ABSTRACT
Human immunodeficiency virus type 1 (HIV-1) is released from infected cells in an immature, noninfectious form in which the structural polyprotein Gag is arranged in a hexameric lattice, forming an incomplete spherical shell. Maturation to the infectious form is mediated by the viral protease, which cleaves Gag at five sites, releasing the CA (capsid) protein, which forms a conical capsid encasing the condensed RNA genome. The pathway of this structural rearrangement is currently not understood, and it is unclear how cone assembly is initiated. RNA represents an integral structural component of retroviruses, and the viral nucleoprotein core has previously been proposed to nucleate mature capsid assembly. We addressed this hypothesis by replacing the RNA-binding NC (nucleocapsid) domain of HIV-1 Gag and the adjacent spacer peptide 2 (SP2) by a leucine zipper (LZ) protein-protein interaction domain [Gag(LZ)] in the viral context. We found that Gag(LZ)-carrying virus [HIV(LZ)] was efficiently released and viral polyproteins were proteolytically processed, though with reduced efficiency. Cryo-electron tomography revealed that the particles lacked a condensed nucleoprotein and contained an increased proportion of aberrant core morphologies caused either by the absence of RNA or by altered Gag processing. Nevertheless, a significant proportion of HIV(LZ) particles contained mature capsids with the wild-type morphology. These results clearly demonstrate that the nucleoprotein complex is dispensable as a nucleator for mature HIV-1 capsid assembly in the viral context.
IMPORTANCE Formation of a closed conical capsid encasing the viral RNA genome is essential for HIV-1 infectivity. It is currently unclear what viral components initiate and regulate the formation of the capsid during virus morphogenesis, but it has been proposed that the ribonucleoprotein complex plays a role. To test this, we prepared virus-like particles lacking the viral nucleocapsid protein and RNA and analyzed their three-dimensional structure by cryo-electron tomography. While most virions displayed an abnormal morphology under these conditions, some particles showed a normal mature morphology with closed conical capsids. These data demonstrate that the presence of RNA and the nucleocapsid protein is not required for the formation of a mature, cone-shaped HIV-1 capsid.
INTRODUCTION
Morphological maturation represents a key event in the replication cycle of retroviruses. Human immunodeficiency virus type 1 (HIV-1) particles are released as immature, noninfectious virions whose main constituents are the structural polyproteins Gag and Gag-Pro-Pol. Cleavage of Gag and Gag-Pro-Pol at multiple sites by the virus genome-encoded PR (protease) releases the mature structural proteins MA (matrix), CA (capsid), and NC (nucleocapsid) as well as the functional viral enzymes PR, reverse transcriptase (RT), and IN (integrase) (1). Proteolytic processing triggers a dramatic rearrangement of the virion architecture. In immature HIV-1, an incomplete sphere made of ∼2,500 radially arranged Gag molecules lines the viral lipid envelope. In contrast, mature, infectious particles are characterized by a cone-shaped capsid assembled from CA molecules. The capsid encases a dense ribonucleoprotein (RNP) core, in which NC condenses the viral RNA genome (2, 3).
Hexagonal arrangements of the CA domain represent the basic structural element of both the immature Gag lattice and the mature capsid. However, contact surfaces involved in CA-CA interactions differ significantly between immature and mature structures, and the spacing of the immature hexagonal lattice (80 Å) is tighter than that of the mature lattice (96 Å) (2, 3). Furthermore, only about 50% of the CA molecules packaged are used for formation of the mature capsid (4). These observations led to the proposition that morphological maturation occurs via dissociation of the immature lattice, followed by new assembly of the mature cone structure. This model is supported by structural analyses of HIV-1 derivatives carrying mutations at individual PR recognition sites in Gag, in particular, those preventing the late step of CA-SP1 processing (5, 6).
Whereas immature HIV-1 particles are very heterogeneous with respect to diameter and the degree of Gag shell completeness (7, 8), the majority of mature particles contain a single capsid core structure and a small number of particles contain two capsid structures (9). It therefore appears to be intuitive that the process of capsid assembly from dissociated CA mono- or oligomers within the confined space of the released particle requires a nucleating event.
Various candidates for a capsid assembly nucleator have been discussed. These include (i) factors at the budding scar where the viral envelope has pinched off from the plasma membrane (10); (ii) the transient processing intermediate CA-SP1 (11); and (iii) the viral RNA genome or the RNP, detected as a single, amorphous, electron-dense structure inside each capsid in electron micrographs and tomograms of wild-type (wt) virus. Larger virus particles, which are thought to contain two RNA genome dimers, often also contain two mature capsids, despite having lower concentrations of CA, lending support to a role for the RNP as a nucleator (9).
The latter possibility seems particularly attractive, since nucleation of the capsid around genomic RNA would ensure that the genome is contained within the capsid, which is important for retroviral particle infectivity. Further, mature capsid formation presumably needs to be coordinated with RNP condensation in order to avoid steric hindrance in the confined intravirion environment. This appears to be achieved by controlled stepwise proteolysis of the Gag precursor to release NC for RNP condensation and CA for capsid formation. Defects in processing of the NC-p6 region of Gag lead to aberrations in capsid morphology (12, 13). Use of the condensed RNP as a nucleator for capsid formation would further facilitate coordination of these processes.
In order to investigate the role of the RNP for mature capsid formation in the viral context, we wanted to analyze the architecture of mature particles devoid of an NC-RNA complex. However, NC-RNA interactions are an important determinant of immature retrovirus assembly; consequently, mutations or deletions within NC that impair RNA binding severely affect the efficiency of HIV-1 particle formation. To circumvent this problem, we made use of the finding that the function of NC-RNA contacts in immature HIV-1 assembly can be replaced by that of a heterologous protein-protein interaction domain. Others have previously reported that replacement of retroviral NC by a leucine zipper (LZ) domain in HIV-1 [HIV-1(LZ)] results in the efficient formation of immature particles (14–18). Since these studies were mainly performed using constructs expressing the modified Gag alone, PR-mediated Gag processing and morphological maturation could not be observed. Here, we have developed a strategy for replacing NC and the adjacent spacer peptide 2 (SP2) by LZ in the full HIV-1 context and used this construct to investigate the role of the NC-RNA complex in mature HIV-1 assembly. Our findings indicate that the RNP is not required for mature HIV-1 capsid formation.
MATERIALS AND METHODS
Cell lines and plasmids.
HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal calf serum (FCS; Biochrom), penicillin (100 IU/ml), streptomycin (100 μg/ml), 4 mM glutamine, and 10 mM HEPES (pH 7.4). Subviral constructs were based on the non-replication-competent pNL4-3 derivative pCHIV (19). A variant of pCHIV in which the gag and pol open reading frames (ORFs) were genetically uncoupled (pCHIVunc) (20) has been previously described. The region encoding NC-SP2, from codon 5 of NC to codon 12 of SP2, was replaced by introducing a unique BstEII restriction site into the respective region of proviral plasmid pNL4-3unc, followed by insertion of a synthetic BstEII/XmaI DNA fragment (Geneart, Regensburg, Germany) encoding an LZ domain from the Saccharomyces cerevisiae GCN4 protein, flanked by flexible glycine-rich linker regions (see Fig. 1A). The modified region was excised and cloned into pCHIV using SwaI restriction sites to yield plasmid pCHIVunc(LZ).
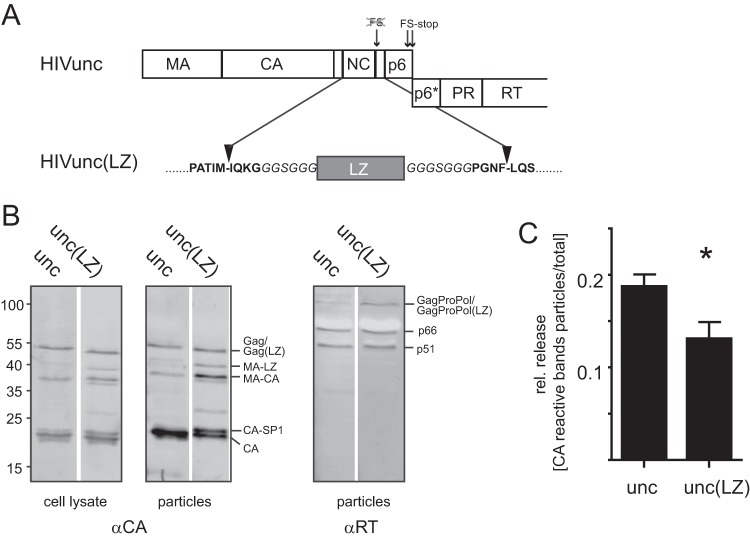
FIG 1.
Characterization of the Gag(LZ)-carrying HIV-1 derivative. (A) Virus derivatives used in this study. The scheme displays the gag coding region of plasmids pCHIVunc and pCHIVunc(LZ). HIV-derived sequences are shown in bold letters, and linker sequences are displayed in italics. See the main text for details. (B) Gag expression and processing. HEK293T cells were transfected with pCHIVunc and pCHIVunc(LZ). At 44 h posttransfection, cell lysates and supernatants were harvested. VLPs were concentrated from the supernatant by ultracentrifugation through a 20% sucrose cushion. Cell lysates and particles were separated by SDS-PAGE and analyzed by quantitative immunoblotting (LI-COR) using antiserum raised against recombinant HIV-1 CA or RT. The positions of molecular mass standards (in kilodaltons) are indicated to the left, and the positions of viral proteins are shown to the right. (C) Relative (rel.) particle release. HEK293T cells were transfected with pCHIVunc or pCHIVunc(LZ). Cell lysates and VLPs were analyzed by immunoblotting, as described in the legend to panel B, and the band intensities of the CA-reactive bands in the respective samples were quantitated using Image Studio Lite software. Relative release efficiencies were determined as the ratio between the total intensity of CA-reactive bands in the particle fraction to the sum of intensities of all anti-CA-reactive bands in cell lysates and particles. The graph shows mean values and SDs from triplicate experiments; significance was calculated using an unpaired Student's t test.
Virus-like particle preparation and characterization.
HEK293T cells were transfected with the plasmids indicated below using polyethyleneimine (PEI; Sigma-Aldrich) following a standard procedure (22). Briefly, at 24 h prior to transfection, 3.5 × 106 cells in 6 ml complete DMEM were seeded per 10-cm petri dish. For transfection, 30 μl of PEI stock (1 mg/ml in double-distilled H2O) was mixed with 10 μg of the plasmid DNA, indicated below, diluted in 1 ml serum-free DMEM. The transfection mixture was incubated for 30 min at room temperature and then added slowly to the petri dish. For the preparation of immature particles, 2 μM the specific HIV-1 PR inhibitor lopinavir (LPV) was added at the time of transfection.
For preparations intended for later RNA purification, medium was removed at 18 h posttransfection, and cells were washed twice with phosphate-buffered saline (PBS) before addition of fresh complete DMEM. At 44 h posttransfection, tissue culture supernatants were harvested and clarified by filtration through 0.45-μm-pore-size nitrocellulose filters. Particles were concentrated from the supernatant by ultracentrifugation through a 20% (wt/wt) sucrose cushion. Particle pellets were resuspended in PBS supplemented with 10 mM HEPES (pH 7.4) and further purified by ultracentrifugation through a preformed iodixanol (OptiPrep; Axis-Shield) density gradient as described by Dettenhofer and Yu (23). The refractive indices of individual fractions were measured and used to calculate the respective densities using the data provided by the reagent's manufacturer (Axis-Shield). Gradient fractions were analyzed by immunoblotting and RNA quantification. Particle-associated RT activity was determined by a Sybr green I PCR-enhanced reverse transcription assay (SG-PERT) (24). Data were analyzed using GraphPad Prism software. For structural analyses, particle-containing fractions from the iodixanol gradient were pooled, and particles were pelleted by ultracentrifugation and resuspended in PBS–10 mM HEPES (pH 7.4). Samples were subjected to mild fixation with 1% paraformaldehyde (PFA; 1 h at room temperature) and stored in aliquots at −80°C.
Immunoblot analyses.
Samples were separated by SDS-PAGE (17.5%; acrylamide-bisacrylamide, 200:1) and transferred to nitrocellulose membranes by semidry blotting. Proteins were detected by polyclonal antisera raised against purified recombinant HIV-1 CA (sheep) or RT (rabbit). Purified recombinant HIV-1 CA (kindly provided by Vanda Lux) was analyzed in parallel as a standard for quantification. Secondary antibodies coupled to Alexa fluorescent dyes were used for detection with an Odyssey infrared imaging system as specified by the manufacturer (LI-COR Biosciences, Lincoln, NE). Band intensities were quantitated using Image Studio Lite software (LI-COR).
RNA quantification.
On the basis of the results from the immunoblot analysis and RT activity measurements of iodixanol gradient fractions, five fractions covering the peak of virus particles were selected from each gradient. Particles from these fractions were pelleted by ultracentrifugation and resuspended in 30 μl of PBS. Samples were divided into aliquots, which were subjected to (i) immunoblotting using antiserum raised against CA and (ii) RNA extraction using an RNeasy Mini Kit (Qiagen). For RNA extraction, the samples were processed according to the manufacturer's instructions; on-column DNase treatment was extended to 30 min.
Total RNA in purified samples was quantified using a Quant-iT RiboGreen RNA assay kit (Invitrogen) according to the manufacturer's instructions. To ensure that values in the linear range of the assay were obtained for all samples, we analyzed two different dilutions (1:1 and 1:10) and used both the high-range (20 ng/ml to 1 μg/ml) and low-range (1 ng/ml to 50 ng/ml) setup provided in the kit. Sample fluorescence was measured using a Tecan Infinite M200Pro fluorescence microplate reader (excitation, 485 nm; emission, 535 nm) and i-control software (Tecan). Values within the linear range of the measurement were used to calculate the total RNA content of the samples.
Determination of HIV-1-specific RNA (using 1:1,000-diluted samples) was performed using an Abbott real-time HIV-1 amplification reagent kit, together with controls prepared using a sample preparation kit (Abbott) and an Abbott m2000sp instrument. Quantitative RT-PCR (qRT-PCR) was performed and the results were analyzed using an Abbott m2000rt instrument and software provided by the manufacturer.
Cryo-electron tomography.
For cryo-electron tomography, samples were mixed with a solution of 10-nm gold beads and vitrified by plunge freezing in liquid ethane. Images were collected on either an FEI Titan Krios or an FEI Tecnai F30 Polara electron microscope, both of which were equipped with a Gatan GIF 2002 postcolumn energy filter and a 2-k by 2-k MultiScan charge-coupled-device (CCD) camera, both operated at 200 keV. Two-dimensional (2D) images were acquired using the SerialEM software package (25) with a total dose of 15 electrons/Å2 at a defocus of −4 or −7 μm. Tomographic tilt series were collected using FEI tomography software (version 4) over an angular range of ±60° with a 3° step at a defocus of −6 μm using a total dose of ∼50 electrons/Å2 for morphological analysis and over an angular range of −45° to +60° with a 3° step at a range of nominal defoci of between −1.5 and −4.5 μm for structural analysis. The nominal magnification for 2D imaging and tomography was ×42,000 (pixel size, 2.02 Å) for the Titan Krios electron microscope and ×34,000 (pixel size, 4.0 Å) for the Polara electron microscope. Tomograms were reconstructed using the IMOD software package (26).
To determine the structure of the immature Gag lattice of HIV-1(LZ) in which the gag and pol open reading frames were genetically uncoupled [HIVunc(LZ)], the set of collected tomograms for structural analysis was split into two halves, each of which contained roughly the same number of virus particles and had a similar distribution of nominal defoci. The image processing of each half data set was carried out in a completely independent manner as described before (27). The two final structures were aligned and compared by using the Fourier shell correlation (FSC) with an estimated resolution of 15.9 at the 0.5 criterion. The two independent references were then averaged into a final reconstruction that was filtered to 16 Å and sharpened with an empirically determined negative B factor of −1,100 Å2.
Morphological analysis.
The 2D images were used to assess the maturation state of intact particles from two independent preparations (42 and 77 particles from the two preparations, respectively), while the three-dimensional (3D) data from tomography were used to describe the core morphology (18 and 36 particles containing 26 and 45 cores, respectively, from the two preparations). Where necessary, to improve efficiency, low-magnification imaging was used to screen the electron microscope grid and identify and select intact particles for tomography.
Each intact particle in the tomographic data sets was classified on the basis of the number of cores, the morphology of the core structure, and whether the core formed a closed shell or was partially open. Morphology was classified as wt (the typical conical or tubular cores found in the wt viruses), mildly aberrant (cores with a slightly deformed structure with some similarity to the conical or tubular ones), or severely aberrant (cores with a spherical or highly disordered core morphology). Core morphology data for the wt preparation were derived from the data set collected for and described by Mattei et al. (28).
RESULTS AND DISCUSSION
Particle assembly and Gag processing.
In order to investigate the role of the NC domain and the viral RNP in HIV-1 maturation, we replaced NC-SP2 with an LZ region in the viral context. Mutagenesis within the C-terminal part of HIV-1 Gag is hampered by the fact that gag and pol open reading frames (ORFs) overlap in the viral genome. Thus, mutations in the NC-SP2-p6 region not only alter the Gag protein but also can affect the Gag-Pro-Pol frameshift efficiency and/or the amino acid sequence of the overlapping region in Pol. Accordingly, replacement of NC by LZ in the viral context resulted in particles containing almost exclusively immature Gag due to inhibition of the Gag-Pro-Pol frameshift (29). In order to allow for the exchange of Gag sequences in the region of the frameshift signal, we employed a virus derivative in which the two ORFs were genetically uncoupled (Fig. 1A). As originally described by Leiherer et al. (30), mutation of the original FS and insertion of a new FS directly upstream of the gag stop codon result in uncoupling of the gag and pol coding regions. This modification impairs neither particle assembly and release nor virus infectivity in tissue culture (20, 30). Accordingly, plasmid pCHIVunc was generated by introducing the respective modifications into the subviral plasmid pCHIV (19), which expresses all proteins of HIV-1 NL4-3 except Nef but is replication deficient due to the lack of long terminal repeat regions. For the generation of pCHIVunc(LZ), most of the NC-SP2 coding region was deleted and replaced by the coding region for a 32-amino-acid long LZ domain from the Saccharomyces cerevisiae yeast GCN4 protein. To allow proteolytic processing of the resulting Gag molecule at the correct positions by the viral PR, codons for amino acids that are part of the N- and C-terminal PR recognition site (i.e., codons 1 to 4 of NC and codons 13 to 16 of SP2) were retained. The resulting construct where the LZ domain was directly inserted into the NC-SP2 coding region exhibited an aberrant Gag processing pattern (data not shown). We therefore introduced flexible glycine-rich linker regions upstream and downstream of the LZ-coding region, yielding plasmid pCHIVunc(LZ) (Fig. 1A).
Immunoblot analysis of cell lysates and virus-like particle (VLP) preparations from HEK293T cells transfected with the respective constructs (Fig. 1B) demonstrated that the modified Gag protein was efficiently expressed and particles were released into the supernatant. The parental construct pCHIVunc showed a pattern of proteolysis products typical of wt HIV-1. Gag with the LZ domain [Gag(LZ)] was also proteolytically processed to the expected products, but comparison with pCHIVunc revealed a slight defect in processing efficiency. Whereas the Gag proteolysis pattern in cell lysates appeared to be similar for both virus derivatives (Fig. 1B, left), immunoblot analysis of HIVunc(LZ) virions revealed a larger amount of CA-containing intermediate products than the control particles did. In particular, we observed larger amounts of an intermediate in which the PR recognition sites between MA and LZ remained uncleaved as well as larger amounts of the late processing intermediate CA-SP1 (Fig. 1B, middle). The mature p66/p51 heterodimer form of the pol-encoded protein reverse transcriptase (RT) was also detected in particle preparations (Fig. 1B, right). However, we noted that the amounts of RT associated with pelletable HIVunc(LZ) particles varied between experiments, ranging from similar levels, as shown here, to significantly reduced levels compared to the amounts of RT associated with HIVunc in some other experiments. A reduced incorporation of wt Gag-Pro-Pol (not carrying an LZ domain) into Gag(LZ) VLPs has been reported earlier (17); it appears that some reduction also occurred in our case, where both Gag and Gag-Pro-Pol carried an LZ domain. Partial impairment of Gag-Pro-Pol recruitment into the virion may explain the reduced Gag processing efficiency observed for HIVunc(LZ).
The relative release of particles was slightly decreased for the LZ-carrying virus (Fig. 1C). This minor defect could be due to the fact that the LZ dimerization domain is also part of the Gag-Pro-Pol polyprotein in the viral context. Gag-Pro-Pol dimer formation is a prerequisite for activation of PR, and enhancement of dimerization can lead to premature intracellular Gag cleavage and decreased virion formation. Accordingly, enhancement of Gag-Pro-Pol dimer formation in the cytoplasm by strong overexpression (31), by addition of nonnucleosidic RT inhibitors that stabilize dimerization of the RT domain (32), or by fusion of an LZ domain C terminal to PR (33) all severely impaired virion formation. In the last case, PR-LZ was overexpressed as a part of Gag by an engineered shift in the Gag reading frame and deletion of downstream pol regions, and no particle release was observed. Here we observed only a subtle decrease of particle formation for HIVunc(LZ), suggesting at most a modest effect of LZ on Gag-Pro-Pol dimer formation in our experimental system.
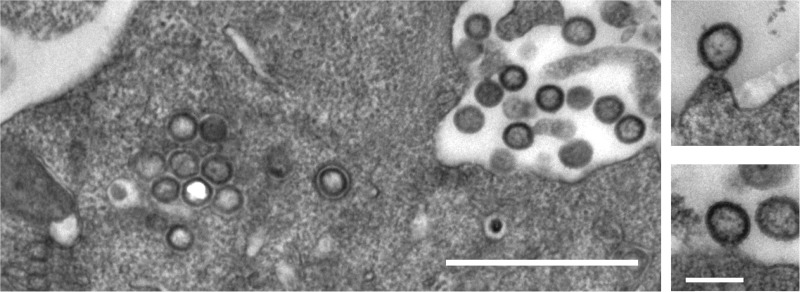
Thin-section electron micrographs of HEK293T cells transfected with pCHIVunc(LZ) revealed numerous particles in the vicinity of the plasma membrane, most of which displayed an immature morphology (Fig. 2). Similar to published data obtained for cells expressing Gag(LZ) alone (15, 29), budding sites detected at the membrane often showed a lollipop-like structure, with an almost complete Gag shell being connected to the plasma membrane by a thin stalk. This phenotype is reminiscent of that of viruses with mutations in the late domain (34), preventing recruitment of the cellular endosomal sorting complex required for transport (ESCRT), which would otherwise mediate virus abscission before formation of a complete Gag shell. It should be noted that late-domain mutants show reduced processing of the CA-SP1 cleavage site, as was also observed for HIVunc(LZ). In some cells, nonenveloped, spherical electron-dense structures with a morphology resembling that of the immature Gag shell were also detected in the cytoplasm (Fig. 2, left). The formation of intracellular spheres has previously been reported for Gag(LZ) variants carrying a mutation of the myristoylation site required for plasma membrane targeting (15, 29). We hypothesize that increased cytoplasmic assembly, a late-domain-like bud phenotype, and reduced proteolytic processing are related phenotypes reflecting an altered kinetic balance between the linked processes of virus assembly, abscission, and PR activation. In the case of HIVunc(LZ), they would all be due to an enhanced assembly propensity of the Gag polyprotein with the LZ substitution and provide an explanation for the slight decrease in particle release observed.
FIG 2.
Thin-section electron micrographs of HIVunc(LZ)-expressing cells. HEK293T cells were transfected with plasmid pCHIVunc(LZ), harvested at 36 h posttransfection, and fixed with 2.5% glutaraldehyde. Postfixation, embedment, and preparation for thin-section electron microscopy were performed as described previously (45). Bars, 1 μm (left) and 200 nm (right).
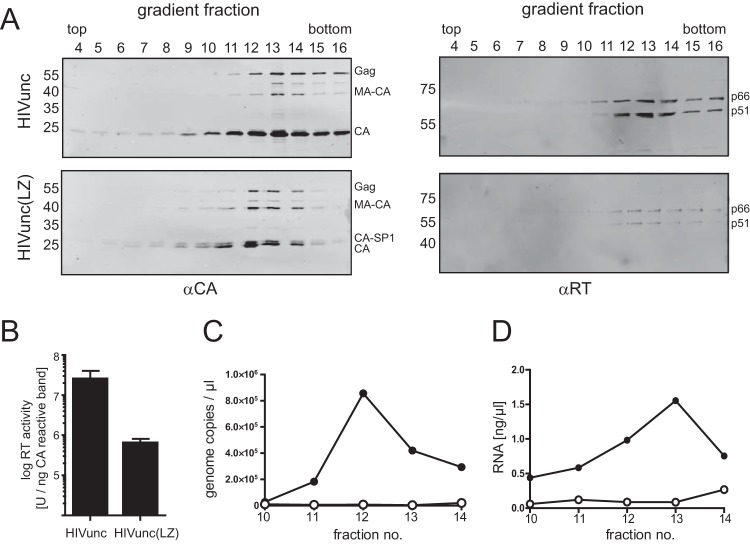
For biochemical characterization, particles were further purified by ultracentrifugation through an iodixanol density gradient. Immunoblot analysis using antiserum raised against CA (Fig. 3A, left) indicated the presence of VLPs in high-density fractions. Similar to what was observed previously for Gag(LZ) particles, HIVunc(LZ) virions displayed a slightly lower density than the corresponding control particles. As is routinely observed in HIV-1 purifications, the peak of CA for the control particles was detected at the transition to the high-density iodixanol cushion (peak fraction 13; refractive index, 1.362). For HIVunc(LZ) particles, the peak was shifted by one fraction toward the top of the gradient (refractive index, 1.359). Although the iodixanol density gradient commonly used for the purification of HIV virions does not separate to complete equilibrium and therefore does not allow the determination of absolute particle densities, the observed differences in sedimentation behavior are consistent with a lower particle density due to the expected lack of RNA in HIVunc(LZ) particles.
FIG 3.
Gradient purification of virus samples. VLPs were concentrated from the supernatant of HEK293T cells transfected with the indicated plasmids by ultracentrifugation through a 20% sucrose cushion and further purified by ultracentrifugation through an iodixanol density gradient. Fractions were collected from the top of the gradients and analyzed by immunoblotting (A), RT activity measurement (B), and RNA quantification (C, D). (A) Samples were separated by SDS-PAGE and transferred to a nitrocellulose membrane. Viral proteins were detected by quantitative immunoblotting (LI-COR) using the indicated antisera. The positions of molecular mass standards (in kilodaltons) are marked at the left, and the positions of viral proteins are shown on the right. (B) The RT activity of gradient fractions was determined using a Sybr green product-enhanced RT assay (24). The values determined for peak fractions [fractions 13 and 12 for HIVunc and HIVunc(LZ), respectively] were normalized to the amount of anti-CA-reactive proteins detected in the respective fractions. The graph shows mean values and SDs from three independent experiments. (C, D) RNA content of peak fractions. RNA was extracted from gradient fractions 10 to 14 as described in Materials and Methods. Samples were analyzed for HIV-1-specific RNA by quantitative RT-PCR (C) or for total RNA content by staining with RiboGreen dye (D). Closed circles, HIVunc; open circles, HIVunc(LZ). Panels A, C, and D show data from one representative experiment out of three independent experiments.
The detection of significant proportions of MA-CA and CA-SP1 in gradient-purified HIVunc(LZ) confirmed the partial processing defect inferred from the sucrose-pelleted preparations (compare Fig. 1B). The mature RT heterodimer (p66/p51) was detected in the same fractions as CA (Fig. 3A, right). Again, the relative RT amounts in gradient-purified HIVunc(LZ) particles varied between preparations but were always decreased compared to those in the control virus particles, in conjunction with notably reduced particle-associated RT activities (Fig. 3B).
The NC-packaging signal interaction is crucial for retroviral genome incorporation. To verify the genomic RNA packaging defect of Gag(LZ)-carrying virus, we tested for the incorporation of virus-specific RNA (Fig. 3C). Particles from iodixanol gradient fractions comprising the CA peak (fractions 10 to 14) were pelleted by ultracentrifugation, treated with DNase, and subjected to RNA extraction. Purified RNA samples were subjected to quantitative RT-PCR to detect HIV-1 genomic RNA (Fig. 3C). A peak of viral RNA associated with control particles was absent in the case of HIVunc(LZ) particles, indicating that they were deficient in viral RNA incorporation.
Retroviruses were found to incorporate large amounts of nonviral RNA in the absence of a packageable genome (35, 36). Furthermore, the MA protein of HIV-1 has been reported to bind to RNA (reviewed in reference 37), and transient binding of host tRNA to MA may be important for regulation of Gag membrane targeting (38). We therefore also analyzed the total RNA content in the same samples by staining with a nucleic acid binding fluorescent dye (Fig. 3D). This analysis revealed a clear peak of nucleic acid associated with the CA peak fractions of control particles. In contrast, a particle-associated RNA peak was absent in HIVunc(LZ) preparations, where only a low background was found in all gradient fractions analyzed. We conclude that HIVunc(LZ) particles are deficient in RNA incorporation.
Particle structure and morphology.
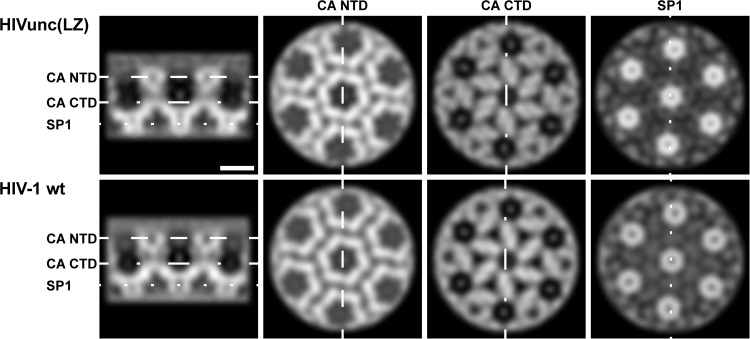
In order to determine whether the Gag(LZ) protein packs normally during immature particle assembly, we produced HIVunc(LZ) particles in the presence of the HIV-1 protease inhibitor lopinavir (LPV), performed a density gradient purification, and imaged the particles by cryo-electron tomography. By applying subtomogram averaging to the purified immature HIVunc(LZ) particles, we determined a low-resolution structure of the CA lattice within these particles and compared it to that determined for immature HIV-1 (EMD-2706 [27]) (Fig. 4). Though we cannot rule out the possibility of small structural changes, we observed no substantial structural differences between the two structures, indicating that replacing NC-SP2 with LZ does not dramatically influence the packing arrangement of CA domains during Gag assembly. Only a blurred, disordered density was observed for the LZ region, suggesting that, as is the case for NC, its arrangement or symmetry does not follow that of the CA layer.
FIG 4.
Comparison of HIVunc(LZ) and HIV-1 wt immature Gag lattice. Orthoslices through the CA-SP1 region of the subtomogram averaging reconstructions of the wild-type and HIVunc(LZ) immature Gag lattices are shown. Both structures were low-pass filtered at a resolution of 16 Å. The dashed lines in the left panel show the positions of the perpendicular orthoslices through the CA N-terminal domains (NTD), CA C-terminal domains (CTD), and SP1 domains. The density is white. Bar, 5 nm.
To analyze the structure of LZ-containing particles after proteolysis, particles were purified by density gradient centrifugation in the absence of the protease inhibitor, vitrified by plunge freezing, and imaged by cryo-electron microscopy. Approximately 50% (60 of 119) of the HIVunc(LZ) particles displayed a characteristically immature morphology (Fig. 5Axii), whereas only ∼1% (4 of 331) of the particles in a typical HIV wt preparation did (Table 1). The reduced level of morphological maturation may be related to the impaired Gag(LZ) cleavage (compare Fig. 1B and 3A), although the increased proportion of immature particles is more pronounced than might be expected from the observed processing defect. Relatively mild processing defects have, however, been previously observed to be associated with a significant reduction of infectivity and morphological aberrations (39, 40). We speculate that the observed defects are caused by reduced PR levels within the virion due to premature intracellular processing and/or a packaging defect of Gag(LZ)-Pro-Pol.
FIG 5.
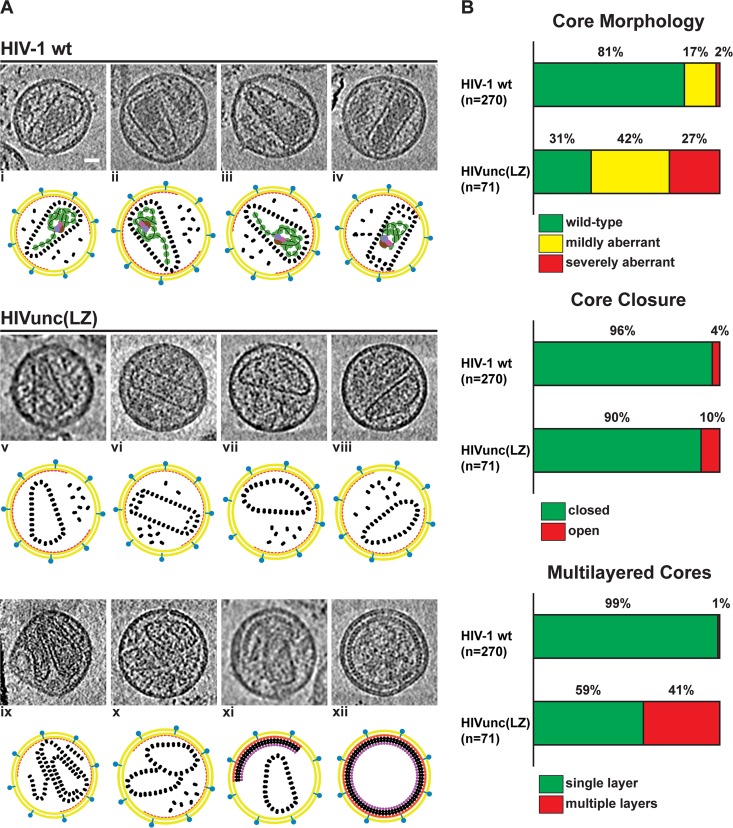
Morphology of HIVunc(LZ) virions analyzed by cryo-electron tomography. (A) Gallery of computational slices through tomographic reconstructions showing a comparison between HIV-1 wt and HIVunc(LZ) particles. (i to iv) HIV-1 wt particles presenting typical conical and tubular cores encasing the condensed RNP density, as described in reference 27. (v to xi) HIVunc(LZ) mature particles showing the representative core morphologies: closed, single-layer, wt cores (v and vi); closed, single-layer, mildly aberrant cores (vii and viii); a closed, multilayer, wt core (ix); closed, single-layer multiple cores with a mildly and severely aberrant morphology (x); a closed, single-layer core in a particle also containing a partial immature lattice (xi). (xii) Immature HIVunc(LZ) particle. Bar, 20 nm. (B) Statistical analysis of the observed phenotypes.
TABLE 1.
Analysis of integrity and maturation state of the samplea
| Sample | Prepn no. | No. (%) of intact particles |
||
|---|---|---|---|---|
| Total | Mature | Immature | ||
| HIV-1 wt | 331 | 327 (99) | 4 (1) | |
| HIVunc(LZ) | 1 | 42 | 23 (55) | 19 (45) |
| HIVunc(LZ) | 2 | 77 | 36 (47) | 41 (53) |
Data are based on 2D analysis of all particles recorded from the indicated virus preparations.
The three-dimensional structure of the mature virus particles was characterized in detail by cryo-electron tomography. Fifty-four mature particles containing a total of 71 cores were analyzed. For comparison, we analyzed the wt preparation described in reference 28. The vast majority of mature HIVunc(LZ) particles contained closed CA assemblies (64 of 71 cores [90%], compared to 259 of 270 cores [96%] in a control sample; Fig. 5B). Furthermore, capsid structures displaying the wt conical or tubular morphology were formed (Fig. 5Av and vi and B). As expected from the absence of RNA and NC in such preparations, we observed no condensed RNP in any of the mature HIVunc(LZ) particles. These observations indicate that wt-like conical capsids can form in the absence of the RNP.
Although capsids with the wt morphology were observed in the HIVunc(LZ) particles, we observed a significantly broader distribution of capsid phenotypes in the HIVunc(LZ) particles than in the wt HIV-1 particles. In a typical HIV-1 wt preparation, approximately 19% of cores showed aberrations in morphology. In contrast, we found that 69% of cores (49 of 71) in the HIVunc(LZ) preparations were aberrant (Fig. 5Avii to viii and x and B). Furthermore, 41% (29 of 71) of HIVunc(LZ) cores displayed a multilayered wall (Fig. 5Aix and B), a feature that was rarely observed in wt particles (3 of 270 cores [∼1%]). A large proportion of mature HIVunc(LZ) particles (30%; 16 of 54) contained more than one capsid (14 contained two capsids, and 2 contained three capsids) (Fig. 5Ax). This feature was less represented in the wt sample, where 19 of 249 mature particles (8%) contained two cores. There was no evident correlation between core morphology and the presence of multiple cores. Interestingly, in 9 of the 54 (17%) analyzed HIVunc(LZ) particles, we observed both a partial immature lattice and a core structure inside the same particle (Fig. 5Axi). Since the number of CA molecules needed to form a mature capsid is smaller than the number of Gag molecules present in an immature HIV-1 particle (4, 41), this phenotype does not require that extra Gag molecules be incorporated during assembly.
This analysis shows that the HIVunc(LZ) particles display a high proportion of aberrant core structures. One explanation for this phenotype would be a direct role of the RNP in nucleating or forming the mature capsid. Mutations of the NC-SP2-p6 cleavage sites lead to defects in capsid architecture, despite the full proteolytic maturation of CA (12, 13), suggesting that RNP condensation influences capsid assembly. The simplest interpretation of the NC-SP2-p6 cleavage defect phenotype, however, is that defects in RNP condensation sterically hinder capsid assembly, and this effect would not be relevant in the absence of any RNP. If the RNP played an essential role in nucleating or forming the mature capsid, HIVunc(LZ) particles should show abolished capsid formation or exclusively aberrant capsid formation. This was, however, not observed.
An alternative possible cause of the higher proportion of aberrant capsids in the HIVunc(LZ) particles are defects in the degree and/or rate of Gag cleavage, due to impaired and variable Gag-Pro-Pol cleavage and incorporation into particles. As shown by us and others (42–44), immature Gag and intermediate processing products (particularly CA-SP1) exert a strong dominant negative effect on morphological maturation and virion infectivity. Furthermore, changes in the rate or degree of processing have been shown to cause severe defects in core morphology; in fact, the morphology defects associated with delayed processing kinetics (28) can be more pronounced than those observed here for the HIVunc(LZ) particles. Based on these previous observations, while we cannot exclude the possibility of a contribution from the absence of the RNP, we argue that the lower relative proportion of mature capsid structures in the case of HIVunc(LZ) could be fully explained by defective Gag processing in these particles.
Taken together, the results presented here show that core nucleation, closure, and the attainment of conical morphology can all be correctly achieved in the absence of RNA. We thus conclude that the presence of RNA and the condensation of the RNP are dispensable for assembly of a mature capsid structure within the HIV-1 virion.
ACKNOWLEDGMENTS
We sincerely thank Angelika Schillak for help with qRT-PCR analysis of viral RNA, Anja Habermann for thin-section electron microscopy, Wim Hagen for support with cryo-electron microscopy, and EMBL IT Services for high-performance computing.
This work was supported by grants from the Deutsche Forschungsgemeinschaft to B.M. (MU 885-5/1), J.A.G.B. (BR 3635/2-1), and H.-G.K. (KR 906/7-1). H.-G.K. and B.M. are investigators of the CellNetworks Cluster of Excellence (EXC81). Work in the J. A. G. Briggs lab is supported by the Chica und Heinz Schaller Stiftung.
REFERENCES
- 1.Sundquist WI, Krausslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harbor Perspect Med 2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briggs JA, Krausslich HG. 2011. The molecular architecture of HIV. J Mol Biol 410:491–500. doi: 10.1016/j.jmb.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 3.Ganser-Pornillos BK, Yeager M, Pornillos O. 2012. Assembly and architecture of HIV. Adv Exp Med Biol 726:441–465. doi: 10.1007/978-1-4614-0980-9_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lanman J, Lam TT, Emmett MR, Marshall AG, Sakalian M, Prevelige PE Jr. 2004. Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange. Nat Struct Mol Biol 11:676–677. doi: 10.1038/nsmb790. [DOI] [PubMed] [Google Scholar]
- 5.de Marco A, Muller B, Glass B, Riches JD, Krausslich HG, Briggs JA. 2010. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS Pathog 6:e1001215. doi: 10.1371/journal.ppat.1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keller PW, Huang RK, England MR, Waki K, Cheng N, Heymann JB, Craven RC, Freed EO, Steven AC. 2013. A two-pronged structural analysis of retroviral maturation indicates that core formation proceeds by a disassembly-reassembly pathway rather than a displacive transition. J Virol 87:13655–13664. doi: 10.1128/JVI.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. 2009. Structure and assembly of immature HIV. Proc Natl Acad Sci U S A 106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson LA, Briggs JA, Glass B, Riches JD, Simon MN, Johnson MC, Muller B, Grunewald K, Krausslich HG. 2008. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe 4:592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briggs JA, Wilk T, Welker R, Krausslich HG, Fuller SD. 2003. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J 22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs JA, Grunewald K, Glass B, Forster F, Krausslich HG, Fuller SD. 2006. The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure 14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 11.England MR, Purdy JG, Ropson IJ, Dalessio PM, Craven RC. 2014. Potential role for CA-SP in nucleating retroviral capsid maturation. J Virol 88:7170–7177. doi: 10.1128/JVI.00309-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coren LV, Thomas JA, Chertova E, Sowder RC II, Gagliardi TD, Gorelick RJ, Ott DE. 2007. Mutational analysis of the C-terminal Gag cleavage sites in human immunodeficiency virus type 1. J Virol 81:10047–10054. doi: 10.1128/JVI.02496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Marco A, Heuser AM, Glass B, Krausslich HG, Muller B, Briggs JA. 2012. Role of the SP2 domain and its proteolytic cleavage in HIV-1 structural maturation and infectivity. J Virol 86:13708–13716. doi: 10.1128/JVI.01704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Accola MA, Strack B, Gottlinger HG. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J Virol 74:5395–5402. doi: 10.1128/JVI.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crist RM, Datta SA, Stephen AG, Soheilian F, Mirro J, Fisher RJ, Nagashima K, Rein A. 2009. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J Virol 83:2216–2225. doi: 10.1128/JVI.02031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Qian H, Love Z, Barklis E. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J Virol 72:1782–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cen S, Niu M, Saadatmand J, Guo F, Huang Y, Nabel GJ, Kleiman L. 2004. Incorporation of Pol into human immunodeficiency virus type 1 Gag virus-like particles occurs independently of the upstream Gag domain in Gag-Pol. J Virol 78:1042–1049. doi: 10.1128/JVI.78.2.1042-1049.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson MC, Scobie HM, Ma YM, Vogt VM. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J Virol 76:11177–11185. doi: 10.1128/JVI.76.22.11177-11185.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lampe M, Briggs JA, Endress T, Glass B, Riegelsberger S, Krausslich HG, Lamb DC, Brauchle C, Muller B. 2007. Double-labelled HIV-1 particles for study of virus-cell interaction. Virology 360:92–104. doi: 10.1016/j.virol.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Radestock B, Morales I, Rahman SA, Radau S, Glass B, Zahedi RP, Muller B, Krausslich HG. 2013. Comprehensive mutational analysis reveals p6Gag phosphorylation to be dispensable for HIV-1 morphogenesis and replication. J Virol 87:724–734. doi: 10.1128/JVI.02162-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reference deleted.
- 22.Swiersy A, Wiek C, Reh J, Zentgraf H, Lindemann D. 2011. Orthoretroviral-like prototype foamy virus Gag-Pol expression is compatible with viral replication. Retrovirology 8:66. doi: 10.1186/1742-4690-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dettenhofer M, Yu XF. 1999. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J Virol 73:1460–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pizzato M, Erlwein O, Bonsall D, Kaye S, Muir D, McClure MO. 2009. A one-step SYBR Green I-based product-enhanced reverse transcriptase assay for the quantitation of retroviruses in cell culture supernatants. J Virol Methods 156:1–7. doi: 10.1016/j.jviromet.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 25.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 27.Schur FK, Hagen WJ, Rumlova M, Ruml T, Muller B, Krausslich HG, Briggs JA. 2015. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 Å resolution. Nature 517:505–508. doi: 10.1038/nature13838. [DOI] [PubMed] [Google Scholar]
- 28.Mattei S, Anders M, Konvalinka J, Krausslich HG, Briggs JA, Muller B. 2014. Induced maturation of human immunodeficiency virus. J Virol 88:13722–13731. doi: 10.1128/JVI.02271-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein KC, Reed JC, Tanaka M, Nguyen VT, Giri S, Lingappa JR. 2011. HIV Gag-leucine zipper chimeras form ABCE1-containing intermediates and RNase-resistant immature capsids similar to those formed by wild-type HIV-1 Gag. J Virol 85:7419–7435. doi: 10.1128/JVI.00288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leiherer A, Ludwig C, Wagner R. 2009. Uncoupling human immunodeficiency virus type 1 Gag and Pol reading frames: role of the transframe protein p6* in viral replication. J Virol 83:7210–7220. doi: 10.1128/JVI.02603-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park J, Morrow CD. 1991. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J Virol 65:5111–5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Figueiredo A, Moore KL, Mak J, Sluis-Cremer N, de Bethune MP, Tachedjian G. 2006. Potent nonnucleoside reverse transcriptase inhibitors target HIV-1 Gag-Pol. PLoS Pathog 2:e119. doi: 10.1371/journal.ppat.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan YY, Wang SM, Huang KJ, Chiang CC, Wang CT. 2012. Placement of leucine zipper motifs at the carboxyl terminus of HIV-1 protease significantly reduces virion production. PLoS One 7:e32845. doi: 10.1371/journal.pone.0032845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci U S A 88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muriaux D, Mirro J, Harvin D, Rein A. 2001. RNA is a structural element in retrovirus particles. Proc Natl Acad Sci U S A 98:5246–5251. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rulli SJ Jr, Hibbert CS, Mirro J, Pederson T, Biswal S, Rein A. 2007. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J Virol 81:6623–6631. doi: 10.1128/JVI.02833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alfadhli A, Barklis E. 2014. The roles of lipids and nucleic acids in HIV-1 assembly. Front Microbiol 5:253. doi: 10.3389/fmicb.2014.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kutluay SB, Zang T, Blanco-Melo D, Powell C, Jannain D, Errando M, Bieniasz PD. 2014. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 159:1096–1109. doi: 10.1016/j.cell.2014.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaplan AH, Zack JA, Knigge M, Paul DA, Kempf DJ, Norbeck DW, Swanstrom R. 1993. Partial inhibition of the human immunodeficiency virus type 1 protease results in aberrant virus assembly and the formation of noninfectious particles. J Virol 67:4050–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore MD, Fu W, Soheilian F, Nagashima K, Ptak RG, Pathak VK, Hu WS. 2008. Suboptimal inhibition of protease activity in human immunodeficiency virus type 1: effects on virion morphogenesis and RNA maturation. Virology 379:152–160. doi: 10.1016/j.virol.2008.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. 2004. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol 11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 42.Checkley MA, Luttge BG, Soheilian F, Nagashima K, Freed EO. 2010. The capsid-spacer peptide 1 Gag processing intermediate is a dominant-negative inhibitor of HIV-1 maturation. Virology 400:137–144. doi: 10.1016/j.virol.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SK, Harris J, Swanstrom R. 2009. A strongly transdominant mutation in the human immunodeficiency virus type 1 gag gene defines an Achilles heel in the virus life cycle. J Virol 83:8536–8543. doi: 10.1128/JVI.00317-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muller B, Anders M, Akiyama H, Welsch S, Glass B, Nikovics K, Clavel F, Tervo HM, Keppler OT, Krausslich HG. 2009. HIV-1 Gag processing intermediates trans-dominantly interfere with HIV-1 infectivity. J Biol Chem 284:29692–29703. doi: 10.1074/jbc.M109.027144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goffinet C, Allespach I, Homann S, Tervo HM, Habermann A, Rupp D, Oberbremer L, Kern C, Tibroni N, Welsch S, Krijnse-Locker J, Banting G, Krausslich HG, Fackler OT, Keppler OT. 2009. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe 5:285–297. doi: 10.1016/j.chom.2009.01.009. [DOI] [PubMed] [Google Scholar]