ABSTRACT
Tomato yellow leaf curl virus (TYLCV) is a begomovirus transmitted exclusively by the whitefly Bemisia tabaci in a persistent, circulative manner. Replication of TYLCV in its vector remains controversial, and thus far, the virus has been considered to be nonpropagative. Following 8 h of acquisition on TYLCV-infected tomato plants or purified virions and then transfer to non-TYLCV-host cotton plants, the amounts of virus inside whitefly adults significantly increased (>2-fold) during the first few days and then continuously decreased, as measured by the amounts of genes on both virus DNA strands. Reported alterations in insect immune and defense responses upon virus retention led us to hypothesize a role for the immune response in suppressing virus replication. After virus acquisition, stress conditions were imposed on whiteflies, and the levels of three viral gene sequences were measured over time. When whiteflies were exposed to TYLCV and treatment with two different pesticides, the virus levels continuously increased. Upon exposure to heat stress, the virus levels gradually decreased, without any initial accumulation. Switching of whiteflies between pesticide, heat stress, and control treatments caused fluctuating increases and decreases in virus levels. Fluorescence in situ hybridization analysis confirmed these results and showed virus signals inside midgut epithelial cell nuclei. Combining the pesticide and heat treatments with virus acquisition had significant effects on fecundity. Altogether, our results demonstrate for the first time that a single-stranded DNA plant virus can replicate in its hemipteran vector.
IMPORTANCE Plant viruses in agricultural crops are of great concern worldwide. Many of them are transmitted from infected to healthy plants by insects. Persistently transmitted viruses often have a complex association with their vectors; however, most are believed not to replicate within these vectors. Such replication is important, as it contributes to the virus's spread and can impact vector biology. Tomato yellow leaf curl virus (TYLCV) is a devastating begomovirus that infects tomatoes. It is persistently transmitted by the whitefly Bemisia tabaci but is believed not to replicate in the insect. To demonstrate that TYLCV is, in fact, propagative (i.e., it replicates in its insect host), we hypothesized that insect defenses play a role in suppressing virus replication. We thus exposed whitefly to pesticide and heat stress conditions to manipulate its physiology, and we showed that under such conditions, the virus is able to replicate and significantly influence the insect's fecundity.
INTRODUCTION
Tomato yellow leaf curl virus (TYLCV) (Begomovirus, Geminiviridae) is the name given to a complex of single-stranded DNA (ssDNA) plant viruses that cause tremendous losses of tomato crops worldwide (1). Begomoviruses are transmitted by several biotypes of the whitefly Bemisia tabaci (2, 3). Begomoviruses have either a single (monopartite) or two (bipartite) circular ssDNA genomes of ∼2,700 nucleotides each. Each genome is encapsidated in an ∼25- by 30-nm geminate particle. TYLCV has a single genomic DNA component, which carries six partially overlapping genes that are bidirectionally organized into two transcriptional units separated by an intergenic region (IR) of ∼300 nucleotides (4, 5). The virus strand is comprised of two genes: V1, which encodes the coat protein (CP) that functions as a nuclear shuttle protein and mediates vector transmission, and V2, which encodes a protein involved in viral movement and in suppression of gene silencing. The virus complementary strand is comprised of four genes: C1 encodes a protein (Rep) that initiates viral replication, C2 is a transcriptional activator that also interferes with transcriptional and posttranscriptional gene silencing, C3 encodes a replication enhancer, and C4 is involved in silencing suppression and has been shown to be a symptom determinant. CP is the only viral protein that has been implicated in vector-mediated transmission of TYLCV (6, 7). In plants, TYLCV, like other begomoviruses, replicates in the infected nucleus via an intermediate double-stranded DNA (dsDNA) replicative form, according to the rolling-circle model (8).
Begomoviruses such as TYLCV are acquired by the whitefly from the plant phloem as intact virions which pass along the food canal in the insect stylet with other phloem components until they reach the esophagus (9). The first tissue through which virions can translocate to the hemocoel is a modification of the digestive system called the filter chamber (10). Based on extensive TYLCV localization studies using fluorescence in situ hybridization (FISH) and transmission electron microscopy, most TYLCV virions have been observed in the filter chamber, suggesting that it is the main site for viral particle translocation to the hemocoel (11–13). Virions are transported through the cytoplasm of the filter chamber epithelial cells in vesicles that fuse with the basal plasma membrane, releasing particles between the membrane and the basal lamina (10, 14, 15). In contrast to the family Luteoviridae, TYLCV is localized in midgut epithelial cells but not in the hindgut (11, 16). Once they cross the midgut cells, the virions take up temporary residence in the hemocoel and then enter the salivary system, from which they are transmitted to plants during feeding (9, 14, 15). It takes a minimum of 8 h (latent period) from the moment that the virus is acquired from infected tomato plants to be effectively transmitted to test plants (9). The vector specificity of geminiviruses is thought to be regulated at the gut interface (17, 18) and by cells around the secretory region of the primary salivary glands (19).
During the transit of begomoviruses in their whitefly vector, CP is the only virus protein exposed to whitefly tissues, and it is hypothesized to interact with insect proteins that have a role in virus movement through insect tissues. TYLCV CP binds to the midgut of B. tabaci, possibly to receptors on epithelial cells (20). Mutational analyses have defined specific regions of begomovirus CPs required for the transmission of begomoviruses by whiteflies (6, 7, 21). Whitefly proteins involved in begomovirus circulative transmission have been intensively sought. A microarray approach examining the response of whiteflies to the acquisition and retention of TYLCV and Squash leaf curl virus (SLCV) showed that heat shock protein 70 (HSP70) interacts with the viruses' CPs (22). It was hypothesized that HSP70 plays a role in protecting whiteflies from the viruses' detrimental effects (23). A small B. tabaci 16-kDa HSP (BtHSP16) was identified by a yeast two-hybrid screen and found to be necessary for the transmission of Tomato yellow leaf curl Sardinia virus (TYLCSV) (24).
Many reports strongly indicate that the interactions between several B. tabaci biotypes and several TYLCV species are not neutral and are reminiscent of an insect-pathogen relationship. TYLCV and other begomoviruses are present in the insect host for most, if not all, of the whitefly's life span (25). The long-term TYLCV-whitefly interaction has been found to have deleterious (23), beneficial (26), or no (27) effects on the insect's fertility and longevity. TYLCV, Tomato yellow leaf curl China virus (TYLCCNV), and TYLCSV have been shown to be transmitted transovarially to progeny (28, 29) and during mating (28, 30, 31). Expression profiling studies examined the transcriptional response of the B. tabaci B biotype to the acquisition and retention of TYLCCNV and identified 1,606 genes involved in 157 biochemical pathways that are differentially expressed in viruliferous versus nonviruliferous whiteflies (32). That study showed that TYLCCNV perturbs the insect's cell cycle and primary metabolism and suppresses some immune responses, such as Toll-like signaling and mitogen-activated protein kinase pathways (32).
The possible replication of begomoviruses in their whitefly vector is still under debate; the dogma is that this group of viruses does not replicate inside their insect vectors, and replication has not been demonstrated experimentally. Some plant RNA viruses have been shown to replicate in their insect vectors, such as nucleorhabdoviruses and cytorhabdoviruses (Rhabdoviridae), which replicate in plant hoppers (33). Similarly, fijiviruses and all phytoreoviruses (Reoviridae) replicate in their plant hopper vectors (34, 35). One of the best-known instances of plant virus replication in an insect vector is that of Tomato spotted wilt virus (TSWV) (Tospovirus, Bunyaviridae) in its thrips vector, Frankliniella occidentalis (36). TSWV was shown to activate genes of the thrips immune system, including those encoding antimicrobial peptides, lectins, and members of the signal transduction pathways activated by Toll-like receptors (37), as was recently shown for the expression of whitefly genes after acquisition and retention of TYLCCNV (32).
Several attempts have been made to test whether TYLCV is able to replicate inside its whitefly vector. In those experiments, the amounts of virus were appraised during whitefly rearing on a non-virus-host plant after a short virus acquisition period on infected plants, under standard laboratory conditions (1, 29, 38, 39). None of those studies was conclusive. However, TYLCV (but not the bipartite begomovirus Tomato mottle virus [ToMoV]) was shown to be actively transcribed in B. tabaci reared on cotton plants following a short acquisition access period (AAP) on infected tomato (40). Interestingly, in those experiments, TYLCV DNA remained stable in the insects, while ToMoV DNA was rapidly degraded, suggesting a steady state of TYLCV DNA synthesis/degradation, whereas degradation prevails for ToMoV. Transcripts of genes located on the viral complementary strand (C3) as well as on the viral strand (V1 and V2) were detected, implying synthesis of the viral complementary strand using the viral strand as the template, to form a dsDNA replicative form.
In this study, we show that following a short AAP on infected tomato plants, the amounts of TYLCV estimated by quantitative real-time PCR (qPCR) increase and then decrease in viruliferous insects reared on cotton plants under standard conditions. Manipulation of the physiological environment of B. tabaci resulted in TYLCV levels that continuously increased and were dynamic under changing stress conditions. Therefore, we propose that TYLCV can replicate in its whitefly vector but that, under normal conditions, the whitefly can prevent virus accumulation using its immune system via an unknown mechanism.
MATERIALS AND METHODS
Maintenance of whiteflies and plants.
The B. tabaci B biotype strains used in this study were reared on cotton seedlings (Gossypium hirsutum cv. Akala) and maintained in insect-proof cages in a growth room under the following standard rearing conditions: temperature of 25°C ± 2°C, 60% relative humidity, and a 14-h-light/10-h-dark photoperiod. The identity of the B biotype population was confirmed by PCR with specific primers amplifying the cytochrome oxidase I gene and the Bem23 microsatellites that differentiate the B and Q biotypes that are present in Israel (41). An Israeli isolate of TYLCV was maintained in infected tomato plants (Solanum lycopersicum cv. Avigail) by insect-mediated transmission. Tomato (host for TYLCV) and cotton (non-TYLCV host) plants were grown in potting mix in 1.5-liter pots under conditions of artificial light and controlled temperature detailed above for insect rearing.
Acquisition of TYLCV from infected tomato plants.
Adult whiteflies, 4 to 5 days after emergence, were used to study the effects of virus acquisition and subsequent heat and pesticide exposure. The insects were caged with the fourth true leaf of TYLCV-infected tomato seedlings for 8-h AAPs, after which the viruliferous whiteflies were collected by aspiration and transferred onto cotton seedlings or were used in further experiments, as detailed below. Samples from 30 insects were collected immediately after transfer to cotton (0 h) and every 24 h thereafter, up to 7 days. Three additional samples were collected every week up to 4 weeks. Midguts were dissected from these whiteflies (collected from days 0 to 7) for FISH and qPCR analyses.
Acquisition of TYLCV from a preparation of purified virions.
TYLCV particles were isolated from young leaves of Nicotiana benthamiana plants ∼4 weeks after whitefly-mediated infection, essentially as described previously (42). In brief, the leaves were homogenized in ice-cold buffer (pH 8.0) containing 100 mM trisodium citrate, 18.5 mM ascorbic acid, 60 mM sodium sulfite, 5 mM EDTA, and 1% (wt/vol) β-mercaptoethanol (2 ml/g tissue). The homogenate was produced in 2.5% (vol/vol) Triton X-100, stirred for 16 h, filtered through cheesecloth, and clarified by a 10-min centrifugation at 8,000 × g. The supernatant was centrifuged for 3 h at 90,000 × g in a Beckman SW27 rotor. The pellet was resuspended in buffered (pH 8.0) CEM buffer (10 mM trisodium citrate, 1 mM EDTA, 0.1% β-mercaptoethanol) and subjected to similar low-speed followed by high-speed centrifugations. Pellets were resuspended in 1 ml 10% (wt/vol) sucrose in CEM buffer and clarified by a 10-min centrifugation at 4,000 × g. The supernatant was loaded onto a 32-ml linear 10 to 50% sucrose gradient in CEM buffer. The sucrose gradient was fractionated (2 ml per fraction) after 14 h of centrifugation at 90,000 × g in an SW27 rotor. The presence of viral particles in the gradient was determined by PCR using specific primers. The positive fractions were diluted with CEM buffer and centrifuged for 3 h at 90,000 × g. The pellet was suspended in 15% sucrose in CEM buffer (without β-mercaptoethanol). The presence of viral particles was confirmed by staining with 2% (wt/vol) aqueous uranyl acetate and observation with an electron microscope (42). Whiteflies were fed the purified virion preparation through membranes (42). Whiteflies collected from the colony were placed into a 3-cm-diameter, 4-cm-high black plastic cylinder (∼50 insects per cylinder) covered with a layer of stretched Parafilm membrane and standing on a black plastic board (insects that died during caging accumulated at the bottom of the cage). About 0.2 ml of a 15% sucrose solution supplemented with yellow food dye and containing ∼50 μg DNA/ml of the virion preparation was deposited onto the membrane and covered with a second layer of stretched membrane. After 8 h, the insects were collected and caged with young leaves of cotton plants by using leaf clip cages. At the specified time intervals, random samples from 30 whiteflies were collected for analyses of TYLCV DNA amounts (using primers for V1) (Table 1) and TYLCV transcript levels (V1 and C3) (Table 1), and 10 insects were collected for virus transmission to tomato test plants (Solanum lycopersicum cv. Daniella) (see below). In these tests, three tomato plants at the 3- to 5-true-leaf stage were used at each time point: for each plant, three insects were caged with a young leaf by using a leaf clip cage. Tomato infection was assessed by observation of symptoms and by PCR using V1-specific primers (Table 1).
TABLE 1.
Primers and probes used in this study
| Gene | Primer or probe | Sequence (5′→3′) | Reference |
|---|---|---|---|
| TYLCV V2 | FV2 | AGTCACGGGCCCTTACA | 30 |
| RV2 | ATACTTGGACACCTAATGGC | ||
| TYLCV V1 | FV1 | CGCCCGTCTCGAAGGTTC | 40 |
| RV1 | GCCATATACAATAACAAGGC | ||
| TYLCV C3 | FC3 | TGAGGCTGTAATGTCGTCCA | 40 |
| RC3 | GCTCCTCAAGCAGAGAATGG | ||
| B. tabaci β-actin | F β-actin | TCTTCCAGCCATCCTTCTTG | 40 |
| R β-actin | CGGTGATTTCCTTCTGCATT | ||
| TYLCV V1 | V1 | GGAACATCAGGGCTTCGATA | 22 |
| TYLCV CV1 | cV1 | GAAGGCTGAACTTCGACAGC | This study |
| TYLCV CC1 | cC1 | GCTCGTAGAGGGTGACGAAG | This study |
| B. tabaci hsp70 | HSP70 | TTCCCGAGGAGATTGTC | 22 |
Exposure of whiteflies to heat stress.
TYLCV genomic DNA and gene transcript levels in whiteflies were quantified after exposure to continuous heat stress at 35°C, as previously described, with slight modifications (43). Whiteflies were allowed an 8-h AAP on TYLCV-infected tomato seedlings as described above, caged with cotton seedlings by using leaf clip cages, and maintained at 35°C for 5 days in an incubator. Following this exposure to heat stress, live individuals were collected for DNA and RNA extractions, and the levels of the TYLCV gene transcript and hsp70 were measured by using reverse transcription-qPCR (qRT-PCR) and qPCR. Dissected midguts from heat-stressed females were subjected to FISH analysis for virus visualization.
Exposure of whiteflies to pesticides.
Four- to five-day-old viruliferous B. tabaci adults (as described above) were reared on pesticide-treated cotton leaves for 5 days. Leaves were exposed to pesticides by using a previously described leaf dip method (44, 45). Briefly, leaves were dipped in the pesticide solution for 20 s and then dried for 2 h under a hood. Insects were confined to the treated leaves for 5 days by using leaf clip cages. Following this exposure, live adults were collected, and the levels of TYLCV were measured by qPCR. Midguts were dissected from these insects, and the localization of the virus was determined by FISH analysis. The pesticides used in these experiments were acetamiprid at a 30% lethal concentration (LC30) of 0.5 mg of active ingredient (a.i.)/liter and imidacloprid, which was applied to the seedlings by drenching (44) at an LC30 of 0.5 mg a.i./liter. Briefly, cotton stems with leaves were kept in the pesticide solution for 24 h, during which the pesticide was systemically absorbed by the stems. After 24 h, the stems were transferred to distilled water for an additional 24 h, after which leaves were detached and used to expose B. tabaci adults to the pesticides. After 5 days, live adults were collected every 24 h for up to 5 days for DNA extraction and FISH analysis.
Exposure of whiteflies to changing conditions.
After an 8-h AAP on TYLCV-infected plants, viruliferous insects were exposed to acetamiprid, imidacloprid, or heat stress as described above. After 48 h, one group of insects was collected for qPCR analysis using the V1, V2, and C3 gene primers. The insects from the remaining three groups (exposure to acetamiprid, imidacloprid, and heat stress) were transferred to cotton plants for 48 h at 25°C. An additional sample from each group was used for qPCR analysis; the remaining insects were reexposed to their respective stress treatments (acetamiprid, imidacloprid, or heat stress at 35°C). These transfers were carried out at 48-h intervals for up to 8 days, and the insects were sampled every 48 h for qPCR analysis. Thus, the relative abundance of TYLCV in the sampled insects was estimated at 0 h and 2, 4, 6, and 8 days.
DNA and RNA extraction and preparation of cDNA.
Genomic DNA was isolated from groups of 30 B. tabaci adults per replicate by using the CTAB (cetyltrimethylammonium bromide) method (46). DNA purity and yield were analyzed by using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). Ten dissected midguts were incubated in 50 μl double-distilled water at 95°C for 10 min to disrupt the cells and then subjected directly to qPCR. Total RNA extractions were carried out by using Tri reagent (Sigma). For each replicate, 30 B. tabaci adults were homogenized in 300 μl Tri reagent containing 1 μl linear polyacrylamide carrier (GenElute LPA; Sigma) and incubated for 5 min at room temperature. Chloroform (60 μl) was added, followed by vigorous mixing and centrifugation at 12,000 × g for 10 min at 4°C. The aqueous phase was transferred to a clean tube, 0.7 volumes of isopropanol were added, and the samples were gently mixed. The samples were then incubated overnight at −20°C for RNA precipitation. The samples were centrifuged at 12,000 × g for 20 min at 4°C. The supernatant was carefully removed. The pellet was washed with 75% ice-cold ethanol, air dried, and dissolved in 50 μl double-distilled water. RNA (100 ng) was used as a template for cDNA synthesis in 25-μl reaction mixtures by using the Verso cDNA kit (Thermo Scientific, Fermentas). RNA integrity and removal of genomic DNA were confirmed by 1% agarose gel electrophoresis. Genomic DNA removal was further performed by using the Turbo DNA-free kit (Ambion).
Quantitation of viral DNA and transcripts.
qRT-PCR and qPCR analyses were used to measure the amounts of viral DNA and the levels of different TYLCV gene transcripts, using the primers listed in Table 1. Amplifications were performed by using Absolute Blue qPCR SYBR green Rox mix (Thermo Scientific). The expression of each gene was tested in three replicates in each of three independent biological experiments. The cycling conditions were 15 min of activation at 95°C; 40 cycles of 15 s at 95°C and 30 s at 58°C (actin and V2 genes), 59°C (C3 gene), or 63°C (V1 gene); and 30 s at 72°C. A melting ramp from 72°C to 95°C was used, with a 1°C increase at each step and a 5-s interval between steps. A Rotor Gene RG-6 machine (Corbett Robotics Pty.), with analysis software, was used for qPCR data normalization and quantification. For each run, standards were loaded onto the same plate to obtain the appropriate standard curve.
Quantification of TYLCV in head, thorax, and abdomen.
After an 8-h AAP as described above, adult females were collected by aspiration and anesthetized with acetone vapors for 2 min. The insects were placed onto a glass slide containing a drop of phosphate-buffered saline (PBS) (4.3 mM Na2HPO4 · 7H2O, 1.4 mM NaH2PO4, 137 mM NaCl, and 1 mM phenylmethylsulfonyl fluoride [pH 7.2]). The insects were then cut with a razor blade under a dissecting microscope into three parts: head, thorax, and abdomen. DNA was extracted from each part as described above. Sixty whiteflies were used for each replicate (three replicates were analyzed for 0 to 7 days).
FISH.
FISH was performed as previously described (47, 48). Briefly, midguts were dissected in PBS (pH 7.2) inside depressed glass wells, fixed in Carnoy's fixative (chloroform-ethanol-glacial acetic acid [6:3:1, vol/vol]) for 5 min, and hybridized overnight in hybridization buffer (20 mM Tris-HCl [pH 8.0], 0.9 M NaCl, 0.01% [wt/vol] sodium dodecyl sulfate, 30% [vol/vol] formamide) containing 10 pmol fluorescent probe/ml. The sequences of the different probes used to localize the genomic viral and cDNA strands are detailed in Table 1. Nuclei in the midgut were stained with 4′,6′-diamidino-2-phenylindole (DAPI) (0.1 mg/ml). The stained samples were mounted whole in hybridization buffer and viewed under an IX81 Olympus FluoView500 confocal laser scanning microscope. For each treatment, 10 to 20 midguts were viewed. Optical confocal sections (100 μm thick) were sometimes prepared from each specimen for better visualization of the signal.
Immunolocalization of TYLCV.
Midguts were dissected on microscopic slides under a dissecting microscope in 1× PBS and fixed in 4% paraformaldehyde for 30 min at room temperature. Permeation of the midguts was then conducted by adding 0.1% Triton X-100 for 30 min at room temperature. The surfactant was then removed, and the midguts were washed three times with PBST (1× PBS with 0.5% [vol/vol] Tween 20), blocked for 1 h at room temperature with blocking buffer (PBST with 1% [wt/vol] bovine serum albumin), and incubated overnight at 4°C with anti-TYLCV CP polyclonal primary antibody raised in rabbit. Midguts were then washed three times with PBST and incubated with goat anti-rabbit secondary antibody conjugated to Cy3 (Sigma-Aldrich) for 1 h at room temperature. Midguts were washed again three times with PBST, mounted in 1× PBS containing 100 μg/ml DAPI, covered with a coverslip, sealed with nail polish, and viewed under a confocal microscope. For each treatment, 10 to 20 midguts were viewed.
Whitefly fecundity under conditions of heat and insecticide stresses.
Healthy 4- to 5-day-old female whiteflies reared on cotton were transferred to new cotton plants and to noninfected and TYLCV-infected tomato plants. After 8 h, the insects were collected from the noninfected and TYLCV-infected tomato plants and placed in leaf clip cages. The cages were attached to (i) cotton leaves for a 48-h exposure to heat stress at 35°C, (ii) acetamiprid-treated cotton leaves (at a concentration of 0.5 mg a.i./liter), or (iii) clean cotton plants. A fourth group was transferred from cotton to cotton as a control. Each replicate consisted of 5 confined adult females in each leaf clip cage; 6 replicates were used for each experiment. The number of eggs laid during this 48-h period was counted.
Statistical analyses.
The significance of the differences between means for all comparisons performed on data from the qPCR, qRT-PCR, and fecundity experiments was determined by replicating experiments at least three times and analyzing the data by using a paired t test with one-way analysis of variance (ANOVA) followed by a Tukey-Kramer honestly significant difference (HSD) test (α = 0.05). JMP7 (SAS Institute) was used for all statistical analyses. Significance (P) values of <0.05 indicate statistically significant differences between the compared means for any specified comparison.
RESULTS
Accumulation of TYLCV DNA and transcripts following virus acquisition from infected plants and retention or administration of purified virions.
After TYLCV acquisition from infected plants and retention on cotton, as described in Materials and Methods, insects were sampled immediately (0 h), every 24 h for up to 7 days, and then every week for 4 weeks. The amount of viral DNA was determined by qPCR using primers specific for the V1, V2, and C3 genes. The insect β-actin gene was used for calibration in this and all other experiments in this study, as this gene did not show any fluctuations under the different treatment conditions. Figure 1A shows that, regardless of the primers used and the gene being tested, the amount of viral DNA peaked 2 days after the insects were transferred to cotton and then decreased sharply, reaching almost undetectable levels by the end of the fourth week. Virus levels on day 2 were significantly higher than those at any other tested time point during the experiment (P = 0.0001 compared to 0 h of retention, as determined by a Tukey HSD test). This increase was likely due to virus replication in its insect vector. To support the replication hypothesis, we eliminated the plant factor by feeding whiteflies for 8 h on preparations of purified virions through membranes. The purity of the virus preparation was evaluated by transmission electron microscopy. Large fields of geminate particles were observed, whereas substantial cell debris was not conspicuous (Fig. 1B). After 8 h, the insects were transferred to cotton plants, and the amounts of viral DNA and V1 and C3 transcripts were measured by qPCR at the specified time intervals (Fig. 1B). The insects that fed on the virus preparation for 8 h were able to infect all of the tomato test plants used in the virus inoculation assays, indicating that the virus preparation contained infectious particles (data not shown). At 0 h, the insects did not contain detectable amounts of V1 or C3 RNA (Fig. 1B). In addition, attempts to prepare cDNA to identify V1 and C3 transcripts in the virion preparation were unsuccessful (not shown). These results indicated that whiteflies do not acquire viral transcripts while feeding on the preparations of viral particles. The amount of viral DNA associated with the whiteflies (using the V1 gene encoding CP as a marker) started to increase between days 1 and 3 and peaked 5 days after transfer to cotton plants (P = 0.0001 compared to 0 h of retention, as determined by a Tukey HSD test). At this time, the amount of viral DNA was ∼7-fold higher than the amount acquired by the insects (0 h), and it decreased thereafter. The accumulation of transcripts of two genes was monitored after the 8-h acquisition of viral particles through membranes: V1, transcribed from the viral genomic strand, and C3, transcribed from the viral complementary strand. The amount of V1 RNA was already at its maximum 1 day after the end of virion acquisition through membranes (P = 0.0001 compared to 0 h of retention, as determined by a Tukey HSD test), and it decreased thereafter. On the other hand, the amount of C3 RNA increased steadily after acquisition of the virions to a peak 5 days later (P = 0.002 compared to 0 h of retention, as determined by a Tukey HSD test), and it then decreased. Since C3 RNA results from transcription of the C3 gene, which is carried solely by the complementary virion genomic strand, the viral genomic strand (which carries the V1 gene) had to serve as the template for synthesis of the complementary virion genomic strand, together forming the dsDNA replicative form.
FIG 1.
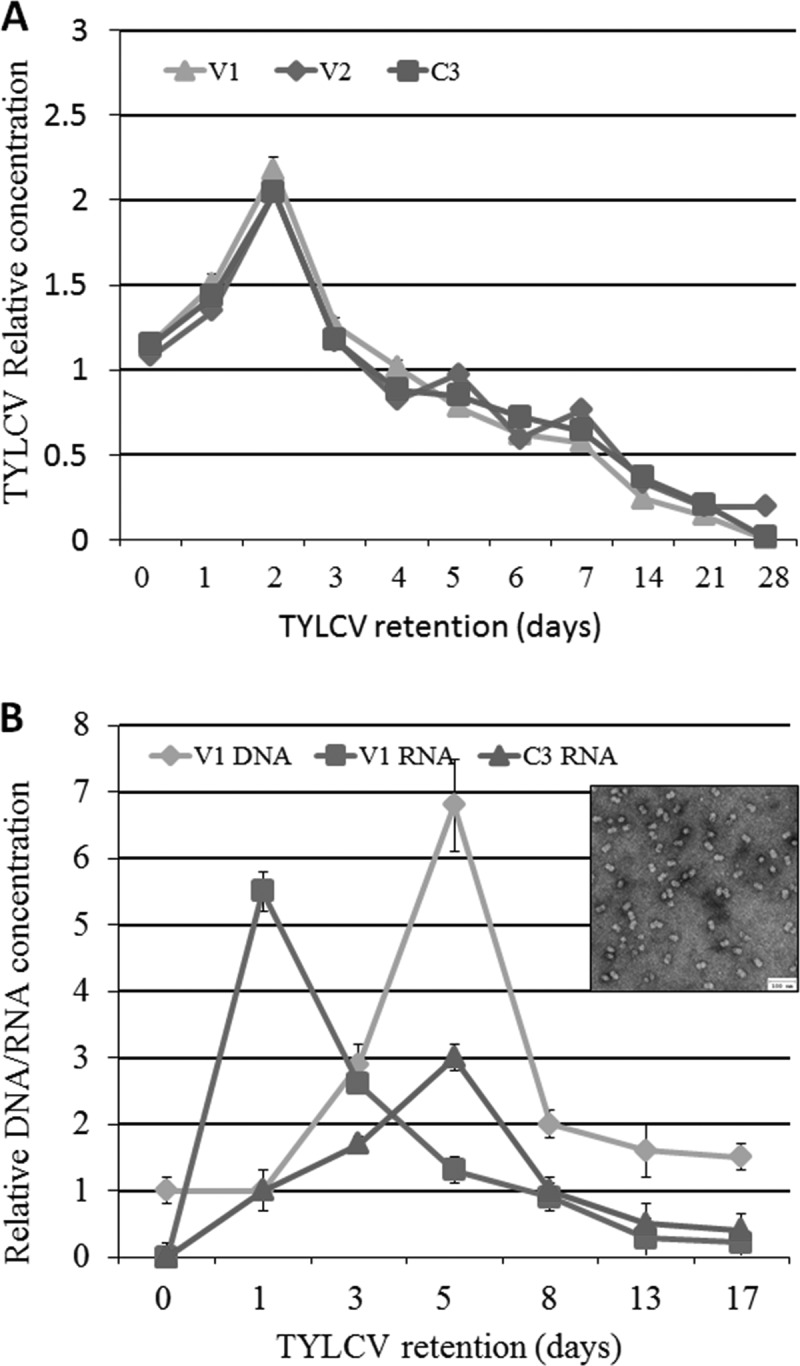
Relative concentrations of TYLCV DNA (A) and RNA (B) obtained by amplifying portions of the V1, V2, and C3 genes using qPCR and qRT-PCR. Adult females were provided an 8-h AAP on TYLCV-infected tomato plants (A) or purified TYLCV virions (B) and then transferred to cotton, a nonhost for TYLCV. Samples were collected at 0 h and on days 1 to 28 for the plant acquisition experiment and at 0 h and on days 1, 3, 5, 8, 13, and 17 days for the purified virion acquisition experiment. Results were normalized to the values for the host nuclear β-actin gene. Each experiment was performed in three replicates and statistically analyzed by a paired t test with an α value of 0.05. The error bars in all graphs represent standard errors of the means. The purified virions used in these experiments are shown in panel B.
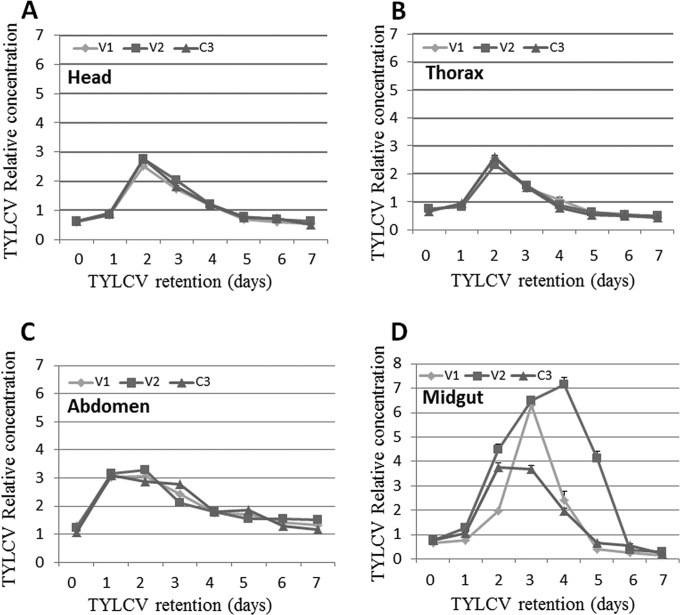
Since the translocation velocity of the virus in the insects depends on the insect organs that it reaches and passes through, the same experiment as the one shown in Fig. 1A was conducted, but the bodies of the insects were divided into three parts, each containing organs involved in the circulative transmission of TYLCV: head (salivary glands), thorax (esophagus), and abdomen (mid- and hindguts). The amount of virus was measured in insects collected every day for 7 days. As shown in Fig. 2, the virus patterns in the three different parts showed a trend similar to that in whole insects (summarized in Fig. 1): the virus levels reached a significant peak ∼2 days after the beginning of whitefly feeding on cotton plants (P = 0.0001 for the three genes in the head and thorax compared to 0 h of retention, as determined by a Tukey HSD test) and then gradually decreased until day 7. It should be noted that the virus amounts in the abdomen reached a significant peak after 1 day of retention, which lasted 2 days, compared to the peak on day 2 in the head and thorax (Fig. 2C, A, and B, respectively). The longer retention in the abdomen might have been related to the midgut, which functions as a major reservoir where virions accumulate during acquisition and are subsequently absorbed into the hemocoel. To test this hypothesis, we collected midguts from adults subjected to the experimental conditions described above. Within 2 to 4 days, the virus associated with the midgut reached levels ca. 8-fold higher than the levels found immediately after feeding on TYLCV-infected tomato plants (0 h), more than double the amount estimated to be present in the abdomen, head, and thorax (Fig. 2D). Furthermore, TYLCV amounts differed profoundly among the three genes tested, V1, V2, and C3, compared to the equal amounts observed when they were measured in the head, thorax, and abdomen. Virus levels reached their peak on day 3, when the amount of V1 was measured (P = 0.001 compared to 0 h of retention, as determined by a Tukey HSD test) and then sharply declined to undetectable levels on day 7. Measurements of V2 levels showed a gradual increase to day 4 (P = 0.023 compared to 0 h of retention, as determined by a Tukey HSD test), while C3 showed the lowest level of increase up to days 3 and 4, about half the amounts seen with V1 and V2; nevertheless, it was still highly significant (P = 0.001 compared to 0 h of retention, as determined by a Tukey HSD test).
FIG 2.
Relative concentration of TYLCV DNA obtained by amplifying portions of the V1, V2, and C3 genes from head (A), thorax (B), abdomen (C), and dissected midguts (D) using qPCR. Adult females were provided an 8-h AAP on TYLCV-infected tomato plants and then transferred to cotton, a nonhost for TYLCV. Adults were collected at 0 h and on days 1 to 7; their bodies were separated into head, thorax, and abdomen sections, or their midguts were dissected; and the virus levels were quantified. Results were normalized to the values for the host nuclear β-actin gene. Each experiment was performed in three replicates and statistically analyzed by using a paired t test with an α value of 0.05. The error bars in all graphs represent standard errors of the means.
TYLCV amounts change upon exposure of B. tabaci to stress.
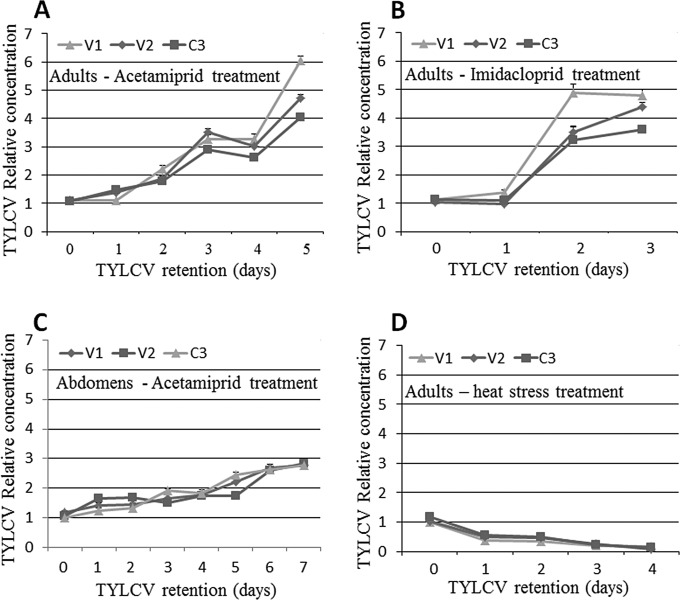
The increases in the amounts of viral DNA and RNA (using gene sequences on the virus genomic and complementary strands) associated with whiteflies following a short acquisition period suggested that TYLCV accumulates in the whitefly (up to 2 days in the whole body and up to 4 days in the midgut). On the other hand, the amounts of viral DNA and RNA decreased sharply thereafter (Fig. 1 and 2). These results suggested that after a short period of virus replication and expression, virus replication is inhibited, possibly by activation of a stress response mechanism in the insect. Imposing further stress on the whitefly, i.e., in addition to the virus (1, 32, 49), might change the dynamics of the virus in the insect. Two types of stress to which whitefly populations in the field are often subjected were applied to whiteflies after virus acquisition: pesticides and heat stress. Following an 8-h AAP on TYLCV-infected tomato, adult B. tabaci females were exposed to cotton plants treated with sublethal concentrations of either acetamiprid or imidacloprid, both neonicotinoid pesticides, for 5 days. An additional group of whiteflies on cotton plants was exposed to heat stress at 35°C for 3 days. As a control, viruliferous whiteflies that had acquired TYLCV for 8 h were reared on untreated cotton plants for 5 days at 25°C. Live adults were acquired from the pesticide-treated, heat-treated, and control groups every 24 h for up to 5 days, starting from 0 h (before exposure to stress). Virus DNA levels were quantified as described above. The use of V1, V2, and C3 gene sequences showed similar virus levels throughout the experiments, with no significant differences (Fig. 3). In the insects exposed to control conditions, the virus copy number peaked after 2 days and then decreased to undetectable levels by day 5, as found in previous experiments (not shown). Interestingly, the virus levels in viruliferous adults that were exposed to acetamiprid, imidacloprid, or heat stress showed different dynamics than those in the controls. Under conditions of acetamiprid exposure, the virus levels (using V1, V2, or C3) increased steadily during the 5 days of the experiment, without the decrease observed after 2 days in controls, reaching significantly higher levels than those at the start of the experiment (P = 0.0001 for the three genes on day 5 compared to 0 h of retention, as determined by a Tukey HSD test). The three genes showed the same pattern and magnitude of increase in virus amounts (Fig. 3A). Similar results and significances were obtained when the insects were exposed to imidacloprid, although the virus levels rapidly increased from day 1 to day 2 and increased only slightly on day 3 (Fig. 3B). To confirm these results, the levels of the virus were measured in abdomens separated from adults treated with acetamiprid for 7 days. Here, the virus levels increased constantly and peaked on day 7, without any decrease in the levels of any of the genes tested (Fig. 3C); their levels were significantly different from those at the start of the experiment (P = 0.0001 for the three genes on day 7 compared to 0 h of retention, as determined by a Tukey HSD test). Interestingly, when the virus levels were measured during 4 days of constant heat stress, a continuous and significant decrease was observed until the virus was no longer detectable, on day 4; none of the gene sequences used showed the typical increase up to 2 days observed in the experiments described above (Fig. 3D) (P = 0.0001 for the three genes on day 4 compared to 0 h of retention, as determined by a Tukey HSD test).
FIG 3.
Relative concentrations of TYLCV based on the gene sequences of V1, V2, and C3 measured by using qPCR, compared to the expression levels of the β-actin housekeeping gene and in a control experiment. (A and B) Relative concentrations of TYLCV in B. tabaci adult females after an 8-h AAP on TYLCV-infected tomato and exposure to 0.5 mg a.i./liter of acetamiprid for 5 days (A) or imidacloprid for 3 days (B). (C) Relative concentrations of TYLCV measured by qPCR based on V1, V2, and C3 sequences in dissected abdomens from adults that had acquired the virus for 8 h and retained it for 7 days. (D) Relative concentrations of TYLCV, based on the three gene sequences, in adults that had acquired the virus for 8 h and retained it for 4 days under heat stress at 35°C. Data shown are the means ± standard errors of the means of data from three independent experiments.
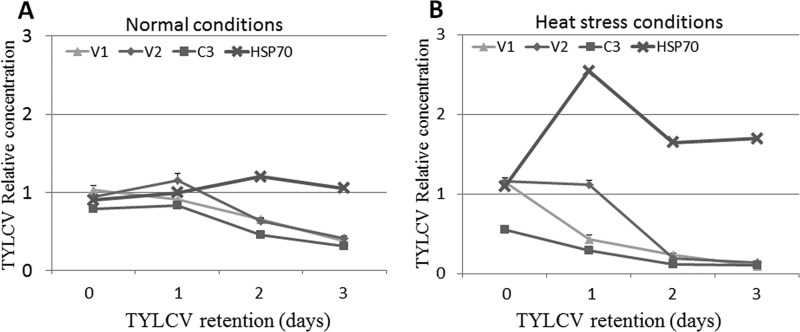
A previous study showed that HSP70 plays a role in TYLCV transmission by B. tabaci and has an inhibitory role against the virus (22). To test whether hsp70 expression changes following heat shock, whiteflies were provided an 8-h AAP on TYLCV-infected plants. The insects were then exposed to a temperature 35°C for 3 days, while control insects were exposed to a temperature of 25°C. The whiteflies were sampled every 24 h for up to 3 days for RNA extraction and qRT-PCR analysis (Fig. 4A). During the first day at 25°C, expression of the V2 and C3 genes increased slightly, while that of the V1 gene showed a slight decrease, although none of these changes were significant. On days 2 and 3, the three genes showed a gradual decrease in expression (Fig. 4A). Under these normal rearing conditions, hsp70 showed a slight increase in expression until day 2 and then a slight decrease on day 3; however, none of these changes was significant across the time points (Fig. 4A) (P = 0.39 for 0 h compared to day 3, as determined by a Tukey HSD test). For the heat stress experiment at 35°C, the results showed a gradual decrease in the expression of the three viral genes to levels that were no longer detectable by the end of day 3 (Fig. 4B). On the other hand, the expression level of hsp70 increased sharply, peaking on the first day of the heat stress; decreased on day 2, although it remained higher than initial levels at the start of the experiment; and remained at this level until the end of day 3 (Fig. 4B). The levels of hsp70 transcript on day 1 were significantly higher than those at any other time point tested (P = 0.002 for hsp70 on day 1 compared to 0 h or 3 days of retention, as determined by a Tukey HSD test). Quantification of HSP70 protein levels by using a Western blot approach showed similar results (not shown).
FIG 4.
Quantification of TYLCV V1, V2, and C3 gene transcripts and hsp70 gene transcripts by qRT-PCR under normal rearing conditions at a temperature of 25°C (A) and heat stress at 35°C (B) for 3 days, relative to the expression level of the housekeeping gene β-actin. Data shown are the means ± standard errors of the means of data from three independent experiments.
Dynamics of TYLCV levels in B. tabaci under changing stress conditions.
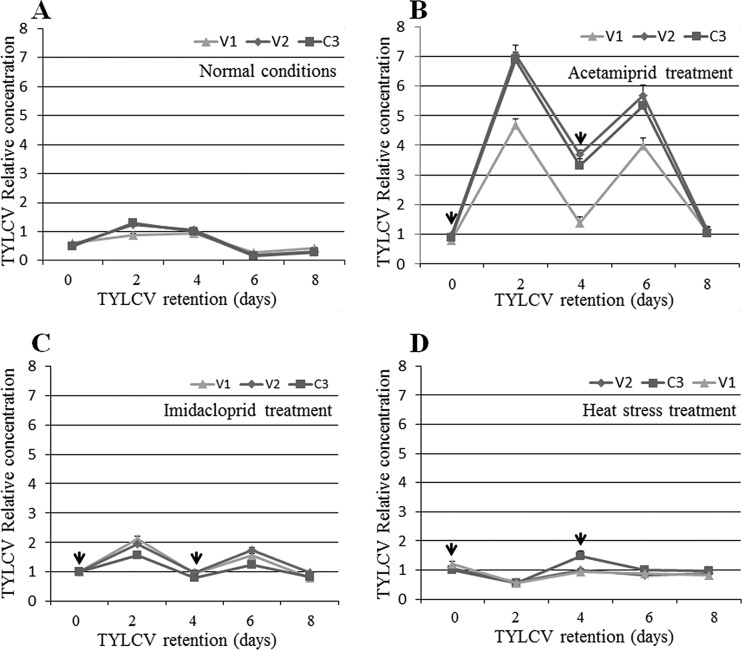
To confirm that stress is a major factor in the increase or decrease in virus levels, we designed experiments in which the insects were switched between stress and normal conditions several times (see Materials and Methods). The results presented in Fig. 5 show that the virus amounts peaked 2 days after retention and then decreased under normal conditions, as observed in the above-described experiments (Fig. 5A). The virus amounts after 2 days were significantly higher than those at the beginning (P = 0.012, as determined by a Tukey HSD test) and end (P = 0.0001, as determined by a Tukey HSD test) of the experiment. When the insects were switched from normal conditions to acetamiprid treatment for 2 days, after they had acquired TYLCV for 8 h, the virus amounts increased ∼7-fold when V2 and C3 levels were measured and ∼5-fold when V1 levels were measured (P = 0.0001 compared to 0 h of retention, as determined by a Tukey HSD test) (Fig. 5B). The virus amounts decreased ∼3-fold when the insects were switched back to untreated cotton for two more days (P = 0.0001 compared to 2 days of retention, as determined by a Tukey HSD test) but increased again significantly when the insects were transferred for a second time to acetamiprid and then decreased significantly to almost undetectable levels when the insects were reared for 2 days without treatment (Fig. 5B). The same significant fluctuations in TYLCV amounts, with similar significance values, were observed when the insects were switched between untreated leaves and leaves treated with imidacloprid; however, in this case, the virus amounts and the fluctuations were lower, albeit still significant, than those observed for the acetamiprid treatment (Fig. 5C). For example, virus amounts were up to 7-fold higher after the first 2 days of treatment with acetamiprid but only ∼2-fold higher when insects were exposed to imidacloprid for the same amount of time (Fig. 5B and C). Hence, upon exposure to heat stress at 35°C during the first 2 days, the virus levels decreased significantly, as observed for the above-described experiments (P = 0.001 compared to 0 h of retention, as determined by a Tukey HSD test); however, the levels significantly increased once the insects were transferred to a temperature of 25°C for 2 days, in direct opposition to the results obtained when insecticides were applied, where the virus levels first increased and then decreased (Fig. 5D). These fluctuations were also observed after switching the temperature from 35°C to 25°C for up to 8 days (Fig. 5D), and the differences in virus amounts were significant.
FIG 5.
Changes in TYLCV levels caused by altering rearing conditions of whiteflies that had acquired TYLCV and were switched between different treatments, as determined by qPCR on viral DNA. (A) Relative abundance of TYLCV measured by using three genes, V1, V2, and C3, in samples obtained from whiteflies after TYLCV acquisition for 8 h and retention for 0 h and 2, 4, 6, and 8 days. (B) Relative abundance of TYLCV in whiteflies after an 8-h AAP (0 h), rearing on acetamiprid-treated cotton leaves (arrows at days 0 and 4 indicate the start of rearing on treated leaves), and then transfer to clean cotton leaves for 48 h (days 2 and 6). (C) Relative concentration of TYLCV in whiteflies after an 8-h AAP (0 h) on TYLCV-infected plants, rearing on imidacloprid-treated cotton leaves (arrows at days 0 and 4 indicate the start of rearing on treated leaves), and then transfer to clean cotton leaves for 48 h (days 2 and 6). (D) Relative amount of TYLCV measured by sequences of three genes, V1, V2, and C3, in B. tabaci adults reared under heat stress (arrows at days 0 and 4 indicate the start of rearing under heat stress) for 2 days and under normal rearing temperatures (days 2 and 6). Data shown are the means ± standard errors of the means of data from three independent experiments.
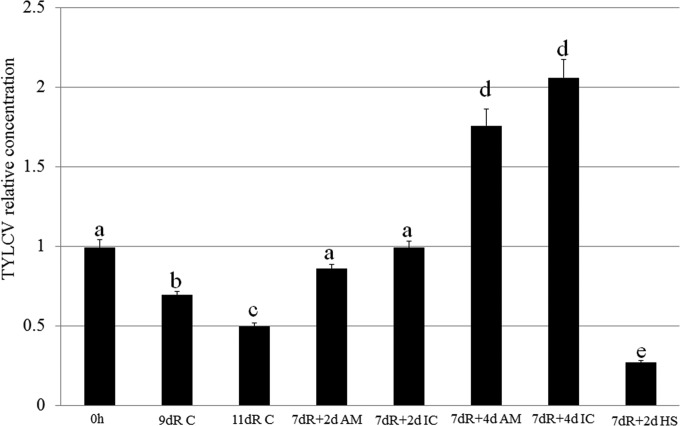
To further investigate the changes in virus amounts following insecticide treatment or heat stress, we measured the virus levels in insects exposed to the insecticide and heat treatments described above after longer retention periods. Whiteflies were transferred to cotton plants after an 8-h AAP on infected tomato plants. After 7 days on cotton, the insects were exposed to acetamiprid, imidacloprid, or heat stress for 2 or 4 days. The results summarized in Fig. 6 showed that following the initial 8-h AAP, retention for 9 or 11 days resulted in a significant decrease in virus levels. When the virus was retained for 7 days and the insects were then transferred to acetamiprid- or imidacloprid-treated leaves for 2 days, the levels of the virus rose significantly again, to levels close to those in the controls at the initial acquisition. When the rearing time on either acetamiprid- or imidacloprid-treated leaves was increased from 2 to 4 days, the virus levels significantly increased and reached higher levels than those for the 2-day treatment. Interestingly, when whiteflies retained TYLCV for 7 days and were then transferred to a temperature of 35°C for 2 days, the virus levels decreased dramatically and were the lowest (significantly so) among all treatments (Fig. 6).
FIG 6.
TYLCV dynamics measured by using the C3 gene sequence under different experimental conditions using qPCR on viral DNA. 0h, TYLCV levels measured immediately after an 8-h AAP; 9dR C, AAP for 8 h and retention on cotton leaves for 9 days; 11dR C, AAP for 8 h and retention on cotton leaves for 11 days; 7dR + 2d AM, AAP for 8 h, retention on cotton leaves for 7 days, and then retention on leaves treated with acetamiprid for an additional 2 days; 7dR + 2d IC, AAP for 8 h, retention on cotton leaves for 7 days, and then retention on leaves treated with imidacloprid for an additional 2 days; 7dR + 4d AM, AAP for 8 h, retention on cotton leaves for 7 days, and then retention on leaves treated with acetamiprid for an additional 4 days; 7dR + 4d IC, AAP for 8 h, retention on cotton leaves for 7 days, and then retention on leaves treated with acetamiprid for an additional 4 days; 7dR + 2d HS, AAP for 8 h, retention on cotton leaves for 7 days, and then retention under heat stress for an additional 2 days.
Localization of viral genomic and complementary viral genomic strands in whitefly midguts.
FISH was applied to visualize the virus genomic strand and its complementary strand (present only on the viral genome replicates), using fluorescent probes designed for different gene sequences on the virus genomic and complementary strands. Probes were designed to visualize V1 (CP) on the virus strand, cV1 for V1 gene sequences on the complementary strand, and cC1 (Rep protein) on the complementary strand. Localization studies were performed with midguts dissected from insects after an 8-h AAP on infected tomatoes (considered 0 h) and then with midguts from whiteflies reared for 2 and 7 days on cotton plants. Midguts dissected at 0 h (after 8 h of TYLCV acquisition) exhibited V1-specific signals in the filter chamber and, to a lesser extent, in other parts of the midgut, including the ceca and descending and ascending midguts (Fig. 7A). The cV1 and cC1 probes, which are complementary to sequences on the replicative form of the virus genomic strand, showed fluorescent signals throughout the midgut (Fig. 7B and C). After the viruliferous insects were reared on cotton plants for 2 days, the intensities of the fluorescent V1, cV1, and cC1 signals increased (Fig. 7D, E, and F, respectively) and then decreased after 7 days on the cotton plants (Fig. 7G, H, and I, respectively). These results were comparable to those obtained by qPCR using genes on the virus genomic strand and on the complementary genome strand (Fig. 1). The FISH results furthermore showed that the observed signals were limited mostly to the filter chamber and ceca and less frequently to the descending and ascending midguts (Fig. 7). Interestingly, probes for sequences on the virus complementary genome strand targeted sites that differed from those illuminated by the V1 viral strand probe and were specifically localized to the nuclei of midgut epithelial cells (Fig. 7E and F).
FIG 7.
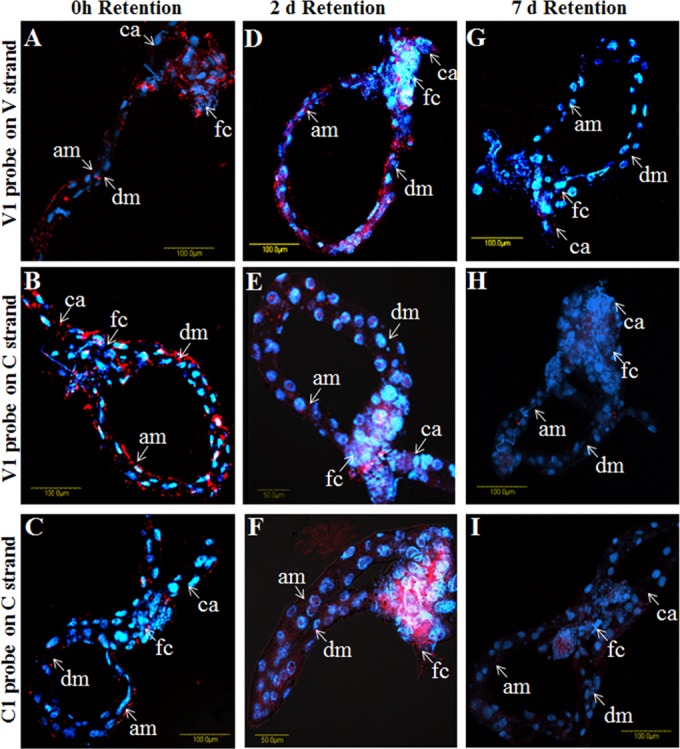
TYLCV localization in midguts dissected from B. tabaci adults that had acquired TYLCV for 8 h and retained it on cotton plants for 0 h (A to C), 2 days (D to F), and 7 days (G to I). Localization assays were performed by using FISH with the following fluorescent probes: V1 for CP on the virus DNA strand (A, D, and G), cV1 for the CP gene sequence on the replicative DNA form of the virus (B, E, and H), and cC1 for the Rep gene sequence on the replicative DNA form of the virus (C, F, and I). All probes were conjugated to Cy3 dye. Blue indicates DAPI staining of nuclei. am, ascending midgut; dm, descending midgut; fc, filter chamber; ca, ceca.
Localization of viral CP and DNA and host hsp70 transcripts in whitefly midgut following insecticide and heat stress.
qPCR and FISH showed that during the rearing of viruliferous whiteflies on cotton plants (following 8 h of acquisition of TYLCV), virus amounts increased to a peak at 2 days and then decreased to almost undetectable levels after 7 days (Fig. 1 and 7). Exposure of viruliferous whiteflies to insecticides and heat stress resulted in an increase and a decrease, respectively, in virus levels. FISH analysis was used to confirm the insecticide and heat stress results. The three probes V1, cV1, and cC1 were used to localize the virus genomic strand and its complementary genomic strand in midguts dissected from whiteflies subjected to the insecticide and heat stress treatments described above.
Following an 8-h AAP on TYLCV-infected plants, the whiteflies were transferred to cotton plants. The insects were sampled at 0 h and after 3 days for immunostaining and FISH analysis with anti-virus CP antibody and the V1 probe on the viral strand. In addition, following an 8-h AAP, additional insects were transferred to a temperature of 35°C and sampled at 0 and 24 h, and their midguts were then dissected and subjected to FISH analysis using the V1 probe and a probe targeting hsp70 mRNA. Midguts dissected at 0 h, before insecticide and heat treatments, showed virus signals mostly in the filter chamber, indicating that the insects had acquired detectable levels of virus (Fig. 8A and D and 9A). After 3 days on the cotton plants, the virus-associated signals decreased compared to those at 0 h (Fig. 8B and E). However, the virus signals drastically increased in midguts dissected from adults exposed to acetamiprid for 3 days, not only in the filter chamber but also along the midgut, including the ceca and descending and ascending midguts (Fig. 8C and F). As mentioned above, TYLCV FISH signals at 0 h, before exposure to heat stress (Fig. 9A), were increased to some extent after 1 day of rearing at 35°C and appeared to be more intense in the nuclei of the epithelial cells (Fig. 9B). The expression of hsp70 was monitored with an hsp70 fluorescent probe. The levels of hsp70 transcript at 0 h were barely detectable (Fig. 9D); however, the signals increased after 24 h on cotton plants (Fig. 9E). The hsp70 transcript was localized throughout the midgut, including within nuclei, and paralleled the presence of the virus (Fig. 9B). At 35°C, the levels of the hsp70 transcript increased, and it was concentrated in the filter chamber and the descending midgut (Fig. 9F). This change in hsp70 expression was accompanied by drastic decreases in TYLCV levels, which were almost undetectable by FISH (Fig. 9C). Taken together, these results suggested that while insecticide treatment increases TYLCV levels, heat stress results in upregulation of hsp70 and a decrease in virus levels.
FIG 8.
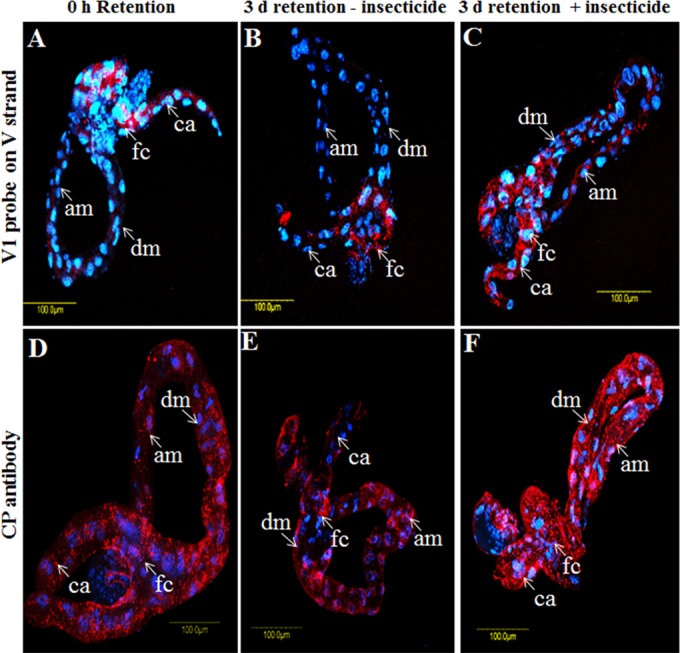
TYLCV (red) localization in midguts dissected from B. tabaci B biotype adults under various treatment conditions. Shown are data from FISH (A to C) using the TYLCV V1 probe and immunolocalization analysis (D to F), using anti-TYLCV CP antibody after virus acquisition for 8 h (A and D) and retention for 3 days without acetamiprid treatment (B and E) or after acetamiprid treatment for 3 days (C and F). The blue signal is DAPI staining of nuclei.
FIG 9.
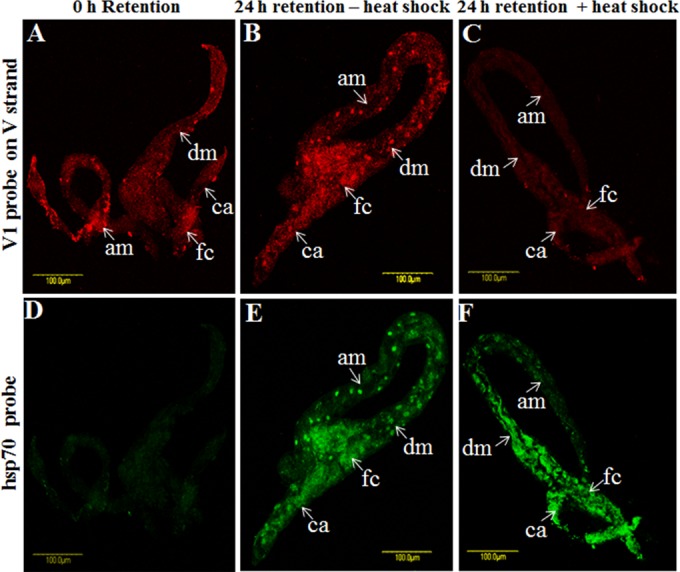
TYLCV (red) (A to C) and hsp70 (green) (D to F) localization in midguts dissected from B. tabaci B biotype adults under various treatment conditions, as determined by FISH analysis. Midguts were probed with virus and hsp70 probes after 8 h of virus acquisition (A and D), after virus retention for 24 h at 25°C without heat shock (B and E), or after virus retention for 24 h with heat shock (C and F).
Influence of TYLCV on whitefly fecundity under normal and stress conditions.
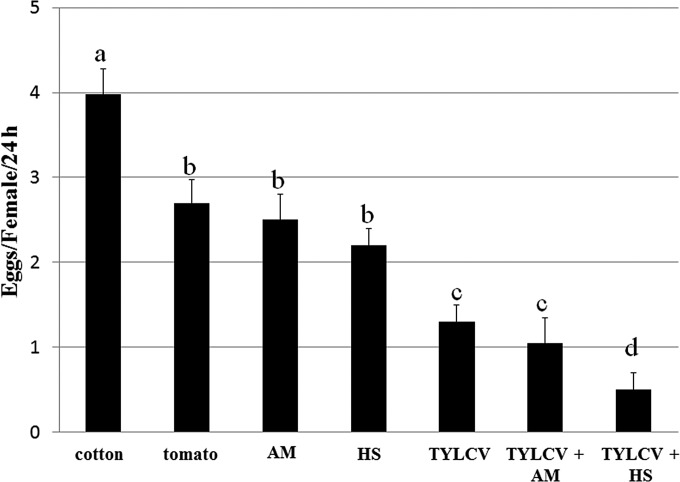
Since we previously reported that TYLCV influences the fecundity and fertility of B. tabaci (23), we tested the hypothesis that virus replication, alone or in combination with stress conditions, influences the insect's fecundity. The mean number of eggs laid by adults on cotton was ∼4 per female per 24 h (Fig. 10). On tomato, this number decreased to 2.7 eggs and was not significantly different from the number laid by whiteflies that were reared on leaves treated with sublethal concentrations of acetamiprid or those reared with a 35°C heat shock (Fig. 10). Interestingly, the number of eggs laid by insects that had acquired TYLCV was 1.3 per female per 24 h, significantly lower than those with previous treatments. When TYLCV was combined with acetamiprid or heat shock, the number of eggs laid decreased to ∼1 per female per 24 h with TYLCV-acetamiprid and 0.5 per female per 24 h with TYLCV-heat shock. The latter combination was the most potent at decreasing oviposition (Fig. 10).
FIG 10.
Effects of TYLCV, insecticide, and heat stress treatments and their combination on whitefly fecundity. Whiteflies were provided an 8-h AAP on TYLCV-infected tomato, retained the virus for 2 days, and were left to lay eggs on cotton leaves for 48 h. After virus retention and before egg laying, the insects were subjected to exposure to acetamiprid treatment (AM) or heat shock (HS). Data shown are the means ± standard errors of the means of data from six independent experiments. Different letters above the columns indicate statistically significant differences.
DISCUSSION
The aim of this study was to investigate whether TYLCV, a begomovirus that belongs to an emerging group of ssDNA plant viruses, is able to replicate and accumulate within its whitefly vector. Several previous reports provided indirect evidence supporting this premise, including accumulation of transcripts of genes located on the complementary virus strand (40), transmission through the egg and during mating (28–31), and deleterious (23) or beneficial (26) effects on fertility and longevity.
Figure 1A shows that following an 8-h AAP on infected tomato plants, the amounts of TYLCV associated with whiteflies increase during the first 2 days of feeding on cotton plants. We hypothesized that this increase was due to replication of the virus in its insect vector. To support this, we eliminated the plant factors (polymerases and primers, etc.) by feeding whiteflies for 8 h on preparations of purified virions through membranes. These insects were then transferred to cotton plants, and the amounts of viral DNA and transcript were measured. As shown in Fig. 1, where the insects acquired TYLCV from infected plants, the amounts of viral DNA associated with the whiteflies increased between days 1 and 3, peaked 5 days after transfer to cotton plants, and decreased thereafter (Fig. 1B). During rearing on cotton, the amounts of V1 and C3 transcripts increased and then decreased (Fig. 1B). The amount of V1 RNA peaked after only 1 day, possibly because large amounts of capsid protein are needed to build the geminate virion. The amounts of C3 RNA increased steadily up to 5 days and decreased thereafter. C3 RNA results from transcription of the C3 gene carried on the virus complementary strand. The viral genomic strand (which carries the V1 gene) serves as the template for complementary-strand synthesis and for the production of the dsDNA replicative form. These results greatly strengthened the replication hypothesis.
Following an increase upon acquisition, the amounts of virus subsequently decreased (Fig. 1 and 2). Inhibition of virus accumulation might be due to cessation of replication, and the decrease in virus amounts might be due to transmission to the cotton plant, destruction by the insect immune system, or some other defense mechanism in the whitefly that is activated against the virus. Indeed, it was previously reported that TYLCCNV is able to manipulate vector genes involved in stress and immune responses (32). The latter report suggested that the immune system or other defenses in the whitefly play a role in suppressing the virus's ability to replicate in B. tabaci. This suppression might also have life history trade-offs, as demonstrated for TYLCV, which had a significant effect on life history parameters in B. tabaci, such as fecundity and fertility (23). The role of the whitefly immune system or other defenses in suppressing virus replication needs to be further investigated, possibly by monitoring the expression dynamics of immune system or defense response genes after virus acquisition and retention. The possible activity of a defense or immune response could be further tested by challenging the insect with additional stresses following TYLCV acquisition and then testing the effect of this exposure on virus dynamics or other biological traits of the insect, because the insect has constraints in responding to multiple stresses. To investigate this, we tested whether the combination of TYLCV and additional stresses would influence the virus's ability to replicate. Imposing additional stress by exposing viruliferous whiteflies to insecticides resulted in continuous virus accumulation (Fig. 3), suggesting that a combination of two stresses impairs the insect's ability to cope with virus accumulation. The insecticides used in this study are neonicotinoids, a class of neuroactive insecticides that specifically bind to acetylcholine receptors in the insect's central nervous system, leading to overstimulation and blocked activity (50). Aside from general stress, this class of insecticides activates molecular responses, such as detoxification responses of P450 enzymes (51), and it has been shown to accelerate the negative impact of pathogens, including viruses, on honeybees by imposing additional stress (52). Thus, the responses caused by the neonicotinoids used in our study might have indirectly impacted the response of B. tabaci to TYLCV, which might in turn have led to the virus's replication. When heat stress was imposed, the opposite response was obtained, with an immediate and continuous decrease in virus amounts to undetectable levels (Fig. 3). The latter response was supported by the fact that imposing heat stress resulted in the accumulation of HSP70 and the hsp70 transcript (Fig. 4 and data not shown for protein levels), which has been shown to inhibit TYLCV transmission (22). It is also known that exposure to abiotic stresses such as heat shock, UV radiation, and chemical pesticides and to biotic stresses such as viruses, bacteria, fungi, and parasitoids may induce the expression of HSP70, which in turn activates the insect's stress response (53). It is therefore likely that in our experiments, exposure to heat led to the activation of a defense mechanism by HSP70, resulting in the inhibition of virus replication (Fig. 4). These results are supported by the increase followed by the decrease in virus levels after acquisition and retention and when the whiteflies were switched between pesticide/heat stress and normal conditions (Fig. 5), by the cumulative influence of exposure to pesticides for 2 and 4 days on virus levels, and by the strong decrease in virus levels after exposure to heat (Fig. 6). This additive influence suggests that the longer the exposure to pesticide stress, the greater the virus's ability to replicate, as illustrated by its levels 2 and 4 days after exposure to each of the pesticides.
The observation that virus accumulation lasted 2 to 3 days longer in the midgut than in the whole body of the whiteflies, and the higher levels of virus in the midgut than in whole insects or their separate parts, suggests that higher levels of virus replication and accumulation occur in midgut cells, as other tissues encountered before the virus reaches the midgut are chitin lined and cannot be penetrated (9). When the increase in virus levels in the midgut is compared with that in the rest of the insect, it seems that the virus increases to significantly high levels and that this increase extends for up to 3 days when targeting the V1 and C3 DNA levels and for up to 4 days when targeting V2 DNA. The differences in the increases in virus gene levels in the midgut might be only technical, as midguts were directly subjected to qPCR analysis without DNA extraction, and the accessibility of the primers to the viral DNA during the amplification reaction is expected not to be uniform. FISH analysis confirmed that the virus reaches high levels in midgut epithelial cells (13). These dynamic levels of TYLCV in the midgut, as observed by using qRT-PCR, qPCR, and environmental manipulation, were confirmed by FISH and immunolocalization experiments using probes targeting the V1, V2, and C3 sequences on the virus strand and the complementary strand and antibody against viral CP. These experiments showed an increase in virus signals after 2 days of retention and a decrease after 7 days (Fig. 7) or fluctuations in the virus levels when the environmental conditions were manipulated by using pesticides and heat stress (Fig. 8 and 9). FISH analysis further confirmed the association between the increase in HSP70 levels and the decrease in virus levels following heat stress (Fig. 9), confirming that heat stress induces HSP70 expression, which results in the inhibition of virus accumulation, possibly via the activation of immune or defense responses. The data from FISH analyses shown in Fig. 9B further demonstrated that the virus localizes to nuclei after 24 h of retention, the exact time when the accumulation of virus is at its peak (Fig. 1 and 2), suggesting that midgut cell nuclei are the sites of virus replication. The effect of the combination of heat stress and pesticides with virus retention on oviposition was remarkable compared to that of each treatment alone (Fig. 10). The effect of virus retention alone was stronger than that of heat stress or acetamiprid, both of which had significantly stronger effects on oviposition than no treatment on cotton or tomato plants. However, the strongest effects were observed when the acetamiprid or heat stress treatment was combined with virus, with the heat stress-virus combination having the strongest effect on the number of eggs laid. These results demonstrate that while pesticide treatment might extract some cost from having to cope with its toxic effects, thus leading to reduced defenses, heat stress causes overexpression of HSP70, which likely results in the activation of defenses, leading to an even stronger effect on oviposition.
REFERENCES
- 1.Czosnek H, Ghanim M, Rubinstein G, Morin S, Fridman V, Zeidan M. 2001. Whiteflies: vectors, and victims(?), of geminiviruses. Adv Virus Res 57:291–322. doi: 10.1016/S0065-3527(01)57006-2. [DOI] [PubMed] [Google Scholar]
- 2.Brown JK. 2010. Phylogenetic biology of the Bemisia tabaci sibling species group, p 31–67. In Stansley PA, Naranjo SE (ed), Bemisia—bionomics and management of a global pest. Springer, New York, NY. [Google Scholar]
- 3.De Barro PJ, Liu SS, Boykin LM, Dinsdale AB. 2011. Bemisia tabaci: a statement of species status. Annu Rev Entomol 56:1–19. doi: 10.1146/annurev-ento-112408-085504. [DOI] [PubMed] [Google Scholar]
- 4.Navot N, Pichersky E, Zeidan M, Zamir D, Czosnek H. 1991. Tomato yellow leaf curl virus: a whitefly-transmitted geminivirus with a single genomic molecule. Virology 185:151–161. doi: 10.1016/0042-6822(91)90763-2. [DOI] [PubMed] [Google Scholar]
- 5.Díaz-Pendón JA, Cañizares MC, Moriones E, Bejarano ER, Czosnek H, Navas-Castillo J. 2010. Tomato yellow leaf curl viruses: ménage à trois between the virus complex, the plant, and the whitefly vector. Mol Plant Pathol 11:441–450. doi: 10.1111/j.1364-3703.2010.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noris E, Vaira AM, Caciagli P, Masenga V, Gronemborn B, Accotto GP. 1998. Amino acids in the capsid protein of tomato yellow leaf curl virus that are crucial for systemic infection, particle formation, and insect transmission. J Virol 72:10050–10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caciagli P, Medina V, Marian D, Vecchiati M, Masenga V, Falcioni T, Noris E. 2009. Virion stability is important for the circulative transmission of Tomato yellow leaf curl Sardinia virus by Bemisia tabaci, but virion access to salivary glands does not guarantee transmissibility. J Virol 83:5784–5795. doi: 10.1128/JVI.02267-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutierrez C. 1999. Geminivirus DNA replication. Cell Mol Life Sci 56:313–329. doi: 10.1007/s000180050433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghanim M, Morin S, Czosnek H. 2001. Rate of Tomato yellow leaf curl virus translocation in the circulative transmission pathway of its vector, the whitefly Bemisia tabaci. Phytopathology 91:188–196. doi: 10.1094/PHYTO.2001.91.2.188. [DOI] [PubMed] [Google Scholar]
- 10.Ghanim M, Rosell RC, Campbell LR, Czosnek H, Brown JK, Ullman DE. 2001. Digestive, salivary, and reproductive organs of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) B type. J Morphol 248:22–40. doi: 10.1002/jmor.1018. [DOI] [PubMed] [Google Scholar]
- 11.Ghanim M, Brumin M, Popovski S. 2009. A simple, rapid and inexpensive method for localization of tomato yellow leaf curl virus and potato leafroll virus in plant and insect vectors. J Virol Methods 159:311–315. doi: 10.1016/j.jviromet.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 12.Ghanim M, Medina V. 2007. Localization of tomato yellow leaf curl virus in its whitefly vector Bemisia tabaci, p 171–183. In Czosnek H. (ed), Tomato yellow leaf curl virus disease management, molecular biology, breeding for resistance. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 13.Skaljac M, Ghanim M. 2010. Tomato yellow leaf curl disease and plant-virus vector interactions. Isr J Plant Sc 58:103–111. doi: 10.1560/IJPS.58.2.103. [DOI] [Google Scholar]
- 14.Cicero JM, Brown JK. 2011. Functional anatomy of whitefly organs associated with squash leaf curl virus (Geminiviridae: Begomovirus) transmission by the B biotype of Bemisia tabaci (Hemiptera: Aleyrodidae). Ann Entomol Soc Am 104:261–279. doi: 10.1603/AN10075. [DOI] [Google Scholar]
- 15.Cicero JM, Brown JK. 2011. Anatomy of accessory salivary glands of the whitefly Bemisia tabaci (Hemiptera: Aleyrodidae) and correlations to begomovirus transmission. Ann Entomol Soc Am 104:280–286. doi: 10.1603/AN10171. [DOI] [Google Scholar]
- 16.Czosnek H, Ghanim M, Ghanim M. 2002. Circulative pathway of begomoviruses in the whitefly vector Bemisia tabaci—insights from studies with Tomato yellow leaf curl virus. Ann Appl Biol 140:215–231. doi: 10.1111/j.1744-7348.2002.tb00175.x. [DOI] [Google Scholar]
- 17.Morin S, Ghanim M, Sobol I, Czosnek H. 2000. The GroEL protein of the whitefly Bemisia tabaci interacts with the coat protein of transmissible and non-transmissible begomoviruses in the yeast two-hybrid system. Virology 276:404–416. doi: 10.1006/viro.2000.0549. [DOI] [PubMed] [Google Scholar]
- 18.Rosell RC, Torres-Jerez I, Brown JK. 1999. Tracing the geminivirus-whitefly transmission pathway by polymerase chain reaction in whitefly extracts, saliva, hemolymph, and honeydew. Phytopathology 89:239–246. doi: 10.1094/PHYTO.1999.89.3.239. [DOI] [PubMed] [Google Scholar]
- 19.Wei J, Zhao JJ, Zhang T, Li FF, Ghanim M, Zhou XP, Ye GY, Liu SS, Wang XW. 2014. Specific cells in the primary salivary glands of the whitefly Bemisia tabaci control retention and transmission of begomoviruses. J Virol 88:13460–13468. doi: 10.1128/JVI.02179-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang LL, Wei XM, Ye XD, Xu HX, Zhou XP, Lu SS, Wang XW. 2014. Expression and functional characterisation of a soluble form of Tomato yellow leaf curl virus coat protein. Pest Manag Sci 70:1624–1631. doi: 10.1002/ps.3750. [DOI] [PubMed] [Google Scholar]
- 21.Hohnle M, Hofer P, Bedford ID, Briddon RW, Markham PG, Frischmuth T. 2001. Exchange of three amino acids in the coat protein results in efficient whitefly transmission of a nontransmissible Abutilon mosaic virus isolate. Virology 290:164–171. doi: 10.1006/viro.2001.1140. [DOI] [PubMed] [Google Scholar]
- 22.Götz M, Popovski S, Kollenberg M, Gorovits R, Brown JK, Cicero J, Czosnek H, Winter S, Ghanim M. 2012. Implication of Bemisia tabaci heat shock protein in begomovirus-whitefly interactions. J Virol 86:13241–13252. doi: 10.1128/JVI.00880-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubinstein G, Czosnek H. 1997. Long-term association of tomato yellow leaf curl virus with its whitefly vector Bemisia tabaci: effect on the insect transmission capacity, longevity and fecundity. J Gen Virol 78:2683–2689. [DOI] [PubMed] [Google Scholar]
- 24.Ohnesorge S, Bejarano ER. 2009. Begomovirus coat protein interacts with a small heatshock protein of its transmission vector (Bemisia tabaci). Insect Mol Biol 18:693–703. doi: 10.1111/j.1365-2583.2009.00906.x. [DOI] [PubMed] [Google Scholar]
- 25.Czosnek H. 2009. Acquisition, circulation and transmission of begomoviruses by their whitefly vectors, p 29–44. In Palombo EA, Kirkwood CD (ed), Viruses in the environment. Research Signpost, Trivandrum, Kerala, India. [Google Scholar]
- 26.Jiu M, Zhou XP, Tong L, Xu J, Yang X, Wan FH, Liu SS. 2007. Vector-virus mutualism accelerates population increase of an invasive whitefly. PLoS One 2:e182. doi: 10.1371/journal.pone.0000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuura S, Hoshino S. 2009. Effect of tomato yellow leaf curl disease on reproduction of Bemisia tabaci Q biotype (Hemiptera: Aleyrodidae) on tomato plants. Appl Entomol Zool 44:143–148. doi: 10.1303/aez.2009.143. [DOI] [Google Scholar]
- 28.Ghanim M, Czosnek H. 2000. Tomato yellow leaf curl geminivirus (TYLCV-Is) is transmitted among whiteflies (Bemisia tabaci) in a sex-related manner. J Virol 74:4738–4745. doi: 10.1128/JVI.74.10.4738-4745.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosco D, Mason G, Accotto GP. 2004. TYLCSV DNA, but not infectivity, can be transovarially inherited by the progeny of the whitefly vector Bemisia tabaci (Gennadius). Virology 323:276–283. doi: 10.1016/j.virol.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 30.Ghanim M, Sobol I, Ghanim M, Czosnek H. 2007. Horizontal transmission of begomoviruses between Bemisia tabaci biotypes. Arthropod Plant Interact 1:195–204. doi: 10.1007/s11829-007-9018-z. [DOI] [Google Scholar]
- 31.Wang J, Zhao H, Liu J, Jiu M, Qian YJ, Liu SS. 2010. Low frequency of horizontal and vertical transmission of two begomoviruses through whiteflies exhibits little relevance to the vector infectivity. Ann Appl Biol 157:125–133. doi: 10.1111/j.1744-7348.2010.00403.x. [DOI] [Google Scholar]
- 32.Luan JB, Li JM, Varela N, Wang YL, Li FF, Bao YY, Zhang CX, Liu SS, Wang XW. 2011. Global analysis of the transcriptional response of whitefly to tomato yellow leaf curl China virus reveals their relationship of coevolved adaptations. J Virol 85:3330–3340. doi: 10.1128/JVI.02507-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Redinbaugh MG, Hogenhout SA. 2005. Plant rhabdoviruses. Curr Top Microbiol Immunol 292:143–163. [DOI] [PubMed] [Google Scholar]
- 34.Nault LR. 1997. Arthropod transmission of plant viruses: a new synthesis. Ann Entomol Soc Am 90:521–541. doi: 10.1093/aesa/90.5.521. [DOI] [Google Scholar]
- 35.Hogenhout SA, Ammar E-D, Whitfield AE, Redinbaugh MG. 2008. Insect vector interactions with persistently transmitted viruses. Annu Rev Phytopathol 46:327–359. doi: 10.1146/annurev.phyto.022508.092135. [DOI] [PubMed] [Google Scholar]
- 36.Ullman DE, German T, Sherwood JL, Wescot DM, Cantone FA. 1993. Immunocytochemical evidence that the nonstructural protein encoded by the S RNA of tomato spotted wilt tospovirus is present in thrips vector cells. Phytopathology 83:456–463. doi: 10.1094/Phyto-83-456. [DOI] [Google Scholar]
- 37.Medeiros RB, De Resende O, De Avila AC. 2004. The plant virus Tomato spotted wilt Tospovirus activates the immune system of its main insect vector, Frankliniella occidentalis. J Virol 78:4976–4982. doi: 10.1128/JVI.78.10.4976-4982.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghanim M, Morin S, Zeidan M, Czosnek H. 1998. Evidence for transovarial transmission of tomato yellow leaf curl virus by its vector, the whitefly Bemisia tabaci. Virology 240:295–303. doi: 10.1006/viro.1997.8937. [DOI] [PubMed] [Google Scholar]
- 39.Su Q, Pan H, Liu B, Chu D, Xie W, Wu Q, Wang S, Xu B, Zhang Y. 2013. Insect symbiont facilitates vector acquisition, retention and transmission of plant virus. Sci Rep 3:1367. doi: 10.1038/srep01367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sinisterra XH, McKenzie CL, Hunter WB, Powell CA, Shatters RG. 2005. Differential transcriptional activity of plantpathogenic begomoviruses in their whitefly vector (Bemisia tabaci, Gennadius: Hemiptera Aleyrodidae). J Gen Virol 86:1525–1532. doi: 10.1099/vir.0.80665-0. [DOI] [PubMed] [Google Scholar]
- 41.Skaljac M, Zanić K, Hrnčić S, Radonjić S, Perović T, Ghanim M. 2013. Diversity and localization of bacterial symbionts in three whitefly species (Hemiptera: Aleyrodidae) from the east coast of the Adriatic sea. Bull Entomol Res 103:48–59. doi: 10.1017/S0007485312000399. [DOI] [PubMed] [Google Scholar]
- 42.Czosnek H, Ber R, Antignus Y, Cohen S, Navot N, Zamir D. 1988. Isolation of the tomato yellow leaf curl virus—a geminivirus. Phytopathology 78:508–512. doi: 10.1094/Phyto-78-508. [DOI] [Google Scholar]
- 43.Brumin M, Kontsedalov S, Ghanim M. 2011. Rickettsia influences thermotolerance in the whitefly Bemisia tabaci B biotype. Insect Sci 18:57–66. doi: 10.1111/j.1744-7917.2010.01396.x. [DOI] [Google Scholar]
- 44.Kontsedalov S, Zchori-Fein E, Chiel E, Gottlieb Y, Inbar M, Ghanim M. 2008. The presence of Rickettsia is associated with increased susceptibility of Bemisia tabaci (Homoptera: Aleyrodidae) to insecticides. Pest Manag Sci 64:789–792. doi: 10.1002/ps.1595. [DOI] [PubMed] [Google Scholar]
- 45.Ghanim M, Kontsedalov S. 2009. Susceptibility to insecticides in the Q biotype of Bemisia tabaci is correlated with bacterial symbiont densities. Pest Manag Sci 65:939–942. doi: 10.1002/ps.1795. [DOI] [PubMed] [Google Scholar]
- 46.Shahjahan RM, Hughes KJ, Leopold RA, DeVault JD. 1995. Lower incubation temperature increases yield of insect genomic DNA isolated by the CTAB method. Biotechniques 19:332–334. [PubMed] [Google Scholar]
- 47.Gottlieb Y, Ghanim M, Chiel E, Gerling D, Portnoy V, Steinberg S, Tzuri GA, Horowitz R, Belausov E, Mozes-Daube N, Kontsedalov S, Gershon M, Gal S, Katzir N, Zchori-Fein E. 2006. Identification and localization of a Rickettsia sp. in Bemisia tabaci (Homoptera: Aleyrodidae). Appl Environ Microbiol 72:3646–3652. doi: 10.1128/AEM.72.5.3646-3652.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skaljac M, Zanic K, Goreta-Ban S, Kontsedalov S, Ghanim M. 2010. Co-infection and localization of secondary symbionts in two whitefly species. BMC Microbiol 10:142. doi: 10.1186/1471-2180-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czosnek H, Ghanim M. 2011. Bemisia tabaci-Tomato yellow leaf curl virus interaction causing worldwide epidemics, p 51–61. In Thompson WMO. (ed), The whitefly, Bemisia tabaci (Homoptera: Aleyrodidae) interaction with geminivirus-infected host plants. Springer Science and Business Media, New York, NY. [Google Scholar]
- 50.Kollmeyer WD, Flattum RF, Foster JP, Powell JE, Schroeder ME, Soloway SB. 1999. Discovery of the nitromethylene heterocycle insecticides, p 71–89. In Yamamoto I, Casida J (ed), Nicotinoid insecticides and the nicotinic acetylcholine receptor. Springer-Verlag, Tokyo, Japan. [Google Scholar]
- 51.Karunker I, Benting J, Lueke B, Ponge T, Nauen R, Roditakis E, Vontas J, Gorman K, Denholm I, Morin S. 2008. Over-expression of cytochrome P450 CYP6CM1 is associated with high resistance to imidacloprid in the B and Q biotypes of Bemisia tabaci (Hemiptera: Aleyrodidae). Insect Biochem Mol Biol 38:634–644. doi: 10.1016/j.ibmb.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 52.Doublet V, Labarussias M, de Miranda JR, Moritz RFA, Paxton RJ. 2015. Bees under stress: sublethal doses of a neonicotinoid pesticide and pathogens interact to elevate honey bee mortality across the life cycle. Environ Microbiol 17:969–983. doi: 10.1111/1462-2920.12426. [DOI] [PubMed] [Google Scholar]
- 53.Moret Y, Schmid-Hempel P. 2001. Immune defence in bumble-bee offspring. Nature 414:506. doi: 10.1038/35107138. [DOI] [PubMed] [Google Scholar]