ABSTRACT
Plant virus species of the family Nanoviridae have segmented genomes with the highest known number of segments encapsidated individually. They thus likely represent the most extreme case of the so-called multipartite, or multicomponent, viruses. All species of the family are believed to be transmitted in a circulative nonpropagative manner by aphid vectors, meaning that the virus simply crosses cellular barriers within the aphid body, from the gut to the salivary glands, without replicating or even expressing any of its genes. However, this assumption is largely based on analogy with the transmission of other plant viruses, such as geminiviruses or luteoviruses, and the details of the molecular and cellular interactions between aphids and nanoviruses are poorly investigated. When comparing the relative frequencies of the eight genome segments in populations of the species Faba bean necrotic stunt virus (FBNSV) (genus Nanovirus) within host plants and within aphid vectors fed on these plants, we unexpectedly found evidence of reproducible changes in the frequencies of some specific segments. We further show that these changes occur within the gut during early stages of the virus cycle in the aphid and not later, when the virus is translocated into the salivary glands. This peculiar observation, which was similarly confirmed in three aphid vector species, Acyrthosiphon pisum, Aphis craccivora, and Myzus persicae, calls for revisiting of the mechanisms of nanovirus transmission. It reveals an unexpected intimate interaction that may not fit the canonical circulative nonpropagative transmission.
IMPORTANCE A specific mode of interaction between viruses and arthropod vectors has been extensively described in plant viruses in the three families Luteoviridae, Geminiviridae, and Nanoviridae, but never in arboviruses of animals. This so-called circulative nonpropagative transmission contrasts with the classical biological transmission of animal arboviruses in that the corresponding viruses are thought to cross the vector cellular barriers, from the gut lumen to the hemolymph and to the salivary glands, without expressing any of their genes and without replicating. By monitoring the genetic composition of viral populations during the life cycle of Faba bean necrotic stunt virus (FBNSV) (genus Nanovirus), we demonstrate reproducible genetic changes during the transit of the virus within the body of the aphid vector. These changes do not fit the view that viruses simply traverse the bodies of their arthropod vectors and suggest more intimate interactions, calling into question the current understanding of circulative nonpropagative transmission.
INTRODUCTION
Plant viruses of the families Luteoviridae, Geminiviridae, and Nanoviridae are transmitted in a circulative (persistent) nonpropagative manner by their insect vectors (1, 2). This transmission mode is characterized by the internalization of the virus within the insect without replication or even transcription of the viral genome. After feeding on an infected plant, the virus is ingested by the vector, along with the sap. The virus is then transported across the insect gut and released into the hemolymph before penetrating into the salivary glands, from where it is injected with saliva during feeding on a new plant. Though not replicating, the virus can remain infectious in the vector for several days and even throughout the vector's life span in some cases. The transcytosis mechanisms by which luteoviruses cross the gut and salivary gland epithelia in aphid vectors have been relatively well described by electron microscopy. The transmitted virus particles are transported across cells into membrane vesicles, preventing any contact between the virus and the insect cell cytoplasm (3). However, no such evidence is available for geminiviruses or nanoviruses, while putative deviations from the canonical circulative nonpropagative transmission model can be suspected from the related literature. For example, several studies have suggested possible replication of the geminivirus Tomato yellow leaf curl virus (TYLCV) within its whitefly vector, although this point is still actively debated (4–7). In another example, in addition to viral particles, an unknown virus-encoded factor has been demonstrated to be mandatory for successful access and/or passage of the nanovirus Faba bean necrotic yellows virus (FBNYV) through the salivary glands of the aphid vectors (8). The involvement of this additional factor indicates at least that the nanovirus-aphid relationship might be more complex than (or different from) that described for luteoviruses.
Virus species of the family Nanoviridae have their genomes segmented into 6 to 8 segments of ∼1 kb each. Each segment is encapsidated as a single copy into individual icosahedral virus particles 18 to 20 nm in diameter (9). Five segments are common to all nanovirus species: the segments C, M, N, R, and S. Segment C encodes a protein interacting with the cell cycle, segment M encodes the movement protein, segment N encodes a nuclear shuttle protein, segment R encodes a protein initiating replication of all the segments, and segment S encodes the capsid protein packaging each segment individually. The other segments, named U1, U2, and U4 for the genus Nanovirus and U3 for the genus Babuvirus, encode proteins with unknown functions (10). Most member species of the family Nanoviridae are aphid transmitted, and the viral form crossing gut and salivary gland barriers has been proposed to be the viral particles (11–13). Though nanovirus particles have never been directly visualized by electron microscopy inside the insect vector, studies using immunofluorescence against the coat protein of Banana bunchy top virus have consistently demonstrated that the virus accumulates within the aphid anterior midgut before being released into the hemolymph and translocated in the principal salivary glands (14).
We have recently shown that each of the eight segments composing the genome of Faba bean necrotic stunt virus (FBNSV) reproducibly accumulates at a specific relative frequency within infected host plants, with the corresponding segment frequency pattern defining the “genome formula” (15). Because in the same study the genome formula appeared to be specific to the host plant species, we questioned whether it could also be affected in a specific way during the transit of the virus within the bodies of aphid vectors. Here, we show that the FBNSV genome formula within aphids is significantly different from that observed within source plants. The observed frequency changes affect primarily the segments N and U2. They are similar in three distinct aphid vector species, Acyrthosiphon pisum, Aphis craccivora, and Myzus persicae, and occur very early during the internalization of the virus within midgut cells with no further changes at later stages of the virus transfer across the aphid body. Although we cannot explain such changes in the viral genome formula at this stage, they suggest the existence of unforeseen intimate interactions between FBNSV and its vectors that are hardly explained by the circulative nonpropagative transmission model.
MATERIALS AND METHODS
Host plants.
Vicia faba (faba bean; var. “Sevilla,” Vilmorin) plants were grown within a P2 restricted-access confinement facility in a growth chamber with a 13-h/11-h day/night photoperiod, a temperature of 26/20°C (day/night), and 70% hygrometry.
Virus isolate.
The FBNSV infectious clone used in this study is derived from an Ethiopian isolate maintained by serial passages on V. faba over 3 years and corresponds to the infectious clone used in previous studies (15–17). Briefly, each of the eight FBNSV genome segments is cloned as a tandem repeat in plasmid pBin19 and transferred into the Agrobacterium tumefaciens strain COR308. To inoculate V. faba plants, eight agrobacterial cultures (each containing one of the eight cloned segments) were grown separately in NZY+ medium (0.1% N-Z-Amine, 0.5% yeast extract, 0.5% NaCl, 12.5 mM MgCl2, 12.5 mM MgSO4, and 0.4% glucose at pH 7.5) before being mixed in equal proportions (equal optical densities at 600 nm [OD600]), centrifuged, and resuspended in a solution containing 10 mM MgCl2 and 150 μM acetosyringone. Young V. faba plants were then needle inoculated in the stem as previously described (15, 16, 18).
Aphid rearing and transmission of FBNSV.
Colonies of the aphid species A. pisum and A. craccivora were reared on V. faba plants, whereas those of M. persicae and Aphis gossypii were maintained on eggplants and zucchini, respectively. All the reared aphids were maintained under similar controlled conditions (25/19°C day/night temperatures and a photoperiod of 13 h/11 h) ensuring reproduction through parthenogenesis.
For transmission of FBNSV, the aphids were allowed an acquisition access period (AAP) of 3 days on infected V. faba plants (at 30 days postinoculation [p.i.]). The aphids were then transferred to two-leaf stage healthy V. faba plants for an inoculation access period (IAP) of three additional days unless otherwise indicated. The aphids were then collected individually or in groups of five for further analysis of their viral content, and the plants were finally treated with the insecticide Pirimor G (Syngenta; 1 g/liter in water). We were not always able to retrieve all the aphids after a 3-day inoculation access period because some aphids ran off the plants or eventually died. This explains the differences observed between the number of aphids used for inoculation and the final number of aphids used in various analyses.
Aphid dissection.
The guts of A. pisum individuals were pulled out of the bodies under a stereomicroscope. Then, the heads containing the salivary glands were severed and set aside. DNA from the guts and heads was extracted with a PureLink Genomic DNA minikit (Invitrogen, Carlsbad, CA, USA) and analyzed by quantitative PCR (Q-PCR).
EDTA-facilitated exudation.
Phloem sap was collected as described previously (19). Apices of infected V. faba plants were severed with a razor blade and immediately rinsed in a solution of 20 mM EDTA (pH 7) to eliminate contamination from wounded cells. The apices were then individually immersed in a tube containing 300 μl of the same solution for 1 h in the dark at 26°C and finally transferred into a second tube containing 300 μl of water for 7 h. DNA from the sap exudates was extracted from both EDTA and water solutions according to the method of Edwards et al. (20), with a longer centrifugation time after isopropanol addition (25 min).
DNA extraction from plants and whole aphids.
DNA from single aphids, or from groups of five, was extracted with a protocol previously described for Bemisia tabaci (21). Briefly, single aphids were ground in 10 μl of extraction buffer containing 50 mM KCl, 10 mM Tris base, pH 8, 0.45% Nonidet P-40, 0.45% Tween 20, and 500 μg/ml proteinase K. Fifteen microliters of extraction buffer was added to each of the extracts, which were then incubated at 65°C for 1 h, followed by incubation at 95°C for 15 min. After final addition of 35 μl of water per extract, they were stored at −20°C until use. The volume of extraction buffer and water was proportionally adjusted when extracting DNA from groups of five aphids.
One leaflet of the last leaf level of V. faba plants was removed at 30 days p.i. and extracted according to the method of Edwards et al. (20), with an additional washing step with 70% ethanol. The DNA of each leaflet was extracted in 400 μl of extraction buffer.
Q-PCR conditions and quantification of genome segments.
All Q-PCRs were carried out using the LightCycler FastStart DNA Master Plus SYBR green I kit (Roche) on the LightCycler 480 thermocycler (Roche), as previously described (15). Primers were used at a final concentration of 0.3 μM and are described in reference 15, supplementary table S6. For DNA extracted from both aphids and plants, Q-PCR analyses were carried out on 10-fold-diluted samples. Fluorescence data were normalized through standard curves and analyzed with the LinRegPCR program (22).
The total viral accumulation was obtained by summing the estimated numbers of copies of the eight segments. The relative frequency of each FBNSV genome segment was calculated as the estimated copy number of a given segment divided by the total viral accumulation. The standardized relative frequency for a given segment (Xi_stand) was calculated as follows: Xi_stand = (Xi − Xs)/Xs, where X is the relative frequency of a given segment and i refers to the aphid and s to the corresponding source plant.
Statistical analysis.
In all the figures, the distributions of the estimated values are shown in the form of Tukey box-and-whisker plot representations. A t test was used to test, for each segment, whether the relative frequencies within aphids were significantly different from those within source plants.
To investigate whether the relationship between the standardized relative frequencies and the duration of the acquisition access period is better explained by a linear model or a function admitting an intermediate maximum, we fitted separately a linear (Xi_stand = a + b × time) and a quadratic model (Xi_stand = a + b × time + c × time2), where Xi_stand corresponds to the standardized relative frequency for a given segment; time to the acquisition access period; and a, b, and c to parameters of the model. The difference in the Akaike information criteria between the two models was then calculated to infer which model best explained the relationship between these traits; we used the maximum-likelihood parameter values for each type of model (see Fig. 2A).
FIG 2.
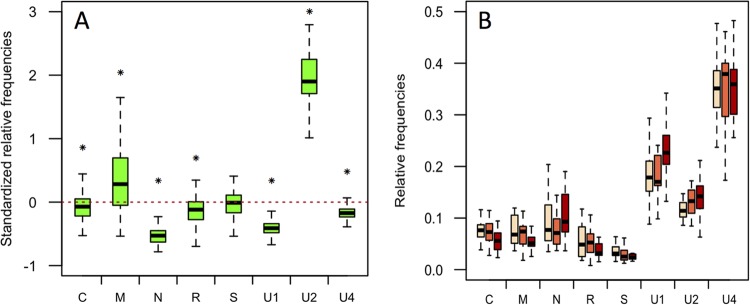
Changes in the relative frequencies of FBNSV segments occur at early stages within aphids. (A) Aphids of the species A. pisum were fed together on a single source plant. Seven groups of five aphids each were removed after AAPs of 6 h (red box plots), 10 h (yellow box plots), 25 h (green box plots), 49 h (blue box plots), and 74 h (purple box plots) and analyzed for their viral contents. The standardized relative frequency of each segment shown here corresponds to the relative frequency of a segment within aphids relative to that within the source plant. The red dashed line represents the “zero line,” where the relative frequencies of a segment are equal in aphids and in the source plant. Deviations in the frequencies of segments in aphids from those in the source plant are evident from the first time point (red box plots), and the apparent additional changes for some of the segments at later time points were statistically analyzed and are described in detail in the text. For each box plot, the bottom and top of the box are the first and third quartiles, the horizontal line within the box plots represents the median value of the distribution, and the whiskers delineate 1.5 times the distance between the first and third quartiles of the distribution. (C) The absolute copy number of each segment in the same aphid pool samples as in panel A, for which the statistical testing of the effect of time on virus accumulation is described in the text. (B) Dissected guts (pink box plots) and heads (blue box plots) of 22 groups of 5 aphids each of the species A. pisum were analyzed for their viral content after a 3-day AAP on an infected plant and a 3-day IAP on a healthy plant. The relative segment frequencies shown here are not standardized and simply compare the FBNSV genome formulas in aphid guts and heads, regardless of that in the source plant. No statistical differences could be detected between the two aphid compartments, and the detailed results of the tests are described in the text. (D) Absolute copy number of each segment in the same head and gut pool samples as in panel B.
A similar approach was used to test whether a linear model or a function admitting an intermediate maximum better explains the relationship between the copy number of each segment in aphids and the duration of the acquisition access period. The segment copy numbers were log transformed to carry out this analysis (see Fig. 2C).
We tested for the effects of the parameters plant extracts and segment and their interaction on the segment relative frequency within plants using a generalized linear model (GLM) (Fig. 1B). We used the glm function of R with a Gaussian distribution as the error structure and an F test to check for their effects. A similar approach was used to test for the effects of the parameters aphid compartment and segment and their interaction on the segment relative frequency within aphids (Fig. 2B), for the effects of the parameters aphid species and segment and their interaction on the standardized relative frequency (Fig. 3 and 4B), and for the effects of the parameters transmission success and segment and their interaction on the standardized relative frequency (Fig. 4A).
FIG 1.
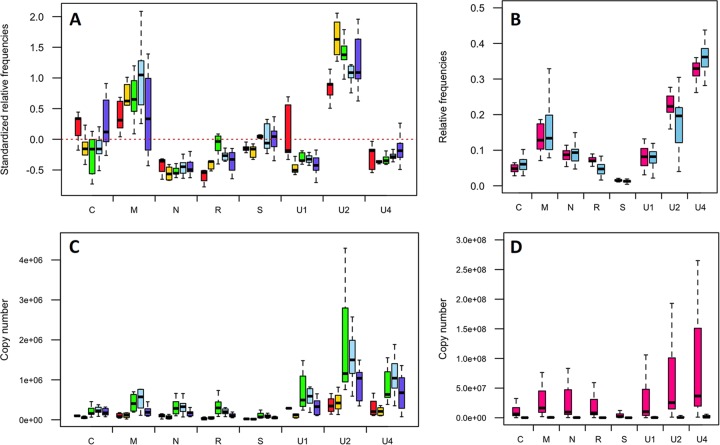
Relative frequencies of FBNSV genome segments in aphids relative to source plants. (A) Aphids of the species A. pisum were fed on an FBNSV-infected V. faba plant before individual transfer onto test plants. This experiment was repeated twice, and the results were pooled. The standardized relative frequency of each segment shown here corresponds to the relative frequency of a segment within aphids relative to that within the corresponding source plants (for details, see Materials and Methods). The box plots summarize data from 117 aphids, and the red dashed line represents the “zero line,” where the relative frequencies of a segment are equal in aphids and plants. For each box plot, the bottom and top of the box are the first and third quartiles, the horizontal line within the box plots represents the median value of the distribution, and the whiskers delineate 1.5 times the distance between the first and third quartiles of the distribution. The segments are shown under the x axis. The asterisks indicate significant differences in the frequencies of a segment in the source plant and in corresponding aphids. Detailed results of statistical tests are given in the text. (B) The relative frequency of each FBNSV genome segment was analyzed specifically within V. faba phloem sap by EDTA-facilitated exudation. After rinsing, the exudates were first collected for 1 h in an EDTA solution (beige box plots) and for 7 additional hours in water (orange box plots). The FBNSV genome formula in these sap exudates did not significantly differ from that in the corresponding apex leaves (brown box plots). The details of the statistical analysis are given in the text.
FIG 3.
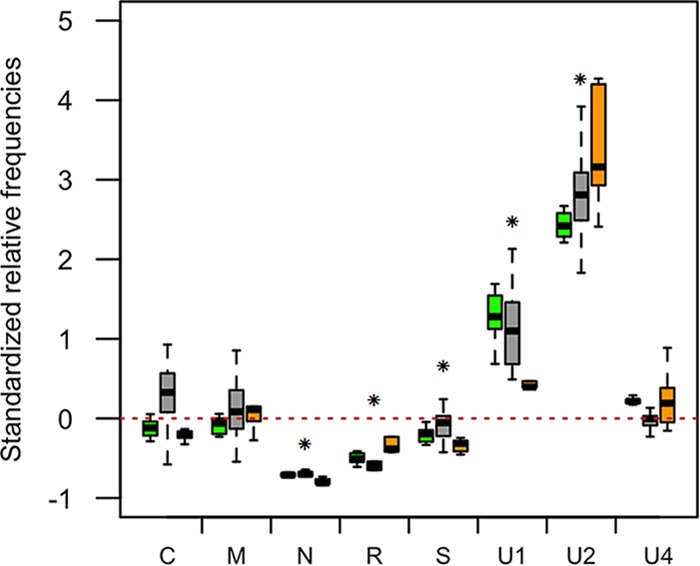
FBNSV genome formulas are similar in distinct aphid vector species. Changes in the frequencies of FBNSV genome segments were estimated in groups of 5 aphids of the species A. pisum (green box plots; n = 11), A. craccivora (gray box plots; n = 13), and M. persicae (orange box plots; n = 8), all caged together and fed on the same source plant. The standardized relative frequency of each segment shown here corresponds to the relative frequency of a segment within aphids relative to that within the source plant. The red dashed line represents the “zero line,” where the relative frequencies of a segment are equal in aphids and in the source plant. For each box plot, the bottom and top of the box are the first and third quartiles, the horizontal line within the box plots represents the median value of the distribution, and the whiskers delineate 1.5 times the distance between the first and third quartiles of the distribution. Significant changes between the source plant and aphids when the three aphid species were pooled are indicated by asterisks.
FIG 4.
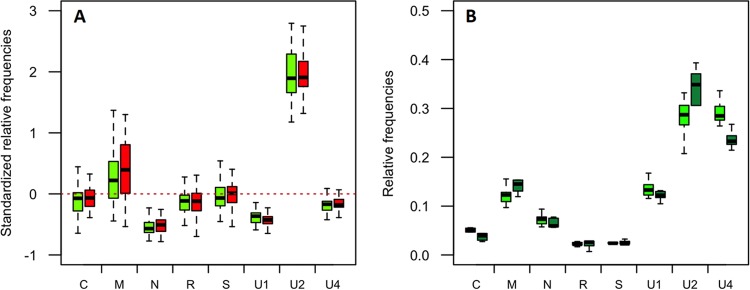
FBNSV genome formula within aphids and transmission success. (A) Comparison of the relative frequencies of FBNSV segments in individual A. pisum aphids that efficiently transmitted the virus (green box plots; n = 54) in the experiment described in the legend to Fig. 1 and in those that did not (red box plots; n = 63). The standardized relative frequency of each segment shown here corresponds to the relative frequency of a segment within aphids relative to that within the source plant. For each box plot, the bottom and top of the box are the first and third quartiles, the horizontal line within the box plots represents the median value of the distribution, and the whiskers delineate 1.5 times the distance between the first and third quartiles of the distribution. The red dashed line represents the “zero line,” where the relative frequencies of a segment are equal in aphids and in the source plant. No significant differences could be detected between the two aphid groups (see the text for details of statistical analysis). (B) Comparison of the relative frequencies of FBNSV segments in groups of 5 aphids of the vector species A. pisum (light-green box plots; n = 10) and of the nonvector species A. gossypii (dark-green box plots; n = 5), all caged and fed together on the same source plant. The relative segment frequencies shown here are not standardized and simply compare the FBNSV genome formulas in vector and nonvector species, regardless of that in the source plant. No statistical differences could be detected between the two aphid species, and the detailed results of the tests are described in the text.
All statistical analyses were carried out with the R software, version 2.12.0 (2011; R Development Core Team), except for the test of the significance of the correlation coefficients, for which JMP10 (SAS) was used. The nature and results of the statistical tests are indicated throughout the text. A P value of <0.05 was used to indicate statistical significance. In each of the analyses where eight tests were conducted (one for each of the eight segments), the significance level was adjusted using the Bonferroni correction.
RESULTS
The relative frequencies of the FBNSV segments changes within aphids.
One hundred aphids of the species A. pisum, an efficient vector of FBNSV (16), were allowed a 3-day AAP on the apex of a symptomatic V. faba source plant at 30 days p.i. At the end of the AAP, aphids were transferred individually onto 100 healthy V. faba test plants for an additional 3-day IAP. Within 10 to 20 days, nearly half of the test plants (38% and 51%) proved to be successfully infected in two repeats of the experiment. The relative frequency of each of the eight FBNSV genome segments was then estimated by Q-PCR in the youngest leaf level of both the source and symptomatic test plants, as well as in individual aphids. Over the two experiment repeats, we could successfully quantify all eight FBNSV genome segments from the 2 source plants, 54 infected test plants, and 117 individual aphids. Consistent with previously reported results, the relative frequencies of all segments were similar in both source and test plants (15), except for segment U1 (results not shown). In contrast, we found that the relative frequencies of the FBNSV genome segments changed significantly within aphids. Figure 1A shows the standardized relative frequency of each segment (standardized as described in Materials and Methods), where deviations from zero indicate a change when the virus population from the source plant was ingested by and internalized within aphids. All segments but segment S had a different relative frequency in aphids (one-sample t test; −41.58 < t < 44.10, df - 116, and P < 0.0016 for all segments except segment S, for which t = −0.58, df = 116, and P = 0.57). In this experiment, we especially observed a sharp aphid-related decrease of N and U1 and a sharp increase of U2.
One could argue that the observed changes in aphids may simply stem from different FBNSV genome formulas in the total cell extract analyzed from the source leaf and in the phloem sap that is specifically ingested by aphids. To evaluate this possibility, we compared the genome formulas found in total leaf extracts and in sap exudates. The apices of 17 infected V. faba plants were used individually to collect phloem sap prior to DNA extraction from each apex. The segment frequencies in the sap exudates were compared to those in the leaves of the corresponding apices (Fig. 1B). The segments' relative frequencies, and thus the genome formulas, in the sap exudates appeared similar to those in the apices, confirming that the differences observed in aphids did not stem from differences in distinct plant compartments (GLM; plant extract effect, F = 0, df = 2, and P = 1; segment effect, F = 153.95, df = 7, and P < 2e−16; interaction between plant extracts and segment, F = 0.85, df = 14, and P = 0.61).
Change of the FBNSV genome formula occurs early within the aphid gut.
We then questioned whether the relative frequencies of the segments gradually change throughout the cycle of the virus within the aphid vector or mainly occur at a precise step. We allowed aphids to continuously feed on the apex of an infected V. faba plant and collected them at different time points. The collected aphids were then purged for at least 4 h on healthy plants before DNA extraction from groups of five individuals and Q-PCR analysis. The genome segments could not be reliably quantified in aphids before a minimum AAP of 6 h, a time at which the frequency changes were in large part already completed (Fig. 2A), with a remarkable drop in the frequencies of N and R and a rise in that of U2 (one-sample t test; t = −6.68, −11.31, and 7.89, respectively; df = 4; P = 0.0026, 0.00035, and 0.0014, respectively). The possible effect of time was further assessed by fitting linear and quadratic models for each segment. It appeared that for segments C, M, and R, a quadratic model better explained the data (Fig. 2A, ΔAIC linear-quadratic = 7.8, 5.7, and 4.2, respectively) with a significant effect of the parameters time (P < 0.021 for all three segments) and time2 (P < 0.017 for all three segments), and thus, for these segments, the standardized relative frequency may present an intermediate maximum. For the segment U1, the data were slightly better explained by a linear model (Fig. 2A, ΔAIC quadratic-linear = 1.7) with a significant effect of time(P = 0.040). For the segments N, S, U2, and U4, no significant effect of time (or of time2) on the standardized relative frequency could be detected (P > 0.052).
To further support the conclusion that most of the changes in the genome formula occur at early stages of the aphid-FBNSV interaction, we compared the viral genome formulas in dissected aphid guts and in the corresponding severed heads, containing the salivary glands. The guts and heads of 110 A. pisum aphids were separated from the rest of the aphid body and analyzed as 22 pools of 5 by Q-PCR. As illustrated in Fig. 2B, the frequency of the FBNSV segments in aphid heads proved very similar to that found within guts (GLM; aphid compartment effect, F = 0, df = 1, and P = 1.00; segment effect, F = 175.78, df = 7, and P < 2.2e−16; interaction between the aphid compartment and segment, F = 3.09, df = 7, and P = 3.64e−3). We thus conclude that the FBNSV genome formula is altered either directly in the gut lumen, during internalization into gut cells, or soon after and that it subsequently remains stable during release into the hemolymph and translocation into the salivary glands.
For the same samples, the absolute copy number of each segment was quantified and is shown in Fig. 2C and D. For viral accumulation within whole aphids during acquisition on infected plants, and for each segment, a quadratic model better explains the data (Fig. 2C, ΔAIC linear-quadratic = 6.9, 21.4,12.9, 22.3,9.6, 12.8, 17.8, and 13.0 for segments C, M, N, R, S, U1, U2, and U4, respectively) with a significant effect of time (P < 5.65e−4 for all segments) and time2 (P < 5.08e−3 for all segments). This result indicates an accumulation of all FBNSV segments over 48 h, which could simply be explained by cumulative internalization of the virus. Surprisingly, the virus load similarly decreased for all 8 segments between 48 and 72 h of AAP, despite continued access of the aphids to infected plants. The reason for this decrease is unknown and will require further investigation. The virus load in the aphids' heads proved much lower than that in the guts (Fig. 2D), indicating that only a small fraction of the virus internalized in the gut transfers to the salivary glands (around 0.2%) (Fig. 2D). Nevertheless, comparison with Fig. 2B suggests that, for each segment, this transfer is proportional to the frequency in the gut.
The genome formula in aphids depends only partly on the initial formula in source plants.
An important follow-up question is whether the FBNSV genome formula, and particularly the frequencies of segments N and U2 (those most reproducibly changing in aphids [Fig. 1, 2A, 3, and 4A]), tends to converge toward a specific endpoint in aphids, whatever the initial situation in the ingested sap. For this, we tested whether the frequencies of these two segments are more constrained within aphids than in the corresponding source plants. Five source plants from different experiments were used for this analysis: the source plants used in Fig. 1 (two source plants) and the one used in Fig. 3 (one source plant) plus two source plants from two additional experiments (not shown). Estimates of the coefficient of variation show that the frequency distributions of segments N and U2 were less dispersed among aphids (39.9% and 18.1%, respectively) than among source plants (81.0% and 39.3%, respectively). This result suggests the existence of unknown aphid-related constraints that tend to induce a low and high relative frequency of N and U2, respectively, whatever the initial situation within source plants.
We also tested for a possible correlation between the decrease in the relative frequency of N and the increase in that of U2, assessing a possible functional link between the two events. No significant correlation could be detected (Pearson's correlation test; ρ = 0.083, P = 0.29), suggesting that changes in the relative frequencies of N and U2 are independent.
The aphid-related FBNSV genome formula is conserved in distinct aphid vector species.
In addition to A. pisum, used in this study, FBNSV has been reported to be transmitted by other aphid species, including A. craccivora (16), which we decided to test for a similar effect on the FBNSV genome formula. In parallel, we tested a third aphid species, M. persicae, because it has been described as a nonvector of the related nanovirus species FBNYV (13). Somewhat to our surprise, M. persicae also proved to be an efficient vector of FBNSV under our experimental conditions. Depending on the experiment, and using five aphids per test plant, A. pisum transmitted FBNSV to 18% to 100% of the test plants, A. craccivora to 81% to 90%, and M. persicae to 31% to 100%.
Over 100 individuals of each of the three aphid species were caged together on the same infected V. faba source plant for a 3-day AAP. Then, groups of five individuals of the same species were each transferred to 16 test plants per species for a 3-day IAP. At the end of the IAP, DNA was extracted from each group of five aphids to determine the FBNSV genome formulas in A. pisum, A. craccivora, and M. persicae and compare them to that in the single source plant (Fig. 3). The overall patterns of the genome formulas were similar across the three aphid species (GLM; F = 1.02, df = 2, and P = 0.36) and quite distinct from that in the source plant. In fact, when the three aphid species results were pooled, the standardized relative frequencies of segments N, R, S, U1, and U2 significantly changed within aphids (one-sample t test; −45.07 < t < 23.15, df = 28, and P < 0.00038), whereas those of segments C, M, and U4 did not. It is interesting that not only the segments, but also the interaction between the segments and the aphid species, proved to have a significant effect on the standardized relative frequency (GLM; F = 305.30, df = 7, and P < 2.2e−16 and F = 5.66, df = 14, and P = 2.94e-09, respectively), suggesting that the small variations of the genome formulas in the three aphid species are significant. Most importantly, however, this experiment confirmed that the more drastic and reproducible changes across the three vector species are a sharp drop and an increase in the frequency of the segments N and U2, respectively.
Relative frequencies of FBNSV segments within aphids and transmission efficiencies.
Finally, we questioned whether the changes in the FBNSV genome formula within aphids play a role in the success of transmission. We first further analyzed the data set presented in Fig. 1A. There, A. pisum aphids had been tested individually for transmission (a single aphid per test plant). It is thus possible to compare the FBNSV genome formula in aphids that actually successfully transmitted the virus (n = 54) and in aphids that failed to transmit (n = 63). Figure 4A shows that the genome formulas are similar in the two aphid sets, indicating that, at least in this experiment, the success or failure of transmission by individual aphids was not related to an altered genome formula of the acquired FBNSV (GLM; F = 0.76, df = 1, and P = 0.38). Also, the interaction between the transmission success and segments had no significant effect on the standardized relative frequency (GLM; F = 1.33, df = 7, and P = 0.23), whereas the segments logically had a significant effect (GLM; F = 858.02 1.33, df = 7, and P < 2e−16). We further compared the total viral loads within aphids by summing up the copy numbers of all eight segments. The viral loads proved similar in the two aphid sets (not shown).
We then tested whether the genome formulas observed in aphid vectors differ in nonvector species. Aphids of the species A. gossypii (here considered a nonvector species) were caged together with A. pisum aphids on the apex of an infected V. faba plant for a 3-day AAP. Groups of five aphids of the same species were then each transferred onto a test plant for a 3-day IAP. In this experimental setting, A. pisum successfully transmitted the virus to 7 out of 23 test plants, whereas A. gossypii failed to transmit to any of the 15 test plants. Despite the fact that A. gossypii proved to be a nonvector or a poor vector species under our experimental conditions, similar amounts of virus accumulated in both A. pisum and A. gossypii (data not shown). Moreover, similar patterns of the genome formula were estimated in the two species (Fig. 4B) (GLM; F = 0, df = 1, and P = 1). It should be noted, however, that both the segments and the interaction between the segments and the aphid species proved to have a significant effect on the relative frequency (GLM; F = 270.31, df = 7, and P < 2.2e−16 and F = 4.54, df = 7, and P = 0.00019, respectively), indicating that although very similar, the formulas show slight differences in the two aphid species.
DISCUSSION
The process underlying the changes in the genome formula of FBNSV within aphids is obscure thus far. One obvious possibility, although contrary to the current view of the nanovirus-aphid interaction, is that some or all FBNSV genome segments could replicate. Because the replication cycles of some double-stranded DNA (dsDNA) viruses can be completed within a few minutes (23), it is currently impossible to exclude a very early and transient phase of FBNSV replication that we cannot see under our experimental conditions and that previous investigations of nanovirus transmission (14, 24) may have overlooked. The observation that the aphid-related genome formula adjusted very early with little further change at later time points (e.g., Fig. 2A and B) would be consistent with this putative replication occurring solely in gut cells and during a limited time. Under this replication hypothesis, mechanisms similar to those we discussed previously (15) for the establishment of the genome formula in host plants could operate similarly in aphids.
Alternatively, the observed changes in aphids could be attributed to a differential decapsidation and degradation of some segments. However, this would involve unknown distinct physical properties (stability) of virus particles, depending on the contained genome segment, with N- and U2-containing particles under this hypothesis being highly labile and stable, respectively. This particle stability hypothesis could explain the fact that segments N and U2 decreased and increased, respectively, in relative frequency in all experimental repeats, but it cannot explain why some segments, for example, M, U1, and U4, behaved in a more erratic way, sometimes decreasing and sometimes increasing relative to the source plant, depending on the experiment, as shown in the figures.
As a third possibility, physicochemical differences of virus particles, depending on the segments contained, could also account for differential interactions with the aphid midgut putative receptor(s). It is widely recognized that the first cellular barrier in insect vectors encountered by circulative viruses is the gut barrier. The time at which a nanovirus passes this barrier has not yet been reported, but geminiviruses and luteoviruses cross it within a few hours—1 h and 12 to 16 h, respectively (25, 26). FBNSV could thus enter the aphid gut cells very rapidly, and differential uptake for particles containing different segments could explain the observed changes in the genome formula. Nevertheless, this hypothesis suffers from the same drawbacks mentioned above for differential degradation: while it could fit the consistent observations for N and U2, it cannot explain the erratic behavior of some other segments.
Unfortunately, at this point, we cannot provide experimental evidence for a correlation between the FBNSV genome formula in aphids and the efficiency of transmission because we did not observe genome formula differences between transmitter and nontransmitter aphids. First, the A. pisum set containing the individuals that transmitted the virus (Fig. 4A) did not yield a genome formula significantly different from that calculated with the set containing individuals that did not transmit the virus. This is not an unexpected result, because it is commonly known that aphid vectors fed with circulative viruses are often able to transmit it through their entire life, but in an “erratic” manner. This means that one individual viruliferous aphid serially transferred onto several test plants will transmit the virus to only some of them, indicating that the transmission success is not 100%. Thus, we believe that all A. pisum individuals in the experiment shown in Fig. 4A were equally able to transmit, irrespective of the fact that they actually did or not, so the sets of transmitters and nontransmitters could be strictly equivalent. Second, all tested aphid vector species induce similar frequency changes in the FBNSV genome formula. The only nonvector species (A. gossypii) tested here accumulates FBNSV in amounts similar to those detected in vector species and induces similar changes in the genome formula. We propose that this observation could be due to compatible early virus-aphid interactions in all cases, followed by transmission blocking at a later step for nonvectors only. For example, late barriers blocking passage across the salivary glands have been demonstrated for some luteovirus- and geminivirus-vector couples (27–29). Further investigation will be required to determine the exact step at which FBNSV is blocked within A. gossypii.
At this stage, we cannot come to a conclusion about a possible functional role of the changes in the FBNSV segment frequencies within aphids, which could also be a corollary of a yet unknown FBNSV-aphid interaction. Further investigation of this point is definitely required, in particular, the localizations and frequencies of segments N and U2 (and of their RNA and protein products) both within plants prior to acquisition and later in the bodies of aphid vectors. This will probably help in unraveling an unforeseen nanovirus-aphid interaction that may not fit into any of the preexisting and longstanding categories of vector transmission in plant viruses.
ACKNOWLEDGMENTS
We are grateful to B. Gronenborn and T. Timchenko for technical help and advice.
A.S., S.B., M.Y., and S.G. acknowledge support from INRA SPE division (project NanoVigs-SPE) and from the Agence National de la Recherche (grant ANR-Nano), Y.M. from CNRS and IRD, J.-L.Z. from IRD, and S.G. from CIRAD.
REFERENCES
- 1.Hogenhout SA, Ammar E-D, Whitfield AE, Redinbaugh MG. 2008. Insect vector interactions with persistently transmitted viruses. Annu Rev Phytopathol 46:327–359. doi: 10.1146/annurev.phyto.022508.092135. [DOI] [PubMed] [Google Scholar]
- 2.Blanc S, Drucker M, Uzest M. 2014. Localizing viruses in their insect vectors. Annu Rev Phytopathol 52:403–425. doi: 10.1146/annurev-phyto-102313-045920. [DOI] [PubMed] [Google Scholar]
- 3.Brault V, Herrbach É Reinbold C. 2007. Electron microscopy studies on luteovirid transmission by aphids. Micron 38:302–312. doi: 10.1016/j.micron.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Czosnek H, Ghanim M, Morin S, Rubinstein G, Fridman V, Zeidan M. 2001. Whiteflies: vectors, and victims (?) of geminiviruses. Adv Virus Res 57:291–322. doi: 10.1016/S0065-3527(01)57006-2. [DOI] [PubMed] [Google Scholar]
- 5.Mehta P, Wyman JA, Nakhla MK, Maxwell DP. 1994. Transmission of tomato yellow leaf curl geminivirus by Bemisia tabaci (Homoptera: Aleyrodidae). J Econ Entomol 87:1291–1297. doi: 10.1093/jee/87.5.1291. [DOI] [PubMed] [Google Scholar]
- 6.Sinisterra XH, McKenzie CL, Hunter WB, Powell CA, Shatters RG Jr. 2005. Differential transcriptional activity of plant-pathogenic begomoviruses in their whitefly vector (Bemisia tabaci, Gennadius: Hemiptera Aleyrodidae). J Gen Virol 86:1525–1532. doi: 10.1099/vir.0.80665-0. [DOI] [PubMed] [Google Scholar]
- 7.Díaz-Pendón JA, Cañizares MC, Moriones E, Bejarano ER, Czosnek H, Navas-Castillo J. 2010. Tomato yellow leaf curl viruses: ménage à trois between the virus complex, the plant and the whitefly vector: Tomato yellow leaf curl viruses. Mol Plant Pathol 11:441–450. doi: 10.1111/j.1364-3703.2010.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franz A, Makkouk KM, Vetten HJ. 1998. Acquisition, retention and transmission of faba bean necrotic yellows virus by two of its aphid vectors, Aphis craccivora (Koch) and Acyrthosiphon pisum (Harris). J Phytopathol 146:347–355. doi: 10.1111/j.1439-0434.1998.tb04703.x. [DOI] [Google Scholar]
- 9.Grigoras I, Ginzo AIDC, Martin DP, Varsani A, Romero J, Mammadov AC, Huseynova IM, Aliyev JA, Kheyr-Pour A, Huss H, Ziebell H, Timchenko T, Vetten H-J, Gronenborn B. 2014. Genome diversity and evidence of recombination and reassortment in nanoviruses from Europe. J Gen Virol 95:1178–1191. doi: 10.1099/vir.0.063115-0. [DOI] [PubMed] [Google Scholar]
- 10.Timchenko T, Bernardi F. 2007. Les nanovirus, petits virus de plantes: similitudes et différences avec les géminivirus. Virologie 11:27–42. [DOI] [PubMed] [Google Scholar]
- 11.Chu PWG, Helms K. 1988. Novel virus-like particles containing circular single-stranded DNAs associated with subterranean clover stunt disease. Virology 167:38–49. doi: 10.1016/0042-6822(88)90052-9. [DOI] [PubMed] [Google Scholar]
- 12.Harding RM, Burns TM, Dale JL. 1991. Virus-like particles associated with banana bunchy top disease contain small single-stranded DNA. J Gen Virol 72:225–230. doi: 10.1099/0022-1317-72-2-225. [DOI] [PubMed] [Google Scholar]
- 13.Katul U, Vetten HJ, Maiss E, Makkouk KM, Lesemann D-E, Casper R. 1993. Characterisation and serology of virus-like particles associated with faba bean necrotic yellows. Ann Appl Biol 123:629–647. doi: 10.1111/j.1744-7348.1993.tb04933.x. [DOI] [Google Scholar]
- 14.Watanabe S, Bressan A. 2013. Tropism, compartmentalization and retention of banana bunchy top virus (Nanoviridae) in the aphid vector Pentalonia nigronervosa. J Gen Virol 94:209–219. doi: 10.1099/vir.0.047308-0. [DOI] [PubMed] [Google Scholar]
- 15.Sicard A, Yvon M, Timchenko T, Gronenborn B, Michalakis Y, Gutierrez S, Blanc S. 2013. Gene copy number is differentially regulated in a multipartite virus. Nat Commun 4:2248. doi: 10.1038/ncomms3248. [DOI] [PubMed] [Google Scholar]
- 16.Grigoras I, Timchenko T, Katul L, Grande-Perez A, Vetten H-J, Gronenborn B. 2009. Reconstitution of authentic nanovirus from multiple cloned DNAs. J Virol 83:10778–10787. doi: 10.1128/JVI.01212-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grigoras I, Timchenko T, Grande-Perez A, Katul L, Vetten H-J, Gronenborn B. 2010. High variability and rapid evolution of a nanovirus. J Virol 84:9105–9117. doi: 10.1128/JVI.00607-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Timchenko T. 2006. Infectivity of nanovirus DNAs: induction of disease by cloned genome components of Faba bean necrotic yellows virus. J Gen Virol 87:1735–1743. doi: 10.1099/vir.0.81753-0. [DOI] [PubMed] [Google Scholar]
- 19.King RW, Zeevaart JAD. 1974. Enhancement of phloem exudation from cut petioles by chelating agents. Plant Physiol 53:96–103. doi: 10.1104/pp.53.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edwards K, Johnstone C, Thompson C. 1991. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delatte H, Reynaud B, Granier M, Thornary L, Lett JM, Goldbach R, Peterschmitt M. 2005. A new silverleaf-inducing biotype Ms of Bemisia tabaci (Hemiptera: Aleyrodidae) indigenous to the islands of the south-west Indian Ocean. Bull Entomol Res 95:29–35. [DOI] [PubMed] [Google Scholar]
- 22.Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37:e45–e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Paepe M, Taddei F. 2006. Viruses' life history: towards a mechanistic basis of a trade-off between survival and reproduction among phages. PLoS Biol 4:e193. doi: 10.1371/journal.pbio.0040193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hafner GJ, Harding RM, Dale JL. 1995. Movement and transmission of banana bunchy top virus DNA component one in bananas. J Gen Virol 76:2279–2285. doi: 10.1099/0022-1317-76-9-2279. [DOI] [PubMed] [Google Scholar]
- 25.Garret A, Kerlan C, Thomas D. 1996. Ultrastructural study of acquisition and retention of potato leafroll luteovirus in the alimentary canal of its aphid vector, Myzus persicae Sulz. Arch Virol 141:1279–1292. doi: 10.1007/BF01718830. [DOI] [PubMed] [Google Scholar]
- 26.Ghanim M, Morin S, Czosnek H. 2001. Rate of Tomato yellow leaf curl virus translocation in the circulative transmission pathway of its vector, the whitefly Bemisia tabaci. Phytopathology 91:188–196. doi: 10.1094/PHYTO.2001.91.2.188. [DOI] [PubMed] [Google Scholar]
- 27.Cohen S. 1989. Acquisition, interference, and retention of cucurbit leaf curl viruses in whiteflies. Phytopathology 79:109. doi: 10.1094/Phyto-79-109. [DOI] [Google Scholar]
- 28.Gildow FE. 1993. Evidence for receptor-mediated endocytosis regulating luteovirus acquisition by aphids. Phytopathology 83:270. doi: 10.1094/Phyto-83-270. [DOI] [Google Scholar]
- 29.Gray S, Gildow FE. 2003. Luteovirus-aphid interactions. Annu Rev Phytopathol 41:539–566. doi: 10.1146/annurev.phyto.41.012203.105815. [DOI] [PubMed] [Google Scholar]