ABSTRACT
Poultry exposure is a major risk factor for human H7N9 zoonotic infections, for which the mode of transmission remains unclear. We studied the transmission of genetically related poultry and human H7N9 influenza viruses differing by four amino acids, including the host determinant PB2 residue 627. A/Silkie chicken/HK/1772/2014 (SCk1772) and A/HK/3263/14 (HK3263) replicated to comparable titers in chickens, with superior oropharyngeal over cloacal shedding; both viruses transmitted efficiently among chickens via direct contact but inefficiently via the airborne route. Interspecies transmission via the airborne route was observed for ferrets exposed to the SCk1772- or HK3263-infected chickens, while low numbers of copies of influenza viral genome were detected in the air, predominantly at particle sizes larger than 4 μm. In ferrets, the human isolate HK3263 replicated to higher titers and transmitted more efficiently via direct contact than SCk1772. We monitored “intrahost” and “interhost” adaptive changes at PB2 residue 627 during infection and transmission of the Sck1772 that carried E627 and HK3263 that carried V/K/E polymorphism at 60%, 20%, and 20%, respectively. For SCk1772, positive selection for K627 over E627 was observed in ferrets during the chicken-to-ferret or ferret-to-ferret transmission. For HK3263 that contained V/K/E polymorphism, mixed V627 and E627 genotypes were transmitted among chickens while either V627 or K627 was transmitted to ferrets with a narrow transmission bottleneck. Overall, our results suggest direct contact as the main mode for H7N9 transmission and identify the PB2-V627 genotype with uncompromised fitness and transmissibility in both avian and mammalian species.
IMPORTANCE We studied the modes of H7N9 transmission, as this information is crucial for developing effective control measures for prevention. Using chicken (SCk1772) and human (HK3263) H7N9 isolates that differed by four amino acids, including the host determinant PB2 residue 627, we observed that both viruses transmitted efficiently among chickens via direct contact but inefficiently via the airborne route. Chicken-to-ferret transmission via the airborne route was observed, along with the detection of viral genome in the air at low copy numbers. In ferrets, HK3263 transmitted more efficiently than SCk1772 via direct contact. During the transmission of SCk1772 that contained E and HK3263 that contained V/K/E polymorphism at PB2 residue 627, positive selections of E627 and K627 were observed in chickens and ferrets, respectively. In addition, PB2-V627 was transmitted and stably maintained in both avian and mammalian species. Our results support applying intervention strategies that minimize direct and indirect contact at the poultry markets during epidemics.
INTRODUCTION
Human zoonotic infections by avian influenza viruses of various subtypes (H5N1, H9N2, H7N9, H6N1, H10N8, and H5N6) are of considerable, but dissimilar, public health concern. The zoonotic and pandemic potential of H7N9 avian influenza virus is evidenced by the rapid surge of human H7N9 disease since 2013 (1). Detection of H7N9 patients through the influenza-like illness surveillance suggests a substantial number of mild or subclinical H7N9 infections in humans (2) and further implies the zoonotic potential of this virus.
The H7N9 virus acquired its surface hemagglutinin (HA) and neuraminidase (NA) glycoproteins from ducks and wild birds and the six internal genes from the H9N2 virus that has been endemic in Asian and Middle Eastern countries for more than a decade (3–6). With expanded geographic distribution, the H7N9 viruses further reassorted with regional H9N2 viruses and increased their genetic diversity in the internal genes (7–9). Genetic comparison of human and poultry H7N9 isolates has identified mammalian adaptive mutations; in particular, many of the human H7N9 isolates contained the well-established mammalian adaptive signature K627 or N701 in the PB2 protein, as opposed to the E627 or D701 signature found in avian H7N9 isolates (7, 8, 10, 11). It is known that the E627K substitution, which confers increased viral replication and transmissibility in mammalian hosts (12–18), may emerge during the viral replication and adaptation inside human hosts, as seen in the Dutch H7N7 human case in 2003 (19, 20). However, the relative fitness of E627 and K627 during the avian-mammalian interspecies transmission of the H7N9 viruses is not fully characterized. In addition to the E627K mutation, V627 has also been identified in the PB2 protein of a human H7N9 isolate (A/Hong Kong/5731/2014; GISAID, accession no. EPI520861). The E627V mutation has been previously reported for H9N2 avian influenza viruses circulating in Hong Kong (A/chicken/Hong Kong/YU158/2011 [GenBank accession no. KF260870] and A/chicken/Hong Kong/JV75/2011 [GenBank accession no. KF260871]), Israel (29 insolates in 2000 to 2006) (21), and Egypt (A/chicken/Egypt/BSU-CU/2011) (22). Valine is nonpolar compared to the positively charged lysine or the negatively charged glutamic acid at residue 627. Applying the minireplicon reporter assay with polymerase proteins derived from the A/Puerto Rico/8/34 virus, the relative polymerase activity of PB2-627V was in between those of PB2-627E and PB2-627K in both human 293T and avian DF-1 cell lines (23).
Epidemiological studies identified exposure to the infected poultry or the contaminated environment at live-poultry markets as one of the major risk factors for human H7N9 infections (24–27). However, approximately 45% of H7N9 patients did not have a definite poultry contact history prior to infection (25). While influenza virus may be transmitted among humans via fomites, droplets, or aerosols (28), the mode of H7N9 transmission remains unclear. Closure of live-poultry markets was effective in temporarily reducing human cases (29–31), but close to 200 human infections were reported during the winter of 2014-2015, with a wider geographic distribution to Xinjiang and Jilin in 2014 (1). While chicken is the most common species in the live-poultry markets harboring the H7N9 virus (6, 32, 33), the predominant mode that facilitates the spread of H7N9 virus among poultry is also not fully understood. Recent studies have established the potential of wild bird species as carriers for H7N9 viruses and the potential transmission to domestic poultry via the contact but not the airborne route (34). To develop effective control measures for H7N9 infections among poultry and from poultry to humans, it is crucial to acquire a comprehensive understanding of the modes of the transmission of the H7N9 virus.
We studied the modes of transmission of a pair of genetically related human and poultry H7N9 viruses in chickens and ferrets. The poultry H7N9 virus (A/SCk/HK/1772-3/14, here referred to as SCk1772) was isolated from Silkie chickens imported from China without apparent clinical signs in January 2014. The isolation of the SCk1772 virus has led to the decision of the Hong Kong government to cull 22,604 poultry. The human H7N9 virus (A/Hong Kong/3263/14, here called HK3263) was isolated from the nasopharyngeal swab of a patient with a nonfatal case. Sanger sequencing results showed that these two viruses only differed by 4 amino acids, including PB2 residue 627. Clonal sequencing confirmed that SCk1772 virus retained the avian signature E, while HK3263 carried V/K/E polymorphism at PB2 residue 627. We thus further investigated the “interhost” and “intrahost” adaptive changes at PB2 residue 627 during interspecies transmission. Our results support the species-specific adaptive changes of E627 and K627 in avian and mammalian hosts, respectively. Furthermore, the V627 genotype, which was transmitted and stably maintained in both avian and mammalian species, may serve as an intermediate genotype and facilitate interspecies jump.
MATERIALS AND METHODS
Viruses and cells.
The avian A/SCk/HK/1772/14 H7N9 virus was isolated from pooled oropharyngeal swabs of Silkie chickens after one passage in the allantoic cavity of embryonated chicken eggs. The virus was further propagated once in eggs prior to the experiments. The A/Hong Kong/3263/14 virus was kindly provided by Department of Health, Hong Kong; it was isolated from the nasopharyngeal swab of a 65-year-old male patient who eventually recovered from the infection. The A/Hong Kong/3263/14 virus has been propagated twice in Madin-Darby canine kidney (MDCK) cells prior to the experiments. All experiments involving live H7N9 viruses were performed in a biosafety level 3 containment facilities at the University of Hong Kong, in compliance with all applicable guidelines. MDCK cells were obtained from the ATCC and were maintained in minimal essential medium (MEM) supplemented with 10% fetal calf serum.
Infectivity.
The 50% tissue culture infectious dose (TCID50) was determined as described previously (35). The 50% egg infectious dose (EID50) was determined in 10-day-old embryonated eggs with 100-μl serially diluted samples; the eggs were incubated at 37°C for 4 days. A hemagglutination assay using 5% turkey or chicken erythrocytes was performed to determine the endpoint of infection; the infectious dose was calculated using the Reed-Muench method.
Ethics statement.
All animal experiments were approved by the Committee on the Use of Live Animals in Teaching and Research (CULATR) at the University of Hong Kong (CULATR number 3231-14), by following the Code of Practice for care and use of animals for experimental purposes established by the Animal Welfare Advisory Group, approved by the Government of the Hong Kong.
Chicken-to-chicken transmission.
The experiments were performed inside walk-in bioisolators with directional airflow (Fig. 1). The airflow rates (mean ± [SD]) were adjusted to 81.3 ± 1.2 ft per min, which led to 42.3 ± 0.2 (mean ± SD) air changes per h for all walk-in bioisolators. White Leghorn chickens were hatched from specific-pathogen-free (SPF) eggs and raised in a clean environment. Nine-week-old SFP chickens were inoculated via a natural route (0.1 ml by ocular route, 0.1 ml intranasally, and 0.3 ml intratracheally) with 5 × 106 EID50s of the SCk1772 or HK3263 viruses in 0.5 ml of phosphate-buffered saline (PBS) and served as donors for the transmission experiment. An additional six chickens were also inoculated with 5 × 106 EID50s of the SCk1772 or HK3263 virus, and two were sacrificed at 2, 4, and 6 days postinoculation (dpi) for pathology examinations (data not shown). At 1 dpi, two direct-contact chickens and three airborne-contact chickens were introduced to evaluate the transmission efficiency of SCk1772 or HK3263. The inoculated chickens were housed together with the direct-contact chickens in one cage, while the airborne-contact chickens were housed in a separate cage 4 cm apart; the two cages were separated by a perforated (0.5-cm holes separated by 1 cm) stainless steel divider (Fig. 1). Oropharyngeal and cloacal swabs were collected every other day from all chickens, and the body weights of the donor and direct-contact chickens were monitored from 2 dpi (1 day postcontact [dpc]) to 14 dpi (13 dpc). Pre- and postcontact sera collected at −7 dpi and 18 dpi (17 dpc) were applied to monitor anti-HA antibody using hemagglutination inhibition assay (HI) or anti-influenza A NP antibody using an ID Screen influenza A virus antibody competition enzyme-linked immunosorbent assay (ELISA) kit (ID.vet) by following the manufacturer's instructions.
FIG 1.
Experiment settings for chicken-to-chicken and chicken-to-ferret transmission. Donor chickens (n = 3; middle shelf) were inoculated via a natural route with 5 ×106 EID50s of the SCk1772 or HK3263 virus. An additional 6 chickens (bottom shelf) were inoculated with SCk1772 or HK3263 virus for pathology examinations at 2, 4, and 6 dpi (data not shown). At 1 dpi, two direct-contact chickens were introduced and cohoused with the three inoculated donors; in addition, three naive airborne-contact chickens were introduced to the adjacent cage to evaluate the airborne-transmission efficiency of SCk1772 or HK3263. The airborne-contact chickens were separated from the donors and direct-contact chickens with a perforated (0.5-cm holes separated by 1 cm) stainless steel divider (dotted blue line). Two naive ferrets were introduced at 1 dpi and were housed separately above the chickens. The two ferrets were separated with a solid stainless steel divider (solid blue line).
H5 vaccination in chickens.
The H5 vaccine (Re-6) was developed by the Harbin Veterinary Research Institute and has been used in China since 2012. White Leghorn SPF chickens were vaccinated intramuscularly in the pectoral muscle with 0.5 ml of the vaccine on days 21 and 42. Sera were collected prior to the experiments from all vaccinated chickens with confirmed HI titers against the vaccine antigen ranging from 768 to 4,096.
Chicken-to-ferret transmission.
Two naive ferrets were introduced at 1 dpi into the isolator where the chicken-to-chicken transmission experiments were performed. The ferrets were housed individually in two separate cages above the chicken cages to prevent any physical contact (Fig. 1). The ferrets were monitored for clinical signs daily, and the nasal washes were collected every other day from 2 to 12dpc to monitor viral shedding. The chickens and ferrets were handled on alternate dates to avoid cross-contamination. Each animal was handled with separate tools, with frequent decontamination in between procedures. Seroconversion was determined using paired sera collected at −3 dpi and 18 dpi (17 dpc) to monitor anti-influenza A virus NP antibody using an ID Screen Influenza A virus antibody competition ELISA kit (ID.vet) by following the manufacturer's instructions.
Ferret-to-ferret transmission.
The experiments were performed as described previously (35). In brief, donor ferrets (n = 2 for each virus) were inoculated intranasally with 105 TCID50s of the SCk1772 or the HK3263 virus in 0.5 ml of medium. At 1 dpi, each donor was cohoused with one naive direct-contact ferret and one respiratory-droplet-contact ferret in the adjacent cage. Clinical signs were monitored and nasal washes were collected daily from 1 to 10 dpc. The naive contact animals were handled first with separate tools, with frequent Virkon decontamination in between procedures. Pre- and postcontact sera collected at 2 dpi and 18 dpi (17 dpc) were applied to monitor anti-influenza A NP antibody using an ID Screen influenza A virus antibody competition ELISA kit (ID.vet).
RNA extraction and qRRT-PCR for viral load quantification in chicken swabs.
Chicken oropharyngeal or cloacal swabs were collected in 1,500 μl of virus transport medium (VTM). RNA was extracted from 200 μl of the medium using NucliSENS easyMAG automated nucleic acid extraction system (bioMérieux, Boxtel, The Netherlands) and resuspended in 50 μl of NucliSENS easyMAG extraction buffer 3. H7 quantitative real-time reverse transcription-PCR (qRRT-PCR) was performed using the LightCycler 480 system (Roche Applied Science, Mannheim, Germany) and the One Step RT-PCR kit (Qiagen, Inc., Hilden, Germany) (36). In each H7 qRRT-PCR run, a standard curve for virus quantification was established with plasmid DNA containing the HA sequence derived from influenza virus A/Anhui/1/2013 (37). Five microliters of the eluted RNA was amplified in a 25-μl reaction mixture, in a fashion similar to that of the plasmid standard. The copy numbers were extrapolated from the standard curve. The limit of detection was determined to be 15.7 copies per reaction or 785 copies/ml of the original VTM.
Air sampling.
NIOSH samplers that are capable of collecting aerosols at three different size ranges (>4, 1 to 4, and <1 μm) were applied to sample the air at the walk-in isolators where the infected chickens were housed. During the transmission experiments when air samplings were performed, the temperatures were recorded at 20 to 21°C and the relative humidity was recorded at 79%. Samplings were performed for 30 min at 3.5 liter air per minute every other day from 2 to 8 dpi. MEM was added to retrieve the particles collected by the NIOSH sampler at each of the three fractions (1 ml per fraction). Viral RNA was extracted (RNeasy; Qiagen) using 400 μl of the medium from each of the three fractions with RNA eluted in 30 μl of nuclease-free water. Influenza virus M gene copy numbers were determined by quantitative real-time RT-PCR (qRRT-PCR) using the LightCycler 480 system (Roche Applied Science, Mannheim, Germany). Experiments were performed in 25-μl reaction mixtures with 5 μl of the eluted RNA and the AgPath-ID One-Step RT-PCR reagents (Life Technology) with primers (forward, 5′-GACCRATCCTGTCACCTCTGAC-3′, and reverse, 5′-AGGGCATTYTGGACAAAKCGTCTA-3′) and probes (5′-6-carboxyfluorescein [FAM]-TGCAGTCCTCGCTCACTGGGCACG-black hole quencher 1 [BHQ1]-3′) reported by the WHO.
Genotyping assay for PB2 residue 627.
A two-step TaqMan single nucleotide polymorphism (SNP) genotyping assay with absolute quantitation was applied to quantify the proportion of V/K/E at PB2 residue 627. The primers (forward, 5′-GGACATTTGACACTGTTCAAATAATAAAGCT-3′, and reverse, 5′-CTCACGTTCACAGTTAGAGAAGAGA-3′) and probes (5′-VIC-CCCGCCGAAGCAGAG-MGB-3′ and 5′-FAM-CCGCCGGAGCAG-MGB-3′) for the first reaction differentiate the proportion of A/G at the first nucleotide of the codon encoding residue 627, which provided an estimate for the copy numbers of K627 (AAG); the primers (forward, 5′-CTATTACCATTTGCAGCAGCCC-3′, and reverse, 5′-CTCACGTTCACAGTTAGAGAAGAGA-3′) and probes (5′-VIC-CGCCGGAGCAGAGT-MGB-3′ and 5′-FAM-CGCCGGTGCAGAGT-MGB-3′) for the second reaction may differentiate the proportion of A/T at the second nucleotide of the codon, which provided an estimate for the copy numbers for V627 (GTG) and E627 (GAA or GAG); the proportion was then calculated based on the copy numbers of each genotype. The specificity of the two-step genotyping assay was evaluated using control PB2 plasmids (PB2 gene derived from A/Shanghai/2/2013) each encoding K627, E627, or V627 at concentrations ranging from 101 to 106 copies per μl. The genotyping assay was only performed using samples containing >100 copies/μl of the PB2 gene.
Full-genome sequencing and clonal sequencing.
Total RNA was extracted from the viral culture supernatants (RNeasy; Qiagen). The full genome was amplified by RT-PCR using the universal primers (38) and subjected to dideoxynucleotide sequencing. To determine the genotype at PB2 residue 627, a 488-bp PCR product (forward, 5′-TGGGNGTRGATGARTAYTC-3′, and reverse, 5′-ATTCCKGAGCCTCTCACATTCACAG-3′) fragment spanning the region of residue 627 was amplified by RT-PCR (Qiagen) and cloned into the pCR4-TOPO vector (Invitrogen). Plasmid DNA was isolated (Miniprep; Qiagen) and subjected to dideoxynucleotide sequencing using the amplification primers.
Statistical analysis.
The total amount of virus shed in infected chickens was estimated by calculating the area under the curve (AUC) values using the viral copy numbers determined at different days postinoculation or postcontact. The AUC values were compared using the Mann Whitney test or the Wilcoxon matched-pair signed-rank test. A P value of <0.05 was considered statistical significant.
Sequence data accession numbers.
The full genome of SCk1772 was submitted to GISAID (accession no. EPI505122, EPI505123, and EPI553197 to EPI553202).
RESULTS
Characterization of the human and chicken H7N9 influenza virus isolates.
The SCk1772 virus was isolated from the pooled oropharyngeal swabs of Silkie chickens in embryonated chicken eggs. The SCk1772 stock virus showed higher infectivity in eggs (7.88 log10 EID50s/ml) than that in MDCK cells (6.46 log10 TCID50s/ml). Similar to the majority of the human and avian H7N9 isolates, SCk1772 contained the mammalian adaptive Q226L mutation at the HA receptor-binding domain; Q226L and G228S have been observed to confer increased binding to alpha2,6-linked sialic acids in human H2N2 and H3N2 influenza viruses (39). In addition, SCk1772 contained the avian-like signatures E627 and D701 in the PB2 protein.
The human H7N9 virus HK3263 was isolated from the nasopharyngeal swab of a 65-year-old male with a history of travel to Guangdong Province, China, prior to disease onset. The virus has been propagated twice in MDCK cells. The HK3263 stock virus showed comparable infectivity in eggs (7.75 log10 EID50s/ml) and MDCK cells (7.68 log10 TCID50s/ml). The genetic sequences of SCk1772 and HK3263 (GISAID; accession no. EPI507080 to EPI507087) were highly homologous and differed by 4 amino acids in the PB2 (E versus V/K/E at residue 627), PB1 (M versus L at residue 372), PA (S versus N at residue 648), and NP (G versus R at residue 485) proteins. Sanger sequencing of the original clinical specimen of HK3263 identified mixed A/G at the first codon and A/T at the second codon for PB2 residue 627. We performed clonal sequencing using the HK3263 stock virus and observed that 6 out of 10 clones contained GTG (V), 2 out of 10 clones contained AAG (K), 1 out of 10 clones contained GAG (E), and 1 out of 10 clones contained GAA (E). In parallel, clonal sequencing for the SCk1772 stock virus at PB2 residue 627 showed homogenous E (GAG) in 17 of 17 clones.
Infectivity and transmissibility of the human and chicken H7N9 viruses in chickens.
It is not known if the human H7N9 viruses with mammalian adaptive changes would transmit among chickens at an efficiency similar to that of the poultry isolates. We evaluated transmissibility of SCk1772 and HK3263 via the direct-contact or airborne route in 9-week old-specific-pathogen-free White Leghorn chickens. Donor chickens (n = 3 per group) were inoculated with 5 × 106 EID50s of the SCk1772 or HK3263 viruses in 0.5 ml of PBS via a natural route (ocular, nasal, or tracheal). An additional six chickens were inoculated for monitoring histopathological changes on 2, 4, and 6 days postinoculation (dpi) (data not shown). At 1 dpi, direct-contact chickens (n = 2) and airborne-contact chickens (n = 3) were introduced (Fig. 1). Transmission was monitored by determining the RNA copy numbers for the HA genes of the SCk1772 and HK3263 viruses from oropharyngeal and cloacal swabs by qRRT-PCR and by monitoring seroconversion.
None of the donor chickens inoculated with SCk1772 or HK3263 viruses showed apparent clinical signs during the course of infection. The mean oropharyngeal peak titers were comparable between chickens inoculated with SCk1772 and HK3263 (5.4 log10 RNA copies/μl for both), and the mean maximum weight losses were detected at 4 dpi (mean ± % SD;5.0 ± 4.1% and 3.7 ± 5.5%, respectively). The area under the curve (AUC) values were calculated to estimate the total amount of virus shed via the oropharyngeal and cloacal routes (Table 1). The SCk1772 and HK3263 viruses replicated to comparable titers in the upper respiratory tract of the inoculated chickens (Fig. 2), with peak oropharyngeal shedding detected at 2 and 4 dpi and comparable mean AUC values (Table 1). Comparable degrees of cloacal shedding were also observed from SCk1772- and HK3263-inoculated chickens, with the mean peak titers detected at 4 and 6 dpi (Fig. 2). Higher viral loads were detected in the oropharyngeal swabs than the cloacal swabs from chickens inoculated with either SCk1772 or HK3263 (P = 0.03, Wilcoxon matched-pair signed-rank test) (Table 1). Overall, the oropharyngeal shedding was superior and generally preceded cloacal shedding for chickens inoculated with SCk1772 or HK3263.
TABLE 1.
Total virus shedding in the oropharyngeal and cloacal swabs, approximated by the AUC values for chickens inoculated or infected by SCk1772 or HK3263 H7N9 virusa
| Swab type | AUC, mean ± SD (95% CI) |
|||||
|---|---|---|---|---|---|---|
| SCk1772 |
HK3263 |
|||||
| Donor (n = 6) | Direct contact (4/4) | Airborne contact (0/6) | Donor (n = 6) | Direct contact (4/4) | Airborne contact (3/6) | |
| Oropharyngeal | 22.4 ± 1.8 (20.5–24.3) | 27.3 ± 3.7 (21.4–33.3) | NA | 20.5 ± 3.2 (17.2–23.8) | 22.8 ± 1.4 (20.5–25.1) | 29.5 ± 1.0b (27.1–32.0) |
| Cloacal | 9.8 ± 4.4 (5.2–14.4) | 14.0 ± 11.7 (−4.7–32.6) | NA | 7.3 ± 5.9 (1.1–13.5) | 4.2 ± 6.8 (−6.6–15.0) | 12.7 ± 0.8b (10.7–14.6) |
The AUC values were calculated to approximate the total virus shed (copies/μl) from the oropharyngeal and cloacal swabs of chickens at 2, 4, 6, 8, 10, 12, and 14 dpi (equivalent to 1, 3, 5, 7, 9, 11, and 13 dpc). CI, confidence interval; NA, not applicable, as no transmission was observed.
Mean AUC ± SD from three infected chickens only.
FIG 2.
Transmission of the chicken SCk1772 and human HK3263 H7N9 isolates in SPF chickens. Donor chickens were inoculated with either 5 × 106 EID50s of SCk1772 or HK3263, and naive direct-contact or airborne-contact chickens were introduced at 1 dpi. Transmission of SCk1772 among chickens was monitored by determining viral RNA copies per microliter in the oropharyngeal swabs or cloacal swabs (A). Transmission of HK3263 among chickens was monitored by determining viral RNA copies per microliter in the oropharyngeal swabs or cloacal swabs (B). The mean viral RNA copy ± SEM is shown, with the solid line representing the limit of detection at 3.14 copies/μl. The proportion of chickens that seroconverted is shown above each group.
Transmission of SCk1772 or HK3263 to naive cage mates via direct contact with shared food and water source (troughs) was rapidly detected on 2 dpi (1 day postcontact [dpc]). Peak oropharyngeal titers were detected from the SCk1772-infected (4.6 to 6.1 log10 RNA copies/μl) or the HK3263-infected (3.9 to 5.1 log10 RNA copies/μl) cage mates at 3 or 5 dpc (Fig. 2). Marginally higher oropharyngeal and cloacal viral shedding was detected for the SCk1772-infected chickens than for those infected with the HK3263 virus, although the differences were not statistically significant (Table 1). Similarly, higher oropharyngeal shedding over the cloacal shedding was observed for the chickens infected by SCk1772 or HK3263 via direct contact (Table 1).
Transmission of SCk1772 or HK3263 virus between chickens via the airborne route was less efficient than by the direct-contact route. There were only low copy numbers of viral RNA (0.6 to 1.3 log10 RNA copies/μl) detected from the SCk1772 airborne-contact chickens without seroconversion at 17 dpc. These SCk1772 airborne-contact chickens were thus considered not infected, and no transmission to the adjacent cage occurred via the airborne route. On the other hand, transmission of HK3263 via the airborne route was observed in one of the two replicate experiments. Viral shedding was first detected from the oropharyngeal swab of one out of three airborne-contact chickens at 3 dpc (4.3 log10 RNA copies/μl), and high copy numbers of viral RNA (5.5 to 5.9 log10 RNA copies/μl) were detected subsequently from its two cage mates at 7 dpc. Viral RNA could be detected in both the oropharyngeal and cloacal samples from the chickens infected by HK3263 via the airborne route, with a higher viral load detected in the oropharyngeal swabs (AUC [mean ± SD] = 29.5 ± 1.0) over the cloacal swabs (AUC = 12.7 ± 0.8). The peak oropharyngeal viral loads were detected at 5 or 7 dpc and ranged from 5.4 to 5.9 log10 RNA copies/μl (Fig. 2); seroconversion was observed for 3 out of 6 exposed chickens at 17 dpc, with HI titers of 128 to 256.
Chickens in China are mandatorily vaccinated against H5N1 virus using the inactivated recombinant Re-6 vaccine derived from a clade 2.3.2.1 H5N1 virus, A/duck/Guangdong/S1322/2010. We evaluated the effect of the two-dose Re-6 vaccination on the replication and transmission of the SCk1772 virus. After challenge, the vaccinated donor chickens shed virus in the oropharyngeal (AUC = 20.4 ± 3.4) and cloacal (AUC = 13.6 ± 7.0) swabs at titers comparable to those from the unvaccinated donor chickens (P = 0.39, Mann-Whitney test) (Table 1). Among the vaccinated chickens, transmission from donors to the direct contacts was as efficient as observed among the nonvaccinated chickens. All chickens in direct contact with the donors started to shed virus in the oropharynx within 1 dpc, with an AUC value comparable to that of the unvaccinated chickens (P = 0.66, Mann-Whitney test) (Table 1). The peak viral titers detected in the oropharyngeal swabs of the Re-6-vaccinated chickens (4.4 to 6.1 log10 RNA copies/μl) (Fig. 3) were also comparable to those of the unvaccinated chickens (Fig. 2).
FIG 3.
Transmission of the SCk1772 virus in Re-6-vaccinated chickens. Chickens were vaccinated with Re-6 H5N1 vaccine. Donor chickens were inoculated with 5 × 106 EID50s of SCk1772, and naive direct-contact chickens were introduced at 1 dpi. Oropharyngeal swabs (A) and cloacal swabs (B) were quantified by H7 qRRT-PCR. The mean viral RNA copy ± SEM is shown, with the solid line representing the limit of detection at 3.14 copies/μl. The proportion of chickens that seroconverted is shown above each group.
Overall, the human (HK3263) and chicken (SCk1772) H7N9 viruses replicated to comparable titers in the inoculated and infected contact chickens, with no apparent clinical signs. We observed rapid transmission of both the SCk1772 and HK3263 among chickens via direct contact with shared food and water sources. Although these chickens were likely infected via fomites, direct or indirect contact, we cannot completely exclude the possibility of close-range airborne transmission under this cohousing setting. Transmission via the airborne route to the adjacent cage was inefficient for HK3263 and was not observed in the two replicate groups of SCk1772. Oropharyngeal shedding was superior to the cloacal shedding for SCk1772- and HK3263-inoculated or -infected chickens. Re-6 vaccine conferred no protection against H7N9 challenge in reducing H7 viral loads or preventing transmission of H7N9 virus among chickens.
Stability of the mammalian adaptive changes at PB2 residue 627 during transmission of the human HK3263 virus in chickens.
The HK3263 stock virus contained polymorphism at PB2 residue 627; clonal sequencing confirmed that V627 (GTG) was the dominant population, with 60% (6/10 clones), followed by K627 (AAG), with 20% (2/10 clones), and E627,with 20% (GAG and GAA at 1/10 clone each). We applied a two-step TaqMan SNP assay to differentiate the proportion of V/K/E at residue 627. The first reaction allows differentiating the proportion of A/G at the first nucleotide of the codon, which provided an estimate for the copy numbers of K627 (AAG); the second reaction may differentiate the proportion of A/T at the second nucleotide of the codon, which provided an estimate for the copy numbers of V627 (GTG); the proportion of E627 was deduced after the two reactions were performed. The specificity of the two-step genotyping assay was evaluated using control PB2 plasmids each encoding K627, E627, or V627 at concentrations from 101 to 106 copies per μl. A minor overestimation for E627 (>1%) was observed when the V627 plasmids were used for genotyping. Using this method, we determined the polymorphism of V/K/E at PB2 residue 627 in stock HK3263 virus as 64.6%, 24.9%, and 10.6%, respectively (Fig. 4), which correlated well with the clonal sequencing data.
FIG 4.
Genotyping of V/K/E at PB2 residue 627 among chickens inoculated or infected with human HK3263 H7N9. Two-step genotyping assays were performed to determine the proportion of V/K/E at PB2 residue 627 from the inoculum of HK3263 or the oropharyngeal swabs of chickens in two separate replicates of the transmission experiment (A and B).
We monitored the proportion of V/K/E at PB2 residue 627 during the transmission of HK3263 in chickens by genotyping the viral RNA present in the oropharyngeal swabs. In donor chickens inoculated with HK3263, we observed that the proportion of K627 was rapidly reduced from 24.9% in the inoculum to less than 10% in 4/6 donors at 2 dpi (Fig. 4) and further reduced to less than 5% in 4/6 donors on 4 dpi. Conversely, the proportion of E627 gradually increased, from 10.6% in the inoculum to 24.2 to 33.2% at 4 dpi. Interestingly, the proportion of V627 remained at 61.0 to 71.2% at 4 dpi and did not change significantly in the donor chickens over time. Among chickens infected by HK3263, V627 was transmitted as the dominant genotype in 3/4 direct contacts at 56.7 to 76.5%, with E627 at 23.5 to 43.3% (Fig. 4); K627 was detected transiently in 2/4 direct-contact chickens. In one out of two replicates, HK3263 was transmitted via the airborne route to at least one naive chicken; both V627 and E627 were transmitted, while the proportion of K627 remained below 1% during the course of infection (Fig. 4A). Interestingly, mixed populations were detected in all infected contact chickens regardless of the route of transmission; this suggests a wide transmission bottleneck during transmission of SCk1772 and HK3263 among chickens. Overall, the genotyping results support the greater fitness of E627 over K627 in chickens and suggest that V627 can be stably maintained and transmitted among chickens.
Detection of influenza viral RNA from the air samples collected during transmission.
To evaluate the potential of transmission of the H7N9 viruses via aerosols, NIOSH samplers (40) were used to collect particles at three different size ranges (>4, 1 to 4, and <1 μm) for 30 min at 3.5 liters of air per minute. The air samplings were performed when chickens were handled to collect oropharyngeal and cloacal swabs, which may generate increased dust and fecal particles in the air. We detected the influenza virus M gene in the air samples from 2 to 8 dpi from the walk-in isolators where the chickens infected with either SCk1772 or HK3263 were housed, although the concentrations (threshold cycle [CT] ranged from 37.28 to 44.51) were mostly below the linear range of quantification (3.71 copies per liter of air at a CT of 38.20) (Table 2). The detection of the M gene was predominantly at particle sizes of >4 μm, which supports the inefficient transmission of the H7N9 virus via the airborne route, as particles of <5 μm are more likely to remain suspended in the air and lead to airborne transmission. The CT values for detection, albeit below the limit of quantification, gradually decreased from 2 to 8 dpi, suggesting an increasing amount of influenza virus particles in the air as the H7N9 virus continued to be transmitted among chickens via the direct-contact route. On two sampling occasions, we detected the M gene copy numbers above the linear range of quantification; these were replicates 2 for SCk1772 and HK3263 at 8 dpi (7 dpc) (Table 2). Examining the data on viral shedding (Fig. 2), we noted that the increased cloacal shedding from the SCk1772 direct-contact chickens at 7 dpc and the transmission of HK3263 to the airborne-contact chickens may have contributed to the increased copy numbers of the M gene detected in the air. There was no apparent difference in the detection rate or the copies of M genes detected in the air from the SCk1772 and HK3263 virus groups. None of the samples with the positive M gene detection yielded positive viral culture in MDCK cells.
TABLE 2.
Detection of influenza virus M gene in the air using the NIOSH air samplera
| No. of dpi | Replicate | Detection of influenza virus M gene (CT value) |
|||||
|---|---|---|---|---|---|---|---|
| SCk1772 |
HK3263 |
||||||
| >4 μm | 1–4 μm | <1 μm | >4 μm | 1–4 μm | <1 μm | ||
| 2 | 1 | − | − | − | − | − | − |
| 2 | − | − | − | + (42.43) | − | − | |
| 4 | 1 | + (41.12) | − | − | + (40.80) | − | + (44.51) |
| 2 | + (44.23) | − | − | + (40.57) | + (40.91) | − | |
| 6 | 1 | + (39.15) | − | − | + (39.27) | − | − |
| 2 | + (39.13) | − | − | + (39.28) | − | − | |
| 8 | 1 | + (39.77) | − | − | − | − | − |
| 2 | + (37.46) | + (36.76) | + (38.97) | + (37.28) | − | + (38.58) | |
Air sampling (30 min at 3.5 liter air per minute) was performed every other day when oropharyngeal and cloacal swabs were collected from the chickens. Viral M gene copy numbers were determined by qRRT-PCR with the linear range limit of quantification at 3.71 copies per liter of air at a CT of 38.20.
Chicken-to-ferret transmission potential of the chicken and human H7N9 isolates.
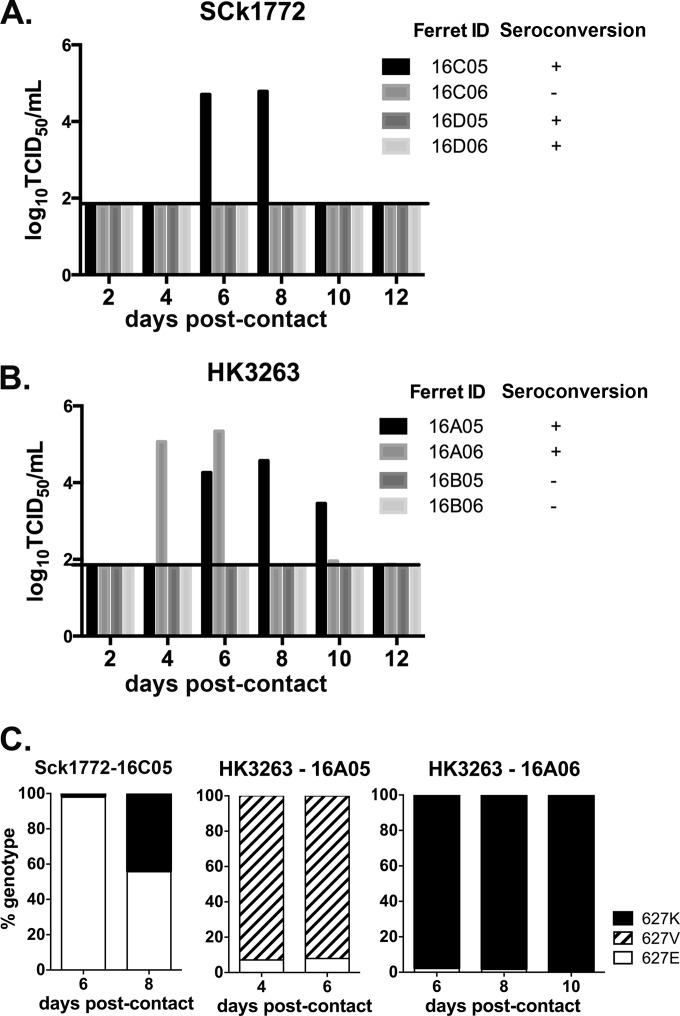
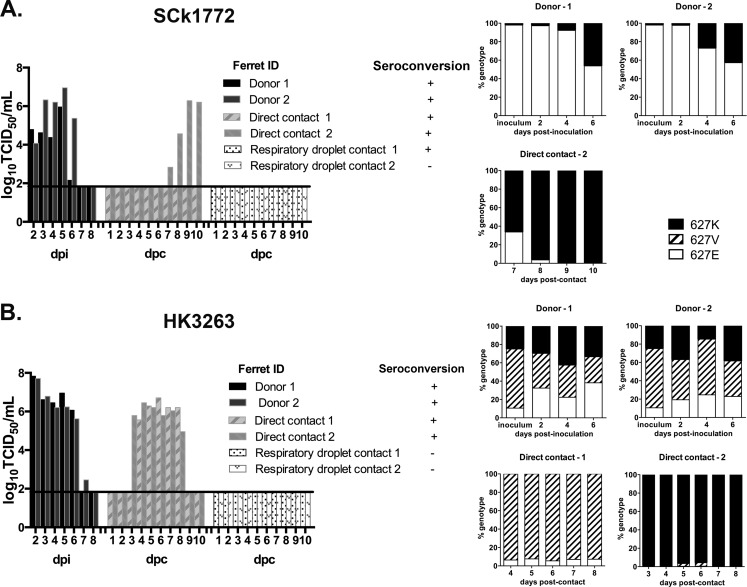
Poultry exposure remains the major risk factor for human zoonotic infections by avian influenza viruses. We therefore evaluated the chicken-to-ferret transmission potential of SCk1772 (Fig. 5A) and HK3263 (Fig. 5B). Naive ferrets were introduced and cohoused with donor and contact chickens at 1 dpi as described (Fig. 1). Ferret nasal washes were collected on alternate dates when the chickens were handled. Viral shedding was detected at 6 dpc in 1/4 ferrets exposed to the SCk1772-infected chickens; however, seroconversion was noted in 3/4 exposed ferrets at 17 dpc (Fig. 5A). Specifically, 1/2 ferrets exposed to SCk1772-infected chickens shed virus in nasal washes in replicate 1, while 2/2 ferrets exposed to SCk1772-infected chickens seroconverted in replicate 2. None of the SCk1772-infected ferrets showed apparent clinical signs over the observation period. The SCk1772 stock virus contained E627 as the dominant genotype. We performed a genotyping assay on viral RNA present in the ferret nasal washes at 6 and 8 dpc to monitor the intrahost adaptive changes at PB2 residue 627. E627 was the dominant genotype transmitted at 6 dpc at 97.9%; however, the proportion of K627 rapidly increased from 1.8% to 44.3% from 6 to 8 dpc (Fig. 5C). This observation suggests that the E627 genotype may transmit from chickens to ferrets but that the mammalian adaptation at PB2 residue 627 may emerge rapidly posttransmission.
FIG 5.
Chicken-to-ferret interspecies transmission efficiency. Naive ferrets were exposed to SPF chickens inoculated or infected with chicken (SCk1772) (A) or human (HK3263) (B) H7N9 virus. Ferret nasal washes were collected every other day and titrated in MDCK cells (log10TCID50 per milliliter) to monitor chicken-to-ferret transmission via the airborne route (A and B). Genotyping of V/K/E at PB2 residue 627 in nasal washes collected from ferrets infected by SCk1772 and HK3263 was also conducted (C).
In comparison, chicken-to-ferret transmission of HK3263 was observed in 2/4 ferrets at 4 and 6 dpc (Fig. 5B) when viral shedding was detected in the nasal washes. The other two contact ferrets remained seronegative at 17 dpc. Specifically, 2/2 ferrets exposed to HK3263-infected chickens shed virus in nasal washes in replicate 1, while 0/2 ferrets exposed to HK3263-infected chickens shed virus or seroconverted in replicate 2. None of the HK3263-infected ferrets showed apparent clinical signs, significant weight loss, or notable temperature change over the observation period. Since HK3263 contained V/K/E polymorphism, we performed the two-step genotyping assay to monitor the interhost and intrahost changes at PB2 residue 627 (Fig. 5C). We observed V627 (92.9%) and K627 (97.9%) being the dominant genotypes transmitted to two separate ferrets, which was distinct from the observation of mixed V/E genotypes being transmitted among chickens (Fig. 4). Furthermore, both the V627 and K627 genotypes were stably maintained within the same host during the course of infection (Fig. 5C). The results suggest that the V627 and K627 genotypes possess higher chicken-to-ferret transmission potential than E627 and suggest a narrow bottleneck during the chicken-to-ferret transmission.
Overall, chicken-to-ferret transmissions via the airborne route were observed for both SCk1772 and HK3263. A previous study that applied the A/Anhui/1/2013 (H7N9) virus showed limited replication in inoculated chickens and poor transmission among chickens and from chickens to ferrets (41). SCk1772 and HK3263 showed efficient replication and transmission to direct-contact chickens, which may contribute to the onward chicken-to-ferret transmission. Interestingly, SCk1772 led to seroconversion without detectable viral shedding in the upper respiratory tracts of 2/3 infected ferrets. From the ferret with detectable virus shedding after SCk1772 infection, we observed initial transmission of the E627 genotype followed by a rapid increase in the proportion of the K627 genotype. From the two ferrets infected with HK3263 containing V/K/E polymorphism, we observed a narrow transmission bottleneck selecting for either the V627 or K627 genotype at transmission; both genotypes were stably maintained within the host during the course of infection.
Transmission efficiency of the human and chicken H7N9 viruses in ferrets.
We further compared the ferret-to-ferret transmissibility of SCk1772 and HK3263. Ferrets inoculated with 105 TCID50s of SCk1772 showed moderately elevated temperature, by 1.1°C and 1.2°C, at 2 dpi, without apparent clinical signs. Transmission of SCk1772 via direct contact was inefficient, as viral shedding was first detected in 1/2 direct-contact ferrets at 7 dpc, with the titer at its peak (6.29 log10 TCID50s/ml) at 9 dpc (Fig. 6A); the AUC value was 10.5. However, seroconversion was observed for both (2/2) direct-contact ferrets. The respiratory-droplet-contact ferrets did not shed detectable virus in the nasal washes, but one (1/2) seroconverted without showing apparent clinical signs (Fig. 6A). A genotyping assay was performed to monitor the changes in PB2 residue 627 from the SCk1772-inoculated donors and the infected direct-contact ferret (Fig. 6A). In both donor ferrets, the proportion of the K627 genotype in the nasal washes gradually increased over time and reached 46.2% and 42.8%, respectively, at 6 dpi. Both the K627 (66.2%) and E627 (33.8%) genotypes were transmitted via direct contact at 7 dpc; however, the K627 genotype rapidly gained dominance (>95%) within the same host (Fig. 6A).
FIG 6.
Transmissibility of the human and chicken H7N9 isolates in ferrets. Donor ferrets were inoculated with 105 TCID50s of SCk1772 (A) or HK3263 (B) intranasally, and naive direct-contact or respiratory-droplet-contact ferrets were introduced at 1 dpi. Nasal washes were collected and titrated in MDCK cells (log10TCID50 per milliliter) from all ferrets to monitor transmission via the direct-contact and airborne routes. Two-step genotyping assays were performed to determine the proportion of V/K/E at PB2 residue 627 from the ferret nasal washes (A and B).
Ferrets inoculated with 105 TCID50s of HK3263 shed higher viral loads in the nasal washes (AUC [mean ± SD] = 27.6 ± 0.5) than did ferrets inoculated with SCk1772 virus (AUC = 17.8 ± 4.8), although the difference was not significant due to the small sample size. Peak titers were also detected earlier for the HK3263 donor ferrets (2 dpi) (Fig. 6B) than for the SCk1772 donors (5 dpi) (Fig. 6A). HK3263 was transmitted more efficiently among ferrets via direct contact than did the SCk1772 virus; viral shedding was detected in the nasal washes of both direct-contact ferrets at 3 and 4 dpc, with peak titers detected at 4 dpc (6.46 log10 TCID50s/ml) and 6 dpc (6.71 log10 TCID50s/ml), respectively (Fig. 6B). The AUC for the two HK3263-infected direct-contact ferrets was 23.3 ± 1.8. Sneezing was noted for the HK3263-inoculated donor ferrets or infected contact ferrets, with moderate temperature elevation (ranging from 0.5 to 1.5°C above baseline). No transmission of HK3263 via the airborne route was observed, as none of the respiratory-droplet-contact ferrets shed virus or seroconverted (Fig. 6B). Genotyping was performed on the nasal washes collected from the HK3263-inoculated donors and infected direct-contact ferrets (Fig. 6B). In both donor ferrets, the proportion of the V/K/E 627 genotypes generally remained stable in the nasal washes over the course of infection, although minor variations were noted. We observed a narrow transmission bottleneck, with V627 (93.6%) being the dominant genotype transmitted to one ferret and K627 (98.8%) being the dominant genotype transmitted to the other ferret. The V627 and K627 genotypes were stably maintained within the infected hosts over the course of infection (Fig. 6B).
Overall, we observed that the human HK3263 isolate transmitted more efficiently among ferrets via direct contact than did the chicken SCk1772 isolate. Genotyping results identified strong positive selection from E to K at PB2 residue 627 among ferrets inoculated or infected with the poultry SCk1772 isolate. For the human HK3263 virus, the V/K/E 627 genotypes remained at comparable proportions in the nasal washes of the inoculated donor ferrets, but the transmission to direct-contact ferrets was dominated by either V627 or K627.
DISCUSSION
We studied the modes of transmission of two genetically related H7N9 viruses isolated in early 2014 from Silkie chickens and a 65-year-old male patient. The chicken (SCk1772) and human (HK3263) H7N9 isolates, which differed by four amino acids in the PB2 (E627V/K/E), PB1 (M372L), PA (S648N), and NP (G485R) proteins, demonstrated efficient direct-contact transmissibility and inefficient airborne transmissibility among chickens. Early and predominant oropharyngeal shedding was noted in chickens infected by SCk1772 or HK3263. Interspecies transmission via the airborne route was observed with ferrets exposed to SCk1772- or HK3263-infected chickens at 4 to 6 dpc. In ferrets, the human isolate HK3263 replicated more efficiently in the upper respiratory tract and transmitted more efficiently to naive direct-contact ferrets than the chicken isolate SCk1772, which predominantly caused asymptomatic infections with seroconversion. Overall, our results identified direct contact as the main mode of transmission for H7N9 viruses. Intervention strategies that minimize poultry-poultry or human-poultry contact frequency should be prioritized during epidemics.
Human and avian H7N9 viruses differ in the adaptive amino acid changes at PB2 residue 627 or 701 (8), suggesting a high selection pressure in favor of K627 and N701 in humans. From the two donor ferrets inoculated with SCk1772, we observed a rapid increase in the proportion of K627 over E627 in the nasal washes during the course of infection. In the two ferrets in which chicken-to-ferret or ferret-to-ferret transmission of SCk1772 was observed, E627 or an E627/K627 mixture was detected in the first positive nasal wash samples, followed by increased proportions of K627 in subsequent nasal washes from both ferrets. While the K627 mutation may enhance viral polymerase activity in mammalian hosts, our results suggest that this adaptive change is not essential for the avian-to-human transmission to occur. The rapid increase in proportion of K627 over E627 would support the contention that the PB2 adaptive change observed in the human H7N9 isolates emerged rapidly within the host after interspecies transmission (19, 20, 42).
The HK3263 virus contained a genetic polymorphism of V (GTG)/K (AAG)/E (GAG and GAA). Considering the avian origin of the H7N9 viruses, the emergence of K (AAG) would require a transition mutation from G to A at the first codon, while the emergence of V (GTG) would require a transversion mutation from A to T at the second codon. Considering the G-to-A and C-to-U mutational bias for influenza in humans (43), it would be more likely to yield the K (AAG) than the V (GTG) adaptive change in a mammalian host. The origin of the V627 genotype observed in the HK3263 isolate remains unknown, but it is likely to be associated with the H9N2-internal gene origin (21, 22). Our results suggest that V627 possesses uncompromised fitness and transmissibility in both chickens and ferrets. This is in agreement with a recent study that applied the mutagenesis approach to explore the plasticity of residue 627 in PB2 (23). The V/K/E polymorphism of HK3263 also allowed us to observe the differences in transmission bottleneck in chickens and ferrets. While mixed V627 and E627 genotypes were transmitted among chickens, only the V627 or K627 genotype was transmitted to four ferrets where chicken-to-ferret or ferret-to-ferret transmission of HK3263 was observed. This observation is in agreement with the recent report of a narrow transmission bottleneck for the H7N9 virus among ferrets (44). However, this narrow transmission bottleneck was not observed for SCk1772, as both E627 and K627 could be detected from one of the two ferrets infected by SCk1772.
Chickens infected with H7N9 viruses portrayed the typical patterns of early onset and predominant oropharyngeal shedding in chickens following low-pathogenic avian influenza virus infection (45) and were in agreement with other H7N9 studies with chickens (11, 41, 46–49). Importantly, chickens inoculated with H9N2 viruses isolated from China also shed in a similar fashion (50, 51). The common tissue tropism shared by the H9N2 and H7N9 viruses would facilitate reassortment, as evidenced by recent surveillance studies (7–9, 52). The internal gene segments 1, 2, 3, 5, and 8 of the SCk1772 and HK3263 viruses we studied were derived from the genetically related clade 3 of the ZJ-HJ/07 lineage of H9N2 viruses and are different from those in the H7N9 viruses reported in the first wave of the epidemic (8). Although the number of ferrets tested was small, the airborne transmissibility of SCk1772 and HK3263 between ferrets was less efficient than that of the A/Shanghai/1/2013 virus, under the same experimental settings (35). Further studies are needed to understand if changes in the internal gene constellation may render the viruses less transmissible in mammals.
We observed limited airborne transmission from chicken to chicken, but interspecies transmission by the airborne route was detected in ferrets exposed to SCk1772- or HK3263-infected chickens. Although we did not determine the minimal infectious dose for SCk1772 and HK3263 in chickens and ferrets, it is possible that chickens and ferrets may differ in their susceptibility for H7N9 viruses by the airborne route. Limited H7N9 studies with chickens support the contention that a high inoculating dose is needed for establishing infection (47, 49). The feasibility of airborne transmission was supported by the detection of low copy numbers of the influenza viral M gene in the air, predominantly at particle sizes of >4 μm. Such large droplets may not remain suspended in the air for a prolonged period and may also confer fomite transmission (28, 53). The air samplings were performed when chickens were handled to collect oropharyngeal and cloacal swabs. It is likely that similar aerosol-generating procedures occur at the live-poultry markets when the birds are handled for inspection by the customers prior to purchase. It is of note that many of the human patients reported no direct physical contact with the market poultry (25), although some of them had visited a market where live poultry were housed; our results would support the possibility that airborne transmission may be one of the modes of zoonotic transmission at live-poultry markets. Reducing use of aerosol generation procedures and improved ventilation at live-poultry markets should be encouraged. Further studies at poultry markets are needed to monitor the quantity and sizes of influenza virus-containing aerosols to assess the airborne-transmission potential of avian influenza viruses.
Rapid transmissions of SCk1772 and HK3263 among chickens via direct contact were observed within 24 h postexposure. Mixing of poultry from different farms allows continuous introduction of susceptible chickens to sustain the chain of H7N9 transmission at live-poultry markets. The lack of apparent clinical signs in H7N9-infected chickens further increases the challenges for outbreak control. Intervention strategies that reduce frequency of direct or indirect contact between poultry from different sources, such as adopting poultry segregation by source farms, slaughtering of unsold chickens at the end of each day to break the infection cycle, and thorough cage cleaning, including change of drinking water and feed, should be considered at live-poultry markets. While closure of live-poultry markets would seem to be the ultimate control measure in area where influenza virus is endemic, practical strategies should be taken in parallel to prevent driving the live-poultry trade underground.
ACKNOWLEDGMENTS
This study was supported by the Health and Medical Research Fund (RRG-01) from the Food and Health Bureau of The Government of Hong Kong, Theme-based Research Scheme (project no. T11-705/14N) from the Research Grants Council of The Government of Hong Kong, the AXA Research Fund, and contract HHSN272201400006C from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA.
We thank Janice Lo at the Department of Health of The Government of Hong Kong for providing the A/HK/3263/2014 virus and Vicky Fang at the School of Public Health, The University of Hong Kong, for the help with statistical analysis. We acknowledge the animal husbandry support from the colleagues from the Laboratory Animal Unit at The University of Hong Kong and the technical support from colleagues at Agriculture, Fisheries and Conservation Department of The Government of Hong Kong.
REFERENCES
- 1.CHP. Avian influenza report, vol 11, week 15.2015 CHP, Hong Kong: http://www.chp.gov.hk/files/pdf/2015_avian_influenza_report_vol11_wk15.pdf. [Google Scholar]
- 2.Yu H, Cowling BJ, Feng L, Lau EH, Liao Q, Tsang TK, Peng Z, Wu P, Liu F, Fang VJ, Zhang H, Li M, Zeng L, Xu Z, Li Z, Luo H, Li Q, Feng Z, Cao B, Yang W, Wu JT, Wang Y, Leung GM. 2013. Human infection with avian influenza A H7N9 virus: an assessment of clinical severity. Lancet 382:138–145. doi: 10.1016/S0140-6736(13)61207-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 4.Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y, Neumann G, Saito T, Kawaoka Y, Tashiro M. 2013. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill 18(15):20453. [PMC free article] [PubMed] [Google Scholar]
- 5.Lam TT, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, Ma C, Lycett SJ, Leung CY, Chen X, Li L, Hong W, Chai Y, Zhou L, Liang H, Ou Z, Liu Y, Farooqui A, Kelvin DJ, Poon LL, Smith DK, Pybus OG, Leung GM, Shu Y, Webster RG, Webby RJ, Peiris JS, Rambaut A, Zhu H, Guan Y. 2013. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 502:241–244. doi: 10.1038/nature12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi JZ, Deng GH, Liu PH, Zhou JP, Guan LZ, Li WH, Li XY, Guo J, Wang GJ, Fan J, Wang JL, Li YY, Jiang YP, Liu LL, Tian GB, Li CJ, Chen HL. 2013. Isolation and characterization of H7N9 viruses from live poultry markets—implication of the source of current H7N9 infection in humans. Chin Sci Bull 58:1857–1863. doi: 10.1007/s11434-013-5873-4. [DOI] [Google Scholar]
- 7.Lu J, Wu J, Zeng X, Guan D, Zou L, Yi L, Liang L, Ni H, Kang M, Zhang X, Zhong H, He X, Monagin C, Lin J, Ke C. 2014. Continuing reassortment leads to the genetic diversity of influenza virus H7N9 in Guangdong, China. J Virol 88:8297–8306. doi: 10.1128/JVI.00630-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lam TT, Zhou B, Wang J, Chai Y, Shen Y, Chen X, Ma C, Hong W, Chen Y, Zhang Y, Duan L, Chen P, Jiang J, Zhang Y, Li L, Poon LL, Webby RJ, Smith DK, Leung GM, Peiris JS, Holmes EC, Guan Y, Zhu H. 11 March 2015. Dissemination, divergence and establishment of H7N9 influenza viruses in China. Nature doi: 10.1038/nature14348. [DOI] [PubMed] [Google Scholar]
- 9.Meng Z, Han R, Hu Y, Yuan Z, Jiang S, Zhang X, Xu J. 2014. Possible pandemic threat from new reassortment of influenza A(H7N9) virus in China. Euro Surveill 19(6):20699. [DOI] [PubMed] [Google Scholar]
- 10.Pasricha G, Mukherjee S, Chakrabarti AK. 2014. Comprehensive sequence analysis of HA protein of H7 subtype avian influenza viruses: an emphasis on mutations in novel H7N9 viruses. Future Virol 9:251–273. doi: 10.2217/fvl.13.132. [DOI] [Google Scholar]
- 11.Zhang Q, Shi J, Deng G, Guo J, Zeng X, He X, Kong H, Gu C, Li X, Liu J, Wang G, Chen Y, Liu L, Liang L, Li Y, Fan J, Wang J, Li W, Guan L, Li Q, Yang H, Chen P, Jiang L, Guan Y, Xin X, Jiang Y, Tian G, Wang X, Qiao C, Li C, Bu Z, Chen H. 2013. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science 341:410–414. doi: 10.1126/science.1240532. [DOI] [PubMed] [Google Scholar]
- 12.Hatta M, Hatta Y, Kim JH, Watanabe S, Shinya K, Nguyen T, Lien PS, Le QM, Kawaoka Y. 2007. Growth of H5N1 influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog 3:1374–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai M, Herfst S, Sorrell EM, Schrauwen EJ, Linster M, De Graaf M, Fouchier RA, Kawaoka Y. 2013. Transmission of influenza A/H5N1 viruses in mammals. Virus Res 178:15–20. doi: 10.1016/j.virusres.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mok CK, Lee HH, Lestra M, Nicholls JM, Chan MC, Sia SF, Zhu H, Poon LL, Guan Y, Peiris JS. 2014. Amino acid substitutions in polymerase basic protein 2 gene contribute to the pathogenicity of the novel A/H7N9 influenza virus in mammalian hosts. J Virol 88:3568–3576. doi: 10.1128/JVI.02740-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steel J, Lowen AC, Mubareka S, Palese P. 2009. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog 5:e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Li X, Guo J, Li L, Chang C, Li Y, Bian C, Xu K, Chen H, Sun B. 2014. The PB2 E627K mutation contributes to the high polymerase activity and enhanced replication of H7N9 influenza virus. J Gen Virol 95:779–786. doi: 10.1099/vir.0.061721-0. [DOI] [PubMed] [Google Scholar]
- 18.Van Hoeven N, Pappas C, Belser JA, Maines TR, Zeng H, García-Sastre A, Sasisekharan R, Katz JM, Tumpey TM. 2009. Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc Natl Acad Sci U S A 106:3366–3371. doi: 10.1073/pnas.0813172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Wit E, Munster VJ, van Riel D, Beyer WE, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA. 2010. Molecular determinants of adaptation of highly pathogenic avian influenza H7N7 viruses to efficient replication in the human host. J Virol 84:1597–1606. doi: 10.1128/JVI.01783-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jonges M, Welkers MR, Jeeninga RE, Meijer A, Schneeberger P, Fouchier RA, de Jong MD, Koopmans M. 2014. Emergence of the virulence-associated PB2 E627K substitution in a fatal human case of highly pathogenic avian influenza virus A(H7N7) infection as determined by Illumina Ultra-Deep sequencing. J Virol 88:1694–1702. doi: 10.1128/JVI.02044-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golender N, Panshin A, Banet-Noach C, Nagar S, Pokamunski S, Pirak M, Tendler Y, Davidson I, Garcia M, Perk S. 2008. Genetic characterization of avian influenza viruses isolated in Israel during 2000–2006. Virus Genes 37:289–297. doi: 10.1007/s11262-008-0272-7. [DOI] [PubMed] [Google Scholar]
- 22.Abdel-Moneim AS, Afifi MA, El-Kady MF. 2012. Isolation and mutation trend analysis of influenza A virus subtype H9N2 in Egypt. Virol J 9:173. doi: 10.1186/1743-422X-9-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chin AW, Li OT, Mok CK, Ng MK, Peiris M, Poon LL. 2014. Influenza A viruses with different amino acid residues at PB2-627 display distinct replication properties in vitro and in vivo: revealing the sequence plasticity of PB2-627 position. Virology 468–470:545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowling BJ, Jin L, Lau EH, Liao Q, Wu P, Jiang H, Tsang TK, Zheng J, Fang VJ, Chang Z, Ni MY, Zhang Q, Ip DK, Yu J, Li Y, Wang L, Tu W, Meng L, Wu JT, Luo H, Li Q, Shu Y, Li Z, Feng Z, Yang W, Wang Y, Leung GM, Yu H. 2013. Comparative epidemiology of human infections with avian influenza A H7N9 and H5N1 viruses in China: a population-based study of laboratory-confirmed cases. Lancet 382:129–137. doi: 10.1016/S0140-6736(13)61171-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu B, Havers F, Chen E, Yuan Z, Yuan H, Ou J, Shang M, Kang K, Liao K, Liu F, Li D, Ding H, Zhou L, Zhu W, Ding F, Zhang P, Wang X, Yao J, Xiang N, Zhou S, Liu X, Song Y, Su H, Wang R, Cai J, Cao Y, Wang X, Bai T, Wang J, Feng Z, Zhang Y, Widdowson MA, Li Q. 2014. Risk factors for influenza A(H7N9) disease—China, 2013. Clin Infect Dis 59:787–794. doi: 10.1093/cid/ciu423. [DOI] [PubMed] [Google Scholar]
- 26.OIE. 2013. World Organisation for Animal Health (OIE) visit to the People's Republic of China to investigate influenza A (H7N9) infections in poultry 25 April–1 May 2013. OIE, Paris, France: http://www.oie.int/fileadmin/Home/eng/Media_Center/docs/pdf/RD_China_H7N9_June2013.pdf. [Google Scholar]
- 27.WHO. 2015. WHO risk assessment of human infections with avian influenza A(H7N9) virus. WHO, Geneva, Switzerland: http://www.who.int/influenza/human_animal_interface/influenza_h7n9/RiskAssessment_H7N9_23Feb20115.pdf?ua=1. [Google Scholar]
- 28.Tellier R. 2009. Aerosol transmission of influenza A virus: a review of new studies. J the Royal Society, Interface/the Royal Society 6 Suppl 6:S783–790. doi: 10.1098/rsif.2009.0302.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He Y, Liu P, Tang S, Chen Y, Pei E, Zhao B, Ren H, Li J, Zhu Y, Zhao H, Pan Q, Gu B, Yuan Z, Wu F. 2014. Live poultry market closure and control of avian influenza A(H7N9), Shanghai, China Emerg Infect Dis 20:1565–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu P, Jiang H, Wu JT, Chen E, He J, Zhou H, Wei L, Yang J, Yang B, Qin Y, Fang VJ, Li M, Tsang TK, Zheng J, Lau EH, Cao Y, Chai C, Zhong H, Li Z, Leung GM, Feng L, Gao GF, Cowling BJ, Yu H. 2014. Poultry market closures and human infection with influenza A(H7N9) virus, China, 2013–14. Emerg Infect Dis 20:1891–1894. doi: 10.3201/eid2011.140556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu H, Wu JT, Cowling BJ, Liao Q, Fang VJ, Zhou S, Wu P, Zhou H, Lau EH, Guo D, Ni MY, Peng Z, Feng L, Jiang H, Luo H, Li Q, Feng Z, Wang Y, Yang W, Leung GM. 2014. Effect of closure of live poultry markets on poultry-to-person transmission of avian influenza A H7N9 virus: an ecological study. Lancet 383:541–548. doi: 10.1016/S0140-6736(13)61904-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.FAO. 2014. Addressing avian influenza A(H7N9) qualitative risk assessment update. FAO, Rome, Italy: http://www.fao.org/3/a-i3813e.pdf. [Google Scholar]
- 33.Zhu H, Wang D, Kelvin DJ, Li L, Zheng Z, Yoon SW, Wong SS, Farooqui A, Wang J, Banner D, Chen R, Zheng R, Zhou J, Zhang Y, Hong W, Dong W, Cai Q, Roehrl MH, Huang SS, Kelvin AA, Yao T, Zhou B, Chen X, Leung GM, Poon LL, Webster RG, Webby RJ, Peiris JS, Guan Y, Shu Y. 2013. Infectivity, transmission, and pathology of human-isolated H7N9 influenza virus in ferrets and pigs. Science 341:183–186. doi: 10.1126/science.1239844. [DOI] [PubMed] [Google Scholar]
- 34.Jones JC, Sonnberg S, Webby RJ, Webster RG. 2015. Influenza A(H7N9) virus transmission between finches and poultry. Emerg Infect Dis 21:619–628. doi: 10.3201/eid2104.141703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yen HL, Zhou J, Choy KT, Sia SF, Teng O, Ng IH, Fang VJ, Hu Y, Wang W, Cowling BJ, Nicholls JM, Guan Y, Peiris JS. 2014. The R292K mutation that confers resistance to neuraminidase inhibitors leads to competitive fitness loss of A/Shanghai/1/2013 (H7N9) influenza virus in ferrets. J Infect Dis 210:1900–1908. doi: 10.1093/infdis/jiu353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slomka MJ, Pavlidis T, Coward VJ, Voermans J, Koch G, Hanna A, Banks J, Brown IH. 2009. Validated RealTime reverse transcriptase PCR methods for the diagnosis and pathotyping of Eurasian H7 avian influenza viruses. Influenza Other Respir Viruses 3:151–164. doi: 10.1111/j.1750-2659.2009.00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong CK, Zhu H, Li OT, Leung YH, Chan MC, Guan Y, Peiris JS, Poon LL. 2013. Molecular detection of human H7N9 influenza A virus causing outbreaks in China. Clin Chem 59:1062–1067. doi: 10.1373/clinchem.2013.208975. [DOI] [PubMed] [Google Scholar]
- 38.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 39.Xu R, de Vries RP, Zhu X, Nycholat CM, McBride R, Yu W, Paulson JC, Wilson IA. 2013. Preferential recognition of avian-like receptors in human influenza A H7N9 viruses. Science 342:1230–1235. doi: 10.1126/science.1243761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blachere FM, Lindsley WG, Pearce TA, Anderson SE, Fisher M, Khakoo R, Meade BJ, Lander O, Davis S, Thewlis RE, Celik I, Chen BT, Beezhold DH. 2009. Measurement of airborne influenza virus in a hospital emergency department. Clin Infect Dis 48:438–440. doi: 10.1086/596478. [DOI] [PubMed] [Google Scholar]
- 41.Ku KB, Park EH, Yum J, Kim HM, Kang YM, Kim JC, Kim JA, Kim HS, Seo SH. 2014. Transmissibility of novel H7N9 and H9N2 avian influenza viruses between chickens and ferrets. Virology 450–451:316–323. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, Liang W, Yang S, Wu N, Gao H, Sheng J, Yao H, Wo J, Fang Q, Cui D, Li Y, Yao X, Zhang Y, Wu H, Zheng S, Diao H, Xia S, Zhang Y, Chan KH, Tsoi HW, Teng JL, Song W, Wang P, Lau SY, Zheng M, Chan JF, To KK, Chen H, Li L, Yuen KY. 2013. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet 381:1916–1925. doi: 10.1016/S0140-6736(13)60903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rabadan R, Levine AJ, Robins H. 2006. Comparison of avian and human influenza A viruses reveals a mutational bias on the viral genomes. J Virol 80:11887–11891. doi: 10.1128/JVI.01414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaraket H, Baranovich T, Kaplan BS, Carter R, Song MS, Paulson JC, Rehg JE, Bahl J, Crumpton JC, Seiler J, Edmonson M, Wu G, Karlsson E, Fabrizio T, Zhu H, Guan Y, Husain M, Schultz-Cherry S, Krauss S, McBride R, Webster RG, Govorkova EA, Zhang J, Russell CJ, Webby RJ. 2015. Mammalian adaptation of influenza A(H7N9) virus is limited by a narrow genetic bottleneck. Nat Commun 6:6553. doi: 10.1038/ncomms7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spackman E, Gelb J Jr, Preskenis LA, Ladman BS, Pope CR, Pantin-Jackwood MJ, McKinley ET. 2010. The pathogenesis of low pathogenicity H7 avian influenza viruses in chickens, ducks and turkeys. Virol J 7:331. doi: 10.1186/1743-422X-7-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalthoff D, Bogs J, Grund C, Tauscher K, Teifke JP, Starick E, Harder T, Beer M. 2014. Avian influenza H7N9/13 and H7N7/13: a comparative virulence study in chickens, pigeons, and ferrets. J Virol 88:9153–9165. doi: 10.1128/JVI.01241-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pantin-Jackwood MJ, Miller PJ, Spackman E, Swayne DE, Susta L, Costa-Hurtado M, Suarez DL. 2014. Role of poultry in the spread of novel H7N9 influenza virus in China. J Virol 88:5381–5390. doi: 10.1128/JVI.03689-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe T, Kiso M, Fukuyama S, Nakajima N, Imai M, Yamada S, Murakami S, Yamayoshi S, Iwatsuki-Horimoto K, Sakoda Y, Takashita E, McBride R, Noda T, Hatta M, Imai H, Zhao D, Kishida N, Shirakura M, de Vries RP, Shichinohe S, Okamatsu M, Tamura T, Tomita Y, Fujimoto N, Goto K, Katsura H, Kawakami E, Ishikawa I, Watanabe S, Ito M, Sakai-Tagawa Y, Sugita Y, Uraki R, Yamaji R, Eisfeld AJ, Zhong G, Fan S, Ping J, Maher EA, Hanson A, Uchida Y, Saito T, Ozawa M, Neumann G, Kida H, Odagiri T, Paulson JC, Hasegawa H, Tashiro M, Kawaoka Y. 2013. Characterization of H7N9 influenza A viruses isolated from humans. Nature 501:551–555. doi: 10.1038/nature12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spackman E, Pantin-Jackwood M, Swayne DE, Suarez DL, Kapczynski DR. 2015. Impact of route of exposure and challenge dose on the pathogenesis of H7N9 low pathogenicity avian influenza virus in chickens. Virology 477:72–81. doi: 10.1016/j.virol.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guan J, Fu Q, Chan M, Spencer JL. 2013. Aerosol transmission of an avian influenza H9N2 virus with a tropism for the respiratory tract of chickens. Avian Dis 57:645–649. doi: 10.1637/10486-010913-Reg.1. [DOI] [PubMed] [Google Scholar]
- 51.Shi H, Ashraf S, Gao S, Lu J, Liu X. 2010. Evaluation of transmission route and replication efficiency of H9N2 avian influenza virus. Avian Dis 54:22–27. doi: 10.1637/8937-052809-Reg.1. [DOI] [PubMed] [Google Scholar]
- 52.Yu X, Jin T, Cui Y, Pu X, Li J, Xu J, Liu G, Jia H, Liu D, Song S, Yu Y, Xie L, Huang R, Ding H, Kou Y, Zhou Y, Wang Y, Xu X, Yin Y, Wang J, Guo C, Yang X, Hu L, Wu X, Wang H, Liu J, Zhao G, Zhou J, Pan J, Gao GF, Yang R, Wang J. 2014. Influenza H7N9 and H9N2 viruses: coexistence in poultry linked to human H7N9 infection and genome characteristics. J Virol 88:3423–3431. doi: 10.1128/JVI.02059-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindsley WG, Blachere FM, Thewlis RE, Vishnu A, Davis KA, Cao G, Palmer JE, Clark KE, Fisher MA, Khakoo R, Beezhold DH. 2010. Measurements of airborne influenza virus in aerosol particles from human coughs. PLoS One 5:e15100. doi: 10.1371/journal.pone.0015100. [DOI] [PMC free article] [PubMed] [Google Scholar]