ABSTRACT
Filoviruses, consisting of Ebola virus (EBOV) and Marburg virus (MARV), are among the most lethal infectious threats to mankind. Infections by these viruses can cause severe hemorrhagic fevers in humans and nonhuman primates with high mortality rates. Since there is currently no vaccine or antiviral therapy approved for humans, there is an urgent need to develop prophylactic and therapeutic options for use during filoviral outbreaks and bioterrorist attacks. One of the ideal targets against filoviral infection and diseases is at the entry step, which is mediated by the filoviral glycoprotein (GP). In this report, we screened a chemical library of small molecules and identified numerous inhibitors, which are known G protein-coupled receptor (GPCR) antagonists targeting different GPCRs, including histamine receptors, 5-HT (serotonin) receptors, muscarinic acetylcholine receptor, and adrenergic receptor. These inhibitors can effectively block replication of both infectious EBOV and MARV, indicating a broad antiviral activity of the GPCR antagonists. The time-of-addition experiment and microscopic studies suggest that GPCR antagonists block filoviral entry at a step following the initial attachment but prior to viral/cell membrane fusion. These results strongly suggest that GPCRs play a critical role in filoviral entry and GPCR antagonists can be developed as an effective anti-EBOV/MARV therapy.
IMPORTANCE Infection of Ebola virus and Marburg virus can cause severe illness in humans with a high mortality rate, and currently there is no FDA-approved vaccine or therapeutic treatment available. The 2013-2015 epidemic in West Africa underscores a lack of our understanding in the infection and pathogenesis of these viruses and the urgency of drug discovery and development. In this study, we have identified numerous inhibitors that are known G protein-coupled receptor (GPCR) antagonists targeting different GPCRs. These inhibitors can effectively block replication of both infectious EBOV and MARV, indicating a broad antiviral activity of the GPCR antagonists. Our results strongly suggest that GPCRs play a critical role in filoviral entry and GPCR antagonists can be developed as an effective anti-EBOV/MARV therapy.
INTRODUCTION
The Filoviridae family consists of Ebola virus (EBOV) and Marburg virus (MARV), which can cause severe hemorrhagic fever in human and nonhuman primates with mortality rates of up to 90%. The viral outbreaks are sporadic and unpredictable and so far have been limited to Africa (1). It is thought that fruit bats are the natural reservoirs of EBOV (2). Although several vaccines and therapeutic options have been developed and shown to be effective in nonhuman primate models (3–5), there is currently no vaccine or treatment approved for filoviral infections in humans. The 2013-2015 West Africa Ebola epidemic, with more than 25,000 people infected and more than 12,000 deaths, underlines the global challenge of treating and controlling the infections and diseases associated with these viruses.
One of the potential targets to block filoviral infection is at the entry step, which is mediated by a single viral glycoprotein (GP). GP is composed of two subunits, GP1, which is responsible for attachment and binding to receptor(s) on susceptible cells, and GP2, which mediates viral and cell membrane fusion. GP on the surface of virions is present as a homotrimer of GP1/GP2 heterodimer (1). Following the initial attachment of virions to the host cell surface, likely through the interaction of GP with heparan sulfate and other closely related glycosaminoglycans (GAGs) (6, 7), virions are believed to enter the cell by a process of endocytosis into the endosome, where fusion of virus and cell membranes occurs. Although numerous other host factors have been implicated in Ebola/Marburg virus entry, the entry mechanism of filovirus is still poorly understood.
In this study, we demonstrate that several classes of G protein-coupled receptor (GPCR) antagonists can effectively inhibit entry of both EBOV and MARV. This finding has important implications in our understanding of the role of GPCRs in filoviral entry and in the development of GPCR antagonists as a potential antifiloviral therapeutic option.
MATERIALS AND METHODS
Cell culture and virus.
Human 293T embryonic kidney cells and human A549 lung epithelial cell lines were cultured in Dulbecco's modified Eagle medium (DMEM; Cellgro) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 μg/ml of streptomycin, and 100 units of penicillin (Invitrogen). Vero E6 cells were maintained in Eagle's minimum essential medium (EMEM, Gibco) supplemented with 10% FBS (Gibco).
The pseudovirions for high-throughput sequencing (HTS) were created by the following plasmids: hemagglutinin (HA) and neuraminidase (NA), isolated from a highly pathogenic avian influenza virus, A/Goose/Qinghai/59/05 (H5N1) strain, Marburg virus glycoprotein, Ebola virus Zaire envelope glycoprotein, and the HIV-1 proviral vector pNL4-3.Luc.R−E−, which was obtained through the NIH AIDS Research and Reference Reagent program. All three types of pseudovirions, HIV/MARV, HIV/AIV, and HIV/EBOV, were produced by transient cotransfection of human 293T cells using a polyethylenimine (PEI)-based transfection protocol. Plasmids encoding MARV GP (MGP), HA/NA, EBOV GP, and replication-defective HIV vector (pNL4-3.Luc.R−E−) were used for transient cotransfection into 293T producing cells. Five hours after transfection, cells were washed with phosphate-buffered saline (PBS), and 40 ml of fresh medium was added to each plate (150 mm). Forty-eight hours posttransfection, the supernatants were collected and filtered through 0.45-μm-pore-size filters (Nalgene). The pseudovirion stocks were stored at 4°C prior to use.
The infectious filoviruses EBOV, enhanced green fluorescent protein (eGFP) Ebola virus (eGFP-EBOV), Sudan virus (SUDV), Marburg virus (MARV/Angola), and Ravn virus (RAVV) were replicated in Vero E6 cells at 90 to 100% confluence. Cells were inoculated with an approximate multiplicity of infection of 0.1 from historical stocks, and the medium was replaced 72 h after inoculation. Cells were monitored for cytopathic effects, and the supernatant was collected once 95 to 100% of the cells had detached from the surface. The cell supernatant was clarified by centrifugation at 1,200 rpm for 10 min at 4°C, and aliquots were placed at −80°C storage until further use. All infectious virus assays were performed at the U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID) at biosafety level 4 (BSL-4) facilities, with personnel wearing positive-pressure protective suits (ILC Dover) fitted with HEPA filters and umbilicated air. USAMRIID is registered with the Centers for Disease Control and Prevention Select Agent Program for the possession and use of biological select agents and has implemented a biological surety program in accordance with U.S. Army regulation AR 50-1 “Biological Surety.”
Compound library and chemicals.
The Prestwick Chemical Library contains 1,200 FDA-approved drugs. The active compounds were selected for high chemical and pharmacological diversity as well as for bioavailability and safety in humans. Three hundred twenty unique compounds were arrayed in a 384-well plate at 10 mM concentration in dimethyl sulfoxide (DMSO), leaving columns 1, 2, 23, and 24 with DMSO. The positive-control drug for this assay, zidovudine (AZT; Sigma), was solubilized at 10 mM in DMSO. The stock solution was diluted to a final concentration of 5 μM for the screens.
Benztropine mesylate, pizotifen malate, and trimeprazine tartrate were purchased from Santa Cruz Biotech; cyproheptadine hydrochloride sequihydrate and heparin were purchased from Sigma-Aldrich; bafilomycin A1 was purchased from Alexis biochemical; serotonin receptors antagonists were purchased from Tocris Bioscience. The compounds were dissolved in DMSO, and aliquots were stored at −80°C until used.
High-throughput screen.
The Prestwick library was screened at 25 μM in duplicate in a 384-well format with a final DMSO concentration of 0.25% to identify a MARV entry inhibitor. Low-passage A549 cells were seeded at the density of 1,000 cells/well in 384-well plates 24 h before infection. In the presence of compounds, A549 cells were infected by HIV/MARV pseudotyped virus, which contains the luciferase reporter gene. Plates were incubated for 48 h, and infection was then quantified by the luciferase activity of infected A549 cells using the neolite reporter gene assay system (PerkinElmer). Virus alone with DMSO was used as a negative control; virus with 5 μM AZT, an HIV reverse transcriptase inhibitor, was used as a positive control. Data were normalized by plate median value, and the criterion of an average >80% inhibition in duplicate wells was applied for picking hits.
The hit compounds were then cherry-picked into 384-well plates and screened against HIV/MARV, HIV/EBOV, or HIV/AIV to validate the primary result and to identify filovirus-specific hits. The cytotoxicity of hit compounds was also examined by the CellTiter-Glo luminescent cell viability assay (PerkinElmer). The signals in the negative-control wells (DMSO) were used to normalize the data.
The hit compounds were serially diluted for 50% inhibitory concentration (IC50) evaluation. IC50s were determined by fitting the dose-response curves against infection of HIV/MARV or HIV/EBOV with four-parameter logistic regression in GraphPad.
Time-of-addition experiment.
A549 cells were incubated with HIV/MARV at 4°C for 1 h to allow virus attachment to the cells. Then, virus was removed and cells were washed with cold PBS two times before fresh medium was added. Temperature was shifted to 37°C to trigger virus entry. At different time points of virus entry, heparin (10 μg/ml), cyproheptadine (25 μM), benztropine (25 μM), bafilomycin A1 (100 nM), or AZT (1 μM) was introduced to assess the impact on virus entry. Triplicate wells were used for each time point. Control-infected cell cultures were treated with drug vehicle (DMSO) only. Virus infection was measured 48 h postinfection as described above.
Microscopy.
Pseudotyped Marburg virus was produced by cotransfection of plasmids encoding MARV GP, replication-defective HIV vector (pNL4-3.Luc.R−E−), and Vpr-GFP. GFP-tagged virions were collected 48 h postinfection, filtered through 0.45-μm-pore-size filters, and then concentrated by ultracentrifugation in a 20% sucrose cushion and suspended in cell culture medium. A549 cells were cultured on glass coverslips and were incubated with GFP-tagged Marburg pseudovirions at 4°C in the presence of benztropine (25 μM), cyproheptadine (25 μM), heparin (10 μg/ml), or DMSO control at 4°C for 15 min. Virus was then removed, and cells were washed twice with cold PBS and cultured for 2 h at 37°C in the continued presence of drugs or control. The cells were washed with cold PBS, fixed, and stained for nuclei using DAPI (4′,6-diamidino-2-phenylindole). All images were taken using a Zeiss laser scanning microscope (LSM) 710 with a 60× objective and analyzed by ImageJ.
Infectious virus assays.
Experiments using live filoviruses were performed in BSL-4 facilities at USAMRIID with personnel wearing positive-pressure protective suits (ILC Dover, Frederica, DE) fitted with HEPA filters and umbilically fed air.
Wild-type-like recombinant EBOV (1976 Mayinga variant)-eGFP and MARV-Angola were used to infect Vero E6 cells. Compounds at a final concentration of 20 μM to 0.08 μM (2-fold serial dilution) were applied together with the viruses, and their antiviral effects were employed to calculate IC50s. Also, the cytotoxicity of these compounds to the Vero E6 cell line was evaluated, and cytotoxicity concentrations (CC50s) were calculated accordingly.
Data analysis.
The screen data were exported as comma-separated-values files and were analyzed using the statistical programming language R (8). An R script was employed to calculate the median of luminescence signals of all samples in each plate, which was used to normalize data. The percentage of inhibition (% inhibition) was calculated as 100 × (1 − normalized signal).
The hit compounds were clustered based on structure similarity using Tanimoto scores calculated from the two-dimensional (2D) structure fingerprint, which was obtained from the online structure clustering service of PubChem. The score matrix was then visualized by an R script.
The mechanism-of-action information of compounds was extracted from the data file from the Prestwick library. The function enrichment analysis of hit compounds was performed based on the information using a Fisher exact test in R.
Disclaimers.
Research was conducted under an IACUC-approved protocol in compliance with the Animal Welfare Act, PHS Policy, and other Federal statutes and regulations relating to animals and experiments involving animals. The facility where this research was conducted is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011.
RESULTS
GPCR antagonists inhibit the EBOV/MARV GP-mediated viral entry.
To develop broad-spectrum antifilovirus entry inhibitors that can block entry of both EBOV and MARV, we developed an HIV-1-based surrogate entry assay. This cell-based assay allows us to quickly screen and identify specific entry inhibitors for filoviruses and other highly pathogenic enveloped viruses such as avian influenza virus H5N1 (AIV) and arenaviruses in a biosafety level 2 (BSL-2) containment with alleviated safety concerns (9). Using this strategy, we screened the Prestwick Chemical Library, which contains 1,200 approved drugs (FDA, European Medicines Agency [EMA], and other agencies), with the MARV/HIV pseudovirions to identify MARV entry-specific inhibitors, with AIV/HIV pseudovirions as specificity controls. In total, 29 drugs were identified to be MARV specific, that is, they exerted more than 80% inhibition on MARV/HIV entry but had little effect on that of AIV/HIV and no observed cytotoxicity at 25 μM. Twenty-four of the 29 drugs were identified as GPCR antagonists, which is interesting considering that there are 74 GPCR agonists (see Table S1 in the supplemental material), 9 GPCR agonists/antagonists (none of them had detectable anti-MARV entry activity; see Table S3 in the supplemental material), and 199 antagonists in the library (see Table S2 in the supplemental material). These results suggested that only GPCR antagonists, but not GPCR agonists, inhibit MARV entry.
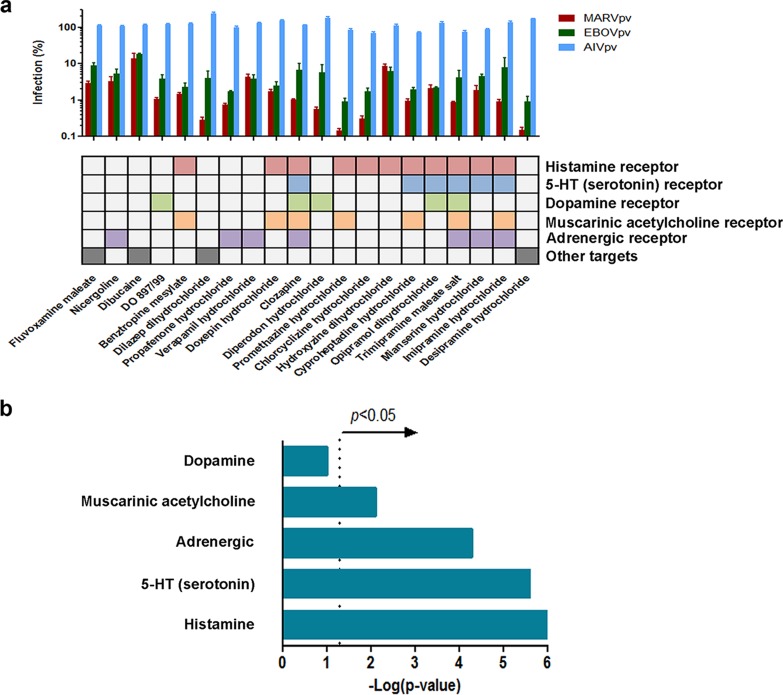
To evaluate whether the identified MARV inhibitors display anti-EBOV activity, each of the 29 drugs was tested on EBOV/HIV pseudovirions (10), and it was found that indeed all 29 drugs blocked EBOV/HIV entry (results not shown), indicating their broad-spectrum anti-filovirus entry property. Shown in Fig. 1 are the 20 most potent drugs that gave more than 80% inhibition on EBOV/HIV and MARV/HIV but did not inhibit AIV/HIV entry at 25 μM. Among them, 16 (80%) are known to be GPCR antagonists, targeting a broad range of GPCR receptors, including histamine receptor, 5-HT (serotonin) receptor, dopamine receptor, muscarinic acetylcholine receptor, and adrenergic receptor (11) (Fig. 1a). Function enrichment analysis using a Fisher exact test (12) revealed that the antagonists of four GPCRs (histamine receptor, 5-HT receptor, muscarinic acetylcholine receptor, and adrenergic receptor; i.e., all five of the above classes with the exception of dopamine receptor) were preferentially enriched by the screening (Fig. 1b), implicating the inhibitory role of these four classes of GPCR antagonists on EBOV and MARV entry.
FIG 1.
Identification of virus entry inhibitors specific to filovirus. (a) The Prestwick library of 1,200 FDA-approved drugs was screened against pseudotyped Marburg virus entry in the A549 cell line. From the primary screen, 20 compounds were identified and validated with pseudotyped H5N1 influenza virus (AIV, blue), Ebola virus (EBOV, green), and Marburg virus (MARV, red). Error bars represent standard deviations (n > 3). The compounds and their respective targeted GPCRs are shown below the graph. (b) Function enrichment analysis of the 20 filovirus entry inhibitors was performed to evaluate if the GPCR antagonism function was enriched by chance. The mechanism of action information of each identified entry inhibitor and every Prestwick library compound was extracted from the drug information sheet provided by the Prestwick library, and data were compared and tested for overrepresentation using a Fisher exact test. The significance threshold is indicated by a dotted line at 1.3, which represents −log(P value) (where P = 0.05).
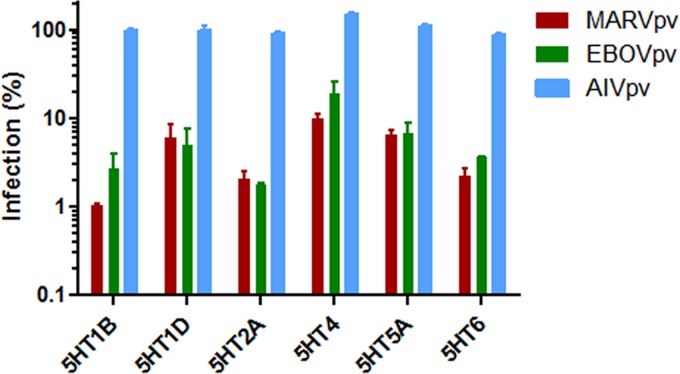
To further demonstrate that GPCR antagonists were effectively able to inhibit the GP-mediated filoviral entry, 11 antagonists targeting different members of 5-HT receptors were purchased and tested using the aforementioned pseudoviral entry assay. Six antagonists were shown to inhibit the MARV and EBOV GP-mediated but not HA-mediated viral entry (Fig. 2). These results are consistent with the notion that GPCR antagonists can specifically block the filoviral GP-mediated entry.
FIG 2.
Serotonin receptor antagonists are able to inhibit cell entry of pseudotyped filoviruses. Antagonists targeting 11 serotonin receptor family members were evaluated for their effect on pseudotyped viruses' entry. Six antagonists exhibited inhibition specific to pseudotyped Marburg virus (MARVpv) and Ebola virus (EBOVpv) but not to pseudotyped H5N1 influenza virus (AIVpv). The antifilovirus effects of the six antagonists are shown at their lowest effective dosages (5HT1D and 5HT6 at 33 μM; 5HT2A at 11 μM; 5HT1B and 5HT4 at 3.7 μM; 5HT5 at 0.41 μM). Error bars represent standard deviations.
GPCR antagonists inhibit replication of infectious EBOV/MARV.
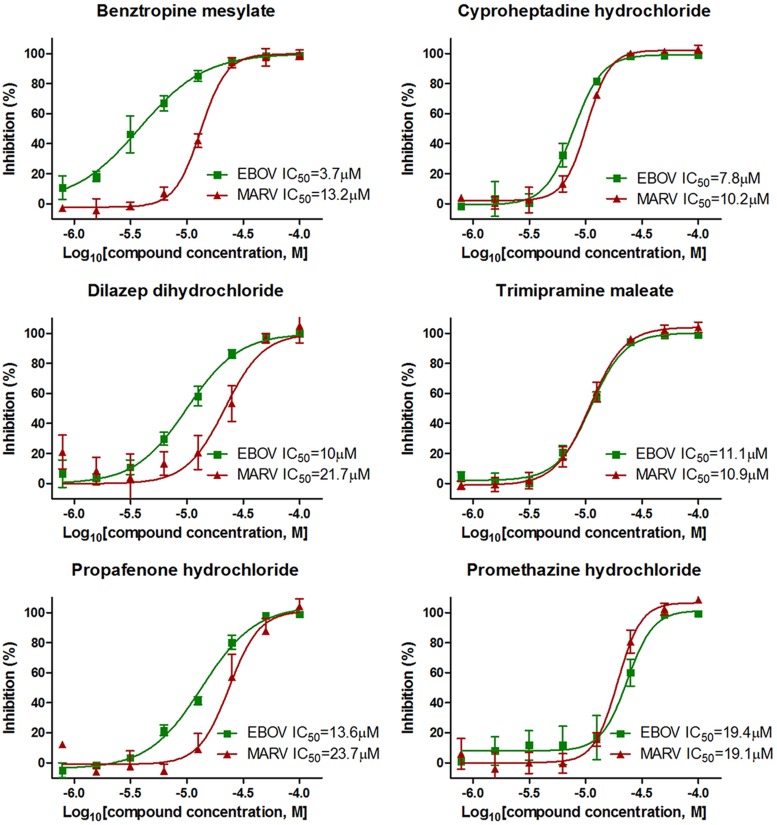
To validate the finding that GPCR antagonists could block both EBOV and MARV infections, six of the most potent drugs, including five GPCR antagonists and dilazep dihydrochloride (which is an adenosine uptake inhibitor with a role in serotonin transport), were evaluated on their ability to inhibit replication of infectious recombinant EBOV (eGFP-EBOV/Zaire) and MARV (MARV/Angola) in tissue-cultured Vero cells under BSL-4 containment conditions. As shown in Fig. 3 (see also Table S4 in the supplemental material), all six drugs displayed dose-dependent inhibition on both EBOV and MARV with potent antifiloviral activity, with 50% inhibitory concentrations (IC50s) ranging from 3.7 μM to 23.7 μM, depending on the drugs and viruses. The most potent drug is benztropine mesylate, which is anticholinergic and an antihistamine and is used clinically in the treatment of Parkinson's disease (13), with IC50s of 3.7 μM and 13.2 μM for EBOV and MARV, respectively. Therefore, we demonstrate that these GPCR antagonists are potent anti-EBOV and -MARV inhibitors.
FIG 3.
In vitro dose-response curves of six filovirus entry inhibitors on infectious EBOV and MARV. Six filovirus entry inhibitors were evaluated in Vero E6 cells against infectious eGFP Ebola virus (eGFP-EBOV) or MARV/Angola infection at 8 2-fold serial dilution concentrations ranging from 0.78 μM to 100 μM. The IC50s were calculated by four-parameter dose-response curve-fitting in GraphPad. Results are from three replicates.
Benztropine displays a broad spectrum of antifilovirus activity.
Benztropine mesylate, because of its high potency, was further evaluated for its broad-spectrum antifilovirus activity using three different isolates of EBOV and two isolates of MARV in Vero cells. The IC50s were determined to be 2.8 μM, 1.7 μM, and 4.9 μM for EBOV Kikwit, Mayinga, and Sudan (Fig. 4a, b, and c), respectively, and 11 μM and 10.6 μM for MARV (Fig. 4d and e), respectively. Under the concentrations used in this study, benztropine mesylate displayed little cytotoxicity to the cells (Fig. 4f). These results demonstrate that benztropine mesylate can potently inhibit both EBOV and MARV strains/isolates.
FIG 4.
Benztropine antagonizes infectious filovirus infection in cell culture. Benztropine was evaluated in Vero E6 cells against infectious EBOV/Kikwit, EBOV/Mayinga, SUDV, MARV/Angola, or RAVV strains (a to e) at 9 2-fold serial dilution concentrations ranging from 0.08 μM to 20 μM. The toxicity of benztropine was also evaluated in Vero E6 cells (f). The IC50s were calculated by four-parameter dose-response curve-fitting in GraphPad. Results are from two replicates.
GPCR antagonists block filoviral entry at a postattachment step.
To investigate if GPCR antagonists blocked the early attachment/binding step or the postbinding steps (e.g., fusion) of MARV, a “time-of-addition experiment” (14) was performed on MARV/HIV pseudovirions with two GPCR antagonists, benztropine mesylate and cyproheptadine (a promiscuous GPCR antagonist), using heparin, bafilomycin A1, and zidovudine (AZT) as controls. As expected, heparin, which was shown by others and us to block the early attachment/binding of filoviruses to the target cells (6, 7), exerted its inhibitory effect at early time points (−1 and 0 h), while bafilomycin A1 (an inhibitor of vacuolar-type H+-ATPase (15) and AZT (an HIV reverse transcriptase inhibitor [16]) were still effective at the late time points (>6 h). In contrast, the GPCR antagonists exerted inhibition at later time points (+2, +3, +4 h) than heparin but at earlier time points than those of bafilomycin A1 and AZT (Fig. 5a). Consistent with this, heparin was shown to be able to block the internalization of the Vpr-GFP-labeled MARV/HIV pseudovirions, while neither benztropine mesylate nor cyproheptadine blocked them (Fig. 5b and c). Together, these results suggest that the GPCR antagonists block infection of EBOV and MARV at a step postbinding but prior to viral/host membrane fusion.
FIG 5.
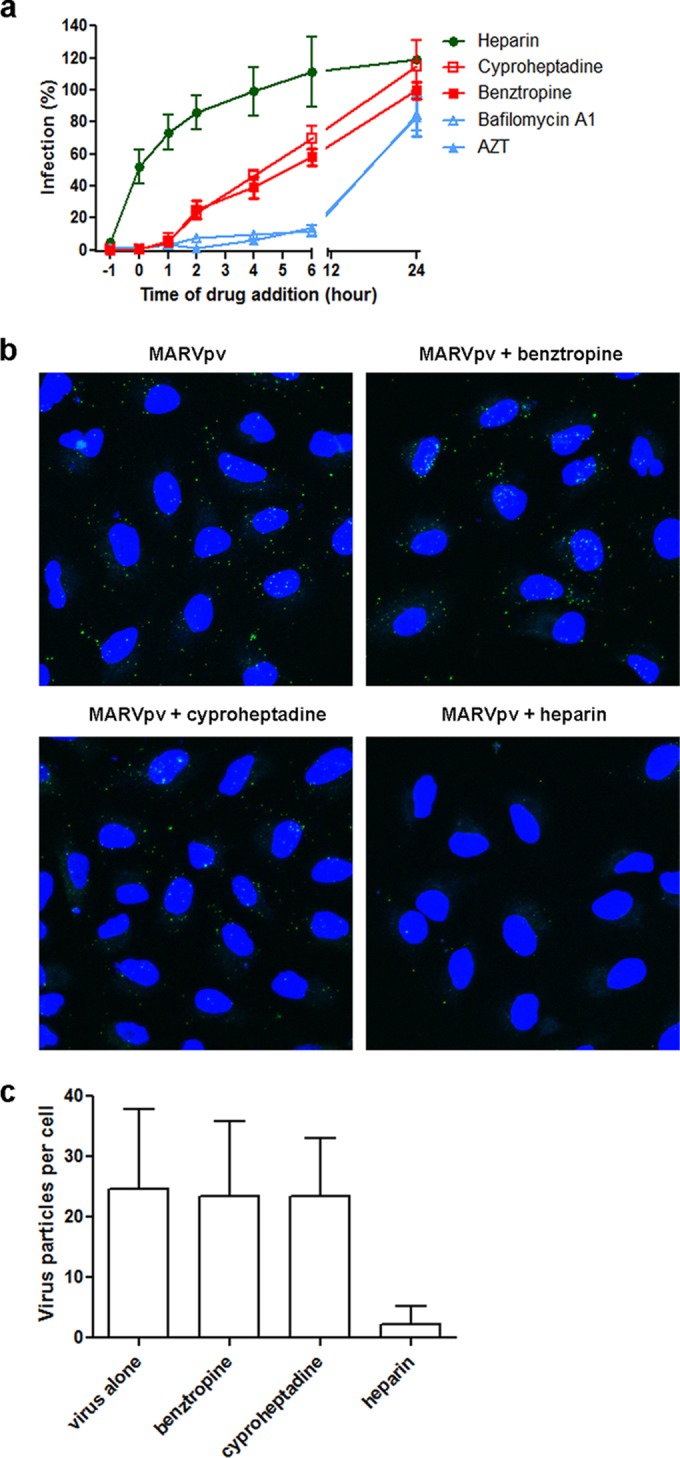
Benztropine and cyproheptadine inhibit pseudotyped Marburg virus at the late-entry stage. (a) Pseudotyped Marburg virus was incubated with A549 cells at 4°C at the −1 h time point. After 1 h of incubation, the virus was removed and temperature was shifted to 37°C to trigger virus internalization. Heparin (10 μg/ml), cyproheptadine (25 μM), benztropine (25 μM), bafilomycin A1 (100 nM), or AZT (1 μM) was introduced at different time points of virus infection, and the drugs' effects on viral infection are shown. (b) Pseudotyped Marburg virus containing Vpr-GFP was incubated with A549 cell in the presence of DMSO (control), benztropine (25 μM), cyproheptadine (25 μM), or heparin (10 μg/ml) at 4°C for 15 min. Virus was then removed, and temperature was shifted to 37°C to trigger virus internalization in the continued presence of drugs or control. After 2 h of incubation, the cells were washed with cold PBS, fixed, stained for nuclei using DAPI, and analyzed by microscopy. Virions inside A549 cells were indicated by green fluorescent foci. (c) Quantification of pseudotyped Marburg virions inside A549 cells. Virion uptake was quantified by counting all the green fluorescent foci inside cells in each image (8 to 14 cells/image). A total of 55 to 68 cells were analyzed for each drug treatment. The data are presented as the average numbers of virions per cell with standard deviations.
DISCUSSION
In this report, we demonstrate that antagonists of the different classes of GPCRs, including histamine receptors, 5-HT (serotonin) receptors, muscarinic acetylcholine receptor, and adrenergic receptor, are potent entry inhibitors of both Ebola and Marburg viruses, indicating a broad antifiloviral activity of these GPCR antagonists. This finding has important implications in elucidating the entry mechanism of filoviruses and for development of antifiloviral therapy.
It is well known that the GPCR superfamily, with roughly 1,000 members identified in the human genome, constitutes the largest group of cell surface proteins that are involved in signal recognition and signal transduction (11, 17), and different viruses use different approaches to usurp the GPCRs and the GPCR-activated signaling pathways (18). Although it is not clear yet how GPCR antagonists exert their effect on EBOV and MARV entry, the time-of-addition experiment and microscopic studies (Fig. 5) suggest that these inhibitors block a postbinding step prior to viral/cell membrane fusion. Our results suggest that GPCRs are directly or indirectly involved in the entry process of EBOV and MARV. One possible mechanism is the binding of filoviral glycoprotein (GP) to one or multiple GPCRs directly, and this interaction is critical for filoviral entry. Since the GPCR antagonists that display potent antiviral activity target different GPCRs, we speculate that multiple GPCRs are likely involved. Another possible mechanism is that Ebola/Marburg virus binding to the hosts can somehow trigger GPCR signaling, and GPCRs are thus indirectly involved in filoviral entry. The involvement of GPCRs and GPCR signaling in filoviral entry provides a plausible explanation for how different host factors and pathways such as the phosphoinoside-3 kinase (PI3K) pathways (19), Tyro3 receptor tyrosine kinase family (20), or integrins (21) are involved in EBOV entry, since these pathways are linked to GPCR signaling and pathways. Considering that numerous host factors such as NPC-1 (22, 23), CatB/L (24), TPCs (25), and EXT1 (6, 7) have been implicated in EBOV and MARV entry, it is clear that EBOV/MARV entry to the host cells is a complex process utilizing multiple host factors and pathways. Insights into these factors/pathways in the entry process should facilitate drug discovery and development to treat and control filovirus infection-associated diseases.
Development of GPCR antagonists as an antifiloviral therapy is an attractive strategy based on the findings in this study. In contrast to previous reports of anti-EBOV inhibitors that target either EBOV glycoprotein (GP) or viral polymerase by a limited number of chemical scaffolds or entities (3, 5, 14, 25), the GPCR antagonists that showed antifiloviral entry activity here represent structurally different scaffolds and target different GPCRs, including histamine receptor, 5-HT (serotonin) receptor, muscarinic acetylcholine receptor, and adrenergic receptor (Fig. 1). Our results are consistent with a recent report that shows that several antagonists of histamine receptors and 5-HT (serotonin) receptors can block entry of EBOV viruslike particles (26). Since it is estimated that overall approximately one-half of all the clinically approved drugs target GPCRs, there is a large number of chemical libraries of small molecules of GPCR antagonists and their derivatives available for screening for potent anti-EBOV/MARV entry inhibitors. Indeed, 6 of 11 5-HT (serotonin) receptor antagonists that were tested using MARV/HIV and EBOV/HIV pseudovirions were shown to be able to block the GP-mediated viral entry (Fig. 2), demonstrating the great promise of developing GPCR antagonists as an effective and broad-spectrum antifilovirus therapy.
Supplementary Material
ACKNOWLEDGMENTS
We thank Thomas Hope at Northwestern University for the Vpr-GFP plasmid and Kiira Ratia at the University of Illinois at Chicago for her assistance with the Prestwick Chemical Library screening.
This research was partially supported by National Institutes of Health (USA) grant AI77767 to L.R. USAMRIID efforts were supported by DTRA project 4.10007_08_RD_B, awarded to G.G.O., and subcontract W81XWH-08-0051, awarded to L.J.
The opinions, interpretations, conclusions, and recommendations are ours and are not necessarily endorsed by the U.S. Army.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01337-15.
REFERENCES
- 1.Feldmann F, Sanchez A, Geisbert JB. 2013. Filoviridae: Marburg and Ebola viruses, p 923–956. In Knipe DM, Howley PM (ed), Fields virology, 6th ed, vol II Wolters Kluwer Lippincott Willliams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Delicat A, Paweska JT, Gonzalez JP, Swanepoel R. 2005. Fruit bats as reservoirs of Ebola virus. Nature 438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 3.Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, Johnson A, Morton J, Swope K, Bohorov O, Bohorova N, Goodman C, Kim D, Pauly MH, Velasco J, Pettitt J, Olinger GG, Whaley K, Xu B, Strong JE, Zeitlin L, Kobinger GP. 2014. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sullivan NJ, Geisbert TW, Geisbert JB, Xu L, Yang ZY, Roederer M, Koup RA, Jahrling PB, Nabel GJ. 2003. Accelerated vaccination for Ebola virus haemorrhagic fever in non-human primates. Nature 424:681–684. doi: 10.1038/nature01876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warren TK, Wells J, Panchal RG, Stuthman KS, Garza NL, Van Tongeren SA, Dong L, Retterer CJ, Eaton BP, Pegoraro G, Honnold S, Bantia S, Kotian P, Chen X, Taubenheim BR, Welch LS, Minning DM, Babu YS, Sheridan WP, Bavari S. 2014. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature 508:402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Hearn A, Wang M, Cheng H, Lear-Rooney CM, Koning K, Rumschlag-Booms E, Varhegyi E, Olinger G, Rong L. 2015. Role of EXT1 and glycosaminoglycans in the early stage of filovirus entry. J Virol 89:5441–5449. doi: 10.1128/JVI.03689-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salvador B, Sexton NR, Carrion R Jr, Nunneley J, Patterson JL, Steffen I, Lu K, Muench MO, Lembo D, Simmons G. 2013. Filoviruses utilize glycosaminoglycans for their attachment to target cells. J Virol 87:3295–3304. doi: 10.1128/JVI.01621-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Team RDC. 2011. R: a language and environment for statistical computing. R Foundation for Statistical computing, Vienna, Austria. [Google Scholar]
- 9.Wang J, Cheng H, Ratia K, Varhegyi E, Hendrickson WG, Li J, Rong L. 2014. A comparative high-throughput screening protocol to identify entry inhibitors of enveloped viruses. J Biomol Screen 19:100–107. doi: 10.1177/1087057113494405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manicassamy B, Wang J, Jiang H, Rong L. 2005. Comprehensive analysis of Ebola virus GP1 in viral entry. J Virol 79:4793–4805. doi: 10.1128/JVI.79.8.4793-4805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroeze WK, Sheffler DJ, Roth BL. 2003. G-protein-coupled receptors at a glance. J Cell Sci 116:4867–4869. doi: 10.1242/jcs.00902. [DOI] [PubMed] [Google Scholar]
- 12.Rivals I, Personnaz L, Taing L, Potier MC. 2007. Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics 23:401–407. doi: 10.1093/bioinformatics/btl633. [DOI] [PubMed] [Google Scholar]
- 13.Gelenberg AJ, Van Putten T, Lavori PW, Wojcik JD, Falk WE, Marder S, Galvin-Nadeau M, Spring B, Mohs RC, Brotman AW. 1989. Anticholinergic effects on memory: benztropine versus amantadine. J Clin Psychopharmacol 9:180–185. [PubMed] [Google Scholar]
- 14.Basu A, Li B, Mills DM, Panchal RG, Cardinale SC, Butler MM, Peet NP, Majgier-Baranowska H, Williams JD, Patel I, Moir DT, Bavari S, Ray R, Farzan MR, Rong L, Bowlin TL. 2011. Identification of a small-molecule entry inhibitor for filoviruses. J Virol 85:3106–3119. doi: 10.1128/JVI.01456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowman EJ, Siebers A, Altendorf K. 1988. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci U S A 85:7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitsuya H, Yarchoan R, Broder S. 1990. Molecular targets for AIDS therapy. Science 249:1533–1544. doi: 10.1126/science.1699273. [DOI] [PubMed] [Google Scholar]
- 17.Rajagopal S, Rajagopal K, Lefkowitz RJ. 2010. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat Rev Drug Discov 9:373–386. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sodhi A, Montaner S, Gutkind JS. 2004. Viral hijacking of G-protein-coupled-receptor signalling networks. Nat Rev Mol Cell Biol 5:998–1012. doi: 10.1038/nrm1529. [DOI] [PubMed] [Google Scholar]
- 19.Saeed MF, Kolokoltsov AA, Freiberg AN, Holbrook MR, Davey RA. 2008. Phosphoinositide-3 kinase-Akt pathway controls cellular entry of Ebola virus. PLoS Pathog 4:e1000141. doi: 10.1371/journal.ppat.1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimojima M, Takada A, Ebihara H, Neumann G, Fujioka K, Irimura T, Jones S, Feldmann H, Kawaoka Y. 2006. Tyro3 family-mediated cell entry of Ebola and Marburg viruses. J Virol 80:10109–10116. doi: 10.1128/JVI.01157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schornberg KL, Shoemaker CJ, Dube D, Abshire MY, Delos SE, Bouton AH, White JM. 2009. Alpha5beta1-integrin controls ebolavirus entry by regulating endosomal cathepsins. Proc Natl Acad Sci U S A 106:8003–8008. doi: 10.1073/pnas.0807578106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Cin PD, Dye JM, Whelan SP, Chandran K, Brummelkamp TR. 2011. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cote M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, Cunningham J. 2011. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakurai Y, Kolokoltsov AA, Chen CC, Tidwell MW, Bauta WE, Klugbauer N, Grimm C, Wahl-Schott C, Biel M, Davey RA. 2015. Ebola virus. Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science 347:995–998. doi: 10.1126/science.1258758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kouznetsova J, Sun W, Martinez-Romero C, Tawa G, Shinn P, Chen CZ, Schimmer A, Sanderson P, McKew JC, Zheng W, Garcia-Sastre A. 2014. Identification of 53 compounds that block Ebola virus-like particle entry via a repurposing screen of approved drugs. Emerg Microbes Infect 3:e84. doi: 10.1038/emi.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.