LETTER
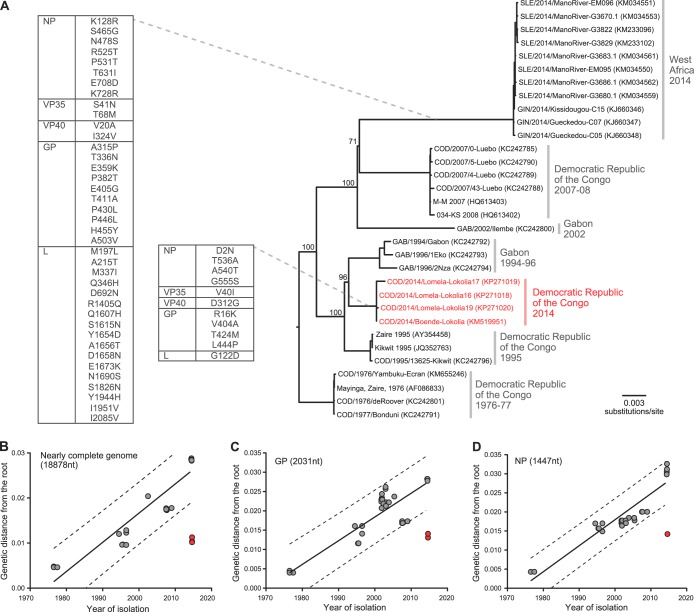
Maganga et al. (1) and Naccache et al. (GenBank numbers KP271018, KP271019, and KP271020) recently reported the genome sequences of the Zaire Ebolavirus (ZEBOV) that caused the 2014 outbreak in the Democratic Republic of the Congo (COD). In contrast to the virus sampled from the ongoing outbreak in West Africa, the sequences from COD (COD/2014/Boende-Lokolia, COD/2014/Lomela-Lokolia16, COD/2014/Lomela-Lokolia17, and COD/2014/Lomela-Lokolia19) are phylogenetically close to the ZEBOVs isolated during the 1995-1996 outbreaks in equatorial Africa (Fig. 1A). Importantly, however, such sequence similarity is far greater than expected given the tree topology and higher rate of ZEBOV evolution determined previously (2, 3). In particular, regression analyses of root-to-tip genetic distances against time of sampling show that the COD sequences deviate from the generally clock-like evolution of other ZEBOVs (Fig. 1B to D). In particular, the COD viruses sampled in 2014 are no further distant from the root of the tree than those from 1994 to 1996, indicating that they have evolved at a lower rate than other ZEBOVs.
FIG 1.
Phylogeny and regression of root-to-tip genetic distances against time of sampling for ZEBOV 1976-2014. (A) Maximum-likelihood phylogeny estimated from the nearly complete genome sequences of 32 ZEBOVs, including those (COD/2014/Boende-Lokolia, COD/2014/Lomela-Lokolia16, COD/2014/Lomela-Lokolia17, and COD/2014/Lomela-Lokolia19; strain names in red) collected from the 2014 outbreak in the Democratic Republic of the Congo (COD). Branch supports (shown at selected nodes) were assessed by bootstrap analysis (1,000 replicates). The boxes on the left show amino acid changes in the NP, VP35, VP40, and GP proteins that have occurred on specific branches, as indicated by dashed lines. Positions are numbered from the first residue of each protein. (B) The genetic distances from the root of the tree to each sequence (tip) are plotted against their year of sampling. Solid and dashed lines are the linear regression fits and the 95% prediction bands for the data points excluding COD/2014 sequences. The COD/2014 ZEBOV sequences (red dots) fall below the expected genetic distance, indicating that they have evolved markedly more slowly. (C and D) Similar observations made using the GP and NP genes, which include more sequences with different time points.
There are several possibilities to account for this markedly slower evolution. Anomalously low evolutionary rates have previously been shown to be artifacts resulting from the inadvertent release of viruses archived in laboratories, with the strain of human influenza A virus that led to the 1977 epidemic as a high-profile example (4). However, such an explanation is clearly implausible in this case, particularly because the COD outbreak occurred in a remote geographic region. More likely is that this lineage of ZEBOV was maintained in an animal host population characterized by a lower replication rate and hence fewer opportunities for mutation, such that the ecology of Ebola is more complex than generally envisaged. Previous PCR and serological studies have suggested that various species of fruit bat are the natural hosts of ZEBOV, with some nonhuman primates potentially acting as intermediate hosts (5–7). It is therefore possible that the COD/2014 lineage is maintained endemically in a different bat species with a lower population that roosts at lower densities or in as-yet-unidentified animal hosts. Finally, it is possible that the COD/2014 lineage replicates and/or mutates at an intrinsically lower rate, which could be conferred by those mutations fixed prior to its emergence (Fig. 1A, smaller box). Further studies of virus genetics and growth kinetics could provide insights into the viability of this hypothesis. Contamination during genome sequencing, which could also generate such data, is unlikely, because similar viral sequences were produced in two different laboratories (1) (GenBank numbers KP271018, KP271019, and KP271020). Given the persisting health threat posed by ZEBOVs, revealing the exact origins of this outbreak clearly merits rigorous investigation.
REFERENCES
- 1.Maganga GD, Kapetshi J, Berthet N, Kebela Ilunga B, Kabange F, Mbala Kingebeni P, Mondonge V, Muyembe JJ, Bertherat E, Briand S, Cabore J, Epelboin A, Formenty P, Kobinger G, Gonzalez-Angulo L, Labouba I, Manuguerra JC, Okwo-Bele JM, Dye C, Leroy EM. 2014. Ebola virus disease in the Democratic Republic of Congo. N Engl J Med 371:2083–2091. doi: 10.1056/NEJMoa1411099. [DOI] [PubMed] [Google Scholar]
- 2.Dudas G, Rambaut A. 2014. Phylogenetic analysis of Guinea 2014 EBOV ebolavirus outbreak. PLoS Curr 6:ecurrents.outbreaks.84eefe5ce43ec9dc0bf0670f7b8b417d. doi: 10.1371/currents.outbreaks.84eefe5ce43ec9dc0bf0670f7b8b417d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gire SK, Goba A, Andersen KG, Sealfon RS, Park DJ, Kanneh L, Jalloh S, Momoh M, Fullah M, Dudas G, Wohl S, Moses LM, Yozwiak NL, Winnicki S, Matranga CB, Malboeuf CM, Qu J, Gladden AD, Schaffner SF, Yang X, Jiang PP, Nekoui M, Colubri A, Coomber MR, Fonnie M, Moigboi A, Gbakie M, Kamara FK, Tucker V, Konuwa E, Saffa S, Sellu J, Jalloh AA, Kovoma A, Koninga J, Mustapha I, Kargbo K, Foday M, Yillah M, Kanneh F, Robert W, Massally JL, Chapman SB, Bochicchio J, Murphy C, Nusbaum C, Young S, Birren BW, Grant DS, Scheiffelin JS, Lander ES, Happi C, Gevao SM, Gnirke A, Rambaut A, Garry RF, Khan SH, Sabeti PC. 2014. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson MI, Viboud C, Simonsen L, Bennett RT, Griesemer SB, St George K, Taylor J, Spiro DJ, Sengamalay NA, Ghedin E, Taubenberger JK, Holmes EC. 2008. Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog 4:e1000012. doi: 10.1371/journal.ppat.1000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leroy EM, Telfer P, Kumulungui B, Yaba P, Rouquet P, Roques P, Gonzalez JP, Ksiazek TG, Rollin PE, Nerrienet E. 2004. A serological survey of Ebola virus infection in central African nonhuman primates. J Infect Dis 190:1895–1899. doi: 10.1086/425421. [DOI] [PubMed] [Google Scholar]
- 6.Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Delicat A, Paweska JT, Gonzalez JP, Swanepoel R. 2005. Fruit bats as reservoirs of Ebola virus. Nature 438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 7.Wittmann TJ, Biek R, Hassanin A, Rouquet P, Reed P, Yaba P, Pourrut X, Real LA, Gonzalez JP, Leroy EM. 2007. Isolates of Zaire ebolavirus from wild apes reveal genetic lineage and recombinants. Proc Natl Acad Sci U S A 104:17123–17127. doi: 10.1073/pnas.0704076104. [DOI] [PMC free article] [PubMed] [Google Scholar]