ABSTRACT
Human bocavirus 1 (HBoV1) is a single-stranded DNA parvovirus that causes lower respiratory tract infections in young children worldwide. In this study, we identified novel splice acceptor and donor sites, namely, A1′ and D1′, in the large nonstructural protein (NS1)-encoding region of the HBoV1 precursor mRNA. The novel small NS proteins (NS2, NS3, and NS4) were confirmed to be expressed following transfection of an HBoV1 infectious proviral plasmid and viral infection of polarized human airway epithelium cultured at an air-liquid interface (HAE-ALI). We constructed mutant pIHBoV1 infectious plasmids which harbor silent mutations (sm) smA1′ and smD1′ at the A1′ and D1′ splice sites, respectively. The mutant infectious plasmids maintained production of HBoV1 progeny virions at levels less than five times lower than that of the wild-type plasmid. Importantly, the smA1′ mutant virus that does not express NS3 and NS4 replicated in HAE-ALI as effectively as the wild-type virus; however, the smD1′ mutant virus that does not express NS2 and NS4 underwent an abortive infection in HAE-ALI. Thus, our study identified three novel NS proteins, NS2, NS3, and NS4, and suggests an important function of the NS2 protein in HBoV1 replication in HAE-ALI.
IMPORTANCE Human bocavirus 1 infection causes respiratory diseases, including acute wheezing in infants, of which life-threatening cases have been reported. In vitro, human bocavirus 1 infects polarized human bronchial airway epithelium cultured at an air-liquid interface that mimics the environment of human lower respiratory airways. Viral nonstructural proteins are often important for virus replication and pathogenesis in infected tissues or cells. In this report, we identified three new nonstructural proteins of human bocavirus 1 that are expressed during infection of polarized human bronchial airway epithelium. Among them, we proved that one nonstructural protein is critical to the replication of the virus in polarized human bronchial airway epithelium. The creation of nonreplicating infectious HBoV1 mutants may have particular utility in vaccine development for this virus.
INTRODUCTION
Human bocavirus 1 (HBoV1) belongs to genus Bocaparvovirus of the Parvoviridae family (1, 2). HBoV1 causes lower respiratory tract infections, especially in infants less than 2 years old (3–7). Severe and deadly cases associated with high viral load, anti-HBoV1 IgM antibody detection, or increased IgG antibody production have been documented (7–9). In vitro, HBoV1 infects polarized primary human airway epithelium cultured at an air-liquid interface (HAE-ALI) (10) and causes damage to the airway epithelium (11–13). Currently, no specific treatments for HBoV1 infection are available for hospitalized infants.
The RNA transcription profiles of three species in the genus Bocaparvovirus, i.e., canine minute virus (CnMV/MVC) (14), bovine parvovirus type 1 (BPV1) (15), and HBoV1 (10), have been studied during virus infection. These species share similarities in their gene expression, with distinguishing features among various parvovirus clades. They use one promoter to transcribe a single precursor mRNA transcript (pre-mRNA), which is both alternatively spliced and polyadenylated, to generate matured mRNA transcripts for encoding viral nonstructural (NS) and structural (viral protein [VP]) proteins (16–19). Members of Bocaparvovirus express one large NS protein (NS1) from the left viral genome, VP proteins from the right side of the viral genome, and at least one small NS protein (NP1) that is encoded by an open reading frame (ORF) located in the middle of the viral genome (11, 16, 17). During MVC infection, we have detected another small NS protein (NS1∼66kd [NS1 protein with an approximate molecular mass of 66 kDa]), which contains the N terminus of the NS1 (18).
The Bocaparvovirus NS1, like the NS1 or Rep78 and Rep68 (Rep78/68) of other parvoviruses, is a multifunctional protein that has a site-specific origin DNA binding domain (OBD) and endonuclease activity at its N terminus (20), ATPase and helicase activities in the middle (21, 22), and a transactivation domain at its C terminus (23, 24). NS1 is essential to viral DNA replication, while NP1 is required for efficient viral DNA replication (11, 17). MVC NP1 protein plays a role in facilitating VP-encoding mRNAs to read through the proximal polyadenylation site that lies in the middle of the viral genome (18). Nothing is known about the functions of the newly identified MVC NS1∼66kd protein. The protein expression profile of HBoV1 has been characterized via transfection using an incomplete HBoV1 genome lacking both the left and right inverted termini (19). Expression of NS1, NP1 and VP from their respective encoding transcripts was identified; however, the identities of the NS proteins with approximate molecular masses of 66 kDa and 34 kDa that were detected by an anti-NS1 C terminus antibody are unknown (19).
In this study, we explored the expression profile of HBoV1 NS proteins in either nonreplicating plasmids or replicating infectious plasmids transfected into HEK293 cells or during HBoV1 infection of HAE-ALI. We identified three novel HBoV1 NS proteins, NS2, NS3, and NS4, both in the HBoV1 genome-replicating HEK293 cell system and in HBoV1 infection of HAE-ALI system. The functions of these proteins were further explored in these two systems.
MATERIALS AND METHODS
Cell culture. (i) Cell line.
Human embryonic kidney 293 (HEK293) cells (CRL-1573) were obtained from American Type Culture Collection (ATCC, Manassas, VA) and were cultured in HyClone Dulbecco's modified Eagle's medium (DMEM; GE Healthcare Bio-Sciences, Piscataway, NJ) with 10% fetal calf serum (FCS) (product number F0926; Sigma-Aldrich, St. Louis, MO).
(ii) Primary human airway epithelium cultures.
Polarized human airway epithelium cultures at an air-liquid interface (ALI), termed HAE-ALI, were generated by growing isolated human bronchial airway epithelial cells on collagen-coated, semipermeable membrane inserts (0.33 cm2, Transwell; Corning, Corning, NY). They were then allowed to differentiate at an air-liquid interface, either in an Ultraser G-containing medium, as described previously (11), or in PneumaCult-ALI medium (StemCell, Vancouver, BC, Canada), in 5% CO2 at 37°C. After 3 to 5 weeks, the polarity of the HAE-ALI cultures was determined based on the transepithelial electrical resistance (TEER); the cultures that had a TEER of over 1,000 Ω · cm2 were used for HBoV1 infection.
DNA constructs. (i) pHBoV1NSCap-based constructs.
The parent pHBoV1NSCap, which harbors HBoV1 NS and Cap genes but lacks the left and right termini (GenBank accession no. DQ000496) (19), was used to construct pHBoV1NS1HACap, in which a hemagglutinin (HA) tag sequence was inserted after the ATG of the NS1-coding sequence. pHBoV1NS1HA65*Cap and pHBoV1NS1HA303*Cap were constructed by inserting a stop codon in the NS1 coding sequence after amino acid residues (aa) 65 and 303, respectively, in pHBoV1NS1HACap. pHBoV1NS1HA296StrepCap was constructed by further introducing a Strep tag after aa 296 of the NS1 coding sequence. pHBoV1NS1HA65*Cap(smA1′) and pHBoV1NS1HA303*Cap(smD1′) were constructed by moving the silent mutations (sm) smA1′ and smD1′, as described below, into pHBoV1NS1HA65*Cap and pHBoV1NS1HA303*Cap, respectively.
(ii) pIHBoV1-based constructs.
pIHBoV1, the infectious plasmid clone of HBoV1, which contains the HBoV1 full-length genome (GenBank accession no. JQ923422), has been described previously (11). pIHBoV1(smA1′) and pIHBoV1(smD1′) were constructed by mutating the pIHBoV1 A1′ acceptor site AG/C at nucleotide (nt) 1015 to TC/G and its D1′ donor site G/GTAGGA at nt 1212 to A/GTTGGC.
(iii) pGEX-4T-3-based constructs.
The NS1-coding sequences from nt 349 to 708 and from nt 349 to 1284 were inserted into pGEX-4T-3 vector (GE Healthcare) to generate pGEX-4T-NS1N and pGEX-4T-NS1NL, respectively.
(iv) pGEM-4Z-based construct.
Plasmid p4Z-PA1′D1′ was constructed by inserting HBoV1 DNA from nt 916 to 1263 into pGEM-4Z (Promega, Madison, WI) via BamHI and HindIII sites.
All nucleotide numbers of the HBoV1 genome refer to the HBoV1 full-length genome (GenBank accession no. JQ923422) unless otherwise specified. Constructs were verified for the HBoV1 sequence by Sanger sequencing at MCLAB (South San Francisco, CA).
Antibody production and antibodies used.
pGEX-4T-3-based plasmids were used to express glutathione S-transferase (GST)-fused proteins comprising aa 1 to 120 (GST-NS1aa1-120) and aa 1 to 313 (GST-NS1aa1-313) of HBoV1 NS1. These proteins were used for the production of antibodies to NS1 N terminus short (anti-NS1N antibody) and NS1 N terminus long (anti-NS1NL antibody), respectively. Purified GST-fused proteins were used to immunize rats, following the antiserum production protocol that was described in our previous publication (17). All animal procedures were approved by the Institutional Animal Care and Use Committee of the University of Kansas Medical Center. Antibodies to NS1 C terminus aa 640 to 781 (anti-NS1C antibody), HBoV1 NP1 (anti-NP1 antibody), and HBoV1 VP (anti-VP antibody) have been described previously (11, 19). Anti-HA (clone HA-7; Sigma), anti-Flag (clone M2; Sigma), anti-Strep (clone 5A9F9; Genscript, Piscataway, NJ), anti-β-actin (product number A5441; Sigma), anti-β-tubulin IV (product number T7941; Sigma), and anti-ZO-1 (catalog number 610966; BD Biosciences) antibodies were purchased.
Transfection.
HEK293 cells grown in 60-mm dishes were transfected with 3 μg of plasmid as indicated in the figures; LipoD293 transfection reagent (SignaGen, Rockville, MD) was used, following the manufacturer's instructions. pBluescript SK plasmid was used as a control.
RNA isolation and RNA analyses. (i) RNA isolation.
At 48 h posttransfection, transfected HEK293 cells from two 60-mm dishes were used to extract total RNA using TRIzol reagent (Life Technologies). At 14 days postinfection (p.i.), at least 2 infected HAE-ALI cultures were used for total RNA extraction using TRIzol reagent, following the manufacturer's instructions.
(ii) Reverse transcription-PCR (RT-PCR).
For identification of A1′ and D1′ splice sites, viral cDNA was synthesized by using a Moloney murine leukemia virus (M-MLV) kit (Life Technologies) and HBoV1-specific primer (HBoV1 nt 2560-2536). PCRs were performed using forward (F) and reverse (R) primers as indicated in Fig. 1A. PCR fragments were sequenced at MCLAB. Oligonucleotides were synthesized at IDT (Coralville, IA).
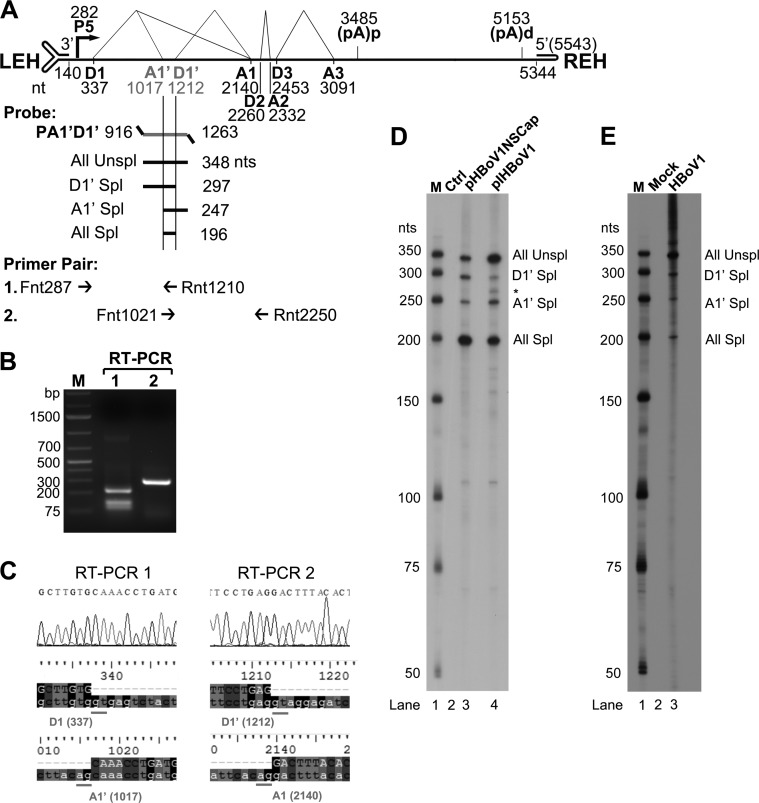
FIG 1.
HBoV1 pre-mRNA is processed at novel A1′ acceptor and D1′ donor sites. (A) Primers and probe used to detect viral mRNAs spliced at the A1′ and D1′ splice sites. The genome of HBoV1 is shown to scale, with transcription landmarks indicated, including the P5 promoter, splice donor (D1, D1′, D2, and D3) and acceptor (A1, A1′, A2, and A3) sites, the internal proximal polyadenylation site [(pA)p], and the distal polyadenylation site [(pA)d]. LEH, left end hairpin; REH, right end hairpin. The RPA probe PA1′D1′ (nt 916 to 1263) is shown, along with the designated bands that are expected to be protected and their predicted sizes (nt). Primers used for RT-PCR are also shown. (B and C) RT-PCR analyses of HBoV1 RNAs. Total RNA was isolated from HEK293 cells transfected with pHBoV1NSCap. cDNA was synthesized and amplified with two pairs of HBoV1-specific primers as shown. (B) Amplified DNA fragments were electrophoresed on 1.6% agarose gel and visualized using ethidium bromide staining. (C) The amplified DNA fragments were excised from the agarose gel and purified. The purified DNA was subjected to sequencing by the Sanger method. The histograms of the sequences at the exon junctions of D1/A1′ and D1′/A1 splice sites, with alignment of the sequences with the HBoV1 genome sequence (at the bottom), are shown. (D and E) Determination of the usage of the D1′ and A1′ splice sites by RPA. Ten micrograms of total RNA isolated at 2 days posttransfection from pBluescript SK (Ctrl) or pHBoV1NSCap- and pIHBoV1-transfected HEK293 cells (D) or at 14 days p.i. from mock- and HBoV1-infected HAE cells (E) was protected by the P1A1′D1′ probe as indicated. Lanes M contain 32P-labeled RNA markers (39), with sizes indicated to the left. The origins of the protected bands in the lanes are indicated to the right. Spl, spliced RNAs; Unspl, unspliced RNAs.
(iii) RPAs.
Probe PA1′D1′ was generated by in vitro transcription of the EcoRI-linearized p4Z-PA1′D1′ plasmid using T7 RNA polymerase. Ten micrograms of the total RNA was used for RNase protection assays (RPAs), following the methods described previously (25). Gels were imaged using a GE Typhoon FLA 9000 and processed with ImageQuant TL 8.1 (GE Healthcare) software for quantification.
DNA isolation and Southern blotting.
Low-molecular-weight (Hirt) DNA was extracted and digested with or without DpnI essentially as previously described (26). DNA samples were run on a 1% agarose gel, and Southern blotting was performed as described previously (27). The blot was hybridized with an HBoV1 NSCap probe (19) and was then processed using a GE Typhoon FLA 9000 and ImageQuant TL 8.1 (GE Healthcare) for quantification.
Immunoprecipitation, SDS-PAGE, and Western blotting.
A Pierce Crosslink IP kit (catalog number 26147; Life Technologies) was used for immunoprecipitation. At 48 h posttransfection, HEK293 cells from a 60-mm dish were collected and lysed for immunoprecipitation with an anti-HA or anti-Strep antibody, following the manufacturer's instructions.
Immunoprecipitated proteins from cell lysates taken either 2 days posttransfection or 14 days p.i. were separated by SDS–10% PAGE, except for those in the blots shown in Fig. 4C and E and Fig. 7C and D, which were separated by SDS–8% PAGE. Proteins were then electrotransferred onto a nitrocellulose membrane. The membrane was first blocked in 5% nonfat milk in 20 mM Tris-HCl, pH 7.6, 150 mM NaCl buffer with 0.1% Tween 20 (TBST) and then incubated with a monoclonal antibody against Flag, HA, or Strep tag, with antisera raised against various regions of HBoV1 NS1 (anti-NS1N, anti-NS1NL, or anti-NS1C antisera), or with anti-NP1, anti-VP, and anti-β-actin antibodies. Thoroughly washed with TBST after each step of incubation, the membrane was incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody (Jackson ImmunoResearch, Inc., West Grove, PA) and then with SuperSignal West Pico chemiluminescent substrate (Life Technologies) for signal development under the Fujifilm imager LAS 4000 (Fujifilm Life Sciences). Images were processed with Multi Gauge version 2.3 software (Fujifilm Life Sciences).
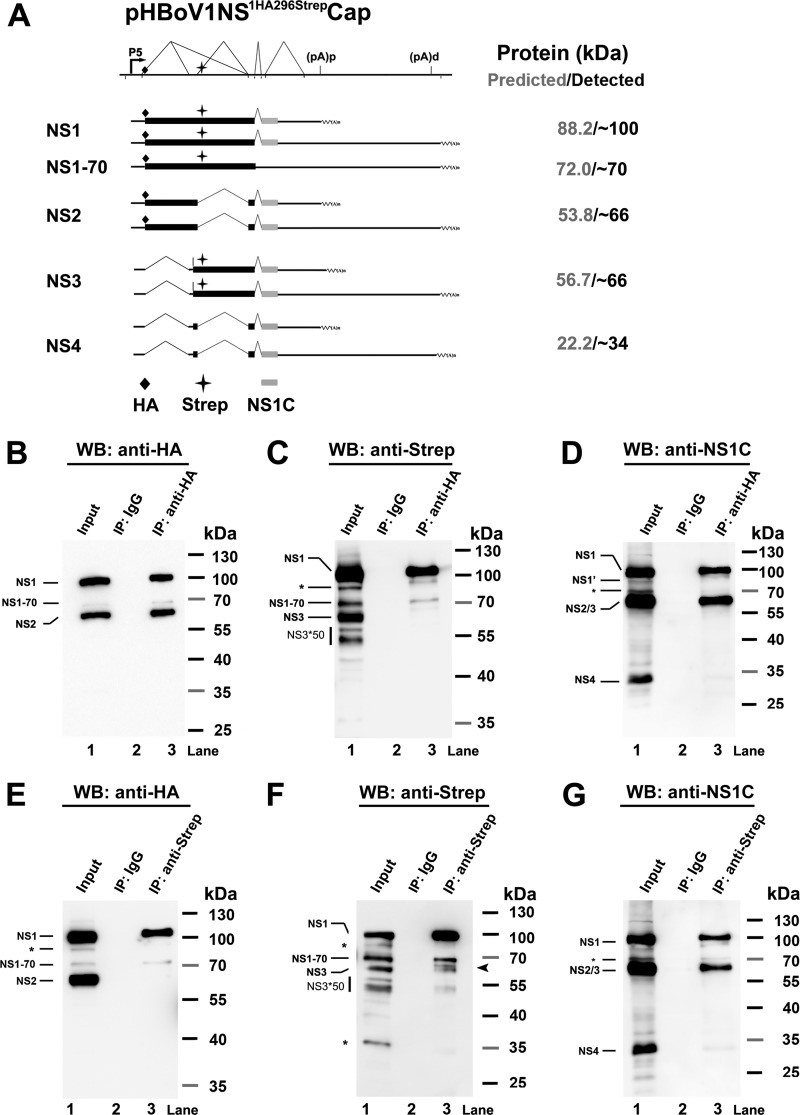
FIG 4.
Detection of HBoV1 NS1, NS2, NS3, and NS4 proteins in transfected HEK293 cells using immunoprecipitation assays. (A) Putative NS expression from transfection of the pHBoV1NS1HA296StrepCap plasmid. A putative expression profile of HBoV1 NS proteins is schematically diagramed, with only HA, Strep, or NS1 C terminus-containing NS-encoding mRNAs shown. The sizes of the putative NS proteins and the sizes actually detected are shown side by side at the right. (B to D) Western blot (WB) analysis of the NS proteins immunoprecipitated (IP) by an anti-HA antibody. HEK293 cells were transfected with pHBoV1NS1HA296StrepCap. At 2 days posttransfection, cells were lysed for immunoprecipitation using the anti-HA antibody or mouse IgG (as a control). Precipitated proteins, together with 10% of the input cell lysate, were analyzed by Western blotting using anti-HA (B), anti-Strep (C), and anti-NS1C (D) antibodies. (E to G) Western blot analysis of the NS proteins immunoprecipitated by an anti-Strep antibody. HEK293 cells were transfected with pHBoV1NS1HA296StrepCap. At 2 days posttransfection, cells were lysed for immunoprecipitation using the anti-Strep antibody or mouse IgG (as a control). Precipitated proteins, together with 10% of the input cell lysate, were analyzed by Western blotting using anti-HA (E), anti-Strep (F), and anti-NS1C (G) antibodies. The identities of detected protein bands are shown with lines to the left of the blot. Asterisks indicate bands that likely originated from degraded, unmodified, or cleaved proteins.
FIG 7.
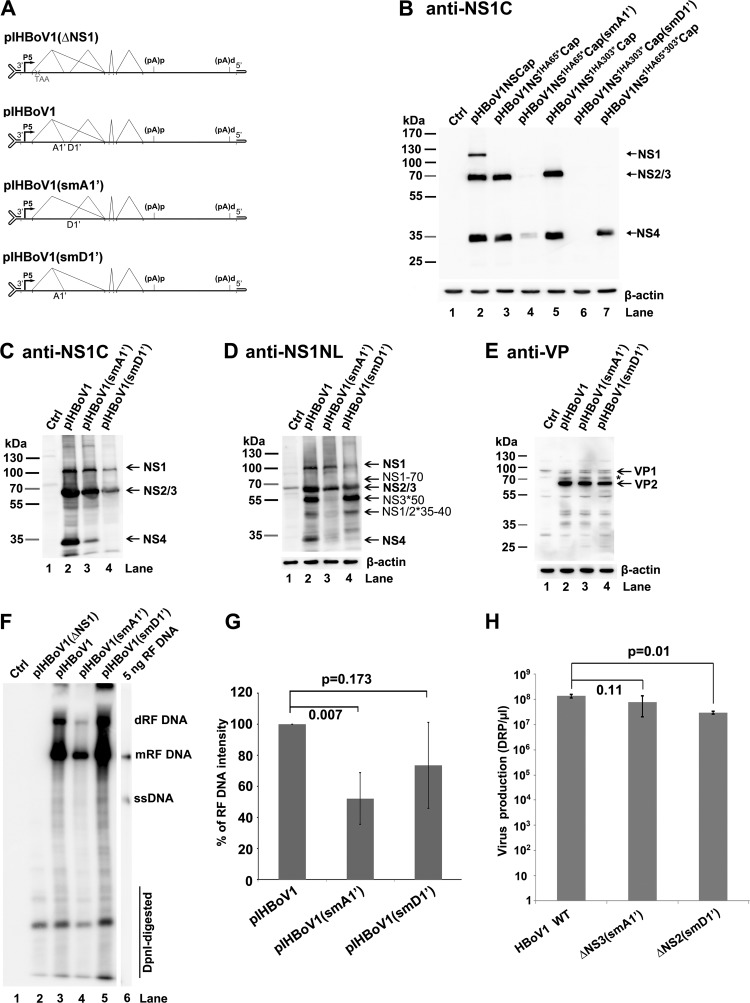
Analyses of virus production from transfection of mutant HBoV1 infectious clones in HEK293 cells. (A) Diagrams of pIHBoV1 and its mutants. pIHBoV1(ΔNS1), which has a stop codon after NS1 aa 65 (11), and pIHBoV1(smA1′) and pIHBoV1(smD1′), in which the A1′ and D1′ splice sites, respectively, are knocked out, are diagramed, with transcription, splicing, and polyadenylation units shown. (B) NS knockdown efficiency from smA1′ and smD1′ mutations. HEK293 cells were transfected with plasmids with smA1′ and smD1′ mutations in the context of pHBoV1NSCap, as indicated. At 2 days posttransfection, cells were lysed for Western blotting using anti-NS1C antibody. (C to E) Western blot analysis of proteins from infectious clones. HEK293 cells were transfected with plasmids as indicated. Cells were harvested and lysed at 2 days posttransfection. The lysates were analyzed by Western blotting using anti-NS1C antibody (C) and reprobed with anti-NS1NL and anti-β-actin antibodies (D) and anti-VP antibody (E). The identities of detected proteins are shown with arrows at the right of the blot. The asterisk indicates a band that is likely a cleaved VP or an unknown VP. (F and G) Southern blot analysis of viral DNA replication. (F) HEK293 cells were transfected with plasmids as indicated. At 2 days posttransfection, cells were harvested for Hirt DNA preparations. The Hirt DNA samples were digested with DpnI and analyzed by Southern blotting with an HBoV1NSCap probe (19). The identities of detected bands are shown. dRF, double replicative form; mRF, monomer replicative form; ssDNA, single-stranded DNA. (G) Quantification of the RF bands, normalized to the level of DpnI-digested DNA, from three independent experiments is shown with means and standard deviations. (H) Quantification of virus production. HEK293 cells were transfected with plasmids as indicated. At 2 days posttransfection, cells were harvested for virus preparation. The final virus preparations (2 ml for each) were quantified for DNase I-resistant particles (DRP) using qPCR and are shown as DRP/μl on a log scale (y axis). Means and standard deviations are calculated from three independent experiments. P values are calculated using the Student t test.
Virus production and infection. (i) Virus production.
HEK293 cells cultured on five 145-mm plates in DMEM–10% FCS were transfected with 30 μg of pIHBoV1 or its mutant per dish using polyethylenimine (PEI) (1 mg/ml, catalog number 23966; Polysciences, Warrington, PA) at a DNA/PEI ratio of 1:2. At 2 days posttransfection, cells were lysed for virus purification as previously described (11).
(ii) Virus infection of HAE-ALI.
Polarized HAE-ALI cultures were apically infected with purified virus at a multiplicity of infection (MOI) of 10 DNase I-resistant particles (DRP) per cell in 50 μl of phosphate-buffered saline (PBS), pH 7.4. After incubation for 2 h, apical medium was aspirated, and the insert was washed 3 times with PBS. Following infection, released virus was collected daily by incubating the apical side of the insert with 100 μl of PBS.
Both purified virus preparations and the viruses released from the apical side were quantified for virus particles as DRP using quantitative PCR (qPCR) as previously described (11).
Immunofluorescence analysis.
At 14 days p.i., HAE-ALI membranes were fixed with 3.7% paraformaldehyde in PBS at room temperature for 15 min. The fixed membranes were cut into small pieces. We then performed immunofluorescence assays with anti-NS1C and anti-β-tubulin IV antibodies or with anti-NS1C and anti-ZO-1 antibodies, as described previously (11).
RESULTS
Novel splice sites, A1′ acceptor and D1′ donor, are used in processing of HBoV1 pre-mRNA.
We have identified a novel splice donor site, 1D′, that lies in the first intron of the pre-mRNA of Bocaparvovirus MVC (18). We suspected the presence of a similar donor site in HBoV1 pre-mRNA (Fig. 1A, D1′). In addition, we previously identified an A1-1 acceptor site in the first intron of HBoV1 pre-mRNA (Fig. 1A, A1′) (19). To confirm the alternative use of these two splice sites in the processing of HBoV1 pre-mRNA, we designed two primer pairs to detect viral mRNA transcripts spliced at D1′, A1′, or both sites in total RNA isolated from pHBoV1NSCap-transfected HEK293 cells. The primer pair F287 and R1210 amplified a DNA fragment of ∼250 bp (Fig. 1B, RT-PCR 1). Sequencing confirmed that the A1′ acceptor site is used at nt 1017 (Fig. 1C, RT-PCR 1). Another primer pair, F1021 and R2250, amplified a band of ∼300 bp (Fig. 1B, RT-PCR 2). Sequencing showed that the D1′ site is used at nt 1212 (Fig. 1C, RT-PCR 2).
Next, using RPAs, we employed an antisense RNA probe spanning nt 916 to 1263 to determine the relative abundance of the HBoV1 mRNAs which were alternatively spliced at the A1′ and D1′ splice sites (Fig. 1A, PA1′D1′). In the RNA samples isolated from transfected cells, we detected 4 protected bands (Fig. 1D), as predicted (Fig. 1A), confirming the involvement of the A1′ and D1′ splice sites in processing HBoV1 pre-mRNA. By quantifying these bands, we found that ∼5% of the protected viral RNAs were spliced at either the A1′ or D1′ splice site in pre-mRNAs generated from either the nonreplicating pHBoV1NSCap or the replicating pIHBoV1 plasmid (Fig. 1D, D1′ and A1′ Spl). However, the replicating pIHBoV1 generated less RNAs spliced at both splice sites than did the nonreplicating pHBoV1NSCap in transfected cells (Fig. 1D, All Spl). The involvement of the A1′ and D1′ splice sites was further validated in the processing of viral pre-mRNA during HBoV1 infection of HAE-ALI (Fig. 1E, lane 3). Notably, virus infection produced much less viral RNA spliced at both the A1′ and D1′ sites than did the pIHBoV1 in transfection (Fig. 1E, lane 3, versus Fig. 1D, lane 4, All Spl).
Since the HBoV1 pre-mRNA is significantly spliced at the A1′ and D1′ splice sites, we searched for potentially encoded small NS proteins spanning the NS1 coding sequence from these alternatively spliced mRNAs. As shown in Fig. 2, R2 short and long (R2S/L) mRNA, in which the D1′-A1 intron is excised, can express NS2 protein; R3 short and long (R3S/L) mRNA, in which the D1-A1′ intron is excised, can translate NS3 protein; and R4 short and long (R4S/L) mRNA, in which both the D1-A1′ and D1′-A1 introns are excised, is expected to have the capability to translate the NS4 protein.
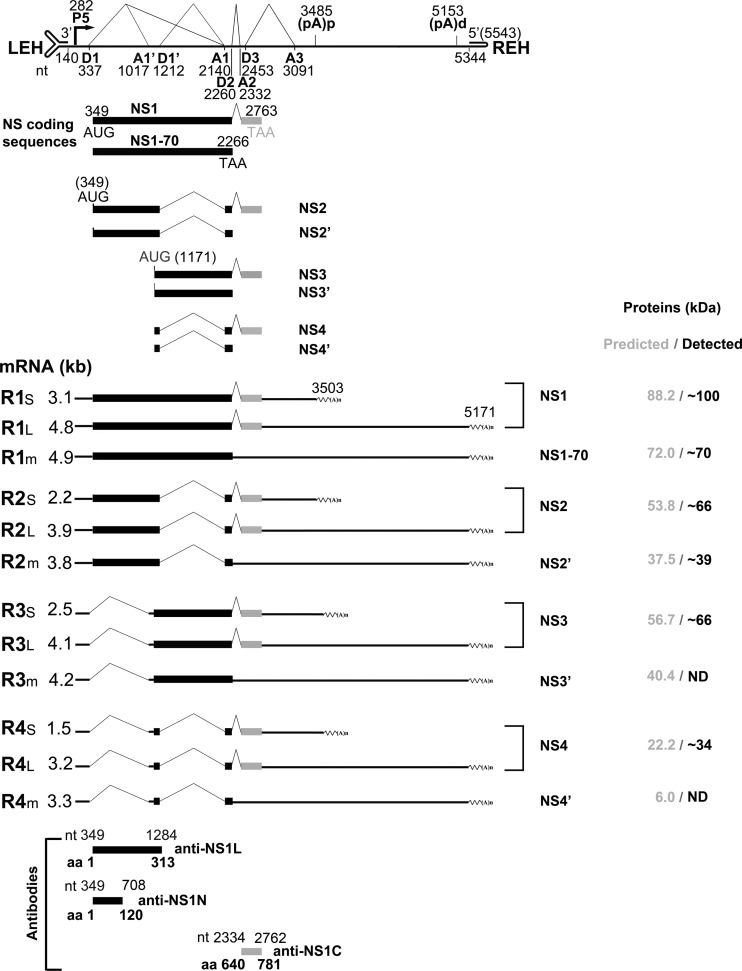
FIG 2.
Putative expression of HBoV1 NS proteins from alternatively processed mRNAs. A putative expression profile of HBoV1 NS proteins, with transcription landmarks and putative NS proteins, is shown. Major species of HBoV1 mRNA transcripts that are alternatively processed at the A1′ and D1′ splice sites and their putatively encoded NS proteins are shown with their relative sizes [minus a poly(A) tail of ∼150 nt]. “m” denotes minor species of the HBoV1 mRNA transcripts. The putative protein sizes and the sizes actually detected in the next experiments are shown side by side at the right. Antibodies raised against various portions of the NS1 protein are diagramed at the bottom.
The predicted sizes of the NS2, NS3, and NS4 proteins are 53.8, 56.7, and 22.2 kDa, respectively. However, in both HBoV1 plasmid-transfected and virus-infected cells, we previously detected two NS protein bands at ∼66 kDa and ∼34 kDa, in addition to the NS1 band at ∼100 kDa, using an anti-NS1C antibody (Fig. 2, α-NS1C) (11, 19). NS1's predicted molecular mass of 88.2 kDa led us to speculate that the C terminus of NS1 is highly posttranslationally modified, which gives an increment of ∼12 kDa in its apparent molecular weight. Since all the predicted NS2, NS3, and NS4 proteins share the C terminus with the NS1 protein, we further postulate that NS2, NS3, and NS4 experience similar posttranslational modification, resulting in detected molecular masses of ∼66, ∼69 and ∼34 kDa, respectively, with an increment of ∼12 kDa from the modification (Fig. 2).
In addition, an in-frame stop codon would terminate NS protein expression in viral mRNAs that retain the D2-A2 intron. NS1 detected at a size of 70 kDa (NS1-70), NS2′, NS3′, and NS4′ could be expressed from the R1m, R2m, R3m, and R4m mRNAs (Fig. 2), which are not abundantly expressed mRNAs, since splicing of the D2-A2 intron is efficient. The ratio of spliced versus unspliced RNA at the D2 site is over 20:1 (19). NS1-70 has been detected previously in HBoV1 plasmid-transfected cells (19).
Detection of NS2, NS3, and NS4 proteins in HEK293 cells transfected with nonreplicating HBoV1 constructs.
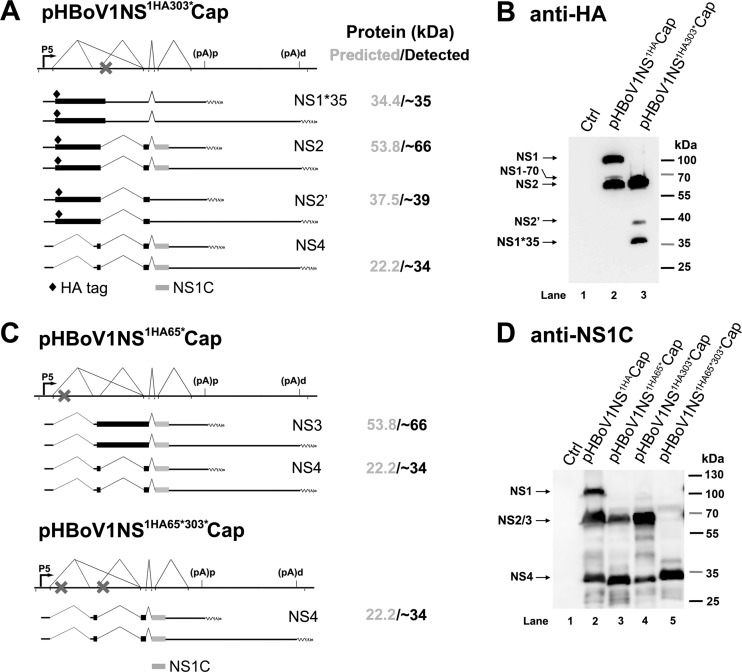
In the following sets of experiments, we aimed to validate the expression of NS2, NS3, and NS4 in HEK293 cells transfected with pHBoV1NSCap-based plasmids. We first introduced a stop codon to experimentally cause early termination of the NS1 and NS3 coding sequences in pHBoV1NS1HACap. Transfection of pHBoV1NS1HA303*Cap produced two major protein bands at ∼66 kDa and ∼35 kDa, as detected by an anti-HA antibody (Fig. 3B, lane 3), supporting the notion that NS1 was truncated to ∼35 kDa. The truncated NS3 should be ∼3 kDa and was undetectable on the blot. We believe that the ∼66-kDa band is NS2, which is assumed to be translated from the NS2 mRNAs that excise the D1′-A1 intron (Fig. 3A, NS2). A weak band at ∼37 kDa that was also detected by the anti-HA antibody fits the size of NS2′ (Fig. 3A, NS2′). Since NS1-70 was detected at a much lower level than the abundant NS1 (Fig. 3B, lane 2), we believe that the NS1-70, NS2′, NS3′, and NS4′ proteins are minimally expressed.
FIG 3.
Detection of HBoV1 NS1, NS2, NS3, and NS4 proteins in HEK293 cells transfected with pHBoV1NSCap-based plasmids. (A and B) Expression of HBoV1 NS2 protein. (A) Expected HA-tagged NS proteins expressed from pHBoV1NS1HA303*Cap. The NS-encoding viral mRNAs, which have the potential to produce HA-tagged NS proteins and C terminus-containing NS proteins, are diagramed. The expected NS proteins and their predicted and detected sizes are shown at the right. (B) HEK293 cells were transfected with pHBoV1NS1HACap or pHBoV1NS1HA303*Cap as indicated. At 2 days posttransfection, cells were lysed for Western blotting using an anti-HA antibody. (C and D) Expression of HBoV1 NS3 and NS4 proteins. (C) Expected NS1 C terminus (NS1C)-containing NS proteins expressed from pHBoV1NS1HA65*Cap and pHBoV1NS1HA65*303*Cap. The NS-encoding viral mRNAs, which have the potential to produce NS1C-containing NS proteins, are diagramed. The expected NS proteins and their predicted and detected sizes are shown at the right. (D) HEK293 cells were transfected with pHBoV1NSCap-based plasmids as indicated. At 2 days posttransfection, cells were lysed for Western blotting using the anti-NS1C antibody.
We next introduced a stop codon after aa 65 of the NS1 coding sequence in pHBoV1NS1HACap, which terminates both the NS1 and NS2 proteins early, but not the NS3 and NS4 proteins. Using an anti-NS1C antibody, we detected protein bands only at ∼66 kDa and ∼34 kDa in pHBoV1NS1HA65*Cap-transfected cells (Fig. 3D, lane 3), which confirms that the NS1 and NS2 proteins are terminated early and suggests that NS3 and NS4 proteins are expressed. On the other hand, only bands at ∼66 kDa and ∼34 kDa were detected in pHBoV1NS1HA303*Cap-transfected cells (Fig. 3D, lane 4), confirming that NS2 and NS4 proteins are expressed. When all the NS1, NS2, and NS3 proteins were terminated early in pHBoV1NS1HA65*303*Cap (Fig. 3C), only NS4 protein was detected in transfected cells (Fig. 3D, lane 5), validating the expression of NS4 from the mRNAs that excised both the D1-A1′ and D1′-A1 introns.
Furthermore, we tagged the NS1 proteins with an HA tag after aa 1 and with a Strep tag after aa 296, which lies in the intron of D1′-A1 in pHBoV1NSCap (Fig. 4A), and used an immunoprecipitation (IP) strategy to confirm the expression of NS2, NS3, and NS4. Anti-HA antibody-conjugated beads were used to pull down HA-tagged NS proteins from the lysate of the HEK293 cells transfected with pHBoV1NS1HA296StrepCap. In the precipitated proteins, we detected NS1 at ∼100 kDa using anti-HA, anti-Strep, or anti-NS1C antibody (Fig. 4B to D, lane 3). However, an NS protein at ∼66 kDa was only detected with an anti-HA or anti-NS1C antibody (Fig. 4B and D, lane 3). These results confirmed that the ∼66-kDa protein pulled down with anti-HA antibody is an NS2 protein that initiates at the HA tag, skips the D1′-A1 intron-encoding region tagged with a Strep tag after aa 295, and ends at the C terminus of the NS1 protein (Fig. 4A, NS2).
Next, a reverse IP using anti-Strep antibody-conjugated beads was performed with the same lysate of the cells transfected with pHBoV1NS1HA296StrepCap. In the precipitated proteins, NS1 was always detected at ∼100 kDa by all three antibodies (Fig. 4E to G, NS1), and an NS protein at ∼66 kDa was detected with the anti-Strep and anti-NS1C antibodies (Fig. 4F and G, lane 3) but not with the anti-HA antibody (Fig. 4E, lane 3), confirming that the ∼66-kDa protein pulled down by the anti-Strep antibody is NS3 protein that spans the D1′-A1 intron-encoding region, is tagged with a Strep tag, and includes the C terminus of the NS1 protein but is not initiated from the HA tag at the N terminus (Fig. 4A, NS3). Protein bands clustered at ∼50 kDa were also detected by the anti-Strep antibody (Fig. 4F, lanes 1 and 3, and Fig. 4C, lane 1, NS3*50). Whether these proteins are cleaved or degraded bands of NS3 or encoded by a new coding sequence remains unclear.
To further understand the NS4 protein expression strategy, we constructed pHBoV1NS275FlagCap, in which a Flag tag is inserted after aa 275 of the NS1 protein, immediately after a putative AUG at nt 1171 and before the D1′ donor site, and pHBoV1NS622FlagCap, in which a Flag tag is inserted after aa 622, which lies in the A1-D2 small exon. An anti-Flag antibody detected an NS protein at ∼34 kDa in cells transfected with either of the plasmids, in addition to the protein bands at ∼100 kDa and ∼66 kDa (Fig. 5, lanes 2 and 3). Together with evidence of the detection of the ∼34-kDa protein with the anti-NS1C antibody, as shown in Fig. 3 and 4, our results confirm that the ∼34-kDa protein is an NS4 protein that initiates at the AUG at aa 275 of the NS1 protein, skips the D1′-A1 intron-encoding region, reads through the A1-D2 exon, and extends to the C terminus of NS1 (Fig. 5, NS4).
FIG 5.
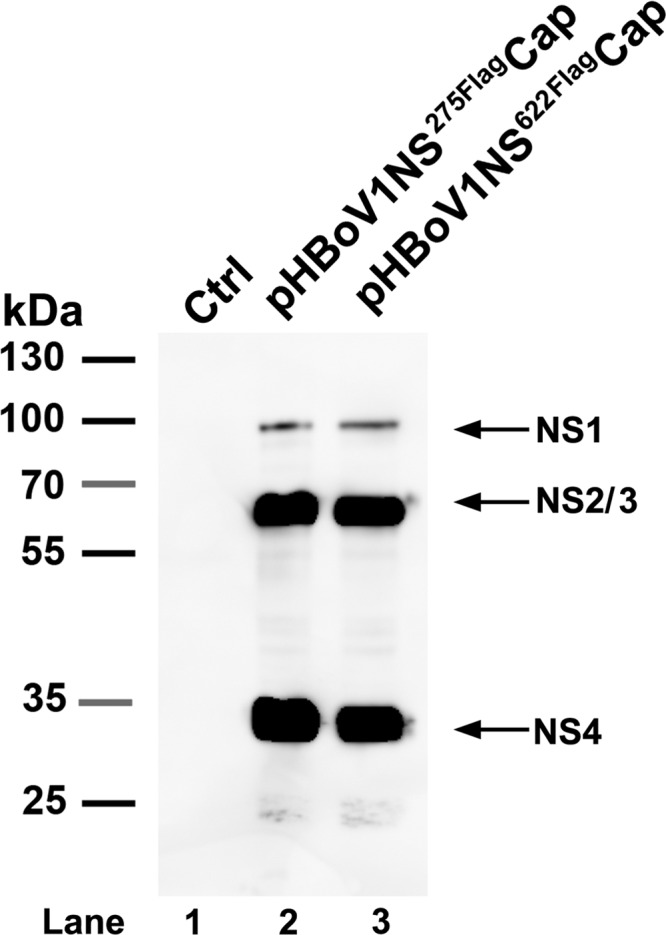
Detection of the HBoV1 NS4 protein in HEK293 cells transfected with plasmid pHBoV1NS275FlagCap or pHBoV1NS622FlagCap. At 2 days posttransfection, cells were lysed for Western blotting using an anti-Flag antibody. The identities of detected proteins are shown with arrows at the right of the blot.
Detection of NS2, NS3, and NS4 during HBoV1 replication in HEK293 cells and HBoV1 infection of HAE-ALI.
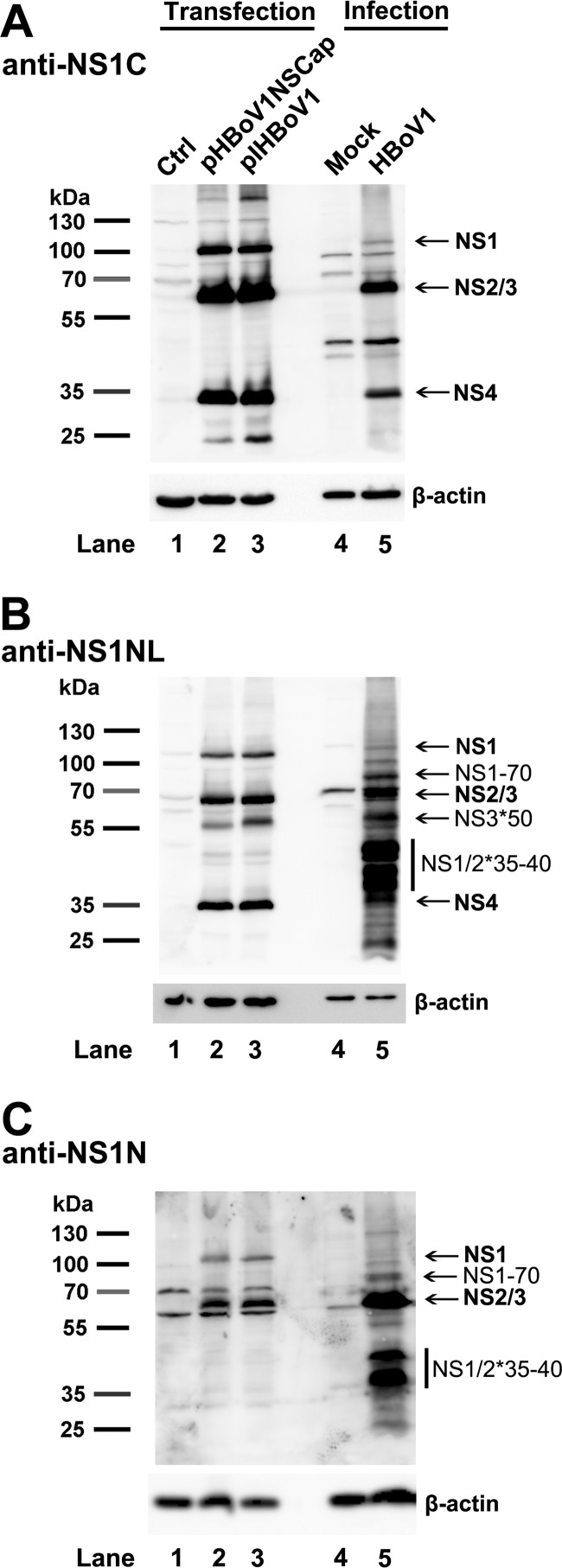
We next sought to detect the NS2, NS3, and NS4 proteins in the HBoV1 reverse genetics system and following HBoV1 infection of HAE-ALI. Using an anti-NS1C antibody, we detected NS1 at ∼100 kDa, a mixed band of NS2 and NS3 at ∼66 kDa, and NS4 at ∼34 kDa in HEK293 cells transfected with the replicating plasmid pIHBoV1 (Fig. 6A, lane 3), as well as in cells of the HAE-ALI infected with HBoV1 (Fig. 6A, lane 5). Using an anti-NS1NL antibody, which was raised against aa 1 to 313 of the NS1, we detected NS1, mixed NS2 and NS3, and NS4 proteins at their expected sizes, both in transfected and infected cells (Fig. 6B, lanes 2, 3, and 5). Additionally, a band at ∼50 kDa was detected by the anti-NS1NL antibody (Fig. 6B, NS3*50), which is similar to the ∼50-kDa protein band detected in the lysate of the cells transfected with pHBoV1NS1HA296StrepCap using the anti-Strep antibody (Fig. 4C and F, lane 1, NS3*50). However, this protein was not detected with an anti-NS1N antibody, which was raised against aa 1 to 120 of NS1 (Fig. 6C, lanes 2, 3, and 5), suggesting that the ∼50-kDa protein is likely a derivative of NS3, named NS3*50.
FIG 6.
Detection of HBoV1 NS2, NS3, and NS4 proteins in pIHBoV1-transfected HEK293 cells and HBoV1-infected HAE cells. HEK293 cells were transfected with pHBoV1NSCap or pIHBoV1. At 2 days posttransfection, cells were lysed. HAE-ALI cultures were infected with HBoV1 at an MOI of 10 (DRP/cell). At 14 days p.i., infected cells in the ALI cultures were lysed. The lysates of both transfected and infected cells, as indicated, were then analyzed by Western blotting using anti-NS1C (A), anti-NS1NL (B), and anti-NS1N (C) antibodies. The identities of detected proteins are shown with arrows at the right of the blot. Blots were reprobed for β-actin as a loading control.
Notably, two significant bands grouped at ∼35 to 40 kDa were detected by both the anti-NS1NL and anti-NS1N antibodies in infected cells but not in transfected cells (Fig. 6B and C, NS1/2*35-40), suggesting that the NS1 and NS2 proteins at 35 to 40 kDa (NS1/2*35-40) are likely cleaved or degraded NS1 and NS2 proteins or, perhaps, that they are NS2′ proteins that are highly expressed in infected cells. As expected, an anti-NS1N antibody detected both ∼100-kDa NS1 and ∼66-kDa NS2 proteins in either transfected or infected cells (Fig. 6C, lanes 2, 3, and 5). As noted, NS1-70 was detected at a higher level in infected cells than in transfected cells (Fig. 6B and C, lane 5 versus lanes 2 and 3). This result may suggest a potential function of NS1-70 during infection. NS4 was clearly detected at ∼34 kDa by the anti-NS1C and anti-NS1NL antibodies (Fig. 6A and B, lanes 2, 3, and 5) but not by the anti-NS1N antibody (Fig. 6C, lane 2, 3, and 5) in both transfected and infected cells.
Taken together, these results confirm the expression of the novel NS2, NS3, and NS4 proteins in HEK293 cells transfected with the infectious clone and, more importantly, in HAE-ALI cells infected with HBoV1. However, only HBoV1 infection demonstrated relatively abundant expression of these short versions of NS proteins (NS1-70, NS1/2*35-40, and NS3*50).
NS2, NS3, and NS4 proteins are dispensable for efficient viral DNA replication and virion production in the HBoV1 reverse genetics system.
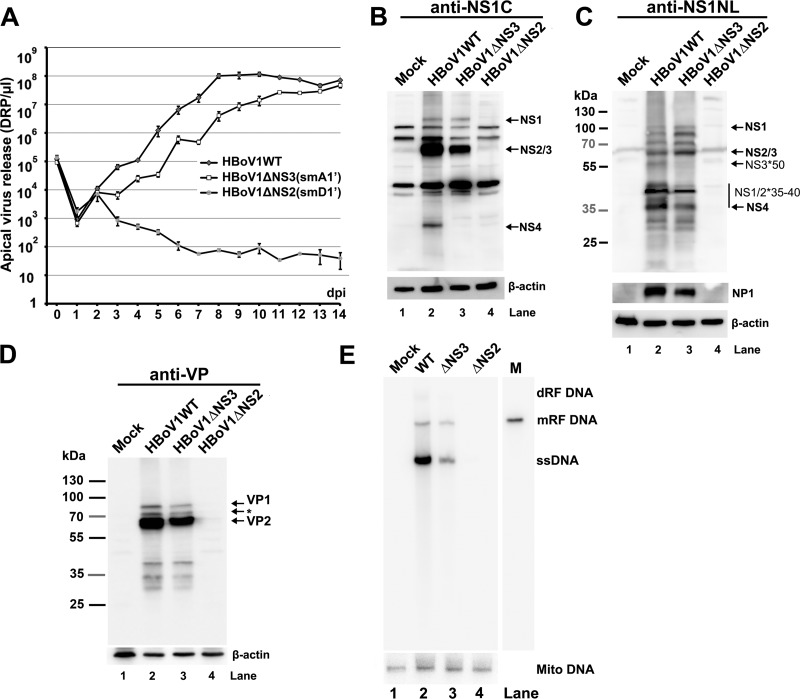
To investigate the functions of NS2, NS3, and NS4 in HBoV1 replication, we constructed pIHBoV1(smA1′) and pIHBoV1(smD1′) by knocking out the A1′ and D1′ splice sites, respectively, in the context of the infectious clone pIHBoV1 (Fig. 7A). Both mutants maintained the encoded NS protein sequences. Analyses of total RNA extracted from HEK293 cells transfected with pIHBoV1(smA1′) or pIHBoV1(smD1′) using RPAs with a homologous PA1′D1′ probe confirmed that splicing at the A1′ or D1′ splice site was abolished (data not shown). In order to examine the knockout efficiencies of NS3 and NS2, we constructed smA1′ and smD1′ mutations in nonreplicating plasmids pHBoV1NS1HA65*Cap and pHBoV1NS1HA303*Cap, respectively. The smA1′ mutation knocked down NS3 expression nearly to the background level of the blot and NS4 expression by ∼10-fold (Fig. 7B, compare lanes 4 and 3). The smD1′ mutation knocked out the expression of both NS2 and NS4 (Fig. 7B, compare lanes 6 and 5). Furthermore, protein analysis of the mutant infectious clones confirmed that pIHBoV1(smD1′) did not produce NS4 (Fig. 7C and D, lane 4) and that pIHBoV1(smA1′) decreased NS3*50 expression nearly to the background level of the blot (Fig. 7D, lane 3) and decreased NS4 by more than 10-fold (Fig. 7C, lane 3). All these mutants produced capsid proteins VP1 and VP2, as well as NP1 (data not shown), at levels similar to their levels in the wild-type (WT) (Fig. 7E).
Next, we tested the replication capability of the mutants in HEK293 cells. pIHBoV1(smA1′) had an ∼2-fold decrease in the production of replicative form (RF) DNA, while pIHBoV1(smD1′) replicated as effectively as the WT (Fig. 7F and G). We then produced mutant viruses ΔNS2(smD1′) and ΔNS3(smA1′) by transfecting HEK293 cells with pIHBoV1(smD1′) and pIHBoV1(smA1′), respectively, and compared viral production against that of the WT. As shown in Fig. 7H, the WT virus was produced at a total yield of 2 × 1011 DRP (2 ml of 1 × 108 DRP/μl) from five 145-mm plates of transfected HEK293 cells, and the mutant ΔNS3 and ΔNS2 viruses were also produced efficiently, with only moderate decreases of 1.7- and 4.5-fold, respectively.
Collectively, these results indicate that NS2, NS3, and NS4 are dispensable for viral DNA replication and for virus production in the HEK293 cell reverse genetics system.
HBoV1 NS2 but not NS3 and NS4 plays an essential role in HBoV1 replication in HAE-ALI.
We finally infected HAE-ALI cultures with the WT, ΔNS2, or ΔNS3 virus. We collected progeny virions produced from the apical side of the ALI cultures for a period of 14 days and quantified their titers. At 2 days p.i., we observed an ∼10-fold increase in virion release from the ALI cultures infected with either WT or mutant viruses (Fig. 8A, 2 dpi), confirming that both the WT and mutant viruses infected HAE-ALI. ΔNS2 virus launched an initial infection at 2 days p.i., as shown by a 1-log increase of apical virus release (Fig. 8A). However, the ΔNS2 virus-infected cultures then showed a gradual decrease in apical virus release, which remained at a level of less than 100 DRP/μl after 6 days p.i. (Fig. 8A, ΔNS2). On the other hand, though ΔNS3 virus-infected cultures produced virions at a level ∼10 times lower than that of the WT-infected cultures from 3 to 9 days p.i., they eventually reached a similar level of apical virus release (less than 2 times difference) at days 13 and 14 p.i. (Fig. 8A, compare ΔNS3 with WT).
FIG 8.
Analyses of virus infection of HAE-ALI cultures. HAE-ALI cultures were prepared in Transwell inserts and infected with HBoV1 WT or its mutants from the apical surface at an MOI of 10 (DRP/cell). (A) Apical virus release. At the indicated days p.i., the apical surface was washed with 100 μl of PBS to collect released virus. Virion particles (DRP) were quantified by qPCR (y axis) and plotted to the days p.i. as shown. Means and standard deviations are shown. (B to D) Western blot analysis of HBoV1-infected HAE-ALI cultures. At 14 days p.i., the cells of the infected HAE-ALI culture were lysed for Western blotting with anti-NS1C (B), anti-NS1NL (C), or anti-VP (D) antibody. The blots were reprobed for β-actin, and the blot in panel C was further reprobed with an anti-NP1 antibody. The identities of detected proteins are shown with arrows at the right of the blot. In panel D, the band indicated by an asterisk is likely a cleaved band of VP1 or a new VP. (E) Southern blot analysis of viral DNA replication. At 14 days p.i., the cells of the infected HAE-ALI cultures were lysed for Hirt DNA preparation. The Hirt DNA samples were analyzed by Southern blotting with an HBoV1 NSCap probe and a mitochondrial DNA (Mito DNA)-specific probe (40). Detected mitochondrial DNA was used for normalization of viral DNA quantification. The identities of detected bands are shown to the right.
At 14 days p.i., we analyzed the NS expression of the infected HAE cells. No NS or capsid (VP) proteins were detectable in ΔNS2 virus-infected cells (Fig. 8B to D, lane 4). ΔNS3 virus-infected cells expressed NS1, NS2 (Fig. 8B and C, lane 3), NS1/2*35-40 (Fig. 8C, lane 3), and VP (Fig. 8D, lane 3) at levels similar to those of the WT counterpart (Fig. 8B to D, lane 2) but did not express NS4 (Fig. 8B, lane 3) and NS3*50 (Fig. 8C, lane 3). This result echoes the NS expression of the ΔNS3 parent infectious clone pIHBoV1(smA1′) (Fig. 7C and D). Southern blot analysis showed that the ΔNS3 virus infection had a decrease in the level of ssDNA but not of mRF DNA (Fig. 8E, compare lanes 3 and 2), suggesting that the ΔNS3 virus replicates at a level similar to the WT in HAE-ALI. We confirmed that there was no obvious viral DNA detected in ΔNS2 virus-infected cells at 14 days p.i. (Fig. 8E, lane 4). Since both the ΔNS2 and ΔNS3 mutants did not express NS4 (Fig. 8B, lanes 3 and 4), these results suggest that NS2 is essential but that NS3 and NS4 are dispensable to HBoV1 replication in HAE-ALI.
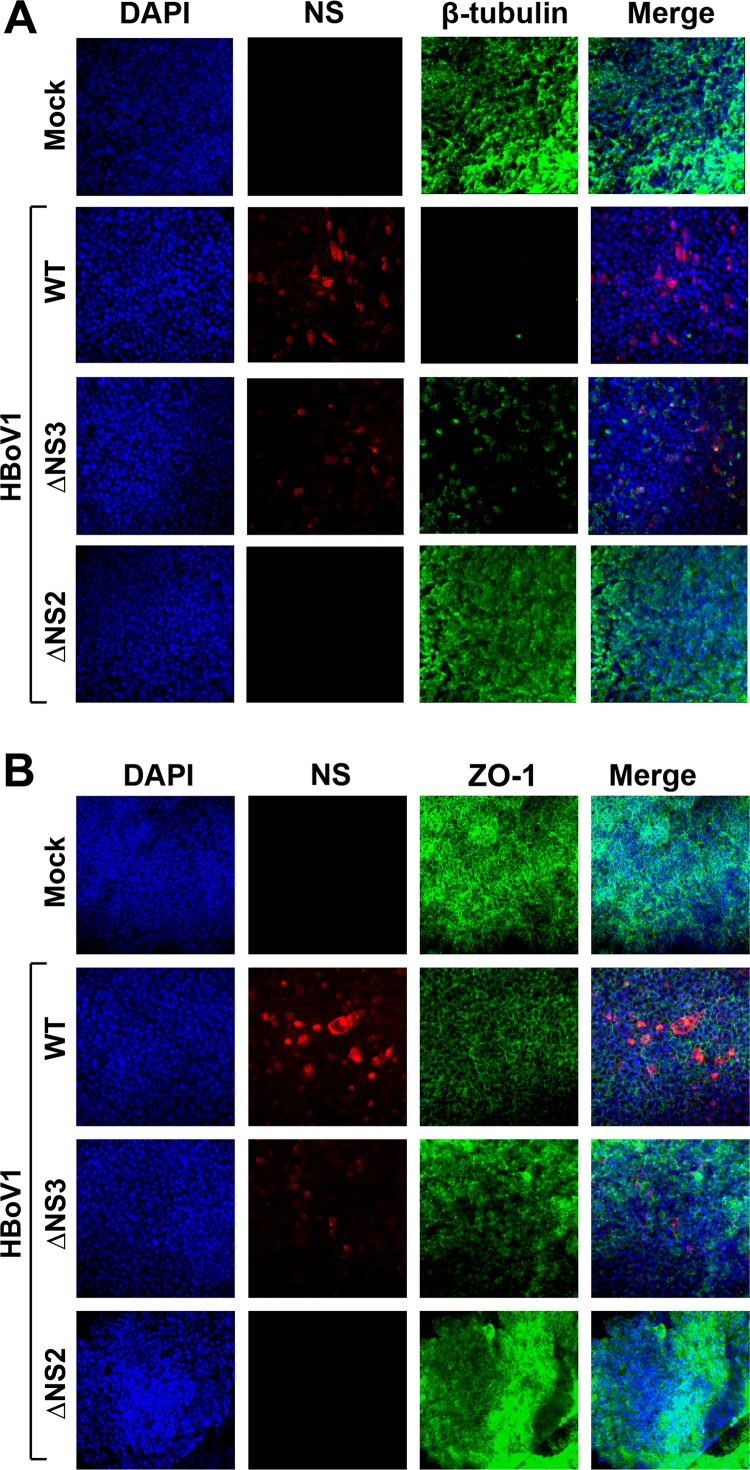
Further analyses of the infected HAE by immunofluorescence assays showed that ΔNS3 virus infection caused loss of cilia on the airway epithelium, as shown by staining of β-tubulin (Fig. 9A, ΔNS3), and redistribution of the tight junction, as shown by staining of ZO-1 (Fig. 9B, ΔNS3), but to a lesser extent than for the WT virus (Fig. 9, WT). This could be due to lower expression of NS proteins as detected by the anti-NS1C antibody during infection. On the other hand, the abortive infection of ΔNS2 virus did not cause any disruption of the tight junction or loss of the cilia, as shown by the ZO-1 and β-tubulin staining, respectively (Fig. 9, ΔNS2), which is consistent with the abortive infection of the ΔNS2 mutant virus.
FIG 9.
Immunofluorescence analysis of the tight junction protein ZO-1 and the cilium marker β-tubulin IV of infected HAE-ALI cultures. At 14 days p.i., mock-, WT-, ΔNS2-, and ΔNS3-infected HAE-ALI cultures were costained with anti-NS1C and anti-β-tubulin IV antibodies (A) or with anti-NS1C and anti-ZO-1 antibodies (B). Confocal images were taken at a magnification of ×40. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) (blue).
DISCUSSION
In this study, we confirmed that two novel splice sites are used to process HBoV1 pre-mRNA, generating mRNA transcripts that are able to translate small NS proteins, NS2, NS3, and NS4. We detected NS2, NS3, and NS4 proteins expressed during HBoV1 DNA replication in HEK293 cells and HBoV1 infection of HAE-ALI cultures. Although NS2, NS3, and NS4 proteins are dispensable for viral DNA replication and virus production in HEK293 cells, NS2 plays a critical role in HBoV1 replication in HAE-ALI cultures. Interestingly, NS2, NS3, and NS4 proteins contain the predicted domains of origin binding/endonuclease and transactivation, helicase and transactivation, and transactivation, respectively, of NS1 (Fig. 10). Importantly, different expression patterns of NS2, NS3, and NS1-70 were found during HBoV1 infection of HAE-ALI compared to those in HBoV1 replication in HEK293 cells, highlighting the importance of these small NS proteins during HBoV1 infection of human airway epithelia.
FIG 10.
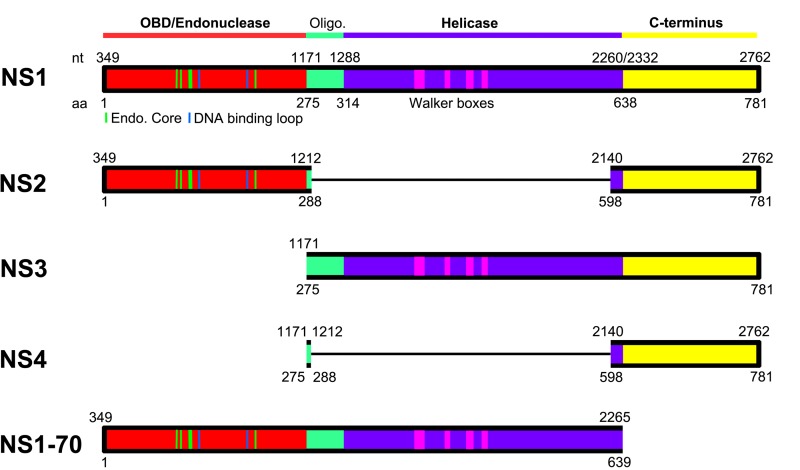
Putative functional domains of HBoV1 NS1, NS2, NS3, NS4, and NS1-70. A schematic diagram of the HBoV1 NS1 protein is shown. The HBoV1 NS1 protein sequence (GenBank accession number AFR53039) is aligned with the AAV5 Rep78 (GenBank accession number AAD13755). The N-terminal origin DNA binding domain (OBD; in red) and helicase domain (in purple) are predicted based on structured regions of the AAV5 Rep78 OBD (41) and AAV2 Rep40 helicase domain (42). Unaligned regions positioned between the OBD and helicase domains (shown in green) are predicted by analogy with AAV and MVM to contain NS oligomerization signals. The C-terminal region (shown in yellow) is predicted to contain a zinc-binding motif as seen in AAV2 Rep78 (23), while in MVM, this region is highly acidic and serves as a potent transcriptional activator (24). The OBD (aa 1 to 271) was initially predicted based on secondary structured and unstructured regions of the NS1 protein. Its structure has now been determined (20). Green and blue lines in the OBD indicate residues that are structured as endonuclease core/DNA binding loop (20), and pink blocks in the helicase domain indicate Walker boxes (43). Domains of the NS2, NS3, NS4, and NS1-70 proteins, deduced from those of the NS1, are diagramed in colored blocks, with thin lines indicating protein sequence excision due to ligation of the neighboring exons of their mRNAs.
Expression strategy of Bocaparvovirus NS proteins.
NS1, or Rep78/68 in adeno-associated virus (AAV), is a multifunctional protein that has site-specific origin DNA binding, endonuclease, ATPase, and helicase activities (Fig. 10, NS1) (21, 22). It is expressed from viral mRNAs transcribed from the promoter near the left end of the genome that are either unspliced or spliced at the small intron that lies in the middle of the genome (28). HBoV1 uses a strategy similar to that of the AAV2 Rep78/68 proteins (28) to express NS1 and NS1-70 from spliced and unspliced mRNAs, respectively, of the pre-mRNA transcribed from the P5 promoter. However, BPV and MVC, also members of the genus Bocaparvovirus, express NS1 at sizes of 100 kDa (16) and 84 kDa (18), respectively.
Parvoviruses also express a number of small NS proteins that are often important for virus replication, one of which is the Bocaparvovirus NP1 (11, 17). However, only a few parvoviruses express small NS proteins that share the amino acid sequences at the N terminus of the NS1. A minute virus of mice (MVM) mRNA excised the large intron, spanned a large portion of the NS1-encoding region, expresses NS2, and, thus, MVM NS2 shares only 85 aa with the NS1 at the N terminus (28, 29). The NS2 and NS3 proteins of Aleutian mink disease virus (AMDV) are expressed by a strategy similar to that of the MVM NS2 (30). They share 60 aa with the AMDV NS1 at the C terminus. Thus, the HBoV1 NS2 is unique in that it shares aa 1 to 288 with NS1, which includes the entire origin DNA binding/endonuclease domain (1 to 271 aa) of NS1 (Fig. 10, NS2) (20). NS2 also contains the C-terminal aa 598 to 781 of NS1. Of note, Bocaparvovirus MVC uses the same strategy to express NS1∼66kd (18), which is equivalent to the HBoV1 NS2. In BPV, we have previously proposed a viral mRNA that has the potential to express such an NS2 protein (16), but its expression has not been confirmed yet. Altogether, our results suggest that NS2 expression is a common feature among members of the genus Bocaparvovirus.
Small NS proteins, such as HBoV1 NS3, that share the helicase activity domain with NS1 have been identified in viruses of the genus Dependoparvovirus, i.e., AAV Rep52/40 (28) and goose parvovirus (GPV) Rep2 (31). But AAV Rep52/40 are expressed from mRNAs that are transcribed from an independent P19 promoter in the middle of the Rep78-coding region (28, 32). GPV Rep2 is expressed from an mRNA that excises the 1D-1A intron lying in the Rep1-encoding region (31, 33). Notably, we previously identified a Bocaparvovirus BPV R3 mRNA that excises an intron (D1-1A1) encoding N-terminal aa 1 to 282 of NS1, and a small NS protein (BPV NS2) is proposed to be expressed from this mRNA at ∼45 kDa (16). In fact, multiple NS bands centering at ∼45 kDa were detected in BPV-infected cell lysates following immunoprecipitation using convalescent-phase sera from BPV-infected calves (34); these may be candidates for the BPV NS2 protein. Therefore, like GPV Rep2 and BPV NS2, HBoV1 NS3 is expressed from the viral mRNA that excises the D1-A1′ intron encoding the origin binding domain (aa 1 to 274) (Fig. 10). However, such an expression strategy of NS3 has not been identified during MVC infection (18).
HBoV1 NS4 is expressed from a viral mRNA that has never been identified in the expression of any parvovirus. The mRNA excises multiple introns (D1-A1′, D1′-A1, and D2-A2). NS4 largely contains the C terminus (aa 598 to 781) of NS1 (Fig. 10). Currently under investigation is whether, in the genus Bocaparvovirus, NS4 protein expression is conserved in MVC and BPV infections.
Functions of the small NS proteins.
Our results suggest that HBoV1 NS2 is essential to virus replication in HAE-ALI but not to viral DNA replication in HEK293 cells. The function of NS2 seems to be cell type dependent, like that of MVM NS2, which is not essential for MVM replication in transformed human cell lines but is required for MVM infection in murine cells (35, 36). HBoV1 NS2 harbors the whole origin DNA binding domain with endonuclease activity (Fig. 10, NS2). Considering that HBoV1 replicates in differentiated/polarized HAE cells that are out of cell cycle (G0 phase) (10, 11), we speculate that the endonuclease feature of NS2 may be required for HBoV1 DNA replication in nondividing cells. The function of the MVC counterpart (NS1∼66kd) in MVC replication is not yet understood (18).
HBoV1 NS3 is dispensable for virus DNA replication and progeny virus production in HEK293 cells, as well as in infection of HAE-ALI. These results suggest that NS3 is not required for viral genome packaging, in contrast to AAV Rep52/40, which are involved in packaging single-stranded viral genomes into capsid structures (37), an activity that is mediated by their helicase activities (38). Since the ΔNS3 mutant virus expresses little NS4, we believe that NS4 is also not essential to virus replication and production in HAE-ALI; however, we cannot rule out functions of NS3 and NS4 that may modulate the host cell innate immune response during infection. Obviously, the function of NS3 and NS4 in the host innate immune response to HBoV1 infection of HAE-ALI warrants further investigation.
NS1-70 and other small NS proteins terminate at a stop codon in mRNAs retaining the D2-A2 intron.
NS1-70 was expressed at a higher level during virus infection of HAE-ALI than in pIHBoV1-transfected HEK293 cells. NS1/2*35-40 were expressed at relatively higher levels than NS1, -2, -3, and -4 during infection, but NS1/2*35-40 are not expressed in cells transfected with the infectious or nonreplicating plasmids. These differences between transfection and infection are not understood but may be due to differences in the types of host cells or to the cell differentiation status between the dividing HEK293 cells and nondividing HAE cells. NS1/2*35-40 are unlikely to be NS2′ proteins (Fig. 2, NS2′), since they were still expressed from infection of a mutant virus that bears the intron D2-A2 deletion (data not shown). Similarly, NS3*50 was expressed in HEK293 cells transfected with a mutant infectious clone that has the D2-A2 intron deleted, indicating that NS3*50 is not the NS3′ protein (Fig. 2, NS3′). We speculate that NS1/2*35-40 and NS3*50 are specifically cleaved products of the NS1/2 and NS3 proteins. Further studies are required to investigate the nature of the NS1/2*35-40 and NS3*50 proteins.
NS1-70 contains the origin DNA binding/endonuclease and helicase domains of NS1 but not the C terminus (Fig. 10, NS1-70). A mutant infectious clone that expresses only NS1-70 still replicates in HEK293 cells but to an extent ∼10 times less than that of the WT (data not shown), suggesting that the C terminus of NS1 plays a helper role in HBoV1 DNA replication. Although NS2′ was detectable in transfected cells (Fig. 3A, lane 3), NS2′ and NS3′ proteins were not obviously detected in infected cells in our study, suggesting that NS4′ is not expressed during infection.
In contrast to the acute infection of most other autonomous parvoviruses, HBoV1 infection is persistent, with the virus replicating in nondividing airway cells. Taken together, our results support the hypothesis that NS2, -3, and -4, NS1-70, NS1/2*35-40, and NS3*50 are required for the virus to balance replication and host cell death during infection of human airway epithelia. The creation of replication-defective HBoV1 mutants may have utility in vaccine development for this virus.
ACKNOWLEDGMENTS
We thank members of the Qiu laboratory for discussions and critical readings of the manuscript.
The study was supported by PHS grants AI070723, AI105543, and AI112803 from NIAID, NIH.
REFERENCES
- 1.Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A 102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, Soderlund-Venermo M, Tattersall P, Tijssen P, Gatherer D, Davison AJ. 2014. The family Parvoviridae. Arch Virol 159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allander T, Jartti T, Gupta S, Niesters HG, Lehtinen P, Osterback R, Vuorinen T, Waris M, Bjerkner A, Tiveljung-Lindell A, van den Hoogen BG, Hyypia T, Ruuskanen O. 2007. Human bocavirus and acute wheezing in children. Clin Infect Dis 44:904–910. doi: 10.1086/512196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meriluoto M, Hedman L, Tanner L, Simell V, Makinen M, Simell S, Mykkanen J, Korpelainen J, Ruuskanen O, Ilonen J, Knip M, Simell O, Hedman K, Soderlund-Venermo M. 2012. Association of human bocavirus 1 infection with respiratory disease in childhood follow-up study, Finland. Emerg Infect Dis 18:264–271. doi: 10.3201/eid1802.111293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nascimento-Carvalho CM, Cardoso MR, Meriluoto M, Kemppainen K, Kantola K, Ruuskanen O, Hedman K, Soderlund-Venermo M. 2012. Human bocavirus infection diagnosed serologically among children admitted to hospital with community-acquired pneumonia in a tropical region. J Med Virol 84:253–258. doi: 10.1002/jmv.22268. [DOI] [PubMed] [Google Scholar]
- 6.Martin ET, Kuypers J, McRoberts JP, Englund JA, Zerr DM. 2015. Human bocavirus-1 primary infection and shedding in infants. J Infect Dis 212:516–524. doi: 10.1093/infdis/jiv044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen A, Nordbo SA, Krokstad S, Rognlien AG, Dollner H. 2010. Human bocavirus in children: mono-detection, high viral load and viraemia are associated with respiratory tract infection. J Clin Virol 49:158–162. doi: 10.1016/j.jcv.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ursic T, Steyer A, Kopriva S, Kalan G, Krivec U, Petrovec M. 2011. Human bocavirus as the cause of a life-threatening infection. J Clin Microbiol 49:1179–1181. doi: 10.1128/JCM.02362-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korner RW, Soderlund-Venermo M, van Koningsbruggen-Rietschel S, Kaiser R, Malecki M, Schildgen O. 2011. Severe human bocavirus infection, Germany. Emerg Infect Dis 17:2303–2305. doi: 10.3201/eid1712.110574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dijkman R, Koekkoek SM, Molenkamp R, Schildgen O, van der Hoek L. 2009. Human bocavirus can be cultured in differentiated human airway epithelial cells. J Virol 83:7739–7748. doi: 10.1128/JVI.00614-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Q, Deng X, Yan Z, Cheng F, Luo Y, Shen W, Lei-Butters DC, Chen AY, Li Y, Tang L, Soderlund-Venermo M, Engelhardt JF, Qiu J. 2012. Establishment of a reverse genetics system for studying human bocavirus in human airway epithelia. PLoS Pathog 8:e1002899. doi: 10.1371/journal.ppat.1002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng X, Yan Z, Luo Y, Xu J, Cheng Y, Li Y, Engelhardt J, Qiu J. 2013. In vitro modeling of human bocavirus 1 infection of polarized primary human airway epithelia. J Virol 87:4097–4102. doi: 10.1128/JVI.03132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng X, Li Y, Qiu J. 2014. Human bocavirus 1 infects commercially available primary human airway epithelium cultures productively. J Virol Methods 195:112–119. doi: 10.1016/j.jviromet.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz D, Green B, Carmichael LE, Parrish CR. 2002. The canine minute virus (minute virus of canines) is a distinct parvovirus that is most similar to bovine parvovirus. Virology 302:219–223. doi: 10.1006/viro.2002.1674. [DOI] [PubMed] [Google Scholar]
- 15.Chen KC, Shull BC, Moses EA, Lederman M, Stout ER, Bates RC. 1986. Complete nucleotide sequence and genome organization of bovine parvovirus. J Virol 60:1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu J, Cheng F, Johnson FB, Pintel D. 2007. The transcription profile of the bocavirus bovine parvovirus is unlike those of previously characterized parvoviruses. J Virol 81:12080–12085. doi: 10.1128/JVI.00815-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y, Chen AY, Cheng F, Guan W, Johnson FB, Qiu J. 2009. Molecular characterization of infectious clones of the minute virus of canines reveals unique features of bocaviruses. J Virol 83:3956–3967. doi: 10.1128/JVI.02569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sukhu L, Fasina O, Burger L, Rai A, Qiu J, Pintel DJ. 2013. Characterization of the non-structural proteins of the bocavirus minute virus of canine (MVC). J Virol 87:1098–1104. doi: 10.1128/JVI.02627-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen AY, Cheng F, Lou S, Luo Y, Liu Z, Delwart E, Pintel D, Qiu J. 2010. Characterization of the gene expression profile of human bocavirus. Virology 403:145–154. doi: 10.1016/j.virol.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tewary SK, Zhao H, Shen W, Qiu J, Tang L. 2013. Structure of the NS1 protein N-terminal origin-recognition/nickase domain from the emerging human bocavirus. J Virol 87:11487–11494. doi: 10.1128/JVI.01770-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cotmore SF, Tattersall P. 2005. A rolling-hairpin strategy: basic mechanisms of DNA replication in the parvoviruses, p 171–181. In Kerr J, Cotmore SF, Bloom ME, Linden RM, Parrish CR (ed), Parvoviruses. Hodder Arnold, London, United Kingdom. [Google Scholar]
- 22.Im DS, Muzyczka N. 1990. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell 61:447–457. doi: 10.1016/0092-8674(90)90526-K. [DOI] [PubMed] [Google Scholar]
- 23.Smith RH, Spano AJ, Kotin RM. 1997. The Rep78 gene product of adeno-associated virus (AAV) self-associates to form a hexameric complex in the presence of AAV ori sequences. J Virol 71:4461–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Legendre D, Rommelaere J. 1994. Targeting of promoters for trans activation by a carboxy-terminal domain of the NS-1 protein of the parvovirus minute virus of mice. J Virol 68:7974–7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu J, Pintel DJ. 2002. The adeno-associated virus type 2 Rep protein regulates RNA processing via interaction with the transcription template. Mol Cell Biol 22:3639–3652. doi: 10.1128/MCB.22.11.3639-3652.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan W, Wong S, Zhi N, Qiu J. 2009. The genome of human parvovirus B19 virus can replicate in non-permissive cells with the help of adenovirus genes and produces infectious virus. J Virol 83:9541–9553. doi: 10.1128/JVI.00702-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu J, Cheng F, Burger LR, Pintel D. 2006. The transcription profile of Aleutian mink disease virus (AMDV) in CRFK cells is generated by alternative processing of pre-mRNAs produced from a single promoter. J Virol 80:654–662. doi: 10.1128/JVI.80.2.654-662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu J, Yoto Y, Tullis GE, Pintel D. 2006. Parvovirus RNA processing strategies, p 253–274. In Kerr J, Cotmore SF, Bloom ME, Linden RM, Parrish CR (ed), Parvoviruses. Hodder Arnold, London, United Kingdom. [Google Scholar]
- 29.Cotmore SF, Tattersall P. 2014. Parvoviruses: small does not mean simple. Annu Rev Virol 1:517–537. doi: 10.1146/annurev-virology-031413-085444. [DOI] [PubMed] [Google Scholar]
- 30.Huang Q, Luo Y, Cheng F, Best SM, Bloom ME, Qiu J. 2014. Molecular characterization of the small nonstructural proteins of parvovirus Aleutian mink disease virus (AMDV) during infection. Virology 452-453:23–31. doi: 10.1016/j.virol.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu J, Cheng F, Yoto Y, Zadori Z, Pintel D. 2005. The expression strategy of goose parvovirus exhibits features of both the Dependovirus and Parvovirus genera. J Virol 79:11035–11044. doi: 10.1128/JVI.79.17.11035-11044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samulski RJ, Muzyczka N. 2014. AAV-mediated gene therapy for research and therapeutic purposes. Annu Rev Virol 1:427–451. doi: 10.1146/annurev-virology-031413-085355. [DOI] [PubMed] [Google Scholar]
- 33.Li L, Pintel DJ. 2012. Splicing of goose parvovirus pre-mRNA influences cytoplasmic translation of the processed mRNA. Virology 426:60–65. doi: 10.1016/j.virol.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lederman M, Cotmore SF, Stout ER, Bates RC. 1987. Detection of bovine parvovirus proteins homologous to the nonstructural NS-1 proteins of other autonomous parvoviruses. J Virol 61:3612–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naeger LK, Cater J, Pintel DJ. 1990. The small nonstructural protein (NS2) of the parvovirus minute virus of mice is required for efficient DNA replication and infectious virus production in a cell-type-specific manner. J Virol 64:6166–6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruiz Z, D'Abramo A Jr, Tattersall P. 2006. Differential roles for the C-terminal hexapeptide domains of NS2 splice variants during MVM infection of murine cells. Virology 349:382–395. doi: 10.1016/j.virol.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 37.Smith RH, Kotin RM. 1998. The Rep52 gene product of adeno-associated virus is a DNA helicase with 3′-to-5′ polarity. J Virol 72:4874–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King JA, Dubielzig R, Grimm D, Kleinschmidt JA. 2001. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J 20:3282–3291. doi: 10.1093/emboj/20.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiu J, Nayak R, Tullis GE, Pintel DJ. 2002. Characterization of the transcription profile of adeno-associated virus type 5 reveals a number of unique features compared to previously characterized adeno-associated viruses. J Virol 76:12435–12447. doi: 10.1128/JVI.76.24.12435-12447.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sowd GA, Li NY, Fanning E. 2013. ATM and ATR activities maintain replication fork integrity during SV40 chromatin replication. PLoS Pathog 9:e1003283. doi: 10.1371/journal.ppat.1003283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hickman AB, Ronning DR, Kotin RM, Dyda F. 2002. Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep. Mol Cell 10:327–337. doi: 10.1016/S1097-2765(02)00592-0. [DOI] [PubMed] [Google Scholar]
- 42.James JA, Escalante CR, Yoon-Robarts M, Edwards TA, Linden RM, Aggarwal AK. 2003. Crystal structure of the SF3 helicase from adeno-associated virus type 2. Structure 11:1025–1035. doi: 10.1016/S0969-2126(03)00152-7. [DOI] [PubMed] [Google Scholar]
- 43.Koonin EV. 1993. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res 21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]