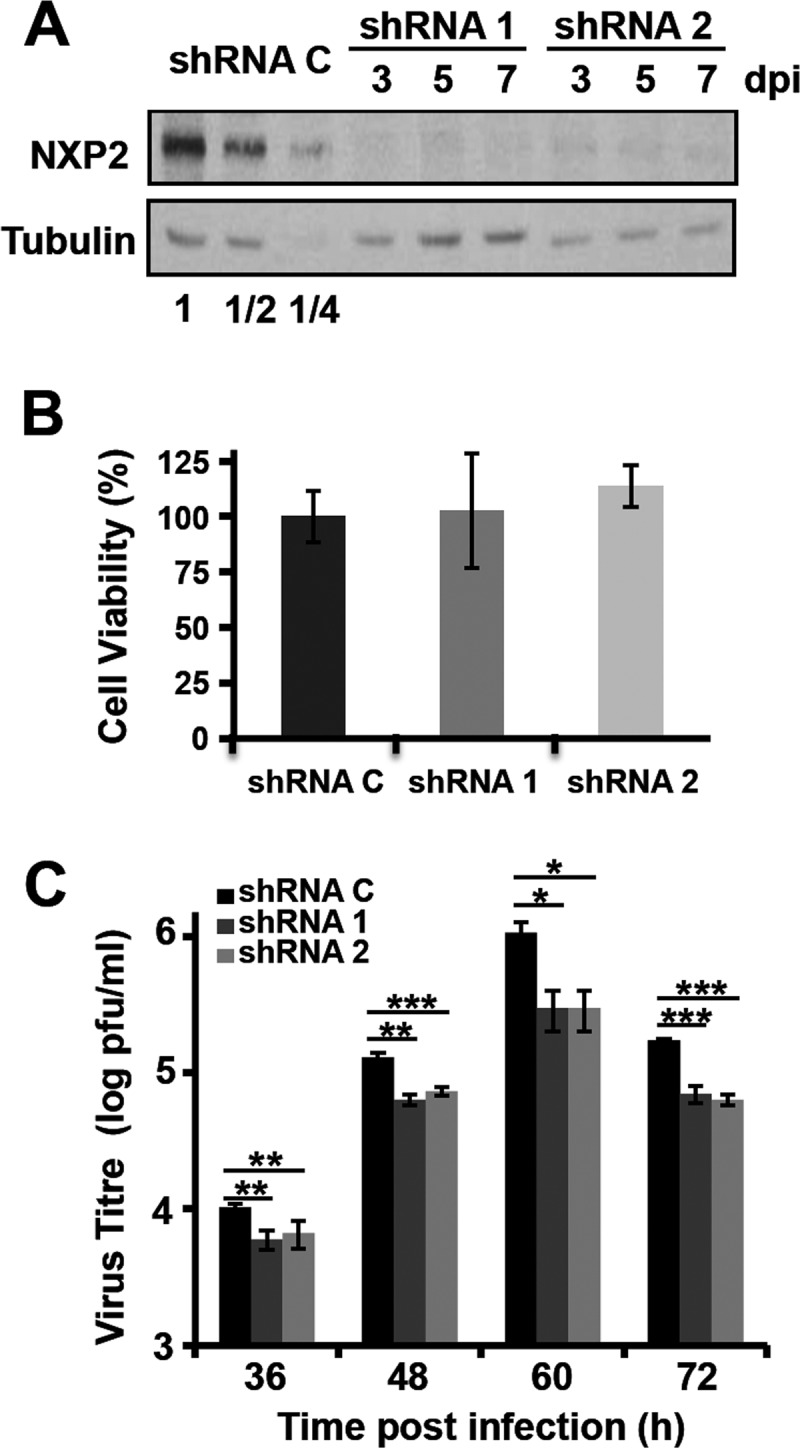
Kinetics of influenza virus multiplication in NXP2/MORC3-downregulated cells. Cultures of A549 cells were transduced with lentiviral vectors expressing an irrelevant short hairpin RNA (shRNA C) or shRNAs targeting the NXP2/MORC3 mRNA (shRNA1 or shRNA2). When the accumulation levels of NXP2/MORC3 were reduced, the cultures were infected with influenza virus WSN at an MOI of 0.001 PFU/cell. (A) Western blot analysis of NXP2/MORC3 at the indicated days post-lentiviral inoculation (dpi), using β-tubulin as a loading control. Serial 2-fold dilutions of control extracts were loaded for improved quantification. (B) The viability of control or downregulated A549 cells was determined using an MTT-based colorimetric assay as described in Materials and Methods. (C) The influenza virus infectivity of the supernatant was determined by plaque assay at the indicated times postinfection. The results presented are representative of three independent experiments. The statistical significance of the differences from the control data set was determined using the Student t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
FIG 4.
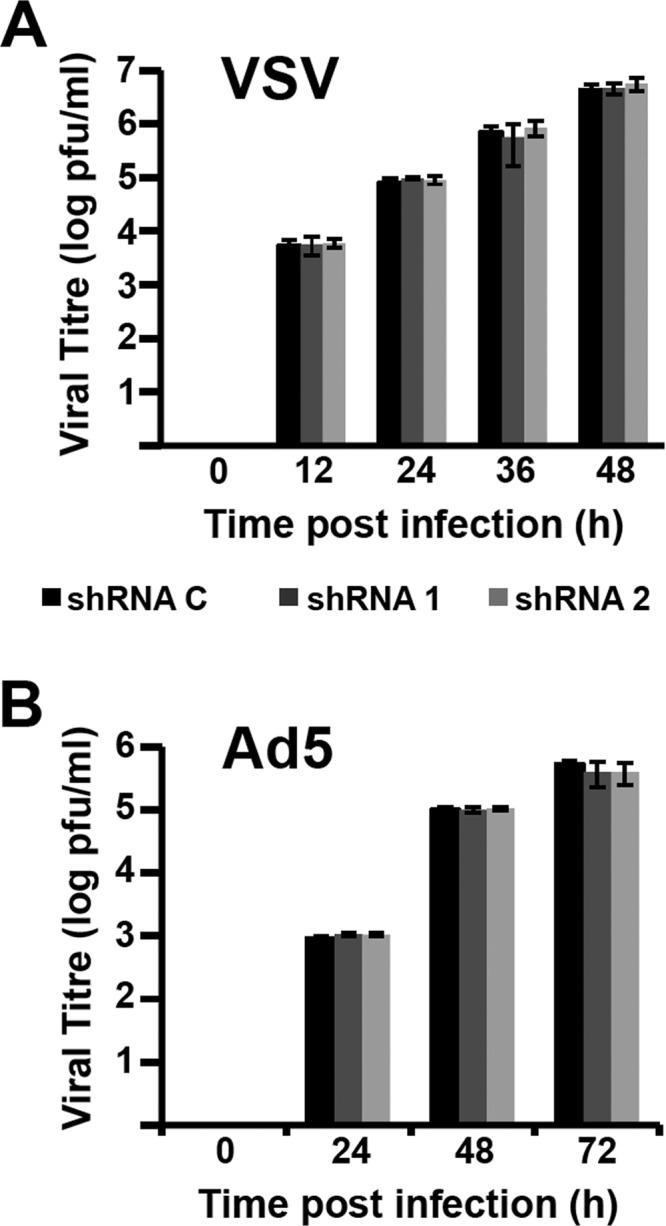
Kinetics of VSV and adenovirus multiplication in NXP2/MORC3-downregulated cells. Cultures of A549 cells were transduced with lentiviral vectors expressing an irrelevant short hairpin RNA (shRNA C) or shRNAs targeting NXP2/MORC3 mRNA (shRNA1 or shRNA2). When the accumulation levels of NXP2/MORC3 were reduced, the cultures were infected with VSV (top; MOI = 0.001 PFU/cell) or Ad5 (bottom; MOI = 5 PFU/cell) as described in Materials and Methods. Virus titration was performed with samples obtained at the indicated times. The results presented are representative of two or three independent experiments.
NXP2/MORC3 is required for virus transcription.
As NXP2/MORC3 is a nuclear matrix protein (15) that associates with viral RNPs, we set out to test whether the synthesis of viral RNAs was affected by NXP2/MORC3 downregulation during infection. Cultures of shRNA C, shRNA1, and shRNA2 cells were infected with influenza virus at an MOI of 3 PFU/cell or mock infected, and total cell RNAs were isolated. The accumulation of virus NP vRNA and mRNA was determined using a strand-specific RT-qPCR technique (45). A reduction of viral RNA accumulation was detected in shRNA1 and shRNA2 cells compared with shRNA C cells (Fig. 5A and B). These results indicate that virus transcription, virus RNA replication, or both are affected by NXP2/MORC3 downregulation. To distinguish between these possibilities, we used a recombinant minireplicon system generated by transfection of HEK293T cells with a mixture of plasmids expressing the viral polymerase subunits, the NP, and a pseudoviral genome carrying the cat gene as a reporter. In this system, virus transcription and RNA replication are uncoupled, as the virus proteins required are provided in trans by cellular RNA Pol II transcription, and the amplification of RNPs and their activity can be measured by the accumulation of CAT protein (43). We first set up the conditions for NXP2/MORC3 downregulation and verified that this silencing did not affect cellular viability. As presented in Fig. 6A, the expression of NXP2/MORC3 was strongly reduced in shRNA1- and shRNA2-transduced cells compared to control shRNA-transduced cells, but such downregulation did not significantly alter cell viability (Fig. 6B). Control- or NXP2/MORC3-silenced cells were then used to generate cat RNPs, and CAT accumulation was determined as a measure of their replication/transcription activity (Fig. 6C). The results showed a 50% reduction in CAT accumulation in NXP2/MORC3-silenced cells compared to control-silenced cells, whereas the expression of RNPs (PA and NP) was not altered by NXP2/MORC3 silencing, as determined by Western blotting (Fig. 6D). Next, the levels of accumulation of positive-polarity and negative-polarity cat RNAs were determined. As shown in Fig. 7A, a small but significant reduction in viral transcription was apparent in NXP2/MORC3-silenced cells compared with control-silenced cells, whereas no significant change in vRNA accumulation was observed (Fig. 7B). These results are consistent with the reduction in viral transcription observed in infected cells and with the decrease in CAT accumulation, and they indicate that NXP2/MORC3 is required for viral transcription but not for virus RNA replication.
FIG 5.
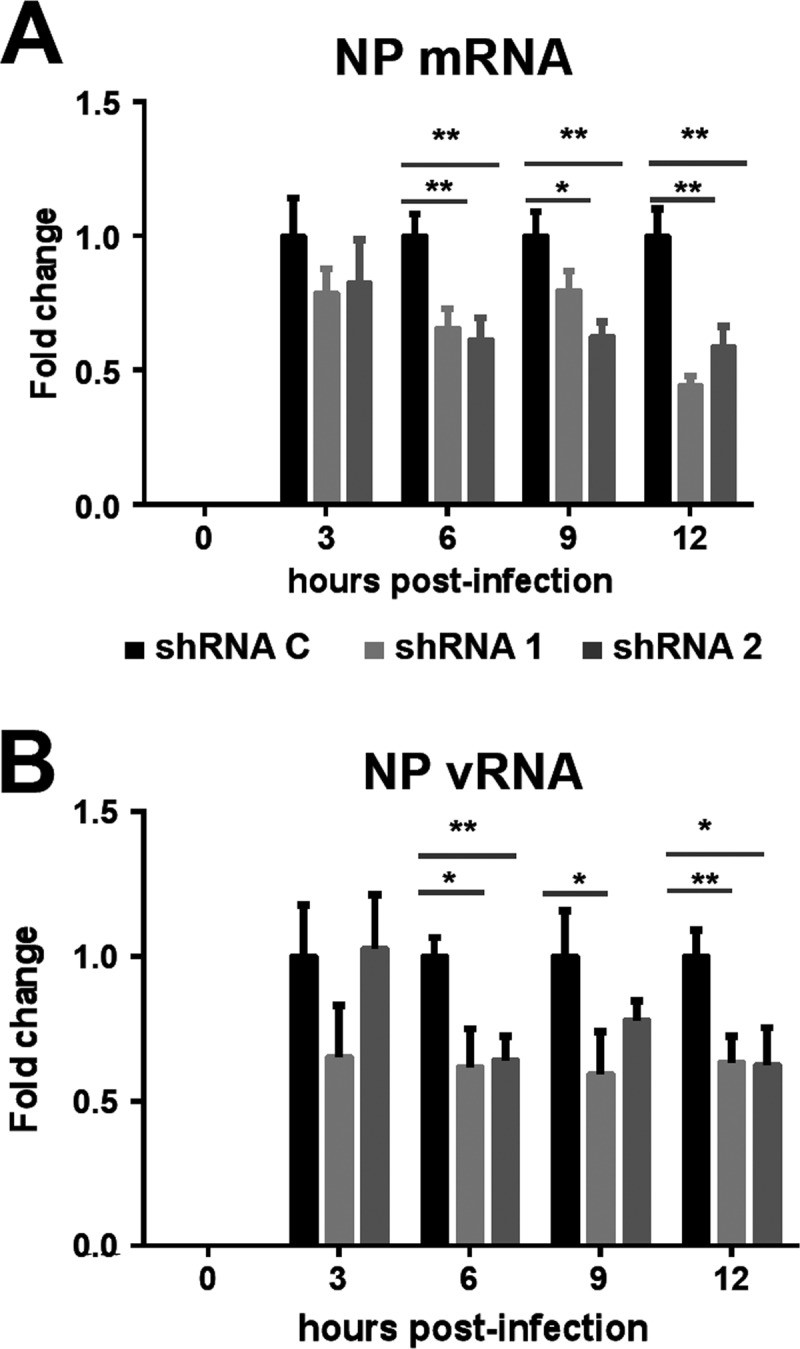
Accumulation of viral genomic RNA and mRNAs during infection of NXP2/MORC3-downregulated cells. Cultures of A549 cells were transduced with lentiviral vectors expressing an irrelevant short hairpin RNA (shRNA C) or shRNAs targeting NXP2/MORC3 mRNA (shRNA1 or shRNA2). When the accumulation levels of NXP2/MORC3 were reduced, the cultures were infected with influenza virus WSN at an MOI of 3 PFU/cell. Total cell RNA was isolated at the indicated times after infection, and the accumulation of NP mRNA (A) and vRNA (B) was determined by RT-qPCR, using the conditions and primers described by Kawakami et al. (45). The values shown are averages and standard deviations for three determinations and have been standardized to the value obtained for control cells at each infection time point. The statistical significance of differences from the data obtained for control cells was determined using the Student t test (*, P < 0.05; **, P < 0.01).
FIG 6.
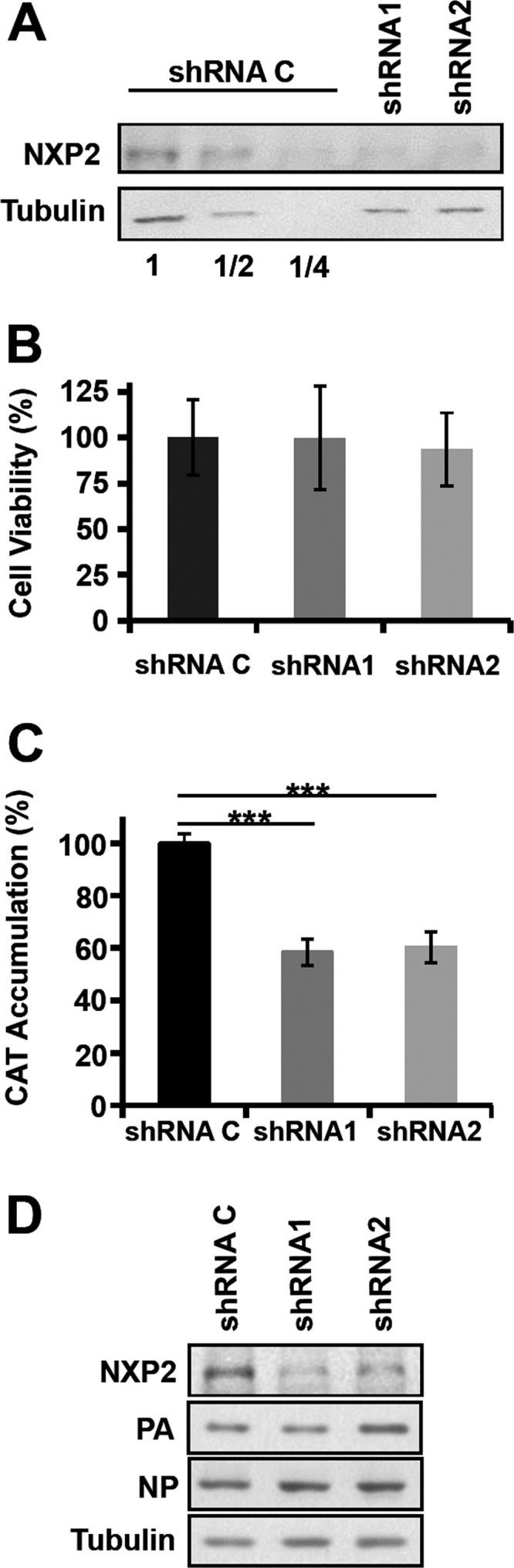
Biological activity of recombinant influenza virus RNPs in NXP2/MORC3-downregulated cells. (A) Cultures of HEK293T cells were transduced with lentiviral vectors expressing an irrelevant short hairpin RNA (shRNA C) or shRNAs targeting NXP2/MORC3 mRNA (shRNA1 or shRNA2), and silencing was monitored in total cell extracts by Western blotting. Serial 2-fold dilutions of control extracts were loaded for improved quantification. The accumulation of β-tubulin was used as a loading control. (B) The viability of control or NXP2/MORC3-downregulated HEK293T cells was tested by using an MTT-based colorimetric assay as described in Materials and Methods. (C) The NXP2/MORC3-downregulated or control HEK293T cells described for panel A were used for in vivo RNP reconstitution. The cultures were transfected with plasmids expressing the polymerase subunits and the NP, as well as with a genomic plasmid expressing a pseudoviral genome containing the cat gene in negative polarity. Twenty-four hours after transfection, the accumulation of the CAT protein in total cell extracts was analyzed by ELISA. The statistical significance of differences from the control data set was determined using the Student t test (***, P < 0.001). (D) Aliquots of the samples used in the experiment whose results are shown in panel C were also analyzed for the presence of RNPs by Western blotting, using β-tubulin as a loading control.
FIG 7.
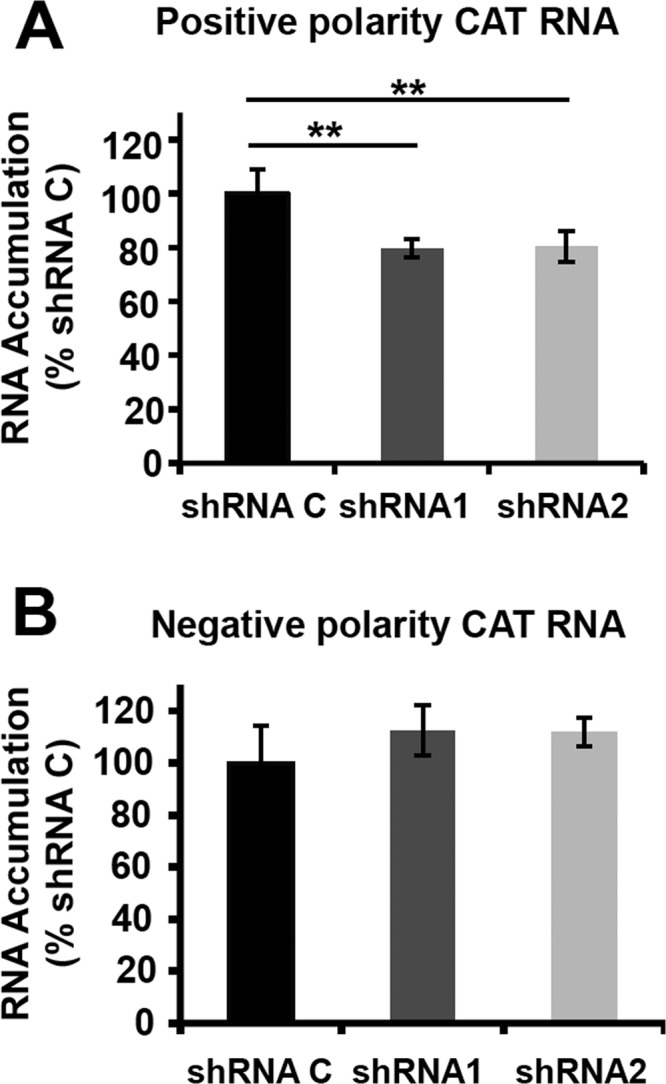
Transcription and replication activity of recombinant influenza virus RNPs in NXP2/MORC3-downregulated cells. Cultures of HEK293T cells were transduced with lentiviral vectors expressing an irrelevant short hairpin RNA (shRNA C) or shRNAs targeting NXP2/MORC3 mRNA (shRNA1 or shRNA2). When the expression of NXP2/MORC3 was downregulated, the cultures were transfected with plasmids expressing the polymerase subunits and the NP, as well as with a genomic plasmid expressing a pseudoviral genome containing the cat gene in negative polarity. At 24 h posttransfection, total cell RNA was isolated. (A) The accumulation of positive-polarity RNA was determined by hybridization with CAT-specific probes and is presented as a percentage of the maximal value obtained for shRNA C-silenced cells. (B) The accumulation of negative-polarity RNA was determined by hybridization with CAT-specific probes and is presented as a percentage of the maximal value obtained for shRNA C-silenced cells. The values represent the averages and standard deviations for three determinations. The statistical significance was determined by the Student t test (**, P < 0.01).
Potential roles of NXP2/MORC3 in influenza virus infection.
Like all viruses, influenza viruses make use of host cell pathways, structures, and specific factors to carry out their replication cycle. Several “-omics” approaches, including two-hybrid, genomic silencing, and proteomic techniques, have revealed a wealth of interactions between the virus and host (48–53). Regarding the RNA transcription and replication steps of infection, proteomic approaches based on the viral polymerase or RNP provided a complex list of interacting host factors (11, 54; reviewed in reference 7), among which we selected NXP2/MORC3 for further analysis. Here we show that NXP2/MORC3 associates with the viral polymerase and colocalizes with viral RNPs during infection (Fig. 1 and 2). Consistent with a slight amount of NXP2/MORC3 overexpression and a partial relocalization during virus infection (Fig. 1 and 2), we further show that NXP2/MORC3 is important for the virus to achieve a proper level of replication fitness (Fig. 3 and 4). In contrast to the results presented here, Bortz et al. reported that NXP2/MORC3 is a negative regulator of influenza virus (53). They reported a 2-fold stimulation of polymerase activity and virus yield by silencing of NXP2/MORC3, while we showed an around 10-fold reduction of virus production under similar conditions. This discrepancy may result from the different approaches used to silence NXP2/MORC3: while Bortz et al. used a combination of three small interfering RNAs (siRNAs), we used stable downregulated cells obtained independently by lentiviral transduction of two alternative shRNAs. It is worth mentioning that in the same study, Bortz et al. reported that SFPQ/PSF is a negative regulator of influenza virus, but we have demonstrated that SFPQ/PSF stimulates influenza virus transcription by promoting virus mRNA polyadenylation (37).
In contrast to most negative-strand RNA viruses, influenza viruses synthesize viral RNAs in the cell nucleus, and virus transcription is coupled to cellular mRNA synthesis. Thus, viral RNPs utilize nascent cell pre-mRNAs to steal capped oligonucleotides and use them as primers for virus transcription (55; reviewed in reference 4). In agreement with this mechanism, associations of the viral polymerase with cellular RNA polymerase II and other transcription-related proteins have been described (46, 47, 56, 57). Because active cell transcriptional complexes are associated with the nuclear matrix (58), it is no surprise that influenza virus RNA synthesis also occurs at specific sites in the nucleus (12), in association with the nuclear matrix (13). The polymerase-associated factor NXP2/MORC3 studied here is an RNA-binding protein connected to the nuclear matrix (15) that may regulate transcription at the epigenetic level (reviewed in reference 14). The potential roles of NXP2/MORC3 in cellular regulation are consistent with the results presented in Fig. 5 to 7, indicating that NXP2/MORC3 is important for viral gene expression at the transcriptional level. However, the lack of any inhibition of virus yield in adenovirus 5-infected cells by NXP2/MORC3 downregulation (Fig. 4) indicates that the reduction in influenza virus transcription is not an indirect consequence of an inhibition of cellular transcription.
In summary, we have shown that NXP2/MORC3, an influenza virus polymerase-binding protein, is relevant for virus infection and is important for efficient virus transcription but not for viral RNA replication.
ACKNOWLEDGMENTS
We thank Pablo Gastaminza, Puri Fortes, Norimitsu Inoue, Francisco J. Iborra, José Antonio Melero, Ariel Rodríguez-Frandsen, and Juan Carlos de la Torre for providing biological reagents and Ariel Rodríguez-Frandsen and Urtzi Garaigorta for their comments on the manuscript. The technical assistance of Silvia Gutiérrez-Erlandsson, Susana Hernández, Yolanda Fernández, and Noelia Zamarreño is gratefully acknowledged.
This work was supported by grants from MICINN (grant BFU2010-17540), FLUPHARM (grant FP7-259751) (European Union), CIBER de Enfermedades Respiratorias (Instituto de Salud Carlos III), and the Fundación Marcelino Botín. S.L.-B. was supported by a predoctoral FPU fellowship (MICINN).
REFERENCES
- 1.Palese P, Shaw M. 2013. Orthomyxoviridae, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Vasin AV, Temkina OA, Egorov VV, Klotchenko SA, Plotnikova MA, Kiselev OI. 2014. Molecular mechanisms enhancing the proteome of influenza A viruses: an overview of recently discovered proteins. Virus Res 185:53–63. doi: 10.1016/j.virusres.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Arranz R, Coloma R, Chichon FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortin J, Martin-Benito J. 2012. The structure of native influenza virion ribonucleoproteins. Science 338:1634–1637. doi: 10.1126/science.1228172. [DOI] [PubMed] [Google Scholar]
- 4.Martin-Benito J, Ortin J. 2013. Influenza virus transcription and replication. Adv Virus Res 87:113–137. doi: 10.1016/B978-0-12-407698-3.00004-1. [DOI] [PubMed] [Google Scholar]
- 5.Resa-Infante P, Jorba N, Coloma R, Ortin J. 2011. The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol 8:207–215. doi: 10.4161/rna.8.2.14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fodor E. 2013. The RNA polymerase of influenza A virus: mechanisms of viral transcription and replication. Acta Virol 57:113–122. doi: 10.4149/av_2013_02_113. [DOI] [PubMed] [Google Scholar]
- 7.Eisfeld AJ, Neumann G, Kawaoka Y. 2015. At the centre: influenza A virus ribonucleoproteins. Nat Rev Microbiol 13:28–41. doi: 10.1038/nrmicro3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuoka Y, Matsumae H, Katoh M, Eisfeld AJ, Neumann G, Hase T, Ghosh S, Shoemaker JE, Lopes TJ, Watanabe T, Watanabe S, Fukuyama S, Kitano H, Kawaoka Y. 2013. A comprehensive map of the influenza A virus replication cycle. BMC Syst Biol 7:97. doi: 10.1186/1752-0509-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naffakh N, Tomoiu A, Rameix-Welti MA, van der Werf S. 2008. Host restriction of avian influenza viruses at the level of the ribonucleoproteins. Annu Rev Microbiol 62:403–424. doi: 10.1146/annurev.micro.62.081307.162746. [DOI] [PubMed] [Google Scholar]
- 10.Nagata K, Kawaguchi A, Naito T. 2008. Host factors for replication and transcription of the influenza virus genome. Rev Med Virol 18:247–260. doi: 10.1002/rmv.575. [DOI] [PubMed] [Google Scholar]
- 11.Jorba N, Juarez S, Torreira E, Gastaminza P, Zamarreno N, Albar JP, Ortin J. 2008. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics 8:2077–2088. doi: 10.1002/pmic.200700508. [DOI] [PubMed] [Google Scholar]
- 12.Jackson DA, Caton AJ, McCready SJ, Cook PR. 1982. Influenza virus RNA is synthesized at fixed sites in the nucleus. Nature 296:366–368. doi: 10.1038/296366a0. [DOI] [PubMed] [Google Scholar]
- 13.Lopez-Turiso JA, Martinez C, Tanaka T, Ortin J. 1990. The synthesis of influenza virus negative-strand RNA takes place in insoluble complexes present in the nuclear matrix fraction. Virus Res 16:325–337. doi: 10.1016/0168-1702(90)90056-H. [DOI] [PubMed] [Google Scholar]
- 14.Li DQ, Nair SS, Kumar R. 2013. The MORC family: new epigenetic regulators of transcription and DNA damage response. Epigenetics 8:685–693. doi: 10.4161/epi.24976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura Y, Sakai F, Nakano O, Kisaki O, Sugimoto H, Sawamura T, Sadano H, Osumi T. 2002. The newly identified human nuclear protein NXP-2 possesses three distinct domains, the nuclear matrix-binding, RNA-binding, and coiled-coil domains. J Biol Chem 277:20611–20617. doi: 10.1074/jbc.M201440200. [DOI] [PubMed] [Google Scholar]
- 16.Dutta R, Inouye M. 2000. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci 25:24–28. doi: 10.1016/S0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 17.Perry J, Zhao Y. 2003. The CW domain, a structural module shared amongst vertebrates, vertebrate-infecting parasites and higher plants. Trends Biochem Sci 28:576–580. doi: 10.1016/j.tibs.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Inoue N, Hess KD, Moreadith RW, Richardson LL, Handel MA, Watson ML, Zinn AR. 1999. New gene family defined by MORC, a nuclear protein required for mouse spermatogenesis. Hum Mol Genet 8:1201–1207. doi: 10.1093/hmg/8.7.1201. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi K, Yoshida N, Murakami N, Kawata K, Ishizaki H, Tanaka-Okamoto M, Miyoshi J, Zinn AR, Shime H, Inoue N. 2007. Dynamic regulation of p53 subnuclear localization and senescence by MORC3. Mol Biol Cell 18:1701–1709. doi: 10.1091/mbc.E06-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mimura Y, Takahashi K, Kawata K, Akazawa T, Inoue N. 2010. Two-step colocalization of MORC3 with PML nuclear bodies. J Cell Sci 123:2014–2024. doi: 10.1242/jcs.063586. [DOI] [PubMed] [Google Scholar]
- 21.Chelbi-Alix MK, Quignon F, Pelicano L, Koken MH, de The H. 1998. Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J Virol 72:1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geoffroy MC, Chelbi-Alix MK. 2011. Role of promyelocytic leukemia protein in host antiviral defense. J Interferon Cytokine Res 31:145–158. doi: 10.1089/jir.2010.0111. [DOI] [PubMed] [Google Scholar]
- 23.Ortin J, Najera R, Lopez C, Davila M, Domingo E. 1980. Genetic variability of Hong Kong (H3N2) influenza viruses: spontaneous mutations and their location in the viral genome. Gene 11:319–331. doi: 10.1016/0378-1119(80)90072-4. [DOI] [PubMed] [Google Scholar]
- 24.Tobita K, Sugiura A, Enomote C, Furuyama M. 1975. Plaque assay and primary isolation of influenza A viruses in an established line of canine kidney cells (MDCK) in the presence of trypsin. Med Microbiol Immunol 162:9–14. doi: 10.1007/BF02123572. [DOI] [PubMed] [Google Scholar]
- 25.Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P. 2006. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J Virol 80:1376–1384. doi: 10.1128/JVI.80.3.1376-1384.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. 1998. A third-generation lentivirus vector with a conditional packaging system. J Virol 72:8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, Weinberg RA, Novina CD. 2003. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falcon AM, Marion RM, Zurcher T, Gomez P, Portela A, Nieto A, Ortin J. 2004. Defective RNA replication and late gene expression in temperature-sensitive influenza viruses expressing deleted forms of the NS1 protein. J Virol 78:3880–3888. doi: 10.1128/JVI.78.8.3880-3888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Llompart CM, Nieto A, Rodriguez-Frandsen A. 2014. Specific residues of PB2 and PA influenza virus polymerase subunits confer the ability for RNA polymerase II degradation and virus pathogenicity in mice. J Virol 88:3455–3463. doi: 10.1128/JVI.02263-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levitz SM, Diamond RD. 1985. A rapid colorimetric assay of fungal viability with the tetrazolium salt MTT. J Infect Dis 152:938–945. doi: 10.1093/infdis/152.5.938. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 32.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barcena J, Ochoa M, de la Luna S, Melero JA, Nieto A, Ortin J, Portela A. 1994. Monoclonal antibodies against influenza virus PB2 and NP polypeptides interfere with the initiation step of viral mRNA synthesis in vitro. J Virol 68:6900–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ochoa M, Barcena J, de la Luna S, Melero JA, Douglas AR, Nieto A, Ortin J, Skehel JJ, Portela A. 1995. Epitope mapping of cross-reactive monoclonal antibodies specific for the influenza A virus PA and PB2 polypeptides. Virus Res 37:305–315. doi: 10.1016/0168-1702(95)00039-S. [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez S, Ortin J. 1999. Characterization of influenza virus PB1 protein binding to viral RNA: two separate regions of the protein contribute to the interaction domain. J Virol 73:631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coloma R, Valpuesta JM, Arranz R, Carrascosa JL, Ortin J, Martin-Benito J. 2009. The structure of a biologically active influenza virus ribonucleoprotein complex. PLoS Pathog 5:e1000491. doi: 10.1371/journal.ppat.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landeras-Bueno S, Jorba N, Perez-Cidoncha M, Ortin J. 2011. The splicing factor proline-glutamine rich (SFPQ/PSF) is involved in influenza virus transcription. PLoS Pathog 7:e1002397. doi: 10.1371/journal.ppat.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nin N, Lorente JA, Sanchez-Rodriguez C, Granados R, Ver LS, Soto L, Hidalgo J, Fernandez-Segoviano P, Ortin J, Esteban A. 2011. Kidney histopathological findings in fatal pandemic 2009 influenza A (H1N1). Intensive Care Med 37:880–881. doi: 10.1007/s00134-011-2183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez A, Perez-Gonzalez A, Nieto A. 2007. Influenza virus infection causes specific degradation of the largest subunit of cellular RNA polymerase II. J Virol 81:5315–5324. doi: 10.1128/JVI.02129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolte S, Cordelieres FP. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 41.Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. 2004. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J 86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Steensel B, van Binnendijk EP, Hornsby CD, van der Voort HT, Krozowski ZS, de Kloet ER, van Driel R. 1996. Partial colocalization of glucocorticoid and mineralocorticoid receptors in discrete compartments in nuclei of rat hippocampus neurons. J Cell Sci 109:787–792. [DOI] [PubMed] [Google Scholar]
- 43.Jorba N, Coloma R, Ortin J. 2009. Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication. PLoS Pathog 5:e1000462. doi: 10.1371/journal.ppat.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wigler M, Pellicer A, Silverstein S, Axel R, Urlaub G, Chasin L. 1979. DNA-mediated transfer of the adenine phosphoribosyltransferase locus into mammalian cells. Proc Natl Acad Sci U S A 76:1373–1376. doi: 10.1073/pnas.76.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawakami E, Watanabe T, Fujii K, Goto H, Watanabe S, Noda T, Kawaoka Y. 2011. Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA. J Virol Methods 173:1–6. doi: 10.1016/j.jviromet.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alfonso R, Lutz T, Rodriguez A, Chavez JP, Rodriguez P, Gutierrez S, Nieto A. 2011. CHD6 chromatin remodeler is a negative modulator of influenza virus replication that relocates to inactive chromatin upon infection. Cell Microbiol 13:1894–1906. doi: 10.1111/j.1462-5822.2011.01679.x. [DOI] [PubMed] [Google Scholar]
- 47.Engelhardt OG, Fodor E. 2006. Functional association between viral and cellular transcription during influenza virus infection. Rev Med Virol 16:329–345. doi: 10.1002/rmv.512. [DOI] [PubMed] [Google Scholar]
- 48.Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Maurer AP, Muller E, Wolff T, Rudel T, Meyer TF. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 49.Konig R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, Garcia-Sastre A, Young JA, Palese P, Shaw ML, Chanda SK. 2010. Human host factors required for influenza virus replication. Nature 463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, Hill DE, Regev A, Hacohen N. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bortz E, Westera L, Maamary J, Steel J, Albrecht RA, Manicassamy B, Chase G, Martinez-Sobrido L, Schwemmle M, Garcia-Sastre A. 2011. Host- and strain-specific regulation of influenza virus polymerase activity by interacting cellular proteins. mBio 2:e00151–11. doi: 10.1128/mBio.00151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayer D, Molawi K, Martinez-Sobrido L, Ghanem A, Thomas S, Baginsky S, Grossmann J, Garcia-Sastre A, Schwemmle M. 2007. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J Proteome Res 6:672–682. doi: 10.1021/pr060432u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plotch SJ, Bouloy M, Ulmanen I, Krug RM. 1981. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 56.Huarte M, Sanz-Ezquerro JJ, Roncal F, Ortin J, Nieto A. 2001. PA subunit from influenza virus polymerase complex interacts with a cellular protein with homology to a family of transcriptional activators. J Virol 75:8597–8604. doi: 10.1128/JVI.75.18.8597-8604.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodriguez A, Perez-Gonzalez A, Nieto A. 2011. Cellular human CLE/C14orf166 protein interacts with influenza virus polymerase and is required for viral replication. J Virol 85:12062–12066. doi: 10.1128/JVI.00684-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mortillaro MJ, Blencowe BJ, Wei X, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R. 1996. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci U S A 93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]