ABSTRACT
During virion maturation, HIV-1 capsid protein assembles into a conical core containing the viral ribonucleoprotein (vRNP) complex, thought to be composed mainly of the viral RNA and nucleocapsid protein (NC). After infection, the viral RNA is reverse transcribed into double-stranded DNA, which is then incorporated into host chromosomes by integrase (IN) catalysis. Certain IN mutations (class II) and antiviral drugs (allosteric IN inhibitors [ALLINIs]) adversely affect maturation, resulting in virions that contain “eccentric condensates,” electron-dense aggregates located outside seemingly empty capsids. Here we demonstrate that in addition to this mislocalization of electron density, a class II IN mutation and ALLINIs each increase the fraction of virions with malformed capsids (from ∼12% to ∼53%). Eccentric condensates have a high NC content, as demonstrated by “tomo-bubblegram” imaging, a novel labeling technique that exploits the susceptibility of NC to radiation damage. Tomo-bubblegrams also localized NC inside wild-type cores and lining the spherical Gag shell in immature virions. We conclude that eccentric condensates represent nonpackaged vRNPs and that either genetic or pharmacological inhibition of IN can impair vRNP incorporation into mature cores. Supplying IN in trans as part of a Vpr-IN fusion protein partially restored the formation of conical cores with internal electron density and the infectivity of a class II IN deletion mutant virus. Moreover, the ability of ALLINIs to induce eccentric condensate formation required both IN and viral RNA. Based on these observations, we propose a role for IN in initiating core morphogenesis and vRNP incorporation into the mature core during HIV-1 maturation.
IMPORTANCE Maturation, a process essential for HIV-1 infectivity, involves core assembly, whereby the viral ribonucleoprotein (vRNP, composed of vRNA and nucleocapsid protein [NC]) is packaged into a conical capsid. Allosteric integrase inhibitors (ALLINIs) affect multiple viral processes. We have characterized ALLINIs and integrase mutants that have the same phenotype. First, by comparing the effects of ALLINIs on several steps of the viral cycle, we show that inhibition of maturation accounts for compound potency. Second, by using cryoelectron tomography, we find that ALLINIs impair conical capsid assembly. Third, by developing tomo-bubblegram imaging, which specifically labels NC protein, we find that ALLINIs block vRNP packaging; instead, vRNPs form “eccentric condensates” outside the core. Fourth, malformed cores, typical of integrase-deleted virus, are partially replaced by conical cores when integrase is supplied in trans. Fifth, vRNA is necessary for ALLINI-induced eccentric condensate formation. These observations suggest that integrase is involved in capsid morphogenesis and vRNP packaging.
INTRODUCTION
The formation of infectious HIV-1 particles starts with the coassembly of the Gag and Gag-Pol polyproteins into a spherical shell, which buds from the cell to produce an immature virion (1). Following budding, the viral protease (PR) is activated and cleaves Gag and Gag-Pol into their component domains, of which the capsid protein CA assembles into a conical capsid shell containing the viral RNA (vRNA) genome, nucleocapsid protein NC, and replication enzymes (we use the term “capsid” to denote the assembled protein shell and “core” for the capsid plus whatever it may contain). This transformation is known as HIV-1 maturation. Drawing mainly on studies by cryoelectron tomography (cryo-ET), a technique that allows three-dimensional (3D) imaging of individual pleomorphic particles with good preservation of native structure (e.g., see reference 2), a number of different models explaining how CA can assemble into conical cores have been proposed (3–6). However, maturation—the process targeted not only by protease inhibitors (PIs) (7) but also by “maturation inhibitors” that act by denying PR access to a key cleavage site (8, 9)—is incompletely understood.
Mutational studies of the viral enzyme integrase (IN) have demonstrated that the effects of some amino acid substitutions, the so-called class II IN mutations, include defects in particle assembly and reverse transcription (compared to class I IN mutations, which affect only integration) (reviewed in reference 10). Class II IN mutant viruses provided an initial clue for a role of IN in the late stages of replication, including maturation. More recently, a group of investigational compounds, 2-(quinolin-3-yl) acetic acid derivatives, which we refer to as allosteric integrase inhibitors (ALLINIs) (11) but which are also called NCINIs for noncatalytic IN inhibitors (12), LEDGINs for lens epithelium-derived growth factor (LEDGF)/p75-IN inhibitors (13), INLAIs for IN-LEDGF/p75 allosteric inhibitors (14), and MINIs for multimeric IN inhibitors (15), were found to inhibit HIV-1 infection (13, 16). ALLINIs engage the interface that is formed by two molecules of the IN catalytic core domain at the binding pocket for the cellular integration cofactor LEDGF/p75 (11, 13–15, 17–19), which itself guides viral DNA integration to active genes (reviewed in reference 20). Inhibition of the LEDGF/p75-IN interaction was initially thought to underlie the mode of ALLINI action (13). However, the compounds were since discovered to affect the late stage of HIV-1 replication in a manner that is independent of LEDGF/p75 expression (12, 15, 18, 21, 22). The compounds are also active during the early stage of HIV-1 replication, but much higher concentrations are required to achieve potencies similar to those when virus producer cells are treated (12, 14, 15, 17, 18, 21, 22). Akin to several class II IN mutant viruses, particles produced in the presence of ALLINIs are reportedly defective for multiple steps of HIV-1 replication, namely, maturation (12, 15, 18, 21), reverse transcription (12, 15, 18, 21), nuclear import of the viral DNA (21), and integration (12–15, 18, 19, 21). The ability of ALLINIs to induce higher-order IN oligomerization during virus production underlies their antiviral activity (11, 12, 14, 15, 17–19, 21, 23, 24).
Transmission electron microscopy of thin sections of plastic embeddings (referred to here as TEM) of HIV-1 particles generated in the presence of ALLINIs revealed an apparent uncoupling of the incorporation of the viral ribonucleoprotein (vRNP) complex, assumed to be composed primarily of vRNA and NC, from assembly of the capsid (12, 15, 18, 21). Instead of a normal electron-dense core, a relatively electron-translucent core was observed, accompanied by a roughly spherical electron-dense feature situated between the viral envelope. We refer to this feature as an “eccentric condensate.” It follows that ALLINIs can be useful probes to help understand how IN contributes to maturation, in a similar way that maturation inhibitors (which prevent the scission of the CA-SP1 junction within Gag, the final proteolytic cleavage that leads to the generation of a mature core [25, 26]), or mutations that prevent the cleavage of the Pol polyprotein (27, 28), have provided structural insights.
In this study, we first analyzed the relationships between core formation, reverse transcription, and infectivity of ALLINI-treated particles and conclude from dose-response curves that inhibition of particle maturation is the process that underlies the potencies of two representative ALLINI compounds. Using cryo-ET, we found that, in addition to affecting vRNP incorporation into the mature core, ALLINIs reduced by a factor of approximately five the efficiency with which conical cores are formed. To demonstrate that the eccentric condensate is indeed composed of NC and RNA, we applied tomo-bubblegram imaging, a novel technique that takes advantage of the acute sensitivity of NC protein to electron irradiation that is expressed in the generation of bubbles as radiation damage products. The same approach confirmed that NC, and by inference the vRNP, is present inside wild-type cores, predominantly at the wide end, and that NC lines the inner part of the thick-walled Gag shell in immature virions. Using Vpr-IN to functionally transcomplement particles that lack IN (ΔIN), we also show that IN promotes conical core formation and HIV-1 infectivity in parallel. Furthermore, we implicate vRNA as a necessary component for ALLINI action during virus production. Together, these results imply that IN has a direct role in nucleating the assembly of conical cores and the incorporation of the vRNP into the mature core and that ALLINI treatment effectively subverts this role(s) of IN during HIV-1 particle maturation.
MATERIALS AND METHODS
Plasmids and antivirals.
HIV-1NL4-3 carrying wild-type (WT) IN was expressed from pNL43/Xmal (29), whereas the V165A IN mutant was expressed from pNL43/XmaI.V165A (30). Single-round derivatives HIV-Luc and HIV-Luc.ΔIN were expressed from pNLX.Luc.R− (31) and pNLX.Luc.R−.ΔIN (18), respectively. The D25A mutation in PR was introduced into pNL43/XmaI using PCR-directed mutagenesis. Gag/ψ-minus virus-like particles (VLPs) were expressed from pRR359 (32) (also known as pCMV55M1-10 [32, 33]) whereas Gag-LeuZip was expressed from pRR546 (32). The HIV-1-based pHP-dI-N/A packaging plasmid (34) was cotransfected with pHI-Luc transfer vector (35) or pGL3-Basic control plasmid (Promega). Vpr fused to the IN protein from HIV-1SG3 (WT or H171T mutant) was expressed from pRL2PVpr-IN (18, 36), whereas the vesicular stomatitis virus G (VSV-G) glycoprotein was expressed using pCG-VSV-G (31).
ALLINIs BI-D and BIB-2 were synthesized as described previously (23, 37). Efavirenz (EFV) was obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (NIH ARRRP).
Cells, viruses, and antiviral assays.
Transformed human embryonic kidney 293T (HEK293T) cells (American Type Tissue Collection item number CRL-3216) were grown in Dulbecco's modified Eagle medium, while human SupT1 T cells (NIH ARRRP item number 100) were maintained in RPMI 1640 medium, both supplemented to contain 10% (vol/vol) fetal bovine serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin. HIV-Luc was produced by cotransfecting HEK293T cells (8 × 105) with 3.5 μg pNLX.Luc.R− and 0.35 μg pCG-VSV-G using PolyJet transfection reagent (SignaGen Laboratories). HIV-1NLX.Luc.R−.ΔIN was transcomplemented with Vpr-IN by cotransfecting HEK293T cells (3 × 106) with 4.5 μg pNLX.Luc.R−.ΔIN, 1.25 μg pRL2P-Vpr-IN containing WT or H171T mutant IN, and 0.5 μg pCG-VSV-G. Dimethyl sulfoxide (DMSO) or ALLINIs were added with fresh medium at ∼17 to 20 h posttransfection, and cell supernatants were collected 24 h later. The cell-free virus concentration, typically 500 to 1,000 ng/ml, was determined using a commercial p24 ELISA kit (Advanced Biosciences Laboratories). For Vpr-IN complementation experiments, SupT1 cells were infected in quadruplicate with 5 ng/ml p24. Luciferase values, expressed in relative light units, were determined 48 h postinfection.
DNA analysis and quantitative PCR.
SupT1 cells (2 × 106) infected with DNase-treated (1 h at 37°C) HIV-Luc (50 ng p24/ml) for 2 h were washed and split into two samples. DNA was isolated from one set of samples 5 h later by use of a DNeasy blood and tissue kit (Qiagen). The other set of samples was seeded in quadruplicate in 96-well plates, and luciferase activity was determined 48 h from the start of infection.
Duplicate PCR mixtures containing 50 ng DNA, 0.2 μM primers (AE2963/AE4422), 0.1 μM probe (AE2965), and 1× QuantiText Probe PCR master mix were incubated at 50°C for 2 min and 95°C for 15 min, followed by 40 cycles of 94°C for 15 s, 59°C for 30 s, and 72°C for 30 s (38). Standards were prepared by endpoint dilution of pNLX.Luc.R− into DNA from uninfected cells. To account for potential residual plasmid carryover from transfection, parallel infections were performed in the presence of 10 μM EFV, and the values obtained from these PCR assays were subtracted from the values from the experimental samples.
RNA isolation and analysis.
Total RNA was extracted from concentrated virus preparations after a 30-min incubation with 0.05 μg/ml proteinase K (Life Technologies) and 0.4% sodium dodecyl sulfate at 56°C. Following phenol-chloroform extraction, RNA was precipitated using isopropanol and GenElute-LPA (Sigma-Aldrich) as the carrier. The RNA concentration was determined by the Ribogreen (Invitrogen) assay, which was completed according to the manufacturer's instructions. PCRs to determine vRNA copy number per nanogram of total RNA were completed as previously described (18), utilizing 2 μl of purified RNA and primer sets AE2963/AE2964 for Fig. 9 samples (excluding HIV-Luc) and AE2961/AE4422 for Fig. 6 samples and HIV-Luc.
FIG 9.
Viral RNA requirement for eccentric condensate formation. (A) Total RNA (in nanograms) of concentrated virion preparations. RNA concentration was standardized by the p24 signal obtained from Western blots of multiple input volumes. Shown are averages (±SD) from four independent experiments reported as percentages of untreated HIV-Luc virions. (B) Average (±SD) copies of vRNA per ng total RNA (n = 4) determined for the samples described for panel A. (C) (Left) Core morphology frequencies in sets of 100 counted virions (average ± SD) from two independent isolates of particles prepared as described for panel A. **, P < 0.01; n.s., not significant (P = 0.37). (Right) Representative images of the mature, eccentric, dual electron density (ED), empty, and immature virions observed in these experiments.
FIG 6.
Tomo-bubblegrams of immature HIV-1 particles. (Ai to Avi) Central tomographic sections (left) and corresponding tomo-bubblegram sections (right). Full tomograms of the virions in panels Aii (Movie S5), Aiii (Movie S6), and Avi (Movie S7) are shown in movies in the supplemental material. The diagrams on the left side of the figure illustrate the composition of each sample. PR− particles contain the Gag and Gag-Pol polyproteins that include NC and incorporate vRNA. The virions are immature due to the inactivating D25A mutation in the PR active site. Gag/ψ-minus particles are also immature, and additionally their RNA lacks the ψ region and therefore vRNA is not incorporated into particles. The Gag-LeuZip particles replace NC with a leucine zipper; these particles lack NC and vRNA. Bar, 50 nm. (B) Total virion RNA from the particles described for panel A. RNA concentration was normalized to the Gag signal obtained from anti-p24 Western blots of multiple input volumes. Shown are the averages (±SD) of total RNA (in nanograms) standardized by Gag signal for four independent experiments, reported as a percentage of PR−. (C) RNA from panel B was utilized to determine the copy number of vRNA. Values, reported as a percentage of PR−, are the averages (±SD) for four independent experiments.
TEM.
Viruses produced from transfected HEK293T cells were pelleted at 30,000 rpm at 4°C for 1.5 h with a Beckman SW41 rotor. Pelleted virus was resuspended in a small volume of phosphate-buffered saline. An equal volume of 4% EM-grade paraformaldehyde (Electron Microscopy Sciences) was added, and samples were incubated at 4°C before being embedded in Epon resin. Thin (75- to 100-nm) sections were applied to 200-mesh carbon-coated copper grids, stained with 0.2% lead citrate, and observed using either a Tecnai-12 (FEI) or JEOL 1200EX microscope equipped with a Gatan or AMT 2k charge-coupled device (CCD) camera, respectively. Images were captured at ×37,500 magnification and were visually scanned to count 100 viral particles per preparation.
Cryo-ET.
HIV-1 carrying the V165A mutation, WT virions produced with the indicated concentration of ALLINIs or DMSO control, PR-defective (PR−) virions, Gag/ψ-minus VLPs, and Gag-LeuZip VLPs were pelleted as described above and were imaged by cryo-ET as described previously (39). In brief, purified fixed virus was mixed (2:1) with a suspension of colloidal gold particles (Electron Microscopy Sciences), applied to Quantifoil R2/2 holey carbon grids (Structure Probe, Inc.), and plunge-frozen in a Vitrobot instrument (FEI). For data acquisition, grids were transferred to a cryo-holder (type 626; Gatan), and single-axis tilt series were recorded on a Tecnai-12 electron microscope equipped with an energy filter (GIF 2002; Gatan) operated at 120 keV in zero-loss mode with an energy slit width of 20 eV. Images were acquired on a 2,048- by 2,048-pixel CCD camera (Gatan) using SerialEM (40). Tilt-series projections were acquired at 2° intervals from ∼−66° to ∼66°, at ×38,500 magnification (0.78-nm/pixel) and a defocus of −4 μm. The electron dose per projection was ∼1.1 e−/Å2, giving a total cumulative dose of ∼75 e−/Å2. Tilt series were reconstructed using the Bsoft package (41), and virions were extracted as subtomograms and denoised by 20 iterations of anisotropic nonlinear diffusion (42).
Bubblegram and tomo-bubblegram imaging.
Dose series of HIV-1 particles were acquired either on a CM200-FEG TEM (FEI), operating at 120 keV with doses of ∼17 e−/Å2 per exposure, or in a Tecnai-12 TEM operating at 120 keV at ∼5 e−/Å2 per exposure. All images were collected on 2,048- by 2,048-pixel CCD cameras. Filled cores and eccentric aggregates bubbled at average doses of 165 ± 40 e−/Å2 (n = 109).
The procedure used to collect and analyze data for tomo-bubblegram imaging is described in Results. The imaging was performed on virions located over holes (and not near their edges) in the Quantifoil grids. The distances from the bubble centers to the center of the viral envelope were measured using Bsoft programs. Briefly, after extraction and centering of a virion, its radius was estimated as follows. First, to reduce noise and to suppress “missing wedge” effects (43), particles were spherically symmetrized (for ease of implementation, we applied icosahedral symmetry, which is not present in HIV-1 virions but gave a close approximation to spherical symmetry and could be applied with the software available). Then the radius was estimated from a density profile of the symmetrized particle. The bubble centers were then selected manually in the tomo-bubblegram, using the BshowX utility, and the distances of the bubbles from the envelope were calculated by subtracting the distance from the center of the bubble to the center of the particle from the particle radius at the middle of the envelope. Isosurface rendering of viral particles was done in Chimera (44).
Bacterially expressed recombinant NC was purified as previously described (45) and lyophilized after the addition of one equivalent Zn2+ per zinc finger. NC was then resuspended in buffer (25 mM HEPES, 30 mM NaCl, 1 mM MgCl2, 3 mM dithiothreitol [DTT], and 10% of glycerol at pH 7.5) at a concentration of 10 μM, aliquoted, and immediately frozen and stored at −80°C. For cryo-EM, thawed aliquots (25 μl) were centrifuged at maximum speed for 5 min in a tabletop centrifuge and the top 20 μl was discarded. The remaining 5 μl was applied to a 400-mesh copper grid covered with a thin carbon film supported by a thick lacy carbon film, washed with buffer (25 mM HEPES, 10 mM NaCl, 1 mM MgCl2, pH 7.5) to remove the glycerol, and plunge-frozen and imaged in a Tecnai-12 EM as described above.
RESULTS
Inhibition of virion maturation underlies ALLINI potency.
ALLINIs have been reported to inhibit multiple aspects of HIV-1 replication, including virion maturation, reverse transcription, nuclear import, and integration (see the introduction), suggesting that it may be the combination of multiple inhibitory processes that underlies ALLINI potency. An alternative scenario is that inhibition may be due to a single dominant or bottleneck step, which in this case would be particle maturation, as it is the first step inhibited from the perspective of the virus producer cell. To investigate this possibility, we compiled dose-response curves measuring the first two steps affected by ALLINIs—maturation and reverse transcription—and compared them to overall antiviral activity as assessed by the expression of the luciferase reporter gene in cells infected with drug-treated HIV-Luc particles. If maturation is a bottleneck step, we anticipated that its inhibition would equate with antiviral potency. If inhibition of multiple steps is involved, then the maturation and reverse transcription inhibitory curves would individually be less potent than overall antiviral activity.
HIV-Luc was produced in the presence of two representative ALLINI compounds, BI-D, with a 50% effective concentration (EC50) for inhibition of HIV-1 infection of 89 nM, and BIB-2 (also known as ALLINI-2), with an EC50 of 630 nM (23, 37) (Fig. 1), at various EC50 doses (1×, 2×, 4×, and 8×). TEM analysis of virus made in the presence of the solvent control DMSO revealed that 87% ± 2% (mean ± SD) of these particles harbored mature cores with internal density. This value decreased by ∼50% when virus was produced with unit EC50s of BI-D or BIB-2, with accompanying increases in particles containing eccentric condensates (Fig. 2A). Higher doses of each compound further reduced the fraction of mature cores and increased the fraction of eccentric condensate-containing particles. The reverse transcription activity and infectivity (luciferase activity) of the particles were measured in SupT1 cells infected with equal amounts of virus, as quantitated by their CA/p24 content. By comparing the results of Fig. 2A and B, we determined that the effect of drug treatment on reverse transcription activity closely paralleled the inhibition of mature core formation (Pearson correlation coefficient [r2] = 0.998 for the two sets of dose-response curves; P < 0.001). Inhibition of reverse transcription, moreover, paralleled overall antiviral activity (Fig. 2C; r2 = 0.903 for these two data sets; P < 0.001).
FIG 1.

Chemical structures of BI-D and BIB-2.
FIG 2.
Inhibition of virion maturation underlies ALLINI potency. (A) (Left) Quantitation of core morphotype frequencies (average ± SD for two independent experiments) for sets of 100 counted HIV-Luc particles; (right) representative TEM images of mature, eccentric, and immature virions produced in these experiments. (B) Quantitation of late reverse transcription (LRT) product formation from infections initiated with samples shown in panel A. (C) Antiviral activity as assessed by bulk luciferase output. Error bars in panels B and C represent the variations obtained from three independent experiments.
ALLINI treatment affects conical core assembly in addition to vRNP incorporation into the mature core.
Having determined that the antiviral activity of two independently tested, representative ALLINIs is due to their inhibition of particle maturation, we performed cryo-ET to further probe the antiviral phenotype. Virions produced with fully inhibitory ALLINI concentrations (10× EC50 for BIB-2; 10 to 100× EC50 for BI-D) (Fig. 2C) were classified according to core morphology (conical, e.g., see Fig. 3A and B; nonconical, e.g., see Fig. 3C and D; or no core, e.g., see Fig. 4A), and the presence (Fig. 3B and D and Fig. 4B) or absence (Fig. 3A and C and Fig. 4A) of an eccentric condensate. The eccentric condensate was previously observed by TEM as a compact, darkly staining body common to class II IN mutant viruses (12, 18, 46, 47) and ALLINI-treated virions (12, 15, 18). We used cryo-ET to extend these observations (Fig. 3B and D). Their compactness and density distinguish condensates from the other material that typically occupies the space between the core and the viral envelope, which we take to be mostly nonpolymerized CA (48, 49) and host cell proteins (50). In BI-D-treated particles, the eccentric condensates have ellipsoidal shapes with major and minor axes of 58 ± 12 nm and 37 ± 12 nm (n = 93), respectively; those present in particles treated with BIB-2 or untreated V165A mutant particles, which was utilized as a class II IN mutant control (18), were similar in size. The incidence of these morphological classes is summarized in Fig. 3E. Eccentric condensates are relatively rare (∼12%) in wild-type (WT) virions but present in ∼75% of particles treated with BI-D or BIB-2 or with the V165A IN mutant. We observed no virions with more than one eccentric condensate.
FIG 3.
Classification of HIV-1 particles observed by cryo-ET. (A to D) Tomographic central slices of representative HIV-1 virions classified according to core morphology (conical or nonconical) and the presence or absence of an eccentric condensate (white arrows). Eccentric condensates appear denser than the material lying between the core and the envelope in particles that lack an eccentric condensate. The cores of virions that have an eccentric condensate are relatively empty (e.g., panel B). Bar, 50 nm. (E) Percentages of HIV-1 virions classified according to their core morphology (conical, nonconical, or no core) and presence or absence of an eccentric condensate. The majority species for each sample is indicated by values in bold. According to the chi-square test, significant differences with P values of <0.00001 were found for comparisons between WT and BIB-2 and between WT and the V165A mutant. The difference between WT and BI-D had a P value of 0.013, whereas the differences between the V165A mutant and BI-D or BIB-II were not significant, with P values of >0.26. The chi-square test was used to test for equal distribution of compartmentalized data but should not be used if more of 20% of the bins contain fewer than 5 counts. Therefore, immature virions were not taken into account and virions with no cores were considered a single category, independent of the presence or absence of an eccentric condensate.
FIG 4.

Examples of relatively rare morphotypes observed in populations of WT virions. (A) Central tomographic slice of a WT particle without a core or eccentric condensate; (B) central tomographic slice (left) and corresponding tomo-bubblegram slice (right) of a WT particle containing an eccentric condensate (marked with an arrow in the left panel). The core is a conical one sampled in an oblique section. While the core is not entirely empty, it lacks sufficient internal density to suggest that it contains a vRNP. This interpretation is confirmed by the bubbling being confined to the eccentric condensate. Bar, 50 nm.
As in earlier studies by cryo-ET (e.g., see references 3, 4, and 25), most WT virions (∼77%) were found to have closed conical cores with internal density (Fig. 3A), noting that the tomograms would not detect small holes if present (51). Most of the eccentric condensate-containing virions from BI-D or BIB-2 treatment or the V165A IN mutant also have cores (Fig. 3E), but there are two striking differences between these cores and WT cores. First, they have a much higher incidence of nonconical cores than the WT (the ratios of nonconical to conical cores were 1.8:1 for BI-D, 0.9:1 for BIB-2, and 1.6:1 for V165A, compared to 1:7.2 for WT). Thus, there is marked impairment of capsid morphogenesis. Second, V165A mutant, BI-D, and BIB-2 conical cores are relatively empty, as exemplified by comparing Fig. 3A and B. With WT cores, internal material is generally taken to consist mainly of the vRNP, although this assignment has not been formally demonstrated and is addressed further below. While V165A mutant, BI-D, and BIB-2 cores have less internal content than the WT ones, they tend to have some internal material (e.g., see Fig. 3D). With this caveat, we estimate that ∼50% of them have little or no content compared to 3% for the WT virions. These data suggest that V165A mutant, BI-D, and BIB-2 virions are deficient in capsid morphogenesis as well as incorporation of the vRNP into the mature core.
Bubblegrams detect radiation-sensitive material in filled cores and eccentric condensates.
It is plausible that eccentric condensates represent NC-vRNA complexes that failed to be incorporated into the mature core. We obtained supporting evidence for this hypothesis by means of a labeling technique called bubblegram imaging (52, 53). Vitrified samples imaged in an electron microscope undergo progressive damage by electron irradiation. This damage is initially expressed as a “smearing” of the macromolecular density, and for this reason the electron dose is kept as low as possible during imaging (54). When proteinaceous specimens are subjected to relatively high levels of electron irradiation in cryo-EM, perhaps 10-fold higher than the dose used in a typical imaging experiment, bubbles of hydrogen gas are generated (55, 56) which are readily visible due to their low density. Different proteins have different bubbling thresholds (N. Cheng, Weimin Wu, Anastasia Aksyuk, J. Fontana, and A. C. Steven, unpublished observations). Moreover, proteins embedded in DNA bubble relatively early because the densely packed DNA impedes the diffusion of radiation products from their sites of origin, accelerating their buildup to a critical concentration for bubbling. Hypothesizing that compact NC-vRNA condensates may have distinctive bubbling behavior, we subjected HIV-1 virions to bubblegram imaging. In dose-series micrographs (not shown), small bubbles first appeared after a cumulative dose of 160 to 200 e−/Å2 and grew in size and number with subsequent exposures. The bubbles coprojected with filled cores and eccentric condensates, and the two structures appeared to have the same bubbling threshold, although in some micrographs the boundaries of these structures were difficult to discern. However, we could not rule out that the bubbles may not be inside these structures but coincidentally lie along the same line-of-sight.
Aiming to eliminate the ambiguity of coprojection, we extended the bubblegram method to 3D imaging by electron tomography. We call this method tomo-bubblegram imaging. First, a regular tilt series is collected from an area of interest and a tomogram is calculated. This involves a total dose of ∼75 e−/Å2, which is below the bubbling threshold for HIV-1. Second, an untilted dose series is recorded, stopping after the first small bubbles begin to develop (another ∼70 e−/Å2). Third, after waiting ∼2 h to allow radiation products to dissipate, a second tilt series is recorded (∼70 e−/Å2) during which larger bubbles appear in the “primed” specimen. These bubbles are visualized in the corresponding tomogram, the tomo-bubblegram. Finally, a comparison of the first tomogram (the “before” state with the virions in a close-to-native state) and the second one (the “after” state) allows the sites in which the bubbles nucleated to be mapped in three dimensions. As illustrated in Fig. 5, Movies S1 to S4 in the supplemental material, and Table 1, the bubbles specifically label the core interiors in WT virions, predominantly toward the wide end of the core. They also label the eccentric condensates of V165A mutant and BI-D or BIB-2 virions. Of note, bubbles were never seen only inside cores (i.e., outside the condensates) of V165A mutant or BI-D or BIB-2 virions that also contained eccentric condensates: in these particles, bubbles were observed either only in the condensate (∼80% of the condensate-containing particles) or in both the condensate and the core (∼20%). In summary, we found that the WT conical core contents and eccentric condensates are more prone to bubbling than the other components of the virion (CA protein, MA protein, envelope glycoprotein) and that they are indistinguishable in this respect. In turn, this correlation strongly supports the idea that they are chemically the same and are indeed NC-vRNA complexes.
FIG 5.
Tomo-bubblegrams of HIV-1 particles. (A to X) Each pair of panels shows a central section from an initial tomogram (left) and the corresponding section from the tomo-bubblegram (right). The types of virions analyzed are indicated on the left of each block. BIB-2 (10×) and BI-D (10× EC50 for panels G, I, and L; 100× EC50 for panels H, J, and K) were used at full inhibitory concentrations. Particles with conical cores are framed in red, and those with nonconical cores are framed in blue; particles lacking eccentric condensates are framed with continuous lines, and particles that have eccentric condensates are framed with dashed lines. Bar, 50 nm. Full tomograms of the virions in panels B (Movie S1), I (Movie S2), R (Movie S3), and X (Movie S4) are shown in the supplemental material.
TABLE 1.
Tomo-bubblegram labeling of HIV-1 virions
| Sample type | % of virions with bubblesa |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Conical core, no condensate |
Nonconical core, no condensate |
Condensate with core (conical or nonconical) |
|||||||
| Inside core | Outside core | Both | Inside core | Outside core | Both | Outside condensate | Inside condensate | Both | |
| WT (n = 78) | 55 | 0 | 19 | 1 | 3 | 3 | 4 | 4 | 9 |
| BI-D (n = 72) | 7 | 0 | 7 | 1 | 1 | 6 | 0 | 56 | 21 |
| BIB-2 (n = 64) | 8 | 0 | 0 | 0 | 3 | 0 | 0 | 77 | 12 |
| V165A mutant (n = 88) | 3 | 0 | 2 | 6 | 0 | 2 | 0 | 67 | 18 |
The data are shown as percentages of virions with bubbles either inside the core, outside the core, or both. The majority species for each sample type is indicated by bold values. Few particles have no bubbles (<3% of the virions observed), and they were not included in this analysis. According to the chi-square test, highly significant differences with P values of <0.00001 were found for comparisons between WT and BIB-2 samples and between WT and V165A mutant samples. The comparison between WT and BI-D samples yielded a P value of 0.009, whereas the differences between the V165A mutant and BI-D or BIB-II samples were not significant, with P values of ∼0.25. To make possible the application of the chi-square test, only four categories were considered: virions with conical or nonconical core without eccentric condensate that were labeled inside the core or inside and outside the core, and virions with condensate and core (conical or nonconical) that were labeled inside the condensate (i.e., eighth column) or inside and outside the condensate (i.e., ninth column).
Bubbles label NC-RNA in immature virions.
To further investigate which component of HIV-1 virions is bubbling, we examined immature (PR-defective [PR−]) virions in which, in the absence of proteolysis, the Gag and Gag-Pol domains are radially ordered, with MA in contact with the viral membrane and CA and NC/vRNA arranged in successive concentric layers. PR− virions (HIV-1NL4-3 with the D25A inactivating mutation in the PR active site) exhibited a similar bubbling threshold as WT virions, although the bubbles appeared in a different location, i.e., within the Gag shell (Fig. 6Ai and ii; see also Movie S5 in the supplemental material). Radially, these bubbles are centered at ∼15 nm (average, 15 ± 5 nm; n = 333) in from the middle of the viral envelope, a position that corresponds to the interface between the CA and NC/vRNA layers (Fig. 7). In the other two dimensions of the Gag lattice, we did not find a preference for bubbling in any particular location.
FIG 7.
Locations of bubbles in PR− virions. (A) Cryo-ET central slice of an immature PR− particle. (B and C) Tomo-bubblegram section corresponding to panel A. In panel C, bubbles are marked with red circles. (D) Surface rendering of a cutaway slice of the particle shown in panels A to C. Most of the bubbles (white) are located between the CA layer (green) and the NC-vRNA layer (yellow). The viral envelope is in cyan, and the internal contents are in red. Bar, 50 nm. (E) Radial density profile of subtomogram averaged Gag from immature virions (from reference 25). (F) Histogram of the distance from the center of the bubbles to the center of the viral envelope (membrane + MA).
Next, immature virions in which vRNA and NC were respectively eliminated from Gag virus-like particles (VLPs) were analyzed to test for the relative contributions of these two components to bubbling phenomena. A VLP construct that lacked the vRNA ψ region (33) reduced the packaging of vRNA relative to total virion RNA by ∼20,000-fold (Fig. 6C). Gag/ψ-minus particles bubbled in the same locations as immature (PR−) virions, thus showing that vRNA is not needed for bubbling (Fig. 6Aiii and iv; see also Movie S6 in the supplemental material). We do, however, note that the bubbling appeared to be slightly reduced with this construct (16 ± 10 bubbles/virion in Gag/ψ-minus particles versus 27 ± 15 bubbles/virion in PR− particles [Table 2]; the differences are significant at a P of <0.01 according to the two-tailed Student t test). The difference, such as it is, might arise from the ∼2.5-fold reduction in overall RNA content in Gag/ψ-minus particles, as the RNA may impede the dispersal of hydrogen gas generated from NC. To eliminate NC but nevertheless retain structurally similar particles, we used a construct, termed Gag-LeuZip, in which the NC domain of Gag was replaced by a leucine zipper (32) but in which Gag otherwise retains its immature conformation (57). The Gag-LeuZip VLPs yielded almost no bubbles (Fig. 6Av and vi; see also Movie S7) (0.3 bubbles/virion versus 16 bubbles/Gag/ψ-minus VLP at the same electron doses), and ∼94% of these particles had 0 or 1 bubble (versus 100% of the control particles that each had two or more bubbles) (Table 2). The relative levels of total RNA and vRNA in Gag-LeuZip VLPs were similar to those in Gag/ψ-minus VLPs (Fig. 6B and C), though we do note that an earlier study failed to detect tangible RNA levels in Gag-LeuZip VLPs (32). Nevertheless, we conclude that apart from a very low background of bubbles from other sources (as illustrated in Fig. 6Av and vi), NC protein is the bubbling component.
TABLE 2.
Bubbles in three kinds of PR− HIV-1 particles
| Particle type | No. of tomograms | Total avg dose (e−/Å2) | No. of virions | No. of bubbles/viriona | % of virions with 2 or more bubbles | % of virions with 1 or no bubbles |
|---|---|---|---|---|---|---|
| PR− | 3 | 215 | 65 | 27 ± 15 | 100 | 0 |
| Gag/ψ-minus | 4 | 221 | 53 | 16 ± 10 | 100 | 0 |
| Gag-LeuZip | 4 | 228 | 53 | 0.3 ± 0.6 | 6 | 94 |
For PR− and Gag/ψ-minus VLPs, the bubbles from 29 and 31 virions, respectively, were counted. For the Gag-LeuZip, bubbles from all particles imaged were counted.
Purified NC exhibits bubbling behavior similar to that observed in mature and immature virions.
Identification of NC protein as the component responsible for bubbling in both mature WT and IN mutant or ALLINI-treated virions and in immature virions led to the prediction that NC protein should also bubble provided that it is suitably concentrated. To test this hypothesis, we examined aggregates of purified NC. Aggregates were similar in diameter to eccentric condensates (average, 51 ± 20 nm [n = 69] versus 48 ± 9 nm [n = 93], respectively) and were found to have a bubbling threshold similar to that of eccentric condensates (average, 154 ± 57 e−/Å2 [n = 69] versus 165 ± 42 e−/Å2 [n = 69], respectively). Their bubbling threshold was somewhat dependent on size, with larger aggregates bubbling earlier than smaller ones (Fig. 8). This is consistent with the greater abundance of NC in larger aggregates providing a richer source of hydrogen gas as a radiation product. Strikingly, bubbling started in the middle of these aggregates and then spread outwards, consistent with the outer layers of molecules impeding the outward diffusion of the gas. Taken together, these observations provide strong evidence that the bubbling component in HIV-1 virions is NC protein and therefore confirm that ALLINI treatment affects incorporation of the vRNP into the core.
FIG 8.
Bubblegram imaging of purified NC protein. (A to E) Cryo-EM dose series of five NC aggregates of different sizes. The images in which bubbles first appear are labeled with a white asterisk in each case (although NC is formally soluble, small aggregates of suitable size were consistently found on EM grids). Bar, 50 nm. (F) Plot of the diameters of NC aggregates versus the cumulative electron dose at which they started bubbling. Points corresponding to the aggregates from panels A to E are shown in red. While the data exhibit some stochastic variability in bubbling threshold, there is a clear trend toward larger aggregates requiring less electron radiation to initiate bubbling. A linear fit is shown for reference (red line).
ALLINI-induced eccentric condensate formation requires vRNA.
To address whether ALLINI-mediated impairment of vRNP incorporation into the mature core is dependent upon the species of RNA within the virus, we cotransfected pHI-Luc, which expresses a minimal HIV-1 transfer vector that harbors ∼45% of the full-length vRNA genome, with pHP-dI-N/A, which expresses the Gag and Pol proteins required to package HI-Luc vRNA in trans. As a transfection control for pHI-Luc, we utilized the promoterless pGL3-basic (GL3-B) construct that carries the luciferase gene but lacks HIV-1-related sequences and the capacity to express RNA. Total RNA levels of pelleted virions nevertheless did not vary significantly between those made in the presence of pHI-Luc or GL3-B, although, as expected, the HIV-1 genome was selectively incorporated into HI-Luc particles (Fig. 9A and B). We also note that HI-Luc total and vRNA levels were similar to those of the HIV-Luc virions characterized in Fig. 2.
TEM revealed that ∼30% of HI-Luc viruses harbored WT-like conical cores, ∼22% harbored cores as well as eccentric aggregates, ∼25% had a previously unseen morphology in which dual electron density (dual ED) appeared both inside and outside the core, ∼15% had empty cores, and ∼15% contained immature-like spherical shells (Fig. 9C). The baseline phenotype of virions made in the presence of GL3-B did not differ dramatically from that of virions that harbored HI-Luc vRNA (Fig. 9C). As expected, BI-D treatment increased the frequency of HI-Luc virions with eccentric aggregates (∼60%), with accompanying decreases in mature (∼5%) and dual ED (∼1%) cores. In contrast, the fraction of virions with eccentric aggregates did not significantly change when sham virus made in the presence of GL3-B was treated with BI-D (Fig. 9C).
Partial recovery of WT core formation by IN in trans.
TEM classification of ΔIN virions revealed that the majority (∼61%) contained eccentric condensates while ∼9% harbored WT-like conical cores and ∼30% were immature (18). To assess the role of IN in HIV-1 maturation and the impact of ALLINI treatment on this role, ΔIN virions were functionally transcomplemented by IN fused to the viral accessory protein Vpr. INH171T, which confers ∼44-fold resistance to BI-D (58), was utilized as a control. Although class II IN mutations such as ΔIN can impede CA/p24 release in a cell type-dependent manner (31, 59), the ΔIN virus used here yielded supernatant p24 values similar to those of the WT control virus (18). Vpr-IN boosted the infectivity of the highly defective ΔIN virions ∼100-fold, to a level equal to ∼5 to 10% of the infectivity of the parental HIV-Luc virus (Fig. 10A). As expected (18), a 10× EC50 dose of BI-D abolished the infectivity of ΔIN virions transcomplemented with Vpr-INWT but not with Vpr-INH171T. Concomitant with the gain in infectivity, the incidence of conical cores rose from ∼9% to ∼30% upon Vpr-IN transcomplementation (Fig. 10B). With Vpr-INWT, ALLINI treatment restored this phenotype to the noncomplemented ΔIN value of ∼9%, whereas virions transcomplemented by Vpr-INH171T harbored ∼30% mature cores, regardless of ALLINI treatment.
FIG 10.

IN in trans stimulates WT core formation in ΔIN virions. (A) Infectivity of HIV-Luc.ΔIN transcomplemented with Vpr-INWT or Vpr-INH171T made in the presence of BI-D or DMSO. Values were normalized to control HIV-Luc virus constructed in the presence of DMSO. (B) (Upper) Quantitation of core morphology frequencies of 100 counted virions (the ΔIN mutation yields more immature particles than does other class II [e.g., missense] mutations [18, 46]). Shown are averages ± SD for two independent experiments for the transcomplemented preparations described for panel A. (Lower) Representative images of the mature, eccentric, and immature virions observed within experiments.
DISCUSSION
In this study, we have utilized the recently discovered antiviral activity of ALLINIs to assess the role of IN in HIV-1 maturation. Of virions assembled in ALLINI-treated cells, the fraction that acquires a closed conical capsid (taken to be a requirement for infectivity [60]) is markedly reduced. Many of these capsids are malformed. Moreover, in most of these virions, the vRNP is not incorporated into the mature core but remains outside the capsid as an “eccentric condensate,” even if a closed conical capsid is assembled. These phenotypes are reproduced by various class II IN mutations, including the V165A point mutation or IN deletion. IN supplied in trans partially rescued the fraction of ΔIN virions that harbored WT cores with internal electron density, a recovery that could be abolished by ALLINI treatment. As vRNA was required for ALLINI-induced eccentric condensate formation, we suggest a model wherein disruption of an IN-vRNA interaction, either by genetic or pharmacological means, results in the inability to properly incorporate the vRNP into the capsid during HIV-1 core morphogenesis.
ALLINI antiviral activity is due to inhibition of HIV-1 particle maturation.
ALLINIs have been shown to inhibit several steps of the HIV-1 replication cycle (see the introduction). However, it has been unclear whether the inhibition of multiple steps combines to determine drug potency or whether ALLINIs inhibit a single bottleneck step in the replication cycle. To address this issue, we correlated dose-response curves from inhibiting maturation and reverse transcription, the first two steps that are inhibited from the perspective of the drug-treated virus-producing cells, to overall antiviral activity. As all three curves (percentage of mature cores, reverse transcription activity, infectivity) effectively superposed with highly significant correlations for two representative inhibitors, we conclude that inhibition of HIV-1 maturation determines the antiviral potency of these compounds.
Similar to PIs (61), ALLINIs display relatively steep dose-response curves (11, 18). Two mutually nonexclusive models for this behavior posit that such compounds inhibit multiple copies of the drug target at the relevant step of the replication cycle (61) or that they inhibit multiple steps in the replication cycle (62). Whereas inhibition of multiple replication steps appears to underlie the cooperative behavior of PIs (62), we conclude from our data that ALLINIs mainly if not exclusively target IN during HIV-1 maturation and, by inference, that multiple copies of IN, if not the full virion complement of the protein, are affected. We accordingly note similar slopes (m) of BI-D dose-response curves for stimulation of purified IN multimerization in vitro (m, ∼2) and inhibition of HIV-1 replication (m, ∼2.8) (18).
Tomo-bubblegram imaging labels NC-containing complexes in 3D.
Cryo-ET and tomo-bubblegram imaging have given a clearer account of the substructure of HIV-1 virions whose maturation was subverted by a class II mutation in IN or by ALLINI treatment. First, we confirmed earlier TEM data (12, 15, 18, 21) that ALLINI treatment yields a high proportion of virions with eccentric condensates. This property recurs in the class II IN mutant control virus. Second, being able to visualize the 3D structures of complete virions in greater detail, we additionally found an adverse effect of the V165A mutation and ALLINIs on capsid morphogenesis.
We now consider the density inside conical capsids of mature WT virions. This density has been interpreted as the vRNP, composed primarily of NC and vRNA. The vRNP also contains the RT and IN enzymes and some other viral and host cell components (63–66). Serendipitously, NC turned out to be highly susceptible to irradiation-induced bubbling, and we exploited this property to demonstrate that NC is indeed a major internal component of WT cores; moreover, the NC is primarily at the wide end of the core (Fig. 5A to F; see also Movie S1 in the supplemental material and reference 67). Because few bubbles are seen elsewhere in virions, it seems that most if not all of the NC protein goes into the core. These data directly support earlier evidence that most or all of NC is in the core, whereby the ratio of NC to CA was found to be significantly higher in isolated cores than in virions (68), in keeping with the property that only about half of the CA present in an immature virion assembles into a mature capsid (48, 49). Similarly, our tomo-bubblegrams have shown that eccentric condensates are rich in NC, with few bubbles seen anywhere else in condensate-containing virions.
The typical dimensions of the material enclosed in the wide end of a WT core are similar to those of an eccentric condensate. It appears therefore that when NC and the attached genomes are released from the Gag shell in a maturing virion, they condense into one or the other of these aggregates. A heuristic calculation supports this inference. If one takes an eccentric condensate to have major axes of 55 by 55 by 35 nm (see above), its volume is ∼55,000 nm3. Assuming ∼50% of this volume to be occupied by protein (a value typical for a protein crystal) and a protein density of 0.78 kDa/nm3, a mass of 21.5 MDa results. Alternatively, an NC copy number of ∼2,000 to 2,500 per virion (1) and a subunit mass of 7.2 kDa give a total mass of ∼14 to 18 MDa. Therefore, the eccentric condensates are large enough to account for all of the NC plus the two copies of the vRNA genome, which remain associated with NC after virion maturation (64, 69).
Our analysis also shows that ∼12% of WT virions contain eccentric condensate: of these, 75% have conical cores and 25% have nonconical cores. An example is shown in Fig. 4B. The existence of such particles demonstrates miscarried vRNP incorporation into mature cores under the otherwise unperturbed WT situation of HIV-1 clade B strain NL4-3 assembly from transfected HEK293T cells. Additional work with other viral clades and cell types would be needed to discern how broadly applicable this observation may be.
Implications for core assembly.
In order to assemble a core, Gag and Gag-Pol must be dissected by PR, with their component domains being separated in a tightly regulated cleavage program (70). Finally, CA is released from the maturing Gag shell into a soluble pool, from where it assembles to form the mature core, leaving about 50% unassembled (4, 26, 48, 49). During core assembly, NC together with the vRNA and IN and RT enzymes becomes situated into the mature core (see above).
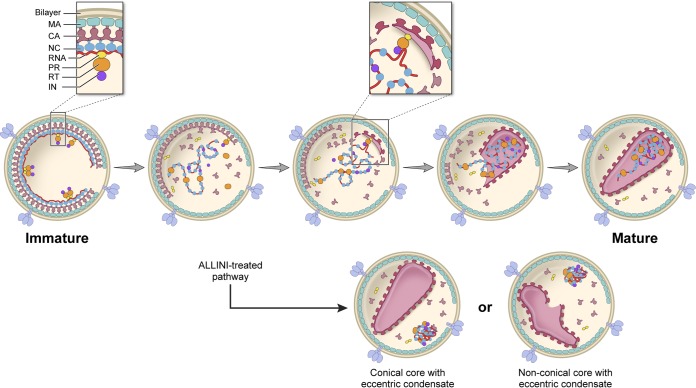
Several models have been proposed for how capsid assembly takes place (3, 4). They differ on whether assembly starts at the wide end (3) or the narrow end (4) and are couched primarily in terms of self-assembly properties attributed to CA protein, since conical capsids similar to the ones observed inside HIV-1 virions can be assembled from purified components in vitro (71). It has also been suggested that the vRNP may help to nucleate assembly (3, 5, 67), ensuring its incorporation into the capsid, and that MA or the inner surface of the viral membrane could direct the closure of the core (4). A related idea for coupling capsid assembly with vRNA packaging was advanced for Rous sarcoma virus in that some unprocessed or incompletely processed copies of Gag in the Gag shell may form a nucleation complex that binds via the NC moieties to a genomic packaging site and, via the CA moieties, nucleates outgrowth of the capsid shell (72). Although IN has been implicated in regulating PR activity during HIV-1 assembly and particle release (59), a role for the viral enzyme in core morphogenesis has not been proposed. Based on our data, we propose an active role for IN in initiating vRNP incorporation into the mature core, which then nucleates capsid formation around the enclosed vRNA-NC complex (Fig. 11). This scenario could in theory apply to precleavage IN as a domain of Gag-Pol or postcleavage IN that has remained in essentially the same position. Supporting a model for an IN-vRNA interaction(s) in core morphogenesis, our data reveal a role for vRNA in ALLINI-induced eccentric condensate formation (Fig. 9) and high-affinity binding of purified HIV-1 IN to RNA has been observed in vitro (73). A linkage between IN and CA changes has, moreover, been noted among strains in drug-naive and drug-experienced AIDS patients (74), indicating a potential biological role for an IN-CA interaction. IN-RNA and IN-CA interactions are two avenues that we are currently pursuing to investigate the mechanistic basis of IN in HIV-1 core formation and vRNP incorporation into the mature core.
FIG 11.
Model for the role of IN in coordinating HIV-1 capsid assembly and vRNP incorporation into the mature core. Maturation starts with activation of the PR and dissection of the Gag and Gag-Pol polyproteins and release of the vRNP or NC from the Gag shell. The key element of this model is that it envisages a molecular complex containing one or several copies of Gag-Pol that initiates both assembly of the capsid by an outgrowth of CA subunits and packaging of the vRNA through binding to IN or to RT as conformationally influenced by IN. Many details remain to be clarified, including, for instance, whether other components are involved in the putative complex, whether the initiation process starts with uncleaved Gag-Pol or whether the polyproteins have been proteolytically processed, with the cleavage products remaining together, and whether capsid assembly begins at the wide or narrow end. We favor the wide end because this narrows down to give a capsid of about the right length, which CA has been shown to be capable of in the absence of a membrane (71). The cartoon shows RT and IN associated with the eccentric condensate for which there is currently no evidence, although they do remain, mostly or totally, inside the wild-type capsid (66). A variant of this model in the Rous sarcoma virus system envisages participation of the cytoplasmic domain of Env (72). The virion and core also contain other components (e.g., Vpr in the core and some host proteins somewhere in the virion), which are not shown.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alan Rein for the generous gift of pRR359 and pRR546 plasmid DNAs, Dennis Winkler and Bernard Heymann for support with resources for EM and computation, Eric Freed for helpful comments on the draft manuscript, and Weifeng Wang for valuable discussion.
This work was funded in part by the Intramural Research Program of NIAMS (A.C.S.), the Intramural AIDS Targeted Antiviral Program (A.C.S.), NIH grants AI070042 (A.N.E.) and GM103368 (A.N.E. and J.R.F.), and funds of the National Cancer Institute under contract HHSN261200800001E with Leidos Biomedical Research, Inc. (R.J.G.).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01522-15.
REFERENCES
- 1.Sundquist WI, Kräusslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med 2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grünewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 3.Benjamin J, Ganser-Pornillos BK, Tivol WF, Sundquist WI, Jensen GJ. 2005. Three-dimensional structure of HIV-1 virus-like particles by electron cryotomography. J Mol Biol 346:577–588. doi: 10.1016/j.jmb.2004.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briggs JA, Grünewald K, Glass B, Forster F, Kräusslich HG, Fuller SD. 2006. The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure 14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 5.Woodward CL, Cheng SN, Jensen GJ. 2015. Electron cryotomography studies of maturing HIV-1 particles reveal the assembly pathway of the viral core. J Virol 89:1267–1277. doi: 10.1128/JVI.02997-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frank GA, Narayan K, Bess JWJ, Del Prete GQ, Wu X, Moran A, Hartnell LM, Earl LA, Lifson JD, Subramaniam S. 2015. Maturation of the HIV-1 core by a non-diffusional phase transition. Nat Commun 6:5854. doi: 10.1038/ncomms6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wlodawer A, Vondrasek J. 1998. Inhibitors of HIV-1 protease: a major success of structure-assisted drug design. Annu Rev Biophys Biomol Struct 27:249–284. doi: 10.1146/annurev.biophys.27.1.249. [DOI] [PubMed] [Google Scholar]
- 8.Li F, Goila-Gaur R, Salzwedel K, Kilgore NR, Reddick M, Matallana C, Castillo A, Zoumplis D, Martin DE, Orenstein JM, Allaway GP, Freed EO, Wild CT. 2003. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc Natl Acad Sci U S A 100:13555–13560. doi: 10.1073/pnas.2234683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou J, Yuan X, Dismuke D, Forshey BM, Lundquist C, Lee KH, Aiken C, Chen CH. 2004. Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J Virol 78:922–929. doi: 10.1128/JVI.78.2.922-929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelman A. 1999. In vivo analysis of retroviral integrase structure and function. Adv Virus Res 52:411–426. doi: 10.1016/S0065-3527(08)60309-7. [DOI] [PubMed] [Google Scholar]
- 11.Kessl JJ, Jena N, Koh Y, Taskent-Sezgin H, Slaughter A, Feng L, de Silva S, Wu L, Le Grice SF, Engelman A, Fuchs JR, Kvaratskhelia M. 2012. Multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors. J Biol Chem 287:16801–16811. doi: 10.1074/jbc.M112.354373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balakrishnan M, Yant SR, Tsai L, O'Sullivan C, Bam RA, Tsai A, Niedziela-Majka A, Stray KM, Sakowicz R, Cihlar T. 2013. Non-catalytic site HIV-1 integrase inhibitors disrupt core maturation and induce a reverse transcription block in target cells. PLoS One 8:e74163. doi: 10.1371/journal.pone.0074163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christ F, Voet A, Marchand A, Nicolet S, Desimmie BA, Marchand D, Bardiot D, Van der Veken NJ, Van Remoortel B, Strelkov SV, De Maeyer M, Chaltin P, Debyser Z. 2010. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat Chem Biol 6:442–448. doi: 10.1038/nchembio.370. [DOI] [PubMed] [Google Scholar]
- 14.Le Rouzic E, Bonnard D, Chasset S, Bruneau JM, Chevreuil F, Le Strat F, Nguyen J, Beauvoir R, Amadori C, Brias J, Vomscheid S, Eiler S, Levy N, Delelis O, Deprez E, Saib A, Zamborlini A, Emiliani S, Ruff M, Ledoussal B, Moreau F, Benarous R. 2013. Dual inhibition of HIV-1 replication by integrase-LEDGF allosteric inhibitors is predominant at the post-integration stage. Retrovirology 10:144. doi: 10.1186/1742-4690-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma A, Slaughter A, Jena N, Feng L, Kessl JJ, Fadel HJ, Malani N, Male F, Wu L, Poeschla E, Bushman FD, Fuchs JR, Kvaratskhelia M. 2014. A new class of multimerization selective inhibitors of HIV-1 integrase. PLoS Pathog 10:e1004171. doi: 10.1371/journal.ppat.1004171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsantrizos YS, Boes M, Brochu C, Fenwick C, Malenfant E, Mason S, Pesant M. 22 November 2007. Inhibitors of human immunodeficiency virus replication. International patent WO 2007/131350 A1.
- 17.Gupta K, Brady T, Dyer BM, Malani N, Hwang Y, Male F, Nolte RT, Wang L, Velthuisen E, Jeffrey J, Van Duyne GD, Bushman FD. 2014. Allosteric inhibition of human immunodeficiency virus integrase: late block during viral replication and abnormal multimerization involving specific protein domains. J Biol Chem 289:20477–20488. doi: 10.1074/jbc.M114.551119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurado KA, Wang H, Slaughter A, Feng L, Kessl JJ, Koh Y, Wang W, Ballandras-Colas A, Patel PA, Fuchs JR, Kvaratskhelia M, Engelman A. 2013. Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation. Proc Natl Acad Sci U S A 110:8690–8695. doi: 10.1073/pnas.1300703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsiang M, Jones GS, Niedziela-Majka A, Kan E, Lansdon EB, Huang W, Hung M, Samuel D, Novikov N, Xu Y, Mitchell M, Guo H, Babaoglu K, Liu X, Geleziunas R, Sakowicz R. 2012. New class of HIV-1 integrase (IN) inhibitors with a dual mode of action. J Biol Chem 287:21189–21203. doi: 10.1074/jbc.M112.347534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kvaratskhelia M, Sharma A, Larue RC, Serrao E, Engelman A. 2014. Molecular mechanisms of retroviral integration site selection. Nucleic Acids Res 42:10209–10225. doi: 10.1093/nar/gku769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desimmie BA, Schrijvers R, Demeulemeester J, Borrenberghs D, Weydert C, Thys W, Vets S, Van Remoortel B, Hofkens J, De Rijck J, Hendrix J, Bannert N, Gijsbers R, Christ F, Debyser Z. 2013. LEDGINs inhibit late stage HIV-1 replication by modulating integrase multimerization in the virions. Retrovirology 10:57. doi: 10.1186/1742-4690-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fadel HJ, Morrison JH, Saenz DT, Fuchs JR, Kvaratskhelia M, Ekker SC, Poeschla EM. 2014. TALEN knockout of the PSIP1 gene in human cells: analyses of HIV-1 replication and allosteric integrase inhibitor mechanism. J Virol 88:9704–9717. doi: 10.1128/JVI.01397-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng L, Sharma A, Slaughter A, Jena N, Koh Y, Shkriabai N, Larue RC, Patel PA, Mitsuya H, Kessl JJ, Engelman A, Fuchs JR, Kvaratskhelia M. 2013. The A128T resistance mutation reveals aberrant protein multimerization as the primary mechanism of action of allosteric HIV-1 integrase inhibitors. J Biol Chem 288:15813–15820. doi: 10.1074/jbc.M112.443390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christ F, Shaw S, Demeulemeester J, Desimmie BA, Marchand A, Butler S, Smets W, Chaltin P, Westby M, Debyser Z, Pickford C. 2012. Small-molecule inhibitors of the LEDGF/p75 binding site of integrase block HIV replication and modulate integrase multimerization. Antimicrob Agents Chemother 56:4365–4374. doi: 10.1128/AAC.00717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller PW, Adamson CS, Heymann JB, Freed EO, Steven AC. 2011. HIV-1 maturation inhibitor bevirimat stabilizes the immature Gag lattice. J Virol 85:1420–1428. doi: 10.1128/JVI.01926-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller PW, Huang RK, England MR, Waki K, Cheng N, Heymann JB, Craven RC, Freed EO, Steven AC. 2013. A two-pronged structural analysis of retroviral maturation indicates that core formation proceeds by a disassembly-reassembly pathway rather than a displacive transition. J Virol 87:13655–13664. doi: 10.1128/JVI.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Marco A, Heuser AM, Glass B, Kräusslich HG, Muller B, Briggs JA. 2012. Role of the SP2 domain and its proteolytic cleavage in HIV-1 structural maturation and infectivity. J Virol 86:13708–13716. doi: 10.1128/JVI.01704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Marco A, Muller B, Glass B, Riches JD, Kräusslich HG, Briggs JA. 2010. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS Pathog 6:e1001215. doi: 10.1371/journal.ppat.1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown HE, Chen H, Engelman A. 1999. Structure-based mutagenesis of the human immunodeficiency virus type 1 DNA attachment site: effects on integration and cDNA synthesis. J Virol 73:9011–9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Limon A, Devroe E, Lu R, Ghory HZ, Silver PA, Engelman A. 2002. Nuclear localization of human immunodeficiency virus type 1 preintegration complexes (PICs): V165A and R166A are pleiotropic integrase mutants primarily defective for integration, not PIC nuclear import. J Virol 76:10598–10607. doi: 10.1128/JVI.76.21.10598-10607.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu R, Limon A, Devroe E, Silver PA, Cherepanov P, Engelman A. 2004. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J Virol 78:12735–12746. doi: 10.1128/JVI.78.23.12735-12746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crist RM, Datta SA, Stephen AG, Soheilian F, Mirro J, Fisher RJ, Nagashima K, Rein A. 2009. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J Virol 83:2216–2225. doi: 10.1128/JVI.02031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider R, Campbell M, Nasioulas G, Felber BK, Pavlakis GN. 1997. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J Virol 71:4892–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang LJ, Urlacher V, Iwakuma T, Cui Y, Zucali J. 1999. Efficacy and safety analyses of a recombinant human immunodeficiency virus type 1 derived vector system. Gene Ther 6:715–728. doi: 10.1038/sj.gt.3300895. [DOI] [PubMed] [Google Scholar]
- 35.Nakajima N, Lu R, Engelman A. 2001. Human immunodeficiency virus type 1 replication in the absence of integrase-mediated DNA recombination: definition of permissive and nonpermissive T-cell lines. J Virol 75:7944–7955. doi: 10.1128/JVI.75.17.7944-7955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu X, Liu H, Xiao H, Conway JA, Hunter E, Kappes JC. 1997. Functional RT and IN incorporated into HIV-1 particles independently of the Gag/Pol precursor protein. EMBO J 16:5113–5122. doi: 10.1093/emboj/16.16.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Jurado KA, Wu X, Shun MC, Li X, Ferris AL, Smith SJ, Patel PA, Fuchs JR, Cherepanov P, Kvaratskhelia M, Hughes SH, Engelman A. 2012. HRP2 determines the efficiency and specificity of HIV-1 integration in LEDGF/p75 knockout cells but does not contribute to the antiviral activity of a potent LEDGF/p75-binding site integrase inhibitor. Nucleic Acids Res 40:11518–11530. doi: 10.1093/nar/gks913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. 2007. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev 21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fontana J, Cardone G, Heymann JB, Winkler DC, Steven AC. 2012. Structural changes in influenza virus at low pH characterized by cryo-electron tomography. J Virol 86:2919–2929. doi: 10.1128/JVI.06698-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 41.Heymann JB, Cardone G, Winkler DC, Steven AC. 2008. Computational resources for cryo-electron tomography in Bsoft. J Struct Biol 161:232–242. doi: 10.1016/j.jsb.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frangakis AS, Hegerl R. 2001. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol 135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- 43.Frank J. 2006. Three-dimensional electron microscopy of macromolecular assemblies. Oxford University Press, New York, NY. [Google Scholar]
- 44.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Carteau S, Gorelick RJ, Bushman FD. 1999. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J Virol 73:6670–6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engelman A, Englund G, Orenstein JM, Martin MA, Craigie R. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol 69:2729–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson BC, Métifiot M, Ferris A, Pommier Y, Hughes SH. 2013. A homology model of HIV-1 integrase and analysis of mutations designed to test the model. J Mol Biol 425:2133–2146. doi: 10.1016/j.jmb.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Briggs JA, Simon MN, Gross I, Kräusslich HG, Fuller SD, Vogt VM, Johnson MC. 2004. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol 11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 49.Lanman J, Lam TT, Emmett MR, Marshall AG, Sakalian M, Prevelige PE Jr. 2004. Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange. Nat Struct Mol Biol 11:676–677. doi: 10.1038/nsmb790. [DOI] [PubMed] [Google Scholar]
- 50.Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW Jr, Sowder RC II, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. 2006. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol 80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu Z, Dobro MJ, Woodward CL, Levandovsky A, Danielson CM, Sandrin V, Shi J, Aiken C, Zandi R, Hope TJ, Jensen GJ. 2013. Unclosed HIV-1 capsids suggest a curled sheet model of assembly. J Mol Biol 425:112–123. doi: 10.1016/j.jmb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu W, Thomas JA, Cheng N, Black LW, Steven AC. 2012. Bubblegrams reveal the inner body of bacteriophage phiKZ. Science 335:182. doi: 10.1126/science.1214120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng N, Wu W, Watts NR, Steven AC. 2014. Exploiting radiation damage to map proteins in nucleoprotein complexes: the internal structure of bacteriophage T7. J Struct Biol 185:250–256. doi: 10.1016/j.jsb.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conway JF, Trus BL, Booy FP, Newcomb WW, Brown JC, Steven AC. 1993. The effects of radiation damage on the structure of frozen hydrated HSV-1 capsids. J Struct Biol 111:222–233. doi: 10.1006/jsbi.1993.1052. [DOI] [PubMed] [Google Scholar]
- 55.Leapman RD, Sun S. 1995. Cryo-electron energy loss spectroscopy: observations on vitrified hydrated specimens and radiation damage. Ultramicroscopy 59:71–79. doi: 10.1016/0304-3991(95)00019-W. [DOI] [PubMed] [Google Scholar]
- 56.Meents A, Gutmann S, Wagner A, Schulze-Briese C. 2010. Origin and temperature dependence of radiation damage in biological samples at cryogenic temperatures. Proc Natl Acad Sci U S A 107:1094–1099. doi: 10.1073/pnas.0905481107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stauffer S, Rahman SA, de Marco A, Carlson LA, Glass B, Oberwinkler H, Herold N, Briggs JA, Muller B, Grunewald K, Krausslich HG. 2014. The nucleocapsid domain of Gag is dispensable for actin incorporation into HIV-1 and for association of viral budding sites with cortical F-actin. J Virol 88:7893–7903. doi: 10.1128/JVI.00428-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fenwick CW, Tremblay S, Wardrop E, Bethell R, Coulomb R, Elston R, Faucher A-M, Mason S, Simoneau B, Tsantrizos Y, Yoakim C. 2011. Resistance studies with HIV-1 non-catalytic site integrase inhibitors. Antivir Ther 16(Suppl 1):A9. [Google Scholar]
- 59.Bukovsky A, Gottlinger H. 1996. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J Virol 70:6820–6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol 76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen L, Peterson S, Sedaghat AR, McMahon MA, Callender M, Zhang H, Zhou Y, Pitt E, Anderson KS, Acosta EP, Siliciano RF. 2008. Dose-response curve slope sets class-specific limits on inhibitory potential of anti-HIV drugs. Nat Med 14:762–766. doi: 10.1038/nm1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rabi SA, Laird GM, Durand CM, Laskey S, Shan L, Bailey JR, Chioma S, Moore RD, Siliciano RF. 2013. Multi-step inhibition explains HIV-1 protease inhibitor pharmacodynamics and resistance. J Clin Invest 123:3848–3860. doi: 10.1172/JCI67399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cochrane AW, McNally MT, Mouland AJ. 2006. The retrovirus RNA trafficking granule: from birth to maturity. Retrovirology 3:18. doi: 10.1186/1742-4690-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Forshey BM, Aiken C. 2003. Disassembly of human immunodeficiency virus type 1 cores in vitro reveals association of Nef with the subviral ribonucleoprotein complex. J Virol 77:4409–4414. doi: 10.1128/JVI.77.7.4409-4414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lyonnais S, Gorelick RJ, Heniche-Boukhalfa F, Bouaziz S, Parissi V, Mouscadet JF, Restle T, Gatell JM, Le Cam E, Mirambeau G. 2013. A protein ballet around the viral genome orchestrated by HIV-1 reverse transcriptase leads to an architectural switch: from nucleocapsid-condensed RNA to Vpr-bridged DNA. Virus Res 171:287–303. doi: 10.1016/j.virusres.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Accola MA, Ohagen A, Gottlinger HG. 2000. Isolation of human immunodeficiency virus type 1 cores: retention of Vpr in the absence of p6(gag). J Virol 74:6198–6202. doi: 10.1128/JVI.74.13.6198-6202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Briggs JA, Wilk T, Welker R, Kräusslich HG, Fuller SD. 2003. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J 22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Welker R, Hohenberg H, Tessmer U, Huckhagel C, Kräusslich HG. 2000. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J Virol 74:1168–1177. doi: 10.1128/JVI.74.3.1168-1177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kutluay SB, Zang T, Blanco-Melo D, Powell C, Jannain D, Errando M, Bieniasz PD. 2014. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 159:1096–1109. doi: 10.1016/j.cell.2014.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee S-K, Potempa M, Swanstrom R. 2012. The choreography of HIV-1 proteolytic processing and virion assembly. J Biol Chem 287:40867–40874. doi: 10.1074/jbc.R112.399444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 72.Butan C, Winkler DC, Heymann JB, Craven RC, Steven AC. 2008. RSV capsid polymorphism correlates with polymerization efficiency and envelope glycoprotein content: implications that nucleation controls morphogenesis. J Mol Biol 376:1168–1181. doi: 10.1016/j.jmb.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allen P, Worland S, Gold L. 1995. Isolation of high-affinity RNA ligands to HIV-1 integrase from a random pool. Virology 209:327–336. doi: 10.1006/viro.1995.1264. [DOI] [PubMed] [Google Scholar]
- 74.Dimonte S, Babakir-Mina M, Aquaro S. 2014. HIV-1 B-subtype capsid protein: a characterization of amino acid's conservation and its significant association with integrase signatures. Virus Genes 48:429–437. doi: 10.1007/s11262-014-1039-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.