Abstract
It has been known for a number of years that integration sites of human immunodeficiency virus type 1 (HIV-1) DNA show a preference for actively expressed chromosomal locations. A number of viral and cellular proteins are implicated in this process, but the underlying mechanism is not clear. Two recent breakthrough publications advance our understanding of HIV integration site selection by focusing on the localization of the preferred target genes of integration. These studies reveal that knockdown of certain nucleoporins and components of nucleocytoplasmic trafficking alter integration site preference, not by altering the trafficking of the viral genome but by altering the chromatin subtype localization relative to the structure of the nucleus. Here, we describe the link between the nuclear basket nucleoporins (Tpr and Nup153) and chromatin organization and how altering the host environment by manipulating nuclear structure may have important implications for the preferential integration of HIV into actively transcribed genes, facilitating efficient viral replication.
INTRODUCTION
HIV is a retrovirus that has the ability to infect nondividing cells. This feature is a consequence of HIV being able to deliver its genome through the nuclear pore. Defining the molecular mechanism of translocation of the HIV genome into the nucleus has been an area of intense focus, revealing a complex series of interactions with alternative pathways and redundancies. We know that multiple viral proteins, such as HIV capsid (CA) and integrase (IN), interact with cellular factors involved in nuclear pore function. Mutations in these proteins have been demonstrated to cause defects in infectivity and decreased nuclear translocation of the viral genome. Likewise, knockdown of nuclear pore components also causes a defect in the nuclear localization of the HIV genome. HIV-1 preferentially integrates within transcribed genes, away from the transcription initiation site through interactions with the cellular factor LEDGF/p75. It is unclear whether transcription directly or indirectly helps integration (for reviews, see references 1 to 4). Interestingly, certain mutations in viral proteins and knockdown of certain nuclear pore components alter the pattern of HIV integration. These observations suggested a continuum of steps where the pathway and efficiency of nuclear translocation are intimately tied to integration site selection. New insights from two publications reveal that the nuclear pore complex (NPC) plays additional roles in organizing nuclear structure (5, 6). By positioning a subset of activated transcription units near the NPC, the produced mRNAs are optimally positioned for efficient nuclear export to the cytoplasm for translation. Therefore, the NPC places the preferred sites of HIV integration in such a way that these actively transcribed genes will be the first host genomic DNA encountered by the incoming viral genome. This new perspective on the relationship between HIV genome nuclear import pathways and integration site selections has broad implications for the interpretation of the current published data set and the design of future studies to determine the mechanisms regarding the ability of HIV to infect dividing cells. To facilitate this discussion, we will review the relevant aspects of the NPC and nuclear import system relating to HIV biology and nuclear structure.
NPC
The NPC is the major gatekeeper of macromolecular traffic between the nucleus and cytoplasm. It is composed of an assembly of ∼30 different proteins called nucleoporins. NPCs penetrate the nuclear envelope, forming a turnstile of ∼100 nm for both active transport of large molecules and diffusion of smaller molecules between the nucleus and the cytoplasm (7). The overall NPC structure is conserved from Saccharomyces cerevisiae to humans and is comprised of a stable membrane-embedded scaffold that delineates a central channel that ranges from ∼9 to 50 nm. It also anchors peripheral structures, such as cytoplasmic filaments (Nup358, Nup214, and Nup88), that extend into the cytoplasm and a nuclear basket (Tpr and Nup153) that extends into the nucleoplasm (Fig. 1A) (7, 8). Nucleoporins play important roles in cellular functions, such as nuclear trafficking, proper chromosome segregation during mitosis, and maintenance of genome integrity (8). Nucleocytoplasmic trafficking is a controlled process that usually involves the small Ras-related GTPase Ran and karyopherin β, employing a crucial RanGTP/RanGDP gradient between the cytoplasmic and nucleoplasmic compartments (7). Cytoplasmic proteins that have a nuclear localization signal are allowed NPC passage in a process mediated by the NPC's phenylalanine-glycine (FG) barrier. Interestingly, mammalian NPCs are disassembled during mitosis; after cytokinesis, NPCs reassemble into the nuclear envelope. Therefore, NPC number and structure are highly coordinated with cell cycle progression (8, 9).
FIG 1.
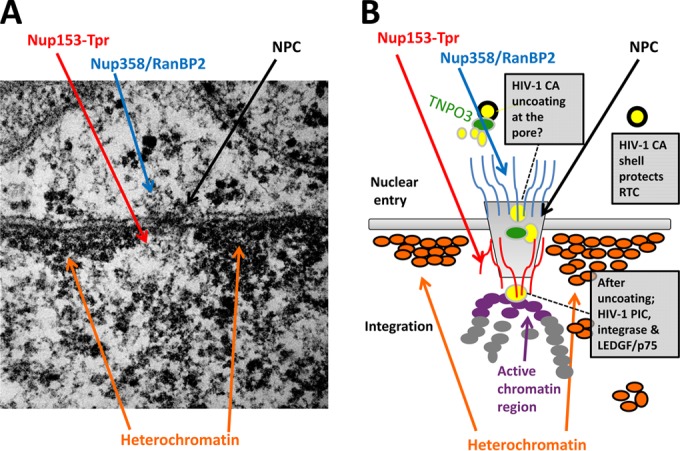
Model for HIV-1 nuclear import and integration. (A) Electron micrograph of the nuclear periphery of HeLa cells. Densely stained heterochromatin (compacted/silenced) regions are visible adjacent to the nucleoplasmic face of the inner nuclear membrane. The lesser-stained euchromatin regions (unfolding/active) are positioned along the nuclear envelop occupied by NPCs. The proposed approximate locations of Nups (Nup358, Nup153, and Tpr) are indicated. (B) Schematic representation summarizing the findings of recent studies into HIV-1 nuclear import and integration during the early steps of HIV-1 infection. Nup358/RanBP2 binds to CA and aids the HIV PIC docking on the NPC; Nup153 binds the PIC and aids nuclear import. The FG repeats from RanBP2 and Nup153 aid PIC nuclear translocation. After the PIC enters the nucleus through the pore, the viral DNA integrates into the active chromatin closest to the nuclear basket Nups (Tpr-Nup153). Tpr ensures that the chromatin environment near the pore is active, thus promoting viral replication.
The nucleus is highly structured to facilitate optimal function, including gene regulation and expression. High-resolution light and electron microscopy of mammalian cells revealed compressed and condensed heterochromatin within the nuclear envelope. The nuclear periphery is normally considered to be a region of gene repression initiated by the presence of heterochromatin and enriched with repressive histone markers. In contrast, active gene regions and euchromatin can be localized in association with the NPC at the nuclear periphery or more centrally in the nucleus (Fig. 1A) (2, 8). In 1985, Günter Blobel proposed the “gene gating hypothesis,” in which gene tethering to the NPC influences mRNA export and production (10). Tethering to the NPC is thought to allow optimal transcriptional activity, which can be regulated by nucleoporins. The nonrandom spatial distribution of chromosomes and genes has been appreciated for decades, although a detailed understanding of the functional role of such positioning remains a mystery (2). Here, we focus on several recent studies on HIV translocation and integration at the nuclear pores, mainly involving the NPC basket nucleoporins Nup153 and Tpr.
Nup153 AND Tpr
Nup153 and Tpr are found on the nuclear basket of the NPC (Fig. 1). The N terminus domain of Nup153 associates with the inner nuclear basket and has been shown to be crucial to the presence of Tpr in the NPC (11). Nup153 has a highly flexible C-terminal domain containing ∼30 FG motif repeats that might be found along the nuclear basket or extending to the cytoplasmic side of the NPC (4). HIV IN was demonstrated to interact with the Nup153 FG/FXFG repeats; an in vitro association with HIV was also found between green fluorescent protein (GFP)-Nup153 and CA in cell lysates (2). Genetics studies of different chimeras between other retroviruses and HIV pointed to Nup153 dependency for determinants in HIV-1 CA (12, 13). However, HIV chimeras expressing IN from murine leukemia virus (MLV) were able to partially restore infection in Nup153-depleted cells, suggesting more than one dependency between the HIV preintegration complex (PIC) and Nup153 (12). The Nup153 FG repeats have been shown to bind to a hydrophobic pocket of HIV-1 CA, which curiously is also the binding pocket for another host protein, CPSF6, and small molecules that inhibit HIV replication, such as PF74 and BI2 (14). Both Nup153 and CPSF6 appear to play a major role in the ability of HIV to infect nondividing cells.
Tpr forms a bridge between the NPC and the underlying chromatin. Mammalian Tpr, which binds directly to Nup153, has been suggested to play a role in nuclear proteins and RNA export. It has also been implicated in multiple additional cellular functions and has been assigned multiple functions, including being a chromatin scaffolding element facilitating transcriptional telomeric chromatin organization by establishing perinuclear heterochromatin exclusion zones. It is also involved in SUMOylation, mitotic spindle bipolarity, centrosome formation, and the control of cellular senescence (8, 15, 16). Since Tpr is localized with an “open-state” euchromatin (decondensed and actively transcribed chromatin), Tpr may play a role in the chromatin target of HIV-1 integration (2). Furthermore, depleting Tpr results in heterochromatin spreading throughout the nuclear periphery in human cells (17).
RECENT INSIGHTS INTO HIV-1 DOCKING AND TRAFFICKING TO THE NUCLEAR PORE
It has become clear that the HIV CA protein contains required determinants of nuclear import (13, 14). HIV CA forms the iconic conical capsid that encases the HIV genome within virions. Eventually, the conical core must dissolve to allow the reverse transcription and interact with nuclear import factors and the nuclear pore through a process called uncoating. There has been much debate about whether that process might take place early, after fusion in the cytoplasm, or much later at the nuclear pore. The point of contention has been that if uncoating happens early, how could CA be a determinant of later events, such as nuclear import and integration site selection. However, a recent report from our laboratory may resolve this issue because a subset of HIV CA proteins can be detected in nuclear HIV complexes (18). The interaction of CA with Nup153 and CPSF6 has been defined in great detail, and studies with drugs that disrupt these interactions with CA can inhibit nuclear import and potentially alter integration site selection (14). The HIV-1 core is transported through the cytoplasm to the nucleus along microtubules, and uncoating is facilitated by dynein and kinesin 1 (19).
RECENT INSIGHTS INTO HIV-1 INTEGRATION INTO ACTIVE GENES AT THE NUCLEAR PORE
As described above, HIV is known to preferentially integrate into active genes, as demonstrated by multiple studies. A recent study by Marini and colleagues examined the nuclear localization of HIV “recurrent integration genes” (RIGs) and found that they were preferentially localized in the periphery of the nucleus (6). This analysis of the three-dimensional architecture of the preferred targets of HIV-1 integration revealed that integration was connected to the transcriptionally active regions of chromatin that are very near the nuclear pores (6). The vast majority of the HIV-1 integration sites mapped to the nuclear periphery within 1 μm of the nuclear membrane. Through chromatin immunoprecipitation followed by sequencing (ChIP-seq) analysis, they also revealed that HIV-1 cDNA insertion sites were connected with transcriptionally active histone markers and with several nucleoporins (Nup62, Nup98, Nup153, and Tpr) but not with known lamin-associated repressive histone markers. Tpr depletion also reduced long terminal repeat-driven gene expression in HIV-1-infected cells (6). Marini et al. suggested that NPC-proximal regions may play a role in HIV-1 gene expression, as proteins within the NPCs were found to be involved in the transcriptional regulation of the viral genome (6, 20).
Another recent report by Lelek and colleagues found that chromatin organization at the nuclear pore favors HIV gene expression and that knockdown of Tpr inhibits the levels of HIV gene expression, with only a minor impact on the nuclear localization and integration of the viral genome. In contrast, they show that the knockdown of Nup153 functions primarily to inhibit the nuclear localization of the HIV genome. These data suggest that Tpr plays a fundamental role in chromatin organization by excluding heterochromatin from the vicinity of NPCs and recruits hyper-transcribed genes near the NPC (5). Tpr depletion alters chromatin organization at the nuclear periphery, resulting in more condensed, transcriptionally silent chromatin near the nuclear basket. The NPC may form a chromatin topology and transcriptional environment encouraging HIV-1 replication. Using superresolution microscopy, they revealed that Tpr depletion caused a reduced density of trimethylated histone H3 Lys36 (H3K36me3) localization in the nuclear periphery beneath NPCs. Moreover, the host cellular factor LEDGF/p75, which is important for HIV-1 integration in active chromatin, is also stabilized at the nuclear periphery when Tpr is overexpressed and vice versa (5).
Together, these two papers provide an important new perspective on the process of HIV nuclear import and integration site preference. It is known that HIV has a propensity to integrate into actively transcribed genes. Subsequent work revealed that amino acid changes in viral proteins and knockdown of certain proteins that are NPC components or that participate in regulated transport through the nuclear pore may alter integration sites. These new results reveal that one mechanism for this targeting is that a function of the nuclear pore is to organize a subset of actively transcribed genes very near the nuclear pores. Therefore, the viral genome will be most likely to encounter these genes immediately after entering the nucleus. In this way, some NPC components are critical for normal integration of the HIV genome, not because the genome utilizes an alternative nuclear import pathway after the nucleoporin knockdown but rather because the structure of the chromatin is altered, which leads to integration into less optimal genes for efficient viral gene expression. Therefore, one consequence of the infection of nondividing cells by translocation through the NPC is that the first chromatin encountered by the PIC after nuclear import is associated with activated genes. The fact that this is the first environment encountered by the HIV-1 PIC ensures high expression of its viral genes by integrating into highly transcribed host DNA regions (2) while simultaneously avoiding silenced chromatin, which is less accessible owing to steric hindrance (Fig. 1B).
IMPLICATIONS
Collectively, these new data from Lelek et al. (5) and Marini et al. (6) highlight that HIV-1 genome integration has a preference for NPC-proximal regions at the nuclear periphery in a nonrandom manner. As usual, new insights lead to new questions. For instance, there are capsids with mutations, such as N74D, that do not infect nondividing cells or utilize the Nup153 import pathway but are still dependent on alternative nucleoporins. This begs the question as to how these HIV derivatives gain access to host DNA. Further study is clearly required to determine if cell division provides access to host chromatin through disruption of the nuclear membrane during cell division or through some alternative mechanism of nuclear import that selects other integration sites. Another area requiring more study is how the knockdown of different nuclear pore components and factors may influence the recruitment of active genes to the NPC. For example, the knockdown of Nup153, which is associated with the nuclear basket of the NPC, is known to have a minor effect on HIV integration site selection and also influences Tpr function, potentially changing access to host cell chromatin (14).
Future studies will provide new insight into how nuclear basket nucleoporins involved in HIV-1 infection might influence contacts between the inner nuclear side of the nuclear pore and the host chromosomal DNA (2). Future studies will also clarify the multiple roles of nucleoporins in the early steps of HIV-1 infection, especially between uncoating and PIC formation, supported by advancements in biochemical, genetic, and microscopic approaches. However, the interpretation of these studies will be complicated by the fact that nucleoporins are interconnected. Distinguishing the specific roles of different HIV proteins and of Nup358, Nup153, and Tpr during nuclear translocation, integration, and transcription is also challenging due to their close structural connection. However, the knowledge that these changes can also alter nuclear structure will facilitate data interpretation and experimental design to advance this work. The use of mutations in viral proteins which interact with transport pathways and the NPC should provide additional insights regarding the interaction between the NPC and HIV integration in different cellular contexts. Ultimately, this knowledge will help us to design novel drugs to inhibit HIV-1 nuclear translocation by disrupting the association between viral proteins and nucleoporins. This knowledge may lead to new therapeutic strategies to prevent, treat, and perhaps cure HIV infection.
ACKNOWLEDGMENTS
Thank you to Ann Carias for assistance with figure development and to Rosemary Clark for assistance with proofreading of the manuscript.
This work was supported by NIH grant P50 GM082545 to T.J.H. This work was also supported by Grants-in-Aid for Challenging Exploratory Research and Grants-in-Aid for Scientific Research (B) from MEXT Japan and by grants from the Takeda Science Foundation to R.W.W.
We apologize to all of our colleagues whose work could not be cited due to length restrictions.
REFERENCES
- 1.Cohen S, Au S, Panté N. 2011. How viruses access the nucleus. Biochim Biophys Acta 1813:1634–1645. doi: 10.1016/j.bbamcr.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 2.Di Nunzio F. 2013. New insights in the role of nucleoporins: a bridge leading to concerted steps from HIV-1 nuclear entry until integration. Virus Res 178:187–196. doi: 10.1016/j.virusres.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Hilditch L, Towers GJ. 2014. A model for cofactor use during HIV-1 reverse transcription and nuclear entry. Curr Opin Virol 4:32–36. doi: 10.1016/j.coviro.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matreyek KA, Engelman A. 2013. Viral and cellular requirements for the nuclear entry of retroviral preintegration nucleoprotein complexes. Viruses 5:2483–2511. doi: 10.3390/v5102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lelek M, Casartelli N, Pellin D, Rizzi E, Souque P, Severgnini M, Di Serio C, Fricke T, Diaz-Griffero F, Zimmer C, Charneau P, Di Nunzio F. 2015. Chromatin organization at the nuclear pore favours HIV replication. Nat Commun 6:6483. doi: 10.1038/ncomms7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marini B, Kertesz-Farkas A, Ali H, Lucic B, Lisek K, Manganaro L, Pongor S, Luzzati R, Recchia A, Mavilio F, Giacca M, Lusic M. 2015. Nuclear architecture dictates HIV-1 integration site selection. Nature 521:227–231. doi: 10.1038/nature14226. [DOI] [PubMed] [Google Scholar]
- 7.Strambio-De-Castillia C, Niepel M, Rout MP. 2010. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol 11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- 8.Ibarra A, Hetzer MW. 2015. Nuclear pore proteins and the control of genome functions. Genes Dev 29:337–349. doi: 10.1101/gad.256495.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cautain B, Hill R, de Pedro N, Link W. 2015. Components and regulation of nuclear transport processes. FEBS J 282:445–462. doi: 10.1111/febs.13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blobel G. 1985. Gene gating: a hypothesis. Proc Natl Acad Sci U S A 82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hase ME, Cordes VC. 2003. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell 14:1923–1940. doi: 10.1091/mbc.E02-09-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matreyek KA, Engelman A. 2011. The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid. J Virol 85:7818–7827. doi: 10.1128/JVI.00325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mamede JI, Sitbon M, Battini J-L, Courgnaud V. 2013. Heterogeneous susceptibility of circulating SIV isolate capsids to HIV-interacting factors. Retrovirology 10:77. doi: 10.1186/1742-4690-10-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matreyek KA, Yücel SS, Li X, Engelman A. 2013. Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity. PLoS Pathog 9:e1003693. doi: 10.1371/journal.ppat.1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi A, Hashizume C, Dowaki T, Wong RW. 2015. Therapeutic potential of mitotic interaction between the nucleoporin Tpr and aurora kinase A. Cell Cycle 14:1447–1458. doi: 10.1080/15384101.2015.1021518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakano H, Funasaka T, Hashizume C, Wong RW. 2010. Nucleoporin translocated promoter region (Tpr) associates with dynein complex, preventing chromosome lagging formation during mitosis. J Biol Chem 285:10841–10849. doi: 10.1074/jbc.M110.105890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krull S, Dörries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC. 2010. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J 29:1659–1673. doi: 10.1038/emboj.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hulme AE, Kelley Z, Foley D, Hope TJ. 2015. Complementary assays reveal a low level of CA associated with viral complexes in the nuclei of HIV-1-infected cells. J Virol 89:5350–5361. doi: 10.1128/JVI.00476-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukic Z, Dharan A, Fricke T, Diaz-Griffero F, Campbell EM. 2014. HIV-1 uncoating is facilitated by dynein and kinesin 1. J Virol 88:13613–13625. doi: 10.1128/JVI.02219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Attar N. 2015. Viral infection: tracking down HIV's hiding place. Nat Rev Microbiol 13:186–187. doi: 10.1038/nrmicro3466. [DOI] [PubMed] [Google Scholar]


