Abstract
We identified two key amino acid residues within human CD134 (hCD134) that are required for its interaction with human herpesvirus 6B (HHV-6B) and for HHV-6B entry into cells. One of the residues (K79) allows access of the HHV-6B ligand to hCD134. Murine CD134 (mCD134) functioned as an HHV-6B receptor when these two amino acid residues were replaced with homologous human residues. This study identifies both the HHV-6B receptor-ligand interaction and the species-specific determinants of hCD134 essential for HHV-6B entry.
TEXT
Human herpesvirus 6B (HHV-6B) is a betaherpesvirus. Initially, HHV-6B and HHV-6A were classified as two variants of HHV-6; however, they are now classified as two virus species due to their different bio-characteristics (1–5). One of the striking differences between these two viruses is receptor ligand usage. The HHV-6A ligand, glycoprotein H (gH)/gL/gQ1/gQ2 complex, binds to human CD46, a ubiquitously expressed molecule that may contribute to the relatively broad cell tropism of HHV-6A (6–8). In contrast, human CD134 (hCD134) is the specific receptor for HHV-6B (9, 10).
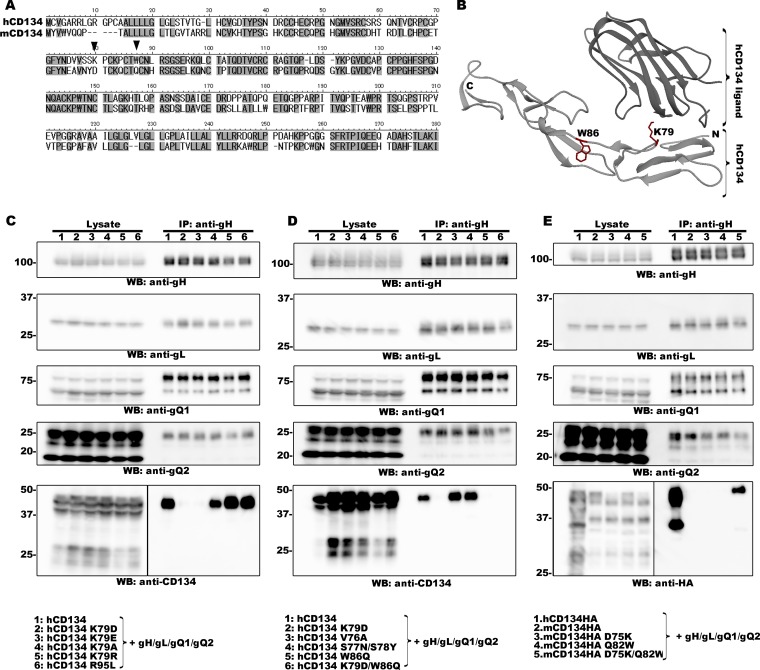
In a previous study, we performed a detailed analysis of the interaction between the HHV-6B ligand and its cellular receptor, hCD134. We found that cysteine-rich domain 2 (CRD 2) of hCD134 is critical for HHV-6B ligand binding and that a key amino acid residue (E127) within HHV-6B gQ1 is required for its function. The E127Q mutation within HHV-6B gQ1 completely abolishes binding to hCD134; however, the homologous mutation (Q135E) in HHV-6A gQ1 does not enable it to bind to CD134, even though these two gQ1s share high amino acid sequence similarity (10). Interestingly, the E (glutamic acid) and Q (glutamine) residues have similar structures in terms of their side chains, although only glutamic acid has a negative electric charge under physiological conditions. These results led us to speculate that the negative electric charge of E might be important for HHV-6B gQ1 function and that there should be a corresponding residue(s) with a positive electric charge in hCD134 (most likely in the CRD 2 domain) that facilitates the interaction between hCD134 and the HHV-6B ligand. To test this, we focused on two amino acid residues in the CRD 2 domain of hCD134 (K79 and R95), both of which have a positive electric charge; neither of these residues is conserved in murine CD134 (mCD134) (Fig. 1A). Also, mCD134 did not interact with the HHV-6B gH/gL/gQ1/gQ2 complex (Fig. 1E).
FIG 1.
Identification of two amino acid residues within CD134 that are required for HHV-6B ligand binding. (A) Amino acid sequence alignment of human and murine CD134. The two amino acid residues of interest are indicated by black triangles. (B) Crystal structure of hCD134 and its ligand (PBD accession number 2HEV). (Bottom panel) Crystal structure of hCD134 (K79 and W86 are shown with their side chains in red); (upper panel) ligand for hCD134. The structure drawing was made by using UCSF Chimera software (23; www.cgl.ucsf.edu/chimera). (C, D, and E) 293T cells were transfected with plasmids expressing the proteins (indicated by a number at the top of each lane and identified at the bottom of the figure) by using Lipofectamine 2000 (Invitrogen). The transfected cells were harvested at day 2 posttransfection and lysed with TNE buffer (10 mM Tris-HCl [pH 7.8], 0.15 M NaCl, 1 mM EDTA, 1% NP-40 [Nacalai Tesque]) containing a cocktail of protease inhibitors (Sigma). The lysates were subjected to ultracentrifugation prior to immunoprecipitation (IP) with an anti-gH antibody. Expression of each protein (Lysate) and the proteins precipitated from the lysates (IP: anti-gH) were confirmed by Western blotting (WB). Numbers at the left of the gels are molecular masses (in kilodaltons).
We replaced these two residues with the homologous residues in mCD134 (K79D and R95L) and then examined the interaction between the CD134 mutants and the HHV-6B gH/gL/gQ1/gQ2 complex by immunoprecipitation with an anti-gH antibody followed by Western blotting (in our previous study, we confirmed that both immunoprecipitation and pulldown assays could be used for the analysis of the HHV-6 receptor-ligand interaction [10, 11]). The interaction between the HHV-6B complex and hCD134 was detected when R95, but not K79, was mutated. Since K79 is located within the loop structure of hCD134 (12) (Fig. 1B), it is highly unlikely that the K79D mutation has a marked effect on the conformational structure of hCD134. Furthermore, to confirm whether the positive electric charge of K79 contributed to the hCD134–HHV-6B ligand interaction, we replaced K79 within hCD134 with the residues containing the negative electric (residue E or D), nonpolar (residue A), or positive (residue R) electric charge and analyzed the interaction with the HHV-6B complex. The K79E and K79D mutations almost completely abolished the interaction with the HHV-6B gH/gL/gQ1/gQ2 complex (Fig. 1C). However, the receptor-ligand interaction was detected after the K79A or K79R mutation. These results indicate that the positive electric charge of hCD134 K79 is responsible for the interaction with the HHV-6B complex. Although the conformational structure of the HHV-6B receptor–ligand has not yet been resolved, based on the effects of changes in the charges of amino acid residues important for the HHV-6B ligand–receptor interaction, it is likely that the positive electric charge of the CD134 residue (K79) contributes to the access of the HHV-6B ligand to hCD134.
Next, we tried to identify the key amino acid residue(s) within hCD134 that contributes to its interaction with the HHV-6B complex. We focused on residues around hCD134 K79 and residues that are not conserved in mCD134. We replaced these residues with the homologous residues of mCD134 (V76A, S77N/S78Y, and W86Q) and analyzed the interaction with the HHV-6B gH/gL/gQ1/gQ2 complex by immunoprecipitation followed by Western blotting as described above. Mutation of V76A and S77N/S78Y had little effect on the interaction between hCD134 and the HHV-6B gH/gL/gQ1/gQ2 complex. In contrast, the W86Q mutation had a marked effect on this interaction (Fig. 1D). Furthermore, we replaced the residue with A (W86A) and performed a similar analysis. The W86A mutation also had a marked effect on the hCD134–HHV-6B complex interaction (data not shown). In addition, W86 is located in the loop structure of hCD134 (12) (Fig. 1B), meaning that mutation of W86 is not likely to have a marked effect on the conformational structure of hCD134. These results indicate that W86 of hCD134 may play a key role in binding to the HHV-6B complex.
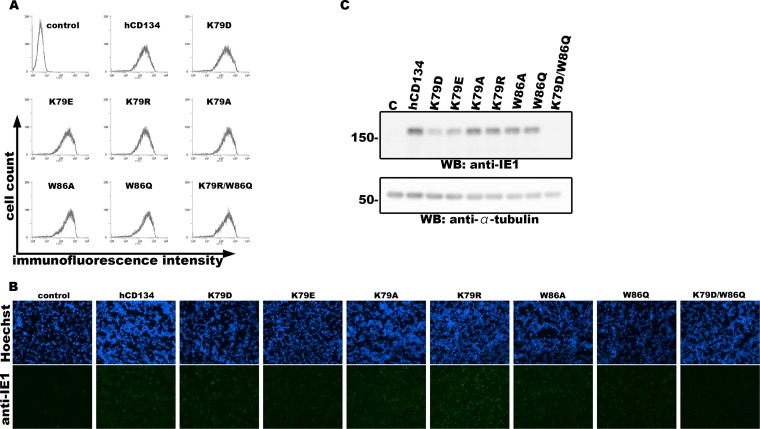
Next, we tested whether these two residues were important for HHV-6B infection by constructing nonreplicative lentiviruses expressing hCD134 mutants in JJHan cells, a cell line rarely permissive for HHV-6B infection, and tested whether these cells could be converted into HHV-6B-permissive cells. Cell surface expression of each CD134 mutant was confirmed by flow cytometry using an anti-CD134 antibody as previously described (10) (Fig. 2A). The cells were then infected with HHV-6B (HST strain) at a multiplicity of infection (MOI) of ∼0.01. HHV-6B used in this study was propagated in cord blood mononuclear cells (CBMCs) as described previously (13), and a portion of the CBMCs were purchased from RIKEN (the Institute of Physical and Chemical Research, Japan). The ethical committee of the Kobe University Graduate School of Medicine approved the use of CBMCs in this study. Viral entry into the cells was evaluated by detecting expression of the HHV-6B IE1 gene by Western blotting and indirect immunofluorescent antibody assay (IFA) as described previously (7, 8). Double mutation of hCD134 (K79D and W86Q) abolished HHV-6B infection of hCD134-expressing JJHan cells, whereas the single mutations (K79 or W86) had only a slight effect (Fig. 2B and C). It is unknown why expression of hCD134K79D, hCD134K79E, hCD134W86A, or hCD134W86Q could also render HHV-6B susceptibility to JJHan cells. It is likely that these hCD134 mutants weakly interact with the HHV-6B ligand, which functions during HHV-6B infection but cannot be detected by immunoprecipitation assay (Fig. 1C and D). There is the other possibility that differences might be seen with shorter adsorption periods or at different temperatures.
FIG 2.
Two amino acid residues within hCD134 are required for HHV-6B infection. (A) JJHan cells were transduced with nonreplicative lentiviruses expressing hCD134 or its mutants for 24 h. A portion of the transduced cells was incubated with anti-CD134 antibody (10) and secondary antibody (FITC-conjugated rabbit anti-mouse IgG; Dako) at 4°C for 1 h. Then, the cells were subjected to surface expression analysis of hCD134 and its mutants by flow cytometry (EC800; Sony). (B and C) The remaining cells were infected with HHV-6B (HST strain). Cells were harvested at 24 h postinfection. A portion of the cells were fixed with acetone-methanol (7:3) and stained with anti-IE1 antibody and secondary antibody for IFA (B). The remaining cells were lysed and examined by Western blotting with anti-IE1 and anti-α-tubulin antibodies (C). JJHan cells transduced with nonreplicative lentiviruses expressing no alien protein were used as a control for these experiments.
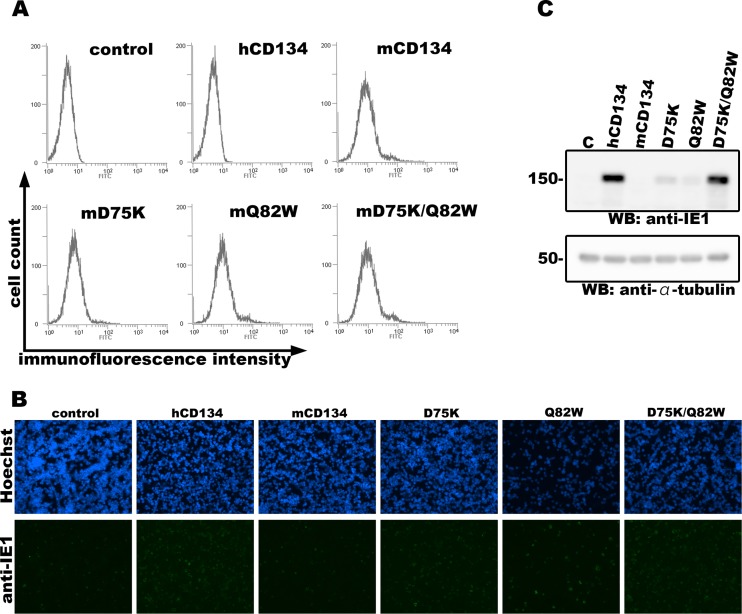
Human and murine CD134s share about 65% amino acid identity; therefore, we examined whether humanization of the homologous residues (D75 and Q82) within mCD134 would enable it to bind the HHV-6B gH/gL/gQ1/gQ2 complex. mCD134 harboring single or double mutations were constructed. Since there is no commercially available anti-mCD134 antibody for Western blotting application, we added a hemagglutinin (HA) tag to the C termini of mCD134 constructs for detection by Western blotting, and their interaction with the HHV-6B complex was analyzed as described above. Comparable interactions were detected when both mCD134 residues were mutated but not when either alone was mutated (Fig. 1E). Next, we constructed nonreplicative lentivirus vectors expressing mCD134 harboring a mutation in the homologous residues within hCD134 as described above (D75K, Q82W, or D75K/Q82W). JJHan cells were transduced with these lentiviruses, and cell surface expression of the mCD134 mutants was confirmed by flow cytometry with a fluorescein isothiocyanate (FITC)-conjugated anti-mCD134 antibody (clone OX-86; Abcam) (Fig. 3A). The transduced cells were then infected with HHV-6B as described above and harvested at 24 h postinfection. Expression of the viral IE1 protein was analyzed by Western blotting and IFA. In agreement with the above results, mutation of either D75K or Q82W had little effect on HHV-6B infection of mCD134-expressing JJHan cells; however, double mutation at these two sites rendered the cells susceptible to HHV-6B infection (Fig. 3B and C). It is likely that mCD134 D75K and mCD134 Q82W interact with HHV-6B ligand weakly (not as strongly as hCD134 or mCD134 D75K/Q82W), which may facilitate HHV-6B infection to some extent, but the interaction could not be confirmed by immunoprecipitation assay (Fig. 1E).
FIG 3.
Cells expressing mCD134 harboring a double amino acid mutation are permissive for HHV-6B infection. (A) JJHan cells were transduced with nonreplicative lentiviruses expressing mCD134 or its mutants for 24 h. A portion of the cells were stained with an FITC-conjugated anti-mCD134 antibody at 4°C for 1 h. Then, the cells were subjected to the surface expression analysis of mCD134 and its mutants by flow cytometry (EC800; Sony). (B and C) The remaining cells were infected with HHV-6B (HST strain) and harvested at 24 h postinfection. A portion of the cells were fixed with acetone-methanol (7:3) and stained with anti-IE1 antibody and secondary antibody for IFA (B). The remaining cells were lysed and examined by Western blotting with anti-IE1 and anti-α-tubulin antibodies (C). JJHan cells transduced with nonreplicative lentiviruses expressing no alien protein were used as a control for these experiments. Numbers at the left of the gels are molecular masses (in kilodaltons).
Receptor usage is but one determinant of species-specific tropism (14, 15). HHV-6B infects only human cells (16–18). Its receptor, hCD134, is expressed on activated human T cells (19), which is one factor that defines the T cell tropism of HHV-6B. Here, we identified two amino acid residues within hCD134 that are important for HHV-6B ligand binding and subsequent viral infection. Interestingly, non-HHV-6B-permissive cells (JJHan cells) became permissive for HHV-6B infection upon expression of mCD134 harboring mutations at these two amino acid residues; thus, these two amino acid residues may determine species-specific HHV-6B entry. In addition, expression of hCD134 or mCD134 D75K/Q82W on the cell surface of a mouse lymphoma cell line (EL4 cells) could not render them susceptible to HHV-6B infection (data not shown), indicating that an additional human molecule(s) is required for HHV-6B infection. No practical animal model of HHV-6 infection exists, in spite of several attempts to devise one (20, 21). Although there are other species-specific determinants for the human cell tropism of HHV-6B, the findings presented herein may contribute to the development of an animal model of HHV-6B infection.
ACKNOWLEDGMENTS
We thank Mitsuhiro Nishimura (Division of Clinical Virology, Kobe University Graduate School of Medicine) for kind discussions about the conformational structure of hCD134 mutants, Jun-Ichi Miyazaki for providing the pCAGGS-MCS plasmid (22), and Hideto Yamada (Department of Obstetrics and Gynecology, Kobe University Graduate School of Medicine) for providing the CBMCs.
This study was supported in part by a Grant-in-Aid for Scientific Research (B) from the Japan Society for the Promotion of Science (JSPS).
REFERENCES
- 1.Ablashi D, Agut H, Alvarez-Lafuente R, Clark DA, Dewhurst S, Diluca D, Flamand L, Frenkel N, Gallo R, Gompels UA, Hollsberg P, Jacobson S, Luppi M, Lusso P, Malnati M, Medveczky P, Mori Y, Pellett PE, Pritchett JC, Yamanishi K, Yoshikawa T. 2014. Classification of HHV-6A and HHV-6B as distinct viruses. Arch Virol 159:863–870. doi: 10.1007/s00705-013-1902-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubin JT, Collandre H, Candotti D, Ingrand D, Rouzioux C, Burgard M, Richard S, Huraux JM, Agut H. 1991. Several groups among human herpesvirus 6 strains can be distinguished by Southern blotting and polymerase chain reaction. J Clin Microbiol 29:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campadelli-Fiume G, Guerrini S, Liu X, Foa-Tomasi L. 1993. Monoclonal antibodies to glycoprotein B differentiate human herpesvirus 6 into two clusters, variants A and B. J Gen Virol 74(Part 10):2257–2262. doi: 10.1099/0022-1317-74-10-2257. [DOI] [PubMed] [Google Scholar]
- 4.Salahuddin SZ, Ablashi DV, Markham PD, Josephs SF, Sturzenegger S, Kaplan M, Halligan G, Biberfeld P, Wong-Staal F, Kramarsky B, Gallo RC. 1986. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science 234:596–601. doi: 10.1126/science.2876520. [DOI] [PubMed] [Google Scholar]
- 5.Wyatt LS, Balachandran N, Frenkel N. 1990. Variations in the replication and antigenic properties of human herpesvirus 6 strains. J Infect Dis 162:852–857. doi: 10.1093/infdis/162.4.852. [DOI] [PubMed] [Google Scholar]
- 6.Santoro F, Kennedy PE, Locatelli G, Malnati MS, Berger EA, Lusso P. 1999. CD46 is a cellular receptor for human herpesvirus 6. Cell 99:817–827. doi: 10.1016/S0092-8674(00)81678-5. [DOI] [PubMed] [Google Scholar]
- 7.Mori Y, Akkapaiboon P, Yang X, Yamanishi K. 2003. The human herpesvirus 6 U100 gene product is the third component of the gH-gL glycoprotein complex on the viral envelope. J Virol 77:2452–2458. doi: 10.1128/JVI.77.4.2452-2458.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akkapaiboon P, Mori Y, Sadaoka T, Yonemoto S, Yamanishi K. 2004. Intracellular processing of human herpesvirus 6 glycoproteins Q1 and Q2 into tetrameric complexes expressed on the viral envelope. J Virol 78:7969–7983. doi: 10.1128/JVI.78.15.7969-7983.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang H, Serada S, Kawabata A, Ota M, Hayashi E, Naka T, Yamanishi K, Mori Y. 2013. CD134 is a cellular receptor specific for human herpesvirus-6B entry. Proc Natl Acad Sci U S A 110:9096–9099. doi: 10.1073/pnas.1305187110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang H, Wang J, Mahmoud NF, Mori Y. 2014. Detailed study of the interaction between human herpesvirus 6B glycoprotein complex and its cellular receptor, human CD134. J Virol 88:10875–10882. doi: 10.1128/JVI.01447-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang H, Hayashi M, Maeki T, Yamanishi K, Mori Y. 2011. Human herpesvirus 6 glycoprotein complex formation is required for folding and trafficking of the gH/gL/gQ1/gQ2 complex and its cellular receptor binding. J Virol 85:11121–11130. doi: 10.1128/JVI.05251-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Compaan DM, Hymowitz SG. 2006. The crystal structure of the costimulatory OX40-OX40L complex. Structure 14:1321–1330. doi: 10.1016/j.str.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 13.Dhepakson P, Mori Y, Jiang YB, Huang HL, Akkapaiboon P, Okuno T, Yamanishi K. 2002. Human herpesvirus-6 rep/U94 gene product has single-stranded DNA-binding activity. J Gen Virol 83:847–854. [DOI] [PubMed] [Google Scholar]
- 14.Douam F, Lavillette D, Cosset FL. 2015. The mechanism of HCV entry into host cells. Prog Mol Biol Transl Sci 129:63–107. doi: 10.1016/bs.pmbts.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Liu D, Liu X, Yan J, Liu WJ, Gao GF. 2009. Interspecies transmission and host restriction of avian H5N1 influenza virus. Sci China C Life Sci 52:428–438. doi: 10.1007/s11427-009-0062-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lusso P, Markham PD, Tschachler E, di Marzo Veronese F, Salahuddin SZ, Ablashi DV, Pahwa S, Krohn K, Gallo RC. 1988. In vitro cellular tropism of human B-lymphotropic virus (human herpesvirus-6). J Exp Med 167:1659–1670. doi: 10.1084/jem.167.5.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mori Y. 2009. Recent topics related to human herpesvirus 6 cell tropism. Cell Microbiol 11:1001–1006. doi: 10.1111/j.1462-5822.2009.01312.x. [DOI] [PubMed] [Google Scholar]
- 18.Tang H, Mori Y. 2010. Human herpesvirus-6 entry into host cells. Future Microbiol 5:1015–1023. doi: 10.2217/fmb.10.61. [DOI] [PubMed] [Google Scholar]
- 19.Croft M. 2003. Costimulation of T cells by OX40, 4-1BB, and CD27. Cytokine Growth Factor Rev 14:265–273. doi: 10.1016/S1359-6101(03)00025-X. [DOI] [PubMed] [Google Scholar]
- 20.Leibovitch E, Wohler JE, Cummings Macri SM, Motanic K, Harberts E, Gaitan MI, Maggi P, Ellis M, Westmoreland S, Silva A, Reich DS, Jacobson S. 2013. Novel marmoset (Callithrix jacchus) model of human herpesvirus 6A and 6B infections: immunologic, virologic and radiologic characterization. PLoS Pathog 9:e1003138. doi: 10.1371/journal.ppat.1003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reynaud JM, Jegou JF, Welsch JC, Horvat B. 2014. Human herpesvirus 6A infection in CD46 transgenic mice: viral persistence in the brain and increased production of proinflammatory chemokines via Toll-like receptor 9. J Virol 88:5421–5436. doi: 10.1128/JVI.03763-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. doi: 10.1016/0378-1119(91)90434-D. [DOI] [PubMed] [Google Scholar]
- 23.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]