Abstract
Background and Aims Accumulation of unfolded proteins caused by inefficient chaperone activity in the endoplasmic reticulum (ER) is termed ‘ER stress’, and it is perceived by a complex gene network. Induction of these genes triggers a response termed the ‘unfolded protein response’ (UPR). If a cell cannot overcome the accumulation of unfolded proteins, the ER-associated degradation (ERAD) system is induced to degrade those proteins. In addition to other factors, reactive oxygen species (ROS) are also produced during oxidative protein-folding in the ER. It has been shown in animal systems that there is a tight association between mitochondrial ROS and ER stress. However, in plants there are no reports concerning how induced ROS production in mitochondria and chloroplasts affects ER stress and if there is a possible role of organelle-originated ROS as a messenger molecule in the unfolded protein response. To address this issue, electron transport in chloroplasts and mitochondria and carnitine acetyl transferase (CAT) activity in peroxisomes were inhibited in wild-type Arabidopsis thaliana to induce ROS production. Expression of UPR genes was then investigated.
Methods Plants of A. thaliana ecotype Col-0 were treated with various H2O2- and ROS-producing agents specific to different organelles, including the mitochondria, chloroplasts and peroxisomes. The expression of ER stress sensor/transducer genes (bZIP28, bZIP17, IRE1A, IRE1B, BiP1, BiP3), genes related to protein folding (CNX, ERO1) and ERAD genes (HRD1, SEL1, DER1, UBC32) were evaluated by qRT-PCR analysis.
Key Results Relatively low concentrations of ROS were more effective for induction of the ER stress response. Mitochondrial and chloroplastic ROS production had different induction mechanisms for the UPR and ER stress responses.
Conclusions Chloroplast- and mitochondria-originated ROS have distinct roles in triggering the ER stress response. In general, low concentrations of ROS induced the transcription of ER stress-related genes, which can be attributed to the roles of ROS as secondary messengers. This is the first time that ROS production in organelles has been shown to affect the ER stress response in a plant system.
Keywords: Oxidative stress, endoplasmic reticulum stress, unfolded protein response, UPR, endoplasmic reticulum-associated degradation, ERAD, reactive oxygen species, ROS, signalling, Arabidopsis thaliana
INTRODUCTION
The endoplasmic reticulum (ER) acts as a hub in the production of secretory proteins in eukaryotic cells. Secretory proteins are modified and folded in the ER, before transfer to their final destination via the secretory pathway (Iwata and Kouzumi, 2012). A series of well-coordinated processes are required for synthesis and secretion of proteins from the ER. First, nascent proteins, synthesized by the ER membrane-bound ribosomes, are moved into ER by a Sec61-like translocon during translation. Then, these proteins are stabilized by HSP40 and HSP70-like proteins such as ERdj3 and binding protein (BiP) in the ER before they are properly modified and fold (Howell, 2013). The interaction of these chaperones with nascent polypeptides prevents aggregation and helps their proper folding. Glycosylated proteins are modified by oligosaccharide transferase in the ER and are folded by a folding apparatus that constitutes the lectin protein calreticulin/calnexin complex and protein disulfide isomerase (PDI). PDI acts as a mediator during formation of disulfide bonds (Kamauchi et al., 2005; Lu and Christopher, 2008). ER oxidoreductase 1 (ERO1) is a glycosylated flavoenzyme that is tightly associated with the luminal face of the ER and catalyses formation of disulfide bonds by oxidizing PDI. In parallel with its role in oxidative protein folding, ERO1 is a significant source of oxidizing equivalents and is responsible for the regulation of ER oxidation state (Dixon et al., 2003; Onda et al., 2009).
Protein load and folding capacity in ER is meticulously controlled. Accumulation of unfolded proteins due to insufficient folding capacity in ER triggers a response called the unfolded protein response (UPR), which induces genes and pathways that facilitate protein folding and degradation of misfolded proteins (Martinez and Chrispeels, 2003; Schröder and Kaufman, 2005; Deng et al., 2011). More detailed information on this subject can be found in a recent excellent review by Howell (2013).
ER stress is sensed by sensors/transducers that are located on the ER membrane. The ER stress response signalling pathway comprises two arms. One arm involves bZIP28, a membrane-associated transcription factor, and the other involves inositol requiring enzyme 1 (IRE1), a membrane-associated dual functioning protein kinase/ribonuclease (Koizumi et al., 2001; Liu and Howell, 2010a, b; Iwata and Koizumi, 2012). Upon accumulation of unfolded proteins in the ER lumen, bZIP28 is mobilized from ER to Golgi bodies, where it is processed by S1P and S2P proteases to release the N-terminal portion of bZIP28 to the cytosol (Liu et al., 2007a, b; Che et al., 2010). Cleaved bZIP28 moves to the nucleus and induces the ER stress response (Tajima et al., 2008). By contrast, upon ER stress, IRE1 splices bZIP60 mRNA and the translated product activates the ER stress response. In addition, bZIP28 and bZIP60 can heterodimerize and these two arms of the pathway may overlap to induce a stress response (Gao et al., 2008; Liu and Howell, 2010a, b).
Another important mechanism in response to ER stress is the removal of unfolded or misfolded proteins from the ER lumen to prevent their aggregation (Su et al., 2011). ER-associated protein degradation (ERAD) effectively removes proteins from the ER and these proteins are degraded by 26S proteasome. Briefly, unfolded glycosylated proteins are recognized by a luminal lectin OS9 which works together with SEL1/HRD3 (a membrane spanning protein) and HRD1 (an E3 ligase). These unfolded proteins are then transferred to cytosol by this complex. After removal from ER lumen, ubiquinated proteins are degraded by 26S proteasome in the cytoplasm (Su et al., 2012).
In a previous study, we investigated the relationship among ER stress, reactive oxygen species (ROS) signalling and antioxidant defence in arabidopsis and showed that treatment with the ER stress agent tunicamycin (Tm) can induce expression of NADPH oxidase encoding genes RBOHD and RBOHF and can induce the activities of ROS scavenging enzymes leading to a change in redox status of the cell (Ozgur et al., 2014). A need for an oxidizing environment in ER for proper disulfide bond formation and results from our previous study clearly imply that ROS signalling and cellular redox status are important players during response to ER stress. Chloroplasts and mitochondria are the major sources of ROS in the plant cell. In animals, mitochondrial ROS production is tightly associated with ER stress (Berridge, 2002; Hotamisligil, 2010). It has been shown that ER stress and mitochondrial dysfunction can induce each other and there is a cross-talk between them via ROS. However, despite the significance of this issue, it is still not known how ROS produced in different compartments of the plant cell such as mitochondria, chloroplasts and peroxisomes affect ER stress response and ER folding machinery.
The aim of this study was to investigate the effects of organellar ROS on different components of the ER stress response such as ER quality control (ERQC), UPR and ERAD at the transcriptional level. For this, different chemicals were used to induce ROS production/accumulation in different cellular compartments: (i) rotenone in mitochondria, (ii) methyl viologen (MV) and 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) in chloroplasts, and (iii) 3-amino-triazole (3-AT) in peroxisomes, in addition to exogenous H2O2 treatments.
Expressions of binding protein 1 (BiP1), BiP3, bZIP28, bZIP17, inositol requiring enzyme1A (IRE1A), IRE1B, calnexin (CNX), endoplasmic reticulum oxidoreductase (ERO1), HRD1, SEL1, UBC32 and DER1 were investigated to elucidate the effects of organellar ROS on ER stress response.
MATERIAL AND METHODS
Plant material, growth conditions and stress treatments
In this study, Arabidopsis thaliana ecotype Col-0 was used as plant material. Plants were grown in a plant growth chamber using a hydroponic system under controlled conditions (23/21 °C day and night temperatures, 60 % relative humidity, 12/12-h light/dark period, 200 µmol photons m–2 s–1 light intensity) with half-strength Hoagland’s solution. Fully expanded rosette leaves of 21-d-old plants were used for the experiments. Leaves were detached and floated on solutions containing H2O2 (10 µm, 100 µm, 1 mm, 10 mm), rotenone (Rot; 1, 10, 25, 50 µm), MV (1, 10, 25, 50 µm), DCMU (1, 10, 25, 50 µm), 3-AT (1 mm) and tunicamycin (0·5 µg mL–1). First, leaves were floated in the dark for 2 h and then lights were turned on for an additional 2 h. Properties of the chemicals used are given in Table 1. ROS staining was done using fresh leaves. For gene expression studies leaves were immediately frozen in liquid nitrogen and stored at –80 °C.
Table 1.
Summary of agents used to induce ROS production/accumulation in specific cellular compartments
| Treatment | Concentration range | Cellular compartment | Function | ROS accumulation |
|---|---|---|---|---|
| H2O2 | 10 µm – 10 mm | – | – | – |
| Rotenone (Rot) | 1–50 µm | Mitochondria | Inhibits NAD(P)H dehydrogenase | O2.– |
| Methyl viologen (MV) | 1–50 µm | Chloroplast | Accepts electrons from PSI and transfers them to O2 | O2.– |
| DCMU | 1–50 µm | Chloroplast | Blocks the plastoquinone binding site of photosystem II | 1O2 |
| 3-Amino-triazole (3-AT) | 1 mm | Peroxisome | Inhibits catalase | H2O2 |
All experiments were done in detached leaves and we tested whether a control detached leaf shows a similar pattern of gene expression compared with an attached leaf (Supplementary Data Fig. S1).
Staining of O2.– and H2O2 with NBT and DAB
In situ staining of O2.– and H2O2 was done according to Dutilleul et al. (2003). For detection of O2.–, after treatments, leaves were vacuum infiltrated (three cycles) in 0·5 mg mL–1 nitro blue tetrazolium (NBT) prepared in 10 mm potassium phosphate buffer (pH 7.8). Samples were incubated for 1 h in the dark at room temperature, and then were cleared in 90 % ethanol at 70 °C until chlorophyll was completely removed. As a control, superoxide dismutase (10 u mL–1) and 10 mm MnCl2 were added to the staining medium before infiltration. For detection of H2O2, leaves were vacuum infiltrated (three cycles) with 1 mg mL–1 3,3′-diaminobnzidine (DAB) in 10 mm sodium acetate buffer (pH 3.8). Samples were incubated in the dark at room temperature for 12 h. After incubation samples were transferred to 3 : 1 : 1 (v/v) ethanol/acetic acid/glycerol and were cleared at 70 °C until complete removal of chlorophyll. Both NBT- and DAB-stained leaves were examined in 70 % glycerol.
Determination of H2O2 content
H2O2 was determined according to Cheeseman (2006) using eFOX reagent. In this assay, 1 % ethanol is added to the reagent, which increases its sensitivity to H2O2 by 50 % (i.e. eFOX). Extraction was carried out using ice-cold acetone containing 25 mm H2SO4. Samples were then centrifuged for 5 min at 3000 g at 4 °C. eFOX reagent [950 µL of 250 µm ferrous ammonium sulfate, 100 µm xylenol orange, 100 µm sorbitol, 1 % ethanol (v/v)] was used for 50 µL of supernatant. Reaction mixtures were incubated at room temperature for 30 min and then absorbance at 550 and 800 nm was measured. H2O2 concentrations were calculated using a standard curve prepared with known concentrations of H2O2.
Quantitative reverse transcriptase PCR (qRT-PCR)
RNA was isolated from 0·1 g of leaf tissue using the Qiagen RNeasy kit according to the manufacturer’s recommendations. Total RNA was treated with DNase I (Fermentas) to remove residual genomic DNA. Then, reverse transcription was performed (1 µg total RNA for each treatment group) using M-MuLV reverse transcriptase (New England Biolabs). These cDNAs were used as templates for qRT-PCR. The amount of RNA in each reaction was normalized to the Arabidopsis thaliana ACTIN8 gene. Power SYBR Green Master Mix was used (Applied Biosystems) to perform the qRT-PCR. Three independent experiments were performed for qRT-PCR assays with an Applied Biosystems StepOne Plus System. The conditions for PCR amplification were as follows: 95 °C for 5 min, and 40 cycles at 94 °C for 15 s, 60 °C for 15 s and 72 °C for 30 s. qRT-PCR data analyses were performed with StepOne Plus software. Non-treated A. thaliana plants were used as a reference point and relative expression levels were calculated with respect to this reference value (set to 1) for genes that were studied.
Expression of bZIP28 (AT3G10800), bZIP17 (AT2G40950), IRE1A (AT2G17520), IRE1B (AT5G24360), BiP1 (AT5G28540), BiP3 (AT1G09080), CNX (AT5G61790), ERO1 (AT1G72280), HRD1 (AT1G65040), SEL1 (AT1G18260), DER1 (AT4G29330) and UBC32 (AT3G17000) were identified by qRT-PCR. The primers were synthesized by Sentromer DNA Technologies.
Primers used in this study are: bZIP28 forward 5′-ATCCTAAGCCTGTCTCGAGTTGTA-3′, reverse 5′-CGCCGACCATTAAAACCCTC-3′; bZIP17 forward 5′-CAAGCTTGTGAAGATAGATGGGA-3′, reverse 5′-TAGAGGCAGTGCAGGGGTAT-3′; IRE1A forward 5′-GCGCTACAGGCGTTACAAATA-3′, reverse 5′-TCGTCGAATCCTTCTGGAACT-3′; IRE1B forward 5′-AGTGGGGAAAAACCAGTTCC-3′, reverse 5′-AACCAAGTCTCGGAAACAGTG-3′; BIP1 forward 5′-TCAGTCCTGAGGAGATTAGTGCT-3′, reverse 5′-TGCCTTTGAGCATCATTGAA-3′; BIP3 forward 5′-CGAAACGTCTGATTGGAAGAA-3′, reverse 5′-GGCTTCCCATCTTTGTTCAC-3′; CNX forward 5′-ATGAGACAACGGCAACTATT-3′, reverse 5′-TTCCTGAGGACGGAGGTACT-3′; ERO1 forward 5′-TGGCGATGGCCTTTAGCGACT-3′, reverse 5′-GGCCAGAATGGGCAGTCACACC-3′; HRD1 forward 5′-TCTCTGTTGGGTTTATCTCTTTGGTT-3′, reverse 5′-CGGACATGAGAGAGCAAAGTCA-3′; SEL1 forward 5′-TGATGGAAGAAGCAGTGGATGA-3′, reverse 5′-CAGCTGCAAATTATGGTGAAG-3′; DER1 forward 5′-CGTAGAAGAGTGGTACAAGCAGATG-3′, reverse 5′-ACCCGACGGTGGTGACTACA-3′; UBC32 forward 5′-CGAGGGCGGGATTTATCATGGG-3′, reverse 5′-GTTGCCAATGCTCAGGGTGGTAG-3′; ACTIN8 forward 5′-TCAGCACTTTCCAGCAGATG-3′, reverse 5′-ATGCCTGGACCTGCTTCAT-3′.
Statistical analysis
The experiments were repeated twice, and each data point was the mean of three replicates (n=6). The results are expressed as mean with error bars indicating standard error of mean (±SEM). Groups were compared using Student’s t-test.
RESULTS
Quantification of ROS (H2O2 and O2.–) under inhibitor treatments
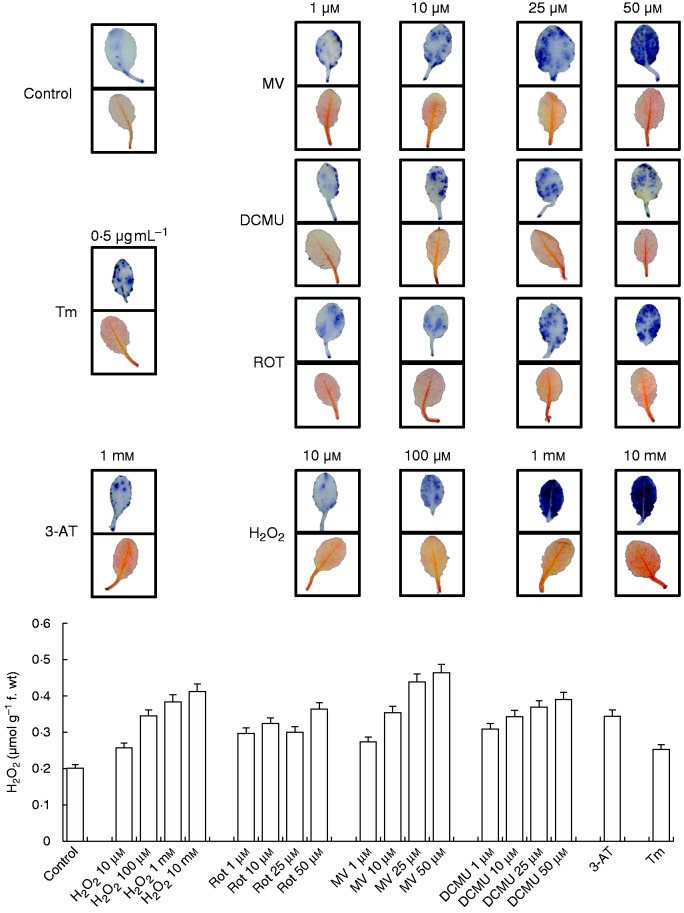
To confirm the effects of different treatments, ROS production in the leaves was evaluated by NBT and DAB staining. Among treatment groups, the highest H2O2 level was observed in H2O2- and MV-treated plants, which can also be seen in H2O2 contents measured spectrophotometrically (Fig. 1).
Fig. 1.
Detection of O2.– and H2O2 in leaves treated with H2O2, Rot, MV, DCMU, 3-AT and Tm by NBT (upper rows) and DAB (lower rows) staining. H2O2 content was also measured using a ferrous oxidation-xylenol orange (FOX) assay (below).
O2.– production was induced by H2O2, rotenone, MV, DCMU and 3-AT treatments (Fig. 1).
Expression levels of ER stress sensors and transducer genes: bZIP28, bZIP17, IRE1A and IRE1B
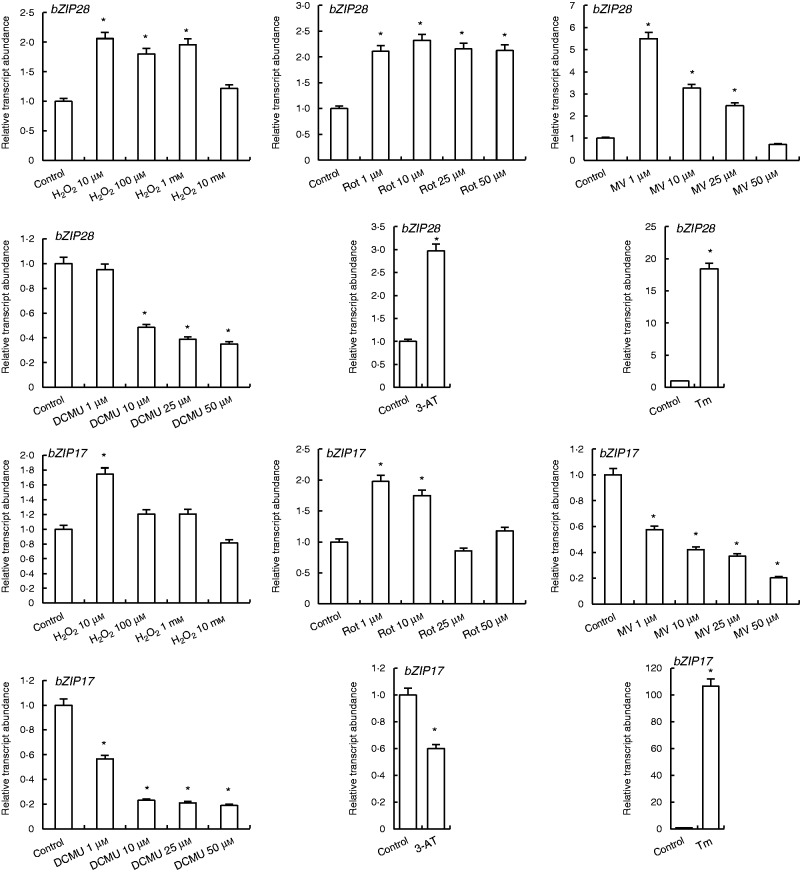
H2O2 and rotenone treatments increased the transcription of bZIP28 except in 10 mm H2O2. While 1 µm MV treatment enhanced bZIP28 expression 5.5-fold, enhanced concentrations of MV (10, 25, 50 µm) gradually decreased its transcription levels to the control levels under 50 µm MV. By contrast, 1 µm DCMU did not have any effect on expression levels of bZIP28, while 10, 25 and 50 µm DCMU decreased the bZIP28 transcript abundance. 3-AT and Tm treatments enhanced expression of this gene by 3 - and 18-fold, respectively (Fig. 2).
Fig. 2.
qRT-PCR analysis of expression of ER stress sensor/transducer genes bZIP28 and bZIP17 in leaves treated with H2O2, Rot, MV, DCMU, 3-AT and Tm. *Significant difference compared with the control group at P < 0·05.
Low concentrations of H2O2 (10 µm) and rotenone (1 and 10 µm) enhanced the bZIP17 transcripts, while other concentrations of these two agents did not affect expression of this gene. MV, DCMU and 3-AT treatments decreased bZIP17 expression. bZIP17 transcripts were highly induced by Tm treatments approx. by 100-fold as compared with control levels (Fig. 2).
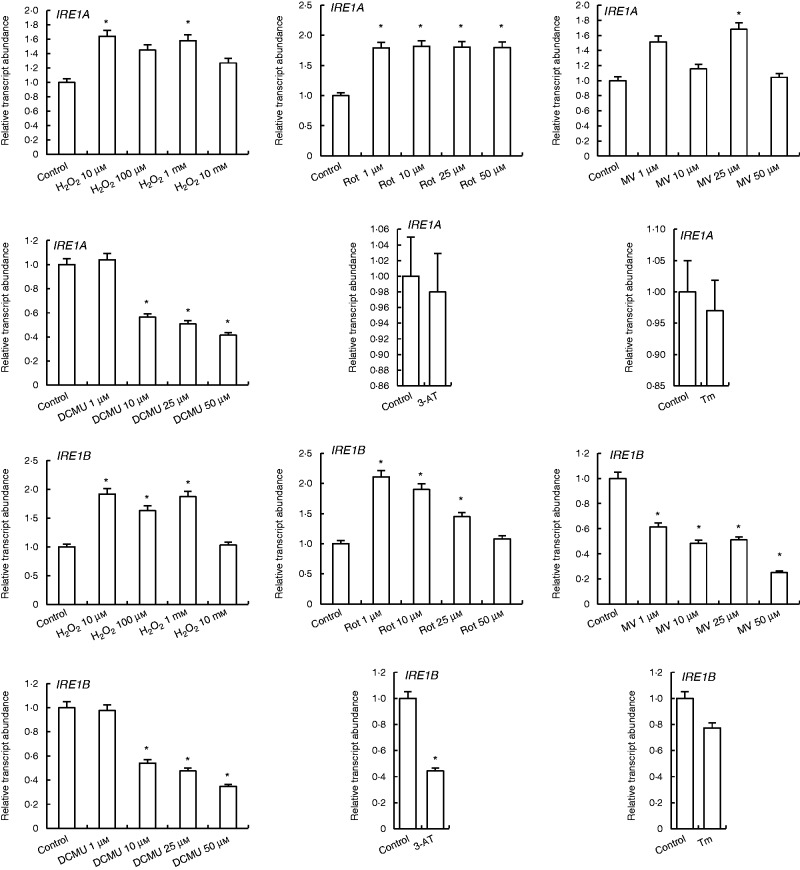
Expression of IRE1A and IRE1B were slightly increased by H2O2 and rotenone treatments as compared with control. By contrast, MV treatments increased the expression of IRE1A but decreased IRE1B expression levels. In the DCMU treatments, neither IRE1A nor IRE1B expression was changed by 1 µm DCMU, while it was decreased by higher concentrations. 3-AT and Tm did not affect the transcription levels of IRE1A and IRE1B (Fig. 3).
Fig. 3.
qRT-PCR analysis of expression of ER stress sensor/transducer genes IRE1A and IRE1B in leaves treated with H2O2, Rot, MV, DCMU, 3-AT and Tm. *Significant difference when compared with the control group at P < 0·05.
Transcription levels of ER stress responsive and protein folding helper BiP1 and BiP3 genes
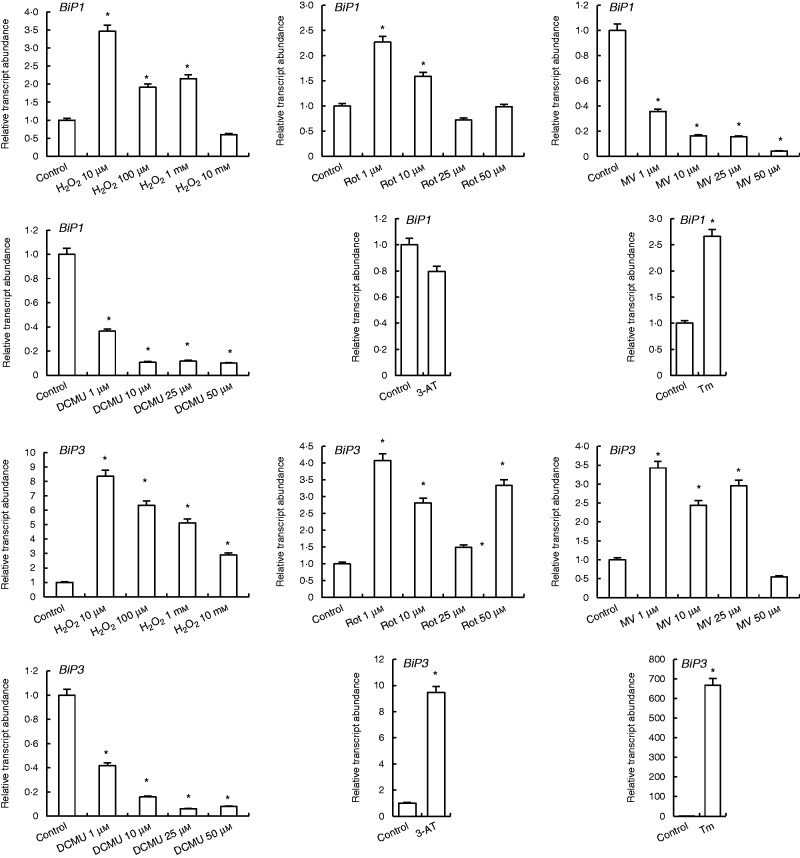
Expression of the BiP1 gene was increased 3 - to 3.5-fold by H2O2 treatments, while the highest concentration of H2O2 (10 mm) decreased it. Rotenone at 1 and 10 µm also increased BiP1 levels, while 25 and 50 µm did not change it, and nor did 3-AT treatment. By contrast, MV and DCMU treatments decreased expression of BiP1, but treatment with the ER stress inducer Tm increased it (Fig. 4).
Fig. 4.
qRT-PCR analysis of expression of ER folding helper/quality control genes BiP1 and BiP3 in leaves treated with H2O2, Rot, MV, DCMU, 3-AT and Tm. *Significant difference when compared with the control group at P < 0·05.
The level of BiP3 transcript was increased by H2O2 treatment. This increase was 8-fold under 10 µm H2O2, and 6 -, 5 - and 3-folds under 100 µm, 1 mm and 10 mm H2O2 treatments, respectively. Rotenone and MV enhanced the levels of BiP3 except with 50 µm MV. Similarly, 3-AT increased BiP3 expression 9-fold, while Tm increased it 660-folds as compared with controls (Fig. 4).
Transcription levels of protein folding machinery elements in the ER: CNX and ERO1
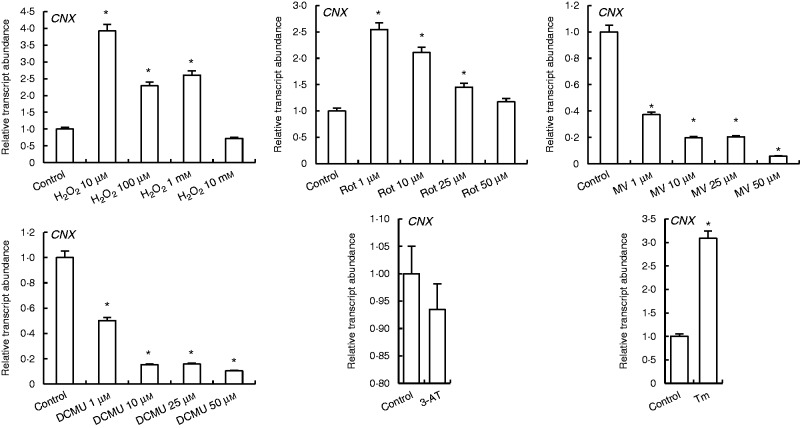
Transcription of CNX was increased by H2O2 and rotenone except with 10 mm H2O2. The highest increases were observed in 10 µm H2O2 and 1 µm rotenone treatments. MV and DCMU treatments decreased CNX expression, while Tm induced it (Fig. 5).
Fig. 5.
qRT-PCR analysis of expression of ER folding helper/quality control gene CNX in leaves treated with H2O2, Rot, MV, DCMU, 3-AT and Tm. *Significant difference when compared with the control group at P < 0·05.
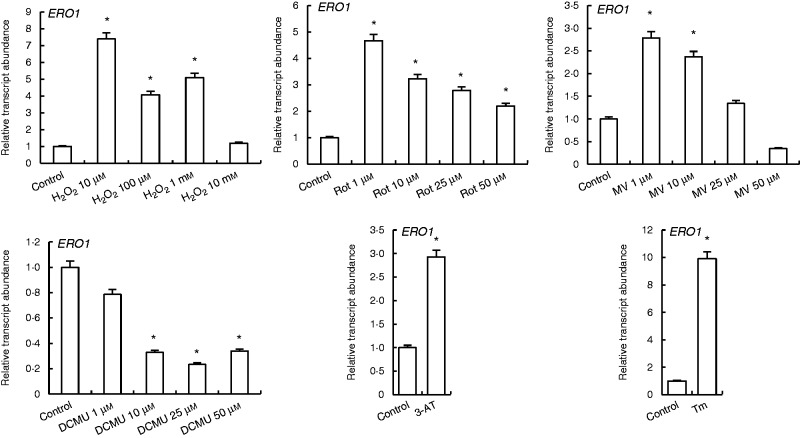
H2O2 at 10 µm enhanced the levels of ERO1 transcripts by 7-fold, while 100 µm and 1 mm H2O2 increased it by 4 - and 5-fold, respectively. By contrast, rotenone treatments increased transcription abundance of ERO1 2 - to 4-fold. MV treatments increased ERO1 expression except in the 50 µm treatment group, while DCMU treatments decreased it. 3-AT and Tm treatments enhanced ERO1 transcript levels 3 - and 10-fold, respectively (Fig. 6).
Fig. 6.
qRT-PCR analysis of expression of ERO1, which is responsible for regulation of the oxidative state of ER, in leaves treated with H2O2, Rot, MV, DCMU, 3-AT and Tm. *Significant difference when compared with the control group at P < 0·05.
Expression of ERAD-related HRD1, SEL1, DER1 and UBC2 genes
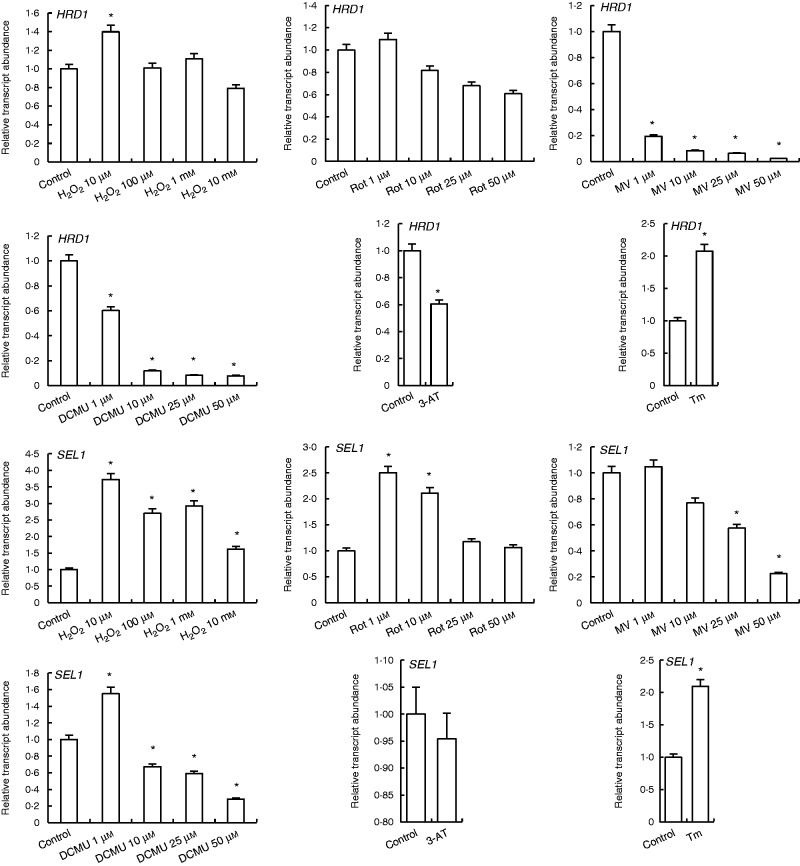
H2O2 at 10 µm slightly increased HRD1 transcript levels, while other concentrations did not affect its expression. HRD1 expression was decreased by rotenone treatments except at 1 µm concentration, which did not change it. By contrast, MV, DCMU and 3-AT treatments decreased HRD1 transcripts, while TM increased it 2-fold (Fig. 7).
Fig. 7.
qRT-PCR analysis of expression of ERAD genes HRD1 and SEL1 in leaves treated with H2O2, Rot, MV, DCMU, 3-AT and Tm. *Significant difference when compared with the control group at P < 0·05.
Transcription of SEL1, another protein degradation related gene, increased 3.7-fold under 10 µm H2O2. SEL1 expression was also increased by 1 µm (2.5-fold) and 10 µm (2-fold) rotenone treatments. By contrast, 10 µm and higher concentrations of MV decreased SEL1 transcription. Moreover, 1 µm DCMU enhanced SEL1 transcription, while other concentrations of DCMU decreased it. Treatment with the ER stress inducer Tm increased SEL1 transcripts 2-fold (Fig. 7).
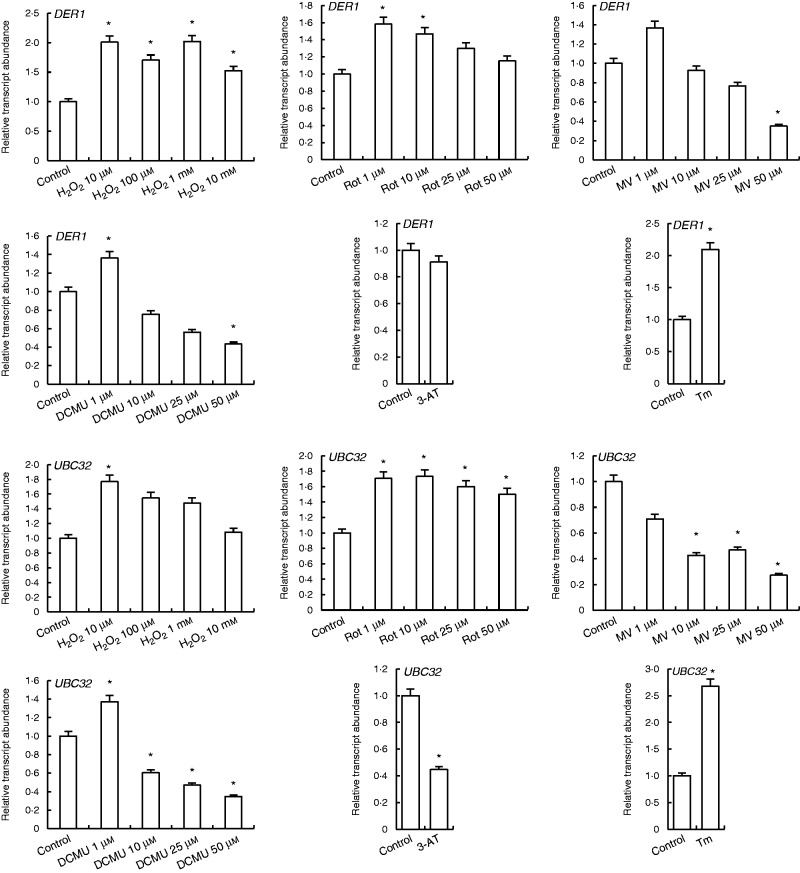
H2O2 and rotenone treatments increased DER1 and UBC32 transcripts. MV at 1 µm increased expression of DER1, while all MV treatments decreased expression of UBC32 genes. Expressions of both DER1 and UBC32 were enhanced by 1 µm DCMU treatments, while higher concentrations decreased them. 3-AT decreased both DER1 and UBC32 transcripts, while Tm treatment had an inducing effect on expression of these genes (Fig. 8).
Fig. 8.
qRT-PCR analysis of expression of ERAD genes DER1 and UBC32 in leaves treated with H2O2, Rot, MV, DCMU, 3-AT and Tm. *Significant difference when compared with the control group at P < 0·05.
DISCUSSION
The main aim of this study was to elucidate the effects of induced organellar (mitochondria, chloroplast and peroxisomes) ROS on ER stress response in Arabidopsis thaliana. As previously mentioned, ER is the centre for protein folding in the cell. Adverse environmental conditions can cause the accumulation of unfolded proteins in the ER, a phenomenon termed ER stress. ER stress can be identified by enhanced expression of unique transcription factors and genes, which are members of the UPR. By inducing UPR, plants can prevent accumulation of the misfolded and unfolded proteins in the ER. By contrast, if the ongoing stress conditions cause accumulation of massive amounts of misfolded or unfolded proteins, expression of ERAD genes can be induced to overcome this issue.
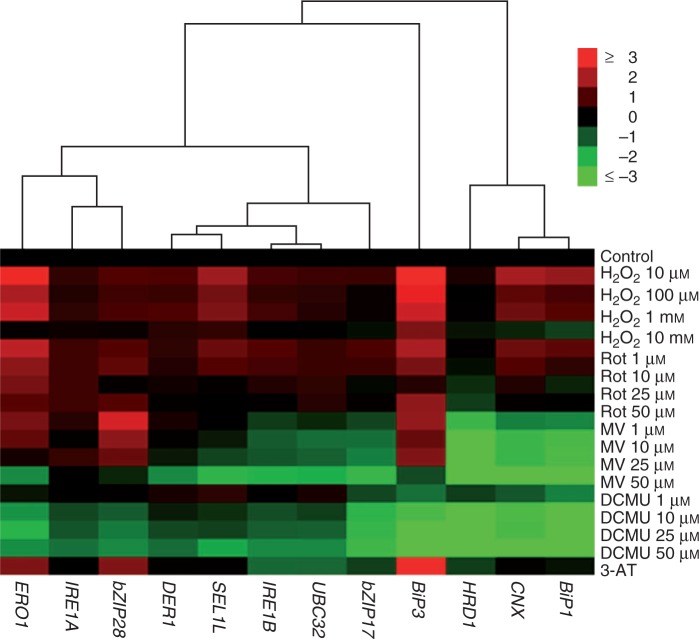
ROS originating from different compartments of the cell have different effects on the cellular response to stress, which was also previously shown in different transcriptomic studies (Gadjev et al., 2006; Laloi et al., 2007; Sewelam et al., 2014). Accordingly, in the present study, 11 of 12 genes were upregulated by rotenone treatment, while 5 of 12 and 3 of 12 genes were upregulated by MV and DCMU treatment, respectively. As can be seen from Fig. 9, there is a clear distinction in the ER stress response against mitochondria (due to Rot treatment), chloroplast (due to MV and DCMU treatments) and peroxisome (due to 3-AT) originated ROS.
Fig. 9.
Hierarchical clustering and heat map of ER stress-related genes in leaves treated with H2O2, Rot, MV, DCMU and 3-AT (average linkage and Euclidean distance as similarity measure). qRT-PCR data for each gene were log2n transformed, so that controls have a value of 0, increased expressions have positive and decreased expressions have negative values. Genes were grouped according to their expression patterns.
Many protein folding and other enzymatic and protein degradation reactions that occur in the ER require ATP (Marzec et al., 2012). However, there is no local source of ATP in the ER and therefore ATP supply depends on import from the cytosol. As mitochondria are the major source of ATP for reactions that take place in the cell, it is inevitable that induction of ROS production in mitochondria causes an imbalance in mitochondrial metabolism and affects the availability of ATP in the cell. HSP70-like proteins BiP1 and BiP3 have ATPase activities and can release their protein ligands only in the presence of ATP (Vitale and Denecke, 1999). Similarly, CDC48, a motor protein that is responsible for removal of proteins from the ER to cytosol for degradation, also shows ATPase activity (Marshall et al., 2008). Therefore, in the present study, the induction of ER stress-related genes by mitochondrial ROS production might be attributed to accumulation of unfolded proteins in the ER due to decreased folding and ERAD capacity caused by insufficient ATP.
Generation of chloroplastic ROS downregulated the expression of genes related to ER stress and protein folding, which might indicate a decrease in the rate of protein synthesis. It was previously shown that protein synthesis can be inhibited under extreme environmental conditions such as drought stress in plants (Dhindsa and Cleland, 1975; Cramer et al., 2011). Like other stresses, drought also causes metabolic imbalance in chloroplasts due to decreased availability of CO2 and increased ROS production. Although response to drought is a complex process, MV and DCMU treatments have some similar consequences to drought in chloroplasts. Therefore, a decrease in the availability of carbon backbone for synthesis of amino acids and proteins might reduce protein synthesis and decrease load on the protein folding machinery.
UPR is initiated by increased levels of ER stress sensor/transducer genes such as bZIP28, bZIP17, IRE1A and IRE1B. Liu et al. (2007a, b) found that salt stress response in arabidopsis requires bZIP17. Activation of bZIP17 upregulates salt stress responsive genes and this suggests that the salt stress response is associated with some of the UPR elements. Moreover, a bZIP17 single mutant was more sensitive to salinity (Liu et al., 2007a, b). Also, expression of the heat stress-induced bZIP17 and bZIP28 (Che et al., 2010) and their single mutants showed higher sensitivity to heat stress (Gao et al., 2008). It is also known that, upregulation of some ER stress-related genes, including BiP3, BiP1 and CNX, in part depends on bZIP28 (Liu and Howell, 2010a). In this study, H2O2 treatment and mitochondrial ROS production induced both bZIP28 and bZIP17, while peroxisomal ROS increased bZIP28 but decreased bZIP17.
Three BiP genes have been identified in A. thaliana (Maruyama et al., 2014). Among them, expression of BiP3 was observed only under ER stress (Noh et al., 2003). Earlier studies showed that overexpression of BiP genes caused osmotic tolerance in tobacco, confirming its link to stress conditions (Alvim et al., 2001). Both BiP1 and BiP3 are key regulators for bZIP28-activated unfolded protein response signalling (Srivastava et al., 2013). BiP3 induction is an important switch for UPR during ER stress conditions and abiotic stress conditions such as heat. Moreover, BiP3 induction is dependent on IRE1B activity (Deng et al., 2011). In this study, exogenous H2O2 application and ROS originating from mitochondria enhanced expression levels of BiP1 and BiP3. Peroxisomal ROS production via 3-AT treatment increased BiP3 but not BiP1. A closer look at chloroplastic ROS production showed that both MV and DCMU application decreased BiP1, while MV enhanced BiP3. These results showed that BiP3 was more responsive than BiP1 to ROS production in the organelles investigated.
ER-resident calnexin (CNX) functions with calreticulin in protein folding, especially of glycoproteins. Several studies have shown that its expression was enhanced by ER stress inducer agents such as Tm and dithiothreitol (Kamauchi et al., 2005). Similarly, various environmental stress conditions changed CNX expression in soybean (Nouri and Komatsu, 2010). Moreover, Sarwat and Naqvi (2013) found that expression of rice CNX was induced by drought in tobacco. Also, Garcia de la Garma et al. (2015) showed that under salinity tobacco BY-2 cells produced ROS in the mitochondria and this probably induced ER stress. Moreover, CNX and calreticulin genes of BY-2 cells were significantly induced by salinity and the ultrastructure of ER was highly altered. However, it is not known how CNX expression is affected directly in response to ROS treatments. In our study, we have shown that CNX expression was increased by H2O2 treatment and mitochondrial ROS production, but not by chloroplastic ROS sources. Induction of CNX by mitochondrial ROS production (rotenone treatment) is consistent with the results obtained by de la Garma et al. (2015).
ERO acts as a regulator of ER redox state and responds to unfolded proteins (Dixon et al., 2003; Onda et al., 2009). Previously, it was shown that expression of ERO1 was enhanced under salt stress (Ozgur et al., 2014). Moreover, ROS-induced ERO expression depends on H2O2 but not on 1O2, as evident from the DCMU-treated plants (Fig. 6).
Remarkably, enhanced expressions of BiP3 and ERO1 were tightly associated with the availability of H2O2 or an ROS that can be converted to H2O2 such as O2.–. Independent of the compartment of H2O2 production, expression of BiP3 and ERO1 increased in this study, except in DCMU-treated groups. ROS produced by DCMU is 1O2, which cannot be enzymatically converted to any other ROS, and damage done by 1O2 is strictly localized to chloroplasts. Due to its inability to diffuse long distances it does not have a signalling role on its own but with its oxidation products. These findings indicate that H2O2 has a signal role to induce ERO1, which is the regulator of redox status of the ER.
Liu et al. (2011) found that salt stress increased ERAD capacity in arabidopsis. Salinity and other environmental conditions increased the accumulation of unfolded and misfolded proteins, which are eventually removed by ERAD (Howell, 2013). A. thaliana plants with a defect in HRD3, an ERAD element, were sensitive to salt stress but not to osmotic stress. These plants also accumulated more ROS upon paraquat application (Liu et al., 2011). SEL1 forms a complex with HRD3 and this complex interacts with DER1 to initiate ERAD (Su et al., 2011). This ERAD mechanism observed in plants is also conserved in organisms through yeast to human (Kaneko and Nomura, 2003). Peroxisomal ROS accumulation did not a have significant effect on the ERAD system. However, ROS produced by DCMU in chloroplasts induced SEL1, DER1 and UBC32. To the best of our knowledge, this is the first report on induction of ERAD via ROS in a plant system.
As evidenced by transcriptomic studies, transcription of UBC32, an important ERAD component, was enhanced under salt, mannitol and drought stresses (Cui et al., 2012). In this study, H2O2 and chloroplastic and mitochondrial ROS increased the transcription level of UBC32.
In conclusion, we have shown how organellar (chloroplast, mitochondria and peroxisome) ROS production affects the ER stress response in a plant system for the first time. Exogenous H2O2 treatments induced transcription of UPR and ERAD genes, but excess amounts of H2O2 (10 mm) did not show a similar effect in the cell. In general, low concentrations of ROS induced the transcription of ER stress-related genes, which might indicate involvement of ROS as secondary messengers during ER stress. ROS originating from mitochondria caused induction of all the investigated genes to some level. Moreover, ERO1 and BiP3 were clearly induced by H2O2 accumulation. In addition, there was a clear distinction between the roles of chloroplastic and mitochondrial ROS in response to the ER stress response.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of Figure S1: relative transcript abundance for genes in attached and detached leaves.
ACKNOWLEDGEMENTS
This work was supported by the Scientific and Technological Research Council of Turkey (TUBITAK, grant no. 212T018) and the Ege University Research Foundation (grant no. 2013/BIL/016).
LITERATURE CITED
- Alvim F, Carolino SMB, Cascardo JCM, et al. 2001. Enhanced accumulation of BiP in transgenic plants confers tolerance to water stress. Plant Physiology 126: 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ. 2002. The endoplasmic reticulum: a multifunctional signaling organelle. Cell Calcium 32: 235–249. [DOI] [PubMed] [Google Scholar]
- Che P, Bussell JD, Zhou W, Estavillo GM, Pogson BJ, Smith SM. 2010. Signaling from the endoplasmic reticulum activates brassinosteroid signaling and promotes acclimation to stress in Arabidopsis. Science Signaling 3: ra69. [DOI] [PubMed] [Google Scholar]
- Cheeseman JM. 2006. Hydrogen peroxide concentrations in leaves under natural conditions. Journal of Experimental Botany 57: 2435–2444. [DOI] [PubMed] [Google Scholar]
- Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K. 2011. Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biology 11: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F, Liu L, Zhao Q, et al. 2012. Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 24: 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Humbert S, Liu JX, Srivastava R, Rothstein SJ, Howell SH. 2011. Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proceedings of the National Academy of Sciences USA 108: 7247–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhindsa RS, Cleland RE. 1975. Water stress and protein synthesis I. Differential inhibition of protein synthesis. Plant Physiology 55: 778–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Van Lith M, Edwards R, Benham A. 2003. Cloning and initial characterization of the Arabidopsis thaliana endoplasmic reticulum oxidoreductins. Antioxidants and Redox Signalling 5: 389–396. [DOI] [PubMed] [Google Scholar]
- Dutilleul C, Garmier M, Noctor G, et al. 2003. Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. The Plant Cell 15: 1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, et al. 2006. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiology 141: 436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Brandizzi F, Benning C, Larkin RM. 2008. A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proceedings of the National Academy of Sciences USA 105: 16398–16403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia de la Garma J, Fernandez-Garcia N, Bardisi E, Pallol B, Asensio-Rubio JS, Bru R, Olmos E. 2015. New insights into plant salt acclimation: the roles of vesicle trafficking and reactive oxygen species signalling in mitochondria and the endomembrane system. New Phytologist205: 216–239. [DOI] [PubMed]
- Hotamisligil GS. 2010. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 140: 900–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell SH. 2013. Endoplasmic reticulum stress responses in plants. Annual Review of Plant Biology 64: 477–499. [DOI] [PubMed] [Google Scholar]
- Iwata Y, Koizumi N. 2012. Plant transducers of the endoplasmic reticulum unfolded protein response. Trends in Plant Science 17: 720–727. [DOI] [PubMed] [Google Scholar]
- Kamauchi S, Nakatani H, Nakano C, Urade R. 2005. Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. Febs Journal 272: 3461–3476. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Nomura Y. 2003. ER signalling in unfolded protein response. Life Sciences 74: 199–205. [DOI] [PubMed] [Google Scholar]
- Koizumi N, Martinez IM, Kimata Y, Kohno K, Sano H, Chrispeels MJ. 2001. Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiology 127: 949–962. [PMC free article] [PubMed] [Google Scholar]
- Laloi C, Stachowiak M, Pers-Kamczyc E, Warzych E, Murgia I, Apel K. 2007. Cross-talk between singlet oxygen-and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proceedings of the National Academy of Sciences USA 104: 672–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH. 2010a. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. The Plant Cell Online 22:782–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH. 2010b. Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. The Plant Cell Online 22: 2930–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH. 2007a. An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. The Plant Cell Online 19: 4111–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH. 2007b. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. The Plant Journal 51: 897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cui F, Li Q, et al. 2011. The endoplasmic reticulum associated degradation is necessary for plant salt tolerance. Cell Research 21: 957–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu DP, Christopher DA. 2008. Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana . Molecular Genetics and Genomics 280: 199–210. [DOI] [PubMed] [Google Scholar]
- Marshall RS, Jolliffe NA, Ceriotti A, et al. 2008. The role of CDC48 in the retro-translocation of non-ubiquitinated toxin substrates in plant cells. Journal of Biological Chemistry 283: 15869–15877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez IM, Chrispeels MJ. 2003. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15: 561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama D, Sugiyama T, Endo T, Nishikawa SI. 2014. Multiple BiP genes of Arabidopsis thaliana are required for male gametogenesis and pollen competitiveness. Plant and Cell Physiology 55: 801–810. [DOI] [PubMed] [Google Scholar]
- Marzec M, Eletto D, Argon Y. 2012. GRP94: an HSP90-like protein specialized for protein folding and quality control in the endoplasmic reticulum. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research 1823: 774–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh SJ, Kwon CS, Oh DH, Moon JS, Chung W I. 2003. Expression of an evolutionarily distinct novel BiP gene during the unfolded protein response in Arabidopsis thaliana. Gene 311: 81–91. [DOI] [PubMed] [Google Scholar]
- Nouri MZ, Komatsu S. 2010. Comparative analysis of soybean plasma membrane proteins under osmotic stress using gel-based and LC MS/MS-based proteomics approaches. Proteomics 10: 1930–1945. [DOI] [PubMed] [Google Scholar]
- Onda Y, Kumamaru T, Kawagoe Y. 2009. ER membrane-localized oxidoreductase Ero1 is required for disulfide bond formation in the rice endosperm. Proceedings of the National Academy of Sciences USA 106, 14156–14161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur R, Turkan I, Uzilday B, Sekmen AH. 2014. Endoplasmic reticulum stress triggers ROS signalling, changes the redox state, and regulates the antioxidant defence of Arabidopsis thaliana. Journal of Experimental Botany 65: 1377–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarwat M, Naqvi AR. 2013. Heterologous expression of rice calnexin (OsCNX) confers drought tolerance in Nicotiana tabacum. Molecular Biology Reporter 40: 5451–5464. [DOI] [PubMed] [Google Scholar]
- Schröder M, Kaufman RJ. 2005. ER stress and the unfolded protein response. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 569: 29–63. [DOI] [PubMed] [Google Scholar]
- Sewelam N, Jaspert N, Van Der Kelen K, et al. 2014. Spatial H2O2 signalling specificity: H2O2 from chloroplasts and peroxisomes modulates the plant transcriptome differentially. Molecular Plant 7: 1191–1210. [DOI] [PubMed] [Google Scholar]
- Srivastava R, Deng Y, Shah S, Rao AG, Howell SH. 2013. BINDING PROTEIN is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis. The Plant Cell 25: 1416–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Liu Y, Xia Y, Hong Z, Li J. 2011. Conserved endoplasmic reticulum-associated degradation system to eliminate mutated receptor-like kinases in Arabidopsis. Proceedings of the National Academy of Sciences USA 108: 870–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Liu Y, Xia Y, Hong Z, Li J. 2012. The Arabidopsis homolog of the mammalian OS-9 protein plays a key role in the endoplasmic reticulum-associated degradation of misfolded receptor-like kinases. Molecular Plant 5: 929–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima H, Iwata Y, Iwano M, Takayama S, Koizumi N. 2008. Identification of an Arabidopsis transmembrane bZIP transcription factor involved in the endoplasmic reticulum stress response. Biochemical and Biophysical Research Communications 374: 242–247. [DOI] [PubMed] [Google Scholar]
- Vitale A, Denecke J. 1999. The endoplasmic reticulum—Gateway of the secretory pathway. The Plant Cell Online 11: 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.