ABSTRACT
Thirty-seven goats carrying different prion protein genotypes (PRNP) were orally infected with a classical scrapie brain homogenate from wild-type (ARQ/ARQ) sheep and then mated to obtain 2 additional generations of offspring, which were kept in the same environment and allowed to be naturally exposed to scrapie. Occurrence of clinical or subclinical scrapie was observed in the experimentally infected goats (F0) and in only one (F1b) of the naturally exposed offspring groups. In both groups (F0 and F1b), goats carrying the R154H, H154H, R211Q, and P168Q-P240P dimorphisms died of scrapie after a longer incubation period than wild-type, G37V, Q168Q-P240P, and S240P goats. In contrast, D145D and Q222K goats were resistant to infection. The immunobiochemical signature of the scrapie isolate and its pathological aspects observed in the sheep donors were substantially maintained over 2 goat generations, i.e., after experimental and natural transmission. This demonstrates that the prion protein gene sequence, which is shared by sheep and goats, is more powerful than any possible but unknown species-related factors in determining scrapie phenotypes. With regard to genetics, our study confirms that the K222 mutation protects goats even against ovine scrapie isolates, and for the first time, a possible association of D145 mutation with scrapie resistance is shown. In addition, it is possible that the sole diverse frequencies of these genetic variants might, at least in part, shape the prevalence of scrapie among naturally exposed progenies in affected herds.
IMPORTANCE This study was aimed at investigating the genetic and pathological features characterizing sheep-to-goat transmission of scrapie. We show that in goats with different prion protein gene mutations, the K222 genetic variant is associated with scrapie resistance after natural and experimental exposure to ovine prion infectivity. In addition, we observed for the first time a protective effect of the D145 goat variant against scrapie. Importantly, our results demonstrate that the phenotypic characteristic of the wild-type sheep scrapie isolate is substantially preserved in goats carrying different susceptible PRNP gene variants, thus indicating that the prion protein gene sequence, which is shared by sheep and goats, plays a fundamental role in determining scrapie phenotypes.
INTRODUCTION
Scrapie is a slowly progressive and ultimately fatal neurodegenerative disease affecting sheep, goats and mouflons. It is part of the family of transmissible spongiform encephalopathies (TSEs) that include similar diseases such as bovine spongiform encephalopathy (BSE) in cattle, Creutzfeldt-Jakob disease (CJD) in humans, and the related TSEs in exotic ungulates and cats (1).
TSEs are caused by an unconventional agent (prion) corresponding largely, if not entirely, to the disease-associated isoform (PrPSc) of the normal host-encoded cellular prion protein (PrPC), which accumulates in the brain and other tissues of sick animals (1).
In sheep, the occurrence of scrapie is strongly associated with certain polymorphisms of the gene region (PRNP) encoding the C-terminal structured domain of the prion protein (2). So far, it is widely accepted that 3 polymorphic codons of the PRNP gene, namely, codons 136, 154, and 171, modulate susceptibility or resistance to classical scrapie in this species (3).
On the basis of this knowledge, the European Community applied a selective breeding plan designed to increase the frequency of the ARR allele, with the objective of controlling scrapie in sheep (4).
Numerous polymorphic codons have also been reported in the open reading frame (ORF) of the goat PRNP gene, which has the ancestral PrPC amino acid sequence seen in wild-type sheep (3), with some dimorphic codons being shared by both species (3, 5). Because of the low frequency of these genetic variants and the small number of the scrapie cases considered in the studies, few associations between scrapie resistance and certain mutations in the goat PRNP gene have been reported. However, the most encouraging, albeit incomplete, data have been reported so far for Q222K, R154H, R211Q, and N146S dimorphisms (6–10). Most importantly, it has yet to be determined whether a correlation between these mutations and scrapie resistance is universally conserved against the different scrapie strains. Indeed, in sheep, the susceptibility to prion infection is strongly influenced by the nature of the TSE strain in addition to PRNP mutations (4, 11). In this respect, it is well known that different TSE agents circulate in small ruminant populations across Europe (11, 12). However, a conclusive representation of this variability in sheep is not available, and there is even less information for goats. In particular, the relationship between goat and sheep scrapie strains is not clear (5).
A recent investigation in Italy showed that goat scrapie isolates share the same pK-resistant PrPSc profile as reported in Italian scrapie sheep cases (6). In addition, after inoculation of bank voles, C57BL mice, and hamsters, Italian sheep and goat isolates showed similar transmission characteristics with regard to incubation time, presence of clinical disease, and attack rate (13).
In the United Kingdom, an immunohistochemical antibody targeting study showed no difference between sheep and goats experimentally infected with SSBP/1, CH1641 scrapie strains, and BSE (14). However, the immunohistochemical analysis of only 4 natural cases of goat scrapie, included in this study, displayed a variability of phenotypes, thus indicating the existence of multiple strains within the goat population.
Taken together, these studies reveal that sheep and goats share many etiopathological aspects of scrapie and suggest that there is apparently no interspecies barrier; however, which factors regulated that transmission remains to be established. In this context, it is worthwhile to know how a scrapie agent of ovine source propagates in goats, particularly in relation to the numerous polymorphic codons of the caprine PRNP gene. This is particularly relevant for understanding whether goats can act as a reservoir of the scrapie agent, in countries in which a selective breeding plan for scrapie resistance is being carried out in sheep populations.
Here, we studied the dynamics of scrapie occurrence with particular emphasis on genetic and phenotypical aspects across two generations in a goat herd in which the parental generation was previously orally challenged with a natural sheep scrapie isolate.
This case study was based in the Sardinia region of Italy. So far, in Sardinia only a few natural cases of scrapie in goats have been observed within the framework of scrapie diagnostic surveillance activity, and all these scrapie-infected goats carried the same PRNP sequence as in wild-type sheep, which is referred to here as the wild-type genotype.
MATERIALS AND METHODS
Experimental design.
Thirty-seven goat kids (F0) were acquired from 10 historically scrapie-free herds located in Sardinia. The PrP genotypes of these kids are detailed in Tables 1 and 2.
TABLE 1.
PRNP genotype, postinfection time, clinical status, and PrPSc detection in goats orally infected with an ovine scrapie brain homogenatea
| Goat | Genotype | Clinical signs | Incubation time (days) | PrPSc detection |
|
|---|---|---|---|---|---|
| Obex | LT | ||||
| 0931 | WT | Yes | 700 | Pos | Pos |
| 0937 | WT | Yes | 613 | Pos | Pos |
| 0947 | WT | Yes | 779 | Pos | Pos |
| 0948 | WT | Yes | 738 | Pos | Pos |
| 0932 | WT | Yes | 681 | Pos | Pos |
| 0946 | G37V | Yes | 766 | Pos | Pos |
| 0941 | G37V | Yes | 723 | Pos | Pos |
| 0903 | G37V | Yes | 688 | Pos | Pos |
| 0908 | G37V-Q222K | No | 1,961 | Neg | Neg |
| 0951 | D145D | No | 1,913 | Neg | Neg |
| 0955 | D145D | No | 1,913 | Neg | Neg |
| 0921 | H154H | Yes | 1,184 | Pos | Pos |
| 0911 | R154H | Yes | 1,101 | Pos | Pos |
| 0926 | R154H | Yes | 955 | Pos | Pos |
| 0902 | R154H-S240P | Yes | 1,030 | Pos | Pos |
| 0904 | R154H-S240P | Yes | 898 | Pos | Pos |
| 0935 | P168Q-P240P | Yes | 1,184 | Pos | Pos |
| 0927 | Q168Q-P240P | Yes | 672 | Pos | Pos |
| 0924 | R211Q | Yes | 2,109 | Pos | Pos |
| 0919 | R211Q-S240P | Yes | 1,347 | Pos | Pos |
| 0928 | R211Q-S240P | Yes | 1,712 | Pos | Pos |
| 0950 | Q222K | No | 2,067 | Neg | Neg |
| 0923 | Q222K-S240P | No | 1,848 | Neg | Neg |
| 0944 | S240P | Yes | 743 | Pos | Pos |
| 0953 | P240P | Yes | 778 | Pos | Pos |
| 0922 | P240P | Yes | 711 | Pos | Pos |
PrPSc was detected via immunohistochemical and Western blotting techniques. LT, lymphoid tissue; Pos, positive; Neg, negative; WT, wild type.
TABLE 2.
Immunohistochemical (IHC) and Western blotting results for PrPSc detection in nervous and lymphoid tissues of goats which were orally challenged with an ovine scrapie isolate and which died of unrelated disease during the course of the studya
| Goat | Genotype | Clinical signs | Incubation time | PrPSc detection |
|
|---|---|---|---|---|---|
| Obex | LT | ||||
| 0917 | WT | No | 205 | Neg | Pos |
| 0929 | R211Q | No | 464 | Neg | Neg |
| 0909 | G145D | No | 185 | Neg | Neg |
| 0914 | G145D-S240P | No | 203 | Neg | Neg |
| 0912 | P240P | No | 196 | Neg | Neg |
| 0906 | Q222K | No | 161 | Neg | Neg |
| 0942 | R211Q | No | 1806 | Pos | Pos |
| 0952 | R211Q-S240P | No | 1036 | Pos | Pos |
| 0910 | G145D-Q222K | No | 1485 | Neg | Neg |
| 0913 | Q222K | No | 1346 | Neg | Neg |
| 0933 | Q222K-S240P | No | 919 | Neg | Neg |
LT, lymphoid tissue; Pos, positive; Neg, negative; WT, wild type.
Kids were weaned at 25 days of age and then were housed together in a barn. Between 60 and 70 days of age, they were infected via the oral route with classical scrapie-infected sheep brain tissue.
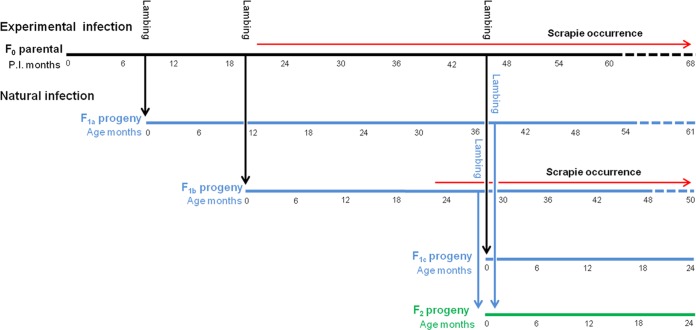
All inoculated animals were mated at 4 months postinfection (p.i.) with 2 external he-goats, carrying, respectively, the wild-type and the Q222K genotype. By using the same he-goats after the first lambing, the same goats were mated a second time at 15 months p.i.; finally, the 4 surviving goats were further mated at 41 months p.i. By doing so we obtained 3 different progenies of the first generation (F1a, F1b, and F1c), which were kept in the same environment with the dams, thus effectively remaining naturally exposed to the scrapie agent. F1a and F1b progenies were in turn allowed to mate at 33 and 22 months of age, respectively, with external wild-type and Q222K he-goats to produce a second generation of progeny (F2a); see Fig. 1 for other details.
FIG 1.
Representation of the experimental scheme. Different progenies of the first generation (F1a, F1b, and F1c) were obtained from 37 goats orally inoculated with scrapie ovine brain homogenate (F0). Subsequently, a second generation of progeny (F2a) was produced from F1a and F1b progenies. All goats were kept in the same environment. The period of occurrence of scrapie in F0 and F1b groups is indicated as months p.i. or months of age of the affected animals.
The goat were intensively managed without access to pasture. In detail, inoculated goats and their offspring were kept together in a single barn with a solid floor of cement connected to an outdoor paddock adequately drained to prevent water and mud buildup. In total, a space allowance of 4 m2 per goat was provided. After birth, kids stayed with the dams for 2 months and then were detached for weaning together with age cohort mates in a separate similar barn. At 3 months of age, the kids returned to be kept in the same barn with the adult goats. For mating and lambing, the goats were temporarily separate in different groups. Goats were fed hay and a concentrate based on cereals adequately enriched with essential minerals and vitamins. The same feeding troughs and drinking troughs were communally available for the goats.
Clinical monitoring of the animals was performed daily. The animals were euthanized after showing severely impaired locomotor capacity indicative of scrapie and after manifesting other incurable pathological conditions.
Inoculum preparation and infection procedure.
The homogenate, in normal saline (10% [wt/vol]), was prepared for inoculation from a pool of brains of wild-type sheep with classical scrapie collected in the framework of the passive surveillance plan and identified as classical scrapie by the Italian National Reference Laboratory for TSE strain typing. Then, 25 ml of this 25% homogenate was slowly injected into the mouths of the kids by using a syringe; it was ascertained that the total inoculum was ingested.
Sampling.
From all the animals included in the experiment, during postmortem examination, the brain, the gut-associated lymphoid tissue (GALT), including tonsils, and several lymph nodes were adequately sampled.
All these samples were fixed in buffered formaldehyde solution (4% [wt/vol]), and one part was stored at −20°C. Each sheep's brain was sagitally divided into two halves; one half was fixed in formaldehyde for immunohistochemical (IHC) investigations, while the other one was frozen at −20°C for immunobiochemical analysis. Finally, placentas from 25 parturient goats were collected and stored at −20°C to be examined for the presence of PrPSc by Western blotting (WB).
Western blotting.
To confirm each scrapie case, frozen nervous and lymphoid tissue samples were thawed and then subjected to a WB protocol for PrPSc detection by using the Prionics check kit (Prionics AG, Switzerland), in accordance with the manufacturer's instructions. The presence of PrPSc in the placentas was evaluated as described elsewhere (15). In short, WB was carried out by means of a modified Prionics check protocol (Prionics AG, Switzerland), with 1 g of placental tissue being first incubated in Tris-buffered saline (TBS) buffer (pH 7.4) containing 1.5 mM CaCl2 and 2.5 mg/ml (final concentration) of type XI collagenase (1.6 U/mg; Sigma, USA) for 2 h at 37°C. P4 monoclonal antibody (MAb; 1:15,000) was used as the primary antibody.
Immunohistochemical technique.
The formalin-fixed tissues were treated for 1 h in 98% formic acid and embedded in paraffin by using conventional protocol. IHC analysis was carried out as described previously (16), with PrPSc immunodetection being performed by using a biotin-streptavidin detection method (Vector Laboratories, Inc., USA), and F99 (1:800) as the primary MAb. Nervous and lymphoid tissue sections from ARR/ARR scrapie-negative sheep were included in each run as controls. Another series of controls was set up by omitting the primary MAb.
Genetic analysis.
Genetic analysis of all goats under study focused on the PRNP gene. Genomic DNA was isolated from the blood with a DNA isolation kit (Qiagen, USA). PCR amplification of the PrP gene was performed using the primers PrP1(+) (5′-ATGGTGAAAAGCCACATAGGCAGT-3′) and PrP2(−) (5′-CTATCCTACTATGAGAAAAATGAG-3′). PrP1(+) and PrP2(-) anneal at the extreme 5′ and 3′ regions of the PrP-coding sequence, respectively. Amplification reactions were performed for 30 cycles of 30 s at 94°C, 30 s at 59°C, and 45 s at 72°C. PrP gene polymorphisms were detected by DNA sequencing on both strands of the PCR products (Life Technology, USA).
Western blot analysis of PrPSc.
Typing of the protease-resistant core of PrPSc (PrPres) was performed by discriminatory immunoblotting, according to the Istituto Superiore di Sanità (ISS) discriminatory WB method (http://www.tse-lab-net.eu/documents/tse-oie-rl-handbook.pdf), as previously described. The ISS discriminatory WB allows an accurate determination of apparent molecular mass of PrPres, obtained by loading Precision Plus streptavidin-tagged molecular markers (Bio-Rad, USA) in three lanes of each gel, and includes the use of two different MAbs in order to increase discriminatory power: SAF84, whose epitope is C terminal (residues 163 to 173 of ovine PrP) and is preserved in BSE and scrapie samples, and P4 (R-Biopharm, Germany), whose epitope (residues 93 to 99 of ovine PrP) is partially lost after PK digestion of BSE samples. Briefly, brain homogenates (20% [wt/vol]) were prepared in 100 mM Tris-HCl with Complete protease inhibitor cocktail (Roche, USA) at pH 7.4. The homogenates were either used directly or stored at −20°C. After addition of an equal volume of 100 mM Tris-HCl containing 4% Sarkosyl, the homogenates were incubated for 30 min at 37°C with gentle shaking. Proteinase K (0.2 mg/ml; Sigma-Aldrich, USA) was added, and then the homogenates were further incubated for 1 h at 37°C with gentle shaking. The reaction was stopped with 3 mM phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, USA). The samples were added with an equal volume of isopropanol-butanol (1:1 [vol/vol]) and centrifuged at 20,000 × g for 5 min; the supernatants were discarded, and the pellets were dissolved in denaturing sample buffer for WB analysis. WB was performed by following the procedures described for ISS discriminatory WB (17, 18), by using 12% bis-Tris polyacrylamide gels (Invitrogen, USA) for electrophoresis. Chemiluminescence was detected with the VersaDoc imaging system (Bio-Rad, USA). All measurements were performed with QuantityOne software (Bio-Rad, USA).
Phenotype of PrPSc deposition in the brain.
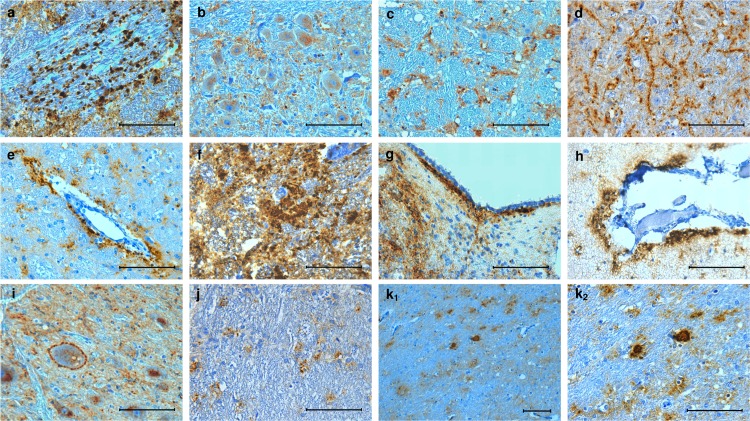
The phenotype of PrPSc deposition in the brain was investigated in experimentally inoculated goats with clinical signs of scrapie, including 3 wild-type, 1 R211Q, 2 R211Q-S240P, 2 R154H, and 1 R154H-S240P goat (see Results). In addition, the brains from 3 wild-type goats of the F1b progeny which had clinical scrapie were examined together with those of 5 naturally scrapie-infected wild-type sheep at the terminal stage of disease, which belonged to the affected flocks that also provided the sheep used as donors of scrapie-infected brain homogenate in this experiment. From the formalin-fixed portions of the brain, 7 distinct coronal sections representative of the frontal cerebral cortex, basal nuclei, thalamus, midbrain, cerebellum, pons, and medulla oblongata (obex) were cut. PrPSc deposition was scored to create a profile by using a method described elsewhere (19). In short, the magnitude of 11 PrPSc accumulation patterns was scored from 0 to 3. The 11 patterns were defined as follows: granular (coarse and punctate), intraneuronal, glial, linear, perivascular, coalescing, subependymal, subpial, perineuronal, intraglial, and plaque-like (Fig. 2). The profiles of PrPSc values represented the respective mean scores of the goats grouped by genotype, as well as of the naturally scrapie-infected sheep group, included for comparison.
FIG 2.
Representative immunolabeling patterns of PrPSc deposition in the brains of the affected goats under study. Of these patterns, magnitude was scored in 7 transverse sections of the brain in order to create an immunohistochemical profile. (a) Granular (coarse and punctate); (b) intraneuronal; (c) glial; (d) linear; (e) perivascular; (f) coalescing; (g) subependymal; (h) subpial; (i) perineuronal; (j) intraglial; (k1 and k2) plaque-like. Bars, 100 μm.
Statistical analysis.
In the case of the 37 F0 goats, survival times were analyzed from the date of inoculation to death, whereas the survival times for both the generations of offspring (41 goats) were calculated from birth. Goats that were not diagnosed as scrapie cases (13 F0 goats and 30 offspring) were included in the analyses as censored at death. Data were stratified by scrapie status, and after a preliminary analysis by genotype, a further stratification was based on a new variable reflecting the level of susceptibility: goats and their genotypes were categorized as resistant, susceptible with long p.i. time, and susceptible with short p.i. time. Differences between survivorship curves were tested using the Kaplan-Meier estimator and the log rank test. Similarly, the potential effect of lag time between the birth of each animal and the scrapie onset in its F0 mother on the offspring's survival times was also evaluated after accounting for the confounding effect of susceptibility. Based on the distribution of the lag times, two scenarios were tested: in the first scenario, animals were divided into two categories based on the first quartile of the distribution (i.e., less than and more than 303 days), whereas in the second scenario, the median of lag times (i.e., 692 days) was used. A stratified log rank test was used with the level of susceptibility as the stratifying variable.
With regard to the phenotype of the PrPSc deposition, statistical analysis were performed by using one-way analysis of variance on the 11 immunohistochemical PrPSc deposition patterns displayed by the different groups of animals. Differences between groups were considered statistically significant if the probability for equality was <0.05 in multiple-comparison tests (Tukey test).
Ethics statement.
The experimental procedures involving the goats investigated herein were officially approved by the Ethics Committee of the Istituto Zooprofilattico Sperimentale of Sardinia (permission n. 1/2013/C.E.), in strict agreement with the guidelines of Italian National Law no. 116/1992.
RESULTS
Experimental oral infection.
The first part of the study aimed to observe which of the polymorphic codons carried by the experimentally inoculated goats were associated with scrapie resistance. With regard to this, we observed that all the wild-type, G37V, Q168Q-P240P, and S/P240P goats had neurological signs indicative of scrapie after a median incubation time of 711 days (95% confidence interval [CI], 672 to 743) (Fig. 3A and Table 1). The goats with R154H, H154H, and R154H-S240P dimorphisms became sick after an ∼1-year-longer incubation time (1,033 days) (Table 1). Similarly, a longer incubation time was observed in the only inoculated P168Q-P240P animal, which developed clinical scrapie at 1,184 days p.i. One R211Q goat had clinical scrapie at 2,109 days postinoculation, while 2 other R211Q goats died of causes unrelated to scrapie at 464 and 1,806 days postinoculation (Table 2). Of these two, only the one that died at 1,806 days postinoculation displayed PrPSc at the level of lymphoid and nervous tissues, with PrPSc deposits being restricted to the nucleus parasympathicus nervi vagi (NPNV) at the level of the obex region of the medulla oblongata. The two remaining R211Q goats both carried the S240P mutation and developed clinical scrapie at an average time of 1,529 days p.i.
FIG 3.
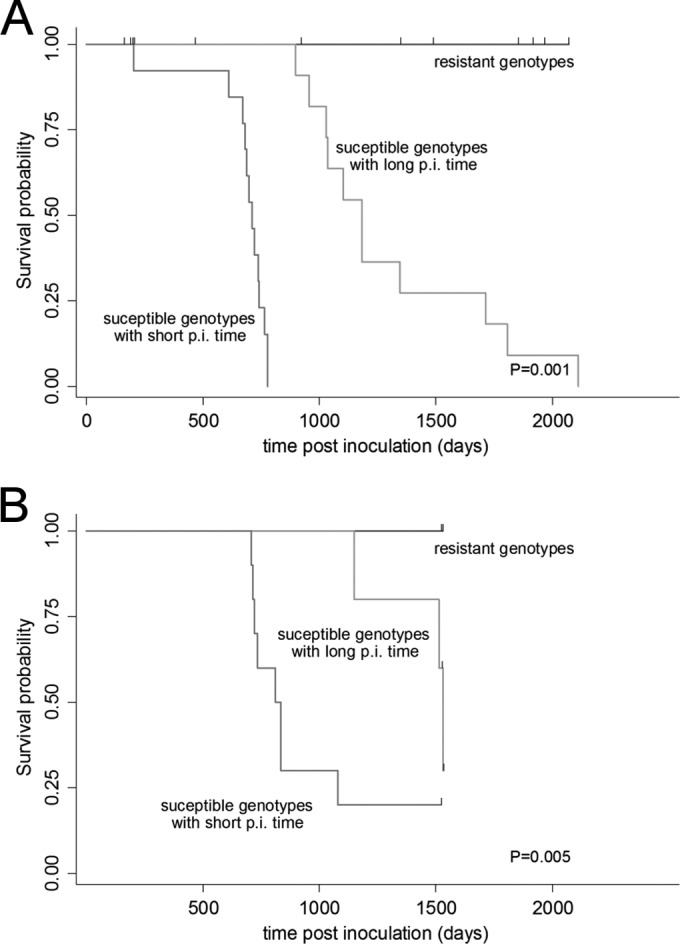
Comparison of goats survivorship by susceptibility status. Goat genotypes were used to categorize goats into three susceptibility levels: resistant (D145D, Q222K, and Q222K-S240P), susceptible with long postinfection time (R/H154H, R154H-S240P, P168Q-P240P, R211Q, and R211Q-S240P), and susceptible with short postinfection time (wild-type, G37V, Q168Q-P240P, and S/P240P). The log rank test was used to compare the three survival curves. Hash marks are used to indicate censoring. (A) Kaplan-Meier survival curves of 37 goats (F0) orally infected with ovine scrapie. Thirteen animals not diagnosed as scrapie cases were included in the analyses as censored at death. (B) Kaplan-Meier survival curves of 20 offspring goats belonging to the F1b group. Nine animals not diagnosed as scrapie cases were included in the analyses as censored at death.
Finally, the goats with homozygous or heterozygous mutations at codon 145 (G/D145D) and with Q222K did not show neurological signs during the course of the study and were found not to have PrPSc in the nervous and lymphoid tissues; they were sacrificed or died at a time ranging from 180 to 2,067 days p.i. (Fig. 3A and Tables 1 and 2).
Based on the above results, goats and their PrP genotypes were categorized (Fig. 3A) as resistant (D145D, Q222K, and Q222K-S240P), susceptible with long p.i. time (R/H154H, R154H-S240P, P168Q-P240P, R211Q and R211Q-S240P; median incubation time to death, 1,184 days; 95% CI, 955 to 1,712 days), or susceptible with short p.i. time (wild type, G37V, Q168Q-P240P, and S/P240P; median incubation time to death, 711 days; 95% CI, 672 to 743 days). The rank test comparing the survivorship between the three levels of susceptibility was highly significant (chi square, 46.9; P = 0.000).
Natural infection in F1 and F2 generations.
With regard to the offspring of the first generation, the goats belonging to the F1a set were sacrificed or died of causes unrelated to scrapie at an average age of 1,755 days without showing neurological signs (Table 3). At necroscopy, no PrPSc was found by means of IHC analysis and WB in the nervous and lymphoid tissues of these goats, including those carrying the wild type, G37V and S/P240P genotypes, which were shown to be susceptible to scrapie in the parental group after experimental oral inoculation.
TABLE 3.
PRNP genotype, clinical status, PrPSc detection, and age at the examination, in offspring from goats naturally exposed to scrapie in a herd where the parental generation was experimentally infected with an ovine scrapie brain homogenatea
| Goat | Progeny | Genotype | Clinical signs | Age (days) | PrPSc detection |
|
|---|---|---|---|---|---|---|
| Obex | LT | |||||
| 806 | F1a | WT | No | 1692 | Neg | Neg |
| 801 | F1a | G37V | No | 1848 | Neg | Neg |
| 802 | F1a | P240P | No | 1854 | Neg | Neg |
| 803 | F1a | S240P | No | 1338 | Neg | Neg |
| 804 | F1a | G145D-P168Q-S240P | No | 1854 | Neg | Neg |
| 805 | F1a | G145D-Q222K | No | 1854 | Neg | Neg |
| 807 | F1a | WT | No | 1848 | Neg | Neg |
| 811 | F1b | WT | Yes | 715 | Pos | Pos |
| 815 | F1b | WT | Yes | 721 | Pos | Pos |
| 819 | F1b | WT | Yes | 707 | Pos | Pos |
| 826 | F1b | WT | Yes | 734 | Pos | Pos |
| 830 | F1b | WT | Yes | 813 | Pos | Pos |
| 824 | F1b | WT | No | 1520 | Neg | Neg |
| 818 | F1b | G37V | Yes | 836 | Pos | Pos |
| 808 | F1b | P168Q-S240P | Yes | 1514 | Pos | Pos |
| 831 | F1b | Q222K | No | 1527 | Neg | Neg |
| 809 | F1b | R154H | Yes | 1149 | Pos | Pos |
| 812 | F1b | R154H | No | 1527 | Neg | Neg |
| 810 | F1b | R211Q | No | 1528 | Pos | Neg |
| 816 | F1b | R211Q | No | 1528 | Neg | Neg |
| 814 | F1b | S240P | Yes | 835 | Pos | Pos |
| 821 | F1b | S240P | No | 1520 | Neg | Neg |
| 825 | F1b | S240P | Yes | 1078 | Pos | Pos |
| 823 | F1b | Q222K | No | 1527 | Neg | Neg |
| 820 | F1b | Q222K | No | 1527 | Neg | Neg |
| 822 | F1b | Q222K | No | 1527 | Neg | Neg |
| 817 | F1b | Q222K | No | 1520 | Neg | Neg |
| 381 | F1c | Q222K | No | 713 | Neg | Neg |
| 379 | F1c | G145D-Q222K | No | 707 | Neg | Neg |
| 375 | F1c | G37V-Q222K | No | 707 | Neg | Neg |
| 389 | F1c | R211Q | No | 707 | Neg | Neg |
| 380 | F2a | Q222K-S240P | No | 714 | Neg | Neg |
| 384 | F2a | G145D- S240P | No | 714 | Neg | Neg |
| 385 | F2a | G145D | No | 714 | Neg | Neg |
| 378 | F2a | G145D | No | 714 | Neg | Neg |
| 383 | F2a | Q222K | No | 714 | Neg | Neg |
| 382 | F2a | K222K | No | 714 | Neg | Neg |
| 386 | F2a | P168Q-S240P | No | 714 | Neg | Neg |
| 387 | F2a | G37V | No | 714 | Neg | Neg |
| 376 | F2a | WT | No | 714 | Neg | Neg |
| 377 | F2a | WT | No | 714 | Neg | Neg |
PrPSc was detected via both immunohistochemical and Western blotting techniques. LT, lymphoid tissue; Pos, positive; Neg, negative; WT, wild type.
In contrast, all but 2 of the susceptible goats with short p.i. times (wild type, G37V, and S240P) belonging to the F1b generation developed clinical scrapie after a median incubation time of 813 days (95% CI, 707 to 1,078 days) (Table 3). In this offspring group, onset of scrapie in the goats carrying the genotypes designated susceptible with long p.i. time (P168Q-S240P, R154H, and R211Q) was observed at an older age (median = 1,528 days) than in the wild-type goat (chi square, 10.64; P = 0.0049) (Fig. 3B and Table 3).
No clinical or subclinical scrapie cases occurred in the goats belonging to group F1c (Table 3), which included no animals suspected of being susceptible, on the basis of the results observed in the previous generations. Finally, in the F2a generation, goats of all genotypes suspected of being susceptible to scrapie were euthanized an average age of 714 days, and no nervous or lymphoid tissues with PrPSc were found (Table 3).
With regard to the goats exposed to natural infection (F1 and F2 generation) and carrying the dimorphism Q/K222 either alone or associated with others, they were found to be clinically healthy throughout the study, and no PrPSc was found in the nervous and lymphoid tissues sampled at necroscopy, which was carried out at 730 to 1,831 days p.i. (Fig. 3B and Table 3).
Finally, it was not possible to identify any statistically effects of the lag time between the birth of each animal and the onset of scrapie in its F0 mother, using either the first quartile (P = 0.065) or the median lag time (P = 0.33).
PrPSc deposition in placentas.
Among the 25 goat placentas examined, only one with the wild-type PRNP genotype was found to harbor PrPSc.
Immunohistochemical profile.
Brains from goats with the wild-type genotype and those carrying the R211Q and the R154H dimorphisms displayed similar immunohistochemical profiles of PrPSc deposition after different incubation times following oral inoculation of scrapie wild-type ovine brain homogenate. In fact, the 11 immunohistochemical patterns did not statistically show significant differences among the groups, although plaque-like deposits were seen in the forebrains of the goats carrying mutations (Fig. 4A). Similar immunohistochemical profiles were observed upon comparison of experimentally inoculated wild-type goats, naturally infected wild-type goats belonging to the F1 offspring, and naturally scrapie-infected wild-type sheep (Fig. 4B).
FIG 4.
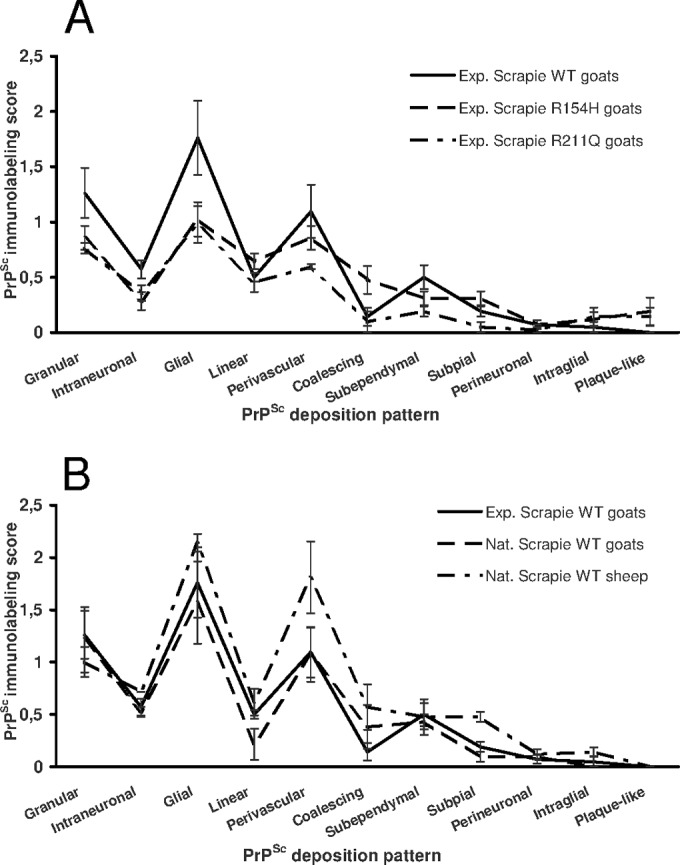
Distribution and magnitude of different PrPSc immunolabeling patterns in the goats under study. (A) Goats with the wild-type genotype or R154H and R211Q dimorphisms experimentally infected with scrapie. (B) Goats experimentally and naturally infected with scrapie under study together with 5 sheep naturally infected with wild-type scrapie and belonging to the flocks that also provided the sheep used as donors of scrapie-infected brain homogenate. Exp, experimentally infected; Nat, naturally infected; WT, wild type.
Western blot analysis of PrPSc.
Medullae oblongatae from F0 and F1b goats of different genotypes (including wild type, R211Q, R211Q-S240P, and R154H) with clinical scrapie were analyzed by discriminatory WB in order to investigate the possible emergence of new PrPSc phenotypes, as reported in previous studies (8). The PrPSc phenotypes in the experimentally infected goats were directly compared with the phenotypes observed in the sheep brain homogenate used for the experimental challenge of F0 and with sheep and goats with natural scrapie obtained from the Italian surveillance program. Overall, the PrPSc phenotype in the experimentally infected goats was indistinguishable from that in the original sheep inoculum, and no variations in molecular weight, SAF84/P4 ratio, or glycotype were observed among goats of F0 and F1b, among goats of different genotypes, or between experimentally and naturally infected goats used as controls (Fig. 5). Given the presence of plaque-like deposits in the forebrains of a subset of experimentally infected mutated goats (see IHC results), we further compared the PrPSc phenotype in brain areas with and without plaque-like deposits. Indeed, the presence of these plaque-like deposits might represent the emergence of a new strain/phenotype of scrapie showing effects on specific brain areas. However, discriminatory WB analysis did not detect any obvious biochemical difference between PrPSc from plaque-free areas and PrPSc from plaques (Fig. 5).
FIG 5.
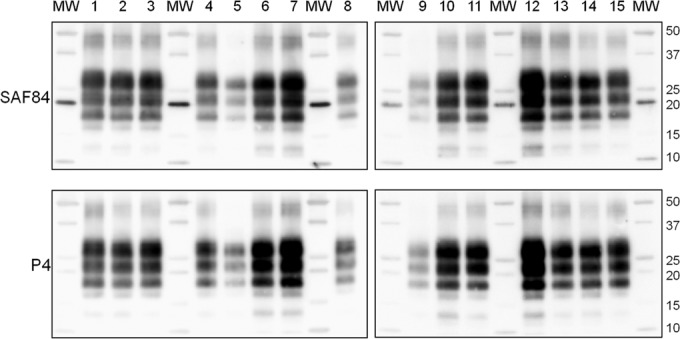
Discriminatory WB in goats with various PrP genotypes experimentally infected with scrapie. Representative WBs show PrPSc from goats experimentally infected with scrapie (lanes 1 to 7 and 11 to 14) compared to PrPSc from the sheep inoculum (lane 8), as well as from goats (lanes 9 and 10) and sheep (lane 15) with natural scrapie. Samples in lanes 1 to 4 and 11 to 14 were from F0 goats, and samples in lanes 5 to 7 were from F1 goats. PrP genotypes of goats were wild type (lanes 1 and 7), R211Q (lanes 2, 5, 11, and 12), R211Q-S240P (lanes 3, 13, and 14), and R154H (lanes 4 and 6). All samples were from medullae oblongatae, with the exception of samples in lanes 12 and 14, which were from the thalamus of the same goats whose medulla samples were loaded in lanes 11 and 13. Replica blots were developed with MAbs SAF84 (top) and P4 (bottom).
DISCUSSION
Field observations (20) and experimental studies (21, 22) suggested that the scrapie agent derived from sheep can infect goats. However, the specific phenotypic traits of the strain involved, the neuropathological aspect displayed in infected goats, and the genetic factors that could have an impact on sheep-to-goat transmission have not been thoroughly explored.
Our results confirm, first, that K222 and D145 mutations protect goats against scrapie under both experimental and natural conditions and, second, that other mutations affect the length of the incubation period under the same conditions. Moreover, by applying WB analysis, we found that the molecular phenotype of a natural wild-type scrapie ovine isolate was mainly preserved across 2 goat generations, by either natural or experimental transmission, and even in animals carrying different susceptible PRNP variants.
In contrast, goats intracerebrally challenged with a natural French caprine isolate displayed diverse PrPSc molecular types based on minimal amino acid differences of the PRNP, thus suggesting that a mixture of scrapie agent strain components was present in that isolate (8).
Our results, coupled with previous findings (6, 23), confirmed that a unique stable scrapie agent strain is circulating in the Italian small ruminant population.
One approach to defining the phenotype of TSE cases is the evaluation of the neuroanatomical distribution and magnitude of given IHC PrPSc patterns in the natural host (24, 25). By this means, we found that IHC phenotype in our goats was similar to that observed in wild-type sheep, and it was highly retained in the experimentally infected goat as well as in naturally exposed ones and their progeny when they expressed the same PRNP genotype. With regard to the goats carrying mutations at PRNP gene codons 154 and 211, their IHC profile was slightly different due to the presence of plaque-like PrPSc deposition in the diencephalic portion of the brain. It is believed that the IHC PrPSc deposition profile results from a composite interaction of various factors, among which is the PRNP host genotype (24, 25). Considering that the same experimental conditions, including the infection source, were used in our study, this finding provides further evidence that minimal amino acid differences in the host PRNP genotype drive the neuropathological phenotype.
Collectively, these results demonstrate that following scrapie wild-type ovine isolate infection, goats mimic the phenotypical aspects of the disease observed in sheep with the absolute absence of an interspecies transmission barrier. Another expression of this resemblance was the incubation time of our wild-type, G37V, and S/P240P goats, which was similar to that which we previously reported for wild-type sheep challenged with the same inoculum (26).
Thus, goats might efficiently act as a reservoir of ovine scrapie strains, sustaining the risk of the disease in a given geographical area where an ovine selective breeding plan for scrapie resistance is carried out.
It is believed that transgenic mice expressing a PrP gene identical to that of the infecting species enhanced susceptibility to prion agent, thus overcoming the species transmission barrier (11). Thus, considering the prion protein gene sequence shared by sheep and goats, our results confirm in natural hosts that the prion protein gene sequence is more powerful than any potential species-related factors in determining the scrapie phenotypes.
Findings from naturally scrapie-infected herds (6, 7) and from goats intracerebrally and orally challenged with scrapie from classical caprine sources (8, 9) as well as from transgenic mice (27) strongly suggest that goats carrying the K222 allele are highly resistant. Our results reinforce these previous findings, as even the wild-type classical ovine scrapie isolate was unable to propagate in Q222K heterozygous goats, which remained uninfected 2,066 days following experimental oral challenging or after 1,505 days of lifetime exposure to a scrapie agent in an infected environment.
The lack of homozygous mutation at codon 222 in our goats exposed to an ovine scrapie isolate prevents our drawing any definite conclusion about its role in resistance to the disease. However, in a recent study (8), the presence of homozygous mutations at certain polymorphic codons, including 222, was found to be associated with a longer survival time than that of heterozygous mutations.
Interestingly, while heterozygous H154 and Q211 goats orally challenged with inoculates from caprine scrapie sources (8) were resistant, in our study, the same genotypes were susceptible to wild-type ovine scrapie isolate, although the onset of the disease was manifested at longer p.i. times than in wild-type goats. These parallel findings led to the conclusion that the results of the interaction between certain PRNP gene polymorphisms and a given TSE strain should not be universally applied to other TSE agents. However, a simple difference in prion infectivity titer between the 2 isolates used in the experiments should also be taken into account.
A D145 allelic variant in Chaouni goats from Morocco was recently reported (28); however, its contingent association with resistance or susceptibility to scrapie has been totally unexplored so far. In our study, 2 goats carrying this dimorphism in homozygous form were found not to have PrPSc after 1,960 days p.i., thus suggesting a possible protective effect comparable to that associated with the K222 allele.
However, the low number of the examined D145 sheep did not allow any definite conclusion, and further experimental and field studies are necessary. With regard to other dimorphisms carried by the goats we studied, an experimentally inoculated P168Q-S240P goat had a longer incubation time than the homozygote Q168Q-P240P goat. Given that the codon at position 240 is deleted after a posttranslational modification (7), we speculate that heterozygous Q168 mutation, and not the homozygous status, may be associated with a longer incubation period. In sheep, the same codon is polymorphic, with a protective effect against experimental BSE in its heterozygous mutated form (29). However, in sheep, the different substitution (proline to leucine) may indicate that the position might play a more important role than the proper type of amino acid change.
In our study, natural scrapie cases were observed only in the second progeny of the first generation (F1b). Since in sheep, horizontal dam-to-offspring transmission of scrapie during the peripartum period is a well-known epidemiological aspect in which the placenta plays a crucial role (30), we assumed that susceptible F1b goats had been more exposed to a scrapie agent because the onset of scrapie in the F0 mother was closer to the time of birth than in the other progenies. However, the lag time between the birth of each F1b animal and the scrapie onset in its F0 mother was found to have no statistically significant effect on scrapie occurrence. Although in our study as well as in another one (31), PrPSc in shed goat placentas was infrequently found, compared to what described in sheep (15, 30), this organ transmits goat scrapie efficiently (32). Moreover, just as in sheep, the dam-to-offspring spread of infection in goats encompasses a variable number of routes and sources of infection governed by numerous interacting conditions. For instance, goat milk can transmit scrapie (33), even though in Sarda wild-type sheep, the presence of chronic inflammatory states of the mammary gland was a condition sine qua non for prion infectivity in milk (34). In light of this knowledge, we suggest that in our experiment, the decreasing frequencies of the susceptible genotypes in the new progenies might have led to reduction in the environmental presence of the scrapie agent.
While a large body of knowledge has already informed the application of a selective breeding plan to eradicate scrapie in sheep (4), only recently available data are encouraging the EU authorities to draw up an equivalent plan for goats. Our results reconfirmed the potential key role of the K222 mutation in developing PRNP genotype selection programs to control and eradicate scrapie in commercial goat populations.
However, in light of the results we obtained for the D145 allele, the large number of caprine polymorphic codons, and their frequency among breeds, as well as the diversity of circulating scrapie agents, a selective breeding plan for resistance to scrapie in goats should not follow a universal strategy but rather should be tailored to the specific characteristics of the local goat populations.
ACKNOWLEDGMENTS
We thank A. Lai, S. Macciocu, and C. Contu for technical assistance as well as S. Sechi for statistical analysis.
This work was supported by RF-2010-2318525, a comprehensive strategy to eradicate prion diseases from the Italian sheep and goat population; EMIDA-ERA-NET, Toward breeding of goats for genetically determined TSEs resistance; and grant RC IZS SA 01/11 from Italian Ministero della Salute.
REFERENCES
- 1.Prusiner SB. 1991. Molecular biology of prion diseases. Science 252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 2.Hunter N. 1997. PrP genetics in sheep and the implications for scrapie and BSE. Trends Microbiol 5:331–334. doi: 10.1016/S0966-842X(97)01081-0. [DOI] [PubMed] [Google Scholar]
- 3.Baylis M, Goldmann W. 2004. The genetics of scrapie in sheep and goats. Curr Mol Med 4:385–396. doi: 10.2174/1566524043360672. [DOI] [PubMed] [Google Scholar]
- 4.Dawson MR, Moore C, Bishop SC. 2008. Progress and limits of PrP gene selection 411 policy. Vet Res 39:25. doi: 10.1051/vetres:2007064. [DOI] [PubMed] [Google Scholar]
- 5.Vaccari G, Panagiotidis CH, Acin C, Peletto S, Barillet F, Acutis P, Bossers A, Langeveld J, van Keulen L, Sklaviadis T, Badiola JJ, Andreéoletti O, Groschup MH, Agrimi U, Foster J, Goldmann W. 2009. State-of-the-art review of goat TSE in the European Union, with special emphasis on PRNP genetics and epidemiology. Vet Res 40:48. doi: 10.1051/vetres/2009031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaccari G, Di Bari MA, Morelli L, Nonno R, Chiappini B, Antonucci G, Marcon S, Esposito E, Fazzi P, Palazzini N, Troiano P, Petrella A, Di Guardo G, Agrimi U. 2006. Identification of an allelic variant of the goat PrP gene associated with resistance to scrapie. J Gen Virol 87:1395–1402. doi: 10.1099/vir.0.81485-0. [DOI] [PubMed] [Google Scholar]
- 7.Acutis PL, Bossers A, Priem J, Riina MV, Peletto S, Mazza M, Casalone C, Forloni G, Ru G, Caramelli M. 2006. Identification of prion protein gene polymorphisms in goats from Italian scrapie outbreaks. J Gen Virol 87:1029–1033. doi: 10.1099/vir.0.81440-0. [DOI] [PubMed] [Google Scholar]
- 8.Lacroux C, Perrin-Chauvineau C, Corbière F, Aron N, Aguilar-Calvo P, Torres JM, Costes P, Brémaud I, Lugan S, Schelcher F, Barillet F, Andréoletti O. 2014. Genetic resistance to Scrapie infection in experimentally challenged goats. J Virol 88:2406–2413. doi: 10.1128/JVI.02872-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acutis PL, Martucci F, D'Angelo A, Peletto S, Colussi S, Maurella C, Porcario C, Iulini B, Mazza M, Dell'atti L, Zuccon F, Corona C, Martinelli N, Casalone C, Caramelli M, Lombardi G. 2012. Resistance to classical scrapie in experimentally challenged goats carrying mutation K222 of the prion protein gene. Vet Res doi: 10.1186/1297-9716-43-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbière F, Perrin-Chauvineau C, Lacroux C, Costes P, Thomas M, Brémaud I, Martin S, Lugan S, Chartier C, Schelcher F, Barillet F, Andreoletti O. 2013. PrP-associated resistance to scrapie in five highly infected goat herds. J Gen Virol 94:241–245. doi: 10.1099/vir.0.047225-0. [DOI] [PubMed] [Google Scholar]
- 11.Beringue V, Vilotte JL, Laude H. 2008. Prion agent diversity and species barrier. Vet Res 39:47. doi: 10.1051/vetres:2008024. [DOI] [PubMed] [Google Scholar]
- 12.Bruce ME. 2003. TSE strain variation. Br Med Bull 66:99–108. doi: 10.1093/bmb/66.1.99. [DOI] [PubMed] [Google Scholar]
- 13.Piening N, Nonno R, Di Bari M, Walter S, Windl O, et al. 2006. Conversion efficiency of bank vole prion protein in vitro is determined by residues 155 and 170, but does not correlate with the high susceptibility of bank voles to sheep scrapie in vivo. J Biol Chem 281:9373–9384. doi: 10.1074/jbc.M512239200. [DOI] [PubMed] [Google Scholar]
- 14.Jeffrey M, Martin S, Gonzàlez LL, Foster J, Langeveld JPM, van Zijderveld FG, Grassi J, Hunter N. 2006. Immunohistochemical features of PrPd accumulation in natural and experimental goat transmissible spongiform encephalopathies. J Comp Path 134:171–181. doi: 10.1016/j.jcpa.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Santucciu C, Maestrale C, Madau L, Attene S, Cancedda MG, Demontis F, Tilocca MG, Saba M, Macciocu S, Carta A, Ligios C. 2010. Association of N176K and L141F dimorphisms of the PRNP gene with lack of pathological prion protein deposition in placentas of naturally and experimentally scrapie-affected wild-type sheep. J Gen Virol 91:2402–2407. doi: 10.1099/vir.0.021188-0. [DOI] [PubMed] [Google Scholar]
- 16.Maestrale C, Di Guardo G, Cancedda MG, Marruchella G, Masia M, Sechi S, Macciocu S, Santucciu C, Petruzzi M, Ligios C. 2013. A lympho-follicular microenvironment is required for pathological prion protein deposition in chronically inflamed tissues from scrapie-affected sheep. PLoS One 8:e62830. doi: 10.1371/journal.pone.0062830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pirisinu L, Migliore S, Di Bari MA, Esposito E, Baron T, D'Agostino C, De Grossi L, Vaccari G, Agrimi U, Nonno R. 2011. Molecular discrimination of sheep bovine spongiform encephalopathy from scrapie. Emerg Infect Dis 17:695–698. doi: 10.3201/eid1704.101215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Migliore S, Esposito E, Pirisinu L, Marcon S, Di Bari M, D'Agostino C, Chiappini B, Conte M, Sezzi E, De Grossi L, Agrimi U, Vaccari G, Nonno R. 2012. Effect of PrP genotype and route of inoculation on the ability of discriminatory Western blot to distinguish scrapie from sheep bovine spongiform encephalopathy. J Gen Virol 93:450–455. doi: 10.1099/vir.0.035469-0. [DOI] [PubMed] [Google Scholar]
- 19.González L, Martin S, Houston FE, Hunter N, Reid HW, Bellworthy SJ, Jeffrey M. 2005. Phenotype of disease-associated PrP accumulation in the brain of bovine spongiform encephalopathy experimentally infected sheep. J Gen Virol 86:827–838. doi: 10.1099/vir.0.80299-0. [DOI] [PubMed] [Google Scholar]
- 20.Hourrigan JL, Klingsporn AL, McDaniel HA, Riemenschneider MN. 1969. Natural scrapie in a goat. J Am Vet Med Assoc 154:538–539. [PubMed] [Google Scholar]
- 21.Pattison IH, Gordon WS, Millson GC. 1959. Experimental production of scrapie in goats. J Comp Pathol 69:300–312. doi: 10.1016/S0368-1742(59)80029-1. [DOI] [PubMed] [Google Scholar]
- 22.Foster J, Goldmann W, Parnham D, Chong A, Hunter N. 2001. Partial dissociation of PrPSc deposition and vacuolation in the brains of scrapie and BSE experimentally affected goats. J Gen Virol 82:267–273. [DOI] [PubMed] [Google Scholar]
- 23.Nonno R, Esposito E, Vaccari G, Conte M, Marcon S, Di Bari M, Ligios C, Di Guardo G, Agrimi U. 2003. Molecular analysis of cases of Italian sheep scrapie and comparison with cases of bovine spongiform encephalopathy (BSE) and experimental BSE in sheep. J Clin Microbiol 41:4127–4133. doi: 10.1128/JCM.41.9.4127-4133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spiropoulos J, Casalone C, Caramelli M, Simmons MM. 2007. Immunohistochemistry for PrPSc in natural scrapie reveals patterns which are associated with the PrP genotype. Neuropathol Appl Neurobiol 33:398–409. doi: 10.1111/j.1365-2990.2007.00800.x. [DOI] [PubMed] [Google Scholar]
- 25.Gonzàlez L, Pitarch JL, Martin S, Thurston L, Simmons H, Acìn C, Jeffrey M. 2014. Influence of polymorphisms in the prion protein gene on the pathogenesis and neuropathological phenotype of sheep scrapie after oral infection. J Comp Pathol 15:57–70. doi: 10.1016/j.jcpa.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Cancedda MG, Di Guardo G, Chiocchetti R, Demontis F, Marruchella G, Sorteni C, Maestrale C, Lai A, Ligios C. 2014. Role of palatine tonsils as a prion entry site in classical and atypical experimental sheep scrapie. J Virol 88:1065–1070. doi: 10.1128/JVI.02750-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguilar-Calvo P, Espinosa JC, Pintado B, Gutiérrez-Adán A, Alamillo E, Miranda A, Prieto I, Bossers A, Andreoletti O, Torres JM. 2014. Role of the goat K222-PrPC polymorphic variant in prion infection resistance. J Virol 88:2670–2676. doi: 10.1128/JVI.02074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serrano C, Hammouchi M, Benomar A, Lyahyai J, Ranera B, Acìn C, el Hamidi M, Monzon M, Badiola JJ, Tligui N, Zaragoza P, Martìn-Burriel I. 2009. PRNP haplotype distribution in Moroccan goats. Anim Genet 40:565–568. doi: 10.1111/j.1365-2052.2009.01873.x. [DOI] [PubMed] [Google Scholar]
- 29.Goldmann W, Houston F, Stewart P, Perucchini M, Foster J, Hunter N. 2006. Ovine prion protein variant A(136)R(154)L(168)Q(171) increases resistance to experimental challenge with bovine spongiform encephalopathy agent. J Gen Virol 87:3741–3745. doi: 10.1099/vir.0.82083-0. [DOI] [PubMed] [Google Scholar]
- 30.Andreoletti O, Lacroux C, Chabert A, Monnereau L, Tabouret G, Lantier F, Berthon P, Eychenne F, Lafond-Benestad S, Elsen JM, Schelcher F. 2002. PrP(Sc) accumulation in placentas of ewes exposed to natural scrapie: influence of foetal genotype and effect on ewe-to-lamb transmission. J Gen Virol 83:2607–2616. [DOI] [PubMed] [Google Scholar]
- 31.O'Rourke KI, Zhuang D, Truscott TC, Yan H, Schneider DA. 2011. Sparse PrP(Sc) accumulation in the placentas of goats with naturally acquired scrapie. BMC Vet Res 7:7. doi: 10.1186/1746-6148-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider DA, Madsen-Bouterse SA, Zhuang D, Truscott TC, Dassanayake RP, O'Rourke KI. 17 April 2015. The placenta shed from goats with classical scrapie is infectious to goat kids and lambs. J Gen Virol doi: 10.1099/vir.0.000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konold T, Simmons HA, Webb PR, Bellerby PJ, Hawkins SA, González L. 2013. Transmission of classical scrapie via goat milk. Vet Rec 172:455. doi: 10.1136/vr.f2613. [DOI] [PubMed] [Google Scholar]
- 34.Ligios C, Cancedda MG, Carta A, Santucciu C, Maestrale C, Demontis F, Saba M, Patta C, DeMartini JC, Aguzzi A, Sigurdson CJ. 2011. Sheep with scrapie and mastitis transmit infectious prions through the milk. J Virol 85:1136–1139. doi: 10.1128/JVI.02022-10. [DOI] [PMC free article] [PubMed] [Google Scholar]